The turnover of mitochondrial outer-membrane proteins is known to be mediated by the cytoplasmic ubiquitin–proteasome pathway. Wu et al. report the unexpected finding that two outer-membrane proteins Tom22 and Om45 are inwardly translocated into mitochondria and degraded by the inner-membrane protease Yme1.
Abstract
Mitochondria are double-membraned organelles playing essential metabolic and signaling functions. The mitochondrial proteome is under surveillance by two proteolysis systems: the ubiquitin–proteasome system degrades mitochondrial outer-membrane (MOM) proteins, and the AAA proteases maintain the proteostasis of intramitochondrial compartments. We previously identified a Doa1–Cdc48-Ufd1-Npl4 complex that retrogradely translocates ubiquitinated MOM proteins to the cytoplasm for degradation. In this study, we report the unexpected identification of MOM proteins whose degradation requires the Yme1-Mgr1-Mgr3 i-AAA protease complex in mitochondrial inner membrane. Through immunoprecipitation and in vivo site-specific photo–cross-linking experiments, we show that both Yme1 adapters Mgr1 and Mgr3 recognize the intermembrane space (IMS) domains of the MOM substrates and facilitate their recruitment to Yme1 for proteolysis. We also provide evidence that the cytoplasmic domain of substrate can be dislocated into IMS by the ATPase activity of Yme1. Our findings indicate a proteolysis pathway monitoring MOM proteins from the IMS side and suggest that the MOM proteome is surveilled by mitochondrial and cytoplasmic quality control machineries in parallel.
Introduction
Mitochondria evolve from endosymbiotic α-proteobacteria (Gray, 2012). Reflecting its evolutionary history, the proteostasis of intramitochondrial compartments are maintained by AAA proteases of prokaryotic origin. These proteases are: Pim1/Lon and ClpXP in the matrix, the m-AAA proteases Afg3 and Yta12, and the i-AAA protease Yme1 embedded in the mitochondrial inner membrane (MIM). The catalytic domains of m-AAA and i-AAA proteases face the matrix and the intermembrane space (IMS), respectively. The mitochondrial AAA proteases form homo- or heterooligomers, which consist of an ATPase ring to extract and unfold substrates as well as a proteolytic chamber to degrade substrates into peptides of 6–20 amino acid residues (Baker et al., 2011; Gerdes et al., 2012).
The mitochondrial outer membrane (MOM) encloses the intramitochondrial compartments and contains protein complexes playing pivotal roles in mitochondrial protein import, fusion/fission dynamics, mitophagy, metabolism, and other biological processes. Consistent with the intimate relationship of MOM with the cytoplasm, the ubiquitin–proteasome system has been shown to mediate the turnover of MOM proteins (Baker et al., 2011; Karbowski and Youle, 2011; Livnat-Levanon and Glickman, 2011; Taylor and Rutter, 2011). In general, ubiquitinated MOM proteins are extracted from membrane by the conserved AAA ATPase Cdc48 in yeast and valosin-containing protein (VCP) in higher eukaryotes and then are degraded by the proteasome. The pathway has been termed mitochondria-associated degradation (MAD). We recently systematically analyzed the turnover of MOM proteins in Saccharomyces cerevisiae, identified four Cdc48 substrates, and revealed Doa1 as a MAD-specific adapter of Cdc48 (Wu et al., 2016; Zhang and Ye, 2016).
During our analysis of MOM protein turnover, we unexpectedly identified two MOM proteins, Tom22 and Om45, whose degradation is independent of the proteasome pathway. In this study, we present experimental evidence that these proteins directly interact with the Yme1-Mgr1-Mgr3 complex, can be translocated into IMS, and are degraded in an Yme1-dependent manner. Our results indicate a proteasome-independent MAD pathway at the IMS side and demonstrate that MOM, which locates at the interface between the cytoplasm and the intramitochondrial compartments, is guarded by protein quality control machineries from both sides.
Results and discussion
The Yme1-Mgr1-Mgr3 complex is required for the degradation of Tom22 and Om45
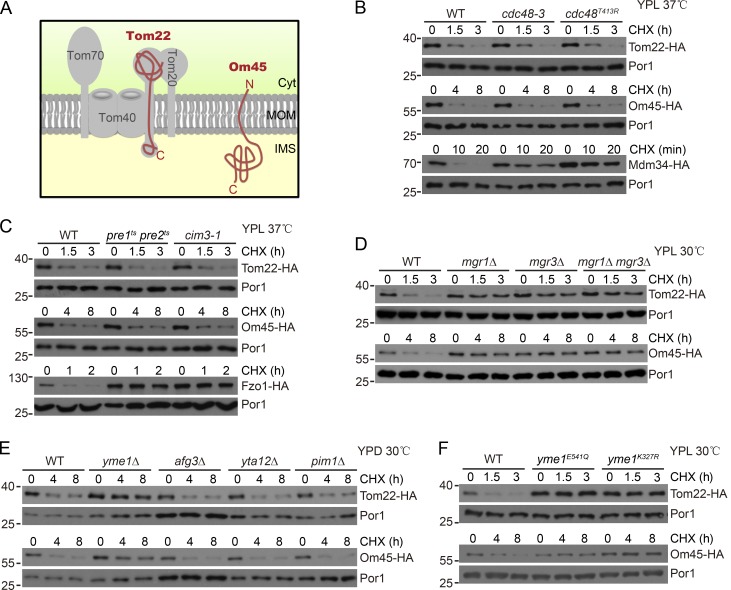
Tom22 and Om45 are two MOM proteins with distinct topology: Tom22 is C-terminally anchored in MOM with a large cytoplasmic domain of 97 amino acid residues and a small IMS domain of 33 residues (Kiebler et al., 1993; Lithgow et al., 1994); Om45 consists of 393 residues with the majority of residues inside the IMS and only five residues in the cytoplasm (Fig. 1 A; Song et al., 2014; Wenz et al., 2014). We chromosomally tagged Tom22 and Om45 with a C-terminal 6× HA tag and monitored their degradation after stopping protein synthesis with cycloheximide (CHX). The degradation of MOM Cdc48 substrates Fzo1-HA and Mdm34-HA was inhibited in mutant strains disrupting the function of Cdc48 and the proteasome (Fig. 1, B and C). However, the degradation of Tom22-HA and Om45-HA was not affected in the same set of mutant strains (Fig. 1, B and C) nor by disrupting Cdc48 adapters (Fig. S1 A). Their degradation was also not inhibited by deleting the MOM ATPase MSP1 (Fig. S1 B; Chen et al., 2014; Okreglak and Walter, 2014). We therefore set up a colony assay monitoring the degradation of Tom22-HA and performed an insertional screen (Wu et al., 2016) for genes required for Tom22-HA degradation.
Figure 1.
Yme1, Mgr1, and Mgr3 are essential for the degradation of Tom22 and Om45. (A) Topology of Tom22 and Om45. Cyt, cytosol. (B) The TOM22-HA, OM45-HA, and MDM34-HA strains in WT, cdc48-3, or cdc48T413R background were grown in lactate media (YPL) to log phase at 25°C and then treated with CHX at 37°C (restrictive temperature). Anti-Por1 blots are shown as loading controls. cdc48-3 and cdc48T413R are two temperature-sensitive (ts) mutations of Cdc48. (C) The TOM22-HA, OM45-HA, and FZO1-HA strains in WT, pre1ts pre2ts, or cim3-1 background were similarly analyzed as in B. pre1ts pre2ts and cim3-1 are ts mutations of the proteasome subunits Pre1, Pre2, and Cim3, respectively. (D–F) The WT and indicated mutant strains expressing either Tom22-HA or Om45-HA were grown in the indicated conditions to log phase and then treated with CHX and collected at the indicated time points. Molecular masses are shown in kilodaltons.
