Cancer therapy reduces tumor burden by killing tumor cells, yet it simultaneously creates tumor cell debris that may stimulate inflammation and tumor growth. Sulciner et al. demonstrate that specific resolvins (RvD1, RvD2, and RvE1) inhibit tumor growth and enhance cancer therapy through the clearance of tumor cell debris.
Abstract
Cancer therapy reduces tumor burden by killing tumor cells, yet it simultaneously creates tumor cell debris that may stimulate inflammation and tumor growth. Thus, conventional cancer therapy is inherently a double-edged sword. In this study, we show that tumor cells killed by chemotherapy or targeted therapy (“tumor cell debris”) stimulate primary tumor growth when coinjected with a subthreshold (nontumorigenic) inoculum of tumor cells by triggering macrophage proinflammatory cytokine release after phosphatidylserine exposure. Debris-stimulated tumors were inhibited by antiinflammatory and proresolving lipid autacoids, namely resolvin D1 (RvD1), RvD2, or RvE1. These mediators specifically inhibit debris-stimulated cancer progression by enhancing clearance of debris via macrophage phagocytosis in multiple tumor types. Resolvins counterregulate the release of cytokines/chemokines, including TNFα, IL-6, IL-8, CCL4, and CCL5, by human macrophages stimulated with cell debris. These results demonstrate that enhancing endogenous clearance of tumor cell debris is a new therapeutic target that may complement cytotoxic cancer therapies.
Introduction
For decades, cancer therapy has focused on killing cancer cells, from broad cytotoxic therapy to the inhibition of specific molecular pathways, in order to reduce tumor burden. However, cancer therapy may inherently be a double-edged sword as radiation-induced apoptotic tumor cells can promote tumor growth (the Révész phenomenon; Révész, 1956; Huang et al., 2011; Chaurio et al., 2013; Ford et al., 2015; Gunjal et al., 2015; da Silva-Jr et al., 2017). Moreover, irradiation and chemotherapy trigger a cytokine storm in the tumor stroma, including the release of tumor-promoting cytokines IL-6 and TNFα (Poth et al., 2010; Reers et al., 2013; Vyas et al., 2014) as well as activation of macrophage production of proinflammatory mediators by apoptotic tumor cells (Ley et al., 2013). Conversely, cell debris can also stimulate antitumor immunity (Casares et al., 2005). Thus, dead and dying tumor cells contribute to an underappreciated component of the tumor microenvironment that may promote tumor progression (Connell and Weichselbaum, 2011; Lauber and Herrmann, 2015; Gregory et al., 2016; Ichim and Tait, 2016). However, the tumor-promoting activity of this treatment byproduct, i.e., tumor cell debris, has not been systematically examined.
In this study, we show that tumor cells killed by chemotherapy or targeted therapy drastically stimulate tumor growth in animal models when coinjected with a subthreshold inoculum of tumor cells that would otherwise not result in macroscopic tumors. Thus, conventional chemotherapy and targeted therapy directly contribute to tumor progression and relapse as tumor cell debris stimulates the survival and growth of living tumor cells. We further demonstrate that chemotherapy-generated tumor cell debris promotes tumorigenesis by stimulating the release of proinflammatory cytokines by macrophages. Overcoming the dilemma between killing tumor cells and debris-induced tumor progression is paramount to preventing tumor recurrence after therapy. In this study, we address this with resolvin D1 (RvD1), RvD2, or RvE1, proresolving lipid autacoids that stimulate the natural debris-clearing process and promote the termination of inflammatory processes (Serhan, 2014). RvD1, RvD2, or RvE1 stimulated the resolution of tumor-promoting inflammation by activating macrophage clearance of cellular debris in tumors.
Results
Chemotherapy-generated or targeted therapy–generated tumor cell debris stimulates primary tumor growth
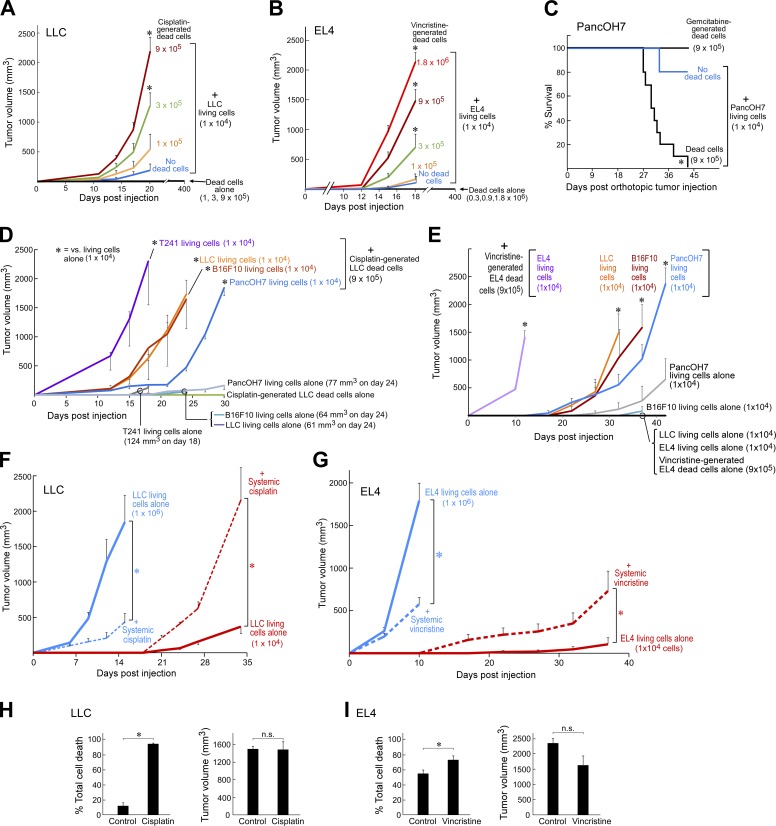
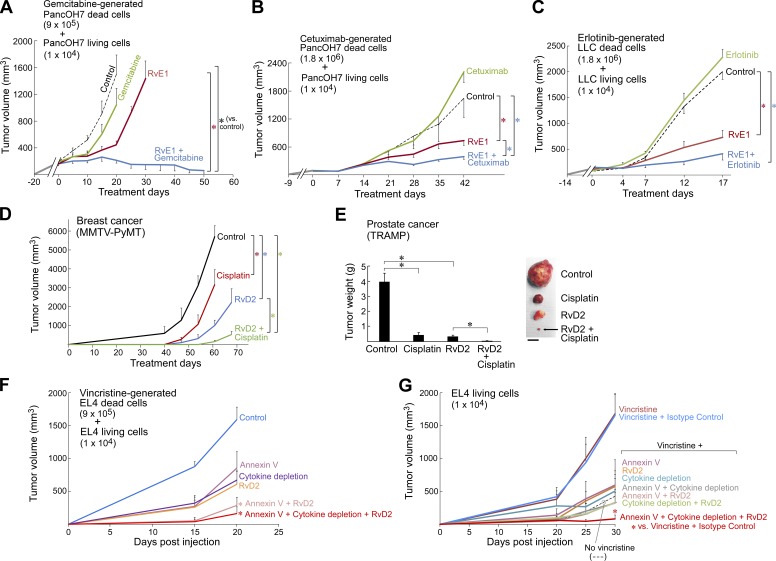
To interrogate the tumor growth–stimulating activity of tumor cell debris, we first developed a mouse debris-stimulated tumor model applicable to many cancer types in which debris generated in vitro can stimulate the growth of grafted tumors from a subthreshold inoculum of tumor cells, which would otherwise not generate a growing tumor. We prepared tumor cell debris in vitro by treating tumor cells with chemotherapy (cisplatin, vincristine, gemcitabine, or docetaxel), targeted therapy (erlotinib or cetuximab), or cycloheximide plus TNFα (a canonical inducer of apoptosis; Niwa et al., 1997; Spite et al., 2009; Chiang et al., 2012). These treatments produced dead cells (apoptotic cells, necrotic cells, and cell fragments; see the Generation of debris by chemotherapy or targeted therapy: General note section of Materials and methods), hereafter referred to as “drug-generated debris” or “debris,” which were collected for coinjection with living tumor cells. In Lewis lung carcinoma (LLC), a widely used mouse tumor model (O’Reilly et al., 1994; Panigrahy et al., 2012), cisplatin-generated LLC debris stimulated LLC tumor growth in a dose-dependent manner up to 100-fold (Fig. 1 A). Increasing the amount of cisplatin-generated LLC debris (105, 3 × 105, or 9 × 105 dead cells) coinjected with a subthreshold inoculum of LLC (104 living cells) resulted in accelerated tumor growth (Figs. 1 A and S1 A). Implantation of a low number of LLC (103 or 104 living cells) mimicked dormancy or minimal growth as these tumor cells survived in the tissue for ≥110 d (Panigrahy et al., 2012). Tumor cell debris alone without living cells did not produce any visible tumors at 400 d after injection. We assessed cell death of drug-generated debris via flow cytometry analysis of annexin V/propidium iodide (PI) and counted the number of dead cell bodies as a surrogate quantity for titrating its tumor-stimulatory potency (Fig. S1, A–K). Using GFP-labeled LLC cells, we verified that debris-stimulated tumors arose from the subthreshold inoculum of living tumor cells (Fig. S1 L). Next, we titrated the number of living tumor cells for a fixed quantity of drug-generated debris (9 × 105 dead cells). LLC debris (9 × 105 dead cells) promoted rapid LLC tumor growth, even from a living tumor cell inoculum as low as 102 cells (Fig. S2 A). LLC alone (102 or 103 living cells) did not result in growing tumors, even at 300 d after injection (Fig. S2 A).
Figure 1.
Chemotherapy-generated tumor cell debris stimulates primary tumor growth. (A and B) Debris-stimulated LLC (A) and EL4 (B) tumor growth from chemotherapy-generated dead cells coinjected with a subthreshold inoculum of 104 living cells. n = 5–15 mice/group. Two-way repeated-measure mixed-effects ANOVAs for tumor growth rates and two-tailed Student’s t test for final tumor measurements were used throughout unless specified; *, P < 0.05 versus 104 living tumor cells alone (“No dead cells;” blue). (C) Percent survival of mice coinjected orthotopically into the pancreas with gemcitabine-generated PancOH7 dead cells and a subthreshold inoculum of PancOH7 living cells. n = 5 mice/group. *, P = 0.004 (Fisher’s exact test). Kaplan-Meier analysis indicated a significantly shortened survival of mice injected with a combination of dead and living cells as depicted by the area under the Kaplan-Meier survival curves (log-rank test = 9.14; *, P < 0.05). (D and E) Debris-stimulated tumor growth from cisplatin-generated LLC dead cells (D) coinjected with 104 LLC, B16F10, T241, or PancOH7 living cells as well as vincristine-generated EL4 dead cells (E) coinjected with 104 EL4, B16F10, LLC, or PancOH7 living cells. n = 5–10 mice/group. *, P < 0.05 versus corresponding living tumor cells alone. (F) Tumor growth from 106 versus 104 LLC living cells with systemic cisplatin. Chemotherapy was initiated on the day of tumor cell injection (dashed lines). n = 5–14 mice/group. *, P < 0.05 versus control (living tumor cells alone; solid lines). (G) Tumor growth from 106 versus 104 EL4 living cells with systemic vincristine. Chemotherapy was initiated on the day of tumor cell injection (dashed lines); n = 5–10 mice/group. *, P < 0.05 versus control (living tumor cells alone; solid lines). (H and I) Flow cytometry analysis for total cell death (sum of percent annexin V+ PI−, annexin V− PI+, and annexin V+ PI+) of comparable sized tumors from 104 LLC (H) or EL4 (I) living cells treated with systemic cisplatin or vincristine versus control (106 LLC or EL4 living cells). n = 4–6 mice/group; *, P < 0.05 versus control. Error bars represent SEM.
To exclude that stimulation of primary tumor growth by chemotherapy-generated tumor cell debris was an idiosyncrasy of the cisplatin and LLC combination, we examined several chemotherapeutics and tumor types. Vincristine-generated lymphoma (EL4) debris (105, 3 × 105, 9 × 105, or 1.8 × 106 dead cells) coinjected with a subthreshold inoculum of EL4 (104 living cells) resulted in progressive acceleration of tumor growth (Fig. 1 B). Debris alone without living cells did not produce visible tumors at 400 d after injection. Gemcitabine-generated debris also shortened survival in an orthotopic pancreatic adenocarcinoma (PancOH7) model (Fig. 1 C). However, debris-stimulated tumor growth was not altered in immunocompromised mouse strains (Fig. S2 B). Human tumor cell debris generated by chemotherapy (e.g., docetaxel, gemcitabine, or cisplatin) also stimulated the growth of a subthreshold inoculum of living tumor cells, including human oral squamous cell carcinoma (HSC-3), pancreatic adenocarcinoma (BxPC3), and prostate carcinoma (PC3M-LN4) xenografts (Fig. S2, C–E).
To determine whether debris-stimulated tumor growth required that both debris and living cells are of the same tumor type, we used LLC or EL4 debris to stimulate the growth of other tumor types. Cisplatin-generated LLC debris (9 × 105 dead cells) stimulated tumor growth from a subthreshold inoculum of 104 LLC, T241 (fibrosarcoma), B16F10 (melanoma), or PancOH7 living cells (Fig. 1 D). Vincristine-generated EL4 debris (9 × 105 dead cells) also stimulated tumor growth from 104 EL4, LLC, B16F10, or PancOH7 living cells (Fig. 1 E). To exclude that debris-stimulated tumor growth was caused by nonspecific cytotoxicity caused by residual chemotherapy in the debris inoculum, we evaluated whether debris generated by the targeted drugs erlotinib or cetuximab (epidermal growth factor receptor inhibitors) could stimulate tumor growth. Erlotinib-generated debris from both therapy-sensitive lung adenocarcinoma (HCC827) or LLC stimulated tumor growth (Fig. S2, F and G). Similarly, cetuximab-generated PancOH7 debris stimulated tumor growth (Fig. S2 H).
We next determined whether cell debris generated by chemotherapies that induce immunogenic cell death (e.g., oxaliplatin or idarubicin; Tesniere et al., 2010; Pol et al., 2015) could stimulate tumor growth when coinjected with a subthreshold inoculum of tumor cells. To compare nonimmunogenic cell debris to immunogenic cell debris, we generated tumor cell debris not only with cisplatin but also with oxaliplatin or idarubicin in MC38 or CT26 colon carcinoma cells (Fig. S1, I and J). Both cisplatin-generated and oxaliplatin-generated MC38 colon cancer cell debris stimulated tumor growth in immunocompetent and immunocompromised mice, including C57BL/6J and RAG1KO (Fig. S2, I and J). In addition, idarubicin-generated CT26 colon cancer cell debris also stimulated tumor growth in immunocompetent BALB/c mice (Fig. S2 K).
We next asked whether systemic chemotherapy could also stimulate tumor growth by generating cell debris in vivo. Systemically administered cisplatin or vincristine inhibited the growth of LLC or EL4 tumors produced by a conventional high inoculum of 106 living cells; in contrast, the same treatment stimulated tumor outgrowth from a subthreshold inoculum of 104 LLC or EL4 living cells (Fig. 1, F and G). Although fast-growing tumors can contain a substantial number of spontaneously dying cells (Kornbluth, 1994; de Jong et al., 2000; Alcaide et al., 2013; Gregory et al., 2016; Ichim and Tait, 2016), analysis of dissociated tumor cells by flow cytometry for annexin V/PI confirmed that systemic chemotherapy indeed increased cell death in tumors derived from a subthreshold inoculum (104 cells) and whose growth was stimulated by the treatment. Tumors of comparable volume, which were established using the inoculum (106 cells) without drug treatment, were used as controls (Fig. 1, H and I).
Debris-stimulated primary tumor growth is phosphatidylserine (PS) dependent
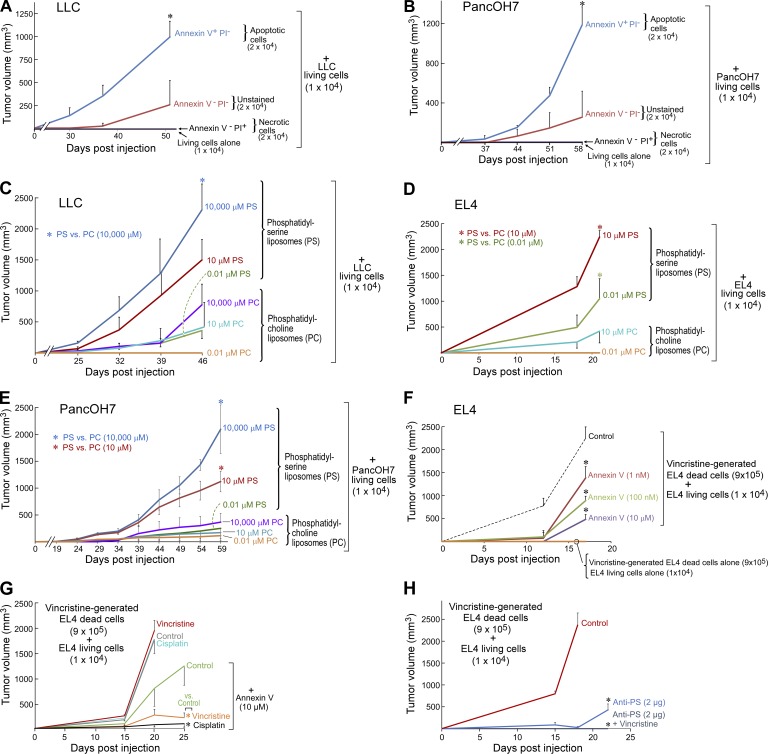
To determine the extent to which either apoptotic or necrotic cells contributed to debris-stimulated tumor growth, we used flow cytometry to cell sort debris into apoptotic (annexin V+ PI−), necrotic (annexin V− PI+), and living (annexin V− PI−) cell populations. Apoptotic cells from cisplatin-generated LLC or gemcitabine-generated PancOH7 debris potently stimulated tumor growth, whereas living cells isolated from debris exhibited minimal tumor-stimulatory activity (Fig. 2, A and B). Necrotic cells alone exhibited no apparent tumor-stimulatory activity (Fig. 2, A and B).
