Li and Springer demonstrate differences between integrins α4β1 and α5β1 in intrinsic affinities and relative free energies of three conformational states. Integrin conformational equilibria are both subunit and cell type specific. The energy landscapes of intact receptors on the cell surface provide a framework for understanding regulation of integrin adhesiveness.
Abstract
Why do integrins differ in basal activity, and how does affinity for soluble ligand correlate with cellular adhesiveness? We show that basal conformational equilibrium set points for integrin α4β1 are cell type specific and differ from integrin α5β1 when the two integrins are coexpressed on the same cell. Although α4β1 is easier to activate, its high-affinity state binds vascular cell adhesion molecule and fibronectin 100- to 1,000-fold more weakly than α5β1 binds fibronectin. Furthermore, the difference in affinity between the high- and low-affinity states is more compressed in α4β1 (600- to 800-fold) than in α5β1 (4,000- to 6,000-fold). α4β1 basal conformational equilibria differ among three cell types, define affinity for soluble ligand and readiness for priming, and may reflect differences in interactions with intracellular adaptors but do not predict cellular adhesiveness for immobilized ligand. The measurements here provide a necessary framework for understanding integrin activation in intact cells, including activation of integrin adhesiveness by application of tensile force by the cytoskeleton, across ligand–integrin–adaptor complexes.
Introduction
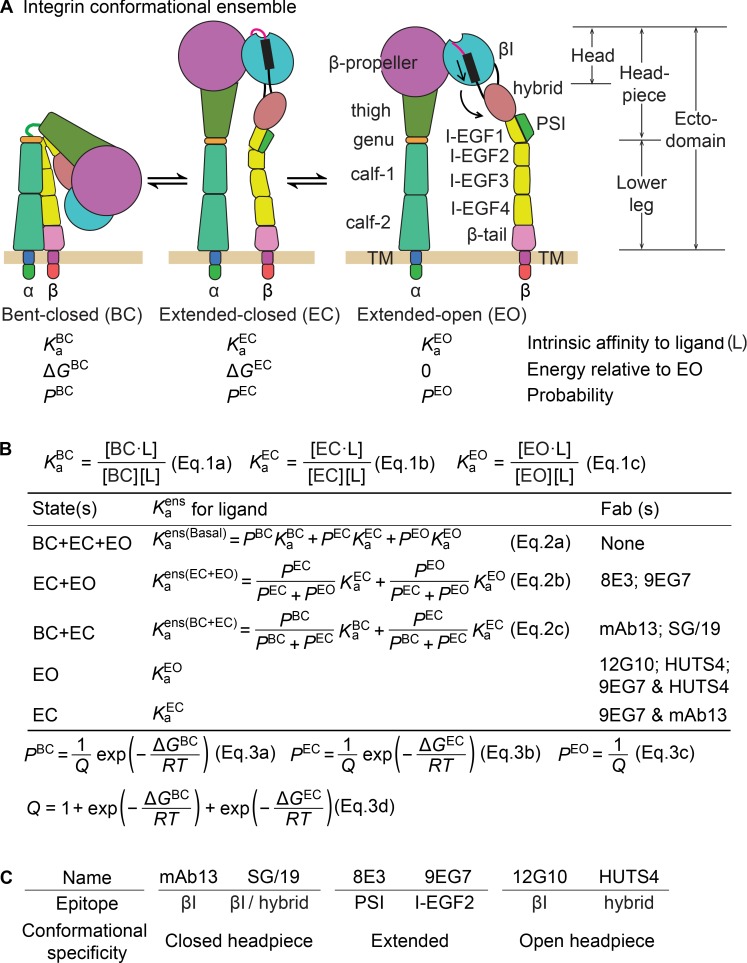
Integrins are cell surface receptors that mediate dynamic cell–cell and cell–matrix adhesion as well as cell migration. Integrins are composed of α and β subunits with long legs that link the ligand-binding head to single-pass transmembrane domains (Fig. 1 A). Crucial to mechanotransduction mediated by integrins (i.e., coordinating cell adhesion and migration with cytoskeletal dynamics) is the modulation of ligand-binding affinity, which is achieved through large-scale conformational changes. Three overall conformational states, termed bent-closed (BC), extended-closed (EC), and extended-open (EO; Fig. 1 A), have been observed in multiple integrins (Luo et al., 2007; Springer and Dustin, 2012).
Figure 1.
Overall integrin conformational states, the equilibria linking them and the strategy to quantify equilibria. (A) The three overall states in an integrin conformational ensemble (Luo et al., 2007) and their thermodynamic parameters. (B) Equations used in this study. (C) Specificities of conformation-specific Fabs.
Many studies have correlated integrin adhesiveness and high affinity for ligand with the EO state (Takagi et al., 2002, 2003; Xiao et al., 2004; Chen et al., 2010; Schürpf and Springer, 2011; Zhu et al., 2013; Su et al., 2016; Li et al., 2017). However, previous integrin affinity measurements, with one recent exception (Li et al., 2017), are for unknown mixtures of integrin states rather than for specific states. It is thought that integrin activation is regulated physiologically by extracellular ligands that preferentially bind to the EO state (termed outside-in signaling), by intracellular adaptors that bind to integrin cytoplasmic tails and regulate their linkage to the actin cytoskeleton (termed inside-out signaling), and by the mechanical force generated by actin retrograde flow (Zhu et al., 2008; Legate and Fässler, 2009; Kim et al., 2011; Nordenfelt et al., 2016; Park and Goda, 2016; Sun et al., 2016). However, the integrin field largely lacks a quantitative framework for understanding these physiological processes. Only if the intrinsic ligand-binding affinity of each conformational state and the conformational equilibria linking them are known under basal conditions can integrin activation be discussed quantitatively.
The work here on integrin α4β1 uses an approach pioneered recently for α5β1 (Li et al., 2017). The affinity intrinsic to each conformational state and the equilibria linking these states were measured using Fab fragments that stabilized specific conformational states (Su et al., 2016). Subsequently, the experimentally determined energy landscape and intrinsic affinities measured for α5β1 were used to thermodynamically evaluate different integrin activation models. It was found that only the combination of cytoskeletal adaptor binding to the integrin cytoplasmic tails and exertion of tensile force by the actin cytoskeleton could provide ultrasensitive regulation of integrin activation (Li and Springer, 2017).
We wondered whether the molecular features that regulate integrin activation and properties, including differences in intrinsic ligand-binding affinity among conformational states, were unique to α5β1 or general. We also wondered whether these properties could be cell type and integrin-subunit specific. Among cell lines, Jurkat was reported to have higher expression than Thp1 of ligand-induced binding site (LIBS) antibody activation epitope on α4β1 (Yednock et al., 1995). Among integrins, α4β1 was found to have the highest expression of a LIBS activation epitope than any other β1 integrin examined on the same cell type, including α5β1 (Bazzoni et al., 1998). Here, we address integrin cell type– and subunit-specific differences in conformational equilibria by comparing α4β1 and α5β1 on different cell types and how these relate to cell adhesion.
Integrin α4β1 binds to acidic motifs within two protein ligands: an IlE-Asp-Ser-Pro motif in vascular cell adhesion molecule (VCAM; Elices et al., 1990; Vonderheide and Springer, 1992; Vonderheide et al., 1994) and a Leu-Asp-Val-Pro (LDVP) motif in the alternatively spliced fibronectin variant containing the type III connecting segment (Fn IIICS; Guan and Hynes, 1990; Mould and Humphries, 1991). In contrast to the RGD motif–binding integrin α5β1, which only mediates firm adhesion to extracellular matrix, integrin α4β1 functions as both a matrix and cell receptor and is capable of mediating both cellular rolling adhesion and firm adhesion (Alon et al., 1995). Lymphocytes and monocytes use α4β1 to migrate from the bloodstream into sites of inflammation and autoimmune diseases (Springer and Cybulsky, 1995; Sixt et al., 2006; Hyun et al., 2009). Antibodies that target α4 integrins are currently used clinically to treat multiple sclerosis, Crohn’s disease, and ulcerative colitis (von Andrian and Engelhardt, 2003; Parikh et al., 2012).
Our results reveal that regulation of integrin conformational equilibria can be both α-subunit–specific and cell type–specific and demonstrate large differences among integrins in intrinsic ligand-binding affinities. By measuring adhesiveness of the same cell types, we also address whether integrin affinity for soluble ligand is sufficient to predict cellular adhesiveness. We find a lack of correlation showing that additional cellular factors are important, consistent with the force-dependent model of integrin activation. Overall, our results establish the framework for understanding the cellular features required for integrin activation, are consistent with a requirement for combination of cytoskeletal adaptors and cytoskeletal force for ultrasensitive regulation of integrin adhesiveness, and suggest that α4β1 can be activated at lower forces or concentrations of adaptors than α5β1.
Results
Approach for measuring intrinsic affinities and energy landscapes of integrin α4β1
Integrin α4β1 adopts multiple conformational states that differ in affinity for ligand (Fig. 1, A and B [Eq. 1, a–c]); therefore, measured affinity depends on the population of each state within the ensemble. Biological conditions that regulate integrin affinity do so by altering the population of conformational states. The apparent affinity of the ensemble in different biological conditions is given by a simple equation: the sum of the products of the affinity of each state multiplied by the population of each state (Fig. 1 B, Eq. 2 a). We measure affinities here by adding different concentrations of ligand and measuring binding. Note that although mixing integrin and ligand can affect the populations of the integrin states by stabilizing and hence increasing the population of the high-affinity state, the apparent affinity that is measured reflects the population of each state in the absence of ligand (Fig. 1 B, Eq. 2). To measure affinities of specific conformational states of α4β1 here, we use saturating concentrations of allosteric, conformation-specific Fabs to convert the three overall conformational states basally present in the integrin α4β1 ensemble into either one or two defined states (Fig. 1 B).
Using these affinities of defined conformational states, we are then able to calculate the population of each state in the ensembles containing two or three states using Eq. 2 (a–c) in Fig. 1 B and supplemental Eqs. S12–S16. The relative population of conformational states is determined by their relative free energies; lower energy states are more populated. The quantitative relationship between population and free energy, known as the Boltzmann distribution, enables the populations to be used to calculate the free energies of the states as shown in Eq. 3 (a–d) in Fig. 1 B.
To stabilize particular conformational states of integrin α4β1, we used Fabs listed in Fig. 1 (B and C). EM studies have directly defined the integrin conformational states that these Fabs stabilize (Su et al., 2016), and the Fabs have previously been used to characterize the intrinsic affinity and conformational equilibria for integrin α5β1 (Li et al., 2017). EM and epitope mapping studies have defined specific regions to which the Fabs bind (Su et al., 2016). Because none of the Fabs compete with ligand or contact both ligand and integrin, they affect only integrin conformational equilibria. To stabilize each conformation, we use two independent Fabs that bind to distinct domains or domain–domain interfaces (Fig. 1 C). As shown below, we measure similar affinities with independent Fabs, as also seen with integrin α5β1 (Li et al., 2017). As previously argued in detail (Li et al., 2017), these results suggest that the Fab-stabilized states are not biased away from native states. To ensure saturable population of target conformations, conformation-specific Fabs were used at concentrations well above the concentration giving half-maximum responses (i.e., their EC50 values; Figs. S1 and S2 and Table S1; Li et al., 2017).
