The TRAPP complexes are nucleotide exchange factors that activate Rab GTPases, and four different versions of TRAPP have been reported. Thomas et al. show that only two versions of TRAPP are detectable in normal cells and demonstrate that the TRAPPIII complex regulates Golgi trafficking in addition to its established role in autophagy.
Abstract
Rab GTPases serve as molecular switches to regulate eukaryotic membrane trafficking pathways. The transport protein particle (TRAPP) complexes activate Rab GTPases by catalyzing GDP/GTP nucleotide exchange. In mammalian cells, there are two distinct TRAPP complexes, yet in budding yeast, four distinct TRAPP complexes have been reported. The apparent differences between the compositions of yeast and mammalian TRAPP complexes have prevented a clear understanding of the specific functions of TRAPP complexes in all cell types. In this study, we demonstrate that akin to mammalian cells, wild-type yeast possess only two TRAPP complexes, TRAPPII and TRAPPIII. We find that TRAPPIII plays a major role in regulating Rab activation and trafficking at the Golgi in addition to its established role in autophagy. These disparate pathways share a common regulatory GTPase Ypt1 (Rab1) that is activated by TRAPPIII. Our findings lead to a simple yet comprehensive model for TRAPPIII function in both normal and starved eukaryotic cells.
Introduction
In eukaryotic cells, virtually every step of membrane transport is mediated by Rab GTPases. Rabs function as molecular switches, cycling between an inactive GDP-bound state and an active GTP-bound state (Barr, 2009; Stenmark, 2009). Rabs are activated by guanine nucleotide exchange factors (GEFs), which catalyze GDP/GTP nucleotide exchange. Activated Rabs anchor to organelle membranes, where they recruit downstream effectors that facilitate vesicular transport. Though Rabs were originally thought to be restricted to specific pathways, it has become increasingly apparent that individual Rabs can coordinate multiple transport pathways by recruiting effectors to different organelles (Lipatova and Segev, 2014).
In budding yeast, the Rab GTPase Ypt1 coordinates several distinct trafficking events including the tethering of COPII vesicles during ER–Golgi transport and membrane expansion during autophagosome formation (Jedd et al., 1995; Lynch-Day et al., 2010). Additional functions have been proposed for Ypt1 at the late Golgi, including endosome–Golgi transport and vesicle formation (Sclafani et al., 2010; McDonold and Fromme, 2014), yet a role for Ypt1 at the late Golgi remains controversial (Lipatova et al., 2013; Kim et al., 2016a). Similarly, the mammalian homologue of Ypt1, Rab1, has conserved functions in ER–Golgi transport, intra-Golgi trafficking, and autophagosome formation (Plutner et al., 1991; Tisdale et al., 1992; Zoppino et al., 2010).
Ypt1 activity is controlled by the transport protein particle (TRAPP) family of multisubunit complexes (Barrowman et al., 2010). TRAPP was originally identified in budding yeast as a complex that copurified with the subunit Bet3 (Sacher et al., 1998). The complex was subsequently delineated into two related complexes, TRAPPI and TRAPPII, with distinct roles in ER–Golgi and late Golgi trafficking, respectively (Sacher et al., 2001). A third autophagy-specific complex, TRAPPIII, was later proposed on the basis that the subunit Trs85 is important for autophagy but apparently dispensable for trafficking in healthy cells (Lynch-Day et al., 2010). More recently, a fourth TRAPP complex was proposed to exist based on synthetic genetic interactions between the genes encoding the Trs85 and Trs33 subunits (Lipatova et al., 2016). All four TRAPP complexes have been implicated as GEFs for Ypt1 (Wang et al., 2000; Lynch-Day et al., 2010), although it remains controversial as to whether TRAPPII activates Ypt1 in vivo (Morozova et al., 2006; Zou et al., 2012; Lipatova et al., 2013; Thomas and Fromme, 2016).
Homologues of all budding yeast TRAPP subunits have been identified in metazoans and assigned to only two distinct TRAPP complexes, TRAPPII and TRAPPIII (Yamasaki et al., 2009; Bassik et al., 2013; Wang et al., 2013; Lamb et al., 2016). Mammalian TRAPPII has a conserved role in late Golgi trafficking, and TRAPPIII has been implicated in both ER–Golgi transport and autophagy (Barrowman et al., 2010; Behrends et al., 2010; Zoppino et al., 2010; Scrivens et al., 2011; Bassik et al., 2013; Brunet and Sacher, 2014; Kim et al., 2016b; Lamb et al., 2016). Differences between the number and subunit composition of TRAPP complexes in yeast and mammalian cells has prevented a clear determination of their distinct functions.
Consistent with a lack of TRAPPI in higher eukaryotes, several studies have suggested that the TRAPPI complex in yeast may be a purification artefact (Brunet et al., 2012, 2013). In fractionation experiments isolating individual TRAPP complexes, the amount of TRAPPI is often very low (Menon et al., 2006; Lynch-Day et al., 2010; Choi et al., 2011). Furthermore, the abundance of TRAPPI increases under high-salt conditions or in TRAPP complex mutants (Montpetit and Conibear, 2009; Choi et al., 2011; Brunet et al., 2012, 2013), indicating that TRAPPI may be a product of destabilized TRAPPII and TRAPPIII in vitro.
In this study, we use two different methods to report that TRAPPII and TRAPPIII are the only detectable TRAPP complexes in WT yeast cells. We show that TRAPPIII-catalyzed nucleotide exchange is an order of magnitude faster than that of TRAPPI in physiological enzyme activity assays. Correspondingly, we find that Ypt1 activation and Golgi trafficking is significantly perturbed in TRAPPIII mutant cells. We propose that only two TRAPP complexes exist in most eukaryotic cells, TRAPPII and TRAPPIII, and that TRAPPIII performs the ER–Golgi trafficking functions previously assigned to TRAPPI.
Our findings support conserved roles for the two TRAPP complexes in both yeast and mammalian cells and indicate that TRAPPIII activates Ypt1/Rab1 to regulate two distinct pathways: normal growth and autophagy.
Results
The TRAPPI complex is virtually absent in WT yeast cells
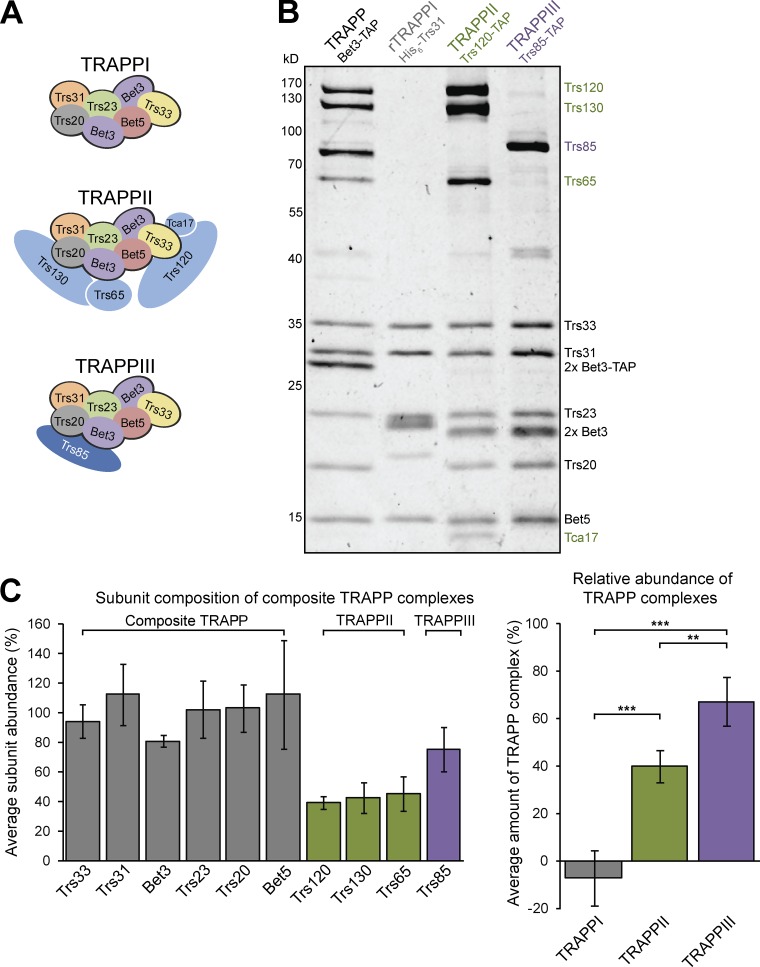
Before 2010, two TRAPP complexes were reported to exist in yeast: TRAPPI contained the subunit Trs85 and facilitated ER–Golgi traffic, and TRAPPII coordinated transport events at the late Golgi (Sacher et al., 2001). The existence of a third TRAPP complex was subsequently suggested based on fractionation of TRAPP complexes purified from yeast, and Trs85 was assigned to an autophagy-specific complex, TRAPPIII, distinct from the TRAPPI complex containing only the six core subunits (Fig. 1 A; Lynch-Day et al., 2010). However, the key fractionation experiments were performed using immunoblots of individual subunits, which did not allow for the quantification of the relative stoichiometry of each subunit in all TRAPP complexes. Furthermore, the amount of TRAPPI in all previous studies appeared to be relatively low, and the abundance of TRAPPI has been shown to increase with high salt or in complex-destabilizing mutants (Montpetit and Conibear, 2009; Lynch-Day et al., 2010; Choi et al., 2011; Brunet et al., 2012, 2013), suggesting that TRAPPI may be an artefact of purification.
Figure 1.
TRAPPII and TRAPPIII are the only TRAPP complexes detected in yeast cells. (A) Subunit composition of TRAPP complexes reported in yeast. (B) Composite TRAPP was purified from log-phase yeast using the shared core subunit Bet3. Distinct TRAPP complexes were purified from yeast or E. coli using the indicated affinity tag. (C) Quantification of the relative abundance of individual TRAPP subunits (left) and TRAPPI, TRAPPII, and TRAPPIII complexes (right) in yeast. Composite TRAPP was purified via Bet3-TAP, and molarities of each complex were determined from complex-specific subunits. Error bars represent 95% confidence intervals (CIs). n = 4 independent purifications. **, P < 0.01; ***, P < 0.001 for one-way ANOVA with Tukey’s test for multiple comparisons.
To obtain a stoichiometric assessment of the TRAPP complexes in cells, we purified “composite” TRAPP from log-phase budding yeast using a tandem affinity purification (TAP)-tag on the core subunit Bet3 and visualized all subunits using colloidal Coomassie (Fig. 1 B). Our buffers contained 300 mM NaCl, the lowest salt concentration that allows for the efficient release of TRAPP complexes from membranes (Choi et al., 2011). For comparison, we purified individual TRAPPII and TRAPPIII complexes using TAP tags on the complex-specific subunits Trs120 and Trs85, respectively. As TRAPPI lacks complex-specific subunits, we purified recombinant TRAPPI (rTRAPPI) by coexpressing the six core subunits in Escherichia coli. Using the colloidal Coomassie staining intensity, we measured the amount of each subunit in the composite TRAPP purification, and abundance of TRAPPII and TRAPPIII were determined from the mean molarity of the complex-specific subunits Trs120, Trs130, and Trs65 (TRAPPII) or Trs85 (TRAPPIII). To determine the relative amount of TRAPPI, we subtracted the molarity of TRAPPII and TRAPPIII from that of the shared core subunits. TRAPPIII accounts for approximately two thirds of composite TRAPP, and TRAPPII makes up the remaining one third (Fig. 1 C). We observed no excess core subunits, indicating a lack of TRAPPI and TRAPPIV in WT yeast cells.
