Significance
Ribosome-targeting macrolide antibiotics can be bacteriostatic or bactericidal. What distinguishes the action of bactericidal macrolides from that of bacteriostatic compounds is unclear. We found that neither the residual translation in the macrolide-treated cells nor the affinity of the inhibitors explain the static/cidal distinction. Instead, bactericidal compounds show a markedly decreased rate of dissociation from the ribosome because of the presence of an extended side chain in their structure. The prolonged translation downtime caused by the slow rate of antibiotic clearance could bring the cell to a point of no return due to depletion of the factors critical for resumption of gene expression. Optimizing the kinetics of binding/dissociation rather than only the affinity could facilitate the development of bactericidal inhibitors of translation.
Keywords: translation, inhibitors, ketolides, solithromycin, erythromycin
Abstract
Antibiotics can cause dormancy (bacteriostasis) or induce death (cidality) of the targeted bacteria. The bactericidal capacity is one of the most important properties of antibacterial agents. However, the understanding of the fundamental differences in the mode of action of bacteriostatic or bactericidal antibiotics, especially those belonging to the same chemical class, is very rudimentary. Here, by examining the activity and binding properties of chemically distinct macrolide inhibitors of translation, we have identified a key difference in their interaction with the ribosome, which correlates with their ability to cause cell death. While bacteriostatic and bactericidal macrolides bind in the nascent peptide exit tunnel of the large ribosomal subunit with comparable affinities, the bactericidal antibiotics dissociate from the ribosome with significantly slower rates. The sluggish dissociation of bactericidal macrolides correlates with the presence in their structure of an extended alkyl-aryl side chain, which establishes idiosyncratic interactions with the ribosomal RNA. Mutations or chemical alterations of the rRNA nucleotides in the drug binding site can protect cells from macrolide-induced killing, even with inhibitor concentrations that significantly exceed those required for cell growth arrest. We propose that the increased translation downtime due to slow dissociation of the antibiotic may damage cells beyond the point where growth can be reinitiated upon the removal of the drug due to depletion of critical components of the gene-expression pathway.
Antibiotics interrupt bacterial growth by interfering with vital cellular functions. Exposure to most antibiotics usually results in rapid cessation of cell growth. However, the fate of the bacterial cells after the removal of the inhibitor can differ dramatically, depending on the nature of the drug. Bacteriostatic antibiotics effectively prevent cell growth but, upon their removal, a significant fraction of the population can resume division. In contrast, only a few cells in the culture, if any, are able to restart growth and proliferation after being exposed to bactericidal drugs. Thus, medically useful antibiotics are operationally classified as bacteriostatic if, following exposure to a fourfold minimally inhibiting concentration (MIC) for 18–24 h, more than 0.1% of bacterial cells can resume growth, or bactericidal, if more than 99.9% of the treated cells never divide again (1). The distinction between bacteriostatic and bactericidal modes of antibiotic action is critically important for the outcome of antimicrobial therapies, especially in immunocompromised patients. However, the molecular mechanisms that differentiate bactericidal inhibitors from structurally related compounds, which act as bacteriostatic inhibitors, are unclear.
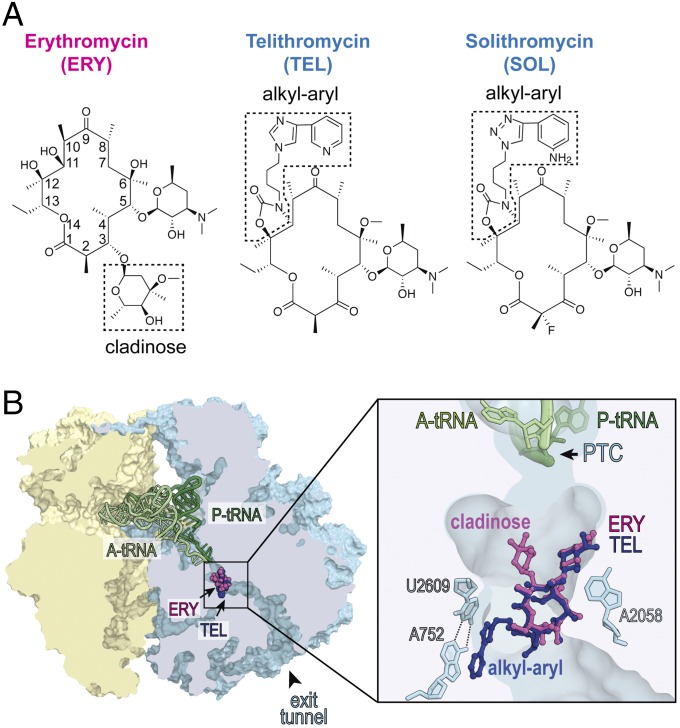
The ribosome-targeting macrolide antibiotics represent a vivid example of this conundrum, because this family of structurally similar drugs includes bacteriostatic as well as bactericidal inhibitors (2–4). Macrolides stop bacterial growth by targeting the ribosome and interfering with protein synthesis (reviewed in refs. 5 and 6). These antibiotics are composed of a macrolactone ring decorated with various substituents (Fig. 1A) and their properties critically depend on the presence and the nature of the side chains. Macrolides bind in the nascent peptide exit tunnel of the large ribosomal subunit and establish specific interactions with several 23S rRNA residues of the tunnel wall (7–11) (Fig. 1B). Erythromycin (ERY), a natural prototype compound of this class composed of a 14-atom macrolactone appended with two sugars (Fig. 1A), is a classic bacteriostatic antibiotic (12). In the newer generation of macrolides known as ketolides, the bulky C3-cladinose of ERY is replaced with a keto group and in addition, an extended alkyl-aryl side chain is attached at the 11,12 carbamate cycle (13, 14) (Fig. 1A). Remarkably, in contrast to bacteriostatic ERY, such ketolides as telithromycin (TEL) or solithromycin (SOL) exhibit moderate to strong cidality against several bacterial species (3, 15–18). The reasons why TEL and SOL, which are structurally related to bacteriostatic macrolides, bind to the same site in the ribosome and exhibit a similar mode of action, are able to kill bacteria, remains unclear.
Fig. 1.
