Weinstein et al. demonstrate that the transcription factors T-bet and STAT4 are necessary for Tfh cell expansion with secretion of IFN-γ and IL-21 and consequent robust germinal center output during acute viral infection.
Abstract
Follicular helper T (Tfh) cells promote germinal center (GC) B cell survival and proliferation and guide their differentiation and immunoglobulin isotype switching by delivering contact-dependent and soluble factors, including IL-21, IL-4, IL-9, and IFN-γ. IL-21 and IFN-γ are coexpressed by Tfh cells during viral infections, but transcriptional regulation of these cytokines is not completely understood. In this study, we show that the T helper type 1 cell (Th1 cell) transcriptional regulators T-bet and STAT4 are coexpressed with Bcl6 in Tfh cells after acute viral infection, with a temporal decline in T-bet in the waning response. T-bet is important for Tfh cell production of IFN-γ, but not IL-21, and for a robust GC reaction. STAT4, phosphorylated in Tfh cells upon infection, is required for expression of T-bet and Bcl6 and for IFN-γ and IL-21. These data indicate that T-bet is expressed with Bcl6 in Tfh cells and is required alongside STAT4 to coordinate Tfh cell IL-21 and IFN-γ production and for promotion of the GC response after acute viral challenge.
Introduction
T follicular helper (Tfh) cells are a functionally and phenotypically distinct subset of CD4+ T helper (Th) cells critical for humoral immunity. Tfh cells reside in B cell follicles and the germinal centers (GCs) of secondary lymphoid organs, therein secreting their canonical cytokine IL-21, which is necessary for GC B cell development and maintenance (Vogelzang et al., 2008). These cells also secrete IFN-γ and IL-4 in type 1 and 2 immune responses, respectively, which are needed for B cell maturation and the Ig isotype switching appropriate to pathogen challenge (Peng et al., 2002; Gerth et al., 2003; Mehta et al., 2003; Ozaki et al., 2004; Kuchen et al., 2007; Reinhardt et al., 2009; Linterman et al., 2010; Zotos et al., 2010; Weinstein et al., 2016), along with IL-9, which promotes B cell memory development (Wang et al., 2017). Defects in either Tfh cell development or function or in antibody production can lead to a failure of viral control (Fahey et al., 2011; Harker et al., 2011; Pallikkuth et al., 2012).
Tfh cell development is initiated in the T cell zone of secondary lymphoid organs when naive T cells are activated by antigen (Ag)-primed dendritic cells in IL-2–limited environments (Baumjohann et al., 2011; Choi et al., 2011; Li et al., 2016), with IL-6 signaling in nascent Tfh cells leading to signal transducer and activator of transcription (STAT) 3 activation and expression of the canonical Tfh cell transcription factor B cell lymphoma 6 (Bcl6; Choi et al., 2013). Dendritic cells also express inducible co-stimulator (ICOS) ligand, which signals through ICOS on developing Tfh cells to transiently inactivate FOXO1, enabling Bcl6-mediated transcriptional regulation (Nurieva et al., 2003; Stone et al., 2015; Weber et al., 2015). The latter represses the transcription factors T box–containing protein expressed in T cells (T-bet) and GATA3, inhibiting differentiation toward Th1 and Th2 pathways, respectively (Yu et al., 2009), while driving the Tfh cell differentiation program (Johnston et al., 2009; Nurieva et al., 2009; Yu et al., 2009), a program also promoted by the transcription factor Ascl2 (Liu et al., 2014).
Bcl6 and Ascl2 regulate expression of surface proteins on Tfh cells, including the chemokine receptor CXCR5, necessary for their migration into the B cell follicle (Schaerli et al., 2000); ICOS, needed for their survival, follicular migration, and support of B cell maturation (Dong et al., 2001; McAdam et al., 2001; Mak et al., 2003; Xu et al., 2013; Liu et al., 2015); and programmed death 1 (PD-1), needed for their GC regulation with the consequent promotion of B cell selection (Good-Jacobson et al., 2010).
A separate subset of CD4+ Th cells, Th1 cells, is critical for protection against challenges by intracellular pathogens (Mosmann and Coffman, 1989). Th1 cells require the expression of the transcription factor T-bet for their development (Szabo et al., 2000). T-bet is up-regulated in CD4+ Th cells upon signaling via the TCR and the IFN-γ receptor, with subsequent engagement and phosphorylation of STAT1 (Mullen et al., 2001; Afkarian et al., 2002; Zhu et al., 2012). IL-12 signaling via STAT4 further stabilizes T-bet and the Th1 cell phenotype (Mullen et al., 2001; Thieu et al., 2008; Schulz et al., 2009; Zhu et al., 2012). T-bet thereupon initiates transcription of the canonical Th1 cell cytokine Ifng and silences the expression of the Th2 cytokine Il4 (Djuretic et al., 2007). Subsequent IFN-γ signaling cements Th1 differentiation via increased STAT1-mediated gene transcription, which, in concert with IL-12–driven STAT4 signaling, perpetuates Tbx21 (gene encoding T-bet) and Ifng expression (Lighvani et al., 2001; Thieu et al., 2008; Wei et al., 2010; Zhu et al., 2012).
Although Tfh and Th1 cells are phenotypically and functionally distinct, they share a transitional developmental stage after T cell activation. In addition to promoting Tbx21 and Ifng expression in Th1 cells, STAT4 drives the expression of Bcl6 and the canonical Tfh cell cytokine Il21 in both mouse and human Tfh cells in vitro (Schmitt et al., 2009; Nakayamada et al., 2011) and binds to and epigenetically regulates Il21 in polarized Th1 cells (Wei et al., 2010). Yet, continued IL-12–driven STAT4 signaling in vitro extinguishes Bcl6 and Il21 expression with the development of repressive chromatin marks, cementing T-bet–driven Th1 cell differentiation at the expense of Tfh cells (Nakayamada et al., 2011). Conversely, if transitional T cells receive signaling via ICOS and IL-6 or TGF-β (Eto et al., 2011; Schmitt et al., 2014; Marshall et al., 2015), Tfh cell differentiation is promoted over that of Th1 cells. The developmental link between Th1 and Tfh cells is not entirely severed with their maturation, however, because the two differentiated subsets do not fully repress the Bcl6 and Tbx21 loci, respectively (Lu et al., 2011; Nakayamada et al., 2011). The two populations also share functional features, as both produce the canonical Th1 cell cytokine IFN-γ and the canonical Tfh cell cytokine IL-21 upon immunization with a nominal Ag (Lüthje et al., 2012; Ray et al., 2014) or after viral infection (Ray et al., 2014; Miyauchi et al., 2016). Moreover, Tfh cells transiently express T-bet and IFN-γ upon Ag immunization or in experimental malaria infection (Lüthje et al., 2012; Carpio et al., 2015).
Yet, the roles of STAT4 and T-bet in promotion of a functional Tfh cell–driven GC response upon type 1 pathogen challenge remain unclear, as does the transcriptional regulation of IFN-γ in Tfh cells. We hypothesized that both of these transcription factors are required for the development of functional Tfh cells after viral challenge and for the secretion of IL-21 and IFN-γ with the consequent humoral immune response. We found that after acute infection with lymphocytic choriomeningitis virus (LCMV), T-bet was coexpressed with Bcl6 in Tfh cells, with the former transcription factor necessary for their expansion and secretion of IFN-γ and with a correspondingly robust GC output. T-bet expression in Tfh cells was STAT4 dependent, as it is in Th1 cells. Although production of IL-21 was independent of T-bet expression, it was dependent on STAT4 signaling. Thus, Tfh cells, like their Th1 counterparts, cosecrete IL-21 and IFN-γ throughout the course of an acute pathogen-induced type 1 immune response, with each cytokine transcriptionally regulated in a distinct manner to properly promote the humoral immune response.
Results
Tfh cells upon acute LCMV infection cosecrete IFN-γ and IL-21
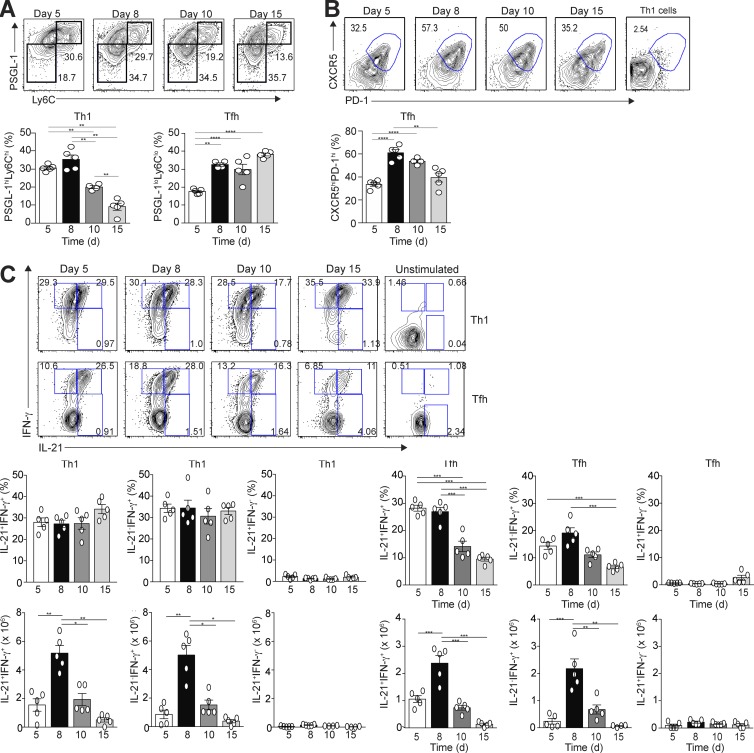
Upon type 1 immune challenge, Tfh cells cosecrete IFN-γ and IL-21; however, the kinetics and mechanism of cytokine cosecretion are not completely understood. To explore these in a system that can be genetically manipulated, we adoptively transferred naive CD4+Thy1.1+ TCR transgenic T cells specific for the LCMV peptide GP66-77 from B6.SMARTA transgenic (Stg) mice into congenically marked Thy1.2+ B6 mice, followed by an LCMV Armstrong challenge and a subsequent assessment of the viral-specific splenic T cell response at days 5, 8, 10, and 15 postinfection (p.i.). Viral-specific Th1 cells were gated as CD4+Thy1.1+CD44hiLy6chiPSGL-1hi, and Tfh cells were gated as CD4+Thy1.1+CD44hiLy6cloPSGL-1lo (Poholek et al., 2010; Marshall et al., 2011). Down-regulation of PSGL-1 is an early event in Tfh cell differentiation that follows their initial activation by dendritic cells before subsequent B cell contact and, hence, is a marker for these cells (Odegard et al., 2008; Poholek et al., 2010; Hale et al., 2013; Weinstein et al., 2014). Th1 cells expanded by day 5 p.i., with the percentage declining at day 8, whereas Tfh cells continued to expand at this time point, remaining stable until day 15 (Fig. 1 A). The Tfh cell population, gated as CD4+Thy1.1+CD44hiLy6cloPSGL-1loCXCR5hiPD-1hi (Crotty, 2011; Lee et al., 2011), also peaked at days 8 and 10 before declining at day 15 (p.i.; Fig. 1 B). For the remainder of our studies, we focused solely on the characterization of this population, gated as CD4+Thy1.1+CD44hiLy6cloPSGL-1loCXCR5hiPD-1hi (designated as Tfh cells in the text), in comparison with Th1 cells, gated as CD4+Thy1.1+CD44hiLy6chiPSGL-1hi.
Figure 1.
Temporal production of IFN-γ and IL-21 by Tfh and Th1 cells after acute LCMV infection. Thy1.1+ SMARTA TCR transgenic (Stg) CD4+ T cells were transferred into Thy1.2+ B6 mice, followed by LCMV Armstrong infection 24 h later. Spleens were harvested at days 5, 8, 10, and 15 p.i. Representative flow cytometry plots of splenic Th1 and Tfh cells, with bar graph summaries, are shown. (A) PSGL-1hiLy6Chi Th1 cells or PSGL-1loLy6Clo Tfh cells with percentages of Th1 and Tfh cells. (B) CXCR5 and PD-1 gated PSGL-1loLy6Clo Tfh cells with cell percentages; gates based on CXCR5 and PD-1 staining of Th1 cells. (C) Representative flow cytometry plots of intracellular IL-21 and IFN-γ staining of Th1 and Tfh cells with percentages and numbers of cytokine-positive cells. Data are representative of three experiments with three to five recipients per group. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by Student’s t test. Error bars represent SEM.
