Significance
Phosphoinositide lipids, produced by phosphorylation of the membrane lipid phosphatidylinositol, play essential roles throughout the cell in defining organelle membrane identity and in initiating and mediating cellular signaling processes. Plasma membrane (PM) phosphoinositides, which reside at the interface between the intracellular and extracellular environments, play especially critical roles. The synthesis of all PM phosphoinositides begins with the conversion of phosphatidylinositol to phosphatidylinositol-4-phosphate by the heterotrimeric PI4KIIIα complex. This study reports the high-resolution structure of the PI4KIIIα complex, revealing a large, intricately structured superassembly whose complexity positions it as a major regulatory junction for PM phosphoinositide synthesis. The structure now provides a framework to guide further efforts to understand its regulation.
Keywords: signaling, phosphoinositides, lipid kinase
Abstract
Plasma membrane (PM) phosphoinositides play essential roles in cell physiology, serving as both markers of membrane identity and signaling molecules central to the cell’s interaction with its environment. The first step in PM phosphoinositide synthesis is the conversion of phosphatidylinositol (PI) to PI4P, the precursor of PI(4,5)P2 and PI(3,4,5)P3. This conversion is catalyzed by the PI4KIIIα complex, comprising a lipid kinase, PI4KIIIα, and two regulatory subunits, TTC7 and FAM126. We here report the structure of this complex at 3.6-Å resolution, determined by cryo-electron microscopy. The proteins form an obligate ∼700-kDa superassembly with a broad surface suitable for membrane interaction, toward which the kinase active sites are oriented. The structural complexity of the assembly highlights PI4P synthesis as a major regulatory junction in PM phosphoinositide homeostasis. Our studies provide a framework for further exploring the mechanisms underlying PM phosphoinositide regulation.
The phosphoinositide lipids at the plasma membrane (PM) play key roles in cell physiology. Phosphatidylinositol-4-phosphate (PI4P) and its metabolite phosphatidylinositol-4,5-bisphosphate [PI(4,5)P2] are determinants of PM identity, specifically recruiting protein effectors to the membrane bilayer to control exo- and endocytic processes and cytoskeletal dynamics (1–3). Further, PI(4,5)P2 and the product of its 3′ phosphorylation, phosphatidylinositol-3,4,5-trisphosphate [PI(3,4,5)P3], have important direct and indirect signaling functions, influencing a wide variety of processes that range from the control of cell shape and cell motility to cell excitability, cell proliferation, and regulation of gene expression, among others (2).
The lipid kinase complex that converts phosphatidylinositol (PI) to PI4P at the PM catalyzes the first committed step in the generation of these phosphoinositides. Conserved in all eukaryotes from yeast to humans, the complex in mammals comprises the lipid kinase PI4KIIIα, also known as PI4KA (and in yeast, Stt4p), and the regulatory subunits TTC7 (Ypp1p in yeast) and FAM126 (a subunit missing in yeast), all of which are recruited to the PM by a fourth protein, EFR3 (4–10). PI4KIIIα is a very large protein (∼240 kDa) in which the catalytic module represents a small portion of the entire molecule, whose structure is unknown. Moreover, nothing is known of how TTC7 and FAM126 interact with PI4KIIIα to modulate its activity.
To better understand its function, we have used cryo-electron microscopy (cryo-EM) to determine the structure of human PI4KIIIα complexed with TTC7 and FAM126 at a nominal resolution of 3.6 Å, finding that two PI4KIIIα/TTC7/FAM126 heterotrimers dimerize via their lipid kinase subunits to form a ∼700-kDa superassembly. Extensive and conserved interfaces between the kinase molecules, between PI4KIIIα and TTC7 and between TTC7 and FAM126, imply an obligate complex. The complex has a flat surface suited for membrane association, with the kinase catalytic domains oriented to interact with PI in the PM bilayer. The N-terminal portion of each kinase forms an α-solenoid protruding from the opposite face of the complex. Interactions with TTC7/FAM126 constrain the conformation of PI4KIIIα and are required for its stability in living cells.
Evident from the reconstruction, the PI4KIIIα assembly is comparable in complexity (although entirely different in structure) to phosphatidylinositol 3-kinase (11), which synthesizes PM PI(3,4,5)P3 and is known to be highly regulated, whereas the architecture of the other major enzymes responsible for PM phosphoinositide homeostasis is much simpler. The structural intricacy of the PI4KIIIα complex underscores the importance of PI4P production as a major regulatory point in PM phosphoinositide homeostasis. Moreover, our structural studies provide a framework guiding further explorations regarding phosphoinositide regulation at the PM.
Results
PI4KIIIα Forms a Stable, Homodimeric ∼700-kDa Complex with TTC7 and FAM126.
