SUMMARY
The immune system is comprised of diverse cell types that coordinate responses to infection and maintain tissue homeostasis. In each of these cells, extracellular cues determine highly specific epigenetic landscapes and transcriptional profiles to promote immunity while maintaining homeostasis. New evidence indicates that long noncoding RNAs (lncRNAs) play critical roles in epigenetic and transcriptional regulation in mammals. Thus, lncRNAs have emerged as key regulatory molecules of immune cell gene expression programs in response to microbial and tissue-derived cues. Herein, we review how lncRNAs control the function and homeostasis of cell populations during immune responses, emphasizing the diverse molecular mechanisms by which lncRNAs tune highly contextualized transcriptional programs. Moreover, we discuss the new challenges faced in interrogating lncRNA mechanisms and function in the immune system.
The regulatory role of the non-coding genome
A longstanding observation in biology is that while the number of protein coding genes has remained relatively stable across multicellular organisms, the genomic content of non-coding DNA increases with the developmental complexity of organisms [1]. This seemingly paradoxical finding, termed the G-value paradox, suggests that the degree of regulatory control exerted by non-coding regions over protein-coding genes determines phenotypic complexity, which has increased throughout evolution [1]. In support of this inference, a number of high-throughput sequencing projects have recently shown that a great proportion of the mammalian non-coding genome comprises an immense network of cis-regulatory elements such as promoters, enhancers, and super-enhancers [2–4]. Strikingly, these consortia also showed that nearly 70% of the human non-coding genome shows evidence of transcriptional activity. The function of the majority of these non-coding transcripts is unknown; nevertheless, several classes of non-coding RNAs have been found to contribute to the transcriptional and epigenetic regulation of gene expression programs. Among these functional non-coding RNAs, lncRNAs are the most abundant and have been extensively studied in recent years. As such, the role of lncRNAs in immune regulation has received a significant amount of attention and will be the focus of this review.
A diverse array of cells from the innate and adaptive immune system orchestrate protective immune responses that clear infections and promote the development of long term immunological memory while preventing the deleterious consequences of prolonged inflammation. The highly-specialized functions of each of these immune cell types are determined by their unique transcriptional programs. While much is known about how transcription factors and different extracellular cues regulate the development and functional specialization of innate and adaptive immune cells, our knowledge regarding the roles of lncRNAs in the immune system is in its infancy. A growing body of evidence indicates that lncRNAs are expressed in a more cell type-specific manner than protein-coding genes and are rapidly induced by unique tissue-derived and environmental signals [5, 6]. As such, these molecules have started to emerge as highly cell type-specific regulators of gene expression programs in the immune system. In the following sections, we will provide an overview of how, through diverse mechanisms of action, lncRNAs tightly control the function and homeostasis of specific cell populations during innate and adaptive immune responses.
MECHANISMS OF ACTION OF LNCRNA LOCI
LncRNAs accomplish a diverse range of biological functions through an equally diverse range of molecular mechanisms [7]. Important recent work has demonstrated that certain ribosome-associated lncRNAs which, by definition, were annotated as non-coding are now reported to code for short peptides with biological function [8–10]. Whether the cryptic translation events of other ribosome-associated lncRNAs in the immune system have meaningful biological function has yet to be determined. Therefore, this type of lncRNA is not discussed further in this review.
LncRNA molecules themselves can directly interact with proteins, DNA, and other RNAs to exert their functions. Nevertheless, not all lncRNA loci that are biologically active depend on the function of the RNA molecule itself. Herein, we distill the biological mechanisms of lncRNAs loci into 3 different groups: (i) lncRNA loci in which the RNA molecule itself is functional, (ii) lncRNA loci in which the act of transcription, but not the lncRNA molecule, has a functional role, and (iii) lncRNAs that represent proxy signals for active cis-regulatory elements. Throughout this section, we aim to exemplify each functional grouping with lncRNA loci described in the immune system, while in the following sections we will describe in detail the biological process they regulate in the context of immune responses.
Functional lncRNAs
The first lncRNAs were characterized by demonstrating direct biological activity of the lncRNA molecule itself [11–13], and this remains the gold standard for characterizing lncRNA loci. Functional lncRNAs can localize to the nucleus or cytosol to specifically interact with proteins, DNA, or other RNAs to perform their molecular functions. The sequence constraints for RNA folding and structure are more lenient than those for protein folding, which has likely helped facilitate the evolution of such mechanistically versatile lncRNA molecules. The recent development of novel RNA-centric tools that directly test lncRNA structure, localization, and interactomes (Box 1) has paved the way for our understanding of the mechanisms of action of lncRNAs. Conceptually, we group functional lncRNAs within two classes: those that serve as inhibitors and those that serve as facilitators of binding partner interactions.
BOX 1. TOOLBOX FOR DETERMINING LNCRNA-INTERACTING MOLECULES AND SECONDARY STRUCTURE.
lncRNAs can associate with DNA, RNA, and proteins to impart an array of functions, many of which are still being discovered. Identifying specific lncRNA-binding partners, in addition to the structural motifs of lncRNAs that might govern these associations, can provide insight into the functional scope of lncRNAs. New RNA-centric tools use in vivo cross-linking methods and high-throughput technologies to discover the binding partners of a particular lncRNA of interest on a global scale. Three methods analogous to ChIP-seq have emerged in recent years to probe lncRNA-biomolecule interactions: ChIRP (chromatin immunoprecipitation by RNA purification), CHART (capture hybridization analysis of RNA targets), and RAP (RNA antisense purification) [66–68]. Although these techniques differ in specific details, they are similar in principle and take advantage of RNA hybridization to pull down a specific lncRNA along with its binding partners. By altering the cross-linking and purification conditions, DNA, RNA and protein partners can be co-purified with a lncRNA and subsequently identified through sequencing and mass spectrometry [69]. These RNA-centric methods for discovering binding partners will be an important component in untangling lncRNA functions.
In addition to identifying lncRNA binding partners, probing the secondary structures of lncRNAs will add to our understanding of lncRNA function. Given that many lncRNA sequences are not well conserved, it has been proposed that their secondary structures could form modular domains that bring an array of binding partners together in a context-specific manner. A new method called PARIS (psoralen analysis of RNA interactions and structures) identifies intra- and inter-molecular RNA-RNA interactions on a genome-wide scale at high resolution, in a manner analogous to 3C for chromatin-chromatin interactions [70]. Unlike ChIRP, CHART, and RAP, this new technology does not probe for a specific lncRNA, and instead analyzes all cross-linked RNAs on a global scale. In addition, PARIS can identify conserved RNA structures through cross-species comparisons, allowing to interrogate the possibility that lncRNA secondary structures are conserved in lieu of their sequences. By first identifying the secondary structures of lncRNAs, researchers can then uncover whether these conformations aid in function and impart binding specificity by introducing point mutations that disrupt these secondary structures. A combination of these approaches will be necessary to characterize lncRNA structure and interactomes in the immune system.
Class I of functional lncRNAs: Inhibitors – keeping things apart
Many protein-protein interactions conform to a classic molecular decoy archetype, wherein the decoy molecule binds to and inhibits one partner in an interaction; a classic example of this is the non-signaling interleukin 1 receptor type 2 (IL1R2), which binds the cytokine IL-1β and prevents it from signaling through its normal receptor [14]. Here we expand this to include lncRNA-mediated processes in which factors are inhibited from interacting with one another (Figure 1A). This class most obviously applies to lncRNAs that act as sinks for microRNAs, such as the lncRNA Phosphatase And Tensin Homolog Pseudogene 1 (PTENP1), whose 3′ UTR binds the same set of miRNAs sequences that normally target the tumor-suppressor gene PTEN [15]. In addition, this class also includes lncRNAs that specifically inhibit classical protein-protein interactions such as the lncRNA lnc-DC [16]. Lnc-DC regulates the differentiation and activation profile of dendritic cells (DCs) in humans by binding STAT3 in the cytoplasm and preventing its dephosphorylation and inactivation by the tyrosine phosphatase SHP1 [16]. Additionally, this inhibitory class includes lncRNAs that block protein-DNA interactions, such as pseudogene-derived lncRNA Lethe [17]. Lethe physically associates with NF-kB p65 (RelA) subunit and directly or indirectly blocks its recruitment to the promoters of certain immune genes such as IL6 and IL8 [17]. Of note, lncRNAs can directly act post-transcriptionally, as their function is not dependent on the process of translation. Thus, by blocking key DNA, RNA, or protein interactions, lncRNAs can exert rapid potent biological outcomes in the immune system.
