Significance
One of the unresolved mysteries of tailless bacteriophages is how they recognize potential targets and translocate their genomes across the periplasmic space of their hosts. In this study, bilayers consisting of lipopolysaccharides (LPS) derived from bacterial cells were found to trigger genome ejection from ΦX174. We investigated the structural response of ΦX174 and showed that the phage binds to LPS through one of its pentameric spikes. Dissociation of the spike, followed by conformational changes in the major capsid proteins, cause DNA ejection through preformed tubes consisting of viral H proteins. This unique infection strategy may give ΦX174 and other members of the Microviridae family an evolutionary advantage by allowing them to protect the DNA conduit until a specific target is identified.
Keywords: bacteriophage, ΦX174, lipopolysaccharide, cryo-EM, DNA ejection
Abstract
Unlike tailed bacteriophages, which use a preformed tail for transporting their genomes into a host bacterium, the ssDNA bacteriophage ΦX174 is tailless. Using cryo-electron microscopy and time-resolved small-angle X-ray scattering, we show that lipopolysaccharides (LPS) form bilayers that interact with ΦX174 at an icosahedral fivefold vertex and induce single-stranded (ss) DNA genome ejection. The structures of ΦX174 complexed with LPS have been determined for the pre- and post-ssDNA ejection states. The ejection is initiated by the loss of the G protein spike that encounters the LPS, followed by conformational changes of two polypeptide loops on the major capsid F proteins. One of these loops mediates viral attachment, and the other participates in making the fivefold channel at the vertex contacting the LPS.
Tailed bacteriophages use their tail complexes for host cell recognition, adsorption, and penetration of bacterial cell walls (1–4). However, it remains unclear how tailless phages like ΦX174 perform tail-associated functions. ΦX174 is a small icosahedral, tailless bacteriophage with a maximum diameter of ∼320 Å. The capsid contains a 5.3-kb circular, single-stranded (ss) DNA genome that encodes four structural proteins: the capsid protein F, the spike protein G, the genome-associated protein J, and the DNA pilot protein H. The ΦX174 structure has T = 1 icosahedral symmetry (5), with 60 F proteins forming the capsid decorated by 12 spikes on the vertices, each containing five G proteins (6).
The major capsid protein F consists of a “jelly roll” fold that has eight antiparallel β strands (B–I) and seven loop insertions. F proteins bind to G proteins through two of the insertions, loops EF (His73-Pro234) and FG (Trp243-Gly262). The N-terminal portion of loop EF forms a “bulge” on the surface of the capsid between the fivefold and twofold axes. On one side of the bulge, Tyr158 interacts with the G protein, whereas the other side of the bulge is mostly exposed. The majority of known mutations affecting assembly are found in these two loops (7). Overall, the F proteins of homologous tailless phages are highly conserved (8), with the exception of these long loop insertions. Before or at the onset of infection, the H proteins assemble into a tube that translocates the genome across the host cell wall (9); however, the trigger that initiates this process is unknown.
Due to their potential to trigger the DNA ejection in vitro, lipopolysaccharides (LPS) have been used to study the mechanisms of infection of many tailed bacteriophages (2, 10). ΦX174-like viruses from Microviridae also use LPS as cellular receptors (11–15). LPS molecules are located on the distal leaflet of the outer membrane of Gram-negative bacteria and consist of three major components: hydrophobic lipid A tails, a hydrophilic polysaccharide core (R core), and repeating O-antigenic polysaccharide chains (O-antigen). The oligosaccharide composition varies among bacterial strains. Proteins G and H are known to interact with the LPS of suitable hosts in a species-specific manner (12, 14). Mutations affecting host cell recognition and/or the kinetics of DNA ejection have been isolated and identified in genes F, G, and H (16–18).
Here we show that the G and F proteins at one of the fivefold vertices recognize and interact with an LPS receptor to initiate DNA ejection. Infectious particles were incubated with LPS to identify the structural changes that led to DNA ejection. In vitro, purified LPS molecules formed membrane-like structures with two parallel layers to which ΦX174 particles were attached. The conformational changes of the ΦX174 particles during this reaction were initially characterized by small-angle X-ray scattering (SAXS). The structures of full and emptied ΦX174, complexed with these LPS bilayers, were then determined to ∼10-Å resolution by single particle cryo-electron microscopy (cryo-EM). In both cases, one of the G spikes was missing, and the virus was interacting with LPS through the exposed EF loops on the F proteins. After the genome had been ejected, the channel formed by the F proteins at the unique fivefold axis remained open.
Results
DNA Ejection Assays.
The loss of infectivity was measured to assess the effectiveness of LPS in triggering a response from the virion. After incubation at 33 °C for 20 min, the infectivity of the group containing LPS derived from Salmonella typhimurium decreased by >90%, while that of the control group containing only buffer decreased by 30%. To evaluate the specificity of this reaction, LPS from a ΦX174-insensitive Escherichia coli strain was tested in a similar manner, but failed to reduce infectivity. In time course experiments, most of the infectivity was lost during the first 5 min (Fig. S1), consistent with the results of previous in vivo studies (17, 19, 20). Both native and LPS-treated ΦX174 particles were examined by negative-stain EM, which showed that >90% of the particles had lost their genomes after a 20-min incubation with LPS (Fig. S1 C and D). These results suggest that S. typhimurium-derived LPS triggers ΦX174 genome ejection and could be used in the subsequent structural studies.
