Abstract
The establishment of a root nodule symbiosis between a leguminous plant and a rhizobium requires complex molecular interactions between the two partners. Compatible interactions lead to the formation of nitrogen-fixing nodules, however, some legumes exhibit incompatibility with specific rhizobial strains and restrict nodulation by the strains. Bradyrhizobium elkanii USDA61 is incompatible with mung bean (Vigna radiata cv. KPS1) and soybean cultivars carrying the Rj4 allele. Here, we explored genetic loci in USDA61 that determine incompatibility with V. radiata KPS1. We identified five novel B. elkanii genes that contribute to this incompatibility. Four of these genes also control incompatibility with soybean cultivars carrying the Rj4 allele, suggesting that a common mechanism underlies nodulation restriction in both legumes. The fifth gene encodes a hypothetical protein that contains a tts box in its promoter region. The tts box is conserved in genes encoding the type III secretion system (T3SS), which is known for its delivery of virulence effectors by pathogenic bacteria. These findings revealed both common and unique genes that are involved in the incompatibility of B. elkanii with mung bean and soybean. Of particular interest is the novel T3SS-related gene, which causes incompatibility specifically with mung bean cv. KPS1.
Keywords: symbiosis, nodulation, Bradyrhizobium elkanii, Vigna radiata, Rj4 soybean
1. Introduction
Symbiotic relationships between leguminous plants and soil bacteria, collectively termed rhizobia, are characterized by the formation of root nodules, which are specialized plant organs that accommodate the rhizobia. In the root nodules, rhizobia fix atmospheric dinitrogen into ammonia, which is utilized by the host legumes. This symbiotic association is of critical importance in agricultural production and the nitrogen cycle. The interactions between legumes and rhizobia exhibit high levels of specificity, which is determined by the exchange of signal molecules. Legume roots excrete flavonoid compounds that interact specifically with the transcriptional regulator NodD in rhizobia [1,2]. The activation of NodD induces the expression of nodulation genes, which are required for the synthesis of lipo-chitooligosaccharides called Nod-factors [1,2]. The Nod factors function as signals for host receptors, which activate signal pathways leading to rhizobial infection and nodule formation [1,2]. In addition, rhizobial factors, such as exopolysaccharides, lipopolysaccharides, and secreted proteins, have also been reported to affect nodulation and host specificity [3,4].
The rhizobium Bradyrhizobium elkanii forms symbiotic relationships with a number of legumes, including soybean, mung bean (Vigna radiata), and groundnuts (Arachys hypogaea). In Brazil, B. elkanii is used as an inoculant for soybean cultivation [5]. The formation of symbiotic root nodules in soybean is controlled by several loci, which are referred to as the rj or Rj loci [6]. Soybean varieties carrying the dominant Rj4 allele, such as the cultivars Hill and BARC2, are incapable of nodulation with B. elkanii USDA61, but are compatible with Bradyrhizobium diazoefficiens USDA110 [7]. These studies revealed that Rj4 incompatibility is strain-specific and cultivar-specific, and appears to resemble gene-for-gene resistance in plant–pathogen interactions [8]. The recent identification of genes that are encoded by the Rj2 and Rj4 genes supports this idea. Rj2 encodes a member of the Toll-interleukin receptor/nucleotide binding site/leucine-rich repeat (TIR-NBS-LRR) class of proteins [9], while Rj4 encodes a thaumatin-like protein [10]. Both of these classes of protein are involved in plant resistance to pathogens.
Mung bean is an important leguminous crop in tropical and subtropical regions. Growth and seed production by V. radiata are highly dependent on symbiosis with nitrogen-fixing rhizobia [11]. The V. radiata cv. KPS1 cultivar is incompatible with B. elkanii USDA61, resulting in ineffective nodulation [12]. In a previous study [12], we mutated two B. elkanii genes, rhcC2 and rhcJ, which are well-conserved components of the type III secretion system (T3SS). The T3SS is a complex protein transport system that is involved in plant and animal pathogenesis by Gram-negative bacteria, and it is also found in many rhizobia [13,14,15,16,17]. The B. elkanii T3SS mutants formed functional nodules on V. radiata cv. KPS1 and Glycine max cv. Hill plants [12]. These results implicated the T3SS in the control of incompatibility between rhizobia and their plant hosts, however, genetic elements involved in the incompatibility between B. elkanii USDA61 and V. radiata cv. KPS1 have not been fully elucidated. Here, we identified five novel B. elkanii genes that contribute to this incompatibility. Four of these genes also control nodulation restriction in Rj4 soybean, suggesting a common mechanism that is underlying both incompatible interactions. The fifth gene encodes a T3SS-related protein with a host-specific function in KPS1. These findings shed light on the diverse and complicated mechanisms underlying the incompatible symbiosis, and suggest the involvement of a novel T3SS-related gene in the nodulation restriction of mung bean.
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
The bacterial strains and plasmids that were used in this study are listed in Table 1. B. elkanii strains were grown at 28 °C in arabinose–gluconate medium [18], supplemented with 50 μg mL−1 polymyxin and 200 μg mL−1 kanamycin. Escherichia coli strains were grown at 37 °C in Luria-Bertani medium (LB) [19] supplemented with 50 μg mL−1 kanamycin.
Table 1.
Bacterial strains, plasmids and oligonucleotides used in this study.
