Significance
The rate of protein synthesis along a messenger RNA is not uniform and can influence the folding of the nascent protein. Using multichannel single-molecule fluorescence microscopy, we simultaneously monitored compositions and conformations of prokaryotic translational complex over translating multiple codons. We show that the dissociation of deacylated tRNA is necessary for conformational changes prior to translocating to the next codon. The deacylated tRNA dissociation kinetics strongly depended on the identity of tRNA, the ionic strength of the solution, and the reaction temperature. These findings suggest that in addition to the tRNA association kinetics, tRNA dissociation kinetics can also alter the dynamics of protein synthesis and may be used as a sensor for different cellular conditions to regulate cotranslational processes.
Keywords: ribosome, protein synthesis, single-molecule FRET, nonfluorescent quencher, E site
Abstract
During protein synthesis, the ribosome simultaneously binds up to three different transfer RNA (tRNA) molecules. Among the three tRNA binding sites, the regulatory role of the exit (E) site, where deacylated tRNA spontaneously dissociates from the translational complex, has remained elusive. Here we use two donor–quencher pairs to observe and correlate both the conformation of ribosomes and tRNAs as well as tRNA occupancy. Our results reveal a partially rotated state of the ribosome wherein all three tRNA sites are occupied during translation elongation. The appearance and lifetime of this state depend on the E-site tRNA dissociation kinetics, which may vary among tRNA species and depends on temperature and ionic strength. The 3-tRNA partially rotated state is not a proper substrate for elongation factor G (EF-G), thus inhibiting translocation until the E-site tRNA dissociates. Our result presents two parallel kinetic pathways during translation elongation, underscoring the ability of E-site codons to modulate the dynamics of protein synthesis.
Translation is carried out by the ribosome, a massive macromolecular machine that coordinates factor compositions and biochemical reactions through its conformation. During translation, each amino acid matching a codon is sequentially added to the nascent peptide through one cycle of elongation. Each elongation cycle can be further divided into two phases. First, during decoding, the cognate aminoacyl-tRNA (aa-tRNA) is selected from the pool of aa-tRNAs to elongate the nascent peptide. Next, during translocation, the translational complex moves to the next codon for decoding. Throughout these processes, the conformation of the ribosome cycles between two major intersubunit conformations, the nonrotated and rotated intersubunit states (also referred to as the classical and hybrid tRNA states, respectively). These states choreograph ligand bindings and dissociations at each elongation phase, facilitating biochemical reactions necessary for translation. Although other global intersubunit conformations with different degrees of intersubunit rotation have been observed during translation initiation (1, 2) and during elongation in the presence of antibiotics (3), their importance during elongation remains unclear.
A detailed model of translation elongation has been elucidated by prior structural, biochemical, and single-molecule studies (4–7). Following a cycle of elongation from the start of decoding, a cognate aminoacyl-tRNA binds to the exposed mRNA codon in the aminoacyl (A) site of the nonrotated state ribosome. After successful accommodation of the aminoacylated 3′ end of the tRNA into the peptidyl-transferase center, the nascent peptide on the tRNA bound to the peptidyl (P) site of the ribosome is transferred to the A-site tRNA. This chemical step alters the energy landscape of the translational complex to allow a 5–7° rotation of the ribosomal subunits with respect to each other to enter the rotated state (8–12). Coupled with the intersubunit rotation, the 3′ ends of two tRNAs in the large ribosomal subunit move from the A and P sites to the P and exit (E) sites, forming A/P and P/E hybrid tRNA binding states. The resulting rotated hybrid state is a proper substrate for elongation factor G (EF-G), which binds near the A site and hydrolyzes GTP to catalyze translocation of the ribosome to the next codon (13). A successful translocation event moves the A-site and P-site tRNAs along with the mRNA codons in the small ribosomal subunit to the P and E sites, resetting the intersubunit conformation of the ribosome back to the nonrotated state. The intersubunit rotation during translocation is coordinated with movements of the L1 stalk, which makes a direct contact with deacylated tRNA to usher it to the E site (12, 14, 15). EF-G (16, 17) and the E-site tRNA (12, 18) then rapidly dissociate in the posttranslocation state.
During elongation, the mechanism and role of E-site tRNA dissociation have been unclear. The occupancy of the E site has been suggested to enhance accuracy of decoding allosterically, possibly through coordinated E-site deacylated tRNA dissociation and the A-site aa-tRNA binding (19–21). Biochemical studies, however, have failed to observe such enhancement of the proofreading efficiency by the presence of the E-site tRNA (18, 22). Further, the allosteric E-site tRNA dissociation mechanism itself has been tested by two key single-molecule studies. First, Uemura et al. (23) observed multiple fluorescently-labeled tRNAs as they bound to an immobilized translational complex. Second, Chen et al. (24) used a clever single-molecule Förster resonance energy transfer (smFRET) assay in an alternating wavelength laser excitation (ALEX) microscope to follow all three tRNAs binding to the translational complex. By monitoring tRNA compositions, both studies revealed spontaneous dissociations of the E-site tRNA during active translation elongation, uncoupled with the A-site occupancy. However, the presence of the E-site tRNA affects the conformational dynamics of the L1 stalk (14), which may correlate with the global ribosomal conformational changes (12, 15). Thus, elucidating the role of the E-site tRNA in modulating ribosomal conformation and elongation rates is essential to our understanding of translation. The presence of E-site tRNA must be signaled to the aa-tRNA or EF-G binding sites, likely via modulating ribosomal conformation, to affect decoding or translocation dynamics, respectively. The conformation of the ribosome is dynamically modulated by both tRNA occupancy and their aminoacylation status (14, 15, 25). As such, simultaneously observing conformational changes of the ribosome and tRNA compositions within the translational complex is a necessary tool to determine the role of the E site.