SAXS.
SAXS was used to study the LPS-induced conformational response of ΦX174 in solution. Model structures aided in the interpretation of features reported by SAXS. For regularly shaped particles with well-defined dimensions, such as full or empty phages, the scattering profiles display a series of maxima and minima, whose positions are determined by particle structure (21). Theoretical SAXS profiles for both types of particle are shown in Fig. 1A. Their differences are characterized by two features: (i) reduced forward scattering intensity [I(q = 0)] of the empty particles relative to full particles, which reflects their lower mass, and (ii) a shift toward lower angles in the positions of the maxima and minima, which reflects an increase in radius of gyration (Rg). Rg is determined by the mean distance between all pairs of the electrons in the sample, and hence is larger for the empty particles. Any loss of rotational symmetry affects the scattering profile by decreasing the depth of the first minimum (without a change in position), as illustrated in Fig. 1B.
Fig. 1.
Time-resolved SAXS data of ΦX174 genome ejection. (A and B) Simulated SAXS profiles of empty capsid, capsid with DNA, and full capsid with asymmetric addition. A illustrates the impact on the scattering profile of increasing the particle’s radius of gyration. B illustrates the impact of losing rotational symmetry by the addition of a model projection at only one vertex. For this model scenario, the mass of the addition is insufficient to significantly alter the overall radius of gyration, and hence the peak position appears unchanged. (C) Experimental time-resolved SAXS data showing the loss of symmetry and fullness of the virion over time. (D and E) Radius of gyration and depth of the first minimum as a function of time. The dotted line represents the noise level in the SAXS data.
Scattering profiles for the full phage (acquired before the addition of LPS; t = 0 data point) and empty phages (acquired using degraded procapsids lacking DNA and the external scaffolding protein) are shown in Fig. 1C. For time-resolved data, LPS was manually mixed with ΦX174 at 4 °C, which allowed attachment but not ejection. The reactions were loaded into a temperature-controlled sample cell held at 33 °C. SAXS profiles collected at various times during the reaction showed changes in the average size, mass, and symmetry of the phage particles (Fig. 1C). The first time-resolved profile, acquired at 45 s after LPS addition, displayed a loss of depth in the first minimum and an increase in intensity at a scattering angle of zero degrees, I(0). These changes indicate a loss of symmetry and increase in particle mass, respectively, which is consistent with LPS binding. Subsequent scattering profiles (acquired after 245 and 545 s) showed a leftward shift and increased first-minimum depth, indicating an increasing Rg and recovery of symmetry. I(0) decreased below that of the initial t = 0 state, reflecting a net loss of particle mass over the reaction’s course. Time courses for the Rg and depth of the first minimum are shown in Fig. 1 D and E, respectively. In summary, the SAXS data show an initial increase in particle mass and loss of symmetry after LPS addition. Viral particles binding LPS at a single vertex would produce this result. At later time points, particle symmetry increased while Rg and particle mass decreased, which is consistent with genome ejection and phage release from the LPS bilayer.
Cryo-EM.
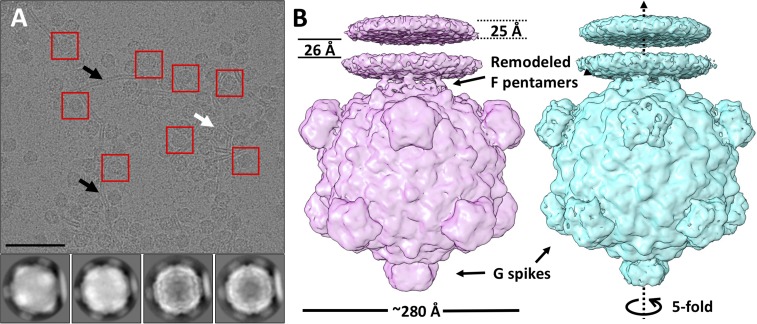
Cryo-EM single-particle analysis was performed to obtain greater structural detail regarding the changes occurring on LPS interaction. Purified ΦX174 particles were mixed with LPS at 33 °C for 1 min and then frozen in vitreous ice. Most of the particles were found attached to LPS bilayers by a single vertex (Fig. 2A, black arrows), but some seemed to be attached by more than one vertex (Fig. 2A, white arrow). The attached particles were “boxed” and subjected to nonreference 2D classification using the RELION software package (22). The selection of particles was dependent on the recognition of LPS bilayers. This gave a preference for selecting particles with their fivefold axis parallel to the plane of the image (Fig. S2).
Fig. 2.
Cryo-EM single-particle processing of ΦX174 complexed with LPS. (A, Top) A raw image of ΦX174 incubated with LPS, with examples of selected particles (red boxes) bound to LPS bilayers (black arrows). Some particles are attached to bilayers at more than one vertex (white arrows). (Scale bar: 100 nm.) (A, Bottom) Examples of 2D classes generated by RELION. Full and emptied particles were well separated. (B) Reconstructions of ΦX174 complexed with a LPS disk before (pink, full) and after (cyan, class A emptied) ejection of the ssDNA genome. These two states of ΦX174 have overall similar architecture, with differences in F proteins at the ΦX174–LPS interface and the G protein spikes.
