Significance
The Hedgehog pathway is critical in development and disease, but how cells respond to the secreted Hedgehog signal remains mysterious. A key step involves the regulation of the seven-transmembrane oncoprotein Smoothened by the 12-pass transporter-like Hedgehog receptor Patched1. We investigate the model that Patched1 is an ion-driven transporter of an endogenous lipidic Smoothened ligand. Whereas Patched–Smoothened regulation has traditionally been studied through indirect, downstream pathway readouts, we developed rapid, direct functional assays to dissect this step in simplified cell-based and in vitro systems. Cholesterol, a major membrane lipid, constitutively activates purified Smoothened by engaging its membrane-spanning region. Patched1 activity depends on extracellular Na+, suggesting that transmembrane Na+ gradients, universal among metazoans, might power Patched1 transporter-like activity in Smoothened regulation.
Keywords: Hedgehog, Patched, Smoothened, cholesterol, Na+
Abstract
Hedgehog signaling specifies tissue patterning and renewal, and pathway components are commonly mutated in certain malignancies. Although central to ensuring appropriate pathway activity in all Hedgehog-responsive cells, how the transporter-like receptor Patched1 regulates the seven-transmembrane protein Smoothened remains mysterious, partially due to limitations in existing tools and experimental systems. Here we employ direct, real-time, biochemical and physiology-based approaches to monitor Smoothened activity in cellular and in vitro contexts. Patched1–Smoothened coupling is rapid, dynamic, and can be recapitulated without cilium-specific proteins or lipids. By reconstituting purified Smoothened in vitro, we show that cholesterol within the bilayer is sufficient for constitutive Smoothened activation. Cholesterol effects occur independently of the lipid-binding Smoothened extracellular domain, a region that is dispensable for Patched1–Smoothened coupling. Finally, we show that Patched1 specifically requires extracellular Na+ to regulate Smoothened in our assays, raising the possibility that a Na+ gradient provides the energy source for Patched1 catalytic activity. Our work suggests a hypothesis wherein Patched1, chemiosmotically driven by the transmembrane Na+ gradient common to metazoans, regulates Smoothened by shielding its heptahelical domain from cholesterol, or by providing an inhibitor that overrides this cholesterol activation.
The Hedgehog (Hh) signaling pathway is a fundamental regulator of embryonic tissue patterning and of postnatal tissue renewal and regeneration (1–5). Loss of Hh pathway activity during development thus gives rise to birth defects, whereas dysregulated pathway activity can directly drive the growth of several malignancies, including basal cell carcinoma, the most common form of skin cancer (6). Hh pathway activity also indirectly restrains growth of certain endodermally derived cancers by causing stromal expression of secreted factors that promote differentiation of epithelial tumor cells (7–12).
The mechanism of Hh response is unusual among signaling cascades for its partitioning of ligand reception and intracellular signal transmission between two distinct multipass transmembrane proteins. Binding of Hh to the transporter-like protein Patched1 (Ptch1) thus releases its constitutive inhibition of the seven-transmembrane (7TM) protein Smoothened (Smo) to initiate coupling to Gli transcription factors within the vertebrate primary cilium (2, 3, 13, 14). Ptch1–Smo regulation plays a central role in ensuring appropriate pathway activity in all Hh-responsive cells, and errors in this process instigate the majority of Hh pathway-driven cancers. However, the molecular mechanism by which Ptch1 communicates with Smo has remained mysterious. A leading model proposes that Ptch1 is an ion-driven transporter that regulates availability of an endogenous lipidic Smo ligand (15). Because Ptch1 and Smo traffic through the primary cilium, it has been speculated that such an endogenous Smo ligand might be highly enriched within the ciliary membrane where Hh pathway regulation takes place (3). This model is based in part on shared sequence homology between Ptch1 and prokaryotic resistance-nodulation-division (RND) transporters that export substrates using a transmembrane chemiosmotic gradient, most commonly a proton-motive force (16). Direct empirical support for these claims, however, is lacking. Indeed, Ptch1 has not been demonstrated to possess intrinsic transport activity, nor does a strong proton gradient exist across metazoan plasma membranes. Thus, the energy source for Ptch1 activity has not been experimentally defined, hindering efforts to evaluate whether the transporter-based model for Ptch1 function is correct.
Furthermore, the identity, subcellular distribution, and mode of action for the endogenous Ptch1-regulated Smo ligand remain unresolved (13, 17). However, among all classes of cellular lipids, sterols have emerged as particularly attractive candidates for mediating Ptch1–Smo communication. Several vertebrate Ptch1-related proteins are functionally linked to sterols, including Niemann-Pick C1 (NPC1), a factor required for egress of LDL-derived cholesterol from lysosomes (18), and Dispatched, which helps to release the cholesterol-modified Hh ligand from Hh-producing cells (19, 20). Hh pathway activity is also highly sensitive to genetic or pharmacological blockade of cholesterol metabolism (21), an effect that maps downstream of Ptch1 and at or upstream of Smo. Supplying exogenous cholesterol to sterol-depleted cells reverses the resulting pathway inhibition (22–24). Furthermore, elevating cellular cholesterol to supraphysiologic levels can overcome Ptch1 inhibitory effects to promote Smo activity, as well as enhance the ability of Hh to derepress Ptch1 (23, 25). These findings raise the possibility that the endogenous Smo ligand might be cholesterol or one of its derivatives, and that Ptch1 might regulate Smo activity by controlling this sterol’s availability. Nevertheless, several fundamental questions remain unanswered. (i) Is Smo activity regulated by cholesterol itself or rather by one of its many metabolic products (such as oxysterols, bile acids, or steroid hormones), all of which are affected by the treatments described above? (ii) Does this sterol act directly on Smo or does it instead target an as yet unidentified pathway component between Ptch1 and Smo? (iii) Is cholesterol merely a cofactor required for efficient Smo activation by another lipid, or is cholesterol sufficient to activate Smo in its own right? (iv) If cholesterol is a Smo ligand, where on Smo is its site of action?
These remaining gaps in our understanding are difficult to resolve by the traditional approaches for measuring Hh pathway activity. Ptch1 and Smo function are typically evaluated using downstream transcriptional readouts or alterations in the ciliary accumulation of pathway components. Such indirect methods measure long-term consequences of changes in Ptch1 and Smo activity in ciliated cells, rendering them unsuitable for experimental manipulations that might disrupt ciliary trafficking or cause toxicity over an extended period. For example, alteration of cellular ion gradients fundamentally perturbs cellular physiology and may exert untoward effects on ciliary trafficking that would confound interpretation of these conventional downstream readouts. A rapid, direct assay of Ptch1 or Smo functional state might circumvent these issues, but such an approach has not been applied to study Ptch1 ionic requirements. Similarly, the outstanding questions regarding cholesterol regulation of Smo are difficult to address in the complex, biochemically undefined environment of a living cell, where as yet unidentified sterols or proteins may contribute to Ptch1–Smo communication. Biochemical reconstitution, on the other hand, might rise to this challenge by enabling a stringent definition of the proteins, lipids, and other cellular factors that suffice to recapitulate Smo regulation in vitro. Unfortunately, existing methods to measure vertebrate Smo activity are indirect and require intact cells with primary cilia. This presents major obstacles for in vitro studies because it is currently not possible to reconstitute ciliary trafficking in cell-free settings. Thus, because conventional Hh pathway assays are long-term and indirect, Ptch1–Smo regulation has remained largely inaccessible.
In the present study, we investigate Smo regulation by developing a set of rapid, robust, and direct Smo activity sensors for cell-based and in vitro studies. We use these assays to recapitulate Ptch1 effects on Smo outside the cilium, demonstrating that the underlying process is unexpectedly dynamic and can proceed in the absence of any cilium-specific proteins or metabolites. We also find that cholesterol is both necessary and sufficient for constitutive activation of purified Smo reconstituted in a defined lipid environment. This activation occurs independently of recently described interactions of cholesterol with the Smo cysteine-rich domain (CRD) (23, 26, 27), an extracellular region that we show is not required for Ptch1–Smo communication. Finally, we show that depletion of extracellular Na+ rapidly and reversibly extinguishes the effects of Ptch1 on Smo. This suggests that Ptch1 might use transmembrane Na+ gradients to power its catalytic cycle and provides evidence that Ptch1 may indeed function as a bona fide ion-driven transporter. Based on these data, we hypothesize that Ptch1 harnesses the energy stored in transmembrane Na+ gradients to either shield the Smo 7TM domain from cholesterol’s activating influence or provide an inhibitory factor that can block Smo even when cholesterol is present. Our study highlights the power of direct conformational sensors in simplified experimental systems to provide insights into Ptch1 and Smo regulation that lie beyond the reach of existing Hh pathway functional assays.
Results
A Cilium-Independent Smo Conformational Biosensor Based on G Protein Coupling.
The cilium might provide a “privileged environment” for Ptch1–Smo regulation by harboring specialized protein and lipid factors that are required for this process. Alternatively, Ptch1–Smo regulation might proceed in the cilium, but the underlying mechanism might use factors that are distributed more ubiquitously throughout the cell, and the ciliary environment may be required mainly for downstream coupling to Gli transcription factors (Fig. S1A). One way to distinguish between these models is to test whether Ptch1–Smo regulation can be recapitulated in a nonciliary location. To this end, we designed a direct Smo conformational assay that takes advantage of Smo’s ability to couple to heterotrimeric G proteins in certain biological settings (28–31). G protein-based assays are ideal in this regard because activated 7TM proteins stimulate downstream coupling by directly binding a G protein heterotrimer, whose conformation changes almost instantaneously without the need for additional cellular components. While it remains controversial whether Smo employs G proteins to relay its “canonical” signal to Gli transcription factors (17, 32), we reasoned that G protein coupling could nevertheless provide a useful direct readout for Smo conformational state (Fig. S1B), allowing us to measure Smo regulation independently of ciliary trafficking.
