Abstract
Magnesium (Mg2+) homeostasis is critical for metabolism. However, the genetic determinants of the renal handling of Mg2+, which is crucial for Mg2+ homeostasis, and the potential influence on metabolic traits in the general population are unknown. We obtained plasma and urine parameters from 9099 individuals from seven cohorts, and conducted a genome-wide meta-analysis of Mg2+ homeostasis. We identified two loci associated with urinary magnesium (uMg), rs3824347 (P=4.4×10−13) near TRPM6, which encodes an epithelial Mg2+ channel, and rs35929 (P=2.1×10−11), a variant of ARL15, which encodes a GTP-binding protein. Together, these loci account for 2.3% of the variation in 24-hour uMg excretion. In human kidney cells, ARL15 regulated TRPM6-mediated currents. In zebrafish, dietary Mg2+ regulated the expression of the highly conserved ARL15 ortholog arl15b, and arl15b knockdown resulted in renal Mg2+ wasting and metabolic disturbances. Finally, ARL15 rs35929 modified the association of uMg with fasting insulin and fat mass in a general population. In conclusion, this combined observational and experimental approach uncovered a gene–environment interaction linking Mg2+ deficiency to insulin resistance and obesity.
Keywords: zebrafish, Genetic determinants, Magnesium homeostasis, Metabolic syndrome, Gene-environment interaction, Tubular transport
Magnesium (Mg2+) is an essential cation for multiple enzymatic reactions, including those involving DNA and protein synthesis and energy metabolism.1 Abnormal Mg2+ levels in serum are associated with common diseases such as diabetes and metabolic disorders.2 Interventional and longitudinal observational studies in humans show that high dietary Mg2+ protects against the risk of developing type 2 diabetes,3–6 and improves glycemic control in patients with diabetes7 as well as in overweight nondiabetic people.8–10 In young adults, high Mg2+ intake is significantly associated with lower incidence of metabolic syndrome.11 Further, there is evidence linking Mg2+ intake to body weight regulation, with low intake potentially impairing lean body mass growth,12,13 whereas Mg2+ supplementation may increase lean body mass and decrease fat mass in overweight women.14
Plasma Mg2+ levels are closely regulated and remain constant throughout life, despite the fact that dietary Mg2+ intake and intestinal absorption decrease with age.15 The control of Mg2+ balance is ensured by a tightly regulated reabsorption of Mg2+ in the distal tubular segments of the kidney. In particular, the filtered Mg2+ is reabsorbed in the proximal tubule and thick ascending loop of Henle (TAL) via paracellular routes, whereas downstream, in the distal convoluted tubule (DCT), Mg2+ is efficiently reabsorbed through transcellular mechanisms involving the transient receptor potential cation channel, subfamily M, member 6 (TRPM6).2 The reabsorption of Mg2+ in the DCT determines the final urinary magnesium (uMg) excretion because no reabsorption of Mg2+ occurs in posterior segments. People harboring inherited or acquired dysfunctions of the renal tubular handling of Mg2+ show inappropriate urinary loss of Mg2+, causing chronic hypomagnesemia and severe, multisystemic manifestations.2,16,17
The genetic component of Mg2+ homeostasis is indicated by a significant heritability (15%–39%) of serum Mg2+18,19 and by rare monogenic disorders disturbing renal tubular transport of Mg2+.20–23 However, most of the regulatory genes of renal Mg2+ channels and transporters remain unknown. A genome-wide association study (GWAS) on serum Mg2+ in European ancestry adults identified six loci, one of which included the gene encoding TRPM6.24 Two more recent studies in children25 and in blacks26 did not reveal additional loci. However, changes in uMg excretion precede changes in circulating serum Mg2+ levels, thus being a more sensitive indicator of any disturbance in Mg2+ homeostasis.27 Further, Mg2+ depletion can be found in individuals with apparently normal total serum Mg2+ levels,28 which underscores the limitations of total serum Mg2+ as a marker of Mg2+ status. Assessment of uMg in GWAS is therefore crucial to elucidate new genes involved in Mg2+ homeostasis. Further, studying the association between the genetic determinants of uMg and metabolic phenotypes linked to disturbed Mg2+ balance (e.g., obesity and diabetes) may provide novel insights into links between Mg2+ and common diseases. Indeed, plasma triglycerides and glucose are major determinants of the Mg2+ balance in patients with diabetes.29
To shed light on the genetic factors and molecular mechanisms linking Mg2+ handling and metabolic disorders, we performed a meta-analysis of GWAS for the renal handling of Mg2+ by combining genetic isolates and population-based studies, and investigating the biologic relevance of the identified loci using cellular systems and model organisms. Given the known relationships of Mg2+ intake with metabolic disorders, we also explored whether the identified loci modified these relationships. These studies establish a novel biologic control of renal Mg2+ handling, and a novel gene–environment interaction sustaining the link between Mg2+ homeostasis, insulin resistance, and obesity.
Results
Meta-Analyses of GWAS for Mg2+ Homeostasis
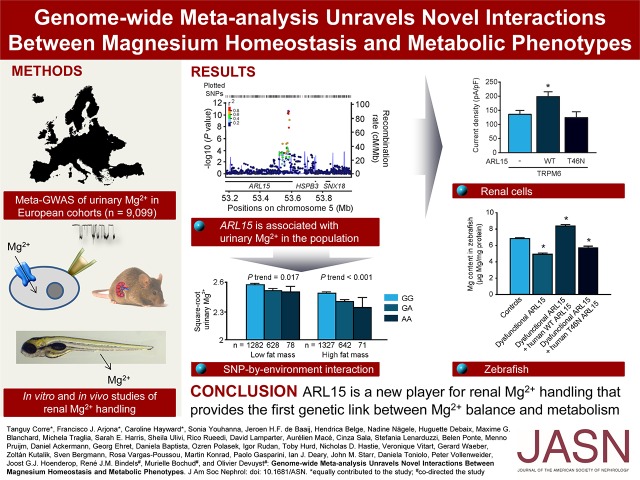
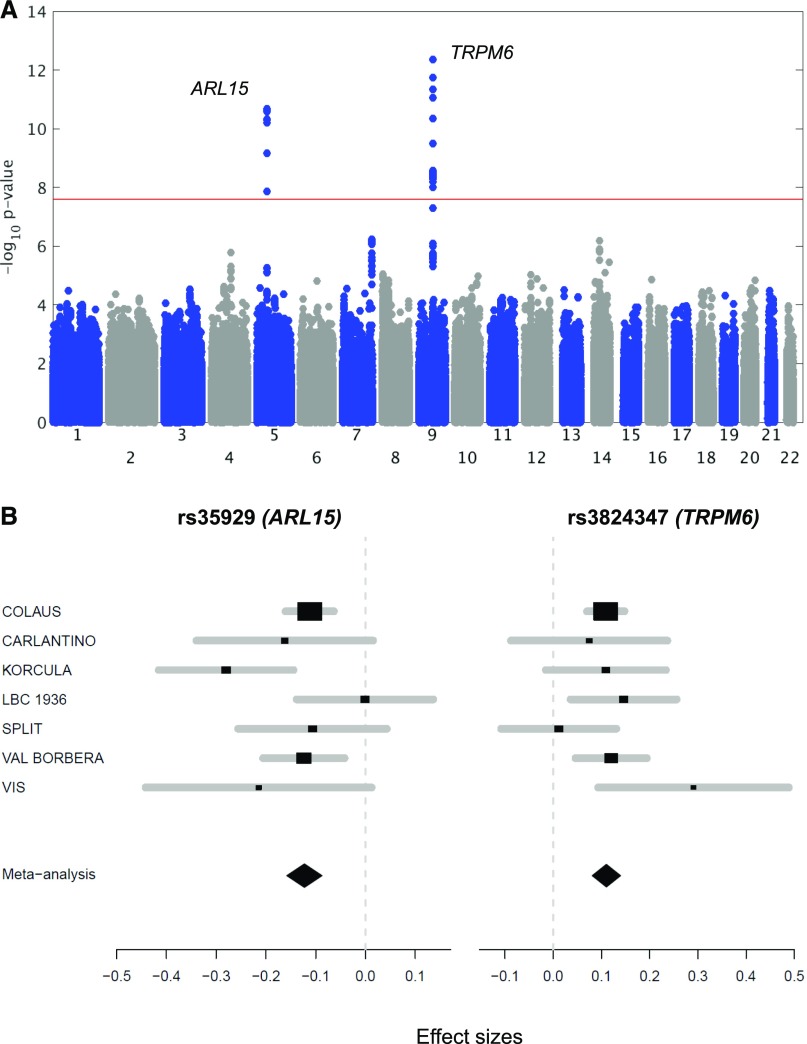
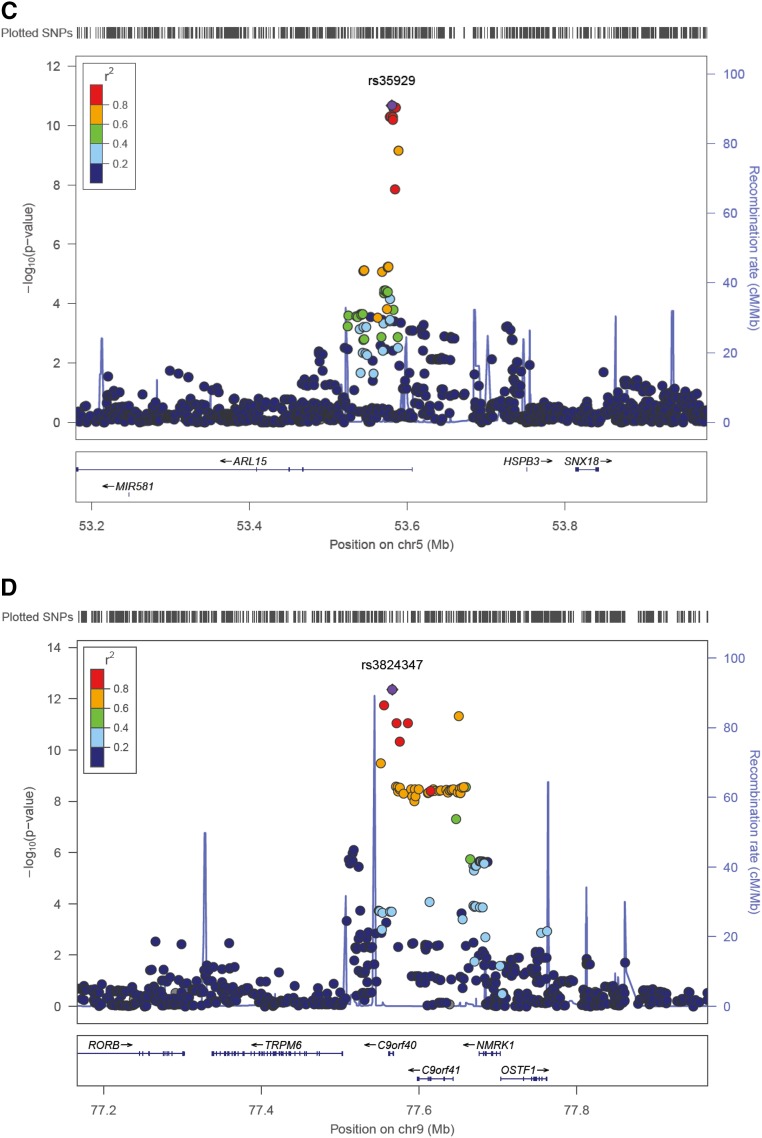
