Hogstad et al. show that the somatic BRAFV600E mutation in myeloid dendritic cell precursors in Langerhans cell histiocytosis promotes lesion formation through impaired dendritic cell migration and resistance to apoptosis, which can be rescued with targeted MAPK pathway inhibition.
Abstract
Langerhans cell histiocytosis (LCH) is an inflammatory myeloid neoplasia characterized by granulomatous lesions containing pathological CD207+ dendritic cells (DCs) with constitutively activated mitogen-activated protein kinase (MAPK) pathway signaling. Approximately 60% of LCH patients harbor somatic BRAFV600E mutations localizing to CD207+ DCs within lesions. However, the mechanisms driving BRAFV600E+ LCH cell accumulation in lesions remain unknown. Here we show that sustained extracellular signal–related kinase activity induced by BRAFV600E inhibits C-C motif chemokine receptor 7 (CCR7)–mediated DC migration, trapping DCs in tissue lesions. Additionally, BRAFV600E increases expression of BCL2-like protein 1 (BCL2L1) in DCs, resulting in resistance to apoptosis. Pharmacological MAPK inhibition restores migration and apoptosis potential in a mouse LCH model, as well as in primary human LCH cells. We also demonstrate that MEK inhibitor-loaded nanoparticles have the capacity to concentrate drug delivery to phagocytic cells, significantly reducing off-target toxicity. Collectively, our results indicate that MAPK tightly suppresses DC migration and augments DC survival, rendering DCs in LCH lesions trapped and resistant to cell death.
Introduction
Langerhans cell histiocytosis (LCH) is characterized by inflammatory lesions that include CD207+ dendritic cells (DCs) with distribution ranging from clinically trivial single bone lesion to life-threatening disease with disseminated lesions that may involve skin, bones, lymph nodes, bone marrow (BM), spleen, liver, lungs, and/or brain (Berres et al., 2015). LCH has an incidence of approximately five cases per million children (similar to pediatric Hodgkin lymphoma), and can also arise de novo in adults (Allen et al., 2015). Current standard of care involves resection or curettage for isolated lesions or/and chemotherapy (vinblastine/prednisone) for disseminated disease. However, approximately half of all LCH patients with multifocal disease relapse or fail to respond to initial therapy (Minkov et al., 2008; Gadner et al., 2013), and treatment failure in LCH with risk-organ involvement is associated with high long-term morbidity and mortality (Haupt et al., 2004). Myelotoxic nucleoside analogues including cytarabine, cladribine, and clofarabine have been effective in salvage therapy strategies, but are associated with concerning rates of treatment-related mortality at higher doses (Allen et al., 2015). Somatic mutations in MAPK pathway genes have been identified in >85% cases of LCH (Chakraborty et al., 2016). However, mechanisms through which ERK activation in myeloid DC precursors mediates LCH pathogenesis are not known. There remains a clear need for improving our understanding of the mechanisms of LCH pathogenesis to develop therapeutic strategies with improved efficacy and minimal toxicity.
DCs are potent professional APCs governing the function of CD4+ and CD8+ T cells in both steady-state and inflammation. They derive from BM-resident myeloid cell hematopoietic progenitors, which give rise to circulating DC precursors differentiating into immature DCs at the tissue site (Hashimoto et al., 2011). While residing in all tissues, immature DCs phagocytize tissue-associated antigens during homeostasis, infection, and inflammation, after which they undergo a terminal maturation step enabling them to migrate to the draining lymph nodes (dLNs). Immature DCs normally persist in tissue with a half-life of 3–7 d and immediately leave the tissue upon maturation. As part of the maturation process, DCs dramatically up-regulate functional maturation-associated markers such as MHC class II (MHCII), C-C motif chemokine receptor 7 (CCR7), CD86, CD80, and CD40. Mature DCs use the up-regulated chemokine receptor CCR7 to migrate along a CCL19/CCL21 chemokine gradient through the afferent lymphatic vessels to the dLN, where they modulate T cells in an antigen-specific manner. After T cell encounters, DCs typically die through apoptosis, and very few DCs are able to exit the LN (Angeli and Randolph, 2006).
A unique feature of DCs in LCH lesions is universal MAPK activation (Badalian-Very et al., 2010; Chakraborty et al., 2014), resulting from mutually exclusive somatic activating mutations in MAPK pathway genes including BRAFV600E (50–65%), MAP2K1 (10–20%), and other less common alterations (e.g., BRAF other than V600E, ARAF, and ERBB3; Badalian-Very et al., 2010; Brown et al., 2014; Chakraborty et al., 2014, 2016; Nelson et al., 2014, 2015).
Depending on the cellular context, the MAPK signaling pathway is critically involved in multifarious cellular functions including cell differentiation, proliferation, cytokine production, and survival as well as cellular arrest and senescence (Cagnol and Chambard, 2010; Cargnello and Roux, 2011). We recently demonstrated that induction of the activating BRAFV600E mutation in myeloid DCs drives an LCH-like phenotype in mice (Berres et al., 2014). However, it remains unclear how genetic alterations of the MAPK pathway in myeloid precursors drive lesion formation. Therefore, our objective was to assess the impact of the somatic BRAFV600E mutation on the key biological functions of DCs including proliferation, differentiation/activation, migration, cytokine production, and apoptosis using mouse experimental models of LCH as well as LCH patient tissue samples.
Identification of MAPK pathway hyperactivation as an essential driver of disease opens a new avenue for therapeutic interventions. Pharmacological BRAF and MEK inhibitors (MEKi’s) have shown clinical promise in some adults with LCH (Haroche et al., 2015; Hyman et al., 2015). However, clinical applicability of these drugs is limited by the narrow therapeutic index because of their toxicity profiles thus far observed in adults with melanoma and other BRAF-mutated tumors due in part to paradoxical MAPK pathway activation in normal cells (Su et al., 2012; Haroche et al., 2015). This limitation might be overcome by targeted delivery of MAPK inhibitors directly to pathogenic cells. In this study, we therefore also assess, as a proof of principle approach, the efficacy and safety of a nanoparticle assisted delivery system of MAPK inhibitors in which the drug concentrates in DCs via phagocytosis in an experimental model of LCH. Finally, given the importance of tissue DC migration to the induction of antitumor and vaccine immunity, our data also reveal a potential role of MEKi’s to influence the induction of therapeutic immunity against cancer and infectious agents.
Results
BRAFV600E-induced DC accumulation at tissue sites is not associated with significantly augmented DC proliferation
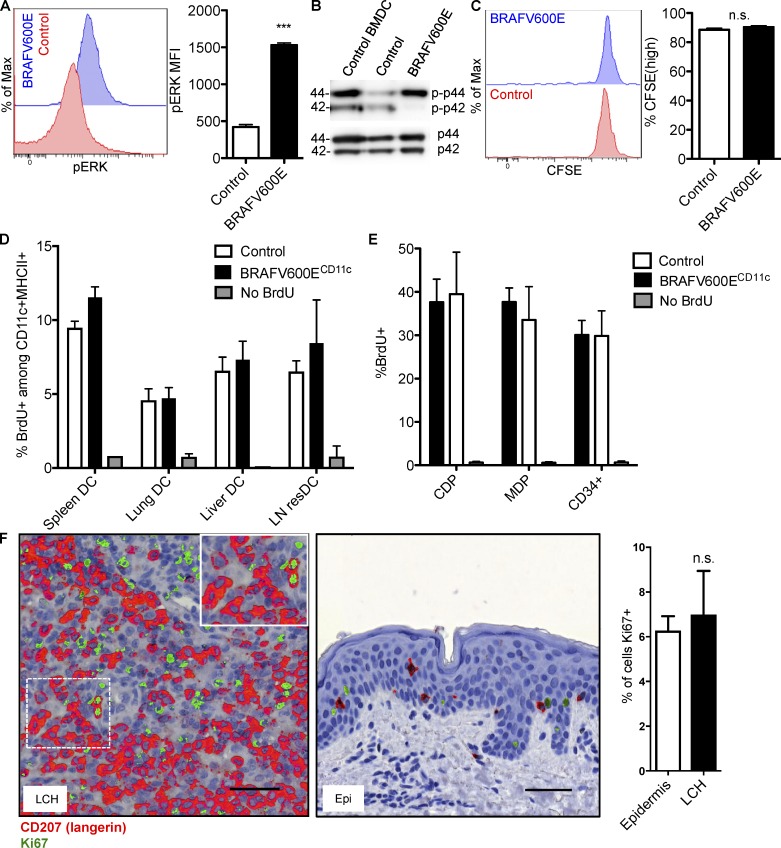
As observed also in other cellular contexts, BRAFV600E causes sustained activation of the MAPK pathways in CD207+ DCs in LCH lesions as indicated by enhanced ERK phosphorylation (Badalian-Very et al., 2010; Chakraborty et al., 2014). In line with these observations, BRAFV600E expression induced increased phosphorylation of ERK in BM-derived DCs (BMDCs; Fig. 1 A) and primary splenic DCs (Fig. 1 B). As LCH is considered a neoplastic disease and ERK signaling has been associated with increased cell proliferation in different settings (Cargnello and Roux, 2011), we first set out to investigate how the BRAFV600E mutation alters DC proliferation potential. For this purpose, we first analyzed the proliferative capacity of BRAFV600E+ DCs from mice in which we conditionally introduced the expression of the constitutively active BRAFV600E allele using Cre-recombinase under the control of the CD11c promoter, a marker enriched in cells of the DC lineage (BRAFV600ECD11c mice; Berres et al., 2014). In these mice, BRAFV600E expression is enforced in BM-resident DC progenitors, circulating precursors, and tissue-resident DCs (Berres et al., 2014). To assess the overall DC proliferation capacity in vivo, we directly compared the proliferation potential of BMDCs isolated from BRAFV600ECD11c mice as well as corresponding controls labeled with CFSE after intranasal adoptive transfer into congenic CD45.1+ mice. 6 d after adoptive transfer, we recovered a significant population of viable CD45.2+ DCs in the lung of recipient animals, suggesting that donor cells engrafted well in the recipient mice. In addition, we found that a large fraction of BRAFV600E-expressing and control BMDCs retained the dye label, indicating that the vast majority of transferred DCs had not undergone cell division at the tissue site after engraftment irrespective of BRAFV600E expression (Fig. 1 C).
Figure 1.
BRAFV600E DCs proliferate at normal DC rates. (A) ERK phosphorylation levels in control and BRAFV600E BMDCs as analyzed by flow cytometry. Histograms show representative ERK MFI expression in control and BRAFV600E BMDC. Bar graph quantification of phospho-ERK MFI expression in control and BRAFV600E BMDCs. Data shown are representative of 5 experiments. Bar graph represents mean of three technical replicates from one experiment ± SEM (***, P < 0.0001; unpaired t test). (B) CD11c+ cells were isolated from the spleen of BRAFV600ECD11c mice and control mice, and whole cell lysates were analyzed for the expression of phospho-ERK (p-p42/44) by Western blot analysis. Representative data of at least three experiments are shown. Molecular mass is indicated in kilodaltons. (C–E) Measuring in vivo proliferation of BRAFV600ECD11c DCs. (C) CFSE dye–labeled BMDCs were transferred intranasally into congenic CD45.1+ C57B6 recipient mice. Proliferation of engrafted BMDCs was analyzed 6 d after transfer. Histograms shown are representative of two experiments. Bar graph represents pooled data from two experiments (n = 3–6 mice per group) ± SEM (P = 0.2769, unpaired t test). (D and E) In vivo BrdU labeling of (D) CD11c+MHCII+ DCs and (E) BM progenitors from control versus BRAFV600ECD11c mice analyzed 5 h after i.p. injection of 1 mg BrdU. Data shown are the mean ± SEM from four experiments (n = 10–12 for spleen, lung and BM; n = 3 for liver, n = 3 for skin dLN). Two-way ANOVA reveals no significant effect of BRAFV600E on overall DC BrdU incorporation (P = 0.1729, ANOVA). (F) Frequency of Ki-67+ cells in LCH lesion from paraffin-embedded tissue sections. LCH lesions and healthy epidermis (Epi) sections were stained serially for CD207 and Ki-67. LCH lesions and Epi were scored for frequency of Ki-67+ nuclei among CD207+ cells ± SEM. Representative images highlight CD207 (red) overlaid with Ki-67 (green) to show staining localization. Scoring is mean of (total Ki-67+CD207+ cells)/(total CD207+ cells) in LCH lesions and Epi obtained from three LCH patients and four healthy donors (P = 0.7068, unpaired t test). Bars, 50 µm.