We screened ∼3,000 colonies and unexpectedly found that mutation of the MGR3 gene blocked Tom22-HA degradation. Mgr3 and its binding partner Mgr1 are transmembrane MIM proteins with a large IMS domain. They associate with the i-AAA protease Yme1 and are required for the degradation of some Yme1 substrates (Dunn et al., 2006, 2008). Deletion of MGR1 and MGR3 significantly inhibited the degradation of Tom22-HA and Om45-HA (Fig. 1 D). Similar effects were observed when YME1, but not other mitochondrial AAA-proteases, was deleted (Fig. 1 E). Yme1 can be inactivated by mutating the Walker-A (K327R) and Walker-B (E381Q) motifs of the AAA-ATPase domain and by mutating the protease domain (E541Q). Inactivation of the ATPase and protease domains of Yme1 also blocked the degradation of the two proteins (Fig. 1 F). Yme1 deletion blocked the degradation of its substrate Nde1-HA (Augustin et al., 2005) but did not affect the degradation of Cdc48 substrate Fzo1-HA (Fig. S1 C), indicating that Yme1 deletion does not affect MOM protein turnover in general.
The majority of Tom22 residues are exposed to the cytoplasm. Yme1 may only proteolyze the IMS domain of Tom22 and leave the remaining domains intact or for proteasomal degradation. We probed cell lysates with an antibody recognizing the cytoplasmic domain of Tom22. No cleavage product of Tom22 was present in WT cells or in cells with defective Cdc48 or proteasome (Fig. S1 D), indicating that Tom22-HA is unlikely to be partially processed by Yme1.
The mitochondrial contact site and cristae organizing system (MICOS) complex in MIM interacts with the TOM40 and SAM complexes in MOM to form intermembrane contact sites (van der Laan et al., 2016). We completely depleted the MICOS complex as reported (Fig. S1 E; Friedman et al., 2015) and found that it had no effect on the turnover of Tom20-HA and Om45-HA (Fig. S1 F).
The IMS domain is critical for the degradation of Tom22 and Om45
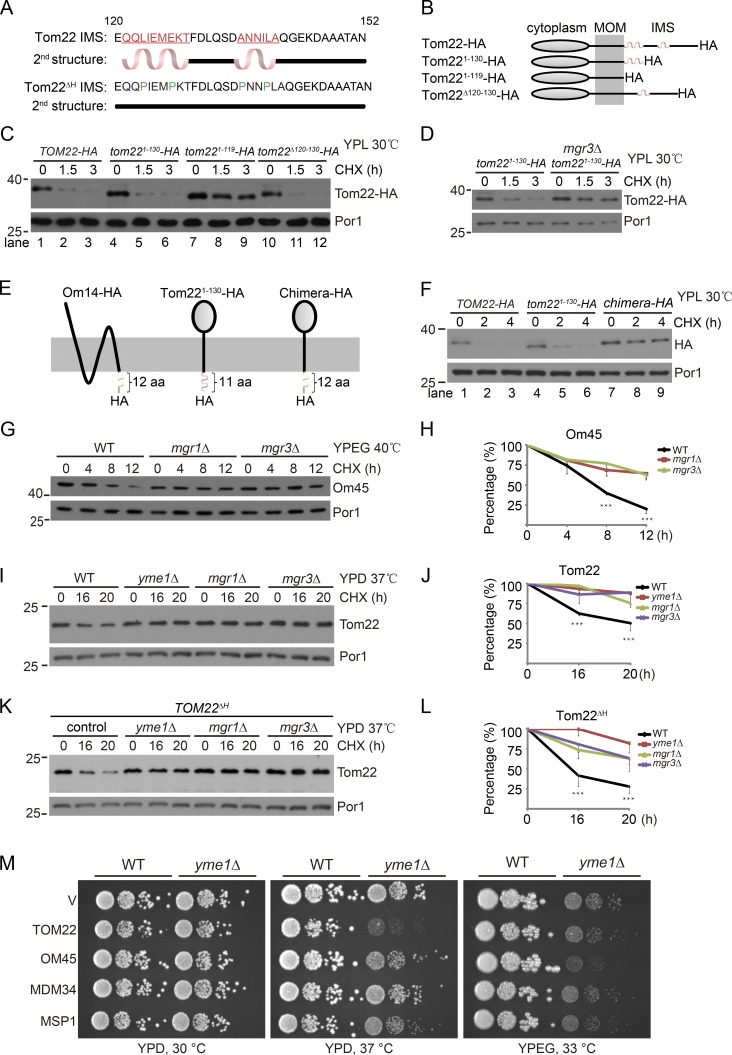
These characterizations suggest a working model that Tom22-HA and Om45-HA are proteolyzed by the Yme1 complex, although they reside in different membranes. A plausible mechanism is that substrate IMS domains mediate the degradation given the topology of both substrates and the Yme1 complex.
Tom22 IMS domain has two putative α-helices (Fig. 2 A). We generated partial and full truncations of Tom22 IMS domain (Fig. 2 B), which did not affect cell viability as previously reported (not depicted; Court et al., 1996; Moczko et al., 1997). The full truncation (Tom221–119-HA) but not partial truncations of the IMS domain inhibited Tom22-HA degradation (Fig. 2 C). Interestingly, Tom221–130-HA, which only retains the first α-helix and the HA tag, was still degraded by the Yme1 complex (Fig. 2 D). During the course of our analysis of MOM protein turnover, we found that Om14-HA, whose 6× HA tag also localizes in the IMS (Fig. 2 E; Sauerwald et al., 2015), is not a Yme1 substrate (Fig. S1 G). We replaced the IMS domain of Tom22 with that of Om14 to generate chimera-HA (Fig. 2 E). Although chimera-HA has an IMS domain of comparable length to that of Tom221–130-HA, it is quite stable (Fig. 2 F). These results indicate that the IMS domain of Tom22 plays a crucial role in its degradation and exclude the possibility that the IMS 6× HA tag itself mediates degradation by Yme1.
Figure 2.
The IMS domain is critical for the degradation of Tom22 and Om45. (A) The predicted secondary structures of the IMS domain of Tom22 and its mutant (Tom22ΔH) using the YASPIN program. (B–D) Schematic illustration (B) and turnover rate analysis (C and D) of the full-length and truncated forms of Tom22-HA. (E and F) Schematic illustration (E) and turnover rate analysis (F) of the indicated mutants. (G and H) The WT and indicated mutant strains were grown in ethanol and glycerol (YPEG) media at 30°C to log phase and then treated with CHX at 40°C. Cell lysates were analyzed by Western blotting with anti-Om45 and anti-Por1 antibodies (G). The Om45/Por1 ratio was measured by ImageJ software and plotted in H. (I and J) The WT and indicated mutant strains were grown in YPD media at 30°C to log phase and then treated with CHX at 37°C. Cell lysates were analyzed by Western blotting with anti-Tom22 and anti-Por1 antibodies (I). The Tom22/Por1 ratio (J) was analyzed as in H. (K and L) The Tom22ΔH mutant was treated and analyzed as in I and J. Data values represent means and SD from three independent experiments. Data were analyzed by two-way ANOVA followed by Bonferroni’s post hoc tests. ***, P < 0.001. Molecular masses are shown in kilodaltons. (M) WT or yme1Δ cells harboring empty vectors (V) or the indicated overexpression 2μ plasmids were grown in glucose media to log phase and then spotted on glucose (YPD) or ethanol and glycerol (YPEG) plates in a 10-fold serial dilution and then were incubated for 2–5 d at the indicated temperature.
