Significance
Leinamycin (LNM) is a promising anticancer drug lead, yet no analog has been isolated since its discovery nearly 30 y ago. By mining bacterial genomes, we discovered 49 potential producers of LNM-type natural products, the structural diversity of which was predicted based on bioinformatics and confirmed by in vitro characterization of selected enzymes and structural elucidation of the guangnanmycins and weishanmycins. These findings demonstrate the power of the discovery-based approach to combinatorial biosynthesis for natural product discovery and structural diversity. New members of the LNM family of natural products should greatly facilitate drug discovery and development. The LNM-type biosynthetic machineries provide outstanding opportunities to dissect and mimic Nature’s strategies for combinatorial biosynthesis and natural product structural diversity.
Keywords: combinatorial biosynthesis, leinamycin, natural products discovery, structural diversity, genome mining
Abstract
Nature’s ability to generate diverse natural products from simple building blocks has inspired combinatorial biosynthesis. The knowledge-based approach to combinatorial biosynthesis has allowed the production of designer analogs by rational metabolic pathway engineering. While successful, structural alterations are limited, with designer analogs often produced in compromised titers. The discovery-based approach to combinatorial biosynthesis complements the knowledge-based approach by exploring the vast combinatorial biosynthesis repertoire found in Nature. Here we showcase the discovery-based approach to combinatorial biosynthesis by targeting the domain of unknown function and cysteine lyase domain (DUF–SH) didomain, specific for sulfur incorporation from the leinamycin (LNM) biosynthetic machinery, to discover the LNM family of natural products. By mining bacterial genomes from public databases and the actinomycetes strain collection at The Scripps Research Institute, we discovered 49 potential producers that could be grouped into 18 distinct clades based on phylogenetic analysis of the DUF–SH didomains. Further analysis of the representative genomes from each of the clades identified 28 lnm-type gene clusters. Structural diversities encoded by the LNM-type biosynthetic machineries were predicted based on bioinformatics and confirmed by in vitro characterization of selected adenylation proteins and isolation and structural elucidation of the guangnanmycins and weishanmycins. These findings demonstrate the power of the discovery-based approach to combinatorial biosynthesis for natural product discovery and structural diversity and highlight Nature’s rich biosynthetic repertoire. Comparative analysis of the LNM-type biosynthetic machineries provides outstanding opportunities to dissect Nature’s biosynthetic strategies and apply these findings to combinatorial biosynthesis for natural product discovery and structural diversity.
Natural products offer unmatched chemical and structural diversity compared with any other small-molecule families (1). Polyketides and nonribosomal peptides, including polyketide–nonribosomal peptide hybrids, are two of the most structurally diverse families of natural products that exhibit broad biological activities, and include some of the most important clinical drugs. While remarkably diverse in structure, the molecular logic for the biosynthesis of these natural products is deceivingly simple, at least conceptually, featuring modular polyketide synthases (PKSs), nonribosomal peptide synthetases (NRPSs), or PKS–NRPS hybrids. The complex and highly functionalized molecular scaffolds of polyketides, nonribosomal peptides, or hybrids thereof are derived from simple building blocks, for example, acyl-CoAs, amino acids, or both, which are activated, incorporated, and further modified as needed by dedicated PKS and NRPS domains and modules within the biosynthetic machinery (2–4). It is the modularity of PKSs and NRPSs that has provided the mechanistic foundation for and inspired the practice of combinatorial biosynthesis to generate polyketide and nonribosomal peptide structural diversity in the past three decades. Today, as an innovative technology, combinatorial biosynthesis has been applied to engineer the biosynthetic machinery of every family of natural products for the production of novel analogs (5–10).
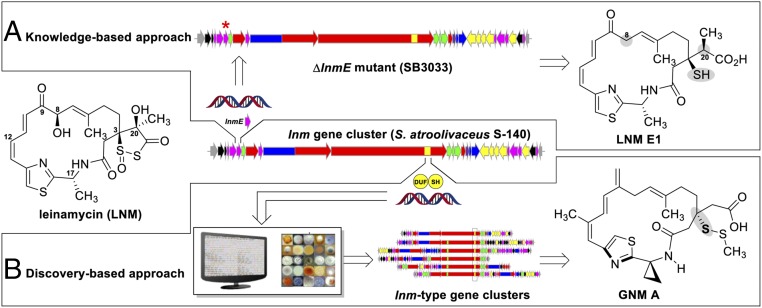
Combinatorial biosynthesis is traditionally defined as the generation of natural product analogs through the use of genetic engineering of biosynthetic pathways (5, 11) (Fig. 1). All existing combinatorial biosynthesis strategies are based on the collective knowledge of the biosynthetic machinery; that is, proteins of known function can be mapped back to their encoding genes, manipulation of which affords the engineered biosynthetic pathways that produce the designer natural products. This “knowledge-based” approach to combinatorial biosynthesis is limited, obviously, by what is known about the biosynthetic machinery and how this knowledge can be exploited to construct designer pathways (5–11). A typical outcome of knowledge-based combinatorial biosynthesis is the modification of one targeted functional moiety at a time, while keeping the rest of the natural product scaffold unchanged (Fig. 1).
Fig. 1.
Two complementary approaches to LNM structural diversity by combinatorial biosynthesis. (A) Knowledge-based approach by inactivating lnmE in S. atroolivaceus S-140, affording the SB3033 mutant strain that specifically produced LNM E1 (28). Shaded groups denote changes resulting from the ΔlnmE mutation. Red * indicates the lnmE gene that has been inactivated. (B) Discovery-based approach by targeting the DUF–SH didomain to mine bacterial genomes, affording strains that are predicted to produce a family of LNM-type natural products as exemplified by GNM A. The shaded group denotes the structural feature targeted by the DUF–SH didomain (27).
Through evolution, Nature has sampled innumerable combinations of genes, proteins, and pathways to create natural product structural diversity and optimize their biosynthesis and production (1, 11–13). To exploit what Nature has already developed, a “discovery-based” approach to combinatorial biosynthesis could be envisaged to search for new natural products, featuring targeted scaffolds or functional groups, by mining Nature’s biosynthetic reservoir using a key gene, or a fragment of it, as a signature probe (Fig. 1). In this context, the discovery-based approach to combinatorial biosynthesis provides an opportunity to explore natural product structural diversity by leveraging the preexisting combinatorial biosynthesis repertoire found in Nature (11, 14–17). Complementing the knowledge-based approach to combinatorial biosynthesis, the discovery-based approach affords natural products with a targeted structural moiety, while allowing the rest of the natural product scaffold to vary (Fig. 1).
Leinamycin (LNM), first isolated from Streptomyces atroolivaceus S-140 in 1989, features a unique 1,3-dioxo-1,2-dithiolane moiety that is spirofused to an 18-membered macrolactam ring (18, 19) (Fig. 1 and SI Appendix, Fig. S1). LNM exhibits potent antitumor activity and is active against tumors that are resistant to clinically important anticancer drugs. Upon reductive activation in the presence of cellular thiols, LNM exerts antitumor activity by an episulfonium ion-mediated DNA alkylation, a mode of action that is unprecedented among all DNA-damaging natural products (20) (SI Appendix, Fig. S1C). Therefore, LNM has been pursued as a promising anticancer drug lead (20).
We have previously cloned and sequenced the lnm gene cluster from S. atroolivaceus S-140 (21, 22) (SI Appendix, Fig. S1 A and B). Characterizations of the LNM biosynthetic machinery have since revealed novel chemistry, enzymology, and molecular logic for natural product biosynthesis. The thiazole-containing 18-membered macrolactam backbone of LNM is synthesized by a hybrid NRPS–PKS, featuring (i) the first experimentally confirmed acyltransferase (AT)-less type I PKS (22, 23), (ii) a novel pathway for β-alkylation in polyketide biosynthesis (24–26), and (iii) an unprecedented domain of unknown function (DUF) and cysteine lyase domain (SH) (i.e., the DUF–SH didomain) for sulfur incorporation into a polyketide backbone (27) (SI Appendix, Fig. S1B). These findings have enabled us to apply knowledge-based combinatorial biosynthesis strategies to the LNM biosynthetic machinery for LNM structural diversity, yielding leinamycin E1 (LNM E1) as the nascent product of the LNM hybrid NRPS–PKS (Fig. 1). Strikingly, LNM E1, complementary to reductive activation of LNM by cellular thiols, can be oxidatively activated by cellular reactive oxygen species (ROS) to generate a similar episulfonium ion, thereby alkylating DNA and leading to eventual cell death (28) (SI Appendix, Fig. S1C). LNM E1 therefore represents a novel anticancer drug lead that exploits the elevated level of ROS as a means to target cancer cells (28).
