Abstract
The advent of nanomedicine has rejuvenated the need for increased understanding of the fundamental physicochemical properties of polymeric amphiphiles. Hyaluronic acid (HA) is a hydrophilic polysaccharide that is frequently conjugated to hydrophobic moieties and then used to entrap dyes and therapeutics. Here, we develop computational models to examine the effects of the hydrophobic modification on supramolecular behavior among three systematically designed HA derivatives substituted with alkyl chains of increasing length. Our simulations coalesce with experimentally obtained results to demonstrate the dependence of supramolecular behavior on intramolecular forces. We show that the formation of clearly defined hydrophobic domains in samples of octadecylamine-modified HA compared to HA conjugates with shorter alkyl chains is a result of more favorable hydrophobic. Trends in hydrodynamic radius and polydispersity are observed in experimental results that coalesce with theoretical calculations, suggesting that supramolecular properties are dependent on the physicochemical characteristics of individual polymer strands.
Keywords: Polymeric nanoparticles, molecular dynamics, structure-property relationship, nanocarrier, self-assembly, computer-aided formulation design
Graphical abstract
1. Introduction
Evolution in the treatment and diagnosis of complicated diseases, such as cancer, has prompted the investigation of more advanced drug or imaging agent formulations to improve prognosis and reduce comorbidity (Elsabahy & Wooley, 2012; Kalepu & Nekkanti, 2015; Livney & Assaraf, 2013; McNeil, 2016). Nanoformulations, such as micelles, liposomes, and hydrogels, have gained popularity as a step toward these goals (Anselmo & Mitragotri, 2016; Livney & Assaraf, 2013). Self-assembled polymeric nanoparticles are particularly attractive as drug delivery platforms due to the low cytotoxicity of the components, highly tuned structures, and increased accumulation in disease sites (Duncan, 2003; Elsabahy & Wooley, 2012; Feng, 2010; Maeda, Nakamura, & Fang, 2013). Among self-assembled polymeric nanoformulations, naturally occurring polymers, including hyaluronic acid, heparin, and chitosan are widely-used for formulation due to innate biocompatibility and ease of chemical modification when compared to peptides or nucleic acids (Mizrahy & Peer, 2012; Ngwuluka, Ochekpe, & Aruoma, 2014; Pushpamalar, Veeramachineni, Owh, & Loh, 2016; Yang, Han, Zheng, Dong, & Liu, 2015; Zhang, Wardwell, & Bader, 2013).
When designing a drug delivery system at the earliest stages, consideration must be given to several key factors that will dictate efficacy as a nanomedicine, including: how the drug or imaging agent will be formulated into the nanoparticle (Hare et al., 2017), how the delivery system will act when administered to cells or in vivo (Beck-Broichsitter, Nicolas, & Couvreur, 2015; Blanco, Shen, & Ferrari, 2015; Livney & Assaraf, 2013), and pharmacokinetic factors such as release kinetics and particle decomposition (Giodini et al., 2017; Wilhelm et al., 2016). Frequently, controlled release and targeted delivery are goals of using a nanoformulation and these outcomes can be influenced or designed through mathematical or computation methods. Furthermore, therapeutic agent formulation through loading into nanoparticles can be accomplished by many means, including covalent attachment (Livney & Assaraf, 2013) and physical entrapment (Hill et al., 2016; Rao & Geckeler, 2011), each requiring different approaches to drug and delivery system compatibility design.
Ultimately, the supramolecular or colloidal behavior of a drug delivery system is derived from its constituent components. In polymer-based delivery systems, the dynamics of inter-particle exchange and the number of polymer chains per particle are a direct result of chemical composition of the polymer chains (Duan & Li, 2013; Lombardo, Kiselev, Magazù, & Calandra, 2015). The design and characterization of these materials can also incorporate strategies for environmentally responsive systems, using factors such as redox potential or pH for controlled release of drugs or specific functions when inside the cell (Such, Yan, Johnston, Gunawan, & Caruso, 2015). Working toward a better understanding of the most fundamental properties and chemical interactions in drug delivery systems will ultimately lead to better drug formulations and patient outcomes (McNeil, 2016).
Recently, the use of mathematical or computational means to expedite and optimize the drug delivery system design process have emerged as promising starting points (Huynh, Neale, Pomès, & Allen, 2012; Peppas, 2013; Peppas & Narasimhan, 2014; Yan & Xie, 2013). While modeling a full-sized drug delivery system at an all-atom or united-atom level remains impractical in most cases, investigations of components or interactions among components has shown value in preclinical studies (Latere Dwan’Isa, Rouxhet, Preat, Brewster, & Arien, 2007; Yongqiang Li, Taulier, Rauth, & Wu, 2006; Liu, Xiao, & Allen, 2004; Soleymani Abyaneh, Vakili, Zhang, Choi, & Lavasanifar, 2015; Youm, Yang, Murowchick, & Youan, 2011). Additionally, computational modeling of polysaccharides presents challenges due to charge, ring systems, or complex geometries, which are not present in many synthetic polymers such as poly(ethylene glycol) (Jiang, Luo, & Nangia, 2015; Ying Li, Abberton, Kröger, & Liu, 2013).
In this work, we present the development of molecular models for amphiphilic hyaluronic acid derivatives by systematically investigating the change in physicochemical properties and supramolecular behavior as a function of hydrophobic modification with alkyl chains of varying length. We support our findings through experimental analysis with dynamic light scattering (DLS), arguing that the trends observed in computational modeling correspond to supramolecular behavior in bulk solution. Furthermore, we observe and describe a relationship between the length of the alkyl chain introduced to HA and the formation of hydrophobic domains, which affects the potential for use of such amphiphilic polymer as a delivery system. Finally, we conclude by placing our work and insights into the context of the theoretical design process for drug delivery systems.
2. Materials and methods
2.1 Materials
N-hydroxysuccinimide (NHS), 1-ethyl-3-(3-dimethylamino)propyl)carbodiimide (EDC), hexylamine, dodecylamine, and octadecylamine were purchased from Sigma-Aldrich (St. Louis, MO). N,N-dimethylformamide (DMF) was purchased from Fisher Scientific (Pittsburgh, PA). Ethanol was purchased from UNMC internal supply. Sodium hyaluronate (HA) was purchased from Lifecore Biomedical (Chaska, MN).
2.2 Molecular Dynamics Simulations
Molecular dynamics simulations were performed using GROMACS 4.6 with the GROMOS 53a6 force field and the SPC water model (Jorgensen, Chandrasekhar, Madura, Impey, & Klein, 1983). The HA model and FF interaction parameters were adopted from the G53A6CARBO united atom FF (Hansen & Hünenberger, 2011; Plazinski, Lonardi, & Hünenberger, 2016), in which the CH, CH2 and CH3 moieties are treated as a single, united interacting site. Each polymer model was simulated for 260 ns. A single polymer chain in a linear extended conformation was placed in a rectangular cell of 20 nm × 15nm × 15nm, filled with approximately 105 water molecules.
To keep physiological ionic strength 0.1 M NaCl was added to aqueous solution. All the MD simulations were carried out at a constant number of particles, constant pressure of P = 1 atm, and constant temperature T = 303 K (the NPT ensemble). The reference temperature of 303 K was kept constant using the velocity rescaling weak coupling scheme (Bussi, Donadio, & Parrinello, 2007) with a coupling constant τ = 0.1 ps. The initial atomic velocities were generated with a Maxwellian distribution at the given absolute temperature. Periodic boundary conditions were applied to all three directions of the simulated box. Electrostatic interactions were simulated with the particle mesh Ewald (PME) approach (Darden, York, & Pedersen, 1993) using the long-range cutoff of 0.8 nm. The cutoff distance of Lennard-Jones interactions was also equal to 0.8 nm. The MD simulation time step was 1–2 fs with the neighbor list updates every 10 fs. All bond lengths in the HA chain were kept constant using the LINCS routine (Hess, 2008; Hess, Bekker, Berendsen, & Fraaije, 1997). The MD simulations were carried out using the GROMACS set of programs, version 4.6 (Van Der Spoel et al., 2005). Molecular graphics and visualization were performed using VMD 1.8.6 (Humphrey, Dalke, & Schulten, 1996).
