Local actin filament formation powers the development of the signal-receiving arbor of neurons. In this study, Izadi et al. demonstrate that Cobl-like, which bears only a single WH2 domain, mediates dendritic branching by coordinating with the F-actin–binding protein Abp1 in a Ca2+/CaM-controlled manner to control actin dynamics.
Abstract
Local actin filament formation powers the development of the signal-receiving arbor of neurons that underlies neuronal network formation. Yet, little is known about the molecules that drive these processes and may functionally connect them to the transient calcium pulses observed in restricted areas in the forming dendritic arbor. Here we demonstrate that Cordon-Bleu (Cobl)–like, an uncharacterized protein suggested to represent a very distantly related, evolutionary ancestor of the actin nucleator Cobl, despite having only a single G-actin–binding Wiskott–Aldrich syndrome protein Homology 2 (WH2) domain, massively promoted the formation of F-actin–rich membrane ruffles of COS-7 cells and of dendritic branches of neurons. Cobl-like hereby integrates WH2 domain functions with those of the F-actin–binding protein Abp1. Cobl-like–mediated dendritic branching is dependent on Abp1 as well as on Ca2+/calmodulin (CaM) signaling and CaM association. Calcium signaling leads to a promotion of complex formation with Cobl-like’s cofactor Abp1. Thus, Ca2+/CaM control of actin dynamics seems to be a much more broadly used principle in cell biology than previously thought.
Introduction
The formation and dynamics of complex neuronal networks involve local Ca2+ and calmodulin (CaM) signals (Rajan and Cline, 1998; Chevaleyre and Castillo, 2002; Fink et al., 2003; Gaudillière et al., 2004; Ohashi et al., 2014) and coincide with transient F-actin formation at sites of dendritic branch induction (Hou et al., 2015).
The recent discovery of the actin nucleator Cordon-Bleu (Cobl; Ahuja et al., 2007) as CaM-controlled component important for early neuronal morphogenesis identified a direct link between Ca2+/CaM signaling and actin filament formation (Hou et al., 2015). Cobl belongs to a more recently identified group of actin nucleators marked by Wiskott–Aldrich syndrome protein Homology 2 (WH2) domains (Chesarone and Goode, 2009; Qualmann and Kessels, 2009; Renault et al., 2013). Cobl contains three G-actin–binding WH2 domains cooperating in actin filament formation and plays a crucial role in dendritic arborization of hippocampal neurons and Purkinje cells of the cerebellum (Ahuja et al., 2007; Haag et al., 2012).
Whereas Cobl is an evolutionary, relatively young gene, its ancestor Cobl-like (Coblr1) already arose with the emergence of bilateria (Schultz and Terhoeven, 2013). Cobl-like mRNA distribution differs from that of its distant relative Cobl during early embryogenesis (Carroll et al., 2003). The Cobl-like gene is linked to diabetes and obesity (Mancina et al., 2013; Sharma et al., 2017) and has been introduced as biomarker of high prognostic value for different types of cancer (Gordon et al., 2003, 2009; Wang et al., 2013; Han et al., 2017). Yet, the cellular functions of Cobl-like and the molecular mechanisms it uses remain completely unknown.
Cobl-like contains a single WH2 domain and shows merely low sequence similarity to Cobl. Single WH2 domains can sequester G-actin (Low and Goldstein, 1982; Paulussen et al., 2009). Suppression of actin filament formation by G-actin sequestering opposes actin nucleation. Balancing neuronal cell shape development by expression of protein isoforms with opposing functions was recently proposed to underlie the formation of the more complex human brain (Charrier and Polleux, 2012). Together, this urgently called for an evaluation of the properties and the functions of the ancestor of the actin nucleator Cobl, Cobl-like.
Here we show that Cobl-like massively promoted the formation of F-actin–rich ruffles in COS-7 cells and of dendritic branches in neurons. Molecular and functional examinations showed that Cobl-like hereby joins the function of its WH2 domain with those of the F-actin–binding protein Abp1—a mechanism and cell biological function controlled by the Ca2+-sensor protein CaM, which modulates the Abp1 interaction of Cobl-like.
Our data therefore suggest that Ca2+/CaM control is not restricted to the actin nucleator Cobl. Instead, powering neuronal morphogenesis by local Ca2+/CaM signaling converging onto actin filament-promoting effectors seems to be a more general principle in cell biology.
Results
Despite its single WH2 domain binding to G-actin, Cobl-like promotes F-actin–rich structures in vivo
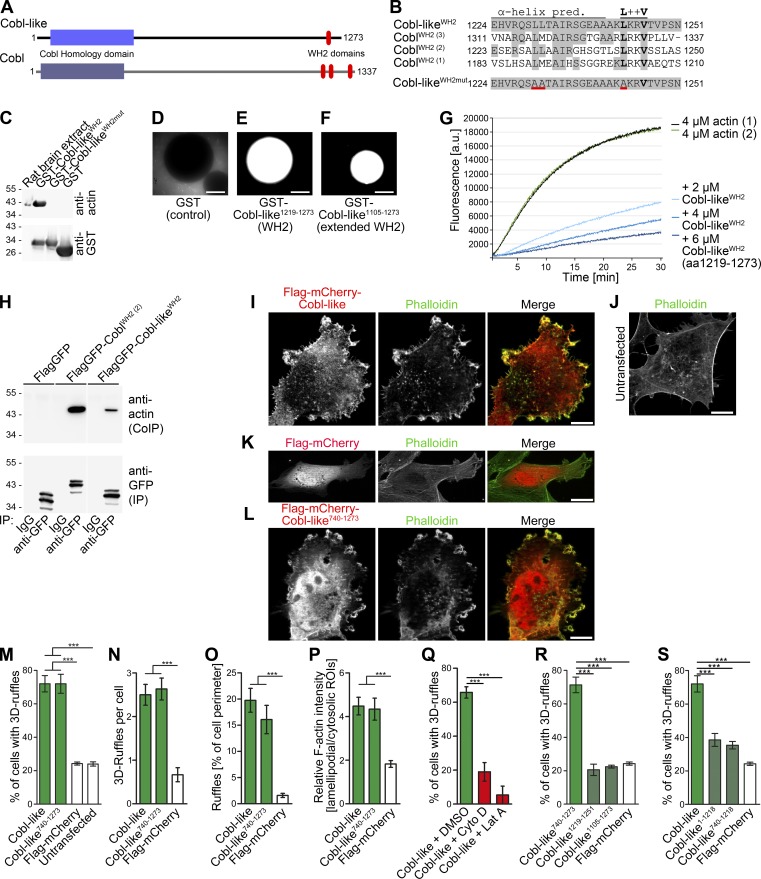
The evolutionary ancestor of the actin nucleator Cobl (Ahuja et al., 2007), Cobl-like, shows only 25% sequence identity to Cobl. This strongly suggested that Cobl-like’s molecular mechanisms and functions fundamentally differ from Cobl’s. Cobl-like contains an N-terminal so-called Cobl Homology domain, which, however, shares only 33% identity with that of Cobl, and a single, C-terminal WH2 domain. Yet, even this small domain shows only 44% conservation (Fig. 1, A and B; Fig. S1 A).
Figure 1.
Cobl-like is a WH2 domain–containing, G-actin–binding protein promoting F-actin–driven shape changes of COS-7 cells. (A) Scheme of murine Cobl-like in comparison to the actin nucleator Cobl. (B) Alignment of the predicted Cobl-like WH2 domain and a mutated (red underlining) version thereof with the three WH2 domains of Cobl. (C) Immunoblotting analyses of precipitations of endogenous actin from rat brain extracts (input) with immobilized Cobl-like WH2 domain, a mutated version of this domain (L1230A, L1231A, and L1243A; WH2mut), and GST as control. (D–F) Beads with immobilized GST-Cobl-like1219–1273 and GST-Cobl-like1105-1273 incubated with rat brain extracts supplemented with fluorescent actin and an energy-regenerating system showing merely a recruitment of fluorescent G-actin (but no formation of F-actin structures at the bead surfaces). Bars, 50 µm. (G) Pyrene-actin assays showing a dose-dependent suppression of spontaneous F-actin formation by Cobl-like1219–1273. (H) Specific coimmunoprecipitations of endogenous actin with Cobl WH2 (2) and the Cobl-like WH2 domain. White line, lanes omitted. (I–L) Maximum intensity projections (MIPs) of ApoTome images of (Flag-tagged) mCherry-Cobl-like (I) and mCherry-Cobl-like740–1273 (L) and F-actin in F-actin–rich membrane-ruffles in COS-7 cells. Untransfected cells (J) and Flag-mCherry–expressing cells (K) are shown as controls. Bars, 10 µm. (M–P) Quantitative evaluations of the Cobl-like and Cobl-like740–1273–induced phenotype of 3D ruffle formation by scoring the percentage of cells with such structures (M) and additional analyses substantiating the phenotype by further quantitative evaluations, such as the number of 3D ruffles per cell (N), ruffles in percentage of cell perimeter (O), and the ratio of lamellipodial versus cytosolic F-actin (P). (Q) Scoring of cells with the Cobl-like–induced 3D ruffles phenotype on latrunculin A (Lat A) and cytochalasin D (Cyto D) incubation, respectively. DMSO, solvent control. (R and S) Quantitative analyses of the molecular requirements of Cobl-like–induced 3D ruffle formation showing that the WH2 domain is not sufficient (R) but critical (S). Data are mean ± SEM, one-way ANOVA plus Tukey’s post test. ***, P < 0.001.
The predicted WH2 domain of Cobl-like specifically precipitated endogenous actin. Mutation of three conserved leucine residues to alanine (L1230A, L1231A, and L1243A; WH2mut) disrupted this actin binding (Fig. 1 C). Thus, the WH2 domain of Cobl-like indeed is functional.
In line with G-actin binding, beads with immobilized Cobl-like WH2 domain and a more extended C-terminal part of Cobl-like showed no induction of F-actin around the beads but only led to recruitment of fluorescent G-actin (Fig. 1, D–F). Consistently, the Cobl-like WH2 domain caused an effective and dose-dependent suppression of spontaneous F-actin assembly in in vitro reconstitutions (Fig. 1 G). The interaction of Cobl-like’s WH2 domain with actin thus is direct and leads to G-actin sequestering.
That Cobl-like also associates with actin in vivo was proven by specific coimmunoprecipitations of endogenous actin with FlagGFP-Cobl-likeWH2 (Fig. 1 H).
G-actin sequestering could represent an interesting mechanism of balancing Cobl functions by opposing its functions. Strikingly, however, subcellular localization studies in COS-7 cells challenged this hypothesis. Instead of reflecting the relatively homogenous distribution of the G-actin pool, Cobl-like was highly enriched at F-actin–rich structures (Fig. 1 I).
In addition, F-actin–rich 3D membrane ruffles were more frequently observed in cells overexpressing Cobl-like than in control cells. The unexpected capability to massively promote the formation of F-actin–rich structures was also displayed by the WH2 domain–containing C-terminal half of Cobl-like (Fig. 1, I–P).
Quantitative analyses showed that >70% of the Cobl-like– or Cobl-like740–1273–expressing cells showed elaborate 3D ruffles, whereas <25% of control cells showed at least some of these F-actin–rich structures. The number of individual ruffles per cell also was strongly increased when compared with control cells (more than threefold). Ruffles of both Cobl-like– or Cobl-like740-1273–expressing cells covered ∼20% of the cell perimeter, whereas data for control cells were 10-fold lower (Fig. 1, M–O).
Cobl-like–induced ruffling was clearly attributable to cortical actin filament formation. The relative F-actin intensity at lamellipodial areas in comparison to the cytosol was more than twofold higher in cells overexpressing Cobl-like or Cobl-like740–1273 when compared to control (Fig. 1 P). Furthermore, both cytochalasin D and latrunculin A incubation fully suppressed the Cobl-like–induced phenotype (Fig. 1 Q).
Cobl requires all three WH2 domains to efficiently nucleate actin in vitro and in mutational in vivo studies (Ahuja et al., 2007). Another study suggested that a combination of a cluster of positively charged residues (“K-region”) and the first WH2 domain nucleates actin and, furthermore, that F-actin severing occurs when a 1:1 molar ratio of actin and a C-terminal Cobl fragment was used in in vitro reconstitutions (Husson et al., 2011). Our in vitro reconstitutions with Cobl-like did not show any severing activity, comparisons of Cobl-like to Cobl’s K-region did not reveal any similarities (our unpublished data; Fig. S1 B), and overexpression of two different C-terminal parts of Cobl-like containing the WH2 domain failed to give rise to any F-actin–rich ruffles (Fig. 1 R; for protein integrity and sufficient expression of functionally negative mutant, see Fig. S1 C). Cobl-like1105–1273 thus is not sufficient for ruffle formation.
Yet, although not sufficient, the WH2 domain was crucial in the context of full-length Cobl-like and in the context of the functionally active C-terminal half, and the respective WH2 domain deletions (Cobl-like1–1218 and Cobl-like740–1218) failed to give rise to the formation of F-actin–rich 3D ruffles (Fig. 1 S and Fig. S1 D).
Cobl-like is expressed in hippocampal and cortical neurons and adopts a dendritic localization
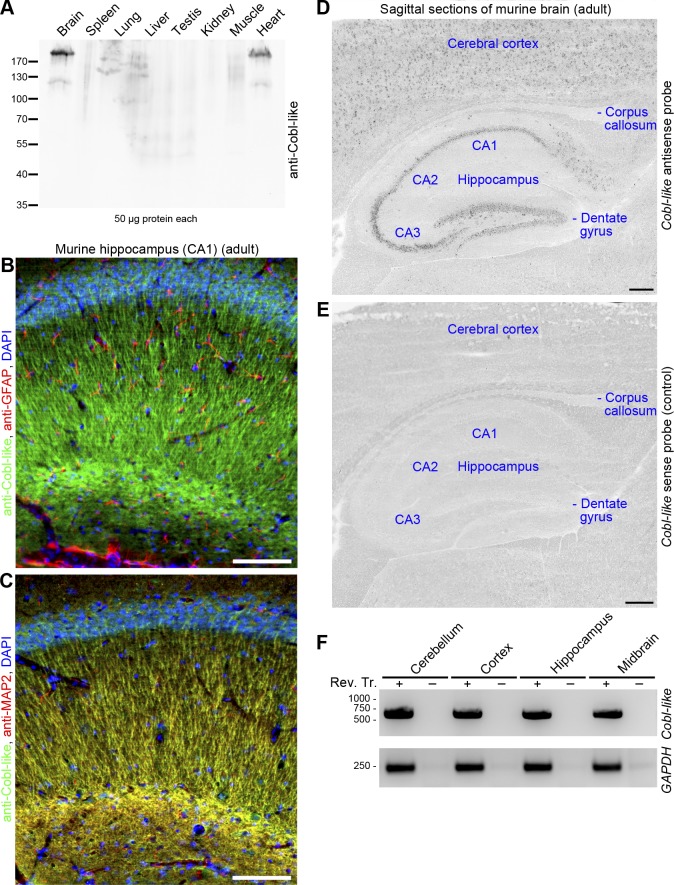
To address whether Cobl-like may have functions overlapping with that of Cobl or not, we raised anti–Cobl-like antibodies. Affinity-purified antibodies recognized Cobl-like with high specificity and good sensitivity (Fig. S1, E and F) and unveiled that, similar to Cobl (Ahuja et al., 2007), Cobl-like is also of relatively low abundance and is a brain-enriched protein (Fig. 2 A).
