Abstract
Phenolic compounds and their derivatives are ubiquitous constituents of numerous synthetic and natural chemicals that exist in the environment. Their toxicity is mostly attributed to their hydrophobicity and/or the formation of free radicals. In a continuation of the study of phenolic toxicity in a systematic manner, we have examined the biological responses of Saccharomyces cerevisiae to a series of mostly monosubstituted phenols utilizing a quantitative structure–activity relationship (QSAR) approach. The biological end points included a growth assay that determines the levels of growth inhibition induced by the phenols as well as a yeast deletion (DEL) assay that assesses the ability of X-phenols to induce DNA damage or DNA breaks. The QSAR analysis of cell growth patterns determined by IC50 and IC80 values indicates that toxicity is delineated by a hydrophobic, parabolic model. The DEL assay was then utilized to detect genomic deletions in yeast. The increase in the genotoxicity was enhanced by the electrophilicity of the phenolic substituents that were strong electron donors as well as by minimal hydrophobicity. The electrophilicities are represented by Brown’s sigma plus values that are a variant of the Hammett sigma constants. A few mutant strains of genes involved in DNA repair were separately exposed to 2,6-di-tert-butyl-4-methyl-phenol (BHT) and butylated hydroxy anisole (BHA). They were subsequently screened for growth phenotypes. BHA-induced growth defects in most of the DNA repair null mutant strains, whereas BHT was unresponsive.
Introduction
Phenolic compounds are ubiquitous in nature and in man-made products. Their role as antioxidants are well-documented, as seen in the case of food preservatives and flavonoids, whose mechanism of action involves a consecutive 2-electron oxidation of the β-ring.1−4 However, under certain conditions such as concentration levels, pH, and presence of metal ions, phenolics have been found to be pro-oxidants by generating an increase in the reactive oxygen species (ROS) in cancerous cells that triggers apoptotic DNA fragmentation.5 Phenol and other benzene-like compounds have been shown to cause statistically significant DNA damage,6,7 whereas other phenols have been shown to protect DNA from damage.7
Clearly, this mixed behavior of simple and complex phenols in eliciting cellular responses has not been clearly delineated and thus warrants further investigation in a systematic manner. In this study, the quantitative structure–activity relationship (QSAR) paradigm8 was used to delineate the biological responses of the yeast, Saccharomyces cerevisiae, in terms of the physicochemical attributes of a series of para-substituted phenolic compounds. Previous work in our laboratory had examined the cytotoxic and apoptotic effects of various mono-, di-, and trisubstituted phenols on mouse leukemia cells.9 Two parameters were found to be of critical importance in determining the overall cytotoxicity of electron-rich substituted phenols: their hydrophobicities and their electron densities.10−12 See eqs 1 and 2.
| 1 |
| 2 |
In these QSAR equations, ID50 represents the molar concentration of X-phenol that results in a 50% inhibition of growth in murine leukemia cells.12 σ+ is Brown’s refinement of the Hammett sigma (σ) constant, which represents the electron-releasing ability of the various substituents, whereas BDE reflects the bond dissociation energy of the O–H bond. log P represents the calculated octanol–water partition coefficient of each phenol. In all models, n represents the number of data points in the study, r is the correlation coefficient, s is the standard deviation of the regression equation, and q2 comprises the cross-validated r2. The coefficient with the hydrophobic term is small and indicates that a surface interaction with a minimally hydrophobic/semipolar receptor is in play. The strong dependence on σ+, as indicated by the coefficient (1.35) also suggests that radical stabilization could be favored by nucleic acids or polar enzymes.13
In this study, two simple, robust, and inexpensive in vivo yeast assays combined with the powerful predicting capabilities of the QSAR analysis were used to determine the variables that define the activity of X-phenols in S. cerevisiae. Because yeast and humans share many orthologues and conserved pathways, molecular mechanisms of toxicity identified in yeast may be relevant to understanding the mechanism of toxicity of the same set of compounds in human cells.14
Two different biological responses pertaining to yeast treated with various X-phenols were measured and assessed. Growth assays were utilized to determine the levels of toxicity induced by a series of X-phenols by measuring the extent of growth inhibition and determining their inhibition concentrations (ICs). This was followed by a genetic assay, that is, the yeast deletion (DEL) assay that was used as an indirect method to assess the capability of X-phenols to induce DNA damage or DNA breaks.15 The DEL assay, developed by Schiestl, measures the induction of deletion events in S. cerevisiae and has been used to detect carcinogenic and clastogenic activities with great success.16,17 The selection of this genetic assay is underscored by three attributes: its sensitivity as confirmed by its ability to detect carcinogens that other short-term genotoxicity assays consistently fail to detect,15 its specificity in distinguishing between noncarcinogenic and carcinogenic structural analogs,18 and its excellent (86%) accuracy at detecting known carcinogens as compared to only 29% accuracy achieved with the Salmonella (Ames) assay.16
To delineate the mechanism of phenol toxicity in yeast, QSAR analysis of ICs from growth inhibition studies as well as deletion frequencies from deletion assays were carried out. By combining the predictive capabilities of QSAR with a simple and reliable in vivo system, this study describes an approach that can be utilized not only to identify potential clastogens but also to address the mechanisms of toxicity and genotoxicity of xenobiotics. Results obtained in this study using yeast as a model system will help identify potential carcinogens in humans and yield insight into the mechanism of the associated chemical toxicity.
Results
Toxicity of X-Phenols in S. cerevisiae
To assess the effects of 4-X-phenols on cell growth and to further examine the cellular response at biologically relevant concentrations, yeast cells are exposed to phenolic compounds, and the extent of growth inhibition is determined. The heterogenous set of X-phenols tested for cytotoxicity includes phenols of varying physicochemical attributes, such as electron density, hydrophobicity, and sterics. In addition, polysubstituted phenols such as butylated hydroxy anisole (BHA) and 2,6-di-tert-butyl-4-methyl-phenol (BHT) are also included in the study.
Although the compounds tested were toxic to S. cerevisiae, there was a wide range in potencies. 4-Sulfonamido-phenol was the least potent with an IC50 value of 318 mM, whereas 4-nonylphenol was the most potent with an IC50 of 0.02 mM (Table S1). These results suggest that the variability of substituents greatly affects the toxicity of the corresponding phenols, as has been previously observed in mammalian cells.9,10,19
Hydrophobicity, a Chemical Parameter Influencing the Toxicity of X-Phenols
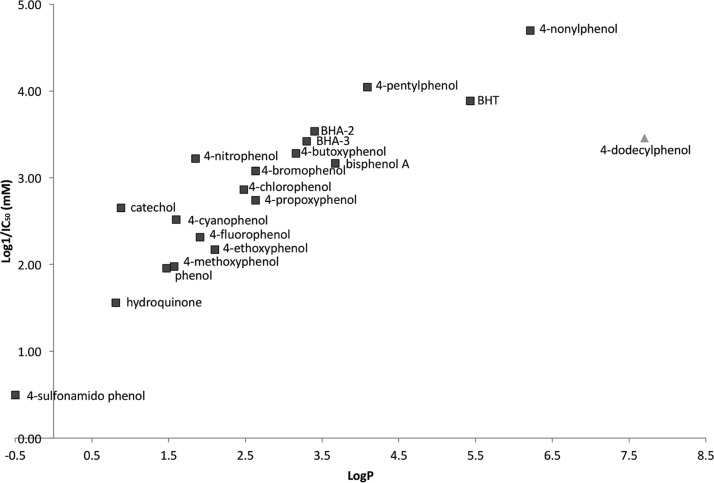
Evaluation of the growth inhibition data indicates that the most significant factor affecting the potency of X-phenol appears to be hydrophobicity, as represented by log P (partition coefficients) in Figure 1.
Figure 1.
Correlation between toxicity and hydrophobicity of phenols used in this study. IC50 values used were obtained for each phenol with the growth assay (square markers, see Table 1). The data for 4-dodecylphenol are an exception; they represent an approximate IC40 value because its poor solubility precluded testing at higher concentrations (triangle marker).
In the present study, the QSAR analysis was performed to delineate the effects of the physicochemical attributes of X-phenols on their associated toxicity activities. From the data in Table 1, a stepwise regression analysis was carried out, and the following QSARs were derived using the Hansch linear and parabolic models.20 Two phenols, 4-nitrophenol and catechol, were omitted from eqs 3–6 and Tables 1 and 2.
| 3 |
| 4 |
Table 1. Inhibition of Growth by X-Phenols (IC50).
