Data demonstrate that short amyloid-β (Aβ) peptides are not toxic in vivo and can partially block toxicity associated with Aβ42 accumulation. Moore et al. further validate the use of γ-secretase modulators that lower Aβ42 and increase short Aβs as potential Alzheimer’s disease therapeutics.
Abstract
Processing of amyloid-β (Aβ) precursor protein (APP) by γ-secretase produces multiple species of Aβ: Aβ40, short Aβ peptides (Aβ37–39), and longer Aβ peptides (Aβ42–43). γ-Secretase modulators, a class of Alzheimer’s disease therapeutics, reduce production of the pathogenic Aβ42 but increase the relative abundance of short Aβ peptides. To evaluate the pathological relevance of these peptides, we expressed Aβ36–40 and Aβ42–43 in Drosophila melanogaster to evaluate inherent toxicity and potential modulatory effects on Aβ42 toxicity. In contrast to Aβ42, the short Aβ peptides were not toxic and, when coexpressed with Aβ42, were protective in a dose-dependent fashion. In parallel, we explored the effects of recombinant adeno-associated virus–mediated expression of Aβ38 and Aβ40 in mice. When expressed in nontransgenic mice at levels sufficient to drive Aβ42 deposition, Aβ38 and Aβ40 did not deposit or cause behavioral alterations. These studies indicate that treatments that lower Aβ42 by raising the levels of short Aβ peptides could attenuate the toxic effects of Aβ42.
Introduction
Alzheimer’s disease (AD) is the most common cause of dementia and the most prevalent neurodegenerative disorder. The defining neuropathological hallmark of AD is the accumulation of amyloid plaques and neurofibrillary tangles (Selkoe and Hardy, 2016). Amyloid plaques contain the ∼4-kD amyloid-β (Aβ) peptide, which is produced through sequential cleavage of the Aβ precursor protein (APP) by the β- and γ-secretases. γ-Secretase cleavage generates numerous Aβ peptides with differing C termini (Wang et al., 1996; Golde et al., 2013). Under physiological conditions, the major species generated are Aβ1–40 (>50% of total Aβ), with Aβ1–37, 1–38, 1–39, and 1–42 generated at lower levels (∼5–20% of total Aβ). In some instances, additional Aβ peptides (e.g., Aβ1–34, 1–36, 1–41, and 1–43) are generated at detectable levels (Seubert et al., 1992; Wang et al., 1996; Golde et al., 2013). Shifts in the relative production of Aβ toward Aβ1–42 or, in rare cases, Aβ1–42 and Aβ1–43 are linked to AD (Iwatsubo et al., 1994; Younkin, 1998; Golde et al., 2000; Saito et al., 2011; Kretner et al., 2016). APP and Presenilin1/2 (PSEN1/2) mutations that elevate the relative level of Aβ1–42/43 as little as 30% deterministically cause early-onset AD (Selkoe, 2001; De Strooper et al., 2010). Further, biochemical studies show that Aβ1–42 aggregates into amyloid fibrils and other assemblies much more readily than Aβ1–40 (Jarrett and Lansbury, 1993), and modeling studies show that AD-associated APP and PSEN mutations that increase Aβ1–42 levels accelerate Aβ deposition (reviewed by Ashe and Zahs, 2010). Other studies using strategies to express Aβ1–42 and Aβ1–40 in the absence of APP overexpression show that Aβ1–42 is required to drive Aβ deposition in mice and that Aβ1–40 may actually inhibit Aβ deposition (McGowan et al., 2005; Kim et al., 2007).
Given the overwhelming evidence that relative increases in Aβ1–42 levels promote aggregation of Aβ into toxic species, there has been great interest in the development of AD therapeutic compounds, referred to as γ-secretase modulators (GSMs), that selectively lower Aβ1–42. GSMs do not affect overall γ-secretase activity but increase the processive cleavages of the APP substrate catalyzed by γ-secretase. Thus, GSMs lower Aβ1–42 but increase the relative production of shorter Aβ peptide (Weggen et al., 2001; Golde et al., 2012; Wagner et al., 2012). First-generation, nonsteroidal anti-inflammatory agent–like, acidic GSMs, did not alter total Aβ production, increase APP C-terminal fragments, or alter cleavage of other γ-secretase substrates, but instead, decreased Aβ1–42 levels and increased Aβ1–38 (Weggen et al., 2001; Golde et al., 2013; Jung et al., 2013). Later, nonacidic GSMs were identified that showed distinct effects on Aβ generation, decreasing both Aβ1–40 and Aβ1–42 and increasing Aβ1–37 and Aβ1–38 (Kounnas et al., 2010). More recently identified triterpenoid GSMs are also distinct, lowering both Aβ1–42 and Aβ1–38 without major effects on other Aβ species (Hubbs et al., 2012).
Although GSMs have been postulated to be inherently safe (Golde et al., 2013), a lingering concern with this therapeutic modality has been whether the shorter Aβ species are potentially harmful. Indeed, an in vitro and cell-culture study suggested that Aβ1–38 aggregated with kinetics similar to Aβ1–42 and that the aggregates were just as neurotoxic as Aβ1–42 (Kuperstein et al., 2010). Further, there is evidence that numerous short Aβ peptides accumulate in the AD brain, suggesting that they could have a role in AD pathogenesis (Moore et al., 2012). Thus, because GSMs are most likely to be efficacious in delaying or preventing the development of AD, it is important to definitively establish whether short Aβ peptides are toxic.
To investigate the potential pathogenic role of short Aβ peptides, we generated PhiC31-based transgenic Drosophila melanogaster expressing Aβ1–36, Aβ1–37, Aβ1–38, Aβ1–39, Aβ1–40, Aβ1–42, and Aβ1–43, fused to the Argos signal peptide and placed under the GAL4-upstream activation sequence (UAS) expression system. In addition, we used recombinant adeno-associated virus (rAAV) vectors to express Aβ1–38 using our BRI2 fusion strategy (Lewis et al., 2001; McGowan et al., 2005) to determine whether the Aβ1–38 peptide deposits on its own, alters cognition, or modulates amyloid deposition in an APP mouse model. Collectively, these studies demonstrate that the short Aβ peptides are not toxic by themselves and have the potential to protect from Aβ1–42 toxicity.
Results
Characterization of transgenic Aβ flies expressing Aβ36, Aβ37, Aβ38, Aβ39, Aβ40, Aβ42, and Aβ43
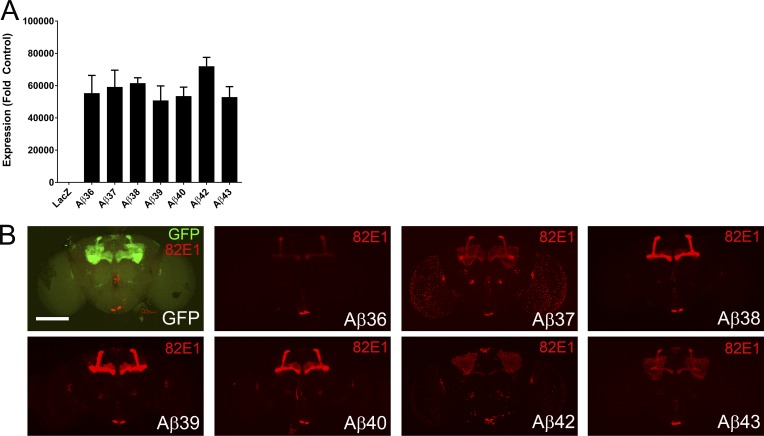
We generated transgenic flies that express a single copy of secreted Aβ through targeted insertion at the attP2 landing site. To initially evaluate expression, we crossed Tg Aβ flies with da-Gal4, isolated RNA, and used real-time quantitative RT-PCR (Q-RT-PCR) to validate the constructs. Q-RT-PCR revealed that the Aβ transgenes were expressed at comparable levels (Fig. 1 A). To confirm that the Aβ peptides were expressed and accumulated at detectable levels, we crossed Tg Aβ flies with OK107-Gal4 flies to direct expression to the mushroom body neurons. Immunostaining with a neo-epitope Aβ1-x antibody (82E1) demonstrated that the Tg Aβ flies expressed Aβ with the authentic N terminus found in mammalian systems (Fig. 1 B). Although RNA expression levels are comparable, the 82E1 immunofluorescence study revealed qualitatively different distributions of Aβ36–Aβ40 peptides and Aβ42 and Aβ43 peptides within the mushroom body (examined in more detail in subsequent experiments).
Figure 1.
Aβ peptide transgenes express in Drosophila at similar levels. (A) Real-time Q-RT-PCR confirms that Aβ transgenes were expressed at relatively similar levels. Data represent expression normalized to two endogenous Drosophila genes that encode β-tubulin and the ribosomal protein L32 relative to LacZ flies, calculated using the comparative CT method. A total of 21 flies were analyzed per transgene, in three pools of n = 7 biological replicates. Data representative of one of two independent experiments. Values represent means ± standard error of the mean. (B) Representative staining with anti-Aβ antibody 82E1 (red) confirms expression of the Aβ peptides. Bar, 250 µm.
