Scally et al. show the molecular, structural, and functional characterization of human antibodies against the C-terminal domain of Plasmodium falciparum (Pf) circumsporozoite (CSP [C-PfCSP]) and reveal that its arrangement on the Pf sporozoite surface and epitope polymorphism contribute to poor C-PfCSP immunogenicity and ineffective humoral responses in volunteers protected against Pf malaria.
Abstract
Antibodies against the central repeat of the Plasmodium falciparum (Pf) circumsporozoite protein (CSP) inhibit parasite activity and correlate with protection from malaria. However, the humoral response to the PfCSP C terminus (C-PfCSP) is less well characterized. Here, we describe B cell responses to C-PfCSP from European donors who underwent immunization with live Pf sporozoites (PfSPZ Challenge) under chloroquine prophylaxis (PfSPZ-CVac), and were protected against controlled human malaria infection. Out of 215 PfCSP-reactive monoclonal antibodies, only two unique antibodies were specific for C-PfCSP, highlighting the rare occurrence of C-PfCSP–reactive B cells in PfSPZ-CVac–induced protective immunity. These two antibodies showed poor sporozoite binding and weak inhibition of parasite traversal and development, and did not protect mice from infection with PfCSP transgenic Plasmodium berghei sporozoites. Structural analyses demonstrated that one antibody interacts with a polymorphic region overlapping two T cell epitopes, suggesting that variability in C-PfCSP may benefit parasite escape from humoral and cellular immunity. Our data identify important features underlying C-PfCSP shortcomings as a vaccine target.
Introduction
The sporozoite stage of the Plasmodium falciparum (Pf) life cycle is a target to elicit sterilizing immunity against malaria by vaccination (Seder et al., 2013; Mordmüller et al., 2017; Sissoko et al., 2017). Circumsporozoite protein (CSP) is the most abundant surface protein of sporozoites (Potocnjak et al., 1982; Swearingen et al., 2016). CSP is encoded by a single gene (Nussenzweig and Nussenzweig, 1989) and is essential for the development of sporozoites in mosquitoes and mammalian hosts (Ménard et al., 1997). CSP has been implicated in the invasion of mosquito salivary glands (Sidjanski et al., 1997) and mediates adhesion of sporozoites to liver cells in the mammalian host (Cerami et al., 1992; Frevert et al., 1993). Anti-CSP antibodies can prevent attachment of sporozoites to hepatocytes, inhibit cell traversal and cell invasion, and thus interfere with the infection (Potocnjak et al., 1980; Yoshida et al., 1980; Charoenvit et al., 1991a,b; Mishra et al., 2012; Sumitani et al., 2013; Foquet et al., 2014). The high abundance of CSP on the parasite surface and its essential roles in invasion—which can be impeded by antibodies—are reasons why CSP is a leading molecular target for the development of a malaria vaccine.
PfCSP consists of a signal peptide, an N-terminal domain, a central region made of tetrapeptide NANP and NVDP repeats, and a C-terminal region (C-PfCSP) that comprises a linker followed by a unique α-thrombospondin type-1 repeat (αTSR) domain (Fig. 1 A; McCutchan et al., 1996; Doud et al., 2012). PfCSP is anchored to the sporozoite plasma membrane by a glycosylphosphatidylinositol (GPI) attachment site at its C terminus (Wang et al., 2005). Three sequence motifs are particularly well conserved in CSP of all Plasmodium species (Dame et al., 1984). The pentapeptide KLKQP, region I, is located immediately upstream of the central repeat; contacts between a lysine cluster N-terminal of this region and heparin sulfate proteoglycans are responsible for sporozoite attachment to hepatocytes (Zhao et al., 2016). An 18–amino acid sequence (EWSXCXVTCGXG[V/I]XXRX[K/R]) within the C-CSP αTSR domain is designated region II–plus (Dame et al., 1984; Kappe et al., 2004), whereas region III is located immediately downstream of the linker to the central repeat and forms an amphipathic α-helix in C-CSP (Fig. 1 A). Sequence elements that confer on PfCSP its many roles in migration, adhesion, and infection have still not been fully elucidated.
Figure 1.
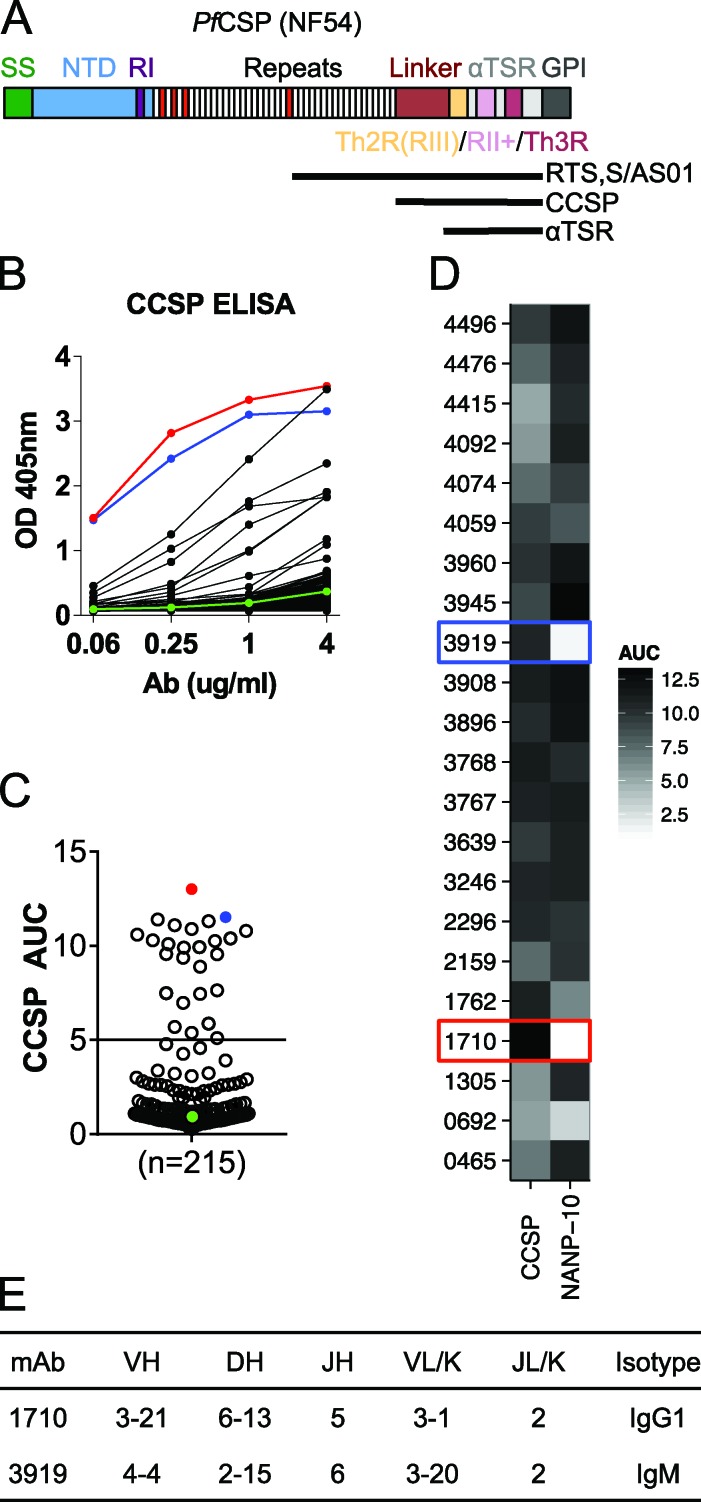
Characterization of antibodies elicited by immunization with live sporozoites under chemoprophylaxis (PfSPZ-CVac) against C-PfCSP. (A) Schematic representation of FL PfCSP (NF54). PfCSP contains a signal sequence (SS), N-terminal domain (NTD), region I (RI), a tandem repeat region consisting of NANP (white) or NVDP (red) repeats, a linker region, and an α1-helix thrombospondin type-1 repeat (αTSR) that contains the largely overlapping region III (RIII)/Th2R, region II+ (RII+) and Th3R, followed by a GPI attachment site. The region corresponding to the RTS,S/AS01 vaccine, the C-PfCSP construct used in ELISA, and the αTSR crystallization construct are indicated by horizontal black lines. (B) Representative ELISA curves of recombinantly expressed mAbs binding to C-PfCSP. Red and blue lines indicate C-PfCSP–binding antibodies 1710 and 3919, respectively. The negative control antibody mGO53 is shown in green. (C) AUCs calculated for 215 PfCSP memory B cell antibodies and the circles colored as in B. (D) Binding of the 22 C-PfCSP antibodies from B and C to a 10mer NANP repeat peptide (NANP-10) indicate most bind to both C-PfCSP and repeat domains. (B–D) Data are representative of two independent experiments. (E) Gene usage and isotype of antibodies 1710 and 3919.
