Significance
Insulin resistance in liver and skeletal muscle are major factors in the pathogenesis of type 2 diabetes; however, the molecular mechanism or mechanisms responsible for this phenomenon have not been established. Recently, an association of a single-nucleotide polymorphism in the human N-acetyltransferase 2 (Nat2) gene with insulin resistance in humans was found. Here, we show that the murine ortholog Nat1 knockout (KO) mice manifested whole-body insulin resistance associated with marked increases in liver and muscle lipid content. Nat1 KO mice also displayed reduced whole-body energy expenditure and reduced mitochondrial activity. Taken together, these studies demonstrate that Nat1 deletion promotes reduced mitochondrial activity and is associated with ectopic lipid-induced liver and muscle insulin resistance.
Keywords: mitochondria, diacylglycerol, protein kinase ε, protein kinase θ, ceramides
Abstract
A single-nucleotide polymorphism in the human arylamine N-acetyltransferase 2 (Nat2) gene has recently been identified as associated with insulin resistance in humans. To understand the cellular and molecular mechanisms by which alterations in Nat2 activity might cause insulin resistance, we examined murine ortholog Nat1 knockout (KO) mice. Nat1 KO mice manifested whole-body insulin resistance, which could be attributed to reduced muscle, liver, and adipose tissue insulin sensitivity. Hepatic and muscle insulin resistance were associated with marked increases in both liver and muscle triglyceride (TAG) and diacylglycerol (DAG) content, which was associated with increased PKCε activation in liver and increased PKCθ activation in skeletal muscle. Nat1 KO mice also displayed reduced whole-body energy expenditure and reduced mitochondrial oxygen consumption in white adipose tissue, brown adipose tissue, and hepatocytes. Taken together, these studies demonstrate that Nat1 deletion promotes reduced mitochondrial activity and is associated with ectopic lipid-induced insulin resistance. These results provide a potential genetic link among mitochondrial dysfunction with increased ectopic lipid deposition, insulin resistance, and type 2 diabetes.
Insulin resistance in liver and skeletal muscle is a major factor in the pathogenesis of type 2 diabetes; however, the molecular mechanism or mechanisms responsible for this phenomenon have not been established. In this context, several cellular mechanisms have been proposed to be responsible for insulin resistance in these tissues, including alterations in circulating adipocytokines resulting from inflammation (1, 2), increased endoplasmic reticulum stress (3, 4), and increases in ectopic lipid [diacylglycerol (DAG) (5–9) and ceramide (10–12)] content.
Recently, a genome-wide association study found an association with a single-nucleotide polymorphism in the human N-acetyltransferase 2 (Nat2) gene associated with insulin resistance in humans (13). This study also found that knockdown of Nat1 caused insulin resistance in murine 3T3-L1 adipocytes and that Nat1 (the murine ortholog of Nat2) knockout (KO) mice displayed decreased glucose tolerance, suggesting whole-body insulin resistance (13). Nevertheless, the mechanisms by which alterations in the Nat1 gene might cause insulin resistance is unknown. To address this question, we performed comprehensive metabolic analyses of regular chow (RC)-fed and high-fat diet (HFD)-fed Nat1 KO mice. Specifically, we examined basal and insulin-stimulated rates of liver, muscle, and white adipose tissue (WAT) glucose metabolism assessed during a hyperinsulinemic-euglycemic clamp combined with [3-3H]glucose, [2H5]glycerol, and [13C16]palmitate infusions. These measurements were combined with assessment of whole-body energy expenditure, activity, food intake, and 1H NMR assessment of lean body and fat mass along with liquid chromatography-tandem mass spectrometry (LC-MS/MS) analyses of liver and muscle lipid intermediates (DAGs, ceramides) that have been implicated in causing insulin resistance in liver and skeletal muscle.
Results
Nat1 Deficiency Causes Whole-Body Insulin Resistance.
The WT and Nat1 KO mice were studied at 12 wk of age and displayed no differences in body weight or body fat between groups when fed either RC or a HFD for 4 wk (Table 1).
Table 1.
General parameters from WT and Nat1 KO mice fed either RC or a HFD at 12 wk of age
| Parameters | WT | KO |
| Body weight (RC), g | 27.5 ± 0.4 | 27.6 ± 0.5 |
| Body fat (RC), g | 1.7 ± 0.1 | 2.0 ± 0.2 |
| Body weight (HFD), g | 34.2 ± 1.3 | 36.5 ± 0.9 |
| Body fat (HFD), g | 7.9 ± 0.8 | 9.9 ± 0.8 |
| Fasting insulin (RC), μU/mL | 5.4 ± 0.7 | 6.9 ± 0.8 |
| Clamped insulin (RC), μU/mL | 34.9 ± 2.6 | 35.9 ± 1.8 |
| Fasting Insulin (HFD), μU/mL | 14.0 ± 3.3 | 37.4 ± 8.5* |
| Clamp insulin (HFD), μU/mL | 97.3 ± 5.2 | 110.5 ± 10.3 |
| Basal NEFA (RC), mmol/L | 1.14 ± 0.07 | 1.12 ± 0.08 |
| Clamp NEFA (RC), mmol/L | 0.34 ± 0.03 | 0.39 ± 0.02 |
| Basal NEFA (HFD), mmol/L | 1.18 ± 0.07 | 1.15 ± 0.05 |
| Clamp NEFA (HFD), mmol/L | 0.70 ± 0.05 | 1.11 ± 0.05** |
| Plasma TAG (RC), mg/dL | 94.5 ± 4.6 | 99.8 ± 5.9 |
| Plasma TAG (HFD), mg/dL | 92.3 ± 3.7 | 91.1 ± 5.0 |
| Plasma FGF21 (RC), pg/mL | 423.2 ± 61.9 | 540.1 ± 80.8 |
Data are represented as mean ± SEM (n = 8–10 per group).