The analysis was performed using GROMACS tools. The radius of gyration, head-to-tail distance, number of polymer-polymer hydrogen bonds, number of polymer-solvent hydrogen bonds, and solvent-accessible surface area were extracted from the trajectory files and plotted using GraphPad Prism 7.0 (GraphPad Software; La Jolla, CA).
2.2 Synthesis of Hyaluronic Acid-Based Amphiphiles
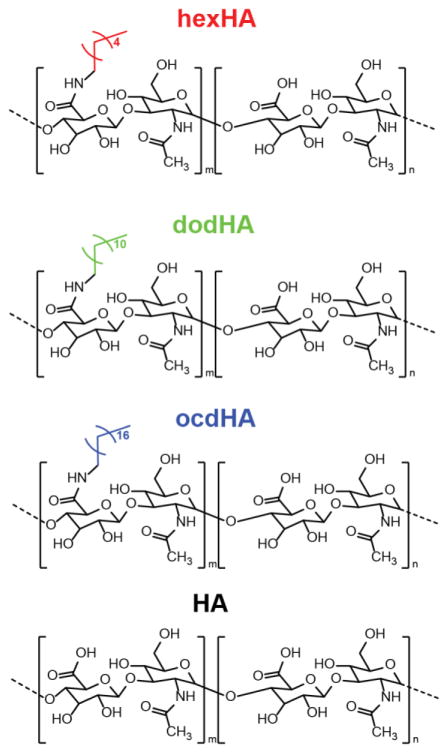
Amphiphilic hyaluronic acid (HA) polymers were synthesized as described in previous reports (Hill et al., 2015). Briefly, 40–45 mg HA (MN = 10–20 kDa, 100 kDa) was dissolved in 1:1 ultrapure water and DMF along with 30 mg of NHS and 30 mg of EDC. After mixing for 30 minutes to activate the HA carboxylic acid groups, 5–10 weight percent of hydrophobic reagent (hexylamine, dodecylamine, or octadecylamine) was added to the HA solution and allowed to react for 24 hours. Samples were then removed and placed in 3500 MWCO dialysis tubing and dialyzed against 1:1 water and ethanol for 4 exchanges over 24 h, then against pure water for 8 exchanges over 48 h to remove any impurities. Finally, samples were frozen and freeze dried for later use. Hereafter, hexylamine-modified HA is referred to as hexHA (Scheme 1A), dodecylamine-modified HA is referred to as dodHA (Scheme 1B), and octadecyl-modified HA is referred to as ocdHA (Scheme 1C).
Scheme 1.
Structures of HA and hydrophobically modified HA derivatives.
2.3 Analysis of HA conjugates
Amphiphilic HA conjugates (hexHA, dodHA, and ocdHA; Scheme 1) were analyzed on a Bruker Avance 600 MHz NMR spectrometer with TXI Cryoprobe at 25 °C with 64 to 128 scans, 8192 to 16 384 data points, and 10–12 s relaxation delay. Samples were analyzed in DMSO-d6 (Cambridge Isotope Laboratories, 99.9% D). Data was processed in Mnova NMR (Escondido, CA).
2.4 Dynamic Light Scattering
To prepare samples for DLS, amphiphilic HA conjugate was dissolved in ultrapure water for a stock solution concentration of 1.0 mg/mL and allowed to equilibrate for at least 24 h, then lower concentrations were prepared from this stock solution and allowed to equilibrate for at least four hours and stored at 4 °C prior to analysis. Samples were prepared in concentrations ranging from 0.008–1.0 mg/mL for DLS analysis using a Malvern ZetaSizer Nano ZS (Malvern Instruments; Malvern, UK). Ten independent measurements were made for each sample concentration at a temperature of 25 °C. Data were exported in CSV format and analyzed in GraphPad Prism 7.0 software (GraphPad Software; La Jolla, CA).
3. Results and Discussion
3.1 Influence of degree of dissociation on HA dynamics
Investigation of HA self-assembly dynamics began with ensuring conformity between experimental and theoretical approaches. Prior to investigating the hydrophobically modified HA derivatives, simulations of HA at different degrees of dissociation were performed to examine differences in polymer dynamics due to protonation of the carboxylic group. Due to the presence of carboxylic groups on HA, the degree of dissociation can potentially affect the polymer-polymer and polymer-solvent interactions through hydrogen bonding.
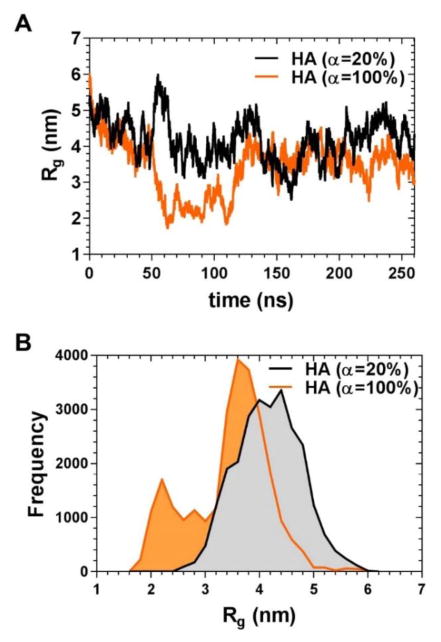
HA with the polymer chain length of 26 units was modeled computationally in the completely dissociated state and with the degree of dissociation of 20% (which, depending on its final concentration, corresponds to the experimental degree of dissociation of aqueous HA with its pKa reported to be 2.9 (Cleland, Wang, & Detweiler, 1982)). Figure S1 visually demonstrates the substitution and deprotonation positions for simulation of HA. Sites of substitution and deprotonation were chosen at random. Since ligands are conjugated to HA through the carboxylic groups on the glucuronic acid moieties and these moieties are active in hydrogen bonding, difference in the degree of dissociation, correlating to the effective pH of the environment, could influence polymer behavior. The radius of gyration was found not to be significantly different between the two samples by performing a Kolmogorov-Smirnov test (p>0.05). Furthermore, the number of hydrogen bonds between the polymer and the surrounding water molecules was not found to be significantly different. These data indicate that the degree of dissociation did not significantly influence the results obtained from simulations of amphiphilic HA.
