Abstract
The MYB gene family constitutes one of the largest transcription factors (TFs) modulating various biological processes in plants. Although genome-wide analysis of this gene family has been carried out in some species, only three MYB members have been functionally characterized heretofore in sesame (Sesamum indicum L.). Here, we identified a relatively high number (287) of sesame MYB genes (SIMYBs) with an uncommon overrepresentation of the 1R-subfamily. A total of 95% of SIMYBs was mapped unevenly onto the 16 linkage groups of the sesame genome with 55 SIMYBs tandemly duplicated. In addition, molecular characterization, gene structure, and evolutionary relationships of SIMYBs were established. Based on the close relationship between sesame and Arabidopsis thaliana, we uncovered that the functions of SIMYBs are highly diverse. A total of 65% of SIMYBs were commonly detected in five tissues, suggesting that they represent key TFs modulating sesame growth and development. Moreover, we found that SIMYBs regulate sesame responses to drought and waterlogging, which highlights the potential of SIMYBs towards improving stress tolerance in sesame. This work presents a comprehensive picture of the MYB gene family in sesame and paves the way for further functional validation of the members of this versatile gene family.
Keywords: abiotic stress, gene expression, MYB, plant growth and development, Sesamum indicum, transcription factors
1. Introduction
Transcription factors (TFs) are important regulators of the expression of functional genes under different biological processes, including development, reproduction, and various environmental conditions [1]. Within the landscape of TFs that are present in the genome, the MYB family constitutes one of the largest and functionally diverse [2]. The MYB proteins are characterized by the MYB domain at the N-terminus which encodes 52 amino acid residues and contains one to four imperfect tandems repeats (R) [2]. According to the numbers of MYB repeat, MYB genes are classified into four different subfamilies, namely MYB –related proteins or 1R–MYB proteins (with 1R), R2R3–MYB proteins (with 2R), R1R2R3-MYB proteins (with 3R), and 4R-MYB proteins (with 4R) [3]. The R1R2R3-MYB proteins are found in animals, but the R2R3-MYB proteins are more frequent in plants [4]. The MYB TFs have been studied in diverse species and particularly in plants. Various numbers of MYBs have been detected suggesting that they have expanded to differing degrees: 177 MYB genes were found in Citrus sinensis [5], 231 in Pyrus bretschneideri [1], 475 in Brassica rapa ssp. pekinensis [6], 524 in Gossypium hirsutum [7], and 104 and 166 in Lotus japonicas and Medicago truncatula, respectively [8].
In plants, MYB proteins are key factors in physiological processes, shoot growth, root formation, organ development, metabolism, hormone signal transduction, and responses to biotic and abiotic stress tolerance [9,10,11,12,13]. For example, expression profiles analysis in Arachis hypogaea identified 30 MYB genes responsive to abiotic stress treatments [14]. Over-expression of SpMYB gene from Solanum pimpinellifolium improved abiotic and biotic stress resistance in tobacco [15]. A total of 44.67% and 47.21% MYB genes were found up- and down-regulated in Arabidopsis under cold stress, respectively [16]. Later on, Zhang et al. [17] reported the gene TaLHY, a 1R-MYB type as a positive regulator of resistance against stripe rust fungus and ear heading in Triticum aestivum. More recently, Xu et al. [18] demonstrated that the R2R3-type MYB gene DcMYB6, is involved in regulating anthocyanin biosynthesis in purple Daucus carota taproots. In the case of drought stress, many MYB genes have been isolated and demonstrated to be involved in drought responses in plants [19]. The transcriptional activation of cuticular wax biosynthesis by MYB96 contributed to drought resistance in Arabidopsis thaliana [20]. A recent study revealed that PbrMYB21 plays a positive role in drought tolerance by modulating polyamine synthesis through the regulation of the ADC expression in Pyrus betulaefolia [21]. Furthermore, Yin et al. [22] showed that OsMYBR1 that was isolated from Oryza sativa improves drought tolerance of transgenic plants through the up-regulation of stress-related genes and the accumulation of osmoprotectants. Similarly, Butt et al. [23] demonstrated that GaMYB85 confers good drought tolerance in Gossypium arboreum, most probably via an ABA-induced pathway. Altogether, these evidences demonstrated the versatility and importance of this gene family in plants.
Sesame (Sesamum indicum L.) is one of the oldest oilseed crops, which is widely grown in tropical and subtropical areas [24]. It has one of the highest oil contents (~55%) among major oilseed crops [25]. Drought and waterlogging represent the most important abiotic stresses that affect sesame plant growth and yield [26]. Sesame is a very sensitive crop to waterlogging [27]. The majority of sesame resources are susceptible to waterlogging damage at the different developmental stages [28], and so far, few studies have been conducted to unravel the genetic basis of waterlogging response in sesame [27,29,30]. In contrast, sesame is moderately tolerant to drought stress [31]. However, severe drought adversely impairs the plant normal growth and development [32], the reproduction and yield [26], and both the quality and quantity of the sesame oil as well [33,34,35]. Therefore, breeding sesame varieties with higher tolerance to these abiotic stresses is drawing great attention from breeders [36].
Given the potential roles of MYB proteins in the regulation of gene expression in response to environmental stresses, it is of the utmost interest to perform a genome-wide survey of this gene family in sesame. In this study, we identified at the genome-wide level 287 MYB genes and revealed that the MYB proteins are highly active in growth and adaptation to the major abiotic stresses namely drought and waterlogging in sesame. We also presented here a comprehensive analysis of the protein characteristics, gene classification and structure, chromosomal distribution, gene duplication, and phylogenetic relationships of sesame MYBs.
2. Materials and Methods
2.1. Identification and Sequence Analysis of MYB Proteins in Sesame
The sesame genome sequence was downloaded from Sinbase (http://ocri-genomics.org/Sinbase/) [37]. The HMM profile of MYB DNA-binding domain (PF00249) was downloaded from Pfam protein families database (http://pfam.sanger.ac.uk/) and was exploited for the identification of MYB genes from Sesamum indicum L. genome, using HMMER3 tool of the software Unipro UGENE (Unipro, Novosibirsk, Russia) [38]. To confirm the presence of MYB domain, the PROSITE (http://prosite.expasy.org/scanprosite/) and the SMART program (http://smart.embl-heidelberg.de/) [39] were used for further analysis. The isoelectric point and protein molecular weight of MYB proteins in sesame were obtained using ExPASy proteomics server (http://www.expasy.ch/tools/protparam.html). In addition, the sequences of 125 Arabidopsis R2R3-MYB proteins were downloaded from The Arabidopsis Information Resource (TAIR; http://www.arabidopsis.org/).
2.2. Chromosomal Location and Gene Duplication of MYB Genes in Sesame
The MYB genes were mapped onto the 16 linkage groups (LGs) of the sesame genome on the basis of their genomic location from Sinbase using the software MapChart 2.3 (Wageningen UR, Wageningen, Netherlands) [40]. The tandemly repeated MYB genes were identified when two sesame MYB genes were separated by three or fewer genes [41].
2.3. Phylogenetic Analysis and Functional Classification of MYB Proteins in Sesame
To characterize the evolutionary features of MYB proteins, we extracted all of the predicted MYB protein sequences from Sinbase (http://ocri-genomics.org/Sinbase/). According to the numbers of the conserved domains, we classified the sesame MYB genes into four subfamilies: R1-(single domain), R1R2-(2 domains), R1R2R3-(3 domains), and Atypical MYB (4 or more domains) [2]. Multiple alignments of their conserved domains were conducted using ClustalW program that was embedded in the MEGA6.0 software (Temple University, Philadelphia, PA, USA) [42], with a gap open penalty of 10 and gap extension penalty of 0.2. Thereafter, a Neighbor-Joining (NJ) tree was constructed with a 1000 bootstrap value. The tree was loaded along with the information of the gene CDS organization into the Gene Structure Display Server (GSDS2.0) (http://gsds.cbi.pku.edu.cn/) [43] to construct the exon-intron map.
Moreover, a Neighbour-Joining (NJ) tree was constructed from ClustalW aligned full-length of 259 R2R3-MYB proteins (125 MYB of Arabidopsis and 134 R2R3 MYB proteins for sesame) using MEGA6.0 software (Temple University, Philadelphia, PA, USA) [42] with the settings described above. Functional classification of the R2R3-MYB genes in sesame was performed according to their phylogenetic relationships with the corresponding Arabidopsis AtMYB genes.
To identify the orthologous genes of the sesame MYBs, we conducted a BLASTp search of their protein sequences against the protein database of Arabidopsis in TAIR. Hits with E-value ≤ 1e−40 and at least 70% similarity were considered significant [44]. The relationships between the orthologous genes of sesame and Arabidopsis MYBs were illustrated using Circos package [45].