Using an unstimulated control as a baseline, we observed three populations of cytokine-secreting Th1 and Tfh cells during infection (IL-21+IFN-γ−, IL-21+IFN-γ+, and IL-21−IFN-γ+; Fig. 1 C). A majority of the cytokine-producing Th1 cells and Tfh cells coproduced IL-21 and IFN-γ or IFN-γ alone, with only a small percentage of single IL-21 producers. These observations were not specific to TCR transgenic T cells, as the endogenous Thy1.2+ Tfh cell population displayed similar kinetics to IL-21 and IFN-γ production (Fig. S1 A). Although the percentage of cytokine double-positive Th1 cells was maintained during the course of infection, their numbers declined alongside a more drastic decrease in the number and percentage of cytokine double-positive Tfh cells (Fig. 1 C). Thus, a majority of the cytokine-expressing Th1 and Tfh cells coproduced IL-21 and IFN-γ during acute LCMV infection, with a decline in numbers of both populations during the later phase of the GC response.
Temporal expression of Bcl6 and T-bet in Th1 and Tfh cells
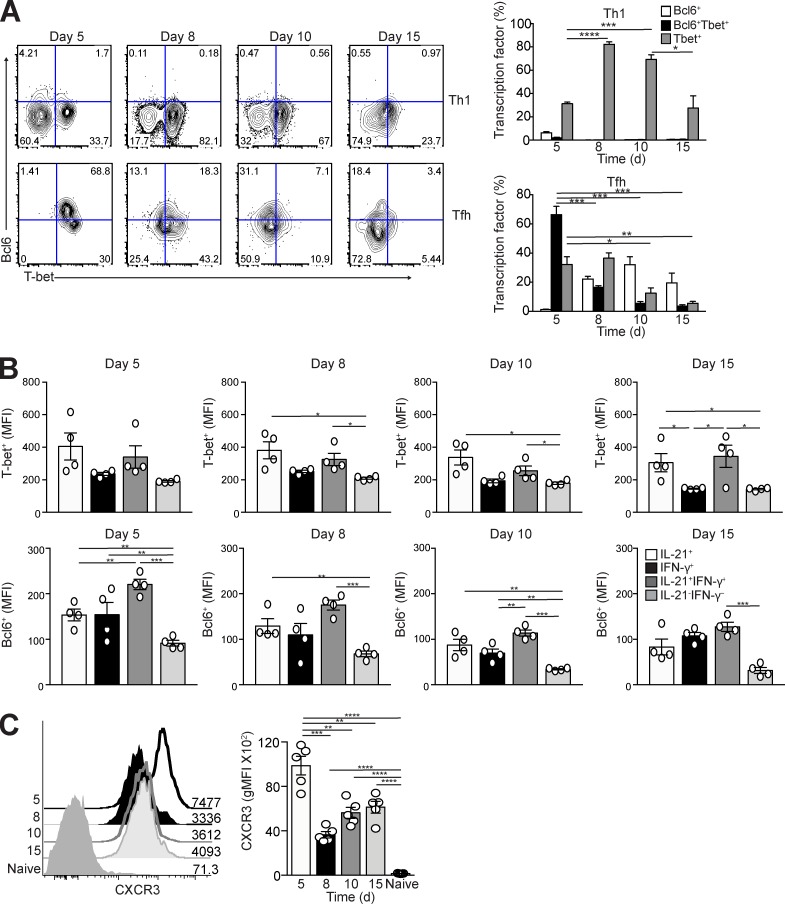
IFN-γ synthesis in Th1 cells is driven by the expression of T-bet (Djuretic et al., 2007), with T-bet and Bcl6 being coexpressed during the initial differentiation of Th1 cells polarized in vitro (Nakayamada et al., 2011). To test whether T-bet and Bcl6 are also coexpressed during Th1 and Tfh cell differentiation in vivo, we again transferred congenically marked naive Stg CD4+ T cells into B6 mice and infected them with LCMV Armstrong, with the expression of the two transcription factors determined in splenic CD4+Thy1.1+CD44hiLy6chiPSGL-1hi Th1 cells and CD4+Thy1.1+CD44hiLy6cloPSGL-1loCXCR5hiPD-1hi Tfh cells at days 5, 8, 10, and 15 p.i. A significant portion of Th1 cells expressed T-bet on day 5 p.i., with a few cells expressing Bcl6 (Fig. 2 A, top). In contrast, the majority of Tfh cells at day 5 p.i. expressed both Bcl6 and T-bet, with a minority expressing T-bet alone (Fig. 2 A, bottom). T-bet expression peaked at day 8 p.i. in Th1 cells, gradually declining by day 15. T-bet expression also decreased in Tfh cells by day 10 p.i, with the remaining transcription factor–positive Tfh cells expressing Bcl6. These observations were not specific to TCR transgenic T cells, as the endogenous Tfh cell population had similar kinetics to transcription factor expression (Fig. S1 B).
Figure 2.
Expression of T-bet and Bcl6 in Tfh and Th1 cells after acute LCMV infection. Thy1.1+ Stg CD4+ T cells were transferred into Thy1.2+ B6 mice followed by LCMV Armstrong infection 24 h later. Spleens were harvested at days 5, 8, 10, and 15 p.i. (A) Representative flow cytometry plots of intracellular Bcl6 and T-bet in splenic Ly6ChiPSGL-1hiCXCR5lo Th1 and Ly6cloPSGL-1loCXCR5hiPD-1hi Tfh cells, with bar graphs that summarize percentages of transcription factor–expressing cells at each time point p.i. (B) Bar graphs that summarize the quantified geometric mean fluorescence intensity (gMFI) of staining of T-bet or Bcl6 from the IL-21– and/or IFN-γ–secreting Tfh cells (as identified in Fig.1 C). (C) CXCR3 expression on naive and Tfh cells, with gMFI. Data are representative of two or three experiments with three to five recipients per group. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by Student’s t test. Error bars represent SEM.
We next assessed T-bet and Bcl6 expression in IL-21– and IFN-γ–secreting Tfh cells. IL-21+IFN-γ+ Tfh cells maintained the greatest T-bet and Bcl6 expression compared with IL-21+IFN-γ− and IL-21−IFN-γ+ populations, despite their temporal decline throughout infection (Fig. 2 B). In contrast, non–cytokine-producing Tfh cells had the least expression of T-bet and Bcl6, indicating that transcription factor expression is associated with cytokine secretion.
To validate the kinetics of T-bet expression in Tfh cells, we assessed staining of the chemokine receptor CXCR3, which is dependent on T-bet transcription (Lord et al., 2005). As for T-bet, CXCR3 was highly expressed at day 5 in Tfh cells, with a reduction by day 8 (Fig. 2 C). These data are consistent with previous in vitro studies demonstrating that polarized Th1 and Tfh cells share a T-bet and Bcl6 intermediate (Nakayamada et al., 2011) while extending these findings to show that in vivo, mature Tfh cells, but not Th1 cells, robustly express both transcription factors early after viral challenge.
T-bet is required for Tfh cell development
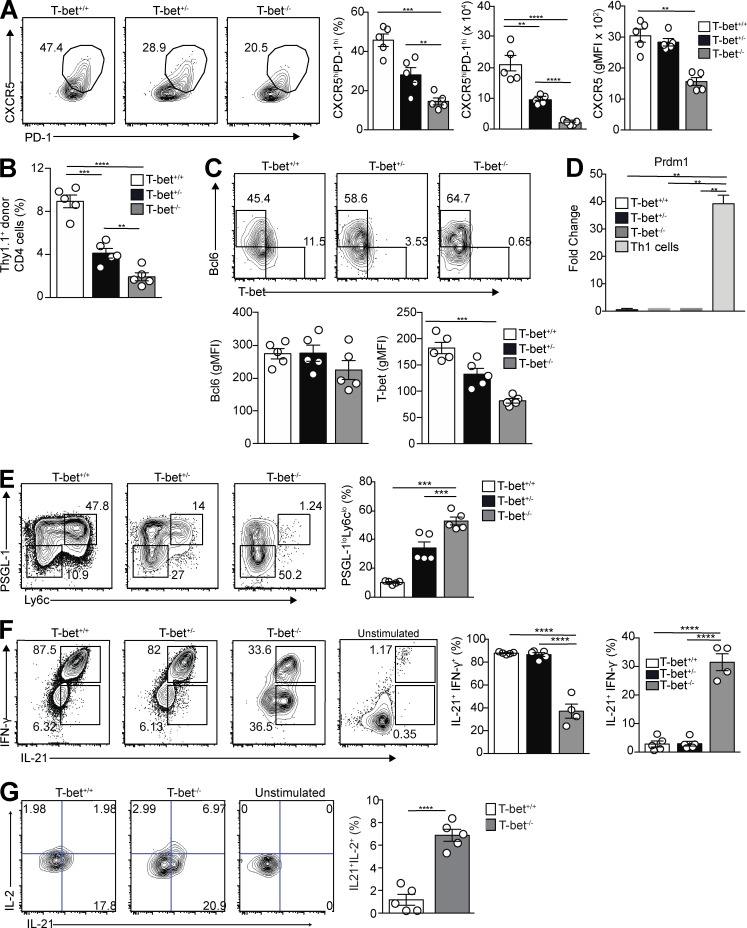
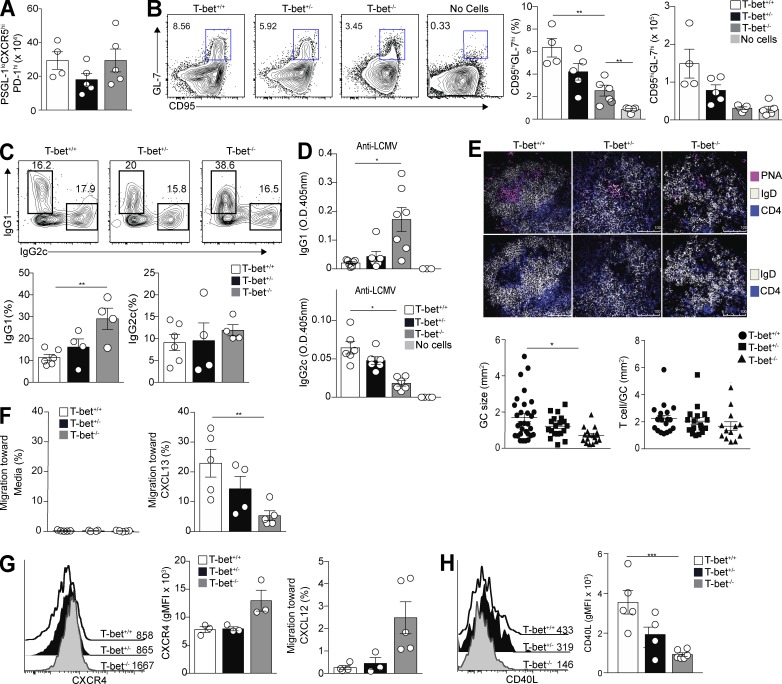
To determine the role of T-bet in regulation of Tfh cell differentiation, congenically marked T-bet–sufficient (T-bet+/+), T-bet–heterozygous (T-bet+/−), and T-bet–deficient (T-bet−/−) Stg CD4+ T cells were transferred into B6 mice before infection with LCMV Armstrong. Splenic Tfh cell development among the donor cells was assessed at day 8 p.i., a time point when T-bet is still expressed in Tfh cells (Fig. 2 A). T-bet−/− cells in comparison with the other two transferred populations had a significant reduction in the percentage and number of Tfh cells (Fig. 3 A), with the percentage of donor Stg cells that differentiated into Tfh cells significantly reduced in a T-bet dose–dependent manner (Fig. 3 B). The reduction in Tfh cells in the absence of T-bet was not a consequence of aberrant Bcl6 expression, which was stable in the T-bet mutant cells compared with T-bet–intact ones (Fig. 3 C), nor was it caused by alterations in Blimp-1, as we observed minimal Prdm1 (the gene encoding Blimp-1) expression in sorted Tfh cells from T-bet+/+, T-bet+/−, and T-bet−/− donors compared with control Th1 cells (Fig. 3 D). In comparison, inhibition of T-bet expression had no effect on Tfh cell development upon challenge with a type 2 immunogen (Fig. S2, A and B). We did observe that after LCMV challenge, a greater percentage of the double-mutant cells, compared with their intact counterparts, down-regulated PSGL-1 (Fig. 3 E), an early event in Tfh cell differentiation (Odegard et al., 2008; Poholek et al., 2010; Weinstein et al., 2014). These findings in aggregate suggest that although the absence of T-bet enhanced initial T cell differentiation along an initial Tfh cell developmental pathway, its presence is required for proper Tfh cell differentiation after type 1 immune challenge as modeled by acute viral infection.
