Abstract
Filarial infections of humans cause some of the most important neglected tropical diseases. The global efforts for eliminating filarial infections by mass drug administration programs may require additional tools (safe macrofilaricidal drugs, vaccines and diagnostic biomarkers). The accurate and sensitive detection of viable parasites is essential for diagnosis and for surveillance programs. Current community-wide treatment modalities do not kill the adult filarial worms effectively; hence, there is a need to identify and develop safe macrofilaricidal drugs. High-throughput sequencing, mass spectroscopy methods and advances in computational biology have greatly accelerated the discovery process. Here, we describe post-genomic developments toward the identification of diagnostic biomarkers and drug targets for the filarial infection of humans.
Recent advances in filarial infections
Parasitic nematodes have a significant impact on human and animal health caused primarily by filarial worms, the common roundworm, hookworms, whipworms and others [1]. Among the eight filarial infections of humans, those caused by Brugia malayi, Wuchereria bancrofti, Loa loa and Onchocerca volvulus are responsible for most of the filarial disease burden. The life cycles of all of the filarial parasites are similar, and each involves an intermediate vector host and long-lived adult parasites, that, depending on the species involved, reside in connective tissues (O. volvulus), the lymphatics (Brugia spp, W. bancrofti) or subcutaneous tissues (L. loa). These adults release microfilariae that circulate in the peripheral circulation (most species) or in the dermal or ocular tissues (O. volvulus) (Figure 1). The human filarial worms (except L. loa and some species of Mansonella) harbor an endosymbiotic bacterium Wolbachia that can affect the viability and sterility of the worms [2].
Figure 1. Life cycle of filarial parasites.
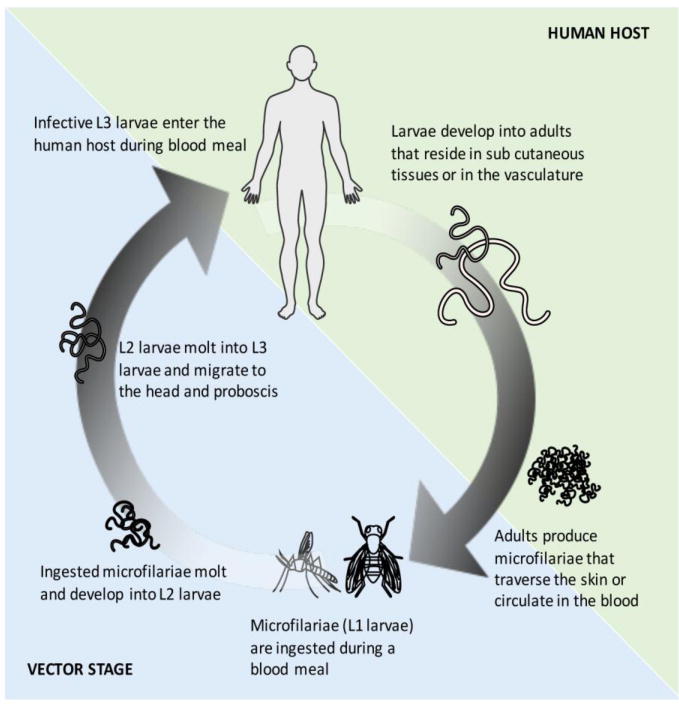
The human filarial parasites are digenetic with a primary human host and an intermediate vector stage. Infection occurs during the blood meal when the infective L3 larvae enter the human host. The larvae develop into adults that reside in subcutaneous tissue or vasculature (blood or lymph). The microfilariae produced by the adults are found in the skin or in the peripheral circulation from which they can be picked up by the vector during a blood meal. In the vector, the ingested microfilariae develop into infective L3 larvae that migrate to the head and proboscis.
During the past decade, advances in sequencing technology, mass spectroscopy as well bioinformatic capabilities have led to a major increase in the number of helminth genomes available. High quality reference genomes provide a platform to investigate the underlying biological makeup and processes by enabling transcriptomics, proteomics, glycomics, metabolomics and other specialized ‘omic’ technologies (Table 1).
Table 1. Post-genomic human filarial datasets.
| Molecule | Life-stages / Samples | Wolbachia | Platform | Accession | Ref | |
|---|---|---|---|---|---|---|
| Brugia malayi | mRNA | Egg; MF; L3; L4; AF; AM | Illumina Genome Analyzer IIx | E-MTAB-811 | [46] | |
| Intra-mosquito host | Illumina Genome Analyzer IIx | GSE53664 | [111] | |||
| 20-Hydroxyecdysone treated AF | Illumina MiSeq | SRP064921 | [112] | |||
| 16-days(L4), 30-,42-, and 120-days post infection | Yes | Illumina HiSeq | SRP090644 | [113] | ||
| MF and MF post-ingestion | Microarray | GSE15017 | [114] | |||
| L3 and L3 post-infection | Microarray | www.nematode.net | [115] | |||
| AM; AF | Microarray | www.nematode.net | [116, 117] | |||
| Doxycycline treated AF | Yes | Microarray | GSE34976 | [118] | ||
| Tetracycline treated adults | Microarray | E-MEXP-2185 | [119] | |||
| AM; MF | Yes | Illumina NextSeq | GSE93139 | [120] | ||
| AF and MF exposed to Heme | Illumina Genome Analyzer IIx | [121] | ||||
| AF exposed to bacteria | Illumina MiSeq | [122] | ||||
| Exosomes | Illumina MiSeq | [123] | ||||
| AF exposed to Flubendazole | Illumina MiSeq | [124] | ||||
| AF exposed to Ivermectin | Illumina MiSeq | GSE75341 | [125] | |||
| Small RNA | Adults | Illumina MiSeq | GSE56651 | [126] | ||
| Exosomes | Illumina MiSeq | [123] | ||||
| AM; AF; MF | Illumina Genome Analyzer IIx | [76] | ||||
| AM; AF; MF | Multiple sequences | [77] | ||||
| Protein | ES proteins from Adults | 2-DE/MALDI-TOF/LC-MS/MS | [50] | |||
| ES proteins from AM; AF; MF | SDS-PAGE/LC-MS/MS | [49] | ||||
| ES proteins from AM; AF; MF; L3; molting L3 | Yes | LC-MS/MS | [51] | |||
| Somatic proteins of L3; MF; UTMF; AF; AM | Yes | LC-MS/MS | [48] | |||
| Body-wall, gut and uterine tissue of AF | Yes | LC-MS/MS | [52] | |||
| 20-Hydroxyecdysone treated AF | LC-MS/MS | [112] | ||||
| AM; AF | 2DE-MALDI-TOF | [98] | ||||
| Exosomes | LC-MS/MS | [123] | ||||
| Tetracycline treated AF | Yes | 2-DE/LC-MS/MS | [127] | |||
| Meta-genomics | Metabolic potential of nematodes | [80] | ||||
| Loa Loa | mRNA | MF | PRJNA60555 | [13] | ||
| Small RNA | Circulating miRNA | [75] | ||||
| Protein | Urine of infected individuals | [44] | ||||
| Onchocerca volvulus | mRNA | Nodular MF; Skin MF; AF; AM; L3; in vitro derived L3 Day1; L3 Day3 | PRJEB2965 | [15, 21] | ||
| Small RNA | Circulating miRNA | [75] | ||||
| Circulating miRNA | [73] | |||||
| Circulating miRNA | [78] | |||||
| Protein | EMB; MF; AF; AM; L3; in vitro derived L3 Day1; L3 Day2; L3 Day3; L4 | Yes | PXD003585 | [21] | ||
| Plasma of infected individuals | [128] | |||||
| Proteome-wide peptide array | [53] |
AF: Adult Female; AM: Adult Male; MF: Microfilariae; L3 and L4: larval stages; EMB: Embryos; UTMF: Uterine Microfilaria; ES: Excretory-secretory proteins. “Yes” in Wolbachia column denotes transcriptome or proteome of Wolbachia was analyzed. Empty fields denote either not applicable or data not available.