Among the 10,000 selected particles, ∼7,000 were empty, while 2,400 still contained DNA (full). The percentage of emptied particles was much higher in LPS-complexed ΦX174 than that measured in the infectivity assay at equivalent time points (Fig. S1), suggesting that binding to LPS bilayers enhanced DNA ejection. Full and emptied particles were each subjected to 3D analysis using RELION (22). A fivefold rotational symmetry was imposed during the reconstruction. All 2,400 full particles were used for the reconstruction. This resulted in a map of DNA-containing ΦX174 with a resolution of 10.2 Å (Fig. 2B, Left and Fig. S3A, orange).
The processing of emptied particles was more complex. The initial reconstruction that included all 7,000 particles had a resolution of 8.2 Å. However, it was not clear whether there were one or two connecting regions of high density per asymmetric unit between the LPS disk and the viral capsid (Fig. S4, black circle). This was followed by two consecutive iterations of masked 3D classification (Materials and Methods) using RELION to separate the particles into two classes, A and B, containing 4,226 and 1,300 particles, respectively. A 3D refinement without any local masking brought the resolution of class A to 8.9 Å (Fig. 2B, Right and Fig. S3A, blue). A similar reconstruction of particles in class B generated a map with a resolution of 10.6 Å. The primary difference between the two maps is that in the class A reconstruction there are two regions of high density connecting the LPS disk to the capsid, whereas in class B there is only one region of connecting density (Fig. S4, red circles).
In the reconstructions, LPS is shown as a bilayer disk. The thickness of each layer is ∼25 Å, separated by ∼26 Å. The greater electron density in each of the bilayers is the consequence of the greater electron scattering power of the heavier phosphate atoms (23). In both maps, there was no density at the interacting fivefold vertex that would account for the G spike, suggesting that the spike dissociated from the rest of the capsid on binding to LPS. The absence of a G spike at the interface leaves the F protein pentamer anchoring the virion on the LPS disk. The LPS disk is connected to ΦX174 through five symmetry-related positions at one of the fivefold vertices of the virus (Fig. 2B).
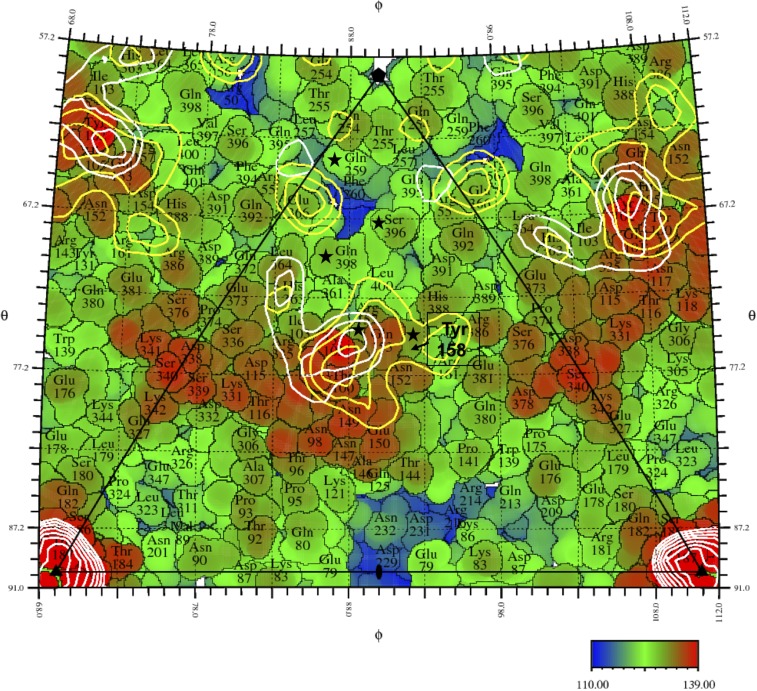
A pentameric F complex, from the crystallographic structure of the virus (6), was then fitted into the cryo-EM maps of the emptied and the full particles at the interacting vertex (Fig. S5A) using EMfit (24). Both maps were low-pass filtered to 10-Å resolution. The other vertex on the fivefold axis, opposite the bound LPS disk, was also fitted with the F pentamer to measure the influence of LPS binding on the conformation of the F proteins. The average density at the atomic positions of the fitted pentamer, sumF (24), was calculated based on only Cα atoms (Table 1). The crystal structure fit better into the EM map of the emptied ΦX174 particles at the vertices that had no contact with LPS. Thus, the binding of LPS caused significant conformational changes in the F proteins at the interacting vertex. Given that fitting into full ΦX174 had a smaller sumF value and lower local resolution at the connecting region compared with emptied ΦX174 (Fig. S3 B and C), it is reasonable to conclude that this conformational change is more significant in full ΦX174 particles with bound LPS compared with emptied particles. A “roadmap” (25) that shows the residues of the F proteins present on the viral surface indicates the position of the density that represents the virus-LPS contact region (Fig. 3). The bulge formed by the EF loop of the F proteins is below the density linking the capsid and the LPS disk. There are positively charged residues Lys118, Arg157 and Lys342 in the contact region, which could form electrostatic interactions with the phosphate group of LPS, while Asn117 and Gln157 may form hydrogen bonds with the hydroxyl group of LPS. This is similar to the crystal structure of a Toll-like receptor molecule complex with LPS in which multiple Lys, Arg, Gln, and hydrophobic residues of the receptor mediate the interaction (23).
