Abstract
Malaria is among the most serious infectious diseases affecting humans, accounting for approximately half a million deaths annually1. Plasmodium falciparum is the causative agent of most life-threatening malaria cases. Acquired immunity to malaria is inefficient, even after repeated exposures to P. falciparum2; immune regulatory mechanisms employed by P. falciparum remain largely unclear. Here, we show that P. falciparum uses immune inhibitory receptors for immune evasion. RIFINs, products of a polymorphic multigene family comprising approximately 150–200 genes per parasite genome3, are expressed on the surface of infected erythrocytes. We found that a subset of RIFINs binds to either leucocyte immunoglobulin-like receptor B1 (LILRB1) or leucocyte-associated immunoglobulin-like receptor 1 (LAIR1). LILRB1-binding RIFINs inhibited activation of LILRB1-expressing B cells and NK cells. Furthermore, interactions between LILRB1 and P. falciparum-infected erythrocytes isolated from malaria patients were associated with severe malaria, although an extended study with larger sample sizes is required to confirm the findings. These results suggest that P. falciparum has acquired multiple RIFINs to evade the host immune system by targeting immune inhibitory receptors.
P. falciparum employs multiple strategies of immune evasion throughout its different developmental stages. Central to parasite survival is the invasion of erythrocytes, within which parasites replicate, while largely protected from immune attack. Blood-stage parasites cause malarial pathology and are the major targets of cellular and acquired antibody-mediated immunity4,5. P. falciparum-infected erythrocytes (IEs) express members of the polymorphic PfEMP1, RIFIN and STEVOR6 protein families on the IE surface. PfEMP1 bind endothelial receptors to sequester IEs from blood circulation, allowing parasites to escape splenic clearance7. RIFIN and STEVOR functions are unclear, although single members of these protein families bind uninfected erythrocytes to form so-called rosettes8 that may facilitate sequestration and evasion of immune recognition in vivo9.
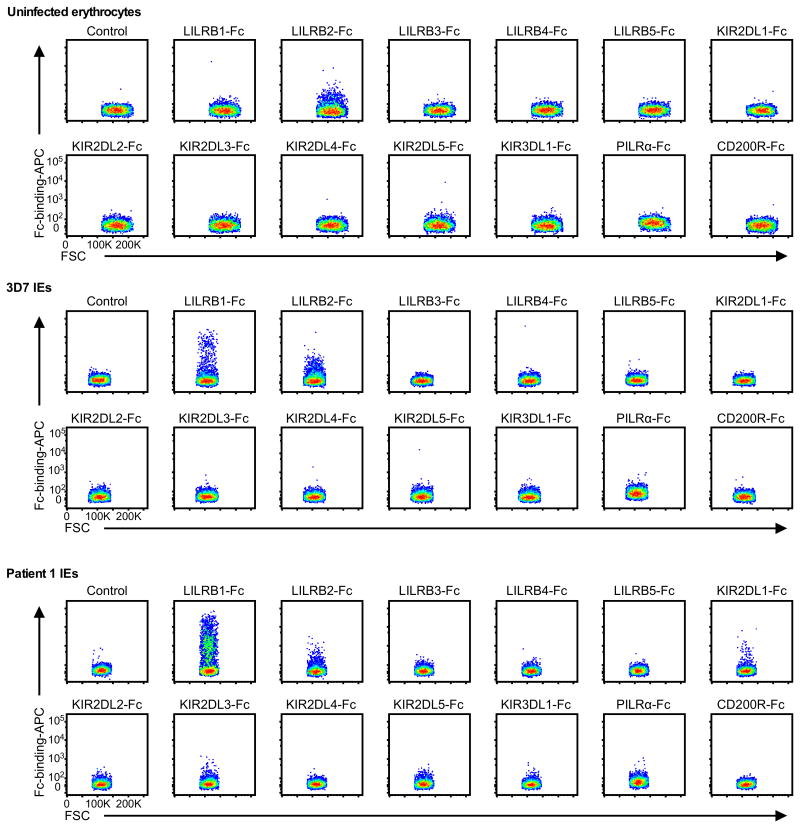
Host immune cells express diverse inhibitory receptors specific for self-antigens to down regulate autoimmune responses. Some viruses exploit these inhibitory receptors for immune evasion by mimicking corresponding ligands10. We hypothesised that P. falciparum similarly exploits inhibitory receptors by expressing parasite-derived ligands on IEs to impair immune responses. We therefore analysed the binding of 13 Fc-fused human inhibitory receptors to erythrocytes infected with the P. falciparum laboratory strain 3D7 or P. falciparum isolated from patients with malaria. In this screen, leucocyte immunoglobulin-like receptor B1 (LILRB1) was the only receptor that exhibited detectable increased binding to a small fraction of IEs (Extended Data Fig. 1).
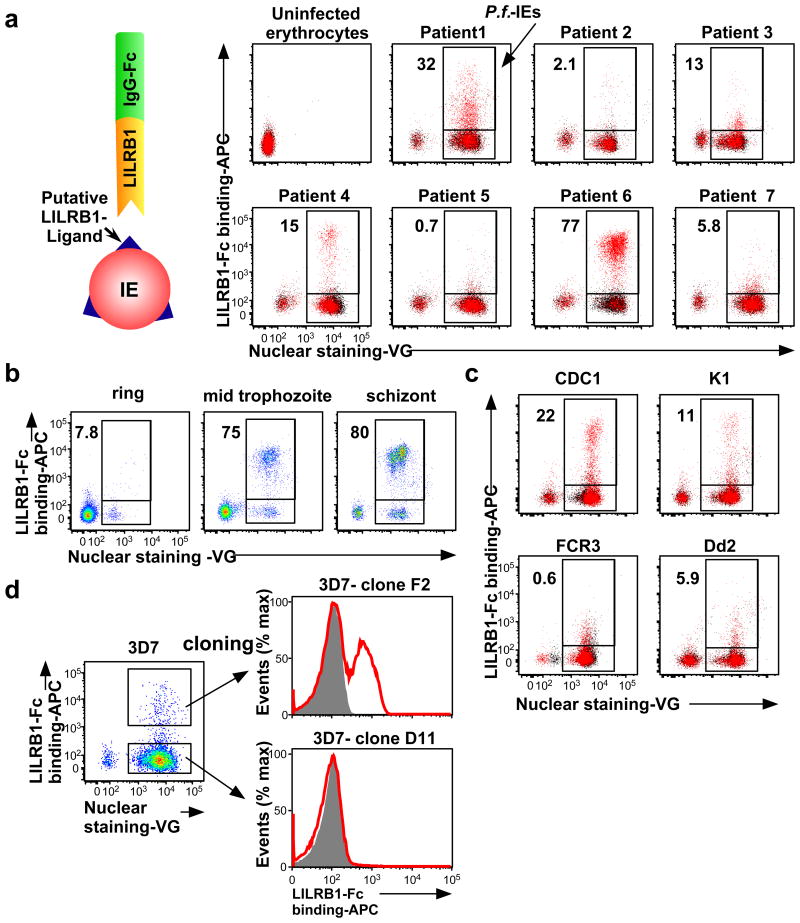
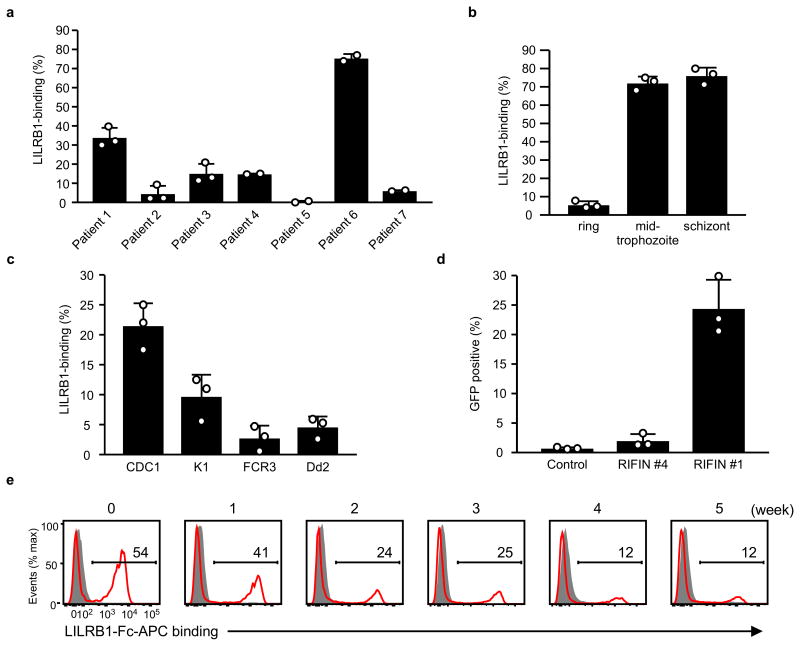
Next, we investigated whether LILRB1 bound erythrocytes infected with P. falciparum isolates acquired from seven Thai patients with malaria. We found that LILRB1 bound a large fraction (77%) of IEs from patient 6 and a small fraction from patients 1 (32%) and 4 (15%) (Fig. 1a and Extended Data Fig. 2a). LILRB1, also known as ILT2, CD85j or LIR-1, is an inhibitory receptor of the LILR family expressed on diverse immune cells and encoded by genes within the leucocyte receptor complex (LRC) region11. LILRB1, which recognises MHC I molecules12, binds the MHC class I-like UL1813 encoded byhuman cytomegalovirus, suggesting that the LILR family coevolved with microbial pathogens11,14. Binding of LILRB1 to IEs was primarily observed at the middle trophozoite and schizont stages of P. falciparum development (Fig. 1b and Extended Data Fig. 2b). LILRB1 bound to IE subpopulations of four P. falciparum laboratory strains (Fig. 1c and Extended Data Fig. 2c). Erythrocytes do not express HLA class I molecules, indicating that a parasite-derived molecule served as an LILRB1 ligand.
Figure 1. Plasmodium falciparum-infected erythrocytes (IEs) are recognised by LILRB1.
a, Analysis of IEs with an LILRB1-Fc fusion protein. Diagram of LILRB1-Fc binding to IEs. Schizont-stage P. falciparum IEs and uninfected erythrocytes were stained with LILRB1-Fc (red dot) and control-Fc (black dot), followed by Vybrant Green (VG) staining. Percentages of LILRB1-binding IEs are shown. b, Different stages of P. falciparum IEs derived from patient 6 were stained with LILRB1-Fc, followed by VG staining. Percentages of LILRB1-positive IEs are shown. c, Schizont-stage IEs from four P. falciparum laboratory strains were stained with LILRB1-Fc (red) and control-Fc (shaded), followed by VG staining. Percentages of LILRB1-binding IEs are shown. d, LILRB1-binding clone 3D7 (F2) and non-binding clone 3D7 (D11). Red and shaded histograms indicate staining with LILRB1-Fc and control-Fc, respectively. Data represent at least three independent experiments and the variabilities of data shown a, b and c are shown in Extended Data Figure 2a, 2b and 2c, respectively.
