Abstract
Antibody responses are essential for protection against influenza virus infection. Humans are exposed to a multitude of influenza viruses throughout their lifetime and it is clear that immune history influences the magnitude and quality of the antibody response. The “original antigenic sin” (OAS) concept refers to the impact of the first influenza virus variant encounter on lifelong immunity. Although this model has been challenged since its discovery, past exposure, and likely one's first exposure, clearly affects the epitopes targeted in subsequent responses. Understanding how previous exposure to influenza virus shape antibody responses to vaccination and infection is critical, especially with the prospect of future pandemics and for the effective development of a universal influenza vaccine.
Antibody responses to influenza
250,000 to 500,000 individuals succumb to influenza virus infection in the world every year. Current seasonal influenza virus vaccines offer protection but are often off-target and they have to be re-formulated and re-administrated every year due to the phenomenon of antigenic drift (see Glossary) [1]. The two surface glycoproteins, hemagglutinin (HA) and neuraminidase (NA) are the main targets of antibody responses. Influenza A viruses are subtyped based on the sequence and antigenic divergence of the HA and NA proteins. A total of 18 HA and 11 NA subtypes have been identified. The classification of influenza viruses into two phylogenetic groups is based on the type of HA expressed on the virus (group 1 includes H1 and H5 and group 2 includes H3 and H7) [2–4]. Influenza B viruses are classified as a single influenza virus type, but two antigenically and genetically distinct lineages circulate, the Victoria-like and the Yamagata-like lineage [5].
Due to antibody pressure, influenza viruses escape the immune system by introducing point mutations, mainly in the immunodominant and highly plastic globular head of HA. In contrast, the more conserved stalk domain of HA does not change as often [6]. Antibodies binding to epitopes on the HA stalk domain are broadly cross-reactive and can neutralize a wide variety of influenza strains (homosubtypic and heterosubtypic neutralization). Unfortunately, the stalk domain is immuno-subdominant and seasonal influenza vaccines do not always induce these broadly neutralizing antibodies [7]. In addition, seasonal vaccines show limited efficacy against novel pandemic influenza virus strains, and producing specific vaccines for these strains in a timely fashion is challenging [8]. Different strategies have been developed to try to induce these broadly neutralizing antibodies, including headless HAs constructs for a better availability of the stalk region and immunization with chimeric HAs made with “exotic” heads [9, 10]. Understanding how immune history affects the production of such antibodies is crucial for the development of new vaccines.
The concept of “original antigenic sin” (OAS) refers to the notion that the first antigenic variant encountered early in life conditions lifelong immunity. This theory has been constantly challenged since its description in the early 1950’s [11, 12]. While it is known that immune memory acquired by past influenza exposure influences the response to subsequent strains, how sequential exposure to antigenically distinct influenza strains shapes the antibody response remains obscure. The terms antigenic seniority or antigen imprinting may more accurately describe such a phenomenon, as these terms encompass both positive and negative impacts of past exposure to vaccine efficacy.
This review focuses on how pre-existing immunity influences the generation and maintenance of broadly cross-reactive antibodies in the context of the development of a “stalk-based” universal influenza virus vaccine.
The concept of Original Antigenic Sin
Around 70 years ago, Thomas Francis Jr. and colleagues made the observation that the antibody response to influenza strains from childhood dominates the anti-influenza virus antibody response over time [12–14]. Even as a person grows older and acquires antibodies to other strains, the original antibodies are maintained at the highest levels at all times. Francis called this phenomenon the OAS, a Biblical reference to how an individual will bear the “sin” of the first influenza virus exposure for the rest of his life. While OAS is most often applied to anti-HA responses, convincing evidence of OAS in anti-NA responses are emerging [15]. The key to understanding the phenomenon of OAS may lie in understanding the nature of the influenza virus itself. When a strain undergoes antigenic drift, some epitopes remain conserved. Pre-existing antibodies to such epitopes cross-react to the drifted strain, thus suppressing the response by reducing antigen levels through Fc-mediated mechanisms and/or epitope masking [16–20]. This reduction in access to antigen would favor recall of memory over de novo activation of naïve B cells. This scenario would therefore boost pre-existing influenza virus antibody responses while the diversity of the overall response is reduced and drifted epitopes are less well targeted [21]. Consistent with the idea of preferential activation of memory B cells at sequential exposure are studies showing that antigen relatedness, but not the length of intervals between exposures, is of great prognostic value for the response to sequential exposure [22–24]. This model is further supported by evidence of how OAS can be alleviated by increasing the available antigen, and/or by shifting antigen-presentation from memory B cells to dendritic cells [25]. The latter can be achieved by using an adjuvanted vaccine, which adds the benefits of an enhanced cellular response [25].