The initial discovery phase consisted of a GWAS (2.5 M markers) performed on the population-based CoLaus cohort (5150 samples) testing for association with urinary Mg2+-to-creatinine ratio. The analysis revealed a single, genome-wide significant signal at rs3824347 (P=3.6×10−8), corresponding to the TRPM6 locus on chromosome 9, and six suggestive loci with P values <10−5 (Supplemental Figure 1). These association results were combined with the genome-wide association scans from six additional European cohorts (LBC1936, CROATIA-Split, CROATIA-Vis, Carlantino, CROATIA-Korcula, and Val Borbera) into a single meta-analysis (9099 samples). As shown on the Manhattan plot (Figure 1A), two signals showed a P value <5×10−8. The QQ plot showed no problematic inflation (λ=1.014, se=3.16×10−5) (Supplemental Figure 2A). Both the forest plots (Figure 1B) and the low I2 values (Table 1) indicated little heterogeneity across cohorts. No secondary signals were detected in the approximate conditional analysis, using the lead single nucleotide polymorphisms (SNPs) as covariates in the regression (Supplemental Figure 2, B and C). The lowest combined P value (4.4×10−13) was observed for the SNP rs3824347 on chromosome 9, at a locus comprising five genes: TRPM6, C9orf40, C9orf41, NMRK1, and OSTF1 (Figure 1C). The second strongest signal maps to chromosome 5, with a P value of 2.1×10−11 for the lead SNP rs35929 (Figure 1D). This SNP and all other genome-wide significant SNPs at this locus lie in the first intron of the ADP ribosylation factor like GTPase 15 (ARL15) gene. None of the two signals showed significant association with urinary creatinine.
Figure 1.
A genome-wide meta-analysis for urinary Mg2+-to-creatinine ratio reveals two signals. (A) Manhattan plot showing −log10(P values) for all SNPs in the genome-wide meta-analysis for normalized urinary Mg2+-to-creatinine ratio in Europeans (n=9099), ordered by chromosomal position. The values correspond to the association of normalized urinary Mg2+-to-creatinine ratio, including age and sex as covariates in the model as well as study-specific covariates if needed. The gene closest to the SNP with the lowest P value is listed at each locus. Two loci reached genome-wide significance (P<5×10−8), indicated by the red horizontal line, at combined analysis (TRPM6, rs3824347 and ARL15, rs35929). (B) Forest plot for rs35929 (ARL15) and rs3824347 (TRPM6) showing effect sizes and 95% confidence intervals across studies as well as the summary meta-analysis results. (C) Regional association plot at the rs3824347 (TRPM6) locus. Regional association plot showing −log10 (P values) for the association of SNPs at the locus of interest ordered by their chromosomal position with normalized urinary Mg2+-to-creatinine ratio. The -log10 (P value) for each SNP is colored according to the correlation of the corresponding SNP, with the SNP showing the lowest P value (index SNP) within the locus using different colors for selected levels of LD (chi-squared). Correlation structures correspond to HapMap 2 CEU. The blue line represents the recombination according to the scale shown on the right-side y-axis. (D) Regional association plot at the rs35929 (ARL15) locus.
Table 1.
Summary statistics for the meta-analysis of urinary Mg2+-to-creatinine ratio
| SNP | Effect Allele | Other Allele | Mean Effect Allele Frequency | Effect Size Discovery (SE) | P Value Discovery | Effect Size Replication (SE) | P Value Replication | Effect Size Combined Analysis (SE) | P Value Combined Analysis | Direction of Effects of Individual Cohorts | I2 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs35929 | a | g | 0.21 | −0.112 | 5.57×10−6 | −0.135 | 5.95×10−7 | −0.1227 | 2.11×10−11 | − − − − − − − | 35.7 |
| (−0.025) | (−0.027) | (−0.183) | |||||||||
| rs3824347 | g | a | 0.41 | −0.109 | 3.61×10−8 | −0.1125 | 2.02×10−6 | −0.1103 | 4.38×10−13 | − − − − − + − | 7 |
| (−0.02) | (−0.024) | (−0.152) |
The effect size sign for each individual cohort in the last but one column is represented by – or +. The cohort displaying a “-” for rs35929 is LBC1936, having an effect close to 0. The last column shows the I2 value, representing the heterogeneity across cohorts.
Results from the versatile gene-based association study (VEGAS analysis)30 of urinary Mg2+-to-creatinine ratio identified C9orf40, C9orf41, and TRPM6 as the only genes with a statistically significant result (P<2.8×10−6); however, these signals are overlapping with the one SNP association in the meta-analysis (rs3824347). None of the discovered SNPs tag copy number variations (CNVs). A pathway analysis was performed using the Magenta algorithm, but no enrichment appeared to be significant after multiple testing correction. Sex-stratified analyses did not reveal additional genome-wide significant loci. No significant eQTL is referenced for rs35929 nor rs3824347 in the GTEx portal.
A meta-GWAS of serum Mg2+ levels replicated the previously published locus on chromosome 1 containing, among others, the gene MUC1 (P=4.45×10−14) (Supplemental Figure 3, Supplemental Table 1).24 Our meta-analysis on fractional excretion of magnesium (FEMg) yielded 12 suggestive loci with a P<10−5 (Supplemental Figure 4), but no genome-wide significant association (the sample size was reduced to cohorts having serum and urine from the same time point, therefore without cohorts LBC1936, Split, and Vis; n=7976). The association of rs35929 (P=2.8×10−4) and rs3824347 (P=1.4×10−5) with FEMg were nominally, but not genome-wide significant.
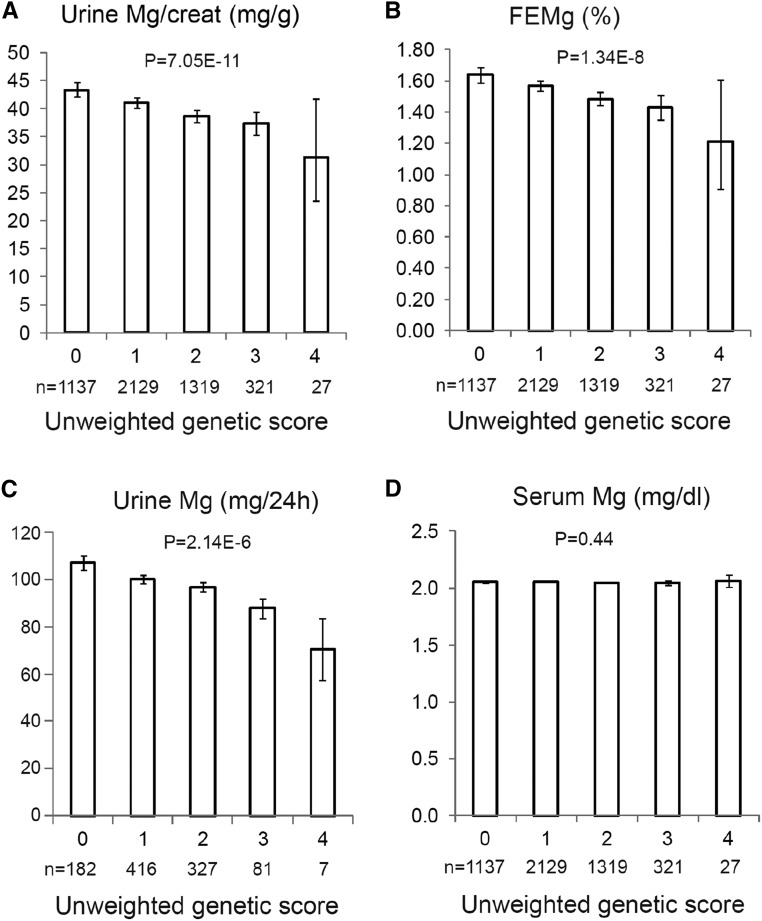
A Genetic Score Including rs35929 and rs3824347 Associates with uMg Traits
Urinary Mg2+-to-creatinine ratio (Figure 2A, CoLaus cohort), FEMg in spot urine (Figure 2B, CoLaus cohort), and uMg excretion in 24-hour urine (Figure 2C, Swiss Kidney Project on Genes in Hypertension [SKIPOGH] cohort), but not serum Mg2+ (Figure 2D, CoLaus) were strongly and linearly associated with an additive unweighted genetic risk score including rs35929-A (ARL15) and rs3824347-G (TRPM6) alleles. The genetic score explained 2.3% of 24-hour uMg excretion in the SKIPOGH study and 1.0% of urinary Mg2+-to-creatinine ratio in CoLaus.