2.4. Analysis of the Expression Patterns of Sesame MYB Genes in Different Tissues, under Drought and Waterlogging Stresses Using RNA-seq Data
To analyze the expression patterns of sesame MYB genes in various sesame tissues, we used RNA-seq data from root, stem seed, capsule, and leaf developed by our group and accessible from sesameFG (http://www.ncgr.ac.cn/SesameFG; [46]). Moreover, we investigated the expression levels of sesame MYB genes in responses to the two major abiotic stresses: waterlogging and drought. Transcriptomic data from a tolerant cultivar (Zhongzhi No. 13) under 3, 9, and 15 h of waterlogging stress were retrieved from GeneBank with the SRA accession number SRR2886790 [30]. Concerning the drought stress, the RNA-seq data of a drought tolerant genotype (ZZM0635) submitted to 3 (d1), 7 (d2) and 11 (d3) days of drought periods [47,48] were downloaded from http://www.ncbi.nlm.nih.gov/bioproject/PRJNA356988. The read per kilobase per million mapped reads (RPKM) values were log2 transformed and used for the construction of heatmaps with the MEV Software (J. Craig Venter Institute Inc., CA, USA) [49].
2.5. Plant Materials and Stress Treatments
To analyze the changes in expression of sesame MYB genes under drought and waterlogging stresses, two experiments were concomitantly conducted. The cultivars ZZM0635 and Zhongzhi No. 13, both obtained from the China National Genebank, Oil Crops Research Institute, Chinese Academy of Agricultural Sciences, were grown in pots for drought and waterlogging experiments, respectively. A three-days water withholding was applied to half of the seedlings when plants entered the early flowering stage with the soil volumetric water content falling from 35% to 10% following descriptions of Dossa et al. [47,48]. For the waterlogging application, the pots were flooded by standing in a plastic bucket filled with tap water to 3 cm above the soil surface and maintained for 9 h according to the experimental descriptions described by Wang et al. [30].
The roots of the seedlings were harvested after stress application for both stressed and control plants. The samples were harvested from three individual plants and were snap-frozen in liquid nitrogen and stored at −80 °C until further use.
2.6. RNA Isolation and Quantitative RT-PCR
Total RNAs from various tissues samples were extracted with Easy Spin RNA kit (Aidlab, Beijing, China). The quantity and quality of RNA samples were assessed by agarose gel electrophoresis and the spectrophotometer (Unico SpectroQuest Model SQ2800) measurement of the A260/A280 ratio. For cDNA synthesis, 1.5 μg of RNA was reverse transcribed using the Superscript III reverse transcription kit (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s instructions. Gene-specific primers were designed by using Primer5.0 [50]. We designed primers to amplify 29 SIMYB genes using Primer Premier 5.0 software (Premier Biosoft International, Palo Alto, CA, USA) (Table S5). The qRT-PCR was conducted on Roche Lightcyler® 480 instrument using SYBR Green Master Mix (Vazyme), according to the manufacturer’s protocol. We performed each reaction using a 20 µL mixture containing 10 µL of 2xChamQ SYBR qPCR Master Mix, 8 µL of nuclease-free water, 2.4 µL of each primer (10μM), and 2 µL of diluted cDNA. All of the reactions were run in 96-well plates and each cDNA was analyzed in triplicate. The reactions were performed with the following cycling profile: 95 °C for 30 s, followed by 40 cycles of 95 °C/10 s, 60 °C/30 s. Each reaction was performed in biological triplicates, and the relative gene expression values were calculated using the 2−ΔΔCt method, as described by [51]. The sesame Actin gene (SIN_1006268) was used as the internal reference gene [52]. For stress related genes, they were considered significantly up- and down-regulated when their relative expression fold change (stress/control) were >2 and <0.5, respectively [53].
3. Results
3.1. Identification of Sesame MYB Genes and Analysis of Their Sequence Features
We identified in the sesame genome, 287 sequences that contain MYB or MYB-like repeats. We followed the united nomenclature for Sesamum indicum genes and used SIMYB (for S. indicum MYB) to represent MYB genes from sesame.
All of the putative MYB genes were further confirmed by SMART and PROSITE searches for the presence of the two MYB repeats. Finally, we confirmed 287 typical MYB genes in the whole genome of sesame, including 134 R2R3-MYB, 5 R1R2R3-MYB, 147 MYB related, and 1 atypical MYB genes. The MYB related genes represent more than half of the total MYB genes, and thus constitute the largest subfamily of MYB genes in sesame (Table 1).
Table 1.
Classification of the MYB gene family and distribution across the linkage groups (LG) of the sesame genome.
| Linkage Groups | R1-MYB | R1R2-MYB | R1R2R3-MYB | Atypical-MYB | Total |
|---|---|---|---|---|---|
| LG01 | 12 | 8 | 1 | 1 | 22 |
| LG02 | 11 | 16 | 27 | ||
| LG03 | 17 | 15 | 1 | 33 | |
| LG04 | 12 | 9 | 21 | ||
| LG05 | 10 | 8 | 1 | 19 | |
| LG06 | 11 | 10 | 1 | 22 | |
| LG07 | 5 | 8 | 13 | ||
| LG08 | 19 | 9 | 28 | ||
| LG09 | 8 | 7 | 15 | ||
| LG10 | 7 | 7 | 14 | ||
| LG11 | 10 | 7 | 17 | ||
| LG12 | 6 | 4 | 10 | ||
| LG13 | 1 | 4 | 5 | ||
| LG14 | 1 | 3 | 4 | ||
| LG15 | 7 | 10 | 17 | ||
| LG16 | 4 | 3 | 7 | ||
| Scaffold | 6 | 6 | 1 | 13 | |
| Total | 147 | 134 | 5 | 1 | 287 |
| Percentage (%) | 51.22 | 46.69 | 1.74 | 0.35 |
The isoelectric point (pI) values for SIMYB proteins ranged from 4.73 (SIMYB3) to 9.91 (SIMYB240). The molecular weight (Mw) was from 9208.63 kDa (SIMYB22) to 73269.43 kDa (SIMYB18) (Table S1).
3.2. Chromosomal Distribution and Duplication Events among Sesame MYB Genes
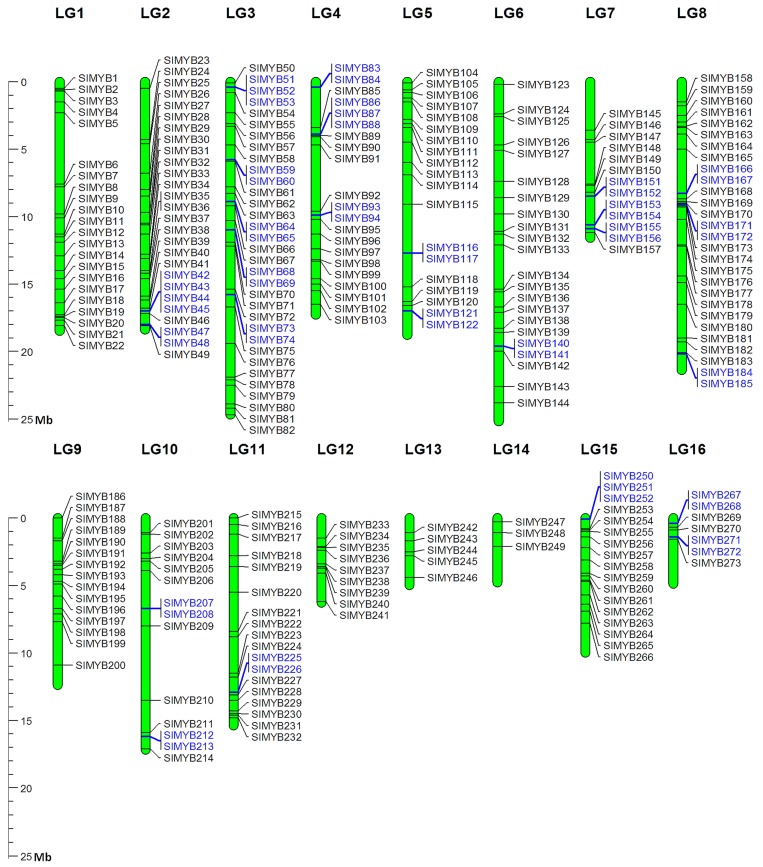
Genome chromosomal location analyses showed that the sesame MYB genes were distributed throughout all of the 16 linkage groups (LG) (Figure 1). In the currently released genome sequences, 273 SIMYB genes (95% of the total SIMYBs) were successfully mapped onto the LGs, whereas 14 genes were remained on as yet unmapped scaffolds. All of the SIMYB genes were named from SIMYB1 to SIMYB287 following their order on the LGs and unanchored sequences. The anchored SIMYB genes were unevenly distributed onto the 16 LGs of the sesame genome. The LG03 encompassed the largest number of SIMYBs (33 genes), followed by 28 genes on LG08. In contrast, only three SIMYB genes were detected on LG14.
Figure 1.
Mapping of sesame MYB genes based on their physical positions. Vertical green bars represent linkage groups (LG) of the sesame genome. The LG numbers are indicated at the top of each LG. The genes colored in blue are the tandemly duplicated SIMYBs. The scale on the left is in megabases (Mb).
We detected 55 genes (20% of the anchored SIMYBs) that were involved in tandem duplicated arrays with sizes ranging from two to four SIMYBs on the LG02, LG03, LG04, LG05, LG06, LG07, LG08, LG10, LG11, LG15, and LG16 (Figure 1).
