Significance
Glycans and proteins are important partners in the regulation of fundamental biological processes such as the immune response, migration, differentiation, morphogenesis, angiogenesis, axon guidance, and response to CNS injury. Understanding glycan–protein interactions is critical to mapping the biological functions of glycans and developing new therapies for diseases such as cancer, autoimmune disorders, and neurodegenerative disorders. It is difficult to extract information about molecular-level interactions from experiments, and theory/computation has not been able to provide definite information to aid the interpretation of experiments. Our glycosaminoglycan (GAG)-Dock methodology addresses this challenge, furthering understanding of GAG–protein interactions by predicting the binding structures and how they depend on glycan structure and predicting precise effects of mutations that can be used to validate and interpret the interactions.
Keywords: docking, chondroitin sulfate, heparin, axonal growth, RPTPσ
Abstract
Cell-surface carbohydrates play important roles in numerous biological processes through their interactions with various protein-binding partners. These interactions are made possible by the vast structural diversity of carbohydrates and the diverse array of carbohydrate presentations on the cell surface. Among the most complex and important carbohydrates are glycosaminoglycans (GAGs), which display varied stereochemistry, chain lengths, and patterns of sulfation. GAG–protein interactions participate in neuronal development, angiogenesis, spinal cord injury, viral invasion, and immune response. Unfortunately, little structural information is available for these complexes; indeed, for the highly sulfated chondroitin sulfate motifs, CS-E and CS-D, there are no structural data. We describe here the development and validation of the GAG-Dock computational method to predict accurately the binding poses of protein-bound GAGs. We validate that GAG-Dock reproduces accurately (<1-Å rmsd) the crystal structure poses for four known heparin–protein structures. Further, we predict the pose of heparin and chondroitin sulfate derivatives bound to the axon guidance proteins, protein tyrosine phosphatase σ (RPTPσ), and Nogo receptors 1–3 (NgR1-3). Such predictions should be useful in understanding and interpreting the role of GAGs in neural development and axonal regeneration after CNS injury.
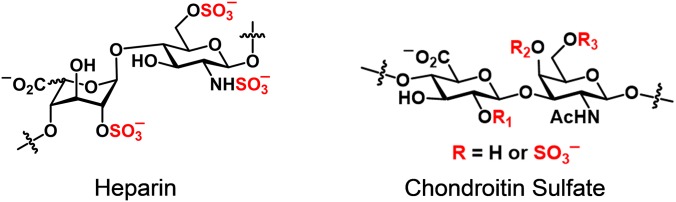
Glycans and proteins are important partners in the regulation of fundamental biological processes such as the immune response, signal transduction, development, and pathogen invasion (1). An understanding of the wide array of glycan–protein interactions is critical to mapping the biological functions of glycans and will pave the way for the development of new therapies that target glycan–protein interactions that contribute to diseases such as cancer and autoimmune and neurodegenerative disorders (2). Glycosaminoglycans (GAGs) are a prototypical example: they are known to interact with more than 300 secreted or membrane-bound proteins and thereby regulate a broad range of phenomena, including cell proliferation, migration, differentiation, morphogenesis, blood coagulation, angiogenesis, axon guidance, and response to CNS injury (3, 4). The GAG family of polysaccharides, which includes heparan sulfate (HS) and chondroitin sulfate (CS), is composed of alternating uronic acid and hexosamine units. The polysaccharides can vary in length, net charge, and the pattern and degree of sulfation (Fig. 1). Recent studies have shown that the biological activity of GAGs is often dependent on their sulfation sequence, with specific, highly sulfated sequences directing interactions with growth factors and other signaling proteins (5–8). Despite the importance of GAG–protein interactions, there is remarkably little structural information about these complexes. This is largely a result of the inherent structural complexity and heterogeneity of GAGs, which makes it difficult to obtain sufficient quantities of oligosaccharides of defined length and sulfation pattern for structural studies. As a result, structural data are available for only a handful of heparin–protein complexes, and no structural information is available for CS-D, CS-E, and HS motifs.
Fig. 1.
Disaccharide representations of the glycosaminoglycans heparin and chondroitin sulfate. Chondroitin sulfate is as follows: CS-A (R1, H; R2, SO3−; R3, H), CS-C (R1, H; R2, H; R3, SO3−), CS-D (R1, SO3−; R2, H; R3, SO3−), and CS-E (R1, H; R2, SO3−; R3, SO3−).
GAGs play critical roles in neuronal growth and axon regeneration after spinal cord and other CNS injuries through their ability to engage transmembrane proteins such as the receptor protein tyrosine phosphatases protein tyrosine phosphatase σ RPTPσ and LAR and Nogo receptors (NgRs) NgR1 and NgR3 (8–11). CS and its associated proteoglycans are the principal inhibitory components of the glial scar, which forms after neuronal damage and acts as a barrier to axon regeneration (12–14). Blocking the interactions of CS using chondroitinase ABC or an antibody specific for the CS-E motif can promote axon regeneration, sprouting, and functional recovery following injury in vivo (8, 15). Intriguingly, RPTPσ engagement by CS and HS can exert opposing effects on neuronal growth, with CS inhibiting and HS promoting axon growth (10). However, it is unclear how GAG binding modulates the activity of RPTPσ and other cell-surface receptors important for axon guidance and regeneration. The lack of structural information on physiologically relevant sulfation motifs has hindered an understanding of GAGs and efforts to develop tools and therapeutic approaches that target GAG-mediated processes.
