Abstract
An abnormal acyl-CoA synthetase/stearoyl-CoA desaturase (ACSL/SCD) lipid network fuels colon cancer progression, endowing cells with invasive and migratory properties. Therapies against this metabolic network may be useful to improve clinical outcomes. Because micro-RNAs (miRNAs/miRs) are important epigenetic regulators, we investigated novel miRNAs targeting this pro-tumorigenic axis; hence to be used as therapeutic or prognostic miRNAs. Thirty-one putative common miRNAs were predicted to simultaneously target the three enzymes comprising the ACSL/SCD network. Target validation by quantitative RT-PCR, Western blotting, and luciferase assays showed miR-544a, miR-142, and miR-19b-1 as major regulators of the metabolic axis, ACSL/SCD. Importantly, lower miR-19b-1 expression was associated with a decreased survival rate in colorectal cancer (CRC) patients, accordingly with ACSL/SCD involvement in patient relapse. Finally, miR-19b-1 regulated the pro-tumorigenic axis, ACSL/SCD, being able to inhibit invasion in colon cancer cells. Because its expression correlated with an increased survival rate in CRC patients, we propose miR-19b-1 as a potential noninvasive biomarker of disease-free survival and a promising therapeutic miRNA in CRC.
Keywords: micro-ribonucleic acid, colorectal tumors, patient relapse, invasiveness, molecular biology, lipid and lipoprotein metabolism, fatty acid/metabolism, clinical studies, acyl-CoA synthetase/stearoyl-CoA desaturase
Colorectal cancer (CRC), one of the most commonly diagnosed cancers, has one of the highest prevalence and mortality rates among malignant tumors (1). Energetic metabolism reprogramming is one of the emerging hallmarks of cancer. Adjustments of energy metabolism are triggered to fuel the uncontrolled growth of cancer cells. Lipid metabolism is altered in cancer, among other metabolic pathways (2, 3), and it represents an important source of energy and structural and biosynthetic resources fundamental for carcinogenic processes (3, 4).
Our group identified cooperative lipid metabolism-related genes involved in CRC (5): two members of the acyl-CoA synthetase (ACSL) family, ACSL1 and ACSL4, and stearoyl-CoA desaturase (SCD). ACSL1 and ACSL4 catalyze the conversion of long chain FAs to acyl-CoA (6) and have been related to carcinogenesis (4, 7). SCD, the rate-limiting enzyme converting saturated FAs into monounsaturated FAs, is also related to malignant transformation, proliferation, or survival (8–10). The ACSL/SCD lipid network fuels migratory and invasive properties through epithelial-mesenchymal transition (EMT) induction and is associated with an increased risk of relapse in CRC patients (5).
Micro-RNAs (miRNAs) are endogenous noncoding small RNAs that have recently emerged as potent epigenetic modulators, which specifically bind to complementary regions within mRNAs to promote their degradation and/or to inhibit translation (11, 12). miRNAs regulate cellular homeostasis and development and also several processes involved in tumorigenesis (12–14). The reported cross-talk between miRNAs and several lipid metabolism processes, like lipogenesis, lipolysis, or lipophagy (13), could contribute to their action as tumor suppressors or as oncomiRNAs.
Here, we characterize novel regulators of the whole ACSL/SCD network through miRNA screening: first by using bioinformatics approaches and then by testing the effect of the strongest miRNA candidates on the ACSL/SCD axis and validating their specificity through the action on their 3′UTR. Finally, we evaluated the expression of the most remarkable regulator in stage II and stage III CRC patients.
MATERIALS AND METHODS
Cell culture and reagents
SW620, DLD-1, LoVo, and HEK-293T cell lines were obtained from ATCC, maintained under standard conditions, and Mycoplasma tested every 2 weeks. Three to four passages (2–3 weeks) for each cell line were used for experiments between collection and thawing. The commercial antibodies used are listed in supplemental Table S3. Antibody against SCD (15) was a gentle contribution from Dr. Jean-Baptiste Demoulin, Université Catholique de Louvain, Brussels, Belgium. Anti-human ACSL4 was generously granted by Dr. Stephen Prescott, University of Utah, Salt Lake, UT and Dr. Diana Stafforini, Huntsman Cancer Institute, University of Utah and was used as indicated (16).
Bioinformatics tools
Four different algorithms were used simultaneously in order to identify miRNA-gene interactions: miRanda, PITA, TargetScan, and PicTar5. miRanda (17) and TargetScan (18) use the degree of sequence complementarity as the primary key parameter to identify miRNA-mRNA interactions. PITA (19) utilizes thermodynamics as the main criterion and miRanda has also included this feature. PicTar5 (20) or the mirSVR scoring model (21) collect different types of features derived from the latest in vivo and in vitro assays and utilize machine learning techniques to find the feature patterns shared by true miRNA-target interactions. A prediction was considered valid whenever it co-occurred in at least two algorithms. For miRNA functional analysis, Genecodis 3 (22) determined overrepresented biological processes from Gene Ontology or overrepresented pathways from the Panther database.
To provide a miRNA-mRNA interaction scoring schema, we used the miRanda/mirSVR scoring schema. The miRanda algorithm provides alignment, thermodynamic, and conservation scores and the mirSVR algorithm uses the machine learning-based score (http://www.microrna.org/). Alignment and conservation scores reflect the level of complementarity between sequences and conservation of the target site among vertebrates, respectively. The seed category represents the number of nucleotides interacting at the seed level. An eight seed-category or eight monomer site (8mer site) means an interaction between all nucleotides from two to nine, a 7mer site between all nucleotides from two to eight, and a 6mer site between all nucleotides from two to seven. However, zero represents a mismatch somewhere along the nucleotides two to seven within the seed, so five or fewer nucleotides interact. The mirSVR score (21) detects experimentally verified noncanonical miRNA-mRNA interactions. The higher the probability of downregulation, the lower the mirSVR score is. Cutoff was set at −0.5 because 15% of all the predicted targets had a score lower than 0.5 and, at such a score, the expected probability of observing a minimal log expression change of −0.1 is approximately 50%.
Quantitative real-time PCR and interaction score method generation
Mimic miRNAs (30 nM) were transfected into HEK-293T cells (Lipofectamine® 2000; Thermo Fisher Scientific). Total RNA was extracted 48 h post transfection using Tri Reagent (Sigma) and reverse-transcribed using the High Capacity RNA-to-cDNA Master Mix system (Life Technologies). Quantitative RT-PCR (RT-QPCR) was performed using VeriQuest SYBR Green qPCR Master Mix (Affymetrix, Santa Clara, CA) and gene-specific primers are listed in supplemental Table S1. miRNA expression in patients was monitored using TaqMan® microRNA reverse transcription kit (Thermo Fisher Scientific) and TaqMan miRNA probes for RT-QPCR (supplemental Table S2). The 2−ΔΔCt method was applied to calculate the relative gene or miRNA expression.
Regarding the interaction scoring method, an individual score (IS) was obtained by subtracting the replica’s mean (2−ΔΔCt) for each miRNA-gene interaction by the global mean (2−ΔΔCt average among the 31 miRNAs for each gene). In this way, we selected only the best interactions for each gene. Consequently, the final score (FS) represents the average of the three ISs of each miRNA for the three target genes, ACSL1, ACSL4, and SCD. The FS gets lower as the probability of downregulation increases. Individual 2−ΔΔCt data (fold change), replica mean, global mean, and IS and FS are listed in supplemental Table S5 for each miRNA-gene interaction.