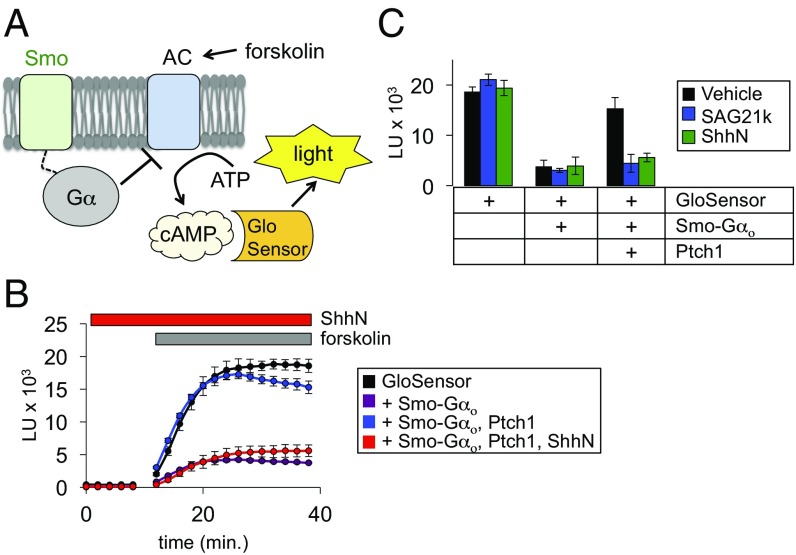
We measured Smo G protein coupling using GloSensor (33), a firefly luciferase variant that emits light only in the presence of cAMP (Fig. 1A). We used HEK293 cells for these studies, as they contain minimal primary cilia (34) and are not transcriptionally responsive to Hh; this minimizes any potentially confounding influences of manipulating cAMP on Gli repressor formation (17) in our experiments. To further ensure that our measurements do not depend on a ciliary pool of Smo, we made use of a direct fusion of a C-terminally truncated Smo to the inhibitory G protein Gαo (Smo–Gαo), thereby eliminating the cytoplasmic sequences required for Smo ciliary localization (35). We transfected the GloSensor cDNA into HEK293 cells, loaded the cells with a luciferase substrate, and raised cellular cAMP levels using the adenylyl cyclase (AC) agonist forskolin, leading rapidly to a large induction of luminescence (Fig. 1B). If Smo is able to couple to the fused inhibitory Gαo protein, we would expect Smo activity to reduce cAMP levels, leading to a decline in GloSensor luminescence. Indeed, cotransfection of Smo–Gαo with GloSensor decreased forskolin-induced luminescence, consistent with Smo–Gαo inhibition of AC (Fig. 1B). When we cotransfected Ptch1 and Smo–Gαo together with GloSensor, luminescence was restored to high levels, showing that Ptch1 inhibits Smo G protein coupling (Fig. 1B and Fig. S2A). Brief pretreatment (10 min) of Ptch1-expressing cells with the recombinant N-terminal fragment of Sonic hedgehog (ShhN) abolished the effect of Ptch1 on Smo–Gαo (Fig. 1B), confirming that our assay detects physiological changes in Ptch1 and Smo activity states.
Fig. 1.
A live-cell assay shows rapid, cilium-independent Ptch1 regulation of Smo. (A) Schematic diagram of the GloSensor cAMP assay. Smo couples to endogenous (or fused) inhibitory Gα proteins, which block forskolin-induced AC activity, thereby reducing cAMP. Increased Smo activity is therefore reflected as a decrease in luminescence. (B) Live-cell luminescence traces from HEK293 cells transfected with the indicated plasmids. Baseline luminescence was recorded for 10 min at 2-min intervals, followed by forskolin treatment and continued monitoring. For ShhN treatment, ShhN was added 10 min before measurement of baseline and forskolin-induced luminescence; ShhN remained present at 200 nM for the entire experiment. (C) Bar graph shows steady state luminescence for HEK293 cells transfected with GloSensor, Smo, and Ptch1 expression plasmids. Cells were preincubated for 10 min with ShhN or SAG21k as indicated in B.
Our control experiments confirmed that Ptch1 inhibition is specific to Smo (Fig. S2B), ruling out artifactual effects of Ptch1 on 7TM protein function in general. Although we used the Smo–Gαo fusion in our cell-based experiments for consistency with the in vitro measurements described below, we observed similar results in the GloSensor assay with non–Gαo-fused Smo, which couples to endogenous pertussis toxin-sensitive G proteins (Fig. 2C). Apart from the influence of Ptch1, Smo activity in Gli transcriptional assays is modulated by a host of exogenous small molecules, including KAAD-cyclopamine, SANT-1, and SAG21k. We found that these molecules all affect Smo G protein coupling (Fig. 1C and Fig. S2C), indicating that the active and inactive conformations sampled by Smo in both Gli and G protein assays are closely related. Thus, cell-based G protein-coupling assays accurately recapitulate coupled Ptch1 and Smo activity with a fast, membrane-proximal real-time readout, even in cell lines such as HEK293 that are poorly ciliated and unable to mount an efficient Gli transcriptional response to Hh. Interestingly, the effects of Smo antagonists are augmented in the presence of Ptch1 in the GloSensor assay (Fig. S2C), whereas such a synergy is usually not seen in Gli-dependent luciferase assays; we further explore this phenomenon in the next section.
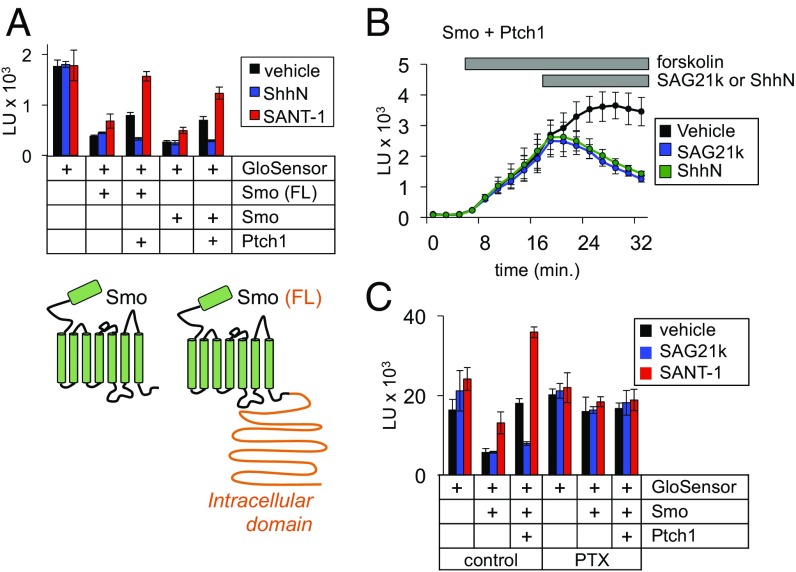
Fig. 2.
Ptch1-mediated regulation of Smo conformation is fast, nonrate-limiting, and occurs independently of Smo ciliary trafficking. (A, Top) Ptch1 blocked the activity of full-length Smo [Smo (FL)] and the cytotail-deleted form (Smo) to similar degrees in the GloSensor assay. (Bottom) A topology diagram of the two Smo expression constructs; the cytoplasmic tail required for transcriptional coupling and ciliary trafficking is indicated in orange. (B) To demonstrate the rapid onset of ShhN or SAG21k effects, cells expressing GloSensor, Smo, and Ptch1 were stimulated with forskolin, and ShhN or SAG21k (vs. a vehicle control) were then added as indicated by the gray bar. (C) Overnight pretreatment with PTX (100 ng/mL) prevents Smo from inhibiting GloSensor luminescence.
The ability of Ptch1 to regulate C-terminally truncated versions of Smo in HEK293 cells suggests that the underlying process does not require a specialized ciliary environment. To more explicitly test this idea, we asked whether restoring ciliary localization signals to our C-terminally truncated Smo altered its ability to couple to G proteins. Both versions of Smo affected GloSensor luminescence to similar extents, further illustrating that ciliary localization signals do not affect Ptch1–Smo regulation in this assay (Fig. 2A).
Our rapid Smo activity sensor shows that Ptch1 can fully shift Smo conformation in a matter of minutes. This measure of the underlying conformational mechanism is faster and more direct than even the earliest known cilium-dependent pathway readout, namely the accumulation of Smo in primary cilia (36, 37). Upon pathway activation, Smo ciliary accumulation requires several hours to reach a steady state, likely because the alterations in Smo ciliary trafficking triggered by the initial changes in Smo conformation involve one or more rate-limiting steps. Because it measures Smo conformation independently of ciliary trafficking, our sensor now provides an opportunity to study the kinetics of Ptch1 action by comparing the effects of a direct Smo agonist to Hh-mediated loss of Ptch1 inhibition. We found that acute addition of ShhN to cells coexpressing Ptch1 and Smo activates G proteins on a similar time scale as SAG21k (Fig. 2B), indicating that the Ptch1 affects Smo conformation in a fast, nonrate-limiting manner. From these data, we calculated a t1/2 for Smo-mediated G protein coupling (the time required for half-maximal inhibition of cAMP to baseline levels, defined by Smo in the absence of Ptch1) as 3.97 min for ShhN and 2.63 min for SAG21k. As predicted, this is markedly faster than Smo ciliary accumulation (t1/2 = ∼2 h) (see ref. 36), a more indirect metric for activity-dependent changes in Smo conformation.
In summary, our rapid cell-based G protein-coupling assay not only faithfully recapitulates the major findings from conventional cell-based transcriptional assays, but also demonstrates that Smo can attain a constitutively active state outside the cilium, likely within the plasma membrane. Thus, even though Ptch1 regulates Smo within the cilium, the underlying mechanism uses factors that are not restricted to this compartment. Furthermore, Ptch1 and Smo change conformation on a time scale significantly faster than is evident from traditional Hh pathway readouts.
Cholesterol Is Necessary and Sufficient for Constitutive Smo Activation in Vitro.
Our preceding experiments establish that G protein coupling serves as a robust measure of Smo activity that accurately recapitulates its fundamental pharmacological properties, providing snapshots of the active and inactive Smo conformational states that are likely involved in physiological coupling to Gli transcription factors. Unlike conventional Hh pathway functional readouts, however, G protein coupling has the distinct advantage that it can be easily measured in vitro. We capitalized on this property, developing cell-free and reconstituted preparations to study how membrane cholesterol and other factors influence Smo activity.