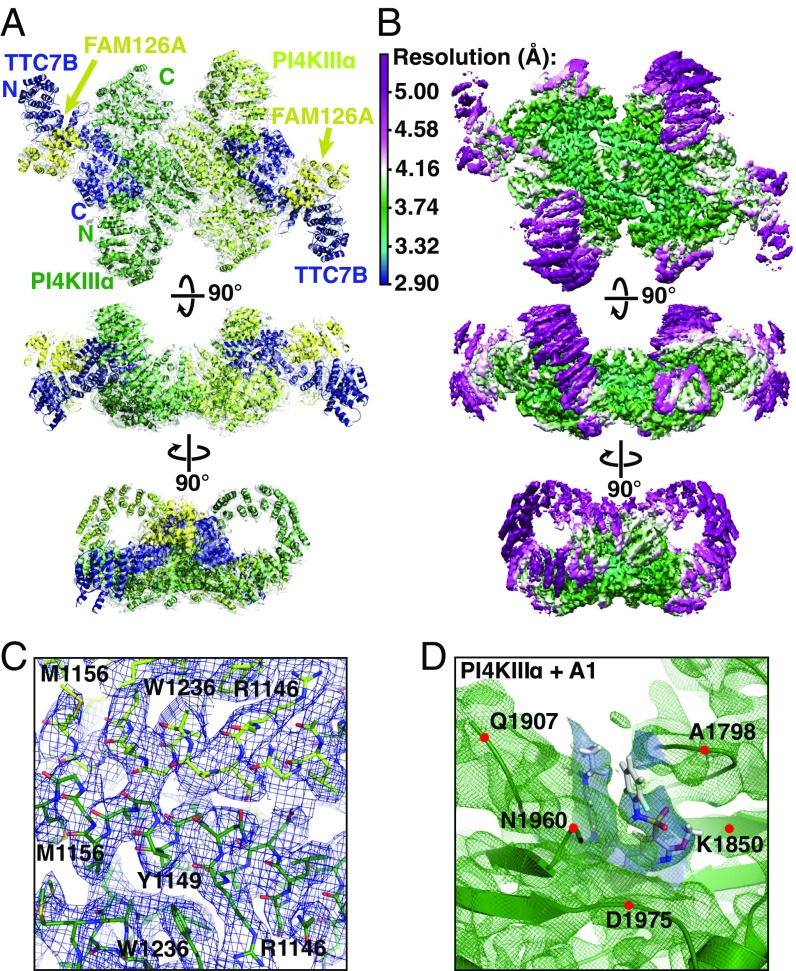
In human cells, PI4KIIIα associates with TTC7 (4, 6) and FAM126 (6), each of which exists in closely related A and B isoforms. Further, the TTC7/FAM126 complex stimulates the enzymatic activity of PI4KIIIα in vitro (6). To obtain structural insights into this biologically active PI4KIIIα assembly, we coexpressed and copurified PI4KIIIα and TTC7B with the ordered N-terminal domain (2–289) of FAM126A in Expi293 cells (Fig. S1), then used cryo-EM and single-particle analysis to obtain a 3D reconstruction of the complex at 3.6-Å resolution (Figs. S2–S4). The complex measures ∼240 × 165 × 85 Å and exhibits twofold symmetry (Fig. 1A), which agrees with the ∼700-kDa size estimated from size exclusion chromatography (Fig. S1A) and is consistent with homodimerization of the ternary complex. Local resolution is highest near the core (3–4 Å; Fig. 1 B and C) and declines toward 5 Å and below at the periphery (Fig. 1 B and D). Existing crystal structures of human TTC7 and FAM126 (6) facilitated identification of structurally congruent map density, into which the crystal structures were first fitted as rigid bodies, then flexibly fitted to match the density. An initial model of the kinase catalytic domain (residues 1,597–2,085) was generated by homology modeling to ensure correct alignment of the catalytic residues, followed by manual rebuilding and flexible fitting to the map density. The core region of the kinase (residues 932–1,596), where the local resolution approaches 3 Å, was built de novo (Fig. 1 B and C). N-terminal portions of the kinase, at lower local resolution, were modeled as a series of polyalanine helices. Due to the poor local resolution, the orientation and connectivity of these helices could not be determined.
Fig. 1.
The PI4KIIIα/TTC7/FAM126 complex forms a ∼700-kDa homodimeric superassembly. (A) Ribbon model of PI4KIIIα/TTC7B/FAM126A complex, colored and labeled by subunit, and superimposed with map density. (B) Single-particle cryo-EM model of PI4KIIIα/TTC7B/FAM126A complex at 3.6-Å resolution, contoured at 8.5 σ. Each view is colored by local resolution according to the scale at Top Left. (C) Model and typical density from the core of the PI4KIIIα map, contoured at 8.5 σ, with resolution approaching 3.0 Å. Prominent residues are labeled for orientation purposes. (D) Ribbon diagram (green) and density (green mesh) from the active site of PI4KIIIa, with the A1 inhibitor in stick representation. Map density used to place the A1 ligand is in blue. The resolution of this region is ∼4–5 Å, with the density contoured at 8.5 σ. Alpha carbons of indicated residues are denoted by red dots for orientation purposes.
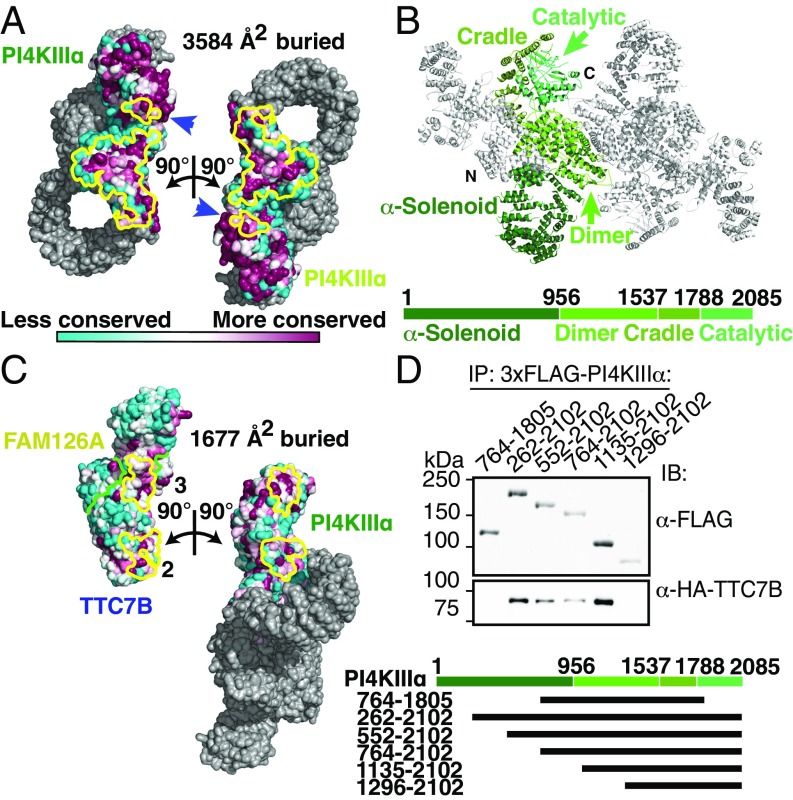
Two molecules of the kinase occupy the center of the complex (Fig. 1A), homodimerizing through an unusually large buried surface area of over 3,500 Å2/monomer (Fig. 2A), which indicates a high-affinity association (interfaces > 1,000 Å2/monomer are considered large). The C-terminal catalytic domains (residues 1,788–2,085) occupy lateral positions opposite the dimerization interface, and each is cradled by a series of ARM repeats (residues 1,537–1,787), which transition into the mostly helical dimerization domain (residues 957–1,536) (Fig. 2B). Both the catalytic domain and the dimerization interface are highly conserved in PI4KIIIα orthologs of other species (Fig. 2A). The N-terminal portion of the kinase comprises a long series of ARM repeats, which form an α-solenoid loop (Fig. 2 A and B) reminiscent of the “horn” reported for the mTOR N terminus (12), although the layouts of the mTOR and PI4KIIIα complexes are otherwise unrelated. The N-terminal tip of this α-solenoid, for which the local resolution was not sufficient for atomic modeling, forms the first of three contacts with TTC7, interacting with its C-terminal helix. Several predicted N-terminal helices (residues 31–43, 75–87, 98–104, and 132–145) of the kinase are conserved and thus are possible candidates to participate in this interface. Two additional contacts with TTC7 occur via the PI4KIIIα dimerization domain and the “cradle” surrounding the catalytic domain, together burying over 1,600 Å2/monomer of solvent-accessible surface area (Fig. 2C). TTC7 and FAM126, based on the crystal structure of the TTC7/FAM126 subcomplex (6), bury over 3,000 Å2/monomer at their interface. The large surface areas buried by each of these interactions suggests that PI4KIIIα, TTC7, and FAM126 form a complex that remains stably associated in the cell.
