Abstract
Compelling evidence indicates that lipid metabolism is in partial control of the circadian system. In this context, it has been reported that the melatonin receptor 1B (MTNR1B) genetic variant influences the dynamics of melatonin secretion, which is involved in the circadian system as a chronobiotic. The objective was to analyze whether the MTNR1B rs10830963 genetic variant was related to changes in lipid levels in response to dietary interventions with different macronutrient distribution in 722 overweight/obese subjects from the POUNDS Lost trial. We did not find a significant association between the MTNR1B genotype and changes in lipid metabolism. However, dietary fat intake significantly modified genetic effects on 2 year changes in total and LDL cholesterol (P interaction = 0.006 and 0.001, respectively). In the low-fat diet group, carriers of the sleep disruption G allele (minor allele) showed a greater reduction of total cholesterol (β ± SE = −5.78 ± 2.88 mg/dl, P = 0.04) and LDL cholesterol (β ± SE = −7.19 ± 2.37 mg/dl, P = 0.003). Conversely, in the high-fat diet group, subjects carrying the G allele evidenced a smaller decrease in total cholesterol (β ± SE = 5.81 ± 2.65 mg/dl, P = 0.03) and LDL cholesterol (β ± SE = 5.23 ± 2.21 mg/dl, P = 0.002). Subjects carrying the G allele of the circadian rhythm-related MTNR1B variant may present a bigger impact on total and LDL cholesterol when undertaking an energy-restricted low-fat diet.
Keywords: clinical trials, diet and dietary lipids, cholesterol, low density lipoprotein, genetics, melatonin receptor 1B, gene-diet interaction, high-fat diet, lipid metabolism, weight-loss intervention
There is scientific evidence that lipid metabolism is partly controlled by the circadian system and exhibits differential 24 h profiles in major metabolic organs in association with sleep/wake, activity/rest, and fast/feeding cycles (1). Plasma lipid concentrations, intestinal absorption, and lipid biosynthesis also show a daily rhythmicity, as has been reported in different models (2, 3). Moreover, it has been found that disruption of the core circadian clock and peripheral clocks leads to a dysregulation of lipid metabolism (2, 4). Interestingly, Clock mutant mice exhibited both hyperlipidemia and obesity phenotypes (5).
Melatonin is a hormone secreted mainly by the pineal gland that plays a major role in the regulation of circadian rhythms (6); melatonin treatment has shown beneficial effects on the lipid profile in humans (7). A genetic variant in the melatonin receptor 1B (MTNR1B) gene, which encodes one of the two high-affinity receptors of melatonin (8, 9), has been associated with altered melatonin rhythm and melatonin signaling (10, 11). Interestingly, the same genetic variant has also been related to the plasma lipid profile (12, 13). Actually, the MTNR1B rs10830963 polymorphism was related to circulating levels of VLDL and triglyceride (TG) (12). In addition, the MTNR1B rs10830963 variant was found to interact with dietary fat on lipid levels in an observational study (13). Specifically, Dashti et al. (13) showed that the relation between the MTNR1B rs10830963 genotype and HDL cholesterol was modified by total fat and MUFA intakes. However, to our knowledge, no study has analyzed the interaction between the MTNR1B rs10830963 polymorphism on long-term changes of lipid metabolism traits in response to dietary interventions. Investigation on such interactions may improve personalized dietary intervention based on the genotype.
The aim of this study was to examine potential interactions between the MTNR1B rs10830963 genotype and weight-loss diets varying in fat content on changes in lipid metabolism traits during 2 years of dietary intervention within the POUNDS Lost trial.
MATERIALS AND METHODS
Study participants
The POUNDS Lost trial is a 2 year randomized clinical trial (clinical trial registration number NCT00072995) designed to compare the effects of four energy-reduced diets with different macronutrient composition on weight loss. The study was conducted at two sites (Harvard School of Public Health and Brigham and Women’s Hospital in Boston, MA and the Pennington Biomedical Research Center of Louisiana State University System, Baton Rouge, LA) from October 2004 through December 2007. The study design and methods have been previously described in detail (14). Briefly, 811 overweight or obese (BMI 25–40 kg/m2) participants were randomly assigned to one of four energy-reduced diets during a 2 year follow-up time. The target percentages of energy derived from fat, protein, and carbohydrate in the four diets were: 20, 15, and 60%; 20, 25, and 55%; 40, 15, and 45%; and 40, 25, and 35%, respectively. In this two-by-two factorial design, two diets were low fat (20%), two diets were high fat (40%), two diets were average in protein (15%), and two diets were high in protein (25%). Major exclusion criteria were the presence of diabetes or unstable cardiovascular disease, the use of medications that affect body weight, and insufficient motivation (14). The study was approved by the human subjects committee at each institution and by a data and safety monitoring board appointed by the National Heart, Lung, and Blood Institute. All participants gave written informed consent.