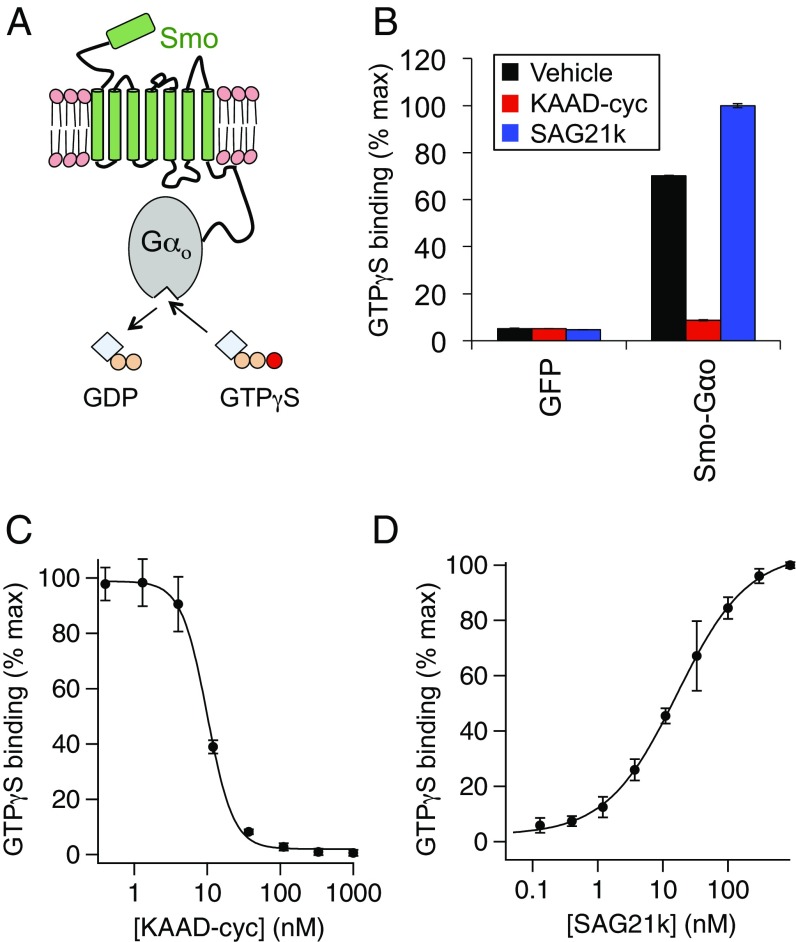
We began our in vitro studies by constructing and characterizing an optimized assay for in vitro Smo G protein coupling. We adapted a previously described approach (30) using the Smo–Gαo fusion, which dramatically increases the efficiency of in vitro measurements compared with nonfused G proteins (38). The fused G protein thus serves as a tethered biosensor that reports on Smo activation state via its nucleotide-exchange capacity, measured as binding of radiolabeled nonhydrolyzable GTP (35S-GTPγS) (Fig. 3A). In membrane fractions derived from transfected HEK293 cells, Smo–Gαo displays a large constitutive activity that is blocked by brief treatment with the Smo inverse agonist KAAD-cyclopamine and is modestly stimulated by the Smo agonist SAG21k (Fig. 3B), each in a dose-dependent fashion (Fig. 3 C and D). A panel of established small-molecule Smo ligands (39) revealed an excellent correlation between assays of G protein coupling and transcriptional regulation (Fig. S3A). Therefore, as with the GloSensor readout, our in vitro pharmacological measurements indicate that the active and inactive conformations sampled by Smo in Gli- and G protein-dependent assays are closely related. Consistent with this idea, a Smo–G protein fusion including the full-length cytotail (SmoFL–Gαo) was able to stimulate Gli-dependent transcription (unlike the C-terminally truncated Smo–Gαo), yet exhibited comparable basal activity and small molecule modulation as Smo–Gαo in the G protein-coupling assay (Fig. S3 B and C).
Fig. 3.
An in vitro assay for Smo conformational state reflects modulation by exogenous small molecules and membrane cholesterol. (A) Diagram of the in vitro Smo–Gαo assay, in which Smo activity promotes GDP dissociation and nucleotide exchange, measured as binding of nonhydrolyzable 35S-GTPγS. (B) Membranes were prepared from HEK293 cells transfected with plasmids encoding GFP or Smo–Gαo, and incubated with the Smo inverse agonist KAAD-cyclopamine (KAAD-cyc, 300 nM), the Smo agonist SAG21k (50 nM), or a DMSO control, followed by 35S-GTPγS binding and scintillation counting. Data were normalized to maximum SAG21k-stimulated binding. (C and D) Concentration-response relationships for KAAD-cyclopamine and SAG21k in the 35S-GTPγS binding assay, revealing a KAAD-cyclopamine IC50 of 9.99 ± 0.36 nM and a SAG21k EC50 of 16.53 ± 1.45 nM, in reasonable agreement with previously published values. Note that D was conducted at high [GDP] (300 µM) to expand the window between basal and SAG21k-induced activity (Materials and Methods).
Curiously, Smo agonists and antagonists displayed weak effects in our in vitro system unless we supplemented the assay buffers with high (10–100 μM) GDP (Fig. S4), which shifts the sensitivity of the G protein into a range that permits efficient responses to Smo conformational changes. This difference provides a plausible explanation for the submaximal effects of Smo antagonists and their synergy with Ptch1 in the GloSensor assay. As GDP levels cannot be effectively altered in cells, the small-molecule effects are similar to in vitro results with low GDP (Fig. S4), but can be augmented with Ptch1, which shifts the Smo equilibrium analogously to high GDP levels. In any case, our in vitro assay recapitulates Smo constitutive activity seen in live cells (Fig. 1) and accurately reflects its sensitivity to small-molecule modulators.
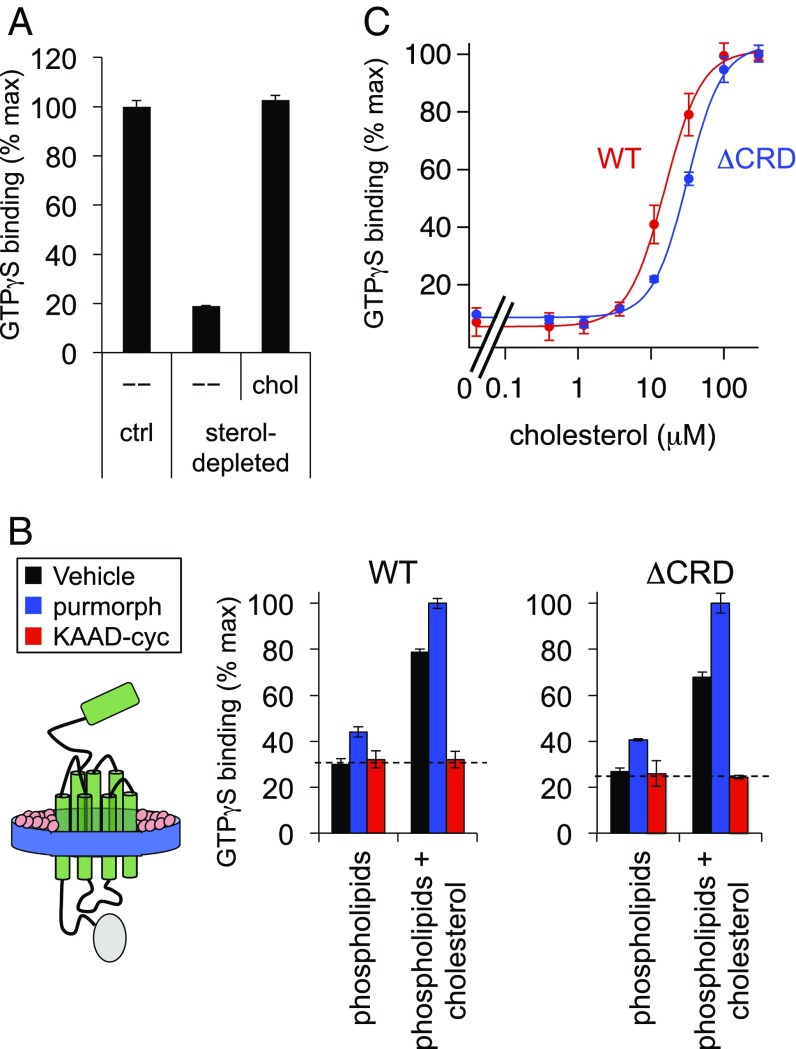
Having characterized our in vitro Smo conformational sensor, we next used this tool to study how cellular lipids, particularly sterols, affect Smo activity. Indeed, several lines of evidence suggest that the endogenous Ptch1-regulated Smo ligand might be cholesterol or one of its derivatives, but the precise roles of cholesterol in Hh pathway regulation remain undefined. For example, cyclodextrin-mediated depletion of cellular sterols inhibits Smo in a manner reversed by addition of exogenous cholesterol, but the mechanism underlying these effects is unclear (21–25, 40). This is because intact cells harbor elaborate sterol biosynthetic machinery that can rapidly transform cholesterol into a host of metabolic products under physiological conditions (41, 42), making it difficult to discriminate using cell-based approaches between roles for cholesterol itself versus one of its many metabolites. In contrast, because extensively washed membranes cannot support sterol metabolic reactions (43, 44), our in vitro Smo functional assay (Fig. 3A) is ideal for distinguishing these possibilities. Indeed, we found that acute (20 min) cyclodextrin extraction of membranes blocked binding of GTPγS to our tethered Smo biosensor (Fig. 4A). This effect was reversed by replenishing the sterol-depleted membranes with cholesterol, mirroring the effects of these treatments on Smo activity in cells (21, 22, 40). Our data therefore indicate that Smo sensitivity to sterol depletion in vivo derives from a requirement for cholesterol itself, and not a cholesterol metabolite. As cholesterol is by far the most-abundant sterol in metazoan plasma membranes (cholesterol:phospholipid ratio = 50%) (45), our in vitro results argue that cholesterol will likely play a similar role in determining Smo activity in cells (21, 22, 24, 40).
Fig. 4.
Smo reconstitution reveals that cholesterol and phospholipids are sufficient for constitutive activity. (A) Membranes from cells expressing Smo–Gαo were depleted of endogenous sterols using methyl-β-cyclodextrin (MβCD) or incubated with buffer alone as a control (ctrl). Sterol-depleted membranes were subsequently incubated with a cholesterol-MβCD complex (chol) or a buffer control (–), then processed for GTPγS binding. Data were normalized to the amount of Smo constitutive GTPγS binding under control (nonsterol-depleted) conditions. (B) Diagram of the Smo–Gαo fusion (green) reconstituted into nanodiscs with a membrane scaffold protein (MSP1D1, blue) and defined lipids (pink). Nanodiscs were formed from purified Smo–Gαo or SmoΔCRD–Gαo (a form lacking the CRD) in combination with synthetic phospholipids alone, or a mixture of phospholipids and 8 mol% cholesterol. Each nanodisc preparation was stimulated with purmorphamine, KAAD-cyc, or a vehicle control, followed by measurement of GTPγS binding. The dashed line represents the baseline level of G protein coupling in this assay, as defined by treatment with KAAD-cyc (a full inverse antagonist). (C) Membranes derived from HEK293-cells transfected with Smo–Gαo or SmoΔCRD–Gαo were stripped of endogenous cholesterol with MβCD and replenished with a cholesterol–MβCD complex as in A. For each data point, the cholesterol concentration in the assay buffer was derived by assuming a 10:1 ratio of cyclodextrin to cholesterol in our saturated cholesterol–MβCD complexes, as defined by previous measurements (67). Data are normalized to GTPγS binding stimulated by the maximal dose of cholesterol–MβCD for each construct. Curve-fitting with the Hill equation revealed EC50s of 15.55 ± 0.83 μM and 31.39 ± 1.73 μM for wild-type and CRD-deleted Smo, respectively.