Bacteriostatic and bactericidal macrolide and ketolide antibiotics bind to the same site in the bacterial ribosome. (A) Chemical structures of bacteriostatic macrolide ERY and bactericidal ketolides TEL and SOL. The C3-cladinose sugar of ERY and the alkyl-aryl side chains in TEL and SOL are boxed. (B) Binding site of macrolides and ketolides in the ribosome. A cross section of the bacterial (Thermus thermophilus) 70S ribosome with ERY (purple) and TEL (blue) bound in the nascent peptide exit tunnel (PDB ID codes: 4V7X and 4V7Z, respectively, from ref. 11). Small subunit is yellow, large subunit is light blue, A-site tRNA is light green, and P-site tRNA is dark green. The zoomed-in image shows interactions of the C5 desosamine of both antibiotics with A2058 and of the alkyl-aryl side chain of TEL with the A752-U2609 base pair in the nascent peptide exit tunnel. The peptidyl-transferase center (PTC) is indicated.
In this paper, we show that cidality strongly correlates with the kinetics of dissociation of the drug from the ribosome. Slowly dissociating macrolides exhibit bactericidal activity, whereas the drugs possessing comparable affinity but faster dissociation rate are bacteriostatic. We present evidence that the slow dissociation of macrolides from the ribosome depends on the presence of the extended alkyl-aryl side chain in the antibiotic molecule. On the basis of our findings, we propose a model where the prolonged inhibition of protein synthesis imposed by slowly dissociating antibiotics depletes one or several proteins essential for gene expression; this brings the cell to a point of no return in which, even upon removal of the inhibitor, restoration of translation and cell growth become no longer possible.
Results
Bactericidal Macrolide Antibiotics Retain Killing Efficiency in the Absence of Residual Translation.
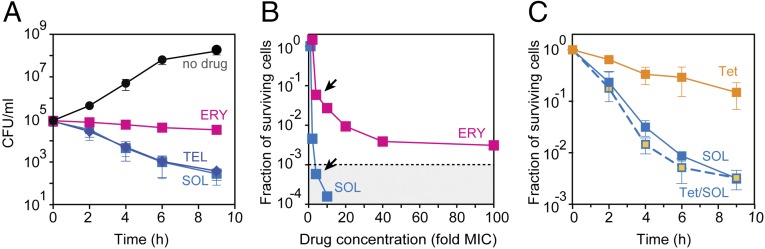
Macrolides are known to be highly active against Gram-positive Streptococcus pneumoniae (3, 15) and MIC testing confirmed that the growth of the S. pneumoniae strain Cp2000 (Table S1) (19) is readily inhibited by a variety of macrolides (Table S2). However, despite showing comparably low MICs, different macrolides varied significantly in their ability to kill bacteria. A large number of S. pneumoniae cells survived after being incubated for 9 h with a fourfold MIC of ERY (Fig. 2A). Even after an 18-h exposure to a 100-fold MIC of ERY, a sizable fraction of the cells were able to form colonies on antibiotic-free agar plates (Fig. 2B). In contrast, SOL exhibited a much more pronounced bactericidal effect at fourfold MIC (Fig. 2A) and essentially no cells survived the 18-h exposure to a 10-fold MIC of SOL (Fig. 2B).
Fig. 2.
Macrolide antibiotics differ significantly in their bactericidal activity. (A) Killing effect of fourfold MIC concentrations of ERY (magenta), TEL (dark blue), or SOL (light blue) against S. pneumoniae cells. The viable cells counts are indicated as colony forming units (CFU). (B) Survival of S. pneumoniae cells exposed for 18 h to various concentrations of ERY (magenta) or SOL (light blue). The experimental points determined at the fourfold MIC concentration of ERY or SOL are indicated by arrows. The dotted line marks the three-orders reduction in CFU, representing the operational definition of the bactericidal activity (1). (C) Survival of cells exposed for the indicated time periods to fourfold MIC of SOL (light blue), 40-fold MIC of Tet (orange), or fourfold MIC of SOL and 40-fold MIC of Tet (light blue/orange). In the latter case, cells were preincubated with Tet for 30 min before the addition of SOL. The error bars in A and C show SD in three independent experiments.
In contrast to antibiotics that indiscriminately inhibit translation, macrolides interfere with the production of specific subsets of cellular proteins (20). Cells exposed to 100-fold MIC of TEL or SOL continue to make a significant number of proteins (Fig. S1 B, D, and E), whereas only a small number of polypeptides are synthesized in S. pneumoniae treated with an equivalently high concentration of ERY (Fig. S1C). We had previously proposed that cidality of TEL or SOL could be related to the continued unbalanced translation of selected proteins (20). Here we noted, however, that the spectra of polypeptides synthesized in cells treated with bactericidal CEM103 or RU69874 (Fig. S1 F and G) resemble more closely those synthesized in the presence of the bacteriostatic ERY than those produced in cells exposed to bactericidal TEL or SOL (Fig. S1 A and C–G). This observation revealed the lack of correlation between cidality and persistent translation of certain proteins.
To test directly whether macrolide cidality requires residual translation, we employed the classic bacteriostatic antibiotic tetracycline (Tet), which interacts with the small ribosomal subunit and, by preventing binding of aminoacyl-tRNA, virtually stops protein synthesis in S. pneumoniae cells at fourfold MIC (Fig. S2) (21). Strikingly, the cidal ability of SOL remained essentially unaffected irrespective of the absence of Tet (when several proteins continued to be synthesized) or its presence (when practically no proteins were produced) (Fig. 2C). This result conclusively demonstrates that continuous synthesis of specific proteins in macrolide-treated cells is irrelevant to the cidality of these antibiotics.
Bactericidal and Bacteriostatic Macrolides Bind to the Ribosome with Comparable Affinities.
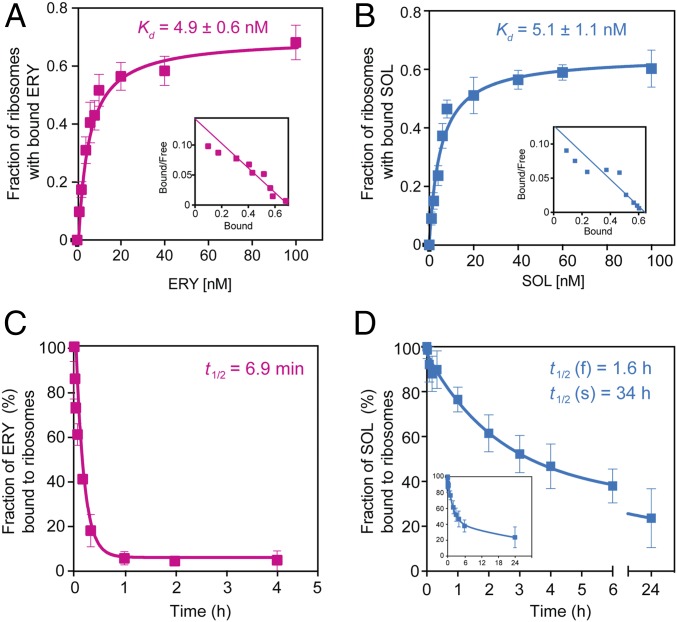
Having eliminated residual translation as an underlying factor determining ketolide cidality, we explored other options. One plausible scenario was that cidality could be defined by a higher affinity of bactericidal macrolides for the ribosome. We tested this possibility by comparing the apparent dissociation constants of bacteriostatic ERY or bactericidal SOL (Fig. 3 A and B). The equilibrium binding data showed that bacteriostatic ERY and bactericidal SOL have comparable affinities [Kd(ERY) = 4.9 ± 0.6 nM and Kd(SOL) = 5.1 ± 1.1 nM]. The determined Kd values were comparable with those previously reported for binding of ERY or SOL to ribosomes from other bacterial species (22–25). From these results we concluded that the drug-induced lethality could not be explained by a higher affinity of bactericidal macrolides for the ribosome.