We then generated antibodies and analyzed the turnover of endogenous untagged Tom22 and Om45. We found that Om45 was rapidly degraded at 40°C under respiratory growth condition (YPEG). The degradation was efficiently blocked by deleting MGR1 or MGR3 (Fig. 2, G and H). yme1Δ cells are unable to grow in YPEG media at 40°C (Campbell and Thorsness, 1998). We thus did not test the function of Yme1. Similarly, Tom22 turnover at 37°C in glucose media (yeast peptone dextrose; YPD) was also blocked by disrupting the Yme1 complex (Fig. 2, I and J).
The turnover of endogenous proteins is slower than the HA-tagged ones. We speculate that the HA tag may have destabilizing effects on IMS domains and thus promotes degradation. If this is true, then we may accelerate substrate degradation by introducing destabilizing mutations into the IMS domain. We replaced four residues of Tom22 IMS domain with proline, a helix breaker, to abolish α-helix formation (Fig. 2 A). The resultant Tom22ΔH mutant exhibited faster degradation, which was also blocked by disrupting the Yme1 complex (Fig. 2, K and L).
If the growth defect of yme1Δ cells is caused by the accumulation of Yme1 substrates, substrate overexpression should exacerbate the growth defect of yme1Δ cells. Overexpression of Tom22 and Om45 had no effect on the growth of WT cells (Fig. 2 M). In contrast, Tom22 overexpression significantly impaired the growth of yme1Δ cells at 37°C in YPD media (Fig. 2 M, middle), under which condition the turnover of Tom22 has been observed (Fig. 2 I). Similarly, Om45 overexpression mildly inhibited the growth of yme1Δ cells at 33°C in YPEG media (Fig. 2 M, right). As negative controls, overexpression of Cdc48 substrates Mdm34 and Msp1 did not impair the growth of WT and yme1Δ cells under the same conditions (Fig. 2 M).
Characterization of the Yme1-Mgr1-Mgr3 complex
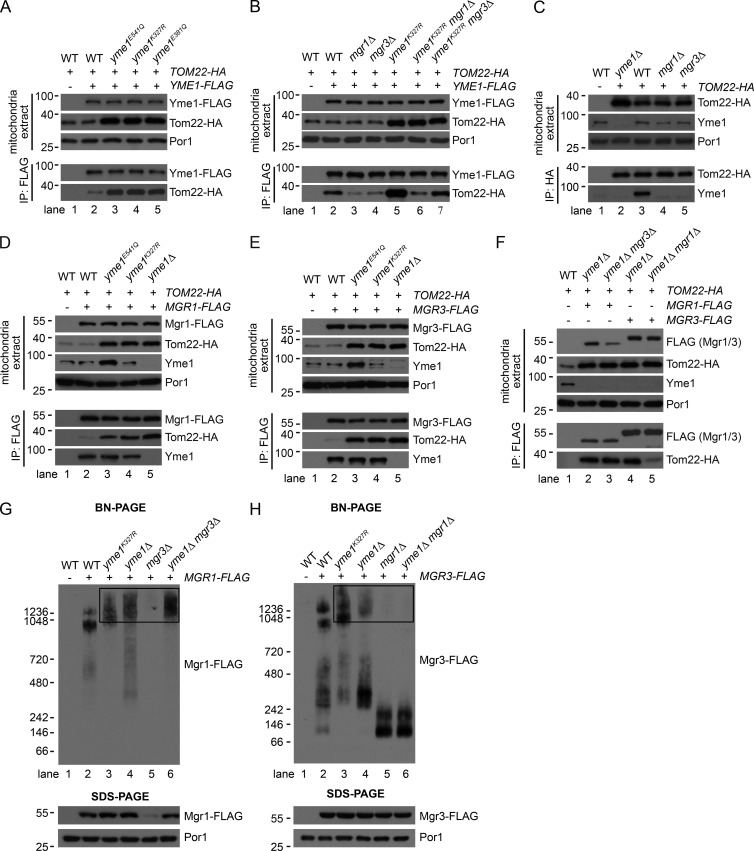
Previous studies of the Yme1-Mgr1-Mgr3 complex have shown that Mgr1 bridges the interaction between Yme1 and Mgr3, that Mgr1 and Mgr3 form a subcomplex in the absence of Yme1, and that both adapters are required for the degradation of Yme1 substrates (Dunn et al., 2006, 2008). We reproduced these results (unpublished data) and further found that Yme1-FLAG and Mgr3-FLAG had stable protein levels (Fig. S2, A and B); however, Mgr1-FLAG is degraded by Yme1 when Mgr3 is absent (Fig. S2 C).
We rescued the protein level of Mgr1-FLAG in mgr3Δ cells by mutating Yme1 (yme1E541Q; Fig. S2 D, lane 5 vs. lane 3). Immunoprecipitation (IP) of Mgr1-FLAG pulled down similar amounts of Yme1 from WT, yme1E541Q, and mgr3Δ yme1E541Q cells (Fig. S2 D, lanes 2, 4, and 5), suggesting that although Mgr3 is essential for Mgr1 stability, it is not required for Mgr1 and Yme1 interaction.
Blue-native (BN)-PAGE analysis revealed two Yme1-containing complexes: the major complex <1,048 kD and a minor complex >1,236 kD (Fig. S2 E, lane 2, white and black arrows). The size of Yme1 complexes was not affected by mutating Yme1 activity or deleting the adapters (Fig. S2 E, lanes 3–6). Mgr1-FLAG and Mgr3-FLAG comigrated with both Yme1 complexes (Fig. S2 F, lanes 4 and 5, white and black arrows) and also existed as Yme1-free forms (Fig. S2 F, lanes 4 and 5, boxed region). Collectively, our characterizations suggest a working model that in the Yme1-Mgr1-Mgr3 complex, Mgr1 bridges complex formation between Yme1 and Mgr3 and that Mgr3 stabilizes Mgr1 but is not required for Mgr1 interaction with Yme1 (Fig. S2 G).
Mgr1 and Mgr3 interact with MOM substrates and facilitate substrate recruitment to Yme1
We next examined the interaction between the Yme1 complex and substrates. Yme1-FLAG had weak interaction with substrates in WT cells but significantly enhanced interaction in Yme1 mutant cells (Figs. 3 A and S2 H). Moreover, deleting the IMS domain abolished the interaction of Tom22-HA with Yme1-FLAG (Fig. S2 I). Yme1-FLAG did not pull down Fzo1-HA or the stable MOM protein Por1 (Fig. S2 J), supporting the specific interaction between Yme1 and its MOM substrates. Mgr1 or Mgr3 deletion greatly reduced the pulldown of Tom22-HA by Yme1-FLAG (Fig. 3 B, lanes 2–4). Reciprocal IP of Tom22-HA and Om45-HA also pulled down endogenous Yme1 in an Mgr1- and Mgr3-dependent manner (lanes 3–5 in Figs. 3 C and S2 K). These results demonstrate that the adapters are required for substrate recruitment to Yme1. However, because deletion of either one of the adapters affects the other, it is not clear whether only one or both adapters participate in substrate recruitment. We thus analyzed substrate recruitment in yme1K327R cells, which preserve the Yme1–Mgr1 subcomplex in the absence of Mgr3. Mgr1 deletion in yme1K327R cells, which also releases Mgr3 from Yme1, greatly reduced Tom22-HA pulldown by Yme1-FLAG (Fig. 3 B, lane 6 vs. lane 5). However, Mgr3 deletion in yme1K327R cells, which preserves the Yme1–Mgr1 subcomplex, partially restored Tom22-HA pulldown by Yme1-FLAG (Fig. 3 B, lane 7 vs. lane 6), but not to the level when both adapters were present (Fig. 3 B, lane 7 vs. lane 5), indicating that Mgr1 alone is able to facilitate substrate recruitment, but both adapters are required for optimal substrate recruitment.
