During crinophagy, secretory granules directly fuse with lysosomes to degrade their contents. Csizmadia et al. show that excess glue granules in Drosophila salivary glands are degraded by crinophagy at the onset of metamorphosis. Glue granule–lysosome fusion requires the HOPS tether, Rab2, Rab7, and a SNARE complex consisting of Syntaxin 13, Snap29, and Vamp7.
Abstract
At the onset of metamorphosis, Drosophila salivary gland cells undergo a burst of glue granule secretion to attach the forming pupa to a solid surface. Here, we show that excess granules evading exocytosis are degraded via direct fusion with lysosomes, a secretory granule-specific autophagic process known as crinophagy. We find that the tethering complex HOPS (homotypic fusion and protein sorting); the small GTPases Rab2, Rab7, and its effector, PLEKHM1; and a SNAP receptor complex consisting of Syntaxin 13, Snap29, and Vamp7 are all required for the fusion of secretory granules with lysosomes. Proper glue degradation within lysosomes also requires the Uvrag-containing Vps34 lipid kinase complex and the v-ATPase proton pump, whereas Atg genes involved in macroautophagy are dispensable for crinophagy. Our work establishes the molecular mechanism of developmentally programmed crinophagy in Drosophila and paves the way for analyzing this process in metazoans.
Introduction
Autophagy refers to several lysosome-mediated self-degradation pathways that occur within eukaryotic cells. Four main types can be distinguished based on how material reaches lysosomes: macroautophagy, microautophagy, chaperone-mediated autophagy, and crinophagy (Marzella et al., 1981; Weckman et al., 2014). In all cases, degradation is performed by lysosomal hydrolases that are active at an acidic pH, which is achieved by the action of the vacuolar/lysosomal proton pump v-ATPase.
Macroautophagy is the best known among these pathways. It starts with the formation of a phagophore cistern, which sequesters various components of the cytoplasm into double-membrane autophagosomes that deliver their cargo to lysosomal degradation (Feng et al., 2014). Autophagosome formation is achieved by the coordinated action of evolutionarily conserved Atg proteins, which form distinct protein complexes. The fusion of autophagosomes with endosomes and lysosomes requires SNAREs, including the autophagosomal Syntaxin 17 and its binding partners, Snap29 and Vamp8 (Vamp7 in Drosophila), together with the small GTPases Rab2, Rab7, and its effectors, PLEKHM1 and the tethering complex HOPS (homotypic fusion and protein sorting; Kimura et al., 2007; Itakura et al., 2012; Takáts et al., 2013, 2014; Jiang et al., 2014; McEwan et al., 2015; Hegedűs et al., 2016; Fujita et al., 2017; Lőrincz et al., 2017).
During microautophagy, part of the lysosomal membrane invaginates and engulfs a portion of the surrounding cytoplasm, which is then degraded within the lysosome (Mijaljica et al., 2011).
The third pathway is chaperone-mediated autophagy, during which the cytoplasmic chaperone Hsc70 recognizes an exposed KFERQ-like amino acid sequence in proteins to be degraded and transports these into the lysosome through a channel formed by the lysosomal membrane protein Lamp2A (Cuervo and Wong, 2014).
During crinophagy, secretory granules directly fuse with lysosomes to ensure the fast breakdown of excess or obsolete secretory material (Weckman et al., 2014). Crinophagy was discovered 50 yr ago by electron microscopical examination of anterior pituitary gland cells in rats (Smith and Farquhar, 1966). This paper provided ultrastructural evidence that the remaining secretory granules fused with lysosomes when secretory activity was suppressed. Similar tightly regulated secretory granule catabolic processes are part of the normal physiology and development of exocrine, endocrine, and neuroendocrine cells to control the secretory granule pool (Weckman et al., 2014). However, the molecular mechanism of crinophagy is unknown.
The salivary gland of fruit flies changes its function during postembryonic development. The gland produces saliva to help food ingestion and intestinal digestion throughout the larval stages. Later on, ∼14 h before puparium formation (−14 h relative to puparium formation [RPF]), the gland starts to synthesize glue proteins. Glue granules are released through a burst of secretory activity at the onset of metamorphosis to attach the pupa to a solid surface. After this wave of glue secretion is over, the remaining cytoplasmic granules may be degraded by crinophagy according to an early ultrastructural study of Drosophila pseudoobscura (Harrod and Kastritsis, 1972). In this work, we characterize crinophagy in the popular animal model Drosophila melanogaster and identify the gene products that are required for developmentally programmed glue granule degradation in salivary gland cells.
Results
Glue granules are degraded via crinophagy in Drosophila salivary gland cells at the onset of metamorphosis
To study glue granule degradation in Drosophila late larval and early prepupal salivary gland cells, we established fly stocks that allow the monitoring of this process by fluorescent microscopy. The first stock expresses two previously described glue granule reporters combined (Glue-Red and Glue-GFP), which are both attached to the glue granule protein Sgs3 expressed by the sgs3 promoter (Biyasheva et al., 2001; Costantino et al., 2008). If glue granules fuse with lysosomes, the fluorescence of GFP is quenched in the acidic, degradative milieu. As DsRed is less sensitive to the low pH of lysosomes, granules undergoing lysosomal degradation lose GFP signal but retain DsRed fluorescence. This “GlueFlux” reporter system thus allows the monitoring of glue granule acidification and degradation, similar to the GFP-RFP-Atg8a autophagic flux reporter that is commonly used to follow the lysosomal degradation of autophagosomes (Kimura et al., 2007; Nezis et al., 2010; Nagy et al., 2015).
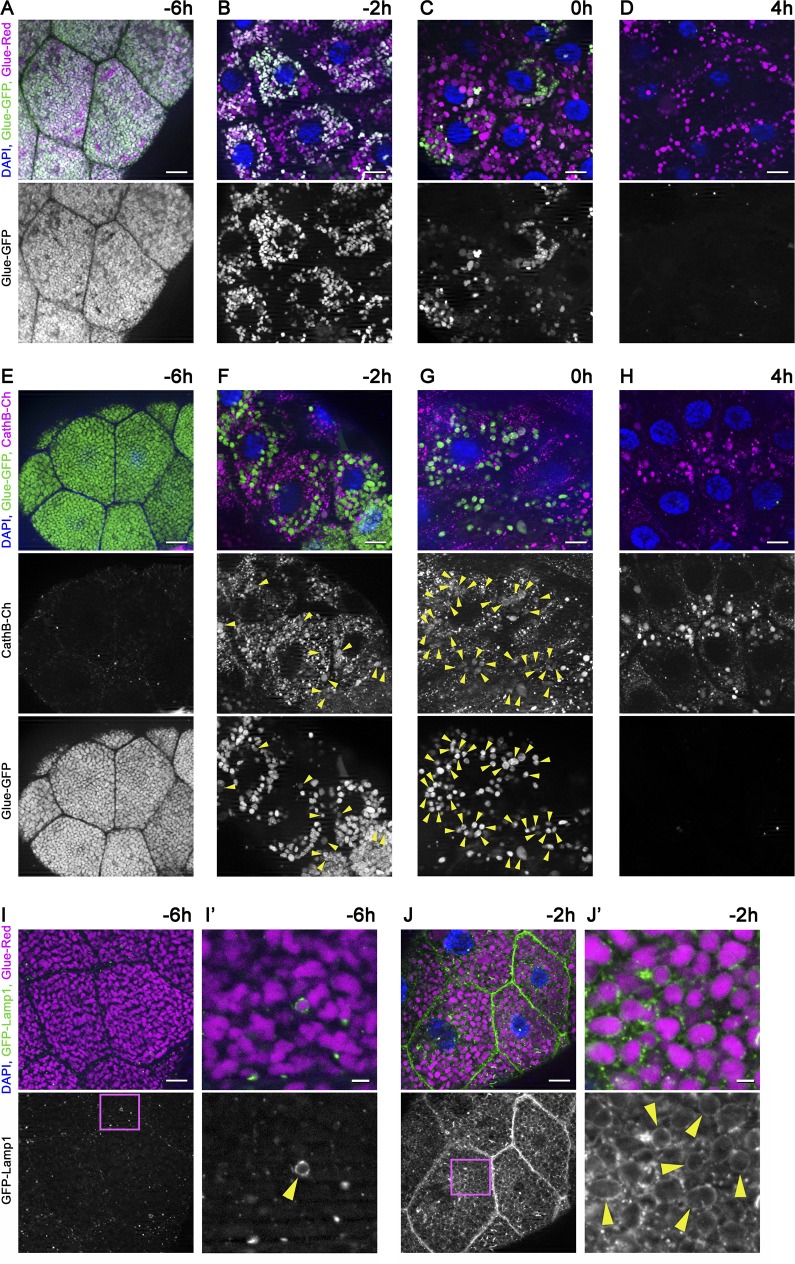
Glue granule biogenesis starts ∼14 h before puparium formation (−14 h RPF; Beckendorf and Kafatos, 1976; Biyasheva et al., 2001; Burgess et al., 2011). The first signs of glue granule degradation were observed as early as in late L3 wandering stage (−6 h RPF), based on the appearance of glue granules that are only positive for DsRed (Figs. 1 A and S1 A). Most of the glue granules remain positive for both DsRed and GFP at this stage, indicating that the majority of these vesicles are intact. The ratio of degrading glue granules readily increased during the next few hours of development, culminating in the complete disappearance of intact granules by 4 h RPF (Fig. 1, B–D; and Fig. S1 A). Acidification of the lysosomal lumen is required for autophagic degradation (Nakamura et al., 1997; Mauvezin et al., 2015). To confirm that the loss of GFP signal is caused by acidification of glue granules, we knocked down VhaSFD in the salivary gland cells, which encodes an essential subunit of the v-ATPase proton pump. As a result, most glue granules remained positive for both GFP and DsRed at the white prepupal stage (0 h RPF; Fig. S1 B), unlike salivary glands in wild-type animals, where only one third of the granules were intact at this stage (Fig. 1 C). Staining glands with Lysotracker red, a dye commonly used for acidic lysosomes, confirmed the acidification defect of glue granules in v-ATPase loss-of-function cells (Fig. S1, C–E).
Figure 1.
Time course of developmentally programmed crinophagy in Drosophila salivary gland cells. (A–D) Glue granule degradation in the salivary gland of animals coexpressing Glue-GFP and Glue-Red reporters (GlueFlux). (A) Wandering L3 stage (−6 h RPF) larval cells contain mostly intact (GFP and DsRed double-positive) secretory granules, and very few degrading glue granules (positive for DsRed only) are seen. (B–D) The number of intact (double positive) glue granules gradually decreases in −2-h (B), 0-h (C), and 4-h (D) old animals, in parallel with the increasing number of DsRed-only crinosomes. (E–H) Glue granules acquire lysosomal Cathepsin B (CathB) 3xmCherry. (E) No colocalization is observed between Glue-GFP granules and CathB at −6 h. (F) CathB structures greatly increase in size and number, and many overlap with Glue-GFP granules (arrowheads) at −2 h. Note that GFP fluorescence often decreases in overlapping structures. (G) The majority of Glue-GFP granules are positive for CathB at 0 h. (H) GFP fluorescence is lost in most granules at 4 h, and the size and number of CathB structures resembles that of Glue-Red in D. (I and J) Glue granule fusion with late endosomes and lysosomes. (I) GFP-Lamp1 localizes to small punctate structures in wandering-stage larvae and very rarely forms rings around Glue-Red granules. (J) GFP-Lamp1 forms prominent rings around Glue-Red granules at −2 h, likely as a result of secretory granule fusion with late endosomes and lysosomes. The boxed regions in I and J are shown enlarged in I′ and J′, with arrowheads pointing to GFP-Lamp1 rings. Green and/or magenta channels of merged images are also shown separately as indicated. Bars: (A–J) 20 µm; (I′ and J′) 3 µm. Please see Fig. S1 (A, F, and G) for quantification of data in A–D, E–H, I, and J, respectively.