Measurements
Body weight was measured in the morning before breakfast at baseline, 6 months, and 2 years of follow-up. Height was measured at baseline. BMI was calculated as weight (in kilograms)/height (in square meters). In the present study, ethnicity was self-reported and grouped as white, black, and others. Fasting blood samples were obtained at routine times in clinical settings at baseline, 6 months, and 2 years. Levels of serum lipids (TG, total cholesterol, and HDL cholesterol) were analyzed in the clinical laboratory at Pennington using the Synchron CX7 (Beckman Coulter). LDL cholesterol was obtained for each participant according to the following equation: total cholesterol – HDL cholesterol – TG/5 (15). However, when TG concentration was >400 mg/dl, LDL cholesterol was measured directly by Synchron CX7 (Beckman Coulter). Dietary intake was assessed in a random sample of 50% of the participants by a review of the 5 day diet record at baseline and by 24 h recall during a telephone interview on three nonconsecutive days at 6 months and 2 years, in order to assess the nutritional adherence across the intervention.
Genotyping
DNA was extracted from the buffy coat fraction of centrifuged blood by the QIAmp blood kit (Qiagen). Genotyping of the previously reported MTNR1B rs10830963 genetic variant was successfully done in 722 of 811 total participants using the Open Array SNP genotyping system (Biotrove) (16). The genotyping success rate was near 90%. The genotype frequency was CC 54.4%, CG 37.8%, and GG 7.8%; the frequency of the minor allele (G allele) was 0.27 in the study population. The genotype distribution was in Hardy-Weinberg equilibrium in both the total population and the major ethnic group (whites) (P > 0.05).
Statistical analysis
In the present study, 722 subjects with baseline genotyped data of the MTNR1B rs10830963 variant were included. Among them, 528 participants completed the intervention after 2 years. The primary outcome of the current study was the change in lipid levels (TG, total cholesterol, LDL cholesterol, and HDL cholesterol) during the 2 years of intervention. To compare baseline characteristics across genotypes, the chi-squared test for categorical variables and general linear models for continuous variables were used. General linear models (model 1: adjusted for age, sex, ethnicity, baseline BMI, the respective baseline variable, and lipid-lowering medication use; model 2: adjusted for model 1 plus weight loss) were performed for comparison of changes from baseline in lipid levels across genotype groups according to low- or high-fat group at 6 months and 2 years intervention. The interaction term (e.g., MTNR1B genotype × high-/low-fat diet group) was included in the models to test gene-diet intervention interactions. Additive genetic models were used in the analysis. Statistical analyses were performed using STATA/SE version 12.0 (StataCorp, College Station, TX). Statistical significance was considered for P < 0.05. Moreover, we used Bonferroni correction to adjust P values for four independent tests (TGs, total cholesterol, LDL cholesterol, and HDL cholesterol). Thus, a P value < 0.012 was considered statistically significant after adjustment of multiple comparisons.
RESULTS
Baseline characteristics of the participants according to the MNTR1B rs10830963 genotype are presented in Table 1. The distribution of the polymorphism was similar by sex and diet groups; meanwhile, statistically significant differences were observed by ethnicity (P < 0.001). No associations of the MNTR1B genotype with baseline lipid levels were observed. Changes in lipid levels at 6 months and 2 years of follow-up were not related to the MTNR1B genotype after adjustment for age, sex, ethnicity, BMI at baseline, baseline value for the respective outcome, lipid-lowering medication use, and diet group (data not shown).
TABLE 1.