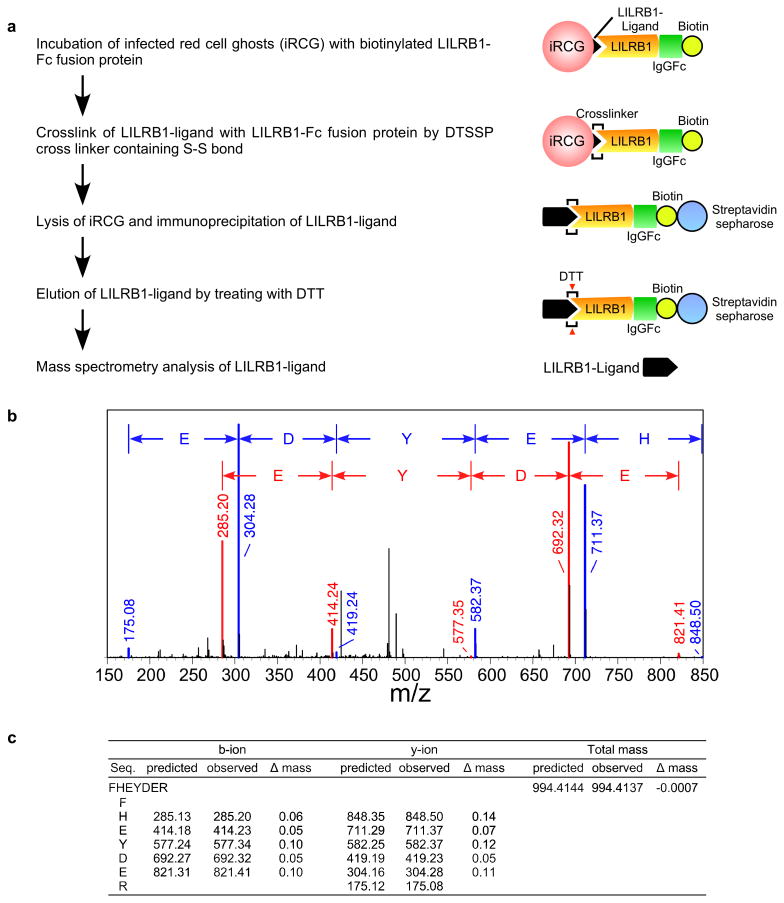
The P. falciparum genome does not encode an MHC class I-like gene15. Thus, to identify putative LILRB1 ligands on IEs, we took advantage of 3D7 parasites, for which whole-genome sequence data are available. LILRB1-Fc bound to a small proportion of 3D7 IEs. We therefore enriched 3D7 IEs for LILRB1-binding using cell sorting and subsequently subcloned 3D7 parasites using limiting dilution to obtain LILRB1-binding (F2) and non-binding (D11) parasite clones (Fig. 1d). Proportions of LILRB1-binding IEs (F2) decreased during passage after cloning (Extended Data Fig. 2e), suggesting that the expression of putative LILRB1 ligands was regulated by transcriptional switching between members of a P. falciparum multigene family. To identify putative LILRB1 ligands on IEs, we conducted mass spectrometric analyses of proteins co-precipitated with LILRB1-Fc from 3D7 F2 and D11 clones (Extended Data Fig. 3a). In two independent experiments, only one peptide (FHEYDER) matching those of RIFINs was specific to F2 immunoprecipitates (Extended Data Fig. 3b, 3c and Supplementary Table 1).
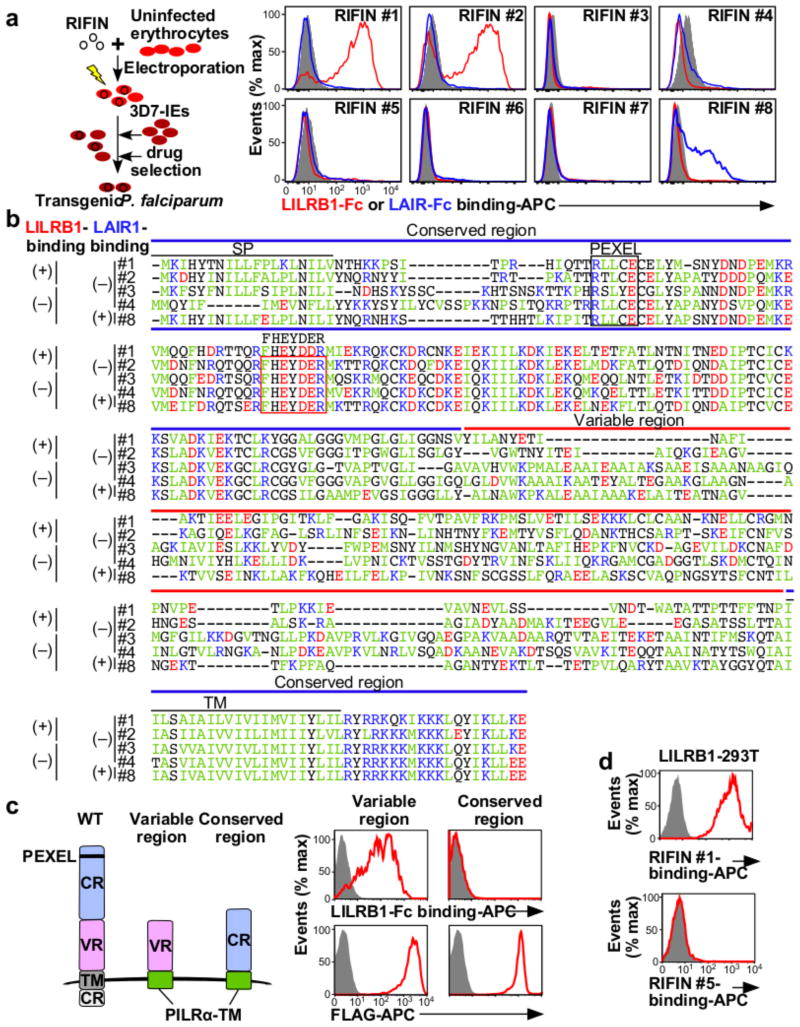
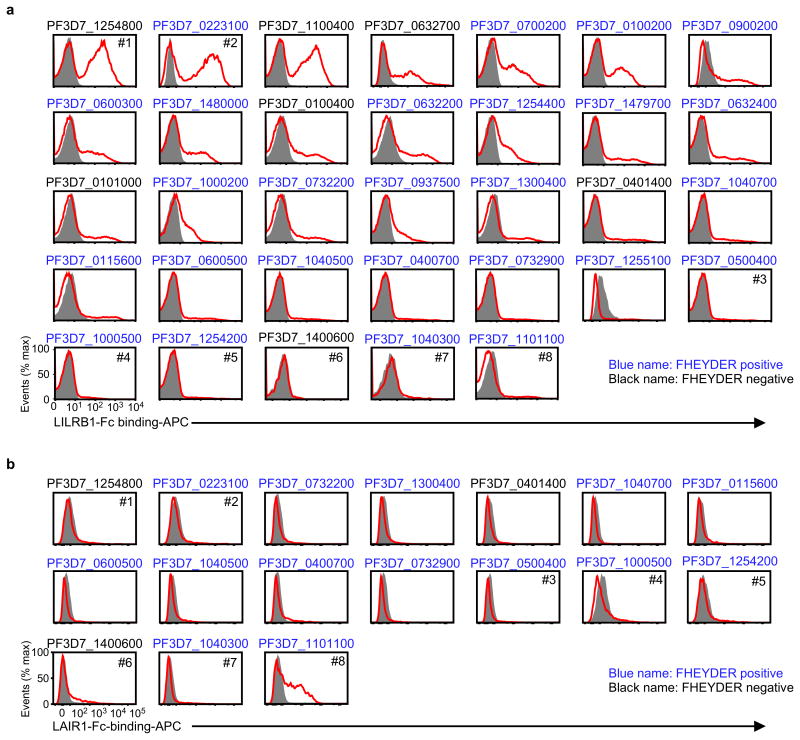
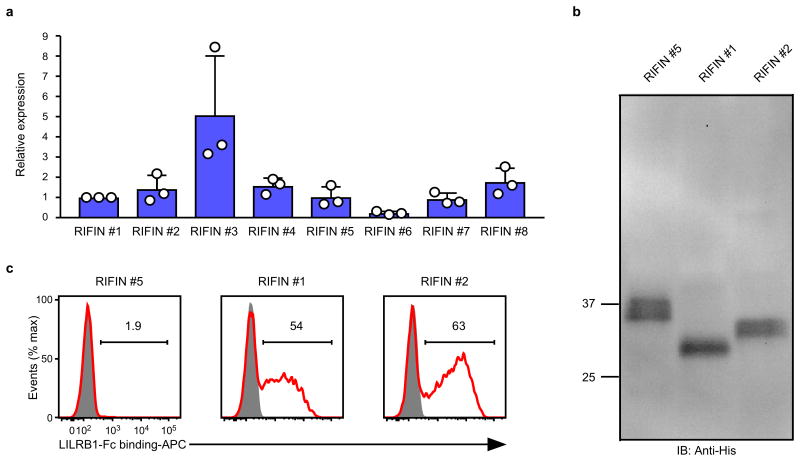
RIFINs are encoded by 150–200 rif genes per parasite genome3. The FHEYDER sequence is shared among 36 different RIFINs in the 3D7 genome. Therefore, cloning of these rif genes was performed to assess RIFIN binding to LILRB1. We cloned 25 and 6 rif genes with or without the FHEYDER sequence, respectively. Erythrocytes infected with 3D7 parasites transfected with each rif gene (Supplementary Table 2) were screened for binding to LILRB1-Fc (Extended Data Fig. 4a). IEs transfected with RIFIN #1–2 (PF3D7_1254800; PF3D7_0223100 )bound strongly to LILRB1-Fc, whereas IEs transfected with RIFINs #3–5 (PF3D7_0500400; PF3D7_1000500; PF3D7_1254200) did not (Fig. 2a and Extended Data Fig. 4a). Presence of the FHEYDER sequence in the transfected RIFINs was not associated with LILRB1-Fc binding (Fig. 2b and Extended Data Fig. 4a). Gene expression levels of LILRB1-Fc non-binding RIFINs (LILRB1− RIFINs) #3–5 were similar or higher than those of LILRB1-Fc-binding RIFINs (LILRB1+ RIFINs) #1 and #2 (Extended Data Fig. 5a). To validate the findings, we generated transgenic parasites expressing C-terminally His-tagged RIFINs #1, #2 or #5 (Extended Data Fig. 5b). Again, IEs transfected with RIFINs #1 and #2, but not RIFIN #5, bound to LILRB1-Fc (Extended Data Fig. 5c). These results indicate that a subset of RIFINs on IEs acts as LILRB1 ligands.
Figure 2. RIFINs are ligands for LILRB1 and LAIR1.
a, Diagram of the method used to generate transgenic P. falciparum. LILRB1-Fc and LAIR1-Fc bind IEs expressing RIFIN-transgenes. RIFIN-transgenic parasites are RIFIN #1: PF3D7_1254800, RIFIN #2: PF3D7_0223100, RIFIN #3: PF3D7_0500400, RIFIN #4: PF3D7_1000500, RIFIN #5: PF3D7_1254200, RIFIN #6: PF3D7_1400600, RIFIN #7: PF3D7_1040300 and RIFIN #8: PF3D7_1101100. Red, blue and shaded histograms indicate staining with LILRB1-Fc, LAIR1-Fc and control-Fc, respectively. b, Amino acid sequence alignments of RIFINs that bind LILRB1, LAIR1 or neither. SP and TM indicate the signal peptide and transmembrane domains, respectively. Green, blue, red and black characters indicate hydrophobic, basic, acidic and neutral amino acid residues, respectively. The FHEYDER sequence is contained in the red box. c, N-terminally FLAG-tagged conserved or variable regions of LILRB1+ RIFIN #1 fused to the PILRα transmembrane region (TM) expressed by 293T cells were tested for LILRB1-Fc binding (upper panel). RIFIN expression was detected using an anti-FLAG antibody (lower panel). Red and shaded histograms indicate RIFIN and mock (GFP) transfectants, respectively. d, LILRB1-transfected 293T cells were stained with His-tagged LILRB1+ RIFIN #1 or LILRB1− RIFIN #5, followed by staining with an anti-His antibody. Data represent at least three independent experiments.
