ABSTRACT
The c-Jun gene encodes a transcription factor that has been implicated in many physiological and pathological processes. c-Jun is a highly unstable protein that is degraded through a ubiquitination/proteasome-dependent mechanism. However, the deubiquitinating enzyme (DUB) that regulates the stability of the c-Jun protein requires further investigation. Here, by screening a DUB expression library, we identified ubiquitin-specific protease 6 (USP6) and showed that it regulates the stability of the c-Jun protein in a manner depending on its enzyme activity. USP6 interacts with c-Jun and antagonizes its ubiquitination. USP6 overexpression upregulates the activity of the downstream signaling pathway mediated by c-Jun/AP-1 and promotes cell invasion. Moreover, many aberrant genes that are upregulated in USP6 translocated nodular fasciitis are great potential targets regulated by c-Jun. Based on our data, USP6 is an enzyme that deubiquitinates c-Jun and regulates its downstream cellular functions.
KEYWORDS: USP6, c-Jun, deubiquitinating enzyme, deubiquitination, nodular fasciitis
INTRODUCTION
The AP-1 complex is a dimeric complex that regulates many cellular processes, including cell proliferation, death, survival, and differentiation. The AP-1 complex comprises members of the JUN, FOS, ATF, and MAF protein families (1). These proteins belong to the basic leucine zipper (bZIP) proteins, which form heterodimers and homodimers through their leucine zipper motifs (2). Among these families, c-Jun is one of most extensively studied proteins.
c-Jun is the proto-oncogene from which v-Jun is derived (3). c-Jun has important functions in embryonic development, inflammation, and malignant transformation (4, 5). Therefore, the expression and activity of c-Jun should be tightly regulated. c-Jun is regulated at the transcriptional, posttranscriptional, translational, and posttranslational levels (2). The posttranslational modifications of c-Jun include phosphorylation, ubiquitination, and acetylation (6–8). Notably, the c-Jun protein is rapidly turned over under normal conditions (9). Based on accumulating evidence, the stability of the c-Jun protein is mainly regulated by a ubiquitination/proteasome-dependent mechanism (10, 11). c-Jun is ubiquitinated by several E3 ubiquitin (Ub) ligases, including F-box and WD repeat domain containing 7 (FBXW7), the E3 ubiquitin-protein ligase RFWD2, cullin 4 (CUL4), itchy E3 ubiquitin protein ligase (ITCH), mitogen-activated protein kinase kinase kinase 1 (MEKK1), and RING-box protein 2 (RNF7) (7, 12–15). However, further investigations are required to determine whether deubiquitinating enzymes (DUB) participate in the regulation of c-Jun stability.
The ubiquitin-specific protease 6 gene (USP6), also known as TRE2 or TRE17, was originally shown to be expressed in a wide variety of human cancer cells but not in human cells from normal tissues (16). Interestingly, USP6 is also expressed at high levels in aneurysmal bone cysts (ABCs) and nodular fasciitis (NF) (17, 18). USP6 is translocated in nearly 70% of ABCs and 90% of NF cases, leading to its transcriptional upregulation (18, 19). Therefore, USP6 has been proposed as a marker of and potential therapeutic target in these diseases. Two downstream effectors of USP6 have recently been discovered (20, 21). However, further studies are required to determine whether other USP6 substrates exist.
We screened an expression library of 68 deubiquitinating enzymes to identify the deubiquitinating enzymes that regulate c-Jun stability. USP6 regulated the stability of the c-Jun protein in a manner dependent on its enzyme activity. USP6 interacts with c-Jun and removes the ubiquitin chain from c-Jun. USP6 regulates signaling downstream of the c-Jun/AP-1 complex and promotes cell migration and invasion. Furthermore, c-Jun is one of the most potent transcription factors that account for the aberrant upregulation of gene expression in USP6 translocation nodular fasciitis disease. Based on our findings, USP6 deubiquitinates c-Jun and regulates its downstream cellular functions.
RESULTS
USP6 regulates c-Jun stability.
Consistent with the results from previous studies (9, 22), the c-Jun protein is rather unstable in many cells, such as MCF7 breast cancer cells and HeLa cervical cancer cells. Pretreatment with the proteasome inhibitor MG132 significantly increased the level of the c-Jun protein (data not shown), indicating that c-Jun underwent ubiquitination-dependent degradation in these cells. We individually transfected 68 DUBs into MCF7 cells and detected the levels of the c-Jun protein to identify the DUBs that regulate the stability of the c-Jun protein (data not shown). Among these DUBs, USP6 significantly increased the level of the c-Jun protein in MCF7 and HeLa cells (Fig. 1A). In contrast, USP6 overexpression only slightly increased the c-Jun mRNA level (Fig. 1B). We also generated USP6 short hairpin RNA (shRNA) expression plasmids. As shown in Fig. 1C and D, USP6 shRNAs significantly reduced both endogenous and exogenous USP6 expression levels. Moreover, knockdown of USP6 reduced the protein levels of c-Jun (Fig. 1E). A similar result was observed in U2OS cells (data not shown). To further determine whether USP6 regulated c-Jun stability, HeLa cells were cotransfected with c-Jun and USP6 expression plasmids and then treated with cycloheximide (CHX). As shown in Fig. 1F, USP6 overexpression enhanced the stability of the cotransfected c-Jun protein. The deubiquitinating enzyme USP28, a binding partner of the E3 ubiquitin ligase FBXW7, regulates the stability of protein substrates of FBXW7, such as c-Myc and c-Jun (23, 24). Therefore, we tested whether USP28 regulates the c-Jun protein level in MCF7 and HeLa cells. As shown in Fig. 1G, USP6, but not USP28, significantly increased the c-Jun protein levels in both MCF7 and HeLa cells. In contrast, USP28 upregulated the c-Myc protein level in HeLa cells (Fig. 1H). Thus, USP6 regulates the stability of the c-Jun protein.
FIG 1.