OAS versus antigenic seniority
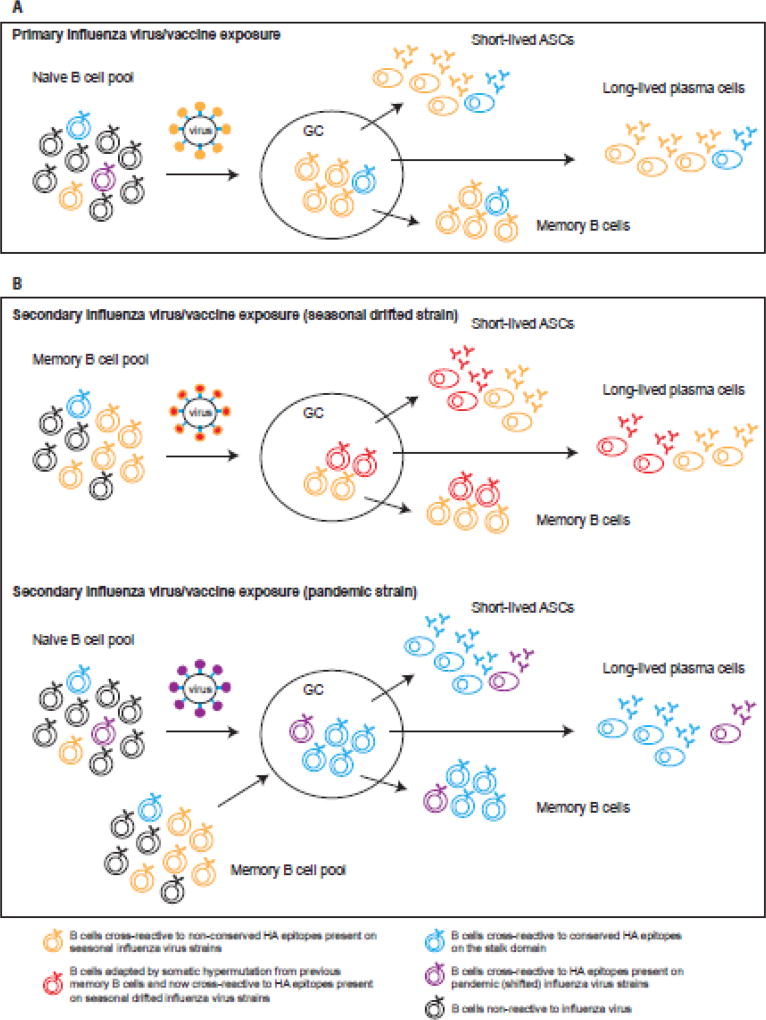
Is it really a “sin” to have immune memory and pre-existing antibodies to the first influenza virus strain an individual is exposed to? It would only be a true sin if nothing good comes from it, and although Francis used this term he also suggested that the concept of OAS could be employed to induce broader immunity [12]. Perhaps the term “antigenic seniority” better describes the phenomenon. This more recent model of OAS dictates that strains from childhood are given a more “senior” antigenic position in our immune repertoire, and that each subsequent strain takes a more junior position in the response [26– 28]. Thus, the relative response to each individual strain will be determined by its hierarchal position, and previously encountered strains will be boosted by encounter with strains of the same subtype. Over time, these responses accumulate, resulting in the highest antibody titers against the strains of childhood. It is also possible that the response to primary exposure is simply larger than that to subsequent exposures [21]. A key distinction of this new model is that every new strain gets a place in the hierarchy, not only the strain of first exposure (figure 1).
Figure 1. Antibody responses to influenza virus and antigenic seniority.
A. Primary exposure to an influenza virus (early in childhood) induces activation of influenza virus-reactive naive B cells. After antigen encounter and T cell help, naïve B cells enter a germinal center (GC) reaction where they class-switch and affinity mature by somatic hypermutation. The majority of the B cells will recognize epitopes on the immunodominant head domain of HA (in orange) but a few might recognize the more conserved stalk region (in blue). After 7–10 days, influenza-reactive short-lived antibody secreting cells (ASCs) or plasmablasts are present in circulation and secrete antibodies. The generation of influenzareactive memory B cells and long-lived plasma cells also occur from the GC and reflects the B cells that have been activated, with the majority of the antibody specificity being directed towards the HA head domain.
B. During secondary exposure to influenza virus/vaccine, memory B cells generated from the first exposure will be reactivated. However different outcomes will happen if the second exposure (infection or vaccination) is a seasonal influenza virus strain or a pandemic strain. In the case of a seasonal virus, antigenic drift happens from year to year resulting in small changes in the globular head. The pool of memory B cells generated from previous exposure will recognize these drifted epitopes on the head, cells will be reactivated and some of them will adapt in the GC by somatic hypermutation to increase binding to the drifted epitope (in red). Short-lived ASCs, long-lived plasma cells and memory B cells will be generated and the antibody specificity will be a mixture of the senior strain and the more junior strain epitopes (antigenic imprinting) but mainly targeting the head. Thus, previously encountered strains will be boosted by encounter with strains of the same subtype and over time, these responses accumulate. The less frequent memory B cells targeting conserved epitopes will not be boosted and will be diluted out. In the OAS model, the reduction in access to antigen because of pre-existing cross-reactive antibodies (epitope masking) is what would favor recall of memory over de novo activation of naïve B cells.
In the case of a pandemic influenza virus (shifted), the majority of the epitopes on the head domain are new (in purple) whereas conserved epitopes on the HA stalk remain the same. Cross-reactive memory B cells specific to the HA stalk (in blue) are being re-activated and now the product of the GC reaction is an antibody response targeting mainly the stalk domain of HA. Naïve responses to the novel epitopes on the head domain are also induced but most likely participate at a lower significance to the overall response.
Evidences supporting or disproving the OAS model
Studying the concept of OAS, or antigenic seniority, in the human population is difficult. While complete vaccination history may be available for a cohort, a comprehensive history of influenza virus exposure is not. In fact, most children have been infected with at least one strain by the age of 3 and adults are re-infected with drifted strains on average every 5–10 years [27, 29]. Nevertheless, numerous human epidemiological studies, as well as experimental animal studies and mathematical modeling, have been conducted on the subject.
There is strong evidence for antigenic seniority, or so-called HA imprinting, where the highest antibody titers are against influenza virus strains from a persons’ childhood [13, 15, 26, 30, 31]. However, whether these titers would have a negative impact on immunological responses to sequential exposure is less certain. The first observations of OAS made by Francis in the mid-20th century found no evidence of deleterious effects of previous exposure on the response to sequential infection with a similar strain [32]. More recent studies provide evidence in favor of a possible impact of pre-existing memory and antibodies on subsequent exposure. Depending on birth year, and thus imprinting, different epitopes on the HA from the 2009 pandemic H1N1 are targeted upon exposure, leading to different levels of protection to infection with later drift variants of the 2009 pandemic H1N1 strain [33–35]. Interestingly, there are indications from studies in mice and ferrets that OAS is stronger when the first exposure is infection rather than vaccination [22, 36]. Another study in mice indicates that antibodies with an OAS “phenotype” share some level of cross-reactivity between priming and recall viral strains and that B cells producing these antibodies can be protective when recalled into secondary immune responses [37]. However, the obvious caveat with studies such as these is the fact that the number of sequential exposures (vaccinations or infections) is limited and does not accurately mimic the situation for an adult human.
