Serebryannyy and Misteli provide a perspective on how protein sequestration at the inner nuclear membrane and nuclear lamina might influence aging.
Abstract
Despite the extensive description of numerous molecular changes associated with aging, insights into the driver mechanisms of this fundamental biological process are limited. Based on observations in the premature aging syndrome Hutchinson–Gilford progeria, we explore the possibility that protein regulation at the inner nuclear membrane and the nuclear lamina contributes to the aging process. In support, sequestration of nucleoplasmic proteins to the periphery impacts cell stemness, the response to cytotoxicity, proliferation, changes in chromatin state, and telomere stability. These observations point to the nuclear periphery as a central regulator of the aging phenotype.
Introduction
Physiological aging is generally defined as a decline in function over time from the cellular to organismal level (López-Otín et al., 2013). As a consequence, with increasing age we are faced with an elevated risk for a spectrum of complex diseases (see text box Premature aging disorders and the nucleus; Chung et al., 2009; Campisi, 2013; Kubben and Misteli, 2017). Deterioration of cellular functions occurs in response to both cell-intrinsic alterations, such as mitochondrial dysfunction and differentiation capacity, and environmental influences, including nutrient availability and endocrine signaling (López-Otín et al., 2013). Although the molecular details that regulate the aging process have remained largely elusive, several characteristics of aging are pervasive. These include changes in nutrient sensing, intercellular communication, proteostasis, mitochondrial dysfunction, cellular senescence, and adult stem cell exhaustion as well as nucleus-specific changes, including altered epigenetic marks, increased genome instability, and telomere attrition (López-Otín et al., 2013). The relative contributions and interdependence of these pathways to the aging process remain largely unclear.
Epidemiological studies have provided key details regarding the risk factors for human longevity (Sebastiani et al., 2012; López-Otín et al., 2013). Nevertheless, given ethical and practical issues in the study of human aging, model systems with shorter lifespans, such as yeast, worms, flies, fish, and mice, have been used extensively to gain molecular insight into evolutionarily conserved mechanisms of longevity and aging (Mitchell et al., 2015). These models have substantiated that aging involves defined cellular pathways that can be experimentally manipulated. A limitation of using animal models, especially non-mammalian systems, to deduce mechanisms of human aging is that the extent they recapitulate the human pathology is highly variable and largely unknown. For example, many short-lived animals used for aging studies such as Caenorhabditis elegans, zebrafish, killifish, mice, and rats, typically die of old age with intact and relatively long telomeres, whereas in aged humans, telomeres are shortened (Blackburn et al., 2015). In addition to their more complex physiology, humans, in contrast to laboratory animals, are exposed to complex environmental influences which in all likelihood contribute to aging. To circumvent some of these limitations, the existence of a wide set of genetic disorders that cause accelerated aging in humans has been exploited (see text box).
Premature aging disorders, or progerias, allow a unique glimpse into the potential mechanisms underlying physiological aging. However, given that no premature aging disorder fully recapitulates all features of human aging, these disorders are designated as segmental progerias (Dreesen and Stewart, 2011; Kubben and Misteli, 2017). Nevertheless, premature and physiological aging share similar features, and the insights gained from premature aging models appear often applicable to physiological aging (Dreesen and Stewart, 2011; López-Otín et al., 2013; Kubben and Misteli, 2017). Conversely, understanding the discrepancies between premature and physiological aging has also proven to be illuminating, such as in highlighting the molecular relationships between oncogenesis and aging (Fernandez et al., 2014; Zane et al., 2014). Intriguingly, a disproportionate number of premature aging disorders have been mapped to defects in nuclear proteins. In this Perspective, we discuss the contribution of nuclear dysfunction to pathological aging, and we specifically posit that the inner nuclear membrane (INM) and the lamina are regulators of the aging phenotype.
The INM
The distinguishing feature of eukaryotes is the nucleus: a segregated organelle that houses the genetic information of the cell. The nucleus is composed of a membrane bilayer: the outer nuclear membrane is contiguous with the ER and links the cytoskeleton to the nucleus, and the INM maintains nuclear architecture and aids in proper function (Hetzer, 2010; Fig. 1). The two nuclear membranes are connected at the numerous nuclear pore complexes that selectively traffic proteins between the cytoplasm and the nucleus (Katta et al., 2014).