Figure 1. Mechanisms of lncRNA action.
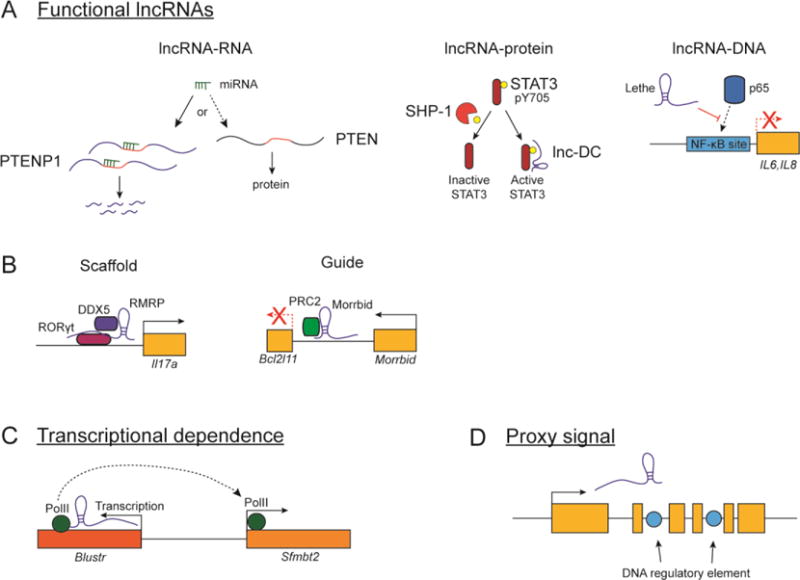
(A) lncRNA mechanisms that depend on the RNA molecule itself include lncRNA-RNA, lncRNA-protein, and lncRNA-DNA-based mechanisms. For example, PTENP1 encodes miRNA target sequences shared by PTEN and acts as a “sponge” for miRNAs targeting PTEN (left). lnc-DC interacts directly with the protein STAT3 and blocks its dephosphorylation by the phosphatase SHP-1 (middle). lncRNA Lethe hinders the recruitment of NF-κB p65 subunit to its target DNA sequences (right).
(B) lncRNAs can activate gene expression by acting as scaffolds for interactions between other proteins, such as lncRNA RMRP (left), or as guides to direct the recruitment of chromatin modifying complexes to neighboring genes, such as Morrbid (right).
(C) The act of lncRNA transcription can intrinsically account for the activity of some lncRNA loci. Transcription across the lncRNA Blustr directly promotes transcription of its neighboring gene Sfmbt2.
(D) Some lncRNAs may not regulate transcription of their target genes, but instead act as a proxy signals for important DNA regulatory elements embedded within the lncRNA locus.
Class II of functional lncRNAs: Facilitators – bringing things together
LncRNAs have been described to form complexes with protein, RNA, and/or DNA, and thus facilitate the interaction of specific binding partners. This class of RNA-dependent mechanism includes lncRNAs that serve as “scaffolds” or “guides” (Figure 1B). “Scaffold” lncRNAs can function as molecular platforms upon which other DNA, RNA, and protein molecules can assemble. For example, the lncRNA RMRP facilitates an interaction between the nuclear receptor RAR-related orphan receptor gamma (RORγt) and DEAD-box protein 5 (DDX5) [18]. This RORγt-DDX5 interaction is necessary for transcriptional co-activation of certain T helper 17 (Th17) genes [18]. On the other hand, lncRNAs that serve as “guides” can bind other DNA, RNA, and protein molecules and target their localization to induce a specific biological function. For example, X-inactive specific transcript (Xist) is, as its name implies, involved in the silencing of the second X chromosome in female placental mammals. Xist is expressed by and coats the inactive X chromosome from which it is transcribed by binding and promoting the recruitment of multiple repressive factors including Polycomb, SHARP, and others [19–21]. Similarly, in the immune system, the lncRNA myeloid RNA regulator of Bim induced death (Morrbid) silences its neighboring pro-apoptotic gene Bcl2l11 (Bim) in an allele specific manner by promoting the enrichment of polycomb repressive complex 2 (PRC2) within the Bcl2l11 promoter [22].
The scaffold and guide classes are closely related. For example, a lncRNA can bind a protein and be required for that protein’s localization to a specific genomic location; yet, this localization can occur through an independent biological effect of the lncRNA. The distinction of the scaffold and guide classes is typically determined by whether the lncRNA’s function is either directly or indirectly dependent on localization. For instance, Xist has the ability to associate with many different protein complexes; nevertheless, its function is predominately dependent on the direct and indirect recruitment of these complexes to the inactive X chromosome. As such, for simplicity, Xist is typically categorized as a lncRNA guide. Of note, the general model of lncRNA direct protein recruitment is complicated by the lack of known protein-binding motifs within lncRNAs and little data on RNA binding specificity (reviewed in [23]). Future studies on lncRNA motifs and structure are needed to better understand how lncRNAs interact with specific proteins and how lncRNAs localize to specific sites.
Transcription across lncRNA has a functional role
As previously discussed, lncRNAs demonstrate exquisite cell type-specific expression and many are responsive to specific extracellular cues. Given the strict regulation of lncRNA biogenesis and that many lncRNAs impact the expression of neighboring genes, it has been suggested that the process of transcription and/or splicing of lncRNAs may impact the expression of spatially proximal genes through the recruitment of specific protein factors [24–26] (Figure 1C). This concept was recently dissected in further detail through the study of the protein coding gene Sfmbt2 and its neighboring lncRNA Blustr. The authors found that Sfmbt2 is upregulated by the splicing and transcription of Blustr, in a mechanism independent of the sequence of Blustr but likely to involve the recruitment of polymerases and chromatin-modifiers [27]. By elegantly inserting a synthetic polyadenylation sequence at different locations within this locus in vitro, these effects were found to be dependent on the length of transcription across the Blustr locus [27]. Additionally, promoter-proximal splice sites are known to enhance transcription, and deletion of this site of Blustr also impacted Sfmbt2 transcription [27]. In parallel, Anderson and colleagues demonstrated in vivo that the heart development regulator Hand2 is controlled in cis by the lncRNA upperhand (Uph), but that this control was independent of the sequence of Uph [28]. Of note, the authors found that transcription of Uph was required to maintain the chromatin state of a cardiac superenhancer-like element within the Uph locus [28]. Although it is not clear mechanistically how transcription and/or splicing recruit critical regulatory factors to coordinate the expression of genes in cis, these studies are highly relevant to the lncRNA field as they clearly demonstrate that lncRNA biogenesis itself can impact proximal genes in a transcript and sequence independent manner. How transcription across lncRNA loci impact immune cells is still yet to be determined.
