Abstract
Background and Objectives:
Enzootic abortion of ewes (EAE) is caused by infection of sheep and goats by Chlamydia abortus bacterium. Chlamydial abortion in bovine could occur by Chlamydia abortus, Chlamydia psittaci and Chlamydia pecorum. C. psittaci is the causative agent of psittacosis or ornithosis disease in humans and birds. It also causes acute pneumonia in cattle and sheep. The present study aimed at surveying the role of chlamydial agents in ruminants abortion.
Materials and Methods:
A total of 117 aborted material samples (Cotyledon, liver, spleen, and abomasal contents of fetus) from 9 cattle and 100 sheep in Shahr-e-Kord and 8 sheep from Bagh-e-Malek were collected from different herds with abortion history during the lambing periods from 2014 to 2016. After DNA extraction, the samples were tested by species-specific PCR to detect C. abortus, C. pecorum and C. psittaci.
Results:
Out of 117 clinical sample (108 sheep and 9 cattle), chlamydial infection was detected in 66 (56.41%) samples by Chlamydiales order-specific primers. A total of 24 (36.36%) and 24 (36.36%) samples indicated positive forms of C. abortus and C. psittasi infections, respectively. Only 1 (1.5%) C. pecorum was identified from cattle using nested PCR during this study. Among 66 Chlamydiales-positive samples, 20 (30.30%) samples with coinfection of C. abortus and C. psittaci were detected, however, infection of 3 species was not detected in the samples.
Conclusion:
Because of the high percentage of chlamydial infection in these regions and probability of coinfection, conducting epidemiological studies on the role of different animals is highly recommended.
Keywords: Abortion, Chlamydia, Goat, PCR, Sheep
INTRODUCTION
The family Chlamydiaceae contains obligate intra-cellular Gram-negative bacteria, with 11 confirmed species (C. trachomatis, C. suis, C. psittaci, C. pneumoniae, C. pecorum, C. muridarum, C. gallinacea, C. felis, C. caviae, C. avium and C. abortus) and candidate species (C. ibidis) relating to single genus of Chlamydia (1–4).
C. abortus is associated with enzootic abortion in ewes (EAE) (5). This is the most common infectious reason for abortion and the birth of weak lambs in many sheep-rearing countries of the world. Abortion usually occurs in the last 2 to 3 weeks of pregnancy. Animals that have been infected before pregnancy show no clinical signs of infection, with the organism arriving into a dormant phase. No clinical signs could be observed in the animals until abortion or delivery of very weak lambs. It was found that the abortion percentage in affected flocks is low in the first year and then reaches 30% and 10% in the second and third years, respectively (5). Hidden infections continuing longer than 3 years have also been described (6). Development of Chlamydiae is highly dependent on nutrient supply and the metabolic status of the host cell (7). Although C. pecorum is frequently isolated from the digestive tract of ruminants with no clinical symptoms, it is a causative agent of fertility disorder, conjunctivitis, arthritis, mastitis, and pulmonary inflammation in sheep, goats and cattle (8). While C. psittaci can cause severe flu-like infections in humans, birds develop largely non-specific, and sometimes, fatal intestinal and respiratory symptoms (9). Moreover, the disease affects goats, and to a lesser degree, cattle, horses, pigs and deer, while little is known about the rate of these infections because of lack of epidemiological evidences (10). Although C. pecorum association in small ruminants abortion incidents was formerly described nearly 20 years ago in south of France (11), its role as an etiological agent of abortion is not well-known in humans. C. psittaci comprises a range of Chlamydia with diverse genetic, serological, and host-tropic properties. By DNA-DNA hybridization examination, 14% to 95% homology was reported among C. psittaci strains (11) and less than 70% among mammalian strains, and avian strains of C. psittaci. Moreover, C. abortus strains are widespread among ruminants and have been related to abortion in horses, rabbits, guinea pigs, mice, pigs and humans (12).