The TRAPP core subunits colocalize completely with TRAPPII- and TRAPPIII-specific subunits
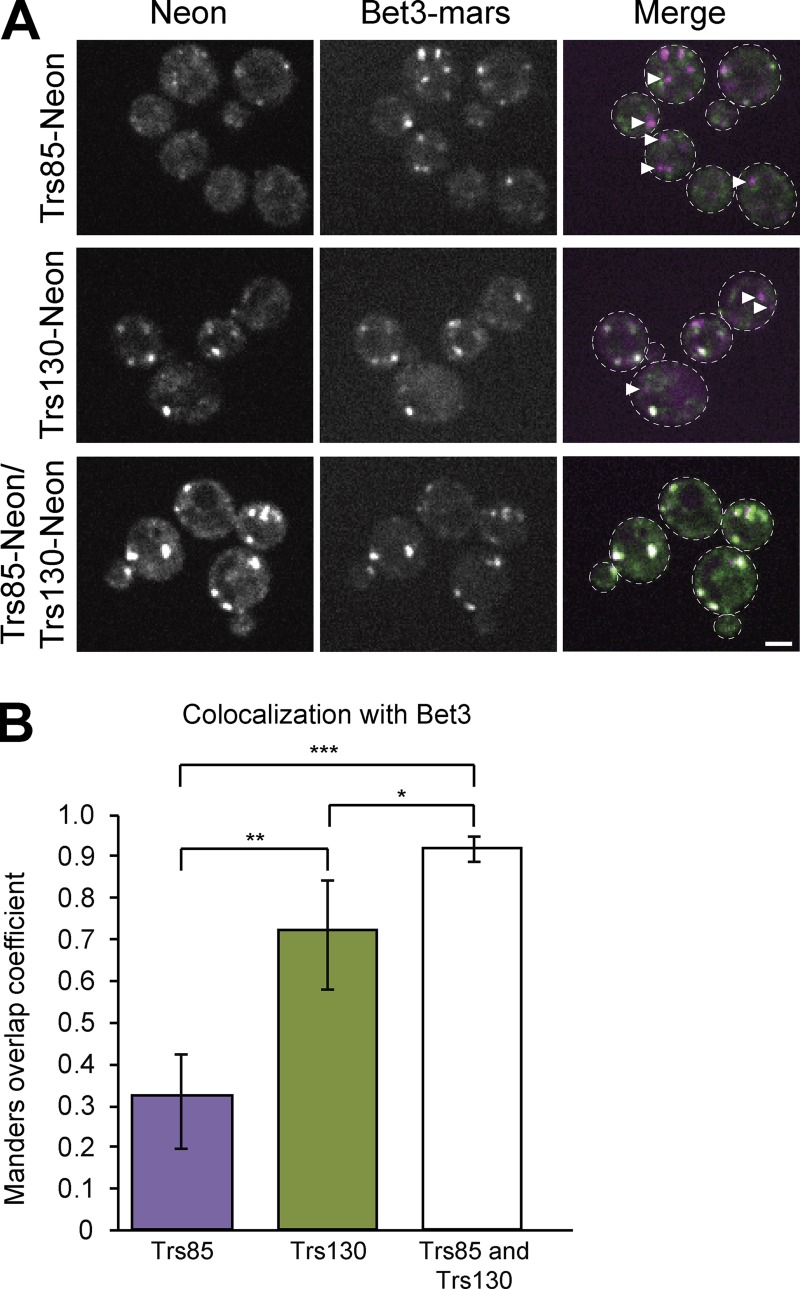
The in vitro analysis of TRAPP subunit ratios presented in the previous section provides strong evidence that yeast contain only two TRAPP complexes. However, it is possible that the relative amounts of each complex were altered during purification. To complement this approach, the distribution of TRAPP subunits was monitored in vivo using fluorescence microscopy of live cells. By endogenously tagging the core subunit Bet3 with an RFP, all TRAPP complexes can be visualized simultaneously. Concurrent tagging of endogenous Trs85 (TRAPPIII-specific) and Trs130 (TRAPPII-specific) with a GFP provided opportunity to monitor each complex individually or together (Fig. 2 A). We reasoned that combining these fusion proteins in a stepwise fashion would allow us to elucidate the balance of distinct TRAPP complexes in vivo.
Figure 2.
The TRAPPI core colocalizes completely with TRAPPII- and TRAPPIII-specific subunits. (A) Single focal planes are shown of live-cell fluorescence microscopy images under normal growth conditions. Cells are expressing endogenously tagged Bet3-3×mRFPmars with Trs85-mNeonGreen (top), Trs130-mNeonGreen (middle), or both (bottom). (B) The amount of overlap between Bet3 and Trs85, Trs130, or both was quantified using the Manders overlap coefficient. Images were cropped to contain between four and nine cells, and then five images for each strain were analyzed to create the mean Manders overlap coefficient. Error bars represent 95% CIs. *, P < 0.05; **, P < 0.01; ***, P < 0.001 for unpaired two-tailed t test with Welch’s correction. Arrowheads designate Bet3 puncta that do not overlap with either Trs85 or Trs130. Dashed lines represent cell boundaries. Bar, 2 µm.
We found that Trs85 and Bet3 colocalize partially but not completely (Fig. 2 A), indicated by a mix of white puncta (corresponding to overlap of the two fluorophores) and magenta puncta (indicating a complex containing Bet3 but not Trs85—likely TRAPPII). Similarly, Trs130 and Bet3 colocalized well, but also not completely. In contrast, the cells containing Trs85-mNeonGreen, Trs130-mNeonGreen, and Bet3-mRFPmars only displayed overlapping white puncta (Fig. 2 A). The Manders Overlap Coefficient was used for quantification of the extent of colocalization and indicated that ∼30% of Bet3 puncta colocalized with Trs85, whereas almost 70% of Bet3 puncta overlapped with Trs130 (Fig. 2 B). Nearly all Bet3 fluorescence overlapped with either Trs85 or Trs130, suggesting that Bet3 is always associated with either TRAPPII- or TRAPPIII-specific subunits. This indicates that distinct TRAPPI or TRAPPIV complexes are not detectable at punctate structures in vivo. Collectively with the results of the biochemical purifications, these data indicate that only two TRAPP complexes, TRAPPII and TRAPPIII, exist in cells.
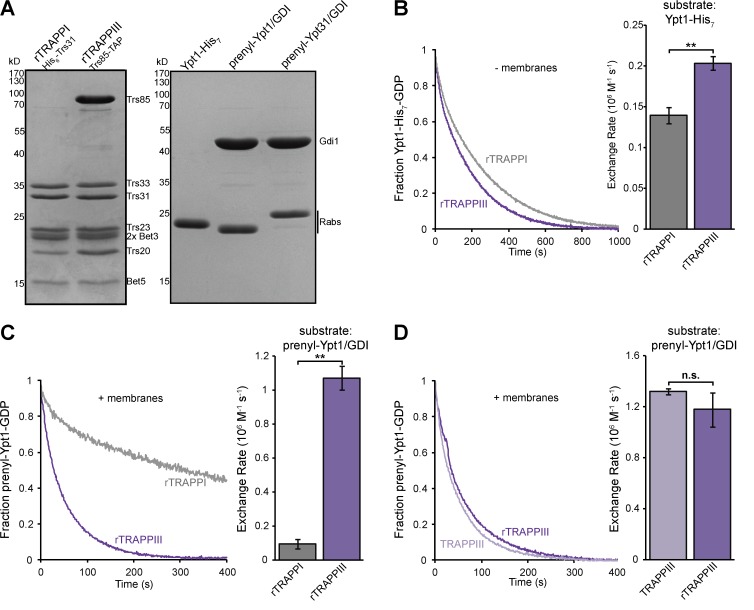
TRAPPIII activates Ypt1 significantly faster than TRAPPI
Previous studies have observed similar nucleotide exchange activity for TRAPPI, TRAPPII, and TRAPPIII, consistent with TRAPPI functioning as a physiological Ypt1 GEF (Sacher et al., 2001; Cai et al., 2008; Lynch-Day et al., 2010). These studies all measured TRAPP-mediated nucleotide exchange on Ypt1 in solution. In agreement with these studies, we found that rTRAPPI activated soluble Ypt1 nearly as fast as TRAPPIII (Fig. 3, A and B). In cells, however, Rab GTPase activation takes place at the membrane surface. We therefore aimed to more closely replicate the in vivo conditions by comparing TRAPPI and TRAPPIII activity with native prenylated Rab/GDP dissociation inhibitor (GDI) substrates in the presence of liposome membranes. With this more physiological assay (Thomas and Fromme, 2016), we found that TRAPPIII was an order of magnitude more active than TRAPPI (Figs. 3 C and S1 A). We also found that recombinant TRAPPIII (rTRAPPIII) purified from E. coli had nearly identical activity to endogenous TRAPPIII purified from yeast (Figs. 3 D and S1 B). Consistent with previous research (Wang et al., 2000), we found that TRAPPIII did not activate the Rab GTPases Ypt31, Ypt6, Sec4, or Vps21 (Fig. S1 C and not depicted), confirming that TRAPPIII is a specific Ypt1 GEF.
Figure 3.
TRAPPIII activates Ypt1 significantly faster than TRAPPI. (A, left) rTRAPPI and TRAPPIII were purified from E. coli using the indicated affinity tag. (A, right) His-tagged and prenylated Rab/GDI substrates used in this study. (B, left) Normalized representative traces showing activation of His-tagged Ypt1 by rTRAPPI and rTRAPPIII in the absence of membranes. (B, right) Rates of rTRAPP-mediated Ypt1 activation determined from the traces at left. Error bars represent 95% CIs. n ≥ 3 reactions. (C, left) Normalized representative traces showing activation of prenylated Ypt1 by rTRAPPI and rTRAPPIII in the presence of synthetic TGN liposomes. (C, right) Rates of rTRAPP-mediated Ypt1 activation from the traces at left. Error bars represent 95% CIs. n = 3 reactions. (D, left) Normalized representative traces showing activation of prenylated Ypt1 by endogenous and rTRAPPIII in the presence of TGN liposomes. (D, right) Rates of TRAPP-mediated Ypt1 activation from the traces at left. Error bars represent 95% CIs. n = 3 reactions. **, P < 0.01 for unpaired two-tailed t test with Welch’s correction.
We observed a fivefold increase in TRAPPIII activity toward the prenylated Ypt1/GDI substrate in the presence of membranes compared with nonprenylated Ypt1 activation in the absence of membranes (compare rates in Fig. 3, B and C). Membranes have been shown to play a key role in GEF-mediated GTPase activation (DiNitto et al., 2007; Richardson et al., 2012; Stalder and Antonny, 2013; Thomas and Fromme, 2016; Peurois et al., 2017), and our data indicate that this is also the case for TRAPPIII.
