Primary cilia are essential for normal development and tissue homeostasis, but the mechanisms that remodel the centriole to promote cilia initiation are not well understood. Agbu et al. report that mouse RSG1, a small GTPase, regulates a late step in cilia initiation, downstream of TTBK2 and the CPLANE protein INTU.
Abstract
Primary cilia, which are essential for normal development and tissue homeostasis, are extensions of the mother centriole, but the mechanisms that remodel the centriole to promote cilia initiation are poorly understood. Here we show that mouse embryos that lack the small guanosine triphosphatase RSG1 die at embryonic day 12.5, with developmental abnormalities characteristic of decreased cilia-dependent Hedgehog signaling. Rsg1 mutant embryos have fewer primary cilia than wild-type embryos, but the cilia that form are of normal length and traffic Hedgehog pathway proteins within the cilium correctly. Rsg1 mother centrioles recruit proteins required for cilia initiation and dock onto ciliary vesicles, but axonemal microtubules fail to elongate normally. RSG1 localizes to the mother centriole in a process that depends on tau tubulin kinase 2 (TTBK2), the CPLANE complex protein Inturned (INTU), and its own GTPase activity. The data suggest a specific role for RSG1 in the final maturation of the mother centriole and ciliary vesicle that allows extension of the ciliary axoneme.
Introduction
Primary cilia are nonmotile microtubule-based organelles that extend from the surface of many eukaryotic cells and serve important mechanosensory and chemosensory functions (Collet et al., 1998; Kindt et al., 2012). Primary cilia are required for all responses to Hedgehog (Hh) family ligands in mice and are therefore required for embryonic development, stem cell maintenance, and Hh-driven tumorigenesis (Huangfu et al., 2003; Goetz and Anderson, 2010). Mice and humans with abnormal primary cilia can exhibit defects in brain patterning, skeletal development, and cardiac morphogenesis, and abnormal primary cilia can cause obesity, polycystic kidney disease, craniofacial defects, and retinal degeneration (Fliegauf et al., 2007; Braun and Hildebrandt, 2017).
The primary cilium assembles onto a modified mother centriole (the basal body), which acts as the template for the nine doublet microtubules of the cilium. A series of steps required for cilia assembly have been defined (Sánchez and Dynlacht, 2016). Maturation of the mother centriole is marked by the appearance of distal and subdistal appendages. Small membrane vesicles fuse to form larger ciliary vesicles (Lu et al., 2015), and the distal appendages mediate association of the mother centriole with the membrane of the ciliary vesicle or the plasma membrane (Tanos et al., 2013). Tau tubulin kinase 2 (TTBK2) controls a rate-limiting step in cilia initiation required for removal of the centriolar capping protein CP110 and recruitment of intraflagellar transport (IFT) proteins that carry cargo in the elongating cilium (Goetz et al., 2012). The microtubule axoneme begins to elongate as the ciliary vesicle and mother centriole are trafficked to the apical surface of the cell (Sorokin, 1962). When the ciliary vesicle docks onto the apical cell surface, it fuses with the plasma membrane to expose the ciliary axoneme to the extracellular environment and produce a functional primary cilium. Despite this information, the mechanisms that remodel centriolar microtubules and the ciliary membrane to allow formation of the ciliary axoneme are poorly understood (Nechipurenko et al., 2017).
The regulation of cilia formation is context-dependent. In many cultured cells, ciliogenesis is initiated after cells have exited the cell cycle (Seeley and Nachury, 2010). In contrast, proliferating cells in the mouse embryo and many adult mouse tissues are ciliated throughout the cell cycle (O’Connor et al., 2013; Bangs et al., 2015), except during M phase when the cilium disassembles and the mother centriole incorporates into one of the two spindle poles. In the early mouse embryo, cilia formation is regulated by cell lineage: all nonmitotic cells of embryonic lineages are ciliated, whereas extraembryonic lineages lack primary cilia at all phases of the cell cycle (Bangs et al., 2015). In adults, tumor progression can be associated with the lack of primary cilia, which could be caused by either cilia disassembly or a failure of cilia initiation (Seeger-Nukpezah et al., 2013; Menzl et al., 2014).
The atypical small GTPase RSG1 was first identified in a human protein interaction screen based on a low-affinity interaction with FUZZY (FUZ), a vertebrate homologue of a Drosophila melanogaster planar polarity effector gene (Gray et al., 2009). Knockdown experiments have shown that FUZ and other homologues of Drosophila planar polarity effector proteins, Inturned (INTU) and Fritz (also called WDPCP), are important for the formation of motile cilia in the multiciliated cells of Xenopus laevis skin (Park et al., 2006; Kim et al., 2010; Toriyama et al., 2016), and mouse mutants have shown that these proteins are also important in primary cilia formation (Heydeck et al., 2009; Zeng et al., 2010; Zhang et al., 2011; Cui et al., 2013). A recent proteomic study defined FUZ, INTU, WDPCP, and JBTS17 as core components of a single protein complex, the CPLANE protein complex, and appear to recruit RSG1 to the complex (Toriyama et al., 2016; Adler and Wallingford, 2017).
Here, we describe a new mouse mutant, pixiebob (pxb), which was identified based on polydactyly and other defects associated with abnormal Hedgehog signaling similar to those seen in ciliopathies. We show that pxb is a strong loss-of-function mutation that inactivates mouse RSG1. Rsg1 null embryos and mouse embryonic fibroblasts (MEFs) make primary cilia at low frequency; however, the few cilia that form are normal in length and function, suggesting a specific role for RSG1 in primary cilia initiation. Although the early steps of cilia initiation proceed normally in Rsg1 mutant cells, mother centrioles fail to extend the microtubule axoneme and fail to move the ciliary vesicle to the cell surface efficiently. We show that recruitment of RSG1 to the mother centriole depends on TTBK2, INTU, and its own GTPase activity, and that the RSG1 GTPase regulates a final, previously unrecognized step in cilia initiation.
Results
Rsg1 mutants arrest during gestation with a ciliopathy-like syndrome of developmental abnormalities
We identified a recessive ethylnitrosourea (ENU)-induced mouse mutation based on completely penetrant polydactyly on all limbs and lethality at embryonic day 12.5 (E12.5), which we named pixiebob (pxb; Fig. 1, A and B). Mapping and next-generation sequencing of exonic DNA from pxb mutants identified a missense mutation that would cause a valine-to-aspartic acid substitution at aa 169 (V169D) of RSG1 (Fig. S1 A and see Materials and methods). RSG1 (REM2- and RAB-like small GTPase 1) has the sequence hallmarks of small GTPases. Although it does not fall within a motif required for GTP or Mg2+ binding, V169 is conserved among vertebrate Rsg1 genes, suggesting that the pxb mutation disrupts gene function (Fig. S1 B).
Figure 1.
Shh signaling phenotypes of Rsg1 embryos. (A and B) Whole-mount images of WT (A) and Rsg1pxb mutant (B) embryonic limbs at E12.5. Note extra preaxial digits in the mutant (arrows). Bar, 500 µm. (C and D) Whole-mount RNA in situ hybridization for Gli1 transcripts in E10.5 WT (C) and Rsg1pxb (D) limbs (n = 6 embryos). Bar, 250 µm. (E) Western blot analysis of GLI3 from extracts of E10.5 limbs, showing the defect in processing of the full-length protein (FL) to the repressor form (R). α-Tubulin was the loading control (n = 3 independent experiments). (F–M) Confocal images of WT, Rsg1pxb, Rsg1pxb/CRISPR, and Rsg1CRISPR lumbar neural tube sections stained for SHH-dependent ventral cell populations marked by FOXA2 (floor plate), NKX2.2 (V3 interneuron progenitors), OLIG2 (motor neuron progenitors), and PAX6 (lateral neural progenitors; n = 3 embryos per genotype). Bars, 70 µm. In contrast to the results shown here, ventral cell fates were specified correctly in the rostral neural tube (not depicted), consistent with a milder defect in SHH response than seen in mutants that completely lack primary cilia.
To confirm that the pxb mutation disrupted the Rsg1 gene, we used the CRISPR/Cas9 system to generate a targeted null allele of the gene (Wang et al., 2013; Romanienko et al., 2016). By targeting the codon for threonine 69, a conserved residue that is essential for GTPase activity (Feig, 1999), we generated the Rsg1CRISPR allele, which contained a 2-bp insertion immediately downstream of codon 69, leading to a frame shift and deletion of essential motifs of the GTPase domain (Fig. S1, A and C; and see Materials and methods). Embryos homozygous for the Rsg1CRISPR allele or trans-heterozygous for Rsg1CRISPR/Rsg1pxb showed the same abnormal morphology at E10.5 and showed polydactyly and lethality at approximately E12.5, demonstrating that the pxb mutation is a strong loss-of-function allele of Rsg1 (Fig. S1, D and E).
The polydactyly of Rsg1 mutant embryos was similar to that seen in mutants with abnormal primary cilia (Goetz and Anderson, 2010), suggesting that the phenotypes of mouse Rsg1 mutants could be caused by disruption of cilia-dependent Hh signaling. Sonic hedgehog (SHH) signaling in the posterior limb bud promotes limb outgrowth and establishes digit number and identity (Harfe et al., 2004; Bouldin et al., 2010). Shh RNA expression was indistinguishable in WT and Rsg1pxb E10.5 limb buds (Fig. S2, A and B), whereas Gli1, a target of SHH signaling in the limb, was expressed at lower levels in the mutant limbs (Fig. 1, C and D), suggesting that RSG1 acts downstream of SHH and upstream of Gli1 in the core Hh signaling pathway. GLI3 is the transcription factor that is primarily responsible for repression of Hh target genes, and Gli3 mutants exhibit polydactyly because they lack the GLI3 repressor (Litingtung et al., 2002; Hill et al., 2009). The proteolytic processing event that produces GLI3 repressor depends on primary cilia (Huangfu and Anderson, 2005; May et al., 2005). As in Gli3 mutants (Aoto et al., 2002; López-Ríos et al., 2012), the domain of expression of Gremlin1, a bone morphogenetic protein inhibitor, was expanded anteriorly and the domain of Fgf8 expression in the apical ectodermal ridge was broadened in E10.5 Rsg1pxb limb buds (Fig. S2, C–F). To analyze GLI3 repressor levels directly, GLI3 protein from Rsg1pxb embryonic limbs was assayed by Western blot (Fig. 1 E). The level of full-length GLI3 was increased, whereas the amount of processed GLI3 was decreased in Rsg1pxb limb buds, consistent with a disruption of cilia-dependent GLI3 processing.