As the specific microenvironment might also impact the proliferation rate of cells, we next assessed the in vivo proliferation of BRAFV600E-expressing DCs in the context of LCH-like disease directly at the lesion site by i.p. injecting a pulse of bromodeoxyuridine (BrdU) into control and BRAFV600ECD11c mice. 5 h after injection, spleen, lung, liver tissues, and LNs were harvested, and BrdU incorporation was assessed using flow cytometry analysis (Fig. 1 D). Consistent with the results obtained upon adoptive transfer, BRAFV600E expression did not significantly impact BrdU incorporation in tissue DCs. ANOVA comparing the effect of BRAFV600E status on BrdU incorporation revealed no significant influence of BRAFV600E. This data collectively indicates that BRAFV600E does not induce augmented proliferation of tissue DCs in vivo.
To test whether the BRAFV600E mutation promoted the proliferation of myeloid progenitors rather than differentiated tissue DCs, we also assessed BrdU incorporation in mononuclear phagocyte progenitors in vivo in BRAFV600ECD11c mice. Similar to the results observed with tissue DCs, we failed to observe increased proliferation of BRAFV600E macrophage and DC precursor, common DC precursors, or CD34+ BM progenitors in BRAFV600ECD11c mice compared with control DC progenitors (Fig. 1 E). In summary, these results suggest that DC accumulation at the tissue site in BRAFV600ECD11c mice did not result from increased proliferation of DCs or their progenitors.
We next correlated our results obtained in the experimental mouse system with tissue samples from LCH patients. To examine the proliferation capacity of BRAFV600E+ in human LCH cells, paraffin-embedded tissue sections were stained for CD207 to identify LCH cells in LCH lesions or Langerhans cells in healthy epidermis and then counterstained with the proliferation marker Ki-67. (Table S1 provides clinical information for LCH patient lesions studied in Fig. 1.) A mean of 6.99% of CD207+ cells coexpressed Ki-67 in the examined BRAFV600E+ LCH lesions (Fig. 1 F), compared with 6.28% of CD207+ cells in healthy epidermis (Fig. 1 F). There was no significant difference between the percentage of CD207+Ki-67+ cells between the LCH lesions and the healthy epidermis (two-tailed unpaired t test, P = 0.7068). These findings align with previous observations reporting no increase in proliferation potential of LCH lesions (Senechal et al., 2007). These data support a model where the BRAFV600E mutation does not augment DC proliferation significantly beyond baseline proliferation rates observed in normal DCs.
BRAFV600E blocks DC emigration potential from nonlymphoid tissues
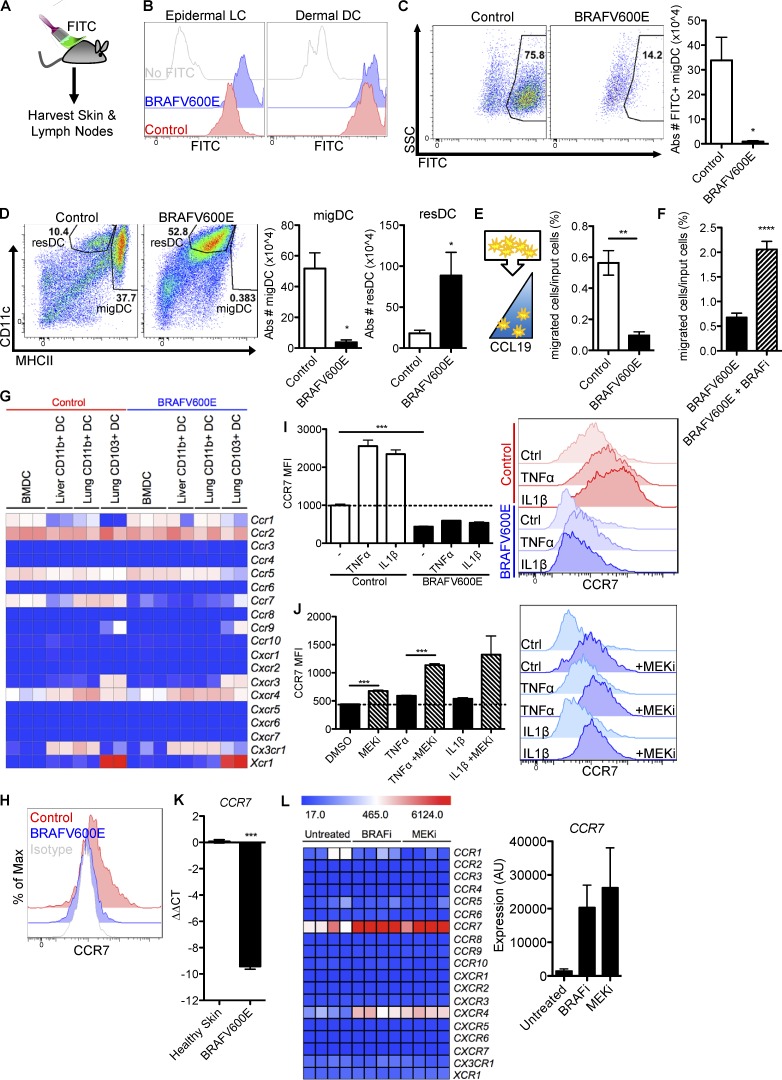
If LCH cells are not hyperproliferative, an alternative mechanism of DC accumulation may be tissue retention. Under homeostatic conditions, DCs constantly egress the tissue to migrate to the LN via the lymphatic vessels. To test whether BRAFV600E alters DC migration potential, we painted the skin of BRAFV600ECD11c or control mice with FITC (Förster et al., 1999; Randolph et al., 2005) to mark and trace migratory DCs (migDCs) in vivo (Fig. 2 A). After skin painting with FITC, BRAFV600E DCs and control DCs were equally able to capture FITC at the tissue site (Fig. 2 B). Despite similar capture efficiency, FITC+ DCs were drastically reduced in the dLNs of BRAFV600ECD11c mice in comparison to control littermates (Fig. 2 C). In fact, the entire population of LN MHCIIhi CD11cintermediate DCs, which phenotypically correspond to tissue migDCs, was nearly absent, whereas LN-resident DCs were greatly expanded (Fig. 2 D) in the dLNs of steady-state BRAFV600ECD11c mice, indicating a specific egress defect of tissue migDCs.
Figure 2.
BRAFV600E abrogates CCR7 function and DC migration and trap DCs in tissues. (A–D) Tracing DC migration to the tissue dLN in BRAFV600ECD11c mice. (A) Cartoon depicts the experimental procedure to trace migDCs. The skin of BRAFV600ECD11c or control mice was painted with FITC to promote the migration of skin FITC+ DCs to the skin dLN. (B) Histograms show FITC uptake by skin DCs including epidermal LC and dermal DCs. (C) Flow cytometry pseudo-color dot plots show the representative frequency and bar graphs show absolute numbers of FITC+CD3−B220−CD11c+MHCIIhigh migDCs that have migrated to the skin dLN in control and BRAFV600ECD11c mice (*, P = 0.0104; unpaired t test). (D) Flow cytometry plots and bar graphs show the quantification of CD11cintMHCIIhigh migDCs (*, P = 0.0104; unpaired t test) and CD11chighMHCIIint lymphoid-resident DCs (P = 0.0328, unpaired t test) in the skin dLN of BRAFV600ECD11c or control mice. Data shown are representative of at least two experiments ± SEM (n = 3–4 per group). (E) Transwell migration assay in which control and BRAFV600E BMDCs were exposed to CCL19 chemokine gradient. Representative data of at least three experiments with three biological replicates are shown ± SEM (**, P = 0.0014; unpaired t test). (F) BRAFV600ECD11c BMDCs were subjected to transwell migration assay as in E ± BRAF inhibitor (****, P < 0.00001; unpaired t test). (G) Heat map summarizes the chemokine receptor expression profile measured by genechip arrays on ex-vivo FACS-sorted DC subsets (CD103+ lung DC, CD11b+ lung DC, and CD11b+ liver DC) and BMDCs from control versus BRAFV600ECD11c mice. (H) CCR7 surface protein levels measured by flow cytometry staining of control (red) and BRAFV600E (blue) BMDCs. Isotype staining control is depicted in gray. Representative of at least five experiments with three technical replicates each. (I and J) CCR7 protein expression levels measured by flow cytometry in control and BRAFV600E BMDCs (***, P < 0.0001; unpaired t test) stimulated overnight with 100 ng/ml TNFα or 100 ng/ml IL-1β. Data representative of at least twp independent experiments with triplicate technical replicates are shown ± SEM. (J) BRAFV600E BMDC unstimulated (***, P < 0.0003; unpaired t test), stimulated with TNFα (***, P < 0.0001; unpaired t test), or stimulated with IL-1β (P = 0.0778, unpaired t test) as in I overnight ± 100 nM GSK1120212 MEKi. (K) Quantitative real-time PCR analysis of CCR7 mRNA expression in BRAF-WT and BRAFV600E lesion relative to healthy skin. CCR7 expression was normalized to CD207 expression in each lesion to normalize for DC numbers. Units are expressed in log2 format to express fold-change relative to healthy skin. Data represent 3 tissue samples per group. (***, P < 0.0001; unpaired t test). (L) Chemokine receptor expression profile analyzed by Affymetrix genechip of purified CD207+ cells isolated from four BRAFV600E+ human LCH lesions untreated or treated with Vemurafenib BRAF inhibitor or Trametinib MEKi for 12 h. Error bars indicate SEM.
Hematopoietic cells depend on chemokine receptor expression for migration. CCR7 expression has been identified as absolutely required for DC migration from peripheral tissues to LN (Förster et al., 1999; Randolph et al., 2005). A transwell migration assay toward a CCL19 chemokine gradient (a CCR7 ligand) revealed a significant defect in migration of BRAFV600E DC toward the CCR7 ligand CCL19 as compared with control cells (Fig. 2 E). To test whether inhibition of BRAFV600E could rescue DC migratory capacity, we subjected mouse BRAFV600E+ BMDC to a CCL19 transwell migration assay in the presence or absence of BRAF inhibitor. As predicted, BRAFV600E inhibition successfully rescued BRAFV600E+ DC migration toward a CCL19 chemokine gradient (Fig. 2 F). These results indicate BRAFV600ECD11c DCs significantly impairs DC migratory potential.
We next assessed the expression profile of chemokine receptors in BMDC cultures and in purified DCs isolated from lung and liver of BRAFV600ECD11c and control mice. In this study, the chemokine receptor Ccr7 transcript was dramatically reduced in BRAFV600ECD11c DCs and was the only chemokine receptor with significantly different expression between BRAFV600ECD11c cells and control DCs (Fig. 2 G).
Reduced Ccr7 mRNA expression in DCs was confirmed by quantitative PCR (qPCR) in BRAFV600ECD11c splenic DCs (Fig. S1 A) and at the protein level using flow cytometry staining for CCR7 on BRAFV600ECD11c BMDC (Fig. 2 H), indicating a cell-intrinsic defect in CCR7 expression associated with BRAFV600E expression. Furthermore, CCR7 was also reduced in FITC+ BRAFV600ECD11c epidermal Langerhans cells and dermal DCs after FITC painting (Fig. S1 B).
Whereas unstimulated tissue DCs migrate at a slow rate to the dLNs, migration is significantly activated in injured tissues. Increased DC migration in inflamed tissues is critically controlled by CCR7 expression induced by inflammatory cytokines such as TNFα or IL-1 (Berthier-Vergnes et al., 2005). To test whether BRAFV600E interferes with inflammation-induced migration as well as steady-state migration, we cultured BRAFV600E and control BMDC in the presence of TNFα or IL-1. Strikingly, BRAFV600E BMDC failed to induce CCR7 expression in response to TNFα or IL-1, whereas CCR7 was highly expressed in control cells upon stimulation (Fig. 2 I). Treatment with MEKi successfully rescued CCR7 expression in BRAFV600E BMDC upon TNFα or IL-1 stimulation (Fig. 2 J). This is consistent with data from human monocyte-derived DC cultures in which ERK inhibition can boost DC maturation and CCR7 expression after TNFα stimulation or serum withdrawal in vitro (Puig-Kröger et al., 2001; Aguilera-Montilla et al., 2013). This suggests that constitutive ERK activation by BRAFV600E exaggerates a physiological role for ERK activity in negatively regulating DC migration.
In line with our results in the mouse setting, CCR7 expression was significantly lower in BRAFV600E+ human LCH lesions compared with healthy control skin (Fig. 2 K). This aligns with our previous array data demonstrating reduced CCR7 expression in CD207+ cells from various LCH lesions compared with CD207+ epidermal Langerhans cells from healthy skin (Allen et al., 2010). (Table S2 provides clinical information for LCH patient lesions studied in Fig. 2.) Genechip expression analysis of primary human LCH lesion CD207+ DCs (BRAFV600E+) cultured in the presence of the BRAF inhibitor (vemurafenib) or MEKi (GSK1120212) revealed that CCR7 was the most highly increased chemokine receptor transcript after both BRAF inhibitor and MEKi treatment (Fig. 2 L), implicating a direct regulatory effect of the BRAF/MEK pathway on CCR7 expression. CXCR4 was also significantly increased in human lesions after BRAF inhibitor and MEKi treatment; however, this expression was ∼10 times lower than CCR7 expression, and this result was not conserved with expression data from the BRAFV600ECD11c mouse model (Fig. 2 G), indicating that CXCR4 is not a conserved functional component between these models. These results suggest that BRAFV600E abrogates DC migration to the dLNs and traps DCs in tissue lesions through inhibition of CCR7 expression.
