The nonreceptor tyrosine kinase Pyk2 is highly expressed in invasive breast cancer, but how it potentiates tumor cell invasiveness is unclear. Genna et al. find that Pyk2 and the closely related kinase FAK modulate breast cancer cell invasiveness by distinct mechanisms and coordinate the balance between focal adhesion–mediated migration and invadopodia-dependent extracellular matrix invasion.
Abstract
The nonreceptor tyrosine kinase Pyk2 is highly expressed in invasive breast cancer, but the mechanism by which it potentiates tumor cell invasiveness is unclear at present. Using high-throughput protein array screening and bioinformatic analysis, we identified cortactin as a novel substrate and interactor of proline-rich tyrosine kinase 2 (Pyk2). Pyk2 colocalizes with cortactin to invadopodia of invasive breast cancer cells, where it mediates epidermal growth factor–induced cortactin tyrosine phosphorylation both directly and indirectly via Src-mediated Abl-related gene (Arg) activation, leading to actin polymerization in invadopodia, extracellular matrix degradation, and tumor cell invasion. Both Pyk2 and the closely related focal adhesion kinase (FAK) regulate tumor cell invasion, albeit via distinct mechanisms. Although Pyk2 regulates tumor cell invasion by controlling invadopodium-mediated functions, FAK controls invasiveness of tumor cells by regulating focal adhesion–mediated motility. Collectively, our findings identify Pyk2 as a unique mediator of invadopodium formation and function and also provide a novel insight into the mechanisms by which Pyk2 mediates tumor cell invasion.
Introduction
Metastatic dissemination of cancer cells from the primary tumor and their spread to distant sites in the body is the leading cause of mortality in breast cancer patients. Metastatic cancer cells must penetrate through several barriers to escape the primary tumor and gain entry into the bloodstream in order to spread to other tissues. Invasive cancer cells penetrate these barriers by forming invadopodia, F-actin–rich protrusions that localize matrix-degrading activity to cell–substrate contact points. Invadopodia represent sites in which cell signaling, proteolytic, adhesive, cytoskeletal, and membrane-trafficking pathways physically converge to execute cell invasion and consequent metastatic dissemination (Weaver, 2006, 2008). Invadopodia proceed through a series of maturation events, beginning with the formation of an invadopodium precursor followed by stabilization and activation of actin polymerization before acquiring the ability to efficiently degrade the ECM (Artym et al., 2006; Oser et al., 2009; Sharma et al., 2013; Beaty and Condeelis, 2014). Invadopodia gain their protrusive ability by combining the physical force generated by actin polymerization with the chemical activity of matrix metalloproteinase (MMP)-mediated ECM degradation (Sibony-Benyamini and Gil-Henn, 2012). Understanding the mechanisms that govern the formation and function of invadopodia could provide insights into the biology, regulation, and potential therapeutic approaches for cancer metastasis.
The cortactin gene CTTN, located in chromosome 11q13, is amplified in various human carcinomas and is correlated with poor patient prognosis (Hui et al., 1997; Ormandy et al., 2003). Cortactin localizes to invadopodia in invasive breast cancer cells, where it regulates both their formation and function (Artym et al., 2006; Clark et al., 2007; Sinha et al., 2016). Cortactin tyrosine phosphorylation is essential for generation of free actin barbed ends, required for actin polymerization in invadopodia (Oser et al., 2009) as well as for efficient ECM degradation (Clark et al., 2007; Ayala et al., 2008; Clark and Weaver, 2008) and metastasis in vivo (Li et al., 2001).
Because cortactin was originally identified as a Src substrate (Wu et al., 1991; Wu and Parsons, 1993) and because Src promotes cortactin phosphorylation when overexpressed in cells (Artym et al., 2006; Oser et al., 2009) and can promote cortactin-mediated actin polymerization in vitro (Tehrani et al., 2007), it has been inferred that Src solely mediates cortactin phosphorylation in invadopodia, thereby controlling invadopodial function. Src has also been shown to phosphorylate the scaffold and Nox organizer protein Tks5, leading to reactive oxygen species production, which induces invadopodia formation and activation (Diaz et al., 2009; Gianni et al., 2009). We have previously shown that the nonreceptor tyrosine kinase Abl-related gene (Arg) can phosphorylate cortactin specifically in invadopodia after its phosphorylation and activation by Src, leading to actin polymerization and consequent tumor cell invasion (Mader et al., 2011).
The focal adhesion kinase (FAK) and its homologous FAK-related proline-rich tyrosine kinase 2 (Pyk2) define a distinct family of nonreceptor tyrosine kinases that exhibits ∼48% amino acid identity, common phosphorylation sites, and a similar domain structure that includes an N-terminal 4.1, exrin, radixin, and moesin (FERM) domain, a kinase domain, proline-rich regions, and a C-terminal focal adhesion-targeting (FAT) domain. Although FAK is expressed in most cells, Pyk2 exhibits a more restricted expression pattern with strongest expression in the central nervous system and in hematopoietic cells (Lev et al., 1995). The differential expression and localization patterns of FAK versus Pyk2 in addition to the distinct binding activities of their nonkinase domains might limit their functional redundancy and may suggest distinct and possibly antagonistic roles in cells.
FAK does not localize to invadopodia but can regulate their formation and activity from a distance by controlling localization of Src kinase. FAK recruits a pool of active Src to mediate localized tyrosine phosphorylation of substrates at focal adhesions. Depletion of FAK releases this pool of active Src to mediate enhanced phosphorylation of substrates in invadopodia, which impairs focal adhesion dynamics while enhancing formation and dynamics of invadopodia. By regulating Src localization, FAK controls the tyrosine phosphorylation balance between invadopodia and focal adhesions (Vitale et al., 2008; Chan et al., 2009; Kolli-Bouhafs et al., 2014). Pyk2 regulates the stabilization and dynamics of podosomes (Gil-Henn et al., 2007), cytoskeletal structures that are closely related to invadopodia, but its role in invadopodia formation or function has never been determined.
To elucidate the molecular mechanism by which Pyk2 regulates tumor cell invasion, we performed high-throughput protein array screens in which 115 novel substrates and interactors of Pyk2 were identified. Additional bioinformatic analysis revealed cortactin as a potential substrate and interactor of Pyk2 in invadopodia. We show in this study that Pyk2 colocalizes with cortactin at invadopodia of invasive breast cancer cells. Pyk2 regulates cortactin tyrosine phosphorylation at invadopodium precursors both directly and indirectly via recruitment and activation of Src and Arg kinases, leading to barbed end generation, ECM degradation, and consequent tumor cell invasion. We also demonstrate that both Pyk2 and FAK regulate tumor cell invasion; however, they do so via distinct mechanisms: although Pyk2 regulates tumor cell invasion by controlling invadopodia-mediated functions, FAK controls tumor cell invasiveness by regulating focal adhesion–mediated motility and functions. Collectively, our findings identify Pyk2 as a unique mediator of invadopodia formation and function in breast cancer cells and provide a novel insight into the mechanisms of Pyk2-mediated tumor cell invasion.
Results
A high-throughput proteomic screen identifies cortactin as a potential substrate and interactor of Pyk2 in invadopodia
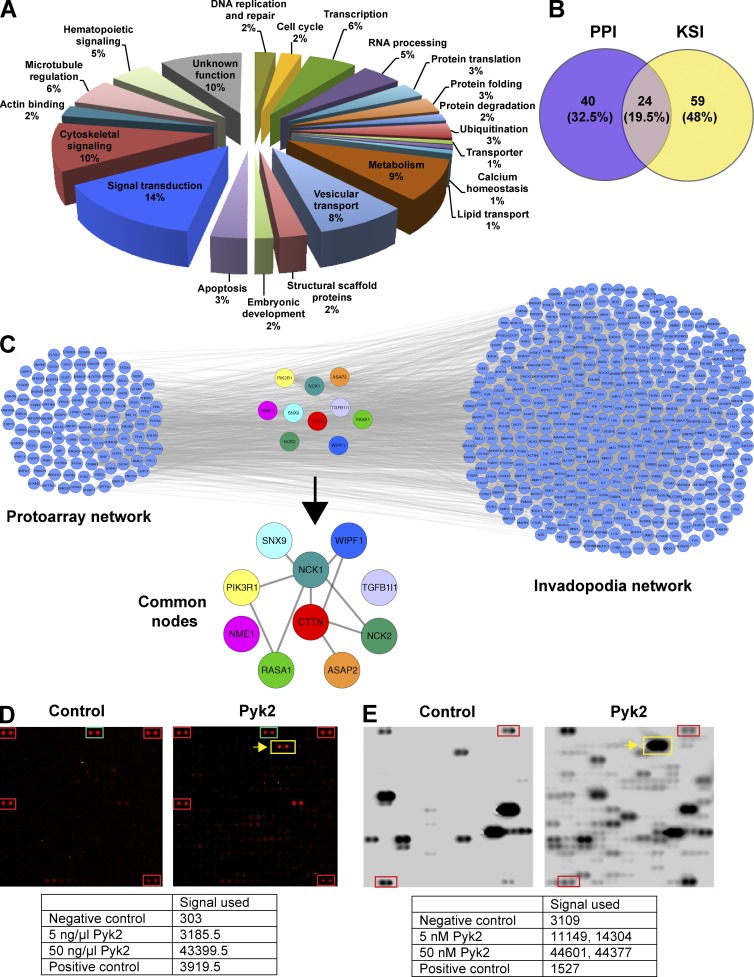
To gain insight into the molecular mechanisms and signaling pathways by which Pyk2 regulates cell invasion, we screened high-throughput protein–protein and kinase–substrate protein microarrays using purified Pyk2 as bait. Using these assays, we have identified a total of 147 candidate Pyk2-interacting proteins (64 candidate interactors and 83 candidate substrates). Detailed comparison with known Pyk2 substrates and interactors that were curated from Ingenuity Pathway Analysis database revealed that 115 proteins out of 147 that are novel substrates or interactors of Pyk2. Classification of identified substrates and interactors of Pyk2 using the GeneCards and UniProt databases revealed that a major fraction of proteins participate in cytoskeletal signaling and regulation as well as in general signal transduction (Fig. 1 A and Table S1). Overlap of proteins from the two screens revealed 24 proteins that were both substrates and interactors of Pyk2 (Fig. 1 B).
Figure 1.
Cortactin is a potential substrate and interactor of Pyk2. (A) Classification of proteins identified in the protein–protein interaction (PPI) and kinase–substrate interaction (KSI) ProtoArray screens based on their GeneCards and UniProt identified function. (B) Venn diagram representing the overlap between proteins that were identified in the protein–protein interaction screen and the kinase–substrate interaction screen. Representation factor = 41.3; P < 5.498e−34. (C) Overlap of known invadopodia proteins (invadopodia network) with Pyk2 substrates and interactors identified by the ProtoArray screens (ProtoArray network). The list of known invadopodia proteins was obtained by literature mining using GLAD4U and ALS. The lists of proteins were overlaid using Cytoscape, and the combined list of common nodes was used to build a physical and functional association network in STRING (gray lines). (D) Representative protein–protein interaction subarray image from control (left) and experimental assay using purified Pyk2 as a bait protein (right). Red and green boxes represent assay controls of biotinylated or Alexa Fluor 647–labeled proteins, and the yellow box and arrow represent the cortactin signal. The table presents the background-subtracted pixel intensity value (signal used) of cortactin in two concentrations of purified Pyk2 that was used for screening compared with the positive and negative controls. (E) Representative kinase–substrate interaction subarray image from control (left) and experimental assay using purified Pyk2 as a bait protein (right). Red boxes represent a positive control of autophosphorylating PKCη, and the yellow box and arrow represent cortactin signal. The table represents the signal used values of cortactin (duplicate) in two concentrations of purified Pyk2 that was used for screening, compared with the positive and negative controls.
Because the closely related FAK is indirectly involved in invadopodia regulation (Chan et al., 2009; Kolli-Bouhafs et al., 2014) and because Pyk2 is involved in regulation of podosomes (cytoskeletal structures that are closely related to invadopodia; Gil-Henn et al., 2007), we hypothesized that Pyk2 might have a role in invadopodia regulation. To test this hypothesis, we performed literature mining using Gene List Automatically Derived for You (GLAD4U) and Agilent Literature Search (ALS) databases for the query invadopodia (Table S2). Overlay of the combined literature mining proteins with protein array data revealed 11 invadopodia-related candidate proteins that directly interact with Pyk2. Out of these candidates, cortactin, which is a major component of invadopodia, was found as both substrate and interactor of Pyk2 (Fig. 1, D and E). Database searches using STRING and Ingenuity Pathway Analysis followed by manual literature validation using PubMed identified cortactin as a novel substrate and interactor of Pyk2.