Here we utilized two donor–quencher pairs to observe two smFRET signals simultaneously, correlating both conformational and compositional changes of a translating ribosome. Our approach leverages the intrinsic redundancy in FRET signals by replacing an acceptor dye with a nonfluorescent quencher that has matching optical properties (26–30), which enabled us to measure multiple smFRET signals simultaneously using common single-molecule optics and photophysically stable fluorophores. This labeling strategy can be readily applied in a standard microscope setup with two optical channels used for smFRET experiments to correlate multiple smFRET signals, without any instrument modifications needed for the existing methods (31–33). Using this technique, we monitored the simultaneous presence of up to three tRNAs within the translational complex, to which we correlated intersubunit conformations during multiple cycles of translation elongation. This revealed a partially rotated conformation of the ribosome in the presence of three tRNAs, which cannot translocate until the E-site tRNA dissociates spontaneously. The E-site tRNA dissociation kinetics depends on multiple factors, such as tRNA identity, temperature, and the buffer ionic strength. Taken together, our data demonstrate that the E-site tRNAs slow protein synthesis, in a manner that is sensitive to the cellular condition as well as the E-site codon sequence.
Results
To correlate two smFRET signals directly, we selected two donor dyes, Cy3 and Cy5, that have the desired photostabilities and quantum yields and chose two quencher dyes, QSY9 and QSY21, as the respective energy transfer quenchers based on the matching of optical spectra (Fig. S1). Using a donor–quencher pair in place of a donor–acceptor pair, changes in FRET efficiency, and underlying structural changes, are detected by the intensity of the donor emission alone encoded in a single fluorescence channel. Although such compression strips a redundancy in the FRET signal and leaves it vulnerable to errors from photophysical artifacts, correlating multiple previously characterized FRET signals strengthens the validity of observed signals and enables probing molecular processes from multiple perspectives. Another restriction arising from the use of donor–quencher pairs is the possibility of cross-talk (27). This is addressed by separating two donor–quencher pairs farther than the dynamic range of FRET (typically 20–80 ). The large size of the ribosome mitigates this issue in our case because the two smFRET labeling pairs within translation complexes are sufficiently distant (>110 ) to minimize interactions between the two donor–quencher pairs. Although the Förster radius (R0) values for Cy3/Cy5 (55.8 ) (34) and Cy3/QSY9 (53 ± 13 ) (27) pairs have been previously reported to calculate the dynamic range of FRET, an R0 value for the Cy5/QSY21 pair has never been reported. We utilized double-stranded DNA as a molecular spacer (27–29) to measure an approximate R0 value for the Cy5/QSY21 pair (49 ± 9 ) (Fig. S2), which is similar to other conventional FRET pairs. These R0 values are sufficient to report on structural changes occurring within the ribosome (Table S1).
Using two characterized donor–quencher pairs, we simultaneously monitored ribosomal intersubunit conformational changes and tRNA–tRNA conformational and compositional changes. To monitor the ribosomal intersubunit rotation, we placed a Cy5 and QSY21 donor–quencher on the small and large ribosomal subunits, respectively. Helix 44 of the small ribosomal subunit (Cy5-30S) and helix 101 of large ribosomal subunits (QSY21-50S) were labeled by annealing dye-labeled DNA oligonucleotides to the mutated ribosomal RNA (35, 36). To monitor the composition and the conformation of tRNAs, we labeled Lys-tRNALys and Phe-tRNAPhe with Cy3 [Lys-(Cy3)-tRNALys] and QSY9 [Phe-(QSY9)-tRNAPhe] at acp3U47 of both tRNAs (37). Our choices of ribosomal labeling sites are known to be insensitive to spontaneous conformational changes (such as L1 stalk fluctuations) and have been used to track productive conformational changes related to decoding and translocation (9, 36). Dye-labeling of tRNA and ribosomes at these sites is well tolerated; it has little effect on the kinetics and dynamics of translation elongation compared with other in vitro assays (9, 37, 38).
Our in vitro translation experiments were prepared by forming a 30S preinitiation complex (30S PIC), using a 5′-biotinylated mRNA, the initiator tRNA (fMet-tRNAfMet), initiation factor 2 (IF2), and Cy5-30S (36). To maximize the number of FRET events between labeled tRNAs per translating molecule, the coding region of the mRNA sequence contained six repeats of phenylalanine–lysine codons followed by a stop codon (Fig. 1C). High concentrations of fluorescently labeled tRNA in solution are required to observe multiple translation–elongation cycles with our labeling scheme. Although this is difficult using conventional total internal reflectance microscopy (TIRFM), which requires lower concentrations of fluorescent ligands in solution, our use of a zero-mode waveguide (ZMW)-based microscope allowed single-fluorophore detection at higher solution background concentrations than in TIRFM by illuminating a smaller volume close to the surface (23, 39). After 30S PIC was surface-tethered within ZMWs, we delivered the required factors to promote translation elongation, which included Lys-(Cy3)-tRNALys ternary complex (TC), Phe-(QSY9)-tRNAPhe TC, QSY21-50S, and EF-G (Fig. 1A), along with GTP and other photostabilizing agents (39). Before delivery, dye-labeled aminoacyl-tRNA TCs were formed with the elongation factor-Tu (EF-Tu) and GTP, in the presence of the elongation factor-Ts (EF-Ts) and a nucleotide regeneration system for GTP. Using the ZMW-based instrument, processive translation elongation of up to 12 codons was observed, driven by a high concentration of Cy3-labeled tRNA (100–150 nM), which resulted in an elongation rate of one codon per 3 s (Fig. S3).
Fig. 1.