Figure 2.
A genetic score including rs35929 and rs3824347 associates with uMg traits. (A–D) Associations of uMg excretion and fractional excretion with an unweighted genetic risk score including rs35929 (ARL15 locus) and rs3824347 (TRPM6 locus). Data are geometric means and whiskers are 95% confidence intervals for Mg2+-related phenotypes in the CoLaus (A, B, D) and SKIPOGH (C) studies. The x-axes represent the unweighted genetic score generated from rs35929 and rs3824347, using as effect allele the one associated with lower urinary Mg2+-to-creatinine ratio, i.e., the A allele for rs35929 and the G allele for rs3824347. P values are from nonparametric trend tests across genetic score. The numbers in each genetic score category are listed in all panels. The y-axis represents urinary Mg2+-to-creatinine ratio (milligrams per gram) (A), FEMg (%) (B), uMg excretion (milligrams per 24 hour) (C), and serum Mg2+ (milligrams per deciliter) (D).
ARL15 Regulates TRPM6 Channel Activity
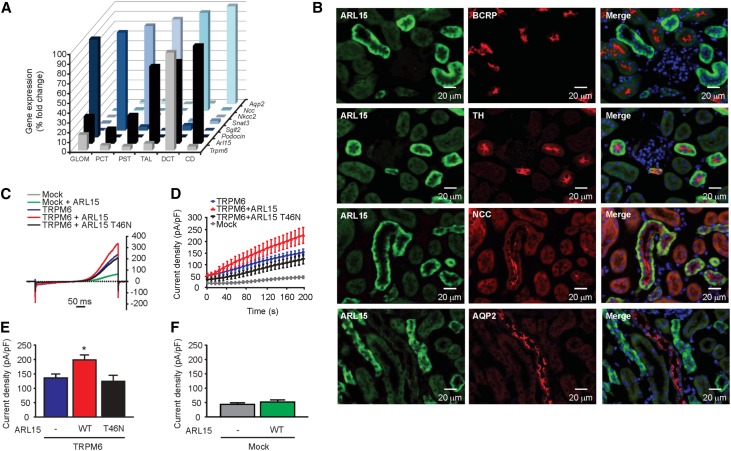
To validate the association between uMg excretion and ARL15 and TRPM6 variants, we first showed that the highest mRNA expression of Arl15 was in distal nephron segments including the TAL and DCT, the DCT also being strongly positive for Trpm6 (Figure 3A). At the protein level, ARL15 is localized predominantly in the TAL and the DCT segments (Figure 3B), which are enriched in the Tamm–Horsfall protein and the Na+-Cl− cotransporter, respectively. Importantly, TRPM6 is also distinctly localized in the DCT segment.31 The spatial correlation between TRPM6 and ARL15 expression in the DCT prompted us to postulate a functional interaction between these two proteins. In addition, functional protein association network analyses for ARL15 (Supplemental Figure 5) showed that ARL15 interacts with key proteins for endocytic trafficking (RAB11 and ARFGEF families), a process that may increase the abundance and channel activity of TRPM6 in the plasma membrane.
Figure 3.
ARL15 localizes in renal DCT regulating TRPM6 channel activity. (A) Gene expression analyses of Trpm6, Arl15, Podocin, Sglt2, Snat3, Nkcc2, Ncc, and Aqp2 in microdissected mouse nephron segments showed coexpression of Arl15 and Trpm6 in DCT. (B) Double immunofluorescence staining of mouse kidney cortex sections for ARL15 (in green) and BCRP (red); TH (red); NCC (red); or AQP2 (red) as markers of the proximal tubule, TAL, DCT, or collecting duct, respectively. For detection, the Alexa Fluor dye was used. (C) Typical current-voltage curves obtained from transfected HEK293 cells 200 seconds after break-in. Outwardly rectifying currents are observed in response to a 500 ms voltage ramp (from −100 to +100 mV) applied 200 seconds after break-in. (D) The average time development of the current density measured at +80 mV is shown (n≥10). The mock plus ARL15 condition is not shown for clarity reasons. (E) WT ARL15 (red, WT, n=47) but not the T46N ARL15 mutant (black, T46N, n=18) increased the whole-cell current density of TRPM6. Asterisk indicates significant difference with respect to the cells transfected with TRPM6 only (blue, “-,” n=44). One-way ANOVA followed by Tukey multiple comparisons post-test; P<0.05. (F) Transfection of HEK293 cells with WT ARL15 did not evoke a significant increase in whole-cell current density when compared with mock-transfected cells (n≥10, unpaired t test).
To assess the potential effect of ARL15 on TRPM6 channel activity, human embryonic kidney 293 (HEK293) cells were transfected with wild-type (WT) TRPM6 in parallel with a mock transfect, WT ARL15, or a dominant negative (T46N) ARL15 mutant. The T46N ARL15 mutant was predicted to be of dominant negative nature by comparative analysis with T27N ARF6, a dominant negative mutant affecting a protein (ARF6) that belongs to the same protein family as ARL15.32 Patch clamp recordings showed slowly developing outwardly rectifying currents, a signature feature of TRPM6 under our recording conditions (Figure 3C and D). A significant increase in TRPM6 channel activity was observed in the presence of WT ARL15, but not the T46N ARL15 mutant (Figure 3E). HEK293 cells express endogenous TRPM7 channels.33 Here, these currents were small (<50 pA/pF) and were not significantly increased by expression of WT ARL15 (Figure 3F).
Physiologic Relevance of ARL15 in Model Organisms
To support the important role of ARL15 in Mg2+ homeostasis, we investigated the regulation of TRPM6 and ARL15 gene expression in mice on exposure to variable Mg2+ diets, known to affect the expression of critical genes in the mouse kidney and intestine,34 where Arl15 is expressed (Supplemental Figure 6A). Metabolic profiling (Supplemental Tables 2 and 3) revealed a remarkable adaptation after exposure to high and low Mg2+ diets, reflected by significant changes in the renal mRNA expression of Trpm6 and Pvalb, contrasting with the stable expression of Arl15 and other segmental markers of the TAL and DCT (Supplemental Figure 6B). A similar effect was observed in the ileum and the cecum, also involved in Mg2+ handling2,34 (Supplemental Figure 6, C and D).
We further tested the biologic relevance of ARL15 by using a zebrafish model, which shows tubular segmentation patterns that are equivalent to the mammalian kidney.35 Two distinct orthologs of mammalian ARL15 were identified in zebrafish (Arl15a and Arl15b), presenting 68% and 83% of amino acid identity respectively, when aligned to the human counterpart (Supplemental Figure 7A). Given the higher amino acid conservation of Arl15b compared with Arl15a, the former was selected to study the function of its mammalian ortholog ARL15. As for mammalian ARL15, arl15b was ubiquitously expressed in all adult zebrafish tissues tested (Supplemental Figure 7B). When adult zebrafish were fed different Mg2+ diets for 3 weeks, arl15b expression patterns reflected compensatory mechanisms to cope with Mg2+ deficiency (kidney and gills) or Mg2+ surplus (gut) (Supplemental Figure 7, C–E).
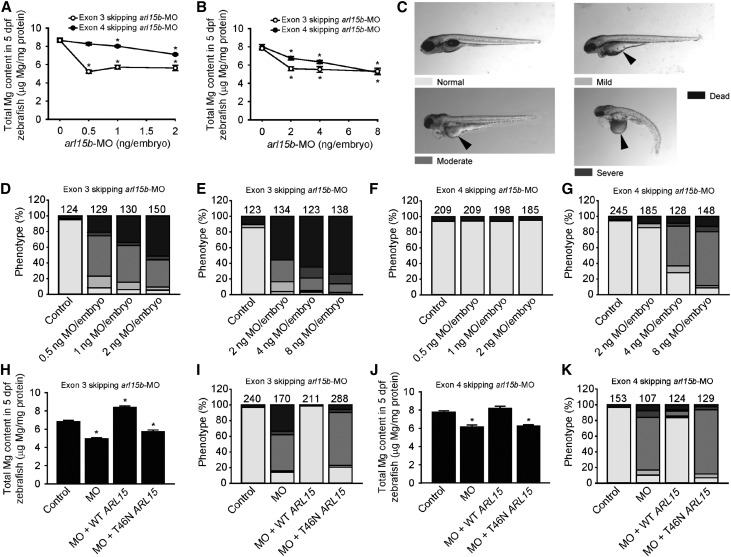
A loss-of-function approach using morpholinos (MOs) against arl15b in zebrafish larvae, where arl15b is expressed in the pronephros (Supplemental Figure 8), was used to relate ARL15 function to renal Mg2+ wasting (Supplemental Figures 9 and 10). Because intestinal Mg2+ absorption does not take place during the time frame where the knockdown is applied, a change in zebrafish larvae Mg2+ content reflects a change in the renal and/or skin (renal-like tissue36) uptake of Mg2+. Total Mg2+ content decreased consistently in arl15b morphants compared with controls, indicating renal Mg2+ wasting because the pronephric kidney is the only extrusion route for Mg2+ (Figure 4, A and B). In parallel, cardiovascular impairments and morphologic phenotypes characterized by a poorly metabolized yolk in comparison with controls, indicative of metabolic disturbances, were also observed (Figure 4, C–G). Coinjection of arl15b-MO with human WT ARL15 cRNA induced a rescue of all phenotypes observed, whereas coinjection with T46N ARL15 cRNA did not result in any rescue (Figure 4, H–K). These rescue experiments demonstrated the specificity of the renal Mg2+ wasting and metabolic defects associated with dysfunctional ARL15.