Protection mediated by CSP-specific immune responses has been firmly established in mouse models and in humans, and is linked to antibodies (Potocnjak et al., 1980; Ockenhouse et al., 2015; Kazmin et al., 2017), CD4 T cells (Tsuji et al., 1990; Migliorini et al., 1993) and CD8 T cells (Schofield et al., 1987; Weiss et al., 1988; Romero et al., 1989). Immunodominant B cell epitopes in CSP are primarily located in the central domain repeat (Godson et al., 1983; Zavala et al., 1983). Indeed, over 90% of sporozoite-specific antibodies recognize CSP repeats in sera from volunteers or animals immunized with sporozoites, or from individuals living in malaria-endemic areas (Nardin et al., 1979; Zavala et al., 1985). Antibodies against the N- and C-terminal domains of CSP have also been characterized (Stüber et al., 1990; Calle et al., 1992; Lopez et al., 1996; Bongfen et al., 2009) Evidence is scarce whether the latter antibodies can inhibit sporozoite infection to the same extent as repeat antibodies. Inhibition studies have been complicated in part by the analysis of polyclonal sera dominated by repeat antibodies (Calle et al., 1992). In cases where functionality has been assessed, antibodies targeting the N-terminal domain of PfCSP were generally associated with better inhibition of sporozoite migration and infection compared with antibodies against C-PfCSP (Bongfen et al., 2009). However, a caveat of previous studies analyzing antibody responses to PfCSP subregions resides in their use of synthetic peptides—and not folded protein, such as the C-PfCSP subdomain—to interrogate reactivity (Doud et al., 2012). Therefore, it remains unclear whether high-affinity antibodies against the three-dimensional C-PfCSP structure can effectively inhibit sporozoite infection.
Here, we screened binding to PfCSP subregions for a panel of 215 human PfCSP-reactive monoclonal memory B cell antibodies derived from individuals immunized with aseptic, purified, cryopreserved live Pf sporozoites (PfSPZ Challenge) under chloroquine prophylaxis (PfSPZ-CVac) who were completely protected against malaria (Mordmüller et al., 2017). We uncovered the molecular basis of antibody recognition of C-PfCSP using binding kinetics experiments and x-ray crystallography. We reveal that PfCSP surface organization on Pf sporozoites and sequence diversity are likely mechanisms used by Pf to impede effective humoral responses against C-PfCSP.
Results and discussion
Antibody responses to C-PfCSP after immunization with PfSPZ-CVac
To understand the mode and functional relevance of the antibody response against C-PfCSP, recombinant mAbs from PfCSP-binding memory B cells of eight European donors who received three PfSPZ-CVac immunizations with 51,200 PfSPZ at 4-wk intervals (Mordmüller et al., 2017) were produced and screened via ELISA for binding to a recombinantly expressed C-PfCSP that includes both the linker and αTSR subdomains (Fig. 1, A and B). In contrast to antibodies that bound the NANP repeat but not to C-PfCSP (193/215), C-PfCSP–binding antibodies were relatively infrequent (22/215). Some of these 22 distinct antibodies belonged to clonally expanded clusters that were diversified through somatic hypermutations. The areas under the ELISA curves (AUCs) of these 22 antibodies illustrate their various degree of binding to C-PfCSP (Fig. 1 C). Only four of the 22 antibodies showed weak polyreactivity, indicating the binding specificity to C-PfCSP for these antibodies (Fig. S1 A). Most of the C-PfCSP–binding antibodies also bound to a peptide with 10 NANP repeats (NANP-10; Fig. 1 D), suggesting their epitopes span the repeat and part of C-PfCSP or there are commonalities associated with these two regions. Antibodies 1710 and 3919 had the highest ELISA reactivity for C-PfCSP (Fig. 1 C) and were the only two antibodies that lacked binding to the NANP repeat, indicating that they exclusively recognized C-PfCSP (Fig. 1 D). Antibody 1710 was cloned from a single IgG1 memory B cell and carried one IGHV and two IGLV mutations, whereas antibody 3919 was cloned from a small cluster of IgM cells that had two clonal relatives and carried one IGHV and two IGKV mutations (Fig. 1 E). Our data demonstrate the poor immunogenicity of C-PfCSP after exposure to three high doses of PfSPZ-CVac with 51,200 PfSPZ, a regimen that was 100% protective at 10 wk after the last vaccine dose (Mordmüller et al., 2017).
C-PfCSP epitope reactivity on recombinant PfCSP
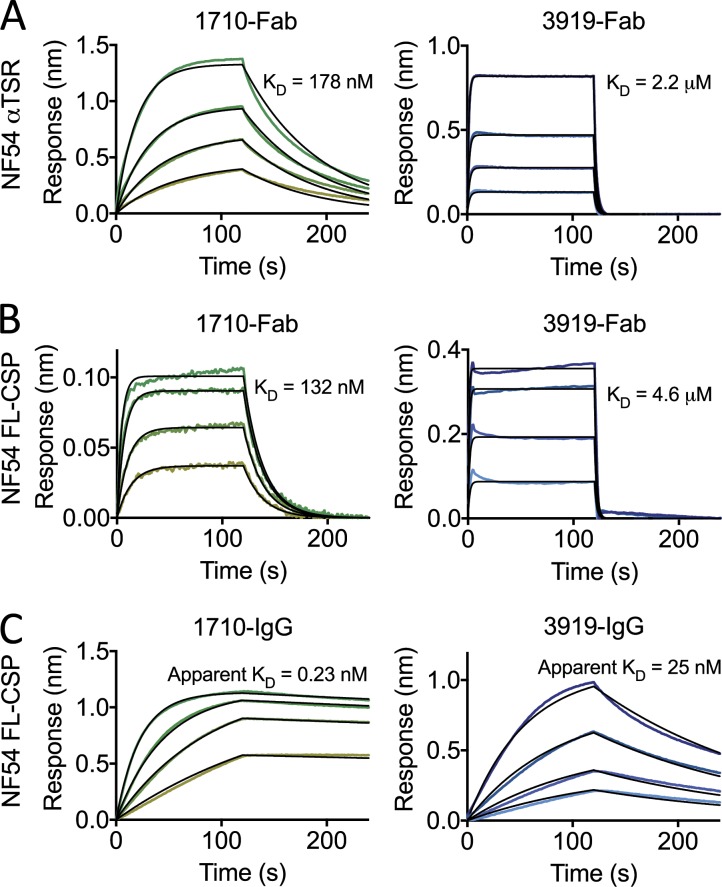
We next determined that the two C-PfCSP–exclusive 1710 and 3919 Fabs bind to the αTSR domain of C-PfCSP with nanomolar and micromolar affinities in biolayer interferometry binding kinetic experiments (178 ± 21 nM and 2.2 ± 0.3 µM, respectively; Fig. 2 A). Binding competition experiments showed that the two antibodies recognize an overlapping epitope on C-PfCSP (Fig. S1 B). In the absence of a full-length (FL) structure of PfCSP, the accessibility of C-PfCSP–embedded epitopes on FL protein and more so on the parasite surface remains unclear. To examine whether the three-dimensional disposition of the N-terminal domain and NANP repeats interfered with C-PfCSP accessibility, we compared the binding kinetics of 1710 and 3919 Fabs to only the αTSR domain of C-PfCSP, and to FL PfCSP. 1710 and 3919 Fabs bound to FL PfCSP with similar affinities as to the αTSR domain, 132 ± 13 nM and 4.6 ± 1.0 µM (Fig. 2 B), respectively, suggesting that the N-terminal domain and the NANP central repeat do not conformationally occlude their epitopes on recombinant PfCSP.
Figure 2.
1710 and 3919 binding to recombinant NF54 PfCSP. (A–C) Representative sensorgrams and 1:1 model best fits (black) for 1710 (green) and 3919 (blue) Fab binding to αTSR (A) and FL PfCSP (B) and 1710 and 3919 IgG binding to FL PfCSP (C). Data are representative of three independent measurements.
In our binding kinetics assays, we observed an ∼500-fold and 200-fold higher apparent KD for 1710 IgG (0.23 nM) and 3919 IgG (25 nM) compared with their Fab counterpart (132 nM, and 4.6 µM, respectively), indicating that these antibodies can effectively use bivalent cross-linking to mediate high-avidity binding to PfCSP (Fig. 2 C). Interestingly, the antibodies were derived from cells expressing IgG1 (1710) and IgM (3919; Fig. 1 E). Pentameric binding of the secreted IgM would likely confer much higher binding avidity to 3919 despite its micromolar binding affinity. In summary, the two exclusive C-PfCSP antibodies recognized the recombinant αTSR domain with high affinity and avidity.
The epitope of antibody 1710 overlaps two T cell epitopes in C-PfCSP
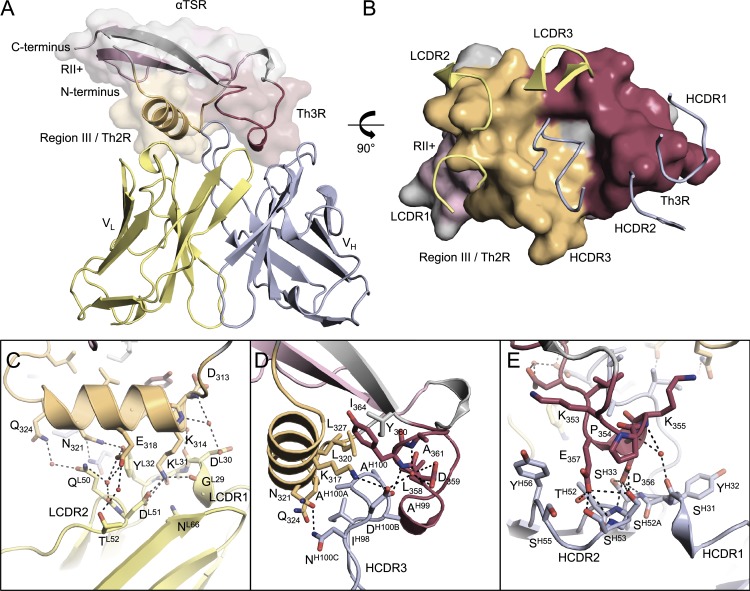
To gain molecular insights into the PfCSP epitope to which 1710 binds with nanomolar affinity, we solved the crystal structure of the complex between 1710 Fab and the αTSR domain of PfCSP with NF54 sequence to 1.95 Å resolution (Fig. 3 A and Table S1). All six 1710 antibody complementarity determining regions (CDRs) contribute to αTSR recognition with a total of 712 Å2 of buried surface area on the antigen: 482 Å2 from the heavy chain and 230 Å2 from the light chain (Fig. 3 B). The importance of the epitope conformation was further corroborated by ELISA experiments with individual C-PfCSP peptides that failed to detect antibody binding (Fig. S1 C).