3.2 Radius of gyration and end-to-end distance analysis
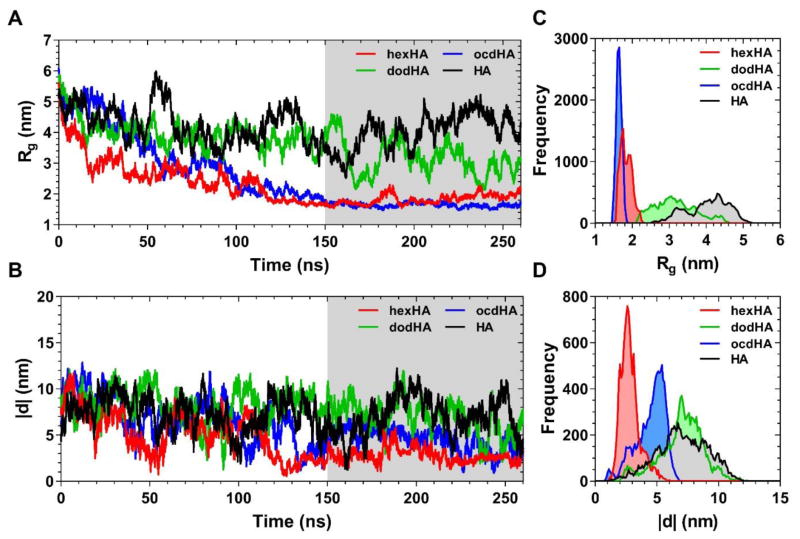
Examining the radius of gyration is the most straight forward method to investigate the effects of hydrophobic modifications on HA. MD simulations were performed for 260 ns to observe the conformational change from linear to random coil conformation of the modified and unmodified HA polymers. Three modified HA polymers were considered: hexHA, dodHA, and ocdHA (Scheme 1, were n=19 and m=7, respectively). As shown in Figure 2A, the radius of gyration for both hexyl- and octadecyl-modified HA reaches a stable radius of gyration of around 2 nm. When compared to unmodified HA (4 nm), these radii are much smaller, demonstrating a more compact conformation as a result of hydrophobically-driven intramolecular interactions. Furthermore, the distributions of 25,000 samplings of Rg taken from 150–260 ns (Figure 2C) indicate a relatively less polydisperse sampling and a narrower distribution than that of unmodified HA. The difference between ocdHA and hexHA is noteworthy in that the ocdHA sample exhibits a smaller distribution of Rg values after collapsing to coil conformation, likely due to the greater influence of hydrophobic packing on driving the compactness of the overall molecule. The much greater length of the hydrophobic chain contributes to the formation of more defined hydrophobic pockets inside the coil, ultimately resulting a more stable conformation.
Figure 2.
Dynamics of single polymer chains of 10 kDa alkyl-substituted HA (degree of substitution 20%): (A) changes of the radius of gyration and (B) head-to-tail distance during 260 ns of simulations. (C) Frequency distribution of the radius of gyration and (D) head-to-tail distance values, respectively, over the equilibrated section of the trajectory (corresponding to the gray shaded region in A and B).
The Rg of dodecyl-modified HA exhibits less pronounced conformational changes over the course of the simulation than the hexyl- or octadecyl-modified samples. While hexHA and ocdHA both reach stable conformations, dodHA continues to fluctuate at a higher Rg and more closely resembles the behavior of unmodified HA. This difference suggests a divergence in the hydrophobic packing of the molecule when compared to ocdHA. When transitioning from linear to random coil conformation, the effect of hydrophobic packing of the alkyl chains associating can cause intramolecular strain. We anticipate that the difference in behavior between dodHA and the other alkyl derivatives of HA is a result of increased strain that is not overcome by hydrophobically-driven collapse. HexHA would have the lowest degree of intramolecular strain, thus being more amenable to hydrophobic collapse, whereas the dodHA polymer would experience significantly more strain than hexHA without the stronger hydrophobic interactions as seen in ocdHA.
The head-to-tail distance is also an important measure of compactness of the polymers in simulation. Similar to the Rg results, the head-to-tail distance is more compact for hexHA and ocdHA, whereas dodHA is more closely comparable to unmodified HA. All modified HA polymers were substituted at the same positions along the polymeric backbone to eliminate any variation due to difference in location of hydrophobic moiteties, and as such, the difference between hexHA, dodHA, and ocdHA in the head-to-tail distance is less pronounced than the Rg as shown in Figure 2B. However, the distribution of the samplings between 150–260 ns clearly demonstrates a narrower distribution of distance for the hexHA and ocdHA samples than the distributions of the dodHA or HA samples as shown in Figure 2D.
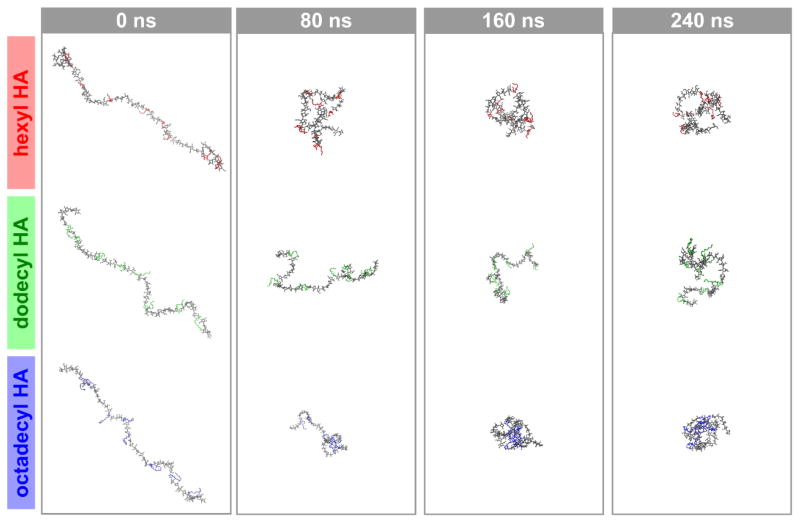
The change in conformation from linear to coil is illustrated with snapshots from the MD simulations in Figure 3. This transition is driven by hydrophobic collapse, ultimately leading to the compact coil formations observed towards the end of the simulations. Furthermore, these snapshots of the polymer amphiphiles conformations help to graphically illustrate the changes in Rg and end-to-end distance explained in this discussion. Of particular note are the final conformations of each molecule; while hexHA and ocdHA form relatively more compact conformations, dodHA exhibits behavior indicative of less favorable collapse and compaction as a result of intramolecular strain. Additional graphical representations to further illustrate the difference in conformations after 260 ns of simulation are shown in Figure S2.
Figure 3.
Snapshots from MD simulations for alkyl-substituted HA derivatives at various time points (water molecules and buffering ions are not shown for clarity). These simulations show that as the polymers transition from linear to random coil conformations, the nature of the hydrophobic moiety contributes to polymer packing and the resulting Rg.
To support and add rigor to the results from our simulations, we performed additional independent simulations to ensure the repeatability of our results. Figure S4 shows the results of such simulations performed using the same starting models used to create Figure 2, as well as self-assembled polymer configurations from the original simulation. Additionally, analyses were performed on the trajectories to evaluate the effect of hydrophobic substitution on the glycosidic dihedral angles (Scheme S1) as shown in Figure S3. Based on the results of our simulations, introduction of alkyl substituents does not cause significant deviation of the glycosidic dihedral angles from the typical value of ±120° (Gargiulo et al., 2010).
3.3 Investigation of solvent-accessible surface area
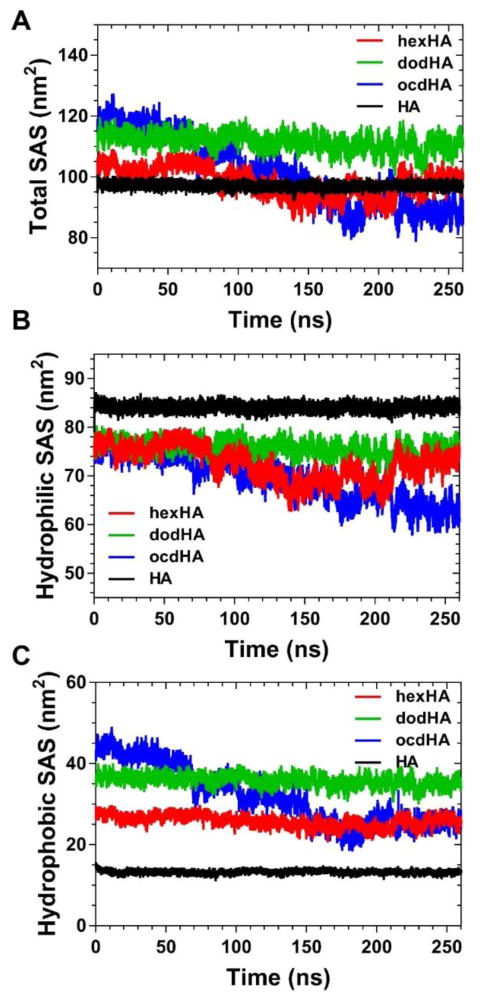
The results illustrated in Figure 3 coincide with changes in the solvent-accessible surface (SAS) area of the polymer stands. The total, hydrobic, and hydrophilic solvent-accessible surface area were extracted to examine their changes as the polymers transmute conformation over the course of the simulation. As the conformation changes, the hydrophobic packing of the alkyl chains results in the formation of hydrophobic domains that are shielded to varying extents by the hydrophilic components of the polymer. Interestingly, dodHA exhibits negligible change in total surface area while both hexHA and ocdHA show an overall decrease in solvent-accessible surface, which is consistant with the trend observed for the radius of gyration in Figure 2. Correlation between Figures 3 and 4 can also be visually inferred by observing that the relative compactness of the polymer at the different timepoints in the simulation correlates strongly with the solvent-accessible surface.