While the current anti-filarial drugs have variable efficacy against the microfilariae (ivermectin (IVM) and diethylcarbamazine (DEC)) or against the adults (albendazole (ALB), DEC, and doxycycline), depending on their mode of action and stage-specificity, they must often be administered once or twice annually for up to 20 years for ivermectin-based MDA for onchocerciasis and for 5-8 years for the standard two-drug regimen of ALB/DEC or IVM/ALB used for lymphatic filariasis elimination programs [3, 4]. Moreover, the development of drug resistance [5, 6] or the induction of severe adverse events following treatment [7-9], or contraindications for DEC in areas endemic for onchocerciasis drives the need for the identification and development of alternative therapeutics, along with sensitive diagnostic tools to assess for efficacy of these alternative treatment strategies [10].
Early genomic studies of the filarial parasites used expressed sequence tags (ESTs) derived from stage-specific cDNA libraries to drive the filarial genome project [11]. Following the elucidation of the first filarial genome (B. malayi) [12], the draft genomes of the human filarial parasites L. loa [13, 14], W. bancrofti [13], O. volvulus [15] and the non-human filarial parasites Brugia pahangi [16], Dirofilaria immitis [17] and Onchocerca ochengi [15] became available. Recent reviews highlight the advances and insights gained from the comparative analyses of the genomes, transcriptomes and proteomes of these filarial nematodes [18, 19].
Although the genomic revolution has been touted as a means of driving vaccine target discovery for filarial parasites, to date, there is currently no vaccine against any of the filarial infections [20], although a number of potential vaccine candidates have been identified for O. volvulus using an “immunonomic” approach [21] or immunoinformatics [22]. Here, we review the implications of the post-genomic developments towards the identification of novel diagnostics and additional drug targets in human filarial infections.
Diagnostic toolbox
The diagnostic toolbox for filarial infections is fairly limited and, until nucleic acid detection and recombinant antigen production became possible, largely relied on classic parasitological methods [23] or serology based on the use of crude parasite extracts [24]. With the advent of recombinant antigen production, polymerase chain reaction (and other methods of nucleic acid detection), based mainly on pre-genomic information [25, 26], sensitive and species-specific diagnostics became available [27-34].
Post-Genomic Protein Biomarker Discovery
When the draft genomes of B. malayi, W. bancrofti, O. volvulus and L. loa became available, they were used quite successfully to identify species-specific antigens [12-15]. An example of this was the identification of Wb123 as a marker of early W. bancrofti infection [35] that was subsequently used for surveillance efforts following mass drug administration (MDA) [36-43]. Nevertheless, there remained a need for new tools with even greater specificity and sensitivity, especially in areas where multiple filarial species were co-endemic.
Having the genome of L. loa allowed for comparative proteomic analysis of both urine and sera from Loa-infected individuals that resulted in the identification of novel proteins (antigens) that could be detected in infected individuals. Among several Loa-derived proteins detected using mass spectrometry and genome-based identification in body fluids of infected patients, we were able to identify LOAG_16297 as a potential biomarker for L. loa microfilarial loads in infected individuals [44]. Likewise, the use of microfilarial RNA-seq data and comparative bioinformatics led to the identification of LOAG_14221 as another quantitative biomarker for Loa-microfilarial density [45].
The genomes of filarial parasites exhibit a range (12,000 – 15,000) of protein-coding genes that can also be found in multiple proteoforms as a result of polymorphisms, post-translational modifications and alternative splicing events [13, 15, 46, 47]. For most genomes (including the filarial genomes), there is a large proportion of uncharacterized or hypothetical genes that have orthologues in other related species. Clearly, most of these hypothetical proteins, whether conserved or unique, are detectable as proteins [21, 48-52] and could be useful biomarkers of infection [21]. The proteins (in the thousands) can be gridded on to high-density arrays. High-throughput protein microarrays, though currently quite expensive, have become a valuable tool in translational research towards discovering and validating potential biomarkers and identifying vaccine candidates. The availability of different protein array formats (protein fragment, peptide, full-length) provides platforms for different queries [53]. For example, the sero-reactivity to a peptide (on a peptide-array) can be MHC- restricted and thus not be recognized by antibody, whereas the full-length protein containing multiple B cell epitopes would be more suitable for protein arrays.
Post-Genomic Nucleotide-based Biomarkers
Nucleic acid-based tools often offer better sensitivity and specificity than parasitological or serological methods. In the peri-genomic era, these nucleic acid-based tools were identified empirically [54-56], and were primarily based on the use of interspersed repeats and internal transcribed spacer sequences [57-62] as genetic markers. Strategies have been designed to utilize qPCR and loop-mediated isothermal amplification (LAMP) methods for the detection of parasite DNA [25]. However, they are not yet applicable in the field and often not deployable in low-resource settings. Isothermal amplification methods circumvent the need for expensive and bulky instrumentation, requiring a simple heat block or water bath. Among the various isothermal-based technologies, loop-mediated isothermal amplification (LAMP) methods have been reported for a variety of filariae in humans [63-67] or in the vectors [66, 68, 69].
The nucleic-acid based tools in the post-genomic era have been largely focused on small RNAs [70, 71]. Among the various small RNAs, microRNAs (miRNA) have been garnering increasing interest not only as important regulators of nematode development but also as diagnostic markers of infection [72-77]. The detection of miRNAs and their potential for use as biomarkers is influenced by: i) sensitivities in depth of coverage, ii) miRNA identification methodology employed, iii) niche of the parasite (blood versus other tissues), and iv) the nature of the miRNA (conserved or unique) itself. For example, while the detection of heartworm specific miRNAs in Dirofilaria-infected (but not from uninfected) dogs demonstrated the utility and specificity of miRNAs [74], the detection of conserved miRNAs in the serum of O. volvulus-infected individuals [73] was not uniform across populations from differing geographic locations [75, 76].