Defective specification of SHH-dependent ventral neural cell types is a hallmark of mutants with abnormal primary cilia (Goetz and Anderson, 2010). In the caudal neural tube of E10.5 embryos, FOXA2+ and NKX2.2+ cell populations, which are specified by the highest level of SHH signaling, were interspersed in Rsg1pxb mutants and clearly reduced in Rsg1CRISPR embryos (Fig. 1, F–I). Motor neuron progenitors, which are specified by an intermediate level of SHH signaling and express OLIG2, reside dorsal to NKX2.2+ V3 interneuron progenitors in WT embryos (Fig. 1 J). Expression of OLIG2 and the more dorsal marker PAX6 shifted ventrally in the Rsg1 mutant embryos, reflecting the loss of ventral neural cell types (Fig. 1, K–M). These phenotypes are consistent with a decreased response to SHH in the Rsg1 neural tube, although the loss of ventral neural cell types is less severe than in mutants that completely lack cilia (Goetz and Anderson, 2010).
Rsg1 embryos survive longer than mouse mutants that lack all cilia and show cilia-related phenotypes that would not be detectable at earlier stages. At E10.5, the domain of Fgf8 expression in craniofacial structures and the brain was expanded in Rsg1pxb mutants (Fig. S2, G and H), similar to the pattern seen in Gli3 mutants (Aoto et al., 2002; Rash and Grove, 2011), which is likely to account for facial clefting seen in E11.5 Rsg1 mutants. The whole-body edema and embryonic lethality at E12.5 suggested that cardiovascular function was compromised in Rsg1pxb embryos. Sections of the E12.5 Rsg1pxb heart showed that inferior and superior atrioventricular endocardial cushions failed to fuse (Fig. S2, I and J). This defect would be sufficient to explain the lethality of the mutants and is consistent with congenital heart defects seen in ciliary mutants (Friedland-Little et al., 2011; Willaredt et al., 2012; Li et al., 2015).
Rsg1 mutant embryos and cells have a reduced number of primary cilia of normal structure
Because of their ciliopathy-like embryonic phenotypes, we examined primary cilia in Rsg1 mutant embryos. Although most cells in the mesenchyme of the limb and adjacent to the neural tube are ciliated in WT, fewer than 30% of mesenchymal cells in Rsg1pxb and Rsg1CRISPR embryos had cilia (85.6 ± 2.0% in WT vs. 26.9 ± 2.9% and 21.0 ± 3.3%, respectively; Fig. 2, A–C and G).
Figure 2.
Reduction of cilia number in Rsg1 mutants. (A–C) Primary cilia in WT (A), Rsg1pxb (B), and Rsg1CRISPR (C) mesenchymal cells in the E10.5 limb. Bar, 3 µm (n = 3 embryos). (D–F) Primary cilia in WT (D), Rsg1pxb (E), and Rsg1CRISPR (F) MEFs. Bar, 3 µm. (G) Quantification of primary cilia formation in limb mesenchyme. Mean ± SD. WT, n = 870 cells; Rsg1pxb, n = 645 cells; Rsg1CRISPR, n = 589 cells. (H) Quantification of primary cilia formation in WT, Rsg1pxb, and Rsg1CRISPR MEFs. Mean ± SD. WT, n = 220 cells; Rsg1pxb, n = 239 cells; Rsg1CRISPR, n = 803 cells. (I and J) Higher-resolution images showing the length of primary cilia in WT (I) and Rsg1pxb (J) MEFs. Bar, 3 µm. (K) Quantification of cilia length in WT and Rsg1pxb MEFs. Mean ± SD. WT, n = 227 cilia; Rsg1pxb, n = 122 cilia. Error bars are SDs. (L and M) Scanning electron micrographs of primary cilia in the WT (L) and Rsg1pxb (M) brachial neural tube. Bar, 1 µm. t test compared with WT, ****, P < 0.0001; ns, no significant difference.
MEFs derived from Rsg1 mutants showed an even stronger defect in cilia formation than seen in the embryos. As visualized by immunostaining for ARL13B, a marker of the ciliary membrane (Caspary et al., 2007), the frequency of cilia formation was decreased four- to fivefold in Rsg1 mutant MEFs (Fig. 2, D–F and H; 18.4 ± 4.0% in Rsg1pxb [n = 239 cells] and 5.0 ± 2.8% in Rsg1CRISPR [n = 803 cells] vs. 76.1 ± 4.7% in WT [n = 220 cells]). Despite their reduced number, the cilia that formed in Rsg1pxb MEFs were normal in length (Fig. 2, I–K). Neural progenitors also had fewer cilia in the mutants: when viewed by scanning electron microscopy, the fraction of Rsg1pxb neural progenitors that were ciliated in the E10.5 neural epithelium was reduced, although the cilia that formed appeared to be of normal length (Fig. 2, L and M).
To test whether ciliary trafficking was affected by the Rsg1pxb mutation, we assayed the relocalization of Hh pathway proteins in response to pathway activation in the few cells that had cilia. After activation of the pathway by the small-molecule Hedgehog activator Smoothened Agonist (SAG), the membrane protein Smoothened (SMO) translocated to primary cilia in both WT and Rsg1pxb MEFs (Fig. 3, A–D). GLI2, the key transcriptional activator of the pathway, localized normally to cilia tips in the mutants and accumulated further after pathway activation by SAG (Fig. 3, E–H). KIF7, a regulator of Shh signaling and of the cilia tip compartment (He et al., 2014), also accumulated further at ciliary tips upon SAG addition in WT and Rsg1pxb MEFs (Fig. 3, I–L).
Figure 3.
Trafficking of Hedgehog pathway proteins is normal in Rsg1 mutant cilia. (A–D) SMO staining in WT (A and B) and Rsg1pxb (C and D) MEF cilia, in the absence of pathway activation (–SAG) or when the pathway was activated by the small molecule SAG (+SAG). (E–H) GLI2 at cilia tips in WT (E and F) and Rsg1pxb (G and H) MEFs, in the presence or absence of SAG. (I–L) KIF7 staining in WT (I and J) and Rsg1pxb (K and L) MEFs, in the presence or absence of SAG. Bars, 1 µm (n = 3 independent experiments). (M and N) Expression of Gli1 and Ptch1 mRNAs in WT, Rsg1pxb, and Rsg1CRISPR MEFs in the absence of pathway activation (–SAG) or when the pathway was activated by SAG (+SAG). Fold change is normalized to WT cells (=1) without SAG treatment; data are presented as mean ± SD, n = 3 experiments. t test *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, no significant difference.
To assay how mutant MEFs respond to Hh activation, we examined expression of two Hh target genes, Gli1 and Ptch1, in MEFs by quantitative PCR. Unlike the robust gene activation in WT MEFs, Gli1 and Ptch1 mRNA failed to be up-regulated in response to SAG in Rsg1pxb MEFs (Fig. 3, M and N). This suggests that normal pathway activation in the small fraction of mutant cells that are ciliated is not sufficient for a normal Hh pathway activation in the cell population, consistent with the embryonic phenotype of Rsg1 mutant embryos.
RSG1 localizes to the mother centriole and the transition zone of the primary cilium
Immunostaining with an antibody that recognizes the human RSG1 protein showed that endogenous RSG1 in human retinal pigment epithelial (RPE1) cells was detected at the transition zone of the primary cilium, between the γ-tubulin–positive basal body and the acetylated α-tubulin–positive axoneme, as well as in small cytoplasmic puncta (Fig. 4 A). Under conditions in which most cells were not ciliated (in the presence of serum), the antibody recognized RSG1 at the mother centriole in ∼30% of unciliated RPE1 cells (Fig. 4 B).
Figure 4.
RSG1 localizes to the mother centriole and the transition zone of the primary cilium. (A and B) Immunolocalization of endogenous RSG1 in human RPE1 cells. (A) In cilia-promoting conditions (absence of serum), RSG1 localizes to the ciliary transition zone, between the γ-tubulin+ mother basal body (pink) and the acetylated α-tubulin+ ciliary axoneme (red). (B) When grown in the presence of serum, the majority of cells lack cilia, and RSG1 was detected at one of the two centrioles in ∼30% of the unciliated cells. (C and D) GFP-RSG1 localization in transiently transfected ciliated (C) and unciliated (D) WT cells (n = 3 independent experiments). (E and F) GFP-RSG1 partially colocalized with CEP164 in transiently transfected ciliated (E) and unciliated (F) WT cells. (G and H) Rescue of cilia formation in Rsg1pxb MEFs by GFP-RSG1. Untransfected Rsg1pxb MEFs (G) and Rsg1pxb MEFs (H) transfected with GFP-RSG1 (n = 3 independent experiments). (I–K) GTPase activity is required for RSG1 localization to the mother centriole. (I) Transfected GFP-RSG1V169D localizes to the transition zone of transfected Rsg1pxb MEFs. (J) In the rare cilia that form after transfection of Rsg1pxb MEFs with GFP-RSG1T69N, GFP was not detected at the base of the cilium (n = 3 independent experiments). (K) Quantification of cilia frequency in MEFs transfected with GFP-RSG1, GFP-RSG1V169D, and GFP-RSG1T69N constructs. WT, n = 70 cells; Rsg1pxb, n = 67 cells; GFP-RSG1, n = 65 cells; GFP-RSG1V169D, n = 63 cells; and GFP-RSG1T69N, n = 55 cells. Mean ± SD. Cilia frequency in GFP-RSG1V169D and GFP-RSG1T69N was significantly reduced compared with WT cells and Rsg1pxb MEFs transfected with WT GFP-RSG1 (t test, *, P < 0.05; ****, P < 0.0001; n = 3 independent experiments). Bars, 2 µm.
To detect the protein in mouse cells, we generated a GFP-RSG1 fusion protein and expressed it in WT MEFs, where it was enriched in the transition zone of primary cilia (Fig. 4 C) and at one of the two centrioles in a subset of unciliated WT MEFs (26.3 ± 7.4%; n = 45 cells; Fig. 4 D). The centriole enriched for GFP-RSG1 also was marked by the distal appendage protein CEP164, indicating that RSG1 specifically associated with the mother centriole (Fig. 4 F). We noted that GFP-RSG1 appeared to localize inside the CEP164 ring and therefore might be closely associated with centriolar microtubules (Fig. 4, E and F). In ciliated cells, GFP-RSG1 was localized distal to CEP164 and proximal to axonemal acetyl-α-tubulin, suggested that it had relocalized to the transition zone (Fig. 4 E). Expression of the GFP-RSG1 fusion protein rescued the ciliation defect of Rsg1pxb MEFs, demonstrating that the fusion protein was functional (15.6 ± 4.7% ciliation in Rsg1pxb vs. 65.0 ± 7.1% in transfected Rsg1pxb MEFs; Fig. 4, G and H). Similarly, although only 1.2% of Rsg1CRISPR cells were ciliated in this experiment (1/75 cells), 27.8 ± 6.6% (16/58 cells) of transfected Rsg1CRISPR had cilia.
RSG1 is required for cilia initiation but not for the early steps of the mother centriole maturation
In previously described mouse mutants that form few primary cilia, the cilia that do form are short (e.g., Gray et al., 2009; Weatherbee et al., 2009; Friedland-Little et al., 2011). In contrast, few Rsg1 mutant cells formed cilia, but those cilia that did form appeared to be normal, which suggested that RSG1 acts specifically during cilia initiation to control the decision of whether to make a cilium. We therefore analyzed whether proteins known to play a role in cilia initiation localized correctly to the mother centriole in Rsg1 mutant MEFs.