Western blot
Mimic miRNAs (60 nM) were transfected into HEK-293T cells (Lipofectamine® 2000; Thermo Fisher Scientific). Cell lysates were separated on SDS-PAGE, transferred onto nitrocellulose membranes, blocked with 5% nonfat dry milk, and probed with the antibodies listed in supplemental Table S3. Signal detection was performed using the Clarity Western ECL substrate (Bio-Rad). Vinculin determination was used as loading control.
Dual-luciferase assays (cloning and cotransfection)
Sequences for ACSL1 (ENST00000281455), ACSL4 (ENST00000348502), and SCD (ENST00000370355) were cloned into psiCheck2 by DNA 2.0. Fifty thousand HEK-293T cells were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s recommendations. Each cotransfection reaction contained 100 ng of either psiCheck2-3′UTR-ACSL1, psiCheck2-3′UTR-ACSL4, or psiCheck2-3′UTR-SCD and 30 nM of each mimic miRNA. The relative luciferase activity (Renilla luminescence/firefly luminescence) was determined after transfection using the Dual-Luciferase Reporter assay system (Promega) representing the translational repression of ACSL1, ACSL4, and SCD upon binding to their 3′UTR of candidate miRNAs.
Patients and samples
Eighty CRC stage II and 46 CRC stage III patients from La Paz University Hospital were enrolled in the study. Formalin-fixed paraffin-embedded samples were obtained with the patients’ authorization and with the approval of the human research ethics review committee of La Paz University Hospital (HULP-PI-1452). Clinical-histopathological data of patients were prospectively collected on clinical history and were confirmed by oncologists of the hospitals implicated in this study (supplemental Table S4).
Invasion assays
SW620 or LoVo cells were nucleotransfected (Neon® transfection system; Thermo Fisher) with 30 nM mimic miR-19b-1 (miR-19b-1) or a control miRNA. For rescue experiments, miRNAs were cotransfected with 300 ng of vectors expressing the coding sequence of ACSL1, ACSL4, and SCD (DNA 2.0; Menlo Park, CA). Cells were seeded in 0.1% FBS-DMEM using Matrigel invasion chambers (Corning). DMEM 10% FBS was used as chemoattractant. After fixation and staining, images were captured (Olympus CKX41) and registered (getIT software, Olympus).
Neutral lipid content distribution
DLD-1 cells were transfected with Lipofectamine 2000 with mimic miRNAs (30 nM) for 48 h. Oleic acid (OA) conjugated with BSA (0.5 mM) was added the day before the analysis to increase the lipid content. FA-free BSA was used as vehicle for the control group.
Triglyceride analysis.
Cell lipids were extracted with chloroform-methanol (23) and analyzed enzymatically using commercial kits (from Spinreact, Sant Esteve de Bas, Spain). Lipid content was expressed as micrograms of triglyceride (TG) per milligram of protein.
BODIPY staining for microscopy.
Cells were treated with 2 μM BODIPY staining solution (BODIPY™ 493/503; Invitrogen) in PBS for 15 min at 37°C and fixed with 4% paraformaldehyde. Images were captured using a Leica DM IL microscope with a 40× Plan Fluotar objective and registered using Leica Application Suite.
Oxygen consumption rate
The oxygen consumption rate (OCR) was monitored as an indicator of mitochondrial respiration with an XF96 extracellular flux analyzer using the XF Cell Mito stress test kit according to the manufacturer’s instructions (Seahorse Biosciences, North Billerica, MA). Cells were seeded into XF96 plates with DMEM 10% FBS at a density of 90,000 cells per well for SW620 cells, as described in (24), and transfected the following day with mimic miRNAs (30 nM) using Lipofectamine 2000. Upon several washes, medium was replaced for 1 h prior to assay with Krebs-Henseleit buffer (111 mM NaCl, 4.7 mM KCl, 1.25 mM CaCl2, 2 mM MgSO4, and 1.2 mM NaH2PO4) supplemented with 2.5 mM glucose, 0.5 mM carnitine, and 5 mM HEPES, as FA oxidation (FAO) medium. Etomoxir (Sigma), a carnitine palmitoyltransferase-1 (CPT-1) inhibitor and therefore a FAO inhibitor, was added to the medium 15 min prior to assay. Once basal rate measurements were taken, mitochondrial respiratory chain drugs were added, following Mito Stress kit specifications. Oligomycin (2 μM) was used to block ATP-linked oxygen consumption, 0.6 μM FCCP as an uncoupling agent to obtain maximal respiration, and 0.5 μM rotenone/antimycin A to inhibit complex I and complex III, stopping all mitochondrial respiration. The OCR was measured three times following injection of each drug and normalized to total protein content. At least six replicates per condition were done for each experiment.
Statistical analysis
Time to relapse was obtained for the analysis of 3 year disease-free survival (DFS), defined from the time of the surgical procedure. The Kaplan-Meier method was used to estimate DFS. Log-rank test for univariate Cox regression analysis was performed to test association between DFS and individual miRNA expression. Hazard ratios (HRs) and 95% CI were calculated from the adjusted Cox regression model (vascular invasion, lymphatic invasion, and perineural invasion, together with age >70). All reported P values were two-sided. Statistical significance was defined as P < 0·05. Statistical analyses were done by R statistical software v3.1 (https://www.r-project.org).
RESULTS
Bioinformatics prediction of common miRNAs regulating the ACSL1/ACSL4/SCD pro-tumorigenic axis
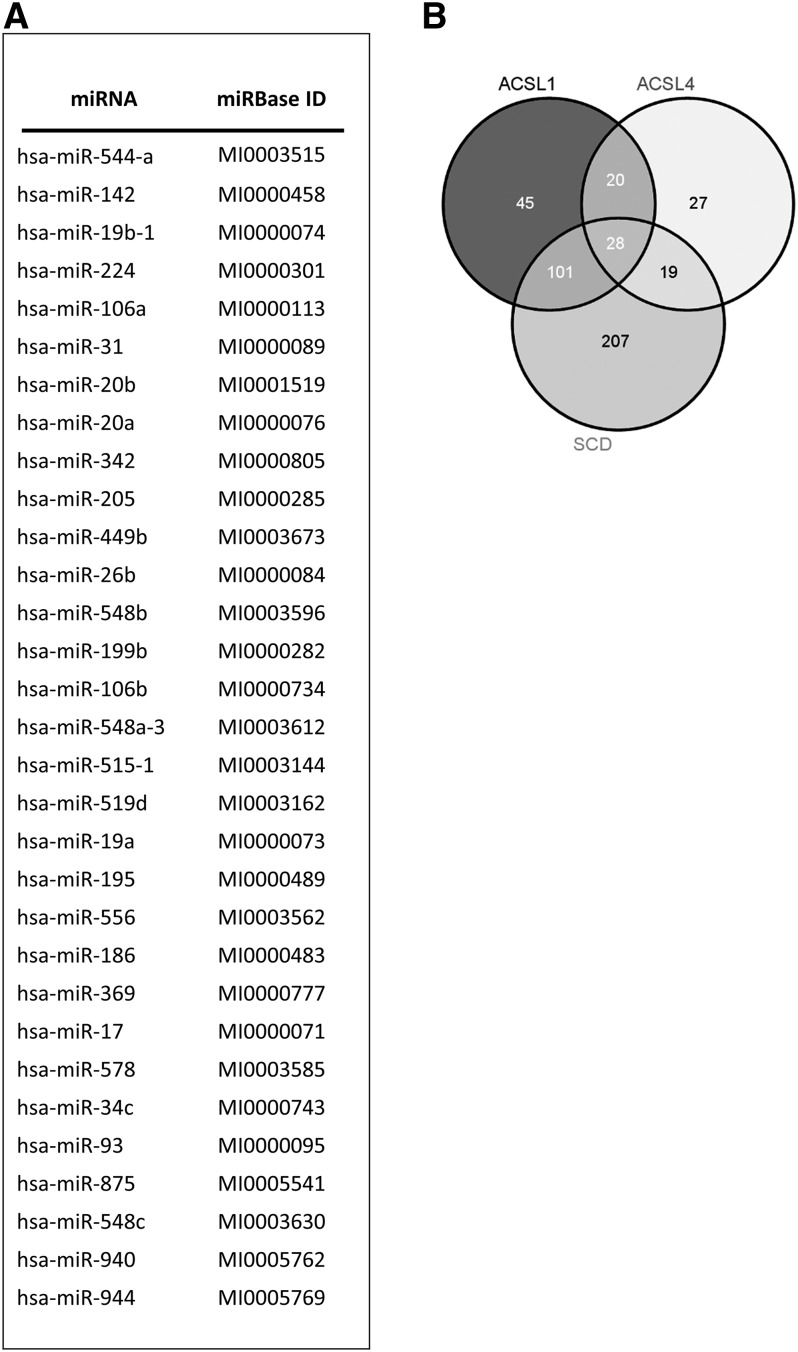
miRanda, PITA, TargetScan, and PICTAR5 were run in order to identify miRNA-gene interactions. A prediction was considered valid whenever it co-occurred in at least two algorithms; we identified 31 putative miRNAs (Fig. 1A) capable of targeting ACSL1, ACSL4, and SCD simultaneously. A more restrictive prediction based on the co-occurrence of at least three algorithms was also performed. A Venn diagram shows the intersection of predictions for 28 common miRNAs of ACSL1/ACSL4/SCD (Fig. 1B). Because the number did not vary significantly, we thus considered the initial 31 candidate miRNAs for further in vitro validation.