An alternative approach to in vitro structural determination is computational modeling of GAG–protein complexes. Modeling GAG–protein interactions is extremely challenging because of the conformational flexibility of GAGs, the high charge density of GAGs and GAG-binding sites, and the weak surface complementarity of GAG–protein interactions. Despite these challenges, we (6) and others (16–18) have used molecular modeling successfully to predict the sites at which GAGs engage their target proteins. Some of these methods have limited accuracy in predicting the bound pose of the ligand or have limited robustness across different systems. Moreover, most of these methods have not been applied to systems other than the known heparin–protein structures.
Herein, we report the GAG-Dock method we developed to accurately model GAG–protein interactions, and we validate this method against known GAG–protein systems. By using GAG-Dock, our predicted heparin binding poses were within 0.70–1.51 Å rmsd of the crystal structures across a diverse set of systems, including FGF1, FGF2, FGF2-FGFR1, and α-antithrombin III (ATIII). We further apply the method to predict the protein-bound pose of various GAGs, including CS-D and CS-E, to systems without known structures. Finally, we demonstrate the utility of these methods to tune the specificity of protein binding, through in silico mutations, to favor a particular GAG sulfation pattern.
Results and Discussion
To validate the GAG-Dock method for such complex ligands and binding sites, we applied it to two sets of systems. The first set consists of the four validation systems for which a crystal structure including the ligand bound to the specific binding site was known. The second set of systems consists of three proteins known to bind to one or more GAG ligands, but for which the specific binding site was not known (although the general region of binding may be known). In each case, we followed the procedure of (i) coarse docking to identify the best regions and (ii) fine docking to identify the best ligand poses. In both cases, the criterion for selection was the predicted binding energy.
Case 1: Validation of Systems for Which There Are X-Ray Structures of the Cocrystal.
Five heparin-protein crystal structures have been solved, providing a means to validate our method. We applied GAG-Dock to four of these cases. We did not consider the fifth system, FGF1-FGFR2 [Protein Data Bank (PDB) ID code 1E0O (19)] because this 10-mer ligand is significantly more demanding computationally, but otherwise is similar to the other validation cases. The rmsd comparison for the predicted and crystal ligands for the validation systems are summarized in SI Appendix, Fig. S1, showing that GAG-Dock reproduces the ligand positions with good accuracy. SI Appendix, Figs. S2–S9, compares nonbond interactions between the ligands and side chains within the binding sites of the validation systems. As can be seen from the plots in SI Appendix, Fig. S10, most of the ligand–side chain interactions were faithfully reproduced. A major source of error in the side chain placement and interaction energies is the lack of waters in our validation systems. For structures without known binding sites, such as RPTPσ and NgR, the placement of waters in an apo-crystal structure cannot be assumed to be correct for a ligand-bound structure, and even that information is lacking if homology modeling is used to generate the protein structure. Therefore, for a realistic assessment of the validation systems, all waters present in the crystal structures were removed. As waters often play a role in ligand binding, removing the waters allows side chains in the protein to interact more strongly with the ligand.
FGF1.
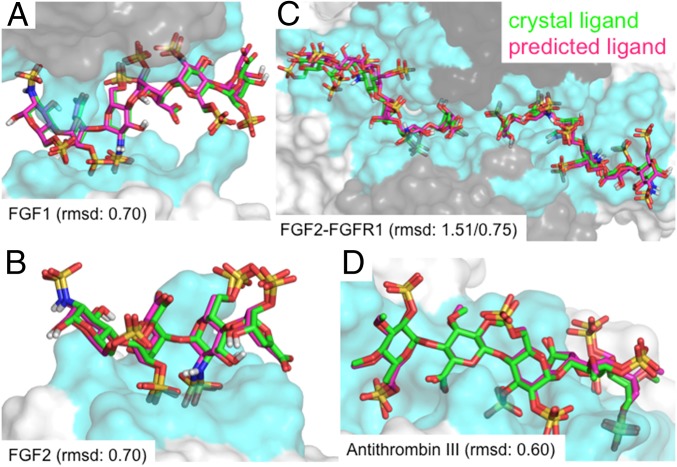
We validated our method by using the crystal structure of the heparin hexasaccharide bound to two molecules of fibroblast growth factor 1 [FGF1; PDB ID code 2AXM (20)]. GAG-Dock correctly identified the binding site, finding that both molecules of FGF1 interact with heparin at the same site, but with different specific residues interacting with the ligand for each protein. The lowest-energy pose was within 0.70 Å rmsd of the crystal structure ligand (Fig. 2A), calculated by comparing all atoms in the docked ligand to all atoms (including added hydrogen atoms) in the X-ray ligand.
Fig. 2.
Comparison of predicted binding sites for heparin (magenta) to the X-ray crystal (green) ligand positions for the four validation systems. The rmsd between predicted and crystal structures of ligand heavy atoms (A) FGF1 (rmsd of 0.70 Å), (B) FGF2 (rmsd of 0.70 Å), (C) FGF2–FGFR1 (rmsds of 1.51 Å, 0.75 Å), and (D) α-antithrombin III (rmsd of 0.60 Å).
As the crystal structure is available, we docked the protein with all side chains in their experimental conformation. In this case, we predict the lowest-energy (i.e., strongest-binding) ligand pose to have an rmsd error of 0.70 Å. Optimizing the ligand and the side chains for the heparin-binding site of the FGF1 molecules, the lowest-energy structure led to an rmsd of 2.08 Å compared with the X-ray structure (SI Appendix, Figs. S2 and S3). We consider that this is a success. Comparing versus the X-ray pose, we find some minor differences in the energy contributions (SI Appendix, Fig. S11). For example, K112 and K113 in chain A and K128 in chain B made stronger Coulomb and hydrogen bonding interactions with the ligand in the docked pose than in the X-ray (probably because the water plays a role in the X-ray structure but not in ours). On the contrary, R119 was positioned farther from the ligand in the docked pose, leading to weaker Coulomb interactions with the ligand. Overall, all interactions found in the crystal structure were recapitulated in the predicted structure. Furthermore, the predicted energy contributions for the ligand interacting with each residue were consistent between the docked and crystal structures, indicating that these energy contributions can be used to understand the relative contributions to binding for each residue of the protein. Our conclusion is that our GAG-Dock methodology accurately predicts the ligand pose and the relative importance of residues toward ligand binding. Our analysis suggests that K112, K113, K118, R122, and K128 make the most important contributions to heparin binding (SI Appendix, Fig. S11).