Measurements on soluble α4β1 headpiece and ectodomain fragments
Ligand-binding affinities of soluble α4β1 fragments (Fig. 2) were measured using fluorescence polarization (FP; Rossi and Taylor, 2011). In FP, a small fluorescent ligand is mixed with a much larger integrin fragment. The fluorescent ligand is excited with plane polarized light, and the intensity (I) of the fluorescence emission is measured in directions parallel (III) and perpendicular (I⊥) to the excitation. If the small ligand is unbound, it tumbles rapidly, and over the ∼3-ns period between excitation and emission, its fluorescence transition dipole becomes unaligned from its orientation during excitation, and III ∼ I⊥. If the small ligand is bound to the much larger integrin during the time period between excitation and emission, the integrin–ligand complex tumbles slowly and the fluorescence transition dipole of the ligand remains largely aligned with the original direction of excitation, and III > I⊥. Thus, FP, defined as (III − I⊥)/(III + I⊥), is maximal when all ligand is bound and minimal when no ligand is bound. FP experiments work similarly to classic saturation ligand-binding assays, but with one twist. Instead of titrating in an excess of ligand, in FP, the receptor is titrated in until it is in excess over the ligand and all ligand is bound. Thus, the concentration of the fluorescent ligand, and total fluorescence intensity, remain constant during receptor titrations. FP is robust, reproducible, and high throughput. Our FP measurements use a well-validated peptidomimetic ligand of α4β1 (Lin et al., 1999; Chigaev et al., 2001), which contains a 2-methylphenylureaphenylacetyl moiety linked to the LDVP motif from the fibronectin IIICS segment conjugated to FITC.
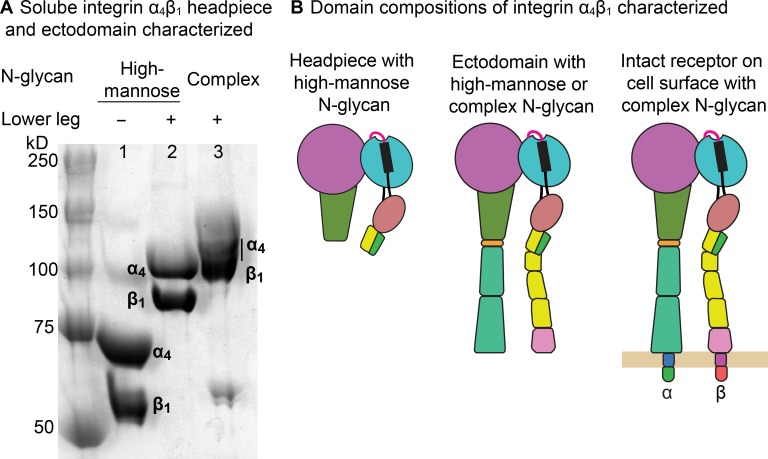
Figure 2.
Integrin α4β1 preparations. (A) SDS 7.5% PAGE of α4β1 headpiece with high-mannose N-glycans (lane 1), unclasped ectodomain with high-mannose N-glycans (lane 2), and unclasped ectodomain with complex N-glycans (lane 3). (B) Domain compositions.
We began FP measurements with an α4β1 headpiece fragment containing high-mannose N-glycans (Fig. 2 A, lane 1). The headpiece lacks the lower integrin legs (Fig. 2 B) and thus has only two conformational states, closed (C) and open (O). For concision here, we use association constants Ka in Eqs. 1 and 2 in Fig. 1 B and dissociation constants (Kd = 1/Ka) for reporting affinities. The affinity of the open headpiece, (0.14 ± 0.05 nM), measured in the presence of HUTS4 Fab was 650-fold higher than the mean affinity of the closed headpiece, (91 ± 21 nM), measured in the presence of two different Fabs, SG/19 and mAb13 (Fig. 3 A). The ensemble affinity, (5.7 ± 1.0 nM), was intermediate between and , showing that both the closed and open conformations were present basally. The populations of the open and closed states (PO and PC) were calculated as the population-weighted contribution from and (Eqs. S15 and S16). The result showed that in the basal ensemble, the α4β1 headpiece is 97.7% in the closed conformation and 2.3% in the open conformation (Fig. 3 D). These population values were then used in Boltzmann distribution equations similar to Eq. 3 (a–d) in Fig. 1 B (Eq. S19) to calculate the relative free energies of the two states. With open state as reference (ΔGO = 0), the free energy of the closed state was found to be ΔGC = −2.2 kcal/mol (Fig. 3 D).
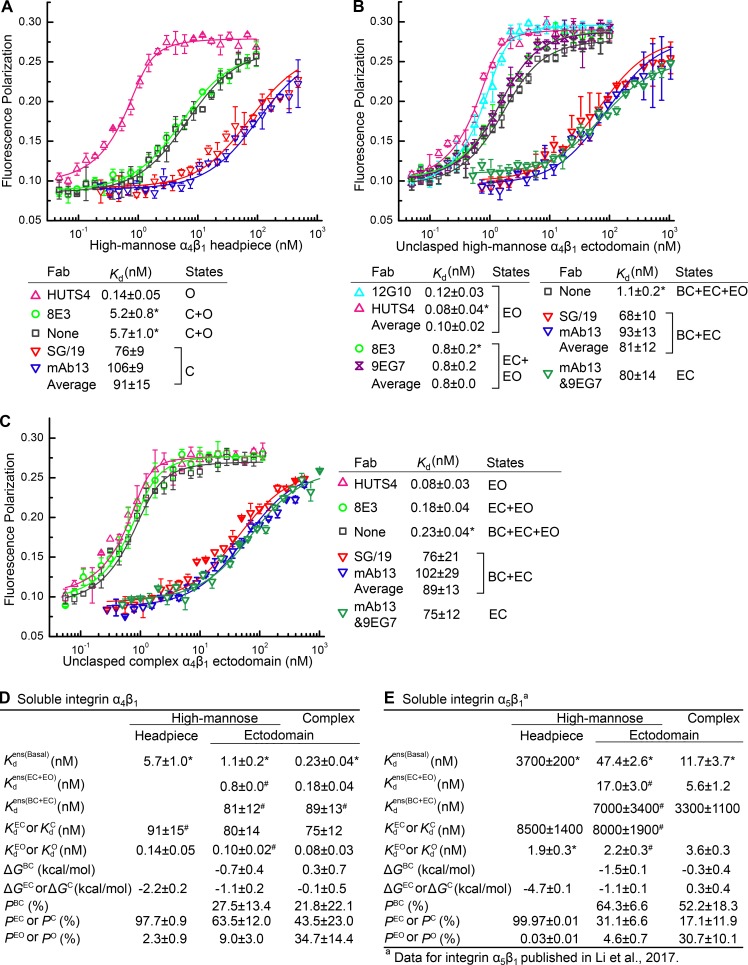
Figure 3.
Intrinsic affinities, ensemble affinities and conformational equilibria of α4β1 headpiece and ectodomain fragments. (A–C) Affinities for FITC-LDVP of α4β1 headpiece with high-mannose N-glycans (A), unclasped ectodomain with high-mannose N-glycans (B) and unclasped ectodomain with complex N-glycans (C) were measured using FP in presence or absence of the indicated Fabs. Errors in plots or inset tables are SD from nonlinear least square fits with triplicates, except values with asterisks are mean and SD from at least three experiments on different days. (D and E) Affinity measurements and calculated thermodynamic parameters for α4β1 (D) and comparison to published (Li et al., 2017) α5β1 measurements (E). Errors are as described for A–C, except values with “#,” which represent mean ± difference from the mean of measurements with distinct Fabs stabilizing the same conformation.
The 8E3 Fab binds the PSI (plexin-semaphorin-integrin) domain, which is present in the headpiece, and induces extension of integrins that contain the lower legs (Su et al., 2016). However, because the headpiece lacks lower legs, we expected 8E3 Fab to have little effect on affinity. Indeed, α4β1 headpiece measured in presence of 8E3 Fab (5.2 ± 0.8 nM) was indistinguishable from measured in absence of Fab (5.7 ± 1.0 nM; Fig. 3 A). This negative control experiment on the headpiece contrasts with the results using 8E3 Fab with the α4β1 ectodomain described in the following paragraph.
We next studied the α4β1 ectodomain bearing high-mannose N-glycans (Fig. 2, A [lane 2] and B). The ectodomain carries the lower legs (Fig. 2 B) and thus can visit three conformational states (Fig. 1 A). The intrinsic affinity of the EO state was = 0.10 ± 0.03 nM (Fig. 3 B), within error of the affinity measured above for the open headpiece of =0.14 ± 0.05 nM. The intrinsic affinity of the EC state was = 80 ± 14 nM. The affinity of an ensemble comprising the two closed states, = 81 ± 17 nM, is indistinguishable from the intrinsic affinity . These results suggest that the intrinsic affinities of the BC and EC states, and , are identical. Moreover, the intrinsic affinities of the closed states of the ectodomain are similar to the intrinsic affinity of the closed headpiece, = 91 ± 21 nM (Fig. 3, A and D). The affinity of the ensemble comprising the two extended states was = 0.8 nM. The affinity of the basal ensemble containing all three states was = 1.1 ± 0.2 nM. Using these affinities, we calculated the population of each state in the basal ectodomain ensemble and in turn the relative free energy of each state using the equations shown in Fig. 1 B (and described in more detail in the supplemental text). The results for the α4β1 ectodomain with high-mannose N-glycans show that among the three integrin conformational states, the EC state is most stable (ΔGEC = −1.1 ± 0.2 kcal/mol, P = 63.5 ± 12%), the BC state is intermediate in stability (ΔGBC = −0.7 ± 0.4 kcal/mol, P = 27.5 ± 13.4%), and the reference EO state is the least stable (ΔGEO = 0, P = 9.0 ± 3%; Fig. 3 D).
To investigate how N-glycosylation regulates α4β1 integrin ectodomain conformational equilibria, we made similar measurements for the α4β1 ectodomain with complex N-glycans (Fig. 2 A, lane 3). The intrinsic affinity of the EO state was = 0.08 ± 0.03 nM. The intrinsic affinity of the EC state was = 75 ± 12 nM. The affinity of the ensemble of the two closed states was = 89 ± 18 nM and thus was similar to that measured for the EC state. These results showed that the intrinsic affinities of the open and closed conformations of the ectodomain with complex carbohydrates were similar to those measured for the headpiece and ectodomain with high-mannose N-glycans (Fig. 3 D). Ensembles containing mixtures of closed and open conformations gave contrasting results. The basal ensemble affinity of the three states of the α4β1 ectodomain with complex N-glycans was = 0.23 ± 0.04 nM, and the ensemble affinity of the two extended states was = 0.18 ± 0.04 nM. These affinities were each significantly higher than for the ectodomain with high-mannose N-glycans, showing that complex N-glycosylation stabilized the high-affinity EO conformation (Fig. 3 D).
Energy landscape of intact α4β1 on the surface of three cell types
We extended our measurements to intact α4β1 on the surface of T lymphocytic Jurkat cells, monocytic Thp1 cells, and erythroleukemia K562 α4 transfectants (KA4; Kassner et al., 1995; Fig. 4). Affinities of intact α4β1 for FITC-LDVP were determined by fluorescent flow cytometry (Chigaev et al., 2001; Dong et al., 2014; Li et al., 2017). Cells were incubated with fluorescent ligand with or without Fabs for 4 h at 22°C to allow binding to reach equilibrium. Then, without any washing, cells were subjected to fluorescent flow cytometry. Background fluorescence corresponding to unbound FITC-LDVP in solution was measured in 10 mM EDTA and subtracted. On Jurkat, Thp1, and KA4 cells, the affinity of the basal α4β1 ensemble, , was 15.0 ± 2.1, 33.7 ± 3.8, and 12.0 ± 2.0 nM, respectively (Fig. 4). The basal affinity on Thp1 cells was significantly lower than that on Jurkat and KA4 cells (P < 0.006 using a two-tailed t test and the Bonferroni correction for multiple comparisons).