Characteristics of the study participants according to MTNR1B rs10830963 genotypes
| CC (n = 393) | CG (n = 273) | GG (n = 56) | P | |
| Age (years) | 50.8 (9.2) | 51.0 (9.1) | 53.0 (10.3) | 0.24 |
| Sex | 0.72 | |||
| Male | 149 (37.9) | 112 (41.0) | 22 (39.3) | |
| Female | 244 (62.1) | 161 (59.0) | 34 (60.7) | |
| Race or ethnic group | <0.001 | |||
| White | 279 (71.0) | 247 (90.5) | 49 (87.5) | |
| Black | 96 (24.4) | 12 (4.4) | 3 (5.4) | |
| Hispanic or other | 18 (4.6) | 14 (5.1) | 4 (7.1) | |
| Diet group | 0.69 | |||
| Low fat | 198 (50.4) | 134 (49.1) | 31 (55.4) | |
| High fat | 195 (49.6) | 139 (50.2) | 25 (44.6) | |
| TG, (mg/dl) | 137.4 (86.8) | 151.4 (85.4) | 151.5 (84.4) | 0.09 |
| Total cholesterol (mg/dl) | 200.0 (37.3) | 204.4 (37.2) | 206.2 (33.9) | 0.22 |
| LDL cholesterol (mg/dl) | 125.0 (32.0) | 125.9 (32.7) | 126.4 (31.2) | 0.92 |
| HDL cholesterol (mg/dl) | 48.2 (13.3) | 48.9 (13.8) | 51.2 (20.3) | 0.31 |
| Body weight (kg) | 93.7 (15.0) | 92.8 (16.0) | 92.0 (17.5) | 0.66 |
| Body weight loss at 2 years (kg) | −3.7 (0.4) | −4.1 (0.5) | −6.0 (1.0) | 0.13a |
| BMI (kg/m2) | 32.9 (3.8) | 32.3 (3.9) | 32.5 (3.7) | 0.11 |
| BMI loss at 2 years (kg/m2) | −1.3 (0.1) | −1.4 (0.2) | −2.1 (0.3) | 0.12a |
Data were calculated by the χ2 test for categorical variables and ANOVA for continuous variables. Data are expressed as n (%) or mean (SD).
Adjusted for age, sex, ethnicity, baseline value for the respective outcome, and diet group.
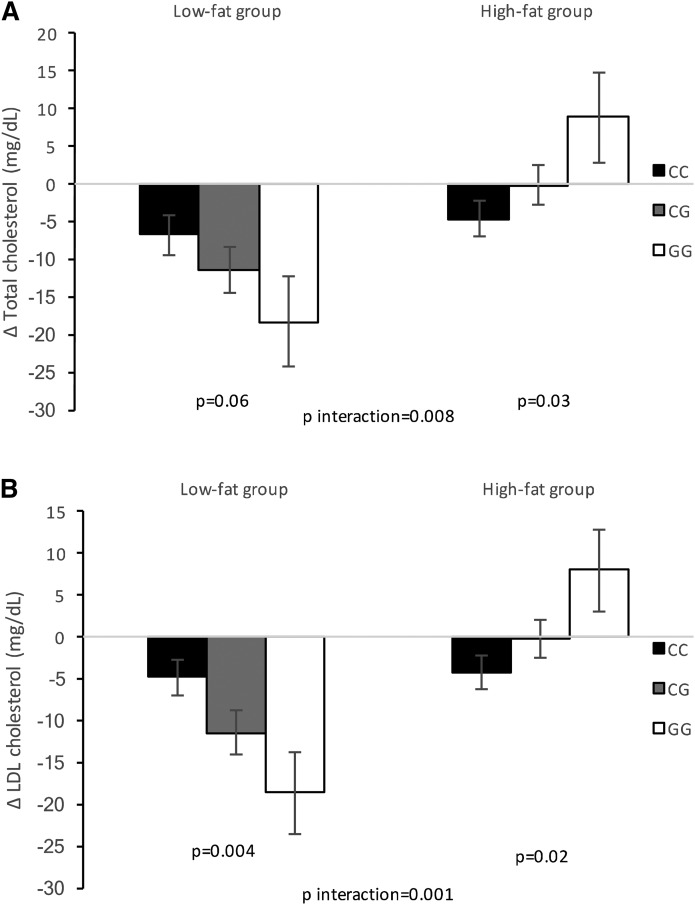
Dietary fat content (high-fat vs. low-fat intake) showed significant differential effects on changes in total cholesterol and LDL cholesterol at 2 years of the intervention depending on the MTNR1B rs10830963 genetic variant, after adjusting for age, sex, ethnicity, BMI at baseline, the respective baseline variable, and lipid-lowering medication use (P interaction = 0.006 and 0.001, respectively) as reported (Table 2; Figs. 1, 2). These genotype-diet interactions were statistically significant after correction for multiple testing (P < 0.012 based on Bonferroni correction for four tests). Within the low-fat diet group, an increasing number of the G allele was associated with greater decreases in total cholesterol (β ± SE = −5.78 ± 2.88 mg/dl per G allele, P = 0.04) and LDL cholesterol (β ± SE = −7.19 ± 2.37 mg/dl per G allele, P = 0.003). Meanwhile, an opposite effect was observed among participants in the high-fat diet group. Thus, carriers of the G allele showed a positive association with increases in total cholesterol (β ± SE = 5.81 ± 2.65 mg/dl per G allele, P = 0.03) and LDL cholesterol (β ± SE = 5.23 ± 2.21 mg/dl per G allele, P = 0.02).