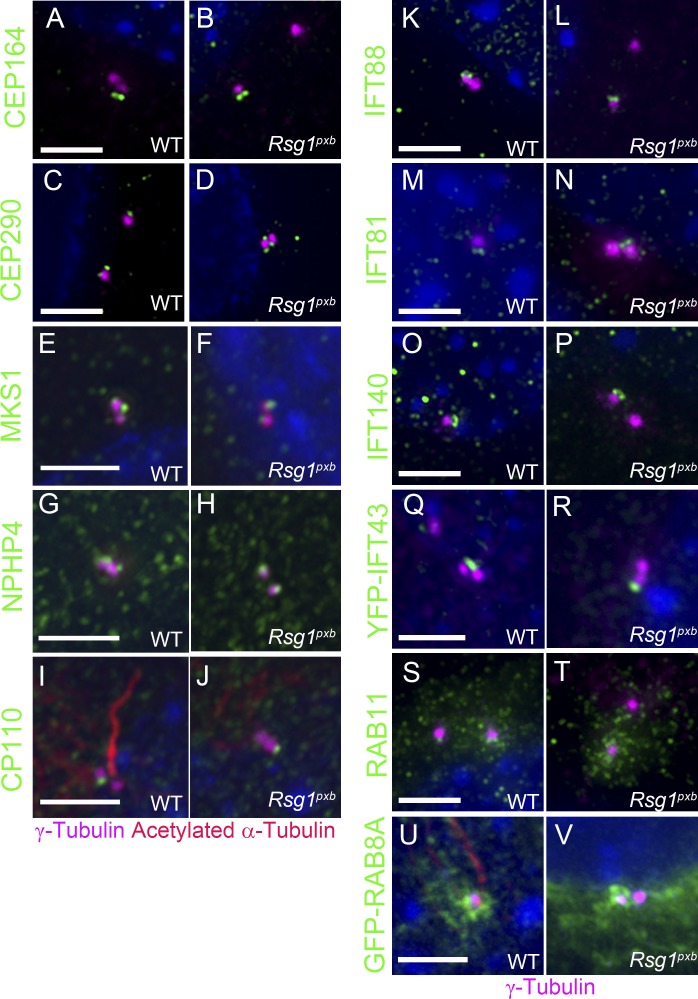
The distal appendages are required for docking of the mother centriole to the membrane (Tanos et al., 2013). CEP164, a distal appendage protein, is required for formation of distal appendages and cilia (Graser et al., 2007; Schmidt et al., 2012; Ye et al., 2014). As in WT, CEP164 associated with Rsg1pxb mother centrioles (Fig. 5, A and B). The ciliary transition zone, which is important for regulating entry and exit of cargo from cilia (Garcia-Gonzalo et al., 2011), forms immediately distal to the distal appendages of the mother centriole. CEP290, a component of the transition zone and centriolar satellites (Gorden et al., 2008; Kim et al., 2008; Garcia-Gonzalo et al., 2011; Stowe et al., 2012), localized to both the mother centriole and the centriolar satellites of WT and Rsg1pxb mutant cells (Fig. 5, C and D). MKS1, a component of the MKS complex that resides at the ciliary transition zone (Cui et al., 2011), localized to Rsg1pxb centrioles (Fig. 5, E and F). NPHP4, a component of a different transition zone complex, the nephronophthisis (NPHP) complex, was also recruited to Rsg1pxb centrioles (Fig. 5, G and H).
Figure 5.
Recruitment of appendage, transition zone, IFT, and cilia-associated RAB proteins to centrioles is normal in Rsg1pxb MEFs. (A and B) CEP164 staining in WT and Rsg1pxb MEFs. As in WT, CEP164 associated with Rsg1pxb mother centrioles (100 ± 0.0% in WT [n = 92 cells] vs. 98.5 ± 1.5% in Rsg1pxb [n = 91 cells]). (C and D) CEP290 localized to mother centrioles in both genotypes (99.5 ± 0.5% in WT [n = 47 cells] vs. 98.6 ± 0.9% in Rsg1pxb [n = 113 cells]). (E and F) MKS1, a transition zone protein, localized normally to Rsg1pxb centrioles (100 ± 0.0% in WT [n = 77 cells] vs. 93.0% ± 2.5% in Rsg1pxb [n = 87 cells]). (G and H) NPHP4, a component of the nephronophthisis (NPHP) complex, was recruited normally to Rsg1pxb centrioles (98.9 ± 1.1% in WT [n = 59 cells] vs. 94.9 ± 1.8% in Rsg1pxb [n = 111 cells]). (I and J) CP110 in WT (I) and Rsg1pxb (J) MEFs (88.8 ± 3.3% in WT [n = 293 cells] vs. 81.1 ± 4.2% in Rsg1pxb [n = 351 cells]). IFT88 (K and L), IFT81 (M and N), IFT140 (O and P), and YFP-IFT43 (Q and R) localization in WT and Rsg1pxb MEFs (IFT88, 98.7 ± 1.0% in WT [n = 134 cells] vs. 94.4 ± 2.1% in Rsg1pxb [n = 381 cells]; IFT81, 99.3 ± 0.4% in WT [n = 133 cells] vs. 96.7% ± 1.2% in Rsg1pxb [n = 178 cells]; IFT43-YFP, 83.6 ± 5.05% in WT [n = 64 cells] vs. 71.4 ± 5.0% in Rsg1pxb [n = 66 cells]). (S and T) RAB11 localization at WT and Rsg1pxb centrosomes after 1 h of serum starvation (46.6 ± 9.2% in WT [n = 34 centrosomes] vs. 56.7 ± 7.8% in Rsg1pxb [n = 57 centrosomes]). n = 3 independent experiments. (U and V) GFP-RAB8A is enriched near the mother centriole in both WT and Rsg1pxb cells. For all experiments, each genotype was examined in three independent experiments. Bars, 2 µm.
Cell-based data suggest that CP110, which localizes to the distal ends of both the daughter and the unciliated mother centriole, is a negative regulator of ciliogenesis and that ciliogenesis cannot begin until CP110 is removed from the mother centriole (Spektor et al., 2007; Tsang and Dynlacht, 2013). Under conditions that promote cilia formation (serum starvation), CP110 localized to only one of the two centrioles in >80% of Rsg1pxb MEFs (Fig. 5, I and J). In the embryo, CP110 also localized to a single centriole in mesenchymal cells of both WT and Rsg1pxb embryos (Fig. S3, A and B). Thus retention of CP110 was not responsible for the failure of cilia formation in the mutants. After CP110 knockdown by RNAi, centrioles elongated in Rsg1pxb MEFs, as previously reported in other cell types (Spektor et al., 2007; Bangs et al., 2015; Fig. S3, C and D), but CP110 knockdown did not increase the percentage of ciliated cells in Rsg1pxb MEFs (0%; 0/53 cells).
IFT-B proteins must localize to mother centrioles to promote axoneme assembly (Pazour et al., 2000). Mouse mutants that lack IFT-B proteins do not form cilia (Huangfu et al., 2003), and the cilia-initiating protein TTBK2 is required to recruit IFT-B proteins to the mother centriole (Goetz et al., 2012). The IFT-B proteins IFT88 and IFT81, which are both required for cilia biogenesis (Haycraft et al., 2007; Bhogaraju et al., 2013), localized to Rsg1pxb mother centrioles at a frequency comparable to WT (Fig. 5, K–N). RSG1 has been implicated in trafficking of IFT-A proteins in Xenopus multiciliated cells (Brooks and Wallingford, 2013; Toriyama et al., 2016). We found that both the IFT-A core complex protein IFT140 and the peripheral IFT-A protein IFT43 localized normally to Rsg1pxb mother centrioles (Fig. 5, O–R). The fluorescence intensity of all four IFT proteins at the mother centriole was comparable in WT and mutant MEFs (Fig. S4, A–D).
The small GTPases RAB11 and RAB8 act sequentially to promote the expansion of the membrane of the ciliary vesicle and the primary cilium (Lu et al., 2015). RAB11 was recruited efficiently to centrosomes in both WT and Rsg1pxb MEFs at 1 h of serum starvation (Fig. 5, S and T). To visualize RAB8, we transfected MEFs with a GFP-RAB8A construct (Westlake et al., 2011). GFP-RAB8A was detected adjacent to the ciliary base of WT, Rsg1pxb, and Rsg1CRISPR MEFs before axoneme formation (Fig. 5, U and V), and endogenous RAB8A was detected in the ciliary membrane of WT and mutant MEFs after 1 h of serum starvation (Fig. S4, E and F). Thus the recruitment of RAB11 and RAB8A, as well as all other proteins examined, to the nascent cilium is independent of RSG1 function.
Rsg1pxb mother centrioles dock on ciliary vesicles but are blocked in axoneme assembly
The data suggest that RSG1 acts at a late step in cilia initiation, after maturation of the mother centriole and recruitment of proteins to the basal body. For a mature mother centriole to form a primary cilium, it must associate with the membrane of the ciliary vesicle, where assembly of axonemal microtubules begins. SMO accumulation has been used as a marker for the ciliary vesicle (Joo et al., 2013; Ye et al., 2014). SMO was not detected at mother centrioles in the presence of serum (Fig. 6 A), but after serum starvation to induce cilia formation, SMO associated with one of the two centrioles (presumably the mother) in WT MEFs that had not yet formed cilia (94.4 ± 1.9%; n = 117 cells; Fig. 6, B and D). Similarly, even though most (Rsg1pxb) or nearly all (Rsg1CRISPR) mutant MEFs were not ciliated, a SMO+ ciliary vesicle associated with one of the two centrioles in serum-starved Rsg1pxb (94.8 ± 1.5%; n = 273 cells; Fig. 6 C) and Rsg1CRISPR MEFs (94.0 ± 1.8%; n = 210 cells; Fig. 6 E), providing evidence that docking of the mother centriole to the ciliary vesicle does not depend on RSG1.
Figure 6.
Mother centrioles associate with ciliary vesicles in the absence of RSG1. (A) SMO is not detected at centrioles of serum-fed unciliated WT MEFs. (B and C) SMO localizes to WT (B) and Rsg1pxb (C) mother centrioles after serum starvation. (D and E) SMO localization to WT (D) and Rsg1CRISPR (E) mother centrioles (n = 3 independent experiments). (A–E) Bars, 3 µm. (F and G) Transmission electron micrographs of ciliary vesicles associated with WT (F) and Rsg1CRISPR (G) mother centrioles from serial sections of the E11.5 neural tube. Bar, 250 nm.