FGF2.
For the complex of a heparin tetrasaccharide with FGF2 [PDB ID code 1BFB (21)], heparin makes contacts primarily with a single molecule of FGF2. However, in the crystal, there are additional contacts with three nearby FGF2 molecules that appear to influence the conformation of the ligand. Thus, we docked the heparin tetrasaccharide to the central protein while including the three nearby FGFs to describe the conditions of the crystal structure. Again, GAG-Dock correctly predicts the binding site and the optimum pose of the crystal ligand (0.70-Å rmsd; Fig. 2B and SI Appendix, Fig. S4).
For FGF2, the side chains of the binding site differ from the X-ray structure by 2.09-Å rmsd. In particular, GAG-Dock predicts conformations of K120, R121, and K130 that lead to stronger hydrogen bond and Coulombic contributions to binding than in the X-ray structure (SI Appendix, Fig. S12). However, no residues had less favorable conformations in the docked structure compared with the crystal structure. This is expected because waters present in the experimental structure generally play a role in binding. Eliminating them usually leads to stronger binding to the residue side chains. Again, GAG-Dock correctly predicts the relative importance of all residues involved in binding, showing that residues K120, R121, K126, K130, and K136 contribute most strongly to heparin binding (SI Appendix, Fig. S12).
FGF2–FGFR1.
Heparin is known to form a ternary complex with FGF and its receptor FGFR2. The crystal structure of the FGF2–FGFR1–heparin complex features a 2:2:2 stoichiometry [PDB ID code 1FQ9 (22)]. In this structure, each heparin molecule (an 8-mer and a 6-mer) binds to the positively charged groove formed at the junction of the proteins, making contacts with an FGF2 molecule and with the D2 domains of both FGFR1 molecules. Interestingly, this structure is very similar to the FGF2–FGFR1 complex without heparin [0.37-Å rmsd (23)], suggesting in this case that GAG-Dock correctly predicts the multimeric protein–receptor–GAG complex. We docked both heparin molecules to regions near the FGF1 molecule and to both FGFR2 molecules. For each heparin molecule, the predicted pose correctly identifies the binding pose found in the crystal structure [with rmsd of 0.75 Å (8-mer) and 1.51 Å (6-mer); Fig. 2C and SI Appendix, Figs. S5–S8]. The rmsds of side chains in the binding site were 1.76 (8-mer) and 2.28 Å (6-mer). The predicted pose accounts for the relative importance of all residues involved in binding, leading to the same pharmacophore identified in the crystal structure (SI Appendix, Figs. S13–S17).
α-Antithrombin III.
The interaction between heparin and ATIII is one of the most studied GAG–protein complexes as a result of its role in blood coagulation (24). The structure of ATIII bound to a heparin analog [PDB ID code 1E03 (25)] provided a more challenging test than the other validation cases. With no other protein species making significant contacts to the ligand, this structure lacked the constraints of the other validation systems. Even without such constraints, GAG-Dock predicts the crystal structure pose with 0.60-Å rmsd (Fig. 2D and SI Appendix, Fig. S8). The protein side chains in the binding site have an rmsd of 1.96 Å compared with the crystal structure. The predicted pose accounts for the relative importance of all residues involved in binding, with residues R13 and K125 contributing more to binding in the docked pose (SI Appendix, Fig. S18).
Case 2: Predictions for Systems for Which No Cocrystal Structure Is Available.
Unlike heparin, no structural information is available for chondroitin sulfate motifs CS-D and CS-E, despite increasing evidence of their biological importance (6, 8, 11). This is because of the difficulty in obtaining CS oligosaccharides that are purely one type (e.g., CS-E) for use in structural studies. The recent identification of RPTPσ and NgR as mediators of CS-induced axon inhibition (9, 11), and the discovery that HS and CS have opposing effects on axon morphology (10), highlight the critical need for structural data to facilitate a mechanistic understanding of GAG function. Interestingly, RPTPσ and NgR bind to polysaccharides enriched in the CS-D, CS-E, or HS epitopes, but not the lower sulfated motifs, such as CS-A (8, 11). Thus, these proteins are ideal first systems to test how consistent our docking predictions are with in vitro binding data. To this end, we predicted docked structures of various GAGs to RPTPσ, NgR1, NgR2, and NgR3.
RPTPσ.