Figure 3.
Mgr1 and Mgr3 interact with MOM substrates and facilitate substrate recruitment to Yme1. (A–F) Digitonin-solubilized mitochondrial extracts from the indicated WT and mutant strains were subject to anti-FLAG or anti-HA IP and analyzed by SDS-PAGE. 5 µl (A, B, and D–F) or 20 µl (C) out of 100 µl immunoprecipitates were loaded for the anti-HA (A, B, and D–F) or anti-Yme1 (C) blots. (G and H) Digitonin-solubilized mitochondrial extracts (10 µg) from the indicated WT and mutant strains were analyzed by BN-PAGE and SDS-PAGE. The boxed regions highlight high MW upshifts of Mgr1-FLAG and Mgr3-FLAG. Molecular masses are shown in kilodaltons.
We then examined whether adapters interact with MOM substrates. Both adapters had weak interaction with substrates in WT cells and significantly enhanced interaction upon inactivation and deletion of Yme1 (lanes 2–5 in Fig. 3 D and E; and Fig. S2 L). Furthermore, Mgr1-FLAG pulled down similar amounts of Tom22-HA in yme1Δ and yme1Δ mgr3Δ cells (Fig. 3 F, lane 2 vs. lane 3), indicating that Mgr1 alone binds substrates as strongly as in the Mgr1–Mgr3 subcomplex. In contrast, Mgr3-FLAG pulled down much less Tom22-HA in yme1Δ mgr1Δ cells than in yme1Δ cells (Fig. 3 F, lane 4 vs. lane 5), suggesting that Mgr3 alone is capable of binding substrates, but the affinity is significantly diminished in the absence of Mgr1.
We further analyzed the adapters by BN-PAGE. Interestingly, Mgr1-FLAG and Mgr3-FLAG formed larger complexes and migrated to higher molecular weight (MW) positions upon inactivation or deletion of Yme1 (lanes 3 and 4 in Fig. 3, G and H). Because the adapters stably associated with substrates in yme1k327R and yme1Δ cells (Fig. 3, D and E; and Fig. S2 L), we hypothesize that these high-MW upshifts (boxed regions in Fig. 3, G and H) represent substrate-bound forms of Mgr proteins. Consistent with the hypothesis, Mgr1-FLAG alone, which has the full capability of binding substrates, formed high-MW upshifts (Fig. 3 G, lane 6), whereas Mgr3-FLAG alone, which has a greatly compromised ability to bind substrates, did not form upshifts and instead dropped to low-MW positions (Fig. 3 H, lanes 5 and 6).
In vivo site-specific photo cross-linking of Tom22 IMS domain to Mgr proteins
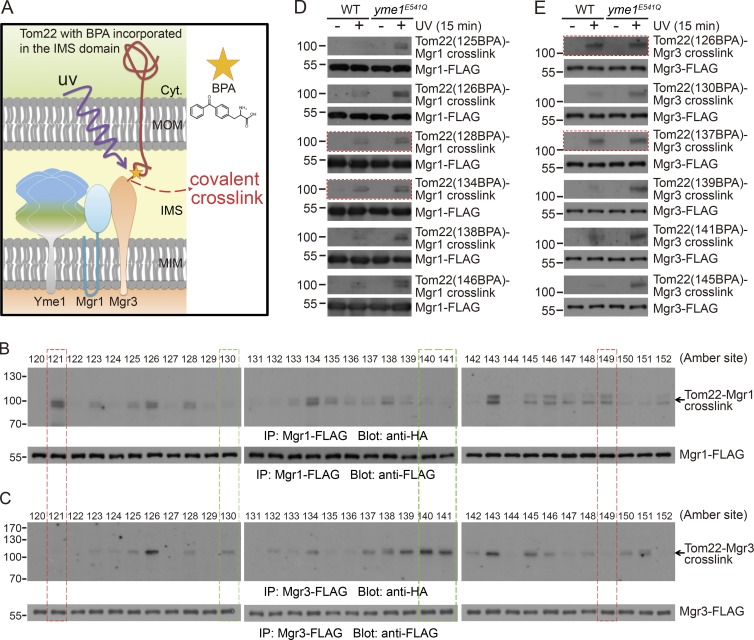
The IP experiments from the previous section were performed under conditions with membrane structures disrupted by detergents. To examine whether the interaction of substrates with the Yme1 complex occurs in intact mitochondria and to obtain further understanding of the interaction, we performed in vivo site-specific photo–cross-linking experiments (Chin et al., 2003; Shiota et al., 2013). The method allowed us to incorporate p-benzoyl-l-phenylalanine (BPA), a photoreactive artificial amino acid, into any residue position of the target protein. BPA can photo cross-link with nearby interacting proteins upon UV irradiation of living cells. Because of the short side chain of BPA, only direct interacting proteins could be crosslinked. Therefore, the method enabled us to detect in vivo, site-specific, and direct interactions.
We incorporated BPA into the Tom22 IMS domain. The incorporation site is specified by an amber codon (Fig. 4 A). To validate our system, we incorporated BPA into positions 124 or 132 (Fig. S3, A and B). Upon UV irradiation, Tom22-(132BPA)-HA clearly cross-linked with Tim50-FLAG, whereas Tom22-(124BPA)-HA did not (Fig. S3 C), reproducing previously published results (Shiota et al., 2011).
Figure 4.
In vivo site-specific photo cross-linking of Tom22 IMS domain to Mgr1 and Mgr3. (A) Cartoon illustration of the in vivo site-specific photo–cross-linking method. (B and C) The MGR1-FLAG yme1E541Q and MGR3-FLAG yme1E541Q strains harboring plasmids for the expression of Tom22-HA with BPA incorporated at the indicated sites were irradiated with UV for 15 min. Whole-cell extracts were prepared and subjected to anti-FLAG IP as described in the Site-specific in vivo photo cross-linking and IP section of Materials and methods. Immunoprecipitates were analyzed by SDS-PAGE. About 30 µl out of 100 µl immunoprecipitates were loaded for the anti-HA blots. Red and green boxes highlight representative residue positions that are only clearly cross-linked to Mgr1-FLAG or Mgr3-FLAG, respectively. (D and E) The MGR1-FLAG strains (D) and MGR3-FLAG strains (E) in WT or yme1E541Q background were transformed with plasmids expressing BPA-incorporated Tom22-HA and analyzed as in B and C. Red boxes highlight Tom22 residue positions showing similar level of cross-linking in WT and yme1E541Q cells. Molecular masses are shown in kilodaltons.