Expression of BRAFV600E suppresses apoptosis in mouse and human DCs via enhanced expression of BCL-XL
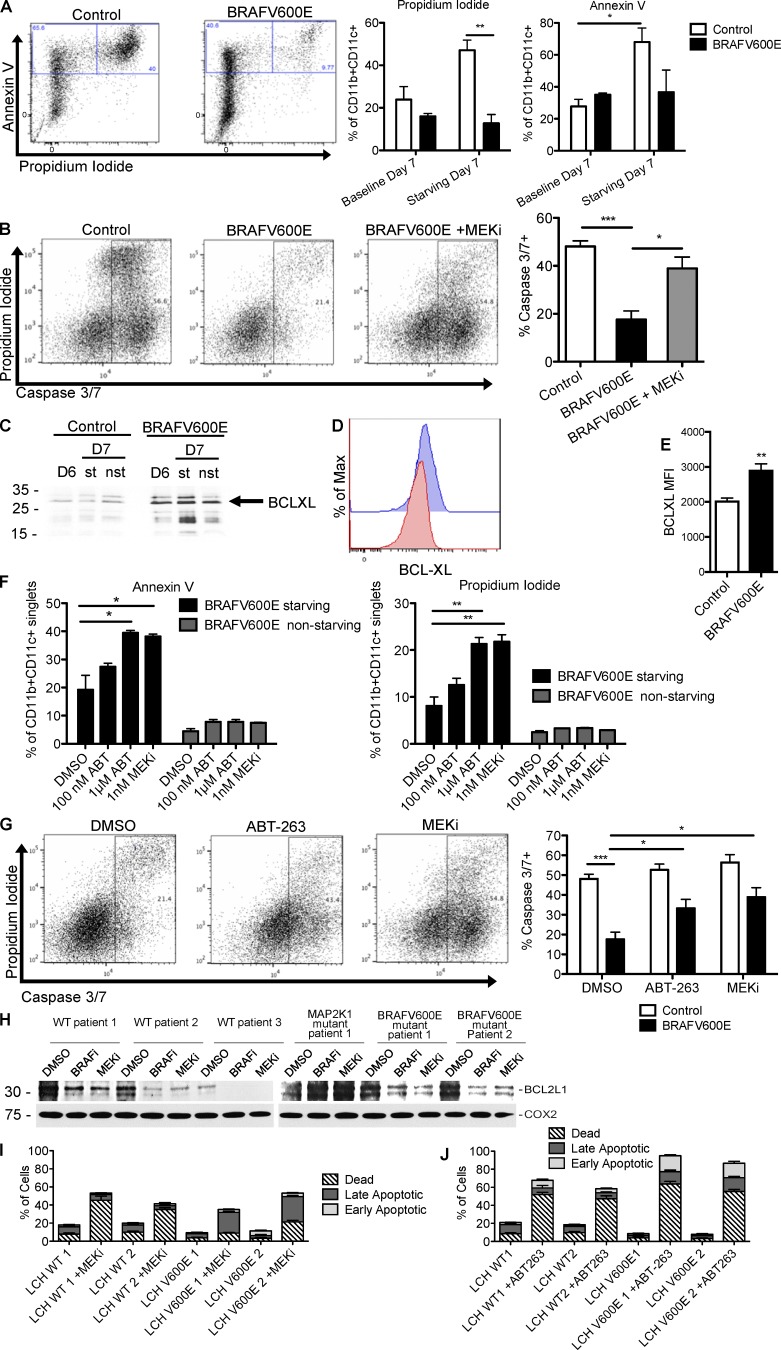
In addition to altered cell migration, another frequent mechanism during tumorigenesis is represented by prolonged cell survival and resistance to apoptosis (Olsson Åkefeldt et al., 2013). Importantly, ERK signaling regulates cell survival in many physiological and neoplastic settings (Michaloglou et al., 2005). Therefore, we set out to investigate if BRAFV600E expression also alters the capacity of DC to undergo apoptosis, thereby contributing to persistence of these cells within the lesions. BRAFV600E and control BMDCs were cultured in vitro in the presence or absence of the DC survival factor GM-CSF for 24 h and then analyzed by flow cytometry analysis for expression of Annexin V+/propidium iodide+ (Fig. 3 A) as general apoptosis marker readouts and caspase 3/7 activation (Fig. 3 B) as marker of the cell-intrinsic pathway of apoptosis activation. Growth factor starvation induced apoptosis in 47% of control DCs compared with only 13% (P = 0.0055) of BRAFV600E+ BMDCs (Fig. 3 A). Resistance to apoptosis was rescued by MEKi treatment during growth factor starvation in BRAFV600E+ BMDCs as measured by increased caspase 3/7 activation (Fig. 3 B), suggesting that increased cell survival is dependent on ERK activation.
Figure 3.
BCL-XL up-regulation via RAF/MEK/ERK signaling contributes to suppressed apoptosis in DCs. (A) Apoptosis measured in control and BRAFV600E BMDC by Annexin V/propidium iodide (PI) staining. On day 6 of culture, GM-CSF cytokine was removed from BMDC culture, and BMDC viability was measured on day 7 by flow cytometry. Dot plots show representative frequency of apoptotic Annexin+PI+ CD11c+CD11b+ BMDC from two experiments. Bar graph show the mean of triplicate samples representative of two experiments ± SEM (n = 3–5; control vs. BRAFV600E starved PI positivity: **, P = 0.0055; unpaired t test; baseline vs. starved control Annexin V positivity: *, P = 0.0419; unpaired t test). (B) Caspase 3/7 activation measured in control and BRAFV600E BMDCs starved of GM-CSF growth factor overnight (***, P = 0.0008; unpaired t test), ±1 nM GSK1120212 (*, P = 0.0161; unpaired t test). Representative samples shown in FACS plots. Bar graphs show the mean of three biological replicates representative of two experiments ± SEM. (C) Bclxl expression was measured by Western blot in BRAFV600ECD11c BMDC starved (st) or not starved (nst) of GM-CSF growth factor in overnight culture and harvested on day 6 (D6) or day 7 (D7) of culture. Representative data of two independent experiments are shown. (D and E) BCL-XL protein levels in control and BRAFV600E BMDCs as measured by flow cytometry. Representative data from at least two experiments with three biological replicates are shown. Bar graph in E represents quantification of triplicate conditions within one experiment ± SEM (**, P = 0.0089; unpaired t test). (F) Percentage of apoptotic BMDCs among control or BRAFV600E BMDC cultured overnight with 100 nM BCL2-family inhibitor ABT-263, 1 µM ABT-263 (Annexin V: *, P = 0.0211; unpaired t test; PI: **, P = 0.0032’ unpaired t test) or 1 nM GSK1120212 MEKi (Annexin V: *, P = 0.0268; unpaired t test; PI: **, P = 0.0030; unpaired t test). BMDCs were starved or nonstarved of GM-CSF growth factor during overnight drug treatment and analyzed for apoptosis using Annexin V/PI staining by flow cytometry. Bar graphs show mean of three biological replicates ± SEM, representative of two independent experiments. (G) Caspase 3/7 activation measuring BRAFV600E BMDC treated with control vehicle (***, P = 0.0004; unpaired t test) or with 1 nM GSK1120212 (*, P = 0.0118; unpaired t test), as shown in B, or in the presence of 1 µM ABT-263 (*, P = 0.0330; unpaired t test) overnight. Bar graphs show the mean results of triplicate conditions from two independent experiments ± SEM. (H) Western blot showing BCL2L1 protein levels in human LCH lesions cultured without serum overnight, then treated with BRAF or MEKi’s for 2 h. (C and H) Molecular mass is indicated in kilodaltons. (I and J) Viability of human LCH lesions cultured overnight without serum, then treated for 2 h with 1 nM GSK1120212 MEKi (I), or 1 µM ABT-263 BCL2-family inhibitor (J). Three patient samples in each treatment group. Data represent means shown ± SEM.
To investigate the mechanism of BRAFV600E-mediated resistance to apoptosis, we measured the expression of pro- and antiapoptotic transcripts by qPCR. We found that transcripts of genes encoding proapoptotic proteins NOXA and BIM, and antiapoptotic BCL2, BCL-XL, and MCL1, were significantly decreased in BRAFV600E+ BMDCs compared with control DCs (Fig. S2 A). ERK signaling can transcriptionally and posttranscriptionally regulate protein levels (Dimmeler et al., 1999; Katz et al., 2009), so we tested protein levels by Western blot and found significantly increased levels of antiapoptotic BCL-XL in BRAFV600E-expressing DCs compared with control DCs (Fig. 3 C), whereas proapoptotic BIM protein was not differentially regulated (Fig. S2 B). Intracellular flow cytometry staining confirmed that BRAFV600E+ BMDCs expressed elevated levels of BCL-XL protein (Fig. 3, D and E). To test relative BCL-XL expression levels, control and BRAFV600E+ BMDCs were stimulated with and without RANK ligand (RANKL), a stimulus known to strongly induce BCL-XL expression and mediate DC survival (Wong et al., 1997). RANKL stimulation boosted the expression levels of BCL-XL protein in control BMDCs; however, BRAFV600E+ BMDCs did not further increase BCL-XL expression, indicating that BRAFV600E BMDCs may already be at maximal expression levels of BCL-XL (Fig. S2 C).
To test whether BCL-XL is important for modulating survival in BMDCs, we treated BRAFV600E+ BMDCs and control BMDCs with ABT-263, a BCL2-family inhibitor with high affinity to BCL-XL (Roberts et al., 2012). Notably, ABT-263 treatment during normal BMDC culture did not induce apoptosis; however, upon GM-CSF growth factor starvation, treatment with ABT-263 led to significantly increased apoptosis in BRAFV600E+ DCs (Fig. 3 F) and increased caspase 3/7 activation (Fig. 3 G), similar to apoptosis levels after treatment with MEKi and growth factor starvation. High levels of the BCL-XL homologue BCL2L1 were also detectable in primary human LCH lesion CD207+ cells by Western blot, and addition of BRAF inhibitor or MEKi dramatically decreased BCL2L1 protein levels (Fig. 3 H) and induced LCH cell death (Fig. 3 I). Accordingly, treatment with BCL2-family inhibitor ABT-263 also induced LCH cell death (Fig. 3 J), recapitulating the observations made in the mouse models. These data indicate that in addition to trapping DCs in tissue, constitutive MAPK pathway activation also prolongs DC survival by increasing the expression of the antiapoptotic molecule BCL-XL.
MEKi’s rescue DC cell accumulation but induce severe side effects
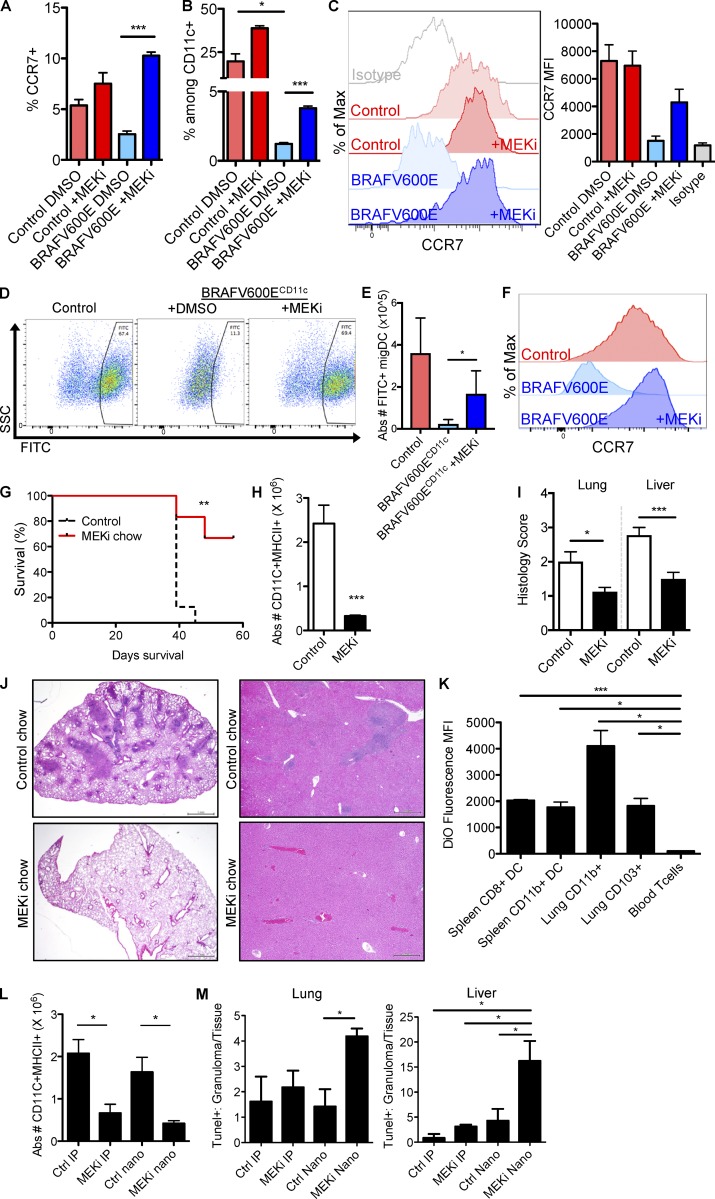
Next we set out to test the therapeutic impact of MEK inhibition in vivo. As BRAFV600E promotes DC accumulation and survival in tissues, we tested the potential for MEK inhibition to rescue DC migration and LCH cell accumulation, and pathogenesis in vivo. In vivo MEKi treatment increased the frequency of CCR7+ lung tissue DCs (Fig. 4 A), increased the frequency of skin dLN migDCs (Fig. 4 B), and rescued expression of CCR7 on skin dLN migDCs (Fig. 4 C). Interestingly, MEKi’s not only rescued the migratory capacity of pathogenic BRAFV600E+ DCs but also increased the migration of WT migDCs (Fig. 4, A and B), suggesting that MEKi’s may influence the function of DCs in relation to tumor immunity.
