RAB-10 is a master regulator of endocytic recycling in polarized epithelial cells. Liu et al. identify LET-413, the Caenorhabditis elegans homolog of Scrib/Erbin, as a RAB-5 effector that is required for the DENN-4–mediated activation of RAB-10 and the control of membrane expansion in the C. elegans intestine.
Abstract
RAB-10/Rab10 is a master regulator of endocytic recycling in epithelial cells. To better understand the regulation of RAB-10 activity, we sought to identify RAB-10(GDP)–interacting proteins. One novel RAB-10(GDP)–binding partner that we identified, LET-413, is the Caenorhabditis elegans homologue of Scrib/Erbin. Here, we focus on the mechanistic role of LET-413 in the regulation of RAB-10 within the C. elegans intestine. We show that LET-413 is a RAB-5 effector and colocalizes with RAB-10 on endosomes, and the overlap of LET-413 with RAB-10 is RAB-5 dependent. Notably, LET-413 enhances the interaction of DENN-4 with RAB-10(GDP) and promotes DENN-4 guanine nucleotide exchange factor activity toward RAB-10. Loss of LET-413 leads to cytosolic dispersion of the RAB-10 effectors TBC-2 and CNT-1. Finally, we demonstrate that the loss of RAB-10 or LET-413 results in abnormal overextensions of lateral membrane. Hence, our studies indicate that LET-413 is required for DENN-4–mediated RAB-10 activation, and the LET-413–assisted RAB-5 to RAB-10 cascade contributes to the integrity of C. elegans intestinal epithelia.
Introduction
In epithelial cells, endocytic transport participates in the formation and/or maintenance of the specialized apical and basolateral plasma membrane domains (Fölsch et al., 2009; Eaton and Martin-Belmonte, 2014). The transport of recycling cargo to the plasma membrane is modulated by a series of regulators, including Rab small GTPases and their effectors (Simonsen et al., 1999; Nielsen et al., 2000; Sasidharan et al., 2012; Murray et al., 2016). RAB-10 was the first GTPase reported to regulate basolateral recycling in the Caenorhabditis elegans intestine, upstream of RME-1 (Chen et al., 2006). In rab-10 mutants, intestinal epithelial cells have fewer basolateral recycling endosomes labeled by RME-1 and accumulate many more RAB-5–labeled early endosomes, some of which are grossly enlarged. Collectively, these results were the first to indicate a regulatory role for RAB-10 at the recycling step from early endosomes to recycling endosomes (Chen et al., 2006). Two previously identified RAB-10 effectors, EHBP-1 and CNT-1, provide insight into RAB-10’s function (Shi et al., 2010, 2012). EHBP-1 promotes RAB-10 endosomal recruitment and bridges recycling endosomes with the actin cytoskeleton (Guilherme et al., 2004; Shi et al., 2010; Wang et al., 2016). Conversely, RAB-10 mediates CNT-1/–ACAP endosomal association, negatively regulating the accumulation of phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2) on endosomes via negative regulation of ARF-6 (Shi et al., 2012).
An important concept in membrane transport regulation revealed over the last decade involves Rab GTPase cascades. The Rab cascade is typically orchestrated via an orderly recruitment of sequentially acting guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs) for Rab GTPases (Hutagalung and Novick, 2011). For example, in the process of early-to-late endosome maturation, Rab5 is replaced by Rab7 through a GEF cascade (Rink et al., 2005). Mon1/SAND-1 functions as GEF to recruit and activate Ypt7p/RAB-7 (Nordmann et al., 2010). Studies of Rab regulation in the genetic model organism C. elegans have also contributed to the understanding of Rab cascades. Several studies indicate that TBC-2 functions as a RAB-5 GAP protein and tbc-2 mutants display endosome abnormalities that are also observed in animals expressing constitutively active RAB-5(Q78L) (Li et al., 2009; Chotard et al., 2010; Sasidharan et al., 2012). In addition to affecting the degradative pathway, TBC-2 has strong effects on recycling transport. RAB-10 helps recruit TBC-2 as early endosomes transition to recycling endosomes, down-regulating RAB-5 in a step required for basolateral recycling (Sun et al., 2012; Liu and Grant, 2015). These effects suggest that TBC-2 acts in the negative feedback loop from RAB-10 to RAB-5, highlighting the importance of Rab cascades as the fundamental determinant of endocytic recycling regulation.
LAP (leucine-rich repeats and PDZ domains) family members have been identified in different systems, including Drosophila melanogaster Scrib, C. elegans LET-413, and mammalian Erbin, Densin-180, and scribble. Specifically, LAP proteins are characterized by the N-terminal 16 leucine-rich repeats (LRRs) and C-terminal PDZ domains. Accumulating evidence indicates that LAP proteins are often functioning as adaptors to participate in polarity and/or receptors targeting (Bryant and Huwe, 2000). In Drosophila, Scrib forms a functional module (Scribble complex) with Discs-large (Dlg) and Lethal giant larvae (Lgl). Scribble complex is reported to regulate the basolateral domain of epithelia by excluding apical regulators such as PAR-3 and PAR-6 (Bilder and Perrimon, 2000; Yamanaka and Ohno, 2008; de Vreede et al., 2014). Mammalian Erbin (Erbb2-interacting protein) also localizes to the basolateral membrane of epithelial cells, where it interacts with the epidermal growth factor receptor family protein ErbB2 and stabilizes ErbB2 to the basolateral membrane (Borg et al., 2000). In human breast cancer cells, Erbin was shown to form a complex with ErbB2 and prevent ErbB2 degradation (Tao et al., 2014). Additionally, the LRR domain of Erbin mediates the interaction with the scaffold protein Sur-8 and disrupts the Sur-8–Ras–Raf complex, inhibiting activation of extracellular ERK1/2 (Kolch, 2003; Dai et al., 2006). C. elegans encodes a single Erbin homologue, LET-413. LET-413 is broadly expressed in multiple epithelial tissues, including the intestine and epidermal seam cells (Legouis et al., 2000). Notably, in let-413 mutant embryos, epidermal cells subapical membranes overextend, forming overlapping membrane extensions (Legouis et al., 2000), which have been proposed to result from the absence of properly positioned apical junctions.
Cell polarity establishment requires endocytic trafficking mediated membrane protein restriction to specific plasma membrane domains. Lgl has been demonstrated to release Rab10 from a Rab10–GDI complex and regulate membrane insertion in growing neurites (Wang et al., 2011). In Drosophila follicle cells and wing imaginal discs, the Scribble complex was reported to regulate retromer-dependent endosome-to-Golgi transport during epithelial polarization (de Vreede et al., 2014). Notably, Scribble1 was suggested to regulate the endocytic recycling of GluN2A via ARF-6 activation (Piguel et al., 2014). A recent study reported that Erbin resides in early endosomes as well, and SARA (SMAD anchor for receptor activation) is required for the endosomal recruitment of Erbin (Sflomos et al., 2011). Nevertheless, the mechanistic role of LET-413 and its homologues in endocytic recycling has not been directly evaluated. Here, we report that LET-413 functions as a RAB-5 effector to boost the affinity of the GEF protein DENN-4 for inactive RAB-10(GDP), therefore promoting RAB-5 to RAB-10 progression in endocytic recycling. Consistent with the premise that LET-413 is essential for RAB-10 activation, loss of LET-413 leads to cytosolic dispersal of RAB-10 effectors TBC-2 and CNT-1. RAB-10 may be a key target for LET-413 in epithelial polarization, because the loss of RAB-10 or EHBP-1 elevated the incidence of abnormal lateral membrane organization. We conclude that LET-413 plays a vital role in DENN-4–mediated RAB-10 activation and that a RAB-5 to RAB-10 cascade contributes to intestinal epithelia integrity.
Results
LET-413 interacts with the active form of RAB-5 and the inactive form of RAB-10
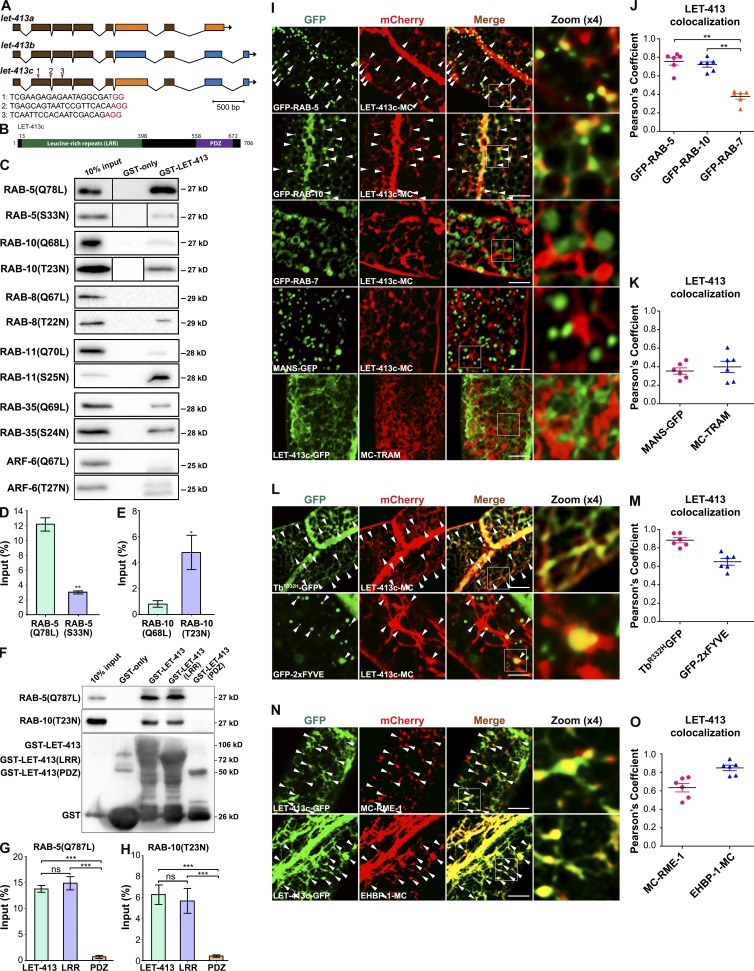
Directed endocytic trafficking is essential to cell polarity establishment, ensuring that specific membrane proteins are restricted to particular membrane domains (de Vreede et al., 2014). Accumulating evidence indicates that many polarity proteins regulate endocytic recycling directly or indirectly, including PAR-6, CDC-42, and PKC-3 (Balklava et al., 2007; Bai and Grant, 2015). As a regulator of epithelial cell polarity, LET-413/Scrib participates in organizing apical junction belts (Bossinger et al., 2001, 2004; Montcouquiol et al., 2003; Yamanaka and Ohno, 2008; Pilipiuk et al., 2009). The N-terminal LRRs of LET-413 have been suggested to mediate interactions with small GTPases (Fig. 1 B), many of which control specific steps in membrane traffic (Legouis et al., 2000, 2003). To determine whether LET-413 interacts with Rab GTPases mediating endocytic recycling, we performed GST pull-down experiments to explore the interaction of LET-413 with RAB-5 and RAB-10. Interestingly, GST-LET-413 interacted strongly with the predicted constitutively active (GTPase-defective) form RAB-5(Q78L) while displaying much less affinity for the GDP-bound mutant form RAB-5(S33N) (Fig. 1, C and D), suggesting that LET-413 preferentially interacts with the active form of RAB-5.
Figure 1.