In addition to DNA-based techniques (polymerase chain reaction and DNA microarray) and RFLP, various diagnosis techniques, such as direct microscopic inspection, culture in embryonated chicken eggs, or in cell cultures, serological exams for protein detection (complement fixation test (CFT), enzyme-linked immunosorbent assay (ELISA) and immunohisto-chemistry and direct immunofluorescence) could be utilized to recognize Chlamydia and Chlamydia in biological samples (13). Conventional and real-time PCR methods have been implemented using PCR, which amplify conserved regions of the chlamydial outer membrane protein genes ompA, omp1, and omp2, the polymorphic membrane gene pmp, genes, or the intergenic space between the 16S and 23S rRNA genes (14, 15). Several studies on C. abortus in sheep and goats by serology (16) and C. psittaci in pigeons (17) by PCR have been documented in Khuzestan province. Considering suspected Chlamydia abortion (last 2–3 weeks of pregnancy) in ruminants (bovine, ovine and goat) in the 2 mentioned provinces and migration of animals to and from these 2 provinces, the aim of this research was the primary study on the presence of important Chlamydia spp. in aborted ruminants with doubtful signs of Chlamydia abortion.
MATERIALS AND METHODS
Preparation of clinical samples.
A total of 117 aborted fetuses were collected from different herds located in southwest of Iran, where abortion had been observed during the lambing periods from 2014 to 2016. A total of 9 cattle and 100 sheep from Saman and Lordegan in Cheharmahal and Bakhtiari province and 8 sheep from Bagh-e-Malek in Khuzestan province were selected. Sampling was targeted, meaning that only aborted fetuses at the last 2 to 3 weeks of gestation were selected and transferred to the laboratory on ice. Sampling was performed in sterile conditions from liver, spleen, and abomasal contents of aborted fetus. Laborious methods were performed to ensure that tissues were collected from the same anatomical location in each animal. Strict aseptic protocols, including the use of new sets of tools, were used to avoid cross-contamination. The samples were stored in sterile microtubes at −20°C till DNA extraction.
DNA extraction.
Genomic DNA was extracted from the tissue samples using a SinaGen Kit (Sina-Gen, Iran), according to the manufacturer’s instructions. Tissue samples were finely chopped using sterile blades prior to extracting DNA. Genomic DNA extracted from each isolate was quantified using the Nano Drop spectrophotometer and stored in −20°C for the next genomic evaluation.
PCR assay.
Precautions were taken to use sterile reagents and conditions; and contamination of reactions by PCR product was avoided by strict separation of working areas. The optimal PCR conditions for C. abortus, C. psittaci and C. pecorum individual amplification were initially determined separately using serial dilutions of respective DNA solution. The PCR reactions were performed in a final volume of 25 μL containing 12.5 microliter of master mix 2× (Ampliquen, Denmark) containing 1× PCR buffer, 200 μM of 4 deoxynucleoside triphosphate (dNTPs), 2 mM MgCl2, and 0.5 U of Taq polymerase, then, 0.5 μM of each primer set and 2 microliter of extracted DNA were added to each reaction. PCR reactions were performed in an Eppendorf thermocycler (Eppendorf, Germany). Thermal conditions for amplification of Chlamydiales specific gene were initial denaturation for 5 minutes at 95°C, 39 one-minute cycles at 94°C, 45 seconds at an annealing temperature of 54°C, and elongation for 45 seconds at 72°C, with a final extension step at 72°C for 5 minutes. The PCR products were subjected to electrophoresis for 1 hour at 70V in 1.5% safe stain containing aga-rose gel, and the results were visualized and photographed under ultraviolet illumination. Detection of C. pecorum infection of samples was conducted by Nested-PCR. The name, sequence and the predicted amplified fragment of studied genes, as well as the annealing temperature are listed in Table 1. The standard strain DNA of C. abortus S26/3 and C. pecorum W73, obtained from Professor Borel (University of Zurich) as a gift, and C. psittaci 6BC, as obtained from Professor Sarryopoglu (University of Turkey) as gift, were used as positive controls for each round of PCR (18–20).
Table 1.