Table 1.
SumF values determined with the EMfit program when fitting the F pentamer into different sites of the maps for the full and emptied particles
| Site fitted with the F pentamer | Emptied | Full |
| Vertex in contact with LPS | 48 | 41 |
| Vertex on the opposite side | 65 | 59 |
Higher values indicate a better agreement between the specific region of the map and the coordinates of the atoms.
Fig. 3.
A roadmap showing the surface of ΦX174 after removal of pentameric G protein spikes, looking down an icosahedral 2-fold axis. The icosahedral asymmetric unit is outlined by a black triangle connecting a fivefold axis in the “north” and two threefold axes along the “equator”. Angles defining the lines of longitude (φ) and latitude (θ) are given along the edge of the diagram. Individual amino acids covering the surface are identified and outlined based on the crystal structure (6, 25). Their distances to the viral center are indicated by color varying from 110 Å (blue) to 139 Å (red). Residues in the F proteins that make contact with the G proteins in the wild-type crystallographic structure are marked with black stars. The yellow and white contours correspond to equal increments of density of the full and emptied particles cryo-EM maps, respectively. The density contours show the highest density connecting the LPS with the virus. A bulge (found mainly in the red region) formed by the long loop insertion between βE and βF of the capsid protein F is located underneath the density that connects the capsid to the LPS disk.
A five-residue-long hydrophobic region was identified by aligning F proteins from Microviridae family members representing the three major evolutionary clades (Fig. S6). This region is located on the bulge formed by the EF loop and buried under the surface of the virus. When the G spike dissociates from the capsid, the interaction that has confined the conformation of the EF loop no longer exists. Thus, this hydrophobic region has the potential to reorient and interact with the lipid component of LPS.
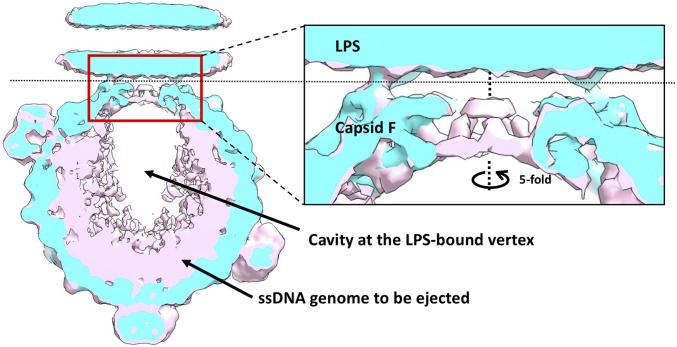
A major difference between the reconstructions of full and emptied ΦX174, both complexed with LPS, lies in the channel along the fivefold axis at the interacting vertex. The other long F protein loop, HI, is located in this region. In the apo state, two gates separate the genome from the outside environment at the fivefold vertices. The first gate is the major spike protein G. The second gate resides 35 Å underneath the first gate and is formed by residues Gly252 to Gln260 of the major capsid protein F. The diameter of the second gate is only ∼4 Å (6). In emptied ΦX174, loop 252–260 is disordered and the gate is more open, with a larger diameter of 30 Å (Fig. S5). However, in the reconstruction of full ΦX174 complexed with LPS, which likely represents an intermediate in the genome delivery pathway, the second gate remains closed, keeping the genome inside the capsid.
The extra electron density inside full ΦX174 particles is not evenly distributed. A radial, cylindrical cavity is found close to the vertex that interacts with the LPS, with a length of ∼110 Å and a diameter of 80 Å (Fig. 4). The dissociation of the G spike protein pentamer and alterations in the structure of the F protein pentamer may result in the release of internal pressure. Consequently, the DNA in this region may be less condensed, which might be the cause of the cavity observed in the reconstruction. Alternatively, a complex between the H protein and the ssDNA genome may form in this region. Since protein has a smaller density than DNA, the electron density in this volume might be somewhat lower.
Fig. 4.
A close look at the differences between full (pink) and emptied (cyan) ΦX174 around the fivefold axis. (Left) Overlaid central slices of the reconstructions of full (pink) and emptied (cyan) ΦX174 along the fivefold axis. (Right) Zoom-in view of the red window. Full ΦX174 has an unevenly distributed internal density, which accounts for the genome and the H proteins. The cavity is very likely a result of averaging less-homogeneous structures.
The full ΦX174 complexed with LPS described here is likely an intermediate during the infection. It has undergone significant structural changes at the interacting vertex but still contains the genome. The portion of ΦX174 particles in this state is relatively low, suggesting that this might be a metastable conformation of ΦX174. The existence of this intermediate implies that ΦX174 pauses transitorily after losing the G spike, so that the ssDNA genome can be rearranged for the subsequent ssDNA ejection.