Figure 2.
Cobl-like is a brain-enriched protein with neuronal and dendritic localization. (A) Anti–Cobl-like immunoblotting of different mouse tissues. (B and C) Immunohistological analyses of sagittal sections of the hippocampus of adult mice with anti–Cobl-like in combination with the glia marker anti–glial fibrillary acidic protein (GFAP; B) and the neuronal and dendritic marker anti-MAP2 (C), respectively. (D and E) In situ hybridizations of sagittal sections of adult murine brain with a probe against Cobl-like mRNA (Cobl-like antisense) and a negative control probe (sense). Bars: (B and C) 100 µm; (D and E) 200 µm. (F) Cobl-like RT-PCR analyses of different brain regions. GAPDH, control. Rev. Tr. + and −, with and without reverse transcription.
Confocal microscopy of sagittal sections of brains of adult mice showed that anti–Cobl-like immunoreactivity was not enriched in glia cells but present in neurons of the hippocampus (Fig. 2, B and C).
Besides in the hippocampus, anti–Cobl-like antibodies also detected Cobl-like in other parts of the central nervous system (Fig. S1 G). The abundance of Cobl-like in different parts of the brain was confirmed by antibody-independent techniques. In situ hybridizations showed Cobl-like expression in the hippocampus and in the cerebral cortex (Fig. 2, D and E). RT-PCR analyses confirmed its expression in the cortex and hippocampus and detected Cobl-like mRNA in the cerebellum and midbrain (Fig. 2 F).
At the subcellular level, Cobl-like adopted a dendritic localization pattern and clearly colocalized with the dendritic marker MAP2, as observed in both hippocampal tissue sections and in cultured hippocampal neurons (Fig. 2, B and C; and Fig. S1 H). Cobl-like thus is present in the dendritic compartment.
Cobl-like massively promotes dendritic arborization of hippocampal neurons and localizes to sites of dendritic branch initiation
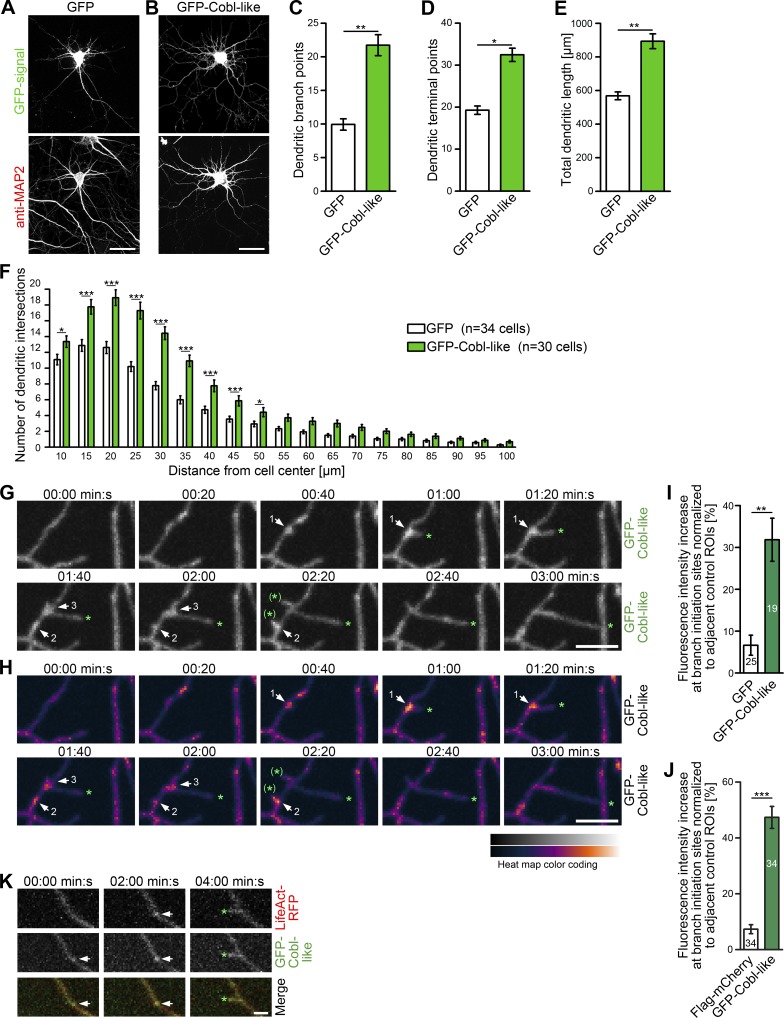
The ability of Cobl-like to promote F-actin–rich ruffles (Fig. 1) and its localization to the dendritic compartment of neurons (Fig. 2) prompted us to test whether Cobl-like shapes neurons. As highlighted by anti-MAP2 immunostaining, an excess of Cobl-like induced a strong increase of dendritic arborization in primary rat hippocampal neurons (Fig. 3, A and B).
Figure 3.
Cobl-like promotes the formation of the dendritic arbor of primary hippocampal neurons. (A and B) Morphology of Cobl-like–overexpressing hippocampal neurons (DIV7). Bars, 20 µm. (C–F) Quantitative determinations of indicated dendritic arbor parameters (C–E) and Sholl analyses (F). (G) MIPs of individual frames of a 3D time-lapse recording of dendrites of DIV7 neurons with GFP-Cobl-like enrichments (distinct sites, white arrows; numbering marks individual events) seconds before protrusion initiation from these sites. Tips of growing dendritic protrusions are marked by a green asterisk. Abandoned protrusive events are marked by a green asterisk in brackets. (H) Heat map representation of data in G. Bars, 5 µm. (I and J) Quantitative evaluations of dynamic Cobl-like enrichment at branch initiation sites 30 s before protrusion formation in comparison to the behavior of GFP used as control (I) and in comparison to directly cotransfected Flag-mCherry firmly excluding theoretical volume artifacts (J), respectively. (K) MIPs of single frames of a 3D time-lapse recording of neurons cotransfected with GFP-Cobl-like and LifeAct-RFP (visualizing F-actin). See the explanation of panel G for the meaning of the markings. Bar, 2 µm. Data are mean ± SEM, two-tailed Student’s t test (C–E, I, and J) and two-way ANOVA plus Bonferroni’s post test (F). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Quantitative evaluations showed an increase of dendritic branch points, terminal points, and the total length of the dendritic arbor (Fig. 3, C–E). These alterations were also visible in Sholl analyses. In both the vicinity of the cell body and in the periphery, the number of Sholl intersections was increased (Fig. 3 F).
3D time-lapse imaging with the use of spinning disk microscopy showed a presence of GFP–Cobl-like during the initiation of protrusions from the existing dendritic arbor. Some of these protrusions were retracted immediately, whereas others grew out longer and developed into stable dendritic branches. Interestingly, Cobl-like accumulated at nascent branch sites during the seconds before the initiation of protrusion formation (Fig. 3, G and H). It also frequently redistributed into the new branch during the first 1–2 min reflecting the first micrometers of growth (≤5 µm; Fig. 3, G and H).
Quantitative evaluations clearly showed that Cobl-like enriches specifically at sites of protrusion initiation (Fig. 3 I). Quantitative examinations of Flag-mCherry and GFP–Cobl-like coexpression experiments firmly excluded that putative volume effects feign the observed accumulations (Fig. 3 J).
Dual 3D time-lapse imaging of GFP–Cobl-like and LifeAct-RFP showed that Cobl-like accumulations are occurring before the build-up of F-actin at nascent branch sites (Fig. 3 K). Collectively, our time-lapse analyses and functional studies thus suggested that Cobl-like may support the initiation of new dendritic protrusions.
Cobl-like is crucial for dendritic arborization
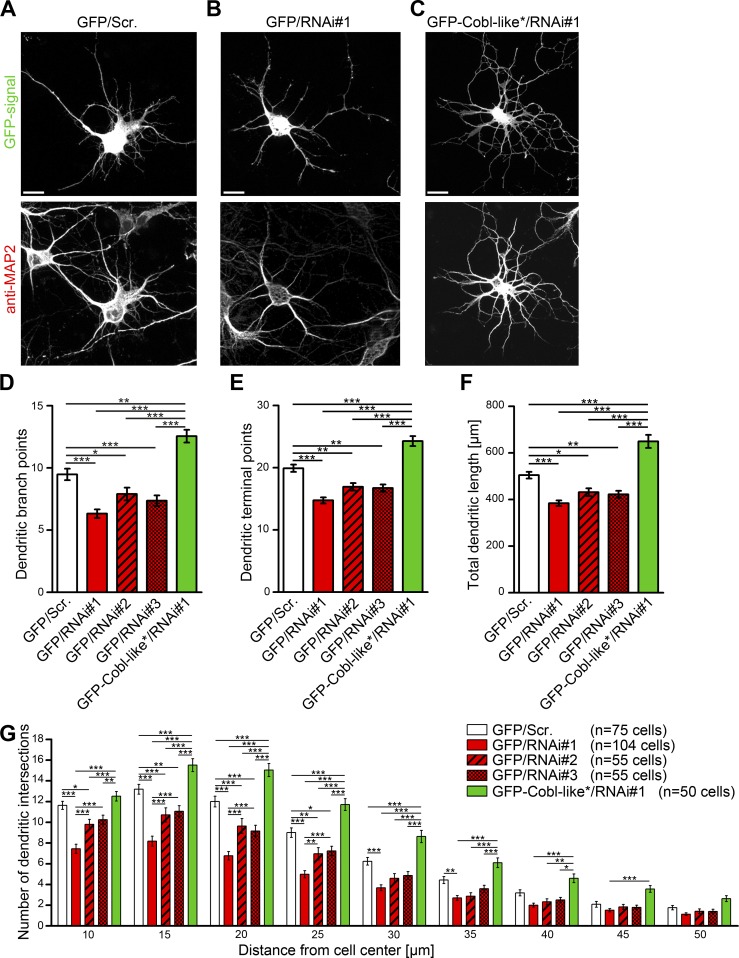
To evaluate whether Cobl-like is crucial for dendritic arbor formation, we next tested three different RNAi sites for efficient knockdown in quantitative immunoblotting analyses. All three RNAi sequences led to Cobl-like knockdown (Fig. S1, I and J).
In hippocampal neurons, all three Cobl-like RNAi plasmids led to consistent reductions of dendritic branches, terminal points, and total dendritic length (Fig. 4, A–F). The phenotypes were dose-dependent. The RNAi#1 yielding the most efficient knockdown (Fig. S1 I–K) also caused the strongest cellular defects (Fig. 4, D–G).
Figure 4.
Cobl-like is critical for dendritic arbor formation. (A–C) Morphology of Cobl-like–depleted neurons (RNAi#1; B) in comparison to control neurons (scrambled RNAi sequence; Scr.; A) and to neurons coexpressing an RNAi#1-insensitive Cobl-like mutant (Cobl-like*; rescue; C). Bars, 10 µm. (D–F) Quantitative determinations of specific defects in dendritic arborization caused by Cobl-like addressing dendritic branch points (D), dendritic terminal points (E), the summarized length of the dendritic arbor (F), and Sholl intersections (G). Data are mean ± SEM, one-way ANOVA Tukey’s post test (D–F) and two-way ANOVA plus Bonferroni’s post test (G). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Fine-structure analyses unveiled reductions of Sholl intersections that were statistically significant over a large distance range. Impairments caused by RNAi#1 remained statistically significant (P < 0.01) up to 35-µm distance. Thus, Cobl-like deficiency led to a reduced complexity of the proximal and peripheral dendritic arbor of neurons (Fig. 4 G).
Importantly, reexpression of RNAi-insensitive Cobl-like (Cobl-like*) fully suppressed all Cobl-like loss-of-function phenotypes and thereby proved their specificity (Fig. 4, D–G).
Cobl-like’s functions in actin filament formation and dendritic arborization require the WH2 domain and a proline-rich region associating with the F-actin–binding protein Abp1
Cobl-like exhibits only a single WH2 domain incapable of promoting F-actin formation in both in vitro and in vivo experimentation. Therefore, Cobl-like’s WH2 domain–dependent capability to induce F-actin–rich ruffles and dendritic branches remained mechanistically unclear. Because we did not observe any hints on putative severing mechanisms (our unpublished data, Fig. S1 B), we next considered mechanisms similar to those suggested for the nucleator Lmod2 (Chereau et al., 2008). However, Cobl-like contains neither an Lmod2-related F-actin–binding leucine-rich repeat nor any other recognizable F-actin–binding domain. In line, the C terminus of Cobl-like did not show F-actin binding in cosedimentation experiments (Fig. S2). Cobl-like thus uses a different mechanism.
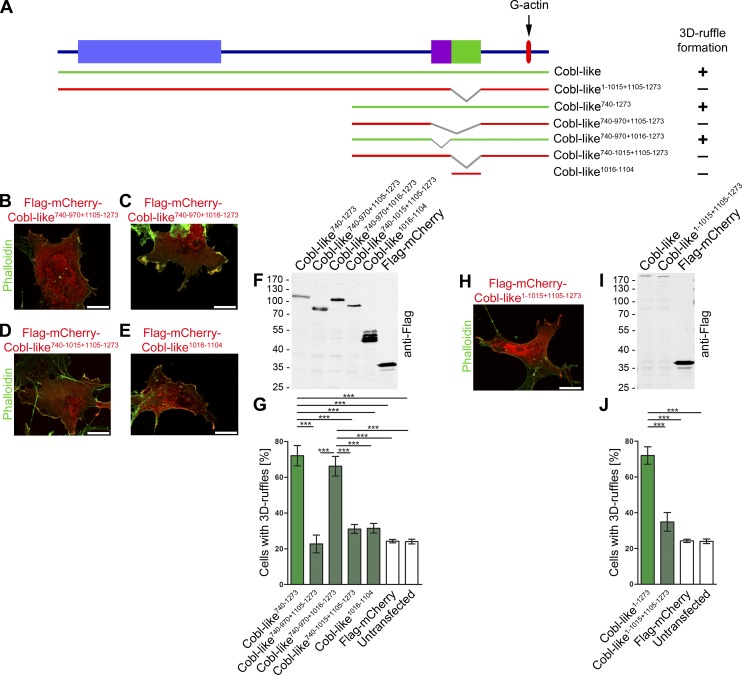
To unveil which parts of Cobl-like are crucially involved in the induction of F-actin–rich ruffles and are cooperating with the WH2 domain, we dissected Cobl-like by deletion screening (Fig. 5, A–J). Strikingly, Cobl-like740–970+1105–1273–expressing cells remained indistinguishable from control whereas the corresponding Cobl-like740–1273 strongly induced 3D ruffling (Fig. 5, A, B, and G).