| log 1/IC50 |
|||||
|---|---|---|---|---|---|
| no. | X | Obsd | Preda | Predb | clog Pc |
| 1 | 4-H | 1.96 | 2.09 | 2.1 | 1.47 |
| 2 | 4-F | 2.32 | 2.36 | 2.43 | 1.91 |
| 3 | 4-CN | 2.52 | 2.17 | 2.2 | 1.6 |
| 4 | 4-Br | 3.08 | 2.79 | 2.92 | 2.63 |
| 5 | 4-Cl | 2.87 | 2.7 | 2.82 | 2.48 |
| 6d | 4-NO2 | 3.22 | 2.32 | 2.38 | 1.85 |
| 7 | 4-OH | 1.56 | 1.69 | 1.58 | 0.81 |
| 8 | 4-SO2NH2 | 0.5 | 0.89 | 0.4 | –0.5 |
| 9 | 4-OCH3 | 1.98 | 2.15 | 2.18 | 1.57 |
| 10 | 4-OC2H5 | 2.17 | 2.47 | 2.56 | 2.1 |
| 11 | 4-OC3H7 | 2.74 | 2.79 | 2.92 | 2.63 |
| 12 | 4-OC4H9 | 3.28 | 3.12 | 3.24 | 3.16 |
| 13 | 4-(CH2)4CH3 | 4.05 | 3.68 | 3.74 | 4.09 |
| 14 | 4-(CH2)8CH3 | 4.7 | 4.97 | 4.54 | 6.21 |
| 15d | 2-OH (catechol) | 2.65 | 1.73 | 1.64 | 0.88 |
| 16 | 2-C(CH3)3, 4-OCH3 | 3.54 | 3.2 | 3.32 | 3.3 |
| 17 | 3-C(CH3)3, 4-OCH3 | 3.42 | 3.26 | 3.38 | 3.4 |
| 18 | BHA | 3.72 | 3.2 | 3.32 | 3.3 |
| 19 | BHT | 3.88 | 4.5 | 4.3 | 5.43 |
| 20 | bisphenol-A | 3.17 | 3.43 | 3.53 | 3.67 |
Table 2. Inhibition of Growth by X-Phenols (IC80).
| log 1/IC80 |
|||||
|---|---|---|---|---|---|
| no. | X | Obsd | Preda | Predb | clog Pc |
| 1 | 4-H | 1.85 | 1.96 | 2.04 | 1.47 |
| 2 | 4-F | 2.21 | 2.22 | 2.35 | 1.91 |
| 3 | 4-CN | 2.26 | 2.04 | 2.13 | 1.6 |
| 4 | 4-Br | 2.96 | 2.65 | 2.82 | 2.63 |
| 5 | 4-Cl | 2.78 | 2.56 | 2.73 | 2.48 |
| 6d | 4-NO2 | 3.09 | 2.19 | 2.31 | 1.85 |
| 7 | 4-OH | 1.39 | 1.58 | 1.51 | 0.81 |
| 8 | 4-SO2NH2 | 0.34 | 0.81 | 0.31 | –0.5 |
| 9 | 4-OCH3 | 1.91 | 2.02 | 2.11 | 1.57 |
| 10 | 4-OC2H5 | 2.1 | 2.33 | 2.48 | 2.1 |
| 11 | 4-OC3H7 | 2.65 | 2.65 | 2.82 | 2.63 |
| 12 | 4-OC4H9 | 3.16 | 2.96 | 3.12 | 3.16 |
| 13 | 4-(CH2)4CH3 | 3.96 | 3.5 | 3.56 | 4.09 |
| 14 | 4-(CH2)8CH3 | 4.4 | 4.75 | 4.14 | 6.21 |
| 15d | 2-OH (catechol) | 2.47 | 1.62 | 1.57 | 0.88 |
| 16 | 2-C(CH3)3, 4-OCH3 | 3.39 | 3.04 | 3.19 | 3.3 |
| 17 | 3-C(CH3)3, 4-OCH3 | 3.31 | 3.1 | 3.24 | 3.4 |
| 18 | BHA | 3.37 | 3.04 | 3.19 | 3.3 |
| 19 | BHT | 3.6 | 4.29 | 3.99 | 5.43 |
| 20 | bisphenol-A | 3.09 | 3.26 | 3.37 | 3.67 |
Equations 3 and 4 both indicate the importance of hydrophobicity in the toxicity of X-phenols. The coefficient of the log P term in eq 4 is close to 1, which suggests that X-phenols are mostly desolvated in the hydrophobic milieu of yeast.20 The statistical parameters of eq 4 (r2, q2, and s) are superior to those of eq 3. The F tests indicate that eq 3 (F1,16 α0.01 = 8.53) and 4 (F1,15 α0.01 = 8.68) are highly significant. The parabolic model is a more robust predictor of toxicity than the linear model. The poor solubility of X-phenols with log P values greater than 6.5 precluded their inclusion in this dataset.
A subsequent QSAR analysis with IC80 values obtained from the growth assays also corroborates the delineation of the parabolic, hydrophobic model for phenol toxicity (see Table 2). Equation 6 is a much better fit and a predictor of toxicity than the linear model (eq 5). The IC80 data yield similar models to those of IC50 models, when one compares eqs 3, 5 and 4, 6. The two models pertaining to the IC80 toxicities are listed below:
| 5 |
| 6 |
The parabolic model is a much better fit with higher r2 (0.950) and q2 values (0.910); it indicates that 95% of the variance in the data can be explained by hydrophobicity alone. On the basis of the QSAR analysis of the data obtained from the growth assays, it is apparent that the overall toxicity induced by X-phenols in this study is highly influenced by their corresponding hydrophobicities. Equations 5 (F1,16 α0.01 = 8.53) and 6 (F1,15 α0.05 = 4.54) are also highly significant. Although the curve appears to taper off around 4-nonylphenol, compounds with greater hydrophobicity, such as 4-dodecylphenol, were subjected to testing, but poor water solubility and high hydrophobicity (clog P = 7.79) precluded further analysis. The dose–response curve with 4-dodecylphenol achieved 40% growth inhibition at 0.35 mM, a value much higher than the IC50 value for 4-nonylphenol at 0.02 mM. This result clearly indicates that 4-dodecylphenol is much less potent than 4-nonylphenol, confirming the predicted downward trend of the parabolic model for compounds of higher hydrophobicity.
In the development of eqs 3–6, there were two outliers: 4-nitrophenol and catechol with toxicities much higher than expected. The substituents on these phenols are reactive entities. In cellular systems, the nitro-moiety is prone to reduction and generation of toxic species such as the nitroso and/or hydroxylamine moieties. 4-Nitrophenol has also consistently been seen to be more toxic than computational models normally predict.21 In the current study, 4-nitrophenol deviates significantly (>3×) when compared to other phenolic congeners, which indicates that other mechanisms are also at play. Catechol, which is easily oxidized, appears to be 4 times more toxic than expected. This may be attributed to its rapid metabolism to other quinones in yeast.22 Electronic effects of the substituents of all phenols were also examined in the QSAR analysis but failed to pass muster.
A toxicity study of a series of multisubstituted phenols on yeast cells was examined by Arnold et al., who utilized a different electro-rotation approach to toxicity.4,23 Electro-rotation is a well-established physical method that involves the noninvasive induction of rotation of yeast by a rotating electric field because the characteristics of its membranes have been shown to affect electro-rotation. An accessible frequency range is utilized to yield the electro-rotation spectra.4 Control yeast cells showed both anti- and cofield rotations (CFRs). However, yeast treated mostly by halogenated phenols behaved differently and yielded a much stronger CFR and practically little or no antifield rotation in this study. The proportion of cells showing the CFR was determined to be a sensitive measure of toxicity.
Using this data that represented the concentrations of X-phenols that yielded 50% CFRs, the following QSAR was developed. See Table 3. Two substituted phenols, 4-nitrophenol and 2-methyl, 4,6-dinitrophenol, were omitted from the development of the QSAR models.
| 7 |
Table 3. Toxicity of X-Phenols in Yeast.
| log 1/ICFR50 |
||||
|---|---|---|---|---|
| no. | X | Obsda | Predb | clog Pc |
| 1 | H | 1.17 | 1.44 | 1.47 |
| 2 | 4-Cl | 2.07 | 2.26 | 2.48 |
| 3 | 2,3-Cl2 | 2.68 | 2.56 | 2.85 |
| 4 | 2,4-Cl2 | 2.77 | 2.65 | 2.97 |
| 5 | 2,6-Cl2 | 2.52 | 2.39 | 2.64 |
| 6 | 2,4,5-Cl3 | 3.26 | 3.16 | 3.6 |
| 7 | 3,4,5-Cl3 | 3.3 | 3.33 | 3.81 |
| 8 | 2,4,6-Cl3 | 3.28 | 2.99 | 3.39 |
| 9 | 2,3,4,5-Cl4 | 3.92 | 3.66 | 4.21 |
| 10 | 2,3,4,5,6-Cl5 | 3.96 | 4.06 | 4.71 |
| 11 | 2-Br | 2.08 | 2.15 | 2.45 |
| 12 | 4-Br | 2.4 | 2.38 | 2.63 |
| 13d | 4-NO2 | 2.85 | 1.75 | 1.85 |
| 14 | 2,4,6-(NO2)3 | 1.74 | 1.55 | 1.64 |
| 15 | 3,5-(OCH3)2 | 1.54 | 1.58 | 1.61 |
| 16 | 4-COOH | 1.75 | 1.51 | 1.56 |
| 17 | 4-Cl, 3,5-(CH3)2 | 2.96 | 3.07 | 3.48 |
| 18 | 2-NH2, 4-CH3 | 1.07 | 1.16 | 1.12 |
| 19 | 4-CH3, 2-NO2 | 2.1 | 2.15 | 2.35 |
| 20d | 2-CH3, 4,6-(NO2)2 | 2.62 | 2.09 | 2.27 |
| 21 | 3-CF3 | 2.31 | 2.58 | 2.88 |
| 22 | 3,5-(C(CH3)3)2 | 3.96 | 4.4 | 5.13 |
| 23 | 2,4,6-(I)3 | 4.3 | 3.96 | 4.58 |
| 24 | 4-I | 2.4 | 2.59 | 2.89 |
| 25 | 2,3,4,5,6-F5 | 2.05 | 2.01 | 2.17 |
In eq 7, the clog P coefficient is close to 0.8 that indicates that X-phenols are mostly desolvated in the biomembrane. With regard to the statistics, the r2 value is 0.953, and the q2 value is 0.937, whereas the F statistic is F1,21 α0.01 = 8.02. Two phenols, 4-nitrophenol and 2-methyl, 4,6-dinitrophenol, were outliers; they are 12 times and 3 times, respectively, more active than predicted.4,23 4-Nitrophenol has a significantly high deviation in the rotation assay, which could be attributed to a strong thru-resonance that enhances uncoupling and inhibits purine transport. Although 2-methyl, 4,6-dinitrophenol also acts as an uncoupler, its overall activity is mitigated by its acidic pKa that enhances ionization and thus marginally enhances the overall cell toxicity. The range in clog P values of this set of phenols extends from 1.12 to 5.13, a spread of only 4 log units, which is inadequate to determine the optimum hydrophobicity of the system. The range in clog P of the first data set runs from −0.50 to 6.21—a spread of 6.7 log units that allows for the determination of clog Po of approximately 8.8.