USP6 regulates the stability of the c-Jun protein. (A) An empty vector or USP6 expression plasmid was transfected into MCF7 or HeLa cells. Thirty-six hours later, the cells were harvested. Whole-cell lysates were subjected to SDS-PAGE and analyzed by immunoblotting (IB) with the indicated antibodies. (B) RNA levels of c-Jun in MCF7 cells transfected with the empty vector and a plasmid expressing USP6. (C) HeLa cells were infected with lentivirus that carried shRNAs targeted to GFP or USP6. Thirty-six hours later, RNA levels of USP6 were determined using RT-PCR. (D) A USP6 expression plasmid was cotransfected with shGFP (shRNA targeted to GFP) or USP6 shRNA plasmids into HeLa cells. Forty-eight hours later, the cells were harvested, subjected to SDS-PAGE, and analyzed by immunoblotting with the indicated antibodies. (E) Cell extracts of HeLa-shGFP or HeLa-USP6 knockdown stable cell lines were harvested and subjected to Western blotting to examine the levels of the indicated proteins. (F) A c-Jun–HA expression plasmid was cotransfected with the empty vector or a USP6 expression plasmid into HeLa cells. Twenty-four hours later, the cells were treated with CHX for the indicated times. Then, cell extracts were subjected to Western blotting to examine the levels of the indicated proteins. The intensity of c-Jun–HA expression for each time point was quantified by densitometry and plotted. The experiment was repeated three times, and a representative experiment is presented. (G) The indicated plasmids were transfected into MCF7 or HeLa cells. Twenty-four hours later, the cells were harvested, subjected to SDS-PAGE, and analyzed by immunoblotting with the indicated antibodies. (H) HeLa cells were transfected with the indicated plasmids; 24 h later, whole-cell lysates were subjected to SDS-PAGE and analyzed by immunoblotting with the indicated antibodies. All experiments were performed at least 3 times. (B, C, and F) The data are presented as means ± standard errors of the mean (SEM) for three triplicate samples and were analyzed by t test. *, P < 0.05.
USP6 regulates the stability of the c-Jun protein in an enzyme activity-dependent manner.
Because USP6 is a deubiquitinating enzyme, we generated an enzyme activity-deficient mutant USP6, C541S (25), in which cysteine-541 is mutated to serine. The enzyme activity of USP6 was required for its ability to regulate c-Jun protein levels in HeLa cells (Fig. 2A). Similar results were observed in MCF7 cells (Fig. 2B). Consistent with this finding, the USP enzyme domain of USP6 (USP6 amino acids [aa] 301 to 1406), but not its TBC domain (USP6 aa 1 to 350), also increased c-Jun protein levels (Fig. 2C). Then, we tested whether a naturally occurring C-terminally truncated isoform of USP6 (USP6-S) that lacks USP activity influenced the c-Jun protein level. As shown in Fig. 2D, USP6, but not USP6-S, increased c-Jun protein levels. Based on these results, the enzyme activity of USP6 is required for its function.
FIG 2.
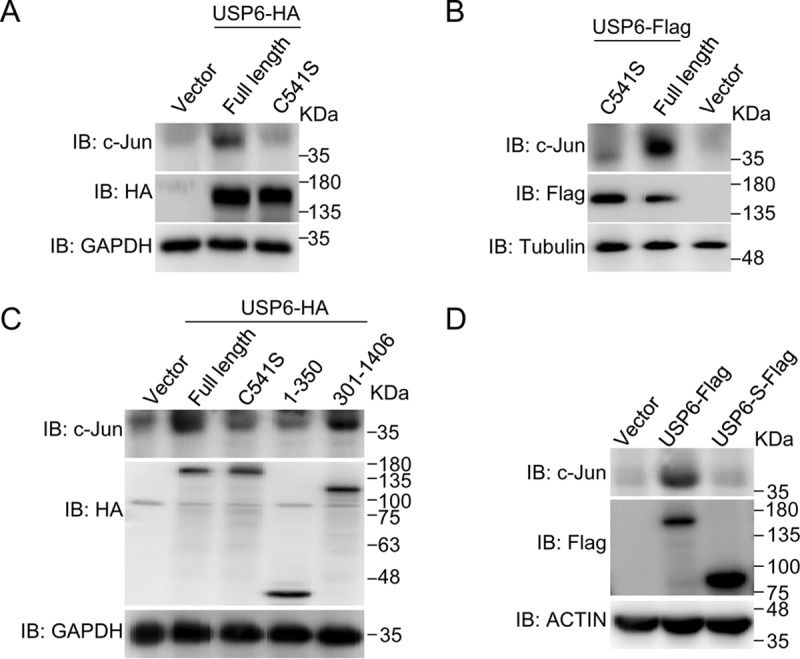
The enzyme activity of USP6 is important for its ability to regulate the stability of the c-Jun protein. (A) An empty vector or a plasmid expressing USP6 or USP6 C541S was transfected into HeLa cells, and the cells were harvested 36 h later. Whole-cell lysates were subjected to SDS-PAGE and analyzed by immunoblotting with the indicated antibodies. (B) The same assay as in panel A, except that the plasmids were transfected into MCF7 cells. (C) The same assay as in panel A, except that the empty vector or USP6-expressing plasmid was transfected into HeLa cells. (D) The indicated plasmids were transfected into MCF7 cells, and the cells were harvested 24 h later. Whole-cell lysates were subjected to SDS-PAGE and analyzed by immunoblotting with the indicated antibodies. All experiments were performed at least 3 times.
USP6 interacts with c-Jun and promotes c-Jun deubiquitination.
We tested whether USP6 interacts with c-Jun to investigate the mechanism by which USP6 regulates the stability of the c-Jun protein. As shown in Fig. 3A and B, USP6 interacts with c-Jun in cotransfected 293T cells in both directions. As c-Jun degradation depends on its ubiquitination, we next investigated whether USP6 influenced c-Jun ubiquitination. As shown in Fig. 3C, USP6, but not the C541S enzyme activity-deficient mutant, deubiquitinated c-Jun. To test whether USP6 is capable of directly deubiquitinating c-Jun in vitro, ubiquitinated c-Jun was immunopurified from 293T cells coexpressing hemagglutinin (HA)–c-Jun and Flag-Ub using anti-HA antibody. The immunoprecipitates were incubated with recombinant USP6, and ubiquitination of c-Jun was detected by anti-Flag immunoblotting. The recombinant catalytic domain of USP6 deubiquitinated c-Jun, while the USP6 C541S mutant and control glutathione S-transferase (GST) protein did not (Fig. 3D). Thus, USP6 regulates the stability of the c-Jun protein through deubiquitination.