The above experiments demonstrate that Smo activity requires cholesterol, and raise several possibilities for its mechanism of action. Cholesterol might function as a permissive factor that promotes Smo activity stimulated by an endogenous ligand, similar to a wide array of canonical ligand-activated G protein-coupled receptors (GPCRs) (46). Alternatively, cholesterol alone might suffice to activate Smo. To more directly examine the role of cholesterol in Smo activation, we assayed Smo conformation in a precisely controlled membrane lipid environment. We purified Smo–Gαo in monodisperse form (Fig. S5) and reconstituted it into apolipoprotein nanodiscs (47, 48) using one of two defined lipid mixtures. When we performed our nanodisc reconstitution using phospholipids alone, Smo–Gαo was devoid of constitutive activity and only weakly stimulated by the agonist purmorphamine (Fig. 4B). In contrast, when we included cholesterol (at 8 mol%) during nanodisc assembly, constitutive and agonist-induced activity increased dramatically (Fig. 4B), mirroring our results from cell-derived membranes (Figs. 3B and 4A). These data indicate that cholesterol is not just necessary, but also sufficient to induce constitutive Smo activity, with no requirement for additional proteins and lipids.
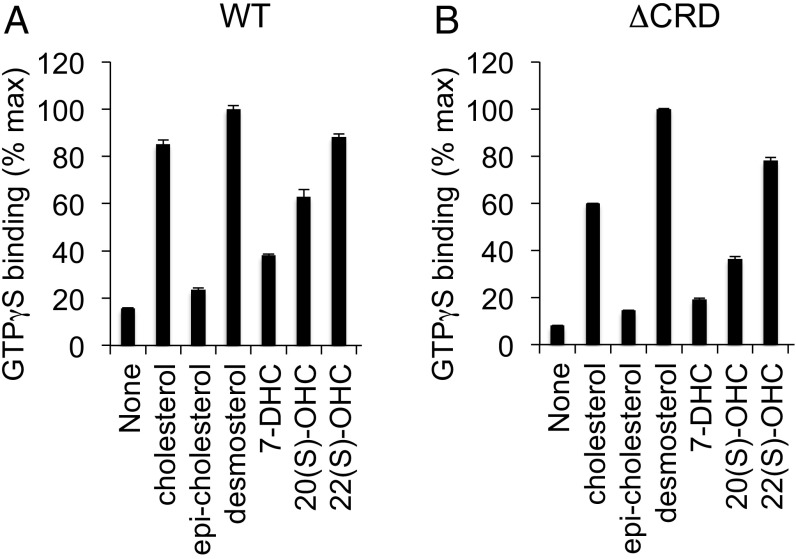
Cholesterol effects in nanodiscs may derive from a direct interaction between cholesterol and Smo. Several papers (23, 25, 27) have reported that cholesterol can interact with the extracellular Smo N-terminal CRD, and recent in vivo labeling studies (26) suggested that cholesterol might become covalently attached to the CRD under some circumstances. We found, however, that CRD-deleted Smo also depends on cholesterol for constitutive activity in nanodiscs (Fig. 4B). Furthermore, cholesterol activates Smo over a nearly identical concentration range both in the presence or absence of its CRD (Fig. 4C). We observed similar results with a CRD point mutant (D99A Y134F, hereafter referred to as DAYF) that cannot bind an alkynyl cholesterol derivative in vitro (Fig. S6) (27; see also ref. 26). Finally, by testing a variety of sterols for their ability to rescue Smo GTPγS binding in cyclodextrin-depleted membranes, we noted a distinct structure–activity relationship from that previously described for CRD-sterol interactions (Fig. 5A); 22(S)-OHC fully rescues the effect of sterol depletion in our assay but does not bind the CRD (49), whereas 20(S)-hydroxycholesterol [20(S)-OHC], which binds the CRD, is less efficient. Furthermore, the effects of sterols in these experiments are strikingly similar for both wild-type and CRD-deleted Smo (compare Fig. 5 A and B), illustrating that the CRD is not required. Taken together, these data indicate that cholesterol can affect Smo activity via its heptahelical bundle. How cholesterol affects the Smo 7TM domain is unclear, but one possibility is that it might engage a specific binding site outside the “cyclopamine pocket” (22, 40).
Fig. 5.
Cholesterol derivatives show a similar structure–activity relationship on wild-type and CRD-deleted Smo in G protein coupling assays. Membranes from HEK293 cells transfected with (A) wild-type or (B) CRD-deleted Smo–Gαo fusions were depleted of endogenous sterols, and then treated with the indicated sterols (in complex with cyclodextrin) as in Fig. 4. Values were normalized to the level of rescue produced by the sterol biosynthesis intermediate desmosterol (which consistently gave slightly higher values of GTPγS binding than cholesterol).
Ptch1 Can Regulate Smo Independently of any CRD–Cholesterol Interactions.
Cholesterol exerts at least two distinct effects on Smo activity, one through the CRD and the other through the 7TM domain (as highlighted by our in vitro studies described above), but the key outstanding question is whether Ptch1 inhibits Smo by altering either of these modes of cholesterol interaction. Several recent studies have provided new data in this regard. Luchetti et al. (25) show that an experimentally induced increase in cellular cholesterol, via loading of cholesterol–cyclodextrin complexes into cholesterol-replete fibroblasts, elicits activation of wild-type Smo but not the DAYF mutant (see also ref. 26). Because Smo DAYF also shows reduced activation of Gli transcription in response to loss of Ptch1, it is tempting to speculate that Ptch1 inhibits Smo by somehow operating directly on cholesterol bound to its CRD (Fig. S7A, model 1). Such a model, however, conflicts with the finding that transfected Ptch1 can suppress the activity of a CRD-deleted Smo mutant (22), implying that Ptch1 can regulate Smo independently of the CRD. These seemingly disparate results might be reconciled if CRD binding to cholesterol instead functions to stabilize the constitutively active Smo conformation attained upon loss of Ptch1 inhibition. In this scenario, Ptch1 does not block Smo by altering CRD-cholesterol binding, but rather acts at a site elsewhere on Smo, while CRD–cholesterol interactions serve an auxiliary role by stabilizing an active Smo conformation (Fig. S7A, model 2).
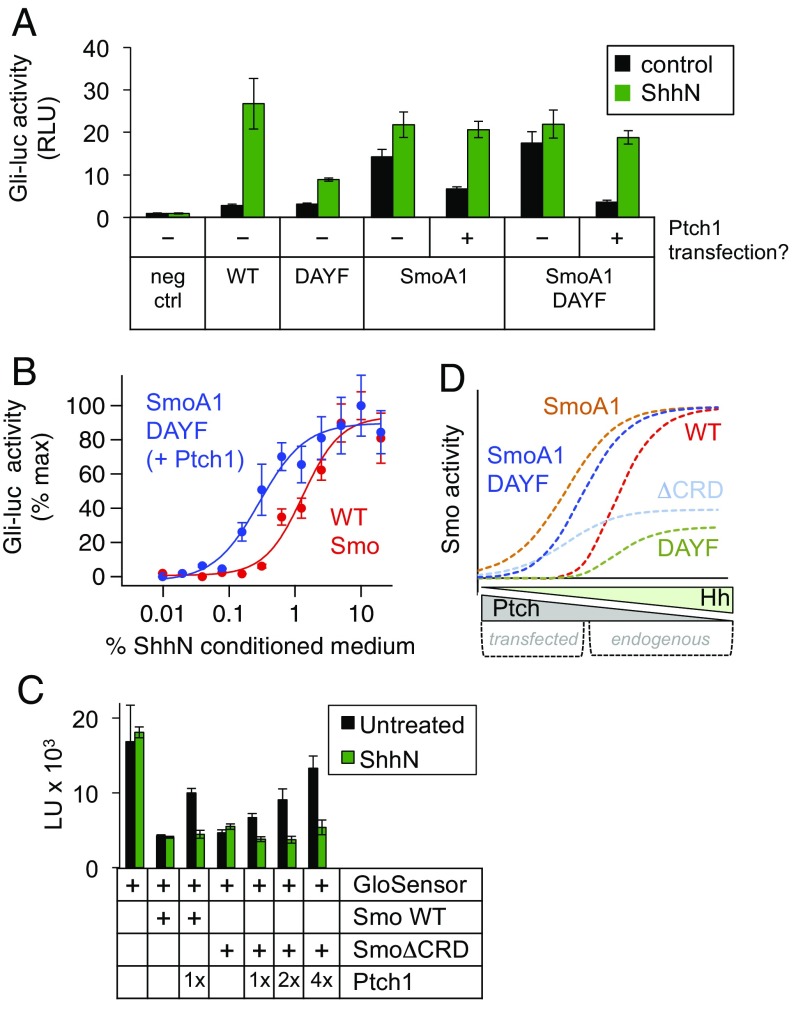
We devised an experiment to distinguish these two models. We combined the DAYF mutation with SmoA1, a 7TM domain mutation that constitutively activates Smo independently of the CRD (15), thereby bypassing the need for any CRD-mediated stabilization of the Smo active state, while still permitting repression by exogenous, transfected Ptch1 (15, 22, 50, 51). If Ptch1 operates directly on a CRD–cholesterol interaction (model 1), Ptch1 transfection should not repress SmoA1–DAYF. In contrast, if the site of Ptch1 action lies outside the CRD, exogenous Ptch1 should block the activity of SmoA1–DAYF (model 2). We found using a Gli transcriptional reporter assay that the constitutive activity of SmoA1–DAYF was dramatically suppressed by Ptch1 transfection (Fig. 6A). This effect closely resembled the action of endogenous Ptch1 on wild-type Smo, as the resulting inhibition was fully reversed by treatment with ShhN (Fig. 6A) in a dose-dependent manner (Fig. 6B). These data indicate that the actions of transfected Ptch1 on Smo in our assays are reflective of true Ptch1 functionality rather than nonspecific consequences of protein overexpression. All of the above results are difficult to reconcile with direct action of Ptch1 via the Smo CRD (model 1), but are readily explained by action of Ptch1 via the Smo 7TM domain, with a critical stabilizing role for the CRD (model 2).
Fig. 6.
Ptch1 can inhibit Smo in the absence of CRD-cholesterol binding. (A) Smo−/− mouse embryonic fibroblasts (MEFs) were transfected with the indicated Smo and Ptch1 expression constructs along with an 8×Gli-luciferase reporter, and stimulated with control (black) or ShhN (green) conditioned medium. Relative luciferase units (RLU) are plotted as a fold-increase over the baseline value, defined as reporter activity from the negative control transfection (no Smo). (B) A similar experiment, in which cells transfected with wild-type Smo (red) or SmoA1 D99A Y134F (SmoA1 DAYF) + Ptch1 (blue) were stimulated with increasing concentrations of ShhN conditioned medium. (C) SmoΔCRD is constitutively active and suppressible by Ptch1. Suppression is evident when Ptch1 is coexpressed according to standard conditions (Materials and Methods), and becomes even more obvious when increasing amounts of Ptch1 cDNA are transfected. The standard 10-min ShhN pretreatment fully reverses the effects of Ptch1 coexpression on both wild-type and CRD-deleted Smo, even at the highest amounts of Ptch1 cDNA transfected. The observed suppression of SmoΔCRD is therefore truly dependent on Ptch1 activity and not simply an artifact of protein overexpression in this assay. (D) Simulation of the functional behavior of wild-type and various CRD-mutated forms of Smo, taking into account observations from Gli transcriptional assays in this and several other studies (22, 27, 68). Smo activity is represented on the y axis, while Ptch1 activity (blocked by binding of Hh) is on the x axis. The possible ranges of Ptch1 activity encompassed by endogenous vs. transfected Ptch are indicated below the x axis.