TRAPPIII plays a major role in Ypt1 activation in vivo
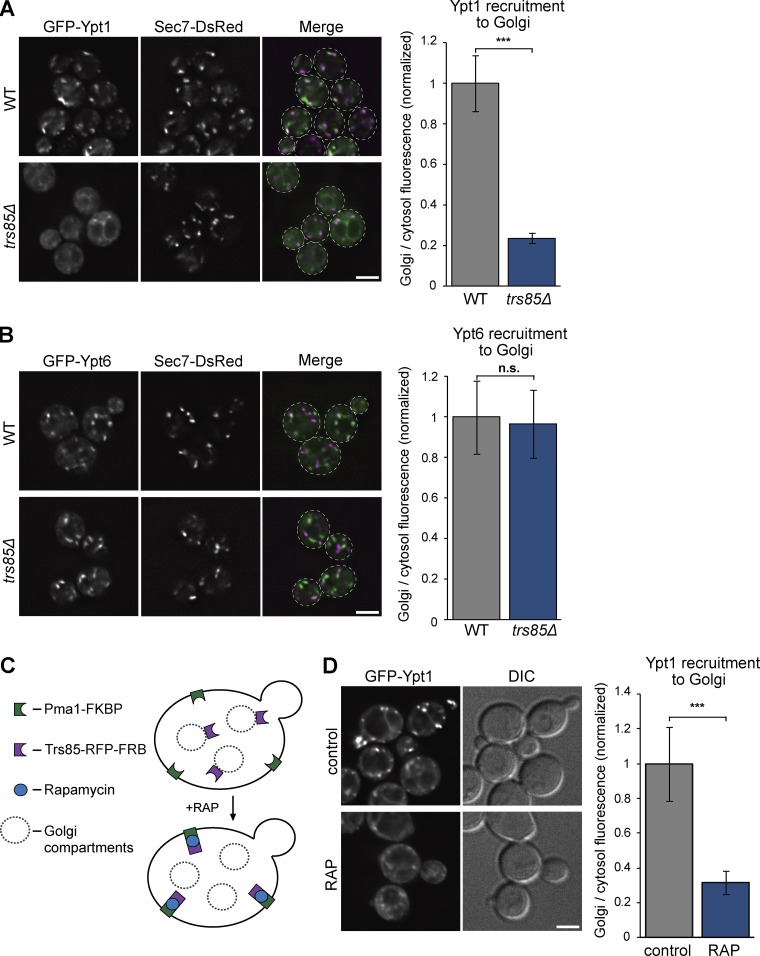
If the TRAPPI complex is a purification artefact and is not biologically relevant in vivo, this raises the question of the identity of the GEF that activates Ypt1 under normal vegetative (nonautophagic) conditions. Several lines of evidence from previous studies suggest that the Trs85-containing TRAPPIII complex activates Ypt1 to mediate ER–Golgi and medial/late Golgi transport. First, Trs85 was originally assigned to TRAPPI (Sacher et al., 2001). Second, multiple studies have shown that trs85Δ cells are impaired in both ER–Golgi trafficking and endocytic recycling (Sacher et al., 2001; Lynch-Day et al., 2010; Zou et al., 2012; Shirahama-Noda et al., 2013). Finally, mammalian TRAPPIII has been reported to play a role in both autophagy and early secretory pathway ricin susceptibility (Zoppino et al., 2010; Scrivens et al., 2011; Bassik et al., 2013; Lamb et al., 2016), raising the possibility that yeast TRAPPIII may activate Ypt1 to coordinate multiple trafficking pathways.
To test whether TRAPPIII is important for Ypt1 activation under normal growth conditions, we examined the localization of GFP-Ypt1 in trs85Δ mutant cells. We used the extent of Ypt1 localization to punctate compartments as a measurement of Rab activation. In WT cells, GFP-Ypt1 localized predominantly to Gea1- and Sec7-labeled early and medial/late Golgi compartments (Figs. 4 A and S2 A). In contrast, in trs85Δ mutant cells, Ypt1 was nearly completely cytosolic. In control experiments, the Rab GTPases Ypt6 and Ypt31, which are not in vitro substrates for TRAPPIII, remained localized to Golgi compartments in trs85Δ cells (Figs. 4 B and S2 B).
Figure 4.
TRAPPIII plays a major role in Ypt1 activation at the Golgi in vivo. (A, left) Localization of plasmid-borne GFP-Ypt1 relative to the late Golgi/TGN marker Sec7-6×DsRed in WT versus trs85Δ mutant cells grown at 30°C. (A, right) Line-trace quantification of Golgi and cytosolic Ypt1 in WT versus trs85Δ cells. Error bars represent 95% CIs. n ≥ 15 cells. (B, left) Localization of plasmid-borne GFP-Ypt6 relative to Sec7-6×DsRed in WT versus trs85Δ mutant cells. (B, right) Line-trace quantification of Golgi and cytosolic Ypt6 in WT versus trs85Δ cells. Error bars represent 95% CIs. n ≥ 15 cells. (C) Schematic depicting the anchor-away method used to conditionally deplete TRAPPIII activity in cells. (D, left) Localization of GFP-Ypt1 in untreated cells versus cells treated with rapamycin to relocalize Trs85 to the PM. (Right) Quantification of Golgi and cytosolic Ypt1 in untreated versus rapamycin-treated cells. Error bars represent 95% CIs. n ≥ 12 cells. ***, P < 0.001 for unpaired two-tailed t test with Welch’s correction. Dashed lines represent cell boundaries. Bars, 2 µm. DIC, differential interference contrast.
To control for the possibility of adaptation artefacts in trs85Δ mutant cells, we used the anchor-away system (Haruki et al., 2008) to trigger acute depletion of the TRAPPIII-specific subunit Trs85. Trs85 was tagged with mRFPmars-FRB in cells expressing the plasma membrane (PM) hook Pma1-2×FKBP (Fig. 4 C). After the addition of rapamycin, Trs85-mRFPmars-FRB was rapidly sequestered at the PM, but a corresponding relocalization of the core subunit Bet3-GFP was not observed (Fig. S2, C and D). Analysis of Bet3-GFP localization in this experiment was confounded by the significant proportion of Bet3 in the TRAPPII complex (Fig. 2); nevertheless, the nearly complete lack of colocalization of Bet3-GFP with the Trs85-mRFPmars-FRB puncta sequestered at the PM suggests that the anchor-away treatment dissociated Trs85 from the rest of the TRAPPIII complex. In comparison with untreated cells, Ypt1 was mislocalized to the cytoplasm after anchor-away of Trs85 (Fig. 4 D). In control experiments, Ypt6 activation was not affected by sequestration of Trs85 at the PM (Fig. S2 E). Collectively, these data indicate that in addition to its previously demonstrated role in autophagy, TRAPPIII is critical for efficient Ypt1 activation at the Golgi in healthy (nonstarved) cells.
TRAPPIII and the Rab–GTPase-activating protein (GAP) Gyp1 control the level of Ypt1 at the Golgi
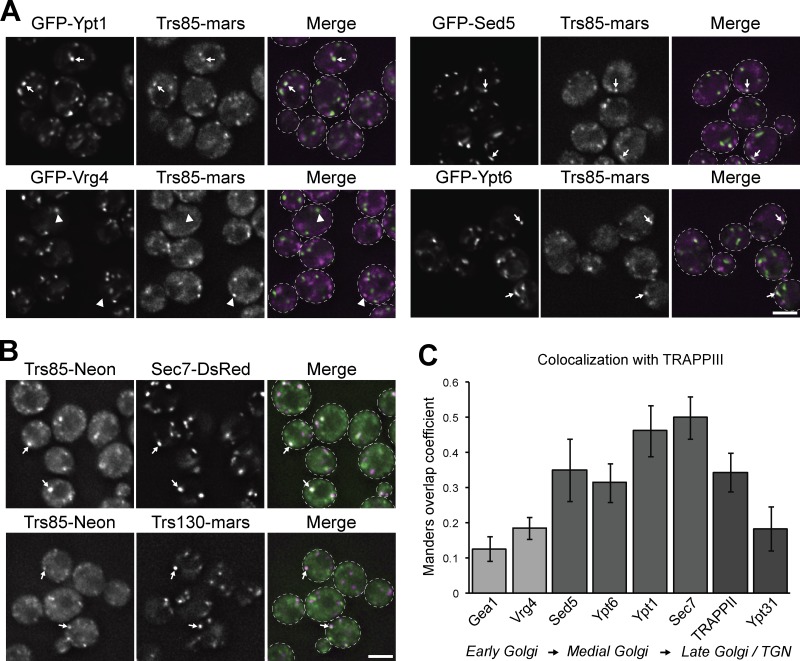
Previous studies have implicated Ypt1 in coordinating endosome–Golgi transport and vesicle formation (Sclafani et al., 2010; McDonold and Fromme, 2014), yet a role for Ypt1 at the late Golgi remains controversial (Lipatova et al., 2013; Kim et al., 2016a). We and others have observed that a significant fraction of Ypt1 colocalizes with the late Golgi/TGN marker Sec7 (Fig. 4 A; Sclafani et al., 2010; Kim et al., 2016a), prompting us to test whether TRAPPIII also localizes to the late Golgi to activate this pool of Ypt1. In agreement with TRAPPIII functioning as a Ypt1 GEF during normal growth, Trs85 colocalized well with Ypt1 at Golgi compartments in nonautophagic cells (Fig. 5, A and C). Trs85 showed limited overlap with the early Golgi proteins Vrg4 and Gea1 (Fig. 5, A and C; and Fig. S3 A), consistent with a previous study that did not observe TRAPP complexes at early Golgi compartments (Montpetit and Conibear, 2009). In contrast, Trs85 colocalized well with markers of the medial Golgi (Sed5 and Ypt6; Fig. 5, A and C) and late Golgi/TGN (Sec7 and Trs130; Fig. 5, B and C). Trs85 overlapped less with Ypt31/32 (Figs. 5 C and S3 A), one of the last proteins to appear at the TGN during secretory vesicle formation, indicating that TRAPPIII localizes to medial/late Golgi compartments and likely leaves the Golgi before terminal vesiculation.
Figure 5.
TRAPPIII localizes to medial/late Golgi compartments. (A) Localization of Trs85-3×mRFPmars relative to GFP-Ypt1, GFP-Vrg4 (early Golgi), GFP-Sed5 (early/medial Golgi), and GFP-Ypt6 (medial/late Golgi) in log-phase cells grown at 30°C. (B) Localization of Trs85-mNeonGreen relative to the late Golgi/TGN markers Sec7-6×DsRed and Trs130-3×mRFPmars. (C) Colocalization analysis of TRAPPIII (Trs85-3×mRFPmars or Trs85-mNeonGreen) with markers of the early, medial, and late Golgi and TGN. Values correspond to the mean overlap of each indicated protein with Trs85, quantified using the Manders overlap coefficient. Error bars represent 95% CIs. n ≥ 60 cells. White arrows and arrowheads denote colocalization, or lack thereof, of visualized proteins at Golgi compartments, respectively. Dashed lines represent cell boundaries. Bars, 2 µm.
Trs85 exhibited the highest degree of colocalization with Sec7, prompting us to test whether TRAPPIII localization is dependent on Sec7 activity. In contrast with the related TRAPPII complex, which is recruited to the Golgi by Sec7-activated Arf1 (Thomas and Fromme, 2016), association of Trs85 with membranes was not impaired after inhibition of Sec7 (Fig. S3 B) or in arf1Δ cells (Fig. S3 C). Instead, Trs85 accumulated at Golgi compartments after Sec7 inhibition, suggesting that the signal for TRAPPIII to dissociate from the Golgi lies downstream of Sec7.
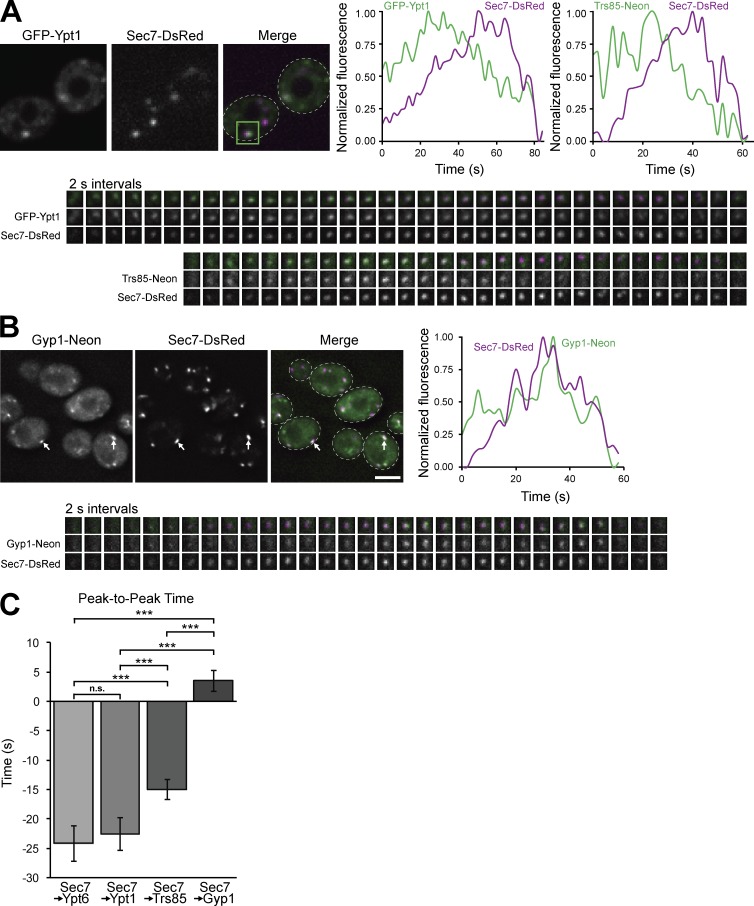
Previous studies have reported that Trs85 colocalizes with Sec7 at late Golgi compartments (Montpetit and Conibear, 2009; Shirahama-Noda et al., 2013), but proposed a role for TRAPPIII in tethering endocytic vesicles. To test whether a major role of TRAPPIII at the medial/late Golgi was activating Ypt1, we used time-lapse imaging to observe the dynamics of TRAPPIII and Ypt1 at Golgi compartments. Consistent with previous studies (Rivera-Molina and Novick, 2009; Suda et al., 2013; McDonold and Fromme, 2014; Kim et al., 2016a; Thomas and Fromme, 2016), we found that Ypt1 recruitment peaked ∼20 s upstream of Sec7, with Ypt6 peaking at a similar time (Fig. 6, A and C; and Fig. S4 A). We observed that Trs85 arrived at the medial/late Golgi before the peak of Ypt1 (Fig. 6, A and C), providing an explanation for the increasing levels of Ypt1 found at this compartment. Curiously, Ypt1 levels began to decline when Trs85 levels were at their peak. We questioned whether the Ypt1 GAP Gyp1 could be responsible for the timing of this decline. Consistent with a previous study (Rivera-Molina and Novick, 2009), we found that Gyp1 began to accumulate directly downstream of Sec7, partially overlapping with Trs85 and coinciding with the decline of Ypt1 at the Golgi (Fig. 6, B and C; and Fig. S4 B). Collectively, these data suggest that opposing activities of TRAPPIII and Gyp1 control the level of active Ypt1 at the medial/late Golgi.
