Abstract
Background:
During lead identification and optimization, the advancement criteria may be driven based on scientific principles, prior experiences, and/or by examining the path paved by approved drugs. However, accessing the discovery data on physicochemical and ADME properties of the approved kinase inhibitors is a monumental task as these are either scattered in the literature or have not been published.
Objective:
Our goals were: 1) To compile the relevant data on all kinase inhibitors approved prior to 2016 for easy access by the biopharmaceutical community, 2) To provide a retrospective analysis to highlight trends and attributes which may have contributed to the “developability” of these drugs, and 3) To ignite focused debates on what constitutes “actionable”, “nice-to-have”, and unnecessary data. Such debates bring about more clarity on stage appropriateness of different types of information and prevent confusion due to abundance of unnecessary data, leading to more efficient and less costly drug discovery programs.
Methods:
A careful and thorough analysis of different bodies of data such as published manuscripts, and available regulatory documents were employed.
Results:
We were able to assemble a large body of data on the first thirty kinase inhibitors approved by US FDA since 2001.
Conclusion:
In conclusion, we have compiled physicochemical and ADME data on the first 30 approved kinase inhibitors and provided our retrospective analysis, which we hope is helpful in constructing advancement criteria in discovery programs. The examination of this data provides an opportunity to develop an opinion on data prioritization and stage appropriateness of assays.
Keywords: Kinase, inhibitor, drug, ADME, physicochemical, discovery, approved, FDA, druggability
1. INTRODUCTION
Protein kinases play pivotal roles in regulating all aspects of cellular function, including differentiation, metabolism, survival, programmed cell death and signal transduction [1]. Protein kinase pioneering research can be traced back to the 1950s [2]. Burnett and Kennedy first characterized protein kinase activities by using isolated rat mitochondria, in 1954 [3]. In 1955, Fischer and Krebs, Sutherland and Wosilai first discovered and characterized a specific type of protein kinase, phosphorylase kinase catalyzing ATP-phosporylation reaction [4-6]. The role of c-AMP, as the second messenger of hormonal signaling, in leading to the activation of phosphorylase was first reported by Sutherland’s group in 1958 [7, 8]. Their research findings led to the discovery of protein kinase A (c-AMP dependent protein kinase), a protein-serine/threonine kinase in late 1960s [9]. As kinase research further advanced, emerging pharmacological research interests were focused on exploiting kinase malfunctions at the genetic level as well as connections between dysregulation of kinase pathways and various types of diseases. In 1960, an abnormal minute chromosome, called Philadelphia Chromosome, was discovered by Peter Nowell and David Hungerford [10, 11]. The Philadelphia Chromosome was the result of reciprocal translocation of chromosomes 9 and 22, generating an elongated chromosome 9 and a truncated chromosome 22. The translocation juxtaposes the Abl1 gene on chromosome 9 to a part of the BCR on chromosome 22 and leads to CML [10, 12]. The translocated Abl1 gene, which encodes a tyrosine kinase, causes deregulated and continual overexpression of kinase activity resulting in tumor development. From this landmark discovery, it became evident that many human malignant diseases were associated with mutations, chromosomal rearrangements and/or overexpression of protein kinases [11, 13-16]. This discovery quickly led to protein kinases becoming well accepted targets for anticancer drug development [17-24]. During late 1980s and early 1990s, tremendous efforts were made to unfold the intracellular signal transduction pathways and aberrations of signaling pathways leading to variety of diseases at the genetic and molecular levels [25-30]. Many extra- and intra-cellularly associated kinases, such as MAPK, ERK, JAK and PI3K, were reported to regulate normal cellular functions [31, 32]. So far, a total of 518 human kinases and 900 human genes encoding for kinase proteins have been revealed [33]. In the meantime, it has been discovered that deregulation and/or over-expression of certain types of kinases lead to changes in the normal cellular functions which further advance to disease states. As evident by a number of marketed drugs and a substantial number of publications, kinase inhibitors can be used for the treatment of various types of cancers and inflammatory diseases (Table 1).
Table 1.
Kinase-targeted inhibitors approved by the FDA from January 2001 to October 2015.
| No. | Drug | Trade Name | Company |
Approval
Date |
Targeted Kinases | Therapeutic Area | Human Efficacious Dose (mg/day) |
|---|---|---|---|---|---|---|---|
| 1 | Imatinib | Gleevec | Novartis | 2001 | Bcr-Abl tyrosine kinase, PDGRR | Chronic Myeloid Leurkemia (CML) | 400-800, QD |
| 2 | Gefitinib | Iressa | AstraZeneca | 2003 | Selective EGFR tyrosine kinase (EGFR-TK1) | Non-small-cell lung cancer (NSCLC) | 250, QD |
| 3 | Erlotinib | Tarceva | OSI | 2004 | EGFR tyrosine kinase (EGFR-TK1) | Non-small-cell-lung cancer (NSCLC) | 150, QD |
| 4 | Sorafenib | Nexavar | Bayer | 2005 | C-RAF, B-RAF, c-KIT, FLT3, VEGFR2, -3, PDGFR-β | Advanced renal cell carcinoma (RCC) and unresectable haptocellular cancinoma (HCC) | 400, BID – 800, QD |
| 5 | Dasatinib | Sprycel | BMS | 2006 | Bcr-Abl, SCR-family kinases, PDGRFβ, c-KIT, ephrin (EPH) receptor kinases | Chronic myeloid leukemia (CML) and Philadelphia Chromosome Positive (Ph+) | 140, BID |
| 6 | Sunitinib | Sutent | Sugen (Pfizer) | 2006 | PDGFRα, -β, VEGFR1, -2, -3, c-KIT, RET, CSF-1R, FLT3 | Gastrointestinal stromal tumor (GIST) and advanced renal cell carcinoma (RCC) | 50, QD |
| 7 | Lapatinib | Tykerb | GSK | 2007 | Erbb2 (HER2) and Erb1 (EGFR) tyrosine kinases | Advanced or metastatic breast cancer | 1250, QD |
| 8 | Nilotinib | Tasigna | Novartis | 2007 | Bcr-Abl, c-KIT, PDGFRα, -β | Chronic Myeloid Leukemia (CML) and Philadelphia Chromosome positive (Ph+) | 600 – 800, QD |
| 9 | Pirfenidone | Glaspira | Marnac | 2008 | Antifibrotic P38 MAP kinase and TGFβ and TNF α synthesis | Idiopathic pulmonary fibrosis (IPF) | 2403, TID |
| 10 | Pazopanib | Votrient | GSK | 2009 | VEGF receptor tyrosine kinase, PDGFR/c-Kit | Advanced cell carcinoma | 200 – 400, QD |
| 11 | Crizotinib | Xalkori | Pfizer | 2011 | Anaplastic lymoma kinase receptor; Ros1 tyrosine kinase receptor | Non-small-cell lung cancer; anaplastic large cell lymphoma | 250, BID |
| 12 | Vemurafenib | Zelboraf | Plexxikon (Daiichi Sankyo) | 2011 | Raf B protein kinase | Thyroid tumor; melanoma | 960, BID |
| 13 | Vandetanib | Zactima | AstraZeneca | 2011 | EGFR family tyrosine kinase receptor, VEGF-2-receptor | Thyroid tumor; Small-cell lung cancer; Multiple myeloma | 300, QD |
| 14 | Ruxolitinib | Jakafi | Incyte | 2011 | Jak1and 2 tyrosine kinases | Multiple myeloma; inflammatory disease | 15 25, BID |
| 15 | Axitinib | INLYTA | Pfizer | 2012 | Selective VEGFR tyrosine kinase, PDGFR, c-Kit | Advanced Renal Cell Carcinoma | 5-10, BID |
| 16 | Bosutinib | Bosulif | Pfizer | 2012 | Tyrosince kinase, specifically an inhibitor of Bcr-Abl and Src-family kinases | Chronic, accelerated or blast phase Ph+ chronic Myelogonous Leukemia | 500, QD |
| 17 | Regorafenib | Stivarga | Bayer | 2012 | EVEGFR, EGF, KIT, RET, RAF, KIT, RET, RAF1, BRAF and BRAFV600E and FGFR1 | Metastatic colorectal cancer | 160, QD |
| No. | Drug | Trade Name | Company |
Approval Date |
Targeted Kinases | Therapeutic Area | Human Efficacious Dose (mg/day) |
| 18 | Tofacitinib | Xeljanz | Pfizer | 2012 | JAK1/JAK3 kinases | Rheumatoid Arthritis (RA) | 5-10, BID |
| 19 | Cabozantinib | Cometriq | Exelixis | 2012 | Multi-targeted inhibitor of receptor tyrosine kinases (RTKs) | Thyroid cancer (MTC) | 140, QD |
| 20 | Ponatinib | Iclusig | Ariad | 2012 | Tyrosine kinase (BCR-ABL), FLT3 | Chronic phase, accelerated phase or blast phase chronic myeloid leukemia (CML) or Ph+ALL acute lymphoblastic leukemia | 45, QD |
| 21 | Trametinib | Mekinist | GSK | 2013 | Mitogen-activated extracellular signal regulated kinase ½ (MEK1 and MEK2) | Metastatic Melanoma | 2, QD |
| 22 | Dabrafenib | Tafinlar | GSK | 2013 | B-raf kinase (BRAF) | Metastatic Melanoma | 150, BID |
| 23 | Afatinib | Gilotrif | Boehringer Ingelheim | 2013 | EGFR, HER2 and HER4 tyrosine kinases | Non-Small Cell Lung Cancer | 40, QD |
| 24 | Ibrutinib | Imbruvica | Janssen | 2013 | Bruton’s tyrosine kinase (BTK) | Non-Hodgkin’s Lymphoma Leukemia, Chronic Lymphatic Leukemia | 560, QD |
| 25 | Ceritinib | Zykadia | Novartis | 2014 | Anaplastic Lymphoma Kinase (ALK) | Non-Small Cell Lung Cancer | 750, QD |
| 26 | Idelalisib | Zydelig | Gilead Sciences | 2014 | Phosphoinositide 3-kinase (PI3Kδ) | Refractory indolent non-Hodginkin’s Lymphoma, Relapsed Chronic Lymphocytic Leukymia, Follicutar Lymphoma | 150, BID |
| 27 | Nintedanib | Ofev | Boehringer Ingelheim | 2014 | Tyrosine kinase, Flt-3, Lck, Lyn and Src kinases | Indiopathic Pulmonary Fibrosis | 100/150, BID |
| 28 | Palbociclib | Ibrance | Pfizer | 2015 | Cyclin-dependent kinases 4 and 6 (CDK4 and CDK6) | Breast Cancer | 125, QD |
| 29 | Lenvatinib | Lenvima | Eisai | 2015 | Multiple receptor tyrosince kinase (RTK) | Thyroid Cancer | 24, QD |
| 30 | Cobimetinib | Cotellic | Genentech | 2015 | MEK tyrosine/serine/threonine protein kinase | Metastatic Melanoma | 60, QD |
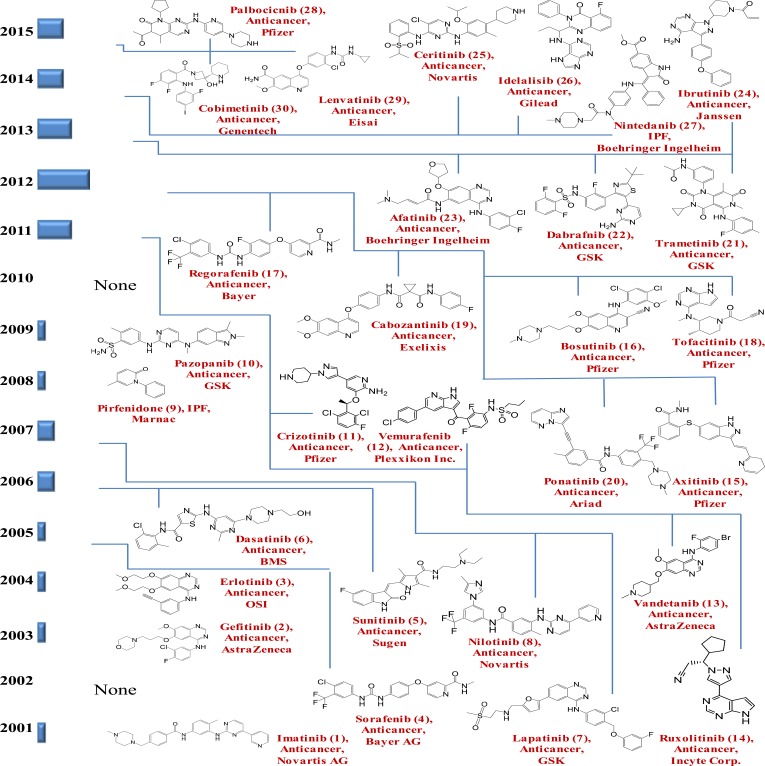
Imatinib was the first kinase inhibitor approved to treat chronic myelogenous leukemia [34, 35]. It was approved in 2001, by the FDA and was marketed by Novartis as Gleevec (USA). Imatinib competes with ATP for the ATP binding site of TK (selective inhibitor of the BCR-Abl TK domain, also inhibits proto-oncogene c-kit and PDGF-R) and has since become the model drug for protein kinase-targeted drug discovery and development [36, 37]. As the understanding of the role of protein kinases in signaling pathways continues to grow, the uses of protein kinase inhibitors are also expanding to include the treatment of inflammation, diabetes, infectious and cardiovascular disease [38]. At present, approximately 150 kinase-targeted compounds are in clinical development and many more are in various stages of preclinical discovery and development [39]. The FDA has approved a total of 30 kinase-targeted drugs between the years 2001 and October 2015 (Table 1 and Fig. 1). Among the 30 approved kinase inhibitors, 27 are used for the treatment of malignancies and 3, pirfenidone (p38 mitogen-activated protein kinase inhibitor), tofacitinib (selective JAK1/JAK3 kinase inhibitor) and nintedanib (multi-kinase inhibitor), are used for the treatment of inflammatory diseases. FDA approval rate for kinase inhibitors was approximately one per year from 2001 to 2010. This increased significantly to approximately five per year from 2011 to 2015. The current trend in kinase-targeted drug development suggests that 1) more efforts will be spent in further exploration of the kinase inhibitor space, 2) the therapeutic indications will expand to cover a broad spectrum of diseases ranging from cancer and inflammation to metabolic, immune, cardiac and CNS disorders, 3) the therapeutic use of combinations of kinase inhibitors and other targeted agents will become the focus for disease treatment and management, 4) there is an urgent need to explore new pharmacophores to diversify the currently available chemical scaffolds for kinase inhibition, 5) multi-kinase inhibitors and single kinase inhibitors with polypharmacology will be needed to produce more efficacious treatment regimens, and 6) major challenges reside in the development of kinase inhibitors with better selectivity, more specificity to driver mutations in diseases, better control of resistance development and less side effects [38-41].