Short Aβ peptides show no toxicity in Drosophila eye
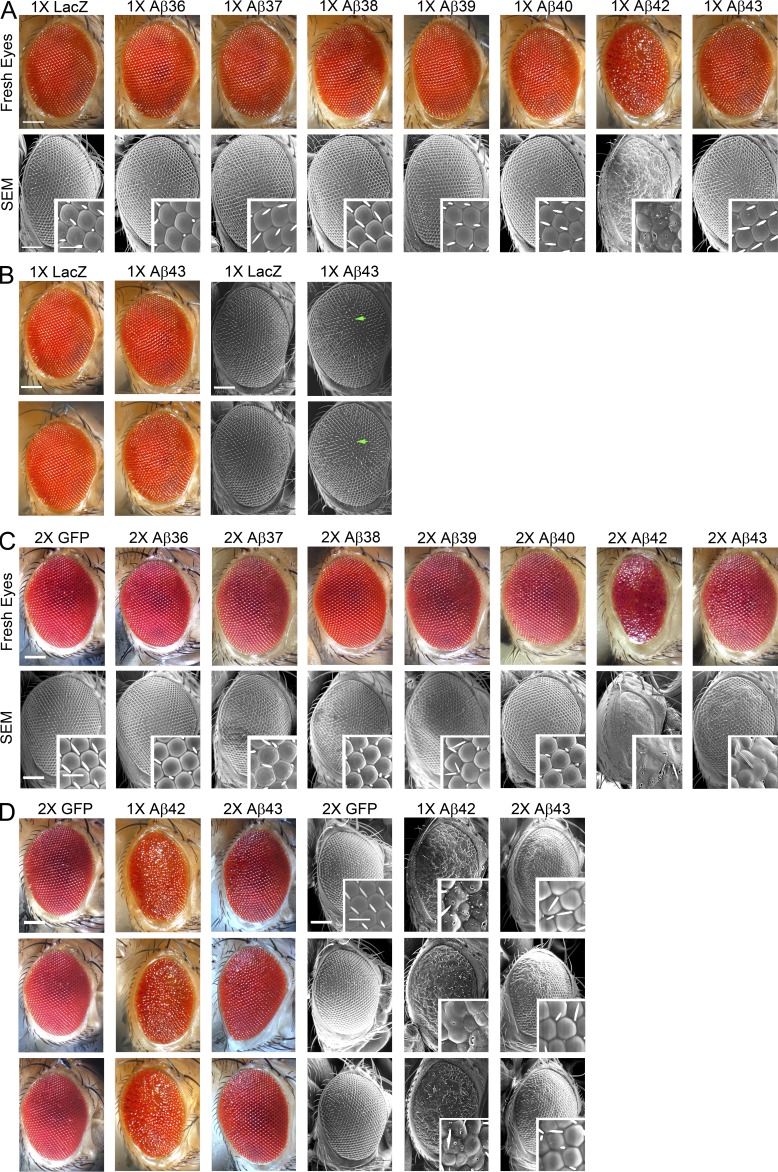
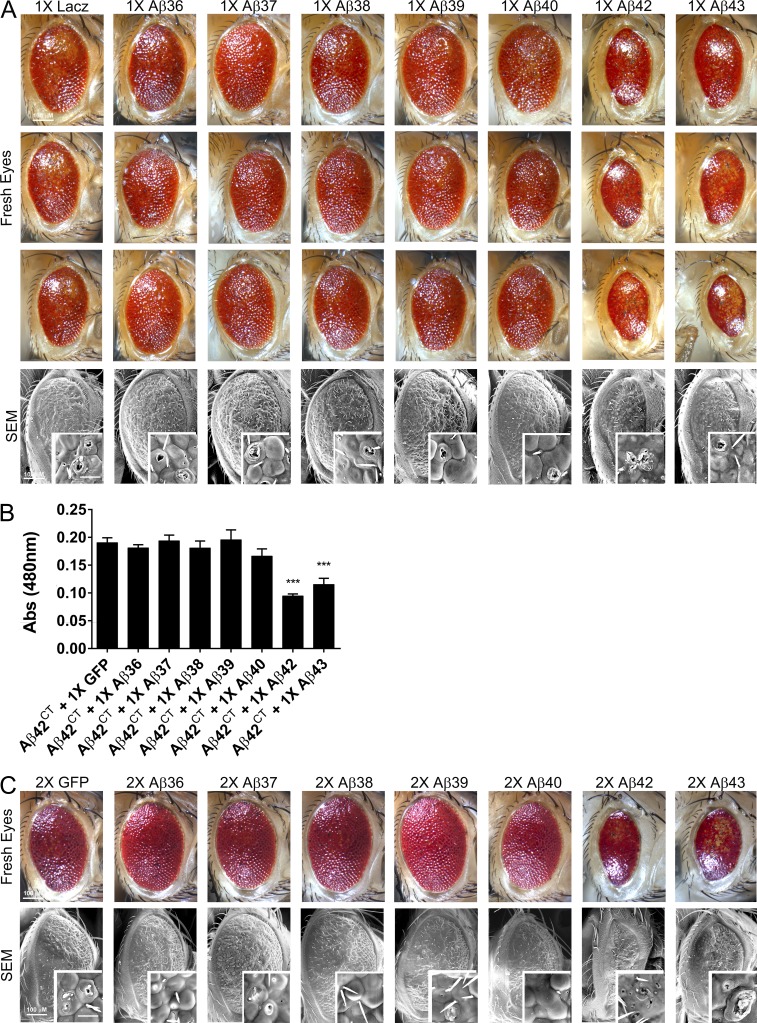
We crossed Tg Aβ flies with GMR-Gal4 to direct expression of the Aβ transgenes to the eye. The eyes of flies expressing one copy (1×) of the shorter Aβ peptides (1× Aβ36–Aβ40) were comparable to control flies (Fig. 2 A); the eyes were composed of a highly organized lattice of ommatidia and an even distribution of bristles located at the vertex of each ommatidium. In contrast, a single copy of the Aβ42 transgene induced a moderate glassy eye phenotype characterized by disorganized and fused ommatidia (Fig. 2 A, fresh eyes), holes in the ommatidia, and missing bristles (Fig. 2 A, scanning electron micrograph [SEM]). These data are similar to previously published results demonstrating that Aβ42 is toxic in flies and induces a smaller, disorganized eye (Iijima and Iijima-Ando, 2008; Casas-Tinto et al., 2011; Burnouf et al., 2015). Aβ43 flies exhibited a detectable, but quite subtle, alteration of phenotype; their eye was oval in shape and contained bristles; however, the ommatidia lattice was slightly distorted at the center of the eye (Fig. 2, A and B). To examine whether higher levels of expression would alter the phenotype, we generated flies containing two copies of the transgene encoding each Aβ peptide (2× Aβ). Eye phenotypes in the 2× Aβ36–Aβ40 flies were again indistinguishable from the control eye phenotype (Fig. 2 C). In contrast, the 2× Aβ42 flies showed markedly exacerbated eye phenotypes: the eyes were much smaller, were elliptical shaped, were composed of disorganized ommatidia that were fused together, and contained necrotic spots (Fig. 2 C). 2× Aβ43 flies showed a clearer disruption of the posterior eye characterized by fusion of the ommatidia compared with the 1× Aβ43 flies; however, the eye phenotype of the 2× Aβ43 flies was still markedly less severe than the 1× Aβ42 eye phenotype (Fig. 2 D).
Figure 2.
Aβ36-Aβ40 do not cause toxicity in the Drosophila eye. Expression of the Aβ peptides in the Drosophila eye. Aβ genotypes are indicated. (A) Representative images, fresh eyes (top), scanning electron micrographs (SEM; bottom), and higher-resolution SEMs (insets), from 1-d-old female Drosophila expressing one copy of the Aβ transgene. (B) Fresh eye images (left) and SEMs (right) of two 1× Aβ43 flies compared with 1× LacZ control. Green arrows indicate disorganized ommatidia in the 1× Aβ43 flies. (C) Representative images, fresh eyes (top), SEM (bottom), and higher-resolution SEMs (inset) from 1-d-old female Drosophila expressing two copies of the Aβ transgene. (D) Comparison of 2× Aβ43 and 1× Aβ42. Fresh eye images (left) and SEMs (right) of Drosophila expressing one copy of Aβ42, two copies of Aβ43, or two copies of GFP. Bars: 100 µm; (inset) 20 µm.
Short Aβ peptides do not induce locomotor dysfunction
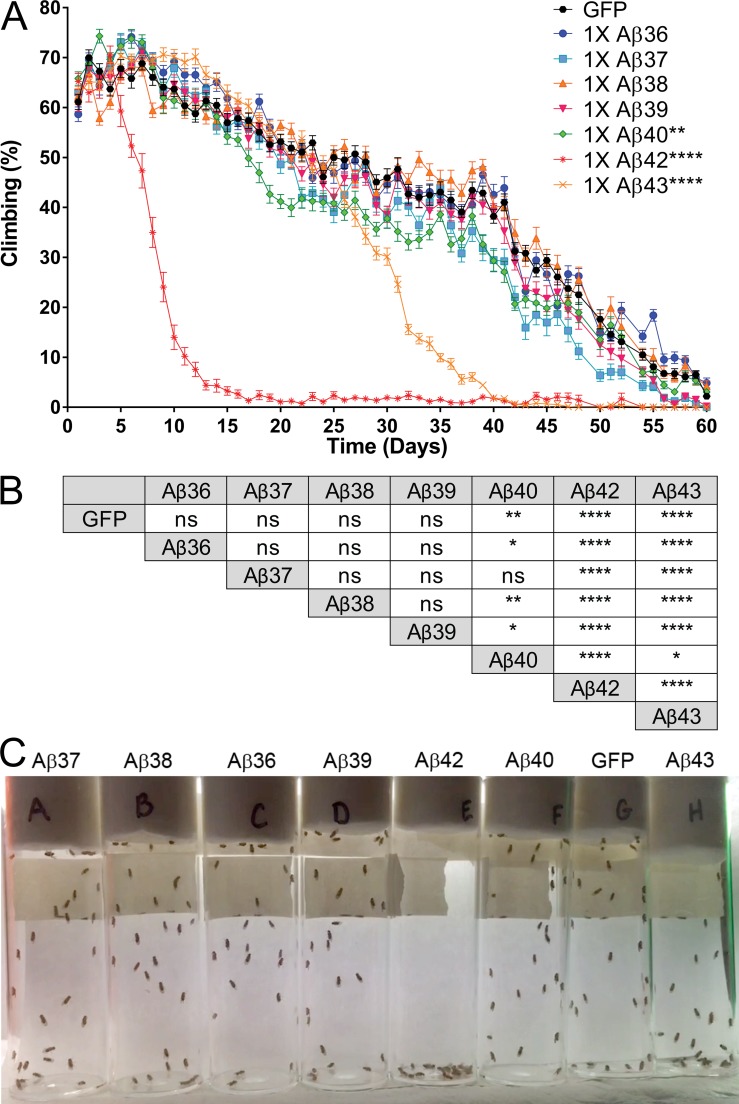
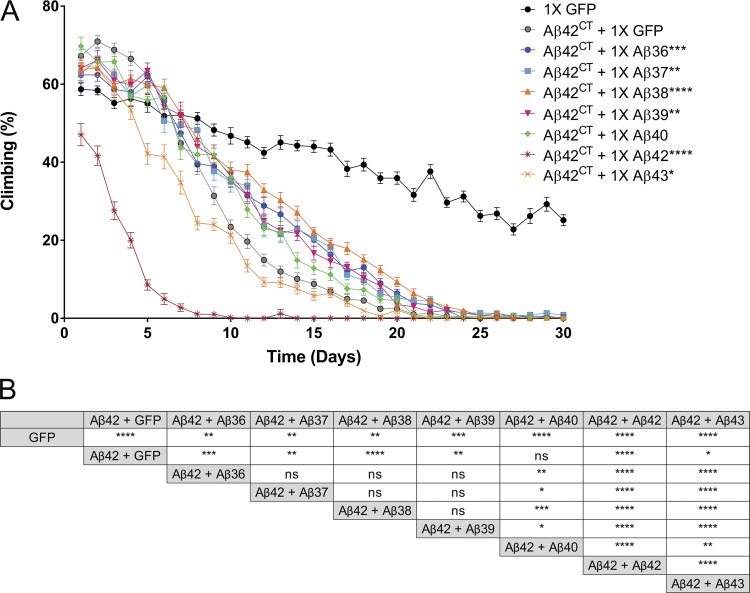
To examine the toxicity of the Aβ peptides more quantitatively, we performed a negative geotaxis assay, which uses the flies’ natural propensity to climb to the top of a vial. As flies age, they show a progressive decrease in their climbing ability and are not able to climb to the top of the vial. For this experiment, we expressed GFP-attP2 or the Aβ peptides pan-neurally under the control of ELAV-Gal4, collected female flies at 1 d of age, and blindly assayed climbing ability daily. To statistically analyze these data, we calculated the climbing index (mean percentage of flies climbing above 5 cm/total flies tested) per day, grouped the data by increments of 10 d to account for the day-to-day variance in climbing, and analyzed by survival curve comparison using the Mantel–Cox test. Flies expressing one copy of the short Aβ peptides, Aβ36 to Aβ39, climbed as well as the control flies expressing 1× GFP; after 7 d, these flies steadily decreased locomotor activity, and after 60 d, they stopped climbing (Fig. 3 A). The climbing ability of flies expressing 1× Aβ42 declined rapidly compared with 1× GFP flies, and they stopped climbing by day 13 (Fig. 3 A). 1× Aβ43 flies performed similarly to control flies until day 27, when their climbing ability decreased rapidly (Fig. 3 A). Locomotor activity was significantly, yet subtly, decreased in flies expressing 1× Aβ40 compared with 1× GFP; however, the magnitude of the effect was clearly distinct from the effect of 1× Aβ42 and 1× Aβ43 (Fig. 3, A and B). To illustrate climbing performance, we imaged climbing activity of a representative replicate from each Aβ transgene on day 40 (Fig. 3 C). At that time, 40–50% of control flies and flies expressing short Aβ peptides were climbing above the 5-cm mark. The 1× Aβ42 flies were localized at the bottom of the vial, moving their wings and legs in an uncoordinated manner resulting from the loss of climbing ability. In the vial containing 1× Aβ43 flies, only one fly of 29 expressing 1× Aβ43 climbed above the 5-cm mark; the remaining were located below the mark and on the bottom of the tube. This was representative of the five replicates of 1× Aβ43 flies.
Figure 3.
Short Aβ peptides do not impair climbing ability. To assess locomotor activity, Aβ transgenic flies were subjected to a climbing assay. (A) Longitudinal study of locomotor activity. The mean percentage of flies that climbed above 5 cm averaged from six trials was plotted over time with flies overexpressing Aβ36 (blue circle), Aβ37 (blue square), Aβ38 (orange triangle), Aβ39 (pink triangle), Aβ40 (green diamond), Aβ42 (dark red asterisk), Aβ43 (orange X symbol), and GFP (black line). n = 5, 25–30 flies/replicate. Statistical analysis of climbing performance of Aβ transgenic flies compared with control flies (GFP; **, P ≤ 0.01; ****, P ≤ 0.0001, Mantel–Cox). Values represent means ± standard error of the mean. (B) Mantel–Cox analysis of climbing performance of Aβ transgenic flies with each other (ns, P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ****, P ≤ 0.0001). (C) Snapshot of locomotor activity assay at day 40, with a representative vial for each transgene.