Figure 4.
Change in (A) total, (B) hydrophilic and (C) hydrophobic solvent accessible surface area of the HA polymers during simulations.
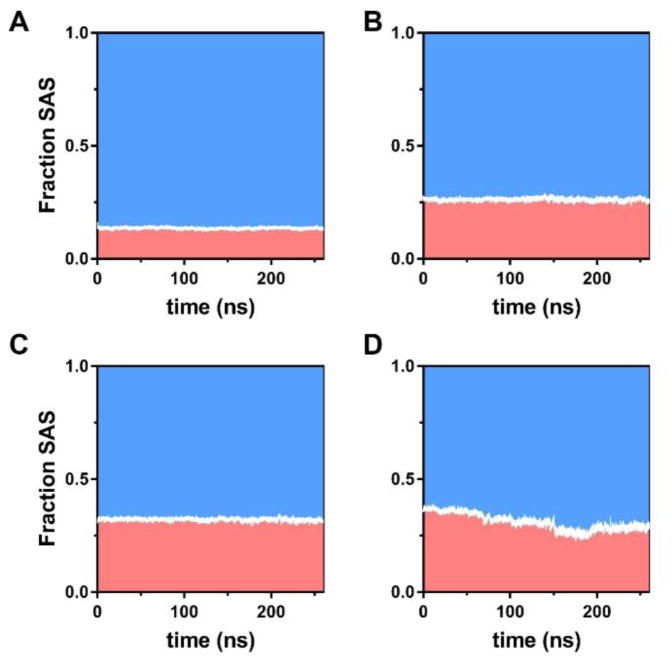
From the discussion of Rg and head-to-tail distance gleaned from Figure 2, the difference in compactness between hexHA and ocdHA is not unambiguous. However, significant difference in dynamics of normalized hydrophobic SAS area between ocdHA and hexHA (Figure 5) better illustrates the tighter packing of the former. The compactness of the hydrophobic pockets and the differences between hexHA and ocdHA are visually apparent in Figure 3. While the Rg for hexHA and ocdHA remain similar throughout the simulation, the interior environment and the hydrophobic domains created differ greatly. As the HA polymers collapse to form hydrophobic pockets, it is thermodynamically favorable and expected that the hydrophobic SAS area would decrease as a fraction of the total SAS area. The hydrophobic SAS of ocdHA decreases from 47% to 39%, whereas its hydrophilic SAS increases from 53% to 61% as seen in Figure 5D.
Figure 5.
Changes of hydrophobic (red) and hydrophilic (blue) solvent accessible surface as fractions of the total solvent accessible surface of HA and alkylated HA derivatives over 260 ns of simulations shows SAS fraction for (A) unmodified HA control, (B) hexHA, (C) dodHA, and (D) ocdHA, which demonstrate the impact in the ratio of hydrophobic and hydrophilic SAS over the course of the MD simulation.
The most insightful observation from the MD analysis of the SAS area for the samples is the difference in hydrophobic contribution to the total SAS when compared to the other modified polymers. Unlike previously discussed results for the radius of gyration in Figure 2, the hexHA and dodHA samples appear to be in accordance whereas the results of MD simulations of ocdHA deviate from the other samples. The trend of decreasing hydrophobic SAS area is very apparent for the ocdHA sample, where it decreases by approximately 20 nm2 while the total SAS area decreases by only approximately 30 nm2. The behavior of ocdHA clearly demonstrates the hydrophobic collapsing and the tighter distribution of Rg when compared to dodHA and hexHA. Despite significantly bulkier hydrophobic moiety, the hydrophobic solvent-accessible surface decreases to the level of that of hexHA. When loading hydrophobic drugs into a nanoparticle drug delivery system, the nature of the hydrophobic pockets in the delivery system dictates loading efficiency and controlled release via passive diffusion of the drug (Beck-Broichsitter et al., 2015; Mu et al., 2014). Thus, when formulating drugs through physical entrapment in polymeric nanoaggregates, an understanding of the fundamental physicochemical properties of the amphiphilic polymer and the hydrophobic pockets created during the collapsing or aggregation process is of paramount importance.
Our experiments only investigate the effects of the alkyl chain length of modified HA using aliphatic hydrocarbons as hydrophobic moieties. The results from the analysis of the change in the Rg and the solvent-accessible surface area demonstrate that the length of the alkyl substitution dictates not only the conformation of the polymer coil, but also the stability of the hydrophobic pockets, and demonstrates the dependence of hydrophobic collapse on intramolecular strain. HexHA, while forming a tightly compact coil structure when compared to dodHA or unmodified HA, does not exhibit enough intramolecular strain to significantly inhibit the formation of hydrophobic pockets inside the coil. Upon moving to the more hydrophobically modified dodHA, one would expect a more compact or more stable conformation than hexHA due to the contribution of additional alkyl chain length. However, a much less defined coil conformation is observed, possibly due to intramolecular (steric) strain limiting the variability conformational contortions. OcdHA, which has the longest alkyl chains and a greater initial hydrophobic surface area, exhibits the outcome of highly favorable hydrophobic collapse, potentially less inhibited by or overcoming the effects of intramolecular strain.
The effect of the degree of alkylation was also investigated computationally. HA derivatives with 50% alkyl chain substitution were constructed and simulated, producing the results shown in Figure S5. The highly-substituted HA systems behaved similarly to those with the experimentally-relevant degree of alkylation as previously discussed. The primary difference was that a higher degree of substitution resulted in reaching conformation equilibrium more quickly. Additionally, the Rg of all samples appears to behave uniformly. Despite the differences in the alkyl chain length, the degree of alkylation overcomes limitations in conformational change, in contrast to the MD simulations with lower conjugation ratios of alkyl chains.
3.4 Dynamic light scattering
The computational simulations we describe are limited to a single polymer strand and do not provide certainty about the behavior of such materials when prepared experimentally in bulk solution where multiple polymer molecules are able to associate with each other. By validating our computational findings with experimentally obtained results from bulk solutions of the modified polymers, we show that in silico methods of designing polymer amphiphiles for drug delivery applications can result in more optimally formulated nanomedicines. Samples of hexHA, dodHA, and ocdHA were prepared and analyzed using dynamic light scattering to observe the behavior of the polymers in bulk solution. As the amphiphilic polymers interact with each other and form nanoparticles, the measurement of the behavior of single polymer strands is not possible, however the behavior of these aggregates can be related back to the simulation results of the individual polymers as presented in Figure 2. The substitution ratio (degree of alkylation) was determined experimentally by NMR. Samples of 10 kDa and 100 kDa hexHA, dodHA, and ocdHA were analyzed to determine the molar ratio of the hydrophobic moiety and are shown in Figure S6–8 and summarized in Table S1.