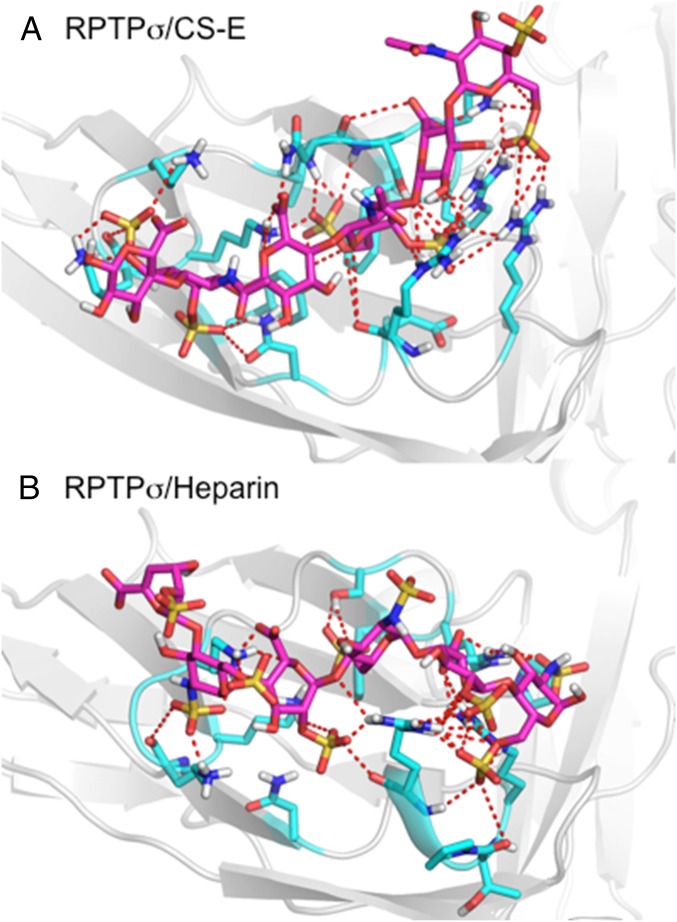
Although structural data for an RPTPσ–GAG complex have not been reported, the GAG binding site on the protein is well understood. A defined GAG-binding site lies on the Ig1 domain of the protein, mediated by the K67, K68, K70, K71, R96, and R99 residues (9). This region forms a shallow electropositive cavity on the surface of the protein between β-strands C-D and E-F (SI Appendix, Fig. S19). The quadruple mutation of K67, K68, K70, and K71 to alanine has been shown to impair binding to CS and HS (11). ELISA binding data to natural GAG polysaccharides indicate that RPTPσ binds strongly to CS-D, CS-E, and heparin, but not to CS-A. To better understand RPTPσ–GAG interactions, we docked CS-D, CS-E, and heparin hexasaccharides to the protein (PDB ID code 2YD2). We also docked CS-A hexasaccharide but did not find significant binding, which is consistent with the lack of binding observed experimentally. The docked CS-E and heparin structures are shown in Fig. 3, with detailed structures shown in SI Appendix, Fig. S20 (CS-E), and SI Appendix, Fig. S21 (heparin).
Fig. 3.
Predicted binding pose for (A) CS-E and (B) heparin bound to the Ig1 domain of RPTPσ, with docked ligand (magenta) and residues within 5 Å shown (cyan). Dashed lines indicate hydrogen bonding and salt bridges between ligand and protein.
Indeed, GAG-Dock predicts that the CS and heparin ligands bind to the previously identified GAG-binding site. CS-E and heparin interacted with the arginine and lysine residues K67, K68, K70, K71, R96, and R99 that line the electropositive cavity found in the Ig1 domain of RPTPσ (SI Appendix, Figs. S22 and S23). Additionally, GAG-Dock identifies significant contributions to CS and heparin binding from R76. We can also gain insights into the relative contribution of these key arginine and lysine residues toward CS-E and heparin binding. The relative contributions to binding for these residues for CS-E is R76, K67, R99, R96, K70, K68, and K71, whereas, for heparin, it is K70, R99, R76, R96, K67, K68, and K71. In addition, a repulsive contribution from D100 and E101 were identified for CS-E but not heparin. GAG-Dock can also identify interactions with polar, noncharged, residues that contribute to GAG binding, such as N73, S74, and Q75. These additional interactions with polar residues would normally not be found from mutational studies, which tend to focus on charged arginine and lysine residues. Compared with mutational studies, GAG-Dock allows us to understand the contribution of all residues in the binding site, structural information that is not readily obtained through traditional experimental methods.
These results are consistent with experimental data, and GAG-Dock predicts similar affinities for CS-E and heparin binding to RPTPσ. However, binding affinity alone cannot explain the opposing effects of CS-E and heparin on neurite outgrowth when interacting with RPTPσ. These differences may arise from the way in which CS-E and heparin ligands engage RPTPσ. For heparin, the predicted structure contains multiple solvent-exposed sulfate groups, whereas the predicted CS-E structure has all sulfate groups oriented toward the GAG binding site of RPTPσ. These differences could allow the heparin–RPTPσ complex to engage an additional RPTPσ through these solvent-exposed sulfate groups. Thus, our GAG-Dock method can shed light mechanistic differences that cannot be explained through binding affinities alone through a more thorough characterization of GAG binding sites.
NgRs.
The NgRs are myelin-associated inhibitors that restrict axonal growth after injury. A recent study demonstrated that NgR1 and NgR3, but not NgR2, are involved in GAG-induced axonal inhibition (11). NgRs are comprised of 8.5 leucine-rich repeat (LRR) domains flanked by N-terminal and C-terminal LRR capping domains (LRR-NT and LRR-CT, respectively) and a C-terminal stalk (CT stalk) that connects the protein to the membrane via a glycosylphosphatidylinositol anchor (26). Compared with RPTPσ, less structural information is known about how NgR binds to GAGs; however, domain deletion studies suggest that the LRR-CT and CT stalk regions are required for GAG binding (11). Unfortunately, available crystal structures of the NgR ectodomain contain the LRR-NT, LRR, and LRR-CT regions but lack the CT stalk region (27).