Observing the translation–elongation cycle using two donor–quencher pairs. (A) Schematic of the experiment. QSY9 and QSY21 are indicated by black circles with green and red outlines, respectively; Cy3 and Cy5 are indicated by green and red stars, respectively; and EF-G is indicated in dark purple. (B) (Top) Translation–elongation cycle over one codon in the mRNA: state i is nonrotated state with one QSY9-labeled tRNA in the P site, state ii is rotated state after accommodation of the Cy3-labeled tRNA, and state ii′ is nonrotated state after translocation with two tRNAs bound. (Bottom) Expected sequence of Cy3/Cy5 signals in each state. (C) The coding region of mRNA construct used in this study. (D) (Left) Representative experimental trace, with state marked as in B. (Right) Postsynchronized plots before and after translocation (marked by a black vertical line) (n = 303). (E) (Left) Representative experimental trace using unlabeled Phe-tRNAPhe, with state marked as in B. (Right) Postsynchronized plots before and after translocation (marked by a black vertical line) (n = 176). Each Cy3 and Cy5 fluorescent intensity has been normalized to the respective unquenched level in postsynchronized plots. White vertical lines have been added to help visualization.
Upon subunit and factor delivery, we observed an expected close correlation between the composition and the conformation of the complex during multiple repeating cycles of elongation (Fig. 1 B and D and Fig. S4). Following one cycle, we observed that binding of Lys-(Cy3)-tRNALys to the A site and peptide bond formation changes the conformation of the ribosome from the nonrotated state (high-quenched state with low Cy5 intensity; state i in Fig. 1 B and D) to the rotated state (low-quenched state with high Cy5 intensity; state ii in Fig. 1 B and D). Transition from the rotated state back to the nonrotated state upon translocation is then followed by E-site tRNA dissociation (state ii′ in Fig. 1 B and D), marked by an increase of the Cy3 signal due to the departure of the quencher-labeled (QSY9)-tRNAPhe (state i in Fig. 1 B and D).
Whereas the donor–quencher smFRET signal for intersubunit conformation has been validated previously (28), we confirmed here the proper behavior of the donor–quencher pair for the tRNA–tRNA FRET signal. We performed control experiments where unlabeled Phe-tRNAPhe was substituted for Phe-(QSY9)-tRNAPhe (Fig. 1E) and observed a slight change of Cy3 fluorescence from Lys-(Cy3)-tRNALys as it transits across tRNA binding sites, which matches previous observations of dye intensity changes in different ribosomal sites (40). As expected, changes of the Cy3 fluorescence intensity from Lys-(Cy3)-tRNALys without the presence of QSY9-labeled tRNA are distinct from changes observed with the quencher present. We further tested our tRNA–tRNA FRET signal over the first elongation cycle, an experimental format consistent with previous studies (25, 37) (Fig. S5). In this experiment, we observed rare, correlated ribosome and tRNA–tRNA conformational changes (Fig. S6). Based on changes of the two FRET efficiencies, this transition is likely to be the intermediate tRNA state, where deacylated tRNA occupies the hybrid P/E site, whereas aa-tRNA occupies the classical A site (wherein a decrease in FRET efficiency is due to an increase in Cy3-QSY9 distance or an increase in Cy3 intensity). This infrequent tRNA movement (observed at least once in 18% of translating molecules, with a mean lifetime of 0.5 s) is coupled to a moderate back-rotation of subunits, similar to conformational changes reported in cryo-EM structures of back-translocating ribosomes (41). Although the cause of this unusual tRNA movement and its relevance during elongation is unclear, we suspect that this may be an artifact that occurs only during the first elongation cycle due to a short nascent dipeptide (42) or incomplete formylation of the initiator fMet amino acid (25).
Comparing our results from two experiments on actively elongating complexes, we noticed a subtle delay between translocation of the ribosome and E-site tRNA dissociation (Fig. 1D). We did not observe this tRNA state after translocation in our control experiment without a quencher labeled tRNA (Fig. 1E). Thus, the decreased in Cy3 intensity is caused by QSY9 quenching, rather than fluorescence intensity changes due to the local tRNA environment. This posttranslocation and E-site occupied state corresponds to the posttranslocation structure of the ribosome by the tRNA and ribosome conformation (43), where the P- and E-site tRNAs are placed closer together after translocation (higher quenching of Cy3 intensity), and the ribosome conformation transitions back to the nonrotated state (higher quenching of Cy5 intensity).
The lifetime of the E-site occupied posttranslocation state was substantially longer with (QSY9)-tRNAPhe than with (Cy3)-tRNALys in the E site (state ii′ in Fig. 1D). This suggested that the deacylated tRNA dissociation kinetics from the E site could be tRNA species dependent and is consistent with previous observations using unlabeled tRNA (15). We compared E-site dissociation kinetics among different tRNA species by measuring the time between translocation, marked by the ribosomal conformation transition and Cy3-labeled E-site tRNA departure in each experiment, without the use of quencher-labeled tRNA. For a quantitative comparison, we used Cy3-labeled tRNAs to avoid possible artifacts that may arise when using different fluorescence dyes that could affect E-site tRNA dissociation kinetics. At physiological Mg2+ ion concentrations, only one (tRNAPhe) out of the three tRNA species tested (tRNAPhe, tRNALys, and tRNAfMet) showed an E-site tRNA dissociation time substantially longer than the time resolution of the experiment (100 ms) (Table 1). Although high concentrations of Mg2+ and polyamines are known to stabilize tRNA in the E site and promote the 3-tRNA occupied state (18, 21, 24), we observed tRNA-specific sensitivities to increasing Mg2+ concentration. At buffer conditions supplemented with an additional 10 mM Mg2+ ion, the E-site residency time for tRNAPhe increased drastically (a lifetime of 10.3 ± 2.3 s, n = 129), whereas the other tRNAs showed modest (a lifetime of 1.8 ± 0.5 s for tRNALys, n = 169) or minor (a lifetime of 0.4 ± 0.1 s for tRNAfMet, n = 129) increases. The source of such correlation between tRNA dissociation kinetics and tRNA identity is likely due to different molecular interactions existing within the E site (15). Further, the exceptional stability of tRNAPhe in the E site at high Mg2+ concentration (15 mM total Mg2+) matches with previous accounts, where its residence lifetime has been estimated to be greater than 22 s (14). These observations suggest that the lifetime of the 3-tRNA occupied state may be determined by the identity of both A-site and E-site tRNAs and their interaction with the translation complex in general.
