Inflammation and tissue regeneration follow tissue damage, but little is known about how these processes are coordinated. Tirone et al. show that alternative redox forms of high mobility group box 1 (HMGB1), the “alarmin” signal released by damaged cells, trigger inflammation or tissue repair after injury by interacting with distinct receptors and that a nonoxidizable HMGB1 mutant promotes regeneration without exacerbating inflammation.
Abstract
Inflammation and tissue regeneration follow tissue damage, but little is known about how these processes are coordinated. High Mobility Group Box 1 (HMGB1) is a nuclear protein that, when released on injury, triggers inflammation. We previously showed that HMGB1 with reduced cysteines is a chemoattractant, whereas a disulfide bond makes it a proinflammatory cytokine. Here we report that fully reduced HMGB1 orchestrates muscle and liver regeneration via CXCR4, whereas disulfide HMGB1 and its receptors TLR4/MD-2 and RAGE (receptor for advanced glycation end products) are not involved. Injection of HMGB1 accelerates tissue repair by acting on resident muscle stem cells, hepatocytes, and infiltrating cells. The nonoxidizable HMGB1 mutant 3S, in which serines replace cysteines, promotes muscle and liver regeneration more efficiently than the wild-type protein and without exacerbating inflammation by selectively interacting with CXCR4. Overall, our results show that the reduced form of HMGB1 coordinates tissue regeneration and suggest that 3S may be used to safely accelerate healing after injury in diverse clinical contexts.
Introduction
Tissue regeneration is a well-orchestrated process that occurs after injury caused by disease, trauma or, in some cases, drug intake. Complete regeneration leads to full restoration to health (restitutio ad integrum), whereas incomplete or defective regeneration can lead to loss of tissue mass or replacement with fibrotic scars, which are associated with impaired functional recovery. Understanding the molecular events underlying the regeneration process and developing agents that aid regeneration is essential for patients with injured tissues in a variety of clinical settings. Because acute inflammation and tissue regeneration follow tissue damage, they are unavoidably entangled, and it is often stated that there can be no regeneration without inflammation (Karin and Clevers, 2016).
High mobility group box 1 (HMGB1) protein is one of the endogenous molecules released on injury and is responsible for triggering inflammation (Scaffidi et al., 2002). HMGB1 is a highly conserved nuclear protein that binds to the minor groove of DNA and acts as a DNA chaperone that helps the binding of transcription factors to DNA and the formation of nucleosomes (Celona et al., 2011). HMGB1 appears to be essential for life because Hmgb1 knockout mice die perinatally (Calogero et al., 1999). Beside its nuclear activities, HMGB1 also functions as a signal of tissue damage or a damage-associated molecular pattern when released passively or actively in the extracellular medium (Scaffidi et al., 2002). Others and we previously demonstrated that sequential oxidation of cysteines modulates and eventually abrogates HMGB1 functions (Venereau et al., 2012, 2013; Yang et al., 2012). Fully reduced HMGB1 (fr-HMGB1) associates with the chemokine CXCL12 and activates the CXCR4 receptor (Schiraldi et al., 2012), acting as a chemoattractant for cells, whereas HMGB1 containing a disulfide bond (ds-HMGB1) is a proinflammatory molecule that interacts with the TLR4 adaptor myeloid differentiation factor-2 (MD-2; Yang et al., 2015); further cysteine oxidation to sulfonates by reactive oxygen species abrogates both activities (Venereau et al., 2012). To dissect the various activities of HMGB1, we created a mutant (3S) in which the cysteines are replaced with serines, which are resistant to oxidation. The 3S mutant behaves as the reduced form of HMGB1 (Venereau et al., 2012, 2013).
We show here that HMGB1, in different redox forms that act on distinct receptors, either promotes inflammation or orchestrates tissue regeneration. Pharmacological treatment with HMGB1, in particular the 3S variant, can accelerate regeneration without exacerbating inflammation in muscle and liver, two paradigms for regenerative medicine and biology.
Results
Fully reduced HMGB1 and 3S promote muscle regeneration after acute muscle injury
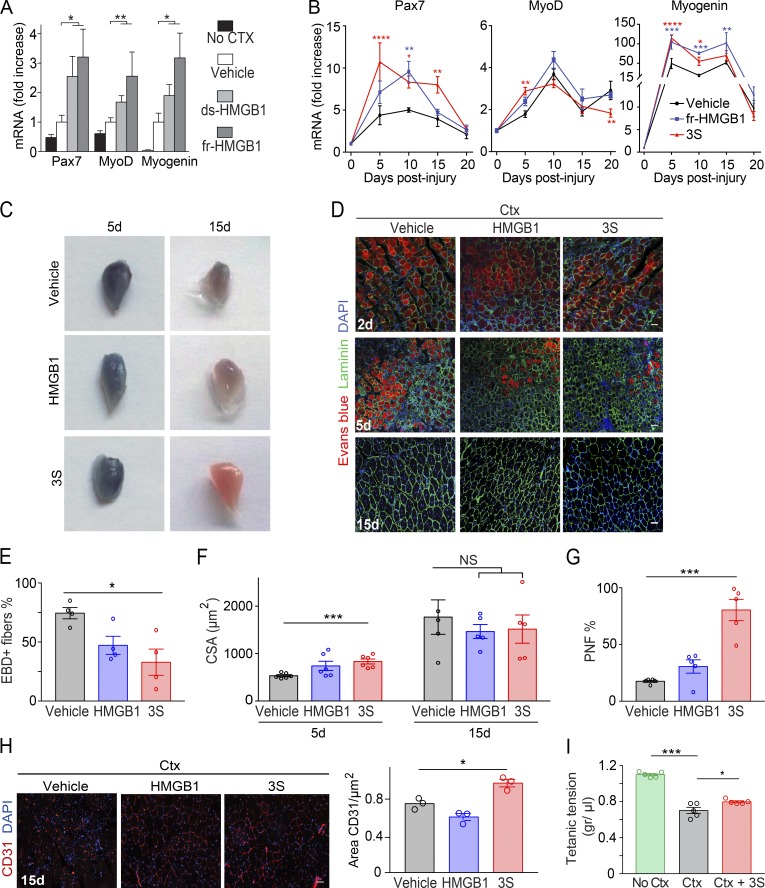
Several studies have suggested a role of HMGB1 in muscle regeneration; in particular, heterozygous Hmgb1+/− mice, which express 50% less HMGB1 when compared with WT mice, have delayed muscle regeneration after acute injury (Dormoy-Raclet et al., 2013). We tested whether providing alternative redox forms of HMGB1 would accelerate muscle regeneration in WT mice. We injured tibialis anterior (TA) muscles with cardiotoxin (Ctx) and simultaneously injected recombinant fully reduced HMGB1 (fr-HMGB1) or disulfide HMGB1 (ds-HMGB1). Both promoted a significant increase in the expression of the satellite cell marker Pax7 and the myogenic factors MyoD and Myogenin (Braun et al., 1989; Seale et al., 2000) compared with vehicle-injected mice (Fig. 1 A). However, HMGB1’s two redox forms can be interconverted in vivo, and their relative contribution cannot be evaluated. We therefore performed similar experiments with 3S, which is resistant to oxidation and functionally similar to fr-HMGB1 (Venereau et al., 2012). Notably, 3S treatment induced a significant increase of Pax7, MyoD, and Myogenin expression at early time points and a faster decrease at late time points (Fig. 1 B). These results indicated that ds-HMGB1 is likely not responsible for the induction of myogenic genes and suggested that 3S treatment might accelerate muscle regeneration. Indeed, the fraction of myofibers positive for staining with Evans blue dye (EBD), a reliable in vivo marker of damage, was significantly lower at day 5 in mice treated with 3S compared with control mice (Fig. 1, C–E). The EBD-negative fibers were centrally nucleated, and their cross-sectional area (CSA; a measure of regeneration) was significantly increased by ∼56% in 3S-treated mice compared with controls (Fig. 1 F). At day 15, the myofiber CSA in 3S-treated mice was comparable to that of control mice but not larger, indicating the absence of any excessive cellular growth or hypertrophy (Fig. 1 F). Finally, we observed a highly significant increase at day 15 in the number of regenerated fibers (myofibers with nuclei at the periphery) in the 3S-treated mice (80 ± 9%) compared with in mice treated with fr-HMGB1 (31 ± 6%) or vehicle (18 ± 1%; Fig. 1 G). In addition, the CD31+ endothelial area was increased in 3S-treated mice (Fig. 1 H), indicating that angiogenesis was also improved. Regenerating muscles of 3S-injected mice exerted a tetanic force significantly greater than muscles of vehicle-injected mice (Fig. 1 I). Overall, these findings clearly demonstrate that one single treatment with 3S considerably accelerates muscle regeneration.
Figure 1.