We generated another reporter line for crinophagy in which Glue-GFP expression is combined with a genomic promoter-driven Cathepsin B (CathB) 3xmCherry transgene expressing a fluorescently tagged lysosomal hydrolase. Only a few small CathB dots were detected at −6 h RPF, and they rarely overlapped with Glue-GFP granules (Figs. 1 E and S1 F). CathB expression strongly increased by −2 h, and structures positive for both CathB and Glue-GFP appeared (Figs. 1 F and S1 F). GFP fluorescence in double-positive granules was often fainter than in intact granules, indicating that the quenching of Glue-GFP starts soon after fusion with lysosomes. Granules positive for both Glue-GFP and CathB-3xmCherry were seen in large numbers at the 0 h stage (Figs. 1 G and S1 F). Finally, GFP signal practically disappeared, and the distribution of CathB-3xmCherry resembled that of Glue-Red at 4 h RPF (Figs. 1 H and S1 F).
Our third crinophagy reporter line consisted of the Glue-Red reporter and a GFP marker linked to a transmembrane protein fragment targeted to late endosomes and lysosomes (GFP-Lamp1), in which GFP faces the luminal side and is rapidly quenched in lysosomes (Pulipparacharuvil et al., 2005). During crinophagy, secretory granules directly fuse with lysosomes to give rise to a new degrading organelle called the crinosome (Ahlberg et al., 1987). In −6 h RPF larvae, although GFP-Lamp1–positive dots did not overlap with glue granules, rarely, a ring was seen surrounding Glue-Red granules (Figs. 1 I and S1 G), indicating that fusions occurred. By −2 h RPF, the majority of glue granules acquired GFP-Lamp1 in the crinosomal membrane (Figs. 1 J and S1 G), as expected based on the other two reporter systems.
An Uvrag-containing Vps34 kinase complex is necessary for proper glue granule degradation
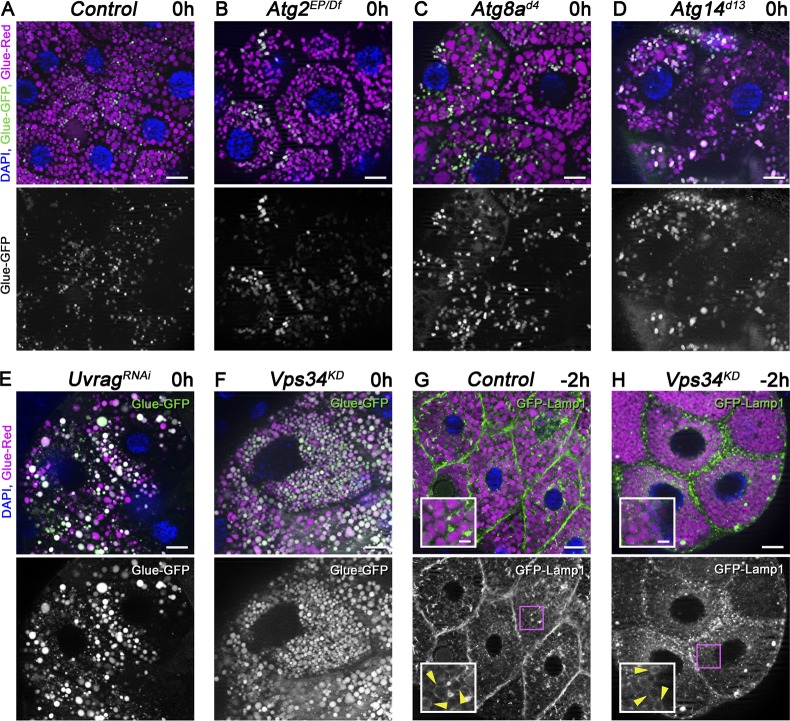
Atg genes are required for the capture of insulin-containing secretory granules into autophagosomes, but their role in crinophagy has not been investigated (Riahi et al., 2016). Interestingly, Atg1, Atg2, Atg3, Atg7, Atg8a, Atg9, Atg14 (the specific subunit of the autophagic Vps34 lipid kinase complex), Atg16, and Atg18a all turned out to be dispensable for secretory granule degradation based on the GlueFlux reporter when compared with controls (Fig. 2, A–D; Fig. S2, A–H; and Fig. S3 A). In contrast, both Vps34 and Uvrag (the specific subunit of the endocytic Vps34 complex) loss of function impaired crinophagy; the majority of granules remained intact in white prepupal gland cells expressing Uvrag RNAi or dominant-negative, kinase-dead Vps34 (Fig. 2, E and F; and Fig. S3 A). Interestingly, GFP-Lamp1 still formed rings around glue granules at −2 h, similar to control cells, and GFP fluorescence was retained within Glue-Red–positive granules in Vps34 loss-of-function cells at 1 h RPF, unlike in control cells (Fig. 2, G and H; Fig. S2, I and J; and Fig. S3 B), indicating that granules fuse with lysosomes, but the subsequent degradation of cargo is impaired in these cells. This is in line with previous studies by others and us showing that the Uvrag–Vps34 complex promotes trafficking of lysosomal enzymes and membrane proteins (Brown et al., 1995; Zeng et al., 2006; Juhász et al., 2008; Lőrincz et al., 2014; Takáts et al., 2014).
Figure 2.
The Uvrag-containing Vps34 kinase complex promotes degradation of glue granules. (A–F) The core macroautophagy proteins Atg2, Atg8a, and Atg14 are dispensable for glue granule degradation, unlike Uvrag and Vps34. Glue-GFP signal is lost in salivary gland cells of white prepupae mutant for Atg2 (B), Atg8a (C), or Atg14 (D), similar to controls (A). In contrast, gland-specific knockdown of Uvrag (E) or expression of dominant-negative, kinase-dead Vps34 (F) prevents the quenching of GFP fluorescence. (G and H) Inhibition of Vps34 does not prevent the formation of GFP-Lamp1 rings (arrowheads) around crinosomes (H) compared with controls (G). The boxed regions of G and H are enlarged in insets, respectively. Green channels are shown separately for A–H. Bars: (A–H) 20 µm; (G and H, insets) 5 µm. Please see Fig. S3 (A and B) for quantification of data.
HOPS tethering complex subunits are required for glue granule degradation in Drosophila
Because the molecular mechanism of glue granule degradation was unknown, we tested candidate genes for involvement in crinophagy. Membrane fusion usually requires tethers, small GTPases, and SNARE proteins. We started our functional analysis with the HOPS tethering complex, because we and others have recently identified it as essential for autophagosome clearance in mammalian and Drosophila cells (Jiang et al., 2014; Takáts et al., 2014).
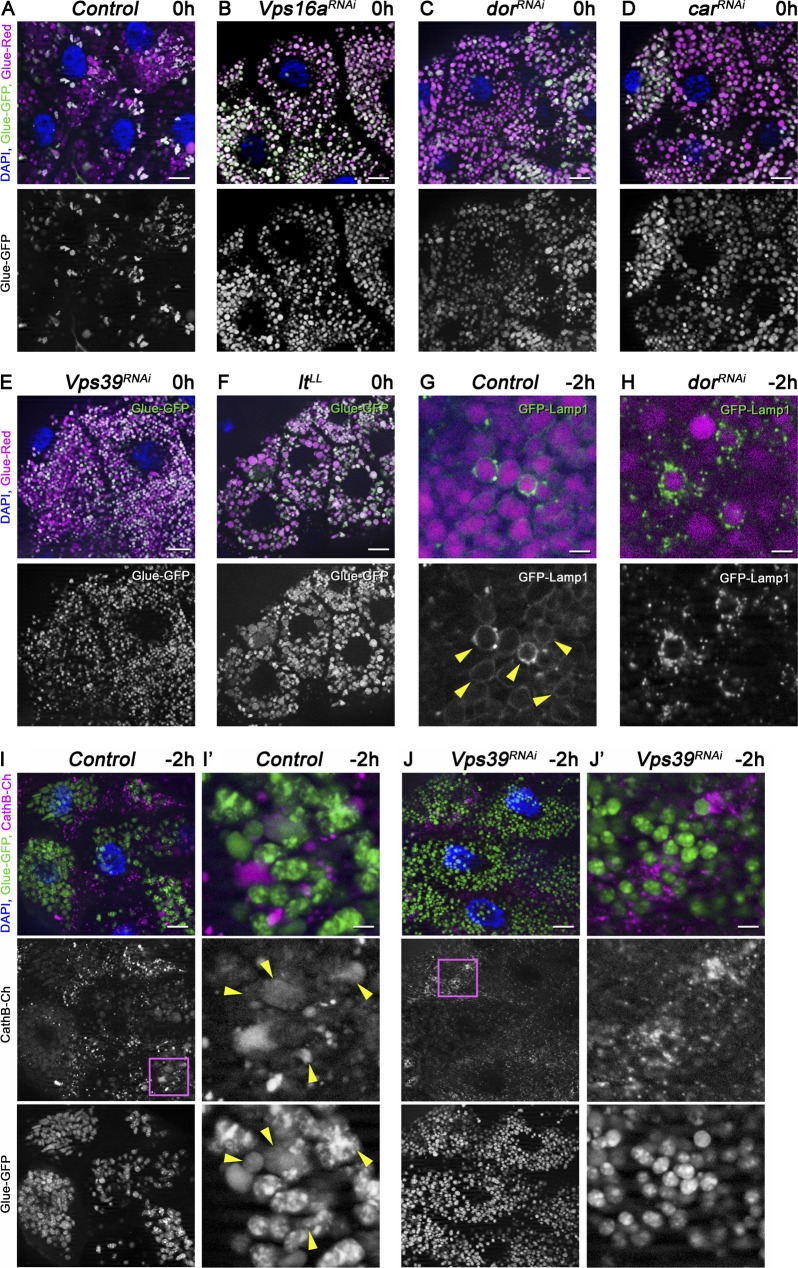
In wild-type control animals, only one third of the glue granules retain GFP fluorescence at the white prepupal stage (Fig. 3 A), indicating ongoing glue degradation. In contrast, most glue granules remain intact at the same developmental stage in the absence of HOPS subunits, that is, in animals with salivary gland–specific RNAi knockdown of Vps16a, dor/Vps18, car/Vps33a, and Vps39, similar to animals mutant for light/Vps41 (Fig. 3, B–F; and Fig. S3 A). Because Vps39 and light are HOPS-specific subunits in Drosophila and are not found in the related tethering complex CORVET (class C core vacuole/endosome tethering; Lőrincz et al., 2016), these data indicate that HOPS is required for crinophagy.
Figure 3.