P < 0.05.
P < 0.001 compared with WT mice fed the same diet.
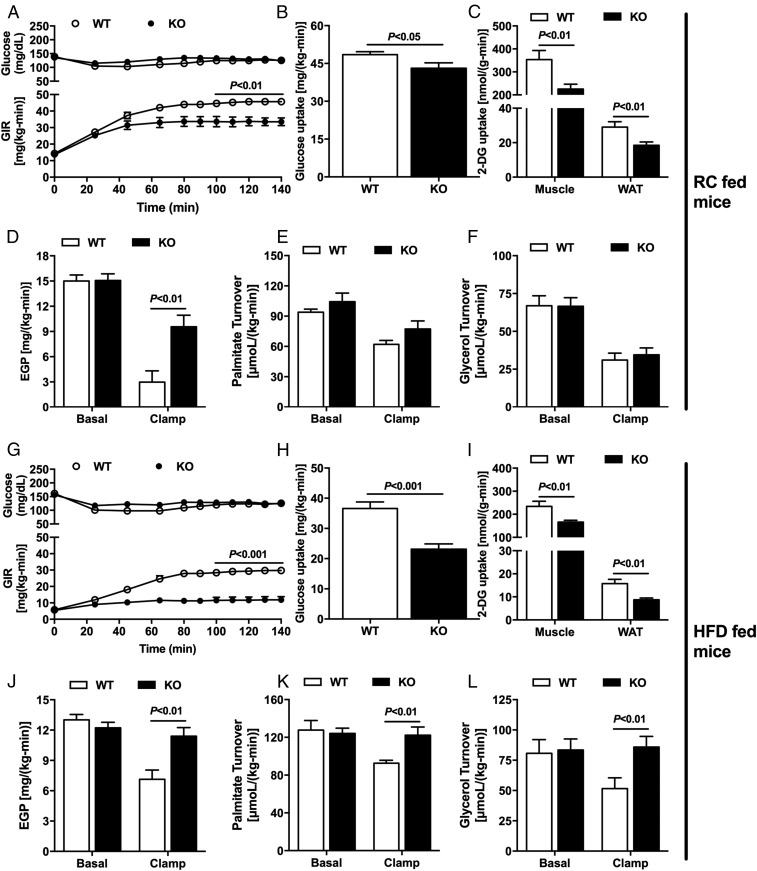
To evaluate whether the Nat1 deficiency would cause insulin resistance and, if so, to determine which tissues were responsible for the insulin resistance, we performed hyperinsulinemic-euglycemic clamp studies combined with radiolabeled glucose in Nat1 KO and WT littermate mice. The deficiency of Nat1 in RC-fed mice caused whole-body insulin resistance, reflected by a marked reduction in the glucose infusion rate required to maintain euglycemia during the hyperinsulinemic-euglycemic clamp (Fig. 1A). This difference in glucose infusion rate was partially accounted for by reduction in peripheral insulin sensitivity in the Nat1 KO mice. The Nat1 KO mice displayed reduced insulin-stimulated muscle and WAT glucose uptake, as well as reduced insulin-stimulated whole-body glucose turnover (Fig. 1 B and C). Nat1 KO mice also manifested hepatic insulin resistance, as reflected by reduced insulin-stimulated suppression of endogenous glucose production during the clamp (Fig. 1D). Although it has been shown that Nat1 deficiency in vitro induced increased lipolysis in 3T3-L1 adipocytes (13), Nat1 KO mice fed RC did not display any alteration in WAT lipolysis measured in vivo, evaluated by nonesterified fatty acid (NEFA) levels (Table 1), palmitate turnover, and glycerol turnover (Fig. 1 E and F).
Fig. 1.
Nat1 KO mice display insulin resistance in liver, muscle, and WAT, which was exacerbated by HFD. Time-course of plasma glucose and glucose infusion rates (GIR) (A) during the hyperinsulinemic-euglycemic clamp of WT and Nat1 KO mice fed RC. (B) Whole-body insulin-stimulated glucose uptake. (C) Insulin-stimulated skeletal muscle and WAT 2-deoxy-glucose uptake. (D) Basal and clamp endogenous glucose production. (E) Basal and clamp palmitate turnover. (F) Basal and clamp glycerol turnover. (G) Time-course of plasma glucose and GIR during the hyperinsulinemic-euglycemic clamp of WT and Nat1 KO mice fed a HFD. (H) Whole-body insulin-stimulated glucose uptake. (I) Insulin-stimulated skeletal muscle and WAT 2-deoxy-glucose uptake. (J) Basal and clamp endogenous glucose production. (K) Basal and clamp palmitate turnover. (L) Basal and clamp glycerol turnover. Data are represented as mean ± SEM (n = 10 per group).
Next, to determine whether Nat1 KO mice would be more prone to HFD-induced insulin resistance, the rodents were fed a HFD for 4 wk before the hyperinsulinemic-euglycemic clamp was performed. Nat1 deficiency induced severe whole-body insulin resistance, observed by a reduction in the glucose infusion rate required to maintain euglycemia during the clamp (Fig. 1G) compared with WT mice. This reduction in glucose infusion rate was accounted for by reduced whole-body glucose turnover during the clamp (Fig. 1H), and consequently, reduction in insulin-stimulated muscle and WAT glucose uptake (Fig. 1I), as well as severe hepatic insulin resistance, as reflected by the lack of suppression in endogenous glucose production during the clamp (Fig. 1J) and a severe defect in insulin-stimulated suppression in WAT lipolysis (Fig. 1 K and L).