We then performed the photo–cross-linking experiments in MGR1-FLAG yme1E541Q and MGR3-FLAG yme1E541Q cells, which have stabilized interactions between Tom22 and the adapters. BPA was introduced to each residue in Tom22 IMS domain (residues 120–152). After UV irradiation, we immunoprecipitated Mgr1-FLAG or Mgr3-FLAG, resolved the immunoprecipitates on SDS-PAGE, and probed with anti-HA antibody to reveal the crosslinked products. Several observations were obtained. First, we detected crosslinked products for both adapters at the expected position of ∼100 kD (black arrows in Fig. 4, B and C). For Mgr1, most cross-linked products had double bands, which were also observed in other cross-linking experiments (Plath et al., 1998; Carvalho et al., 2010). A conceivable explanation is that BPA has multiple accessible residues on the target protein, and the cross-linked products are different in mobility on SDS-PAGE. Second, both adapters cross-linked to BPA at a variety of positions widely distributed throughout Tom22 IMS domain. This result was consistent with the observation that Tom22-HA degradation was inhibited only upon deletion of the whole IMS domain (Fig. 2 C). Last, we found that Mgr1 and Mgr3 cross-linked to BPA at common and, more interestingly, different positions of Tom22 (highlighted by colored boxes in Fig. 4, B and C), indicating that Mgr1 and Mgr3 have overlapped but also distinct interaction surfaces on the same substrate. During photo cross-linking, Tom22-(BPA)-HA is continuously expressed and imported. We added CHX to stop protein synthesis before cross-linking and found that the cross-linking efficiency was similar in the presence or absence of CHX (Fig. S3 D). Therefore, cross-linking mainly occurs to preexisting proteins.
We then selected several IMS residue positions of Tom22 and compared their cross-linking with Yme1 adapters in WT and yme1E541Q cells. BPA incorporated at most positions showed no or much weaker cross-linking in WT cells as compared with yme1E541Q cells (Fig. 4, D and E). Interestingly, we also observed positions exhibiting similar cross-linking efficiency in WT and yme1E541Q cells (red boxes in Fig. 4, D and E). These residues may be most easily exposed and recognized by Yme1 adapters.
Photo cross-linking of Tom22 cytoplasmic domain to Mgr3 in yme1E541Q cells
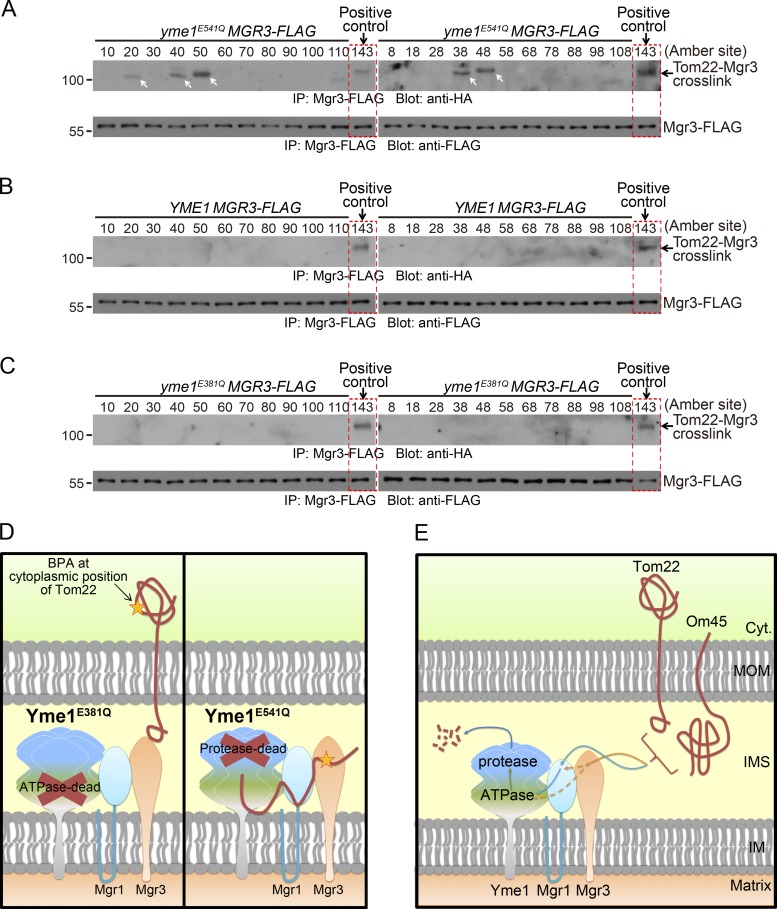
The ATPase domain of AAA proteases can function independently of the protease activity to drive protein translocation across the membrane (Rainey et al., 2006; Tatsuta et al., 2007) and remain associated with unfolded substrates when the protease domain is inactivated (Van Melderen and Gottesman, 1999; Singh et al., 2000). To examine whether Tom22 cytoplasmic domain can be dislocated into the IMS, we incorporated BPA into 22 randomly chosen positions in Tom22 cytoplasmic and transmembrane domains and observed that positions 20, 38, 40, 48, and 50 showed clear cross-linking with Mgr3 in yme1E541Q (ATPase-active and protease-dead) cells (Fig. 5 A, white arrows). In contrast, no cross-linking with Mgr3 was observed in WT and yme1E381Q (ATPase dead) cells (Fig. 5, B and C). The cross-linking in yme1E541Q cells was not affected by CHX treatment (Fig. S3 E). The differential cross-linking of Yme1 mutants with Tom22 cytoplasmic positions suggest that the Tom22 cytoplasmic domain can be dislocated into IMS by the ATPase activity of Yme1 (Fig. 5 D).
Figure 5.
In vivo site-specific photo cross-linking of the Tom22 cytoplasmic domain to Mgr3. (A–C) The MGR3-FLAG strains in yme1E541Q (A), WT (B), or yme1E381Q (C) background were transformed with plasmids expressing Tom22-HA with BPA incorporated at the indicated sites and analyzed as in Fig. 4 C. Red boxes highlight a positive control, which is the cross-linking of Tom22 (143 BPA) with Mgr3 in yme1E541Q background. White arrows highlight cross-linked bands. Molecular masses are shown in kilodaltons. (D) Cartoon illustration showing that the Yme1E541Q mutant (protease dead but ATPase active) but not the Yme1E381Q mutant (ATPase dead but protease active) can cross-link to Tom22 cytosolic domain after the dislocation of the entire protein into IMS. (E) Working model for the proteolysis of Tom22 and Om45 by the Yme1-Mgr1-Mgr3 complex.
Concluding remarks
Our data suggest a working model that Yme1 adapters Mgr1 and Mgr3 recognize the IMS domains of Tom22 and Om45 and recruit them to Yme1. After engaging the IMS domain of substrates, the ATPase activity of Yme1 powers substrate unfolding and dislocation into IMS for proteolysis (Fig. 5 E). Our model is based on the following observations: (A) Tom22 and Om45 are degraded in an Yme1-dependent manner (Figs. 1 and 2); (B) the substrate IMS domain and Yme1 adapters are required for substrate recruitment to Yme1 and degradation (Figs. 1, 2, and 3); (C) Substrate IMS domains directly interact with the Yme1 adapters (Figs. 3 and 4); and (D) The cytoplasmic domain of Tom22 can be dislocated into IMS by the ATPase activity of Yme1 (Fig. 5).
Yme1 can access substrates through its middle ATPase and C-terminal protease domains (Graef et al., 2007) and also through the Mgr1/3 adapter (Dunn et al., 2006, 2008; this study). These substrate-binding sites may work independently for different substrates or cooperatively/sequentially for the same substrate. Further studies are required to examine the potential role of other substrate-binding sites in the processing of MOM substrates.
To this end, we have not found other proteins required for the degradation of MOM substrates by Yme1. But we cannot exclude the potential involvement of cytoplasmic and mitochondrial factors in the degradation of Tom22 and Om45. These factors may help separate substrates from their interacting partners and participate in the inward translocation of substrates.