Fig. 3.
Thermodynamic and kinetic parameters of interaction of bacteriostatic and bactericidal macrolides with the bacterial ribosome. (A and B) Equilibrium binding of [14C]-ERY (A) or [14C]-SOL (B) to S. pneumoniae ribosomes. Ribosomes were mixed with varying concentrations of the radiolabeled drugs and incubated for 2 h at 37 °C before determining the amount of bound antibiotic (see Materials and Methods for details). Insets show the Scatchard plots for ERY and SOL equilibrium binding. (C and D) Kinetics of dissociation of [14C]-ERY (C) or [14C]-SOL (D) from the ribosome. The Inset in D shows the complete curve. Ribosomes were preequilibrated with the [14C]-labeled antibiotics and after addition of an excess of the respective nonlabeled drug the amount of ribosome-associated radioactivity was monitored over time. The data were fitted to a one-phase (ERY) or two-phase (SOL) exponential functions that yielded dissociation rate constants of (10 ± 1.4) × 10−2 min−1 for ERY, (0.72 ± 0.25) × 10−2 min−1 for the faster (f) phase of SOL, and (0.034 ± 0.02) × 10−2 min−1 for the slower (s) phase of SOL. The values of dissociation rate constants were used for calculating the half-life (t1/2) of the complexes (shown in the figure). All experiments were performed in triplicates. Error bars represent standard deviation (SD).
Slow Dissociation from the Ribosome Distinguishes Bactericidal SOL from Bacteriostatic ERY.
Because the equilibrium dissociation constant reflects the ratio of the on- and off- rates of the ligand binding, inhibitors with comparable affinities can display significantly different kinetics of association and dissociation from the target. Therefore, we asked whether instead of the drug affinity, it is the difference in the rates of dissociation of ERY and SOL from the ribosome that underlies the difference in their killing potential. To measure the drug dissociation rates, ribosomes immobilized on DEAE magnetic beads were preequilibrated with [14C]-ERY or [14C]-SOL and, after addition of an excess of unlabeled drug, the displacement of the bound radiolabeled antibiotic was monitored (Fig. 3 C and D). In agreement with published data (22, 23), the dissociation of ERY from the ribosome occurred relatively fast (half-life 6.9 min) and could be described by a single exponential function. In contrast, [14C]-SOL dissociated from the ribosome with biphasic kinetics. Even the faster component of SOL dissociation exhibited a half-life of 1.6 h, which was much slower compared with that of ERY, whereas the half-life of the second component (34 h) was nearly 300 times longer in comparison with ERY. Hence, the dramatically slower dissociation rates distinguished the bactericidal SOL from the bacteriostatic ERY.
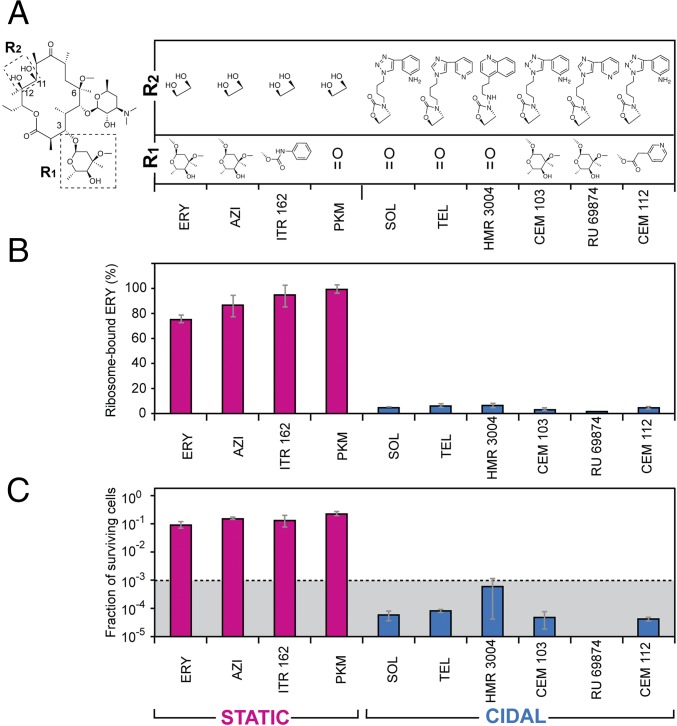
We tested whether the correlation between slow dissociation rate and cidality observed for SOL holds true for a broader range of macrolide compounds. Therefore, we compared the dissociation kinetics of a range of macrolide compounds from the ribosome and in parallel analyzed the extent of their bactericidal activity. Two key structural features distinguish bactericidal SOL and TEL from bacteriostatic ERY: the lack of C3-cladinose and the presence of an extended alkyl-aryl side chain (Fig. 1A). Therefore, we selected a number of macrolide compounds that vary in these two structural elements (Fig. 4A) and determined how rapidly they dissociate from the ribosome. Ribosomes were preequilibrated with the selected antibiotics and then, upon removal of the free inhibitor and addition of an excess of [14C]-ERY, the extent of replacement of prebound compound with ERY after 30-min incubation was measured. Strikingly, all tested antibiotics that lacked the 11,12 side chain dissociated rapidly and were efficiently replaced with [14C]-ERY (Fig. 4B). In contrast, almost no [14C]-ERY bound to the ribosomes that were preincubated with the 11,12 side chain-containing antibiotics (TEL, SOL, HMR3004, RU69874, CEM103, and CEM112) (Fig. 4B), indicating that a large fraction of these antibiotics remained associated to their ribosomal binding site.
Fig. 4.