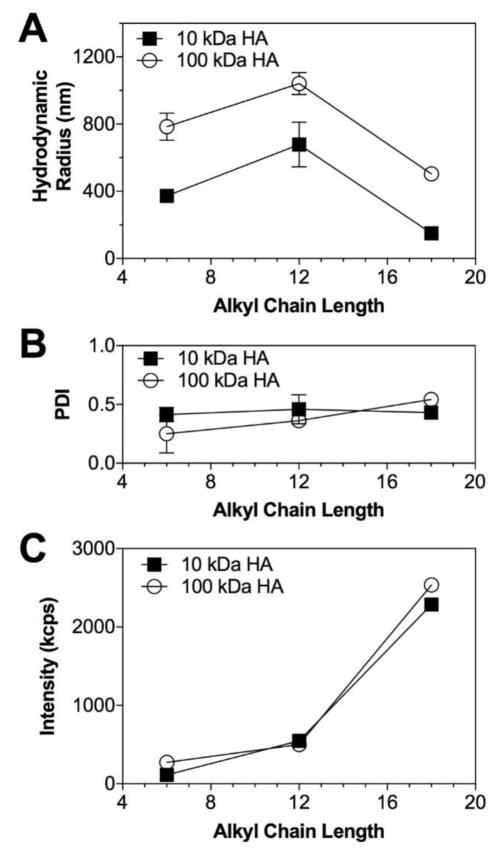
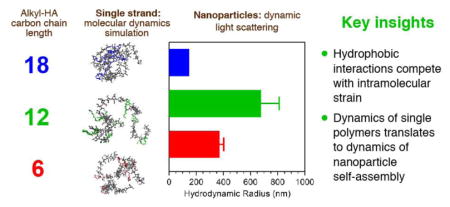
The behavior of amphiphilic HA conjugates in bulk solution closely resembles the results obtained from simulation studies of the individual polymer components as shown in Figure 6. The first explicit correlation observed is the trend of nanoparticle mean size and standard deviation when compared to the computationally determined Rg of the constituent amphiphile. In accord with the relative stability of the Rg for both hexHA and ocdHA, the standard deviation in mean nanoparticle size (385 ± 30 nm and 175 ± 15 nm, respectively) for each of these experimental samples (Figure 6A) exhibits trends parallel to those observed for both Rg (1.84 ± 0.16 nm and 1.64 ± 0.07 nm, respectively) and head-to-tail distance observed in silico. Assent in behavior between two different polymer lengths prepared experimentally reinforces the similarity in trends observed between the theoretical and experimental results. Two sets of amphiphilic conjugates were prepared—10 kDa and 100 kDa—to determine if the polymer length affected the behavior observed after hydrophobic modification. Although simulations were performed using only 10 kDa polymer chains due to computational limitations, the effects of hydrophobic modification on the stability and size are distinguished for the 100 kDa samples prepared experimentally (Figure 6) and align well with the observations of 10 kDa samples. In particular, the standard deviation from the mean hydrodynamic radius observed for the most compact ocdHA samples was 15 nm and 20 nm for 10 kDa and 100 kDa variants, respectively.
Figure 6.
Dependence of the (A) nanoparticle size, (B) polydispersity index and (C) scattering intensity upon the hydrophobic ligand chain length for 10 kDa and 100 kDa HA nanoparticles in water at a concentration of 0.5 mg/mL.
The experimental results obtained for ocdHA samples highlight the dependence of the formation and stability of hydrophobic pockets on the physicochemical properties of the hydrophobic moiety. The small mean hydrodynamic diameter and narrow size distribution also clarifies our investigation of the formation of hydrophobic pockets within the aggregates and relates strongly to our computational results. When comparing the ocdHA and hexHA samples, the Rg (1.84 vs. 1.64 nm), normalized hydrophobic solvent-accessible surface area (0.43 vs. 0.39), and mean hydrodynamic radius (385 nm vs. 175 nm) suggest a distinct difference between the hydrophobic pockets formed inside the aggregates, contending that the ocdHA samples produce more well-defined and stable hydrophobic regions than the hexHA or dodHA.
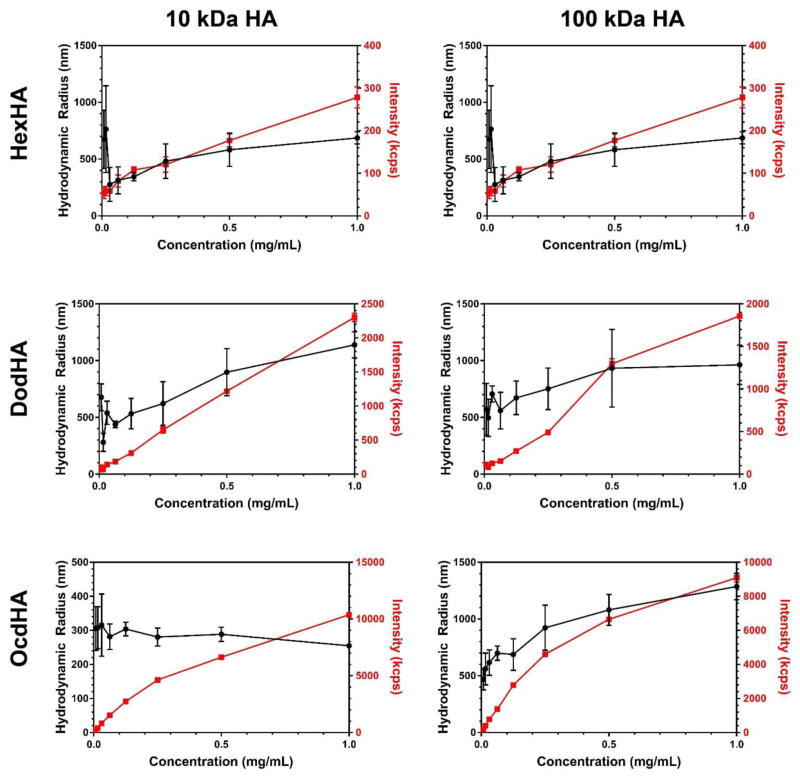
Supramolecular behavior as a function of individual strand properties is supported through the concentration-dependent intensities obtained from DLS for the HA derivatives shown in Figure 7. The scattering intensity depends on the size, shape, and number of particles in solution, and therefore the DLS data obtained shows that the ocdHA samples produce smaller, more tightly aggregated nanoparticles in bulk solution. Figure 7 details the scattering profiles as a function of concentration for both 100 kDa and 10 kDa variants of the three amphiphilic HA derivatives. When comparing HA derivatives of the same concentration, the scattering intensities differ greatly, with ocdHA producing the highest scattering intensities. Since scattering intensity in DLS is proportional to the Brownian motion of the particles in solution, the small standard deviations suggest better colloidal stability and more uniform particles than the dodHA or hexHA samples.
Figure 7.
Dynamic light scattering of the HA amphiphiles derived from (left column) 10 kDa HA and (right column) 100 kDa HA. Hydrodynamic radius and scattering intensity are reported as a function of concentration to demonstrate differences in aggregation and colloidal stability.
The correlations observed between our theoretical and experimental work are supported by previously published insights (Cho et al., 2011; Lee, Choi, & Park, 2006; Ramezanpour et al., 2016; Sun & Mao, 2016) and expand the understanding of the self-assembly dynamics of HA-based materials. Where others have investigated the computer-aided formulation design (Jha & Larson, 2014; Latere Dwan’Isa et al., 2007; Patel, Lavasanifar, & Choi, 2008; Rostamizadeh, Vahedpour, & Bozorgi, 2012), including HA-based systems (Sun & Mao, 2016), our work is the first example of systematically investigating the effects of hydrophobic modification on single chain and supramolecular dynamics. As new drug delivery systems based on polysaccharides are developed, computational methods for design and characterization will be useful to shorten development time.