Fig. 1.
Bioinformatics prediction of common miRNAs regulating ACSL1, ACSL4, and SCD. A: miRanda, PITA, TargetScan, and PICTAR5 algorithms were run to predict 31 common miRNAs putatively regulating ACSL1, ACSL4, and SCD. A prediction was considered valid whenever co-occurring in at least two algorithms. MiRbase identification (ID) for all the candidates is also shown. B: Venn diagram showing the intersection of predictions for ACSL1, ACSL4, and SCD based on the co-occurrence of at least three predicting algorithms.
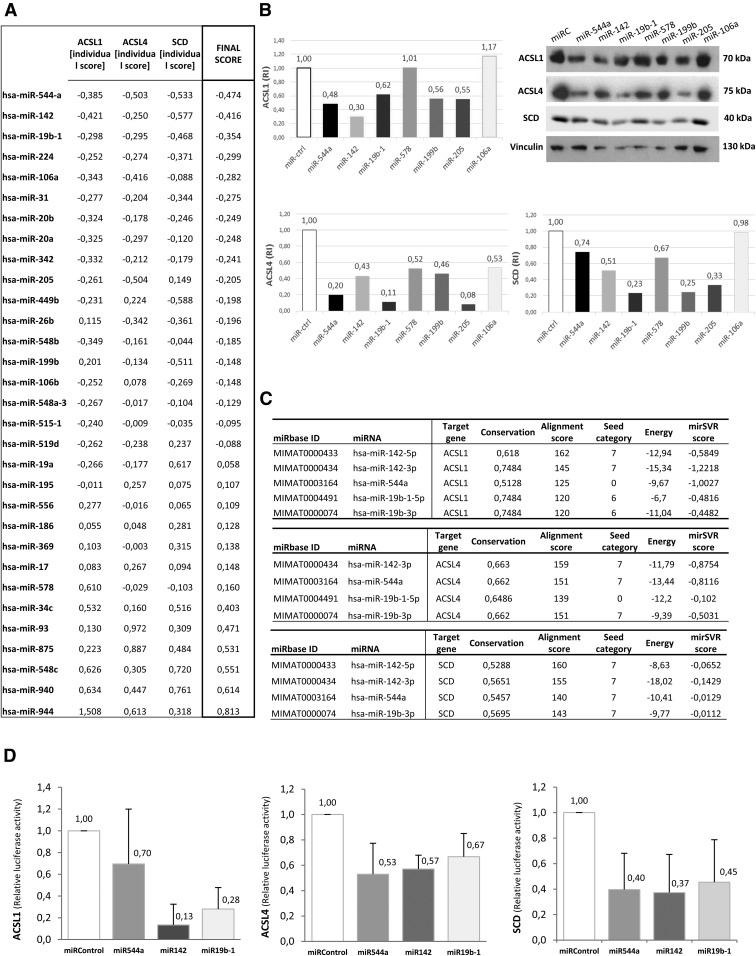
Screening of the predicted miRNAs shows miR-544a, miR-142, and miR-19b-1 as regulators of the ACSL1/ACSL4/SCD network
Because in vitro screening and validation are essential to avoid false predictions, HEK-293T cells were transfected with miRNA mimics and the ACSL1/ACSL4/SCD protein expression levels were measured. We next developed an interaction scoring method based on RT-QPCR data to select stronger miRNA-gene interactions based on better mRNA depletions. Figure 2A shows ISs and FSs from miRNA-gene interactions (the lower the scores are, the higher the target’s depletion is). We defined a FS cutoff of −0.35, which selects miR-544a (FS = −0.474), miR-142 (FS = −0.416), and miR-19b-1 (FS = −0.354) as the best candidates.
Fig. 2.
Validation of the predicted miRNAs shows evidence of three selected miRNAs: miR-142, miR-544a, and miR-19b-1. A: Interaction scoring method. ISs and FSs from miRNA-gene interactions upon performing RT-QPCR. The IS was obtained by subtracting the RT-QPCR replica’s mean (2−ΔΔCt) for each interaction from the global mean (2−ΔΔCt average among the 31 miRNAs for each gene). The FS represents the average of the three ISs. B: Western blotting showing ACSL1, ACSL4, and SCD protein depletion upon treatment with mimic miRNAs for selected candidates from previous RT-QPCR experiments: miR-578, miR-199-b, miR-205, miR-106a, miR-142, miR-544a, and miR-19b-1. A negative mimic miRNA control without any known target was used as a control. RI, the relative intensity of protein bands compared with controls. C: miRanda/mirSVR scoring schema of mRNA-miRNA interactions: ACSL1, ACSL4, and SCD and the selected miRNAs [miR-544a, miR-142 (miR-142-3p and miR-142-5p), and miR-19b-1 (miR-19b-1-5p and miR-19b-3p)], based on three different scores generated by miRanda (alignment, thermodynamic, and conservation scores) and the mirSVR machine learning-based score. D: The relative luciferase activity of ACSL1 (left graph), ACSL4 (middle graph), or SCD (right graph) 3′UTR psiCHECK™-2 vectors upon treatment with the selected miRNAs (miR-142, miR-544a, and miR-19b-1). The relative luciferase activity (Renilla luminescence/firefly luminescence) was determined 48 h after transfection, representing the translational repression of the proteins upon binding to the 3′UTR of candidate miRNAs. A miRNA with no predictive target was used as a negative control. Results represent the fold-change mean ± SD (n = 3).