Figure 5.
Identification of an additional part of Cobl-like that, besides the WH2 domain, is critical for Cobl-like–mediated promotion of F-actin–rich ruffles. (A) Scheme of Cobl-like and mutants thereof used to identify the Cobl-like regions required for the formation of F-actin–rich 3D ruffles in COS-7 cells (green, inducing 3D ruffles; red, not inducing 3D ruffles; graphical summary on the right side). (B–J) Functional analyses in the context of the C-terminal half (B–G) and in full-length (H–J). (B–E and H) Representative examples of phalloidin-stained cells overexpressing the Cobl-like deletion mutants indicated. Bars, 10 µm. (F and I) Anti-Flag immunoblottings of Cobl-like and mutants thereof ensuring that all proteins are intact and that proteins negative in functional assays were expressed at least as high as the active proteins. (G and J) Quantitative analyses of ruffle formation in Cobl-like mutants-overexpressing COS-7 cells in comparison to positive and negative controls (datasets from Fig. 1). Cobl-like1016-1104 is critical for 3-D ruffle formation (yet, not sufficient); all Cobl-like mutants lacking this sequence are not sufficient to induce ruffles and are not statistically different from mCherry and untransfected cells (controls). Data are mean ± SEM, one-way ANOVA plus Tukey’s post test (G and J). ***, P < 0.001.
Dissecting the identified critical part of Cobl-like even further revealed that aa 1016–1104 were indispensable (but alone not sufficient), whereas the more N-terminal part (aa 971–1015) was not required (Fig. 5, A–G). Consistently, also in the full-length context, deletion of aa 1016–1104 rendered Cobl-like incapable of promoting F-actin–rich ruffles (Fig. 5, A and H–J).
Alignments of Cobl-like proteins from different species revealed that Cobl-like1016–1104 contains an unusual, extended, and in some cases multiplied PxxP motif with the consensus PAEtSPPPvAPKPxxl (Fig. 6 A). It seemed possible that this motif, located more than 100 amino acids N-terminal of the WH2 domain, represents a binding site for some cofactor and that only in complex with this functional partner Cobl-like is capable of promoting the formation of F-actin.
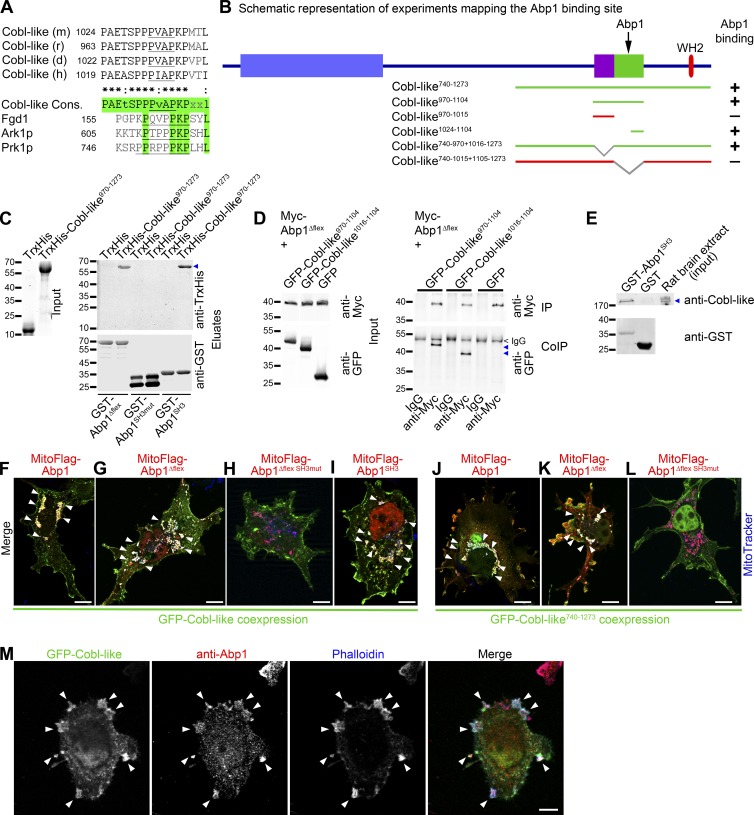
Figure 6.
The Cobl-like region that together with the WH2 domain is critical for promotion of F-actin formation is an Abp1-binding site. (A) Alignment and identification of proline-rich sequences of mouse (m; gi:74201419), rat (r; gi: 672016899), dolphin (d; gi:1131285576), and human (h; gi:510937001) Cobl-like proteins and of related motifs in the Abp1-binding proteins Fgd1 (mouse; BC011462), Ark1p (yeast; gi:285814631), and Prk1p (yeast; gi:285812559). Green shows similarities between the Cobl-like consensus and the other proteins. PxxP motifs are underlined. (B) Scheme of Cobl-like and mutants thereof used to identify the Abp1-binding region (binding, green lines, +; not binding, red lines, −; for original data see Fig. S3, C and D). Right side shows additional graphical summary (+ vs. −). (C) In vitro reconstitutions of complex formation of purified TrxHis-Cobl-like970–1273 (blue arrowhead) versus TrxHis control with immobilized GST-Abp1Δflex, GST-Abp1SH3, and a mutant thereof incapable of binding PxxP motifs (Abp1SH3mut). Input is shown by Coomassie (left). (D) Immunoblotting analyses showing specific coimmunoprecipitations of GFP-Cobl-like970–1104 and GFP-Cobl-like1016–1104 (blue arrowheads) with Myc-Abp1Δflex. (E) Anti-GSTgp and anti–Cobl-like immunoblottings of endogenous Cobl-like (blue arrowhead) specifically precipitated from rat brain extracts with immobilized GST-Abp1SH3. (F–L) MIPs of merged images of successful complex reconstitutions (arrowheads mark examples) of GFP-Cobl-like (F–I) and GFP-Cobl-like740–1273 (J–L) with mitochondrially targeted Abp1 (MitoFlag-Abp1; F and J), Abp1Δflex (G and K), and the Abp1 SH3 domain (I) in intact COS-7 cells costained with MitoTracker (blue). In contrast, H and L show unsuccessful reconstitutions with MitoFlag-Abp1Δflex SH3mut. For single channels of the merged images shown here, see Fig. S4. (M) Colocalization of endogenous Abp1 (red) with GFP-Cobl-like–induced, phalloidin-stained F-actin–rich 3D ruffles (shown in blue; arrowheads) in COS-7 cells. Bars, 10 µm.
Database analysis did not reveal any other proteins containing such extended consensus sequences. However, somewhat related sequences containing the core PKPxxL in combination with further PxxP motifs were found in faciogenital dysplasia 1 (Fgd1) and in the actin cytoskeleton-regulating kinases Ark1p and Prk1p in yeast (Fig. 6 A). Interestingly, these three proteins were reported to interact with the SH3 domain-containing protein Abp1 (Fazi et al., 2002; Hou et al., 2003). Abp1 is a phylogenetically old F-actin–binding protein that is highly expressed in the brain and especially in the hippocampus (Kessels et al., 2000; Qualmann et al., 2004). Our analyses indeed unveiled that Abp1’s SH3 domain specifically interacted with the C-terminal part of Cobl-like (Fig. 6 B; and Fig. S3, A and B). Further coprecipitations showed that both Cobl-like970–1104 and Cobl-like1024–1104, which contained the extended PAEtSPPPvAPKPxxl motif, were sufficient for Abp1 SH3 association (Fig. 6 B and Fig. S3 C).
Consistently, Cobl-like740–970+1016–1273, which was cell biologically active in our ruffling assays, associated with Abp1’s SH3 domain, whereas the inactive mutant Cobl-like740–1015+1105–1273 did not (Fig. 6 B and Fig. S3 D).
Reconstitutions with the use of purified TrxHis-Cobl-like970–1273 in combination with immobilized GST-Abp1Δflex (a proteolytically more stable Abp1 version), GST-Abp1SH3, and a P422L,G425R mutation-carrying SH3 domain incapable of associating with PxxP motifs (Abp1SH3mut; Kessels et al., 2001), respectively, demonstrated that the discovered Abp1 interaction with Cobl-like reflects a direct complex formation of Cobl-like with Abp1SH3 (Fig. 6 C).
Coimmunoprecipitations of both proteins showed that the Cobl-like/Abp1 interaction also is of relevance in vivo (Fig. 6 D). These results were further corroborated by the fact that Abp1SH3 specifically precipitated endogenous Cobl-like from rat brain extracts (Fig. 6 E).
Cobl-like/Abp1 complexes could also be directly visualized in intact cells. Both GFP-Cobl-like and GFP-Cobl-like740-1273 were specifically recruited by Abp1, Abp1Δflex, and Abp1SH3 targeted to defined cellular sites (mitochondrial surfaces). Mutation of the Abp1 SH3 domain abolished the Cobl-like recruitment (Fig. 6, F–L; and Fig. S4).
Consistent with the Abp1-binding region identified in coprecipitation and coimmunoprecipitation studies, additional experiments showed that two different Cobl-like mutants lacking the aa 1016–1104 interface were not recruited (Fig. S5, A–C).
Immunofluorescence analyses furthermore showed that endogenous Abp1 is enriched in Cobl-like–induced membrane ruffles and that Abp1 and Cobl-like colocalized at these F-actin–rich sites (Fig. 6 M).
Cobl-like–mediated dendrite branching relies on Cobl-like’s WH2 domain and complex formation with Abp1
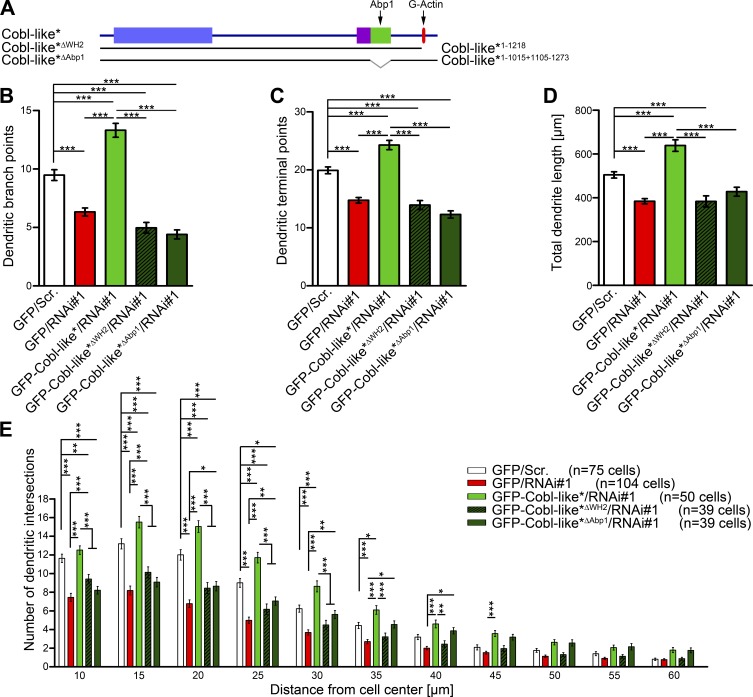
Cobl-like’s functions in promoting F-actin–rich ruffles in COS-7 cells relied on the WH2 domain and on an Abp1-associating interface. We next addressed the relevance of these findings for Cobl-like’s physiological functions in developing hippocampal neurons. Whereas reexpression of WT Cobl-like fully rescued all Cobl-like RNAi phenotypes, an RNAi-insensitive Cobl-like mutant lacking the WH2 domain (GFP-Cobl-like*ΔWH2) failed to rescue. Quantitative evaluations demonstrated that the numbers of dendritic branch points and terminal points as well as the total dendrite length remained as low as for Cobl-like–deficient cells. Also, Sholl analyses demonstrated that without the WH2 domain no rescue of the Cobl-like loss-of-function phenotypes was obtained (Fig. 7, A–E).
Figure 7.
Cobl-like functions in dendritogenesis strongly rely on the actin-associating WH2 domain and on the Abp1 interaction capability of Cobl-like. (A) Scheme of Cobl-like mutants used for functional analyses. (B–E) Functional analyses testing Cobl-like*ΔWH2 (Cobl-like*1-1218) and Cobl-like*ΔAbp1 (Cobl-like*1-1015+1105-1273) mutants reexpressed in neurons deficient for Cobl-like for their ability to restore Cobl-like–mediated dendritic branch formation (Scrambled control [Scr.], Cobl-like RNAi, and WT Cobl-like rescue data; see Fig. 4 for comparison). Data are mean ± SEM, one-way ANOVA plus Tukey’s post test (B–D) and two-way ANOVA plus Bonferroni’s post test (E). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Also, GFP-Cobl-like*ΔAbp1 failed to rescue the Cobl-like RNAi phenotypes in dendritic branching. The number of dendritic branch points and terminal points as well as the total length of the dendritic arbor was as low as in Cobl-like RNAi cells (Fig. 7, A–D). Thus, Cobl-like’s ability to interact with G-actin via its WH2 domain and to interact with the F-actin–binding protein Abp1 are both required for Cobl-like’s functions in dendritogenesis. Also, Sholl analyses confirmed that rescue attempts with GFP-Cobl-like*ΔAbp1 failed. Especially in the proximity of the cell bodies, Sholl intersections remained statistically significantly fewer than control, as also observed for Cobl-like loss-of-function (Fig. 7 E).