| Strain, Plasmid or Oligonucleotide | Characteristics or Sequence a | Reference or Source |
|---|---|---|
| Bacterial strains | ||
| Bradyrhizobium elkanii | ||
| USDA61 | Wild-type strain, Polr | Keyser b |
| BErhcJ | USDA61 derivative harboring insertion in rhcJ region, Polr, Kmr, Tcr | [12] |
| BE5 | Tn5 mutant of USDA61, Polr, Kmr | This study |
| BE53 | Tn5 mutant of USDA61, Polr, Kmr | This study |
| BE85 | Tn5 mutant of USDA61, Polr, Kmr | This study |
| BE103 | Tn5 mutant of USDA61, Polr, Kmr | This study |
| BE168 | Tn5 mutant of USDA61, Polr, Kmr | This study |
| BE53S | USDA61 derivative containing an insertion of plasmid pSUPSCAKm:int innB, Polr, Kmr | This study |
| Escherichia coli | ||
| HB101 | recA, hsdR, hsdM, pro, Smr | Invitrogen, Carlsbad, CA, USA |
| S17-1 | pro recA RP4-2(Tcs:Mu) (Kms:Tn7); Mob+ | [20] |
| DH10B | Cloning strain | Invitrogen |
| Plasmids | ||
| pUTKm | Transposon delivery vector; Apr, Kmr | [21] |
| pRK2013 | Helper plasmid, ColE1 replicon carrying RK2 transfer genes; Kmr, tra | [22] |
| pSUPSCAKm | Derivative of pSUPPOL2SCA [23] with a kanamycin resistance gene in the DraI site, oriT of RP4, Tcr, Kmr | This study |
| pSUPSCAKm:int innB | pSUPSCAKm carrying a 0.5-kb DNA fragment containing internal sequence of innB, Kmr | This study |
| Oligonucleotides | ||
| Linker 1 | 5′-TTTCTGCTCGAATTCAAGCTTCTAACGATGTACGGGGACACATG-3′ | [24] |
| Linker 2 | 5′-TGTCCCCGTACATCGTTAGAACTACTCGTACCATCCACAT-3′ | [24] |
| Y-linker primer | 5′-CTGCTCGAATTCAAGCTTCT-3′ | [24] |
| Tn5 primer | 5′-GGCCAGATCTGATCAAGAGA-3′ | [24] |
| Psuppol-F | 5′-ATAAACCAGCCAGCCGGAA-3′ | This study |
| Psuppol-R | 5′-TTCTGACAACGATCGGAGGA-3′ | This study |
| BE5-F | 5′-TCATGCAGGTGAATGTCGAT-3′ | This study |
| BE5-R | 5′-CTATCCGCAGGAGTTGAACG-3′ | This study |
| BE53-F | 5′-AGATTGATGTTGCCGAGGAC-3′ | This study |
| BE53-R | 5′-TGAAAAAGCTCCGTGAGGTC -3′ | This study |
| BE85-F | 5′-GCGCGGATATTGACATTGAT-3′ | This study |
| BE85-R | 5′-AGGCCGTCGATCTCTATCAC-3′ | This study |
| BE103-F | 5′-ACAAGAAGATGTCGGCCAAG-3′ | This study |
| BE103-R | 5′-TGCTCGCAGAATACAACTGC-3′ | This study |
| BE168-F | 5′-TCGAAAGCGCACTAGATTGA-3′ | This study |
| BE168-R | 5′-AGCCGTAAATATCGGACAGC-3′ | This study |
| InnBXbaI-intF | 5′-CGGTGGCGGCGGCCGCTCTAGAAAAATGCGCAACTGGAAGAT-3′ | This study |
| InnBEcoRI-intR | 5′-CGATAAGCTTGATATCGAATTCATCTGCTCACCAAGCCAATC-3′ | This study |
a Polr: Polymyxin resistant; Kmr: Kanamycin resistant; Smr: Streptomycin resistant; Spr: Spectinomycin resistant; Tcr: Tetracycline resistant; Apr: Ampicillin resistant; inn: incompatible nodulation. b United States Department of Agriculture, Beltsville, MD, USA.
2.2. Transposon Mutagenesis of B. elkanii USDA61
The transposon mini-Tn5 was introduced into the B. elkanii USDA61 strain by conjugation, as described by Krause et al. [23], using E. coli strain S17-1 [20] carrying the plasmid pUTKm [21], and E. coli HB101 (pRK2013) [22]. Strains containing transposons were selected on arabinose–gluconate agar plates, supplemented with 50 μg mL−1 polymyxin and 200 μg mL−1 kanamycin.
2.3. Plant Cultivation, Mutant Screening, and Inoculation Tests
Seeds of V. radiata cv. KPS1 and G. max cv. BARC2 were surface sterilized and germinated on wet sterile paper, then the seedlings were transferred to sterile vermiculite, using procedures described previously [8]. One day after transplantation, 216 V. radiata KPS1 seedlings were inoculated with approximately 36,000 Tn5 insertion strains of B. elkanii USDA61 (1 mL of 107 cells per mL on each plant). Plants were cultivated in a plant growth cabinet (LPH-410SP; NK Systems Co. Ltd., Osaka, Japan) at 25 °C and 70% humidity under a day/night regimen of 16/8 h. After 30 days of cultivation, nodules that formed on the V. radiata KPS1 plants were sampled, and the B. elkanii mutants were isolated, as described previously [8]. These isolates were used to re-inoculate V. radiata KPS1 plants in order to confirm the nodulation phenotypes. G. max cv. BARC2 seedlings were also inoculated with the identified mutants in order to determine if the mutants could form nodules on this G. max cultivar. Inoculation tests were performed, as described previously [8].
2.4. Nucleotide Sequence Analysis of Tn5-Flanking Regions
Genomic DNA was isolated from B. elkanii strains using the Wizard Genomic DNA Purification Kit (Promega, Madison, WI, USA). Transposon-flanking sequences were amplified using the Y-linker method [24]. The genomic DNA was digested with NlaIII or SphI, and ligated with the Y-linker. PCR was carried out using the GoTaq Green Master Mix (Promega), Y-linker primer and Tn5 primer (Table 1). The thermal cycling conditions were as described previously [8]. The PCR fragments were sequenced directly using the Tn5 or Y-linker primers. Homology searches were carried out using BLASTN at the National Center for Biotechnology Information and an in-house database for the B. elkanii USDA61 genome [25]. For the identification of tts box (TCGTCAG[CGT]TTNTCG[TA][CA]AGCTN(8)[TC]A) motif, the 2 kb upstream sequences of each gene were searched with the program “fuzznuc” of the EMBOSS package [26]. The consensus sequences from Rhizobium sp. strain NGR234 [27] and B. diazoefficiens USDA110 [23] were compared to select the tts box motif sequence (data not shown). In order to confirm that the observed phenotypes of the mutants were caused by transposon insertion and not by a secondary mutation independent of the transposon, innB identified in BE53 was mutated via single-crossover recombination. The internal region of innB was amplified by PCR using the primers InnBXbaI-intF and InnBEcoRI-intR (Table 1). The PCR product was cloned into the vector pSUPSCAKm and was mobilized into B. elkanii USDA61 via conjugation, as described previously [12]. The mutant generated by integration of the plasmids was selected on PSY medium containing polymyxin and kanamycin. Integration of the plasmids was confirmed by PCR using the primer set Psuppol-F and Psuppol-R (Table 1).
2.5. Microscopy
Nodules were excised from V. radiata cv. KPS1 and G. max cv. BARC2 and were fixed, sectioned, and photographed under a stereoscopic microscope, as described previously [8].
2.6. Nucleotide Sequence Accession Numbers
The sequences of the innA, innB, innC, innD, and innE genes have been submitted to GenBank and may be found under accession numbers KX499540, KX499541, KX499542, KX499543, and KX499544, respectively.