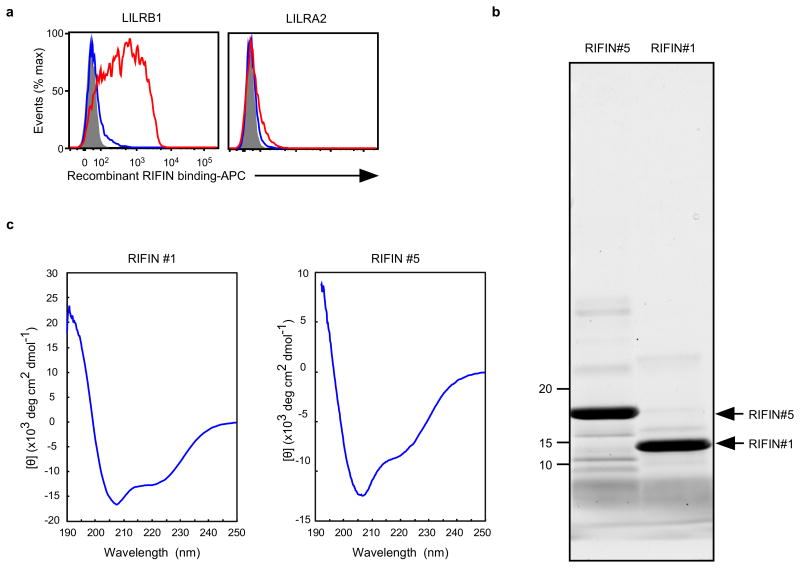
The extracellular domain of RIFINs comprises a relatively conserved N-terminal region containing the FHEYDER sequence and a highly variable C-terminal region (Fig. 2b).LILRB1-Fc bound to the variable and not the conserved region of LILRB1+ RIFIN #1 expressed on 293T cells (Fig. 2c). Further, recombinant LILRB1+ RIFIN #1 protein, but not LILRB1− RIFIN #5, bound to LILRB1 expressed on transfected 293T cells (Fig. 2d and Extended Data Fig. 6). These results indicate that RIFINs interact directly with LILRB1 through specific amino acids or conformations within their variable region.
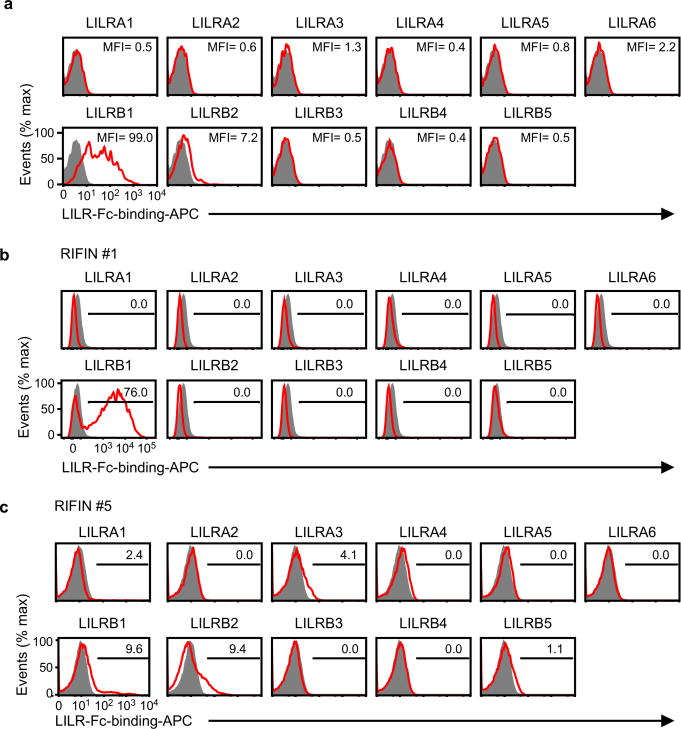
The LILR family comprises five activating, five inhibitory and one soluble form. LILRB1 shares >80% sequence identity with the activating LILRA2 receptor that recognises microbially cleaved immunoglobulins14. LILRA2 and other LILRs did not bind LILRB1+ RIFIN #1 expressed on 293T cells (Extended Data Fig. 7a). Similarly, LILRB1 was the only LILR family member that bound erythrocytes infected with RIFIN #1-transgenic parasites (Extended Data Fig. 7b). No LILR bound erythrocytes infected with RIFIN #5-transgenic parasites (Extended Data Fig. 7b). These data suggest that certain RIFINs evolved to specifically interact with LILRB1. Natural ligands for LILRB1 are HLA class I molecules12. However, LILRB1+ RIFINs or LILRB1− RIFINs do not share amino acid sequence similarities or predicted structural homology with HLA class I molecules as indicated by results of a search using Phyre2. Further, we could not identify amino acid sequence motifs characteristic of LILRB1+ RIFINs. Similar to the recent determination of the protein structure of PfEMP1-host receptor interactions16,17, solving the structure of the LILRB1–RIFIN interaction may clarify molecular constraints that confer receptor-binding specificity and antigenic diversity of RIFINs.
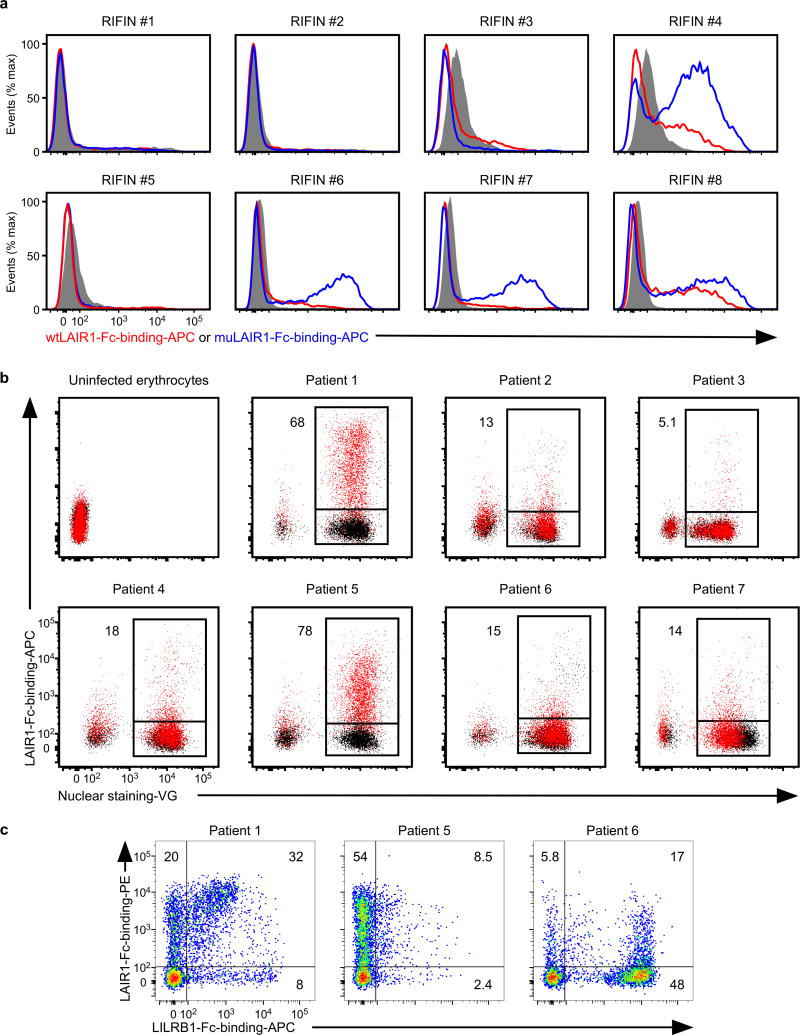
LAIR1 is an inhibitory receptor for collagens encoded within the LRC, similar to LILRB1, suggesting that these genes are evolutionarily related18. Unique antibodies isolated from individuals living in a malaria-endemic region contained a mutated LAIR1 insert that bound RIFIN #6 (PF3D7_1400600) and RIFIN #7 (PF3D7_1040300), whereas binding of wild-type LAIR1 to these RIFINs was undetectable or weak19,20. Three amino acid substitutions in the mutated LAIR1 (T67L/N69S/A77T) are responsible for its increased binding to RIFINs. Consistent with previous observations19,20, mutated LAIR1 (T67L/N69S/A77T)-Fc bound IEs from parasites transfected with RIFINs #6 and #7, but not wild-type LAIR1-Fc (Extended Data Fig. 8a). However, when analysing the binding of wild-type LAIR1 to IEs from 15 RIFIN-transgenic parasite lines (Extended Data Fig. 4b),we found that LAIR1-Fc bound to IEs from parasites transfected with LILRB1−RIFIN #8 (PF3D7_1101100) (Fig. 2a). Moreover, wild-type LAIR1-Fc bound 5%–78% of erythrocytes infected with parasites from Thai malaria patients (Extended Data Fig. 8b).
When IE binding of both LILRB1-Fc and LAIR1-Fc was analysed using parasites isolated from three patients, IEs derived from patient 1 predominantly bound LILRB1-Fc and LAIR1-Fc, whereas IEs derived from patients 5 and 6 predominantly bound LAIR1-Fc or LILRB1-Fc, respectively (Extended Data Fig. 8c). Similar to LILRB1+ RIFINs, structural homology to natural ligands for LAIR1 (collagens)21 was not predicted from our analysis of the LAIR1-binding (LAIR1+) RIFIN #8 sequence. Consistent with previous studies of RIFIN expression22,these results suggest that RIFINs are expressed on IE cell surface in a variegated manner and that P. falciparum evades immune attack through the interaction of RIFIN with different inhibitory receptors.