Short Aβ peptides do not accumulate as amyloid in Drosophila
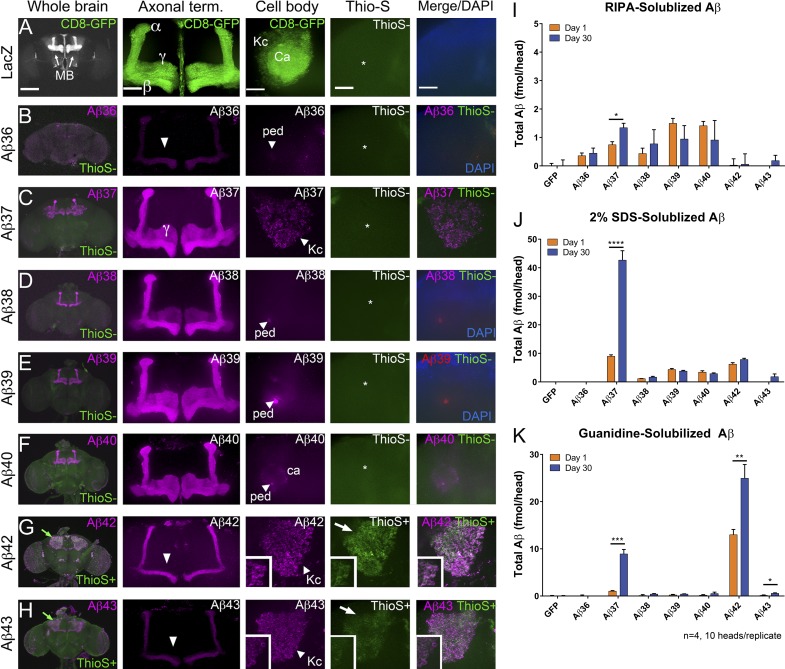
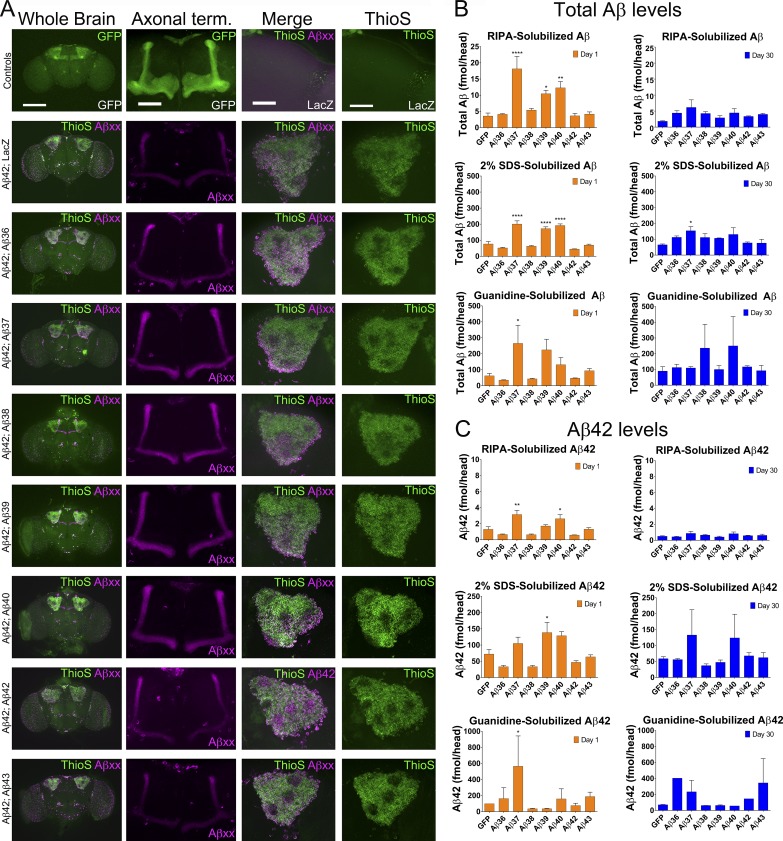
To further investigate the relationship between toxicity of Aβ peptides and accumulation, we histochemically and biochemically analyzed Tg Aβ flies in which the Aβ transgenes were expressed in the mushroom bodies (Fig. 4). 1× Aβ36 was weakly detected by immunofluorescence methods, suggesting that it may be more rapidly turned over than other short Aβs. 1× Aβ38, 1× Aβ39, and 1× Aβ40 displayed similar immunofluorescence patterns, with strong immunofluorescence in the axonal terminals but very little Aβ in the cell bodies and no evidence for Thioflavin-S (Thio-S) fluorescence (Fig. 4, D–F). In contrast to these other short Aβs, 1× Aβ37 was detected in the cell body, similar to 1× Aβ42 and 1× Aβ43, but in the 1× Aβ37 flies these accumulations were not Thio-S positive (Fig. 4, C and G). As noted in the original 82E1 immunofluorescent study (Fig. 1 B), 1× Aβ42 and 1× Aβ43 showed markedly less staining of the axons and axon terminals than 1× Aβ37–Aβ40; however, prominent cell body staining was observed with 1× Aβ42 and 1× Aβ43, and these accumulations were Thio-S positive, although the Thio-S fluorescence was stronger in 1× Aβ42 flies. To further evaluate Aβ accumulation, Aβ levels from 1- and 30-d-old flies were measured from fly heads sequentially extracted with radioimmunoprecipitation assay (RIPA) buffer, 2% SDS, or guanidinium (Fig. 4, I–K). Our expectation in these studies was that there might be significant increases over time in levels of Aβ, and especially detergent-insoluble Aβ. Although there are several significant differences in the levels of Aβ between Aβ transgenic flies and control flies at days 1 and 30 (Table S1), the a priori expectation that there might be exponential increases in detergent-insoluble Aβ levels between day 1 and day 30 was, in general, not supported by the data. Higher levels of Aβ37 (RIPA, 2% SDS, and guanidine fractions), Aβ42 (guanidine fraction), and Aβ43 (guanidine fraction) were observed at day 30 than at day 1, but these modest increases were not typical of the exponential accumulations often observed in rodent models of Aβ accumulation. Notably, the partitioning of Aβ37 was distinct from Aβ42, with the majority of Aβ37 peptide present in the SDS fraction as opposed to the guanidine fractions. This differential partitioning likely reflects a difference in the aggregation state of Aβ37 in the flies, which is also reflected by the lack of Thio-S positivity.
Figure 4.
Differential distribution of Aβ peptides in Drosophila brain. Expression of Aβ peptides or LacZ in the mushroom bodies with the promoter OK107-Gal4; UAS-CD8-GFP followed by immunohistochemistry, Thio-S staining, and biochemical analysis. (A–H) Representative brain sections (whole brain, axonal terminals, and cell body) of 1-d-old females expressing Aβ peptides, as annotated, stained with anti-Aβ mAb 33.1.1 (magenta) and Thio-S (green). MB, mushroom bodies; Kc, Kenyon cells; Ca, Calyx (dendritic terminals); Ped, pedunculus (axonal projections). Thio-S only accumulates in the cell bodies of flies expressing Aβ42 and Aβ43 (G and H, arrows). Expression of Aβ36-Aβ40 does not result in Thio-S signal (B–F, asterisks). Aβ36-Aβ40 accumulate mainly in the axons (B and D–F, arrowheads) and axonal terminals of the mushroom body neurons with high levels in the γ lobes (C–F). Aβ36, Aβ42, and Aβ43 accumulate at low levels in the γ lobes (B, G, and F, arrowheads). Aβ37, Aβ42, and Aβ43 accumulate at high levels in the cell bodies of the Kenyon cells (B, G, and F, arrowheads). Bars: (whole brain) 200 µm; (axonal term., cell body, Thio-S, and merge/DAPI) 50 µm. (I–K) Sequentially extracted total Aβ levels from 1- and 30-d-old females. Data are presented as femtomoles/head. (I) RIPA-extracted Aβ. (J) 2% SDS-extracted Aβ. (K) Guanidinium-extracted Aβ. A total of 40 fly heads were analyzed per transgenic fly, in four pools of n = 10 biological replicates. Statistical analysis of Aβ levels between two time points of the transgenic flies is displayed (*, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001, unpaired t test). Values represent means ± standard error of the mean. Statistical comparison of Aβ levels between transgenic flies is presented in Table S1.
Because the biophysical properties of short Aβ peptides have not been studied as extensively as Aβ42, 43, and 40, we evaluated how the shorter Aβ peptides aggregate in vitro. Notably, under conditions in which synthetic Aβ42, 43, and 40 aggregate into Thioflavin T–positive structures that demonstrate a β-pleated sheet conformation as assessed by circular dichroism spectroscopy, under the conditions and times studied, synthetic Aβ36, 37, and 38 do not form Thioflavin T–positive assemblies in vitro and maintain random coil conformation during conditions favorable for aggregation (Fig. S1). In these assays, Aβ39 did show a slight increase in Thioflavin T fluorescence over time and also an intermediate structure between random coil and β-pleated sheet.
Short Aβ peptides partially rescue the Aβ42-induced eye phenotype
Having established that short Aβ peptides are not toxic in the fly models when individually expressed, we generated Aβ combinations to determine whether the short Aβ peptides could alter the Aβ42-induced eye phenotype. Tg 1× Aβ flies were crossed with flies expressing Aβ42 from a bicistronic construct, as described by Casas-Tinto et al. (2011) (Aβ42CT), to coexpress one copy of the shorter Aβ peptides with high levels of Aβ42. Flies containing 1× Aβ36–Aβ40 with Aβ42CT displayed eyes with subtly improved eye phenotype relative to those expressing Aβ42CT and LacZ (Fig. 5 A). The eyes were larger and oval shaped and composed of a defined ommatidia subunit; however, the ommatidia lacked organization. Coexpression of Aβ42CT with 1× Aβ42 exacerbated the eye phenotype. These eyes appeared smaller and elliptical in shape; the ommatidia structure was deteriorated; and the bristles were distributed irregularly (Fig. 5 A). Coexpression of Aβ42CT with 1× Aβ43 also enhanced the degenerative eye phenotype, but not to the same degree as the 1× Aβ42 cross (Fig. 5 A). To quantify the effects of the Aβ peptides on the toxicity of Aβ42 in the eye, we extracted eye pigments and quantified the red pigments as an indirect measure of the eye size and organization. Although this assay was not sensitive enough to distinguish between controls expressing Aβ42CT and LacZ or GFP versus the short Aβ peptides, we detected a dramatic reduction in the amount of red pigments in flies coexpressing Aβ42CT and 1× Aβ42 or 1× Aβ43 (Fig. 5 B).
Figure 5.
Aβ36-Aβ40 partially rescue Aβ42 induced eye toxicity. Coexpression of Aβ42CT with Aβ peptides in 1-d-old Drosophila eye. (A) Three images of eyes containing Aβ42CT with 1× Aβ peptides, fresh eyes (top three panels), scanning electron micrographs (SEMs; bottom), and higher-resolution SEMs (inset). (B) Relative quantification of eye pigments from Drosophila coexpressing Aβ42CT and Aβ peptides. Eye pigments were significantly decreased in Aβ42 and Aβ43 flies compared with control flies, Aβ42CT + 1× GFP. Data represent mean absorbance ± standard error of the mean. 15 fly heads per transgene were analyzed in three groups of five biological replicates. (***, P < 0.001, one-way ANOVA with Dunnett’s multiple comparison test). (C) Representative images of eye containing Aβ42CT with 2× Aβ peptides, fresh eyes (top), SEMs (bottom), and higher-resolution SEMs (inset). Bars: 100 µm; (inset) 20 µm.
To further examine the effects of the shorter Aβ peptides, we generated transgenic flies that coexpress two copies of the Aβ peptides with Aβ42CT. 2× Aβ36–Aβ40 further improved the Aβ42CT eye phenotype (Fig. 5 C). Coexpression of Aβ42CT with either 2× Aβ42 or 2× Aβ43 further worsened the eye phenotype compared with the control (Fig. 5 C); eyes were even smaller and elliptical in shape, and the ommatidia subunits were fused together, with depigmentation (Fig. 5 C). In this case, the difference between the 2× Aβ42 and 2× Aβ43 crosses was more easily observed, with 2× Aβ42 showing a more severe phenotype.
Short Aβ peptides protect against Aβ42-induced locomotor dysfunction
To evaluate whether shorter Aβ peptides altered Aβ42 toxicity in the negative geotaxis assay, we crossed 1× Aβ transgenic flies with Aβ42CT and measured climbing ability. Coexpression of 1× Aβ36–Aβ39 with Aβ42CT significantly improved locomotor activity (Fig. 6, A and B). Alternatively, expression of Aβ42CT with 1× Aβ42 induced rapid locomotor dysfunction; only 50% of flies were climbing at day 1, and by day 8 they had stopped climbing altogether (Fig. 6 A). Flies coexpressing Aβ42CT and 1× Aβ43 also showed accelerated locomotor dysfunction relative to controls, but not as accelerated as observed with 1× Aβ42 (Fig. 6 A). The addition of 1× Aβ40 did not alter the climbing activity of Aβ42CT; there was no significant difference between flies coexpressing Aβ42CT + 1× Aβ40 and control (Aβ42CT + 1× GFP; Fig. 6, A and B).
Figure 6.
