Abstract
Combining fluorescence microscopy with electron cryo-tomography allows, in principle, spatial localization of tagged macromolecular assemblies and structural features within the cellular environment. To allow precise localization and scale integration between the two disparate imaging modalities, accurate alignment procedures are needed. Here, we describe a marker-free method for aligning images from light or cryo-light fluorescence microscopy and from electron cryo-microscopy that takes advantage of sample support features, namely the holes in the carbon film. We find that the accuracy of this method, as judged by prediction errors of the hole center coordinates, is better than 100 nm.
Introduction
Correlative imaging of samples using fluorescence markers to identify macromolecular assemblies and structural features in images and volumes obtained by electron cryo-microscopy approaches has become an important tool for biomedical research in recent years. In electron cryo-microscopy (cryo-EM), samples are vitrified by fast freezing, resulting in the biological samples being embedded in a layer of amorphous ice, preserved in their native state (Dubochet et al., 1988). Recent hardware and software developments have revolutionized the field, allowing now to obtain structural information at near atomic resolution for a wide variety of macromolecular assemblies (Subramaniam et al., 2016). The same advances also enable imaging of unperturbed cellular landscapes at high resolution (Asano et al., 2015). Although some large macromolecular assemblies are identifiable in such cellular reconstructions (Asano et al., 2015; Mahamid et al., 2016; Woodward et al., 2015; Fukuda et al., 2017) the majority of the components are not identifiably without specific labeling, presenting a major challenge in crowded cellular environments.
The advantages of fluorescence microscopy are complementary to those of electron cryo-microscopy. The fluorescence microscopy approach allows live cell imaging and to follow specific fluorescently labeled molecules in real time. The larger field of view also facilitates the identification and localization of sparse cellular events in a larger context. The combination of the two microscopy approaches potentially allows identifying regions associated with specific biological events by fluorescence microscopy and then investigating the underlying structural features at high resolution by cryo-EM.
For correlative imaging, vitrification of the sample can be either performed before or after fluorescence imaging. The advantages of room temperature fluorescence imaging include the use of optimized equipment such as high-powered, oil-immersion lenses with large numerical apertures and that dynamic processes can be characterized and followed. Even if precautions are being made to avoid imaging induced damage related to buffer and anti-oxidative agents, the disadvantage of this procedure is that structures may change or that the biological process of interest may be completed in the time required to perform the vitrification. To avoid this problem, cells can be fixed with carefully chosen fixation protocols to minimize artifacts that might be caused by cross-linking of cellular components. For fluorescence imaging after vitrification, specialized cryo-stages that allow fluorescence imaging at cryo-temperatures are required (Sartori et al., 2007; Briegel et al., 2010; Faas et al., 2013; Hampton et al., 2017; Arnold et al., 2016) and the resolution is restricted to about 400 nm or less owing to lack of immersion cryo-objectives. In addition, cryo-fluorescence imaging loses the capability of capturing the dynamics of the biological process. Correlative imaging using super-resolution techniques and electron cryo-microscopy are also under development (Wolff et al., 2016; Nahmani et al., 2017).
For all these correlative techniques, it is necessary to find an accurate alignment between the images from the two imaging modalities. The general workflow for this type of correlative imaging involves acquisition of fluorescence images and acquisition of cryo-EM images at increasing magnifications towards a final high magnification image or tilt series for tomographic reconstruction. The coordinate transform for the first step, i.e. correlating the fluorescence image with a low-magnification cryo-EM image, generally involves in-plane rotation, translation, and scaling. In some instances, when there are significant beam-induced changes to the sample (Brilot et al., 2012) or the sample support is tilted significantly in respect to the second imaging modality, further correction may be necessary. The subsequent steps are less complex for several reasons: (i) There is no change in rotation between the subsequent cryo-EM images; (ii) the difference in magnification is well defined, (iii) because the imaging modality does not change, features correlate very well between the successive images because one is simply a lower-resolution version of the other. Highly accurate methods for alignment of cryo-EM images based on fiducial markers (Kremer et al., 1996) or correlation (Winkler and Taylor, 2006) are well established in the cryo-EM field.
Early applications of correlative fluorescence/cryo-EM imaging took advantage of large cellular features to correlate fluorescence images with low-magnification cryo-EM images, which allowed a straightforward relative orientation of the sample (van Driel et al., 2009). To increase the accuracy of the alignment and to allow determination of accuracy estimates, the use of fluorescence beads as fiducial markers for alignment was introduced (Kukulski et al., 2011; Schorb and Briggs, 2014; Schellenberger et al., 2014). Here, we propose a marker-free approach using features of the sample support to provide alignment between images from fluorescence or cryo-fluorescence microscopy and cryo-EM images. The alignment quality of this approach is comparable to that previously reported for marker-based alignment and does not involve modification of the experimental procedures.