3.3. Phylogenetic Relationships, Classification and Exon-Intron Structures of SIMYB Gene Family in Sesame
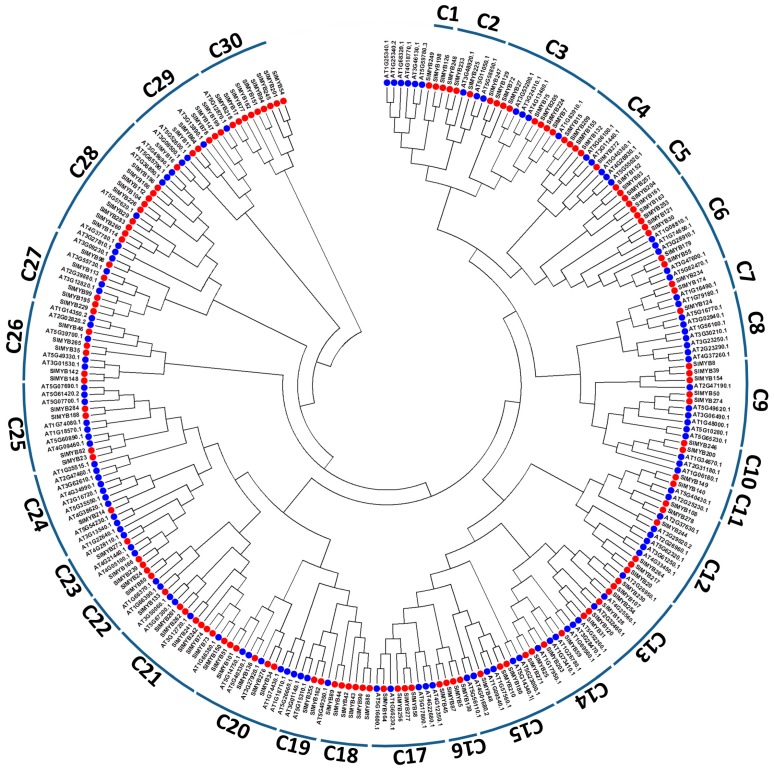
Using the Arabidopsis R2R3-MYB proteins as reference for classification, we subdivided the SIMYB members into 30 subgroups (designated as C1–C30 in this study) according to the sequence similarity and topology (Figure 2; Table S2). Most of the subgroups in our classification were in accordance with those in Arabidopsis, and only seven subgroups (C1, C5, C11, C16, C18, C29, and C30) were not retrieved in the phylogenetic tree of Arabidopsis MYB proteins. According to the functions described for Arabidopsis R2R3-MYB subgroups [2], we therefore inferred the functions of the corresponding subgroups of R2R3-MYB in sesame (Table S2).
Figure 2.
Phylogenetic relationships of the R2R3-MYB subfamily between sesame and Arabidopsis. The subgroups were designated as C1 to C30 following the classification in Arabidopsis [2]. Sesame SIMYBs are represented by the red dot whereas Arabidopsis MYB are labeled by a blue dot.
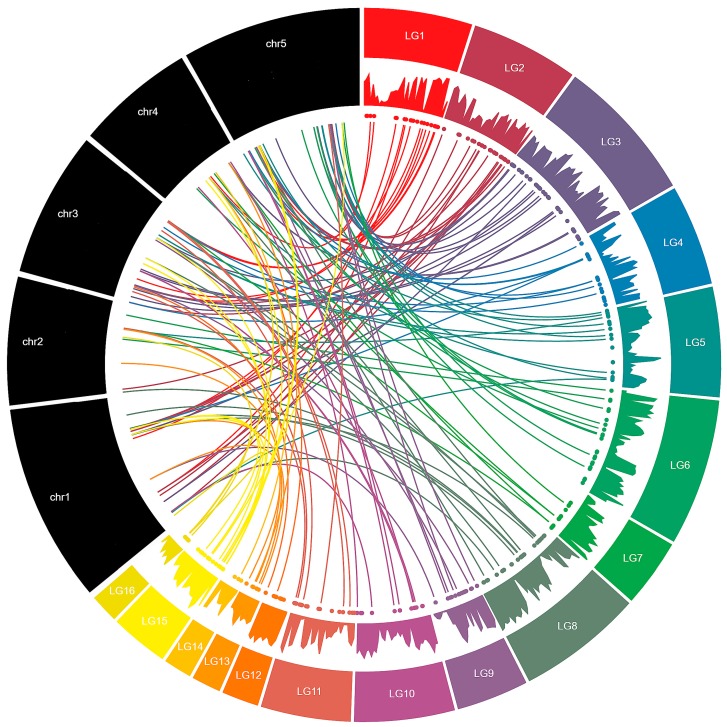
Synteny analysis of SIMYB genes when compared to those of Arabidopsis could provide more functional insight. Comparative orthologous mapping of MYB genes between the two species revealed that 133 SIMYBs are orthologs to Arabidopsis MYB genes across the 16 LGs of the sesame genome. The LG03 and LG02 exhibited the highest numbers of orthologous genes (16 and 15 gene pairs, respectively) while only one gene pair was detected on LG14. Moreover, Arabidopsis MYB genes retained from 1 to 6 copies of SIMYBs while conversely, no gene in sesame has conserved more than a single copy in Arabidopsis genome (Figure 3). Based on the functions described for the Arabidopsis MYB genes [1,2], we assigned various functions to the orthologous SIMYBs (Table S3).
Figure 3.
Syntenic relationships between MYB genes in sesame and Arabidopsis genomes. The black bars represent the chromosomes of Arabidopsis whereas the colored bars represent the linkage groups of the sesame genome. The numbers LG01–16 represent linkage groups within the sesame genome and the five Arabidopsis chromosomes are labeled Chr1–Chr5. The area chart displays the gene density along the LGs in the sesame genome. The colored dots represent the locations of the SIMYB genes along the LGs. Colored lines indicate orthologous genes in sesame and Arabidopsis.
The exon/intron structure analysis for the 287 SIMYBs indicated that most of the coding sequences were disrupted by introns, with exception of 16 genes (Figure S1). The numbers of exon of the 287 SIMYB genes ranged from 1 to 31. In addition, we identified five genes that were possessing high numbers of exons from 16 to 31 (SIMYB70, SIMYB83, SIMYB187, SIMYB197, and SIMYB135). The clustering pattern of SIMYBs was not obviously consistent with the exon/intron structures. However, in few cases, some of the genes in the same cluster exhibited a similar exon/intron structure. For example, the intronless genes SIMYB119, SIMYB32, SIMYB77, SIMYB190, and SIMYB182, all clustered together on the phylogenetic tree. Moreover, clustering patterns of SIMYB genes did not strictly follow their subfamily’s classification (Figure S1).
3.4. Expression Patterns of SIMYB Genes in Different Tissues of Sesame
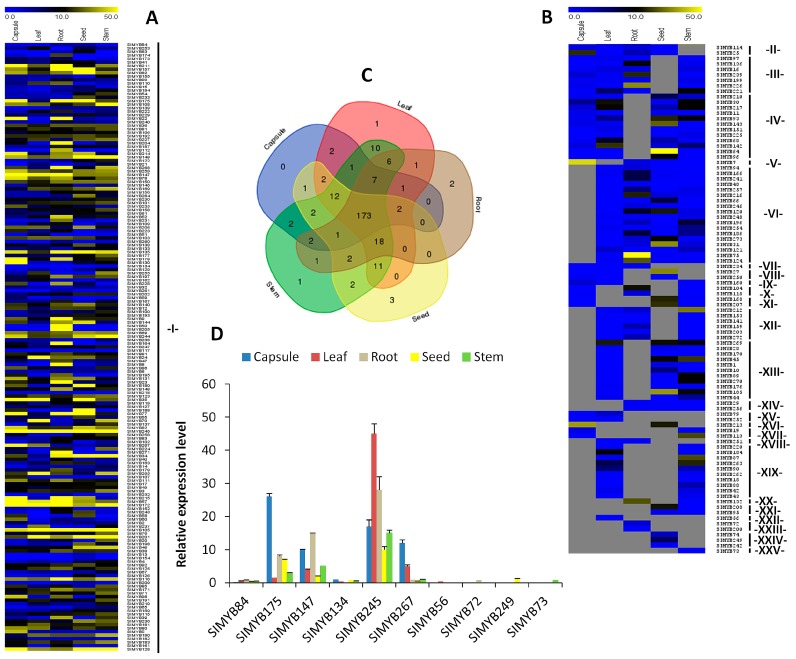
The gene expression pattern can provide important clues for its function. To explore the expression pattern of MYB gene family in sesame development and growth, we investigated the relative expression level of SIMYBs in five tissues, including root, capsule, leaf, stem, and seed. The results revealed that SIMYB genes have a variety of expression patterns in different tissues of sesame (Figure 4), and could be classified into 25 groups (I to XXV) according to their tissue-specificity expression (Table S4). A total of 268 SIMYB genes were expressed (RPKM > 1) in the different tissues, although their transcript abundance was highly varied according to the tissues. Of these expressed genes, 173 SIMYBs (~65%) were commonly detected in all of the tissues, suggesting that they regulate sesame development and growth (Figure 4a). We found some highly expressed genes (SIMYB69, SIMYB106, SIMYB171, SIMYB175, SIMYB245, etc.) in the tissues, some low expressed genes (SIMYB84, SIMYB134, etc.), and also found some tissue-specific SIMYBs. For instance, the gene SIMYB73 was only expressed in the stem; two genes, including SIMYB72 and SIMYB200 were exclusively expressed in the sesame root; one gene in the leaf (SIMYB56), three genes (SIMYB74, SIMYB242, and SIMYB249) were expressed in the seed and one gene (SIMYB19) was exclusively expressed in the capsule (Figure 4b,c; Table S4). To further validate the gene expression profile from RNA-seq data, we performed qRT-PCR analysis with ten selected genes, including six genes with high or low expression levels from the group I and one tissue-specific gene each from the groups XXII, XXIII, XIV, and XXV. For all of the genes tested, qRT-PCR results confirmed well the RNA-seq expression profiles, proving the accuracy of the RNA-seq dataset that was used in this study (Figure 4d).