Nat1 Deficiency Leads to Ectopic Lipid Accumulation in Liver and Muscle and Reduced Insulin Signaling in Liver and Skeletal Muscle.
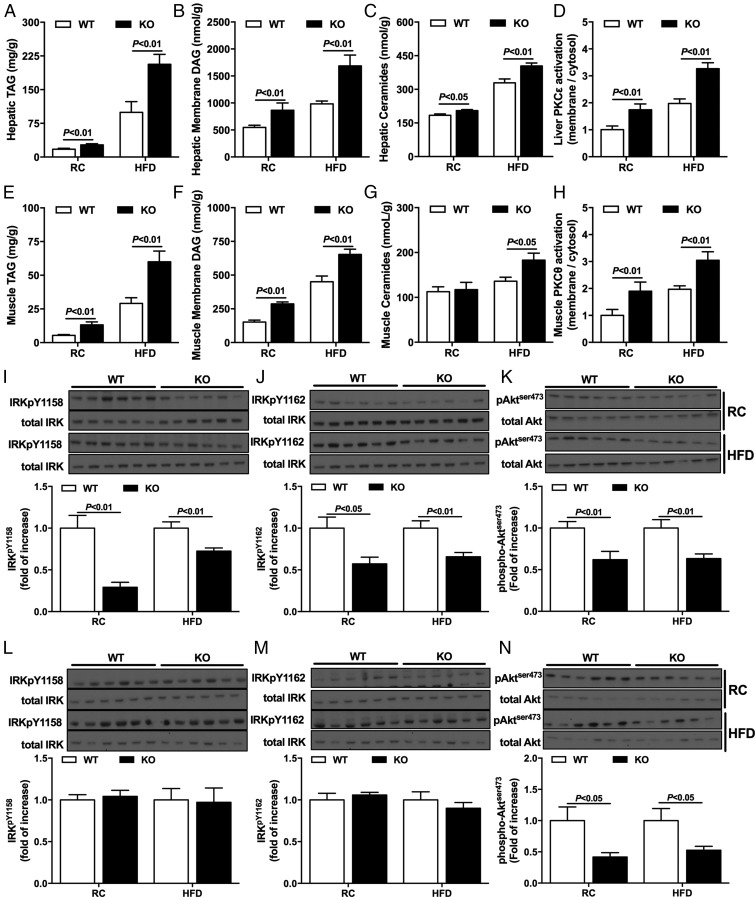
Given the strong causal relationship between ectopic lipid accumulation and hepatic and muscle insulin resistance shown in previous studies (4, 14–16), we measured DAG and ceramide content as well as translocation (activation) of PKCε in liver and PKCθ in skeletal muscle in RC and HFD rodents. Hepatic TAG, DAG, and ceramides were increased in KO mice in either the RC or HFD condition (Fig. 2 A–C and Tables S1 and S3). Consistent with increased DAG content in liver, Nat1 KO mice fed either RC or a HFD displayed increased PKCε activation (Fig. 2D), suggesting the increased hepatic DAG content led to hepatic insulin resistance through activation of PKCε, as previously demonstrated (5, 17–19).
Fig. 2.
Nat1 KO mice display increased ectopic lipid content and reduced insulin signaling in liver and skeletal muscle. Hepatic TAG (A), hepatic membrane DAG (B), and hepatic ceramides (C) content in WT and KO mice fed either RC or a HFD. Hepatic PKCε activation (D) in WT and KO mice fed either RC or a HFD. Muscle TAG (E), muscle membrane DAG (F), and muscle ceramides (G) content in WT and KO mice fed either RC or a HFD. Muscle PKCθ activation (H) in WT and KO mice fed either RC or a HFD. Western blot images and quantification for insulin-stimulated IRK phosphorylation pY1158 (I) and pY1162 (J), and Akt phosphorylation (K) in liver. Western blot images and quantification for insulin-stimulated IRK phosphorylation pY1158 (L) and pY1162 (M) and Akt phosphorylation (N) in muscle. Data are represented as mean ± SEM (n = 8–12 per group).
Ectopic lipid content was also evaluated in skeletal muscle. TAG and DAG content were increased in muscle from Nat1 KO mice fed either RC or a HFD (Fig. 2 E and F and Tables S2 and S4). In contrast to what was observed in liver, ceramide content was not increased in skeletal muscle from RC-fed mice, despite the presence of muscle insulin resistance (Fig. 2G). Nat1 KO mice fed a HFD displayed increased muscle DAG (Fig. 2F) and ceramide content (Fig. 2G), and the increased DAG content in muscle was associated with increased PKCθ activation (Fig. 2H).
Increased DAG content in liver and skeletal muscle has been shown to play a causal role in liver and muscle insulin resistance by activation of PKCε in liver and PKCθ in skeletal muscle, leading to inhibition of insulin signaling at the level of the insulin receptor kinase (IRK) in liver (17–19) and at the level of insulin receptor substrate 1 tyrosine phosphorylation in skeletal muscle (5, 6, 8, 9). Consistent with this hypothesis, increased DAG content in liver in Nat1 KO mice was associated with reduced insulin-stimulated IRK tyrosine phosphorylation and Akt phosphorylation in liver (Fig. 2 I–K), which provides further support of recent studies demonstrating that DAG activation of PKCε in liver promotes increased phosphorylation of a critical threonine (threonine1160 human IRK, threonine1150 mouse IRK) in the catalytic subunit of the IRK leading to decreased IRK tyrosine phosphorylation and reduced IRK activity (19). Consistent with DAG activation of PKCθ inhibiting insulin signaling downstream of the IRK, Nat1 KO mice displayed reduced insulin-stimulated Akt phosphorylation in skeletal muscle without any changes in IRK tyrosine phosphorylation (9) (Fig. 2 L–N).