Materials and methods
Sample preparation
GFP:paxillin-transfected mouse embryonic fibroblast and Chinese hamster ovary cells (Bachir et al., 2014) were cultured on electron microscopy grids as previously described (Anderson et al., 2016). Streptococcus pneumoniae samples were prepared as previously described (Durand et al., 2015). Grids were manually plunge frozen in liquid ethane at liquid nitrogen temperature either after room temperature fluorescence imaging and mild fixation and/or before cryo-fluorescence imaging.
Correlative fluorescence and electron cryo-microscopy
We tested the alignment protocols using samples on commonly used electron microscopy grids (Quantifoil Grids with R1.2/1.3, R1/4 or R5/20 hole patterns). Fluorescence modalities used for correlated imaging included FRET, confocal, and cryo-fluorescence imaging with mCherry or GFP, recorded with a variety of objectives and camerapixel sizes. Cryo-fluorescence imaging was performed using a CorrSight CLEM microscope (FEI Company), equipped with a motorized stage, a broad-band Xenon light source and motorized fluorescent filter sets with details previously described (Arnold et al., 2016). The CorrSight contains a 40 × objective and the camera pixel size is 161 nm. The vitrified grids were mounted into grid support rings (Auto-gridTEM sample holder, FEI) and transferred into the cartridge holder of a CorrSight CLEM microscope for imaging. The clipped grids are then transferred into the Titan Krios electron microscope so no manual grid handling between cryo-fluorescence and cryo-EM imaging is necessary. Cryo-EM images were acquired with an FEI Titan Krios transmission electron cryo-microscope (FEI Company) equipped with an extra-high brightness field emission gun and operated at 300 kV using a back-thinned 4k × 4k FEI Falcon II direct detection device. Images used for hole identification were taken at magnifications between 500 × (pixel size 28 nm) and 1500 × (pixel size 9.3 nm) depending on the magnification of the correlated fluorescence images and the density of the hole-pattern on the grid. The defocus was individually adjusted to emphasize the edges of the holes as much as possible.
Identification of holes in the carbon film
To obtain estimates for the center positions of the holes in the images we applied the following multi-step procedure.
An iterative median filter (van der Heide et al., 2007) was applied to suppress background noise while preserving the sharpness of the hole edges. We used 20 iterations, which is usually enough to reach the stationary fixed point (Tyan, 1981). This step is fully automatic.
The optimal threshold for binarization was determined interactively.
The Canny edge detector (Canny, 1986) was applied to the binarized image. This step is automatic.
An initial estimate of the hole-radius was obtained interactively either from the binarized image or from the edge image.
The circular Hough transform (Kimme et al., 1975) was run on the edge image with variable radius around the initial radius to determine the optimal radius for detecting the holes. High values in the circular Hough transform image correspond to centers of circular objects in the transformed image.
The circular Hough transform was re-run with the optimized radius.
The optimal number of peaks was determined interactively using a simplified peak search. The determined number of peaks was extracted using a full-blown, high-quality peak search algorithm. These peaks correspond to the center coordinates of the detected circles.
Alignment using the circle center coordinates
To allow the use of the circle center coordinates for alignment, we first needed to determine the mapping of the same holes in the two different imaging domains. The first step is to pick a small number of correspondences interactively (a minimum of two). This is achieved by giving each peak a unique label, rotating the fluorescence image approximately in the same orientation as the cryo-EM image and then displaying the two labeled images next to each other and to the corresponding reference images. Picking a small number of correspondences is then straightforward. Once initial correspondences were determined, these were used to automatically find all other correspondences between the two images. Initial scales were automatically determined using all distances between all holes and their correspondences. Using this scale estimate, initial estimates of the in-plane rotation and the relative translation were obtained using simplex optimizations (Nelder and Mead, 1964) every 45° and picking the one that resulted in the lowest root-mean-square deviation (rmsd) between the reference center coordinates and the corresponding transformed coordinates in the other imaging domain.
As a second step, all parameters i.e. scale, in-plane rotation, and relative translation are refined by minimizing the rmsd using the simplex optimizer. The entire workflow has been implemented in the pyCoAn framework, a python-based extension of the CoAn package (Volkmann and Hanein, 1999; Volkmann, 2009). A complete run takes about 5–10 minutes on a modern workstation. The software is freely available and can be accessed, together with documentation and tutorials, at the pyCoAn website (coan.burnham.org).