To better understand the role of the CT stalk region in GAG binding, we generated models of the entire ectodomain, including the previously uncrystallized CT stalk, of NgR isoforms 1–3 by using Rosetta software (28). We carried out 5 ns of molecular dynamics (MD) in the presence of explicit water and counter ions to allow the five models per isoform to relax. We then selected the structure closest to the average conformation for each model, minimized it, and selected the lowest-energy structure for each isoform to use for docking. The electrostatic potential surfaces of these homology models of the extracellular domain of NgR isoforms 1–3 suggest an electrostatic basis for the difference in activity between NgR2 and NgRs 1 and 3 (SI Appendix, Fig. S24). For NgR1 and NgR3, we solvated the GAG–protein complex and carried out 5 ns of MD at 298K. We did not observe the formation of any new interactions between NgR and the GAG ligand that were not identified using GAG-Dock. However, we did observe small changes in protein side-chain conformation that improved binding scores. Unlike the GAG-binding isoforms, NgR2 lacks significant regions of electropositive potential, and, in fact, its surface is quite electronegative. Our binding energies from coarse-level docking with a CS-E tetrasaccharide to NgR2 predict much weaker interactions (−297.67 kcal/mol), relative to NgR1 and 3 (−641.27 and −985.46 kcal/mol, respectively), consistent with experimental findings.
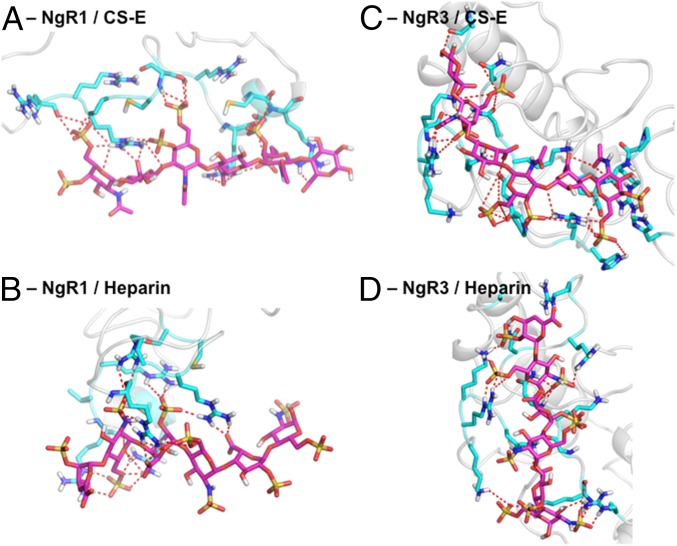
Based on fine-level docking with CS-A, -D, -E, and heparin hexasaccharides, followed by 5 ns MD relaxation in a full water box with counter ions, we predict that GAGs bind to regions of electropositive potential on the CT stalk of NgR1 and NgR3 (Fig. 4). GAG-Dock studies predict that the GAG-binding domains of NgR1 and NgR3 are on different faces of the CT stalk, although this could be the result of the structural flexibility of this region of the protein and discrepancies between the model and the natural state of the protein. We predict that the GAGs make polar or electrostatic contacts with residues R399, R414, R415, R416, R421, K422, R424, R426, and R430 on NgR1 and with residues R346, R350, K354, N355, N358, R360, K364, K399, R400, K401, K403, and R406 on NgR3. Many of these residues, particularly residues 414–426 on NgR1 and 399–406 on NgR3, were shown by mutagenesis studies to be important for GAG binding (11). Together, these results validate that GAG-Dock can be used to understand the structural basis for extreme differences in GAG-binding activity between related proteins and to identify reliably the pharmacophore even in cases in which the protein structure is ill-defined. Detailed structures for CS-A, CS-D, CS-E, and heparin bound to NgR1 are shown in SI Appendix, Figs. S25–S30. Detailed structures for those ligand bound to NgR3 are shown in SI Appendix, Figs. S31–S36.
Fig. 4.
Predicted structures (magenta) for CS-E and heparin bound to NgR1 and NgR3. Residues within 5 Å of GAG ligand are shown (cyan) with hydrogen bonding and salt bridges between ligand and protein displayed (dashed lines). (A) CS-E and (B) heparin bound to NgR1. (C) CS-E and (D) heparin bound to NgR3.
Tuning GAG Binding Through in Silico Mutations.
To identify mutations that increase or decrease GAG binding, we applied an in silico mutations for our predicted CS-A, CS-D, CS-E, and heparin binding sites for RPTPs, NgR1, and NgR3. The common strategy of probing the binding site by mutation of arginine or lysine to alanine leads to a drastic change in character that might result in significant disruption of the system beyond direct effects on binding. Instead, we employed subtler mutations to asparagine or glutamine, which allows the possibility of maintaining some polar contact with the ligand, but without the benefit of strong charged interactions. A second common strategy in probing the binding site is to identify only mutations that decrease binding, which we consider to be ambiguous, as binding can be lost for many reasons. Therefore, we sought to identify mutations we expect might increase binding of the ligand to the protein.
We first employed single-residue mutations of each of the residues within the binding sites to asparagine or glutamine while simultaneously optimizing the remaining side-chain conformations in the binding site by using SCREAM, followed by 50 steps of conjugate gradient energy minimization. From these calculations, we identified mutations that increased or lost hydrogen bonding to the ligands. Based on these individual mutations, we identified sets of mutations to enhance or reduce ligand binding for each GAG–protein complex. We should note that, even though some mutations of arginine or lysine may still lead to increased hydrogen bonding, there would generally be a net loss of overall binding energy as a result of the lost Coulomb interactions. Therefore, we considered only mutations of arginine or lysine to asparagine or glutamine for our loss-of-binding mutations sets.
For RPTPs, we identified three sets of mutations that improve binding to CS-A, CS-D, or CS-E, but, interestingly, we found none to heparin (SI Appendix, Figs. S37 and S38). Perhaps RPTPσ is already optimized for heparin binding, but not for CS binding. Mutation set “G1” is specific for CS-E, whereas “G2” is specific for CS-D and “G3” is nonspecific with the exception of decreasing heparin binding. Based on single-residue mutations, we generated four sets of mutations to decrease binding. As all of these mutations affect the key arginine and lysine residues, they all result in significant reductions in binding energy, as would be expected.