Cytotoxicity
Our results indicate that there is a strong correlation between hydrophobicity and toxicity in yeast cells (Figure 1 and eqs 3–6). However, when similar QSAR studies were conducted in murine leukemia cells, phenols with electron-donating groups (EDGs) and electron-withdrawing groups (EWGs) behaved differently. The cytotoxicity of phenols with EWGs correlated with hydrophobicity, whereas phenols with EDGs presented a very low correlation with hydrophobicity.9,12,24 The drastic mechanistic difference between yeast and murine leukemia cells may be attributed to the cell wall of yeast cells that present some type of barrier that is not existent in mammalian cells. This barrier contains mannoproteins and β-glucans, which are abundant components of yeast cell walls with some hydrophobic characteristics.25 They could readily enhance transport of hydrophobic X-phenols in yeast cells.
When studying the cytotoxicity of a series of para-substituted phenols in murine leukemia cells, Selassie et al. predicted that substituents with EDGs would result in DNA damage through a phenoxy radical mechanism.9 Hence, yeast was used as a model system to test this hypothesis because the yeast DEL assay is a well-defined and consistent method for measuring the presence of DNA breaks in the cell.17 The results obtained were analyzed by QSAR, and the prediction that X-phenols with EDGs would interact with DNA to cause damage was confirmed.
Genomic Rearrangements Induced by 4-X-Phenols
The toxicity indices induced by the phenols could be attributed to several different cellular targets. The focus was on identifying and predicting which phenols would result in DNA damage, particularly in deletion formation, a type of genomic rearrangement that in higher eukaryotic cells can result in neoplastic transformation.26−28 Genomic deletions in yeast cells induced by chemicals can be readily detected by the DEL (deletion) assay. Compounds that cause single- and double-strand breaks or oxidative damage will lead to a significant increase in the deletion events17 and genomic instability.29 These qualities of the DEL assay are known to correctly identify carcinogenic compounds with a greater success than other short-term genotoxicity tests.16
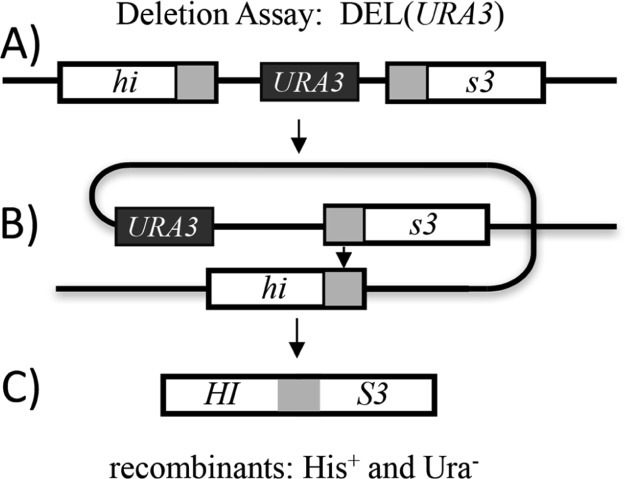
The yeast DEL assay is a simple method in which the frequency of deletion events in the yeast genome is calculated after treating cells with a suspected damaging agent. The deletion events result from an intrachromosomal recombination between two truncated his3 alleles that share approximately 400 bp of homologous sequences flanking a genetic marker. One allele is truncated at its 3′ end and the other at its 5′ end resulting in a histidine auxotroph mutant. A homologous recombination between the his3 direct repeats results in deletion of the intervening sequence and restoration of a functional HIS3 gene, giving rise to histidine prototrophs (Figure 2). The frequency of deletion formation is determined by dividing the number of recombinant cell colonies by the total number of cells. The total number of cells is calculated by plating cells onto histidine-containing media, whereas the recombinant cells give origin to colonies growing in agar medium lacking histidine. In this study, the recombination frequencies of cells exposed to IC80 concentrations of phenol will be compared to the frequency of recombination in a negative control consisting of cells exposed only to the solvent [1–3% dimethylsulfoxide (DMSO)]. The effect of a phenolic compound is considered to be positive for inducing recombination when the treatment of cells results in deletion frequencies 2-fold or higher than that of the negative control.16,30 The DEL assay will therefore not only identify which phenols induce DNA damage in S. cerevisiae but also identify potential carcinogenic compounds and provide a biological descriptor to be used in a QSAR analysis to provide insight into the mechanism of phenolic toxicity. Testing structurally related compounds with different substituents that confer different physicochemical properties will help elucidate a potential mechanism of phenol toxicity.
Figure 2.
Deletion Assay. Diagram of the DEL(URA3) assay. (A) At the HIS3 locus, yeast strains carry a construct with a functional URA3 gene flanked on both sides by his3 sequences. The upstream his3 allele lacks its 3′ end and the downstream his3 allele lacks its 5′ end and both contain his3 direct repeated sequences of 415 bp (indicated in diagram as gray shaded boxes). (B) Direct repeats can recombine by homologous recombination by several different mechanisms, not discussed in figure, resulting in deletion of all intervening sequences. (C) A deletion event by homologous recombination results in a functional HIS3 gene.
The first growth and deletion assays were performed using the yeast DEL(LEU2) strain kindly provided by Robert Schiestl (RS112). Subsequent experiments were performed using DEL(URA3) provided by Adam Bailis that yielded similar results. We continued our analysis with the DEL(URA3) assay system as our laboratory strains are isogenic to those of the Bailis laboratory, facilitating our genetic analysis. This strain consists of HIS3 repeats flanking a URA3 marker inserted at the HIS3 locus31 instead of the LEU2 marker used in RS112. Growth assays were repeated with several of the phenolic compounds tested with RS112, and the same or very similar IC values were obtained. In addition, included in the phenolic derivatives tested in this study for deletion formation, hydroquinone and catechol had been tested by Schiestl’s group with the DEL(LEU2) assay;22 we obtained similar results with our modified deletion assay. Having a different DEL assay strain resulted in reproducible and similar findings to those obtained by Schiestl’s group.
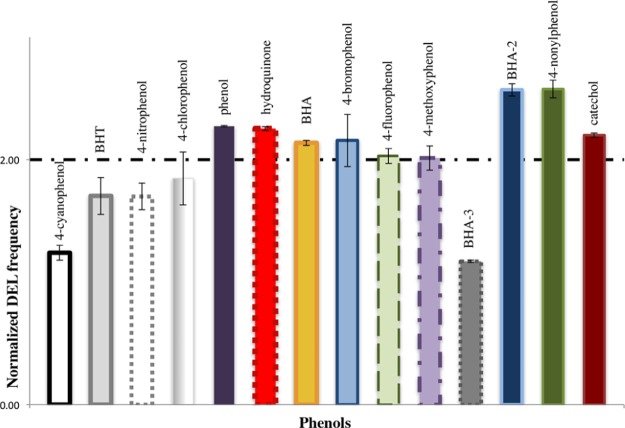
To gain a comprehensive view of phenol toxicity and its ability to induce DNA damage, the DEL(URA3) assay was performed on a select group of compounds containing different chemical properties. DEL assays were successfully conducted at IC80 concentrations of the following 14 phenols: 4-nitrophenol, 4-methoxyphenol, 4-hydroquinone, phenol, BHA, BHT, 4-cyanophenol, 4-chlorophenol, 4-flourophenol, 4-bromophenol, 4-nonylphenol, catechol, 2-tert-butyl-4-methoxy-phenol (BHA-2), and 3-tert-butyl-4-methoxy-phenol (BHA-3). The IC80 concentration is used to ensure that the assay is carried out under conditions that will not cause more than 90% growth inhibition, and the growing conditions used are the same as those used for determining the IC values (growth assay). Of the 14 compounds tested, 9 resulted in recombination frequencies that are 2-fold or higher than the frequency obtained for control cells treated only with the solvent (2% DMSO) (Figure 3 and Supporting Information Table S2). The percent viability of treated cells was determined by counting the colony-forming units. Treatments with IC80 values resulted in an average of 30% viability.
Figure 3.
Deletion formation induced by phenolic compounds. A series of phenolic compounds were tested for their ability to induce deletion formation in yeast cells carrying the DEL(URA3) assay. Cells were treated with an IC80 concentration of the indicated phenols for 17 h. Appropriate dilutions of treated cells were plated onto SC-Ura plates to determine the number of viable cells and onto SC-His to determine the number of cells that have undergone deletion formation by homologous recombination. The frequency of deletion formation (His+ cells) is calculated by dividing the total number of His+ cells by the total number of viable cells. Untreated cells also were run through this assay to determine the baseline frequency of deletion formation in untreated cells (for details, see the deletion assay in the Methods Section). The plot of the DEL frequency is determined by the ratio of the deletion frequency of the treated sample to the deletion frequency of the untreated control. The horizontal line drawn at a value of 2 clearly identifies those phenolic compounds that induced a deletion frequency that was 2-fold or higher than the untreated control.