Fig. (1).
Thirty FDA approved kinase inhibitors from January 2001 to October 2015.
In today’s fast-paced and competitive environment, there is an ever-increasing need for more effective drug design by fully balancing potency, selectivity, molecular properties and appropriate ADME properties early in drug discovery. However, incorporating drug-like properties at the onset is often a challenging task. Understanding the molecular and ADME properties of successful drugs provides helpful insight in defining parameters to facilitate this process. In 2013, we published our detailed analysis on molecular and ADME properties of the first 14 kinase inhibitors approved by FDA between 2001-2011 [42]. Herein, we add the next 16 approved kinase inhibitors and summarize our systematic analysis of the common physicochemical and ADME properties of all 30 marketed kinase inhibitors. Although the dataset is still limited, we believe this retrospective analysis provides a valuable insight into what attributes are more crucial to the success of kinase inhibitor discovery and development programs. Furthermore, this work allows us to determine whether the new generation of kinase inhibitors is significantly different than the first set.
2. Retrospective analysis of available data on thirty marketed kinase inhibitors
2.1. Structural and Physicochemical Properties
2.1.1. MW, Log P, H-bond Acceptors/Donors/ Rotations, and PSA
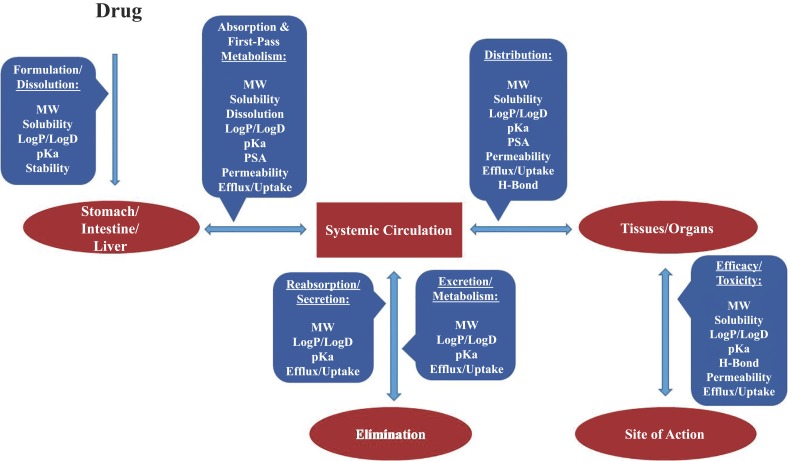
Physicochemical properties considered for lead optimization include hydrogen bonding, lipophilicity, molecular weight, pKa, PSA, shape and reactivity. Fig. (2) depicts the relationship and interplay of physicochemical/biochemical properties and in vivo drug pharmacokinetic and dynamic processes. One way to isolate the more impactful physicochemical factors in drug discovery is to examine the marketed drugs and their attributes.
Fig. (2).
Relationship and interplay of physicochemical properties and in vivo drug kinetic/dynamic processes following oral administration.
Christopher Lipinski et al. published the analysis of 2245 marketed drugs and drug candidates in clinical trials and their guidelines in 1997 [43]. This guideline, commonly referred to as the Lipinski “Rule of Five” (RO5), states that ideally an orally active drug has a MW<500 daltons, Log P≤5, H-bond donors ≤5, and H-bond acceptors≤10. Lipinski RO5 was used to enable the selection of compounds more likely to become orally bioavailable drugs based on early discovery data. Daniel Veber et al. examined over 1100 drug candidates at SmithKline Beecham Pharmaceuticals (now GlaxoSmithKline) and analyzed the relationship between physicochemical properties and rat bioavailabilities [44]. Veber found that compounds with total hydrogen bonds ≤ 12, rotatable bonds ≤ 10 and PSA ≤ 140 tend to have oral bioavailability ≥ 20% in rats. This analysis gave rise to Veber’s rules, which complement Lipinski’s RO5 and enhance drug discovery efficiency. Verber’s analysis was based only on compounds with rat bioavailability data. Mark Wenlock [45] reported on the limitation inherent in the compound collections used to give rise to Lipinski’s and Verber’s rules and suggested that following these rules was likely to lead to lead-like molecules rather than drug-like molecules. Furthermore, they concluded that the mean MW and lipophilicity of orally administered compounds decreased with NCE’s that progressed further down discovery/development path and eventually converge towards the mean values of marketed drugs.
We performed a trend analysis on the physicochemical properties of 30 FDA approved kinase inhibitors and identified the commonality of their structural properties. For the 30 FDA approved kinase inhibitors, structural properties were either measured or predicted using ACD software (Advanced Chemistry Development, Inc., Toronto, Ontario, Canada) and GastroPlus
ADMET Predictor software (Simulation Plus Inc., Lancaster, CA).
2.1.1.1. Analysis
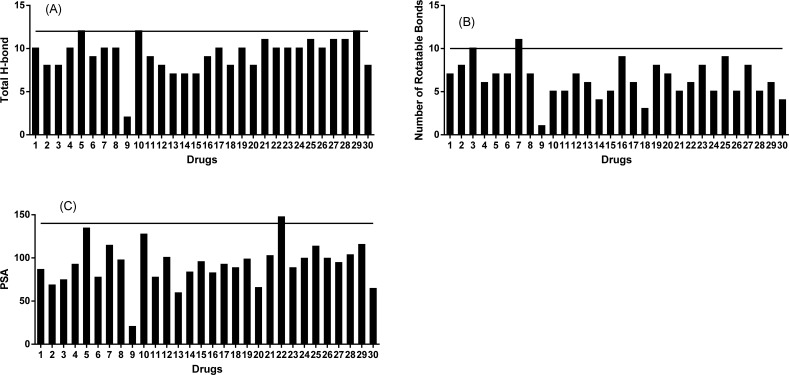
Of the 30 kinase inhibitors ~30% violated Lipinski’s RO5 with molecular weights slightly over 500 daltons (Fig. 3A). The measured lipophilicity (LogP values) was not available for eight of the drugs. For the remaining ones, ~20% violated RO5 with LogP>5 (Fig. 3B). The same level of violation was observed when using predicted LogP for all 30 drugs (Fig. 3C). Overall, ~80% of the drugs had LogP values between 1-5. While all thirty followed RO5 by having ≤5 H-bond donors, ~97% actually had ≤3 H-bond donors (Fig. 3D). While all 30 followed RO5 and had ≤ 10 H-bond acceptors, ~85% actually had ≤8 H-bond acceptors (Fig. 3E).
Fig. (3).
Structural properties of the 30 approved kinase inhibitors follow Lipinski’s Rule of Five. Each kinase inhibitor is shown on the X-axis with the numbering system used in table 1. (A) 30 out of 30 had molecular weight (MW) <600 Daltons while 20 out 30 had MW ≤ 500 Daltons; (B) 19 out of 22 kinase inhibitors had measured lipophilicity LogP values ≤ 5.0; (C) 17 out of 22 kinase inhibitors had measured lipophilicity LogP values ≤ 5.0; (D) 29 out 29 had total number of hydrogen donor ≤ 5 while 28 out of 29 had ≤ 3.0 ; (E) all 30 kinase inhibitors had a total number of hydrogen acceptor ≤ 10. * symbolize the kinase inhibitors without reported values.
As shown in (Fig. 4), all of the drugs followed Veber’s rule by having ≤ 12 total hydrogen bonds. With one exception, all drugs had ≤ 10 rotatable bonds. With regards to PSA, 29 drugs followed Veber’s rule and had PSA ≤ 140, however, ~73% of them actually had PSA ≤100 (Fig. 4C).
Fig. (4).
Structural properties of 30 approved kinase inhibitors follow Veber’s Rule. Each kinase inhibitor is shown on the X-axis with the numbering system used in table 1. (A) all kinase inhibitors had a total H-bond ≤12 while 23 out of 30 ≤ 10; (B) 30 out 30 had a total number of rotatable bonds ≤ 12 while 29 out of 30 ≤ 10; (C) Twenty-nine kinase inhibitors had polar surface area (PSA) values ≤140 while 22 out of 30 ≤100.
2.1.2. pKa
pKa is an important parameter, which has not received adequate attention from Lipinski and Weber. pKa is a measurement of the ionization potential of a compound and depends on ionizability of different functional groups within a molecule. Based on the pKa values on the strongest ionizable groups, NCEs can be classified into acidic, neutral and basic compounds. In vivo, whether NCEs remain in their neutral state or ionized form is influenced by the physiological pH in a given environment. For a given compound, solubility consists of both intrinsic solubility in the neutral state and apparent solubility of its ionized form at the local physiological pH. Oral absorption is affected by dissolution rate, apparent solubility in gastrointestinal tract, and permeability and stability throughout different regions of GI tract. Because pKa determines the ionization state of an NCE, it is a critical factor affecting the rate and extent of oral absorption from the GI tract therefore influencing its bioavailability. A highly-ionized molecule tends to have higher solubility and dissolution, but because ions cannot easily pass through membrane lipid bilayer, that same trait might compromise the ability of that molecule to pass through gut cells and into circulation; resulting in low bioavailability.
Tissue distribution of basic molecules has been in part related to their pKa values. Since the mid-1970s, many researchers have investigated the in vivo distribution profiles of basic molecules, including their whole body and intracellular distributions, and their interactions with factors leading to pharmacology and toxicity [46-50]. Their findings suggest that strongly basic amines tend to be sequestered in acidic organelles such
as lysosomes and bind to tissue membrane phospholipids. These result in high volumes of distribution, highly localized intracellular concentrations, and possible alterations of PK behavior and potential toxicity [46, 47, 50]. In addition, molecules with strong basicity tend to dissolve in gastric fluid but precipitate under intestinal environment with more basic pH; thereby resulting in low absorption owing to low solubility and slow dissolution rates at the site of absorption.
Together with LogP, plasma protein binding and blood-to-plasma partition ratio, the pKa value can be a predictor of intracellular distribution profiles of NCEs. These factors directly affect absorption, distribution, metabolism, elimination and toxicity (ADMET) properties and, in turn, systemic and target tissue exposures (Fig. 2) [46-51].
2.1.2.1. Analysis
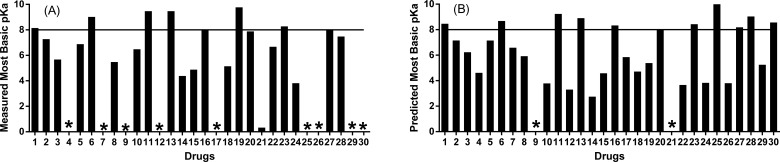
A wide range of weak to strong basicity was observed for the approved kinase inhibitors. We summarized the measured and predicted pKa values for the 30 kinase inhibitors (Fig. 5). Measured pKa values were not available in 30% of compounds. GastroPlus software was used to predict the most basic pKa values for all 30 drugs. The most basic pKa of pirfenidone and trametinib could not be determined because they lacked basic nitrogens. The predicted pKa’s were in good agreement with the measured values when available. Examination of the predicted values revealed that ~64% of these drugs had a pKa ≤8, ~29% had pKa values within 8-9, and ~ 7% with pKa values >9 (Fig. 5).
Fig. (5).
Structural properties of 30 approved kinase inhibitors (predicted using GastroPlus software and measured most basic pKa values). Each kinase inhibitor is shown on the X-axis with the numbering system used in table 1. (A) 15 out 21 kinase inhibitors had measured most basic pKa values ≤ 8.0 while 18 out of 21 ≤ 9.0 (B) 18 out of 28 kinase inhibitors had predicted most basic pKa values ≤ 8.0 while 26 out of 28 ≤ 9.0. * symbolize the inhibitors without reported values
2.1.3. Solubility, Permeability, and Efflux
Following oral administration, successful delivery of a drug to the site of action requires adequate absorption from the GI tract into the systemic blood circulation. Solubility, permeability, and efflux potential are the primary properties which impact the rate and the extent of oral absorption. These parameters have been used to categorize NCE’s according to BCS guidelines provided by US FDA [52]. Chemical and metabolic stability are also important factors which are beyond the scope of this review.
In brief, BCS classification is based primarily on solubility and permeability. Drugs can be grouped into four categories. Class I and II refer to high permeability drugs with high and low solubility, respectively. Class III and IV refer to low permeability drugs with high and low solubility.
2.1.3.1. Analysis
Twenty-nine drugs had reported values for in vitro Caco-2 permeability (Papp A-B) and solubility (Table 2). For these, BCS characterization data revealed that ~10% fell into BCS Class I and exhibited high solubility and high permeability, ~52% fell into BCS Class II and exhibited low solubility but high permeability, ~7% fell into BCS Class III with high solubility but low permeability, while ~27% had low solubility and low to moderate permeability (BCS Class IV). Only one drug, lenvatinib, had inclusive classification and could fall into either Class II or IV category [53-89, 109, 127].
Table 2.