Figure 3.
T-bet is necessary for T cell development and their robust IFN-γ expression after LCMV challenge. T-bet+/+, T-bet+/−, or T-bet−/− Thy1.1+ Stg CD4+ T cells were transferred to Thy1.2+ B6 mice, followed by LCMV Armstrong infection 24 h later. Spleens were harvested 8 d p.i. (A) Representative flow cytometry plots of CXCR5 and PD-1 gating on PSGL-1loLy6Clo Tfh cells with bar graphs that summarize cell percentages, cell numbers, and CXCR5 MFI. (B) Percentage of donor cells that differentiated into Tfh cells in transfer recipients. (C) Representative intracellular Bcl6 and T-bet expression in PSGL-1loCXCR5hiPD-1hi Tfh cells with gMFI for both. (D) Quantitative RT-PCR for Prdm1 expression from sorted Tfh cells and control T-bet+/+ Th1 cells. (E) Representative flow cytometry plots of T-bet+/+, T-bet+/−, and T-bet−/− CD4+Thy1.1+PSGL-1loLy6Clo splenic Tfh cells from recipients, with a bar graph that summarizes percentages of cells. (F) Representative flow cytometry plots of intracellular IL-21 and IFN-γ staining of T-bet+/+, T-bet+/−, and T-bet−/− cells from recipient spleens, with bar graphs that summarize percentages of cells. (G) Representative flow cytometry plots of intracellular IL-21 and IL-2 staining of T-bet+/+ and T-bet−/− cells from recipient spleens, with a bar graph that summarizes percentages of cells. Data are representative of three experiments with three to five recipients per group. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by Student’s t test. Error bars represent SEM.
We next asked whether T-bet expression in Tfh cells is required for their IFN-γ production, as in Th1 cells (Szabo et al., 2000). Intracellular cytokine staining was performed on donor T-bet+/+, T-bet+/−, or T-bet−/− Stg CD4+ splenic Tfh cells at 8 d p.i. T-bet−/− Tfh cells had a significant reduction in the percentage of double-positive IL-21+IFN-γ+ cells compared with that of T-bet+/+ and T-bet+/− Tfh cells, a finding in contrast to the significant increase of single IL-21+ producers (Fig. 3 F). Because Th2 cell development is increased in the absence of T-bet (Usui et al., 2006), we also assessed IL-4 production in donor T-bet+/+, T-bet+/−, or T-bet−/− that became Tfh cells after transfer and viral infection; however, we observed no differences in production of this cytokine (Fig. S2 C). These data indicate that T-bet promotes expansion and IFN-γ production by Tfh cells after viral infection, as in Th1 cells, but that it does not regulate their IL-21 or IL-4 production.
Because T-bet is a negative regulator of IL-2 production, which in turn negatively regulates Bcl-6 and promotes Blimp-1 expression (Oestreich et al., 2012), we also assessed IL-2 synthesis by Tfh cells, finding enhanced secretion alongside that of IL-21 in the mutant cells (Fig. 3 G). Despite increased IL-2 expression in mutant cells compared with T-bet–intact cells, this did not alter their expression of Bcl6 or Prdm1 (Fig. 3, C and D). Thus, while promoting IFN-γ production in Tfh cells during acute viral infection, T-bet inhibits that of IL-2, helping to cement the Tfh cell phenotype (Nurieva et al., 2012; Locci et al., 2016).
T-bet–intact and T-bet–deficient Tfh cells are transcriptionally distinct
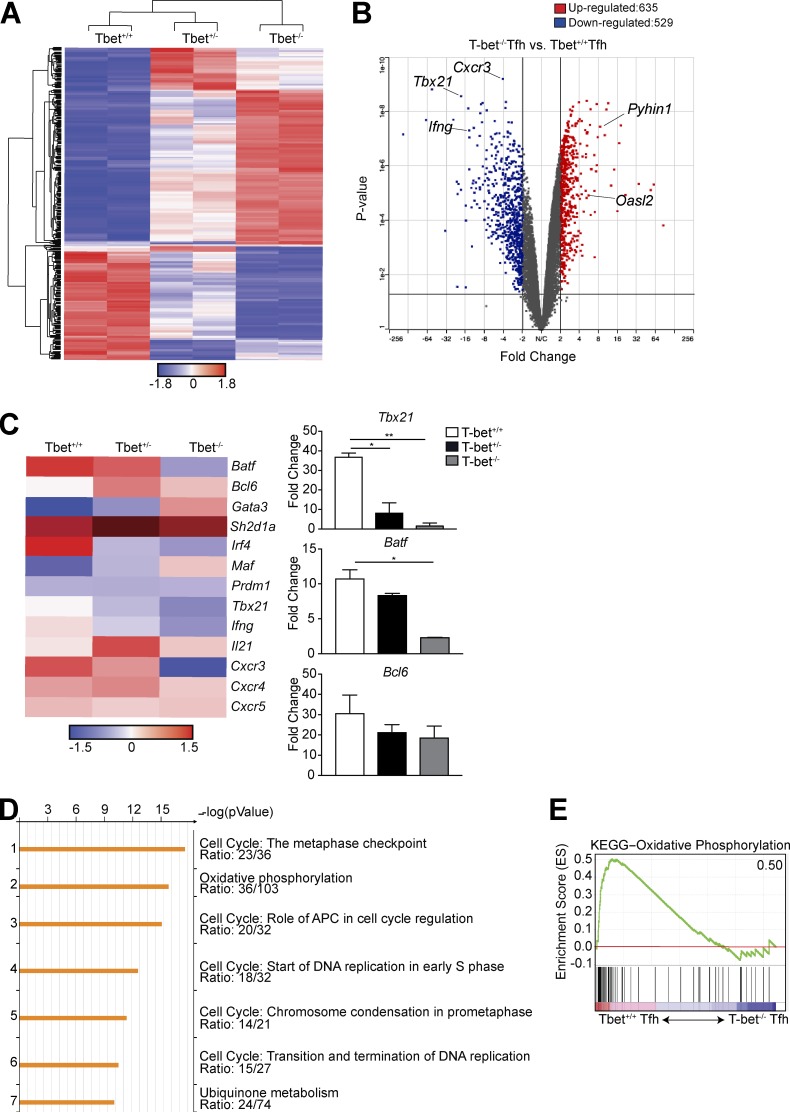
We next performed a transcriptome analysis of splenic Tfh cells after adoptive transfer of T-bet+/+, T-bet+/−, or T-bet−/− Stg Thy1.1+CD4+ T cells 8 d p.i. with LCMV Armstrong. We found 837 genes differentially expressed between T-bet+/− and T-bet+/+ Tfh cells, with 1,164 genes differentially expressed between the T-bet+/+ and T-bet−/− populations (Fig. 4 A). In the absence of T-bet, 635 genes were up-regulated, including type I IFN–stimulated genes known to be repressed by T-bet in Th1 cells (including Pyhin1 and Oasl2; Iwata et al., 2017), with 529 genes down-regulated, including those emblematic of Th1 cell differentiation, including Ifng and Cxcr3 (Fig. 4 B), as well as Tbx21. To further analyze the transcriptional differences among the Tfh cell populations, we examined a curated set of genes previously described to be up- or down-regulated in Tfh cells compared with other CD4+ Th subsets (Fig. 4 C; Bauquet et al., 2009; Eddahri et al., 2009; Ise et al., 2011). Although the three Tfh cell populations shared expression of Tfh cell–defining genes including Bcl6, Cxcr5, and Sh2d1a (Fig. 4 C), as well as reduction in that of Prdm1 (Fig. 3 E), the T-bet−/− population had reduced expression of the Tfh cell–associated genes Irf4 and Batf. Pathway enrichment analysis of the down-regulated genes in T-bet−/− compared with T-bet+/+ Tfh cells revealed down-regulation of multiple cell cycle pathways and oxidative phosphorylation (Fig. 4 D), the latter supported by gene set enrichment analysis (Fig. 4 E). Thus, T-bet has a role in regulating the cytokine, cell division, and metabolic processes of Tfh cells in a manner similar to that in Th1 cells (Iwata et al., 2017).
Figure 4.
T-bet is required for proper transcriptional development of Tfh cells. T-bet+/+, T-bet+/−, or T-bet−/− Thy1.1+ Stg CD4+ T cells were transferred into Thy1.2+ B6 mice with LCMV Armstrong infection 24 h later. Spleens were harvested 8 d p.i. (A) Heat map of significantly differentially expressed genes (rows) in the three populations (columns). FDR, α < 0.05. (B) Volcano plot of gene expression comparing T-bet+/+ with T-bet−/− Ly6cloPSGL-1loCXCR5hiPD-1hi Tfh cells to identify differentially expressed genes with a cutoff p-value of <0.05, fold change >2, comparing T-bet+/+ or T-bet−/− cells. (C) Heat map of selected T cell–related genes. FDR-adjusted q < 0.05 comparing T-bet+/+ or T-bet−/− Tfh cells. Quantitative RT-PCR analysis of selected T cell–related genes, normalized to results obtained for the control gene Hprt. Data from two independent sorts using 10 mice each; fragments per kilobase of transcript per million mapped reads (FPKM). (D) Enrichment pathways analysis of the down-regulated genes in T-bet−/− Tfh cells, with the top seven pathways listed, including the enrichment p-value and the number of genes per total genes in a pathway. (E) Enrichment of gene signatures related to activation of the Kyoto Encyclopedia of Genes and Genomes oxidatve phosphorylation pathway in T-bet+/+ versus T-bet−/− Tfh cells. Number in top-right corner is the enrichment score; all p-values and FDRs = 0. *, P < 0.05; **, P < 0.01 by Student’s t test. Error bars represent SEM.
T-bet is required for Tfh cell function
To evaluate the requirement for T-bet in the Tfh cell function, we transferred naive T-bet+/+, T-bet+/−, or T-bet−/− Stg CD4+ T cells into TCR-β−/− mice that lacked T cells; thus, only transferred cells were capable of Tfh cell differentiation and the promotion of a GC response. At day 13 after LCMV infection (the height of the GC response), the reduction in T-bet expression in transferred T cells corresponded with a decrease in the percentage that developed a Tfh phenotype (Fig. S3 A). To account for the Ag-induced expansion defect in T-bet−/− T cells upon adoptive transfer (Marshall et al., 2011), we transferred four times as many T-bet−/− as T-bet+/− and T-bet+/+ naive CD4+ T cells into recipients and found equivalent numbers of Tfh cells among the recipients of the three transferred populations (Fig. 5 A). Yet, similar to results from transfers into T cell–intact recipients (Fig. 3 F), T-bet−/− Tfh cells arising from cells transferred into TCR-β−/− mice had defects in IFN-γ production, with reduced numbers of IL-21+IFN-γ+ double-positive cells compared with T-bet+/+ and T-bet+/− donors (Fig. S3 B). In contrast, numbers of IL-21 single-positive cells were equivalent. Despite Tfh cell production of IL-21, and despite equivalent numbers of Tfh cells after transfer, recipients of T-bet−/− cells had a significant reduction of B220+IgDloCD95hiGL-7hi GC B cells compared with those of T-bet+/+ and T-bet+/− recipients (Fig. 5 B). Ablation of T-bet in Tfh cells also influenced Ig isotype usage, with GC B cells in recipients of T-bet–deficient transfers having increased intracellular IgG1 expression compared with animals receiving T-bet–sufficient CD4+ T cells (Fig. 5 C), consistent with the increase in IgG1 production by GC B cells in IFN-γ−/− mice infected with LCMV (Fig. S3 C). In contrast to the increased IgG1 in recipients of T-bet mutant T cells, intracellular B cell and serum IgG2c was decreased (Fig. 5, C and D), providing additional evidence that T-bet is required by Tfh cells to ensure the proper maturation of GC B cells appropriate to viral challenge.
Figure 5.