Intramuscular injection of fr-HMGB1 or 3S accelerates muscle regeneration. (A) Muscle acute injury was induced by injection of Ctx in triceps muscles. Vehicle (PBS) or HMGB1 (fr-HMGB1 or ds-HMGB1) was injected at the same time as Ctx. Quantitative PCR of Pax7, MyoD, and Myogenin (Myog) mRNA in triceps was performed at day 5 after injury (fold increase vs. vehicle). The significance of the difference in gene expression between mice treated with HMGB1 (fr-HMGB1 or ds-HMGB1) and vehicle-treated mice was assessed with Mann-Whitney tests. *, P < 0.05; **, P < 0.01. n = 9 mice per group, three independent experiments. (B–I) Ctx was injected together with vehicle (PBS) or WT fr-HMGB1 (HMGB1) or 3S in TA and/or triceps muscles. Muscle regeneration was assessed at days 2, 5, 10, 15, and 20 after injury. (B) Quantitative PCR of Pax7, MyoD, and Myog mRNA in triceps at days 5, 10, 15, and 20 after injury (fold increase vs. uninjured muscles). n ≥ 5 mice per group, at least two independent experiments. (C) TA muscles, stained with EBD, at days 5 and 15 after injury. (D) EBD, laminin, and DAPI staining of TA muscle sections at days 2, 5, and 15 after injury. Bars, 50 µm. (E) Percentage of EBD-positive myofibers in regenerating TA muscles at day 5 after injury. n = 4 mice per group, two independent experiments. (F) CSA of fibers in TA muscles at days 5 and 15 after injury. n ≥ 5 mice per group, at least two independent experiments. (G) Quantification of myofibers with nuclei at the periphery (PNF) in TA muscles at day 15 after injury. n = 5 mice per group, two independent experiments. (H) CD31 and DAPI staining on TA muscle sections at day 15 after injury and relative quantification of CD31+ area. n = 3 mice per group of three independent experiments. Bars, 50 µm. (I) Tetanic force of TA muscles: uninjured (No Ctx), cardiotoxin-injured (Ctx), Ctx-injured treated with 3S (Ctx + 3S), at day 10 after injury. n = 5 mice per group, two independent experiments. Differences between groups in B and E–I were assessed with one-way ANOVA plus Dunnett’s post-test; data are means ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
HMGB1 directly acts on muscle stem cells to support muscle repair
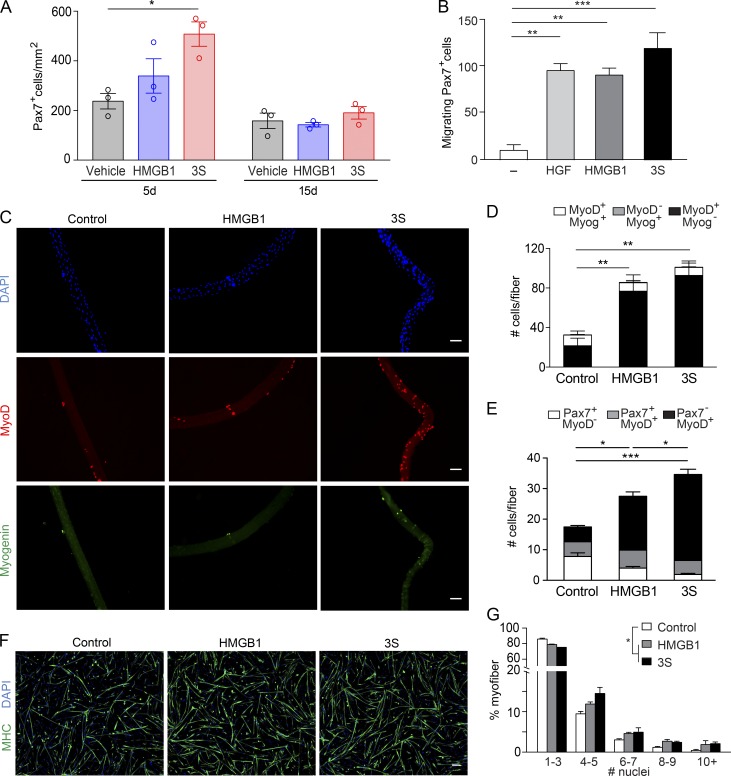
The number of Pax7+ cells (quiescent/activated satellite cells) was increased by ∼77% in 3S-treated mice at day 5 after injury compared with vehicle-treated mice (Fig. 2 A), suggesting that the regenerative properties of HMGB1 were mediated by muscle stem cells. HMGB1 forms a heterocomplex with the chemokine CXCL12 to induce cell migration via CXCR4, in particular fibroblasts and leukocytes (Schiraldi et al., 2012). Ex vivo migration experiments showed that both fr-HMGB1 and 3S were chemoattractant for Pax7+ cells and as effective as the conventional positive control human growth factor (Bischoff, 1997; Fig. 2 B).
Figure 2.
fr-HMGB1/3S promotes skeletal muscle regeneration by acting on satellite cells. (A) TA muscle acute injury was induced by injection of Ctx; vehicle (PBS) or WT fr-HMGB1 (HMGB1) or 3S was injected together with Ctx. Pax7+ cells were quantified in regenerating TA muscles at days 5 and 15 after injury. n = 3 mice per group, two independent experiments. (B) Ex vivo migration of primary mouse Pax7+ cells toward 40 nM HMGB1 (fully reduced) or 3S. Hepatocyte growth factor (HGF) was used as the positive control. n = 3 independent experiments. (C–E) Single myofibers isolated from mouse muscles were cultured in proliferation medium for 72 (C and D) or 16 h (E) with or without 40 nM fr-HMGB1 or 3S. Relative numbers of MyoD+ and/or Myogenin+ cells at 72 h (D) and of Pax7+ and/or MyoD+ cells at 16 h (E) on single fibers. n = 3 mice, 10 fibers/mouse, three independent experiments. Bars, 50 µm. (F and G) Representative images of mouse primary myoblasts cultured for 48 h in differentiating medium, with or without 40 nM fr-HMGB1 or 3S, and stained with DAPI and anti–myosin heavy chain (MHC) antibody (F), and distribution of myofiber size (G). Bar, 100 µm. The differences between control and treatments are statistically significant (P < 0.0001, χ2 test). Data are representative of two independent experiments with biological triplicates. Data are means ± SEM. Statistical significance was assessed with one-way ANOVA plus Dunnett’s post-test in A, B, D, and E. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
On injury, satellite cells get activated, proliferate, differentiate, and fuse to form new myofibers. To further investigate the effects of HMGB1 on such processes, we isolated single myofibers and analyzed the expression of specific markers: Pax7 (quiescent/activated satellite cells), MyoD (activated/proliferating myoblasts), and Myogenin (differentiating myoblasts). After 72 h of culture in the presence of fr-HMGB1 or 3S, the total number of myoblasts was significantly higher relative to the control, in particular the number of MyoD-positive cells, suggesting that treatment with HMGB1/3S promoted activation and proliferation of myoblasts (Fig. 2, C and D). Indeed, the total number of myoblasts was already significantly increased at 16 h, in particular the number of activated myoblasts (Pax7−MyoD+ cells), whereas the number of quiescent satellite cells (Pax7+MyoD− cells) was significantly decreased (Fig. 2 E). Both fr-HMGB1 and 3S induced the formation of larger myotubes from primary myoblasts (Fig. 2, F and G). Overall, these results identify HMGB1 as an activator of satellite cell myogenic output; 3S is even more effective.
fr-HMGB1 and 3S establish a tissue-healing microenvironment
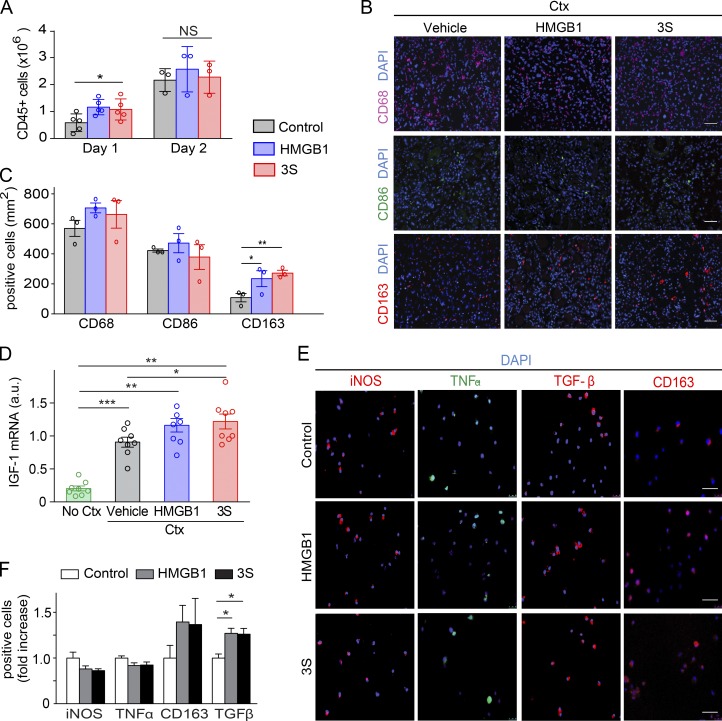
In the extracellular environment, HMGB1 is one of the main mediators in both acute and chronic inflammation. To investigate whether treatment with fr-HMGB1 or 3S might modulate the inflammatory state of the regenerating muscles, we examined the infiltration of leukocytes. We previously showed that endogenous fr-HMGB1 forms a heterocomplex with the chemokine CXCL12 that recruits leukocytes into injured muscles (Schiraldi et al., 2012). Only a slight increase of CD45+ cells was observed at day 1 after injury in muscles injected with fr-HMGB1 or 3S, suggesting that endogenous HMGB1 is already sufficient to promote optimal leukocyte recruitment (Fig. 3 A).
Figure 3.
fr-HMGB1/3S modulates macrophage polarization toward a tissue-healing phenotype. (A–D) TA muscles were injected with Ctx alone or Ctx plus WT fr-HMGB1 (HMGB1) or 3S. (A) Quantification of CD45+ cells isolated from injured muscles at days 1 (n = 5 mice per group of three independent experiments) and 2 (n = 3 mice per group of three independent experiments) after injury. (B and C) Representative immunofluorescence staining for CD68, CD86, and CD163 on sections of TA muscles at day 5 after injury (B) and quantification of CD68+ (n = 3 mice per group), CD68+CD86+, and CD68+CD163+ cells (n = 5 mice per group) in sections of TA muscles (C), two independent experiments. Bars, 50 µm. (D) Quantitative PCR of IGF-1 mRNA in triceps at day 5 after injury. n = 9 mice per group of three independent experiments. (E and F) Mouse bone marrow–derived macrophages were cultured for 7 d in DMEM conditioned by L929 cells (enriched in CSF-1) and polarized for 3 d toward a proinflammatory phenotype by stimulation with IFNγ (50 ng/ml) with or without 40 nM fr-HMGB1 or 3S. Representative images of macrophages (E) and quantification of cells positive for iNOS, TNFα, TGFβ, and CD163 expression, analyzed by immunofluorescence staining (F; fold increase vs. control; at least three independent experiments). Bars, 50 µm. Data are means ± SEM. Statistical significance was assessed with one-way ANOVA plus Dunnett’s post-test in A, C, D, and F. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
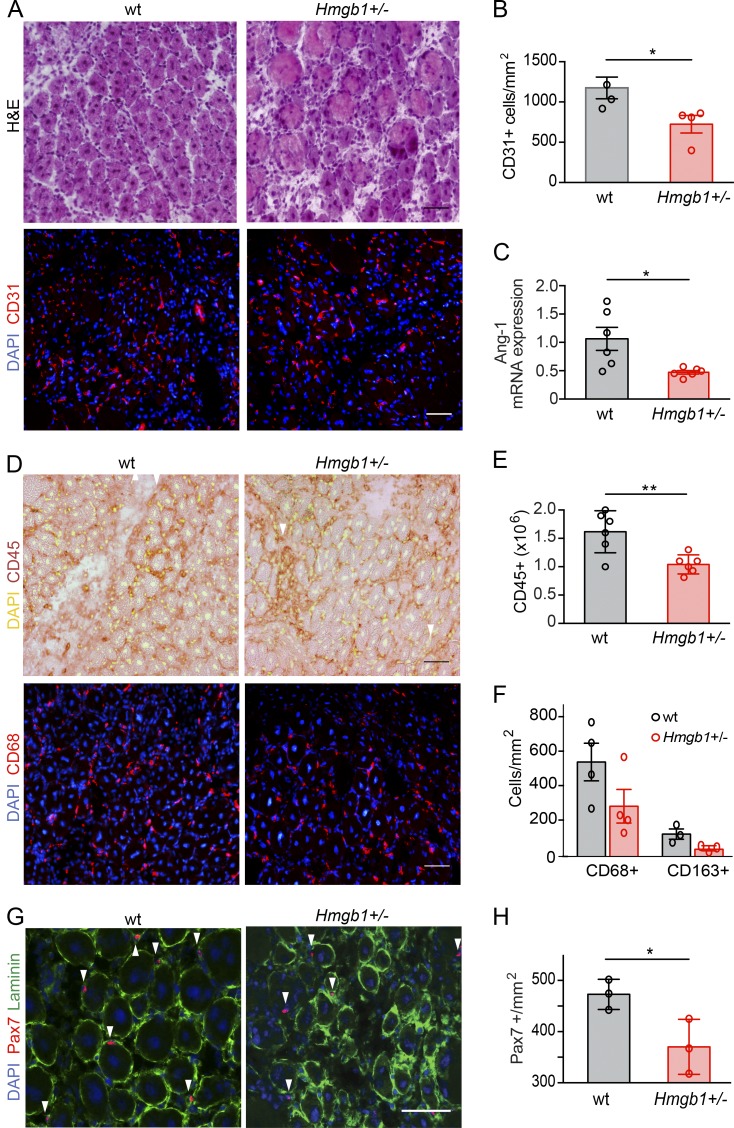
Recent work showed that HMGB1 modulates the polarization of macrophages (Schaper et al., 2016; Son et al., 2016). The switch from a proinflammatory to a tissue-healing phenotype in macrophages is an essential step in muscle regeneration to limit the inflammatory response and favor myogenesis (Arnold et al., 2007; Cohen and Mosser, 2013; Saclier et al., 2013; Wang et al., 2014). Thus, we analyzed the expression of specific macrophage surface markers at day 5 after injury: CD68 for total macrophages, CD86 for proinflammatory, and CD163 for tissue-healing macrophages (Saclier et al., 2013). Treatment with fr-HMGB1 or 3S increased, respectively, by ∼80% and 97% the number of CD163+ macrophages in the muscle relative to controls (Fig. 3, B and C). Expression of insulin-like growth factor-1 (IGF-1), a potent enhancer of tissue regeneration that promotes macrophage polarization toward the tissue-healing phenotype (Lefaucheur and Sébille, 1995; Lu et al., 2011; Tonkin et al., 2015), was also significantly increased in 3S-treated mice at day 5 after injury compared with vehicle controls (Fig. 3 D). Similarly, treatment of bone marrow–derived macrophages with fr-HMGB1 or 3S, in proinflammatory polarizing conditions, increased the expression of markers of the tissue-healing phenotype, such as CD163 and TGF-β (Fig. 3, E and F). Collectively, these data reveal that fr-HMGB1 and 3S act on myoblasts and macrophages to promote muscle regeneration rather than inflammation. Conversely, the marked delay of muscle regeneration in Hmgb1+/− mice after acute injury was associated with a defect in angiogenesis and in mobilization of satellite cells and leukocytes (Fig. 4); in particular both CD163+ macrophages and Pax7+ cells were sparser in injured muscles of Hmgb1+/− mice compared with WT (Fig. 4, F–H).