We further confirmed their action on their targets by Western blot (WB) (Fig. 2B). The FS (−0.474) for miR-544a is in accordance with a reduction of the target’s protein expression (52, 80, and 26% of protein downregulation of ACSL1, ACSL4, and SCD, respectively). miR-142 obtained a FS of −0.416 (−0.421/−0.25/−0.577 as IS for ACSL1/ACSL4/SCD, respectively), which is in agreement with 70, 57, and 49% of protein downregulation of ACSL1, ACSL4, and SCD, respectively. Finally, miR-19b-1 (FS = −0.354) showed 38, 89, and 77% of protein downregulation of ACSL1, ACSL4, and SCD, respectively; also in agreement with previous ISs (−0.298/−0.295/−0.468 for ACSL1/ACSL4/SCD, respectively). Additionally, two miRNAs not preselected, miR-578 and miR-199b, with higher FSs and therefore less predicted capacity to downregulate the targets, were also able to downregulate the three target proteins in the case of miR-199b (44, 54, and 75% of protein downregulation of ACSL1, ACSL4, and SCD, respectively) and ACSL4 and SCD proteins in the case of miR-578 (0, 48, and 33% of protein downregulation of ACSL1, ACSL4, and SCD, respectively). We also analyzed miR-205, whose prognostic value in CRC has been described (14), and miR-106, characterized as a biomarker in CRC (25, 26). Although miR-205 and miR-106-a were good candidates globally (FS −0.205 and −0.282, respectively), only miR-205 presented significant protein downregulation (45, 92, and 67% of ACSL1, ACSL4, and SCD protein downregulation, respectively).
Even though miR-205 and miR-199b presented significant downregulation capacity at the protein level, we decided to continue studying miR-544a, miR-142, and miR-19b-1 as the most robust candidates downregulating both RNA and protein levels of the ACSL/SCD pro-tumorigenic axis.
To complement WB results, those mRNA-miRNA interactions were quantified based on the miRanda/mirSVR scoring schema. Regarding ACSL1 (Fig. 2C, upper table), selected miRNA alignment scores were quite low because 120 is the minimum score for a target to be considered as valid. It is remarkable that four out of five interactions were canonical at the seed level. mirSVR scores, which get lower as downregulation probability increases, were better for 7mer sites. Curiously, miR-544a had a high probability to downregulate ACSL1, stated by a mirSVR score of −1.0027, demonstrating this score’s ability to detect noncanonical miRNA-mRNA interactions. Thus, miR-142-3p, which had a canonical 7mer interaction site together with good alignment, energy, and conservation scores, and miR-544a, which was not a canonical interaction, but presented a significantly low mirSVR score, may be among the best candidates. Regarding miR-19b-1 mature forms [miR-19b-1-5p (MIMAT0004491) and miR-19b-3p (MIMAT0000074)], further in vitro assessment is needed because they presented the minimal alignment score to be considered as valid candidates and the mirSVR did not reflect a high downregulation probability. In the case of ACSL4 (Fig. 2C, middle table), the highest alignment scores corresponded to 7mer canonical sites, which, in addition, had acceptable mirSVR scores based on our cutoff. The miR-19b-1-5p interaction did not account for a canonical site type, and the mirSVR score did not reflect a potential noncanonical interaction. Regarding SCD (Fig. 2C, lower table), the alignment and energy scores were strong and all the interaction sites were canonical with a seven nucleotide interaction along the seed, with the exception of miR-19b-1-5p, whose interaction was not potent enough to overcome the detection threshold. This was also the case for the ACSL4-miR-142-5p interaction. Considering the above-mentioned results, we reaffirmed the decision to consider miR-544a, miR-142, and miR-19b-1 as the best candidates to regulate the ACSL1/ACSL4/SCD pro-tumorigenic axis for more detailed analysis.
miR-142, miR-544a, and miR-19b-1 target the 3′UTR of ACSL1, ACSL4, and SCD
To validate the direct interaction of these miRNAs to their targets, we performed dual-luciferase assays (Fig. 2D). In agreement with previous results, miR-544a, miR-142, and miR-19b-1 downregulated the activity of the three reporter constructions reflecting their binding on the corresponding 3′UTRs. Hence, we assumed miR-544a, miR-142, and miR-19b-1 as validated miRNAs regulating the ACSL/SCD pro-tumorigenic axis.
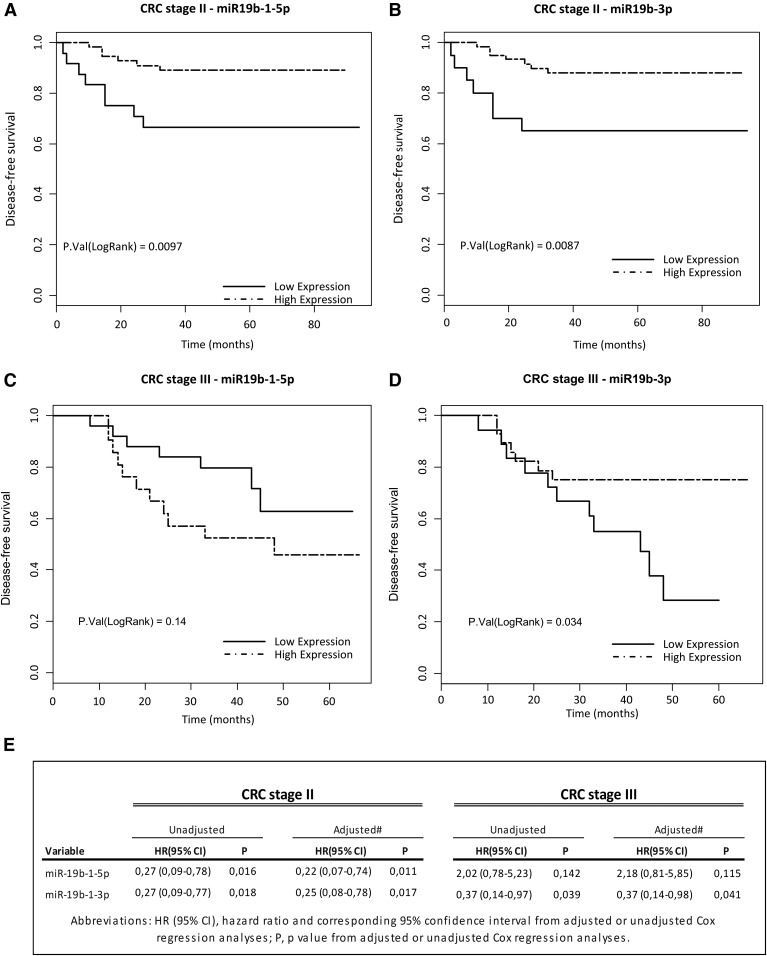
Low expression of miR-19b-1 correlates with a poorer prognosis in CRC stage II and stage III patients
In order to confirm the potency of these miRNAs as regulators of the ACSL/SCD axis involved in CRC prognosis, their expression levels were evaluated in CRC patients. For both mature forms of miR-142 [miR-142-3p (MIMAT0000434) and miR-142-5p (MIMAT0000433)] and for miR-544a, significant correlation with DFS was not found (data not shown). However, both miR-19b mature forms significantly predicted DFS in CRC stage II patients. miR-19b-5p displayed a HR(95% CI) of 0.27(0.09–0.78), log-rank P = 0.0097 (Fig. 3A) and miR-19b-3p displayed a HR(95% CI) of 0.27(0.09–0.77), log-rank P = 0.0087 (Fig. 3B). Furthermore, the miR-19b-3p prognostic value was also observed in stage III CRC patients’ DFS [HR(95% CI) of 0.37(0.14–0.97), log-rank P = 0.034] (Fig. 3C, D). Finally, to evaluate whether miR-19b-1 might constitute independent prognostic classifiers, clinical and histopathological data were included in both univariate and multivariate Cox regression analyses (Fig. 3E) revealing that both miR-19b-1 forms were DFS-independent prognostic biomarkers in stage II CRC patients [miR-19b-1-5p: HR(95% CI) of 0.22(0.07–0.74), P = 0.0111; miR-19b-3p: HR(95% CI) of 0.25(0.08–0.78), P = 0.0173] (Fig. 3E). Again, only miR-19b-3p [HR(95% CI) of 0.37(0.14–0.98), P = 0.041] constituted a DFS-independent prognostic biomarker in stage III CRC patients (Fig. 3E). Hence, miR-19b-1 could be considered as a potential miRNA with predictive value related to good prognosis in CRC patients through the inhibition of the ACSL/SCD network activation.