For NgR1, we identified four sets of mutations to increase binding by combining the single-mutation information (SI Appendix, Figs. S39 and S40). Set G1 improved CS-A and CS-D binding, but not CS-E or heparin binding. Set G2 improved CS-D and heparin binding, and set G4 improved binding for every ligand except CS-E. However, surprisingly, set G3 did not show any improvement in binding. It is again interesting that none of the mutation sets improved CS-E binding. Similarly, for NgR3, we identified four sets of mutations that increase ligand binding (SI Appendix, Figs. S41 and S42). Set G1 improved CS-A, CS-E, and heparin binding but not CS-D binding. Set G2 modestly improved CS-A and CS-E binding but not CS-D or heparin binding. Set G3 is the only set to improve binding for all ligands, and set G4 improved binding for CS-A and heparin only. As with RPTPs, the four loss-of-binding mutation sets identified for NgR1 and NgR3 were all effective in reducing ligand binding, but were nonspecific for any ligand. Importantly, the methods outlined here could be employed to engineer GAG binding sites to tune their affinity for a particular GAG structure.
Conclusions
Predicting the binding sites of highly charged GAG ligands with multiple independent charge sites and numerous possible conformations is a formidable challenge. The very large number of charged sites on the ligands and in the binding site likely leads to redistributions of the water and ions in the solvent, making polarization likely of great importance. Nevertheless, we show here, for eight independent systems, that the simple GAG-Dock modifications of the DarwinDock general docking approach accounts well for the enormous importance of electrostatic interactions and leads to plausible structures and relative binding energies that help distinguish the strength of binding for various GAG ligands to a wide variety of receptors likely to play essential roles in axonal growth. Given the difficulty of obtaining high-quality cocrystals for X-ray studies, this simple GAG-Dock computational methodology may provide the best means for predicting the structure with sufficient accuracy to help design experimental probes to elucidate the mechanisms by which GAGs modulate important processes such as axon growth and regeneration.
Summary of the GAG-Dock Method
Unlike small-molecule ligands often docked successfully with various techniques (29–33), even the truncated GAGs are large (a CS-A 4-mer has 60 heavy atoms and a net charge of −4; a CS-E 8-mer has 137 heavy atoms and a net charge of −12). Additionally, they bind to protein surfaces rather than in pockets and engage proteins primarily through electrostatic interactions.
Our GAG-Dock method is based on the DarwinDock/GenDock methodology (29, 30) with modifications to accommodate bulky, highly charged, surface-binding ligands characteristic of GAGs. Whereas our previous approach predicted GAG-binding regions within proteins (6), the method reported here accurately predicts the binding pose, giving deeper insights into the specific GAG–protein pairwise interactions critical for recognition. The GAG-binding site is generally not known; hence, it is necessary to systematically examine all possible binding regions. To do this, we conduct two rounds of docking. For “coarse docking,” we dock a single GAG conformation to the entire protein surface to identify likely binding sites. Docking to the “alanized” structure (see System Preparation) allows us to quickly scan the entire protein for putative GAG binding sites by optimizing the long range Coulomb interactions first. For the “fine-grained” approach, we redock to the best coarse regions to identify specific, strongly bound poses. In this step, we are more completely sampling conformations and the intrinsic flexibility of the GAG ligand by allowing rotation about bonds. Final structures in the fine-grained approach are subjected to minimization before scoring to identify the top docked structure for each GAG–protein complex. These methods are detailed further in the subsequent sections and in the SI Appendix.
DarwinDock/GenDock.
The DarwinDock/GenDock docking method applied here has been applied recently to predict ligand binding sites for GPCRs such as OR1G1 (31), AA3R (32), and 5HT2b-R (33). Briefly, it consists of four parts:
i) System preparation.
Starting with target protein structures (usually with no hydrogen atoms), we prepare the systems as follows: (i) add hydrogens to various heavy atoms using standard bond distances and hydrogen binding criteria, (ii) assign partial charges to all protein atoms based on general force-field criteria and to all heteroatoms based on Mulliken charges from quantum mechanics calculations, (iii) optimize the protein structure using the force field to minimize the energy, (iv) replace the seven bulky, nonpolar residues (V, L, I, M, F, Y, and W) with alanine (i.e., alanization) to allow more complete sampling of the binding site, and (v) generate and select sphere regions defining the space to be sampled by the ligand.
Generally, the conformations of the protein side chains at the ligand binding site depend on the location and the conformation of the ligand (the pose), whereas the location and conformation of the ligand depends on the side-chain conformations. Our solution to this “chicken/egg” problem is to alanize the bulky, nonpolar side chains in step iv (above) to allow the ligand to fully sample available sites on the protein surface in the presence of the polar interactions. After selecting the best poses, the original nonpolar side chains are replaced and reoptimized for each pose by using the SCREAM side-chain optimization method (34) in a process we call “dealanization.” This allows a different set of protein side chains for each ligand pose.
To select poses that are close enough to the protein to interact favorably, but not too close to clash with protein atoms, we generate spheres to describe the space available for the ligand. This is done with the sphgen program (35), modified to work with protein surfaces. The spheres are partitioned into overlapping boxes (“sphere regions”) for docking.
ii) Generation of a complete set of poses.
Before evaluating interaction energies between the ligand and protein, we wanted to sample the complete set of all possible poses. We do this by iteratively generating poses and then clustering them into Voronoi-like families using rmsd as the distance metric. This is continued until the number of families stops changing as additional poses are added. For the cases considered here, we used an rmsd criterion of 2 Å in defining families, which generally leads to ∼50,000 poses partitioned into ∼2,000 families, for each of which we select the “family head” as the central pose. During the pose-generation process, no energies are calculated. To choose the best binding region, a quick but systematic coarse docking is first done by using 10,000 poses without attempting the iterative, complete sampling.
iii) Scoring.