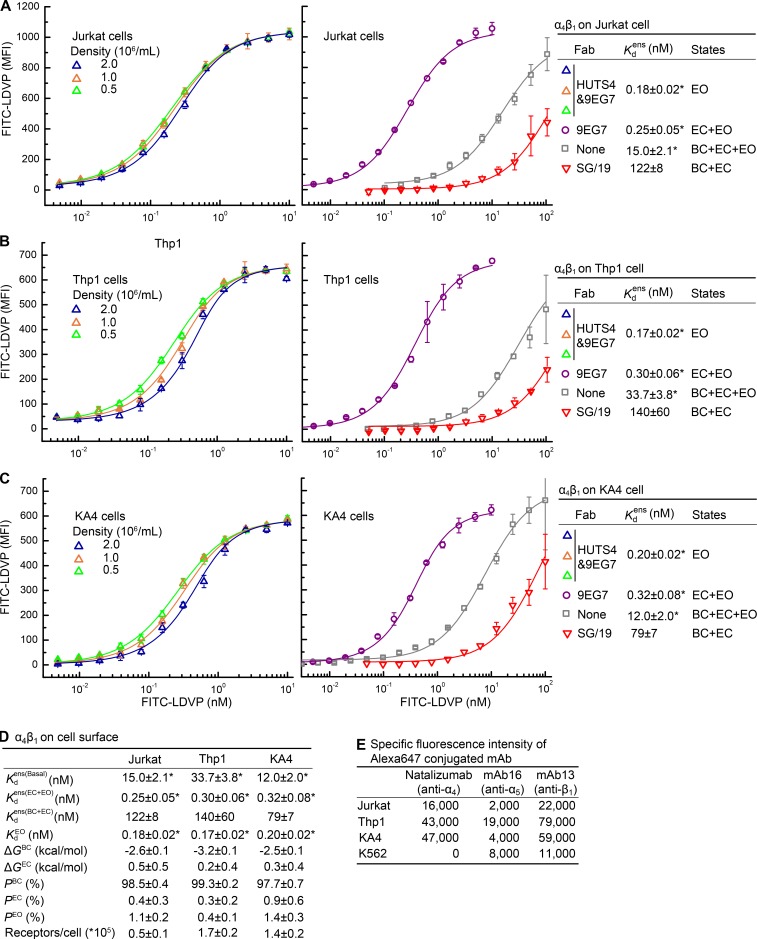
Figure 4.
Intrinsic and ensemble affinities of α4β1 on the surface of three cell types and deduced conformational equilibria. (A–C) Affinities for FITC-LDVP of intact α4β1 on Jurkat (A), Thp1 (B), and KA4 cells (C) in the presence of the indicated Fabs and cell concentrations. (D) Tabulation of results from A–C. Errors in each panel and inset table are as described in the Fig. 3 legend. (E) Specific fluorescence intensity of cells stained with 25 nM Alexa647-conjugated mAb.
The affinity for FITC-LDVP measured for open, soluble α4β1 was ∼0.1 nM, which approached the concentration range of integrins in the cell suspension. Under these conditions, binding to cells can significantly deplete ligand from solution, and the total concentration of FITC-LDVP could not be used as an approximation of its free concentration. Therefore, cells were used at three different densities in the assay (Fig. 4, A–C, left) so that the fitted value of the α4β1 concentration in the cell suspension could be used to correct for the effect of ligand depletion (Eq. S8). The affinity intrinsic to the EO conformation on cells was thus measured as = 0.18 ± 0.02 nM, 0.17 ± 0.02 nM, and 0.20 ± 0.02 nM on Jurkat, Thp1, and KA4 respectively (Fig. 4 D). These values are within each other’s error range and close to intrinsic affinities measured with purified, soluble α4β1 (Fig. 3 D). Fits to Eq. S8 also yielded the number of FITC-LDVP–binding sites (α4β1 receptors) per cell. Jurkat, Thp1 and KA4 cells expressed 50,000, 170,000, and 140,000 α4β1 receptors per cell, respectively (Fig. 4 D). These site numbers were in agreement with the approximately threefold higher expression of α4β1 on Thp1 and KA4 cells than Jurkat cells measured by fluorescent staining with Alexa647-conjugated α4 antibody (natalizumab; Fig. 4 E).
We further measured and (Fig. 4, A–D). The latter was less accurately measured because of high backgrounds from free FITC-LDVP in solution (Fig. 4, A–C, middle); however, measured on the cell surface was similar to that measured with soluble α4β1 fragments (Fig. 3 D), and its uncertainty had little impact on energy landscape calculations (Fig. S3).
Using these affinity measurements, and assuming that the affinities of the BC and EC states were identical as we had found with soluble α4β1 preparations (Fig. 3 D), we calculated the population of each state, and in turn the relative free energy of each state, in basal cell surface ensembles. On the surface of all three cell types, the BC state was the most stable, and its free energy ranged from −3.2 to −2.5 kcal/mol (Fig. 4 D). Thus, the BC state is much more stable in intact, cell surface α4β1 than in the soluble α4β1 ectodomain with similar complex N-glycosylation (Fig. 3 D). Correspondingly, the high-affinity EO state of α4β1 is rare on cell surfaces, with a population of only 1.1 ± 0.2%, 1.4 ± 0.3%, and 0.4 ± 0.1% on Jurkat, KA4, and Thp1 cells, respectively. Thus, PEO values on Jurkat and Thp1 cells were similar and significantly higher than on Thp1 cells (Fig. 4 D).
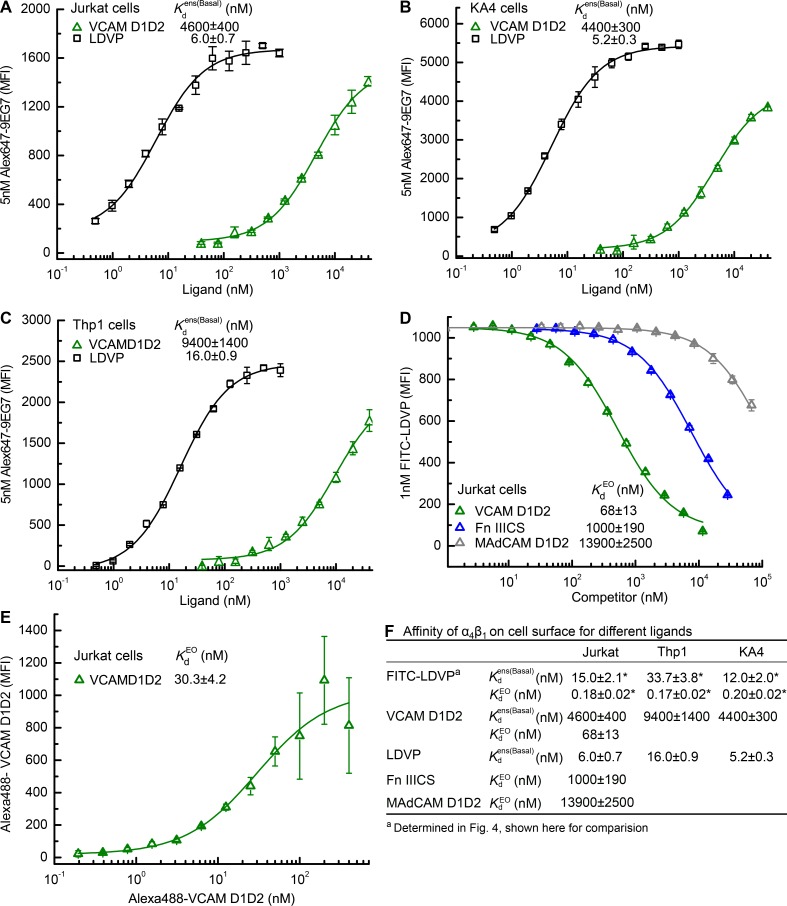
To independently validate the basal ensemble affinity differences on the three cell types, we measured binding affinity of different ligands by titrating in ligand and measuring enhancement of binding of a fixed concentration of Alexa647-labeled 9EG7 Fab, which is specific for the extended conformations of β1 integrins (Su et al., 2016; Fig. 5, A–C). A fragment of VCAM containing domains 1 and 2 (VCAM D1D2) bound with of 4,600 ± 400 nM, 4,400 ± 300 nM, and 9,400 ± 1,400 nM to Jurkat, KA4, and Thp1 cells, respectively (Fig. 5, A–C and F). The basal ensemble affinities on Jurkat and KA4 cells were each significantly higher than on Thp1 cells (P < 0.014 using a two-tailed t test and Bonferroni correction). Similarly, the affinity of LDVP, the nonfluorescent analogue of FITC-LDVP, was significantly higher at 6.0 ± 0.7 nM and 5.2 ± 0.3 nM on Jurkat and KA4 cells, respectively, than on Thp1 cells at 16.0 ± 0.9 nM (Fig. 5, A–C and F; P < 0.0006, using a two-tailed t test and the Bonferroni correction). Importantly, these results showed concordance among the three cell types in their relative basal affinities for VCAM D1D2, FITC-LDVP, and LDVP (Fig. 5 F).
Figure 5.
Affinities of α4β1 on cell surface for different ligands. (A–C) Basal ensemble affinity of α4β1 for LDVP and VCAM D1D2 on Jurkat (A), KA4 (B), and Thp1 (C) cells determined by binding of 5nM Alexa647-9EG7 Fab. (D) Intrinsic affinity of α4β1 EO conformation for VCAM D1D2, Fn IIICS, and MAdCAM D1D2 measured on Jurkat cells by inhibition of binding of 1nM FITC-LDVP in presence of EO-stabilizing Fabs 9EG7&HUTS4. (E) Intrinsic affinity of α4β1 EO conformation in presence of Fabs 9EG7&HUTS4 measured by saturation binding of Alexa488-VCAM D1D2 using fluorescence flow cytometry. (F) Tabulation of results. Errors in each panel and inset table are as described in Fig. 3 legend.
Comparisons among biological ligands of affinity for α4β1
VCAM and Fn IIICS are biological ligands of integrin α4β1. Additionally, α4β1 binds mucosal addressin cell adhesion molecule (MAdCAM), although with lower affinity than integrin α4β7 (Newham et al., 1997). In the presence of Fabs 9EG7 and HUTS4 to stabilize the EO conformation, competition with FITC-LDVP showed that VCAM D1D2, Fn IIICS, and MAdCAM D1D2 bind with intrinsic affinities of 68 ± 13, 1,000 ± 190, and 13,900 ± 2,500 nM, respectively (Fig. 5 D). Direct measurement of the intrinsic affinity of Alexa488-VCAM D1D2 for the α4β1 EO state on Jurkat cells in presence of Fabs 9EG7 and HUTS4 yielded = 30.3 ± 4.3 nM (Fig. 5 E). At higher Alexa488-VCAM D1D2 concentrations, the background of free Alexa488-VCAM D1D2 created uncertainty in determination of bound Alexa488-VCAM D1D2 (Fig. 5 E). Furthermore, the presence of the Alexa488 label may alter the affinity of VCAM D1D2 as similarly seen with the difference in affinity between FITC-LDVP and LDVP (Fig. 5 F). Nonetheless, the =30.3 ± 4.3 nM measured for Alexa488-VCAM D1D2 differed only by twofold from that measured for unlabeled VCAM of 68 ± 13 nM in Fig. 5 D.