Nat1 Deficiency Was Associated with Decreased Energy Expenditure and Mitochondrial Dysfunction.
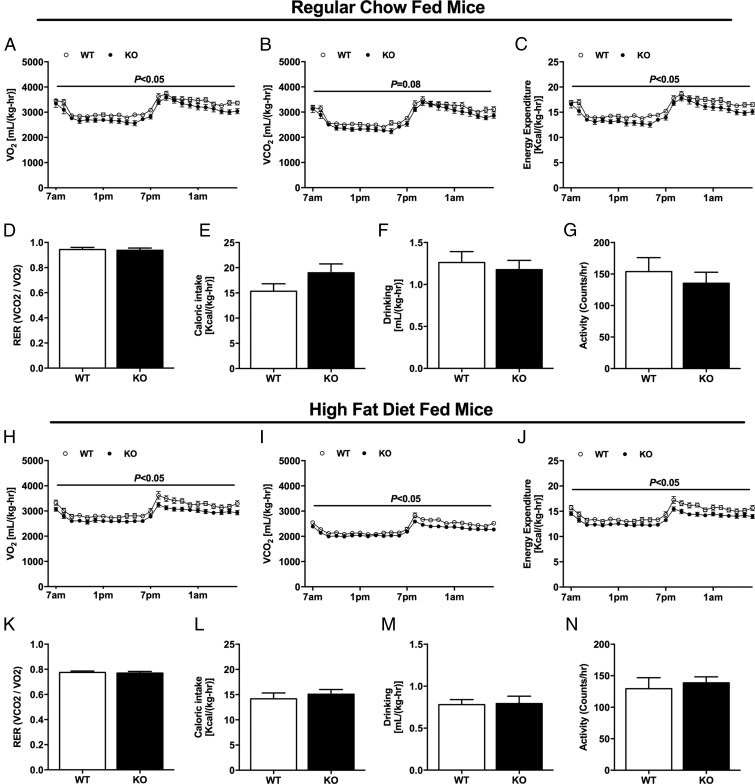
Previous studies in humans have found a strong relationship between increased ectopic lipid content in liver and skeletal muscle and insulin resistance, and hypothesized that reduced mitochondrial activity in these organs may be a predisposing factor for increased ectopic lipid accumulation and insulin resistance in these organs (20–23). To evaluate whether whole-body oxygen consumption and energy expenditure were altered in these Nat1 KO mice, these parameters were assessed by metabolic cages. Whole-body oxygen consumption, carbon dioxide production, and energy expenditure were reduced in Nat1 KO mice fed RC compared with WT mice (Fig. 3 A–C), without any difference between groups regarding quotient respiratory, caloric intake, drinking, and activity (Fig. 3 D–G). These effects were also observed in Nat1 KO mice fed HFD, displaying reduced whole-body oxygen consumption, carbon dioxide production, and energy expenditure (Fig. 3 H–J). In contrast, there was no difference among groups regarding quotient respiratory, caloric intake, drinking, and activity (Fig. 3 K–N).
Fig. 3.
Nat1 KO mice display reduced whole-body energy expenditure. Whole-body oxygen consumption (A), carbon dioxide production (B), energy expenditure (C), respiratory exchange ratio (D), caloric intake (E), drinking (F), and daily activity (G) in WT and KO mice fed RC. Whole-body oxygen consumption (H), carbon dioxide production (I), energy expenditure (J), respiratory exchange ratio (K), caloric intake (L), drinking (M), and daily activity (N) in WT and KO mice fed a HFD. Data are represented as mean ± SEM (n = 10 per group).
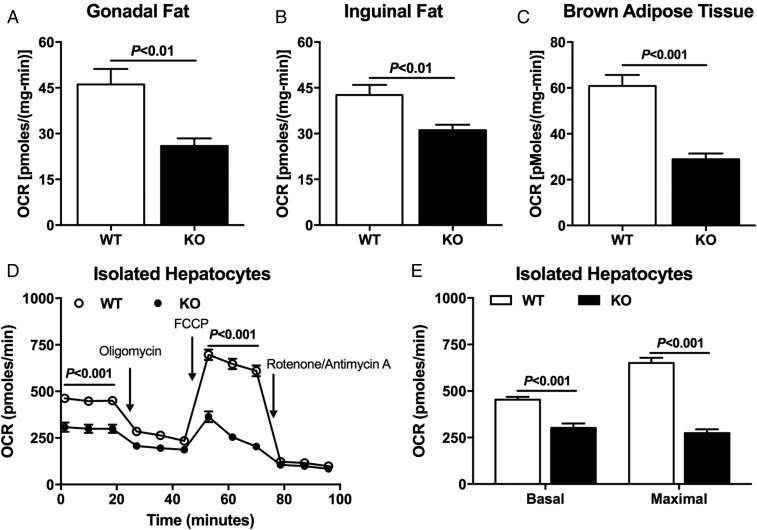
We also performed an evaluation of oxygen consumption in hepatocytes, WAT, and brown adipose tissue (BAT) isolated from Nat1 KO and WT mice by Seahorse analyses to determine whether these tissues were contributing to the reduced whole-body energy expenditure in the Nat1 KO mice. In vitro oxygen consumption in WAT and BAT were reduced in Nat1 KO mice compared with WT mice (Fig. 4 A–C). Hepatocytes from Nat1 KO mice also displayed reduced oxygen consumption in vitro compared with WT mice (Fig. 4 D and E).
Fig. 4.