3. Results
3.1. Isolation of the Transposon Mutants of B. elkanii
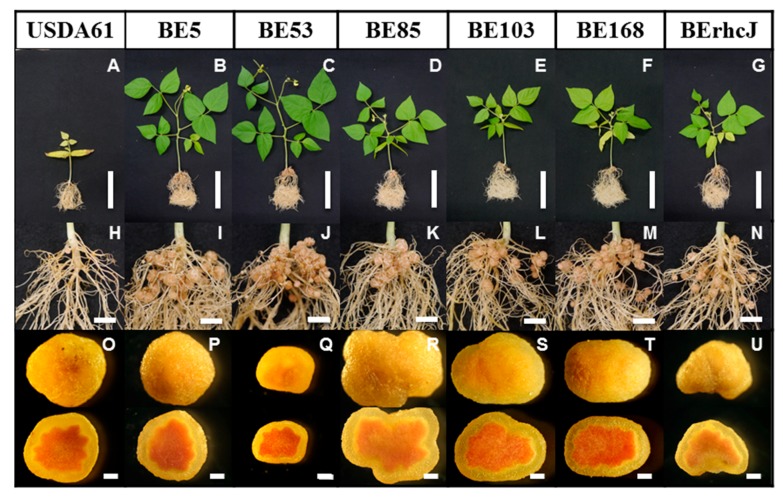
To investigate the molecular mechanisms underlying the incompatibility of V. radiata cv. KPS1 with B. elkanii USDA61, we created about 36,000 Tn5 transposon insertion mutants of USDA61, and then inoculated 216 KPS1 seedlings with the mutants. Tn5 mutants that formed nodules on KPS1 were isolated and used to re-inoculate KPS1 plants to confirm the nodulation phenotypes. Five mutants (BE5, BE53, BE85, BE103, and BE168) were confirmed to form nodules on KPS1 (Figure 1). The wild-type strain USDA61 formed only a small number of nodules on KPS1 plants, and these plants exhibited nitrogen deficiency symptoms (Figure 1A,H). In contrast, all of the Tn5 mutants were able to form mature nodules and promote the growth of KPS1, indicating efficient nitrogen-fixing activity (Figure 1B–F,I–M). The B. elkanii strain BErhcJ, a mutant defective in the type III protein secretion system [12], also formed nodules and promoted growth of KPS1 plants (Figure 1G,N). Nodules formed by all the plants showed a red color in the central tissue due to the presence of leghemoglobin, which is an indicator of nitrogenase activity (Figure 1O–U).
Figure 1.
Vigna radiata (L.) cv. KPS1 plants, roots, and nodules inoculated with Bradyrhizobium elkanii USDA61, Tn5 mutants and type III secretion system-deficient mutant BErhcJ. Plants were photographed at 40 days post-inoculation. Scale bars: (A–G) 10 cm, (H–N) 1 cm, and (O–U) 1 mm.
3.2. Symbiotic Phenotypes of the Transposon Mutants on V. radiata cv. KPS1 Plants
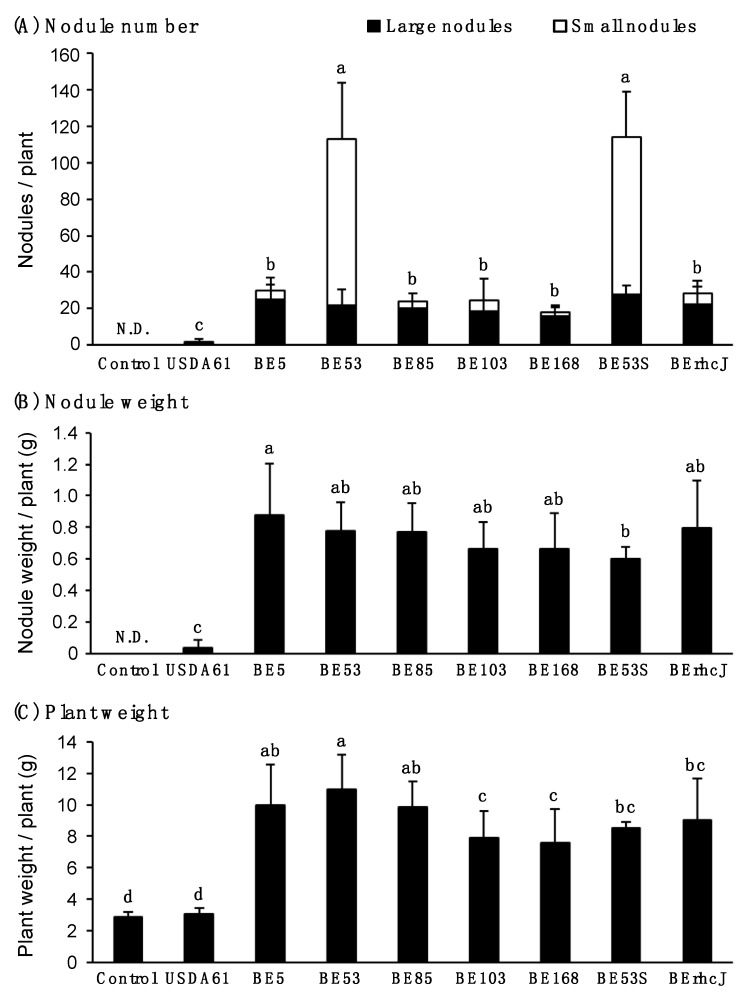
To characterize the symbiotic phenotypes of the mutants, we measured the nodule numbers, nodule weights, and plant fresh weights of inoculated KPS1 plants at 40 days after inoculation (Figure 2). For this analysis, we classified the nodules into two types by size: <2 mm and ≥2 mm in diameter. The Tn5 mutants exhibited significantly higher nodule numbers (about 18 to 114 nodules per plant) than the wild-type USDA61 strain (1.8 nodules per plant) (Figure 2A). The number of nodules and the total weight of nodules per plant were highest in plants that were inoculated with BE53 (Figure 2A,B). These plants also showed the highest fresh weights. Interestingly, BE53 induced many small nodules: 80% of total nodules (Figure 2A). In contrast, the mutants BE5, BE85, BE103, and BE168 formed mainly large nodules, and the numbers of large nodules and nodule weights per plant were similar to those of BErhcJ (Figure 2A,B). Apart from plants that were inoculated with BE53, the fresh weights of plants inoculated with the other mutants were similar to one another, and correlated with the numbers of large nodules and total nodule weights. These correlations were probably due to similar nitrogen fixation levels. As we reported previously, BErhcJ formed large nodules on KPS1 plants and promoted plant growth (Figure 2C).
Figure 2.
Symbiotic properties of V. radiata (L.) cv. KPS1 inoculated with B. elkanii USDA61, Tn5 mutants and type III secretion system-deficient mutant BErhcJ. (A) Nodule number, (B) nodule weight and (C) plant weight measured at 40 days post inoculation. Open bars and closed bars in (A) show the numbers of small nodules (<2 mm) and large nodules (≥2 mm), respectively. Nodulation tests were performed at least five times, the values are means of seven mung bean plants, and the error bars indicate standard deviations. Statistical analysis (Fisher’s method) was performed comparing the total nodule numbers induced by USDA61, BErhcJ and Tn5 mutants. Means followed by the same letters are not significantly different at 5% level of significance by Fisher’s test. N.D.: Not determined.