LET-413 preferentially associated with the active RAB-5 and the inactive RAB-10. (A) let-413 open reading frames encoding let-413a, let-413b, and let-413c isoforms. Red arrowheads above let-413c indicate three single guide RNAs target locations in let-413(ycx23) CRISPR/Cas9 intestine somatic mutants. (B) LET-413c contains N-terminal LRRs and a C-terminal PDZ domain; amino acid numbers are indicated. (C) Glutathione beads loaded with GST and GST-LET-413 were incubated with in vitro–expressed HA-tagged RAB-5(Q78L), RAB-10(Q68L), RAB-8(Q67L), RAB-11(Q70L), RAB-35(Q69L), and ARF-6(Q67L) and inactive RAB-5(S33N), RAB-10(T23N), RAB-8(T22N), RAB-11(S25N), RAB-35(S24N), and ARF-6(T27N). Eluted proteins were separated on SDS-PAGE gel and analyzed by Western blotting using anti–HA antibody. Input lanes contain in vitro–expressed HA-tagged proteins in the binding assays (10%). (D) The binding affinity of GST-LET-413 was fourfold higher for RAB-5(Q78L) than for RAB-5(S33N). The SEMs from three independent experiments are shown. Asterisks indicate significant differences (**, P < 0.01; one-tailed Student’s t test). (E) The binding affinity of GST-LET-413 was sixfold lower for RAB-10(Q68L) than for RAB-10(T23N). The SEMs from three independent experiments are shown. Asterisks indicate significant differences (*, P < 0.05; one-tailed Student’s t test). (F–H) Glutathione beads loaded with GST, GST-LET-413, GST-LET-413(LRRs), and GST-LET-413(PDZ) were incubated with HA-tagged RAB-5(Q78L) and RAB-10(T23N). Bound proteins were eluted and analyzed by Western blotting using anti–HA or anti–GST (bottom) antibodies. The SEMs from three independent experiments are shown. Asterisks indicate significant differences (***, P < 0.001; ns, not significant; one-tailed Student’s t test). (I–K) LET-413c-mCherry colocalized well with GFP-RAB-5– or GFP-RAB-10–labeled endosomes. However, LET-413c-mCherry displayed little overlap with the late endosome marker GFP-RAB-7, the Golgi marker MANS-GFP, and the ER marker mCherry-TRAM. (L and M) TbR332H-GFP colocalized with LET-413c-mCherry significantly, whereas GFP-2xFYVE partially overlapped with LET-413c-mCherry. (N and O) LET-413c-GFP colocalized extensively with mCherry-RME-1 or EHBP-1-mCherry on tubules and puncta. Arrowheads indicate positive overlap. Pearson’s correlation coefficients for GFP and mCherry signals are calculated (n = 6 animals). Error bars represent SEM; **, P < 0.01. Bars, 10 µm.
We next examined whether LET-413 can interact with RAB-10. Surprisingly, GST-LET-413 pulled down inactive RAB-10(T23N), but not active RAB-10(Q68L) (Fig. 1, C and E). Consistent with this result, LET-413 strongly interacted with the nucleotide-free mutant RAB-10(N122I) (Fig. S1 A). Binding of the GDP-bound mutant form RAB-10(T23N) to LET-413 suggested a functional link between the activation of RAB-10 and LET-413 during endocytic recycling. To characterize the specificity of LET-413’s interaction with Rabs, we examined additional endocytic GTPases mutant forms, including RAB-8(Q67L) and RAB-8(T22N), RAB-11(Q70L) and RAB-11(S25N), RAB-35(Q69L) and RAB-35(S24N), and ARF-6(Q67L) and ARF-6(T27N). We failed to observe the interaction of LET-413 with either the active and inactive forms of ARF-6. However, RAB-8(T22N), RAB-11(S25N), and RAB-35(S24N) showed interactions with LET-413 (Figs. 1 C and S1 B). In C. elegans, RAB-8 and RAB-10 are the closely related paralogs and function redundantly in the nonpolarized germ cells while maintaining distinct functions in polarized cells like the intestine and neurons (Shi et al., 2010). RAB-11 and RAB-35 have also reported being a recycling regulator in C. elegans and mammals (Kouranti et al., 2006; Sato et al., 2008; Sakaguchi et al., 2015).
To determine the region within LET-413 that interacts with Rabs, we tested fragments containing either the LRR or the PDZ domain with RAB-5(Q78L) and RAB-10(T23N). Our experiments indicated that LRRs (aa 1–400) are necessary and sufficient to mediate the interaction with both RAB-5 and RAB-10, whereas the PDZ domain–containing segment (aa 500–706) failed to display any interaction (Fig. 1, F–H). These results are in agreement with previous studies in which LRRs were suggested to be involved in the interaction with small GTPases (Legouis et al., 2000, 2003).
LET-413 colocalizes with RAB-5 and RAB-10 on sorting endosomes
LET-413 is broadly expressed in multiple epithelial tissues of C. elegans, including the intestine, epidermal seam cells, pharynx, rectum, vulva, uterus, and spermatheca (Legouis et al., 2000). To determine the precise subcellular localization of LET-413, we expressed the longest isoform of LET-413 (isoform c) in the intestinal epithelia (Fig. 1 A). We found that LET-413c-GFP is enriched on the basolateral tubular and punctate meshwork resembling that previously identified in the studies of RAB-10 effector EHBP-1, suggesting that LET-413 is resident on endosomes (Shi et al., 2010; Wang et al., 2016; Fig. S1 D).
The endocytosed cargo proteins are sorted in early endosomes for delivery to several distinct destinations (Maxfield and McGraw, 2004; Grant and Donaldson, 2009). Rab5/RAB-5 is a master regulator of early endosomes, directing virtually all aspects of early endosome function through a host of effectors (Simonsen et al., 1999; Nielsen et al., 2000; Grant and Donaldson, 2009; Murray et al., 2016). RAB-10 partially overlaps with RAB-5 on endosomes and regulates endosome-to-plasma membrane recycling transport (Chen et al., 2006; Shi et al., 2010). To determine whether LET-413 also resides on early and/or basolateral recycling endosomes, we compared the subcellular localization of LET-413 to RAB-5, RAB-10, and other organelle markers in living animals. Live-cell imaging revealed significant colocalization of LET-413c-mCherry with GFP-RAB-5 and GFP-RAB-10 on tubular network–associated puncta (Fig. 1, I and J). We also observed extensive colocalization of LET-413c-mCherry on punctate aspects of endosomes labeled by GFP-RAB-8, which is known to colocalize with RAB-10 (Fig. S1 C; Shi et al., 2010). Previous work showed that basolateral recycling tubules could be visualized in the intestine using probes for the lipid PI(4,5)P2, whereas early endosomes are enriched in phosphatidylinositol 3-phosphate. We found that the functional LET-413c-mCherry colocalized extensively with PI(4,5)P2 marker TbR332H-GFP on intracellular puncta and tubules (Quinn et al., 2008; Hardie et al., 2015), and with phosphatidylinositol 3-phosphate marker GFP-2xFYVE on puncta, consistent with the residence of LET-413 on both early and basolateral recycling endosomes in the intestine (Fig. 1, L and M). Indeed, LET-413c-GFP colocalizes well with mCherry-tagged RME-1 and EHBP-1, additional markers of the tubular basolateral recycling endosomes (Fig. 1, N and O). To confirm this interpretation, we assayed LET-413c-GFP localization in mutants that specifically disrupt basolateral recycling tubule architecture. We found that LET-413c-GFP appeared aggregated and less tubular in arf-6(tm1447) and rme-1(b1045) mutants (Fig. S1, D–G).
We failed to detect obvious overlap between LET-413 and the late endosome marker RAB-7, the Golgi marker MANS-GFP, or the ER marker mCherry-TRAM (Fig. 1, I and K), suggesting a specific association between LET-413 and intestinal early and basolateral recycling endosomes.
Loss of LET-413 leads to reduced endosomal TBC-2 and increased endosomal RAB-5
If LET-413 is involved in activating RAB-10, then the loss of LET-413 should lead to specific phenotypes known to result from the loss of RAB-10. Previous work showed that RAB-10 is required to recruit the RAB-5 GAP TBC-2 to endosomes (Fig. S1, J and K), inactivating RAB-5 as part of a RAB-5/RAB-10 countercurrent GTPase cascade (Sasidharan et al., 2012; Liu and Grant, 2015). To better characterize the function of LET-413 in the recycling process, we developed a transgenic strain, let-413(ycx23), an intestine-specific CRISPR/Cas9 somatic mutant (Fig. 1 A and Fig. S1, H and I; Li et al., 2015). Importantly, loss of LET-413 resulted in the loss of GFP-TBC-2 from endosomal puncta. Instead, GFP-TBC-2 displayed a diffusive distribution in the intestinal cells of let-413(ycx23) or let-413(RNAi) animals (Fig. 2, A and B; and Fig. S1, J and K). let-413(ycx23) animals also displayed greatly increased intensity of GFP-RAB-5 labeling on endosomes, as would be expected if TBC-2 fails to be recruited to endosomes (Fig. 2, C and D). In contrast, there were no changes in the intensity and number of GFP-RAB-7–labeled late endosomes or GFP-RAB-11–labeled apical recycling endosomes upon loss of LET-413, indicating the specificity of the phenotype (Fig. S1, N–P).
Figure 2.
The absence of LET-413 resulted in cytoplasmic dispersion of TBC-2 and CNT-1, as well as RAB-5-labeled endosome accumulation and increased PI(4,5)P2 levels. (A and B) Compared with wild-type animals, the labeling of GFP-TBC-2 on the endosomes was significantly reduced by ∼79% and showed cytoplasmic diffusion in let-413(ycx23) mutants. (C and D) GFP-RAB-5–labeled puncta intensity significantly increased (by ∼65%) in let-413(ycx23) mutants. (E and F) In let-413(ycx23) mutants, CNT-1-GFP labeled puncta intensity was significantly reduced (by ∼75%), and CNT-1-GFP appeared diffusive in the cytoplasm. (G and H) TbR332H-GFP basolateral tubules labeling increased by approximately twofold in let-413(ycx23) mutants. (I and J) GFP-RME-1 labels basolateral recycling endosomal meshwork. In let-413(ycx23) animals, the GFP-RME-1–labeled meshwork was disrupted, and GFP-RME-1 accumulated in punctate structures in the middle focal plane. Asterisks in the panels indicate intestinal lumen. Error bars represent SEM (n = 18 each); asterisks indicate significant differences (***, P < 0.001; one-tailed Student’s t test). Bars, 10 µm.
Loss of LET-413 leads to cytosolic dispersal of CNT-1 and accumulation of endosomal PI(4,5)P2
RAB-10 also recruits CNT-1 to intestinal endosomes (Fig. S1, L and M). CNT-1 acts as an ARF-6 GAP, modulating ARF-6 activity and maintaining levels of the lipid PI(4,5)P2 on these endosomes (Shi et al., 2012; Shi and Grant, 2013). Consistent with a loss of RAB-10 activation, we found that CNT-1-GFP labeling became diffusive in let-413(ycx23) or let-413(RNAi) animals, with a distinct loss of CNT-1 from punctate endosomes (Fig. 2, E and F; and Fig. S1, L and M).
We also found that let-413(ycx23) animals have increased levels of endosomal PI(4,5)P2, as would be expected from the loss of endosomal CNT-1. We measured PI(4,5)P2 levels on endosomes using the TbR332H-GFP reporter (Quinn et al., 2008; Hardie et al., 2015). let-413(ycx23) mutant animals displayed an increased TbR332H-GFP signal on basolateral tubules, similar to that previously observed in rab-10 and cnt-1 mutants (Fig. 2, G and H; approximately twofold increase). The level of the PI(4,5)P2-binding protein RME-1 on endosomes also increased in let-413(ycx23) mutants, with a strong disruption in the normal tubularity of RME-1–labeled structures (Fig. 2, I and J).