A recent study [78] points out the feasibility and diagnostic power of using plasma-derived miRNA as diagnostic biomarkers. Among the suggested ways to optimize the detection of miRNAs were to use: a) a higher volume of plasma; b) a pre-amplification step to boost sensitivity; c) miRNAs from fractions enriched in exosomes. While each of these options are likely to enhance detection, the practical feasibility of handling larger volumes of plasma and/or purification of exosomes in the field or even in a centralized regional laboratory need to be considered. This is especially important because small amounts of parasite-derived miRNAs were detected in plasma in individuals infected with the skin-tissue dwelling O. volvulus, when compared to individuals infected with blood-tissue dwelling L. loa, W. bancrofti or B. malayi [74]. Nevertheless, adopting small RNA-based biomarkers for inexpensive assays for applications in the field or at point-of-care still requires technological advancements.
Post-Genomic Metabolite-based Biomarkers
Prior to the genomic era, metabolite mapping was used to understand the composition of the filarial worms [79]. Metabolic reconstructions at the genome-level utilizing flux balance analysis (FBA) of the genomes of B. malayi, L. loa, O. volvulus and the relative contribution of Wolbachia highlighted potential metabolic chokepoints [15, 80]. Metabolomic approaches identified urine-derived N-acetyltyramine-O-b-glucuronide (NATOG) as a potential biomarker for O. volvulus [81], though the utility of NATOG measurements is somewhat controversial [82, 83].
Using a slightly different approach, metabolite profiling of O. volvulus-infected individuals resulted in the identification of many unknown metabolites as potential biomarkers of infection [84]. Because metabolite profiling of the parasites is largely limited by availability of live worms, it would be important to leverage the metabolic composition identified empirically to fill the holes in the metabolic maps based on genomic information. Furthermore, because filarial worms source their nutrients from their host(s) or from the Wolbachia endosymbiont, the significantly depleted levels of metabolites in the infected individuals suggests the likely dependence of the filarial worms on specific metabolites [84].
Therapeutic targets
Current control measures, as part of the MDA campaigns, are largely based on annual or bi-annual distribution of ivermectin (IVM) (onchocerciasis endemic areas) or diethylcarbamazine citrate (DEC; areas non-endemic of onchocerciasis) either alone or in combination with albendazole. At the individual patient level, a number of drugs, including DEC, albendazole, ivermectin, and doxycycline, are in use and have demonstrated efficacy against a number of the filarial infections [85, 86]. However, there have been substantial efforts in the past 10 years to identify more effective macrofilaricides [87]. Although biochemical extracts from medicinal plants have been explored for bioactive compounds with filaricidal activity [88], current efforts are largely focused on repurposing approved drugs that could accelerate more effective therapies.
Post-Genomic Drug Targets
Comparative genomics of the parasite(s) and human host provide a dataset of potential targets. The process of comparing the genomes and their inferred biochemical pathways yields features that are evolutionarily conserved and features that are divergent between organisms. Currently, considering the logistics and length of time between drug discovery and market introduction, the use of existing natural, synthetic or semi-synthetic compounds with known target and safety profiles are being identified and explored as potential alternatives. When targeting metabolic networks present in both the parasite and the host, selectivity for the parasite is the most important property.
Wolbachia appear to be essential for the development and survival of the filarial parasites that harbor them. Depletion of Wolbachia by antibiotics (e.g., the tetracyclines) disrupts embryogenesis, microfilariae (mf) development and worm survival [2], but, despite the loss of Wolbachia, filarial worms can survive for long periods of time [89]. This longevity, the length of time needed for antibiotic treatment (4-6 weeks) and contraindications in children under 8 years of age and in pregnant women, has led to the search for novel anti-Wolbachia agents [90]. Repurposing drugs such as minocycline [91] and rifampicin [92] as anti-Wolbachia therapy could potentially reduce the treatment duration from 6 weeks to 1-2 weeks, allowing for more ease in delivering these regimens on a community-wide basis.
Post-Genomic Drug Targets: High-Throughput Screens
Using a yeast-based model, screening of 400 drugs available from the Malaria Box project [93] yielded filarial-specific active compounds that affected adult females of B. pahangi (used as surrogate for human infections) in vitro and did not affect the corresponding human homologues [94]. Repurposing of approved drugs also led to the identification of auranofin, approved for the treatment of rheumatoid arthritis, as a lead candidate for treating lymphatic filariasis and onchocerciasis, whose likely target is thioredoxin reductase (TrxR) [95]. It is likely that auranofin treatment renders the parasite susceptible to oxidative damage allowing for subsequent clearance of the parasite by the host immune system. Likewise, screening for developmental inhibitors of filarial parasites identified closantel, a known anthelmintic drug that acts like a proton ionophore, and targets L3-expressed chitinase [96, 97]. Further, proteomic studies resulted in the identification of 62 gender-associated proteins expressed during embryogenesis or spermatogenesis [98]. Most of these gender-associated proteins have homologues in C. elegans with severe RNAi phenotypes and, hence, might be targets for new drugs or vaccines. High-throughput drug screening for repurposing approved drugs targeting Wolbachia yielded effective compounds from the tetracycline, fluoroquinolone and rifamycin classes [99].
Post-Genomic Drug Targets: Genome-wide Screens
Predictive genome-wide screening of drugs approved for human use with the potential use as anthelmintics led to the identification of 16 O. volvulus proteins (predominantly enzymes and proteins involved in ion transport and neurotransmission) as likely targets [15]. Information gained from the filarial genome projects identified other protein kinases in the filarial genomes —EGFR, Src, Raf/Raf, FRAP and AGC/DMPK/ROCK —for which there are orally available small molecule inhibitors on the commercial market [13]. Aside from kinases, other targets found based on genome mining of the O. volvulus genome, that link to FDA-approved drugs with likely activity include well-known and commonly used drugs such as metformin, baclofen, acetaminophen, and sertraline [15].
Proteins that are unique to nematodes and sufficiently distinct from homologous human proteins are particularly attractive as drug targets. Comparison of filarial and non-filarial nematodes revealed the presence of filarial-nematode specific kinases which could be targeted using existing approved drugs for human use. Of the 205 conventional and 10 atypical protein kinases encoded in the B. malayi genome, 142 are deemed essential based on the RNAi phenotype in C. elegans [12]. Similarly, 168 kinases of O. volvulus have no significant human matches [15]. One of the post-genomic insights from L. loa genome was the existence of a tyrosine kinase c-Abl like protein in filarial worms. Moving from a theoretical basis, the applicability of repurposing drugs based on L. loa genome, was tested with the tyrosine kinase inhibitors (TKI) imatinib, nilotinib and dasatinib [100]. The TKIs were able to effectively affect all life-stages of B. malayi at concentrations that are physiologically achievable. Given the structural similarity of these c-Abl like proteins in the filarial nematodes, and the conserved binding site of the TKI, it is likely to be successful in targeting the parasites that cause lymphatic filariasis, loiasis and onchocerciasis [100]. Because the expression of c-abl-like protein localized predominantly to the reproductive organs, muscle and intestine of the adult B. malayi worms, an effort to perform clinical trials in humans (e.g., NCT02644525) with drugs such as these TKIs that are effective as both micro- and macrofilaricides might greatly shorten the length of MDA treatment and significantly boost elimination efforts [101].