Figure 3.
Crystal structure of the PfCSP αTSR domain in complex with 1710 Fab. (A) The 1710 Fab variable regions bound to PfCSP αTSR. Region III/Th2R, region II+, and Th3R are orange, pink, and maroon, respectively. (B) CDR loops interacting with the Th2R and Th3R regions of PfCSP αTSR. LCDRs predominantly interact with the Th2R peptide (orange), whereas HCDRs predominantly interact with the hydrophobic core and the Th3R peptide (maroon). (C–E) PfCSP αTSR interactions with LCDRs (C), HCDR3 (D), and HCDR1/2 (E) loops. Hydrogen bonds are denoted as black dashed lines.
Interestingly, the 1710 epitope is primarily located in peptide regions that correspond to Th2R and Th3R epitopes. Light-chain CDR1 (LCDR1) and LCDR2 interact closely with the α1-helix (residues 313–324, corresponding to region III or Th2R), forming a total of 12 hydrogen-bonds (H-bonds), most of which are water-mediated (Fig. 3 C). The predominantly hydrophobic 16-residue long heavy-chain CDR3 (HCDR3) inserts in a conserved hydrophobic groove on PfCSP formed at the interface of the α1-helix and loop 355–364 (Fig. 3 D). AspH100B bridges the two structural elements through a salt bridge with PfCSP residue K317 and H-bonds with residues D359, Y360, and A361 (Fig. 3 D). PfCSP residues 353–357 (peptide corresponding to Th3R) are contacted by HCDR1 and HCDR2 primarily through intimate H-bonding (Fig. 3 E).
The PfCSP antigenic surface recognized by 1710 is made of basic and acidic patches on either side of a recessed central hydrophobic groove (Fig. S2 A). The shape and charge complementarity of the 1710 paratope indicates an optimally evolved molecular recognition (Fig. S2 B). This is highlighted by the key basic residue K314 from the Th2R epitope occupying an acidic pocket formed by LCDR1, and acidic residues D356 and E357 of the Th3R peptide occupying a basic pocket formed by HCDR1 and HCDR2 (Fig. S2, C and D). Compared with the αTSR-bound antibody structure, unliganded 1710 Fab (Fig. S2 E and Table S1) shows high conformational similarity across all CDRs (root mean square deviation [r.m.s.d.] of 0.57 Å), thus indicating a fine-tuned lock-and-key interaction of 1710 with its antigen.
Previous studies have described O-fucosylation in the conserved CSVTCGNG sequence in PfCSP expressed from HEK 293T cells (Doud et al., 2012) or from the sporozoite surface (Swearingen et al., 2016; Lopaticki et al., 2017). We observed partial evidence of O-linked glycosylation in the electron density map of our αTSR construct expressed from HEK 293F cells. ESI/TOF mass spectrometry analysis of our recombinant αTSR construct revealed three major species (Fig. S2 F). This included a predominant nonglycosylated species (9,168 Da), and two species, 146 Da and 308 Da heavier, likely indicating an O-fucose (146 Da) and an O-glucosylfucose disaccharide (308 Da) glycosylation at T337. Differential O-fucosylation likely led to the partial electron density we observe in the crystal structure, as crystal packing would not preclude an O-fucose from being accommodated. In any case, the O-fucosylation site is located distally from the 1710 epitope, and this posttranslational modification is unlikely to affect recognition.
Our co-complex structure reveals that the overall fold of 1710-bound αTSR has a low r.m.s.d. of 1.34 Å (Cα’s) compared with the unliganded antigen (Fig. S2 G; Doud et al., 2012). Minor differences are observed in the N- and C-terminal regions of the crystal structures, likely because of differences in crystal packing. This high similarity indicates that the αTSR-fold previously described for PfCSP (Doud et al., 2012) is biologically relevant, and seen by the immune system after repeated exposure to PfSPZ-CVac. Unique structural elements of the PfCSP αTSR (compared with other TSR folds) map to the hydrophilic faces of the α1-helix and neighboring loop, arranging to form a central hydrophobic groove (Doud et al., 2012). Thus, it is not unexpected that these features are the ones recognized by sporozoite-elicited antibodies in humans. Importantly, our structural delineation of 1710 binding to αTSR uncovered that the B cell epitope overlaps two T cell epitopes, and corresponds to C-PfCSP regions with highest sequence variation between Pf isolates (Weedall et al., 2007; Bailey et al., 2012; Gandhi et al., 2012). No C-PfCSP specific Abs were isolated against the highly conserved C-PfCSP region II–plus, which is likely less immunogenic or less exposed on PfCSP on the surface of sporozoites. Low C-PfCSP immunogenicity in the context of whole Pf sporozoites immunizations is in line with the results of our studies on healthy Gabonese adults with lifelong exposure to Pf, where out of 208 mAbs cloned from PfCSP-reactive memory B cells, no antibodies were found to be solely C-PfCSP reactive (Triller et al., 2017).
Strain specificity of the antibody response against C-PfCSP
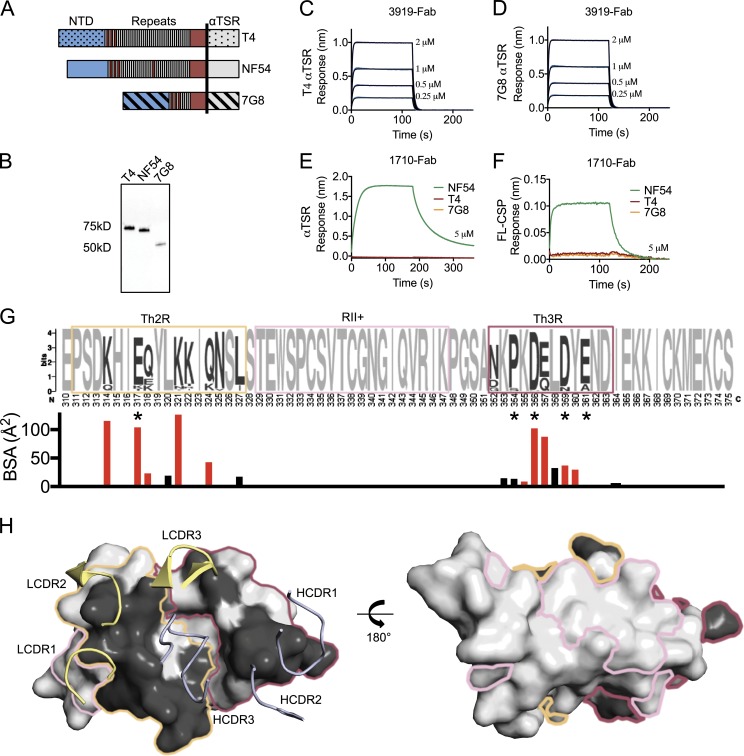
We next tested whether 1710 and 3919 can react broadly against PfCSP of different genotypes, or are specific to the NF54 PfCSP against which they were elicited. PfCSP constructs from three Pf genotypes (NF54, T4, and 7G8) were generated as FL and αTSR PfCSP constructs (Fig. 4, A and B). Whereas 3919 reacted weakly with αTSR constructs from all three strains, 1710 was specific to NF54 PfCSP constructs (Fig. 4, C–F). T4 and 7G8 constructs failed to show any binding at the highest 1710 concentration tested (5 µM; Fig. 4, E and F). Inspection of the three C-PfCSP sequences revealed key polymorphisms in the 1710 epitope that explain its strain-specific binding reactivity (Fig. S2 J). Whereas the centrally located αTSR hydrophobic groove is conserved across the three genotypes, surrounding epitope regions that comprise PfCSP residues 317, 318, 321, 324, and 361 differ significantly in sequence between NF54, T4, and 7G8, and generally across Pf strains (Fig. 4, G and H). In NF54, K317 forms a salt bridge with AspH100B of the HCDR3 (Fig. 3 D). A K317E polymorphism in the T4 and 7G8 strains would likely introduce electrostatic repulsion in this region. Additionally, N321 is located within a pocket formed by residues of the LCDR1/2 and HCDR3 and forms H-bonds with AsnH100C and TyrL32 (Fig. 3 C). Thus, the N321K polymorphism would likely create a steric clash with this region of the 1710 paratope; in this scenario, AsnH100C and TyrL32 would prevent K321 from adopting an alternate conformation. Swapping the αTSR domain in FL T4 and 7G8 PfCSP with that of NF54 resulted in similar binding affinities to FL NF54 PfCSP, thereby attributing 1710 strain specificity entirely to the NF54 αTSR domain (Fig. S2, H and I). Thus, the 1710 paratope likely cannot accommodate either the K317E or N321K polymorphisms present in the T4 and 7G8 genotypes.
Figure 4.