Figure 6.
TRAPPIII and the Rab GAP Gyp1 regulate Ypt1 activation at the medial/late Golgi. (A) Time-lapse imaging series (2-s intervals) of single Golgi compartments (green boxed region of the Merge panel) in cells expressing Sec7-6×DsRed with GFP-Ypt1 or Trs85-mNeonGreen. (A, top right) Normalized quantification of fluorescent signal from the boxed regions. (B, top left) Localization of Gyp1-mNeonGreen relative to Sec7-6×DsRed. (B, top right and bottom) Time-lapse imaging series and normalized quantification of Gyp1-mNeonGreen and Sec7-6×DsRed at a single Golgi compartment. (C) Quantification of peak-to-peak times for each indicated protein relative to Sec7-6×DsRed. t = 0 corresponds to peak Sec7 recruitment to the Golgi. Error bars represent 95% CIs. n ≥ 16 series. ***, P < 0.001 for one-way ANOVA with Tukey’s test for multiple comparisons. White arrows denote colocalization of Gyp1 and Sec7 at Golgi compartments. Dashed lines represent cell boundaries. Bars, 2 µm. Regions of interest for time-lapse imaging are 0.7 × 0.7 µm.
TRAPPIII functions in normal trafficking through the Golgi complex
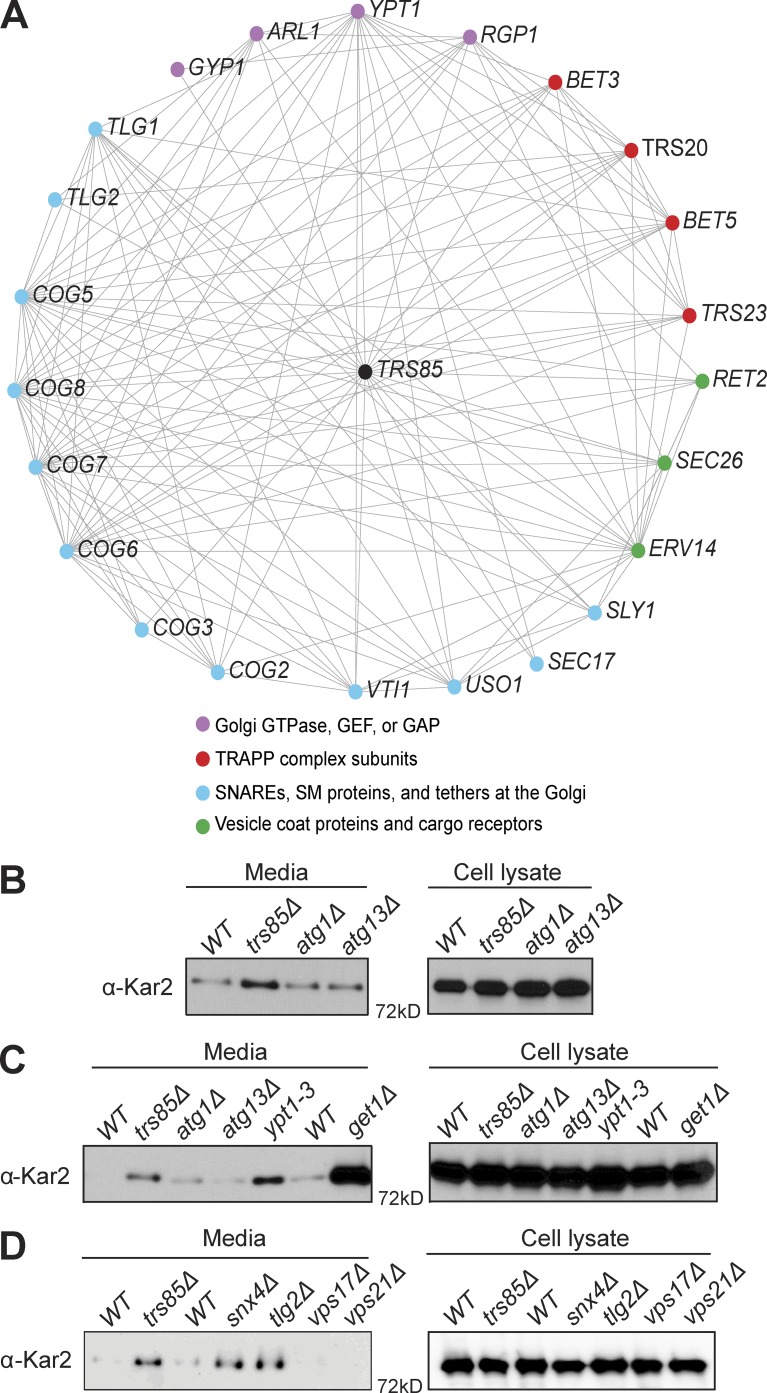
Previous studies have firmly established a role for Trs85 in autophagy; however, the role of Trs85 in Golgi trafficking has been less clear (Sacher et al., 2001; Meiling-Wesse et al., 2005; Lynch-Day et al., 2010; Zou et al., 2012; Shirahama-Noda et al., 2013). As an approach to uncover functional information from genetic network analysis, we generated a profile similarity network (PSN) map (Usaj et al., 2017) for TRS85 (Fig. 7 A). PSN analysis compares the set of genetic interactions of a particular gene with the genetic interactions sets for all other genes. If the interaction sets of two genes are similar, it can be inferred that the two genes have similar or related functions. The PSN indicated that TRS85 is functionally related to several genes known to mediate transport events at the Golgi complex. These genes can be grouped into four categories: (A) small GTPases, GEFs, and GAPs that function at the Golgi complex, (B) SNAREs, Sec1/Munc18 (SM) proteins, and protein tethers at the Golgi complex, (C) Golgi vesicle coat proteins and cargo receptors, or (D) other TRAPP subunits. Importantly, these genes include USO1 and subunits of the conserved oligomeric Golgi (COG) complex, vesicle tethers that are effectors of Ypt1 (Suvorova et al., 2002) and essential components of the secretory pathway. This PSN analysis provides strong support for an important role of Trs85 in normal secretory trafficking through the Golgi complex.
Figure 7.
TRAPPIII mediates Golgi trafficking. (A) The PSN of TRS85, showing all genes with similar genetic interactions, using a similarity cutoff of 0.4. Note that Uso1 and the COG complex mediate vesicle tethering at the Golgi complex and are known effectors of Ypt1, which is activated by TRAPPIII. (B) Western blot for Kar2 of TCA-precipitated cell and media fractions under normal growth conditions showing aberrant Kar2 secretion in trs85Δ cells but not atg1Δ or atg13Δ autophagy mutants. (C) Western blot comparing Kar2 secretion in trs85Δ and ypt1-3 mutants with that of get1Δ cells, a mutant previously shown to significantly disrupt ER–Golgi transport (Schuldiner et al., 2005). (D) Western blot comparing Kar2 secretion in a trs85Δ mutant with that of snx4Δ and tlg2Δ endosome-recycling mutants or vps17Δ and vps21Δ VPS mutants. Blots are representative of three independent experiments.
As a functional test of the role of Trs85 in regulating Golgi trafficking, we monitored the secretion of the ER-resident protein Kar2. Under normal growth conditions in WT cells, Kar2 is localized to the ER via constant recycling from the Golgi complex back to the ER. Previous studies have shown that when ER quality control or trafficking at the Golgi complex is disrupted, Kar2 will be aberrantly secreted from the cell (Schuldiner et al., 2005, 2008). Thus, secretion of Kar2 can be used as an indicator for improper trafficking at the Golgi complex. A genomewide screen determined that trs85Δ mutants secrete Kar2 at levels above those of WT cells (Copic et al., 2009). In agreement with this study, we found that trs85Δ cells exhibit a significant level of Kar2 secretion compared with WT cells (Fig. 7 B). To determine whether this phenomenon is the result of Trs85 function in autophagy, we examined Kar2 secretion in known autophagy mutants (atg1Δ and atg13Δ). These autophagy mutants did not secrete significant amounts of Kar2 (Fig. 7 B), indicating that the Kar2 secretion phenotype of trs85Δ is independent of the role of Trs85 in the autophagy pathway.
To assess the magnitude of the Kar2 secretion phenotype of a trs85Δ mutant, we also examined Kar2 secretion in get1Δ cells, a mutant known to exhibit significant Golgi trafficking defects (Schuldiner et al., 2008), as well as in ypt1-3 mutant cells at the permissive temperature (Fig. 7 C). Kar2 secretion in trs85Δ and ypt1-3 cells was similar but lower than that of the get1Δ mutant cells. These results indicate that loss of Trs85 compromises Golgi trafficking but that in the absence of Trs85, the resulting TRAPPI complex (present only in trs85Δ mutant cells) is sufficient to provide a minimal level of activated Ypt1 required for growth.
To gain insight into whether Kar2 secretion can arise through defects in endosome-to-Golgi trafficking, we examined cells lacking Snx4, Tlg2, Vps17, or Vps21/Ypt51 (Fig. 7 D). Neither vps17Δ nor vps21/ypt51Δ mutants exhibited Kar2 secretion, indicating that the vacuolar protein sorting (VPS) pathway is not required for proper Kar2 retention. However, both snx4Δ and tlg2Δ mutants secreted Kar2 at a level similar to that of trs85Δ. Given the role of Tlg2 as a SNARE involved in fusion of endosome-derived vesicles at the Golgi (Lewis et al., 2000) and the role of Snx4 in proper recycling of SNAREs to the Golgi (Hettema et al., 2003), we interpret these results to mean that proper Golgi homeostasis is required for normal Kar2 retention, as previously reported (Schuldiner et al., 2005, 2008; Copic et al., 2009), and that VPS function is not required.
Finally, we wished to test the functional effects of acute Trs85 depletion via anchor-away. The Kar2 secretion assay requires a long time course, so instead we monitored trafficking of the cargo GFP-Snc1. Snc1 is a v/R-SNARE protein that functions in the fusion of Golgi-derived secretory vesicles with the PM and is recycled back to the Golgi via endocytosis. Anchor-away of Trs85 resulted in a reduction in the amount of GFP-Snc1 observed at the PM (Fig. S5, A and B), similar to a more dramatic phenotype observed in trs85Δ cells at elevated temperature (Fig. S5 C; Zou et al., 2012) and consistent with a role for Trs85 in proper function of the secretory and/or PM recycling pathways that transit through the Golgi in budding yeast. Collectively, our findings demonstrate that Trs85 functions in normal Golgi trafficking in addition to its role in autophagy.