Aβ36-Aβ40 partially rescue Aβ42 induced locomotor dysfunction. Climbing ability of female flies coexpressing Aβ42CT and 1× Aβ peptides was measured daily. (A) The mean percentage of flies climbing above 5 cm from six trials was plotted over time with five replicates of flies coexpressing Aβ42CT and GFP (gray circle), Aβ36 (dark blue circle), Aβ37 (blue square), Aβ38 (orange triangle), Aβ39 (pink triangle), Aβ40 (green diamond), Aβ42 (dark red asterisk), Aβ43 (orange asterisk), and GFP alone (black circle). n = 5, 15–25 flies/replicate. Statistical analysis of climbing performance of Aβ transgenic flies compared with control flies (Aβ42H + GFP; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001, Mantel–Cox). Values represent means ± standard error of the mean. (B) Mantel–Cox analysis of climbing performance of Aβ transgenic flies with each other (ns, P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001).
Lack of correlation between Aβ deposition and toxicity in the crossed flies
To try to gain insight into the protective effect of these shorter peptides, we evaluated coexpression of Aβ42CT with 1× Aβ36–Aβ40 in mushroom body neurons, which enabled us to assess the distribution and aggregation of Aβ. Coexpression of short peptides did not dramatically alter the distribution of Aβ assessed by either immunofluorescence or Thio-S staining (Fig. 7 A). Biochemical studies on flies aged 1 or 30 d showed that many of the crosses resulted in altered distribution of Aβ in the various biochemical fractions (Fig. 7, B and C). Despite these biochemically detectable alterations in Aβ accumulation, there was no strong correlation between Aβ deposition and toxicity. This lack of correlation is easily illustrated by comparing the effects of expression of Aβ36 and Aβ37. Both peptides were equally protective in functional assays, but expression of Aβ36 did not alter Aβ deposition, whereas expression of Aβ37 had the most dramatic effect on Aβ accumulation (Fig. 7, B and C).
Figure 7.
Aβ36-Aβ40 do not alter neuronal distribution of Aβ42 and have variable effects on Aβ42 accumulation in the mushroom bodies. Coexpression of Aβ42CT with 1× Aβ peptides or LacZ, as illustrated, in the mushroom bodies with the promoter OK107-Gal4; UAS-CD8-GFP. (A) Representative brain sections (whole brain, axonal terminals, and cell bodies) of 1-d-old females imaged by GFP, stained with anti-Aβ mAb 33.1.1 (magenta) and Thio-S (green). Bars: (whole brain) 200 µm; (axonal term., merge, and Thio-S) 50 µm. (B and C) Sequentially extracted total Aβ (B) or Aβ42 (C) levels from 1- and 30-d-old female flies. Data are presented as femtomoles/head; the y-axis is scaled differently in the RIPA-solubilized total Aβ graphs. Data from 1- and 30-d-old flies are presented. Aβ42 data are presented as femtomoles/head with different y-axis scaling based on extraction (C). Data from 1- and 30-d-old flies are presented. 40 fly heads per transgenic fly were analyzed in four groups of n = 10 biological replicates. Statistical analyses of Aβ levels in transgenic flies were compared with GFP flies (ns [not significant], P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ****, P ≤ 0.0001, one-way ANOVA with Dunnett’s multiple comparison test). Values represent means ± standard error of the mean.
Aβ38 does not deposit when expressed in the brain of mice
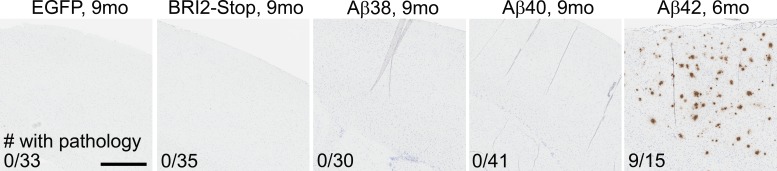
To examine whether short Aβ peptides might produce a phenotype in mice, we generated rAAV2/1 vectors encoding EGFP, BRI2-Aβ38, BRI2-Aβ40, BRI2-Aβ42, and a BRI2-Stop truncated at the furin cleavage site. We had previously used the BRI2-Aβ fusion system to produce transgenic animals that express either Aβ40 or Aβ42 and shown that rAAV2/1 vectors expressing BRI2-Aβ42 could drive amyloid deposition in the brain of nontransgenic rats (Lawlor et al., 2007). We first confirmed that the BRI2-Aβ38 vectors generate only Aβ38 (Fig. S2 A), then proceeded to inject a large number of postnatal day 0 (P0) mice with the various BRI2 constructs and appropriate controls. Overexpression of Aβ42 for only 6 mo resulted in amyloid deposition in nine of 15 NTg mice (Fig. 8). Conversely, despite higher mean expression levels of BRI2-Aβ38 or BRI2-Aβ40 transgenes as assessed by measurement of the BRI2 fusion protein (Fig. S2, B and C), expression of Aβ38 or Aβ40 did not result in accumulation of Aβ deposits (Fig. 8). χ2 testing showed a strong relationship between Aβ peptide length and pathology (χ2 [4, n = 154] = 154.0; P < 0.001).
Figure 8.
Aβ38 does not deposit. Neonatal P0 CRND8 mice were injected with either rAAV-Aβ38, Aβ40, BRI2-Stop (control), or EGFP (control) in the cerebral ventricles and analyzed after 9 mo. Non-Tg mice injected with rAAV-BRI2-Aβ42 were analyzed at 6 mo of age. (A) Representative cortex sections of 9-mo-old NTg littermates overexpressing EGFP, BRI2-Stop, Aβ38, and Aβ40 and 6-mo-old NTg mice overexpressing Aβ42 stained for Aβ plaque pathology with Aβ mAb 33.1.1. Number of mice with pathology relative to total number of mice analyzed is reported. Bar, 250 µm.
Aβ38 lowers plaque load and does not alter behavioral phenotypes in non-Tg and APP mice
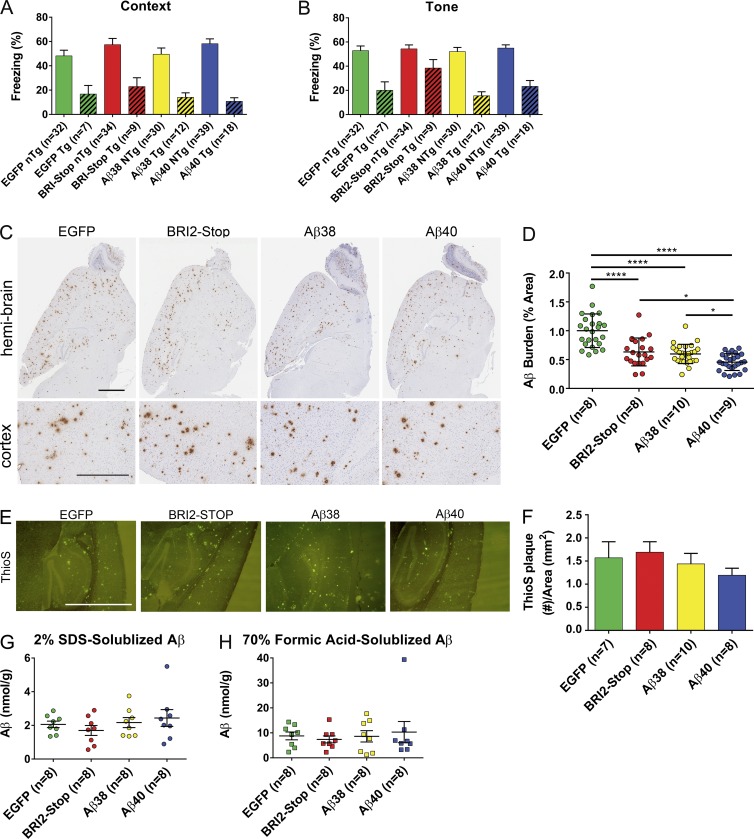
We had previously shown that rAAV-mediated expression of BRI2-Aβ40 from P0 to 3 mo of age reduced amyloid loads in TgCRND8 mice (Kim et al., 2007). To examine whether Aβ38 expressed from rAAV vectors encoding BRI2-Aβ38 altered phenotypes in TgCRND8 mice, we expressed rAAV-BRI2-Aβ38, rAAV-BRI2-Aβ40, rAAV-BRI2-Stop, and rAAV-EGFP vectors from P0 to 9 mo in TgCRND8 mice and non-Tg (NTg) littermates. At 9 mo of age, the mice were behaviorally tested in a fear-conditioning paradigm, as 9-mo-old CRND8 mice exhibit robust deficits in contextual fear memory or tone-conditioned fear memory (Hanna et al., 2012). Control groups included mice transduced with rAAV-BRI2-stop, rAAV-BRI2-Aβ40, and rAAV-EGFP. The analysis of the conditioned context fear memories and tone fear memories of TgCRND8 and NTg littermate mice revealed that Aβ38 and Aβ40 had no effect on behavior in either TgCRND8 mice or their NTg littermates (Fig. 9 A). Although expression of BRI2-Stop resulted in a trend toward increased freezing in the tone test of TgCRND8 mice, this effect was not robust enough to be significant given the correction for multiple group testing (Fig. 9 B). Because of the large group sizes in the NTg mice, it is clear that expression of Aβ38 and Aβ40 had no effect on fear conditioning–based learning and memory.
Figure 9.
Aβ38 reduces Aβ deposition but has no effect on Aβ levels or behavior in TgCRND8 mice. (A and B) CRND8 mice injected with rAAV-Aβ38, Aβ40, BRI2-Stop (control), or EGFP (control) were subjected to contextual fear conditioning at 9 mo of age. Mean percentage freezing ± standard error of the mean exhibited by TgCRND8 and NTg littermate overexpression of EGFP, BRI2-Stop, Aβ38, or Aβ40. (A) Context paradigm. (B) Tone test. n = 7–18/Tg, n = 30–39/NTg mice per group. (C) Representative brain sections (hemi-brain [top] and cortex [bottom]) of TgCRND8 mice overexpressing EGFP, BRI2-Stop, Aβ38, and Aβ40 stained with anti–Aβ mAb 33.1.1. Bars: 500 µm (hemi-brain); 200 µm (cortex). (D) Quantification of Aβ plaque deposits immunostained with anti–Aβ Ab5. Data represent mean ± standard error of the mean; n = 7–10 mice per group (*, P < 0.05; ****, P < 0.0001, one-way ANOVA with Dunnett’s multiple comparison test). (E) Representative brain sections of TgCRND8 mice overexpressing EGFP, BRI2-Stop, Aβ38, and Aβ40 stained with Thio-S. Bar, 2.00 mm. n = 7–10 mice per group. (F) Quantification of Thio-S plaques per area measured. Data represent mean ± standard error of the mean; n = 7–10 mice per group. (G and H) Biochemical analysis of sequentially extracted Aβ42 and Aβ40 levels by end-specific sandwich ELISA. (G) 2% SDS-extracted Aβ. (H) 70% formic acid–extracted Aβ. Data represent mean ± standard error of the mean (n = 7–19 per group).
After behavioral experiments, mice were killed, and brain Aβ burden was examined in a subset of mice by immunohistochemical (IHC; Fig. 9 C) and biochemical methods (Fig. 9, G and H). NTg mice from these groups were included in the analysis described above (Fig. 8) and showed no deposition phenotype. Blinded quantification of Aβ IHC burden revealed subtle differences between the groups, with BRI2-Aβ40 showing an almost 60% decrease in Aβ IHC burden (P < 0.001) and BRI2-stop and BRI2-Aβ38 showing 33% and 35% reductions in Aβ IHC burdens, respectively (P < 0.05; Fig. 9 D). In contrast to the histochemical burden analyses, we saw no significant difference in Aβ40 or Aβ42 levels in the 2% SDS–solubilized or 70% formic acid–solubilized Aβ from mice overexpressing short Aβ peptides or BRI2-stop (Fig. 9, G and H, respectively). Given this unexpected discrepancy, we evaluated Thio-S plaque load, which measures only cored plaque. In this blinded analysis, we saw no difference in Thio-S plaque loads (Fig. 9, E and F). Collectively, the presence of an effect on Aβ IHC burdens coupled with a lack of effect on biochemical loads and Thio-S burden would be consistent with an effect of BRI2-Stop, BRI2-Aβ38, and BRI2-Aβ40 on more diffuse Aβ deposits that do not contribute significantly to the overall amount of Aβ either detected biochemically or deposited as amyloid.
We did conduct studies to explore the feasibility of using rAAV-BRI2 fusion protein system to express Aβ36, 37, and 39. Unfortunately, despite multiple attempts, we were unable to achieve sufficient expression levels of the transgenes from these rAAV-BRI2-short Aβ vectors to draw any firm conclusions on the effects of these peptides in mouse brains.