Figure 4.
MEK inhibition rescues DC migration and pathological DC accumulation. (A–C) BRAFV600ECD11c and control mice were injected with two doses of 1 mg/kg MEKi GSK1120212 and sacrificed 30 h after the first injection (5 h after the second injection). (A) Frequency of CCR7+ DCs among live CD11c+MHCII+F480− lung DCs is shown (***, P < 0.0001; unpaired t test). (B) Frequency of CD11cintMHCIIhigh mDCs and resident CD11chighMHCIIint DCs among live MHCII+CD11c+CD3−B220− from skin dLN (*, P < 0.0132; ***, P = 0.0002, unpaired t test). Flow cytometry plots show representative samples, and bar graph shows the mean ± SEM (n = 3). (C) Histogram shows CCR7 surface protein levels CD11cintMHCIIhigh migDCs from skin dLN. (D–F) BRAFV600ECD11c and control mice were injected with 1 mg/kg MEKi GSK1120212 or control vehicle 16 h before FITC painting challenge, injected with drug or control vehicle immediately after FITC painting, and then sacrificed 16 h later to harvest LNs for flow cytometry analysis. Representative of three experiments with two to three mice per group. (D) Representative frequency of FITC+ migDCs within the skin dLN. (E) Graph represents quantification of absolute numbers of FITC+ migDCs (*, P = 0.0213; unpaired t test). (F) CCR7 expression on skin dLN migDCs ±MEKi treatment. (G–J) BRAFV600ECD11c chimeras treated with control or MEKi chow for 3 wk. (G) Survival of BRAFV600ECD11c chimeric mice treated with either control chow or PD0325901 MEKi chow as described in the methods (**, P = 0.0014; Mantel-Cox test). Data shown are the mean survival of six to eight mice per group. (H) Absolute number of CD11c+MHCII+ DCs in the lung of BRAFV600ECD11c chimeras (***, P = 0.0005; unpaired t test) after 3 wk of treatment with PD0325901 MEKi or control chow (n = 8–9 mice/treatment group). (I) Histological scores of LCH lesions in lungs (*, P = 0.0178; unpaired t test) and livers (***, P = 0.0006; unpaired t test) of PD0325901 MEKi or control chow treated BRAFV600ECD11c chimeras. (J) Representative images of treated lung and liver lesions. Bars, 500 µm. (K) Fluorescent nanoparticle labeling of DCs in BRAFV600ECD11c chimeric mice injected i.v. with a single dose of DiO-HDL nanoparticles. Tissues were harvested 48 h after injection and analyzed for fluorescent labeling by flow cytometry. Data represent mean, n = 2 mice. Representative of two experiments (***, P = 0.0005; *, P < 0.05; unpaired t test). (L and M) Efficacy of i.p. injected GSK1120212 MEKi and GSK1120212–loaded nanoparticles. BRAFV600ECD11c chimeras were treated with i.p. injection of control vehicle (Ctrl IP) or MEKi (1 mg/kg GSK1120212; MEKi IP), i.v. injection of empty HDL nanoparticle (Ctrl nano), or i.v. 1 mg/kg GSK1120212-loaded nanoparticle (MEKi nano) two times per week for 2 wk (four total injections). (L) Bar graphs show the absolute numbers of CD11+MHCII+ DCs in the lungs of treated BRAFV600ECD11c chimeras (*, P < 0.05; unpaired t test). Data represent mean ± SEM (n = 3–4 mice per treatment group). (M) TUNEL staining in the liver (*, P = 0.0102; unpaired t test) and lung (*, P = 0.0193; unpaired t test) of treated BRAFV600ECD11c chimeras. Bar graphs show the ratio of TUNEL+ nuclei within granuloma divided by TUNEL+ nuclei in total tissue (*, P < 0.05; unpaired t test). Data represent mean ± SEM (n = 3–4 mice per treatment group).
MEKi treatment 16 h before FITC skin painting challenge made a dramatic impact, restoring the frequency (Fig. 4 D) and absolute number of FITC+ migDC (Fig. 4 E) in the skin dLN of BRAFV600ECD11c mice. Furthermore, MEKi treatment also restored CCR7 surface protein expression in the migDC compartment after FITC painting challenge (Fig. 4 F).
Mice reconstituted with BM cells isolated from BRAFV600ECD11c mice developed a rapid, fatal LCH-like syndrome with progressive tissue infiltration of CD11c+MHCII+ DCs (Fig. 4 G). BRAFV600ECD11c chimeric mice treated with MEKi–supplemented chow (estimated at 1 mg/kg daily dosing) survived significantly longer than mice on control chow (Fig. 4 G). MEKi chow significantly reduced tissue DC accumulation by absolute numbers (Fig. 4 H). Accordingly, histological analysis revealed significant improvement of LCH lesions, with reduced histiocytic infiltration of alveoli and peribronchiolar stroma in lungs (Fig. 4 I) and significantly reduced sinusoidal distension and periportal cellular infiltration in livers (Fig. 4 I) of MEKi–treated mice compared with controls. Whereas overall survival and lesion severity improved with MEKi chow, nearly all MEKi–treated mice developed dermatitis between 2–3 wk of treatment (Fig. S3). These side effects are reminiscent of rashes commonly seen in human patients receiving MEKi therapy (Falchook et al., 2012).
Nanoparticles loaded with MEKi’s deliver targeted therapy to LCH cells
Methods directly targeting MAPK inhibition toward LCH cells would be desirable in the clinical setting to reduce detrimental off-target effects. High-density lipoprotein (HDL)–based nanoparticle platforms have the potential to deliver highly insoluble drugs to phagocytic cells (Duivenvoorden et al., 2014; Sanchez-Gaytan et al., 2015; Tang et al., 2015), thereby reducing systemic off-target side effects in nonphagocytic cells. To demonstrate phagocytic cell targeting in the BRAFV600ECD11c LCH mouse model, we injected HDL nanoparticles loaded with the hydrophobic fluorescent dye DiO as a surrogate to hydrophobic drug and assessed their uptake into DC subsets by flow cytometry. DiO nanoparticle labeling was enriched in DCs at levels 20–40 times higher than T cells (Fig. 4 K), demonstrating phagocyte-specific targeting within the hematopoietic compartment.
We next generated a HDL nanoparticle loaded with the MEKi GSK1120212 (trametinib). The trametinib-loaded nanoparticles display a diameter of ∼20 nm and a discoid morphology (not depicted), similar to the natural HDL. To compare the effects of systemic i.p. delivery to nanoparticle delivery of MEKi, we generated a cohort of chimeric mice reconstituted with BM cells isolated from BRAFV600ECD11c mice or control mice. Starting at 3 wk after transplant, chimeric mice were treated with i.p. injection of 1 mg/kg MEKi (MEKi IP), i.p. injection of vehicle (Ctrl IP), i.v. injection of 1 mg/kg MEKi-loaded nanoparticles (MEKi nano), or i.v. injection of empty nanoparticles (Ctrl nano). At 2 wk of treatment, both MEKi delivery approaches had significantly reduced DC numbers in tissues (Fig. 4 L). However, MEKi-loaded nanoparticles induced a higher ratio of apoptosis within LCH lesions relative to those in surrounding healthy tissue as a surrogate marker of off-target apoptosis and cell death induction quantified by TUNEL staining (Fig. 4 M). These results demonstrate that pharmacological targeting of MEKi’s to DCs has the potential to rescue DC migratory capacity and resistance to apoptosis in LCH lesions. Further, MEKi-loaded HDL nanoparticles show potential to increase the therapeutic index of MEKi’s by reducing off-target toxicity.
Discussion
The findings of this study elucidate the mechanisms driving lesion formation and persistence in LCH driven by constitutive MAPK pathway activation in myeloid DC precursor cells. There has been long-standing historical debate of LCH as a neoplastic versus reactive immune disorder. Discovery of BRAFV600E in LCH lesions supported an oncogenic model, but lack of obvious proliferation was puzzling (Arceci et al., 1998; Badalian-Very et al., 2010; Berres et al., 2013). BRAFV600E mutation is commonly mutated in melanoma, yet BRAFV600E alone is not sufficient to drive nevus cells into uncontrolled proliferation. Indeed, BRAFV600E is involved in induction of cellular senescence, and the process by which nevi overcome growth arrest is through accumulation of further mutations. However, unlike melanoma, where BRAFV600E is one of hundreds of somatic mutations (Alexandrov et al., 2013), LCH is characterized by an overall very low rate of mutations (median of 1 exomic mutation per patient; Chakraborty et al., 2014). Lesion clonality has been detected in patients’ lesions through nonrandom X-inactivation patterns of CD1a+ LCH lesion cells, suggesting clonal expansion of LCH cells (Willman et al., 1994; Yu et al., 1994). However, other studies have shown that CD1a+ LCH lesion cells have a low proliferative index (Senechal et al., 2007; Sahm et al., 2012), suggesting that DCs may not be aberrantly proliferating in situ at the lesion site. Our study demonstrates that BRAFV600E mutant tissue DCs and control DCs have similar proliferation rates, which affirms the notion that the BRAFV600E does not drive augmented proliferation of the CD207+ cell within the LCH lesion. Furthermore, BRAFV600E progenitors in BM in patients with high-risk LCH (with hematopoietic organ involvement) are typically <1% of total peripheral blood cells or BM aspirate (Berres et al., 2014), making a hematopoietic reservoir of hyperproliferative precursor cells unlikely.
Although there is no statistically significant effect of BRAFV600E expression in driving proliferation when using ANOVA to compare all DCs from various organs, it is true that there is a P < 0.05 significance specifically in the spleen CD11c+MHCII+ compartment (Fig. 1 D). However, whereas a P value of 0.05 is considered statistically significant, it is a small difference of ∼9% versus 12% in WT and BRAFV600ECD11c spleen DCs, respectively. This small difference in proliferation does not necessarily suggest BRAFV600E-driven proliferation. As the spleen is a hematopoietic organ, there may be an indirect 3% increase in extramedullary hematopoiesis because of systemic inflammation. We feel that the ANOVA is a more appropriate statistical analysis for this study as we are testing for the overall effect of the BRAFV600E mutation in regards to DC proliferation, regardless of tissue origin. Thus, although there is a very slight increase in splenic DC BrdU incorporation in BRAFV600ECD11c mice, the overall picture suggests that BRAFV600E does not significantly increase DC proliferation. Furthermore, although there is a 3% increase in BrdU incorporation in the spleen, DCs still accumulate in the other tissues despite the absence of augmented proliferation. These data demonstrate that augmented proliferation is not a prerequisite for lesion formation.
LCH lesions include clonal CD207+ DCs, which can give an appearance of clonal proliferation that drives tumor formation in most malignant conditions. However, according to results from this study, we hypothesize that lesions result from the accumulation of CD207+ cells derived from clonal hematopoietic stem cells (HSCs) or myeloid progenitor cells bearing the BRAFV600E mutation. These LCH cells are trapped in lesion tissue sites and resistant to cell death (whereas WT DCs migrate away and turn over), resulting in a concentration of clonal populations of differentiated CD207+ cells derived from the same mutation-bearing precursors.