TABLE 2.
Effect of the MTNR1B rs10830963 genetic variant on changes in lipid metabolism traits in response to a low-/high-fat diet at 2 years of diet intervention
| Low Fat (n = 270) | High Fat (n = 258) | P Interaction | |||
| β (SE) | P | β (SE) | P | ||
| Model 1 | |||||
| Δ TG (mg/dl) | 5.37 (5.51) | 0.33 | 0.65 (5.44) | 0.90 | 0.65 |
| Δ Total cholesterol (mg/dl) | −5.78 (2.88) | 0.04 | 5.81 (2.65) | 0.03 | 0.006 |
| Δ LDL cholesterol (mg/dl) | −7.19 (2.37) | 0.003 | 5.23 (2.21) | 0.02 | 0.001 |
| Δ HDL cholesterol (mg/dl) | 0.71 (0.67) | 0.29 | 0.70 (0.74) | 0.34 | 0.90 |
| Model 2 | |||||
| Δ TG (mg/dl) | 8.62 (5.30) | 0.10 | 0.51 (5.18) | 0.92 | 0.44 |
| Δ Total cholesterol (mg/dl) | −5.41 (2.90) | 0.06 | 5.81 (2.66) | 0.03 | 0.008 |
| Δ LDL cholesterol (mg/dl) | −6.85 (2.38) | 0.004 | 5.23 (2.21) | 0.02 | 0.001 |
| Δ HDL cholesterol (mg/dl) | 0.23 (0.63) | 0.71 | 0.74 (0.69) | 0.28 | 0.76 |
Data were calculated by using linear regression models. The interaction term was included in the models to test gene-diet interactions. The β represents changes in outcomes for the increasing number of the G allele of the rs10830963 variant. Model 1: adjusted for age, sex, ethnicity, BMI at baseline, the respective baseline variable, and lipid-lowering medication use. Model 2: adjusted for age, sex, ethnicity, BMI at baseline, the respective baseline variable, lipid-lowering medication use, and body weight loss at each intervention time.
Fig. 1.
Interaction between the MTNR1B rs10830963 genetic variant and dietary fat intervention on changes in total cholesterol (A) and LDL cholesterol (B) at 2 years of diet intervention. Data are means (SE) after being adjusted for age, sex, ethnicity, BMI at baseline, the value for the respective outcome trait at baseline, lipid-lowering medication use, and body weight loss. Low-fat group sample sizes: CC n = 146, CG n = 97, and GG n = 27. High-fat group sample sizes: CC n = 137, CG n = 100, and GG n = 21.
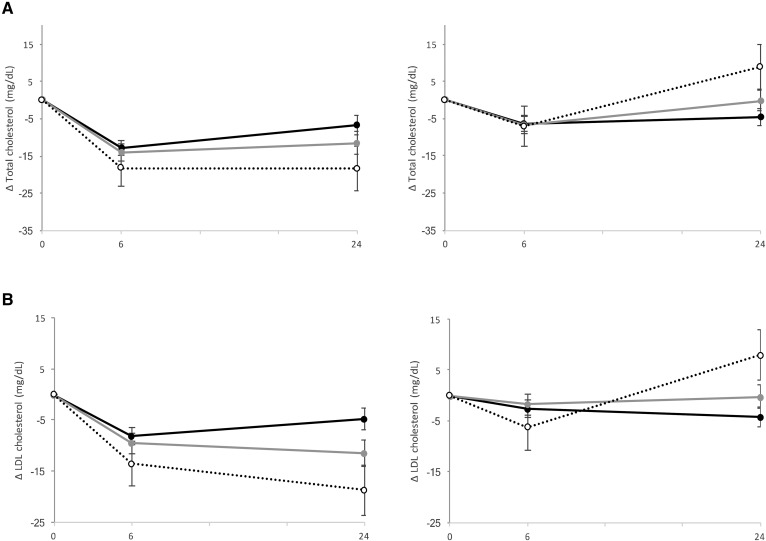
Fig. 2.