The concept of MAD originally refers to a molecular pathway that MOM proteins are ubiquitinated and retrogradely translocated by the Cdc48/VCP complex to the cytoplasm for proteasomal degradation (Karbowski and Youle, 2011; Livnat-Levanon and Glickman, 2011; Taylor and Rutter, 2011). In this study, we report an unexpected pathway in which MOM substrates can be inwardly translocated into mitochondria and degraded by the Yme1-Mgr1-Mgr3 complex. We therefore propose to expand the concept of MAD to include all the MOM-associated protein turnover pathways: the Doa1–Cdc48-Ufd1-Npl4 complex recognizes the ubiquitinated cytoplasmic domains (Wu et al., 2016) and the Yme1-Mgr1-Mgr3 complex recognizes the IMS domains of MOM proteins. The Cdc48- and Yme1-mediated pathways represent MAD pathways at the cytoplasmic and the IMS sides respectively. It remains to be determined whether there exist specific pathways monitoring the transmembrane domains of MOM proteins.
Materials and methods
Yeast strains and cell culture
The yeast strains used in this study are listed in Table S1. Strain transformation was performed using the lithium acetate method, selected on appropriate media, and confirmed by PCR and gene expression as needed. All deletions were generated by PCR-based homologous recombination to replace the entire ORF with appropriate selection cassettes (Longtine et al., 1998; Gueldener et al., 2002). All the C-terminal tags were generated by PCR-based homologous recombination to replace the endogenous stop codon with cassettes containing appropriate tags and selectable markers (Longtine et al., 1998). The ΔMICOS strain was made by using the Cre–loxP system as described previously (Gueldener et al., 2002; Friedman et al., 2015). In brief, the entire ORF of each MICOS gene was replaced by homologous recombination with the loxP–his5+–loxP cassette (amplified from pUG27). The resulting strain was transformed with pSH47, which carries the URA3 selection cassette and expresses the Cre recombinase under the control of a repressible GAL1 promoter. To remove the selection cassette, cells were inoculated in glucose (YPD) media for 1 d to allow the leaky expression of Cre to occur. Individual histidine auxotrophic clones were isolated and cultured in nonselective YPD media for 1 d. During this period, the random loss of pSH47 plasmid would occur. Individual histidine and uracil auxotrophic clones were then isolated and verified by PCR. This process was repeated sequentially to knock out all the MICOS genes.
Media used in this study included: YPD (1% yeast extract, 2% peptone, and 2% glucose), YPL (1% yeast extract, 2% peptone, and 2% lactate), YPEG (1% yeast extract, 2% peptone, 3% ethanol, and 3% glycerol), SCD (0.67% yeast nitrogen base without amino acids, 0.079% complete supplement mixture, and 2% glucose), SCEG (0.67% yeast nitrogen base without amino acids, 0.079% complete supplement mixture, 3% ethanol, and 3% glycerol), SCD-LEU (0.67% yeast nitrogen base without amino acids, 0.069% Leu dropout supplement, and 2% glucose), and SCD-TRP-URA (0.67% yeast nitrogen base without amino acids, 0.072% Trp/Ura dropout supplement, and 2% glucose). Yeast strains were grown at 30°C if not otherwise indicated.
Antibodies and chemicals
The following antibodies were from Sigma-Aldrich: G6PDH produced in rabbit (A9521), HA-peroxidase (H6533), and FLAG M2 produced in mouse (F1804). The antibody for Por1 produced in mouse (459500) was from Thermo Fisher Scientific. The synthetic peptides RKDIGDDKPK (amino acid residues 731–740 of Yme1), ENETLLDRIVALKD (amino acid residues 54–67 of Tom22), and KEDALSLKDALLGV (amino acids residues 64–77 of Om45) were used for generating Yme1, Tom22, and Om45 antibodies, respectively.
Yeast extract, peptone, and yeast nitrogen base without amino acids were from BD. Yeast complete supplement mixture was from MP Biomedicals. Yeast amino acid dropout supplements were from Takara Bio Inc. CHX was from Amresco. Other chemicals or reagents were from Sigma-Aldrich if not otherwise indicated.
Plasmids and primers
The 2μ plasmids for overexpressing Tom22, Om45, Mdm34, and Msp1 were made as follows: the coding sequence of each gene and the flanking endogenous promoter (∼1,000 bp) and terminator (∼500 bp) sequences were subcloned to pRS42N by Gibson assembly (Gibson et al., 2009).
The 2μ plasmid pYES2-TOM22-HA was generated as follows: the coding sequence of TOM22 together with a C-terminal 6× HA tag was amplified using the TOM22-HA strain as the template, and then they were cloned into pYES2 (Thermo Fisher Scientific) at KpnI–EcoRI sites. The amber (TAG) codon was introduced to specific positions by QuikChange site-directed mutagenesis (Agilent Technologies).
Primers used for plasmid construction and introducing amber codons to TOM22 are listed in Table S1.
Colony screen assay
To randomly generate insertion mutants by homologous recombination (Burns et al., 1994), the founder strain TOM22-HA, leu2Δ in CEN.PK background, was transformed with the yeast genomic mini-Tn3::lacZ::LEU2 transposon insertion library. Approximately 3,000 mutants were generated and tested by colony assay. On day 0, the transformed yeast cells were grown on selective glucose (SCD-LEU) plates. On day 3, we replica plated the cells from the SCD-LEU plates to lactate (YPL) plates. On day 4, these cells were replica plated onto nitrocellulose membranes that were placed on YPL plates. On day 5, the nitrocellulose membranes were placed on new YPL plates supplemented with 50 µg/ml CHX. After 8 h of treatment, we lysed the yeast cells by placing the membranes on filter paper soaked with colony lysis buffer (0.1% SDS, 0.2 M NaOH, 0.5% β-mercaptoethanol; Knop et al., 1996). After incubation for 1 h at room temperature, we cleaned the membranes by rinsing off the cell debris with water. The membranes were then subject to HA immunoblotting. The colonies with relatively stronger HA signals were collected onto a new plate and subsequently reexamined for protein degradation by colony assay and Western blotting. Finally, the insertion sites were mapped using the Vectorette PCR method (Riley et al., 1990).
Yeast whole-cell extract preparation
Cell pellets were resuspended in 300 µl yeast lysis buffer (50 mM NaCl, 50 mM NaF, 100 mM Tris-HCl, pH 7.5, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 10% glycerol, 14 mM 2-mercaptoethanol, 2 mM PMSF, 5 µM pepstatin A, and 10 µM leupeptin). After adding ∼80 µl of glass beads, cells were lysed via three rounds of bead beating (40 s beating followed by 1 min of cooling on ice). For the degradation assay, CHX was used at 50 µg/ml.
Isolation of the mitochondria-enriched fraction and IP
Mitochondria were isolated following a previously described method (Diekert et al., 2001) with some modifications. In brief, ∼200 ml of cells grown in YPL media to late log phase (OD ∼3) were collected. Cells were washed once with water and incubated in TD buffer (10 mM DTT and 100 mM Tris-SO4, pH 9.4) for 15 min at 30°C. Cells were then washed once with SP buffer (1.2 M sorbitol and 20 mM potassium phosphate, pH 7.4) and treated with zymolyase 20T/100T (MP Biomedicals) for 40 min at 30°C to generate spheroplasts. After two times of washes with SP buffer, the spheroplasts were resuspended in SHE buffer (0.6 M sorbitol, 20 mM Hepes-KOH, pH 7.4, 1 mM EGTA, pH 8, and 2 mM MgCl2) supplemented with protease inhibitors and homogenized by a French press (EmulsiFlex-C3, AVESTIN Inc.) at pressures in the range of 1,000 to 1,500 psi. The mitochondria-enriched fraction was obtained by differential centrifugation, flash frozen by liquid nitrogen, and stored at −80°C until use.