Cidality of macrolides correlates with slow dissociation and the presence of an extended alkyl-aryl side chain. (A) Chemical structures of the C3 or C11, C12 side chains (boxed in the depicted ERY structure) of the macrolide and ketolide antibiotics. Note that ERY, AZI, and PKM carry a C6 hydroxyl instead of the shown metoxy group present in the rest of the compounds. (B) Displacement of nonlabeled macrolides bound to S. pneumoniae ribosomes by [14C]-ERY. After pre-equilibration with the corresponding antibiotic, ribosomes were incubated for 30 min at 37 °C with a 20-fold molar excess of [14C]-ERY and the amount of labeled ERY replacing the prebound inhibitor was measured. Binding of [14C]-ERY to the vacant drug-free ribosomes was taken as 100%. The blue bars represent slow-dissociating antibiotics that after 30-min incubation remain bound to more than 90% of the ribosomes; the fast-dissociating inhibitors that vacate more than 90% of the ribosomes over a 30-min incubation time are indicated by the magenta bars. (C) Surviving of S. pneumoniae cells after 18-h exposure to the corresponding fourfold MIC concentration of the indicated antibiotics. The dotted line indicates the three-orders reduction in the viable cell count representing the operational definition of the bactericidal activity. Error bars indicate SD in three independent experiments. The graph bars representing bactericidal antibiotics are blue and those corresponding to bacteriostatic inhibitors are magenta.
We then tested the cidality of the selected compounds by exposing cells for 18 h to fourfold MIC concentrations of these drugs. Replacement of the C3 keto group of SOL or TEL with cladinose (CEM103 and HMR69874, respectively) did not eliminate the bactericidal activity (Fig. 4C), suggesting that the nature of a chemical group at the C3 atom of the macrolactone ring is inconsequential for cidality. In contrast, all of the tested antibiotics that carried an alkyl-aryl side chain exhibited bactericidal activity. Appending such side chains to an erythromycin-like scaffold (CEM103, RU69874, or HMR004) or even to the minimalist ketolide PKM (thereby, essentially converting it into TEL), transformed the largely bacteriostatic drugs into strongly bactericidal compounds (Fig. 4C).
These results revealed two critical pieces of information: (i) the rate of antibiotic clearance from the ribosome likely determines the ability of the inhibitor to irreversibly abolish cell proliferation; and (ii) cidality of macrolides correlates with the presence in their structure of the extended alkyl-aryl side chain, which slows their dissociation, presumably due to the additional idiosyncratic interactions that this structure establishes with the ribosome (10, 26, 27).
Alterations in the Drug Binding Site Alleviates Cidality of the Macrolide Inhibitors.
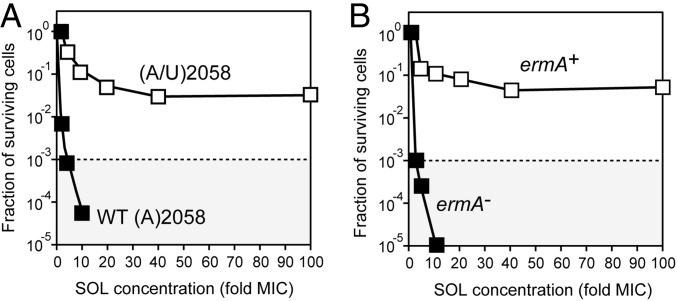
To extend our understanding of the mechanism of macrolide cidality, we selected S. pneumoniae mutants that, while retaining the general sensitivity toward an originally bactericidal macrolide, gained the ability to restore their growth upon the removal of the antibiotic. For that, cells were exposed for 6 h to a 100-fold MIC of TEL and then surviving cells were allowed to proliferate on an antibiotic-free agar plate. After repeating the selection cycle six times, eight of the randomly picked colonies were characterized. In all of the selected clones, two of the four rRNA operons acquired the same mutation in the 23S rRNA gene: nucleotide A2058 located in the macrolide binding site (Fig. 1B) was replaced with U. Consistently, approximately half of the ribosomes in the mutant cells carried the A2058U mutation (Fig. S3A). These mutants remained highly susceptible to SOL and TEL even though the MIC increased ∼10-fold compared with WT cells (Table S2). Remarkably, the cidality of SOL and TEL toward the mutant cells was dramatically reduced: while only a negligible fraction of the WT cells were able to resume growth after exposure to 100-fold MIC of SOL (Fig. 2 A and B), a significant fraction of the mutant cells survived the exposure to the adequately adjusted 100-fold MIC of this drug (Fig. 5A). The most straightforward explanation of these results is that the alterations in the macrolide binding site, which changed the interactions of the antibiotic with the mutant ribosomes in the cells, alleviated the cidal effect of the antibiotic. Interestingly, the overall level of residual translation in the WT and mutant cells at the respective 100-fold MIC of SOL remained essentially the same (Fig. S4). Therefore, the most likely explanation of why the otherwise bactericidal antibiotic lost the ability to kill the mutant cells is that the mutation of the rRNA residue that forms the key contacts with the antibiotic accelerated the kinetics of drug dissociation.
Fig. 5.
Alterations of the ribosomal drug-binding site protect bacteria from killing by bactericidal antibiotics. (A) Survival of the WT S. pneumoniae strain Cp2000 and the killing-resistant mutant upon exposure for 18 h to varying concentrations of SOL. All four 23S rRNA gene alleles in the WT cells carry A2058, whereas in the mutant cells the same 23S rRNA position in two of the alleles has been mutated to U. (B) Survival of the parental (ermA−) S. pneumoniae strain CPM1 and its ermA+ variant (Cp1290) upon exposure for 18 h to varying concentrations of SOL. In both panels the dotted line indicates the three-orders reduction in the viable cell count. The concentrations of SOL for the treatment of WT and altered strains were adjusted according to the respective MIC values (Table S2).
We further tested the correlation between cidality and the parameters of the drug binding by using S. pneumoniae cells expressing ErmA rRNA methyltransferase that modifies residue A2058. In the S. pneumoniae Cp1290 strain carrying the ermA gene (Table S1) (28), A2058 is modified in ∼94% of the ribosomes; of these, 77% of ribosomes carry dimethylated A2058 and in the remaining 17% the same nucleotide is monomethylated (Fig. S3B). Despite the A2058 modifications, the ermA(+) Cp1290 cells remained sensitive to SOL, even though the MIC was ∼100-fold higher compared with the parental strain lacking the methylase gene (Table S2). However, when the ermA(+) Cp1290 cells were exposed to 40 µg/mL of SOL (100-fold MIC), the drug showed dramatically reduced cidality compared with its effect upon ermA− cells at the respective MIC-fold value (Fig. 5B). Because of the overall reduced affinity of SOL to the ErmA-modified ribosome, it was not feasible to directly measure the kinetics of its binding or dissociation. However, we noted that, similar to the effect observed for the selected A2058U mutant, translation in the ErmA+ cells was efficiently inhibited at 100-fold MIC concentration of SOL (Fig. S4). These results suggest that the main reason for the improved survival of the ErmA-expressing cells is not the continued protein synthesis, but the altered kinetics of the drug binding.