Effect of the MTNR1B rs10830963 genetic variant and fat diets on changes in total cholesterol (A) and LDL cholesterol (B) at 6 months and 2 years of diet intervention (black circle and solid line, CC genotype; gray circle and gray solid line, CG genotype; white circle and dotted line, GG genotype). Data are means (SE) after being adjusted for age, sex, ethnicity, BMI at baseline, the value for the respective outcome trait at baseline, lipid-lowering medication use, and body weight loss. Low-fat group sample sizes at 6 months: CC n = 167, CG n = 121, and GG n = 27. High-fat group sample sizes at 6 months: CC n = 165, CG n = 115, and GG n = 23. Low-fat group sample sizes at 24 months: CC n = 146, CG n = 97, and GG n = 27. High-fat group sample sizes at 2 years: CC n = 137, CG n = 100, and GG n = 21.
In order to detect whether the observed effects were mediated by weight loss, the analyzed models were further adjusted for body weight loss (model 2) (Table 2). We found that the gene-diet interactions on changes in total cholesterol and LDL cholesterol remained significant (P for interaction = 0.008 and 0.001, respectively) and passed the threshold of P < 0.012 after correcting for multiple testing. Only in the low-fat diet group was the association between total cholesterol and the genotype attenuated (P = 0.063). When we analyzed the data among Caucasians, similar results were found (P interaction = 0.025 and 0.003 in model 2 for total cholesterol and LDL cholesterol, respectively) (supplemental Table S1).
There were no significant interactions between the genotype and the low-/high-fat diet on changes in HDL cholesterol and TG at 2 years of follow-up (both P for interaction >0.05). In addition, the MTNR1B rs10830963 genetic variant did not interact with dietary protein intake on changes in lipid levels (data not shown).
DISCUSSION
In one of the largest available randomized dietary intervention trials on weight loss, we, for the first time, report a significant interaction between the circadian rhythm-related MTNR1B rs10830963 genetic variant and dietary fat intake on changes in total cholesterol and LDL cholesterol. Our results indicate that an increasing number of the G allele was associated with greater decreases in total cholesterol and LDL cholesterol in response to the low-fat diet, whereas an opposite effect was found in the high-fat diet group.
Compelling evidence has shown that the circadian system plays an important role in coordinating lipid metabolic pathways through rhythmic activation or repression of genes involved in lipid metabolism, either directly or indirectly by controlling other transcription factors (17). In addition, the disruption of the core molecular clock results in abnormal lipid metabolism, including altered fat storage and lipid transport and deficits in absorption of dietary lipids (1, 2). In this sense, melatonin is one of the chronobiotics used by the central master clock to synchronize circadian rhythms (6). Regarding lipid metabolism, it has been demonstrated that melatonin treatment can improve dyslipidemia in both animal and human studies (7). In humans, daily administration of melatonin for 2 months significantly improved LDL cholesterol among subjects with features of the metabolic syndrome (18). Moreover, treatment with melatonin and zinc decreased the levels of TG, total cholesterol, and LDL cholesterol and increased the levels of HDL cholesterol in type 2 diabetic patients poorly controlled with metformin (19). Despite the fact that the effect of melatonin on lipid profiles has been widely studied, the mechanisms by which the MTNR1B rs10830963 affects lipid metabolism remains unknown. However, the MTNR1B genetic variant has recently been associated with melatonin levels and melatonin signaling (10, 11). On the one hand, Lane et al. (10) reported that rs10830963 G allele carriers showed a disruption of melatonin rhythm, because subjects presented a later melatonin offset and a longer duration of elevated melatonin levels. Given that melatonin appears to be involved in various lipid phenotypes, it can be speculated that the effect of the MTNR1B genetic variant on dynamics of melatonin expression thereby could influence lipid levels. On the other hand, Tuomi et al. (11) reported that subjects carrying one or two MTNR1B rs10830963 G alleles showed a 2- and 4-fold increase in MTNR1B mRNA expression in human pancreatic islets, respectively, compared with subjects carrying the CC genotype. Although the results by Tuomi et al. (11) did not show a direct impact on lipid levels, such findings suggest that MTNR1B rs10830963 might affect MTNR1B mRNA expression in other cell types related to lipid metabolism.