FIG 3.
USP6 interacts with c-Jun and mediates c-Jun deubiquitination. (A) 293T cells were transfected with the indicated plasmids. The cell lysates were immunoprecipitated with an anti-Flag antibody. The lysates (TCL) and immunoprecipitates were immunoblotted. (B) 293T cells were transfected with the indicated plasmids. The cell lysates were immunoprecipitated with an anti-HA antibody. The lysates and immunoprecipitates were immunoblotted. (C) The empty vector or a plasmid expressing USP6 or USP6 C541S was cotransfected with c-Jun–HA and Flag-ubiquitin expression plasmids into 293T cells. The cells were treated with MG132 (10 μM) for 4 h, and then the cells were lysed in RIPA buffer and the cell lysates were immunoprecipitated with anti-HA antibody. The lysates and immunoprecipitates were immunoblotted with the indicated antibodies. (D) 293T cells were cotransfected with HA–c-Jun and Flag-Ub. After 4 h of MG132 treatment, c-Jun was purified using anti-HA antibody and then incubated with GST or GST-tagged recombinant USP6 or USP6-C541S. c-Jun ubiquitylation was detected by immunoblotting with anti-FLAG. All experiments were performed three times.
USP6 regulates signaling downstream of the AP-1 complex through c-Jun.
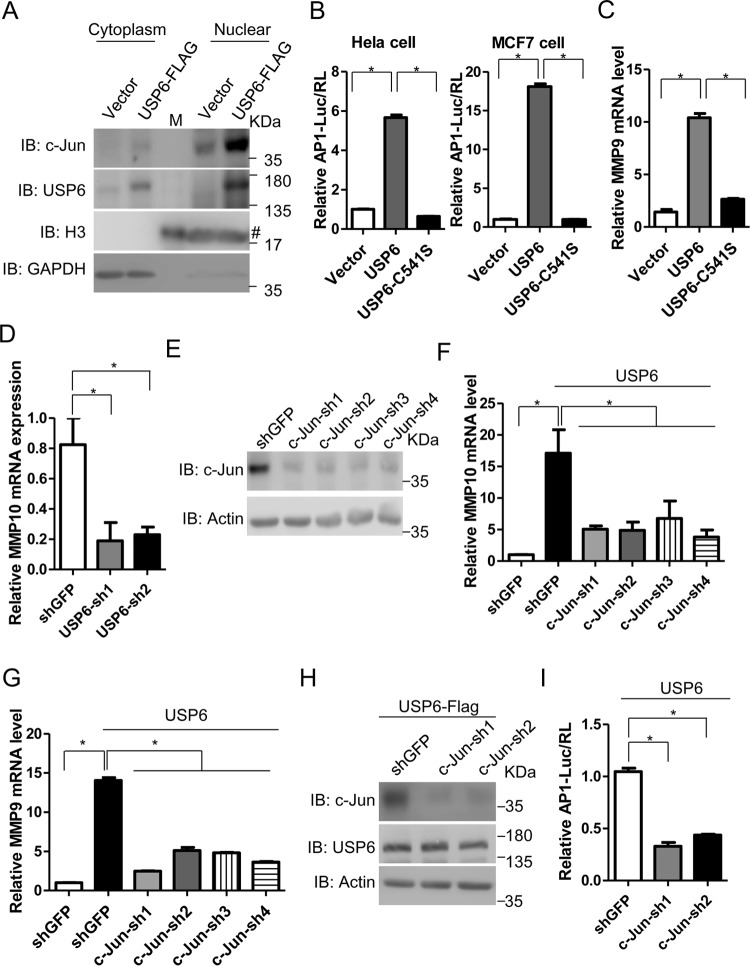
The nuclear localization of c-Jun is important for its function. We found that c-Jun in the nuclear fraction was difficult to detect when diluted to a volume equal to that of the cytoplasmic fraction. Therefore, the volumes of nuclear fraction samples we prepared were about 1/8 of those of cytoplasmic fraction samples. As shown in Fig. 4A, USP6 significantly increased the levels of the c-Jun protein in the nuclear fraction compared to the control sample. As one of the important components of the AP-1 complex, c-Jun regulates many downstream effectors. We investigated whether USP6 regulated the transcriptional activity of the AP-1 complex. As shown in Fig. 4B, USP6 increased AP-1 reporter activation, whereas the enzyme activity-deficient mutant of USP6 abolished this activation. Matrix metalloproteinases (MMPs) are important downstream effectors of USP6 (25). Consistent with previous reports, USP6, but not the USP6 C541S mutant, increased the RNA levels of MMP9 and MMP10 (Fig. 4C and data not shown). In contrast, knockdown of USP6 downregulated the expression of MMP10 (Fig. 4D). In order to evaluate the roles of c-Jun in the USP6-induced upregulation of MMP expression and AP-1 activity, we generated c-Jun shRNA plasmids and found that these shRNAs significantly knocked down c-Jun protein levels (Fig. 4E). Then, we generated c-Jun knockdown stable cell lines using c-Jun shRNAs. As shown in Fig. 4F and G, USP6-induced MMP10 and MMP9 expression was abolished in the c-Jun knockdown cell lines. Moreover, c-Jun shRNAs also attenuated c-Jun protein levels and AP-1 activity induced by USP6 (Fig. 4H and I). Thus, USP6-mediated regulation of AP-1 downstream signaling depends on USP6 enzyme activity and c-Jun.
FIG 4.