Discussion
We have examined the pathobiological activity of shorter Aβ peptides in Drosophila. These data establish that the short Aβ peptides, Aβ36, Aβ37, Aβ38, and Aβ39, are not toxic in Drosophila and partially protect, in a dose-dependent fashion, against Aβ42 toxicity. Although Aβ40 does not accumulate by itself and protects from Aβ42 toxicity, its effects are statistically distinguishable from those of the other short Aβ peptides. Aβ40 expression slightly impairs climbing performance by itself and has a less protective effect on Aβ42-induced climbing dysfunction than the shorter Aβ40 peptides. These findings are reminiscent of a recent study showing a very subtle effect on survival, but not climbing, of Aβ40 expressed neuronally (Jonson et al., 2015). Our finding that Aβ43 expression is less toxic than Aβ42 is somewhat unexpected given recent studies with select PSEN mutations (e.g., PSEN1 L435F, R278I, C410Y) that increased Aβ43 and produced rather dramatic amyloid phenotypes in mice and humans (Saito et al., 2011; Kretner et al., 2016; Veugelen et al., 2016); however, the data are consistent with the study by Burnouf et al. (2015), who also found Aβ42 to be more toxic than Aβ43. Because Aβ43 does seem to synergistically increase Aβ42 toxicity in Drosophila, one possible explanation for these studies is that the PSEN mutations L435F and R278I still do produce Aβ42. Thus, the dramatic amyloid phenotype in patients could be attributed to the combination of the two peptides depositing in some synergistic fashion. Future studies looking at effects of Aβ43 expressed by itself or in combination with Aβ42, in mice using the BRI2 fusion strategy, could help to clarify this discrepancy.
In contrast to the other short Aβ peptides, expression of Aβ37 did result in Aβ accumulation that was detectable by biochemical methods and by immunofluorescence. These deposits were not Thio-S positive, were more SDS soluble than Aβ42 deposits, and were not toxic. Collectively, these studies and those from others suggest that expression of Aβ peptides with an enhanced propensity to form β-pleated sheet aggregates in vitro and the ability to form Thio-S–positive deposits is, to some degree, associated with toxicity in the Drosophila model (Iijima and Iijima-Ando, 2008; Casas-Tinto et al., 2011; Burnouf et al., 2015). Further, it is clear that the in vitro studies of pure synthetic Aβ do not capture the complex in vivo interactions that may modulate toxicity in any model system. For example, we find that although synthetic Aβ40 can form amyloid structures in vitro, in vivo it appears to have minimal toxicity and does protect from Aβ42. Nevertheless, when expressed by itself, Aβ40 slightly impairs climbing ability relative to several of the shorter Aβs and is also less protective from Aβ42 than the other short Aβs. Biochemical solubility measures of the deposited Aβ along with Thio-S staining appear to show that propensity to form amyloid is a good surrogate for toxicity in the flies expressing only a single species of Aβ. However, such measures are insufficient to provide insight into why the shorter Aβ peptides partially protect from Aβ42 toxicity. Indeed, we conducted standard studies designed to explore why the short Aβ peptides attenuate Aβ42 toxicity by examining how expression of the short peptides altered Aβ accumulation. Unfortunately, these studies were not particularly informative. Distribution and Thio-S positivity of Aβ42 are not altered to any appreciable degree by expression of the shorter peptides. Biochemical analysis did reveal that the presence of the short Aβ peptides increased total Aβ and Aβ42 in multiple biochemical fractions in some of the short Aβ + Aβ42 combinations; however, altered biochemical levels did not correlate with alterations in protective effects. Given these data and the overall lack of precise mechanistic insight into Aβ toxicity in Drosophila models (Shulman et al., 2003), we can only speculate that short Aβ directly interact with Aβ42 in some subtle way to alter either the aggregate structure or the kinetics of aggregation to reduce toxicity.
Despite such limitations, these observations support a growing body of literature that subtle differences in the structure of Aβ assemblies can influence the biological activity of the aggregate (Tycko, 2015; Selkoe and Hardy, 2016). Further, it is clear that despite more than two decades of intensive study, large gaps in our knowledge remain regarding precise correlations between structure and toxicity of Aβ aggregates.
Selective expression of Aβ38 or Aβ40 from the BRI2 fusion protein in the brains of mice did not lead to amyloid deposition. Transgene expression from these rAAV vectors was, on the mean, equal or higher to the expression of Aβ42 in mice injected with AAV-BRI2-Aβ42. Further, the BRI2-Aβ42 were analyzed at 6 mo of age, whereas the mice expressing Aβ38 were analyzed at 9 mo of age. These data suggest that in mammalian systems, Aβ38 does not deposit on its own when expressed at levels sufficient to drive Aβ42 deposition.
The effects of Aβ38 from the rAAV-expressed BRI2 fusion proteins in APP TgCRND8 mice are somewhat more complex. First, the data suggest that BRI2 expression itself can have a subtle effect on Aβ deposition in vivo. Expression of the BRI2 containing constructs in the brain resulted in lower Aβ IHC burden. As Thio-S–positive amyloid plaque load or biochemical Aβ loads were not altered, we interpret this data to mean that the effect was restricted to diffuse Aβ deposits detectable by IHC that do not contribute significantly to biochemical or Thio-S plaque loads. These findings are consistent with publications suggesting that the extracellular BRICHOS domain of BRI2 can alter Aβ aggregation dynamics (Peng et al., 2010; Willander et al., 2012; Hermansson et al., 2014; Biverstål et al., 2015; Poska et al., 2016). Second, the additional lowering of Aβ IHC burden by BRI2-Aβ40, relative to BRI2-Stop or BRI2-Aβ38, would be consistent with our previous data showing that Aβ40 can inhibit Aβ42 accumulation in both Tg2576 and 3-mo-old TgCRND8 mice (Kim et al., 2007, 2008). Third, although these effects on reduction of Aβ IHC burden are intriguing, they did not result in any alteration in memory and learning as assessed in a fear-conditioning paradigm. Overall, these data suggest that expression of Aβ38 in an aggressive model of amyloid deposition has little effect on the phenotype at the time AD-like plaque loads and robust behavioral deficits are observed.
These studies are therapeutically relevant, as GSMs that increase short Aβ peptides remain in both clinical and preclinical development for AD (Wagner et al., 2012; Golde et al., 2013). As emerging clinical data continue to reinforce the hypothesis that Aβ production inhibitors are likely to be most effective as prophylactic therapies to prevent development of AD, safety of the inhibitor becomes of paramount importance (Golde et al., 2011; Golde, 2016). These studies provide clear-cut preclinical data that short Aβ peptides are not themselves toxic and in certain circumstances may also be protective. These data further reinforce the assertion that the GSM mechanism of action is inherently safe and suggest that, at least in prevention settings, increasing short Aβ peptides might be beneficial. It is not clear that an optimal GSM that avoids off-target toxicity has yet been developed (Golde et al., 2013). However, given that target-based toxicity remains a potential safety concern for BACE1 inhibitors (Willem et al., 2009; Barão et al., 2016), these and other data support continued efforts to both evaluate current best-in-class GSMs (Wagner et al., 2014; Brendel et al., 2015; Blain et al., 2016) and develop novel GSM for AD prophylaxis.
Materials and methods
Generation of transgenic flies expressing Aβ36–Aβ40, Aβ42, and Aβ43
To generate transgenic flies expressing comparable levels of Aβ, cDNA fragments encoding human Aβ1–36, Aβ1–37, Aβ1–38, Aβ1–39, Aβ1–40, Aβ1–42, and Aβ1–43 peptides fused to the Argos signal peptide for secretion were cloned under the control of UAS in the Drosophila pJFRC-MHU vector carrying an attB site for site-directed integration. pJFRC-MUH was a gift from G. Rubin (plasmid #26213; Addgene; Pfeiffer et al., 2010). The resulting constructs were microinjected into yellow white (yw) embryos at Rainbow Transgenics (Camarillo, CA) and targeted to the same genomic location, the attP2 site, to achieve similar expression levels in vivo. At least two transgenic lines for each Aβ construct were established. The flies were raised and maintained at 25°C in regular media. To express the Aβ constructs, we combined these transgenic lines with several Gal4 drivers, including daughterless (da)-Gal4 (ubiquitous), glass multimer reporter (GMR)-Gal4 (all eye cells), OK107-Gal4 (mushroom bodies), and ELAV-Gal4 (pan-neural; Fig. S3). We also used flies expressing high levels of Aβ42 (Aβ42CT) from a construct carrying two copies of Aβ42 that we described previously (Casas-Tinto et al., 2011). As controls, we used the reporters UAS-LacZ and UAS-GFP-attP2 from the Bloomington Drosophila Stock Center. To induce high levels of the Aβ peptides, all crosses were incubated at 28°C, and adult females were collected at day 1 and either processed or aged at 28°C, unless otherwise noted.
RNA isolation and analysis
To examine mRNA expression levels from the Aβ constructs, we crossed the transgenic flies with da-Gal4 at 25°C. Adult female flies were collected at day 1 and snap-frozen for RNA extraction. Total RNA from 21 flies, in three pools of seven biological replicates, was purified using RNeasy Mini kit (Qiagen) and reverse transcribed using Superscript III (Thermo Fisher Scientific). Real-time Q-RT-PCR was performed using a custom primer/probe mix (Table S2), amplifying the Aβ peptides with the Argos probe and β-tubulin and the ribosomal protein L32 as internal controls. Probes (Roche Universal Probe library) were labeled at the 5′ end with fluorescein (FAM) and at the 3′ end with dark blue quencher. Target-specific primer sequences were ordered from IDT. The PCR (initial denaturation cycle 95°C/30 s, followed by 39 amplification cycles of 95°C/5 s and 60°C/5 s) was done using SSoFast EvaGreen Supermix (Bio-Rad Laboratories).
Drosophila eye imaging
We crossed all Aβ transgenic flies with GMR-Gal4 or GMR-Gal4;Aβ42 at 25°C for 2 d; the progeny were raised at 28°C, and we collected females at day 1. To image fresh eyes, we froze the flies at −80°C for at least 24 h and collected images as z-stacks with a Leica Z16 APO using a 2× Plan-Apo objective. Flattened in-focus images were produced with the Montage Multifocus module of the Leica Application Software. For scanning electron microscopy, flies were serially dehydrated in ethanol, air-dried in hexamethyldisilazane (Electron Microscope Sciences), and metal-coated for observation in a Jeol 1500 SEM.
Negative geotaxis assay
To assess the function of the Aβ peptides on behavior, the Aβ transgenic flies were crossed with ELAV-Gal4 flies to direct expression of the transgene to the neurons. The crosses were initially placed at 25°C for 2 d; the progeny were raised at 26°C, and female flies were collected at 1 d of age. The progeny were subjected to daily climbing assays that used the natural propensity of flies to exhibit negative geotaxic response (Bainton et al., 2000; Feany and Bender, 2000; Friggi-Grelin et al., 2003; Rival et al., 2004; Fernandez-Funez et al., 2009, 2016; Sofola et al., 2010; Zhang et al., 2014). In brief, each day, 25–30 flies were placed into empty vials (9.5 cm high, 1.7 cm in diameter) with flat bottoms, and their subsequent climb to the top of the vial was analyzed. Five replicates (vials containing 25–30 flies) were prepared for each construct. At the beginning of each test session, the flies were forced to the bottom of a vial by firmly tapping the vial against the bench surface. Eight seconds after the final tap, the number of flies that climbed up the walls of a vial above the 5-cm mark was recorded. Each session consisted of six trials repeated with the interval of 15 s. Scores recorded were the mean number of flies climbing to the criterion during each daily session. During the test, the observer was not aware of the genotype of each tested batch of flies.
Immunofluorescence
To examine the distribution of the Aβ peptides in the mushroom body neurons, we crossed Aβ transgenic flies with flies expressing CD8-GFP under the control of OK107-Gal4. We also combined flies expressing Aβ42 in the mushroom bodies (Aβ42; OK107-Gal4) with LacZ (negative control) or the Aβ peptides. All these crosses were placed at 28°C to maximize expression of the transgenes. We collected adult flies at day 1 post-eclosion and aged them for 30 d. We then imaged the distribution of Aβ and GFP at days 1 and 30 by dissecting brains, which were fixed in 4% formaldehyde, washed with PBS, blocked with 3% BSA, and mounted as described previously (Fernandez-Funez et al., 2000). To detect each Aβ peptide, we used the Aβ antibody 82E1 (IBL) followed by anti–mouse Cy3 (Molecular Probes) at 1:300. After washing the secondary antibody, tissues were mounted on Vectashield antifade (Vector).