PfCSP sequence specificity of C-PfCSP–reactive antibodies. (A) Schematic representation of the FL PfCSP from NF54, T4, and 7G8 Pf genotypes. The vertical black line represents the boundary for the αTSR construct. (B) Western blot of T4, NF54, and 7G8 FL PfCSP constructs expressed recombinantly, and probed with an anti-(His)6x tag antibody. (C and D) 3919 Fab binds both T4 αTSR (C) and 7G8 αTSR (D) constructs weakly as measured by biolayer interferometry. (E and F) 1710 Fab binds NF54 constructs, but not T4 and 7G8 constructs as measured by biolayer interferometry for αTSR (E) and FL PfCSP (F). Data are representative of two independent measurements. (G) αTSR sequence diversity from the NCBI database in Weblogo representation (Crooks et al., 2004). Th2R, RII+, and Th3R residues are shown in orange, pink, and maroon boxes, respectively. Invariant residues are light gray, whereas polymorphic residues are dark gray. Asterisks indicates residues associated with efficacy in the RTS,S/AS01 vaccine in sieve analysis when parasites were matched to the vaccine sequence (Neafsey et al., 2015). Below is the surface area contribution of each αTSR residue buried by antibody 1710 as determined by PISA (Krissinel and Henrick, 2007). Residues that form a salt bridge or H-bond with 1710 are red, whereas those that provide van der Waals interactions are black. (H) Polymorphism mapped onto the αTSR surface colored using the scheme in (E). Polymorphisms are constrained to the Th2R and Th3R peptides and occupy one face of αTSR, which is the recognition site of antibody 1710. The invariant region II+ presides on the opposite face, which is likely less exposed on PfCSP on the surface of sporozoites. NTD, N-terminal domain.
Together our data provide a molecular basis for understanding how antigenic diversity in C-PfCSP might limit the breadth of antibody responses against this subregion (Calle et al., 1992) in addition to the previously described evasion from cellular immunity (Udhayakumar et al., 1997; Gilbert et al., 1998; Bonelo et al., 2000).
Inhibition of parasite traversal, development, and infection
To investigate to what extent epitopes were accessible to C-PfCSP–reactive antibodies in the context of whole Pf sporozoites, we performed live immunofluorescence and flow cytometry experiments (Fig. 5, A and B). In both assays, C-PfCSP–reactive antibodies that also have high affinity to a NANP-5 peptide in SPR (3908: 54 ± 5 nM and 2296: 201 nM) bound to a NF54 PfCSP transgenic Plasmodium berghei (Pb-PfCSP) parasite line (Triller et al., 2017) to a similar level as the NANP-specific positive control (2A10: 2.2 nM; Fig. 5, A and B). In contrast, antibody 1710 showed weak binding at 100 µg/ml and no binding at 1 µg/ml. This observation was confirmed in IFA with fixed NF54 Pf sporozoites (Fig. S3 A). Presumably because of its lower αTSR affinity compared with 1710, antibody 3919 failed to bind in either of the Pb-PfCSP assays even at high concentration (Fig. 5, A and B).
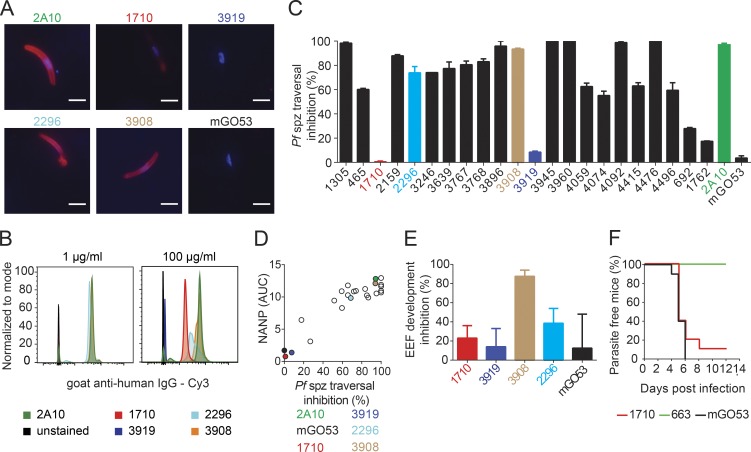
Figure 5.
Functional activity of C-PfCSP–reactive antibodies. (A and B) Binding of the indicated mAbs to live Pb-PfCSP transgenic sporozoites at either 1 µg/ml or 100 µg/ml as determined by immunofluorescence assay (A) and flow cytometry (B). Bars, 5 µm. (C) Percent inhibition of Pf sporozoite traversal activity as measured in hepatocyte traversal assay in vitro. (D) Pf sporozoite traversal inhibition correlates with NANP cross-reactivity of C-PfCSP–binding antibodies. C-PfCSP–specific antibodies 1710 and 3919 are colored as in Fig. 1 B, whereas the positive control, 2A10, and negative control, mGO53, are shown in green and black, respectively. Data are representative of two independent experiments. (E) Percent inhibition of parasite liver stage development by C-PfCSP antibodies. 3908 and 2296 are C-PfCSP binding antibodies that cross-react with the NANP repeat (see Fig. 1). (F) Percentage of parasite-free mice after passive immunization of antibody 1710 and infection with Pb-PfCSP sporozoites. Antibodies 663 and mGO53 were used as positive and negative controls, respectively. (A–D) Data are representative of two independent experiments. (E and F) Data are pooled from two independent experiments with n = 5 mice per group. Error bars indicate standard error of the mean.
We next tested the potency of C-PfCSP–reactive antibodies in functional assays. Inhibition of Pf sporozoite traversal was directly correlated with the ability of C-PfCSP–reactive antibodies to also bind the NANP repeat (Fig. 5, C and D). Consequently, 1710 and 3919, which bind C-PfCSP exclusively, showed poor Pf sporozoite traversal inhibition (Figs. 1 D and 5 C). The lack of protective efficacy was confirmed using Pb-PfCSP sporozoites. Both antibodies failed to inhibit Pb-PfCSP sporozoite development into exoerythrocytic forms (EEFs) in hepatoma cells in vitro, compared with the anti–C-PfCSP antibodies 2296 and 3908 with intermediate and high repeat reactivity, respectively (Fig. 5 E). Our findings were further validated in vivo (Fig. 5 F and Fig. S3 C). Passive immunization with 400 µg of 1710 failed to protect WT mice from Pb-PfCSP sporozoite infection and the development of blood-stage parasites, demonstrating that the anti-αTSR antibodies lacked protective efficacy compared with the positive control anti-NANP repeat antibody 663, which protected all mice from the infection (Triller et al., 2017).
Our data suggest that a unique PfCSP disposition on the surface of sporozoites might mitigate the accessibility of C-PfCSP antibodies to their epitopes, negatively impacting their inhibitory activity. A model where PfCSP is densely packed on sporozoites preventing antibodies targeting the αTSR by steric hindrance has previously been proposed (Coppi et al., 2011). Such molecular crowding might be particularly relevant for antibodies that bind epitopes located near the GPI anchor at the C terminus, as described here for 1710. Despite an ability for 1710 and 3919 antibodies to cross-link two adjacent recombinant PfCSP molecules to achieve high-avidity binding (0.23 nM and 25 nM, respectively), we note that only one C-PfCSP antibody can bind per PfCSP, whereas multiple anti-repeat antibodies can bind PfCSP (Fisher et al., 2017), perhaps contributing to differences in overall antibody binding and protective activity. Whether conformation and epitope accessibility of recombinant PfCSP is representative of that on the surface of sporozoites also remains to be determined. Our data suggest that antibody efficiency to bind PfCSP epitopes and epitope accessibility on sporozoites both contribute to antibody immune effectiveness.
Concluding remarks
Our work provides insights into human B cell–mediated immunity to a non-immunodominant and largely polymorphic C-PfCSP epitope on sporozoites. Antibody recognition of this αTSR region in recombinant proteins is linked to inefficient inhibition of sporozoite traversal, development, and infection, which corroborates previous findings of the poor functionality of antibody responses against this PfCSP subregion (Calle et al., 1992; Lopez et al., 1996; Bongfen et al., 2009). Our molecular characterization of a C-PfCSP–reactive antibody with weak inhibitory characteristics is important for iterative vaccine design. It gives clues as to which regions should be altered or removed in subunit immunogens to decrease undesired immune responses, as has previously been done in HIV (de Taeye et al., 2015) and influenza (Impagliazzo et al., 2015) vaccine development. Antibodies against the N-terminal domain and the central repeat region have been shown to be associated with broader and more potent inhibitory effects (Potocnjak et al., 1980; Godson et al., 1983; Zavala et al., 1983; Charoenvit et al., 1991a,b; Bongfen et al., 2009), and thus we propose that their re-elicitation should be prioritized in vaccination strategies. Approaches to develop a broadly effective PfCSP-based subunit malaria vaccine should therefore carefully evaluate whether displaying αTSR in immunogens is the best approach to providing Pf B and T cell epitopes. Instead, immunogen design strategies that can elicit superior responses focused on conserved and accessible PfCSP regions might improve on current vaccine efficacy.
Materials and methods
Flow cytometric isolation and sorting of PfCSP memory B cells
Inoculation of European volunteers with Sanaria PfSPZ Challenge in the Sanaria PfSPZ-CVac clinical trial (TÜCHMI-2) was previously reported (Richie et al., 2015; Mordmüller et al., 2017). The study was approved by the ethics committee of the medical faculty and the university clinics of the University of Tübingen and strictly adhered to Good Clinical Practice and the principles of the Declaration of Helsinki. The clinical trial is registered at ClinicalTrials.gov (NCT02115516). The study was performed under FDA IND 15862 and with approval of the Paul Ehrlich Institute. Peripheral blood mononuclear cells (PBMCs) were isolated from the blood sample using Ficoll gradient density and frozen down in liquid nitrogen for long-term storage. Before sorting, the cells were thawed and twice washed in RPMI (Gibco). Recombinantly expressed PfCSP (Protein Potential, LLC), labeled with Alexa 647 (Molecular Probes), was used for the isolation of PfCSP-reactive B cells. PBMCs were incubated with the labeled PfCSP and following labeled antibodies and dead cell marker at the mentioned dilutions: CD20-APC-H7 (2H7; 1:20), CD19-BV786 (SJ25C1; 1:10), CD27-PE (M-T271; 1:5), CD38-FITC (HIT2; 1:5), CD138-BV421 (MI15; 1:20), IgG-BV510 (G18-145; 1:20; all BD Biosciences), CD21-PE-Cy7 (Bu32; 1:20; BioLegend), and 7AAD (1:400; Invitrogen). 7-AAD−CD19+CD27+CSP+ single cells were sorted using a FACS AriaII (Becton Dickinson) with FACSDiVa software version 8.0.1 (Becton Dickinson). Data were analyzed using FlowJo V.10.0.8 (Tree Star).