In contrast, a study on responses to seasonal vaccination in humans showed no evidence of OAS, as pre-existing antibody titers were not prognostic of the post-vaccination response [18]. These findings are also supported by studies from our laboratory, where single B cell clones were investigated rather than total serum antibodies [38]. In addition, the emergence of a new pandemic H1N1 strain in 2009 provided new opportunities to test the OAS hypothesis in humans. Following the model of OAS rather than antigenic seniority, if the first strain of childhood was another H1N1 strain, exposure to variants of the new pandemic strain would lead to a boosted response to old strains, while response to the pandemic strain would be minimal. However neither human epidemiological studies nor experimental studies in ferrets could fully prove this hypothesis [39].
Is OAS harmful?
Paradoxically, the phenomenon of OAS may be both beneficial and harmful at the same time. Perhaps the biggest impact of both the dangers and the benefits of OAS lies in the ever-lurking threat of emerging pandemic and highly-pathogenic zoonotic influenza virus strains. These are novel to the human population, and whenever such a strain emerges it can be deadly for those who do not have some level of pre-existing cross-protective immunity. However, HA imprinting with a strain from the same phylogenetic group may confer protection against severe infection. Indeed, the decreased susceptibility of the elderly population both to the 1918 and the 2009 H1N1 pandemics has been attributed to cross protection from antibodies generated to strains encountered in childhood [7, 40–42]. Furthermore, heterosubtypic protection to highly pathogenic avian strains might depend on birth year, and thus which strain dominated during childhood [30]. Further supporting this notion are studies from our laboratory showing that seasonal, or 2009 pandemic H1N1, vaccination of persons born after 1968, when H3N2 (group 2) strains dominated, can induce antibodies that can neutralize group 2 H7N9 avian strains [43]. In another series of studies we showed that people targeted conserved, frequently HA-stalk, epitopes on the 2009 pandemic strain [44, 45], but upon subsequent exposures the response was to immunodominant and more specific head epitopes [7]. Additionally, OAS can also negatively affect responses to seasonal strains. Pre-existing memory and antibodies can, through epitope masking, force the response to an antigenically similar strain to be focused to a single epitope [46]. This may indeed be harmful if that epitope experiences antigenic drift, whereby all capability of recognizing the drift-variant is lost. It is possible that the influenza virus can use this aspect of OAS as a potential way of escaping from the host’s immune system [22].
Towards a universal influenza vaccine: targeting conserved epitopes
Most protective antibodies generated in response to influenza target the HA protein, a trimeric surface glycoprotein comprised of the HA1 and HA2 domains. The majority of the HA1 chain forms the globular head, which contains the receptor-binding site (RBS) while the HA stalk (or stem) is predominantly comprised of the HA2 domain (plus the N- and C-terminus of HA1) [47]. Despite the fact that the HA head is highly plastic and subjected to mutations, cross-reactive antibodies targeting conserved epitopes on the head have been described [48–54]. Nonetheless the majority of broadly neutralizing antibodies target epitopes on the stalk domain, as the level of conservation across influenza virus strains is much higher [6, 55–57]. The breadth of cross-reactivity observed is diverse; antibodies targeting protective epitopes conserved within group 1 or within group 2 strains have been described [45, 55, 56, 58–62]. Additionally, protective epitopes on influenza A virus conserved between group 1 and 2 [43, 63–65] and even between influenza A and B strains have been reported as well [49].
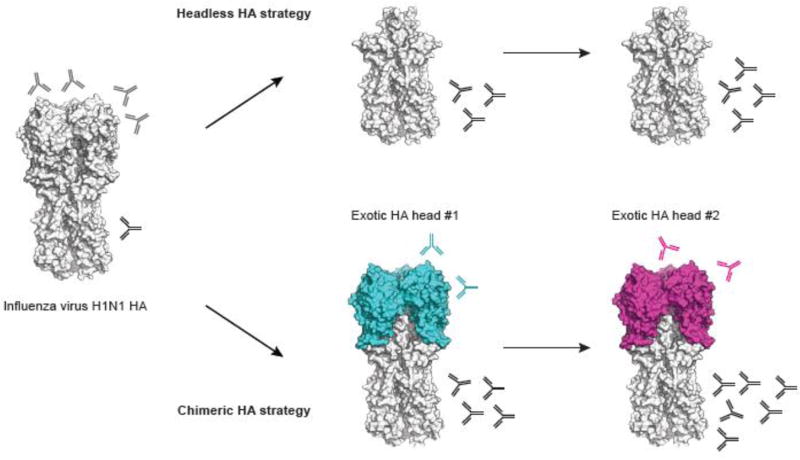
Broadly neutralizing stalk antibodies are induced following natural influenza infection [45, 66–68]. Although it was previously thought that seasonal influenza virus vaccines rarely induced such antibodies, more recent studies demonstrated that they were not uncommon and that their generation is boosted upon exposure to antigenically divergent HA head domains [43, 69–73]. Based on this, two strategies to induce stalk broadly neutralizing antibodies have been developed (figure 2). The first focuses on removal of the entire head domain to construct headless HAs while the second is based on chimeric HAs [74, 75]. While the first strategy seemed an obvious solution, headless HAs may lack important conformational epitopes, thus inducing antibodies that may be unable to bind pre-fusion HA and so will not be able to neutralize viral infectivity [57, 75]. Several constructs based on the stalk domain have been developed and vaccination with recombinantly expressed protein provided protection against viral challenge in a mouse model [76–79]. More recently two studies improved the stabilization of the stalk domain and increased immunogenicity [80, 81]. Both constructs induced stalk-reactive antibodies in animal models and protected from viral challenge, but neutralizing antibody titers were low (using homosubtypic viruses) or almost undetectable (using heterosubtypic viruses). A new study describing the use of a group 1 HA mini-stem showed protection against both group 1 and group 2 viruses [82].