Measured permeability and solubility of approved kinase-targeted inhibitors.
| No. | Drug | Papp A-Ba (x 10-6 cm/sec) | Solubilityb (mg/mL at pH) | BCS Class | References |
|---|---|---|---|---|---|
| 1 | Imatinib | 0.95 at 1µM, 7.9 at 50 µM | Very soluble | I | [53] |
| 2 | Gefitinib | 15.5 at 3 µM | 1 at pH 1.2 - 4.0, 0.43-0.01 at pH 5.0 - 6.0, < 0.01 at pH 6.8-8.0 | II | [54] |
| 3 | Erlotinib | 34 at 10 µM | 0.4 at pH 2.0, pH dependent solubility | II | [55] |
| 4 | Sorafenib | 16.4 at 0.1 µM, 33.5 at 1 µM | Low solubility | II | [56, 127] |
| 5 | Dasatinib | 10.2 at 50 µM | ~18 at pH 2.6, <0.001 at pH 7.0 | II | [57] |
| 6 | Sunitinib | 2.20 at 1 µM, 3.81 at 10 µM | 25 at pH 1.0-6.8, low solubility at pH > 6.8 | IV | [58] |
| 7 | Lapatinib | Low permeability | 0.007 in water, 0.001 at pH 1.1 | IV | [59, 60] |
| 8 | Nilotinib | Moderate permeability | Insoluble in buffer solutions of pH ≥ 4.5, pH dependent solubility | IV | [61] |
| 9 | Pirfenidone | NA | Sparingly soluble in water at any pH | NA | [62] |
| 10 | Pazopanib | 17.6 at 1 µM, 14.9 at 3 µM, 18.1 at 10 µM | Slightly soluble at pH 1.0, Insoluble above pH 4.0 | II | [63, 64] |
| 11 | Crizotinib | Low permeability, <LOQ at 1-10 µM | Low solubility, pH dependent solubility | IV | [65] |
| 12 | Vemurafenib | 2.9 | Insoluble in aq. media (<0.0005) across the pH range of 1 to 7.5 | IV | [69] |
| 13 | Vandetanib | 17.9 at 1 µM, 23.8 at 10 µM | Highly soluble at pH ≤ 6.0, Low solubility at pH > 6.0 | II | [68] |
| 14 | Ruxolitinib | 21.5 at 10 µM | 0.52 at pH ≤3.3, 0.15 at pH=7.5 | I | [66, 67] |
| 15 | Axitinib | 13.8 | 1.8 at pH1.1, 0.0002 at pH ≥ 6.0 | II | [70] |
| 16 | Bosutinib | 2.08 at 1µM, 2.96 at 10 µM | 11.03 at pH1.0, 0.02 at pH 6.8 | IV | [71] |
| 17 | Regorafenib | 12.4 | Low solubility across the pH range of 1 to 7.5 | II | [72] |
| 18 | Tofacitinib | Moderate permeability | 3.48 - >28 at pH 1.0-3.9, 0.20 – 0.59 at pH 4.53 – 8.0 | III | [73, 74] |
| 19 | Cabozantinib | Highly permeable | Low solubility | II | [75, 109] |
| 20 | Ponatinib | 4.4 | Highly soluble at pH<1.7, Slightly soluble at pH 1.7-2.7, Insoluble > pH 2.7 | IV | [76] |
| 21 | Trametinib | 16.2-59.5 at pH 7.4, MDCKII-MDR1 | Low solubility at pH 1.0 – 8.0 | II | [77] |
| 22 | Dabrafenib | High permeability - MDR1-MDCK | 0.043 at pH 1.2, 0.068 at pH4.9 FeSSIF, 0.0062 at pH6.3 FaSSIF | II | [78] |
| 23 | Afatinib | Higher permeability | >50 at pH less than 6.0, >1 at pH 6-7, 0.04 at pH>7.0 | I | [79] |
| 24 | Ibrutinib | 57.9 | 2 at pH 1.2, 0.003 at pH 6.0 | II | [80] |
| 25 | Ceritinib | Low permeability | 11.9 at pH 1.2, 0.01 at pH6.8, pH dependent solubility | IV | [81, 82] |
| 26 | Idelalisib | High permeability | at pH1.2, <0.1 at pH7.7 | II | [83] |
| 27 | Nintedanib | High permeability | >1 at acidic media, <0.001 at pH≥7.0 | II | [84, 85] |
| 28 | Palbociclib | High permeability | High solubility at pH<4.0, Solubility significantly reduced at pH>4.0 | II | [86, 87] |
| 29 | Lenvatinib | NA | pH dependent solubility | II, IV | [88] |
| 30 | Cobimetinib | 18.5-5.1, MDCK moderate-high permeability | 48 at pH 1.9, 0.78 at pH 6.8 | III | [89] |
a Apparent permeability was determined using Caco-2 assays at pH 7.4. Permeability was categorized in comparison to the permeability values obtained from low and high permeability standards recommended by FDA, which were run in parallel with the corresponding drugs in each study to calibrate permeability boundary of each drug substance.
b Solubility was categorized based on FDA guideline: a drug substance is considered highly soluble when the highest dose strength is soluble in 250 mL or less aqueous media at pH range of 1 to 7.5.
Uptake and efflux transporters can also play a role. Six major uptake and efflux transporters are P-gp, BCRP, BSEP, OATP, OAT and OCT. The interaction of these transporters with the approved kinase inhibitors was evaluated. In general, it was determined that ~80% of these drugs were substrates for one or more efflux transporters with the majority interacting with P-gp and BCRP efflux transporters (Table 3) [53-90, 109, 116, 125, 140].
Table 3.
Enzymes and transporters involved in pharmacokinetics of kinase inhibitors.
| No. | Drug | Major CYP450 enzymes responsible for kinase inhibitor metabolism | Major transporters responsible for efflux transport | References |
|---|---|---|---|---|
| 1 | Imatinib | CYP3A4 | P-gp and BCRP | [53, 138] |
| 2 | Gefitinib | CYP3A4 and CYP2D6 | P-gp and BCRP | [54] |
| 3 | Erlotinib | CYP3A4/5 and CYP1A1 | P-gp and BCRP | [55] |
| 4 | Sorafenib | CYP3A4 | BCRP | [56, 90, 140] |
| 5 | Dasatinib | CYP3A4 | P-gp and BCRP | [57] |
| 6 | Sunitinib | CYP3A4 | P-gp and BCRP | [58] |
| 7 | Lapatinib | CYP3A4/5 | P-gp and BCRP | [59, 60] |
| 8 | Nilotinib | CYP3A4 and CYP2C8 | P-gp and BCRP | [61] |
| 9 | Pirfenidone | CYP1A2 | None | [62] |
| 10 | Pazopanib | CYP3A4 | P-gp | [63, 64] |
| 11 | Crizotinib | CYP3A4/5 | P-gp | [65] |
| 12 | Vemurafenib | CYP3A4 | P-gp and BCRP | [69] |
| 13 | Vandetanib | CYP3A4 | BCRP | [68] |
| 14 | Ruxolitinib | CYP3A4 | None | [67] |
| 15 | Axitinib | CYP3A4 | P-gp and BCRP | [70] |
| 16 | Bosutinib | CYP3A4 | P-gp, BCRP and MRPs | [71] |
| 17 | Regorafenib | CYP3A4 and UGT1A19 | None | [72] |
| 18 | Tofacitinib | CYP3A4 and CYP2C9 | P-gp | [73, 74] |
| 19 | Cabozantinib | CYP3A4, CYP2C9, UGT, ST and amidase | None | [75, 109] |
| 20 | Ponatinib | CYP3A4, CYP2C8, CYP2D6 and CYP3A5 | P-gp and BCRP | [76] |
| 21 | Trametinib | Primarily non-CYP mediated metabolism, CYP3A4 plays minor role | None | [77] |
| 22 | Dabrafenib | CYPs 2C8, 3A4 and 2C9 | P-gp and BCRP | [78] |
| 23 | Afatinib | Mainly CYP3A4, FMO and UGT | P-gp | [79] |
| 24 | Ibrutinib | CYP3A4 | None | [80] |
| 25 | Ceritinib | CYP3A4/5 | P-gp and BCRP | [81, 82] |
| 26 | Idelalisib | CYP3A4 and UGT1A4 | P-gp and BCRP | [83, 125] |
| 27 | Nintedanib | Esterase, CYP3A4 and UGT | P-gp and OCT-1 | [84, 85, 116] |
| 28 | Palbociclib | CYP3A and SULT1A1 | P-gp and BCRP | [86, 87] |
| 29 | Lenvatinib | AO, CYP3A and GST | P-gp, MDR1 and BCRP | [88] |
| 30 | Cobimetinib | CYP3A4 and UGT2B7 | None | [89] |
The above data can be summarized as follows: 1) ~65% were Class I and II drugs with high permeability, 2) ~83% showed poor solubility, and 3) 80% were substrates of P-gp and/or BCRP efflux transporters. It is noteworthy that 30% of these drugs had poor solubility and poor permeability. While formulation attempts to combat poor solubility/dissolution to improve exposure levels have had some success, similar attempts aimed at improving permeability without damaging intestinal epithelial membranes have proven challenging. It is also important to point out that in vitro Caco-2 and MDCK system may underestimate in vivo permeability through membranes due to their intrinsic flaws of tighter junction and lack of adequate expression of uptake transporters. Finally, efflux transporters can be saturated in the GI tract at higher dose level or with high apparent intestinal solubility. These factors might explain satisfactory exposure levels of these drugs despite their low solubility and poor permeability data from in vitro assays.
3. Pharmacokinetics
3.1. Absorption
As eluted to previously, the overall oral exposures usually reflect an interplay of systemic CL (metabolism in gastrointestinal tract and liver, and other organs in addition to excretion), permeability, solubility, efflux/uptake transporters, and dissolution rate in small intestine as well as chemical stability. During lead optimization, systemic CL is the most important guides for compound selection industry-wide [91, 128]. This parameter is always judged against liver blood flow (LBF) to be binned as rapid, moderate, or slow CL. It is customary to characterize CL values as rapid if >70% LBF. LBF values have been reported to be 90 mL/min/kg in the mouse, 60 mL/min/kg in the rat, 31 mL/min/kg in the dog, 44 mL/min/kg in the monkey [92-94]. As a general trend, kinase inhibitors with acceptable F% tend to have slow to moderate CL (Tables 4 and 5). However, ~26% of these drugs were cleared rapidly in one or more preclinical species, nonetheless the oral plasma exposures for the majority of these drugs appeared to be reasonable with F% values ≥ 20% (Tables 4 and 5). Nevertheless, it is critical to emphasize that CL is still a critical factor in advancement of compounds in discovery as it is ultimately a determinant of dose.
In general, these kinase inhibitors reached Cmax relatively rapidly (Tmax between 0.5 to 4 hr) across all species (Table 4). About 76% of these inhibitors had F% values greater than 20% across all reported species. High solubility and/or high permeability in GI tract might overwhelm intestinal and even hepatic first-pass clearance mechanisms. This might off-set the negative impact of rapid clearance to oral exposures and salvage bioavailability normally impaired by the rapid clearance. This could be the case for kinase inhibitors in BCS class I (high permeability and high solubility) and II (high permeability and low solubility) (Table 2). The available data cannot offer a good explanation for bosutinib (BCS Class IV), which exhibited poor permeability, poor solubility, and moderate to rapid CL (Tables 2, 4 and 5).
Table 4.
Oral pharmacokinetic parameters of the kinase inhibitors.
|
Drug
(Dose (mg/kg) in Mouse/Rat/ Dog/Monkey) |
Cmax
(ng/mL) |
Tmax
(hr) |
AUC
(ng.hr/mL) |
Bioavailability
(%) |
Reference | |||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ms | Rt | Dg | Mnk | Ms | Rt | Dg | Mnk | Ms | Rt | Dg | Mnk | Ms | Rt | Dg | Mnk | |||||||||||||||||||
| Imatinib (50/10/10/3) |
6990 | 870 | 60-1410 | 40 | NA | 2.0 | 2-4 | 2.5 | 12820 | 4030 | 250-12400 | 216 | 28 | 53 | 29-68 | 27 | [96] | |||||||||||||||||
| Gefitinib (NA/5/5/NA) |
NA | 164 | 357-510 | NA | NA | 2.0 | 2-4 | NA | NA | 1440 | 3380-3882 | NA | NA | 44 | 49-64 | NA | [142] | |||||||||||||||||
| Erlotinib (NA/NA/10/NA) |
NA | NA | 650 | NA | NA | NA | 1.0 | NA | NA | NA | 4792 | NA | NA | NA | 45 - 88 | NA | [90] | |||||||||||||||||
| Sorafenib (NA) |
NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 79 | 79 | 60 | NA | [146] | |||||||||||||||||
| Dasatinib (15/10/3/5) |
156 | 239 | 146 | 166 | 2.0 | 2.3 | 0.75 | 0.60 | 586 | 1854 | 683 | 366 | 14 | 27 | 34 | 15 | [103] | |||||||||||||||||
| Sunitinib (NA) |
NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 100 | NA | 41 | [104] | |||||||||||||||||
| Lapatinib (10/10/10/NA) |
504 | 288 | 555 | NA | 0.5 | 0.5 | 4.0 | NA | 1735 | 861 | 5916 | NA | 50 | 24 | 42 | NA | [143] | |||||||||||||||||
| Nilotinib (25/20/10/NA) | 7910 | 1740 | 518 | NA | 0.5 | 4.0 | NA | 2.7 | 36100 | 26100 | 3880 | NA | 43 | 34 | NA | 24 | [144] | |||||||||||||||||
| Pirfenidone (NA/50/18/NA) |
NA | 45100 | 11700 | NA | NA | 0.35 | 0.25 | NA | NA | 1706000 | 711000 | NA | NA | 52 | 81 | NA | [62] | |||||||||||||||||
| Pazopanib (10/10/1/50) | 19500 | 17267 | 810 | 30310 | 6.0 | 0.8 | 3.5 | 0.9 | 220200 | 70429 | 5332 | 141017 | NA | 72 | 47 | 30 | [145] | |||||||||||||||||
| Crizotinib (NA/10/10/NA) |
NA | 220 | 938 | NA | NA | 8.0 | 6.0 | NA | NA | 3160 | 17600 | NA | NA | 26-63 | 38-66 | 44 | [99, 100] | |||||||||||||||||
| Vemurafenib (NA/30/ 24.5/NA) | NA | NA | NA | NA | NA | NA | NA | NA | NA | 70.5-172 | 62.1 | NA | NA | 18-43 | 40 | NA | [102] | |||||||||||||||||
| Vandetanib (NA/10/20/NA) | NA | 326 | 267 | NA | NA | 2.0 | 3.3 | NA | NA | 5298 | 4930 | NA | NA | 55 | 56 | NA | [101] | |||||||||||||||||
| Ruxolitinib (NA/50/10/NA) | NA | 970 | 3522 | NA | NA | NA | 2.0 | NA | NA | 2184 | 15730 | NA | NA | 29 | 57 | NA | [67] | |||||||||||||||||
| Axitinib (NA) |
NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 16 | 3-31 | 10-59 | 3 | [105] | |||||||||||||||||
| Bosutinib (50/50/5/NA) |
1509 | 224 | 206 | NA | 4.0 | 3.0 | 1.3 | NA | 11677 | 1507 | 3091 | NA | 53 | 23 | 50 | NA | [106] | |||||||||||||||||
| Regorafenib (NA/0.5/1.0/NA) |
NA | 277 | 369 | NA | NA | 6.0 | 2.67 | NA | NA | 2830 | 2460 | NA | NA | 85 | 67 | NA | [107, 141] | |||||||||||||||||
| Tofacitinib (NA/10/5/5) |
NA | 261-670 | 1020 | 791 | NA | 0.5 | 0.5 | 1.1 | NA | 462-1138 | 2330 | 2280 | NA | 12-17 | 43 | 48 | [108] | |||||||||||||||||
| Cabozantinib (NA/5/3/3) |
NA | 4584-7623 | 4278-6118 | 20.2-491 | NA | 4.0 | 4.0 | 2.0-3.0 | NA | 76731-144435 | 51154-69208 | 547-3175 | NA | 66-90 | 51-55 | 13-73 | [109] | |||||||||||||||||
| Ponatinib (NA/15/NA/2-3) |
NA | 453 | NA | 96 | NA | 6.0 | NA | 4.0 | NA | 8320 | NA | 942 | NA | 54 | NA | 21 | [110] | |||||||||||||||||
| Trametinib (3/3/0.3/NA) |
1662 | 289 | 80 | NA | NA | NA | NA | NA | 14462 | 3754 | 1723 | NA | 111 | 42 | 86 | NA | [111] | |||||||||||||||||
| Debrafenib (10/4.0/0.7/1.0) |
NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 70 | 77 | 82 | 46 | [131] | |||||||||||||||||
| Afatinib (NA) |
NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 38 | 81 | 42 | [113] | |||||||||||||||||
| Ibrutinib (NA) |
NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 9.4 | 11 | 0.33 | [113] | |||||||||||||||||
| Ceritinib (20/25/NA/60) |
388 | 363 | NA | 947 | 7.0 | 12 | NA | 13 | 6864 | 8390 | NA | 45300 | 55 | 48 | NA | 56 | [114] | |||||||||||||||||
| Idelalisib (NA/3/1/NA) |
NA | 129 | 209 | NA | NA | 3.0 | 1.0 | NA | NA | 141 | 671 | NA | NA | 39 | 48 | NA | [115] | |||||||||||||||||
| Nintedanib (50/50/NA/40) |
547 | 105 | NA | 175 | NA | NA | NA | NA | 2720 | 375 | NA | 2390 | NA | 12 | NA | 13 | [116] | |||||||||||||||||
| Palbociclib (NA/5/20/2.66) |
NA | 178 | 664 | 86 | NA | 3.5 | 8.7 | 2.7 | NA | 1140 | 17400 | 768 | NA | 53 | 37 | 23 | [117] | |||||||||||||||||
| Lenvatinib (3/3/3/3) |
1965 | 6167 | 1272 | 2501 | 0.5 | 0.5 | 2.0 | 2.0 | 5596 | 20697 | 5481 | 10272 | 64 | 69 | 70 | 78 | [118] | |||||||||||||||||
| Cobimetinib (3/3/3/3) |
0.184 | 0.193 | 0.843 | 0.050 | 0.25 | 3.5 | 2.7 | 2.0 | 1.51 | 1.59 | 11.9 | 0.64 | 54 | 69 | 69 | 20 | [132] | |||||||||||||||||
Ms = Mouse, Rt = Rat, Dg = Dog, Mnk = Monkey, Hmn = Human, NA = Not Available.