Among the eight compounds that caused an increase in the DEL recombination of at least 2-fold over the control, there is an overrepresentation of phenolic compounds with electron-donating substituents (Figure 3).
QSAR of the DNA Deletion Frequency
A QSAR analysis of the DNA deletion frequency was performed to elucidate the mechanism of this chemical–biological interaction in yeast and determine if there is a relationship between the physicochemical properties of X-phenols and the ability to induce deletion events in yeast cells. It is well-established that many radical cations of substituted phenols are correlated by σ+, the Brown variant of the Hammett electronic parameter.32 Thus, this descriptor and hydrophobicity were utilized for the QSAR analysis. The biological descriptor DNA-R represents the increase in the deletion frequency compared to control cells. See Table 4.
Table 4. Induction of Deletion by X-Phenols.
| no. | X | Obsda | Predb | σ+ | clog Pc |
|---|---|---|---|---|---|
| ld | 4-H | 2.27 | 1.84 | 0 | 1.47 |
| 2 | 4-F | 2.03 | 1.92 | –0.07 | 1.91 |
| 3 | 4-CN | 1.25 | 1.6 | 0.66 | 1.6 |
| 4 | 4-Br | 2.16 | 1.92 | 0.15 | 2.63 |
| 5 | 4-Cl | 1.85 | 1.92 | 0.11 | 2.48 |
| 6 | 4-NO2 | 1.7 | 1.57 | 0.79 | 1.85 |
| 7 | 4-OH | 2.25 | 2.13 | –0.92 | 0.81 |
| 8 | 4-OCH3 | 2.01 | 2.16 | –0.78 | 1.57 |
| 9 | 4-(CH2)8CH3 | 2.58 | 2.51 | –0.29 | 6.21 |
| 10 | 2-OH (catechol) | 2.26 | 2.14 | –0.92 | 0.88 |
| 11 | 2-C(CH3)3, 4-OCH3 | 2.57 | 2.47 | –1.04 | 3.3 |
| 12d | 3-C(CH3)3, 4-OCH3 | 1.17 | 2.42 | –0.88 | 3.4 |
| 13 | BHA | 2.14 | 2.47 | –1.04 | 3.3 |
| 14d | BHT | 1.7 | 2.63 | –0.83 | 5.43 |
Fold increase over untreated control is presented. Fold increase in deletion formation was determined by dividing the frequency of recombination of cells treated with X-phenol (average of three samples) by the frequency of recombination of cells treated only with the solvent (untreated control), as explained in the experimental procedures.
clog P version 1.6.
Using this data, the following QSAR models, 8 and 9, were formulated.
| 8 |
| 9 |
Equation 9 indicates that 52% of the variance in the data can be attributed primarily to the electrophilicity of the EDG substituents, whereas hydrophobicity accounts for 22% of the variance in the data. The range in the deletion frequency is 1.55 units, which is limiting. The correlation between DEL formation and electrophilicity plus hydrophobicity yields a suitable fit of the data with an r2 value of 0.74, a cross-validated q2 value of 0.49, and a standard deviation of 0.22. The small number of compounds in this study (N = 14) and the limited range in activity could account for the low r2 value in this equation and therefore can potentially be improved with the addition of more data points (see Table 4).
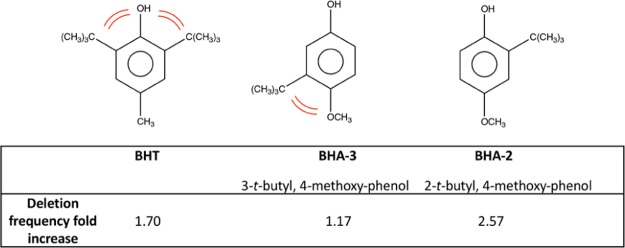
There are three outliers in this dataset: phenol, BHA-3, and BHT. The latter two are highly hydrophobic, bulky, and sterically hindered by interactions between the t-butyl substituent and the methoxy group in the 3-t-butyl-4-methoxy-phenol and the crowded “flanking” of the hydroxyl group by the t-butyl groups in BHT. See Figure 4.
Figure 4.
Steric hindrance in BHT and BHA-3. Deletion frequency fold increase values, from Table 4, are indicated for each compound.
The steric repulsions can be described by the size of the critical substituents involved in the negative interactions. Molar refraction (MR) of a substituent is an appropriate reflection of its size and can be used to delineate the interactions between the substituents on a molecule. The hydroxyl group (MR = 0.28) of BHT is closely flanked by two bulky tert-butyl groups (MR = 1.96) that would impede easy accessibility to DNA, hence its low deletion index of 1.70. With BHA-3, the methoxy group (MR = 0.79) is right up against the bulky t-butyl group that would distort the planarity of the phenyl ring and reduce its ability to interact with DNA. This behavior contrasts sharply with that on BHA-2, whereby the smaller hydroxyl group (MR = 0.28) is ortho to the t-butyl group and has a higher deletion frequency. Apparently, interactions with DNA are impaired by phenols with high hydrophobicities and bulky substituents. Phenol is also an outlier and an anomaly. Triplicate tests were run, but it always yielded the same outcome. It behaved well in the cytotoxicity tests but not the deletion assay. Studies have shown that phenol can inhibit DNA synthesis and replication in HeLa cells.33 Thus, it is possible that phenol could disrupt the DNA synthesis in yeast, resulting in a higher recombination frequency.
Genetic Pathways Involved in the Response to Phenol Treatment
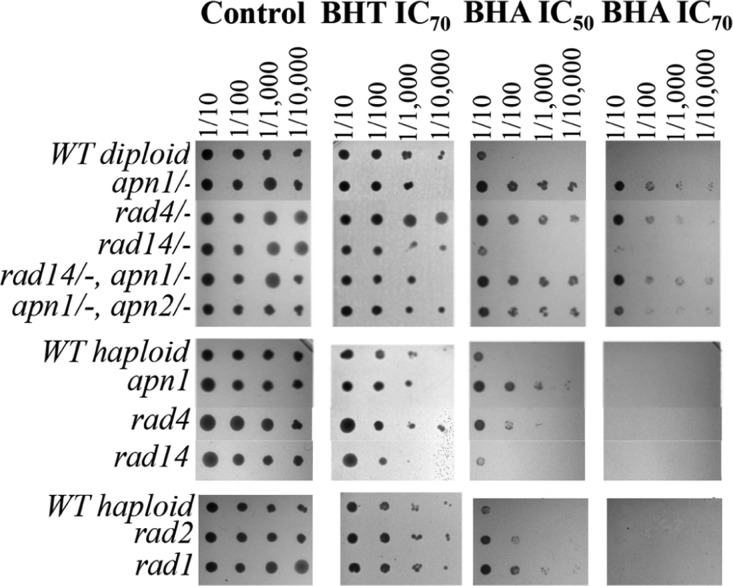
The induction of deletion formation by phenol treatment is indirect evidence that DNA damage is occurring and is a consequence of the treatment of cells with a small set of these phenolic compounds. If cells are accumulating damaged DNA, the cell must respond and try to repair this damage. A genetic approach was utilized in which several null mutant strains of genes involved in DNA repair were treated with phenolic compounds and screened for growth phenotypes. The test included BHA, a compound that resulted in greater than 2-fold deletion formation (DEL+), and BHT, that did not increase deletion formation over 2-fold (DEL–). Serial dilutions of each strain were plated using a 96-pin replicator onto synthetic complete (SC) plates with or without BHA or BHT. A comparison of the level of growth against the WT strain to the growth observed for each strain in the untreated control plate (no phenolic compound + DMSO) clearly identified strains that are resistant, sensitive, or nonresponsive (Figure 5). These results indicate that DNA repair proteins are an integral part of the cellular response to treatment with phenols that elicit a genotoxic effect. Clearly, BHA causes growth defects in most of the DNA repair null mutant strains tested, whereas the response to BHT was negligible.
Figure 5.
Growth phenotype of DNA repair null mutant yeast strains exposed to phenol treatment. The indicated yeast strains were grown to saturation, and serial dilutions were plated using a 96-pin replicator onto SC 1% DMSO, SC IC50 BHA in 1% DMSO, SC IC70 BHA in 1% DMSO, or SC IC70 BHT in 1% DMSO. The first panel consists of homozygous diploid null mutant yeast strains (name of deleted gene/-). The second and third panels consist of haploid null mutant strains.
Discussion
Cytotoxicity in Yeast Cells
To delineate the effects of phenol toxicity in yeast cells, a kill curve was first established, and various specific concentrations were determined for this study. Yeast cells were grown in liquid media and exposed to different concentrations of a particular compound in a 96-well plate format, which allows for rapid screening of many compounds at the same time and the subsequent generation of dose–response curves. From these curves, IC50 and IC80 values were determined for each compound tested. This ensured that in yeast cells treated with IC50, IC80, or in between concentrations, growth inhibition would not exceed 90%, and viability measured by colony forming units did not go below 10% (see Table S1). This approach avoided the use of concentrations of compounds tested that may have resulted in acute effects that are not biologically relevant and are not a focus of this study.