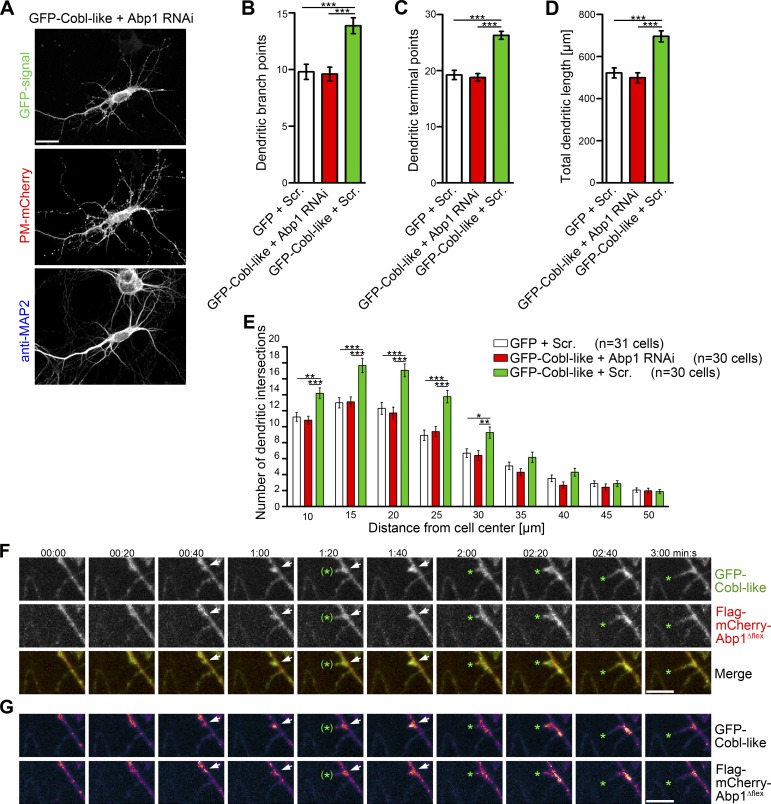
Next, we asked whether the Cobl-like gain-of-function phenotype in dendritic arborization indeed explicitly relies on Abp1. Abp1 RNAi completely suppressed both Cobl-like–mediated branching and growth of the dendritic arbor of developing hippocampal neurons (Fig. 8, A–E; and Fig. S5, D and E).
Figure 8.
Cobl-like–mediated dendritic arborization relies on Abp1 and both proteins coincide at nascent branch induction sites. (A) MIPs of primary hippocampal neurons transfected with GFP-Cobl-like and Abp1 RNAi. Bars, 20 µm. (B–E) Quantitative analyses of indicated dendritic parameters addressing whether the Cobl-like gain-of-function phenotypes are Abp1-dependent. Data are mean ± SEM, one-way ANOVA plus Tukey’s post test (B–D) and two-way ANOVA plus Bonferroni’s post test (E). (F and G) Individual frames (MIPs) of 3D time lapse recordings of GFP-Cobl-like and Flag-mCherry-Abp1Δflex in a dendrite of a hippocampal neuron. Labeling and heat map representation (G) are explained in Fig. 3. Bars, 2 µm. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
3D time-lapse recordings showed that Cobl-like accumulations at nascent branch initiation sites, which seconds later gave rise to protrusions, coincided with Abp1Δflex and that spatial dynamics of Abp1 and Cobl-like were largely synchronous (Fig. 8, F and G).
Cobl-like’s role in dendritic branch induction is controlled by Ca2+/CaM signaling
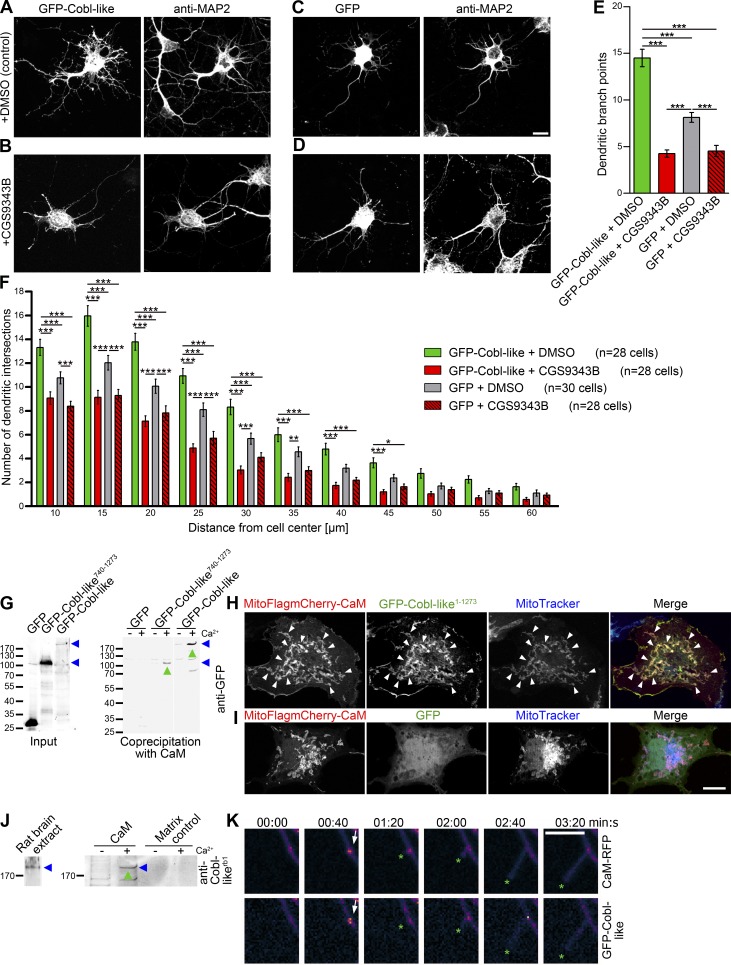
We have recently shown that the actin nucleator Cobl is controlled by transient, local Ca2+/CaM signaling (Hou et al., 2015). Our work thus far had unveiled that Cobl-like also plays a critical role in dendritic arbor formation, albeit by using different molecular mechanisms. We therefore next asked whether Cobl-like–mediated dendritic arborization may also be dependent on Ca2+/CaM signaling. Strikingly, application of the CaM inhibitor CGS9343B to GFP-Cobl-like–transfected neurons resulted in a complete block of Cobl-like–mediated functions. The number of branch points per cell dropped to the levels of untransfected cells incubated with inhibitor (Fig. 9, A–E). Sholl analysis showed that CaM inhibition affected Cobl-like–mediated branching in the entire dendritic arbor (Fig. 9 F).
Figure 9.
Cobl-like–mediated dendritogenesis is suppressed by CaM inhibition and Cobl-like associates with CaM and dynamically coenriches with CaM at nascent branch sites. (A–D) Hippocampal neurons transfected as indicated and treated with DMSO (A and C) and CaM inhibitor CGS9343B (B and D), respectively. Bar, 10 µm. (E and F) Quantitative morphological examinations (dendritic branch point [E] and Sholl analyses [F]). Data are mean ± SEM, one-way ANOVA plus Tukey’s post test (E) and two-way ANOVA plus Bonferroni’s post test (F). (G) Anti-GFP immunoblotting analyses of coprecipitation experiments with immobilized CaM and GFP-Cobl-like, GFP-Cobl-like740–1273 (both marked by blue arrowheads) and GFP, respectively, in absence (−) and presence (+) of Ca2+ (500 µM). Green upward arrowheads highlight Ca2+-dependent associations of Cobl-like and Cobl-like740–1273 with CaM. (H and I) Reconstitutions of CaM/Cobl-like complexes with mitochondrially targeted CaM (MitoFlagmCherry-CaM) in combination with GFP-Cobl-like (H) and GFP (I), respectively, in intact COS-7 cells (arrowheads mark examples of successful reconstitutions of protein complexes). Bar, 10 µm. (J) Immunoblotting analyses of Ca2+-dependent (2 µM Ca2+; green upward arrowheads) coprecipitation analyses with immobilized CaM and endogenous Cobl-like from rat brain extracts (blue arrowheads). (K) Individual frames (MIPs) of heat maps of fluorescence intensities of 3D time-lapse recordings of GFP-Cobl-like and CaM-RFP in DIV7 hippocampal neurons. Arrows mark accumulations of both proteins at a nascent branch initiation site. Green asterisks mark the protrusion tip. Bar, 5 µm. Data are mean ± SEM, one-way ANOVA plus Tukey’s post test (B–D) and two-way ANOVA plus Bonferroni’s post test (E). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
In addition, the inhibitor already resulted in some reduction of dendritic branch formation in GFP-expressing control cells when compared with untreated control cells (Fig. 9, C–F). Thus, both dendritic branching in general and Cobl-like–mediated dendritic branching is controlled by Ca2+/CaM signaling.
CaM associates with Cobl-like and accumulates at Cobl-like–enriched branch initiation sites
Because Cobl-like functions are controlled by Ca2+/CaM signaling and CaM can modulate the activity of target proteins by direct, physical association, we next asked whether CaM interacts with Cobl-like and if so, whether such interactions would be Ca2+-dependent and involve the Abp1- and actin-binding C terminus. CaM indeed precipitated both Cobl-like full-length and C terminus in a Ca2+-specific manner (Fig. 9 G). Furthermore, specific recruitments of Cobl-like to CaM-enriched sites in COS-7 cells showed that the Cobl-like/CaM interaction is of relevance in intact cells (Fig. 9, H and I). These findings were corroborated further by the fact that immobilized CaM precipitated endogenous Cobl-like from rat brain extracts (Fig. 9 J).
3D time-lapse analyses demonstrated a dynamic coenrichment of Cobl-like and CaM at nascent branch sites. Similar to the accumulation of Cobl-like, CaM also accumulated at branching sites during the seconds before protrusion initiation (Fig. 9 K).
A CaM association site adjacent to Cobl-like’s Abp1-binding region is required for dendritogenesis
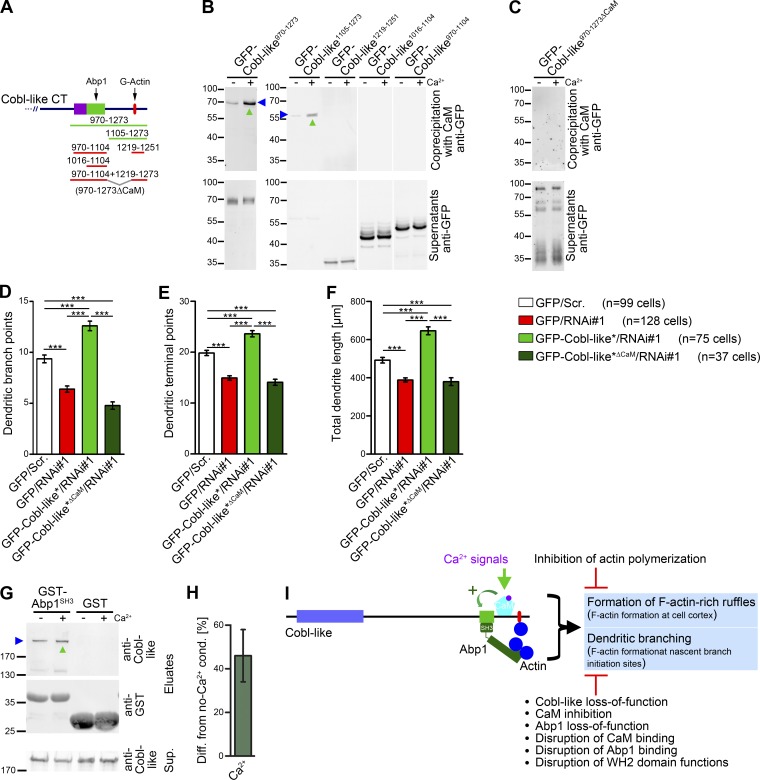
To obtain first insights into how Cobl-like’s role in dendritic branching may be regulated by Ca2+/CaM signaling, we next conducted detailed mutational analyses. Interestingly, these experiments unveiled a site critical for CaM binding that is located between the Abp1-binding region and the WH2 domain (Fig. 10, A and B).
Figure 10.
Cobl-like’s role in dendritic arbor formation relies on a C-terminal–binding site for the Ca2+-sensor CaM, and Ca2+ promotes the association of Cobl-like’s crucial cofactor Abp1. (A) Scheme of C-terminal Cobl-like deletion mutants used for mapping of CaM-binding (binding, green lines; not binding, red lines). (B and C) Coprecipitation studies with GFP-Cobl-like deletion mutants (B) as well as with Cobl-like970–1273ΔCaM (Cobl-like970–1104+1219–1273; C) and immobilized CaM (±Ca2+; 500 µM). (D–F) Functional analyses in primary hippocampal neurons unveiling that reexpression of Cobl-like*ΔCaM fails to rescue the indicated Cobl-like loss-of-function phenotypes (i.e., data not statistically significantly different from Cobl-like RNAi data). Data are mean ± SEM, one-way ANOVA plus Tukey’s post test. (G) Anti–Cobl-like immunoblottings unveiling that 2 µM Ca2+ increases the ability of endogenous Cobl-like from rat brain extracts to associate with Abp1SH3 (marked by upward green arrowhead). (H) Quantitative analyses of Cobl-like/Abp1 complex formation show an increase of 46 ± 12% on Ca2+ presence (n = 3 assays; data are mean ± SEM). (I) Model summarizing the cell biological functions of Cobl-like and the molecular mechanisms it uses. ***, P < 0.001.
To address whether indeed CaM/Cobl-like complex formation is required for dendritic branching, we generated a deletion mutant lacking the identified site (Cobl-like970–1104+1219–1273; Cobl-like970–1273ΔCaM). Coprecipitation experiments confirmed that this Cobl-like mutant did not associate with CaM anymore (Fig. 10, A and C).
An RNAi-insensitive version (Cobl-like*ΔCaM) indeed failed to rescue the Cobl-like loss-of-function phenotypes in dendritic branching. Dendritic branch points, terminal points, and the total dendrite length of primary hippocampal neurons expressing an RNAi-insensitive version of Cobl-like*ΔCaM together with Cobl-like RNAi all remained as affected as on Cobl-like RNAi, i.e., none of the three parameters was rescued by the Cobl-like mutant lacking the CaM-binding site (Fig. 10, D–F).
The ability of Ca2+/CaM signaling to converge on the cytoskeletal component Cobl-like thus is of critical importance for Cobl-like’s function in dendrite branching.
Ca2+/CaM signaling promotes the functionally crucial Abp1/Cobl-like complex formation
Because we discovered that Ca2+/CaM signaling as well as Abp1 presence and Abp1 complex formation with Cobl-like are crucial for Cobl-like’s functions, we next asked whether these aspects may be mechanistically interconnected. Strikingly, coprecipitation assays unveiled that under conditions of CaM association, the Abp1 SH3 domain interacted more effectively with endogenous Cobl-like from rat brain compared with conditions of inhibited Ca2+/CaM signaling (Fig. 10 G). Cobl-like/Abp1SH3 interactions in the presence of 2 µM Ca2+ were 46 ± 12% above control levels (Fig. 10 H).
Ca2+ signals thus lead to increased complex formation of Cobl-like with its crucial cofactor Abp1, and our studies strongly suggest that this molecular mechanism is a prerequisite for proper dendritic branch formation by Cobl-like.