Discussion
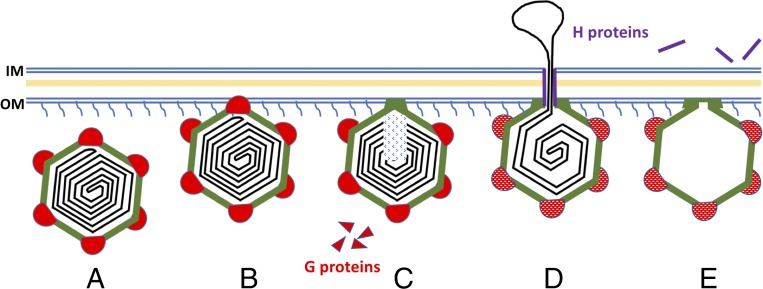
Our present findings, together with results from previous studies (9, 12–14, 17, 19, 26, 27), suggest a pathway for the ΦX174 infection (Fig. 5). The initial contact with the host cell is made by one of the 12 G protein spikes. This spike then dissociates from the capsid, leaving a structurally altered F protein pentamer to maintain viral attachment with LPS. When the G proteins dissociate from the capsid, the F proteins on this vertex gain more conformational freedom, particularly the EF and FG loops. The change in the bulge formed by the EF loop enables formation of a stable contact between the capsid and the host cell’s membrane. Meanwhile, the change in the FG loop opens the gate at the special vertex and prepares the virus for DNA translocation. At the same time, changes in the conformations of the H proteins and the ssDNA presumably occur inside the capsid to facilitate genome ejection through the vertex that is in contact with LPS.
Fig. 5.
Proposed model for ΦX174 DNA injection. (A and B) Native ΦX174 recognizes its host carrying specific LPS on the surface hypothetically through one of the G spikes (red). (C) The interaction destabilizes the G spike and causes its dissociation, leaving the loops on the surface of F proteins (green) to maintain the interaction with the cell wall of the host. (D) The dissociation of G proteins causes subsequent conformational changes in the F proteins (green) and H proteins (purple). The F and H proteins cooperate in translocating the genome (black) across the cell wall of the host. (E) The exit used by DNA remains open after ejection. H proteins are ejected along with the genome. F proteins still interact with the outer membrane.
In a crystallographic study, McKenna et al. (6) found that the G proteins make minimal contact with the capsid. Thus, it is likely that LPS can induce dissociation of the G spike by disrupting the interaction between the G spikes and the F proteins. Another question is how ΦX174 penetrates the peptidoglycan and the inner membrane of the host after dissociation of the G proteins. A partial answer is that the H proteins may form a tube-like conduit that serves to translocate the ssDNA genome (9, 28). It may be relevant that ΦX174 particles congregate at membrane adhesions sites, or Bayer’s patches, where the inner and outer membranes appear to merge (29). Although the H tube is not seen in this structure, we may expect it to be immediately behind the G spike in contact with the LPS, but as long as the H tube is still mixed up with the genome, we are unlikely to recognize it in any reconstruction, as has been suggested previously (9) (Fig. S2). Furthermore, it is unknown whether the H proteins assemble into tubes at the time of viral assembly or are assembled later during ejection. After the genome is ejected, the H tube disappears into the cytoplasm (9). During the infection, the H proteins and the DNA are internalized, while the rest of the capsid remains outside the host cell (30). These actions have been confirmed by the reconstruction of emptied ΦX174 in this study. Therefore, no tubular structure should be observed associated with the emptied capsid, as is the case.
Materials and Methods
Amplification and Purification of ΦX174.
Between 6 and 8 L of E. coli C122 were grown in TK broth (1.0% tryptone, 0.5% KCl) to 1.0 × 108 cells/mL at 37 °C. Before infection at multiplicity of infection of 10−5, MgCl2 and CaCl2 were added to respective concentrations of 10 mM and 5 mM. Infections were incubated for ∼5 h until a clear majority of cells had lysed. Cultures were refrigerated overnight to allow phage to attach to cell debris, which was concentrated by centrifugation. The resulting pellet was resuspended in 32 mL of 50 mM Na2B4O7/3.0 mM EDTA and then shaken at 4 °C overnight to elute attached particles. Debris was removed by centrifugation, and the supernatant was layered atop CsCl gradients made in 50 mM Na2B4O7/3.0 mM EDTA and spun as described previously (31). Virion bands were pulled and dialyzed against 100 mM NaCl, 5.0 mM EDTA, 6.4 mM Na2HPO4, and 3.3 mM KH2PO4 (pH 7.0), and then concentrated down to 0.8 mL. Then 0.2 mL was loaded atop a 5–30% sucrose (wt/vol) gradient made in 100 mM NaCl, 5.0 mM EDTA, 6.4 mM Na2HPO4, and 3.3 mM KH2PO4 (pH 7.0) and spun at 192,000 × g for 1 h. Gradients were divided into ∼40 125-µL fractions and analyzed by UV spectroscopy (OD280).
ΦX174 DNA Ejection Assay.