Loss of LET-413 results in intracellular accumulation of endocytic recycling cargo in intestinal epithelia
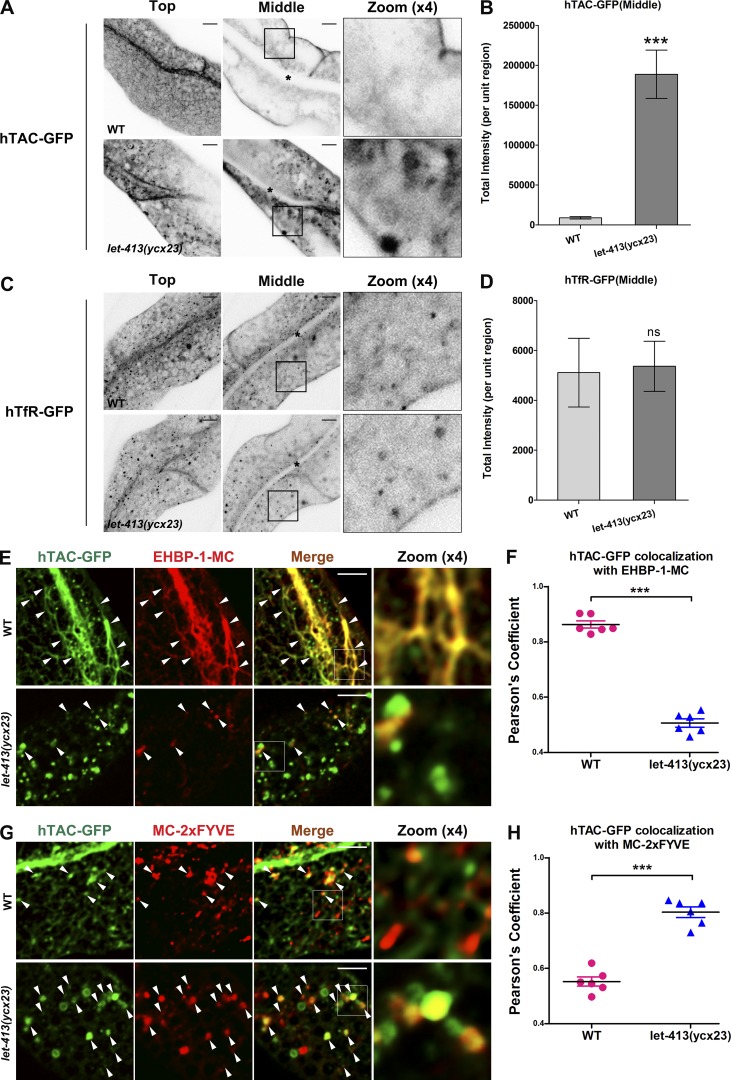
If LET-413 functions in the RAB-10–mediated recycling pathway, then we would expect to observe defects in recycling in let-413 mutants similar to those previously described in rab-10 mutants (Chen et al., 2006; Shi et al., 2010). rab-10 mutant effects are cargo specific, trapping clathrin-independent recycling cargo human IL-2 receptor α-chain (hTAC) GFP, but not clathrin-dependent recycling cargo human transferrin receptor (hTfR) GFP (Chen et al., 2006). In contrast, there was no hTAC-GFP recycling aberration in RAB-11– or RAB-35–depleted cells (Fig. S2, A and B). We found that in let-413(ycx23) mutants, hTAC-GFP strongly accumulated in intracellular vacuoles and enlarged puncta in the cytoplasm (Fig. 3 A) in a manner very similar to rab-10 mutants (Fig. S2, A and B). Quantification indicated that hTAC-GFP–labeled structure fluorescence intensity increased by more than 20-fold in let-413(ycx23) mutants (Fig. 3 B). In the wild-type background, hTAC-GFP mainly colocalizes with the recycling endosome marker EHBP-1 and, to a lesser extent, the early endosome marker EEA-1-2xFYVE (Wang et al., 2016; Fig. 3, E and G). In let-413(ycx23) mutants, the amount of hTAC-GFP in recycling endosomes decreased, and the amount in early endosomes increased, further indicating a rab-10–like defect impairing early endosome to recycling endosome transport (Fig. 3, F and H). Similar defects were observed in let-413(ycx23) mutants expressing transgenic DAF-4-GFP (type II TGF-β receptor), another RAB-10–dependent recycling cargo (Fig. S2, C and D; Gleason et al., 2014).
Figure 3.
Loss of LET-413 caused endocytic recycling defects in C. elegans intestine. (A–D) Confocal images of the worm intestinal cells expressing GFP-tagged recycling cargo proteins, the IL-2 receptor α chain (hTAC-GFP), and the hTfR-GFP. Asterisks indicate the location of the intestinal lumen. (A) In the top focal plane, hTAC-GFP–labeled tubular structures were disrupted in let-413(ycx23) mutants. In the middle focal plane, hTAC-GFP accumulated significantly on the cytosolic endosomal structures, compared with wild-type animals. An approximately 20-fold increase of total hTAC-GFP intensity was observed in let-413(ycx23) animals (B). Error bars represent SEM (n = 18 each). Asterisks indicate significant differences (***, P < 0.001; one-tailed Student’s t test). (C and D) hTfR-GFP showed no obvious intensity difference in wild-type and let-413(ycx23) animals. Error bars represent SEM (n = 18 each). ns, not significant (one-tailed Student’s t test). Asterisks in C indicate intestinal lumen. (E–H) The subcellular localization of the accumulated hTAC-GFP in let-413(ycx23) mutants. In let-413(ycx23) animals, the overlap between hTAC-GFP and EHBP-1-mCherry was reduced by ∼41.8%, whereas the colocalization between hTAC-GFP and mCherry-2xFYVE was increased by ∼45.4%. Arrowheads indicate positive overlap. Pearson’s correlation coefficients for GFP and mCherry signals under wild-type and mutant conditions are calculated (n = 6 animals). Error bars represent SEM. ***, P < 0.001. Bars, 10 µm.
Importantly, the subcellular localization of the clathrin-dependent cargos hTfR-GFP, MIG-14-GFP (a retromer-dependent cargo), or GFP-CD4-LL (a nonrecycling cargo) was not perturbed in let-413(ycx23) mutants, indicating that LET-413 has the same cargo specificity as RAB-10 (Fig. 3, C and D; and Fig. S2, E and F; Shi et al., 2009; Gu et al., 2013; Bai and Grant, 2015). These results also suggest that LET-413 does not affect clathrin-dependent uptake from the basolateral plasma membrane.
We confirmed the specificity of our intestine-specific CRISPR/Cas9 let-413(ycx23) mutants by expressing a CRISPR/Cas9 editing–resistant form of LET-413c-mCherry in the intestine, finding that it fully rescued the hTAC-GFP accumulation phenotypes of let-413(ycx23) mutants in vivo (Fig. S2, G and H). These results also indicate that the requirement for LET-413 in endocytic recycling is cell autonomous.
LET-413 functions independently of DLG-1 and LGL-1 to promote recycling
In Drosophila, the Scribble module (Scrib, Dlg, and Lgl) localizes to the basolateral plasma membrane and acts to confine apical proteins distribution (Yamanaka and Ohno, 2008; de Vreede et al., 2014). In other organisms, Lgl is thought to have a regulatory role in transmembrane protein targeting (Wirtz-Peitz and Knoblich, 2006), and Lgl1 was reported to release Rab10 from the Rab10–GDI complex to regulate membrane insertion in growing mammalian neurites (Wang et al., 2011). However, we did not find any significant changes in the intracellular localization of hTAC-GFP in dlg-1(RNAi) or lgl-1(tm2616) animals (Fig. S3, A and B). We further analyzed the subcellular distribution of RAB-5 and RAB-10 upon loss of DLG-1 or LGL-1. Consistently, the subcellular localization of GFP-RAB-5 or GFP-RAB-10 was not disturbed in dlg-1(RNAi) or lgl-1(tm2616) animals (Fig. S3, C and D). Together, our results suggested that LET-413 functions independently of DLG-1 and LGL-1 to promote recycling in the intestinal epithelia of C. elegans.
To this end, our results suggested that LET-413 exists in two distinct functional modules, participating in RAB-10–mediated transport and Scribble module–mediated apical protein confinement. To better assess the localization of LET-413, we performed a colocalization assay to detect the overlap level of LET-413 or RAB-10 with DLG-1 and LGL-1, respectively. These experiments demonstrated that LET-413 has a significant colocalization with DLG-1 or LGL-1, whereas the coexistence of RAB-10 with DLG-1 or LGL-1 is very limited (Fig. S3, E–H). Collectively, the data indicate that LET-413 is localized in at least two independent functional modules. These results also explain why dlg-1(RNAi) and lgl-1(tm2616) animals have no significant hTAC-GFP overaccumulation in the cytosol (Fig. S3, A and B).
LET-413 functions to prevent the basolateral diffusion of apical junction proteins, including PAR-3 and PAR-6 (Bossinger et al., 2001). To determine whether the recycling defects in let-413(ycx23) animals were caused by perturbed epithelial polarity, we assayed PAR-3 and PAR-6 distribution (Cox and Hardin, 2004; de Vreede et al., 2014; Waaijers et al., 2016). PAR-3-GFP and PAR-6-GFP are both normally localized on the apical plasma membrane in let-413(ycx23) animals (Fig. S4, A and B), indicating that the let-413(ycx23) animals in our studies maintains proper epithelial polarity, likely because our method removes LET-413 after intestinal polarity is already established. Expression of the vha-6 promoter used in the CRISPR/Cas9 somatic mutagenesis begins in the intestine at the relatively late twofold embryo stage (>500 cells; Pujol et al., 2001), whereas polarity establishment occurs much earlier, during the 16-cell stage (Leung et al., 1999).
Erbin depletion disturbs endocytic recycling of TAC-GFP in mammalian cells
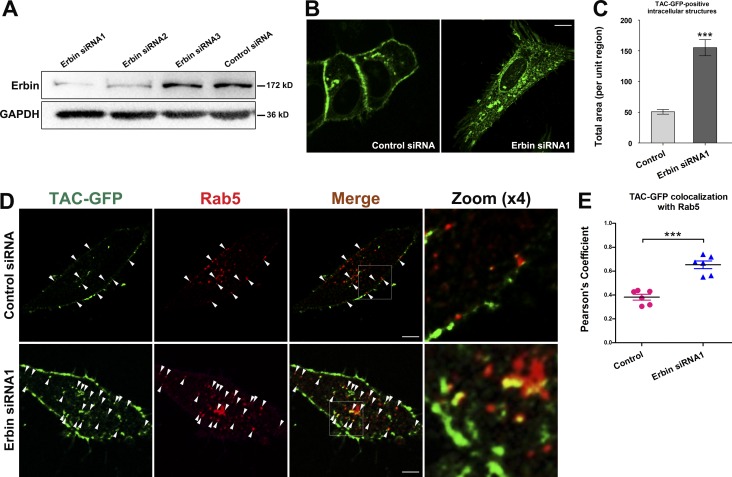
To determine whether the LET-413 human homologue Erbin is also required for the recycling of clathrin-independent cargo, we analyzed the ARF-6–dependent recycling cargo TAC in HeLa cells (Radhakrishna and Donaldson, 1997). Cells transfected with Erbin siRNA1 were compared with cells transfected with a control vector (Fig. 4 A). At steady state, TAC-GFP localized mostly to the plasma membrane of the cells, with a subpopulation found in intracellular puncta juxtaposed to the nucleus (Fig. 4 B). In contrast, TAC-GFP overaccumulated in intracellular tubules and puncta in Erbin siRNA1 cotransfected cells (Fig. 4, B and C), suggesting that the requirement for Erbin/LET-413 in the recycling pathway is conserved from C. elegans to mammals. Further analyses demonstrated that the colocalization of TAC-GFP with endogenous Rab5 in endosomal structures was considerably increased upon loss of Erbin (Fig. 4, D and E). Erbin siRNA1 cotransfected cells showed the prominent overlap of TAC-GFP with Rab5 (Pearson’s correlation coefficient, ∼67%) when compared with cells cotransfected with siRNA control plasmid only (Pearson’s correlation coefficient, ∼37%), indicating the overaccumulation of TAC-GFP in Rab5-positive endosomes.
Figure 4.
Erbin knockdown leads to TAC-GFP cytosolic accumulation in HeLa cells. (A) The knockdown efficiency of three Erbin siRNAs was analyzed by Western blotting using anti–Erbin antibody. (B and C) TAC-GFP subcellular localization in cells cotransfected with Erbin siRNA1 was compared with cells cotransfected with a control vector. Asterisks indicate significant differences (***, P < 0.001; one-tailed Student’s t test); n = 18 cells. Error bars represent SEM. (D and E) In control siRNA cells, some of the intracellular Tac-GFP was observed in Rab5-positive puncta (∼37%). However, the colocalization of Tac-GFP with Rab5 was significantly increased upon loss of Erbin (∼67%). Arrowheads indicate positive overlap. Pearson’s correlation coefficients for GFP and mCherry signals are calculated (n = 6 cells). Error bars represent SEM. ***, P < 0.001. Bars, 10 µm.
LET-413 LRRs and the PDZ domain are indispensable for endocytic recycling
The functions of LET-413 homologue domains have been studied in different organisms, but their precise roles remain vague. The hErbin PDZ expressed without the LRRs localizes in the cytosol and nucleus, whereas the LRRs expressed without the PDZ mainly associate with the basolateral membrane (Legouis et al., 2003). To characterize the roles of LRRs and the PDZ domain in endosome association, we examined the localization of GFP-tagged LET-413 truncations in the intestinal cells of C. elegans (Fig. S4, C and D). Consistent with studies in other systems, truncated fragments containing the C-terminal PDZ domain (aa 500–706) but lacking LRRs localized diffusively in the cytosol and nucleus (Fig. S4 C). However, the LRRs (aa 1–400) displayed a punctate subcellular localization (Fig. S4 C). This was not fully in agreement with previous studies in C. elegans epithelia, where a similar LRR-containing construct was reported to localize to the basolateral membrane (Legouis et al., 2003). Notably, we found that a fragment containing LRRs and central region (aa 1–499) resembles full-length LET-413 subcellular localization, labeling basolateral tubular and punctate structures (Fig. S4 C). These results suggested that LRRs are responsible for membrane targeting, whereas the central region facilitates the specificity of basolateral membrane association.