Table 1.
E-site tRNA dissociation kinetics for different tRNA species at 20 °C
| E-site tRNA | Mg2+ concentration, mM | E-site tRNA dissociation, s | n |
| Cy3-tRNAPhe | 3.5 | 0.1 ± 0.1 | 272 |
| Cy3-tRNAPhe | 5 | 0.4 ± 0.1 | 143 |
| Cy3-tRNAPhe | 10 | 6.8 ± 1.0 | 212 |
| Cy3-tRNAPhe | 15 | 10.3 ± 2.3 | 129 |
| Cy3-tRNALys | 5 | 0.1 ± 0.1 | 166 |
| Cy3-tRNALys | 15 | 1.8 ± 0.5 | 169 |
| Cy3-tRNAfMet | 5 | 0.1 ± 0.1 | 103 |
| Cy3-tRNAfMet | 15 | 0.4 ± 0.1 | 129 |
To obtain estimates of activation energies involved in the E-site tRNA dissociation, we additionally measured the E-site tRNAPhe residence time over a temperature range from 20 to 35 °C (at 10 mM Mg2+) using the temperature control ability of our instrument (Fig. S7). At 20 °C, tRNAPhe exhibited a slow dissociation rate of 7.6 ± 1.0 s (n = 212). The E-site tRNA residence time precipitously decreased as temperature was increased by 5 °C steps, down to 0.4 ± 0.1 s at 35 °C (n = 158). From these data, we calculated the activation entropy (T∆S‡ = 70 ± 60 kJ⋅mol−1 at 20 °C or 293 K) and the activation enthalpy (∆H‡ = 140 ± 60 kJ⋅mol−1) of E-site tRNA dissociation using Arrhenius plots and the Eyring equation, resulting in a ∆G‡ of 70 ± 80 kJ⋅mol−1 via the Gibbs–Helmholtz equation. Although the effect of temperature on E-site tRNA dissociation kinetics exhibited a clear trend, our fitting introduced a large margin of error in calculating its thermodynamic parameters. However, considering that ∆Gdiss of a single base pair within an eight-base-pair–long RNA duplex ranges from 10 to 40 kJ⋅mol−1 at high salt concentrations (44, 45), our estimates are consistent with the expected enthalpic and entropic barriers of a three-base-pair dissociation from the E site.
The presence of the third tRNA at the E site could potentially inhibit translation elongation because the dissociation of deacylated tRNA is necessary for the ribosome to translocate to the next codon. To test this hypothesis, we repeated our elongation experiments using two donor–quencher pairs in buffer containing 15 mM Mg2+ to further stabilize tRNA in the E site. Our two donor–quencher pair method allowed us to observe up to three tRNAs bound to the ribosome via the Cy3/QSY9 signal due to the alternating binding of labeled tRNAs, while simultaneously monitoring the conformation of the ribosome using the Cy5/QSY21 signal (Fig. 2A). As expected, we observed a prolonged 3-tRNA bound state (40 ± 10 s, n = 94), which could not translocate while a tRNA occupied the E site (Fig. 2B). The 3-tRNA bound state was mainly seen while tRNAPhe resided in the E site, where its E-site dissociation lifetimes were much longer than that of tRNALys and tRNAfMet. The 3-tRNA bound state exhibited a markedly different ribosome conformation than both rotated and nonrotated states; we detected an intermediate quenching between Cy5 and QSY21 compared with those of the rotated and nonrotated states. This intermediate signal suggests the existence of a partial rotation of the ribosomal subunits relative to each other, and we term this conformation the “partially rotated state” (Fig. 2B). At 5 mM Mg2+, and higher tRNA and EF-G concentrations, we still observed the partially rotated state, suggesting that the state persists in more physiologically relevant conditions (Fig. S8).
Fig. 2.
Partially rotated states observed using different labeling schemes. (A) Translation–elongation cycle via 3-tRNA state (marked as iii). (B–D) Data from experiments using different dye labeling schemes. For each panel: (Left) dye labeling schemes for the expected 3-tRNA state, (Right) a representative experimental trace showing a partial rotation of the ribosome on state iii, and (Bottom) postsynchronized plots for the start and the end of the 3-tRNA state iii. (B) Cy5/QSY21 pair on the ribosome and Cy3/QSY9 pair on tRNA (n = 94). NR, PR, and R denote nonrotated, partially rotated, and rotated states, respectively. (C) Cy5/QSY21 pair on the ribosome and Cy3 on tRNA (n = 66). (D) Cy3B/QSY7 pair on the ribosome and Cy5 on tRNA (n = 215).
To test whether the appearance of the partially rotated state is due to the presence of the bulky quencher moieties on the tRNA, we performed the same experiments by substituting Phe-(QSY9)-tRNAPhe and Lys-(Cy3)-tRNALys with Phe-(Cy3)-tRNAPhe and Lys-tRNALys, and preserving the labeling of the ribosome (Fig. 2C). With this labeling scheme, we also identified the partially rotated state of the ribosomal subunits when it is simultaneously bound by three tRNAs. Although this approach did not track the presence of a P-site tRNA, we inferred its presence by monitoring A-site and E-site tRNA bound states. To test whether the change in intersubunit fluorescence intensity arises from potential FRET from the tRNA to the labeled ribosomal subunits, we employed the Cy3B/QSY7 pair (with optical spectra nearly identical to Cy3/QSY9 pair) to monitor intersubunit conformations and substituted Phe-(Cy3)-tRNAPhe with Phe-(Cy5)-tRNAPhe. We again observed the partially rotated state when three tRNAs are bound to the ribosome (Fig. 2D). Together, the results from these donor–quencher pair permutation experiments support the existence of the partially rotated state, excluding the possibility that it is a photophysical artifact from, for example, interchannel cross-talk.