Discussion
Establishment and adaptations of the elaborate shapes of neurons underlie information processing as well as regeneration processes in the brain. Both processes are expected to involve signals that are converted into locally restricted and transient activity of force-generating cytoskeletal effectors at the cell cortex (Lohmann, 2009). In neurons, Ca2+ triggers important locally restricted signaling cascades. Yet, thus far, only very few direct links of Ca2+ signaling to the actin cytoskeleton have been discovered in developing neurons. However, neither the described CaM associations with brain-enriched spectrin isoforms (Riederer et al., 1986) nor competitive binding of the F-actin–bundling protein α-actinin and CaM to N-methyl-d-aspartic acid–type glutamate receptors and L-type Ca2+ channels (Wyszynski et al., 1997; Hall et al., 2013) offered any obvious mechanisms that could power dendritic arborization in response to the transient Ca2+ signals that are observable in dendritic trees of developing neurons. These Ca2+ signals, which were shown to be mediated by N-methyl-d-aspartic acid receptors, voltage-gated calcium channels, and ryanodine receptors (Rajan and Cline, 1998; Chevaleyre and Castillo, 2002; Ohashi et al., 2014), seem to be sensed and transmitted by the Ca2+ sensor protein CaM, because inhibition of CaM-dependent kinases caused impairments in dendritogenesis (Fink et al., 2003; Gaudillière et al., 2004).
Our examinations unveil that the thus far uncharacterized gene product Cobl-like is a Ca2+/CaM-responsive, G-actin–binding protein that promotes dendritogenesis of developing neurons in a CaM-dependent manner. Strikingly, Cobl-like thereby seems to functionally resemble its distant relative, the actin nucleator Cobl (Ahuja et al., 2007; Hou et al., 2015), although sequence conservation is only 24.8% and neither conserved CaM-binding sites nor Cobl’s C-terminal domain topology can be found in Cobl-like. In contrast to Cobl, which uses a combination of three WH2 domains to nucleate actin (Ahuja et al., 2007), Cobl-like has only a single WH2 domain. We demonstrated by in vitro and in vivo experimentation that this WH2 domain is functional. Cobl-like thus effectively associates with G-actin. Consistently, in vitro reconstitutions showed that the WH2 domain–containing C terminus of Cobl-like sequesters G-actin. Strikingly, however, Cobl-like did not display the disperse G-actin distribution in cells but instead colocalized with F-actin. Also, Cobl-like did not seem to quench G-actin in vivo but instead massively promoted the formation of F-actin–rich 3D ruffles in COS-7 cells. It is possible that this Cobl-like–mediated F-actin formation relates to findings linking the Cobl-like gene to different types of cancer (Gordon et al., 2003, 2009; Wang et al., 2013; Han et al., 2017), because changed cytoskeletal organization and dynamics are key characteristics of tumor cells.
In developing neurons, an excess of Cobl-like led to a dramatic increase of new dendritic branches protruding from F-actin–rich sites in the dendritic arbor. This Cobl-like gain-of-function effect indeed reflected the physiological role of Cobl-like because, conversely, Cobl-like RNAi led to a strong impairment in dendritic arborization. The numbers of dendritic branch points, terminal points, and the total length of the dendritic arbor were all strongly affected by Cobl-like RNAi. Sholl analyses confirmed these findings. Cobl-like thus is a critical component in dendritic arbor formation.
In line with the identified critical cell biological function of Cobl-like in dendritogenesis, Cobl-like was predominantly expressed in the brain. It could be detected in all parts of the central nervous system analyzed and occurred in neurons. These similarities with Cobl in adult mice are in contrast to the description that, at least in very early development, Cobl-like (Coblr) and Cobl mRNA can be observed in unrelated patterns (Carroll et al., 2003).
Mutational analyses clearly demonstrated that the cell-shaping, cytoskeletal functions of Cobl-like were WH2 domain–dependent, because Cobl-like mutants lacking the WH2 domain were neither able to promote the formation of F-actin–rich ruffles in COS-7 cells nor able to rescue the Cobl-like loss-of-function phenotypes in the dendritic branching of neurons. The WH2 domain sequestered G-actin in in vitro reconstitutions, and removal of G-actin from the pool of actin monomers available for filament formation would oppose the formation of actin filaments. Therefore, G-actin binding of the WH2 domain could only be one important aspect of the molecular mechanisms underlying Cobl-like’s functions in vivo and some additional mechanism had to be involved (Fig. 10 I). Indeed, our mutational analyses in both COS-7 cells and primary hippocampal neurons unveiled that Cobl-like functions critically relied on the WH2 domain and a region in some distance from the WH2 domain. Although this finding seemed somewhat reminiscent of the actin nucleator Lmod2, which uses its single WH2 domain in combination with an F-actin–binding domain to promote actin filament formation in vitro (Chereau et al., 2008), we did not observe any direct F-actin binding of the respective part of Cobl-like. Instead, we identified this region in Cobl-like to be an SH3 domain-binding site for the F-actin–binding protein, Abp1. Thus, Cobl-like seems to use a mechanism that involves indirect F-actin association via its interaction partner Abp1 and G-actin association via its own WH2 domain to form F-actin–rich, protrusive structures in cells.
Consistently, a Cobl-like mutant lacking the ability to associate with Abp1 (Cobl-like*ΔAbp1) was neither able to elicit the formation of F-actin–rich 3D ruffles in COS-7 cells nor able to rescue the Cobl-like loss-of-function phenotypes in dendritic branching. That indeed specifically Abp1 as binding partner is required for Cobl-like’s functions in dendritic branching was demonstrated by the fact that Abp1 RNAi completely suppressed Cobl-like–mediated dendritogenesis.
Thus, Cobl-like functions in dendritogenesis rely on both the G-actin–binding WH2 domain and on association with the F-actin–binding protein Abp1 to a site located more N-terminally from the WH2 domain (Fig. 10 I). The identification of such a combinatory mechanism for effective promotion of F-actin formation in different cells suggests that it may be useful to revisit and carefully analyze the plethora of single WH2 domain–containing proteins for related mechanisms at both the molecular and the functional level.
From an evolutionary point of view, it is interesting that also the actin nucleator Cobl, which had been proposed to have emerged from the ancestor protein Cobl-like millions of years later, i.e., with the emergence of vertebrates (Schultz and Terhoeven, 2013), is an Abp1-binding protein (Haag et al., 2012). Yet, the mechanisms that Cobl and Cobl-like use seem to be different. The Abp1-binding regions of both proteins do not show obvious sequence conservation. Furthermore, they are located at distinct positions, i.e., in the N terminus of Cobl versus in the C terminus of Cobl-like.
It is also interesting that both proteins are regulated by Ca2+/CaM signaling. Ca2+/CaM signaling modulates the actin binding of the WH2 domains of Cobl (Hou et al., 2015). In the case of Cobl-like, we unveiled that the crucial complex formation with Abp1 is regulated by Ca2+/CaM signaling. This mechanism of regulation correlated with the identification of a CaM-binding site adjacent to the Abp1-binding region. In line with an important role of Ca2+/CaM signaling in Cobl-like–mediated dendritic arborization, Cobl-like and CaM both accumulated specifically at nascent dendritic branch points during the seconds before dendritic branch induction, and Cobl-like–mediated dendritic branching was completely suppressed by the CaM inhibitor CGS9343B. Furthermore, a Cobl-like mutant lacking the identified CaM-binding site was unable to rescue the defects in dendritic arbor formation caused by Cobl-like RNAi (Fig. 10 I).
The observation that Ca2+/CaM-mediated mechanisms control Cobl-like’s activity in neurons unveils how local Ca2+ signals steer and power branch initiation during early arborization of nerve cells. Also, the formin INF2 has recently been identified as a CaM-binding protein (Wales et al., 2016). This strongly suggests that Ca2+/CaM-controlled actin filament formation, which was originally identified for the actin nucleator Cobl (Hou et al., 2015), may be a thus-far unrecognized, general principle in cell biology that is of much more wide-spread use for cytoskeletal control and for shaping cells than initially thought.
Materials and methods
DNA constructs
Plasmids encoding for GFP-, FlagGFP- and Flag-mCherry-Cobl-like and parts thereof were generated by PCR by using the EST clone UniProtID Q3UMF0 as template, by using an internal EcoRI site for subclonings and fusing N-terminal and C-terminal parts and by subcloning into pEGFP (Clontech) and into FlagGFP and Flag-mCherry derivatives of a pCMV vector, respectively. Cobl-like fragments were generated by using internal restriction sites (at aa 456/457, aa 536/537, aa 740/741, and aa 1250/1251) and/or by PCRs combining the following primers—aa 1 forward: 5′-AATTAGATCTATGGACCGCAGCGTCCCGGATCC-3′, aa 261 forward: 5′-AAAGATCTGATATCAGCAGAGAG-3′, aa 537 (BglII) forward: 5′-AAAGATCTAAGGATCCTGATTCAGC-3′, aa 740 forward: 5′-GCCTCAAGAGAATTCAGG-3′, aa 970 forward: 5′-AAGAATTCGTCCACACTGCCCCTG-3′, aa 1016 forward: 5′-AAGATATCGCTGATGATGATATAATTC-3′, aa 1024 forward: 5′-AAGGATCCCCTGCCGAGACCTCTC-3′, aa 1105 forward: 5′-AAGATATCGGTAGTCAGGGAAC-3′, or aa 1219 forward: 5′-AAGGATCCCAGAGCTCTGACCCAGAG-3′ with aa 741 reverse: 5′-TCCTGAATTCTCTTGAGG-3′, aa 970 reverse: 5′-TTGATATCGACGCTCTTAGCAGC-3′, aa 1015 reverse: 5′-TTGATATCTTTTGAAATATTGG-3′, aa 1104 reverse: 5′-TTTGTCGACAGGGAGAGCCACAGG-3′, aa 1218 reverse: 5′-AAGAATTCGAATGTTAGAGAACTCTG-3′, or aa 1251 reverse: 5′-TTGAATTCTTAATTCGATGGAACAGTAAC-3′.
Cobl-like deletion mutants, such as ΔAbp1 and ΔCaM binding as well as ΔWH2 domain, were generated by combining the respective parts of Cobl-like by introducing an EcoRV site.
Cobl-likeΔAbp1 mutants were generated by combining a PCR product (primers, aa 740 forward and aa 1015 reverse) with a second PCR product (primers, aa 1105 forward and aa 1273 reverse). The fused sequence encoding for a mutated C-terminal half was then fused with a sequence encoding for the Cobl-like N-terminal half to generate a full-length version of the mutant.
Cobl-likeΔCaM mutants were generated by fusing a PCR product (primers, aa 740 forward and aa 1104 (EcoRV) reverse: 5′-TTGATATCAGGGAGAGCCACAGG-3′) with a second PCR product (primers, aa 1219 (EcoRV) forward: 5′-AAGATATCCAGAGCTCTGACCCAG-3′, and aa 1273 reverse). Fusion of the resulting product with a sequence encoding for the Cobl-like N terminus by using the internal EcoRI site at aa 740/741 then resulted in the respective mutated full-length protein.
Cobl-likeΔWH2 was generated by PCR (primers, aa 737 forward and aa 1218 reverse) and fusing the resulting PCR product to the sequence encoding for the Cobl-like N terminus.
Plasmids encoding for GST fusion proteins of Cobl-like were generated by PCR using the primers described in the previous section and by subcloning into pGEX-5X-1 and pGEX-4T-2 (GE Healthcare), respectively.
Plasmids encoding for TrxHis-Cobl-like proteins were generated by PCR using the primers described in the previous section and by subcloning into pET-32 (Novagen).
RNAi constructs that are directed against mouse and rat Cobl-like and are coexpressing GFP were generated according to procedures described previously (Ahuja et al., 2007). In brief, phosphorylated primers for RNAi#1 (788–808 bp; target sequence 5′-GCAGAGAGTCCTGCCAAATAT-3′), RNAi#2 (947–967 bp; target sequence 5′-GGTCCAACACCATTTCCAAAC-3′) and RNAi#3 (569–587 bp; target sequence 5′-CTGTGAGAGTAGTGATCAA-3′) were annealed, and the products were subcloned into pRNAT H1.1-GFP. RNAi#1 was also inserted into a pRNAT vector coexpressing farnesylated mCherry (mCherry-F; pRNAT-mCherryF). Corresponding pRNAT vectors expressing a scrambled RNAi sequence served as controls (Pinyol et al., 2007).
For quantitative RNAi validations, GFP was replaced by GFP-Cobl-like1–457 in scrambled RNAi and RNAi#1–3 vectors.
An RNAi-insensitive GFP-Cobl-like (GFP-Cobl-like*) was generated by introducing several silent mutations into the RNAi#1 site to render the mRNA resistant to Cobl-like RNAi#1. The primers used were 5′-GATATCTCACGCGAAAGCTGTCAGATCAGTCACAAC-3′ as the forward and 5′-GTTGTGACTGATCTGACAGCTTTCGCGTGAGATATC-3′ as the reverse primer. Cobl-like RNAi constructs and scrambled RNAi constructs coexpressing RNAi-insensitive GFP-Cobl-like full-length as well as mutants thereof lacking the WH2 domain, Abp1-binding site and CaM-binding site, respectively, as well as related constructs in a scrambled RNAi vector were constructed by cloning into the respective pRNAT vectors by using NheI and SmaI restriction sites.
LifeAct-RFP was a gift from K. Murk (Charité Universitätsmedizin Berlin, Berlin, Germany) and described previously (Riedl et al., 2008). A vector expressing PM-targeted (farnesylated) mCherry was provided by M. Korte (Technische Universität Braunschweig, Braunschweig, Germany).
Plasmids encoding for GFP- and GST-tagged Abp1 full-length and SH3 domain, respectively, as well as for a P422L,G425R-mutated SH3 (Abp1SH3mut) mutant were as described (Kessels et al., 2000, 2001; Haag et al., 2012).
The more stable Abp1 derivative Abp1Δflex was generated by PCR amplification of the ADF-H and helical domains (aa 1–281) and the SH3 domain (aa 371–443) and combining them by an introduced EcoRI site (primers, aa 1 forward: 5′-CCCGGGATCCATGGCGGTGAACCTGAGCC-3′, aa 281 reverse: 5′-CTCGGAATTCTCACGAGCTGGTGACAGAGGTGG-3′, aa 371 forward: 5′-CTCGGAATTCGGCTTCAGTGGACAAGGGC-3′, and aa 433 reverse: 5′-TATGCCAAGCTTATCACTCTATGAGCTCCACGTAG-3′). A version comprising a mutated SH3 domain was generated accordingly by using Abp1SH3mut as PCR template.
The resulting Abp1Δflex mutant was cloned into pGAT (Kessels et al., 2000) and pEGFP vectors. Myc-Abp1Δflex was generated by subcloning into pRK5. MitoFlag-Abp1 and MitoFlag-Abp1SH3 were as described previously (Pinyol et al., 2007). MitoFlag-Abp1Δflex and MitoFlag-Abp1Δflex SH3mut were generated by subcloning.
Flag-mCherry-Abp1Δflex was generated by subcloning into a Flag-mCherry vector (Hou et al., 2015). Plasma membrane-targeted mCherry-coexpressing Abp1 RNAi (181–201 bp) vector and the corresponding control pRNAT vector were described previously (Pinyol et al., 2007; Haag et al., 2012).