Figure 4.
High expression of HMGB1 is required for optimal skeletal muscle regeneration. Muscle acute injury was induced by injection of Ctx in TA and/or triceps muscles of WT or Hmgb1+/− mice, and regeneration was assessed at day 5 after injury. (A) H&E and immunofluorescence staining (DAPI and CD31) of TA muscle sections from WT and Hmgb1+/− mice at day 5 after injury. Bars, 50 µm. (B) Quantification of CD31-positive cells/mm2 in TA muscles of WT and Hmgb1+/− mice at day 5 after injury. n = 4 mice per group of three independent experiments. (C) Quantitative PCR analysis of mRNA levels of angiopoietin-1 (Ang-1) in triceps at day 5 after injury. n = 6 mice per group of two independent experiments. (D) Representative immunostaining (DAPI and CD45, top; DAPI and CD68, bottom) of TA muscle sections from WT and Hmgb1+/− mice at day 5 after injury. Bars, 50 µm. (E) Quantification of CD45-positive cells isolated with immunobeads from injured muscles. n = 6 mice per group of two independent experiments. (F) Quantification of CD68-positive cells (n = 4 mice) and CD68/CD163-positive macrophages (n = 3 mice) in TA muscle sections from WT and Hmgb1+/− mice at day 5 after injury, two independent experiments. (G) Representative immunofluorescence staining for DAPI, laminin, and Pax7 in TA muscles from WT and Hmgb1+/− mice at day 5 after injury (Pax7-positive cells indicated with white arrowheads). Bars, 50 µm. (H) Quantification of Pax7-positive cells in TA muscles of WT and Hmgb1+/− mice at day 5 after injury. n = 3 mice per group of two independent experiments. In all panels, data are means ± SEM, and statistical significance was assessed with Student’s t test. *, P < 0.05; **, P < 0.01.
Alternative redox forms of HMGB1 contribute to muscle repair or inflammation by binding to different receptors
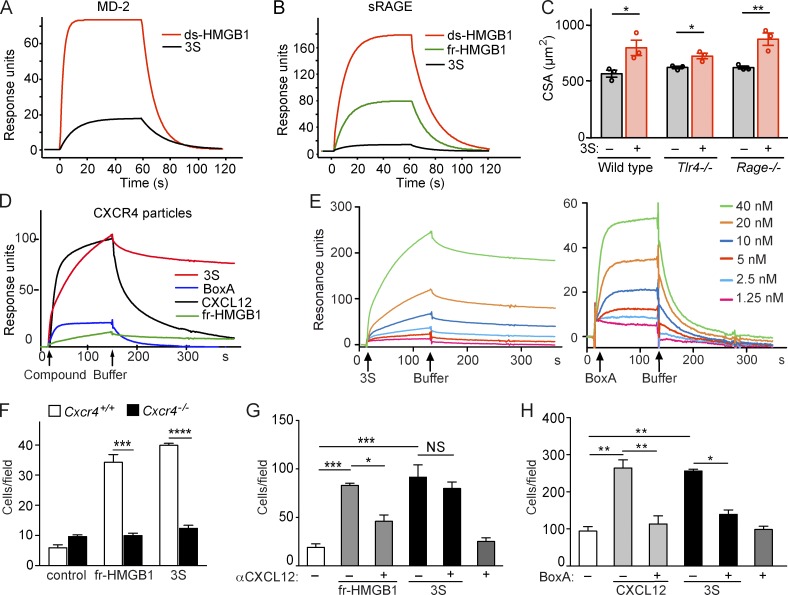
Our results indicate that the regenerative properties of HMGB1 are dictated by its redox state, and we next asked which HMGB1 receptor or receptors are involved in muscle regeneration. ds-HMGB1 binds TLR4 via the adaptor protein MD-2, and cysteine 106 is essential (Yang et al., 2010, 2015). Surface plasmon resonance (SPR) experiments showed that 3S, in which cysteine 106 is mutated to serine, binds to MD-2 with a very low affinity compared with ds-HMGB1 (Fig. 5 A), in agreement with its inability to induce cytokine and chemokine expression in macrophages (Venereau et al., 2012). The receptor for advanced glycation end products (RAGE) also preferentially bound to ds-HMGB1 (Fig. 5 B), and 3S bound inefficiently. These results suggested that the regenerative properties of fr-HMGB1 and 3S were not mediated by TLR4/MD-2 or RAGE, contrary to the inflammatory activities of ds-HMGB1. Indeed, 3S promoted muscle regeneration when injected in the muscles of Ctx-injured WT, Tlr4−/−, and Rage−/− mice, with no significant differences between the different genotypes (Fig. 5 C).
Figure 5.
3S and BoxA directly bind to CXCR4. (A and B) SPR sensorgrams representing the binding of HMGB1 isoforms to immobilized MD-2 (A) or sRAGE (B). ds-HMGB1 and 3S bind to MD-2 with an apparent KD of 0.35 µM and 2 µM, respectively (A). Binding to sRAGE: apparent KD of 1 µM for ds-HMGB1 and 3.5 µM for fr-HMGB1 and 3S (B). Data are representative of three independent experiments. (C) TA muscles of WT, Tlr4−/− and Rage−/− mice were injected with Ctx and vehicle (PBS) or 3S, and the CSA of centronucleated fibers was measured at day 5 after injury. n = 3 mice per group, two independent experiments. Data are means ± SEM. Student t tests within each genotype: *, P < 0.05; **, P < 0.01. (D) Sensorgrams representing the binding of 40 nM fr-HMGB1 or 3S, 50 nM CXCL12, and 40 nM BoxA to immobilized lentiviral particles with membrane-bound CXCR4. (E) Sensorgrams representing the binding of different concentrations of 3S (left) or BoxA (right) to immobilized lentiviral particles with membrane-bound CXCR4. Curves derived from these assays were analyzed by fitting to a simple one-site interaction model with BIA Evaluation 4.1 software (GE Healthcare). Data are representative of three independent experiments. (F) Migration of WT (Cxcr4+/+) or Cxcr4−/− MEFs toward 0.4 nM fr-HMGB1 or 3S. Data are representative of three independent experiments and are means ± SEM. Statistical significance between genotypes was calculated with Student’s t test. ***, P < 0.001; ****, P < 0.0001. (G and H) Migration of myoblasts toward 40 nM fr-HMGB1 and 3S with or without 10 nM anti-CXCL12 antibody (G) or toward 40 nM 3S and 30 nM CXCL12 with or without 250 nM BoxA (H). Data are means ± SEM of triplicates, two independent experiments. Differences between treatments were assessed with one-way ANOVA plus Tukey’s post-test. *, P < 0.05; **, P < 0.01, ***, P < 0.001.