Fig. 2.
PI4KIIIα homodimerizes and interacts stably with TTC7 via large conserved surfaces. (A) Solvent-accessible surface representation of PI4KIIIα/TTC7B/FAM126A complex, with the PI4KIIIα surface (excluding the α-solenoid) colored by conservation. Each heterotrimer is rotated by ±90° as indicated to reveal the homodimerization interface. PI4KIIIα surface residues participating in the dimerization are outlined in yellow. Blue arrows indicate regions of the kinase catalytic domains that contact the opposite copy of PI4KIIIα. (B) Ribbon representation of the PI4KIIIα complex, with one copy of PI4KIIIα colored by domain. Boundary residue numbers for each domain are indicated on the schematic below. (C) Solvent-accessible surface representation of PI4KIIIα/TTC7B/FAM126A complex, with the TTC7/FAM126 complex and PI4KIIIα surfaces colored by conservation, as in A. PI4KIIIα and the TTC7B/FAM126A complex are each rotated by ±90° as indicated to reveal the PI4KIIIα/TTC7B dimerization interface. Interacting residues are outlined in yellow, while the TTC7B/FAM126A boundary is indicated by a green line. TTC7 surfaces 2 and 3 interact with the dimerization and cradle domains of PI4KIIIα, respectively. The additional contact between TTC7B and the tip of the PI4KIIIα α-solenoid, which was not sufficiently resolved to model by a Cα trace, is neither shown nor included in the surface area calculation. (D) Truncation constructs of 3xFLAG-tagged PI4KIIIα were overexpressed alongside HA-TTC7B and EGFP-FAM126A(2–289) in Expi293 cells. α-FLAG-immunoprecipitated samples were resolved by SDS/PAGE and immunoblotted with anti-FLAG and anti-HA antibodies to probe coprecipitation of TTC7B with the different PI4KIIIα constructs. Boundaries of PI4KIIIα truncation constructs are indicated above the blot and in the schematic at bottom.
To test our model and to evaluate the importance of the interfaces for complex formation, we coexpressed a series of N- and C-terminal truncations of the kinase with TTC7 and FAM126 in Expi293 cells and monitored complex formation by immunoprecipitation (Fig. 2D). Constructs with deletions of up to 1,134 amino acids from the N terminus retain interaction with the TTC7/FAM126 subcomplex, indicating that loss of the N-terminal contact does not disrupt complex formation. A more severe deletion of the first 1,295 residues, which additionally destabilizes the second interface, abolishes interaction completely, as does disruption of the first and third interfaces in a PI4KIIIα construct comprising residues 764–1,805. Thus, none of the contact points individually is sufficient to maintain a stable complex, likely due to the smaller buried surface areas (675 Å2 for interface “2” and 1,002 Å2 for interface “3”; Fig. 2C). The second and third interfaces together are, however, sufficient for interaction.
PI4KIIIα Interactions with TTC7 Are Required for Stability of the Complex Components in Vivo.
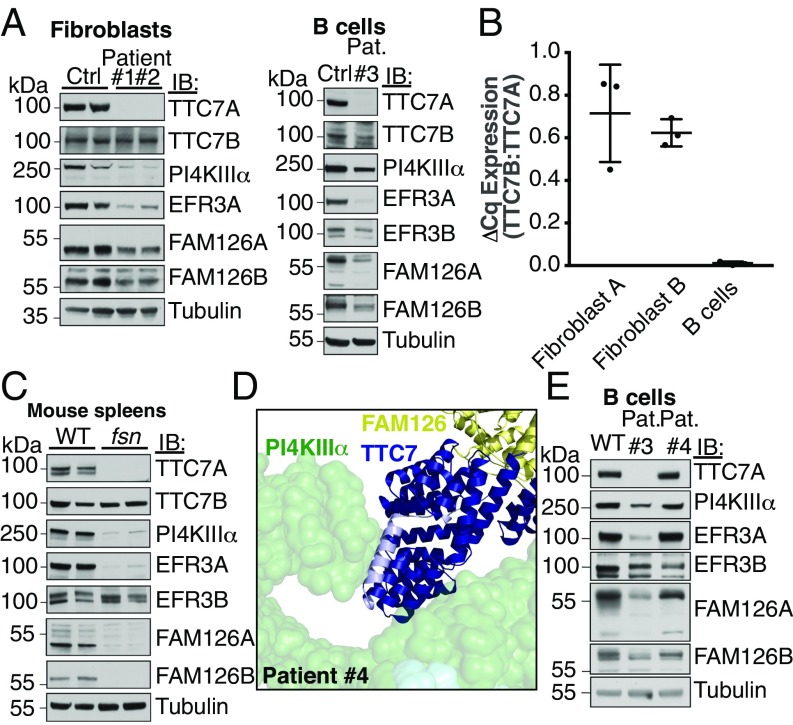
Mutations in TTC7A, one of the two genes encoding TTC7 homologs, have been associated with the rare hereditary human disease, combined immunodeficiency with multiple intestinal atresias (CID-MIA) (13–16), and with various mouse models, including the flaky skin (fsn) mouse, which involves pleiotropic abnormalities, including immune system dysfunction (17, 18). To better understand the role of TTC7 in kinase complex formation and stability, we examined the PI4KIIIα complex in patient cells and fsn mouse tissue by Western blotting. Three CID-MIA cell lines, in which TTC7A was undetectable (patients 1–3: from two homozygous patients and one compound heterozygous patient), exhibited reduced levels of the other complex components (PI4KIIIα and FAM126) compared with cells of healthy individuals, suggesting loss of protein stability in the absence of complex formation (Fig. 3A). As TTC7B was still present in these cells, it most likely is a minor contributor to the complex in these cells, possibly because of its lower levels of expression (Fig. 3B). Absence of TTC7 in the fsn mouse produced a similar dramatic loss of other complex components (Fig. 3C), indicating that PI4KIIIα, TTC7, and FAM126 likely form a constitutive core complex. Levels of EFR3, which is not part of the catalytic complex, were also affected in these CID-MIA patients, consistent with the requirement of PI4KIIIα, possibly via its impact on the phospholipid composition of the PM, in the efficient targeting to and possibly stabilization of EFR3 at the PM (7).