3.3. Symbiotic Phenotypes of the KPS1-Nodulating Mutants on Rj4 Soybean
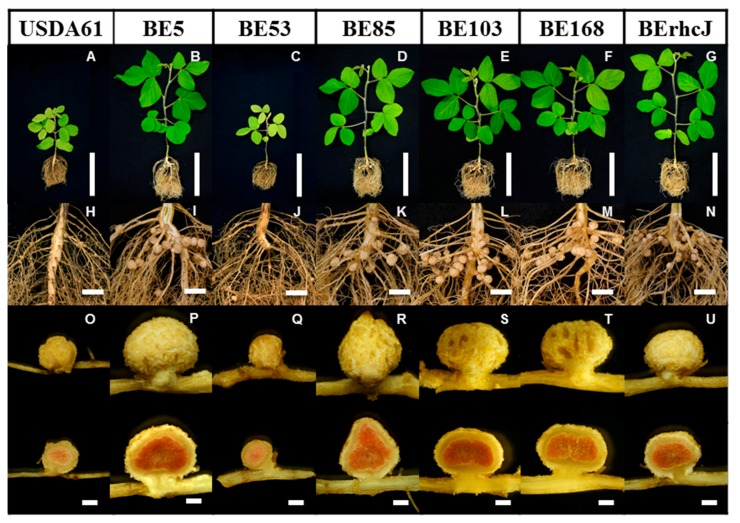
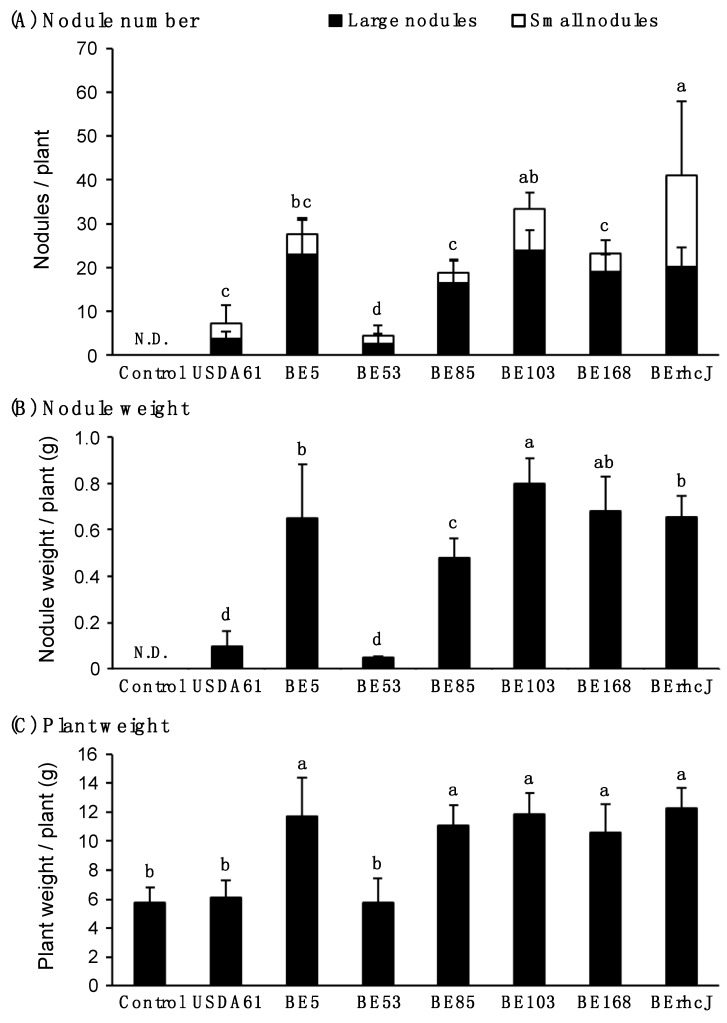
We also tested the symbiotic phenotypes of the Tn5 mutants on soybean cultivar BARC2 (Rj4/Rj4), which is known to restrict nodulation by B. elkanii [8,12]. Wild-type USDA61 induced only a few nodules on BARC2 plants, and the plants exhibited retarded growth due to nitrogen deficiency (Figure 3A,H). Interestingly, the four mutants BE5, BE85, BE103, and BE168 induced numerous nodules on the BARC2 roots (Figure 3B,I,D–F,K–M). More than 75% of the nodules that were formed by these mutants were large (Figure 4A,B), and they enhanced plant growth (Figure 4C). In contrast, BE53, which formed numerous nodules on V. radiata KPS1, formed only a few nodules on soybean BARC2 (Figure 3C,J). The nodules formed by BE53 were small and irregular in shape, and were similar to those that were formed by USDA61 (Figure 3Q). The BARC2 plants inoculated with BE53 showed nitrogen deficiency symptoms, indicating that the small nodules were ineffective.
Figure 3.
Glycine max cv. BARC2 (Rj4Rj4) plants, roots, and nodules inoculated with wild-type B. elkanii USDA61, Tn5 mutants and type III secretion system-deficient mutant BErhcJ. Plants were photographed at 30 days after inoculation. Scale bars: (A–G) 10 cm, (H–N) 1 cm, and (O–U) 1 mm.
Figure 4.
Symbiotic properties of G. max cv. BARC2 (Rj4Rj4) inoculated with B. elkanii USDA61, Tn5 mutants and type III secretion system-deficient mutant BErhcJ. (A) Nodule number, (B) nodule weight and (C) plant weight measured at 30 days post inoculation. Open bars and closed bars in (A) show the numbers of small nodules (<2 mm) and large nodules (≥2 mm), respectively. Nodulation tests were performed at least two times, the values are means of six soybean plants, and the error bars indicate standard deviations. Statistical analysis (Fisher’s method) was performed comparing the total nodule numbers induced by USDA61, BErhcJ and Tn5 mutants. Means followed by the same letters are not significantly different at 5% level of significance by Fisher’s test. N.D.: Not determined.
3.4. B. elkanii Genes Responsible for the Symbiotic Incompatibility
To identify the genes that were disrupted by the Tn5 insertions, Y-linker PCR was performed using genomic DNA from the Tn5 mutants. All of the mutant genomes contain the Tn5 fragment, and the PCR fragment sizes differ among strains, indicating that they are independent mutations. The genome of B. elkanii USDA61 has been completely sequenced [25], and the Tn5 flanking regions all contain sequences that are found in the genome (Table 2). The location map and genome context of the identified genes indicated that these genes are located outside T3SS gene cluster (Figures S1 and S2). BE5 contains a Tn5 insertion in a 1212 bp open reading frame (ORF), designated as innA, that encodes a cytosine deaminase, which is similar to those in rhizobia and other Gram-negative bacteria. BE85 contains a Tn5 insertion in a 504 bp ORF (designated as innC) that encodes a tellurite resistance protein (TerB). The gene products are similar with those in other rhizobia and pathogenic bacteria. BE103 has an insertion in a 1089 bp ORF (innD) that encodes an ABC-transporter substrate-binding protein, whose homologues were well conserved among rhizobia as well as species of Afipia, Rhodopseudomonas, Azospirillum, and Agrobacterium. One of its homologues in B. diazoefficiens USDA110, blr3743, has been reported as a secreted protein [28]. BE168 has an insertion in a 2292 bp ORF (innE) encoding GTP pyrophosphokinase (Table 2). The innE product has 88% and 60% identity with the products of relA genes in rhizobia and spoT genes in other bacteria including Rhodopseudomonas palustris, Xanthomonas campestris, Pseudomonas fluorescens, and Salmonella enterica.