Concentrated ΦX174 in 0.06 M NH4Cl, 0.09 M NaCl, 0.1 M KCl, 0.1 M Tris⋅HCl pH 7.4, 1.0 mM MgSO4, and 1.0 mM CaCl2 was mixed with 5 mg/mL stock LPS dissolved in Tris⋅HCl pH 8.0 buffer to achieve the desired concentrations. CaCl2 was added to a concentration of 5.0 mM. ΦX174 and Tris⋅HCl buffer without LPS served as a negative control. The final concentration of ΦX174 was ∼108–109 pfu/mL. The mixtures including the negative control were then kept at 33 °C for 20 min. After 1, 3, 5, 8, 12, or 20 min, the mixtures containing LPS were diluted into 3.0 mM EDTA to terminate the DNA ejection of the particles. Diluted samples were titered as described previously (31). The dilution procedure was optimized for each batch of sample so that the number of plaques on each plate was between 10 and 500. The plaque counts of three replicates were averaged for each condition to establish the relative infectivity.
SAXS.
SAXS data were collected using 11.18-keV X-rays at the Cornell High-Energy Synchrotron Source G1 station. Scattering profiles (0.006–0.26 Å−1) were collected with a sample-to-detector distance of 2.05 m (measured using an Ag-behenate standard) onto a Pilatus 200K detector (Dectris). SAXS profiles were normalized using the transmitted beam through a semitransparent molybdenum beamstop. All samples were oscillated during data collection to reduce the effects of radiation damage inside a quartz capillary (2- mm diameter, 10-μm walls; Hampton Research) mounted in a custom-built temperature-controlled holder. Multiple 10-s exposures were acquired and averaged to improve the signal-to-noise ratio. All SAXS images were analyzed using MATLAB (MathWorks). Scattering profiles were azimuthally averaged about the beam center. Buffer scattering was measured before and after each sample and averaged before being subtracted from the sample scattering. For time-resolved experiments, profiles are shown at the indicated time after the addition of LPS to the sample. Sample conditions were as follows: 30-µL samples at 0.05–0.1 mg/mL ΦX174, 0.15 mg/mL LPS, 0.06 M NH4Cl2, 0.09 M NaCl, 0.1 M KCl, 1 mM MgS04, 1 mM CaCl2, and 0.1 M Tris⋅HCl pH 7.4 at 33 °C. The same trends were observed but with faster rates at elevated temperatures (37 °C) and slower rates at lower LPS concentrations (0.01 and 0.05 mg/mL).
Cryo-EM.
Cryo-EM samples were prepared using the Cryoplunge 3 system (Gatan). Here 5 µL of purified ΦX174 at 1011 pfu/mL was incubated with 1 µL of 1 mg/mL LPS from Salmonella enterica TV119 (Sigma-Aldrich) at 33 °C for 1 min. Then 3-µL aliquots of the mixture were loaded onto lacey carbon grids (400 mesh; Ted Pella). The grids were blotted by filter paper for 5 s and then plunged into liquid ethane for cryo-EM inspection. Forty consecutive frames of ΦX174 reacting with LPS embedded in vitreous ice were recorded using a Titan Krios transmission electron microscope (FEI) operated at 300 kV and equipped with a Gatan K2 Summit direct electron detector (3,838 × 3,710). All of the frames were collected automatically in counting mode using Leginon (32) at a magnification of 18,000× and with a defocus range of 1–3 μm, which generated a pixel size of 1.62 Å. For each frame stack, the total electron dose was 25 e−/Å2.
Image Processing.
A total of 2,908 frame stacks were collected. Beam-induced motion was corrected by realigning the frames within one stack using the MotionCorr program (33). The version of MotionCorr used here had been modified by Wen Jiang to take three consecutive frames instead of one to calculate the correlation coefficient. Contrast transfer function values were estimated for each motion-corrected image using CTFFIND3 (34). From the 2,908 images, 11,845 particles were manually selected using e2boxer.py in the EMAN2 package (35). The particle images were 2× binned and subjected to nonreference 2D classification using RELION (22). The particles in the classes that failed to generate a good average projection of ΦX174 were discarded, leaving 7,002 emptied particles and 2,384 full particles. Two iterations of masked 3D classifications were then used (Fig. S4) to select particles more homogeneous in the region where the LPS disk is connected to ΦX174. The first iteration was done with a large mask covering the top half of the map, while the second iteration was done with a smaller mask containing mainly the connecting region. The masks were generated using Chimera (36) and relion_mask_create from RELION to identify the region of interest. The 3D classification and subsequent refinement were done in RELION. The initial model was generated using jspr (37) from a smaller dataset containing 800 particles. The initial model used for that reconstruction was a 50-Å low-pass–filtered map of native icosahedral ΦX174.
For the reconstruction of emptied ΦX174, after two rounds of 3D classification, a subset of 4,226 particles (Fig. S4, class A) was selected, and a map at 8.9-Å resolution was achieved. If all 7,025 particles from the 2D classification were used, the final resolution would be 8.2 Å, but with less continuous density in the connecting region. This suggests that the connecting region is more flexible and heterogeneous. However, 3D classification did not improve the map of full ΦX174, likely due to the smaller number of particles to start with. All 2,384 full particles selected based on 2D classification were used for the final reconstruction, which generated a map with a resolution of 10.2 Å judged by the gold standard criterion. The map of the full particles has been deposited in the EMDataBank (accession code EMD-8862), as have the map of class A emptied particles (Fig. S4) and the masks used in the classification (accession code EMD-7033).