The HOPS tethering complex is required for glue granule degradation. (A–F) Glue granule degradation in white prepupal (0 h) salivary glands. (A) The majority of glue granules lack GFP signal in control cells. (B–F) Loss of HOPS function leads to persisting GFP signal at the same developmental stage upon salivary gland-specific knockdown of Vps16a (B), dor/Vps18 (C), car/Vps33a (D), Vps39 (E), and in lt/Vps41 mutant animals (F). (G and H) Glue granule fusion with late endosomes and lysosomes at −2 h. (G) GFP-Lamp1 is seen as rings (arrowheads) around DsRed-positive glue granules, indicating ongoing crinophagy in control gland cells. (H) In contrast, no rings are seen and small GFP-Lamp1–positive lysosomes often accumulate near Glue-Red granules in dor/Vps18 RNAi cells because a block of fusion. (I and J) Presence of lysosomal cathepsin in glue granules at −2 h. Glue-GFP granules acquire lysosomal CathB in control cells (I; arrowheads in I′), unlike in Vps39 knockdown cells (J). The boxed regions of I and J are shown enlarged in I′ and J′, respectively. Green and/or magenta channels of merged images are also shown separately as indicated. Bars: (A–F, I, and J) 20 µm; (G and H) 3 µm; (I′ and J′) 5 µm. Please see Fig. S3 (A–C) for quantification of data.
We used the GFP-Lamp1 reporter to test whether the lack of HOPS indeed prevents the fusion of glue granules with lysosomes. GFP-Lamp1, which is normally seen as a ring around Glue-Red granules in −2 h glands, failed to form rings and often accumulated as small dots around granules upon salivary gland–specific knockdown of dor/Vps18 (Fig. 3, G and H; and Fig. S3 B). Similarly, CathB failed to reach Glue-GFP granules in Vps39 RNAi cells, unlike in controls (Fig. 3, I and J; and Fig. S3 C), in line with a block of fusion.
Ultrastructure of glue granule crinophagy
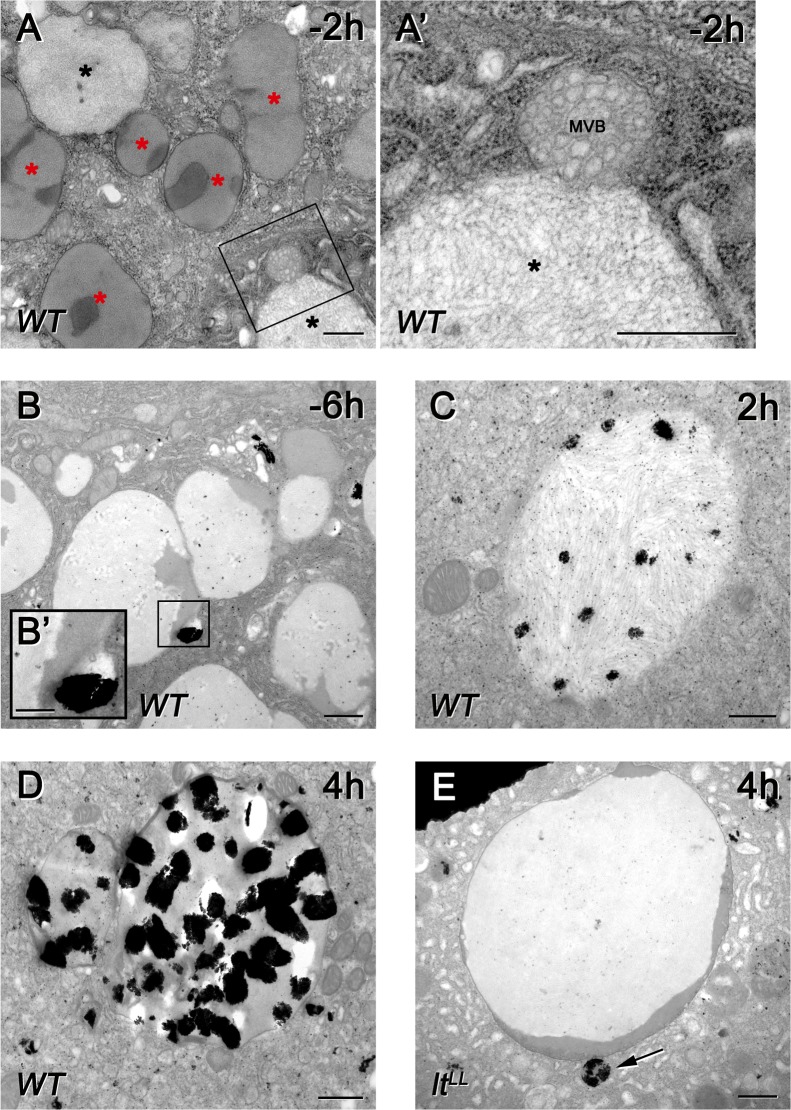
We sought to confirm our fluorescence microscopy data using ultrastructural analysis. Transmission EM of late wandering stage larval glands revealed lots of intact glue granules as well as granules with loose inner structure, which likely represent crinosomes (Fig. 4 A). In line with this, fusion of multivesicular endosomes with glue granules could be detected (Fig. 4, A and A′). The identity of glue granules (Glue-GFP positive), lysosomes (CathB positive), and crinosomes (double positive) were further confirmed by immuno-EM analysis (Fig. S4).
Figure 4.
Ultrastructure of crinophagy. (A) Both intact (red asterisks) and degrading, more translucent (black asterisks) glue granules are seen in salivary gland cells at −2h RPF. The boxed region of A is enlarged in A′, showing the fusion of a degrading glue granule (crinosome) with a late endosome (MVB, multivesicular body). (B–E) Acid phosphatase enzyme reaction (black precipitates) reveals the sites of lysosomal activity in gland cells. (B) Intact glue granules are separate from acid phosphatase positive lysosomes in wandering larvae (−6 h RPF), with the boxed region shown enlarged in B′ to highlight the ongoing fusion of a lysosome with a glue granule. (C) Black precipitate is observed within granules/crinosomes of 2-h prepupal cells, indicating crinophagy. (D) Crinosomes display very high acid phosphatase activity by 4 h RPF. (E) In contrast, intact glue granules lacking acid phosphatase activity are still seen at 4 h RPF in homozygous mutants of lt/Vps41 encoding a HOPS-specific subunit. The arrow points to an acid phosphatase positive lysosome next to the glue granule. Bars: (A–E and A′) 500 nm; (B′) 250 nm.
Salivary gland cells contain acid phosphatase–positive vesicles representing primary lysosomes (Harrod and Kastritsis, 1972). We could indeed capture the fusion of acid phosphatase positive lysosomes with glue granules (Fig. 4 B). The granules that evade secretion were becoming increasingly more positive for acid phosphatase in older animals (Fig. 4, C and D). In contrast, enlarged glue granules remained negative for acid phosphatase in animals lacking the HOPS subunit light/Vps41 even at 4 h RPF (Fig. 4 E). These results support our model that glue granules in salivary gland cells are degraded by crinophagy, which requires the HOPS-dependent direct fusion of secretory granules with late endosomes and lysosomes.
The small GTPases Rab2 and Rab7 and the SNAREs Syntaxin 13 (Syx13), Snap29, and Vamp7 are required for glue granule to lysosome fusion
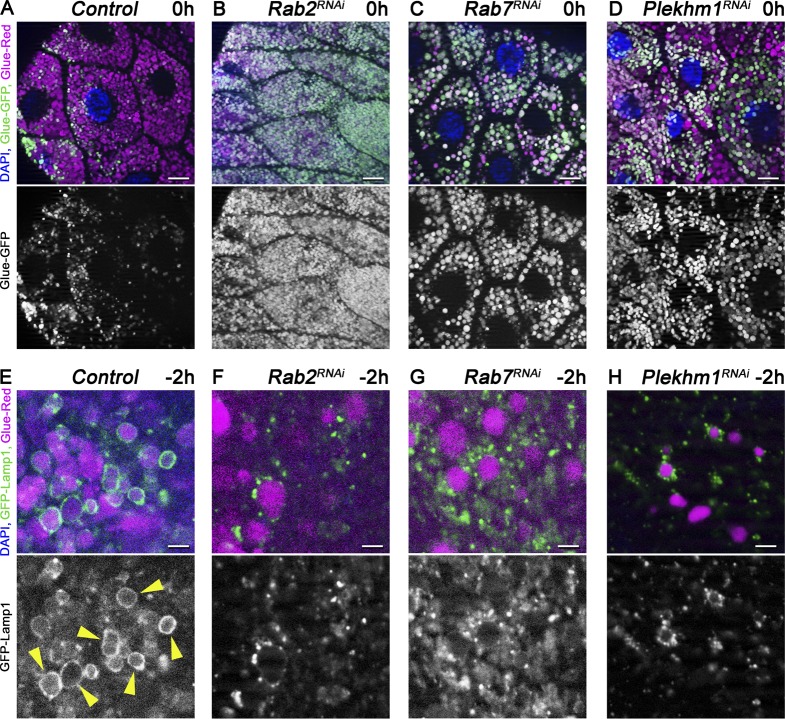
Small GTPases play critical roles in vesicular trafficking processes (Stenmark, 2009). We thus tested the small GTPases Rab2 and Rab7 and its binding partner, PLEKHM1, which are involved in autophagosome–lysosome fusion (McEwan et al., 2015; Hegedűs et al., 2016; Fujita et al., 2017; Lőrincz et al., 2017). Gland-specific knockdown of either Rab2 or Rab7 or Plekhm1 caused a glue granule degradation defect, as most granules remained intact at 0 h RPF, unlike in control cells (Fig. 5, A–D; and Fig. S3 A). Moreover, loss of Rab2, Rab7, or PLEKHM1 resulted in the accumulation of Lamp1-positive lysosomes near glue granules instead of ring formation, as in control cells (Fig. 5, E–H; and Fig. S3 B).
Figure 5.
Rab2, together with Rab7 and its binding partner, PLEKHM1, is required for glue granule–lysosome fusion. (A–D) Glue granule degradation in white prepupal (0 h) salivary glands. The number of intact glue granules retaining GFP signal greatly increases upon knockdown of Rab2 (B), Rab7 (C), and Plekhm1 (D) compared with control cells (A). (E and H) Glue granule fusion with late endosomes and lysosomes at −2 h. GFP-Lamp1 is seen as rings (arrowheads) around Glue-Red granules, indicating ongoing crinophagy in control cells (E). In contrast, no rings are seen and small GFP-Lamp1 dots often accumulate near Glue-Red granules in Rab2, Rab7, and Plekhm1 RNAi cells (F–H) because of a block of fusion. Green channels are shown separately for A–H. Bars: (A–D) 20 µm; (E–H) 3 µm. Please see Fig. S3 (A and B) for quantification of data.
