Abstract
Sirtuin 3 (SIRT3) is a major regulator of oncometabolism. Its activity or lack thereof significantly affects cellular oxidative stress, glycolytic gene expression, and tumorigenic potential. Thus, a system to accurately measure the level of SIRT3 transcriptional activation could lead to the identification of new cancer therapies and may have diagnostic applications. Here, we describe the development of a luciferase-based plasmid reporter system to measure activation of the human SIRT3 promoter. We detail the steps involved in construction of the system, including primer design, promoter fragment amplification, cloning, bacterial transformation, and mutagenesis. We validate this system in human 293T cells using the known activation of the SIRT3 promoter by the transcription factor estrogen-related receptor α, and we further show novel activation by the small molecule rapamycin, which has been used as a calorie restriction mimetic. Finally, we give an overview of the complementary molecular biology techniques that may be used to verify the results of this system.
Keywords: SIRT3, oncometabolism, promoter analysis, luciferase reporter plasmid, rapamycin
1. Introduction
Sirtuins are mammalian homologs of the yeast enzyme silent information regulator 2 (SIR2), a regulator of chromatin packing and transcription which may also be involved in the regulation of replicative lifespan (Lin, Defossez, & Guarente, 2000). The seven mammalian sirtuins possess NAD+-dependent activities including deacetylation, deacylation, and ADP-ribosylation (as reviewed in Houtkooper, Pirinen, & Auwerx, 2012). Their best-known biological functions involve helping the cell adapt to nutrient challenges and stress. For example, SIRT1 and SIRT3 levels are upregulated during calorie restriction to promote fatty acid oxidation (Cohen et al., 2004; Chakrabarti et al., 2011; Hallows et al., 2011; Hirschey et al., 2010). Recently, the sirtuins have come under close study by cancer biologists because several of them – at least SIRT3, SIRT4, and SIRT6 – function as tumor suppressors through their regulation of cellular metabolism (e.g. Kim et al., 2010; Finley et al., 2011; Jeong et al., 2013; Sebastián et al., 2012).
SIRT3 in particular seems to lie at the nexus of metabolism and tumor suppression. In addition to promoting fatty acid metabolism, SIRT3 activates superoxide dismutase 2 (SOD2), one of the cell’s main defenses against reactive oxygen species (ROS) (Tao et al., 2010). By fighting ROS, SIRT3 plays dual roles: first, it helps to promote genomic integrity (Kim et al., 2010), and second, it maintains hypoxia inducible factor 1α (HIF1α) in a destabilized state, preventing it from inducing the glycolytic gene expression and Warburg effect which are typical of cancer cells (Finley et al., 2011; Bell, Emerling, Ricoult, & Guarente, 2011). When SIRT3 activity is low, tumor formation is more likely – in fact, one copy of SIRT3 is deleted in approximately 20% of all human tumors (Finley et al., 2011). Because SIRT3 plays such an important role in metabolism and tumor suppression, it is important to develop quantitative and sensitive methods to monitor its expression.
This chapter details the construction of a luciferase-based reporter system for assaying the activity of the SIRT3 promoter. The system was inspired by previous work (Bellizzi et al., 2007) but greatly expands and improves upon it. The first part of the protocol describes cloning the SIRT3 promoter into an otherwise promoter-less luciferase reporter plasmid. The construct can then be mutagenized site-specifically to test the effects of ablating certain sequence elements. Assaying SIRT3 promoter activation with the construct involves transfecting it into cells under different conditions and measuring the activity of the luciferase reporter gene; we present two example applications to illustrate validation and use of the system, including the novel upregulation of SIRT3 expression by rapamycin, a small molecule inhibitor of the nutrient-responsive kinase mammalian target of rapamycin (mTOR) (Zoncu, Efeyan, & Sabatini, 2011). The protocol concludes by describing verification of reporter results by quantitative PCR and western blot. Though tailored specifically for SIRT3 in this chapter, these same methods could be applied to any target gene of interest.
2. Protocol for construction of reporter plasmid
2.1. Primer design
Perhaps the most crucial step in creating a reporter construct is to decide which DNA fragment to clone. Because this fragment will be bounded by the primers used to amplify it, the first step is therefore a decision about where to place the primers.
2.1.1. Place the reverse (downstream) primer
The reverse primer is more straightforward to place than the forward (upstream) primer. In most cases, place the reverse primer in the region after the transcriptional start site for the gene of interest but before the translation start codon. This will allow the construct to respond to the transcriptional machinery that normally controls the gene but instead produce the reporter enzyme after translation. The location of a gene’s transcriptional and translational start sites can be read from DNA sequence information annotated by the National Center for Biotechnology Information (NCBI), displayed via the NCBI’s own website (searchable by gene at http://www.ncbi.nlm.nih.gov/gene) or the University of California, Santa Cruz Genome Browser (http://genome.ucsc.edu/index.html).
We followed these guidelines and placed our downstream primer between the SIRT3 transcription start site and translation start codon (Table 1).
Table 1.
Location of salient features within the SIRT3-PSMD13 promoter (using the NM_001017524.2 transcript annotation for SIRT3 and NM_002817.3 transcript annotation for PSMD13 from the National Center for Biotechnology Information).
| Feature | Location relative to Sirt3 transcription start site |
|---|---|
|
| |
| PSMD13 start codon | −687 to −685 |
| First homologous base of forward primer | −682 |
| Last base of forward primer | −663 |
| PSMD13 transcription start site | −446 |
| First base of reverse primer | +8 |
| Last homologous base of reverse primer | +29 |
| Sirt3 start codon | +35 to +37 |
| First base of Bellizzi reverse primer | +150 |
| Last homologous base of Bellizzi reverse primer | +169 |
2.1.2. Place the forward (upstream) primer
Placement of the forward (upstream) primer depends on the size of the promoter and the locations of known upstream regulatory sequences. Use DNA sequence analysis algorithms such as MAPPER (http://genome.ufl.edu/mapper/) or TFSearch (http://www.cbrc.jp/research/db/TFSEARCH.html) to identify upstream regions containing potential transcription factor binding sites, and combine this information with any regulatory sequences already known to exist in the promoter. When possible, place the primer to include these regions in the amplified fragment. For genes with lengthy promoters or vague promoter boundaries, the number of kilobases between the forward and reverse primers may depend on how large a region you are able to amplify via PCR. Approximately one in ten human genes has a bidirectional promoter – that is, a short (generally less than 1 kb) stretch of non-coding DNA between the 5’ ends of two genes on opposite strands that may be functionally related and co-regulated (Lin et al., 2007). This situation establishes more definite bounds for the promoter region.