Simulated data
A hole pattern was created in the GNU Image Manipulation Program (www.gimp.com). The rest of the operations were performed within the pyCoAn framework. A collection of large interspersed Gaussian spheres of different sizes and weights were created and projected to emulate a cell-like density. The resulting image was overlaid with the hole pattern. The image was scaled down by a factor of six, rotated by 255° and shifted to provide the background of the simulated fluorescence image. Sixteen small but intense Gaussian peaks were created in the simulated cell area to emulate localized fluorescence signals. The centers of these Gaussians served as the point-like features to be located in the simulated cryo-EM images. The contrast between the background and the signals was adjusted to match the appearance of a typical experimental fluorescence image. Spatially correlated mixed Gaussian and impulse noise was added and adjusted to match the noise in the experimental fluorescence test images. To generate a simulated cryo-EM image, the original image of the cell-like density on the hole-pattern was inverted. A series of edge-enhancements and convolution steps were performed to add line-like features inside the simulated cell. A number of smaller Gaussian densities were added in random locations to generate some higher-resolution texture. The pixel size was set to 30 nm and a defocus of 50 microns was computationally applied to the image. Spatially correlated mixed Gaussian and impulse noise was added and adjusted to match the noise in the experimental cryo-EM test images. The simulated data are distributed as part of the tutorial on the pyCoAn website (coan.burnham.org).
Alignment accuracy estimation
To obtain an estimate of the alignment accuracy, we averaged over individual prediction errors following a similar procedure previously developed for fiducial-based alignments (Schellenberger et al., 2014; Kukulski et al., 2011). Briefly, for the calculation of individual prediction errors, one hole center coordinate from the fluorescence image was excluded from the alignment calculation. This coordinate was treated as the object of interest and its position predicted within the cryo-EM image. The predicted position was compared to the actual position in the cryo-EM image to estimate the individual prediction error. This operation was repeated for all hole centers in the fluorescence images. The estimate of the alignment accuracy was then calculated as the average of the individual prediction errors.
Results
We developed a method for accurate alignment of fluorescence images from light microscopy with images obtained by cryo-EM. The approach is based on the visibility of holes in the carbon support in both imaging modalities. The holes are readily identifiable in the cryo-EM images (Figure 1A) as long as the ice is not too thick. In the fluorescence images the holes are not always immediately apparent (Figure 1B). However, there is always a subtle difference between the background fluorescence in the holes versus background fluorescence on the carbon that becomes visible when increasing the contrast to maximum. The resulting images tend to be rather noisy, which makes accurate detection of the hole boundaries difficult. Application of iterative median filtering (van der Heide et al., 2007) up to the stationary fixed point improves the situation considerably and most holes become identifiable in the fluorescence images as well (Figure 1C). For images with very high noise levels, it can be beneficial to apply more sophisticated noise reduction algorithms such as non-local means (Buades et al., 2005), bilateral filtering (Jiang et al., 2003) or anisotropic nonlinear diffusion (Frangakis and Hegerl, 2001; Fernandez and Li, 2003). The optimal application of these algorithms usually involves a significant amount of parameter tweaking. It is also possible to use light microscopy bright-field images to assist with the hole identification with the caveat that there might be an additional uncertainty in the alignment between the fluorescence and bright-field images. The cryo-EM images also benefit from iterative median filtering, which suppresses the background variations, especially inside the holes. Cryo-EM images from samples with thicker ice, where the signal-to-noise ratio is reduced, may require the application of more sophisticated noise reduction algorithms that can be better at suppressing noise.
Figure 1. Correlative imaging.
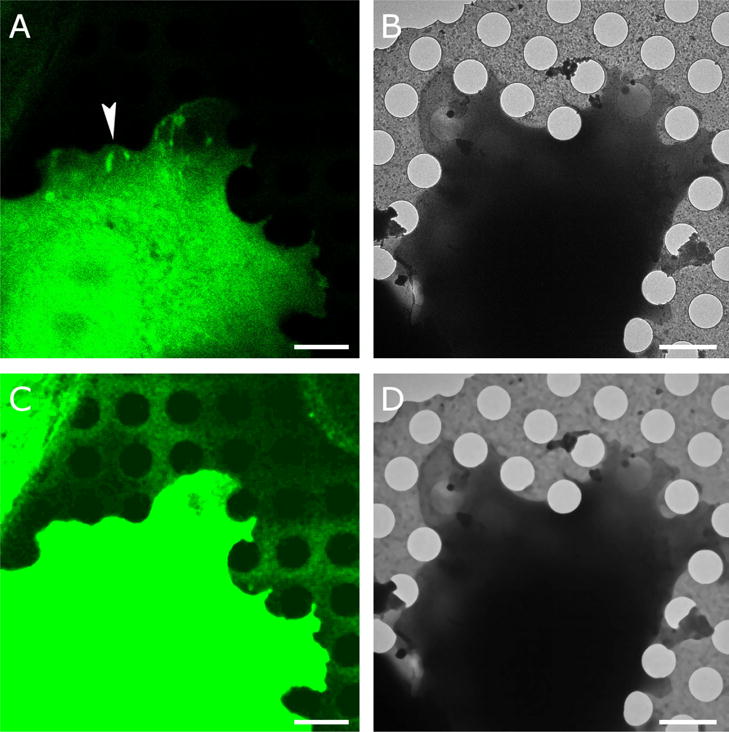
A. An Olympus FV1000 confocal microscope equipped with a 60 × 1.35 UPlanSApo oil objective was used to capture Paxillin-GFP fluorescence (green) with a pixel size of 81 nm. Fluorescence was obtained at room temperature and the cell was plunge frozen after mild fixation (Anderson et al. 2016). B. Cryo-EM image of the same cell at 1500 × magnification resulting in 9.3-nm pixel size. The arrowhead in (A) points at a region with elongated spots of high paxillin fluorescence, indicative of focal adhesions, an actin-based assembly involved in cell adhesion to extracellular substrates. C. The holes in the carbon film are not visible in (A) but become visible after contrast adjustment and noise suppression with iterative median filtering. D. Iterative median filtering also suppresses background fluctuations in the cryo-EM image, allowing more accurate determination of the hole centers. Bars are 2 microns.