Primers Used to Detect Chlamydia Bacterium in Aborted Fetus
| Gene | Sequences | Segment (bp) | Ref. |
|---|---|---|---|
| Chlamydiales (16s–23s spacer region) | F: 5-CAAGGTGAGGCTGATGAC-3 | 352 | (18) |
| R: 5-TCGCCTKTCAATGCCAAG-3 | |||
| C. abortus (16srRNA) | F: 5′-TGG TAT TCTTGC CGA TGA C-3′ | 479 | (19) |
| R: 5′-GAT CGT AAC TGC TTA ATA AAC CG-3′ | |||
| C. psittaci (pmp gene) | F: 5′-ATG AAA CAT CCA GTC TAC TGG-3′ | 300 | (13) |
| R: 5′-TTG TGT AGT AAT ATT ATC AAA-3′ | |||
| C. pecorum (momp) | F: 5-GCICTITGGGAATGCGGITGCGCIAC-3 | 576–597 | (20) |
| R: 5-TTAGAAICGGAATTGIGCATTIACGTGIGCICG-3 | |||
| F: 5-CCAATACGCACAATCGAAACCTCGC-3 | 426–441 | ||
| R:5-CCACAAAATTTTCTAGACTTCAACTTGTTAAT-3 |
RESULTS
The samples were tested by conventional PCR to identify specific 16S rRNA and pmp genes of C. abortus and C. psittaci, respectively. As expected, PCR amplification of DNA for C. abortus produced 222bp fragment and produced 300 bp fragments for C. psittaci. The annealing temperature of 54°C and 48°C were used for these PCR experiments, respectively (Figs. 1, 3).
Fig 1.
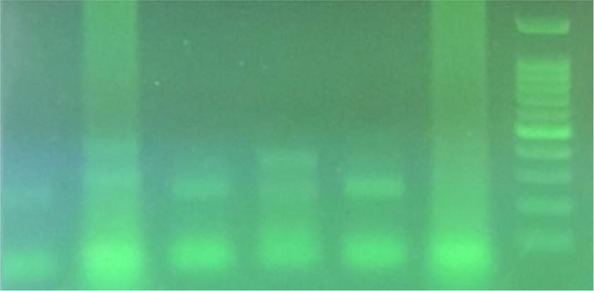
Result of PCR using C. psittaci specific primers: right to left: 100 bp DNA ladder; negative control; positive control (300 bp); 4 samples
Fig. 3.
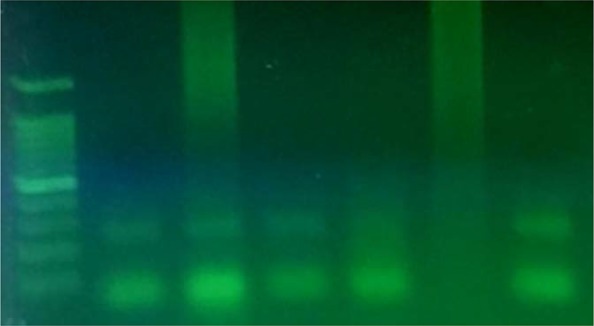
Agarose gel electrophoresis of C. abortus S26/3 species-specific conventional PCR: left to right: 100 bp DNA ladder; 3 positive samples; 1 negative sample; negative control; positive control (222bp).
Out of 117 doubtful chlamydial clinical samples taken from the infected animals (108 sheep and 9 cow), 66 (56.41 %) samples were detected by either one of the 3 pathogens. A total of 24 (36.36%) and 24 (36.36%) sheep samples were positive for C. abortus and C. psittasi, respectively. In this study, only 1 (1.5%) C. pecorum was identified from cattle by producing a 576–597 and 426–441 bp fragment using Nested-PCR. Annealing temperatures used in the first and second stages were 52°C and 50°C, respectively (Fig. 2).
Fig. 2.

Agarose gel electrophoresis of C. pecorum species-specific Nested-PCR: left to right: 100 bp DNA ladder; seven negative samples; positive control (426–441 bp); negative control (distilled water).
The specificity of the PCR experiments using these primers were checked on genomic DNA samples from unrelated bacteria. None of the DNA samples from non-chlamydial bacteria created a measurable PCR bands in these experiments. No PCR product was produced using water instead of target DNA. The results are demonstrated in Table 2.
Table 2.