Figure 2. Strategies to boost broadly neutralizing stalk antibodies.
The first strategy focuses on headless HAs, with the removal of the entire head domain to make the stalk domain more “available” and thus induce antibody responses against the stalk domain.
The second strategy uses chimeric HAs consisting of the stalk domain from H1, H3 or influenza B viruses (here H1N1 as an example) in combination with head domains of exotic influenza virus strains (avian strains for the majority). Sequential exposure with such chimeric HAs elicits boosted responses against the conserved stalk domain.
Structures are based on PDB ID 4M4Y and visualized using PyMOL.
The second strategy uses sequential exposure with chimeric HAs consisting of the stalk domain from H1, H3 or influenza B viruses in combination with head domains of exotic influenza virus strains (mostly avian strains) [75]. Multiple studies in mice and ferrets have demonstrated that it is feasible to concentrate the immune response toward the stalk domain by using sequential immunization with chimeric HAs that have different head domains but the same stalk domain [83–86]. Moreover, different adjuvants and routes of immunization were proven effective in boosting anti-stalk antibodies and enhancing the longevity of the response in mice using chimeric HAs [87].
Universal influenza virus vaccines that target the conserved stalk domain by using chimeric HAs are in late preclinical development. Additionally, vaccines that target other conserved regions of the influenza virus including the ectodomain of the M2 ion channel or the internal matrix and nucleoproteins are in clinical development [88].
Pre-existing immunity and broadly neutralizing stalk antibodies
Both strategies to enhance stalk-antibody responses have so far been tested only in animal models. A major difference between ferrets/mice and humans is that humans have pre-existing immunity to influenza viruses. Humans are continuously exposed to antigenically distinct influenza virus strains (by infection or vaccination), containing both novel and immune experienced epitopes [27, 29]. How stalk-reactive antibodies are maintained over time and to which extent they can be boosted upon sequential exposures to distinct HA subtypes is of major interest. The direct analysis of human serological data presents an opportunity to assess and understand immune responses in the context of pre-existing immunity. A longitudinal analysis of antibody titers against various pandemic and seasonal strains of influenza virus spanning a 20-year period showed that titers of stalk cross-reactive antibodies rose modestly over time [28]. These results are consistent with another study reporting an increase in serum titers of broadly neutralizing hemagglutinin stalk-reactive antibodies with age [89]. However, both studies pointed out the relative weakness of group 2 anti-HA stalk antibodies compared to group 1, mainly due to the absence of major antigenic shifts in group 2 viruses. Another argument for this weakness is the glycosylation on the Asn38 residue on HA of group 2 viruses, which could affect the development of broadly neutralizing stalk antibodies [56, 59, 63, 64]. In addition, recent work reconstructing influenza A virus exposure by year of birth highlighted the persistence of group 1 imprinting in older adults, despite decades of natural exposure to H3N2 after 1968. This suggests that HA exposures later in life do not substantially change broadly protective responses in individuals already imprinted to a particular HA group [30]. However, there is some evidence suggesting that if an already imprinted individual is exposed to a new pandemic strain before the age of approximately 20 years, the hierarchy can be “re-set” to that strain [90]. This leads to a question concerning timing of the delivery: should a universal influenza virus vaccine be administered to children before natural infection? Would a universal influenza virus vaccine change the individual long-term protection if given before natural infection? What about individuals that are already primed?
Another key aspect of influenza immune history is the reliance on memory B cells (figure 1). A secondary exposure to an antigen results in the activation and differentiation of memory B cells. It is well accepted that the B cell response to influenza virus in adults is principally a secondary immune response mediated through activation of memory B cells [38, 44, 45]. In fact we found that at the single cell level, almost half of the plasmablasts recalled over two consecutive years were from the same clonal progenitors [7]. Understanding how an evolving pathogen such as influenza virus shapes the memory B cell repertoire is central to designing a universal influenza virus vaccine and boosting stalk-reactive antibodies in primed individuals. A recent longitudinal analysis of the plasmablasts, memory and serological response upon vaccination in individuals between 2006 and 2013 demonstrated that B cells specific for more variant strains are preferentially activated while revaccination with similar strains produces a relatively weak response [91]. In accordance with this, we reported that only individuals with low preexisting serological levels of 2009 pandemic H1N1-specific antibodies generated a broadly neutralizing plasmablast response directed toward the HA stalk domain. But repeated vaccination with the pandemic H1N1 strain resulted in head-specific responses [7]. These studies demonstrate how serological antibody levels allow exposure to various influenza virus strains to diversify the memory B cell precursor pool and provide overall improved protection to evolving pathogens such as influenza viruses. Additional evidence comes from clinical studies using H5 or H7 subtype vaccines (novel head domain epitopes combined with conserved stalk domain epitopes), to which individuals are naive. They all showed significantly boosted anti-stalk antibody titers [92, 93] and the reactivation of stalk-reactive memory B cells [73, 94].
Concluding remarks
The concept of antigenic seniority or antigenic imprinting is a more adequate description for the hierarchical nature of antibody responses to previously encountered influenza virus strains rather than the OAS doctrine. Understanding the impact of pre-existing immunity on antibody responses to conserved influenza virus epitopes is critical for the induction of broadly stalk-reactive antibodies and their sustainability over time. Multiple strategies are currently being explored to boost such responses and ongoing clinical trials with vaccine candidates in humans will demonstrate how feasible a universal influenza virus vaccine is (see Outstanding Questions). Further, this concept has potential implications for other viruses, such as dengue and HIV.
Outstanding questions.
What is the best strategy for preferentially boosting and maintaining HA stalk-reactive and broadly neutralizing antibodies over time?
Are there other (better?) conserved epitopes, besides those on the HA stalk, that can be targeted to induce sustainable broadly neutralizing responses?
What is the ideal timing for administering a universal influenza virus vaccine? Should it be given to children before first exposure by natural infection?