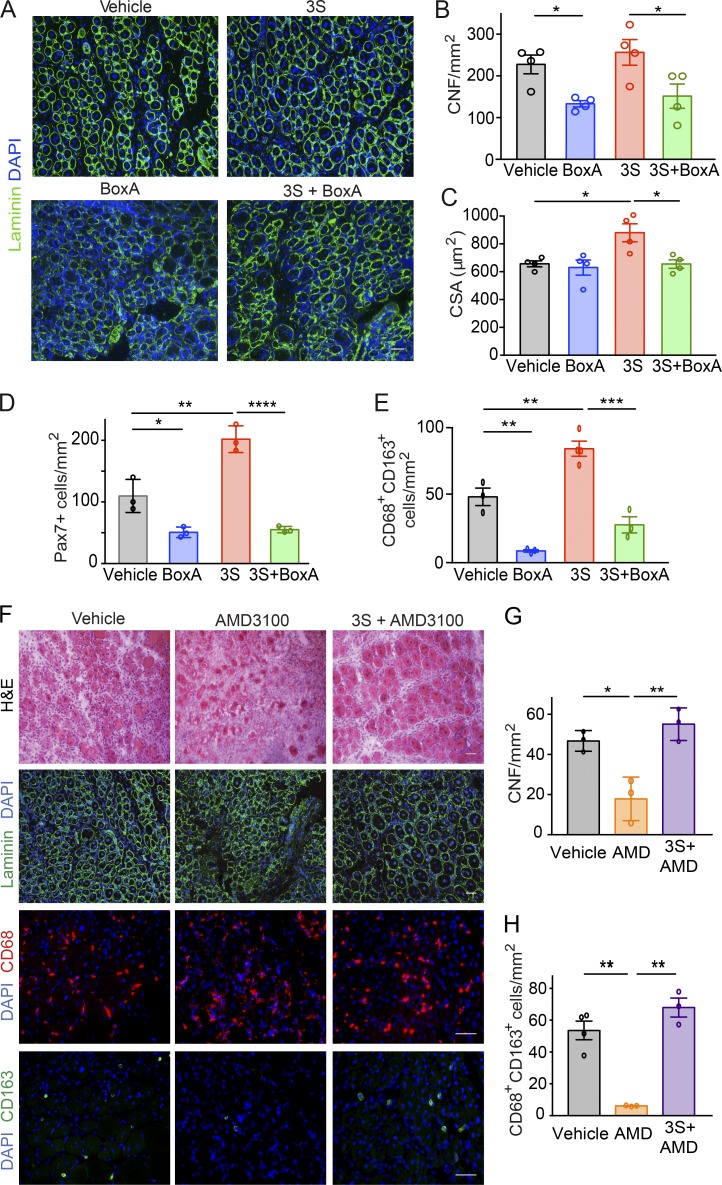
HMGB1 forms a heterocomplex with the chemokine CXCL12 to induce cell migration via CXCR4 (Schiraldi et al., 2012). By analogy, we explored the involvement of the CXCL12/CXCR4 axis in HMGB1-mediated regeneration. To analyze the binding of 3S to CXCR4, we performed SPR experiments with immobilized lentiviral particles expressing CXCR4 on their surface (Vega et al., 2011). As reported previously (Schiraldi et al., 2012), fr-HMGB1 failed to bind to CXCR4 in the absence of CXCL12 (Fig. 5 D). Surprisingly, we found that 3S directly bound to CXCR4, with an affinity 10-fold higher than that of CXCL12 itself (KD of 6.44 vs. 76.9 nM; Fig. 5, D and E; and Table 1). We also found that BoxA, a truncated form of HMGB1 used as an antagonist of HMGB1, was able to bind to CXCR4 with high affinity (KD, 6.66 nM) in the absence of CXCL12 (Fig. 5, D and E; and Table 1). Accordingly, 3S-induced migration is mediated by CXCR4, because Cxcr4−/− cells did not migrate toward 3S (Fig. 5 F) and anti-CXCL12 antibodies inhibited the migration of myoblasts toward fr-HMGB1 but had no effect on migration toward 3S (Fig. 5 G). BoxA inhibited myoblast migration in response to 3S or CXCL12, demonstrating that BoxA is a CXCR4 inhibitor (Fig. 5 H). Consistently, BoxA abrogated the beneficial effects of 3S (Fig. 6, A–C), in particular the expansion of Pax7+ cells and CD163+ cells (Fig. 6, D and E). Interestingly, BoxA also interfered with the unaided muscle regeneration process (Fig. 6, A–E) and so did treatment with AMD3100/Plerixafor, the most widely used CXCR4 antagonist (Fig. 6, F–H). AMD3100 binds to CXCR4 with an affinity similar to that of CXCL12 (Rosenkilde et al., 2004), whereas 3S has a 10-fold higher affinity (Table 1). Accordingly, 3S was able to restore muscle regeneration in AMD3100-treated mice (Fig. 6, F–H). Overall, our findings demonstrate that CXCR4 plays a central role in muscle regeneration and that 3S can bypass the need for CXCL12 by directly interacting with CXCR4.
Table 1. Kinetic constants of CXCL12, 3S, and BoxA binding to CXCR4.
| Ligand | Ka | Kd | KD |
|---|---|---|---|
| 1/Ms | 1/s | nM | |
| CXCL12 | 1.5 × 105 | 5.83 × 10−2 | 76.9 |
| 3S | 6.81 × 105 | 1.04 × 10−3 | 6.4 |
| BoxA | 5.75 × 106 | 2.3 × 10−2 | 6.7 |
Curves derived from binding of CXCL12, 3S, and BoxA to immobilized lentiviral particles with membrane-bound CXCR4 were used to generate kinetic constants (Ka, Kd, KD) by using a simple one-site interaction model with BIA Evaluation 4.1 software (GE Healthcare). Data are representative of three independent experiments.
Figure 6.
HMGB1 supports muscle regeneration via CXCR4. (A–E) Comparison of C57BL/6 mice treated with Ctx plus vehicle versus Ctx plus BoxA and Ctx plus 5 mg/kg 3S with or without 10 mg/kg BoxA. (A) Laminin and DAPI staining of TA muscle sections at day 5 after injury. Bar, 50 µm. Number (B) and CSA of centronucleated fibers (CNFs; C) in TA muscles at day 5 after injury. n = 4 mice per group, two independent experiments. (D) Quantification of Pax7+ cells in regenerating TA muscles at day 5 after injury. n = 3 mice per group, two independent experiments. (E) Quantification of CD68+CD163+ cells in sections of TA muscles at day 5 after injury. n = 4 mice per group, two independent experiments. (F–H) Comparison of muscles from mice receiving one single intramuscular injection of Ctx plus vehicle (PBS) versus Ctx plus 5 mg/kg AMD3100 with or without 5 mg/kg 3S. (F) Representative images of H&E and immunofluorescence staining for DAPI and laminin or CD68/CD163. Bars, 50 µm. (G) Number of CNFs in TA muscles at day 5 after injury. n = 3 mice per group, two independent experiments. (H) Quantification of CD68+CD163+ cells on sections of TA muscles at day 5 after injury. n = 3 mice per group, two independent experiments. Differences between groups were assessed with one-way ANOVA plus Tukey’s post-test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
3S promotes liver regeneration without exacerbating inflammation
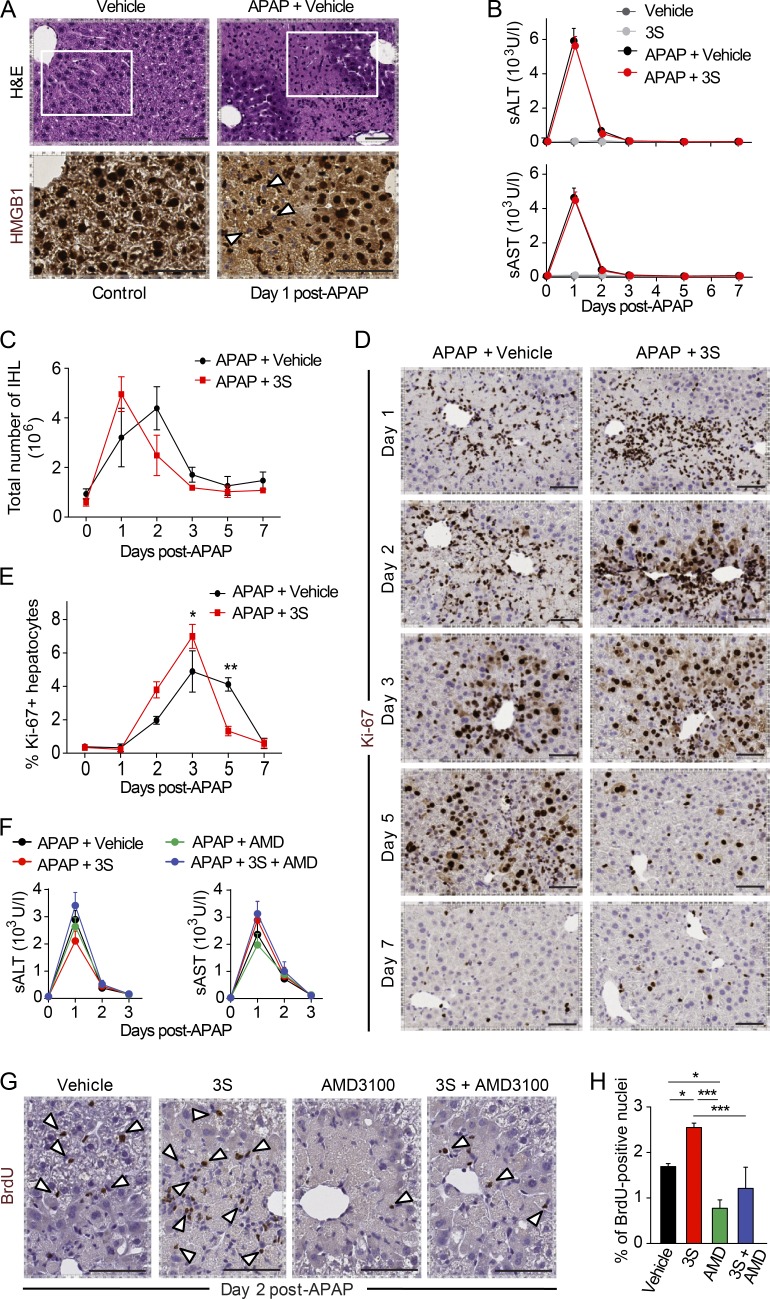
We then asked whether 3S promotes regeneration in other tissues. We examined liver regeneration, which is one of the most studied models of tissue regeneration. Previous studies have demonstrated that endogenous HMGB1 is a mediator of drug-induced hepatoxicity by promoting inflammation via its interaction with TLR4/MD-2 (Antoine et al., 2012; Yang et al., 2015). Hence, the drug-induced liver injury (DILI) model represents an ideal system to evaluate the regenerative and noninflammatory properties of 3S. We first analyzed HMGB1 expression in healthy and acetaminophen (APAP)-intoxicated livers (Fig. 7 A). Notably, hepatocytes abutting the APAP-induced injury had HMGB1-negative nuclei, indicating that they secreted HMGB1. We administered 3S systemically 2 h after APAP injection. Vehicle- and 3S-treated mice had similar levels of serum alanine aminotransferase (sALT) and serum aspartate aminotransferase (sAST; Fig. 7 B). Thus, 3S does not modulate APAP-induced liver injury. Accordingly, flow cytometry quantification of intrahepatic subpopulations of leukocytes showed no significant difference between vehicle- or 3S-treated mice (Fig. 7 C).
Figure 7.