To determine the functional contributions of LRRs and the PDZ domain, we examined the hTAC-GFP recycling defects in let-413(ycx23) animals coexpressing CRISPR/Cas9 editing–resistant mCherry-tagged LET-413 truncations. Expression of fragments containing the LRRs and the central region, or just the PDZ domain, were not sufficient to relieve hTAC-GFP intracellular accumulation (Fig. S4, E and F), indicating that LRRs and the PDZ domain are both required for endocytic recycling in the intestinal epithelia.
Loss of LET-413 leads to cytosolic diffusion of RAB-10
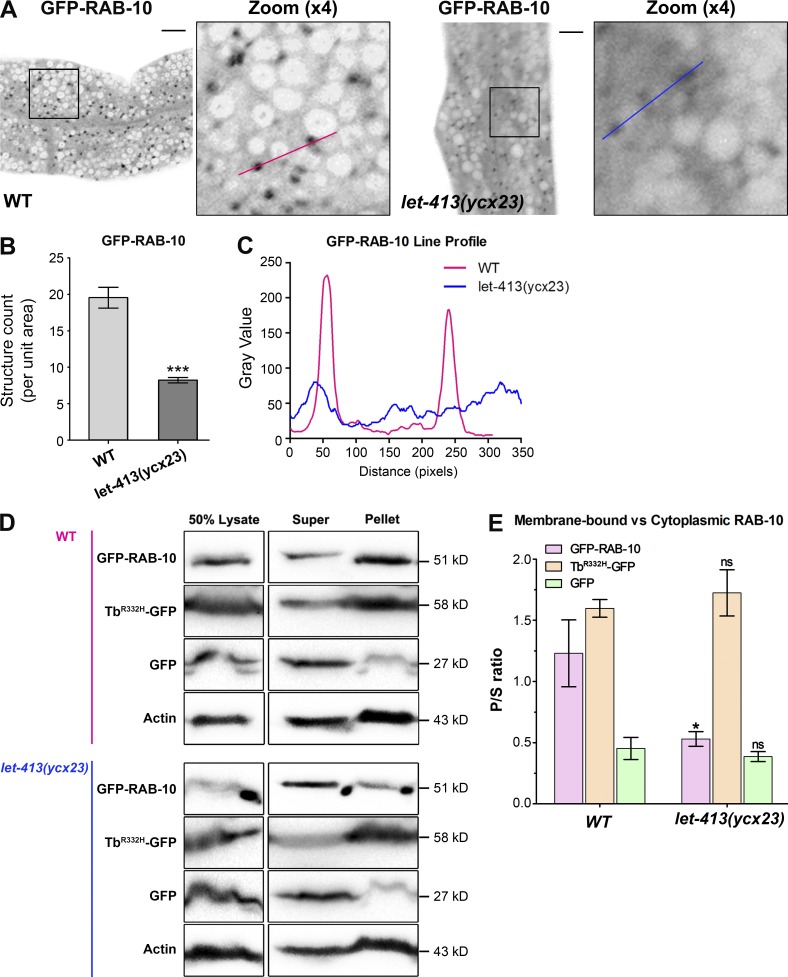
The aforementioned results clearly linked LET-413 to RAB-10 related recycling, and LET-413 possesses the affinity for a GDP-bound mutant form, RAB-10(T23N). These results suggested that LET-413 might be important for the activation of RAB-10. Because activation of Rabs to the GTP-bound state is generally associated with their recruitment to membranes, we examined RAB-10 subcellular distribution in let-413(ycx23) or let-413(RNAi) animals. Loss of LET-413 significantly decreased GFP-RAB-10–labeled puncta number (Fig. 5, A and B; and Fig. S5, A and B), with a diffusive redistribution of GFP-RAB-10 in the cytosol (Fig. 5 C). We further measured the membrane association of RAB-10 biochemically, separating cytosol from the membrane in worm lysates by ultracentrifugation and assaying intestinal GFP-RAB-10 distribution by Western blot (Liu and Grant, 2015). Consistent with LET-413 playing a role in the endosomal association of RAB-10, the membrane-to-cytosol ratio of GFP-RAB-10 in let-413(ycx23) mutants significantly declined (by ∼57%; Fig. 5, D and E). As a control, we assayed the membrane association of the PI(4,5)P2 biosensor TbR332H-GFP, which specifically associates with PI(4,5)P2-enriched membranes. In the meantime, we deployed GFP as a cytosolic protein control. It is worth noting that the membrane-to-cytosol ratio of membrane-associated TbR332H-GFP or cytosolic GFP was not significantly affected in let-413(ycx23) mutants (Fig. 5, D and E). Together, these data could reflect an effect on the RAB-10(GTP) level, perhaps by affecting RAB-10’s interaction with the GEF DENN-4. Alternatively, LET-413 may affect RAB-10 endosomal recruitment more directly.
Figure 5.
Loss of LET-413 resulted in the cytosolic dispersal of RAB-10. (A and B) Compared with wild-type animals, the lack of LET-413 resulted in a reduction of ∼60% of the number of RAB-10–labeled puncta and cytosolic diffusion of GFP-RAB-10. (C) Line-scan profile of fluorescence intensity of GFP-RAB-10–labeled structures in wild-type (red) and let-413(ycx23) animals (blue). Error bars represent SEM (n = 18 each). Asterisks indicate significant differences (***, P < 0.001; one-tailed Student’s t test). Bars, 10 µm. (D) The membrane-to-cytosol ratio of RAB-10 decreased in let-413(ycx23) mutants. Membrane structures were separated from the cytosol of wild-type and let-413(ycx23) worm lysates by ultracentrifugation at 100,000 g for 1 h. GFP-RAB-10, TbR332H-GFP, and GFP in the supernatants and pellets were analyzed by Western blotting using anti–GFP antibody. Loading control was blotted by the anti–actin antibody. (E) Quantification of the membrane-to-cytosol ratio (P/S) of GFP-RAB-10, TbR332H-GFP, and GFP in wild-type and let-413(ycx23) backgrounds. The SEMs from three independent experiments are shown (*, P < 0.05; ns, not significant).
LET-413 facilitates the interaction between DENN-4 and RAB-10(GDP) and promotes the GEF activity of DENN-4 toward RAB-10
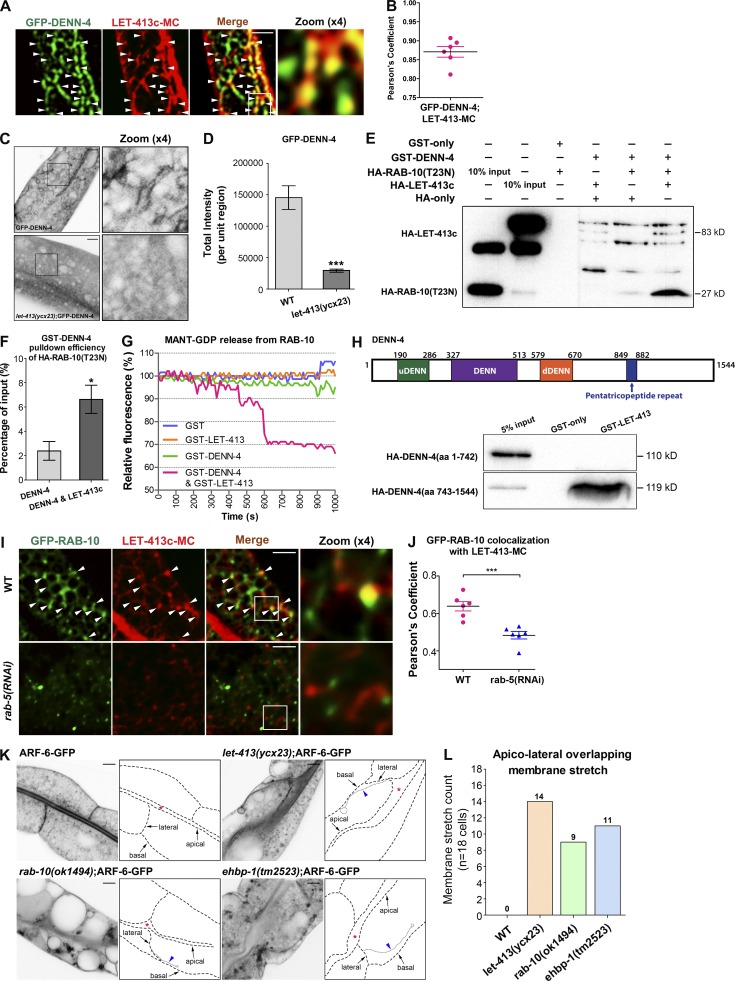
Given the biochemical evidence that LET-413 preferentially interacts with the inactive form RAB-10(GDP), LET-413 could participate in the process of RAB-10’s conversion to a GTP-bound form. Recently, DENND4 was identified as a Rab10 GEF in adipocytes that is required for the Rab10-dependent translocation of glucose transporter GLUT4 upon insulin stimulation (Yoshimura et al., 2010; Sano et al., 2011). Consistently, the number of GFP-RAB-10–labeled puncta was significantly reduced upon loss of DENN-4, the C. elegans homologue of DENND4, and GFP-RAB-10 showed a prominent cytosolic diffusion (Fig. S5, C and D). Interestingly, we found that DENN-4 colocalized with LET-413 on intestinal endosomes, suggesting that LET-413 might act through DENN-4 (Fig. 6, A and B). In our GST pull-down assay, GST-DENN-4 interacted with HA-LET-413 directly (Fig. S5 E). Furthermore, depletion of LET-413 impaired the labeling of GFP-DENN-4 at tubules and puncta and caused cytosolic dispersion of DENN-4 (Fig. 6, C and D). We thus hypothesized that LET-413 could act as a cofactor to enhance the activity of DENN-4 toward RAB-10.
Figure 6.
LET-413 facilitated the interaction of DENN-4 with RAB-10(GDP), and the intestinal cells’ lateral membrane organization was distorted in let-413 and recycling regulator mutants. (A and B) GFP-DENN-4 and LET-413c-mCherry displayed significant colocalization on punctate structures. Pearson’s correlation coefficient for GFP and mCherry signals is calculated (n = 6 animals). (C and D) Loss of LET-413 led to the cytosolic dispersal of GFP-DENN-4. Error bars represent SEM (n = 18 each). Asterisks indicate significant differences (***, P < 0.001; one-tailed Student’s t test). (E) In the presence of LET-413, the affinity of GST-DENN-4 for HA-RAB-10(T23N) was significantly elevated. (F) GST-DENN-4 pull-down efficiency of HA-RAB-10(T23N) with LET-413c was increased by approximately threefold. The SEMs from three independent experiments are shown. Asterisks indicate significant differences (*, P < 0.05; one-tailed Student’s t test). (G) In vitro GEF assay. MANT-GDP release from RAB-10 was measured by adding GST-LET-413, GST-DENN-4, and GST-DENN-4&GST-LET-413. (H) LET-413c lacks binding to the fragment containing DENN domain (aa 1–742) but has a significant interaction with the C-terminal fragment of DENN-4 (aa 743–1544). (I and J) The colocalization of LET-413c-mCherry with GFP-RAB-10 in wide-type (∼64% overlap) and rab-5(RNAi) (∼48% overlap) animals was compared. Pearson’s correlation coefficients for GFP and mCherry signals are calculated (n = 6 animals). Error bars represent SEM. ***, P < 0.001. (K) Intestinal cells’ lateral membranes (labeled by ARF-6-GFP) were distorted in let-413, rab-10, and enbp-1 mutants. Arrows indicate apical, basal, and lateral plasma membranes. Arrowheads indicate the overextended lateral membrane. (L) Overextended lateral membranes were quantified. Numbers of lateral membrane overextensions in 18 intestinal cells (total of six animals per each genotype) were quantified and plotted. Bars, 10 µm.