Discussion
In this study we identified a major difference in the mode of action of bactericidal and bacteriostatic macrolide antibiotics targeting the ribosome. We show that pretreatment of cells with Tet, which completely stops cellular protein synthesis, does not affect the rate with which bactericidal macrolides reduce the number of viable cells; therefore, neither the residual translation of specific proteins (20), nor the reported miscoding activity of some macrolides (29, 30) could account for cidality. We further show that there is no major distinction in the general affinity of bacteriostatic and bactericidal macrolide compounds for the ribosome. Instead, our findings provide strong evidence that the bactericidal action of certain macrolide drugs could be explained by the slow rate of their dissociation from the ribosome.
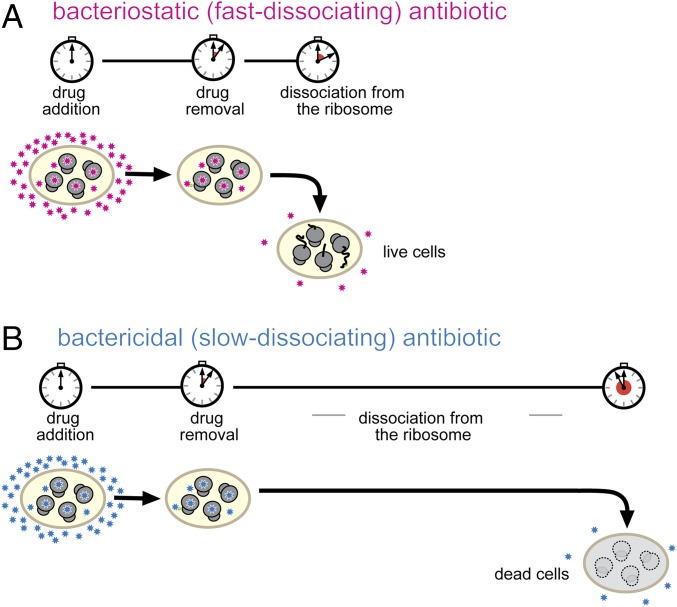
We propose a simple model that satisfactorily accounts for our experimental observations (Fig. 6). With fast-dissociating antibiotics, the extent of translation downtime is defined essentially by the duration of the antibiotic treatment (Fig. 6A). Soon after omission of the fast-dissociating inhibitor from the medium, translation resumes and the cell can recover from the antibiotic-induced dormancy. In contrast, slowly dissociating drugs continue to block protein synthesis long after antibiotic withdrawal (Fig. 6B). When the inhibitor eventually vacates a sufficient fraction of ribosomes, translation could be potentially resumed but only if enough ribosomes, translation factors, aminoacyl-tRNA synthetases, and all other direct or accessory participants of the gene-expression pathway remain available and functional. However, if the cellular content of at least one of the essential components falls below a critical level, protein synthesis cannot be restarted even when ribosomes become drug-free.
Fig. 6.
Prolonged inhibition of protein synthesis leads to cell death. (A) With fast-dissociating inhibitors (magenta stars), the duration of protein synthesis shut-down is determined primarily by the timing of exposure to the antibiotic. Translation readily resumes upon the removal of the drug and cells can restart their growth. (B) With slowly dissociating antibiotics (blue stars), the translation downtime is prolonged due to the slow off-rate of the drug. During the extended translation down time depletion of critical cellular components prevents the resumption of translation and of cell growth.
The steady-state level of functional biomolecules in the cell is determined by the rates of their production and degradation (31). Once translation is interrupted, the decay will prevail and eventually the concentration of a protein, RNA or other components will fall below a critical threshold, making resumption of protein synthesis impossible. Our findings are also compatible with the possibility that interrupted translation depletes cells of proteins involved in detoxification of harmful chemicals, exposure to which could cause irreparable damage (32). Currently, we do not know which key component gets depleted first when cells are deprived of active translation. It is conceivable that overproduction of a limiting factor could prolong cell survival in the absence of protein synthesis, while its depletion before antibiotic treatment could increase the antibiotic cidality. Identification of the factors determining cell persistence in the absence of active translation could be the subject of future studies.
One of the predictions that follows from the proposed model of macrolide cidality is that resumption of cell growth should not be possible if exposure to a normally bacteriostatic inhibitor is prolonged for an excessive period of time. Indeed, we observed a steady drop over time in the number of viable cells upon exposure of S. pneumoniae to either the macrolide (ERY) or the unrelated Tet (Fig. S5), both classic bacteriostatic protein synthesis inhibitors. Our model of macrolide cidality was corroborated further by the finding that changing the structure, and thus the properties, of the macrolide binding site in the ribosome dramatically improves cell survival upon treatment with an excess of bactericidal macrolides. Because such alterations do not increase residual translation in the presence of the excess of antibiotic, we believe that the alterations of the binding site result in an accelerated dissociation of the drug from the modified ribosome leading to a faster restoration of translation upon antibiotic removal.
Although we originally hypothesized that the cidality of ketolides is defined by the lack of C3 cladinose (20), here we showed that it is rather the presence of an alkyl-aryl side chain that converts a bacteriostatic inhibitor into a bactericidal one by slowing down the rate of its dissociation from the ribosome (Fig. 4B). In agreement with this conclusion, the clinical macrolide candidates carrying extended alkyl-aryl side chains, exhibit a prolonged postantibiotic effect: that is, delayed regrowth of bacteria following exposure to antibiotic (3, 33–35). Previous binding studies have suggested that TEL, an example of a drug with an extended side chain (Fig. 1A), binds to the Escherichia coli ribosome in a two-step process, with the slower step gated by formation of a strong stacking interaction between the alkyl-aryl side chain of the drug and the base pair U2609:A752 of 23S rRNA (Fig. 1B) (27). While the exact position of the alky-aryl side chain in the S. pneumoniae ribosome is unknown, it is conceivable that the slow off-rate of bactericidal macrolides could be delimited by breaking the interactions of the alkyl-aryl moiety of the drug with the target. Different placements of the side chain in the ribosome or the presence of structurally distinct populations of ribosomes in our preparation could account for the biphasic nature of dissociation of bactericidal antibiotics.