Fig. 3.
The interaction between TTC7 and PI4KIIIα is required for PI4KIIIα stability in vivo. (A) Patient fibroblasts or B cells were isolated and immunoblotted with antibodies against components of the PI4KIIIα complex. The levels of other complex subunits are reduced in the absence of intact TTC7A. (B) TTC7B mRNA is less abundant than TTC7A mRNA in human fibroblasts and nearly absent in B cells. cDNA from control human fibroblasts and B cells used in C was analyzed by qRT-PCR with TTC7A- and TTC7B-specific primer pairs. The ratio of TTC7B expression to that of TTC7A is shown for each cell type. Error bars represent SD (n = 3). (C) Spleens isolated from “flaky skin” mice (17, 18) were immunoblotted for PI4KA complex subunits. Loss of TTC7A caused reduction in the levels of other complex components. (D) CID-MIA patient 4 carries a mutation that truncates TTC7A at residue 823 (corresponding to residue 809 of TTC7B). The corresponding truncated residues are indicated in light blue on the structure of TTC7B in the context of the kinase complex. This mutation likely abolishes the interaction between TTC7 and the tip of the PI4KIIIα α-solenoid, causing the pathology of CID-MIA. (E) CID-MIA patient 4 carries an intact, though mildly truncated version of TTC7A. Immunoblots for other PI4KA complex components indicate little change in protein levels. Immunoblots against B cells from patient 3 are shown for comparison. Ctrl, Control; IB, immunoblot; Pat., Patient.
Cells from a fourth CID-MIA patient carried a mutation in TTC7A that leaves the protein largely intact, exhibiting near wild-type expression. This patient carries one copy of TTC7A from which the encoded protein is truncated at residue 600, likely resulting in a degraded, nonfunctional protein. The second copy, however, carries a c.2839+1insC frameshift that alters its amino acid sequence starting only 35 residues from its C terminus. For TTC7B, this corresponds to deviation from the wild-type sequence starting at position 809 and loss of its C-terminal helix (Fig. 3D, light blue; Fig. S7). The other proteins in the PI4KIIIα complex are expressed at near wild-type levels, as assessed by Western blot (Fig. 3E), but the alteration is sufficient to cause the disease, suggesting that the interaction between the C-terminal tip of TTC7 and the N-terminal tip of PI4KIIIα, while not required for complex assembly, is critical for kinase function. One possibility is that the large α-solenoid of PI4KIIIα forms an interaction surface for additional factors that regulate the complex, and that its particular conformation, determined by the interaction with TTC7 at its tip, is sensitive to perturbation.
The PI4KIIIα Complex Interacts with the PM via a Broad Conserved Surface, Orienting Its Active Sites Toward Substrate.
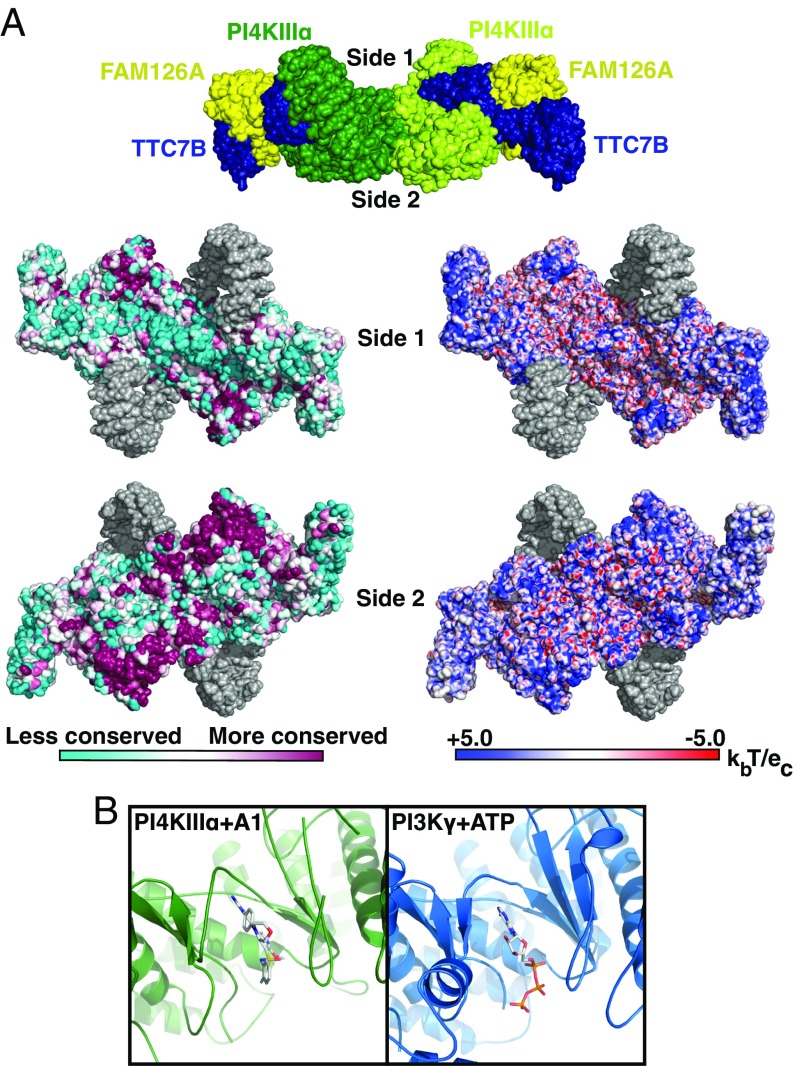
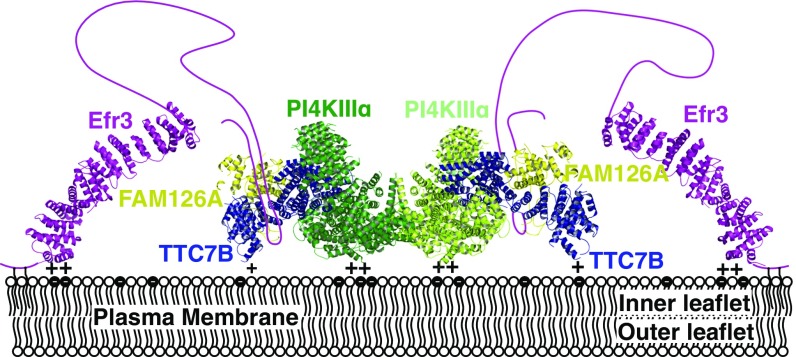
The mode of kinase complex interaction with the membrane has thus far been unknown. Our structural model reveals the complex as a flattened structure with two broad surfaces (Fig. 4A). One surface, denoted as “side 1,” is poorly conserved, with acidic residues near the dimer interface and peripheral hotspots of basic residues in the kinase catalytic domain and the TTC7 N terminus. This surface is uneven, largely because of the two kinase α-solenoids (depicted in gray) that protrude over it. The “side 2” surface is flatter and is highly conserved (Fig. 4A), comprising chiefly the catalytic and dimerization domains of the kinase, with further contributions from the N-terminal tips of the TTC7 molecules, which curve gently toward this face. The most highly conserved portions of this surface form plateaus (Fig. S7 A and B, outlined in yellow), which, while not strongly basic, have an excess of exposed basic residues compared with acidic residues (18 R/K vs. 10 D/E residues; Fig. S7 B and C), making this region suitable for interaction with the acidic inner leaflet of the PM. While interaction of the PI4KIIIα/TTC7/FAM126 complex with the PM is known to require EFR3 in vivo (4, 5, 7), the flat, basic surface may rather serve to help orient the complex with respect to the membrane upon recruitment by EFR3.