Interestingly, we found that dietary fat intake modified the effect of the MTNR1B rs10830963 genetic variant on changes in total cholesterol and LDL cholesterol, which provides suggestive implications in preventive medicine and clinical practice. In this context, a meta-analysis found nominal significant interactions between the MTNR1B genotype and fat intake (total fat and MUFA) on HDL cholesterol levels (13). However, the results did not pass the prespecified Bonferroni-corrected significance level. Although the mechanisms underlying the observed MTNR1B rs10830963 gene-dietary fat interaction are unknown, there is evidence that a high-fat diet could alter the expression and the rhythmic mRNA expression levels of circadian-clock genes and circadian clock-controlled lipogenic genes (20–22). For example, Sun et al. (22) observed a rhythmic expression of the clock-controlled output gene, Ppar-α, and downstream lipid metabolism genes (Srebp-1c, Fas, and Acc1) in normally fed mice. Meanwhile, when mice were fed with a high-fat diet, the rhythmic expression in the liver of such genes was significantly altered. Moreover, the effects of the genotype on changes in lipid metabolism traits showed opposite trends in participants with low-fat versus high-fat intake. The results are in line with the “differential susceptibility hypothesis,” which proposes that vulnerability genes or risk alleles may function like plasticity genes because genetic risk can be modified by environmental exposures, including dietary factors (23–26). In other words, some individuals might be more responsive to environmental influences in a “for-better-and-for-worse” manner because of the genetic background (24). Consistent with this hypothesis, we observed that carriers of the risk allele might function as either a protective or a detrimental factor, depending on the differences in dietary fat intake. In the present study, we failed to ascertain a genotype-diet interaction on changes in TGs and HDL cholesterol. The reason for this differential genetic effect on various lipid phenotypes is unclear. However, it can be hypothesized that different mechanisms might drive the reported gene-diet interactions for changes on different lipid components.
Our data indicate that the gene-diet interaction on changes in total cholesterol and LDL cholesterol remained statistically significant after additional adjustment for weight loss, although a slight attenuation of the effect was observed on changes in total cholesterol. These results suggest that the effects on changes in total cholesterol and LDL cholesterol concentrations might be independent of weight loss, although weight reduction has been shown to induce beneficial effects on lipid profile, mainly on total cholesterol and LDL cholesterol (27). However, in our study, weight loss was correlated with TG levels and HDL cholesterol, but not with total cholesterol and LDL cholesterol, at 2 years of the intervention (28).
To the best of our knowledge, this is the first study to analyze interactions between a MTNR1B genetic variant and dietary fat intake on changes in plasma lipids in a large and long-term dietary intervention trial. These findings add novel insights into the role of the circadian rhythm-related MTNR1B genetic variant on metabolic responses (16). However, the present study has some limitations that should be addressed. We did not measure circulating melatonin levels in the study population, which prevented the potential analysis of the relationship between the genetic variant and circulating melatonin levels. Nonetheless, according to the Mendelian randomization principle, a genetic variant could be a surrogate for the biomarker in causal inference, because it is less likely to be affected by confounding and reverse causation (29). In addition, it was difficult to determine which macronutrient played the key role of the observed interactions because the low-fat intake was characterized by high-carbohydrate intake and vice versa, to maintain energy balance. Finally, the results should be replicated in order to be extended to other ethnic groups because most of the participants were whites (around 80%), and to rule out the possibility of false positive findings.
In summary, these results suggest that carriers of the G allele of the circadian rhythm-related MTNR1B rs10830963 genetic variant may benefit more in the improvement of their lipid profile by choosing a low-fat diet instead of a high-fat diet. These findings may lend support to personalized dietary interventions in improvement of lipid metabolism.