T-bet in Tfh cells is required for proper GC B cell output after viral infection. T-bet+/+, T-bet+/−, or T-bet−/− Thy1.1+ Stg CD4+ T cells were transferred to TCR-β−/− mice. Spleens were harvested 13 d p.i. (A) Number of T-bet+/+, T-bet+/−, or T-bet−/− donor cells that differentiated into Tfh cells in spleens of recipients. (B) Representative flow cytometry plots of CD4−B220+IgDloCD95hiGL-7hi GC B cells in recipients of T-bet+/+, T-bet+/−, or T-bet−/− donor cells with cell percentages and numbers. (C) Representative flow cytometry plots of intracellular IgG1 and IgG2c staining of GC B cells with percentages of isotype-staining cells. (D) Anti-LCMV IgG2c and IgG1 antibodies in sera of recipient mice. (E) Splenic B cell follicles of TCR-β−/− recipient mice stained with anti-IgD, anti-CD4, and peanut agglutinin (PNA), with the numbers of T cells per GC size (bottom left) and GC sizes (bottom right) quantified. (F and G) Total splenocytes from mice 12 d p.i., co-cultured with CXCL13, CXCL12, or media to assess percentages of Tfh cells migrating to each (F and G) or stained for CXCR4 (G). (H) Staining of Tfh cells in recipient spleens for CD40L. Data are representative of three experiments with three to five recipients per group. *, P < 0.05; **, P < 0.01; ***, P < 0.001 by Student’s t test. Error bars represent SEM.
Even though we observed equivalent numbers of Tfh cells after infection, as assessed by surface phenotype upon transfer (Fig. 5 A), we wondered whether the reduction in GC output among recipients of the double-mutant population was a consequence of defective GC migration, especially given their reduced expression of CXCR5 (Fig. 3 A and Fig. S3 A). The number of Tfh cells per GC area was similar to that of recipients of all three populations; however, TCR-β−/− mice infected after receipt of T-bet−/− cells had smaller GCs 13 d p.i. (Fig. 5 E). Consistent with their reduced CXCR5 expression, T-bet−/− cells migrated less efficiently in vitro to the GC light zone ligand CXCL13 than their T-bet+/+ and T-bet+/− counterparts (Fig. 5 F). In contrast, cell surface expression of the chemokine receptor CXCR4 was increased in T-bet−/− Tfh cells compared with T-bet+/+ and T-bet+/− cells, consistent with their increased ability to migrate to the dark zone ligand CXCL12 (Fig. 5 G; Weinstein et al., 2016). In addition, surface expression of CD40L (Koguchi et al., 2007), which via CD40 signaling promotes isotype switching and regulates GC B cell differentiation, was significantly reduced on T-bet−/− Tfh cells compared with T-bet+/+ cells (Fig. 5 H). Thus, even though numbers of T-bet−/− Tfh cells were not substantially different than those of transcription factor–sufficient cells after transfer, their decreased expression of CXCR5 and migration to CXCL13, despite increased expression of CXCR4 and migration to CXCL12, suggested the T-bet−/− cells had defective follicular migration. This, along with a reduction in their expression of IFN-γ and that of CD40L, hindered their ability to promote GC expansion and the isotype switching appropriate to the nature of pathogen challenge. Together, these data indicate that T-bet is required in a dosage-dependent fashion for Tfh expansion, migration, and function, including maximal secretion of IFN-γ and proper GC output after type 1 immune challenge.
STAT4 is necessary for Tfh cell development and cytokine production after acute LCMV infection
Because Ag-specific T-bet–deficient Tfh cells produced IL-21 equivalent to that of T-bet–intact cells, we next examined the transcriptional regulation of Il21 by STAT4, given that this factor binds to the Il21 locus in Th1 cells and its deletion results in ablation of activating histone marks (H3K4me3) within that locus (Wei et al., 2010). STAT4 expression is also shared with that of T-bet in in vitro–polarized Th1 cells that have a Tfh cell intermediate (Nakayamada et al., 2011). We accordingly measured STAT4 phosphorylation (pSTAT4) ex vivo after IL-12 or IFN-β stimulation, both of which are present in viral infections and promote pSTAT4 expression in Th1 cells (Cho et al., 1996; Morinobu et al., 2002). Naive Stg CD4+ T cells were transferred into B6 mice, and splenocytes from days 5, 10, and 15 p.i. were cultured with IL-12 or IFN-β. In response to IL-12 stimulation, Tfh cells taken from recipient mice had significantly increased phosphorylated STAT4 at day 5 compared with those from days 10 and 15 p.i. (Fig. S4 A). pSTAT1 was also increased at day 5 p.i. in response to IL-12 stimulation, consistent with previous studies (Choi et al., 2013), affirming a role for STAT1 phosphorylation in Tfh cell development (Fig. S4 A; Nakayamada et al., 2014). In response to IFN-β stimulation, pSTAT4 was also increased in Tfh cells p.i., with pSTAT1 similarly up-regulated (Fig. S4 B). In contrast, Th1 cells had increased stimulated pSTAT4 to IL-12 and IFN-β at later time points p.i. (Fig. S4 C). In response to IFN-β stimulation, pSTAT1-positive Tfh cells were also pSTAT4 positive (Fig. S4 D), suggesting that signaling by these two STATs is involved in their development. Thus, IL-12 and IFN-β activate STAT4 and STAT1 in Tfh cells early after viral infection, corresponding to the kinetics of their T-bet expression (Fig. 2 A).
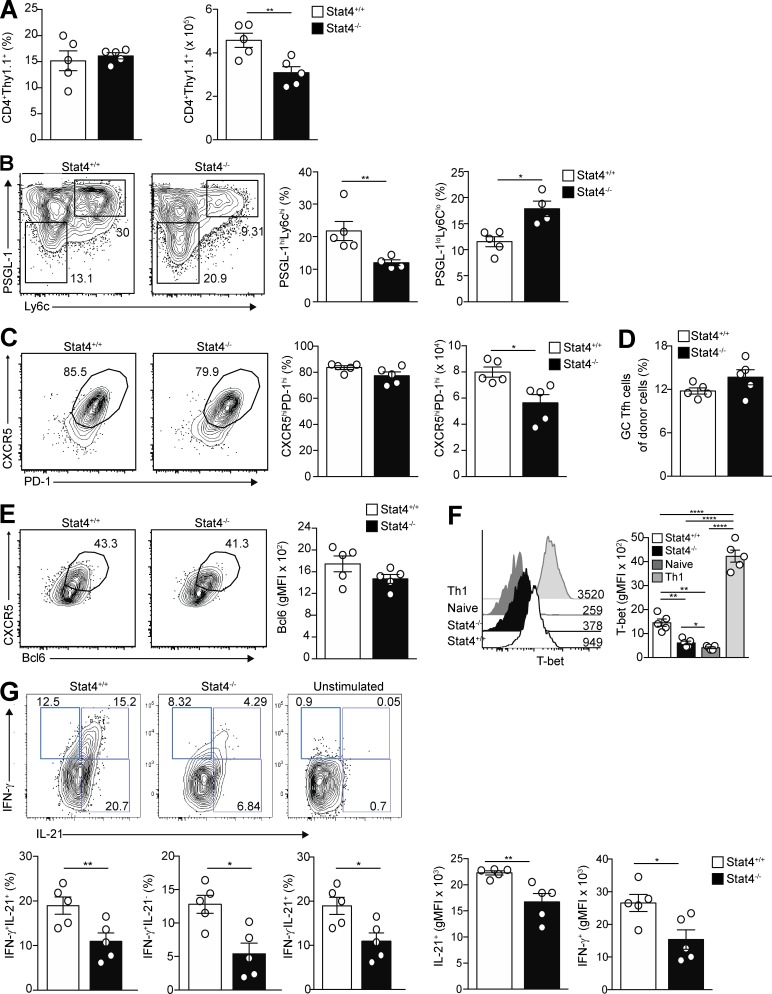
To explore whether pSTAT4 is necessary for Tfh cell differentiation, we transferred congenically marked STAT4+/+ or STAT4−/− Stg CD4+ T cells into B6 mice and infected them with LCMV. Because pSTAT4 was increased at day 5 p.i in Tfh cells (Fig. S4, A and B), we examined their differentiation in the recipient mice at this time point, finding that donor STAT4+/+ and STAT4−/− cells comprised similar percentages of CD4+ T cells in recipients, although STAT4−/− cells had reduced expansion (Fig. 6 A), similar to T-bet−/− donors. Donor T cells were stimulated with IL-12 to reconfirm the presence or absence of pSTAT4 at the time of sacrifice (Fig. S5 A). Th1 cells were decreased in the STAT4−/− donor population compared with those from the STAT4+/+ cells (Fig. 6 B), consistent with the fact that STAT4 is required for Th1 development (Thieu et al., 2008). We observed a corresponding increase in the CD4+Thy1.1+CD44hiLy6cloPSGL-1lo Tfh cell population, similar to that seen with T-bet−/− donors (Fig. 6 B and Fig. 3 E). However, unlike T-bet−/− donor cells, the STAT4−/− Ly6cloPSGL-1lo cells did not have a reduced CXCR5 expression (Fig. 6 C), although the number of Tfh cells was reduced compared with the STAT4+/+ donors, presumably a consequence of the expansion defect of the mutants (Fig. 6 A). Nonetheless, a similar percentage of naive STAT4-deficient and -sufficient donor cells became Tfh cells (Fig. 6 D). Moreover, the percentage of Bcl6hi Tfh cells and degree of Bcl6 expression among STAT4-deficient donor cells were comparable with those from STAT4-sufficient cells (Fig. 6 E). STAT4−/− donor cells also produced similar percentages of Tfh cells with equivalent Bcl6 expression compared with STAT4+/+ Tfh cells at a later, day 8 p.i. time point (Fig. S5 C).
Figure 6.
Tfh cells require STAT4 for cytokine production but not formation. STAT4+/+ or STAT4−/− Thy1.1+ Stg CD4+ T cells were transferred into Thy1.2+ B6 mice followed by infection with LCMV Armstrong 24 h later, and spleens were harvested at day 5 p.i. (A) Summary of the percentages and numbers of donor CD4+ T cells in recipient mice. (B) Representative flow cytometry plots of PSGL-1hiLy6Chi Th1 or PSGL-1loLy6Clo Tfh cells from STAT4+/+ or STAT4−/− donors with percentages of cells summarized. (C) Representative flow cytometry plots of CXCR5 and PD-1 gating of PSGL-1loLy6Clo Tfh cells from STAT4+/+ and STAT4−/− donors with cell percentages and numbers. (D) Percentage of STAT4+/+ and STAT4−/− donor CD4+ T cells that differentiated into Tfh cells. (E) Intracellular and surface staining for Bcl6 and CXCR5, respectively, in CD4+Thy1.1+Ly6cloPSGL-1lo STAT4+/+ and STAT4−/− Tfh cells in recipients of transferred cells with gMFI of Bcl6 staining. (F) Intracellular T-bet staining in STAT4+/+ and STAT4−/− Thy1.1+ PSGL-1loCXCR5hiPD-1hi Tfh and in Th1 and naive CD4+ T cells with their gMFI. (G) Representative flow cytometry plots of intracellular IL-21 and IFN-γ expression in STAT4+/+ or STAT4−/− Tfh cells of recipient mice with summary of cell percentages and MFI of staining of Tfh cells. Data are representative of three experiments with five recipients per group. *, P < 0.05; **, P < 0.01; ****, P < 0.0001 by Student’s t test. Error bars represent SEM.
We next assessed whether STAT4 regulates T-bet expression in Tfh cells, as it does in Th1 cells (Thieu et al., 2008). We found a significant reduction in T-bet expression in the absence of STAT4 in Tfh cells at day 5 p.i. (Fig. 6 F). This reduction did not affect Bcl6 expression, consistent with our finding that T-bet–deficient Tfh cells also had no such alterations (Fig. 3 C). T-bet expression in STAT4−/− Tfh cells was not completely ablated and was higher than in naive CD4+ T cells, indicating that additional pathways such as STAT1 can separately promote expression of this transcription factor (Afkarian et al., 2002). Hence, similar to Th1 cells, signaling through STAT4 promotes increased T-bet expression in Tfh cells.
We also asked whether STAT4 is required for Tfh cell production of IL-21, finding that mutant Tfh cells 5 d p.i., compared with STAT4+/+ cells, had a reduction in the percentage of IFN-γ+ and IL-21+IFN-γ+ cells, but not IL-21+IFN-γ− cells (Fig. 6 G). Moreover, total IFN-γ and IL-21 expression was decreased in STAT4−/− Tfh cells compared with STAT4+/+ cells. Thus, Tfh cells require STAT4 to promote IL-21 and T-bet expression, with the latter driving their IFN-γ production.