Whereas the experiments presented above involved overexpression of Ptch1 along with a compound SmoA1 mutant, we also observed that endogenous Ptch1 can regulate Smo DAYF even without the activating SmoA1 mutation (Fig. S7B; see also figure 3F in ref. 27); as predicted, the maximal extent of Hh-induced activity is less than its wild-type counterpart because the mutations compromise the CRD’s ability to stabilize active Smo. In contrast, Smo DAYF is completely insensitive to the oxysterol 20(S)-OHC (Fig. S7B), confirming that these mutations disrupt CRD-sterol binding and providing further evidence that Ptch1 effects are separable from CRD–lipid interactions. We also note that whereas wild-type Smo and the DAYF mutant are activated to similar extents by the 7TM agonist SAG (Fig. S7B) (see also refs. 25 and 27), this finding does not rule out model 2 because SAG does not require the CRD to stabilize an active Smo conformation. Finally, Ptch1 was able to repress SmoΔCRD (Fig. 6C) and Smo DAYF (Fig. S7C) in our GloSensor assay, consistent with transcriptional data presented here and in our previous study (22).
We conclude from these experiments that Ptch1 is capable of acting outside of the Smo CRD, likely within the 7TM domain at a site that remains to be mapped. One reason that other studies may not have reached these conclusions is that the Ptch1 responsiveness of CRD-mutated Smo alleles becomes particularly obvious after manipulating Ptch1 levels over a broad range (graphically summarized in Fig. 6D). This is only possible by introducing exogenous Ptch1 via transfection as in the present study (see also refs. 22 and 51), whereas other investigations (23, 25, 27) have sampled a relatively narrow range of Ptch1 activity by relying exclusively on endogenous Ptch1 protein, which is expressed at low levels.
Ptch1 Regulation of Smo G Protein Coupling Depends on Extracellular Na+.
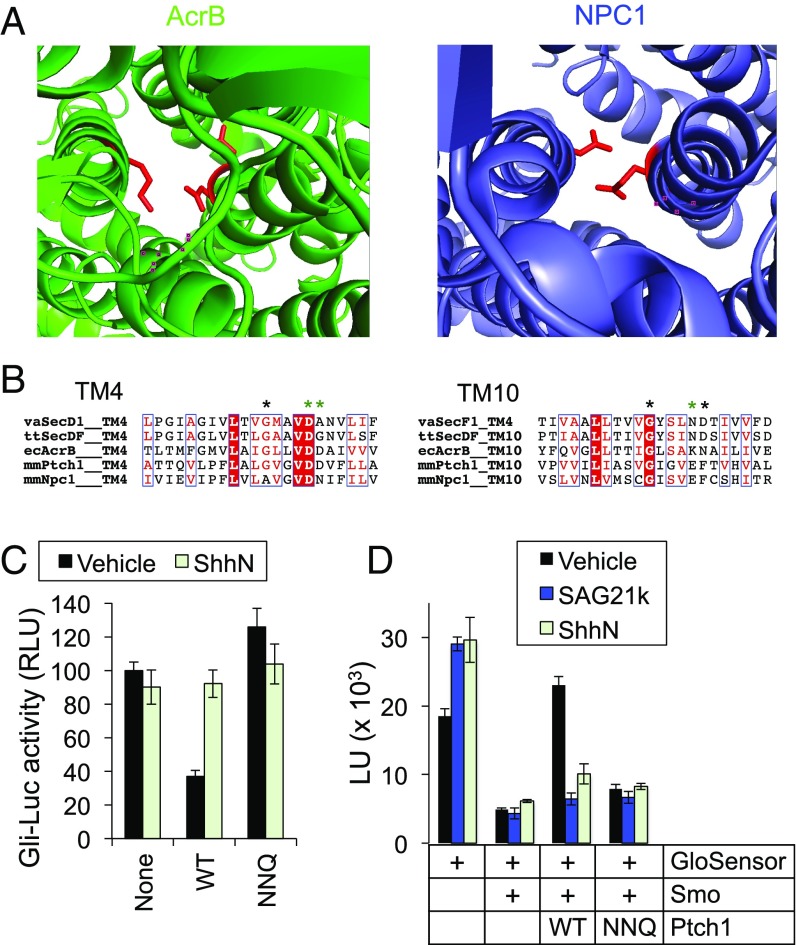
Smo constitutive activity in our GloSensor experiments is dramatically reduced when Ptch1 is cotransfected (Fig. 1). In contrast, we failed to observe any Ptch1-mediated reduction in Smo GTPγS binding in membrane fractions (Fig. S8A) or detergent-permeabilized cells (Fig. S8B). This result suggests that Ptch1 activity is not recapitulated in these preparations, and must somehow be lost during the process of cell disruption. The lack of Ptch1 activity following cell breakage might reflect, for example, requirements for intact cellular structure, diffusible cytoplasmic factors, or transmembrane ion gradients in Ptch1 function. In this regard, it is noteworthy that Ptch1 is predicted to function as a transmembrane transporter based on its homology to prokaryotic H+ gradient-driven RND small-molecule efflux pumps (15). In fact, a recent crystal structure of NPC1 (52), a putative cholesterol transporter and the closest vertebrate Ptch1 relative, reveals that the overall fold and the positions of key amino acids mediating transmembrane H+ transfer in prokaryotic RNDs (53, 54) are highly conserved (Fig. 7A). Neutralizing the analogous Ptch1 residues (D499N, D500N, E1081Q, hereafter referred to as NNQ) (Fig. 7B) inactivates Ptch1 in transcriptional (Fig. 7C) and G protein-coupling assays (Fig. 7D). These findings are consistent with previous reports implying that Ptch1, like its bacterial counterparts, is powered by an ion gradient (15). But while bacterial RNDs rely primarily on a large H+ gradient between cytoplasm and periplasm (16, 55), no such pH difference exists across the plasma membrane of most metazoan cells. Thus, the energy source for Ptch1 activity is still unclear, and any active-transport–based models for Ptch1 function remain largely speculative.
Fig. 7.
Ptch1 activity depends on conserved residues that mediate intramembrane ion flux in bacterial RNDs. (A) Top-down views of the prototypical bacterial RND AcrB (green, PDB ID code 2DHH) and NPC1 (blue, PDB ID code 5U73), with conserved charged residues in TM4 and TM10 highlighted in red. (B) Sequence alignment of transmembrane helices 4 and 10 of several RND family proteins. ecAcrB, Escherichia coli AcrB; mmNpc1, Mus musculus (mouse) NPC1; mmPtch1, Mus musculus (mouse) Ptch1; ttSecDF, Thermus thermophiles secDF; vaSecD1, Vibrio alginolyticus secD-1; vaSecF1, Vibrio alginolyticus secF-1. SecD/F in certain organisms, including E. coli and V. alginolyticus, is a heterodimer composed of D + F, with each subunit corresponding to either the first or last six-transmembrane helices of the RND family. Asterisks indicate key residues for ion flux, with those neutralized by the Ptch1 “NNQ” mutation colored green. The activity of different Ptch1 constructs was tested in a Gli-luciferase transcriptional reporter assay in Ptch1−/− cells (C) or the GloSensor assay in HEK293 cells (D). In both assays, wild-type Ptch1 suppressed Smo activity and the suppression was reversed by addition of ShhN or SAG21k, while the NNQ mutation diminished Ptch1 activity.
We hypothesized that a different ion might drive Ptch1 enzymatic activity. One of the most fundamental chemiosmotic differences across animal cell membranes is low Na+/high K+ inside and high Na+/low K+ outside the cell (56). We asked whether Ptch1 is inactivated when these gradients are eliminated by bathing cells acutely in a low Na+/high K+ solution, such that extracellular ionic strength is maintained but Na+ is low and K+ is high on both sides of the cell membrane. Initially, we attempted to measure loss of Ptch1 activity via a conventional Smo activity assay, namely accumulation of endogenous Smo in primary cilia (36, 37). We reasoned that disrupting the Ptch1 energy source should induce Smo ciliary accumulation, similar to that observed by treating cells with either Shh or the Smo agonist SAG21k. Instead, the low Na+/high K+ solution abolished SAG21k-induced Smo accumulation (Fig. S9). Because SAG21k binds Smo directly, thereby acting downstream of the Ptch1–Smo regulatory step, our results indicate that alterations in Na+ and K+ concentrations nonspecifically inhibit Smo ciliary accumulation during these experiments. Indeed, disrupting Na+ and K+ gradients likely exerts a multitude of pleiotropic effects on cellular metabolism and physiology, any of which could interrupt the complex, multicomponent ciliary trafficking process (14).
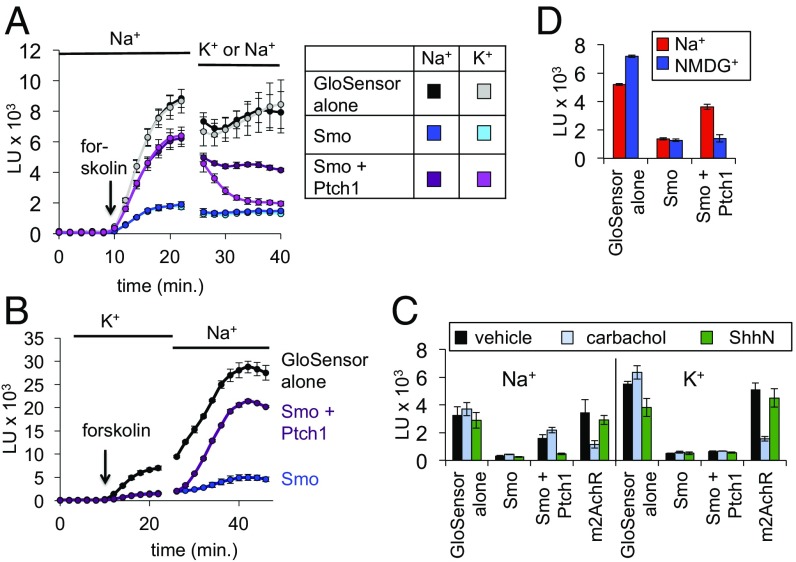
To circumvent this issue, we attempted to assess the effect of manipulating Na+ and K+ concentrations on Ptch1 in the rapid, membrane-proximal GloSensor assay. Remarkably, we found that changing the extracellular bath to a low Na+/high K+ solution reversed Ptch1-mediated Smo inhibition within minutes (Fig. 8A). Conversely, Ptch1 inhibition was briskly restored upon switching from a low Na+/high K+ bath to a physiological high Na+/low K+ bath (Fig. 8B and Fig. S10A). Thus, Ptch1 regulation of Smo requires normal extracellular Na+ and K+ concentrations. In similar experiments, we found that replacement of Na+ with Li+ in the bath also eliminated Ptch1 inhibitory effects (Fig. S10B). We observed little or no effect of changing the ionic composition of the bath in cells expressing Smo alone (Smo suppression of GloSensor activity remained), in cells expressing GloSensor alone, or in cells expressing the muscarinic acetylcholine receptor, a control GPCR that also affects cAMP levels (Fig. 8C). These control experiments (Fig. 8) also indicate that the GloSensor readout remains robust when transmembrane ion gradients are acutely manipulated, in marked contrast to the Smo ciliary accumulation assay (Fig. S9). A low Na+/high K+ extracellular solution will affect both transmembrane Na+ and K+ gradients as well as depolarize the plasma membrane (56) (cytoplasm typically −30 to −60 mV with respect to extracellular space, largely set by the K+ equilibrium potential). Substituting Na+ with the organic cation N-methyl-d-glucamine (NMDG+), which maintains transmembrane K+ gradients and perturbs membrane potential only minimally (57), also inactivated Ptch1 (Fig. 8D). Taken together, these data suggest that among monovalent cations, Ptch1 stringently requires extracellular Na+. Furthermore, the rapid (<5 min) changes in Ptch1 activity resulting from extracellular ion substitution imply that Ptch1 acts at the cell surface, or else in an endocytic compartment that fully exchanges its contents with the extracellular space over these short times.