What is the most optimal vaccination strategy for already influenza virus-primed individuals? Can OAS/antigenic seniority be overcome by e.g. increasing the antigen amount in the vaccine?
Would long-term protection by antigenic imprinting be altered if the first exposure were by vaccination rather than by natural infection?
How are T cell responses affected by OAS/antigenic seniority? What role does “T cell OAS” play in the shaping of the antibody response?
Trends.
The concept of antigenic seniority or antigenic imprinting is a more adequate model compared to the Original Antigenic Sin (OAS) theory.
Understanding how pre-existing immunity shapes the antibody response to conserved epitopes on influenza virus is critical for the development of new influenza virus vaccines.
Different strategies are being developed to boost and sustain the production of broadly neutralizing stalk-reactive antibodies over time.
Clinical trials with vaccine candidates targeting conserved regions on influenza virus are ongoing in humans.
Acknowledgments
We thank Dr. Charles L. Dulberger, the master of PyMOL, for help with the HA structure and Karlynn E. Neu for critical review of the manuscript. This project was funded in part from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under CEIRS contract HHSN272201400006C (PW) and HHSN272201400008C (FK), and grant numbers: U19AI109946-01 (PW), U19AI082724 (PW), P01AI097092-03 (PW), U19AI057266-11 (PW), 1R01AI117287 (FK), 1R01AI128821 (FK) and U19AI109946 (FK).
Glossary
- Antigenic drift
The introduction of point mutations, mainly in the globular head domain of HA, which allows the influenza virus to continuously evolve into new strains.
- Antigenic seniority
The model by which the first influenza virus strain a person is exposed to as a child takes the most “senior” antigenic position in the immune repertoire. This “senior” response will be continuously boosted upon sequential exposure to other strains, which in turn will take progressively more “junior” positions. Thus, the more “senior” a strain is, the more boosted it will be upon infection and/or vaccination.
- Antigenic shift
The reassortment of gene segments between human and/or zoological influenza viruses, which leads to the emergence of novel strains.
- Broadly neutralizing antibody
An antibody that binds to conserved epitopes on the influenza virus, thus being capable of neutralizing multiple strains.
- Epitope masking
The direct blocking or steric hindrance of an epitope due to pre-existing antibodies, which will result in reduced access to antigen.
- Hemagglutinin (HA)
Influenza virus surface glycoprotein that mediates viral entry by binding sialic acid on host cells. HA is a trimer arranged into a membrane-distal globular head domain and a membrane-proximal stalk domain.
- Heterosubtypic strains
Influenza viruses belonging to different subtypes (for example H1N1 or H3N2 are subtypes of influenza A viruses).
- Homosubtypic strains
Influenza viruses belonging to the same subtype.
- Immune history
The accumulation of all previous influenza virus exposures (infection and vaccination).
- Immunodominance
The phenomenon of preferential induction of immune responses to certain epitopes due to the unequal immunogenicity between different antigens, or between different epitopes on the same antigen.
- Neuraminidase (NA)
Tetrameric influenza virus surface glycoprotein that cleaves sialic acid, thus allowing release of virions from host cells.
- Original Antigenic Sin
The notion that the first variant of an influenza virus encountered early in life will dictate lifelong immunity to all subsequently encountered antigenic variants of that virus.
- Pandemic strain
A novel influenza strain capable of causing a global disease outbreak.
- Pre-existing immunity
The memory cells, long-lived plasma cells, and circulating antibodies that were generated in a previous immune response to influenza virus.
- Zoonotic strain
An influenza strain that is transmissible from animals to humans.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gerdil C. The annual production cycle for influenza vaccine. Vaccine. 2003;21(16):1776–9. doi: 10.1016/s0264-410x(03)00071-9. [DOI] [PubMed] [Google Scholar]
- 2.Air GM. Sequence relationships among the hemagglutinin genes of 12 subtypes of influenza A virus. Proc Natl Acad Sci U S A. 1981;78(12):7639–43. doi: 10.1073/pnas.78.12.7639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tong S, et al. New world bats harbor diverse influenza A viruses. PLoS Pathog. 2013;9(10):e1003657. doi: 10.1371/journal.ppat.1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu X, et al. Hemagglutinin homologue from H17N10 bat influenza virus exhibits divergent receptor-binding and pH-dependent fusion activities. Proc Natl Acad Sci U S A. 2013;110(4):1458–63. doi: 10.1073/pnas.1218509110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rota PA, et al. Cocirculation of two distinct evolutionary lineages of influenza type B virus since 1983. Virology. 1990;175(1):59–68. doi: 10.1016/0042-6822(90)90186-u. [DOI] [PubMed] [Google Scholar]
- 6.Krystal M, et al. Evolution of influenza A and B viruses: conservation of structural features in the hemagglutinin genes. Proc Natl Acad Sci U S A. 1982;79(15):4800–4. doi: 10.1073/pnas.79.15.4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andrews SF, et al. Immune history profoundly affects broadly protective B cell responses to influenza. Sci Transl Med. 2015;7(316):316ra192. doi: 10.1126/scitranslmed.aad0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krammer F, Palese P. Advances in the development of influenza virus vaccines. Nat Rev Drug Discov. 2015;14(3):167–82. doi: 10.1038/nrd4529. [DOI] [PubMed] [Google Scholar]
- 9.Krammer F, Palese P. Influenza virus hemagglutinin stalk-based antibodies and vaccines. Curr Opin Virol. 2013;3(5):521–30. doi: 10.1016/j.coviro.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krammer F, et al. Advances in universal influenza virus vaccine design and antibody mediated therapies based on conserved regions of the hemagglutinin. Curr Top Microbiol Immunol. 2015;386:301–21. doi: 10.1007/82_2014_408. [DOI] [PubMed] [Google Scholar]