While there are a few examples of FDA-approved nanomedicines (Havel et al., 2016; Schütz, Juillerat-Jeanneret, Mueller, Lynch, & Riediker, 2013), more research into the fundamental science of nanomedicine will help new nanoformulations translate into clinical use (McNeil, 2016; Moghimi & Farhangrazi, 2014; Satalkar, Elger, Hunziker, & Shaw, 2016; Wicki, Witzigmann, Balasubramanian, & Huwyler, 2015). Empirical and computational methods to evaluate nanomedicine formulations theoretically, before experimental investigation, are gaining in popularity, but still require further development to achieve widespread use. Our work contributes to a growing body of research supporting the use of theoretical methods to predict the self-assembly properties of nanomaterials for formulation.
4. Conclusions
We report the fundamental investigation of hydrophobically modified HA derivatives and the physicochemical properties that influence their collapse into coil conformations when simulated in silico, as well as experimentally observed aggregation of multiple polymer strands into nanoparticles. Using a series of alkyl-modified HA, we investigate in detail the influence on alkyl chain length on the radius of gyration, head-to-tail distance, and solvent-accessible surface area, ultimately relating theoretical insights to behavior of real polymers in bulk solution.
Molecular dynamics simulations revealed a distinct correlation between alkyl chain lengths and HA conjugate stability in the form of Rg and solvent-accessible surface area when individual amphiphilic polymers solvated in water were modeled. When compared to unmodified HA, which is innately hydrophilic, amphiphilic HA conjugates quickly transitioned to random coil conformations driven by hydrophobic collapse and the formation of hydrophobic domains. The definition of the hydrophobic domains depends heavily on the properties of the polymer, where conformational transitions are driven by the alkyl chain length, but inhibited by forces such as intramolecular strain. The delicate balance between hydrophobicity and steric hindrance calls to attention a new factor for consideration in the design of supramolecular assemblies for drug delivery applications.
Our computationally obtained insights are substantiated through experimental results. Samples of each HA derivative were prepared in two polymer chain lengths to elucidate any dependence of aggregation or conformational trends for polymer molecules of different sizes. The behavior observed for experimental samples—analysis in bulk solution rather than individual strands—correlated to the trends exposed in silico; where Rg and relative hydrophobic solvent-accessible surface area correlate to compactness and stability of coil conformation, nanoaggregates in bulk demonstrate stability and degree of hydrophobic domain definition to be dependent on the properties of the individual polymer components. Ultimately, these results support our hypothesis that the dynamics of supramolecular assemblies composed of polymeric amphiphiles mirror the characteristics of its components.
Our work is a fundamental investigation aiming to improve the understanding of factors that should be considered when designing a drug delivery system from amphiphilic polymers. Elucidating the properties of structures that are poorly understood and difficult to picture will aid in the optimization of formulations for a variety of aspects, and the insights gained herein are a step forward in creating a paradigm for physicochemical design of nanomedicines.
Supplementary Material
Figure 1.
Dynamics of a single 10 kDa HA polymer chain of 20% and 100% dissociation in water. (A) Changes in the radius of gyration during 260 ns of simulations; (B) frequency distribution of the radius of gyration values over the full trajectory shown in A.
Highlights.
Colloidal properties of HA-based nanomaterials result from single strand dynamics.
Results of MD simulations are supported experimentally.
Trends observed in silico are also observed in bulk solution experiments.
Acknowledgments
Funding Sources
This work was funded in part by National Institutes of Health [grant numbers R01 EB019449, P20 GM103480, P30 CA036727 (Fred & Pamela Buffett Cancer Center), 1S10RR17846, and 1S10RR027940], and the Nebraska Research Initiative. Computational aspects of this work were completed utilizing the Holland Computing Center at the University of Nebraska.
Abbreviations
- HA
hyaluronic acid
- CAC
critical aggregation concentration
- DLS
dynamic light scattering
- MD
molecular dynamics
- Rg
radius of gyration
- NMR
nuclear magnetic resonance
- FF
force field
- SAS
solvent accessible surface
Footnotes
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
MD simulation data for the alkylated HA derivatives with the 50% degree of substitution, parallel independent simulations for alkHA derivatives, analysis of glycosidic torsion angles dynamics, and NMR data with degree of substitution quantification for experimental hydrophobically modified HA samples are provided in the Supporting Information, which is available free of charge.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anselmo AC, Mitragotri S. Nanoparticles in the clinic. Bioengineering & Translational Medicine. 2016;1(1):10–29. doi: 10.1002/btm2.10003. http://doi.org/10.1002/btm2.10003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck-Broichsitter M, Nicolas J, Couvreur P. Design attributes of long-circulating polymeric drug delivery vehicles. European Journal of Pharmaceutics and Biopharmaceutics. 2015;97:304–317. doi: 10.1016/j.ejpb.2015.03.033. http://doi.org/10.1016/j.ejpb.2015.03.033. [DOI] [PubMed] [Google Scholar]
- Blanco E, Shen H, Ferrari M. Principles of nanoparticle design for overcoming biological barriers to drug delivery. Nature Biotechnology. 2015;33(9):941–951. doi: 10.1038/nbt.3330. http://doi.org/10.1038/nbt.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussi G, Donadio D, Parrinello M. Canonical sampling through velocity rescaling. The Journal of Chemical Physics. 2007;126(1):14101. doi: 10.1063/1.2408420. http://doi.org/10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- Cho HJ, Yoon HY, Koo H, Ko SH, Shim JS, Lee JH, … Kim D-D. Self-assembled nanoparticles based on hyaluronic acid-ceramide (HA-CE) and Pluronic® for tumor-targeted delivery of docetaxel. Biomaterials. 2011;32(29):7181–7190. doi: 10.1016/j.biomaterials.2011.06.028. http://doi.org/10.1016/j.biomaterials.2011.06.028. [DOI] [PubMed] [Google Scholar]
- Cleland RL, Wang JL, Detweiler DM. Polyelectrolyte properties of sodium hyaluronate. 2. Potentiometric titration of hyaluronic acid. Macromolecules. 1982;15(2):386–395. [Google Scholar]
- Darden T, York D, Pedersen L. Particle mesh Ewald: An N log(N) method for Ewald sums in large systems. The Journal of Chemical Physics. 1993;98(12):10089. http://doi.org/10.1063/1.464397. [Google Scholar]