Strikingly sluggish dissociation of the alkyl-aryl–equipped macrolides from the vacant ribosomes observed in in vitro experiments could be exacerbated even further in the living cell. While some short-growing peptide chains can dislodge the macrolide inhibitor from its binding site (36, 37), many nascent chains bypass the antibiotic molecule in the exit tunnel without displacing it (20, 30). The growing protein is elongated at a fast rate of ∼20 residues per second (38); therefore, if the drug fails to dissociate rapidly at the early stages of translation, it would be trapped in its binding site by the elongated nascent polypeptide. Subsequent arrest of the translation at the sequence problematic for the macrolide-bound ribosome (20, 39–41) could then make dissociation of the drug particularly slow.
Noteworthy, the placement of the macrolide’s alkyl-aryl chain may vary when the drug binds to ribosomes from different species (9–11, 42–45). The idiosyncratic interactions of the side chain with the target could explain varying bactericidal activity of the macrolides against different bacteria (46, 47). Our results suggest that optimizing such interactions with the purpose of reducing the off-rate of the inhibitor rather than its general affinity could be a way to improve the bactericidal activity and possibly species-specificity of macrolide antibiotics.
Materials and Methods
Antibiotics and bacterial strains used in the study are listed in SI Materials and Methods.
Determination of the MIC.
All S. pneumoniae strains were cultivated in the casein-tryptone (CAT) medium, as described in ref. 48. Exponentially growing S. pneumoniae cells were diluted to the final culture density of A600 = 0.001, and cultivated in 96-well plates (100 µL per well) in the presence of increasing concentrations of antibiotics. After overnight incubation at 37 °C, the presence of live cells was detected by positive staining with AlamarBlue dye.
Kinetics and Concentration Dependence of Macrolide-Induced Cell Killing.
The cidality assay was carried out according to National Committee for Clinical Laboratory Standards guidelines (1). Specifically, exponential culture of S. pneumoniae was diluted to 105 CFU/mL in 5 mL of CAT media supplemented with fourfold MIC of an antibiotic and the mixture was incubated at 37 °C without shaking. Aliquots of the culture (0.1 mL) were withdrawn at different times of incubation, diluted 50- and 2,500-times in antibiotic-free CAT media, and embedded in agar plates (5 mL of CAT media mixed with 5 mL of 1.5% CAT agar) (48). Colonies were counted after 36- to 48-h incubation at 37 °C.
The concentration dependence of antibiotic cidality was examined by incubating S. pneumoniae cells (105 CFU/mL) in 5 mL of CAT media with 2-, 4-, 10-, 20-, 40-, and 100-fold MIC of antibiotics at 37 °C without shaking. After 18 h of incubation, 0.5-mL aliquots of the cultures were passed by centrifugation (4,000 × g for 1 min) through the Spin-X filters (Costar) to remove antibiotic. Cells retained on the filter were resuspended in 0.1 mL CAT, and the dilutions were plated on CAT agar plates as described above. Colonies were counted after 36–48 h incubation at 37 °C.
Equilibrium Binding of Antibiotics to the Ribosome.
S. pneumoniae ribosomes, purified according to ref. 49, were diluted to 3 nM and combined with varying concentrations of [14C]-ERY or [14C]-SOL in 1.5 mL of binding buffer (20 mM Tris⋅HCl, pH 7.6, 10 mM MgCl2, 150 NH4Cl, and 6 mM β-mercaptoethanol). After a 2-h incubation at 37 °C, 10 µL of a 50 mg/mL suspension of DEAE magnetic beads (BioClone) was added to the reactions and incubation continued for 15 min at room temperature. The beads with immobilized ribosomes were captured using a magnetic stand (Invitrogen), the supernatant was aspirated and beads were rapidly washed twice with 0.7 mL of ice-cold binding buffer. Ribosomes was released by resuspending the magnetic beads in 100 µL of 10 mM EDTA and incubating for 10 min at room temperature. Beads were recaptured and ribosome-containing supernatant was transferred to scintillation vials. After quantifying the ribosome-bound radioactivity, the Kd values were calculated using Prism software (GraphPad).
The Kinetics of Dissociation of Antibiotics from the Ribosome.
Ribosomes (225 pmol), preimmobilized on 7.5 mg of DEAE magnetic beads, were incubated for 1 h at 37 °C with 225 pmol of [14C]-ERY or [14C]-SOL in 15 mL of binding buffer. After addition of a 50-fold molar excess of the respective nonradioactive antibiotic solution, incubation continued at 37 °C and 1.5-mL aliquots of the reaction suspension were transferred to an Eppendorf tube at the specified time points. Magnetic beads were captured, the supernatant was removed, and the ribosomes were eluted using 10 mM EDTA solution, as described above. The amount of the ribosome-associated radioactivity was quantified in a scintillation counter. The experimental data points were fitted to one-phase (ERY) or two-phase (SOL) exponential functions using Prism software (GraphPad).
Displacement of Ribosome-Bound Macrolides by [14C]- ERY.
Ribosomes (15 pmol) immobilized on 0.5 mg of DEAE magnetic beads were preincubated with 15 pmol of different label-free macrolide antibiotics in 1.5 mL of binding buffer for 1 h at 37 °C. The reactions were then supplemented with 300 pmol of [14C]-ERY and incubation continued at 37 °C for 30 min. The beads with immobilized ribosomes were captured, washed twice with ice-cold binding buffer, and treated with 10 mM EDTA as described above. Eluted radioactivity was quantified in a scintillation counter.
Supplementary Material
Acknowledgments
We thank Donald Morrison (University of Illinois at Chicago) for providing the bacterial strains and the invaluable advice on their manipulation; Prabha Fernandes (Cempra, Inc), Andre Bryskier (Aventis Pharma), and Scott Franzblau (Institute of Tuberculosis Research, University of Illinois at Chicago) for providing various macrolide antibiotics; Prabha Fernandes for advising us on using S. pneumoniae for this study; and Elizabeth Woods for her help in proofreading the manuscript. This work was supported by the NIH Grant R01 GM106386.
Footnotes
Conflict of interest statement: In the previous years, A.S.M. had grants from Cempra, Inc. and Melinta Therapeutics, which were involved in the development of macrolide antibiotics.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1717168115/-/DCSupplemental.