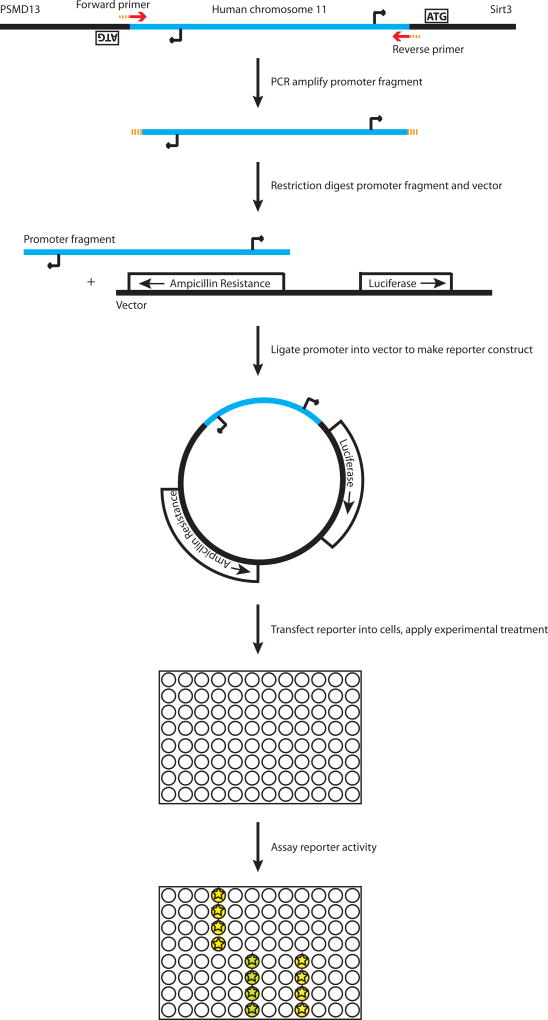
SIRT3 shares a bidirectional promoter with the proteasome subunit gene PSMD13. The intergenic sequence is not prohibitively long (only 721bp between start codons for PSMD13 and the “canonical” SIRT3 isoform, using the NCBI NC_000011.9 human chromosome 11 sequence and UniProt.org for isoform information). We therefore placed the forward primer between the PSMD13 translation start codon and transcription start site (Figure 1, Table 1). This symmetric design allowed for the amplified fragment to be used in assaying either SIRT3 or PSMD13 promoter activity depending on its orientation in the reporter vector.
Figure 1.
Overview of promoter fragment amplification, cloning into reporter plasmid, and experimental use.
2.1.3. Design the exact primer sequences
Having selected the rough locations, design the exact primer sequences using the NCBI Primer Blast tool (http://www.ncbi.nlm.nih.gov/tools/primer-blast/). This tool designs primer pairs that have minimal sequence similarity to other regions in the genome while maintaining desirable estimated hybridization properties to a given input sequence. To enable subsequent cloning, add the recognition site for a restriction enzyme to the 5’ ends of the primers, as well as an overhang of three bases to allow the restriction enzyme to bind. The forward and reverse primers may optionally be designed to have recognition sites for different restriction enzymes, which will direct the insert to adopt a definite orientation during its later ligation into the vector (ensure that the order of recognition sites in the vector’s multiple cloning site matches the planned directionality of the insert). Check that the recognition sequences do not occur in the region you plan to amplify.
For SIRT3, the NCBI Primer Blast algorithm identified a primer pair which fit the desired regions. We added a recognition site (ggtacc) for the restriction enzyme KpnI on the 5’ end of each primer, as well as a three-base (ctc) overhang. The specific primers we used for the reversible insert were thus:
Forward: 5’-ctcggtaccCAGGAAGACCCCCGGCACAG-3’
Reverse: 5’-ctcggtaccGCGCAGTCCAAGGAGTCCTCCG-3’
Their positions within the SIRT3-PSMD13 promoter are given in Table 1
We additionally made a SIRT3 reporter using the reverse primer from Bellizzi et al. (2007), which is 5’-ctcggtaccATCGTCCCTGCCGCCAAGCA-3’. This primer falls after the SIRT3 translation start codon. Transcription factor binding sites or other regulatory elements may in some instances overlap the start codon (Xi, Yu, Fu, Foley, Halees, & Weng, 2007), necessitating its inclusion in the amplified promoter fragment. In this scenario, it is important to design the reverse primer so that the reporter gene encoded by the construct remains in frame.
2.2. Promoter fragment amplification
Following primer design, the next step is to amplify the promoter fragment. This process involves generating a starting material to use as template, amplifying the promoter fragment via PCR, and verifying correct amplification.
2.2.1. Extract template genomic DNA
A single 10 cm dish or T75 flask of cultured cells that is near confluence will provide ample DNA for use as template. Collect the cells and proceed to DNA extraction. We extracted DNA from a pellet of human embryonic kidney 293T cells using a DNEasy kit (Qiagen, cat. # 69504), following manufacturer’s instructions.
2.2.2. Amplify promoter fragment via PCR
Amplify the promoter fragment using the primers designed above and the template DNA extracted in the previous step. We used a FastStart PCR Master kit (Roche, cat. # 04710444001) with 50 ng template DNA in 20 µl total volume, following manufacturer’s instructions. The recommended length of the elongation step is 1 minute for every 1 kb of DNA to be amplified (up to 3 minutes); because the SIRT3-PSMD13 intergenic promoter sequence is slightly less than 1 kb, we used a 1 minute elongation step.