To determine if various structural changes in phenols could affect the toxicity to yeast cells, a systematic approach that included a variation of substituents in the size, hydrophobicity, electronic attributes, and position was examined. The first biological response to be analyzed was cytotoxicity. QSAR analysis identified a strong correlation between cytotoxicity and hydrophobicity of the compounds tested. Cytotoxicity increased with the hydrophobicity of the compounds tested, up to a certain point (clog P 7.5), after which the increase in hydrophobicity resulted in a downward toxicity trajectory.
The correlation between hydrophobicity and toxicity appears to be a strong relationship observed in yeast cells. When similar QSAR studies were done in murine leukemia cells, looking at the toxicity of para-substituted phenols, those with EDGs and EWGs behaved differently. The cytotoxicity of phenols with EWGs correlated with hydrophobicity, whereas phenols with EDGs showed a very low correlation with hydrophobicity.9,12,34 The reason for this discrepancy observed between yeast and mammalian cells may be attributed to differences in how and how much of the tested phenols can penetrate and accumulate within cells. Cells have two main mechanisms to control the actual amount of a particular chemical that is found inside the cell: the presence of a physical barrier and mechanisms of detoxification. The plasma membrane acts as a barrier between the outside and inside of a cell, and in the case of yeast cells, there is in addition, a cell wall. The detoxifying mechanisms present in yeast and mammalian membranes are not completely understood but they are known to present a wide range of specificity and redundancy in terms of having overlapping specificities. Because of the wide range of transporters available in both systems, we do not believe detoxifying mechanisms cause the difference observed between mammalian and yeast cells.35 This critical difference may be attributed to the cell wall of yeast cells.36,37
Mechanism of Phenol Toxicity
Phenols have the ability to quench the ROS by donating hydrogen atoms to superoxide or hydroxyradicals in mammalian cells. The phenoxy radical formed is a less reactive molecule that will stop the propagation of radical reactions preventing damage to macromolecules within the cell as well as overall detrimental effects to cells.9,38,39 Phenols with substituents that are EDGs in the para position will help stabilize the phenoxy radical formed inside the cell and propagate an oxidative stress state by being more reactive and inducing the formation of superoxide, hydroxyradicals, hydrogen peroxide, or oxidation products of the phenoxy radical that can react with DNA, causing strand breaks. The strong negative dependence on Brown’s sigma-plus constant suggests that the electron-donating capability of the phenols enhances DNA damage. This result is significant because it also indicates that a radical mechanism may be at play, implicating a phenoxy radical in the generation of DNA damage. Electron-withdrawing substituents in the para position would decrease the likelihood of a phenol causing an increase in deletion formation, whereas electron-donating substituents would increase its likelihood to be more damaging. Selassie et al. studied the toxicity of a similar set of X-phenols in a murine cell line and proposed that a radical mechanism was at play.9 Our current study using yeast as a model system supports this hypothesis; it suggests the formation of a phenoxy radical as the culprit for cell toxicity and identifies DNA as one of the putative end targets.
In yeast, previous results obtained by Schiestl match our prediction that phenolic compounds with EDGs will result in greater genotoxicity than phenols with EWGs. When assaying aniline metabolites, 4-aminophenol caused a greater than 2-fold increase in deletion frequencies over untreated control cells, whereas 4-acetamidophenol, another metabolite with a moderate electron donor substituent, did not show a significant increase.15 The amino group of 4-aminophenol is a much more potent electron donor (σ+ = −1.30) than the acetomido substituent of 4-acetamidophenol (σ+ = −0.60). In the latter case, the electrons are more delocalized over the nitrogen and oxygen atoms.
Among the compounds tested, a couple of more complex phenols, such as BHA and BHT, were tested for toxicity and genotoxicity. Even though these two compounds have similar structures, BHA-treated cells resulted in a significant increase in deletion events, whereas BHT showed no significant genotoxicity. Because BHA is a mixture of two isomers, BHA-2 and BHA-3, the DEL assay was performed with cells treated with each individual isomer. Cells treated with BHA-2 phenol resulted in a greater than 2-fold deletion formation, whereas the BHA-3 isomer had a minimal effect. It must be noted that hydroquinone, catechol, and phenol, all produce a similar increase in the DEL frequency but their potencies are different. Catechol is an outlier in the potency data (Tables 1 and 2), but its predicted genotoxicity based on eq 8 is in line with the prediction. It is well-known that catechol is easily oxidized in air and in vivo because of interaction with heavy metals or cell metabolism that results in redox reactions.11,40 However, phenol turns out to be an outlier in the DEL assay based on its σ+ value and hydrophobicity.
Shadnia and Wright propose that after the phenoxyradical is formed, phenols with EDGs can rapidly form a quinone intermediate, and quinone is responsible for reacting with DNA.11 Prevention of the formation of quinone would thus render the phenol intermediates less genotoxic. A mechanism to reduce quinone formation is to block the ortho position of the phenolic ring by bulky substituents or to have EWGs on the phenol ring. Our results indicate that phenols with EWGs do not express genotoxicity in addition to the more complex phenolic compounds with blocked ortho positions that do not increase the deletion formation in yeast (i.e., BHT and BHA-3).
Genetic Control of Phenol Response
To gain insight into the molecular mechanism of toxicity due to phenol exposure, a genetic approach was taken. Yeast strains with gene deletions in important DNA repair genes were exposed to phenols, and their growth was assessed in a spot assay. The mutant strains that were screened lack genes that play a role in the nucleotide excision repair (NER) and base excision repair (BER) pathways. NER deals mainly with bulky DNA lesions or those that distort the DNA backbone such as damage caused by UV light. Growing evidence suggests that NER is involved in the repair of other types of DNA lesions such as those generated by oxidative damage,41,42 which are handled mainly by BER. In Figure 5, null mutant strains for NER and BER genes all show some level of growth defect when exposed to BHA, for example, Δrad4 and Δrad14 strains. The Rad4 protein is involved in one of the first steps in the NER pathway, identifying DNA damage and recruiting other NER proteins to the site.43,44 The Rad14 protein is involved in recruiting one of the endonucleases, Rad1, to the site of DNA damage,45 possibly also playing a role in recruiting the rest of the NER proteins and other roles that are still unknown.46 Volker et al. showed in mammalian cells that XPA (Rad14 homolog) is needed to recruit XPF (Rad1 homolog).47 Under our experimental conditions, the Δrad4 strain is more resistant to BHA than the WT strain, which appeared to be an anomaly. This suggests that NER processing may actually cause the formation of an intermediate that affects the viability or that the processing of the damage by NER may be detrimental under some circumstances.48,49 On the other hand, the Δrad14 strain shows increased sensitivity in the presence of BHA. A possible explanation for what seems a contradictory result is as follows: in this mutant, NER can be initiated by Rad4, but the whole repair process cannot be completed in the absence of Rad14 that eventually leads to a loss of viability.50,51 This assay only measures the ability of yeast cells to grow in the presence of phenolic compounds and does not yield information on other consequences such as accumulation of mutations, genomic rearrangements, and so forth. Exposure of the same strains to BHT did not cause clear growth defects as observed with BHA, indicating that cellular targets are not the same. This preliminary screen using a specific subset of DNA repair null mutant strains confirms that phenols can cause damage to DNA, and perhaps more than one DNA repair pathway may be involved in the cellular response.
Experimental Procedures
Chemicals
Phenol, 4-fluorophenol, 4-cyanophenol, 4-bromophenol, 4-ethoxyphenol, 4-pentylphenol, 4-nonylphenol, catechol, 2-t-butyl-4-methoxy phenol, 3-t-butyl-4-methoxy phenol, and butylated hydroxytoluene were purchased from Sigma-Aldrich. 4-Chlorophenol, 4-nitrophenol, 4-methoxyphenol, 4-propoxyphenol, 4-butoxyphenol, and bisphenol A were purchased from Aldrich. Butylated hydroxyanisole hydroquinone, and DMSO were purchased from Sigma, and 4-sulfonamido phenol was purchased from Acros.
Culture Conditions and Media
All S. cerevisiae strains were maintained and grown in YPD (yeast nitrogen base, peptone, dextrose), synthetic minimal media (SC), or the appropriate synthetic drop-out medium (SC-X) (1.7 g/L yeast nitrogen base without amino acids and ammonium sulfate, 0.5 g/L serine, 0.2 g/L aspartate, 0.17 g/L tryptophan, 0.12 g/L adenine, 0.1 g/L leucine, 0.02 g/L tyrosine, and 0.05 g/L of histidine, uracil, lysine, alanine, phenylalanine, tyrosine, isoleucine, valine, and tyrosine) with 2% dextrose. SC-X media was made by leaving out the corresponding amino acid or base. For example, SC-Ura is SC media lacking uracil.