Fitting of the Pentameric F Protein Crystal Structure into the EM Maps.
One F protein pentamer was selected from the crystal structure of the whole virus (Protein Data Bank ID code 2BPA). The cryo-EM maps of full and class A emptied ΦX174 were both low-pass filtered to 10-Å resolution. EMfit (24) was then used to fit the pentamer into different vertices of these low-pass–filtered maps. The initial coordinates for placing the center of the F pentamer were inherited from a preliminary fitting by Chimera. Then a search for translation and rotation was done using EMfit. SumF values (a measurement of quality of the fit) were calculated based on only the Cα atoms.
Data Deposition.
The cryo-EM map of the full ΦX174 particles has been deposited in the EMDataBank (accession code EMD-8862), as have the map of the emptied ΦX174 particles and the 3D masks used in the classifications (accession code EMD-7033). Raw SAXS data have been deposited in the Small Angle Scattering Biological Data Bank (accession codes ASDCT9, SASDCU9, SASDCV9, SASDCW9, and SASDCX9).
Supplementary Material
Acknowledgments
We thank Yue Liu and Lei Sun for helpful discussions, Seong ha Park for assistance in interpreting SAXS data, and Valorie Bowman (Purdue Electron Microscopy Facility), Weifeng Shang and Srinivas Chakravarthy [Advanced Photon Source (APS) sector 18], and Richard Gillilan and Arthur Woll [Cornell High-Energy Synchrotron Source (CHESS)] for help and advice. We also thank Sheryl Kelly for help in preparing this manuscript. Preliminary data were acquired at APS, a US Department of Energy (DOE) Office of Science user facility operated by Argonne National Laboratory, supported by the US DOE under Contract DE-AC02-06CH11357. CHESS is supported by the National Science Foundation (NSF) (DMR01332208) and the National Institutes of Health/National Institute of General Medical Sciences. This research was supported by NSF Grant MCB-1515260 (to M.G.R.), NSF Grant MCB-1408217 (to B.A.F.), and US Department of Agriculture Hatch Funds awarded to the University of Arizona and the BIO5 Institute (to B.A.F.). A.M. was supported by an NSF Graduate Research Fellowship under Grant DGE-1650441.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The cryo-EM map of the full ΦX174 particles has been deposited in the EMDataBank (accession no. EMD-8862), as has the map of the emptied ΦX174 particles and the three-dimensional masks used in the classifications (accession no. EMD-7033). Raw SAXS data have been deposited in the Small Angle Scattering Biological Data Bank, https://www.sasbdb.org (accession codes SASDCT9, SASDCU9, SASDCV9, SASDCW9, and SASDCX9).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1716614114/-/DCSupplemental.
References
- 1.Yap ML, et al. Role of bacteriophage T4 baseplate in regulating assembly and infection. Proc Natl Acad Sci USA. 2016;113:2654–2659. doi: 10.1073/pnas.1601654113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.González-García VA, et al. Conformational changes leading to T7 DNA delivery upon interaction with the bacterial receptor. J Biol Chem. 2015;290:10038–10044. doi: 10.1074/jbc.M114.614222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taylor NMI, et al. Structure of the T4 baseplate and its function in triggering sheath contraction. Nature. 2016;533:346–352. doi: 10.1038/nature17971. [DOI] [PubMed] [Google Scholar]
- 4.Hu B, Margolin W, Molineux IJ, Liu J. The bacteriophage T7 virion undergoes extensive structural remodeling during infection. Science. 2013;339:576–579. doi: 10.1126/science.1231887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caspar DL, Klug A. Physical principles in the construction of regular viruses. Cold Spring Harb Symp Quant Biol. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- 6.McKenna R, et al. Atomic structure of single-stranded DNA bacteriophage ΦX174 and its functional implications. Nature. 1992;355:137–143. doi: 10.1038/355137a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ilag LL, Incardona NL. Structural basis for bacteriophage ΦX174 assembly and eclipse as defined by temperature-sensitive mutations. Virology. 1993;196:758–768. doi: 10.1006/viro.1993.1533. [DOI] [PubMed] [Google Scholar]
- 8.Roux S, Krupovic M, Poulet A, Debroas D, Enault F. Evolution and diversity of the Microviridae viral family through a collection of 81 new complete genomes assembled from virome reads. PLoS One. 2012;7:e40418. doi: 10.1371/journal.pone.0040418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun L, et al. Icosahedral bacteriophage ΦX174 forms a tail for DNA transport during infection. Nature. 2014;505:432–435. doi: 10.1038/nature12816. [DOI] [PubMed] [Google Scholar]
- 10.Andres D, Baxa U, Hanke C, Seckler R, Barbirz S. Carbohydrate binding of Salmonella phage P22 tailspike protein and its role during host cell infection. Biochem Soc Trans. 2010;38:1386–1389. doi: 10.1042/BST0381386. [DOI] [PubMed] [Google Scholar]