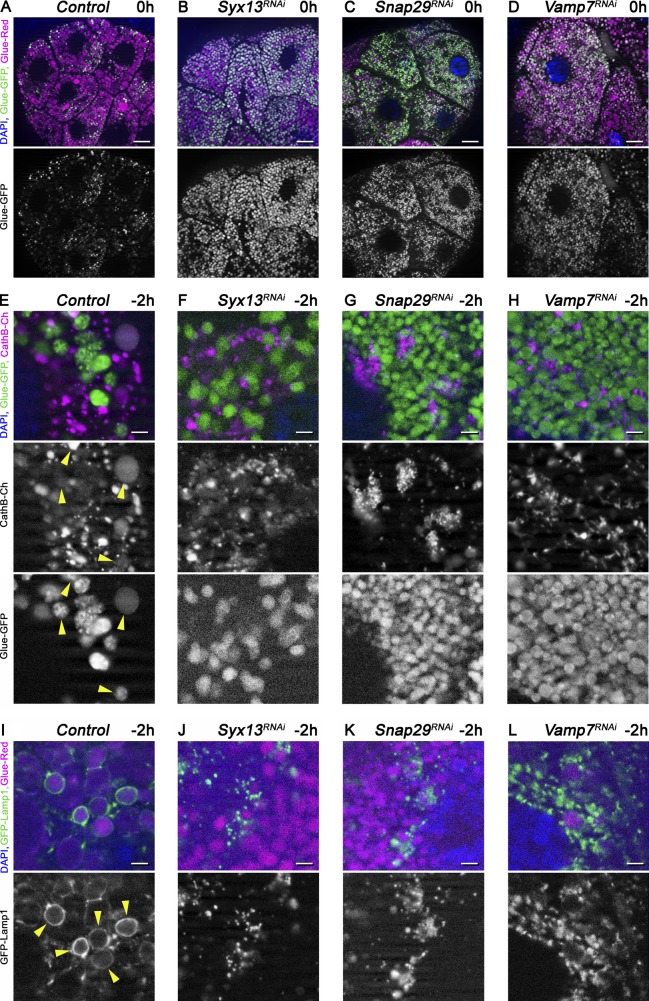
SNARE proteins mediate the majority of membrane fusions within cells, likely including glue granule fusion with lysosomes. We thus tested the role of the SNAREs involved in autophagosome–lysosome fusion (Itakura et al., 2012; Takáts et al., 2013). Syntaxin 17 turned out to be dispensable for glue granule degradation based on the GlueFlux reporter; the GFP signal was readily quenched in Syntaxin 17–null mutant animals, similar to controls (Fig. S5, A and B; and Fig. S3 A). In contrast, the GFP signal persisted in the granules in Snap29 and Vamp7 RNAi cells, respectively (Fig. 6, C and D; and Fig. S3 A). We next analyzed the Drosophila SNAREs that are similar to Syntaxin 17 (Qa SNAREs that have a glutamine in the zero ionic layer of the assembled complex; Table S1). Loss of Syx13 caused a glue granule degradation defect similar to Snap29 and Vamp7 compared with controls (Fig. 6, A and B; and Fig. S3 A). Furthermore, delivery of CathB to Glue-GFP granules was reduced (Fig. 6, E–H; and Fig. S3 C), and Lamp1-positive lysosomes accumulated near glue granules in Syx13, Snap29 and Vamp7 RNAi animals, unlike labeling crinosome membranes as in controls (Fig. 6, I–L; and Fig. S3 B).
Figure 6.
The SNAREs Syx13, Snap29 and Vamp7 are required for crinophagy. Loss of Syx13, Snap29, and Vamp7 results in a glue granule degradation defect (A–D). Salivary gland-specific knockdown of Syx13 (B), Snap29 (C), or Vamp7 (D) inhibits the quenching of GFP within glue granules compared with control cells (A) at 0 h RPF. Glue granules fail to properly acquire lysosomal CathB in the absence of these SNAREs (E–H). Numerous Glue-GFP granules overlap with CathB-3xmCherry in control cells (E, arrowheads), which decreases upon silencing of Syx13 (F), Snap29 (G), or Vamp7 (H) in −2-h gland cells. Impaired glue granule fusion with late endosomes and lysosomes in SNARE RNAi cells (I–L). Formation of GFP-Lamp1 rings (arrowheads) around Glue-Red granules that is seen in control cells (I) is impaired in cells undergoing Syx13 (J), Snap29 (K), or Vamp7 (L) RNAi, indicating a fusion defect. Green and/or magenta channels are shown separately as indicated. Bars: (A–D) 20 µm; (E–L) 3 µm. Please see Fig. S3 (A–C) for quantification of data.
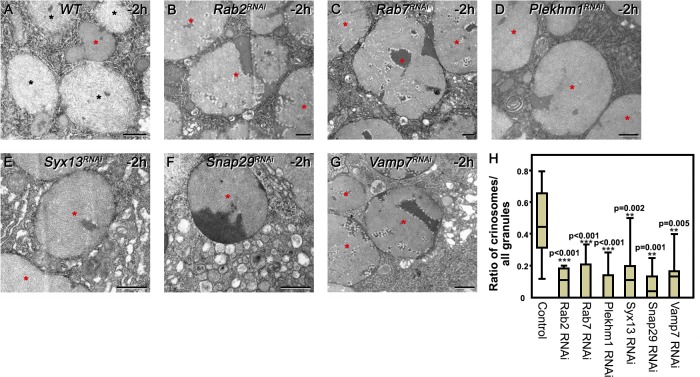
Indeed, most glue granules retained an intact ultrastructure in Rab2, Rab7, Plekhm1, Syx13, Snap29, and Vamp7 RNAi cells, unlike in controls (Fig. 7). Collectively, these findings suggest that Rab2, Rab7 (and its binding partner, PLEKHM1), and the SNAREs Syx13, Snap29, and Vamp7 are all required for glue granule–lysosome fusion.
Figure 7.
Intact glue granules persist in Rab2, Rab7, Plekhm1, Syx13, Snap29, and Vamp7 knockdown cells. Ultrastructural analysis of glue granules and crinosomes (A–H). Both intact glue granules with highly ordered contents and characteristic dark regions (red asterisk) and crinosomes containing loose filamentous contents (black asterisks) are obvious in wild-type gland cells at −2 h RPF (A). Loss of Rab2 (B), Rab7 (C), PLEKHM1 (D), Syx13 (E), Snap29 (F), or Vamp7 (G) impairs crinosome formation; mostly intact granules (red asterisks) are seen in these cells. (H) Quantification of data from A–G, n = 8–21 cells, Kruskal–Wallis tests with post-hoc U test. In the box plots, bars show the data ranging between the upper and lower quartiles, and median is indicated as a horizontal black line within the box. Whiskers plot the smallest and largest observations. Bars, 500 nm.
Syx13, Snap29, and Vamp7 form a SNARE complex and may associate with HOPS
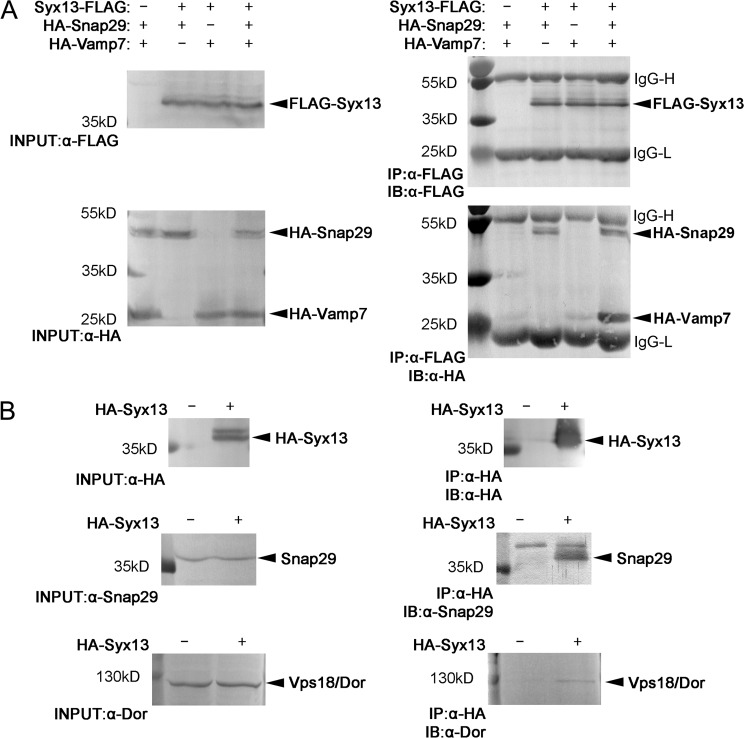
Our loss-of-function data raised the possibility that Syx13, Snap29, and Vamp7 function as part of the same SNARE complex. Syx13 is a Qa SNARE, Snap29 has two SNARE domains (Qb and Qc), and Vamp7 is an R SNARE (having an arginine in the zero ionic layer), which fits well with the rule of SNARE complex assembly. Indeed, FLAG-tagged Syx13 readily coprecipitated both HA-tagged Snap29 and Vamp7 in cultured Drosophila cells (Fig. 8 A). Interestingly, the amount of Vamp7 bound to beads dramatically increased when all three SNARE proteins were coexpressed, similar to what we have observed previously for the Syx17-containing autophagosomal SNARE complex (Takáts et al., 2013). This phenomenon suggested that Syx13 and Snap29 together bind more efficiently to Vamp7 than Syx13 does alone. We further tested protein interactions in animal lysates. HA-Syx13 coprecipitated both endogenous Snap29 and endogenous Dor/Vps18, a HOPS subunit (Fig. 8 B), raising the possibility that HOPS and these SNAREs act together during crinophagy.
Figure 8.
Syx13 forms a SNARE complex with Snap29 and Vamp7. (A) Coimmunoprecipitation experiments in cultured cells reveal that FLAG-Syx13 strongly binds to HA-Snap29 and weakly to HA-Vamp7. Note that the amount of Vamp7 that coprecipitates with Syx13 is dramatically increased by the presence of Snap29. (B) HA-Syx13 coprecipitates both endogenous Snap29 and endogenous Dor/Vps18 from animal lysates. H, heavy chain; IB, immunoblot; IP, immunoprecipitation; L, light chain.