2.2.3. Verify amplification
Check for successful amplification by running a small amount of PCR product (2 µl of the reaction, plus 2 µl of DNA loading dye, New England Biolabs, cat. # B7021S) on a 2% agarose gel with SYBR Safe DNA Gel Stain (Life Technologies, cat. # S33102) incorporated into the gel at a 1:10,000 dilution. Include control lanes (e.g. 5 µl of 100bp DNA ladder alongside 5 µl of 1kb DNA ladder, New England Biolabs, cat. #s N3231S and N3232S) for size comparison. Bands may be visualized by exciting the gel with blue or ultraviolet light (if using UV light, use proper shielding to limit personal UV exposure). If the PCR bands are clean and the expected size, proceed to cloning the promoter fragment into the vector.
2.2.4. Troubleshoot
If the PCR results are not clean, try one or more troubleshooting strategies. To optimize the reaction, use annealing temperatures five degrees warmer or cooler than recommended, or design entirely new primers. To control for template quality, include reactions with a pair of primers known to amplify well. If these steps do not lead to improved PCR results, then it may help to use as template a bacterial artificial chromosome (BAC) with the segment of the genome that includes the promoter of interest. BACs generally contain only a few hundred kilobases, greatly increasing the chances that primers will encounter the desired region for amplification rather than a different genomic location of similar sequence.
Though our PCR results with genomic template were usable, we found that our reactions were cleaner when using BACs as a template. We tested two BACs which contain SIRT3: CTD-2344F1 (a clone with human DNA from sperm, developed by the California Institute of Technology, available from Life Technologies) and RP11-652O18 (a clone with human male DNA from white blood cells, developed by the Roswell Park Cancer Institute, available from Life Technologies).
2.3. Inserting the promoter into the vector
After verifying that a band of the desired length is the major PCR product, insert it into the reporter vector. This process involves digesting both the PCR products and the vector plasmid with a restriction enzyme. The vector is additionally treated with alkaline phosphatase to prevent re-circularization prior to the ligation reaction, which inserts the amplified promoter fragment into the vector.
2.3.1. Purify PCR product
Remove excess primer from the PCR product by prepping the reactions with a PCR purification kit (Qiagen, cat. # 28104), eluting into 40 µl of water in the final step. This typically gives PCR products with concentrations of 100–200 ng/µl, of which only ~1% is residual template.
2.3.2. Digest PCR product with restriction enzyme
Cleave the PCR products with the restriction enzyme corresponding to the recognition sites that were added to the primers designed above. To digest the PCR products and create the insert for our SIRT3 reporter construct, we used 35.5 µl of each product and 0.5 µl of the restriction enzyme KpnI-HF (New England Biolabs, cat. # R3142S) with 4 µl of NEBuffer 4. We then incubated in a 37°C water bath for 1.5 hours. The 0.5 µl of restriction enzyme was 10 units, which was sufficient to cut 10 µg of DNA in one hour at that temperature.
2.3.3. Similarly cleave the vector
As with the PCR products, cleave the vector. We used the vector pGL3 basic (Promega, cat. # E1751). A similar vector may also be used, provided it is a promoterless vector with a luciferase reporter gene immediately following the recognition site(s) for the restriction enzyme(s) used in amplifying the promoter fragment. We cleaved with KpnI-HF, again using 10 units of enzyme with approximately 10 µg of DNA in a 37°C water bath for 1.5 hours.
2.3.4. Treat the vector with alkaline phosphatase
Treat the vector with alkaline phosphatase (from calf intestine, New England Biolabs, cat. # M0290S) to ensure that it does not self-ligate and re-circularize after digestion. An efficient way to do this is to prep the post-digest vector DNA using the same purification kit as above (Qiagen, cat. # 28104) and elute with 40 µl of water directly into a microtube containing 5 µl of the 10× buffer, 2 µl of water, and 3 µl of alkaline phosphatase (or approximately 0.5 units per µg of DNA). Then mix and incubate in a 37°C water bath for 1 hour.
2.3.5. Gel extract the PCR products and vector
To prepare pure DNA samples for ligation, run the cleaved fragments (PCR product and vector alike) on a 2% agarose gel with 1:10,000 SYBR Safe. Under blue or UV light, carefully excise the desired DNA bands (if using UV light, work quickly to avoid excessive DNA crosslinking). Extract the DNA from the bands with a gel extraction kit (Qiagen, cat. # 28704), eluting into 40 µl of water. Yields are typically ~20 ng/µl for the PCR products.
2.3.6. Perform ligation
Ligate the fragment into the vector using T4 DNA ligase (New England Biolabs, cat. # M0202S). Each reaction is 20 µl total volume and should contain 1 µl of a 1:10 dilution of the ligase and 2 µl of the 10× buffer. Use 50 ng of vector DNA with a 5-to-1 molar ratio of insert to vector and incubate at 16°C in a thermal cycler for 30 minutes. Include two control ligation reactions: one with no DNA, and one with vector DNA but no ligase. These will be important for troubleshooting in the next section. (Note that molarity of DNA need not be calculated explicitly. To calculate the ng of insert to use per ng of vector, multiply the desired 5-to-1 molar ratio by the insert/vector length ratio. In our case, the promoter fragment was only 0.13 times the length of the vector, so we used approximately 0.65 ng insert per ng vector DNA.)
2.4. Transforming the bacteria with the reporter construct
At this point, the reporter construct may be finished, but usable quantities of it must be produced in order to verify proper incorporation of the insert. This is accomplished by electroporating and growing bacteria, followed by extracting DNA from individual colonies. This section follows standard biological procedures with bacteria.
2.4.1. Electroporate
Transform electrocompetent bacteria with the reporter construct by electroporation. To do this, add 1 µl of ligation product to 60 µl of DH5α E. coli bacteria, place into an electroporation cuvette (Bio-Rad, cat. #165-2089), and electroporate with a Gene Pulser (Bio-Rad) or similar instrument. Resuspend the bacteria in 600 µl LB medium (Sigma, cat. # L7275) and incubate for one hour at 37°C. Next, spread 200 µl of each onto a 10 cm dish containing LB-agar (Sigma, cat. # L2897) and 100 µg/mL ampicillin (EMD Millipore, cat. # 171254) and at grow overnight at 37°C. Include control plates which contain bacteria treated with the no-DNA and the no-ligase control reactions from the previous step.