USP6 regulates c-Jun downstream signaling. (A) Cells transfected with the empty vector or USP6 expression plasmid were harvested, and nuclear and cytoplasmic fractions were generated as described in Materials and Methods. In order to detect c-Jun protein, the nuclear fraction was not diluted as much as the cytoplasmic fraction. The final volumes of the nuclear fraction samples were about 1/8 those of the cytoplasmic fraction samples. M, protein marker; #, the histone 3 antibody or the goat anti-rabbit secondary antibody might cross-react with marker proteins (lane 3). (B) Luciferase activity of HeLa or MCF7 cells transfected with AP-1 luciferase reporter and a Renilla luciferase plasmid, together with the empty vector or a plasmid expressing USP6 or USP6 C541S. (C) Levels of MMP9 mRNA in HeLa cells transfected with the empty vector or a plasmid expressing USP6 or USP6 C541S determined using RT-PCR. (D) HeLa cells were infected with lentivirus carrying shGFP or USP6 shRNAs. Forty-eight hours later, the RNA levels of MMP10 were determined using RT-PCR. (E) HeLa cells were infected with lentivirus carrying shGFP or c-Jun shRNAs. Forty-eight hours later, the cells were harvested, subjected to SDS-PAGE, and analyzed by immunoblotting with the indicated antibodies. (F) The empty vector or a plasmid expressing USP6 was transfected into MCF7-shGFP or c-Jun knockdown stable cell lines. The RNA levels of MMP10 were determined using RT-PCR. (G) Levels of the MMP9 mRNAs in MCF7-shGFP and c-Jun knockdown stable cell lines transfected with the empty vector or a plasmid expressing USP6 were determined using RT-PCR. (H) A plasmid expressing USP6-Flag was transfected into MCF7-shGFP or c-Jun knockdown stable cell lines; 36 h later, the cells were harvested, subjected to SDS-PAGE, and analyzed by immunoblotting with the indicated antibodies. (I) A plasmid expressing USP6 or an AP-1 luciferase reporter and a Renilla luciferase plasmid were transfected into MCF7-shGFP or c-Jun knockdown stable cell lines, and 24 h later, luciferase activities were determined. All experiments were performed at least 3 times. (B to D and F to I) The data are presented as means and SEM for three triplicate samples and were analyzed by t test. *, P < 0.05.
USP6 promotes cell migration and invasion.
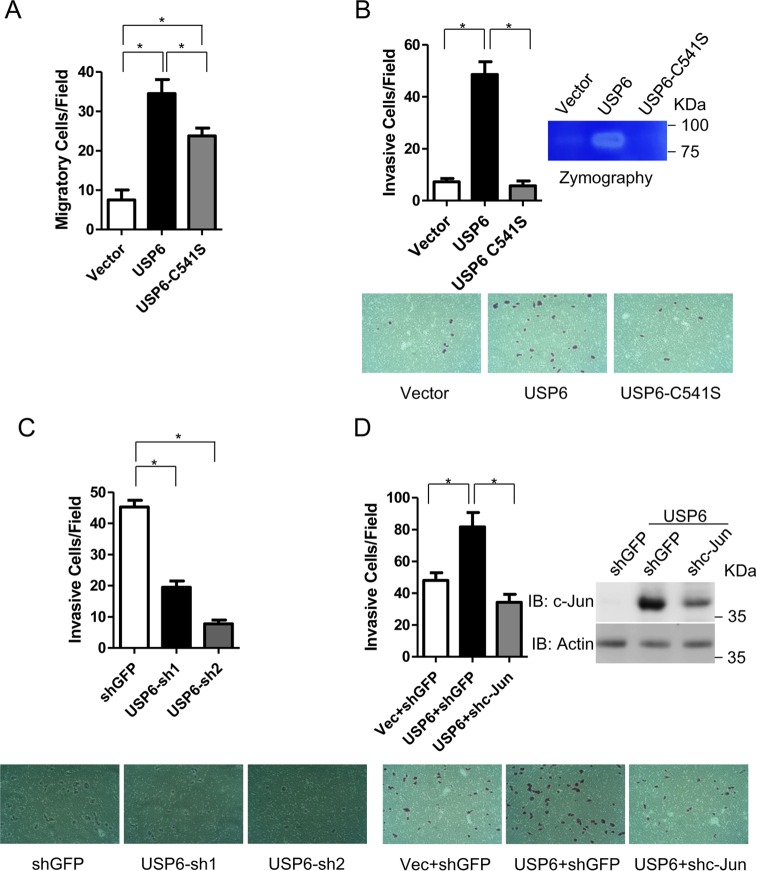
As USP6 has been implicated in various human tumors, we decided to investigate the role of USP6 in cell migration and invasion. We generated USP6 correlate stable cell lines. As shown in Fig. 5A, overexpression of USP6 significantly enhanced the migratory activity of HeLa cells. Interestingly, the enzymatic USP6 mutant C541S also enhanced the migratory activity. In contrast, USP6, but not USP6 C541S, promoted cell invasion (Fig. 5B). Consistently, a zymography assay revealed that USP6 significantly enhanced the gelatinolytic activity of HeLa cells. Moreover, knockdown of USP6 inhibited invasion of HeLa cells (Fig. 5C). Then, we tested whether c-Jun is involved in USP6-dependent cell invasion. As shown in Fig. 5D, c-Jun knockdown abolished USP6-dependent cell invasion.
FIG 5.
USP6 promotes cell migration and invasion. (A) HeLa cells were infected with the indicated lentivirus to generate stable cell lines, and 2 × 104 HeLa cells were seeded into the upper chamber of each Transwell. Twenty-four hours later, the cells were subjected to transwell assays to measure cell migration. (B) Approximately 2 × 104 of the indicated HeLa cells were seeded into the upper chamber of a Transwell containing Matrigel. Twenty-four hours later, the cells were subjected to an in vitro invasion assay. A zymography assay was performed to determine MMP activity as described in Materials and Methods. (C) HeLa cells were infected with lentivirus carrying shGFP or USP6 shRNAs to generate stable cell lines, and 2 × 104 of the HeLa cells were seeded into the upper chamber of a Transwell containing Matrigel. Thirty-six hours later, the cells were subjected to an in vitro invasion assay. (D) The indicated HeLa stable cell lines were infected with lentivirus carrying shGFP or c-Jun shRNAs; 48 h later, approximately 2 × 104 of the HeLa cells were seeded into the upper chamber of a Transwell containing Matrigel. Thirty-six hours later, the cells were subjected to an in vitro invasion assay. All experiments were performed 3 times. In all panels data are presented as means and SEM for three triplicate samples and were analyzed by t test. *, P < 0.05.
c-Jun is potentially involved in USP6 translocation nodular fasciitis disease.