Nat1 KO mice have lower tissue oxygen consumption. Oxygen consumption rates (OCR) measured in gonadal fat (A), inguinal fat (B), BAT (C), and isolated primary hepatocytes (D and E) from WT and KO mice. Data are represented as mean ± SEM (n = 5 per group).
Discussion
A single-nucleotide polymorphism in the human Nat2 gene has recently been identified to be associated with insulin resistance in humans. However, the mechanism by which alterations in the Nat2 gene might cause insulin resistance is unknown. To address this question, we performed a comprehensive metabolic analysis of murine ortholog Nat1 KO mice fed either RC or a HFD. Specifically, we examined basal and insulin-stimulated rates of liver, muscle, and WAT glucose metabolism assessed by a hyperinsulinemic-euglycemic clamp study. These measurements were combined with assessment of whole-body energy expenditure, activity, food intake, and 1H NMR measurements of lean body and fat mass, as well as LC-MS/MS analyses of liver and muscle lipid intermediates (ceramides, DAGs), which have been implicated as causal factors in liver and muscle insulin resistance. Using this approach, we find that Nat1 KO mice displayed whole-body insulin resistance on a RC diet, with reduced insulin-stimulated glucose uptake in skeletal muscle and reduced insulin-stimulated suppression of endogenous glucose production during a hyperinsulinemic-euglycemic clamp. Interestingly, insulin resistance in liver and skeletal muscle was further exacerbated when the Nat1 KO mice were fed a HFD. In addition to the reduced insulin-stimulated muscle glucose uptake and decreased suppression of hepatic glucose production, Nat1 KO mice also displayed reduced insulin-stimulated suppression of WAT lipolysis, which may have contributed to the severe metabolic phenotype in these mice (24).
Ectopic lipid deposition in liver and skeletal muscle has been shown to be strongly associated with insulin resistance in liver and muscle (9, 25, 26), and increases in DAGs (4, 6, 7, 9, 14, 27) and ceramides (10–12) have both been implicated at the molecular triggers in this process. Increased DAG content in liver has been shown to promote increase PKCε translocation to the plasma membrane, which in turn has been shown to phosphorylate threonine1160 (threonine1150 mouse) in the IRK catalytic domain, leading to inhibition of IRK activity (17–19). In contrast, increases in muscle DAG content have been shown to increase PKCθ translocation, leading to decreased insulin signaling at the level of IRS-1 tyrosine phosphorylation (28–30). These findings have recently been translated to humans, in that increased DAG content and PKCε translocation have been shown to be strongly associated with hepatic insulin resistance in obese individuals undergoing bariatric surgery (31, 32), and increased DAG content and PKCθ translocation were found to be strongly associated with muscle insulin resistance in obese subjects and patients with type 2 diabetes (33). Consistent with this mechanism, we observed an increase in liver and muscle DAG content in Nat1 KO mice that was associated with an increase in PKCε and PKCθ in liver and muscle, respectively. In contrast, we saw only small increases in hepatic ceramide content in Nat1 KO mice and no increase in muscle ceramide content in Nat1 KO mice, despite the presence of muscle insulin resistance.
To further understand the underlying mechanism responsible for the increased ectopic lipid content in Nat1 KO mice, we performed comprehensive metabolic cage studies for 4 consecutive days to evaluate energy expenditure and feeding behavior. We observed that the Nat1 KO mice displayed reduced whole-body energy expenditure. Taken together, these data suggest that decreased energy expenditure may be responsible for the increased hepatic and muscle ectopic lipid accumulation and insulin resistance in Nat1 KO mice. Furthermore, these results are consistent with prior studies demonstrating that decreased energy expenditure in rodents leads to reduced insulin sensitivity associated with increased ectopic lipid content (15), and that increased energy expenditure resulting from increased mitochondrial activity in liver and/or skeletal muscle (27, 28, 34, 35) can protect mice from HFD-induced insulin resistance in liver and skeletal muscle.
To understand which tissues might be responsible for the reduced whole-body energy expenditure in Nat1 KO mice, we assessed in vitro oxygen consumption in WAT, BAT, and primary hepatocytes obtained from Nat1 KO and age- and weight-matched littermate WT mice. Using this approach, we found that oxygen consumption was markedly reduced in all these tissues, suggesting Nat1 deficiency may lead to global reductions in mitochondrial activity in all tissues. Indeed, it was recently shown that Nat1 KO mice display mitochondrial dysfunction (36), corroborating our data. This manuscript showed that mitochondria from Nat1 KO mice displayed increased superoxide production and reduction in PGC-1α expression, as well as mitochondrial genes (such as Nrf1, Tfam, Cyscs, and Atp5). These alterations resulted in reduced mitochondria oxygen consumption, reduced whole-body energy expenditure, and reduced exercise tolerance. The authors in this study performed the in vivo experiments in 16-wk-old mice, which resulted in differences in body weight between groups. Knowing that differences in body weight can profoundly affect the analysis of whole-body energy metabolism (37), we extended these findings of in vivo energy metabolism in Nat1 KO mice by performing all the experiments in body weight-matched mice (12-wk-old mice) and examining these same parameters in HFD-fed Nat1 KO mice. Moreover, most of the in vitro oxygen measurement data by Chennamsetty et al. (36) were collected from immortalized cell lines with knockdown of Nat1 protein. Here, we expanded and corroborated these findings by measuring oxygen consumption in WAT and BAT as well as primary hepatocytes from WT and Nat1 KO mice, showing that all these tissues displayed reduced oxygen consumption.