- 11.Brown DT, MacKenzie JM, Bayer ME. Mode of host cell penetration by bacteriophage ΦX174. J Virol. 1971;7:836–846. doi: 10.1128/jvi.7.6.836-846.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inagaki M, et al. Characterization of the binding of spike H protein of bacteriophage ΦX174 with receptor lipopolysaccharides. J Biochem. 2000;127:577–583. doi: 10.1093/oxfordjournals.jbchem.a022643. [DOI] [PubMed] [Google Scholar]
- 13.Kawaura T, et al. Recognition of receptor lipopolysaccharides by spike G protein of bacteriophage ΦX174. Biosci Biotechnol Biochem. 2000;64:1993–1997. doi: 10.1271/bbb.64.1993. [DOI] [PubMed] [Google Scholar]
- 14.Inagaki M, et al. Different contributions of the outer and inner R-core residues of lipopolysaccharide to the recognition by spike H and G proteins of bacteriophage ΦX174. FEMS Microbiol Lett. 2003;226:221–227. doi: 10.1016/S0378-1097(03)00601-3. [DOI] [PubMed] [Google Scholar]
- 15.Suzuki R, et al. Specific interaction of fused H protein of bacteriophage ΦX174 with receptor lipopolysaccharides. Virus Res. 1999;60:95–99. doi: 10.1016/s0168-1702(98)00145-2. [DOI] [PubMed] [Google Scholar]
- 16.Bull JJ, et al. Exceptional convergent evolution in a virus. Genetics. 1997;147:1497–1507. doi: 10.1093/genetics/147.4.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Young LN, Hockenberry AM, Fane BA. Mutations in the N terminus of the ΦX174 DNA pilot protein H confer defects in both assembly and host cell attachment. J Virol. 2014;88:1787–1794. doi: 10.1128/JVI.03227-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Incardona NL. Mechanism of adsorption and eclipse of bacteriophage ΦX174. 3: Comparison of the activation parameters for the in vitro and in vivo eclipse reactions with mutant and wild-type virus. J Virol. 1974;14:469–478. doi: 10.1128/jvi.14.3.469-478.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Incardona NL, Tuech JK, Murti G. Irreversible binding of phage ΦX174 to cell-bound lipopolysaccharide receptors and release of virus-receptor complexes. Biochemistry. 1985;24:6439–6446. doi: 10.1021/bi00344a020. [DOI] [PubMed] [Google Scholar]
- 20.Doore SM, Fane BA. The kinetic and thermodynamic aftermath of horizontal gene transfer governs evolutionary recovery. Mol Biol Evol. 2015;32:2571–2584. doi: 10.1093/molbev/msv130. [DOI] [PubMed] [Google Scholar]
- 21.Svergun DI, Koch MHJ. Small-angle scattering studies of biological macromolecules in solution. Rep Prog Phys. 2003;66:1735–1782. [Google Scholar]
- 22.Scheres SHW. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park BS, et al. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature. 2009;458:1191–1195. doi: 10.1038/nature07830. [DOI] [PubMed] [Google Scholar]
- 24.Rossmann MG, Bernal R, Pletnev SV. Combining electron microscopic with x-ray crystallographic structures. J Struct Biol. 2001;136:190–200. doi: 10.1006/jsbi.2002.4435. [DOI] [PubMed] [Google Scholar]
- 25.Xiao C, Rossmann MG. Interpretation of electron density with stereographic roadmap projections. J Struct Biol. 2007;158:182–187. doi: 10.1016/j.jsb.2006.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Incardona NL, Selvidge L. Mechanism of adsorption and eclipse of bacteriophage ΦX174. II: Attachment and eclipse with isolated Escherichia coli cell wall lipopolysaccharide. J Virol. 1973;11:775–782. doi: 10.1128/jvi.11.5.775-782.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Inagaki M, Wakashima H, Kato M, Kaitani K, Nishikawa S. Crucial role of the lipid part of lipopolysaccharide for conformational change of minor spike H protein of bacteriophage ΦX174. FEMS Microbiol Lett. 2005;251:305–311. doi: 10.1016/j.femsle.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 28.Sun L, Rossmann MG, Fane BA. High-resolution structure of a virally encoded DNA-translocating conduit and the mechanism of DNA penetration. J Virol. 2014;88:10276–10279. doi: 10.1128/JVI.00291-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bayer ME, Starkey TW. The adsorption of bacteriophage ΦX174 and its interaction with Escherichia coli: A kinetic and morphological study. Virology. 1972;49:236–256. doi: 10.1016/s0042-6822(72)80026-6. [DOI] [PubMed] [Google Scholar]
- 30.Jazwinski SM, Marco R, Kornberg A. The gene H spike protein of bacteriophages ΦX174 and S13. II: Relation to synthesis of the parenteral replicative form. Virology. 1975;66:294–305. doi: 10.1016/0042-6822(75)90199-3. [DOI] [PubMed] [Google Scholar]
- 31.Fane BA, Hayashi M. Second-site suppressors of a cold-sensitive prohead accessory protein of bacteriophage ΦX174. Genetics. 1991;128:663–671. doi: 10.1093/genetics/128.4.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suloway C, et al. Automated molecular microscopy: The new Leginon system. J Struct Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 33.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 35.Tang G, et al. EMAN2: An extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 36.Pettersen EF, et al. UCSF Chimera–A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 37.Guo F, Jiang W. Single particle cryo-electron microscopy and 3-D reconstruction of viruses. Methods Mol Biol. 2014;1117:401–443. doi: 10.1007/978-1-62703-776-1_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.