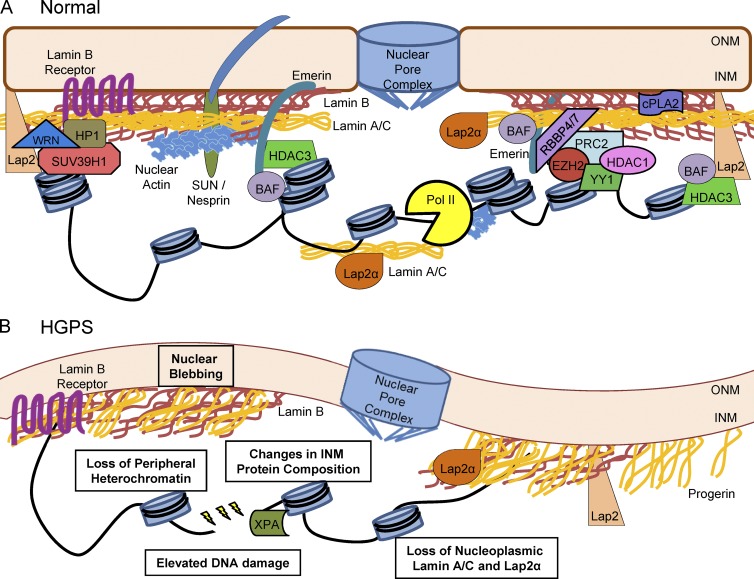
Figure 1.
The INM and lamina in normal and HGPS nuclei. (A) The INM is composed of an array of proteins responsible for maintaining nuclear function and an associated protein meshwork composed of tetrameric lamin filaments (Turgay et al., 2017). The nuclear lamina is composed of B-type lamins that are integrated into the INM and proximal but segregated lamin A/C filaments (Delbarre et al., 2006; Shimi et al., 2008, 2015; Xie et al., 2016). The structural integrity of the nucleus is also influenced by the SUN/Nesprin LINC complex that links the nucleus to the cytoskeleton (Zhang et al., 2005; Chang et al., 2015). Furthermore, integral INM proteins such as lamin B receptor, emerin, and Lap2 isoforms maintain heterochromatin at the periphery by interacting with chromatin remodeling complexes WRN, HP1/SUV39H1, PRC/NURD, and HDAC3/BAF (Makatsori et al., 2004; Somech et al., 2005; Shumaker et al., 2006; Holaska and Wilson, 2007; Montes de Oca et al., 2009; Pegoraro et al., 2009; Demmerle et al., 2012; Laugesen and Helin, 2014; Cesarini et al., 2015; Zhang et al., 2015b). Additionally, peripheral and nucleoplasmic lamin A/C may have different roles in transcription and replication via regulation of Lap2α, nuclear actin, and polymerases (Pol II; Dechat et al., 2000; Nili et al., 2001; Pekovic et al., 2007; Simon et al., 2010; de Lanerolle and Serebryannyy, 2011; Vidak et al., 2015). Through these interactions, the INM maintains proper DNA replication, DNA repair, proliferation, proinflammatory signaling via cytosolic phospholipase A2 (cPLA2), and differentiation (Dittmer and Misteli, 2011; Burke and Stewart, 2013; Enyedi et al., 2016). (B) Progerin expression and incorporation into the INM disrupts nuclear organization, causing misshapen nuclei (Goldman et al., 2004; Dahl et al., 2006; Verstraeten et al., 2008). HGPS models exhibit reduced expression of integral INM proteins such as lamin B1 and isoforms of Lap2 (Scaffidi and Misteli, 2005) and a corresponding loss of peripheral heterochromatin (Goldman et al., 2004; Scaffidi and Misteli, 2005; Shumaker et al., 2006; McCord et al., 2013). Not only is the composition of the periphery altered, but nucleoplasmic protein populations are also lost (i.e., lamin A/C and Lap2α; Dechat et al., 2000; Chojnowski et al., 2015; Vidak et al., 2015). These defects correlate with increased DNA damage, abnormal DNA damage repair, and impaired proliferation (Decker et al., 2009; Benson et al., 2010; Cao et al., 2011a; Musich and Zou, 2011; Chojnowski et al., 2015).