- 11.Cobey S, Hensley SE. Immune history and influenza virus susceptibility. Curr Opin Virol. 2017;22:105–111. doi: 10.1016/j.coviro.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Francis T., Jr On the Doctrine of Original Antigenic Sin. Proceedings of the American Philosophical Society. 1960;104(6):572–578. [Google Scholar]
- 13.Davenport FM, et al. EPIDEMIOLOGIC AND IMMUNOLOGIC SIGNIFICANCE OF AGE DISTRIBUTION OF ANTIBODY TO ANTIGENIC VARIANTS OF INFLUENZA VIRUS. The Journal of Experimental Medicine. 1953;98(6):641–656. doi: 10.1084/jem.98.6.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davenport FM, et al. Epidemiology of influenza; comparative serological observations in England and the United States. Lancet. 1955;269(6888):469–74. doi: 10.1016/s0140-6736(55)93328-6. [DOI] [PubMed] [Google Scholar]
- 15.Rajendran M, et al. Analysis of Anti-Influenza Virus Neuraminidase Antibodies in Children, Adults, and the Elderly by ELISA and Enzyme Inhibition: Evidence for Original Antigenic Sin. MBio. 2017;8(2) doi: 10.1128/mBio.02281-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zarnitsyna VI, et al. Masking of antigenic epitopes by antibodies shapes the humoral immune response to influenza. Philos Trans R Soc Lond B Biol Sci. 2015;370(1676) doi: 10.1098/rstb.2014.0248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bergstrom JJ, et al. Epitope-Specific Suppression of IgG Responses by Passively Administered Specific IgG: Evidence of Epitope Masking. Front Immunol. 2017;8:238. doi: 10.3389/fimmu.2017.00238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gulati U, et al. Amount and avidity of serum antibodies against native glycoproteins and denatured virus after repeated influenza whole-virus vaccination. Vaccine. 2005;23(11):1414–25. doi: 10.1016/j.vaccine.2004.08.053. [DOI] [PubMed] [Google Scholar]
- 19.DiLillo DJ, et al. Broadly neutralizing anti-influenza antibodies require Fc receptor engagement for in vivo protection. J Clin Invest. 2016;126(2):605–10. doi: 10.1172/JCI84428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henry Dunand CJ, et al. Both Neutralizing and Non-Neutralizing Human H7N9 Influenza Vaccine-Induced Monoclonal Antibodies Confer Protection. Cell Host Microbe. 2016;19(6):800–13. doi: 10.1016/j.chom.2016.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fonville JM, et al. Antibody landscapes after influenza virus infection or vaccination. Science. 2014;346(6212):996–1000. doi: 10.1126/science.1256427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim JH, et al. Original antigenic sin responses to influenza viruses. J Immunol. 2009;183(5):3294–301. doi: 10.4049/jimmunol.0900398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fazekas de St G, Webster RG. Disquisitions of Original Antigenic Sin. I. Evidence in man. J Exp Med. 1966;124(3):331–45. doi: 10.1084/jem.124.3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Virelizier JL, et al. Antibody responses to antigenic determinants of influenza virus hemagglutinin. II. Original antigenic sin: a bone marrow-derived lymphocyte memory phenomenon modulated by thymus-derived lymphocytes. J Exp Med. 1974;140(6):1571–8. doi: 10.1084/jem.140.6.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim JH, et al. Strategies to alleviate original antigenic sin responses to influenza viruses. Proc Natl Acad Sci U S A. 2012;109(34):13751–6. doi: 10.1073/pnas.0912458109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lessler J, et al. Evidence for antigenic seniority in influenza A (H3N2) antibody responses in southern China. PLoS Pathog. 2012;8(7):e1002802. doi: 10.1371/journal.ppat.1002802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kucharski AJ, et al. Estimating the life course of influenza A(H3N2) antibody responses from cross-sectional data. PLoS Biol. 2015;13(3):e1002082. doi: 10.1371/journal.pbio.1002082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller MS, et al. Neutralizing antibodies against previously encountered influenza virus strains increase over time: a longitudinal analysis. Sci Transl Med. 2013;5(198):198ra107. doi: 10.1126/scitranslmed.3006637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bodewes R, et al. Prevalence of antibodies against seasonal influenza A and B viruses in children in Netherlands. Clin Vaccine Immunol. 2011;18(3):469–76. doi: 10.1128/CVI.00396-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gostic KM, et al. Potent protection against H5N1 and H7N9 influenza via childhood hemagglutinin imprinting. Science. 2016;354(6313):722–726. doi: 10.1126/science.aag1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nachbagauer R, et al. Defining the antibody cross-reactome directed against the influenza virus surface glycoproteins. Nat Immunol. 2017;18(4):464–473. doi: 10.1038/ni.3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Francis T, Jr, et al. Experience with vaccination against influenza in the spring of 1947; a preliminary report. Am J Public Health Nations Health. 1947;37(8):1013–6. doi: 10.2105/ajph.37.8.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y, et al. Immune history shapes specificity of pandemic H1N1 influenza antibody responses. J Exp Med. 2013;210(8):1493–500. doi: 10.1084/jem.20130212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Linderman SL, et al. Potential antigenic explanation for atypical H1N1 infections among middle-aged adults during the 2013–2014 influenza season. Proc Natl Acad Sci U S A. 2014;111(44):15798–803. doi: 10.1073/pnas.1409171111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petrie JG, et al. Antibodies Against the Current Influenza A(H1N1) Vaccine Strain Do Not Protect Some Individuals From Infection With Contemporary Circulating Influenza A(H1N1) Virus Strains. J Infect Dis. 2016;214(12):1947–1951. doi: 10.1093/infdis/jiw479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bodewes R, et al. Vaccination against seasonal influenza A/H3N2 virus reduces the induction of heterosubtypic immunity against influenza A/H5N1 virus infection in ferrets. J Virol. 2011;85(6):2695–702. doi: 10.1128/JVI.02371-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Linderman SL, Hensley SE. Antibodies with 'Original Antigenic Sin' Properties Are Valuable Components of Secondary Immune Responses to Influenza Viruses. PLoS Pathog. 2016;12(8):e1005806. doi: 10.1371/journal.ppat.1005806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wrammert J, et al. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature. 2008;453(7195):667–71. doi: 10.1038/nature06890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Donnell CD, et al. Humans and ferrets with prior H1N1 influenza virus infections do not exhibit evidence of original antigenic sin after infection or vaccination with the 2009 pandemic H1N1 influenza virus. Clin Vaccine Immunol. 2014;21(5):737–46. doi: 10.1128/CVI.00790-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Viboud C, et al. Age- and sex-specific mortality associated with the 1918–1919 influenza pandemic in Kentucky. J Infect Dis. 2013;207(5):721–9. doi: 10.1093/infdis/jis745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hancock K, et al. Cross-reactive antibody responses to the 2009 pandemic H1N1 influenza virus. N Engl J Med. 2009;361(20):1945–52. doi: 10.1056/NEJMoa0906453. [DOI] [PubMed] [Google Scholar]