Fig. 3.
Prognostic value of miR-19b-1 in CRC stage II patients. Kaplan-Meier survival curves for CRC patients showing correlation between DFS in stage II patients and the expression levels of miR19b-1-5p (A) or miR19-b-3p (B), or in stage III patients and the expression levels of miR19b-1-5p (C) or miR19-b-3p (D). E: Univariate and multivariate Cox regression analyses. (CRC stage II patients n = 80, 17.50% of recurrence; CRC stage III patients n = 46, 39.13% of recurrence).
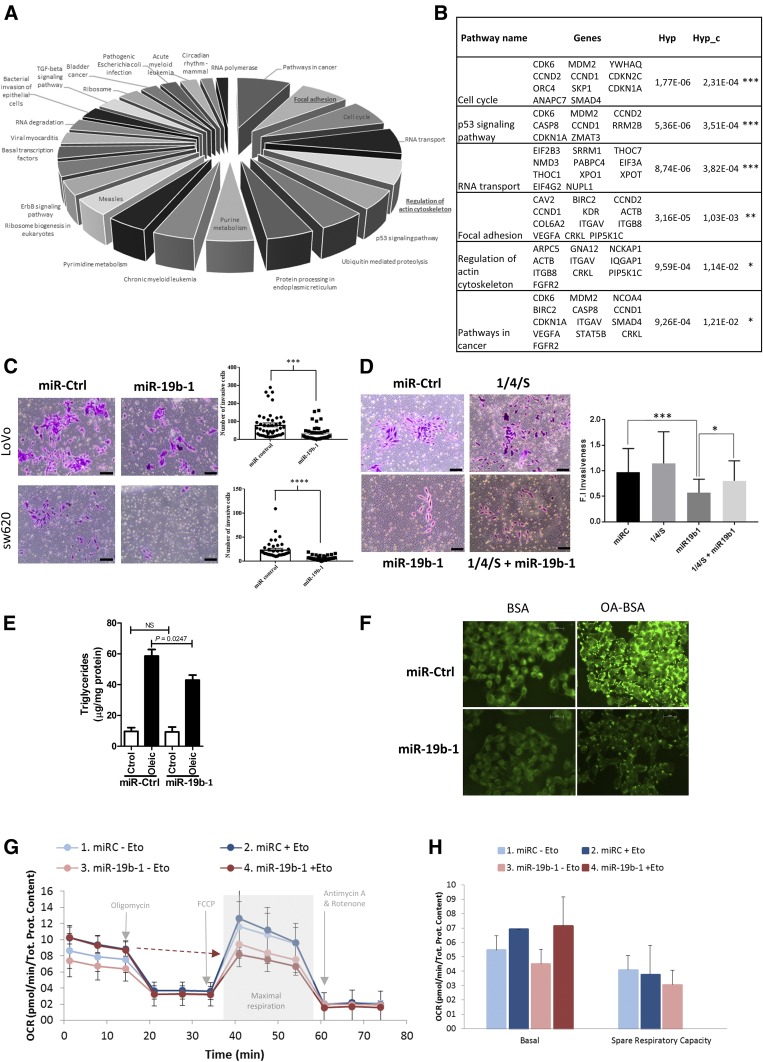
miR-19b-1 blocks cell invasion and regulates lipid metabolism in CRC cells
To further validate miR-19b-1 interference with CRC cell invasiveness mediated by the ACSL/SCD axis, we performed an in silico functional analysis, elucidating putative miRNA biological effects. Figure 4A shows biological processes ordered according to the increasing number of genes involved in each statistically significant process (Hyp_c < 0.05); while in Fig. 4B, they are ordered by corrected hypergeometric test (Hyp_c). Interestingly, focal adhesions (Hyp_c = 0.001) and actin cytoskeleton regulation (Hyp_c = 0.011), both related to invasion and migration (27, 28), are putatively elicited by miR-19b-1, in accordance with the reported ACSL/SCD effect promoting invasion and migration (5). To confirm the miR-19b-1 implication in these processes, we performed a Matrigel-based invasion assay in two well-known invasive CRC cell lines, SW620 and LoVo (Fig. 4C). Remarkably, in both cases, a clear decrease in the number of invasive cells was observed upon miR-19b-1 electroporation. Therefore, miR-19b-1 blocked CRC cell invasion, which was in agreement with the opposite effect exerted by its targets, ACSL1, ACSL4, and SCD (5). To demonstrate whether the effect of miR-19b-1 in CRC cell invasion is regulated directly by the suppression of ACSL1, ACSL4, and SCD, we performed rescue experiments by reintroducing ACSL1, ACSL4, and SCD in miR-19b-1-transfected cells. As shown in Fig. 4D, the invasive phenotype of CRC cells was significantly recovered when miR-19b-1 transfection was combined with ACSL/SCD expression vectors lacking the 3′UTR and, thus, was not affected by miR-19b-1 regulation. This demonstrates that re-expression of the ACSL/SCD axis is able to recover the lost invasion upon miR-19b-1 treatment of SW620 cells; therefore, it evidences the specific role of the miR-19b-1/ACSL/SCD regulatory axis in controlling CRC invasion.
Fig. 4.