LncRNAs as proxy signals for cis-regulatory elements
The promoters of many lncRNA loci are conserved across species and have enhancer chromatin signatures, suggesting that certain lncRNA promoters may function as cis-acting enhancer elements that result in bystander noncoding transcripts [27, 29]. Given that lncRNA loci frequently contain or are in close proximity to important regulatory DNA elements, the production of lncRNAs from these loci can delineate critical cis-regulatory DNA elements within these loci (Figure 1D). For example, mapping of RNA polymerase II using Chromatin Interaction Analysis with Paired-End-Tag sequencing (ChIA-PET) revealed strong genome-wide promoter-centered intergenic interactions, suggesting that promoters of coding and non-coding genes may have dual functions as enhancers [30]. Therefore, certain lncRNA transcripts may arise as non-functional by-products, yet they may serve as proxy signals of the activity of important regulatory elements. For example, deletion of the lncRNA down-stream of Cdkn1b (Lockd) locus was found to impact the transcription of its neighboring gene Cdkn1b; however, pre-mature polyadenylation of Lockd had no impact on Cdkn1b transcription [31]. The Lockd promoter was found to have many enhancer-like features suggesting that this lncRNA locus may indeed function through DNA elements within its promoter and that the RNA transcript produced from this locus marks an important regulatory element [31]. Additionally, we recently showed that the lncRNA Rroid is specifically expressed in group 1 innate lymphoid cells (ILCs), and marks a critical cis-regulatory element important for the maturation, function, and lineage identify of these cells while being dispensable for the development and homeostasis of closely related ILC2s and ILC3s (Mowel et al. 2017), suggesting that other cell-type specific lncRNAs will demarcate critical cis-regulatory elements that control highly specialized processes in immune cells.
BIOLOGICAL FUNCTIONS OF LNCRNAS IN THE IMMUNE SYSTEM
LNCRNAS IN THE INNATE IMMUNE SYSTEM
In biological immune responses, the cells of the innate immune system – macrophages, dendritic cells, neutrophils, basophils, eosinophils, and ILCs – must undergo rapid changes in their gene expression programs to immediately respond to infection, tissue damage, or other changes in environmental conditions. As discussed above, one of the key advantages of lncRNA-mediated gene regulation is that existing pools of transcription factors can be utilized to promote expression of target lncRNAs, which can then rapidly affect expression of both individual genes or sets of genes within minutes without requiring new protein synthesis. As such, it is not surprising that many of the initial discoveries of lncRNAs in the immune system were focused on the regulation of inflammation in innate immune cells.
lncRNAs regulate inflammatory gene expression in innate immune cells
One of the first lncRNAs described in the immune system, lincRNA-Cox2, was discovered in bone marrow-derived dendritic cells stimulated with lipopolysaccharide (LPS), an agonist of TLR4 [32]. Strikingly, lincRNA-Cox2 is rapidly induced within 30 minutes in RAW264.6 macrophages in response to inflammatory cues [33]. Carpenter et al. showed that this lncRNA regulates a large set of inflammatory response genes, as knockdown of lincRNA-Cox2 resulted in increased expression of a gene set including Ccl5 and Cx3cl1 [34] (Figure 2A). lincRNA-Cox2 interacts with hnRNP-A/B and hnRNP-A2/B1 in vitro, and knockdown of both hnRNP-A/B and hnRNP-A2/B1 results in de-repression of Ccl5 [34]. In contrast, other work has shown that lincRNA-Cox2 in immortalized macrophage cell lines interacts with the SWI/SNF chromatin remodeling complex to promote its recruitment to the Ccl5 promoter and activate transcription [33]. lincRNA-Cox2 has also been shown to interact with and recruit the Mi2/NuRD repressive complex to the Il12b promoter in response to TNF stimulation in intestinal epithelial cell lines to silence Il12b expression [35].
Figure 2. lncRNAs in the innate immune system.
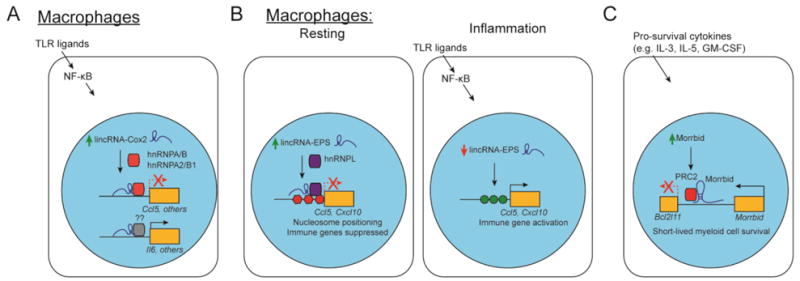
(A) LincRNA-Cox2 is induced by TLR ligands and associates with hnRNPA/B and hnRNPA2/B1 to suppress transcription of target genes, such as Ccl5, and promotes the expression of genes such as Il6 through a yet unknown mechanism.
(B) Resting macrophages express high levels lincRNA-EPS, which interacts with hnRNPL to suppress target genes. Upon stimulation, expression of lincRNA-EPS is reduced, and repression of these targets is released.
(C) In short-lived myeloid cells, pro-survival cytokines induce expression of Morrbid, which acts as a guide to target the repressive PRC2 complex to the Bcl2l11 locus (which encodes the pro-apoptotic molecule Bim), to promote the survival of these cells.
This type of mechanism, wherein lncRNAs act as guides or scaffolds to rapidly regulate the recruitment chromatin modifying complexes to their target genes, is likely widespread. Indeed, a recent study identified lnc13, which is downregulated in response to LPS, as a repressor of genes involved in inflammatory pathways such as Myd88, Stat1, Stat3, and Il1ra. This effect is mediated through the regulation of HDAC1 and hnRNPD recruitment to their target genes in an RNA-dependent mechanism [36]. Additionally, lincRNA-EPS suppresses genes such as Cxcl10 and Ccl5 by interacting with hnRNPL and regulating nucleosomal positioning at these target loci [37] (Figure 2B). In vivo, mice deficient for lincRNA-EPS exhibit significant increases in circulating proinflammatory cytokines and are extremely susceptible to LPS challenge.
Constitutively expressed transcription factors in innate immune cells transduce signals within minutes of TLR ligation, promoting inflammatory gene expression. lncRNAs regulate this process not only by directing the recruitment of chromatin modifying complexes to target genes, but can also directly control the ability of transcription factors to form complexes with other proteins or bind their target loci. As described above, one example of this activity is lnc-DC, which is highly expressed in human DCs and directly promotes STAT3 activity by preventing its dephosphorylation [16]. Functionally, this results in reduced expression of the costimulatory molecules CD80 and CD86, as well as MHC-II [16]. Thus, lnc-DC and upstream signals regulating its expression may play an important role in CD4+ T cell priming during in vivo immune responses. Similarly, the lncRNA Lethe interacts with the p65 subunit of NF-κB to inhibit its binding to the TNF promoter, negatively regulating TNF production in mouse macrophages, while the lncRNA PACER blocks the inhibitory NF-κB p50 subunit from binding the COX2 promoter, resulting in increased COX2 expression in human macrophages in vitro [17, 38]. Altogether, these studies show that lncRNAs utilize a variety of mechanisms to rapidly regulate inflammatory gene expression programs in innate immune cells in response to microbial-derived cues, representing a new and potent regulatory layer in inflammatory responses.
lncRNAs in the development and homeostasis of innate immune cells
While several lncRNAs regulating inflammatory processes have been described in the literature, very few are yet known to regulate the development or homeostasis of innate cells. HOTAIRM1 is one such example that is highly expressed in human granulocytes and induced by retinoic acid signaling and PU.1 during myeloid differentiation [39, 40]. Knockdown of HOTAIRM1 results in reduced expression of CD11b and CD18 as well as impaired granulocyte differentiation in cell lines [39]. However, the role of HOTAIRM1 in the differentiation of human granulocytes in vivo still unknown.