Pyk2 colocalizes with cortactin at invadopodia of invasive breast cancer cells
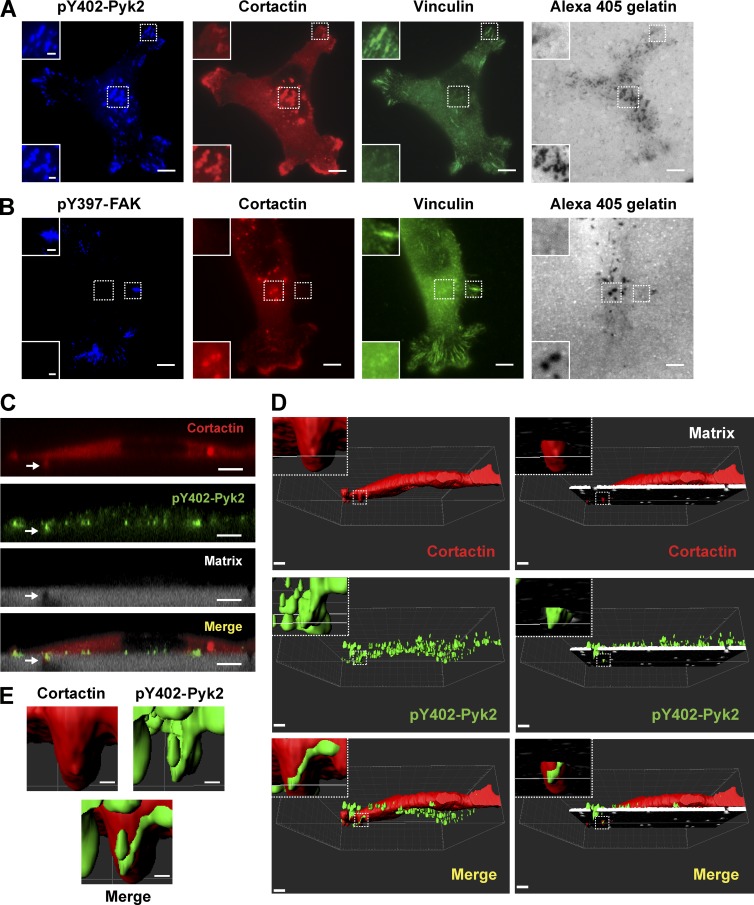
Based on our protein arrays data, we sought to determine whether Pyk2 localizes to invadopodia in invasive breast cancer cells. Immunofluorescent labeling of endogenous activated Pyk2 (pY402-Pyk2) in MDA–MB-231 cells plated on a fluorescent ECM substrate showed colocalization with cortactin and with degradation puncta, markers for mature invadopodia, and with vinculin at focal adhesions (Fig. 2 A). Labeling of the closely related activated FAK (pY397-FAK) showed colocalization with vinculin at focal adhesions only, as previously reported (Fig. 2 B; Bowden et al., 2006; Chan et al., 2009; Kolli-Bouhafs et al., 2014). Similar results were observed using GFP-Pyk2 and GFP-FAK constructs in MDA–MB-231 cells (Fig. S1, A and B). To verify localization of Pyk2 to matrix-degrading invadopodia, we plated MDA–MB-231 cells on 1-µm transwell filters overlaid with fluorescent ECM mixture. As previously demonstrated (Fig. S1; Schoumacher et al., 2010; Magalhaes et al., 2011), only F-actin–rich protrusions containing invadopodial marker proteins can protrude through the 1-µm pores by actively degrading the ECM overlay. X–Z confocal images and 3D reconstruction clearly indicated colocalization of activated Pyk2 with cortactin to ECM-degrading invadopodial protrusions (Fig. 2, C–E). These data suggest that Pyk2 may regulate invadopodial formation or function in invasive breast cancer cells.
Figure 2.
Pyk2 colocalizes with cortactin at invadopodia. (A and B) MDA–MB-231 cells expressing cortactin-TagRFP were plated on Alexa Fluor 405 gelatin matrix, fixed, and labeled for vinculin and pY402-Pyk2 (A) or pY397-FAK and vinculin (B). Insets depict magnification of boxed areas showing either focal adhesions or invadopodia. Bars: (main images) 10 µm; (insets) 1 µm. (C) Representative X–Z confocal images showing colocalization of cortactin (red) and pY402-Pyk2 (green) to an invadopod (arrow) projecting into the Matrigel/Alexa Fluor 405 gelatin matrix (gray) that was plated in the top chamber of a 1-µm transwell filter. Bar, 5 µm. (D and E) Representative 3D reconstruction images showing colocalization of cortactin (red) and pY402-Pyk2 (green) to an invadopod (box) projecting into the Matrigel/Alexa Fluor 405 gelatin matrix (white, right images in D) that was plated in the top chamber of a 1-µm transwell filter. Bars: (main images) 5 µm; (insets and E) 1 µm.
Pyk2 binds to cortactin at invadopodium precursors
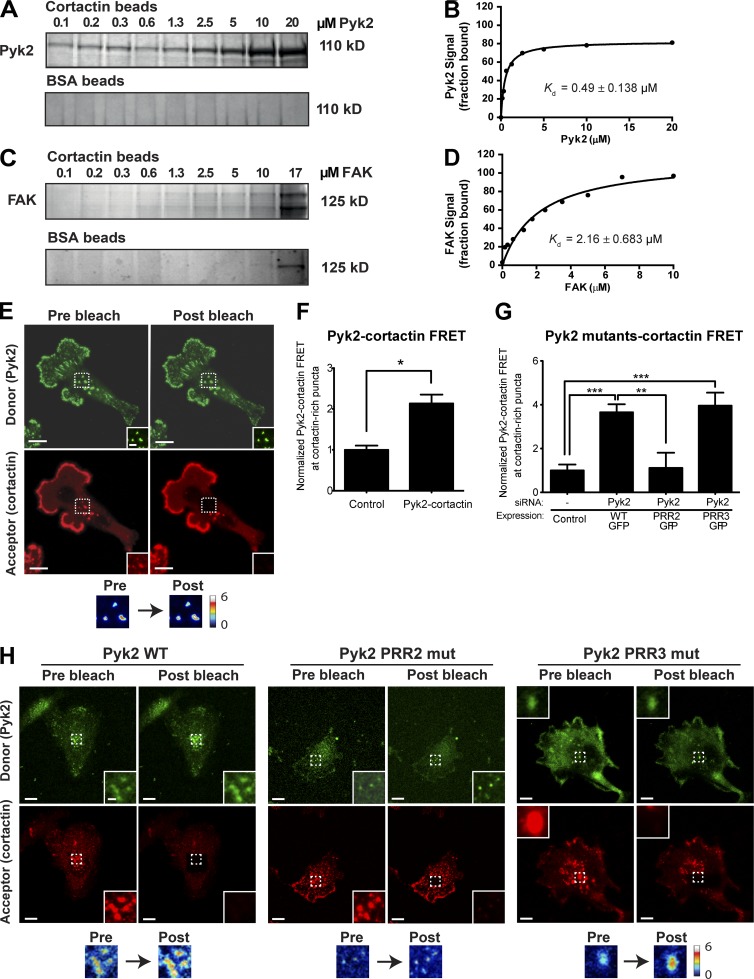
To examine whether Pyk2 binds cortactin directly, we performed an in vitro pulldown assay using purified proteins. Increasing concentrations of purified Pyk2 were incubated with constant concentration of cortactin, and the amount of bound Pyk2 was quantified. As shown in Fig. 3 (A and B), cortactin binds to Pyk2 with a Kd of 0.49 ± 0.138 µM. Previous studies demonstrated that the closely related FAK binds cortactin at focal adhesions (Wang et al., 2011; Tomar et al., 2012). We therefore sought to determine whether FAK binds cortactin in our in vitro system and thus compared the FAK–cortactin interaction to Pyk2–cortactin binding. As demonstrated in Fig. 3 (C and D), FAK binds to cortactin with a Kd of 2.16 ± 0.683 µM. These experiments suggest that Pyk2 binds cortactin directly in vitro with higher affinity than FAK.
Figure 3.
Cortactin binds to Pyk2. (A–D) Increasing concentrations of recombinant His-tagged Pyk2 (0–20 µM; A and B) or FAK (0–17 µM; C and D) were incubated with 1 µM cortactin covalently bound to AminoLink beads or to BSA control beads only. Pyk2 bound to cortactin with a Kd of 0.49 ± 0.138 µM (B), whereas FAK bound cortactin with a Kd of 2.16 ± 0.683 µM (D). n = 3 independent pulldown experiments with either Pyk2 or FAK in solution and cortactin beads, and n = 2 independent pulldown experiments with Pyk2 or FAK with BSA control beads. (E) Representative Pyk2-cortactin FRET efficiency images of an MDA–MB-231 cell labeled for Pyk2 (green) and cortactin (red). Insets (left) and box (right) indicate the bleached area. (F) Quantification of FRET between pY402-Pyk2 and cortactin at cortactin-rich puncta in locations where invadopodium precursors are observed. n = 23 invadopods from three independent experiments. (G and H) Representative images and quantification of FRET between GFP-tagged Pyk2 WT, PRR2 mutant, or PRR3 mutant and cortactin at cortactin-rich puncta in locations where invadopodium precursors are observed. n = 43–52 invadopods per group from three independent experiments. Bars: (main images) 10 µm; (insets) 2 µm. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Error bars represent SEM.
To determine whether Pyk2 and cortactin interact at cortactin-rich puncta in locations where invadopodium precursors are observed, we performed Förster resonance energy transfer (FRET) acceptor photobleaching experiments. MDA–MB-231 cells were plated on gelatin matrix, fixed, and stained with antibodies against Pyk2 (donor) and cortactin (acceptor). The results of this experiment indicate that Pyk2 and cortactin interact at cortactin-containing puncta (Fig. 3, E and F). To identify the region in Pyk2 that binds to cortactin, we performed similar experiments using Pyk2 WT or mutants of Pyk2 in which the proline-rich region 2 or 3 have been mutated (PRR2 and PRR3). As demonstrated in Fig. 3 (G and H), only mutation in the second proline-rich region of Pyk2 eliminated binding to cortactin. To confirm that the FRET interaction between Pyk2 and cortactin is specific, we performed acceptor photobleaching FRET between cortactin and Tks5, two invadopodial proteins that do not interact with each other. As expected, an interaction between cortactin and Tks5 was not observed in invadopodia using FRET (Fig. S1 E). Collectively, these data demonstrate that Pyk2 can bind cortactin in vitro as well as in cortactin-positive puncta of breast cancer cells in locations where invadopodium precursors are observed.
Invadopodium precursor formation depends on Pyk2
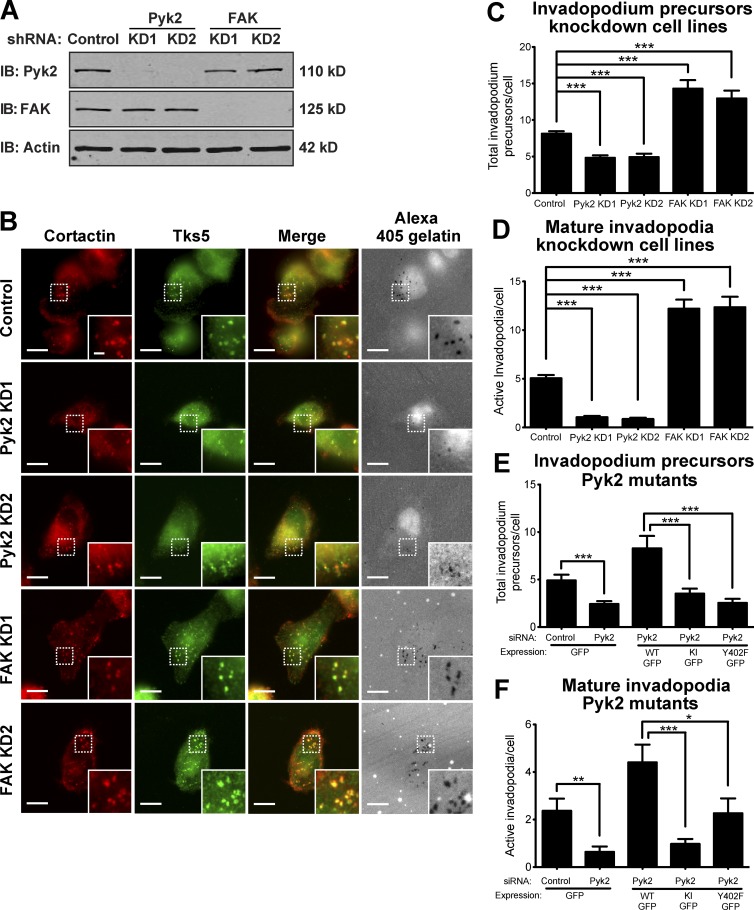
Invadopodia initially form as punctate structures called precursors, enriched in F-actin, cortactin, Tks5, and the Arp2/3 complex, which mature to acquire matrix-degrading capabilities (Artym et al., 2006; Bowden et al., 2006; Oser et al., 2009). To examine whether Pyk2 regulates the initial assembly of invadopodium precursors, MDA–MB-231 stably knocked down for Pyk2 or FAK were plated on fluorescently labeled gelatin matrix and labeled for Tks5 and cortactin (Fig. 4, A–D). Pyk2-knockdown cells showed a significant decrease in Tks5- and cortactin-positive invadopodium precursors as well as a decrease in active matrix-degrading invadopodia. In accordance with previous studies (Chan et al., 2009; Kolli-Bouhafs et al., 2014), cells knocked down for the closely related FAK showed increases in both invadopodium precursors and active invadopodia numbers.
Figure 4.
Pyk2 and FAK oppositely regulate invadopodium precursor formation in breast cancer cells. (A) Representative immunoblots (IBs) of MDA–MB-231 cells stably expressing control, Pyk2, or FAK shRNA. Whole-cell lysates were blotted for either Pyk2 or FAK and for actin as loading controls. (B) MDA–MB-231 cells stably expressing control, Pyk2 shRNA, or FAK shRNA were plated on Alexa Fluor 405 gelatin, fixed, and labeled for cortactin (red) and Tks5 (green). Boxed regions and insets depict colocalization of cortactin and Tks5 as markers for invadopodium precursors with Alexa Fluor 405 gelatin as a marker for mature invadopodia. Bars: (main images) 10 µm; (insets) 2 µm. (C and D) Quantification of invadopodium precursors defined by colocalization of cortactin and Tks5 (C) as well as mature (active) invadopodia, defined by colocalization of cortactin and Tks5 with degradation regions (D). n = 44–262 cells per group from three independent experiments. (E and F) Quantification of invadopodium precursors (E) and mature invadopodia (F) from cells expressing WT Pyk2-GFP, kinase-inactive (KI) Pyk2-GFP, or Y402F-Pyk2 GFP. n = 42–73 cells per group from three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Error bars represent SEM.
To elucidate the mechanism by which Pyk2 regulates invadopodia formation, we rescued Pyk2-knockdown cells with a kinase-inactive (Pyk2-KI) or autophosphorylation and Src recruitment mutant (Pyk2-Y402F) of Pyk2. As demonstrated in Fig. 4 (E and F), both invadopodium precursor formation and mature invadopodia numbers were significantly reduced in cells expressing either mutant of Pyk2.
Similar results were obtained using MTLn3 rat adenocarcinoma cells, U2OS human osteosarcoma cells, and A431 epidermoid carcinoma cells knocked down for either Pyk2 or FAK (Fig. S2, C–F; and Fig. S3). Altogether, these data suggest that both Pyk2 and FAK regulate invadopodium precursor formation and invadopodia maturation and activation in several different cancer cell lines, but they do so in distinct or possibly opposite mechanisms.