Fig. 4.
PI4KIIIα complex interacts with the plasma membrane via a large conserved surface that orients the catalytic domains toward substrate. (A, Top) Surface representation of PI4KIIIα complex, colored by subunit, as in Fig. 1A. Surfaces designated side 1 and side 2 are indicated by labels. (A, Lower Left) Side 1 and side 2 surfaces of PI4KIIIα complex, colored by conservation. Regions built as polyalanine helices are indicated in gray. (A, Lower Right) Side 1 and side 2 surfaces of PI4KIIIα complex, colored by surface charge. Regions built as polyalanine helices are indicated in gray. (B) Active site structures of PI4KIIIα + A1 inhibitor and PI3Kγ + ATP (PDB ID code 1E8X) are shown after structural alignment for comparison and oriented such that the putative membrane-binding surface of PI4KIIIα lies directly below the indicated view. In this orientation, the PI-binding site of each molecule lies directly below the bound ligand in each molecule.
Alignment of the PI4KIIIα catalytic domain with the crystal structure of ATP-bound PI3Kγ identifies the active site and reveals the likely disposition of ATP within it. In our structure, density in the ATP-binding site likely corresponds to the specific PI4KIIIα inhibitor A1 (19) (Fig. 4B, Left), which was added to the sample before grid preparation. Based on the homology model of the PI4KIIIα catalytic domain, the walls of the active site in contact with the A1 compound are lined primarily with hydrophobic residues, so that the A1 binding interactions are similar to those of other lipid kinase inhibitors, such as wortmannin (20). The ATP-binding and active sites are accessible from the side 2 surface of the PI4KIIIα complex. The disposition of the kinase catalytic domains, the flatness and conservation of side 2, and its overall positive charge suggest that the complex interacts with the acidic PM via this side (Fig. 5), placing the catalytic domains in close proximity to the PI substrate, whose head groups would be properly oriented for reaction with ATP.
Fig. 5.
A model for PI4KIIIα complex recruitment to the plasma membrane. EFR3, for which the structure from Saccharomyces cerevisiae (PDB ID code 4N5A) is shown, localizes to the membrane via a basic patch at its N terminus as well as N-terminal palmitoylation (33). The PI4KIIIα complex is recruited to the plasma membrane by an interaction between TTC7/FAM126 and the EFR3 unstructured C terminus (5, 6). The PI4KIIIα/TTC7/FAM126 complex interacts with the acidic inner leaflet of the plasma membrane via a flat, basic surface, orienting its active sites optimally for reaction with phosphatidylinositol in the membrane.
Discussion
The structure reported here reveals the intricate organization of the enzyme complex with the most upstream role in the PM phosphoinositide signaling cascade. Each catalytic domain of PI4KIIIα is ∼35 kDa (residues 1,788–2,085), raising questions as to the function of the remaining ∼600 kDa of the PI4KIIIα complex, including the N-terminal portions of the kinase (residues 1–1,787) and the TTC7/FAM126 heterodimer. The structure of the complex indicates that PI4KIIIα both homodimerizes and interacts with TTC7/FAM126 by way of extensive buried surfaces. One major role of these interactions may be to conformationally constrain PI4KIIIα, allowing for formation of the complex’s membrane-interacting surface, as well as for orientation of the PI4KIIIα active sites toward this surface. Indeed, the PI4KIIIα dimerization interface and the PI4KIIIα/TTC7 interface include contacts with the catalytic and cradle domains, respectively, that may help to position the active site. Such a role would account for the stimulatory effect of TTC7/FAM126 on PI4KIIIα catalytic activity observed in vitro. Further, via its interaction with EFR3, TTC7 is critical in recruiting the PI4KIIIα complex to the PM (13), and we speculate that a conserved surface in TTC7/FAM126 lateral to the PI4KIIIα membrane interaction surface (Fig. S8) may serve as a docking site for EFR3. The contact between the C terminus of TTC7 and the N-terminal tip of the PI4KIIIα α-solenoid positions the latter, and the mutations in CID-MIA patient 4 suggest that this contact is important for PI4KIIIα activity in vivo.
Given its crucial role in phosphoinositide homeostasis and hence signaling, PI4KIIIα activity is likely to be tightly regulated by a variety of inputs. The structural complexity of the PI4KIIIα superassembly is consistent with the occurrence of multiple regulatory mechanisms. By analogy to numerous other examples of α-solenoids that function as protein–protein interaction modules, the N-terminal α-solenoid of PI4KIIIα may similarly serve as an interaction site for regulators of this complex. Elucidation of the structure of PI4KIIIα complexed with its two major accessory factors now opens the possibility of gaining fresh mechanistic insights into the regulation of phosphoinositide homeostasis at the PM.
Materials and Methods
Antibodies and Reagents.
Antibodies for immunoblots were obtained from the following commercial sources: Roche (anti–HA-HRP 3F10), AbGent (rabbit anti-FLAG), Thermo (goat anti-rabbit DyLight 800), Sigma (EFR3A Ab2; TTC7B; tubulin B-5-1-2), Novus Biologicals (FAM126B), Proteus Biosciences (GAPDH 1D4), and Cell Signaling Technology (PI4KIIIα). Antibodies to EFR3B, mouse TTC7A and TTC7B, and human FAM126A were reported previously (6, 7). Other reagents were obtained from Sigma-Aldrich, American Bioanalytical, or Fisher Scientific if not otherwise indicated.
Plasmid Construction.