Comparisons of energy landscapes between integrins α4β1 and α5β1 on the same cell
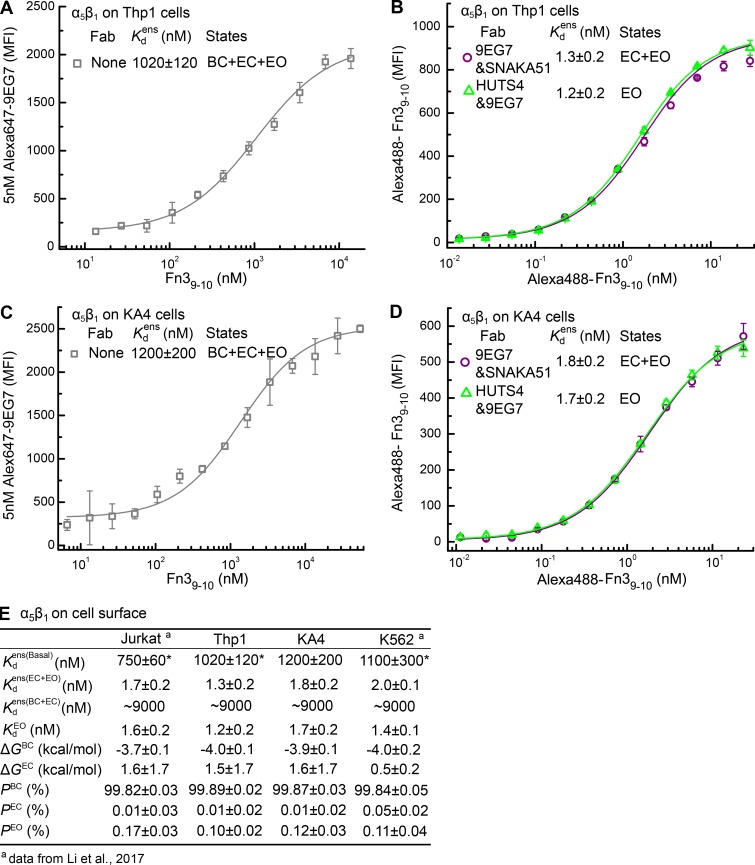
We supplemented previous α5β1 energy landscape measurements on Jurkat and K562 cells (Li et al., 2017) with measurements here of , , and on Thp1 and KA4 cells (Fig. 6). We used previous measurements of values for closed conformations (Li et al., 2017). The populations and free energies of α5β1 conformational states on the four cell types are summarized in Fig. 6 E. ΔGBC is well determined and similar on the four cell types with a mean value of −3.9 and range of −4.0 to −3.7 kcal/mol (Fig. 6 E). The ensemble affinity, the most important parameter for accurate calculations of P and ΔG values, was also well determined and ranged from 750 nM on Jurkat to 1,200 nM on KA4 cells (Fig. 6 E). The values for and were close to one another, showing that PEC is very small. Because of the closeness of the and values, ΔGEC was not well determined (Fig. 6 E); however, this has little effect on conclusions in Discussion about and ΔGBC differences among integrins and cell types, as shown in Fig. S3 and (Li et al., 2017).
Figure 6.
Intrinsic and ensemble affinities of α5β1 on the surface of Thp1 and KA4 cells and deduced conformational equilibria for comparison with α4β1. (A and C) Affinity of α5β1 on Thp1 (A) and KA4 (C) cells for Fn39-10 by enhancement of 5 nM Alexa647-9EG7 Fab binding. (B and D) Affinity of α5β1 on Thp1 (B) and KA4 (D) cells for Alexa488-Fn39-10 in presence of indicated Fabs. (E) Tabulation of results. Data for α5β1 on Jurkat and K562 cell surfaces are published (Li et al., 2017) and shown for comparison. Errors in each panel are as described in Fig. 3 legends.
Measurements of cellular adhesiveness
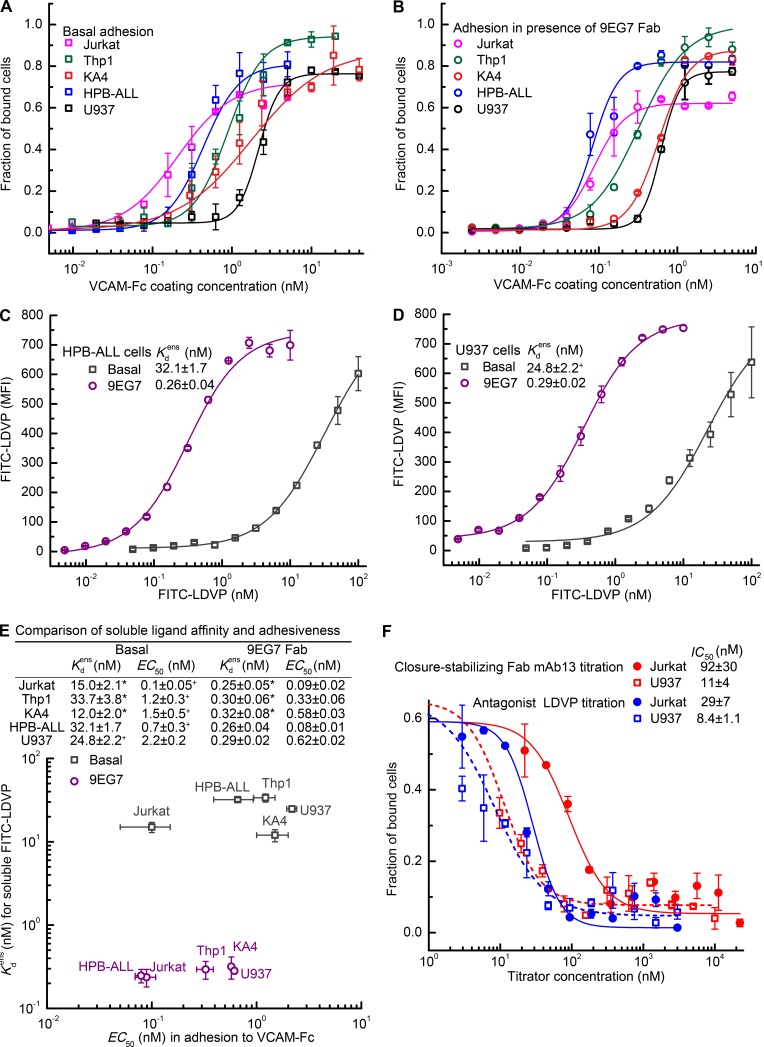
To test whether cellular adhesiveness correlated with cellular affinity for soluble ligand, we tested multiple cell lines for adhesion to VCAM (Fig. 7). In addition to the three α4β1 integrin-bearing cell lines characterized above, we chose HPB-ALL and U937 cells for their high and low adhesiveness (Masumoto and Hemler, 1993). To facilitate quantitative comparisons, we used 96-well plates absorbed with Protein A, added 100 μl VCAM-Fc over a >1,000-fold range of concentrations, washed out unbound VCAM-Fc, added cells, and measured the fraction of adherent cells after 1 h at 37°C. Under basal conditions, half-maximal adherence was given at VCAM-Fc coating concentrations (EC50) of 0.1 ± 0.05, 0.7 ± 0.3, 1.2 ± 0.3, 1.5 ± 0.5, and 2.2 ± 0.2 nM for Jurkat, HPB-ALL, Thp1, KA4, and U937 cells, respectively (Fig. 7 A).
Figure 7.
Lack of correlation between affinity of cell surface integrin α4β1 for soluble ligand and cellular adhesiveness. (A and B) Cell adhesion to VCAM-Fc under basal conditions (A) or in presence of 8 µM 9EG7 Fab (B). (C and D) Ensemble affinity of α4β1 on HPB-ALL (C) and U937 (D) cells for FITC-LDVP under basal conditions and in presence of 8 µM 9EG7 Fab to stabilize extension. (E) Comparison of ensemble affinity for FITC-LDVP on five cell types to adhesiveness measured as VCAM-Fc EC50 values under basal or extension-stabilizing conditions. (F) Jurkat or U937 cells were mixed with LDVP or closure-stabilizing Fab mAb13 and assayed for adhesion to 5 nM VCAM-Fc bound to protein A–coated plates as described in Results. Lines in A, B, and F show fits to dose–response curves. Errors in each panel are as described in Fig. 3 legend, except values with “+,” which represent mean and deviation from the mean from two experiments on different days.
We next tested the effect of inducing extension with a saturating concentration of 9EG7 Fab. The free energies of the EC and EO conformations of α4β1 are similar (Fig. 4 D), so that after Fab-stabilized extension, approximately half of the cell surface α4β1 integrins are in each of these conformations, which still provides some scope for cellular regulation of whether integrins are in the low-affinity EC or high-affinity EO conformations. All cell types showed increased adhesiveness in presence of 9EG7 Fab (Fig. 7 B), with a lowering of VCAM-Fc EC50 values compared with basal conditions (Fig. 7 A and E). Furthermore, the VCAM-Fc EC50 values became more compressed, with a 7.8-fold range of variation in presence of 9EG7 Fab compared with a 22-fold range of variation basally (Fig. 7 E).
To enable comparison of adhesion to affinity for soluble ligand, we extended measurements of soluble ligand affinity to HPB-ALL and U937 cells (Fig. 7 C and D). Log scale comparisons showed that the EC50 of VCAM-Fc required to support adhesion correlated neither with nor with binding affinities for soluble ligand (Fig. 7 E).
We further compared the most and least adhesive cells for inhibition of cell adhesion by the closure-stabilizing Fab mAb13 and by the VCAM antagonist LDVP. Cells were preincubated with inhibitors and then allowed to adhere to VCAM-Fc coated at 5 nM. Higher concentrations of inhibitors were required to inhibit Jurkat cell adhesion than U937 cell adhesion, with the fold difference in IC50 (50% inhibitory concentration) values between cell types being higher for the closure-stabilizing Fab (8.4-fold) than the antagonist (3.4-fold). Saturating concentrations of antagonist and closure-stabilizing Fab completely inhibited cell adhesion, demonstrating the complete dependence of adhesiveness on binding to VCAM of integrin α4β1 and its high-affinity, EO conformation.
Discussion
A long-standing discussion point in the integrin field has been differences among integrins and cell types in their basal adhesiveness and readiness for activation. Studies with K562 cells, in which different integrin α-subunits were transfected to pair with the endogenous β1 subunit, showed marked differences among β1 integrins in basal expression of LIBS epitopes and induction of these epitopes with Mn2+ or ligand (Bazzoni et al., 1998). Furthermore, differences in basal activity were found for the same integrin expressed on different cell types; for example, higher LIBS expression on α4β1 was found on Jurkat than Thp1 cells (Masumoto and Hemler, 1993; Yednock et al., 1995; Shu et al., 2002). The mechanistic basis for these differences has been unclear; indeed, an underlying conceptual framework and quantitative methods for understanding differences among integrins and their activity on different cell types have become available only recently. Here, we have filled gaps in understanding by evaluating differences among both integrins and cell types in terms of the intrinsic affinities of integrin conformational states for ligand and the population of these states (their energy landscape) on the surface of cells. This section first addresses our overall findings on integrin intrinsic ligand-binding affinities, integrin equilibria, and similarities and differences among integrins in how these equilibria are regulated. We then discuss differences among cells in integrin equilibria set points, the potential regulators of the equilibria set points, and relevance to the concept of integrin priming. Finally, we discuss the limitations of what measurements on binding of soluble ligands to cells in suspension can teach us about integrin adhesiveness and the important framework that our findings provide for understanding how other factors, including cytoskeletal forces transmitted through integrins, are required to regulate cellular adhesiveness by integrins.