Figure 4.
ARL15 dysfunction results in renal Mg2+ wasting and metabolic disturbances. (A and B) Knockdown of arl15b by 0.5–2 (A) and 2–8 (B) ng per embryo of the two arl15b-MOs used (exon skipping 3 and 4 arl15b-MOs) resulted in a dose-dependent decrease of the total Mg2+ content of zebrafish arl15b morphants, reflecting renal Mg2+ wasting. The zero dose represents injection with control-MO (2 [A] and 8 [B] ng MO per embryo). (C) Morphologic phenotypes distinguished in zebrafish larvae (5 days postfertilization) after treatment with arl15b-MO or control-MO (25× magnification). A complete description of each phenotype is detailed in the Supplemental Material. Metabolic defects (poor metabolization of the yolk) are indicated by arrowheads. (D–G) Distribution of morphologic phenotypes in zebrafish larvae injected with 0.5–2 (D, F) and 2–8 (E, G) ng per embryo of exon skipping 3 (D, E) arl15b-MO, exon skipping 4 (F, G) arl15b-MO, or control-MO (2 [D, F] and 8 [E, G] ng MO per embryo). (H–K) Rescue of renal Mg2+ wasting (H, J) and of metabolism defects (I, K) in morphant zebrafish by coinjection of exon skipping 3 arl15b-MO (0.5 ng MO per embryo [H, I]) or exon skipping 4 arl15b-MO (8 ng MO per embryo [J, K]) with cRNA encoding human WT ARL15 (50 pg cRNA per embryo). Coinjection with cRNA encoding human T46N ARL15 mutant (50 pg cRNA per embryo) did not rescue renal Mg2+ wasting or the defects in metabolism. (D–G, I, K) Numbers on top of the bars indicate the number of animals in each experiment. (A, B, H, J) Data are presented as mean±SEM (n=10, except for [B], where n=6–10). (A, B, H, J) Asterisks indicate significant differences respect to the control condition (one-way ANOVA followed by Tukey multiple comparisons post-test; P<0.05).
Genetic Background Influences the Link between uMg and Metabolism Phenotypes
The functional relevance of ARL15 for Mg2+ homeostasis, as demonstrated in this study, in combination with the association between ARL15 and obesity and insulin biology,37 urged us to investigate a possible link between genetic background, renal Mg2+ handling, and metabolic traits in general population cohorts.
Urinary Mg2+-to-creatinine ratio was positively associated with circulating fasting adiponectin (P<0.001) and glucose levels (P=0.001), and negatively with body mass index (BMI) (P=0.001), homocysteine (P<0.001), and γ-glutamyl transeptidase (GGT) (P<0.001), independently of serum Mg2+, in CoLaus (the largest cohort available). In CoLaus, the positive associations with adiponectin and fasting glucose, as well as the negative associations with BMI, GGT, and homocysteine, remained significant (P<0.05) upon adjustment for all the other metabolic phenotypes. In SKIPOGH, 24-hour uMg excretion was negatively associated with fasting serum insulin (P<0.001), even after adjustment for other metabolic covariates (P=0.003). The negative associations of 24-hour uMg excretion with fasting triglycerides or glucose disappeared upon adjustment for fasting insulin; we found no association of 24-hour uMg excretion with BMI nor with fat mass (P>0.40).
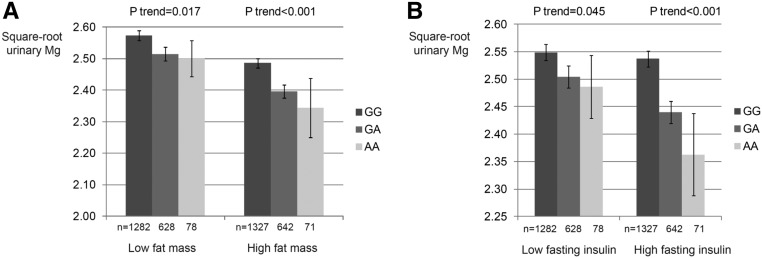
In CoLaus, the negative association of the ARL15 rs35929 variant with uMg excretion was stronger in overweight people (P<0.01 for interaction between BMI and rs35929 for their effect on uMg concentration in spot urine), in people with high fat mass (P interaction =0.02) (Figure 5A), and in people with high fasting insulin levels (P interaction =0.01) (Figure 5B), highlighting an SNP-by-environment interaction. In SKIPOGH, the negative association of daytime uMg excretion with fasting insulin was stronger in carriers of the ARL15 rs35929 A allele (P interaction <0.04), which is consistent with results in CoLaus in that rs35929 appears to modify the association between uMG excretion and fasting insulin. The corresponding P values for interaction were 0.14 and 0.14 when replacing fasting insulin with BMI or fat mass, respectively.
Figure 5.
Genotypes at rs35929 influence the link between uMg and metabolism related phenotypes. (A) Effect modification of fat mass on the association of uMg concentration with rs35929 (ARL15) genotypes. Data represent adjusted square-root-transformed uMg levels by rs35929 genotypes and fat mass strata in CoLaus. The model was adjusted for age, sex, height, lean mass, CKD-EPI, urinary creatinine (square root), serum Mg2+, serum and urinary Ca2+ (square root), and menopausal status. P interaction =0.02. Fat mass strata are cut by sex-specific medians (n=4729). (B) Effect modification of fasting insulin on the association of uMg levels with rs35929 (ARL15) genotypes. Data represent adjusted square-root-transformed uMg levels by rs35929 genotypes and fasting insulin strata in CoLaus. The model was adjusted for age, sex, height, lean mass, CKD-EPI, urinary creatinine (square root), serum Mg2+, serum and urinary Ca2+ (square root), and menopausal status. P interaction =0.01. Fasting insulin strata are cut by sex-specific medians (n=4729).
Discussion
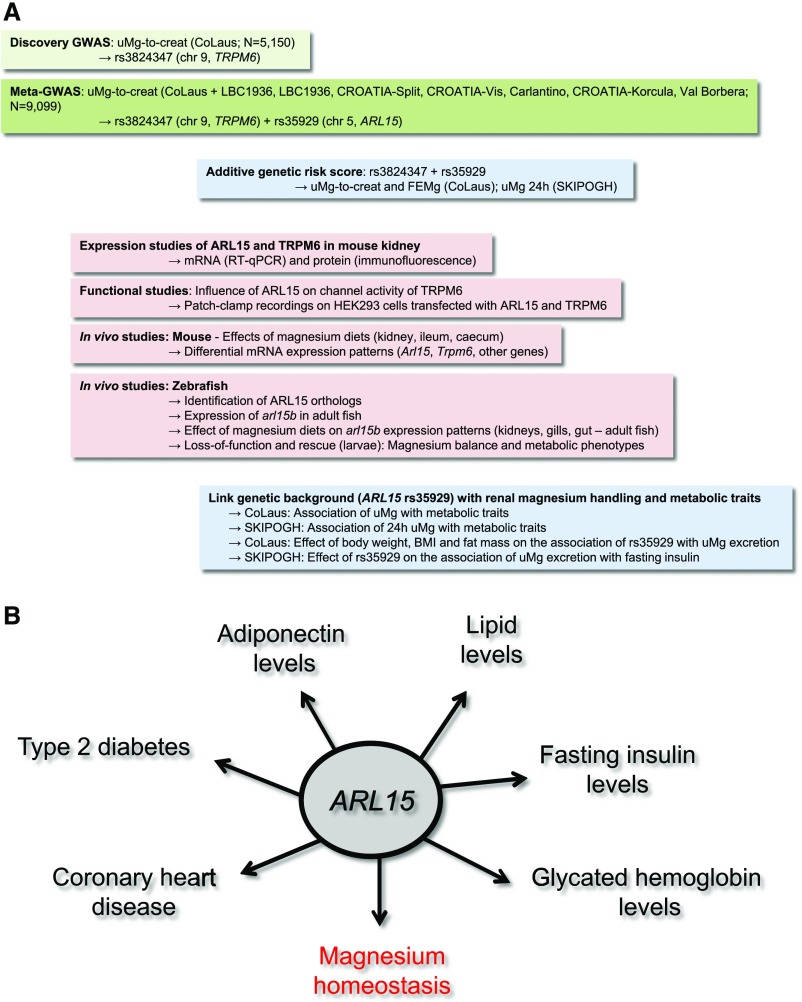
In this multistep study (Figure 6), we conducted the first genome-wide meta-analysis for uMg levels using data from seven population-based studies of European descent. We identified two genome-wide significant loci associated with urinary Mg2+-to-creatinine ratio: (1) rs3824347 located on chromosome 9, near the TRPM6 gene coding for a Mg2+ channel; and (2) rs35929 located on chromosome 5, an intronic variant of the ARL15 gene known for its association with adiponectin38 and lipid levels,39,40 type 2 diabetes,40 fasting insulin levels,41 and coronary heart disease,38 but without a prior physiologic link to Mg2+ homeostasis (Supplemental Table 4). Next we provide robust evidence that ARL15 influences renal Mg2+ reabsorption through regulation of the Mg2+ channel, TRPM6. An additive genetic score including the rs35929 A (ARL15) and rs3824347 G (TRPM6) alleles is strongly associated with 24-hour uMg excretion, a physiologic test of Mg2+ status, which integrates both intestinal absorption and renal wasting.42 In addition, the association of ARL15 rs35929 with uMg levels is modified by the metabolic status, and the association of uMg excretion with metabolic phenotypes is modified by ARL15 rs35929 in the general adult population. Altogether, these findings suggest a gene–diet interaction relating Mg2+ homeostasis to metabolic disorders in the general population. If replicated in independent populations, the observed genetic effect sizes are sufficiently large to be of clinical and public health relevance.