In the absence of augmented proliferation, accumulation and persistence of pathological DCs is a plausible mechanism of lesion formation. There have been contradictory studies on expression of CCR6 and CCR7 of LCH DCs (Annels et al., 2003; Fleming et al., 2003). This study demonstrates that MAPK pathway activation inhibits CCR7 expression, effectively trapping LCH CD207+ cells within the lesion. CCR7 is required for constitutive steady-state migration as well as migration of DCs in response to tissue injury (Förster et al., 2008; Braun et al., 2011). CXCR4 was also up-regulated in BRAF inhibitor– and MEKi-treated human LCH lesion cells. In addition to its role in HSC homing to the BM niche, CXCR4 up-regulation has been implicated in promoting tissue DC egress into the lymphatics. Thus, BRAF-mediated suppression of CXCR4 expression may also contribute to impaired LCH cell migration and tissue accumulation (Kabashima et al., 2007; Stutte et al., 2010). However, it is noteworthy that the BRAFV600E-mediated suppression of CXCR4 expression does not appear to be conserved between mouse and human, according to our expression data. Conversely, ERK-mediated suppression of CCR7 expression and resulting paralysis of DC migration toward CCR7 ligands to the LN is conserved between human and mouse LCH cells and is rescued with MAPK pathway signaling blockade.
Overnight treatments of BRAFV600ECD11c DC both in vitro and in vivo with BRAF or MEKi’s were able to restore DC migratory capacity (Figs. 2 F and 4 D), arguing that the cells are DCs with stunted migratory capacity that can be readily rescued when constitutive BRAFV600E activity is interrupted. This argues that BRAF activity is regulating DC migratory function rather than generating systematic differentiation bias toward macrophages in the cultures. In the case of the human LCH lesions, this model is also supported as BRAF and MEK inhibition rescues CCR7 expression in characteristic CD207+CD163− LCH DCs isolated from the lesions (Fig. 2 L).
Corbí’s group demonstrated that ERK activity modulates human DC maturation and CCR7 expression in normal human DCs in vitro (Puig-Kröger et al., 2001; Aguilera-Montilla et al., 2013). The presence of serum in cell culture medium activates ERK phosphorylation and suppresses the maturation of human monocyte–derived DCs. When serum was removed from culture, ERK phosphorylation was reduced and maturation marker expression was boosted in response to maturation stimulus (TNFα; Puig-Kröger et al., 2001). Furthermore, treatment with MEKi increased the DC maturation in response to TNFα stimulation (Puig-Kröger et al., 2001). Their microarray study revealed that MEKi treatment in normal human DCs induced increased CCR7 mRNA expression (Aguilera-Montilla et al., 2013). These data align with our observations, thereby suggesting that BRAFV600E is locking on and exaggerating a normal function of BRAF in DCs. Whereas WT BRAF activity can be turned off in response to changes in cellular environment, BRAFV600E activity sustains ERK activity independent of extracellular signals, thereby constantly and potently suppressing DC migration. ERK activity suppresses CCR7 expression and DC migration, and down-regulation of ERK activity appears to be a necessary step for DC tissue egress and steady-state migration to the LN.
In addition to impaired mobility, inhibition of apoptosis appears to be a significant contributor to LCH cell accumulation. LCH cells are resistant to apoptosis, and treatment with MEKi induces apoptosis in both mouse BRAFV600ECD11c and human LCH lesion cells. MEKi treatment induced cell death in human LCH lesions bearing both BRAFV600E mutation and WT BRAF (Fig. 3 I). From our previous study, in patient samples where no MAPK pathway gene mutations were detected, MEK1 phosphorylation was variable, but ERK1/2 phosphorylation levels remained universally high (Chakraborty et al., 2014). Patient samples where no MAPK pathway gene mutations were detected exhibited variable responses to BRAF and MEK inhibition, in contrast to predicted responses from BRAFV600E and MAP2K1 mutant samples. Thus, response to MEKi is highly variable in patients with no detected mutation (and uncertain mechanisms of MEK and/or ERK activation), and these results are consistent with what we have shown before (Chakraborty et al., 2014, 2016).
Treatment with MEKi induces apoptosis in LCH cells, and we found that this resistance to apoptosis is at least partly mediated by the antiapoptotic BCL-XL protein as treatment with BCL-2 family inhibitor ABT-263–induced apoptosis in serum-starved BRAFV600ECD11c DCs. Whereas ABT-263 inhibits both BCL2 and BCL-XL, we believe that BCL-XL is the key player in this case as BCL2-transgenic mice do not demonstrate any accumulation of classical DCs. Rather, BCL2-transgenic mice accumulate plasmacytoid DCs in excess (Carrington et al., 2015).
Although our results show that BRAFV600E+ DCs are tissue-restricted and evade apoptosis, resulting in accumulation in the tissue, the signals that initially recruit these cells are still unknown. Mechanisms of LCH tumor dissemination remain uncertain, but our mouse BM chimera data (Fig. 4 G–J) support the hypothesis that BM-derived LCH cell precursors seed tissues to drive lesion formation. The inflammatory cytokine milieu within the lesions may play a role in recruitment of DC progenitors to the lesion site. If increased recruitment was the only mechanism at play, normal DC migration and apoptosis should clear the lesions. Yet we see that upon in vivo challenge with FITC painting, the cells are not able to leave the tissue. Thus, although there may be additional factors at play, we feel the migration and survival defects are critical factors to contributing to lesion formation. Upon DC precursor recruitment to the lesion site, we hypothesize that the precursors differentiate into immature DCs that are incapable of up-regulating CCR7, thereby becoming restricted within the tissue. We hypothesize that the local granuloma formation and cytokine storm results from dysfunctional and sustained inflammatory signaling between the paralyzed LCH cells and reactive immune cells.
A limitation of our model is that in our model, ∼95% of the conventional DCs will express the BRAFV600E mutation (Caton et al., 2007), whereas in human LCH there will be only a minor fraction of lesional cells positive for BRAFV600E. This might explain the aggressive and disseminated phenotype of the BRAFV600ECD11c mouse model. Cre-mediated recombination is detected in more than 95% of conventional CD11chigh DCs both from lymphoid organs and from nonlymphoid tissues such as lung and epidermis, and in 50–80% of plasmacytoid DCs in mice. Moreover, relatively low amounts of recombination are detected in lymphocytes (<10%), natural killer cells (12%), and other myeloid cells such as granulocytes (<1%). No increase of recombination frequency was observed in CD11clow-activated T cells (Caton et al., 2007).
To more closely mimic the clinical scenario of human LCH in which BRAFV600E-mutated cells are present alongside nonmutated DCs in the same individual, we tried to adjust our model by generating mixed chimeric mice. To this end, we reconstituted C57BL6/J mice after lethal irradiation with BM cells derived from BRAFV600ECD11c mice, littermate control mice, or both. Engraftment and chimerism were assessed by expression of congenic markers of the donors and host by flow cytometry analysis. However, as depicted in Fig. S4, we thereby revealed a severe competitive engraftment disadvantage of BRAFV600ECD11c mice–derived progenitors as compared with control BM progenitors (4 out of 5 mice with less than 5% cells of BRAFV600ECD11c origin), whereas there was no engraftment failure of BRAFV600ECD11c BM in mice receiving BRAFV600ECD11c BM in a noncompetitive setting. This engraftment disadvantage in the mixed environment precludes further investigations on the interfering effect of mutated and nonmutated myeloid cells in this mouse model of LCH. This is reminiscent of the engraftment disadvantage seen in BM from enflamed donor mice injected with TLR ligands. BM from LPS-challenged mice demonstrates a MYD88- and TRIF-dependent engraftment disadvantage when mixed with healthy BM from untreated mice during a competitive engraftment experiment (Zhang et al., 2016). Direct sensing of the inflammatory environment in the donor mouse induces changes in HSCs resulting in engraftment disadvantage relative to HSCs derived from normal BM. This may be what we are seeing in this situation of competitive engraftment with BRAFV600ECD11c BM as BRAFV600ECD11c mice display severe systemic inflammation.
One possible reason a significant fraction of patients relapse after cessation of vinblastine/prednisone therapy despite initial responses (Minkov et al., 2008; Gadner et al., 2013) might be transient benefits from disrupting the supportive inflammatory lesion microenvironment while sparing the resistant DC precursor cells. Therapies with activity in other myeloid malignancies, such as nucleoside analogues cytarabine and clofarabine, may more effectively inhibit myelopoiesis, thereby reducing recruitment of new progenitors to differentiate at the lesion site (Abla et al., 2010). However, as these chemotherapy strategies all carry increasing toxicity with increasing doses, optimal therapy regimens remain to be defined.
Besides providing insight in disease initiation and progression, the discovery of mutations in the MAPK signaling pathway have additionally opened up opportunities to generate novel therapies. Indeed, adult LCH patients have already been treated with BRAF inhibitors with promising response rates (Haroche et al., 2015; Hyman et al., 2015). However, BRAF inhibitors also carry significant risks from toxicity, including keratoacanthoma and squamous cell carcinoma caused by paradoxical activation of the MAPK pathway in nonmutated cells (Su et al., 2012). Among MAPK pathway inhibitors, MEKi’s have the advantage of being able to target a broader range of mutations than agents specific for BRAF monomers such as vemurafenib. However, MEKi’s have a very narrow therapeutic index in vivo. MEK is required for cellular growth and homeostasis downstream of various growth factors in many cell types, which may explain the frequent toxicities of these treatments including severe rash, leukopenia, and diarrhea. Indeed, we demonstrated that DC accumulation can be rescued in BRAFV600ECD11c chimeric mice treated with MEKi chow; however, these mice experienced severe skin toxicity. Moreover, as LCH is largely a pediatric disease, the impact of prolonged therapy on cellular growth and survival on children is uncertain. Recently, efforts have been made to combine BRAF and MEK inhibition in the treatment of MAPK-mutated solid cancers, e.g., melanoma, to reduce the off-target effect initiated by paradoxical pathway activation in nonmutated cells, but it remains to be proven that these regimens will also substantially reduce other long-term side effects and increase efficacy and safety (Long et al., 2014).
ABT-263 is a BCL-2 family inhibitor that is being clinically tested for treatment of various cancers (Tse et al., 2008). ABT-263 inhibits the antiapoptotic proteins Bcl-2, Bcl-w, and Bcl-xL (Tse et al., 2008). ABT-263 treatment did not induce apoptosis in BRAFV600E+ DCs under normal culture conditions with growth factor GM-CSF present; however, removal of growth factor potentiated apoptosis upon ABT-263 treatment. As platelets require BCL-XL for survival, thrombocytopenia is a dose-limiting effect of ABT-263 treatment. In the future, the combination of ABT-263 with MEKi may present opportunities to lower dosages of each individual drug, but further studies will be required to find optimal combined in vivo dosing.
In LCH mouse models in this study, treatment with MEKi demonstrated significant response as well as significant toxicity. We therefore took advantage of the phagocytic ability of DCs to test an innovative drug delivery strategy: MEKi-loaded nanoparticles were preferentially taken up by DCs in BRAFV600ECD11c chimeras, decreasing tumor burden and prolonging survival with substantially reduced side effects as compared with free drug delivery. With this proof of concept experiment, we set the stage for further strategies aiming to directly target pathogenic LCH cells within the lesions, maximizing the effects of the drugs on the pathogenic cells while minimizing their impact on bystander and healthy cells and thereby limiting side effects. Modified nanoparticles loaded with BRAF, MEK, or BCL-XL inhibitors may therefore have therapeutic potential for treating LCH and other histiocytic diseases. Although optimization for tissue targeting and nanoparticle stability will be required, this study provides proof-of-concept evidence that nanoparticle targeting of pathogenic histiocytes may be a viable approach for future histiocytosis treatments.
Specifically targeting DC migration and survival may spare patients from extreme immunosuppression and toxicity involved in targeting overall myelopoiesis through chemotherapeutic use of nucleoside analogues. Persistent MAPK pathway activation in LCH lesion CD207+ cells arrests the DC life cycle at an immature stage without significant CCR7 expression and with inhibition of apoptosis. Blocking MAPK signaling restored DC migration and apoptosis stages rather than inhibiting overall myeloid cell production. Further understanding of the chemokines, cytokines, and growth factors involved in the suppression of DC migration and prolonging of DC survival may reveal new drug targets to allow development of novel clinical treatments for LCH. Our results have been demonstrated using the BRAFV600ECD11c mouse model and human lesions with MAPK pathway activation as a result of several different somatic gene mutations. ERK activation appears to be universal in LCH DCs (Badalian-Very et al., 2010; Chakraborty et al., 2014), and we suspect that MAPK-mediated regulation of CCR7 and apoptosis are common mechanisms across all LCH lesions regardless of the specific driver mutation.