Ig PCR amplification, sequencing, cloning, and expression
Ig heavy and light chain genes were amplified from sorted single B cells using published protocols (Busse et al., 2014; Murugan et al., 2015). In brief, cDNA libraries from single B cells were prepared using random hexamers, and Ig heavy and light chain genes were amplified by barcoded gene specific primers using a high-throughput platform. Amplicons were sequenced using the 454 (Roche) or Illumina (Life Technologies) sequencing strategy, and the reads were analyzed using a previously described pipeline (Imkeller et al., 2016). Specific V and J primers tagged with restriction enzyme sites were used to amplify and clone Ig genes using published protocols (Tiller et al., 2008). Vectors with paired Ig heavy and light chain genes were co-transfected in HEK293T (ATCC, no. CRL-11268) or HEK293F (Thermo Fisher Scientific) cells for expression of mAbs.
ELISA
Protein G Sepharose 4 Fast Flow (GE Healthcare) was used for purification of mAbs, and concentrations were measured (Tiller et al., 2008). High-binding polystyrene ELISA plates were coated with the antigens: recombinantly expressed C-PfCSP, NANP-10, or streptavidin at 200 ng/well, 50 ng/well, or 50 ng/well, respectively, overnight at 4°C. Wells were blocked with 1% BSA in PBS (wt/vol). For peptide-ELISA, biotinylated peptides were coated at 50 ng/well. Binding of mAbs to the coated antigen was characterized by incubation with four serial dilutions (1 in 4 dilution) of each mAb starting with 4 µg/ml. Anti–human IgG-HRP (Jackson ImmunoResearch) used at 1:1,000 dilution in blocking buffer, and further absorbance measurement of the enzymatic reaction in One-step ABTS substrate (Roche), were used to detect the bound antibodies. mGO53 was used as an isotype control (Wardemann et al., 2003). Prism 6.07 (GraphPad) was used to calculate AUCs for the serially diluted mAbs.
1710 and 3919 Fab production
1710 and 3919 VL/K and VH regions were cloned into expression vectors upstream of human Igλ/Igκ and Igγ1 constant regions, respectively, as previously described (Tiller et al., 2008). Additionally, 3919 VL and VH regions were cloned into a pcDNA3.4 TOPO expression vector immediately upstream of human Igκ and Igγ1-CH1 domains, respectively. 1710 IgG, 3919 IgG, and 3919 Fab were transiently expressed in HEK293F cells and purified via protein A or KappaSelect affinity chromatography (GE Healthcare). 1710 Fab was generated by papain digestion and purified via an additional protein A chromatography step followed by cation exchange chromatography (MonoS, GE Healthcare). 1710 and 3919 Fab were further purified by size exclusion chromatography (Superdex 200 Increase 10/300 GL, GE Healthcare).
αTSR expression and purification
A construct based on the previously determined crystal structure (PDB ID: 3VDJ; Doud et al., 2012) of the Pf αTSR domain of PfCSP with NF54 genotype sequence was cloned into the pHLsec vector via AgeI and XhoI restriction sites. αTSR was transiently expressed in HEK293F cells and purified via a Histrap FF column (GE Healthcare), followed by size exclusion chromatography (Superdex 200 Increase 10/300 GL, GE Healthcare). Intact protein samples were subjected to electrospray ionization mass spectrometry to determine their molecular weight (The Scripps Research Institute).
PfCSP construct design for binding studies
Multiple constructs were used for binding studies. This includes FL PfCSP constructs from strains representative of Pf infection across three continents with a high prevalence of malaria: NF54 (UniProt accession no. P19597, residues 20–384, Africa), T4 (accession no. P13814, residues 20–411, Asia) and 7G8 (accession no. EUR80924, residues 20–278, South America). Potential N-linked glycosylation sites were mutated to glutamine. αTSR constructs were also generated for all three strains. The αTSR sequences of the T4 and 7G8 PfCSP strains were replaced with the NF54 αTSR sequence to generate T4-CSP-NF54-αTSR and 7G8-CSP-NF54-αTSR chimeric constructs. FL PfCSP constructs and C-terminal constructs were cloned in pHLsec for transient expression in HEK293F cells. All samples were purified via HisTrap FF (GE Healthcare) and size exclusion chromatography (Superdex 200 Increase 10/300 GL, GE Healthcare) before binding studies. An anti-His antibody (Santa Cruz Biotechnology) and an anti–mouse HRP secondary antibody (Abcam) were used in Western blot experiments according to the manufacturer’s protocols.
Biolayer interferometry binding studies
Biolayer interferometry (Octet RED96, FortéBio) experiments were conducted to determine the specificity and binding kinetics of 1710 and 3919 for multiple PfCSP constructs. CSPs diluted to 10 µg/ml in kinetics buffer (PBS, pH 7.4, 0.01% [wt/vol] BSA, 0.002% [vol/vol] Tween-20) were immobilized onto Ni-NTA (NTA) biosensors (FortéBio). After establishment of a stable baseline with loaded ligand in kinetics buffer, biosensors were dipped into wells containing twofold dilution series of 1710 or 3919 Fab or IgG. Tips were then dipped back into kinetics buffer to monitor the dissociation rate. Kinetics data were analyzed using FortéBio’s Data Analysis software 9.0, and curves were fitted to a 1:1 binding model. Mean kinetic constants reported are the results of three independent experiments. In competition binding studies, αTSR (NF54) was diluted to 10 µg/ml in kinetics buffer and immobilized onto Ni-NTA (NTA) biosensors (FortéBio). After a 30-s baseline step, biosensors were dipped into wells containing the first antibody (10 µg/ml) for 600 s, followed by another 30-s baseline, and then dipped into wells containing the second antibody for 300 s.
Crystallization and structure determination
Purified 1710 Fab and PfCSP αTSR were mixed in a 1:3 molar ratio, and excess αTSR was purified away by size exclusion chromatography (Superdex 200 Increase 10/300 GL, GE Healthcare). The 1710 Fab/αTSR complex was then concentrated to 25 mg/ml and mixed 1:1 with 200 mM MgCl2, 20% (wt/vol) PEG 3350. Crystals appeared after ∼30 d. 1710 Fab crystals were grown at 10 mg/ml and mixed 1:1 with 17% (wt/vol) PEG 400, 15% (vol/vol) glycerol, 8.5% (vol/vol) isopropanol, and 85 mM Hepes pH 7.5. The 3919 Fab/αTSR co-complex was generated similarly to the 1710 Fab/αTSR co-complex. Extensive crystallization trials were performed, without yielding crystals, likely because of a relatively weak affinity for the complex. 1710 Fab/αTSR co-complex crystals were cryoprotected in 15% (vol/vol) glycerol before being flash-frozen in liquid nitrogen. Data were collected at the 08ID-1 beamline at the Canadian Light Source, processed, and scaled using XDS (Kabsch, 2010). The structures were determined by molecular replacement using Phaser (McCoy et al., 2007). Refinement of the structures was performed using phenix.refine (Adams et al., 2010) and iterations of refinement using Coot (Emsley et al., 2010). The crystal structures have been deposited in the Protein Data Bank (PDB IDs 6B0S and 6B0W).
Sporozoite production
Pf NF54 sporozoites were isolated from salivary glands of infected Anopheles coluzzii mosquitoes (Ngousso strain). NF54 PfCSP transgenic P. berghei (Pb-PfCSP) sporozoites (Triller et al., 2017) were isolated from salivary glands of infected immuno-compromised Anopheles gambiae 7b mosquitoes (Pompon and Levashina, 2015).
Traversal assay
Traversal assays were performed as described (Triller et al., 2017). In brief, HC-04 cells of a human hepatocyte cell line (MRA-975, deposited by Jetsumon Sattabongkot) were seeded at 6 × 104 cells/well in a 96-well plate (Greiner) and allowed to grow at 37°C and 5% CO2 for 24 h to attain 70% confluency. 105 Pf salivary gland sporozoites were incubated with 100 µg/ml of mAb for 30 min and added to the cells along with dextran-rhodamine (Molecular Probes) at 0.5 mg/ml. The maximum traversal efficiency, as measured in the wells with non–antibody-treated sporozoites, was used for normalization. Background noise, as detected in the wells where the dextran-rhodamine was added without sporozoites and antibodies, was subtracted for a given plate. mGO53 and the chimeric version of 2A10 (with the mouse 2A10 variable region and human heavy and kappa constant regions) were used as negative and positive controls, respectively (Zavala et al., 1983; Wardemann et al., 2003; Triller et al., 2017). All controls were set up in each plate as duplicates. The plates were centrifuged at 3,000 rpm at room temperature for 10 min without brake and incubated at 37°C and 5% CO2 for 2 h. Cells were thrice washed with PBS, treated with trypsin (Sigma) for 10 min, resuspended in 10% (vol/vol) FCS in PBS, centrifuged at 1,400 g for 5 min, and resuspended in 150 µl of 1% (vol/vol) PFA (Alfa Alsar) in PBS. Sporozoite traversal activity as detected by dextran positivity was measured in a FACS LSR II instrument (Becton Dickinson). Data were analyzed using FlowJo V.10.0.8 (Tree Star).