Yeast Strains
Yeast strains used in this study are presented in Table 5. Preliminary data were obtained with the yeast strain RS112 kindly provided by Dr. Robert Schiestl.52 All other strains used were derived in our research lab or provided by Dr. Adam Bailis because they are isogenic to our strains. The apn1, apn2, rad4, and rad14 null mutant alleles were constructed using the one-step gene disruption method and standard yeast genetic techniques.53
Table 5. Genotypes of Yeast Strains Used in This Study.
| strain | genotype | source |
|---|---|---|
| RS112 | MATa/α, ura3-52/ura3-52, leu2-3,112/leu2-Δ98, trp5-27/TRP5, ade2-40/ade2-101, ilv1-92/ILV1, HIS3::pRS6/his3-Δ200, LYS2/lys2-801 | Schiestl |
| ABX195-14D | Mata, ade2-1, can1-100, leu2-3, 112, ura3-1, trp1-1, his3::URA3::his3 | Bailis |
| ABX731-13C | Matα, ade2-1, can1-100, LEU2, ura3-l, trp1-1, his3-Δ200 | Bailis |
| TNX70 | MATa/α, ade2-1/-, can1-100/-, his3-Δ200/his3::URA3::his3, LEU2/leu2-3, 112, ura3-1/-, trp1-1/-, RAD5/+ | Negritto |
| TNX71 | MATa/α, ade2-1/-, can1-100/-, his3-Δ200/his3::URA3::his3, LEU2/leu2-3, 112, ura3-1/-, trp1-1/-, RAD5/+ | Negritto |
| TNX72 | MATa/α, ade2-1/-, can1-100/-, his3-Δ200/his3::URA3::his3, LEU2/leu2-3, 112, ura3-1/-, trp1-1/-, RAD5/+ | Negritto |
| TNX90 | MATa/α, ade2-1/-, can1-100/-, his3-Δ200/his3::URA3::his3, LEU2/leu2-3, 112, ura3-1/-, trp1-1/-, RAD5/+ | Negritto |
| TNX84 | MATa/α, ade2-1/-, can1-100/-, leu2-3, 112/-, trp1-1/-, ura3-1/-, his3::ura3::LEU2/HIS3, apn1::TRP1/-, RAD5/+ | Negritto |
| TNX13 | MATa/α, ade2-1/-, can1-100/-, leu2-3, 112/-, trp1-1/-, ura3-1/-, HIS3/his3::ura3::LEU2, rad4::TRP1/- | Negritto |
| TNX78 | MATa/α, ade2-1/-, can1-100/-, leu2-3, 112/-, trp1-1/-, ura3-1/-, rad14::KNMX/-, HIS3/his3::ura3::LEU2, RAD5/+ | Negritto |
| TNX82 | MATa/α, ade2-1/-, can1-100/-, leu2-3, 112/-, HIS3/his3::ura3::LEU2, rad14::KNMX/-, apn1::TRP1/-, RAD5/+ | Negritto |
| TNX87 | MATa/α, ade2-1/-, can1-100/-, leu2-3,112/-, ura3-1/-, trp1-1/-, his3::ura3::LEU2/HIS3, apn1::TRP1/-, apn2::KNMX/-, RAD5/+ | Negritto |
| TNX88 | MATa/α, ade2-1/-, can1-100/-, leu2-3,112/-, ura3-1/-, trp1-1/-, his3::ura3::LEU2/HIS3, apn1::TRP1/-, apn2::KNMX/-, RAD5/+ | Negritto |
| TNX89 | MATa/α, ade2-1/-, can1-100/-, leu2-3,112/-, ura3-1/-, trp1-1/-, his3::ura3::LEU2/HIS3, apn1::TRP1/-, apn2::KNMX/-, RAD5/+ | Negritto |
| ABX129-3D | MATa, ade2-1, can1-100, HIS3, leu2-3, 112, trp1-1, ura3-1, rad1::LEU2 | Bailis |
| ABX135-2B | MATa, ade2-1, can1-100, HIS3, leu2-3, 112, trp1-1, ura3-1, rad2::TRP1 | Bailis |
| TNT6 | MATa, ade2-1, can1-100, leu2-3, 112, trp1-1, ura3-1, HIS3, rad4::TRP1 | Negritto |
| TNX73-3B | MAT?, ade2-1, can1-100, leu2-3, 112trp1-1, ura3-1, his3::URA3::his3(405bp), apn1::TRP1, RAD5 | Negritto |
| TNX74-5B | MAT?, ade2-1, can1-100, leu2-3, 112, trp1-1, ura3-1, his3::URA3::his3(405bp), rad14::KNMX, RAD5 | Negritto |
| TNX69-5A | MATα, ade2-1, can1-100, his3::URA3::his3, leu 2-3, 112, ura3-1, trp1-1, RAD5 | Negritto |
Growth Inhibition Assay Adapted for 96-Well Microplate Format
The IC values for the different compounds used in our studies were determined with a cell growth inhibition assay. A 10 mL overnight starter culture was prepared in liquid SC media inoculated with a single colony of a strain carrying the deletion assay and grown for a 17 h overnight incubation at 30 °C on a rotating wheel. The cells were then counted using a hemacytometer, and aliquots of 2 × 106 cells were placed into 1.7 mL sterile Eppendorf tubes with varied volumes of a stock solution of the phenolic compound in 100% DMSO to obtain different concentrations of the compound to be tested. The total amount of DMSO in the final culture was always between 1 and 3% DMSO. SC media were then added to bring the final volume of each tube to 1 mL. We adapted the growth assay to grow cells in a 96-well microplate in small volumes (200 μL). In a typical growth assay, two different phenolic compounds were run at the same time; 12 different concentrations were each tested in quadruplicates. The 1 mL culture was then aliquoted into 96-well plates (microplate sterile 96K U-form Greiner Bio-One, USA scientific) such that there were four replicates for each concentration (200 μL aliquots in each well). To prevent contamination and possibly excessive evaporation, microtiter plates were covered with a sealing film (TempPlate sealing film sterile, USA Scientific) and incubated at 30 °C clipped to a rotating drum to maintain constant agitation. After 17 h, the plate was scanned at 655 nm using a Bio-Rad microplate reader. The average background absorbance of the solvent control samples (SC media, DMSO) was subtracted from the average absorbance measured for each treated sample and plotted against the log of the phenol concentrations used. A regression was fit to the linear region of the resulting sigmoidal curve, and the concentrations of the compound that induced 20%, 50%, and 80% inhibition of yeast cell growth were interpolated from the curve (IC20, IC50, and IC80 values, respectively) (Supporting Information Table S1 and Figure S1).
DEL Recombination Assay
The S. cerevisiae strains used for the DEL assay were diploid strains TNX70, TNX71, and TNX90 (Table 5) and were constructed by crossing a haploid strain containing the his3::URA3::his3 allele that consists of a URA3 gene disrupting the HIS3 gene. The his3 sequences on either side lack the 3′ end and 5′ end of the HIS3 sequence, respectively, and they share 415 bp of the homologous sequence in direct orientation such that if they recombine, it results in a functional HIS3 allele. The other haploid strain carries the his3-Δ200 allele that consists of a deletion of the HIS3 open reading frame. The his3-Δ200 and his3::URA3::his3 alleles were kindly provided by Adam Bailis, and the latter was built as described in Maines et al. 1998.31
To determine the effect of different phenolic compounds on deletion formation in yeast, the strains were first streaked and grown on SC-Ura plates at 30 °C to isolate single colonies. Drop-out media were used to keep under selection the his3::URA3::his3 allele to prevent spontaneous direct repeat recombination. A single colony was transferred to 5 mL of liquid SC-Ura media and grown on a rotator at 30 °C for 17 h. Cells were counted and new cultures for untreated and treated samples were prepared by inoculating with an appropriate volume of overnight culture to obtain 2 × 106 cells/mL. The control (untreated) samples consisted of 2 × 106 cells/mL in SC-Ura media with 2% DMSO in a final volume of 2 mL. The phenol-treated samples contained 2 × 106 cells/mL, the appropriate amount of phenol stock solution prepared in 100% DMSO that would result in a final phenol concentration equivalent to IC20, IC50, or IC80 values (previously determined by the growth assay), and DMSO was then added to ensure that the final concentration of DMSO in the treated sample matched the % DMSO in the control sample (% DMSO used never exceeded 3% of the final volume).
Cells were then grown following the same method used for growing cells in 96-well plates, as described in the growth assay with slight modifications. Four 220 μL aliquots of each sample were transferred to a 0.5 mL sterile 96-well plate and grown on a rotator for 17 h at 30 °C. After the 17 h incubation, 200 μL from each aliquot were pooled into a sterilized Eppendorf tube. Cells were pelleted in a microcentrifuge and washed three times with sterile distilled water and resuspended in sterile water. Cells were then diluted appropriately and plated onto five SC-Ura agar plates to determine the total number of viable cells and onto eight SC-HIS agar plates to determine the number of His+ recombinants (DEL events). All plates were grown at 30 °C for 3 days, and the colonies were counted. The recombination frequency (DEL frequency) for each sample was calculated by dividing the total number of recombinants by the total number of viable cells. Frequencies from three different biological samples were then averaged and presented as the deletion frequency with SD (Supporting Information Table S2). Statistical significance of recombination frequencies between controls and treated samples were determined by the Student’s t test. The fold increase in deletion formation was calculated by dividing the deletion frequencies of treated versus deletion frequencies of untreated samples. Fold deletion formation values were then analyzed by QSAR. The percent viability after a phenolic compound treatment for a deletion assay was calculated by dividing the number of viable cells (colony forming units in SC-Ura) in the treated sample by the number of viable cells in the untreated sample (Supporting Information Table S2).