We used transmission electron microscopy (TEM) to investigate docking of the mother centriole onto the membrane at high resolution in the embryonic neural epithelium (Fig. 6, F and G; and Fig. S5). As shown in Fig. 2 M, the mutant neural epithelium has few primary cilia on its surface. Quantitation of TEM images showed that 40% of the mother centrioles (defined by the presence of distal appendages) were associated with primary cilia at the cell surface of the WT neural epithelium (28/70 mother centrioles), whereas only ∼17% of mother centrioles (13/76) were associated with the surface in the Rsg1CRISPR neural epithelium. In contrast to the decreased number of mother centrioles at the plasma membrane, the majority of mother centrioles away from the surface of the neural epithelium docked onto ciliary vesicles in both WT and mutant: 76 ± 25% in WT (29/38 mother centrioles) and 54 ± 27% in Rsg1CRISPR (18/33 mother centrioles). These values are not statistically different, which suggests that the loss of cilia in the Rsg1 mutant neural plate is not caused by a defect in docking of the mother centriole onto the ciliary vesicle.
Inside most WT ciliary vesicles located away from the plasma membrane, microtubule-containing axonemes extended from the mother centriole (29/33; Fig. S5, A, C, E, and G). In contrast, the majority of Rsg1CRISPR mother centrioles docked onto ciliary vesicles failed to bud into the ciliary vesicle: less than half (13/27) of the Rsg1CRISPR mother centrioles extended microtubules into the ciliary vesicle (Fig. S5, B, D, F, and H), and the remainder had short axonemes that were a mean of half the length of WT axonemes in ciliary vesicles (424.3 ± 48.2 nm in WT [n = 33 ciliary vesicles] vs. 201.8 ± 35.0 nm [n = 27 ciliary vesicles] in Rsg1CRISPR). Thus the data suggest that RSG1 activity promotes assembly of the microtubule axoneme after the mother centriole has docked onto the ciliary vesicle and that RSG1 may also accelerate the rate of elongation of the young axoneme.
GTPase activity is required for RSG1 localization and function
RSG1 is a member of the RAS superfamily of small GTPases. Mutation of a Ser/Thr residue required for guanine nucleotide binding (position 69 in RSG1) to asparagine interferes with GTPase activity in other RAS family proteins (Feig, 1999), and overexpression of RSG1T69N interferes with ciliogenesis in the Xenopus epidermis (Gray et al., 2009). A transfected mouse GFP-RSG1T69N construct failed to localize to the ciliary base in transfected MEFs (Fig. 4 J). This construct also failed to rescue cilia formation in Rsg1pxb MEFs: the number of ciliated cells after expression of the GFP-RSG1T69N construct (23.2% ± 7.1%; n = 55 cells) was not significantly different from that of untransfected Rsg1pxb MEFs (Fig. 4 K; 15.6 ± 4.7%; n = 67 cells; P = 0.39); in the same experiments, 92.9% ± 2.9% of WT MEFs were ciliated. These data suggest that GTPase activity is required for RSG1 localization, and that localization to the ciliary base is required for RSG1 to promote cilia formation. In contrast, a GFP-RSG1V169D construct, which should mimic the product of the Rsg1pxb allele, localized correctly to the ciliary base (Fig. 4 I). Thus the V169D mutation does not prevent correct protein localization and instead is likely to interfere with the function of the correctly localized protein.
TTBK2 is required for recruitment of RSG1 to the mother centriole
TTBK2 is a serine/threonine kinase that, like RSG1, is essential for cilia initiation in vivo (Goetz et al., 2012). Using 3D structured illumination microscopy (SIM), both TTBK2 and GFP-RSG1 were visualized in a ring around the ciliary base in transfected MEFs (Fig. 7, A and B), a localization similar to the patterns of CEP164 and other proteins that reside at the base of primary cilia (Yang et al., 2015). In double-labeling experiments, TTBK2 and RSG1 were present in overlapping domains at the transition zone (Fig. 7 C).
Figure 7.
RSG1 localization to the mother centriole depends on TTBK2. (A–C) 3D-SIM image of GFP-RSG1 (A) and TTBK2 (B) localization at the ciliary base of a transfected WT MEF. Merged image is shown in C. Bar, 0.1 µm. (D and E) TTBK2 localization in serum-starved WT (D) and Rsg1pxb (E) MEFs. (F and G) GFP-RSG1 localization in serum-starved WT (F) and Ttbk2bby (G) MEFs. GFP-RSG1 never localized to the mother centriole in Ttbk2bby mutant MEFs (n = 60). n = 3 independent experiments. (H and I) TTBK2 localization in serum-starved WT (H) and Intu−/− (I) MEFs. (J and K) GFP-INTU localization in serum-starved WT (J) and Ttbk2bby (K) MEFs. GFP-INTU never localized to the mother centriole in Ttbk2bby mutant MEFs (n = 75). (D–K) Bars, 3 µm. n = 3 independent experiments.
In unciliated WT MEFs, TTBK2 localizes to one of two centrioles (presumably the mother; Goetz et al., 2012; Fig. 7 D). TTBK2 localized to one centriole in Rsg1pxb MEFs, indicating that TTBK2 recruitment to the mother centriole does not depend on RSG1 (Fig. 7 E). In contrast, although GFP-RSG1 localized to the mother centriole in a subset of unciliated WT MEFs (Fig. 7 F; 26.3 ± 7.4%; n = 45 cells), GFP-RSG1 never localized to the mother centriole in Ttbk2bby MEFs (0%; n = 60 cells; Fig. 7 G).
RSG1 acts downstream of the CPLANE complex protein INTU
INTU is a core component of the CPLANE complex that binds and recruits RSG1 to the basal bodies of multiciliated cells (Toriyama et al., 2016). Like Rsg1 mutants, mouse Intu null mutant embryos show phenotypes characteristic of partial loss of Hh activity, including polydactyly and a partial loss of ventral neural cell types (Zeng et al., 2010). Intu null cells have fewer cilia, and the cilia that do form are short, in contrast to the normal length of Rsg1 mutant cilia.
Although GFP-RSG1 localized to the mother centriole or transition zone in ∼90% of WT MEFs, GFP-RSG1 was never detected at the mother centriole or cilium of Intu null MEFs (Fig. 8, E and F), indicating that INTU is required for recruitment of RSG1 to the mother centriole. By use of methanol fixation, it was possible to detect GFP-tagged INTU at the mother centriole or ciliary transition zone in >90% of WT and Intu−/− MEFs (Fig. 8, A and B). GFP-INTU was also detected at the mother centriole or transition zone in ∼30% of Rsg1pxb MEFs (Fig. 8, C and D), indicating that INTU can be recruited to the base of the cilium in the absence of RSG1.
Figure 8.
RSG1 localization to the mother centriole depends on INTU. (A and B) Transfected GFP-INTU localizes to the mother centriole (A) of unciliated Intu−/− cells and the ciliary transition zone of Intu−/− ciliated MEFs (B). (C and D) GFP-INTU localization to the centriole (C) and cilium (D) was normal in Rsg1pxb mutant MEFs. (E and F) In contrast, GFP-RSG1 failed to localize to the centriole (E) and cilium (F) of Intu null MEFs. Bars, 2 µm. (G) Expression of GFP-RSG1 partially rescued the ciliogenesis defect of Intu−/− cells. GFP-RSG1 rescues the cilia ratio of Intu−/− cells to 20.0%, whereas expression of GFP-INTU does not increase the fraction of ciliated Rsg1pxb cells. (H) Expression of GFP-RSG1 does not increase the length of Intu−/− cilia, whereas expression of GFP-INTU rescues cilium length in Intu−/− cells; ****, P < 0.0001; ns, no significant difference. Error bars are the SD. (I) Working model for the network of interactions that controls cilia initiation.
We observed that ∼3% of Intu null mutant cells were ciliated, and the mutant cilia were ∼30% shorter than WT cilia (Fig. 8, G and H). Expression of the GFP-RSG1 construct partially overcame the ciliogenesis defect: 20% of Intu mutant cells were ciliated after GFP-RSG1 expression (Fig. 8 G), although the cilia that formed were the same short cilia as seen in Intu−/− MEFs (Fig. 8 H). Thus overexpression of RSG1 partially bypassed the requirement for INTU in cilia initiation but not the requirement for INTU in the control of cilia length. In contrast, expression of GFP-INTU did not increase the number of ciliated Rsg1pxb cells (6.7 ± 2.5% ciliated cells after expression of GFP-INTU, vs. 7.0 ± 1.9% in control cells). GFP-INTU was never associated with the centriole in Ttbk2 null cells (Fig. 7, J and K; n = 0/75 cells), whereas TTBK2 localized normally without INTU (Fig. 7, H and I). We therefore suggest that TTBK2 recruits INTU to the mother centriole, and that INTU then facilitates the recruitment of RSG1 (Fig. 8 I).
Discussion
The data show that the small GTPase RSG1 is required for embryonic development: embryos that lack RSG1 have a ciliopathy-like syndrome of defects that include polydactyly, loss of ventral neural cell types, craniofacial defects, and heart defects that are likely to be responsible for death before E12.5 of gestation. Two additional ENU-induced alleles of Rsg1 were identified in an independent screen and appear to have phenotypes that overlap those of the null alleles described here (http://www.informatics.jax.org/allele/summary?markerId=MGI:1923416&alleleType=Chemically%20induced%20). Although no mutations in human RSG1 have yet been identified, we predict that mutations in the human gene would cause a classic ciliopathy phenotype.
The Rsg1 phenotypes are milder than those caused by the complete absence of primary cilia (Huangfu et al., 2003; Goetz and Anderson, 2010) but are similar to those of other mutants that make short or abnormal primary cilia, including hypomorphic alleles of IFT-B genes (e.g., Ift88fxo; Huangfu et al., 2003); mutations that disrupt the ciliary transition zone (e.g., Tctn1; Garcia-Gonzalo et al., 2011); and mutations that disrupt Fuzzy, Inturned, or Wdpcp, which encode components of the CPLANE complex (Heydeck et al., 2009; Zeng et al., 2010; Zhang et al., 2011; Cui et al., 2013). Rsg1 is the only gene identified to date in which null mutations cause the production of fewer primary cilia of normal length and without a detectable defect in ciliary trafficking, suggesting that the function of RSG1 is dedicated to increasing the efficiency of primary cilia initiation.
The roles of RSG1 and other CPLANE proteins in apical docking of basal bodies were first defined in the multiciliated cells of the Xenopus skin (Gray et al., 2009; Brooks and Wallingford, 2013; Adler and Wallingford, 2017). Proteomic analysis suggests that RSG1 is a peripheral component of the CPLANE complex, whereas INTU, FUZ, WDPCP, and JBTS17 are core components (Toriyama et al., 2016; Adler and Wallingford, 2017). The knockdown phenotypes of these genes in the Xenopus multiciliated cells are similar, but there are differences in the primary cilia-dependent phenotypes of mouse mutants that lack components of the CPLANE complex. Null mutations in Wdpcp allow survival until birth (Zhang et al., 2011; Cui et al., 2013), whereas Fuz, Intu, and Rsg1 mutants die at midgestation with more severe developmental phenotypes (Heydeck et al., 2009; Zeng et al., 2010; Tabler et al., 2013). The cellular phenotypes are also distinct: Fuz, Intu, and Wdpcp mutant mouse embryos have short primary cilia (Zeng et al., 2010; Seo et al., 2011; Zhang et al., 2011; Cui et al., 2013), whereas the few cilia that are present in Rsg1 null mutants are normal in length and function. Thus the CPLANE complex proteins appear to have some shared and some distinct functions.