Figure 2.
Debris-stimulated primary tumor growth is PS-dependent. (A and B) Growth of apoptotic (annexin V+ PI−), late apoptotic/necrotic (annexin V+ PI+), necrotic (annexin V− PI+), and living (annexin V− PI−; unstained; A) cisplatin-generated LLC and gemcitabine-generated PancOH7 (B) debris-stimulated tumors. n = 4–10 mice/group. One-factor ANOVA for tumor growth rates and two-tailed Student’s t test for final tumor measurements were used throughout unless specified; *, P < 0.05 versus 104 living tumor cells alone. (C–E) PS or PC liposomes coinjected with a subthreshold inoculum of 104 LLC (C), EL4 (D), or PancOH7 (E) living cells. n = 5–15 mice/group. *, P < 0.05 versus PC liposomes. (F) Debris-stimulated EL4 tumor growth from annexin V–treated vincristine-generated EL4 debris coinjected with a subthreshold inoculum of 104 EL4 living cells. Debris was pretreated with annexin V recombinant protein (10 µM) or vehicle. n = 5 mice/group. *, P < 0.05 versus control. (G) Debris-stimulated EL4 tumors with systemic chemotherapy (vincristine or cisplatin). Chemotherapy was initiated on the day of tumor cell injection. Debris was pretreated with annexin V recombinant protein (10 µM) or vehicle. n = 5–10 mice/group. *, P < 0.05 versus control (green). (H) Debris-stimulated EL4 tumor growth with systemic vincristine. Chemotherapy was initiated on the day of tumor cell injection. Debris was pretreated with anti-PS antibody (2 µg) or control IgG. n = 5 mice/group. *, P < 0.05 versus control. Error bars represent SEM.
Because apoptotic debris stimulated tumor growth, we asked whether PS, which is presented on the surface of apoptotic cells and is detected by annexin V, could be a molecular mediator of tumor stimulation by the debris. Coinjection of PS liposomes (Hosseini et al., 2015) in lieu of debris with a subthreshold inoculum of 104 living tumor cells, i.e., LLC, EL4, or PancOH7, stimulated tumor growth in a dose-dependent manner in comparison with phosphatidylcholine (PC) liposomes (Fig. 2, C–E). Additionally, blocking PS in the debris with an annexin V–recombinant protein or an anti-PS neutralizing antibody drastically, albeit not completely, suppressed debris-stimulated tumor growth in a dose-dependent manner (Fig. 2, F–H). Intriguingly, chemotherapy alone did not exhibit therapeutic activity in debris-stimulated EL4 tumors (Fig. 2 G). In contrast, debris-stimulated tumors in which debris was treated with annexin V or anti-PS neutralizing antibody before injection were responsive to chemotherapy (cisplatin or vincristine; Fig. 2, G and H).
Chemotherapy-generated debris stimulates primary tumor growth via proinflammatory cytokines
To further evaluate the potential mechanism or mechanisms by which drug-generated debris stimulates tumor growth, we next measured the extent to which debris promotes an inflammatory infiltrate, which is known to propagate tumor growth and progression (Mantovani et al., 2008). Debris-stimulated tumors exhibited an increased proportion of infiltrating leukocytes (CD45+), specifically macrophages (CD45+ F4/80+), compared with nondebris tumors as assessed by flow cytometry analysis of cells from dissociated tumors (Fig. S3 A). In contrast, the number of infiltrating myeloid-derived suppressor cells (CD11b+Gr1+) and neutrophils (CD11b+Gr1+Ly6G+) was not altered compared with tumors derived from living cells alone (Fig. S3, B and C). We also found that conditioned media from RAW264.7 mouse macrophages cocultured with tumor cell debris stimulated the proliferation of tumor and endothelial cells while not altering tumor cell viability or apoptosis (Fig. S3, D–F).
Proinflammatory cytokines released by activated immune cells in the tumor stroma mediate the tumor-promoting activity of inflammatory infiltrates (Mantovani et al., 2008). Cytokine array screening of conditioned media from human monocyte–derived macrophages revealed an increase in proinflammatory cytokine release by macrophages cocultured with tumor cell debris (HSC-3 or PC3M-LN4), including IL-6, IL-8, TNFα, CCL2, CCL3, CCL4, CCL5, and GROα, compared with macrophages alone (Fig. S3 G). This was independent of tumor type and the treatment used to generate debris. PS liposomes but not PC liposomes stimulated macrophages to release the same series of proinflammatory cytokines as drug-generated debris, including IL-6, CCL4, and CCL5 (Fig. S3 H). Intriguingly, treatment with annexin V recombinant protein inhibited the production of proinflammatory cytokines, such as CCL4, by debris-stimulated mouse macrophages (RAW264.7), consistent with the PS-dependent activity of debris (Fig. S3 I). Further, liquid chromatography–tandem mass spectrometry (MS; LC-MS-MS)–based profiling of plasma from debris-stimulated tumor-bearing mice revealed elevated inflammatory and tumor-promoting mediators, including prostaglandin E2 (PGE2; Wang and DuBois, 2010) and leukotriene B4 (LTB4; Fig. S3, J and K). Immunohistochemistry studies showed that debris-stimulated (PC3M-LN4) tumors exhibited increased levels of proinflammatory cytokines IL-6, IL-8, and TNFα compared with nondebris tumors generated from only living tumor cells (Fig. S3 L).
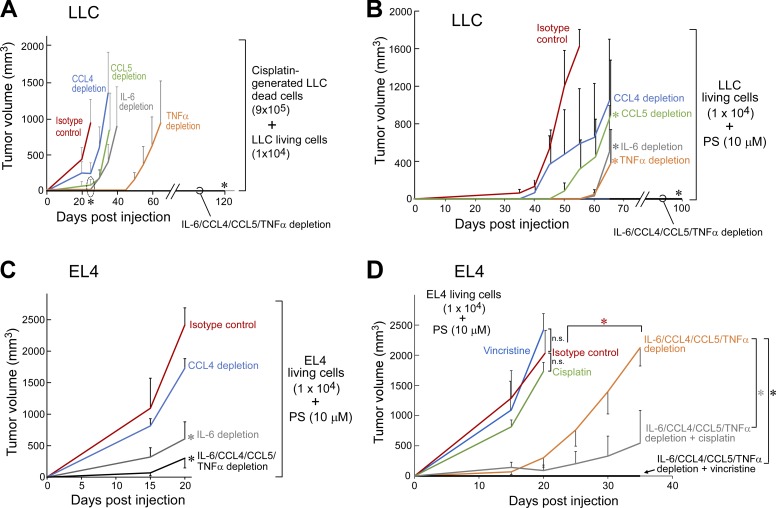
To determine whether these proinflammatory cytokines were critical for the tumor-promoting activity of debris and/or PS liposomes, we depleted these cytokines in debris-stimulated and PS liposome–stimulated tumor models using neutralizing antibodies. The cytokines CCL4, CCL5, IL-6, and TNFα were chosen for in vivo depletion as these cytokines were consistently released by macrophages when exposed to tumor cell debris and/or PS liposomes (Fig. S3, G–I). Although depletion of a single cytokine (CCL4, CCL5, IL-6, or TNFα) only delayed debris- and PS liposome–stimulated tumor growth, the simultaneous depletion of all four cytokines (IL-6/CCL4/CCL5/TNFα) not only prevented debris- and PS liposome–stimulated growth of LLC tumors but also potently suppressed the growth of EL4 tumors stimulated by PS compared with mice administered isotype control antibodies (Fig. 3, A–C). Systemic cytokine depletion also sensitized PS liposome–stimulated tumors to chemotherapy (cisplatin or vincristine; Fig. 3 D).
Figure 3.
Chemotherapy-generated debris stimulates primary tumor growth via proinflammatory cytokines. (A–C) Debris-stimulated LLC tumor growth (A) and PS liposome-stimulated LLC and EL4 tumor growth (B and C) in mice systemically depleted of CCL4, CCL5, IL-6, TNFα, or IL-6/CCL4/CCL5/TNFα versus isotype control. Cytokine depletion was initiated on the day of tumor cell injection. n = 5–10 mice/group. (D) PS liposome-stimulated EL4 tumor growth in mice with or without multiple cytokine depletion (IL-6/CCL4/CCL5/TNFα) versus isotype control. Systemic chemotherapy (vincristine or cisplatin) and/or cytokine depletion were initiated on the day of tumor cell injection. n = 5 mice/group. *, P < 0.05 versus control or IL-6/CCL4/CCL5/TNFα depletion alone. Two-way repeated-measures mixed-effects ANOVAs for tumor growth rates and two-tailed Student’s t test for final tumor measurements were used throughout. *, P < 0.05 versus isotype control. Error bars represent SEM.
Resolvins, chemotherapy, and anti-inflammatories exhibit differential tumor-inhibitory activity on debris-stimulated versus nondebris tumor models
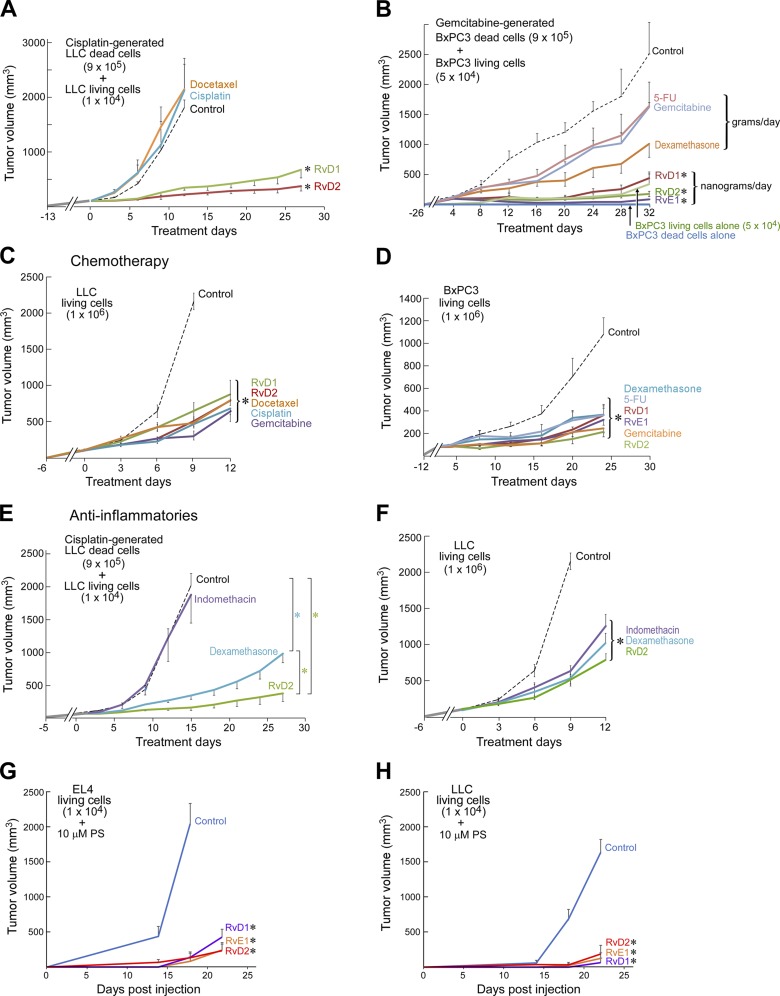
We reasoned that if drug-generated debris promotes tumor growth, clearance of debris may mitigate this growth. Resolvins are endogenous proresolving and antiinflammatory mediators that stimulate the resolution of inflammation by increasing macrophage phagocytosis of debris and countering proinflammatory molecules (Serhan et al., 2002; Serhan, 2014). Therefore, we examined whether resolvins (RvD1, RvD2, or RvE1; Spite et al., 2009; Chiang et al., 2012) could accelerate removal of drug-generated debris. Indeed, RvD1, RvD2, and RvE1 each delayed the onset of debris-stimulated tumor growth of a variety of tumors (LLC, BxPC3, PancOH7, or PC3M-LN4), achieving sustained suppression after 12–70 d of treatment (Fig. 4, A and B; and Fig. S4, A–C). In contrast, chemotherapy exhibited less therapeutic activity than resolvins in debris-stimulated tumors (Fig. 4, A and B; and Fig. S4 C). In the corresponding nondebris tumor models (106 LLC, BxPC3, PancOH7, or PC3M-LN4 living cells), resolvins exhibited antitumor activity equivalent to chemotherapy (docetaxel, cisplatin, gemcitabine, or 5-fluorouracil [5-FU]; Fig. 4, C and D; and Fig. S4, D and E).
Figure 4.
Resolvins, chemotherapy, and antiinflammatories exhibit differential tumor inhibitory activity on debris-stimulated versus nondebris tumor models. (A–F) Debris-stimulated or nondebris LLC or BxPC3 tumors treated with systemic resolvins (RvD1, RvD2, or RvE1), docetaxel, cisplatin, gemcitabine, 5-FU, dexamethasone, or indomethacin. Treatment was initiated once tumors reached 100–200 mm3 throughout unless otherwise specified. n = 4–10 mice/group. Two-way repeated-measures mixed-effects ANOVAs assessed by the F test for tumor growth rates and post hoc Tukey comparisons for final tumor measurements were used throughout unless specified. *, P < 0.05 versus control in A–F; additionally, *, P < 0.05 RvD2 versus dexamethasone in E. (G and H) Growth of PS liposome–stimulated EL4 or LLC tumors treated systemically with resolvins (RvD1, RvD2, or RvE1). Treatment was initiated on the day of tumor cell injection. n = 5 mice/group. *, P < 0.05 versus control. Error bars represent SEM.
Resolvins differ from classic anti-inflammatories in that they stimulate, as agonists, the resolution of inflammation, act at significantly lower doses, and are not immunosuppressive (Serhan, 2014; Fullerton and Gilroy, 2016). To assess the contribution of resolution in the observed tumor suppression by resolvins, we compared their tumor-inhibitory activity to anti-inflammatories (dexamethasone and indomethacin). Resolvins, and to a lesser extent dexamethasone, inhibited debris-stimulated growth of BxPC3, LLC, and PancOH7 tumors (Fig. 4, B and E; and Fig. S4 B). Dexamethasone, indomethacin, and resolvins (RvD1, RvD2, or RvE1) also inhibited nondebris tumor growth (BxPC3, LLC, or PancOH7 living cells), albeit not completely (Fig. 4, D and F; and Fig. S4 F). This is consistent with the known activity of dexamethasone to stimulate the resolution of inflammation, including macrophage phagocytosis of apoptotic cells (Maderna et al., 2005). However, dexamethasone (2 mg/kg/d) required a 1,000-fold higher dose compared with resolvins (6 µg/kg/d) to inhibit debris-stimulated tumor growth. Importantly, resolvins do not exhibit the immunosuppressive actions associated with dexamethasone (Serhan, 2014). Resolvins (RvD1, RvD2, or RvE1) also inhibited PS liposome–stimulated tumor growth (EL4 and LLC; Fig. 4, G and H). Furthermore, resolvins inhibited the growth of orthotopic (PC3M-LN4) and spontaneous tumors (genetically engineered mouse models, transgenic adenocarcinoma of the mouse prostate [TRAMP], and mouse mammary tumor virus [MMTV]-PyMT; Fig. S4, G–I).
Using a well-established model in which resection of a primary tumor reproducibly stimulates development of distant metastasis 14–17 d after resection (O’Reilly et al., 1994; Panigrahy et al., 2012), we investigated whether resolvins could inhibit spontaneous metastatic growth. Resolvins suppressed metastasis in the lung, as measured by lung weight and number of surface lung metastases, compared with control mice (Fig. S4 J). To determine whether the inhibitory activity of resolvins was limited to the LLC model, we injected B16F10 melanoma cells into the tail vein, a common (nonspontaneous) hematogenic metastasis model in which B16F10 cells exclusively colonize the lung and produce pulmonary metastases (Parhar and Lala, 1987). Administration of RvD1 or RvD2 inhibited B16F10 lung metastasis (Fig. S4 K).
Given the unique ability of resolvins to stimulate clearance of debris (Serhan, 2014), we next treated debris-stimulated tumors with resolvins in combination with the debris-generating chemotherapy or targeted therapy. Chemotherapy (gemcitabine or cisplatin) or targeted therapy (cetuximab or erlotinib) in combination with resolvins resulted in pancreatic tumor regression in debris-stimulated tumors (PancOH7) and had additive antitumor activity in debris-stimulated LLC and spontaneous (MMTV-PyMT and TRAMP) tumor models over a treatment period of 17–84 d (Fig. 5, A–E). The combination of resolvins and cisplatin treatment delayed MMTV-PyMT cancer onset and growth (Fig. 5 D). Furthermore, the antitumor activity of immunogenic chemotherapy (e.g., oxaliplatin) was improved when combined with resolvins in debris-stimulated tumors (Fig. S4 L). Finally, treatment with a combination of annexin V recombinant protein, cytokine depletion, and resolvins had the most potent activity in suppressing both debris-stimulated and chemotherapy-stimulated EL4 tumor growth (Fig. 5, F and G). Whereas systemic vincristine stimulated the outgrowth of a subthreshold inoculum of EL4 (104 living cells), the combination of vincristine with annexin V, cytokine depletion, and/or resolvins inhibited tumor growth (Fig. 5 G).
Figure 5.