3S accelerates liver regeneration via CXCR4. DILI was induced by i.p. injection of 300 mg/kg (body weight) APAP. 2 h later, mice received one single i.p. injection of vehicle (NaCl), 500 µg/mouse 3S, or 200 µg/mouse AMD3100 with or without 500 µg/mouse 3S. Serum collection and necroscopy were performed at different time points, and hepatic regeneration was assessed by i.p. injection of 1 mg/mouse BrdU 16 h before liver collection. (A) Representative images of H&E staining and HMGB1 immunostaining of liver in control mice on day 1 after DILI. HMGB1 negative nuclei (white arrowheads) are visible at higher magnification of the areas identified by rectangles in APAP-treated mice. Bars, 50 µm. (B) sALT and sAST in serum after APAP injection in mice treated with vehicle or 3S. n = 6 mice per group, two independent experiments. (C) Quantification of intrahepatic leukocytes (IHL) at days 1, 2, 3, 5, and 7 after APAP injection in control mice and after DILI in mice treated with vehicle or 3S. n = 4 mice per group, two independent experiments. Data are means ± SEM, and differences between groups were assessed with two-way ANOVA plus Bonferroni post-tests. The interaction between treatment and time is not significant. (D and E) Representative images of Ki-67 immunostaining (D) and quantification of Ki-67–positive hepatocytes (E) in livers from mice treated with vehicle or 3S at days 1, 2, 3, 5, and 7 after APAP injection. Bars, 50 µm. n = 4 mice per group, two independent experiments. Data are means ± SEM, and differences between groups were assessed with two-way ANOVA plus Bonferroni post-tests. The interaction between treatment and time is significant (P = 0.0003). (F) sALT and sAST after APAP injection in mice treated with vehicle, 3S, or AMD3100 with or without 3S. n = 4 mice per group, two independent experiments. Differences between groups in B and F were assessed with one-way ANOVA plus Dunnett’s post-test. (G and H) Representative images of BrdU immunostaining (G; BrdU-positive hepatocytes indicated with white arrowheads) and quantification of BrdU-positive hepatocytes (H) in livers from mice treated with vehicle, 3S, or AMD3100 with or without 3S at day 2 after APAP injection. Bars, 50 µm. n = 4 mice per group. Data are means ± SEM, and differences between groups were assessed with one-way ANOVA plus Bonferroni post-tests. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
In contrast to muscle, liver regeneration does not rely on stem cells but on differentiated hepatocytes, which proliferate in a highly regulated manner after loss of liver mass. Interestingly, hepatocyte proliferation occurred significantly earlier in 3S-treated mice: the number of Ki67-positive hepatocytes was higher at days 2 and 3 after APAP injection compared with control mice and declined already at day 5 (Fig. 7, D and E). To investigate the role of CXCR4 in the DILI model, we administered 3S, AMD3100, or a combination of both, 2 h after APAP injection. All groups of mice had similar APAP-induced liver injury, indicating that the different treatments did not cause further liver damage (Fig. 7 F). At day 2 after APAP injection, when liver regeneration was starting, AMD3100-treated mice had a lower number of BrdU-positive hepatocytes compared with control mice, indicating that CXCR4 blockade inhibits hepatocyte proliferation (Fig. 7, G and H). Overall, our findings indicate that 3S is noninflammatory in a DILI model in contrast to endogenous HMGB1 and conversely promotes liver regeneration through the CXCR4 receptor.
Discussion
In this study we have shown that HMGB1 coordinates tissue repair and inflammation by switching among alternative redox forms. Specifically, fr-HMGB1 promotes muscle and liver repair through several cell-specific responses mediated by CXCR4, whereas ds-HMGB1, which triggers and sustains inflammation via the TLR4/MD-2 and RAGE receptors, is not involved in regeneration. HMGB1 needs to form a complex with CXCL12 to interact with CXCR4 (Schiraldi et al., 2012) and appears to be a limiting factor in physiological conditions, because the reduction of HMGB1 content in Hmgb1+/− mice delays muscle repair (Dormoy-Raclet et al., 2013), whereas the injection of exogenous fr-HMGB1 sizably accelerates it. The 3S variant is a nonoxidizable form of HMGB1, which cannot be converted to ds-HMGB1 or be inactivated by sulfonylation. Notably, 3S binds inefficiently to TLR4/MD-2 and RAGE, the receptors mediating the inflammatory properties of HMGB1, but it directly binds to CXCR4 with high affinity, bypassing the requirement for CXCL12. Accordingly, 3S is more effective than the WT protein in promoting regeneration and does not exacerbate inflammation.
HMGB1 is a ubiquitous protein that is released by all cell types upon injury, and consequently it might contribute to regeneration in multiple tissues. Indeed, HMGB1 appears to exert beneficial effects under several pathological conditions by recruiting different types of stem or progenitor cells and promoting their proliferation (Palumbo et al., 2004; Limana et al., 2005; Chavakis et al., 2007; Deneault et al., 2009; Tamai et al., 2011). Similarly, the CXCL12/CXCR4 axis has been reported to activate the body’s own tissue repair pathways, and different strategies aimed at increasing CXCL12 availability have been tried to facilitate tissue repair (Lau and Wang, 2011). We now show that the critical ligand (or coligand) of CXCR4 in muscle and liver regeneration is HMGB1. Muscle and liver have very different tissue architectures and rely on different cellular mechanisms for regeneration: muscle relies on local stem cells, the satellite cells, and liver relies on the proliferation capacities of hepatocytes. Based on these extreme differences, and the common regeneration responses to 3S, we suggest that the HMGB1/CXCR4 axis may be involved in the repair and regeneration of most tissues.
Since the discovery of its extracellular release, most attention has been paid to the contribution of HMGB1 to inflammation. Our data demonstrate that fr-HMGB1 and 3S are devoid of proinflammatory activities, indicating that the HMGB1 redox form responsible for inflammation is ds-HMGB1, acting via RAGE and TLR4 signaling. Accordingly, ds-HMGB1 has been reported to be the isoform responsible for DILI, neuroinflammation, epilepsy, thrombosis, and pain (Agalave et al., 2014; Balosso et al., 2014; Yang et al., 2015; Stark et al., 2016; Lian et al., 2017). Conversely, mice deficient in HMGB1 in specific cell types—intestinal epithelial cells, pancreatic cells, hepatocytes, or monocytes—display worsened phenotypes of inflammatory bowel disease, pancreatitis, liver damage after ischemia/reperfusion, and sepsis, respectively (Yanai et al., 2013; Huang et al., 2014; Kang et al., 2014; Zhu et al., 2015). Although the beneficial role of HMGB1 in these models has been mostly attributed to its intracellular functions, it is equally possible that the worsened outcome after insult is due to a defective repair of the damaged tissue. Therapeutic strategies aiming at the prevention of HMGB1 oxidation, rather than HMGB1 neutralization, might be useful in diverse clinical settings to fine-tune the pharmacological control of inflammation and to support tissue regeneration. Accordingly, the current therapy for APAP intoxication is the administration of the antioxidant N-acetylcysteine (NAC). Based on our results, the efficacy of NAC could be explained, at least in part, by the shift in balance between deleterious ds-HMGB1 and beneficial fr-HMGB1.
Several strategies have been developed so far to counteract the extracellular activities of HMGB1. A truncated form of HMGB1, BoxA, has been extensively used in vivo to inhibit HMGB1, and its protective effects have been demonstrated in multiple mouse models of pathological conditions (Venereau et al., 2016). However, its mechanism of action has not been fully elucidated yet. Notably, we show that BoxA interacts directly with CXCR4 and suppresses the activity of both 3S and CXCL12, indicating that BoxA is an antagonist of CXCR4. Hence, some effects of BoxA might be attributed to the inhibition of the CXCL12/CXCR4 axis, in particular in the context of cell recruitment and cancer (Guo et al., 2016).
More generally, this study provides an example in which the organism uses the same molecule in alternative redox forms to orchestrate sequential physiological processes after tissue injury—signaling damage, triggering inflammation, and eventually promoting regeneration. Of most direct utility, we identified 3S as a promising drug candidate to promote tissue repair without exacerbating inflammation. In agreement with our findings, 3S has been recently demonstrated to exert no deleterious effects in neuroinflammation, excitotoxicity, and thrombosis (Balosso et al., 2014; Stark et al., 2016; Lian et al., 2017), contrary to WT HMGB1. The ability to safely accelerate regeneration with a single administration of 3S (locally in the muscle or systemically in the liver) opens up promising therapeutic opportunities for regenerative medicine.
Materials and methods
Mice and injury models
8-wk-old C57BL/6 mice were purchased from Charles River Laboratories. Hmgb1+/−, Tlr4−/−, Rage−/−, and Pax7-ZsGreen mice (C57BL/6), were bred in the animal facility at San Raffaele Scientific Institute. All mice were housed under standard or specific pathogen–free conditions and allowed access to food and water ad libitum. All experimental protocols were approved by the San Raffaele Institutional Animal Care and Use Committee (IACUC 590 and 663) in accordance with Italian law. All efforts were made to minimize suffering. Male and female mice were used and always sex-matched in each experiment.
In the acute muscle injury model, animals were anaesthetized by i.p. injection of Avertin (T48402, 2,2,2-Tribromoethanol 97%; Sigma-Aldrich), and sterile injury was induced by injection of 50 µl 15-µM Ctx (C9759 Sigma-Aldrich) in TA and triceps muscles. Treatment with vehicle or HMGB1 (5 nmol WT fr-HMGB1 or 3S; HMGBiotech) was administered together with Ctx. Mice were sacrificed 2, 5, 15, or 20 d after Ctx injection, and muscles were collected and either sectioned for histological analysis or subjected to RNA extraction.
For the DILI model, 8-wk-old C57BL/6 males were fasted 16 h before i.p. injection of 300 mg/kg (body weight) APAP (Sigma-Aldrich) dissolved in sterile warm saline. 2 h after APAP intoxication, mice received vehicle (NaCl) or 3S (500 µg/mouse), AMD3100 (200 µg/mouse; Sigma-Aldrich), or 3S and AMD3100 by i.p. injection. Transaminases (sALT and sAST) were quantified in serums at days 1, 2, 3, 5, and 7 after APAP administration with an International Federation of Clinical Chemistry and Laboratory Medicine–optimized kinetic UV method in an ILab Aries chemical analyzer (Instrumentation Laboratory), and they are expressed as U/liter. To assess hepatocyte proliferation, 16 h before sacrifice mice received 1 mg BrdU (Sigma-Aldrich) by i.p. injection.
Morphometry
Morphometric analyses were performed on sections collected from similar regions of each TA muscle. CSA and central nucleation analyses were performed on 500–750 fibers/TA muscle after laminin and DAPI immunostaining by using ImageJ software (http://rsbweb.nih.gov/ij/).
Histology and immunofluorescence
Muscles were collected and embedded in OCT (Bio-Optica) or directly frozen in liquid nitrogen–cooled isopentane. Serial muscle sections, 8-µm thick, were stained with hematoxylin and eosin (H&E; Sigma-Aldrich) according to standard procedures. Immunofluorescence on frozen sections of muscles was performed as previously described (Brunelli et al., 2007) by using the following primary antibodies: chicken anti-laminin (1:500; Sigma-Aldrich), rat anti-CD68 (1:100; AbD Serotec, Bio Rad), rat anti-CD86 (1:100; BD-PharMingen), rabbit anti-CD163 (1:200; Santa Cruz Biotech), rat anti-CD31 (1:2, clone MEC13.3; gift from E. Dejana, Uppsala University, Uppsala, Sweden). Appropriate Alexa Fluor (Alexa 488 or Alexa 546)–conjugated antibodies (1:500; Invitrogen) were used as second-step reagent. Nuclei were counterstained with DAPI (1:1,000; Sigma Aldrich), and coverslips were mounted with Fluorescence Mounting Medium (Dako).
For Pax7 staining, 8-µm serial muscle sections were permeabilized with 0.2% Triton, 1% BSA in PBS for 30 min at room temperature and then blocked in 10% serum, 1% BSA solution in PBS for 30 min before incubation with the primary antibody (Pax7; Developmental Studies Hybridoma Bank), after an antigen retrieval step in sodium citrate 10 mM, pH 6.0, for 10 min between fixation and blocking steps (2 h).
Livers were collected, fixed in zinc-formalin, and then embedded in paraffin and stained with H&E, anti–Ki-67 (1:100, clone SP6; Neomarkers) and anti-HMGB1 (1:200; BD PharMingen) and anti-BrdU (1:100, Abcam) antibodies.