In addition to defects in apical docking of basal bodies, knockdown of Rsg1 in Xenopus epidermal multiciliated cells reduces the length of motile cilia and disrupts IFT within cilia (Brooks and Wallingford, 2013; Toriyama et al., 2016). Unlike the data from Xenopus multiciliated cells, it appears that RSG1 does not regulate IFT in primary cilia in the mouse, as we did not detect defects in cilia length, cilia diameter, or ciliary trafficking in the rare cilia that formed in the mutants. We suggest that INTU regulates two aspects of primary cilia formation: it both recruits RSG1 and regulates IFT, as the Intu mutant cilia rescued by expression of RSG1 are still short. In contrast, we propose that the only function of RSG1 in primary cilia is the regulation of cilia initiation.
Our data represent the first high-resolution analysis of the role of a CPLANE-associated protein in the events of primary cilia initiation. We find that distal appendage proteins and transition zone proteins associate with the Rsg1 mother centriole, the mutant centriole associates a morphologically normal ciliary vesicle, and the cilia-initiating kinase TTBK2 is recruited to the mother centriole, where it promotes removal of CP110 and recruitment of IFT proteins to the basal body. The Rsg1 mutant mother centriole docks successfully onto the membrane of the ciliary vesicle but fails to efficiently extend the axoneme and move to the apical surface. Thus RSG1 acts at a late step in cilia initiation.
TTBK2 was shown to regulate two aspects of cilia initiation: removal of CP110 from the mother centriole and recruitment of IFT proteins (Goetz et al., 2012). The data presented here define a new function for TTBK2 in cilia initiation: recruitment of INTU to the mother centriole, and INTU is then required for recruitment of RSG1 to the mother centriole. RSG1 recruitment to the mother centriole also depends on its own GTPase activity; once it is at the basal body, we infer that interaction of RSG1 with other unidentified proteins promotes cilia initiation (Fig. 8 I).
The downstream function of RSG1 remains enigmatic. GTPase activity is required for RSG1 recruitment to mother centriole; once recruited, the protein is then required for efficient axoneme extension and apical trafficking of the mature ciliary vesicle. The protein most closely related in sequence to RSG1 is REM2, another member of a small atypical branch of the RAS family, the RGK family (Finlin et al., 2000; Wennerberg et al., 2005). REM2 and other RGK proteins regulate the activity of specific voltage-dependent calcium channels (Finlin et al., 2003; Flynn et al., 2008; Magyar et al., 2012; Manning et al., 2013; Xu et al., 2015), REM2 has also been reported to regulate the pattern of dendritic branching in cortical neurons (Ghiretti et al., 2014), and the Drosophila RGK protein GEM binds the kinesin KIF9 (Piddini et al., 2001). Unlike most RAS superfamily proteins, RSG1 and other RGK proteins lack motifs that would mediate membrane association by prenylation or myristoylation (Finlin et al., 2000). RSG1 also lacks the polybasic C-terminal domain found in REM2 and other RGK proteins that mediates its membrane association through phosphatidylinositols (Correll et al., 2008), suggesting that RSG1 may not directly associate with membranes.
Because the GTPase domain of RSG1 also has amino acid substitutions that distinguish it from other RGK proteins, sequence comparisons do not provide clear hypotheses about the downstream activities of RSG1. Based on the Rsg1 mutant phenotype and the requirement for dynamic remodeling of centriolar microtubules during cilia initiation, we hypothesize that RSG1 interacts with regulators of microtubule dynamics and ciliary vesicle trafficking, and that RSG1 assists in remodeling axonemal microtubules to promote axoneme assembly.
CP110, KIF24, and interacting proteins have been implicated in remodeling of centriolar microtubules (Sánchez and Dynlacht, 2016), and the phenotype of a null mutation in mouse CP110 suggests that the protein has complex roles in cilia initiation (Yadav et al., 2016). It will be worthwhile to determine whether RSG1 modulates the function rather than localization of these proteins during the process of cilia initiation.
Materials and methods
Mouse strains and phenotypic analysis
All animal experiments were performed in accordance with institutional animal care and welfare guidelines of Memorial Sloan Kettering Cancer Center (MSKCC). The pxb allele was isolated from an ENU screen for recessive mutations that disrupt mouse embryonic development (García-García et al., 2005; unpublished data). For recombination mapping, genomic DNA was extracted with the Qiagen/Gentra PureGene kit and analyzed with a single nucleotide polymorphism panel (Illumina Mouse Medium Density Linkage Panel) at the Hospital for Sick Children (Toronto, ON, Canada), placing the pxb mutation between single nucleotide polymorphism markers rs13478002 and rs13478089 on chromosome 4. Subsequent single nucleotide polymorphism–based positional mapping localized the pxb mutation to a 1.5-Mbp region. For whole-exome sequencing, pxb genomic DNA was amplified and used for in-solution exon capture (Agilent SureSelect). Whole-exome sequencing was performed with 30× coverage on the Illumina HiSeq system as 2× 100 paired-end reads. Exome sequencing identified a single nucleotide change in the interval, an adenine-to-thymine transversion in exon 5 of the Rsg1 gene. For genotyping, a 175-bp fragment was PCR amplified from genomic DNA and digested with AccI. The pxb allele created an AccI restriction site, whereas the WT allele lacked this restriction site.
The Rsg1CRISPR mice were generated by the MSKCC Mouse Genetics Core Facility. The sgRNA sequences 5′-GTGTCCGGGAAGAGTGGTGTGGG-3′ and 5′-TGGTGTGGGCAAGACAGCACTGG-3′ were coinjected with Cas9 RNA into the male pronucleus of one-cell-stage embryos. Embryos were subsequently transplanted into foster females, and pups were genotyped at postnatal day 11 (P11) to P14 via the T7 endonuclease assay. Sanger sequencing confirmed and identified genetic mutations at the Cas9 cut site, and genotyping was performed by amplification of a 150-bp fragment from genomic DNA followed by restriction digestion with HpyCH4V. The CRISPR allele created an HpyCH4V restriction site, whereas the WT allele lacked HpyCH4V restriction sites.
Cell culture
MEFs were created by dissociation of E11.5 embryos in DMEM supplemented with 10% FBS, l-glutamine, and penicillin/streptomycin. MEFs were cultured at 5% CO2 at 37°C for five to six passages. MEFs were subsequently immortalized by serial passaging and used for transfection experiments. For cilia analysis, MEFs were serum-starved for 48 h in serum-free media and harvested. To induce Shh pathway activity, MEFs were serum-starved for 48 h in serum-free media and treated with 100 nM Smoothened agonist (SAG) for 24 h. Ttbk2 null (Ttbk2bby) and Intu−/− MEFs have been described (Zeng et al., 2010; Goetz et al., 2012).
RNAi
MEFs were transfected with 10 µM siRNA using Lipofectamine RNAiMax (Life Technologies). CP110 knockdown was performed with siGENOME SMARTpool (Thermo Fisher Scientific); the four primers in the pool were 5′-GCAAUAACGUCACUGUUGA-3′, 5′-GACAAACCAAGCCUUAAUA-3′, 5′-CAAGUAAUGUUUCUGCUAC-3′, and 5′-GCAUGUCAGAUCCUUAUAA-3′.
Cell transfection
MEFs were plated onto eight-well chamber slides, and each well was transfected with 0.2 µg DNA at 85–90% confluence. Transfections were done with Lipofectamine 2000 (Life Technologies) and Opti-MEM according to the manufacturer’s instructions. Media was changed 24 h posttransfection, and gene expression was visualized 48 h posttransfection.
Constructs
The YFP-IFT43 plasmid was a gift from R. Roepman (Radboud University Nijmegen Medical Centre, Nijmegen, Netherlands; Arts et al., 2011), and GFP-Rab8A was a gift from C. Westlake (National Cancer Institute, Bethesda, MD; Westlake et al., 2011). The full-length Rsg1 cDNA was prepared by reverse transcription of RNA from WT MEFs and cloned into pEGFP-C1, which was overexpressed under the control of the CMV promoter. Mutant primers were designed according to the QuikChange Site-Directed Mutagenesis kit (Agilent), and the V169D and T69N constructs were generated using the WT construct as a template according to the manufacturer’s instructions. Protein expression was verified by Western blot using transfected MEF lysates. GFP-INTU was previously described (Zeng et al., 2010).
Phenotypic analysis
For RNA in situ hybridization, dissected E10.5 embryos were fixed overnight in 4% PFA and dehydrated through a methanol series. Embryos were then rehydrated through a methanol series before whole-mount RNA in situ hybridization with digoxigenin-labeled riboprobes was performed as described (Belo et al., 1997). The Gremlin1 probe was a gift from S. Weatherbee (Yale University, New Haven, CT) and R. Harland (University of California, Berkeley, Berkeley, CA).
Hematoxylin and eosin staining
Embryonic tissue was fixed in 4% PFA for 48 h and dehydrated through an ethanol series. Embryonic tissue was then washed three times in Histoclear and three times in paraffin (Leica). Tissue was incubated in paraffin-filled plastic molds overnight at 60°C. Embryonic tissue was sectioned on a microtome at a thickness of 7 µm. For hematoxylin and eosin staining, sections were warmed at 37°C for 10–15 min before three washes in Histoclear and dehydrated through a graded ethanol series. Sections were washed in water, Harris hematoxylin, acid alcohol, sodium bicarbonate, 95% ethanol, and alcoholic eosin. Sections were then washed three times in 100% ethanol and twice in Histoclear. Mounting was performed in Permount (Thermo Fisher Scientific), and images were acquired using an Axioplan 2 imaging wide-field microscope (Zeiss) and a 10× 0.5-NA objective at room temperature. Images were obtained with a wide-field monochrome camera (AxioCam MR) using Axiovision software (Zeiss).
Microscopy and image acquisition
Imaging was performed with an image restoration system (DeltaVision, GE Healthcare) using a wide-field microscope (IX71; Olympus) and a 100× 1.4-NA objective at room temperature. Samples were mounted in ProlongGold (Life Technologies), and a cooled charge-coupled device camera (CoolSNAP HQ2; Photometrics) was used. Images were subsequently processed by iterative constrained deconvolution (softWoRx; GE Healthcare) and corrected for chromatic aberrations. The Volocity software package was used to produce extended-view images (PerkinElmer). Neural tube images were acquired on a Leica TCS Confocal SP5-II confocal microscope with a 20× 0.7-NA water objective, and samples were mounted in ProlongGold. For superresolution microscopy, samples were mounted in ProlongDiamond (Life Technologies). Superresolution imaging was performed at room temperature on a DeltaVision OMX Blaze 3D-SIM superresolution microscope (Applied Precision) with a 100× 1.4-NA UPLSAPO oil objective (Olympus) and three-evolve electron-multiplying CCD cameras (Photometrics) for sequential acquisition. Images were subsequently processed with softWoRx software (GE Healthcare) and Imaris (Bitplane).