Flag-GFP-CoblWH2 (2) and GST-CoblWH2 were as described previously (Ahuja et al., 2007).
GST-CaM, TrxHis-CaM, and MitoFlagmCherry-CaM were as described (Hou et al., 2015). CaM-RFP was generated by PCR and subcloning into an RFP-derivative of pEGFP-N1 (by using BglII and BamHI site insertion into BamHI; primers, aa 1 forward: 5′-AAAGATCTGCCACCATGGCTGATCAG-3′, and aa 149 reverse: 5′-CCGGATCCCATTTTGCAGTCATCATC-3′.
Correct cloning by PCR was verified by sequencing in all cases.
Antibodies, reagents, and proteins
Rabbit anti–Cobl-like antibodies were raised against a combination of two GST-Cobl-like fusion proteins (GST-Cobl-like537–741 and GST-Cobl-like740–1015). Terminal bleeds were collected at day 100 and tested for immunoreactivity. Rabbit #1 showed a strong immune response. Affinity purifications of serum were done on immobilized TrxHis-Cobl-like537–1015 according to procedures described previously (Qualmann et al., 1999; Kessels et al., 2000). In brief, purified TrxHis-tagged Cobl-like537–1015 was subjected to preparative SDS-PAGE and transferred to nitrocellulose. After blocking with BSA, blot pieces with the immobilized antigen were incubated with anti–Cobl-like sera, washed, and eluted with 5 mM glycine, pH 2.3, in 500 mM NaCl, 0.5% Tween 20, and 0.01% BSA. Eluates were neutralized immediately and characterized.
Rabbit anti-Abp1 and anti-GST as well as anti-TrxHis antibodies were described previously (Qualmann et al., 1999; Kessels et al., 2000; Fenster et al., 2003; Braun et al., 2005).
Polyclonal rabbit anti-GFP and anti-MAP2 antibodies were from Abcam (ab290 and ab32454). Monoclonal mouse anti-GFP antibodies (JL-8) were from Clontech (632381). Monoclonal mouse (M2) and polyclonal rabbit anti-Flag antibodies were from Sigma (F3165 and F7425). Monoclonal anti–β-actin, anti-MAP2 (HM-2), anti–β-tubulin, and anti–glial fibrillary acidic protein antibodies also were from Sigma (A5441, M4403, T4026, and G3893). Monoclonal anti-Myc antibodies (9E10) and anti-SMI312 antibodies were from Covance (MMS-150P and SMI-32P). Polyclonal rabbit anti-CaM antibodies were from Cell Signaling Technology (4830).
Alexa Fluor 488– and Alexa Fluor 647–conjugated phalloidin and MitoTracker Deep Red 633 were from Molecular Probes.
Secondary antibodies used included Alexa Fluor 488– and Alexa Fluor 568–labeled goat anti–guinea pig antibodies (A-11073 and A-11075), Alexa Fluor 488– and Alexa Fluor 568–labeled donkey anti–mouse antibodies (R37114 and A10037), Alexa Fluor 647– and Alexa Fluor 680–labeled goat anti–mouse antibodies (A-21236 and A32723), Alexa Fluor 488–labeled donkey anti–rabbit (A-21206), Alexa Fluor 568–, Alexa Fluor 647–, and Alexa Fluor 680–labeled goat anti–rabbit antibodies (A-11036, A-21245, and A-21109; Thermo Fisher Scientific) as well as DyLight800-conjugated goat anti–rabbit and anti–mouse antibodies (35571 and 35521; Thermo Fisher Scientific). Donkey anti–guinea pig antibodies coupled to IRDye680 and IRDye800, respectively were from LI-COR Bioscience (925-32411 and 926-32411). Goat anti–rabbit, anti–guinea pig, and anti–mouse-peroxidase antibodies were from Dianova (711-035-152, 106-036-003, and 115-036-003; Jackson ImmunoResearch), and alkaline phosphatase-conjugated anti-Dig-Fab fragments used for in situ hybridizations were from Roche (11 093 274910).
Rabbit skeletal muscle actin and pyrene-labeled rabbit skeletal muscle actin were from Cytoskeleton. Alexa Fluor 488–labeled actin was from Thermo Fisher Scientific. Sepharose 4B-coupled CaM was from GE Healthcare. GST- and TrxHis-tagged fusion proteins were purified from Escherichia coli as described previously (Qualmann and Kelly, 2000; Schwintzer et al., 2011). In brief, both GST and TrxHis proteins were expressed in E. coli BL21. GST fusion proteins were purified from bacterial lysates on glutathione-agarose or -sepharose (Sigma; GenScript) and eluted with 20 mM glutathione in 120 mM NaCl and 50 mM Tris, pH 8.0. TrxHis fusion proteins were purified by using Talon metal affinity resin (Clontech) and eluted with 250 mM imidazole, 300 mM NaCl, and 50 mM Na2HPO4, pH 7.0. After purification, fusion proteins were dialyzed against PBS, characterized by SDS-PAGE, and either snap-frozen and stored at −80°C or stored in glycerol at −20°C.
In vitro reconstitutions of direct protein–protein interactions
Direct protein–protein interactions were demonstrated by coprecipitations with combinations of recombinant TrxHis- and GST-tagged fusion proteins purified from E. coli. Complex formation of TrxHis-Cobl-like970–1273 with Abp1 fusion proteins was demonstrated in 10 mM Hepes, pH 7.4, 300 mM NaCl, 0.1 mM MgCl2, and 1% (vol/vol) Triton X-100 supplemented with EDTA-free protease inhibitor cocktail. Eluted proteins were analyzed by SDS-PAGE, transferred to polyvinylidene fluoride membranes by either tank or semi-dry blotting and then subjected to immunodetection with anti-TrxHis and anti-GST antibodies by using a Licor Odyssey System.
In vitro actin assays
Actin bead assays were conducted with Alexa Fluor 588–labeled actin, rat brain extract, and an energy-regenerating mix as well as immobilized GST-tagged fusion proteins as described previously (Ahuja et al., 2007; Pinyol et al., 2007). In brief, GST-fusion proteins were immobilized on glutathione sepharose 4B beads (Amersham Bioscience) and incubated with rat brain lysates in 10 mM Hepes, pH 7.0, 0.5 mM DTT, 1 mM ATP, and EDTA-free protease inhibitor cocktail (Roche) that were supplemented 1:10 with 150 mM creatine phosphate, 20 mM ATP, and 20 mM MgCl2 as the energy-regenerating system and Alexa Fluor 488–coupled G-actin to visualize recruited actin and putative F-actin structures formed. After preincubation on ice (5 min), the reactions were initiated by the temperature shift caused by transfer onto glass slides and viewed at RT by fluorescence microscopy.
Pyrene actin assays were performed with rabbit skeletal muscle actin (4 µM final) and pyrene-labeled rabbit skeletal muscle actin (0.4 µM final) according to procedures described previously (Ahuja et al., 2007; Pinyol et al., 2007). In brief, polymerization reactions with 4 µM G-actin and 0.4 µM pyrene-actin in G-buffer (10 mM Tris-HCl, pH 8.0, 100 mM KCl, 1 mM MgCl2, 0.1 mM EDTA, and 10 mM DTT) were started with a 10× polymerization initiation mix containing 10 mM KCl, 10 mM MgCl2, and 10 mM ATP in G-buffer. The change in the fluorescence of pyrene-actin was measured at 407 nm in a fluorescence spectrometer (excitation 365 nm; 5-s time increments; 1-s integration time; Fluoromax 3; Jobin Yvon, Horiba Group).
F-actin–pelleting assays were conducted with rabbit muscle actin and purified, recombinant GST-Cobl-like970–1273 and GST, respectively, largely as described previously (Kessels et al., 2000). In brief, F-actin formation was started by adding initiation mix (10×; composed of 500 mM KCl, 20 mM MgCl2, and 5 mM ATP) to 10 µM G-actin in G-actin buffer (5 mM Tris-HCl, pH 7.5, 0.2 mM DTT, 0.2 mM CaCl2, and 0.2 mM ATP). The F-actin was then incubated with proteins (30 min, RT). Subsequent to ultracentrifugation (352,000 g; 90,000 rpm; TLA100 rotor; Beckmann Instruments), equivalent portions of pellet (F-actin and bound protein) and supernatant (unbound protein) were analyzed by SDS-PAGE and Coomassie staining (actin) and immunoblotting (GST and GST-Cobl-like970–1273) with anti-GST antibodies, respectively.
Preparation of HEK293 cell lysates
24–48 h after transfection, HEK293 cells were washed with PBS, harvested, and subjected to sonification for 10 s and/or lysed by incubation in lysis buffer (10 mM Hepes, pH 7.4, 0.1 mM MgCl2, 1 mM EGTA, and 1% Triton X-100) containing 150 mM NaCl and EDTA-free protease inhibitor Complete (Roche) for 20–30 min at 4°C (Kessels and Qualmann, 2006). Cell lysates were obtained as supernatants from centrifugations at 20,000 g (20 min at 4°C).
Coprecipitation of proteins from HEK293 cell lysates
Coprecipitation experiments with extracts from HEK293 cells expressing different GFP-Cobl-like proteins were essentially done as described before (Qualmann et al., 1999; Schwintzer et al., 2011). In brief, HEK293 cell lysates were incubated for 3 h at 4°C with purified, recombinant GST-fusion proteins immobilized on glutathione sepharose beads (GenScript). The reactions were washed several times with lysis buffer containing 150 mM NaCl and EDTA-free protease inhibitor Complete. Bound protein complexes were eluted either with 20 mM reduced glutathione, 120 mM NaCl, and 50 mM Tris-HCl, pH 8.0 (30 min RT), or by boiling the beads in 4× SDS sample buffer.
For coprecipitations with CaM, HEK293 cell lysates were prepared in an EGTA-free lysis buffer containing 150 mM NaCl, EDTA-free protease inhibitor cocktail, and 200 µM calpain I inhibitor. Cell lysates were supplemented with 1 mM EGTA or 500 µM CaCl2. After incubation with 25 µl CaM-sepharose 4B (GE Healthcare) for 3 h at 4°C and washing, bound proteins were isolated by boiling in SDS sample buffer.
Lysates, supernatants, and eluates were analyzed by immunoblotting using anti-GST and anti-GFP antibodies, respectively.
Coimmunoprecipitation analyses
For coimmunoprecipitations of endogenous actin together with GFP-tagged Cobl-like deletion mutants, protein A agarose beads were coated for ∼4 h with rabbit anti-GFP antibodies and nonimmune IgG, respectively, and were then washed with PBS and with EGTA-free lysis buffer containing 100 mM NaCl. The antibody-coated beads were incubated overnight with lysates of HEK293 cells transfected with FlagGFP-Cobl-likeWH2 (Cobl-like1105–1273), flagGFP-CoblWH2 (2), and FlagGFP, respectively. The lysates were made by using EGTA-free lysis buffer containing 100 mM NaCl. After washing, immunoprecipitated GFP proteins and coimmunoprecipitated proteins were eluted by boiling with SDS-PAGE sample buffer. Eluates were immunoblotted with anti-GFP and anti-actin antibodies and fluorescently labeled secondary antibodies and analyzed by using a Licor Odyssey System.
Heterologous coimmunoprecipitations addressing Cobl-like/Abp1 interactions were done with lysates of HEK293 cells transfected with GFP-Cobl-like fusion proteins and GFP, respectively, in combination with Myc-Abp1Δflex. The cell lysates were incubated with 5 µg rabbit anti-Myc antibody and nonimmune IgG, respectively, in lysis buffer containing 60 mM NaCl and protease inhibitor cocktail for 3 h at 4°C. Antibody-associated protein complexes were isolated by 2-h incubation with protein A–agarose (Santa Cruz Biotechnology) at 4°C. The immunoprecipitates were washed with lysis buffer containing 60 mM NaCl and eluted from the matrix by boiling in a mix of 2 M (final) urea and SDS-sample buffer. The eluates were immunoblotted with anti-GFP and anti-Myc antibodies and analyzed quantitatively by using fluorescently labeled secondary antibodies and a Licor Odyssey System.
Mouse and rat tissue extracts
Tissue extracts were prepared from mice sacrificed by cervical dislocation (for tissue distribution analyses) and from rats sacrificed by CO2 inhalation (for dissection of brain regions and for different coprecipitation experiments). Extracts were prepared in lysis buffer supplemented with 100 mM NaCl and 200 µM calpain inhibitor I by using an ultra turrax and were cleared from cell debris by centrifugation at 1,000 g for 15 min.
Precipitations of endogenous Cobl-like from rat brain extracts and quantitative analyses of Abp1/Cobl-like complex formation
Endogenous Cobl-like was precipitated from rat brain extracts by immobilized GST-Abp1SH3 by incubation for 3 h at 4°C. The precipitates were washed with lysis buffer and eluted with 20 mM of reduced glutathione, 120 mM NaCl, and 50 mM Tris-HCl, pH 8.0.
Precipitates of endogenous Cobl-like with CaM immobilized on sepharose 4B (GE Healthcare) were obtained by incubations with rat brain extracts in EGTA-free lysis buffer supplemented with 150 mM NaCl and 200 µM calpain inhibitor I in the presence (2 µM CaCl2) or absence (1 mM EGTA) of calcium. Bound proteins were eluted by boiling the CaM-sepharose matrix with 4× SDS sample buffer.
Lysates, supernatants, and eluates were analyzed by immunoblotting. Endogenous Cobl-like was detected by immunoblotting with rabbit anti–Cobl-like antibodies.
For quantitative evaluations of Cobl-like/Abp1 complex formation on presence (2 µM CaCl2) and absence of Ca2+ (1 mM EGTA), the amounts of precipitated endogenous Cobl-like were quantified based on the detection of fluorescent antibody signals by using a Licor Odyssey System and normalized to the amounts of immobilized Abp1 fusion protein that were immunodetected on the same blot by using the second fluorescence channel of the Licor Odyssey System. Data were expressed as the percentage of difference from Ca2+-free conditions. Statistical analyses were performed by using one-way ANOVA with Tukey’s post test.
Culture, transfection, immunostaining, and evaluations of HEK293 and COS-7 cells
Culturing of HEK293 and COS-7 cells, phalloidin-staining, and immunolabeling were essentially as described (Kessels et al., 2001). HEK293 and COS-7 cells were transfected by using TurboFect (Thermo Fisher Scientific).
For subcellular recruitment assays, mitochondria of COS-7 cells were stained with 0.2 µM MitoTracker Deep Red 633 in medium at 37°C for 1 h, and cells were subsequently fixed with 4% PFA for 7 min.