2.4.2. Pick and grow colonies
The next day, pick colonies from the plates to seed mini (~7 mL) LB-ampicillin cultures (final concentration of ampicillin 100 µg/mL, as in the agar plates). The control plate with no DNA should not grow colonies unless the ampicillin selection is faulty, and the control with no ligase should not grow more than a few colonies unless the vector was able to re-circularize itself efficiently, which would indicate a problem with alkaline phosphatase treatment. After seeding the cultures, grow them overnight (with shaking) at 37°C.
2.4.3. Extract DNA
After growing overnight, set aside ~500 µl of each culture at 4°C for later use. Prep the rest using a Wizard Plus SV Miniprep DNA Purification kit (Promega, cat. # A1460). Eluting each sample into 50 µl of water typically gives a yield of about 5 µg DNA, which is sufficient to test for incorporation of the insert into the clone.
2.4.4. Verify incorporation of insert by restriction digest
First, test for incorporation of the insert by cleaving 1 µg of each clone with restriction enzyme, following manufacturer’s instructions as above. Run on a 2% agarose gel (with SYBR Safe, adding loading dye to the DNA and running ladder lanes as above) and visualize. If the insert has been incorporated, the lane will have one band for the insert and one band for the vector. If the insert is not present in the clone, the vector will be the only visible band.
2.4.5. Verify proper insert sequence and orientation by sequencing
For the clones which contain an insert, verify proper sequence and proper insert orientation by sequencing. The forward and reverse cloning primers may be used as sequencing primers; additional internal primers may also be designed to obtain better read coverage of the entire insert. Because the construct will be used as template, primers can be designed with tools such as Primer3 (http://bioinfo.ut.ee/primer3/) without worrying about potential homology to other locations in the genome.
We designed and used multiple internal primers to verify the sequence of our SIRT3 constructs:
5’-GTGGGCGCCTGTGGTCGAAC-3’ (starts at position -441)
5’-TCACCGCCATCCGGGTTGAA-3’ (starts at position -277)
5’-AGGTTTGACCTCCGGGGCGA-3’ (starts at position -221)
Our sequencing runs were performed by the Dana-Farber / Harvard Cancer Center DNA Sequencing Facility and required 1 µg of template per reaction.
2.4.6. Grow larger amounts of correct construct(s)
After identifying which constructs contain the correct insert in the desired orientation, take 50 µl of the saved aliquot from these samples and seed larger (~50 mL) LB-ampicillin cultures. Grow them overnight (with shaking) at 37°C, then prep the DNA from each. We used Plasmid Plus Midi Kits (Qiagen, cat. # 12945) because the vacuum manifold allowed for quick sample preparation (although it was necessary to clear the bacterial lysate by centrifugation prior to the syringe filtering step in order to prevent the DNA-binding column from becoming overloaded with debris). We then eluted into 200 µl of elution buffer. Yields were on the order of 100 µg DNA.
2.5. Mutagenesis
To disrupt specific sequence elements by introducing site-specific mutations into the promoter construct, use the QuikChange II Site-Directed Mutagenesis Kit (Agilent, cat. # 200523). This kit uses mutagenized primers to amplify a new version of the construct in a PCR reaction followed by cleaving methylated template DNA with the restriction enzyme DpnI, leaving behind only the newly synthesized mutagenized strands which can then be used to transform bacteria.
2.5.1. Design primers
As with the original construct design, primer design is important. This depends on identifying the specific sequence element to alter and deciding upon the specific mutation(s) to introduce. Once these decisions are made, use the Agilent mutagenesis primer design tool (http://www.genomics.agilent.com/primerDesignProgram.jsp) to design primer pairs which introduce the desired mutations while adhering to the parameters of the kit.
The SIRT3 promoter contains a binding site for the transcription factor estrogen-related receptor α (ERRα), and this site plays a role in mediating induction of SIRT3 expression by the transcription factor peroxisome proliferator-activated receptor gamma coactivator 1-α (PGC-1α) (Kong et al., 2010). We created a version of our plasmid with an abrogated ERRα element by mutating the native binding site (TGCCATTG) to TGTAATTG (as Kong et al. did with murine SIRT3). We used the following mutagenic primers (with the binding site underlined):
Forward ERRα mutagenic primer: 5’-ccgcgcacttggctgtgtaattgaggcgttaaagag-3’
Reverse ERRα mutagenic primer: 5’-ctctttaacgcctcaattacacagccaagtgcgcgg-3’
2.5.2. Amplify mutagenized construct
Create a mutagenized construct by using the mutagenic primers in a PCR reaction. The manufacturer’s instructions can largely be followed, though we slightly modified the PCR protocol to better handle a large amplification product. In our mutagenesis reactions, we used a 7-minute elongation step and repeated the denaturation-annealing-elongation cycle 18 times. We then added 1 µl of DpnI restriction enzyme directly to each amplification reaction and incubated at 37°C for 1 hour to degrade the original template, and we subsequently checked for presence of amplified plasmid by running 5 µl of each reaction (mixed with 5 µl of DNA sample buffer) on a 2% agarose gel (with SYBR Safe). We ran an equal volume of a 1:50 template dilution to use as comparison.
2.5.3. Transform bacteria
To introduce the mutagenized vector into bacteria, we found that electroporation of DH5α electrocompetent cells worked more efficiently than the heat shock system described in the manufacturer’s protocol. We electroporated and then grew these bacteria in the same manner as described above for cloning the construct. We also verified introduction of the correct mutation by sequencing. Note that the internal sequencing primers designed above are especially important when high-quality sequences are needed to examine a few specific base pairs.
3. Assaying SIRT3 promoter activity with the construct
3.1. Assay protocol
The completed constructs may be used to assay promoter activity when transfected into cells growing in an opaque tissue culture plate. The assay involves transfecting the cells with the construct along with a second, constitutively expressed plasmid (to control for transfection efficiency), applying one or more treatments to the cells, waiting 24–48 hours, and measuring luciferase activity after addition of a substrate for the luciferase reporter. The resulting data indicate which treatments stimulate activity from the promoter fragment cloned into the reporter construct.