Resolvins exhibit additive antitumor activity in combination with chemotherapy or targeted therapy in debris-stimulated and genetically engineered mouse tumor models. (A) Debris-stimulated PancOH7 primary tumor growth with RvE1 and/or gemcitabine. n = 5 mice/group. Treatment initiated once tumors reached 100–200 mm3 throughout unless otherwise specified. Two-way repeated-measures mixed-effects ANOVAs for tumor growth rates and two-tailed Student’s t test for final tumor measurements used throughout unless specified. *, P < 0.05 versus control or RvE1 alone. (B) Debris-stimulated PancOH7 primary tumor growth with RvE1 and/or cetuximab. n = 5 mice/group. *, P < 0.05 versus control or RvE1 alone. (C) Debris-stimulated LLC tumor growth with RvE1 and/or erlotinib. n = 5–10 mice/group. *, P < 0.05 versus control. (D) MMTV-PyMT tumor growth with RvD2 and/or cisplatin. Treatment initiated when mice were 8 wk of age. Tumor volume represents the sum tumor volume of all visible tumors per mouse. n = 5–10 mice/group. *, P < 0.05. (E) Tumor burden in TRAMP mice treated with RvD2 and/or cisplatin. Treatment initiated when mice were 8 wk of age for a duration of 84 d. n = 5–9 mice/group. *, P < 0.05. Images show representative tumors after 84 d of treatment. Bar, 1 cm. (F) Debris-stimulated EL4 tumor growth from in vitro annexin V recombinant protein–treated vincristine-generated EL4 debris coinjected with a subthreshold inoculum of 104 EL4 living cells. Debris was pretreated with annexin V recombinant protein (10 µM) or vehicle. RvD2 treatment and cytokine depletion (IL-6/CCL4/CCL5/TNFα) was initiated on the day of tumor cell injection versus isotype control. n = 5 mice/group. *, P < 0.05 versus control. (G) Tumor growth from 104 EL4 living cells treated with systemic vincristine. Chemotherapy was initiated on the day of tumor cell injection; systemic treatment was initiated on the day of tumor cell injection with annexin V recombinant protein, RvD2, or cytokine depletion (IL-6/CCL4/CCL5/TNFα) versus isotype control. n = 5–10 mice/group. *, P < 0.05 versus vincristine and isotype control. Error bars represent SEM.
Antitumor activity of resolvins is receptor dependent
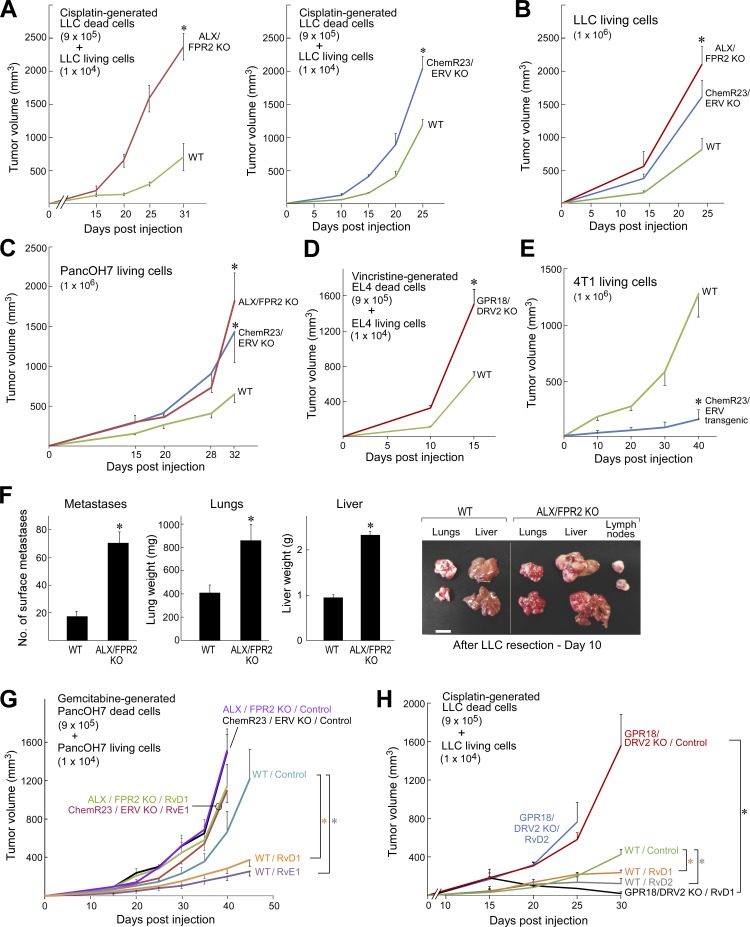
To confirm the specificity of resolvin action, we generated debris-stimulated and nondebris tumors in mice with a genetic deletion (KO) of the resolvin D1 receptor (ALX/FPR2; Dufton et al., 2010), resolvin E1 receptor (ChemR23/ERV; Arita et al., 2007), or resolvin D2 receptor (GPR18/DRV2; Chiang et al., 2015). Debris-stimulated LLC, EL4, and PancOH7 tumors exhibited accelerated growth in ALX/FPR2 KO, ChemR23/ERV KO, and GPR18/DRV2 KO mice in comparison with WT mice (Fig. 6, A, D, G, and H). Nondebris LLC and PancOH7 tumors (106 living cells) also displayed accelerated growth in ALX/FPR2 KO and ChemR23/ERV KO mice (Fig. 6, B and C). In ChemR23/ERV–overexpressing transgenic mice (high expression of the RvE1 receptor in BALB/c background; Gao et al., 2013), mammary tumor growth (4T1) was inhibited compared with WT mice (Fig. 6 E). Lung, liver, and lymph node metastases after primary tumor resection (O’Reilly et al., 1994; Panigrahy et al., 2012) were enhanced in ALX/FPR2 KO mice compared with WT mice (Fig. 6 F). These findings suggest that endogenous resolvins may restrict growth of both debris-stimulated and nondebris tumors. Indeed, measurement of resolvin production by macrophages revealed that debris stimulates resolvin production (Fig. S5, A and B). The antitumor activity of administered RvD1, RvE1, or RvD2 was lost in RvD1 receptor (ALX/FPR2), RvE1 receptor (ChemR23/ERV), and RvD2 receptor (GPR18/DRV2) KO mice, respectively (Fig. 6, G and H).
Figure 6.
Primary tumor growth and metastasis are stimulated in genetically engineered resolvin receptor KO mice. (A and B) Debris-stimulated (A) and nondebris (B) LLC tumor growth in resolvin D1 receptor (ALX/FPR2) KO and resolvin E1 receptor (ChemR23/ERV) KO mice compared with WT mice. n = 4–10 mice/group. Two-way repeated-measures mixed-effects ANOVAs for tumor growth rates and two-tailed Student’s t test for final tumor measurements were used throughout unless specified. (C) Nondebris PancOH7 tumor growth in ALX/FPR2 KO and ChemR23/ERV KO mice compared with WT mice. n = 3–10 mice/group. (D) Debris-stimulated EL4 tumor growth in resolvin D2 receptor (GPR18/DRV2) KO mice compared with WT mice. n = 4–5 mice/group. (E) Nondebris 4T1 tumor growth in ChemR23/ERV transgenic (overexpressed) mice compared with WT mice. n = 3–5 mice/group. (F) Spontaneous LLC lung, liver, and lymph node metastasis in ALX/FPR2 KO mice 10 d after removal of the primary tumor (LLC resection). n = 5–10 mice/group. *, P < 0.05 versus WT. Images show representative lung, liver, and lymph node metastases 10 d after primary tumor resection. Bar, 1 cm. (G and H) Debris-stimulated PancOH7 tumor growth in ALX/FPR2 KO and ChemR23/ERV KO mice (G) as well as debris-stimulated LLC tumor growth in GPR18/DRV2 KO mice vs. WT mice (H). Systemic resolvin treatment was initiated on the day of tumor cell injection. n = 4–5 mice/group. *, P < 0.05 versus WT control, and *, P < 0.05 of GPR18/DRV2 KO / RvD1 (treated with RvD1) versus GPR18/DRV2 KO control. Error bars represent SEM.
Resolvins stimulate macrophage phagocytosis of tumor cell debris and counterregulate macrophage secretion of protumorigenic cytokines
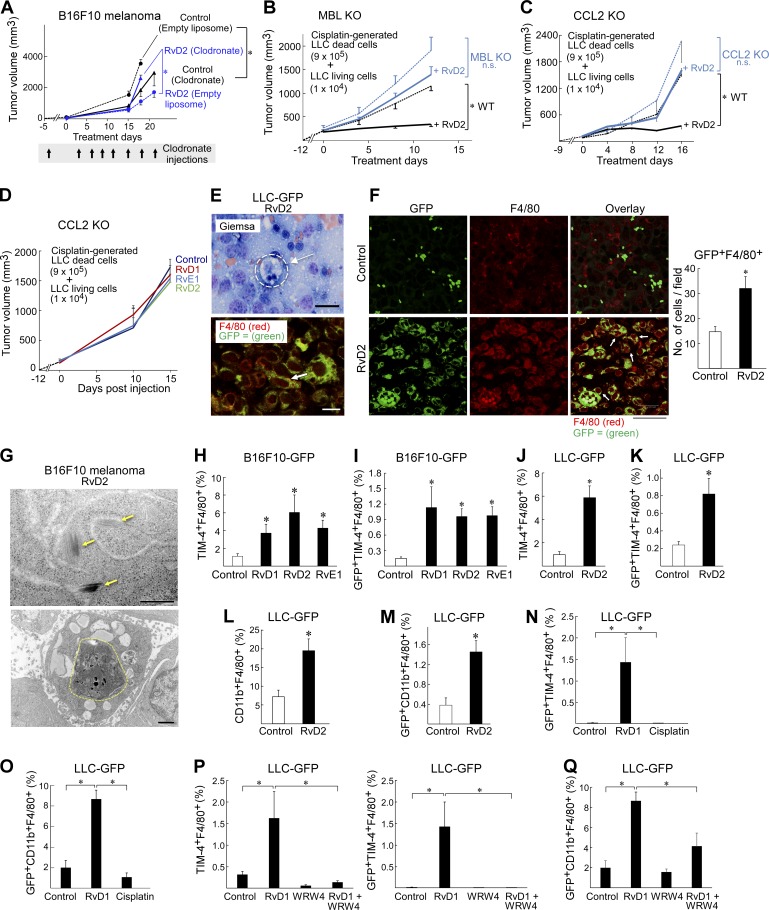
A critical function of resolvins is to stimulate nonphlogistic macrophage phagocytosis of debris (Serhan, 2014). To establish whether the antitumor activity of resolvins is macrophage dependent, we depleted macrophages in mice via clodronate liposomes (Zeisberger et al., 2006). As expected, clodronate liposomes inhibited tumor growth in animal tumor models, because of the known tumor-promoting activity of macrophage infiltration (Qian and Pollard, 2010) compared with control mice administered empty (control) liposomes (Fig. 7 A). Macrophage depletion further abrogated the tumor-inhibitory actions of RvD2, resulting in faster growing tumors than RvD2-treated mice administered empty liposomes (Fig. 7 A). Neither RvD1, RvD2, nor RvE1 inhibited tumor growth in mannose-binding lectin (MBL)–deficient (MBL KO; Stuart et al., 2005; Stienstra et al., 2014) or CCL2 KO (Lu et al., 1998) mice, two genetically engineered models with impaired macrophage phagocytosis and chemotaxis, respectively, consistent with defective clearance (Fig. 7, B–D). Thus, the presence of functional macrophages was necessary for maximal tumor inhibition by resolvins.
Figure 7.
Antitumor activity of resolvins is macrophage dependent and induces a prophagocytic macrophage phenotype. (A) B16F10 (106 living cells) tumor growth after macrophage depletion with clodronate liposomes and RvD2 or vehicle treatment. n = 5–10 mice/group. Systemic RvD2 treatment was initiated on the day of tumor cell injection. Clodronate was administered 3 d before tumor cell injection and every 3 d thereafter for 21 d. Two-way repeated-measures mixed-effects ANOVAs for tumor growth rates and two-tailed Student’s t test for final tumor measurements were used throughout unless specified. *, P < 0.05. (B and C) Debris-stimulated LLC tumor growth in MBL KO (B) and CCL2 KO (C) mice with systemic RvD2 treatment. n = 5–7 mice/group. Resolvin treatment was initiated once tumors reached 100–200 mm3 throughout unless otherwise specified. *, P < 0.05 versus WT control (black dashed lines). (D) Debris-stimulated LLC tumor growth in resolvin or vehicle-treated CCL2 KO mice. n = 5–10 mice/group. (E) Giemsa staining of RvD2-treated LLC-GFP tumor (top); macrophage ingesting tumor cell (dashed circle). Immunofluorescent double-staining (bottom) for tumor cell marker GFP (green) and macrophage marker F4/80 (red) or colocalization of macrophages and tumor cells (yellow, indicated by arrows). (F) Immunofluorescent double staining for tumor cell marker GFP (green, left) and macrophage marker F4/80 (red, center) or colocalization of macrophages and tumor cells (macrophage phagocytosis indicated by arrows, right) in RvD2- or vehicle-treated LLC-GFP tumors on day 14 of treatment. Macrophage phagocytosis quantified by GFP+F4/80+ cells/field. Bars, 10 µm. *, P < 0.05 versus control. Images represent five sections each of four different samples. (G) Top: electron microscopy of RvD2-treated B16F10 tumors showing melanosomes (yellow arrows). Bottom: macrophage ingesting melanoma tumor cells (dashed circle). Bars: (top) 500 nm; (bottom) 1 µm. (H and I) Flow cytometry analysis of resolvin (RvD1, RvD2, or RvE1) versus vehicle-treated B16F10-GFP tumors for efferocytic macrophages (percent TIM-4+F4/80+) and macrophage efferocytosis of tumor debris (percent GFP+TIM-4+F4/80+). GFP+ cells were gated on TIM-4+F4/80+ cell populations. Macrophage efferocytosis of tumor debris quantified as the proportion of GFP+TIM-4+F4/80+ cells in each tumor sample. n = 4 mice/group. *, P < 0.05 versus control. (J and K) Flow cytometry analysis of efferocytic macrophages (TIM-4+F4/80+) in RvD2 versus vehicle-treated LLC-GFP tumors. GFP+ cells were gated on TIM-4+F4/80+ cell populations. Macrophage phagocytosis of tumor debris quantified as the proportion of GFP+TIM-4+F4/80+ cells in each tumor sample. n = 4 mice/group. *, P < 0.05 versus control. (L) Flow cytometry analysis of phagocytic macrophages (CD11bhighF4/80+) in RvD2- versus vehicle-treated LLC-GFP tumors. n = 6 mice/group. *, P < 0.05 versus control. (M) Macrophage phagocytosis of tumor debris quantified as the proportion of GFP+CD11b+F4/80+ cells in each tumor sample. GFP+ cells were gated on CD11b+F4/80+ cell populations. n = 3–6 mice/group. *, P < 0.05 versus control. (N) Flow cytometry analysis of LLC-GFP tumors treated systemically with RvD1 or cisplatin. GFP+ cells were gated on TIM-4+F4/80+ cell populations. Macrophage efferocytosis of tumor debris quantified as the proportion of GFP+TIM-4+F4/80+ cells in each tumor sample. n = 4–5 mice/group. *, P < 0.05 versus control. (O) Flow cytometry analysis of LLC-GFP tumors treated systemically with RvD1 or cisplatin. Macrophage phagocytosis of tumor debris quantified as the proportion of GFP+CD11b+F4/80+ cells in each tumor sample. n = 4–5 mice/group. *, P < 0.05 versus control. (P) Flow cytometry analysis of efferocytic macrophages (TIM-4+F4/80+; left) in LLC-GFP tumors treated systemically with RvD1 and/or WRW4. GFP+ cells were gated on TIM-4+F4/80+ cell populations. Macrophage efferocytosis of tumor debris quantified as the proportion of GFP+TIM-4+F4/80+ cells in each tumor sample (right). n = 4–5 mice/group. *, P < 0.05 versus control. (Q) Flow cytometry analysis of phagocytic macrophages (GFP+CD11b+F4/80+) in LLC-GFP tumors treated systemically with RvD1 and/or WRW4. Macrophage phagocytosis of tumor debris quantified as the proportion of GFP+CD11b+F4/80+ cells in each tumor sample. n = 4–5 mice/group. *, P < 0.05 versus control or RvD1. Data are representative of two biological repeats. Error bars represent SEM.
To demonstrate clearance of tumor debris by phagocytosis in vivo, we performed immunohistochemical analysis of GFP-labeled tumors in animals treated with resolvins. Sections showed macrophages (identified by Giemsa or F4/80 stain) containing GFP signals, indicative of phagocytosed tumor cells (Fig. 7, E and F). Moreover, electron microscopy of B16F10 melanoma tumors revealed melanosomes (a distinct electron-dense tumor cell marker; Drochmans, 1960) in the cytoplasm of macrophages in RvD2-treated tumors (Fig. 7 G). To confirm the localization of tumor cell debris in macrophages was caused by resolvin-stimulated phagocytosis, we used flow cytometry to detect macrophages in GFP-labeled tumors that (a) were positive for TIM-4, a PS-mediated macrophage efferocytosis marker (Kobayashi et al., 2007; Miyanishi et al., 2007; Wong et al., 2010), or CD11b, which distinguishes between phagocytic (CD11bhigh) and “satiated” (CD11blow) macrophage subtypes (Schif-Zuck et al., 2011); and (b) contained tumor cell material (GFP+), indicative of phagocytosis. Systemic treatment with resolvins (RvD1, RvD2, or RvE1) increased the proportion of efferocytic double-positive (TIM-4+ F4/80+) macrophages compared with vehicle-treated tumors. Resolvins further increased the fraction of macrophages that were triple-positive (GFP+TIM-4+F4/80+), indicating ingestion of tumor cells (LLC-GFP or B16F10-GFP) by efferocytosis, by fourfold compared with vehicle-treated tumors (Fig. 7, H–K). Similarly, resolvin-treated tumors exhibited more than double the proportion of double-positive (CD11b+F4/80+) phagocytic macrophages compared with vehicle-treated tumors and almost fourfold more macrophages were triple-positive (GFP+CD11b+F4/80+), indicative of phagocytosis of GFP-labeled tumor cells (Fig. 7, L and M). In contrast, chemotherapy (e.g., cisplatin) did not stimulate macrophage phagocytosis or efferocytosis of GFP-labeled tumor cells (Fig. 7, N and O). The RvD1 receptor antagonist (WRW4) inhibited resolvin-stimulated macrophage tumor cell phagocytosis and efferocytosis, albeit not completely (Fig. 7, P and Q).