MiR-19b-1 implication in cell invasion and lipid metabolism. A: Biological processes elicited by miR-19b-1 obtained from GeneCodis 3 (29). The chart is ordered according to the increasing number of genes involved in each process (Hyp_c < 0.05). The Hyp_c the P value obtained from the hypergeometric test determined the significance of the biological process. For miRNA hsa-miR-19b-1, due to its promiscuity, targets were limited to the ones experimentally validated and were retrieved from the TarBase database. B: Genes involved in each biologically predicted process elicited by miR-19b-1 and ordered by Hyp_c C: Boyden chamber transwell assay of invasion through Matrigel. miR-19b-1 or a control miRNA with no predicting binding site in the 3′UTR of ACSL1, ACSL4, or SCD was electroporated into SW620 or LoVo cells. D: Boyden chamber transwell assay for invasion through Matrigel. As a rescue experiment, vectors expressing ACSL1, ACSL4, and SCD were transfected together with mimic miR-19b-1 (miR-19b-1) or a control miRNA (miR-Ctrl) in SW620 cells. The positive invasion controls, miR-Ctrl alone and (1/4/S), were also included. In both cases (C, D), after 72 h with chemoattractant (10% FBS), inserts were fixed and stained with crystal violet. Nonmigrated cells were removed using cotton swabs and images were captured using an Olympus CKX41 microscope (Olympus, Tokyo, Japan) with a 20× LCAch objective and registered using analysis getIT software (Olympus). Scale bars, 50 μm. Results represent the number of migrated cells found in six random microscope fields in two independent inserts in three independent experiments ± SEM (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. E: TG analysis content in DLD-1 cells transfected with mimic miR-19b-1 or a mimic miRNA control upon a 0.5 mM OA-BSA or FA-free BSA (vehicle control) input. Lipid content is expressed in micrograms of TG per milligram of protein in two independent experiments ± SEM (n = 2). *P < 0.05. F: Representative immunofluorescence images of the distribution of the fluorophore, BODIPY, in DLD-1 cells transfected with mimic miR-19b-1 or a mimic miRNA control, upon a 0.5 mM OA-BSA or FA-free BSA (vehicle control) input. Images were captured using a Leica DM IL microscope from a representative experiment, with a 40× Plan Fluotar objective and registered using Leica Application Suite (n = 2). G: Bioenergetic profile (OCR) of cells transfected with miR-19b-1 or a mimic miRNA control with or without the addition of etomoxir. Injection schema (oligomycin, FCCP, antimycin A/rotenone) and key parameters assayed (basal respiration, maximal respiration, and spare capacity) are also indicated. H: Basal respiration and spare respiratory capacity quantification of cells transfected with miR-19b-1 or a mimic miRNA control with or without the addition of etomoxir. Both results show a representative experiment of two independent experiments with at least six replicates per condition ± SEM (n = 2).
To further assess the impact of our miRNA candidate in lipid metabolism, we measured the TG levels in miR-19b-1-transfected cells, as shown in Fig. 4E. Because CRC cells do not normally store substantial amounts of neutral lipids, cells were exposed to OA to favor lipid storage. Under these conditions, cells transfected with miR-19b-1 presented a clear reduction in the cellular TG content, indicating that miR-19b-1 diminishes neutral lipid accumulation in the cells. As a complementary analysis, we also used the BODIPY fluorophore, a specific dye for cellular neutral lipid droplets. Figure 4F shows again that cells transfected with miR-19b-1 are less prone to neutral lipid droplet accumulation upon OA supplementation (OA-BSA), compared with the vehicle control alone (BSA).
To get further insight into the metabolic role of miR-19b-1 on CRC cells, we performed bioenergetics experiments using the Seahorse XF bioanalyzer. Figure 4G shows that miR-19b-1 did not only trigger an overall effect on lipid droplet maintenance or mobilization, but also at the bioenergetic level. Upon treatment with a β-oxidation inhibitor (etomoxir), which inhibits FA transport to mitochondria and therefore respiration dependent on FA, miR-19b-1-transfected cells presented a lower maximal OCR compared with miR control cells. As a consequence of this, the spare capacity (measured as the difference between maximal respiration and the correspondent basal respiration) was null in cells transfected with miR-19b-1, differently from cells transfected with miR control. This spare respiratory capacity indicates the presence of a reserve that is accessible for the cells in case of a bioenergetic demand or a stressful situation. These results suggest that miR-19b-1 compromises respiratory capacity of cells, leading to an even more dramatic effect upon FAO inhibition, eliminating the spare respiratory capacity of cells.
DISCUSSION
The ACSL/SCD network is a clear example of how cancer cells reprogram their metabolism to promote cell invasion and poorer disease prognosis (5). Hence, it is crucial to unveil this network’s regulating mechanisms in order to design effective cancer therapies. In this sense, regulatory miRNAs emerge as promising therapeutic tools and noninvasive biomarkers because they can be detected in body fluids.
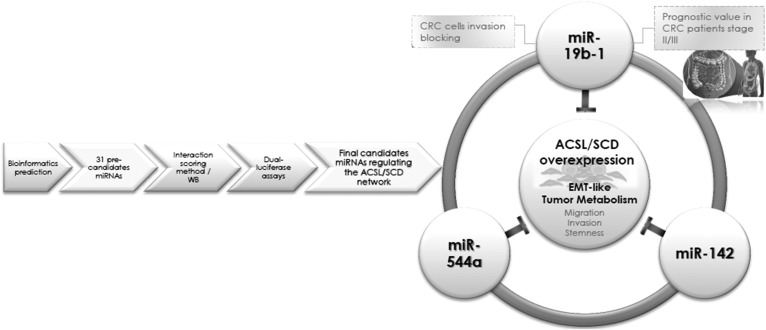
Combined bioinformatics tools led us to a miRNA library with 31 candidates (Fig. 1) that were screened by analyzing their effect on their targets’ ACSL1/ACSL4/SCD expression using a novel interaction scoring method (Fig. 2A). WB, miRanda/mirSVR scoring schema, and dual-luciferase assays (Fig. 2) validated miR-544a, miR-142, and miR-19b-1 as ACSL/SCD regulators. Importantly, miR-19b-1’s lower expression correlated with a poorer prognosis in stage II and stage III CRC patients (Fig. 3), suggesting a protective role for this miRNA in CRC, very likely through its involvement in cell invasion and lipid metabolism regulation (Fig. 4). The overall workflow to obtain the tumorigenic axis ACSL/SCD candidate miRNAs is clearly represented in Fig. 5.
Fig. 5.
Graphical abstract representing the overall procedure to obtain the tumorigenic axis ACSL/SCD candidate miRNAs.
Here we have used novel scoring approaches for more convenient managing and screening of bioinformatics data. For instance, the interaction scoring method enabled us to select the best miRNA candidates for a combination of targets. Furthermore, the combination of two important bioinformatic tools in the defined miRanda/mirSVR scoring schema allowed us to detect canonical and noncanonical interaction sites, which are often filtered out during the prediction process by most algorithms, such as miRanda, making it hard to detect experimentally verified noncanonical interactions. This was the case for the miR-544a-ACSL1 interaction, whose mirSVR score of −1.0027 represented a high downregulation probability, which was further validated despite not having a canonical interaction (17).
miR-19b-1 is a member of the miR-17-92 cluster frequently recognized as an oncomiR (29). This cluster has been associated with CRC (30), among other cancers (31). However, recent studies describe miR-19 to be decreased in gastric cancer (32, 33), suggesting that the role of the miR-17-92 cluster needs to be further investigated. Probably these miRNAs’ roles and expression may be tissue- and tumor-stage dependent (34). This study opens a new avenue for miR-17-92 cluster involvement in CRC regulation; particularly, miR-19b-1, because its potential role as a tumor suppressor is described here.
An important finding was miR-19b-1’s ability to inhibit CRC cell invasion. In silico analysis indicated this miRNA’s implication in processes like focal adhesions and regulation of the actin cytoskeleton. Focal adhesions, which regulate cellular behaviors, such as cell proliferation, migration, and invasion (35), also connect ECM and, thus, the tumor microenvironment to the actin cytoskeleton, which is also involved in cancer cell migration and invasion (28). The fact that miR-19b-1 may be able to inhibit these processes is in agreement with previous findings describing the ACSL/SCD metabolic signature to promote invasion and migration (5).