Electron microscopy
For scanning electron microscopy, E10.5 embryos were fixed in 2% PFA and 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer (Electron Microscopy Services). Samples were dehydrated through an ethanol series, critical point-dried, and coated with gold particles. Scanning electron microscopy was performed using a field emission microscope (Supra 25; Zeiss) at room temperature. For TEM, E11.5 embryos were fixed in 2% PFA, 2.5% glutaraldehyde, and 2 mM calcium chloride in 0.075 M sodium cacodylate buffer, pH 7.4. Embryonic tissue was then treated with 0.1% tannic acid and washed in sodium cacodylate buffer, postfixed with 1% osmium tetroxide for 1 h, and stained en bloc with uranyl acetate for 30 min. Tissue was then dehydrated in a graded series of ethanol, washed with acetone, and embedded in resin (Eponate 12; Electron Microscopy Sciences). After polymerization at 60°C for 48 h, ultrathin serial sections were cut, poststained with 2% uranyl acetate and 1% lead citrate, and imaged in a TEM (100CX; Jeol) with a digital imaging system at room temperature (XR41-C; AMT). Images were acquired at 80 kV. To improve clarity, some images were adjusted in Photoshop (Adobe Systems). In every case, all pixels in the image were adjusted uniformly, and all panels within a figure were adjusted with identical settings.
Western blotting
E10.5 limbs and MEFs were lysed in general lysis buffer (50 mM Tris buffer, 150 mM NaCl, 0.5% NP-40, and 1× protease inhibitor; Roche) for 20 min, run through a 10-G needle (BD), and spun in a centrifuge for 5 min at 4°C. Protein lysates were quantified via BCA Assay (Life Technologies) and separated on an SDS-PAGE gel. Proteins were transferred onto PVDF membranes (Millipore), blocked with 5% milk/TBST, and blotted with primary and secondary antibodies diluted in 5% milk/TBST. Donkey anti–goat and sheep anti–mouse HRP-conjugated secondary antibodies (Amersham) were used. Signal was detected via chemiluminescence using the ECL Prime detection kit (GE Healthcare).
Immunostaining
Cells were fixed with 2% PFA for 10 min at room temperature followed by 100% methanol at −20°C for 5 min. Cells were permeabilized with 0.5% Triton X-100/PBS, washed in 0.3% Triton X-100/PBS, and blocked in 0.3% Triton X-100/2% BSA/5% goat serum in 1× PBS. For NPHP4, CEP290, and MKS1 protein staining, antigen retrieval was performed with 1× Pipes/Hepes/EGTA/MgCl2 (PHEM)/0.5% Triton X-100 buffer for 30 s after methanol fixation, which was followed by permeabilization with 1% Triton X-100/PBS and three washes in 0.3% Triton X-100/PBS. For RAB11 analysis, washes were performed with PBS/0.1% saponin. For RAB8A and Rab8A-GFP staining, cells were pretreated with 0.5% Triton X-100/PHEM buffer for 30 s followed by PFA fixation and the rest of the immunostaining protocol. For YFP-IFT43 staining, cells were fixed with 4% PFA followed by methanol fixation for 5 min at −20°C. For GFP-INTU, cells were fixed in methanol for 10 min at −20°C, permeabilized with 0.5% Triton X-100/PBS for 10 min, washed, and incubated with an anti-GFP antibody. For analysis of neural patterning, E10.5 embryos were fixed in 2% PFA overnight, cryoprotected in 30% sucrose, embedded in Tissue-Tek OCT (Sakura), and cryosectioned at 10 µm. The sections were then stained for ventral cell types in the neural tube. All sections were stained with DAPI to mark nuclei.
Antibodies used were rabbit α-FOXA2 (1:1,000; Abcam), mouse α-NKX2.2 (1:10; Developmental Studies Hybridoma Bank), rabbit α-OLIG2 (1:200; Millipore); mouse α-PAX6 (1:10; Developmental Studies Hybridoma Bank), rabbit α-ARL13B (1:1,000; gift of T. Caspary, Emory University, Atlanta, GA), mouse α-acetylated α-tubulin (1:2,000; Sigma-Aldrich), mouse α-γ-tubulin (1:1,000; Sigma Aldrich), rabbit α-Smoothened (1:1,000; Ocbina and Anderson, 2008), rabbit α-CP110 (1:500; Proteintech), rabbit α-IFT88 (1:500; Proteintech), rabbit α-IFT81 (1:500; Proteintech), rabbit α-IFT140 (1:1,000; gift of G. Pazour, University of Massachusetts Medical School, Worcester, MA), rabbit α-TTBK2 (1:1,000; Sigma Aldrich), guinea pig α-GLI2 (1:1,000; gift of J. Eggenschwiler, University of Georgia, Athens, GA), goat anti-GLI2 (1:3,000), goat anti-GLI3 (1:3,000), rabbit α-KIF7 (1:1,000; He et al., 2014), rabbit α-RAB11 (1:100; Cell Signaling Technology), rabbit α-RAB8A (1:5,000; Proteintech), rabbit α-CEP164 (1:500; Sigma-Aldrich), rabbit α-CEP290 (1:500; Proteintech), rabbit α-MKS1 (1:300; Proteintech), rabbit α-NPHP4 (1:100; Proteintech), chicken α-GFP (1:1,000; Abcam), and rabbit α-GFP (1:1,000; Life Technologies). Goat Alexa Fluor 405–, 488–, 594–, and 647–conjugated secondary antibodies were used (Life Technologies). Double antibody staining with rabbit antibodies was performed in series using Dylight 488-Fab fragment goat anti-rabbit H+L (Jackson ImmunoResearch Laboratories).
Quantitative RT-PCR
For Gli1 and Ptch1 mRNA expression analysis, WT, Rsg1pxb, and Rsg1CRISPR MEFs were grown on six-well dishes and serum-starved for 48 h, and then grown with or without 100 nM Hh agonist SAG for 24 h. Total RNA extraction was performed using the RNeasy Mini kit with the RNase-Free DNase set to perform on-column DNase digestion. For RT-PCR, 1–2 µg RNA was used with random hexamer primers to synthesize cDNA using the SuperScript III First-Strand Synthesis System (Invitrogen). cDNA was diluted fivefold for quantitative PCR. Quantitative PCR was performed using Thermo Fisher SYBR Green PCR Master Mix (Life Technologies) for detecting products on a LightCycler 480 II Real-Time PCR instrument (Roche). All reactions were done at 60° annealing temperature, with an extension time of 15 s for all the primers: Gli1, 5′-GAAGGAATTCGTGTGCCATT-3′ and 5′-GCAACCTTCTTGCTCACACA-3′; Ptch1, 5′- CCATTTGAGGACAGGACTGG-3′ and 5′-CATGCTGAGAATTGCAGGAA-3′; and Gapdh, 5′- GCAATGCATCCTGCACCACCA-3′ and 5′-TTCCAGAGGGGCCATCCACA-3′ (Eguether et al., 2014). All reactions were done in triplicate and accompanied by control reactions using cDNA synthesized without reverse transcription (–RT controls). The success of reactions was confirmed by analysis of amplification curves, melting curves, and electrophoresis of representative amplification products on agarose gels.
LightCycler 480 Software was used to quantify products by absolute quantification analysis using the second-derivative maximum method. All crossing point values were normalized to the mean crossing point value obtained for the triplicate Gapdh reactions, to get relative values. The mean relative value represents the relative amount of product in any given experiment (cell line and condition). To obtain fold change values, the mean relative value for each experiment was normalized to the mean of that obtained for WT MEFs without SAG treatment.
Statistics
Comparisons between two conditions were performed with the two-tailed Student’s t test, and comparisons between three or more conditions were performed with one-way ANOVA and Tukey’s multiple comparison test (Prism). P < 0.05 was considered to be significant.
Online supplemental material
Fig. S1 is a schematic of the Rsg1 alleles and shows that the two mutant alleles fail to complement. Fig. S2 shows limb, craniofacial, and heart phenotypes of the Rsg1pxb allele. Fig. S3 shows that CP110 removal from the mother centriole in embryonic tissues is not affected by the Rsg1pxb mutation. Fig. S4 shows IFT intensity and RAB8A localization in Rsg1pxb MEFs. Fig. S5 shows WT and Rsg1CRISPR ciliary vesicles and microtubule axonemes visualized by TEM.
Supplementary Material
Acknowledgments
We thank Heather Alcorn for assistance with exome sequencing and Kunihiro Uryu (Rockefeller Electron Microscopy Core) and Nina Lampen (MSKCC Electron Microscopy Core) for assistance with transmission and scanning electron microscopy, respectively. We also thank the Rockefeller Bio-Imaging Resource Center and MSKCC Molecular Cytology Facility (Y. Romin, S. Fujisawa, and V. Boyko) for assistance with wide-field and confocal microscopy, respectively. We thank Peter Romanienko (MSKCC Mouse Genetics Core) for assistance with generating CRISPR mice and the Genomics Core Facility and Nicholas Socci (MSKCC Bioinformatics Core) for assistance in processing whole-exome sequencing data. We thank Ronald Roepman and Christopher Westlake for tagged constructs. We thank Devanshi Jain (MSKCC) and Greg Pazour (University of Massachusetts) for the help with qPCR experiments. We are grateful to Angela Parrish, Rocío Hernández-Martinez, and Benjamin Cyge for comments on the manuscript.
The work was supported by the National Institutes of Health (R37 HD03455 and R01 NS044385 to K.V. Anderson), a Memorial Sloan Kettering Cancer Center Support Grant (P30 CA008748), and a National Institutes of Health training grant (T32 HD060600) to S.O. Agbu.
The authors declare no competing financial interests.
Author contributions: S.O. Agbu, Y. Liang, and K.V. Anderson designed and carried out the experiments, analyzed the data, and wrote the paper. A. Liu provided reagents.