- Duan X, Li Y. Physicochemical characteristics of nanoparticles affect circulation, biodistribution, cellular internalization, and trafficking. Small. 2013;9(9–10):1521–1532. doi: 10.1002/smll.201201390. http://doi.org/10.1002/smll.201201390. [DOI] [PubMed] [Google Scholar]
- Duncan R. The dawning era of polymer therapeutics. Nature Reviews Drug Discovery. 2003;2(5):347–360. doi: 10.1038/nrd1088. http://doi.org/10.1038/nrd1088. [DOI] [PubMed] [Google Scholar]
- Elsabahy M, Wooley KL. Design of polymeric nanoparticles for biomedical delivery applications. Chemical Society Reviews. 2012;41(7):2545–2561. doi: 10.1039/c2cs15327k. http://doi.org/10.1039/c2cs15327k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X. Polymeric Conjugates for Anti-cancer Drug Delivery. Polymer. 2010:50–52. [Google Scholar]
- Gargiulo V, Morando MA, Silipo A, Nurisso A, Perez S, Imberty A, … De Castro C. Insights on the conformational properties of hyaluronic acid by using NMR residual dipolar couplings and MD simulations. Glycobiology. 2010;20(10):1208–1216. doi: 10.1093/glycob/cwq067. http://doi.org/10.1093/glycob/cwq067. [DOI] [PubMed] [Google Scholar]
- Giodini L, Re F Lo, Campagnol D, Marangon E, Posocco B, Dreussi E, Toffoli G. Nanocarriers in cancer clinical practice: a pharmacokinetic issue. Nanomedicine: Nanotechnology, Biology, and Medicine. 2017;13(2):583–599. doi: 10.1016/j.nano.2016.07.012. http://doi.org/10.1016/j.nano.2016.07.012. [DOI] [PubMed] [Google Scholar]
- Hansen HS, Hünenberger PH. A reoptimized GROMOS force field for hexopyranose-based carbohydrates accounting for the relative free energies of ring conformers, anomers, epimers, hydroxymethyl rotamers, and glycosidic linkage conformers. Journal of Computational Chemistry. 2011;32(6):998–1032. doi: 10.1002/jcc.21675. http://doi.org/10.1002/jcc.21675. [DOI] [PubMed] [Google Scholar]
- Hare JI, Lammers T, Ashford MB, Puri S, Storm G, Barry ST. Challenges and strategies in anti-cancer nanomedicine development: An industry perspective. Advanced Drug Delivery Reviews. 2017;108:25–38. doi: 10.1016/j.addr.2016.04.025. http://doi.org/10.1016/j.addr.2016.04.025. [DOI] [PubMed] [Google Scholar]
- Havel H, Finch G, Strode P, Wolfgang M, Zale S, Bobe I, … Liu M. Nanomedicines: From Bench to Bedside and Beyond. The AAPS Journal. 2016;18(6):1373–1378. doi: 10.1208/s12248-016-9961-7. http://doi.org/10.1208/s12248-016-9961-7. [DOI] [PubMed] [Google Scholar]
- Hess B. P-LINCS: A Parallel Linear Constraint Solver for Molecular Simulation. Journal of Chemical Theory and Computation. 2008;4(1):116–122. doi: 10.1021/ct700200b. http://doi.org/10.1021/ct700200b. [DOI] [PubMed] [Google Scholar]
- Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. LINCS: A linear constraint solver for molecular simulations. Journal of Computational Chemistry. 1997;18(12):1463–1472. http://doi.org/10.1002/(SICI)1096-987X(199709)18:12<1463::AID-JCC4>3.0.CO;2-H. [Google Scholar]
- Hill TK, Abdulahad A, Kelkar SS, Marini FC, Long TE, Provenzale JM, Mohs AM. Indocyanine Green-Loaded Nanoparticles for Image-Guided Tumor Surgery. Bioconjugate Chemistry. 2015;26(2):294–303. doi: 10.1021/bc5005679. http://doi.org/10.1021/bc5005679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill TK, Davis AL, Wheeler FB, Kelkar SS, Freund EC, Lowther WT, … Mohs AM. Development of a Self-Assembled Nanoparticle Formulation of Orlistat, Nano-ORL, with Increased Cytotoxicity against Human Tumor Cell Lines. Molecular Pharmaceutics. 2016;13(3):720–728. doi: 10.1021/acs.molpharmaceut.5b00447. http://doi.org/10.1021/acs.molpharmaceut.5b00447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. Journal of Molecular Graphics. 1996;14(1):33–38. doi: 10.1016/0263-7855(96)00018-5. http://doi.org/10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Huynh L, Neale C, Pomès R, Allen C. Computational approaches to the rational design of nanoemulsions, polymeric micelles, and dendrimers for drug delivery. Nanomedicine: Nanotechnology, Biology and Medicine. 2012;8(1):20–36. doi: 10.1016/j.nano.2011.05.006. http://doi.org/10.1016/j.nano.2011.05.006. [DOI] [PubMed] [Google Scholar]
- Jha PK, Larson RG. Assessing the Efficiency of Polymeric Excipients by Atomistic Molecular Dynamics Simulations. Molecular Pharmaceutics. 2014;11(5):1676–1686. doi: 10.1021/mp500068w. http://doi.org/10.1021/mp500068w. [DOI] [PubMed] [Google Scholar]
- Jiang W, Luo J, Nangia S. Multiscale Approach to Investigate Self-Assembly of Telodendrimer Based Nanocarriers for Anticancer Drug Delivery. Langmuir. 2015;31(14):4270–4280. doi: 10.1021/la503949b. http://doi.org/10.1021/la503949b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. The Journal of Chemical Physics. 1983;79(2):926. http://doi.org/10.1063/1.445869. [Google Scholar]
- Kalepu S, Nekkanti V. Insoluble drug delivery strategies: Review of recent advances and business prospects. Acta Pharmaceutica Sinica B. 2015;5(5):442–453. doi: 10.1016/j.apsb.2015.07.003. http://doi.org/10.1016/j.apsb.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latere Dwan’Isa JP, Rouxhet L, Preat V, Brewster ME, Arien A. Prediction of drug solubility in amphiphilic di-block copolymer micelles: the role of polymer-drug compatibility. Die Pharmazie. 2007;62(7):499–504. http://doi.org/10.1691/ph.2007.7.6273. [PubMed] [Google Scholar]
- Lee H, Choi SH, Park TG. Direct Visualization of Hyaluronic Acid Polymer Chain by Self-Assembled One-Dimensional Array of Gold Nanoparticles. Macromolecules. 2006;39(1):23–25. http://doi.org/10.1021/ma051929c. [Google Scholar]
- Li Y, Abberton B, Kröger M, Liu W. Challenges in Multiscale Modeling of Polymer Dynamics. Polymers. 2013;5(2):751–832. http://doi.org/10.3390/polym5020751. [Google Scholar]
- Li Y, Taulier N, Rauth AM, Wu XY. Screening of lipid carriers and characterization of drug-polymer-lipid interactions for the rational design of polymer-lipid hybrid nanoparticles (PLN) Pharmaceutical Research. 2006;23(8):1877–1887. doi: 10.1007/s11095-006-9033-2. http://doi.org/10.1007/s11095-006-9033-2. [DOI] [PubMed] [Google Scholar]
- Liu J, Xiao Y, Allen C. Polymer-Drug Compatibility: A Guide to the Development of Delivery Systems for the Anticancer Agent, Ellipticine. Journal of Pharmaceutical Sciences. 2004;93(1):132–143. doi: 10.1002/jps.10533. http://doi.org/10.1002/jps.10533. [DOI] [PubMed] [Google Scholar]
- Livney YD, Assaraf YG. Rationally designed nanovehicles to overcome cancer chemoresistance. Advanced Drug Delivery Reviews. 2013;65(13–14):1716–1730. doi: 10.1016/j.addr.2013.08.006. http://doi.org/10.1016/j.addr.2013.08.006. [DOI] [PubMed] [Google Scholar]
- Lombardo D, Kiselev MA, Magazù S, Calandra P. Amphiphiles Self-Assembly: Basic Concepts and Future Perspectives of Supramolecular Approaches. Advances in Condensed Matter Physics. 2015;2015:1–22. http://doi.org/10.1155/2015/151683. [Google Scholar]
- Maeda H, Nakamura H, Fang J. The EPR effect for macromolecular drug delivery to solid tumors: Improvement of tumor uptake, lowering of systemic toxicity, and distinct tumor imaging in vivo. Advanced Drug Delivery Reviews. 2013;65(1):71–79. doi: 10.1016/j.addr.2012.10.002. http://doi.org/10.1016/j.addr.2012.10.002. [DOI] [PubMed] [Google Scholar]