Pyk2 regulates invadopodium precursor assembly by recruiting Src kinase
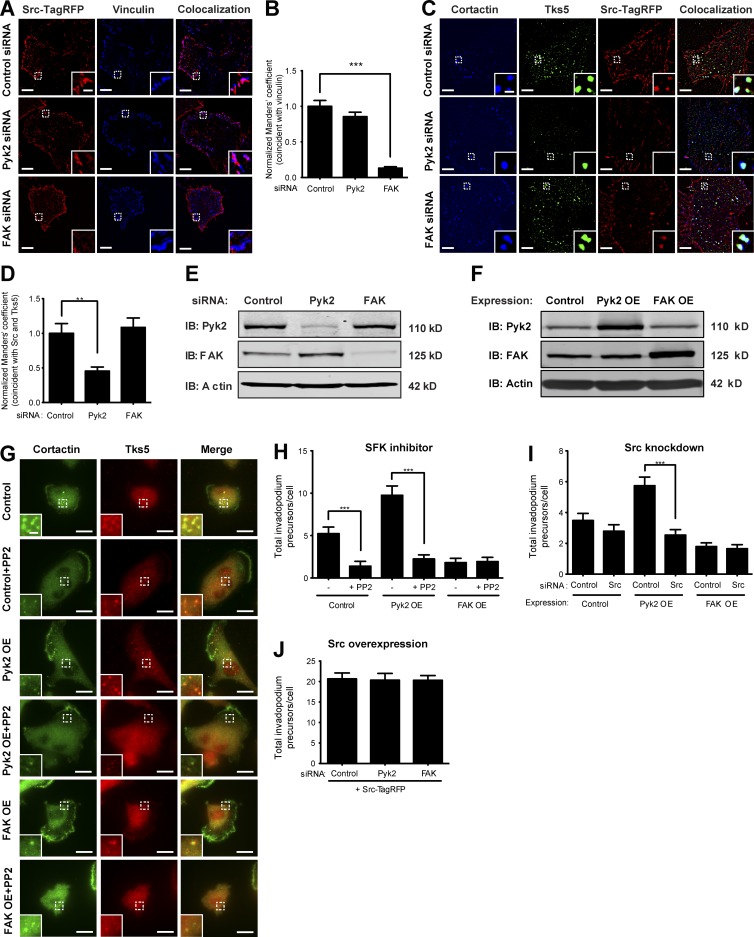
Overexpression of WT or activated mutants of Src in MDA–MB-231 cells or head-and-neck squamous cell carcinoma (HNSCC) cells produces a significant increase in the number of invadopodium precursors (Artym et al., 2006; Chan et al., 2009; Oser et al., 2009), whereas inhibition of Src family kinases by PP2 reduces the number of invadopodia and consequent matrix degradation (Chan et al., 2009; Liu et al., 2010). Previous publications suggested that FAK indirectly regulates invadopodium precursor formation by sequestering Src to focal adhesions (Chan et al., 2009; Kolli-Bouhafs et al., 2014), but the identity of the protein that localizes Src to invadopodium precursors has never been determined. To investigate whether Pyk2 and FAK coordinate the recruitment of Src kinase to invadopodium precursors and focal adhesions, respectively, we examined colocalization of fluorescently labeled Src to cortactin and Tks5-enriched invadopodium precursors and with vinculin at focal adhesions in Pyk2- or FAK-depleted cells. Depletion of Pyk2 from MDA–MB-231 breast cancer cells significantly reduced localization of Src to invadopodium precursors and only mildly changed its distribution to vinculin-containing focal adhesions, whereas depletion of FAK significantly reduced Src localization to vinculin-containing focal adhesions (Fig. 5, A–E). These observations indicate that Pyk2 localizes Src to invadopodium precursors and suggest that Pyk2 and FAK coordinate the balance between focal adhesions and invadopodia by controlling Src kinase localization.
Figure 5.
Pyk2 regulates invadopodium precursor assembly by recruiting Src kinase. (A) MDA–MB-231 cells stably expressing Src-TagRFP and knocked down for control, Pyk2, or FAK were plated on gelatin, fixed, and labeled for vinculin as a marker for focal adhesions. Colocalization masks are shown for Src with vinculin at focal adhesions. (B) Quantification of colocalization (Manders’ coefficient) of Src and vinculin at focal adhesions. n = 14–25 cells per group from three independent experiments. (C) MDA–MB-231 cells stably expressing Src-TagRFP and knocked down for control, Pyk2, or FAK were plated on gelatin, fixed, and labeled for Tks5 and cortactin as markers for invadopodium precursors. Colocalization masks are shown for Src with Tks5 and cortactin at invadopodium precursors. (D) Quantification of colocalization of Src with Tks5 and cortactin at invadopodium precursors of control, Pyk2 knockdown, or FAK knockdown cells. n = 22–30 cells per group from three independent experiments. (E) Representative immunoblot of MDA–MB-231 cells transiently transfected with control, Pyk2, or FAK siRNA. (F) Immunoblot of MDA–MB-231 cells overexpressing Pyk2, FAK, or control. Whole-cell lysates were blotted for either Pyk2 or FAK and for actin as loading control. (G) Control, Pyk2 overexpression (OE), and FAK overexpression cells were treated with the Src family kinase (SFK) inhibitor PP2, plated on gelatin-coated plates, and immunolabeled for cortactin and Tks5 as markers of invadopodium precursors. Bars: (main images) 10 µm; (insets) 2 µm. (H) Quantification of invadopodium precursors defined by colocalization of cortactin and Tks5. n = 44–61 cells per group from three independent experiments. (I) Quantification of invadopodium precursors defined by colocalization of cortactin and Tks5 in control, Pyk2 overexpression, or FAK overexpression cells that were treated with either control siRNA or Src siRNA. n = 70–93 cells per group from three independent experiments. (J) Quantification of invadopodium precursor formation in MDA–MB-231 cells stably expressing Src-TagRFP and treated with control, Pyk2 siRNA, or FAK siRNA, plated on gelatin-coated plates and labeled for cortactin and Tks5. n = 34–46 cells per group from three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Error bars represent SEM.
To further determine whether the opposing regulation of invadopodium precursor formation by Pyk2 and FAK is mediated via Src kinase, we examined the ability of the Src family inhibitor PP2 to alter the assembly of invadopodium precursors by Pyk2- and FAK-overexpressing cells. Overexpression of Pyk2 in MDA–MB-231 cells significantly increased the assembly of Tks5- and cortactin-rich invadopodium precursors, which was reduced by Src inhibition with PP2. As expected, overexpression of FAK significantly reduced the assembly of these structures, which was not changed by Src inhibition with PP2 (Fig. 5, G and H). Similar results were obtained using Src-specific knockdown (Fig. 5 I).
To examine whether Pyk2 and FAK function upstream or downstream of Src in regulating invadopodium precursor formation, we examined precursor formation in MDA–MB-231 cells stably overexpressing Src-TagRFP. Depletion of either Pyk2 or FAK in Src-overexpressing cells did not change precursor formation (Fig. 5 J). Together with the complementary experiment above in which we overexpressed Pyk2 or FAK and knocked down or inhibited Src and in accordance with previous publications (Chan et al., 2009; Kolli-Bouhafs et al., 2014), this result suggests that Src functions downstream of Pyk2 to regulate the assembly of invadopodium precursors in MDA–MB-231 breast cancer cells.
Pyk2 is required for cortactin tyrosine phosphorylation at invadopodium precursors
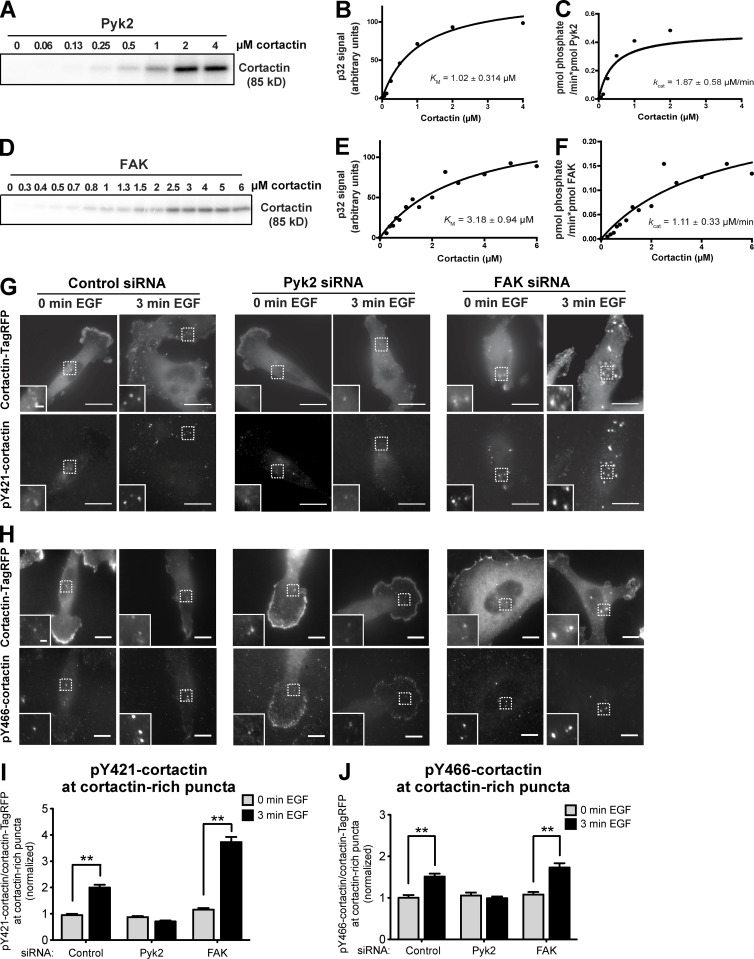
In the advanced stages of invadopodia assembly and maturation, kinases are activated and phosphorylate cortactin on tyrosine residues. These phosphorylations mediate invadopodia maturation and activation of invadopodial functions such as actin polymerization, matrix degradation, and tumor cell invasion. To examine whether Pyk2 phosphorylates cortactin, we performed the in vitro kinase assay using purified recombinant Pyk2 and cortactin. As shown in Fig. 6 (A–C), Pyk2 directly phosphorylated cortactin (KM = 1.02 ± 0.314 µM and kcat = 1.87 ± 0.58 µM/min). As previously described (Wang et al., 2011; Tomar et al., 2012), FAK was also able to phosphorylate cortactin in vitro, albeit with higher constants than Pyk2 (KM = 3.18 ± 0.94 µM and kcat = 1.11 ± 0.33 µM/min). These data suggest that Pyk2 is more adept in phosphorylating cortactin in vitro.
Figure 6.
Pyk2 phosphorylates cortactin in vitro and in breast cancer cells. (A–F) Increasing concentrations of recombinant His-tagged WT cortactin were incubated with constant concentrations of Pyk2 (10 nM; A–C) or FAK (10 nM; D–F) in the presence of [γ32]P-ATP. Kinase reactions were permitted to proceed for 5 min (Pyk2) or 30 min (FAK) and then quenched by adding Laemmli sample buffer and boiling for 5 min. Reactions were run on an SDS-PAGE gel and exposed to a PhosphorImager screen. Shown are representative phosphoimages and graphs from three independent experiments each. (G and H) MDA–MB-231 cells stably expressing cortactin-TagRFP were treated with control, Pyk2, or FAK siRNA, plated on FN/gelatin matrix, starved, and stimulated with EGF. Cells were fixed and labeled for phosphorylated cortactin (anti-pY421 in G or anti-pY466 in H) before (0 min) or after (3 min) EGF stimulation. Bars: (main images) 10 µm; (insets) 2 µm. (I) Quantification of pY421-cortactin/cortactin-TagRFP signal at cortactin-rich puncta. n = 489–660 puncta per group from three independent experiments. (J) Quantification of pY466 cortactin/cortactin-TagRFP signal at cortactin-rich puncta. n = 77–196 puncta per group from three independent experiments. **, P < 0.01. Error bars represent SEM.
To investigate which tyrosine of cortactin is phosphorylated by Pyk2, we used purified cortactin that was mutated in tyrosine 421 (Y421F), tyrosine 466 (Y466F), or a mutant in which the three tyrosines Y421, Y466, and Y482 were mutated to phenylalanine (3YF). Mutation of tyrosine Y421 in cortactin increased the KM to 8.47 ± 1.32 µM (Fig. S4, A and B), whereas the reaction using mutated tyrosine 466 did not reach saturation (Fig. S4, C and D). Mutation of the three tyrosines together eliminated the phosphorylation signal almost completely (Fig. S4 E). In conclusion, these data suggest that Pyk2 phosphorylates cortactin on tyrosine residues Y421, Y466, and Y482.
Given that cortactin phosphorylation on tyrosines Y421 and Y466 mediates mature invadopodial functions such as actin polymerization and matrix degradation (Oser et al., 2010) and based on our data above showing that Pyk2 can directly phosphorylate cortactin in vitro, we hypothesized that Pyk2 may phosphorylate cortactin in invadopodium precursors. To test this hypothesis, control, Pyk2-knockdown, and FAK-knockdown cells were treated with EGF to induce invadopodia formation and labeled with phosphorylation-specific antibodies for cortactin tyrosine Y421 or Y466 (Fig. 6, G–I). Cortactin phosphorylation was increased on both tyrosines in EGF-stimulated control cells, whereas no cortactin phosphorylation was detected on either tyrosine in Pyk2-depleted cells. Interestingly, and in accordance with our above data demonstrating increased invadopodium precursor formation and Src localization to invadopodia in FAK-knockdown cells, cortactin phosphorylation on tyrosine Y421 was significantly increased in FAK-depleted cells, whereas only a mild increase above control level was observed for tyrosine Y466 (Fig. 6, G and I). Altogether, these data suggest that both Pyk2 and FAK control cortactin phosphorylation in invadopodia in an opposite manner: although Pyk2 increases cortactin phosphorylation, FAK decreases the phosphorylation of cortactin in invadopodium precursors.