Image acquisition and analyses
Fluorescent and phase contrast images were taken by using Nikon Eclipse E600 or Zeiss Imager M2 microscopes. Image acquisition was done by using the Nikon digital camera DXM1200 or AxioCam MRc5 camera and the acquisition software Nikon ACT-1 (Plan Fluor lenses: X4/0.13, X10/0.33, X20/0.50, X40/0.75) or AxioVision. Images showing double fluorescence were first acquired separately by using appropriate filters, and then the different layers were merged by using Adobe Photoshop CS6. Histological quantifications were done by counting at least 20 fields (20× and/or 40×) for each data point.
For the quantification of CD31-positive staining, the red signal from CD31 immunostaining was filtered by using the same parameters for all samples, and regions on the tissue sections were marked to outline areas to be quantified. A morphological image processing operation, flood-fill, was used to identify the pixels within the image that belonged to the regions of interest. CD31-stained pixels were identified by thresholding red-to-blue channel intensity. An empirically determined low threshold of 1.1 was used to segment out CD31-positive regions.
All images of liver immunohistochemical staining were acquired by using the Aperio Scanscope CS2 system (Leica Biosystems). Quantifications of Ki-67– and BrdU-positive hepatocytes were performed by automated image analysis software through dedicated macros of the ImageScope program (Leica Biosystems) on liver sections of an area between 5 and 30.4 mm2, as previously described (Catarinella et al., 2016).
Muscle force
TA muscles were dissected from mouse legs under a stereomicroscope (×10 to ×60) and transferred to the myograph to be mounted between the hooks of a force transducer (AME 801; Aksjeselkapet Mikkroelektronik). Preparations were placed in an organ bath filled with Krebs solution (120 mM NaCl, 2.4 mM KCl, 2.5 mM CaCl2, 1.2 mM MgSO4, 5.6 mM glucose, 1.2 mM KH2PO4, and 24.8 mM NaHCO3, pH 7.4), bubbled with 95% O2 and 5% CO2 at the constant temperature of 22°C, and attached to a force transducer (FT-03; AstroNova). On both sides of the perfusion bath, at a distance of ∼2 mm from the preparations, plate platinum electrodes connected with a Grass stimulator allowed field electrical stimulation (S48 stimulator; AstroNova). The preparations were stretched to Lo (the length at which the maximal twitch force is observed), and their response to electrical stimulation was tested. If a response could be elicited, the preparation was stimulated for ∼30 min with supramaximal, low-frequency (0.03 Hz) stimuli. Tetanic isometric contractions were evoked (110 Hz, 500 ms, supramaximal amplitude) at Lo. Normalized tetanic force was expressed as maximal tetanic force/muscle volume (gr/µl). The output of the tension transducer was stored in a personal computer after analogue-to-digital conversion and was recalled for analysis. A 1401 A/D converter (Cambridge Electronic Design) and CEA Spike2 software (Cambridge Electronic Design) were used.
Quantitative PCR
Total RNA was isolated from triceps by using the TRIzol reagent (15596–026; Ambion) according to manufacturer’s instructions. Extracted RNA was quantified with NanoDrop (ND-1000 Spectrophotometer; EuroClone). cDNA was obtained by retrotranscription with Oligo(dT) primers (Invitrogen) and SuperScript II Reverse transcription (Invitrogen). Quantitative real-time PCR was performed in duplicates by using SYBR Green I master mix and LightCycler480 (Roche Molecular Diagnostics). ΔCt were calculated by using RSP29 as normalizer. The following primers were used (Tm 60°C): angiopoietin-1: 5′-GACAGTAATACAACACCGGGAAGA-3′ (forward), 5′-CAAAACCCATTTTATACTCCTTCCA-3′ (reverse); CD31: 5′-AGGGGACCAGCTGCACATTAGG-3′ (forward), 5′-AGGCCGCTTCTCTTGACCACTT-3′ (reverse); IGF-1: 5′-GAATGTTCCCCCAGCTGTTTC-3′ (forward), 5′-CATTGCGCAGGCTCTATCTG-3′ (reverse); MyoD: 5′-ACGGCTCTCTCTGCTCCTTT-3′ (forward), 5′-GTAGGGAAGTGTGCGTGCT-3′ (reverse); Myogenin: 5′-GACATCCCCCTATTTCTACCA-3′ (forward), 5′-GTCCCCAGTCCCTTTTCTTC-3′ (reverse); Pax7: 5′-GACTCGGCTTCCTCCATCTC-3′ (forward), 5′-AGTAGGCTTGTCCCGTTTCC-3′ (reverse); and RSP29: 5′-AGTCACCCACGGAAGTTCG-3′ (forward), 5′-AGCATGATCGGTTCCACTTG-3′ (reverse).
Quantification of leukocytes
Muscles were dissociated in RPMI 1640 containing 0.2% collagenase B (Roche Diagnostics) at 37°C for 1 h. CD45-positive cells were purified by magnetic cell sorting by using anti-CD45 beads (Miltenyi Biotec) according to the manufacturer’s instructions and quantified by Countess (Invitrogen).
Blood samples from WT and Hmgb1+/− mice were collected from the tail vein, and circulating leukocytes were measured by Sysmex Kx21N according to the manufacturer’s instructions (Sismex Inc.). Leukocyte isolation from the liver was performed as previously described (Sitia et al., 2007).
Myoblast isolation, differentiation, and migration assays
15-d-old CD1 mice were sacrificed and muscles were cut in fine pieces and digested twice in collagenase and dispase medium (Gibco by Life Technologies) at 37°C for 20 min under strong agitation in a water bath. Pellets were resuspended in DMEM 10% FBS and passed through 70-µm and 40-µm filters. After incubation with red blood cell lysis buffer (Z314; Promega), cells were preplated on one 150-mm uncoated dish for 2 h. Myoblasts in suspension were centrifuged, resuspended in proliferation medium (IMDM; 1% glutamine, 1% penicillin/streptomycin, 0.1% gentamicin, 20% FBS, and 3% chicken embryo extract), and plated in collagen-coated 150-mm dishes. The purity of cells was assessed by FACS analysis by using SMC/2.6-biotin (a gift from S.-i. Fukada, Graduate School of Pharmaceutical Sciences, Osaka University, Osaka, Japan) and Streptavidin-APC (405207; Biolegend) as previously described (Fukada et al., 2004). For the differentiation assay, cells were plated after 4 d in culture on Matrigel-coated slides in 12-well plates (105 cells/well) in differentiating medium (IMDM; 1% glutamine, 1% penicillin/streptomycin, 0.1% gentamicin, 2% horse serum, and 1% chicken embryo extract) with or without 40 nM WT fr-HMGB1 or 40 nM 3S. After 48 h, cells were fixed with 4% paraformaldehyde in PBS and stained with DAPI and anti–myosin heavy chain (clone MF20; DHSB).
For migration assays, 8-µm transwell filters (Corning) were first coated with collagen. Myoblasts were isolated as described above in the previous paragraph, after 2 h of preplating the suspended myoblasts were counted, and 5 × 104 cells were transferred onto the upper side of the transwell chamber in proliferation medium alone or with 150 pM human growth factor or 40 nM HMGB1 (WT fully reduced form or 3S mutant). After 16 h the cells were fixed with 4% paraformaldehyde, and Pax7 immunostaining (Pax7; Developmental Studies Hybridoma Bank) was performed. Alternatively, Pax7-positive cells were isolated by cell sorting from Pax7-ZsGreen mice and tested with 40 nM fr-HMGB1, 40 nM 3S, or 30 nM CXCL12 with or without 10 nM anti-CXCL12 or 250 nM BoxA. Pax7-positive cells were counted in 10 random fields on the lower face of the transwell membrane at 10× magnification.
Single myofibers were prepared from the extensor digitorum longus and TA muscles of 6-to 12-wk-old mice as previously described (Pasut et al., 2013). Individual myofibers were plated on 35-mm dishes in proliferating medium (20% FBS and 3% chicken embryo extract in DMEM) in the presence or not of 40 nM fr-HMGB1 or 3S for 16 and 72 h. Fibers were fixed with 4% PFA and stained with DAPI, anti-Pax7 mAb (Hybridoma Bank), anti–MyoD mAb (DakoCytomation), and anti–Myogenin mAb (Hybridoma Bank). 10 fibers/condition were counted in biological triplicate (n = 3 mice, 30 fibers in total per condition).
Bone marrow–derived macrophage polarization
Macrophages were obtained from bone marrow precursor cells. In brief, bone marrow was obtained from mice by flushing the femur and tibia with DMEM. Cells were cultured for 7 d in DMEM conditioned by L929 cells (enriched in CSF-1). Macrophages were polarized toward a proinflammatory phenotype with 50 ng/ml IFN-γ in DMEM containing 10% FBS and were incubated with or without 40 nM fr-HMGB1 or 3S for 3 d. Macrophages were labeled with primary antibodies against iNOS (ab3523; Abcam), TNFα (ab34839; Abcam), CD163 (sc-33560; Santa Cruz), TGFβ1 (ab64715, Abcam), and Cy3- or FITC-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories).
SPR
The analysis of the binding of HMGB1 forms to the MD-2 or soluble RAGE (sRAGE) was conducted by using a Biacore T200 instrument (GE Healthcare Bio-Sciences). Binding reactions were done in HBS-EP buffer (Biacore) containing 10 mM Hepes, 150 mM NaCl, 3 mM EDTA, pH 7.4, and 0.05% polyoxyethylenesorbitan. MD-2 or sRAGE proteins were coated on the surface of CM5 dextran sensor chips by direct immobilization. MD-2 or sRAGE proteins were diluted to a concentration of 15 µg/ml in 10 mM acetate buffer (pH 5.5 and pH 5.0, respectively). A 1:1 mixture of N-hydroxysuccinimide and N-ethyl-N-(dimethylaminopropyl) carbodiimide was used to activate 2 flow-cells of the CM5 chip. One flow-cell was used as a reference and thus immediately blocked on activation by 1 M ethanolamine (pH 8.5). The sample flow-cell was injected with the diluted MD-2 or sRAGE at a flow rate of 10 µl/min. MD-2 or sRAGE injections were stopped when the SPR reached ∼1,200 or 200 resonance units (RU). For binding assays, the analytes (fr-HMGB1, ds-HMGB1, or 3S) were diluted to 1 µM in HBS-EP buffer (filtered, 0.22 µm) and injected over the chips at a flow rate of 30 µl/min for 60 s at 25°C; the dissociation time was set to 1 min. Binding experiments were performed at least 3 times. Equilibrium dissociation constants (KD) were obtained by using the BIA Evaluation 2.0 software (GE Healthcare) and assuming a 1:1 binding ratio.