Ligand-binding affinities intrinsic to integrin conformational states and differences among integrins and ligands
We found that when integrin α4β1 was stabilized in the open conformation, its affinity for ligand was essentially indistinguishable, whether measured for headpiece or ectodomain fragments, for different glycoforms, or for intact integrins on the surface of different cell types. Similarly, when stabilized in the closed conformation, the affinity of integrin α4β1 for ligand showed little dependence on type of N-glycans or presence or absence of lower integrin legs or display on the cell surface. Furthermore, BC and EC conformations had similar affinities. Thus, affinity for ligand is a property that is intrinsic to each conformational state of α4β1, and only the conformation of the head, which is the moiety that binds ligand, is important. Similar results were previously found for integrin α5β1 (Li et al., 2017).
The importance of integrin conformational change was reemphasized here by the 600- to 800-fold higher intrinsic affinity of the open than the closed conformations of α4β1. The importance of conformational change and affinity regulation for integrin adhesiveness, which has often been debated in the integrin field, was further emphasized by the finding that stabilizing α4β1 in the closed conformation completely abolished cell adhesion.
In contrast to the 600- to 800-fold increase in affinity seen here with α4β1, integrin α5β1 shows an ∼5,000-fold increase in affinity upon opening (Li et al., 2017). The magnitude of the difference in affinity between the open and closed states is clearly lesser for α4β1 than for α5β1. The exact mechanism by which the α subunit modulates the magnitude of the affinity increase given by β subunit opening is an interesting question for further study.
Large differences were found among ligands in intrinsic affinity for open α4β1. Both VCAM and fibronectin are considered important biological ligands of α4β1. The intrinsic affinities of open α4β1 for VCAM D1D2 and the IIICS segment of fibronectin were found to be 70 and 1,000 nM, respectively. Interestingly, fibronectin can costimulate T cell responses as efficiently as VCAM (Lehnert et al., 1998), despite its 14-fold-lower affinity. MAdCAM is preferred by α4β7 over α4β1, whereas the reverse is true for VCAM. We found an intrinsic open affinity of α4β1 for MADCAM D1D2 of 14,000 nM. Thus, α4β1 binds VCAM with 200-fold higher affinity than MAdCAM.
Intrinsic affinities of open α4β1 were markedly lower than the affinity of open α5β1 of 1.5 nM for its biological ligand, fibronectin, which it binds at Fn3 domains 9 and 10. All fibronectin isoforms include Fn3 domains 9 and 10, whereas the IIICS segment, recognized by integrin α4β1, is alternatively spliced. It is quite remarkable that integrins α4β1 and α5β1 can efficiently use the same ligand despite binding to different domains with affinities that differ by 700-fold. Could the lower affinity of α4β1 for biological ligands, or its more compressed difference in affinity between closed and open conformations compared with α5β1, relate to the ability of α4β1 to mediate both rolling and firm adhesion (Alon et al., 1995)? These are examples of new concepts that the findings reported here now open up for future testing. It is also possible that the lower intrinsic affinity of the open conformation of α4β1 for biological ligands is tuned to its higher basal activity on cell surfaces compared with α5β1 (Fig. 8, A–C).
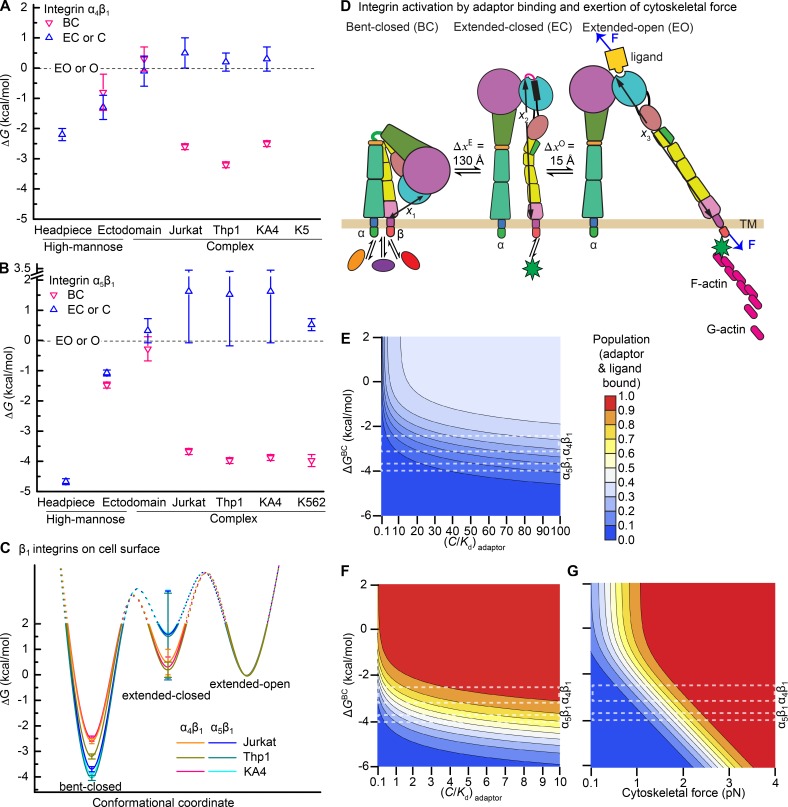
Figure 8.
Regulation of integrin activation by intracellular and extracellular signaling. (A and B) ΔG of each integrin conformational state for integrin headpiece or ectodomain preparations or intact cell surface integrins measured here for α4β1 (A) and here and previously for α5β1 (B; Li et al., 2017). (C) Energy landscape comparisons for intact α5β1 and α4β1 on Jurkat, Thp1, and KA4 cells. (D) Schematic illustration of integrin activation regulated by intracellular protein binding to integrin cytoplasmic tails and application of cytoskeletal force. Ovals represent inhibitors and stars represent adaptors. F represents tensile force exerted across ligand–integrin–adaptor complexes by the cytoskeleton and resisted by immobilized ligand. Distances in the force-bearing pathway between the ligand-binding site and the C terminus of the β-tail domain known from structures and molecular dynamics are shown with arrows (x1, x2, and x3). (E–G) Population of integrin states that mediate cell adhesion in absence (E) or presence of force (F and G). Colors in the key encode the population of integrin states that can mediate cell adhesion (i.e., the sum of adaptor and ligand bound states over all three integrin conformational states). >99% of such adaptor- and ligand-bound integrins are in the EO conformation (Li and Springer, 2017). Ligands are used at concentrations equal to . Adaptors are assumed to bind to EC and EO and not BC states and are used at a concentration equal to the adaptor Kd in G. Rectangles with white dashed lines show the range of ΔG values found for integrins α4β1 and α5β1 on different cell types. (E) Induction of cell adhesion by an increase in adaptor concentration is insensitive to adaptor concentration. (F) Presence of a moderate 1.5 pN cytoskeletal force gives sensitive integrin activation by variation in adaptor concentration. (G) Variation in force with a fixed adaptor concentration gives ultrasensitive regulation of integrin adhesiveness. Population of adaptor and ligand bound states was calculated according to Eq. S20 as previously described (Li and Springer, 2017).
Molecular features modulating integrin conformational equilibria and differences among integrins
Differences in the relative free energies of the states present in different types of integrin preparations gave rise to large differences in basal ensemble affinities. Comparing the high-mannose glycoforms of the α4β1 ectodomain and headpiece, the presence of the lower legs stabilized the open conformation by 1.1 kcal/mol, resulting in a fivefold increase in basal ensemble affinity of the ectodomain compared with the headpiece. Complex compared with high-mannose N-glycans raised the energies of the BC and EC ectodomain conformations relative to the EO conformation by 1 kcal/mol and consequently increased basal ensemble affinity by an additional fivefold. In the BC and EC conformations, the integrin α and β-subunit knees and lower legs are much closer together than in the open conformation (Fig. 1 A). The results on α4β1 are consistent with repulsive or crowding interactions between the lower legs and N-glycans that favor the open headpiece conformation as found with integrin α5β1 (Li et al., 2017). Because the upper and lower legs are further away in the EO than in the EC and BC conformations, charge repulsion and steric repulsion between N-glycans can explain the higher energies of the EC and BC conformations relative to the EO conformation in complex compared with high-mannose ectodomain preparations. The amount of stabilization of the EO state relative to the BC and EC states by complex compared with high-mannose glycans was similar for α4β1 and α5β1 (∼1 and ∼1.3 kcal/mol, respectively). In contrast, the lower legs stabilized the EO state relative to the EC state much more in α5β1 than in α4β1 (i.e., by 3.6 compared with 0.9 kcal/mol) and with an 80-fold increase in basal ensemble affinity in α5β1 compared with a fivefold increase in α4β1 (Li et al., 2017). These results show that although similar qualitative features regulated conformational equilibria in α4β1 and α5β1, their quantitative extents differed, and emphasize the importance of α-subunit–specific differences in regulating integrin conformational equilibria.
We found that on cell surfaces, the BC conformation of α4β1 was much more stable than in ectodomain preparations, as found for α5β1 (Li et al., 2017; Fig. 8, A and B). Thus, in the context of the cell surface, the integrin transmembrane and cytoplasmic domains and their potential interactions with other proteins contributed to the stability of the BC conformation. Structures in intact cells and detergent bicelles show that integrin α and β-subunit transmembrane domains associate over a large interface extending through the membrane bilayer (Lau et al., 2009; Zhu et al., 2009). The strong stabilization of the BC state relative to both the EC and EO states by integrin transmembrane and cytoplasmic domains in the context of the cell surface suggest that the transmembrane and cytoplasmic domains stabilize the BC, but not the EC and EO states, and hence that the transmembrane and cytoplasmic domains participate in intersubunit interactions in the BC but not the EC or EO states. In other words, transmembrane and cytoplasmic domain separation occurs during the BC-EC transition rather than the EC-EO transition, and on cell surfaces, integrin extension is associated with separation between the α and β-subunit transmembrane and cytoplasmic domains (Fig. 1 A).
Subunit- and cell type–specific integrin conformational equilibria set points on cell surfaces and integrin priming
Our thermodynamic measurements revealed the mechanistic basis for the long-standing observation that integrin α4β1 has higher basal activity than α5β1 (Bazzoni et al., 1998): on three cell types, the difference in energy between the BC and EO conformations was less for α4β1 than for α5β1 (Fig. 8, A–C). In basal integrin ensembles on the cell types, the high-affinity EO conformation comprised 0.3 to 1.4% of cell surface α4β1 compared with only 0.12 to 0.17% of cell surface α5β1. Although β1 integrins have commonly been considered to be constitutively active, only a small proportion of α4β1 and α5β1 are in their high-affinity state. α4β1 and α5β1 are more active than α2β1, α6β1, and α3β1 integrins (Bazzoni et al., 1998), which suggests that the latter integrins will basally exhibit an even lower proportion of the EO state on cell surfaces. Although the readiness of integrin α4β1 for activation and its presence on microvilli may each correlate with its ability to mediate leukocyte rolling, microvilli localization does not correlate with integrin activity, because integrin α6β1 also localizes to microvilli (Abitorabi et al., 1997).