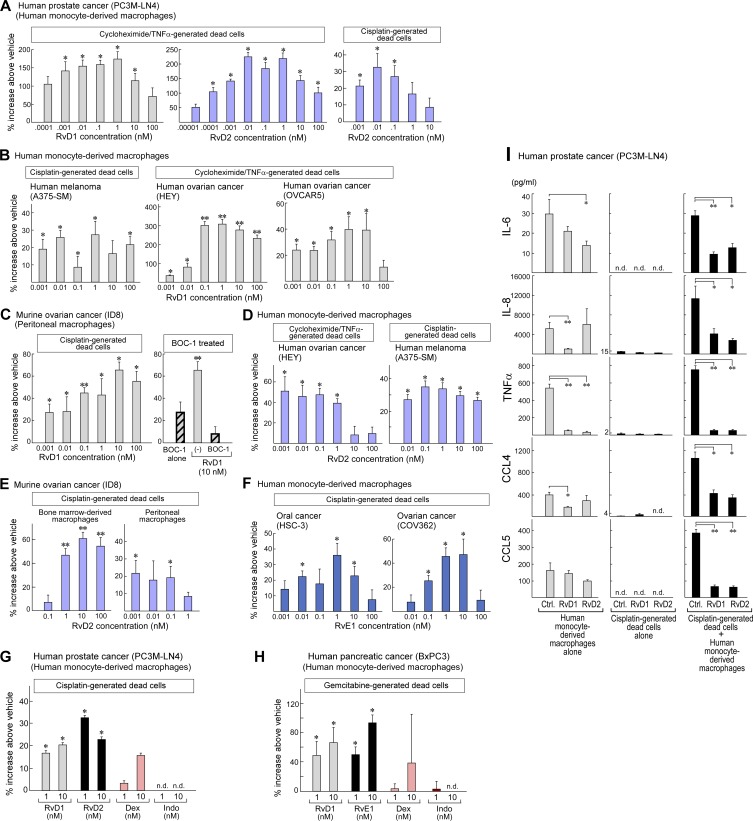
Consistent with the observed resolvin-stimulated phagocytosis of tumor debris in vivo (Fig. 7, E–Q), treatment of human monocyte–derived and mouse macrophages in vitro with RvD1, RvD2, or RvE1 at 1 pM to 100 nM stimulated phagocytosis of debris from several human and mouse tumor cell lines (PC3M-LN4, BxPC3, COV362, A375-SM, HEY, OVCAR5, HSC-3, and ID8) generated by cisplatin, gemcitabine, or cycloheximide plus TNFα (Fig. 8, A–H). A biphasic dose-response curve with diminishing activity at doses >1 nM has also been observed for resolvin-stimulated phagocytosis of neutrophils (Spite et al., 2009; Chiang et al., 2012). Moreover, the biological activity of resolvins is mediated by G protein–coupled receptors, known to display characteristic bell-shaped dose responses when activated by their cognate ligands (Perretti et al., 2002; Krishnamoorthy et al., 2010). The bell-shaped dose response is consistent with many other studies in which resolvins stimulate macrophage phagocytosis of cell debris in noncancer settings, including sepsis (Spite et al., 2009), infection (Chiang et al., 2012), clot remodeling (Elajami et al., 2016), and obesity (Titos et al., 2011). Stimulation of phagocytosis by resolvins was receptor specific, as the selective RvD1 receptor (ALX/FPR2) antagonist, BOC-1, neutralized RvD1-stimulated phagocytosis of debris by peritoneal macrophages (Fig. 8 C). In contrast, the antiinflammatory drugs dexamethasone and indomethacin did not significantly stimulate macrophage phagocytosis of debris (Fig. 8, G and H).
Figure 8.
Resolvins stimulate macrophage phagocytosis of tumor cell debris and counterregulate the secretion of protumorigenic cytokines by macrophages exposed to tumor cell debris. (A–H) Macrophage phagocytosis of CFDA-labeled tumor cell debris measured as RFUs after resolvin treatment. RFUs are displayed as percent increase above vehicle throughout. Data are representative of four biological repeats. (A) Human macrophage phagocytosis of cycloheximide plus TNFα– or cisplatin-generated dead cells (PC3M-LN4) after RvD1 (gray bars) or RvD2 (purple bars) treatment. n = 6/group. *, P < 0.05 versus vehicle. (B and C) Human macrophage phagocytosis of cisplatin-generated dead cells (A375-SM) or cycloheximide plus TNFα–generated dead cells (HEY and OVCAR5) after RvD1 treatment (B) and mouse peritoneal macrophage phagocytosis of cisplatin-generated dead cells (mouse ovarian cancer; ID8) treated with the RvD1 receptor (ALX/FPR2) antagonist BOC-1 and/or RvD1 (C). n = 6/group. (D) Human macrophage phagocytosis of cycloheximide plus TNFα–generated dead cells (HEY) or cisplatin-generated dead cells (A375-SM) after RvD2. n = 6/group *, P < 0.05 versus vehicle. (E) Mouse bone marrow–derived macrophage and peritoneal macrophage phagocytosis of cisplatin-generated ID8 dead cells after RvD2 treatment. n = 6/group. *, P < 0.05; **, P < 0.0001 versus vehicle. (F) Human macrophage phagocytosis of cisplatin-generated dead cells (HSC-3 and COV362) after RvE1 treatment. n = 6/group (left) or n = 12/group (right). *, P < 0.05 versus vehicle. (G and H) Human macrophage phagocytosis of tumor cell debris (cisplatin-generated PC3M-LN4 or gemcitabine-generated BxPC3) after RvD1 (gray bars), RvD2 or RvE1 (black bars), dexamethasone (pink bars), or indomethacin (red bars) treatment. n = 6/group. *, P < 0.05 versus vehicle. Nondetectable RFUs are labeled n.d. (I) ELISA quantification of IL-6, IL-8, TNFα, CCL4, and CCL5 production by resolvin-treated human monocyte–derived macrophages coincubated with (black bars) or without (gray bars) tumor cell debris (cisplatin-generated PC3M-LN4) or by tumor cell debris alone (cisplatin-generated dead cells alone). n = 5–6/group. Data are representative of three biological repeats (three human peripheral blood monocyte donors). *, P < 0.05; **, P < 0.0001 versus control. Error bars represent SEM.
Stimulation of the resolution of inflammation by resolvins not only enhances macrophage phagocytosis of debris, but it also counterregulates proinflammatory cytokine production and halts leukocyte infiltration (Serhan, 2014). RvD1 or RvD2 (1 nM) inhibited the release of proinflammatory tumor-promoting cytokines by tumor debris–activated macrophages including IL-6, IL-8, TNFα, CCL4, and CCL5 by up to 10–40% compared with vehicle-treated macrophages (Fig. 8 I). As only traces of cytokines were found in tumor cell debris alone, these mediators were products of macrophages and not of debris (Fig. 8 I). To quantify leukocyte infiltration in vivo after resolvin treatment, we mimicked the tumor microenvironment using a Matrigel plug assay (Benton et al., 2009). Systemic administration of RvD1 or RvD2 inhibited the proportion of infiltrating leukocytes (CD45+) found in the dissociated implant, whereas RvD2 also decreased the proportion of infiltrating macrophages (CD45+F4/80+; Fig. S5, C and D). RvD2 also inhibited the proportion of infiltrating macrophages in LLC-GFP tumors (Fig. S5 E). Systemic resolvins also decreased blood vessel formation (as quantified by the number of CD31+ cells coinciding with vessel structures; Fig. S5 F).
It is not likely that the antitumor activity of resolvins was mediated by direct antiproliferative or antimigratory activity on tumor cells. Neither RvD1 nor RvD2 (≤1 µM) inhibited proliferation of B16F10, LLC, or PC3M-LN4 cells in vitro (Fig. S5 G). In addition, neither RvD1 nor RvD2 inhibited tumor cell migration in vitro (Fig. S5 H). Also, Western blot analysis did not detect expression of the RvD1 receptor ALX/FPR2 in six different mouse tumor cell lines including LLC, B16F10, T241, pancreatic β cell tumor (BTC), sarcoma (MS-180), and hemangioendothelioma (EOMA; Fig. S5 I). Immunohistochemical analysis of clinical prostate carcinoma specimens showed that the human RvD1 receptor GPR32/DRV1 was expressed on tumor-infiltrating inflammatory cells but not on tumor cells (Fig. S5 J). Similarly, histological staining for ALX/FPR2 in sections of LLC tumors revealed expression only in stromal cells, including tumor-associated inflammatory cells and endothelial cells (Fig. S5 K). This was confirmed by double staining of LLC tumors for ALX/FPR2 and the macrophage marker F4/80 or the endothelial cell marker MECA-32 (Fig. S5, L and M). Collectively, these results indicate that the antitumor activity of resolvins may be mediated by cells in the tumor stroma and not by direct action on tumor cells.
Discussion
In this study, we demonstrate that tumor cell debris can stimulate tumor growth, which has pivotal implications for the treatment of cancer patients. One central mechanism of debris-stimulated tumor growth, in which debris is generated by chemotherapy or targeted therapy in vitro or in situ in grafted tumor models, is via stimulating macrophage release of proinflammatory cytokines. Resolvins promote nonphlogistic clearance of debris by TIM4+ and CD11b+ phagocytic macrophages and suppress therapy-induced tumor growth. Current antiinflammatory cancer therapies have focused on suppressing proinflammatory mediators, (i.e., cytokines and prostaglandins; Wang and DuBois, 2010); however, they possess limited therapeutic efficacy. In this study, we demonstrate that endogenous clearance of inflammation (caused by tumor cell debris) mediated by resolvins contributes to the suppression of tumor growth (Fig. S5 N). Notably, resolvins (RvD1, RvD2, or RvE1) inhibited tumor growth at doses 10,000 times lower than their substrates (eicosapentaenoic acid and docosahexaenoic acid; Grenon et al., 2013) or other antiinflammatory agents such as aspirin and other nonsteroidal antiinflammatory drugs (NSAIDs; Rothwell et al., 2012; Fullerton and Gilroy, 2016).
Debris-stimulated tumor growth is likely relevant to many types of current cancer therapy, including chemotherapy, radiation, and targeted therapy. During cytotoxic tumor treatment, some tumor cells inevitably survive (Pisco and Huang, 2015). Thus, in the process of reducing tumor burden via cytotoxic mechanisms, continuous production of chemotherapy-generated apoptotic cell debris in tumors perpetuates tumor growth via tumor-promoting cytokines released by macrophages (Fig. S5 N). The failure to clear apoptotic cells in a timely manner and the accumulation of apoptotic cells within tissue can stimulate an inflammatory response (Birge et al., 2016). Given the large debris to living cell ratio used for tumor implantation in the debris-stimulated tumor models (30–90:1 debris cells/living cells), the mass accumulation of debris and defective clearance may promote an inflammatory response that in turn contributes to debris-stimulated tumor progression. We show that chemotherapy-generated tumor cell debris stimulates living tumor cells, acting as a “feeder,” consistent with previous research on radiation-generated apoptotic tumor cells (Huang et al., 2011). Thus, cytotoxic cancer treatment designed to kill tumor cells may be a double-edged sword. Our preclinical studies in a large variety of tumor models demonstrate that the growth-stimulating activity of therapy-generated cell debris may contribute to the inherent limitation of cancer treatment in general. Indeed, we confirmed that established living cell tumors in mice contain cell debris that can be stimulated in situ by systemic administration of chemotherapy, supporting the pathophysiological relevance of the debris-stimulated tumor models. Thus, debris-stimulated tumor growth may have clinical relevance. Our studies on chemotherapy-generated debris are consistent with previous observations on radiation-generated tumor cell debris by Révész (1956), in which the stimulation of tumor growth had been attributed to the production of diffusible factors that conditioned the tumor microenvironment (Révész phenomenon; Révész, 1956; Seelig and Revesz, 1960; van den Brenk et al., 1977). The Révész phenomenon has been confirmed in followup studies of radiation-induced cell death (Huang et al., 2011; Chaurio et al., 2013; Donato et al., 2014; Ford et al., 2015; Yu et al., 2016; da Silva-Jr et al., 2017).
Our studies also have implications for tumor immunotherapy. Specifically, we demonstrate that debris-stimulated tumor growth is in part mediated by PS, which accumulates in the tumor microenvironment and may antagonize adaptive tumor immunity (Birge et al., 2016). In support of our data, an apoptotic response to therapy has been shown to generate a PS-mediated immunosuppressive environment (Mochizuki et al., 2003; Birge et al., 2016), a response in which prostaglandin release has recently been implicated (Hangai et al., 2016). Our data are also consistent with an alternative mechanism in which annexin V, a naturally occurring specific PS ligand, suppresses the tumorigenicity of dead tumor cells by promoting antitumor immunity (Frey et al., 2009). In addition, annexin V–coupled irradiated cells induced the regression of growing tumors (Bondanza et al., 2004), and administration of radiation and anti-PS antibody induced tumor immunity in a glioblastoma model (He et al., 2009). Moreover, PS-targeting antibodies augment the antitumor activity of immunotherapy by enhancing immune activation (Gray et al., 2016).
The concept of immunogenic cell death postulates that debris generated by certain (but not all) chemotherapeutic agents (e.g., doxorubicin, anthracyclines, oxaliplatin, or bleomycin) would stimulate the uptake of cell debris by dendritic cells for antigen presentation or contribute to adaptive immunity by acting as an adjuvant (Casares et al., 2005; Obeid et al., 2007). However, debris stimulated tumor growth in the subthreshold inoculum model presented in this study. This may be a result of the response to tumor cell debris by the innate immune system and inflammation, which may overwhelm and counteract a potential immunizing effect of immunogenic cell death. However, other studies demonstrate that tumor cell debris generated by radiation, the Herpes simplex virus thymidine kinase/ganciclovir (HSVtk/GCV) system, photodynamic therapy, or radiofrequency ablation (e.g., ultrasound) can inhibit tumor growth, an effect that had been attributed to induction of antitumor immunity (Melcher et al., 1998; Todryk et al., 1999; Gough et al., 2001; Akazawa et al., 2004; Bondanza et al., 2004; Casares et al., 2005; Apetoh et al., 2007; Korbelik et al., 2007; Obeid et al., 2007; Dromi et al., 2009; Deng et al., 2010; Garg et al., 2010, 2012; Unga and Hashida, 2014; Shan et al., 2015; Kindy et al., 2016). Our studies suggest that the generation of therapy-generated tumor cell death may be a doubled-edged sword. Future studies are required to establish the specific conditions in which therapy-generated cell debris suppresses or activates antitumor immunity.
We also demonstrate in this study that chemotherapy-generated tumor cell debris triggers a “cytokine storm” that stimulates tumor growth that is resistant to chemotherapy. Immunotherapy, including chimeric antigen receptor (CAR)–T cell therapy, may lead to the release of toxic levels of cytokines (DeFrancesco, 2014). This therapy-induced cytokine release may be particularly relevant in patients with dormant or small residual tumors as modeled by the subthreshold inoculum tumor models, and it may provide a potential mechanism whereby chemotherapy paradoxically harbors the potential to stimulate or induce tumor initiation, growth, and/or metastasis (de Ruiter et al., 1979; Ormerod et al., 1986; Orr et al., 1986; Poth et al., 2010; Abubaker et al., 2013; Volk-Draper et al., 2014; Vyas et al., 2014; Gunjal et al., 2015; Chang et al., 2017; Karagiannis et al., 2017). We show in the subthreshold inoculum model that systemic chemotherapy stimulated small or dormant tumors to grow, instead of suppressing the tumor. However, the therapeutic activity of chemotherapy was restored in the debris-stimulated tumor models if the source of this inflammatory cascade, namely the PS presented by apoptotic cells, was neutralized with annexin V or anti-PS antibodies (Fig. 2, G and H). Collectively, these findings suggest a critical role for PS in debris-stimulated tumors.
Therapy-generated tumor cell debris activates macrophages to secrete proinflammatory and protumorigenic cytokines, thus sustaining an inflammatory tumor microenvironment, which in turn promotes tumor growth (Fig. S5 N). This protumorigenic activity could fuel a positive feedback loop that is difficult to overcome with more aggressive cytotoxic therapy. Sterile inflammation is sustained by the presence of cell debris, as apoptotic cells release inflammation-initiating “danger signals” (Kornbluth, 1994; Chan et al., 2012). However, resolvins can polarize these protumorigenic and proinflammatory macrophages to a prophagocytic state, inhibiting further proinflammatory cytokine secretion. Consistently, clearance of tumor cell debris disrupts debris-dependent tumor growth. Thus, resolvins (i.e., RvE1, RvD1, and RvD2) represent a novel mechanism to suppress tumor progression, growth, and recurrence. Unlike the majority of antiinflammatory agents, including NSAIDs, resolvins are endogenous, nonimmunosuppressive, and nontoxic inhibitors of inflammation (Serhan et al., 2002). We show that the specific resolvins RvD1, RvD2, and RvE1 promote the clearance of tumor debris and subsequent inhibition of tumor growth by stimulating macrophage phagocytosis of tumor cell debris and by counterregulating the release of critical proinflammatory protumorigenic cytokines/chemokines.
We showed that adding resolvins to existing chemotherapy or targeted therapy regimens induced sustained regression of primary tumors by blocking the tumor-stimulatory activity of therapy-generated tumor cell debris produced by cytotoxic therapy. Although both aspirin and omega-3 fatty acids reduce cancer risk (Greene et al., 2011; Rothwell et al., 2012), they only weakly trigger the production of resolvins by the human body (Sun et al., 2007). Resolvins have entered clinical development as novel therapeutic approaches for inflammatory diseases including keratoconjunctivitis sicca, periodontal disease, eczema, and various neurodegenerative diseases. Current antiinflammatory agents, including NSAIDs, have potentially severe side effects, such as stomach and brain bleeding, as well as cardiovascular and kidney toxicity. In contrast, targeting the resolvin pathways provides an entirely new, nontoxic, and nonimmunosuppressive approach to cancer therapy by increasing the body’s natural production of endogenous proresolving and antiinflammatory mediators.