3.2. Distribution
Distribution of drugs into the body can be studied by various methods. It can be estimated by calculating volume of distribution at steady state (Vss), during PK modeling, tissue extraction followed by quantitation of the drug, or by using tissue imaging techniques such as MALDI-TOF using non-labeled drugs, or whole body autoradiography using radiolabeled drugs. Vss is a commonly used theoretical parameter to reflect the extent of drug distribution throughout the body. It is devoid of actual physiological meaning, but provides an overall understanding of distribution. In relation to total body water volume (~ 0.7 L/kg), Vss can be classified into three categories: small (<0.7 L/kg), moderate (0.7 – 3.5 L/kg) and large (> 3.5 L/kg) [95].
All 30 kinase inhibitors were well distributed into various tissues, including liver, kidney, lung, gastrointestinal tract and glandular tissues. Also, the majority exhibited limited CNS penetration [53-63, 65-69, 90, 96, 97, 99-118]. Their Vss values ranged from 0.28 L/kg to 43.6 L/kg (Table 5). In rodents, majority of these drugs fell into small to large Vss category, ranging from less than total body water volume (~ 0.7 L/kg) to greater than 10 L/kg. Our limited data analysis (Tables 4 and 5) suggested that basic kinase inhibitors (~33%) with the most basic pKa > 9 tend to have Vss > 10 L/kg, which may pose a risk for undesirable effects due to higher tendency for tissue accumulation, ie. in the heart. Lipophilicity (logP) and plasma protein binding may also be indicators of large Vss. However, in the overall evaluation of pKa, logP and PPB and their relationship to Vss it was observed that pKa was a greater correlate to Vss.
Table 5.
Disposition parameters of kinase-targeted inhibitors in mouse, rat, dog and monkey.
| Drug | Most Basic pKa (Predicted/measured) |
Plasma Clearancea
(mL/min/kg) |
Vssb
(L/kg) |
MRT
(hr) |
References | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ms | Rt | Dg | Mnk | Ms | Rt | Dg | Mnk | Ms | Rt | Dg | Mnk | |||
| Imatinib | 8.4/8.1 | 18 | 9.5 | 52 | 38 | 5.2 | 3.3 | 10.4 | 11 | 4.9 | 5.8 | 3.3 | 4.8 | [96] |
| Gefitinib | 7.1/7.2 | NA | 34 | 13 | NA | NA | 9.4 | 2.1 | NA | NA | 5.3 | 2.5 | NA | [142] |
| Erlotinib | 6.5/5.6 | NA | ~70 | 34 | NA | NA | NA | 2.6 | NA | NA | NA | 1.3 | NA | [90] |
| Sorafenib | 4.5/NA | 2.5 | 0.67 | 2.2 | NA | 0.68 | 0.65 | 0.74 | NA | 4.5 | 16 | 5.7 | NA | [146] |
| Dasatinib | 7.1/6.8 | 62 | 26 | 25 | 34 | 4.2 | 6.3 | 4.7 | 3.5 | 1.1 | 4.1 | 3.2 | 1.7 | [103] |
| Sunitinib | 8.6/8.9 | 77 | 30 | 41 | 31 | 8.1 | 5.6 | 21 | 17 | 1.8 | 4.7 | 8.5 | 14 | [104] |
| Lapatinib | 6.5/NA | 48 | 23 | 13 | NA | 9.6 | 1.8 | 5.5 | NA | 3.3 | 2.1 | 7.8 | NA | [143] |
| Nilotinib | 5.9/5.4 | 5.0 | 4.3 | NA | 11 | 0.52 | 7.9 | NA | 0.67 | 1.7 | 31 | NA | 1.0 | [144] |
| Pirfenidone | NA/NA | 100 | 29 | 20 | NA | 0.71 | NA | NA | NA | 0.11 | NA | NA | NA | [62] |
| Pazopanib | 3.7/6.4 | NA | 1.8 | 1.4 | 1.5 | NA | 0.45 | 0.30 | 0.28 | NA | 4.7 | 3.5 | 2.9 | [145] |
| Crizotinib | 9.2/9.4 | NA | 35 | 7.9 | 18 | NA | 24 | 12 | 13 | NA | 11 | 25 | 12 | [99, 100] |
| Vemurafenib | 3.2/NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | [102] |
| Vandetanib | 8.8/9.4 | NA | 17.1 | 34.7 | NA | NA | 27.2 | 43.6 | NA | NA | 27 | 21 | NA | [101] |
| Ruxolitinib | 2.7/4.3 | NA | 157 | 8.0 | NA | NA | 3.8 | 1.1 | NA | NA | 0.40 | 2.3 | NA | [67] |
| Axitinib | 4.5/4.8 | 25 | 397 | 12 | 11.2 | 1.67 | 32.3 | 1.17 | 0.8 | 1.1 | 1.4 | 1.6 | 1.2 | [105] |
| Bosutinib | 8.3/7.9 | 37.5 | 128 | 15.2 | NA | 11.5 | 15.2 | 13.5 | NA | 5.1 | 2.0 | 15 | NA | [106] |
| Regorafenib | 5.8/NA | NA | 2.42 | 4.53 | NA | NA | 0.88 | 1.89 | NA | NA | 6.1 | 7.0 | NA | [107, 141] |
| Tofacitinib | 4.6/5.1 | NA | 62 | 19.4 | 18.2 | NA | 2.6 | 1.8 | 1.7 | NA | 0.7 | 1.5 | 1.6 | [108] |
| Cabozantinib | 5.3/NA | NA | 0.50 | 7.18 | 1.07 | NA | 0.41 | NA | 2.67 | NA | 13-16 | NA | 42 | [109] |
| Ponatinib | 8.0/7.8 | NA | 26.5 | NA | 8.7 | NA | 17.7 | NA | 2.6 | NA | 11.1 | NA | 5.0 | [110] |
| Trametinib | NA/0.25 | 3.44 | 5.41 | 2.41 | NA | 0.9 | 2.9 | 3.0 | NA | 4.4 | 8.9 | 21 | NA | [111] |
| Debrafenib | 3.6/6.6 | 43.5 | 17.6 | 3.6 | 22.5 | 1.0 | 1.0 | 0.4 | 0.5 | 0.38 | 0.95 | 1.9 | 0.37 | [131] |
| Afatinib | 8.4/8.2 | NA | 90 | 64 | 25 | NA | NA | NA | NA | NA | NA | NA | NA | [113] |
| Ibrutinib | 3.8/3.7 | NA | 34 | 54 | 28 | NA | NA | NA | NA | NA | NA | NA | NA | [113] |
| Ceritinib | 10/9.7 | 26.6 | 24.8 | NA | 13 | 9.7 | 19.9 | NA | 13.5 | 6.1 | 13 | NA | 17 | [114] |
| Idelalisib | 3.7/3.4 | NA | 47.8 | 11.6 | NA | NA | 2.49 | 1.23 | NA | NA | 0.87 | 1.8 | NA | [115] |
| Nintedanib | 8.1/7.9 | NA | 202 | NA | 37.5 | NA | 41.2 | NA | 8.64 | NA | 3.4 | NA | 3.8 | [116] |
| Palbociclib | 9.0/7.4 | NA | 38 | 7.22 | 13.4 | NA | 5.7 | 6.2 | 5.1 | NA | 2.5 | 14 | 6.3 | [117] |
| Lenvatinib | 5.2/NA | 5.76 | 1.67 | 6.14 | 3.96 | 0.714 | 0.391 | 1.61 | 0.794 | 2.1 | 3.9 | 4.4 | 3.3 | [118] |
| Cobimetinib | 8.5/NA | 33.5 | 37.8 | 5.53 | 29.6 | 9.4 | 34.6 | 3.99 | 29.6 | 4.7 | 15 | 12 | 17 | [132] |
a LBF of mouse, rat, dog and monkey are: 90, 60, 31 and 44 mL/min/kg, respectively.
b Total body water volume of moue, rat, dog and monkey are: 0.725, 0.668, 0.604 and 0.693 L/kg, respectively.
MRT describes the sojourn of drug in the body and correlates with t1/2. The MRT values reported here were estimated using clearance and steady-state volumes of distribution. MRT values ranged from 0.11 to 42 hr with majority of the drugs having MRTs around 2-5 hr in mice, rats, dogs and monkeys (Table 5). The trend analysis on Vss and MRT for these kinase inhibitors revealed that Vss ≤ 10 L/kg and MRT ≥ 2 hr in mice, rats, dogs and monkeys were prevalent in this class of drugs. However, because ~30% of these approved drugs have Vss > 10 L/kg, it should be considered that in absence of preclinical adverse events large Vss should be tolerated. In our view, Vss cannot constitute a selection criteria in compound advancement in discovery.
Reviewing the PPB levels across mice, rats, dogs, monkeys, and human, revealed that about 30% of these drugs had extremely high PPB values (≥ 99%) and less than 10% of them exhibited low PPB (<70%). In between the two extremes, roughly 50% of these drugs had high PPB (90-99%) and less than 17% of them had moderate PPB (70-90%) (Table 6). Overall, ~80% of approved kinase inhibitors are highly protein bound with PPB values ≥90%. Therefore, in our view PPB should not be used as a screening criteria during lead optimization or as a determinant for drug candidacy.
Table 6.
Plasma protein binding of the kinase inhibitors.
| Drug | Plasma Protein Binding (%) | References | ||||
|---|---|---|---|---|---|---|
| Ms | Rt | Dg | Mnk | Hmn | ||
| Imatinib | 98 | 95 | 81 | 90 | 93 | [96] |
| Gefitinib | 91 | 87 | 91 | NA | 90 | [142] |
| Erlotinib | 89 | 85 | 85 | NA | 94 | [90] |
| Sorafenib | >99 | >99 | 99 | NA | >99 | [146] |
| Dasatinib | 92 | 97 | 96 | 97 | 94 | [103] |
| Sunitinib | 94 | 98 | 95 | 95 | 95 | [104] |
| Lapatinib | >99 | >99 | >99 | >99 | >99 | [143] |
| Nilotinib | 97 | 99 | 98 | 99 | 98 | [144] |
| Pirfenidone | 30 | 30 | 30 | NA | 50 | [62] |
| Pazopanib | >99 | >99 | >99 | >99 | >99 | [145] |
| Crizotinib | 96 | 94 | 96 | 93 | 91 | [99, 100] |
| Vemurafenib | NA | NA | NA | NA | NA | [102] |
| Vandetanib | >99 | >99 | >99 | >99 | >99 | [101] |
| Ruxolitinib | 97 | 82 | 90 | NA | 97 | [67] |
| Axitinib | 97 | 98 | 98 | 96-99 | 99 | [105] |
| Bosutinib | 93-95 | 93-94 | 95-96 | NA | 93-94 | [106] |
| Regorafenib | >99 | >99 | >99 | >99 | >99 | [141] |
| Tofacitinib | 33 | 6-31 | 20 | 35 | 39 | [108] |
| Cabozantinib | >99 | >99 | >99 | >99 | >99 | [109] |
| Ponatinib | >99 | >99 | NA | >99 | >99 | [110] |
| Trametinib | 95 | 96 | 97 | 98 | 97 | [111] |
| Debrafenib | 93 | 99 | 95 | NA | 98-99 | [131] |
| Afatinib | 94 | 91-93 | 93 | 92 | 90-95 | [113] |
| Ibrutinib | 99 | 97 to >99 | 96-98 | 97 | 97-98 | [113] |
| Ceritinib | NA | 98 | 98 | 95 | 97 | [114] |
| Idelalisib | 80 | 81 | 79 | NA | 86 | [115] |
| Nintedanib | 97 | 98 | NA | 93 | 98 | [116] |
| Palbociclib | 81-87 | 87-88 | 55-61 | NA | 84-86 | [117] |
| Lenvatinib | 96-97 | 98 | 90-92 | 96 | 98-99 | [118] |
| Cobimetinib | 96-97 | 97 | 99 | 95-96 | 94-95 | [132] |
Ms = Mouse, Rt = Rat, Dg = Dog, Mnk = Monkey, Hmn = Human, NA = Not Available.
3.3. Potential Drug-drug Interactions (DDI) for Transporters and CYP450 Enzymes
Inhibition of transporters and metabolizing enzymes by drugs could pose potential DDI effects leading to altered PK for co-administered drugs (or standalone). The reason why DDI has attracted so much attention is that patients may be treated with multiple drugs simultaneously (polypharmacy) and their interactions might result in adverse drug reaction (ADR). In a study done in United States outpatient setting, rates of ADRs due to DDI ranges from 2% to 50% [119, 120].