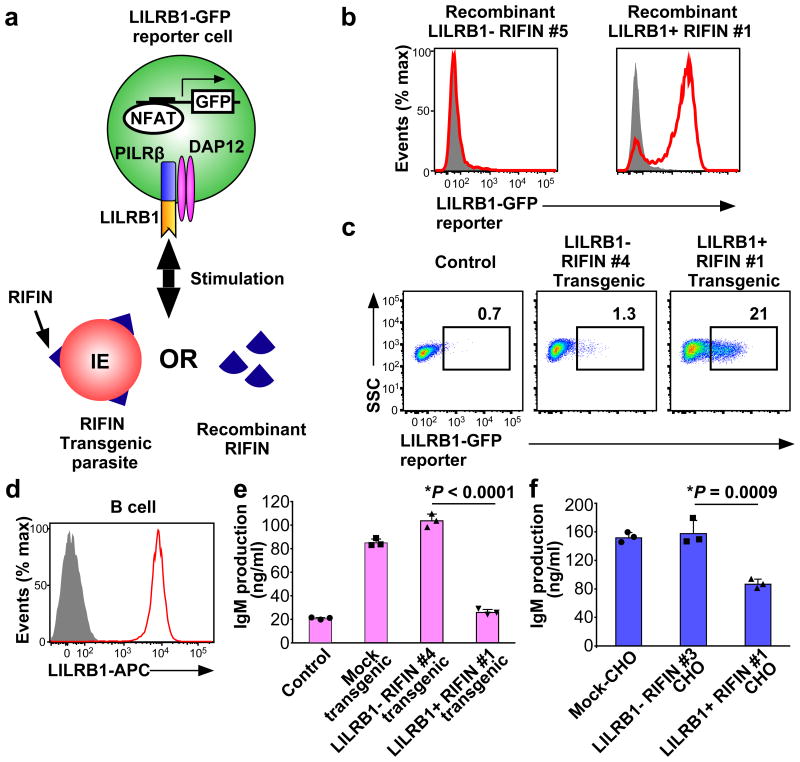
Next, we investigated whether RIFINs induced LILRB1- or LAIR1-mediated cell signalling using the nuclear factor of activated Tcells (NFAT)-GFP reporter system wherein GFP expression is induced upon ligand interaction with LILRB1 or LAIR114,23,24 (Fig. 3a). When the LILRB1-reporter cells were co-cultured with immobilised recombinant RIFINs, the cells were activated, and GFP expression was induced by LILRB1+ RIFIN #1 but not by LILRB1− RIFIN #5 (Fig. 3b). Moreover, when LILRB1-reporter cells were co-cultured with erythrocytes infected with RIFIN-transgenic parasites, LILRB1-reporter cells were activated by IEs expressing LILRB1+ RIFIN #1 but not by IEs expressing LILRB1− RIFIN #4 (Fig. 3c and Extended Data Fig. 2d). In contrast, IEs expressing LAIR1+ RIFIN #8 did not significantly stimulate LAIR1-reporter cells, whereas collagen, a host ligand for LAIR1, significantly stimulated the reporter cells (Extended Data Fig. 9a). Similarly, IEs derived from patient 1 and LAIR1+ IEs enriched from patient 4 IEs using cell sorting did not significantly stimulate LAIR1-reporter cells (Extended Data Fig.9b and 9c). These results suggest that the LAIR1–RIFIN interaction was not sufficiently strong to deliver inhibitory signals that were detectable in this assay. However, the LAIR1–RIFIN interaction may transduce an inhibitory signal via LAIR1 in other in vivo situations.
Figure 3. Inhibition of LILRB1-expressing B cells by RIFIN.
a, Diagram of the LILRB1 NFAT-GFP reporter assay. b, GFP expression in LILRB1-reporter cells upon treatment with recombinant RIFIN. Red histograms indicate treatment with recombinant LILRB1+ RIFIN#1 or LILRB1− RIFIN#5. Shaded histogram areas represent medium alone. c, GFP expression in LILRB1-reporter cells upon treatment with IEs expressing the LILRB1+ RIFIN #1 or LILRB1− RIFIN #4 transgene. Percentages of GFP-expressing cells are shown. SSC: Side-scattered light. d, Red and shaded histograms indicate staining of primary human B cells from a healthy donor with an anti-LILRB1 antibody and control, respectively. e, Inhibition of human immunoglobulin M (IgM) production in PBMCs by IEs. Human PBMCs were co-cultured with IEs, and IgM was measured in culture supernatants (mean ± s.d.). Transgenic malarial parasites expressing LILRB1+ RIFIN #1, LILRB1− RIFIN #4 or mock (GFP) are shown. Control indicates PBMCs alone. n = 3 technically independent samples. f, Inhibition of human IgM production by RIFIN-transfected CHO cells. PBMCs were co-cultured with CHO cells expressing LILRB1+ RIFIN #1, LILRB1− RIFIN #3 and an unrelated gene (MDA5). Data represent the mean ± s.d. (n = 3 technically independent samples). *P< 0.05 (one-way ANOVA with Tukey's post hoc test). Data represent at least three independent experiments, and the variability of data presented in c is shown in Extended Data Fig.2d.
LILRB1 is expressed on various immune cell types, including T cells, B cells, natural killer (NK) cells and monocytes. LILRB1 expressed by primary human B cells is involved in suppressing B cell responses25. We therefore examined the effects of RIFINs on B cells expressing LILRB1 (Fig. 3d). IEs expressing LILRB1+ RIFIN #1, but not LILRB1− RIFIN #4, inhibited IE-induced immunoglobulin M (IgM) production26 of primary human B cells present in preparations of peripheral blood mononuclear cells (PBMCs) (Fig. 3e). Similarly, CHO cells expressing LILRB1+ RIFIN #1, but not LILRB1− RIFIN #3, inhibited IgM production by PBMCs (Fig. 3f). We next investigated the effects of LILRB1+ RIFIN #1 and LILRB1− RIFIN #5 on the cytolytic function of an LILRB1-expressing human NK cell line, NKL (Extended Data Fig. 9d). K562 cells, often used as targets for NK cells, stably transfected with LILRB1+ RIFIN #1 were more resistant to NK-mediated killing than parental or K562 cells stably transfected with LILRB1−RIFIN #5, suggesting that NK cell response is down regulated by the LILRB1–RIFIN interaction (Extended Data Fig. 9e and 9f). These results indicate that malaria parasites use RIFINs to down regulate immune cells and impair anti-malarial immunity.
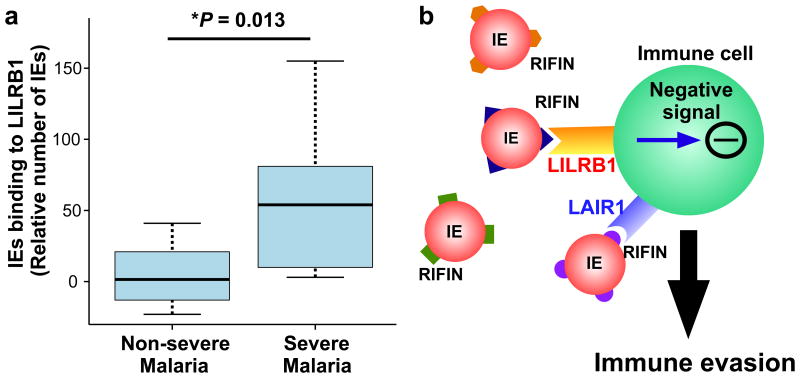
These data led us to hypothesise that IE binding to LILRB1 contributes to parasite survival during infection and is potentially associated with development of severe malaria. To evaluate this hypothesis, we analysed LILRB1 binding to IEs from Tanzanian patients with malaria. A significantly higher number of IEs from patients with severe malaria (cerebral malaria or severe malarial anaemia) bound to recombinant LILRB1-Fc compared with IEs from patients without these severe forms of malaria (Fig. 4a).The median number of IEs binding to the LILRB1-Fc from non-severe malaria patients was close to zero when compared to the control-Fc. This finding indicates that IEs from patients with severe malaria more often express LILRB1+ RIFINs and that LILRB1-binding IEs contribute to the pathogenesis of severe malaria. However, the sample size in this study was small, and further studies are required to clarify LILRB1's role in the pathogenesis of malaria.
Figure 4. Binding of Plasmodium falciparum-infected erythrocytes to LILRB1 is associated with severe malaria.
a, Binding of IEs from Tanzanian patients with malaria diagnosed with cerebral malaria, severe anaemia or both (severe malaria, n = 9) or non-cerebral or non-severe anaemia (non-severe malaria, n = 30) to LILRB1-Fc, relative to control-Fc (median, 75 and 95 percentiles and two-sided Mann–Whitney U test). b, Diagram of the suggested mechanism of immune evasion. P. falciparum induces the expression of RIFINs on the surface of IEs. Individual RIFINs may have evolved to target host inhibitory receptors, thus facilitating escape from host immune systems, which may lead to inefficient development of immunity.
Variation of P. falciparum surface antigens in vivo is governed by the host's immune status27,28. In areas with high P. falciparum transmission, immunity to severe malaria is acquired early in life and before acquisition of clinical immunity2. Thus, antibodies against RIFIN subsets associated with severe malaria may be acquired early and before antibodies to other RIFINs. Therefore, we analysed IgG reactivity with recombinant proteins derived from the RIFIN-variable regions of LILRB1+ RIFIN #1 and LILRB1− RIFIN #5 in plasma from 222 Tanzanians. IgG acquisition was similar for the two RIFINs, and as previously reported for another RIFIN29, with a very early and marked increase in the proportion of individuals responding to the RIFINs one year after birth, plateauing thereafter (Extended Data Fig. 10). Though antibody acquisition was similar for the two RIFINs, the rapid acquisition of antibodies suggests that RIFINs play an active role in early infections and may serve as important targets of protective immunity.
RIFIN-binding antibodies with LAIR1 inserted into their variable regions19, appear to be produced in response to P. falciparum infection20, potentially to inhibit the interaction between RIFINs and native LAIR1. These findings and data presented here suggest that RIFINs evolved to bind inhibitory receptors to facilitate an escape from the host immune system (Fig. 4b). RIFINs are unique to P. falciparum but belong to the Plasmodium interspersed repeats (pir) protein super family encoded by multigene families present in all human- and rodent-specific species of Plasmodium (e.g., vir of P. vivax, kir of P. knowlesi and cir/bir/yir of rodent malaria parasites30). Studies of immune receptor interactions with these polymorphic protein families may further identify crucial host–parasite interactions in the regulation of malarial infections.
Methods
Cells
Human embryonic kidney 293T and Chinese hamster ovary (CHO) cells were obtained from and authenticated by the RIKEN Cell Bank. NKL cells31 were a gift from Dr. Lanier (University of California, San Francisco). The K562 cell line was obtained from the Cell Resource Centre for Biomedical Research, Institute of Development, Ageing and Cancer, Tohoku University. PBMCs were separated from the blood of healthy donors using Ficoll density gradient centrifugation (GE Healthcare, Pittsburgh, PA, USA). Each cell line was tested regularly for Mycoplasma contamination using PCR.
Collection of P. falciparum parasites from Thai patients with malaria
All clinical P. falciparum isolates were acquired from patients living in the Mae Sariang district, Mae Hong Son Province in Thailand according to the ethical guidelines described below. Clonal parasite lines were established from field-isolated parasites using limiting dilution. All parasite strains were cultivated with human erythrocytes (type O blood, haematocrit [Ht] 2%, the Japanese Red Cross Blood Centre) in complete medium, which consisted of RPMI-1640 medium containing 20% Albu MAX I (Life Technologies), 25 mM HEPES, 0.225% sodium bicarbonate and 0.38 mM hypoxanthine supplemented with 10g/ml gentamicin. Strains were incubated in an atmosphere containing 90% N2, 5% CO2 and 5% O2.
P. falciparum culture
The P. falciparum strains 3D732, CDC133, K134, FCR334, Dd232 and parasites from Thai patients were cultured with human erythrocytes in RPMI-1640 containing 10% human serum. Transgenic parasites were maintained in RPMI-1640 containing 10% human serum and 25 ng/ml pyrimethamine (Sigma-Aldrich). Parasites were synchronised at the ring stage using 5% D-sorbitol, and trophozoite- and schizont-stage parasites were enriched using Percoll density gradient centrifugation (GE Healthcare). Parasite cultures were tested routinely for Mycoplasma contamination using PCR.