We found cell-specific differences in integrin α4β1 basal ensemble affinity. Affinity was significantly lower on Thp1 cells than on Jurkat or KA4 cells. Our results agree with the finding of higher LIBS expression on α4β1 on Jurkat cells than on Thp1 cells (Yednock et al., 1995). Priming is a term that has been introduced to describe conformational changes in integrins induced by perturbations such as Mn2+ ions or LIBS antibodies that shift integrins toward, but not fully to, their ligand-occupied state (Humphries, 2004). We propose to define priming as the constellation of factors that regulate integrin ensemble affinity for soluble ligand. Posttranslational modifications of integrins, physicochemical differences in the environment of the integrin outside or inside the cell, and differences in the concentrations of other proteins with which integrins interact, either outside or inside the cell, may all modulate the relative stability of the three overall integrin conformational states.
Intracellular factors that regulate integrin priming are of particular interest. The activating adaptor talin appears to bind to the integrin β-subunit cytoplasmic domain only when the integrin α and β-subunit transmembrane domains are not associated (Kim et al., 2003). Silencing talin in Jurkat cells was found to decrease soluble VCAM binding to α4β1 (Manevich et al., 2007). Evidence here that integrin α and β-subunit transmembrane domains are associated in the BC and not in the EC and EO conformations suggests that talin should prime integrins by increasing the population of the EC and EO states relative to the BC state. Inhibitory proteins that bind to integrin cytoplasmic tails include SHARPIN, ICAP1, and filamin (Bouvard et al., 2013; Iwamoto and Calderwood, 2015). Inhibitors may decrease ensemble affinity by directly stabilizing closed conformations or by antagonizing binding of activators. SHARPIN binds a motif at the junction between the integrin α-subunit transmembrane and cytoplasmic domains but also blocks talin binding to the β subunit (Bouvard et al., 2013). ICAP1 binds a distal NXXY motif of β1 integrin, to which the activating adaptor kindlin also binds (Bouvard et al., 2013). Structural data show that filamin binds to a complex between the integrin α and β-subunit cytoplasmic domains (Liu et al., 2015). Adaptors and inhibitors may also influence integrin adhesiveness by regulating clustering and enhancing localized binding to multimeric integrin ligands. For example, talin is a dimer, and actin is a filament.
Our thermodynamic results suggest that the integrin α and β-subunit transmembrane and cytoplasmic domains are associated in the BC, but not the EC and EO, conformations (Fig. 1 A). In turn, this would suggest that filamin, and any other inhibitor that binds both the integrin α and β-subunit cytoplasmic domains, should decrease ensemble affinity by stabilizing the BC conformation. Do such inhibitors contribute to the much lower ensemble affinity measured here for cell-surface integrins than for integrin ectodomain fragments? The free energy of association of integrin α and β-subunit transmembrane domains measured in bicelles of −4.8 kcal/mol (Schmidt et al., 2016) is sufficient to explain our finding that the BC conformation is ∼3 kcal/mol lower in energy on cell surfaces that in ectodomain preparations. A definitive answer to the question of whether integrin inhibitors make an important contribution to the low energy of the BC conformation of intact integrins in cells would require comparisons to purified, intact integrins reconstituted into an artificial membrane environment.
Comparisons here between cellular integrin ensemble affinity for soluble ligand and integrin-dependent cell adhesion using a panel of five cell lines suggest the overriding importance of cellular regulatory mechanisms that only come into play when integrins bind immobilized ligands on substrates. Although cells varied by up to 2.8-fold in basal affinity and 22-fold in the EC50 of VCAM-Fc required for substrate adhesiveness, affinity for soluble ligand and adhesiveness to VCAM substrates did not correlate (Fig. 7). Addition of 9EG7 Fab to stabilize the EC and EO states raised ensemble affinity for soluble ligand by ∼100-fold and resulted in essentially identical affinities on all five cell types (Fig. 7 E). Nonetheless, the effectiveness of VCAM in supporting adhesion increased by only 1.1 to 8.2-fold, and there was little change in the rank order of EC50 values for the five cell types. Stabilizing integrin extension gave the largest enhancement of adhesiveness with the least adhesive cell types, whereas little effect was seen with the most adhesive cell type, Jurkat. Moreover, the number of α4β1 integrins per cell had little effect, because the most adhesive cell, Jurkat, expressed approximately threefold less α4β1 than less adhesive KA4 and Thp1 cells (Fig. 4, D and E). The high-affinity, EO conformation of α4β1 was absolutely required for cell adhesion, because stabilizing the closed conformations of α4β1 completely blocked cell adhesion. These results suggest significant variation among cells in the machinery that enables integrins to be stabilized in the high-affinity, EO conformation on substrates bearing VCAM. The similar adhesiveness of Jurkat cells in presence and absence of extension-stabilizing Fab suggests that cellular mechanisms that come into play on ligand-bearing substrates are quite effective in inducing the EO conformation and thus exogenous stimulators have little effect on Jurkat cells. In contrast, the less adhesive cell types appeared to have less effective cellular machinery for inducing the EO conformation of integrin α4β1 on VCAM substrates, because boosting the population of the EO conformation from ∼1% to ∼50% gave a 2.6- to 8.8-fold enhancement of adhesiveness (Fig. 7 F). Thus the measurements here of intrinsic ligand-binding affinities and relative free energies of the three integrin conformational states on cell surfaces do not predict adhesiveness directly. Instead, they provide a necessary framework for understanding how additional factors, including dissociation of inhibitors, binding of activating adaptors to integrin cytoplasmic domains, and application of force by the cytoskeleton that can be resisted by substrate-bound integrin ligands (and not by soluble ligands) can stabilize the integrin EO conformation to mediate cell adhesiveness (Fig. 8 D).
A framework for understanding integrin activation in cell adhesion and migration
To illustrate how our results provide a framework for understanding cellular regulation of integrin adhesiveness, we used our measurements of affinities and free energies to calculate the ability of intracellular adaptors that are selective for specific integrin conformational states and application of force by the cytoskeleton to stabilize the EO conformation of integrin α4β1 and α5β1. Calculations were as previously described for integrin α5β1 binding to fibronectin (Li and Springer, 2017). Using thermodynamics and the concentrations of conformation-specific adaptors such as talin relative to their Kd, we calculate how adaptors shift the populations of the three integrin conformational states. Similarly, using the Kd of VCAM and fibronectin for the BC, EC, and EO states of α4β1 and α5β1, respectively, we calculate how the concentration of ligands shifts the population of integrin states. We calculate the population of each unbound integrin state, each state bound to adaptor only, each state bound to ligand only, and each state bound to both adaptor and ligand. Additionally, for states bound to both adaptor and ligand, we calculate the effect of tensile force (F) on stabilizing extension and headpiece opening, given the increase in distance (Δx) along the force-bearing axis of 130 Å in extension and 15 Å in opening, and the change in energy (E) of E = −F × Δx (Fig. 8 D). Tensile force would be applied to the integrin through the adaptor by actin polymerization or actin-myosin contractility that is resisted by a substrate-bound ligand.
We compared different scenarios. In the absence of force, with an adaptor that is specific for the EC + EO states, large increases in free adaptor concentrations from 0.1- to 100-fold above Kd give proportionally much smaller increases in integrin activation (Fig. 8 E). In contrast, regulation is highly sensitive when a force of 1.5 pN is applied (Fig. 8 F), which is within the range measured on ligands and within integrins (Chang et al., 2016; Nordenfelt et al., 2016; Sun et al., 2016). In this case, activation is highly sensitive to adaptor concentration, with complete activation occurring with a concentration of adaptor within several fold of its Kd (Fig. 8 F). Alternatively, with a fixed concentration of an adaptor equal to its Kd, we calculated the effect of applying variable force. With the range of free energies of the BC conformation measured for α4β1 and α5β1 on cell surfaces, the population of the EO conformation went from <10% to >90% with only a two- to threefold change in force (Fig. 8 G). Force-dependent activation of integrin adhesiveness is thus ultrasensitive, because it shows all or none regulation over only a twofold range of biological signal input. Interestingly, ATP hydrolysis drives actin polymerization and actin-myosin contractility, similarly to MAP kinase cascades and chemotaxis that exemplify ultrasensitive biological regulation (Kuriyan and Wemmer, 2012).
What are the consequences of cell type–specific and integrin-specific differences in conformational equilibria set points for integrin activation? For regulation by adaptor-binding alone or adaptor binding in presence of a force of 1.5 pN, a much wider range of population of the EO conformation is accessible to α4β1 than to α5β1; furthermore, among the three cell types studied here, Thp1 cells have less access to the EO state of α4β1 than the other two cell types (Fig. 8, E and F). In contrast, for the model in which force can vary, all cell types can modulate the population of the EO state for both α4β1 and α5β1 from rarely to fully populated over a narrow range of forces, from 1 to 3 pN. In a remarkable concordance, measurements on adherent cells show that among ligands on substrates bound to α5β1, 70% experience forces in the 1 to 3 pN range (Chang et al., 2016). We conclude that the ΔGBC values and ΔGEC values measured here on multiple cell types for α4β1 and α5β1 are well suited to give ultrasensitive integrin activation in the range of forces exerted on integrins and their ligands in live cells (Chang et al., 2016; Nordenfelt et al., 2016; Sun et al., 2016). Our measurements provide information that is essential to building realistic models of how integrin adhesiveness is regulated in living cells. In the future, similar measurements on the β2 and β3 integrins, which are thought to be more resistant to activation than β1 integrins, and on the αV integrins, which have a diverse range of β subunits, will be important to explore the diversity of mechanisms that have evolved to regulate integrin adhesiveness.
Materials and methods
Fabs
Sources of hybridomas for 12G10, HUTS4, 8E3, 9EG7 (Askari et al., 2010), and SG/19 (Luo et al., 2004) were as described previously. mAb13 (Akiyama et al., 1989) hybridoma was a gift of K. Yamada (National Institutes of Health, Bethesda, MD). IgG was purified by protein G affinity; Fabs were prepared with papain digestion in PBS with 10 mM EDTA and 10 mM cysteine and papain/IgG mass ratio of 1:500 for 8 h at 37°C, followed by Hi-Trap Q chromatography in Tris-HCl, pH 9.0, with a gradient in the same buffer to 0.5 M NaCl.
Integrin α4β1 soluble preparations
Integrin α4β1 headpiece (α4 F1 to F587 with R558A mutation and β1 Q1 to E481) and ectodomain (α4 F1 to T943 with R558A mutation and β1 Q1 to D708) with secretion peptide, purification tags, and C-terminal clasp (Takagi et al., 2001) were produced by cotransfecting the pcDNA3.1/Hygro(−) vector coding the α-subunit and pIRES vector coding the β-subunit into HEK 293 cells (for protein with complex N-glycans) or HEK 293S GnTI−/− (N-acetylglucosaminyl transferase I deficient) cells (for protein with high-mannose N-glycans). Stable transfectants were selected with 100 µg/ml hygromycin and 1 mg/ml G418, and proteins were purified from culture supernatants by His-tag affinity chromatography and Superdex S200 gel filtration (Li et al., 2017).