Thioflavin-S staining
We generated flies expressing LacZ alone, each Aβ peptide alone, or combined with Aβ42 under the control of OK107-Gal4 at 25°C. Then, we fixed 1- and 30-d-old brains, incubated them with a 0.03% solution of freshly prepared and filtered Thio-S (Sigma-Aldrich) in 50% ethanol/PBS for 10 min, washed, and mounted in Vectashield.
Microscopy and image processing
We collected fluorescent images with AxioVision (Zeiss) in an Axio-Observer Z1 microscope (Zeiss) by optical sectioning using ApoTome (structured light microscopy) with 10× NA: 0.45 (air), 20× NA: 0.7 (air), 40× NA: 1.4 (oil), and 63× NA: 1.4 (oil) objectives. Maximum-intensity projection images (Aβ distribution) were created in AxioVision (geometric processing, orthoview) from z-stacks containing complete structures. Representative single plane images of Kenyon cells (Thio-S) were extracted from z-stacks. Image processing was minimal but included brightness/contrast adjustment to whole images for optimal viewing and printing.
Aβ extraction and quantification by ELISA (Drosophila)
In brief, for ELISA, 1- and 30-d-old females were frozen, and a total of 40 heads were cut and aliquoted to four tubes containing 10 heads per transgene. The tissue was sequentially extracted in protease inhibitor cocktail (Roche) containing 50 µl RIPA buffer, 2% SDS, and 5 M guanidinium/50 mM Tris-HCL (Gn-HCl), as previously described (Kawarabayashi et al., 2001). RIPA-, 2% SDS–, and Gn-HCl–extracted samples were diluted appropriately and used for sandwich ELISAs as described previously (Kim et al., 2008). Total Aβ was captured with mAb Ab9 (human Aβ1-16 specific; T.E. Golde) and detected by HRP-conjugated mAb 4G8 (Covance). Aβ42 was captured with mAb 2.1.3 (human Aβ35-42 specific; T.E. Golde) and detected by HRP-conjugated mAb 4G8 (Covance).
Aβ aggregation assay
Aβ peptides, Aβ36–40, Aβ42, and Aβ43 (Anaspec) were pretreated. In brief, they were solubilized in hexafluoro-2-propanol (Sigma), dried by SpeedVac, stored at −20°C, and used within 2 wk. Reactions were initiated in siliconized Eppendorf tubes by adding 100 µM monomeric Aβ36–40, Aβ42, or Aβ43 to reaction buffer (20 mM Tris-HCl and 150 mM NaCl, pH 8.0) and incubated with shaking at 37°C. Aggregation was monitored by Thioflavin T fluorescence. In brief, at specific time points, an aliquot of the Aβ reaction mixture was diluted 15-fold in buffer containing 5 mM Thioflavin T and 5 mM Tris HCl, pH 8.0, and fluorescence (excitation 415 nm, emission 487 nm) was measured (FlexStation3; Molecular Devices; Rangachari et al., 2007). Circular dichroism spectra were obtained using an Aviv Model 430 circular dichroism spectrometer (AVIV biomedical). Scans were performed at 25°C using a cuvette with a path length of 0.1 cm. Instrument optics and lamp chamber were purged with nitrogen gas at a rate sufficient to maintain oxygen levels less than 7 ppm. Each scan was performed in the far-UV range of 260–190 nm, with readings recorded at 1-nm intervals. Data were analyzed with CAPITO and K2D program (Perez-Iratxeta and Andrade-Navarro, 2008; Wiedemann et al., 2013).
Eye pigment extraction
To quantify red pigments, 1-d-old females were immediately frozen. A total of 15 heads per genotype were analyzed, in three groups of five biological replicates. Five heads per transgene were placed into a vial (in triplicate) and eye pigments were extracted by homogenization and overnight incubation in 200 µl of 30% ethanol, pH 2.0 (Falcón-Pérez et al., 2007). The next day, the relative quantification of the red pigments was determined by measuring their absorption at 480 nm (FlexStation3).
Animal models and AAV injection
All animal procedures were approved by the Institutional Animal Care and Use Committee in accordance with NIH guidelines. TgCRND8 mice overexpressing amyloid precursor protein (APP; Chishti et al., 2001) were bred in-house. Recombinant rAAV2/1 expressing BRI2del244-266 (BRI2-Stop), BRI2-Aβ38, BRI2-Aβ40, BRI2-Aβ42, and EGFP were generated as previously described (Kim et al., 2013). The genomic titer of each virus was quantitated as 2.7 × 1012 to 3.9 × 1013 genome particles. Neonatal intracerebroventricular injections of rAAV2/1 were performed as described previously (Chakrabarty et al., 2010). Mice were aged 9 mo; after behavioral examination, the mice were killed, and brain tissue was collected for immunohistochemical and biochemical analysis.
Immunoprecipitation/mass spectrometry
Fusion constructs encoding the first 243 amino acids of BRI2 protein followed by Aβ peptides encompassing various Aβ species were generated as previously described (Kim et al., 2007). The fragments were ligated into the expression vector pAG3. Sequences were verified by DNA sequencing. Overexpression was performed by transiently transfecting human embryonic kidney (HEK 293T) cells. Cells were grown in DMEM supplemented with 10% FBS (Hyclone) and 1% penicillin/streptomycin (Life Technologies). In brief, 2.7 µg DNA was applied to a 75% confluent six-well plate (Corning) using the polycation polyethylenimine transfection method. Cells were incubated with transfection reagent for 12–16 h, after which the growth medium was replaced with fresh medium. 24 h later, the medium was collected for assay by immunoprecipitation, followed by mass spectrometry, as previously described (Ran et al., 2014). In brief, 50 µl magnetic sheep anti–mouse IgG beads (Invitrogen) were incubated with 4.5 µg Ab5 antibody for 30 min at room temperature with constant shaking. The beads were then washed with PBS and incubated with 1–10 ml of conditioned medium containing 0.1% Triton X-100 for 60 min. Bound beads were washed sequentially with 0.1% and 0.05% octyl glucoside (Sigma-Aldrich) followed by water. Samples were eluted with 10 µl of 0.1% TFA (Thermo Scientific) in water. 2 µl of elute was mixed with an equal volume of saturated α-cyano-4-hydroxycinnamic acid (Sigma) solution in 60% acetonitrile, 40% methanol. 1 µl of sample mixture was loaded to α-cyano-4-hydroxycinnamic acid–pretreated MSP 96 target plates (Bruker Daltonics). The samples were analyzed with a Microflex (Bruker Daltonics) mass spectrometer.
Western blotting
2% SDS brain lysates were heated at 70°C for 5 min in the presence of denaturing sample buffer, separated on a 10% Bis-Tris gel (Bio-Rad) in 1× 2-(N-morpholino)ethanesulfonic acid (MES) running buffer (Bio-Rad), and transferred onto 0.2 µm PVDF (EMD Millipore). Membranes were blocked in casein blocking buffer and incubated overnight with primary antibodies, Ab5 (human Aβ1-16; T.E. Golde) and β-tubulin (Covance), and detected with Alexa Fluor anti–mouse 680 and Alexa Fluor anti–rabbit 800 (Life Technologies). Images were developed using an Odyssey infrared scanner (Li-Cor Biosciences) and analyzed using the densitometric feature for semiquantitative analysis using the Odyssey Infrared Imaging System software v.3.0.21 (Li-Cor Biosciences). Bands were manually selected, and background readings of the BRI2-Stop were subtracted.
Contextual fear conditioning
Mice were aged to 9 mo and subjected to contextual fear conditioning as described previously (Hanna et al., 2012; Chakrabarty et al., 2015). The conditioning procedure was performed in four identical chambers (25.3 cm length × 29.5 cm width × 29.5 cm height; Coulbourn Institute). The total floor area of each chamber was 746 cm2. The chambers were constructed from aluminum (sidewalls and ceiling) and Plexiglas (rear and front walls). They were placed individually in sound-attenuated cabinets with black inside walls (interior dimensions: 43.3 cm length × 55.3 cm width × 58.5 cm height; Coulbourn Institute), which were located in a dedicated room. A ventilation fan in each cabinet provided 50 dB background noise, and a 24V DC white light, mounted on a wall of each chamber, provided illumination (65 lux at the floor level). A speaker mounted in the wall opposite to the light delivered an acoustic conditioned stimulus (CS). The floor of each chamber, which consisted of 26 stainless steel rods (3 mm in diameter) spaced 11 mm center to center, was wired to a precision-regulated shocker (H13-15; Coulbourn Institute). A camera mounted above the chamber recorded mouse activity. Conditioning was assessed by the analysis of fear response expressed as freezing behavior with the aid of FreezeFrame program (v.3.06; Actimetrics). Freezing was defined as the cessation of all movements other than respiratory activity (Fanselow, 1980).
Mice were exposed to the context of a training chamber and a tone, both initially novel and neutral stimuli, in one training session. They were transported in squads of 4-in. individual containers filled with home cage bedding and placed singly in the conditioning chamber. During training, the mice received two pairings between a tone (80 dB, pulse [6 c.p.s.], 30-s duration) and a 0.45-mA foot shock (2-s duration, coterminated with a tone). The first CS-US pairing was delivered at the end of 120 s of the initial exploration of the chamber, and the second after a 60-s interval. After the second CS-US pairing, the mice were given a 60-s posttraining period. The total duration of the training session was 300 s. After a day of recovery, the mice were returned to their respective conditioning chambers and tested for fear-induced freezing to the context in which they received foot shocks. The test, performed in an extinction mode with no shock administered, lasted 300 s. The next day, the mice were tested for the association between the tone and the foot-shock in a modified chamber. The floor and the walls of the chamber were replaced with plastic inserts (opaque white for the floor, and semi-transparent white at the front and opaque green at the back for the walls), which also eliminated corners in the chamber. The total floor area of the modified chamber was ∼671 cm2. A Petri dish containing a drop of Pure Vanilla Extract (McCormick) was placed underneath the floor of each chamber to provide a distinct novel odor in the chamber. These modifications did not change the light intensity in the chamber. The tone test lasted 360 s. During the first 180 s, the mice were allowed to explore the new environment, and during the second 180 s a tone, with the same characteristics as the tone used during training, was delivered. Mice activity was recorded during all tests.
Immunohistochemical imaging and image processing
After behavioral testing, we analyzed a random subset of mice by IHC. Mice were killed, brain tissue was harvested, and the right hemisphere was fixed in formalin, embedded in paraffin, sectioned, and stained with a biotinylated pan-Aβ antibody Ab5 (1:500, human Aβ1-16 specific; T.E. Golde). 1% Thio-S (Sigma-Aldrich) staining was performed on paraffin-embedded brain sections using established protocols. Immunohistochemically and fluorescently stained sections were captured using the Scansope XT image scanner (Aperio; Leica Biosystems) or BX 60 (Olympus) and analyzed using ImageScope program. Aβ plaque burden was calculated using the Positive Pixel Count program (Aperio), as previously described (Chakrabarty et al., 2015). In brief, at least three sections per sample, at least 30 µm apart, were calculated blindly and then the mean was taken to determine plaque burden. For Thio-S quantification, one section per sample was used by a blinded observer to calculate the number of cored plaques per area using ImageJ (Schneider et al., 2012).
Aβ ELISA (mouse experiments)
After tissue harvesting, the left hemisphere was flash-frozen in isopentane. The frozen cortex was sequentially extracted with protease inhibitor cocktail (Roche) containing Tris-buffered saline, RIPA buffer, 2% SDS, and 70% formic acid (FA) as described previously at a concentration of 150 mg/ml (Moore et al., 2012). Aβ levels from the 2% SDS– and 70% FA–extracted samples were quantified using end-specific sandwich ELISA as previously described (Moore et al., 2012). Aβ40 was captured with mAb 13.1.1 (human Aβ35–40 specific; T.E. Golde) and detected by HRP-conjugated mAb 33.1.1 (human Aβ1–16; T.E. Golde). Aβ42 was captured with mAb 2.1.3 (human Aβ35–42 specific; T.E. Golde) and detected by HRP-conjugated mAb 33.1.1 (human Aβ1–16; T.E. Golde). ELISA results were analyzed using SoftMax Pro software (Molecular Devices).