The INM contains a number of integral scaffolding proteins (e.g., emerin, LBR, Lap2β, SUN, and MAN1; Schirmer and Gerace, 2005) and is supported by an associated 3.5-nm protein meshwork composed of tetrameric lamin filaments, referred to as the nuclear lamina (Turgay et al., 2017; Fig. 1 A). In mammals, there are three differentially expressed lamin genes: LMNA, LMNB1, and LMNB2 (Dittmer and Misteli, 2011). LMNA is alternatively spliced into two major isoforms, lamin A and C. B-type lamins are incorporated into the INM, whereas an associated lamin A network is thought to remain proximal but largely segregated (Delbarre et al., 2006; Goldberg et al., 2008; Shimi et al., 2008, 2015; Xie et al., 2016). In addition to the lamina, the INM is physically connected to the cytoskeleton via the linker of nucleoskeleton and cytoskeleton complex (LINC; Lombardi and Lammerding, 2011; Chang et al., 2015). In this way, the nuclear periphery integrates both signaling and mechanical cues to regulate gene expression and gene positioning, as well as DNA replication, DNA repair, proliferation, proinflammatory signaling, and differentiation (Dittmer and Misteli, 2011; Burke and Stewart, 2013; Enyedi et al., 2016).
Proteomic analyses have highlighted the complexity of the INM. Earlier studies found that the INM is composed of ∼70 proteins involved in protein scaffolding, maintenance of structural stability, and genomic regulation (Dreger et al., 2001; Schirmer et al., 2003; Schirmer and Gerace, 2005). More recent research describes several hundred to several thousand proteins enriched at the nuclear envelope in a tissue-specific manner (Korfali et al., 2010, 2012; Wilkie et al., 2011; Smoyer et al., 2016; Thul et al., 2017). These discrepancies may be explained by a model wherein ER proteins that pass through the nuclear pore complex can diffuse along the INM, but only a subpopulation is retained (Ungricht et al., 2015; Smoyer et al., 2016). Evidence relating INM composition with function has come from INM-associated protein mutations that map to numerous pathologies including Charcot-Marie-Tooth disease, limb-girdle and Emery–Dreifuss muscular dystrophies, several myopathies, and most strikingly, premature aging disorders (Schirmer et al., 2003; Somech et al., 2005; Dittmer and Misteli, 2011).
Premature aging and the nuclear periphery
Strong evidence for a role for regulatory events at the nuclear periphery in aging comes from the premature aging disorder Hutchinson–Gilford progeria syndrome (HGPS; Fig. 1 B). This rare childhood disease, which occurs at a rate of ∼1 in 4 million births, is caused by a silent single-base mutation (classically 1824C > T) in the LMNA gene, and its symptoms include abnormal dentition, hair loss, joint contractures, growth retardation, lipodystrophy, osteoporosis and bone hypoplasia, sclerodermatous skin, skeletal muscle atrophy, and premature atherosclerosis with resultant cardiac failure in the early teens (De Sandre-Giovannoli et al., 2003; Eriksson et al., 2003; Merideth et al., 2008). The disease-causing mutation introduces a cryptic splice site in LMNA exon 11, resulting in heterozygous expression of a truncated form of lamin A, termed progerin (De Sandre-Giovannoli et al., 2003; Eriksson et al., 2003). Lamin A undergoes several processing steps including C-terminal farnesylation and subsequent cleavage by the endoprotease Zmpste24 (FACE-1), as well as tail methylation by isoprenylcysteine carboxylmethyltransferase (Barrowman and Michaelis, 2009; Davies et al., 2009; Ibrahim et al., 2013). Progerin, however, fails to undergo complete processing because it lacks the Zmpste24 cleavage site as a consequence of aberrant splicing in LMNA exon 11 caused by the disease mutation. Progerin incorporation into the nuclear lamina is thought to disrupt the lamin A and B networks, contributing to both structural and signaling dysfunction (Delbarre et al., 2006; Lee et al., 2016). In addition to classical HGPS, other mutations in lamin A can lead to atypical progerias that have symptoms similar to those of HGPS with varying severity (Chen et al., 2003; Csoka et al., 2004; Plasilova et al., 2004; Verstraeten et al., 2006; Liang et al., 2009; Doubaj et al., 2012). Mutations in ZMPSTE24 have also been documented, resulting in the accumulation of a farnesylated lamin A (prelamin A) and leading to restrictive dermopathy (RD), a usually more severe form of HGPS (Moulson et al., 2005; Navarro et al., 2005, 2014; Wang et al., 2016).