The availability of stage-specific transcriptional and proteomic data, coupled to comparative genomics identified potential genes and pathways of Wolbachia that can be targeted. This focus has largely arisen from work on the metabolic pathways of the parasites [102] that are perceived to be complemented by the Wolbachia-containing filarial parasite and its endosymbiont [15]. However, given that the genetic makeup of L. loa (filarial parasite devoid of Wolbachia) is the same as other filarial worms, the nature of the metabolic provisioning in the symbiotic relationship remains enigmatic [13]. Further, the observed heterogeneity and extreme variations in Wolbachia copy numbers within and between populations in O. volvulus raises the issue of how metabolic provisioning by Wolbachia is balanced, and a complicating factor in finding an efficacious antibiotic for MDA programs [103]. Another genome-based approach relies on the prediction of essential Wolbachia genes that have experimental evidence or verified genes in other bacterial taxa and phylogenetically conserved in Rickettsiales [104]. Incidentally, many of the potential inhibitors identified by the A-WOL screens target these essential genes/gene products [105].
From the endosymbiont perspective, Filobase [106] — a database derived from post-genomic insights from B. malayi and its Wolbachia endosymbiont (wBm) using a combination of bioinformatic tools, pathway analyses and data mining -- provides a list of essential proteins that could be potential drug-targets for the parasites that cause lymphatic filariasis. This approach, however, requires de novo drug development, a time consuming and costly endeavor with uncertainty about safety.
Based on a curated set of ‘essential genes’ across bacteria, archaea and eukaryotes [107] and sequence homology to humans, a subset of B. malayi predicted genes have been postulated as potential drug targets [108]. This strategy is, however, limited to genes that exhibit sequence homology with genes of known functional characteristics; they are not applicable to the gene families restricted to filarial parasites that are probably essential [21, 46, 48].
Concluding Remarks
Comparative genomic and post-genomic investigations have provided a vast amount of molecular information. Through these studies, sets of genes, proteins or critical pathways have been identified and can be further exploited to develop improved diagnostic and therapeutic tools. Moreover, the availability of stage-specific expression data allows for the identification of parasite-derived biomarkers that can reflect different periods of parasite development. These in turn can be exploited for various point-of-care tools that may be needed for support of intervention strategies aimed at control and elimination of filarial infections. For example, we now have the ability to identify markers of: 1) early infection or of recrudescence (L3-L4 based); 2) active patent infection (adults and microfilariae based); or 3) ongoing transmission (vector stages). Whatever the approach, good point-of-care tools are needed for aiding and evaluating the priorities towards intervention strategies aimed at control and elimination of filarial infections [109, 110].
Rational design-based approaches toward the identification of biomarkers and toward the development of rapid format assays is now of primary importance if the goals of the MDA programs are to be achieved. Moreover, it is important to have the political will to provide to the global health community the resources needed for testing and validating these biomarkers and therapeutic targets along with models that can be used to predict drug efficacy in human infection (see Outstanding Questions).
Trends.
Current diagnostic methods are not sufficient to reliably detect active filarial infection once transmission is controlled.
Accurate detection of viable filarial parasites requires diagnostic tools with better specificity and sensitivity to detect low to very low-level infections.
Diagnostic tools should preferentially be point-of-care, be able to distinguish among related filarial species or detect the presence of multiple infections.
Current treatment strategies are not adequately effective against the adult filarial worms. Triple drug therapy looks promising.
Identification and development of novel and safe macrofilaricides is largely focused on repurposing existing approved drugs and those that target metabolic chokepoint reactions.
Outstanding Questions.
What is the in vivo efficacy of the FDA-approved drugs identified through post-genomic approaches in human filarial infection?
Can POC tests for filarial infections be made more sensitive and specific?
Can we develop an animal model to test drug efficacy reflective of human responses?
-
Can there be community wide resources such as:
banked material that includes sera/plasma/urine from filarial-infected individuals pre- and post-treatment
parasite material for genomics/transcriptomics for population biology and drug resistance studies
arrayed proteins or glycans or metabolites for screening for discovery and validation of promising biomarkers?
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hotez PJ, et al. Helminth infections: the great neglected tropical diseases. Journal of Clinical Investigation. 2008;118(4):1311–1321. doi: 10.1172/JCI34261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Foster J, et al. The molecular biology, immunology and chemotherapy of Wolbachia bacterial endosymbionts of filarial nematodes. In: Kennedy M, Harnett W, editors. Parasitic nematodes: Molecular Biology, Biochemistry and Immunology. CABI; 2013. [Google Scholar]
- 3.Fischer PU, et al. Potential Value of Triple Drug Therapy with Ivermectin, Diethylcarbamazine, and Albendazole (IDA) to Accelerate Elimination of Lymphatic Filariasis and Onchocerciasis in Africa. PLoS Negl Trop Dis. 2017;11(1):e0005163. doi: 10.1371/journal.pntd.0005163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thomsen EK, et al. Efficacy, Safety, and Pharmacokinetics of Coadministered Diethylcarbamazine, Albendazole, and Ivermectin for Treatment of Bancroftian Filariasis. Clin Infect Dis. 2016;62(3):334–341. doi: 10.1093/cid/civ882. [DOI] [PubMed] [Google Scholar]
- 5.Schwab AE, et al. Detection of benzimidazole resistance-associated mutations in the filarial nematode Wuchereria bancrofti and evidence for selection by albendazole and ivermectin combination treatment. Am J Trop Med Hyg. 2005;73(2):234–8. [PubMed] [Google Scholar]
- 6.Osei-Atweneboana MY, et al. Phenotypic evidence of emerging ivermectin resistance in Onchocerca volvulus. PLoS Negl Trop Dis. 2011;5(3):e998. doi: 10.1371/journal.pntd.0000998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boussinesq M, et al. Three probable cases of Loa loa encephalopathy following ivermectin treatment for onchocerciasis. Am J Trop Med Hyg. 1998;58(4):461–9. doi: 10.4269/ajtmh.1998.58.461. [DOI] [PubMed] [Google Scholar]