Recently, we showed that the lncRNA Morrbid controls the homeostasis of eosinophils, neutrophils, and Ly6Chi monocytes in vivo by regulating the pro-apoptotic molecule Bcl2l11, also known as Bim [22]. Morrbid is induced in these short-lived myeloid cells in response to pro-survival cytokines and represses Bcl2l11 transcription in an allele-specific manner by promoting the enrichment of the repressive PRC2 complex within the Bcl2l11 promoter (Figure 2C). In the absence of Morrbid, Bcl2l11 is de-repressed in these cells, resulting in their apoptosis [22]. This example illustrates a novel and critical pathway used to precisely regulate the lifespan of these highly inflammatory cell populations. Indeed, expression of MORRBID is highly upregulated in eosinophils collected from patients with hypereosinphilic syndrome (HES), a group of disorders characterized by an altered eosinophil lifespan [22]. Taken together, these data suggest that dysregulation of the Morrbid-Bcl2l11 axis may be an important element in HES and other disorders of dysregulated myeloid lifespan such as autoinflammation and cancer.
Finally, the lncRNA biogenesis and proxy signal-based mechanisms of action are an emerging field, and as such have not yet been studied in detail in the context of the immune system. Interestingly, a recent study identified linc1405, a lncRNA expressed upstream of the Eomes gene in mouse embryonic stem cells [27]. Deletion of this locus or its promoter reduced Eomes expression in these cells. However, Eomes-dependent immune cells, such as natural killer (NK) cells were not examined. We have recently shown that a lncRNA, Rroid, marks a cis-regulatory element that is critical for promoting the homeostasis and function of group 1 ILCs, but not ILC2s or ILC3s (Mowel CITATION). The Rroid locus, but not the RNA itself, regulates ILC1 homeostasis by promoting open chromatin and STAT5 deposition at the promoter of Id2, a key gene required for ILC homeostasis. Although the Rroid locus forms a physical interaction with the Id2 locus, the exact mechanisms of action are still unknown. It is intriguing to speculate that other lncRNAs expressed near key lineage-specific transcription factors or cytokines may mark cis-regulatory elements critical for the development, homeostasis, and function of varied immune cell subsets.
LNCRNAS IN THE ADAPTIVE IMMUNE SYSTEM
Composed of B and T lymphocytes, the adaptive immune system must generate an effective antigen-specific effector response and form long-term immunological memory while avoiding autoimmunity and chronic inflammation. Unbiased studies have shown that lncRNAs are differentially expressed by many, if not most subsets of B and T cells [41–43]. Mechanistic description of lncRNA function in developing lymphocytes and mature B cells remains a nascent area of inquiry, but lncRNAs are emerging as key regulators of the defining steps of helper T cell-mediated immunity. Herein we describe recent work on lncRNA function in the activation of naive CD4+ T cells, their differentiation into specific effector subsets, and production of subset-specific cytokines.
lncRNAs in T cell activation
Upon activation, naive T cells undergo transcriptional and epigenetic changes as they proliferate and differentiate into specific effector subsets. One canonical example of this process is the production of IL-2 and the switch from a low-affinity to a high-affinity IL-2 receptor, which promote naive T cell proliferation and survival. Production of IL-2 requires the transcription factors NFAT1 (nuclear factor of activated T cells), AP-1, and NF-κB. Of these, the function of NFAT1 in T cells is regulated by the lncRNA NRON (ncRNA repressor of NFAT) [44]. In resting T cells, inactive NFAT1 is sequestered in the cytoplasm. As T cells are activated, NFAT1 is dephosphorylated by the phosphatase calcineurin, and translocated to the nucleus [45]. NRON serves as a scaffold for the interaction of NFAT1 with IQGAP and the three inhibitory NFAT1 kinases CKe, GSKb, and DYRK, which leads to the retention of NFAT1 in the cytoplasm [44, 46] (Figure 3A). Importantly, CD8+ T cells from IQGAP knockout mice produce significantly increased levels of IFN-γ, a known NFAT1 target, suggesting that an NRON-IQGAP axis may be a key factor in regulating NFAT-dependent cytokine production in activated CD8+ T cells [46].
Figure 3. lncRNAs in the adaptive immune system.
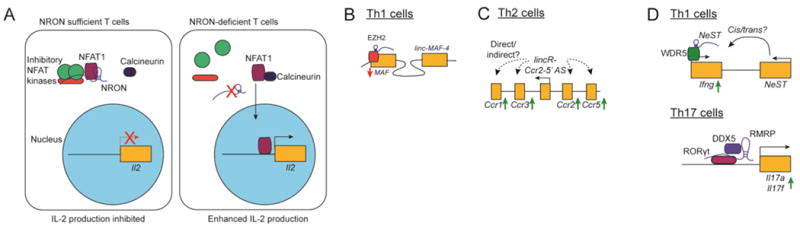
(A) The lncRNA NRON represses T cell NFAT1 activity by promoting its association with inhibitory factors in the cytoplasm. In the absence of NRON, the phosphatase Calcineurin is able to dephosphorylate NFAT1, resulting in its nuclear translocation and increased cytokine production.
(B) lncRNAs promote Th1 cell polarization. linc-MAF-4 promotes the recruitment of EZH2 to the promoter of the Th2-specific transcription factor c-MAF to repress its expression.
(C) LincR-Ccr2-5′AS promotes expression of Th2 genes, including nearby chemokine receptors such as Ccr1, Ccr2, Ccr3, and Ccr5. Whether this regulation is direct or indirect, and the specific mechanisms through which LincR-Ccr2-5′AS regulate these genes is unknown.
(D) lncRNAs promote T cell effector function. NeST promotes WDR5 recruitment to the Ifng locus to activate its transcription (above), while RMRP promotes Th17 cytokine production by acting as a scaffold for the association of the Th17-specific transcription factor RORγt and the helicase DDX5 at the Il17a and Il17f loci.
lncRNAs in CD4+ T cell polarization
Following their activation by cognate antigen, naive CD4+ T cells further differentiate into different lineages of helper and regulatory T cells with specific immunological functions. While environmental cues shape CD4+ T cell fate, the major lineage-specifying determinants are cytokines that activate networks of transcription factors leading to characteristic transcriptional profiles and functions. Th1 cells require the master transcription factor T-bet for their differentiation, Th2 cells require GATA3, and Th17 cells require RORγt. Recent work has shown that linc-MAF-4 plays a key role in regulating the expression of the transcription factors which control the Th1/Th2 fate decision [47, 48](Figure 3B). Ranzani et al. showed that linc-MAF-4 expression is high in Th1 cells but considerably reduced under Th2-polarizing conditions, and that knockdown of linc-MAF-4 in naive CD4+ cells results in expression of the Th2-specific genes MAF, GATA3, and IL-4 [47]. The authors went on to demonstrate physical interaction between linc-MAF-4 and the MAF promoter, increased H3K27 trimethylation at the MAF promoter, and association of linc-MAF-4 and the methyltransferases EZH2 and LSD1 [47]. Together, these observations suggest a model in which linc-MAF-4 associates with the MAF promoter via DNA looping, where it recruits or acts as a scaffold for a repressive chromatin-modifying complex.