Pyk2 mediates direct and indirect tyrosine phosphorylation of cortactin in invadopodia
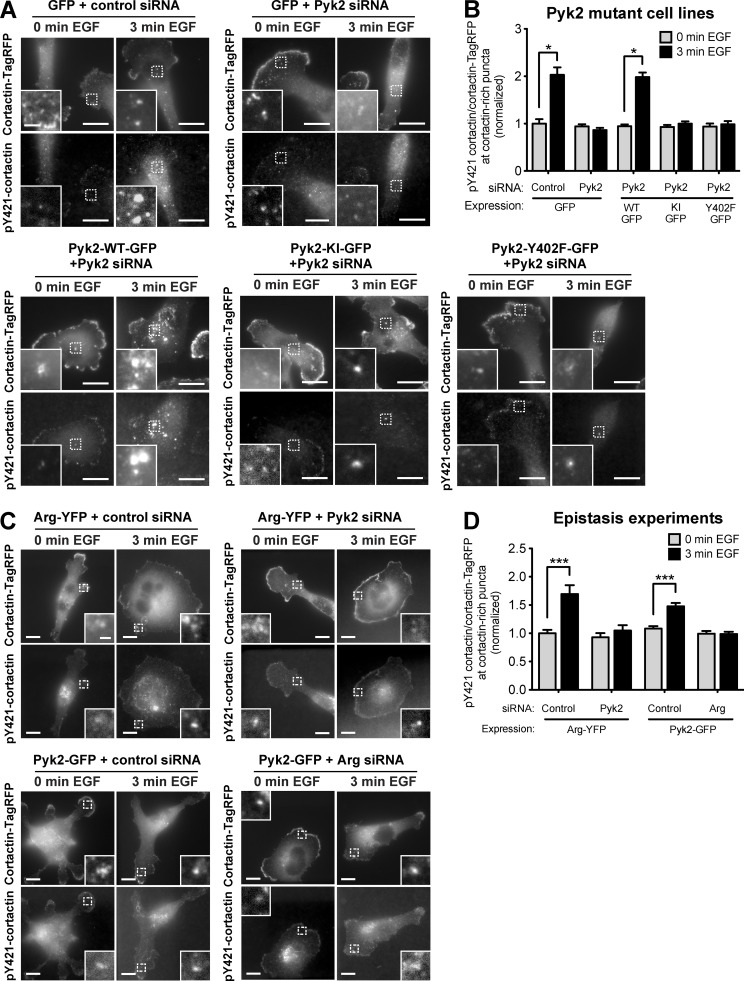
We have previously shown that Arg phosphorylates cortactin in invadopodia of breast cancer cells after activation by Src kinase (Mader et al., 2011). To determine whether Pyk2 phosphorylates cortactin directly or indirectly via Src recruitment and consequent Arg activation, we rescued Pyk2-knockdown cells by reexpressing either a kinase-inactive mutant (Pyk2-KI) or an autophosphorylation and Src-binding mutant of Pyk2 (Pyk2-Y402F). Interestingly, both Pyk2-KI and Pyk2-Y402F were unable to rescue cortactin phosphorylation at invadopodia of Pyk2-depleted cells, suggesting that Pyk2-mediated tyrosine phosphorylation of cortactin in invadopodia is dependent on both the kinase activity of Pyk2 as well as on the recruitment and activation of Src kinase (Fig. 7, A and B).
Figure 7.
Pyk2 mediates cortactin phosphorylation in invadopodia by activation of the Src–Arg pathway. (A) MDA–MB-231 cells stably expressing cortactin-TagRFP and either GFP or Pyk2-GFP WT and mutants were treated with control or Pyk2 siRNA, plated on FN/gelatin matrix, starved, and stimulated with EGF. Cells were fixed and labeled for phosphorylated cortactin (anti-pY421) before (0 min) or after (3 min) EGF stimulation. Bars: (main images) 10 µm; (insets) 2 µm. (B) Quantification of pY421-cortactin/cortactin-TagRFP signal at cortactin-rich puncta. n = 97–211 puncta per group from three independent experiments. KI, kinase inactive. (C) MDA–MB-231 cells stably expressing cortactin-TagRFP were treated with control siRNA, Arg siRNA, or Pyk2 siRNA and transfected with either Arg-YFP or Pyk2-GFP, plated on FN/gelatin matrix, starved, and stimulated with EGF. Cells were fixed and labeled for phosphorylated cortactin (anti-pY421) before (0 min) or after (3 min) EGF stimulation. Bars: (main images) 10 µm; (insets) 1 µm. (D) Quantification of pY421-cortactin/cortactin-TagRFP signal at cortactin-rich puncta. n = 65–118 puncta per group from three independent experiments. *, P < 0.05; ***, P < 0.001. Error bars represent SEM.
To confirm the direct and indirect effects of Pyk2 on cortactin phosphorylation, we used cells overexpressing Arg-YFP and treated with Pyk2 siRNA or a nonsilencing siRNA. Cells overexpressing Arg-YFP and treated with control siRNA showed an ∼1.7-fold increase in cortactin phosphorylation, whereas knockdown of Pyk2 in these cells eliminated most of the phosphorylation of cortactin, suggesting that overexpression of Arg cannot compensate for Pyk2 depletion. Similarly, overexpression of Pyk2-GFP could not rescue cortactin phosphorylation in invadopodia of cells that were knocked down for Arg, confirming that the two kinases work in the same signaling pathway for cortactin phosphorylation in addition to direct phosphorylation of cortactin by Pyk2 (Fig. 7, C and D).
Pyk2 is required for actin barbed end generation at invadopodia-rich cellular regions
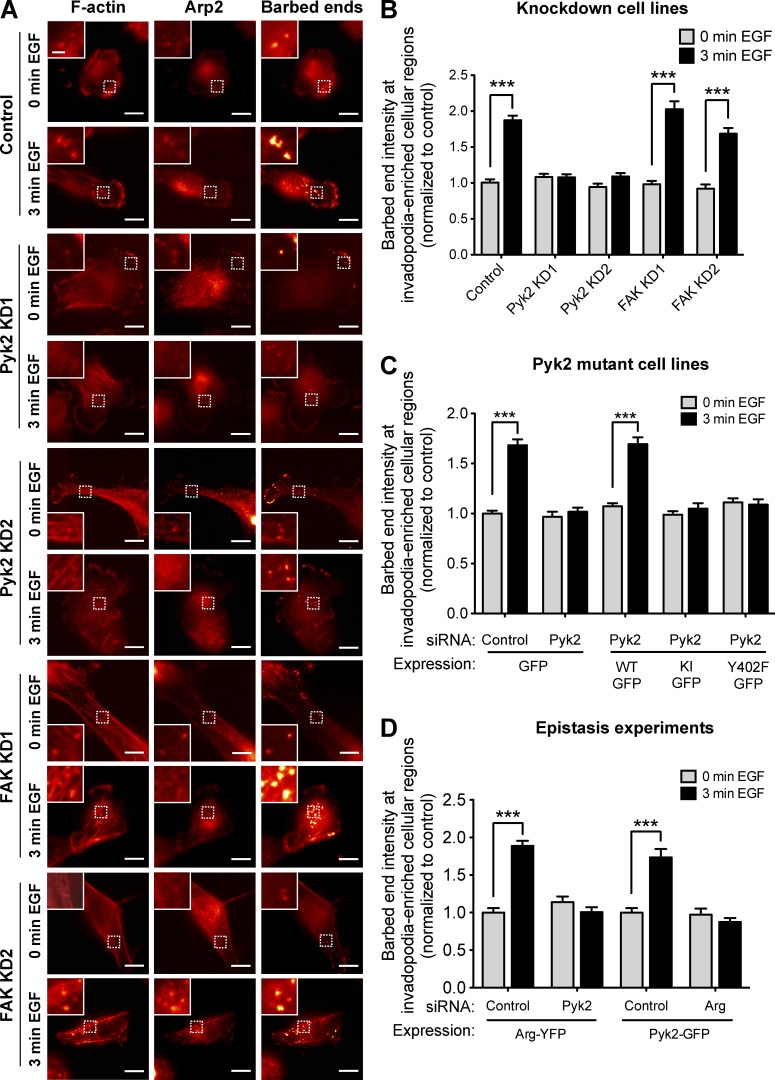
Free actin barbed end generation and consequent actin polymerization in invadopodia are induced by EGF-mediated tyrosine phosphorylation of cortactin (Yamaguchi et al., 2005; Oser et al., 2009). Because Pyk2 is required for cortactin tyrosine phosphorylation in invadopodia in response to EGF, we hypothesized that Pyk2 might regulate barbed end generation leading to actin polymerization in these structures. To test this hypothesis, we stimulated control, Pyk2-knockdown, or FAK-knockdown cells with EGF to synchronize the generation of free actin barbed ends within regions of cells rich in invadopodia (Fig. 8 A). We then quantified free actin barbed end generation specifically within these structures after EGF stimulation. Whereas control and FAK-knockdown cells generated approximately twofold increase in barbed end intensity in response to EGF stimulation, knockdown of Pyk2 completely disrupted generation of barbed ends at regions of cells rich in invadopodia (Fig. 8 B). Interestingly, and in accordance with these data, overexpression of Pyk2 in MDA–MB-231 cells generated an increase in barbed end intensity in EGF-stimulated cells, whereas overexpression of FAK in these cells completely disrupted this process (Fig. S4, G and H).
Figure 8.
Pyk2 regulates actin polymerization at regions of cells rich in invadopodia. (A) MDA–MB-231 cells stably expressing control, Pyk2 shRNA, or FAK shRNA were left untreated (0 min EGF) or stimulated with EGF for 3 min (3 min EGF). Cells were fixed and labeled for F-actin and Arp2 as invadopodia markers and for biotin–actin as a marker for newly formed barbed ends. Bars: (main images) 10 µm; (insets) 2 µm. (B) Quantification of free barbed ends as measured by mean biotin–actin intensity at stimulated invadopodia containing F-actin and Arp2. n = 40–101 invadopods per group from three independent experiments. (C) Quantification of free barbed ends at stimulated invadopodia of cells stably expressing either GFP or Pyk2-GFP WT, Pyk2-GFP kinase inactive (KI), or Pyk2-GFP Y402F and treated with control siRNA or Pyk2 siRNA. n = 54–126 invadopods per group from three independent experiments. (D) Quantification of free barbed ends at stimulated invadopodia of cells that were treated with control siRNA, Arg siRNA, or Pyk2 siRNA and transfected with either Arg-YFP or Pyk2-GFP. n = 22–59 invadopods per group from three independent experiments. ***, P < 0.001. Error bars represent SEM.
In agreement with our cortactin phosphorylation data above, a kinase-inactive mutant of Pyk2 or an autophosphorylation and Src-binding mutant of Pyk2 were unable to rescue barbed end formation at invadopodia of Pyk2-depleted cells, suggesting that Pyk2-mediated regulation of actin polymerization in invadopodia is dependent on both the kinase activity of Pyk2 and on the recruitment and activation of Src kinase (Fig. 8 C).
To determine whether Pyk2 regulates barbed end formation in invadopodia both directly and indirectly via activation of Src and Arg kinases, we used cells overexpressing Arg-YFP treated with Pyk2 siRNA or a nonsilencing control and cells overexpressing Pyk2-GFP and treated with Arg siRNA or a nonsilencing control. In accordance with cortactin phosphorylation in invadopodia, both combinations eliminated barbed end formation in invadopodia, suggesting that polymerization of actin in invadopodia is controlled both directly by Pyk2 and indirectly by Pyk2-mediated recruitment and activation of Src and Arg kinases. Collectively, our data suggest that Pyk2 is essential both directly and indirectly for EGF-mediated actin-free barbed end generation at regions of cells rich in invadopodia, whereas FAK may be inhibiting this process.
Knockdown of Pyk2 reduces MMP secretion and ECM degradation by breast cancer cells
At their final stage of maturation, invadopodia acquire the ability to focally degrade the ECM via recruitment and activation of MMPs (Artym et al., 2006; Ayala et al., 2008; Oser et al., 2009). This process allows the cells to escape through the basement membrane surrounding the primary tumor, to invade through the dense stromal ECM, and to penetrate through the endothelial basement membrane into blood vessels (Beaty and Condeelis, 2014).
To examine the ability of Pyk2 and FAK to regulate MMP-mediated ECM degradation by mature invadopodia, control, Pyk2-knockdown, and FAK-knockdown cells were plated on fluorescent fibronectin (FN)/gelatin matrix and allowed to degrade (Fig. 9 A). Quantification of the degradation area revealed a significant decrease in the ability of Pyk2-depleted cells to degrade the matrix. Although Pyk2-depleted cells exhibited a decreased ability to degrade the FN/gelatin matrix compared with control cells, FAK-knockdown cells showed significantly increased matrix degradation (Fig. 9 B). Similar results were obtained using control, Pyk2-knockdown, and FAK-knockdown MTLn3 rat adenocarcinoma cell lines (Fig. S2, C–F) as well as human osteosarcoma (U2OS) and epidermoid carcinoma (A431) cell lines (Fig. S3, D and H). Interestingly, and in agreement with our knockdown results, overexpression of Pyk2 in MDA–MB-231 cells significantly increased ECM degradation, whereas overexpression of FAK resulted in a substantial decrease in the area degraded by the cells (Fig. S4, I and J).
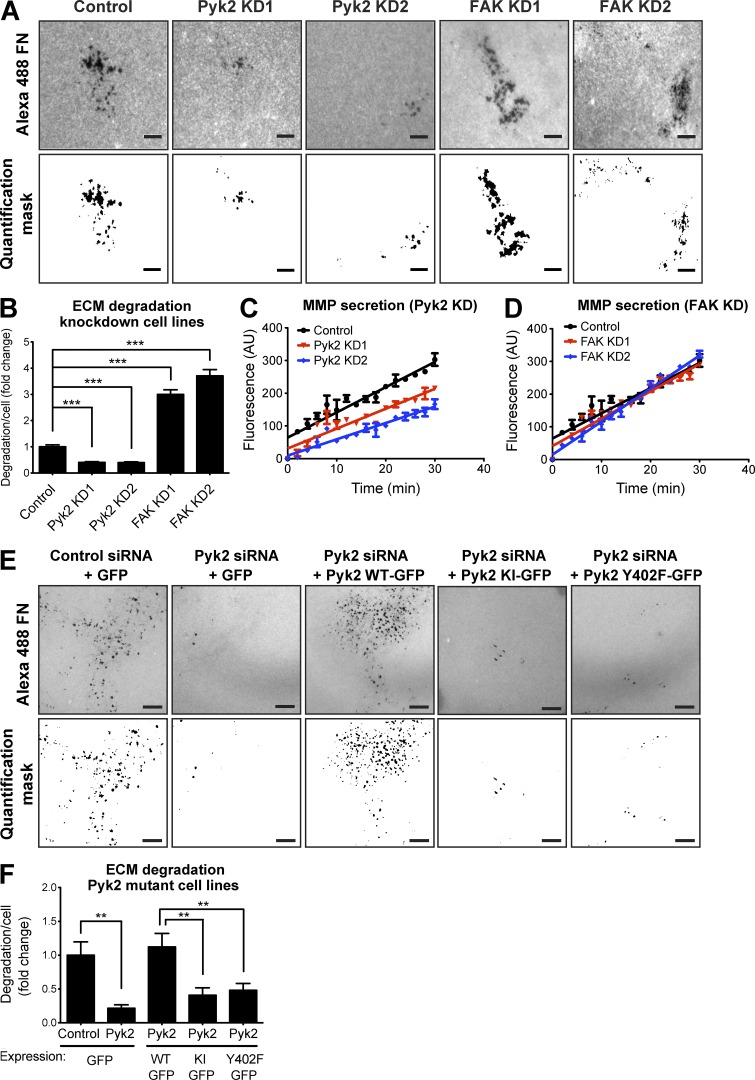
Figure 9.