Insulin resistance associated with mitochondrial dysfunction is a notable finding, as this relationship has been described in rodents (27, 38, 39), as well as in humans (20–23), where reductions in mitochondrial oxidative-phosphorylation activity assessed by in vivo NMR spectroscopy was found to be associated with increased ectopic lipid content and insulin resistance in muscle of lean healthy elderly people and insulin-resistant offspring of patients with type 2 diabetes who are prone to develop diabetes later in life (20–23, 40). The results complement previous studies demonstrating a key role for alterations in mitochondrial dysfunction in the pathogenesis of insulin resistance, which found that aging-associated reduction in mitochondrial activity and aging-associated ectopic lipid (DAG-novel PKC)-induced insulin resistance in liver and skeletal muscle can be prevented by targeting catalase to the mitochondria (27).
In summary, these data demonstrate that deletion of the Nat1 gene in mice caused reduced mitochondrial activity, which resulted in decreased whole-body energy expenditure and increased ectopic lipid accumulation in liver and skeletal muscle. Increased DAG content in liver and skeletal muscle was associated with increased PKCε (liver) and PKCθ (muscle) activity and insulin resistance in liver and skeletal muscle when mice were fed a RC diet. These effects were further exacerbated when Nat1 KO mice were challenged with a HFD.
These data provide insights into how variants in the Nat2 gene may predispose humans to insulin resistance and type 2 diabetes and provide a potential genetic link between mitochondrial dysfunction with increased ectopic lipid deposition and insulin resistance.
Methods
Animal Procedures.
Nat1 KO mice were obtained from Jackson Labs. Mice were then generated for experiments breeding heterozygous × heterozygous mice to obtain WT and KO littermates mice. The animals were individually housed under controlled temperature (23 °C) and lighting (12:12 h light/dark cycle, lights on at 7:00 AM) conditions, with free access to water and food. Mice were fed either RC or a HFD (D12492; Research Diets) for 4 wk. Body composition was assessed by 1H magnetic resonance spectroscopy, using a Bruker Minispec analyzer (Bruker BioSpin). All experimental procedures were approved by and conducted in accordance with the Institutional Animal Care and Use Committee guidelines of Yale University School of Medicine.
Hyperinsulinemic-Euglycemic Clamp Studies.
Hyperinsulinemic-euglycemic clamps were performed as previously described (6). Briefly, a catheter was implanted in the jugular vein 7 d before the experiments. After overnight fasting, conscious mice were infused with [3-3H]-glucose (HPLC purified; Perkin-Elmer Life Sciences) at a rate of 0.05 μCi/min for 120 min for basal glucose turnover measurement. After the basal infusion, hyperinsulinemic-euglycemic clamps were conducted for 140 min with a 3-min primed infusion of insulin [6.0 mU/(kg-min)] and [3-3H]-glucose (0.24 μCi/min), followed by a continuous infusion of insulin [2.5 mU/(kg-min)] and [3-3H]-glucose (0.1 μCi/min), and a variable infusion of 20% dextrose to maintain euglycemia (∼120 mg/dL). After 85 min, a bolus of 2-deoxy-d-[1-14C]glucose (PerkinElmer) (10 μCi) was injected to evaluate insulin-stimulated tissue glucose uptake. At the end of the clamps, mice were anesthetized with a sodium pentobarbital injection (150 mg/kg), and liver, WAT, and skeletal muscle (gastrocnemius + soleus) were taken and snap-frozen in liquid nitrogen and stored at −80 °C for subsequent analyses.
Comprehensive Animal Metabolic Monitoring System.
Comprehensive animal metabolic monitoring system (Columbus Instruments) was used to evaluate O2 consumption, CO2 production, energy expenditure, activity, and food consumption. Drinking was assessed by a computer system counting consumed water droplets.
Lipid Measurements.
Tissue TAGs were extracted using the method of Folch et al. (41) and measured using a DCL TAG reagent (Diagnostic Chemicals). For DAG extraction, livers and muscles were homogenized in a buffer solution (20 mM Tris⋅HCl at pH 7.4, 1 mM EDTA, 0.25 mM EGTA, 250 mM sucrose) containing a protease inhibitor mixture (Roche), and samples were centrifuged at 100,000 × g for 1 h. The supernatants containing the cytosolic fraction and the pellet containing the membrane fraction were collected. DAG and ceramide concentrations were measured by LC-MS/MS, as previously described (30). Total cytosolic and membrane DAG and ceramide content are expressed as the sum of individual species. All lipid measurements were made from tissues harvested from 6-h fasted mice.
Oxygen Consumption Measurements.
Primary hepatocytes from WT and KO mice were isolated at the Yale Liver Center. Cells were washed three times with recovery media (DMEM with high glucose plus 10% FBS), and an equal amount of cells (12,000) was seeded in each well of a Seahorse XF24 cell culture plate (Seahorse Bioscience). These experiments were repeated six times under the same conditions as previously described (14). Briefly, cells were kept in recovery media for 4–6 h and then washed with DMEM media (low glucose plus 10% FBS) and incubated overnight. The following morning, cells were washed with prewarmed (∼37 °C) XF24 Assay media. XF24 Assay media (525 µL) was then added to each well. Immediately before measurements, cells with assay media were placed in an unbuffered, humidified incubator at 37 °C for 1 h to allow temperature and pH equilibration. Three measurements of oxygen consumption rate were taken, and the average of three measurements was used for analysis. These experiments were repeated six times using six different mice per group under the same conditions.
In addition, freshly isolated mouse gonadal or inguinal s.c. WAT or BAT were rinsed with XF-DMEM (containing 25 mM Hepes) and cut into small pieces (∼10 mg). After extensive washing, one piece of tissue was placed in each well of a XF24 Islet Capture Microplate (Seahorse Bioscience) and covered with the islet capture screen that allows free perfusion while minimizing tissue movement. XF Assay Medium (500 μL) was added, and samples were analyzed in the XF24 Analyzer.