Statistical analyses
For Drosophila Q-RT-PCR, one-way ANOVA with Tukey’s multiple comparison test, was used for statistical comparison (Prism 5; GraphPad Software). For the climbing assay, for each Aβ transgene, the climbing index, expressed as percentage of flies climbing above the criterion of 5-cm mark (no. flies above 5-cm mark/total no. of flies × 100), was calculated. The climbing index was then averaged from six trials of five replicates for each day and normalized. To account for day-to-day variability in climbing, we averaged the climbing index over 10- and 3-d (Fig. 3 and Fig. 6, respectively) increments, plotted as a survival curve and analyzed by Mantel–Cox using Prism 6 (GraphPad Software). Final images were created using Photoshop CS5 (Adobe Systems). All values in the text and figures represent means ± standard error of the mean. For Drosophila ELISA data, t test with Prism 6 was used to compare 1- and 30-d-old flies, and one-way ANOVA with Dunnett’s multiple comparison test was used for statistical comparison of Aβ transgenic flies to each other (Prism 6). One-way ANOVA with Dunnett’s multiple comparison test (Prism 6) was used to analyze mammalian data unless otherwise noted. For mouse memory tests, the overall analyses was by factorial ANOVA with genotype/construct as between-subject factor and age/time or pre- and poststimulus stage of a test as repeated measure (within-subject) factor. When necessary, degrees of freedom were adjusted by Greenhouse–Geisser epsilon correction for the heterogeneity of variance. In analyses requiring multiple comparisons between means, the Bonferroni adjustment of α level minimizing type I (family-wise) error rate was used (Howell, 1992). A priori comparisons were performed using Bonferroni t test (MODLSD), and post hoc of multiple pairwise comparisons were done using Student-Newman-Keuls test (Howell, 1992). All statistical analyses were done using Statistical Package for Social Sciences (SPSS) v.23 for Macintosh. Comparisons between two independent groups were done using Student’s t test. In mouse studies, freezing responses of mice during context and tone tests were expressed as percentage of total duration of a test. In the case of tone test, which encompassed pretone phase of exploration of a modified chamber, followed by the tone test, the freezing shown during tone phase was analyzed using analysis of covariance with freezing during pretone phase as covariate to control for the increased freezing in the modified chamber because of generalization of conditional response (Stevens, 1990). Consequently, the freezing rates in response to tone stimulus were adjusted for the freezing rates during pretone phase and presented in Fig. 9 B.
Online supplemental material
Fig. S1 shows the aggregation of Aβ36–40, Aβ42, and Aβ43. Fig. S2 shows expression of Aβ38, Aβ40, and Aβ42 in nontransgenic mice. Fig. S3 shows the Drosophila drivers used in this study. Table S1 shows statistical analysis of Aβ levels from Drosophila expressing Aβ peptides. Table S2 details Q-RT-PCR primers and probes.
Supplementary Material
Acknowledgments
This research was supported by National Institutes of Health (NIH) grants U01AG046139, P50AG047266, R01AG18454, and P01AG020206. L. de Mena is a Howard Hughes Medical Institute fellow of the Life Sciences Research Foundation. J.J. Kurian is partially supported by an NIH T32 Basic Microbiology and Infectious Diseases Training Grant (5T32AI007110-34) managed by Dr. David Bloom at the University of Florida.
The authors declare no competing financial interests.
Author contributions: B.D. Moore, J. Martin, L. de Mena, P.E. Cruz, C. Ceballos-Diaz, Y. Ran, Y. Levites, T.L. Kukar, J.J. Kurian, D. Rincon-Limas, and P. Fernandez-Funez performed experiments. B.D. Moore, L. de Mena, J. Sanchez, E.H. Koo, D.R. Borchelt, C. Janus, D. Rincon-Limas, P. Fernandez-Funez, and T.E. Golde planned experiments. B.D. Moore, L. de Mena, Y. Levites, T.L. Kukar, C. Janus, D. Rincon-Limas, P. Fernandez-Funez, and T.E. Golde analyzed data. B.D. Moore and T.E. Golde wrote the manuscript. B.D. Moore, L. de Mena, T.B. Ladd, Y. Levites, T.L. Kukar, R. McKenna, E.H. Koo, D.R. Borchelt, C. Janus, D. Rincon-Limas, P. Fernandez-Funez, and T.E. Golde edited the manuscript.
References
- Ashe K.H., and Zahs K.R.. 2010. Probing the biology of Alzheimer’s disease in mice. Neuron. 66:631–645. 10.1016/j.neuron.2010.04.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bainton R.J., Tsai L.T., Singh C.M., Moore M.S., Neckameyer W.S., and Heberlein U.. 2000. Dopamine modulates acute responses to cocaine, nicotine and ethanol in Drosophila. Curr. Biol. 10:187–194. 10.1016/S0960-9822(00)00336-5 [DOI] [PubMed] [Google Scholar]
- Barão S., Moechars D., Lichtenthaler S.F., and De Strooper B.. 2016. BACE1 physiological functions may limit its use as therapeutic target for Alzheimer’s disease. Trends Neurosci. 39:158–169. 10.1016/j.tins.2016.01.003 [DOI] [PubMed] [Google Scholar]
- Biverstål H., Dolfe L., Hermansson E., Leppert A., Reifenrath M., Winblad B., Presto J., and Johansson J.. 2015. Dissociation of a BRICHOS trimer into monomers leads to increased inhibitory effect on Aβ42 fibril formation. Biochim. Biophys. Acta. 1854:835–843. 10.1016/j.bbapap.2015.04.005 [DOI] [PubMed] [Google Scholar]
- Blain J.F., Bursavich M.G., Freeman E.A., Hrdlicka L.A., Hodgdon H.E., Chen T., Costa D.E., Harrison B.A., Kapadnis S., Murphy D.A., et al. 2016. Characterization of FRM-36143 as a new γ-secretase modulator for the potential treatment of familial Alzheimer’s disease. Alzheimers Res. Ther. 8:34 10.1186/s13195-016-0199-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendel M., Jaworska A., Herms J., Trambauer J., Rötzer C., Gildehaus F.J., Carlsen J., Cumming P., Bylund J., Luebbers T., et al. 2015. Amyloid-PET predicts inhibition of de novo plaque formation upon chronic γ-secretase modulator treatment. Mol. Psychiatry. 20:1179–1187. 10.1038/mp.2015.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnouf S., Gorsky M.K., Dols J., Grönke S., and Partridge L.. 2015. Aβ43 is neurotoxic and primes aggregation of Aβ40 in vivo. Acta Neuropathol. 130:35–47. 10.1007/s00401-015-1419-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas-Tinto S., Zhang Y., Sanchez-Garcia J., Gomez-Velazquez M., Rincon-Limas D.E., and Fernandez-Funez P.. 2011. The ER stress factor XBP1s prevents amyloid-beta neurotoxicity. Hum. Mol. Genet. 20:2144–2160. 10.1093/hmg/ddr100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarty P., Jansen-West K., Beccard A., Ceballos-Diaz C., Levites Y., Verbeeck C., Zubair A.C., Dickson D., Golde T.E., and Das P.. 2010. Massive gliosis induced by interleukin-6 suppresses Abeta deposition in vivo: Evidence against inflammation as a driving force for amyloid deposition. FASEB J. 24:548–559. 10.1096/fj.09-141754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarty P., Li A., Ceballos-Diaz C., Eddy J.A., Funk C.C., Moore B., DiNunno N., Rosario A.M., Cruz P.E., Verbeeck C., et al. 2015. IL-10 alters immunoproteostasis in APP mice, increasing plaque burden and worsening cognitive behavior. Neuron. 85:519–533. 10.1016/j.neuron.2014.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chishti M.A., Yang D.S., Janus C., Phinney A.L., Horne P., Pearson J., Strome R., Zuker N., Loukides J., French J., et al. 2001. Early-onset amyloid deposition and cognitive deficits in transgenic mice expressing a double mutant form of amyloid precursor protein 695. J. Biol. Chem. 276:21562–21570. 10.1074/jbc.M100710200 [DOI] [PubMed] [Google Scholar]
- De Strooper B., Vassar R., and Golde T.. 2010. The secretases: Enzymes with therapeutic potential in Alzheimer disease. Nat. Rev. Neurol. 6:99–107. 10.1038/nrneurol.2009.218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcón-Pérez J.M., Romero-Calderón R., Brooks E.S., Krantz D.E., and Dell’Angelica E.C.. 2007. The Drosophila pigmentation gene pink (p) encodes a homologue of human Hermansky-Pudlak syndrome 5 (HPS5). Traffic. 8:154–168. 10.1111/j.1600-0854.2006.00514.x [DOI] [PubMed] [Google Scholar]
- Fanselow M.S. 1980. Conditioned and unconditional components of post-shock freezing. Pavlov. J. Biol. Sci. 15:177–182. [DOI] [PubMed] [Google Scholar]
- Feany M.B., and Bender W.W.. 2000. A Drosophila model of Parkinson’s disease. Nature. 404:394–398. 10.1038/35006074 [DOI] [PubMed] [Google Scholar]
- Fernandez-Funez P., Nino-Rosales M.L., de Gouyon B., She W.C., Luchak J.M., Martinez P., Turiegano E., Benito J., Capovilla M., Skinner P.J., et al. 2000. Identification of genes that modify ataxin-1-induced neurodegeneration. Nature. 408:101–106. 10.1038/35040584 [DOI] [PubMed] [Google Scholar]
- Fernandez-Funez P., Casas-Tinto S., Zhang Y., Gómez-Velazquez M., Morales-Garza M.A., Cepeda-Nieto A.C., Castilla J., Soto C., and Rincon-Limas D.E.. 2009. In vivo generation of neurotoxic prion protein: Role for hsp70 in accumulation of misfolded isoforms. PLoS Genet. 5:e1000507 10.1371/journal.pgen.1000507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Funez P., Sanchez-Garcia J., de Mena L., Zhang Y., Levites Y., Khare S., Golde T.E., and Rincon-Limas D.E.. 2016. Holdase activity of secreted Hsp70 masks amyloid-β42 neurotoxicity in Drosophila. Proc. Natl. Acad. Sci. USA. 113:E5212–E5221. 10.1073/pnas.1608045113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friggi-Grelin F., Coulom H., Meller M., Gomez D., Hirsh J., and Birman S.. 2003. Targeted gene expression in Drosophila dopaminergic cells using regulatory sequences from tyrosine hydroxylase. J. Neurobiol. 54:618–627. 10.1002/neu.10185 [DOI] [PubMed] [Google Scholar]
- Golde T.E. 2016. Overcoming translational barriers impeding development of Alzheimer’s disease modifying therapies. J. Neurochem. 139(Suppl 2):224–236. 10.1111/jnc.13583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golde T.E., Eckman C.B., and Younkin S.G.. 2000. Biochemical detection of Abeta isoforms: Implications for pathogenesis, diagnosis, and treatment of Alzheimer’s disease. Biochim. Biophys. Acta. 1502:172–187. 10.1016/S0925-4439(00)00043-0 [DOI] [PubMed] [Google Scholar]
- Golde T.E., Schneider L.S., and Koo E.H.. 2011. Anti-aβ therapeutics in Alzheimer’s disease: The need for a paradigm shift. Neuron. 69:203–213. 10.1016/j.neuron.2011.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golde T.E., Ran Y., and Felsenstein K.M.. 2012. Shifting a complex debate on γ-secretase cleavage and Alzheimer’s disease. EMBO J. 31:2237–2239. 10.1038/emboj.2012.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golde T.E., Koo E.H., Felsenstein K.M., Osborne B.A., and Miele L.. 2013. γ-Secretase inhibitors and modulators. Biochim. Biophys. Acta. 1828:2898–2907. 10.1016/j.bbamem.2013.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna A., Iremonger K., Das P., Dickson D., Golde T., and Janus C.. 2012. Age-related increase in amyloid plaque burden is associated with impairment in conditioned fear memory in CRND8 mouse model of amyloidosis. Alzheimers Res. Ther. 4:21 10.1186/alzrt124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermansson E., Schultz S., Crowther D., Linse S., Winblad B., Westermark G., Johansson J., and Presto J.. 2014. The chaperone domain BRICHOS prevents CNS toxicity of amyloid-β peptide in Drosophila melanogaster. Dis. Model. Mech. 7:659–665. 10.1242/dmm.014787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell D.C. 1992. Statistical methods for psychology. Duxbury Press, Belmont, California. [Google Scholar]
- Hubbs J.L., Fuller N.O., Austin W.F., Shen R., Creaser S.P., McKee T.D., Loureiro R.M., Tate B., Xia W., Ives J., and Bronk B.S.. 2012. Optimization of a natural product-based class of γ-secretase modulators. J. Med. Chem. 55:9270–9282. 10.1021/jm300976b [DOI] [PubMed] [Google Scholar]