In this vein, miR-19b-1’s implication in lipid metabolism agrees with the lipid metabolism enzyme condition of its targets ACSL1, ACSL4, and SCD. The use of two different tracers of lipid trafficking pointed toward the role of this miRNA in the maintenance of lipid droplets, either causing decreased intake, impaired incorporation of the FAs into neutral lipids, or increased mobilization. miR-19b-1 could act by inhibiting de novo lipogenesis by targeting FA-activating enzymes (ACSLs and SCD) and, therefore, the previous steps to TG synthesis, leading to less accumulation of lipid droplets. Furthermore, Seahorse analysis suggested that miR-19b-1 compromises the respiratory capacity of cells, potentiating the effect of the β-oxidation inhibitor, etomoxir. Again, by inhibiting ACSL-mediated FA activation, miR-19b-1 would lead to diminished β-oxidation and to a limited maximal mitochondrial respiration and impaired spare capacity. Nevertheless, further studies will help to elucidate the mechanisms underlying the miRNA’s fine-tuning control of lipid metabolism fueling cancer energy.
Considering miR-19b-1 interacting together with the other selected candidates, miR-142 and miR-544a, a Panther analysis designated the Wnt route as the most represented biological pathway common to these three miRNAs (supplemental Fig. S1). Wnt activation requires avoiding β-catenin degradation upon GSK3β inhibitory phosphorylation leading to invasion gene transcription (36). ACSL/SCD increases GSK3β phosphorylation, activating Wnt signaling and EMT (5); therefore, the downregulation elicited by these miRNAs over this network suggests a role for them in Wnt regulation. Although with an opposite role, miR-544a has been involved in Wnt pathway actions in gastric and lung cancer (37, 38). miR-142-3p has been reported to target cancer stem cell markers, such as the Wnt target, LGR5, in CRC cells (39), in agreement with its action on the ACSL/SCD network, which is also involved in cancer stem cell feature generation (5). Furthermore, reported miR-142-3p downregulation in CRC patients (40) supports our bioinformatics analysis. All of this data supports the idea of miRNA networks controlling functional gene interactions, such as ACSL/SCD.
Finally, we propose miR-19b-1 as a potential noninvasive biomarker, given its strong association with a better prognosis in CRC patients and as a promising therapeutic miRNA inhibiting CRC cell invasion. miR-19b-1 could be used as a biomarker of CRC prognosis, which could be done either by directly evaluating its expression in biopsy tissues, as described here, or its circulating levels in serum, as previously described for other types of cancer (33). From a therapeutic point of view, miRNA replacement therapy could be an alternative to increase the expression of this particular miRNA in colorectal tissue, as exemplified by the use of miR-34 for other types of human neoplasias (41). Other alternatives, such as the dietary modulation of miRNAs could also be a possibility, because miR-19b-1, induced by low protein diet in piglets has been recently reported (42). However, more in vivo research to test efficacy and safety issues is needed before we could benefit from therapeutic approaches to increase miRNA levels in humans.
Supplementary Material
Acknowledgments
The authors thank Dr. Jean-Baptiste Demoulin for the anti-SCD1 antibody and Dr. Stephen Prescott and Dr. Diana Stafforini for anti-ACSL4 antibody.
Footnotes
Abbreviations:
- ACSL
- acyl-CoA synthetase
- CRC
- colorectal cancer
- DFS
- disease-free survival
- EMT
- epithelial-mesenchymal transition
- FAO
- FA oxidation
- FS
- final score
- HR
- hazard ratio
- Hyp_c
- corrected hypergeometric test
- IS
- individual score
- miRNA/miR
- micro-RNA
- OA
- oleic acid
- OCR
- oxygen consumption rate
- RT-QPCR
- quantitative RT-PCR
- SCD
- stearoyl-CoA desaturase
- TG
- triglyceride
- WB
- Western blot
This work was supported by Secretaría de Estado de Investigación, Desarrollo e Innovación Grants MINECO AGL2013-48943-C2 and AGL2016-76736-C3, Consejería de Educación, Juventud y Deporte, Comunidad de Madrid Grant P2013/ABI-2728, ALIBIRD-CM, and EU Structural Funds. The authors declare no potential conflicts of interest.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Siegel R. L., Miller K. D., and Jemal A.. 2016. Cancer statistics, 2016. CA Cancer J. Clin. 66: 7–30. [DOI] [PubMed] [Google Scholar]
- 2.Cairns R. A., Harris I. S., and Mak T. W.. 2011. Regulation of cancer cell metabolism. Nat. Rev. Cancer. 11: 85–95. [DOI] [PubMed] [Google Scholar]
- 3.Carracedo A., Cantley L. C., and Pandolfi P. P.. 2013. Cancer metabolism: fatty acid oxidation in the limelight. Nat. Rev. Cancer. 13: 227–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mashima T., Seimiya H., and Tsuruo T.. 2009. De novo fatty-acid synthesis and related pathways as molecular targets for cancer therapy. Br. J. Cancer. 100: 1369–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sánchez-Martínez R., Cruz-Gil S., Gómez de Cedrón M., Álvarez-Fernández M., Vargas T., Molina S., García B., Herranz J., Moreno-Rubio J., Reglero G., et al. . 2015. A link between lipid metabolism and epithelial-mesenchymal transition provides a target for colon cancer therapy. Oncotarget. 6: 38719–38736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coleman R. A., Lewin T. M., Van Horn C. G., and Gonzalez-Baró M. R.. 2002. Do long-chain acyl-CoA synthetases regulate fatty acid entry into synthetic versus degradative pathways? J. Nutr. 132: 2123–2126. [DOI] [PubMed] [Google Scholar]
- 7.Gaisa N. T., Reinartz A., Schneider U., Klaus C., Heidenreich A., Jakse G., Kaemmerer E., Klinkhammer B. M., Knuechel R., and Gassler N.. 2013. Levels of acyl-coenzyme A synthetase 5 in urothelial cells and corresponding neoplasias reflect cellular differentiation. Histol. Histopathol. 28: 353–364. [DOI] [PubMed] [Google Scholar]
- 8.Mason P., Liang B., Li L., Fremgen T., Murphy E., Quinn A., Madden S. L., Biemann H-P., Wang B., Cohen A., et al. . 2012. SCD1 inhibition causes cancer cell death by depleting mono-unsaturated fatty acids. PLoS One. 7: e33823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roongta U. V., Pabalan J. G., Wang X., Ryseck R. P., Fargnoli J., Henley B. J., Yang W. P., Zhu J., Madireddi M. T., Lawrence R. M., et al. . 2011. Cancer cell dependence on unsaturated fatty acids implicates stearoyl-CoA desaturase as a target for cancer therapy. Mol. Cancer Res. 9: 1551–1561. [DOI] [PubMed] [Google Scholar]
- 10.von Roemeling C. A., Marlow L. A., Wei J. J., Cooper S. J., Caulfield T. R., Wu K., Tan W. W., Tun H. W., and Copland J. A.. 2013. Stearoyl-CoA desaturase 1 is a novel molecular therapeutic target for clear cell renal cell carcinoma. Clin. Cancer Res. 19: 2368–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahmad N., Haider S., Jagannathan S., Anaissie E., and Driscoll J. J.. 2014. MicroRNA theragnostics for the clinical management of multiple myeloma. Leukemia. 28: 732–738. [DOI] [PubMed] [Google Scholar]