Rapid tumor growth is invariably linked to apoptotic cell death because of unfavorable conditions such as hypoxia (Holmgren et al., 1995; de Jong et al., 2000; Alcaide et al., 2013; Gregory et al., 2016; Ichim and Tait, 2016), and continuous production of apoptotic cell debris sustains inflammation that can stimulate tumor growth (Kornbluth, 1994). Thus, natural apoptotic cell death can also contribute to tumor progression (Reiter et al., 1999), and this underappreciated source of tumor stimulation is further enhanced by treatment-induced apoptosis. High levels of spontaneous apoptotic cell death in tumors of patients with cancer have also been shown to correlate with poor prognosis in several cancer types and may be causatively involved in tumor growth (Wyllie, 1985; Kornbluth, 1994; de Jong et al., 2000; Naresh et al., 2001; Jalalinadoushan et al., 2004; Sun et al., 2006; Gregory and Pound, 2011; Alcaide et al., 2013; Gregory et al., 2016; Ichim and Tait, 2016). This may explain the moderate antitumor activity of resolvins in the nondebris tumor models. Moreover, cytotoxic treatment in cancer patients without evidence of progressive cancer could be a double-edged sword as therapy-generated debris could inadvertently stimulate proliferation of dormant tumor cells or small tumors. Although generation of tumor cell debris throughout treatment may explain an inherent therapeutic limit to chemotherapy, targeted therapy, and any cytocidal therapy, stimulating the clearance of such tumor cell debris via specialized proresolving mediators such as resolvins represents a novel approach to prevent tumor growth and recurrence.
Materials and methods
Tumor debris, xenograft, and metastasis studies
Reporting of the following animal experiments abided by the Animal Research: Reporting of In Vivo Experiments (ARRIVE) guidelines (Kilkenny et al., 2010). All animal studies were reviewed and approved by the Animal Care and Use Committees of Boston Children’s Hospital and Beth Israel Deaconess Medical Center. Animals at each institution were housed up to five mice/cage in a pathogen-free facility. Mice had unlimited access to sterile water and chow. Throughout each animal experiment, daily welfare evaluations were performed, and animal sacrifice guidelines were followed per institutional committee guidelines.
Experiments involving human blood cells (deidentified) at Brigham and Women’s Hospital are under protocol 1999P001297, and discarded human materials are under protocol 1999P001279. Both protocols are approved by the Partners Human Research Committee.
Generation of debris by chemotherapy or targeted therapy: General note
Cell debris for the debris-stimulated tumor models was generated in vitro by incubating cells in fresh media containing the drug at the indicated dose for the indicated time (detailed for each tumor type below). For annexin V/PI flow cytometry characterization of debris, only the media containing floating cells (debris) from drug-treated cell cultures was assessed. This was compared with vehicle-treated (control) cell cultures in which media was collected and combined with trypsinized cells (Fig. S1, A–J). The representative FACS analyses for control groups included all cells taken from a single culture, and both adhered and floating cells were collected. For mouse tumor injections, living cells were collected by trypsinization of adherent cells only from untreated cell cultures, whereas debris was collected for injection by aspirating floating cells from drug-treated cell cultures. Surrogate measurement for quantification of debris involved counting whole-cell bodies in the floating cell population after the specified treatment (except for the suspension cultures, i.e., EL4 cells, see the Vincristine-generated EL4 debris tumors section), indicated as number of dead cells (Fig. S1 K). This quantitation provides a tool for standardization and comparison of various tumor cell lines. Protocols for debris generation (drug dose and length of treatment) were adjusted such that at 900,000 dead cells in all tumor cell lines used, robust debris-stimulated tumor growth was observed when coinjected with a subthreshold inoculum of living tumor cells. This subthreshold inoculum was separately determined by injection of living cells at a (typical) series of 106, 105, 104, and 103 cells, and assessment of tumor take was observed for 200–400 d. Once the quantitative parameters for each drug and cell line combination used were established, routine generation of debris for tumor studies followed a strict protocol regarding treatment, cell collection, washing, and handling before injection. Dead cells (apoptotic and necrotic) and living cells were counted by hemocytometer. Pelleted dead and living cells were resuspended at desired concentrations. Experimental groups were then prepared by mixing equal volumes of dead cell bodies with living cells. The specifics are listed below.
Cisplatin-generated LLC debris tumors
Cisplatin-generated LLC debris was prepared by treating 75–80% confluent T150 flasks with complete media with 10% FBS plus 50 µM cisplatin (Sigma-Aldrich) and incubating for 24 h. Dead cell bodies (apoptotic and necrotic cells) were counted by hemocytometer. Pelleted cells were resuspended at 1.8 × 107 dead cells/ml in PBS. Untreated LLC cells were trypsinized, pelleted, and resuspended at 2 × 105 living cells/ml in PBS. Experimental groups were prepared by mixing equal volumes of dead cell bodies with living cells. Cisplatin-generated LLC debris (9 × 105, 3 × 105, or 105 dead cells) and/or LLC (102, 103, or 104 living cells) were coinjected into C57BL/6J, RAG1 KO mice (The Jackson Laboratory), and SCID mice (Charles River). All tumor debris and/or living tumor cells were injected subcutaneously into the mid-dorsum of 6–8-wk-old male mice at 100 µl/mouse with a 30-G needle unless specifically noted. Cisplatin-generated LLC debris (9 × 105 dead cells) and/or T241, B16F10, LLC, or PancOH7 (104 living cells) were coinjected into C57BL/6J mice. LLC (104 or 106 living cells) were injected into C57BL/6J mice, and systemic cisplatin was initiated on the day of tumor injection (see the Tumor inhibition studies section).
Gemcitabine-generated PancOH7 debris tumors
Gemcitabine-generated PancOH7 debris was prepared by treating 75–80% confluent T150 flasks with complete media with 10% FBS plus 40 µM gemcitabine (Sigma-Aldrich) and incubating for 72 h. Dead cell bodies were counted and prepared as described in the previous section. Gemcitabine-generated PancOH7 debris (9 × 105 dead cells) and/or PancOH7 (104 living cells) were coinjected into C57BL/6J, RAG1 KO, and SCID mice. For orthotopic tumors, gemcitabine-generated PancOH7 debris (9 × 105 dead cells) and/or PancOH7 (104 living cells) were coinjected directly into the pancreas of C57BL/6J mice with a 30-G needle.
Vincristine-generated EL4 debris tumors
Vincristine-generated EL4 debris was prepared by treating confluent T150 flasks (7 × 106 cells) with complete media with 10% horse serum plus 40 nM vincristine (Sigma-Aldrich) and incubating for 72 h. Dead cell bodies were isolated via Ficoll gradient (Enzo Life Sciences) and resuspended in PBS at 1.8 × 107 cells/ml. Untreated EL4 cells were pelleted and resuspended in PBS at 2 × 105 cells/ml. Experimental groups were prepared as described in the General note and injected into C57BL/6J mice. Vincristine-generated EL4 debris (9 × 105 dead cells) and/or EL4, LLC, B16F10, or PancOH7 (104 living cells) were prepared as described in the General note and coinjected into C57BL/6J mice. EL4 (104 or 106 living cells) were injected into C57BL/6J mice, and systemic vincristine (Sigma-Aldrich) was initiated on the day of tumor injection (see the Tumor inhibition studies section).
Docetaxel-generated HSC-3 debris tumors
Docetaxel-generated HSC-3 debris was prepared by treating 75–80% confluent T150 flasks with complete media with 10% FBS plus 10 nM docetaxel (Sigma-Aldrich) and incubating for 48 h. Dead cell bodies were counted and prepared as described in the General note. Untreated HSC-3 cells were trypsinized, pelleted, and resuspended at 106 living cells/ml in PBS. Subsequent groups were made by serial dilutions and combining equal volumes of dead cell bodies and living cells. Docetaxel-generated HSC-3 debris (9 × 105, 3 × 105, or 105 dead cells) and/or HSC-3 (5 × 104 living cells) were coinjected into SCID mice.
Erlotinib-generated human and mouse lung debris tumor
Erlotinib-generated HCC827 or LLC debris was prepared by treating 75–80% confluent T150 flasks with complete media with 10% FBS plus 10 µM erlotinib (SelleckChem) and incubated for 72 h. Dead cell bodies were counted and prepared as described in the General note and resuspended in PBS at 3.6 × 107 dead cells/ml. Untreated HCC827 and untreated LLC were trypsinized, pelleted, and resuspended in PBS at 106 living cells/ml and 2 × 105 living cells/ml, respectively. Subsequent groups were made by serial dilutions and combining equal volumes of dead cell bodies and living cells. Erlotinib-generated HCC827/LLC debris (9 × 105 or 1.8 × 106 dead cells) and/or HCC827/LLC (5 × 104 or 104 living cells, respectively) were coinjected into SCID mice or C57BL/6J mice, respectively.
Cetuximab-generated PancOH7 debris tumors
Cetuximab-generated PancOH7 debris was prepared by treating 75–80% confluent T150 flasks with complete media with 10% FBS plus 343 nM cetuximab and incubating for 72 h. Dead cell bodies were counted and prepared as described in the General note and resuspended in PBS at 3.6 × 107 dead cells/ml. Untreated PancOH7 cells were trypsinized, pelleted, and resuspended in PBS at 2 × 105 living cells/ml. Experimental groups were prepared by mixing equal volumes of dead cell bodies with living cells as described in the General note and coinjected into C57BL/6J mice.
Gemcitabine-generated BxPC3 debris tumors
Gemcitabine-generated BxPC3 debris was prepared by treating 75–80% confluent T150 flasks with complete media with 10% FBS plus 40 µM gemcitabine and incubating for 72 h. Dead cell bodies were counted and prepared as described in the General note and then resuspended in PBS at 1.8 × 106 dead cells/ml. Untreated BxPC3 cells were trypsinized, pelleted, and resuspended in PBS at 106 or 107 living cells/ml. Experimental groups were prepared by mixing equal volumes of dead cell bodies with living cells as described in the General note. Gemcitabine-generated BxPC3 debris (9 × 105 dead cells) and/or BxPC3 (5 × 104 living cells) were coinjected into SCID mice.
Cisplatin-generated PC3M-LN4 debris tumors
Cisplatin-generated PC3M-LN4 debris was prepared by treating 75–80% confluent T150 flasks with complete media with 10% FBS plus 50 µM cisplatin and incubating for 24 h. Dead cell bodies were counted and prepared as described in the General note and resuspended in PBS at 1.8 × 106 dead cells/ml. Untreated PC3M-LN4 tumor cells (provided by I.J. Fidler, University of Texas MD Anderson Cancer Center, Houston, TX) were trypsinized, pelleted, and resuspended in PBS at 107 living cells/ml. Experimental groups were prepared by mixing equal volumes of dead cell bodies with living cells as previously described. PC3M-LN4 debris (9 × 105 dead cells) and/or PC3M-LN4 (5 × 104 living cells) were coinjected into SCID mice. Treatment of debris-stimulated (9 × 105 dead cells with 5 × 104 living cells/mouse) or nondebris (106 living cells/mouse) PC3M-LN4 tumors with cisplatin or resolvins was initiated on the day of tumor injection.
Oxaliplatin-generated MC38 debris tumors
Oxaliplatin-generated MC38 debris was prepared by treating 75–80% confluent T150 flasks with complete media with 10% FBS plus 50 µM oxaliplatin (MedChem Express) and incubating for 48 h. Dead cell bodies were counted and prepared as described in the General note. Oxaliplatin-generated MC38 debris (9 × 105 dead cells) and/or MC38 (104 living cells) were coinjected into C57BL/6J mice, and systemic oxaliplatin was initiated on the day of tumor injection (see the Tumor inhibition studies section).
Cisplatin-generated MC38 debris tumors
Cisplatin-generated MC38 debris was prepared by treating 75–80% confluent T150 flasks with complete media with 10% FBS plus 50 µM cisplatin (Sigma-Aldrich) and incubating for 48 h. Dead cell bodies were counted and prepared as described in the General note. Cisplatin-generated MC38 debris (9 × 105 dead cells) and/or MC38 (104 living cells) were coinjected into C57BL/6J mice.
Idarubicin-generated CT26 debris tumors
Idarubicin-generated CT26 debris was prepared by treating 75–80% confluent T150 flasks with complete media with 10% FBS plus 1 µM idarubicin (Sigma-Aldrich) and incubating for 24 h. Dead cell bodies were counted and prepared as described in the General note. Idarubicin-generated CT26 debris (9 × 105 dead cells) and/or CT26 (104 living cells) were coinjected into BALB/c mice.
Nondebris tumors (living cells only)
For nondebris tumors, untreated tumor cells (LLC, LLC-GFP, EL4, PancOH7, B16F10, B16F10-GFP, BxPC3, and PC3M-LN4) were collected and resuspended in PBS at 107 living cells/ml. LLC or EL4 (104, 105 or 106 living cells) and LLC-GFP, PancOH7, B16F10, or B16F10-GFP (106 living cells) were injected subcutaneously into C57BL/6J mice, and BxPC3 and PC3M-LN4 (106 living cells) were injected into SCID mice. For orthotopic human prostate tumors, PC3M-LN4 (2 × 105 living cells) were injected directly into the prostate of SCID mice. RvD1, RvD2, or RvE1 (15 ng/d; Cayman Chemical) or vehicle miniosmotic pumps (Alzet Inc.) were implanted intraperitoneally on the day of injection and changed once at 14 d after injection. For the genetically engineered TRAMP or MMTV-PyMT mice (The Jackson Laboratory), RvD2 (15 ng/d) or vehicle was administered via miniosmotic pump and/or cisplatin (5 mg/kg q 5 d) initiated when mice were 8 wk of age (the miniosmotic pumps were changed every 28 d for 2 or 3 mo).
Metastasis studies
For LLC metastasis studies, 106 LLC living cells were injected subcutaneously into 6-wk-old male C57BL/6J mice (The Jackson Laboratory). Tumors were resected once they had reached a size of 2 cm3, and miniosmotic pumps were implanted on the day of resection. B16F10 (2.5 × 105 living cells) were injected into 6-wk-old male C57BL/6J mice (The Jackson Laboratory) intravenously via the tail vein.
Macrophage depletion studies
Macrophages were depleted by clodronate liposomes (a gift from R. Schwendener, University of Zurich, Zurich, Switzerland) in tumor-bearing mice (106 B16F10 living cells), and depletion was confirmed by flow cytometry. Initial clodronate dose was administered intraperitoneally at 2 mg/20 g mouse body weight followed by 1 mg/20 g mouse body weight every 3 d. RvD2 (15 ng/d) or vehicle was administered via miniosmotic pump beginning on the day of tumor injection. Cisplatin-generated LLC debris (9 × 105 dead cells) and/or LLC (104 or 106 living cells) were injected into male MBL-deficient (MBL KO) and/or CCL2 KO mice (The Jackson Laboratory). Treatment with RvD1, RvD2, RvE1 (15 ng/d; Cayman Chemical), or vehicle via miniosmotic pump was initiated when tumors reached 100–200 mm3.
Resolvin receptor KO mice and transgenic studies
Cisplatin-generated LLC or gemcitabine-generated PancOH7 debris (9 × 105 dead cells) and/or LLC or PancOH7 living tumor cells (104 or 106 cells) were injected into ALX/FPR2 KO and ChemR23/ERV KO mice (ChemR23/ERV KO mice were provided by B. Zabel [Stanford University, Stanford, CA] and E. Butcher [Stanford School of Medicine, Stanford, CA]). Vincristine-generated EL4 or cisplatin-generated LLC debris (9 × 105 dead cells) and EL4 or LLC living cells (104) were coinjected into GPR18/DRV2 KO mice, and mice were treated systemically with resolvins (see the Tumor inhibition studies section). For metastasis studies, LLC tumors were resected 14 d after injection. 4T1 (106 living cells) were collected and prepared as described in the General note and injected into ChemR23-overexpressing transgenic and WT mice.
FACS cell sorting of debris
Cisplatin-generated LLC or gemcitabine-generated PancOH7 debris was prepared as described in the Cisplatin-generated LLC debris tumors and Gemcitabine-generated PancOH7 debris tumors sections and resuspended in annexin V binding buffer at a concentration of 106 cells/ml according to FITC Annexin V/Dead Cell Apoptosis kit protocol (Thermo Fisher Scientific). Cells were sorted using Sorter BD FACSAria Ilu SORP UV (DFCI; Jimmy Fund Flow Cytometry Core) as follows: annexin V single stain, PI single stain, annexin V/PI double stain, and unstained. Dead cell bodies were counted by hemocytometer, pelleted, and resuspended in PBS at 4 × 105 cells/ml. Untreated LLC or PancOH7 cells were trypsinized, pelleted, and resuspended at 2 × 105 cells/ml in PBS. Experimental groups were prepared by mixing equal volumes of dead cell bodies with living LLC or PancOH7 cells, and combinations were injected subcutaneously into C57BL/6J mice.
PS liposome–stimulated tumors
PS and PC liposomes (Avanti Polar Lipids) were filtered through a 20-µm filter and resuspended in PBS at the concentrations of 104, 101, and 10−2 μM. LLC, EL4, or PancOH7 cells were pelleted and resuspended in PBS for a final concentration of 2 × 105 cells/ml. Cells were resuspended in PS or PC liposomes for a final concentration of 105 cells/ml liposomes. Combination of liposomes and tumor cells were injected into C57BL/6J mice. Systemic treatment with resolvins (RvD1, RvD2, or RvE1) was initiated on the day of tumor cell injection and administered via miniosmotic pumps (Alzet).
Tumor inhibition studies
Treatment with chemotherapy, targeted therapy, antiinflammatory drugs, and/or resolvins was initiated once tumors reached 100–200 mm3 unless otherwise noted. Resolvins (RvD1, RvD2, or RvE1; 15 ng/d; Cayman Chemical) or vehicle were administered intraperitoneally via miniosmotic pumps (Alzet); docetaxel (25 mg/kg q 10 d; Sigma-Aldrich), cisplatin (5 mg/kg q 5 d; Sigma-Aldrich), gemcitabine (50 mg/kg q 4 d; Sigma-Aldrich), 5-FU (10 mg/kg q 5 d; Sigma-Aldrich), dexamethasone (2 mg/kg/d; Sigma-Aldrich), cetuximab (40 mg/kg/d), oxaliplatin (6 mg/kg q 4 d; MedChem Express), or WRW4 (1 mg/kg/d; Sigma-Aldrich) were administered intraperitoneally; and erlotinib (40 mg/kg/d) or indomethacin (3 mg/kg/d; Sigma-Aldrich) were administered by gavage. Tumor size was measured by caliper (width2 × length × 0.52 = mm3). Tumor experiments were terminated per protocol when tumor sizes reached a mean of 2,000–2,500 mm3. Number of mice per group was determined by minimum number required to achieve statistical significance (in collaboration with a statistician). Experiments were performed at least three times with similar results unless otherwise specified. For annexin V recombinant protein studies, vincristine-generated EL4 tumor cell debris (9 × 105 dead cells) was prepared as previously described and treated in vitro with recombinant annexin V protein (1 nM, 100 nM, or 10 µM; eBioscience) for 1 h before coinjection with EL4 (104 living cells) into C57BL/6J mice. Annexin V recombinant protein (4 µg/kg/d) was administered in vivo to mice injected with 104 living EL4 tumors via miniosmotic pumps. For cytokine depletion studies, C57BL/6J mice were treated intraperitoneally with combinations of anti–IL-6, anti-CCL4, anti-CCL5, and/or anti-TNFα neutralizing antibodies beginning on the day of tumor cell injection (20 µg each q 4 d; R&D Systems). Systemic chemotherapy, cytokine depletion, annexin V, and/or resolvin treatment initiated on the day of tumor cell injection.