Discussion
The lack of obvious secretory pathway defects in previous research involving trs85Δ mutants was the primary rationale for the assignment of Trs85 to an autophagy-specific TRAPP complex (Lynch-Day et al., 2010). However, previously reported TRAPP complex fractionations resulted in the isolation of relatively low amounts of TRAPPI, which increased under high-salt conditions or in complex-destabilizing mutants (Montpetit and Conibear, 2009; Choi et al., 2011; Brunet et al., 2012, 2013). For example, levels of TRAPPI and TRAPPII increased and decreased, respectively, in tca17Δ or trs33Δ mutants that likely destabilize TRAPPII to produce TRAPPI (Montpetit and Conibear, 2009; Choi et al., 2011). A previous study also reported that the amount of TRAPPI increases in trs85Δ cells, presumably caused by a loss of TRAPPIII (Choi et al., 2011). Our new biochemical, functional, and localization analyses lead us to conclude that yeast possess only two TRAPP complexes: TRAPPII and TRAPPIII, akin to mammalian cells. The TRAPPI complex represents the core subunits of each but does not appear to be present as a distinct complex at detectable levels in normal yeast or mammalian cells.
We found that TRAPPIII is required for activating the bulk of Ypt1 at the Golgi and also for normal Golgi trafficking. A subset of Ypt1 remained at Golgi compartments in trs85Δ cells, implying that the TRAPPI core in trs85Δ mutants activates sufficient levels of Ypt1 during ER–Golgi transport to support viability. In contrast, trs85Δ mutants exhibit more pronounced autophagy phenotypes, perhaps because Trs85 physically interacts with several proteins involved in autophagy (Kakuta et al., 2012; Lipatova et al., 2012; Wang et al., 2013). We cannot completely rule out the possibility that small, transient amounts of TRAPPI function in ER–Golgi trafficking in WT cells. However, the simplest explanation of our data is that TRAPPIII is the primary activator of Ypt1 in the secretory pathway and that TRAPPIII performs the ER–Golgi transport functions previously assigned to TRAPPI.
Although the role of Ypt1 in ER–Golgi transport has been well established, a function for Ypt1 at the medial/late Golgi remains more controversial (Lipatova et al., 2013). Multiple groups have reported significant colocalization of Ypt1 with the late Golgi/TGN marker Sec7 (Sclafani et al., 2010; Kim et al., 2016a), yet a recent study proposed that these are “transitional” compartments rather than true late Golgi compartments (Kim et al., 2016a). Given the broad distribution of Sec7 across the medial/late Golgi, it is likely that the transitional compartment corresponds to a medial Golgi compartment that has just begun to acquire Sec7. A major argument against a role for Ypt1 at the medial/late Golgi is the controversy regarding TRAPPII as a GEF for Ypt1 (Morozova et al., 2006; Zou et al., 2012; Thomas and Fromme, 2016). We previously determined that TRAPPII can activate Ypt1 in vitro, but it appears to have only a minor role in Ypt1 activation in vivo (Thomas and Fromme, 2016). Given the significant loss of Ypt1 localization in trs85Δ cells and our previous observation that TRAPPII arrives far downstream of Ypt1 (Thomas and Fromme, 2016), we conclude that TRAPPIII is the primary Ypt1 GEF at the Golgi. Consistent with a previous study (Shirahama-Noda et al., 2013), we also observed that TRAPPIII localizes to medial/late Golgi compartments, arriving at the medial/late Golgi just before peak Ypt1 activation (Figs. 5, 6, S3, and S4). Previous studies have reported that endosome–Golgi transport is compromised in both Ypt1 and Trs85 mutants (Sclafani et al., 2010; Shirahama-Noda et al., 2013); furthermore, our PSN analysis suggests that TRAPPIII functions in the same or similar pathway or pathways as the COG complex, a Ypt1 effector that mediates intra-Golgi trafficking and endosome–Golgi transport (Ungar et al., 2006). Collectively, these observations suggest that TRAPPIII-activated Ypt1 contributes to endocytic recycling and thereby strongly support a function for Ypt1 at the medial/late Golgi. In future studies, it will be important to determine the mechanism by which TRAPPIII is recruited to medial/late Golgi compartments.
Our observation that Ypt1 is present at the Golgi complex before the arrival of Trs85 was surprising. We postulate that this occurs because a pool of Ypt1 remains associated with the Golgi complex after arrival via fusion of Ypt1-laden COPII vesicles, a step that is regulated by the direct action of TRAPPIII. In addition to its partial localization to the medial/late Golgi, we observed that Trs85 localized to multiple smaller compartments that were not labeled with Golgi markers (Fig. 5, A and B; and Fig. S3 A) and were not affected by inhibition of Sec7 activity (Fig. S3 B) or the ypt1-3 temperature-sensitive mutant that disrupts the Golgi (not depicted), indicating that TRAPPIII localizes to compartments in addition to the Golgi. Fluorescently tagged Trs85 appears as a diffuse haze throughout the cell in addition to its punctate localization. Cellular fractionation studies have shown that the majority of TRAPPIII is associated with membranes (Sacher et al., 1998; Lynch-Day et al., 2010), and studies have shown that TRAPPIIII associates directly with ER-derived COPII vesicles through an interaction between the core TRAPP subunit Bet3 and the COPII coat subunit Sec23 (Sacher et al., 2001; Yu et al., 2006; Cai et al., 2007; Lord et al., 2011; Bassik et al., 2013; Tan et al., 2013; Zhao et al., 2017). Additionally, Trs85 and Ypt1 physically associate with Atg9 vesicles, an essential membrane source for autophagosome formation (Lynch-Day et al., 2010; Kakuta et al., 2012; Lipatova et al., 2012; Shirahama-Noda et al., 2013). We therefore predict that the small Trs85-labeled compartments and cytoplasmic haze corresponds to TRAPPIII bound to highly mobile COPII and Atg9 vesicles. As the essential function of Ypt1 appears to be ER–Golgi transport rather than endocytic recycling at the medial/late Golgi, we assume that a significant fraction of TRAPPIII is devoted to activating Ypt1 on COPII vesicles. Combining our fluorescent and biochemical data, we estimate that <20% of the TRAPPIII in cells is localized to medial/late Golgi puncta, whereas >80% of the TRAPPIII in cells is localized diffusely to transport vesicles.
Previous models have proposed that Ypt1 activity is controlled by distinct GEFs in the biosynthetic pathway (TRAPPI) and autophagy (TRAPPIII; Lynch-Day et al., 2010; Davis et al., 2017), but there is also a precedent for the regulation of both normal and autophagic trafficking by the same GEF: the Rab GTPases Ypt7 and Vps21 are activated by the same GEFs (Mon1/Ccz1 and Vps9, respectively) under both normal trafficking conditions and during autophagy (Wang et al., 2002; Chen et al., 2014). Furthermore, Ypt7 and Vps21 recruit the same effectors to mediate both endolysosomal transport and autophagy. Similarly, Ypt1 recruits the tethering complex COG as an effector to capture incoming vesicles at the Golgi (Suvorova et al., 2002). COG has also been implicated in autophagy, and COG subunits localize to the phagophore assembly site/preautophagosomal structure (PAS; Yen et al., 2010), supporting a model in which GEF-Rab–effector modules may play similar roles in both healthy and starved cells.
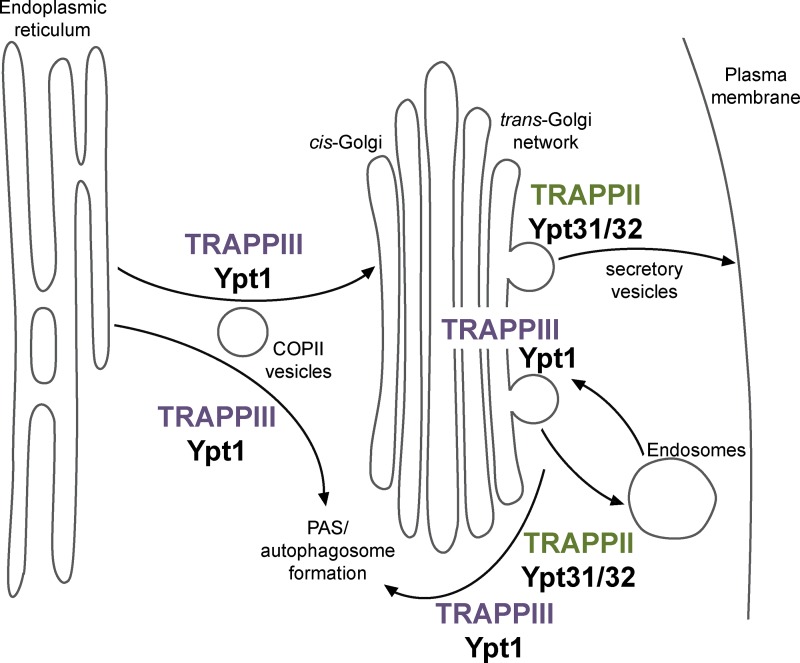
In light of the extensive literature on the role of the TRAPP complexes, our findings lead to a new model in which TRAPPIII and Ypt1 control membrane trafficking events under normal growth conditions and during autophagy. In this model, the COPII coat subunit Sec23 recruits TRAPPIII to COPII vesicles once they bud from the ER in both normal and starved cells (Fig. 8). TRAPPIII then activates Ypt1, either directly on COPII vesicles or in trans at the cis-Golgi or PAS, to mediate vesicle tethering and fusion. TRAPPIII likely dissociates from membranes once COPII vesicles completely uncoat, explaining why Ypt1 localizes to early Golgi compartments but TRAPPIII does not. TRAPPIII is also recruited to medial/late Golgi compartments, where it activates additional Ypt1 to facilitate vesicle formation (via its role in Sec7 recruitment) as well as endosome–Golgi transport in both normal and starved cells. Finally, TRAPPIII also activates Ypt1 at the PAS to facilitate membrane expansion during autophagosome formation. Therefore, TRAPPIII is responsible for activating Ypt1 at multiple locations to coordinate multiple distinct membrane transport pathways in both normal growth and autophagy.
Figure 8.
TRAPPIII activates Ypt1 (Rab1) to mediate ER–Golgi transport, medial/late Golgi trafficking, and autophagy. Model for TRAPPIII controlling Ypt1 activation in both the secretory pathway and autophagy. TRAPPIII is the primary activator of Ypt1 (Rab1), whereas TRAPPII is the primary activator of Ypt31/32 (Rab11). In addition to its established role in autophagy, TRAPPIII activates Ypt1 on COPII vesicles to facilitate normal ER–Golgi transport and at the medial/late Golgi to mediate normal endosome–Golgi trafficking. TRAPPII activates Ypt31/32 at the TGN to coordinate secretory vesicle formation and trafficking. TRAPPII and Ypt31/32 have also been reported to play a role in autophagy (Zou et al., 2013). PAS, phagophore assembly site/preautophagosomal structure.
Materials and methods
Strains and plasmids
All yeast strains and plasmids were constructed using standard techniques and are described in Tables S1 and S2. Several plasmids and strains were obtained from R. Collins and S. Emr (Cornell University, Ithaca, NY), S. Ferro-Novick (University of California, San Diego, La Jolla, CA), B. Glick (University of Chicago, Chicago, IL), K. Reinisch (Yale University, New Haven, CT), N. Segev (University of Illinois at Chicago, Chicago, IL), A. Spang (Biozentrum, Basel, Switzerland), and C. Ungermann (University of Osnabrück, Osnabrück, Germany).