Results of Chlamydial Infection of the Studied Samples
| Total of samples | Positive number of Chlamydiales order | Cp. abortus | Cp. psittasi | Cp. pecorum | Coinfection (Cl. abortus+Cl. psittasi) | Coinfection (Cl. abortus+Cl. psittasi + Cl. pecorum) |
|---|---|---|---|---|---|---|
| 117 | 66 | 24 | 24 | 1 | 20 | 0 |
| (9 cattle +108 sheep) | (56.41%) | (36.36%) | (36.36%) | (1.5%) | (30.30%) | (0%) |
DISCUSSION
Chlamydiaceae family is considered as one of the main bacterium related to abortion in ruminants, such as sheep, goats, and cattle (21). Abortion is economically important in many herds of sheep and goats in Europe, North America, Africa, and Iran. The bacteria causes premature birth, reproductive disorders in ruminants, inflammation of the epididymis, pneumonia, arthritis, and conjunctivitis in the feces of healthy sheep and goats (22); also, it is a zoonotic risk for numerous pregnant women. It is reported that C. abortus can be spread in human placenta (9). This bacterial family is remarkably important. Thus, many studies have been conducted to identify and recognize these bacteria. For example, the prevalence of infection with this bacterium was reported to be 8.9% in a serological study by ELISA in sheep of Ahvaz, Iran (16). In house ELISA kit, based on rPOMP-90-3, 4 and 3+4 antigens were designed by Bakhtiari et al. to prevent available cross-reaction between C. abortus and C. pecorum in commercial kits (23). Moreover, in Mahzouniyeh et al. research (2014), C. abortus contamination in Shahr-e-Kord was reported to be 52% using Nested-PCR (24). In 2009, Pantchev et al. detected C. psittaci and C. abortus based on ompA gene from tissue samples using real-time PCR (25). Regular methods, such as bacterial culturing and staining, are slightly more sensitive in detecting Chlamydia bacterium in field samples. However, these methods are uncertain in most situations and are more difficult. The new development of different PCR assays has been described to detect Chlamydia bacterium in samples from the aborted fetuses (26). PCR provides a rapid diagnosis without the need for a culture or identifying species and strains with more similarity. Also, PCR detection is not affected by the lack of viability of the microorganism and is more sensitive than culture in detecting nonfeasible organisms and cellular DNA. Previous results have revealed that the PCR amplification of 16S rRNA genes is a good target for identifying Chlamydia spp. (15). Although there are different sets of primers that allow the identification of all species of the Chlamydiaceae family, PCR assays that amplify segments of the 16S rRNA genes present high sensitivity and specificity (27). Based on present results, C. abortus, C. psittaci and C. pecorum can be differentiated by PCR products obtained with species-specific primers to 16S rRNA, pmp and momp gene. The specificity of those primers allows the differentiation of C. abortus and C. pecorum using a conventional PCR. The fact that a considerable proportion of sheep samples (20 of 57 positive samples) were contaminated with 2 chlamydial agents is in line with previous study. The clinical features of abortion caused by C. abortus and C. psittaci are highly similar and such mixed infections have been proposed to be a common incidence in sheep and goat herds (28). Investigation of a large panel of diagnostic samples revealed an interesting epidemiological aspect, which was the occurrence of 2 chlamydial species in 1 sample. This was in agreement with previous findings (29) that reported the same species in pigs suffering from respiratory symptoms or fertility problems. Moreover, infections caused by C. suis, C. abortus, C. pecorum and C. psittaci were reported (30, 31). The existing data suggest that the sheep seem to be a host mainly susceptible to co-infections. In the present study, combinations