Figure 4.
Expression profile analysis of MYB genes in five sesame tissues. The SIMYB genes expressed (RPKM > 1) in leaf, stem, root, seed and capsule were represented according to their tissue specificity: (A) SIMYBs commonly expressed in the five tissues; (B) SIMYBs expressed in only some tissues; (C) Venn diagram exhibiting the numbers of common and unique SIMYB genes expressed in the various sesame tissues; and, (D) Validation through qRT-PCR of the expression patterns of ten selected genes. Gray cells represent genes not expressed. -I- to -XXV- represent the groups of genes according to their tissue specificity (Table S4).
3.5. Expression Profiles of SIMYB Genes in Response to Waterlogging Stress
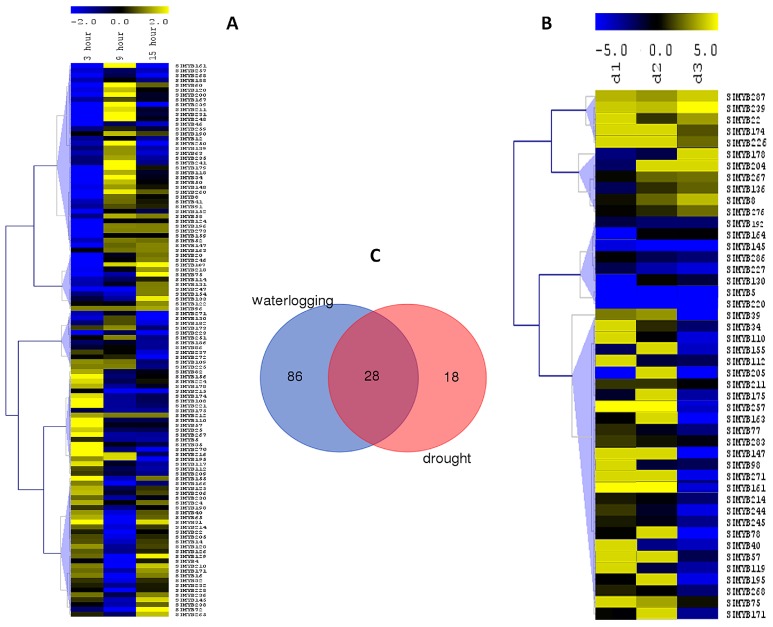
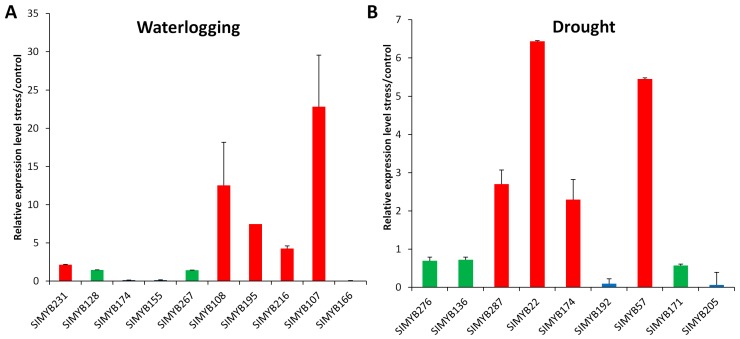
The expression of SIMYB genes under waterlogging stress was examined at 3, 9, and 15 h after treatment. In total, we identified 114 SIMYBs as differentially expressed genes (DEGs) with various expression patterns across the three time points of waterlogging stress application (Figure 5a). The observed expression patterns revealed that SIMYB genes could be categorized into five main groups. The group I was composed of 39 genes, which were strongly induced at 9 h of stress application. The group II gathered together 12 genes which were induced when the stress became more intense (15 h). The group III encompassed 12 genes induced at 3 and 9 h, but their expression levels were significantly decreased at 15 h under stress. The group IV included 22 genes that were up-regulated at the beginning of the stress application (3 h), but their expression levels decreased with an increasing level of stress. Finally, the group V was composed of 29 down-regulated genes with their expression levels significantly down-regulated at 9 h under stress but induced at the beginning of the stress (3 h) and under severe stress (15 h). The RNA-seq data was further verified by qRT-PCR experiment which was performed on root samples of a waterlogging-tolerant cultivar Zhongzhi No.13 based on ten selected SIMYB genes differentially expressed (Figure 6a). A total of five genes were detected up-regulated with the gene SIMYB107, the most induced one (>22-fold increase in gene expression). In contrast, the genes SIMYB166, SIMYB155, and SIMYB174 were down-regulated under waterlogging, as revealed by the RNA-seq data.
Figure 5.
Transcriptome profiling of MYB gene expression under waterlogging and drought stress. (A) Heatmap displaying expression changes of differentially expressed MYB genes in sesame plants stressed for 3 h, 9 h and 15 h of waterlogging compared with the control; (B) Heatmap displaying expression changes of differentially expressed MYB genes in sesame plants stressed for 3 days (d1), 7 days (d2), and 11 days (d3) of drought compared with the control; and, (C) Venn diagram depicting the shared and stress-specific differentially expressed genes between waterlogging and drought.
Figure 6.
MYB gene induction rates in sesame roots during abiotic stress treatments. (A) 9 h of waterlogging; (B) 3 days of drought. Transcripts abundance was quantified through qRT-PCR and the experimental values were normalized using sesame Actin as reference gene. The histograms represent the relative expression values of induction rates (stress/control). The green bars represent the no significantly regulated genes, the red bars represent the genes significantly up-regulated (induction rate > 2) and the blue bars represent the significantly down-regulated genes (induction rate < 2).
3.6. Expression Profiles of SIMYB Genes in Response to Drought Stress
Previously developed RNA-seq data under drought stress [42,43] allowed for us to analyze the expression profiles of SIMYBs under this important abiotic stress. First, we retrieved all SIMYBs from the DEGs at d1, d2, and d3 when compared to control (period prior to water stress application). A total of 46 SIMYBs were found as DEGs across the different drought application periods, and their expression profiles were subsequently summarized in the heatmap represented in the Figure 5b.
The results showed that SIMYBs regulating drought responses could be assembled into four clusters: The first group (I) represented five genes constitutively up-regulated under drought stress with a striking induction at d3. The group II gathered together six genes with increased activity under severe stresses (d2 and d3). The group III was composed of eight genes constitutively down-regulated under drought stress. Finally, the group IV encompassed 27 genes highly induced at d1 or d2, but down-regulated under severe drought stress (d3).
To further confirm the transcriptomic expression patterns of SIMYBs, we selected nine SIMYBs and performed qRT-PCR analysis in the drought tolerant genotype after three days of drought stress. Among these genes, we found four SIMYBs, including SIMYB22, SIMYB57, SIMYB174, and SIMYB287, which were significantly up-regulated in response to drought, with SIMYB22 being the most induced gene. The genes SIMYB192 and SIMYB205, in contrast, were down-regulated in response to drought stress exactly, as shown in the RNA-seq results. However, we found three SIMYB genes (SIMYB136, SIMB171, and SIMYB267) that were not significantly regulated under drought stress, as their expression levels in the stressed plants when compared to the control ones remained unaffected (Figure 6b).
By crossing the significantly regulated SIMYB genes under drought and waterlogging stresses, we found that a total of 132 genes (46% of the total SIMYB genes) were implicated in these major abiotic stresses in sesame. Moreover, we identified some stress-specific genes while 28 genes were commonly involved in drought and waterlogging stresses (Figure 5c). Most of these shared genes displayed contrasting expression patterns under both stresses. For example, the gene SIMYB174 was highly down-regulated under waterlogging, but was found strongly up-regulated under drought stress (Figure 6).