References
- 1.Clinical and Laboratory Standards Institute . Methods for Determining Bactericidal Activity of Antimicrobial Agents; Approved Guideline. NCCLS Document M26-A. NCCLS; Wayne, PA: 1999. [Google Scholar]
- 2.Goldstein FW, Emirian MF, Coutrot A, Acar JF. Bacteriostatic and bactericidal activity of azithromycin against Haemophilus influenzae. J Antimicrob Chemother. 1990;25(Suppl A):25–28. doi: 10.1093/jac/25.suppl_a.25. [DOI] [PubMed] [Google Scholar]
- 3.Woosley LN, Castanheira M, Jones RN. CEM-101 activity against Gram-positive organisms. Antimicrob Agents Chemother. 2010;54:2182–2187. doi: 10.1128/AAC.01662-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kosowska K, et al. Activities of two novel macrolides, GW 773546 and GW 708408, compared with those of telithromycin, erythromycin, azithromycin, and clarithromycin against Haemophilus influenzae. Antimicrob Agents Chemother. 2004;48:4113–4119. doi: 10.1128/AAC.48.11.4113-4119.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaynor M, Mankin AS. Macrolide antibiotics: Binding site, mechanism of action, resistance. Curr Top Med Chem. 2003;3:949–961. doi: 10.2174/1568026033452159. [DOI] [PubMed] [Google Scholar]
- 6.Dinos GP. The macrolide antibiotic renaissance. Br J Pharmacol. 2017;174:2967–2983. doi: 10.1111/bph.13936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moazed D, Noller HF. Chloramphenicol, erythromycin, carbomycin and vernamycin B protect overlapping sites in the peptidyl transferase region of 23S ribosomal RNA. Biochimie. 1987;69:879–884. doi: 10.1016/0300-9084(87)90215-x. [DOI] [PubMed] [Google Scholar]
- 8.Schlünzen F, et al. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature. 2001;413:814–821. doi: 10.1038/35101544. [DOI] [PubMed] [Google Scholar]
- 9.Tu D, Blaha G, Moore PB, Steitz TA. Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance. Cell. 2005;121:257–270. doi: 10.1016/j.cell.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 10.Dunkle JA, Xiong L, Mankin AS, Cate JH. Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action. Proc Natl Acad Sci USA. 2010;107:17152–17157. doi: 10.1073/pnas.1007988107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bulkley D, Innis CA, Blaha G, Steitz TA. Revisiting the structures of several antibiotics bound to the bacterial ribosome. Proc Natl Acad Sci USA. 2010;107:17158–17163. doi: 10.1073/pnas.1008685107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garrod LP, Waterworth PM. Behaviour in vitro of some new antistaphylococcal antibiotics. BMJ. 1956;2:61–65. doi: 10.1136/bmj.2.4984.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bryskier A, Denis A. Ketolides: Novel antibacterial agents designed to overcome resistance to erythromycin A within gram-positive cocci. In: Schönfeld W, Kirst HA, editors. Macrolide Antibiotics. Birkhäuser; Basel: 2002. pp. 97–140. [Google Scholar]
- 14.Fernandes P, Martens E, Bertrand D, Pereira D. The solithromycin journey—It is all in the chemistry. Bioorg Med Chem. 2016;24:6420–6428. doi: 10.1016/j.bmc.2016.08.035. [DOI] [PubMed] [Google Scholar]
- 15.Hamilton-Miller JM, Shah S. Comparative in-vitro activity of ketolide HMR 3647 and four macrolides against gram-positive cocci of known erythromycin susceptibility status. J Antimicrob Chemother. 1998;41:649–653. doi: 10.1093/jac/41.6.649. [DOI] [PubMed] [Google Scholar]
- 16.Pankuch GA, Visalli MA, Jacobs MR, Appelbaum PC. Susceptibilities of penicillin- and erythromycin-susceptible and -resistant pneumococci to HMR 3647 (RU 66647), a new ketolide, compared with susceptibilities to 17 other agents. Antimicrob Agents Chemother. 1998;42:624–630. doi: 10.1128/aac.42.3.624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malathum K, Coque TM, Singh KV, Murray BE. In vitro activities of two ketolides, HMR 3647 and HMR 3004, against gram-positive bacteria. Antimicrob Agents Chemother. 1999;43:930–936. doi: 10.1128/aac.43.4.930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Credito KL, Ednie LM, Jacobs MR, Appelbaum PC. Activity of telithromycin (HMR 3647) against anaerobic bacteria compared to those of eight other agents by time-kill methodology. Antimicrob Agents Chemother. 1999;43:2027–2031. doi: 10.1128/aac.43.8.2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weng L, Piotrowski A, Morrison DA. Exit from competence for genetic transformation in Streptococcus pneumoniae is regulated at multiple levels. PLoS One. 2013;8:e64197. doi: 10.1371/journal.pone.0064197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kannan K, Vázquez-Laslop N, Mankin AS. Selective protein synthesis by ribosomes with a drug-obstructed exit tunnel. Cell. 2012;151:508–520. doi: 10.1016/j.cell.2012.09.018. [DOI] [PubMed] [Google Scholar]
- 21.Wilson DN. The A-Z of bacterial translation inhibitors. Crit Rev Biochem Mol Biol. 2009;44:393–433. doi: 10.3109/10409230903307311. [DOI] [PubMed] [Google Scholar]
- 22.Pestka S. Binding of [14C]erythromycin to Escherichia coli ribosomes. Antimicrob Agents Chemother. 1974;6:474–478. doi: 10.1128/aac.6.4.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lovmar M, et al. Erythromycin resistance by L4/L22 mutations and resistance masking by drug efflux pump deficiency. EMBO J. 2009;28:736–744. doi: 10.1038/emboj.2009.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dinos GP, Kalpaxis DL. Kinetic studies on the interaction between a ribosomal complex active in peptide bond formation and the macrolide antibiotics tylosin and erythromycin. Biochemistry. 2000;39:11621–11628. doi: 10.1021/bi000811f. [DOI] [PubMed] [Google Scholar]