We determined whether the reduction in protein expression of transcription factors and cytokines in the absence of STAT4 occurred at the chromatin or transcriptional level. Because STAT4 has been shown to be necessary for IL-12–induced chromatin remodeling of the CD25 locus (Shin et al., 2005), we asked whether it alters the genomic landscape in Tfh cells. We characterized chromatin accessibility via assay for transposase-accessible chromatin using sequencing (ATAC-seq) in STAT4+/+ and STAT4−/− Tfh cells at 5 d p.i. Principal component analysis and clustering analyses performed on called peaks revealed that STAT4+/+ and STAT4−/− Tfh cells did not form distinct groups (Fig. S5 F). Comparison of differentially called peaks in the two replicates of STAT4+/+ versus STAT4−/− cells identified by the DiffBind package with DEseq normalization revealed only 12 differentially expressed peaks from the 35,330 merged regions (false discovery rate [FDR] <0.01). No significant differences were seen in chromatin accessibility at the Bcl6, Il21, Tbx21, and Ifng gene loci (Fig. S5 G). A parallel analysis performed to detect differential ATAC peaks using the csaw package yielded similar results (Lun and Smyth, 2016). These findings suggest that the regulation of differential gene expression in STAT4+/+ and STAT4−/− Tfh cells is not mediated at the level of chromatin accessibility (Table S3).
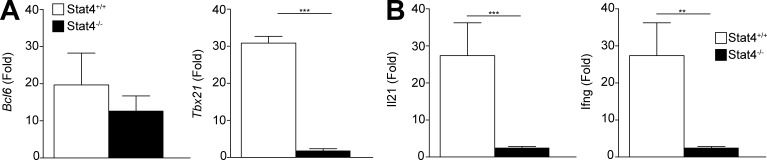
We also examined whether these genes were transcriptionally repressed in the absence of STAT4. Bcl6 expression was similar between STAT4+/+ and STAT4−/− Tfh cells, although Tbx21 expression was significantly reduced in the latter at 5 d p.i. (Fig. 7 A). Il21 and Ifng expression was significantly reduced in the mutant compared with STAT4+/+ cells at 5 d p.i. (Fig. 7 B). The transcriptional expression of these genes was consistent with the pattern of observed protein expression. Thus, Bcl6, Tbx21, Il21, and Ifng expression in Tfh cells is regulated at the transcriptional level and not via changes in chromatin accessibility mediated by STAT4.
Figure 7.
Influence of STAT4 deletion on gene expression in Tfh cells. Thy1.2+ B6 mice were infected with LCMV a day after receiving in transfer naive Thy1.1+STAT4+/+ or Thy1.1+STAT4−/− Stg CD4+ T cells. (A and B) Quantitative PCR was performed on sorted splenic Thy1.1+PSGL-1loLy6cloCXCR5hiPD-1hi Tfh cells to assess expression of Tbx21 and Bcl6 (A) and expression of Il21 and Ifng. (B) Data are representative of two experiments with five recipients per group. **, P < 0.01; ***, P < 0.001 by Student’s t test. Error bars represent SEM.
Discussion
We found that Tfh and Th1 cells coproduce IFN-γ and IL-21 after acute LCMV infection. Tfh cells required T-bet for IFN-γ secretion and for proliferation, robust CXCR5 expression with proper follicular migration, and CD40L expression—all necessary for GC formation and humoral output. In contrast, IL-21 secretion was not dependent on T-bet but on the presence of STAT4. STAT4 signaling was also necessary for T-bet expression in Tfh cells, as it is in Th1 cells (Thieu et al., 2008), and thus is also required for expansion of the former and for GC function upon viral challenge.
Previous studies have examined the divergent roles of T-bet and Bcl6 in the differentiation of Th1 and Tfh cells, largely in in vitro analyses. Initiation of Tbx21 expression with Th1 cell differentiation in vitro is dependent on IL-12–driven STAT4 signaling, which also promotes Bcl6 expression (Nakayamada et al., 2011). IL-2R signaling similarly promotes Th1 cell differentiation via expression of Blimp-1 (Nakayamada et al., 2011; Johnston et al., 2012; Oestreich et al., 2012), a repressor of transcription of Bcl6; yet, the latter locus in Th1 cells is not fully repressed because T-bet in concert with Bcl6 in Th1 cells is necessary to repress alternative Th cell programs (Nakayamada et al., 2011; Oestreich et al., 2011). We primarily took an in vivo approach, finding that T-bet–expressing Th1 cells expressed little Bcl6 after viral infection of mice. In contrast, Bcl6-expressing Tfh cells expressed T-bet, which was critical for their development and function and transcriptionally required for proper Tfh cell programming and for pathways of cell cycle and oxidative phosphorylation, as assessed by transcriptome analysis. The finding that T-bet is essential for Tfh cell development is at odds with previous in vitro experiments, in which naive polyclonal T-bet−/− CD4+ T cells developed into Tfh cells when transferred into RAG2−/− mice, followed by infection with Toxoplasma gondii (Nakayamada et al., 2011). Because naive CD4+ T cells and T cells with genetic defects in activation undergo homeostatic proliferation upon transfer into RAG2−/− mice (Min et al., 2004; Sena et al., 2013), it is possible that naive T-bet−/− cells in these experiments overcame the expansion defect that we have observed (Marshall et al., 2011). Likewise, the absence of B cells in RAG-deficient recipients may have contributed to Tfh cell expansion in the toxoplasma infection experiments (Nakayamada et al., 2011), given the role of B cell PD-L1 in regulation of Tfh cell numbers after immune stimulation (Good-Jacobson et al., 2010). Our experiments also used Ag-specific Tfh cells, which may have additional requirements for expansion in comparison with polyclonal cells analyzed in the earlier transfer into and toxoplasma infection of RAG-deficient recipients.
We found that T-bet and STAT4 expression in Tfh cells was similar to that in Th1 cells, as both transcription factors were expressed early after LCMV infection. Also akin to Th1 cells, Tbx21 and T-bet expression in Tfh cells was significantly reduced, albeit not ablated, in the absence of STAT4, suggesting that STAT4-independent signaling, likely via STAT1, promotes T-bet expression in Tfh cells, in accordance with its actions in Th1 cells (Afkarian et al., 2002). Although STAT1 signaling is not known to drive T-bet expression in Tfh cells, it does promote that of Bcl6 (Choi et al., 2013; Nakayamada et al., 2014). Nonetheless, robust Tfh cell expansion was dependent on STAT4 and T-bet. In contrast, Il21 transcription was dependent on STAT4, and not T-bet, as in Th1 cells (Nakayamada et al., 2011). The dichotomous roles of STAT4 and T-bet in normal Tfh cell function are consistent with the previous observation that subsets of genes depend on the presence of STAT4 or T-bet or of both for their expression in Th1 cells (Thieu et al., 2008).
Our finding that STAT4 in Tfh cells did not lead to changes in chromatin configuration is similar to that seen during Th cell development (Yu et al., 2008), in which changes in chromatin configuration distinguishing the induced state in Th1 cells from the repressed state in Th2 cells were STAT4 independent. STAT4 is required for recruitment of the histone acetyltransferase P300 to active enhancers in Th1 cells (Yu et al., 2008). If a similar mechanism is operative in Tfh cells, it is likely that many of the changes in gene expression we observed are driven by differences in histone acetylation, rather than by global rearrangement in chromatin occupancy.
Coordinated secretion of Tfh cell cytokines is necessary for promotion of intact humoral immunity and mediation of host effector function. Mouse IgG2a (or, more precisely, IgG2c in the B6 background used herein) antibodies dependent on Tfh cell T-bet expression and IFN-γ production, as we observed, are protective upon viral infection compared with IgG1 (Schlesinger and Chapman, 1995). The Fc region of secreted IgG2a interacts with complement components and activates critical antiviral Fc receptor–mediated effector functions including the stimulation of antibody-dependent, cell-mediated cytotoxicity and opsonization by macrophages (Kipps et al., 1985; Takai et al., 1994). Conversely, in the absence of T-bet, we found that the cytokine production by Tfh cells shifted away from IFN-γ toward IL-21, with enhanced IgG1 anti-LCMV responses. Although well suited for neutralizing secreted proteins produced by invading extracellular pathogens such as helminths, the Fc portion of IgG1 antibodies mediates a lower-affinity interaction with Fc receptors in comparison with IgG2c, with less effective receptor stimulation and consequent inflammatory responses (Hewitson et al., 2015). Our data also are consistent with recent studies demonstrating that in B cells, IFN-γ, and not IL-21, drives T-bet expression and subsequent IgG2a transcription along with gene expression programs necessary for proliferation and proper B cell localization (Barnett et al., 2016; Naradikian et al., 2016). These findings and ours explain why T-bet expression is required for IFN-γ synthesis by Tfh cells and clearance of experimental malaria infection (Carpio et al., 2015), with IFN-γ–driven T-bet expression in GC B cells being necessary for proper isotype switching and function (Barnett et al., 2016; Naradikian et al., 2016), whereas IL-21 is needed for full GC expansion and Ig affinity maturation (Weinstein et al., 2016).
We show that T-bet and STAT4 are essential for Tfh cell development after acute viral infection and their regulation of IFN-γ and IL-21 production and GC responses. Expression of T-bet is required for Tfh cell expansion and IFN-γ production and viral-specific GC output. Activated STAT4 promotes T-bet expression, as in Th1 cells (Nakayamada et al., 2011; Oestreich et al., 2012), along with IL-21. STAT4 and T-bet expression is driven by IL-12 and IFN-β. Both are needed to transcriptionally regulate Tfh cell expansion and coproduction of IL-21 and IFN-γ and, along with Bcl6, enable a proper GC response upon type 1 immune response. The loss of T-bet expression in Tfh cells during a chronic infection, such as experimental malaria (Carpio et al., 2015), accordingly leads to the loss of IFN-γ production, but not of IL-21. This imbalance results in improper GC B cell class switching from IgG2c to IgG1 and the failure of pathogen clearance. Conversely, our findings potentially explain the persistence of GCs and production of pathogenic IgG2a autoantibodies in chronic autoimmune diseases, such as systemic lupus erythematosus, which are driven by aberrant production of IL-12 and type I IFNs (Luzina et al., 2001; Kirou et al., 2005; Harigai et al., 2008), with downstream STAT4- and T-bet–mediated transcriptional activation in Tfh cells.
Materials and methods
Mice
Mice were housed in pathogen-free conditions at the Yale School of Medicine, New Haven, CT. C57BL/6J (B6), B6.Tcratm1Mom, B6.129S6-Tbx21tm1Glm/J, and C.129S2-Stat4tm1Gru/J animals were purchased from the Jackson Laboratory. B6.Tg(TcrLCMV)1Aox (SMARTA; Stg) mice (Oxenius et al., 1998) were provided by S. Kaech (Yale University, New Haven, CT). All animals were used at 6–8 wk of age, with approval for procedures given by the Institutional Animal Care and Use Committee of Yale University.
Cell transfers and viral infections
2.5 × 104 naive Stg TCR transgenic CD4 T cells were transferred to recipient mice via retroorbital injection. Mice were infected i.p. with 2 × 105 PFU LCMV Armstrong 24 h after transfer. Animals were sacrificed at different time points p.i., and harvested spleens were processed for flow cytometry.
ELISA for antibodies to LCMV
Anti-LCMV antibodies were measured by ELISA using sonicated cell lysate from LCMV-infected BHK-21 cells as capture Ag. 96-well microtiter plates (Polysorp; Nunc) were coated overnight with lysate in PBS. The antibodies used are listed in Table S1. ODs were read at 405 nm (Softmax Pro 3.1 software; Molecular Devices).
Flow cytometry and cell sorting
Tissues were homogenized by crushing with the head of a 1-ml syringe in a Petri dish followed by straining through a 40-µm nylon filter. Ammonium–chloride–potassium buffer was used for red blood cell lysis, and remaining cells were counted. Antibodies used for flow cytometry staining are listed in Table S1. Staining for CXCR5 was performed at room temperature (25°C) with 30 min of incubation. Intracellular staining for cytokines was performed using Cytofix/Cytoperm kits (BD Biosciences) according to the manufacturer’s protocol. Stained and rinsed cells were analyzed using a multilaser cytometer (LSRII; BD Biosciences). We performed a CD40L surface mobilization as previously described (Koguchi et al., 2007). In brief, cells were blocked with anti–Fc-γRII/III antibody, and 10 µg/ml APC-labeled anti-CD40L was introduced into the cell culture immediately before stimulation with 50 ng/ml PMA and 1 µg/ml ionomycin (Sigma-Aldrich), followed by 30-min incubation at 37°C. After three washes, cells were surface stained to identify subsets of CD4+ T cells. For sorting, CD4+ T cells were enriched using a biotin-based magnetic separation kit (EasySep; StemCell Technologies) before cell surface staining, with specific populations sorted using a cell sorter (FACSAria; BD Biosciences).