Supplementary Material
Acknowledgments
The authors thank all participants of the study for their dedication and contribution to the research.
Footnotes
This work was supported by National Heart, Lung, and Blood Institute Grants HL071981, HL034594, and HL126024; National Institute of Diabetes and Digestive and Kidney Diseases Grants DK091718, DK100383, and DK078616; Boston Obesity Nutrition Research Center Grant DK46200; and United States-Israel Binational Science Foundation Grant 2011036. L.G. is a recipient of predoctoral and mobility grants from the Spanish Ministry of Education, Culture, and Sport. Y.H. is a recipient of a Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science. L.Q. was a recipient of American Heart Association Scientist Development Award 0730094N. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Kumar Jha P., Challet E., and Kalsbeek A.. 2015. Circadian rhythms in glucose and lipid metabolism in nocturnal and diurnal mammals. Mol. Cell. Endocrinol. 418: 74–88. [DOI] [PubMed] [Google Scholar]
- 2.Gooley J. J. 2016. Circadian regulation of lipid metabolism. Proc. Nutr. Soc. 75: 440–450. [DOI] [PubMed] [Google Scholar]
- 3.Bailey S. M., Udoh U. S., and Young M. E.. 2014. Circadian regulation of metabolism. J. Endocrinol. 222: R75–R96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Obayashi K., Saeki K., Iwamoto J., Okamoto N., Tomicka K., Nezu S., Ikada Y., and Kurumatani N.. 2013. Exposure to light at night, nocturnal urinary melatonin secretion, and obesity/dyslipidemia in the elderly: a cross-sectional analysis of the HEIJO-KYO study. J. Clin. Endocrinol. Metab. 98: 337–344. [DOI] [PubMed] [Google Scholar]
- 5.Turek F. W., Joshu C., Kohsaka A., Lin E., Ivanova G., McDearmon E., Laposky A., Losee-Olson S., Easton A., Jensen D. R., et al. 2005. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 308: 1043–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cipolla-Neto J., Amaral F. G., Afeche S. C., Tan D. X., and Reiter R. J.. 2014. Melatonin, energy metabolism, and obesity: a review. J. Pineal Res. 56: 371–381. [DOI] [PubMed] [Google Scholar]
- 7.Sun H., Huang F., and Qu S.. 2015. Melatonin: a potential intervention for hepatic steatosis. Lipids Health Dis. 14: 75–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dubocovich M. L., Delagrange P., Krause D. N., Sugden D., Cardinali D. P., and Olcese J.. 2010. International Union of Basic and Clinical Pharmacology. LXXV. Nomenclature, classification, and pharmacology of G protein-coupled melatonin receptors. Pharmocol. Rev. 62: 343–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gamble K. L., Berry R., Frank S. J., and Young M. E.. 2014. Circadian clock control of endocrine factors. Nat. Rev. Endocrinol. 10: 466–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lane J. M., Chang A. M., Bjonnes A. C., Aeschbach D., Anderson C., Cade B. E., Cain S. W., Czeisler C. A., Gharib S. A., Gooley J. J., et al. 2016. Impact of common diabetes risk variant in MTNR1B on sleep, circadian, and melatonin physiology. Diabetes. 65: 1741–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tuomi T., Nagorny C. L. F., Singh P., Bennet H., Yu Q., Alenkvist I., Isomaa B., Östman B., Söderström J., Pesonen A. K., et al. 2016. Increased melatonin signaling is a risk factor for type 2 diabetes. Cell Metab. 23: 1067–1077. [DOI] [PubMed] [Google Scholar]
- 12.DeMenna J., Puppala S., Chittoor G., Schneider J., Kim J. Y., Shaibi G. Q., Mandarino L. J., Duggirala R., and Coletta D. K.. 2014. Association of common genetic variants with diabetes and metabolic syndrome related traits in the Arizona insulin resistance registry: a focus on Mexican American families in the southwest. Hum. Hered. 78: 47–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dashti H. S., Follis J. L., Smith C. E., Tanaka T., Garaulet M., Gottlieb D. J., Hruby A., Jacques P. F., Kiefte-De Jong J. C., Lamon-Fava S., et al. 2015. Gene-environment interactions of circadian-related genes for cardiometabolic traits. Diabetes Care. 38: 1456–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sacks F. M., Bray G. A., Carey V. J., Smith S. R., Ryan D. H., Anton S. D., McManus C., Champagne C. M., Bishop L. M., Laranjo N., et al. 2009. Comparison of weight-loss diets with different compositions of fat, protein, and carbohydrates. N. Engl. J. Med. 360: 859–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedewald W. T., Levy R. I., and Fredrickson D. S.. 1972. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin. Chem. 18: 499–502. [PubMed] [Google Scholar]