Cloning of P. falciparum 3D7 parasites
The 3D7 parasite cultures enriched for LILRB1-binding IEs were generated by sorting cells that bound LILRB1-Fc fusion protein pre-mixed with anti-IgG Fc-APC antibodies. These enriched IEs were then subjected to limiting dilution to obtain a single clone. Briefly, the enriched parasites were synchronised at the ring stage using 5% D-sorbitol. After 48 h, parasites at the late schizont stage were purified using 63% (v/v) Percoll (Amersham Pharmacia Biotech) density gradient centrifugation. The parasites in culture media (10% O-type human serum in RPMI 1640; GIBCO BRL) were mixed with freshly prepared human erythrocytes and cultured in a T-75 flask (parasitemia, 1%; haematocrit, 2%). One hour after incubation at 37°C in an atmosphere containing 5% CO2, 5% O2 and 90% N2 (CO2-O2-N2 incubator BNP-110, TABAI ESPEC Corp), the cap of the flask was tightly sealed, and the flask was shaken on an orbital shaker at 100 rpm to avoid multiple infections of a single erythrocyte. The parasite stage was monitored every 2 h using Giemsa staining, and after most parasites proceeded to the ring stage, the parasite culture was diluted using fresh culture media with erythrocytes (3% haematocrit). The diluted parasites (0.5 parasite/0.2 ml) were plated in 96-well flat bottom plates, and 50% of the culture medium was changed every 48 h. After 2 weeks in culture, parasites were detected in approximately 20 wells per plate, and LILRB1-binding positive and negative wells were screened using flow cytometric analysis. LILRB1+ F2 and LILRB1− D11 clones were obtained using this procedure.
Plasmids
Plasmids for paired immunoglobulin-like type 2 receptor (PILR)α-Fc, CD200R-Fc and LILR-Fc fusion protein production were constructed as previously described14,35,36. Plasmids for LAIR1 (GenBank: AF013249.1, amino acid residues 23-165), KIR2DL1*003 (amino acid residues 22-245), KIR2DL2*001 (amino acid residues 22-245), KIR2DL3*001 (amino acid residues 22-245), KIR2DL4*001 (amino acid residues 24-240), KIR2DL5*001 (amino acid residues 22-239) and KIR3DL1*001 (amino acid residues 22-339) were constructed in the same manner. Fc fusion proteins of these inhibitory receptors were used for analyses of binding to IEs. A plasmid encoding a mutated LAIR1 (T67L/N69S/A77T) Fc-fusion protein was generated using site-directed mutagenesis of the wild-type LAIR1 and a QuikChange Multi Site-Directed Mutagenesis Kit (Agilent Technologies). To produce a biotinylated LILR-Fc fusion protein, C-terminally AviTagged-LILR-Fc plasmids were constructed via insertion into a pCAGGS expression vector. To generate transgenic parasites, full-length genome sequence of RIFINs, including an intron, were obtained using PCR amplification of the 3D7 genome with primer sets (Supplementary Table 3) and inserted into the PfCEN5 expression vector. Each RIFIN-PfCEN5 expression vector was cloned in Escherichia coli and subjected to Sanger sequencing (Supplementary Sequence Chromatogram). Each rif sequence was compared with the 3D7 genome version 3 (release 32, PlasmoDB, http://plasmodb.org). For RIFINs that were unexpectedly cloned because of sequence similarity their 5′ and 3′ termini, nucleic acid sequences of primer annealing sites were replaced with primer sequences. However, sequences of conserved and variable regions were identical to the reference sequences. Silent and missense mutations in the cloned RIFINs are summarised in Supplementary Table 2. As a mock control, we used a PfCEN5 expression vector containing GFP. To confirm that the first intron in the rif transgenes was correctly spliced out of their IE-transcripts, cDNAs synthesised from RIFIN #1∼5 and #8-transgenic IEs were amplified using PCR with the primers (5′-TTATCCTTATTTTTTAATAACTGCC-3′ and 5′-GTTCGTGGCATTCCAC-3′), followed by direct sequencing of the amplicons. As shown in Supplementary Sequence Chromatogram, the first intron was correctly spliced out of their transcripts, resulting in in-frame transcripts. A structural homology search of RIFINs was conducted using Phyre2 (http://www.sbg.bio.ic.ac.uk/phyre2). C-terminal His-tagged RIFINs #1, #2 and #5 were constructed based on these cloned RIFIN-PfCEN5 expression vectors. Full-length RIFIN #6 (PF3D7_1400600) and RIFIN #7 (PF3D7_1040300) were synthesised by IDT and GenScript, respectively, and cloned into the PfCEN5 expression vector. For cell surface expression of RIFINs on 293T and CHO cells, the extracellular domains of RIFIN #1 (amino acid residues 166–275) and RIFIN #3 (amino acid residues 166–331) linked to the transmembrane and cytoplasmic regions of PILRα (amino acid residues 196–256)37 were cloned into a pME18s expression vector37. N-terminal conserved and C-terminal variable regions of RIFIN #1 corresponding to amino acid residues 39–139 and 166–275, respectively, were expressed with the transmembrane and cytoplasmic regions of PILRα using the pME18s expression vector, respectively. To generate recombinant RIFIN #1 (amino acid residues 166–275) and #5 (amino acid residues 168-309) using a wheat germ cell-free protein expression system, C-terminal His-tagged RIFINs were constructed by synthesising codon-optimised versions of the rif gene (https://www.eurofinsgenomics.com/en/home.aspx), followed by insertion into a pEU-E01 expression vector (CellFree Sciences). To generate recombinant RIFIN #1 (amino acid residues 166–275) and #5 (amino acid residues 168–309) in E. coli, N-terminal His-tagged RIFINs were constructed by synthesising codon-optimised versions of rif genes (IDT), followed by insertion into the pET-15b vector (N-terminal His-tagged cassette).
Transfection
The transient transfection of plasmids into 293T and CHO cells was conducted using PEI Max (Polysciences Inc.). For 293T cell surface expression, plasmids encoding target molecules and GFP were used to co-transfect 293T cells, and GFP-positive cells were analysed as previously described38. To express RIFINs on the cell surface of CHO cells, plasmids encoding fusion proteins comprising RIFINs and the transmembrane and cytoplasmic domains of mouse PILRα or mock (human melanoma differentiation-associated protein 5 (MDA5) were used to co-transfect CD8 into CHO cells. Stable transfectants of K562-RIFIN #1 (amino acid residues 166–275) and #5 (amino acid residues 168-309) were generated using retrovirus-mediated transduction with the pMxs retroviral expression vector and PLAT-E retroviral packaging cells transfected with an amphotropic envelope as previously described39.
Flow cytometry
The binding of human receptor-Fc fusion proteins to IEs was examined by staining IEs with Fc fusion proteins premixed with an allophycocyanin (APC)-conjugated anti-human IgG Fc antibody (109-136-098, Jackson Immuno Research Laboratories). For PILRα-Fc fusion protein, cells were mixed with PILRα-Fc fusion protein followed by APC conjugated anti-human IgG Fc antibody to decrease non-specific staining. For double staining of IEs with LAIR1-Fc and LILRB1-Fc fusion proteins, the LAIR1-Fc or LILRB1-Fc fusion protein was mixed with an APC-conjugated anti-human IgG Fc antibody or an phycoerythrin (PE)-conjugated anti-human IgG Fc antibody (109-116-098, Jackson Immuno Research Laboratories), respectively, followed by blocking these complexes with purified human IgG (Cappel). Thereafter, IEs were stained with these complexes. The LILRA2-Fc fusion protein was used as a control. The binding of His-tagged recombinant RIFINs to LILR-transfected 293T cells was analysed by staining with recombinant His-tagged RIFINs, followed by detection with an anti-His antibody (clone 28-75; WAKO) or an anti-DYKDDDDK (FLAG) antibody (clone L5; Biolegend). Nuclei of IEs were stained using Vybrant Green (Thermo). Populations of IEs or reporter cells were gated using FSC and SSC parameters (Supplementary gating strategy for flow cytometry analysis). Flow cytometric analyses were conducted using BD FACS Calibur, LSR II and FACS Verse flow cytometers (BD Biosciences). Data were analysed using FlowJo software (FlowJo, LLC).
Preparation of infected red cell ghosts
P. falciparum-infected red cells were incubated for 15 min at 4 °C in 40 volumes of hypotonic solution (RPMI-1640 diluted 5-fold in water). Ghost cells were collected by centrifugation at 15,000 rpm for 15 min and washed three times with a hypotonic solution.
Biotinylation of LILRB1-Fc protein and immunoprecipitation
Purified AviTag-LILRB1-Fc was biotinylated using BirA biotin-protein ligase (Avidity) (Extended Data Fig. 3a). Red cell ghosts obtained from erythrocytes infected with the LILRB1+ F2 or LILRB1− D11 clone at the schizont-stage were incubated with biotinylated LILRB1-Fc fusion protein and subsequently treated with 0.25 mM 3,3-dithiobis (sulfosuccinimidyl propionate) (DTSSP, Thermo Scientific) to generate crosslinks. Red cell ghosts treated without LILRB1-Fc were used as controls. The red cell ghosts were then washed in phosphate-buffered saline, solubilised by boiling in sample buffer in the absence of 2-mercaptoethanol and immunoprecipitated using streptavidin Sepharose (GE Healthcare). The immunoprecipitated products were eluted using 50 mM dithiothreitol (DTT) and analysed using a liquid chromatography-tandem mass spectrometer (LC-MS/MS, LTQ Orbitrap, Thermo Scientific) after tryptic digestion. All MS/MS spectra data were analysed using Mascot software (Matrix Science) with the 3D7 sequence database version 3 (PlasmoDB, http://plasmodb.org). Proteins detected in control precipitates from F2 and D11 clones and LILRB1-Fc precipitates from D11 clone were considered non-specific (Extended Data Fig. 3b, 3c and Supplementary Table 1).
Generation of transgenic parasites
Transgenic parasites were generated as previously described40. Briefly, fresh erythrocytes were transfected with the RIFIN-PfCEN5 plasmid via electroporation and subsequently infected with 3D7 parasites. Four days later, the parasites were cultured in RPMI-1640 containing pyrimethamine. Transcription of the rif transgenes was controlled by the promoter for the P. berghei elongation factor-α. These RIFIN-PfCEN5 plasmids can be maintained stably in an episomal form in the parasite during multiple rounds of cell division.
Quantitative PCR
Trophozoite-stage P. falciparum-infected erythrocytes were cultivated and subjected to RNA extraction using TRIzol (Thermo Fisher Scientific) according to the manufacturer's instructions. Contaminating genomic DNA was removed from extracted RNA using the TURBO DNA-free kit (Thermo Fisher Scientific) according to the manufacturer's instructions. The absence of detectable DNA in extracted RNA was confirmed using PCR with a primer set specific for the seryl-tRNA synthetase gene as previously described41. The cDNA was synthesised using SuperScript III (Thermo Fisher Scientific) and random hexamers according to the manufacturer's instructions. Quantitative PCR was performed for the rif transgene transcripts using Power SYBR Green Master Mix (Thermo Fisher Scientific) with primers (5′-AGGAATAAAGTGAATGGAGTCGA-3′ and 5′-AGCTTCGATCAAATGTGGTGG-3′) located within the 3′ UTR of the P. berghei heat shock protein, located downstream of the rif transgenes. The seryl-tRNA synthetase gene was used as internal control. Relative expression was calculated using theΔCT method. Briefly, the expression level of each transgene was normalised to that of the internal control using their threshold cycle (Ct) values, and then the relative expression levels of eight different transgenes were calculated, defining the average of the RIFIN#1 expression level = 1.