- 8.Chippaux JP, et al. Severe adverse reaction risks during mass treatment with ivermectin in loiasis-endemic areas. Parasitol Today. 1996;12(11):448–50. doi: 10.1016/0169-4758(96)40006-0. [DOI] [PubMed] [Google Scholar]
- 9.Gardon J, et al. Serious reactions after mass treatment of onchocerciasis with ivermectin in an area endemic for Loa loa infection. Lancet. 1997;350(9070):18–22. doi: 10.1016/S0140-6736(96)11094-1. [DOI] [PubMed] [Google Scholar]
- 10.WHO. Investing to overcome the global impact of neglected tropical diseases: Third WHO report on negelected tropical diseases 2016 [Google Scholar]
- 11.Williams SA. Deep within the filarial genome: progress of the filarial genome project. Parasitol Today. 1999;15(6):219–24. doi: 10.1016/s0169-4758(99)01454-4. [DOI] [PubMed] [Google Scholar]
- 12.Ghedin E, et al. Draft genome of the filarial nematode parasite Brugia malayi. Science. 2007;317(5845):1756–1760. doi: 10.1126/science.1145406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desjardins CA, et al. Genomics of Loa loa, a Wolbachia-free filarial parasite of humans. Nature Genetics. 2013;45(5):495–500. doi: 10.1038/ng.2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tallon LJ, et al. Single molecule sequencing and genome assembly of a clinical specimen of Loa loa, the causative agent of loiasis. BMC Genomics. 2014;15:788. doi: 10.1186/1471-2164-15-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cotton JA, et al. The genome of Onchocerca volvulus, agent of river blindness. Nat Microbiol. 2016;2:16216. doi: 10.1038/nmicrobiol.2016.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lau YL, et al. Draft genome of Brugia pahangi: high similarity between B. pahangi and B. malayi. Parasites & Vectors. 2015;8 doi: 10.1186/s13071-015-1064-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Godel C, et al. The genome of the heartworm, Dirofilaria immitis, reveals drug and vaccine targets. Faseb Journal. 2012;26(11):4650–4661. doi: 10.1096/fj.12-205096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grote A, et al. Lessons from the genomes and transcriptomes of filarial nematodes. Mol Biochem Parasitol. 2017;(17):S0166–6851. 30012–9. doi: 10.1016/j.molbiopara.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lustigman S, et al. The role of ‘omics’ in the quest to eliminate human filariasis. PLoS Negl Trop Dis. 2017;11(4):e0005464. doi: 10.1371/journal.pntd.0005464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morris CP, et al. A comprehensive, model-based review of vaccine and repeat infection trials for filariasis. Clin Microbiol Rev. 2013;26(3):381–421. doi: 10.1128/CMR.00002-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bennuru S, et al. Stage-Specific Transcriptome and Proteome Analyses of the Filarial Parasite Onchocerca volvulus and Its Wolbachia Endosymbiont. MBio. 2016;7(6) doi: 10.1128/mBio.02028-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Terry FE, et al. Time for T? Immunoinformatics addresses vaccine design for neglected tropical and emerging infectious diseases. Expert Rev Vaccines. 2015;14(1):21–35. doi: 10.1586/14760584.2015.955478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eberhard ML, Lammie PJ. Laboratory diagnosis of filariasis. Clin Lab Med. 1991;11(4):977–1010. [PubMed] [Google Scholar]
- 24.Lal RB, et al. Circulating parasite antigen(s) in lymphatic filariasis: use of monoclonal antibodies to phosphocholine for immunodiagnosis. J Immunol. 1987;138(10):3454–60. [PubMed] [Google Scholar]
- 25.Alhassan A, et al. Expanding the MDx toolbox for filarial diagnosis and surveillance. Trends Parasitol. 2015;31(8):391–400. doi: 10.1016/j.pt.2015.04.006. [DOI] [PubMed] [Google Scholar]
- 26.McCarthy JS, et al. A research agenda for helminth diseases of humans: diagnostics for control and elimination programmes. PLoS Negl Trop Dis. 2012;6(4):e1601. doi: 10.1371/journal.pntd.0001601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lobos E, et al. An immunogenic Onchocerca volvulus antigen: a specific and early marker of infection. Science. 1991;251(5001):1603–5. doi: 10.1126/science.2011741. [DOI] [PubMed] [Google Scholar]
- 28.Weil GJ, et al. A rapid-format antibody card test for diagnosis of onchocerciasis. J Infect Dis. 2000;182(6):1796–9. doi: 10.1086/317629. [DOI] [PubMed] [Google Scholar]
- 29.Steel C, et al. Rapid Point-of-Contact Tool for Mapping and Integrated Surveillance of Wuchereria bancrofti and Onchocerca volvulus Infection. Clin Vaccine Immunol. 2015;22(8):896–901. doi: 10.1128/CVI.00227-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lipner EM, et al. Field applicability of a rapid-format anti-Ov-16 antibody test for the assessment of onchocerciasis control measures in regions of endemicity. J Infect Dis. 2006;194(2):216–21. doi: 10.1086/505081. [DOI] [PubMed] [Google Scholar]
- 31.Lammie PJ, et al. Recombinant antigen-based antibody assays for the diagnosis and surveillance of lymphatic filariasis - a multicenter trial. Filaria J. 2004;3(1):9. doi: 10.1186/1475-2883-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klion AD, et al. Serum immunoglobulin G4 antibodies to the recombinant antigen, Ll-SXP-1, are highly specific for Loa loa infection. J Infect Dis. 2003;187(1):128–33. doi: 10.1086/345873. [DOI] [PubMed] [Google Scholar]
- 33.More SJ, Copeman DB. A highly specific and sensitive monoclonal antibody-based ELISA for the detection of circulating antigen in bancroftian filariasis. Trop Med Parasitol. 1990;41(4):403–6. [PubMed] [Google Scholar]
- 34.Weil GJ, Liftis F. Identification and partial characterization of a parasite antigen in sera from humans infected with Wuchereria bancrofti. J Immunol. 1987;138(9):3035–41. [PubMed] [Google Scholar]
- 35.Kubofcik J, et al. Identification of Wb123 as an early and specific marker of Wuchereria bancrofti infection. PLoS Negl Trop Dis. 2012;6(12):e1930. doi: 10.1371/journal.pntd.0001930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steel C, et al. Rapid Wuchereria bancrofti-specific antigen Wb123-based IgG4 immunoassays as tools for surveillance following mass drug administration programs on lymphatic filariasis. Clin Vaccine Immunol. 2013;20(8):1155–61. doi: 10.1128/CVI.00252-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harris JR, Wiegand RE. Detecting infection hotspots: Modeling the surveillance challenge for elimination of lymphatic filariasis. PLoS Negl Trop Dis. 2017;11(5):e0005610. doi: 10.1371/journal.pntd.0005610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilson NO, et al. Evaluation of Lymphatic Filariasis and Onchocerciasis in Three Senegalese Districts Treated for Onchocerciasis with Ivermectin. PLoS Negl Trop Dis. 2016;10(12):e0005198. doi: 10.1371/journal.pntd.0005198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coulibaly YI, et al. Dynamics of antigenemia and transmission intensity of Wuchereria bancrofti following cessation of mass drug administration in a formerly highly endemic region of Mali. Parasit Vectors. 