Crude mitochondria were solubilized with 1% digitonin buffer (50 mM Hepes-KOH, pH 7.4, 50 mM KOAc, 2 mM Mg(OAc)2, 1 mM CaCl2, 200 mM sorbitol, 1 mM NaF, and 1% [wt/vol] digitonin) supplemented with 1 mM DTT, 2 mM ATP, and protease inhibitors for 45–60 min at 4°C. The lysates were cleared by centrifugation at 17,000 g for 10 min at 4°C. The supernatant was then mixed with anti-FLAG or anti-HA agarose beads (Sigma-Aldrich) and incubated at 4°C for 5–6 h (anti-FLAG) or 8–18 h (anti-HA). The agarose beads were then washed five times with 0.1% digitonin buffer and eluted overnight with FLAG or HA peptide (ChinaPeptides Co. Ltd.) at 4°C.
Site-specific in vivo photo cross-linking and IP
Experiments were preformed following a previously described method (Shiota et al., 2013) with specific modifications as follows. In brief, strains in W303 background were transformed with two high-copy plasmids, one expressing Tom22-HA with an amber (TAG) codon at a specific site under the control of the repressible GAL1 promoter and the other one expressing amber suppressor tRNA and a modified aminoacyl-tRNA synthetase that specifically charges the amber suppressor tRNA with the photoreactive unnatural amino acid BPA (a gift from P.G. Schultz, The Scripps Research Institute, Jupiter, FL). Cells were grown in selective glucose media to log phase and then switched to selective lactate media containing 0.05% galactose and 0.2 mM BPA for 16–18 h to induce the expression of BPA-incorporated Tom22-HA. About 100 OD600 U of cells in late log phase were then split into two halves. One half was kept on ice and used as the control. The other half was subjected to UV irradiation for 15 min. To prepare whole-cell extracts, cells were resuspended in 0.1 M NaOH and incubated at room temperature for 5–10 min. Cell pellets were then resuspended in SDS buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 2% [wt/vol] SDS, and 4% [vol/vol] β-mercaptoethanol) and boiled at 98°C for 10 min. The lysates were cleared by centrifugation at 17,000 g for 10 min. The supernatant was then diluted with 15 volumes of Triton X-100 buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 1% [vol/vol] Triton X-100), mixed with anti-FLAG agarose beads, and incubated at 4°C for 5–6 h. The agarose beads were then washed four times with Triton X-100 buffer and eluted overnight with FLAG peptide at 4°C.
Statistical analysis
For Fig. 2 (H, J, and L), protein bands were quantified using ImageJ software (National Institutes of Health). Data were then processed in Excel (Microsoft) and analyzed in Prism (GraphPad Software) using two-way ANOVA followed by Bonferroni’s post hoc tests.
Online supplemental material
Fig. S1 shows that the degradation of Tom22-HA and Om45-HA is not affected by inhibiting the function of the Doa1–Cdc48-Ufd1-Npl4 complex or the proteasome, by deletion of MSP1, or by deletion of the MICOS complex. Fig. S2 characterizes the Yme1-Mgr1-Mgr3 complex and analyzes the interaction between substrates and the Yme1 complex. Fig. S3 validates the photo cross-linking between Tom22 and Tim50 and analyzes the photo cross-linking of Tom22 (BPA)-HA with Mgr1 and Mgr3 in the presence or absence of CHX. Table S1 lists the strains and primers used in this study.
Supplementary Material
Acknowledgments
We thank Dr. Peter G. Schultz (The Scripps Research Institute) for providing the plasmid expressing BpaRS and amber suppressor tRNA.
The research is supported by the National Basic Research Program of China 973 (2012CB837503) and the municipal government of Beijing. X. Wu is supported by the Beijing Postdoctoral Research Foundation and China Postdoctoral Science Foundation.
The authors declare no competing financial interests.
Author contributions: Lanlan Li generated strains and plasmids; Xi Wu and Lanlan Li performed CHX chasing experiments; Xi Wu did growth tests, IP, and in vivo site-specific photo–cross-linking experiments; Hui Jiang performed the BN gel experiments; and Xi Wu and Hui Jiang conceived the project, designed the experiments, analyzed data, prepared the figures, and wrote the manuscript.
Footnotes
Abbreviations used:
- BN
- Blue-native
- BPA
- p-benzoyl-l-phenylalanine
- CHX
- cycloheximide
- IMS
- intermembrane space
- IP
- immunoprecipitation
- MAD
- mitochondria-associated degradation
- MICOS
- mitochondrial contact site and cristae organizing system
- MIM
- mitochondrial inner membrane
- MOM
- mitochondrial outer membrane
- MW
- molecular weight
- ts
- temperature sensitive
References
- Augustin S., Nolden M., Müller S., Hardt O., Arnold I., and Langer T.. 2005. Characterization of peptides released from mitochondria: evidence for constant proteolysis and peptide efflux. J. Biol. Chem. 280:2691–2699. 10.1074/jbc.M410609200 [DOI] [PubMed] [Google Scholar]
- Baker M.J., Tatsuta T., and Langer T.. 2011. Quality control of mitochondrial proteostasis. Cold Spring Harb. Perspect. Biol. 3:a007559 10.1101/cshperspect.a007559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns N., Grimwade B., Ross-Macdonald P.B., Choi E.Y., Finberg K., Roeder G.S., and Snyder M.. 1994. Large-scale analysis of gene expression, protein localization, and gene disruption in Saccharomyces cerevisiae. Genes Dev. 8:1087–1105. 10.1101/gad.8.9.1087 [DOI] [PubMed] [Google Scholar]
- Campbell C.L., and Thorsness P.E.. 1998. Escape of mitochondrial DNA to the nucleus in yme1 yeast is mediated by vacuolar-dependent turnover of abnormal mitochondrial compartments. J. Cell Sci. 111:2455–2464. [DOI] [PubMed] [Google Scholar]
- Carvalho P., Stanley A.M., and Rapoport T.A.. 2010. Retrotranslocation of a misfolded luminal ER protein by the ubiquitin-ligase Hrd1p. Cell. 143:579–591. 10.1016/j.cell.2010.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.C., Umanah G.K., Dephoure N., Andrabi S.A., Gygi S.P., Dawson T.M., Dawson V.L., and Rutter J.. 2014. Msp1/ATAD1 maintains mitochondrial function by facilitating the degradation of mislocalized tail-anchored proteins. EMBO J. 33:1548–1564. 10.15252/embj.201487943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin J.W., Cropp T.A., Anderson J.C., Mukherji M., Zhang Z., and Schultz P.G.. 2003. An expanded eukaryotic genetic code. Science. 301:964–967. 10.1126/science.1084772 [DOI] [PubMed] [Google Scholar]
- Court D.A., Nargang F.E., Steiner H., Hodges R.S., Neupert W., and Lill R.. 1996. Role of the intermembrane-space domain of the preprotein receptor Tom22 in protein import into mitochondria. Mol. Cell. Biol. 16:4035–4042. 10.1128/MCB.16.8.4035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diekert K., de Kroon A.I.P.M., Kispal G., and Lill R.. 2001. Isolation and subfractionation of mitochondria from the yeast Saccharomyces cerevisiae. Methods Cell Biol. 65:37–51. 10.1016/S0091-679X(01)65003-9 [DOI] [PubMed] [Google Scholar]