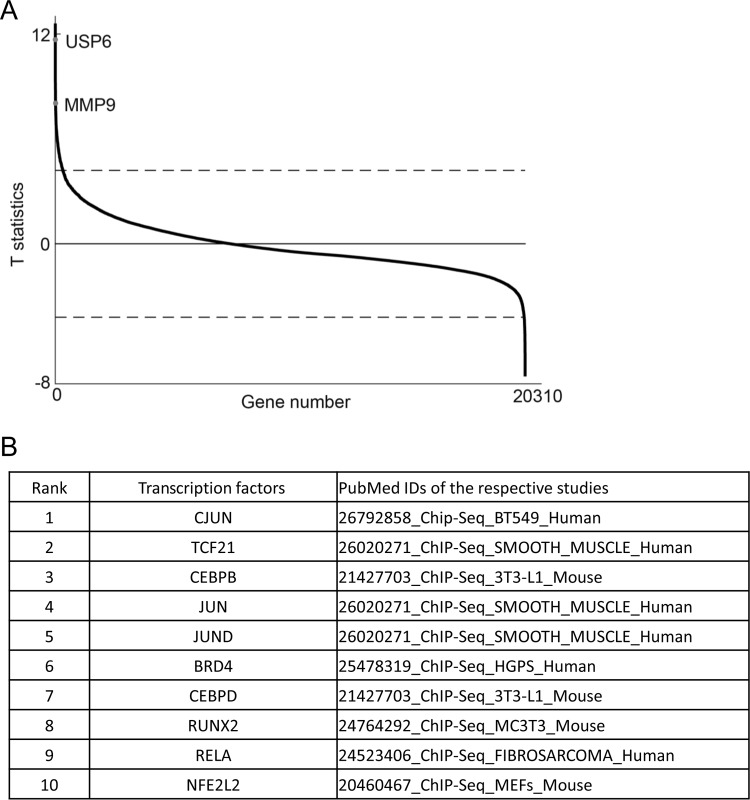
USP6 has been demonstrated to have important roles in NF and ABCs, in which the expression of USP6 is upregulated due to gene translocation (18). Using nodular fasciitis transcriptome data from a Gene Expression Omnibus (GEO) database, we sought to identify genes that were upregulated in NF samples. As shown in Table S1 in the supplemental material, more than 300 genes were upregulated in NF samples. Among these genes, USP6 was one of most significant (ranked no. 2) (Fig. 6A; see Table S1 in the supplemental material), consistent with previous reports showing that the translocation of USP6 drives its overexpression in NF (18). We performed an enrichment analysis of the ChIP-X Enrichment Analysis (ChEA) gene set library (26) using Enrichr (27, 28) to characterize the putative transcription factors that control the aberrant expression of these genes. Among the transcription factors in the database, c-Jun is ranked as one of the most potent transcription factors involved in regulating the expression of these genes (Fig. 6B; see Table S2 in the supplemental material).
FIG 6.
c-Jun is potentially involved in USP6 translocation nodular fasciitis disease. (A) The T statistics (y axis) of all the genes measured (x axis) in the data set were plotted. The dashed lines represent the threshold (FDR < 0.01) used to identify DE genes. DE genes above or below the dashed lines were upregulated or downregulated in the nodular fasciitis group compared with the “other tumors” group. The points representing two genes, USP6 and MMP9, are labeled. T statistics reflect the degree of expression difference of a gene between two groups of samples. The larger the absolute value of a gene's T statistic is, the greater the expression difference is. (B) Chromatin immunoprecipitation (ChIP) enrichment analysis of aberrantly upregulated genes. The aberrantly upregulated genes in a USP6 translocation nodular fasciitis sample from the ChEA 2016 gene set library with FDR values of <0.01 were analyzed using Enrichr and the combined-score ranking method. The top 10 ranked (by P value) transcription factors are shown (see Tables S1 and S2 in the supplemental material).
DISCUSSION
Using a deubiquitinating enzyme expression library, we showed that USP6 regulates the stability of the c-Jun protein. USP6 interacts with and promotes the deubiquitination of c-Jun. Therefore, USP6 stabilized the c-Jun protein and potentiated c-Jun-dependent downstream signaling and cell invasion. Based on our analysis of nodular fasciitis transcriptome data, the interaction between USP6 and c-Jun might be important in nodular fasciitis disease.
Posttranslational modifications are important regulators of c-Jun function. The stability of the c-Jun protein is tightly regulated in a ubiquitination/proteasome-dependent manner. Several E3 ligases were reported to promote the ubiquitination and degradation of c-Jun. In contrast, the deubiquitinating enzyme(s) of c-Jun required further investigation. Our work identified USP6 as an enzyme that deubiquitinates c-Jun. By regulating the stability of the c-Jun protein, USP6 promoted c-Jun-dependent downstream signaling and cell invasion. We found that USP6, but not USP28, increased c-Jun protein levels in MCF7 and HeLa cells in our study. USP28, a binding partner of the E3 ubiquitin ligase FBXW7, regulates the stability of protein substrates of FBXW7. It is possible that in our systems c-Jun was mainly ubiquitinated by other E3 ligases instead of FBXW7. Our studies have improved our knowledge about the diverse mechanisms regulating c-Jun.
USP6 translocation frequently occurs in ABC and NF. Therefore, studies identifying the downstream effectors of USP6 will improve our knowledge of and ability to cure these diseases. Actually, two novel genes, the Frizzled and JAK1 genes, were recently identified as USP6 substrates (20, 21). Using a DUB library, we identified the transcription factor c-Jun as a novel substrate of USP6. USP6 regulates the stability of the c-Jun protein and its downstream effectors in an enzyme activity-dependent manner. According to the results of mechanistic studies, USP6 interacts with c-Jun and mediates c-Jun deubiquitination. Furthermore, by analyzing genes that are upregulated in USP6 translocation NF samples, we identified many genes that are predicted targets of c-Jun. Thus, c-Jun is a novel substrate of USP6 and might play important roles in NF.