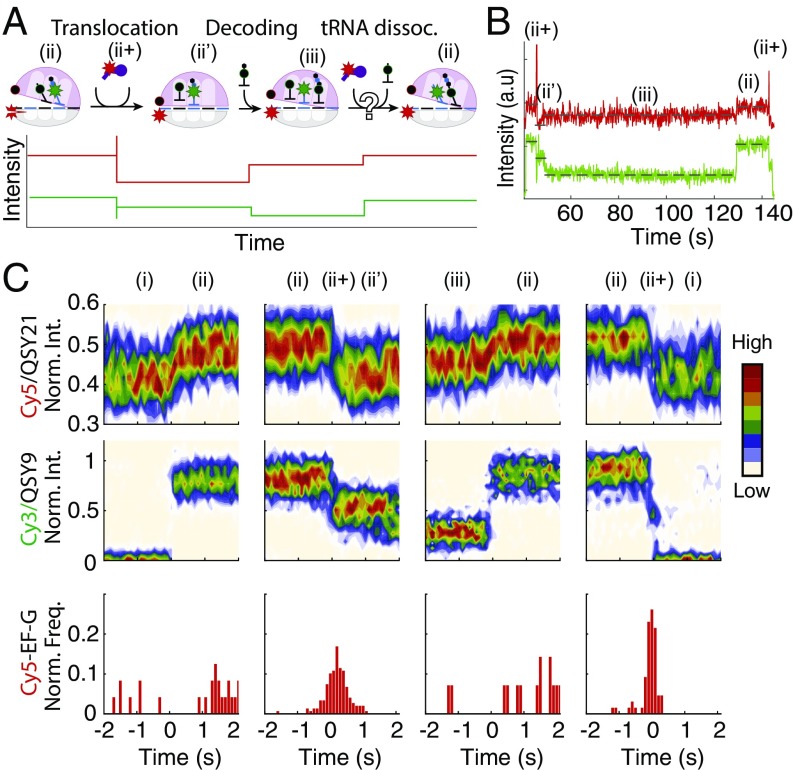
We considered two possible roles for the partially rotated state during translation. First, it may be a substrate for EF-G, which attempts to catalyze translocation and induces E-site tRNA dissociation. Second, the partially rotated conformation may be resolved through spontaneous E-site tRNA dissociation under our experimental conditions. To distinguish between these two possibilities, we monitored FRET between Cy3-labeled tRNA and a Cy5-labeled EF-G (Cy5-EF-G) (40, 46) (Fig. 3A). The distinctively anticorrelated features of the tRNA-EF-G FRET signal allowed us to monitor specific binding events of EF-G, while simultaneously monitoring tRNA composition using a Cy3/QSY9 pair and intersubunit conformation using a Cy5/QSY21 pair. As expected, EF-G bound to the rotated state, as marked by a sharp increase in Cy5 intensity and an anticorrelated decrease of Cy3 intensity due to FRET. These transient EF-G binding events triggered an intersubunit rotation to the nonrotated state and populated the E site with deacylated tRNA (Fig. 3 B and C). In contrast, we did not observe binding of EF-G to the partially rotated state at a time resolution of 100 ms. Out of 86 instances, we witnessed zero EF-G binding events, which would have been marked by sharp increase of Cy5 intensity that were correlated with E-site tRNA dissociation events. In contrast, 85 ± 4% of the rotated state to nonrotated state transition events correlated with EF-G binding, which matched the labeling efficiency of EF-G (83% labeled based on optical measurements of Cy5 and protein concentrations). We therefore conclude that the presence of the E-site tRNA impedes EF-G–catalyzed translocation, and its dissociation is independent of EF-G bindings. Our data also eliminate the possibility that the partially rotated state rapidly fluctuates between two states because EF-G would have captured the rotated state leading to translocation upon E-site tRNA dissociation.
Fig. 3.
EF-G does not resolve the partially rotated state. (A) (Top) Translation–elongation cycle via 3-tRNA state, with ii+ marking the EF-G bound state. (Bottom) Expected 3-tRNA state with two donor–quencher pairs reporting on tRNA composition and ribosome conformation. (B) Representative experimental trace showing EF-G binding to the rotated state but not to the partially rotated state. (C) Postsynchronized plots synchronized on four events: (Cy3)-tRNALys binding (first column), translocation (second column), (QSY9)-tRNAPhe dissociation from the E site (third column), and translocation (fourth column). Top and Middle show normalized Cy3 and Cy5 intensities, whereas Bottom shows frequency of Cy5-EF-G bound state appearances (n = 94).
Discussion
We have identified a partially rotated state intermediate of the ribosome when it is occupied by three tRNAs during translation elongation. The use of two donor–quencher pairs to simultaneously track and correlate tRNA occupancy and ribosomal conformation was essential in identifying the partially rotated state, given its transient nature at low Mg2+ concentration and its subtle difference in the fluorescence signal. Overlaying two smFRET signal based on donor–quencher pairs with a conventional smFRET between two fluorophores, we successfully monitored the composition of a single translational complex in real time with up to six key factors (3-tRNAs, two ribosomal subunits, and EF-G) as well as their global conformations (tRNA–tRNA and ribosome intersubunit conformations).