Footnotes
Abbreviations used:
- E
- embryonic day
- ENU
- ethylnitrosourea
- IFT
- intraflagellar transport
- MEF
- mouse embryonic fibroblast
- SHH
- Sonic hedgehog
- SIM
- structured illumination microscopy
- TEM
- transmission electron microscopy
References
- Adler P.N., and Wallingford J.B.. 2017. From planar cell polarity to ciliogenesis and back: The curious tale of the PPE and CPLANE proteins. Trends Cell Biol. 27:379–390. 10.1016/j.tcb.2016.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoto K., Nishimura T., Eto K., and Motoyama J.. 2002. Mouse GLI3 regulates Fgf8 expression and apoptosis in the developing neural tube, face, and limb bud. Dev. Biol. 251:320–332. 10.1006/dbio.2002.0811 [DOI] [PubMed] [Google Scholar]
- Arts H.H., Bongers E.M., Mans D.A., van Beersum S.E., Oud M.M., Bolat E., Spruijt L., Cornelissen E.A., Schuurs-Hoeijmakers J.H., de Leeuw N., et al. 2011. C14ORF179 encoding IFT43 is mutated in Sensenbrenner syndrome. J. Med. Genet. 48:390–395. 10.1136/jmg.2011.088864 [DOI] [PubMed] [Google Scholar]
- Bangs F.K., Schrode N., Hadjantonakis A.K., and Anderson K.V.. 2015. Lineage specificity of primary cilia in the mouse embryo. Nat. Cell Biol. 17:113–122. 10.1038/ncb3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belo J.A., Bouwmeester T., Leyns L., Kertesz N., Gallo M., Follettie M., and De Robertis E.M.. 1997. Cerberus-like is a secreted factor with neutralizing activity expressed in the anterior primitive endoderm of the mouse gastrula. Mech. Dev. 68:45–57. 10.1016/S0925-4773(97)00125-1 [DOI] [PubMed] [Google Scholar]
- Bhogaraju S., Čajánek L., Fort C., Blisnick T., Weber K., Taschner M., Mizuno N., Lamla S., Bastin P., Nigg E.A., and Lorentzen E.. 2013. Molecular basis of tubulin transport within the cilium by IFT74 and IFT81. Science. 341:1009–1012. 10.1126/science.1240985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouldin C.M., Gritli-Linde A., Ahn S., and Harfe B.D.. 2010. Shh pathway activation is present and required within the vertebrate limb bud apical ectodermal ridge for normal autopod patterning. Proc. Natl. Acad. Sci. USA. 107:5489–5494. 10.1073/pnas.0912818107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun D.A., and Hildebrandt F.. 2017. Ciliopathies. Cold Spring Harb. Perspect. Biol. 9:a028191 10.1101/cshperspect.a028191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks E.R., and Wallingford J.B.. 2013. The small GTPase Rsg1 is important for the cytoplasmic localization and axonemal dynamics of intraflagellar transport proteins. Cilia. 2:13 10.1186/2046-2530-2-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspary T., Larkins C.E., and Anderson K.V.. 2007. The graded response to Sonic Hedgehog depends on cilia architecture. Dev. Cell. 12:767–778. 10.1016/j.devcel.2007.03.004 [DOI] [PubMed] [Google Scholar]
- Collet J., Spike C.A., Lundquist E.A., Shaw J.E., and Herman R.K.. 1998. Analysis of osm-6, a gene that affects sensory cilium structure and sensory neuron function in Caenorhabditis elegans. Genetics. 148:187–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correll R.N., Botzet G.J., Satin J., Andres D.A., and Finlin B.S.. 2008. Analysis of the Rem2-voltage dependant calcium channel beta subunit interaction and Rem2 interaction with phosphorylated phosphatidylinositide lipids. Cell. Signal. 20:400–408. 10.1016/j.cellsig.2007.10.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui C., Chatterjee B., Francis D., Yu Q., SanAgustin J.T., Francis R., Tansey T., Henry C., Wang B., Lemley B., et al. 2011. Disruption of Mks1 localization to the mother centriole causes cilia defects and developmental malformations in Meckel-Gruber syndrome. Dis. Model. Mech. 4:43–56. 10.1242/dmm.006262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui C., Chatterjee B., Lozito T.P., Zhang Z., Francis R.J., Yagi H., Swanhart L.M., Sanker S., Francis D., Yu Q., et al. 2013. Wdpcp, a PCP protein required for ciliogenesis, regulates directional cell migration and cell polarity by direct modulation of the actin cytoskeleton. PLoS Biol. 11:e1001720 10.1371/journal.pbio.1001720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguether T., San Agustin J.T., Keady B.T., Jonassen J.A., Liang Y., Francis R., Tobita K., Johnson C.A., Abdelhamed Z.A., Lo C.W., and Pazour G.J.. 2014. IFT27 links the BBSome to IFT for maintenance of the ciliary signaling compartment. Dev. Cell. 31:279–290. 10.1016/j.devcel.2014.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feig L.A. 1999. Tools of the trade: Use of dominant-inhibitory mutants of Ras-family GTPases. Nat. Cell Biol. 1:E25–E27. 10.1038/10018 [DOI] [PubMed] [Google Scholar]
- Finlin B.S., Shao H., Kadono-Okuda K., Guo N., and Andres D.A.. 2000. Rem2, a new member of the Rem/Rad/Gem/Kir family of Ras-related GTPases. Biochem. J. 347:223–231. 10.1042/bj3470223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlin B.S., Crump S.M., Satin J., and Andres D.A.. 2003. Regulation of voltage-gated calcium channel activity by the Rem and Rad GTPases. Proc. Natl. Acad. Sci. USA. 100:14469–14474. 10.1073/pnas.2437756100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fliegauf M., Benzing T., and Omran H.. 2007. When cilia go bad: Cilia defects and ciliopathies. Nat. Rev. Mol. Cell Biol. 8:880–893. 10.1038/nrm2278 [DOI] [PubMed] [Google Scholar]
- Flynn R., Chen L., Hameed S., Spafford J.D., and Zamponi G.W.. 2008. Molecular determinants of Rem2 regulation of N-type calcium channels. Biochem. Biophys. Res. Commun. 368:827–831. 10.1016/j.bbrc.2008.02.020 [DOI] [PubMed] [Google Scholar]
- Friedland-Little J.M., Hoffmann A.D., Ocbina P.J.R., Peterson M.A., Bosman J.D., Chen Y., Cheng S.Y., Anderson K.V., and Moskowitz I.P.. 2011. A novel murine allele of Intraflagellar Transport Protein 172 causes a syndrome including VACTERL-like features with hydrocephalus. Hum. Mol. Genet. 20:3725–3737. 10.1093/hmg/ddr241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-García M.J., Eggenschwiler J.T., Caspary T., Alcorn H.L., Wyler M.R., Huangfu D., Rakeman A.S., Lee J.D., Feinberg E.H., Timmer J.R., and Anderson K.V.. 2005. Analysis of mouse embryonic patterning and morphogenesis by forward genetics. Proc. Natl. Acad. Sci. USA. 102:5913–5919. 10.1073/pnas.0501071102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gonzalo F.R., Corbit K.C., Sirerol-Piquer M.S., Ramaswami G., Otto E.A., Noriega T.R., Seol A.D., Robinson J.F., Bennett C.L., Josifova D.J., et al. 2011. A transition zone complex regulates mammalian ciliogenesis and ciliary membrane composition. Nat. Genet. 43:776–784. 10.1038/ng.891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiretti A.E., Moore A.R., Brenner R.G., Chen L.F., West A.E., Lau N.C., Van Hooser S.D., and Paradis S.. 2014. Rem2 is an activity-dependent negative regulator of dendritic complexity in vivo. J. Neurosci. 34:392–407. 10.1523/JNEUROSCI.1328-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz S.C., and Anderson K.V.. 2010. The primary cilium: A signalling centre during vertebrate development. Nat. Rev. Genet. 11:331–344. 10.1038/nrg2774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz S.C., Liem K.F. Jr., and Anderson K.V.. 2012. The spinocerebellar ataxia-associated gene Tau tubulin kinase 2 controls the initiation of ciliogenesis. Cell. 151:847–858. 10.1016/j.cell.2012.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorden N.T., Arts H.H., Parisi M.A., Coene K.L., Letteboer S.J., van Beersum S.E., Mans D.A., Hikida A., Eckert M., Knutzen D., et al. 2008. CC2D2A is mutated in Joubert syndrome and interacts with the ciliopathy-associated basal body protein CEP290. Am. J. Hum. Genet. 83:559–571. 10.1016/j.ajhg.2008.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graser S., Stierhof Y.-D., Lavoie S.B., Gassner O.S., Lamla S., Le Clech M., and Nigg E.A.. 2007. Cep164, a novel centriole appendage protein required for primary cilium formation. J. Cell Biol. 179:321–330. 10.1083/jcb.200707181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray R.S., Abitua P.B., Wlodarczyk B.J., Szabo-Rogers H.L., Blanchard O., Lee I., Weiss G.S., Liu K.J., Marcotte E.M., Wallingford J.B., and Finnell R.H.. 2009. The planar cell polarity effector Fuz is essential for targeted membrane trafficking, ciliogenesis and mouse embryonic development. Nat. Cell Biol. 11:1225–1232. 10.1038/ncb1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe B.D., Scherz P.J., Nissim S., Tian H., McMahon A.P., and Tabin C.J.. 2004. Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell. 118:517–528. 10.1016/j.cell.2004.07.024 [DOI] [PubMed] [Google Scholar]
- Haycraft C.J., Zhang Q., Song B., Jackson W.S., Detloff P.J., Serra R., and Yoder B.K.. 2007. Intraflagellar transport is essential for endochondral bone formation. Development. 134:307–316. 10.1242/dev.02732 [DOI] [PubMed] [Google Scholar]
- He M., Subramanian R., Bangs F., Omelchenko T., Liem K.F. Jr., Kapoor T.M., and Anderson K.V.. 2014. The kinesin-4 protein Kif7 regulates mammalian Hedgehog signalling by organizing the cilium tip compartment. Nat. Cell Biol. 16:663–672. 10.1038/ncb2988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heydeck W., Zeng H., and Liu A.. 2009. Planar cell polarity effector gene Fuzzy regulates cilia formation and Hedgehog signal transduction in mouse. Dev. Dyn. 238:3035–3042. 10.1002/dvdy.22130 [DOI] [PubMed] [Google Scholar]
- Hill P., Götz K., and Rüther U.. 2009. A SHH-independent regulation of Gli3 is a significant determinant of anteroposterior patterning of the limb bud. Dev. Biol. 328:506–516. 10.1016/j.ydbio.2009.02.017 [DOI] [PubMed] [Google Scholar]
- Huangfu D., and Anderson K.V.. 2005. Cilia and Hedgehog responsiveness in the mouse. Proc. Natl. Acad. Sci. USA. 102:11325–11330. 10.1073/pnas.0505328102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D., Liu A., Rakeman A.S., Murcia N.S., Niswander L., and Anderson K.V.. 2003. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature. 426:83–87. 10.1038/nature02061 [DOI] [PubMed] [Google Scholar]
- Joo K., Kim C.G., Lee M.-S., Moon H.-Y., Lee S.-H., Kim M.J., Kweon H.S., Park W.Y., Kim C.H., Gleeson J.G., and Kim J.. 2013. CCDC41 is required for ciliary vesicle docking to the mother centriole. Proc. Natl. Acad. Sci. USA. 110:5987–5992. 10.1073/pnas.1220927110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Krishnaswami S.R., and Gleeson J.G.. 2008. CEP290 interacts with the centriolar satellite component PCM-1 and is required for Rab8 localization to the primary cilium. Hum. Mol. Genet. 17:3796–3805. 10.1093/hmg/ddn277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.K., Shindo A., Park T.J., Oh E.C., Ghosh S., Gray R.S., Lewis R.A., Johnson C.A., Attie-Bittach T., Katsanis N., and Wallingford J.B.. 2010. Planar cell polarity acts through septins to control collective cell movement and ciliogenesis. Science. 329:1337–1340. 10.1126/science.1191184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindt K.S., Finch G., and Nicolson T.. 2012. Kinocilia mediate mechanosensitivity in developing zebrafish hair cells. Dev. Cell. 23:329–341. 10.1016/j.devcel.2012.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Klena N.T., Gabriel G.C., Liu X., Kim A.J., Lemke K., Chen Y., Chatterjee B., Devine W., Damerla R.R., et al. 2015. Global genetic analysis in mice unveils central role for cilia in congenital heart disease. Nature. 521:520–524. 10.1038/nature14269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litingtung Y., Dahn R.D., Li Y., Fallon J.F., and Chiang C.. 2002. Shh and Gli3 are dispensable for limb skeleton formation but regulate digit number and identity. Nature. 418:979–983. 10.1038/nature01033 [DOI] [PubMed] [Google Scholar]