3.1.1. Seed cells
Grow cells on an opaque 96-well plate (Corning, cat. # 3917). Seeding densities will vary by cell line; for 293T cells, use 10,000 cells per well. It may be helpful to simultaneously grow cells on a clear plate (Corning, cat. # 3595) at an identical density to allow for visual inspection of population confluence and health through a normal light microscope.
3.1.2. Transfect cells
Prepare the transfection mixture by adding 0.2–0.4 µl per well of FuGene 6 transfection reagent (Promega, cat. # E2691) to 4 µl per well of serum-free medium, then adding 0.03–0.06 µg per well of construct and 0.01–0.03 µg per well of a pRL renilla control vector (used to control for cell number and transfection efficiency, Promega, cat. # E2261). The exact amounts of these two plasmids should be empirically determined for each assay in order to obtain optimal luciferase and renilla signals. (Note that the constitutive promoter of the renilla vector is quite strong and, in some cases, a larger construct/renilla ratio may be necessary than the typical values given here.) Mix according to the manufacturer’s instructions, then add to cells.
3.1.3. Add chemicals or apply other treatment
Depending on the hypothesis being tested, add chemicals, transfect additional plasmids, or apply another treatment to the cells.
3.1.4. Measure luciferase activity
24 to 48 hours after the initial transfection, assay luciferase activity using a Dual-Luciferase Reporter Assay kit (Promega, cat. # E1910). Begin by aspirating the medium from the cells, then add 25 µl per well of the included passive lysis buffer (dilute the 5× stock to a 1× working solution with distilled water). Place the cells on a shaker with gentle movement for at least 20 minutes to ensure lysis.
Following lysis, add 100 µl of the provided luciferase assay reagent to each well. Use a multichannel pipettor to do this quickly, as the luminescence signal degrades with time. Measure the luminescence of each well in a luminescence-equipped plate reader such as a Cary Varian Eclipse fluorescence spectrophotometer with a gate time of 200 ms and an emission slit of 2.5 nm around a target emission wavelength of 560 nm (the first two of these parameters may be adjusted to optimize signal capture). Next, add 100 µl per well of the provided “Stop & Glo” reagent to quench the luciferase reaction and begin the renilla luminescence. Measure the luminescence of each well again, changing the target emission wavelength from 560 nm to 500 nm.
For data analysis, divide the first (luciferase) luminescence reading by the second (renilla) luminescence reading. This provides a signal value for each well that is normalized by transfection efficiency. Average by condition to obtain a readout of the relative activity of the luciferase promoter construct under different treatments.
3.2. Example application: Importance of ERRα for SIRT3 promoter activity in 293T cells
We used the reversible SIRT3-PSMD13 promoter construct to verify the effect of ERRα binding on SIRT3 promoter activity in 293T cells. To do this, we generated both forward and reverse SIRT3 promoter reporter constructs, making versions for each with either wildtype sequence or a mutated ERRα binding site, as described above. We also used Gateway cloning techniques to generate an HA-tagged ERRα overexpression plasmid (starting from the HsCD00079872 ORF in a pDONR221 entry clone from the PlasmID database of the Dana-Farber / Harvard Cancer Center DNA Resource Core).
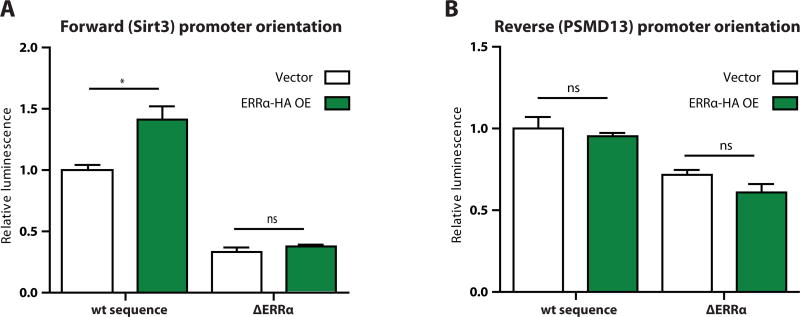
We then seeded 32 wells of an opaque 96-well plate with 10,000 293T cells per well. Cells were grown in Dulbecco’s Modified Eagle Medium (DMEM) (Life Technologies, cat. # 11995) with 10% fetal bovine serum (FBS) (HyClone, cat. # SH30910.03) and 1% penicillin-streptomycin supplement (Life Technologies, cat. #15140). The next day, we mixed and applied to each well 4 µl DMEM, 0.2 µl FuGene6 transfection reagent, 0.03 µg renilla plasmid, and 0.06 µg of a reporter plasmid construct. We additionally combined and added 4 µl DMEM, 0.2 µl FuGene6, and 0.02 µg ERRα-HA expression plasmid or vector control. Forty-eight hours later, we assayed luciferase activity as described above and saw that ERRα overexpression increased promoter activity from the wildtype sequence promoter construct in the SIRT3 direction (Figure 2A), but not from the mutated promoter in the SIRT3 direction (Figure 2A) or from either sequence variant in the PSMD13 direction (Figure 2B). This verified that the previously reported activation of the SIRT3 promoter by ERRα was exhibited by our system.
Figure 2.
Luciferase reporter results after transfection of wildtype sequence or mutated estrogen-related receptor α (ERRα) reporter constructs plus vector or ERRα-HA overexpression constructs in human 293T cells. Promoters were oriented in the A) forward or B) reverse orientation relative to SIRT3. N = 4 wells per condition.