3D membrane ruffling of phalloidin-stained COS-7 cells transfected with Flag-mCherry, Flag-mCherry-Cobl-like, and different mutants thereof was determined according to Ahuja et al. (2007). 200–250 cells from three independent assays were analyzed for each condition. The Cobl-like–induced phenotype of 3D ruffle formation was confirmed by blinded, quantitative datasets from several independent investigators.
In a subset of experiments, a final concentration of 2 µM latrunculin A and 2.5 µM cytochalasin D was applied (in 1% and 0.0025% DMSO, respectively) for 20 min before fixation. These quantitative analyses of 3D ruffles were performed for at least 150 cells from two independent assays.
In addition to the established procedure (Ahuja et al., 2007), the Cobl-like–induced phenotype furthermore was corroborated by three different quantitative assessments. First, the number of 3D ruffle areas per transfected cell was determined. Second, the lateral extensions of these ruffles were measured and expressed as the percentage of the cell perimeter, which was obtained by using the thresholding function of ImageJ (Fiji). Third, ApoTome fluorescent microscopy image stacks were processed by using ImageJ, and the averaged F-actin content of the two brightest lamellipodial or lamellipodial-related areas of Z-projections was put in relation to the F-actin content measured in the cytosol. For each of these, three further validations and examinations of the Cobl-like–induced 3D ruffles phenotype 30 cells (Flag-mCherry-Cobl-like; Flag-mCherry-Cobl-like740–1273) and 33 cells (Flag-mCherry control), respectively, (from three transfections) per experimental condition were analyzed.
Immunolabeling of mouse brain sections
Sagittal sections of brains of adult male mice were immunolabeled as described previously (Haag et al., 2012). In brief, brains were removed, postfixed in 4% PFA for 16 h at 4°C, rinsed, and placed in 30% (wt/vol) sucrose in PBS at 4°C until saturation. Brains were sliced sagittally on a sliding microtome (SM 2000R; Leica). Obtained 55-µm sections were washed with phosphate buffer (PB; 77.4 mM Na2HPO4 and 22.6 mM NaH2PO4) and then permeabilized and blocked by incubation with 5% (vol/vol) goat serum, 0.25% (vol/vol) Triton X-100 in PB (block solution) for 2 h at RT. Antibody labelings were done in block solution (primary: 48 h, 4°C; secondary: 16 h at 4°C). Slices were then DAPI stained (30 min, 1:1,000 in PB), transferred into PBS, and mounted onto HistoBond slides with Fluoromount-G (Southern Biotech).
Microscopy
Confocal images were recorded by using a TCS SP5 microscope (equipped with 40×/0.75dry and 63×/1.4 oil objectives, LAS AF software; Leica), an LSM Meta 510, or an AxioObserver.Z1 microscope (Zeiss) equipped with an ApoTome. Both Zeiss microscopes were equipped with Plan-Apochromat 100×/1.4, 63×/1.4, 40×/1.3, and 20×/0.5 objectives and an AxioCam MRm CCD camera (Zeiss).
Digital images from Zeiss microscopes were recorded by ZEN2012 or AxioVision Software (Vs40 4.8.2.0). Image processing was done by Adobe Photoshop.
In situ hybridizations
In situ hybridization probes were generated by PCR amplification of nucleotides 2827–3391 of the coding sequence of the murine Cobl-like gene. The primers used were 5′-GAGAGCTCCAGGCCAAACCAAGTTCATT-3′ (forward) and 5′-GAGGTACCAGGGGCCAGAACTGGAGTAT-3′ (reverse). The PCR fragment was subcloned into pBluescript II-SK (Stratagene). DIG-labeled antisense and sense RNA probes were prepared by in vitro transcription by using 1 µg linearized plasmid and a 10× DIG-RNA labeling mix containing the ribonucleotides (Roche) as well as T3 and T7 RNA polymerases, respectively, for 2 h at 37°C. The probes were purified by LiCl/ethanol precipitation (final concentrations, 200 mM LiClDT, 75% ethanol).
Cryostat sagittal sections (16 µm) from WT C57BL/6J mice were postfixed in 4% PFA for 10 min at RT, rinsed in PBSDT, placed for 10 min into 0.2 M HClDT, and then incubated with 5 mM acetic anhydride in 0.1 M triethanolamine/HClDT (10 min, RT).
The sections were hybridized overnight at 68.5°C with 4 ng DIG-labeled Cobl-like sense and antisense probes in 150 µl hybridization solution. The hybridization solution was composed of 50% formamide, 0.075 mM sodium citrate/HCl buffer, pH 7.0, containing 750 mM NaCl (5× SSC), 5× Denhardt's reagent, 0.4 mg/ml torula yeast tRNA, and 0.1 mg/ml tRNA. The slides were rinsed with 5×SSC at RT and then washed for 1 h in 3 µM sodium citrate/HCl, pH 7.0, and 30 mM NaCl (0.2× SSC) at 73°C and another 15 min in 0.2× SSC at RT. The samples were then blocked in 2% Blocking Reagent (Roche) in 0.1 M maleic acid/NaOH buffer, pH 7.5, containing 0.15 M NaCl (MaBS) for 1 h. Alkaline phosphatase-conjugated anti-DIG antibodies in 2% Blocking Reagent in MaBS were added to the samples for 30 min at RT. After washing with MaBS (three times), endogenous alkaline phosphatase activity was blocked by tetramisol addition (5 min, RT), and the samples were placed in a moisture chamber and incubated with 0.1 M TRIS-HCl buffer, pH 9.5, containing 100 mM NaCl and 3.75 µl/ml 5-Bromo-4-chloro-3-indoxylphosphate (Roche) and 5 µl/ml nitro blue tetrazolium (Roche; 500 µl/each slide) for 24 h. After being washed with PBS (5 min), the sections on the slides were mounted with Moviol (Calbiochem).
Images were recorded by using an AxioObserver.Z1 microscope equipped with an ApoTome and a Plan-Apochromat 20×/0.5 objective by using the tile function of the ZEN2012 software to generate larger tissue overviews.
RT-PCR analyses of Cobl-like expression in the brain
Total RNA was extracted from the cortex, the cerebellum, the hippocampus, and the midbrain with phenol-chloroform. cDNA was prepared by using equal amounts of RNA with a ReverdAid H Minus First Strand cDNA Synthesis kit (Thermo Fisher Scientific). The pairs of exon-spanning PCR primers were as follows—Cobl-like: forward 5′-CTCCGCAGGAAAAAGGAGGTT-3′, reverse 5′-GCTCCTGAGCCTGGTAATCC-3′; and GAPDH (control): forward 5′-ATTGACCTCAACTACATGGTCTACA-3′, reverse 5′-CCAGTAGACTCCACGACATACTC-3′. The PCR product amplified by the Cobl-like–specific primers from the hippocampal cDNA was cloned into pGEMT (Seqlab), and its identity was verified by sequencing.
Spinning-disc live microscopy of developing neurons
Primary hippocampal neurons were transiently transfected with Lipofectamine 2000 at day in vitro (DIV) 6. For imaging, the culture medium was replaced by 20 mM Hepes, pH 7.4, 0.8 mM MgCl2, 1.8 mM CaCl2, 5 mM KCl, 140 mM NaCl, and 5 mM d-glucose (live-imaging buffer) adjusted to isoosmolarity by using a freezing point osmometer (Osmomat 3000; Gonotec).
Live imaging was conducted at 37°C 16–28 h after transfection in an open coverslip holder placed into a temperature- and CO2-controlled incubator built around a spinning-disc microscope. The microscope was a motorized Axio Observer combined with a spinning-disc unit CSU-X1A 5000 and equipped with a 488-nm/100-mW optically pumped semiconductor laser, a 561-nm/40-mW diode laser, and a QuantEM 512SC EMCCD camera (Zeiss).
Images were taken as Z-stacks (stacks of 7–17 images at Z-intervals of 0.31 µm depending on cellular morphology) at time intervals of 10 s with exposure times of 50–200 ms and 3–12% laser power by using a C-Apochromat objective (63×/1.20W Korr M27; Zeiss). Image processing was done by using ZEN2012, Imaris, ImageJ, and Photoshop software.
Quantitative evaluations of dynamic protein enrichments at dendritic branch initiation sites were done according to Hou et al. (2015). In brief, a circular region of interest (ROI) covering the dendrite diameter was placed at the branch initiation site and at a reference site at the same dendrite (adjacent to the branch initiation site; distance, two ROI diameters). At both sites, the fluorescence signal intensities of GFP and GFP-Cobl-like, respectively, were measured 30 s before initiation of protrusion and expressed as increases at branch initiation site relative to the corresponding adjacent control ROI (GFP-Cobl-like, n = 19; GFP, n = 25). Additionally, similar measurements were conducted with neurons transfected with both GFP-Cobl-like and Flag-mCherry to firmly exclude putative volume artifacts (which, however, were technically prevented by the use of spinning-disk microscopy providing excellent X,Y but also Z resolution; GFP-Cobl-like, n = 34; Flag-mCherry, n = 34).
Culturing, transfection, and immunostaining of primary rat hippocampal neurons
Primary rat hippocampal neuronal cultures for immunofluorescence analyses were prepared, maintained, and transfected as described previously (Qualmann et al., 2004; Pinyol et al., 2007; Schwintzer et al., 2011). In brief, neurons prepared from hippocampi of E18 rats were seeded at densities of ∼200,000/well (12-well plate) and 60,000/well (24-well plate), respectively. Cells were cultured in Neurobasal medium containing 2 mM l-glutamine, 1× B27, and 1 µm/ml penicillin/streptomycin and were maintained at 37°C with 90% humidity and 5% CO2. Transfections were done by using 2 µl Lipofectamine 2000 and 1 µg DNA per well in antibiotic-free medium. After 4 h, the transfection medium was replaced by the conditioned medium and neurons were cultured further.
Fixation was done in 4% (wt/vol) PFA in PBS, pH 7.4, at RT for 4–6 min. Permeabilization and blocking were done with 10% (vol/vol) horse serum and 5% (wt/vol) BSA in PBS with 0.2% (vol/vol) Triton X-100 (blocking solution). Phalloidin stainings and antibody incubations were done in the same buffer without Triton X-100 according to Kessels et al. (2001) and Pinyol et al. (2007). In brief, neurons were incubated with primary antibodies for 1 h at RT, washed three times with blocking solution, and then incubated with secondary antibodies (1 h, RT). Finally, the coverslips were washed with blocking solution, PBS, and water and mounted onto coverslips by using Moviol.
Quantitative assessment of RNAi efficiency in primary hippocampal neurons
DIV4 hippocampal neurons were transfected with Abp1 RNAi and control vectors, respectively, fixed and immunostained with anti-Abp1 and anti-MAP2 antibodies ∼48 h later at DIV6. Transfected cells were imaged by systematic sweeps over the coverslips and evaluated for anti-Abp1 staining intensities in ImageJ by using constant microscope and imaging parameters. ROIs represent the entire cell body. The mean intensities of Z-projections of Abp1 RNAi cells were compared with the values of neighboring, untransfected cells on the same image. In case two untransfected cells could be evaluated in the vicinity of a transfected cell, then these control data were averaged (8 of 16 images). Anti-Abp1 immunofluorescence levels in Abp1 RNAi neurons versus untransfected neurons were presented as mean ± SEM (Abp1 RNAi, n = 16; untransfected, n = 16).
Analyses of dendrites of primary hippocampal neurons
Cobl-like overexpression phenotypes were analyzed 27–30 h after transfection at DIV5. For loss-of-function analyses and the corresponding rescue experiments, as well as for suppressions of Cobl-like overexpression phenotypes, DIV4 hippocampal neurons were transfected with RNAi and control vectors, respectively, and fixed and immunostained ∼34 h later at DIV6. Two to six independent coverslips per condition per assay and neurons of at least two independent neuronal preparations were analyzed based on the anti-MAP2 immunostaining of transfected neurons.
For inhibitor studies, neurons were transfected at DIV4 and the CaM inhibitor CGS9343B (1,3-dihydro-1-[1-[(4-methyl-4H,6H-pyrrolo[1,2-a][4,1]-benzoxazepin-4-yl)methyl]-4-piperidinyl]-2H-benzimidazol-2-one(1:1) maleate; Tocris) was applied 12 h after transfection at a final concentration of 10 µM and 0.1% DMSO for 18 h. Neurons were then processed for anti-MAP2 immunostaining.
In all quantitative assays, transfected neurons were sampled systematically on each coverslip. Morphometric measurements were based on anti-MAP2 immunolabeling of transfected neurons. With the use of Imaris 7.6 software, the number of dendritic branching points, dendritic terminal points, and dendritic filament length were determined, and Sholl analyses (Sholl, 1953) were conducted.
For each neuron, one filament was drawn by Imaris software with the use of the following settings: largest diameter, cell body diameter; thinnest diameter, 0.2 µm; start seed point, 1.5× cell body diameter; disconnected points, 2 µm; minimum segment size, 10 µm. Immunopositive areas, which were artificially spliced by Imaris or protrusions belonging to different cells, as well as filament branch points that the software erroneously placed inside the cell body were manually removed from the filament. Parameters determined by Imaris 7.6 were saved as Excel files.
Statistical analyses
All quantitative data shown represent mean ± SEM. Tests for normal data distribution and statistical significance analyses were done by using GraphPad Prism 5 and Prism 6 software (SCR_002798).
All morphological analyses of COS-7 cell ruffling were subjected to statistical significance calculations by using one-way ANOVA and Tukey’s post test (Figs. 1 and 5).
Data evaluating the dendritic arbor formation were analyzed for statistical significance by using one-way ANOVA and Tukey’s post test throughout (Figs. 4, 7, 8, 9, and 10).
The comparison of dendritic arborization on GFP-Cobl-like versus GFP expression was evaluated by two-tailed Student’s t test (Fig. 3).
All Sholl analyses were tested by two-way ANOVA and Bonferroni’s post test (Figs. 3, 4, 7, 8, 9, and10).
Dynamic GFP-Cobl-like enrichments at branch initiation sites in comparison to independent GFP controls and to cotransfected mCherry, respectively, were evaluated by two-tailed Student’s t test (Fig. 3).
Quantitative determinations of knock-down efficiencies were analyzed by one-way ANOVA with Tukey’s post test (Cobl-like RNAi) and unpaired Student’s t test (Abp1 RNAi), respectively.
Statistical significance was marked by *, P < 0.05; **, P < 0.01; and ***, P < 0.001 throughout.