4. Discussion
The function of MYB genes has been extensively investigated in numerous plants species, but, to date, no comprehensive analysis of the MYB gene family members has been reported in sesame and their functions are still largely unknown. In this study, we comprehensively studied this important gene family based on bioinformatic tools and expression profiling. We identified 287 SIMYB genes, which is quite higher than the 198 MYB genes reported in Arabidopsis, 183 in rice [16], 247 in soybean [54], 177 in sweet orange [5], 125 in Jatropha curcas [55], 231 in Pyrus bretschneideri [1], and 104 and 166 in Lotus japonicus and Medicago truncatula, respectively [8]. To date, only two species, namely, cotton and Chinese cabbage had much higher MYB genes than sesame, which could be attributed to their bigger genome sizes [6,7]. Hence, according to the previous reports related to the expansion of gene families in sesame [44,53,56,57], we speculated that sesame has retained and expanded during the evolutionary process, the MYB family members that may play essential roles in various biological processes. Similarly as reported in cotton [7], soybean [54], rice, and Arabidopsis [16], tandem duplication events have contributed to the expansion of the MYB gene family in sesame. More importantly, we deduced that tandem duplication events have also contributed to the functional divergence of SIMYBs since some tandem duplicated SIMYB genes were predicted distinct functions.
One surprising feature of the SIMYB gene family is the over-representation of the 1R-MYB subfamily. The 2R-MYB subfamily has been reported to be the largest subfamily of MYB family, followed by the 1R-MYB in many plant species [7,16,58,59]. Contrarily to the 2R-MYB, the 1R-MYB subfamily is less-studied and their functions were assigned for circadian and light regulation, cell differentiation, and telomeric DNA binding [9]. In sesame, we have found its members to be highly active in abiotic stress response, suggesting a novel role of this subfamily. Nonetheless, the abundance of 1R-MYB subfamily members in the sesame genome is intriguing and may deserve in-depth functional investigations.
To date, only three MYB genes (SIMYB186 (SIN_1010473), SIMYB261 (SIN_1025617), and SIMYB257 (SIN_1004921)) have been functionally characterized as being involved in the corolla and petiole pigmentation in sesame [52]. Wherefore, revealing the functions of the members of this important gene family should be the focus of future studies in sesame. The R2R3-MYB genes are known to be involved in plant specific processes, such as control of secondary metabolism or cellular morphogenesis, defense, pigmentation, and root formation [60]. Structurally similar MYB proteins within species were found to be functionally orthologous. For example, the genes ZmMYBC1, ZmMYBPL from maize and PhMYBAN2 from Petunia are structurally related and play the same function: control of anthocyanin synthesis [7]. Hence, we performed an evolutionary relationship analysis of the R2R3-MYB subfamily in sesame when compared with the well-studied species Arabidopsis. Out of 30 sesame clades, 23 clades were consistent with those in Arabidopsis [1,2], providing an excellent reference to explore the functions of sesame R2R3-MYB genes [61]. For example, SIMYB50, SIMYB74, SIMYB242, and SMYB261 were assembled together with Arabidopsis AtMYB044 and AtMYB077 into the clade 21, referring to abiotic stress response and anthocyanin biosynthesis [62]. The genes SIMYB24 and SIMYB80 were grouped into the clade 22 with two Arabidopsis genes AtMYB90 and AtMYB113, representing the functional clade of anthocyanin biosynthesis [63]. The genes SIMYB72 and SIMYB200 were found in the groups C3 and C10, respectively, related to root development. Accordingly, based on tissue RNA-seq analysis, we uncovered that these genes were exclusively expressed in sesame roots. Seven R2R3-SIMYB subgroups of sesame have no representative in Arabidopsis, suggesting that these proteins might have specialized roles that were either lost in Arabidopsis or gained after divergence from the last common ancestor [2]. Furthermore, the close relationship between sesame and Arabidopsis helped to identify some homolog MYB genes and allowed for us to predict the functions of SIMYB genes [53].
Few MYB genes have been reported to be involved in plant developmental processes [6]. In sesame, 65% of SIMYB genes were expressed in all of the tissues, indicating that these genes might contribute in various aspects of growth and developmental processes. In contrast, we detected few tissue-specific SIMYBs that may play major role in their respective tissues [2]. To cite an instance, the gene SIMYB200, which is exclusively expressed in the sesame root, is the homolog of the gene AtMYB93 which regulates lateral root development in Arabidopsis [64]. Until now, the sesame root system architecture has not been yet examined at the molecular level [36]. Therefore, SIMYB200 could be a potential gene to be targeted for functional characterization in the sesame root. In addition, the homolog of the sesame seed-specific genes SIMYB74, SIMYB242, and SIMYB249 in Arabidopsis (AtMYB113) was found to be involved in anthocyanin biosynthesis [65]. We suspected that these genes may be involved in seed coat coloration in sesame.
Waterlogging is a common adverse environmental condition that limits sesame plant growth and reproduction [30]. Similarly, drought stress constitutes the second major abiotic stress in sesame production [26]. Unfortunately, advances in developing abiotic stress tolerant sesame varieties have been hampered by the lack of functional gene resources [36]. Increasing evidences have shown that MYB TFs are implicated in drought response in various plant species, including Arabidopsis thaliana, Zea mays, Gossypium herbaceum, Vitis vinifera, Oryza sativa, Solanum tuberosum, Triticum aestivum, and Glycine max [66,67,68,69,70,71,72]. In sesame, 16% of the total SIMYB genes were significantly active in drought stress responses, and most of their homologs in Arabidopsis are described as abiotic stress responsive genes. By way of illustration, the homologs of the sesame drought responsive genes SIMYB204 (AtMYB015), SIMYB77, SIMYB245, SIMYB78 (AtMYB073), and SIMYB8 (AtMYB2) were proved to be implicated in drought responses in Arabidopsis. This suggests a conservation of gene function across the two species, implying that these identified genes could be regarded as candidate gene resources for drought tolerance improvement in sesame [1,2].
On the other hand, little is known concerning the role of MYB genes in the waterlogging response in plants. It was shown that AtMYB2 is induced by hypoxia, with mRNA levels peaking after 2–4 h under hypoxic conditions [73]. The gene TaMyb1 (ortholog of AtMYB2) exhibited a dramatic increase in transcripts under waterlogging stress, indicating its positive involvement in alleviating the damages in common wheat root [74]. The gene SIMYB8, which is the homolog of AtMYB2, was also highly induced after 9 h waterlogging stress in sesame, indicating a functional conservation. Recently, based on a transcriptome profiling under waterlogging stress in kiwifruit plants, Zhang et al. [75] found that the MYB TFs were abundant in the differentially expressed genes. In the present study, 40% of SIMYBs was significantly associated with waterlogging stress responses, representing potential genes for enhancing sesame endurance under this important abiotic stress. The higher number of SIMYBs regulated under waterlogging as compared to drought confirms sesame’s greater susceptibility to waterlogging stress [30].
SIMYBs commonly involved in drought and waterlogging responses are of a great interest as they could be targeted for improvement towards tolerance to both stresses simultaneously. Our results are in agreements with previous reports which showed that some MYB gene members are key regulators of several abiotic stresses, including cold, osmotic stress, salt, etc. [76,77,78]. However, it appears that SIMYBs may function differentially under drought and waterlogging stresses as shown by their contrasting expression patterns under these stresses. Therefore, in-depth analysis of these particular genes is needed to get insight into the mechanisms of co-regulation of drought and waterlogging stresses in sesame.
5. Conclusions
In the present study, we analyzed the characteristics of MYB gene structures, phylogenetic relationships, chromosomal locations, and their functional diversification in sesame. For the first time in sesame, we identified 287 SIMYB genes that were unevenly distributed on the 16 LGs. SIMYBs are quite abundant in sesame probably due to tandem duplication events with an over-representation of the 1R-MYB subfamily. Phylogenetic-based prediction of the SIMYB functions indicated that they are involved in diverse biological processes. Expression profiling in various tissues suggested that SIMYBs are highly active in modulating sesame growth and development. Moreover, by integrating RNA-seq data and qRT-PCR analysis, we demonstrated that SIMYBs are key transcription factors regulating sesame responses to drought and waterlogging stresses. Altogether, we provide here a firm groundwork for the future functional characterization of SIMYBs.
Acknowledgments
This work was financially supported by the China Agriculture Research System (CARS-1), the Agricultural Science and Technology Innovation Project of the Chinese Academy of Agricultural Sciences (CAAS-ASTIP-2013-OCRI), and Fundamental Research Funds for Central Non-profit Scientific Institution. The first two authors are grateful to the DAAD scholarship.
Supplementary Materials
The following are available online at www.mdpi.com/2073-4425/8/12/362/s1. Table S1: Full details of the sesame SIMYB genes, subfamily, ORF length, number of CDS, genome location and protein characteristics. Table S2: Classification and functional prediction of R2R3- SIMYB genes based on Arabidopsis functional clades [1,2,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99]. Table S3: Function prediction of SIMYBs based on sesame and Arabidopsis orthology. Table S4: List of the SIMYB genes expressed in five tissues of sesame. Table S5: List of gene specific primers used for qRT-PCR expression analysis of MYB genes in sesame under drought, waterlogging and various tissues. Figure S1: Gene structures of MYB proteins detected in sesame. Exon and intron are represented by blue box and black line, respectively.