- McNeil SE. Evaluation of nanomedicines: stick to the basics. Nature Reviews Materials. 2016;1(10):16073. http://doi.org/10.1038/natrevmats.2016.73. [Google Scholar]
- Mizrahy S, Peer D. Polysaccharides as building blocks for nanotherapeutics. Chem Soc Rev. 2012;41(7):2623–2640. doi: 10.1039/c1cs15239d. http://doi.org/10.1039/C1CS15239D. [DOI] [PubMed] [Google Scholar]
- Moghimi SM, Farhangrazi ZS. Just so stories: The random acts of anti-cancer nanomedicine performance. Nanomedicine: Nanotechnology, Biology, and Medicine. 2014;10(8):1661–1666. doi: 10.1016/j.nano.2014.04.011. http://doi.org/10.1016/j.nano.2014.04.011. [DOI] [PubMed] [Google Scholar]
- Mu Q, Jiang G, Chen L, Zhou H, Fourches D, Tropsha A, Yan B. Chemical basis of interactions between engineered nanoparticles and biological systems. Chemical Reviews. 2014;114(15):7740–7781. doi: 10.1021/cr400295a. http://doi.org/10.1021/cr400295a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngwuluka NC, Ochekpe NA, Aruoma OI. Naturapolyceutics: The science of utilizing natural polymers for drug delivery. Polymers. 2014;6(5):1312–1332. http://doi.org/10.3390/polym6051312. [Google Scholar]
- Patel S, Lavasanifar A, Choi P. Application of Molecular Dynamics Simulation To Predict the Compatability between Water-Insoluble Drugs and Self-Associating Poly(ethylene oxide)- b -poly(ε-caprolactone) Block Copolymers. Biomacromolecules. 2008;9(11):3014–3023. doi: 10.1021/bm800320z. http://doi.org/10.1021/bm800320z. [DOI] [PubMed] [Google Scholar]
- Peppas NA. Historical perspective on advanced drug delivery: How engineering design and mathematical modeling helped the field mature. Advanced Drug Delivery Reviews. 2013;65(1):5–9. doi: 10.1016/j.addr.2012.09.040. http://doi.org/10.1016/j.addr.2012.09.040. [DOI] [PubMed] [Google Scholar]
- Peppas NA, Narasimhan B. Mathematical models in drug delivery: How modeling has shaped the way we design new drug delivery systems. Journal of Controlled Release. 2014;190:75–81. doi: 10.1016/j.jconrel.2014.06.041. http://doi.org/10.1016/j.jconrel.2014.06.041. [DOI] [PubMed] [Google Scholar]
- Plazinski W, Lonardi A, Hünenberger PH. Revision of the GROMOS 56A6(CARBO) force field: Improving the description of ring-conformational equilibria in hexopyranose-based carbohydrates chains. Journal of Computational Chemistry. 2016;37(3):354–65. doi: 10.1002/jcc.24229. http://doi.org/10.1002/jcc.24229. [DOI] [PubMed] [Google Scholar]
- Pushpamalar J, Veeramachineni AK, Owh C, Loh XJ. Biodegradable Polysaccharides for Controlled Drug Delivery. ChemPlusChem. 2016;81(6):504–514. doi: 10.1002/cplu.201600112. http://doi.org/10.1002/cplu.201600112. [DOI] [PubMed] [Google Scholar]
- Ramezanpour M, Leung SSW, Delgado-Magnero KH, Bashe BYM, Thewalt J, Tieleman DP. Computational and experimental approaches for Investigating Nanoparticle-Based Drug Delivery Systems. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2016;1858(7):1688–1709. doi: 10.1016/j.bbamem.2016.02.028. http://doi.org/10.1016/j.bbamem.2016.02.028. [DOI] [PubMed] [Google Scholar]
- Rao JP, Geckeler KE. Polymer nanoparticles: Preparation techniques and size-control parameters. Progress in Polymer Science. 2011;36(7):887–913. http://doi.org/10.1016/j.progpolymsci.2011.01.001. [Google Scholar]
- Rostamizadeh K, Vahedpour M, Bozorgi S. Synthesis, characterization and evaluation of computationally designed nanoparticles of molecular imprinted polymers as drug delivery systems. International Journal of Pharmaceutics. 2012;424(1–2):67–75. doi: 10.1016/j.ijpharm.2011.12.054. http://doi.org/10.1016/j.ijpharm.2011.12.054. [DOI] [PubMed] [Google Scholar]
- Satalkar P, Elger BS, Hunziker P, Shaw D. Challenges of clinical translation in nanomedicine: A qualitative study. Nanomedicine: Nanotechnology, Biology, and Medicine. 2016;12(4):893–900. doi: 10.1016/j.nano.2015.12.376. http://doi.org/10.1016/j.nano.2015.12.376. [DOI] [PubMed] [Google Scholar]
- Schütz CA, Juillerat-Jeanneret L, Mueller H, Lynch I, Riediker M. Therapeutic nanoparticles in clinics and under clinical evaluation. Nanomedicine. 2013;8(3):449–467. doi: 10.2217/nnm.13.8. http://doi.org/10.2217/nnm.13.8. [DOI] [PubMed] [Google Scholar]
- Soleymani Abyaneh H, Vakili MR, Zhang F, Choi P, Lavasanifar A. Rational design of block copolymer micelles to control burst drug release at a nanoscale dimension. Acta Biomaterialia. 2015;24:127–139. doi: 10.1016/j.actbio.2015.06.017. http://doi.org/10.1016/j.actbio.2015.06.017. [DOI] [PubMed] [Google Scholar]
- Such GK, Yan Y, Johnston APR, Gunawan ST, Caruso F. Interfacing Materials Science and Biology for Drug Carrier Design. Advanced Materials. 2015;27(14):2278–2297. doi: 10.1002/adma.201405084. http://doi.org/10.1002/adma.201405084. [DOI] [PubMed] [Google Scholar]
- Sun Y, Mao S. Computational approach to the design of hyaluronic acid copolymer based doxorubicin micelles. Asian Journal of Pharmaceutical Sciences. 2016;11(1):62–63. http://doi.org/10.1016/j.ajps.2015.10.045. [Google Scholar]
- Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC. GROMACS: Fast, flexible, and free. Journal of Computational Chemistry. 2005;26(16):1701–1718. doi: 10.1002/jcc.20291. http://doi.org/10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- Wicki A, Witzigmann D, Balasubramanian V, Huwyler J. Nanomedicine in cancer therapy: Challenges, opportunities, and clinical applications. Journal of Controlled Release. 2015;200:138–157. doi: 10.1016/j.jconrel.2014.12.030. http://doi.org/10.1016/j.jconrel.2014.12.030. [DOI] [PubMed] [Google Scholar]
- Wilhelm S, Tavares AJ, Dai Q, Ohta S, Audet J, Dvorak HF, Chan WCW. Analysis of nanoparticle delivery to tumours. Nature Reviews Materials. 2016;1(5):16014. http://doi.org/10.1038/natrevmats.2016.14. [Google Scholar]
- Yan LT, Xie XM. Computational modeling and simulation of nanoparticle self-assembly in polymeric systems: Structures, properties and external field effects. Progress in Polymer Science. 2013;38(2):369–405. http://doi.org/10.1016/j.progpolymsci.2012.05.001. [Google Scholar]
- Yang J, Han S, Zheng H, Dong H, Liu J. Preparation and application of micro/nanoparticles based on natural polysaccharides. Carbohydrate Polymers. 2015;123:53–66. doi: 10.1016/j.carbpol.2015.01.029. http://doi.org/10.1016/j.carbpol.2015.01.029. [DOI] [PubMed] [Google Scholar]
- Youm I, Yang XY, Murowchick JB, Youan BBC. Encapsulation of docetaxel in oily core polyester nanocapsule intended for breast cancer therapy. Nanoscale Research Letters. 2011;6(1):630. doi: 10.1186/1556-276X-6-630. http://doi.org/10.1186/1556-276X-6-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Wardwell PR, Bader RA. Polysaccharide-based micelles for drug delivery. Pharmaceutics. 2013;5(2):329–352. doi: 10.3390/pharmaceutics5020329. http://doi.org/10.3390/pharmaceutics5020329. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.