Once the hole center locations are determined, a one-to-one correspondence mapping between the holes in one versus the other imaging modality is determined (Figure 2C–D). The coordinates can then be used to determine the rotation, translation, and relative scale between the fluorescence and cryo-EM images by minimizing the root-mean-square deviation between the reference coordinates and the transformed coordinates of the other imaging domain. To estimate the alignment accuracy, we calculated averages of the prediction errors of individual circle centers (Schellenberger et al., 2014; Kukulski et al., 2011). Previous studies (Schellenberger et al., 2014) distinguish between three different error terms contributing to the prediction error.
Figure 2. Hole detection. This is the same cell as in Fig. 1.
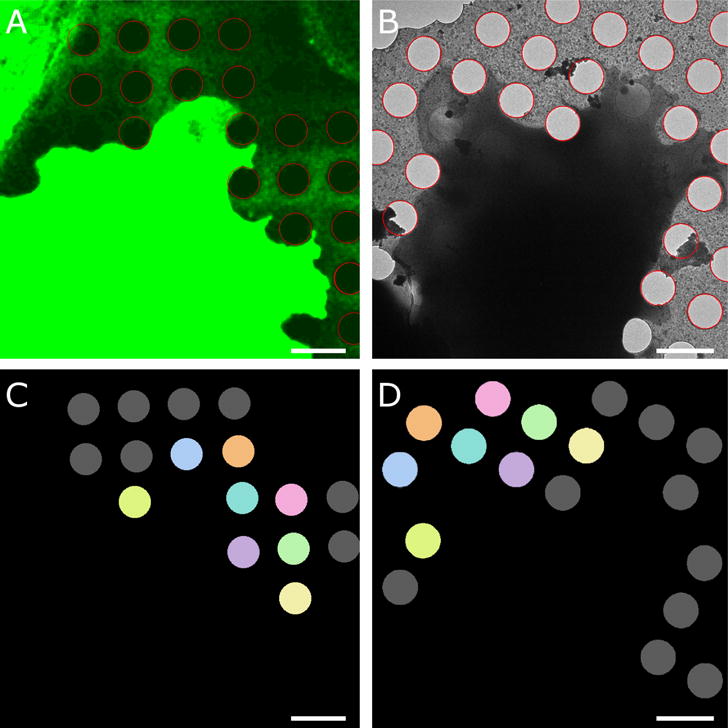
A. Application of edge detection and circular Hough transform allows localization of the hole boundaries (red circles) in the enhanced fluorescence image. B. Hole boundaries (red) in electron cryo-microscopy image of the same cell. C–D. Correspondence mapping of the holes in the respective images. Identical colors refer to the same hole, holes marked in grey do not have a corresponding hole in the other imaging modality. Bars are 2 microns.
(i) an error associated to the alignment of fluorescence channels; (ii) an error associated with the alignment between the fiducials markers in the two imaging modalities; and (iii) an error associated with the localization accuracy of point-like fiducials in the fluorescence images. In our case, the first term is not present because we deal with single fluorescence channels. The third term is also absent because we detect circles and not point-like features. Therefore, our prediction error corresponds most closely to the second term above but also accounts for some uncertainty in the determination of the hole centers.
We find that the average Euclidian distance between the predicted coordinates and the coordinates based on the full set was between 59.3 and 89.4 nm, depending on the actual correlative image pair (Table 1). Application of the coordinate transform to the fluorescence images shows excellent visual overlap between the hole boundaries with those in the cryo-EM images (Figure 4A) and allows accurate overlay of features of interest in the fluorescence image with the cryo-EM image of the same region for room-temperature (Figure 3B) as well as cryo-fluorescence images (Figure 4).
Table 1. Alignment statistics for various CLEM data sets.