Antibodies and inhibitors
The anti-Kar2 rabbit polyclonal antibody was a gift from E. Miller (Medical Research Council Laboratory of Molecular Biology, Cambridge, England, UK) and was used at a 1:10,000 dilution. The anti-GFP mouse monoclonal antibody was purchased from Santa Cruz Biotechnology, Inc. (sc-9996) and was used at a dilution of 1:500. The anti–glucose-6-phosphate dehydrogenase (G6PDH) rabbit polyclonal antibody was purchased from Sigma-Aldrich (SAB2100871) and was used at a concentration of 1:30,000.
To test whether Sec7 activity plays a role in TRAPPIII recruitment to Golgi compartments, 40 µM 6-methyl-5-nitro-2-(trifluoromethyl)-4H-chromen-4-one (MNTC; MolPort) was added to growth media 20 min before imaging. To deplete TRAPPIII from the Golgi in anchor-away experiments, cells expressing Pma1-2×FKBP and Trs85-mRFPmars-FRB were treated with 1 µg/ml rapamycin (LC Laboratories) for 1 h before imaging. The rapamycin growth plates shown in Fig. S2 D contained 1 µg/ml rapamycin.
TRAPP purification from yeast
Endogenous TRAPP complexes were purified from 12 liters of log-phase yeast expressing Trs120-TAP (TRAPPII), Trs85-TAP (TRAPPIII), or Bet3-TAP (composite TRAPP). The TAP tag used in this study consisted of a calmodulin-binding peptide separated from a protein A tag by a tobacco etch virus (TEV) protease site. Yeast cells were lysed using a BeadBeater (Biospec) or freezer mill (SPEX SamplePrep) in lysis buffer (20 mM Hepes, pH 7.4, 300 mM NaCl, 5% glycerol, 1% CHAPS, 2 mM MgCl2, 1 mM DTT, 50 mM NaF, 0.1 mM Na3VO4, 1 mM PMSF, and 1× protease inhibitor cocktail [Roche]). The lysate was cleared by centrifugation and incubated with Sepharose 6B (Sigma-Aldrich) for 30 min to remove any proteins that bind nonspecifically to Sepharose. The cleared lysate was then incubated with IgG Sepharose (GE Healthcare) for 3 h at 4°C to isolate protein A–tagged TRAPP complexes. The resin was then washed with IPP300 buffer (25 mM Tris-HCl, pH 8.0, 300 mM NaCl, 5% glycerol, 0.1% CHAPS, and 1 mM DTT) followed by TEV cleavage buffer (IPP300 with 0.5 mM EDTA), and TRAPP complexes were cleaved off the resin by overnight treatment with TEV protease at 4°C. Cleaved protein was diluted in calmodulin-binding buffer (25 mM Tris-HCl, pH 8.0, 300 mM NaCl, 5% glycerol, 0.1% CHAPS, 1 mM magnesium acetate, 1 mM imidazole, 2 mM CaCl2, and 1 mM DTT) and incubated with calmodulin Sepharose (GE Healthcare) for 2–3 h at 4°C. The calmodulin resin was then washed with calmodulin binding buffer, and TRAPP complexes were eluted using calmodulin elution buffer (25 mM Tris-HCl, pH 8.0, 300 mM NaCl, 5% glycerol, 0.1% CHAPS detergent, 1 mM magnesium acetate, 1 mM imidazole, 20 mM EGTA, and 1 mM DTT).
Samples from each “composite” TRAPP preparation (purified via Bet3-TAP) were run on a 15% SDS-PAGE gel, stained for total protein using colloidal Coomassie (Bio-Rad Laboratories), and imaged with an Odyssey instrument (LI-COR Biosciences). For each purification, the molarities of all individual TRAPP subunits were measured using ImageJ (National Institutes of Health). The amount of total TRAPP complexes in each preparation was then determined by averaging the molarities of the shared core subunits Trs33, Trs31, Bet3, Trs23, Trs20, and Bet5. Individual TRAPP complex preparations contained approximately stoichiometric amounts of all core subunits with two copies of Bet3.
To determine the mean abundance of individual subunits (Fig. 1 C, left), the measured molarity of each subunit was divided by that calculated for total TRAPP complexes within the same preparation. Amounts of TRAPPII and TRAPPIII complexes present (Fig. 1 C, right) were calculated from the mean molarities of the complex-specific subunits Trs65, Trs120, and Trs130 or Trs85, respectively (Tca17 was excluded from the analysis because its signal was too close to the background). The amount of TRAPPI present was then determined by subtracting the amount of TRAPPII and TRAPPIII from that of total TRAPP within the same preparation.
Recombinant protein purification
All recombinant proteins were purified using Rosetta2 cells (Novagen). rTRAPPI was expressed from a single pCOLADuet-1 vector containing the six core genes (TRS33, TRS31, TRS23, BET3, TRS20, and BET5; pLT14) with a cleavable N-terminal His6 tag on TRS31. The expression vector was transformed into Rosetta2 E. coli and grown in 1–4 liters terrific broth (TB) to an OD600 of ∼3.0. Protein expression was then induced with 300 µM IPTG for 12–16 h at 18°C. Cells were lysed by sonication in lysis buffer (40 mM Tris-HCl, pH 8.0, 300 mM NaCl, 10% glycerol, 10 mM imidazole, 2 mM MgCl2, 5 mM β-mercaptoethanol [BME], and 1 mM PMSF), and the lysate was cleared by centrifugation. His-tagged rTRAPPI was affinity purified using Ni-NTA resin (QIAGEN), and complexes were eluted using elution buffer (lysis buffer with 250 mM imidazole). Eluted protein was treated with TEV protease overnight at 4°C to remove the His6 tag. Stoichiometric complexes were isolated using gel filtration with a Superdex 200 10/300 GL column (GE Healthcare); for consistency across endogenous and recombinant TRAPP complexes, calmodulin elution buffer was used for gel filtration.
rTRAPPIII was purified by coexpressing the following two plasmids in Rosetta2 cells: a rTRAPPI plasmid with the His6 tag removed from TRS31 (pLT21) and pETDuet-1 containing TRS85 with a C-terminal TAP tag (pLT92). rTRAPPIII was purified from 4–8 liters of cells using the same protocol as for endogenous TRAPP complexes, with a final gel filtration step used to isolate stoichiometric complexes.
His-tagged Mrs6 and Bet2–Bet4 complexes were purified from 1–8 liters TB in the same manner as rTRAPPI complexes without TEV treatment to remove the His6 tags. Prenylation buffer (20 mM Hepes, pH 7.4, 150 mM NaCl, 2 mM MgCl2, and 1 mM DTT) was used for gel filtration.
Rab GTPases and Gdi1 were expressed with cleavable N-terminal GST tags in the pGEX-6P vector backbone. GST-Rab constructs were purified with and without C-terminal His7 tags; the His-tagged Rabs were designed so that the His7 tag replaced the C-terminal cysteines to allow for anchoring on membranes with Ni2+-DOGS. After protein expression, cells grown in 1–4 liters TB were lysed in lysis buffer (1× PBS, 2 mM MgCl2, 5 mM BME, and 1 mM PMSF). The cleared lysate was incubated with Glutathione resin (G-Biosciences) for 2–3 h at 4°C to isolate GST-tagged proteins. The resin was washed in lysis buffer, resuspended in PreScission cleavage buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 2 mM MgCl2, and 1 mM DTT), and treated overnight with PreScission (3C) protease (GE Healthcare) at 4°C to remove the GST tag. Cleavage of the GST tag eluted the proteins from the resin.
Purified Rab GTPases were loaded with mantGDP (Biolog Life Science Institute) in order to measure nucleotide exchange in GEF activity assays. To generate prenylated Rab/GDI substrates, the mantGDP-loaded Rab, Gdi1, His6-Mrs6, and His6-Bet2/Bet4 were mixed in a 10:10:1:1 molar ratio with a sixfold excess of geranylgeranyl pyrophosphate (Cayman) in prenylation buffer with 20 µM mantGDP added to maintain loading, and incubated at 37°C for 1 h to allow prenylation. After prenylation, imidazole was added for a final concentration of 10 mM. A 1/10th volume of Ni-NTA resin was then added, and the mixture was incubated at 4°C for 1 h to deplete His6-Mrs6 and His6-Bet2/Bet4. Prenylated Rab/GDI complexes were further purified using gel filtration to isolate stoichiometric complexes.
Liposome preparation
Synthetic TGN liposomes were prepared using a mixture of lipids that approximates the lipid composition of the TGN (Klemm et al., 2009). Lipids were combined, vacuum dried, rehydrated in HK buffer (20 mM Hepes, pH 7.4, and 150 mM KOAc), and extruded through 100-nm filters (Whatman) to generate liposomes. See Table S3 for the composition of TGN and phosphatidylcholine liposomes.
GEF activity assays
For assays with prenylated Rab substrates, nucleotide exchange was measured by sequentially adding 333 µM liposomes, 200 µM nonfluorescent GTP, and 250 nM mantGDP-labeled Rab to HK buffer with 2 mM MgCl2 at 30°C. The mixture was then incubated at 30°C for 2 min to allow the Rab to begin partitioning into the membrane, and nucleotide exchange was initiated by the addition of 20 nM TRAPP. After the addition of TRAPP, mantGDP fluorescence was measured (365-nm excitation and 440-nm emission) for 20 min to obtain the exchange trace. To determine exchange rates, traces of fluorescence versus time were fit to a single exponential curve with an additional linear drift term using Prism (GraphPad Software). The resulting rate constants were divided by the GEF concentration to obtain the exchange rate.
Assays with soluble Rab-His7 substrates were performed using identical conditions but without the addition of liposomes. To control for the intrinsic nucleotide exchange of soluble substrates, mock reactions were performed with buffer added instead of TRAPP complexes. Rate constants were obtained for replicate mock reactions and averaged, and the mean mock rate constant was subtracted from that of TRAPP-mediated reactions before dividing by the GEF concentration.
Kar2 secretion assay and GFP-Snc1 Western blot
Cells with endogenous mutations were grown to an OD600 = 0.85 − 1 in YPD media. 1.2 ml of culture was transferred to an Eppendorf tube and spun for 5 min at 21,000 g. 1 ml of the supernatant fraction was transferred to a new tube and spun a second time for 5 min at 21,000 g, and then the entire supernatant was transferred to a clean tube. The cell pellet was washed once and then resuspended in 1 ml cold distilled H2O. Both the supernatant and cell pellet fractions were then subjected to TCA precipitation including acetone washes. Samples were resolved by SDS-PAGE and transferred to a polyvinylidene fluoride membrane before immunoblotting using anti-Kar2 primary antibody at 1:10,000 dilution and anti–rabbit secondary antibody at 1:20,000 dilution.
To test whether total GFP-Snc1 levels were altered by anchor-away of Trs85, cells expressing GFP-Snc1, Trs85-mRFPmars-FRB, and Pma1-2×FKBP (CFY3144) were grown to log phase and treated with 1 µg/ml rapamycin or DMSO as a control for 20 or 60 min. 10-OD units of cells were collected for each treatment and subjected to TCA precipitation. Levels of GFP-Snc1 were measured by immunoblotting with anti-GFP; anti-G6PDH was used as a loading control.
Microscopy
Cells were grown in synthetic dropout media at 30°C and imaged at room temperature in log phase on glass coverslips or in glass-bottomed dishes. Each image shown is a single focal plane. All images were processed using ImageJ, adjusting only the min/max brightness levels for clarity with identical processing of all images within a single figure panel. Time-lapse imaging series in Figs. 6 and S4 were generated by imaging each channel every 2 s for 2–4 min.
Images shown in Figs. 2, 6, and S4 A were captured with a CSU-X spinning-disk confocal microscope system (Intelligent Imaging Innovations) using a DMI6000 microscope (Leica Microsystems) outfitted with a CSU-X1 spinning-disk confocal unit (Yokogawa Electric Corporation) with a QuantEM 512SC (Photometrics). The objective was a 100× 1.46 NA Plan Apochromat oil immersion lens (Leica Microsystems). Additional components included a laser stack and mSAC (spherical aberration correction; Intelligent Imaging Innovations). SlideBook software was used to control the system (Intelligent Imaging Innovations).