Our studies revealed a novel link between USP6 and c-Jun. As USP6 has been shown to have important roles in ABC and NF diseases, inhibition of c-Jun and other USP6-related genes, such as NF-κB, JAK1, and Frizzled genes, would benefit people who suffer from these diseases. Meanwhile, considering the important roles of c-Jun in cancer, inflammation, and development, the contributions of USP6 to these pathological and physiological processes still require further investigation.
MATERIALS AND METHODS
Cell culture.
HEK293T, MCF7, and HeLa cell lines were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, 4 mM l-glutamine, 100 IU penicillin, and 100 mg/ml streptomycin at 37°C in a humidified incubator containing 5% CO2.
Reagents.
Mouse anti-HA (F-7), mouse anti-GAPDH (anti-glyceraldehyde-3-phosphate dehydrogenase) (6C5), and mouse anti-β-actin (C4) antibodies were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). The mouse anti-Flag antibody (M2), USP6 antibody, and protease inhibitor cocktail were obtained from Sigma (Saint Louis, MO). The c-Jun antibody (D155181) was obtained from Sangon Biotech. Histone 3 (17168-1-AP), tubulin (11224-1-AP), c-Myc (10828-1-AP), and ubiquitin (10201-2-AP) antibodies were obtained from the Proteintech Company. The Dual-Luciferase reagent was obtained from Promega. TRIzol reagent was obtained from Invitrogen. Top Green quantitative PCR (qPCR) SuperMix was obtained from Transgen Biotech.
Immunoprecipitation.
Protein interactions were assessed using immunoprecipitation, as previously described (29). Briefly, cells were washed once with cold phosphate-buffered saline (PBS) and lysed in cold lysis buffer containing protease inhibitors. The supernatants of the cell lysates were isolated by centrifugation at 20,000 × g for 30 min at 4°C. Immunoprecipitation was performed using anti-Flag M2 beads or anti-HA beads as previously described (29). For the ubiquitin immunoprecipitation assay, cells were lysed in radioimmunoprecipitation assay (RIPA) buffer containing protease inhibitors. The anti-HA beads were washed with RIPA buffer. The other steps were similar to those in typical immunoprecipitation assays of protein interactions.
Plasmids.
The gene expression clones and shRNA clones were constructed as previously described (29). The genes of interest were amplified by PCR using specific primers and cloned into the BamHI and SmaI sites of the pLV-EF1α-MCS-IRES-Puro vector (Biosettia, San Diego, CA) or the BamHI and XhoI sites of the pBOBE vector using the ExoIII-assisted ligase-free cloning method, as described previously (30). For lentivirus-based shRNA expression vectors, DNA oligonucleotides containing shRNA sequences were designed and cloned into the pLV-H1-EF1α-puro expression vector using the single-oligonucleotide RNA interference (RNAi) technology developed by Biosettia, as described previously (31). All lentiviral shRNA vectors were constructed according to the manufacturer's protocol. cDNA sequence encoding aa 500 to 1406 of USP6 and its enzymatic-activity mutant was subcloned into pGEX-4T1 to produce a GST recombinant USP6 (aa 500 to 1406) or USP6 C541S (aa 500 to 1406) proteins.
RNA extraction and real-time PCR analysis.
RNA extractions and real-time PCR analyses were performed as previously described (29). Total RNA was isolated from cells using TRIzol reagent, according to the manufacturer's instructions. Approximately 1 μg of each RNA sample was used to prepare cDNAs by reverse transcription using oligo(dT)15 primers. RNA expression levels were normalized to an internal control, actin. Real-time PCR (RT-PCR) was performed in a Stratagene Mx3000P system (Agilent Technologies).
Fractionation of nuclear and cytoplasmic extracts.
Approximately 3 × 106 cells were collected to generate nuclear and cytoplasmic fractions. Cells were isolated after one wash with PBS, scraped into 800 μl of nuclear extract buffer (NEB; 0.01 M Tris-HCl, pH 7.4, 0.01 M NaCl, 0.003 M MgCl2, 0.03 M sucrose, and 0.5% NP-40), and then transferred to 1.5-ml Eppendorf tubes. The tubes were immediately rotated at 4°C for 10 min and then centrifuged at 1,500 × g for 10 min at 4°C. The supernatants were collected as the cytoplasmic fraction. The pellets were washed 3 times with 1 ml of NEB buffer to obtain the nuclear proteins. The pellets were collected as the nuclear fraction. GAPDH and histone 3 were used as markers of the cytoplasm and nucleus, respectively.
Determination of MMP activity.
Determination of MMP activity generally followed the gelatin zymography protocol presented on the Abcam website. Cells were transfected with the indicated plasmids. Thirty-six hours later, the cells were starved in serum-free medium for 36 h prior to harvesting. Conditioned media were collected and centrifuged to eliminate dead cells. Then, the conditioned media were concentrated using an Amicon Ultra-0.5 centrifugal filter. The conditioned-medium samples were fractionated on 7.5% gels containing 4 mg/ml gelatin (Sigma). The gels were washed with washing buffer (2.5% Triton X-100, 50 mM Tris-HCl, 5 mM CaCl2, 1 μM ZnCl2, pH 7.5), incubated in incubation buffer (1% Triton X-100, 50 mM Tris-HCl, 5 mM CaCl2, 1 μM ZnCl2, pH 7.5) at 37°C for 36 h, and then stained with Coomassie brilliant blue.
Ubiquitylation assays.