Fig. 8.
A role for extracellular Na+ in Ptch1–Smo regulation. (A) HEK293 cells were transfected with GloSensor + the indicated plasmids and stimulated with forskolin in physiological saline (Na+). The bath was replaced at the indicated time with a low Na+/high K+ buffer (K+), in which all Na+ was replaced by K+, and vice versa. A control buffer replacement maintained physiological Na+-based saline for the entire experiment. (B) Cells were transfected with GloSensor alone (black), or in the additional presence of Smo (blue), or Smo and Ptch1 (purple). After loading with luciferin in K+ buffer, cells were switched at the indicated time to physiological Na+ buffer. While Ptch1 failed to suppress Smo in the K+ buffer (note the overlapping blue and purple traces), Ptch1 activity increased almost immediately after the buffer change and plateaued within 20 min. See Fig. S10A for quantification. (C) Steady-state luminescence in HEK293 cells expressing GloSensor + the indicated plasmids (m2AchR, m2 muscarinic acetylcholine receptor, stimulated by its ligand carbachol). Cells were bathed in Na+ (Left) vs. K+ (Right) for all steps following substrate loading, and forskolin-induced luminescence was measured as in Fig. 1C. (D) Steady-state forskolin-induced luminescence in cells bathed in saline solution based on Na+ or the organic cation NMDG+, revealing that NMDG+ cannot sustain Ptch1 activity.
One appealing hypothesis suggested by these data is that Ptch1 uses a transmembrane Na+ gradient to power its transport cycle. Alternatively, extracellular Na+ might somehow act as a cofactor required to maintain Ptch1 or Smo in active or inactive conformations, respectively. Several control experiments argue against the latter possibility. First, GTPγS binding of Smo in cell-free membrane fractions or purified nanodiscs, normally measured in a high Na+/low K+ buffer, is no greater in a high K+/low Na+ buffer (Fig. S10C), and SANT-1 can still block Smo G protein coupling when cells are bathed in high K+/low Na+ (Fig. S10D). Indeed, while ligand binding to some but not all class A GPCRs is modulated by changes in Na+ binding near the GPCR orthosteric site, the critical residues involved in Na+ coordination are not conserved in the Smoothened/Frizzled subfamily (58), consistent with the lack of Na+ effects in the experiments described above. These data demonstrate that depletion of extracellular Na+ is unlikely to activate Smo directly. Finally, Ptch1 can still bind Hh when cells are bathed in high K+/low Na+, arguing that these ion substitutions do not affect Ptch1 folding or stability (Fig. S10E).
The above data are consistent with the idea that Ptch1 activity relies on transmembrane sodium gradients (Fig. 9). Our results, however, do not exclude the possibility that Ptch–Smo regulation requires extracellular Na+ in a manner that is unrelated to transmembrane ion gradients. Definitive proof that Na+ gradients provide the energy source for Ptch1 function awaits demonstration of Ptch1-mediated transmembrane Na+ flux, which will require the development of a direct Ptch1 transport assay. Nevertheless, our findings should provide a useful starting point to investigate Ptch1 using cellular physiology-based approaches.
Fig. 9.
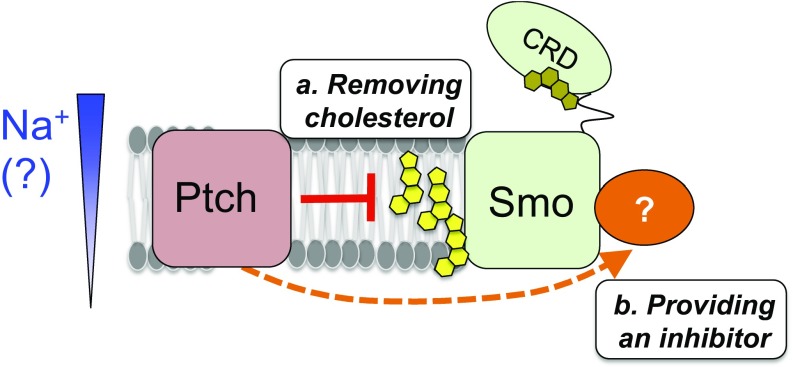
Model for the roles of membrane cholesterol in the Hh pathway. When the pathway is on, following Hh binding to and inactivation of Ptch1, membrane cholesterol activates Smo by binding to an as yet unidentified site within the Smo heptahelical bundle (or by modulating the membrane’s properties). In the “off” state of the pathway, Ptch1 could repress Smo by either: (a) altering cholesterol availability to Smo (yellow); or (b) providing a negative regulator (orange) that overrides the cholesterol activating effect on Smo. In either case, the modulatory influences of Ptch1 activity and membrane cholesterol on Smo function are distinct from the CRD-cholesterol interactions highlighted in previous structural studies (see main text). Based on the observed Ptch1 requirement for extracellular Na+, we hypothesize that Ptch1 depends on the transmembrane Na+ gradient to power its activity. See Discussion for more details.
Discussion
Genetic and cell biological studies clearly support a role for primary cilia in the vertebrate Hh cascade (14), but the precise function of this organelle has remained speculative. We find that Ptch1–Smo regulation can be studied using simple, direct assays that do not depend on cilia. Our measurements show that Ptch1 can switch Smo conformational state within minutes. This time scale is faster than cilium-dependent assays such as the activity-dependent accumulation of Smo in primary cilia (36, 37), as ciliary accumulation not only reflects Smo conformational state but also incorporates downstream, rate-limiting alterations in ciliary trafficking following the initial Smo conformational changes.
The cilium is likely the subcellular location where Ptch1 regulates Smo conformation to modulate Gli transcriptional coupling, as all three proteins localize to this compartment and blockade of ciliary biogenesis or trafficking inhibits transcriptional activation. However, our observation that Ptch1 can regulate Smo G protein coupling independently of cilia implies that the underlying mechanism does not require a specialized ciliary environment, and instead must use factors present both inside and outside cilia. Rather than providing an obligate, privileged setting for Ptch1–Smo regulation, the cilium may instead be uniquely required as a “meeting place” to concentrate Ptch1 and Smo with downstream pathway components, thereby permitting efficient coupling to transcriptional effectors. Nevertheless, the ciliary compartment does possess a distinct lipid repertoire (59–61), and cilium-specific lipids might play physiologically relevant roles in fine-tuning the core Ptch1–Smo regulatory step that we have recapitulated in our experiments. Our Smo nanodisc reconstitution system may allow direct testing of this hypothesis in the future. In the long term, it will be important to extend our findings to the endogenous Hh pathway in living cells by developing biochemical and imaging-based tools to directly interrogate Smo conformation in real-time within its native ciliary environment. It may also be worthwhile to extend our studies to other model systems such as the Drosophila Hh pathway, which functions independently of primary cilia, once a reliable short-term readout for Drosophila Smo activity has been developed.
A potential limitation of our approach is that it relies on G protein coupling as a Smo conformational readout, and the contribution of G proteins to Smo regulation under physiological conditions remains a matter of debate. Nevertheless, the G protein-based assays in our study reliably reflect all of Smo’s hallmark functional properties (modulation by established small molecules, inhibition by cholesterol depletion, and sensitivity to Ptch1), arguing that our findings represent a useful, valid framework for future investigations of Smo regulation.
Previously, it was not known whether Ptch1 inhibits Smo by removing an activator or providing an inhibitor, as either scenario is consistent with established genetic relationships. However, our nanodisc reconstitution (Fig. 4) shows that cholesterol is not merely a permissive factor that facilitates the action of another endogenous agonist, but is sufficient in its own right to stimulate Smo activity. This stimulation does not require the CRD, and so cannot be mediated by cholesterol bound to this domain, highlighting the importance of the 7TM region in cholesterol regulation of Smo. Furthermore, Smo shows similar constitutive activity in cell-derived membrane fractions (Fig. 3) and intact cells lacking exogenous Ptch1 (Figs. 1 and 2), even though both of these systems possess an extensive repertoire of lipid species besides cholesterol that could in principle influence Smo activity. Our reconstitution studies thus nominate cholesterol as the most likely substrate for Ptch1 action, if Ptch1 indeed functions by removing a Smo activator. Our in vitro experiments complement prior studies in living systems demonstrating that cholesterol biosynthesis is essential for Smo-mediated signaling (21, 22, 24, 40), and that addition of exogenous cholesterol to cells can promote Smo activity and enhance the ability of Hh to derepress Ptch1 (23, 25). In this regard, a salient feature of in vitro reconstitution is its ability to rigorously establish which components are truly sufficient for a biological process. This overcomes an inherent limitation of previous studies performed within the complex, incompletely defined environment of living cells.
How might Ptch1 act on cholesterol to influence Smo activity? Ptch1 might directly affect an interaction between membrane cholesterol and the Smo 7TM domain (Fig. 9), thereby coopting a near-universal mediator of GPCR stability (46) to allow for robust Hh-mediated regulation of Smo conformation. How Ptch1 could operate to efficiently deplete such a ubiquitous lipid is not obvious, but colocalization of Ptch1 and Smo within a specific membrane domain or a transient Ptch1–Smo interaction might allow Ptch1 to adjust cholesterol concentrations or the inner vs. outer leaflet distribution of cholesterol (62) selectively within the Smo vicinity. Alternatively, Ptch1 might catalyze the accumulation of an as yet unidentified negative regulator that blocks Smo activity by overriding the positive effects of membrane cholesterol (Fig. 9). Future studies using the approaches described here may help to fully define the molecular mechanism of Ptch1–Smo communication, including the identity of any endogenous Ptch1-regulated ligands.