To gain insight into the mechanism of LET-413 function, we examined the impact of LET-413 on the affinity between DENN-4 and RAB-10(T23N) (Fig. 6, E and F; and Fig. S5 F). Surprisingly, the interaction of DENN-4 with RAB-10(T23N) was relatively weak. However, after addition of HA-LET-413 to the reaction, the ability of GST-DENN-4 to pull down HA-RAB-10(T23N) increased by approximately threefold (Fig. 6 F). Furthermore, we found that DENN-4 itself has no significant RAB-10 GEF activity in vitro (Fig. 6 G). Notably, when we added LET-413 to the reaction, the RAB-10-GEF activity of DENN-4 was initiated, and preloaded MANT-GDP (2′-/3′-O-(N′-methylanthraniloyl) guanosine-5′-O-diphosphate), a fluorescent GDP analogue, was gradually released (Fig. 6 G). This result brought us two important messages. First, in agreement with the results in adipocytes, DENN-4 is a bona fide GEF protein of RAB-10 in C. elegans. Second, the activation of the GEF activity of DENN-4 toward RAB-10 requires the assistance of LET-413. DENN-4 itself cannot effectively promote GDP to GTP conversion on RAB-10. A recent study showed that DENND3 takes an autoinhibited conformation (Xu and McPherson, 2017). DENN-4 may adopt a similar self-regulatory configuration. The interaction between LET-413 and the DENN-4 C terminus could release the N-terminal DENN domain from the self-inhibiting conformation, thereby enhancing the affinity of DENN domain to RAB-10(GDP). In agreement with the model, LET-413 did not bind to the fragment containing the DENN domain but had a strong interaction with the C terminus of the DENN-4 containing the pentatricopeptide repeat (Fig. 6 H). We then assayed the distribution of GFP-RAB-11 in denn-4(RNAi) animals (Fig. S5, G and H). We note that the subcellular labeling of RAB-11 did not show significant abnormalities upon loss of DENN-4, indicating that DENN-4 does not possess GEF activity toward RAB-11 in intestinal cells. Collectively, our in vivo and in vitro results strongly suggested that LET-413 specifically promotes the gaining of active RAB-10(GTP) through strengthening the physical interaction of RAB-10(GDP) with DENN-4.
Membrane trafficking routes are often mediated through Rab GTPase cascades (Hutagalung and Novick, 2011). Our evidence suggested that in C. elegans intestinal epithelia, LET-413 is a putative RAB-5 effector and plays a crucial role in activating RAB-10. To test this idea, we assayed the subcellular localization of LET-413 in rab-5(RNAi) animals and found that LET-413c-GFP labeling on tubules significantly decreased (Fig. S5, I and J). We then compared the colocalization of LET-413 with RAB-10 in rab-5(RNAi) animals. In the wild-type background, LET-413 colocalizes well with RAB-10 at the endosomes. Silencing of RAB-5 significantly impaired the overlap between LET-413 and RAB-10. GFP-RAB-10 displayed more diffusive localization in the cytoplasm when RAB-5 was depleted (Fig. 6, I and J, insets). To further characterize the functional position of LET-413 in RAB-5-to-RAB-10 cascade, we examined the subcellular localization of LET-413 in rab-10(ok1494) and ehbp-1(tm2523) null mutants. We observed that knockout of RAB-10 or EHBP-1 induced the accumulation of LET-413 in aggregates and vacuoles (Fig. S5, K and L). This phenotype could be explained by the loss of endosomal recruitment of TBC-2/RAB-5-GAP in RAB-10– or EHBP-1–deficient animals, resulting in the increased amount of active RAB-5. Together, our data suggested that LET-413 functions together with DENN-4 to mediate RAB-5 to RAB-10 progression in endocytic recycling.
Lateral membrane organization of intestinal epithelia is impaired in let-413 and recycling regulators mutants
In let-413 mutant embryonic intestine and epidermis, extended and discontinuous adherens junctions were observed. Apical membranes of epidermal cells abnormally extended and formed overlapping membrane extensions (Legouis et al., 2000). These phenotypes were thought to be the consequence of the absence of properly positioned apical junctions. Likewise, in C. elegans young adult intestines, close inspection using plasma membrane marker ARF-6 revealed the impaired lateral membrane organization in let-413 mutants. In wild-type animals, ARF-6-GFP–labeled lateral membranes are short and mostly vertical to the apical lumen (Fig. 6 K). However, in let-413(ycx23) mutants, lateral membranes were often disorganized and overextended into the cytosol (Fig. 6 K), which is comparable to the abnormalities observed in let-413 mutant embryos epidermis (Legouis et al., 2000). To determine whether this is also the case in recycling regulators mutants, we examined the lateral membranes organization in rab-10(ok1494) and ehbp-1(tm2523) null mutants. We found that loss of either RAB-10 or its effector, EHBP-1, caused an elevated incidence of lateral membrane overextension (Fig. 6, K and L), suggesting a requirement for RAB-10–mediated recycling transport in the proper organization and positioning of apical junction proteins. We next examined the localization of apical junction protein HMP-1(α-catenin) in intestinal epithelia (Cox and Hardin, 2004; Segbert et al., 2004). HMP-1-GFP normally resides in the subapical contact zone between cells, displaying a punctate labeling at the apicolateral membrane junction. Nevertheless, in the absence of LET-413, HMP-1-GFP subcellular distribution was disturbed and abnormally appeared in lateral plasma membranes (55.6% defective; Fig. S5, M and N). A similar phenotype was also observed in rab-10 mutant animals (28.6% defective; Fig. S5, M and N). These studies highlight the importance of the LET-413–RAB-10 module as an intrinsic determinant of correct targeting of apical junctions.
Discussion
Cell polarity regulators have been implicated in endocytic trafficking. For example, Lgl regulates transmembrane protein targeting (Yamanaka and Ohno, 2008; Wang et al., 2011). Scribble1 has been postulated to function in concert with Arf-6 during the endocytic recycling of GluN2A (Piguel et al., 2014). However, the mechanisms that link LET-413, Erbin, and Scrib in endocytic recycling were not fully examined in these previous studies. Here, we identified LET-413 as a new RAB-5 effector. LET-413 facilitates the interaction of the RAB-10 GEF protein DENN-4 with RAB-10(GDP). Loss of LET-413 impaired the localization of DENN-4 on endosomes and resulted in the cytosolic diffusion of RAB-10 effectors TBC-2 and CNT-1, indicating that RAB-10 is not well activated in the absence of LET-413.
Specifically, our data suggest that LET-413 and DENN-4 jointly mediate a RAB-5 to RAB-10 cascade in the recycling pathway. Rab GTPase cascades are characterized by a transition from an upstream Rab to a downstream Rab. During Rab switching on endosomes, the upstream Rab recruits a GEF as its effector (GEF cascade) to activate the downstream Rab. Studies in C. elegans suggest that the Mon1 homologue SAND-1 functions as the relevant RAB-7 GEF (Nordmann et al., 2010; Poteryaev et al., 2010). Interestingly, as a Rab5 effector, Mon1 forms a complex with Ccz1 and CCZ-1 to exhibit Rab7-GEF activity (Kinchen and Ravichandran, 2010). In a similar manner, we identified LET-413 as a previously unrecognized RAB-5 effector potentiating a GEF cascade via interaction with RAB-10-GEF DENN-4. Our data suggest that LET-413 is involved in the regulation of RAB-10 activity by promoting the RAB-10(GDP) affinity of DENN-4. Impairment of this process results in mislocalization of RAB-10 and the intracellular trapping of hTAC-GFP. It is possible that DENN-4 possesses an autoinhibited conformation, similar to that reported for the Rab12 exchange factor DENND3 (Xu and McPherson, 2017). Access of RAB-10(GDP) to the DENN domain of DENN-4 may require relief from a DENN-4 intramolecular interaction, and the interaction between DENN-4 and LET-413 could contribute to this conformational alteration.
LET-413–mediated recycling transport, as previously found for RAB-10–mediated transport, is cargo specific, strongly affecting clathrin-independent cargo, whereas clathrin-dependent cargo was not affected. A previous study identified a requirement of rat Scribble1 for the internalization of the GluN2A subunit via AP2-mediated clathrin-dependent uptake (Piguel et al., 2014). Conversely, in Drosophila Scribble mutants, the endocytosis of the AP2-dependent cargo Notch was not disturbed (de Vreede et al., 2014). Instead, the endosome-to-TGN trafficking of the retromer-dependent cargo Wntless was abnormal in imaginal discs cells. We analyzed the well-established retrograde cargo MIG-14-GFP in let-413 mutants (Shi et al., 2009) and did not observe any differences from the results reported in Drosophila. The study in mammals also revealed the implication of LAP proteins in the regulation of various aspects of intracellular transport. Erbin was found to localize to the basolateral membrane and interacts with the epidermal growth factor receptor family protein ErbB2, thereby restricting ErbB2 to the basolateral domain of epithelia (Borg et al., 2000). Moreover, we noticed that loss of Erbin caused the accumulation of recycling cargo TAC-GFP on Rab5-positive endosomes in HeLa cells, suggesting that Erbin is required for the recycling transport in mammals. Indeed, a recent study also showed that Erbin resides in the EEA1-positive early endosomes (Sflomos et al., 2011). Another mammalian LAP member, scribble, was found to interact with AP2 to facilitate the endocytic uptake of N-methyl-d-aspartate receptor (Piguel et al., 2014). In contrast, scribble has also been proposed to play a role in limiting the endocytosis of clathrin-dependent cargo E-cadherin in polarized MDCK cells (Georgiou et al., 2008; Lohia et al., 2012). This inconsistency implies that the role of mammalian scribble is differentiated in distinct cellular context. An alternative explanation is that the scribble functionality in endocytosis is cargo-specific. It is noteworthy that previous studies on scribble homologues in Drosophila and yeast suggest that Scrib could act to regulate the polarized targeting of Golgi-derived apical proteins (Grindstaff et al., 1998; Bilder and Perrimon, 2000;). The junction complex could serve as a sorting site for a subset of the Golgi-derived vesicular carrier (Yeaman et al., 1999), where Scrib might interact with the exocyst and regulate the polarized sorting of cargo proteins (Grindstaff et al., 1998; Bilder and Perrimon, 2000). Together, these findings indicate that LET-413 and its homologues, including scribble and Erbin, are implicated in the different routes of endocytic transport. In addition to participating in the regulation of the endocytic uptake at the plasma membrane, scribble may also work with other secretory regulators to coordinate the flow of exocytic vesicles. More work will be required to understand the functional diversity of LET-413 homologues in endocytic transport in metazoans.
In addition, our GST pull-down results suggest that LET-413 may also regulate RAB-11 activation. Nevertheless, the depletion of RAB-11 failed to disturb the subcellular localization of hTAC-GFP (Fig. S2, A and B). Moreover, loss of LET-413 did not affect subcellular localization of RAB-11 in C. elegans intestinal epithelia (Fig. S1, N and P). Consistently, we also noticed that the GEF activity of DENN-4 appears to be explicitly directed against RAB-10 rather than RAB-11 (Fig. S5, G and H). One plausible explanation is that Rab regulatory multiplicity of LET-413 can be mediated through distinct GEF partners in various tissues, contributing to the tissue-specific requirements for transport regulators like RAB-11 and RAB-10. Extensive experiments need to be executed to determine whether LET-413 modulates RAB-11 activation in a similar manner in other cell types.
Our study showed that loss of either RAB-10 or LET-413 induced disorganized lateral membranes in live animals, suggesting the likely role of endocytic trafficking in the maintenance of epithelial membrane integrity. Small GTPases have been described to regulate epithelial membrane integrity. In MDCK cells, Cdc42 localizes at the apical membrane, interacting with the PAR complex and cytoskeletal regulators to coordinate endocytosis of cadherin (Johansson et al., 2000; Lin et al., 2000; Garrard et al., 2003; West and Harris, 2016). Rab8/RAB-8 has also been reported to sustain polarized traffic in intestinal epithelia (Sato et al., 2007). Endocytosed Cdc42 colocalizes with Sec4, the yeast homologue of Rab8, on recycling endosomes, suggesting the functional role of Sec4 in the recycling of Cdc42 (Watson et al., 2014). In the mammalian neuron, Rab10 promotes polarized membrane trafficking and axonal development. Lgl1 acts as a positive regulator of the Rab10 in such neurons (Wang et al., 2011; Liu et al., 2013). During Drosophila tracheal terminal cell branching morphogenesis, Rab10 was found to mediate polarized branch outgrowth of terminal cells (Jones et al., 2014). Our current findings show that the proper GEF-mediated activation of RAB-10 via LET-413 is required for the integrity of lateral membranes, suggesting a model in which LET-413 regulates lateral membrane composition by influencing sorting steps and cargo transfer between early endosomes and recycling endosomes, regulating the recycling itinerary of key apical junction proteins.