A second lncRNA, LincR-Ccr2-5′AS, coordinates the expression of a Th2-specific gene program downstream of GATA3 signaling. Located in the cluster of genes encoding Ccr1, Ccr3, Ccr2, and Ccr5, LincR-Ccr2-5′AS regulates the expression of these genes and others by an unknown mechanism, independent of modulating H3K4 methylation or RNA pol II accessibility [49](Figure 3C). These studies and others suggest that lncRNAs influence T helper cell development by regulating the expression of a range of lineage-controlling transcription factors and other subset-specific genes.
lncRNAs control the function of adaptive immune cells
In addition to their master transcription factors, effector CD4+ T cell lineages are defined functionally by the cytokines they produce; Th1 cells are characterized by production of IFN-γ, Th2 by IL-4, and Th17 cells by the production of IL-17A and IL-17F. It has recently been shown that lineage-specific lncRNAs are preferentially located adjacent to or intergenic with cytokine coding genes, and act downstream of linage-specific transcription factors to regulate their expression [49]. A well-characterized example of this is the lncRNA NeST, which is located in the cluster of genes encoding Ifng and II22 and requires the Th1-specific transcription factors T-bet and STAT4 for its expression [50, 51]. NeST interacts with the methyltransferase subunit WDR5, and expression of NeST is correlated with increased levels of transcriptionally permissive H3K4 trimethylation at the Ifng locus [52, 53] (Figure 3D). The lncRNA Th2-LCR appears to regulate cytokine production in Th2 cells by a similar mechanism. Th2-LCR is located in the IL-4, IL-5, and IL-13 gene cluster, co-precipitates with WDR5, and its knockdown in vitro reduced both H3K4 methylation and WDR5 binding of the IL-4 and IL-13 promoters [54, 55].
Th17 cells, a third lineage of CD4+ T cells, are characterized by production of the cytokines IL-17A, IL-17F, and IL-22 downstream of the transcription factor RORγt [56]. As mentioned above, Huang et al showed that the lncRNA RMRP is required for the association of RORγt and DDX5 [18]. Proper assembly of the DDX5-RMRP complex is not required for Th17 cell differentiation, but is required for RORγt-mediated expression of Th17-specific cytokines (Figure 3D). Significantly, a single G>T point mutation in the human RMRP gene is associated with cartilage-hair hypoplasia (CHH), a disorder characterized by immunodeficiency among a number of other symptoms. Mice bearing the CHH-associated RMRP allele harbor Th17 cells with a defective effector program, and this remains one of the only studies directly linking altered lncRNA function to a human immunological disease [18]. Another lncRNA, Flicr, is expressed in mouse and human Foxp3+ Tregs and functions to negatively regulate Foxp3 expression and Treg function, as in vivo deletion of Flicr significantly protected mice from autoimmune diabetes [57]. The exact mechanisms through which Flicr regulates Foxp3 expression are unknown, but appear to work in part through the control of chromatin accessibility at the CNS3/AR5 region within the Foxp3 gene [57]. Together, these studies demonstrate that lncRNAs act in conjunction with lineage-specific transcription factors to control expression of cell type-specific effector molecules in the adaptive immune system.
LNCRNAS IN HUMAN INFLAMMATORY DISORDERS
A longstanding observation in human GWAS studies is that more than 90% of disease-related SNPs are associated with noncoding elements of the genome, and it is tempting to speculate that mutations in lncRNAs may explain some of these phenotypes [58]. Thus far, much of the lncRNA-related research that has been done in the immune system has focused on delineating the function of lncRNAs in mouse and human primary cells and cell lines. However, examples have begun to emerge that demonstrate the importance of lncRNAs in human inflammatory disorders. As we discuss above, the lncRNA MORRBID is highly upregulated in eosinophils collected from patients with HES, suggesting the MORRBID-BCL2L11 axis may contribute to this disease [22]. lnc13, a highly expressed lncRNA in the healthy human gut, suppresses expression of a subset of inflammatory disease related genes and is significantly downregulated in patients with celiac disease, suggesting that dysregulated lnc13 expression may contribute to inflammation in this disease [36]. Indeed, celiac disease-associated SNPs have been found in lnc13 [36]. Additionally, linc-MAF-4 has been shown to be dysregulated in multiple sclerosis, and as discussed above, mutations in RMRP are associated with CHH [18, 48]. Together, the in vivo and in vitro data regarding lncRNAs in the immune system suggest that these molecules play key roles in mediating both protection and susceptibility to inflammatory diseases. In the future, it is critical that robust in vivo models are developed and utilized to test the physiological consequences of lncRNAs in inflammatory disorders such as inflammatory bowel disease, diabetes, allergies, asthma, and cancer, among many others.
TECHNICAL CHALLENGES IN DISSECTING LNCRNA MECHANISM IN VIVO
Although high-throughput sequencing has allowed for great strides to be made in profiling immune cell lncRNA expression patterns, the functional characterization of these lncRNAs in vivo has lagged far behind (Figure 4A). Much of this delay can be attributed to the difficulty in separating the role of the genomic lncRNA loci from that of its RNA products. For example, many groups have deleted full lncRNA loci to test lncRNA functionality [22, 59, 60] (Figure 4B). While this deletion approach is a rapid way to initially screen functional lncRNA loci, especially with the efficiency of the CRISPR/Cas9 system, it does not separate any of the 3 lncRNA mechanism categories discussed above: functional RNAs, transcription-dependent, and proxy signals for cis-regulatory elements. As such, additional complementary experimental approaches need to be taken to disentangle the functionality of these lncRNAs [61].
Figure 4. Testing lncRNA function in vivo.
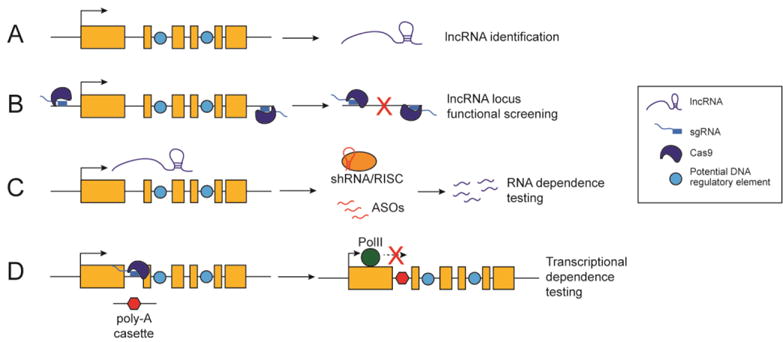
In vivo functional testing of lncRNAs remains challenging but is critical to understanding their relevance in the immune system.
(A) Cell-type specific lncRNA expression profiles can be generated from RNA-seq on purified cell populations or publicly available datasets.
(B) The functional relevance of these loci encoding these lncRNAs can be quickly and easily assessed by generating genomic deletions using the CRISPR/Cas9 system.
(C) RNA dependence of functional lncRNA loci can be assessed using shRNAs and ASOs.
(D) Transcriptional dependence can be determined by insertion of a premature poly-A cassette using CRISPR/Cas9.
RNA interference (RNAi)-based techniques, in particular short-hairpin RNAs (shRNAs), are an important addition to the experimental arsenal of lncRNA biologists as they allow for stable targeting of specific RNAs (Figure 4C). Of note, nuclear RNAi targeting is less well understood [62, 63]; thus, RNAi techniques are most well suited for testing the RNA-dependence of cytoplasmic lncRNAs. Antisense oligonucleotides (ASOs), provide an alternative post-transcriptional RNA targeting method [64] and their nuclear RNA targeting is significantly better characterized, yet they cannot be used to generate stable transgenic lines (Figure 4C). Moreover, if the lncRNA is suggestive to have functional roles in trans, complementation ‘rescue’ studies are a direct method to address their mechanism of action. To test transcription-dependent lncRNA function, additional genetic approaches can be taken to insert transcriptional terminator sequences [27, 28, 65] (Figure 4D). Targeted deletion of DNA elements within the lncRNA locus, such as the lncRNA promoter, may help to expose the lncRNA as a proxy for these important cis-regulatory elements. As with all genetic approaches, investigators must be vigilant and control for disruption of important DNA regulatory elements and inadvertent impact of inserted DNA elements. Importantly, it is not possible to predict lncRNA function from expression levels and sequence composition with current bioinformatics tools, especially with little known about conserved lncRNA motifs and secondary structures. Thus, a combination of the empiric approaches above must be taken and interpreted in the context of each lncRNA to accurately characterize its functionality.