- Iijima K., and Iijima-Ando K.. 2008. Drosophila models of Alzheimer’s amyloidosis: The challenge of dissecting the complex mechanisms of toxicity of amyloid-beta 42. J. Alzheimers Dis. 15:523–540. 10.3233/JAD-2008-15402 [DOI] [PubMed] [Google Scholar]
- Iwatsubo T., Odaka A., Suzuki N., Mizusawa H., Nukina N., and Ihara Y.. 1994. Visualization of A beta 42(43) and A beta 40 in senile plaques with end-specific A beta monoclonals: Evidence that an initially deposited species is A beta 42(43). Neuron. 13:45–53. 10.1016/0896-6273(94)90458-8 [DOI] [PubMed] [Google Scholar]
- Jarrett J.T., and Lansbury P.T. Jr. 1993. Seeding “one-dimensional crystallization” of amyloid: A pathogenic mechanism in Alzheimer’s disease and scrapie? Cell. 73:1055–1058. 10.1016/0092-8674(93)90635-4 [DOI] [PubMed] [Google Scholar]
- Jonson M., Pokrzywa M., Starkenberg A., Hammarstrom P., and Thor S.. 2015. Systematic Aβ analysis in Drosophila reveals high toxicity for the 1–42, 3-42 and 11–42 peptides, and emphasizes N- and C-terminal residues. PLoS One. 10:e0133272 10.1371/journal.pone.0133272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J.I., Ladd T.B., Kukar T., Price A.R., Moore B.D., Koo E.H., Golde T.E., and Felsenstein K.M.. 2013. Steroids as γ-secretase modulators. FASEB J. 27:3775–3785. 10.1096/fj.12-225649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawarabayashi T., Younkin L.H., Saido T.C., Shoji M., Ashe K.H., and Younkin S.G.. 2001. Age-dependent changes in brain, CSF, and plasma amyloid (beta) protein in the Tg2576 transgenic mouse model of Alzheimer’s disease. J. Neurosci. 21:372–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Onstead L., Randle S., Price R., Smithson L., Zwizinski C., Dickson D.W., Golde T., and McGowan E.. 2007. Abeta40 inhibits amyloid deposition in vivo. J. Neurosci. 27:627–633. 10.1523/JNEUROSCI.4849-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Miller V.M., Levites Y., West K.J., Zwizinski C.W., Moore B.D., Troendle F.J., Bann M., Verbeeck C., Price R.W., et al. 2008. BRI2 (ITM2b) inhibits Abeta deposition in vivo. J. Neurosci. 28:6030–6036. 10.1523/JNEUROSCI.0891-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Chakrabarty P., Hanna A., March A., Dickson D.W., Borchelt D.R., Golde T., and Janus C.. 2013. Normal cognition in transgenic BRI2-Aβ mice. Mol. Neurodegener. 8:15 10.1186/1750-1326-8-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kounnas M.Z., Danks A.M., Cheng S., Tyree C., Ackerman E., Zhang X., Ahn K., Nguyen P., Comer D., Mao L., et al. 2010. Modulation of gamma-secretase reduces beta-amyloid deposition in a transgenic mouse model of Alzheimer’s disease. Neuron. 67:769–780. 10.1016/j.neuron.2010.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretner B., Trambauer J., Fukumori A., Mielke J., Kuhn P.H., Kremmer E., Giese A., Lichtenthaler S.F., Haass C., Arzberger T., and Steiner H.. 2016. Generation and deposition of Aβ43 by the virtually inactive presenilin-1 L435F mutant contradicts the presenilin loss-of-function hypothesis of Alzheimer’s disease. EMBO Mol. Med. 8:458–465. 10.15252/emmm.201505952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuperstein I., Broersen K., Benilova I., Rozenski J., Jonckheere W., Debulpaep M., Vandersteen A., Segers-Nolten I., Van Der Werf K., Subramaniam V., et al. 2010. Neurotoxicity of Alzheimer’s disease Aβ peptides is induced by small changes in the Aβ42 to Aβ40 ratio. EMBO J. 29:3408–3420. 10.1038/emboj.2010.211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawlor P.A., Bland R.J., Das P., Price R.W., Holloway V., Smithson L., Dicker B.L., During M.J., Young D., and Golde T.E.. 2007. Novel rat Alzheimer’s disease models based on AAV-mediated gene transfer to selectively increase hippocampal Abeta levels. Mol. Neurodegener. 2:11 10.1186/1750-1326-2-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis P.A., Piper S., Baker M., Onstead L., Murphy M.P., Hardy J., Wang R., McGowan E., and Golde T.E.. 2001. Expression of BRI-amyloid beta peptide fusion proteins: A novel method for specific high-level expression of amyloid beta peptides. Biochim. Biophys. Acta. 1537:58–62. 10.1016/S0925-4439(01)00054-0 [DOI] [PubMed] [Google Scholar]
- McGowan E., Pickford F., Kim J., Onstead L., Eriksen J., Yu C., Skipper L., Murphy M.P., Beard J., Das P., et al. 2005. Abeta42 is essential for parenchymal and vascular amyloid deposition in mice. Neuron. 47:191–199. 10.1016/j.neuron.2005.06.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore B.D., Chakrabarty P., Levites Y., Kukar T.L., Baine A.M., Moroni T., Ladd T.B., Das P., Dickson D.W., and Golde T.E.. 2012. Overlapping profiles of Aβ peptides in the Alzheimer’s disease and pathological aging brains. Alzheimers Res. Ther. 4:18 10.1186/alzrt121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng S., Fitzen M., Jörnvall H., and Johansson J.. 2010. The extracellular domain of Bri2 (ITM2B) binds the ABri peptide (1-23) and amyloid beta-peptide (Abeta1–40): Implications for Bri2 effects on processing of amyloid precursor protein and Abeta aggregation. Biochem. Biophys. Res. Commun. 393:356–361. 10.1016/j.bbrc.2009.12.122 [DOI] [PubMed] [Google Scholar]
- Perez-Iratxeta C., and Andrade-Navarro M.A.. 2008. K2D2: Estimation of protein secondary structure from circular dichroism spectra. BMC Struct. Biol. 8:25 10.1186/1472-6807-8-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer B.D., Ngo T.T., Hibbard K.L., Murphy C., Jenett A., Truman J.W., and Rubin G.M.. 2010. Refinement of tools for targeted gene expression in Drosophila. Genetics. 186:735–755. 10.1534/genetics.110.119917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poska H., Haslbeck M., Kurudenkandy F.R., Hermansson E., Chen G., Kostallas G., Abelein A., Biverstål H., Crux S., Fisahn A., et al. 2016. Dementia-related Bri2 BRICHOS is a versatile molecular chaperone that efficiently inhibits Aβ42 toxicity in Drosophila. Biochem. J. 473:3683–3704. 10.1042/BCJ20160277 [DOI] [PubMed] [Google Scholar]
- Ran Y., Cruz P.E., Ladd T.B., Fauq A.H., Jung J.I., Matthews J., Felsenstein K.M., and Golde T.E.. 2014. γ-Secretase processing and effects of γ-secretase inhibitors and modulators on long Aβ peptides in cells. J. Biol. Chem. 289:3276–3287. 10.1074/jbc.M113.512921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangachari V., Moore B.D., Reed D.K., Sonoda L.K., Bridges A.W., Conboy E., Hartigan D., and Rosenberry T.L.. 2007. Amyloid-beta(1–42) rapidly forms protofibrils and oligomers by distinct pathways in low concentrations of sodium dodecylsulfate. Biochemistry. 46:12451–12462. 10.1021/bi701213s [DOI] [PubMed] [Google Scholar]
- Rival T., Soustelle L., Strambi C., Besson M.T., Iché M., and Birman S.. 2004. Decreasing glutamate buffering capacity triggers oxidative stress and neuropil degeneration in the Drosophila brain. Curr. Biol. 14:599–605. 10.1016/j.cub.2004.03.039 [DOI] [PubMed] [Google Scholar]
- Saito T., Suemoto T., Brouwers N., Sleegers K., Funamoto S., Mihira N., Matsuba Y., Yamada K., Nilsson P., Takano J., et al. 2011. Potent amyloidogenicity and pathogenicity of Aβ43. Nat. Neurosci. 14:1023–1032. 10.1038/nn.2858 [DOI] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., and Eliceiri K.W.. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 9:671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selkoe D.J. 2001. Alzheimer’s disease: Genes, proteins, and therapy. Physiol. Rev. 81:741–766. [DOI] [PubMed] [Google Scholar]
- Selkoe D.J., and Hardy J.. 2016. The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Mol. Med. 8:595–608. 10.15252/emmm.201606210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seubert P., Vigo-Pelfrey C., Esch F., Lee M., Dovey H., Davis D., Sinha S., Schlossmacher M., Whaley J., Swindlehurst C., et al. 1992. Isolation and quantification of soluble Alzheimer’s beta-peptide from biological fluids. Nature. 359:325–327. 10.1038/359325a0 [DOI] [PubMed] [Google Scholar]
- Shulman J.M., Shulman L.M., Weiner W.J., and Feany M.B.. 2003. From fruit fly to bedside: Translating lessons from Drosophila models of neurodegenerative disease. Curr. Opin. Neurol. 16:443–449. 10.1097/01.wco.0000084220.82329.60 [DOI] [PubMed] [Google Scholar]
- Sofola O., Kerr F., Rogers I., Killick R., Augustin H., Gandy C., Allen M.J., Hardy J., Lovestone S., and Partridge L.. 2010. Inhibition of GSK-3 ameliorates Abeta pathology in an adult-onset Drosophila model of Alzheimer’s disease. PLoS Genet. 6:e1001087 10.1371/journal.pgen.1001087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens J. 1990. Intermediate statistics: A modern approach. Lawrence Erlbaum Associates, Inc., Hillsdale, New Jersey. [Google Scholar]
- Tycko R. 2015. Amyloid polymorphism: Structural basis and neurobiological relevance. Neuron. 86:632–645. 10.1016/j.neuron.2015.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veugelen S., Saito T., Saido T.C., Chávez-Gutiérrez L., and De Strooper B.. 2016. Familial Alzheimer’s disease mutations in presenilin generate amyloidogenic Aβ peptide seeds. Neuron. 90:410–416. 10.1016/j.neuron.2016.03.010 [DOI] [PubMed] [Google Scholar]
- Wagner S.L., Tanzi R.E., Mobley W.C., and Galasko D.. 2012. Potential use of γ-secretase modulators in the treatment of Alzheimer disease. Arch. Neurol. 69:1255–1258. 10.1001/archneurol.2012.540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner S.L., Zhang C., Cheng S., Nguyen P., Zhang X., Rynearson K.D., Wang R., Li Y., Sisodia S.S., Mobley W.C., and Tanzi R.E.. 2014. Soluble γ-secretase modulators selectively inhibit the production of the 42-amino acid amyloid β peptide variant and augment the production of multiple carboxy-truncated amyloid β species. Biochemistry. 53:702–713. 10.1021/bi401537v [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Sweeney D., Gandy S.E., and Sisodia S.S.. 1996. The profile of soluble amyloid beta protein in cultured cell media. Detection and quantification of amyloid beta protein and variants by immunoprecipitation-mass spectrometry. J. Biol. Chem. 271:31894–31902. 10.1074/jbc.271.50.31894 [DOI] [PubMed] [Google Scholar]
- Weggen S., Eriksen J.L., Das P., Sagi S.A., Wang R., Pietrzik C.U., Findlay K.A., Smith T.E., Murphy M.P., Bulter T., et al. 2001. A subset of NSAIDs lower amyloidogenic Abeta42 independently of cyclooxygenase activity. Nature. 414:212–216. 10.1038/35102591 [DOI] [PubMed] [Google Scholar]
- Wiedemann C., Bellstedt P., and Görlach M.. 2013. CAPITO—A web server-based analysis and plotting tool for circular dichroism data. Bioinformatics. 29:1750–1757. 10.1093/bioinformatics/btt278 [DOI] [PubMed] [Google Scholar]
- Willander H., Presto J., Askarieh G., Biverstål H., Frohm B., Knight S.D., Johansson J., and Linse S.. 2012. BRICHOS domains efficiently delay fibrillation of amyloid β-peptide. J. Biol. Chem. 287:31608–31617. 10.1074/jbc.M112.393157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willem M., Lammich S., and Haass C.. 2009. Function, regulation and therapeutic properties of beta-secretase (BACE1). Semin. Cell Dev. Biol. 20:175–182. 10.1016/j.semcdb.2009.01.003 [DOI] [PubMed] [Google Scholar]
- Younkin S.G. 1998. The role of A beta 42 in Alzheimer’s disease. J. Physiol. Paris. 92:289–292. 10.1016/S0928-4257(98)80035-1 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Casas-Tinto S., Rincon-Limas D.E., and Fernandez-Funez P.. 2014. Combined pharmacological induction of Hsp70 suppresses prion protein neurotoxicity in Drosophila. PLoS One. 9:e88522 10.1371/journal.pone.0088522 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.