Immunofluorescence assay
8-well microscopy slides (Medco) were pre-coated with 3% BSA in RPMI (wt/vol) for 15 min at 37°C. For the analysis of fixed Pf sporozoites, 4 × 104 salivary gland Pf NF54 sporozoites were added per well and air-dried overnight at room temperature. Fixation was performed with 4% PFA (Alfa Alsar) in PBS (vol/vol) for 10 min at room temperature and then blocked with 10% (vol/vol) FCS in PBS for 15 min at 37°C. Pf sporozoites were stained with mAbs (100 µg/ml) in 10% (vol/vol) FCS in PBS for 45 min at 37°C. mAbs were detected using a Cy3-conjugated goat anti–human IgG (1:1,000, Jackson ImmunoResearch). Slides were washed three times for 2 min with 1% (vol/vol) FCS in PBS after each staining step. For the analysis of live sporozoites, 2 × 105 Pb-PfCSP salivary gland sporozoites were incubated in siliconized tubes with 100 µg/ml of mAbs in a total volume of 100 µl PBS with 1% (vol/vol) FCS in PBS for 30 min on ice. Sporozoites were washed with 1.5 ml of 1% (vol/vol) FCS in PBS by centrifuging at 9,300 g for 4 min at 4°C. The sporozoite pellet was resuspended in 100 µl 1% (vol/vol) FCS in PBS containing goat anti–human IgG-Cy3 (1:1,000, Jackson ImmunoResearch) and incubated for 30 min on ice. The suspension was washed once again, and the sporozoite pellet was resuspended in 40 µl PBS. 20 µl sporozoite suspension was added per well and allowed to settle for 2.5 h at room temperature. Excess liquid was removed, and sporozoites were fixed with 20 µl of 4% (wt/vol) PFA in PBS per well for 15 min at room temperature. After fixation, the wells were washed twice. Vectashield Mounting Medium with DAPI (Vector Laboratories) was used to mount all microscopy slides and to visualize sporozoite nuclei. Images were acquired on an AxioObserver Z1 fluorescence microscope equipped with an Apotome module (Zeiss) using the 63× objective. Images were analyzed using the AxioVision ZEN 2012 software (Zeiss), ImageJ (Schneider et al., 2012), and Fiji (Schindelin et al., 2012).
Live sporozoite flow cytometry
105 Pb-PfCSP salivary gland sporozoites were incubated in siliconized tubes with 100 µg/ml or 1 µg/ml of mAbs in a total volume of 60 µl PBS with 1% (vol/vol) FCS for 30 min on ice. The sporozoites were washed with 1.5 ml of 1% (vol/vol) FCS in PBS by centrifuging at 9,300 g for 4 min at 4°C. The sporozoite pellet was resuspended in 100 µl of 1% (vol/vol) FCS in PBS containing goat anti–human IgG-Cy3 (1:1,000; Jackson ImmunoResearch) and incubated for 30 min on ice. The sporozoite suspension was washed once more, and the pellet was resuspended in 200 µl of 1% (vol/vol) FCS in PBS. Antibody-stained sporozoites were detected by intrinsic GFP expression and Cy3 staining in a FACS LSR II instrument (Becton Dickinson). Data were analyzed using FlowJo V.10.0.8 (Tree Star).
Pf liver stage developmental assay
Developmental assays were performed as described previously (Triller et al., 2017). In brief, 104 HC-04 cells/well were seeded into a 96-well plate with transparent bottom (Nalgene International) and pre-treated with rat collagen (BD Biosciences). 104 Pb-PfCSP salivary gland sporozoites were incubated with 100 µg/ml of mAb for 30 min on ice and then added in technical triplicates to the cells. Untreated sporozoites were used as positive infection control. The plate was centrifuged at 110 g for 1 min at room temperature and incubated at 37°C for 2 h. After incubation, the wells were washed thrice with 2% penicillin/streptomycin solution with 5 µg/ml fungizone to remove extracellular sporozoites and once without fungizone. After 2 d of incubation at 37°C, the cells were fixed with 4% (wt/vol) PFA for 15 min at room temperature and permeabilized with 50 µM NH4Cl2 (Sigma-Aldrich), 3% (vol/vol) FBS, and 0.3% (vol/vol) Triton (Sigma-Aldrich) for 1 h at 37°C. EEFs were detected using rabbit anti-GFP antibody (1:1,000, Abcam) diluted in permeabilization buffer for 1 h at 37°C, and anti–rabbit IgG antibody conjugated to Alexa Fluor 555 (1:1,000, Molecular Probes). DAPI (1.25 µg/ml, Molecular Probes) was used to stain cell nuclei. An AxioObserver Z1 fluorescence microscope equipped with an Apotome module (10× objective, Zeiss) and AxioVision ZEN 2012 software (Zeiss) was used to acquire images of each well. EEF development was determined in comparison to the untreated sporozoite control, which was set to 0% inhibition. mAb mGO53 was used as a negative control.
In vivo Plasmodium infections
In vivo infection experiments were performed as described previously (Triller et al., 2017) and were approved by LAGeSo (H0027/12). In brief, female C57BL/6 mice (5 per group) were passively immunized by i.p. injection with 400 µg of monoclonal human anti-PfCSP antibody 1710 or the positive and negative control antibodies 663 (Triller et al., 2017) and mGO53 (Wardemann et al., 2003), respectively, in 250 µl of PBS. 1 d later, mice were inoculated with 5 × 103 Pb-PfCSP sporozoites by s.c. injection at the tail base. Blood parasitemia was determined daily by Giemsa-stained thin blood smears starting on day 3 after the infection. At least 100 microscopic fields were counted. Mice were sacrificed 4 d after the detection of blood stage parasites.
Online supplemental material
Fig. S1 shows the characterization of C-PfCSP binding to C-PfCSP–reactive antibodies. Fig. S2 shows the structural and biophysical analysis of 1710 and the PfCSP αTSR. Fig. S3 shows Pb-PfCSP sporozoite binding and functional evaluation of C-PfCSP antibodies. Table S1 shows the data collection and refinement statistics for 1710 Fab-αTSR and unliganded 1710 Fab crystal structures.
Supplementary Material
Acknowledgments
We thank the vaccine trial participants for their contribution and commitment to vaccine research, C. Kreschel from the Vector Biology Unit, Max Planck Institute for Infection Biology, Berlin, for her support in Pf sporozoite production, as well as L. Spohr and M. Andres for mosquito rearing and infections. We thank the Sanaria and Protein Potential teams for the conception, manufacture, and shipping of investigational products, PfSPZ Challenge and diluents, regulatory, quality, and clinical site activities, and legal and administrative support. Pb-PfCSP parasites were provided by Drs. S.M. Khan and C.J. Janse (Leiden University Medical Center, Leiden, Netherlands). The following reagents were obtained from BEI Resources, National Institute of Allergy and Infectious Diseases, National Institutes of Health: hybridoma 2A10 anti-PfCSP, MRA-183, contributed by Elizabeth Nardin; and HC-04, hepatocyte (human), MRA-975, contributed by Jetsumon Sattabongkot Prachumsri. We appreciate the expert reviews of the Safety Monitoring Committee (W. Chen, P. Coyne, and P. Zanger). Research described in this paper was performed using beamline 08ID-1 at the Canadian Light Source, which is supported by the Canada Foundation for Innovation, Natural Sciences and Engineering Research Council of Canada, the University of Saskatchewan, the Government of Saskatchewan, Western Economic Diversification Canada, the National Research Council Canada, and the Canadian Institutes of Health Research.
R. Murugan was supported by the International Max Planck Research School for Infectious Diseases and Immunology (IMPRS-IDI), and G. Triller was supported by the IMPRS-IDI and the German National Academic Foundation. The clinical trial was funded by the German Federal Ministry of Education and Research (BMBF) through the German Center for Infection Research (DZIF). Manufacture of investigational product by Sanaria was supported in part by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under SBIR award nos. 5R44AI058375 and 5R44AI055229.
The authors declare no competing financial interests.
Author contributions: S.W. Scally, R. Murugan, B. Mordmüller, P.G. Kremsner, B.K.L. Sim, S.L. Hoffman, E.A. Levashina, H. Wardemann, and J.-P. Julien conceived and designed the research. S.W. Scally, R. Murugan, A. Bosch, G. Triller, and G. Costa performed the research. S.W. Scally, R. Murugan, A. Bosch, G. Triller, G. Costa, H. Wardemann, and J.-P. Julien analyzed data. S.W. Scally, R. Murugan, H. Wardemann, and J.-P. Julien wrote the paper.