Figure 6.
ARL15 was identified as a key player for metabolism and renal homeostasis. The figure shows the multistep approach (A) used to investigate the genetic determinants of the renal handling of Mg2+ and their influence on metabolic traits in the general population (B).
In contrast to TRPM6, little is known about the function of the ARL15 gene, except its association with adiponectin and lipid levels, type 2 diabetes, and higher fasting insulin levels in humans.38–40,43 We found that ARL15 is highly expressed in the TAL and DCT, where Mg2+ reabsorption is mostly regulated and the main bulk of Mg2+ is reabsorbed. Particularly, we show that ARL15 is enriched along with the Mg2+ channel TRPM6 in the DCT, and that ARL15 increases TRPM6-mediated currents in renal epithelial cells. Because TRPM6 is the gatekeeper in transepithelial Mg2+ transport, its regulation by ARL15 supports a key role of the latter in the transepithelial Mg2+ transport in the distal part of the renal tubule. ARL15 is structurally similar to Ras-related GTP-binding proteins, which regulate intracellular vesicle trafficking.44 It interacts with proteins of the RAB11 and ARFGEF families, which are involved in endocytic traffic, suggesting that ARL15 regulates the trafficking of vesicular TRPM6 to the plasma membrane, thereby influencing Mg2+ reabsorption in the kidney. In this sense, our in vivo data substantiates ARL15 as a key protein for the maintenance of Mg2+ homeostasis. This conclusion is supported by the high sensitivity of ARL15 expression to dietary Mg2+, as demonstrated by the regulation of the highly conserved zebrafish ortholog of ARL15, arl15b, in the kidney and gills of fish exposed to different Mg2+ diets—as already observed for the zebrafish ortholog of TRPM6.45 Conversely, Arl15 expression in intestine and kidney remained unchanged in mice fed different Mg2+ diets. This discrepancy between the mouse and the zebrafish models might be due to the fact that, in zebrafish and freshwater fish in general, active transcellular Mg2+ reabsorption is more prominent than in terrestrial vertebrates, as freshwater fish are hyperosmotic with respect to the surrounding aquatic medium.46,47 Thus, the dietary Mg2+ challenge performed with mice in this study may not be sufficiently severe to evoke regulation of Arl15 expression.
The association of the ARL15 rs35929 variant with uMg excretion in the present GWAS predicted a physiologic relevance of ARL15 in Mg2+ homeostasis. In agreement with these findings, knockdown experiments in zebrafish recognized ARL15 as a relevant key protein that determines renal Mg2+ excretion. In agreement with previous GWAS linking ARL15 variants with risk of congestive heart disease,38 cardiovascular impairments (i.e., deficient tail blood circulation) was recognized in zebrafish arl15b morphants. Further, previous links between ARL15 and lipid metabolism37–39,43 were functionally supported in zebrafish, where arl15b morphants showed a poor mobilization of yolk metabolite reserves. In zebrafish larvae, the yolk serves mainly as a lipid store,48,49 therefore the deficiency in yolk absorption observed in arl15b morphants revealed poor lipid mobilization. The zebrafish arl15b morphants also accumulated fluid in the pericardial cavity and in the pronephros in the form of cysts showing kidney dysfunction. However, when lowering the intensity of the arl15b knockdown (by using doses of 1 and 2 ng exon 4–skipping arl15b-MO per embryo), renal Mg2+ wasting was the only phenotype observed in the fish morphants. Therefore, the disturbances in Mg2+ balance observed in arl15b morphants are a primary effect of ARL15 dysfunction and not a secondary disturbance to the kidney/metabolic/cardiovascular phenotypes observed. The phenotype rescue experiments performed with normal or mutant versions of human ARL15 confirmed that renal Mg2+ wasting and metabolic defects were directly due to ARL15 knockdown. Altogether, the ARL15 functions unraveled in this study illustrate an unprecedented case of shared pathophysiologic genetics of renal Mg2+ wasting, cardiovascular disease, and metabolic disorders, and demonstrate the pleiotropic nature of ARL15, inferred from the present and published GWAS. The importance of ARL15 is also supported by recent genetic evidence for its positive selection among adaptive loci identified in cattle.50 Thus far, no mutations in ARL15 have been associated with a monogenic disorder, including unidentified forms of renal Mg2+ wasting (data not shown).
Could the metabolic traits linked to ARL15 variants be connected? By integrating intestinal absorption and renal wasting, uMg excretion represents a physiologic test of Mg2+ status more suitable than total serum Mg2+ levels.42 Urinary Mg2+-to-creatinine ratio was associated with multiple metabolic phenotypes, independently of serum Mg2+, in the population-based CoLaus study, providing insights into the metabolic consequences of Mg2+ deficiencies. Our results are in line with longitudinal studies in humans suggesting that self-reported dietary Mg2+ intake may protect against type 2 diabetes, insulin resistance,51 and metabolic syndrome.52 Growing experimental evidence in favor of a beneficial role of Mg2+ intake on type 2 diabetes is accumulating in humans. Mg2+ supplements were found to significantly improve glucose tolerance and insulin sensitivity in diabetic and nondiabetic people.53 Whether such benefit would persist over years is currently unknown. The public health relevance of dietary Mg2+ intake is further underscored by an association with all-cause mortality and risk of cardiovascular disease.51 The novelty of our results is that we benefit from an objective measure of Mg2+ intake (namely uMg excretion) that, unlike circulating Mg2+, was found to be associated with risk of ischemic heart disease.54
In population-based studies, the association between uMg and metabolic phenotypes (i.e., BMI, fat mass, fasting insulin levels) tended to differ by ARL15 rs35929 variants. These results are in line with human studies showing that Mg2+ intake exerts more protective effects on type 2 diabetes in overweight compared with nonoverweight people.3 Such gene–diet interaction has already been observed for type 2 diabetes, for which the increased risk conferred by the rs7903146 TCF7L2 genotype was only observed in the absence of lifestyle intervention.55 Therefore, the ARL15 gene may modulate the beneficial effect of Mg2+ intake on insulin resistance in humans, offering a novel paradigm in nutrigenetics. Further, the beneficial effect of high Mg2+ intake on metabolic disorders, such as type 2 diabetes and obesity, may not be uniform across individuals.
In conclusion, the translational data described in this study offer novel insights for the understanding of Mg2+ homeostasis and its relation to metabolic phenotypes, and identify ARL15 as a key player for these processes.
Concise Methods
Genome-Wide Meta-Analyses
Seven European population-based cohorts with Caucasian ethnicity (CoLaus, CROATIA-Vis, CROATIA-Korcula, CROATIA-Split, Lothian Birth Cohort 1936, INGI-Val Borbera, and INGI-Carlantino, described in the Supplemental Material) participated in the study, with a total of 9099 individuals. A variety of electrolytes including Mg2+ as well as creatinine, were measured in both urine and serum (methods and mean values described in the Supplemental Methods and Supplemental Table 5).
All cohorts were whole genome genotyped before imputation using the HapMap CEU panel as reference (Supplemental Tables 6 and 7). Serum Mg2+ measurements and FEMg were subjected to quantile-quantile normalization in order to reach normality before performing linear regression. uMg measurements expressed in milligrams per deciliter were first standardized to urinary creatinine, then corrected for age-, sex-, and study-specific covariates (such as ancestry principal components, study center) before undergoing a quantile-quantile normalization. Approximately 2.5 M SNPs passing quality control checks were subjected to a linear regression of the residuals for each phenotype, and association summary statistics were collected.
Even though quantile-quantile plots of the individual GWAS showed minimal inflation, ruling out the presence of population substructure, a genomic-control correction on the basis of the λ factor was applied to the association P values. Subsequently, a meta-analysis was conducted using the inverse variant weighting method implemented in METAL.56 A new genomic-control correction was applied to combined statistics.
SKIPOGH Cohort
SKIPOGH is a population-based, family-based, multicenter study focusing on BP regulation and renal function.57 Each participant collected 24-hour urine. The study was approved by the institutional ethical committees of each participating university hospital and participants signed written informed consent.
VEGAS Analysis
The SNP association P values from the meta-analysis of uMg were analyzed using VEGAS,30 a program for performing gene-based tests for association using the summary statistics from genetic association studies. VEGAS assigns SNPs to genes and combines their association P values into a gene-based test statistic. Permutations are used to calculate the null distribution of the test statistic for each gene in order to derive an empirical gene-wise P value. The gene-based approach also reduces the multiple-testing problem of GWAS by only considering statistical tests for 17,787 genes, giving a Bonferroni-corrected threshold of P<2.8E−6.
Pathway Analysis
We used Magenta58 to check enrichment for pathways in either the BIOCARTA KEGG REACTOME databases, which were contained in the msigdb version 4.0 (http://www.broadinstitute.org/gsea/msigdb/collections.jsp#C2).
Metabolome Analysis
Nuclear magnetic resonance spectroscopy data obtained from urine samples was binned and normalized to produce metabolome features.59 These features were then associated with the SNPs of interest, including relevant covariates among standard covariates (age, sex, genotype principal components) and lifestyle factors (smoking, caffeine use, dietary intake, etc.).