- 16.Mirzaei K., Xu M., Qi Q., De Jonge L., Bray G. A., Sacks F., and Qi L.. 2014. Variants in glucose- and circadian rhythm-related genes affect the response of energy expenditure to weight-loss diets: the POUNDS LOST trial. Am. J. Clin. Nutr. 99: 392–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gooley J. J., and Chua E. C.. 2014. Diurnal regulation of lipid metabolism and applications of circadian lipidomics. J. Genet. Genomics. 41: 231–250. [DOI] [PubMed] [Google Scholar]
- 18.Koziróg M., Poliwczak A. R., Duchnowicz P., Koter-Michalak M., Sikora J., and Broncel M.. 2011. Melatonin treatment improves blood pressure, lipid profile, and parameters of oxidative stress in patients with metabolic syndrome. J. Pineal Res. 50: 261–266. [DOI] [PubMed] [Google Scholar]
- 19.Kadhim H. M., Ismail S. H., Hussein K. I., Bakir I. H., Sahib A. S., Khalaf B. H., and Hussain S. A. R.. 2006. Effects of melatonin and zinc on lipid profile and renal function in type 2 diabetic patients poorly controlled with metformin. J. Pineal Res. 41: 189–193. [DOI] [PubMed] [Google Scholar]
- 20.Yanagihara H., Ando H., Hayashi Y., Obi Y., and Fujimura A.. 2006. High-fat feeding exerts minimal effects on rhythmic mRNA expression of clock genes in mouse peripheral tissues. Chronobiol. Int. 23: 905–914. [DOI] [PubMed] [Google Scholar]
- 21.Hsieh M. C., Yang S. C., Tseng H. L., Hwang L. L., Chen C. T., and Shieh K. R.. 2010. Abnormal expressions of circadian-clock and circadian clock-controlled genes in the livers and kidneys of long-term, high-fat-diet-treated mice. Int. J. Obes. (Lond). 34: 227–239. [DOI] [PubMed] [Google Scholar]
- 22.Sun L., Wang Y., Song Y., Cheng X. R., Xia S., Rahman M. R. T., Shi Y., and Le G.. 2015. Resveratrol restores the circadian rhythmic disorder of lipid metabolism induced by high-fat diet in mice. Biochem. Biophys. Res. Commun. 458: 86–91. [DOI] [PubMed] [Google Scholar]
- 23.Belsky J., Jonassaint C., Pluess M., Stanton M., Brummett B., and Williams R.. 2009. Vulnerability genes or plasticity genes? Mol. Psychiatry. 14: 746–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Belsky J., and Hartman S.. 2014. Gene-environment interaction in evolutionary perspective: differential susceptibility to environmental influences. World Psychiatry. 13: 87–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hartman S., and Belsky J.. 2016. An evolutionary perspective on family studies: differential susceptibility to environmental influences. Fam. Process. 55: 700–712. [DOI] [PubMed] [Google Scholar]
- 26.Dalle Molle R., Fatemi H., Dagher A., Levitan R. D., Silveira P. P., and Dubé L.. 2017. Gene and environment interaction: is the differential susceptibility hypothesis relevant for obesity? Neurosci. Biobehav. Rev. 73: 326–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poobalan A., Aucott L., Smith W. C. S., Avenell A., Jung R., Broom J., and Grant A.. 2004. Effects of weight loss in overweight/obese individuals and long-term lipids outcomes a systematic review. Obes. Rev. 5: 43–50. [DOI] [PubMed] [Google Scholar]
- 28.Zhang X., Qi Q., Bray G. A., Hu F. B., Sacks F. M., and Qi L.. 2012. APOA5 genotype modulates 2-y changes in lipid profile in response to weight-loss diet intervention: the Pounds Lost trial. Am. J. Clin. Nutr. 96: 917–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qi L. 2009. Mendelian randomization in nutritional epidemiology. Nutr. Rev. 67: 439–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.