Since its discovery nearly 30 y ago (18, 19), LNM has remained the only member known for this family of natural products. Considering the potent antitumor activities of LNM and LNM E1, their fascinating modes of action, and the promise of LNM and LNM E1 as novel anticancer drug leads, as well as the rich chemistry and enzymology associated with the LNM biosynthetic machinery, we set out to search for novel members of the LNM family of natural products by exploring Nature’s combinatorial biosynthesis repertoire. We report here the use of the discovery-based approach to combinatorial biosynthesis by using the DUF–SH didomain, specific for sulfur incorporation from the LNM biosynthetic machinery, as a probe to target LNM analogs that feature the characteristic sulfur-containing moiety (Fig. 1). By mining the genomes from both public databases [National Center for Biotechnology Information (NCBI) and Joint Genome Institute (JGI)] and the actinomycetes strain collection at The Scripps Research Institute (TSRI), we discovered 49 potential producers that could be grouped into 18 distinct clades based on phylogenetic analysis. Further analysis of the representative genomes from each of the clades identified 28 lnm-type gene clusters. Structural diversity encoded by the LNM-type biosynthetic machineries was first predicted based on bioinformatics analysis and subsequently confirmed by isolation and structural elucidation of the guangnanmycins (GNMs) and the weishanmycins (WSMs), members of the LNM family of natural products, and in vitro scanning of the substrate specificities of adenylation proteins from representative biosynthetic machineries. These findings demonstrate the power of the discovery-based approach to combinatorial biosynthesis for natural product discovery and structural diversity (11–17).
Results
Genome Mining of Public Databases Identifying 19 Potential Producers of the LNM Family of Natural Products.
To explore Nature’s combinatorial biosynthesis repertoire for the LNM family of natural products, we first carried out a virtual survey of all bacterial genomes in public databases (as of March 2017, ∼48,780 bacterial genomes, including ∼12,000 actinobacterial genomes, are available in the NCBI and JGI online databases) using the DUF–SH didomain from the LNM biosynthetic machinery as a probe, identifying 19 lnm-type gene clusters from 19 different strains (SI Appendix, Fig. S2 and Table S1). Bioinformatics analysis revealed that each of the gene clusters featured a hybrid NRPS–AT–less type I PKS that consisted of two NRPS modules and six PKS modules and was predicted to assemble an LNM-like 18-membered macrolactam scaffold (Fig. 2 and SI Appendix, Fig. S3). Notably, among the 19 strains identified, 18 of them belong to the order Actinomycetales (SI Appendix, Fig. S2 and Table S1).
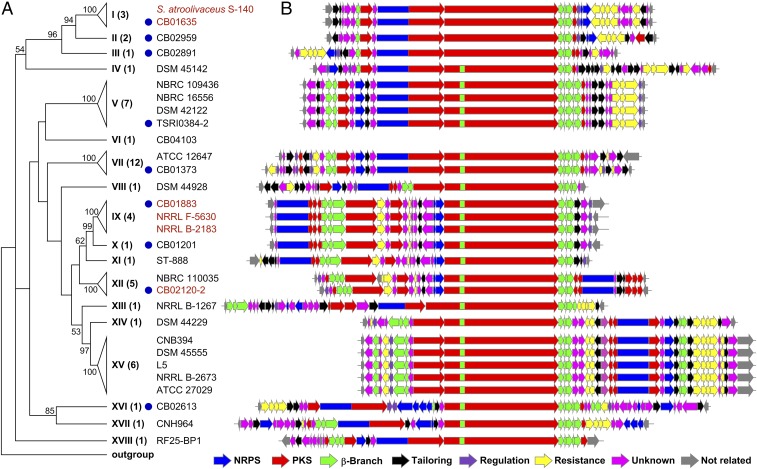
Fig. 2.
Survey of bacterial genomes (∼48,780) available from public databases and strains (∼5,000) from the actinomycetes collection at TSRI (including ∼100 from the Naicons collection and ∼500 from Myongji University), identifying 49 potential producers for LNM-type natural products by targeting the DUF–SH didomain. (A) Phylogenetic analysis of the 49 producers, based on the translated 1.2-kb internal fragment of DUF–SH didomains (27) and with S. atroolivaceus S-140 as a reference (21), affording 18 distinct clades (clades I to XVIII) when subjected to ∼70% amino acid identity cutoff (also see SI Appendix, Fig. S3B). CalE6 (AAM94792) from Micromonospora echinospora was used as the outgroup. Numbers in parentheses are the hits identified from each of the clades. Representative hits from each of the clades that have been genome-sequenced are listed. Blue dots indicate the nine hits from TSRI collection whose genomes have been sequenced. The strains from which the production of LNM-type natural products has been confirmed are highlighted in red. (B) The 28 lnm-type gene clusters from 17 of the 18 clades, in comparison with the lnm gene cluster (clade I), highlighting the rich structural diversities of the encoded family of LNM-type natural products. Clusters within the same clades are highly homologous, indicative of producing highly similar natural products. Genes are color-coded based on their proposed functions (see SI Appendix, Tables S4–S36 for annotations).
Genome Survey of 5,000 Actinomycetes at TSRI Identifying an Additional 30 Potential Producers of the LNM Family of Natural Products.
Inspired by the accuracy and specificity of the virtual genome mining, we next screened the actinomycetes strain collection at TSRI (17) for additional producers of the LNM family of natural products. We first adapted our recently developed high-throughput real-time PCR method for strain prioritization (14) (SI Appendix, Fig. S2), using degenerate primers that targeted a 0.6-kb internal fragment of the DUF–SH didomain-coding region (27) (SI Appendix, Tables S2 and S3). Upon surveying 5,000 strains from TSRI actinomycetes collection, we identified 72 hits, the identities of which were confirmed by sequencing a 1.2-kb internal fragment of the DUF–SH didomain-coding region; the translated amino acid sequences show 36 to 99% identity to the 19 DUF–SH didomains identified from public databases. The 72 initial hits were then dereplicated to 30 distinct hits on the basis of their 1.2-kb DNA sequences, taxonomy, and geographic locations where they were isolated (SI Appendix, Fig. S2 and Table S1). Finally, we sequenced the genomes of 9 representative hit strains, revealing that each strain contained one distinct lnm-type gene cluster (Fig. 2 and SI Appendix, Table S1).
Bioinformatics Analysis Revealing Remarkable Potential of Structural Diversity for the LNM Family of Natural Products.
We next subjected the 49 hit strains (19 from public databases and 30 from TSRI strain collection), with the known LNM producer S. atroolivaceus S-140 as a reference (21, 22), to phylogenetic analysis, using the amino acid sequences translated from the 1.2-kb internal fragments of the DUF–SH didomain-coding regions (27) (SI Appendix, Fig. S3B). The 49 hits could be grouped into 18 distinct clades when subjected to 70% amino acid identity cutoff (Fig. 2A). Modeled on the LNM biosynthetic machinery, biosynthesis of the LNM family of natural products could be divided into two stages: (i) biosynthesis of the nascent 18-membered macrolactam intermediates, such as LNM E1, from the acyl-CoA and amino acid building blocks by the hybrid NRPS–AT–less type I PKS; and (ii) tailoring of the nascent NRPS–PKS intermediates into the final natural products, such as LNM (27, 28) (SI Appendix, Fig. S1 A and B). Thus, using the lnm gene cluster as a reference (21, 22) (SI Appendix, Table S23), we annotated 28 representative lnm-type gene clusters from the total of 49 identified hit strains (Fig. 2B), including 19 from public databases (SI Appendix, Tables S4–S22) and 9 from TSRI strain collection (SI Appendix, Tables S23–S31). The 28 lnm-type gene clusters represented hits from 17 of the 18 clades (clade VI was the only exception for which the Streptomyces sp. CB04103 strain has not been sequenced) (Fig. 2A). In analogy to LNM, the 28 biosynthetic machineries all feature an eight-module hybrid NRPS–AT–less type I PKS biosynthetic machinery, including two NRPS modules (loading module and module-2), six PKS modules (module-3 to module-8), and enzymes for β-alkylation, all of which together are responsible for the biosynthesis of a nascent 18-membered macrolactam intermediate. Close examination of the domain compositions for each of the modules among the different biosynthetic machineries further revealed that while NRPS module-2 and PKS module-3, -4, and -8 were highly conserved, indicative of structural similarity, distinct features could be readily found within the NRPS loading module and PKS module-5, -6, and -7, indicative of structural variations among the resultant 18-membered macrolactam scaffolds (SI Appendix, Fig. S3). It is also worth noting that while the hits from different clades yield distinct lnm-type gene clusters, the hits from the same clade afford highly homologous lnm-type gene clusters, as exemplified by the multiple clusters from clades I, V, VII, IX, XII, and XV, respectively (Fig. 2 and SI Appendix, Tables S23 and S32–S36). These findings support our strategy of using DUF–SH didomain sequence phylogeny as a reference to further streamline genome mining of the hits for novel members of the LNM family of natural products.
NRPS loading module.