The partial rotation of ribosomal subunits suggests that the peptidyl transfer reaction has occurred to allow ribosomal intersubunit rotations (10), yet transition to a fully rotated state is obstructed by the presence of the E-site tRNA. Our findings very likely explain why existing ribosome structures containing three tRNAs are present in the nonrotated state: either deacylated tRNAs are used to form the complex (47) or the peptidyl transfer reaction has been thwarted by the presence of antibiotics such as erythromycin (48). The absence of an effect of E-site tRNA occupancy on A-site tRNA binding and potentially peptidyl transfer observed here agrees with previous reports (18, 22). The 3-tRNA, partially rotated state is a functionally distinct state from both the nonrotated and rotated states because it is a postdecoding complex and not a proper substrate for EF-G. Although our experiment does not preclude a transient binding (<100 ms) of EF-G to the partially rotated state, it shows that EF-G catalyzed translocation does not occur until E-site tRNA dissociates in a manner independent of EF-G.
As an idling state that is limited by slow tRNA dissociation kinetics, the E-site occupied partially rotated state may have different regulatory roles during protein synthesis. For example, E-site tRNA dissociation may directly sense cellular stress conditions, such as low temperature or high Mg2+ concentrations, to modulate translation elongation rates. Given that the average elongation rate ranges from 5 to 20 cycles per second (49), a bound lifetime of E-site tRNA on the order of 0.1–1 s in vivo (Table 1) would represent a substantial slowdown of the protein synthesis. The sensitivity of E-site tRNA dissociation kinetics to Mg2+ concentration and temperature suggests a role in extreme conditions, where elongation can be switched between two tRNA occupancy pathways (24) to result in different protein synthesis dynamics (Fig. 4). The lengthening of the partially rotated state at different ionic conditions may add another explanation to the high sensitivity of translocation rate to the Mg2+ concentration reported by Borg and Ehrenberg (50), along with the suggested effect of Mg2+ ions in stabilizing the interaction between ribosomal RNA monitoring bases and the minor groove of the codon–anticodon helix during translocation.
Fig. 4.
A model for the kinetic pathways of translation elongation through either the 3-tRNA state (iii) or the 1-tRNA state (i). The combination of the A-site tRNA–mRNA interaction, the E-site tRNA–ribosome interaction, and the ionic strength directs translation elongation via two pathways.
The partially rotated state may be further controlled by other translation factors that facilitate E-site tRNA dissociation. In fact, eukaryotic elongation factor 3 (eEF3), an essential fungal-specific elongation factor, has been proposed to facilitate departure of E-site tRNA during each round of elongation (51, 52). A similar role could be performed by its bacterial homolog, elongation factor 4 (EF4, also referred as LepA), that binds to the posttranslocation state (53, 54). Although the physiological role of EF4 during translation elongation has been debated in recent reports (55, 56), it has been shown that EF4 aids cell proliferation under stress conditions, such as high Mg2+ concentration or low temperature (53, 55), both of which have been shown here to prolong E-site tRNA residency time.
In summary, our findings suggest a mechanism by which the occupancy of the ribosomal E site modulates the ribosomal conformation and controls translation–elongation dynamics. The presence of E-site tRNA is manifested in the conformation of the ribosome, which allosterically limits its ability to translocate by blocking EF-G binding after a successful decoding. The appearance of the 3-tRNA bound state depends on the identities of the codons in the A site and E site, as well as cellular conditions such as ionic strength and temperature.
Materials and Methods
Reagents and buffers were prepared as previously reported (9, 37) (SI Materials and Methods). The synthetic biotinylated mRNA was purchased from GE Dharmacon, and tRNAs were purchased from Sigma–Aldrich and Chemical Block Ltd. and labeled with Cy3, Cy5, or QSY9 as previously described (9, 37). Details on buffer composition as well as specific concentration used can be found in SI Materials and Methods. Briefly, small and large ribosomal subunits were annealed with their respective fluorescently labeled DNA oligonucleotide for each experiment. Next, the small subunit was mixed with biotinylated mRNA, initiation factor 2, and aminoacylated formyl-methionine tRNA in the presence of GTP. Formed complex was diluted and incubated in the zero-mode waveguide chip for immobilizing. The chip was washed to remove unbound complexes before imaging. At the same time, tRNA ternary complex was formed by incubating tRNA with EF-Tu•GTP with GTP and delivered to the chip along with EF-G, large ribosomal subunits in the presence of GTP within the zero-mode waveguide instrument. Resulting movies were analyzed using in-house–written MATLAB (MathWorks) scripts, as previously described (39) (SI Materials and Methods).
Supplementary Material
Acknowledgments
We thank the members of the J.D.P. laboratory, especially Alexey Petrov, Rosslyn Grosely, Arjun Prabhakar, Christopher Lapointe, Alex Johnson, and Michael Lawson for their input. This work was supported by US National Institutes of Health Grant GM51266 (to J.D.P.) and a Stanford Bio-X graduate fellowship (to J.C.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1719592115/-/DCSupplemental.