To reduce computational cost, we wanted to minimize the number of poses for which an energy must be evaluated. Thus, scoring of the poses is broken into two steps. First, the protein–ligand interaction energy of each family head is calculated, and the families are ranked. Then, 90% of the families are eliminated based on the energy of the family head. Finally, the binding energies are calculated for all of the family members (i.e., “children”) in these 10% best families, and the poses are ranked with only the best 100 poses selected for further analysis. This hierarchical scoring procedure allows for a majority of the poses from the complete set (∼50,000) to be eliminated without energy calculations.
iv) Optimization and refinement.
The 100 best poses from step iii are further optimized and refined to identify the best poses. The first step is to dealanize, i.e., replace and reoptimize the alanized residues with the full hydrophobic side chains. Simultaneously, all side chains in the binding site are reoptimized (i.e., “SCREAMed”) by using SCREAM (34) in the presence of the specific ligand pose. Thus, we end up with 100 distinct sets of side-chain conformations, one for each of the 100 ligand poses. Then, each of these 100 systems is energy-minimized for 10 conjugate gradient steps. At this point, the 100 poses are rescored and 50% are eliminated. Then, another 50 steps of minimization are performed for these 50, with the poses again rescored. This final round of minimization is skipped during coarse docking.
GAG-Dock Modifications.
The small-molecule docking methodology (DarwinDock/GenDock) was adapted to GAG structures through the following changes. Sphere generation for flat protein surfaces requires alterations to the standard sphgen procedure (35). First, all spheres are generated with the “dotlim” parameter in sphgen set to −0.9, which allows spheres to be generated for flat surfaces. Second, to prevent the generation of deeply buried spheres that would be inaccessible to GAG ligands, a second set of spheres is generated by using a probe radius of 2.8 Å instead of the normal 1.4 Å. The normal (1.4-Å probe radius) set of spheres is compared with the restricted (2.8-Å set), and only spheres within 2.8 Å of the restricted set sphere are kept. This procedure generates spheres focused on the protein surface while preventing them from being so close to the surface to cause a large number of clashes with the protein during pose generation.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health (NIH) Grants R01 GM084724 (to L.C.H.-W.) and 5T32 GM07616 (to C.J.R. and G.M.M.). Initial support for this work was from NIH Grants R01-NS071112, R01-NS073115, and R01-AI040567 (to A.R.G., R.A., and W.A.G.), and support from National Science Foundation (NSF) Emerging Frontiers in Research and Innovation (EFRI)-Origami Design for Integration of Self-assembling Systems for Engineering Innovation (ODISSEI) Grant 1332411 was used to complete the project. The computers used in this research were funded by grants from the Defense University Research Instrument Program (to W.A.G.) and from NSF Materials Research Science and Engineering Center (MRSEC), Center for the Science and Engineering of Materials (CSEM) (equipment part of the NSF-MRSEC-CSEM).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1715093115/-/DCSupplemental.
References
- 1.Varki A. Biological roles of glycans. Glycobiology. 2017;27:3–49. doi: 10.1093/glycob/cww086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dube DH, Bertozzi CR. Glycans in cancer and inflammation–Potential for therapeutics and diagnostics. Nat Rev Drug Discov. 2005;4:477–488. doi: 10.1038/nrd1751. [DOI] [PubMed] [Google Scholar]
- 3.Sharma K, Selzer ME, Li S. Scar-mediated inhibition and CSPG receptors in the CNS. Exp Neurol. 2012;237:370–378. doi: 10.1016/j.expneurol.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu D, Esko JD. Demystifying heparan sulfate-protein interactions. Annu Rev Biochem. 2014;83:129–157. doi: 10.1146/annurev-biochem-060713-035314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gama CI, et al. Sulfation patterns of glycosaminoglycans encode molecular recognition and activity. Nat Chem Biol. 2006;2:467–473. doi: 10.1038/nchembio810. [DOI] [PubMed] [Google Scholar]
- 6.Rogers CJ, et al. Elucidating glycosaminoglycan-protein-protein interactions using carbohydrate microarray and computational approaches. Proc Natl Acad Sci USA. 2011;108:9747–9752. doi: 10.1073/pnas.1102962108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Handel TM, Johnson Z, Crown SE, Lau EK, Proudfoot AE. Regulation of protein function by glycosaminoglycans–As exemplified by chemokines. Annu Rev Biochem. 2005;74:385–410. doi: 10.1146/annurev.biochem.72.121801.161747. [DOI] [PubMed] [Google Scholar]
- 8.Brown JM, et al. A sulfated carbohydrate epitope inhibits axon regeneration after injury. Proc Natl Acad Sci USA. 2012;109:4768–4773. doi: 10.1073/pnas.1121318109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen Y, et al. PTPsigma is a receptor for chondroitin sulfate proteoglycan, an inhibitor of neural regeneration. Science. 2009;326:592–596. doi: 10.1126/science.1178310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coles CH, et al. Proteoglycan-specific molecular switch for RPTPσ clustering and neuronal extension. Science. 2011;332:484–488. doi: 10.1126/science.1200840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dickendesher TL, et al. NgR1 and NgR3 are receptors for chondroitin sulfate proteoglycans. Nat Neurosci. 2012;15:703–712. doi: 10.1038/nn.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Silver J, Miller JH. Regeneration beyond the glial scar. Nat Rev Neurosci. 2004;5:146–156. doi: 10.1038/nrn1326. [DOI] [PubMed] [Google Scholar]