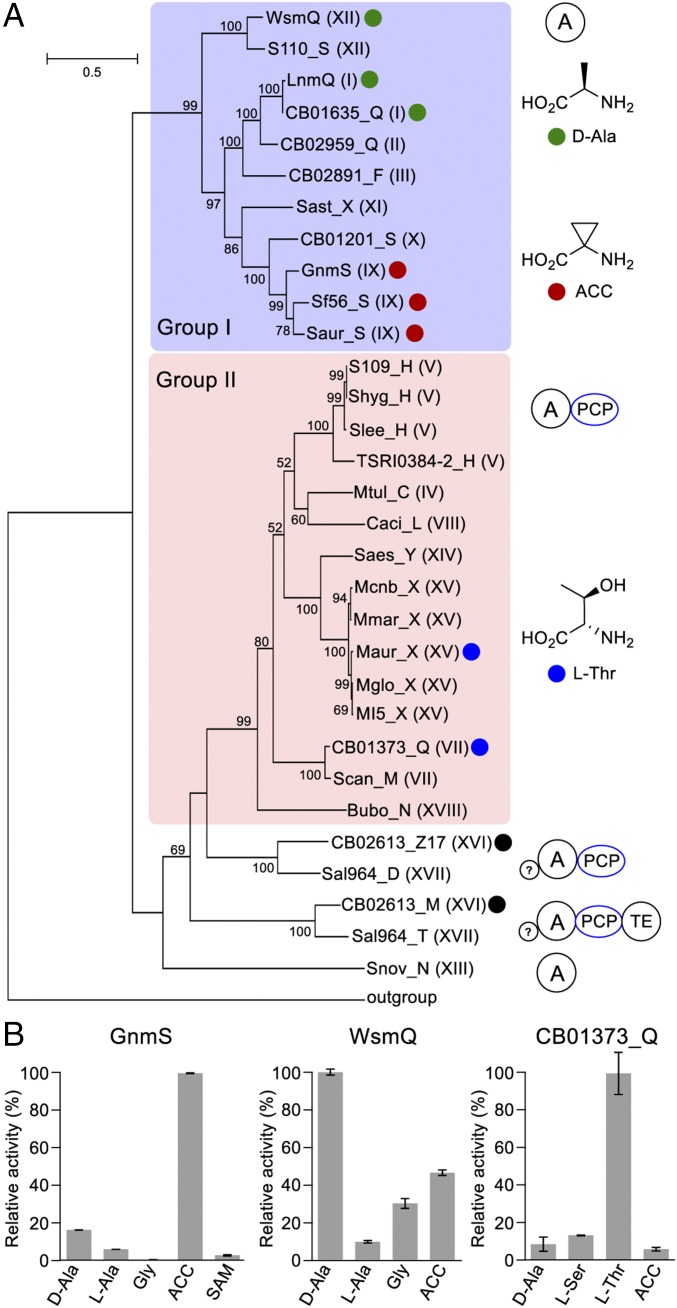
The NRPS loading module of the LNM biosynthetic machinery consists of an adenylation (A) protein LnmQ and a peptidyl carrier protein (PCP) LnmP, which are responsible for the selection, activation, and incorporation of the starting amino acid d-Ala into LNM, hence determining the substitution pattern at the C17 position for the LNM family of natural products (SI Appendix, Fig. S1B). Phylogenetic analysis of the A proteins, including LnmQ and its homologs from all of the LNM-type pathways, revealed two major groups (Figs. 2B and 3A). Homologs in the form of discrete A proteins (clades I to III and IX to XII) tend to cluster with LnmQ (group I), while homologs in the form of A domains, which are fused with their PCP counterparts (i.e., as an A–PCP didomain protein) (clades IV and V, VII and VIII, and XIV, XV, and XVIII), form group II (Figs. 2B and 3A). In group I, LnmQ is the only functionally characterized A protein that is specific for d-Ala (29), and the other members, with the exception of CB01635_Q, do not match any of the specificity-conferring codes for known A proteins (30) (SI Appendix, Table S37), indicating the potential for the activation of other amino acids. The A domains from group II are predicted to use l-Thr as a substrate according to the specificity-conferring codes for known A domains (30) (SI Appendix, Table S37). Furthermore, the LNM-type biosynthetic machineries encoded by CB02613 (clade XVI) and Salinispora arenicola CNH964 (clade XVII) each contains two putative A proteins that fell out of the two main groups based on phylogenetic analysis (Figs. 2B and 3A). In addition to the A–PCP didomain, CB02613_Z17 and Sal964_D each contains an extra N-terminal region (∼200 residues), while CB02613_M and Sal964_T each contains a similar N-terminal region and an additional C-terminal thioesterase (TE)-like domain. Amino acid substrate specificity of these putative A domains could not be predicted bioinformatically (30) (SI Appendix, Table S37).
Fig. 3.
Functional diversity of the A proteins or domains from LNM-type biosynthetic machineries. (A) Phylogenetic analysis of the A proteins from the 28 LNM-type machineries, in comparison with LnmQ, which specifies d-Ala (29), revealing two major groups. AfsK (BAA08229) from Streptomyces coelicolor was used as the outgroup. Roman numerals in parentheses refer to the corresponding clades shown in Fig. 2A. The architectures of A proteins from different groups are shown (Right), with A as a discrete protein (group I), A-PCP didomain (group II), and A-PCP accompanied by an extra N-terminal sequence (?) with/without an additional C-terminal thioesterase domain (the rest). The colored dots denote A proteins whose substrate specificities have been confirmed experimentally or deduced from the isolated natural products: green, d-Ala; red, ACC; blue, l-Thr; black, preferred substrate not detected among the 22 amino acids tested (also see SI Appendix, Table S37 for substrate specificities predicted based on the NRPS codes). (B) In vitro assay of representative A proteins to determine their substrate specificities, as exemplified by GnmS, WsmQ, and CB01373_Q that specify ACC, d-Ala, and l-Thr, respectively (also see SI Appendix, Fig. S29). Error bars are generated from three replicates.
PKS module-5, -6, and -7.
Different domain compositions of PKS module-5, -6, and -7 have been found in most of the LNM-type biosynthetic machineries (SI Appendix, Fig. S3C). While the collinear model of modular PKS allows correlations between domain organizations and the resulting structural moieties in the nascent polyketide products, in analogy to the LNM biosynthetic machinery (SI Appendix, Fig. S1B), the insertion of the enoyl-CoA hydratase (ECH2) domain in module-5, the lack of the acyl carrier protein (ACP6-1), methyltransferase (MT), or dehydratase (DH) domain in module-6, the missing of the DH domain in module-7, or any combination thereof, would suggest structural variations for the corresponding moieties of the nascent polyketide products (SI Appendix, Fig. S3).
β-Alkyl branches.
β-Alkylation of the growing polyketide intermediates imbues additional structural variations to the nascent polyketide products (24–26, 31). A β-alkyl branch is typically installed by the hydroxymethylglutaryl-CoA synthase (HCS)-catalyzed condensation of an acyl-S-ACP with a β-keto group of the growing ACP-tethered polyketide intermediate (31). The resultant β-hydroxyacyl-S-ACP intermediate would further undergo dehydration and/or decarboxylation catalyzed by ECH1 and ECH2, respectively (SI Appendix, Fig. S4). Thus, in analogy to the LNM biosynthetic machinery (21–29), potential variations that would lead to the diverse structural outcomes for the LNM family of natural products, including varying β-alkylation at C3 and C9, acetyl- or propionyl-S-ACP as the preferred substrates by HCSs, and different modifications after condensation by ECH1 alone or in combination with ECH2, have all been found within PKS module-5 or module-8 in the identified biosynthetic machineries (SI Appendix, Figs. S3–S5).
DUF–SH didomains.
While the domain organization of PKS module-8 is absolutely conserved among the identified biosynthetic machineries (SI Appendix, Fig. S3), close examination of the associated HCSs, which catalyze β-alkylation at the C3 position of the full-length β-ketoacyl-S-ACP intermediates attached to PKS module-8, revealed that they could be grouped into two clusters according to acetyl- or propionyl-S-ACP as the preferred substrates (SI Appendix, Figs. S3–S5). Since β-alkylation is the prerequisite for installing the SH group into the polyketide backbone by the DUF–SH didomain, as exemplified by the LNM biosynthetic machinery (27, 28) (SI Appendix, Fig. S1B), we wondered if there would be a correlation between β-alkylation and sulfur incorporation chemistry. Indeed, phylogenetic analysis of either the DUF or the SH domains yielded similar clustering as HCSs (SI Appendix, Fig. S5 vs. SI Appendix, Fig. S6), suggesting potentially distinct chemistry for the DUF–SH didomains to install the SH group into the polyketide backbones bearing varying β-branches at the C3 position.
Tailoring enzymes.
While previous studies of LNM biosynthesis allowed the correlation of the hybrid NRPS–AT–less type I PKS to the structural features of the nascent 18-membered macrolactam intermediate LNM E1 (21–29), the tailoring enzymes responsible for converting LNM E1 to LNM were poorly understood (27, 28) (SI Appendix, Fig. S1 A and B). To probe the novel tailoring enzymes encoded in the lnm-type gene clusters, we constructed a genome neighborhood network (GNN) (12, 17) to facilitate the overall analysis of conservation and variation of proteins from all of the LNM-type biosynthetic machineries (SI Appendix, Fig. S7). The GNN analysis revealed that homologs of LnmDEHXZ′ are fairly conserved, indicating their common functions in the biosynthesis of the LNM family of natural products. Notably, the biosynthetic machineries are diverse and rich in new chemistries, featuring enzymes that are unprecedented compared with those of the LNM biosynthetic machinery, such as methyltransferase, aminotransferase, hydrolase, FAD-dependent oxidoreductase, halogenase, and enzymes without predictable functions, hence suggesting tailoring modifications that would lead to additional structural diversity for the LNM family of natural products (Fig. 1 and SI Appendix, Fig. S7).