QSAR Analysis
The comparative quantitative structure–activity relationship suite of programs (BioByte Inc., Claremont, CA) was used to derive the various models.20,54P represents the partition coefficients of the phenols. In this study, calculated log P (clog P) values were utilized to represent hydrophobicity. Sigma plus (σ+) is Brown’s refinement of the Hammett electronic constant, sigma (σ).55 In all models, n represents the number of data points in the study, r is the correlation coefficient, s is the standard deviation of the regression equation, and q2 comprises the cross-validated r2. The 95% confidence intervals for the terms in the equations are listed in parenthesis. The cross-validated r2 designated by q2 is obtained by using the leave-one-out procedure.56
Pinning Assay
Single colonies were inoculated into liquid SC media and grown to saturation at 30 °C. One hundred microliters of each saturated culture were aliquoted into a 96-well plate and serially diluted (1:10, 1:100, 1:1000, and 1:10 000). Using a 96-well floating-pin replicator, 3 μL was transferred onto SC agar plates with 2% DMSO and IC50, or IC80 values of BHA, BHT, or no phenol as the control. The plates were imaged after 3 days of growth at 30 °C with an LAS 500 imager (Figure 5).
Acknowledgments
We thank R. Schiestl and Adam Bailis for kindly providing yeast strains.
Glossary
Abbreviations
- BDE
bond dissociation energy
- BHA
butylated hydroxyl anisole
- BHT
2,6-di-tert-butyl-4-methyl-phenol
- BHA-2
2-tert-butyl-4-methoxy-phenol
- BHA-3
3-tert-butyl-4-methoxy-phenol
- CFR
co-field rotations
- CQSAR
comparative quantitative structure–activity relationship
- DEL
yeast deletion assay
- DMSO
dimethyl sulfoxide
- EDG
electron donating groups
- EWG
electron withdrawing groups
- IC
inhibitory concentration
- log P
partition coefficient
- clog P
calculated log P
- MR
molar refraction
- OD
optical density
- q2
cross validated r2
- QSAR
quantitative structure activity relationship
- s
standard deviation of the regression
- SC
synthetic complete, synthetic minimal media (defined yeast media)
- SC-X
synthetic drop-out medium
- YPD
rich yeast media contains yeast nitrogen base, peptone, dextrose
- σ
Hammet sigma constant
- σ+
Brown’s sigma constant
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.7b01200.
IC values for each compound used and equation from linear dose-dependent survival curve; graph with dose-dependent survival curves for each compound used; degree of fold increase in deletion formation over the untreated control; and percent viability for each compound tested (ZIP)
Author Present Address
§ Department of Neurology, Northwestern University, 3510 N Springfield, Unit 3N Chicago, IL 60634 (C.V.).
Author Present Address
∥ 9961 Sierra Ave, Kaiser Fontana, Fontana CA 92335 (J.S.).
Author Present Address
⊥ Boston Children’s Hospital, 300 Longwood Ave. Boston, MA 02115 (C.R.).
This work was supported by funding from the National Institutes of Health grant (1R15-ESO14812), Pomona College Research funds, and Student Summer Research fellowships from HHMI, the Rose Hills Foundation, and the Merck and Pomona College.
The authors declare no competing financial interest.
Supplementary Material
References
- Hadi S. M.; Bhat S. H.; Azmi A. S.; Hanif S.; Shamim U.; Ullah M. F. Oxidative breakage of cellular DNA by plant polyphenols: a putative mechanism for anticancer properties. Semin. Cancer Biol. 2007, 17, 370–376. 10.1016/j.semcancer.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Williams G. M.; Iatropoulos M. J.; Whysner J. Safety assessment of butylated hydroxyanisole and butylated hydroxytoluene as antioxidant food additives. Food Chem. Toxicol. 1999, 37, 1027–1038. 10.1016/s0278-6915(99)00085-x. [DOI] [PubMed] [Google Scholar]
- Jovanovic S. V.; Steenken S.; Tosic M.; Marjanovic B.; Simic M. G. Flavonoids as Antioxidants. J. Am. Chem. Soc. 1994, 116, 4846–4851. 10.1021/ja00090a032. [DOI] [Google Scholar]
- Arnold W. M.; Zimmermann U.; Pauli W.; Benzing M.; Niehns C.; Ahlers J. The comparative influence of substituted phenols (especially chlorophenols) on yast cells assayed by electro-rotation and other methods. Biochim. Biophys. Acta, Biomembr. 1988, 942, 83–95. 10.1016/0005-2736(88)90277-5. [DOI] [PubMed] [Google Scholar]
- Oikawa S.; Nishino K.; Oikawa S.; Inoue S.; Mizutani T.; Kawanishi S. Oxidative DNA damage and apoptosis induced by metabolites of butylated hydroxytoluene. Biochem. Pharmacol. 1998, 56, 361–370. 10.1016/s0006-2952(98)00037-9. [DOI] [PubMed] [Google Scholar]
- Klein P. J.; Van Vleet T. R.; Hall J. O.; Coulombe R. A. Jr. Effects of dietary butylated hydroxytoluene on aflatoxin B1-relevant metabolic enzymes in turkeys. Food Chem. Toxicol. 2003, 41, 671–678. 10.1016/s0278-6915(02)00332-0. [DOI] [PubMed] [Google Scholar]
- Stich H. F. The Beneficial and Hazardous Effects of Simple Phenolic Compounds. Mutat. Res., Genet. Toxicol. 1991, 259, 307–324. 10.1016/0165-1218(91)90125-6. [DOI] [PubMed] [Google Scholar]
- Hansch C.; Maloney P. P.; Fujita T.; Muir R. M. Correlation of Biological Activity of Phenoxyacetic Acids with Hammett Substituent Constants and Partition Coefficients. Nature 1962, 194, 178–180. 10.1038/194178b0. [DOI] [Google Scholar]
- Selassie C. D.; DeSoyza T. V.; Rosario M.; Gao H.; Hansch C. Phenol toxicity in leukemia cells: a radical process?. Chem.-Biol. Interact. 1998, 113, 175–190. 10.1016/s0009-2797(98)00027-1. [DOI] [PubMed] [Google Scholar]
- Melzig M. F.; Tran G. D.; Henke K.; Selassie C. D.; Verma R. P. Inhibition of neutrophil elastase and thrombin activity by caffeic acid esters. Pharmazie 2005, 60, 869–873. [PubMed] [Google Scholar]
- Wright J. S.; Shadnia H. Computational modeling of substituent effects on phenol toxicity. Chem. Res. Toxicol. 2008, 21, 1426–1431. 10.1021/tx800085a. [DOI] [PubMed] [Google Scholar]
- Selassie C. D.; Shusterman A. J.; Kapur S.; Verma R. P.; Zhang L. T.; Hansch C. On the toxicity of phenols to fast growing cells. A QSAR model for a radical-based toxicity. J. Chem. Soc., Perkin Trans. 2 1999, 2729–2733. 10.1039/a905764a. [DOI] [Google Scholar]
- Núñez M. E.; Hall D. B.; Barton J. K. Long-range oxidative damage to DNA: effects of distance and sequence. Chem. Biol. 1999, 6, 85–97. 10.1016/s1074-5521(99)80005-2. [DOI] [PubMed] [Google Scholar]
- Gaytán B. D.; Vulpe C. D. Functional toxicology: tools to advance the future of toxicity testing. Front. Genet. 2014, 5, 110. 10.3389/fgene.2014.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan R. J.; Schiestl R. H. Aniline and its metabolites generate free radicals in yeast. Mutagenesis 1997, 12, 215–220. 10.1093/mutage/12.4.215. [DOI] [PubMed] [Google Scholar]
- Brennan R. J.; Schiestl R. H.. Detecting carcinogens with the yeast DEL assay. Genetic Recombination; Methods in Molecular Biology; Springer, 2004; Vol. 262, pp 111–124. [DOI] [PubMed] [Google Scholar]
- Kirpnick Z.; Homiski M.; Rubitski E.; Repnevskaya M.; Howlett N.; Aubrecht J.; Schiestl R. H. Yeast DEL assay detects clastogens. Mutat. Res., Genet. Toxicol. Environ. Mutagen. 2005, 582, 116–134. 10.1016/j.mrgentox.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Brennan R. J.; Schiestl R. H. The aromatic amine carcinogens o-toluidine and o-anisidine induce free radicals and intrachromosomal recombination in Saccharomyces cerevisiae. Mutat. Res., Fundam. Mol. Mech. Mutagen. 1999, 430, 37–45. 10.1016/s0027-5107(99)00118-9. [DOI] [PubMed] [Google Scholar]
- Verma R. P.; Kapur S.; Barberena O.; Shusterman A.; Hansch C. H.; Selassie C. D. Synthesis, cytotoxicity, and QSAR analysis of X-thiophenols in rapidly dividing cells. Chem. Res. Toxicol. 2003, 16, 276–284. 10.1021/tx020103q. [DOI] [PubMed] [Google Scholar]
- Hansch C.; Leo A.. Exploring QSAR. Fundamentals and Applications in Chemistry and Biology, 1st ed.; Heller S. R., Ed.; American Chemical Society: Washington, DC, 1995; p 557. [Google Scholar]
- Selassie C. D.; Garg R.; Kapur S.; Kurup A.; Verma R. P.; Mekapati S. B.; Hansch C. Comparative QSAR and the radical toxicity of various functional groups. Chem. Rev. 2002, 102, 2585–2605. 10.1021/cr940024m. [DOI] [PubMed] [Google Scholar]