To determine c-Jun ubiquitylation in vivo, 293T cells were cotransfected with c-Jun–HA and Flag-Ub, with or without USP6 plasmids. After lysis with RIPA buffer, c-Jun was immunoprecipitated with anti-HA beads. The immunoprecipitated c-Jun–HA was subjected to SDS-PAGE, and the immunoblots were probed with anti-Flag antibody. For in vitro deubiquitination assays, 293T cells were cotransfected with c-Jun–HA and Flag-Ub. The cells were treated with MG132 for 4 h before being harvested. Cell lysates were immunoprecipitated with anti-HA beads and washed with lysis buffer. Then, the samples were incubated with GST, GST-USP6 (aa 500 to 1406), or GST-USP6 C541S (aa 500 to 1406) for 4 h at 37°C. Samples were washed with lysis buffer and then immunoblotted with Flag.
Cell migration and invasion assay.
Cell invasion was determined using 24-well-formatted Transwells (migration) or 24-well-formatted Matrigel-coated Transwells (invasion), according to the manufacturer's instructions. Briefly, 2 × 104 HeLa cells in 200 μl of serum-free medium were added to the upper chamber of each Transwell and allowed to migrate or invade for 24 or 36 h. Cells in the upper chambers were removed with cotton swabs, and cells that had invaded the lower surface of the chamber were stained with crystal violet. The numbers of migratory or invasive cells were determined by counting the stained cells under a microscope.
Data sources and preprocessing.
The three data sets (GSE52252, GSE78991, and GSE18229) used in this study were downloaded from GEO (http://www.ncbi.nlm.nih.gov/geo/) (21, 32–34). Differentially expressed (DE) genes were identified and compared among 9 nodular fasciitis tumors with USP6 translocation in data set GSE78991 and 27 other tumors in data set GSE52252. Detailed information about the samples is provided in a study by L. Quick et al. (21). Student's t test was used to select DE genes between two groups of samples from data sets GSE78991 and GSE52252. The P values were adjusted using the Benjamini and Hochberg procedure to control the false-discovery rate (FDR). Gene expression was considered significantly different between two groups when the FDR was <0.01.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by grants from the National Natural Science Foundation of China (grants no. 31501120, 81602738, and 81602498); Joint Funds for the Innovation of Science and Technology, Fujian Province (grant no. 2016Y9040); the Natural Science Foundation of Fujian Province (grants no. 2015J05065 and 2015J05157); the Health and Family Planning Commission of Fujian Province (grants no. 2016-ZQN-65, 2014-1-38, and 2016-1-40); the Educational Scientific Research Project of Young Teachers of Fujian Province (grants no. JA14131, JA14140, and JAT160216); the Outstanding Youth Scientific Research Personnel Training Program at Fujian Province University (grant no. 2016B023); the Scientific Research Project of Young Teachers of Fujian Medical University (grants no. 2014MP002 and 2014MP001); and the Setting Sail Foundation of Fujian Medical University (grant no. 2016QH006).
We declare that we have no conflicts of interest with the contents of this article.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00320-17.
REFERENCES
- 1.Angel P, Karin M. 1991. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim Biophys Acta 1072:129–157. [DOI] [PubMed] [Google Scholar]
- 2.Shaulian E, Karin M. 2002. AP-1 as a regulator of cell life and death. Nat Cell Biol 4:E131–E136. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- 3.Vogt PK. 2002. Fortuitous convergences: the beginnings of JUN. Nat Rev Cancer 2:465–469. doi: 10.1038/nrc818. [DOI] [PubMed] [Google Scholar]
- 4.Mechta-Grigoriou F, Gerald D, Yaniv M. 2001. The mammalian Jun proteins: redundancy and specificity. Oncogene 20:2378–2389. doi: 10.1038/sj.onc.1204381. [DOI] [PubMed] [Google Scholar]
- 5.Johnson RS, van Lingen B, Papaioannou VE, Spiegelman BM. 1993. A null mutation at the c-jun locus causes embryonic lethality and retarded cell growth in culture. Genes Dev 7:1309–1317. doi: 10.1101/gad.7.7b.1309. [DOI] [PubMed] [Google Scholar]
- 6.Meng Q, Xia Y. 2011. c-Jun, at the crossroad of the signaling network. Protein Cell 2:889–898. doi: 10.1007/s13238-011-1113-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nateri AS, Riera-Sans L, Da Costa C, Behrens A. 2004. The ubiquitin ligase SCFFbw7 antagonizes apoptotic JNK signaling. Science 303:1374–1378. doi: 10.1126/science.1092880. [DOI] [PubMed] [Google Scholar]
- 8.Vries RG, Prudenziati M, Zwartjes C, Verlaan M, Kalkhoven E, Zantema A. 2001. A specific lysine in c-Jun is required for transcriptional repression by E1A and is acetylated by p300. EMBO J 20:6095–6103. doi: 10.1093/emboj/20.21.6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jariel-Encontre I, Salvat C, Steff AM, Pariat M, Acquaviva C, Furstoss O, Piechaczyk M. 1997. Complex mechanisms for c-fos and c-jun degradation. Mol Biol Rep 24:51–56. doi: 10.1023/A:1006804723722. [DOI] [PubMed] [Google Scholar]
- 10.Treier M, Staszewski LM, Bohmann D. 1994. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell 78:787–798. doi: 10.1016/S0092-8674(94)90502-9. [DOI] [PubMed] [Google Scholar]
- 11.Westermarck J. 2010. Regulation of transcription factor function by targeted protein degradation: an overview focusing on p53, c-Myc, and c-Jun. Methods Mol Biol 647:31–36. doi: 10.1007/978-1-60761-738-9_2. [DOI] [PubMed] [Google Scholar]
- 12.Gao M, Labuda T, Xia Y, Gallagher E, Fang D, Liu YC, Karin M. 2004. Jun turnover is controlled through JNK-dependent phosphorylation of the E3 ligase Itch. Science 306:271–275. doi: 10.1126/science.1099414. [DOI] [PubMed] [Google Scholar]