Rab2, Rab7, Syx13, and Vamp7 localize to the membrane of crinosomes
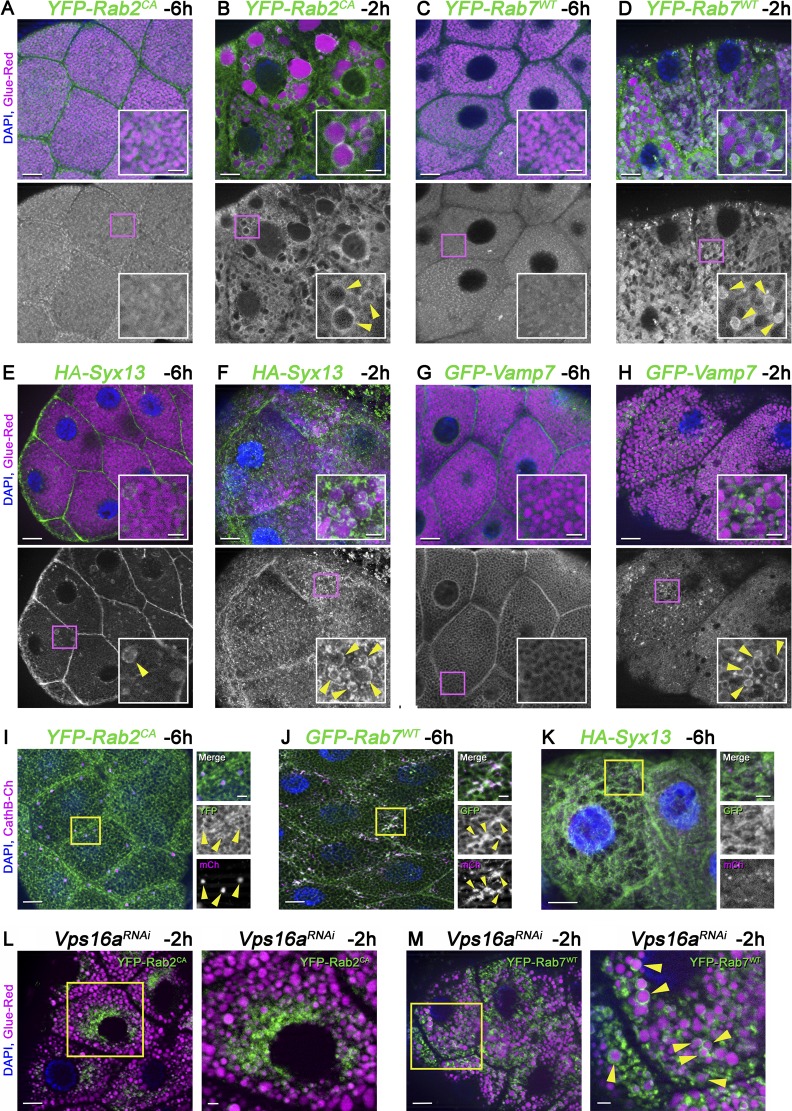
To further support our functional analyses, we determined the localization of Rab2, Rab7, and Syx13 in salivary gland cells. All of these proteins are membrane bound, because small GTPases of the Rab family have lipid anchors, and Syx13 and Vamp7 have a transmembrane domain, respectively. We could rarely detect the wild-type form of Rab2 in the limiting membrane of glue granules/crinosomes (Fig. S5, C and D), whereas the constitutively active (CA), Q65L mutant form that is thought to promote a GTP-locked state was clearly present in the membrane of crinosomes at −2 h RPF (Fig. 9 B). It is worth noting that expression of Rab2-CA resulted in a striking increase of crinosome size (Fig. 9 B), suggesting that it may alter the rate or degree of crinophagy, and perhaps even influence membrane identity. Rab2-CA exhibited a punctate localization and rarely surrounded Glue-Red granules in −6 h glands (Fig. 9 A). Wild-type Rab7 exhibited a localization pattern resembling that of Rab2-CA; it formed rings around crinosomes at −2 h RPF, but not at −6 h RPF (Fig. 9, C and D), similar to Vamp7 (Fig. 9, G and H). Interestingly, Syx13 appeared to form pronounced rings around a subset of glue granules already at −6 h RPF, which was observed more frequently at −2 h RPF (Fig. 9, E and F).
Figure 9.
The active forms of Rab2, wild-type Rab7, Syx13, and Vamp7 associate with the membrane of crinosomes. Rab2, Rab7, Syx13, and Vamp7 are present in the limiting membrane of crinosomes but absent from the membrane of the majority of intact glue granules (A–H). (A and B) The CA, supposedly GTP-locked form of Rab2 does not localize to Glue-Red granules at −6 h RPF (A), whereas it forms rings around enlarged crinosomes at −2 h (B, arrowheads in inset). (C and D) The wild-type form of Rab7 displays punctate localization distinct from Glue-Red granules at −6 h RPF (C), but it is present in the crinosome membrane at −2 h (D, arrowheads in inset). (E and F) HA-Syx13 already forms obvious rings around a subset of Glue-Red granules at −6 h RPF (E, arrowhead in inset), and it is detected in the limiting membrane of most granules at −2 h (F, arrowheads in inset). (G and H) GFP-Vamp7 signal fills the spaces between Glue-Red granules at −6 h (G), whereas it also forms striking rings around crinosomes at −2 h (H). Green channels are shown separately and boxed regions are enlarged in insets in A–H, respectively. (I–K) Lysosomal localization of selected factors at −6 h RPF. (I) Rab2-CA forms clear rings around CathB-positive lysosomes. (J) GFP-Rab7 signal overlaps with CathB-3xmCherry. (K) HA-Syx13 does not colocalize with CathB. Boxed regions are shown enlarged in I–K, respectively. (L and M) Rab2-CA and Rab7 localization in Vps16a RNAi cells at −2 h RPF. (L) Knockdown of Vps16a prevents the formation of large Glue-Red structures (compare with B) and causes a dramatic redistribution of the active form of Rab2; it is mostly seen in the perinuclear region instead of forming rings around Glue-Red granules. Note that Rab2-CA is present on a subset of small perinuclear Glue-Red structures, the identity of which is not known. (M) Loss of HOPS does not prevent Rab7 recruitment to the membrane of Glue-Red granules. Boxed regions in L and M are shown enlarged in L′ and M′, respectively, and arrowheads mark Rab7 rings around glue granules in M′. Bars: (A–M) 20 µm; (A–K, L′, and M′, insets) 3 µm.
Rab2 is known as a Golgi Rab, but its active form was recently shown to relocalize to lysosomes, where it directly binds to the HOPS subunit Vps39 to promote fusions (Gillingham et al., 2014; Lőrincz et al., 2017). Rab7 is also known to localize to late endosomes, lysosomes, and autophagosomes (Hegedűs et al., 2016). Rab2-CA clearly formed rings around CathB-positive lysosomes in −6 h gland cells, as expected (Fig. 9 I), and Rab7 also localized to lysosomes (Fig. 9 J). Finally, no overlap between Syx13 and CathB was detected at this stage (Fig. 9 K), raising the possibility that Syx13 is recruited to glue granules before fusion.
To further study the dynamics of protein localizations, we analyzed Rab2 and Rab7 distribution in Vps16a RNAi cells, where loss of HOPS prevents fusion. Interestingly, the distribution of Rab2-CA dramatically changed in HOPS knockdown cells; it exhibited a perinuclear localization and was absent from the membrane of most granules, whereas Rab7 was still recruited to granules on this genetic background (Fig. 9, L and M). These data suggest that Rab7 is recruited to glue granules to promote fusion with Rab2-positive lysosomes, similar to our model of Rab2 and Rab7 action during autophagosome–lysosome fusion (Lőrincz et al., 2017).
Crinophagy is dispensable for glue secretion
Because lysosomes are also secretory organelles in several cell types (Luzio et al., 2014), we tested whether the fusion of lysosomes with glue granules is necessary for their secretion. Glue-GFP signal was readily detected in the gland lumen in animals with salivary gland–specific loss of Vps39/HOPS, Rab2, or Snap29, unlike in EcR RNAi animals that failed to secrete glue (Fig. S5, E–I), because this event is triggered by ecdysone (Biyasheva et al., 2001; Rousso et al., 2016). Thus, crinophagy does not seem to be required for glue granule exocytosis.
Discussion
Our body contains numerous types of secretory cells, including exocrine, endocrine, and neuroendocrine cells, whose main function is to either continuously or temporarily produce, store, and exocytose secretory material into the extracellular space. The amount of the released material is tightly controlled according to the needs of the organism. Thus, usually only part of the secretory granule pool is released, and the remaining vesicles are often degraded via crinophagy (Marzella et al., 1981).
Developmentally programmed crinophagy likely plays important roles during postembryonal development in Drosophila. Salivary gland cells produce much more glue than it is necessary to fix the pupa to a solid surface at the onset of metamorphosis. This likely ensures a practically unlimited amount of glue to prevent the pupa from falling down and dying in a wet place. Another possible reason for excess glue production is the storage function of polyploid larval tissues including salivary gland cells. Unused glue that is produced by these cells is recycled via crinophagy, and the building blocks may be released during metamorphosis: either actively by these cells, or passively at the time of larval salivary gland regression.
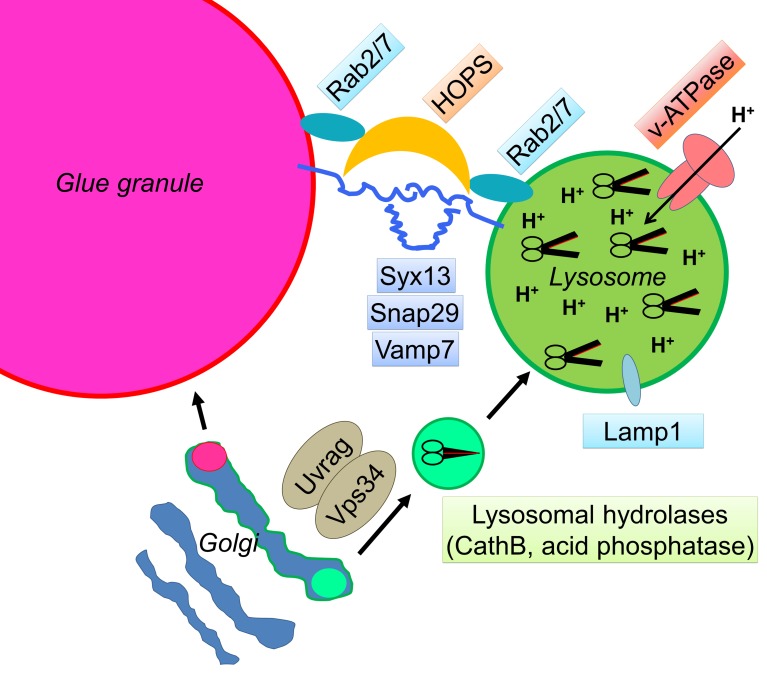
Surprisingly, the molecular mechanism of crinophagy is practically unknown. Here, we identified the SNAREs, tethering complexes, and small GTPases that are essential for programmed elimination of obsolete glue granules in Drosophila salivary gland cells (Fig. 10). This process requires the direct fusion of these secretory granules with lysosomes and late endosomes. Our data reveal that there is a significant overlap between the factors involved in autophagosome–lysosome fusion and crinophagy. Several known players functioning in autophagosome clearance, namely, the HOPS tethering complex (Jiang et al., 2014; Takáts et al., 2014), its direct binding partner, Rab2 (Gillingham et al., 2014; Fujita et al., 2017; Lőrincz et al., 2017), Rab7 (Kimura et al., 2007; Hegedűs et al., 2016), and its effector, PLEKHM1 (McEwan et al., 2015), are required for both processes. Similarly, the SNARE proteins Snap29 (Qbc) and Vamp7 (R) also function in both types of vesicle fusions, but they seem to form a functional complex with different Qa SNAREs: Syx17 in case of autophagosome–lysosome fusion (Itakura et al., 2012; Takáts et al., 2013) and Syx13 for glue granule–lysosome fusion, respectively. Thus, Qa SNAREs may specify which organelles can undergo fusion with lysosomes. Interestingly, Syx13 has been identified as a genetic modifier of frontotemporal dementia, possibly acting via promoting autophagic flux (Lu et al., 2013). In contrast, Syx17 appears to be specific for autophagosome–lysosome fusion, because its loss did not impair crinophagic glue granule degradation in our experiments.
Figure 10.
Model of crinophagy. During crinophagy, excess secretory glue granules fuse with lysosomes, which requires the tethering complex HOPS (and its binding partner, PLEKHM1 [not depicted]), the activity of two small GTPases (Rab2 and Rab7), and a SNARE complex composed of Syx13 (Qa), Snap29 (Qbc), and Vamp7 (R). The degradation of secretory material also depends on proper lysosomal functioning (i.e., adequate amounts of lysosomal enzymes and an acidic pH), which is supported by the activity of the Uvrag-containing Vps34 lipid kinase complex and the v-ATPase proton (H+) pump, respectively.