Guangnanmycins from Streptomyces sp. CB01883 and Weishanmycins from Streptomyces sp. CB02120-2 Confirming the Hits as Producers of the LNM Family of Natural Products.
All 49 hits were subjected to fermentation optimization and prioritized for natural product dereplication based on their metabolite profiles. Combined with structural novelties predicted according to the lnm-type gene clusters, S. sp. CB01883 and S. sp. CB02120-2 were selected to showcase the isolation and structural elucidation of new members of the LNM family of natural products.
GNMs from S. sp. CB01883.
Strain S. sp. CB01883 in clade IX contains the gnm gene cluster that differs from the lnm gene cluster in both genetic organization and encoded enzymes (Figs. 2 and 4A and SI Appendix, Fig. S7 and Table S28). To facilitate the identification of the gnm cluster-specific natural products, we first constructed the mutant strain SB21001 (i.e., ΔgnmB) (SI Appendix, Fig. S8), in which the gnmB gene encoding NRPS module-2 was deleted (SI Appendix, Fig. S9), and its complementation recombinant strain SB21002 (i.e., SB21001/pBS21005), in which the deleted gnmB gene was provided in trans by pBS21005 that constitutively expressed gnmB under the control of the strong promoter kasO* (SI Appendix, Tables S2 and S3). The CB01883 wild-type strain was then fermented under the same condition for LNM production by S. atroolivaceus S-140 (21, 22), with both SB21001 and SB21002 as controls. HPLC analysis of the fermentation revealed two distinct metabolites from the CB01883 wild-type strain, whose production was completely abolished in SB21001 and partially restored in SB21002, confirming that these metabolites are encoded by the gnm gene cluster (Fig. 4B, I–III). S. sp. CB01883 was isolated from a soil sample collected in Guangnan County, Yunnan Province, China, and we therefore named the metabolites from CB01883 guangnanmycins.
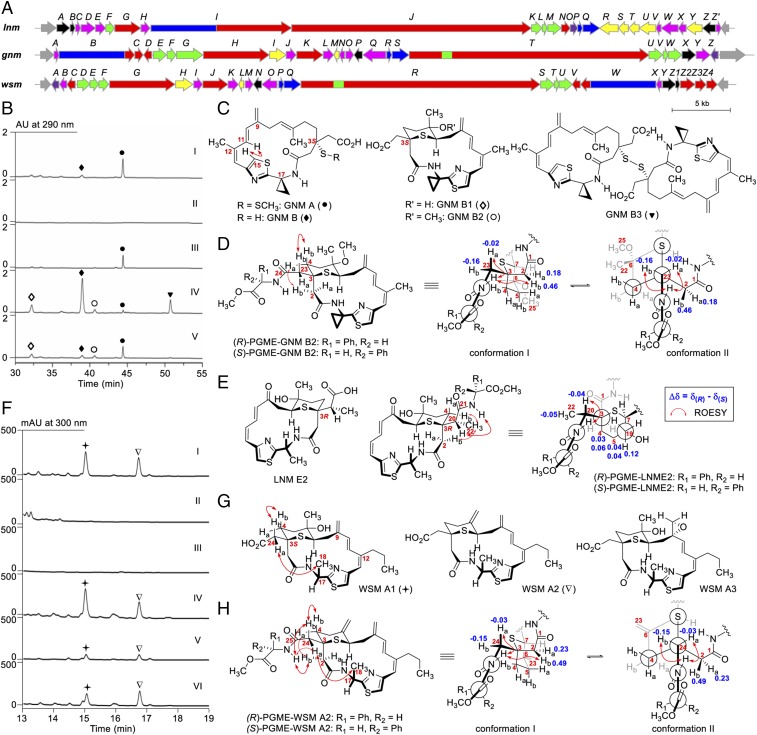
Fig. 4.
Discovery of GNMs and WSMs exemplifying the rich structural diversity of the LNM family of natural products. (A) Genetic organizations of the gnm and wsm gene clusters in comparison with the lnm gene cluster. (B) HPLC analysis of fermentations of the S. sp. CB01883 wild-type (I), SB21001 (i.e., ΔgnmB) (II), SB21002 (i.e., ΔgnmB/pBS21005) (III), SB21003 (i.e., ΔgnmO) (IV), and SB21004 (i.e., ΔgnmO/pBS21007) (V) mutant strains. (C) Structures of GNMs isolated from the S. sp. CB01883 wild-type (GNM A and B) and SB21003 mutant (GNM B, B1, B2, and B3). (D) Determination of the absolute configuration of GNMs at C3 to be S as shown based on the differences of the chemical shifts in 1H NMR of H2 and H23 between (R)- and (S)-PGME derivatives of GNM B2. Two major conformations are shown based on the analysis of their ROESY correlation signals (see SI Appendix, Fig. S14 for details). (E) Confirmation of the absolute configuration of LNM E2 at C3 to be R as shown based on the differences of the chemical shifts in 1H NMR of H4, H5, and H22 between the (R)- and (S)-PGME derivatives of LNM E2 (see SI Appendix, Fig. S16 for details). (F) HPLC analysis of fermentations of the S. sp. CB02120-2 wild-type (I), SB22001 (i.e., ΔwsmW) (II), SB22002 (i.e., ΔwsmZ3) (III), SB22003 (i.e., ΔwsmZ3/pBS22006) (IV), SB22004 (i.e., ΔwsmZ4) (V), and SB22005 (i.e., ΔwsmZ4/pBS22008) (VI) mutant strains. (G) Structures of WSMs isolated from the S. sp. CB02120-2 wild-type strain. (H) Determination of the absolute configuration of WSMs at C3 to be S as shown based on the differences of the chemical shifts in 1H NMR of H2 and H24 between (R)- and (S)-PGME derivatives of WSM A2 (see SI Appendix, Fig. S24 for details). Ha and Hb denote one of the two geminal hydrogens appearing at lower and higher field, respectively, in 1H NMR. GNM A, ●; GNM B, ◆; GNM B1, ◇; GNM B2, ○; GNM B3, ▼; WSM A1, ✦; WSM A2, ∇.
The two major metabolites from CB01883 were isolated and named GNM A (0.5 mg/L) and B (0.1 mg/L) (Fig. 4 B and C). The structures of GNM A and GNM B were established on the basis of high-resolution mass spectrometry (HRMS) and one-dimensional (1D) and 2D NMR spectroscopy (SI Appendix, Figs. S10–S12 and Tables S38 and S39). On the basis of their common biosynthesis (SI Appendix, Fig. S9), we proposed that GNM A and B share the same absolute configuration at C3, which was determined to be 3S, opposite to that found in LNM, by analyzing the differences of the chemical shifts in 1H NMR of H2 and H23 between the (R)- and (S)-phenylglycine methyl ester (PGME) derivatives of GNM B2 (32) (Fig. 4D and SI Appendix, Figs. S13–S15 and Table S40). This method, based on the anisotropic effect of the auxiliary PGME group to determine the absolute stereochemistry at C3, was first validated with LNM E2 as a positive control (Fig. 4E and SI Appendix, Figs. S13, S16, and S17 and Table S41), whose absolute configuration at C3 has been unambiguously established previously (28). In addition, several interesting structural features, including a cyclopropane ring at C17, methyl substitution at C12, terminal double bond at C9, carboxymethyl group at C3, and methyldithiol (or thiol) group at C3, are found in GNM A (or GNM B), all of which differ from those of LNM (Figs. 1 and 4C) and could be partially correlated with the variations predicted for NRPS module-1 and PKS module-3, -5, and -8, as well as the tailoring enzymes in the GNM biosynthetic machinery (SI Appendix, Figs. S1 and S9). Furthermore, the incorporation of a small strained cyclopropane ring into GNM A and B pushed the thiazole ring to a direction with its H15 pointing to the inside of the 18-membered macrolactam ring, which could be observed from the ROESY correlation between H15 and H11 (Fig. 4C and SI Appendix, Fig. S10). The spirofusion of a cyclopropane ring to the 18-membered macrolactam ring also changed the physicochemical property of the secondary N-cyclopropyl amide, and an increased ratio (∼2:1) of two rotamers was detected in DMSO-d6 solution at room temperature (33) (SI Appendix, Figs. S10 and S11).
WSMs from S. sp. CB02120-2.