Recombinant proteins
Fc fusion proteins were produced in 293T cells using transient transfection and purified using protein A Sepharose (GE Healthcare) as previously described14. Recombinant RIFINs were produced in E. coli BL21 (DE3) via IPTG induction, followed by TALON metal affinity chromatography (Takara) purification and subsequent protein refolding. Briefly, inclusion bodies were obtained from IPTG-induced cells by disruption using a sonicator and were washed with 0.5% Triton X-100. The inclusion bodies were then solubilised with 6 M guanidinium hydrochloride and subjected to TALON metal affinity chromatography to purify the denatured RIFINs, which were refolded as previously described42. Final purification of the recombinant LILRB1+ RIFIN #1 (PF3D7_1254800) and LILRB1− RIFIN #5 (PF3D7_1254200) was achieved using gel filtration chromatography through a Superdex 200 Increase 10/300 GL column (GE Healthcare) equilibrated with phosphate-buffered saline. Both proteins eluted as monodisperse peaks, indicating the lack of misfolded aggregates. These proteins were analysed using SDS-PAGE and Oriole staining (Bio-Rad, Extended Data Fig. 6b and Supplementary Data). The structural integrity of the refolded RIFINs was further evaluated according to their circular dichroism (CD) spectra. Briefly, proteins were diluted to 0.2 mg/ml with 20 mM phosphate buffer (pH 7.4), and far-UV CD spectra (190–250 nm) were recorded using a Jasco J-820 spectropolarimeter (Jasco Co., Ltd.) with a 1-mm path length quartz cell at 25 °C. The spectra are expressed as the mean residue ellipticity after subtracting the solvent background. Both #1 and #5 RIFINs exhibited CD spectra typical for well-folded proteins with α-helical(208+222 nm) and β-sheet (215 nm) structures (Extended Data Fig. 6c). Prediction of the secondary structures for each RIFIN using the BeStSel server (http://bestsel.elte.hu/index.php) yielded α/β values of ∼30%/∼10% and ∼30%/∼20% for RIFINs #1 and #5, respectively. Recombinant RIFINs from the wheat germ cell-free protein expression system (CellFree Sciences) were produced as previously described43.
Reporter Assay
The LILRB1- and LAIR1- reporter cells were mouse T cell hybridomas that were stably transfected with NFAT-GFP, FLAG-tagged DAP12 and a fusion of the extracellular domain of LILRB1 or LAIR1 with the transmembrane and intracellular domains of paired immunoglobulin-like receptor β (PILRβ) as previously described24. The transmembrane and cytoplasmic domains of PILRβ were used to induce GFP via the DAP12 adaptor molecule in the reporter cells. LILRB1- and LAIR1-reporter cells were immobilised on a 96-well plate at 1 × 105 cells per well and cultured with stimulants for 16 h. Recombinant RIFINs (10 μg/ml) or Type IV collagen (10 μg/ml, Sigma-Aldrich) were immobilised on the plate before stimulation. GFP expression was analysed using flow cytometry.
IgM production by RIFIN-stimulated B cells
For stimulation with IEs, PBMCs were co-cultured with IEs at 1:100 for 16 h, washed and then cultured for 3 days. IgM levels in the supernatants were determined using an enzyme-linked immunosorbent assay (ELISA). For stimulation with RIFIN-expressing CHO cells, CD8- and RIFIN-positive CHO cells were separated using an auto MACS Pro Separator (Miltenyi Biotec) as previously described44 and co-cultured with PBMCs at 1:2. After 24 h, the PBMCs were stimulated with K3 CpG (2 μg/ml). After 3 days, IgM levels in the culture supernatants were analysed using the ELISA. MDA5 was used as an unrelatedgene (mock control).
Cytotoxicity assay
The human erythroleukemia cell line K562, which serves as a target cell for NK cell killing assays, was stably transfected with LILRB1+ RIFIN #1 or LILRB1–RIFIN #5 fused with transmembrane and cytoplasmic regions of PILR α and labelled with 15 μM Calcein AM (Thermo Fisher Scientific) in complete medium (RPMI-1640 without phenol red, supplemented with heat-inactivated 10% FCS) for 30 min at 37°C. After washing twice with complete medium, cells were adjusted to 5 × 103/100 μl. The NKL effector cells were washed twice with complete medium and mixed with target parental or rif-transfected K562 cells in a round-bottom 96-well plate with E:T ratios ranging from 50:1 to 12.5:1 in triplicate. The assay was also performed for spontaneous and maximum release (target cells in 2% Triton X-100-containing complete medium). Cells were centrifuged at 100×g for 5 min and incubated at 37°C in an atmosphere containing 5% CO2 atmosphere for 4 h. After incubation, cells were centrifuged at 1500 rpm for 2 min, and the fluorescence of the supernatants were measured using a TriStar LB941 (Berthold Technologies). Percent-specific lysis was calculated as follows, [experimental release - spontaneous release]/[maximum release - spontaneous release]× 100.
Binding assay
Parasites collected from Tanzanian paediatric patients with malaria were isolated and cultured as previously described45. Binding assays were performed using freshly isolated parasites within 24 h of culturing in petri dishes coated with LILRB1-Fc or LILRA2-Fc (as a control) as previously described46. Unbound parasites were washed away, while bound parasites were fixed with glutaraldehyde, stained with Giemsa and counted. Cerebral malaria and severe anaemia were defined according to a Blantyre coma score <3 and blood haemoglobin <5 g/dl, respectively, as previously described47. A statistical method was not used to specify sample size.
Serological analyses
For this study, 222 plasma samples from children and adults, 6 months to 60 years, were collected in Mkokola, Tanzania in 2004. Frozen samples were thawed and analysed for the presence of IgG reacting with recombinant RIFIN as previously described48. Antibody reactivity was measured in a bead-based Luminex assay, using beads coated with the variable regions of LILRB1+ RIFIN #1 (PF3D7_1254800), LILRB1− RIFIN #5 (PF3D7_1254200) and GLURP R2, which was used as a marker of exposure. The cut-off was determined according to mean reactivity +2 SD of 43 European donors never exposed to malaria.
Ethics approvals
Ethics approvals for the collection of parasite-infected blood from Thai patients were granted by the Research Ethics Committees of the Faculty of Medicine, Chiang Mai University (permission number: 187/2554) and the Department of Medicine, Mie University, Japan (permission number: 1312). The work was conducted in compliance with all relevant ethical standards and regulations governing research involving human samples. Written informed consent was obtained from all patients or the parents or guardians of paediatric patients. Erythrocytes were obtained from the Japanese Red Cross (research ID: 25J0143). Written informed consent was obtained from the parents or legal guardians of the paediatric Tanzanian patients. All patients received treatment according to national guidelines. The study protocols were reviewed and approved by the National Health Research Ethics Committee (NatHREC), hosted by the National Institute for Medical Research (NIMR) of Tanzania (reference no. NIMR/HQ/R.8c/Vol.II/436), and the methods used in this study were performed in accordance with the approved guidelines.
Data availability
Source data for Figures 3-4 and Extended Data Figures 2, 5-6, 9-10 are provided with the paper. All data are available from the corresponding author upon reasonable request.
Extended Data
Extended Data Figure 1. Binding of Fc fusion proteins of inhibitory receptors to IEs.
Erythrocytes infected with 3D7 (3D7 IEs), P. falciparum obtained from patient 1 (Patient 1 IEs), or uninfected erythrocytes were stained with Fc fusion proteins of inhibitory receptors. As a control, IEs were stained with APC-labelled anti-human IgG Fc Abalone (Control). FSC: Forward-scattered light. The experiments were replicated twice.
Extended Data Figure 2. Variability and stability of LILRB1 binding to IEs and LILRB1 reporter activity.
a, LILRB1 binding to schizont-stage P. falciparum-infected erythrocytes from patients with malaria in Figure 1a. b, LILRB1 binding to P. falciparum-infected erythrocytes at the ring, mid-trophozoite and schizont stages of P. falciparum-infected erythrocytes derived from patient 6 in Figure 1b. c, LILRB1 binding to schizont-stage P. falciparum-infected erythrocytes from the laboratory strains CDC1, K1, FCR3 and Dd2 shown in Figure 1c. d, GFP expression in LILRB1-expressing reporter cells upon stimulation with P. falciparum-infected erythrocytes in Figure 3c. Data represent the mean ± s.d. of three independent experiments. e, Proportions of LILRB1-Fc- binding erythrocytes infected with clone 3D7-F2 were analysed during 5 weeks of culture. The experiment was performed once.
Extended Data Figure 3. Identification of the LILRB1-ligand.
a, Diagram of LILRB1-ligand identification. A putative LILRB1 ligand was immunoprecipitated from IE ghosts using an LILRB1-Fc fusion protein and was identified using mass spectrometry analysis. b, Mass spectrometry of LILRB1-Fc immunoprecipitates. The observed m/z values of b-ions (red) and y-ions (blue) in the MS/MS spectra of the peptide FHEYDER present in RP-HPLC fractions of trypsin-digests of LILRB1 precipitates from IEs infected with F2 clones. The experiments were replicated twice. c, The observed m/z values of b-ions and y-ions in the MS/MS spectra of the peptide FHEYDER present in RP-HPLC fractions of trypsin-digests of LILRB1 precipitates. The predicted m/z values are shown for comparison. The differences between the m/z values for observed ions and the predicted values are shown.
Extended Data Figure 4. Screening of RIFINs that bound the LILRB1-Fc or LAIR1-Fc fusion protein.
a, IEs of 3D7 carrying RIFIN-transgenes were stained with the LILRB1-Fc fusion protein. RIFIN-transgenes are indicated in the figure. Red and shaded histograms indicate staining with LILRB1-Fc and control-Fc fusion proteins, respectively. b, IEs of 3D7 carrying RIFIN-transgenes were stained with the LAIR1-Fc fusion protein. RIFIN-transgenes are indicated in the figure. Red and shaded histograms indicate staining with LAIR1-Fc and control-Fc fusion proteins, respectively. Presence of the FHEYDER sequence in each RIFIN is indicated in the figure. Representative data from independent analyses are shown. Therefore, the proportions of IEs bound to Fc-fusion proteins and the levels of Fc-fusion protein binding IEs may not be comparable among different RIFINs. All experiments were replicated twice.