Table 2.
Results of BLAST and tts box searches of Tn5-flanking sequences.
| Gene with Tn5 Insertion | ||||||
|---|---|---|---|---|---|---|
| Strains | Nodulation on Rj4 Soybean a | Designated Symbol/GenBank Accession No. b | Length (bp) | Deduced Gene Products | tts Box c | Strain and Locus Tag/Gene (GenBank Accession No., % Identity by BLASTP Analysis) |
| BE5 | + | innA/KX499540 | 1212 | Cytosine deaminase | - |
|
| BE53 | - | innB/KX499541 | 2280 | Hypothetical protein | + |
|
| BE85 | + | innC/KX499542 | 504 | TerB family tellurite resistance protein | - |
|
| BE103 | + | innD/KX499543 | 1089 | ABC transporter substrate-binding protein | - |
|
| BE168 | + | innE/KX499544 | 2292 | GTP pyrophosphokinase | - |
|
a Nodulation on G. max cv. BACR2 (Rj4Rj4) soybean; +: Positive; −: Negative. b inn: incompatible nodulation. c Presence of tts box in upstream regions of the genes; +: Present; −: Absent.
BE53 mutant carries a Tn5 insertion in the 2280 bp ORF (innB), which encodes a hypothetical protein that is conserved among rhizobia as well as Xanthomonas and Rhodopseudomonas. Notably, a tts box motif (5′-TATGGGACCTAGCTTTCGAAAAGCTGACGA-3′) is present at 96 bp upstream of the innB ORF. The tts box motifs are often located in the promoter regions of genes encoding structural components of the T3SS and secreted proteins. Furthermore, a homologue of innB in Mesorhizobium loti, mlr6327 (GenBank accession No. BAB52639.1), is located in the cluster of genes that are encoding the T3SS [29].
In order to confirm that the observed phenotypes of the mutants were caused by transposon insertion and not by a secondary mutation independent of the transposon, we reintroduced the mutation into the wild-type background. The innB gene was mutated via single-crossover recombination in wildtype USDA61 and the newly constructed mutant, designated BE53S, was used to inoculate V. radiata cv. KPS1 plants, and its symbiotic phenotypes were examined. As a result, the BE53S formed numerous mature nodules similar to the transposon mutant (BE53) (Figure 2A–C), confirming that transposon insertion in BE53 was responsible for the altered nodulation phenotype.
4. Discussion
In fields, the application of high nitrogen-fixing rhizobia to their target legume crops often fails to increase the crop yields. One of the major causes of this phenomenon is that more competitive indigenous rhizobial strains, with low nitrogen fixation activity, occupy most of the nodules. This problem is known as the competition problem, and it is very important in the cultivation of legumes. Besides the competitiveness of different rhizobial strains, compatibility between leguminous plants and rhizobia also affect nodule occupancy. In soybean, Rj genes control nodulation: plants carrying specific Rj genes cannot form nodules when inoculated with specific rhizobial strains [6]. This compatibility/incompatibility might be reminiscent of host–pathogen interactions. Consistent with this observation, the Rj2 and Rj4 genes encode TIR-NBS-LRR and thaumatin-like proteins, respectively [10], and these proteins are involved in plant resistance to pathogens.
Similarly, host-controlled nodulation occurs in mung beans, and the compatibility is dependent on both the host cultivar and the rhizobial strain. In a previous report, we showed that two genes that encode components of the T3SS, rhcC2 and rhcJ, are necessary for the incompatibility between V. radiata cv. KPS1 and B. elkanii USDA61 [12]. The T3SS is involved in plant and animal pathogenesis, and many rhizobial strains possess a functional T3SS [13,14,15,16,17]. The secreted proteins, called nodulation outer proteins (Nops), affect symbiosis positively or negatively, depending on the host plant [23,30,31]. The expression of the rhizobial T3SS is controlled by flavonoids that are derived from the host legume. Flavonoids perceived by the NodD protein activate the expression of ttsI, a transcriptional activator of the type III secretion gene cluster (ttsI). TtsI subsequently activates the ttsI gene cluster, and proteins are secreted via the T3SS. A conserved motif, called the tts box, is located in the promoter region of genes encoding structural components and secreted effector proteins [23,32,33].
In the present study, we explored additional genetic determinants of B. elkanii, which are involved in the nodulation incompatibility between V. radiata KPS1 and B. elkanii USDA61. We performed Tn5 transposon mutagenesis and identified five Tn5 insertion mutants of B. elkanii USDA61, which were able to form functional nodules on V. radiata KPS1 and enhance the growth of the plants.
The BE5 mutant contains a Tn5 insertion in the innA gene, which encodes cytosine deaminase. Cytosine deaminase deaminates cytosine to form uracil and ammonia [34]. Although the mechanism for the involvement of cytosine deaminase in nodulation restriction remains unclear, the BE5 mutant efficiently nodulated and promoted the growth of both KPS1 and Rj4 plants. The disruption of innA might cause the inactivation of the T3SS, which abolishes the secretion of effector proteins that are responsible for nodulation restriction in both Rj4 soybean and V. radiata KPS1.
The BE53 mutant carries transposon in the innB gene, which encodes a hypothetical protein with an unknown function. The innB gene is preceded by the consensus tts box, and the innB homologue in M. loti, mlr6327, occurs in the cluster of genes encoding the T3SS [28]. Intriguingly, the innB-deficient mutant formed nodules on V. radiata KPS1 plants, but not on Rj4 soybean. This suggests that innB encodes a protein with a host-specific function in KPS1. One possibility is that innB encodes an effector protein that is recognized specifically in V. radiata KPS1. It is likely that the rhizobial effector proteins that are recognized by KPS1 and Rj4 soybean differ. The InnB might be recognized specifically by a resistance (R) protein in KPS1, causing nodulation restriction in KPS1, but not in Rj4 soybean.
The BE85 mutant contains a Tn5 insertion in innC, encoding TerB family tellurite resistance protein that is conserved among the Bradyrhizobiaceae. It has been reported that the Tellurium resistance proteins of Frankia spp. are induced by seed phenolic compounds that are produced by their actinorhizal host plants [35]. Considering that the disruption of innC induced nodulation on both KPS1 and Rj4 plants, the gene might be involved in the T3SS. However, the function of these proteins in rhizobial symbiosis remains unknown.