For CXCR4 binding assays, lentiviral particles were produced, characterized, and titrated as previously described (Vega et al., 2011) by JetPei cotransfection of HEK293T cells with the LVTHM/GFP, PAX2, and VSVG plasmids (Tronolab) at a 1:1:1 ratio and plasmids for CXCR4 or siRNA for CXCR4 when needed. Lentiviral particles with a similar titration index were aliquoted and stored at −80°C. To activate carboxymethylated dextran, equal volumes of 0.1 M N-hydroxysuccinimide and 0.4 M 1-ethyl-3-(3-dimethylamino-propyl) carbodiimide were mixed and injected (5 µl/min, 7 min, RT) over the surface of a CM5 sensor chip (GE Healthcare). Hepes-buffered saline (HBS-P: 10 mM Hepes, pH 7.4, 0.15 M NaCl, 0.005% polyoxyethylenesorbitan) was used as an immobilization running buffer. Lentiviral particles (107/ml) diluted in 10 mM sodium acetate buffer (pH 4.0) were injected over the activated surfaces (5 µl/min, 7 min, RT), followed by 1 M ethanolamine (pH 8.5, 5 µl/min, 7 min, RT) to deactivate remaining active carboxyl groups. We usually detected 6,000 RU of coupled lentiviral particles. All determinations were performed by using a Biacore 3000 (GE Healthcare). Fr-HMGB1, 3S, and BoxA (1.25–40 nM) diluted in PBS-P buffer (137 mM NaCl, 10 mM Na2HPO4, 1.76 mM KH2PO4, 2.7 mM KCl, and 0.005% polyoxyethylenesorbitan, pH 7.4) were injected over the immobilized viral particles (30 µl/min, 2 min, 25°C; association phase), followed by a 2-min injection period of PBS-P buffer alone over the surface (dissociation phase). Sensorgrams were corrected for signals obtained in two reference flow channels, a control chamber without viral particles, and another with viral particles obtained from HEK293 cells transfected with siRNA for CXCR4 that were activated and deactivated in parallel. All kinetic assays were followed by injection of 5 mM HCl to dissociate remaining ligand from the virions (regeneration phase). All steps were performed by using the system’s automated robotics; all phases were followed in real time as a change in signal expressed in RU. Curves derived from these assays were used to generate kinetic constants, which were analyzed by fitting to a simple one-site interaction model with BIA Evaluation 4.1 software (GE Healthcare).
Statistical analysis
Every experiment was replicated at least twice and was performed at least in biological triplicate. Animals were assigned randomly to experimental groups (save for the matching between males and females), and no animals were excluded from the study. According to the 3R rules, a power calculation analysis was previously performed. The evaluator was blinded to the identity of the specific sample as far as the nature of the experiment allowed it. Bars represent the mean ± SEM. Statistical significance was assessed by using the tests indicated in the figure legends (Prism 5; GraphPad Software). In brief, t tests and Mann-Whitney tests were used for pairwise comparisons, and ANOVA and Kruskal-Wallis tests were used for multiple comparisons. P-values <0.05 were considered statistically significant.
Acknowledgments
We thank A. Fiocchi and M. Raso from the Ospedale San Raffaele mouse clinic for immunohistochemistry analyses of liver sections and serum biochemical measurements, respectively. We thank D. Gabellini and R. Piccirillo for constructive discussions and for careful reading of the manuscript. We also thank M.-V. Neguembor, R. Caccia, S. Pedrotti, and S. Samadi Shams for technical assistance and productive discussions.
This work was supported by grants from the Ministero della Salute (GR-2011-02351814 to E. Vénéreau and G. Messina, GR-2010-2312693 to A. Raucci, and RF-2011-02346754 to G. Sitia), the Associazione Italiana per la Ricerca sul Cancro (IG-18468 to G. Sitia and IG-14233 to M.E. Bianchi), 7th Framework Programme Endostem (241440 to S. Brunelli, B. Chazaud, and HMGBiotech), Ministero dell’Istruzione, dell’Università e della Ricerca (PRIN 2010–11, 20108YB5W3_007 to S. Brunelli), Università Milano Milano-Bicocca (FAR-QC2014 to S. Brunelli), Fondazione Cariplo (2013-0583 to S. Brunelli), and National Institutes of Health (S10 RR033072-01 to Y. Al-Abed). G. Messina also thanks the ERC Starting Grants 2011 (RegeneratioNfix 280611), the Italian Ministry for University and Research (MIUR-Futuro in Ricerca 2010), and the AFM-Téléthon (20002). M. Mellado and C. Santiago were supported by a grant from the Spanish Ministry of Economy and Competitiveness (SAF 2014-53416-R). N.L. Tran was supported by a Swiss National Science Foundation Fellowship (P2GEP3_171976). S. Di Maggio was supported by Centro Cardiologico Monzino-IRCCS (Ricerca Corrente 2015-2017).
M.E. Bianchi, S. Brunelli, M. Casalgrandi, and E. Vénéreau have an indirect financial interest as inventors on a patent application based partially on the data reported in this paper. M.E. Bianchi is founder and part owner of HMGBiotech, a company that provides goods and services related to HMGB proteins, and M. Casalgrandi, A. Preti, and E. Vénéreau were partially supported by HMGBiotech. The other authors declare no competing financial interests.
Author contributions: All contributing authors have agreed to submission of this manuscript for publication. M. Tirone designed, carried out most of the experiments in the muscle, and analyzed data. N.L. Tran and G. Sitia designed, carried out experiments in the liver, and analyzed data. C. Ceriotti, A. Gorzanelli, G. Careccia, and V. Conti designed, carried out some experiments in the muscle, and analyzed data. M. Canepari and R. Bottinelli designed and carried out muscle force experiments. A. Raucci, S. Di Maggio, C. Santiago, and M. Mellado designed and carried out SPR experiments for CXCR4 binding and analyzed data. M. He and Y. Al-Abed designed and carried out SPR experiments for MD-2 and sRAGE binding and analyzed data. F. De Marchis performed cell migration assays. S. François and G. Messina designed and carried out experiments on isolated single myofibers and myoblasts and analyzed data. M. Saclier, S. Ben Larbi, S. Cuvellier, and B. Chazaud designed and performed experiments of macrophage polarization and analyzed data. M. Casalgrandi and A. Preti produced recombinant proteins. S. Brunelli and M.E. Bianchi discussed results and provided important advice on experimental design. E. Vénéreau designed experiments, directed the project, and wrote the manuscript with comments from all authors.
References
- Agalave N.M., Larsson M., Abdelmoaty S., Su J., Baharpoor A., Lundbäck P., Palmblad K., Andersson U., Harris H., and Svensson C.I.. 2014. Spinal HMGB1 induces TLR4-mediated long-lasting hypersensitivity and glial activation and regulates pain-like behavior in experimental arthritis. Pain. 155:1802–1813. 10.1016/j.pain.2014.06.007 [DOI] [PubMed] [Google Scholar]
- Antoine D.J., Jenkins R.E., Dear J.W., Williams D.P., McGill M.R., Sharpe M.R., Craig D.G., Simpson K.J., Jaeschke H., and Park B.K.. 2012. Molecular forms of HMGB1 and keratin-18 as mechanistic biomarkers for mode of cell death and prognosis during clinical acetaminophen hepatotoxicity. J. Hepatol. 56:1070–1079. 10.1016/j.jhep.2011.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Arnold L., Henry A., Poron F., Baba-Amer Y., van Rooijen N., Plonquet A., Gherardi R.K., and Chazaud B.. 2007. Inflammatory monocytes recruited after skeletal muscle injury switch into antiinflammatory macrophages to support myogenesis. J. Exp. Med. 204:1057–1069. 10.1084/jem.20070075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balosso S., Liu J., Bianchi M.E., and Vezzani A.. 2014. Disulfide-containing high mobility group box-1 promotes N-methyl-D-aspartate receptor function and excitotoxicity by activating Toll-like receptor 4-dependent signaling in hippocampal neurons. Antioxid. Redox Signal. 21:1726–1740. 10.1089/ars.2013.5349 [DOI] [PubMed] [Google Scholar]
- Bischoff R. 1997. Chemotaxis of skeletal muscle satellite cells. Dev. Dyn. 208:505–515. [DOI] [PubMed] [Google Scholar]
- Braun T., Bober E., Buschhausen-Denker G., Kohtz S., Grzeschik K.H., and Arnold H.H.. 1989. Differential expression of myogenic determination genes in muscle cells: possible autoactivation by the Myf gene products. EMBO J. 8:3617–3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunelli S., Sciorati C., D’Antona G., Innocenzi A., Covarello D., Galvez B.G., Perrotta C., Monopoli A., Sanvito F., Bottinelli R., et al. 2007. Nitric oxide release combined with nonsteroidal antiinflammatory activity prevents muscular dystrophy pathology and enhances stem cell therapy. Proc. Natl. Acad. Sci. USA. 104:264–269. 10.1073/pnas.0608277104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calogero S., Grassi F., Aguzzi A., Voigtländer T., Ferrier P., Ferrari S., and Bianchi M.E.. 1999. The lack of chromosomal protein Hmg1 does not disrupt cell growth but causes lethal hypoglycaemia in newborn mice. Nat. Genet. 22:276–280. 10.1038/10338 [DOI] [PubMed] [Google Scholar]
- Catarinella M., Monestiroli A., Escobar G., Fiocchi A., Tran N.L., Aiolfi R., Marra P., Esposito A., Cipriani F., Aldrighetti L., et al. 2016. IFNα gene/cell therapy curbs colorectal cancer colonization of the liver by acting on the hepatic microenvironment. EMBO Mol. Med. 8:155–170. 10.15252/emmm.201505395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celona B., Weiner A., Di Felice F., Mancuso F.M., Cesarini E., Rossi R.L., Gregory L., Baban D., Rossetti G., Grianti P., et al. 2011. Substantial histone reduction modulates genomewide nucleosomal occupancy and global transcriptional output. PLoS Biol. 9:e1001086 10.1371/journal.pbio.1001086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavakis E., Hain A., Vinci M., Carmona G., Bianchi M.E., Vajkoczy P., Zeiher A.M., Chavakis T., and Dimmeler S.. 2007. High-mobility group box 1 activates integrin-dependent homing of endothelial progenitor cells. Circ. Res. 100:204–212. 10.1161/01.RES.0000257774.55970.f4 [DOI] [PubMed] [Google Scholar]