Strain S. sp. CB02120-2 in clade XII harbors the wsm gene cluster with different genetic organization and encoded proteins from both the lnm and gnm gene clusters (Figs. 2 and 4A and SI Appendix, Fig. S7 and Table S30). To facilitate the discovery of wsm gene cluster-encoded natural products, we also made a mutant strain, SB22001 (i.e., ΔwsmW) (SI Appendix, Fig. S18), in which the wsmW gene encoding NRPS module-2 was deleted, hence inactivating the WSM biosynthetic machinery (SI Appendix, Fig. S19). Both the CB02120-2 wild-type and the SB22001 mutant strains were fermented under the same condition for LNM production by S. atroolivaceus S-140 (21, 22). Comparison of the metabolic profiles between the CB02120-2 wild-type and the SB22001 mutant strains by HPLC analysis revealed two distinct metabolites whose biosynthesis could be readily correlated to the wsm gene cluster (Fig. 4F, I and II). S. sp. CB02120-2 was isolated from a soil sample collected in Weishan County, Yunnan Province, China, and we therefore named the metabolites from CB02120-2 weishanmycins.
The two metabolites, together with a third metabolite produced in significantly lower titer that eluded initial HPLC analysis of the fermentation, were isolated from CB02120-2 and named WSM A1 (1.4 mg/L), A2 (1.5 mg/L), and A3 (0.2 mg/L). The structures of WSMs were similarly established based on extensive HRMS and 1D and 2D NMR analysis (Fig. 4G and SI Appendix, Figs. S20–S23 and Table S42). WSM A1, A2, and A3 were assumed to have the same absolute configuration at C3 based on their common biosynthesis (SI Appendix, Fig. S19), which was similarly determined to be 3S, the same as that found in GNMs, by analyzing the differences of the chemical shifts in 1H NMR of H2 and H24 between the (R)- and (S)-PGME derivatives of WSM A2 (Fig. 4H and SI Appendix, Figs. S13, S24, and S25 and Table S43). The 3S assignment was further supported by the NOE correlation between 24-Ha and 18-CH3 (Fig. 4H), which was attached to C17 with its absolute configuration being defined by the incorporation of d-Ala by the WSM NRPS loading module (i.e., WsmQP) (Fig. 3B and SI Appendix, Fig. S19). WSM A1, A2, and A3 all featured the characteristic tetrahydrothiopyran ring system, which could be envisaged deriving from WSM A, the proposed nascent 18-membered macrolactam intermediate of the WSM biosynthetic machinery, via a similar oxidative mechanism in analogy to LNM and LNM E1 (28) (SI Appendix, Figs. S1C and S19). However, all attempts to detect WSM A throughout the time course of CB02120-2 fermentation were unsuccessful.
Other than the new structural features already found in the GNMs, the presence of a propyl side chain at C12 of the WSMs is noteworthy, suggesting a unique propylmalonyl-derived extender unit being incorporated by WSM PKS module-3 (Fig. 4G). While no obvious differences in WSM PKS module-3 could be readily observed bioinformatically (SI Appendix, Fig. S3), we noticed the presence of a three-gene operon, wsmZ2–wsmZ3–wsmZ4, near the boundary of the wsm gene cluster (Fig. 4A and SI Appendix, Fig. S19), which encodes a crotonyl-CoA carboxylase/reductase (CCR), ketoacyl-acyl carrier protein synthase III (KAS III), and 3-hydroxyacyl-CoA dehydrogenase (HCDH). CCR and associated enzymes are known to mediate the formation of a variety of substituted malonyl-CoA derivatives that could be incorporated as extender units by PKSs into polyketide natural products (34). To investigate the role the WsmZ2/WsmZ3/WsmZ4 cassette may play in WSM biosynthesis, we deleted wsmZ3 or wsmZ4 in CB02120-2 to afford the mutant strains SB22002 (i.e., ΔwsmZ3) or SB22004 (i.e., ΔwsmZ4), respectively (SI Appendix, Fig. S26). We also prepared their corresponding complementation recombinant strains SB22003 (i.e., ΔwsmZ3/pBS22006) and SB22005 (i.e., ΔwsmZ4/pBS22008). All strains were fermented under the same condition as for WSM production, with the CB02120-2 wild-type strain as a control. HPLC analysis of fermentation showed that production of WSMs was completely abolished in SB22002 (i.e., ΔwsmZ3) or significantly reduced in SB22004 (i.e., ΔwsmZ4), and was restored to levels comparable to that of the wild type in SB22003 and SB22005, respectively (SI Appendix, Fig. S26). These findings support the proposal that the WSM biosynthetic machinery utilizes propylmalonyl-CoA as an extender unit to install the propyl group at C12 (SI Appendix, Fig. S19).
Streptomyces sp. CB01635 as an Alternative LNM E1 Producer and Streptomyces sp. NRRL F-5630 and Streptomyces aureofaciens NRRL B-2183 as Alternative GNM Producers.
It has been noted that strains within the same clade tend to harbor highly homologous gene clusters (Fig. 2). For example, S. sp. CB01635 fell into the same clade as the known LNM and LNM E1 producer S. atroolivaceus S-140 and harbored a gene cluster with identical genetic organization and ∼97%/98% identity of DNA–amino acid sequences to the lnm cluster from S. atroolivaceus S-140 (Fig. 2, clade I and SI Appendix, Table S23). Similarly, S. sp. NRRL F-5630 and S. aureofaciens NRRL B-2183 fell into the same clade as the GNM producer of S. sp. CB01883, and the three strains harbored a highly homologous gene cluster with identical genetic organization and ∼75%/72% identity of DNA–amino acid sequences (Fig. 2, clade IX and SI Appendix, Table S34). We subjected S. sp. CB01635, S. sp. NRRL F-5630, and S. aureofaciens NRRL B-2183 to fermentation under the same condition as for LNM production (21, 22), with S. atroolivaceus and S. sp. CB01883 as controls. LNM E1 production by S. sp. CB01635 (SI Appendix, Fig. S27) and GNM production by S. sp. NRRL F-5630 and S. aureofaciens NRRL B-2183 (SI Appendix, Fig. S28) were confirmed by HPLC analysis in comparison with authentic standards of LNM E1, GNM A, and GNM B, as well as HRMS analysis. These findings provided experimental evidence supporting the DUF–SH didomain sequence-based phylogeny to further streamline dereplication of the hits for the discovery of novel members of the LNM family of natural products. LNM was discovered originally from S. atroolivaceus S-140, isolated from a soil sample collected in Natori-shi, Miyagi, Japan (18, 19) and rediscovered recently from S. atroolivaceus THS-44, isolated from a soil sample collected in Baghdad, Iraq (35), while CB01635 was isolated from a soil sample collected in Guangnan County, Yunnan Province, China (SI Appendix, Table S1). For the three GNM producers, CB01883, NRRL F-5630, and NRRL B-2183 were isolated from soil samples collected in Guangnan County, Yunnan Province, China, an unknown location, and Missouri, United States, respectively (SI Appendix, Fig. S2 and Table S1). While CB01635 closely resembles S. atroolivaceus S-140 based on selected housekeeping genes, CB01883, NRRL F-5630, and NRRL B-2183 are quite distinct (SI Appendix, Fig. S3A). It is fascinating that varying strains from such geographically distant locations are found to harbor nearly identical gene clusters for the production of the same natural products. Multiple producers with varying growth characteristics and genetic amenability present great opportunities for yield improvement, structural diversity, or both by applying combinatorial biosynthesis and synthetic biology strategies and methods.
Characterization of the Adenylation Proteins with Varying Amino Acid Specificities at the NRPS Loading Modules Supporting Additional Structural Diversity for the LNM Family of Natural Products.
Bioinformatics analysis of the A proteins or domains of the NRPS loading modules, according to the specificity-conferring codes for known A domains, suggested varying amino acids priming the biosynthesis of the LNM family of natural products (29, 30) (Fig. 3A and SI Appendix, Table S37). To provide experimental evidence supporting these predictions, we took advantage of the hydroxylamine-trapping assay to quickly scan the amino acid substrate specificities of the A proteins or domains (36). Six representative A proteins or domains from different groups, including WsmQ and GnmS from group I, Maur_X and CB01373_Q from group II, as well as CB02613_M and CB02613_Z17, were overproduced as N-His6–tagged fusion proteins and purified (Fig. 3A and SI Appendix, Fig. S29). The six proteins were tested against an array of 22 different amino acids, including the 20 proteinogenic amino acids, d-Ala, and 1-aminocyclopropane-1-carboxylic acid (ACC), for substrate specificities. d-Ala and ACC were found to be the preferred substrates of WsmQ and GnmS, respectively, thus showing the presence of substrate variations of the A proteins in group I (Fig. 3 and SI Appendix, Fig. S29). These findings are consistent with the structures of WSMs and GNMs as encoded by the wsm and gnm gene clusters, respectively (Figs. 2 and 4). S-adenosyl methionine (SAM) was also tested for GnmS but was found not to be a substrate (Fig. 3B), indicative of a distinct pathway for ACC biosynthesis between GNM and colibactin (37). Both Maur_X and CB01373_Q from group II were confirmed to use l-Thr as their preferred substrate as predicted by the bioinformatics analysis, while CB02613_M and CB02613_Z17 did not show any obvious activities toward any of the 22 amino acids tested, implying their utilization of other unnatural amino acids (Fig. 3 and SI Appendix, Fig. S29 and Table S37).