Majority of oncology drugs are cleared via CYP450 metabolism pathways. Therefore, it is critical to understand DDI potential for kinase inhibitors, particularly in oncology, to better manage polypharmacy. While gathering the data, it became evident that a variety of formats were used to report transporter inhibition and CYP inhibition/induction. This posed a challenge in organizing the data. As a result, we have simplified the available information into Tables 7 and 8 for ease of discussion. More detailed information is available in the references cited in these tables.
3.3.1. Transporter Inhibition
Transporter inhibition data were reported on ABC and SLC transporter proteins, such as P-gp, BCRP, OATP, OCT and OAT (Table 7). About 53% of the kinase inhibitors inhibited P-gp at some level. Other transporters such as BCRP, OATP,
Table 7.
Transporter inhibition potential for kinase inhibitors.
| Drug | Transporters | References | |||||
|---|---|---|---|---|---|---|---|
| P-gp | BCRP | BSEP | OAT | OATP | OCT | ||
| Imatinib | Y | Y | NA | NA | NA | NA | [39, 53, 96, 133, 139] |
| Gefitinib | NA | NA | NA | NA | NA | NA | [39, 54, 134] |
| Erlotinib | NA | NA | NA | NA | NA | NA | [39, 55, 90, 136] |
| Sorafenib | Y | NA | NA | NA | NA | NA | [39, 56, 127, 134, 140] |
| Dasatinib | N | NA | NA | NA | NA | NA | [39, 57, 97, 103, 134, 137] |
| Sunitinib | NA | NA | NA | NA | NA | NA | [39, 58, 98, 104, 134] |
| Lapatinib | Y | Y | NA | Y | Y | NA | [39, 59, 134] |
| Nilotinib | Y | NA | NA | NA | NA | NA | [39, 61, 134] |
| Pirfenidone | N | NA | NA | NA | NA | NA | [62] |
| Pazopanib | N | NA | NA | NA | NA | NA | [39, 63, 134] |
| Crizotinib | Y | Y | NA | NA | NA | NA | [39, 65, 99, 100, 134] |
| Vemurafenib | Y | Y | NA | NA | NA | NA | [39, 69, 102, 134, 135] |
| Vandetanib | N | NA | NA | NA | NA | NA | [39, 68, 101, 134] |
| Ruxolitinib | N | N | NA | NA | NA | NA | [39, 66, 67, 134] |
| Axitinib | Y | NA | NA | NA | NA | NA | [70, 77] |
| Bosutinib | Y | NA | NA | NA | NA | NA | [71] |
| Regorafenib | Y | Y | NA | NA | NA | NA | [72] |
| Tofacitinib | Y | NA | NA | NA | Y | Y | [73, 74] |
| Cabozantinib | Y | NA | NA | NA | NA | NA | [75] |
| Ponatinib | Y | NA | NA | NA | NA | NA | [76] |
| Trametinib | N | N | NA | NA | N | NA | [77] |
| Debrafenib | N | Y | NA | NA | Y | NA | [78] |
| Afatinib | Y | NA | NA | NA | NA | NA | [79] |
| Ibrutinib | Y | NA | NA | NA | NA | NA | [80] |
| Ceritinib | N | N | NA | Y | Y | Y | [81, 82] |
| Idelalisib | N | N | NA | N | N | N | [83] |
| Nintedanib | Y | Y | NA | NA | NA | Y | [85] |
| Palbociclib | Y | Y | NA | Y | Y | Y | [86, 87] |
| Lenvatinib | N | N | Y | Y | Y | Y | [88] |
| Cobimetinib | N | N | NA | NA | NA | NA | [89] |
Y = Yes, N = No, NA = Not Available
Efflux transporter inhibition: A decrease in net flux ratio of probe substrate of P-gp/BCRP/BSEP in presence of investigational kinase drug.
Uptake transporter inhibition: A decrease in uptake of model substrate of OATP/OAT/OCT in presence of investigational kinase drug.
OCT, OAT, and BSEP were inhibited by about 27%, 20%, 17%, 13%, and 3% of these approved drugs, respectively. As can be observed, transporter inhibition is prevalent among kinase inhibitors.
3.3.2. Metabolic Enzyme Profiling
After examining metabolic pathways and enzymes responsible for the clearance of these 30 kinase inhibitors, our analysis suggested that ~90% of kinase inhibitors were cleared by CYP3A4 (Table 3). Also, CYP2D6, CYP1A1, CYP2C8, UGT1A19, CYP2C9, UGT, SULT1A1 as well as hydrolytic enzymes play a role in metabolizing ~30% of these drugs.
3.3.3. CYP450 Inhibition and Induction
The in vitro CYP450 inhibition and induction data are summarized in Table 8. Some clinical DDI data are also briefly touched on here for simple comparison between in vitro and in vivo results.
Table 8.
CYP450 inhibition and induction potentials for kinase inhibitors.
| Drug | CYP450 Inhibition | CYP450 Induction | References | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1A2 | 2A6 | 2B6 | 2C8 | 2C9 | 2C19 | 2D6 | 3A4 | 1A2 | 2B6 | 3A4 | ||
| Imatinib | N | N | NA | NA | N | N | L | L and TDI | NA | NA | NA | [39, 53, 96, 123, 134] |
| Gefitinib | L | NA | NA | NA | L | L | L | L and TDI | N | N | N | [39, 54, 123, 134] |
| Erlotinib | L | NA | NA | L | L | L | L | L and TDI | NA | NA | Y | [39, 55, 90, 123, 136] |
| Sorafenib | NA | NA | M | M | L | L | L | L | N | NA | N | [39, 56, 123, 134] |
| Dasatinib | L | L | N | M | N | N | N | L and TDI | N | N | N | [39, 57, 97, 123, 134] |
| Sunitinib | M | L | N | N | N | N | M | L and TDI | Y | Y | Y | [39, 58, 98, 104, 123, 134] |
| Lapatinib | NA | NA | NA | M | NA | NA | NA | M and TDI | N | N | N | [39, 59, 123, 134] |
| Nilotinib | NA | NA | NA | H | H | M | M | H and TDI | Y | Y | Y | [39, 61, 123, 134] |
| Pirfenidone | N | N | N | N | N | N | N | N | N | N | N | [62] |
| Pazopanib | M | NA | M | M | M | M | M | M and TDI | N | Y | Y | [39, 63, 123, 134] |
| Crizotinib | L | NA | L | L | L | L | L | M and TDI | NA | NA | Y (mRNA) | [39, 65, 99, 100, 134] |
| Vemurafenib | L | N | NA | NA | M | L | L | N | NA | NA | Y (mRNA) | [39, 69, 102, 134, 135] |
| Vandetanib | N | N | N | N | N | N | N | N | Y | NA | Y | [39, 68, 101, 134] |
| Ruxolitinib | L | NA | L | L | L | L | L | L | N | N | N | [39, 66, 67, 123, 134] |
| Axitinib | M | NA | NA | NA | M | NA | NA | NA | N | NA | N | [70, 105] |
| Bosutinib | N | N | N | N | N | N | N | N | N | N | N | [71] |
| Regorafenib | NA | NA | L | M | L | L | NA | L | N | N | N | [72] |
| Tofacitinib | N | N | N | N | N | N | N | N | N | N | N | [73, 74] |
| Cabozantinib | L | NA | NA | M | M | M | L | L | Y | N | N | [75] |
| Ponatinib | L | NA | M | M | NA | M | L | L to M | N | N | N | [76] |
| Trametinib | L | L | L | H | M | M | L | L | N | N | Y | [77] |
| Debrafenib | N | N | N | M | M | L | N | L | N | Y (mRNA) | Y (mRNA) | [78] |
| Afatinib | N | N | N | N | N | N | N | N | N | N | N | [79] |
| Ibrutinib | N | NA | M | M | M | M | M | M | N | N | N | [80] |
| Ceritinib | N | M | M | L | M | N | L | H | N | N | Y (mRNA) | [81, 82] |
| Idelalisib | N | NA | L | L | N | N | N | N | N | Y (mRNA) | Y (mRNA) | [83] |
| Nintedanib | N | N | N | N | N | N | N | N | N | N | N | [84, 85] |
| Palbociclib | L | L | L | L | L | L | L | L and TDI | N | N | N | [86, 87] |
| Lenvatinib | N | N | N | M | N | N | N | N and TDI | N | N | Y (mRNA) | [88] |
| Cobimetinib | N | NA | N | N | N | N | M | L to M and TDI | N | N | Y (mRNA) | [89] |
For CYP450 inhibition: L = Low Risk Potential (IC50: 10-50 µM), M = Moderate Risk (IC50: 1-10 µM), H = High Risk (IC50<1 µM), N = No Risk Potential (IC50>50 µM), NA = Not available, TDI = Time Dependent Inhibition, For Ki values: High Risk Potential (<0.5 µM), Moderate Risk Potential (0.5-5 µM), Low Risk Potential (5-25 µM), No Risk Potential (>25 µM)
For CYP450 induction: Y = Yes, N = No, ≥40% positive control response is considered as inductive potential.
Ki, IC50 and % inhibition at defined inhibitor concentrations are commonly used to describe the inhibitory potentials of drugs toward an enzyme [39, 53-69, 90, 96-102, 121, 122], while kinact, Ki and kinact/Ki ratio are often used to describe the potentials for the time-dependent inhibition (TDI) or mechanism based inhibi-tion of an enzyme. TDI is due to reactive metabolites of a drug irreversibly or quasi-irreversibly binding to an enzyme or the heme iron of the CYP’s in a manner that deactivates that enzyme. In general, the impact of TDI could be more significant compared to reversible inhibition due to its irreversible nature of deactivation of the enzymes. Therefore, TDI effect continues even after cessation of dosing and elimination of the drugs, until newly expressed enzymes are available. We have organized the data according to each drug’s ability to inhibit enzymes. More detailed discussions can be found in the references cited in Table 8.
Over 66% of the kinase inhibitors inhibited CYP3A4 with a third of them falling in the “Moderate” to “High” risk categories. Additionally, 40% of these drugs inhibited other CYP450’s tested in “Moderate” to “High” risk categories (Table 8).
Kenny et al. published in vitro time-dependent CYP3A4 inhibition profile of 26 marketed oncology drugs, of which, dasatinib, erlotinib, gefitinib, imatinib, lapatinib, nilotinib, pazopanib, sorafenib and sunitinib were kinase inhibitors [123]. It was indicated that all nine kinase inhibitors were moderate to strong inhibitors of CYP3A4 capable of time-dependent inhibition. This information alerted the investigators to the possibility of DDI in the clinic. Surprisingly, the available clinical DDI data did not validate this concern (dasatinib, erlotinib, gefitinib, imatinib, nilotinib, pazopanib and sorafenib as perpetrators [123]), brining preclinical prediction of DDI into question. According to FDA guidance [124], perpetrators can be classified into weak, moderate and strong inhibitors based on the AUC ratio of victim drug in the presence and absence of perpetrator drug (CYP inhibitor) after oral administration of the two interacting drugs. The DDI is considered weak, moderate, or strong if the ratio of victim drug’s exposure with and without co-administered perpetrator is ≥5, 5-2, 2-1.25-fold, respectively [125]. The clinical data showed that with the exception of imatinib, the observed AUC ratios for 6 victim drugs in the presence and absence of these 7 kinase drugs with potential for TDI to be less than 1.5-fold; suggesting an overestimation of clinical DDI by in vitro inhibition assays used in preclinical setting.
3.3.4. CYP450 Induction
CYP450 expression is inducible by certain compounds after multiple doses and continuous exposure. This results in increased levels of CYP’s leading to enhanced drug metabolism and reduced exposure levels in vivo. Metabolizing enzyme induction is of concern because it might lead to loss of efficacy. It has become increasingly attractive for discovery scientists to assess induction potential of NCEs early in the process in order to alleviate potential DDI risks. The most commonly used in vitro systems are gene reporter (PXR, AhR and CAR) and cultured human hepatocyte assays. In vivo, transgenic mouse and monkey models are also known as appropriate to recapitulate human enzyme induction potential of interested drugs. Herein, we simply report the in vitro CYP450 induction potential of approved kinase inhibitors as “Yes” and “No” based on criteria set by FDA (≥40% positive control response is considered inductive potential). Due to lack of availability of a full data packages, a comprehensive list of could not be assembled. Available information on in vitro hepatocyte induction potential of kinase inhibitors for CYP1A2, CYP2B6, and CYP3A4 was organized in Table 8. This data set suggested that at least 40% of the approved kinase inhibitors were in vitro inducers of one or more of the three CYP’s mentioned above. It is important to point out that to this date a correlation between the in vitro and clinical CYP450 induction data has not been observed. This may in part be due to the dual inhibitory and inductive effects observed in many drugs which results in net lessening of these effects and manifestation of a minor inhibition or induction in the clinic.
4. DISCUSSION
It has been 15 years since the discovery and launch of the first kinase drug, imatinib. Imatinib marked a significant breakthrough in cancer treatment. This opened a new era for exploring targeted therapy for oncology indications with better therapeutic effectiveness and reduced adverse effects compared to conventional cancer therapy. From 2001 to October 2015, a total of 30 kinase inhibitors have been approved by FDA and numerous others have entered in various stages of clinical development. An extraordinary level of attention has been paid to the discovery and development of tyrosine kinase inhibitors, which covers the majority of the approved drugs. Despite all the efforts to advance high quality kinase inhibitors into the clinic, the clinical attrition rate is still rising [126]. Determination of what constitutes the right balance of attributes for an inhibitor in early discovery is crucial, yet challenging. This manuscript summarizes the physicochemical and ADME properties of the first 30 kinase inhibitor drugs that have entered the market. Retrospective analysis of the attributes for these kinase inhibitors can provide insights as to what the minimally acceptable criteria might be to advance the candidate molecules through lead optimization efforts in discovery. Furthermore, availability of this dataset can better guide discussions between medicinal chemists and DMPK scientists in deciding the significance of different parameters during lead optimization. In absence of such information, an overwhelming body of low impact and expensive data is generated at the expense of productivity and efficiency.
With the advent of automation, high throughput screening processes have been devised to rapidly and indiscriminately generate huge volumes of data in in vitro ADME assays for an excessively large number of compounds. Often, the misconception that more data provides better clarity rules the philosophy of drug discovery groups, in particular in organizations with the means to capitalize such efforts. The net result in the creation of large sets of data which might not be necessary or stage appropriate is that it leads to confusion and distraction in teams.