3.3. Example application: Effect of rapamycin on SIRT3 promoter activity in 293T cells
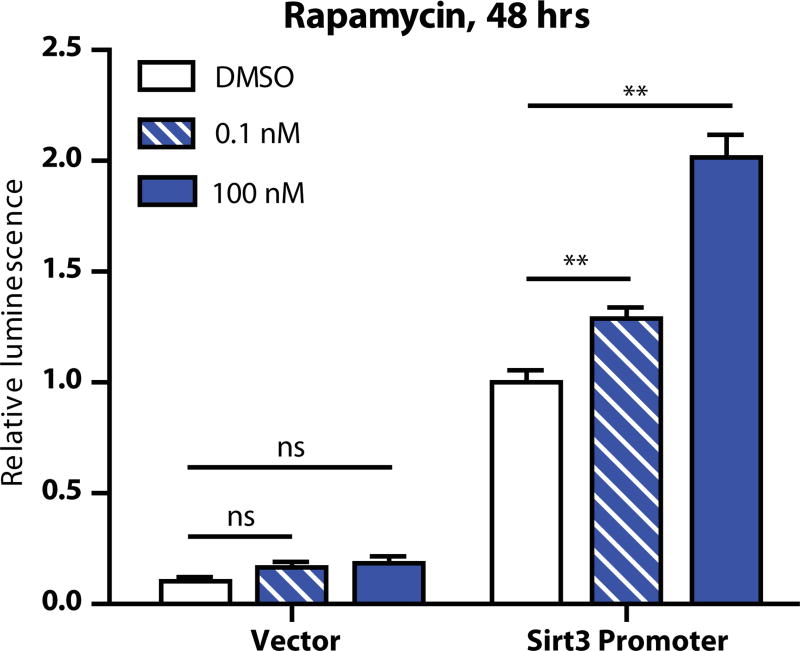
Because SIRT3 expression is known to be upregulated by calorie restriction, we also tested the responsiveness of our longer SIRT3 promoter construct to rapamycin (Sigma, cat. # R0395), a small molecule that inhibits the nutrient-sensing TOR pathway and has been used as a calorie restriction mimetic (Harrison et al., 2009; Zoncu et al., 2011). To do this, we seeded 293T cells into 24 wells of a 96-well plate at 10,000 cells per well. The next day, we mixed and applied to each well 4 µl DMEM, 0.4 µl FuGene6 transfection reagent, 0.03 µg renilla plasmid, and 0.06 µg of a reporter plasmid construct or promoterless vector control. The following day, we treated with DMSO or one of two concentrations of rapamycin, to final concentrations of 0, 0.1, or 100 nm of rapamycin in 0.01% DMSO. Forty-eight hours later we assayed luciferase activity as described above and observed that increasing rapamycin dosage led to increased activity from the SIRT3 promoter construct (Figure 3).
Figure 3.
Luciferase reporter results after transfection of the longer-form SIRT3 reporter construct or vector control plus DMSO or the indicated concentration of rapamycin. N = 4 wells per condition.
4. Verifying reporter results with complementary methods
Multiple molecular biology methods may be used to verify the results of the luciferase reporter system. Quantitative polymerase chain reaction (qPCR) is conceptually similar, as it assays promoter activity by measuring the amount of RNA transcript detected by a pair of primers specific to the gene of interest. A standard western blot can also be used to estimate sirtuin expression by measuring protein level.
4.1. Quantitative PCR
4.1.1. Quantitative PCR protocol
Begin by extracting RNA with an RNEasy Mini kit (Qiagen, cat. #74104). One well of a six-well plate (or 10 mg of tissue) is sufficient for each biological replicate. Reverse transcribe RNA into cDNA with an iScript cDNA synthesis kit (Bio-Rad, cat. # 170-8891), using 1 µg of RNA as starting material.
Quantitative PCR may be performed using 2× LightCycler 480 SYBR Green I Master mix (Roche, cat. #04707516001) on a LightCycler 480 thermal cycler (Roche). Halve the 20 µl reaction volume called for by the manufacturer’s protocol and use 5 µl of master mix, 0.5 µl each of the forward and reverse primers (at 10 µM stock concentration), and 4 µl of a 1:50 working cDNA dilution for each qPCR reaction. Run two technical replicates for each biological replicate. In addition, prepare a set of six standards, beginning with a 1:4 dilution of pooled cDNA and serially diluting 1:4 five times to create a basis for software estimation of the relative concentrations of target RNA in each sample. Run each plate through 45 amplification cycles, following manufacturer’s instructions, and end with a melting curve to verify homogeneity of the amplified DNA.
Before concentration estimates for the gene of interest in each sample can be compared to one another, they must be normalized by the levels of a reference gene within the same sample. We found that β2 microglobulin (B2M), ribosomal protein S16 (RPS16), and peptidylprolyl isomerase A (PPIA) were generally stable in expression across different treatments in human 293T cells and made good reference genes. In cases where detection of small perturbations in target gene level was important, we ran all three and used the geometric mean of their expression as reference.
4.1.2. Verification of ERRα results by qPCR
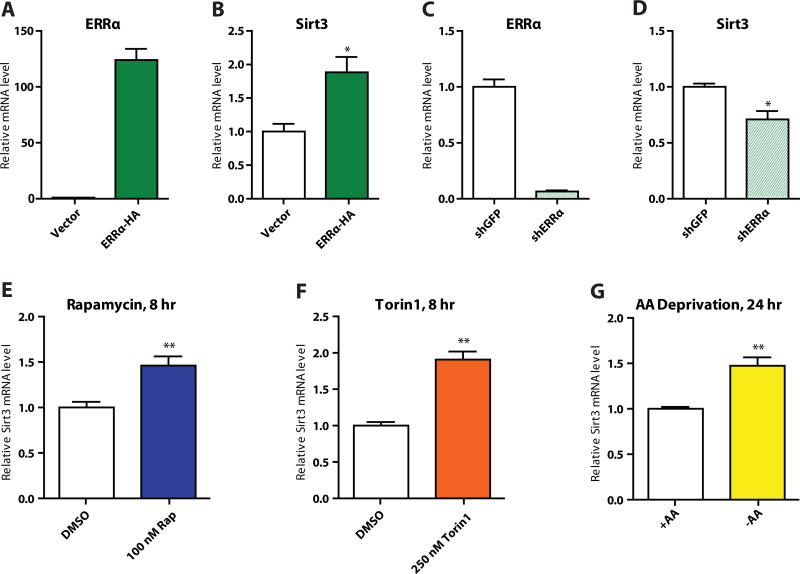
To assay SIRT3 expression in 293T cells following overexpression of the ERRα-HA construct described above, we seeded 100,000 cells per well in a six-well plate. The next day, we treated each well with a mix of 8 µl FuGene6 and 0.5 µg ERRα-HA or vector. Two days later, we extracted RNA, synthesized cDNA, and analyzed SIRT3 expression by qPCR. Consistent with our luciferase data, we saw that SIRT3 mRNA was upregulated with ERRα overexpression (Figure 4A–B). For this data, n = 3 and B2M was used as the reference gene.