Author Contributions
M.A.M., K.D., M.O.S., N.C. and X.Z. conceived and designed the experiments; M.A.M., K.D., R.Z., Y.W. performed the experiments; L.W. and K.D. provided transcriptome data. M.A.M., K.D., M.O.S., N.C. and L.W. analyzed the data; R.Z. and X.Z. contributed reagents/materials/analysis tools; M.A.M. wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Li X., Xue C., Li J., Qiao X., Li L., Yu L., Huang Y., Wu J. Genome-wide identification, evolution and functional divergence of MYB transcription factors in Chinese white Pear (Pyrus bretschneideri) Plant Cell Physiol. 2016;57:824–847. doi: 10.1093/pcp/pcw029. [DOI] [PubMed] [Google Scholar]
- 2.Dubos C., Stracke R., Grotewold E., Weisshaar B., Martin C., Lepiniec L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010;15:573–581. doi: 10.1016/j.tplants.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 3.Geethalakshmi S., Barathkumar S., Prabu G. The MYB transcription factor family genes in sugarcane (Saccharum sp.) Plant Mol. Biol. Rep. 2015;33:512–531. doi: 10.1007/s11105-014-0768-3. [DOI] [Google Scholar]
- 4.Lipsick J.S. One billion years of Myb. Oncogene. 1996;13:223–235. [PubMed] [Google Scholar]
- 5.Hou X.J., Li S.B., Liu S.R., Hu C.G., Zhang J.Z. Genome-wide classification and evolutionary and expression analyses of Citrus MYB transcription factor families in Sweet orange. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0112375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saha G., Park J.I., Ahmed N.U., Kayum M.A., Kang K.K., Nou I.S. Characterization and expression profiling of MYB transcription factors against stresses and during male organ development in Chinese cabbage (Brassica rapa ssp. pekinensis) Plant Physiol. Biochem. 2016;104:200–215. doi: 10.1016/j.plaphy.2016.03.021. [DOI] [PubMed] [Google Scholar]
- 7.Salih H., Gong W., He S., Sun G., Sun J., Du X. Genome-wide characterization and expression analysis of MYB transcription factors in Gossypium hirsutum. BMC Genet. 2016;17:129. doi: 10.1186/s12863-016-0436-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jin J., Tian F., Yang D.C., Meng Y.Q., Kong L., Luo J., Gao G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017;45:1040–1045. doi: 10.1093/nar/gkw982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin H., Martin C. Multifunctionality and diversity within the plant MYB-gene family. Plant Mol. Biol. 1999;41:577–585. doi: 10.1023/A:1006319732410. [DOI] [PubMed] [Google Scholar]
- 10.Wang G.P., Hui Z., Li F., Zhao M.-R., Zhang J., Wang W. Improvement of heat and drought photosynthetic tolerance in wheat by over accumulation of glycine betaine. Plant Biotechnol. Rep. 2010;4:213–222. doi: 10.1007/s11816-010-0139-y. [DOI] [Google Scholar]
- 11.Liu C., Wang X., Xu Y., Deng X., Xu Q. Genome-wide analysis of the R2R3-MYB transcription factor gene family in sweet orange (Citrus sinensis) Mol. Biol. Rep. 2014;41:6769–6785. doi: 10.1007/s11033-014-3563-1. [DOI] [PubMed] [Google Scholar]
- 12.Ambawat S., Sharma P., Yadav N.R., Yadav R.C. MYB transcription factor genes as regulators for plant responses: An overview. Physiol. Mol. Biol. Plant. 2013;19:307–321. doi: 10.1007/s12298-013-0179-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roy S. Function of MYB domain transcription factors in abiotic stress and epigenetic control of stress response in plant genome. Plant Signal. Behav. 2016;11 doi: 10.1080/15592324.2015.1117723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen N., Yang Q., Pan L., Chi X., Chen M., Hu D., Yang Z., Wang T., Wang M., Yu S. Identification of 30 MYB transcription factor genes and analysis of their expression during abiotic stress in peanut (Arachis hypogaea L.) Genes. 2014;533:332–345. doi: 10.1016/j.gene.2013.08.092. [DOI] [PubMed] [Google Scholar]
- 15.Li J., Luan Y., Yin Y. SpMYB overexpression in tobacco plants leads to altered abiotic and biotic stress responses. Gene. 2014;547:145–151. doi: 10.1016/j.gene.2014.06.049. [DOI] [PubMed] [Google Scholar]
- 16.Katiyar A., Smita S., Keshari S., Ravi L., Chinnusamy R.V., Bansal K.C. Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. BMC Genom. 2012;13:544. doi: 10.1186/1471-2164-13-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Z., Chen J., Su Y., Liu H., Chen Y., Luo P., Du X., Wang D., Zhang H. TaLHY, a 1R-MYB transcription factor, plays an important role in disease resistance against stripe rust fungus and ear heading in wheat. PLoS ONE. 2015;10 doi: 10.1371/journal.pone.0127723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu Z.S., Feng K., Que F., Wang F., Xiong A.S. A MYB transcription factor, DcMYB6, is involved in regulating anthocyanin biosynthesis in purple carrot taproots. Sci. Rep. 2017;7 doi: 10.1038/srep45324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baldoni E., Genga A., Cominelli E. Plant MYB transcription factors: Their role in drought response mechanisms. Int. J. Mol. Sci. 2015;16:15811–15851. doi: 10.3390/ijms160715811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seo P.J., Lee S.B., Suh M.C., Park M.J., Go Y.S., Park C.M. The MYB96 transcription factor regulates cuticular wax biosynthesis under drought conditions in Arabidopsis. Plant Cell. 2011;23:1138–1152. doi: 10.1105/tpc.111.083485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li K., Xing C., Yao Z., Huang X. PbrMYB21, a novel MYB protein of Pyrus betulaefolia, functions in drought tolerance and modulates polyamine levels by regulating arginine decarboxylase gene. Plant Biotechnol. J. 2017;15:1186–1203. doi: 10.1111/pbi.12708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yin X., Cui Y., Wang M., Xia X. Overexpression of a novel MYB-related transcription factor, OsMYBR1, confers improved drought tolerance and decreased ABA sensitivity in rice. Biochem. Biophys. Res. Commun. 2017;490:1355–1361. doi: 10.1016/j.bbrc.2017.07.029. [DOI] [PubMed] [Google Scholar]
- 23.Butt H.I., Yang Z., Gong Q., Chen E., Wang X., Zhao G., Ge X., Zhang X., Li F. GaMYB85 an R2R3 MYB gene, in transgenic Arabidopsis plays an important role in drought tolerance. BMC Plant Biol. 2017;17:142. doi: 10.1186/s12870-017-1078-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bedigian D. Evolution of sesame revisited: Domestication, diversity and prospects. Genet. Resour. Crop Evol. 2003;50:779–787. [Google Scholar]
- 25.Pathak N., Rai A.K., Kumari R., Thapa A., Bhat K.V. Sesame crop: An underexploited oilseed holds tremendous potential for enhanced food value. Agric. Sci. 2014;5:519–529. doi: 10.4236/as.2014.56054. [DOI] [Google Scholar]
- 26.Sun J., Rao Y., Le M., Yan T., Yan X., Zhou H. Effects of drought stress on sesame growth and yield characteristics and comprehensive evaluation of drought tolerance. Chin. J. Oil Crop Sci. 2010;32:525–533. [Google Scholar]
- 27.Wang L., Zhang Y., Qi X., Li D., Wei W., Zhang X. Global gene expression responses to waterlogging in roots of sesame (Sesamum indicum L.) Acta Physiol. Plant. 2012;34:2241–2249. doi: 10.1007/s11738-012-1024-9. [DOI] [Google Scholar]
- 28.Wei W.L., Li D.H., Wang L.H., Ding X., Zhang Y.X., Gao Y., Zhang X.R. Morpho-anatomical and physiological responses to waterlogging of sesame (Sesamum indicum L.) Plant Sci. 2013;208:102–111. doi: 10.1016/j.plantsci.2013.03.014. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y., Wang L., Li D., Gao Y., Lu H., Zhang X. Mapping of sesame waterlogging tolerance QTL and identification of excellent waterlogging tolerant germplasm. Sci. Agric. Sin. 2014;47:422–430. [Google Scholar]
- 30.Wang L., Li D., Zhang Y., Gao Y., Yu J., Wei X., Zhang X. Tolerant and susceptible sesame genotypes reveal waterlogging stress response patterns. PLoS ONE. 2016;11 doi: 10.1371/journal.pone.0149912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eskandari H., Zehtab-Salmasi S., Ghassemi-Golezani K., Gharineh M.H. Effects of water limitation on grain and oil yields of sesame cultivars. J. Food Agric. Environ. 2009;7:339–342. [Google Scholar]
- 32.Anjum A.H., Xie X.Y., Wang L.C., Saleem M.F., Man C., Le W. Morphological, physiological and biochemical responses of plants to drought stress. Afr. J. Agric. Res. 2011;6:2026–2032. [Google Scholar]
- 33.Kim K.S., Ryu S.N., Chung H.G. Influence of drought stress on chemical composition of sesame seed. Korean J. Crop Sci. 2006;51:73–80. [Google Scholar]
- 34.Ozkan A., Kulak M. Effects of water stress on growth, oil yield, fatty acid composition and mineral content of Sesamum indicum. J. Anim. Plant Sci. 2013;23:1686–1690. [Google Scholar]