2016;9(1):628. doi: 10.1186/s13071-016-1911-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lau CL, et al. Lymphatic Filariasis Elimination in American Samoa: Evaluation of Molecular Xenomonitoring as a Surveillance Tool in the Endgame. PLoS Negl Trop Dis. 2016;10(11):e0005108. doi: 10.1371/journal.pntd.0005108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moss DM, et al. Serological Responses to Filarial Antigens in Malian Children Attending Elementary Schools. Am J Trop Med Hyg. 2017;96(1):229–232. doi: 10.4269/ajtmh.16-0560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lau CL, et al. Seroprevalence and spatial epidemiology of Lymphatic Filariasis in American Samoa after successful mass drug administration. PLoS Negl Trop Dis. 2014;8(11):e3297. doi: 10.1371/journal.pntd.0003297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hamlin KL, et al. Longitudinal monitoring of the development of antifilarial antibodies and acquisition of Wuchereria bancrofti in a highly endemic area of Haiti. PLoS Negl Trop Dis. 2012;6(12):e1941. doi: 10.1371/journal.pntd.0001941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Drame PM, et al. Identification and Validation of Loa loa Microfilaria-Specific Biomarkers: a Rational Design Approach Using Proteomics and Novel Immunoassays. Mbio. 2016;7(1) doi: 10.1128/mBio.02132-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Drame PM, et al. Discovery of specific antigens that can predict microfilarial intensity in Loa loa infection. J Clin Microbiol. 2017 doi: 10.1128/JCM.00513-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Choi YJ, et al. A deep sequencing approach to comparatively analyze the transcriptome of lifecycle stages of the filarial worm, Brugia malayi. PLoS Negl Trop Dis. 2011;5(12):e1409. doi: 10.1371/journal.pntd.0001409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Choi YJ, et al. Genomic diversity in Onchocerca volvulus and its Wolbachia endosymbiont. Nat Microbiol. 2016;2:16207. doi: 10.1038/nmicrobiol.2016.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bennuru S, et al. Stage-specific proteomic expression patterns of the human filarial parasite Brugia malayi and its endosymbiont Wolbachia. Proc Natl Acad Sci U S A. 2011;108(23):9649–54. doi: 10.1073/pnas.1011481108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moreno Y, Geary TG. Stage- and gender-specific proteomic analysis of Brugia malayi excretory-secretory products. PLoS Negl Trop Dis. 2008;2(10):e326. doi: 10.1371/journal.pntd.0000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hewitson JP, et al. The secretome of the filarial parasite, Brugia malayi: proteomic profile of adult excretory-secretory products. Mol Biochem Parasitol. 2008;160(1):8–21. doi: 10.1016/j.molbiopara.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 51.Bennuru S, et al. Brugia malayi excreted/secreted proteins at the host/parasite interface: stage- and gender-specific proteomic profiling. PLoS Negl Trop Dis. 2009;3(4):e410. doi: 10.1371/journal.pntd.0000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morris CP, et al. A Proteomic Analysis of the Body Wall, Digestive Tract, and Reproductive Tract of Brugia malayi. PLoS Negl Trop Dis. 2015;9(9):e0004054. doi: 10.1371/journal.pntd.0004054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lagatie O, et al. Identification of three immunodominant motifs with atypical isotype profile scattered over the Onchocerca volvulus proteome. PLoS Negl Trop Dis. 2017;11(1):e0005330. doi: 10.1371/journal.pntd.0005330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Erttmann KD, et al. A DNA sequence specific for forest form Onchocerca volvulus. Nature. 1987;327(6121):415–7. doi: 10.1038/327415a0. [DOI] [PubMed] [Google Scholar]
- 55.Williams SA, et al. Species-specific oligonucleotide probes for the identification of human filarial parasites. Mol Biochem Parasitol. 1988;28(2):163–9. doi: 10.1016/0166-6851(88)90064-3. [DOI] [PubMed] [Google Scholar]
- 56.Klion AD, et al. Cloning and characterization of a species-specific repetitive DNA sequence from Loa loa. Mol Biochem Parasitol. 1991;45(2):297–305. doi: 10.1016/0166-6851(91)90098-q. [DOI] [PubMed] [Google Scholar]
- 57.Jimenez M, et al. Detection and discrimination of Loa loa, Mansonella perstans and Wuchereria bancrofti by PCR-RFLP and nested-PCR of ribosomal DNA ITS1 region. Exp Parasitol. 2011;127(1):282–6. doi: 10.1016/j.exppara.2010.06.019. [DOI] [PubMed] [Google Scholar]
- 58.Morales-Hojas R, et al. Characterisation of nuclear ribosomal DNA sequences from Onchocerca volvulus and Mansonella ozzardi (Nematoda: Filarioidea) and development of a PCR-based method for their detection in skin biopsies. Int J Parasitol. 2001;31(2):169–77. doi: 10.1016/s0020-7519(00)00156-9. [DOI] [PubMed] [Google Scholar]
- 59.Nuchprayoon S, et al. Detection and differentiation of filarial parasites by universal primers and polymerase chain reaction-restriction fragment length polymorphism analysis. Am J Trop Med Hyg. 2005;73(5):895–900. [PubMed] [Google Scholar]
- 60.Fischer P, et al. Detection of DNA of nocturnally periodic Brugia malayi in night and day blood samples by a polymerase chain reaction-ELISA-based method using an internal control DNA. Am J Trop Med Hyg. 2000;62(2):291–6. doi: 10.4269/ajtmh.2000.62.291. [DOI] [PubMed] [Google Scholar]
- 61.Rao RU, et al. Detection of Brugia parasite DNA in human blood by real-time PCR. J Clin Microbiol. 2006;44(11):3887–93. doi: 10.1128/JCM.00969-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rao RU, et al. A real-time PCR-based assay for detection of Wuchereria bancrofti DNA in blood and mosquitoes. Am J Trop Med Hyg. 2006;74(5):826–32. [PMC free article] [PubMed] [Google Scholar]
- 63.Fernandez-Soto P, et al. Development of a highly sensitive loop-mediated isothermal amplification (LAMP) method for the detection of Loa loa. PLoS One. 2014;9(4):e94664. doi: 10.1371/journal.pone.0094664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Drame PM, et al. Loop-mediated isothermal amplification for rapid and semiquantitative detection of Loa loa infection. J Clin Microbiol. 2014;52(6):2071–7. doi: 10.1128/JCM.00525-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Poole CB, et al. Diagnosis of brugian filariasis by loop-mediated isothermal amplification. PLoS Negl Trop Dis. 2012;6(12):e1948. doi: 10.1371/journal.pntd.0001948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Takagi H, et al. Development of loop-mediated isothermal amplification method for detecting Wuchereria bancrofti DNA in human blood and vector mosquitoes. Parasitol Int. 2011;60(4):493–7. doi: 10.1016/j.parint.2011.08.018. [DOI] [PubMed] [Google Scholar]