Figure 4.
A) Verification of ERRα overexpression in 293T cells and B) effect on SIRT3 expression; C) verification of ERRα knockdown in 293T cells and D) effect on SIRT3 expression; induction of SIRT3 expression by E) 8-hour 100 nM rapamycin treatment, F) 8-hour 250 nM Torin1 treatment, and G) 24-hour amino acid deprivation.
We also created an ERRα knockdown 293T cell line using the shRNA construct TRCN0000330191 from the RNAi consortium (via the Dana-Farber / Harvard Cancer Center RNAi Core facility). We conducted qPCR on these cells and saw the complementary result, that ERRα knockdown led to lower SIRT3 transcript levels (Figure 4C–D). For this data, n = 3 and B2M was used as the reference gene. Primer sequences used are given in Table 2; the ERRα pair is from Schreiber, Knutti, Brogli, Uhlmann, & Kralli (2003).
Table 2.
Quantitative PCR primer sequences used in this study.
| qPCR Primer | Sequence |
|---|---|
|
| |
| Sirt3 Forward | 5’-AGCCCTCTTCATGTTCCGAAGTGT-3’ |
| Sirt3 Reverse | 5’-TCATGTCAACACCTGCAGTCCCTT-3’ |
|
| |
| ERRα Forward | 5’-AAGACAGCAGCCCCAGTGAA-3’ |
| ERRα Reverse | 5’-ACACCCAGCACCAGCACCT-3’ |
|
| |
| B2M Forward | 5’-AGATGAGTATGCCTGCCGTGTGAA-3’ |
| B2M Reverse | 5’-TGCTGCTTACATGTCTCGATCCCA-3’ |
|
| |
| RPS16 Forward | 5’-AGATCAAAGACATCCTCATCCAG-3’ |
| RPS16 Reverse | 5’-TGAGTTTTGAGTCACGATGGG-3’ |
|
| |
| PPIA Forward | 5’-AGCATACAGGTCCTGGCATCTTGT-3’ |
| PPIA Reverse | 5’-CAAAGACCACATGCTTGCCATCCA-3’ |
4.1.3. Verification of rapamycin results by qPCR
To verify the rapamycin luciferase results, we inhibited the TOR pathway in three different ways. We inhibited by treatment with rapamycin for 8 hours at 100 nM (Figure 4E); by treatment with the small molecule Torin1, which inhibits the catalytic site of mTOR (Thoreen et al., 2009), for 8 hours at 250 nM (Figure 4F); and by 24-hour amino acid deprivation (Figure 4G). In all three cases, inhibition of the TOR pathway led to greater expression of SIRT3, validating our luciferase results. In this case, n = 4, and we used the geometric mean of B2M, RPS16, and PPIA expression as a reference. Primer sequences used are given in Table 2.
4.2. Western blot
Additional verification may be performed by western blot, which looks at protein rather than mRNA levels. We used this to further verify the upregulation of SIRT3 expression by rapamycin treatment because it represented a novel finding.
4.2.1. Western blot protocol
Briefly, obtain whole cell lysate by applying 1% NP40 buffer (1% NP40 detergent with 150 mM NaCl, 50 mM Tris [pH 8], 1 mM dithiothreitol, plus 1× Roche cOmplete Mini protease inhibitor tablets [cat. # 11836170001] and 1% each of Sigma phosphatase inhibitor cocktail #s 2 [cat. # P5726] and 3 [cat. # P0044]) to a pellet of collected cells. Use 150 µl of lysis buffer for mostly confluent cells in a 35 mm well, or 300 µl for cells in a 10 cm dish (providing a more concentrated lysate). Separate the proteins by running the lysate on a polyacrylamide gel (Bio-Rad, cat. # 345-0043), transfer to nitrocellulose membrane (Bio-Rad, cat. # 162-0112), and blot for the protein of interest using standard molecular biology techniques.
4.2.2. Verification of rapamycin results by western blot
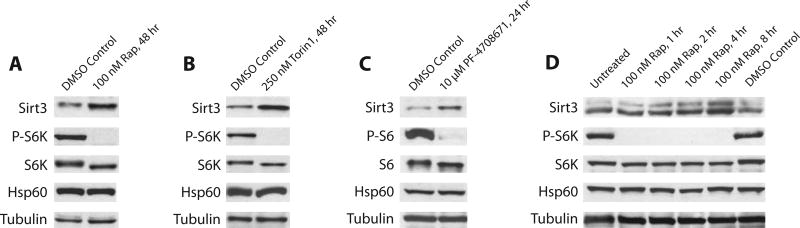
For detection of human SIRT3, we used Cell Signaling C73E3 (cat. # 2627) as the primary antibody at a dilution of 1:1000 in 3% BSA with 0.02% sodium azide (note that this antibody detects human but not mouse SIRT3, while Cell Signaling D22A3 [cat. # 5490] detects both). We observed that our western blot reproduced at the protein level the induction of SIRT3 expression that we saw at the RNA level. In 293T cells grown in DMEM with 10% FBS and 1% penicillin-streptomycin, inhibition of the TOR pathway by 100 nM rapamycin for 48 hours (Figure 5A) or 250 nM Torin1 for 48 hours (Figure 5B) led to an increase in SIRT3 expression, as did inhibition of the mTOR target p70-S6 kinase (S6K) via the S6K inhibitor PF-4708671 (Pearce et al., 2010) at 10 µM for 24 hours (Figure 5C). An 8-hour time course of 100 nM rapamycin treatment displayed gradual SIRT3 induction (Figure 5D). In these experiments we used phosphorylation of S6K (Cell Signaling, cat. # 9234, 1:1000) as a marker of mTOR pathway activation (except in the case of direct S6K inhibition, where we used phosphorylation of ribosomal protein S6 [Cell Signaling, cat. # 4856, 1:1000]). HSP60 (Abcam, cat. # ab3080, 1:5000) was used as a control for total amount of mitochondria, and the loading controls were total S6K (Cell Signaling, cat. # 9202, 1:1000) or total S6 (Cell Signaling, cat. # 2317, 1:1000) and α-tubulin (Santa Cruz Biotechnology, cat. # sc-8035, 1:5000).