of C. abortus and C. psittasi (35.08%) were regularly identified in sheep samples. A certain preference of C. abortus and C. psittasi to perform in concert with another chlamydial agent has already been reported (32). How does a bacterium that causes systemic disease in birds transform into an organism of mammalian abortion? The response will offer important visions into the mechanisms of chlamydial virulence and can finally be answered by genome sequence comparison. Until then, our capability to differentiate C. psittaci and C. abortus will remain to rely on ecological alterations, mAbs, and genetic data (16S or 23S rRNA signature sequences), and ompA, cysteine-rich proteins (27, 33, 34). In this study, PCR-amplification of momp gene, using specie-specific primers by nested-PCR, identified C. pecorum strain in cattle. Another study revealed that C. pecorum was more widespread in cattle than C. abortus and that the bacteria were frequently detected in vaginal swabs and fecal samples (35). Earlier data on C. pecorum involvement in abortion in Tunisia and Morocco indicated that C. pecorum may cause abortion in small ruminants in North African countries. Several studies have indicated that C. pecorum can also be a possible reason of abortion in ewes and goats (36). Clinically unclear intestinal infections produced by C. pecorum have already been reported in both abortion-affected and unaffected ruminant flocks (37). Also, the mixed infection of C. pecorum with C. abortus related to abortion in water buffalo in south of Italy (38) suggests that C. pecorum could also be associated with abortion in large ruminants. Consequently, differentiating the 2 species in abortion material is highly necessary. Nevertheless, it is still unknown whether C. pecorum-related abortion is a consequence of C. pecorum alone or is due to a development of its pathogenesis mediated by the coinfection with C. abortus; its pathogenicity may be related to a lack of nutrients or parasitic invasions, which frequently occur in these countries. It could also be considered that no pathogenic C. pecorum strains might be spread from the intestine through the blood circulation and reach the placenta, where they cause abortion due to some unidentified physiopathologic situations. The presence of 1 C. pecorum among 66 samples included in our study suggests that abortion by C. pecorum is rare in the region. Also, migration of the flocks toward Baghe-Malek and Shahr-e-Kord during winter and summer can cause co-contamination and simultaneous infection in these areas. Thus, as co-infections are not rare events, the combination of various specific diagnostic tests is crucial for epidemiological studies.
REFERENCES
- 1.Kuo CC, Stephens RS, Bavoil PM, Kaltenboeck B. Genus I. Chlamydia. In: Krieg N, Staley J, Brown D, Hedlund B, Paster B, Ward W, et al. editors. Bergey’s Manual of Systematic Bacteriology. 2nd ed vol 4. New York, USA: Springer-Verlag; 2011. pp. 846–865. [Google Scholar]
- 2.Sachse K, Laroucau K, Riege K, Wehner S, Dilcher M, Creasy HH, et al. Evidence for the existence of two new members of the family Chlamydiaceae and proposal of Chlamydia avium sp. nov. and Chlamydia gallinacea sp. nov. Syst Appl Microbiol 2014; 37:79–88. [DOI] [PubMed] [Google Scholar]
- 3.Vorimore F, Hsia RC, Huot-Creasy H, Bastian S, Deruyter L, Passet A, et al. Isolation of a new Chlamydia species from the feral Sacred Ibis (Threskiornis aethiopicus): Chlamydia ibidis. PLoS One 2013; 8(9): e74823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siarkou VI, Vorimore F, Vicari N, Magnino S, Rodolakis A, Pannekoek Y, et al. Diversification and distribution of ruminant Chlamydia abortus clones assessed by MLST and MLVA. PLoS One 2015; 10: e0126433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Victoria S, Alexandros FL, Sofia Ch, Kotsis A, Papadopoulos O, et al. Subspecies variation in Greek strains of Chlamydophila abortus. Vet Microbiol 2002; 85: 145–157. [DOI] [PubMed] [Google Scholar]