- Dunn C.D., Lee M.S., Spencer F.A., and Jensen R.E.. 2006. A genomewide screen for petite-negative yeast strains yields a new subunit of the i-AAA protease complex. Mol. Biol. Cell. 17:213–226. 10.1091/mbc.E05-06-0585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn C.D., Tamura Y., Sesaki H., and Jensen R.E.. 2008. Mgr3p and Mgr1p are adaptors for the mitochondrial i-AAA protease complex. Mol. Biol. Cell. 19:5387–5397. 10.1091/mbc.E08-01-0103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J.R., Mourier A., Yamada J., McCaffery J.M., and Nunnari J.. 2015. MICOS coordinates with respiratory complexes and lipids to establish mitochondrial inner membrane architecture. eLife. 4:e07739 10.7554/eLife.07739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes F., Tatsuta T., and Langer T.. 2012. Mitochondrial AAA proteases--towards a molecular understanding of membrane-bound proteolytic machines. Biochim. Biophys. Acta. 1823:49–55. 10.1016/j.bbamcr.2011.09.015 [DOI] [PubMed] [Google Scholar]
- Gibson D.G., Young L., Chuang R.Y., Venter J.C., Hutchison C.A. III, and Smith H.O.. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 6:343–345. 10.1038/nmeth.1318 [DOI] [PubMed] [Google Scholar]
- Graef M., Seewald G., and Langer T.. 2007. Substrate recognition by AAA+ ATPases: distinct substrate binding modes in ATP-dependent protease Yme1 of the mitochondrial intermembrane space. Mol. Cell. Biol. 27:2476–2485. 10.1128/MCB.01721-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray M.W. 2012. Mitochondrial evolution. Cold Spring Harb. Perspect. Biol. 4:a011403 10.1101/cshperspect.a011403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueldener U., Heinisch J., Koehler G.J., Voss D., and Hegemann J.H.. 2002. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 30:e23 10.1093/nar/30.6.e23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbowski M., and Youle R.J.. 2011. Regulating mitochondrial outer membrane proteins by ubiquitination and proteasomal degradation. Curr. Opin. Cell Biol. 23:476–482. 10.1016/j.ceb.2011.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiebler M., Keil P., Schneider H., van der Klei I.J., Pfanner N., and Neupert W.. 1993. The mitochondrial receptor complex: a central role of MOM22 in mediating preprotein transfer from receptors to the general insertion pore. Cell. 74:483–492. 10.1016/0092-8674(93)80050-O [DOI] [PubMed] [Google Scholar]
- Knop M., Finger A., Braun T., Hellmuth K., and Wolf D.H.. 1996. Der1, a novel protein specifically required for endoplasmic reticulum degradation in yeast. EMBO J. 15:753–763. [PMC free article] [PubMed] [Google Scholar]
- Lithgow T., Junne T., Suda K., Gratzer S., and Schatz G.. 1994. The mitochondrial outer membrane protein Mas22p is essential for protein import and viability of yeast. Proc. Natl. Acad. Sci. USA. 91:11973–11977. 10.1073/pnas.91.25.11973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livnat-Levanon N., and Glickman M.H.. 2011. Ubiquitin-proteasome system and mitochondria - reciprocity. Biochim. Biophys. Acta. 1809:80–87. 10.1016/j.bbagrm.2010.07.005 [DOI] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A. III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., and Pringle J.R.. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961. [DOI] [PubMed] [Google Scholar]
- Moczko M., Bömer U., Kübrich M., Zufall N., Hönlinger A., and Pfanner N.. 1997. The intermembrane space domain of mitochondrial Tom22 functions as a trans binding site for preproteins with N-terminal targeting sequences. Mol. Cell. Biol. 17:6574–6584. 10.1128/MCB.17.11.6574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okreglak V., and Walter P.. 2014. The conserved AAA-ATPase Msp1 confers organelle specificity to tail-anchored proteins. Proc. Natl. Acad. Sci. USA. 111:8019–8024. 10.1073/pnas.1405755111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plath K., Mothes W., Wilkinson B.M., Stirling C.J., and Rapoport T.A.. 1998. Signal sequence recognition in posttranslational protein transport across the yeast ER membrane. Cell. 94:795–807. 10.1016/S0092-8674(00)81738-9 [DOI] [PubMed] [Google Scholar]
- Rainey R.N., Glavin J.D., Chen H.W., French S.W., Teitell M.A., and Koehler C.M.. 2006. A new function in translocation for the mitochondrial i-AAA protease Yme1: import of polynucleotide phosphorylase into the intermembrane space. Mol. Cell. Biol. 26:8488–8497. 10.1128/MCB.01006-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley J., Butler R., Ogilvie D., Finniear R., Jenner D., Powell S., Anand R., Smith J.C., and Markham A.F.. 1990. A novel, rapid method for the isolation of terminal sequences from yeast artificial chromosome (YAC) clones. Nucleic Acids Res. 18:2887–2890. 10.1093/nar/18.10.2887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauerwald J., Jores T., Eisenberg-Bord M., Chuartzman S.G., Schuldiner M., and Rapaport D.. 2015. Genome-Wide Screens in Saccharomyces cerevisiae Highlight a Role for Cardiolipin in Biogenesis of Mitochondrial Outer Membrane Multispan Proteins. Mol. Cell. Biol. 35:3200–3211. 10.1128/MCB.00107-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiota T., Mabuchi H., Tanaka-Yamano S., Yamano K., and Endo T.. 2011. In vivo protein-interaction mapping of a mitochondrial translocator protein Tom22 at work. Proc. Natl. Acad. Sci. USA. 108:15179–15183. 10.1073/pnas.1105921108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiota T., Nishikawa S., and Endo T.. 2013. Analyses of protein-protein interactions by in vivo photocrosslinking in budding yeast. Methods Mol. Biol. 1033:207–217. 10.1007/978-1-62703-487-6_14 [DOI] [PubMed] [Google Scholar]
- Singh S.K., Grimaud R., Hoskins J.R., Wickner S., and Maurizi M.R.. 2000. Unfolding and internalization of proteins by the ATP-dependent proteases ClpXP and ClpAP. Proc. Natl. Acad. Sci. USA. 97:8898–8903. 10.1073/pnas.97.16.8898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Tamura Y., Yoshihisa T., and Endo T.. 2014. A novel import route for an N-anchor mitochondrial outer membrane protein aided by the TIM23 complex. EMBO Rep. 15:670–677. 10.1002/embr.201338142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsuta T., Augustin S., Nolden M., Friedrichs B., and Langer T.. 2007. m-AAA protease-driven membrane dislocation allows intramembrane cleavage by rhomboid in mitochondria. EMBO J. 26:325–335. 10.1038/sj.emboj.7601514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor E.B., and Rutter J.. 2011. Mitochondrial quality control by the ubiquitin-proteasome system. Biochem. Soc. Trans. 39:1509–1513. 10.1042/BST0391509 [DOI] [PubMed] [Google Scholar]
- van der Laan M., Horvath S.E., and Pfanner N.. 2016. Mitochondrial contact site and cristae organizing system. Curr. Opin. Cell Biol. 41:33–42. 10.1016/j.ceb.2016.03.013 [DOI] [PubMed] [Google Scholar]
- Van Melderen L., and Gottesman S.. 1999. Substrate sequestration by a proteolytically inactive Lon mutant. Proc. Natl. Acad. Sci. USA. 96:6064–6071. 10.1073/pnas.96.11.6064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenz L.S., Opaliński L., Schuler M.H., Ellenrieder L., Ieva R., Böttinger L., Qiu J., van der Laan M., Wiedemann N., Guiard B., et al. . 2014. The presequence pathway is involved in protein sorting to the mitochondrial outer membrane. EMBO Rep. 15:678–685. 10.1002/embr.201338144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Li L., and Jiang H.. 2016. Doa1 targets ubiquitinated substrates for mitochondria-associated degradation. J. Cell Biol. 213:49–63. 10.1083/jcb.201510098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., and Ye Y.. 2016. Doa1 is a MAD adaptor for Cdc48. J. Cell Biol. 213:7–9. 10.1083/jcb.201603078 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.