Figure 5.
SIRT3 protein level after treatment with a) rapamycin, b) Torin1, or c) PF-4708671, an inhibitor of p70-S6 kinase (S6K); d) rapamycin-induced increase in SIRT3 protein level over 8 hours.
5. Summary
In this chapter we have detailed the development of a luciferase-based plasmid reporter system to measure activation of the human SIRT3 promoter, we have validated its activity, and we have shown that SIRT3 expression is upregulated by inhibition of the nutrient-sensing TOR pathway in 293T cells. Not only can this system be used with cell lines in culture, as described above, but it could also see future utilization as a diagnostic tool for use with primary tumor cell lines to assay the level of SIRT3 activation. Furthermore, future studies using such a system may identify small molecule compounds and proteins that activate or inhibit SIRT3 expression. Because SIRT3 exerts a range of metabolic effects on tumors, including fighting against the Warburg effect, the knowledge gained by implementations of this system could have important diagnostic, prognostic, and therapeutic consequences.
Acknowledgments
F. K. S. was supported by NIH Training Grant No. T32 DK007260. M. C. H. was supported by an American Cancer Society New Scholar Award and the Glenn Foundation for Medical Research.
References
- Bell EL, Emerling BM, Ricoult SJ, Guarente L. SIRT3 suppresses hypoxia inducible factor 1α and tumor growth by inhibiting mitochondrial ROS production. Oncogene. 2011;30(26):2986–96. doi: 10.1038/onc.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellizzi D, Dato S, Cavalcante P, Covello G, Di Cianni F, Passarino G, et al. Characterization of a bidirectional promoter shared between two human genes related to aging: SIRT3 and PSMD13. Genomics. 2007;89(1):143–50. doi: 10.1016/j.ygeno.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Chakrabarti P, English T, Karki S, Qiang L, Tao R, Kim J, et al. SIRT1 controls lipolysis in adipocytes via FOXO1-mediated expression of ATGL. Journal of Lipid Research. 2011;52(9):1693–701. doi: 10.1194/jlr.M014647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen HY, Miller C, Bitterman KJ, Wall NR, Hekking B, Kessler B. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305(5682):390–2. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- Finley LW, Carracedo A, Lee J, Souza A, Egia A, Zhang J, et al. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1α destabilization. Cancer Cell. 2011;19(3):416–28. doi: 10.1016/j.ccr.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallows WC, Yu W, Smith BC, Devries MK, Ellinger JJ, Someya S, et al. SIRT3 promotes the urea cycle and fatty acid oxidation during dietary restriction. Molecular Cell. 2011;41(2):139–49. doi: 10.1016/j.molcel.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460(7253):392–5. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB, et al. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464(7285):121–5. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper RH, Pirinen E, Auwerx J. Sirtuins as regulators of metabolism and healthspan. Nature Reviews Molecular Cell Biology. 2012;13(4):225–38. doi: 10.1038/nrm3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong SM, Xiao C, Finley LW, Lahusen T, Souza AL, Pierce K, et al. SIRT4 has tumor-suppressive activity and regulates the cellular metabolic response to DNA damage by inhibiting mitochondrial glutamine metabolism. Cancer Cell. 2013;23(4):450–63. doi: 10.1016/j.ccr.2013.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Patel K, Muldoon-Jacobs K, Bisht KS, Aykin-Burns N, Pennington JD, et al. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell. 2010;17(1):41–52. doi: 10.1016/j.ccr.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X, Wang R, Xue Y, Liu X, Zhang H, Chen Y, et al. Sirtuin 3, a new target of PGC-1alpha, plays an important role in the suppression of ROS and mitochondrial biogenesis. PLoS One. 2010;5(7):e11707. doi: 10.1371/journal.pone.0011707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JM, Collins PJ, Trinklein ND, Fu Y, Xi H, Myers RM, et al. Transcription factor binding and modified histones in human bidirectional promoters. Genome Research. 2007;17(6):818–27. doi: 10.1101/gr.5623407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289(5487):2126–8. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- Pearce LR, Alton GR, Richter DT, Kath JC, Lingardo L, Chapman J, et al. Characterization of PF-4708671, a novel and highly specific inhibitor of p70 ribosomal S6 kinase (S6K1) Biochemical Journal. 2010;431(2):245–55. doi: 10.1042/BJ20101024. [DOI] [PubMed] [Google Scholar]
- Sebastián C, Zwaans BM, Silberman DM, Gymrek M, Goren A, Zhong L, et al. The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell. 2012;151(6):1185–99. doi: 10.1016/j.cell.2012.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber SN, Knutti D, Brogli K, Uhlmann T, Kralli A. The transcriptional coactivator PGC-1 regulates the expression and activity of the orphan nuclear receptor estrogen-related receptor alpha (ERRalpha) Journal of Biological Chemistry. 2003;278(11):9013–8. doi: 10.1074/jbc.M212923200. [DOI] [PubMed] [Google Scholar]
- Tao R, Coleman MC, Pennington JD, Ozden O, Park SH, Jiang H, et al. SIRT3-mediated deacetylation of evolutionarily conserved lysine 122 regulates MnSOD activity in response to stress. Molecular Cell. 2010;40(6):893–904. doi: 10.1016/j.molcel.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, et al. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. Journal of Biological Chemistry. 2009;284(12):8023–32. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi H, Yu Y, Fu Y, Foley J, Halees A, Weng Z. Analysis of overrepresented motifs in human core promoters reveals dual regulatory roles of YY1. Genome Research. 2007;17(6):798–806. doi: 10.1101/gr.5754707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nature Reviews Molecular Cell Biology. 2011;12(1):21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]