Microscopy
Spleens were snap frozen in optimum cutting temperature tissue-freezing solution and stored at −80°C. Tissues were cut into 8-µm sections and processed as described previously (Odegard et al., 2008). Reagents used to stain sections are listed in Table S1. Images were obtained from a laser-scanning confocal microscope (510 META; Carl Zeiss) at 25× magnification. ImageJ software (National Institutes of Health) was used for the measurement of GC and B cell follicle size, distance measurements, and for T cell counting. The latter analyses were performed in a blinded manner.
Transwell migration assays
Chemotaxis assays were performed as described previously (Beck et al., 2014). 5 × 105 enriched CD4+ splenic T cells were incubated for 1 h with 1× DMEM containing 0.5% fatty acid–free BSA (EMD Biosciences), 5% antibiotics, l-glutamine (Cellgro), and Hepes. Cells were then allowed to migrate through 5-µm-pore–sized transwells (Corning) toward soluble CXLC13 (R&D Systems) or media alone for 3 h at 37°C. Cells were collected, stained, and resuspended in 45 µl of staining buffer and analyzed by flow cytometry.
Retroviral transduction
Transductions were done as previously described (Ray et al., 2015). In brief, 1 mg LMP and T-bet shRNA vectors with 0.5 mg EcoHelp plasmid was used to transfect HEK293T viral packaging cells using X-tremeGENE 9 DNA transfection reagent (Roche) overnight. The media were then replaced, and viruses were grown for another 24 h. 6 × 106 splenocytes were activated for 24 h using CD3 plus CD28 stimulation and spin transduced with the supernatant of the transfection with polybrene. After transduction, 5 × 105 T cells were transferred via retroorbital injection into infected B6 recipients that were subsequently infected with LCMV Armstrong or Nippostrongylus brasiliensis plus OVA administration (Weinstein et al., 2016).
Quantitative PCR
Real-time PCR was set up using Brilliant II SYBR Green Master Mix and performed on an thermal cycler (MX4005P) according to the manufacturer’s protocols (Agilent Technologies), using the primers as noted in Table S2. Expression was calculated with the ΔΔxp method normalized to Hprt, and all measurements were performed in triplicate.
RNA-seq and analysis
Tfh cell populations were sorted by flow cytometry, with two separate sorts performed on different days, pooling spleens of 10 mice each day. Quality verification, library preparation, and sequencing were performed at the Yale Center for Genomic Analysis. Samples were sequenced on a high-throughput sequencing system (HiSeq 2500; Illumina) using 75-bp paired-end reads. RNA-seq was performed using Partek Flow (Partek Inc.). Reads were aligned to the mm9 mouse genome using STAR version 2.41d; unaligned reads were then aligned using TopHat version 2.1.0, and differential expression of the combined reads was then computed with Quantify to the annotation model (Partek E/M). Differentially expressed genes were filtered by keeping transcripts with at least one read from each population significant at the FDR-adjusted p-value of <0.05 between two of the populations, and fragments per kilobase of transcript per million mapped reads were summed across isoforms to obtain data for 668 significant genes. Clustering was done using hierarchical clustering in Partek by the average method or the partitioning around medoids method and Euclidean distance metrics. For heat maps, expression values were normalized per gene. Pathway enrichment analysis was performed using MetaCore (version 6.31 build). Enrichment analysis was performed using GSEA software v3.0 (Broad Institute), with the following settings: 1,000 gene set permutations, data collapsed to gene symbols using max probe mode, enrichment statistic = weighted, and ranking metric = Log2ratio of classes.
ATAC-seq
Sorted cells were lysed in buffer (10 mM Tris-HCl, 10 mM NaCl, 3 mM MgCl2, and 0.1% octylphenoxypolyethoxyethanol for 15 min at 4°C and then centrifuged and resuspended with transposase reaction mix (2× tagmented DNA buffer, transposase [Ilumina Nextera], and nuclease-free water) and incubated for 30 min at 37°C. Cells were then added to a MinElute column (Qiagen) and PCR amplified in KAPA HiFi 2× mix (Kapa Biosystems) with barcoding primers (Buenrostro et al., 2013). Amplification was conducted for 45 s at 98°C, followed by five cycles of denaturing at 98°C for 15 s, annealing at 63°C for 30 s, extension at 72°C for 30 s, and a final extension of 72°C for 1 min in a thermal cycler (MX4005P). PCR products were cleaned with MinElute columns, and size exclusion was performed with magJet NGS Cleanup and Size Selection kit (Thermo Fisher). Quantitative PCR library amplification test and PCR library amplification were performed as previously described (Buenrostro et al., 2015). DNA processing and high-throughput sequencing were performed as described in the previous section. Sequenced reads were mapped to the mouse genome (mm10 NCBI Build 38) using the Burrows-Wheeler Aligner version 0.7.9a alignment program. Reads mapping to the ENCODE project blacklist of repetitive regions and mitochondrial regions were removed (ENCODE Project Consortium, 2012). MACS2 program version 2.1.0.20150420 was used to identify peaks using parameters nomodel shift −100 extsize 200 with a q value of <0.05. Read counts for open chromatin regions in all samples were obtained using the DiffBind version 2.0.7 package (Ross-Innes et al., 2012) with DESEQ2 normalization using read counts in analyzed regions (parameter bFullLibrarySize = FALSE). Statistically significant differentially accessible regions were identified by DiffBind using the DESEQ2 method with default parameters. Peaks were annotated to mouse genome sequence features using the annotatePeak function in the Chipseeker version 1.8.9 package (Yu et al., 2015) and the University of California, Santa Cruz, mm10 knownGene database using parameter tss Region from −1,000 to +1,000 bp relative to the transcription start site.
Statistics
Data were analyzed using the Student’s t test or Fisher’s exact test with Prism 6 (GraphPad Software). The number of asterisks represents the degree of significance with respect to p-value, with the exact value presented within each figure legend.
Deposition of data
RNA sequencing and ATAC-seq data have been deposited into the GEO database (accession nos. GSE105806, GSE105807, and GSE105808).
Online supplemental material
Fig. S1 shows that endogenous Tfh cells express T-bet and coproduce IFN-γ and IL-21. Fig. S2 demonstrates that T-bet is not required for Tfh cell differentiation in type 2 infection. Fig. S3 demonstrates that T-bet expression in Tfh cells is necessary for proper GC development. Fig. S4 shows pSTAT4 in Tfh cells during viral infection. Fig. S5 reveals that STAT4 is required for cytokine production by Tfh cells, but not their development. Table S1 lists the antibodies used for flow cytometry, ELISA, and microscopy. Table S2 lists the primer sets used for quantitative PCR. Table S3 shows statistically significant differentially accessible regions from ATAC-seq.
Supplementary Material
Acknowledgments
The authors acknowledge members of our departments for critical review of the manuscript.
J.S. Weinstein was supported in part by National Institutes of Health (NIH) grants K01AR067892-02, UL1 TR001863, and NIH P30 AR053495. Other support came from NIH grants R37 AR40072 and P30 AR053495 (both to J. Craft), the Alliance for Lupus Research (to J. Craft), and NIH grants R01 AR068994 (to J. Craft and P.G. Gallagher) and U54DK106857 (to P.G. Gallagher).
The authors declare no competing financial interests.
Author contributions: J.S. Weinstein designed and performed experiments and wrote the manuscript. B.J. Laidlaw contributed to the experimental design and performed experiments. Y. Lu performed experiments. J.K. Wang contributed to the design and performed experiments. V.P. Schulz, E.I. Herman, and N. Li helped with ATAC-seq analysis. S.M. Kaech helped design experiments. P.G. Gallagher and J. Craft helped design experiments and wrote the manuscript. All authors read and approved the manuscript.
References
- Afkarian M., Sedy J.R., Yang J., Jacobson N.G., Cereb N., Yang S.Y., Murphy T.L., and Murphy K.M.. 2002. T-bet is a STAT1-induced regulator of IL-12R expression in naïve CD4+ T cells. Nat. Immunol. 3:549–557. 10.1038/ni794 [DOI] [PubMed] [Google Scholar]
- Barnett B.E., Staupe R.P., Odorizzi P.M., Palko O., Tomov V.T., Mahan A.E., Gunn B., Chen D., Paley M.A., Alter G., et al. 2016. Cutting Edge: B Cell-Intrinsic T-bet Expression Is Required To Control Chronic Viral Infection. J. Immunol. 197:1017–1022. 10.4049/jimmunol.1500368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumjohann D., Okada T., and Ansel K.M.. 2011. Cutting Edge: Distinct waves of BCL6 expression during T follicular helper cell development. J. Immunol. 187:2089–2092. 10.4049/jimmunol.1101393 [DOI] [PubMed] [Google Scholar]
- Bauquet A.T., Jin H., Paterson A.M., Mitsdoerffer M., Ho I.C., Sharpe A.H., and Kuchroo V.K.. 2009. The costimulatory molecule ICOS regulates the expression of c-Maf and IL-21 in the development of follicular T helper cells and TH-17 cells. Nat. Immunol. 10:167–175. 10.1038/ni.1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck T.C., Gomes A.C., Cyster J.G., and Pereira J.P.. 2014. CXCR4 and a cell-extrinsic mechanism control immature B lymphocyte egress from bone marrow. J. Exp. Med. 211:2567–2581. 10.1084/jem.20140457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro J.D., Giresi P.G., Zaba L.C., Chang H.Y., and Greenleaf W.J.. 2013. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 10:1213–1218. 10.1038/nmeth.2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro J.D., Wu B., Chang H.Y., and Greenleaf W.J.. 2015. ATAC-seq: A method for assaying chromatin accessibility genome-wide. In Current Protocols in Molecular Biology. Ausubel F.M., Brent R., Kingston R.E., Moore D.D., Seidman J.G., and Struhl K., editors. 109:21.29.1–9. 10.1002/0471142727.mb2129s109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpio V.H., Opata M.M., Montañez M.E., Banerjee P.P., Dent A.L., and Stephens R.. 2015. IFN-γ and IL-21 Double Producing T Cells Are Bcl6-Independent and Survive into the Memory Phase in Plasmodium chabaudi Infection. PLoS One. 10:e0144654 10.1371/journal.pone.0144654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S.S., Bacon C.M., Sudarshan C., Rees R.C., Finbloom D., Pine R., and O’Shea J.J.. 1996. Activation of STAT4 by IL-12 and IFN-alpha: evidence for the involvement of ligand-induced tyrosine and serine phosphorylation. J. Immunol. 157:4781–4789. [PubMed] [Google Scholar]
- Choi Y.S., Kageyama R., Eto D., Escobar T.C., Johnston R.J., Monticelli L., Lao C., and Crotty S.. 2011. ICOS receptor instructs T follicular helper cell versus effector cell differentiation via induction of the transcriptional repressor Bcl6. Immunity. 34:932–946. 10.1016/j.immuni.2011.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y.S., Eto D., Yang J.A., Lao C., and Crotty S.. 2013. Cutting edge: STAT1 is required for IL-6-mediated Bcl6 induction for early follicular helper cell differentiation. J. Immunol. 190:3049–3053. 10.4049/jimmunol.1203032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S. 2011. Follicular helper CD4 T cells (TFH). Annu. Rev. Immunol. 29:621–663. 10.1146/annurev-immunol-031210-101400 [DOI] [PubMed] [Google Scholar]
- Djuretic I.M., Levanon D., Negreanu V., Groner Y., Rao A., and Ansel K.M.. 2007. Transcription factors T-bet and Runx3 cooperate to activate Ifng and silence Il4 in T helper type 1 cells. Nat. Immunol. 8:145–153. 10.1038/ni1424 [DOI] [PubMed] [Google Scholar]
- Dong C., Juedes A.E., Temann U.A., Shresta S., Allison J.P., Ruddle N.H., and Flavell R.A.. 2001. ICOS co-stimulatory receptor is essential for T-cell activation and function. Nature. 409:97–101. 10.1038/35051100 [DOI] [PubMed] [Google Scholar]