- 13.Miller GM, Hsieh-Wilson LC. Sugar-dependent modulation of neuronal development, regeneration, and plasticity by chondroitin sulfate proteoglycans. Exp Neurol. 2015;274:115–125. doi: 10.1016/j.expneurol.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bartus K, James ND, Bosch KD, Bradbury EJ. Chondroitin sulphate proteoglycans: Key modulators of spinal cord and brain plasticity. Exp Neurol. 2012;235:5–17. doi: 10.1016/j.expneurol.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 15.Bradbury EJ, et al. Chondroitinase ABC promotes functional recovery after spinal cord injury. Nature. 2002;416:636–640. doi: 10.1038/416636a. [DOI] [PubMed] [Google Scholar]
- 16.Bitomsky W, Wade RC. Docking of glycosaminoglycans to heparin-binding proteins: Validation for aFGF, bFGF, and antithrombin and application to IL-8. J Am Chem Soc. 1999;121:3004–3013. [Google Scholar]
- 17.Forster M, Mulloy B. Computational approaches to the identification of heparin-binding sites on the surfaces of proteins. Biochem Soc Trans. 2006;34:431–434. doi: 10.1042/BST0340431. [DOI] [PubMed] [Google Scholar]
- 18.Takaoka T, Mori K, Okimoto N, Neya S, Hoshino T. Prediction of the structure of complexes comprised of proteins and glycosaminoglycans using docking simulation and cluster analysis. J Chem Theory Comput. 2007;3:2347–2356. doi: 10.1021/ct700029q. [DOI] [PubMed] [Google Scholar]
- 19.Pellegrini L, Burke DF, von Delft F, Mulloy B, Blundell TL. Crystal structure of fibroblast growth factor receptor ectodomain bound to ligand and heparin. Nature. 2000;407:1029–1034. doi: 10.1038/35039551. [DOI] [PubMed] [Google Scholar]
- 20.DiGabriele AD, et al. Structure of a heparin-linked biologically active dimer of fibroblast growth factor. Nature. 1998;393:812–817. doi: 10.1038/31741. [DOI] [PubMed] [Google Scholar]
- 21.Faham S, Hileman RE, Fromm JR, Linhardt RJ, Rees DC. Heparin structure and interactions with basic fibroblast growth factor. Science. 1996;271:1116–1120. doi: 10.1126/science.271.5252.1116. [DOI] [PubMed] [Google Scholar]
- 22.Schlessinger J, et al. Crystal structure of a ternary FGF-FGFR-heparin complex reveals a dual role for heparin in FGFR binding and dimerization. Mol Cell. 2000;6:743–750. doi: 10.1016/s1097-2765(00)00073-3. [DOI] [PubMed] [Google Scholar]
- 23.Plotnikov AN, Schlessinger J, Hubbard SR, Mohammadi M. Structural basis for FGF receptor dimerization and activation. Cell. 1999;98:641–650. doi: 10.1016/s0092-8674(00)80051-3. [DOI] [PubMed] [Google Scholar]
- 24.Bourin M-C, Lindahl U. Glycosaminoglycans and the regulation of blood coagulation. Biochem J. 1993;289:313–330. doi: 10.1042/bj2890313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCoy AJ, Pei XY, Skinner R, Abrahams J-P, Carrell RW. Structure of β-antithrombin and the effect of glycosylation on antithrombin’s heparin affinity and activity. J Mol Biol. 2003;326:823–833. doi: 10.1016/s0022-2836(02)01382-7. [DOI] [PubMed] [Google Scholar]
- 26.Fournier AE, GrandPre T, Strittmatter SM. Identification of a receptor mediating Nogo-66 inhibition of axonal regeneration. Nature. 2001;409:341–346. doi: 10.1038/35053072. [DOI] [PubMed] [Google Scholar]
- 27.He XL, et al. Structure of the Nogo receptor ectodomain: A recognition module implicated in myelin inhibition. Neuron. 2003;38:177–185. doi: 10.1016/s0896-6273(03)00232-0. [DOI] [PubMed] [Google Scholar]
- 28.Kim DE, Chivian D, Baker D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 2004;32(suppl 2):W526–W531. doi: 10.1093/nar/gkh468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cho AE, et al. The MPSim-Dock hierarchical docking algorithm: Application to the eight trypsin inhibitor cocrystals. J Comput Chem. 2005;26:48–71. doi: 10.1002/jcc.20118. [DOI] [PubMed] [Google Scholar]
- 30.Floriano WB, Vaidehi N, Zamanakos G, Goddard WA., 3rd HierVLS hierarchical docking protocol for virtual ligand screening of large-molecule databases. J Med Chem. 2004;47:56–71. doi: 10.1021/jm030271v. [DOI] [PubMed] [Google Scholar]
- 31.Charlier L, et al. How broadly tuned olfactory receptors equally recognize their agonists. Human OR1G1 as a test case. Cell Mol Life Sci. 2012;69:4205–4213. doi: 10.1007/s00018-012-1116-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim SK, Riley L, Abrol R, Jacobson KA, Goddard WA., 3rd Predicted structures of agonist and antagonist bound complexes of adenosine A3 receptor. Proteins. 2011;79:1878–1897. doi: 10.1002/prot.23012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim S-K, Li Y, Abrol R, Heo J, Goddard WA., 3rd Predicted structures and dynamics for agonists and antagonists bound to serotonin 5-HT2B and 5-HT2C receptors. J Chem Inf Model. 2011;51:420–433. doi: 10.1021/ci100375b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tak Kam VW, Goddard WA., 3rd Flat-bottom strategy for improved accuracy in protein side-chain placements. J Chem Theory Comput. 2008;4:2160–2169. doi: 10.1021/ct800196k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moustakas DT, et al. Development and validation of a modular, extensible docking program: DOCK 5. J Comput Aided Mol Des. 2006;20:601–619. doi: 10.1007/s10822-006-9060-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.