- 67.Poole CB, et al. Colorimetric tests for diagnosis of filarial infection and vector surveillance using non-instrumented nucleic acid loop-mediated isothermal amplification (NINA-LAMP) Plos One. 2017;12(2) doi: 10.1371/journal.pone.0169011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Alhassan A, et al. A simple isothermal DNA amplification method to screen black flies for Onchocerca volvulus infection. PLoS One. 2014;9(10):e108927. doi: 10.1371/journal.pone.0108927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kouassi BL, et al. Assessing the presence of Wuchereria bancrofti in vector and human populations from urban communities in Conakry, Guinea. Parasit Vectors. 2015;8:492. doi: 10.1186/s13071-015-1077-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Britton C, et al. Application of small RNA technology for improved control of parasitic helminths. Vet Parasitol. 2015;212(1-2):47–53. doi: 10.1016/j.vetpar.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Quintana JF, et al. Small RNAs and extracellular vesicles in filarial nematodes: From nematode development to diagnostics. Parasite Immunol. 2017;39(2) doi: 10.1111/pim.12395. [DOI] [PubMed] [Google Scholar]
- 72.Buck AH, et al. Exosomes secreted by nematode parasites transfer small RNAs to mammalian cells and modulate innate immunity. Nat Commun. 2014;5:5488. doi: 10.1038/ncomms6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Quintana JF, et al. Extracellular Onchocerca-derived small RNAs in host nodules and blood. Parasit Vectors. 2015;8:58. doi: 10.1186/s13071-015-0656-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tritten L, et al. Detection of circulating parasite-derived microRNAs in filarial infections. PLoS Negl Trop Dis. 2014;8(7):e2971. doi: 10.1371/journal.pntd.0002971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tritten L, et al. Loa loa and Onchocerca ochengi miRNAs detected in host circulation. Mol Biochem Parasitol. 2014;198(1):14–7. doi: 10.1016/j.molbiopara.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 76.Poole CB, et al. Diversity and expression of microRNAs in the filarial parasite, Brugia malayi. PLoS One. 2014;9(5):e96498. doi: 10.1371/journal.pone.0096498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Poole CB, et al. Cloning and bioinformatic identification of small RNAs in the filarial nematode, Brugia malayi. Mol Biochem Parasitol. 2010;169(2):87–94. doi: 10.1016/j.molbiopara.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 78.Lagatie O, et al. Plasma-derived parasitic microRNAs have insufficient concentrations to be used as diagnostic biomarker for detection of Onchocerca volvulus infection or treatment monitoring using LNA-based RT-qPCR. Parasitol Res. 2017;116(3):1013–1022. doi: 10.1007/s00436-017-5382-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shukla-Dave A, et al. Metabolite mapping of human filarial parasite, Brugia malayi with nuclear magnetic resonance. Magn Reson Imaging. 1999;17(10):1503–9. doi: 10.1016/s0730-725x(99)00091-0. [DOI] [PubMed] [Google Scholar]
- 80.Tyagi R, et al. Pan-phylum Comparison of Nematode Metabolic Potential. PLoS Negl Trop Dis. 2015;9(5):e0003788. doi: 10.1371/journal.pntd.0003788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Globisch D, et al. Onchocerca volvulus-neurotransmitter tyramine is a biomarker for river blindness. Proc Natl Acad Sci U S A. 2013;110(11):4218–23. doi: 10.1073/pnas.1221969110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lagatie O, et al. Evaluation of the diagnostic potential of urinary N-Acetyltyramine-O,beta-glucuronide (NATOG) as diagnostic biomarker for Onchocerca volvulus infection. Parasit Vectors. 2016;9(1):302. doi: 10.1186/s13071-016-1582-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Globisch D, et al. Validation of onchocerciasis biomarker N-acetyltyramine-Oglucuronide (NATOG) Bioorg Med Chem Lett. 2017 doi: 10.1016/j.bmcl.2017.05.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bennuru S, et al. Metabolite profiling of infection-associated metabolic markers of onchocerciasis. Mol Biochem Parasitol. 2017 doi: 10.1016/j.molbiopara.2017.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Melrose WD. Chemotherapy for lymphatic filariasis: progress but not perfection. Expert Rev Anti Infect Ther. 2003;1(4):571–7. doi: 10.1586/14787210.1.4.571. [DOI] [PubMed] [Google Scholar]
- 86.Geary TG. Are new anthelmintics needed to eliminate human helminthiases? Curr Opin Infect Dis. 2012;25(6):709–17. doi: 10.1097/QCO.0b013e328359f04a. [DOI] [PubMed] [Google Scholar]
- 87.Sharma OP, et al. Drug targets for lymphatic filariasis: a bioinformatics approach. J Vector Borne Dis. 2013;50(3):155–62. [PubMed] [Google Scholar]
- 88.Al-Abd NM, et al. Recent advances on the use of biochemical extracts as filaricidal agents. Evid Based Complement Alternat Med. 2013;2013:986573. doi: 10.1155/2013/986573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hoerauf A, et al. Doxycycline in the treatment of human onchocerciasis: Kinetics of Wolbachia endobacteria reduction and of inhibition of embryogenesis in female Onchocerca worms. Microbes Infect. 2003;5(4):261–73. doi: 10.1016/s1286-4579(03)00026-1. [DOI] [PubMed] [Google Scholar]
- 90.Taylor MJ, et al. Anti-Wolbachia drug discovery and development: safe macrofilaricides for onchocerciasis and lymphatic filariasis. Parasitology. 2014;141(1):119–127. doi: 10.1017/S0031182013001108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sharma R, et al. Minocycline as a re-purposed anti-Wolbachia macrofilaricide: superiority compared with doxycycline regimens in a murine infection model of human lymphatic filariasis. Sci Rep. 2016;6:23458. doi: 10.1038/srep23458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Aljayyoussi G, et al. Short-Course, High-Dose Rifampicin Achieves Wolbachia Depletion Predictive of Curative Outcomes in Preclinical Models of Lymphatic Filariasis and Onchocerciasis. Sci Rep. 2017;7(1):210. doi: 10.1038/s41598-017-00322-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Spangenberg T, et al. The open access malaria box: a drug discovery catalyst for neglected diseases. PLoS One. 2013;8(6):e62906. doi: 10.1371/journal.pone.0062906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bilsland E, et al. Yeast-Based High-Throughput Screens to Identify Novel Compounds Active against Brugia malayi. PLoS Negl Trop Dis. 2016;10(1):e0004401. doi: 10.1371/journal.pntd.0004401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bulman CA, et al. Repurposing auranofin as a lead candidate for treatment of lymphatic filariasis and onchocerciasis. PLoS Negl Trop Dis. 2015;9(2):e0003534. doi: 10.1371/journal.pntd.0003534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Garner AL, et al. Design, synthesis, and biological activities of closantel analogues: structural promiscuity and its impact on Onchocerca volvulus. J Med Chem. 2011;54(11):3963–72. doi: 10.1021/jm200364n. [DOI] [PubMed] [Google Scholar]