The BE103 mutant carries a Tn5 insertion in the gene innD, which encodes a substrate-binding protein in the ABC transporter family. These substrate-binding proteins are located in the periplasm, and are involved in the active transport of substrates into the cytoplasm [36,37,38]. Although the function of the innD protein remains unclear, its homologue in B. diazoefficiens USDA110 (blr3743, BAC49008.1) is secreted [28], and is upregulated in bacteroids in nodules [39,40]. Clearly, functional analyses are needed to determine the contribution of these transporters to the compatibility/incompatibility between rhizobia and their hosts. Since the inactivation of the innD gene by the Tn5 insertion canceled the nodulation restriction in both KPS1 and Rj4 plants, the innD product might be involved in the T3SS. Further molecular experiments will be performed to explore this possibility.
The BE168 mutant contains a Tn5 insertion in innE, which encodes a GTP pyrophosphokinase (ppGpp synthetase) that is conserved among various rhizobia and pathogenic bacteria. innE shares similarity with the relA genes of B. diazoefficens USDA110 (BAC50330.1) and other bacteria. It has been proposed that ppGpp synthetase binds to, and alters the ability of, RNA polymerase to initiate and elongate transcription. ppGpp synthetase activates genes that function in the biosynthesis of amino acids and transcription factors that are responsible for stress responses [41,42]. In addition, ppGpp synthetase plays important roles in the virulence and the persistence of phytopathogens within host plants [43]. In Rhizobium etli, the relA gene regulates gene expression in bacteroid cells [44]. A relA mutant of Sinorhizobium meliloti shows a non-nodulation phenotype on alfalfa [45]. These findings suggest that the mutation in innE affects cellular ppGpp synthetase levels and leads to changes in cell functions that affect the strain specific incompatibility between B. elkanii USDA61 and mung bean and soybean. It should be noted that, except for the innB gene, which was confirmed by the newly constructed mutant BE53S, the other genes also need to be confirmed by mutational and/or complementation analysis to unequivocally associate the gene with the phenotype.
In addition to the genes and gene products discussed above, rhizobial factors have been reported to be involved in incompatible symbioses, including the type IV secretion system [46], extracellular polysaccharides [47], and peptidase that cleaves host-derived signaling peptides [48]. T3SSs in other rhizobia involved in incompatible symbioses have been reported for the following interactions: Rhizobium sp. NGR234 with Crotalaria juncea [49], M. loti MAFF303099 with Lotus halophilus [28], Sinorhizobium fredii USDA257 with G. max cv. McCall [15], B. elkanii USDA61 with Rj4 soybean [8], Bradyrhizobium japonicum USDA122 with Rj2 soybean [50], and B. diazoefficiens USDA110 with V. radiata KPS2 [51]. The type III-secreted proteins NopE1 and NopE2 of B. diazoefficiens USDA110 were reported as negative effectors of compatibility with V. radiata KPS2 [51]. In our preliminary results, B. diazoefficiens USDA110 formed nodules on V. radiata KPS1 and promoted plant growth efficiently (data not shown), indicating that NopE1 and NopE2 do not control incompatibility with KPS1, and that B. elkanii might possess other effectors that are recognized by KPS1. These findings suggest that the molecular mechanisms that are controlling incompatibility between various rhizobia and legumes differ, although the T3SS appears to underlie at least three types of nodulation restriction. Clearly, further study is necessary to define the roles of the T3SS components in both innate-immunity and rhizobial nodulation.
5. Concluding Remarks
In conclusion, we have identified novel genes of B. elkanii that are involved in incompatibility with V. radiata cv. KPS1. Four of the genes (innA, innC, innD, and innE) are also involved in incompatibility with Rj4 soybean, suggesting that they participate in a common mechanism underlying nodulation restriction in both of the legumes. In contrast, innB is involved in nodulation restriction with V. radiata cv. KPS1, but not with Rj4 soybean. The innB gene is preceded by a tts box, which is a promoter element in type III secretion system-related genes, suggesting its T3SS-related function. These findings shed light on the diverse and complicated mechanisms underlying the incompatible symbiosis, and suggest the involvement of a novel T3SS-related gene in the nodulation restriction of mung bean.
Acknowledgments
We thank K. Suzuki for her kind helps. This work was supported in part by a grant-in-aid (No. 25450082) and JSPS Bilateral Joint Research Project to S.O.
Supplementary Materials
The following are available online at www.mdpi.com/2073-4425/8/12/374/s1. Figure S1: The location map of the identified genes innA, innB, innC, innD, and innE in B. elkanii USAD61 genome, Figure S2: Genome context of the identified genes innA, innB, innC, innD, and innE.
Author Contributions
H.P.N. and S.O. designed the research; H.P.N. and H.M. performed the experiments; H.M., T.K. and S.S. contributed with new analytic tools; H.P.N., H.M. and S.O. analyzed the data; and H.P.N and S.O. wrote the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Cullimore J.V., Ranjeva R., Bono J.-J. Perception of lipo-chitooligosaccharidic Nod factors in legumes. Trends Plant Sci. 2001;6:24–30. doi: 10.1016/S1360-1385(00)01810-0. [DOI] [PubMed] [Google Scholar]