Pyk2 and FAK oppositely regulate ECM degradation by breast cancer cells. (A) MDA–MB-231 cells stably expressing control, Pyk2 shRNA, or FAK shRNA were plated on Alexa Fluor 488 FN/gelatin matrix and allowed to degrade for 24 h. Shown are representative images (left) and quantification masks (right) of degradation areas. (B) Quantification of matrix degradation by control and knockdown (KD) cells. n = 280–475 fields per group from three independent experiments. (C and D) MDA–MB-231 cells expressing control, Pyk2 shRNA, or FAK shRNA were incubated in serum-free medium in the presence of a fluorogenic substrate peptide, and their MMP activity was measured over 30 min. Shown are linear regression graphs. n = 8–10 samples per group from three independent experiments. (E and F) Representative images (E) and quantification (F) of matrix degradation by cells expressing WT Pyk2-GFP, kinase-inactive (KI) Pyk2-GFP, or Y402F-Pyk2 GFP and treated with control or Pyk2 siRNA. Bars, 10 µm. n = 42–73 cells per group from three independent experiments. **, P < 0.01; ***, P < 0.001. Error bars represent SEM.
To investigate whether decreased or increased matrix degradation in Pyk2- and FAK-knockdown cells results from changes in MMP secretion and activation, we performed a fluorogenic substrate-based MMP assay (Knight et al., 1992) using serum-free medium conditioned by control, Pyk2-knockdown, or FAK-knockdown cells. Depletion of Pyk2 from MDA–MB-231 cells significantly reduced MMP secretion, whereas FAK depletion showed no change in MMP secretion and activation despite a significant increase in ECM degradation (Fig. 9, C and D). In accordance with our above invadopodium precursor formation and cortactin phosphorylation data, rescue of Pyk2-depleted cells by either kinase-inactive or autophosphorylation and Src-binding mutants significantly reduced matrix degradation (Fig. 9, E and F). Altogether, these data indicate that Pyk2 controls MMP-mediated ECM degradation, whereas FAK negatively regulates this function.
Pyk2 and FAK regulate tumor cell invasion via distinct mechanisms
Invadopodia gain their protrusive ability by combining the physical force generated by actin polymerization with the chemical activity of MMP-mediated ECM degradation (Sibony-Benyamini and Gil-Henn, 2012). As Pyk2 regulates cortactin phosphorylation–mediated actin polymerization and ECM degradation in breast cancer cells, we hypothesized that it might also regulate the ability of cells to invade through an ECM barrier. To test this hypothesis, we examined the ability of serum-starved control, Pyk2-, and FAK-knockdown cells to invade through Matrigel-coated membranes toward complete medium. As demonstrated in Fig. 10 (A and B), Pyk2-knockdown cells exhibited a dramatic reduction in proteolysis-dependent invasion through Matrigel-coated membranes. Interestingly, despite the fact that FAK depletion increases MMP-dependent ECM degradation (Fig. 9), FAK-knockdown cells showed lower ECM invasion.
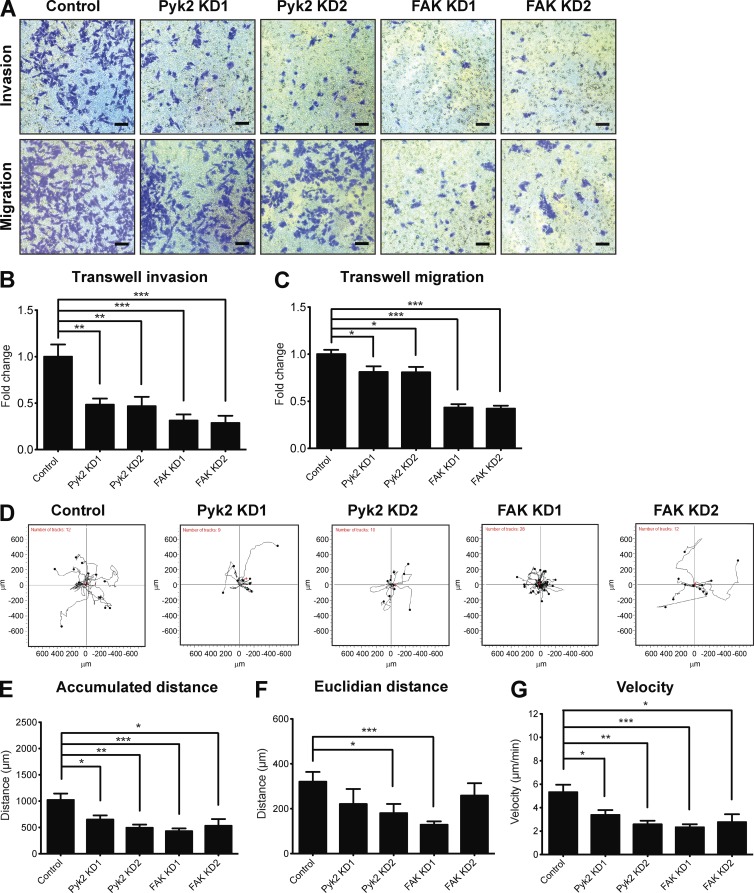
Figure 10.
Pyk2 and FAK regulate migration and invasion via distinct mechanisms. (A) MDA–MB-231 cells stably expressing control, Pyk2 shRNA, or FAK shRNA were plated on Matrigel-coated (top) or uncoated (bottom) membranes, allowed to invade or migrate for 24 h, fixed, and stained. Representative images of cells that migrated or invaded to the lower part of the membranes are shown. Bars, 5 µm. (B) Quantification of cell invasion through Matrigel-coated membranes. n = 12–20 fields per group from three independent experiments. (C) Quantification of cell migration through uncoated membranes. n = 24 (control, Pyk2 KD1, Pyk2 KD2, and FAK KD1) or 20 (FAK KD2) fields from three independent experiments. (D) Trajectory plots of single-cell migration experiments of control, Pyk2-knockdown, and FAK-knockdown cell lines. (E–G) Quantification of accumulated distance (total cell path length; E), Euclidian distance (the shortest distance between the beginning and end points; F), and velocity (G) of migrating cells. n = 9–28 cells per group from three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Error bars represent SEM.
FAK plays a critical role in 2D cell migration and has been shown to coordinate lamellipodial formation as well as focal adhesion dynamics. Moreover, focal adhesion proteins including FAK have been shown to regulate migration of cells through ECM by affecting protrusion activity and matrix deformation (Fraley et al., 2010). To investigate whether decreased ability of FAK-knockdown cells to invade results from other mechanisms such as reduced focal adhesion–dependent motility, we performed the transwell migration assay using uncoated membranes. As demonstrated in Fig. 10 C, FAK-knockdown cells exhibited a dramatic reduction in migration through uncoated membranes toward complete medium, whereas Pyk2-knockdown cells exhibited a slight decrease in migration. To further examine the ability of Pyk2- and FAK-depleted cells to migrate in 2D, control or knockdown cells were plated on FN and allowed to randomly migrate over it. As demonstrated in Fig. 10 (D–G), both Pyk2-depleted and FAK-depleted cells showed reduced 2D migration over an ECM substrate.
In line with these results, cells overexpressing Pyk2 showed increased ECM invasion as well as transwell and 2D migration. Surprisingly, overexpression of FAK significantly decreased transwell invasion and migration while also increasing the velocity and distance traveled by these cells in 2D (Fig. S5). Collectively, these results suggest that Pyk2 or FAK depletion results a significant decrease in cell motility and invasion. Although the decrease in ECM invasion in Pyk2-knockdown cells may be explained by reduction in cortactin phosphorylation–dependent actin polymerization and matrix degradation, the phenotype of FAK-knockdown cells is mainly attributed to a defect in focal adhesion–dependent functions. Lack of motility in FAK-depleted cells could also provide an explanation to their increased matrix degradation capability despite no significant increase in MMP secretion.
Discussion
It has been established that tyrosine kinase signaling has an important role in regulating invadopodial functions such as matrix degradation (Bowden et al., 2006; Clark et al., 2007; Ayala et al., 2008) and actin polymerization (Oser et al., 2009, 2010; Mader et al., 2011); however, little is known about the mechanisms of regulation or the role of specific kinases in these processes. In this study, we identified the nonreceptor tyrosine kinase Pyk2 as a novel regulator of both invadopodium precursor formation as well as invadopodium maturation and function. Pyk2 mediates invadopodium maturation and function by controlling cortactin tyrosine phosphorylation and consequent downstream signaling. Pyk2 is critical for regulating invadopodial activation in response to EGF by promoting the generation of free actin barbed ends. Maturation of invadopodium precursors into ECM-degrading invadopodia is dependent on Pyk2, which mediates this function by controlling focal MMP secretion and activation in invadopodia. By coordinating invadopodial actin polymerization with focal MMP-dependent ECM degradation, Pyk2 controls tumor cell invasion. To our knowledge, this is the first documentation of Pyk2 as a critical regulator of cortactin in invadopodia.
Pyk2 controls cortactin tyrosine phosphorylation in invadopodia by a combination of direct and indirect mechanisms
Src has been shown to localize to invadopodia, where it regulates their formation by inducing reactive oxygen species production (Diaz et al., 2009; Gianni et al., 2009); however, the identity of the protein that recruits Src to invadopodia has not been determined to date. We show here, for the first time, that Pyk2 recruits Src kinase into invadopodium precursors. Moreover, we demonstrate that Pyk2 mediates the assembly of these structures by coordinating with FAK on the spatial regulation of Src kinase in breast cancer cells.
We have previously shown that invadopodial Src activates Arg kinase to induce cortactin phosphorylation in invadopodia as well as their consequent maturation and activation (Mader et al., 2011). In this study, we showed that Pyk2 controls cortactin phosphorylation in invadopodia by both direct binding and phosphorylation of cortactin as well as by indirect recruitment and activation of Src and Arg kinases. Our previous and current data indicate that, although Arg and Pyk2 cannot compensate for each other, simultaneous activation of both the direct and Src-Arg–mediated pathways is necessary for efficient cortactin phosphorylation in invadopodia.
ECM invasion is mediated by both invadopodia- and focal adhesion–dependent mechanisms
Our experiments demonstrated that both Pyk2 and FAK are essential for invasion through the ECM. Regulation of tumor cell invasion by Pyk2 could be attributed to its role in regulating Src-mediated invadopodium precursor formation as well as to its role in controlling EGF-mediated cortactin phosphorylation and consequent actin polymerization and MMP-mediated ECM degradation. FAK depletion showed decreased tumor cell invasion despite elevated cortactin phosphorylation, actin polymerization, and MMP-mediated ECM degradation. The increase in invadopodial functions could be explained by relocalization of Src kinase to invadopodium precursors and consequent formation and activation of these structures in FAK-depleted cells (Chan et al., 2009; Kolli-Bouhafs et al., 2014). However, despite the increase in invadopodial functions, FAK-knockdown cells showed a significant reduction in ECM invasion. Additional investigation revealed a significant reduction in the ability of FAK-depleted cells to migrate toward serum-containing medium or randomly over an ECM substrate. FAK plays a critical role in 2D cell migration and has been shown to coordinate lamellipodial formation as well as focal adhesion turnover and dynamics. Moreover, focal adhesion proteins including FAK have been shown to regulate 3D migration of cells through ECM by affecting protrusion activity and matrix deformation (Fraley et al., 2010).
We suggest a model by which both Pyk2 and FAK regulate tumor cell invasion albeit via different mechanisms. Although Pyk2 regulates invasion mostly by controlling cortactin phosphorylation–dependent maturation and activation of invadopodia, FAK controls ECM invasion by regulating focal adhesion–dependent motility and functions. Whether Pyk2 and FAK use these distinct mechanisms for regulating in vivo invasion and metastatic dissemination is a subject for future investigation.
Pyk2 regulates both migration and invasion in breast cancer cells
Pyk2 localizes to invadopodium precursors in MDA–MB-231 breast cancer cells, but it can also colocalize with FAK to vinculin-rich focal adhesions. Although Pyk2 localization was described as mainly cytosolic or perinuclear, it was also suggested to localize to focal adhesions in certain cancer cell lines (Litvak et al., 2000; Selitrennik and Lev, 2015). Indeed, breast cancer cells that were depleted of Pyk2 showed a significant decrease in 2D motility in addition to their decrease in invadopodial-mediated functions and consequent matrix invasion.
An emerging question is why both Pyk2 and FAK localize to focal adhesions in breast cancer cells. One possible explanation is that focal adhesions fulfill an indispensable role in cancer invasiveness and metastasis. Continuing this line, Pyk2 and FAK may be able to compensate for each other to fulfill the same function in focal adhesions in the case where one of the proteins is disabled. Indeed, it was previously shown that FAK-mediated focal adhesion turnover and consequent cell motility is regulated by a FAK–cortactin signaling complex (Wang et al., 2011; Tomar et al., 2012). Cortactin is a common substrate and interactor of both Pyk2 and FAK; however, it is left to be determined whether Pyk2 directly interacts with cortactin at focal adhesions. Our random migration experiments indicate that deletion of either Pyk2 or FAK ablates the motility of breast cancer cells, suggesting that Pyk2 and FAK cannot compensate for each other in 2D cell migration.