References
- Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66:213–221. 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey J.A., Mvalo T., Aragam N., Weiser M., Congdon S., Kamwendo D., Martinson F., Hoffman I., Meshnick S.R., and Juliano J.J.. 2012. Use of massively parallel pyrosequencing to evaluate the diversity of and selection on Plasmodium falciparum csp T-cell epitopes in Lilongwe, Malawi. J. Infect. Dis. 206:580–587. 10.1093/infdis/jis329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonelo A., Valmori D., Triponez F., Tiercy J.M., Mentha G., Oberholzer J., Champagne P., Romero J.F., Esposito F., Nebié I., et al. 2000. Generation and characterization of malaria-specific human CD8(+) lymphocyte clones: effect of natural polymorphism on T cell recognition and endogenous cognate antigen presentation by liver cells. Eur. J. Immunol. 30:3079–3088. [DOI] [PubMed] [Google Scholar]
- Bongfen S.E., Ntsama P.M., Offner S., Smith T., Felger I., Tanner M., Alonso P., Nebie I., Romero J.F., Silvie O., et al. 2009. The N-terminal domain of Plasmodium falciparum circumsporozoite protein represents a target of protective immunity. Vaccine. 27:328–335. 10.1016/j.vaccine.2008.09.097 [DOI] [PubMed] [Google Scholar]
- Busse C.E., Czogiel I., Braun P., Arndt P.F., and Wardemann H.. 2014. Single-cell based high-throughput sequencing of full-length immunoglobulin heavy and light chain genes. Eur. J. Immunol. 44:597–603. 10.1002/eji.201343917 [DOI] [PubMed] [Google Scholar]
- Calle J.M., Nardin E.H., Clavijo P., Boudin C., Stüber D., Takacs B., Nussenzweig R.S., and Cochrane A.H.. 1992. Recognition of different domains of the Plasmodium falciparum CS protein by the sera of naturally infected individuals compared with those of sporozoite-immunized volunteers. J. Immunol. 149:2695–2701. [PubMed] [Google Scholar]
- Cerami C., Frevert U., Sinnis P., Takacs B., Clavijo P., Santos M.J., and Nussenzweig V.. 1992. The basolateral domain of the hepatocyte plasma membrane bears receptors for the circumsporozoite protein of Plasmodium falciparum sporozoites. Cell. 70:1021–1033. 10.1016/0092-8674(92)90251-7 [DOI] [PubMed] [Google Scholar]
- Charoenvit Y., Collins W.E., Jones T.R., Millet P., Yuan L., Campbell G.H., Beaudoin R.L., Broderson J.R., and Hoffman S.L.. 1991a Inability of malaria vaccine to induce antibodies to a protective epitope within its sequence. Science. 251:668–671. 10.1126/science.1704150 [DOI] [PubMed] [Google Scholar]
- Charoenvit Y., Mellouk S., Cole C., Bechara R., Leef M.F., Sedegah M., Yuan L.F., Robey F.A., Beaudoin R.L., and Hoffman S.L.. 1991b Monoclonal, but not polyclonal, antibodies protect against Plasmodium yoelii sporozoites. J. Immunol. 146:1020–1025. [PubMed] [Google Scholar]
- Coppi A., Natarajan R., Pradel G., Bennett B.L., James E.R., Roggero M.A., Corradin G., Persson C., Tewari R., and Sinnis P.. 2011. The malaria circumsporozoite protein has two functional domains, each with distinct roles as sporozoites journey from mosquito to mammalian host. J. Exp. Med. 208:341–356. 10.1084/jem.20101488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks G.E., Hon G., Chandonia J.M., and Brenner S.E.. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188–1190. 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dame J.B., Williams J.L., McCutchan T.F., Weber J.L., Wirtz R.A., Hockmeyer W.T., Maloy W.L., Haynes J.D., Schneider I., Roberts D., et al. 1984. Structure of the gene encoding the immunodominant surface antigen on the sporozoite of the human malaria parasite Plasmodium falciparum. Science. 225:593–599. 10.1126/science.6204383 [DOI] [PubMed] [Google Scholar]
- de Taeye S.W., Ozorowski G., Torrents de la Peña A., Guttman M., Julien J.P., van den Kerkhof T.L., Burger J.A., Pritchard L.K., Pugach P., Yasmeen A., et al. 2015. Immunogenicity of Stabilized HIV-1 Envelope Trimers with Reduced Exposure of Non-neutralizing Epitopes. Cell. 163:1702–1715. 10.1016/j.cell.2015.11.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doud M.B., Koksal A.C., Mi L.Z., Song G., Lu C., and Springer T.A.. 2012. Unexpected fold in the circumsporozoite protein target of malaria vaccines. Proc. Natl. Acad. Sci. USA. 109:7817–7822. 10.1073/pnas.1205737109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., and Cowtan K.. 2010. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66:486–501. 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher C.R., Sutton H.J., Kaczmarski J.A., McNamara H.A., Clifton B., Mitchell J., Cai Y., Dups J.N., D’Arcy N.J., Singh M., et al. 2017. T-dependent B cell responses to Plasmodium induce antibodies that form a high-avidity multivalent complex with the circumsporozoite protein. PLoS Pathog. 13:e1006469 10.1371/journal.ppat.1006469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foquet L., Hermsen C.C., van Gemert G.J., Van Braeckel E., Weening K.E., Sauerwein R., Meuleman P., and Leroux-Roels G.. 2014. Vaccine-induced monoclonal antibodies targeting circumsporozoite protein prevent Plasmodium falciparum infection. J. Clin. Invest. 124:140–144. 10.1172/JCI70349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frevert U., Sinnis P., Cerami C., Shreffler W., Takacs B., and Nussenzweig V.. 1993. Malaria circumsporozoite protein binds to heparan sulfate proteoglycans associated with the surface membrane of hepatocytes. J. Exp. Med. 177:1287–1298. 10.1084/jem.177.5.1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi K., Thera M.A., Coulibaly D., Traoré K., Guindo A.B., Doumbo O.K., Takala-Harrison S., and Plowe C.V.. 2012. Next generation sequencing to detect variation in the Plasmodium falciparum circumsporozoite protein. Am. J. Trop. Med. Hyg. 86:775–781. 10.4269/ajtmh.2012.11-0478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert S.C., Plebanski M., Gupta S., Morris J., Cox M., Aidoo M., Kwiatkowski D., Greenwood B.M., Whittle H.C., and Hill A.V.. 1998. Association of malaria parasite population structure, HLA, and immunological antagonism. Science. 279:1173–1177. 10.1126/science.279.5354.1173 [DOI] [PubMed] [Google Scholar]
- Godson G.N., Ellis J., Svec P., Schlesinger D.H., and Nussenzweig V.. 1983. Identification and chemical synthesis of a tandemly repeated immunogenic region of Plasmodium knowlesi circumsporozoite protein. Nature. 305:29–33. 10.1038/305029a0 [DOI] [PubMed] [Google Scholar]
- Imkeller K., Arndt P.F., Wardemann H., and Busse C.E.. 2016. sciReptor: analysis of single-cell level immunoglobulin repertoires. BMC Bioinformatics. 17:67 10.1186/s12859-016-0920-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Impagliazzo A., Milder F., Kuipers H., Wagner M.V., Zhu X., Hoffman R.M., van Meersbergen R., Huizingh J., Wanningen P., Verspuij J., et al. 2015. A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science. 349:1301–1306. 10.1126/science.aac7263 [DOI] [PubMed] [Google Scholar]
- Kabsch W. 2010. XDS. Acta Crystallogr. D Biol. Crystallogr. 66:125–132. 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappe S.H., Buscaglia C.A., and Nussenzweig V.. 2004. Plasmodium sporozoite molecular cell biology. Annu. Rev. Cell Dev. Biol. 20:29–59. 10.1146/annurev.cellbio.20.011603.150935 [DOI] [PubMed] [Google Scholar]
- Kazmin D., Nakaya H.I., Lee E.K., Johnson M.J., van der Most R., van den Berg R.A., Ballou W.R., Jongert E., Wille-Reece U., Ockenhouse C., et al. 2017. Systems analysis of protective immune responses to RTS,S malaria vaccination in humans. Proc. Natl. Acad. Sci. USA. 114:2425–2430. 10.1073/pnas.1621489114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E., and Henrick K.. 2007. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372:774–797. 10.1016/j.jmb.2007.05.022 [DOI] [PubMed] [Google Scholar]
- Lopaticki S., Yang A.S.P., John A., Scott N.E., Lingford J.P., O’Neill M.T., Erickson S.M., McKenzie N.C., Jennison C., Whitehead L.W., et al. 2017. Protein O-fucosylation in Plasmodium falciparum ensures efficient infection of mosquito and vertebrate hosts. Nat. Commun. 8:561 10.1038/s41467-017-00571-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez J.A., Roggero M.A., Duombo O., Gonzalez J.M., Tolle R., Koita O., Arevalo-Herrera M., Herrera S., and Corradin G.. 1996. Recognition of synthetic 104-mer and 102-mer peptides corresponding to N- and C-terminal nonrepeat regions of the Plasmodium falciparum circumsporozoite protein by sera from human donors. Am. J. Trop. Med. Hyg. 55:424–429. 10.4269/ajtmh.1996.55.424 [DOI] [PubMed] [Google Scholar]
- McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., and Read R.J.. 2007. Phaser crystallographic software. J. Appl. Cryst. 40:658–674. 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutchan T.F., Kissinger J.C., Touray M.G., Rogers M.J., Li J., Sullivan M., Braga E.M., Krettli A.U., and Miller L.H.. 1996. Comparison of circumsporozoite proteins from avian and mammalian malarias: biological and phylogenetic implications. Proc. Natl. Acad. Sci. USA. 93:11889–11894. 10.1073/pnas.93.21.11889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ménard R., Sultan A.A., Cortes C., Altszuler R., van Dijk M.R., Janse C.J., Waters A.P., Nussenzweig R.S., and Nussenzweig V.. 1997. Circumsporozoite protein is required for development of malaria sporozoites in mosquitoes. Nature. 385:336–340. 10.1038/385336a0 [DOI] [PubMed] [Google Scholar]