- 2.Radutoiu S., Madsen L.H., Madsen E.B., Jurkiewicz A., Fukai E., Quistgaard E.M.H., Albrektsen A.S., James E.K., Thirup S., Stougaard J. LysM domains mediate lipochitin-oligosaccharide recognition and Nfr genes extend the symbiotic host range. EMBO J. 2007;26:3923–3935. doi: 10.1038/sj.emboj.7601826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lerouge P., Roche P., Faucher C., Maillet F., Truchet G., Promé J.C., Dénarié J. Symbiotic host-specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature. 1990;344:781–784. doi: 10.1038/344781a0. [DOI] [PubMed] [Google Scholar]
- 4.Okazaki S., Kaneko T., Sato S., Saeki K. Hijacking of leguminous nodulation signaling by the rhizobial type III secretion system. Proc. Natl. Acad. Sci. USA. 2013;110:17131–17136. doi: 10.1073/pnas.1302360110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rumjanek N.G., Dobert R.C., van Berkum P., Triplett E.W. Common soybean inoculant strains in Brazil are members of Bradyrhizobium elkanii. Appl. Environ. Microbiol. 1993;59:4371–4373. doi: 10.1128/aem.59.12.4371-4373.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hayashi M., Saeki Y., Haga M., Harada K., Kouchi H., Umehara Y. Rj (rj) genes involved in nitrogen-fixing root nodule formation in soybean. Breed. Sci. 2012;61:544–553. doi: 10.1270/jsbbs.61.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sadowsky M.J., Cregan P.B. The Soybean Rj4 Allele Restricts Nodulation by Bradyrhizobium japonicum Serogroup 123 Strains. Appl. Environ. Microbiol. 1992;58:720–723. doi: 10.1128/aem.58.2.720-723.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faruque O.M., Miwa H., Yasuda M., Fujii Y., Kaneko T., Sato S., Okazaki S. Identification of Bradyrhizobium elkanii Genes Involved in Incompatibility with Soybean Plants Carrying the Rj4 Allele. Appl. Environ. Microbiol. 2015;81:6710–6717. doi: 10.1128/AEM.01942-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang S., Tang F., Gao M., Krishnan H.B., Zhu H. R gene-controlled host specificity in the legume-rhizobia symbiosis. Proc. Natl. Acad. Sci. USA. 2010;107:18735–18740. doi: 10.1073/pnas.1011957107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang F., Yang S., Liu J., Zhu H. Rj4, a Gene Controlling Nodulation Specificity in Soybeans, Encodes a Thaumatin-Like Protein But Not the One Previously Reported. Plant Physiol. 2016;170:26–32. doi: 10.1104/pp.15.01661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loh J., Stacey G. Nodulation gene regulation in Bradyrhizobium japonicum: A unique integration of global regulatory circuits. Appl. Environ. Microbiol. 2003;69:10–17. doi: 10.1128/AEM.69.1.10-17.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okazaki S., Zehner S., Hempel J., Lang K., Göttfert M. Genetic organization and functional analysis of the type III secretion system of Bradyrhizobium elkanii. FEMS Microbiol. Lett. 2009;295:88–95. doi: 10.1111/j.1574-6968.2009.01593.x. [DOI] [PubMed] [Google Scholar]
- 13.De Lyra Mdo C., Lopez-Baena F.J., Madinabeitia N., Vinardell J.M., Espuny Mdel R., Cubo M.T., Belloguin R.A., Ruiz-Sainz J.E., Ollero F.J. Inactivation of the Sinorhizobium fredii HH103 rhcJ gene abolishes nodulation outer proteins (Nops) secretion and decreases the symbiotic capacity with soybean. Int. Microbiol. 2006;9:125–133. [PubMed] [Google Scholar]
- 14.Göttfert M., Röthlisberger S., Kündig C., Beck C., Marty R., Hennecke H. Potential symbiosis-specific genes uncovered by sequencing a 410-kilobase DNA region of the Bradyrhizobium japonicum chromosome. J. Bacteriol. 2001;183:1405–1412. doi: 10.1128/JB.183.4.1405-1412.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krishnan H.B., Lorio J., Kim W.S., Jiang G., Kim K.Y., DeBoer M., Pueppke S.G. Extracellular proteins involved in soybean cultivar-specific nodulation are associated with pilus-like surface appendages and exported by a type III protein secretion system in Sinorhizobium fredii USDA257. Mol. Plant Microbe Interact. 2003;16:617–625. doi: 10.1094/MPMI.2003.16.7.617. [DOI] [PubMed] [Google Scholar]
- 16.Viprey V., Del Greco A., Golinowski W., Broughton W.J., Perret X. Symbiotic implications of type III protein secretion machinery in Rhizobium. Mol. Microbiol. 1998;28:1381–1389. doi: 10.1046/j.1365-2958.1998.00920.x. [DOI] [PubMed] [Google Scholar]
- 17.Kaneko T., Nakamura Y., Sato S., Asamizu E., Kato T., Sasamoto S., Watanabe A., Idesawa K., Ishikawa A., Kawashima K., et al. Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. DNA Res. 2000;7:331–338. doi: 10.1093/dnares/7.6.331. [DOI] [PubMed] [Google Scholar]
- 18.Sadowsky M.J., Tully R.E., Cregan P.B., Keyser H.H. Genetic Diversity in Bradyrhizobium japonicum Serogroup 123 and Its Relation to Genotype-Specific Nodulation of Soybean. Appl. Environ. Microbiol. 1987;53:2624–2630. doi: 10.1128/aem.53.11.2624-2630.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Green M.R., Sambrook J. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY, USA: 2012. [Google Scholar]
- 20.Simon R., Priefer U., Pühler A. A Broad Host Range Mobilization System for In Vivo Genetic Engineering: Transposon Mutagenesis in Gram Negative Bacteria. Nat. Biotechnol. 1983;1:784–791. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]
- 21.Herrero M., de Lorenzo V., Timmis K.N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Figurski D.H., Helinski D.R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krause A., Doerfel A., Göttfert M. Mutational and transcriptional analysis of the type III secretion system of Bradyrhizobium japonicum. Mol. Plant Microbe Interact. 2002;15:1228–1235. doi: 10.1094/MPMI.2002.15.12.1228. [DOI] [PubMed] [Google Scholar]
- 24.Kwon Y.M., Ricke S.C. Efficient amplification of multiple transposon-flanking sequences. J. Microbiol. Methods. 2000;41:195–199. doi: 10.1016/S0167-7012(00)00159-7. [DOI] [PubMed] [Google Scholar]
- 25.Kaneko T. (Faculty of Life Sciences, Kyoto Sangyo University, Motoyama, Kamigamo, Kita-Ku, Kyoto, Japan). Complete genome sequence of Bradyrhizobium elkanii USDA61. 2015. Unpublished work.