CNV Analysis
In-house datasets were used to call CNVs and to check their correlation with the SNPs of interest. The CNVs call was done using pennCNV software.60 An SNP-by-sample matrix with the copy number status was created. Then the square correlation (Pearson correlation) between the value of each SNP of interest and the SNPs copy number status in a ±100 kb region has been calculated. For the SNPs of interest for which no correspondence has been found in the datasets, they have been replaced by the closest SNPs in high linkage disequilibrium (LD) and present in the datasets. LD between the SNPs of interest and a list of SNPs tagging CNVs from the Genetic Investigation of ANthropometric Traits (GIANT) consortium has also been calculated. The SNPs from the GIANT list are in LD >0.8 with their corresponding CNV.60
Laboratory Measurements
Electrolytes, hematology parameters, and glycaemia were dosed in the biochemical platform of the University of Zürich using standard clinical laboratory methods. Creatinine was measured using Jaffe kinetic compensated method (Roche Diagnostics, Switzerland; intra-assay variability 0.7%–2.9%). The CKI Epidemiology Collaboration (CKD-EPI) formula was used to calculate the eGFR. In CoLaus, urinary uromodulin, creatinine, electrolytes, and osmolality concentrations were measured in morning spot urine samples.
All urinary biochemical parameters were measured from samples stored at −80°C, using the same biochemical platform UniCel DxC 800 Synchron Clinical System (Beckman Coulter, Nyon, Switzerland) at the University of Zürich. Appropriate controls and sets of calibration standards were used before running each sample batch. All cohorts were subjected to the same measurement protocol, in the same laboratory.
Tissue Distribution and Localization of ARL15
The tissue distribution of Arl15 and of its highly conserved zebrafish ortholog arl15b was studied in adult mouse, adult zebrafish, and zebrafish larvae, as described in the Supplemental Material (for information on the primer sequences used for the RT-qPCR measurements, see Supplemental Table 8). Segmental expression of ARL15 in the mouse (C57BL/6J) kidney was studied at both protein and mRNA level. Five-micrometer sections of fixed frozen kidney samples were costained for ARL15 and specific segment markers: breast cancer resistance protein for the proximal tubule, Tamm–Horsfall protein for the TAL, Na+-Cl− cotransporter for the DCT, and aquaporin-2 for the collecting duct. For mRNA expression, kidneys were dissected and minced, before isolation of well characterized nephron segments. For more details of the immunohistochemistry, microdissection studies, and mRNA analyses in the nephron segments, see the Supplemental Material.
Functional Studies in Cells and Animal Models
To study the regulation of TRPM6 channel activity by ARL15, HEK293 cells were transfected with 1 µg of human TRPM6 cDNA33 and 250 ng of empty vector (mock), human WT ARL15, or a human dominant negative (T46N) ARL15 mutant as negative control. Whole-cell patch clamp recordings were performed to determine the electrophysiological properties of the cells transfected with the constructs mentioned above (see details in the Supplemental Material).
The regulation of ARL15 gene expression by dietary Mg2+ was studied in C57BL/6J mice and Tupfel long-fin zebrafish (details in the Supplemental Material. Primer information for RT-qPCR determinations is indicated in Supplemental Table 8). These studies were followed by a loss-of-function approach in zebrafish larvae using two nonoverlapping, splice-site blocking MOs: 5′-AAACACTGAAAGACGGGACAAAGAC-3′ and 5′-GTTAAGCGAGTATTAGGTTACCTCT-3′ (Gene Tools, Philomath, OR), designated as exon skipping 3 and 4 arl15b-MO, respectively. These antisense oligonucleotides were used to knockdown the highly conserved ARL15 ortholog in the zebrafish, arl15b. Phenotype rescue experiments were performed by coinjecting arl15b-MOs with human ARL15 cRNA as previously described,61 and detailed in the Supplemental Material. The use of two nonoverlapping arl15b-MOs and the combination with rescue experiments with human ARL15 cRNA served to rule out potential off-target effects in our knockdown approach.
Statistical Analyses in Experiments Performed in Cellular and Animal Models
All results are depicted as mean±SEM. Statistical analyses were conducted by one-way ANOVA followed by the Tukey multiple comparison post-test. When only two experimental groups were affected by the factor of variance, an unpaired t test was used. Statistical significance was set at P˂0.05. The datasets and summary statistics are available in the IUMSP Research Data Repository, under the link https://data.iumsp.ch/index.php/catalog/6 (identification number: CHE-GWAS-V01; created on July 12, 2017).
Metabolic Factors Associated with uMg Excretion
We used multiple linear regression to explore the association between square-root-transformed urinary Mg2+-to-creatinine ratio in spot urine and several metabolic markers (i.e., fasting triglycerides, total cholesterol, HDL cholesterol, glucose, insulin, homocysteine, adiponectin, GGT, and BMI), taken individually as the dependent variable, and also adjusting for serum Mg2+ and other potential confounders in SKIPOGH and CoLaus cohorts. In SKIPOGH, we used multivariate linear mixed effect regression to explore the association of fasting insulin with 24-hour uMg excretion, with adjustment for potential confounders. We used a conservative P value (0.05/9=0.0056) threshold to consider the associations of uMg with metabolic phenotypes as statistically significant.
SNP-by-Environment Interaction on uMg Levels
We explored the role of metabolic phenotypes on the association of ARL15 rs35929 with uMg levels in spot urine among CoLaus participants. We conducted multiple linear regression to explore the modifying effect of BMI, fat mass, and fasting insulin levels on the association of the ARL15 rs35929 with square-root-transformed uMg concentration, including age, sex, height, lean mass, CKD-EPI, urinary creatinine (square root), serum Mg2+, serum and urine Ca2+ (square-root), and menopausal status. The P values for interaction were not corrected for multiple testing because we tested a single global hypothesis (does the association between rs35929 and uMg differ by metabolic status?), guided by prior knowledge.
Disclosures
None.
Supplementary Material
Acknowledgments
We would like to acknowledge the invaluable contributions of the recruitment team in Korcula and Split, the administrative teams in Croatia and Edinburgh, and the people of Korcula and Split. The SNP genotyping for the CROATIA-Korcula cohort was performed in Helmholtz Zentrum München, Neuherberg, Germany. The SNP genotyping for the CROATIA-Split cohort was performed by AROS Applied Biotechnology, Aarhus, Denmark. We thank the LBC1936 (Lothian Birth Cohort 1936) participants, the LBC1936 team for data collection and collation, and the staff at the Wellcome Trust Clinical Research Facility for bio-sample collection and genotyping. We thank Dr. Sjoerd Verkaart, Anke Hoefnagels and Sami G. Mohammed for their excellent technical support during the performance of the functional analyses, and acknowledge Dr. Erwin van Wijk and Dr. Gert Flik for providing the facilities for zebrafish experimentation. For INGI (Italian Network on Genetic Isolates)-Carlantino, we thank Anna Morgan and Angela D’Eustacchio for technical support; the municipal administrators for their collaboration on the project and for logistic support; and all participants of the study.
This work was supported by the Long-Term Fellowship from the European Renal Association European Dialysis and Transplant Association (ERA LTF 175-2014) and the Impulsion Grant from the ERA-EDTA Working Group on Inherited Kidney Disorders to F.J.A; and by the grant from The Netherlands Organization for Scientific Research (NWO; VICI 016.130.668) to J.G.J.H. J.H.F.d.B. is supported by a grant from NWO (Rubicon 825.14.021) and R.J.M.B. is supported by the EURenOmics project from the European Union Seventh Framework Programme (FP7/2007–2013, agreement no 305608). Z.K. received financial support from the Leenaards Foundation, the Swiss National Science Foundation (31003A-143914), and SystemsX.ch (51RTP0_151019). M.B. and T.C. are supported by the Swiss National Centre of Competence in Research Kidney Control of Homeostasis (NCCR Kidney.C.H.) program. O.D. is supported by the European Community’s Seventh Framework Programme (305608 EURenOmics), the Swiss National Centre of Competence in Research Kidney Control of Homeostasis (NCCR Kidney.C.H.) program, the Swiss National Science Foundation (31003A_169850), and the Rare Disease Initiative Zurich (radiz), a clinical research priority program of the University of Zurich, Switzerland. The SKIPOGH (Swiss Kidney Project on Genes in Hypertension) study is supported by a grant from the Swiss National Science Foundation (FN 33CM30-124087). The CoLaus (Cohorte Lausannoise) study is supported by research grants from GlaxoSmithKline, the Faculty of Biology and Medicine of Lausanne, Lausanne University, and the Swiss National Science Foundation (grants 33CSCO-122661, 33CS30-139468, and 33CS30-148401). The CROATIA-Korcula and CROATIA-Split studies were funded by grants from the Medical Research Council (UK), European Commission Framework six-project EUROSPAN (European Special Populations Research Network) (contract no. LSHG-CT-2006-018947), and Republic of Croatia Ministry of Science, Education and Sports research grants to I.R. (108-1080315-0302). For the INGI-Val Borbera study, the research was supported by funds from Compagnia di San Paolo, Torino, Italy; Fondazione Cariplo, Italy; and Ministry of Health, Ricerca Finalizzata 2008 to D.T. Phenotype collection in LBC1936 was supported by Age UK (The Disconnected Mind project). Genotyping was funded by the BBSRC (Biotechnology and Biological Sciences Research Council) (BB/F019394/1). The work was undertaken by the University of Edinburgh Centre for Cognitive Ageing and Cognitive Epidemiology, part of the crosscouncil Lifelong Health and Wellbeing Initiative (MR/K026992/1). Funding from the BBSRC and Medical Research Council is gratefully acknowledged.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2017030267/-/DCSupplemental.