- 42.Worobey M, et al. Genesis and pathogenesis of the 1918 pandemic H1N1 influenza A virus. Proc Natl Acad Sci U S A. 2014;111(22):8107–12. doi: 10.1073/pnas.1324197111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Henry Dunand CJ, et al. Preexisting human antibodies neutralize recently emerged H7N9 influenza strains. J Clin Invest. 2015;125(3):1255–68. doi: 10.1172/JCI74374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li GM, et al. Pandemic H1N1 influenza vaccine induces a recall response in humans that favors broadly cross-reactive memory B cells. Proc Natl Acad Sci U S A. 2012;109(23):9047–52. doi: 10.1073/pnas.1118979109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wrammert J, et al. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J Exp Med. 2011;208(1):181–93. doi: 10.1084/jem.20101352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang KY, et al. Focused antibody response to influenza linked to antigenic drift. J Clin Invest. 2015;125(7):2631–45. doi: 10.1172/JCI81104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Skehel JJ, Wiley DC. Influenza haemagglutinin. Vaccine. 2002;20(Suppl 2):S51–4. doi: 10.1016/s0264-410x(02)00131-7. [DOI] [PubMed] [Google Scholar]
- 48.Iba Y, et al. Conserved neutralizing epitope at globular head of hemagglutinin in H3N2 influenza viruses. J Virol. 2014;88(13):7130–44. doi: 10.1128/JVI.00420-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dreyfus C, et al. Highly conserved protective epitopes on influenza B viruses. Science. 2012;337(6100):1343–8. doi: 10.1126/science.1222908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schmidt AG, et al. Preconfiguration of the antigen-binding site during affinity maturation of a broadly neutralizing influenza virus antibody. Proc Natl Acad Sci U S A. 2013;110(1):264–9. doi: 10.1073/pnas.1218256109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsibane T, et al. Influenza human monoclonal antibody 1F1 interacts with three major antigenic sites and residues mediating human receptor specificity in H1N1 viruses. PLoS Pathog. 2012;8(12):e1003067. doi: 10.1371/journal.ppat.1003067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ekiert DC, et al. Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature. 2012;489(7417):526–32. doi: 10.1038/nature11414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee PS, et al. Heterosubtypic antibody recognition of the influenza virus hemagglutinin receptor binding site enhanced by avidity. Proc Natl Acad Sci U S A. 2012;109(42):17040–5. doi: 10.1073/pnas.1212371109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Whittle JR, et al. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc Natl Acad Sci U S A. 2011;108(34):14216–21. doi: 10.1073/pnas.1111497108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ekiert DC, et al. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009;324(5924):246–51. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sui J, et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol. 2009;16(3):265–73. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Graves PN, et al. Preparation of influenza virus subviral particles lacking the HA1 subunit of hemagglutinin: unmasking of cross-reactive HA2 determinants. Virology. 1983;126(1):106–16. doi: 10.1016/0042-6822(83)90465-8. [DOI] [PubMed] [Google Scholar]
- 58.Wang TT, et al. Broadly protective monoclonal antibodies against H3 influenza viruses following sequential immunization with different hemagglutinins. PLoS Pathog. 2010;6(2):e1000796. doi: 10.1371/journal.ppat.1000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ekiert DC, et al. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science. 2011;333(6044):843–50. doi: 10.1126/science.1204839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Friesen RH, et al. A common solution to group 2 influenza virus neutralization. Proc Natl Acad Sci U S A. 2014;111(1):445–50. doi: 10.1073/pnas.1319058110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tan GS, et al. Characterization of a broadly neutralizing monoclonal antibody that targets the fusion domain of group 2 influenza A virus hemagglutinin. J Virol. 2014;88(23):13580–92. doi: 10.1128/JVI.02289-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tan GS, et al. A pan-H1 anti-hemagglutinin monoclonal antibody with potent broad-spectrum efficacy in vivo. J Virol. 2012;86(11):6179–88. doi: 10.1128/JVI.00469-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Corti D, et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science. 2011;333(6044):850–6. doi: 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]