Several recent studies (23, 25–27) demonstrated an intriguing capacity of cholesterol to interact with the Smo CRD, raising the possibility that Ptch1 might regulate Smo activity by changing this extracellular domain’s cholesterol occupancy. However, transcriptional reporter assays, both here (Fig. 6) and in prior studies (22, 24, 27), along with our reconstitution (Fig. 4) and GloSensor (Fig. 6) data, all demonstrate that the Smo CRD is not absolutely required for sensitivity to either cellular cholesterol or Ptch1 action. Instead, we suggest that any effects of Ptch1 on Smo-associated cholesterol proceed via a site within the Smo 7TM domain. The location of this site is unknown, but may be revealed through future mutagenesis, crystallographic studies of active Smo, or computational prediction of ligand binding surfaces (63). Whereas the CRD is not required to achieve Ptch1 regulation of Smo, CRD interaction with cholesterol may help stabilize the active conformation of Smo to an extent that is critical for in vivo pathway function, as suggested by the severe developmental phenotype of a Smo D99N mutant mouse (26). In addition, our data do not exclude the possibility that Ptch1 directly operates on both the CRD and 7TM domains simultaneously, or that the CRD renders Ptch1 action on the 7TM domain more efficient. Nevertheless, our study highlights a direct role for the Smo heptahelical domain as a critical mediator of Ptch1 action. To fully understand how endogenous metabolites regulate Smo, it will be essential going forward to distinguish between the “primary” sites on which Ptch1 directly operates versus “secondary” sites that are required to stabilize active Smo (which either constitutively bind a cofactor or whose occupancy changes as a downstream consequence of Ptch1 activity on its primary site). By developing direct assays for Ptch activity that do not depend on Smo for a functional readout, it may be possible to resolve this dilemma in the future.
Our study shows that cellular Ptch1 function requires extracellular Na+, leading us to hypothesize that transmembrane Na+ gradients, universally present in metazoans, may supply the energy for Ptch1 action. We cannot at present rule out that the ion substitution effects in our GloSensor assay might reflect a Na+ requirement for Ptch1-Smo regulation that is independent of transmembrane Na+ gradients, in which case Ptch1 activity would rely on some other cellular energy source that remains to be identified. Nevertheless, our data raise the possibility that Ptch1 activity might depend on an ion gradient, implying that the long-hypothesized transporter model for Ptch1 function might be correct. The reliance of Ptch1 on Na+ gradients may seem unusual considering that most RND transporters are H+-driven (16), but Na+-driven RND transporters have been identified in halophilic organisms. Vibrio alginolyticus, a marine-estuarine bacterium, encodes two sets of RND transporters responsible for peptide secretion (SecD/F-1 and SecD/F-2), the first of which is Na+-dependent (64). Like Ptch1, both H+- and Na+-dependent SecDF proteins are sensitive to charge-neutralizing mutations in transmembrane helices 4 and 10 (65), illustrating that the core ion-conducting circuit within the RND fold can be evolutionarily rewired to accommodate at least two different types of chemiosmotic gradients. Further efforts to systematically mutagenize transmembrane residues could define the permeation pathway in Ptch1 and other Na+-dependent RND transporters in greater detail. Apart from Ptch1, our direct cell-based and in vitro approaches may also shed light on other mechanistically mysterious Ptch1-related proteins for which sensitive live-cell approaches are not yet available, including the putative Hh exporter Dispatched (19, 20) and the NPC1 protein required to release LDL-derived cholesterol from lysosomes (18). Given that the lysosomal lumen contains high concentrations of both Na+ and H+ (66), it will be interesting to determine whether the NPC1 protein functions by harnessing the energy from a Na+ gradient.
Seminal advances in many areas of membrane signaling, such as the regulation of GPCRs and ion channels by hormones and neurotransmitters, have relied on rapid, direct, real-time functional readouts in living and cell-free systems. In the past, the question of Ptch1–Smo regulation has not benefited from such approaches. By measuring Smo activity with direct assays in simplified cell-based and in vitro settings, our work now renders the upstream Hh pathway accessible to a variety of experimental treatments that would be impractical with conventional cilium-dependent readouts. This includes manipulations that, while potentially informative, might kill cells or compromise ciliary trafficking over longer time scales. In addition, our cell-free readouts may allow us to more deeply interrogate Ptch–Smo regulation using in vitro systems where proteins, lipids, and ionic compositions can be rigorously and precisely controlled. We anticipate that such strategies will be generally useful in unraveling the mechanism of Ptch1–Smo communication, potentially revealing fundamental aspects of an enduring mystery in development, cancer, and regeneration.
Materials and Methods
Culture and transfection of Sf9 cells, HEK293 fibroblasts, NIH 3T3 fibroblasts, and Smo−/− fibroblasts is described in SI Materials and Methods. G protein coupling in live HEK293 cells expressing Smo or Ptch1 was masured using a cAMP-dependent luciferase (GloSensor 22F). Membrane fractions derived from suspension HEK293 cells were analyzed for G protein coupling via 35S-GTPγS binding and scintillation proximity assay. An N-terminally SBP-tagged Smo–Gαo fusion was purified from BacMam-infected suspension HEK293 cells in DDM/CHS detergent, and reconstituted into MSP1D1 nanodiscs, and analyzed for G protein coupling in a similar manner. In experiments studying effects of cholesterol depletion, membrane cholesterol was first depleted from membranes using MβCD, and subsequently restored using various various sterol:MβCD complexes. Di-isoleucine-initiated Shh was purified from bacteria as previously described. Detailed information is provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank J. K. Chen, L. Feng, A. Manglik, R. Mann, and W. Weis for providing critical feedback on our manuscript; J. Belani and S. Rychnovsky for their generous gift of ent-cholesterol; and the Vincent Coates Foundation Mass Spectrometry Laboratory at Stanford University for mass spectrometry assistance. Portions of this work were funded by NIH Grant 5R01GM102498-04. B.R.M. was a fellow of the Damon Runyon Cancer Research Foundation. L.N. was supported by the Stanford Bio-X Undergraduate Summer Research Program. Y.Z. is supported by California Institute for Regenerative Medicine Training Grant TG2-01159. K.J.R. is supported by a National Science Foundation Graduate Research Fellowship, and by the Gerhard Casper Stanford Graduate Fellowship. P.A.B. is an investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1717891115/-/DCSupplemental.
References
- 1.Briscoe J, Thérond PP. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat Rev Mol Cell Biol. 2013;14:416–429. doi: 10.1038/nrm3598. [DOI] [PubMed] [Google Scholar]
- 2.Pak E, Segal RA. Hedgehog signal transduction: Key players, oncogenic drivers, and cancer therapy. Dev Cell. 2016;38:333–344. doi: 10.1016/j.devcel.2016.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu F, Zhang Y, Sun B, McMahon AP, Wang Y. Hedgehog signaling: From basic biology to cancer therapy. Cell Chem Biol. 2017;24:252–280. doi: 10.1016/j.chembiol.2017.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shin K, et al. Hedgehog/Wnt feedback supports regenerative proliferation of epithelial stem cells in bladder. Nature. 2011;472:110–114. doi: 10.1038/nature09851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao C, et al. Stromal Gli2 activity coordinates a niche signaling program for mammary epithelial stem cells. Science. 2017;356:eaal3485. doi: 10.1126/science.aal3485. [DOI] [PubMed] [Google Scholar]
- 6.Teglund S, Toftgård R. Hedgehog beyond medulloblastoma and basal cell carcinoma. Biochim Biophys Acta. 2010;1805:181–208. doi: 10.1016/j.bbcan.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 7.Lee JJ, et al. Stromal response to Hedgehog signaling restrains pancreatic cancer progression. Proc Natl Acad Sci USA. 2014;111:E3091–E3100. doi: 10.1073/pnas.1411679111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rhim AD, et al. Stromal elements act to restrain, rather than support, pancreatic ductal adenocarcinoma. Cancer Cell. 2014;25:735–747. doi: 10.1016/j.ccr.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shin K, et al. Hedgehog signaling restrains bladder cancer progression by eliciting stromal production of urothelial differentiation factors. Cancer Cell. 2014;26:521–533. doi: 10.1016/j.ccell.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee JJ, et al. Control of inflammation by stromal Hedgehog pathway activation restrains colitis. Proc Natl Acad Sci USA. 2016;113:E7545–E7553. doi: 10.1073/pnas.1616447113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerling M, et al. Stromal Hedgehog signalling is downregulated in colon cancer and its restoration restrains tumour growth. Nat Commun. 2016;7:12321. doi: 10.1038/ncomms12321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Özdemir BC, et al. Depletion of carcinoma-associated fibroblasts and fibrosis induces immunosuppression and accelerates pancreas cancer with reduced survival. Cancer Cell. 2014;25:719–734. doi: 10.1016/j.ccr.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arensdorf AM, Marada S, Ogden SK. Smoothened regulation: A tale of two signals. Trends Pharmacol Sci. 2016;37:62–72. doi: 10.1016/j.tips.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goetz SC, Anderson KV. The primary cilium: A signalling centre during vertebrate development. Nat Rev Genet. 2010;11:331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taipale J, Cooper MK, Maiti T, Beachy PA. Patched acts catalytically to suppress the activity of Smoothened. Nature. 2002;418:892–897. doi: 10.1038/nature00989. [DOI] [PubMed] [Google Scholar]
- 16.Tseng TT, et al. The RND permease superfamily: An ancient, ubiquitous and diverse family that includes human disease and development proteins. J Mol Microbiol Biotechnol. 1999;1:107–125. [PubMed] [Google Scholar]
- 17.Mukhopadhyay S, Rohatgi R. G-protein-coupled receptors, Hedgehog signaling and primary cilia. Semin Cell Dev Biol. 2014;33:63–72. doi: 10.1016/j.semcdb.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carstea ED, et al. Niemann-Pick C1 disease gene: Homology to mediators of cholesterol homeostasis. Science. 1997;277:228–231. doi: 10.1126/science.277.5323.228. [DOI] [PubMed] [Google Scholar]