Notably, our study revealed that MEK inhibition not only rescued the migration of BRAFV600E+ DCs but also enhanced the migration of tissue-resident WT DCs to the tissue dLN. Given the critical contribution of tissue-resident DC migration to the induction of therapeutic immunity against tumor antigens, our results suggest that MEKi’s may influence the treatment of BRAFV600E tumors not only though a direct antitumor effect but also through modulation of DC function during the orchestration of antitumor immunity. MEKi’s may also modulate DC migration and function in other disease settings.
In summary, this study provides a novel insight into the underlying driving mechanisms of LCH pathology and support the rational development of novel therapies that target DC migration and survival rather than cellular proliferation. Targeted MAPK inhibition delivered through nanoparticles could enhance accessibility of MAPK pathway inhibitors in LCH where development of these promising agents has been challenged by a concerning toxicity profile for a largely pediatric population.
These studies also reveal some basic aspects about the role of the MAPK pathway in regulating DC migration and survival in general. We speculate that growth factors required for tissue DC homeostasis (such as CSF2 [King et al., 2010; Greter et al., 2012] and TGFβ [Kaplan et al., 2007; Kel et al., 2010]) may contribute to tissue DC tissue retention and homeostasis in part through ERK activation, thereby suppressing CCR7 while also enhancing DC survival through suppression of apoptosis. Our data also suggest the potential benefits from understanding the effects of MAPK inhibition in modulating DC migration and DC survival in other therapeutic settings beyond LCH, such as antitumor and vaccine immunity.
Materials and methods
Mouse models
All animal experiments performed in this study were approved by the Institutional Animal Care and Use Committee of Mount Sinai School of Medicine. BRAFV600ECD11c mice were created by crossing BRAFV600Eca/wt or BRAFV600Eca/ca mice (provided by M.W. Bosenberg, Yale University, New Haven, CT; Dankort et al., 2007) with mice expressing cre recombinase under the control of the CD11c promoter (C57BL/6 background; The Jackson Laboratory). BRAFV600ECD11c mice with one BRAFV600ECA allele and one CD11c-Cre allele develop a lethal LCH-like syndrome, as described (Berres et al., 2014). All animals were housed under specific pathogen-free conditions and sacrificed at the indicated time points. All experiments were controlled using littermates negative for the cre recombinase transgene construct or cre-positive littermates negative for the BRAFV600Eca construct.
BRAFV600ECD11c chimeras were generated by transplantation of 1–3 × 10^6 whole BM cells flushed from the long bones of 5–8 wk old BRAFV600ECD11c mice into lethally irradiated CD45.1 mice age 8–12 wk (C57BL/6 background; Charles River Laboratory). Mice were allowed to recover for 3 wk after transplantation before initiation of drug treatments.
Statistics
Statistical analysis was performed using unpaired two-tailed Student’s t tests, and error bars indicate SEM. Statistical significance is indicated by *, P < 0.05; **, P < 0.01; and ***, P < 0.001. Two-way ANOVA was performed for Fig. 1 (D and E), to measure the impact of BRAFV600E on proliferation between different cell types.
Human subject approvals
Human tissues used in these studies were collected, stored, and processed according to protocols approved by the Baylor College of Medicine Internal Review Board.
Reagents
Recombinant mouse GM-CSF, TNFα, and IL-1β were purchased from Peprotech. MEKi chow PD0325901 at 7 ppm (estimated 1 mg/kg daily dosing) was kindly provided by Plexxikon. GSK1120212, ABT-263, and Vemurafenib (PLX4720) were obtained from Selleck Chem.
Histology and immunohistochemistry analysis
For the immunohistochemistry analysis, 4-µm tissue sections were deparaffinized and rehydrated through xylene/absolute alcohol. Endogenous peroxidase activity was blocked by incubating the sections in methanol with 0.6% hydrogen peroxide for 10 min at room temperature. Heat-induced antigen retrieval was performed with citrate buffer, pH 6.0, in a steamer for 15 min. To block nonspecific staining, Rodent Block M (Biocare Medical) was incubated with the section for 1 h at room temperature. Sections were stained with anti-CD3 (Abcam), anti–MHC II (BD), anti-FOXP3 (BioLegend), or anti-CD68 (Thermo Fisher Scientific) mAbs overnight at 4°C. Application of the primary antibodies was followed by a 30-min incubation with a Rabbit on Rodent/Mouse on Mouse/Rat on Rodent Polymer-HRP (BioCare Medical) and visualized with DAB (Diagnostics BioSystems) as a chromogen with CAT hematoxylin counterstaining (BioCare Medical). For immunohistochemistry on human specimens, 5-µm-thick formalin-fixed and paraffin-embedded tissue sections were deparaffinized before incubation in antigen retrieval solution (S2369 and S2367, Dako) at 95°C for 30 min. Tissue sections were incubated in 3% hydrogen peroxide and in serum-free protein block solution (X0909, Dako) before adding the primary antibodies. After signal amplification using biotinylated-secondary antibody and streptavidin-HRP, chromogenic revelation was performed and slides counterstained with hematoxylin. Slides were scanned for digital imaging and quantification (Olympus whole-slide scanner with Olyvia software). After scanning, the slide was bleached to remove the chromogen stain and subjected to the next round of staining (MICSSS method; Remark et al., 2016). The following Abs were used for LCH cell analysis: anti-CD207 (clone 12D6) and anti-Ki-67 (clone 30–9). After image acquisition, each stain was artificially attributed a color code, and images were overlaid using Adobe Photoshop CS6.
Generation of BMDCs
Bones were flushed with RPMI + 10% FBS media, and single cell BM suspensions were incubated with RBC lysis buffer (BioLegend) for 2 min at room temperature before plating at a concentration of 1 × 106 cells/ml with 20 ng/ml GM-CSF growth factor (PeproTech). Media were replaced on days 3 and 6 of culture. Cells were treated for harvest between days 6 to 8 of culture. DCs were analyzed by FACS analysis. BMDCs were gated as MHCII+CD11b+CD11c+. Recombinant TNFα (PeproTech) or IL-1β (PeproTech) was added to indicated cultures at 10 ng/ml or 100 ng/ml.
Adoptive transfer of DCs
BRAFV600E and control BMDCs were labeled with CFSE for 10 min, then extensively washed, and 600,000 cells were resuspended in 25 µl PBS before being administered dropwise to the nose of mice anesthetized with ketamine/xylazine. Lungs were harvested 6 d after cell transfer and processed into single-cell suspensions. Adoptively transferred cells were gated as live CD45.2+CD11c+CD11b+ DCs and analyzed for CFSE dilution as a measure of proliferation.
In vivo BrdU labeling
BRAFV600ECD11c mice aged 8 wk were injected i.p. with 1 mg BrdU. Tissues were harvested 5 h later and processed into single cell suspension for BrdU incorporation by flow cytometry analysis. Tissue preparation and BrdU staining were performed according to the BD PharMingen FITC BrdU Flow kit.
FITC painting
The ventral abdomen and thorax were shaved and stained with 400 µl FITC (5 mg/ml dissolved in equal volumes of dibutyl phthalate and acetone). After 24 h, cell suspensions of single LNs (inguinal, brachial) were analyzed by flow cytometry. 25 µl of FITC in dibutyl phthalate and acetone was applied to each ear. Epidermis and dermis were prepared by separating dorsal and ventral ear flaps and incubating with 4 Mandl units/ml dispase at 37°C for 90 min, then separating epidermis and dermis, dicing with scissors, digesting with collagenase D for 45 min at 37°C, and finally filtering through a 70-µM cell strainer to obtain a single cell suspension.
Flow cytometry analysis
Single cell suspension was obtained from indicated tissues after digestion with Roche collagenase D (Roche) at 37°C for 45 min. For liver tissues, nonparenchymal cells were enriched by density gradient centrifugation with 40/70 Percoll (GE Healthcare) for 30 min at 1,100 rcf. BM and blood single cell suspensions were incubated with RBC lysis buffer (BioLegend) for 2 min at room temperature before staining. mAbs specific to mouse CD45 (clone 30F11; BioLegend), MHCII (I-A/I-E; clone M5/114.15.2; BioLegend), CD11c (clone N418; eBioscience), CD103 (clone 2E7; eBioscience), CD11b (clone M1/70; BioLegend), F4/80 (clone CI: A3-1; BioLegend), CD3 (clone 145-2C11; BioLegend), CD4 (clone L3T4; BioLegend), NK1.1 (clone PK136; eBioscience), FoxP3 (clone FJK-16s; eBioscience), Sca-1 (clone D7; eBioscience), CD115 (clone AFS98; eBioscience), CD34 (clone RAM34; eBioscience), pERK (clone 20A, eBioscience), CD40 (clone 20A, BD), and CCR7 (clone 4B12; BioLegend) were purchased from the indicated vendors. For intracellular FoxP3 staining, cells were fixed overnight in fixation/permeabilization buffer (eBioscience). Before acquisition, cells were resuspended in PBS/BSA 0.5%/EDTA (2 mM) solution with 1 µg/ml of DAPI to exclude dead cells. For FoxP3 staining, cells were stained with a live/dead fixable dead cell stain kit (Invitrogen) before fixation to assess viability. For phospho-ERK flow staining, cells were fixed with 2% paraformaldehyde for 10 min, then washed with PBS and resuspended in 95% −80°C chilled methanol, and stored at −80°C until ready for analysis. Cells were rehydrated in 1:1 (PBS/methanol), then stained with anti-pERK antibody for 1 h at room temperature. Multiparameter analysis was performed on the LSR II (BD) and analyzed with FlowJo software.
Apoptosis assays
Apoptosis induction was measured using the Annexin V and propidium iodide staining kit from BioLegend. Cells were stained for surface markers, then washed and stained in calcium-containing binding buffer according to the manufacturer’s directions. To measure Caspase 3/7 activation, cells were stained for surface markers, then stained for caspase-3 and -7 activation according to the Vybrant FAM caspase-3 and -7 activation kit manufacturer’s instructions. Human LCH lesion apoptosis was measured using the Annexin V & Dead Cell Assay kit (Millipore) on a Muse Cell Analyzer (Millipore) according to the manufacturer’s recommendation. In brief, 100 µl of cells in suspension was incubated with 100 μl Muse Annexin V & Dead Cell Reagent for 20 min at room temperature before being analyzed on the Muse Cell Analyzer.
Trametinib-HDL nanoparticle synthesis
In brief, trametinib (GSK1120212, ApexBio), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), and 1-palmitoyl-2-hexadecyl-sn-glycero-3-phosphocholine (PHPC; Avanti Polar Lipids) were mixed in a ratio of 1:3:1 and dissolved in chloroform/methanol (4:1 by volume) solvent and dried to form a thin film. Human apolipoprotein A1 (APOA1) proteins, separated from human plasma, were added to the film, and the solution was incubated at 37°C until the film was hydrated and a homogeneous solution was formed. The solution was sonicated, and aggregates were removed by centrifugation to yield small trametinib-loaded nanoparticles (Tra-HDL). For fluorescence detection purposes, DiO (Invitrogen) was incorporated when flow cytometry techniques were applied. The nanoparticle solution was washed extensively with sterilized PBS by using a 100-kD filter (Vivaspin, Vivaproducts) and filtered through a 0.22-µm nylon filter before being administered to animals. Trametinib incorporation efficiency was determined by HPLC (Shimadzu), and the final yield of the compound in the nanoparticle was ∼50%.
Immunoblot analysis
LCH patient biopsy cell suspensions were lysed using a buffer containing 25 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP-40, and 5% glycerol supplemented with Halt protease inhibitor cocktail (Thermo Fisher Scientific), phosphatase inhibitor cocktails 2 and 3 (Sigma-Aldrich), and 1 mM sodium orthovanadate (Sigma-Aldrich). 10 µg of whole cell lysates were resolved on a Criterion TGX 4–20%, transferred to an Immobilon PVDF membrane (Millipore), and probed with antibodies recognizing Bcl-xL (BCL2L1; clone: 54H6; no. 2764, Cell Signaling Technology). All blots were subsequently stripped and reprobed with COXIV antibody (no. 4844, Cell Signaling Technology) to confirm equivalent loading across the lanes.
RNA purification and cDNA amplification
Unsorted LCH lesion single-cell suspensions were treated with drug for 12 h. Then cells were sorted as previously described (Chakraborty et al., 2014). Total RNA was isolated from sorted cells by using the Arcturius Picopure RNA isolation kit (Applied Biosystems). cDNA amplification was performed with the Ovation WTA system (NuGen) according to the manufacturer’s protocol.
Human transcriptome data analysis
Transcriptional profiles were generated by using Affymetrix Human Transcriptome Array 2.0 Genechips. The raw data obtained after scanning the arrays was processed using the Affymetrix Expression Console. A signal space transformation in conjunction with the regular robust multiple-array average normalization method (RMA) was applied before obtaining gene expression signal values. The dataset was then checked for quality control in Expression Console by inspecting array metrics, signal distribution box plots, correlation heat maps, and principal component analysis. QC-passed samples were used to generate heat maps using the TM4 MultiExperiment viewer (MeV).