- 13.Wertz IE, O'Rourke KM, Zhang Z, Dornan D, Arnott D, Deshaies RJ, Dixit VM. 2004. Human De-etiolated-1 regulates c-Jun by assembling a CUL4A ubiquitin ligase. Science 303:1371–1374. doi: 10.1126/science.1093549. [DOI] [PubMed] [Google Scholar]
- 14.Xia Y, Wang J, Xu S, Johnson GL, Hunter T, Lu Z. 2007. MEKK1 mediates the ubiquitination and degradation of c-Jun in response to osmotic stress. Mol Cell Biol 27:510–517. doi: 10.1128/MCB.01355-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu Q, Bowden GT, Normolle D, Sun Y. 2007. SAG/ROC2 E3 ligase regulates skin carcinogenesis by stage-dependent targeting of c-Jun/AP1 and IkappaB-alpha/NF-kappaB. J Cell Biol 178:1009–1023. doi: 10.1083/jcb.200612067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakamura T, Hillova J, Mariage-Samson R, Onno M, Huebner K, Cannizzaro LA, Boghosian-Sell L, Croce CM, Hill M. 1992. A novel transcriptional unit of the tre oncogene widely expressed in human cancer cells. Oncogene 7:733–741. [PubMed] [Google Scholar]
- 17.Oliveira AM, Hsi BL, Weremowicz S, Rosenberg AE, Dal Cin P, Joseph N, Bridge JA, Perez-Atayde AR, Fletcher JA. 2004. USP6 (Tre2) fusion oncogenes in aneurysmal bone cyst. Cancer Res 64:1920–1923. doi: 10.1158/0008-5472.CAN-03-2827. [DOI] [PubMed] [Google Scholar]
- 18.Erickson-Johnson MR, Chou MM, Evers BR, Roth CW, Seys AR, Jin L, Ye Y, Lau AW, Wang X, Oliveira AM. 2011. Nodular fasciitis: a novel model of transient neoplasia induced by MYH9-USP6 gene fusion. Lab Invest 91:1427–1433. doi: 10.1038/labinvest.2011.118. [DOI] [PubMed] [Google Scholar]
- 19.Oliveira AM, Perez-Atayde AR, Dal Cin P, Gebhardt MC, Chen CJ, Neff JR, Demetri GD, Rosenberg AE, Bridge JA, Fletcher JA. 2005. Aneurysmal bone cyst variant translocations upregulate USP6 transcription by promoter swapping with the ZNF9, COL1A1, TRAP150, and OMD genes. Oncogene 24:3419–3426. doi: 10.1038/sj.onc.1208506. [DOI] [PubMed] [Google Scholar]
- 20.Madan B, Walker MP, Young R, Quick L, Orgel KA, Ryan M, Gupta P, Henrich IC, Ferrer M, Marine S, Roberts BS, Arthur WT, Berndt JD, Oliveira AM, Moon RT, Virshup DM, Chou MM, Major MB. 2016. USP6 oncogene promotes Wnt signaling by deubiquitylating Frizzleds. Proc Natl Acad Sci U S A 113:E2945–E2954. doi: 10.1073/pnas.1605691113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quick L, Young R, Henrich IC, Wang X, Asmann YW, Oliveira AM, Chou MM. 2016. Jak1-STAT3 signals are essential effectors of the USP6/TRE17 oncogene in tumorigenesis. Cancer Res 76:5337–5347. doi: 10.1158/0008-5472.CAN-15-2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shao J, Teng Y, Padia R, Hong S, Noh H, Xie X, Mumm JS, Dong Z, Ding HF, Cowell J, Kim J, Han J, Huang S. 2013. COP1 and GSK3beta cooperate to promote c-Jun degradation and inhibit breast cancer cell tumorigenesis. Neoplasia 15:1075–1085. doi: 10.1593/neo.13966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Popov N, Wanzel M, Madiredjo M, Zhang D, Beijersbergen R, Bernards R, Moll R, Elledge SJ, Eilers M. 2007. The ubiquitin-specific protease USP28 is required for MYC stability. Nat Cell Biol 9:765–774. doi: 10.1038/ncb1601. [DOI] [PubMed] [Google Scholar]
- 24.Diefenbacher ME, Popov N, Blake SM, Schulein-Volk C, Nye E, Spencer-Dene B, Jaenicke LA, Eilers M, Behrens A. 2014. The deubiquitinase USP28 controls intestinal homeostasis and promotes colorectal cancer. J Clin Invest 124:3407–3418. doi: 10.1172/JCI73733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ye Y, Pringle LM, Lau AW, Riquelme DN, Wang H, Jiang T, Lev D, Welman A, Blobel GA, Oliveira AM, Chou MM. 2010. TRE17/USP6 oncogene translocated in aneurysmal bone cyst induces matrix metalloproteinase production via activation of NF-kappaB. Oncogene 29:3619–3629. doi: 10.1038/onc.2010.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lachmann A, Xu H, Krishnan J, Berger SI, Mazloom AR, Ma'ayan A. 2010. ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics 26:2438–2444. doi: 10.1093/bioinformatics/btq466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma'ayan A. 2013. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, McDermott MG, Monteiro CD, Gundersen GW, Ma'ayan A. 2016. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li L, Chen W, Liang Y, Ma H, Li W, Zhou Z, Li J, Ding Y, Ren J, Lin J, Han F, Wu J, Han J. 2014. The Gbetagamma-Src signaling pathway regulates TNF-induced necroptosis via control of necrosome translocation. Cell Res 24:417–432. doi: 10.1038/cr.2014.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li C, Evans RM. 1997. Ligation independent cloning irrespective of restriction site compatibility. Nucleic Acids Res 25:4165–4166. doi: 10.1093/nar/25.20.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang DW, Shao J, Lin J, Zhang N, Lu BJ, Lin SC, Dong MQ, Han J. 2009. RIP3, an energy metabolism regulator that switches TNF-induced cell death from apoptosis to necrosis. Science 325:332–336. doi: 10.1126/science.1172308. [DOI] [PubMed] [Google Scholar]
- 32.Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, He X, Perou CM. 2010. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res 12:R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edgar R, Domrachev M, Lash AE. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang X, Bledsoe KL, Graham RP, Asmann YW, Viswanatha DS, Lewis JE, Lewis JT, Chou MM, Yaszemski MJ, Jen J, Westendorf JJ, Oliveira AM. 2014. Recurrent PAX3-MAML3 fusion in biphenotypic sinonasal sarcoma. Nat Genet 46:666–668. doi: 10.1038/ng.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.