Coding sequences for PI4KIIIα (full-length and truncation fragments, as described in the text), TTC7B, and FAM126A (2–289) were amplified by PCR from human cDNA. PI4KIIIα PCR products were ligated into pcDNA3.1–3×FLAG, TTC7B PCR product was ligated into pCMV6-HA, and FAM126A (2–289) PCR product was ligated into pEGFP-C1 (the latter modified to include a cleavage site for PreScission protease).
Protein Expression and Purification.
For cryo-EM sample preparation, constructs for expression of human EGFP-FAM126A(2–289), human HA-TTC7B(1–843), and 3×FLAG-PI4KIIIα(1–2102) were cotransfected into Expi293F cells according to manufacturer instructions. Expression was carried out for 72 h, after which cells were harvested by centrifugation and either frozen at −80 °C or processed immediately. Cells were solubilized by resuspension in lysis buffer (50 mM Tris⋅HCl, pH 8.0, 200 mM NaCl, 10% glycerol, 1 mM TCEP-HCl, 1× Roche Complete EDTA-free protease inhibitor mixture, 0.5% Triton X-100) and the lysate was incubated on ice for 10 min. Insoluble debris was pelleted at 17,600 × g for 40 min. Clarified lysate was applied to GFP-Trap_A beads (Chromotek) preequilibrated in lysis buffer, then incubated at 4 °C for 2 h to capture intact complex. Beads were washed with 3 × 10 bed volumes of wash buffer (lysis buffer lacking Triton X-100), then incubated in 10 bed volumes of wash buffer supplemented with 1 mM ATP and 1 mM MgCl2 for 12–16 h to remove chaperone. The ATP/Mg2+ wash was then removed and the beads were washed in 3 × 10 bed volumes of wash buffer containing 5% glycerol, then resuspended in 0.4 bed volumes of 5% glycerol wash buffer supplemented with 0.1 mg PreScission protease and incubated overnight to cleave the complex from the beads. Supernatant was carefully removed from beads and transferred to an equal volume of preequilibrated amylose beads (New England Biolabs) to bind and remove PreScission protease. The supernatant (totaling 100–200 μL) was recovered as completely as possible from the beads. Protein concentration was determined by absorbance at 280 nm. A1, a competitive inhibitor of PI4KIIIα activity (19), was added from a 1-mM stock (in DMSO) to a 2.5-fold molar excess over PI4KIIIα. A small aliquot was diluted to 1% glycerol and preassessed by negative-stain EM. Well-behaved samples were used directly for cryo-EM grid preparation.
EM Specimen Preparation and Data Collection.
The homogeneity of the purified complex was examined by negative staining with 0.7% (wt/vol) uranyl formate as described (21). A total of 58 images of negatively stained specimens were collected on a Philips CM10 electron microscope (FEI) operated at 100 kV. The images were recorded at a defocus of −1.5 μm on an XR16L-ActiveVu charge-coupled device camera (AMT) at a nominal magnification of 52,000× (calibrated pixel size of 2.62 Å on the specimen level) (Fig. S2A).
Two types of grids were used to prepare specimens for cryo-EM: C-flat 300 mesh 1.2/1.3 Cu grids (Protochips) overlaid with a thin layer of home-made carbon film (∼10-nm thickness) and Quantifoil 300 mesh 1.2/1.3 Cu grids. The grids were glow discharged for 30 s immediately before use. Freshly purified sample was centrifuged at 13,000 × g for 2 min to remove potential protein aggregates. The protein concentration of the supernatant was measured with a NanoDrop spectrophotometer (Thermo Fisher Scientific) and adjusted to 0.25 mg/mL. Specimens were frozen with a Vitrobot Mark VI (FEI) set at 4 °C and 90% humidity. A 4-μL sample aliquot (containing 5% glycerol) was applied to a glow-discharged grid. After 10 s, grids were blotted for 2 s with a blot force of −2 and plunged into liquid ethane cooled to liquid-nitrogen temperature.
First, a small dataset of 569 image stacks was collected with a 200 kV Talos Arctica electron microscope (FEI) at a nominal magnification of 28,000× (calibrated pixel size of 1.5 Å on the specimen level) using a sample prepared with a carbon-covered C-flat 1.2/1.3 Cu grid (Fig. S3A). A larger dataset of 3,280 image stacks was then collected with a 300 keV Titan Krios electron microscope (FEI) at a nominal magnification of 22,500× (calibrated pixel size of 1.3 Å on the specimen level) using specimens prepared on Quantifoil 1.2/1.3 holy carbon grids (Fig. S4A).
All of the images were recorded with a K2 Summit direct detector camera (Gatan) in superresolution counting mode. Exposures of 10 s were dose fractionated into 40 frames (250-ms exposure per frame), with a dose rate of 8 electrons/pixel/s (∼1.18 electrons/Å2/frame) and a total dose of 47 electrons/Å2. Images were collected using a defocus range from −2 μm to −3.5 μm. Data were acquired automatically using SerialEM (22).
Image Processing.
For the negative-stain EM dataset, 18,473 particles were picked with EMAN2 (23) and windowed into 156 × 156 pixel boxes. The particle images were reduced to 64 × 64 pixels, centered, and subjected to classification with the iterative stable alignment and clustering (ISAC) algorithm (24), specifying 100 images per group and a pixel error threshold of 0.7. After 10 generations, 257 averages were obtained (Fig. S2B) and used to calculate an initial model with the validation of individual parameter reproducibility (VIPER) algorithm implemented in SPARX (25) (Fig. S2C).
For the cryo-EM datasets, the dose-fractionated image stacks collected in superresolution counting mode were motion corrected, dose weighted, and binned twice in Motioncorr2 (26). The CTF was determined and corrected with Gctf (27).
For the Arctica dataset, ∼6,000 particles were automatically and template-free picked with Gautomatch (www.mrc-lmb.cam.ac.uk/kzhang/Gautomatch/) and subjected to 2D classification in Relion 1.4 (28). Four of the class averages were selected as templates for automated picking in Gautomatch, and 138,285 particles from 569 micrographs were windowed into 280 × 280 pixel images. All subsequent image processing was carried out in Relion 1.4 unless noted otherwise. After 2D classification (Fig. S3B) and removal of classes creating poor averages, the remaining 83,705 particles were subjected to 3D classification into four classes with C1 symmetry (Fig. S3C). The density map obtained with VIPER from the negative-stain EM dataset was used as the reference map. The resulting map with the most structural detail showed clear C2 symmetry. This map, containing 46,122 particles, was refined with C2 symmetry and, after postprocessing, reached a resolution of 6.4 Å using gold-standard Fourier shell correlation (FSC) and a cutoff of 0.143 (Fig. S3C).