All other images were acquired using a DeltaVision RT widefield deconvolution microscope (Applied Precision Ltd.), a 100× 1.35 NA objective oil immersion lens, and a CoolSNAP HQ camera (Photometrics) and deconvolved (conservative setting; six cycles) using SoftWoRx software (Applied Precision Ltd.).
Image analysis
Line-trace analysis was performed using ImageJ to quantify the amount of cytosolic versus Golgi-localized Ypt1, Ypt6, and TRAPPIII. Traces were chosen to pass through the image background as well as the cell cytoplasm and brightest Golgi compartments to allow for simultaneous quantification of each. For each cell, the background value was subtracted from those obtained for the cytosol and Golgi. The background-subtracted Golgi fluorescence was then divided by that of the cytosol. Recruitment of Ypt31 to the TGN was measured by quantifying the ratio of GFP-Ypt31 to mRFPmars-Sec7–labeled TGN compartments as previously described (Thomas and Fromme, 2016). A similar line-trace analysis was also used to quantitate the extent of GFP-Snc1 localization to the PM. For each cell, the trace was chosen to pass through the PM of the daughter cell as well as the brightest Golgi compartment, and then background-subtracted PM fluorescence was divided by that of the Golgi compartment.
Manders analysis was used to quantify the colocalization of fluorescent signals at Golgi compartments. Images were cropped to contain between four and nine cells. Five crops of each strain were analyzed to calculate a mean Manders coefficient of colocalization using the JACoP plugin in ImageJ (Bolte and Cordelières, 2006). Images were thresholded to only quantify signals at puncta (therefore avoiding any cytoplasmic background). All of the red channel images were thresholded to the same level across all strains, whereas the green threshold varied for each strain.
Time-lapse imaging was analyzed using SlideBook 5.0 software to quantify peak-to-peak recruitment times. The fluorescence intensity for each visualized protein at a single Golgi compartment was tracked over the lifetime of the compartment. Fluorescence signal was normalized for each channel, and the time point where normalized fluorescence equaled 1.0 was taken as the peak recruitment time for each protein. Only compartments that remained in the focal plane and distinct from other compartments were analyzed.
PSN analysis
The PSN was generated using an internet-based tool (thecellmap.org) developed by the Boone and Andrews laboratories (Usaj et al., 2017). A threshold of 0.4 was used as a cutoff.
Statistical tests
For Figs. 1 C and 6 C, statistical significance was determined using a one-way ANOVA with Tukey’s test for multiple comparisons. For all other figures, significance was determined using an unpaired two-tailed t test with Welch’s correction. Data distribution was assumed to be normal, but this was not formally tested.
Online supplemental material
Fig. S1 demonstrates that TRAPPIII is a specific GEF for Ypt1. Fig. S2 shows that Trs85, but not TRAPP core subunits, is relocalized to the PM during anchor-away and that Ypt1 is specifically affected by loss of Trs85 at the Golgi. Fig. S3 shows that TRAPPIII localizes to medial/late Golgi compartments independent of Sec7 activity and Arf1. Fig. S4 summarizes the timing of GEF, GAP, and Rab recruitment to the medial/late Golgi. Fig. S5 shows that Snc1 trafficking is perturbed by loss of Trs85 at the Golgi. Tables S1 and S2 describe the plasmids and strains used in this study, respectively. Table S3 lists the lipid composition of liposomes used in GEF activity assays.
Supplementary Material
Acknowledgments
We thank N. Navarro and E. Miller for the Kar2 antibody and advice regarding the Kar2 secretion assay. We thank R. Gingras and A. Bretscher for assistance with microscopy. We thank R. Collins, S. Emr, S. Ferro-Novick, B. Glick, K. Reinsich, N. Segev, A. Spang, and C. Ungermann for reagents. We thank S. Suzuki and B. Richardson for feedback on the manuscript and members of the Fromme laboratory for helpful discussions.
This work was supported by National Institutes of Health grant R01GM116942 to J.C. Fromme, by an Alfred P. Sloan Foundation Fellowship and National Science Foundation Graduate Research Fellowships Program grant DGE-1144153 (any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation) to A.M.N. Joiner, and by a National Institutes of Health training grant T32GM007273 to L.L. Thomas.
The authors declare no competing financial interests.
Author contributions: conceptualization and methodology, L.L. Thomas, A.M.N. Joiner, and J.C. Fromme; investigation, visualization, and writing (original draft), L.L. Thomas and A.M.N. Joiner; writing (review and editing), L.L. Thomas, A.M.N. Joiner and J.C. Fromme; funding acquisition and project administration, J.C. Fromme.
Footnotes
Abbreviations used:
- BME
- β-mercaptoethanol
- CI
- confidence interval
- COG
- conserved oligomeric Golgi
- G6PDH
- glucose-6-phosphate dehydrogenase
- GAP
- GTPase-activating protein
- GDI
- GDP dissociation inhibitor
- GEF
- guanine nucleotide exchange factor
- MNTC
- 6-methyl-5-nitro-2-(trifluoromethyl)-4H-chromen-4-one
- PM
- plasma membrane
- PSN
- profile similarity network
- TAP
- tandem affinity purification
- TB
- terrific broth
- TEV
- tobacco etch virus
- TRAPP
- transport protein particle
- VPS
- vacuolar protein sorting
References
- Barr F.A. 2009. Rab GTPase function in Golgi trafficking. Semin. Cell Dev. Biol. 20:780–783. 10.1016/j.semcdb.2009.03.007 [DOI] [PubMed] [Google Scholar]
- Barrowman J., Bhandari D., Reinisch K., and Ferro-Novick S.. 2010. TRAPP complexes in membrane traffic: convergence through a common Rab. Nat. Rev. Mol. Cell Biol. 11:759–763. 10.1038/nrm2999 [DOI] [PubMed] [Google Scholar]
- Bassik M.C., Kampmann M., Lebbink R.J., Wang S., Hein M.Y., Poser I., Weibezahn J., Horlbeck M.A., Chen S., Mann M., et al. 2013. A systematic mammalian genetic interaction map reveals pathways underlying ricin susceptibility. Cell. 152:909–922. 10.1016/j.cell.2013.01.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrends C., Sowa M., Gygi S., and Harper J.W.. 2010. Network organization of the human autophagy system. Nature. 466:68–76. 10.1038/nature09204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte S., and Cordelières F.P.. 2006. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224:213–232. 10.1111/j.1365-2818.2006.01706.x [DOI] [PubMed] [Google Scholar]
- Brunet S., and Sacher M.. 2014. In sickness and in health: the role of TRAPP and associated proteins in disease. Traffic. 15:803–818. 10.1111/tra.12183 [DOI] [PubMed] [Google Scholar]
- Brunet S., Noueihed B., Shahrzad N., Saint-Dic D., Hasaj B., Guan T.L., Moores A., Barlowe C., and Sacher M.. 2012. The SMS domain of Trs23p is responsible for the in vitro appearance of the TRAPP I complex in Saccharomyces cerevisiae. Cell. Logist. 2:28–42. 10.4161/cl.19414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet S., Shahrzad N., Saint-Dic D., Dutczak H., and Sacher M.. 2013. A trs20 mutation that mimics an SEDT-causing mutation blocks selective and non-selective autophagy: a model for TRAPP III organization. Traffic. 14:1091–1104. 10.1111/tra.12095 [DOI] [PubMed] [Google Scholar]
- Cai H., Yu S., Menon S., Cai Y., Lazarova D., Fu C., Reinisch K., Hay J.C., and Ferro-Novick S.. 2007. TRAPPI tethers COPII vesicles by binding the coat subunit Sec23. Nature. 445:941–944. 10.1038/nature05527 [DOI] [PubMed] [Google Scholar]
- Cai Y., Chin H.F., Lazarova D., Menon S., Fu C., Cai H., Sclafani A., Rodgers D.W., De La Cruz E.M., Ferro-Novick S., and Reinisch K.M.. 2008. The structural basis for activation of the Rab Ypt1p by the TRAPP membrane-tethering complexes. Cell. 133:1202–1213. 10.1016/j.cell.2008.04.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Zhou F., Zou S., Yu S., Li S., Li D., Song J., Li H., He Z., Hu B., et al. 2014. A Vps21 endocytic module regulates autophagy. Mol. Biol. Cell. 25:3166–3177. 10.1091/mbc.E14-04-0917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi C., Davey M., Schluter C., Pandher P., Fang Y., Foster L.J., and Conibear E.. 2011. Organization and assembly of the TRAPPII complex. Traffic. 12:715–725. 10.1111/j.1600-0854.2011.01181.x [DOI] [PubMed] [Google Scholar]
- Copic A., Dorrington M., Pagant S., Barry J., Lee M.C.S., Singh I., Hartman J.L. IV, and Miller E.A.. 2009. Genomewide analysis reveals novel pathways affecting endoplasmic reticulum homeostasis, protein modification and quality control. Genetics. 182:757–769. 10.1534/genetics.109.101105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis S., Wang J., and Ferro-Novick S.. 2017. Crosstalk between the secretory and autophagy pathways regulates autophagosome formation. Dev. Cell. 41:23–32. 10.1016/j.devcel.2017.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiNitto J.P., Delprato A., Gabe Lee M.-T., Cronin T.C., Huang S., Guilherme A., Czech M.P., and Lambright D.G.. 2007. Structural basis and mechanism of autoregulation in 3-phosphoinositide-dependent Grp1 family Arf GTPase exchange factors. Mol. Cell. 28:569–583. 10.1016/j.molcel.2007.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruki H., Nishikawa J., and Laemmli U.K.. 2008. The anchor-away technique: rapid, conditional establishment of yeast mutant phenotypes. Mol. Cell. 31:925–932. 10.1016/j.molcel.2008.07.020 [DOI] [PubMed] [Google Scholar]
- Hettema E.H., Lewis M.J., Black M.W., and Pelham H.R.B.. 2003. Retromer and the sorting nexins Snx4/41/42 mediate distinct retrieval pathways from yeast endosomes. EMBO J. 22:548–557. 10.1093/emboj/cdg062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedd G., Richardson C., Litt R., and Segev N.. 1995. The Ypt1 GTPase Is Essential for the First Two Steps of the Yeast Secretory Pathway. J. Cell Biol. 131:583–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakuta S., Yamamoto H., Negishi L., Kondo-Kakuta C., Hayashi N., and Ohsumi Y.. 2012. Atg9 vesicles recruit vesicle-tethering proteins Trs85 and Ypt1 to the autophagosome formation site. J. Biol. Chem. 287:44261–44269. 10.1074/jbc.M112.411454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.J., Lipatova Z., Majumdar U., and Segev N.. 2016a Regulation of Golgi Cisternal Progression by Ypt/Rab GTPases. Dev. Cell. 36:440–452. 10.1016/j.devcel.2016.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.J., Lipatova Z., and Segev N.. 2016b TRAPP Complexes in Secretion and Autophagy. Front. Cell Dev. Biol. 4:20 10.3389/fcell.2016.00020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm R.W., Ejsing C.S., Surma M.A., Kaiser H.J., Gerl M.J., Sampaio J.L., de Robillard Q., Ferguson C., Proszynski T.J., Shevchenko A., and Simons K.. 2009. Segregation of sphingolipids and sterols during formation of secretory vesicles at the trans-Golgi network. J. Cell Biol. 185:601–612. 10.1083/jcb.200901145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb C.A., Nühlen S., Judith D., Frith D., Snijders A.P., Behrends C., and Tooze S.A.. 2016. TBC1D14 regulates autophagy via the TRAPP complex and ATG9 traffic. EMBO J. 35:281–301. 