Extended Data Figure 5. Expression of RIFINs in transgenic malaria parasites.
a, The rif transgene transcript levels normalised to the internal control gene. The average of RIFIN #1 transcript levels was defined as 1. Data represent the mean ± s.d. (n = 3 technically independent samples). RIFIN #6 expression was lowest among the transgenes. However, cell surface expression of RIFIN #6 was detected using a mutated LAIR1-Fc fusion protein (Extended Data Fig. 8a), indicating that all the transgenes were sufficiently expressed at the transcript level. b, Western blot analysis of the expression of transfected C-terminally His-tagged RIFINs transfected in malaria parasites using an anti-His-tag mAb. The His-tagged RIFINs were detected at approximately equal levels (Supplementary Data). The expected molecular masses are 31.7 (RIFIN #1), 32.9 (RIFIN #2) and 34.8 (RIFIN #5) kDa. The experiment was performed once. c, P. falciparum-infected erythrocytes expressing C-terminally His-tagged RIFIN-transgenes RIFIN #1: PF3D7_1254800, RIFIN #2: PF3D7_0223100 and RIFIN #5: PF3D7_1254200 were stained with LILRB1-Fc (red) and control-Fc (shaded). The experiments were replicated at least twice.
Extended Data Figure 6. Recombinant RIFINs.
a, Binding of recombinant RIFINs to LILRB1 that was produced using a wheat germ cell-free protein expression system. 293T cells expressing transfected LILRB1 or LILRA2 were stained with recombinant His-tagged RIFINs that were produced using a wheat germ cell-free protein expression system. LILRA2 is an activating counterpart of LILRB1 and was used as a control. Red and blue histograms indicate binding of LILRB1+RIFIN #1 and LILRB1−RIFIN #5, respectively. The shaded histogram represents an unstained control. The experiments were replicated at least twice. b, Production of recombinant RIFINs in Escherichia coli.N-terminal-His-tagged variable regions of RIFINs were expressed in E. coli and purified using TALON metal-affinity chromatography. Recombinant RIFINs were analysed using SDS-PAGE and Oriole staining. LILRB1+RIFIN #1 and LILRB1−RIFIN #5 are shown on the right and left, respectively. The experiments were replicated at least twice. c, Circular dichroism (CD) spectra of recombinant RIFINs. Refolded and purified recombinant RIFIN #1 and #5 were subjected to CD spectral analysis. The spectra are shown as the mean residue ellipticity after subtracting the solvent background. RIFINs #1 and #5 exhibited CD spectra typical for well-folded proteins with α-helix (208 nm +222 nm) and β-sheet (215 nm) structures. Prediction of the secondary structures of each RIFIN using the BeStSel server (http://bestsel.elte.hu/index.php) yielded α/β values of ∼30%/∼10% and ∼30%/∼20% for RIFINs #1 and #5, respectively. The experiment was performed once.
Extended Data Figure 7. LILRB1-binding RIFIN did not bind to other LILRs.
a, The sequence encoding the variable region of LILRB1+ RIFIN #1 was transfected into 293T cells, and the transfectants were stained with LILR-Fc fusion proteins. The levels of LILR-Fc-binding are indicated in the figureas mean fluorescence intensities (MFIs). Control indicates fluorescence of cells reacted only with the secondary antibody. The experiment was performed once. b and c, Binding of LILR-Fc fusion proteins to IEs. RIFIN #1- and RIFIN #5-transgenic IEs were stained with 11 LILR-Fc fusion proteins and analysed using flow cytometry. Control indicates fluorescence of cells reacted only with the secondary antibody. The proportions of IEs stained with LILR-Fc fusion proteins are shown. The experiment in b was replicated twice and experiment in c was performed once.
Extended Data Figure 8. Binding of wild-type and mutated LAIR1 to IEs.
a, IEs from the RIFIN-transgenic parasites RIFIN #1: PF3D7_1254800, RIFIN #2: PF3D7_0223100, RIFIN #3: PF3D7_0500400, RIFIN #4: PF3D7_1000500, RIFIN #5: PF3D7_1254200, RIFIN #6: PF3D7_1400600, RIFIN #7: PF3D7_1040300 and RIFIN #8: PF3D7_1101100 were stained with wild-type LAIR1-Fc (wtLAIR1-Fc, red), mutated LAIR1-Fc (muLAIR1-Fc, blue), and control-Fc (shaded histogram) fusion proteins. The experiments were replicated twice. b, LAIR1-Fc bound to erythrocytes infected with P. falciparum derived from Thai patients with malaria. Schizont-stage erythrocytes infected with P. falciparum from patients with malaria and uninfected erythrocytes were stained with LAIR1-Fc (red dot) and control-Fc (black dot) fusion proteins, followed by Vybrant Green(VG) staining. Percentages of LAIR1-ligand-expressing IEs are shown. The experiments were replicated at least twice. c, Patterns of LILRB1 and LAIR1 binding to IEs derived from Thai patients with malaria. Schizont-stage IEs derived from Thai patients with malaria were stained with LAIR1-Fc (vertical) and LILRB1-Fc (horizontal) fusion proteins, followed by VG staining. VG-positive cells were analysed. The percentages of LAIR1-ligand single-positive, LILRB1-ligand single-positive and LAIR- and LILRB1-ligand double-positive IEs. The experiments were replicated twice.
Extended Data Figure 9. Functional analysis of cells expressing LAIR1 and LILRB1.
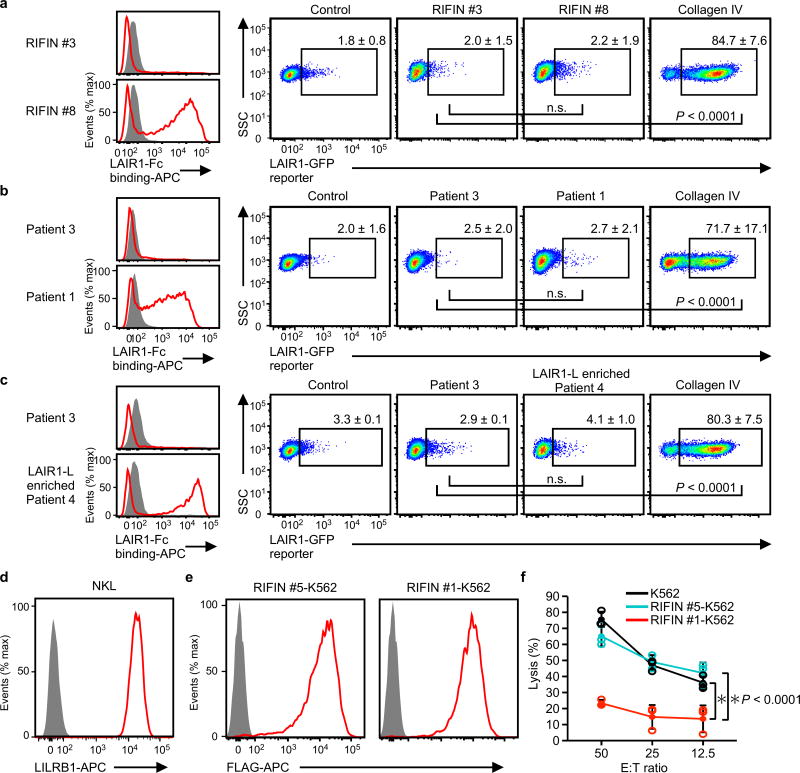
a–c, Erythrocytes infected with LAIR1-Fc-binding parasites (RIFIN #8 transgenic parasites [a], parasites from Thai patient 1 [b], LAIR1-Fc-binding parasites enriched by cell sorting from Thai patient isolate 4 [LAIR1-L enriched patient 4, c]) or erythrocytes infected with parasites that did not bind LAIR1-Fc (RIFIN #3 transgenic parasites [a], parasites from Thai malaria patient 3 [b, c]) were prepared (left histogram). LAIR1-reporter cells were co-cultured with these IEs and their expression levels in reporter cells were analysed using flow cytometry (right dot plots). Immobilised collagen IV served as a positive control for LAIR1-reporter activation. Proportions of GFP-expressing cells are shown as the mean ± s.d. (n = 3 biologically independent samples, one-way ANOVA with Tukey's post hoc test).d, LILRB1 expressed by the NK cell line NKL. e, FLAG-tagged RIFINs were expressed by K562 cells stably transfected with LILRB1+ RIFIN #1 or LILRB1− RIFIN #5. f, RIFIN-K562 cells or parental K562 cells were used as targets for NKL. The E:T ratio indicates the Effector: Target ratio. Data represent the mean ± s.d. (n = 3 technically independent samples). *P < 0.05, two-way ANOVA with Tukey's post hoc test. SSC: Side-scattered light. The experiments in d and e were replicated at least twice.
Extended Data Figure 10. Age dependence of the antibody response to LILRB1+ and LILRB1− RIFINs.
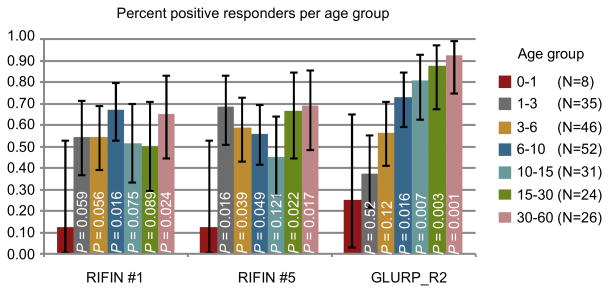
Plasma IgG positivity for the recombinant proteins comprising the variable regions of LILRB1+ RIFIN #1 and LILRB1− RIFIN #5 as well as GLURP_R2 in 222 Tanzanian individuals divided into age groups. Error bars represent 95% CI.P values were calculated using logistic regression comparing percent responders among children aged 0-1 years to children of the other age groups.
Supplementary Material
Acknowledgments
We thank T. Mitamura for discussions, K. Saito (DNA-chip Development Centre for Infectious Diseases, RIMD, Osaka University) for mass spectrometry analysis, M. Matsumoto, S. Matsuoka for technical assistance, the Thai and Tanzanian donors and the Japanese Red Cross Society for providing human erythrocytes and human plasma. This work was partly supported by the Platform Project for Supporting Drug Discovery and Life Science Research (J.T.) and Japanese Initiative for Progress of Research on Infectious Disease for global Epidemic (H.A.) funded by the Japan Agency for Medical Research and Development (AMED), JSPS KAKENHI Grant Numbers 16K08839 (K.H.), 16H05195 (T.S.), 15K08531 (M.K.), MEXT KAKENHI Grant Number JP26117714 (H.A.), JP23117008 (T.T.), JP24115005 (H.A.), Senri Life Science Foundation (K.H.), Kato Memorial Bioscience Foundation (K.H.), Danish Council for Independent Research grants 1333-00220 (C.W.W.) and 4004-00624B (T.L.) and the United States National Institutes of Health (NIH R01HL130678, T.L.). F.S was supported by the Taniguchi Memorial Fellowship program.
Footnotes
Supplementary Information is available in the online version of the paper.
Author Information: The authors declare no competing financial interests.
Author Contributions: F.S. and K.H. performed most of the binding and functional experiments, analysed and discussed the data. T. Satoh performed limiting dilution experiments and assisted with ligand identification. C.W.W. performed binding and serological analyses in Tanzania. J.L. assisted with experiments in Tanzania. T.A. performed CD spectra analysis. K.S. prepared recombinant plasmids. N.M.Q.P. assisted with P. falciparum culture and cloning. S. Itagaki assisted with P. falciparum culture. S. Iwanaga assisted with transgenic parasite experiments and provided Thai P. falciparum isolates. E.T. prepared wheat germ cell-free proteins. T.T. designed wheat germ cell-free protein expression system and discussed the data. M.K. assisted with functional experiments. T. Suenaga assisted with ligand identification. M.C. assisted with LILR analyses. J.T. assisted with protein experiments and discussed the data. T.L. assisted with experiments in Tanzania and discussed the data. T.H. assisted with P. falciparum experimental design and discussed the data. H.A. designed the study and analysed the data. All authors contributed to the writing of the manuscript.