Peptidomimetic and macromolecule fragments
FITC-conjugated α4β1 specific probe, 4-((N′-2-methylphenyl)ureido)-phenylacetyl-l-leucyl-l-aspartyl-l-valyl-l-prolyl-l-alanyl-l-alanyl-l-lysine (FITC-LDVP) and its unlabeled version, LDVP, were from Tocris Bioscience. Human VCAM-Fc was from R&D Systems. Human VCAM D1D2 (mature residues F1 to T202; Yu et al., 2013) and human MAdCAM D1D2 (mature residues Q1 to S231; Tan et al., 1998) were expressed and purified from HEK 293S GnTI−/− and CHO Lec.3.2.8.1 cell line supernatants, respectively, by affinity chromatography and gel filtration as described previously. The alternatively spliced connecting segment III of human fibronectin (Fn IIICS; mature residues D1992 to G2071) with an N-terminal His tag and TEV linker was expressed in Escherichia coli BL21 (DE3) cells. Protein codon optimized for E. coli was synthesized as gblocks from IDT and ligated into BamHI and NdeI sites of the pET11c vector. Fn IIICS from E. coli lysate was purified by His-tag affinity and Superdex 75 gel filtration.
FP
Each 10-µl sample contained 150 mM NaCl, 1 mM CaCl2, 1 mM MgCl2, 1 nM FITC-LDVP, the indicated concentration of α4β1, and indicated Fabs at concentrations shown in Table S1 in 20 mM Tris buffer, pH 7.4. The mixture was allowed to equilibrate at 22°C for 2 h before recording FP on a Synergy NEO HTS multimode microplate reader (Biotek).
Quantitative fluorescent flow cytometry
Jurkat, Thp1, K562, KA4, HPB-ALL, and U937 cells (106 cells/ml in RPMI-1640 medium and 10% FBS) were washed twice with assay medium (Leibovitz’s L-15 medium and 10 mg/ml BSA) containing 5 mM EDTA, twice with assay medium alone, and resuspended in assay medium. For saturation binding experiments, each 50-µl sample contained 5 × 105 cells/ml unless otherwise specified, indicated concentrations of FITC-LDVP, Alexa488-Fn39-10, or Alexa488-VCAM D1D2, and indicated Fabs at concentrations shown in Table S1 in assay medium. For competitive binding experiments, each 50-µl sample contained 5 × 105 cells/ml, 1 nM FITC-LDVP, varying concentrations of competitors, and indicated Fabs in assay medium. For Fab titration experiments, each 50-µl sample contained 5 × 105 cells/ml, 1 nM FITC-LDVP (for extension- and opening-stabilizing Fabs), or 30 nM FITC-LDVP (for closure-stabilizing Fabs) and indicated concentrations of Fabs. For LIBS antibody epitope exposure assay, each 50-µl sample contained 5 × 105 cells/ml, 5 nM Alexa647-9EG7, and indicated concentrations of ligands. The mixture was allowed to equilibrate at 22°C for 4 h before flow cytometry (BD FACSCanto II) without washing (Chigaev et al., 2001; Dong et al., 2014; Li et al., 2017). Binding was measured as mean fluorescence intensity (MFI); background MFI for FITC-LDVP, Alexa488-VCAM D1D2, and Alexa647-9EG7 in the presence of 10 mM EDTA was subtracted; and background MFI for α5β1 ligand Alexa488-Fn39-10 in the presence of a 100-fold-higher concentration of α5 specific blocking antibody mAb16 (Akiyama et al., 1989; Burrows et al., 1999) at each Alexa488-Fn39-10 concentration was subtracted.
Cell adhesion
VCAM-Fc at the indicated concentrations was absorbed to protein A–coated 96-well plates (ThermoFisher Scientific) in PBS for 16 h at 4°C. Plates were washed and blocked for 1 h at 37°C with PBS containing 30 mg/ml BSA. Cells were prelabeled with 2′,7′-bis-(2-carboxyethyl)-5-(and-6)-carboxyfluorescein, acetoxymethyl ester (BCECF AM; ThermoFisher Scientific) at 37°C for 1 h in assay medium (Leibovitz’s L-15 medium and 10 mg/ml BSA) and washed with assay medium containing 5 mM EDTA. Cells were washed once more in assay medium, resuspended (7.5 ×105 cells/ml, 100 µl) in assay medium, and added to VCAM-Fc–coated plates either directly or after preincubation for 0.5 h at 37°C with 8 µM 9EG7 Fab or indicated concentrations of LDVP or mAb13 Fab. Plates were incubated at 37°C for 1 h and washed thrice. Adherence was measured as the ratio of fluorescence intensity after and before washing.
Calculations
Probabilities of each conformation in the basal ensemble and their free energies relative to that of the extended open conformation (ΔGBC and ΔGEC; ΔGEO = 0) were calculated from intrinsic and ensemble affinities. Detailed derivations and equations used for data fitting and thermodynamic parameter calculation were exactly as described previously (Li et al., 2017). Final equations for fitting and calculations are also shown in the supplemental text. In brief, FP saturation binding data (shown in Fig. 3) were fit to Eq. S1 for affinities; saturation binding data on cell surface by FACS (Figs. 4 and 6) were fit to Eq. S8; competitive binding data on cell surface (Fig. 5 D) were fit to Eqs. S10 and S11; probability and free energy of each conformational state under basal condition (Fig. 3, D and E; Fig. 4 D; Fig. 6 E; and Fig. 8, A–C) were calculated according to Eqs. S12–S16 and Eqs. S17–S19, respectively; populations of the adaptor- and ligand-bound states in the presence of intracellular and extracellular signaling events (Fig. 8, E–G) were calculated according to Eq. S20; and affinity of ligands for integrin determined by LIBS antibody exposure assay (Fig. 5, A–C) were fitted from Eq. S22.
Online supplemental material
Supplementary text includes equations S1–S22 for data fitting and calculations. Figs. S1 and Fig. S2 show the EC50 values of conformation-specific Fabs for soluble α4β1 preparations and intact α4β1 receptors on cell surfaces, respectively, assayed by their influence on FITC-LDVP binding. Table S1 summarizes EC50 values of Fabs used on different preparations of integrin α4β1 and α5β1, Fab concentrations used in ligand-binding affinity measurements, and the percentage of Fab-bound integrins.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (grants P01-HL-103526 and R01-HL-131729) and the Susan G. Komen Breast Cancer Foundation (fellowship PDF16381021).
The authors declare no competing financial interests.
Author contributions: J. Li and T.A. Springer designed research. J. Li performed research. J. Li and T.A. Springer analyzed data and wrote the manuscript.
Footnotes
Abbreviations used:
- BC
- bent-closed
- EC
- extended-closed
- EO
- extended-open
- FP
- fluorescence polarization
- LDVP
- Leu-Asp-Val-Pro
- LIBS
- ligand-induced binding site
- MAdCAM
- mucosal addressin cell adhesion molecule
- MFI
- mean fluorescence intensity
- VCAM
- vascular cell adhesion molecule
References
- Abitorabi M.A., Pachynski R.K., Ferrando R.E., Tidswell M., and Erle D.J.. 1997. Presentation of integrins on leukocyte microvilli: A role for the extracellular domain in determining membrane localization. J. Cell Biol. 139:563–571. 10.1083/jcb.139.2.563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama S.K., Yamada S.S., Chen W.T., and Yamada K.M.. 1989. Analysis of fibronectin receptor function with monoclonal antibodies: Roles in cell adhesion, migration, matrix assembly, and cytoskeletal organization. J. Cell Biol. 109:863–875. 10.1083/jcb.109.2.863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alon R., Kassner P.D., Carr M.W., Finger E.B., Hemler M.E., and Springer T.A.. 1995. The integrin VLA-4 supports tethering and rolling in flow on VCAM-1. J. Cell Biol. 128:1243–1253. 10.1083/jcb.128.6.1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askari J.A., Tynan C.J., Webb S.E., Martin-Fernandez M.L., Ballestrem C., and Humphries M.J.. 2010. Focal adhesions are sites of integrin extension. J. Cell Biol. 188:891–903. 10.1083/jcb.200907174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzoni G., Ma L., Blue M.-L., and Hemler M.E.. 1998. Divalent cations and ligands induce conformational changes that are highly divergent among β1 integrins. J. Biol. Chem. 273:6670–6678. 10.1074/jbc.273.12.6670 [DOI] [PubMed] [Google Scholar]
- Bouvard D., Pouwels J., De Franceschi N., and Ivaska J.. 2013. Integrin inactivators: balancing cellular functions in vitro and in vivo. Nat. Rev. Mol. Cell Biol. 14:430–442. 10.1038/nrm3599 [DOI] [PubMed] [Google Scholar]
- Burrows L., Clark K., Mould A.P., and Humphries M.J.. 1999. Fine mapping of inhibitory anti-alpha5 monoclonal antibody epitopes that differentially affect integrin-ligand binding. Biochem. J. 344:527–533. [PMC free article] [PubMed] [Google Scholar]
- Chang A.C., Mekhdjian A.H., Morimatsu M., Denisin A.K., Pruitt B.L., and Dunn A.R.. 2016. Single molecule force measurements in living cells reveal a minimally tensioned integrin state. ACS Nano. 10:10745–10752. 10.1021/acsnano.6b03314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Xie C., Nishida N., Li Z., Walz T., and Springer T.A.. 2010. Requirement of open headpiece conformation for activation of leukocyte integrin αXβ2. Proc. Natl. Acad. Sci. USA. 107:14727–14732. 10.1073/pnas.1008663107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chigaev A., Blenc A.M., Braaten J.V., Kumaraswamy N., Kepley C.L., Andrews R.P., Oliver J.M., Edwards B.S., Prossnitz E.R., Larson R.S., and Sklar L.A.. 2001. Real time analysis of the affinity regulation of α4-integrin. The physiologically activated receptor is intermediate in affinity between resting and Mn(2+) or antibody activation. J. Biol. Chem. 276:48670–48678. 10.1074/jbc.M103194200 [DOI] [PubMed] [Google Scholar]
- Dong X., Hudson N.E., Lu C., and Springer T.A.. 2014. Structural determinants of integrin β-subunit specificity for latent TGF-β. Nat. Struct. Mol. Biol. 21:1091–1096. 10.1038/nsmb.2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elices M.J., Osborn L., Takada Y., Crouse C., Luhowskyj S., Hemler M.E., and Lobb R.R.. 1990. VCAM-1 on activated endothelium interacts with the leukocyte integrin VLA-4 at a site distinct from the VLA-4/fibronectin binding site. Cell. 60:577–584. 10.1016/0092-8674(90)90661-W [DOI] [PubMed] [Google Scholar]
- Guan J.L., and Hynes R.O.. 1990. Lymphoid cells recognize an alternatively spliced segment of fibronectin via the integrin receptor α4β1. Cell. 60:53–61. 10.1016/0092-8674(90)90715-Q [DOI] [PubMed] [Google Scholar]
- Humphries M.J. 2004. Monoclonal antibodies as probes of integrin priming and activation. Biochem. Soc. Trans. 32:407–411. 10.1042/bst0320407 [DOI] [PubMed] [Google Scholar]