Materials and methods
General methods and strains
All C. elegans strains used in this study were derived from Bristol strain N2. C. elegans cultures, and genetic crosses were performed by standard methods (Brenner, 1974). All strains were grown at 20°C on nematode growth media plates seeded with the Escherichia coli strain OP50. A complete list of strains used in this study can be found in Table S1.
C. elegans RNAi-mediated interference was performed using a previously described feeding protocol (Timmons and Fire, 1998). Feeding constructs in this study were either from the Ahringer library (Kamath and Ahringer, 2003) or prepared by PCR from cDNA templates followed by subcloning into the RNAi vector L4440 (Timmons and Fire, 1998). For most experiments, synchronized L1- or L3-stage animals were treated for 48–72 h and scored as adults.
Antibodies
The antibodies used in this study were rabbit anti–actin polyclonal antibody (sc-1616-R; Santa Cruz Biotechnology), rabbit anti–HA monoclonal antibody (C29F4; Cell Signaling Technology), rabbit anti–GST monoclonal antibody (91G1; Cell Signaling Technology), rabbit anti–GFP polyclonal antibody ChIP grade (ab290; Abcam), rabbit anti–Erbin polyclonal antibody (ab55930; Abcam), rabbit anti–Rab5 monoclonal antibody (3547; Cell Signaling Technology), and anti–rabbit IgG-Alexa Fluor 594 conjugate (8889; Cell Signaling Technology).
Somatic CRISPR/Cas9 mutant strains
CRISPR/Cas9 constructs were prepared according to a previously described method (Shen et al., 2014; Li et al., 2015). The somatic CRISPR/Cas9 expression vectors were constructed by replacing the eft-3 promoter in pDD162 (47549; Addgene) with the C. elegans intestine-specific promoter Pvha-6 through overlapping PCR by Phusion high-fidelity DNA polymerase (Thermo Scientific). The CRISPR design tool (http://crispr.mit.edu) was used to select the specific targets. Three let-413 target sequences were chosen: 5′-TCGAAGAGAGAATAGGCGATGG-3′, 5′-TGAGCAGTAATCCGTTCACAAGG-3′, and 5′-TCAATTCCACAATCGACAGAGG-3′. The somatic CRISPR/Cas9 vectors were transformed into DH5α competent cells. The clones were confirmed to contain the target sequences via sequencing. CRISPR/Cas9 constructs transgenic C. elegans were generated by microinjection of DNA plasmids at 50 ng/µl and a selection marker, Podr-1::rfp, into an N2 hermaphrodite germline (Zhou et al., 2016).
Cell cultures and transfection
Human cervical adenocarcinoma cells (HeLa) were cultured in DMEM supplemented with 10% heat-inactivated FBS, 100 U/ml penicillin, and 100 U/ml streptomycin. Exponentially growing cells were trypsinized; solutions with 50,000 cells/ml were prepared in 90% medium (no phenol red or antibiotics) with 10% vol/vol of the appropriate serum. Plates with an appropriate volume of cell stock were incubated at 37°C for 2 h before siRNA transfection. 12 µl X-tremeGENE siRNA Transfection Reagent (Sigma), siRNA nontargeting control or siRNA1 plasmid (2 µg per siRNA; Ribobio Inc.), and 2 µg TAC-GFP plasmid were added into 100 µl OPTI-MEM I (Gibco) in sterile microcentrifuge tubes. The tubes were incubated at room temperature for 5 min and then mixed for 20 min. 24 h after transfection, cells were fixed with 5% paraformaldehyde for 5 min at room temperature. 0.1% Triton X-100 was used for 15 min to permeate cell membrane. 5% BSA was added to the cells to block nonspecific antigen recognition for 30 min. Rab5 rabbit mAb was then used to detect endogenous Rab5 at a dilution of 1:200. Cells were plated at 4°C overnight. Anti–rabbit IgG Alexa Fluor 594 conjugate was used to detect the primary antibody with a dilution of 1:200 for 2 h at room temperature before confocal imaging. siRNA oligonucleotides were synthesized with the following sequences: si-hErbin001, 5′-GCAACTAAGTGGATTGAAA-3′; si-hErbin002, 5′-GAACAAGTACTTCGACATA-3′; and si-hErbin003, 5′-CAGATGAGCTTAAGAATAT-3′ (Ribobio Inc.).
Protein expression and GST pull-down assays
rab-5(Q78L), rab-5(S33N), rab-8(Q67L), rab-8(T22N), rab-10(Q68L), rab-10(T23N), rab-10(N122I), rab-11.1(Q70L), rab-11.1(S25N), rab-35(Q69L), rab-35(S24N), arf-6(Q67L), arf-6(T27N), denn-4, denn-4(1-742aa), and denn-4(743-1544aa) cDNAs were transferred into an in-house–modified vector (pcDNA3.1(+); Invitrogen) with a 2xHA epitope tag and Gateway cassette (Invitrogen) for in vitro transcription and translation. Equivalent let-413c, let-413(1-400aa), and let-413(500-706aa) PCR products were introduced in-frame into vector pGEX-2T (GE Healthcare Life Sciences) modified with a Gateway cassette. The N-terminally HA-tagged proteins were synthesized in vitro using the TNT-coupled transcription/translation system (Promega). GST-only, GST-LET-413, GST-LET-413(1-400aa), and GST-LET-413(500-706aa) fusion proteins were expressed in the ArcticExpress strain of E. coli (Stratagene). Bacterial pellets were lysed in 50 ml lysis buffer (50 mM Hepes, pH 7.5, 400 mM NaCl, 1 mM DTT, and 1 mM PMSF) with Complete Protease Inhibitor Mixture Tablets (Sigma). Extracts were cleared by centrifugation, and supernatants were collected and incubated with glutathione Sepharose 4B beads (GE Healthcare Life Sciences) at 4°C for 3 h. Beads were washed six times with cold STET buffer (10 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, and 0.1% [vol/vol] Tween-20). In vitro synthesized HA-tagged protein (15 µl TNT mix diluted in 500 µl STET) was added to the beads and allowed to bind at 4°C overnight. After six additional washes in STET, the proteins were eluted by boiling in 30 µl of 2× SDS-PAGE sample buffers. Eluted proteins were separated on SDS-PAGE (12% [wt/vol] polyacrylamide) blotted to nitrocellulose. After blocking with 5% milk and washing with STET, the blot was probed with the anti–HA antibody and then stripped and reprobed with the anti–GST antibody.
Membrane fractionation assay
Wild-type and let-413(ycx23) mutant C. elegans expressing intestinal GFP or GFP fusion proteins were synchronized and cultured on nematode growth media. Mixed stage animals were washed off with M9 buffer, and the worm pellet was resuspended in 500 µl lysis buffer (50 mM Tris-HCl, pH 8.0, 20% sucrose, 10% glycerol, 2 mM DTT, and protease inhibitor). The worms were then disrupted using an automatic grinding machine (Jingxin Inc.). Carcasses and nuclei were cleared by centrifugation at 1,000 g for 5 min at 4°C. 200 µl of the post-cleared lysate was centrifuged at 100,000 g for 1 h. Supernatants were collected as samples, and pellets were reconstituted in the same volume of lysis buffer as that of the supernatant. Equal volume supernatants and pellets were subjected to immunoblotting using anti-actin and anti–GFP antibodies.
Plasmids and transgenic strains
For let-413(ycx23) mutant rescue assays, corresponding single guide RNA–resistant plasmids were constructed. We introduced one silent mutation in or around the PAM and three silent mutations in the codon of each target sequence (5′-TCGAAGAGAAAACAGACGATGA-3′, 5′-TGAGCAGTAACCCTTTTACTAGG-3′, and 5′-TCAATTCAACGATTGACAAAGG-3′). To construct GFP or mCherry fusion transgenes specifically expressed in C. elegans intestine, previously described vha-6 promoter-driven vectors modified with a Gateway cassette inserted upstream or downstream of the GFP or mCherry coding region were used. The sequences of let-413, let-413(1-400aa), let-413(1-499aa), let-413(500-706aa), Tubby-PH(R332H) (the faithful reporter of PI(4,5)P2; Quinn et al., 2008; Hardie et al., 2015), par-3, par-6, 2xFYVE(eea-1), and denn-4 lacking a stop codon or a start codon were cloned into entry vector pDONR221 by PCR and BP reaction and then transferred into intestinal expression vectors by LR reaction to generate fusion plasmids (Chen et al., 2006). Transgenic lines for these plasmids were obtained using standard microinjection techniques; plasmids were coinjected with the selection markers Podr-1::gfp, Podr-1::rfp into N2 or let-413(ycx23) hermaphrodite germlines.
Nucleotide exchange assay
The GEF assay was performed as described previously (Sakaguchi et al., 2015), with some modifications. RAB-10 tagged with C-terminal His6 was purified as described previously (Ebine et al., 2014). 50 µM MANT-GDP (Invitrogen) was incubated with 500 pmol RAB-10-His6 in the preloading buffer at 25°C for 60 min, and 10 mM MgCl2 was added to stop the reaction. 200 pmol preloaded Rab-His6 was incubated with 100 pmol of the indicated GST-tagged proteins in GEF buffer (50 mM Tris, pH 8.0, 150 mM NaCl, and 0.5 mM MgCl2) at 25°C for 100 s The nucleotide exchange reaction was started by adding the hydrolysis-resistant GTP analogue GMP-PNP (guanylyl-imidodiphosphate; Calbiochem) at a final concentration of 0.1 mM. The nucleotide exchange reaction was recorded using a Synergy 2 modular multimode reader (BioTek) at an excitation wavelength of 366 nm and an emission wavelength of 443 nm.
Confocal microscopy and imaging analysis
Live C. elegans animals were mounted on 2% agarose pads with 100 mM levamisole. Multiwavelength fluorescence images (GFP, mCherry, and DAPI channels) were obtained at 20°C using a C2 laser scanning confocal microscope (Nikon) equipped with a 100× NA 1.2 oil-immersion objective and captured using NIS-Elements AR 4.40.00 software. Monofluorescence images (GFP channel) were obtained at 4°C using the C2 laser scanning confocal microscope equipped with a 100× NA 1.2 oil-immersion objective and captured using NIS-Elements AR 4.40.00 software. Z-series of optical sections were acquired using a 0.5 µm step size.
Fluorescence data from GFP channel were analyzed by Metamorph software version 7.8.0.0 (Universal Imaging). The “Integrated Morphometry Analysis” function of Metamorph was used to measure the fluorescent structures that are significantly brighter than the background and to measure fluorescence intensity (total intensity), puncta number (structure count), and fluorescence area (total area) within unit regions. From a total of six animals of each genotype, “total intensity,” “structure count,” and “total area” were sampled in three different unit regions of each intestine defined by a 100 × 100 (pixel2) box positioned at random (n = 18 each). In most cases, total area was used to compare tubularity, because the normal endosomal tubule meshwork covers much more area than when the network collapses into puncta. Another parameter, structure count, was also used to assay this aspect, where the structure count increases as the network breaks down into puncta. Quantification of colocalization images was performed using the open source Fiji (ImageJ) software (Schindelin et al., 2012). Pearson’s correlation coefficients for GFP and mCherry signals were calculated, and six animals for each genotype were analyzed.
To establish a quantitative index for the extended lateral membranes phenotype, total lateral membranes overextension numbers were quantified in three intestinal cells of six let-413(ycx23), rab-10(ok1494), and ehbp-1(tm2523) mutant animals (n = 18 per genotype) using endosome visualization marker ARF-6-GFP.
Statistical analysis
All statistical analyses were performed and plotted using Prism software version 5.01 (GraphPad Software).
Online supplemental material
Fig. S1 demonstrates the endosomal labeling of GFP-TBC-2 or CNT-1-GFP is disrupted in rab-10(ok1494) or let-413(RNAi) animals. However, GFP-RAB-7 and GFP-RAB-11 subcellular localizations are intact in let-413(ycx23) mutants. Fig. S2 shows that hTAC-GFP overaccumulates in rab-10(ok1494) mutants, but not in rab-11(RNAi) and rab-35(RNAi) animals. Similarly, cargo protein DAF-4-GFP accumulates in let-413(ycx23) mutants. Fig. S3 shows that DLG-1 and LGL-1 colocalize well with LET-413, but not with RAB-10. Fig. S4 shows that the apical domain of plasma membrane remains normal in the let-413(ycx23) background. Fig. S5 shows that loss of LET-413 or DENN-4 affects RAB-10 puncta labeling. Moreover, in let-413(ycx23) or rab-10(ok1494) mutants, HMP-1-GFP mislocalizes in lateral plasma membranes. Table S1 lists transgenic and mutant strains and is included as a Word file.