Mouse transcriptome data analysis
RNA isolation and Affymetrix gene analysis were performed as previously described (Miller et al., 2012). RNA was prepared from sorted cell populations from C57BL/6J mice using TRIzol reagent. RNA was amplified and hybridized on the Affymetrix Mouse Gene 1.0 ST array according to the manufacturer’s procedures. Raw data for all populations were preprocessed and normalized using the RMA algorithm implemented in the “Expression File Creator” module in the GenePattern suite. All datasets have been deposited at the National Center for Biotechnology Information/Gene Expression Omnibus under accession no. GSE108251.
Quantitative real-time PCR
Quantitative real-time PCR reactions were performed with TaqMan Gene Expression Assays (Applied Biosystems). Total RNA was isolated and cDNA was generated as described in the "RNA purification and cDNA amplification" method section above. Each quantitative real-time PCR reaction included 20 ng cDNA. The TaqMan Fast Universal PCR Master Mix (Applied Biosystems) was used in 25-µl reactions in 96-well plates on a CFX96 Touch Real-time PCR detection system (Bio-Rad Laboratories). Assays were performed in triplicate. Thermal cycling conditions were set at 10 min at 95°C and then 40 cycles of 95°C for 15 s and 60°C for 1 min. TaqMan probe sets used were CCR7: Hs01013469_m1 and GAPDH: Hs02758991_g1. Data were normalized to GAPDH expression and analyzed by the −ΔΔCt method relative to CD207 expression. Data are presented as mean ± SEM except where otherwise stated.
Online supplemental material
Fig. S1 shows reduced CCR7 expression in BRAFV600ECD11c spleen DC by qPCR and FITC-painted skin DCs at the protein level. Fig. S2 shows expression of pro- and antiapoptotic proteins in control and BRAFV600E DCs. Fig. S3 shows skin ulcerative lesions in mice treated with MEKi. Fig. S4 shows blood myeloid cell chimerism 4 wk after lethal irradiation, and reconstitution by BM progenitors derived from BRAFV600ECD11c mice and littermate controls reveals a competitive engraftment disadvantage of BRAFV600ECD11c mice–derived progenitors as compared with control BM progenitors. Table S1 shows clinical information for human LCH lesions studied in Fig. 1. Table S2 shows clinical information for LCH lesions studied in Fig. 2.
Supplementary Material
Acknowledgments
This study was supported in part by funding from the HistioCure Foundation (TXCH Histiocytosis Program). Research reported in this publication was supported by a grant from the National Heart, Lung, and Blood Institute of the National Institutes of Health (no. F31HL126484 to B. Hogstad). Support includes grants from the following institutions: National Institutes of Health (no. 2R01CA154947-06A1 to B. Hogstad and M. Merad), National Institutes of Health R01 (no. CA154489 to C.E. Allen, K. McClain, and T.-K. Man), National Institutes of Health SPORE in Lymphoma (no. P50CA126752 to C.E. Allen), National Institutes of Health R01 (nos. CA154947, AI10008, and AI089987 to M. Merad), the German Research Association (Deutsche Forschungsgemeinschaft, nos. BE 4818/1-1 and SFBTRR57 P07 to M.-L. Berres), the St Baldrick’s Foundation (North American Consortium for Histiocytosis to C.E. Allen, K. McClain, and M. Merad), the Alex’s Lemonade Stand Foundation Young Investigator Grant (to R. Chakraborty), the American Society of Hematology Scholar Award (to R. Chakraborty), and the Howard Hughes Medical Institute to the Baylor College of Medicine Med into Grad Initiative (to K.P.H. Lim). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We also appreciate the support of shared resources by a Dan L. Duncan Cancer Center support grant (no. P30CA125123).
The authors declare no competing financial interests.
Author contributions: B. Hogstad, M.-L. Berres, R. Chakraborty, designed the studies, performed the experiments, and wrote the manuscript. C.E. Allen and M. Merad coordinated the studies, analyzed data, and wrote the manuscript. J. Tang, S. Baxter, and W.J.M. Mulder designed and produced the nanoparticles. C. Bigenwald, M. Lebouef, E. Brandt, R. Remark, K.P.H. Lim, T.-K. Man, V. Kana, S. Jordan, W.-h. Kwan, and H. Salmon performed research and provided critical feedback. H. Lin performed bioinformatic analyses. Z. Karoulia and P. Poulikakos designed experiments and analyzed data regarding the RAF/MEK/ERK pathway. M. Serasinghe designed and performed apoptotic protein studies. J. Chipuk analyzed data regarding apoptotic protein studies. K. McClain analyzed human LCH specimen studies.
References
- Abla O., Egeler R.M., and Weitzman S.. 2010. Langerhans cell histiocytosis: Current concepts and treatments. Cancer Treat. Rev. 36:354–359. 10.1016/j.ctrv.2010.02.012 [DOI] [PubMed] [Google Scholar]
- Aguilera-Montilla N., Chamorro S., Nieto C., Sánchez-Cabo F., Dopazo A., Fernández-Salguero P.M., Rodríguez-Fernández J.L., Pello O.M., Andrés V., Cuenda A., et al. 2013. Aryl hydrocarbon receptor contributes to the MEK/ERK-dependent maintenance of the immature state of human dendritic cells. Blood. 121:e108–e117. 10.1182/blood-2012-07-445106 [DOI] [PubMed] [Google Scholar]
- Alexandrov L.B., Nik-Zainal S., Wedge D.C., Aparicio S.A., Behjati S., Biankin A.V., Bignell G.R., Bolli N., Borg A., Børresen-Dale A.L., et al. ICGC PedBrain . 2013. Signatures of mutational processes in human cancer. Nature. 500:415–421. 10.1038/nature12477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen C.E., Li L., Peters T.L., Leung H.C., Yu A., Man T.K., Gurusiddappa S., Phillips M.T., Hicks M.J., Gaikwad A., et al. 2010. Cell-specific gene expression in Langerhans cell histiocytosis lesions reveals a distinct profile compared with epidermal Langerhans cells. J. Immunol. 184:4557–4567. 10.4049/jimmunol.0902336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen C.E., Ladisch S., and McClain K.L.. 2015. How I treat Langerhans cell histiocytosis. Blood. 126:26–35. 10.1182/blood-2014-12-569301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angeli V., and Randolph G.J.. 2006. Inflammation, lymphatic function, and dendritic cell migration. Lymphat. Res. Biol. 4:217–228. 10.1089/lrb.2006.4406 [DOI] [PubMed] [Google Scholar]
- Annels N.E., Da Costa C.E., Prins F.A., Willemze A., Hogendoorn P.C., and Egeler R.M.. 2003. Aberrant chemokine receptor expression and chemokine production by Langerhans cells underlies the pathogenesis of Langerhans cell histiocytosis. J. Exp. Med. 197:1385–1390. 10.1084/jem.20030137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arceci R.J., Brenner M.K., and Pritchard J.. 1998. Controversies and new approaches to treatment of Langerhans cell histiocytosis. Hematol. Oncol. Clin. North Am. 12:339–357. 10.1016/S0889-8588(05)70514-1 [DOI] [PubMed] [Google Scholar]
- Badalian-Very G., Vergilio J.A., Degar B.A., MacConaill L.E., Brandner B., Calicchio M.L., Kuo F.C., Ligon A.H., Stevenson K.E., Kehoe S.M., et al. 2010. Recurrent BRAF mutations in Langerhans cell histiocytosis. Blood. 116:1919–1923. 10.1182/blood-2010-04-279083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berres M.L., Allen C.E., and Merad M.. 2013. Pathological consequence of misguided dendritic cell differentiation in histiocytic diseases. Adv. Immunol. 120:127–161. 10.1016/B978-0-12-417028-5.00005-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berres M.L., Lim K.P., Peters T., Price J., Takizawa H., Salmon H., Idoyaga J., Ruzo A., Lupo P.J., Hicks M.J., et al. 2014. BRAF-V600E expression in precursor versus differentiated dendritic cells defines clinically distinct LCH risk groups. J. Exp. Med. 211:669–683. 10.1084/jem.20130977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berres M.L., Merad M., and Allen C.E.. 2015. Progress in understanding the pathogenesis of Langerhans cell histiocytosis: back to Histiocytosis X? Br. J. Haematol. 169:3–13. 10.1111/bjh.13247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthier-Vergnes O., Bermond F., Flacher V., Massacrier C., Schmitt D., and Péguet-Navarro J.. 2005. TNF-alpha enhances phenotypic and functional maturation of human epidermal Langerhans cells and induces IL-12 p40 and IP-10/CXCL-10 production. FEBS Lett. 579:3660–3668. 10.1016/j.febslet.2005.04.087 [DOI] [PubMed] [Google Scholar]
- Braun A., Worbs T., Moschovakis G.L., Halle S., Hoffmann K., Bölter J., Münk A., and Förster R.. 2011. Afferent lymph-derived T cells and DCs use different chemokine receptor CCR7-dependent routes for entry into the lymph node and intranodal migration. Nat. Immunol. 12:879–887. 10.1038/ni.2085 [DOI] [PubMed] [Google Scholar]
- Brown N.A., Furtado L.V., Betz B.L., Kiel M.J., Weigelin H.C., Lim M.S., and Elenitoba-Johnson K.S.. 2014. High prevalence of somatic MAP2K1 mutations in BRAF V600E-negative Langerhans cell histiocytosis. Blood. 124:1655–1658. 10.1182/blood-2014-05-577361 [DOI] [PubMed] [Google Scholar]
- Cagnol S., and Chambard J.C.. 2010. ERK and cell death: mechanisms of ERK-induced cell death--apoptosis, autophagy and senescence. FEBS J. 277:2–21. 10.1111/j.1742-4658.2009.07366.x [DOI] [PubMed] [Google Scholar]
- Cargnello M., and Roux P.P.. 2011. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol. Mol. Biol. Rev. 75:50–83. 10.1128/MMBR.00031-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington E.M., Zhang J.G., Sutherland R.M., Vikstrom I.B., Brady J.L., Soo P., Vremec D., Allison C., Lee E.F., Fairlie W.D., et al. 2015. Prosurvival Bcl-2 family members reveal a distinct apoptotic identity between conventional and plasmacytoid dendritic cells. Proc. Natl. Acad. Sci. USA. 112:4044–4049. 10.1073/pnas.1417620112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caton M.L., Smith-Raska M.R., and Reizis B.. 2007. Notch-RBP-J signaling controls the homeostasis of CD8- dendritic cells in the spleen. J. Exp. Med. 204:1653–1664. 10.1084/jem.20062648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty R., Hampton O.A., Shen X., Simko S.J., Shih A., Abhyankar H., Lim K.P., Covington K.R., Trevino L., Dewal N., et al. 2014. Mutually exclusive recurrent somatic mutations in MAP2K1 and BRAF support a central role for ERK activation in LCH pathogenesis. Blood. 124:3007–3015. 10.1182/blood-2014-05-577825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty R., Burke T.M., Hampton O.A., Zinn D.J., Lim K.P., Abhyankar H., Scull B., Kumar V., Kakkar N., Wheeler D.A., et al. 2016. Alternative genetic mechanisms of BRAF activation in Langerhans cell histiocytosis. Blood. 128:2533–2537. 10.1182/blood-2016-08-733790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dankort D., Filenova E., Collado M., Serrano M., Jones K., and McMahon M.. 2007. A new mouse model to explore the initiation, progression, and therapy of BRAFV600E-induced lung tumors. Genes Dev. 21:379–384. 10.1101/gad.1516407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimmeler S., Breitschopf K., Haendeler J., and Zeiher A.M.. 1999. Dephosphorylation targets Bcl-2 for ubiquitin-dependent degradation: a link between the apoptosome and the proteasome pathway. J. Exp. Med. 189:1815–1822. 10.1084/jem.189.11.1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duivenvoorden R., Tang J., Cormode D.P., Mieszawska A.J., Izquierdo-Garcia D., Ozcan C., Otten M.J., Zaidi N., Lobatto M.E., van Rijs S.M., et al. 2014. A statin-loaded reconstituted high-density lipoprotein nanoparticle inhibits atherosclerotic plaque inflammation. Nat. Commun. 5:3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falchook G.S., Lewis K.D., Infante J.R., Gordon M.S., Vogelzang N.J., DeMarini D.J., Sun P., Moy C., Szabo S.A., Roadcap L.T., et al. 2012. Activity of the oral MEK inhibitor trametinib in patients with advanced melanoma: a phase 1 dose-escalation trial. Lancet Oncol. 13:782–789. 10.1016/S1470-2045(12)70269-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming M.D., Pinkus J.L., Fournier M.V., Alexander S.W., Tam C., Loda M., Sallan S.E., Nichols K.E., Carpentieri D.F., Pinkus G.S., and Rollins B.J.. 2003. Coincident expression of the chemokine receptors CCR6 and CCR7 by pathologic Langerhans cells in Langerhans cell histiocytosis. Blood. 101:2473–2475. 10.1182/blood.V101.7.2473 [DOI] [PubMed] [Google Scholar]