For the Krios dataset, 508,504 particles were automatically picked from 3,280 micrographs with Gautomatch using the same templates used for the Arctica dataset but scaled and windowed into 320 × 320 pixel images. After 2D classification, classes yielding poor averages were removed. Since 2D classification revealed both dimer and monomer classes (Fig. S4B), the remaining 361,684 particles were subjected to supervised 3D classification. The full density map and half of the density map obtained with the Arctica dataset were used as references for the dimer and monomer (Fig. S4C). The 119,673 particles assigned to the dimer class were further classified into four classes by unsupervised 3D classification. The two classes that showed the best fine structure detail were combined (64,104 particles) and the resulting map after refinement and postprocessing, imposing C2 symmetry, reached a resolution of 3.6 Å (Figs. S4C and S5A). Local resolution variations were estimated from two half data maps using Relion. Further processing of the monomer class revealed that the particles adopted preferred orientations, resulting in a poor density map (Fig. S4C).
Model Building and Refinement.
Coordinates from Protein Data Bank (PDB) ID code 5DSE, chains C and D (TTC7B/FAM126A) were extracted and rigid-body fitted into congruent density in the final cryo-EM map using University of California San Francisco (UCSF) Chimera (29). Structures were flexibly fitted to the map using phenix.real_space_refine from the Phenix software suite (30). The PI4KIIIα cradle and catalytic domains (residues 1,598–2,102) were modeled using I-TASSER with PDB ID code 1E7U as template (similar to ref. 20). The resulting top hit homology model was rigid-body fitted into the map using UCSF Chimera then locally fitted to the density by hand using Coot (31). A model of A1 PI4KIIIα inhibitor was constructed in Coot, with restraints generated using Elbow (32) as implemented in Phenix. The remaining N-terminal portion of PI4KIIIα (residues 1–1,597) was hand built in Coot by first building helices into strong helical density. A single short sequence anchor was registered using the “Dock Sequence” extension in Coot against these primitive helices in the map. The remaining sequence and connectivity (spanning residues 932–1,597) were identified and built into the density manually using Coot and refined iteratively. The model for this portion was merged with the catalytic domain and the model was refined against Relion half-map 1 using phenix.real_space_refine. FSC curves were calculated between the refined model and (i) half map 1, which was used in refinement; (ii) half map 2, which was not used in refinement; and (iii) the combined map (Fig. S5B). The portion of the map corresponding to PI4KIIIα residues 1–931, the α-solenoid horn, was not at sufficient resolution to trace, and is currently modeled as polyalanine helices.
PI4KIIIα Truncation Immunoprecipitations.
The 3×FLAG-tagged PI4KIIIα truncation constructs were coexpressed with EGFP-FAM126A(2–289) and HA-TTC7B(1–843) in Expi293 cells. Cells were lysed as for cryo-EM sample preparation and clarified lysate was applied to anti-FLAG M2 Affinity Beads (Sigma) and incubated for 2 h at 4 °C. After washing, beads were incubated overnight in wash buffer with 1 mM each ATP and MgCl2 to remove chaperone, then resuspended in 1 bed volume of wash buffer. Bead slurry samples were resolved by SDS/PAGE and bands were wet transferred to a nitrocellulose membrane, which was immunoblotted separately with rat anti-HA HRP and rabbit anti-FLAG (with goat anti-rabbit DyLight 800 secondary) antibodies. HA immunoblot was developed with Pierce enhanced chemiluminescence substrate and imaged using a GE ImageQuant LAS 4000. FLAG immunoblot was imaged using a LI-COR Odyssey imager.
Cell Lines and Cell Culture.
Patient and control cells were obtained upon informed consent using institutional review board-approved protocols in use at Boston Children’s Hospital (protocol 04-09-113R), and stored at the National Institutes of Health (protocol 16-I-N139). Following ethical guidelines, samples were distributed for analysis and storage with written informed consent. Patient TTC7A mutations are as follows: patient 1 (c.1000delAAGT; c.A1817G), patient 2 (c.1190delCA; c.1190delCA), patient 3 (c.315_318delTCTA; c.2355+5G > A), and patient 4 (c.2170+3G > C; c2839+1insC). Some of these mutations have been previously described (13). Fibroblasts were cultured as described previously (6). B cells were grown at 37 °C at 5% CO2 in culture medium [Roswell Park Memorial Institute medium (RPMI)-1640 with 10% FBS, MEM-NEAA, l-glutamine, 0.1 mM 2-mercaptoethanol, and penicillin/streptomycin].
Immunoblotting of Patient Samples.
Patient and control cells were harvested and resuspended in lysis buffer: 150 mM NaCl, 20 mM Tris at pH 7.4, 1 mM EDTA, 1% Triton X-100, supplemented with protease inhibitors [cOmplete, EDTA-free (Roche)]. Organs from fsn mice (17, 18) were homogenized in lysis buffer plus benzonase. Lysates and homogenates were then sonicated briefly and centrifuged for 10 min at 16,000 × g. The supernatant was collected and analyzed by SDS/PAGE and Western blot with chemiluminescence.
Quantitative RT-PCR.
Total RNA from control human fibroblast and B cell lines (described above) was isolated using the RNeasy Plus Mini kit (Qiagen). Yield and purity were determined by absorbance at 230 nm, 260 nm, and 280 nm. RNA integrity was monitored by electrophoresis on a bleach-treated agarose gel (33). Equal amounts (700 ng each) of total RNA were used for cDNA synthesis using the iScript cDNA Synthesis kit (Bio-Rad). Quantitative PCR reactions were performed in triplicate using the iTaq Universal SYBR Green Supermix (Bio-Rad) with the corresponding validated PrimePCR intron-spanning primer pairs for detection of human TTC7A and TTC7B, with primers against S26 ribosomal protein cDNA as a control.
Supplementary Material
Acknowledgments
We thank M. Ebrahim and J. Sotiris (Evelyn Gruss Lipper Cryo-Electron Microscopy Resource Center, The Rockefeller University) for assistance in data collection; Drs. S. Pelsue and J. Walker (University of Southern Maine) for the gift of tissue samples from flaky skin (fsn) mutant mice and corresponding controls; M. Messa (Yale University) for assistance with RT-PCR experiments; T. Balla (National Institute of Child Health and Human Development) for the gift of the A1 inhibitor; and Brynn Wainstein, John Ziegler, Paul Gray (Sydney Children’s Hospital), and Dr. Waleed Al-Herz (Kuwait University) for the generous gift of fibroblast cell lines from TTC7A-mutated patients. J.M.B. was supported by NIH Award GM110121. L.D.N. and K.D. were partially supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, NIH. P.D.C. was supported in part by the NIH (Awards NS036251 and DK082700) and the Kavli Foundation. T.W. and K.M.R. were supported by NIH Award GM114068. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does the mention of trade names, commercial products, or organizations imply endorsement by the U.S. government.