RT: room temperature; accuracy: estimated accuracy calculated from prediction errors; std: standard deviation for the estimated accuracy; cryo LM: obtained by the CorrSight CLEM microscope under cryo conditions.
| grid type | FM pixel | EM pixel | accuracy | |
|---|---|---|---|---|
| RT confocal | R1.2/1.3 | 81 nm | 9.3 nm | 62.4 nm |
| RT confocal | R 1/4 | 81 nm | 14 nm | 76.3 nm |
| RT confocal | R 5/20 | 81 nm | 14 nm | 75.2 nm |
| RT FRET | R 5/20 | 330 nm | 28 nm | 89.4 nm |
| RT FRET | R 5/20 | 61 nm | 28 nm | 63.6 nm |
| cryo LM | R 1.2/1.3 | 161 nm | 28 nm | 59.3 nm |
| cryo LM | R 1.2/1.3 | 161 nm | 28 nm | 79.9 nm |
Figure 4. Correlating cryo-fluorescence and electron cryo-microscopy images.
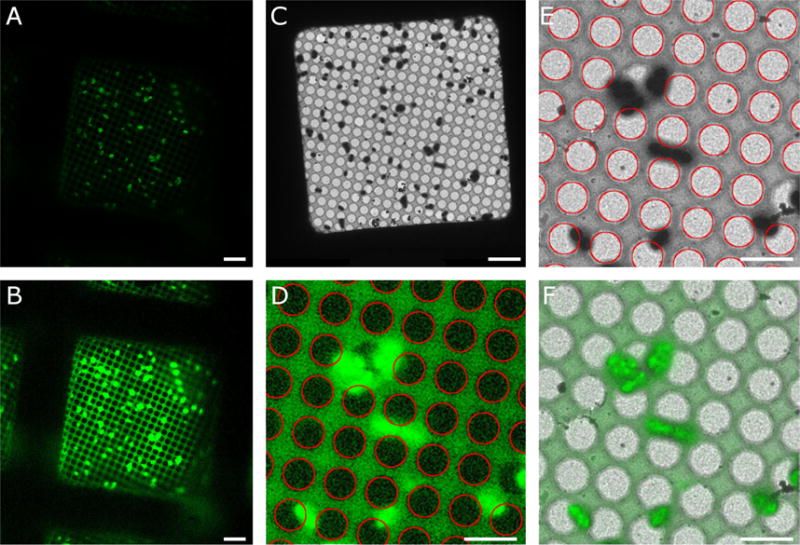
A. Correlated R3045 ComGA:GFP fluorescence (green) of expressing Streptococcus pneumoniae recorded with a CorrSight cryo-microscope (FEI company). A 40 × objective was used with a camera pixel size of 161 nm. B. Enhanced cryo-fluorescence image showing the detectability of the holes in the carbon film. C. Cryo-EM image of the same field of view. The magnification is 500 × with 28-nm pixel size. D. Enlarged region of enhanced cryo-fluorescence with Hough transform circles (red) overlaid. E. Enlarged region of cryo-EM image with Hough circles (red) overlaid. F. Alignment of cryo-fluorescence (green) with cryo-EM image (grayscale) based on the centers of corresponding Hough circles. Bars are 5 microns.
Figure 3. Overlay of Paxilin-GFP fluorescence image (green) with electron cryomicroscopy image (greyscale) after applying the transform determined using the carbon hole center correspondences. Same cell as in Figures 1 and 2.
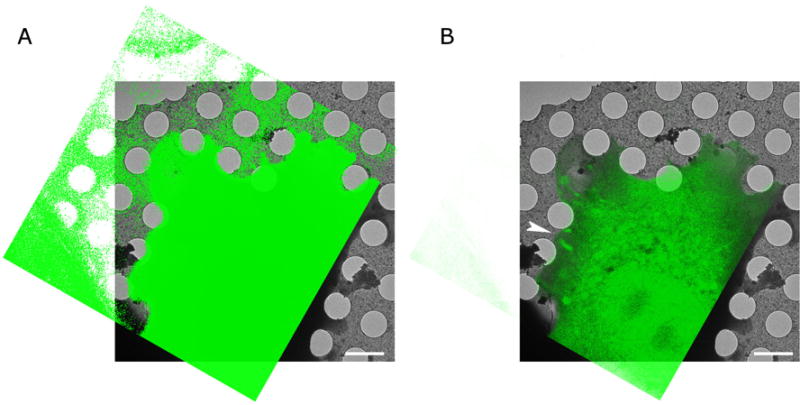
A. Is shown at high contrast to show the excellent correspondence between the holes. B. Is shown at lower contrast to show the overlay of fluorescence features with the electron cryo-microscopy image. The arrowhead points at the same focal adhesion region as the arrowhead in Figure 1. The bars are 2 microns.
It was shown previously using fluorescing objects visible at the EM level such as virus particles or microtubule ends that the estimated alignment accuracy using prediction errors correlates well with the localization accuracy of the individual fluorescing objects (Kukulski et al., 2011; Kukulski et al., 2012; Schellenberger et al., 2014; Schorb and Briggs 2014; Schorb et al., 2017). Here, we used simulated images to investigate not only the correspondence between the estimated accuracy and the actual localization accuracy in more detail but also to investigate the influence of local deformations that can be adequately approximated by a rotation around an arbitrary axis in the imaging plane (tilt) within the field of view. Because the locations of point-like features are known for simulated images before and after the coordinate transform, we can determine a meaningful relationship between estimated and true accuracy. The accuracy estimate based on prediction errors correlates very well (R = 0.98) with the actual localization accuracy over the entire range of accuracies tested (Figure 5A). The error introduced by tilt rotation between the two imaging modalities is essentially negligible below an angle of 5° and, depending on the pixel size at the cryo-EM level, significantly higher angles might be tolerable (Figure 5B). For example, at 9.3 nm pixel size, an angular offset of 13° would result in less than 30 nm mean displacement.