- López-Ríos J., Speziale D., Robay D., Scotti M., Osterwalder M., Nusspaumer G., Galli A., Holländer G.A., Kmita M., and Zeller R.. 2012. GLI3 constrains digit number by controlling both progenitor proliferation and BMP-dependent exit to chondrogenesis. Dev. Cell. 22:837–848. 10.1016/j.devcel.2012.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q., Insinna C., Ott C., Stauffer J., Pintado P.A., Rahajeng J., Baxa U., Walia V., Cuenca A., Hwang Y.S., et al. 2015. Early steps in primary cilium assembly require EHD1/EHD3-dependent ciliary vesicle formation. Nat. Cell Biol. 17:228–240. 10.1038/ncb3109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magyar J., Kiper C.E., Sievert G., Cai W., Shi G.X., Crump S.M., Li L., Niederer S., Smith N., Andres D.A., and Satin J.. 2012. Rem-GTPase regulates cardiac myocyte L-type calcium current. Channels (Austin). 6:166–173. 10.4161/chan.20192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning J.R., Yin G., Kaminski C.N., Magyar J., Feng H.-Z., Penn J., Sievert G., Thompson K., Jin J.-P., Andres D.A., and Satin J.. 2013. Rad GTPase deletion increases L-type calcium channel current leading to increased cardiac contraction. J. Am. Heart Assoc. 2:e000459 10.1161/JAHA.113.000459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- May S.R., Ashique A.M., Karlen M., Wang B., Shen Y., Zarbalis K., Reiter J., Ericson J., and Peterson A.S.. 2005. Loss of the retrograde motor for IFT disrupts localization of Smo to cilia and prevents the expression of both activator and repressor functions of Gli. Dev. Biol. 287:378–389. 10.1016/j.ydbio.2005.08.050 [DOI] [PubMed] [Google Scholar]
- Menzl I., Lebeau L., Pandey R., Hassounah N.B., Li F.W., Nagle R., Weihs K., and McDermott K.M.. 2014. Loss of primary cilia occurs early in breast cancer development. Cilia. 3:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechipurenko I.V., Berciu C., Sengupta P., and Nicastro D.. 2017. Centriolar remodeling underlies basal body maturation during ciliogenesis in Caenorhabditis elegans. eLife. 6:e25686 10.7554/eLife.25686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor A.K., Malarkey E.B., Berbari N.F., Croyle M.J., Haycraft C.J., Bell P.D., Hohenstein P., Kesterson R.A., and Yoder B.K.. 2013. An inducible CiliaGFP mouse model for in vivo visualization and analysis of cilia in live tissue. Cilia. 2:8 10.1186/2046-2530-2-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocbina P.J.R., and Anderson K.V.. 2008. Intraflagellar transport, cilia, and mammalian Hedgehog signaling: Analysis in mouse embryonic fibroblasts. Dev. Dyn. 237:2030–2038. 10.1002/dvdy.21551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park T.J., Haigo S.L., and Wallingford J.B.. 2006. Ciliogenesis defects in embryos lacking inturned or fuzzy function are associated with failure of planar cell polarity and Hedgehog signaling. Nat. Genet. 38:303–311. 10.1038/ng1753 [DOI] [PubMed] [Google Scholar]
- Pazour G.J., Dickert B.L., Vucica Y., Seeley E.S., Rosenbaum J.L., Witman G.B., and Cole D.G.. 2000. Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene Tg737, are required for assembly of cilia and flagella. J. Cell Biol. 151:709–718. 10.1083/jcb.151.3.709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piddini E., Schmid J.A., de Martin R., and Dotti C.G.. 2001. The Ras-like GTPase Gem is involved in cell shape remodelling and interacts with the novel kinesin-like protein KIF9. EMBO J. 20:4076–4087. 10.1093/emboj/20.15.4076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rash B.G., and Grove E.A.. 2011. Shh and Gli3 regulate formation of the telencephalic-diencephalic junction and suppress an isthmus-like signaling source in the forebrain. Dev. Biol. 359:242–250. 10.1016/j.ydbio.2011.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanienko P.J., Giacalone J., Ingenito J., Wang Y., Isaka M., Johnson T., You Y., and Mark W.H.. 2016. A vector with a single promoter for in vitro transcription and mammalian cell expression of CRISPR gRNAs. PLoS One. 11:e0148362 10.1371/journal.pone.0148362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez I., and Dynlacht B.D.. 2016. Cilium assembly and disassembly. Nat. Cell Biol. 18:711–717. 10.1038/ncb3370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt K.N., Kuhns S., Neuner A., Hub B., Zentgraf H., and Pereira G.. 2012. Cep164 mediates vesicular docking to the mother centriole during early steps of ciliogenesis. J. Cell Biol. 199:1083–1101. 10.1083/jcb.201202126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger-Nukpezah T., Little J.L., Serzhanova V., and Golemis E.A.. 2013. Cilia and cilia-associated proteins in cancer. Drug Discov. Today Dis. Mech. 10:e135–e142. 10.1016/j.ddmec.2013.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley E.S., and Nachury M.V.. 2010. The perennial organelle: assembly and disassembly of the primary cilium. J. Cell Sci. 123:511–518. 10.1242/jcs.061093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo J.H., Zilber Y., Babayeva S., Liu J., Kyriakopoulos P., De Marco P., Merello E., Capra V., Gros P., and Torban E.. 2011. Mutations in the planar cell polarity gene, Fuzzy, are associated with neural tube defects in humans. Hum. Mol. Genet. 20:4324–4333. 10.1093/hmg/ddr359 [DOI] [PubMed] [Google Scholar]
- Sorokin S. 1962. Centrioles and the formation of rudimentary cilia by fibroblasts and smooth muscle cells. J. Cell Biol. 15:363–377. 10.1083/jcb.15.2.363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spektor A., Tsang W.Y., Khoo D., and Dynlacht B.D.. 2007. Cep97 and CP110 suppress a cilia assembly program. Cell. 130:678–690. 10.1016/j.cell.2007.06.027 [DOI] [PubMed] [Google Scholar]
- Stowe T.R., Wilkinson C.J., Iqbal A., and Stearns T.. 2012. The centriolar satellite proteins Cep72 and Cep290 interact and are required for recruitment of BBS proteins to the cilium. Mol. Biol. Cell. 23:3322–3335. 10.1091/mbc.E12-02-0134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabler J.M., Barrell W.B., Szabo-Rogers H.L., Healy C., Yeung Y., Perdiguero E.G., Schulz C., Yannakoudakis B.Z., Mesbahi A., Wlodarczyk B., et al. 2013. Fuz mutant mice reveal shared mechanisms between ciliopathies and FGF-related syndromes. Dev. Cell. 25:623–635. 10.1016/j.devcel.2013.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanos B.E., Yang H.-J., Soni R., Wang W.-J., Macaluso F.P., Asara J.M., and Tsou M.F.B.. 2013. Centriole distal appendages promote membrane docking, leading to cilia initiation. Genes Dev. 27:163–168. 10.1101/gad.207043.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toriyama M., Lee C., Taylor S.P., Duran I., Cohn D.H., Bruel A.L., Tabler J.M., Drew K., Kelly M.R., Kim S., et al. University of Washington Center for Mendelian Genomics . 2016. The ciliopathy-associated CPLANE proteins direct basal body recruitment of intraflagellar transport machinery. Nat. Genet. 48:648–656. 10.1038/ng.3558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang W.Y., and Dynlacht B.D.. 2013. CP110 and its network of partners coordinately regulate cilia assembly. Cilia. 2:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Yang H., Shivalila C.S., Dawlaty M.M., Cheng A.W., Zhang F., and Jaenisch R.. 2013. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 153:910–918. 10.1016/j.cell.2013.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weatherbee S.D., Niswander L.A., and Anderson K.V.. 2009. A mouse model for Meckel syndrome reveals Mks1 is required for ciliogenesis and Hedgehog signaling. Hum. Mol. Genet. 18:4565–4575. 10.1093/hmg/ddp422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wennerberg K., Rossman K.L., and Der C.J.. 2005. The Ras superfamily at a glance. J. Cell Sci. 118:843–846. 10.1242/jcs.01660 [DOI] [PubMed] [Google Scholar]
- Westlake C.J., Baye L.M., Nachury M.V., Wright K.J., Ervin K.E., Phu L., Chalouni C., Beck J.S., Kirkpatrick D.S., Slusarski D.C., et al. 2011. Primary cilia membrane assembly is initiated by Rab11 and transport protein particle II (TRAPPII) complex-dependent trafficking of Rabin8 to the centrosome. Proc. Natl. Acad. Sci. USA. 108:2759–2764. 10.1073/pnas.1018823108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willaredt W.A., Gorgas K., Gardner H.A., and Tucker K.L.. 2012. Multiple essential roles for primary cilia in heart development. Cilia. 11:1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Zhang F., Zamponi G.W., and Horne W.A.. 2015. Solution NMR and calorimetric analysis of Rem2 binding to the Ca2+ channel β4 subunit: A low affinity interaction is required for inhibition of Cav2.1 Ca2+ currents. FASEB J. 29:1794–1804. 10.1096/fj.14-264499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav S.P., Sharma N.K., Liu C., Dong L., Li T., and Swaroop A.. 2016. Centrosomal protein CP110 controls maturation of the mother centriole during cilia biogenesis. Development. 143:1491–1501. 10.1242/dev.130120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T.T., Su J., Wang W.J., Craige B., Witman G.B., Tsou M.F.B., and Liao J.C.. 2015. Superresolution pattern recognition reveals the architectural map of the ciliary transition zone. Sci. Rep. 5:14096 10.1038/srep14096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X., Zeng H., Ning G., Reiter J.F., and Liu A.. 2014. C2cd3 is critical for centriolar distal appendage assembly and ciliary vesicle docking in mammals. Proc. Natl. Acad. Sci. USA. 111:2164–2169. 10.1073/pnas.1318737111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H., Hoover A.N., and Liu A.. 2010. PCP effector gene Inturned is an important regulator of cilia formation and embryonic development in mammals. Dev. Biol. 339:418–428. 10.1016/j.ydbio.2010.01.003 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Wlodarczyk B.J., Niederreither K., Venugopalan S., Florez S., Finnell R.H., and Amendt B.A.. 2011. Fuz regulates craniofacial development through tissue specific responses to signaling factors. PLoS One. 6:e24608 10.1371/journal.pone.0024608 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.