CONCLUDING REMARKS
LncRNAs are critical epigenetic and transcriptional regulators that allow cells to specifically adapt their gene expression profiles to cues within their extracellular environment. It is becoming clear that the diverse cell types of the innate and adaptive immune system are no exception, and that lncRNAs are key regulatory molecules of immune cell gene expression programs in response to microbial and tissue-derived cues. Although much progress has been made in cataloging lncRNAs expressed in the immune system and in assessing their functional impact, there still exists a large void in our understanding of how these lncRNAs function at the molecular level in the context of immune responses. The development of new experimental and bioinformatic technologies will be paramount in filling this gap in lncRNA biology. In particular, novel techniques directed towards predicting and interrogating the structure of lncRNAs will be essential as they will help to uncover lncRNA motifs and structures that ultimately dictate the localization and interactomes of these molecules. Additionally, novel techniques to address lncRNA impact on higher order 3D chromatin organization will help uncover parallels between certain lncRNAs and potentially new principles of lncRNA function. Ultimately, better understanding of lncRNA function in the immune system will expose new means to augment protective immunity, quell excess inflammation, and promote tissue homeostasis.
Table 1.
lncRNAs with identified immune functions
| LncRNAs in innate immunity | ||||
|---|---|---|---|---|
| lncRNA | Species | Phenotype/function | Proposed mechanism | Reference |
| Lethe | Ms | Pseudogene lncRNA activated by TNFα. Negatively regulates NF-κB. | Binds RelA to inhibit DNA binding | [17] |
| Linc-Cox2 | Ms | Induced in BMDC by LPS (TLR4) and Pam3CSK (TLR2). Regulates inflammatory gene sets. | Interactions with hnRNP-A/B, hnRNP-A2/B1, and SWI/SNF | [32–34] |
| PACER | Hu | Antisense RNA upstrem of COX-2 gene. Promotes COX2 by inducing epigenetic changes at COX2 promoter. | Interacts w/NFkB p50 subunit, prevents it from binding & repressing COX2 | [38] |
| THRIL | Hu | Decreased upon Pam3CSK stimulation and promotes TNFa. Associates with hnRNPL located at TNFa promoter | hnRNPL | [71] |
| AS-IL-1β | Ms | Highly upregulated following LPS stimulation in RAW and peritoneal macrophages. Suppresses IL1b expression. | Regulates RNApolII recruitment to II-1b locus | [72] |
| IL1β-eRNA & IL1 β-RBT46 | Hu | eRNA and site of bidirectional transcription near Il-1b locus. Knockdown attenuates Il-1b and CXCL8 transcription/release. | Unknown | [73] |
| AS-IL-1α | Ms | Induced by Pam3CSK, LPS, polyIC in BMDM to regulate levels of IL-1a | Regulates recruitment of PolII to IL-1a promoter | [74] |
| lnc-DC | Hu | Highly expressed in all DC populations. Promotes CD40, CD80, HLA-DR expression on DCs. | Blocks dephosphorylation of STAT3 | [16] |
| Lnc13 | Hu,Ms | Decreased in inflammation & active celiac. Overlaps 3′ end of Il18rap Levels depend on levels of Dcp2 decapping enzyme. Shown to interact with Hdac1 and hnRNPD. Celiac risk alleles reduce association with hnRNPD | Localized to TSS of targets along with hnRNPD and Hdac1 ; likely a scaffold | [36] |
| lincRNA-EPS | Ms | Downregulated by TLR stimulation. Full deletion results in increased inflammatory genes basally and during inflammation. | Interacts w/hnRNPL through specific RNA motif; loss alters positioning of nucleosomes at target genes | [37] |
| Morrbid | Hu,Ms | Induced by prosurvival cytokines in short-lived myeloid cells (neutrophils, eosinophils, Ly6Chi monocytes). Inhibition results in increased short-lived myeloid cell death. | Interacts with repressive PRC2 complex to target it to Bcl2l11 promoter | [22] |
| lncHSC-1/2 | Ms | Promote HSC function in vitro and in vivo; altered T/B/myeloid reconstitution upon knockdown. | At least partially by promoting recruitment of E2A to target genes | [75] |
| NRAV | Hu | Expressed basally & reduced by infection w/virus. Mouse phenotype is via transgenic overexpression. Negatively regulates ISG expression. | Increases H3K27me3 modifications | [76] |
| iNOS AS | Rat | Antisense RNA in 3′ UTR of iNOS. Promotes expression of iNOS mRNA. | Interactions with 3′ UTR of iNOS mRNA | [77] |
| HOTAIRM1 | Hu | Knockdown results in reduced CD11B and CD18 expression; reduced granulocytic differentiation of cell lines | Activated downstream of RA signaling; mechanism unknown | [39] |
| NKILA | Hu | Induced in response to TNF and IL-1b to regulate NF-kB signaling in breast cancer cell lines. Function in immune cells is unknown. | Blocks IkB phosphorylation | [78] |
| NEAT1 | Hu,Ms | Regulates IL-8 synthesis following immune stimulation | Alters localization of repressor protein SFPQ | [79] |
| lncRHOXF1 | Hu | Suppresses expression of viral response genes in trophoblast progenitors. Viral infection induces lncRHOXF1 expression. | Unknown | [80] |
| lncKdm2b | Ms | Regulates ILC3 homeostasis by promoting ILC3 proliferation | Interacts with Satb1 | [81] |
| Rroid | Ms | Promotes Id2 expression specifically in NK/ILC1 to control their homeostasis | Promotes STAT5 deposition and chromatin accessibility at Id2. RNA-independent | [MOWEL citation] |
| LncRNAs in adaptive immunity | ||||
| NeST / Tmevpg1 | Ms,Hu | Promotes expression of Ifng in Th1 cells downstream of Stat4, Tbet, NFkB, Ets1 | Recruits H3K4 methyltransferases at Ifng promoter | [51–53] |
| NRON | Ms,Hu | Regulates expression of IL-2 in Th1 cells | Maintaints NFAT1 in cytoplasm by providing scaffold for inhibitory NFAT kinases | [44, 46] |
| linc-MAF-4 | Hu | Represses Th2 differentiation by regulating levels of the master Th2 transcription factor c-MAF | Recruits LDS1 and EZH2 to the MAF locus | [47, 48] |
| Rmrp | Ms,Hu | Loss decreases production of IL-17A and IL-17F by Th17 cells. SNP associated with the genetic disease cartilage hair hypoplasia | Required for the interaction of RORγt with DDX5 | [18] |
| Th2-LCR | Ms,Hu | Regulates expression of the IL-4, IL-5, and IL-13 gene cluster in Th2 cells | Associates with WDR5, promotes H3K4 methylation at the II4 and II13 promoters | [54, 55] |
| LincR-Ccr2-5′AS | Ms | Controls a Th2-specific gene expression program including Ccr1, Ccr2, Ccr3, and Ccr5 | Unknown | [49] |
| Fas-AS1 | Ms,Hu | Increases Fas/FasL-mediated apoptosis in B cells, T cells, and other cell types | Binds RBM5, reduces exon skipping in Fas | [82] |
| Gata3-AS | Hu | Coexpressed with GATA3 under Th2-polarizing conditions | Unknown | [49, 83] |
| Flicr | Ms | Partially overlaps FoxP3, specifically expressed in Tregs. Deletion reduces the number of FoxP3lo Tregs and protects against diabetes in NOD mice. Negatively regulated by IL-2 | May alter chromatin structure and accessability at the CNS3/AR5 FoxP3 regulatory region | [57] |
TRENDS.
Immune cells exhibit a wide variety of functions that are controlled by cell type-specific epigenetic and gene expression profiles. Long non-coding RNAs (lncRNAs) have recently emerged as key epigenetic and transcriptional regulators of immune cells.