- 26.Rice P., Longden I., Bleasby A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000 doi: 10.1016/S0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 27.Marie C., Deakin W.J., Ojanen-Reuhs T., Diallo E., Reuhs B., Broughton W.J., Perret X. TtsI, a Key Regulator of Rhizobium Species NGR234 Is Required for Type III-Dependent Protein Secretion and Synthesis of Rhamnose-Rich Polysaccharides. Mol. Plant Microbe Interact. 2004;958:958–966. doi: 10.1094/MPMI.2004.17.9.958. [DOI] [PubMed] [Google Scholar]
- 28.Hempel J., Zehner S., Göttfert M., Patschkowski T. Analysis of the secretome of the soybean symbiont Bradyrhizobium japonicum. J. Biotechnol. 2009;140:51–58. doi: 10.1016/j.jbiotec.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 29.Okazaki S., Okabe S., Higashi M., Shimoda Y., Sato S., Tabata S., Hashiguchi M., Akashi R., Gottfert M., Saeki K. Identification and functional analysis of type III effector proteins in Mesorhizobium loti. Mol. Plant Microbe Interact. 2010;23:223–234. doi: 10.1094/MPMI-23-2-0223. [DOI] [PubMed] [Google Scholar]
- 30.Marie C., Deakin W.J., Viprey V., Kopciñska J., Golinowski W., Krishnan H.B., Perret X., Broughton W.J. Characterization of Nops, nodulation outer proteins, secreted via the type III secretion system of NGR234. Mol. Plant Microbe Interact. 2003;16:743–751. doi: 10.1094/MPMI.2003.16.9.743. [DOI] [PubMed] [Google Scholar]
- 31.Meinhardt L.W., Krishnan H.B., Balatti P.A., Pueppke S.G. Molecular cloning and characterization of a sym plasmid locus that regulates cultivar-specific nodulation of soybean by Rhizobium fredii USDA257. Mol. Microbiol. 1993;9:17–29. doi: 10.1111/j.1365-2958.1993.tb01665.x. [DOI] [PubMed] [Google Scholar]
- 32.Wassem R., Kobayashi H., Kambara K., Le Quéré A., Walker G.C., Broughton W.J., Deakin W.J. TtsI regulates symbiotic genes in Rhizobium species NGR234 by binding to tts boxes. Mol. Microbiol. 2008;68:736–748. doi: 10.1111/j.1365-2958.2008.06187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zehner S., Schober G., Wenzel M., Lang K., Göttfert M. Expression of the Bradyrhizobium japonicum type III secretion system in legume nodules and analysis of the associated tts box promoter. Mol. Plant Microbe Interact. 2008;21:1087–1093. doi: 10.1094/MPMI-21-8-1087. [DOI] [PubMed] [Google Scholar]
- 34.Danielsen S., Kilstrup M., Barilla K., Jochimsen B., Neuhard J. Characterization of the Escherichia coli codBA operon encoding cytosine permease and cytosine deaminase. Mol. Microbiol. 1992;6:1335–1344. doi: 10.1111/j.1365-2958.1992.tb00854.x. [DOI] [PubMed] [Google Scholar]
- 35.Bagnarol E., Popovici J., Alloisio N., Maréchal J., Pujic P., Normand P., Fernandez M.P. Differential Frankia protein patterns induced by phenolic extracts from Myricaceae seeds. Physiol. Plant. 2007;130:380–390. doi: 10.1111/j.1399-3054.2007.00875.x. [DOI] [Google Scholar]
- 36.Higgins C.F. ABC transporters: Physiology, structure and mechanism—An overview. Res. Microbiol. 2001;152:205–210. doi: 10.1016/S0923-2508(01)01193-7. [DOI] [PubMed] [Google Scholar]
- 37.Abouhamad W.N., Manson M.D. The dipeptide permease of Escherichia coli closely resembles other bacterial transport systems and shows growth-phase-dependent expression. Mol. Microbiol. 1994;14:1077–1092. doi: 10.1111/j.1365-2958.1994.tb01340.x. [DOI] [PubMed] [Google Scholar]
- 38.Doeven M.K., van den Bogaart G., Krasnikov V., Poolman B. Probing receptor-translocator interactions in the oligopeptide ABC transporter by fluorescence correlation spectroscopy. Biophys. J. 2008;94:3956–3965. doi: 10.1529/biophysj.107.120964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang W.-S., Franck W.L., Cytryn E., Jeong S., Joshi T., Emerich D.W., Sadowsky M.J., Xu D., Stacey G. An Oligonucleotide Microarray Resource for Transcriptional Profiling of Bradyrhizobium japonicum. Mol. Plant Microbe Interact. 2007 doi: 10.1094/MPMI-20-10-1298. [DOI] [PubMed] [Google Scholar]
- 40.Pessi G., Ahrens C.H., Rehrauer H., Lindemann A., Hauser F., Fischer H.-M., Hennecke H. Genome-Wide Transcript Analysis of Bradyrhizobium japonicum Bacteroids in Soybean Root Nodules. Mol. Plant Microbe Interact. 2007;20:1353–1363. doi: 10.1094/MPMI-20-11-1353. [DOI] [PubMed] [Google Scholar]
- 41.Vogt S.L., Green C., Stevens K.M., Day B., Erickson D.L., Woods D.E., Storey D.G. The stringent response is essential for Pseudomonas aeruginosa virulence in the rat lung agar bead and Drosophila melanogaster feeding models of infection. Infect. Immun. 2011;79:4094–4104. doi: 10.1128/IAI.00193-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Calderón-Flores A., Du Pont G., Huerta-Saquero A., Merchant-Larios H., Servín-González L., Durán S. The stringent response is required for amino acid and nitrate utilization, nod factor regulation, nodulation, and nitrogen fixation in Rhizobium etli. J. Bacteriol. 2005;187:5075–5083. doi: 10.1128/JB.187.15.5075-5083.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chatterji D., Kumar Ojha A. Revisiting the stringent response, ppGpp and starvation signaling. Curr. Opin. Microbiol. 2001;4:160–165. doi: 10.1016/S1369-5274(00)00182-X. [DOI] [PubMed] [Google Scholar]
- 44.Moris M., Braeken K., Schoeters E., Verreth C., Beullens S., Vanderleyden J., Michiels J. Effective symbiosis between Rhizobium etli and Phaseolus vulgaris requires the alarmone ppGpp. J. Bacteriol. 2005;187:5460–5469. doi: 10.1128/JB.187.15.5460-5469.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wells D.H., Long S.R. Mutations in rpoBC suppress the defects of a Sinorhizobium meliloti relA mutant. J. Bacteriol. 2003;185:5602–5610. doi: 10.1128/JB.185.18.5602-5610.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hubber A., Vergunst A.C., Sullivan J.T., Hooykaas P.J.J., Ronson C.W. Symbiotic phenotypes and translocated effector proteins of the Mesorhizobium loti strain R7A VirB/D4 type IV secretion system. Mol. Microbiol. 2004;54:561–574. doi: 10.1111/j.1365-2958.2004.04292.x. [DOI] [PubMed] [Google Scholar]
- 47.Kelly S.J., Muszyński A., Kawaharada Y., Hubber A.M., Sullivan J.T., Sandal N., Carlson R.W., Stougaard J., Ronson C.W. Conditional Requirement for Exopolysaccharide in the Mesorhizobium-Lotus Symbiosis. Mol. Plant Microbe Interact. 2013;26:319–329. doi: 10.1094/MPMI-09-12-0227-R. [DOI] [PubMed] [Google Scholar]
- 48.Price P.A., Tanner H.R., Dillon B.A., Shabab M., Walker G.C., Griffitts J.S. Rhizobial peptidase HrrP cleaves host-encoded signaling peptides and mediates symbiotic compatibility. Proc. Natl. Acad. Sci. USA. 2015 doi: 10.1073/pnas.1417797112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dai W.-J., Zeng Y., Xie Z.-P., Staehelin C. Symbiosis-Promoting and Deleterious Effects of NopT, a Novel Type 3 Effector of Rhizobium sp. Strain NGR234. J. Bacteriol. 2008;190:5101–5110. doi: 10.1128/JB.00306-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsukui T., Eda S., Kaneko T., Sato S., Okazaki S., Kakizaki-Chiba K., Itakura M., Mitsui H., Yamashita A., Terasawa K., et al. The type III secretion system of Bradyrhizobium japonicum USDA122 mediates symbiotic incompatibility with Rj2 soybean plants. Appl. Environ. Microbiol. 2013;79:1048–1051. doi: 10.1128/AEM.03297-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wenzel M., Friedrich L., Göttfert M., Zehner S. The type III-secreted protein NopE1 affects symbiosis and exhibits a calcium-dependent autocleavage activity. Mol. Plant Microbe Interact. 2010;23:124–129. doi: 10.1094/MPMI-23-1-0124. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.