- Hyun Y.M., Chung H.L., McGrath J.L., Waugh R.E., and Kim M.. 2009. Activated integrin VLA-4 localizes to the lamellipodia and mediates T cell migration on VCAM-1. J. Immunol. 183:359–369. 10.4049/jimmunol.0803388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto D.V., and Calderwood D.A.. 2015. Regulation of integrin-mediated adhesions. Curr. Opin. Cell Biol. 36:41–47. 10.1016/j.ceb.2015.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassner P.D., Alon R., Springer T.A., and Hemler M.E.. 1995. Specialized functional properties of the integrin α4 cytoplasmic domain. Mol. Biol. Cell. 6:661–674. 10.1091/mbc.6.6.661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C., Ye F., and Ginsberg M.H.. 2011. Regulation of integrin activation. Annu. Rev. Cell Dev. Biol. 27:321–345. 10.1146/annurev-cellbio-100109-104104 [DOI] [PubMed] [Google Scholar]
- Kim M., Carman C.V., and Springer T.A.. 2003. Bidirectional transmembrane signaling by cytoplasmic domain separation in integrins. Science. 301:1720–1725. 10.1126/science.1084174 [DOI] [PubMed] [Google Scholar]
- Kuriyan J.K.B., and Wemmer D.. 2012. The Molecules of Life: Physical and Chemical Principles. Garland Science, New York. 1008 pp. [Google Scholar]
- Lau T.L., Kim C., Ginsberg M.H., and Ulmer T.S.. 2009. The structure of the integrin αIIbβ3 transmembrane complex explains integrin transmembrane signalling. EMBO J. 28:1351–1361. 10.1038/emboj.2009.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legate K.R., and Fässler R.. 2009. Mechanisms that regulate adaptor binding to β-integrin cytoplasmic tails. J. Cell Sci. 122:187–198. 10.1242/jcs.041624 [DOI] [PubMed] [Google Scholar]
- Lehnert K., Print C.G., Yang Y., and Krissansen G.W.. 1998. MAdCAM-1 costimulates T cell proliferation exclusively through integrin α4β7, whereas VCAM-1 and CS-1 peptide use α4β1: evidence for “remote” costimulation and induction of hyperresponsiveness to B7 molecules. Eur. J. Immunol. 28:3605–3615. [DOI] [PubMed] [Google Scholar]
- Li J., and Springer T.A.. 2017. Integrin extension enables ultrasensitive regulation by cytoskeletal force. Proc. Natl. Acad. Sci. USA. 114:4685–4690. 10.1073/pnas.1704171114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Su Y., Xia W., Qin Y., Humphries M.J., Vestweber D., Cabañas C., Lu C., and Springer T.A.. 2017. Conformational equilibria and intrinsic affinities define integrin activation. EMBO J. 36:629–645. 10.15252/embj.201695803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin K., Ateeq H.S., Hsiung S.H., Chong L.T., Zimmerman C.N., Castro A., Lee W.C., Hammond C.E., Kalkunte S., Chen L.L., et al. . 1999. Selective, tight-binding inhibitors of integrin α4β1 that inhibit allergic airway responses. J. Med. Chem. 42:920–934. 10.1021/jm980673g [DOI] [PubMed] [Google Scholar]
- Liu J., Das M., Yang J., Ithychanda S.S., Yakubenko V.P., Plow E.F., and Qin J.. 2015. Structural mechanism of integrin inactivation by filamin. Nat. Struct. Mol. Biol. 22:383–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo B.-H., Strokovich K., Walz T., Springer T.A., and Takagi J.. 2004. Allosteric β1 integrin antibodies that stabilize the low affinity state by preventing the swing-out of the hybrid domain. J. Biol. Chem. 279:27466–27471. 10.1074/jbc.M404354200 [DOI] [PubMed] [Google Scholar]
- Luo B.-H., Carman C.V., and Springer T.A.. 2007. Structural basis of integrin regulation and signaling. Annu. Rev. Immunol. 25:619–647. 10.1146/annurev.immunol.25.022106.141618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manevich E., Grabovsky V., Feigelson S.W., and Alon R.. 2007. Talin 1 and paxillin facilitate distinct steps in rapid VLA-4-mediated adhesion strengthening to vascular cell adhesion molecule 1. J. Biol. Chem. 282:25338–25348. 10.1074/jbc.M700089200 [DOI] [PubMed] [Google Scholar]
- Masumoto A., and Hemler M.E.. 1993. Multiple activation states of VLA-4. Mechanistic differences between adhesion to CS1/fibronectin and to vascular cell adhesion molecule-1. J. Biol. Chem. 268:228–234. [PubMed] [Google Scholar]
- Mould A.P., and Humphries M.J.. 1991. Identification of a novel recognition sequence for the integrin α4β1 in the COOH-terminal heparin-binding domain of fibronectin. EMBO J. 10:4089–4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newham P., Craig S.E., Seddon G.N., Schofield N.R., Rees A., Edwards R.M., Jones E.Y., and Humphries M.J.. 1997. α4 integrin binding interfaces on VCAM-1 and MAdCAM-1. Integrin binding footprints identify accessory binding sites that play a role in integrin specificity. J. Biol. Chem. 272:19429–19440. 10.1074/jbc.272.31.19429 [DOI] [PubMed] [Google Scholar]
- Nordenfelt P., Elliott H.L., and Springer T.A.. 2016. Coordinated integrin activation by actin-dependent force during T-cell migration. Nat. Commun. 7:13119 10.1038/ncomms13119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh A., Leach T., Wyant T., Scholz C., Sankoh S., Mould D.R., Ponich T., Fox I., and Feagan B.G.. 2012. Vedolizumab for the treatment of active ulcerative colitis: a randomized controlled phase 2 dose-ranging study. Inflamm. Bowel Dis. 18:1470–1479. 10.1002/ibd.21896 [DOI] [PubMed] [Google Scholar]
- Park Y.K., and Goda Y.. 2016. Integrins in synapse regulation. Nat. Rev. Neurosci. 17:745–756. 10.1038/nrn.2016.138 [DOI] [PubMed] [Google Scholar]
- Rossi A.M., and Taylor C.W.. 2011. Analysis of protein-ligand interactions by fluorescence polarization. Nat. Protoc. 6:365–387. 10.1038/nprot.2011.305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt T., Ye F., Situ A.J., An W., Ginsberg M.H., and Ulmer T.S.. 2016. A Conserved Ectodomain-Transmembrane Domain Linker Motif Tunes the Allosteric Regulation of Cell Surface Receptors. J. Biol. Chem. 291:17536–17546. 10.1074/jbc.M116.733683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schürpf T., and Springer T.A.. 2011. Regulation of integrin affinity on cell surfaces. EMBO J. 30:4712–4727. 10.1038/emboj.2011.333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu F., Holzmann B., Seibold F., Erle D., and Kearney J.F.. 2002. Activated α4 integrins are preferentially expressed on immature thymocytes and activated T cells. Dev. Immunol. 9:73–84. 10.1080/1044667021000024229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sixt M., Bauer M., Lämmermann T., and Fässler R.. 2006. β1 integrins: zip codes and signaling relay for blood cells. Curr. Opin. Cell Biol. 18:482–490. 10.1016/j.ceb.2006.08.007 [DOI] [PubMed] [Google Scholar]
- Springer T.A., and Cybulsky M.I.. 1995. Traffic signals on endothelium for leukocytes in health, inflammation, and atherosclerosis. In Atherosclerosis and coronary artery disease. Fuster V., Ross R., and Topol E.J., eds. Lippincott-Raven Publishers, Philadelphia: 511–537. [Google Scholar]
- Springer T.A., and Dustin M.L.. 2012. Integrin inside-out signaling and the immunological synapse. Curr. Opin. Cell Biol. 24:107–115. 10.1016/j.ceb.2011.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y., Xia W., Li J., Walz T., Humphries M.J., Vestweber D., Cabañas C., Lu C., and Springer T.A.. 2016. Relating conformation to function in integrin α5β1. Proc. Natl. Acad. Sci. USA. 113:E3872–E3881. 10.1073/pnas.1605074113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z., Guo S.S., and Fässler R.. 2016. Integrin-mediated mechanotransduction. J. Cell Biol. 215:445–456. 10.1083/jcb.201609037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi J., Erickson H.P., and Springer T.A.. 2001. C-terminal opening mimics ‘inside-out’ activation of integrin α5β1. Nat. Struct. Biol. 8:412–416. 10.1038/87569 [DOI] [PubMed] [Google Scholar]
- Takagi J., Petre B.M., Walz T., and Springer T.A.. 2002. Global conformational rearrangements in integrin extracellular domains in outside-in and inside-out signaling. Cell. 110:599–11. 10.1016/S0092-8674(02)00935-2 [DOI] [PubMed] [Google Scholar]
- Takagi J., Strokovich K., Springer T.A., and Walz T.. 2003. Structure of integrin α5β1 in complex with fibronectin. EMBO J. 22:4607–4615. 10.1093/emboj/cdg445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan K., Casasnovas J.M., Liu J.-H., Briskin M.J., Springer T.A., and Wang J.-H.. 1998. The structure of immunoglobulin superfamily domains 1 and 2 of MAdCAM-1 reveals novel features important for integrin recognition. Structure. 6:793–801. 10.1016/S0969-2126(98)00080-X [DOI] [PubMed] [Google Scholar]
- von Andrian U.H., and Engelhardt B.. 2003. α4 integrins as therapeutic targets in autoimmune disease. N. Engl. J. Med. 348:68–72. 10.1056/NEJMe020157 [DOI] [PubMed] [Google Scholar]
- Vonderheide R.H., and Springer T.A.. 1992. Lymphocyte adhesion through VLA-4: Evidence for a novel binding site in the alternatively spliced domain of VCAM-1 and an additional α4 integrin counter-receptor on stimulated endothelium. J. Exp. Med. 175:1433–1442. 10.1084/jem.175.6.1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonderheide R.H., Tedder T.F., Springer T.A., and Staunton D.E.. 1994. Residues within a conserved amino acid motif of domains 1 and 4 of VCAM-1 are required for binding to VLA-4. J. Cell Biol. 125:215–222. 10.1083/jcb.125.1.215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao T., Takagi J., Coller B.S., Wang J.H., and Springer T.A.. 2004. Structural basis for allostery in integrins and binding to fibrinogen-mimetic therapeutics. Nature. 432:59–67. 10.1038/nature02976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yednock T.A., Cannon C., Vandevert C., Goldbach E.G., Shaw G., Ellis D.K., Liaw C., Fritz L.C., and Tanner L.I.. 1995. α4β1 integrin-dependent cell adhesion is regulated by a low affinity receptor pool that is conformationally responsive to ligand. J. Biol. Chem. 270:28740–28750. 10.1074/jbc.270.48.28740 [DOI] [PubMed] [Google Scholar]
- Yu Y., Schürpf T., and Springer T.A.. 2013. How natalizumab binds and antagonizes α4 integrins. J. Biol. Chem. 288:32314–32325. 10.1074/jbc.M113.501668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Luo B.H., Xiao T., Zhang C., Nishida N., and Springer T.A.. 2008. Structure of a complete integrin ectodomain in a physiologic resting state and activation and deactivation by applied forces. Mol. Cell. 32:849–861. 10.1016/j.molcel.2008.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Luo B.H., Barth P., Schonbrun J., Baker D., and Springer T.A.. 2009. The structure of a receptor with two associating transmembrane domains on the cell surface: integrin αIIbβ3. Mol. Cell. 34:234–249. 10.1016/j.molcel.2009.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Zhu J., and Springer T.A.. 2013. Complete integrin headpiece opening in eight steps. J. Cell Biol. 201:1053–1068. 10.1083/jcb.201212037 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.