Manipulation of GNM Biosynthesis in S. sp. CB01883 Further Enriching Structural Diversity for the LNM Family of Natural Products.
Since we can manipulate GNM biosynthesis in S. sp. CB01883, as exemplified by the construction of the ΔgnmB mutant SB21001, we wished to manipulate the GNM biosynthetic machinery to further enrich structural diversity for the LNM family of natural products. GnmO, a homolog of LnmX, is predicted to be an N-acetylglucosamine deacetylase belonging to the LmbE family, and its homologs are highly conserved among all of the LNM-type biosynthetic machineries (SI Appendix, Fig. S7 and Table S28). Thus, we constructed both the ΔgnmO mutant strain SB21003 (SI Appendix, Figs. S9 and S30) and its complementation recombinant strain SB21004 (i.e., SB21003/pBS21007) (SI Appendix, Table S2). Both SB21003 and SB21004 were fermented under the same condition as for GNM production, with the CB01883 wild-type strain as a control. HPLC analysis of the fermentation revealed that GNM A production in SB21003 was nearly abolished, with a concomitant production of GNM B (2.6 mg/L) as the major metabolite, together with three additional new metabolites. GNM A production in SB21004 was restored to a level comparable to the CB01883 wild type (Fig. 4B, I, IV, and V). These findings supported GnmO playing a critical role in the conversion of GNM B, the nascent 18-membered macrolactam intermediate, into GNM A, the final product of the GNM biosynthetic machinery (SI Appendix, Fig. S9).
GNM B, together with the three minor metabolites, were subsequently isolated from SB21003, and the structures of the three minor metabolites, named GNM B1, B2, and B3, were established on the basis of HRMS and 1D and 2D NMR analysis (Fig. 4C and SI Appendix, Figs. S10 and S31–S33 and Tables S39 and S44). GNM B features the same SH group at C3 as LNM E1, which is known to exert its DNA alkylation activity via an episulfonium ion intermediate upon oxidative activation and, in the absence of DNA, could undergo facile rearrangements to afford various products bearing the characteristic tetrahydrothiopyran ring system (28). The coisolation of GNM B1, B2, and B3, together with GNM B as the major metabolite from SB21003, therefore suggested that they could be derived from adventitious oxidation of GNM B during the fermentation or isolation process. Thus, to confirm the intermediacy of GNM B in the formation of GNM B1, B2, and B3, we treated GNM B with H2O2 in acetone and indeed found the formation of GNM B1 and B3 as major products (SI Appendix, Fig. S34 A and B). When the reaction was performed in methanol, GNM B2 was detected as the major product, together with GNM B1 and B3 (SI Appendix, Fig. S34 A and C). The similar reactivity toward oxidants and product distribution thereof between GNM B and LNM E1 would support that GNM B, like LNM E1 (SI Appendix, Fig. S1C), could be similarly activated upon exposure to ROS, hence serving as a promising anticancer drug lead targeting cancer cells known to be under high cellular oxidative stress (28).
Discussion
Nature’s ability to generate diverse natural products from simple building blocks, by evolving modular biosynthetic machineries in a combinatorial fashion, has provided inspiration for natural product biosynthesis (2–11). By manipulating Nature’s biosynthetic machineries, the knowledge-based approach to combinatorial biosynthesis has allowed the production of designer natural products and their analogs by rational metabolic pathway engineering and microbial fermentation (5–11). While successful, structural alterations engineered into the parent natural product scaffolds are often limited and very minor, and the designer natural products and their analogs are often produced in compromised titers (38, 39). This should not come as a surprise, given the fact that the biosynthetic machinery has been optimized by evolution to specifically produce the parent natural product. In this context, the discovery-based approach to combinatorial biosynthesis complements the knowledge-based approach by exploring the vast combinatorial biosynthesis repertoire found in Nature (10–17). The power of combinatorial biosynthesis by discovery is therefore the ability to probe Nature for a targeted structural motif while allowing the rest of the natural product scaffolds to change. Depending on the structural motifs targeted, the discovery-based approach could result in the discovery of natural products with similar core scaffolds but varying modifications or peripheral moieties, as exemplified by the LNM family of natural products reported in this study, or completely distinct scaffolds that share only the targeted motifs (11, 14–17). Since these biosynthetic machineries are the result of Nature’s combinatorial biosynthesis, they have been evolved to produce the specific natural products efficiently. They therefore provide outstanding opportunities to dissect biosynthetic strategies and learn from Nature how to efficiently practice combinatorial biosynthesis for natural product structural diversity.
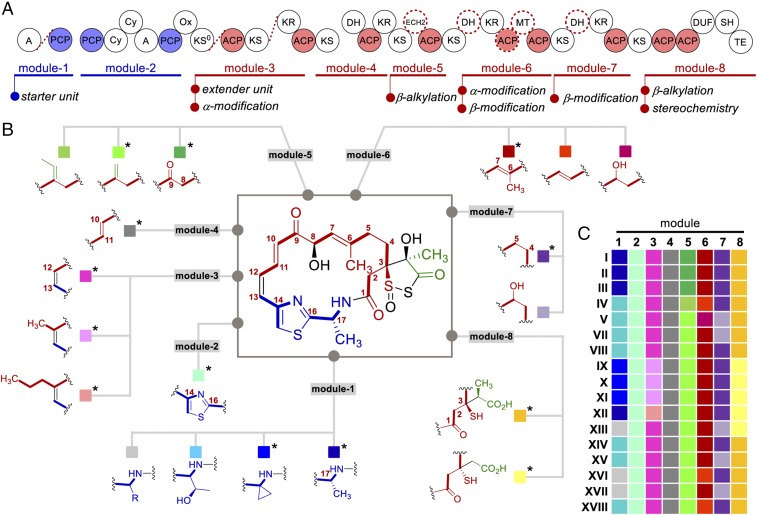
LNM was discovered by the traditional grind-and-find approach nearly 30 y ago (18, 19). With the exception of an alternative LNM producer (35), no LNM analog has been discovered in the past three decades. The unique molecular architecture of LNM remained unprecedented in any other natural products until the current study as exemplified by the GNMs and WSMs (Figs. 1 and 4). The lnm gene cluster was cloned and sequenced from S. atroolivaceus S-140 in 2003 (21, 22), and significant progress has been made since in characterizing the LNM biosynthetic machinery (23–29). However, despite great efforts, the knowledge-based approach to combinatorial biosynthesis for LNM structural diversity by manipulating the LNM machinery has met with limited success to date (25, 28). It is therefore remarkable that the discovery-based approach in this study by exploiting Nature’s combinatorial biosynthesis repertoire has resulted in the discovery of a family of LNM-type natural products (Fig. 1). Modeled on LNM, GNM, and WSM biosynthesis (SI Appendix, Figs. S1, S9, and S19), one could now start to piece together parts from the LNM-type biosynthetic machineries to construct a combinatorial biosynthetic platform to account for how Nature generates structural diversity for the LNM family of natural products (Fig. 5). Thus, the modularity of the LNM-type biosynthetic machineries sets the stage for Nature to practice combinatorial biosynthesis. Each module has its own evolutionary route to generate variants, as exemplified by changing the substrate specificities (e.g., starter unit for NRPS module-1 or extender unit for PKS module-3) or by varying modifications of the growing polyketide intermediates (e.g., α- or β-modifications for PKS module-3 and -6 or PKS module-5, -6, -7, and -8, respectively) (Fig. 5A). Combinations of the varying modules afford the biosynthesis of the LNM family of natural products, as exemplified by LNMs, GNMs, and WSMs (Figs. 1 and 5B). It is clear that Nature’s full combinatorial biosynthetic potential, in a Lego analogy (Fig. 5C), has just begun to be appreciated (23, 40, 41). Innovative strategies and technologies are now needed to discover these novel natural products.
Fig. 5.
Nature’s combinatorial biosynthesis for the LNM family of natural products. (A) The LNM-type biosynthetic machineries, featuring a hybrid NRPS–AT–less type I PKS with varying substrate specificity and modification domains, to account for the structural diversity found within the LNM family of natural products. Domains marked with red dotted circles vary among the machineries. Gaps between domains denote protein boundaries, with red dotted lines denoting that the two domains (enzymes) are fused in some of the machineries. (B) A composite structure depicting varying features of the LNM family of natural products that could be correlated with different modules shown with colored squares. Those marked with asterisks denote the structural motifs that have been discovered from the structures of LNMs, GNMs, and WSMs. (C) A mosaic view of the structural diversity of the LNM family of natural products, highlighting Nature’s intrinsic use of combinatorial biosynthesis. The roman numerals (I–V, VII–XVIII) represent the 17 different clades of potential producers of LNM-type natural products (Fig. 2A).