- 19.Burke R, et al. Dispatched, a novel sterol-sensing domain protein dedicated to the release of cholesterol-modified hedgehog from signaling cells. Cell. 1999;99:803–815. doi: 10.1016/s0092-8674(00)81677-3. [DOI] [PubMed] [Google Scholar]
- 20.Ma Y, et al. Hedgehog-mediated patterning of the mammalian embryo requires transporter-like function of dispatched. Cell. 2002;111:63–75. doi: 10.1016/s0092-8674(02)00977-7. [DOI] [PubMed] [Google Scholar]
- 21.Cooper MK, et al. A defective response to Hedgehog signaling in disorders of cholesterol biosynthesis. Nat Genet. 2003;33:508–513. doi: 10.1038/ng1134. [DOI] [PubMed] [Google Scholar]
- 22.Myers BR, et al. Hedgehog pathway modulation by multiple lipid binding sites on the smoothened effector of signal response. Dev Cell. 2013;26:346–357. doi: 10.1016/j.devcel.2013.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang P, et al. Cellular cholesterol directly activates smoothened in Hedgehog signaling. Cell. 2016;166:1176–1187.e14. doi: 10.1016/j.cell.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blassberg R, Macrae JI, Briscoe J, Jacob J. Reduced cholesterol levels impair Smoothened activation in Smith-Lemli-Opitz syndrome. Hum Mol Genet. 2016;25:693–705. doi: 10.1093/hmg/ddv507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luchetti G, et al. Cholesterol activates the G-protein coupled receptor Smoothened to promote hedgehog signaling. eLife. 2016;5:e20304. doi: 10.7554/eLife.20304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiao X, et al. Cholesterol modification of Smoothened is required for Hedgehog signaling. Mol Cell. 2017;66:154–162.e10. doi: 10.1016/j.molcel.2017.02.015. [DOI] [PubMed] [Google Scholar]
- 27.Byrne EFX, et al. Structural basis of Smoothened regulation by its extracellular domains. Nature. 2016;535:517–522. doi: 10.1038/nature18934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DeCamp DL, Thompson TM, de Sauvage FJ, Lerner MR. Smoothened activates Galphai-mediated signaling in frog melanophores. J Biol Chem. 2000;275:26322–26327. doi: 10.1074/jbc.M004055200. [DOI] [PubMed] [Google Scholar]
- 29.Riobo NA, Saucy B, Dilizio C, Manning DR. Activation of heterotrimeric G proteins by Smoothened. Proc Natl Acad Sci USA. 2006;103:12607–12612. doi: 10.1073/pnas.0600880103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen F, Cheng L, Douglas AE, Riobo NA, Manning DR. Smoothened is a fully competent activator of the heterotrimeric G protein G(i) Mol Pharmacol. 2013;83:691–697. doi: 10.1124/mol.112.082511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masdeu C, et al. Identification and characterization of Hedgehog modulator properties after functional coupling of smoothened to G15. Biochem Biophys Res Commun. 2006;349:471–479. doi: 10.1016/j.bbrc.2006.07.216. [DOI] [PubMed] [Google Scholar]
- 32.Ayers KL, Thérond PP. Evaluating Smoothened as a G-protein-coupled receptor for Hedgehog signalling. Trends Cell Biol. 2010;20:287–298. doi: 10.1016/j.tcb.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 33.Binkowski BF, et al. A luminescent biosensor with increased dynamic range for intracellular cAMP. ACS Chem Biol. 2011;6:1193–1197. doi: 10.1021/cb200248h. [DOI] [PubMed] [Google Scholar]
- 34.Gerdes JM, et al. Disruption of the basal body compromises proteasomal function and perturbs intracellular Wnt response. Nat Genet. 2007;39:1350–1360. doi: 10.1038/ng.2007.12. [DOI] [PubMed] [Google Scholar]
- 35.Kim J, et al. The role of ciliary trafficking in Hedgehog receptor signaling. Sci Signal. 2015;8:ra55. doi: 10.1126/scisignal.aaa5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rohatgi R, Milenkovic L, Scott MP. Patched1 regulates hedgehog signaling at the primary cilium. Science. 2007;317:372–376. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- 37.Kim J, Kato M, Beachy PA. Gli2 trafficking links Hedgehog-dependent activation of smoothened in the primary cilium to transcriptional activation in the nucleus. Proc Natl Acad Sci USA. 2009;106:21666–21671. doi: 10.1073/pnas.0912180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seifert R, Wenzel-Seifert K, Kobilka BK. GPCR-Galpha fusion proteins: Molecular analysis of receptor-G-protein coupling. Trends Pharmacol Sci. 1999;20:383–389. doi: 10.1016/s0165-6147(99)01368-1. [DOI] [PubMed] [Google Scholar]
- 39.Sharpe HJ, Wang W, Hannoush RN, de Sauvage FJ. Regulation of the oncoprotein Smoothened by small molecules. Nat Chem Biol. 2015;11:246–255. doi: 10.1038/nchembio.1776. [DOI] [PubMed] [Google Scholar]
- 40.Sever N, et al. Endogenous B-ring oxysterols inhibit the Hedgehog component Smoothened in a manner distinct from cyclopamine or side-chain oxysterols. Proc Natl Acad Sci USA. 2016;113:5904–5909. doi: 10.1073/pnas.1604984113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Russell DW. Oxysterol biosynthetic enzymes. Biochim Biophys Acta. 2000;1529:126–135. doi: 10.1016/s1388-1981(00)00142-6. [DOI] [PubMed] [Google Scholar]
- 42.Russell DW. Fifty years of advances in bile acid synthesis and metabolism. J Lipid Res. 2009;50:S120–S125. doi: 10.1194/jlr.R800026-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pikuleva IA. Cholesterol-metabolizing cytochromes P450. Drug Metab Dispos. 2006;34:513–520. doi: 10.1124/dmd.105.008789. [DOI] [PubMed] [Google Scholar]
- 44.Nebert DW, Wikvall K, Miller WL. Human cytochromes P450 in health and disease. Philos Trans R Soc Lond B Biol Sci. 2013;368:20120431. doi: 10.1098/rstb.2012.0431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Meer G, Voelker DR, Feigenson GW. Membrane lipids: Where they are and how they behave. Nat Rev Mol Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gimpl G. Interaction of G protein coupled receptors and cholesterol. Chem Phys Lipids. 2016;199:61–73. doi: 10.1016/j.chemphyslip.2016.04.006. [DOI] [PubMed] [Google Scholar]
- 47.Denisov IG, Sligar SG. Nanodiscs for structural and functional studies of membrane proteins. Nat Struct Mol Biol. 2016;23:481–486. doi: 10.1038/nsmb.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vélez-Ruiz GA, Sunahara RK. Reconstitution of G protein-coupled receptors into a model bilayer system: Reconstituted high-density lipoprotein particles. Methods Mol Biol. 2011;756:167–182. doi: 10.1007/978-1-61779-160-4_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nachtergaele S, et al. Oxysterols are allosteric activators of the oncoprotein Smoothened. Nat Chem Biol. 2012;8:211–220. doi: 10.1038/nchembio.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taipale J, et al. Effects of oncogenic mutations in Smoothened and patched can be reversed by cyclopamine. Nature. 2000;406:1005–1009. doi: 10.1038/35023008. [DOI] [PubMed] [Google Scholar]
- 51.Sweeney RT, et al. Identification of recurrent SMO and BRAF mutations in ameloblastomas. Nat Genet. 2014;46:722–725. doi: 10.1038/ng.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li X, et al. 3.3 Å structure of Niemann-Pick C1 protein reveals insights into the function of the C-terminal luminal domain in cholesterol transport. Proc Natl Acad Sci USA. 2017;114:9116–9121. doi: 10.1073/pnas.1711716114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guan L, Nakae T. Identification of essential charged residues in transmembrane segments of the multidrug transporter MexB of Pseudomonas aeruginosa. J Bacteriol. 2001;183:1734–1739. doi: 10.1128/JB.183.5.1734-1739.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Murakami S, Nakashima R, Yamashita E, Yamaguchi A. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature. 2002;419:587–593. doi: 10.1038/nature01050. [DOI] [PubMed] [Google Scholar]
- 55.Seeger MA, et al. Engineered disulfide bonds support the functional rotation mechanism of multidrug efflux pump AcrB. Nat Struct Mol Biol. 2008;15:199–205. doi: 10.1038/nsmb.1379. [DOI] [PubMed] [Google Scholar]
- 56.Hille B. Ionic Channels of Excitable Membranes. Sinauer Associates Incorporated; Sunderland, MA: 1992. [Google Scholar]
- 57.Reuss L, Grady TP. Effects of external sodium and cell membrane potential on intracellular chloride activity in gallbladder epithelium. J Membr Biol. 1979;51:15–31. doi: 10.1007/BF01869341. [DOI] [PubMed] [Google Scholar]
- 58.Katritch V, et al. Allosteric sodium in class A GPCR signaling. Trends Biochem Sci. 2014;39:233–244. doi: 10.1016/j.tibs.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vieira OV, et al. FAPP2, cilium formation, and compartmentalization of the apical membrane in polarized Madin-Darby canine kidney (MDCK) cells. Proc Natl Acad Sci USA. 2006;103:18556–18561. doi: 10.1073/pnas.0608291103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Garcia-Gonzalo FR, et al. Phosphoinositides regulate ciliary protein trafficking to modulate Hedgehog signaling. Dev Cell. 2015;34:400–409. doi: 10.1016/j.devcel.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chávez M, et al. Modulation of ciliary phosphoinositide content regulates trafficking and sonic Hedgehog signaling output. Dev Cell. 2015;34:338–350. doi: 10.1016/j.devcel.2015.06.016. [DOI] [PubMed] [Google Scholar]
- 62.Liu SL, et al. Orthogonal lipid sensors identify transbilayer asymmetry of plasma membrane cholesterol. Nat Chem Biol. 2017;13:268–274. doi: 10.1038/nchembio.2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ruat M, Hoch L, Faure H, Rognan D. Targeting of Smoothened for therapeutic gain. Trends Pharmacol Sci. 2014;35:237–246. doi: 10.1016/j.tips.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 64.Ishii E, et al. Nascent chain-monitored remodeling of the Sec machinery for salinity adaptation of marine bacteria. Proc Natl Acad Sci USA. 2015;112:E5513–E5522. doi: 10.1073/pnas.1513001112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tsukazaki T, et al. Structure and function of a membrane component SecDF that enhances protein export. Nature. 2011;474:235–238. doi: 10.1038/nature09980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang X, et al. TPC proteins are phosphoinositide- activated sodium-selective ion channels in endosomes and lysosomes. Cell. 2012;151:372–383. doi: 10.1016/j.cell.2012.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Klein U, Gimpl G, Fahrenholz F. Alteration of the myometrial plasma membrane cholesterol content with beta-cyclodextrin modulates the binding affinity of the oxytocin receptor. Biochemistry. 1995;34:13784–13793. doi: 10.1021/bi00042a009. [DOI] [PubMed] [Google Scholar]
- 68.Nedelcu D, Liu J, Xu Y, Jao C, Salic A. Oxysterol binding to the extracellular domain of Smoothened in Hedgehog signaling. Nat Chem Biol. 2013;9:557–564. doi: 10.1038/nchembio.1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.