Footnotes
The authors declare no conflict of interest.
Data deposition: The cryo-EM maps have been deposited to EMDataBank, www.emdatabank.org/ (accession no. EMD-7129). The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 6BQ1).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1718471115/-/DCSupplemental.
References
- 1.Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 2.Balla T. Phosphoinositides: Tiny lipids with giant impact on cell regulation. Physiol Rev. 2013;93:1019–1137. doi: 10.1152/physrev.00028.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tan J, Brill JA. Cinderella story: PI4P goes from precursor to key signaling molecule. Crit Rev Biochem Mol Biol. 2014;49:33–58. doi: 10.3109/10409238.2013.853024. [DOI] [PubMed] [Google Scholar]
- 4.Baird D, Stefan C, Audhya A, Weys S, Emr SD. Assembly of the PtdIns 4-kinase Stt4 complex at the plasma membrane requires Ypp1 and Efr3. J Cell Biol. 2008;183:1061–1074. doi: 10.1083/jcb.200804003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu X, et al. Structural insights into assembly and regulation of the plasma membrane phosphatidylinositol 4-kinase complex. Dev Cell. 2014;28:19–29. doi: 10.1016/j.devcel.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baskin JM, et al. The leukodystrophy protein FAM126A (hyccin) regulates PtdIns(4)P synthesis at the plasma membrane. Nat Cell Biol. 2016;18:132–138. doi: 10.1038/ncb3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakatsu F, et al. PtdIns4P synthesis by PI4KIIIα at the plasma membrane and its impact on plasma membrane identity. J Cell Biol. 2012;199:1003–1016. doi: 10.1083/jcb.201206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balla A, Balla T. Phosphatidylinositol 4-kinases: Old enzymes with emerging functions. Trends Cell Biol. 2006;16:351–361. doi: 10.1016/j.tcb.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 9.Nakagawa T, Goto K, Kondo H. Cloning, expression, and localization of 230-kDa phosphatidylinositol 4-kinase. J Biol Chem. 1996;271:12088–12094. doi: 10.1074/jbc.271.20.12088. [DOI] [PubMed] [Google Scholar]
- 10.Audhya A, Foti M, Emr SD. Distinct roles for the yeast phosphatidylinositol 4-kinases, Stt4p and Pik1p, in secretion, cell growth, and organelle membrane dynamics. Mol Biol Cell. 2000;11:2673–2689. doi: 10.1091/mbc.11.8.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baretić D, Williams RL. PIKKsThe solenoid nest where partners and kinases meet. Curr Opin Struct Biol. 2014;29:134–142. doi: 10.1016/j.sbi.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 12.Aylett CHS, et al. Architecture of human mTOR complex 1. Science. 2016;351:48–52. doi: 10.1126/science.aaa3870. [DOI] [PubMed] [Google Scholar]
- 13.Chen R, et al. Whole-exome sequencing identifies tetratricopeptide repeat domain 7A (TTC7A) mutations for combined immunodeficiency with intestinal atresias. J Allergy Clin Immunol. 2013;132:656–664.e17. doi: 10.1016/j.jaci.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ngan B, et al. Mutations in tetratricopeptide repeat domain 7A (TTC7A) are associated with combined immunodeficiency with dendriform lung ossification but no intestinal atresia. LymphoSign J. 2014;01:10–26. [Google Scholar]
- 15.Yang W, et al. Compound heterozygous mutations in TTC7A cause familial multiple intestinal atresias and severe combined immunodeficiency. Clin Genet. 2015;88:542–549. doi: 10.1111/cge.12553. [DOI] [PubMed] [Google Scholar]
- 16.Samuels ME, et al. Exome sequencing identifies mutations in the gene TTC7A in French-Canadian cases with hereditary multiple intestinal atresia. J Med Genet. 2013;50:324–329. doi: 10.1136/jmedgenet-2012-101483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Helms C, et al. The Tetratricopeptide repeat domain 7 gene is mutated in flaky skin mice: A model for psoriasis, autoimmunity, and anemia. Exp Biol Med (Maywood) 2005;230:659–667. doi: 10.1177/153537020523000908. [DOI] [PubMed] [Google Scholar]
- 18.White RA, et al. Positional cloning of the Ttc7 gene required for normal iron homeostasis and mutated in hea and fsn anemia mice. Genomics. 2005;85:330–337. doi: 10.1016/j.ygeno.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 19.Bojjireddy N, et al. Pharmacological and genetic targeting of the PI4KA enzyme reveals its important role in maintaining plasma membrane phosphatidylinositol 4-phosphate and phosphatidylinositol 4,5-bisphosphate levels. J Biol Chem. 2014;289:6120–6132. doi: 10.1074/jbc.M113.531426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Balla A, et al. Design of drug-resistant alleles of type-III phosphatidylinositol 4-kinases using mutagenesis and molecular modeling. Biochemistry. 2008;47:1599–1607. doi: 10.1021/bi7017927. [DOI] [PubMed] [Google Scholar]
- 21.Ohi M, Li Y, Cheng Y, Walz T. Negative staining and image classification: Powerful tools in modern electron microscopy. Biol Proced Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 23.Tang G, et al. EMAN2: An extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 24.Yang Z, Fang J, Chittuluru J, Asturias FJ, Penczek PA. Iterative stable alignment and clustering of 2D transmission electron microscope images. Structure. 2012;20:237–247. doi: 10.1016/j.str.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hohn M, et al. SPARX, a new environment for Cryo-EM image processing. J Struct Biol. 2007;157:47–55. doi: 10.1016/j.jsb.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 26.Zheng SQ, et al. MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang K. Gctf: Real-time CTF determination and correction. J Struct Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scheres SHW. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pettersen EF, et al. UCSF Chimera: A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 30.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moriarty NW, Grosse-Kunstleve RW, Adams PD. electronic Ligand Builder and Optimization Workbench (eLBOW): A tool for ligand coordinate and restraint generation. Acta Crystallogr D Biol Crystallogr. 2009;65:1074–1080. doi: 10.1107/S0907444909029436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aranda PS, LaJoie DM, Jorcyk CL. Bleach gel: A simple agarose gel for analyzing RNA quality. Electrophoresis. 2012;33:366–369. doi: 10.1002/elps.201100335. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.