References
- 1.Feeney KA, Hansen LL, Putker M, Olivares-Yañez C, Day J, Eades LJ, Larrondo LF, Hoyle NP, O’Neill JS, van Ooijen G: Daily magnesium fluxes regulate cellular timekeeping and energy balance. Nature 532: 375–379, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Baaij JHF, Hoenderop JGJ, Bindels RJM: Magnesium in man: Implications for health and disease. Physiol Rev 95: 1–46, 2015 [DOI] [PubMed] [Google Scholar]
- 3.Dong JY, Xun P, He K, Qin LQ: Magnesium intake and risk of type 2 diabetes: Meta-analysis of prospective cohort studies. Diabetes Care 34: 2116–2122, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hruby A, Meigs JB, O’Donnell CJ, Jacques PF, McKeown NM: Higher magnesium intake reduces risk of impaired glucose and insulin metabolism and progression from prediabetes to diabetes in middle-aged americans. Diabetes Care 37: 419–427, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lopez-Ridaura R, Willett WC, Rimm EB, Liu S, Stampfer MJ, Manson JE, Hu FB: Magnesium intake and risk of type 2 diabetes in men and women. Diabetes Care 27: 134–140, 2004 [DOI] [PubMed] [Google Scholar]
- 6.Schulze MB, Schulz M, Heidemann C, Schienkiewitz A, Hoffmann K, Boeing H: Fiber and magnesium intake and incidence of type 2 diabetes: A prospective study and meta-analysis. Arch Intern Med 167: 956–965, 2007 [DOI] [PubMed] [Google Scholar]
- 7.Song Y, He K, Levitan EB, Manson JE, Liu S: Effects of oral magnesium supplementation on glycaemic control in Type 2 diabetes: A meta-analysis of randomized double-blind controlled trials. Diabet Med 23: 1050–1056, 2006 [DOI] [PubMed] [Google Scholar]
- 8.Chacko SA, Sul J, Song Y, Li X, LeBlanc J, You Y, Butch A, Liu S: Magnesium supplementation, metabolic and inflammatory markers, and global genomic and proteomic profiling: A randomized, double-blind, controlled, crossover trial in overweight individuals. Am J Clin Nutr 93: 463–473, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mooren FC, Krüger K, Völker K, Golf SW, Wadepuhl M, Kraus A: Oral magnesium supplementation reduces insulin resistance in non-diabetic subjects - a double-blind, placebo-controlled, randomized trial. Diabetes Obes Metab 13: 281–284, 2011 [DOI] [PubMed] [Google Scholar]
- 10.Guerrero-Romero F, Tamez-Perez HE, González-González G, Salinas-Martínez AM, Montes-Villarreal J, Treviño-Ortiz JH, Rodríguez-Morán M: Oral magnesium supplementation improves insulin sensitivity in non-diabetic subjects with insulin resistance. A double-blind placebo-controlled randomized trial. Diabetes Metab 30: 253–258, 2004 [DOI] [PubMed] [Google Scholar]
- 11.He K, Liu K, Daviglus ML, Morris SJ, Loria CM, Van Horn L, Jacobs DR Jr., Savage PJ: Magnesium intake and incidence of metabolic syndrome among young adults. Circulation 113: 1675–1682, 2006 [DOI] [PubMed] [Google Scholar]
- 12.Bertinato J, Lavergne C, Rahimi S, Rachid H, Vu NA, Plouffe LJ, Swist E: Moderately low magnesium intake impairs growth of lean body mass in obese-prone and obese-resistant rats fed a high-energy diet. Nutrients 8: E253, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ried JS, Jeff M J, Chu AY, Bragg-Gresham JL, van Dongen J, Huffman JE, Ahluwalia TS, Cadby G, Eklund N, Eriksson J, et al. : A principal component meta-analysis on multiple anthropometric traits identifies novel loci for body shape. Nat Commun 7: 13357, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moslehi N, Vafa M, Sarrafzadeh J, Rahimi-Foroushani A: Does magnesium supplementation improve body composition and muscle strength in middle-aged overweight women? A double-blind, placebo-controlled, randomized clinical trial. Biol Trace Elem Res 153: 111–118, 2013 [DOI] [PubMed] [Google Scholar]
- 15.Barbagallo M, Belvedere M, Dominguez LJ: Magnesium homeostasis and aging. Magnes Res 22: 235–246, 2009 [DOI] [PubMed] [Google Scholar]
- 16.Glaudemans B, Knoers NVAM, Hoenderop JGJ, Bindels RJM: New molecular players facilitating Mg(2+) reabsorption in the distal convoluted tubule. Kidney Int 77: 17–22, 2010 [DOI] [PubMed] [Google Scholar]
- 17.Devuyst O, Knoers NV, Remuzzi G, Schaefer F; Board of the Working Group for Inherited Kidney Diseases of the European Renal Association and European Dialysis and Transplant Association : Rare inherited kidney diseases: Challenges, opportunities, and perspectives. Lancet 383: 1844–1859, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hunter DJ, Lange M, Snieder H, MacGregor AJ, Swaminathan R, Thakker RV, Spector TD: Genetic contribution to renal function and electrolyte balance: A twin study. Clin Sci (Lond) 103: 259–265, 2002 [DOI] [PubMed] [Google Scholar]
- 19.Moulin F, Ponte B, Pruijm M, Ackermann D, Bouatou Y, Guessous I, Ehret G, Bonny O, Pechère-Bertschi A, Staessen JA, Paccaud F, Martin PY, Burnier M, Vogt B, Devuyst O, Bochud M: A population-based approach to assess the heritability and distribution of renal handling of electrolytes. Kidney Int doi: 10.1016/j.kint.2017.06.020, 2017 [DOI] [PubMed] [Google Scholar]
- 20.Simon DB, Lu Y, Choate KA, Velázquez H, Al-Sabban E, Praga M, Casari G, Bettinelli A, Colussi G, Rodríguez-Soriano J, McCredie D, Milford D, Sanjad S, Lifton RP: Paracellin-1, a renal tight junction protein required for paracellular Mg2+ resorption. Science 285: 103–106, 1999 [DOI] [PubMed] [Google Scholar]
- 21.Konrad M, Schaller A, Seelow D, Pandey AV, Waldegger S, Lesslauer A, Vitzthum H, Suzuki Y, Luk JM, Becker C, Schlingmann KP, Schmid M, Rodriguez-Soriano J, Ariceta G, Cano F, Enriquez R, Juppner H, Bakkaloglu SA, Hediger MA, Gallati S, Neuhauss SC, Nurnberg P, Weber S: Mutations in the tight-junction gene claudin 19 (CLDN19) are associated with renal magnesium wasting, renal failure, and severe ocular involvement. Am J Hum Genet 79: 949–957, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schlingmann KP, Weber S, Peters M, Niemann Nejsum L, Vitzthum H, Klingel K, Kratz M, Haddad E, Ristoff E, Dinour D, Syrrou M, Nielsen S, Sassen M, Waldegger S, Seyberth HW, Konrad M: Hypomagnesemia with secondary hypocalcemia is caused by mutations in TRPM6, a new member of the TRPM gene family. Nat Genet 31: 166–170, 2002 [DOI] [PubMed] [Google Scholar]
- 23.Walder RY, Landau D, Meyer P, Shalev H, Tsolia M, Borochowitz Z, Boettger MB, Beck GE, Englehardt RK, Carmi R, Sheffield VC: Mutation of TRPM6 causes familial hypomagnesemia with secondary hypocalcemia. Nat Genet 31: 171–174, 2002 [DOI] [PubMed] [Google Scholar]
- 24.Meyer TE, Verwoert GC, Hwang SJ, Glazer NL, Smith AV, van Rooij FJ, Ehret GB, Boerwinkle E, Felix JF, Leak TS, Harris TB, Yang Q, Dehghan A, Aspelund T, Katz R, Homuth G, Kocher T, Rettig R, Ried JS, Gieger C, Prucha H, Pfeufer A, Meitinger T, Coresh J, Hofman A, Sarnak MJ, Chen YD, Uitterlinden AG, Chakravarti A, Psaty BM, van Duijn CM, Kao WH, Witteman JC, Gudnason V, Siscovick DS, Fox CS, Köttgen A; Genetic Factors for Osteoporosis Consortium; Meta Analysis of Glucose and Insulin Related Traits Consortium : Genome-wide association studies of serum magnesium, potassium, and sodium concentrations identify six Loci influencing serum magnesium levels. PLoS Genet 6: e1001045, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang X, Li J, Guo Y, Wei Z, Mentch FD, Hou C, Zhao Y, Qiu H, Kim C, Sleiman PM, Hakonarson H: Genome-wide association study of serum minerals levels in children of different ethnic background. PLoS One 10: e0123499, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tin A, Köttgen A, Folsom AR, Maruthur NM, Tajuddin SM, Nalls MA, Evans MK, Zonderman AB, Friedrich CA, Boerwinkle E, Coresh J, Kao WH: Genetic loci for serum magnesium among African-Americans and gene-environment interaction at MUC1 and TRPM6 in European-Americans: The Atherosclerosis Risk in Communities (ARIC) study. BMC Genet 16: 56, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fleming CR, George L, Stoner GL, Tarrosa VB, Moyer TP: The importance of urinary magnesium values in patients with gut failure. Mayo Clin Proc 71: 21–24, 1996 [DOI] [PubMed] [Google Scholar]
- 28.Barbagallo M, Dominguez LJ: Magnesium and type 2 diabetes. World J Diabetes 6: 1152–1157, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kurstjens S, de Baaij JHF, Bouras H, Bindels RJM, Tack CJ, Hoenderop JGJ: Determinants of hypomagnesemia in patients with type 2 diabetes mellitus. Eur J Endocrinol 176: 11–19, 2017 [DOI] [PubMed] [Google Scholar]