10.15252/embj.201592695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis M.J., Nichols B.J., Prescianotto-Baschong C., Riezman H., and Pelham H.R.. 2000. Specific retrieval of the exocytic SNARE Snc1p from early yeast endosomes. Mol. Biol. Cell. 11:23–38. 10.1091/mbc.11.1.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipatova Z., and Segev N.. 2014. Ypt/Rab GTPases regulate two intersections of the secretory and the endosomal/lysosomal pathways. Cell. Logist. 4:e954870 10.4161/21592780.2014.954870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipatova Z., Belogortseva N., Zhang X.Q., Kim J., Taussig D., and Segev N.. 2012. Regulation of selective autophagy onset by a Ypt/Rab GTPase module. Proc. Natl. Acad. Sci. USA. 109:6981–6986. 10.1073/pnas.1121299109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipatova Z., Shah A.H., Kim J.J., Mulholland J.W., and Segev N.. 2013. Regulation of ER-phagy by a Ypt/Rab GTPase module. Mol. Biol. Cell. 24:3133–3144. 10.1091/mbc.E13-05-0269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipatova Z., Majumdar U., and Segev N.. 2016. Trs33-containing TRAPP IV: A novel autophagy-specific Ypt1 GEF. Genetics. 204:1117–1128. 10.1534/genetics.116.194910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord C., Bhandari D., Menon S., Ghassemian M., Nycz D., Hay J., Ghosh P., and Ferro-Novick S.. 2011. Sequential interactions with Sec23 control the direction of vesicle traffic. Nature. 473:181–186. 10.1038/nature09969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch-Day M.A., Bhandari D., Menon S., Huang J., Cai H., Bartholomew C.R., Brumell J.H., Ferro-Novick S., and Klionsky D.J.. 2010. Trs85 directs a Ypt1 GEF, TRAPPIII, to the phagophore to promote autophagy. Proc. Natl. Acad. Sci. USA. 107:7811–7816. 10.1073/pnas.1000063107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonold C.M., and Fromme J.C.. 2014. Four GTPases differentially regulate the Sec7 Arf-GEF to direct traffic at the trans-Golgi network. Dev. Cell. 30:759–767. 10.1016/j.devcel.2014.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiling-Wesse K., Epple U.D., Krick R., Barth H., Appelles A., Voss C., Eskelinen E.-L., and Thumm M.. 2005. Trs85 (Gsg1), a component of the TRAPP complexes, is required for the organization of the preautophagosomal structure during selective autophagy via the Cvt pathway. J. Biol. Chem. 280:33669–33678. 10.1074/jbc.M501701200 [DOI] [PubMed] [Google Scholar]
- Menon S., Cai H., Lu H., Dong G., Cai Y., Reinisch K., and Ferro-Novick S.. 2006. mBET3 is required for the organization of the TRAPP complexes. Biochem. Biophys. Res. Commun. 350:669–677. 10.1016/j.bbrc.2006.09.096 [DOI] [PubMed] [Google Scholar]
- Montpetit B., and Conibear E.. 2009. Identification of the novel TRAPP associated protein Tca17. Traffic. 10:713–723. 10.1111/j.1600-0854.2009.00895.x [DOI] [PubMed] [Google Scholar]
- Morozova N., Liang Y., Tokarev A.A., Chen S.H., Cox R., Andrejic J., Lipatova Z., Sciorra V.A., Emr S.D., and Segev N.. 2006. TRAPPII subunits are required for the specificity switch of a Ypt-Rab GEF. Nat. Cell Biol. 8:1263–1269. 10.1038/ncb1489 [DOI] [PubMed] [Google Scholar]
- Peurois F., Veyron S., Ferrandez Y., Ladid I., Benabdi S., Zeghouf M., Peyroche G., and Cherfils J.. 2017. Characterization of the activation of small GTPases by their GEFs on membranes using artificial membrane tethering. Biochem. J. 474:1259–1272. 10.1042/BCJ20170015 [DOI] [PubMed] [Google Scholar]
- Plutner H., Cox A.D., Pind S., Khosravi-Far R., Bourne J.R., Schwaninger R., Der C.J., and Balch W.E.. 1991. Rab1b regulates vesicular transport between the endoplasmic reticulum and successive Golgi compartments. J. Cell Biol. 115:31–43. 10.1083/jcb.115.1.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson B.C., McDonold C.M., and Fromme J.C.. 2012. The Sec7 Arf-GEF is recruited to the trans-Golgi network by positive feedback. Dev. Cell. 22:799–810. 10.1016/j.devcel.2012.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Molina F.E., and Novick P.J.. 2009. A Rab GAP cascade defines the boundary between two Rab GTPases on the secretory pathway. Proc. Natl. Acad. Sci. USA. 106:14408–14413. 10.1073/pnas.0906536106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacher M., Jiang Y., Barrowman J., Scarpa A., Burston J., Zhang L., Schieltz D., Yates J.R. III, Abeliovich H., and Ferro-Novick S.. 1998. TRAPP, a highly conserved novel complex on the cis-Golgi that mediates vesicle docking and fusion. EMBO J. 17:2494–2503. 10.1093/emboj/17.9.2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacher M., Barrowman J., Wang W., Horecka J., Zhang Y., Pypaert M., and Ferro-Novick S.. 2001. TRAPP I implicated in the specificity of tethering in ER-to-Golgi transport. Mol. Cell. 7:433–442. 10.1016/S1097-2765(01)00190-3 [DOI] [PubMed] [Google Scholar]
- Schuldiner M., Collins S.R., Thompson N.J., Denic V., Bhamidipati A., Punna T., Ihmels J., Andrews B., Boone C., Greenblatt J.F., et al. 2005. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 123:507–519. 10.1016/j.cell.2005.08.031 [DOI] [PubMed] [Google Scholar]
- Schuldiner M., Metz J., Schmid V., Denic V., Rakwalska M., Schmitt H.D., Schwappach B., and Weissman J.S.. 2008. The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell. 134:634–645. 10.1016/j.cell.2008.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sclafani A., Chen S., Rivera-Molina F., Reinisch K., Novick P., and Ferro-Novick S.. 2010. Establishing a role for the GTPase Ypt1p at the late Golgi. Traffic. 11:520–532. 10.1111/j.1600-0854.2010.01031.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scrivens P.J., Noueihed B., Shahrzad N., Hul S., Brunet S., and Sacher M.. 2011. C4orf41 and TTC-15 are mammalian TRAPP components with a role at an early stage in ER-to-Golgi trafficking. Mol. Biol. Cell. 22:2083–2093. 10.1091/mbc.E10-11-0873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirahama-Noda K., Kira S., Yoshimori T., and Noda T.. 2013. TRAPPIII is responsible for vesicular transport from early endosomes to Golgi, facilitating Atg9 cycling in autophagy. J. Cell Sci. 126:4963–4973. 10.1242/jcs.131318 [DOI] [PubMed] [Google Scholar]
- Stalder D., and Antonny B.. 2013. Arf GTPase regulation through cascade mechanisms and positive feedback loops. FEBS Lett. 587:2028–2035. 10.1016/j.febslet.2013.05.015 [DOI] [PubMed] [Google Scholar]
- Stenmark H. 2009. Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 10:513–525. 10.1038/nrm2728 [DOI] [PubMed] [Google Scholar]
- Suda Y., Kurokawa K., Hirata R., and Nakano A.. 2013. Rab GAP cascade regulates dynamics of Ypt6 in the Golgi traffic. Proc. Natl. Acad. Sci. USA. 19:18976–18981. 10.1073/pnas.1308627110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suvorova E.S., Duden R., and Lupashin V.V.. 2002. The Sec34/Sec35p complex, a Ypt1p effector required for retrograde intra-Golgi trafficking, interacts with Golgi SNAREs and COPI vesicle coat proteins. J. Cell Biol. 157:631–643. 10.1083/jcb.200111081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan D., Cai Y., Wang J., Zhang J., Menon S., Chou H.T., Ferro-Novick S., Reinisch K.M., and Walz T.. 2013. The EM structure of the TRAPPIII complex leads to the identification of a requirement for COPII vesicles on the macroautophagy pathway. Proc. Natl. Acad. Sci. USA. 110:19432–19437. 10.1073/pnas.1316356110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas L.L., and Fromme J.C.. 2016. GTPase cross talk regulates TRAPPII activation of Rab11 homologues during vesicle biogenesis. J. Cell Biol. 215:499–513. 10.1083/jcb.201608123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisdale E.J., Bourne J.R., Khosravi-Far R., Der C.J., and Balch W.E.. 1992. GTP-binding mutants of Rab1 and Rab2 are potent inhibitors of vesicular transport from the endoplasmic reticulum to the Golgi complex. J. Cell Biol. 119:749–761. 10.1083/jcb.119.4.749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungar D., Oka T., Krieger M., and Hughson F.M.. 2006. Retrograde transport on the COG railway. Trends Cell Biol. 16:113–120. 10.1016/j.tcb.2005.12.004 [DOI] [PubMed] [Google Scholar]
- Usaj M., Tan Y., Wang W., VanderSluis B., Zou A., Myers C.L., Costanzo M., Andrews B., and Boone C.. 2017. TheCellMap.org: A web-cccessible database for visualizing and mining the global yeast genetic interaction network. G3: Genes. Genomes. Genetics. 7:1539–1549. 10.1534/g3.117.040220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.W., Stromhaug P.E., Shima J., and Klionsky D.J.. 2002. The Ccz1-Mon1 protein complex is required for the late step of multiple vacuole delivery pathways. J. Biol. Chem. 277:47917–47927. 10.1074/jbc.M208191200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Menon S., Yamasaki A., Chou H.-T., Walz T., Jiang Y., and Ferro-Novick S.. 2013. Ypt1 recruits the Atg1 kinase to the preautophagosomal structure. Proc. Natl. Acad. Sci. USA. 110:9800–9805. 10.1073/pnas.1302337110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Sacher M., and Ferro-Novick S.. 2000. TRAPP stimulates guanine nucleotide exchange on Ypt1p. J. Cell Biol. 151:289–296. 10.1083/jcb.151.2.289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki A., Menon S., Yu S., Barrowman J., Meerloo T., Oorschot V., Klumperman J., Satoh A., and Ferro-Novick S.. 2009. mTrs130 is a component of a mammalian TRAPPII complex, a Rab1 GEF that binds to COPI-coated vesicles. Mol. Biol. Cell. 20:4205–4215. 10.1091/mbc.E09-05-0387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen W.L., Shintani T., Nair U., Cao Y., Richardson B.C., Li Z., Hughson F.M., Baba M., and Klionsky D.J.. 2010. The conserved oligomeric Golgi complex is involved in double-membrane vesicle formation during autophagy. J. Cell Biol. 188:101–114. 10.1083/jcb.200904075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S., Satoh A., Pypaert M., Mullen K., Hay J.C., and Ferro-Novick S.. 2006. mBet3p is required for homotypic COPII vesicle tethering in mammalian cells. J. Cell Biol. 174:359–368. 10.1083/jcb.200603044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., Li C.M., Luo X.M., Siu G.K.Y., Gan W.J., Zhang L., Wu W.K.K., Chan H.C., and Yu S.. 2017. Mammalian TRAPPIII Complex positively modulates the recruitment of Sec13/31 onto COPII vesicles. Sci. Rep. 7:43207 10.1038/srep43207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoppino F.C.M., Militello R.D., Slavin I., Álvarez C., and Colombo M.I.. 2010. Autophagosome formation depends on the small GTPase Rab1 and functional ER exit sites. Traffic. 11:1246–1261. 10.1111/j.1600-0854.2010.01086.x [DOI] [PubMed] [Google Scholar]
- Zou S., Liu Y., Zhang X.Q., Chen Y., Ye M., Zhu X., Yang S., Lipatova Z., Liang Y., and Segev N.. 2012. Modular TRAPP complexes regulate intracellular protein trafficking through multiple Ypt/Rab GTPases in Saccharomyces cerevisiae. Genetics. 191:451–460. 10.1534/genetics.112.139378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou S., Chen Y., Liu Y., Segev N., Yu S., Liu Y., Min G., Ye M., Zeng Y., Zhu X., et al. 2013. Trs130 participates in autophagy through GTPases Ypt31/32 in Saccharomyces cerevisiae. Traffic. 14:233–246. 10.1111/tra.12024 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.