The LNM-type biosynthetic machineries also provide opportunities to unveil new chemistry and enzymology for natural product biosynthesis and to exploit the LNM family of natural products for drug discovery. For example, the GNMs feature ACC (Fig. 4C), a rare building block found in microbial natural products. While ACC biosynthesis in plants has been well-characterized (42), little is known about ACC biosynthesis in bacteria. A recent study revealed a PCP-tethered SAM as a key intermediate for biosynthesis of the ACC moiety of colibactin in Escherichia coli (37). The fact that ACC is directly activated and incorporated into GNMs by GnmS (Fig. 3B) would argue for a distinct pathway for ACC biosynthesis in S. sp. CB01883 (SI Appendix, Fig. S9). The WSMs feature a propyl branch at C12 (Fig. 4G), indicative that the WSM type I AT-less PKS is capable of selectively incorporating both malonyl- and propylmalonyl-CoAs as extender units in WSM biosynthesis. The WSM type I PKS joins a growing family of AT-less PKSs that are capable of incorporating multiple extender units in polyketide biosynthesis, whose utility in combinatorial biosynthesis has just begun to be appreciated (22, 23, 43). Most intriguing is the finding that both GNMs and WSMs feature a 3S configuration (Fig. 4 D and H), which is opposite to the 3R configuration of LNM E2 (28) (Fig. 4E). Discovery of the DUF–SH didomain from the LNM biosynthetic machinery unveiled a new family of PKS domains, expanding the chemistry and enzymology of PKSs (27). Comparative characterization of the DUF–SH didomains from the GNM, WSM, and LNM biosynthetic machineries has now promised to reveal new insights into the stereochemistry for β-thiol branching in polyketide biosynthesis. Finally, both LNM and LNM E1 have been pursued as promising anticancer drug leads, with distinct modes of action (20, 28), yet few analogs of LNM or LNM E1 are available for structure–activity relationship studies (44–47). The LNM family of natural products discovered in the current study therefore sets the stage to investigate their structure–activity relationship by exploring the new natural products directly or applying combinatorial biosynthetic strategies to the biosynthetic machineries to further enrich their structural diversity.
Materials and Methods
Materials, methods, and detailed experimental procedures are provided in SI Appendix. SI Appendix tables include: the summary of taxonomy, geographic origin, and genome sequence information of the 49 hit strains (SI Appendix, Table S1); strains, plasmids, and primers used (SI Appendix, Tables S2 and S3); annotation of the 19 lnm-type gene clusters from public databases (SI Appendix, Tables S4–S22); the 9 lnm-type gene clusters from the strain collection at TSRI (SI Appendix, Tables S23–S31); comparison of the lnm-type gene clusters within the same clades (SI Appendix, Tables S32–S36); amino acid substrate specificity for the A proteins or domains predicted according to the NRPS codes (SI Appendix, Table S37); and 1H and 13C NMR data of GNMs and WSMs, as well as the PGME derivatives of GNM B2, WSM A1, and LNM E1 (SI Appendix, Tables S38–S44). SI Appendix figures include: the proposed biosynthetic pathway and modes of action for LNM and LNM E1 (SI Appendix, Fig. S1); scheme depicting the discovery-based approach to the LNM family of natural products by genome mining of the DUF–SH didomains (SI Appendix, Fig. S2); phylogenetic analysis of the hits based on the DNA sequences of selected housekeeping genes, amino acid sequences of their DUF–SH domains, and organization and the proposed or identified nascent 18-membered macrolactams of LNM-type biosynthetic machineries from 17 distinct clades (SI Appendix, Fig. S3); proposed pathways for β-alkylation in the biosynthesis of the LNM family of natural products (SI Appendix, Fig. S4); predicting substrate specificity of HCSs by phylogenetic analysis (SI Appendix, Fig. S5); correlation between DUF–SH domains and the substrates for β-alkylation at C3 by phylogenetic analysis (SI Appendix, Fig. S6); GNN predicting novel chemistry for the biosynthesis of the LNM family of natural products (SI Appendix, Fig. S7); construction of the ΔgnmB mutant strain SB21001 in S. sp. CB01883 (SI Appendix, Fig. S8); proposed biosynthetic pathway for the GNMs (SI Appendix, Fig. S9); 1H and 13C NMR spectra and key correlations supporting structural elucidation of the GNMs and determination of their absolute stereochemistry at C3 in comparison with LNM E1 (SI Appendix, Figs. S10–S17); construction of the ΔwsmW mutant strain SB22001 in S. sp. CB02120-2 (SI Appendix, Fig. S18); proposed biosynthetic pathway for the WSMs (SI Appendix, Fig. S19); 1H and 13C NMR spectra and key correlations supporting structural elucidation of the WSMs and determination of their absolute stereochemistry at C3 in comparison with GNM B2 and LNM E2 (SI Appendix, Figs. S20–S25); construction of the ΔwsmZ3 mutant strain SB22002 and the ΔwsmZ4 mutant strain SB22004 in S. sp. CB02120-2 (SI Appendix, Fig. S26); confirmation of S. sp. CB01635 as an alternative LNM E1 producer (SI Appendix, Fig. S27) and S. sp. NRRL F-5630 and S. aureofaciens NRRL B-2183 as alternative GNM producers (SI Appendix, Fig. S28); in vitro characterization of selected A proteins or domains (SI Appendix, Fig. S29); construction of the ΔgnmO mutant strain SB21003 in S. sp. CB01883 (SI Appendix, Fig. S30); and 1H and 13C NMR spectra of GNM B1, B2, B3 and conversion of GNM B to GNM B1, B2, and B3 upon oxidation (SI Appendix, Figs. S31–S34).
Supplementary Material
Acknowledgments
We thank Kyowa Hakko Kogyo Co. Ltd. for the S. atroolivaceus S-140 wild-type strain, Drs. Paolo Monciardini and Stefano Donadio, Naicons Srl for selected strains from the Naicons strain collection, and the Next Generation Sequencing core and NMR core facilities at TSRI for genome sequencing and 1D and 2D NMR analysis, respectively. This work was supported in part by Chinese Ministry of Education 111 Project B08034 (to Y.D.), National High Technology Joint Research Program of China Grant 2011ZX09401-001 (to Y.D.), National High Technology Research and Development Program of China Grant 2012AA02A705 (to Y.D.), Cooperative Research Program for Agriculture Science and Technology Development, Rural Development Administration, Korea, Project PJ01128901 (to J.-W.S.), and NIH Grant CA106150 (to B.S.). Z.G. was supported in part by the Institute of Tropical Bioscience and Biotechnology, Chinese Academy of Tropical Agricultural Sciences and a scholarship from the China Scholarship Council (201403260013). This is manuscript 29599 from The Scripps Research Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the National Center for Biotechnology Information database. See SI Appendix, Table S1 for a summary of all accession numbers.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1716245115/-/DCSupplemental.