Based on our initial localization data, we hypothesize that first Syx13 is recruited to glue granules (because it was clearly present as rings surrounding a subset of granules at −6 h RPF; Fig. 9 E), and granules can also acquire Rab7 independent of fusion (because Rab7 still forms rings around most granules in the absence of HOPS; Fig. 9 M). Glue granules may then tether Rab2-positive vesicles, including lysosomes, by recruiting HOPS, which promotes SNARE assembly and fusion. Rab proteins most likely influence SNARE assembly indirectly (Grosshans et al., 2006), for example via HOPS that binds to both Rabs and SNAREs (Balderhaar and Ungermann, 2013; Takáts et al., 2014; Lőrincz et al., 2017).
Lysosomal degradation of secretory cargo also requires the Uvrag-containing Vps34 lipid kinase complex, a known regulator of trafficking to lysosomes (Brown et al., 1995; Juhász et al., 2008). Importantly, large-scale accumulation of glue granules has recently been reported in Vps34-null mutant salivary gland cells in 4 h RPF prepupae of Drosophila, although it was attributed to a secretory defect (Shravage et al., 2013). Autophagy is known to play a role in the noncanonical secretion of cytosolic proteins (Subramani and Malhotra, 2013), but glue granules are produced in the secretory pathway followed by their exocytosis, so it is not clear how autophagy would promote that process. Indeed, our results indicate that the persistence of glue in the absence of Vps34 kinase function is primarily caused by a failure of crinophagic degradation within lysosomes. This is different from the defective fusion of granules with lysosomes and late endosomes, which is seen upon loss of Rab2, Rab7, HOPS, PLEKHM1, Syx13, Snap29, and Vamp7. Interestingly, both Rab2 and HOPS are also known to promote the proper trafficking of lysosomal hydrolases, and these may function together in protein sorting at the Golgi in addition to lysosomal fusions, which is also supported by our recent identification of direct binding between Rab2 and the HOPS subunit Vps39 (Takáts et al., 2014; Lőrincz et al., 2017). Several of these factors are phosphoinositide effectors, including HOPS and Rab7 via its activator, the Ccz1–Mon1 complex (Stroupe et al., 2006; Cabrera et al., 2014; Hegedűs et al., 2016). It is thus possible that the product of Vps34, phosphatidylinositol 3-phosphate, also contributes to the regulation of glue granule–lysosome fusion, but loss of Vps34 may be compensated by other classes of phosphatidylinositol 3-kinases or other phosphoinositide species during this process, unlike in the case of lysosomal protein trafficking.
The mechanisms of membrane fusion are highly conserved among metazoans, suggesting that the mammalian orthologues of these Drosophila factors may play similar roles in crinophagy. This process is a major degradation route for insulin-containing secretory granules in β cells, and it is strongly up-regulated upon starvation (Orci et al., 1984; Uchizono et al., 2007; Goginashvili et al., 2015). The continuous synthesis, release, and degradation of these granules ensure a steady pool of vesicles. This balance of synthesis, exocytosis, and breakdown is necessary for β cell homeostasis. Degradation of insulin granules occurs via multiple routes: either by the simple and fast direct fusion of secretory granules with lysosomes (crinophagy) or by the capturing of these small granules into autophagosomes (macroautophagy) or lysosomes (microautophagy; Marsh et al., 2007; Weckman et al., 2014). It has been shown that Rab3A−/− β cells are defective in exocytosis, which leads to enhanced secretory granule degradation mainly through crinophagy (Marsh et al., 2007). Crinophagy thus likely plays an important role in controlling insulin secretion.
Crinophagy also occurs in the exocrine pancreas, but its relevance has not been functionally tested because of a lack of insight into its molecular mechanism. Interestingly, one of the major causes of pancreatitis is the premature, intracellular activation of normally secreted digestive hydrolase precursors, including trypsinogen, by lysosomal proteases within a hybrid organelle (Logsdon and Ji, 2013). This triggers the death of acinar cells, leading to inflammation and digestive enzyme insufficiency. Crinophagy is a possible route that may give rise to such hybrid organelles via direct fusion of secretory vesicles with lysosomes. Consistently, morphological analysis suggested that ethionine treatment induces necrosis of pancreatic acinar cells because of a blockade of exocytosis followed by premature intracellular activation of digestive hydrolase precursors at least in part via crinophagy (Koike et al., 1982).
Similarly, inhibition of exocytosis in liver cells by vinblastine treatment up-regulates crinophagy (Ahlberg et al., 1987). Isolation and subsequent biochemical characterization of crinosomes confirmed that these organelles contain both secretory material and active lysosomal hydrolases (Glaumann et al., 1989).
These descriptive studies of the liver and the exocrine and endocrine pancreas clearly indicate that the basal rate of crinophagy strongly increases in response to certain stresses. Activation of crinophagy likely protects the cells and the organism by degrading and recycling unnecessary secretory material. However, it may also be detrimental, for example, because of premature activation of intestinal enzymes within acinar cells of the exocrine pancreas.
Collectively, we demonstrate that HOPS, Rab2, Rab7, PLEKHM1, and the SNAREs Syx13, Snap29, and Vamp7 are required for glue protein degradation by promoting the fusion of Lamp1-positive lysosomes with glue granules, whereas the loss of either the v-ATPase proton pump or the UVRAG-containing Vps34 lipid kinase complex leads to a glue granule degradation defect downstream of fusion caused by lysosomal dysfunction (Fig. 10). Our study thus paves the way for functional analyses of crinophagy in metazoan cells.
Materials and methods
Fly stocks
The following fly stocks were obtained from the Bloomington Drosophila Stock Center: Sgs3 (Glue)-GFP (Biyasheva et al., 2001), da-Gal4, UAS-YFP-Rab2, UAS-YFP-Rab2Q65L, UAS-YFP-Rab7 (Lőrincz et al., 2017), Df(3L)BSC119, and RNAi stocks generated by the Transgenic RNAi Project (UAS-EcRJF02538, UAS-Vps16aHMS01716, UAS-Syx13JF01920, UAS-Snap29JF01883, UAS-Vamp7GL01524, UAS-UvragHMS01357, UAS-Plekhm1HMS23559, and UAS-Atg7HMS01358; Ni et al., 2009). RNAi lines obtained from the Vienna Drosophila Resource Center included UAS-VhaSFDGD8795 (Mauvezin et al., 2015), UAS-dorKK107053, UAS-carGD1397, UAS-Vps39GD12152 (Lőrincz et al., 2016), UAS-Rab2GD11158, UAS-Rab7GD11800 (Lőrincz et al., 2017), UAS-Atg1GD16133, UAS-Atg3KK101368, UAS-Atg9GD10045, UAS-Atg16GD25652, UAS-Atg17KK104864, and UAS-Atg18aKK105366 (Pircs et al., 2012). Additional fly lines included UAS-GFP-Lamp1 (Pulipparacharuvil et al., 2005), Sgs3-DsRed (Glue-Red, provided by A. Andres, University of Nevada, Las Vegas, NV; Costantino et al., 2008), fkh-Gal4 (provided by E. Baehrecke, University of Massachusetts Medical School, Worcester, MA; Berry and Baehrecke, 2007), UAS-Vps34KD (Juhász et al., 2008), UAS-HA-Syx13 (provided by F.-B. Gao, University of Massachusetts Medical School, Worcester, MA; Lu et al., 2013), and CathB-3xmCherry (this study; generated by cloning a genomic PCR fragment amplified with primers 5′-TATACTGAATTCCAGCTTGGGCAGACCCGCCGAAAT-3′ and 5′-GCATGCTGCATGCGTGCCTGTATCGCTTGGGCATC-3′ into our 3xmCherry vector [Hegedűs et al., 2016] and standard embryo injections). Mutant lines included ltLL07138 (Lőrincz et al., 2016), Atg2EP3697, Syx17LL06330 (Takáts et al., 2013), Atg8ad4 (Pircs et al., 2012), and Atg14d13 (Hegedűs et al., 2016).
Fluorescent microscopy
Salivary glands were dissected from control, mutant, and RNAi animals at the indicated developmental stages, fixed for 5 min in 4% paraformaldehyde in PBS, and covered with PBS/glycerin (9:1) containing DAPI. HA-Syx13 was detected essentially as described previously (Takáts et al., 2013). In brief, salivary glands were dissected in ice-cold PBS then fixed with 4% formaldehyde in PBTX (0.1% Triton X-100 in PBS for overnight at 4°C). Samples were extensively washed with PBTX (3 × 15 min at RT), and then incubated in blocking solution (5% FCS in PBTX for 30 min at RT). Samples were then incubated with rat anti-HA (clone 3F10; Roche) diluted 1:100 in the blocking solution overnight at 4°C. Salivary glands were then washed (3 × 15 min in PBTX at RT) and incubated in blocking solution again for 30 min at RT, followed by incubation with DyLight 488–conjugated goat anti–rat (SA5-10018; Thermo Fisher Scientific) diluted 1:600 in blocking solution for 3 h at RT. Washing steps were repeated, and samples were mounted with PBS/glycerol (9:1) containing DAPI. Images were taken at RT using an AxioImager Z1 epifluorescent microscope (Zeiss) equipped with an Apotome grid confocal unit and a HBO 100 mercury lamp, using AxioCam MRm camera and EC Plan-Neofluar 40× NA = 0.75 or EC Plan-Neofluar 10× NA = 0.3 air objectives (all Zeiss). Images of DAPI, GFP, YFP, DsRed, and Dylight 488 fluorescence were acquired in AxioVision SE64 Rel. 4.9.1 (Zeiss) and processed in Photoshop CS3 Extended (Adobe).
Transmission EM
Dissected salivary glands were fixed in 3.2% paraformaldehyde, 0.5% glutaraldehyde, 1% sucrose, and 0.028% CaCl2 in 0.1 N sodium cacodylate buffer, pH 7.4, for overnight at 4°C. Samples were postfixed in 0.5% osmium tetroxide for 1 h and in half-saturated aqueous uranyl acetate for 30 min, dehydrated in a graded series of ethanol, and embedded into Durcupan (Fluka) according to the manufacturer’s recommendations. 70-nm sections were stained in Reynold’s lead citrate and viewed on a transmission electron microscope (JEM-1011; JEOL) equipped with a digital camera (Morada; Olympus) using iTEM software (Olympus).
Acid phosphatase cytochemistry
Acid phosphatase reaction was performed essentially as described previously (Lőrincz et al., 2014). In brief, dissected salivary glands were fixed with 2% formaldehyde, 2% glutaraldehyde, 3 mM CaCl2, and 1% sucrose in 0.1 M sodium-cacodylate, pH 7.4, overnight. Subsequently, the buffer was replaced with 0.05 M sodium-acetate buffer (pH 5.0, three 5-min washes). Next, the samples were incubated in Gömöri’s medium (5 mM sodium-β-glycerophosphate and 4 mM lead nitrate) dissolved in acetate buffer for three 5-min incubations and processed as described for transmission EM. Ultrathin sections were analyzed unstained. Substrate-free medium was used for control experiments.