References
- 1.Sprink T, et al. Structures of ribosome-bound initiation factor 2 reveal the mechanism of subunit association. Sci Adv. 2016;2:e1501502. doi: 10.1126/sciadv.1501502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ling C, Ermolenko DN. Initiation factor 2 stabilizes the ribosome in a semirotated conformation. Proc Natl Acad Sci USA. 2015;112:15874–15879. doi: 10.1073/pnas.1520337112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang L, et al. Allosteric control of the ribosome by small-molecule antibiotics. Nat Struct Mol Biol. 2012;19:957–963. doi: 10.1038/nsmb.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Voorhees RM, Ramakrishnan V. Structural basis of the translational elongation cycle. Annu Rev Biochem. 2013;82:203–236. doi: 10.1146/annurev-biochem-113009-092313. [DOI] [PubMed] [Google Scholar]
- 5.Frank J, Gonzalez RL., Jr Structure and dynamics of a processive Brownian motor: The translating ribosome. Annu Rev Biochem. 2010;79:381–412. doi: 10.1146/annurev-biochem-060408-173330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Korostelev A, Ermolenko DN, Noller HF. Structural dynamics of the ribosome. Curr Opin Chem Biol. 2008;12:674–683. doi: 10.1016/j.cbpa.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen J, et al. The molecular choreography of protein synthesis: Translational control, regulation, and pathways. Q Rev Biophys. 2016;49:e11. doi: 10.1017/S0033583516000056. [DOI] [PubMed] [Google Scholar]
- 8.Frank J, Agrawal RK. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- 9.Aitken CE, Puglisi JD. Following the intersubunit conformation of the ribosome during translation in real time. Nat Struct Mol Biol. 2010;17:793–800. doi: 10.1038/nsmb.1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agirrezabala X, et al. Visualization of the hybrid state of tRNA binding promoted by spontaneous ratcheting of the ribosome. Mol Cell. 2008;32:190–197. doi: 10.1016/j.molcel.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunkle JA, et al. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science. 2011;332:981–984. doi: 10.1126/science.1202692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ning W, Fei J, Gonzalez RL., Jr The ribosome uses cooperative conformational changes to maximize and regulate the efficiency of translation. Proc Natl Acad Sci USA. 2014;111:12073–12078. doi: 10.1073/pnas.1401864111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spirin AS. Ribosomal translocation: Facts and models. Prog Nucleic Acid Res Mol Biol. 1985;32:75–114. doi: 10.1016/s0079-6603(08)60346-3. [DOI] [PubMed] [Google Scholar]
- 14.Fei J, Kosuri P, MacDougall DD, Gonzalez RL., Jr Coupling of ribosomal L1 stalk and tRNA dynamics during translation elongation. Mol Cell. 2008;30:348–359. doi: 10.1016/j.molcel.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 15.Fei J, et al. Allosteric collaboration between elongation factor G and the ribosomal L1 stalk directs tRNA movements during translation. Proc Natl Acad Sci USA. 2009;106:15702–15707. doi: 10.1073/pnas.0908077106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peske F, Matassova NB, Savelsbergh A, Rodnina MV, Wintermeyer W. Conformationally restricted elongation factor G retains GTPase activity but is inactive in translocation on the ribosome. Mol Cell. 2000;6:501–505. doi: 10.1016/s1097-2765(00)00049-6. [DOI] [PubMed] [Google Scholar]
- 17.Chen J, Petrov A, Tsai A, O’Leary SE, Puglisi JD. Coordinated conformational and compositional dynamics drive ribosome translocation. Nat Struct Mol Biol. 2013;20:718–727. doi: 10.1038/nsmb.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Semenkov YP, Rodnina MV, Wintermeyer W. The “allosteric three-site model” of elongation cannot be confirmed in a well-defined ribosome system from Escherichia coli. Proc Natl Acad Sci USA. 1996;93:12183–12188. doi: 10.1073/pnas.93.22.12183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sergiev PV, et al. Function of the ribosomal E-site: A mutagenesis study. Nucleic Acids Res. 2005;33:6048–6056. doi: 10.1093/nar/gki910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nierhaus KH. The allosteric three-site model for the ribosomal elongation cycle: Features and future. Biochemistry. 1990;29:4997–5008. doi: 10.1021/bi00473a001. [DOI] [PubMed] [Google Scholar]
- 21.Burkhardt N, Jünemann R, Spahn CM, Nierhaus KH. Ribosomal tRNA binding sites: Three-site models of translation. Crit Rev Biochem Mol Biol. 1998;33:95–149. doi: 10.1080/10409239891204189. [DOI] [PubMed] [Google Scholar]
- 22.Petropoulos AD, Green R. Further in vitro exploration fails to support the allosteric three-site model. J Biol Chem. 2012;287:11642–11648. doi: 10.1074/jbc.C111.330068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uemura S, et al. Real-time tRNA transit on single translating ribosomes at codon resolution. Nature. 2010;464:1012–1017. doi: 10.1038/nature08925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen C, et al. Allosteric vs. spontaneous exit-site (E-site) tRNA dissociation early in protein synthesis. Proc Natl Acad Sci USA. 2011;108:16980–16985. doi: 10.1073/pnas.1106999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Munro JB, Altman RB, O’Connor N, Blanchard SC. Identification of two distinct hybrid state intermediates on the ribosome. Mol Cell. 2007;25:505–517. doi: 10.1016/j.molcel.2007.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marras SAE, Kramer FR, Tyagi S. Efficiencies of fluorescence resonance energy transfer and contact-mediated quenching in oligonucleotide probes. Nucleic Acids Res. 2002;30:e122. doi: 10.1093/nar/gnf121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Le Reste L, Hohlbein J, Gryte K, Kapanidis AN. Characterization of dark quencher chromophores as nonfluorescent acceptors for single-molecule FRET. Biophys J. 2012;102:2658–2668. doi: 10.1016/j.bpj.2012.