- 97.Gloeckner C, et al. Repositioning of an existing drug for the neglected tropical disease Onchocerciasis. Proc Natl Acad Sci U S A. 2010;107(8):3424–9. doi: 10.1073/pnas.0915125107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jiang DJ, et al. Multiplex proteomics analysis of gender-associated proteins in Brugia malayi. International Journal for Parasitology. 2012;42(9):841–850. doi: 10.1016/j.ijpara.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Johnston KL, et al. Repurposing of approved drugs from the human pharmacopoeia to target Wolbachia endosymbionts of onchocerciasis and lymphatic filariasis. Int J Parasitol Drugs Drug Resist. 2014;4(3):278–86. doi: 10.1016/j.ijpddr.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.O'Connell EM, et al. Targeting Filarial Abl-like Kinases: Orally Available, Food and Drug Administration-Approved Tyrosine Kinase Inhibitors Are Microfilaricidal and Macrofilaricidal. J Infect Dis. 2015;212(5):684–93. doi: 10.1093/infdis/jiv065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.O'Connell EM, et al. Defining the target and the effect of imatinib on the filarial c-Abl homologue. PLoS Negl Trop Dis. 2017;11(7):e0005690. doi: 10.1371/journal.pntd.0005690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mukherjee S, et al. Metabolic Inhibitors as Antiparasitic Drugs: Pharmacological, Biochemical and Molecular Perspectives. Curr Drug Metab. 2016;17(10):937–970. doi: 10.2174/1389200217666161004143152. [DOI] [PubMed] [Google Scholar]
- 103.Armoo S, et al. Significant heterogeneity in Wolbachia copy number within and between populations of Onchocerca volvulus. Parasit Vectors. 2017;10(1):188. doi: 10.1186/s13071-017-2126-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Holman AG, et al. Computational prediction of essential genes in an unculturable endosymbiotic bacterium, Wolbachia of Brugia malayi. BMC Microbiol. 2009;9:243. doi: 10.1186/1471-2180-9-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Johnston KL, et al. Overcoming the challenges of drug discovery for neglected tropical diseases: the A.WOL experience. J Biomol Screen. 2014;19(3):335–43. doi: 10.1177/1087057113511270. [DOI] [PubMed] [Google Scholar]
- 106.Sharma OP, Kumar MS. Essential proteins and possible therapeutic targets of Wolbachia endosymbiont and development of FiloBase--a comprehensive drug target database for Lymphatic filariasis. Sci Rep. 2016;6:19842. doi: 10.1038/srep19842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Luo H, et al. DEG 10, an update of the database of essential genes that includes both protein-coding genes and noncoding genomic elements. Nucleic Acids Res. 2014;42(Database issue):D574–80. doi: 10.1093/nar/gkt1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hande S, et al. Exploring apposite therapeutic target for apoptosis in filarial parasite: a plausible hypothesis. Med Hypotheses. 2014;82(3):356–61. doi: 10.1016/j.mehy.2013.12.023. [DOI] [PubMed] [Google Scholar]
- 109.Prichard RK, et al. A research agenda for helminth diseases of humans: intervention for control and elimination. PLoS Negl Trop Dis. 2012;6(4):e1549. doi: 10.1371/journal.pntd.0001549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Boatin BA, et al. A research agenda for helminth diseases of humans: towards control and elimination. PLoS Negl Trop Dis. 2012;6(4):e1547. doi: 10.1371/journal.pntd.0001547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Choi YJ, et al. Dual RNA-seq of parasite and host reveals gene expression dynamics during filarial worm-mosquito interactions. PLoS Negl Trop Dis. 2014;8(5):e2905. doi: 10.1371/journal.pntd.0002905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mhashilkar AS, et al. Phenotypic and molecular analysis of the effect of 20-hydroxyecdysone on the human filarial parasite Brugia malayi. Int J Parasitol. 2016;46(5-6):333–41. doi: 10.1016/j.ijpara.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Grote A, et al. Defining Brugia malayi and Wolbachia symbiosis by stage-specific dual RNA-seq. PLoS Negl Trop Dis. 2017;11(3):e0005357. doi: 10.1371/journal.pntd.0005357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Griffiths KG, et al. Use of microarray hybridization to identify Brugia genes involved in mosquito infectivity. Parasitol Res. 2009;106(1):227–35. doi: 10.1007/s00436-009-1655-y. [DOI] [PubMed] [Google Scholar]
- 115.Li BW, et al. Transcriptomes and pathways associated with infectivity, survival and immunogenicity in Brugia malayi L3. BMC Genomics. 2009;10:267. doi: 10.1186/1471-2164-10-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Li BW, et al. Gender-associated genes in filarial nematodes are important for reproduction and potential intervention targets. PLoS Negl Trop Dis. 2011;5(1):e947. doi: 10.1371/journal.pntd.0000947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Li BW, et al. Transcription profiling reveals stage- and function-dependent expression patterns in the filarial nematode Brugia malayi. BMC Genomics. 2012;13:184. doi: 10.1186/1471-2164-13-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Rao RU, et al. Effects of doxycycline on gene expression in Wolbachia and Brugia malayi adult female worms in vivo. J Biomed Sci. 2012;19:21. doi: 10.1186/1423-0127-19-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ghedin E, et al. Brugia malayi gene expression in response to the targeting of the Wolbachia endosymbiont by tetracycline treatment. PLoS Negl Trop Dis. 2009;3(10):e525. doi: 10.1371/journal.pntd.0000525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Luck AN, et al. Removing the needle from the haystack: Enrichment of Wolbachia endosymbiont transcripts from host nematode RNA by Cappable-seq. PLoS One. 2017;12(3):e0173186. doi: 10.1371/journal.pone.0173186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Luck AN, et al. Heme acquisition in the parasitic filarial nematode Brugia malayi. FASEB J. 2016;30(10):3501–3514. doi: 10.1096/fj.201600603R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Libro S, et al. Characterization of innate immunity genes in the parasitic nematode Brugia malayi. Symbiosis. 2016;68:145–155. doi: 10.1007/s13199-015-0374-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zamanian M, et al. Release of Small RNA-containing Exosome-like Vesicles from the Human Filarial Parasite Brugia malayi. PLoS Negl Trop Dis. 2015;9(9):e0004069. doi: 10.1371/journal.pntd.0004069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.O'Neill M, et al. Profiling the macrofilaricidal effects of flubendazole on adult female Brugia malayi using RNAseq. Int J Parasitol Drugs Drug Resist. 2016;6(3):288–296. doi: 10.1016/j.ijpddr.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ballesteros C, et al. The Effect of In Vitro Cultivation on the Transcriptome of Adult Brugia malayi. PLoS Negl Trop Dis. 2016;10(1):e0004311. doi: 10.1371/journal.pntd.0004311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sarkies P, et al. Ancient and novel small RNA pathways compensate for the loss of piRNAs in multiple independent nematode lineages. PLoS Biol. 2015;13(2):e1002061. doi: 10.1371/journal.pbio.1002061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Dangi A, et al. Tetracycline treatment targeting Wolbachia affects expression of an array of proteins in Brugia malayi parasite. Proteomics. 2009;9(17):4192–208. doi: 10.1002/pmic.200800324. [DOI] [PubMed] [Google Scholar]
- 128.McNulty SN, et al. An Integrated Multiomics Approach to Identify Candidate Antigens for Serodiagnosis of Human Onchocerciasis. Mol Cell Proteomics. 2015;14(12):3224–33. doi: 10.1074/mcp.M115.051953. [DOI] [PMC free article] [PubMed] [Google Scholar]