Supplementary Material
Acknowledgments
We are grateful to Dr. Barth D. Grant (Rutgers University, New Brunswick, NJ), Dr. Guangshuo Ou (Tsinghua University, Beijing, China), and Dr. Roger C. Hardie (University of Cambridge, Cambridge, England, UK) for important reagents. We thank Xin Zhang, Sophia Z. Tao, Xin Fu, and Zhenrong Yang for the technical assistance.
This work was supported by the National Natural Science Foundation of China (grants 31771570, 31571466, and 81371418), the Program for HUST Interdisciplinary Innovation Team and the Fundamental Research Funds for the Central University (2016JCTD108), the Specialized Research Fund for the Doctoral Program of Higher Education (grant 20130142110071), the Program for New Century Excellent Talents in University (grant NCET-13-0234), and the Junior Thousand Talents Program of China (to A. Shi).
The authors declare no competing financial interests.
Author contributions: H. Liu and A. Shi conceived and designed the study. H. Liu, S. Wang, W. Hang, W. Zhang, C. Yang, and Z. Cheng performed the experiments. J. Gao, J. He, and J. Zhou contributed the reagents. H. Liu, J. Chen, and A. Shi analyzed and interpreted the data. H. Liu and A. Shi wrote the paper with inputs and final approval from all authors.
Footnotes
Abbreviations used:
- GAP
- GTPase-activating protein
- GEF
- guanine nucleotide exchange factor
- hTAC
- human IL-2 receptor α-chain
- hTfR
- human transferrin receptor
- LRR
- leucine-rich repeat
- PI(4,5)P2
- phosphatidylinositol 4,5-bisphosphate
References
- Bai Z., and Grant B.D.. 2015. A TOCA/CDC-42/PAR/WAVE functional module required for retrograde endocytic recycling. Proc. Natl. Acad. Sci. USA. 112:E1443–E1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balklava Z., Pant S., Fares H., and Grant B.D.. 2007. Genome-wide analysis identifies a general requirement for polarity proteins in endocytic traffic. Nat. Cell Biol. 9:1066–1073. 10.1038/ncb1627 [DOI] [PubMed] [Google Scholar]
- Bilder D., and Perrimon N.. 2000. Localization of apical epithelial determinants by the basolateral PDZ protein Scribble. Nature. 403:676–680. 10.1038/35001108 [DOI] [PubMed] [Google Scholar]
- Borg J.P., Marchetto S., Le Bivic A., Ollendorff V., Jaulin-Bastard F., Saito H., Fournier E., Adélaïde J., Margolis B., and Birnbaum D.. 2000. ERBIN: a basolateral PDZ protein that interacts with the mammalian ERBB2/HER2 receptor. Nat. Cell Biol. 2:407–414. 10.1038/35017038 [DOI] [PubMed] [Google Scholar]
- Bossinger O., Klebes A., Segbert C., Theres C., and Knust E.. 2001. Zonula adherens formation in Caenorhabditis elegans requires dlg-1, the homologue of the Drosophila gene discs large. Dev. Biol. 230:29–42. 10.1006/dbio.2000.0113 [DOI] [PubMed] [Google Scholar]
- Bossinger O., Fukushige T., Claeys M., Borgonie G., and McGhee J.D.. 2004. The apical disposition of the Caenorhabditis elegans intestinal terminal web is maintained by LET-413. Dev. Biol. 268:448–456. 10.1016/j.ydbio.2004.01.003 [DOI] [PubMed] [Google Scholar]
- Brenner S. 1974. The genetics of Caenorhabditis elegans. Genetics. 77:71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant P.J., and Huwe A.. 2000. LAP proteins: what’s up with epithelia? Nat. Cell Biol. 2:E141–E143. 10.1038/35019616 [DOI] [PubMed] [Google Scholar]
- Chen C.C., Schweinsberg P.J., Vashist S., Mareiniss D.P., Lambie E.J., and Grant B.D.. 2006. RAB-10 is required for endocytic recycling in the Caenorhabditis elegans intestine. Mol. Biol. Cell. 17:1286–1297. 10.1091/mbc.E05-08-0787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotard L., Mishra A.K., Sylvain M.A., Tuck S., Lambright D.G., and Rocheleau C.E.. 2010. TBC-2 regulates RAB-5/RAB-7-mediated endosomal trafficking in Caenorhabditis elegans. Mol. Biol. Cell. 21:2285–2296. 10.1091/mbc.E09-11-0947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox E.A., and Hardin J.. 2004. Sticky worms: adhesion complexes in C. elegans. J. Cell Sci. 117:1885–1897. 10.1242/jcs.01176 [DOI] [PubMed] [Google Scholar]
- Dai P., Xiong W.C., and Mei L.. 2006. Erbin inhibits RAF activation by disrupting the sur-8-Ras-Raf complex. J. Biol. Chem. 281:927–933. 10.1074/jbc.M507360200 [DOI] [PubMed] [Google Scholar]
- de Vreede G., Schoenfeld J.D., Windler S.L., Morrison H., Lu H., and Bilder D.. 2014. The Scribble module regulates retromer-dependent endocytic trafficking during epithelial polarization. Development. 141:2796–2802. 10.1242/dev.105403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton S., and Martin-Belmonte F.. 2014. Cargo sorting in the endocytic pathway: a key regulator of cell polarity and tissue dynamics. Cold Spring Harb. Perspect. Biol. 6:a016899 10.1101/cshperspect.a016899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebine K., Inoue T., Ito J., Ito E., Uemura T., Goh T., Abe H., Sato K., Nakano A., and Ueda T.. 2014. Plant vacuolar trafficking occurs through distinctly regulated pathways. Curr. Biol. 24:1375–1382. 10.1016/j.cub.2014.05.004 [DOI] [PubMed] [Google Scholar]
- Fölsch H., Mattila P.E., and Weisz O.A.. 2009. Taking the scenic route: biosynthetic traffic to the plasma membrane in polarized epithelial cells. Traffic. 10:972–981. 10.1111/j.1600-0854.2009.00927.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrard S.M., Capaldo C.T., Gao L., Rosen M.K., Macara I.G., and Tomchick D.R.. 2003. Structure of Cdc42 in a complex with the GTPase-binding domain of the cell polarity protein, Par6. EMBO J. 22:1125–1133. 10.1093/emboj/cdg110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiou M., Marinari E., Burden J., and Baum B.. 2008. Cdc42, Par6, and aPKC regulate Arp2/3-mediated endocytosis to control local adherens junction stability. Curr. Biol. 18:1631–1638. 10.1016/j.cub.2008.09.029 [DOI] [PubMed] [Google Scholar]
- Gleason R.J., Akintobi A.M., Grant B.D., and Padgett R.W.. 2014. BMP signaling requires retromer-dependent recycling of the type I receptor. Proc. Natl. Acad. Sci. USA. 111:2578–2583. 10.1073/pnas.1319947111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant B.D., and Donaldson J.G.. 2009. Pathways and mechanisms of endocytic recycling. Nat. Rev. Mol. Cell Biol. 10:597–608. 10.1038/nrm2755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grindstaff K.K., Yeaman C., Anandasabapathy N., Hsu S.C., Rodriguez-Boulan E., Scheller R.H., and Nelson W.J.. 1998. Sec6/8 complex is recruited to cell-cell contacts and specifies transport vesicle delivery to the basal-lateral membrane in epithelial cells. Cell. 93:731–740. 10.1016/S0092-8674(00)81435-X [DOI] [PubMed] [Google Scholar]
- Gu M., Liu Q., Watanabe S., Sun L., Hollopeter G., Grant B.D., and Jorgensen E.M.. 2013. AP2 hemicomplexes contribute independently to synaptic vesicle endocytosis. eLife. 2:e00190 10.7554/eLife.00190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilherme A., Soriano N.A., Furcinitti P.S., and Czech M.P.. 2004. Role of EHD1 and EHBP1 in perinuclear sorting and insulin-regulated GLUT4 recycling in 3T3-L1 adipocytes. J. Biol. Chem. 279:40062–40075. 10.1074/jbc.M401918200 [DOI] [PubMed] [Google Scholar]
- Hardie R.C., Liu C.H., Randall A.S., and Sengupta S.. 2015. In vivo tracking of phosphoinositides in Drosophila photoreceptors. J. Cell Sci. 128:4328–4340. 10.1242/jcs.180364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutagalung A.H., and Novick P.J.. 2011. Role of Rab GTPases in membrane traffic and cell physiology. Physiol. Rev. 91:119–149. 10.1152/physrev.00059.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson A., Driessens M., and Aspenström P.. 2000. The mammalian homologue of the Caenorhabditis elegans polarity protein PAR-6 is a binding partner for the Rho GTPases Cdc42 and Rac1. J. Cell Sci. 113:3267–3275. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Nikolova L.S., Schjelderup A., and Metzstein M.M.. 2014. Exocyst-mediated membrane trafficking is required for branch outgrowth in Drosophila tracheal terminal cells. Dev. Biol. 390:41–50. 10.1016/j.ydbio.2014.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath R.S., and Ahringer J.. 2003. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 30:313–321. 10.1016/S1046-2023(03)00050-1 [DOI] [PubMed] [Google Scholar]
- Kinchen J.M., and Ravichandran K.S.. 2010. Identification of two evolutionarily conserved genes regulating processing of engulfed apoptotic cells. Nature. 464:778–782. 10.1038/nature08853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolch W. 2003. Erbin: sorting out ErbB2 receptors or giving Ras a break? Sci. STKE. 2003:pe37. [DOI] [PubMed] [Google Scholar]
- Kouranti I., Sachse M., Arouche N., Goud B., and Echard A.. 2006. Rab35 regulates an endocytic recycling pathway essential for the terminal steps of cytokinesis. Curr. Biol. 16:1719–1725. 10.1016/j.cub.2006.07.020 [DOI] [PubMed] [Google Scholar]
- Legouis R., Gansmuller A., Sookhareea S., Bosher J.M., Baillie D.L., and Labouesse M.. 2000. LET-413 is a basolateral protein required for the assembly of adherens junctions in Caenorhabditis elegans. Nat. Cell Biol. 2:415–422. 10.1038/35017046 [DOI] [PubMed] [Google Scholar]
- Legouis R., Jaulin-Bastard F., Schott S., Navarro C., Borg J.P., and Labouesse M.. 2003. Basolateral targeting by leucine-rich repeat domains in epithelial cells. EMBO Rep. 4:1096–1102. 10.1038/sj.embor.7400006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung B., Hermann G.J., and Priess J.R.. 1999. Organogenesis of the Caenorhabditis elegans intestine. Dev. Biol. 216:114–134. 10.1006/dbio.1999.9471 [DOI] [PubMed] [Google Scholar]
- Li W., Zou W., Zhao D., Yan J., Zhu Z., Lu J., and Wang X.. 2009. C. elegans Rab GTPase activating protein TBC-2 promotes cell corpse degradation by regulating the small GTPase RAB-5. Development. 136:2445–2455. 10.1242/dev.035949 [DOI] [PubMed] [Google Scholar]
- Li W., Yi P., and Ou G.. 2015. Somatic CRISPR-Cas9-induced mutations reveal roles of embryonically essential dynein chains in Caenorhabditis elegans cilia. J. Cell Biol. 208:683–692. 10.1083/jcb.201411041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D., Edwards A.S., Fawcett J.P., Mbamalu G., Scott J.D., and Pawson T.. 2000. A mammalian PAR-3-PAR-6 complex implicated in Cdc42/Rac1 and aPKC signalling and cell polarity. Nat. Cell Biol. 2:540–547. 10.1038/35019592 [DOI] [PubMed] [Google Scholar]
- Liu O., and Grant B.D.. 2015. Basolateral Endocytic Recycling Requires RAB-10 and AMPH-1 Mediated Recruitment of RAB-5 GAP TBC-2 to Endosomes. PLoS Genet. 11:e1005514 10.1371/journal.pgen.1005514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Xu X.H., Chen Q., Wang T., Deng C.Y., Song B.L., Du J.L., and Luo Z.G.. 2013. Myosin Vb controls biogenesis of post-Golgi Rab10 carriers during axon development. Nat. Commun. 4:2005. [DOI] [PubMed] [Google Scholar]