Immunoblot Analysis.
Tissues were homogenized in RIPA lysis buffer supplemented with protease inhibitor mixture (Roche) for protein isolation. Proteins from homogenized liver or skeletal muscle (100 μg protein extracts) were electrophoretically separated by 4–12% SDS/PAGE (Invitrogen) and then transferred to polyvinylidene difluoride membranes (Millipore), using a semidry transfer cell (Bio-Rad) for 120 min. After blockade of nonspecific sites with 5% nonfat dry milk TBST (10 mM Tris, 100 mM NaCl, 0.1% Tween 20) solution, membranes were incubated overnight at 4 °C with the following primary antibodies: total IRK, tyrosine pY1158 IRK, tyrosine pY1162 IRK, total Akt, and Akt phosphorylation Ser473 (Santa Cruz Biotechnology, Inc.), PKCε (BD Transduction Laboratories), PKCθ (BD Transduction Laboratories), or GAPDH (Santa Cruz Biotechnology, Inc.). After washing with TBST, membranes were incubated with peroxidase-conjugated anti-rabbit, or anti-mouse. Membranes were thoroughly washed, and immune complexes were detected using an enhanced luminol chemiluminescence system (Thermo Scientific) and subjected to photographic films. Signals on the immunoblot were quantified by optical densitometry (Scion Image Software).
Statistics.
All data are expressed as mean ± SEM. Results were assessed using two-tailed unpaired Student’s t test or two-way ANOVA (GraphPad Prism 5). A P value less than 0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank Ali Nasiri, Mario Kahn, and Gina Butrico for their skilled technical assistance. This study was funded by grants from the US Public Health Service (R01 DK40936, R01 AG-23686, P30 DK059635). Kasper Faarkrog was supported with a Danish scholarship from Direktør Jacob Madsen og hustru Olga Madsens Fond.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1716990115/-/DCSupplemental.
References
- 1.Hotamisligil GS, Budavari A, Murray D, Spiegelman BM. Reduced tyrosine kinase activity of the insulin receptor in obesity-diabetes. Central role of tumor necrosis factor-alpha. J Clin Invest. 1994;94:1543–1549. doi: 10.1172/JCI117495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hotamisligil GS, Shargill NS, Spiegelman BM. Adipose expression of tumor necrosis factor-alpha: Direct role in obesity-linked insulin resistance. Science. 1993;259:87–91. doi: 10.1126/science.7678183. [DOI] [PubMed] [Google Scholar]
- 3.Ozcan L, Tabas I. Calcium signalling and ER stress in insulin resistance and atherosclerosis. J Intern Med. 2016;280:457–464. doi: 10.1111/joim.12562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Samuel VT, Shulman GI. Mechanisms for insulin resistance: Common threads and missing links. Cell. 2012;148:852–871. doi: 10.1016/j.cell.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Samuel VT, Shulman GI. The pathogenesis of insulin resistance: Integrating signaling pathways and substrate flux. J Clin Invest. 2016;126:12–22. doi: 10.1172/JCI77812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Camporez JP, et al. Cellular mechanism by which estradiol protects female ovariectomized mice from high-fat diet-induced hepatic and muscle insulin resistance. Endocrinology. 2013;154:1021–1028. doi: 10.1210/en.2012-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jurczak MJ, et al. Dissociation of inositol-requiring enzyme (IRE1α)-mediated c-Jun N-terminal kinase activation from hepatic insulin resistance in conditional X-box-binding protein-1 (XBP1) knock-out mice. J Biol Chem. 2012;287:2558–2567. doi: 10.1074/jbc.M111.316760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee HY, et al. Mitochondrial-targeted catalase protects against high-fat diet-induced muscle insulin resistance by decreasing intramuscular lipid accumulation. Diabetes. 2017;66:2072–2081. doi: 10.2337/db16-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shulman GI. Ectopic fat in insulin resistance, dyslipidemia, and cardiometabolic disease. N Engl J Med. 2014;371:1131–1141. doi: 10.1056/NEJMra1011035. [DOI] [PubMed] [Google Scholar]
- 10.Holland WL, et al. Receptor-mediated activation of ceramidase activity initiates the pleiotropic actions of adiponectin. Nat Med. 2011;17:55–63. doi: 10.1038/nm.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xia JY, et al. Targeted induction of ceramide degradation leads to improved systemic metabolism and reduced hepatic steatosis. Cell Metab. 2015;22:266–278. doi: 10.1016/j.cmet.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xia JY, Morley TS, Scherer PE. The adipokine/ceramide axis: Key aspects of insulin sensitization. Biochimie. 2014;96:130–139. doi: 10.1016/j.biochi.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knowles JW, et al. RISC (Relationship between Insulin Sensitivity and Cardiovascular Disease) Consortium; EUGENE2 (European Network on Functional Genomics of Type 2 Diabetes) Study; GUARDIAN (Genetics UndeRlying DIAbetes in HispaNics) Consortium; SAPPHIRe (Stanford Asian and Pacific Program for Hypertension and Insulin Resistance) Study Identification and validation of N-acetyltransferase 2 as an insulin sensitivity gene. J Clin Invest. 2015;125:1739–1751. doi: 10.1172/JCI74692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Camporez JP, et al. Cellular mechanisms by which FGF21 improves insulin sensitivity in male mice. Endocrinology. 2013;154:3099–3109. doi: 10.1210/en.2013-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Camporez JP, et al. Hepatic insulin resistance and increased hepatic glucose production in mice lacking Fgf21. J Endocrinol. 2015;226:207–217. doi: 10.1530/JOE-15-0136. [DOI] [PubMed] [Google Scholar]