- Eddahri F., Denanglaire S., Bureau F., Spolski R., Leonard W.J., Leo O., and Andris F.. 2009. Interleukin-6/STAT3 signaling regulates the ability of naive T cells to acquire B-cell help capacities. Blood. 113:2426–2433. 10.1182/blood-2008-04-154682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium 2012. An integrated encyclopedia of DNA elements in the human genome. Nature. 489:57–74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eto D., Lao C., DiToro D., Barnett B., Escobar T.C., Kageyama R., Yusuf I., and Crotty S.. 2011. IL-21 and IL-6 are critical for different aspects of B cell immunity and redundantly induce optimal follicular helper CD4 T cell (Tfh) differentiation. PLoS One. 6:e17739 10.1371/journal.pone.0017739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahey L.M., Wilson E.B., Elsaesser H., Fistonich C.D., McGavern D.B., and Brooks D.G.. 2011. Viral persistence redirects CD4 T cell differentiation toward T follicular helper cells. J. Exp. Med. 208:987–999. 10.1084/jem.20101773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerth A.J., Lin L., and Peng S.L.. 2003. T-bet regulates T-independent IgG2a class switching. Int. Immunol. 15:937–944. 10.1093/intimm/dxg093 [DOI] [PubMed] [Google Scholar]
- Good-Jacobson K.L., Szumilas C.G., Chen L., Sharpe A.H., Tomayko M.M., and Shlomchik M.J.. 2010. PD-1 regulates germinal center B cell survival and the formation and affinity of long-lived plasma cells. Nat. Immunol. 11:535–542. 10.1038/ni.1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale J.S., Youngblood B., Latner D.R., Mohammed A.U., Ye L., Akondy R.S., Wu T., Iyer S.S., and Ahmed R.. 2013. Distinct memory CD4+ T cells with commitment to T follicular helper- and T helper 1-cell lineages are generated after acute viral infection. Immunity. 38:805–817. 10.1016/j.immuni.2013.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harigai M., Kawamoto M., Hara M., Kubota T., Kamatani N., and Miyasaka N.. 2008. Excessive production of IFN-gamma in patients with systemic lupus erythematosus and its contribution to induction of B lymphocyte stimulator/B cell-activating factor/TNF ligand superfamily-13B. J. Immunol. 181:2211–2219. 10.4049/jimmunol.181.3.2211 [DOI] [PubMed] [Google Scholar]
- Harker J.A., Lewis G.M., Mack L., and Zuniga E.I.. 2011. Late interleukin-6 escalates T follicular helper cell responses and controls a chronic viral infection. Science. 334:825–829. 10.1126/science.1208421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitson J.P., Filbey K.J., Esser-von Bieren J., Camberis M., Schwartz C., Murray J., Reynolds L.A., Blair N., Robertson E., Harcus Y., et al. 2015. Concerted activity of IgG1 antibodies and IL-4/IL-25-dependent effector cells trap helminth larvae in the tissues following vaccination with defined secreted antigens, providing sterile immunity to challenge infection. PLoS Pathog. 11:e1004676 10.1371/journal.ppat.1004676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ise W., Kohyama M., Schraml B.U., Zhang T., Schwer B., Basu U., Alt F.W., Tang J., Oltz E.M., Murphy T.L., and Murphy K.M.. 2011. The transcription factor BATF controls the global regulators of class-switch recombination in both B cells and T cells. Nat. Immunol. 12:536–543. 10.1038/ni.2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata S., Mikami Y., Sun H.W., Brooks S.R., Jankovic D., Hirahara K., Onodera A., Shih H.Y., Kawabe T., Jiang K., et al. 2017. The Transcription Factor T-bet Limits Amplification of Type I IFN Transcriptome and Circuitry in T Helper 1 Cells. Immunity. 46:983–991.e4. 10.1016/j.immuni.2017.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston R.J., Poholek A.C., DiToro D., Yusuf I., Eto D., Barnett B., Dent A.L., Craft J., and Crotty S.. 2009. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 325:1006–1010. 10.1126/science.1175870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston R.J., Choi Y.S., Diamond J.A., Yang J.A., and Crotty S.. 2012. STAT5 is a potent negative regulator of TFH cell differentiation. J. Exp. Med. 209:243–250. 10.1084/jem.20111174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipps T.J., Parham P., Punt J., and Herzenberg L.A.. 1985. Importance of immunoglobulin isotype in human antibody-dependent, cell-mediated cytotoxicity directed by murine monoclonal antibodies. J. Exp. Med. 161:1–17. 10.1084/jem.161.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirou K.A., Lee C., George S., Louca K., Peterson M.G., and Crow M.K.. 2005. Activation of the interferon-alpha pathway identifies a subgroup of systemic lupus erythematosus patients with distinct serologic features and active disease. Arthritis Rheum. 52:1491–1503. 10.1002/art.21031 [DOI] [PubMed] [Google Scholar]
- Koguchi Y., Thauland T.J., Slifka M.K., and Parker D.C.. 2007. Preformed CD40 ligand exists in secretory lysosomes in effector and memory CD4+ T cells and is quickly expressed on the cell surface in an antigen-specific manner. Blood. 110:2520–2527. 10.1182/blood-2007-03-081299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchen S., Robbins R., Sims G.P., Sheng C., Phillips T.M., Lipsky P.E., and Ettinger R.. 2007. Essential role of IL-21 in B cell activation, expansion, and plasma cell generation during CD4+ T cell-B cell collaboration. J. Immunol. 179:5886–5896. 10.4049/jimmunol.179.9.5886 [DOI] [PubMed] [Google Scholar]
- Lee S.K., Rigby R.J., Zotos D., Tsai L.M., Kawamoto S., Marshall J.L., Ramiscal R.R., Chan T.D., Gatto D., Brink R., et al. 2011. B cell priming for extrafollicular antibody responses requires Bcl-6 expression by T cells. J. Exp. Med. 208:1377–1388. 10.1084/jem.20102065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Lu E., Yi T., and Cyster J.G.. 2016. EBI2 augments Tfh cell fate by promoting interaction with IL-2-quenching dendritic cells. Nature. 533:110–114. 10.1038/nature17947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lighvani A.A., Frucht D.M., Jankovic D., Yamane H., Aliberti J., Hissong B.D., Nguyen B.V., Gadina M., Sher A., Paul W.E., and O’Shea J.J.. 2001. T-bet is rapidly induced by interferon-γ in lymphoid and myeloid cells. Proc. Natl. Acad. Sci. USA. 98:15137–15142. 10.1073/pnas.261570598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linterman M.A., Beaton L., Yu D., Ramiscal R.R., Srivastava M., Hogan J.J., Verma N.K., Smyth M.J., Rigby R.J., and Vinuesa C.G.. 2010. IL-21 acts directly on B cells to regulate Bcl-6 expression and germinal center responses. J. Exp. Med. 207:353–363. 10.1084/jem.20091738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Xu H., Shih C., Wan Z., Ma X., Ma W., Luo D., and Qi H.. 2015. T-B-cell entanglement and ICOSL-driven feed-forward regulation of germinal centre reaction. Nature. 517:214–218. 10.1038/nature13803 [DOI] [PubMed] [Google Scholar]
- Liu X., Chen X., Zhong B., Wang A., Wang X., Chu F., Nurieva R.I., Yan X., Chen P., van der Flier L.G., et al. 2014. Transcription factor achaete-scute homologue 2 initiates follicular T-helper-cell development. Nature. 507:513–518. 10.1038/nature12910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locci M., Wu J.E., Arumemi F., Mikulski Z., Dahlberg C., Miller A.T., and Crotty S.. 2016. Activin A programs the differentiation of human TFH cells. Nat. Immunol. 17:976–984. 10.1038/ni.3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord G.M., Rao R.M., Choe H., Sullivan B.M., Lichtman A.H., Luscinskas F.W., and Glimcher L.H.. 2005. T-bet is required for optimal proinflammatory CD4+ T-cell trafficking. Blood. 106:3432–3439. 10.1182/blood-2005-04-1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K.T., Kanno Y., Cannons J.L., Handon R., Bible P., Elkahloun A.G., Anderson S.M., Wei L., Sun H., O’Shea J.J., and Schwartzberg P.L.. 2011. Functional and epigenetic studies reveal multistep differentiation and plasticity of in vitro-generated and in vivo-derived follicular T helper cells. Immunity. 35:622–632. 10.1016/j.immuni.2011.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun A.T., and Smyth G.K.. 2016. csaw: a Bioconductor package for differential binding analysis of ChIP-seq data using sliding windows. Nucleic Acids Res. 44:e45 10.1093/nar/gkv1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüthje K., Kallies A., Shimohakamada Y., Belz G.T., Light A., Tarlinton D.M., and Nutt S.L.. 2012. The development and fate of follicular helper T cells defined by an IL-21 reporter mouse. Nat. Immunol. 13:491–498. 10.1038/ni.2261 [DOI] [PubMed] [Google Scholar]
- Luzina I.G., Atamas S.P., Storrer C.E., daSilva L.C., Kelsoe G., Papadimitriou J.C., and Handwerger B.S.. 2001. Spontaneous formation of germinal centers in autoimmune mice. J. Leukoc. Biol. 70:578–584. [PubMed] [Google Scholar]
- Mak T.W., Shahinian A., Yoshinaga S.K., Wakeham A., Boucher L.M., Pintilie M., Duncan G., Gajewska B.U., Gronski M., Eriksson U., et al. 2003. Costimulation through the inducible costimulator ligand is essential for both T helper and B cell functions in T cell-dependent B cell responses. Nat. Immunol. 4:765–772. 10.1038/ni947 [DOI] [PubMed] [Google Scholar]
- Marshall H.D., Chandele A., Jung Y.W., Meng H., Poholek A.C., Parish I.A., Rutishauser R., Cui W., Kleinstein S.H., Craft J., and Kaech S.M.. 2011. Differential expression of Ly6C and T-bet distinguish effector and memory Th1 CD4(+) cell properties during viral infection. Immunity. 35:633–646. 10.1016/j.immuni.2011.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall H.D., Ray J.P., Laidlaw B.J., Zhang N., Gawande D., Staron M.M., Craft J., and Kaech S.M.. 2015. The transforming growth factor beta signaling pathway is critical for the formation of CD4 T follicular helper cells and isotype-switched antibody responses in the lung mucosa. eLife. 4:e04851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAdam A.J., Greenwald R.J., Levin M.A., Chernova T., Malenkovich N., Ling V., Freeman G.J., and Sharpe A.H.. 2001. ICOS is critical for CD40-mediated antibody class switching. Nature. 409:102–105. 10.1038/35051107 [DOI] [PubMed] [Google Scholar]
- Mehta D.S., Wurster A.L., Whitters M.J., Young D.A., Collins M., and Grusby M.J.. 2003. IL-21 induces the apoptosis of resting and activated primary B cells. J. Immunol. 170:4111–4118. 10.4049/jimmunol.170.8.4111 [DOI] [PubMed] [Google Scholar]
- Min B., Foucras G., Meier-Schellersheim M., and Paul W.E.. 2004. Spontaneous proliferation, a response of naive CD4 T cells determined by the diversity of the memory cell repertoire. Proc. Natl. Acad. Sci. USA. 101:3874–3879. 10.1073/pnas.0400606101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyauchi K., Sugimoto-Ishige A., Harada Y., Adachi Y., Usami Y., Kaji T., Inoue K., Hasegawa H., Watanabe T., Hijikata A., et al. 2016. Protective neutralizing influenza antibody response in the absence of T follicular helper cells. Nat. Immunol. 17:1447–1458. 10.1038/ni.3563 [DOI] [PubMed] [Google Scholar]
- Morinobu A., Gadina M., Strober W., Visconti R., Fornace A., Montagna C., Feldman G.M., Nishikomori R., and O’Shea J.J.. 2002. STAT4 serine phosphorylation is critical for IL-12-induced IFN-gamma production but not for cell proliferation. Proc. Natl. Acad. Sci. USA. 99:12281–12286. 10.1073/pnas.182618999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosmann T.R., and Coffman R.L.. 1989. TH1 and TH2 cells: different patterns of lymphokine secretion lead to different functional properties. Annu. Rev. Immunol. 7:145–173. 10.1146/annurev.iy.07.040189.001045 [DOI] [PubMed] [Google Scholar]
- Mullen A.C., High F.A., Hutchins A.S., Lee H.W., Villarino A.V., Livingston D.M., Kung A.L., Cereb N., Yao T.P., Yang S.Y., and Reiner S.L.. 2001. Role of T-bet in commitment of TH1 cells before IL-12-dependent selection. Science. 292:1907–1910. 10.1126/science.1059835 [DOI] [PubMed] [Google Scholar]