We have offered our opinion on efficiency in drug discovery elsewhere [91, 128-130]. Our analysis suggests that not all ADME data are equally critical in early discovery. One conclusion might be that by using solubility, plasma protein binding, CYP450 inhibition and induction, permeability and efflux data as selection criteria in early discovery one could have inadvertently excluded some of the approved kinase inhibitors from advancement into the clinic. These data can only be valuable when put in the context of targetted disease, in vivo efficacy, efficacious exposure, and dosing regimen, which are all late stage considerations. Therefore, we recommend reserving judgment based on such criteria until early development. Based on this work and our internal experience, we propose focusing on in vitro metabolic stability and in vivo rodent disposition properties as primary screening assays. Moreover, we recommend against using permeability/efflux data as gate keeper in early screening stage. Instead, this data can be effective as a diagnostic assay to explain poor oral exposure or as a way to salvage compounds with rapid rodent plasma clearance, if they exhibit other redeeming qualities.
Oral systemic exposure of an NCE is a result of the interplay of structural features, apparent solubility/dissolution rate, permeability and efflux in the GI tract, disposition properties and food effects. Based on our analysis, Lipinski’s and Veber’s rules should be followed closely with regards to the number of H-acceptors, total number of H-bonds, and rotatable bonds. The recommendations on the number of H-donors (5) and PSA (140) can be followed also, however, it is noteworthy that most kinase inhibitors seem to follow a stricter trend of H-donor ≤3 and PSA≤100, respectively (Fig. 5). There seems to be some tolerance for violating the recommendations on MW and LogP. Kinase inhibitors with MW and logP as high as 600 and 6, respectively, have reached the market (Fig. 4). One parameter for which a guideline has not been reported is the most basic pKa. Our recommendation is to maintain this parameter below 9 in order to improve PK behavior. If rat IV PK is a primary screen in early discovery, we recommend prioritizing compounds with plasma CL ≤ 70% liver blood flow. However, our analysis revealed that plasma CL > 70% LBF criteria may be tolerated only if the higher species plasma CL is in line with LBF and there are other exquisite and compelling attributes such as potency (Table 5). The majority of kinase inhibitors had bioavailability above 20% in all preclinical species (Table 4), and therefore, prioritizing compounds with that level of bioavailability is warranted. The exception to that recommendations may apply to compounds with exquisite potency and selectivity, treatment rationale for unmet medical needs, and other positive attributes.
Based on the current information, in vitro data such as solubility, PPB, Caco-2 (or MDCK) permeability/efflux, transporter (uptake/efflux), CYP450 inhibition/induction, preclinical DDI do not appear to be gating, and therefore, one should carefully consider where and when to place these on the critical path for compound optimization. As shown, many negative attributes related to these data have been tolerated in the first 30 approved kinase inhibitors. Going forward these negative attributes may be minimized or eliminated to provide a competitive edge and yield higher quality drugs with better safety/efficacy profiles for patients; particularly as investigators move into areas beyond oncology.
An example of an ongoing challenge regarding the relevance of a preclinical assay to the clinic might be in vitro DDI. The in vitro DDI data are often over-interpreted likely because the in vitro systems designed for investigating potential for DDI are over-simplified and devoid of all the proper in vivo interactions. In the closed in vitro system, the interaction between the drug and CYPs can be exaggerated due to proximity and lack of compartmentalization. Also, it is nearly impossible to conduct the in vitro DDI studies at clinically relevant drug concentrations while in discovery mode, simply due to lack of accurate information on human efficacious dose/exposure levels early in the process. Moreover, the extent of decline in circulating concentrations of drugs over time due to in vivo clearance mechanisms can be more extensive than in isolated in vitro systems. Finally, in vivo DDI is a dynamic process and can be an interplay of multiple factors such as inhibition and induction, compensating metabolic pathways, contribution of CYPs to the entire metabolic pathways, etc. In vitro inhibition and induction studies are by nature closed system and more static. Therefore, in vitro DDI data can only become meaningful when combined with factors such as profiles of co-administered drugs, dosing route, dosing regimen, elimination pathways, enzyme phenotyping profile, efficacious exposure concentrations, concentration-time profile, and therapeutic target in the clinic.
A glance at the attributes of the approved kinase inhibitors between 2001-2011 compared to the ones approved from 2012-2015 reveals the following about the new generation kinase inhibitors: 1) more diverse types of kinases have been targeted, expanding beyond oncology, 2) metabolism of these new drugs is no longer predominantly through CYP3A4, 3) while rat CL has become more rapid, this parameter has been better optimized in the higher species, 4) Pgp/BCRP inhibition is more prevalent, 5) CYP450 induction and inhibition appears to be less of a concern.
CONCLUSION
Herein, we have organized a large body of physicochemical and ADME data on the first 30 approved kinase inhibitors. A detailed examination of this data can reveal what criteria may be acceptable/tolerable or in need of further optimization. It is clear that discovery teams should continue to strive for continuous improvement of NCE’s based on rigorous analysis of what is proven to add value, and not necessarily the conventional thinking. We hope that our analysis offers a baseline for attributes of kinase inhibitors and constitutes a platform for improving quality and provides insights into what “actionable” and “stage-appropriate” data might be. This can help to minimize generation of unnecessary data; thereby preventing “data pollution” and confusion during lead optimization.
ACKNOWLEDGEMENTS
Declared none.
LIST OF ABBREVIATIONS
- ABC
ATP-Binding Cassette
- ADME
Absorption, Distribution, Metabolism and Elimination
- ADR
Adverse Drug Reaction
- AUC
Area Under the Curve
- BCR-Abl
Breakpoint Cluster Region-Abelson murine leukemia viral oncogene homolog 1
- BCRP
Breast Cancer Resistance Protein
- BCS
Biopharmaceutics Classification System
- BSEP
Bile Salt Export Pump
- Caco-2
Human colorectal adenocarcinoma
- c-AMP
Cyclic AMP
- CL
Clearance
- Cmax
Maximum Concentration
- CML
Chronic Myelogenous Leukemia
- CNS
Central Nervus System
- CYP450
Cytochrome P450
- DDI
Drug-Drug Interaction
- DMPK
Drug Metabolism and Pharmacokinetics
- ERK
Extracellular Signal-regulated Kinase
- F%
Bioavailability
- FaSSIF
Fasted-state Simulated Small Intestinal Fluid
- FDA
Food and Drug Administration
- FeSSIF
Fed-state Simulated Small Intestinal Fluid
- GI
Gastro-Intestinal
- IV
Intravenous
- Ki
Inhibitory Constant
- Kinact
Maximal Rate of Enzyme Inactivation
- LBF
Liver Blood Flow
- MDCK
Madin-Darby Canine Kidney
- MDR
Multi-Drug Resistant
- MRP
Multidrug resistance-associated protein
- MRT
Mean Residence Time
- MW
Molecular Weight
- NA
Not Available
- NCE
New Chemical Entities
- OAT
Organic Anion Transporter
- OATP
Organic Anion-Transporting Polypeptide
- OCT
Organic Cation Transporter
- Papp A-B
Apparent Permeability from A-B
- PDGF-R
Proto-oncogen c-kit and platelet-derived Growth Factor Receptor
- Pgp
P-glycoprotein
- PI3K
Phosphoinositide 3-Kinase
- PK
Pharmacokinetics
- PO
Per Os (Oral)
- PPB
Plasma Protein Binding
- PS
Phosphatidylserine
- PSA
Polar Surface Area
- SGF
Simulated Gastric Fluid
- SLC
Solute Carrier
- SULT
Sulfotranserase
- TDI
Time Dependent Inhibition
- TK
Tyrosine Kinases
- Tmax
Time at maximum concentration
- UGT
UDP Glucuronosyltransferases
- Vss
Steady State Volume of Distribution
CONSENT FOR PUBLICATION
Not applicable.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Manning G., Whyte D.B., Martinez R., Hunter T., Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298(5600):1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 2.Wu P., Nielsen T.E., Clausen M.H. FDA-approved small-molecule kinase inhibitors. Trends Pharmacol. Sci. 2015;36(7):422–439. doi: 10.1016/j.tips.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 3.Burnett G., Kennedy E.P. The enzymatic phosphorylation of proteins. J. Biol. Chem. 1954;211(2):969–980. [PubMed] [Google Scholar]
- 4.Krebs E.G., Fischer E.H. Phosphorylase activity of skeletal muscle extracts. J. Biol. Chem. 1955;216(1):113–120. [PubMed] [Google Scholar]
- 5.Fischer E.H., Krebs E.G. Conversion of phosphorylase b to phosphorylase a in muscle extracts. J. Biol. Chem. 1955;216(1):121–132. [PubMed] [Google Scholar]
- 6.Sutherland E.W., Jr, Wosilait W.D. Inactivation and activation of liver phosphorylase. Nature. 1955;175(4447):169–170. doi: 10.1038/175169a0. [DOI] [PubMed] [Google Scholar]
- 7.Rall T.W., Sutherland E.W. Formation of a cyclic adenine ribonucleotide by tissue particles. J. Biol. Chem. 1958;232(2):1065–1076. [PubMed] [Google Scholar]
- 8.Sutherland E.W., Rall T.W. Fractionation and characterization of a cyclic adenine ribonucleotide formed by tissue particles. J. Biol. Chem. 1958;232(2):1077–1091. [PubMed] [Google Scholar]
- 9.Walsh D.A., Perkins J.P., Krebs E.G. An adenosine 3′,5′-monophosphate-dependant protein kinase from rabbit skeletal muscle. J. Biol. Chem. 1968;243(13):3763–3765. [PubMed] [Google Scholar]
- 10.Nowell P.C., Hungerfor D.A. A minute chromosome in human chronic granulocytic leukemia. Science. 1960;132:1497. [Google Scholar]
- 11.Rudkin C.T., Hungerford D.A., Nowell P.C. Nowell PC. DNA contents of chromosome PH1 and chromosome 21 in human chronic granulocytic leukemia. Science. 1964;144(3623):1229–1231. doi: 10.1126/science.144.3623.1229. [DOI] [PubMed] [Google Scholar]
- 12.Blume-Jensen P., Hunter T. Oncogenic kinase signalling. Nature. 2001;411(6835):355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- 13.Bardelli A., Parsons D.W., Silliman N., Ptak J., Szabo S., Saha S., Markowitz S., Willson J.K., Parmigiani G., Kinzler K.W., Vogelstein B., Velculescu V.E. Mutational analysis of the tyrosine kinome in colorectal cancers. Science. 2003;300(5621):949. doi: 10.1126/science.1082596. [DOI] [PubMed] [Google Scholar]
- 14.Greenman C., Stephens P., Smith R., Dalgliesh G.L., Hunter C., Bignell G., Davies H., Teague J., Butler A., Stevens C., Edkins S., O’Meara S., Vastrik I., Schmidt E.E., Avis T., Barthorpe S., Bhamra G., Buck G., Choudhury B., Clements J., Cole J., Dicks E., Forbes S., Gray K., Halliday K., Harrison R., Hills K., Hinton J., Jenkinson A., Jones D., Menzies A., Mironenko T., Perry J., Raine K., Richardson D., Shepherd R., Small A., Tofts C., Varian J., Webb T., West S., Widaa S., Yates A., Cahill D.P., Louis D.N., Goldstraw P., Nicholson A.G., Brasseur F., Looijenga L., Weber B.L., Chiew Y.E., DeFazio A., Greaves M.F., Green A.R., Campbell P., Birney E., Easton D.F., Chenevix-Trench G., Tan M.H., Khoo S.K., Teh B.T., Yuen S.T., Leung S.Y., Wooster R., Futreal P.A., Stratton M.R. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446(7132):153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thomas R.K., Baker A.C., Debiasi R.M., Winckler W., Laframboise T., Lin W.M., Wang M., Feng W., Zander T., MacConaill L., Lee J.C., Nicoletti R., Hatton C., Goyette M., Girard L., Majmudar K., Ziaugra L., Wong K.K., Gabriel S., Beroukhim R., Peyton M., Barretina J., Dutt A., Emery C., Greulich H., Shah K., Sasaki H., Gazdar A., Minna J., Armstrong S.A., Mellinghoff I.K., Hodi F.S., Dranoff G., Mischel P.S., Cloughesy T.F., Nelson S.F., Liau L.M., Mertz K., Rubin M.A., Moch H., Loda M., Catalona W., Fletcher J., Signoretti S., Kaye F., Anderson K.C., Demetri G.D., Dummer R., Wagner S., Herlyn M., Sellers W.R., Meyerson M., Garraway L.A. High-throughput oncogene mutation profiling in human cancer. Nat. Genet. 2007;39(3):347–351. doi: 10.1038/ng1975. [DOI] [PubMed] [Google Scholar]
- 16.Zhang J., Yang P.L., Gray N.S. Targeting cancer with small molecule kinase inhibitors. Nat. Rev. Cancer. 2009;9(1):28–39. doi: 10.1038/nrc2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rowley J.D. Letter: A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243(5405):290–293. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- 18.Mayall B.H., Carrano A.V., Moore D.H., II, Rowley J.D. Quantification by DNA-based cytophotometry of the 9q+/22q-chromosomal translocation associated with chronic myelogenous leukemia. Cancer Res. 1977;37(10):3590–3593. [PubMed] [Google Scholar]