Alternative debris generation for in vitro assays
Cycloheximide plus TNFα–generated debris was prepared by treating 75–80% confluent T150 flasks of PC3M-LN4, HEY, HSC-3, or OVCAR5 with complete media with 10% FBS plus 4 ng/ml TNFα and 5 µM cycloheximide and incubating for 18 h. Dead cell bodies were counted by hemocytometer, pelleted, and then resuspended in PBS to give a final desired concentration. See the Flow cytometry, Human macrophage phagocytosis assays, and Mouse phagocytosis assays sections.
Cisplatin-generated COV362, ID8 (provided by J. Lawler, Beth Israel Deaconess Medical Center, Boston, MA), or A375-SM debris was prepared by treating 75–80% confluent T150 flasks of respective cells with complete media with 10% FBS plus 50 µM cisplatin (Sigma-Aldrich) and incubating for 24 h. Dead cell bodies were counted by hemocytometer. Pelleted cells were resuspended in PBS at the desired concentration. See the Flow cytometry, Human macrophage phagocytosis assays, and Assays sections.
Immunohistochemistry
Tumor samples were processed, and immunohistochemical staining was performed according to standard protocol. Giemsa and 3-3’-diaminobenzidine (DAB) staining were performed according to standard protocol. Sections were deparaffinized with xylene and rehydrated in graded ethanol. Sections were microwaved in 10 mM sodium citrate and then incubated with IL-6 (1:100; Abcam), IL-8 (1:100; Abcam), TNFα (1:100; Abcam), F4/80 (1:100; Bio-Rad Laboratories), GPR32 (1:200; GeneTex), ALX/FPR2 (1:100; GeneTex), or CD31 antibodies (1:250; BD). MECA-32 and CD31 stains were amplified using Tyramide signal amplification direct and indirect kits (NEN Life Science Products Inc.). Human prostate sections were obtained from M. Loda (Dana-Farber Cancer Institute, Boston, MA). Histological sections of tumors were analyzed for vessel density as previously described (Panigrahy et al., 2012). Immunohistochemistry and localization of fluorescently labeled cells were analyzed using a confocal SP2 microscope (Leica Microsystems).
Western blotting
Protein was extracted from cell lysates, and immunoblotting was performed based on standard protocols to measure ALX/FPR2 (GeneTex) in LLC, B16F10 (melanoma), T241 (fibrosarcoma), BTC, MS-180 (sarcoma), and EOMA. Protein from mouse spleen was used as a positive control.
Flow cytometry
For flow cytometry of in vitro debris cultures, vincristine-generated EL4 debris was isolated via Ficoll gradient (GE Healthcare). For all other chemotherapy- and targeted therapy–generated debris, cells were treated and collected as described in the General note. Cells were pelleted and resuspended in annexin V binding buffer at 106 cells/ml and double stained with Annexin V and PI according to FITC Annexin V/Dead Cell Apoptosis kit protocol. Staining was assessed using a FACSCanto II (BD) or LSR Fortessa (BD) and analyzed using FlowJo software (Tree Star).
For flow cytometry analysis of dissociated cells from whole tumors, tumors were removed when they reached ≥1,500–2,000 mm3, and single-cell suspensions were prepared by enzymatic digestion with Liberase (40 min at 37°C; Roche). Digested tissue was filtered through a 40-µm cell strainer and resuspended in PBS. For nondebris LLC or EL4 tumors treated with and without systemic cisplatin or vincristine, respectively, cell death was assessed via annexin V/PI staining as described in the FACS cell sorting of debris. For LLC-GFP– and LLC debris–stimulated tumors, GFP levels were assessed using FACSCanto II or LSR Fortessa and analyzed using FlowJo software. For flow cytometry analysis of resolvin-treated LLC-GFP or B16F10-GFP tumors, CD11b-PE and F4/80-APC (Roche) were assessed using FACS Calibur and CellQuest software (BD) and analyzed with WinMDI 2.8 software. For flow cytometry analysis of tumor cell phagocytosis and efferocytosis in the resolvin-, cisplatin-, and/or WRW4-treated LLC-GFP tumors, TIM-4–Alexa Fluor 647 (BioLegend), CD11b–Alexa Fluor 405 (R&D Systems), and F4/80-PE (Miltenyi Biotec) were assessed using LSR Fortessa and analyzed with FlowJo software. TIM-4+F4/80+ cells were gated on whole-tumor populations, whereas GFP+ cells were gated on TIM-4+F4/80+ cells to reflect macrophage efferocytosis (GFP+TIM-4+F4/80+). Likewise, CD11b+F4/80+ cells were gated on whole-tumor populations, and GFP+ cells were gated on CD11b+F4/80+ cells to reflect macrophage phagocytosis (GFP+CD11b+F4/80+).
In tumors generated from 106 LLC living cells and cisplatin-generated LLC debris–stimulated tumors, CD45-PE and F4/80-APC (Bio-Rad Laboratories) were assessed using LSR Fortessa and analyzed with FlowJo software. For examining immune cell types in tumors generated from 106 LLC or EL4 living cells and cisplatin-generated LLC debris or vincristine-generated EL4 debris-stimulated tumors, F4/80-PE (Miltenyi Biotec), CD11b–Alexa Fluor 405 (R&D Systems), Gr1-APC (BioLegend), and Ly6G–Alexa Fluor 488 (Novus Biologicals) were assessed using LSR Fortessa and analyzed with FlowJo software. The percentage of CD45+ cells referred to the proportion of leukocytes (CD45+) within the whole tumor lysate included the double-stained CD45+F4/80+ population (macrophages) as well as CD45+F4/80− (leukocytes excluding macrophages).
For flow cytometry of Matrigel plugs (100 mg/ml; BD), plugs were injected into both flanks, resected after 7 d, and digested as described above with Liberase. CD45-PE (Bio-Rad Laboratories) and F4/80-APC (Roche) were assessed using FACSCanto II and analyzed with FlowJo software.
Electron microscopy and light microscopy
Electron microscopic thin sections were examined on a FEI/Phillips EM 208S (FEI Electron Optics BV) equipped with a digital camera (Advanced Microscopy Techniques). Light microscopic images were taken on an Eclipse E600 microscope (Nikon) with a 100× 0.30 NA oil immersion lens and an RT Slider SPOT 2.3.1 camera (Diagnostic Instruments) using SPOT Advanced software (3.5.9; SPOT Imaging). Tumor cell debris was prepared as described in the General note. Debris was collected and resuspended in PBS. Cells were placed on glass slides. Light microscopic images were taken on an Axiophot (ZEISS) with a 20× dry objective lens using SPOT Advanced software.
Human monocyte–derived macrophages
Human peripheral blood mononuclear cells from healthy human volunteers from the Children’s Hospital Boston blood bank were isolated by density-gradient Histopaque-1077 (Sigma-Aldrich). Macrophages were differentiated using RPMI media plus 10 ng/ml GM-CSF (R&D Systems) for 7 d (37°C at 5% CO2).
Macrophage-secreted cytokines
For human tumor cell debris, human monocyte–derived macrophages were plated in six-well plates at 2 × 106 cells per well. Cells were incubated in complete RPMI medium with 10% FBS and 10 ng/ml GM-CSF for 7 d. Cisplatin-generated PC3M-LN4, cisplatin-generated HSC-3, and cycloheximide plus TNFα–generated PC3M-LN4 debris were prepared as described in the General note. Macrophage plates were rinsed with PBS and then were treated with vehicle, RvD1 (1 nM), or RvD2 (1 nM) for 30 min at 37°C. Collected tumor cell debris was added to six-well plates at 4 × 106 cells per well. PC3M-LN4 or HSC-3 debris was added to empty six-well plates as a control. Plates were incubated for 1 h at 37°C. Plates were rinsed with PBS and then refed with 5 ml complete RPMI medium with 10% FBS. Plates were incubated for 24 h at 37°C. Conditioned media from plates was collected and used according to the protocol of R&D Systems Proteome Profiler: Human Cytokine Array Panel A. Array control allows for comparison between membranes. For ELISA analysis, human monocyte–derived macrophage-conditioned media was prepared as described above. Media was used according to the protocols of each ELISA kit (R&D Systems). Human monocyte–derived macrophage-conditioned media collection was repeated using three individual patient donations of peripheral blood monocytes, and debris collection was repeated for a total of three separate experiments.
For mouse tumor cell debris, mouse RAW264.7 macrophages were plated in six-well plates at 5 × 105 cells per well and incubated for 2 h in Dulbecco’s PBS at 37°C. Cisplatin-generated LLC debris was prepared as described in the Cisplatin-generated LLC debris tumors section and pretreated with annexin V recombinant protein (10 µM; eBioscience) for 1 h. Collected tumor cell debris was added to six-well plates at 2 × 106 cells per well. Plates were incubated for 1 h at 37°C. Plates were rinsed with PBS, refed with 3 ml/well serum-free DMEM media, and incubated for 24 h at 37°C. Conditioned media from plates was collected and used according to the protocol of R&D Systems Proteome Profiler: Mouse Cytokine Array kit, Panel A. Array control allows for comparison between membranes.
For PS and PC liposome studies, RAW264.7 macrophages were plated in six-well plates at 106 cells per well. PS or PC liposomes (Avanti Polar Lipids) were resuspended in PBS for a final concentration of 100 µM. Macrophages were incubated with PS/PC liposomes for 2 h at 37°C, refed with serum-free DMEM media, and incubated for 24 h at 37°C. Conditioned media was collected and used according to the protocols for each ELISA kit (R&D Systems).
Human monocyte–derived macrophage phagocytosis assays
Human primary monocyte–derived macrophages were plated in 96-well plates at 5 × 104 cells per well in complete RPMI medium with 10% FBS for 18–24 h. Dead cell bodies were collected and prepared as described in the General note and then were fluorescently stained with carboxyfluorescein diacetate (CFDA). Macrophages were treated with vehicle, RvD1 RvD2, or RvE1 (0.001–100 nM), dexamethasone (1 nM or 10 nM), or indomethacin (1 nM or 10 nM) for 30 min at 37°C. Collected tumor cells were added to 96-well plates at a 1:2 (PC3M-LN4, A375-SM, and BxPC3) or 1:4 (COV362, HEY, OVCAR5, and HSC-3) monocyte-derived macrophage/dead cell body ratio, and plates were incubated for 1 h at 37°C. Plates were quenched with trypan blue, and fluorescence was measured using a Spectra Max M5 plate reader (Molecular Devices). Relative fluorescence units (RFUs) were used to measure phagocytosis compared with control monocyte-derived macrophages. Experiments were performed three times with similar results. We performed macrophage phagocytosis assays using nontumorigenic macrophages (e.g., human monocyte–derived macrophages, mouse peritoneal macrophages, and mouse bone marrow–derived macrophages).
Mouse phagocytosis assays
Mouse resident peritoneal and bone marrow–derived macrophages
Mouse resident peritoneal macrophages were collected by peritoneal lavage of C57BL/6J male mice using sterile PBS. Macrophages were plated in 96-well plates at 5 × 104 cells per well and incubated in PBS for 1–2 h at 37°C. Mouse bone marrow–derived macrophages were extracted from C57BL/6J male mice and differentiated by incubation in RPMI containing mouse CSF for 7 d (37°C at 5% CO2). Bone marrow–derived macrophages were plated in 96-well plates at 5 × 104 cells per well and incubated in DMEM supplemented with 10% FBS and 30% l-929 medium overnight. ID8 cells were treated with cisplatin (Sigma-Aldrich) and collected as described in the General note. After 24 h, dead cell bodies were fluorescently stained with CFDA. Macrophage wells were treated with vehicle, RvD1, or RvD2 (0.001–100 nM), and/or BOC-1 (10 µM; MP Biomedicals) for 30 min at 37°C. CFDA-stained dead tumor cell bodies were added to 96-well plates at a 1:4 macrophage/dead cell body ratio, and plates were incubated for 1 h at 37°C. Plates were quenched with trypan blue, and fluorescence was measured using a Spectra Max M5 plate reader. RFUs were used to measure phagocytosis compared with control macrophages.
RvD1 and RvD2 levels in vitro (ELISA)
For analysis of RvD1 and RvD2 in tissue culture samples, mouse RAW264.7 macrophages were plated in six-well plates at 2 × 106 macrophages per well in PBS and incubated for 2 h at 37°C. Cisplatin-generated LLC debris was counted and prepared as described in the General note. Debris was added to applicable plates at a 1:2 macrophage/dead cell body ratio. Plates were incubated for 1 h at 37°C. Conditioned media from each well was collected according to RvD1 or RvD2 ELISA kit instructions (Cayman Chemical). Samples were run in triplicates (RvD1) and duplicates (RvD2) per kit instructions. n = 4 per group.
Proliferation assays
For tumor cell (B16F10, LLC, and PC3M-LN4) proliferations assays, B16F10 cells were plated at an initial 5 × 103 cells/well in 24-well plates. LLC cells were plated at an initial 7.5 × 103 cells/well in 24-well plates. PC3M-LN4 cells were plated at an initial 104 cells/well in 24-well plates. B16F10 and LLC cells were treated with RvD1 (10 nM and 1 µM), RvD2 (10 nM and 1 µM), or vehicle for 48 h. PC3M-LN4 cells were treated with RvD1 (10 nM and 1 µM), RvD2 (10 nM and 1 µM), or vehicle for 24 h. After 1 or 2 d, B16F10, LLC, and PC3M-LN4 cells were counted with a Coulter Counter. n = 4–8/group, three biological repeats.
Additional proliferation assays used Cell Proliferation kit 1 (MTT; Roche), according to recommended protocol to quantify proliferation. PancOH7 or MS1 cells were plated at an initial 5 × 103 cells/well in 96-well plates with complete media with 10% FBS and incubated overnight. Cells were refed with conditioned media from RAW264.7 mouse macrophages or RAW264.7 macrophages exposed to gemcitabine-generated PancOH7 debris, prepared as described in the Gemcitabine-generated PancOH7 debris tumors. After a 24-h incubation at 37°C with the macrophage-conditioned media, 10 µl of MTT reagent (Roche) was added to each well and incubated for 4 h at 37°C. Next, 100 µl of solubilizing solution (Roche) was added to each well and incubated overnight at 37°C. VersaMax microplate reader (Molecular Devices) was used to quantify the results of the assay.
Tumor cell viability
Conditioned media from RAW264.7 mouse macrophages or RAW264.7 macrophages exposed to gemcitabine-generated PancOH7 debris was collected as described in the Macrophage secreted cytokines section. PancOH7 cells were plated at 5 × 105 and incubated overnight in complete media plus 10% FBS. The next day, PancOH7 cells were refed with conditioned media from macrophages and incubated overnight. Cells were stained according to the FITC Annexin V/Dead Cell Apoptosis kit protocol (Thermo Fisher Scientific), and cell viability was assessed via flow cytometry analysis.
Migration studies
RvD1, RvD2, or control was added to serum-free media at a final concentration of 10 nM. In transwell permeable supports with an 8.0-µm polycarbonate membrane (Costar), 10 nM RvD1, 10 nM RvD2, or vehicle was added to the bottom chamber, and 5 × 105 LLC in serum-free media was added to the top chamber. Cells were incubated at 37°C at 5% CO2 for 5 h. After the incubation, remaining cells were removed from the top chamber with a cotton-tipped applicator. Cells were fixed with methanol and stained using Giemsa stain (Sigma-Aldrich) for 40 min at room temperature. Excess Giemsa stain was removed with water. Migration was quantified using an EVOS microscope (Thermo Fisher Scientific) and counting the number of cells per field (four fields/transwell) at 10× magnification.
LC-MS-MS
Plasma from nontumor-bearing and tumor-bearing mice was analyzed by LC-MS-MS by LC-20AD HPLC and a SIL-20AC autoinjector (Shimadzu Corp.) paired with a QTrap 6500 (ABSciex). Mouse tumors were placed in 1 ml of methanol, gently homogenized using a glass dounce, and kept at −20°C to allow for protein precipitation. Lipid mediators were extracted using solid-phase extraction (Colas et al., 2014). In brief, before sample extraction, a deuterated internal standard (d5-LXA4) representing the region of interest in the chromatographic analysis (500 pg) was added to facilitate quantification. Extracted samples were analyzed by a LC-MS-MS system, QTrap 6500 (AB Sciex) equipped with a SIL-20AC autoinjector and LC-20AD binary pump (Shimadzu Corp.). An Eclipse Plus C18 column (100 × 4.6 mm × 1.8 µm; Agilent Technologies) was used with a gradient of methanol/water/acetic acid of 55:45:0.01 (vol/vol/vol) that was ramped to 85:15:0.01 (vol/vol/vol) over 10 min and then to 98:2:0.01 (vol/vol/vol) for the next 8 min. This was subsequently maintained at 98:2:0.01 (vol/vol/vol) for 2 min. The flow rate was maintained at 0.4 ml/min. To monitor and quantify the levels of lipid mediators, a multiple reaction monitoring method was developed with signature ion fragments (m/z) for each molecule monitoring the parent ion (Q1) and a characteristic daughter ion (Q3). Identification was conducted using published criteria where a minimum of six diagnostic ions were used (Colas et al., 2014). Calibration curves were determined using a mixture of lipid mediators obtained via total organic synthesis. Linear calibration curves for each compound were obtained with r2 values ranging from 0.98–0.99. Detection limit was ∼0.1 pg. Quantification was performed as described by Colas et al. (2014).