Figure 5. Evaluation of accuracy estimates using simulated images.
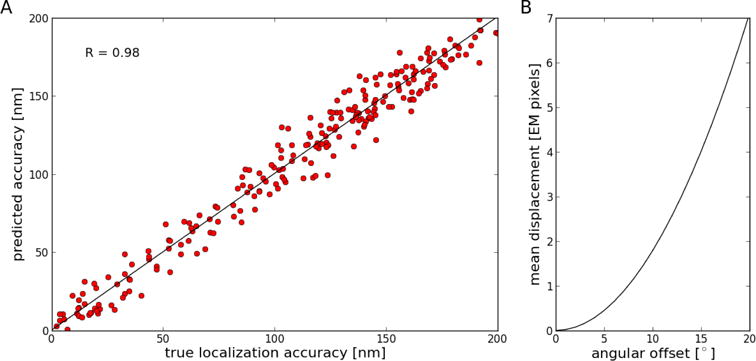
A. Correlation between true localization accuracy and accuracy estimated from prediction errors. Linear regression shows that the data is highly correlated (R = 0.98; slope = 1.002; intercept = 0.21). Accuracy is defined here as the mean Euclidean distance between corresponding point-like features. B. Effect of angular offset between images on mean displacement between locations of point-like features in the reference sample versus in the tilted sample. An angular offset of about 7.5° results in a one-pixel mean displacement.
Discussion
Fiducial-less alignment based on features of the electron-microscopy sample support has several advantages over marker-based alignment methods. First, no additional material needs to be added to the sample. Markers need to be explicitly introduced and may obscure features of interest. In contrast, the sample support is always present. Second, the distribution of the holes is regular and covers the entire grid in predictable ways with excellent coverage. Marker concentrations need to be optimized. Too many markers can interfere with the analysis of the sample; too few markers may not be enough for high-quality alignment. In addition, markers can spatially cluster, which can also degrade alignment quality. For example, if ten markers form a small cluster in the lower right-hand corner, then alignment between features in the top left-hand corner may be off significantly even though the precision estimate using the ten marker coordinates will not give any indication of the issue. Because the grid is regular by nature, clustering of hole positions is much less likely. Third, the large size of the holes makes them amenable for aligning over a larger range of magnifications than point-like fiducial markers such as fluorescent beads.
The estimated alignment accuracy of our method is comparable to accuracy estimates previously provided for marker-based alignment of fluorescence and cryo-EM images (Schellenberger et al., 2014; Schorb and Briggs, 2014). For bead-based alignment, it is assumed that the bead is essentially a point-like object at the light-microscopy level and a two-dimensional Gaussian is fitted to get a precise, sub-pixel estimate of the center position of the bead (Schellenberger et al., 2014). When using holes for alignment, a circle must be fitted to the rim of the hole to determine the precise center position. On one hand, this can be an advantage because more image pixels are available for fitting the circle than are normally available for fitting a two-dimensional Gaussian to a bead position. On the other hand, because of the relatively large size of the circles, small errors in center estimates might not be as apparent as an offset Gaussian would be. The cumulative effect does not appear to make a significant difference because the overall estimated alignment accuracy of bead-based and hole-based alignments is comparable.
The use of the holes in the carbon film of the cryo-EM sample support for alignment is a viable alternative to bead-based alignment techniques in terms of alignment accuracy and does not require any modification of the sample. It is thus less invasive than bead-based methods. The regular and predictable distribution of holes ensures adequate coverage of reference points for alignment in any given area on the grid and the regularity will also help to detect and correct for distortions and relative tilt of the sample support between imaging modalities.