- 19.Konopka J.B., Watanabe S.M., Witte O.N. An alteration of the human c-abl protein in K562 leukemia cells unmasks associated tyrosine kinase activity. Cell. 1984;37(3):1035–1042. doi: 10.1016/0092-8674(84)90438-0. [DOI] [PubMed] [Google Scholar]
- 20.Heisterkamp N., Stam K., Groffen J., de Klein A., Grosveld G. Structural organization of the bcr gene and its role in the Ph’ translocation. Nature. 1985;315(6022):758–761. doi: 10.1038/315758a0. [DOI] [PubMed] [Google Scholar]
- 21.Stam K., Heisterkamp N., Grosveld G., de Klein A., Verma R.S., Coleman M., Dosik H., Groffen J. Evidence of a new chimeric bcr/c-abl mRNA in patients with chronic myelocytic leukemia and the Philadelphia chromosome. N. Engl. J. Med. 1985;313(23):1429–1433. doi: 10.1056/NEJM198512053132301. [DOI] [PubMed] [Google Scholar]
- 22.Grosveld G., Hermans A., De Klein A., Bootsma D., Heisterkamp N., Groffen J. The role of the Philadelphia translocation in chronic myelocytic leukemia. Ann. N. Y. Acad. Sci. 1987;511:262–269. doi: 10.1111/j.1749-6632.1987.tb36254.x. [DOI] [PubMed] [Google Scholar]
- 23.Naldini L., Stacchini A., Cirillo D.M., Aglietta M., Gavosto F., Comoglio P.M. Phosphotyrosine antibodies identify the p210c-abl tyrosine kinase and proteins phosphorylated on tyrosine in human chronic myelogenous leukemia cells. Mol. Cell. Biol. 1986;6(5):1803–1811. doi: 10.1128/mcb.6.5.1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Comoglio P.M., Di Renzo M.F., Gaudino G. Protein tyrosine kinases associated with human malignancies. Ann. N. Y. Acad. Sci. 1987;511:256–261. doi: 10.1111/j.1749-6632.1987.tb36253.x. [DOI] [PubMed] [Google Scholar]
- 25.Anderson N.G., Maller J.L., Tonks N.K., Sturgill T.W. Requirement for integration of signals from two distinct phosphorylation pathways for activation of MAP kinase. Nature. 1990;343(6259):651–653. doi: 10.1038/343651a0. [DOI] [PubMed] [Google Scholar]
- 26.Fujioka A., Terai K., Itoh R.E., Aoki K., Nakamura T., Kuroda S., Nishida E., Matsuda M. Dynamics of the Ras/ERK MAPK cascade as monitored by fluorescent probes. J. Biol. Chem. 2006;281(13):8917–8926. doi: 10.1074/jbc.M509344200. [DOI] [PubMed] [Google Scholar]
- 27.Roskoski R., Jr The ErbB/HER family of protein-tyrosine kinases and cancer. Pharmacol. Res. 2014;79:34–74. doi: 10.1016/j.phrs.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 28.Roskoski R., Jr MEK1/2 dual-specificity protein kinases: structure and regulation. Biochem. Biophys. Res. Commun. 2012;417(1):5–10. doi: 10.1016/j.bbrc.2011.11.145. [DOI] [PubMed] [Google Scholar]
- 29.Roskoski R., Jr ERK1/2 MAP kinases: structure, function, and regulation. Pharmacol. Res. 2012;66(2):105–143. doi: 10.1016/j.phrs.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 30.Roskoski R., Jr RAF protein-serine/threonine kinases: structure and regulation. Biochem. Biophys. Res. Commun. 2010;399(3):313–317. doi: 10.1016/j.bbrc.2010.07.092. [DOI] [PubMed] [Google Scholar]
- 31.Cohen P. The origins of protein phosphorylation. Nat. Cell Biol. 2002;4(5):E127–E130. doi: 10.1038/ncb0502-e127. [DOI] [PubMed] [Google Scholar]
- 32.Steelman L.S., Pohnert S.C., Shelton J.G., Franklin R.A., Bertrand F.E., McCubrey J.A. JAK/STAT, Raf/MEK/ ERK, PI3K/Akt and BCR-ABL in cell cycle progression and leukemogenesis. Leukemia. 2004;18(2):189–218. doi: 10.1038/sj.leu.2403241. [DOI] [PubMed] [Google Scholar]
- 33.Pennisi E. Genomics. ENCODE project writes eulogy for junk DNA. Science. 2012;337(6099):1159–1161. doi: 10.1126/science.337.6099.1159. [DOI] [PubMed] [Google Scholar]
- 34.Fabbro D., Fendrich G., Guez V., Meyer T., Furet P., Mestan P., Griffin J.D., Manley P.W., Cowan-Jacob S.W. Targeted therapy with imatinib: An exception or a rule? Handbook of Experimental Pharmacology, Inhibitors of Protein Kinases and Protein Phosphoates., . 2005;167:361–389. [Google Scholar]
- 35.Druker B.J., Talpaz M., Resta D.J., Peng B., Buchdunger E., Ford J.M., Lydon N.B., Kantarjian H., Capdeville R., Ohno-Jones S., Sawyers C.L. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N. Engl. J. Med. 2001;344(14):1031–1037. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- 36.van Oosterom A.T., Judson I., Verweij J., Stroobants S., Donato di Paola E., Dimitrijevic S., Martens M., Webb A., Sciot R., Van Glabbeke M., Silberman S., Nielsen O.S., European Organisation for Research and Treatment of Cancer Soft Tissue and Bone Sarcoma Group Safety and efficacy of imatinib (STI571) in metastatic gastrointestinal stromal tumours: a phase I study. Lancet. 2001;358(9291):1421–1423. doi: 10.1016/s0140-6736(01)06535-7. [DOI] [PubMed] [Google Scholar]
- 37.Food and Drug Administration Sorafenib (Nexavar), Application number: NDA 21-923, Pharmacology Review(s). Available at https://www. accessdata.fda.gov/drugsatfda_docs/nda/2005/021923_s000_ Nexavar_PharmR.pdf
- 38.Fabbro D., Cowan-Jacob S.W., Möbitz H., Martiny-Baron G. Targeting cancer with small-molecular-weight kinase inhibitors. Methods Mol. Biol. 2012;795:1–34. doi: 10.1007/978-1-61779-337-0_1. [DOI] [PubMed] [Google Scholar]
- 39.van Erp N.P., Gelderblom H., Guchelaar H-J. Clinical pharmacokinetics of tyrosine kinase inhibitors. Cancer Treat. Rev. 2009;35(8):692–706. doi: 10.1016/j.ctrv.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 40.Rask-Andersen M., Masuram S., Schiöth H.B. The druggable genome: Evaluation of drug targets in clinical trials suggests major shifts in molecular class and indication. Annu. Rev. Pharmacol. Toxicol. 2014;54:9–26. doi: 10.1146/annurev-pharmtox-011613-135943. [DOI] [PubMed] [Google Scholar]
- 41.Fabbro D. 25 years of small molecular weight kinase inhibitors: potentials and limitations. Mol. Pharmacol., 2015 May; 87(5), 766-775, and Fischer, P.M., Approved and experimental small-molecule oncology kinase inhibitor drugs. Med. Res. Rev. 2017;37(2):314–367. doi: 10.1002/med.21409. [DOI] [PubMed] [Google Scholar]
- 42.O’Brien Z., Fallah Moghaddam M. Small molecule kinase inhibitors approved by the FDA from 2000 to 2011: a systematic review of preclinical ADME data. Expert Opin. Drug Metab. Toxicol. 2013;9(12):1597–1612. doi: 10.1517/17425255.2013.834046. [DOI] [PubMed] [Google Scholar]
- 43.Lipinski C.A., Lombardo F., Dominy B.W., Feeney P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001;46(1-3):3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 44.Veber D.F., Johnson S.R., Cheng H.Y., Smith B.R., Ward K.W., Kopple K.D. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002;45(12):2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- 45.Wenlock M.C., Austin R.P., Barton P., Davis A.M., Leeson P.D. A comparison of physiochemical property profiles of development and marketed oral drugs. J. Med. Chem. 2003;46(7):1250–1256. doi: 10.1021/jm021053p. [DOI] [PubMed] [Google Scholar]
- 46.Kaufmann A.M., Krise J.P. Lysosomal sequestration of amine-containing drugs: analysis and therapeutic implications. J. Pharm. Sci. 2007;96(4):729–746. doi: 10.1002/jps.20792. [DOI] [PubMed] [Google Scholar]
- 47.Murakami T., Yumoto R. Role of phosphatidylserine binding in tissue distribution of amine-containing basic compounds. Expert Opin. Drug Metab. Toxicol. 2011;7(3):353–364. doi: 10.1517/17425255.2011.548805. [DOI] [PubMed] [Google Scholar]
- 48.Arbuckle W., Baugh M., Belshaw S., Bennett D.J., Bruin J., Cai J., Cameron K.S., Claxton C., Dempster M., Everett K., Fradera X., Hamilton W., Jones P.S., Kinghorn E., Long C., Martin I., Robinson J., Westwood P. 1H-imidazo[4,5-c]pyridine-4-carbonitrile as cathepsin S inhibitors: separation of desired cellular activity from undesired tissue accumulation through optimization of basic nitrogen pka. Bioorg. Med. Chem. Lett. 2011;21(3):932–935. doi: 10.1016/j.bmcl.2010.12.065. [DOI] [PubMed] [Google Scholar]
- 49.Meanwell N.A. Improving drug candidates by design: a focus on physicochemical properties as a means of improving compound disposition and safety. Chem. Res. Toxicol. 2011;24(9):1420–1456. doi: 10.1021/tx200211v. [DOI] [PubMed] [Google Scholar]
- 50.Manallack D.T. The pKa distribution of drugs: application to drug discovery. Perspect. Medicin. Chem. 2007;1:25–38. [PMC free article] [PubMed] [Google Scholar]
- 51.Wan H., Ulander J. High-throughput pKa screening and prediction amenable for ADME profiling. Expert Opin. Drug Metab. Toxicol. 2006;2(1):139–155. doi: 10.1517/17425255.2.1.139. [DOI] [PubMed] [Google Scholar]
- 52.Varma M.V., Gardner I., Steyn S.J., Nkansah P., Rotter C.J., Whitney-Pickett C., Zhang H., Di L., Cram M., Fenner K.S., El-Kattan A.F. pH-Dependent solubility and permeability criteria for provisional biopharmaceutics classification (BCS and BDDCS) in early drug discovery. Mol. Pharm. 2012;9(5):1199–1212. doi: 10.1021/mp2004912. [DOI] [PubMed] [Google Scholar]
- 53.Food and Drug Administration, Washington (USA): Imatinib (Gleevec), Application number: NDA 21-335, Clinical Pharmacology and Biopharmaceutics Review(s). Available at: https://www.accessdata.fda.gov/drugsat-fda_docs/nda/2003/21-335S003_Gleevec_BioPharmr.pdf
- 54.Food and Drug Administration, Washington (USA): Gefitinib (Iressa), Application number: 21-339, Clinical Pharmacology and Biopharmaceutics Review. Available at: https://www.accessdata.fda.gov/drugsatfda_docs/nda/2003/21-399_IRESSA_Clinr.pdf
- 55.Food and Drug Administration, Washington (USA): Erlotinib (Tarceva), Application number: 21-743, Clinical Pharmacology and Biopharmaceutics Review. Available at: https://www.accessdata.fda.gov/drugsatfda_docs/nda/2004/21-743_Tarceva_biopharmr.PDF
- 56.Food and Drug Administration, Washington (USA): Sorafenib (Nexavar), Application number: NDA 21-923, Clinical Pharmacology and Biopharmaceutics Review. Available at: https://www.accessdata.fda.gov/drugsatfda_docs/nda/2005/021923_s000_Nexavar_BioPharmR.pdf
- 57.Food and Drug Administration, Washington (USA): Dasatinib (Sprycel), Application number: 21-986 & 22-072,Clinical Pharmacology & Biopharmaceutics Review(s).Available at: https://www.accessdata.fda.gov/drugsatfda_ docs/nda/2006/021986s000_Sprycel__ClinPharmR.pdf
- 58.Food & Drug Administration, Washington (USA): Sunitinib (Sutent), Application number: NDA 21-938 (GIST) & NDA 21-968 (MRCC), Clinical Pharmacology and Biopharmaceutics Review. Available at: https://www. accessdata.fda.gov/drugsatfda_docs/nda/2006/021938_S000_Sutent_BioPharmR.pdf
- 59.Food and Drug Administration https://www.accessdata.fda.gov/drugsatfda_docs/nda/ 2007/022059s000_ClinPharmR.pdf
- 60.Novartis Pharmaceuticals Canada Inc
- 61.Food and Drug Administration https://www.accessdata.fda.gov/drugsatfda_docs/nda/ 2007/022068s000_ClinPharmR.pdf
- 62.European Medicines Agency http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_Public_assessment_report/human/002154/WC500103073.pdf
- 63.Food and Drug Administration https://www.accessdata.fda.gov/drugsatfda_docs/nda/ 2009/022465s000_ClinPharmR.pdf
- 64.Food and Drug Administration https://www.accessdata.fda.gov/ drugsatfda_docs/label/2009/022465lbl.pdf
- 65.Food and Drug Administration https://www.accessdata.fda.gov/ drugsatfda_docs/nda/2011/202570Orig1s000ClinPharmR.pdf
- 66.Food and Drug Administration https://www.accessdata.fda.gov/ drugsatfda_docs/nda/2011/202192Orig1s000ClinPharmR.pdf
- 67.Food and Drug Administration https://www. accessdata.fda.gov/drugsatfda_docs/nda/2011/ 202192Orig1s-000PharmR.pdf
- 68.Food and Drug Administration https://www.accessdata.fda.gov/drugsatfda_ docs/nda/2011/022405Orig1s000ClinPharmR.pdf
- 69.Food and Drug Administration https://www.accessdata.fda.gov/drugsatfda_docs/nda/2011/ 202429Orig1s000ClinPharmR.pdf
- 70.Food and Drug Administration https://www.accessdata.fda.gov/drugsatfda_docs/ nda/2012/202324Orig1s000ClinPharmR.pdf
- 71.Food and Drug Administration https://www.accessdata.fda.gov/ drugsatfda_docs/nda/2012/203341Orig1s000ClinPharmR.pdf
- 72.Food and Drug Administration https://www. accessdata.fda.gov/ drugsatfda_docs/nda/2012/203085Orig 1s000ClinPharmR. pdf
- 73.Food and Drug Administration https://www.accessdata.fda.gov/drugsatfda_ docs/nda/2012/203214Orig1s000ClinPharmR.pdf
- 74.European Medicines Agency http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_Public_assessment_report/human/002542/WC500154697.pdf
- 75.Food and Drug Administration https://www.accessdata.fda.gov/ drugsatfda_docs/nda/2012/203756Orig1s000ClinPharmR. pdf
- 76.Food and Drug Administration https://www.accessdata.fda.gov/drugsatfda_ docs/nda/2012/203469Orig1s000ClinPharmR.pdf
- 77.Food and Drug Administration http://datastore. opentrials.net/documents/fda/0766877a666a938ae849897942db79dc40210de6.pdf