- 6.Schiller I, Koesters R, Weilenmann R, Thoma R, Kaltenboeck B, Heitz P, et al. Mixed infections with porcine Chlamydia trachomatis/pecorum and infections with ruminant Chlamydia psittaci serovar 1 associated with abortions in swine. Vet Microbiol 1997; 58: 251–260. [DOI] [PubMed] [Google Scholar]
- 7.Wang C, Gao D, Kaltenboeck B. Acute Chlamydia pneumonia re-infection accelerates the development of insulin resistance and diabetes in obese C57BL6 mice. J Infect Dis 2009; 200: 279–287. [DOI] [PubMed] [Google Scholar]
- 8.Reinhold P, Jaeger J, Liebler-Teneorio E, Berndt A, Bachmann R, Schubert E, et al. Imapct of latent infections with Chlamydophila species in young cattle. Vet J 2008; 175: 202–211. [DOI] [PubMed] [Google Scholar]
- 9.Buxton D, Anderson IE, Longbottom D, Livingstone M, Wattegedera S, Entrican G. Ovine chlamydial abortion: characterization of the inflammatory immune response in placental tissues. J Comp Pathol 2002;127:133–141. [DOI] [PubMed] [Google Scholar]
- 10.Longbottom D, Coulter LJ. Animal Chlamydioses and zoonotic implications. J Comp Pathol 2003;128:217–244. [DOI] [PubMed] [Google Scholar]
- 11.Rodolakis A, Salinas J, Papp J. Recent advances on ovine chlamydial abortion. Vet Res 1998; 29: 275–288. [PubMed] [Google Scholar]
- 12.Fukushi H, Hirai K. Genetic diversity of avian and mammalian Chlamydia psittaci strains and relation to host origin. J Bacteriol 1989; 171: 2850–2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laroucau K, Trichereau A, Vorimore F, Mahé AM. A pmp genes based PCR as a valuable tool for the diagnosis of avian Chlamydiosis. Vet Microbiol 2007; 121: 150–157. [DOI] [PubMed] [Google Scholar]
- 14.Everett KD. Chlamydial and Chlamydiales: more than meet the eye. Vet Microbiol 2000; 75: 109–126. [DOI] [PubMed] [Google Scholar]
- 15.Madico G, Quinn TC, Boman J, Gaydos CA. Touch-down enzyme time release-PCR for detection and identification of Chlamydia trachomatis, Chlamydia pneumoniae and Chlamydia psittaci: using the 16S-23S spacer rRNA genes. J Clin Microbiol 2000;38:1085–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghorbanpoor M, Goraninejad D, Heydari R. Serological study on enzootic abortion of ewes in Ahvaz, Iran. Anim Vet Adv 2007; 6: 1194–1196. [Google Scholar]
- 17.Ghorbanpoor M, Moori Bakhtiari N, Mayahi M, Hana Moridveisi. Detection of Chlamydophila psittaci from pigeons by polymerase chain reaction in Ahvaz. Iran J Microbiol 2015; 7: 18–22. [PMC free article] [PubMed] [Google Scholar]
- 18.Sachse K, Laroucau K, Vorimore F, Magnino S, Feige J, Müller W, et al. DNA microarray-based genotyping of Chlamydophila psittaci strains from culture and clinical samples. Vet Microbiol 2009; 135: 22–30. [DOI] [PubMed] [Google Scholar]
- 19.Longbottom D, Fairley S, Chapman T, Psarrou E, Vretou E, Livingstone M. Serological diagnosis of ovine enzootic abortion by enzyme-linked immunosorbent assay with a recombinant protein fragment of the Polymorphic outer membrane protein POMP90 of Chlamydophila abortus. J Clin Microbiol 2002; 40(11): 4235–4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sachse K, Hotzel H. Detection and differentiation of Chlamydiae by Nested PCR. Detection of microbial pathogens. Methods Mol Biol 2003; 216:123–136. [DOI] [PubMed] [Google Scholar]
- 21.DeGraves FJ, Gao D, Hehnen HR, Schlapp T, Kaltenboeck B. Quantitative Detection of Chlamydia psittaci and C. pecorum by high-sensitivity real-time PCR reveals high prevalence of vaginal infection in Cattle. J Clin Microbiol 2003; 41: 1726–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Creelan JL, McCullough SJ. Evaluation of strain-speciec primer sequences from an abortifacient strain of ovine Chlamydophila abortus (Chlamydia psittaci) for the detection of EAE by PCR. FEMS Microbiol Lett 2000; 190(1):103–108. [DOI] [PubMed] [Google Scholar]