- 35.Kadkhodaie A., Razmjoo J., Zahedi M., Pessarakli M. Oil content and composition of sesame (Sesamum indicum L.) genotypes as affected by irrigation regimes. J. Am. Oil Chem. Soc. 2014;91:1737–1744. doi: 10.1007/s11746-014-2524-0. [DOI] [Google Scholar]
- 36.Dossa K., Diouf D., Wang L., Wei X., Yu J., Niang M., Fonceka D., Yu J., Mmadi M.A., Yehouessi L.W., et al. The emerging oilseed crop Sesamum indicum enters the “Omics” era. Front. Plant Sci. 2017;8:1154. doi: 10.3389/fpls.2017.01154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang L., Yu J., Li D., Zhang X. Sinbase: An integrated database to study genomics, genetics and comparative genomics in Sesamum indicum. Plant Cell Physiol. 2014;56 doi: 10.1093/pcp/pcu175. [DOI] [PubMed] [Google Scholar]
- 38.Okonechnikov K., Golosova O., Fursov M. UGENE team Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics. 2012;28:1166–1167. doi: 10.1093/bioinformatics/bts091. [DOI] [PubMed] [Google Scholar]
- 39.Letunic I., Doerks T., Bork P. SMART: Recent updates, new developments and status in 2015. Nucleic Acids Res. 2015;43 doi: 10.1093/nar/gku949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Voorrips R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002;93:77–78. doi: 10.1093/jhered/93.1.77. [DOI] [PubMed] [Google Scholar]
- 41.Sharoni A.M., Nuruzzaman M., Satoh K., Shimizu T., Kondoh H., Sasaya T., Choi I.-R., Omura T., Kikuchi S. Gene structures, classification and expression models of the AP2/EREBP transcription factor family in rice. Plant Cell Physiol. 2011;52:344–360. doi: 10.1093/pcp/pcq196. [DOI] [PubMed] [Google Scholar]
- 42.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu B., Jin J.P., Guo A.Y., Zhang H., Luo J.H., Gao G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics. 2015;31:1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dossa K., Diouf D., Cissé N. Genome wide investigation of Hsf genes in sesame reveals their segmental duplication expansion and their active role in drought stress response. Front. Plant Sci. 2016;7:1522. doi: 10.3389/fpls.2016.01522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wei X., Gong H., Yu J., Liu P., Wang L., Zhang Y., Zhang X. Sesame FG: An integrated database for the functional genomics of sesame. Sci. Rep. 2017;7:2342. doi: 10.1038/s41598-017-02586-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dossa K., Li D., Wang L., Zheng X., Yua J., Wei X., Fonceka D., Diouf D., Liao B., Cisse N., et al. Dynamic transcriptome landscape of Sesame (Sesamum indicum L.) under progressive drought and after rewatering. Genom. Data. 2017;11:122–124. doi: 10.1016/j.gdata.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dossa K., Li D., Wang L., Zheng X., Liu A., Yu J., Wei X., Zhou R., Fonceka D., Diouf D., et al. Transcriptomic, biochemical and physio-anatomical investigations shed more light on responses to drought stress in two contrasting sesame genotypes. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-09397-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saeed A.I., Bhagabati N.K., Braisted J.C., Liang W., Sharov V., Howe E.A., Li J.W., Thiagarajan M., White J.A., Quackenbush J. TM4 microarray software suite. Method Enzymol. 2006;411:134–193. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- 50.Lalitha S. Primer premier 5. Biotechnol. Softw. Internet Rep. 2000;1:270–272. doi: 10.1089/152791600459894. [DOI] [Google Scholar]
- 51.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real time quantitative PCR and the 2−ΔΔCt method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 52.Wei X., Liu K., Zhang Y., Feng Q., Wang L., Zhao Y., Li D., Zhao Q., Zhu X., Zhu X., et al. Genetic discovery for oil production and quality in sesame. Nat. Commun. 2015;6 doi: 10.1038/ncomms9609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dossa K., Wei X., Li D., Fonceka D., Zhang Y., Wang L., Yu J., Liao B., Diouf D., Cissé N., et al. Insight into the AP2/ERF transcription factor superfamily in sesame and expression profiling of DREB subfamily under drought stress. BMC Plant Biol. 2016;16:171. doi: 10.1186/s12870-016-0859-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Du H., Yang S.S., Liang Z., Feng B.R., Liu L., Huang Y.B., Tang Y.X. Genome-wide analysis of the MYB transcription factor superfamily in soybean. BMC Plant Biol. 2012;12:106. doi: 10.1186/1471-2229-12-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou C., Chen Y., Wu Z., Lu W., Han J., Wu P., Chen Y., Li M., Jiang H., Wu G. Genome-wide analysis of the MYB gene family in physic nut (Jatropha curcas L.) Gene. 2015;572:63–71. doi: 10.1016/j.gene.2015.06.072. [DOI] [PubMed] [Google Scholar]
- 56.Wang L., Yu S., Tong C., Zhao Y., Liu Y., Song C., Zhang Y., Zhang X., Wang Y., Hua W., et al. Genome sequencing of the high oil crop sesame provides insight into oil biosynthesis. Genome Biol. 2014;15 doi: 10.1186/gb-2014-15-2-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu J., Wang L., Guo H., Liao B., King G., Zhang X. Genome evolutionary dynamics followed by diversifying selection explains the complexity of the Sesamum indicum genome. BMC Genom. 2017;18:257. doi: 10.1186/s12864-017-3599-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cao Z.H., Zhang S.Z., Wang R.K., Zhang R.F., Hao Y.J. Genome wide analysis of the apple MYB transcription factor family allows the identification of MdoMYB121 gene conferring abiotic stress tolerance in plants. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0069955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Z., Tang J., Hu R., Wu P., Hou X.L., Song X.M., Xiong A.S. Genome-wide analysis of the R2R3-MYB transcription factor genes in Chinese cabbage (Brassica rapa ssp. pekinensis) reveals their stress and hormone responsive patterns. BMC Genom. 2015;16:17. doi: 10.1186/s12864-015-1216-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ramachandran S., Hiratsuka K., Chua N.H. Transcription factors in plant growth and development. Curr. Opin. Genet. Dev. 1994;4:642–646. doi: 10.1016/0959-437X(94)90129-Q. [DOI] [PubMed] [Google Scholar]
- 61.Zhang L., Zhao G., Jia J., Liu X., Kong X. Molecular characterization of 60 isolated wheat MYB genes and analysis of their expression during abiotic stress. J. Exp. Bot. 2012;63:203–214. doi: 10.1093/jxb/err264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jung C., Seo J.S., Han S.W., Koo Y.J., Kim C.H., Song S.I., Nahm B.H., Choi Y.D., Cheong J.J. Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol. 2008;146:623–635. doi: 10.1104/pp.107.110981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stracke R., Werber M., Weisshaar B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001;4:447–456. doi: 10.1016/S1369-5266(00)00199-0. [DOI] [PubMed] [Google Scholar]
- 64.Gibbs D., Voss U., Harding S., Fannon J., Moody L., Yamada E., Swarup K., Nibau C., Bassel G., Choudhary A., et al. AtMYB93 is a novel negative regulator of lateral root development in Arabidopsis. New Phytol. 2014;203:1194–1207. doi: 10.1111/nph.12879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li T., Jia K.P., Lian H.L., Yang X., Li L., Yang H.Q. Jasmonic acid enhancement of anthocyanin accumulation is dependent on phytochrome A signaling pathway under far-red light in Arabidopsis. Biochem. Biophys. Res. Commun. 2014;454:78–83. doi: 10.1016/j.bbrc.2014.10.059. [DOI] [PubMed] [Google Scholar]
- 66.Jia J., Fu J., Zheng J., Zhou X., Huai J., Wang J., Wang M., Zhang Y., Chen X., Zhang J. Annotation and expression profile analysis of full-length cDNAs from stress-induced maize (Zea mays L.) seedlings. Plant J. 2006;48:710–727. doi: 10.1111/j.1365-313X.2006.02905.x. [DOI] [PubMed] [Google Scholar]
- 67.Davey M.W., Graham N.S., Vanholme B., Swennen R., May S.T., Keulemans J. Heterologous oligonucleotide microarrays for transcriptomics in a non-model species; a proof-of-concept study of drought stress in Musa. BMC Genom. 2009;10:436. doi: 10.1186/1471-2164-10-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Golldack D., Lüking I., Yang O. Plant tolerance to drought and salinity: Stress regulating transcription factors and their functional significance in the cellular transcriptional network. Plant Cell Rep. 2011;30:1383–1391. doi: 10.1007/s00299-011-1068-0. [DOI] [PubMed] [Google Scholar]