- Lohia M., Qin Y., and Macara I.G.. 2012. The Scribble polarity protein stabilizes E-cadherin/p120-catenin binding and blocks retrieval of E-cadherin to the Golgi. PLoS One. 7:e51130 10.1371/journal.pone.0051130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxfield F.R., and McGraw T.E.. 2004. Endocytic recycling. Nat. Rev. Mol. Cell Biol. 5:121–132. 10.1038/nrm1315 [DOI] [PubMed] [Google Scholar]
- Montcouquiol M., Rachel R.A., Lanford P.J., Copeland N.G., Jenkins N.A., and Kelley M.W.. 2003. Identification of Vangl2 and Scrb1 as planar polarity genes in mammals. Nature. 423:173–177. 10.1038/nature01618 [DOI] [PubMed] [Google Scholar]
- Murray D.H., Jahnel M., Lauer J., Avellaneda M.J., Brouilly N., Cezanne A., Morales-Navarrete H., Perini E.D., Ferguson C., Lupas A.N., et al. 2016. An endosomal tether undergoes an entropic collapse to bring vesicles together. Nature. 537:107–111. 10.1038/nature19326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen E., Christoforidis S., Uttenweiler-Joseph S., Miaczynska M., Dewitte F., Wilm M., Hoflack B., and Zerial M.. 2000. Rabenosyn-5, a novel Rab5 effector, is complexed with hVPS45 and recruited to endosomes through a FYVE finger domain. J. Cell Biol. 151:601–612. 10.1083/jcb.151.3.601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordmann M., Cabrera M., Perz A., Bröcker C., Ostrowicz C., Engelbrecht-Vandré S., and Ungermann C.. 2010. The Mon1-Ccz1 complex is the GEF of the late endosomal Rab7 homolog Ypt7. Curr. Biol. 20:1654–1659. 10.1016/j.cub.2010.08.002 [DOI] [PubMed] [Google Scholar]
- Piguel N.H., Fievre S., Blanc J.M., Carta M., Moreau M.M., Moutin E., Pinheiro V.L., Medina C., Ezan J., Lasvaux L., et al. 2014. Scribble1/AP2 complex coordinates NMDA receptor endocytic recycling. Cell Reports. 9:712–727. 10.1016/j.celrep.2014.09.017 [DOI] [PubMed] [Google Scholar]
- Pilipiuk J., Lefebvre C., Wiesenfahrt T., Legouis R., and Bossinger O.. 2009. Increased IP3/Ca2+ signaling compensates depletion of LET-413/DLG-1 in C. elegans epithelial junction assembly. Dev. Biol. 327:34–47. 10.1016/j.ydbio.2008.11.025 [DOI] [PubMed] [Google Scholar]
- Poteryaev D., Datta S., Ackema K., Zerial M., and Spang A.. 2010. Identification of the switch in early-to-late endosome transition. Cell. 141:497–508. 10.1016/j.cell.2010.03.011 [DOI] [PubMed] [Google Scholar]
- Pujol N., Bonnerot C., Ewbank J.J., Kohara Y., and Thierry-Mieg D.. 2001. The Caenorhabditis elegans unc-32 gene encodes alternative forms of a vacuolar ATPase a subunit. J. Biol. Chem. 276:11913–11921. 10.1074/jbc.M009451200 [DOI] [PubMed] [Google Scholar]
- Quinn K.V., Behe P., and Tinker A.. 2008. Monitoring changes in membrane phosphatidylinositol 4,5-bisphosphate in living cells using a domain from the transcription factor tubby. J. Physiol. 586:2855–2871. 10.1113/jphysiol.2008.153791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishna H., and Donaldson J.G.. 1997. ADP-ribosylation factor 6 regulates a novel plasma membrane recycling pathway. J. Cell Biol. 139:49–61. 10.1083/jcb.139.1.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rink J., Ghigo E., Kalaidzidis Y., and Zerial M.. 2005. Rab conversion as a mechanism of progression from early to late endosomes. Cell. 122:735–749. 10.1016/j.cell.2005.06.043 [DOI] [PubMed] [Google Scholar]
- Sakaguchi A., Sato M., Sato K., Gengyo-Ando K., Yorimitsu T., Nakai J., Hara T., Sato K., and Sato K.. 2015. REI-1 Is a Guanine Nucleotide Exchange Factor Regulating RAB-11 Localization and Function in C. elegans Embryos. Dev. Cell. 35:211–221. 10.1016/j.devcel.2015.09.013 [DOI] [PubMed] [Google Scholar]
- Sano H., Peck G.R., Kettenbach A.N., Gerber S.A., and Lienhard G.E.. 2011. Insulin-stimulated GLUT4 protein translocation in adipocytes requires the Rab10 guanine nucleotide exchange factor Dennd4C. J. Biol. Chem. 286:16541–16545. 10.1074/jbc.C111.228908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasidharan N., Sumakovic M., Hannemann M., Hegermann J., Liewald J.F., Olendrowitz C., Koenig S., Grant B.D., Rizzoli S.O., Gottschalk A., and Eimer S.. 2012. RAB-5 and RAB-10 cooperate to regulate neuropeptide release in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 109:18944–18949. 10.1073/pnas.1203306109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M., Sato K., Liou W., Pant S., Harada A., and Grant B.D.. 2008. Regulation of endocytic recycling by C. elegans Rab35 and its regulator RME-4, a coated-pit protein. EMBO J. 27:1183–1196. 10.1038/emboj.2008.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Mushiake S., Kato Y., Sato K., Sato M., Takeda N., Ozono K., Miki K., Kubo Y., Tsuji A., et al. 2007. The Rab8 GTPase regulates apical protein localization in intestinal cells. Nature. 448:366–369. 10.1038/nature05929 [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segbert C., Johnson K., Theres C., van Fürden D., and Bossinger O.. 2004. Molecular and functional analysis of apical junction formation in the gut epithelium of Caenorhabditis elegans. Dev. Biol. 266:17–26. 10.1016/j.ydbio.2003.10.019 [DOI] [PubMed] [Google Scholar]
- Sflomos G., Kostaras E., Panopoulou E., Pappas N., Kyrkou A., Politou A.S., Fotsis T., and Murphy C.. 2011. ERBIN is a new SARA-interacting protein: competition between SARA and SMAD2 and SMAD3 for binding to ERBIN. J. Cell Sci. 124:3209–3222. 10.1242/jcs.062307 [DOI] [PubMed] [Google Scholar]
- Shen Z., Zhang X., Chai Y., Zhu Z., Yi P., Feng G., Li W., and Ou G.. 2014. Conditional knockouts generated by engineered CRISPR-Cas9 endonuclease reveal the roles of coronin in C. elegans neural development. Dev. Cell. 30:625–636. 10.1016/j.devcel.2014.07.017 [DOI] [PubMed] [Google Scholar]
- Shi A., and Grant B.D.. 2013. Interactions between Rab and Arf GTPases regulate endosomal phosphatidylinositol-4,5-bisphosphate during endocytic recycling. Small GTPases. 4:106–109. 10.4161/sgtp.23477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi A., Sun L., Banerjee R., Tobin M., Zhang Y., and Grant B.D.. 2009. Regulation of endosomal clathrin and retromer-mediated endosome to Golgi retrograde transport by the J-domain protein RME-8. EMBO J. 28:3290–3302. 10.1038/emboj.2009.272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi A., Chen C.C., Banerjee R., Glodowski D., Audhya A., Rongo C., and Grant B.D.. 2010. EHBP-1 functions with RAB-10 during endocytic recycling in Caenorhabditis elegans. Mol. Biol. Cell. 21:2930–2943. 10.1091/mbc.E10-02-0149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi A., Liu O., Koenig S., Banerjee R., Chen C.C., Eimer S., and Grant B.D.. 2012. RAB-10-GTPase-mediated regulation of endosomal phosphatidylinositol-4,5-bisphosphate. Proc. Natl. Acad. Sci. USA. 109:E2306–E2315. 10.1073/pnas.1205278109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen A., Gaullier J.M., D’Arrigo A., and Stenmark H.. 1999. The Rab5 effector EEA1 interacts directly with syntaxin-6. J. Biol. Chem. 274:28857–28860. 10.1074/jbc.274.41.28857 [DOI] [PubMed] [Google Scholar]
- Sun L., Liu O., Desai J., Karbassi F., Sylvain M.A., Shi A., Zhou Z., Rocheleau C.E., and Grant B.D.. 2012. CED-10/Rac1 regulates endocytic recycling through the RAB-5 GAP TBC-2. PLoS Genet. 8:e1002785 10.1371/journal.pgen.1002785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y., Shen C., Luo S., Traoré W., Marchetto S., Santoni M.J., Xu L., Wu B., Shi C., Mei J., et al. 2014. Role of Erbin in ErbB2-dependent breast tumor growth. Proc. Natl. Acad. Sci. USA. 111:E4429–E4438. 10.1073/pnas.1407139111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L., and Fire A.. 1998. Specific interference by ingested dsRNA. Nature. 395:854 10.1038/27579 [DOI] [PubMed] [Google Scholar]
- Waaijers S., Muñoz J., Berends C., Ramalho J.J., Goerdayal S.S., Low T.Y., Zoumaro-Djayoon A.D., Hoffmann M., Koorman T., Tas R.P., et al. 2016. A tissue-specific protein purification approach in Caenorhabditis elegans identifies novel interaction partners of DLG-1/Discs large. BMC Biol. 14:66 10.1186/s12915-016-0286-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Liu H., Wang Y., Liu O., Zhang J., Gleason A., Yang Z., Wang H., Shi A., and Grant B.D.. 2016. RAB-10 Promotes EHBP-1 Bridging of Filamentous Actin and Tubular Recycling Endosomes. PLoS Genet. 12:e1006093 10.1371/journal.pgen.1006093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Liu Y., Xu X.H., Deng C.Y., Wu K.Y., Zhu J., Fu X.Q., He M., and Luo Z.G.. 2011. Lgl1 activation of rab10 promotes axonal membrane trafficking underlying neuronal polarization. Dev. Cell. 21:431–444. 10.1016/j.devcel.2011.07.007 [DOI] [PubMed] [Google Scholar]
- Watson L.J., Rossi G., and Brennwald P.. 2014. Quantitative analysis of membrane trafficking in regulation of Cdc42 polarity. Traffic. 15:1330–1343. 10.1111/tra.12211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West J.J., and Harris T.J.. 2016. Cadherin trafficking for tissue morphogenesis: control and consequences. Traffic. 17:1233–1243. 10.1111/tra.12407 [DOI] [PubMed] [Google Scholar]
- Wirtz-Peitz F., and Knoblich J.A.. 2006. Lethal giant larvae take on a life of their own. Trends Cell Biol. 16:234–241. 10.1016/j.tcb.2006.03.006 [DOI] [PubMed] [Google Scholar]
- Xu J., and McPherson P.S.. 2017. Regulation of DENND3, the exchange factor for the small GTPase Rab12 through an intramolecular interaction. J. Biol. Chem. 292:7274–7282. 10.1074/jbc.M116.772434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka T., and Ohno S.. 2008. Role of Lgl/Dlg/Scribble in the regulation of epithelial junction, polarity and growth. Front. Biosci. 13:6693–6707. 10.2741/3182 [DOI] [PubMed] [Google Scholar]
- Yeaman C., Grindstaff K.K., and Nelson W.J.. 1999. New perspectives on mechanisms involved in generating epithelial cell polarity. Physiol. Rev. 79:73–98. [DOI] [PubMed] [Google Scholar]
- Yoshimura S., Gerondopoulos A., Linford A., Rigden D.J., and Barr F.A.. 2010. Family-wide characterization of the DENN domain Rab GDP-GTP exchange factors. J. Cell Biol. 191:367–381. 10.1083/jcb.201008051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Zeng J., Ouyang C., Luo Q., Yu M., Yang Z., Wang H., Shen K., and Shi A.. 2016. A novel bipartite UNC-101/AP-1 μ1 binding signal mediates KVS-4/Kv2.1 somatodendritic distribution in Caenorhabditis elegans. FEBS Lett. 590:76–92. 10.1002/1873-3468.12043 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.