Immunogold labeling
Progressive lowering temperature embedding and subsequent immunolabeling were performed as previously described (Lőrincz et al., 2014). In brief, salivary glands from Glue-GFP, CathB-3xmCherry animals were dissected in PBS and fixed with 4% formaldehyde, 0.05% glutaraldehyde, and 0.2% tannic acid in phosphate buffer (PB; 0.1 M, pH 7.4) overnight at 4°C. Samples were then washed extensively with PB, and free aldehyde groups were quenched with 50 mM glycine and 50 mM NH4Cl in PB. Salivary glands were then postfixed in 1% uranyl acetate in 0.05 M maleate buffer (3 h at RT). Samples were then dehydrated in a graded series of ethanol as follows: 25% EtOH (10 min, 0°C), 50% EtOH (10 min, 0°C), 70% EtOH (10 min, −20°C), 96% EtOH (20 min, −20°C), and absolute EtOH (2 × 60 min, −20°C). Next, salivary glands were infiltrated with pure LR White (Sigma-Aldrich) containing 2% benzoyl peroxide as catalyst (24 h, −20°C). Curing was performed using a homemade UV chamber (equipped with two 2 × 6-W UV lamps) for 48 h at −20°C. Ultrathin sections (80–90 nm) were cut and collected on nickel grids. All immunoreactions were performed at RT. The following incubations were performed: (1) 5% H2O2 for 1 min; (2) bi-distilled water for 3 × 5 min; (3) 0.1% NaBH4 in TBS, pH 7.4, for 10 min; (4) 50 mM glycine in TBS for 30 min; (5) TBS for 3 × 5 min; (6) 10% FCS, 5% skimmed milk powder, and 1% BSA in TBS for 30 min; (7) primary antibodies diluted in 5% FCS, 2.5% skimmed milk powder, and 1% BSA in TBS overnight at 4°C; (8) 2% FCS, 1.25% skimmed milk powder, and 1% BSA in TBS for 3 × 5 min; (9) secondary antibodies in 2% FCS, 1.25% skimmed milk powder, and 1% BSA in TBS for 90 min; (10) 3 × 5 min TBS; (11) 1% glutaraldehyde in TBS for 10 min; and (12) extensive wash with bi-distilled water. Ultrathin sections were then stained with uranyl acetate. The following primary and secondary antibodies were used: rat anti-mCherry (1:50; Takáts et al., 2013), chicken anti-GFP (1:50; A10262; Life technologies), 18 nm gold-conjugated goat anti–rat (1:50; 112-215-167; Jackson ImmunoResearch Laboratories), and 10 nm gold-conjugated rabbit anti–chicken (1:50; EM.RCHL10; BBI Solutions). Imaging was done as for transmission EM analysis.
Statistics
Fluorescence structures from original, unmodified single focal planes were quantified manually. Three to five cells were randomly selected for counting from pictures of control, RNAi, or mutant salivary glands from three to seven animals. In CathB-3xmCherry Glue-GFP experiments, both GFP- and GFP-negative CathB structures >1 µm in diameter were counted as crinosomes, because especially in the later stages the GFP signal was quenched in most crinosomes. In GFP-Lamp1 experiments, GFP-Lamp1 rings around the Glue-Red granules were counted as double positive. For EM analyses, 8–21 cells from two different animals per genotype were evaluated manually.
We used SPSS17 (IBM) for data analysis. Mann–Whitney U tests were used for comparing two samples, and Kruskal–Wallis tests with post-hoc U test were used for comparing multiple samples, as in all cases, at least one variable showed non-Gaussian data distribution (determined by Kolmogorov–Smirnov test of normality). In the box plots, bars show the data ranging between the upper and lower quartiles; median is indicated as a horizontal black line within the box. Whiskers plot the smallest and largest observations. p-values for the relevant comparisons are shown in the panels.
Cell culture, immunoprecipitation, and Western blots
Syx13 (amino acids 1–259) was amplified from the EST LD27581 using the primers 5′-ATGTCCAAGGCCTTGAACAATCCC-3′ and 5′-GATCTTGCGCCGGTAGCTCT-3′ and cloned into pUAST-3xFLAG vector. Transfections and immunoprecipitations were performed in D.mel-2 cells using the plasmids pMT-Gal4, UAS-HA-Vamp7, and UAS-HA-Snap29 (Takáts et al., 2013). For in vivo experiments, UAS-HA-Syx13 was expressed systematically using da-Gal4. 100-mg animals of mixed life stages were collected in 1 ml lysis buffer containing 1% Triton X-100, and homogenized using an Ultra-Turret T10 (IKA) with S10N-5G disperser for 2 × 10 s on ice. Both cultured cell and animal lysates were spun at 30,130 g for 2 × 10 min at 4°C. Immunoprecipitation was performed using anti-HA or anti-FLAG agarose beads (A2095 and A2220, respectively; Sigma-Aldrich) according to the manufacturer’s instructions. Finally, beads were boiled in 30 µl Laemmli buffer, followed by Western blot analysis using mouse anti-FLAG (1:2,000; F1804; Sigma-Aldrich), rat anti-HA (1:2,000; clone 3F10; Roche), rat anti-Snap29 (1:3,000; Takáts et al., 2013), and rabbit anti-Dor (1:1,000; Pulipparacharuvil et al., 2005) primary and alkaline phosphatase–conjugated goat anti–rabbit, rabbit anti–mouse, and rabbit anti–rat (all 1:5,000; A3812, A4312, and A6066, respectively; Sigma-Aldrich). Signal was developed using nitroblue tetrazolium chloride and 5-bromo-4-chloro-3-indolylphosphate (Sigma-Aldrich) in 100 mM Trizma base, 100 mM NaCl, 5 mM MgCl2, and 0.05% Tween 20, pH adjusted to 9.5 with HCl. Blots were dried and then scanned on a Perfection 4990 Photo scanner (Epson), followed by processing in Photoshop CS3 Extended.
Online supplemental material
Fig. S1 illustrates the effect of v-ATPase loss on glue degradation and statistical analysis of developmental crinophagy progression. Fig. S2 shows that core macroautophagy genes are dispensable for the degradation of glue granules. Fig. S3 shows quantification of crinophagy in different genetic backgrounds. Fig. S4 shows the identification of intact glue granules, lysosomes and crinosomes in salivary gland cells using immunogold labeling. Fig. S5 contains additional Syntaxin 17, YFP-Rab2, and glue secretion data. Table S1 shows the effect of Qa SNARE inhibitions on the quenching of Glue-GFP.
Supplementary Material
Acknowledgments
We thank Eduard Burghardt, Sarolta Pálfia, and Mónika Truszka for skillful technical assistance and the public repositories and colleagues listed in Materials and methods for reagents.
This work was supported by the Hungarian Academy of Sciences (grant Momentum LP-2014/2 to G. Juhász), the Wellcome Trust (grant 087518/Z/08/Z to G. Juhász), and the National Research, Development, and Innovation Office of Hungary (grants GINOP-2.3.2-15-2016-00032 and -00006, K119842 to G. Juhász and grant PD112632 to K. Hegedűs).
The authors declare no competing financial interests.
Author contributions: T. Csizmadia and G. Juhász designed research. T. Csizmadia, P. Lőrincz, K. Hegedűs, S. Széplaki, and P. Lőw performed experiments. T. Csizmadia, P. Lőrincz, and G. Juhász evaluated data. T. Csizmadia, P. Lőw, and G. Juhász wrote the paper with comments from all authors.
Footnotes
Abbreviations used:
- CA
- constitutively active
- CathB
- Cathepsin B
- RPF
- relative to puparium formation
- Syx13
- Syntaxin 13
References
- Ahlberg J., Beije B., Berkenstam A., Henell F., and Glaumann H.. 1987. Effects on in vivo and in vitro administration of vinblastine on the perfused rat liver--identification of crinosomes. Exp. Mol. Pathol. 47:309–326. 10.1016/0014-4800(87)90016-5 [DOI] [PubMed] [Google Scholar]
- Balderhaar H.J., and Ungermann C.. 2013. CORVET and HOPS tethering complexes - coordinators of endosome and lysosome fusion. J. Cell Sci. 126:1307–1316. 10.1242/jcs.107805 [DOI] [PubMed] [Google Scholar]
- Beckendorf S.K., and Kafatos F.C.. 1976. Differentiation in the salivary glands of Drosophila melanogaster: characterization of the glue proteins and their developmental appearance. Cell. 9:365–373. 10.1016/0092-8674(76)90081-7 [DOI] [PubMed] [Google Scholar]
- Berry D.L., and Baehrecke E.H.. 2007. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell. 131:1137–1148. 10.1016/j.cell.2007.10.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biyasheva A., Do T.V., Lu Y., Vaskova M., and Andres A.J.. 2001. Glue secretion in the Drosophila salivary gland: a model for steroid-regulated exocytosis. Dev. Biol. 231:234–251. 10.1006/dbio.2000.0126 [DOI] [PubMed] [Google Scholar]
- Brown W.J., DeWald D.B., Emr S.D., Plutner H., and Balch W.E.. 1995. Role for phosphatidylinositol 3-kinase in the sorting and transport of newly synthesized lysosomal enzymes in mammalian cells. J. Cell Biol. 130:781–796. 10.1083/jcb.130.4.781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess J., Jauregui M., Tan J., Rollins J., Lallet S., Leventis P.A., Boulianne G.L., Chang H.C., Le Borgne R., Krämer H., and Brill J.A.. 2011. AP-1 and clathrin are essential for secretory granule biogenesis in Drosophila. Mol. Biol. Cell. 22:2094–2105. 10.1091/mbc.E11-01-0054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera M., Nordmann M., Perz A., Schmedt D., Gerondopoulos A., Barr F., Piehler J., Engelbrecht-Vandré S., and Ungermann C.. 2014. The Mon1-Ccz1 GEF activates the Rab7 GTPase Ypt7 via a longin-fold-Rab interface and association with PI3P-positive membranes. J. Cell Sci. 127:1043–1051. 10.1242/jcs.140921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantino B.F., Bricker D.K., Alexandre K., Shen K., Merriam J.R., Antoniewski C., Callender J.L., Henrich V.C., Presente A., and Andres A.J.. 2008. A novel ecdysone receptor mediates steroid-regulated developmental events during the mid-third instar of Drosophila. PLoS Genet. 4:e1000102 10.1371/journal.pgen.1000102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo A.M., and Wong E.. 2014. Chaperone-mediated autophagy: roles in disease and aging. Cell Res. 24:92–104. 10.1038/cr.2013.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., He D., Yao Z., and Klionsky D.J.. 2014. The machinery of macroautophagy. Cell Res. 24:24–41. 10.1038/cr.2013.168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N., Huang W., Lin T.H., Groulx J.F., Jean S., Nguyen J., Kuchitsu Y., Koyama-Honda I., Mizushima N., Fukuda M., and Kiger A.A.. 2017. Genetic screen in Drosophila muscle identifies autophagy-mediated T-tubule remodeling and a Rab2 role in autophagy. eLife. 6:e23367 10.7554/eLife.23367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillingham A.K., Sinka R., Torres I.L., Lilley K.S., and Munro S.. 2014. Toward a comprehensive map of the effectors of rab GTPases. Dev. Cell. 31:358–373. 10.1016/j.devcel.2014.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaumann H., Ahlberg J., Hultenby K., Jansson H., Mengarelli-Widholm S., and Motakefi A.M.. 1989. Isolation and characterization of crinosomes--a subclass of secondary lysosomes. Exp. Mol. Pathol. 50:167–182. 10.1016/0014-4800(89)90028-2 [DOI] [PubMed] [Google Scholar]