- Nakayamada S., Kanno Y., Takahashi H., Jankovic D., Lu K.T., Johnson T.A., Sun H.W., Vahedi G., Hakim O., Handon R., et al. 2011. Early Th1 cell differentiation is marked by a Tfh cell-like transition. Immunity. 35:919–931. 10.1016/j.immuni.2011.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayamada S., Poholek A.C., Lu K.T., Takahashi H., Kato M., Iwata S., Hirahara K., Cannons J.L., Schwartzberg P.L., Vahedi G., et al. 2014. Type I IFN induces binding of STAT1 to Bcl6: divergent roles of STAT family transcription factors in the T follicular helper cell genetic program. J. Immunol. 192:2156–2166. 10.4049/jimmunol.1300675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naradikian M.S., Myles A., Beiting D.P., Roberts K.J., Dawson L., Herati R.S., Bengsch B., Linderman S.L., Stelekati E., Spolski R., et al. 2016. Cutting Edge: IL-4, IL-21, and IFN-γ Interact To Govern T-bet and CD11c Expression in TLR-Activated B Cells. J. Immunol. 197:1023–1028. 10.4049/jimmunol.1600522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurieva R.I., Mai X.M., Forbush K., Bevan M.J., and Dong C.. 2003. B7h is required for T cell activation, differentiation, and effector function. Proc. Natl. Acad. Sci. USA. 100:14163–14168. 10.1073/pnas.2335041100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurieva R.I., Chung Y., Martinez G.J., Yang X.O., Tanaka S., Matskevitch T.D., Wang Y.H., and Dong C.. 2009. Bcl6 mediates the development of T follicular helper cells. Science. 325:1001–1005. 10.1126/science.1176676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurieva R.I., Podd A., Chen Y., Alekseev A.M., Yu M., Qi X., Huang H., Wen R., Wang J., Li H.S., et al. 2012. STAT5 protein negatively regulates T follicular helper (Tfh) cell generation and function. J. Biol. Chem. 287:11234–11239. 10.1074/jbc.M111.324046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odegard J.M., Marks B.R., DiPlacido L.D., Poholek A.C., Kono D.H., Dong C., Flavell R.A., and Craft J.. 2008. ICOS-dependent extrafollicular helper T cells elicit IgG production via IL-21 in systemic autoimmunity. J. Exp. Med. 205:2873–2886. 10.1084/jem.20080840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oestreich K.J., Huang A.C., and Weinmann A.S.. 2011. The lineage-defining factors T-bet and Bcl-6 collaborate to regulate Th1 gene expression patterns. J. Exp. Med. 208:1001–1013. 10.1084/jem.20102144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oestreich K.J., Mohn S.E., and Weinmann A.S.. 2012. Molecular mechanisms that control the expression and activity of Bcl-6 in TH1 cells to regulate flexibility with a TFH-like gene profile. Nat. Immunol. 13:405–411. 10.1038/ni.2242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxenius A., Bachmann M.F., Zinkernagel R.M., and Hengartner H.. 1998. Virus-specific major MHC-class II-restricted TCR-transgenic mice: effects on humoral and cellular immune responses after viral infection. Eur. J. Immunol. 28:390–400. [DOI] [PubMed] [Google Scholar]
- Ozaki K., Spolski R., Ettinger R., Kim H.P., Wang G., Qi C.F., Hwu P., Shaffer D.J., Akilesh S., Roopenian D.C., et al. 2004. Regulation of B cell differentiation and plasma cell generation by IL-21, a novel inducer of Blimp-1 and Bcl-6. J. Immunol. 173:5361–5371. 10.4049/jimmunol.173.9.5361 [DOI] [PubMed] [Google Scholar]
- Pallikkuth S., Parmigiani A., Silva S.Y., George V.K., Fischl M., Pahwa R., and Pahwa S.. 2012. Impaired peripheral blood T-follicular helper cell function in HIV-infected nonresponders to the 2009 H1N1/09 vaccine. Blood. 120:985–993. 10.1182/blood-2011-12-396648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng S.L., Szabo S.J., and Glimcher L.H.. 2002. T-bet regulates IgG class switching and pathogenic autoantibody production. Proc. Natl. Acad. Sci. USA. 99:5545–5550. 10.1073/pnas.082114899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poholek A.C., Hansen K., Hernandez S.G., Eto D., Chandele A., Weinstein J.S., Dong X., Odegard J.M., Kaech S.M., Dent A.L., et al. 2010. In vivo regulation of Bcl6 and T follicular helper cell development. J. Immunol. 185:313–326. 10.4049/jimmunol.0904023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray J.P., Marshall H.D., Laidlaw B.J., Staron M.M., Kaech S.M., and Craft J.. 2014. Transcription factor STAT3 and type I interferons are corepressive insulators for differentiation of follicular helper and T helper 1 cells. Immunity. 40:367–377. 10.1016/j.immuni.2014.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray J.P., Staron M.M., Shyer J.A., Ho P.C., Marshall H.D., Gray S.M., Laidlaw B.J., Araki K., Ahmed R., Kaech S.M., and Craft J.. 2015. The Interleukin-2-mTORc1 Kinase Axis Defines the Signaling, Differentiation, and Metabolism of T Helper 1 and Follicular B Helper T Cells. Immunity. 43:690–702. 10.1016/j.immuni.2015.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt R.L., Liang H.E., and Locksley R.M.. 2009. Cytokine-secreting follicular T cells shape the antibody repertoire. Nat. Immunol. 10:385–393. 10.1038/ni.1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Innes C.S., Stark R., Teschendorff A.E., Holmes K.A., Ali H.R., Dunning M.J., Brown G.D., Gojis O., Ellis I.O., Green A.R., et al. 2012. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 481:389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaerli P., Willimann K., Lang A.B., Lipp M., Loetscher P., and Moser B.. 2000. CXC chemokine receptor 5 expression defines follicular homing T cells with B cell helper function. J. Exp. Med. 192:1553–1562. 10.1084/jem.192.11.1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger J.J., and Chapman S.. 1995. Neutralizing F(ab’)2 fragments of protective monoclonal antibodies to yellow fever virus (YF) envelope protein fail to protect mice against lethal YF encephalitis. J. Gen. Virol. 76:217–220. 10.1099/0022-1317-76-1-217 [DOI] [PubMed] [Google Scholar]
- Schmitt N., Morita R., Bourdery L., Bentebibel S.E., Zurawski S.M., Banchereau J., and Ueno H.. 2009. Human dendritic cells induce the differentiation of interleukin-21-producing T follicular helper-like cells through interleukin-12. Immunity. 31:158–169. 10.1016/j.immuni.2009.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt N., Liu Y., Bentebibel S.E., Munagala I., Bourdery L., Venuprasad K., Banchereau J., and Ueno H.. 2014. The cytokine TGF-β co-opts signaling via STAT3-STAT4 to promote the differentiation of human TFH cells. Nat. Immunol. 15:856–865. 10.1038/ni.2947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz E.G., Mariani L., Radbruch A., and Höfer T.. 2009. Sequential polarization and imprinting of type 1 T helper lymphocytes by interferon-gamma and interleukin-12. Immunity. 30:673–683. 10.1016/j.immuni.2009.03.013 [DOI] [PubMed] [Google Scholar]
- Sena L.A., Li S., Jairaman A., Prakriya M., Ezponda T., Hildeman D.A., Wang C.R., Schumacker P.T., Licht J.D., Perlman H., et al. 2013. Mitochondria are required for antigen-specific T cell activation through reactive oxygen species signaling. Immunity. 38:225–236. 10.1016/j.immuni.2012.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H.J., Park H.Y., Jeong S.J., Park H.W., Kim Y.K., Cho S.H., Kim Y.Y., Cho M.L., Kim H.Y., Min K.U., and Lee C.W.. 2005. STAT4 expression in human T cells is regulated by DNA methylation but not by promoter polymorphism. J. Immunol. 175:7143–7150. 10.4049/jimmunol.175.11.7143 [DOI] [PubMed] [Google Scholar]
- Stone E.L., Pepper M., Katayama C.D., Kerdiles Y.M., Lai C.Y., Emslie E., Lin Y.C., Yang E., Goldrath A.W., Li M.O., et al. 2015. ICOS coreceptor signaling inactivates the transcription factor FOXO1 to promote Tfh cell differentiation. Immunity. 42:239–251. 10.1016/j.immuni.2015.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo S.J., Kim S.T., Costa G.L., Zhang X., Fathman C.G., and Glimcher L.H.. 2000. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 100:655–669. 10.1016/S0092-8674(00)80702-3 [DOI] [PubMed] [Google Scholar]
- Takai T., Li M., Sylvestre D., Clynes R., and Ravetch J.V.. 1994. FcR gamma chain deletion results in pleiotrophic effector cell defects. Cell. 76:519–529. 10.1016/0092-8674(94)90115-5 [DOI] [PubMed] [Google Scholar]
- Thieu V.T., Yu Q., Chang H.C., Yeh N., Nguyen E.T., Sehra S., and Kaplan M.H.. 2008. Signal transducer and activator of transcription 4 is required for the transcription factor T-bet to promote T helper 1 cell-fate determination. Immunity. 29:679–690. 10.1016/j.immuni.2008.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usui T., Preiss J.C., Kanno Y., Yao Z.J., Bream J.H., O’Shea J.J., and Strober W.. 2006. T-bet regulates Th1 responses through essential effects on GATA-3 function rather than on IFNG gene acetylation and transcription. J. Exp. Med. 203:755–766. 10.1084/jem.20052165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelzang A., McGuire H.M., Yu D., Sprent J., Mackay C.R., and King C.. 2008. A fundamental role for interleukin-21 in the generation of T follicular helper cells. Immunity. 29:127–137. 10.1016/j.immuni.2008.06.001 [DOI] [PubMed] [Google Scholar]
- Wang Y., Shi J., Yan J., Xiao Z., Hou X., Lu P., Hou S., Mao T., Liu W., Ma Y., et al. 2017. Germinal-center development of memory B cells driven by IL-9 from follicular helper T cells. Nat. Immunol. 18:921–930. 10.1038/ni.3788 [DOI] [PubMed] [Google Scholar]
- Weber J.P., Fuhrmann F., Feist R.K., Lahmann A., Al Baz M.S., Gentz L.J., Vu Van D., Mages H.W., Haftmann C., Riedel R., et al. 2015. ICOS maintains the T follicular helper cell phenotype by down-regulating Krüppel-like factor 2. J. Exp. Med. 212:217–233. 10.1084/jem.20141432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L., Vahedi G., Sun H.W., Watford W.T., Takatori H., Ramos H.L., Takahashi H., Liang J., Gutierrez-Cruz G., Zang C., et al. 2010. Discrete roles of STAT4 and STAT6 transcription factors in tuning epigenetic modifications and transcription during T helper cell differentiation. Immunity. 32:840–851. 10.1016/j.immuni.2010.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein J.S., Bertino S.A., Hernandez S.G., Poholek A.C., Teplitzky T.B., Nowyhed H.N., and Craft J.. 2014. B cells in T follicular helper cell development and function: separable roles in delivery of ICOS ligand and antigen. J. Immunol. 192:3166–3179. 10.4049/jimmunol.1302617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein J.S., Herman E.I., Lainez B., Licona-Limón P., Esplugues E., Flavell R., and Craft J.. 2016. TFH cells progressively differentiate to regulate the germinal center response. Nat. Immunol. 17:1197–1205. 10.1038/ni.3554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Li X., Liu D., Li J., Zhang X., Chen X., Hou S., Peng L., Xu C., Liu W., et al. 2013. Follicular T-helper cell recruitment governed by bystander B cells and ICOS-driven motility. Nature. 496:523–527. 10.1038/nature12058 [DOI] [PubMed] [Google Scholar]
- Yu G., Wang L.G., and He Q.Y.. 2015. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 31:2382–2383. 10.1093/bioinformatics/btv145 [DOI] [PubMed] [Google Scholar]
- Yu Q., Chang H.C., Ahyi A.N., and Kaplan M.H.. 2008. Transcription factor-dependent chromatin remodeling of Il18r1 during Th1 and Th2 differentiation. J. Immunol. 181:3346–3352. 10.4049/jimmunol.181.5.3346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q., Sharma A., Oh S.Y., Moon H.-G., Hossain M.Z., Salay T.M., Leeds K.E., Du H., Wu B., Waterman M.L., et al. 2009. T cell factor 1 initiates the T helper type 2 fate by inducing the transcription factor GATA-3 and repressing interferon-gamma. Nat. Immunol. 10:992–999. 10.1038/ni.1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Jankovic D., Oler A.J., Wei G., Sharma S., Hu G., Guo L., Yagi R., Yamane H., Punkosdy G., et al. 2012. The transcription factor T-bet is induced by multiple pathways and prevents an endogenous Th2 cell program during Th1 cell responses. Immunity. 37:660–673. 10.1016/j.immuni.2012.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zotos D., Coquet J.M., Zhang Y., Light A., D’Costa K., Kallies A., Corcoran L.M., Godfrey D.I., Toellner K.-M., Smyth M.J., et al. 2010. IL-21 regulates germinal center B cell differentiation and proliferation through a B cell-intrinsic mechanism. J. Exp. Med. 207:365–378. 10.1084/jem.20091777 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.