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen J, Tsai A, Petrov A, Puglisi JD. Nonfluorescent quenchers to correlate single-molecule conformational and compositional dynamics. J Am Chem Soc. 2012;134:5734–5737. doi: 10.1021/ja2119964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwartz JJ, Quake SR. Single molecule measurement of the “speed limit” of DNA polymerase. Proc Natl Acad Sci USA. 2009;106:20294–20299, and erratum (2010) 107:1254. doi: 10.1073/pnas.0907404106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tyagi S, Kramer FR. Molecular beacons: Probes that fluoresce upon hybridization. Nat Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 31.Hohng S, Joo C, Ha T. Single-molecule three-color FRET. Biophys J. 2004;87:1328–1337. doi: 10.1529/biophysj.104.043935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DeRocco V, Anderson T, Piehler J, Erie DA, Weninger K. Four-color single-molecule fluorescence with noncovalent dye labeling to monitor dynamic multimolecular complexes. Biotechniques. 2010;49:807–816. doi: 10.2144/000113551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee J, et al. Single-molecule four-color FRET. Angew Chem Int Ed Engl. 2010;49:9922–9925. doi: 10.1002/anie.201005402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dietrich A, Buschmann V, Müller C, Sauer M. Fluorescence resonance energy transfer (FRET) and competing processes in donor-acceptor substituted DNA strands: A comparative study of ensemble and single-molecule data. J Biotechnol. 2002;82:211–231. doi: 10.1016/s1389-0352(01)00039-3. [DOI] [PubMed] [Google Scholar]
- 35.Dorywalska M, et al. Site-specific labeling of the ribosome for single-molecule spectroscopy. Nucleic Acids Res. 2005;33:182–189. doi: 10.1093/nar/gki151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marshall RA, Dorywalska M, Puglisi JD. Irreversible chemical steps control intersubunit dynamics during translation. Proc Natl Acad Sci USA. 2008;105:15364–15369. doi: 10.1073/pnas.0805299105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blanchard SC, Gonzalez RL, Kim HD, Chu S, Puglisi JD. tRNA selection and kinetic proofreading in translation. Nat Struct Mol Biol. 2004;11:1008–1014. doi: 10.1038/nsmb831. [DOI] [PubMed] [Google Scholar]
- 38.Choi J, et al. N(6)-methyladenosine in mRNA disrupts tRNA selection and translation-elongation dynamics. Nat Struct Mol Biol. 2016;23:110–115. doi: 10.1038/nsmb.3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen J, et al. High-throughput platform for real-time monitoring of biological processes by multicolor single-molecule fluorescence. Proc Natl Acad Sci USA. 2014;111:664–669. doi: 10.1073/pnas.1315735111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Munro JB, Wasserman MR, Altman RB, Wang L, Blanchard SC. Correlated conformational events in EF-G and the ribosome regulate translocation. Nat Struct Mol Biol. 2010;17:1470–1477. doi: 10.1038/nsmb.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fischer N, Konevega AL, Wintermeyer W, Rodnina MV, Stark H. Ribosome dynamics and tRNA movement by time-resolved electron cryomicroscopy. Nature. 2010;466:329–333. doi: 10.1038/nature09206. [DOI] [PubMed] [Google Scholar]
- 42.Wasserman MR, Alejo JL, Altman RB, Blanchard SC. Multiperspective smFRET reveals rate-determining late intermediates of ribosomal translocation. Nat Struct Mol Biol. 2016;23:333–341. doi: 10.1038/nsmb.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Korostelev A, Trakhanov S, Laurberg M, Noller HF. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell. 2006;126:1065–1077. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 44.Steinert HS, Rinnenthal J, Schwalbe H. Individual basepair stability of DNA and RNA studied by NMR-detected solvent exchange. Biophys J. 2012;102:2564–2574. doi: 10.1016/j.bpj.2012.03.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rinnenthal J, Klinkert B, Narberhaus F, Schwalbe H. Direct observation of the temperature-induced melting process of the Salmonella fourU RNA thermometer at base-pair resolution. Nucleic Acids Res. 2010;38:3834–3847. doi: 10.1093/nar/gkq124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen C, et al. Elongation factor G initiates translocation through a power stroke. Proc Natl Acad Sci USA. 2016;113:7515–7520. doi: 10.1073/pnas.1602668113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yusupov MM, et al. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 48.Arenz S, et al. A combined cryo-EM and molecular dynamics approach reveals the mechanism of ErmBL-mediated translation arrest. Nat Commun. 2016;7:12026. doi: 10.1038/ncomms12026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Young R, Bremer H. Polypeptide-chain-elongation rate in Escherichia coli B/r as a function of growth rate. Biochem J. 1976;160:185–194. doi: 10.1042/bj1600185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Borg A, Ehrenberg M. Determinants of the rate of mRNA translocation in bacterial protein synthesis. J Mol Biol. 2015;427:1835–1847. doi: 10.1016/j.jmb.2014.10.027. [DOI] [PubMed] [Google Scholar]
- 51.Triana-Alonso FJ, Chakraburtty K, Nierhaus KH. The elongation factor 3 unique in higher fungi and essential for protein biosynthesis is an E site factor. J Biol Chem. 1995;270:20473–20478. doi: 10.1074/jbc.270.35.20473. [DOI] [PubMed] [Google Scholar]
- 52.Buskirk AR, Green R. Ribosome pausing, arrest and rescue in bacteria and eukaryotes. Philos Trans R Soc Lond B Biol Sci. 2017;372:20160183. doi: 10.1098/rstb.2016.0183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pech M, et al. Elongation factor 4 (EF4/LepA) accelerates protein synthesis at increased Mg2+ concentrations. Proc Natl Acad Sci USA. 2011;108:3199–3203. doi: 10.1073/pnas.1012994108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qin Y, et al. The highly conserved LepA is a ribosomal elongation factor that back-translocates the ribosome. Cell. 2006;127:721–733. doi: 10.1016/j.cell.2006.09.037. [DOI] [PubMed] [Google Scholar]
- 55.Balakrishnan R, Oman K, Shoji S, Bundschuh R, Fredrick K. The conserved GTPase LepA contributes mainly to translation initiation in Escherichia coli. Nucleic Acids Res. 2014;42:13370–13383. doi: 10.1093/nar/gku1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu H, et al. The conserved protein EF4 (LepA) modulates the elongation cycle of protein synthesis. Proc Natl Acad Sci USA. 2011;108:16223–16228. doi: 10.1073/pnas.1103820108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.