Acknowledgments
This work was supported by NIGMS grants R01 GM115972 and P01 GM121203, CCSG Pilot Project funding (P30 CA030199). This work was also supported by NIH grants S10 OD12372 and GM098412-S1 for the Titan Krios and Falcon II direct detector. We thank Drs. Rémi Fronzes and Esther Marza for providing the R3045 ComGA:GFP Streptococcus pneumoniae samples.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson KL, Page C, Swift MF, Suraneni P, Janssen ME, Pollard TD, Li R, Volkmann N, Hanein D. Nano-scale actin-network characterization of fibroblast cells lacking functional Arp2/3 complex. J Struc Biol. 2016;197:312–321. doi: 10.1016/j.jsb.2016.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold J, Mahamid J, Lucic V, de Marco A, Fernandez JJ, Laugks T, Mayer T, Hyman AA, Baumeister W, Plitzko JM. Site-Specific Cryo-focused Ion Beam Sample Preparation Guided by 3D Correlative Microscopy. Biophys J. 2016;110:860–869. doi: 10.1016/j.bpj.2015.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano S, Engel BD, Baumeister W. In Situ Cryo-Electron Tomography: A Post-Reductionist Approach to Structural Biology. J Mol Biol. 2015;428:332–343. doi: 10.1016/j.jmb.2015.09.030. [DOI] [PubMed] [Google Scholar]
- Asano S, Fukuda Y, Beck F, Aufderheide A, Förster F, Danev R, Baumeister W. Proteasomes. A molecular census of 26S proteasomes in intact neurons. Science. 2015;347:439–442. doi: 10.1126/science.1261197. [DOI] [PubMed] [Google Scholar]
- Bachir AI, Zareno J, Moissoglu K, Plow EF, Gratton E, Horwitz AR. Integrin-associated complexes form hierarchically with variable stoichiometry in nascent adhesions. Curr Biol. 2014;24:1845–1853. doi: 10.1016/j.cub.2014.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briegel A, Chen S, Koster AJ, Plitzko JM, Schwartz CL, Jensen GJ. Correlated light and electron cryo-microscopy. Methods Enzymol. 2010;481:317–341. doi: 10.1016/S0076-6879(10)81013-4. [DOI] [PubMed] [Google Scholar]
- Brilot AF, Chen JZ, Cheng A, Pan J, Harrison SC, Potter CS, Carragher B, Henderson R, Grigorieff N. Beam-induced motion of vitrified specimen on holey carbon film. J Struc Biol. 2012;177:630–637. doi: 10.1016/j.jsb.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buades A, Coll B, Morel JM. A non-local algorithm for image denoising. IEEE Comput Soc Conf Comp Vis Pattern Recog. 2005;2:60–65. [Google Scholar]
- Canny J. A computational approach to edge detection. IEEE Trans Pattern Anal Mach Intell. 1986;8:679–698. [PubMed] [Google Scholar]
- Dubochet J, Adrian M, Chang JJ, Homo JC, Lepault J, McDowall AW, Schultz P. Cryo-electron microscopy of vitrified specimens. Q Rev Biophys. 1988;21:129–228. doi: 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
- Durand E, Nguyen VS, Zoued A, Logger L, Péhau-Arnaudet G, Aschtgen MS, Spinelli S, Desmyter A, Bardiaux B, Dujeancourt A, Roussel A, Cambillau C, Cascales E, Fronzes R. Biogenesis and structure of a type VI secretion membrane core complex. Nature. 2015;523:555–560. doi: 10.1038/nature14667. [DOI] [PubMed] [Google Scholar]
- Faas FG, Bárcena M, Agronskaia AV, Gerritsen HC, Moscicka KB, Diebolder CA, van Driel LF, Limpens RW, Bos E, Ravelli RB, Koning RI, Koster AJ. Localization of fluorescently labeled structures in frozen-hydrated samples using integrated light electron microscopy. J Struc Biol. 2013;181:283–290. doi: 10.1016/j.jsb.2012.12.004. [DOI] [PubMed] [Google Scholar]
- Fernandez JJ, Li S. An improved algorithm for anisotropic nonlinear diffusion for denoising cryo-tomograms. J Struct Biol. 2003;144:152–161. doi: 10.1016/j.jsb.2003.09.010. [DOI] [PubMed] [Google Scholar]
- Frangakis AS, Hegerl R. Noise reduction in electron tomographic reconstructions using nonlinear anisotropic diffusion. J Struct Biol. 2001;135:239–250. doi: 10.1006/jsbi.2001.4406. [DOI] [PubMed] [Google Scholar]
- Fukuda Y, Beck F, Plitzko JM, Baumeister W. In situ structural studies of tripeptidyl peptidase II (TPPII) reveal spatial association with proteasomes. Proc Natl Acad Sci U S A. 2017;114:4412–4417. doi: 10.1073/pnas.1701367114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton CM, Strauss JD, Ke Z, Dillard RS, Hammonds JE, Alonas E, Desai TM, Marin M, Storms RE, Leon F, Melikyan GB, Santangelo PJ, Spearman PW, Wright ER. Correlated fluorescence microscopy and cryo-electron tomography of virus-infected or transfected mammalian cells. Nat Protoc. 2017;12:150–167. doi: 10.1038/nprot.2016.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Baker ML, Wu Q, Bajaj C, Chiu W. Applications of a bilateral denoising filter in biological electron microscopy. J Struct Biol. 2003;144:114–122. doi: 10.1016/j.jsb.2003.09.028. [DOI] [PubMed] [Google Scholar]