Online supplemental material
Fig. S1 provides more information on the Cobl-like WH2 domain and C terminus, displays Cobl-like’s wide expression in the nervous system and dendritic localization, and shows the validation and effectiveness of anti–Cobl-like antibodies and RNAi constructs. Fig. S2 shows that the C terminus of Cobl-like does not directly associate with F-actin. Fig. S3 shows the experimental data underlying the schematic summary of data presented in Fig. 6 B and additional experiments revealing that the region of Cobl-like identified to be functionally critical in addition to the WH2 domain in F-actin-driven alterations of cell morphology in COS-7 cells is an interface for SH3 domain association of the F-actin–binding protein Abp1. Fig. S4 shows the fluorescence microscopy signals of all three channels used to generate the merged images shown in Fig. 6 (F–L) visualizing the successful reconstitution of the Abp1 SH3 domain-dependent formation of Cobl-like/Abp1 protein complexes in intact (COS-7) cells by recruitments of Cobl-like and GFP-Cobl-like740–1273 by using a set of mitochondrially anchored Abp1 and mutants thereof. Fig. S5 confirms by subcellular recruitment studies that Cobl-like interacts with the SH3 domain of Abp1 via the part of the proline-rich domain that was identified as functionally critical in cell-ruffling assays conducted with COS-7 cells (Cobl-like1016–1104) and furthermore corroborates by quantitative analyses again that endogenous Abp1 in primary hippocampal neurons is knocked down by RNAi (compare quantitative data already presented by Pinyol et al. (2007) for the same cell system).
Supplementary Material
Acknowledgments
We thank A. Kreusch and B. Schade for excellent technical support.
This work was supported by Deutsche Forschungsgemeinschaft grants KE685/3-2 and KE685/4-2 to M.M. Kessels and QU116/5-2 to B. Qualmann.
The authors declare no competing financial interests.
Author contributions: M. Izadi, D. Schlobinski, M. Lahr, and L. Schwintzer generated data. M. Izadi, D. Schlobinski, M. Lahr, L. Schwintzer, B. Qualmann, and M.M. Kessels analyzed the data. M. Izadi, B. Qualmann, and M.M. Kessels wrote the manuscript. B. Qualmann and M.M. Kessels designed the project.
References
- Ahuja R., Pinyol R., Reichenbach N., Custer L., Klingensmith J., Kessels M.M., and Qualmann B.. 2007. Cordon-bleu is an actin nucleation factor and controls neuronal morphology. Cell. 131:337–350. 10.1016/j.cell.2007.08.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun A., Pinyol R., Dahlhaus R., Koch D., Fonarev P., Grant B.D., Kessels M.M., and Qualmann B.. 2005. EHD proteins associate with syndapin I and II and such interactions play a crucial role in endosomal recycling. Mol. Biol. Cell. 16:3642–3658. 10.1091/mbc.E05-01-0076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll E.A., Gerrelli D., Gasca S., Berg E., Beier D.R., Copp A.J., and Klingensmith J.. 2003. Cordon-bleu is a conserved gene involved in neural tube formation. Dev. Biol. 262:16–31. 10.1016/S0012-1606(03)00323-3 [DOI] [PubMed] [Google Scholar]
- Charrier C., and Polleux F.. 2012. [How human-specific SRGAP2 gene duplications control human brain development]. [In French.] Med. Sci. (Paris). 28:911–914. 10.1051/medsci/20122811003 [DOI] [PubMed] [Google Scholar]
- Chereau D., Boczkowska M., Skwarek-Maruszewska A., Fujiwara I., Hayes D.B., Rebowski G., Lappalainen P., Pollard T.D., and Dominguez R.. 2008. Leiomodin is an actin filament nucleator in muscle cells. Science. 320:239–243. 10.1126/science.1155313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesarone M.A., and Goode B.L.. 2009. Actin nucleation and elongation factors: mechanisms and interplay. Curr. Opin. Cell Biol. 21:28–37. 10.1016/j.ceb.2008.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevaleyre V., and Castillo P.E.. 2002. Assessing the role of Ih channels in synaptic transmission and mossy fiber LTP. Proc. Natl. Acad. Sci. USA. 99:9538–9543. 10.1073/pnas.142213199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazi B., Cope M.J., Douangamath A., Ferracuti S., Schirwitz K., Zucconi A., Drubin D.G., Wilmanns M., Cesareni G., and Castagnoli L.. 2002. Unusual binding properties of the SH3 domain of the yeast actin-binding protein Abp1: Structural and functional analysis. J. Biol. Chem. 277:5290–5298. 10.1074/jbc.M109848200 [DOI] [PubMed] [Google Scholar]
- Fenster S.D., Kessels M.M., Qualmann B., Chung W.J., Nash J., Gundelfinger E.D., and Garner C.C.. 2003. Interactions between Piccolo and the actin/dynamin-binding protein Abp1 link vesicle endocytosis to presynaptic active zones. J. Biol. Chem. 278:20268–20277. 10.1074/jbc.M210792200 [DOI] [PubMed] [Google Scholar]
- Fink C.C., Bayer K.U., Myers J.W., Ferrell J.E. Jr., Schulman H., and Meyer T.. 2003. Selective regulation of neurite extension and synapse formation by the beta but not the alpha isoform of CaMKII. Neuron. 39:283–297. 10.1016/S0896-6273(03)00428-8 [DOI] [PubMed] [Google Scholar]
- Gaudillière B., Konishi Y., de la Iglesia N., Yao G., and Bonni A.. 2004. A CaMKII-NeuroD signaling pathway specifies dendritic morphogenesis. Neuron. 41:229–241. 10.1016/S0896-6273(03)00841-9 [DOI] [PubMed] [Google Scholar]
- Gordon G.J., Jensen R.V., Hsiao L.L., Gullans S.R., Blumenstock J.E., Richards W.G., Jaklitsch M.T., Sugarbaker D.J., and Bueno R.. 2003. Using gene expression ratios to predict outcome among patients with mesothelioma. J. Natl. Cancer Inst. 95:598–605. 10.1093/jnci/95.8.598 [DOI] [PubMed] [Google Scholar]
- Gordon G.J., Dong L., Yeap B.Y., Richards W.G., Glickman J.N., Edenfield H., Mani M., Colquitt R., Maulik G., Van Oss B., et al. 2009. Four-gene expression ratio test for survival in patients undergoing surgery for mesothelioma. J. Natl. Cancer Inst. 101:678–686. 10.1093/jnci/djp061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag N., Schwintzer L., Ahuja R., Koch N., Grimm J., Heuer H., Qualmann B., and Kessels M.M.. 2012. The actin nucleator Cobl is crucial for Purkinje cell development and works in close conjunction with the F-actin binding protein Abp1. J. Neurosci. 32:17842–17856. 10.1523/JNEUROSCI.0843-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall D.D., Dai S., Tseng P.Y., Malik Z., Nguyen M., Matt L., Schnizler K., Shephard A., Mohapatra D.P., Tsuruta F., et al. 2013. Competition between α-actinin and Ca2+-calmodulin controls surface retention of the L-type Ca2+ channel Ca(V)1.2. Neuron. 78:483–497. 10.1016/j.neuron.2013.02.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S.H., Kim S.H., Kim H.J., Lee Y., Choi S.Y., Park G., Kim D.H., Lee A., Kim J., Choi J.M., et al. 2017. Cobll1 is linked to drug resistance and blastic transformation in chronic myeloid leukemia. Leukemia. 31:1532–1539. 10.1038/leu.2017.72 [DOI] [PubMed] [Google Scholar]
- Hou P., Estrada L., Kinley A.W., Parsons J.T., Vojtek A.B., and Gorski J.L.. 2003. Fgd1, the Cdc42 GEF responsible for Faciogenital Dysplasia, directly interacts with cortactin and mAbp1 to modulate cell shape. Hum. Mol. Genet. 12:1981–1993. 10.1093/hmg/ddg209 [DOI] [PubMed] [Google Scholar]
- Hou W., Izadi M., Nemitz S., Haag N., Kessels M.M., and Qualmann B.. 2015. The actin nucleator Cobl is controlled by calcium and calmodulin. PLoS Biol. 13:e1002233 10.1371/journal.pbio.1002233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husson C., Renault L., Didry D., Pantaloni D., and Carlier M.F.. 2011. Cordon-Bleu uses WH2 domains as multifunctional dynamizers of actin filament assembly. Mol. Cell. 43:464–477. 10.1016/j.molcel.2011.07.010 [DOI] [PubMed] [Google Scholar]
- Kessels M.M., and Qualmann B.. 2006. Syndapin oligomers interconnect the machineries for endocytic vesicle formation and actin polymerization. J. Biol. Chem. 281:13285–13299. 10.1074/jbc.M510226200 [DOI] [PubMed] [Google Scholar]
- Kessels M.M., Engqvist-Goldstein A.E.Y., and Drubin D.G.. 2000. Association of mouse actin-binding protein 1 (mAbp1/SH3P7), an Src kinase target, with dynamic regions of the cortical actin cytoskeleton in response to Rac1 activation. Mol. Biol. Cell. 11:393–412. 10.1091/mbc.11.1.393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessels M.M., Engqvist-Goldstein A.E.Y., Drubin D.G., and Qualmann B.. 2001. Mammalian Abp1, a signal-responsive F-actin-binding protein, links the actin cytoskeleton to endocytosis via the GTPase dynamin. J. Cell Biol. 153:351–366. 10.1083/jcb.153.2.351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann C. 2009. Calcium signaling and the development of specific neuronal connections. Prog. Brain Res. 175:443–452. 10.1016/S0079-6123(09)17529-5 [DOI] [PubMed] [Google Scholar]
- Low T.L., and Goldstein A.L.. 1982. Chemical characterization of thymosin beta 4. J. Biol. Chem. 257:1000–1006. [PubMed] [Google Scholar]
- Mancina R.M., Burza M.A., Maglio C., Pirazzi C., Sentinelli F., Incani M., Montalcini T., Pujia A., Congiu T., Loche S., et al. 2013. The COBLL1 C allele is associated with lower serum insulin levels and lower insulin resistance in overweight and obese children. Diabetes Metab. Res. Rev. 29:413–416. 10.1002/dmrr.2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi R., Sakata S., Naito A., Hirashima N., and Tanaka M.. 2014. Dendritic differentiation of cerebellar Purkinje cells is promoted by ryanodine receptors expressed by Purkinje and granule cells. Dev. Neurobiol. 74:467–480. 10.1002/dneu.22139 [DOI] [PubMed] [Google Scholar]
- Paulussen M., Landuyt B., Schoofs L., Luyten W., and Arckens L.. 2009. Thymosin beta 4 mRNA and peptide expression in phagocytic cells of different mouse tissues. Peptides. 30:1822–1832. 10.1016/j.peptides.2009.07.010 [DOI] [PubMed] [Google Scholar]
- Pinyol R., Haeckel A., Ritter A., Qualmann B., and Kessels M.M.. 2007. Regulation of N-WASP and the Arp2/3 complex by Abp1 controls neuronal morphology. PLoS One. 2:e400 10.1371/journal.pone.0000400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qualmann B., and Kelly R.B.. 2000. Syndapin isoforms participate in receptor-mediated endocytosis and actin organization. J. Cell Biol. 148:1047–1062. 10.1083/jcb.148.5.1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qualmann B., and Kessels M.M.. 2009. New players in actin polymerization--WH2-domain-containing actin nucleators. Trends Cell Biol. 19:276–285. 10.1016/j.tcb.2009.03.004 [DOI] [PubMed] [Google Scholar]
- Qualmann B., Roos J., DiGregorio P.J., and Kelly R.B.. 1999. Syndapin I, a synaptic dynamin-binding protein that associates with the neural Wiskott-Aldrich syndrome protein. Mol. Biol. Cell. 10:501–513. 10.1091/mbc.10.2.501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qualmann B., Boeckers T.M., Jeromin M., Gundelfinger E.D., and Kessels M.M.. 2004. Linkage of the actin cytoskeleton to the postsynaptic density via direct interactions of Abp1 with the ProSAP/Shank family. J. Neurosci. 24:2481–2495. 10.1523/JNEUROSCI.5479-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan I., and Cline H.T.. 1998. Glutamate receptor activity is required for normal development of tectal cell dendrites in vivo. J. Neurosci. 18:7836–7846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault L., Deville C., and van Heijenoort C.. 2013. Structural features and interfacial properties of WH2, β-thymosin domains and other intrinsically disordered domains in the regulation of actin cytoskeleton dynamics. Cytoskeleton (Hoboken). 70:686–705. 10.1002/cm.21140 [DOI] [PubMed] [Google Scholar]
- Riederer B.M., Zagon I.S., and Goodman S.R.. 1986. Brain spectrin(240/235) and brain spectrin(240/235E): Two distinct spectrin subtypes with different locations within mammalian neural cells. J. Cell Biol. 102:2088–2097. 10.1083/jcb.102.6.2088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl J., Crevenna A.H., Kessenbrock K., Yu J.H., Neukirchen D., Bista M., Bradke F., Jenne D., Holak T.A., Werb Z., et al. 2008. Lifeact: a versatile marker to visualize F-actin. Nat. Methods. 5:605–607. 10.1038/nmeth.1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J., and Terhoeven N.. 2013. The bilaterian roots of cordon-bleu. BMC Res. Notes. 6:393 10.1186/1756-0500-6-393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwintzer L., Koch N., Ahuja R., Grimm J., Kessels M.M., and Qualmann B.. 2011. The functions of the actin nucleator Cobl in cellular morphogenesis critically depend on syndapin I. EMBO J. 30:3147–3159. 10.1038/emboj.2011.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma V., Sharma I., Sethi I., Mahajan A., Singh G., Angural A., Bhanwer A.J.S., Dhar M.K., Singh V., Rai E., and Sharma S.. 2017. Replication of newly identified type 2 diabetes susceptible loci in Northwest Indian population. Diabetes Res. Clin. Pract. 126:160–163. 10.1016/j.diabres.2017.02.013 [DOI] [PubMed] [Google Scholar]
- Sholl D.A. 1953. Dendritic organization in the neurons of the visual and motor cortices of the cat. J. Anat. 87:387–406. [PMC free article] [PubMed] [Google Scholar]
- Wales P., Schuberth C.E., Aufschnaiter R., Fels J., García-Aguilar I., Janning A., Dlugos C.P., Schäfer-Herte M., Klingner C., Wälte M., et al. 2016. Calcium-mediated actin reset (CaAR) mediates acute cell adaptations. eLife. 5:e19850 10.7554/eLife.19850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Yan Z., Zhang B., Rao Z., Zhang Y., Liu J., Yu L., Zhao Y., Yang B., Wu T., and Gao J.. 2013. Identification of a 5-gene signature for clinical and prognostic prediction in gastric cancer patients upon microarray data. Med. Oncol. 30:678–688. 10.1007/s12032-013-0678-5 [DOI] [PubMed] [Google Scholar]
- Wyszynski M., Lin J., Rao A., Nigh E., Beggs A.H., Craig A.M., and Sheng M.. 1997. Competitive binding of α-actinin and calmodulin to the NMDA receptor. Nature. 385:439–442. 10.1038/385439a0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.