LncRNAs control the development and function of specific immune cell-types through a variety of mechanisms including interactions with other RNAs, DNA, and proteins. LncRNAs are frequently induced downstream of cytokine, innate and adaptive receptors, which allows immune cells to rapidly modulate their gene expression programs in response to stimuli.
In innate immune cells, lncRNAs have been shown to directly regulate several biological functions, including cytokine production, development, and cell survival.
In adaptive immune cells, lncRNAs have primarily been shown to regulate the polarization and effector function of CD4+ T cells.
OUTSTANDING QUESTIONS.
There is growing appreciation that transcription of lncRNA loci, rather than the lncRNA itself, is required to mediate the biological functions of lncRNAs. What is the role of transcription across lncRNA loci in protective immunity and immune cell development?
Secondary structures are likely to be critical for lncRNA function. What lncRNA motifs and secondary structures are important for their function in the immune system?
How do lncRNAs impact nuclear organization and 3D chromatin architecture of immune cells?
The majority of known immune acting lncRNAs have been characterized in vitro. What are the in vivo functions of lncRNAs in the immune system?
What are the roles of lncRNAs in human inflammatory disorders? What are the functional consequences of disease-associated SNPs that fall within lncRNA loci?
Acknowledgments
The work in this manuscript was supported by funds from NIH R21AI128060 and R01HL136572 (J.H.M.); NIH F30HL138739-01A1 (J.J.K.).
GLOSSARY
- lncRNAsLong
non-coding RNAs are defined as being RNA species that are greater than 200nt in length, capped, spliced, and polyadenylated. As their name implies, lncRNAs lack strong Kozak consensus sequences and therefore are not predicted to encode proteins.
- Cis-regulatory element
A region of non-coding DNA which impacts the transcription of a neighboring or proximally located gene.
- Cis-acting lncRNA
A lncRNA that directly impacts the transcription of genes located proximally to the lncRNA loci.
- Trans-acting lncRNA
A lncRNA that directly impacts the transcription of distally located genes and is not confined to acting on the chromosome from which it is transcribed
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Taft RJ, et al. The relationship between non-protein-coding DNA and eukaryotic complexity. Bioessays. 2007;29(3):288–99. doi: 10.1002/bies.20544. [DOI] [PubMed] [Google Scholar]
- 2.Encode Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Derrien T, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775–89. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hnisz D, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155(4):934–47. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cabili MN, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25(18):1915–27. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guttman M, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol. 2010;28(5):503–10. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:14566. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anderson DM, et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. 2015;160(4):595–606. doi: 10.1016/j.cell.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bazzini AA, et al. Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J. 2014;33(9):981–93. doi: 10.1002/embj.201488411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nelson BR, et al. A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science. 2016;351(6270):271–275. doi: 10.1126/science.aad4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Penny GD, et al. Requirement for Xist in X chromosome inactivation. Nature. 1996;379(6561):131–7. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- 12.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–23. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pandey RR, et al. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32(2):232–46. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 14.Colotta F, et al. Interleukin-1 type II receptor: a decoy target for IL-1 that is regulated by IL-4. Science. 1993;261(5120):472–5. doi: 10.1126/science.8332913. [DOI] [PubMed] [Google Scholar]
- 15.Poliseno L, et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465(7301):1033–8. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang P, et al. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science. 2014;344(6181):310–3. doi: 10.1126/science.1251456. [DOI] [PubMed] [Google Scholar]
- 17.Rapicavoli NA, et al. A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. Elife. 2013;2:e00762. doi: 10.7554/eLife.00762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang W, et al. DDX5 and its associated lncRNA Rmrp modulate TH17 cell effector functions. Nature. 2015 doi: 10.1038/nature16193. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Zhao J, et al. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322(5902):750–6. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McHugh CA, et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015;521(7551):232–6. doi: 10.1038/nature14443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cerase A, et al. Xist localization and function: new insights from multiple levels. Genome Biol. 2015;16:166. doi: 10.1186/s13059-015-0733-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kotzin JJ, et al. The long non-coding RNA Morrbid regulates Bim and short-lived myeloid cell lifespan. Nature. 2016;537(7619):239–243. doi: 10.1038/nature19346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davidovich C, Cech TR. The recruitment of chromatin modifiers by long noncoding RNAs: lessons from PRC2. RNA. 2015;21(12):2007–22. doi: 10.1261/rna.053918.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ebisuya M, et al. Ripples from neighbouring transcription. Nat Cell Biol. 2008;10(9):1106–13. doi: 10.1038/ncb1771. [DOI] [PubMed] [Google Scholar]
- 25.Sleutels F, et al. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415(6873):810–3. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- 26.Swiezewski S, et al. Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature. 2009;462(7274):799–802. doi: 10.1038/nature08618. [DOI] [PubMed] [Google Scholar]
- 27.Engreitz JM, et al. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature. 2016;539(7629):452–455. doi: 10.1038/nature20149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anderson KM, et al. Transcription of the non-coding RNA upperhand controls Hand2 expression and heart development. Nature. 2016;539(7629):433–436. doi: 10.1038/nature20128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Necsulea A, Kaessmann H. Evolutionary dynamics of coding and non-coding transcriptomes. Nat Rev Genet. 2014;15(11):734–48. doi: 10.1038/nrg3802. [DOI] [PubMed] [Google Scholar]
- 30.Li G, et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148(1–2):84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paralkar Vikram R, et al. Unlinking an lncRNA from Its Associated cis Element. Molecular Cell. 2016 doi: 10.1016/j.molcel.2016.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458(7235):223–7. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu G, et al. LincRNA-Cox2 Promotes Late Inflammatory Gene Transcription in Macrophages through Modulating SWI/SNF-Mediated Chromatin Remodeling. J Immunol. 2016 doi: 10.4049/jimmunol.1502146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carpenter S, et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science. 2013;341(6147):789–92. doi: 10.1126/science.1240925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tong Q, et al. LincRNA-Cox2 modulates TNF-alpha-induced transcription of Il12b gene in intestinal epithelial cells through regulation of Mi-2/NuRD-mediated epigenetic histone modifications. FASEB J. 2016;30(3):1187–97. doi: 10.1096/fj.15-279166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Castellanos-Rubio A, et al. A long noncoding RNA associated with susceptibility to celiac disease. Science. 2016;352(6281):91–5. doi: 10.1126/science.aad0467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Atianand Maninjay K, et al. A Long Noncoding RNA lincRNA-EPS Acts as a Transcriptional Brake to Restrain Inflammation. Cell. 2016;165(7):1672–1685. doi: 10.1016/j.cell.2016.05.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krawczyk M, Emerson BM. p50-associated COX-2 extragenic RNA (PACER) activates COX-2 gene expression by occluding repressive NF-kappaB complexes. Elife. 2014;3:e01776. doi: 10.7554/eLife.01776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang X, et al. A myelopoiesis-associated regulatory intergenic noncoding RNA transcript within the human HOXA cluster. Blood. 2009;113(11):2526–34. doi: 10.1182/blood-2008-06-162164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wei S, et al. PU.1 controls the expression of long noncoding RNA HOTAIRM1 during granulocytic differentiation. J Hematol Oncol. 2016;9(1):44. doi: 10.1186/s13045-016-0274-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pang KC, et al. Genome-wide identification of long noncoding RNAs in CD8+ T cells. J Immunol. 2009;182(12):7738–48. doi: 10.4049/jimmunol.0900603. [DOI] [PubMed] [Google Scholar]