- Kimme C, Ballard D, Sklansky J. Finding circles by an array of accumulators. Communications of the ACM. 1975;18:120–122. [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Kukulski W, Schorb M, Welsch S, Picco A, Kaksonen M, Briggs JA. Correlated fluorescence and 3D electron microscopy with high sensitivity and spatial precision. J Cell Biol. 2011;192:111–119. doi: 10.1083/jcb.201009037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukulski W, Schorb M, Welsch S, Picco A, Kaksonen M, Briggs JA. Precise, correlated fluorescence microscopy and electron tomography of lowicryl sections using fluorescent fiducial markers. Methods Cell Biol. 2012;111:235–257. doi: 10.1016/B978-0-12-416026-2.00013-3. [DOI] [PubMed] [Google Scholar]
- Mahamid J, Pfeffer S, Schaffer M, Villa E, Danev R, Cuellar LK, Förster F, Hyman AA, Plitzko JM, Baumeister W. Visualizing the molecular sociology at the HeLa cell nuclear periphery. Science. 2016;351:969–972. doi: 10.1126/science.aad8857. [DOI] [PubMed] [Google Scholar]
- Nahmani M, Lanahan C, DeRosier D, Turrigiano GG. High-numerical-aperture cryogenic light microscopy for increased precision of superresolution reconstructions. Proc Natl Acad Sci U S A. 2017;114:3832–3836. doi: 10.1073/pnas.1618206114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelder JA, Mead R. A simplex method for function minimization. Comp J. 1964;7:308–313. [Google Scholar]
- Sartori A, Gatz R, Beck F, Rigort A, Baumeister W, Plitzko JM. Correlative microscopy: bridging the gap between fluorescence light microscopy and cryo-electron tomography. J Struct Biol. 2007;160:135–145. doi: 10.1016/j.jsb.2007.07.011. [DOI] [PubMed] [Google Scholar]
- Schellenberger P, Kaufmann R, Siebert CA, Hagen C, Wodrich H, Grünewald K. High-precision correlative fluorescence and electron cryo microscopy using two independent alignment markers. Ultramicroscopy. 2014;143:41–51. doi: 10.1016/j.ultramic.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorb M, Briggs JA. Correlated cryo-fluorescence and cryo-electron microscopy with high spatial precision and improved sensitivity. Ultramicroscopy. 2014;143:24–32. doi: 10.1016/j.ultramic.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorb M, Gaechter L, Avinoam O, Sieckmann F, Clarke M, Bebeacua C, Bykov YS, Sonnen AF, Lihl R, Briggs JA. New hardware and workflows for semi-automated correlative cryo-fluoresence and cryo-electron microscopy/tomography. J Struct Biol. 2017;197:83–93. doi: 10.1016/j.jsb.2016.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam S, Earl LA, Falconieri V, Milne JL, Egelman EH. Resolution advances in cryo-EM enable application to drug discovery. Curr Opin Struct Biol. 2016;41:194–202. doi: 10.1016/j.sbi.2016.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyan SG. Median filtering: deterministic properties. In: Huang TS, editor. Two-dimensional digital signal processing II. Springer; New York: 1981. pp. 197–217. [Google Scholar]
- van der Heide P, Xu XP, Marsh BJ, Hanein D, Volkmann N. Efficient automatic noise reduction of electron tomographic reconstructions based on iterative median filtering. J Struct Biol. 2007;158:196–204. doi: 10.1016/j.jsb.2006.10.030. [DOI] [PubMed] [Google Scholar]
- van Driel LF, Valentijn JA, Valentijn KM, Koning RI, Koster AJ. Tools for correlative cryo-fluorescence microscopy and cryo-electron tomography applied to whole mitochondria in human endothelial cells. Eur J Cell Biol. 2009;88:669–684. doi: 10.1016/j.ejcb.2009.07.002. [DOI] [PubMed] [Google Scholar]
- Volkmann N. Confidence intervals for fitting of atomic models into low-resolution densities. Acta Crystallogr D Biol Crystallogr. 2009;65:679–689. doi: 10.1107/S0907444909012876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkmann N, Hanein D. Quantitative fitting of atomic models into observed densities derived by electron microscopy. J Struc Biol. 1999;125:176–184. doi: 10.1006/jsbi.1998.4074. [DOI] [PubMed] [Google Scholar]
- Winkler H, Taylor KA. Accurate marker-free alignment with simultaneous geometry determination and reconstruction of tilt series in electron tomography. Ultramicroscopy. 2006;106:240–254. doi: 10.1016/j.ultramic.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Wolff G, Hagen C, Grünewald K, Kaufmann R. Towards correlative super-resolution fluorescence and electron cryo-microscopy. Biol Cell. 2016;108:245–258. doi: 10.1111/boc.201600008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward CL, Mendonça LM, Jensen GJ. Direct visualization of vaults within intact cells by electron cryo-tomography. Cell Mol Life Sci. 2015;72:3401–3409. doi: 10.1007/s00018-015-1898-y. [DOI] [PMC free article] [PubMed] [Google Scholar]