- 25.Llano-Sotelo B, et al. Binding and action of CEM-101, a new fluoroketolide antibiotic that inhibits protein synthesis. Antimicrob Agents Chemother. 2010;54:4961–4970. doi: 10.1128/AAC.00860-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garza-Ramos G, Xiong L, Zhong P, Mankin A. Binding site of macrolide antibiotics on the ribosome: New resistance mutation identifies a specific interaction of ketolides with rRNA. J Bacteriol. 2001;183:6898–6907. doi: 10.1128/JB.183.23.6898-6907.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kostopoulou ON, Petropoulos AD, Dinos GP, Choli-Papadopoulou T, Kalpaxis DL. Investigating the entire course of telithromycin binding to Escherichia coli ribosomes. Nucleic Acids Res. 2012;40:5078–5087. doi: 10.1093/nar/gks174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo P, Morrison DA. Transient association of an alternative sigma factor, ComX, with RNA polymerase during the period of competence for genetic transformation in Streptococcus pneumoniae. J Bacteriol. 2003;185:349–358. doi: 10.1128/JB.185.1.349-358.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson J, Pratt CA, Dahlberg AE. Effects of a number of classes of 50S inhibitors on stop codon readthrough during protein synthesis. Antimicrob Agents Chemother. 2004;48:4889–4891. doi: 10.1128/AAC.48.12.4889-4891.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gupta P, Kannan K, Mankin AS, Vázquez-Laslop N. Regulation of gene expression by macrolide-induced ribosomal frameshifting. Mol Cell. 2013;52:629–642. doi: 10.1016/j.molcel.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levin BR, et al. A numbers game: Ribosome densities, bacterial growth, and antibiotic-mediated stasis and death. MBio. 2017;8:e02253-16. doi: 10.1128/mBio.02253-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dwyer DJ, Collins JJ, Walker GC. Unraveling the physiological complexities of antibiotic lethality. Annu Rev Pharmacol Toxicol. 2015;55:313–332. doi: 10.1146/annurev-pharmtox-010814-124712. [DOI] [PubMed] [Google Scholar]
- 33.Boswell FJ, Andrews JM, Wise R. Pharmacodynamic properties of HMR 3647, a novel ketolide, on respiratory pathogens, enterococci and Bacteroides fragilis demonstrated by studies of time-kill kinetics and postantibiotic effect. J Antimicrob Chemother. 1998;41:149–153. doi: 10.1093/jac/41.2.149. [DOI] [PubMed] [Google Scholar]
- 34.Jacobs MR, Bajaksouzian S, Appelbaum PC. Telithromycin post-antibiotic and post-antibiotic sub-MIC effects for 10 Gram-positive cocci. J Antimicrob Chemother. 2003;52:809–812. doi: 10.1093/jac/dkg437. [DOI] [PubMed] [Google Scholar]
- 35.Cao Z, et al. Ribosome affinity and the prolonged molecular postantibiotic effect of cethromycin (ABT-773) in Haemophilus influenzae. Int J Antimicrob Agents. 2004;24:362–368. doi: 10.1016/j.ijantimicag.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 36.Tripathi S, Kloss PS, Mankin AS. Ketolide resistance conferred by short peptides. J Biol Chem. 1998;273:20073–20077. doi: 10.1074/jbc.273.32.20073. [DOI] [PubMed] [Google Scholar]
- 37.Lovmar M, et al. The molecular mechanism of peptide-mediated erythromycin resistance. J Biol Chem. 2006;281:6742–6750. doi: 10.1074/jbc.M511918200. [DOI] [PubMed] [Google Scholar]
- 38.Bremer H, Dennis P. Modulation of chemical composition and other parameters of the cell by growth rate. In: Neidhardt FC, et al., editors. Escherichia coli and Salmonella: Cellular and Molecular Biology. 2nd Ed. Vol 2. ASM Press; Washington, DC: 1996. pp. 1553–1569. [Google Scholar]
- 39.Davis AR, Gohara DW, Yap MN. Sequence selectivity of macrolide-induced translational attenuation. Proc Natl Acad Sci USA. 2014;111:15379–15384. doi: 10.1073/pnas.1410356111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kannan K, et al. The general mode of translation inhibition by macrolide antibiotics. Proc Natl Acad Sci USA. 2014;111:15958–15963. doi: 10.1073/pnas.1417334111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sothiselvam S, et al. Binding of macrolide antibiotics leads to ribosomal selection against specific substrates based on their charge and size. Cell Rep. 2016;16:1789–1799. doi: 10.1016/j.celrep.2016.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiong L, Shah S, Mauvais P, Mankin AS. A ketolide resistance mutation in domain II of 23S rRNA reveals the proximity of hairpin 35 to the peptidyl transferase centre. Mol Microbiol. 1999;31:633–639. doi: 10.1046/j.1365-2958.1999.01203.x. [DOI] [PubMed] [Google Scholar]
- 43.Hansen LH, Mauvais P, Douthwaite S. The macrolide-ketolide antibiotic binding site is formed by structures in domains II and V of 23S ribosomal RNA. Mol Microbiol. 1999;31:623–631. doi: 10.1046/j.1365-2958.1999.01202.x. [DOI] [PubMed] [Google Scholar]
- 44.Schlünzen F, et al. Structural basis for the antibiotic activity of ketolides and azalides. Structure. 2003;11:329–338. doi: 10.1016/s0969-2126(03)00022-4. [DOI] [PubMed] [Google Scholar]
- 45.Eyal Z, et al. Structural insights into species-specific features of the ribosome from the pathogen Staphylococcus aureus. Proc Natl Acad Sci USA. 2015;112:E5805–E5814. doi: 10.1073/pnas.1517952112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Al-Lahham A, Appelbaum PC, van der Linden M, Reinert RR. Telithromycin-nonsusceptible clinical isolates of Streptococcus pneumoniae from Europe. Antimicrob Agents Chemother. 2006;50:3897–3900. doi: 10.1128/AAC.00057-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kays MB, Lisek CR, Denys GA. Comparative in vitro and bactericidal activities of telithromycin against penicillin-nonsusceptible, levofloxacin-resistant, and macrolide-resistant Streptococcus pneumoniae by time-kill methodology. Int J Antimicrob Agents. 2007;29:289–294. doi: 10.1016/j.ijantimicag.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 48.Tovpeko Y, Morrison DA. Competence for genetic transformation in Streptococcus pneumoniae: Mutations in σA bypass the comW requirement. J Bacteriol. 2014;196:3724–3734. doi: 10.1128/JB.01933-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ohashi H, Shimizu Y, Ying BW, Ueda T. Efficient protein selection based on ribosome display system with purified components. Biochem Biophys Res Commun. 2007;352:270–276. doi: 10.1016/j.bbrc.2006.11.017. [DOI] [PubMed] [Google Scholar]
- 50.Almutairi MM, et al. Resistance to ketolide antibiotics by coordinated expression of rRNA methyltransferases in a bacterial producer of natural ketolides. Proc Natl Acad USA. 2015;112:12956–12961. doi: 10.1073/pnas.1512090112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cato A, Guild WR. Transformation and DNA size. I. Activity of fragments of defined size and a fit to a random double cross-over model. J Mol Biol. 1968;37:157–178. doi: 10.1016/0022-2836(68)90080-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.