A second explanation for localization of both Pyk2 and FAK to focal adhesions could be that they have complementary or opposite roles that are executed by distinct downstream interactors and signaling pathways. Careful examination of protein–protein interactions and specific substrates that are unique to either Pyk2 or FAK in focal adhesions should resolve this enigma.
Materials and methods
Protein microarrays
Protein–protein and kinase–substrate interaction profiling were performed by Invitrogen. In both screens, the ProtoArray human protein microarray (5.0) containing 9,480 purified human proteins was used as a platform. For the protein–protein interaction screen, purified biotinylated Pyk2 was used to probe the microarrays at concentrations of 5 ng/µl and 50 ng/µl. As a positive control, 50 ng/µl of calmodulin kinase 1 was probed with the microarray that contained calmodulin, a known binding partner of calmodulin kinase 1. The results from this assay were compared with a negative control assay in which the Pyk2 probe protein was excluded. All interactions of the probe protein with the immobilized proteins on the microarrays were evaluated by five specified statistical threshold criteria within the array: z score (normalized fluorescent signal), z factor (background-subtracted pixel intensity value), signal used (background-subtracted pixel intensity value), replicate spot coefficient of variations, and confidence interval p-value (the probability that a signal is derived from the distribution of signals arising from a set of defined negative controls). The obtained values were compared with interactions observed on the negative control array. A total of 64 human proteins on the array were identified as candidate interactors for Pyk2.
For the kinase substrate interaction screen, purified Pyk2 was used to probe the microarrays at 5 nM and 50 nM in duplicate in the presence of [γ33]P-ATP. As a positive control, 100 nM of MAPK14/p38α was probed with the microarray. The results from this assay were compared with a negative control assay in which Pyk2 kinase was excluded. Phosphorylation of all possible kinase substrates on the experimental arrays was evaluated within the array by the five statistical criteria as above (z score, z factor, signal used, replicate spot coefficient of variations, and confidence interval p-value) and then compared with autophosphorylation of corresponding proteins observed on the negative control assay. A total of 86 human proteins on the array were identified as candidate substrates of Pyk2.
Bioinformatic analysis and literature mining
The GeneCards and UniProt databases were used to classify proteins from the protein–protein interaction screen and the kinase substrate interaction screen by manual curation. The novelty of substrates and interactors that were identified in the protein–protein and kinase–substrate interaction screens was examined by overlapping data from the two screens with known Pyk2 substrates and interactors that were identified by Ingenuity Pathway Analysis (http://www.ingenuity.com; QIAGEN).
Overlap between proteins from the protein–protein interaction and kinase–substrate interaction screens was calculated using Venny 2.1 (Oliveros, 2015). The statistical significance of overlap between the two groups of proteins was calculated using hypergeometric distribution.
Comparative literature mining was performed using two different automated literature-mining tools, GLAD4U and ALS (Cytoscape; Agilent Labs). The GLAD4U search was performed using the query “invadopodia” and was limited to human context with a threshold of 0.01. The ALS search was performed through Cytoscape using the query “invadopodia” and was limited to Homo sapiens with limited interaction lexicon. The combined lists of proteins identified by GLAD4U and ALS (a total of 425 nonredundant proteins) and the list of proteins identified in both of the ProtoArray screens (a total of 126 nonredundant proteins) were used to build networks of physical and functional associations in the STRING database. Networks were overlaid using Cytoscape (Shannon et al., 2003), and overlapped proteins were validated by manual literature search in PubMed.
Cell culture
MDA–MB-231 human breast adenocarcinoma cells, U2OS human osteosarcoma cells, and A431 human epidermoid carcinoma of the skin were obtained from the American Type Culture Collection. MDA–MB-231 and U2Os were cultured in DMEM/10% FBS, and A431 was cultured in DMEM/10% FBS and supplemented with sodium pyruvate. MTLn3 rat mammary adenocarcinoma cells (provided by G. Nicolson, Institute for Molecular Medicine, Huntington Beach, CA; Neri and Nicolson, 1981) were cultured in DMEM/5% FBS. For EGF stimulation experiments, MDA–MB-231 cells were starved for 16 h in 0.5% FBS/0.8% BSA in DMEM and then for 10 min in L15 media/0.345% BSA. Cells were then stimulated with 2.5 nM EGF (Invitrogen) for 3 min.
For Src inhibition experiments, MDA–MB-231 cells were pretreated with 2 µM PP2 (Sigma-Aldrich) for 18 h, detached, and plated on gelatin-coated plates for additional 4 h in presence of PP2. Cells were then fixed and immunofluorescently labeled for invadopodium precursor markers.
DNA constructs, RNA interference, and transfection
Human Pyk2 and FAK were cloned from human cDNA library into the retroviral pBABE plasmid. Human Tks5-GFP was cloned from N1-Tks5-GFP (provided by S. Courtneidge, Oregon Health and Science University, Portland, OR) into the retroviral plasmid pQCTK. pLXSN-Arg-YFP retroviral plasmid containing murine Arg has been previously described (Mader et al., 2011).
For shRNA-mediated knockdown in MDA–MB-231 cells, the following sequences were cloned into the pLKO.2 retroviral plasmid: Pyk2 KD1, 5′-GGTGGTGGTACCAGTAGAT-3′; Pyk2 KD2, 5′-GGATCATCATGGAATTGTA-3′; FAK KD1, 5′-GGGCATCATTCAGAAGATA-3′; and FAK KD2, 5′-TAGTACAGCTCTTGCATAT-3′.
For shRNA-mediated knockdown in MTLn3 cells, the following sequences were cloned into the pSUPER-Retro plasmid (Oligoengine): Pyk2 KD1, 5′-GGTGGTGGTACCAGTAGAT-3′; Pyk2 KD2, 5′-GGATCATCATGGAATTGTA-3′; FAK KD1, 5′-GGGAATCATTCAGAAGATA-3′; and FAK KD2, 5′-TAGTACAGCACTCGCGTAT-3′.
For siRNA-mediated knockdown, control nonsilencing siRNA SMARTpool (D-001810-10), Pyk2 siRNA SMARTpool (L-003165-00), FAK siRNA SMARTpool (L-003164-00), Src siRNA SMARTpool (L-003175-00; Mader et al., 2011), and Arg siRNA SMARTpool (L-003101-00; Mader et al., 2011) were obtained from GE Healthcare.
Pyk2-WT-GFP, Pyk2-K457A-GFP (kinase-inactive mutant), and Pyk2-Y402F-GFP (autophosphorylation and Src SH2 domain–binding mutant) were generated by subcloning WT and mutant Pyk2 cDNAs (Gil-Henn et al., 2007) into the pQCTK-GFP retroviral plasmid. Pyk2-PRR-GFP constructs containing mutations in proline-rich region 2 (PRR2; P714A, P717A, and P720A) or proline-rich region 3 (PRR3; P856A and P859A) of Pyk2 were generated from pQCTK-Pyk2-WT-GFP by the QuikChange site-directed mutagenesis kit (Agilent Technologies). Endogenous Pyk2 was knocked down in stable cell lines expressing WT or mutant Pyk2 using the Pyk2 KD1 siRNA sequence 5′-GGTGGTGGTACCAGTAGAT-3′ (GE Healthcare). siRNA transfections were performed by resuspending 106 MDA–MB-231 cells in 100 µl Ingenio electroporation solution (Mirus Bio LLC) with 2 µM siRNA 72 h before each experiment.
Cell line generation
Stable knockdown or overexpressing cell lines were generated by transfecting pLKO.2 or pBABE viral expression vectors carrying shRNA sequences or overexpression constructs into HEK293T- or GP2-packaging cells, respectively. Viral supernatants were used to infect MDA–MB-231 or MTLn3 cells, which were then selected with puromycin. Knockdown or overexpression levels were verified by Western blotting. Control cell lines were generated by infection with viral supernatants containing empty pLKO.2 (control for knockdown cell lines) or empty pBABE (control for Pyk2- and FAK-overexpressing cell lines).
MDA–MB-231 cells stably expressing cortactin-TagRFP and Src-Tag-RFP were previously described (Oser et al., 2010; Mader et al., 2011). MDA–MB-231 cells stably expressing both cortactin-TagRFP and Tks5-GFP were generated by infecting the cortactin-TagRFP cell line with viral supernatant that was collected from GP2 cells that were transfected with pQCTK-Tks5-GFP. Stable cell lines expressing GFP, Pyk2-WT-GFP, and GFP-tagged mutants of Pyk2 were generated by transfecting pQCTK-Pyk2-GFP plasmids into GP2-packaging cells. Viral supernatants were used to infect MDA–MB-231 cells or cortactin-TagRFP cells, which were then selected with hygromycin. Expression levels of GFP-tagged Pyk2 were compared with endogenous protein levels in noninfected MDA–MB-231 cells and in cells expressing empty GFP control plasmid by Western blotting.
Antibodies and reagents
For immunofluorescence, anti-cortactin (ab-33333) was obtained from Abcam; anti–pY402-Pyk2 and anti–pY397-FAK were obtained from Invitrogen; anti-Arp2 (H-84; SC-15389) and anti-Tks5 (FISH M-300; SC-30122) were obtained from Santa Cruz Biotechnology, Inc.; and antivinculin (clone hVIN1; V9131), anti–pY421-cortactin (C0739), and anti–pY466-cortactin (C0864) were obtained from Sigma-Aldrich. Rhodamine-labeled phalloidin and Alexa Fluor–conjugated secondary antibodies were obtained from Molecular Probes. For Western blotting, anticortactin (clone 4F11; 05-180) and antiphosphotyrosine (clone 4G10; 05-321) were obtained from EMD Millipore; anti-Pyk2 (3480 and 3292) was obtained from Cell Signaling Technology; anti-FAK (clone 77; 610088) was obtained from BD; anti-GFP (clones 7.1 and 13.1; 11814460001) was obtained from Roche; and anti–β-actin (clone AC-15; A5441) was obtained from Sigma-Aldrich. Secondary antibodies (goat anti–mouse 680LT and goat anti–rabbit 800CW) were obtained from LI-COR Biosciences.
2.5D invadopodia localization assay
The 2.5D invadopodia localization assay was adapted from Magalhaes et al. (2011). In brief, growth factor–reduced Matrigel (2.5 mg/ml; BD) was mixed with 0.2 mg/ml Alexa Fluor 405 gelatin (1:20 dilution) and applied to the top chamber of a 1.0-µm transwell support (Corning). Excess Matrigel/gelatin solution was removed, and both chambers were allowed to equilibrate in plain DMEM at 37°C for 1 h. After equilibration, the bottom chamber was filled with 750 µl of DMEM/10% FBS. 30,000 cells were resuspended in 200 µl of DMEM/0.5% FBS, plated in the top chamber, and allowed to invade for 24 h. Cells were fixed in 3.7% paraformaldehyde and immunofluorescently labeled for cortactin and pY402-Pyk2. Membranes were imaged using an inverted laser-scanning confocal microscope (LSM780; 60× 1.4 NA with oil objective with ZEN black edition acquisition software; ZEISS). 3D invadopodia images were reconstructed using Imaris 8.0.0 (Bitplane).
Protein purification
High Five cells (Invitrogen) expressing His-tagged Pyk2, His-tagged FAK, and His-tagged cortactin were lysed in lysis buffer containing 50 mM Hepes, pH 7.25, 500 mM NaCl, 5% glycerol, 1% Triton X-100, 20 mM imidazole, 1 mM DTT, 0.5 mM EDTA, and protease inhibitors. The extract was clarified by ultracentrifugation of 100,000 g for 45 min at 4°C. His-tagged proteins were loaded on an Ni-NTA column (QIAGEN), washed with 10 column volumes of wash buffer A (20 mM Hepes, pH 7.25, 500 mM NaCl, 5% glycerol, 0.01% Triton X-100, 20 mM imidazole, 1 mM DTT, and 0.5 mM EDTA), then with 10 column volumes of wash buffer B (20 mM Hepes, pH 7.25, 1 M NaCl, 5% glycerol, 0.01% Triton X-100, 20 mM imidazole, 1 mM DTT, and 0.5 mM EDTA), and again in 10 column volumes of wash buffer A. Proteins were eluted from the column using elution buffer (20 mM Hepes, pH 7.25, 500 mM NaCl, 5% glycerol, 0.01% Triton X-100, 100 mM imidazole, 1 mM DTT, and 0.5 mM EDTA). Before use in assays, all proteins were buffer-exchanged into assay buffer containing 25 mM Hepes buffer, pH 7.25, 100 mM NaCl, 0.01% Triton X-100, 5% glycerol, and 1 mM DTT using a G-25 Sephadex column (GE Healthcare).
Quantitative in vitro binding assay
Quantitative in vitro binding assay was performed as described previously by Lapetina and Gil-Henn (2017). In brief, purified cortactin was buffer exchanged into 3.65× PBS buffer using a G-25 Sephadex column by gravity flow and covalently coupled to AminoLink beads (Thermo Fisher Scientific) according to the manufacturer’s instructions to a final concentration of 1 µM. The remaining active sites on protein-linked beads were blocked in 1 M Tris-HCl, pH 7.25, and 100 mg/ml BSA. Recombinant Pyk2 or FAK were buffer exchanged into binding buffer (25 mM Hepes buffer, pH 7.25, 100 mM NaCl, 0.01% Triton X-100, 5% glycerol, and 1 mM DTT) using a G-25 Sephadex column. 490 µl of serial dilutions of Pyk2 (0–20 µM) and of FAK (0–17 µM) were incubated with 10 µl of cortactin-linked beads or 10 µl of BSA control beads for 1 h at 4°C with rotation. After incubation, the supernatant was removed, beads were briefly washed, and bound material was recovered by boiling in Laemmli sample buffer and then separated on 10% SDS-PAGE. Gel bands were resolved using Coomassie blue silver stain, and their densities were quantified using ImageJ software (National Institutes of Health). For measurements of binding affinities (Kd), band densities were plotted against concentration of the free solution protein, and binding isotherms were set using Prism (GraphPad Software) using the one site-specific binding equation: Y = Bmax × X/X + Kd. In this equation, Y equals the specific binding signal, X equals the concentration of Pyk2 or FAK added to cortactin beads, and Bmax is the maximum specific binding. The equilibrium constant Kd is solved as the value of X when Y equals 50% of Bmax. To ensure saturation of the curves, Pyk2 or FAK maximal concentrations of at least five times the Kd were used in the assay.