- Förster R., Schubel A., Breitfeld D., Kremmer E., Renner-Müller I., Wolf E., and Lipp M.. 1999. CCR7 coordinates the primary immune response by establishing functional microenvironments in secondary lymphoid organs. Cell. 99:23–33. 10.1016/S0092-8674(00)80059-8 [DOI] [PubMed] [Google Scholar]
- Förster R., Davalos-Misslitz A.C., and Rot A.. 2008. CCR7 and its ligands: balancing immunity and tolerance. Nat. Rev. Immunol. 8:362–371. 10.1038/nri2297 [DOI] [PubMed] [Google Scholar]
- Gadner H., Minkov M., Grois N., Pötschger U., Thiem E., Aricò M., Astigarraga I., Braier J., Donadieu J., Henter J.I., et al. Histiocyte Society . 2013. Therapy prolongation improves outcome in multisystem Langerhans cell histiocytosis. Blood. 121:5006–5014. 10.1182/blood-2012-09-455774 [DOI] [PubMed] [Google Scholar]
- Greter M., Helft J., Chow A., Hashimoto D., Mortha A., Agudo-Cantero J., Bogunovic M., Gautier E.L., Miller J., Leboeuf M., et al. 2012. GM-CSF controls nonlymphoid tissue dendritic cell homeostasis but is dispensable for the differentiation of inflammatory dendritic cells. Immunity. 36:1031–1046. 10.1016/j.immuni.2012.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haroche J., Cohen-Aubart F., Emile J.F., Maksud P., Drier A., Tolédano D., Barete S., Charlotte F., Cluzel P., Donadieu J., et al. 2015. Reproducible and sustained efficacy of targeted therapy with vemurafenib in patients with BRAF(V600E)-mutated Erdheim-Chester disease. J. Clin. Oncol. 33:411–418. 10.1200/JCO.2014.57.1950 [DOI] [PubMed] [Google Scholar]
- Hashimoto D., Miller J., and Merad M.. 2011. Dendritic cell and macrophage heterogeneity in vivo. Immunity. 35:323–335. 10.1016/j.immuni.2011.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupt R., Nanduri V., Calevo M.G., Bernstrand C., Braier J.L., Broadbent V., Rey G., McClain K.L., Janka-Schaub G., and Egeler R.M.. 2004. Permanent consequences in Langerhans cell histiocytosis patients: a pilot study from the Histiocyte Society-Late Effects Study Group. Pediatr. Blood Cancer. 42:438–444. 10.1002/pbc.20021 [DOI] [PubMed] [Google Scholar]
- Hyman D.M., Puzanov I., Subbiah V., Faris J.E., Chau I., Blay J.Y., Wolf J., Raje N.S., Diamond E.L., Hollebecque A., et al. 2015. Vemurafenib in Multiple Nonmelanoma Cancers with BRAF V600 Mutations. N. Engl. J. Med. 373:726–736. 10.1056/NEJMoa1502309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabashima K., Shiraishi N., Sugita K., Mori T., Onoue A., Kobayashi M., Sakabe J., Yoshiki R., Tamamura H., Fujii N., et al. 2007. CXCL12-CXCR4 engagement is required for migration of cutaneous dendritic cells. Am. J. Pathol. 171:1249–1257. 10.2353/ajpath.2007.070225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan D.H., Li M.O., Jenison M.C., Shlomchik W.D., Flavell R.A., and Shlomchik M.J.. 2007. Autocrine/paracrine TGFβ1 is required for the development of epidermal Langerhans cells. J. Exp. Med. 204:2545–2552. 10.1084/jem.20071401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz S.I., Zhou L., Chao G., Smith C.D., Ferrara T., Wang W., Dicker D.T., and El-Deiry W.S.. 2009. Sorafenib inhibits ERK1/2 and MCL-1(L) phosphorylation levels resulting in caspase-independent cell death in malignant pleural mesothelioma. Cancer Biol. Ther. 8:2406–2416. 10.4161/cbt.8.24.10824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kel J.M., Girard-Madoux M.J., Reizis B., and Clausen B.E.. 2010. TGF-beta is required to maintain the pool of immature Langerhans cells in the epidermis. J. Immunol. 185:3248–3255. 10.4049/jimmunol.1000981 [DOI] [PubMed] [Google Scholar]
- King I.L., Kroenke M.A., and Segal B.M.. 2010. GM-CSF-dependent, CD103+ dermal dendritic cells play a critical role in Th effector cell differentiation after subcutaneous immunization. J. Exp. Med. 207:953–961. 10.1084/jem.20091844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long G.V., Stroyakovskiy D., Gogas H., Levchenko E., de Braud F., Larkin J., Garbe C., Jouary T., Hauschild A., Grob J.J., et al. 2014. Combined BRAF and MEK inhibition versus BRAF inhibition alone in melanoma. N. Engl. J. Med. 371:1877–1888. 10.1056/NEJMoa1406037 [DOI] [PubMed] [Google Scholar]
- Michaloglou C., Vredeveld L.C., Soengas M.S., Denoyelle C., Kuilman T., van der Horst C.M., Majoor D.M., Shay J.W., Mooi W.J., and Peeper D.S.. 2005. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 436:720–724. 10.1038/nature03890 [DOI] [PubMed] [Google Scholar]
- Miller J.C., Brown B.D., Shay T., Gautier E.L., Jojic V., Cohain A., Pandey G., Leboeuf M., Elpek K.G., Helft J.; Immunological Genome Consortium, et al. 2012. Deciphering the transcriptional network of the dendritic cell lineage. Nat. Immunol. 13:888–899. 10.1038/ni.2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minkov M., Steiner M., Potschger U., Arico M., Braier J., Donadieu J., Grois N., Henter J.I., Janka G., McClain K., et al. International LCH Study Group . 2008. Reactivations in multisystem Langerhans cell histiocytosis: data of the international LCH registry. J. Pediatr. 153:700–705. [DOI] [PubMed] [Google Scholar]
- Nelson D.S., Quispel W., Badalian-Very G., van Halteren A.G., van den Bos C., Bovée J.V., Tian S.Y., Van Hummelen P., Ducar M., MacConaill L.E., et al. 2014. Somatic activating ARAF mutations in Langerhans cell histiocytosis. Blood. 123:3152–3155. 10.1182/blood-2013-06-511139 [DOI] [PubMed] [Google Scholar]
- Nelson D.S., van Halteren A., Quispel W.T., van den Bos C., Bovée J.V., Patel B., Badalian-Very G., van Hummelen P., Ducar M., Lin L., et al. 2015. MAP2K1 and MAP3K1 mutations in Langerhans cell histiocytosis. Genes Chromosomes Cancer. 54:361–368. 10.1002/gcc.22247 [DOI] [PubMed] [Google Scholar]
- Olsson Åkefeldt S., Ismail M.B., Valentin H., Aricò M., Henter J.I., and Delprat C.. 2013. Targeting BCL2 family in human myeloid dendritic cells: a challenge to cure diseases with chronic inflammations associated with bone loss. Clin. Dev. Immunol. 2013:701305 10.1155/2013/701305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig-Kröger A., Relloso M., Fernández-Capetillo O., Zubiaga A., Silva A., Bernabéu C., and Corbí A.L.. 2001. Extracellular signal-regulated protein kinase signaling pathway negatively regulates the phenotypic and functional maturation of monocyte-derived human dendritic cells. Blood. 98:2175–2182. 10.1182/blood.V98.7.2175 [DOI] [PubMed] [Google Scholar]
- Randolph G.J., Angeli V., and Swartz M.A.. 2005. Dendritic-cell trafficking to lymph nodes through lymphatic vessels. Nat. Rev. Immunol. 5:617–628. 10.1038/nri1670 [DOI] [PubMed] [Google Scholar]
- Remark R., Merghoub T., Grabe N., Litjens G., Damotte D., Wolchok J.D., Merad M., and Gnjatic S.. 2016. In-depth tissue profiling using multiplexed immunohistochemical consecutive staining on single slide. Sci. Immunol. 1:aaf6925 10.1126/sciimmunol.aaf6925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A.W., Seymour J.F., Brown J.R., Wierda W.G., Kipps T.J., Khaw S.L., Carney D.A., He S.Z., Huang D.C., Xiong H., et al. 2012. Substantial susceptibility of chronic lymphocytic leukemia to BCL2 inhibition: results of a phase I study of navitoclax in patients with relapsed or refractory disease. J. Clin. Oncol. 30:488–496. 10.1200/JCO.2011.34.7898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahm F., Capper D., Preusser M., Meyer J., Stenzinger A., Lasitschka F., Berghoff A.S., Habel A., Schneider M., Kulozik A., et al. 2012. BRAFV600E mutant protein is expressed in cells of variable maturation in Langerhans cell histiocytosis. Blood. 120:e28–e34. 10.1182/blood-2012-06-429597 [DOI] [PubMed] [Google Scholar]
- Sanchez-Gaytan B.L., Fay F., Lobatto M.E., Tang J., Ouimet M., Kim Y., van der Staay S.E., van Rijs S.M., Priem B., Zhang L., et al. 2015. HDL-mimetic PLGA nanoparticle to target atherosclerosis plaque macrophages. Bioconjug. Chem. 26:443–451. 10.1021/bc500517k [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senechal B., Elain G., Jeziorski E., Grondin V., Patey-Mariaud de Serre N., Jaubert F., Beldjord K., Lellouch A., Glorion C., Zerah M., et al. 2007. Expansion of regulatory T cells in patients with Langerhans cell histiocytosis. PLoS Med. 4:e253 10.1371/journal.pmed.0040253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutte S., Quast T., Gerbitzki N., Savinko T., Novak N., Reifenberger J., Homey B., Kolanus W., Alenius H., and Förster I.. 2010. Requirement of CCL17 for CCR7- and CXCR4-dependent migration of cutaneous dendritic cells. Proc. Natl. Acad. Sci. USA. 107:8736–8741. 10.1073/pnas.0906126107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su F., Viros A., Milagre C., Trunzer K., Bollag G., Spleiss O., Reis-Filho J.S., Kong X., Koya R.C., Flaherty K.T., et al. 2012. RAS mutations in cutaneous squamous-cell carcinomas in patients treated with BRAF inhibitors. N. Engl. J. Med. 366:207–215. 10.1056/NEJMoa1105358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J., Lobatto M.E., Hassing L., van der Staay S., van Rijs S.M., Calcagno C., Braza M.S., Baxter S., Fay F., Sanchez-Gaytan B.L., et al. 2015. Inhibiting macrophage proliferation suppresses atherosclerotic plaque inflammation. Sci. Adv. 1:e1400223 10.1126/sciadv.1400223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse C., Shoemaker A.R., Adickes J., Anderson M.G., Chen J., Jin S., Johnson E.F., Marsh K.C., Mitten M.J., Nimmer P., et al. 2008. ABT-263: a potent and orally bioavailable Bcl-2 family inhibitor. Cancer Res. 68:3421–3428. 10.1158/0008-5472.CAN-07-5836 [DOI] [PubMed] [Google Scholar]
- Willman C.L., Busque L., Griffith B.B., Favara B.E., McClain K.L., Duncan M.H., and Gilliland D.G.. 1994. Langerhans’-cell histiocytosis (histiocytosis X)--a clonal proliferative disease. N. Engl. J. Med. 331:154–160. 10.1056/NEJM199407213310303 [DOI] [PubMed] [Google Scholar]
- Wong B.R., Josien R., Lee S.Y., Sauter B., Li H.L., Steinman R.M., and Choi Y.. 1997. TRANCE (tumor necrosis factor [TNF]-related activation-induced cytokine), a new TNF family member predominantly expressed in T cells, is a dendritic cell-specific survival factor. J. Exp. Med. 186:2075–2080. 10.1084/jem.186.12.2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu R.C., Chu C., Buluwela L., and Chu A.C.. 1994. Clonal proliferation of Langerhans cells in Langerhans cell histiocytosis. Lancet. 343:767–768. 10.1016/S0140-6736(94)91842-2 [DOI] [PubMed] [Google Scholar]
- Zhang H., Rodriguez S., Wang L., Wang S., Serezani H., Kapur R., Cardoso A.A., and Carlesso N.. 2016. Sepsis Induces Hematopoietic Stem Cell Exhaustion and Myelosuppression through Distinct Contributions of TRIF and MYD88. Stem Cell Reports. 6:940–956. 10.1016/j.stemcr.2016.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.