- 42.Casero D, et al. Long non-coding RNA profiling of human lymphoid progenitor cells reveals transcriptional divergence of B cell and T cell lineages. Nat Immunol. 2015 doi: 10.1038/ni.3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brazao TF, et al. Long noncoding RNAs in B-cell development and activation. Blood. 2016;128(7):e10–9. doi: 10.1182/blood-2015-11-680843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Willingham AT, et al. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. 2005;309(5740):1570–3. doi: 10.1126/science.1115901. [DOI] [PubMed] [Google Scholar]
- 45.Okamura H, et al. Concerted dephosphorylation of the transcription factor NFAT1 induces a conformational switch that regulates transcriptional activity. Mol Cell. 2000;6(3):539–50. doi: 10.1016/s1097-2765(00)00053-8. [DOI] [PubMed] [Google Scholar]
- 46.Sharma S, et al. Dephosphorylation of the nuclear factor of activated T cells (NFAT) transcription factor is regulated by an RNA-protein scaffold complex. Proc Natl Acad Sci U S A. 2011;108(28):11381–6. doi: 10.1073/pnas.1019711108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ranzani V, et al. The long intergenic noncoding RNA landscape of human lymphocytes highlights the regulation of T cell differentiation by linc-MAF-4. Nat Immunol. 2015;16(3):318–25. doi: 10.1038/ni.3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang F, et al. Linc-MAF-4 regulates Th1/Th2 differentiation and is associated with the pathogenesis of multiple sclerosis by targeting MAF. FASEB J. 2017;31(2):519–525. doi: 10.1096/fj.201600838R. [DOI] [PubMed] [Google Scholar]
- 49.Hu G, et al. Expression and regulation of intergenic long noncoding RNAs during T cell development and differentiation. Nat Immunol. 2013;14(11):1190–8. doi: 10.1038/ni.2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vigneau S, et al. Tmevpg1, a candidate gene for the control of Theiler’s virus persistence, could be implicated in the regulation of gamma interferon. Journal of Virology. 2003;77(10):5632–5638. doi: 10.1128/JVI.77.10.5632-5638.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Collier SP, et al. Cutting edge: influence of Tmevpg1, a long intergenic noncoding RNA, on the expression of Ifng by Th1 cells. J Immunol. 2012;189(5):2084–8. doi: 10.4049/jimmunol.1200774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gomez JA, et al. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-gamma locus. Cell. 2013;152(4):743–54. doi: 10.1016/j.cell.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Collier SP, et al. Regulation of the Th1 genomic locus from Ifng through Tmevpg1 by T-bet. J Immunol. 2014;193(8):3959–65. doi: 10.4049/jimmunol.1401099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koh BH, et al. Th2 LCR is essential for regulation of Th2 cytokine genes and for pathogenesis of allergic asthma. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(23):10614–10619. doi: 10.1073/pnas.1005383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spurlock CF, 3rd, et al. Expression and functions of long noncoding RNAs during human T helper cell differentiation. Nat Commun. 2015;6:6932. doi: 10.1038/ncomms7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Korn T, et al. IL-17 and Th17 Cells. Annu Rev Immunol. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- 57.Zemmour D, et al. Flicr, a long noncoding RNA, modulates Foxp3 expression and autoimmunity. Proc Natl Acad Sci U S A. 2017;114(17):E3472–E3480. doi: 10.1073/pnas.1700946114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maurano MT, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337(6099):1190–5. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Eissmann M, et al. Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA Biol. 2012;9(8):1076–87. doi: 10.4161/rna.21089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li L, et al. Targeted disruption of Hotair leads to homeotic transformation and gene derepression. Cell Rep. 2013;5(1):3–12. doi: 10.1016/j.celrep.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bassett AR, et al. Considerations when investigating lncRNA function in vivo. Elife. 2014;3:e03058. doi: 10.7554/eLife.03058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Castel SE, Martienssen RA. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat Rev Genet. 2013;14(2):100–12. doi: 10.1038/nrg3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gagnon KT, et al. Analysis of nuclear RNA interference in human cells by subcellular fractionation and Argonaute loading. Nat Protoc. 2014;9(9):2045–60. doi: 10.1038/nprot.2014.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dias N, Stein CA. Antisense oligonucleotides: basic concepts and mechanisms. Mol Cancer Ther. 2002;1(5):347–55. [PubMed] [Google Scholar]
- 65.Levitt N, et al. Definition of an efficient synthetic poly(A) site. Genes Dev. 1989;3(7):1019–25. doi: 10.1101/gad.3.7.1019. [DOI] [PubMed] [Google Scholar]
- 66.Chu C, et al. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44(4):667–78. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Simon MD, et al. The genomic binding sites of a noncoding RNA. Proc Natl Acad Sci U S A. 2011;108(51):20497–502. doi: 10.1073/pnas.1113536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Engreitz JM, et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 2013;341(6147):1237973. doi: 10.1126/science.1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chu C, et al. Systematic discovery of xist RNA binding proteins. Cell. 2015;161(2):404–16. doi: 10.1016/j.cell.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lu Z, et al. RNA Duplex Map in Living Cells Reveals Higher-Order Transcriptome Structure. Cell. 2016;165(5):1267–79. doi: 10.1016/j.cell.2016.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li Z, et al. The long noncoding RNA THRIL regulates TNFalpha expression through its interaction with hnRNPL. Proc Natl Acad Sci U S A. 2014;111(3):1002–7. doi: 10.1073/pnas.1313768111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lu J, et al. A potential suppressive effect of natural antisense IL-1beta RNA on lipopolysaccharide-induced IL-1beta expression. J Immunol. 2013;190(12):6570–8. doi: 10.4049/jimmunol.1102487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ilott NE, et al. Long non-coding RNAs and enhancer RNAs regulate the lipopolysaccharide-induced inflammatory response in human monocytes. Nat Commun. 2014;5:3979. doi: 10.1038/ncomms4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chan J, et al. Cutting Edge: A Natural Antisense Transcript, AS-IL1alpha, Controls Inducible Transcription of the Proinflammatory Cytokine IL-1alpha. J Immunol. 2015;195(4):1359–63. doi: 10.4049/jimmunol.1500264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Luo M, et al. Long Non-Coding RNAs Control Hematopoietic Stem Cell Function. Cell Stem Cell. 2015;16(4):426–38. doi: 10.1016/j.stem.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ouyang J, et al. NRAV, a long noncoding RNA, modulates antiviral responses through suppression of interferon-stimulated gene transcription. Cell Host Microbe. 2014;16(5):616–26. doi: 10.1016/j.chom.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Matsui K, et al. Natural antisense transcript stabilizes inducible nitric oxide synthase messenger RNA in rat hepatocytes. Hepatology. 2008;47(2):686–97. doi: 10.1002/hep.22036. [DOI] [PubMed] [Google Scholar]
- 78.Liu B, et al. A cytoplasmic NF-kappaB interacting long noncoding RNA blocks IkappaB phosphorylation and suppresses breast cancer metastasis. Cancer Cell. 2015;27(3):370–81. doi: 10.1016/j.ccell.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 79.Imamura K, et al. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol Cell. 2014;53(3):393–406. doi: 10.1016/j.molcel.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 80.Penkala I, et al. lncRHOXF1, a Long Noncoding RNA from the X Chromosome That Suppresses Viral Response Genes during Development of the Early Human Placenta. Mol Cell Biol. 2016;36(12):1764–75. doi: 10.1128/MCB.01098-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu B, et al. Long noncoding RNA lncKdm2b is required for ILC3 maintenance by initiation of Zfp292 expression. Nat Immunol. 2017;18(5):499–508. doi: 10.1038/ni.3712. [DOI] [PubMed] [Google Scholar]
- 82.Sehgal L, et al. FAS-antisense 1 lncRNA and production of soluble versus membrane Fas in B-cell lymphoma. Leukemia. 2014;28(12):2376–87. doi: 10.1038/leu.2014.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang H, et al. Profiling of human CD4+ T-cell subsets identifies the TH2-specific noncoding RNA GATA3-AS1. J Allergy Clin Immunol. 2013;132(4):1005–8. doi: 10.1016/j.jaci.2013.05.033. [DOI] [PubMed] [Google Scholar]