- 16.Camporez JP, et al. ApoA5 knockdown improves whole-body insulin sensitivity in high-fat-fed mice by reducing ectopic lipid content. J Lipid Res. 2015;56:526–536. doi: 10.1194/jlr.M054080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Samuel VT, et al. Mechanism of hepatic insulin resistance in non-alcoholic fatty liver disease. J Biol Chem. 2004;279:32345–32353. doi: 10.1074/jbc.M313478200. [DOI] [PubMed] [Google Scholar]
- 18.Samuel VT, et al. Inhibition of protein kinase Cepsilon prevents hepatic insulin resistance in nonalcoholic fatty liver disease. J Clin Invest. 2007;117:739–745. doi: 10.1172/JCI30400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petersen MC, et al. Insulin receptor Thr1160 phosphorylation mediates lipid-induced hepatic insulin resistance. J Clin Invest. 2016;126:4361–4371. doi: 10.1172/JCI86013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petersen KF, et al. Mitochondrial dysfunction in the elderly: Possible role in insulin resistance. Science. 2003;300:1140–1142. doi: 10.1126/science.1082889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petersen KF, Dufour S, Befroy D, Garcia R, Shulman GI. Impaired mitochondrial activity in the insulin-resistant offspring of patients with type 2 diabetes. N Engl J Med. 2004;350:664–671. doi: 10.1056/NEJMoa031314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petersen KF, et al. Effect of aging on muscle mitochondrial substrate utilization in humans. Proc Natl Acad Sci USA. 2015;112:11330–11334. doi: 10.1073/pnas.1514844112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Befroy DE, et al. Impaired mitochondrial substrate oxidation in muscle of insulin-resistant offspring of type 2 diabetic patients. Diabetes. 2007;56:1376–1381. doi: 10.2337/db06-0783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perry RJ, et al. Hepatic acetyl CoA links adipose tissue inflammation to hepatic insulin resistance and type 2 diabetes. Cell. 2015;160:745–758. doi: 10.1016/j.cell.2015.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shulman GI. Cellular mechanisms of insulin resistance. J Clin Invest. 2000;106:171–176. doi: 10.1172/JCI10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Samuel VT, Petersen KF, Shulman GI. Lipid-induced insulin resistance: Unravelling the mechanism. Lancet. 2010;375:2267–2277. doi: 10.1016/S0140-6736(10)60408-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee HY, et al. Targeted expression of catalase to mitochondria prevents age-associated reductions in mitochondrial function and insulin resistance. Cell Metab. 2010;12:668–674. doi: 10.1016/j.cmet.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choi CS, et al. Overexpression of uncoupling protein 3 in skeletal muscle protects against fat-induced insulin resistance. J Clin Invest. 2007;117:1995–2003. doi: 10.1172/JCI13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim JK, et al. PKC-theta knockout mice are protected from fat-induced insulin resistance. J Clin Invest. 2004;114:823–827. doi: 10.1172/JCI22230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu C, et al. Mechanism by which fatty acids inhibit insulin activation of insulin receptor substrate-1 (IRS-1)-associated phosphatidylinositol 3-kinase activity in muscle. J Biol Chem. 2002;277:50230–50236. doi: 10.1074/jbc.M200958200. [DOI] [PubMed] [Google Scholar]
- 31.Kumashiro N, et al. Cellular mechanism of insulin resistance in nonalcoholic fatty liver disease. Proc Natl Acad Sci USA. 2011;108:16381–16385. doi: 10.1073/pnas.1113359108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ter Horst KW, et al. Hepatic diacylglycerol-associated protein kinase Cε translocation links hepatic steatosis to hepatic insulin resistance in humans. Cell Rep. 2017;19:1997–2004. doi: 10.1016/j.celrep.2017.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szendroedi J, et al. Role of diacylglycerol activation of PKCθ in lipid-induced muscle insulin resistance in humans. Proc Natl Acad Sci USA. 2014;111:9597–9602. doi: 10.1073/pnas.1409229111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perry RJ, et al. Reversal of hypertriglyceridemia, fatty liver disease, and insulin resistance by a liver-targeted mitochondrial uncoupler. Cell Metab. 2013;18:740–748. doi: 10.1016/j.cmet.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perry RJ, Zhang D, Zhang XM, Boyer JL, Shulman GI. Controlled-release mitochondrial protonophore reverses diabetes and steatohepatitis in rats. Science. 2015;347:1253–1256. doi: 10.1126/science.aaa0672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chennamsetty I, et al. Nat1 deficiency is associated with mitochondrial dysfunction and exercise intolerance in mice. Cell Rep. 2016;17:527–540. doi: 10.1016/j.celrep.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tschöp MH, et al. A guide to analysis of mouse energy metabolism. Nat Methods. 2011;9:57–63. doi: 10.1038/nmeth.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bonnard C, et al. Mitochondrial dysfunction results from oxidative stress in the skeletal muscle of diet-induced insulin-resistant mice. J Clin Invest. 2008;118:789–800. doi: 10.1172/JCI32601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang D, et al. Mitochondrial dysfunction due to long-chain Acyl-CoA dehydrogenase deficiency causes hepatic steatosis and hepatic insulin resistance. Proc Natl Acad Sci USA. 2007;104:17075–17080. doi: 10.1073/pnas.0707060104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morino K, et al. Reduced mitochondrial density and increased IRS-1 serine phosphorylation in muscle of insulin-resistant offspring of type 2 diabetic parents. J Clin Invest. 2005;115:3587–3593. doi: 10.1172/JCI25151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.