In vitro kinase assay
Purified Pyk2 or FAK (10 nM) were preincubated with increasing concentrations of cortactin (0–4 µM for Pyk2 or 0–6 µM for FAK) for 5 min at 32°C. The reactions were started by addition of 5 µM cold ATP and 0.75 µCi of radiolabeled [γ32]P-ATP (BLU502H; PerkinElmer) in kinase buffer (25 mM Hepes, 100 mM NaCl, 5% glycerol, 5 mM MgCl2, 5 mM MnCl2, 1 mM Na3VO4, and 1 mM DTT) and incubated at 32°C for an additional 5 min (Pyk2) or 30 min (FAK). All reactions were quenched with Laemmli sample buffer, boiled, and separated on 10% SDS-PAGE. The gels were exposed to a PhosphorImaging screen overnight and scanned using a Personal Molecular Imager (Bio-Rad Laboratories), and then band densities were quantified using the Quantity One software (Bio-Rad Laboratories). The values for each concentration series were analyzed by Prism software and fitted to Michaelis-Menten isotherms, Y = Vmax * X/(KM + X), where Y is the enzyme velocity, Vmax is the maximum enzyme velocity, X is the substrate concentration, and Km is the substrate concentration needed to achieve a half-maximal enzyme velocity.
After exposure and phosphorimaging scanning, gels were stained with Coomassie blue silver to visualize cortactin bands. Specific and negative control bands were cut out of the gel and quantified by a scintillation counter along with 2 µl sample from the kinase assay. The number of counts per minute was calculated and corrected to integrated density from ImageJ software, and kcat values were determined as previously described (Boyle and Koleske, 2007).
Immunoprecipitation and Western blotting
Cells were washed twice in ice-cold PBS and lysed in modified radioimmunoprecipitation assay buffer (50 mM Tris, pH 7.2, 150 mM NaCl, 1% NP-40, 0.5% deoxycholate, 0.1% SDS, 1 mM EDTA, 2 mM NaF, 1 mM Na3VO4, and protease inhibitors). Total protein concentration was determined using DC Protein Assay (Bio-Rad Laboratories), and equal amounts were loaded on SDS-PAGE, transferred to a nitrocellulose membrane, blocked in Odyssey blocking buffer, incubated with primary and secondary antibodies, and imaged using the Odyssey CLx imaging system (LI-COR Biosciences). Cells were lysed in lysis buffer (25 mM Hepes, pH 7.4, 120 mM NaCl, 1 mM EGTA, 1% Triton X-100, 10% glycerol, 0.75 mM MgCl2, 2 mM NaF, 1 mM Na3VO4, and protease inhibitors). 1 mg of protein was incubated with 1 µg of antiphosphotyrosine antibodies and 30 µl of protein A/G agarose beads for 4 h in 4°C and washed with washing buffer (25 mM Hepes, pH 7.4, 120 mM NaCl, 1 mM EGTA, 0.1% Triton X-100, 10% glycerol, and 0.75 mM MgCl2). Proteins were released from beads by boiling the sample in Laemmli sample buffer, separated on SDS-PAGE, and subjected to Western blot analysis as above.
Invadopodium precursor formation assay
The invadopodium precursor formation assay was performed as previously described (Beaty et al., 2013). In brief, gelatin was conjugated to Alexa Fluor 405 dye (Thermo Fisher Scientific). MatTek dishes (MatTek Corporation) were treated with 1 N HCl and coated with 50 µg/ml poly-l-lysine. A 0.2% gelatin solution was prepared in PBS, and a 1:40 mixture of Alexa Fluor 405–labeled gelatin/unlabeled gelatin was warmed to 37°C before addition to the poly-l-lysine–coated plates. Gelatin was crosslinked with 0.01% glutaraldehyde followed by quenching with 5 mg/ml sodium borohydride. 150,000 MDA–MB-231 cells or 75,000 MTLn3 cells were plated on Alexa Fluor 405–labeled gelatin plates for 4 h (MDA–MB-231) or overnight (MTLn3) and fixed in 3.7% paraformaldehyde. Cells were permeabilized with 0.1% Triton X-100, blocked with 1% FBS and 1% BSA in PBS, and then labeled with anti-Tks5 and anticortactin. Images were acquired using an inverted fluorescent microscope (AF6000; 63× 1.3 NA with oil objective with LAS AF acquisition software; Leica Microsystems) equipped with an ORCA-Flash 4.0 V2 digital CMOS camera (Hamamatsu Photonics). Invadopodium precursors were identified as Tks5 and cortactin-rich punctate structures that do not colocalize with degradation areas, whereas mature invadopodia were identified as Tks5 and cortactin-rich puncta that colocalize with degradation areas. Total invadopodia includes both degrading and nondegrading Tks5 and cortactin-rich punctate structures.
Colocalization experiments
MDA–MB-231 cells stably knocked down for either Pyk2 or FAK or MDA–MB-231 cells stably expressing Src-TagRFP and knocked down with Pyk2 or FAK siRNA were plated on gelatin for 4 h, fixed using 3.7% paraformaldehyde, and fluorescently labeled for vinculin or cortactin and Tks5. Images were acquired using an inverted fluorescent microscope (AF6000; 63× 1.3 NA with oil objective). Images were processed using the ImageJ spot enhancer filter 2D, and threshold levels were set to select only invadopodium precursors or focal adhesions. Images were then analyzed for colocalization (Manders’ coefficient) using the JACoP colocalization plugin for ImageJ.
Matrix degradation assay
FN/gelatin matrix was prepared as described previously (Chen, 1989). In brief, MatTek dishes were treated with 2.5% gelatin/2.5% sucrose, crosslinked with 0.5% glutaraldehyde, treated with 10 µg/ml of fluorescently labeled FN (Alexa Fluor 568; Invitrogen), and then treated with 1 mg/ml NaBH4 in PBS. 125,000 MDA–MB-231 cells or 100,000 MTLn3 cells were plated on the FN/gelatin matrix for 24 h or 16 h, respectively. Cells were then fixed in 3.7% paraformaldehyde, and random fields were imaged using an inverted fluorescent microscope (AF6000; 40× 1.3 NA with oil objective; Leica Microsystems). Matrix degradation was analyzed by quantifying the mean degraded area in pixels per field using ImageJ and then normalized to the number of cells per field.
MMP activity assay
MDA–MB-231 cells (10,000 cells per well) were seeded in a glass-bottomed 96-well plate (black walls; Greiner Bio-One), allowed to adhere overnight, washed three times in serum-free DMEM without phenol red, and incubated at 37°C in 150 µl of the same medium. After 4 h, the fluorogenic substrate Mca-Lys-Pro-Leu-Gly-Leu-Dpa-Ala-Arg-NH2 (BML-P276-0001; Enzo Life Sciences; Knight et al., 1992) was added at 5 µM final concentration with or without the MMP inhibitor GM6001 (Enzo Life Sciences) at 100 µM final concentration. Fluorescence intensity was measured every 1 min for 30 min using the Infinite M1000 Pro plate reader (excitation, 328 nm; emission, 400 nm; Tecan). To determine specific activity, the fluorescence of samples containing the MMP inhibitor GM6001 was subtracted from the total fluorescence at each time point. Averaged data were used to generate a trend line by linear regression.
Transwell migration/invasion assay
To measure invasion, the upper surfaces of 8.0-µm transwell supports (Greiner Bio-One) were coated with 20 µl of growth factor–reduced Matrigel (2.5 mg/ml) for 1 h at 37°C. Excess Matrigel was removed, and both chambers were allowed to equilibrate in plain DMEM at 37°C for 1 h. After equilibration, the bottom chamber was filled with 750 µl of DMEM/10% FBS. 50,000 cells were resuspended in 250 µl of DMEM/0.5% FBS, plated in the upper chamber, and allowed to invade for 24 h. Before fixation, cells that did not invade were removed from the upper surface of the membranes using a cotton swab. Membranes were fixed in ice-cold methanol for 10 min, stained with cell stain solution (Cell Biolabs), washed extensively with DDW, and allowed to dry overnight. Intact membranes were imaged using an Eclipse TS100 inverted microscope (10× 0.25 NA air objective; Nikon). To measure tumor cell migration, cells were plated in the upper chamber of transwell supports without Matrigel coating and allowed to migrate for 24 h followed by fixation and staining as above.
Random migration assay
μ-Slide 8-well chambered coverslips (Ibidi) were coated with 10 µg/ml FN and blocked with 1% BSA. 2,000 cells per well were plated in each well in DMEM/10% FBS and allowed to adhere for 12–16 h. Before imaging, medium was replaced to DMEM/10% FBS/10 mM Hepes. Cells were placed in a 37°C heated chamber, and images were collected using the AF6000 inverted microscope (40× 0.60 NA with air objective). Phase images were acquired every 5 min for 8 h. Trajectory plots, accumulated distance (total cell path length), Euclidian distance (the shortest distance between the starting point and end point of migration), and velocity were calculated using the Chemotaxis and Migration Tool (Ibidi).
Immunofluorescence analysis of cortactin tyrosine phosphorylation at invadopodium precursors
72 h before each experiment, Pyk2 or FAK were transiently knocked down using siRNA in MDA–MB-231 cells stably expressing mouse cortactin-TagRFP. 24 h before each experiment, cells were plated on FN/gelatin-unlabeled matrix, starved overnight, and stimulated with 2.5 nM EGF for 3 min or left unstimulated (0 min EGF). Cells were fixed and stained with mouse phosphospecific cortactin antibodies (pY421 or pY466). The intensity (mean gray value [mgv] minus background) of cortactin-TagRFP, pY421-cortactin, or pY466-cortactin at invadopodium precursors was quantified, and the pY421-cortactin/cortactin-TagRFP or pY466-cortactin/cortactin-TagRFP ratios were calculated as measures of tyrosine-phosphorylated cortactin at invadopodium precursors. Data were normalized to resting cells (0 min EGF) and presented as relative fold change in cortactin tyrosine phosphorylation.
Barbed end formation assay
The barbed end assay was performed using biotin-conjugated actin as previously described (Chan et al., 1998). In brief, cells were starved, stimulated with 2.5 nM EGF, and permeabilized with a permeabilization buffer (20 mM Hepes, pH 7.5, 138 mM KCl, 4 mM MgCl2, 3 mM EGTA, 0.2 mg/ml saponin, 1 mM ATP, and 1% BSA) containing 0.4 µM biotin-conjugated muscle actin (AB07; Cytoskeleton Inc.) for 1 min at 37°C. Cells were fixed in 3.7% paraformaldehyde for 5 min, blocked in PBS containing 1% FBS, 1% BSA, and 3 µM unlabeled phalloidin (P3457; Molecular Probes), and then labeled with FITC antibiotin (200-092-211, Jackson ImmunoResearch Laboratories, Inc.) to visualize barbed ends, and with rhodamine-phalloidin (R415; Molecular Probes) and Arp2 to identify regions of cells rich in invadopodia. The barbed end intensity at invadopodia-rich regions was quantified by measuring mgv at invadopodia-rich regions minus mgv of the background. Data were normalized to the control condition for each experiment.
Acceptor photobleaching FRET
Acceptor photobleaching experiments were performed as previously described (Oser et al., 2009; Beaty et al., 2013). In brief, MDA–MB-231 cells were plated on gelatin, fixed, and immunostained with pY402-Pyk2/Alexa Fluor 488 goat anti–rabbit (donor) and cortactin/Alexa Fluor 555 goat anti–mouse (acceptor). As control, cells were stained with Tks5/Alexa Fluor 488 goat anti–rabbit (donor) and cortactin/Alexa Fluor 555 goat anti–mouse (acceptor). Colocalization of cortactin with degradation holes in Alexa Fluor 405–labeled gelatin was used to identify invadopodia.
For all FRET experiments, cells were imaged in PBS at room temperature on a laser-scanning inverted confocal microscope (LSM780; 60× 1.4 NA with oil objective). A region of interest surrounding invadopodia was bleached using 100% 561-nm laser power (acceptor channel), and images were acquired in both the 488-nm and the 561-nm channels before and after bleaching. FRET efficiency was calculated as the E = 1 − (donor before/donor after) in background-subtracted images and was corrected for fluctuations in laser power and donor bleaching in ImageJ.
Statistical analysis
Statistical significance was calculated using an unpaired, two-tailed Student’s t test. Values were considered statistically significant if the p-value was <0.05. For all figures: *, P < 0.05; **, P < 0.01; ***, P < 0.001. Error bars represent SEM.
Online supplemental material
Fig. S1 shows MDA–MB-231 expressing Pyk2-GFP or FAK-GFP, X–Z sections and 3D reconstruction images of cortactin and Tks5 colocalizing to matrix degrading invadopodia, and FRET control experiments between Tks5 and cortactin at degrading invadopodia. Fig. S2 presents Western blots of WT and mutant Pyk2-GFP constructs used for rescue experiments and total invadopodium precursors, active invadopodia, and matrix degradation experiments in MTLn3 cell lines knocked down for Pyk2 or FAK. Fig. S3 shows total invadopodium precursors, active invadopodia, and matrix degradation experiments in U2OS and A431 cell lines knocked down for Pyk2 or FAK. Fig. S4 shows in vitro phosphorylation the of cortactin mutants cortactin Y421F, cortactin Y466F, and cortactin 3YF by Pyk2, tyrosine phosphorylation blots of Pyk2- and FAK-overexpressing cell lines, and barbed end formation and matrix degradation experiments in these cells. Fig. S5 shows transwell migration, transwell invasion, and single-cell migration experiments in Pyk2- and FAK-overexpressing cell lines. Table S1 is an Excel spreadsheet containing the protein–protein and kinase–substrate interaction data obtained in the ProtoArray screens with description and classification of all proteins. Table S2 is an Excel spreadsheet containing lists of invadopodia-associated proteins identified by GLAD4U and ALS as well as a list of invadopodia-related publications identified by ALS.