- 69.Galbiati M., Matus J.T., Francia P., Rusconi F., Cañón P., Medina C., Conti L., Cominelli E., Tonelli C., Johnson P.A. The grapevine guard cell-related VvMYB60 transcription factor is involved in the regulation of stomatal activity and is differentially expressed in response to ABA and osmotic stress. BMC Plant Biol. 2011;11:142. doi: 10.1186/1471-2229-11-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang A., Dai X., Zhang W.H. A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J. Exp. Bot. 2012;63:2541–2556. doi: 10.1093/jxb/err431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bi H., Luang S., Li Y., Bazanova N., Morran S., Song Z., Perera M.A., Hrmova M., Borisjuk N., Lopato S. Identification and characterization of wheat drought-responsive MYB transcription factors involved in the regulation of cuticle biosynthesis. J. Exp. Bot. 2016;67 doi: 10.1093/jxb/erw298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang N., Zhang W., Qin M., Li S., Qiao M., Liu Z., Xiang F. Drought tolerance conferred in soybean (Glycine max. L) by GmMYB84, a novel R2R3-MYB transcription factor. Plant Cell Physiol. 2017;58:1764–1776. doi: 10.1093/pcp/pcx111. [DOI] [PubMed] [Google Scholar]
- 73.Hoeren F.U., Dolferus R., Wu Y., Peacock W.J., Dennis E.S. Evidence for a role for AtMYB2 in the induction of the Arabidopsis alcohol dehydrogenase (ADH1) gene by low oxygen. Genetics. 1998;149:479–490. doi: 10.1093/genetics/149.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee T.G., Jang C.S., Kim J.Y., Kim D.S., Park J.H., Kim D.Y., Seo Y.W. A Myb transcription factor (TaMyb1) from wheat roots is expressed during hypoxia: Roles in response to the oxygen concentration in root environment and abiotic stresses. Physiol. Plant. 2007;129:375–385. doi: 10.1111/j.1399-3054.2006.00828.x. [DOI] [Google Scholar]
- 75.Zhang J.Y., Huang S.N., Mo Z.H., Xuan J.P., Jia X.D., Wang G., Guo Z.R. De novo transcriptome sequencing and comparative analysis of differentially expressed genes in kiwifruit under waterlogging stress. Mol. Breed. 2015;35:208. doi: 10.1007/s11032-015-0408-0. [DOI] [Google Scholar]
- 76.Peng X.J., Liu H., Wang D., Shen S.H. Genome-wide identification of the Jatropha curcas MYB family and functional analysis of the abiotic stress responsive gene JcMYB2. BMC Genom. 2016;17:251. doi: 10.1186/s12864-016-2576-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Guo H., Wang Y., Wang L., Hu P., Wang Y., Jia Y., Zhang C., Zhang Y., Zhang Y., Wang C., Yang C. Expression of the MYB transcription factor gene BplMYB46 affects abiotic stress tolerance and secondary cell wall deposition in Betula platyphylla. Plant Biotechnol. J. 2017;15:107–121. doi: 10.1111/pbi.12595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Duan M., Huang P., Yuan X., Chen H., Huang J., Zhang H. CMYB1 encoding a MYB transcriptional activator is involved in abiotic stress and circadian rhythm in rice. Sci. World J. 2014;2014 doi: 10.1155/2014/178038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang Y., Cao G., Qu L., Gu H. Involvement of an R2R3-MYB transcription factor gene AtMYB118 in embryogenesis in Arabidopsis. Plant Cell Rep. 2009;28:337–346. doi: 10.1007/s00299-008-0644-4. [DOI] [PubMed] [Google Scholar]
- 80.Mu R.L., Cao Y.R., Liu Y.F., Lei G., Zou H.F., Liao Y., Wang H.W., Zhang W.K., Ma B., Du J.Z., et al. An R2R3-type transcription factor gene AtMYB59 regulates root growth and cell cycle progression in Arabidopsis. Cell Res. 2009;19:1291–1304. doi: 10.1038/cr.2009.83. [DOI] [PubMed] [Google Scholar]
- 81.Millar A.A., Gubler F. The Arabidopsis GAMYB-Like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell. 2005;17:705–721. doi: 10.1105/tpc.104.027920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cominelli E., Galbiati M., Vavasseur A., Conti L., Sala T., Vuylsteke M., Leonhardt N., Dellaporta S.L., Tonelli C. A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Curr. Biol. 2005;15:1196–1200. doi: 10.1016/j.cub.2005.05.048. [DOI] [PubMed] [Google Scholar]
- 83.Zhou J., Lee C., Zhong R., Ye Z. MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell. 2009;21:248–266. doi: 10.1105/tpc.108.063321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Scoville R., Barnett L.L., Roels S.B., Kelly J.K., Hileman L.C. Differential regulation of a MYB transcription factor is correlated with transgenerational epigenetic inheritance of trichome density in Mimulus guttatus. New Phytol. 2011;191:251–263. doi: 10.1111/j.1469-8137.2011.03656.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Abe H., Urao T., Ito T., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell. 2003;15:63–78. doi: 10.1105/tpc.006130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Devaiah B.N., Madhuvanthi R., Karthikeyan A.S., Raghothama K.G. Phosphate starvation responses and gibberellic acid biosynthesis are regulated by the MYB62 transcription factor in Arabidopsis. Mol. Plant. 2009;2:43–58. doi: 10.1093/mp/ssn081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yang S.W., Jang I.C., Henriques R., Chua N.H. FAR-RED ELONGATED HYPOCOTYL1 and FHY1-LIKE associate with the Arabidopsis transcription factors LAF1 and HFR1 to transmit phytochrome A signals for inhibition of hypocotyl elongation. Plant Cell. 2009;21:1341–1359. doi: 10.1105/tpc.109.067215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Newman L.J., Lisa J., Perazza D.E., Juda L., Campbell M.M. Involvement of the R2R3-MYB, AtMYB61, in the ectopic lignification and dark-photomorphogenic components of the det3 mutant phenotype. Plant J. 2004;37:239–250. doi: 10.1046/j.1365-313X.2003.01953.x. [DOI] [PubMed] [Google Scholar]
- 89.Patzlaff A., Newman L.J., Dubos C., Whetten R.W., Smith C., McInnis S., Bevan M.W., Sederoff R.R., Campbell M.M. Characterization of PtMYB1, an R2R3-MYB from pine xylem. Plant Mol. Biol. 2003;53:597–608. doi: 10.1023/B:PLAN.0000019066.07933.d6. [DOI] [PubMed] [Google Scholar]
- 90.Zhao Q., Dixon R.A. Transcriptional networks for lignin biosynthesis: More complex than we thought? Trends Plant Sci. 2011;16:227–233. doi: 10.1016/j.tplants.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 91.Mandaokar A., Thines B., Shin B., Lange B.M., Choi G., Koo Y.J., Yoo Y.J., Choi Y.D., Choi G., Browse J. Transcriptional regulators of stamen development in Arabidopsis identified by transcriptional profiling. Plant J. 2006;46:984–1008. doi: 10.1111/j.1365-313X.2006.02756.x. [DOI] [PubMed] [Google Scholar]
- 92.Kirik V., Lee M.M., Wester K., Herrmann U., Zheng Z., Oppenheimer D., Schiefelbein J., Hulskamp M. Functional diversification of MYB23 and GL1 genes in trichome morphogenesis and initiation. Development. 2005;132:1477–1485. doi: 10.1242/dev.01708. [DOI] [PubMed] [Google Scholar]
- 93.Gonzalez A., Mendenhall J., Huo Y., Lloyd A. TTG1 complex MYBs, MYB5 and TT2, control outer seed coat differentiation. Dev. Biol. 2009;325:412–421. doi: 10.1016/j.ydbio.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 94.Borevitz J.O., Xia Y., Blount J., Dixon R.A., Lamb C. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell. 2000;12:2383–2394. doi: 10.1105/tpc.12.12.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dubos C., Le Gourrierec J., Baudry A., Huep G., Lanet E., Debeaujon I., Routaboul J.M., Alboresi A., Weisshaar B., Lepiniec L. MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J. 2008;55:940–953. doi: 10.1111/j.1365-313X.2008.03564.x. [DOI] [PubMed] [Google Scholar]
- 96.Celenza J.L., Quiel J.A., Smolen G.A., Merrikh H., Silvestro A.R., Normanly J., Bender J. The Arabidopsis ATR1 Myb transcription factor controls indolic glucosinolate homeostasis. Plant Physiol. 2005;137:253–262. doi: 10.1104/pp.104.054395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Stracke R., Ishihara H., Huep G., Barsch A., Mehrtens F., Niehaus K., Weisshaar B. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 2007;50:660–677. doi: 10.1111/j.1365-313X.2007.03078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sun P., Zhu X., Huang X., Liu J.H. Overexpression of a stress responsive MYB transcription factor of Poncirus trifoliata confers enhanced dehydration tolerance and increases polyamine biosynthesis. Plant Physiol. Biochem. 2014;78:71–79. doi: 10.1016/j.plaphy.2014.02.022. [DOI] [PubMed] [Google Scholar]
- 99.Feng C., Andreasson E., Maslak A., Mock P.H., Mattsson O., Mundy J. Arabidopsis MYB68 in development and responses to environmental cues. Plant Sci. 2004;167:1099–1107. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.