References
- 1.World Health Organization. World malaria report 2016. World Health Organization; 2016. [Google Scholar]
- 2.Langhorne J, Ndungu FM, Sponaas AM, Marsh K. Immunity to malaria: more questions than answers. Nat Immonol. 2008;9:725–732. doi: 10.1038/ni.f.205. [DOI] [PubMed] [Google Scholar]
- 3.Joannin N, Abhiman S, Sonnhammer EL, Wahlgren M. Sub-grouping and sub-functionalization of the RIFIN multi-copy protein family. BMC Genomics. 2008;9:19. doi: 10.1186/1471-2164-9-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gomes PS, Bhardwaj J, Rivera-Correa J, Freire-De-Lima CG, Morrot A. Immune Escape Strategies of Malaria Parasites. Front Microbiol. 2016;7:1617. doi: 10.3389/fmicb.2016.01617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Renia L, Goh YS. Malaria Parasites: The Great Escape. Front Immunol. 2016;7:463. doi: 10.3389/fimmu.2016.00463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kyes S, Horrocks P, Newbold C. Antigenic variation at the infected red cell surface in malaria. Annu Rev Microbiol. 2001;55:673–707. doi: 10.1146/annurev.micro.55.1.673. [DOI] [PubMed] [Google Scholar]
- 7.Miller LH, Baruch DI, Marsh K, Doumbo OK. The pathogenic basis of malaria. Nature. 2002;415:673–679. doi: 10.1038/415673a. [DOI] [PubMed] [Google Scholar]
- 8.Goel S, et al. RIFINs are adhesins implicated in severe Plasmodium falciparum malaria. Nat Med. 2015;21:314–317. doi: 10.1038/nm.3812. [DOI] [PubMed] [Google Scholar]
- 9.Yam XY, Niang M, Madnani KG, Preiser PR. Three Is a Crowd - New Insights into Rosetting Plasmodium falciparum. Trends Parasitol. 2017 doi: 10.1016/j.pt.2016.12.012. in press. [DOI] [PubMed] [Google Scholar]
- 10.Arase H, Lanier LL. Specific recognition of virus-infected cells by paired NK receptors. Rev Med Virol. 2004;14:83–93. doi: 10.1002/rmv.422. [DOI] [PubMed] [Google Scholar]
- 11.Hirayasu K, Arase H. Functional and genetic diversity of leukocyte immunoglobulin-like receptor and implication for disease associations. J Hum Genet. 2015;60:703–708. doi: 10.1038/jhg.2015.64. [DOI] [PubMed] [Google Scholar]
- 12.Colonna M, et al. A common inhibitory receptor for major histocompatibility complex class I molecules on human lymphoid and myelomonocytic cells. J Exp Med. 1997;186:1809–1818. doi: 10.1084/jem.186.11.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cosman D, et al. A novel immunoglobulin superfamily receptor for cellular and viral MHC class I molecules. Immunity. 1997;7:273–282. doi: 10.1016/s1074-7613(00)80529-4. [DOI] [PubMed] [Google Scholar]
- 14.Hirayasu K, et al. Microbially cleaved immunoglobulins are sensed by the innate immune receptor LILRA2. Nat Microbiol. 2016;1:16054. doi: 10.1038/nmicrobiol.2016.54. [DOI] [PubMed] [Google Scholar]
- 15.Gardner MJ, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lau CK, et al. Structural conservation despite huge sequence diversity allows EPCR binding by the PfEMP1 family implicated in severe childhood malaria. Cell Host Microbe. 2015;17:118–129. doi: 10.1016/j.chom.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsieh FL, et al. The structural basis for CD36 binding by the malaria parasite. Nat Commun. 2016;7:12837. doi: 10.1038/ncomms12837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trowsdale J, Jones DC, Barrow AD, Traherne JA. Surveillance of cell and tissue perturbation by receptors in the LRC. Immunol Rev. 2015;267:117–136. doi: 10.1111/imr.12314. [DOI] [PubMed] [Google Scholar]
- 19.Tan J, et al. A LAIR1 insertion generates broadly reactive antibodies against malaria variant antigens. Nature. 2016;529:105–109. doi: 10.1038/nature16450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pieper K, et al. Public antibodies to malaria antigens generated by two LAIR1 insertion modalities. Nature. 2017;548:597–601. doi: 10.1038/nature23670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lebbink RJ, et al. Collagens are functional, high affinity ligands for the inhibitory immune receptor LAIR-1. J Exp Med. 2006;203:1419–1425. doi: 10.1084/jem.20052554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petter M, et al. Variant proteins of the Plasmodium falciparum RIFIN family show distinct subcellular localization and developmental expression patterns. Mol Biochem Parasitol. 2007;156:51–61. doi: 10.1016/j.molbiopara.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 23.Arase H, Mocarski ES, Campbell AE, Hill AB, Lanier LL. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science. 2002;296:1323–1326. doi: 10.1126/science.1070884. [DOI] [PubMed] [Google Scholar]
- 24.Shiroishi M, et al. Efficient leukocyte Ig-like receptor signaling and crystal structure of disulfide-linked HLA-G dimer. J Biol Chem. 2006;281:10439–10447. doi: 10.1074/jbc.M512305200. [DOI] [PubMed] [Google Scholar]
- 25.Naji A, et al. Binding of HLA-G to ITIM-bearing Ig-like transcript 2 receptor suppresses B cell responses. J Immunol. 2014;192:1536–1546. doi: 10.4049/jimmunol.1300438. [DOI] [PubMed] [Google Scholar]
- 26.Donati D, et al. Identification of a polyclonal B-cell activator in Plasmodium falciparum. Infect Immun. 2004;72:5412–5418. doi: 10.1128/IAI.72.9.5412-5418.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Warimwe GM, et al. Plasmodium falciparum var gene expression is modified by host immunity. Proc Natl Acad Sci USA. 2009;106:21801–21806. doi: 10.1073/pnas.0907590106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lavstsen T, et al. Plasmodium falciparum erythrocyte membrane protein 1 domain cassettes 8 and 13 are associated with severe malaria in children. Proc Natl Acad Sci USA. 2012;109:E1791–1800. doi: 10.1073/pnas.1120455109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mwakalinga SB, et al. Expression of a type B RIFIN in Plasmodium falciparum merozoites and gametes. Malar J. 2012;11:429. doi: 10.1186/1475-2875-11-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Janssen CS, Phillips RS, Turner CM, Barrett MP. Plasmodium interspersed repeats: the major multigene superfamily of malaria parasites. Nucleic Acids Res. 2004;32:5712–5720. doi: 10.1093/nar/gkh907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robertson MJ, et al. Characterization of a cell line, NKL, derived from an aggressive human natural killer cell leukemia. Exp Hematol. 1996;24:406–415. [PubMed] [Google Scholar]
- 32.Hanada K, et al. Plasmodium falciparum phospholipase C hydrolyzing sphingomyelin and lysocholinephospholipids is a possible target for malaria chemotherapy. J Exp Med. 2002;195:23–34. doi: 10.1084/jem.20010724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitamura T, Hanada K, Ko-Mitamura EP, Nishijima M, Horii T. Serum factors governing intraerythrocytic development and cell cycle progression of Plasmodium falciparum. Parasitol Int. 2000;49:219–229. doi: 10.1016/s1383-5769(00)00048-9. [DOI] [PubMed] [Google Scholar]
- 34.Tanabe K, et al. Allelic dimorphism-associated restriction of recombination in Plasmodium falciparum msp1. Gene. 2007;397:153–160. doi: 10.1016/j.gene.2007.04.033. [DOI] [PubMed] [Google Scholar]
- 35.Satoh T, et al. PILRα is a herpes simplex virus-1 entry coreceptor that associates with glycoprotein B. Cell. 2008;132:935–944. doi: 10.1016/j.cell.2008.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shiratori I, et al. Down-regulation of basophil function by human CD200 and human herpesvirus-8 CD200. J Immunol. 2005;175:4441–4449. doi: 10.4049/jimmunol.175.7.4441. [DOI] [PubMed] [Google Scholar]
- 37.Jiang Y, et al. Transport of misfolded endoplasmic reticulum proteins to the cell surface by MHC class II molecules. Int Immunol. 2013;25:235–246. doi: 10.1093/intimm/dxs155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jin H, et al. Autoantibodies to IgG/HLA class II complexes are associated with rheumatoid arthritis susceptibility. Proc Natl Acad Sci USA. 2014;111:3787–3792. doi: 10.1073/pnas.1401105111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morita S, Kojima T, Kitamura T. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene therapy. 2000;7:1063–1066. doi: 10.1038/sj.gt.3301206. [DOI] [PubMed] [Google Scholar]
- 40.Iwanaga S, Kato T, Kaneko I, Yuda M. Centromere plasmid: a new genetic tool for the study of Plasmodium falciparum. PLoS One. 2012;7:e33326. doi: 10.1371/journal.pone.0033326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang CW, et al. Evidence for in vitro and in vivo expression of the conserved VAR3 (type 3) plasmodium falciparum erythrocyte membrane protein 1. Malar J. 2012;11:129. doi: 10.1186/1475-2875-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Furukawa A, et al. Structural analysis for glycolipid recognition by the C-type lectins Mincle and MCL. Proc Natl Acad Sci USA. 2013;110:17438–17443. doi: 10.1073/pnas.1312649110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arumugam TU, et al. Application of wheat germ cell-free protein expression system for novel malaria vaccine candidate discovery. Expert Rev Vaccines. 2014;13:75–85. doi: 10.1586/14760584.2014.861747. [DOI] [PubMed] [Google Scholar]
- 44.Suenaga T, Kohyama M, Hirayasu K, Arase H. Engineering large viral DNA genomes using the CRISPR-Cas9 system. Microbiol Immunol. 2014;58:513–522. doi: 10.1111/1348-0421.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mkumbaye SI, et al. Cellulose filtration of blood from malaria patients for improving ex vivo growth of Plasmodium falciparum parasites. Malar J. 2017;16:69. doi: 10.1186/s12936-017-1714-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Magistrado PA, et al. High efficacy of anti DBL4varepsilon-VAR2CSA antibodies in inhibition of CSA-binding Plasmodium falciparum-infected erythrocytes from pregnant women. Vaccine. 2011;29:437–443. doi: 10.1016/j.vaccine.2010.10.080. [DOI] [PubMed] [Google Scholar]
- 47.Turner L, et al. Severe malaria is associated with parasite binding to endothelial protein C receptor. Nature. 2013;498:502–505. doi: 10.1038/nature12216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Turner L, et al. IgG antibodies to endothelial protein C receptor-binding cysteine-rich interdomain region domains of Plasmodium falciparum erythrocyte membrane protein 1 are acquired early in life in individuals exposed to malaria. Infect Immun. 2015;83:3096–3103. doi: 10.1128/IAI.00271-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data for Figures 3-4 and Extended Data Figures 2, 5-6, 9-10 are provided with the paper. All data are available from the corresponding author upon reasonable request.