- Cohen H.B., and Mosser D.M.. 2013. Extrinsic and intrinsic control of macrophage inflammatory responses. J. Leukoc. Biol. 94:913–919. 10.1189/jlb.0413236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deneault E., Cellot S., Faubert A., Laverdure J.P., Fréchette M., Chagraoui J., Mayotte N., Sauvageau M., Ting S.B., and Sauvageau G.. 2009. A functional screen to identify novel effectors of hematopoietic stem cell activity. Cell. 137:369–379. 10.1016/j.cell.2009.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormoy-Raclet V., Cammas A., Celona B., Lian X.J., van der Giessen K., Zivojnovic M., Brunelli S., Riuzzi F., Sorci G., Wilhelm B.T., et al. 2013. HuR and miR-1192 regulate myogenesis by modulating the translation of HMGB1 mRNA. Nat. Commun. 4:2388 10.1038/ncomms3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukada S., Higuchi S., Segawa M., Koda K., Yamamoto Y., Tsujikawa K., Kohama Y., Uezumi A., Imamura M., Miyagoe-Suzuki Y., et al. 2004. Purification and cell-surface marker characterization of quiescent satellite cells from murine skeletal muscle by a novel monoclonal antibody. Exp. Cell Res. 296:245–255. 10.1016/j.yexcr.2004.02.018 [DOI] [PubMed] [Google Scholar]
- Guo F., Wang Y., Liu J., Mok S.C., Xue F., and Zhang W.. 2016. CXCL12/CXCR4: a symbiotic bridge linking cancer cells and their stromal neighbors in oncogenic communication networks. Oncogene. 35:816–826. 10.1038/onc.2015.139 [DOI] [PubMed] [Google Scholar]
- Huang H., Nace G.W., McDonald K.A., Tai S., Klune J.R., Rosborough B.R., Ding Q., Loughran P., Zhu X., Beer-Stolz D., et al. 2014. Hepatocyte-specific high-mobility group box 1 deletion worsens the injury in liver ischemia/reperfusion: a role for intracellular high-mobility group box 1 in cellular protection. Hepatology. 59:1984–1997. 10.1002/hep.26976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang R., Zhang Q., Hou W., Yan Z., Chen R., Bonaroti J., Bansal P., Billiar T.R., Tsung A., Wang Q., et al. 2014. Intracellular Hmgb1 inhibits inflammatory nucleosome release and limits acute pancreatitis in mice. Gastroenterology. 146:1097–1107. 10.1053/j.gastro.2013.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M., and Clevers H.. 2016. Reparative inflammation takes charge of tissue regeneration. Nature. 529:307–315. 10.1038/nature17039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau T.T., and Wang D.A.. 2011. Stromal cell-derived factor-1 (SDF-1): homing factor for engineered regenerative medicine. Expert Opin. Biol. Ther. 11:189–197. 10.1517/14712598.2011.546338 [DOI] [PubMed] [Google Scholar]
- Lefaucheur J.P., and Sébille A.. 1995. Muscle regeneration following injury can be modified in vivo by immune neutralization of basic fibroblast growth factor, transforming growth factor beta 1 or insulin-like growth factor I. J. Neuroimmunol. 57:85–91. 10.1016/0165-5728(94)00166-L [DOI] [PubMed] [Google Scholar]
- Lian Y.J., Gong H., Wu T.Y., Su W.J., Zhang Y., Yang Y.Y., Peng W., Zhang T., Zhou J.R., Jiang C.L., and Wang Y.X.. 2017. Ds-HMGB1 and fr-HMGB induce depressive behavior through neuroinflammation in contrast to nonoxid-HMGB1. Brain Behav. Immun. 59:322–332. 10.1016/j.bbi.2016.09.017 [DOI] [PubMed] [Google Scholar]
- Limana F., Germani A., Zacheo A., Kajstura J., Di Carlo A., Borsellino G., Leoni O., Palumbo R., Battistini L., Rastaldo R., et al. 2005. Exogenous high-mobility group box 1 protein induces myocardial regeneration after infarction via enhanced cardiac C-kit+ cell proliferation and differentiation. Circ. Res. 97:e73–e83. 10.1161/01.RES.0000186276.06104.04 [DOI] [PubMed] [Google Scholar]
- Lu H., Huang D., Saederup N., Charo I.F., Ransohoff R.M., and Zhou L.. 2011. Macrophages recruited via CCR2 produce insulin-like growth factor-1 to repair acute skeletal muscle injury. FASEB J. 25:358–369. 10.1096/fj.10-171579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palumbo R., Sampaolesi M., De Marchis F., Tonlorenzi R., Colombetti S., Mondino A., Cossu G., and Bianchi M.E.. 2004. Extracellular HMGB1, a signal of tissue damage, induces mesoangioblast migration and proliferation. J. Cell Biol. 164:441–449. 10.1083/jcb.200304135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasut A., Jones A.E., and Rudnicki M.A.. 2013. Isolation and culture of individual myofibers and their satellite cells from adult skeletal muscle. J. Vis. Exp. (73):e50074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenkilde M.M., Gerlach L.O., Jakobsen J.S., Skerlj R.T., Bridger G.J., and Schwartz T.W.. 2004. Molecular mechanism of AMD3100 antagonism in the CXCR4 receptor: transfer of binding site to the CXCR3 receptor. J. Biol. Chem. 279:3033–3041. 10.1074/jbc.M309546200 [DOI] [PubMed] [Google Scholar]
- Saclier M., Cuvellier S., Magnan M., Mounier R., and Chazaud B.. 2013. Monocyte/macrophage interactions with myogenic precursor cells during skeletal muscle regeneration. FEBS J. 280:4118–4130. 10.1111/febs.12166 [DOI] [PubMed] [Google Scholar]
- Scaffidi P., Misteli T., and Bianchi M.E.. 2002. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature. 418:191–195. 10.1038/nature00858 [DOI] [PubMed] [Google Scholar]
- Schaper F., de Leeuw K., Horst G., Bootsma H., Limburg P.C., Heeringa P., Bijl M., and Westra J.. 2016. High mobility group box 1 skews macrophage polarization and negatively influences phagocytosis of apoptotic cells. Rheumatology (Oxford). 55:2260–2270. 10.1093/rheumatology/kew324 [DOI] [PubMed] [Google Scholar]
- Schiraldi M., Raucci A., Muñoz L.M., Livoti E., Celona B., Venereau E., Apuzzo T., De Marchis F., Pedotti M., Bachi A., et al. 2012. HMGB1 promotes recruitment of inflammatory cells to damaged tissues by forming a complex with CXCL12 and signaling via CXCR4. J. Exp. Med. 209:551–563. 10.1084/jem.20111739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P., Sabourin L.A., Girgis-Gabardo A., Mansouri A., Gruss P., and Rudnicki M.A.. 2000. Pax7 is required for the specification of myogenic satellite cells. Cell. 102:777–786. 10.1016/S0092-8674(00)00066-0 [DOI] [PubMed] [Google Scholar]
- Sitia G., Iannacone M., Müller S., Bianchi M.E., and Guidotti L.G.. 2007. Treatment with HMGB1 inhibitors diminishes CTL-induced liver disease in HBV transgenic mice. J. Leukoc. Biol. 81:100–107. 10.1189/jlb.0306173 [DOI] [PubMed] [Google Scholar]
- Son M., Porat A., He M., Suurmond J., Santiago-Schwarz F., Andersson U., Coleman T.R., Volpe B.T., Tracey K.J., Al-Abed Y., and Diamond B.. 2016. C1q and HMGB1 reciprocally regulate human macrophage polarization. Blood. 128:2218–2228. 10.1182/blood-2016-05-719757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark K., Philippi V., Stockhausen S., Busse J., Antonelli A., Miller M., Schubert I., Hoseinpour P., Chandraratne S., von Brühl M.L., et al. 2016. Disulfide HMGB1 derived from platelets coordinates venous thrombosis in mice. Blood. 128:2435–2449. 10.1182/blood-2016-04-710632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamai K., Yamazaki T., Chino T., Ishii M., Otsuru S., Kikuchi Y., Iinuma S., Saga K., Nimura K., Shimbo T., et al. 2011. PDGFRalpha-positive cells in bone marrow are mobilized by high mobility group box 1 (HMGB1) to regenerate injured epithelia. Proc. Natl. Acad. Sci. USA. 108:6609–6614. 10.1073/pnas.1016753108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonkin J., Temmerman L., Sampson R.D., Gallego-Colon E., Barberi L., Bilbao D., Schneider M.D., Musarò A., and Rosenthal N.. 2015. Monocyte/Macrophage-derived IGF-1 Orchestrates Murine Skeletal Muscle Regeneration and Modulates Autocrine Polarization. Mol. Ther. 23:1189–1200. 10.1038/mt.2015.66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega B., Muñoz L.M., Holgado B.L., Lucas P., Rodríguez-Frade J.M., Calle A., Rodríguez-Fernández J.L., Lechuga L.M., Rodríguez J.F., Gutiérrez-Gallego R., and Mellado M.. 2011. Technical advance: Surface plasmon resonance-based analysis of CXCL12 binding using immobilized lentiviral particles. J. Leukoc. Biol. 90:399–408. 10.1189/jlb.1010565 [DOI] [PubMed] [Google Scholar]
- Venereau E., Casalgrandi M., Schiraldi M., Antoine D.J., Cattaneo A., De Marchis F., Liu J., Antonelli A., Preti A., Raeli L., et al. 2012. Mutually exclusive redox forms of HMGB1 promote cell recruitment or proinflammatory cytokine release. J. Exp. Med. 209:1519–1528. 10.1084/jem.20120189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venereau E., Schiraldi M., Uguccioni M., and Bianchi M.E.. 2013. HMGB1 and leukocyte migration during trauma and sterile inflammation. Mol. Immunol. 55:76–82. 10.1016/j.molimm.2012.10.037 [DOI] [PubMed] [Google Scholar]
- Venereau E., De Leo F., Mezzapelle R., Careccia G., Musco G., and Bianchi M.E.. 2016. HMGB1 as biomarker and drug target. Pharmacol. Res. 111:534–544. 10.1016/j.phrs.2016.06.031 [DOI] [PubMed] [Google Scholar]
- Wang H., Melton D.W., Porter L., Sarwar Z.U., McManus L.M., and Shireman P.K.. 2014. Altered macrophage phenotype transition impairs skeletal muscle regeneration. Am. J. Pathol. 184:1167–1184. 10.1016/j.ajpath.2013.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai H., Matsuda A., An J., Koshiba R., Nishio J., Negishi H., Ikushima H., Onoe T., Ohdan H., Yoshida N., and Taniguchi T.. 2013. Conditional ablation of HMGB1 in mice reveals its protective function against endotoxemia and bacterial infection. Proc. Natl. Acad. Sci. USA. 110:20699–20704. 10.1073/pnas.1320808110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Hreggvidsdottir H.S., Palmblad K., Wang H., Ochani M., Li J., Lu B., Chavan S., Rosas-Ballina M., Al-Abed Y., et al. 2010. A critical cysteine is required for HMGB1 binding to Toll-like receptor 4 and activation of macrophage cytokine release. Proc. Natl. Acad. Sci. USA. 107:11942–11947. 10.1073/pnas.1003893107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Lundbäck P., Ottosson L., Erlandsson-Harris H., Venereau E., Bianchi M.E., Al-Abed Y., Andersson U., Tracey K.J., and Antoine D.J.. 2012. Redox modification of cysteine residues regulates the cytokine activity of high mobility group box-1 (HMGB1). Mol. Med. 18:250–259. 10.2119/molmed.2011.00389 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Yang H., Wang H., Ju Z., Ragab A.A., Lundbäck P., Long W., Valdes-Ferrer S.I., He M., Pribis J.P., Li J., et al. 2015. MD-2 is required for disulfide HMGB1-dependent TLR4 signaling. J. Exp. Med. 212:5–14. 10.1084/jem.20141318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Messer J.S., Wang Y., Lin F., Cham C.M., Chang J., Billiar T.R., Lotze M.T., Boone D.L., and Chang E.B.. 2015. Cytosolic HMGB1 controls the cellular autophagy/apoptosis checkpoint during inflammation. J. Clin. Invest. 125:1098–1110. 10.1172/JCI76344 [DOI] [PMC free article] [PubMed] [Google Scholar]