- 23.Moori Bakhtiari N, Seifi M, Ghorbanpour M, Gooraninejad S. Cloning and expression segment of the POMP90 gene Chlamydophila abortus strain S26? 3 in E. coli. Iran Vet J 2011; 7: 74–80. [Google Scholar]
- 24.Mahzouniyeh M, Golboui daghdari SH, Pourahmad R. Detection of Chlamydophila abortus abortions in sheep in the Chaharmahal-va-Bakhtiyari province, using Nested PCR. Vet J 2014; 2: 80–74. [Google Scholar]
- 25.Pantchev A, Sting R, Bauerfeind R. New real-time PCR tests for species-specific detection of Chlamydophila psittaci and Chlamydophila abortus from tissue samples. Vet J 2009; 181: 145–150. [DOI] [PubMed] [Google Scholar]
- 26.Pelletier C, Chartier S, Berthillier J, Spohr H, Carvalho Lima BAD, Negrão FJ, et al. Validation of an internal method for the diagnosis of infections with Chlamydophila abortus and Coxiella burnetii by real-time multiplex PCR. Dev Biol (Basel) 2006; 126: 219–226. [PubMed] [Google Scholar]
- 27.Meijer A, Kwakke GJ, de Vries A, Schouls LM, Ossewaarde JM. Species identification of Chlamydia isolates by analyzing restriction fragment length polymorphism of the 16S-23S rRNA spacer region. J Clin Microbiol 1997; 35:1179–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aitken ID, Clarkson MJ, Linklater K. Enzootic abortion of ewes. Vet Rec 1990; 126: 136–138. [DOI] [PubMed] [Google Scholar]
- 29.Hoelzle LE, Steinhausen G, Wittenbrink MM. PCR-based detection of chlamydial infection in swine and subsequent PCR-coupled genotyping of chlamydial ompA-Gene amplicons by DNA-hybridization, RFLP analysis, and nucleotide sequence analysis. Epidemiol Infect 2000; 125: 427–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Teankum K, Pospischil A, Janett F, Burgi E, Brugnera E, Hoelzle K, et al. Detection of chlamydiae in boar semen and genital tracts. Vet Microbiol 2006; 116(1–3):149–157. [DOI] [PubMed] [Google Scholar]
- 31.Kauffold J, Melzer F, Henning K, Schulze K, Leiding C, Sachse K. Prevalence of chlamydiae in boars and semen used for artificial insemination. Theriogenology 2006; 65: 1750–1758. [DOI] [PubMed] [Google Scholar]
- 32.Pantchev A, Sting R, Bauerfeind R, Tyczka J, Sachse K. Detection of all Chlamydophila and Chlamydia spp. of veterinary interest using species-specific real-time PCR assays. Comp Immunol Microbiol Infect Dis 2010; 33: 473–484. [DOI] [PubMed] [Google Scholar]
- 33.Herrmann B, Pettersson B, Everett KD, Mikkelsen NE, Kirsebom LA. Characterization of the rnpB gene and the RNase P RNA in the order Chlamydiales. Int J Syst Evol Microbiol 2000; 50 Pt 1:149–158. [DOI] [PubMed] [Google Scholar]
- 34.Bush RM, Everett KDE. Molecular evolution of the Chlamydiaceae. Int J Syst Evol Microbiol 2001; 51(Pt 1):203–220. [DOI] [PubMed] [Google Scholar]
- 35.Berri M, Rekiki A, Boumedine KS, Rodolakis A. Simultaneous differential detection of Chlamydophila abortus, Chlamydophila pecorum and Coxiella burentii from aborted ruminant’s clinical samples using Multiplex PCR. BMC Microbiol 2009; 9: 130–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rekiki A, Bouakane A, Hammami S, El Idrissi AH, Bernard F, Rodolakis A. Efficacy of live Chlamydophila abortus vaccine 1B in protecting mice placentas and foetuses against strains of Chlamydophila pecorum isolated from cases of abortion. Vet Microbiol 2004; 99:295–299. [DOI] [PubMed] [Google Scholar]
- 37.Kaltenboeck B, Hehnen HR, Vaglenov A. Bovine Chlamydophila spp. Infection: Do we underestimate the impact on fertility? Vet Res Commun 2005; 29 Suppl 1:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Greco G, Corrente M, Buonavoglia D, Campanile G, Di Palo R, Martella V, et al. Epizootic abortion related to infections by Chlamydophila abortus and Chlamydophila pecorum in water buffalo (Bubalus bubalis). Theriogenology 2008; 69: 1061–1069. [DOI] [PubMed] [Google Scholar]