Statistics
For in vivo experiments, Student’s t test and ANOVA were used. Student’s t test was used to evaluate significance of in vitro experiments. The Kolmogorov-Smirnov goodness-of-fit statistic was used to test the assumption of normality of the tumor volume measurements and other continuous variables, and no significant departures from a Gaussian-shaped distribution were detected. Therefore, summary data are reported as mean values and SEM. Longitudinal tumor growth data were analyzed using two-factor repeated-measures mixed effects ANOVA with the Greenhouse–Geisser F test to assess overall group differences followed by Tukey post hoc comparisons, where treatment was considered the between subjects factor, and serial tumor measurements were considered the within subjects or repeated factor (Liu et al., 2010). In addition, one-factor ANOVA was used to compare treatment and control groups with respect to cytokines and biomarker variables, and the Student’s t test was used to compare the percentage of total cell death between cisplatin and control. Survival after orthotopic injection was analyzed using the Kaplan-Meier product-limit model with the log-rank test to evaluate survival differences over time after tumor injection between treatment groups for chemotherapy-generated dead cells and PancOH7 living cells versus living cells alone. P-values <0.05 were considered statistically significant.
Online supplemental material
Fig. S1 characterizes the chemotherapy- and targeted therapy–generated debris used in our debris tumor models, describes how the debris was quantified, and confirms that debris-stimulated tumors indeed arose from the living tumor cells. Fig. S2 provides additional titrations of subthreshold inoculums of various tumor types with debris generated by different therapies in immunocompromised versus immunocompetent mouse models. Fig. S3 highlights and characterizes the role of macrophages and inflammation within the tumor microenvironment in debris-stimulated tumors. Fig. S4 demonstrates the role of resolvins in human xenograft mouse models, genetically engineered mouse models, spontaneous tumor models, and metastasis models. Fig. S5 further elucidates that the antitumor activity of resolvins is stromal and provides a schematic that summarizes our model.
Supplementary Material
Acknowledgments
We gratefully acknowledge and thank Steve Moskowitz (Advanced Medical Graphics) for photography and preparation of the figures; we thank Dr. Sesquile Ramon, Dr. William Aird, Thad Vickery, John Daley, Catherine Butterfield, Dr. Howard Mulhern, Dr. Ofra Benny, Dr. Jo-Anne Vergilio, Donna Vatnick, and Chantal Barksdale for excellent technical assistance; Dr. Isaiah J. Fidler for PC3M-LN4 cells; Dr. Jack Lawler for ID8 cells; Dr. Reto Schwendener for clodronate liposomes; and Dr. Massimo Loda for human prostate sections. The resolvin E1 receptor (ChemR23) KO mice were provided by Dr. Brian Zabel (supported by National Institutes of Health grant R01 AI079320) and Professor Eugene Butcher (supported by National Institutes of Health grant CA169354).
This work was supported by the National Cancer Institute grants RO1 01CA170549-02 (to D. Panigrahy and C.N. Serhan), ROCA148633-01A4 (to D. Panigrahy), and GM095467 (to C.N. Serhan); the Stop and Shop Pediatric Brain Tumor Fund (to M.W. Kieran); the CJ Buckley Pediatric Brain Tumor Fund (to M.W. Kieran); Alex Lemonade Stand (to M.W. Kieran); Molly’s Magic Wand for Pediatric Brain Tumors (to M.W. Kieran); the Markoff Foundation Art-In-Giving Foundation (to M.W. Kieran); the Kamen Foundation (to M.W. Kieran); Jared Branfman Sunflowers for Life (to M.W.K.); and The Wellcome Trust program 086867/Z/08 (to M. Perretti).
The authors declare no competing financial interests.
Author contributions: conceptualization, M.L. Sulciner, C.N. Serhan, M.W. Kieran, S. Huang, and D. Panigrahy; data curation, M.L. Sulciner and D. Panigrahy; formal analysis, M.L. Sulciner, M.M. Gilligan, D. Zurakowski, C.N. Serhan, J. Dalli, S. Huang, and D. Panigrahy; funding acquisition, C.N. Serhan, M.W. Kieran, V.P. Sukhatme, and D. Panigrahy; investigation, M.L. Sulciner, C.N. Serhan, M.M. Gilligan, D.K. Mudge, J. Chang, A. Gartung, K.A. Lehner, D.R. Bielenberg, B. Schmidt, E.R. Greene, J. Dalli, Y. Gus-Brautbar, J. Piwowarski, M Perretti, T. Mammoto, V.P. Sukhatme, and D. Panigrahy; methodology, M.L. Sulciner, C.N. Serhan, M.M. Gilligan, J. Dalli, M. Perretti, A. Kaipainen, M.W. Kieran, S. Huang, and D. Panigrahy; project administration, C.N. Serhan, M.W. Kieran, S. Huang, and D. Panigrahy; resources, C.N. Serhan. M.W. Kieran, and D. Panigrahy; supervision, C.N. Serhan, M.W. Kieran, S. Huang, and D. Panigrahy; visualization, M.L. Sulciner, C.N. Serhan, M.M. Gilligan, D.M. Mudge, and D. Panigrahy; writing (original draft), M.L. Sulciner, C.N. Serhan, A. Kaipainen, M.W. Kieran, S. Huang, and D. Panigrahy; writing (review and editing), M.L. Sulciner, C.N. Serhan, M.M. Gilligan, D.M. Mudge, J. Dalli, V.P. Sukhatme, A. Kaipainen, M.W. Kieran, S. Huang, and D. Panigrahy.
References
- Abubaker K., Latifi A., Luwor R., Nazaretian S., Zhu H., Quinn M.A., Thompson E.W., Findlay J.K., and Ahmed N.. 2013. Short-term single treatment of chemotherapy results in the enrichment of ovarian cancer stem cell-like cells leading to an increased tumor burden. Mol. Cancer. 12:24 10.1186/1476-4598-12-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akazawa T., Masuda H., Saeki Y., Matsumoto M., Takeda K., Tsujimura K., Kuzushima K., Takahashi T., Azuma I., Akira S., et al. 2004. Adjuvant-mediated tumor regression and tumor-specific cytotoxic response are impaired in MyD88-deficient mice. Cancer Res. 64:757–764. 10.1158/0008-5472.CAN-03-1518 [DOI] [PubMed] [Google Scholar]
- Alcaide J., Funez R., Rueda A., Perez-Ruiz E., Pereda T., Rodrigo I., Coveñas R., Muñoz M., and Redondo M.. 2013. The role and prognostic value of apoptosis in colorectal carcinoma. BMC Clin. Pathol. 13:24 10.1186/1472-6890-13-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apetoh L., Ghiringhelli F., Tesniere A., Obeid M., Ortiz C., Criollo A., Mignot G., Maiuri M.C., Ullrich E., Saulnier P., et al. 2007. Toll-like receptor 4-dependent contribution of the immune system to anticancer chemotherapy and radiotherapy. Nat. Med. 13:1050–1059. 10.1038/nm1622 [DOI] [PubMed] [Google Scholar]
- Arita M., Ohira T., Sun Y.P., Elangovan S., Chiang N., and Serhan C.N.. 2007. Resolvin E1 selectively interacts with leukotriene B4 receptor BLT1 and ChemR23 to regulate inflammation. J. Immunol. 178:3912–3917. 10.4049/jimmunol.178.6.3912 [DOI] [PubMed] [Google Scholar]
- Benton G., George J., Kleinman H.K., and Arnaoutova I.P.. 2009. Advancing science and technology via 3D culture on basement membrane matrix. J. Cell. Physiol. 221:18–25. 10.1002/jcp.21832 [DOI] [PubMed] [Google Scholar]
- Birge R.B., Boeltz S., Kumar S., Carlson J., Wanderley J., Calianese D., Barcinski M., Brekken R.A., Huang X., Hutchins J.T., et al. 2016. Phosphatidylserine is a global immunosuppressive signal in efferocytosis, infectious disease, and cancer. Cell Death Differ. 23:962–978. 10.1038/cdd.2016.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondanza A., Zimmermann V.S., Rovere-Querini P., Turnay J., Dumitriu I.E., Stach C.M., Voll R.E., Gaipl U.S., Bertling W., Pöschl E., et al. 2004. Inhibition of phosphatidylserine recognition heightens the immunogenicity of irradiated lymphoma cells in vivo. J. Exp. Med. 200:1157–1165. 10.1084/jem.20040327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casares N., Pequignot M.O., Tesniere A., Ghiringhelli F., Roux S., Chaput N., Schmitt E., Hamai A., Hervas-Stubbs S., Obeid M., et al. 2005. Caspase-dependent immunogenicity of doxorubicin-induced tumor cell death. J. Exp. Med. 202:1691–1701. 10.1084/jem.20050915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.K., Roth J., Oppenheim J.J., Tracey K.J., Vogl T., Feldmann M., Horwood N., and Nanchahal J.. 2012. Alarmins: awaiting a clinical response. J. Clin. Invest. 122:2711–2719. 10.1172/JCI62423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y.S., Jalgaonkar S.P., Middleton J.D., and Hai T.. 2017. Stress-inducible gene Atf3 in the noncancer host cells contributes to chemotherapy-exacerbated breast cancer metastasis. Proc. Natl. Acad. Sci. USA. 114:E7159–E7168. 10.1073/pnas.1700455114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaurio R., Janko C., Schorn C., Maueröder C., Bilyy R., Gaipl U., Schett G., Berens C., Frey B., and Munoz L.E.. 2013. UVB-irradiated apoptotic cells induce accelerated growth of co-implanted viable tumor cells in immune competent mice. Autoimmunity. 46:317–322. 10.3109/08916934.2012.754433 [DOI] [PubMed] [Google Scholar]
- Chiang N., Fredman G., Bäckhed F., Oh S.F., Vickery T., Schmidt B.A., and Serhan C.N.. 2012. Infection regulates pro-resolving mediators that lower antibiotic requirements. Nature. 484:524–528. 10.1038/nature11042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang N., Dalli J., Colas R.A., and Serhan C.N.. 2015. Identification of resolvin D2 receptor mediating resolution of infections and organ protection. J. Exp. Med. 212:1203–1217. 10.1084/jem.20150225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colas R.A., Shinohara M., Dalli J., Chiang N., and Serhan C.N.. 2014. Identification and signature profiles for pro-resolving and inflammatory lipid mediators in human tissue. Am. J. Physiol. Cell Physiol. 307:C39–C54. 10.1152/ajpcell.00024.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connell P.P., and Weichselbaum R.R.. 2011. A downside to apoptosis in cancer therapy? Nat. Med. 17:780–782. 10.1038/nm0711-780 [DOI] [PubMed] [Google Scholar]
- da Silva-Jr I.A. Jr., Chammas R., Lepique A.P., and Jancar S.. 2017. Platelet-activating factor (PAF) receptor as a promising target for cancer cell repopulation after radiotherapy. Oncogenesis. 6:e296 10.1038/oncsis.2016.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFrancesco L. 2014. CAR-T cell therapy seeks strategies to harness cytokine storm. Nat. Biotechnol. 32:604 10.1038/nbt0714-604 [DOI] [PubMed] [Google Scholar]
- de Jong J.S., van Diest P.J., and Baak J.P.. 2000. Number of apoptotic cells as a prognostic marker in invasive breast cancer. Br. J. Cancer. 82:368–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J., Zhang Y., Feng J., and Wu F.. 2010. Dendritic cells loaded with ultrasound-ablated tumour induce in vivo specific antitumour immune responses. Ultrasound Med. Biol. 36:441–448. 10.1016/j.ultrasmedbio.2009.12.004 [DOI] [PubMed] [Google Scholar]
- de Ruiter J., Cramer S.J., Smink T., and van Putten L.M.. 1979. The facilitation of tumour growth in the lung by cyclophosphamide in artificial and spontaneous metastases models. Eur. J. Cancer. 15:1139–1149. 10.1016/0014-2964(79)90130-0 [DOI] [PubMed] [Google Scholar]
- Donato A.L., Huang Q., Liu X., Li F., Zimmerman M.A., and Li C.Y.. 2014. Caspase 3 promotes surviving melanoma tumor cell growth after cytotoxic therapy. J. Invest. Dermatol. 134:1686–1692. 10.1038/jid.2014.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drochmans P. 1960. Electron microscope studies of epidermal melanocytes, and the fine structure of melanin granules. J. Biophys. Biochem. Cytol. 8:165–180. 10.1083/jcb.8.1.165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dromi S.A., Walsh M.P., Herby S., Traughber B., Xie J., Sharma K.V., Sekhar K.P., Luk A., Liewehr D.J., Dreher M.R., et al. 2009. Radiofrequency ablation induces antigen-presenting cell infiltration and amplification of weak tumor-induced immunity. Radiology. 251:58–66. 10.1148/radiol.2511072175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufton N., Hannon R., Brancaleone V., Dalli J., Patel H.B., Gray M., D’Acquisto F., Buckingham J.C., Perretti M., and Flower R.J.. 2010. Anti-inflammatory role of the murine formyl-peptide receptor 2: ligand-specific effects on leukocyte responses and experimental inflammation. J. Immunol. 184:2611–2619. 10.4049/jimmunol.0903526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elajami T.K., Colas R.A., Dalli J., Chiang N., Serhan C.N., and Welty F.K.. 2016. Specialized proresolving lipid mediators in patients with coronary artery disease and their potential for clot remodeling. FASEB J. 30:2792–2801. 10.1096/fj.201500155R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford C.A., Petrova S., Pound J.D., Voss J.J., Melville L., Paterson M., Farnworth S.L., Gallimore A.M., Cuff S., Wheadon H., et al. 2015. Oncogenic properties of apoptotic tumor cells in aggressive B cell lymphoma. Curr. Biol. 25:577–588. 10.1016/j.cub.2014.12.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey B., Schildkopf P., Rödel F., Weiss E.M., Munoz L.E., Herrmann M., Fietkau R., and Gaipl U.S.. 2009. AnnexinA5 renders dead tumor cells immunogenic--implications for multimodal cancer therapies. J. Immunotoxicol. 6:209–216. 10.3109/15476910903204058 [DOI] [PubMed] [Google Scholar]
- Fullerton J.N., and Gilroy D.W.. 2016. Resolution of inflammation: a new therapeutic frontier. Nat. Rev. Drug Discov. 15:551–567. 10.1038/nrd.2016.39 [DOI] [PubMed] [Google Scholar]
- Gao L., Faibish D., Fredman G., Herrera B.S., Chiang N., Serhan C.N., Van Dyke T.E., and Gyurko R.. 2013. Resolvin E1 and chemokine-like receptor 1 mediate bone preservation. J. Immunol. 190:689–694. 10.4049/jimmunol.1103688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg A.D., Nowis D., Golab J., Vandenabeele P., Krysko D.V., and Agostinis P.. 2010. Immunogenic cell death, DAMPs and anticancer therapeutics: an emerging amalgamation. Biochim. Biophys. Acta. 1805:53–71. 10.1016/j.bbcan.2009.08.003 [DOI] [PubMed] [Google Scholar]
- Garg A.D., Krysko D.V., Verfaillie T., Kaczmarek A., Ferreira G.B., Marysael T., Rubio N., Firczuk M., Mathieu C., Roebroek A.J., et al. 2012. A novel pathway combining calreticulin exposure and ATP secretion in immunogenic cancer cell death. EMBO J. 31:1062–1079. 10.1038/emboj.2011.497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough M.J., Melcher A.A., Ahmed A., Crittenden M.R., Riddle D.S., Linardakis E., Ruchatz A.N., Emiliusen L.M., and Vile R.G.. 2001. Macrophages orchestrate the immune response to tumor cell death. Cancer Res. 61:7240–7247. [PubMed] [Google Scholar]
- Gray M.J., Gong J., Hatch M.M., Nguyen V., Hughes C.C., Hutchins J.T., and Freimark B.D.. 2016. Phosphatidylserine-targeting antibodies augment the anti-tumorigenic activity of anti-PD-1 therapy by enhancing immune activation and downregulating pro-oncogenic factors induced by T-cell checkpoint inhibition in murine triple-negative breast cancers. Breast Cancer Res. 18:50 10.1186/s13058-016-0708-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene E.R., Huang S., Serhan C.N., and Panigrahy D.. 2011. Regulation of inflammation in cancer by eicosanoids. Prostaglandins Other Lipid Mediat. 96:27–36. 10.1016/j.prostaglandins.2011.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory C.D., and Pound J.D.. 2011. Cell death in the neighbourhood: direct microenvironmental effects of apoptosis in normal and neoplastic tissues. J. Pathol. 223:177–194. 10.1002/path.2792 [DOI] [PubMed] [Google Scholar]
- Gregory C.D., Ford C.A., and Voss J.J.. 2016. Microenvironmental Effects of Cell Death in Malignant Disease. Adv. Exp. Med. Biol. 930:51–88. 10.1007/978-3-319-39406-0_3 [DOI] [PubMed] [Google Scholar]
- Grenon S.M., Owens C.D., Alley H., Chong K., Yen P.K., Harris W., Hughes-Fulford M., and Conte M.S.. 2013. n-3 Polyunsaturated fatty acids supplementation in peripheral artery disease: the OMEGA-PAD trial. Vasc. Med. 18:263–274. 10.1177/1358863X13503695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunjal P.M., Schneider G., Ismail A.A., Kakar S.S., Kucia M., and Ratajczak M.Z.. 2015. Evidence for induction of a tumor metastasis-receptive microenvironment for ovarian cancer cells in bone marrow and other organs as an unwanted and underestimated side effect of chemotherapy/radiotherapy. J. Ovarian Res. 8:20 10.1186/s13048-015-0141-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hangai S., Ao T., Kimura Y., Matsuki K., Kawamura T., Negishi H., Nishio J., Kodama T., Taniguchi T., and Yanai H.. 2016. PGE2 induced in and released by dying cells functions as an inhibitory DAMP. Proc. Natl. Acad. Sci. USA. 113:3844–3849. 10.1073/pnas.1602023113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J., Yin Y., Luster T.A., Watkins L., and Thorpe P.E.. 2009. Antiphosphatidylserine antibody combined with irradiation damages tumor blood vessels and induces tumor immunity in a rat model of glioblastoma. Clin. Cancer Res. 15:6871–6880. 10.1158/1078-0432.CCR-09-1499 [DOI] [PubMed] [Google Scholar]