- Goginashvili A., Zhang Z., Erbs E., Spiegelhalter C., Kessler P., Mihlan M., Pasquier A., Krupina K., Schieber N., Cinque L., et al. . 2015. Insulin secretory granules control autophagy in pancreatic β cells. Science. 347:878–882. 10.1126/science.aaa2628 [DOI] [PubMed] [Google Scholar]
- Grosshans B.L., Ortiz D., and Novick P.. 2006. Rabs and their effectors: achieving specificity in membrane traffic. Proc. Natl. Acad. Sci. USA. 103:11821–11827. 10.1073/pnas.0601617103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrod M.J., and Kastritsis C.D.. 1972. Developmental studies in Drosophila. II. Ultrastructural analysis of the salivary glands of Drosophila pseudoobscura during some stages of development. J. Ultrastruct. Res. 38:482–499. 10.1016/0022-5320(72)90086-X [DOI] [PubMed] [Google Scholar]
- Hegedűs K., Takáts S., Boda A., Jipa A., Nagy P., Varga K., Kovács A.L., and Juhász G.. 2016. The Ccz1-Mon1-Rab7 module and Rab5 control distinct steps of autophagy. Mol. Biol. Cell. 27:3132–3142. 10.1091/mbc.E16-03-0205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E., Kishi-Itakura C., and Mizushima N.. 2012. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 151:1256–1269. 10.1016/j.cell.2012.11.001 [DOI] [PubMed] [Google Scholar]
- Jiang P., Nishimura T., Sakamaki Y., Itakura E., Hatta T., Natsume T., and Mizushima N.. 2014. The HOPS complex mediates autophagosome-lysosome fusion through interaction with syntaxin 17. Mol. Biol. Cell. 25:1327–1337. 10.1091/mbc.E13-08-0447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhász G., Hill J.H., Yan Y., Sass M., Baehrecke E.H., Backer J.M., and Neufeld T.P.. 2008. The class III PI(3)K Vps34 promotes autophagy and endocytosis but not TOR signaling in Drosophila. J. Cell Biol. 181:655–666. 10.1083/jcb.200712051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S., Noda T., and Yoshimori T.. 2007. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 3:452–460. 10.4161/auto.4451 [DOI] [PubMed] [Google Scholar]
- Koike H., Steer M.L., and Meldolesi J.. 1982. Pancreatic effects of ethionine: blockade of exocytosis and appearance of crinophagy and autophagy precede cellular necrosis. Am. J. Physiol. 242:G297–G307. [DOI] [PubMed] [Google Scholar]
- Logsdon C.D., and Ji B.. 2013. The role of protein synthesis and digestive enzymes in acinar cell injury. Nat. Rev. Gastroenterol. Hepatol. 10:362–370. 10.1038/nrgastro.2013.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lőrincz P., Lakatos Z., Maruzs T., Szatmári Z., Kis V., and Sass M.. 2014. Atg6/UVRAG/Vps34-containing lipid kinase complex is required for receptor downregulation through endolysosomal degradation and epithelial polarity during Drosophila wing development. BioMed Res. Int. 2014:851349 10.1155/2014/851349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lőrincz P., Lakatos Z., Varga Á., Maruzs T., Simon-Vecsei Z., Darula Z., Benkő P., Csordás G., Lippai M., Andó I., et al. . 2016. MiniCORVET is a Vps8-containing early endosomal tether in Drosophila. eLife. 5:e14226 10.7554/eLife.14226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lőrincz P., Tóth S., Benkő P., Lakatos Z., Boda A., Glatz G., Zobel M., Bisi S., Hegedűs K., Takáts S., et al. . 2017. Rab2 promotes autophagic and endocytic lysosomal degradation. J. Cell Biol. 216:1937–1947. 10.1083/jcb.201611027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Zhang Z., Sun D., Sweeney S.T., and Gao F.B.. 2013. Syntaxin 13, a genetic modifier of mutant CHMP2B in frontotemporal dementia, is required for autophagosome maturation. Mol. Cell. 52:264–271. 10.1016/j.molcel.2013.08.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzio J.P., Hackmann Y., Dieckmann N.M., and Griffiths G.M.. 2014. The biogenesis of lysosomes and lysosome-related organelles. Cold Spring Harb. Perspect. Biol. 6:a016840 10.1101/cshperspect.a016840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh B.J., Soden C., Alarcón C., Wicksteed B.L., Yaekura K., Costin A.J., Morgan G.P., and Rhodes C.J.. 2007. Regulated autophagy controls hormone content in secretory-deficient pancreatic endocrine beta-cells. Mol. Endocrinol. 21:2255–2269. 10.1210/me.2007-0077 [DOI] [PubMed] [Google Scholar]
- Marzella L., Ahlberg J., and Glaumann H.. 1981. Autophagy, heterophagy, microautophagy and crinophagy as the means for intracellular degradation. Virchows Arch. B Cell Pathol. Incl. Mol. Pathol. 36:219–234. 10.1007/BF02912068 [DOI] [PubMed] [Google Scholar]
- Mauvezin C., Nagy P., Juhász G., and Neufeld T.P.. 2015. Autophagosome-lysosome fusion is independent of V-ATPase-mediated acidification. Nat. Commun. 6:7007 10.1038/ncomms8007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwan D.G., Popovic D., Gubas A., Terawaki S., Suzuki H., Stadel D., Coxon F.P., Miranda de Stegmann D., Bhogaraju S., Maddi K., et al. . 2015. PLEKHM1 regulates autophagosome-lysosome fusion through HOPS complex and LC3/GABARAP proteins. Mol. Cell. 57:39–54. 10.1016/j.molcel.2014.11.006 [DOI] [PubMed] [Google Scholar]
- Mijaljica D., Prescott M., and Devenish R.J.. 2011. Microautophagy in mammalian cells: revisiting a 40-year-old conundrum. Autophagy. 7:673–682. 10.4161/auto.7.7.14733 [DOI] [PubMed] [Google Scholar]
- Nagy P., Varga Á., Kovács A.L., Takáts S., and Juhász G.. 2015. How and why to study autophagy in Drosophila: it’s more than just a garbage chute. Methods. 75:151–161. 10.1016/j.ymeth.2014.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N., Matsuura A., Wada Y., and Ohsumi Y.. 1997. Acidification of vacuoles is required for autophagic degradation in the yeast, Saccharomyces cerevisiae. J. Biochem. 121:338–344. 10.1093/oxfordjournals.jbchem.a021592 [DOI] [PubMed] [Google Scholar]
- Nezis I.P., Shravage B.V., Sagona A.P., Lamark T., Bjørkøy G., Johansen T., Rusten T.E., Brech A., Baehrecke E.H., and Stenmark H.. 2010. Autophagic degradation of dBruce controls DNA fragmentation in nurse cells during late Drosophila melanogaster oogenesis. J. Cell Biol. 190:523–531. 10.1083/jcb.201002035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J.-Q., Liu L.-P., Binari R., Hardy R., Shim H.-S., Cavallaro A., Booker M., Pfeiffer B.D., Markstein M., Wang H., et al. . 2009. A Drosophila resource of transgenic RNAi lines for neurogenetics. Genetics. 182:1089–1100. 10.1534/genetics.109.103630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orci L., Ravazzola M., Amherdt M., Yanaihara C., Yanaihara N., Halban P., Renold A.E., and Perrelet A.. 1984. Insulin, not C-peptide (proinsulin), is present in crinophagic bodies of the pancreatic B-cell. J. Cell Biol. 98:222–228. 10.1083/jcb.98.1.222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pircs K., Nagy P., Varga A., Venkei Z., Erdi B., Hegedus K., and Juhasz G.. 2012. Advantages and limitations of different p62-based assays for estimating autophagic activity in Drosophila. PLoS One. 7:e44214 10.1371/journal.pone.0044214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulipparacharuvil S., Akbar M.A., Ray S., Sevrioukov E.A., Haberman A.S., Rohrer J., and Krämer H.. 2005. Drosophila Vps16A is required for trafficking to lysosomes and biogenesis of pigment granules. J. Cell Sci. 118:3663–3673. 10.1242/jcs.02502 [DOI] [PubMed] [Google Scholar]
- Riahi Y., Wikstrom J.D., Bachar-Wikstrom E., Polin N., Zucker H., Lee M.S., Quan W., Haataja L., Liu M., Arvan P., et al. . 2016. Autophagy is a major regulator of beta cell insulin homeostasis. Diabetologia. 59:1480–1491. (published erratum appears in Diabetologia. 2016. 59:1575-1576) 10.1007/s00125-016-3868-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousso T., Schejter E.D., and Shilo B.Z.. 2016. Orchestrated content release from Drosophila glue-protein vesicles by a contractile actomyosin network. Nat. Cell Biol. 18:181–190. 10.1038/ncb3288 [DOI] [PubMed] [Google Scholar]
- Shravage B.V., Hill J.H., Powers C.M., Wu L., and Baehrecke E.H.. 2013. Atg6 is required for multiple vesicle trafficking pathways and hematopoiesis in Drosophila. Development. 140:1321–1329. 10.1242/dev.089490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R.E., and Farquhar M.G.. 1966. Lysosome function in the regulation of the secretory process in cells of the anterior pituitary gland. J. Cell Biol. 31:319–347. 10.1083/jcb.31.2.319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H. 2009. Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 10:513–525. 10.1038/nrm2728 [DOI] [PubMed] [Google Scholar]
- Stroupe C., Collins K.M., Fratti R.A., and Wickner W.. 2006. Purification of active HOPS complex reveals its affinities for phosphoinositides and the SNARE Vam7p. EMBO J. 25:1579–1589. 10.1038/sj.emboj.7601051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramani S., and Malhotra V.. 2013. Non-autophagic roles of autophagy-related proteins. EMBO Rep. 14:143–151. 10.1038/embor.2012.220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takáts S., Nagy P., Varga Á., Pircs K., Kárpáti M., Varga K., Kovács A.L., Hegedűs K., and Juhász G.. 2013. Autophagosomal Syntaxin17-dependent lysosomal degradation maintains neuronal function in Drosophila. J. Cell Biol. 201:531–539. 10.1083/jcb.201211160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takáts S., Pircs K., Nagy P., Varga Á., Kárpáti M., Hegedűs K., Kramer H., Kovács A.L., Sass M., and Juhász G.. 2014. Interaction of the HOPS complex with Syntaxin 17 mediates autophagosome clearance in Drosophila. Mol. Biol. Cell. 25:1338–1354. 10.1091/mbc.E13-08-0449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchizono Y., Alarcón C., Wicksteed B.L., Marsh B.J., and Rhodes C.J.. 2007. The balance between proinsulin biosynthesis and insulin secretion: where can imbalance lead? Diabetes Obes. Metab. 9(Suppl 2):56–66. 10.1111/j.1463-1326.2007.00774.x [DOI] [PubMed] [Google Scholar]
- Weckman A., Di Ieva A., Rotondo F., Syro L.V., Ortiz L.D., Kovacs K., and Cusimano M.D.. 2014. Autophagy in the endocrine glands. J. Mol. Endocrinol. 52:R151–R163. 10.1530/JME-13-0241 [DOI] [PubMed] [Google Scholar]
- Zeng X., Overmeyer J.H., and Maltese W.A.. 2006. Functional specificity of the mammalian Beclin-Vps34 PI 3-kinase complex in macroautophagy versus endocytosis and lysosomal enzyme trafficking. J. Cell Sci. 119:259–270. 10.1242/jcs.02735 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.