- 12.Bartel D. P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 116: 281–297. [DOI] [PubMed] [Google Scholar]
- 13.Gómez de Cedrón M., and Ramírez de Molina A.. 2016. Microtargeting cancer metabolism: opening new therapeutic windows based on lipid metabolism. J. Lipid Res. 57: 193–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Orang A. V., Safaralizadeh R., Hosseinpour Feizi M. A., and Somi M. H.. 2014. Diagnostic and prognostic value of miR-205 in colorectal cancer. Asian Pac. J. Cancer Prev. 15: 4033–4037. [DOI] [PubMed] [Google Scholar]
- 15.Demoulin J. B., Ericsson J., Kallin A., Rorsman C., Rönnstrand L., and Heldin C. H.. 2004. Platelet-derived growth factor stimulates membrane lipid synthesis through activation of phosphatidylinositol 3-kinase and sterol regulatory element-binding proteins. J. Biol. Chem. 279: 35392–35402. [DOI] [PubMed] [Google Scholar]
- 16.Cao Y., Murphy K. J., McIntyre T. M., Zimmerman G. A., and Prescott S. M.. 2000. Expression of fatty acid-CoA ligase 4 during development and in brain. FEBS Lett. 467: 263–267. [DOI] [PubMed] [Google Scholar]
- 17.John B., Enright A. J., Aravin A., Tuschl T., Sander C., and Marks D. S.. 2004. Human microRNA targets. PLoS Biol. 2: e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friedman R. C., Farh K. K., Burge C. B., and Bartel D. P.. 2009. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19: 92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kertesz M., Iovino N., Unnerstall U., Gaul U., and Segal E.. 2007. The role of site accessibility in microRNA target recognition. Nat. Genet. 39: 1278–1284. [DOI] [PubMed] [Google Scholar]
- 20.Krek A., Grün D., Poy M. N., Wolf R., Rosenberg L., Epstein E. J., MacMenamin P., da Piedade I., Gunsalus K. C., Stoffel M., et al. . 2005. Combinatorial microRNA target predictions. Nat. Genet. 37: 495–500. [DOI] [PubMed] [Google Scholar]
- 21.Betel D., Koppal A., Agius P., Sander C., and Leslie C.. 2010. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 11: R90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tabas-Madrid D., Nogales-Cadenas R., and Pascual-Montano A.. 2012. GeneCodis3: a non-redundant and modular enrichment analysis tool for functional genomics. Nucleic Acids Res. 40: W478–W483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Folch J., Lees M., and Sloane Stanley G. H.. 1957. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226: 497–509. [PubMed] [Google Scholar]
- 24.Sánchez-Martínez R., Cruz-Gil S., García-Álvarez M. S., Reglero G., and Ramirez de Molina A.. 2017. Complementary ACSL isoforms contribute to a non-Warburg advantageous energetic status characterizing invasive colon cancer cells. Sci. Rep. 7: 11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen W-Y., Zhao X-J., Yu Z-F., Hu F-L., Liu Y-P., Cui B-B., Dong X-S., and Zhao Y-S.. 2015. The potential of plasma miRNAs for diagnosis and risk estimation of colorectal cancer. Int. J. Clin. Exp. Pathol. 8: 7092–7101. [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang L., Meng L., Fan Z., Liu B., Pei Y., and Zhao Z.. 2014. [Expression of plasma miR-106a in colorectal cancer and its clinical significance]. Nan Fang Yi Ke Da Xue Xue Bao. 34: 354–357. Chinese. [PubMed] [Google Scholar]
- 27.Nagano M., Hoshino D., Koshikawa N., Akizawa T., and Seiki M.. 2012. Turnover of focal adhesions and cancer cell migration. Int. J. Cell Biol. Accessed October 2, 2017, at https://www.hindawi.com/journals/ijcb/2012/310616/. [DOI] [PMC free article] [PubMed]
- 28.Yamaguchi H., and Condeelis J.. 2007. Regulation of the actin cytoskeleton in cancer cell migration and invasion. Biochim. Biophys. Acta. 1773: 642–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mo M-H., Chen L., Fu Y., Wang W., and Fu S. W.. 2012. Cell-free circulating miRNA biomarkers in cancer. J. Cancer. 3: 432–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diosdado B., van de Wiel M. A., Terhaar Sive Droste J. S., Mongera S., Postma C., Meijerink W. J. H. J., Carvalho B., and Meijer G. A.. 2009. MiR-17-92 cluster is associated with 13q gain and c-myc expression during colorectal adenoma to adenocarcinoma progression. Br. J. Cancer. 101: 707–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kandalam M. M., Beta M., Maheswari U. K., Swaminathan S., and Krishnakumar S.. 2012. Oncogenic microRNA 17-92 cluster is regulated by epithelial cell adhesion molecule and could be a potential therapeutic target in retinoblastoma. Mol. Vis. 18: 2279–2287. [PMC free article] [PubMed] [Google Scholar]
- 32.Yang O., Huang J., and Lin S.. 2014. Regulatory effects of miRNA on gastric cancer cells. Oncol. Lett. 8: 651–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang J., Song Y., Zhang C., Zhi X., Fu H., Ma Y., Chen Y., Pan F., Wang K., Ni J., et al. . 2015. Circulating MiR-16–5p and MiR-19b-3p as two novel potential biomarkers to indicate progression of gastric cancer. Theranostics. 5: 733–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reid G., Kirschner M. B., and van Zandwijk N.. 2011. Circulating microRNAs: association with disease and potential use as biomarkers. Crit. Rev. Oncol. Hematol. 80: 193–208. [DOI] [PubMed] [Google Scholar]
- 35.Wozniak M. A., Modzelewska K., Kwong L., and Keely P. J.. 2004. Focal adhesion regulation of cell behavior. Biochim. Biophys. Acta. 1692: 103–119. [DOI] [PubMed] [Google Scholar]
- 36.Clevers H. 2006. Wnt/beta-catenin signaling in development and disease. Cell. 127: 469–480. [DOI] [PubMed] [Google Scholar]
- 37.Yanaka Y., Muramatsu T., Uetake H., Kozaki K., and Inazawa J.. 2015. miR-544a induces epithelial-mesenchymal transition through the activation of WNT signaling pathway in gastric cancer. Carcinogenesis. 36: 1363–1371. [DOI] [PubMed] [Google Scholar]
- 38.Mo X. M., Li H. H., Liu M., and Li Y. T.. 2014. Downregulation of GSK3β by miR-544a to maintain self-renewal ability of lung caner stem cells. Oncol. Lett. 8: 1731–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen W. W., Zeng Z., Zhu W. X., and Fu G. H.. 2013. MiR-142–3p functions as a tumor suppressor by targeting CD133, ABCG2, and Lgr5 in colon cancer cells. J. Mol. Med. (Berl.). 91: 989–1000. [DOI] [PubMed] [Google Scholar]
- 40.Ghanbari R., Mosakhani N., Asadi J., Nouraee N., Mowla S. J., Yazdani Y., Mohamadkhani A., Poustchi H., Knuutila S., and Malekzadeh R.. 2015. Downregulation of plasma MiR-142–3p and MiR-26a-5p in patients with colorectal carcinoma. Iran. J. Cancer Prev. 8: e2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beg M. S., Brenner A. J., Sachdev J., Borad M., Kang Y-K., Stoudemire J., Smith S., Bader A. G., Kim S., and Hong D. S.. 2017. Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Invest. New Drugs. 35: 180–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun R. P., Xi Q. Y., Sun J. J., Cheng X., Zhu Y. L., Ye D. Z., Chen T., Wei L. M., Ye R. S., Jiang Q. Y., et al. . 2016. In low protein diets, microRNA-19b regulates urea synthesis by targeting SIRT5. Sci. Rep. 6: 33291. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.