- Holmgren L., O’Reilly M.S., and Folkman J.. 1995. Dormancy of micrometastases: balanced proliferation and apoptosis in the presence of angiogenesis suppression. Nat. Med. 1:149–153. 10.1038/nm0295-149 [DOI] [PubMed] [Google Scholar]
- Hosseini H., Li Y., Kanellakis P., Tay C., Cao A., Tipping P., Bobik A., Toh B.H., and Kyaw T.. 2015. Phosphatidylserine liposomes mimic apoptotic cells to attenuate atherosclerosis by expanding polyreactive IgM producing B1a lymphocytes. Cardiovasc. Res. 106:443–452. 10.1093/cvr/cvv037 [DOI] [PubMed] [Google Scholar]
- Huang Q., Li F., Liu X., Li W., Shi W., Liu F.F., O’Sullivan B., He Z., Peng Y., Tan A.C., et al. 2011. Caspase 3-mediated stimulation of tumor cell repopulation during cancer radiotherapy. Nat. Med. 17:860–866. 10.1038/nm.2385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichim G., and Tait S.W.. 2016. A fate worse than death: apoptosis as an oncogenic process. Nat. Rev. Cancer. 16:539–548. 10.1038/nrc.2016.58 [DOI] [PubMed] [Google Scholar]
- Jalalinadoushan M., Peivareh H., and Azizzadeh Delshad A.. 2004. Correlation between Apoptosis and Histological Grade of Transitional Cell Carcinoma of Urinary Bladder. Urol. J. 1:177–179. [PubMed] [Google Scholar]
- Karagiannis G.S., Pastoriza J.M., Wang Y., Harney A.S., Entenberg D., Pignatelli J., Sharma V.P., Xue E.A., Cheng E., D’Alfonso T.M., et al. 2017. Neoadjuvant chemotherapy induces breast cancer metastasis through a TMEM-mediated mechanism. Sci. Transl. Med. 10.1126/scitranslmed.aan0026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilkenny C., Browne W.J., Cuthill I.C., Emerson M., and Altman D.G.. 2010. Improving bioscience research reporting: the ARRIVE guidelines for reporting animal research. PLoS Biol. 8:e1000412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindy M.S., Yu J., Zhu H., Smith M.T., and Gattoni-Celli S.. 2016. A therapeutic cancer vaccine against GL261 murine glioma. J. Transl. Med. 14:1 10.1186/s12967-015-0757-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi N., Karisola P., Peña-Cruz V., Dorfman D.M., Jinushi M., Umetsu S.E., Butte M.J., Nagumo H., Chernova I., Zhu B., et al. 2007. TIM-1 and TIM-4 glycoproteins bind phosphatidylserine and mediate uptake of apoptotic cells. Immunity. 27:927–940. 10.1016/j.immuni.2007.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbelik M., Stott B., and Sun J.. 2007. Photodynamic therapy-generated vaccines: relevance of tumour cell death expression. Br. J. Cancer. 97:1381–1387. 10.1038/sj.bjc.6604059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornbluth R.S. 1994. The immunological potential of apoptotic debris produced by tumor cells and during HIV infection. Immunol. Lett. 43:125–132. 10.1016/0165-2478(94)00149-9 [DOI] [PubMed] [Google Scholar]
- Krishnamoorthy S., Recchiuti A., Chiang N., Yacoubian S., Lee C.H., Yang R., Petasis N.A., and Serhan C.N.. 2010. Resolvin D1 binds human phagocytes with evidence for proresolving receptors. Proc. Natl. Acad. Sci. USA. 107:1660–1665. 10.1073/pnas.0907342107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber K., and Herrmann M.. 2015. Tumor biology: with a little help from my dying friends. Curr. Biol. 25:R198–R201. 10.1016/j.cub.2015.01.040 [DOI] [PubMed] [Google Scholar]
- Ley S., Weigert A., Weichand B., Henke N., Mille-Baker B., Janssen R.A., and Brune B.. 2013. The role of TRKA signaling in IL-10 production by apoptotic tumor cell- activated macrophages. Oncogene. 32:631–640. 10.1038/onc.2012.77 [DOI] [PubMed] [Google Scholar]
- Liu C., Cripe T.P., and Kim M.O.. 2010. Statistical issues in longitudinal data analysis for treatment efficacy studies in the biomedical sciences. Mol. Ther. 18:1724–1730. 10.1038/mt.2010.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B., Rutledge B.J., Gu L., Fiorillo J., Lukacs N.W., Kunkel S.L., North R., Gerard C., and Rollins B.J.. 1998. Abnormalities in monocyte recruitment and cytokine expression in monocyte chemoattractant protein 1-deficient mice. J. Exp. Med. 187:601–608. 10.1084/jem.187.4.601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maderna P., Yona S., Perretti M., and Godson C.. 2005. Modulation of phagocytosis of apoptotic neutrophils by supernatant from dexamethasone-treated macrophages and annexin-derived peptide Ac(2-26). J. Immunol. 174:3727–3733. 10.4049/jimmunol.174.6.3727 [DOI] [PubMed] [Google Scholar]
- Mantovani A., Allavena P., Sica A., and Balkwill F.. 2008. Cancer-related inflammation. Nature. 454:436–444. 10.1038/nature07205 [DOI] [PubMed] [Google Scholar]
- Melcher A., Todryk S., Hardwick N., Ford M., Jacobson M., and Vile R.G.. 1998. Tumor immunogenicity is determined by the mechanism of cell death via induction of heat shock protein expression. Nat. Med. 4:581–587. 10.1038/nm0598-581 [DOI] [PubMed] [Google Scholar]
- Miyanishi M., Tada K., Koike M., Uchiyama Y., Kitamura T., and Nagata S.. 2007. Identification of Tim4 as a phosphatidylserine receptor. Nature. 450:435–439. 10.1038/nature06307 [DOI] [PubMed] [Google Scholar]
- Mochizuki T., Kuge Y., Zhao S., Tsukamoto E., Hosokawa M., Strauss H.W., Blankenberg F.G., Tait J.F., and Tamaki N.. 2003. Detection of apoptotic tumor response in vivo after a single dose of chemotherapy with 99mTc-annexin V. J. Nucl. Med. 44:92–97. [PubMed] [Google Scholar]
- Naresh K.N., Lakshminarayanan K., Pai S.A., and Borges A.M.. 2001. Apoptosis index is a predictor of metastatic phenotype in patients with early stage squamous carcinoma of the tongue. Cancer. 91:578–584. [DOI] [PubMed] [Google Scholar]
- Niwa M., Hara A., Kanamori Y., Kohno K., Yoshimi N., Mori H., and Uematsu T.. 1997. Comparison of susceptibility to apoptosis induced by rhTNF-alpha and cycloheximide between human circulating and exudated neutrophils. Life Sci. 61:205–215. 10.1016/S0024-3205(97)00375-5 [DOI] [PubMed] [Google Scholar]
- O’Reilly M.S., Holmgren L., Shing Y., Chen C., Rosenthal R.A., Moses M., Lane W.S., Cao Y., Sage E.H., and Folkman J.. 1994. Angiostatin: a novel angiogenesis inhibitor that mediates the suppression of metastases by a Lewis lung carcinoma. Cell. 79:315–328. 10.1016/0092-8674(94)90200-3 [DOI] [PubMed] [Google Scholar]
- Obeid M., Tesniere A., Ghiringhelli F., Fimia G.M., Apetoh L., Perfettini J.L., Castedo M., Mignot G., Panaretakis T., Casares N., et al. 2007. Calreticulin exposure dictates the immunogenicity of cancer cell death. Nat. Med. 13:54–61. 10.1038/nm1523 [DOI] [PubMed] [Google Scholar]
- Ormerod E.J., Everett C.A., and Hart I.R.. 1986. Enhanced experimental metastatic capacity of a human tumor line following treatment with 5-azacytidine. Cancer Res. 46:884–890. [PubMed] [Google Scholar]
- Orr F.W., Adamson I.Y., and Young L.. 1986. Promotion of pulmonary metastasis in mice by bleomycin-induced endothelial injury. Cancer Res. 46:891–897. [PubMed] [Google Scholar]
- Panigrahy D., Edin M.L., Lee C.R., Huang S., Bielenberg D.R., Butterfield C.E., Barnés C.M., Mammoto A., Mammoto T., Luria A., et al. 2012. Epoxyeicosanoids stimulate multiorgan metastasis and tumor dormancy escape in mice. J. Clin. Invest. 122:178–191. 10.1172/JCI58128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parhar R.S., and Lala P.K.. 1987. Amelioration of B16F10 melanoma lung metastasis in mice by a combination therapy with indomethacin and interleukin 2. J. Exp. Med. 165:14–28. 10.1084/jem.165.1.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perretti M., Chiang N., La M., Fierro I.M., Marullo S., Getting S.J., Solito E., and Serhan C.N.. 2002. Endogenous lipid- and peptide-derived anti-inflammatory pathways generated with glucocorticoid and aspirin treatment activate the lipoxin A4 receptor. Nat. Med. 8:1296–1302. 10.1038/nm786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisco A.O., and Huang S.. 2015. Non-genetic cancer cell plasticity and therapy-induced stemness in tumour relapse: ‘What does not kill me strengthens me’. Br. J. Cancer. 112:1725–1732. 10.1038/bjc.2015.146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pol J., Vacchelli E., Aranda F., Castoldi F., Eggermont A., Cremer I., Sautès-Fridman C., Fucikova J., Galon J., Spisek R., et al. 2015. Trial Watch: Immunogenic cell death inducers for anticancer chemotherapy. OncoImmunology. 4:e1008866 10.1080/2162402X.2015.1008866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poth K.J., Guminski A.D., Thomas G.P., Leo P.J., Jabbar I.A., and Saunders N.A.. 2010. Cisplatin treatment induces a transient increase in tumorigenic potential associated with high interleukin-6 expression in head and neck squamous cell carcinoma. Mol. Cancer Ther. 9:2430–2439. 10.1158/1535-7163.MCT-10-0258 [DOI] [PubMed] [Google Scholar]
- Qian B.Z., and Pollard J.W.. 2010. Macrophage diversity enhances tumor progression and metastasis. Cell. 141:39–51. 10.1016/j.cell.2010.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reers S., Pfannerstill A.C., Rades D., Maushagen R., Andratschke M., Pries R., and Wollenberg B.. 2013. Cytokine changes in response to radio-/chemotherapeutic treatment in head and neck cancer. Anticancer Res. 33:2481–2489. [PubMed] [Google Scholar]
- Reiter I., Krammer B., and Schwamberger G.. 1999. Cutting edge: differential effect of apoptotic versus necrotic tumor cells on macrophage antitumor activities. J. Immunol. 163:1730–1732. [PubMed] [Google Scholar]
- Révész L. 1956. Effect of tumour cells killed by x-rays upon the growth of admixed viable cells. Nature. 178:1391–1392. 10.1038/1781391a0 [DOI] [PubMed] [Google Scholar]
- Rothwell P.M., Wilson M., Price J.F., Belch J.F., Meade T.W., and Mehta Z.. 2012. Effect of daily aspirin on risk of cancer metastasis: a study of incident cancers during randomised controlled trials. Lancet. 379:1591–1601. 10.1016/S0140-6736(12)60209-8 [DOI] [PubMed] [Google Scholar]
- Schif-Zuck S., Gross N., Assi S., Rostoker R., Serhan C.N., and Ariel A.. 2011. Saturated-efferocytosis generates pro-resolving CD11b low macrophages: modulation by resolvins and glucocorticoids. Eur. J. Immunol. 41:366–379. 10.1002/eji.201040801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seelig K.J., and Revesz L.. 1960. Effect of lethally damaged tumour cells upon the growth of admixed viable cells in diffusion chambers. Br. J. Cancer. 14:126–138. 10.1038/bjc.1960.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serhan C.N. 2014. Pro-resolving lipid mediators are leads for resolution physiology. Nature. 510:92–101. 10.1038/nature13479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serhan C.N., Hong S., Gronert K., Colgan S.P., Devchand P.R., Mirick G., and Moussignac R.L.. 2002. Resolvins. J. Exp. Med. 196:1025–1037. 10.1084/jem.20020760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan C.C., Shi L.R., Ding M.Q., Zhu Y.B., Li X.D., Xu B., Jiang J.T., and Wu C.P.. 2015. Cytokine-induced killer cells co-cultured with dendritic cells loaded with the protein lysate produced by radiofrequency ablation induce a specific antitumor response. Oncol. Lett. 9:1549–1556. 10.3892/ol.2015.2977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spite M., Norling L.V., Summers L., Yang R., Cooper D., Petasis N.A., Flower R.J., Perretti M., and Serhan C.N.. 2009. Resolvin D2 is a potent regulator of leukocytes and controls microbial sepsis. Nature. 461:1287–1291. 10.1038/nature08541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stienstra R., Dijk W., van Beek L., Jansen H., Heemskerk M., Houtkooper R.H., Denis S., van Harmelen V., Willems van Dijk K., Tack C.J., and Kersten S.. 2014. Mannose-binding lectin is required for the effective clearance of apoptotic cells by adipose tissue macrophages during obesity. Diabetes. 63:4143–4153. 10.2337/db14-0256 [DOI] [PubMed] [Google Scholar]
- Stuart L.M., Takahashi K., Shi L., Savill J., and Ezekowitz R.A.. 2005. Mannose-binding lectin-deficient mice display defective apoptotic cell clearance but no autoimmune phenotype. J. Immunol. 174:3220–3226. 10.4049/jimmunol.174.6.3220 [DOI] [PubMed] [Google Scholar]
- Sun B., Sun Y., Wang J., Zhao X., Wang X., and Hao X.. 2006. Extent, relationship and prognostic significance of apoptosis and cell proliferation in synovial sarcoma. Eur. J. Cancer Prev. 15:258–265. 10.1097/01.cej.0000198896.02185.68 [DOI] [PubMed] [Google Scholar]
- Sun Y.P., Oh S.F., Uddin J., Yang R., Gotlinger K., Campbell E., Colgan S.P., Petasis N.A., and Serhan C.N.. 2007. Resolvin D1 and its aspirin-triggered 17R epimer. Stereochemical assignments, anti-inflammatory properties, and enzymatic inactivation. J. Biol. Chem. 282:9323–9334. 10.1074/jbc.M609212200 [DOI] [PubMed] [Google Scholar]
- Tesniere A., Schlemmer F., Boige V., Kepp O., Martins I., Ghiringhelli F., Aymeric L., Michaud M., Apetoh L., Barault L., et al. 2010. Immunogenic death of colon cancer cells treated with oxaliplatin. Oncogene. 29:482–491. 10.1038/onc.2009.356 [DOI] [PubMed] [Google Scholar]
- Titos E., Rius B., González-Périz A., López-Vicario C., Morán-Salvador E., Martínez-Clemente M., Arroyo V., and Clària J.. 2011. Resolvin D1 and its precursor docosahexaenoic acid promote resolution of adipose tissue inflammation by eliciting macrophage polarization toward an M2-like phenotype. J. Immunol. 187:5408–5418. 10.4049/jimmunol.1100225 [DOI] [PubMed] [Google Scholar]
- Todryk S., Melcher A.A., Hardwick N., Linardakis E., Bateman A., Colombo M.P., Stoppacciaro A., and Vile R.G.. 1999. Heat shock protein 70 induced during tumor cell killing induces Th1 cytokines and targets immature dendritic cell precursors to enhance antigen uptake. J. Immunol. 163:1398–1408. [PubMed] [Google Scholar]
- Unga J., and Hashida M.. 2014. Ultrasound induced cancer immunotherapy. Adv. Drug Deliv. Rev. 72:144–153. 10.1016/j.addr.2014.03.004 [DOI] [PubMed] [Google Scholar]
- van den Brenk H.A., Crowe M.C., and Stone M.G.. 1977. Reactions of the tumour bed to lethally irradiated tumour cells, and the Révész effect. Br. J. Cancer. 36:94–104. 10.1038/bjc.1977.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk-Draper L., Hall K., Griggs C., Rajput S., Kohio P., DeNardo D., and Ran S.. 2014. Paclitaxel therapy promotes breast cancer metastasis in a TLR4-dependent manner. Cancer Res. 74:5421–5434. 10.1158/0008-5472.CAN-14-0067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas D., Laput G., and Vyas A.K.. 2014. Chemotherapy-enhanced inflammation may lead to the failure of therapy and metastasis. Onco Targets Ther. 7:1015–1023. 10.2147/OTT.S60114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., and DuBois R.N.. 2010. Eicosanoids and cancer. Nat. Rev. Cancer. 10:181–193. 10.1038/nrc2809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong K., Valdez P.A., Tan C., Yeh S., Hongo J.A., and Ouyang W.. 2010. Phosphatidylserine receptor Tim-4 is essential for the maintenance of the homeostatic state of resident peritoneal macrophages. Proc. Natl. Acad. Sci. USA. 107:8712–8717. 10.1073/pnas.0910929107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyllie A.H. 1985. The biology of cell death in tumours. Anticancer Res. 5:131–136. [PubMed] [Google Scholar]
- Yu Y., Tian L., Feng X., Cheng J., Gong Y., Liu X., Zhang Z., Yang X., He S., Li C.Y., and Huang Q.. 2016. eIF4E-phosphorylation-mediated Sox2 upregulation promotes pancreatic tumor cell repopulation after irradiation. Cancer Lett. 375:31–38. 10.1016/j.canlet.2016.02.052 [DOI] [PubMed] [Google Scholar]
- Zeisberger S.M., Odermatt B., Marty C., Zehnder-Fjällman A.H., Ballmer-Hofer K., and Schwendener R.A.. 2006. Clodronate-liposome-mediated depletion of tumour-associated macrophages: a new and highly effective antiangiogenic therapy approach. Br. J. Cancer. 95:272–281. 10.1038/sj.bjc.6603240 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.