- 78.Food and Drug Administration https://www.accessdata.fda.gov/ drugsatfda_docs/nda/2013/202806Orig1s000ClinPharmR. pdf
- 79.Australian Government, Department of Health, Therapeutic Goods Administration, Symonston 2014 https://www.tga.gov.au/sites/default/files/auspar-afatinib-dimaleate-140414.pdf
- 80.Food and Drug Administration https://www.accessdata.fda.gov/drugsatfda_ docs/ nda/2013/205552Orig1s000ClinPharmR.pdf
- 81.Food and Drug Administration https://www.accessdata.fda.gov/drugsatfda_ docs/nda/2014/205755Orig1s000ClinPharmR.pdf
- 82.Food and Drug Administration https://www.ac-cessdata.fda.gov/drugsatfda_docs/nda/2014/205755Orig1s-000PharmR.pdf
- 83.Food and Drug Administration https://www.accessdata.fda.gov/drugsatfda_ docs/nda/2014/206545Orig1s000ClinPharmR.pdf
- 84.Food and Drug Administration https://www.accessdata.fda.gov/drugsatfda_ docs/nda/2014/205832Orig1s000ClinPharmR.pdf
- 85.Boehringer Ingelheim (Canada) Ltd 2016 https://www.boehringer-ingelheim.ca /sites/ca/files/documents/ofevpmen.pdf
- 86.Ruiz-Garcia A., Plotka A., Pawlak S., O’Gorman M., Kosa M., Nidadavolu S., Philips S., Wang D.D. Effect of food on the bioavailability of palbociclib 125 mg capsules in healthy volunteers. Ann. Oncol. 2014;25(Suppl. 4):iv146–iv164. [Google Scholar]
- 87.Food and Drug Administration https://www.accessdata.fda.gov/drugsatfda_ docs/nda/2015/207103Orig1s000ClinPharmR.pdf
- 88.Food and Drug Administration https://www.accessdata.fda.gov/drugsatfda_ docs/nda/2015/206947Orig1s000ClinPharmR.pdf
- 89.Food and Drug Administration https://www.accessdata.fda.gov/ drugsatfda_docs/nda/2015/206192Orig1s000ClinPharmR. pdf
- 90.Government of Canada
- 91.Moghaddam M.F., Tang Y., O’Brien Z., Richardson S.J., Bacolod M., Chaturedi P., Apuy J., Kulkarni A. A proposed screening paradigm for discovery of covalent inhibitor drugs. Drug Metab. Lett. 2014;8(1):19–30. doi: 10.2174/1872312808666140317151735. [DOI] [PubMed] [Google Scholar]
- 92.Davies B., Morris T. Physiological parameters in laboratory animals and humans. Pharm. Res. 1993;10(7):1093–1095. doi: 10.1023/a:1018943613122. [DOI] [PubMed] [Google Scholar]
- 93.Di L., Atkinson K., Orozco C.C., Funk C., Zhang H., McDonald T.S., Tan B., Lin J., Chang C., Obach R.S. In vitro-in vivo correlation for low-clearance compounds using hepatocyte relay method. Drug Metab. Dispos. 2013;41(12):2018–2023. doi: 10.1124/dmd.113.053322. [DOI] [PubMed] [Google Scholar]
- 94.Jolivette L.J., Ward K.W. Extrapolation of human pharmacokinetic parameters from rat, dog, and monkey data: Molecular properties associated with extrapolative success or failure. J. Pharm. Sci. 2005;94(7):1467–1483. doi: 10.1002/jps.20373. [DOI] [PubMed] [Google Scholar]
- 95.Evans C.A., Jolivette L.J., Nagilla R., Ward K.W. Extrapolation of preclinical pharmacokinetics and molecular feature analysis of “discovery-like” molecules to predict human pharmacokinetics. Drug Metab. Dispos. 2006;34(7):1255–1265. doi: 10.1124/dmd.105.006619. [DOI] [PubMed] [Google Scholar]
- 96.Food and Drug Administration https://www. accessdata.fda.gov/drugsatfda_docs/nda/2001/21-335_Glee-vec_pharmr_P1.pdf
- 97.Government of Canada
- 98.Government of Canada
- 99.Food and Drug Administration https://www. accessdata.fda.gov/drugsatfda_docs/nda/2011/202570Orig1s-000PharmR.pdf
- 100.Government of Canada
- 101. Food and Drug Administration, Washington (USA): Vandetinib (Zactima), Application number: 022405- Orig1s000, Pharmacology Review(s). Available at: https://www.accessdata.fda.gov/drugsatfda_docs/nda/2011/022405Orig1s000PharmR.pdf
- 102.European Medicines Agency http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_Public_assessment_report/human/002409/WC500124400.pdf
- 103.Kamath A.V., Wang J., Lee F.Y., Marathe P.H. Preclinical pharmacokinetics and in vitro metabolism of dasatinib (BMS-354825): a potent oral multi-targeted kinase inhibitor against SRC and BCR-ABL. Cancer Chemother. Pharmacol. 2008;61(3):365–376. doi: 10.1007/s00280-007-0478-8. [DOI] [PubMed] [Google Scholar]
- 104.Speed B., Bu H.Z., Pool W.F., Peng G.W., Wu E.Y., Patyna S., Bello C., Kang P. Pharmacokinetics, distribution, and metabolism of[14C]sunitinib in rats, monkeys, and humans. Drug Metab. Dispos. 2012;40(3):539–555. doi: 10.1124/dmd.111.042853. [DOI] [PubMed] [Google Scholar]
- 105.Australian Government, Department of Health, Therapeutic Goods Administration, Symonston 2013.
- 106.Food and Drug Administration https://www. accessdata.fda.gov/drugsatfda_docs/nda/2012/203341Orig-1s000PharmR.pdf
- 107.Ministry of Health, Labour and Welfare. Tokyo: Pharmaceutical and Food Safety Bureau; 2013. [Google Scholar]
- 108.European Medicines Agency 2013 http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_Public_assessment_report/human/002542/WC500154697.pdf
- 109.European Medicines Agency 2013 http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_Public_assessment_report/human/002542/WC500154697.pdf
- 110.Food and Drug Administration https://www.accessdata.fda.gov/drugsatfda_ docs/nda/2012/203469Orig1s000PharmR.pdf
- 111.Food and Drug Administration
- 112.Denton C.L., Minthorn E., Carson S.W., Young G.C., Richards-Peterson L.E., Botbyl J., Han C., Morrison R.A., Blackman S.C., Ouellet D. Concomitant oral and intravenous pharmacokinetics of dabrafenib, a BRAF inhibitor, in patients with BRAF V600 mutation-positive solid tumors. J. Clin. Pharmacol. 2013;53(9):955–961. doi: 10.1002/jcph.127. [DOI] [PubMed] [Google Scholar]
- 113.Shibata Y., Chiba M. The role of extrahepatic metabolism in the pharmacokinetics of the targeted covalent inhibitors afatinib, ibrutinib, and neratinib. Drug Metab. Dispos. 2015;43(3):375–384. doi: 10.1124/dmd.114.061424. [DOI] [PubMed] [Google Scholar]
- 114.Food and Drug Administration https://www. accessdata.fda.gov/drugsatfda_docs/nda/2014/205755Orig-1s000PharmR.pdf
- 115.Food and Drug Administration https://www.accessdata.fda.gov/drugsatfda_ docs/nda/2014/205858Orig1s000PharmR.pdf
- 116.Food and Drug Administration https://www.accessdata.fda.gov/ drugsatfda_docs/nda/2014/205832Orig1s000PharmR.pdf
- 117.Food and Drug Administration https://www.accessdata.fda.gov/ drugsatfda_docs/nda/2015/207103Orig1s000PharmR.pdf
- 118.Food and Drug Administration https://www.accessdata.fda.gov/ drugsatfda_docs/nda/2015/206947Orig1s000PharmR.pdf
- 119.Bates D.W., Spell N., Cullen D.J., Burdick E., Laird N., Petersen L.A., Small S.D., Sweitzer B.J., Leape L.L., Adverse Drug Events Prevention Study Group The costs of adverse drug events in hospitalized patients. JAMA. 1997;277(4):307–311. [PubMed] [Google Scholar]
- 120.Nolan L., O’Malley K. Prescribing for the elderly. Part I: Sensitivity of the elderly to adverse drug reactions. J. Am. Geriatr. Soc. 1988;36(2):142–149. doi: 10.1111/j.1532-5415.1988.tb01785.x. [DOI] [PubMed] [Google Scholar]
- 121.Giannoudis A., Davies A., Lucas C.M., Harris R.J., Pirmohamed M., Clark R.E. Effective dasatinib uptake may occur without human organic cation transporter 1 (hOCT1): implications for the treatment of imatinib-resistant chronic myeloid leukemia. Blood. 2008;112(8):3348–3354. doi: 10.1182/blood-2007-10-116236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.AstraZeneca Canada Inc https://www.astrazeneca.ca/ content/dam/az-ca/downloads/productinformation/I
- 123.Kenny J.R., Mukadam S., Zhang C., Tay S., Collins C., Galetin A., Khojasteh S.C. Drug-drug interaction potential of marketed oncology drugs: In vitro assessment of time-dependent cytochrome P450 inhibition, reactive metabolite formation and drug-drug interaction prediction. Pharm. Res. 2012;29(7):1960–1976. doi: 10.1007/s11095-012-0724-6. [DOI] [PubMed] [Google Scholar]
- 124.FDA Guidance for Industry 2012.
- 125.2017 http:/www.fda/gov/Drugs/Development/Approval/Process/DevelopmentResourses/DrugInteractions-Labeling/ucom080499.html
- 126.Lacombe D., Liu Y. The future of clinical research in oncology: where are we heading to? Linchuang Zhongliuxue Zazhi. 2013;2(1):9–16. doi: 10.3978/j.issn.2304-3865.2012.11.14. [DOI] [PubMed] [Google Scholar]
- 127.Gnoth M.J., Sandmann S., Engel K., Radtke M. In vitro to in vivo comparison of the substrate characteristics of sorafenib tosylate toward P-glycoprotein. Drug Metab. Dispos. 2010;38(8):1341–1346. doi: 10.1124/dmd.110.032052. [DOI] [PubMed] [Google Scholar]
- 128.Kulkarni A., Riggs J., Phan C., Bai A., Calabrese A., Shi T., Moghaddam M.F. Proposing advancement criteria for efficient DMPK triage of new chemical entities. Future Med. Chem. 2014;6(2):131–139. doi: 10.4155/fmc.13.190. [DOI] [PubMed] [Google Scholar]
- 129.Richardson S., Bai A., Katz J., Kulkarni A., Moghaddam M.F. Efficiency in Drug Discovery: Using S9 Fractions as a Gating DMPK Drug Discovery Screen. Drug Metab. Lett. 2016;10:83–90. doi: 10.2174/1872312810666160223121836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Manna J.D., Richardson S.J., Moghaddam M.F. Efficiency in Drug Discovery: Development and Implementation of an Ultra Fast Mass Spectrometric (UFast-MS) Metabolic Stability Assay Using Exact Mass TOF-MS. Bioanalysis. 9(4) doi: 10.4155/bio-2016-0187. [DOI] [PubMed] [Google Scholar]
- 131.Food and Drug Administration https://www.accessdata.fda.gov/drugsatfda_docs/nda/2013/202806Orig1s000PharmR.pdf
- 132.Choo E.F., Belvin M., Boggs J., Deng Y., Hoeflich K.P., Ly J., Merchant M., Orr C., Plise E., Robarge K., Martini J.F., Kassees R., Aoyama R.G., Ramaiya A., Johnston S.H. Preclinical disposition of GDC-0973 and prospective and retrospective analysis of human dose and efficacy predictions. Drug Metab. Dispos. 2012;40(5):919–927. doi: 10.1124/dmd.111.043778. [DOI] [PubMed] [Google Scholar]
- 133.Levêque D., Maloisel F. Clinical pharmacokinetics of imatinib mesylate. In Vivo. 2005;19(1):77–84. [PubMed] [Google Scholar]
- 134.Azzariti A., Porcelli L., Simone G.M., Quatrale A.E., Colabufo N.A., Berardi F., Perrone R., Zucchetti M., D’Incalci M., Xu J.M., Paradiso A. Tyrosine kinase inhibitors and multidrug resistance proteins: interactions and biological consequences. Cancer Chemother. Pharmacol. 2010;65(2):335–346. doi: 10.1007/s00280-009-1039-0. [DOI] [PubMed] [Google Scholar]
- 135.Mittapalli R.K., Vaidhyanathan S., Sane R., Elmquist W.F. Impact of P-glycoprotein (ABCB1) and breast cancer resistance protein (ABCG2) on the brain distribution of a novel BRAF inhibitor: vemurafenib (PLX4032). J. Pharmacol. Exp. Ther. 2012;342(1):33–40. doi: 10.1124/jpet.112.192195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kodaira H., Kusuhara H., Ushiki J., Fuse E., Sugiyama Y. Kinetic analysis of the cooperation of P-glycoprotein (P-gp/Abcb1) and breast cancer resistance protein (Bcrp/Abcg2) in limiting the brain and testis penetration of erlotinib, flavopiridol, and mitoxantrone. J. Pharmacol. Exp. Ther. 2010;333(3):788–796. doi: 10.1124/jpet.109.162321. [DOI] [PubMed] [Google Scholar]
- 137.Chen Y., Agarwal S., Shaik N.M., Chen C., Yang Z., Elmquist W.F. P-glycoprotein and breast cancer resistance protein influence brain distribution of dasatinib. J. Pharmacol. Exp. Ther. 2009;330(3):956–963. doi: 10.1124/jpet.109.154781. [DOI] [PubMed] [Google Scholar]
- 138.Breedveld P., Pluim D., Cipriani G., Wielinga P., van Tellingen O., Schinkel A.H., Schellens J.H. The effect of Bcrp1 (Abcg2) on the in vivo pharmacokinetics and brain penetration of imatinib mesylate (Gleevec): implications for the use of breast cancer resistance protein and P-glycoprotein inhibitors to enable the brain penetration of imatinib in patients. Cancer Res. 2005;65(7):2577–2582. doi: 10.1158/0008-5472.CAN-04-2416. [DOI] [PubMed] [Google Scholar]
- 139.Hamada A., Miyano H., Watanabe H., Saito H. Interaction of imatinib mesilate with human P-glycoprotein. J. Pharmacol. Exp. Ther. 2003;307(2):824–828. doi: 10.1124/jpet.103.055574. [DOI] [PubMed] [Google Scholar]
- 140.Agarwal S., Sane R., Ohlfest J.R., Elmquist W.F. The role of the breast cancer resistance protein (ABCG2) in the distribution of sorafenib to the brain. J. Pharmacol. Exp. Ther. 2011;336(1):223–233. doi: 10.1124/jpet.110.175034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Food and Drug Administration https://www.accessdata. fda.gov/drugsatfda_docs/nda/2012/203085Orig1s000PharmR.pdf
- 142.European Medicines Agency http://www.ema.europa. eu/docs/en_GB/document_library/EPAR_-_Public_assess-ment_report/human/001016/WC500036361.pdf
- 143.Food and Drug Administration https://www.accessdata.fda. gov/drugsatfda_docs/nda/2007/022059s000TOC.cfm
- 144.Food and Drug Administration https://www.accessdata. fda.gov/drugsatfda_docs/nda/2007/022068TOC.cfm
- 145.Food and Drug Administration https://www.accessdata. fda.gov/drugsatfda_docs/nda/2009/022465s000_PharmR. pdf
- 146.Food and Drug Administration https://www.accessdata. fda.gov/drugsatfda_docs/nda/2005/021923_s000_Nexavar_ PharmR.pdf