- 64.Kallewaard NL, et al. Structure and Function Analysis of an Antibody Recognizing All Influenza A Subtypes. Cell. 2016;166(3):596–608. doi: 10.1016/j.cell.2016.05.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Joyce MG, et al. Vaccine-Induced Antibodies that Neutralize Group 1 and Group 2 Influenza A Viruses. Cell. 2016;166(3):609–23. doi: 10.1016/j.cell.2016.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moody MA, et al. H3N2 influenza infection elicits more cross-reactive and less clonally expanded anti-hemagglutinin antibodies than influenza vaccination. PLoS One. 2011;6(10):e25797. doi: 10.1371/journal.pone.0025797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Margine I, et al. H3N2 influenza virus infection induces broadly reactive hemagglutinin stalk antibodies in humans and mice. J Virol. 2013;87(8):4728–37. doi: 10.1128/JVI.03509-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu L, et al. Induction of Broadly Cross-Reactive Stalk-Specific Antibody Responses to Influenza Group 1 and Group 2 Hemagglutinins by Natural H7N9 Virus Infection in Humans. J Infect Dis. 2017;215(4):518–528. doi: 10.1093/infdis/jiw608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Miller MS, et al. 1976 and 2009 H1N1 influenza virus vaccines boost anti-hemagglutinin stalk antibodies in humans. J Infect Dis. 2013;207(1):98–105. doi: 10.1093/infdis/jis652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sui J, et al. Wide prevalence of heterosubtypic broadly neutralizing human anti-influenza A antibodies. Clin Infect Dis. 2011;52(8):1003–9. doi: 10.1093/cid/cir121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sangster MY, et al. B cell response and hemagglutinin stalk-reactive antibody production in different age cohorts following 2009 H1N1 influenza virus vaccination. Clin Vaccine Immunol. 2013;20(6):867–76. doi: 10.1128/CVI.00735-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thomson CA, et al. Pandemic H1N1 Influenza Infection and Vaccination in Humans Induces Cross-Protective Antibodies that Target the Hemagglutinin Stem. Front Immunol. 2012;3:87. doi: 10.3389/fimmu.2012.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ellebedy AH, et al. Induction of broadly cross-reactive antibody responses to the influenza HA stem region following H5N1 vaccination in humans. Proc Natl Acad Sci U S A. 2014;111(36):13133–8. doi: 10.1073/pnas.1414070111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Krammer F. The Quest for a Universal Flu Vaccine: Headless HA 2.0. Cell Host Microbe. 2015;18(4):395–7. doi: 10.1016/j.chom.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 75.Krammer F. Novel universal influenza virus vaccine approaches. Curr Opin Virol. 2016;17:95–103. doi: 10.1016/j.coviro.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sagawa H, et al. The immunological activity of a deletion mutant of influenza virus haemagglutinin lacking the globular region. J Gen Virol. 1996;77(Pt 7):1483–7. doi: 10.1099/0022-1317-77-7-1483. [DOI] [PubMed] [Google Scholar]
- 77.Steel J, et al. Influenza virus vaccine based on the conserved hemagglutinin stalk domain. MBio. 2010;1(1) doi: 10.1128/mBio.00018-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wohlbold TJ, et al. Vaccination with soluble headless hemagglutinin protects mice from challenge with divergent influenza viruses. Vaccine. 2015;33(29):3314–21. doi: 10.1016/j.vaccine.2015.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mallajosyula VV, et al. Influenza hemagglutinin stem-fragment immunogen elicits broadly neutralizing antibodies and confers heterologous protection. Proc Natl Acad Sci U S A. 2014;111(25):E2514–23. doi: 10.1073/pnas.1402766111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yassine HM, et al. Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat Med. 2015;21(9):1065–70. doi: 10.1038/nm.3927. [DOI] [PubMed] [Google Scholar]
- 81.Impagliazzo A, et al. A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science. 2015;349(6254):1301–6. doi: 10.1126/science.aac7263. [DOI] [PubMed] [Google Scholar]
- 82.Valkenburg SA, et al. Stalking influenza by vaccination with pre-fusion headless HA mini-stem. Sci Rep. 2016;6:22666. doi: 10.1038/srep22666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Krammer F, et al. Chimeric hemagglutinin influenza virus vaccine constructs elicit broadly protective stalk-specific antibodies. J Virol. 2013;87(12):6542–50. doi: 10.1128/JVI.00641-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Krammer F, et al. H3 stalk-based chimeric hemagglutinin influenza virus constructs protect mice from H7N9 challenge. J Virol. 2014;88(4):2340–3. doi: 10.1128/JVI.03183-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Margine I, et al. Hemagglutinin stalk-based universal vaccine constructs protect against group 2 influenza A viruses. J Virol. 2013;87(19):10435–46. doi: 10.1128/JVI.01715-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nachbagauer R, et al. Hemagglutinin Stalk Immunity Reduces Influenza Virus Replication and Transmission in Ferrets. J Virol. 2015;90(6):3268–73. doi: 10.1128/JVI.02481-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Goff PH, et al. Adjuvants and immunization strategies to induce influenza virus hemagglutinin stalk antibodies. PLoS One. 2013;8(11):e79194. doi: 10.1371/journal.pone.0079194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nachbagauer R, Krammer F. Universal influenza virus vaccines and therapeutic antibodies. Clin Microbiol Infect. 2017;23(4):222–228. doi: 10.1016/j.cmi.2017.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nachbagauer R, et al. Age Dependence and Isotype Specificity of Influenza Virus Hemagglutinin Stalk-Reactive Antibodies in Humans. MBio. 2016;7(1):e01996–15. doi: 10.1128/mBio.01996-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gagnon A, et al. Is antigenic sin always "original?" Re-examining the evidence regarding circulation of a human H1 influenza virus immediately prior to the 1918 Spanish flu. PLoS Pathog. 2015;11(3):e1004615. doi: 10.1371/journal.ppat.1004615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Andrews SF, et al. High preexisting serological antibody levels correlate with diversification of the influenza vaccine response. J Virol. 2015;89(6):3308–17. doi: 10.1128/JVI.02871-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Krammer F, et al. An H7N1 influenza virus vaccine induces broadly reactive antibody responses against H7N9 in humans. Clin Vaccine Immunol. 2014;21(8):1153–63. doi: 10.1128/CVI.00272-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nachbagauer R, et al. Induction of broadly reactive anti-hemagglutinin stalk antibodies by an H5N1 vaccine in humans. J Virol. 2014;88(22):13260–8. doi: 10.1128/JVI.02133-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Halliley JL, et al. High-Affinity H7 Head and Stalk Domain-Specific Antibody Responses to an Inactivated Influenza H7N7 Vaccine After Priming With Live Attenuated Influenza Vaccine. J Infect Dis. 2015;212(8):1270–8. doi: 10.1093/infdis/jiv210. [DOI] [PMC free article] [PubMed] [Google Scholar]