Supplementary Material
Acknowledgments
We are grateful to Dr. Anthony J. Koleske, Dr. Mark A. Simpson, Dr. Massimiliano Baldassarre, and Dr. Robert Eddy for technical advice and assistance, to the Gruss Lipper Biophotonics Center and Analytical Imaging Facility at Albert Einstein College of Medicine for their assistance in imaging, and to Dr. Gilgi Friedlander from the Israel National Center for Personalized Medicine at Weizmann Institute for her kind assistance and advice in bioinformatic analysis.
This work was funded by the Israel Science Foundation (grants 996/12 and 1749/12), the Israel Cancer Research Fund (grant 12-709-RCDA), the Israel Cancer Association (grant 20141043), a European Commission Marie Curie Career Integration Grant (grant 321556), the Helmsley Charitable Trust Fund (grant 2012PG-ISL013 to H. Gil-Henn), and the National Institutes of Health (grant CA150344 to J.S. Condeelis).
The authors declare no competing financial interests.
Author contributions: A. Genna and S. Twafra performed invadopodia experiments and analysis. S. Lapetina performed in vitro binding and phosphorylation assays. N. Lukic performed cell migration assays. T. Meirson and H. Gil-Henn performed bioinformatic analysis. H. Gil-Henn designed and supervised experiments and wrote the manuscript. J.S. Condeelis and V.P. Sharma advised and assisted in designing and supervising experiments and edited the manuscript.
Footnotes
Abbreviations used:
- ALS
- Agilent Literature Search
- Arg
- Abl-related gene
- FAK
- focal adhesion kinase
- FN
- fibronectin
- FRET
- Förster resonance energy transfer
- GLAD4U
- Gene List Automatically Derived for You
- mgv
- mean gray value
- MMP
- matrix metalloproteinase
References
- Artym V.V., Zhang Y., Seillier-Moiseiwitsch F., Yamada K.M., and Mueller S.C.. 2006. Dynamic interactions of cortactin and membrane type 1 matrix metalloproteinase at invadopodia: defining the stages of invadopodia formation and function. Cancer Res. 66:3034–3043. 10.1158/0008-5472.CAN-05-2177 [DOI] [PubMed] [Google Scholar]
- Ayala I., Baldassarre M., Giacchetti G., Caldieri G., Tetè S., Luini A., and Buccione R.. 2008. Multiple regulatory inputs converge on cortactin to control invadopodia biogenesis and extracellular matrix degradation. J. Cell Sci. 121:369–378. 10.1242/jcs.008037 [DOI] [PubMed] [Google Scholar]
- Beaty B.T., and Condeelis J.. 2014. Digging a little deeper: the stages of invadopodium formation and maturation. Eur. J. Cell Biol. 93:438–444. 10.1016/j.ejcb.2014.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaty B.T., Sharma V.P., Bravo-Cordero J.J., Simpson M.A., Eddy R.J., Koleske A.J., and Condeelis J.. 2013. beta1 integrin regulates Arg to promote invadopodial maturation and matrix degradation. Mol. Biol. Cell. 24:1661–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden E.T., Onikoyi E., Slack R., Myoui A., Yoneda T., Yamada K.M., and Mueller S.C.. 2006. Co-localization of cortactin and phosphotyrosine identifies active invadopodia in human breast cancer cells. Exp. Cell Res. 312:1240–1253. 10.1016/j.yexcr.2005.12.012 [DOI] [PubMed] [Google Scholar]
- Boyle S.N., and Koleske A.J.. 2007. Use of a chemical genetic technique to identify myosin IIb as a substrate of the Abl-related gene (Arg) tyrosine kinase. Biochemistry. 46:11614–11620. 10.1021/bi701119s [DOI] [PubMed] [Google Scholar]
- Chan A.Y., Raft S., Bailly M., Wyckoff J.B., Segall J.E., and Condeelis J.S.. 1998. EGF stimulates an increase in actin nucleation and filament number at the leading edge of the lamellipod in mammary adenocarcinoma cells. J. Cell Sci. 111:199–211. [DOI] [PubMed] [Google Scholar]
- Chan K.T., Cortesio C.L., and Huttenlocher A.. 2009. FAK alters invadopodia and focal adhesion composition and dynamics to regulate breast cancer invasion. J. Cell Biol. 185:357–370. 10.1083/jcb.200809110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W.T. 1989. Proteolytic activity of specialized surface protrusions formed at rosette contact sites of transformed cells. J. Exp. Zool. 251:167–185. 10.1002/jez.1402510206 [DOI] [PubMed] [Google Scholar]
- Clark E.S., and Weaver A.M.. 2008. A new role for cortactin in invadopodia: regulation of protease secretion. Eur. J. Cell Biol. 87:581–590. 10.1016/j.ejcb.2008.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark E.S., Whigham A.S., Yarbrough W.G., and Weaver A.M.. 2007. Cortactin is an essential regulator of matrix metalloproteinase secretion and extracellular matrix degradation in invadopodia. Cancer Res. 67:4227–4235. 10.1158/0008-5472.CAN-06-3928 [DOI] [PubMed] [Google Scholar]
- Diaz B., Shani G., Pass I., Anderson D., Quintavalle M., and Courtneidge S.A.. 2009. Tks5-dependent, nox-mediated generation of reactive oxygen species is necessary for invadopodia formation. Sci. Signal. 2:ra53 10.1126/scisignal.2000368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraley S.I., Feng Y., Krishnamurthy R., Kim D.H., Celedon A., Longmore G.D., and Wirtz D.. 2010. A distinctive role for focal adhesion proteins in three-dimensional cell motility. Nat. Cell Biol. 12:598–604. 10.1038/ncb2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianni D., Diaz B., Taulet N., Fowler B., Courtneidge S.A., and Bokoch G.M.. 2009. Novel p47(phox)-related organizers regulate localized NADPH oxidase 1 (Nox1) activity. Sci. Signal. 2:ra54 10.1126/scisignal.2000370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil-Henn H., Destaing O., Sims N.A., Aoki K., Alles N., Neff L., Sanjay A., Bruzzaniti A., De Camilli P., Baron R., and Schlessinger J.. 2007. Defective microtubule-dependent podosome organization in osteoclasts leads to increased bone density in Pyk2(-/-) mice. J. Cell Biol. 178:1053–1064. 10.1083/jcb.200701148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui R., Campbell D.H., Lee C.S., McCaul K., Horsfall D.J., Musgrove E.A., Daly R.J., Seshadri R., and Sutherland R.L.. 1997. EMS1 amplification can occur independently of CCND1 or INT-2 amplification at 11q13 and may identify different phenotypes in primary breast cancer. Oncogene. 15:1617–1623. 10.1038/sj.onc.1201311 [DOI] [PubMed] [Google Scholar]
- Knight C.G., Willenbrock F., and Murphy G.. 1992. A novel coumarin-labelled peptide for sensitive continuous assays of the matrix metalloproteinases. FEBS Lett. 296:263–266. 10.1016/0014-5793(92)80300-6 [DOI] [PubMed] [Google Scholar]
- Kolli-Bouhafs K., Sick E., Noulet F., Gies J.P., De Mey J., and Rondé P.. 2014. FAK competes for Src to promote migration against invasion in melanoma cells. Cell Death Dis. 5:e1379 10.1038/cddis.2014.329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapetina S., and Gil-Henn H.. 2017. Aguide to simple, direct, and quantitative in vitro binding assays. J. Biol. Methods. 4:e62 10.14440/jbm.2017.161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lev S., Moreno H., Martinez R., Canoll P., Peles E., Musacchio J.M., Plowman G.D., Rudy B., and Schlessinger J.. 1995. Protein tyrosine kinase PYK2 involved in Ca(2+)-induced regulation of ion channel and MAP kinase functions. Nature. 376:737–745. 10.1038/376737a0 [DOI] [PubMed] [Google Scholar]
- Li Y., Tondravi M., Liu J., Smith E., Haudenschild C.C., Kaczmarek M., and Zhan X.. 2001. Cortactin potentiates bone metastasis of breast cancer cells. Cancer Res. 61:6906–6911. [PubMed] [Google Scholar]
- Litvak V., Tian D., Shaul Y.D., and Lev S.. 2000. Targeting of PYK2 to focal adhesions as a cellular mechanism for convergence between integrins and G protein-coupled receptor signaling cascades. J. Biol. Chem. 275:32736–32746. 10.1074/jbc.M004200200 [DOI] [PubMed] [Google Scholar]
- Liu S., Yamashita H., Weidow B., Weaver A.M., and Quaranta V.. 2010. Laminin-332-beta1 integrin interactions negatively regulate invadopodia. J. Cell. Physiol. 223:134–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mader C.C., Oser M., Magalhaes M.A., Bravo-Cordero J.J., Condeelis J., Koleske A.J., and Gil-Henn H.. 2011. An EGFR-Src-Arg-cortactin pathway mediates functional maturation of invadopodia and breast cancer cell invasion. Cancer Res. 71:1730–1741. 10.1158/0008-5472.CAN-10-1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magalhaes M.A., Larson D.R., Mader C.C., Bravo-Cordero J.J., Gil-Henn H., Oser M., Chen X., Koleske A.J., and Condeelis J.. 2011. Cortactin phosphorylation regulates cell invasion through a pH-dependent pathway. J. Cell Biol. 195:903–920. 10.1083/jcb.201103045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neri A., and Nicolson G.L.. 1981. Phenotypic drift of metastatic and cell-surface properties of mammary adenocarcinoma cell clones during growth in vitro. Int. J. Cancer. 28:731–738. 10.1002/ijc.2910280612 [DOI] [PubMed] [Google Scholar]
- Oliveros J.C. 2015. Venny. An interactive tool for comparing lists with Venn's diagrams. http://bioinfogp.cnb.csic.es/tools/venny/index.html (accessed February, 2017).
- Ormandy C.J., Musgrove E.A., Hui R., Daly R.J., and Sutherland R.L.. 2003. Cyclin D1, EMS1 and 11q13 amplification in breast cancer. Breast Cancer Res. Treat. 78:323–335. 10.1023/A:1023033708204 [DOI] [PubMed] [Google Scholar]
- Oser M., Yamaguchi H., Mader C.C., Bravo-Cordero J.J., Arias M., Chen X., Desmarais V., van Rheenen J., Koleske A.J., and Condeelis J.. 2009. Cortactin regulates cofilin and N-WASp activities to control the stages of invadopodium assembly and maturation. J. Cell Biol. 186:571–587. 10.1083/jcb.200812176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oser M., Mader C.C., Gil-Henn H., Magalhaes M., Bravo-Cordero J.J., Koleske A.J., and Condeelis J.. 2010. Specific tyrosine phosphorylation sites on cortactin regulate Nck1-dependent actin polymerization in invadopodia. J. Cell Sci. 123:3662–3673. 10.1242/jcs.068163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoumacher M., Goldman R.D., Louvard D., and Vignjevic D.M.. 2010. Actin, microtubules, and vimentin intermediate filaments cooperate for elongation of invadopodia. J. Cell Biol. 189:541–556. 10.1083/jcb.200909113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selitrennik M., and Lev S.. 2015. PYK2 integrates growth factor and cytokine receptors signaling and potentiates breast cancer invasion via a positive feedback loop. Oncotarget. 6:22214–22226. 10.18632/oncotarget.4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., and Ideker T.. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13:2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma V.P., Eddy R., Entenberg D., Kai M., Gertler F.B., and Condeelis J.. 2013. Tks5 and SHIP2 regulate invadopodium maturation, but not initiation, in breast carcinoma cells. Curr. Biol. 23:2079–2089. 10.1016/j.cub.2013.08.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibony-Benyamini H., and Gil-Henn H.. 2012. Invadopodia: the leading force. Eur. J. Cell Biol. 91:896–901. 10.1016/j.ejcb.2012.04.001 [DOI] [PubMed] [Google Scholar]
- Sinha S., Hoshino D., Hong N.H., Kirkbride K.C., Grega-Larson N.E., Seiki M., Tyska M.J., and Weaver A.M.. 2016. Cortactin promotes exosome secretion by controlling branched actin dynamics. J. Cell Biol. 214:197–213. 10.1083/jcb.201601025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tehrani S., Tomasevic N., Weed S., Sakowicz R., and Cooper J.A.. 2007. Src phosphorylation of cortactin enhances actin assembly. Proc. Natl. Acad. Sci. USA. 104:11933–11938. 10.1073/pnas.0701077104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomar A., Lawson C., Ghassemian M., and Schlaepfer D.D.. 2012. Cortactin as a target for FAK in the regulation of focal adhesion dynamics. PLoS One. 7:e44041 10.1371/journal.pone.0044041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale S., Avizienyte E., Brunton V.G., and Frame M.C.. 2008. Focal adhesion kinase is not required for Src-induced formation of invadopodia in KM12C colon cancer cells and can interfere with their assembly. Eur. J. Cell Biol. 87:569–579. 10.1016/j.ejcb.2008.04.002 [DOI] [PubMed] [Google Scholar]
- Wang W., Liu Y., and Liao K.. 2011. Tyrosine phosphorylation of cortactin by the FAK-Src complex at focal adhesions regulates cell motility. BMC Cell Biol. 12:49 10.1186/1471-2121-12-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver A.M. 2006. Invadopodia: specialized cell structures for cancer invasion. Clin. Exp. Metastasis. 23:97–105. 10.1007/s10585-006-9014-1 [DOI] [PubMed] [Google Scholar]
- Weaver A.M. 2008. Invadopodia. Curr. Biol. 18:R362–R364. 10.1016/j.cub.2008.02.028 [DOI] [PubMed] [Google Scholar]
- Wu H., and Parsons J.T.. 1993. Cortactin, an 80/85-kilodalton pp60src substrate, is a filamentous actin-binding protein enriched in the cell cortex. J. Cell Biol. 120:1417–1426. 10.1083/jcb.120.6.1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Reynolds A.B., Kanner S.B., Vines R.R., and Parsons J.T.. 1991. Identification and characterization of a novel cytoskeleton-associated pp60src substrate. Mol. Cell. Biol. 11:5113–5124. 10.1128/MCB.11.10.5113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H., Lorenz M., Kempiak S., Sarmiento C., Coniglio S., Symons M., Segall J., Eddy R., Miki H., Takenawa T., and Condeelis J.. 2005. Molecular mechanisms of invadopodium formation. J. Cell Biol. 168:441–452. 10.1083/jcb.200407076 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.