References
- 1.Newman DJ, Cragg GM. Natural products as sources of new drugs from 1981 to 2014. J Nat Prod. 2016;79:629–661. doi: 10.1021/acs.jnatprod.5b01055. [DOI] [PubMed] [Google Scholar]
- 2.Shen B. Polyketide biosynthesis beyond the type I, II and III polyketide synthase paradigms. Curr Opin Chem Biol. 2003;7:285–295. doi: 10.1016/s1367-5931(03)00020-6. [DOI] [PubMed] [Google Scholar]
- 3.Fischbach MA, Walsh CT. Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: Logic, machinery, and mechanisms. Chem Rev. 2006;106:3468–3496. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- 4.Walsh CT. The chemical versatility of natural-product assembly lines. Acc Chem Res. 2008;41:4–10. doi: 10.1021/ar7000414. [DOI] [PubMed] [Google Scholar]
- 5.Cane DE, Walsh CT, Khosla C. Harnessing the biosynthetic code: Combinations, permutations, and mutations. Science. 1998;282:63–68. doi: 10.1126/science.282.5386.63. [DOI] [PubMed] [Google Scholar]
- 6.Van Lanen SG, Shen B. Progress in combinatorial biosynthesis for drug discovery. Drug Discov Today Technol. 2006;3:285–292. doi: 10.1016/j.ddtec.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 7.Kim E, Moore BS, Yoon YJ. Reinvigorating natural product combinatorial biosynthesis with synthetic biology. Nat Chem Biol. 2015;11:649–659. doi: 10.1038/nchembio.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen B. A new golden age of natural products drug discovery. Cell. 2015;163:1297–1300. doi: 10.1016/j.cell.2015.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smanski MJ, et al. Synthetic biology to access and expand nature’s chemical diversity. Nat Rev Microbiol. 2016;14:135–149. doi: 10.1038/nrmicro.2015.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katz L, Baltz RH. Natural product discovery: Past, present, and future. J Ind Microbiol Biotechnol. 2016;43:155–176. doi: 10.1007/s10295-015-1723-5. [DOI] [PubMed] [Google Scholar]
- 11.Rudolf JD, Cronovčić I, Shen B. The role of combinatorial biosynthesis in natural products discovery. In: Newman DJ, Cragg GM, Grothaus P, editors. Chemical Biology of Natural Products. Taylor and Francis; Boca Raton, FL: 2017. pp. 87–125. [Google Scholar]
- 12.Rudolf JD, Yan X, Shen B. Genome neighborhood network reveals insights into enediyne biosynthesis and facilitates prediction and prioritization for discovery. J Ind Microbiol Biotechnol. 2016;43:261–276. doi: 10.1007/s10295-015-1671-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pye CR, Bertin MJ, Lokey RS, Gerwick WH, Linington RG. Retrospective analysis of natural products provides insights for future discovery trends. Proc Natl Acad Sci USA. 2017;114:5601–5606. doi: 10.1073/pnas.1614680114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hindra, et al. Strain prioritization for natural product discovery by a high-throughput real-time PCR method. J Nat Prod. 2014;77:2296–2303. doi: 10.1021/np5006168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Owen JG, et al. Multiplexed metagenome mining using short DNA sequence tags facilitates targeted discovery of epoxyketone proteasome inhibitors. Proc Natl Acad Sci USA. 2015;112:4221–4226. doi: 10.1073/pnas.1501124112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ju KS, et al. Discovery of phosphonic acid natural products by mining the genomes of 10,000 actinomycetes. Proc Natl Acad Sci USA. 2015;112:12175–12180. doi: 10.1073/pnas.1500873112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan X, et al. Strain prioritization and genome mining for enediyne natural products. MBio. 2016;7:e02104-16. doi: 10.1128/mBio.02104-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hara M, et al. DC 107, a novel antitumor antibiotic produced by a Streptomyces sp. J Antibiot (Tokyo) 1989;42:333–335. doi: 10.7164/antibiotics.42.333. [DOI] [PubMed] [Google Scholar]
- 19.Hara M, et al. Leinamycin, a new antitumor antibiotic from Streptomyces: Producing organism, fermentation and isolation. J Antibiot (Tokyo) 1989;42:1768–1774. doi: 10.7164/antibiotics.42.1768. [DOI] [PubMed] [Google Scholar]
- 20.Viswesh V, Gates K, Sun D. Characterization of DNA damage induced by a natural product antitumor antibiotic leinamycin in human cancer cells. Chem Res Toxicol. 2010;23:99–107. doi: 10.1021/tx900301r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang GL, Cheng YQ, Shen B. Leinamycin biosynthesis revealing unprecedented architectural complexity for a hybrid polyketide synthase and nonribosomal peptide synthetase. Chem Biol. 2004;11:33–45. doi: 10.1016/j.chembiol.2003.12.014. [DOI] [PubMed] [Google Scholar]
- 22.Cheng YQ, Tang GL, Shen B. Type I polyketide synthase requiring a discrete acyltransferase for polyketide biosynthesis. Proc Natl Acad Sci USA. 2003;100:3149–3154. doi: 10.1073/pnas.0537286100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lohman JR, et al. Structural and evolutionary relationships of “AT-less” type I polyketide synthase ketosynthases. Proc Natl Acad Sci USA. 2015;112:12693–12698. doi: 10.1073/pnas.1515460112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu T, Huang Y, Shen B. Bifunctional acyltransferase/decarboxylase LnmK as the missing link for β-alkylation in polyketide biosynthesis. J Am Chem Soc. 2009;131:6900–6901. doi: 10.1021/ja9012134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang Y, et al. Characterization of the lnmKLM genes unveiling key intermediates for β-alkylation in leinamycin biosynthesis. Org Lett. 2011;13:498–501. doi: 10.1021/ol102838y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lohman JR, Bingman CA, Phillips GN, Jr, Shen B. Structure of the bifunctional acyltransferase/decarboxylase LnmK from the leinamycin biosynthetic pathway revealing novel activity for a double-hot-dog fold. Biochemistry. 2013;52:902–911. doi: 10.1021/bi301652y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma M, Lohman JR, Liu T, Shen B. C-S bond cleavage by a polyketide synthase domain. Proc Natl Acad Sci USA. 2015;112:10359–10364. doi: 10.1073/pnas.1508437112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang SX, et al. Leinamycin E1 acting as an anticancer prodrug activated by reactive oxygen species. Proc Natl Acad Sci USA. 2015;112:8278–8283. doi: 10.1073/pnas.1506761112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang GL, Cheng YQ, Shen B. Chain initiation in the leinamycin-producing hybrid nonribosomal peptide/polyketide synthetase from Streptomyces atroolivaceus S-140. Discrete, monofunctional adenylation enzyme and peptidyl carrier protein that directly load D-alanine. J Biol Chem. 2007;282:20273–20282. doi: 10.1074/jbc.M702814200. [DOI] [PubMed] [Google Scholar]
- 30.Stachelhaus T, Mootz HD, Marahiel MA. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem Biol. 1999;6:493–505. doi: 10.1016/S1074-5521(99)80082-9. [DOI] [PubMed] [Google Scholar]
- 31.Calderone CT. Isoprenoid-like alkylations in polyketide biosynthesis. Nat Prod Rep. 2008;25:845–853. doi: 10.1039/b807243d. [DOI] [PubMed] [Google Scholar]
- 32.Yabuuchi T, Kusumi T. Phenylglycine methyl ester, a useful tool for absolute configuration determination of various chiral carboxylic acids. J Org Chem. 2000;65:397–404. doi: 10.1021/jo991218a. [DOI] [PubMed] [Google Scholar]
- 33.González-de-Castro Á, Broughton H, Martínez-Pérez JA, Espinosa JF. Conformational features of secondary N-cyclopropyl amides. J Org Chem. 2015;80:3914–3920. doi: 10.1021/acs.joc.5b00236. [DOI] [PubMed] [Google Scholar]
- 34.Wilson MC, Moore BS. Beyond ethylmalonyl-CoA: The functional role of crotonyl-CoA carboxylase/reductase homologs in expanding polyketide diversity. Nat Prod Rep. 2012;29:72–86. doi: 10.1039/c1np00082a. [DOI] [PubMed] [Google Scholar]
- 35.Hassani HH, Kadhim TA, Al-Shimary AM. Cytotoxic and apoptotic activity of leinamycin produced by Streptomyces atroolivaceus THS-44 isolate from Iraqi soil. Eur J Exp Biol. 2013;3:301–306. [Google Scholar]
- 36.Kadi N, Challis GL. Siderophore biosynthesis: A substrate specificity assay for nonribosomal peptide synthetase-independent siderophore synthetases involving trapping of acyl-adenylate intermediates with hydroxylamine. Methods Enzymol. 2009;458:431–457. doi: 10.1016/S0076-6879(09)04817-4. [DOI] [PubMed] [Google Scholar]
- 37.Zha L, et al. Colibactin assembly line enzymes use S-adenosylmethionine to build a cyclopropane ring. Nat Chem Biol. 2017;13:1063–1065. doi: 10.1038/nchembio.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McDaniel R, et al. Multiple genetic modifications of the erythromycin polyketide synthase to produce a library of novel “unnatural” natural products. Proc Natl Acad Sci USA. 1999;96:1846–1851. doi: 10.1073/pnas.96.5.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xue Q, Ashley G, Hutchinson CR, Santi DV. A multiplasmid approach to preparing large libraries of polyketides. Proc Natl Acad Sci USA. 1999;96:11740–11745. doi: 10.1073/pnas.96.21.11740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nguyen T, et al. Exploiting the mosaic structure of trans-acyltransferase polyketide synthases for natural product discovery and pathway dissection. Nat Biotechnol. 2008;26:225–233. doi: 10.1038/nbt1379. [DOI] [PubMed] [Google Scholar]
- 41.Nett M, Ikeda H, Moore BS. Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat Prod Rep. 2009;26:1362–1384. doi: 10.1039/b817069j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harpaz-Saad S, Yoon GM, Mattoo AK, Kieber JJ. The formation of ACC and competition between polyamines and ethylene for SAM. Annu Plant Rev. 2012;44:53–81. [Google Scholar]
- 43.Helfrich EJN, Piel J. Biosynthesis of polyketides by trans-AT polyketide synthases. Nat Prod Rep. 2016;33:231–316. doi: 10.1039/c5np00125k. [DOI] [PubMed] [Google Scholar]
- 44.Kanda Y, et al. Synthesis and antitumor activity of leinamycin derivatives: Modifications of C-8 hydroxy and C-9 keto groups. Bioorg Med Chem Lett. 1998;8:909–912. doi: 10.1016/s0960-894x(98)00133-4. [DOI] [PubMed] [Google Scholar]
- 45.Kanda Y, et al. Synthesis and antitumor activity of novel thioester derivatives of leinamycin. J Med Chem. 1999;42:1330–1332. doi: 10.1021/jm9900366. [DOI] [PubMed] [Google Scholar]
- 46.Kanda Y, Ashizawa T, Kawashima K, Ikeda S, Tamaoki T. Synthesis and antitumor activity of novel C-8 ester derivatives of leinamycin. Bioorg Med Chem Lett. 2003;13:455–458. doi: 10.1016/s0960-894x(02)00949-6. [DOI] [PubMed] [Google Scholar]
- 47.Liu T, et al. Synthesis and evaluation of 8,4′-dideshydroxy-leinamycin revealing new insights into the structure-activity relationship of the anticancer natural product leinamycin. Bioorg Med Chem Lett. 2015;25:4899–4902. doi: 10.1016/j.bmcl.2015.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.