- 30.Liu JZ, McRae AF, Nyholt DR, Medland SE, Wray NR, Brown KM, Hayward NK, Montgomery GW, Visscher PM, Martin NG, Macgregor S; AMFS Investigators : A versatile gene-based test for genome-wide association studies. Am J Hum Genet 87: 139–145, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nijenhuis T, Vallon V, van der Kemp AWCM, Loffing J, Hoenderop JGJ, Bindels RJM: Enhanced passive Ca2+ reabsorption and reduced Mg2+ channel abundance explains thiazide-induced hypocalciuria and hypomagnesemia. J Clin Invest 115: 1651–1658, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lawrence JTR, Birnbaum MJ: ADP-ribosylation factor 6 delineates separate pathways used by endothelin 1 and insulin for stimulating glucose uptake in 3T3-L1 adipocytes. Mol Cell Biol 21: 5276–5285, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Voets T, Nilius B, Hoefs S, van der Kemp AW, Droogmans G, Bindels RJM, Hoenderop JGJ: TRPM6 forms the Mg2+ influx channel involved in intestinal and renal Mg2+ absorption. J Biol Chem 279: 19–25, 2004 [DOI] [PubMed] [Google Scholar]
- 34.Groenestege WM, Hoenderop JGJ, van den Heuvel L, Knoers N, Bindels RJM: The epithelial Mg2+ channel transient receptor potential melastatin 6 is regulated by dietary Mg2+ content and estrogens. J Am Soc Nephrol 17: 1035–1043, 2006 [DOI] [PubMed] [Google Scholar]
- 35.Wingert RA, Davidson AJ: The zebrafish pronephros: A model to study nephron segmentation. Kidney Int 73: 1120–1127, 2008 [DOI] [PubMed] [Google Scholar]
- 36.Hwang P-P, Chou M-Y: Zebrafish as an animal model to study ion homeostasis. Pflugers Arch 465: 1233–1247, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shungin D, Winkler TW, Croteau-Chonka DC, Ferreira T, Locke AE, Mägi R, Strawbridge RJ, Pers TH, Fischer K, Justice AE, et al.; ADIPOGen Consortium; CARDIOGRAMplusC4D Consortium; CKDGen Consortium; GEFOS Consortium; GENIE Consortium; GLGC; ICBP; International Endogene Consortium; LifeLines Cohort Study; MAGIC Investigators; MuTHER Consortium; PAGE Consortium; ReproGen Consortium : New genetic loci link adipose and insulin biology to body fat distribution. Nature 518: 187–196, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Richards JB, Waterworth D, O’Rahilly S, Hivert MF, Loos RJ, Perry JR, Tanaka T, Timpson NJ, Semple RK, Soranzo N, Song K, Rocha N, Grundberg E, Dupuis J, Florez JC, Langenberg C, Prokopenko I, Saxena R, Sladek R, Aulchenko Y, Evans D, Waeber G, Erdmann J, Burnett MS, Sattar N, Devaney J, Willenborg C, Hingorani A, Witteman JC, Vollenweider P, Glaser B, Hengstenberg C, Ferrucci L, Melzer D, Stark K, Deanfield J, Winogradow J, Grassl M, Hall AS, Egan JM, Thompson JR, Ricketts SL, König IR, Reinhard W, Grundy S, Wichmann HE, Barter P, Mahley R, Kesaniemi YA, Rader DJ, Reilly MP, Epstein SE, Stewart AF, Van Duijn CM, Schunkert H, Burling K, Deloukas P, Pastinen T, Samani NJ, McPherson R, Davey Smith G, Frayling TM, Wareham NJ, Meigs JB, Mooser V, Spector TD; GIANT Consortium : A genome-wide association study reveals variants in ARL15 that influence adiponectin levels. PLoS Genet 5: e1000768, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S, Ganna A, Chen J, Buchkovich ML, Mora S, et al.; Global Lipids Genetics Consortium : Discovery and refinement of loci associated with lipid levels. Nat Genet 45: 1274–1283, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mahajan A, Go MJ, Zhang W, Below JE, Gaulton KJ, Ferreira T, Horikoshi M, Johnson AD, Ng MC, Prokopenko I, et al.; DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium; Asian Genetic Epidemiology Network Type 2 Diabetes (AGEN-T2D) Consortium; South Asian Type 2 Diabetes (SAT2D) Consortium; Mexican American Type 2 Diabetes (MAT2D) Consortium; Type 2 Diabetes Genetic Exploration by Nex-generation sequencing in muylti-Ethnic Samples (T2D-GENES) Consortium : Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat Genet 46: 234–244, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scott RA, Lagou V, Welch RP, Wheeler E, Montasser ME, Luan J, Mägi R, Strawbridge RJ, Rehnberg E, Gustafsson S, et al.; DIAbetes Genetics Replication and Meta-analysis (DIAGRAM) Consortium : Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nat Genet 44: 991–1005, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Swaminathan R: Magnesium metabolism and its disorders. Clin Biochem Rev 24: 47–66, 2003 [PMC free article] [PubMed] [Google Scholar]
- 43.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, et al.: Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466: 707–713, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gillingham AK, Munro S: The small G proteins of the Arf family and their regulators. Annu Rev Cell Dev Biol 23: 579–611, 2007 [DOI] [PubMed] [Google Scholar]
- 45.Arjona FJ, Chen Y-X, Flik G, Bindels RJM, Hoenderop JGJ: Tissue-specific expression and in vivo regulation of zebrafish orthologues of mammalian genes related to symptomatic hypomagnesemia. Pflugers Arch 465: 1409–1421, 2013 [DOI] [PubMed] [Google Scholar]
- 46.Bijvelds MJC, Velden JA, Kolar ZI, Flik G: Magnesium transport in freshwater teleosts. J Exp Biol 201: 1981–1990, 1998 [DOI] [PubMed] [Google Scholar]
- 47.Kersten S, Arjona FJ: Ion transport in the zebrafish kidney from a human disease angle: Possibilities, considerations, and future perspectives. Am J Physiol Renal Physiol 312: F172–F189, 2017 [DOI] [PubMed] [Google Scholar]
- 48.Miyares RL, de Rezende VB, Farber SA: Zebrafish yolk lipid processing: A tractable tool for the study of vertebrate lipid transport and metabolism. Dis Model Mech 7: 915–927, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Q, Dalman M, Chen Y, Akhter M, Brahmandam S, Patel Y, Lowe J, Thakkar M, Gregory A-V, Phelps D, Riley C, Londraville RL: Knockdown of leptin A expression dramatically alters zebrafish development. Gen Comp Endocrinol 178: 562–572, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boitard S, Boussaha M, Capitan A, Rocha D, Servin B: Uncovering adaptation from sequence data: Lessons from genome resequencing of four cattle breeds. Genetics 203: 433–450, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fang X, Wang K, Han D, He X, Wei J, Zhao L, Imam MU, Ping Z, Li Y, Xu Y, Min J, Wang F: Dietary magnesium intake and the risk of cardiovascular disease, type 2 diabetes, and all-cause mortality: A dose-response meta-analysis of prospective cohort studies. BMC Med 14: 210, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sarrafzadegan N, Khosravi-Boroujeni H, Lotfizadeh M, Pourmogaddas A, Salehi-Abargouei A: Magnesium status and the metabolic syndrome: A systematic review and meta-analysis. Nutrition 32: 409–417, 2016 [DOI] [PubMed] [Google Scholar]
- 53.Simental-Mendía LE, Sahebkar A, Rodríguez-Morán M, Guerrero-Romero F: A systematic review and meta-analysis of randomized controlled trials on the effects of magnesium supplementation on insulin sensitivity and glucose control. Pharmacol Res 111: 272–282, 2016 [DOI] [PubMed] [Google Scholar]
- 54.Joosten MM, Gansevoort RT, Mukamal KJ, van der Harst P, Geleijnse JM, Feskens EJ, Navis G, Bakker SJ; PREVEND Study Group : Urinary and plasma magnesium and risk of ischemic heart disease. Am J Clin Nutr 97: 1299–1306, 2013 [DOI] [PubMed] [Google Scholar]
- 55.Florez JC, Jablonski KA, Bayley N, Pollin TI, de Bakker PI, Shuldiner AR, Knowler WC, Nathan DM, Altshuler D; Diabetes Prevention Program Research Group : TCF7L2 polymorphisms and progression to diabetes in the diabetes prevention program. N Engl J Med 355: 241–250, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Willer CJ, Li Y, Abecasis GR: METAL: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26: 2190–2191, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ponte B, Pruijm M, Ackermann D, Vuistiner P, Eisenberger U, Guessous I, Rousson V, Mohaupt MG, Alwan H, Ehret G, Pechere-Bertschi A, Paccaud F, Staessen JA, Vogt B, Burnier M, Martin PY, Bochud M: Reference values and factors associated with renal resistive index in a family-based population study. Hypertension 63: 136–142, 2014 [DOI] [PubMed]
- 58.Segrè AV, Groop L, Mootha VK, Daly MJ, Altshuler D; DIAGRAM Consortium; MAGIC investigators : Common inherited variation in mitochondrial genes is not enriched for associations with type 2 diabetes or related glycemic traits. PLoS Genet 6: e1001058, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rueedi R, Ledda M, Nicholls AW, Salek RM, Marques-Vidal P, Morya E, Sameshima K, Montoliu I, Da Silva L, Collino S, Martin FP, Rezzi S, Steinbeck C, Waterworth DM, Waeber G, Vollenweider P, Beckmann JS, Le Coutre J, Mooser V, Bergmann S, Genick UK, Kutalik Z: Genome-wide association study of metabolic traits reveals novel gene-metabolite-disease links. PLoS Genet 10: e1004132, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang K, Li M, Hadley D, Liu R, Glessner J, Grant SF, Hakonarson H, Bucan M: PennCNV: An integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res 17: 1665–1674, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Arjona FJ, de Baaij JHF, Schlingmann KP, Lameris ALL, van Wijk E, Flik G, Regele S, Korenke GC, Neophytou B, Rust S, Reintjes N, Konrad M, Bindels RJM, Hoenderop JGJ: CNNM2 mutations cause impaired brain development and seizures in patients with hypomagnesemia. PLoS Genet 10: e1004267, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.