- Migliorini P., Betschart B., and Corradin G.. 1993. Malaria vaccine: immunization of mice with a synthetic T cell helper epitope alone leads to protective immunity. Eur. J. Immunol. 23:582–585. 10.1002/eji.1830230245 [DOI] [PubMed] [Google Scholar]
- Mishra S., Nussenzweig R.S., and Nussenzweig V.. 2012. Antibodies to Plasmodium circumsporozoite protein (CSP) inhibit sporozoite’s cell traversal activity. J. Immunol. Methods. 377:47–52. 10.1016/j.jim.2012.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mordmüller B., Surat G., Lagler H., Chakravarty S., Ishizuka A.S., Lalremruata A., Gmeiner M., Campo J.J., Esen M., Ruben A.J., et al. 2017. Sterile protection against human malaria by chemoattenuated PfSPZ vaccine. Nature. 542:445–449. 10.1038/nature21060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murugan R., Imkeller K., Busse C.E., and Wardemann H.. 2015. Direct high-throughput amplification and sequencing of immunoglobulin genes from single human B cells. Eur. J. Immunol. 45:2698–2700. 10.1002/eji.201545526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardin E., Gwadz R.W., and Nussenzweig R.S.. 1979. Characterization of sporozoite surface antigens by indirect immunofluorescence: detection of stage- and species-specific antimalarial antibodies. Bull. World Health Organ. 57(Suppl 1):211–217. [PMC free article] [PubMed] [Google Scholar]
- Neafsey D.E., Juraska M., Bedford T., Benkeser D., Valim C., Griggs A., Lievens M., Abdulla S., Adjei S., Agbenyega T., et al. 2015. Genetic Diversity and Protective Efficacy of the RTS,S/AS01 Malaria Vaccine. N. Engl. J. Med. 373:2025–2037. 10.1056/NEJMoa1505819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussenzweig V., and Nussenzweig R.S.. 1989. Circumsporozoite proteins of malaria parasites. Bull. Mem. Acad. R. Med. Belg. 144:493–504. [PubMed] [Google Scholar]
- Ockenhouse C.F., Regules J., Tosh D., Cowden J., Kathcart A., Cummings J., Paolino K., Moon J., Komisar J., Kamau E., et al. 2015. Ad35.CS.01-RTS,S/AS01 Heterologous Prime Boost Vaccine Efficacy against Sporozoite Challenge in Healthy Malaria-Naïve Adults. PLoS One. 10:e0131571 10.1371/journal.pone.0131571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pompon J., and Levashina E.A.. 2015. A New Role of the Mosquito Complement-like Cascade in Male Fertility in Anopheles gambiae. PLoS Biol. 13:e1002255 10.1371/journal.pbio.1002255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potocnjak P., Yoshida N., Nussenzweig R.S., and Nussenzweig V.. 1980. Monovalent fragments (Fab) of monoclonal antibodies to a sporozoite surface antigen (Pb44) protect mice against malarial infection. J. Exp. Med. 151:1504–1513. 10.1084/jem.151.6.1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potocnjak P., Zavala F., Nussenzweig R., and Nussenzweig V.. 1982. Inhibition of idiotype--anti-idiotype interaction for detection of a parasite antigen: a new immunoassay. Science. 215:1637–1639. 10.1126/science.6122269 [DOI] [PubMed] [Google Scholar]
- Richie T.L., Billingsley P.F., Sim B.K., James E.R., Chakravarty S., Epstein J.E., Lyke K.E., Mordmüller B., Alonso P., Duffy P.E., et al. 2015. Progress with Plasmodium falciparum sporozoite (PfSPZ)-based malaria vaccines. Vaccine. 33:7452–7461. 10.1016/j.vaccine.2015.09.096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero P., Maryanski J.L., Corradin G., Nussenzweig R.S., Nussenzweig V., and Zavala F.. 1989. Cloned cytotoxic T cells recognize an epitope in the circumsporozoite protein and protect against malaria. Nature. 341:323–326. 10.1038/341323a0 [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., and Eliceiri K.W.. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 9:671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield L., Villaquiran J., Ferreira A., Schellekens H., Nussenzweig R., and Nussenzweig V.. 1987. Gamma interferon, CD8+ T cells and antibodies required for immunity to malaria sporozoites. Nature. 330:664–666. 10.1038/330664a0 [DOI] [PubMed] [Google Scholar]
- Seder R.A., Chang L.J., Enama M.E., Zephir K.L., Sarwar U.N., Gordon I.J., Holman L.A., James E.R., Billingsley P.F., Gunasekera A., et al. VRC 312 Study Team . 2013. Protection against malaria by intravenous immunization with a nonreplicating sporozoite vaccine. Science. 341:1359–1365. 10.1126/science.1241800 [DOI] [PubMed] [Google Scholar]
- Sidjanski S.P., Vanderberg J.P., and Sinnis P.. 1997. Anopheles stephensi salivary glands bear receptors for region I of the circumsporozoite protein of Plasmodium falciparum. Mol. Biochem. Parasitol. 90:33–41. 10.1016/S0166-6851(97)00124-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sissoko M.S., Healy S.A., Katile A., Omaswa F., Zaidi I., Gabriel E.E., Kamate B., Samake Y., Guindo M.A., Dolo A., et al. 2017. Safety and efficacy of PfSPZ Vaccine against Plasmodium falciparum via direct venous inoculation in healthy malaria-exposed adults in Mali: a randomised, double-blind phase 1 trial. Lancet Infect. Dis. 17:498–509. 10.1016/S1473-3099(17)30104-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stüber D., Bannwarth W., Pink J.R., Meloen R.H., and Matile H.. 1990. New B cell epitopes in the Plasmodium falciparum malaria circumsporozoite protein. Eur. J. Immunol. 20:819–824. 10.1002/eji.1830200416 [DOI] [PubMed] [Google Scholar]
- Sumitani M., Kasashima K., Yamamoto D.S., Yagi K., Yuda M., Matsuoka H., and Yoshida S.. 2013. Reduction of malaria transmission by transgenic mosquitoes expressing an antisporozoite antibody in their salivary glands. Insect Mol. Biol. 22:41–51. 10.1111/j.1365-2583.2012.01168.x [DOI] [PubMed] [Google Scholar]
- Swearingen K.E., Lindner S.E., Shi L., Shears M.J., Harupa A., Hopp C.S., Vaughan A.M., Springer T.A., Moritz R.L., Kappe S.H., and Sinnis P.. 2016. Interrogating the Plasmodium Sporozoite Surface: Identification of Surface-Exposed Proteins and Demonstration of Glycosylation on CSP and TRAP by Mass Spectrometry-Based Proteomics. PLoS Pathog. 12:e1005606 10.1371/journal.ppat.1005606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller T., Meffre E., Yurasov S., Tsuiji M., Nussenzweig M.C., and Wardemann H.. 2008. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J. Immunol. Methods. 329:112–124. 10.1016/j.jim.2007.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triller G., Scally S.W., Costa G., Pissarev M., Kreschel C., Bosch A., Marois E., Sack B.K., Salman A.M., Janse C.J., et al. 2017. Protective human anti-malarial antibodies induced by natural exposure. Immunity. 10.1016/j.immuni.2017.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji M., Romero P., Nussenzweig R.S., and Zavala F.. 1990. CD4+ cytolytic T cell clone confers protection against murine malaria. J. Exp. Med. 172:1353–1357. 10.1084/jem.172.5.1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udhayakumar V., Ongecha J.M., Shi Y.P., Aidoo M., Orago A.S., Oloo A.J., Hawley W.A., Nahlen B.L., Hoffman S.L., Weiss W.R., and Lal A.A.. 1997. Cytotoxic T cell reactivity and HLA-B35 binding of the variant Plasmodium falciparum circumsporozoite protein CD8+ CTL epitope in naturally exposed Kenyan adults. Eur. J. Immunol. 27:1952–1957. 10.1002/eji.1830270819 [DOI] [PubMed] [Google Scholar]
- Wang Q., Fujioka H., and Nussenzweig V.. 2005. Mutational analysis of the GPI-anchor addition sequence from the circumsporozoite protein of Plasmodium. Cell. Microbiol. 7:1616–1626. 10.1111/j.1462-5822.2005.00579.x [DOI] [PubMed] [Google Scholar]
- Wardemann H., Yurasov S., Schaefer A., Young J.W., Meffre E., and Nussenzweig M.C.. 2003. Predominant autoantibody production by early human B cell precursors. Science. 301:1374–1377. 10.1126/science.1086907 [DOI] [PubMed] [Google Scholar]
- Weedall G.D., Preston B.M., Thomas A.W., Sutherland C.J., and Conway D.J.. 2007. Differential evidence of natural selection on two leading sporozoite stage malaria vaccine candidate antigens. Int. J. Parasitol. 37:77–85. 10.1016/j.ijpara.2006.09.001 [DOI] [PubMed] [Google Scholar]
- Weiss W.R., Sedegah M., Beaudoin R.L., Miller L.H., and Good M.F.. 1988. CD8+ T cells (cytotoxic/suppressors) are required for protection in mice immunized with malaria sporozoites. Proc. Natl. Acad. Sci. USA. 85:573–576. 10.1073/pnas.85.2.573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida N., Nussenzweig R.S., Potocnjak P., Nussenzweig V., and Aikawa M.. 1980. Hybridoma produces protective antibodies directed against the sporozoite stage of malaria parasite. Science. 207:71–73. 10.1126/science.6985745 [DOI] [PubMed] [Google Scholar]
- Zavala F., Cochrane A.H., Nardin E.H., Nussenzweig R.S., and Nussenzweig V.. 1983. Circumsporozoite proteins of malaria parasites contain a single immunodominant region with two or more identical epitopes. J. Exp. Med. 157:1947–1957. 10.1084/jem.157.6.1947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavala F., Tam J.P., Hollingdale M.R., Cochrane A.H., Quakyi I., Nussenzweig R.S., and Nussenzweig V.. 1985. Rationale for development of a synthetic vaccine against Plasmodium falciparum malaria. Science. 228:1436–1440. 10.1126/science.2409595 [DOI] [PubMed] [Google Scholar]
- Zhao J., Bhanot P., Hu J., and Wang Q.. 2016. A Comprehensive Analysis of Plasmodium Circumsporozoite Protein Binding to Hepatocytes. PLoS One. 11:e0161607 10.1371/journal.pone.0161607 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.