- Sommers C. H.; Schiestl R. H. Effect of benzene and its closed ring metabolites on intrachromosomal recombination in Saccharomyces cerevisiae. Mutat. Res., Fundam. Mol. Mech. Mutagen. 2006, 593, 1–8. 10.1016/j.mrfmmm.2005.06.026. [DOI] [PubMed] [Google Scholar]
- Arnold W. M.; Zimmermann U. Rotation of an isolated cell in a rotating electric field. Naturwissenschaften 1982, 69, 297–298. 10.1007/bf00396446. [DOI] [PubMed] [Google Scholar]
- Selassie C. D.; Kapur S.; Verma R. P.; Rosario M. Cellular apoptosis and cytotoxicity of phenolic compounds: a quantitative structure-activity relationship study. J. Med. Chem. 2005, 48, 7234–7242. 10.1021/jm050567w. [DOI] [PubMed] [Google Scholar]
- Orlean P. Architecture and biosynthesis of the Saccharomyces cerevisiae cell wall. Genetics 2012, 192, 775–818. 10.1534/genetics.112.144485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop A. J. R.; Schiestl R. H. Role of homologous recombination in carcinogenesis. Exp. Mol. Pathol. 2003, 74, 94–105. 10.1016/s0014-4800(03)00010-8. [DOI] [PubMed] [Google Scholar]
- Stratton M. R.; Campbell P. J.; Futreal P. A. The cancer genome. Nature 2009, 458, 719–724. 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosato V.; Grüning N.-M.; Breitenbach M.; Arnak R.; Ralser M.; Bruschi C. V. Warburg effect and translocation-induced genomic instability: two yeast models for cancer cells. Front. Oncol. 2013, 2, 212. 10.3389/fonc.2012.00212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettier G.; Feng Q.; de Mayolo A. A.; Erdeniz N.; Reid R. J. D.; Lisby M.; Mortensen U. H.; Rothstein R. The role of DNA double-strand breaks in spontaneous homologous recombination in S. cerevisiae. PLoS Genet. 2006, 2, e194 10.1371/journal.pgen.0020194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan R. J.; Swoboda B. E. P.; Schiestl R. H. Oxidative mutagens induce intrachromosomal recombination in yeast. Mutat. Res., Fundam. Mol. Mech. Mutagen. 1994, 308, 159–167. 10.1016/0027-5107(94)90151-1. [DOI] [PubMed] [Google Scholar]
- Maines S.; Negritto M. C.; Wu X.; Manthey G. M.; Bailis A. M. Novel mutations in the RAD3 and SSL1 genes perturb genome stability by stimulating recombination between short repeats in Saccharomyces cerevisiae. Genetics 1998, 150, 963–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansch C.; Gao H. Comparative QSAR: Radical Reactions of Benzene Derivatives in Chemistry and Biology. Chem. Rev. 1997, 97, 2995–3060. 10.1021/cr9601021. [DOI] [PubMed] [Google Scholar]
- Michalowicz J.; Duda W. Phenols—Sources and toxicity. Pol. J. Environ. Stud. 2007, 16, 347–362. [Google Scholar]
- Zhang L.; Gao H.; Hansch C.; Selassie C. D. Molecular orbital parameters and comparative QSAR in the analysis of phenol toxicity to leukemia cells. J. Chem. Soc., Perkin Trans. 2 1998, 2553–2556. 10.1039/a801684d. [DOI] [Google Scholar]
- Piecuch A.; Obłąk E. Yeast ABC proteins involved in multidrug resistance. Cell. Mol. Biol. Lett. 2014, 19, 1–22. 10.2478/s11658-013-0111-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M.; Hanna M.; Li J.; Butcher S.; Dai H.; Xiao W. Creation of a hyperpermeable yeast strain to genotoxic agents through combined inactivation of PDR and CWP genes. Toxicol. Sci. 2010, 113, 401–411. 10.1093/toxsci/kfp267. [DOI] [PubMed] [Google Scholar]
- Zhang M.; Liang Y.; Zhang X.; Xu Y.; Dai H.; Xiao W. Deletion of yeast CWP genes enhances cell permeability to genotoxic agents. Toxicol. Sci. 2008, 103, 68–76. 10.1093/toxsci/kfn034. [DOI] [PubMed] [Google Scholar]
- Valko M.; Rhodes C. J.; Moncol J.; Izakovic M.; Mazur M. Free radicals, metals and antioxidants in oxidative stress-induced cancer. Chem.-Biol. Interact. 2006, 160, 1–40. 10.1016/j.cbi.2005.12.009. [DOI] [PubMed] [Google Scholar]
- Forester S. C.; Lambert J. D. The role of antioxidant versus pro-oxidant effects of green tea polyphenols in cancer prevention. Mol. Nutr. Food Res. 2011, 55, 844–854. 10.1002/mnfr.201000641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreto G.; Madureira D.; Capani F.; Aon-Bertolino L.; Saraceno E.; Alvarez-Giraldez L. D. The role of catechols and free radicals in benzene toxicity: An oxidative DNA damage pathway. Environ. Mol. Mutagen. 2009, 50, 771–780. 10.1002/em.20500. [DOI] [PubMed] [Google Scholar]
- Chalissery J.; Jalal D.; Al-Natour Z.; Hassan A. H. Repair of Oxidative DNA Damage in Saccharomyces cerevisiae. DNA Repair 2017, 51, 2–13. 10.1016/j.dnarep.2016.12.010. [DOI] [PubMed] [Google Scholar]
- Shafirovich V.; Geacintov N. E. Removal of oxidatively generated DNA damage by overlapping repair pathways. Free Radical Biol. Med. 2017, 107, 53–61. 10.1016/j.freeradbiomed.2016.10.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugasawa K.; Hanaoka F. Sensing of DNA damage by XPC/Rad4: one protein for many lesions. Nat. Struct. Mol. Biol. 2007, 14, 887–888. 10.1038/nsmb1007-887. [DOI] [PubMed] [Google Scholar]
- Guzder S. N.; Sung P.; Prakash L.; Prakash S. Nucleotide excision repair in yeast is mediated by sequential assembly of repair factors and not by a pre-assembled repairosome. J. Biol. Chem. 1996, 271, 8903–8910. 10.1074/jbc.271.15.8903. [DOI] [PubMed] [Google Scholar]
- Mardiros A.; Benoun J. M.; Haughton R.; Baxter K.; Kelson E. P.; Fischhaber P. L. Rad10-YFP focus induction in response to UV depends on RAD14 in yeast. Acta Histochem. 2011, 113, 409–415. 10.1016/j.acthis.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzder S. N.; Sommers C. H.; Prakash L.; Prakash S. Complex formation with damage recognition protein Rad14 is essential for Saccharomyces cerevisiae Rad1-Rad10 nuclease to perform its function in nucleotide excision repair in vivo. Mol. Cell. Biol. 2006, 26, 1135–1141. 10.1128/mcb.26.3.1135-1141.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volker M.; Moné M. J.; Karmakar P.; van Hoffen A.; Schul W.; Vermeulen W.; Hoeijmakers J. H. J.; van Driel R.; van Zeeland A. A.; Mullenders L. H. F. Sequential assembly of the nucleotide excision repair factors in vivo. Mol. Cell 2001, 8, 213–224. 10.1016/s1097-2765(01)00281-7. [DOI] [PubMed] [Google Scholar]
- Swanson R. L.; Morey N. J.; Doetsch P. W.; Jinks-Robertson S. Overlapping specificities of base excision repair, nucleotide excision repair, recombination, and translesion synthesis pathways for DNA base damage in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999, 19, 2929–2935. 10.1128/mcb.19.4.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva C. R.; Almeida G. S.; Caldeira-de-Araújo A.; Leitão A. C.; de Pádula M. Influence of Ogg1 repair on the genetic stability of ccc2 mutant of Saccharomyces cerevisiae chemically challenged with 4-nitroquinoline-1-oxide (4-NQO). Mutagenesis 2016, 31, 107–114. 10.1093/mutage/gev062. [DOI] [PubMed] [Google Scholar]
- Boiteux S.; Guillet M. Abasic sites in DNA: repair and biological consequences in Saccharomyces cerevisiae. DNA Repair 2004, 3, 1–12. 10.1016/j.dnarep.2003.10.002. [DOI] [PubMed] [Google Scholar]
- de Graaf B.; Clore A.; McCullough A. K. Cellular pathways for DNA repair and damage tolerance of formaldehyde-induced DNA-protein crosslinks. DNA Repair 2009, 8, 1207–1214. 10.1016/j.dnarep.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiestl R. H.; Gietz R. D.; Mehta R. D.; Hastings P. J. Carcinogens induce intrachromosomal recombination in yeast. Carcinogenesis 1989, 10, 1445–1455. 10.1093/carcin/10.8.1445. [DOI] [PubMed] [Google Scholar]
- Amberg D. C.; Burke D.; Strathern J. N.. Methods in yeast genetics. A Cold Spring Harbor Laboratory Course Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor NY, 2000. [Google Scholar]
- Hansch C.; Hoekman D.; Leo A.; Zhang L. T.; Li P. The Expanding Role of Quantitative Structure-Activity Relationships (Qsar) in Toxicology. Toxicol. Lett. 1995, 79, 45–53. 10.1016/0378-4274(95)03356-p. [DOI] [PubMed] [Google Scholar]
- Selassie C.; Verma R. P.. History of Quantitative Structure–Activity Relationships. Burger’s Medicinal Chemistry and Drug Discovery; John Wiley & Sons, Inc., 2010; pp 1–95. [Google Scholar]
- Cramer R. D.; Patterson D. E.; Bunce J. D. Comparative molecular field analysis (CoMFA). 1. Effect of shape on binding of steroids to carrier proteins. J. Am. Chem. Soc. 1988, 110, 5959–5967. 10.1021/ja00226a005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.