At the cellular level, progerin-expressing cells exhibit misshapen and lobulated nuclei both in vitro and in vivo, with some cell types affected more than others (De Sandre-Giovannoli et al., 2003; Goldman et al., 2004; Navarro et al., 2004; Dahl et al., 2006; Verstraeten et al., 2008). HGPS-induced changes in the nuclear lamina correlate with reduced expression of lamin B1 and some isoforms of the chromatin-binding Lap2 proteins (Scaffidi and Misteli, 2005). Lamin B1 loss has been observed in multiple models of cellular senescence and has been linked to changes in chromatin organization (Dreesen et al., 2013; Shah et al., 2013; Zhang et al., 2016). Furthermore, HGPS cells show a loss of peripheral heterochromatin (Goldman et al., 2004); repressive histone marks (H3K9me3 and H3K27me3; Scaffidi and Misteli, 2005; Shumaker et al., 2006; McCord et al., 2013; Zane et al., 2014); and association with heterochromatin protein 1 (HP1), enhancer of zeste homologue 2 (EZH2; Shumaker et al., 2006), and barrier to autointegration factor (BAF; Capanni et al., 2010; Loi et al., 2016). Intriguingly, BAF gene mutations result in Nestor–Guillermo progeria syndrome, a disease similar to, but less severe than, HGPS (Cabanillas et al., 2011; Puente et al., 2011). Along with changes in organization at the periphery, HGPS animal models also exhibit loss of the nucleoplasmic populations of lamin A/C and Lap2α (Naetar et al., 2017). These nucleoplasmic pools have been implicated in maintaining chromatin organization and gene expression and depletion of these nucleoplasmic pools may exacerbate the aging phenotype.
HGPS models exhibit defects in DNA replication, the cell cycle, and proliferation (Goldman et al., 2004; Dechat et al., 2007; Musich and Zou, 2011; Tang et al., 2012). These deficiencies correlate with increased DNA damage, abnormal DNA damage repair, and telomere dysfunction (Decker et al., 2009; Benson et al., 2010; Cao et al., 2011a; Musich and Zou, 2011; Chojnowski et al., 2015; Fig. 1 B). Progerin expression also leads to extranuclear defects such as inflammation (Di Micco et al., 2016; Tran et al., 2016), increased mitochondrial dysfunction (Peinado et al., 2011; Richards et al., 2011; Rivera-Torres et al., 2013; Kubben et al., 2016; Xiong et al., 2016), altered autophagy activation (Mariño et al., 2008), and changes to the ECM (Csoka et al., 2004; Hernandez et al., 2010; de la Rosa et al., 2013; Vidak and Foisner, 2016). Globally, proteomic and yeast two-hybrid screens have identified 35 and 225 proteins, respectively, that differentially interact with progerin versus wild-type lamin A (Kubben et al., 2010; Dittmer et al., 2014). Furthermore, RNA profiling has found drastic changes in the transcription program of HGPS models (Ly et al., 2000; Csoka et al., 2004; Scaffidi and Misteli, 2008; Marji et al., 2010; Kubben et al., 2012; Wang et al., 2015; Chen et al., 2017), which may be a consequence of the role of the lamina in gene positioning (Meaburn, 2016; Shachar and Misteli, 2017) and transcription regulation (Kumaran et al., 2002; Spann et al., 2002).
In normal human fibroblasts, progerin expression can be induced as a consequence of telomere shortening or uncapping (Cao et al., 2011a), as well as UV damage (Takeuchi and Rünger, 2013), strongly suggesting that aging may directly influence nuclear integrity. In support, several studies have reported increased sensitivity to or accumulation of progerin or prelamin A in physiological aging models (Cao et al., 2007; McClintock et al., 2007; Scaffidi and Misteli, 2008; Rodriguez et al., 2009; Marji et al., 2010; Olive et al., 2010; Ragnauth et al., 2010; Luo et al., 2013; Lattanzi et al., 2014; Petrini et al., 2017). Furthermore, single nucleotide polymorphisms in LMNA have been correlated with long-lived individuals (Conneely et al., 2012), and treatments that increase human longevity (i.e., rapamycin, metformin, statins) appear to alleviate symptoms of HGPS, at least in vitro (Varela et al., 2008; Cao et al., 2011b; Egesipe et al., 2016). Conversely, some HIV protease inhibitor treatments impair Zmpste24 function and lead to HGPS-like defects (Barrowman and Michaelis, 2009; Mehmood et al., 2016). Whereas the accumulation of progerin has a dose-dependent effect on cellular dysfunction (Moulson et al., 2007; Hisama et al., 2011; Chojnowski et al., 2015), the depletion of progerin by reversion of HGPS-derived fibroblasts into induced pluripotent stem cells reverses nuclear defects until progerin is reexpressed upon differentiation into mesenchymal stem cells (MSCs; Liu et al., 2011; Zhang et al., 2011b; Miller et al., 2013; Chen et al., 2017). Together, these studies implicate lamin A variants in HGPS and, most likely, in physiological aging.
Sequestration at the nuclear periphery in the aging process
The observation that HGPS and RD are caused by mutations that directly affect nuclear lamina proteins points to the nuclear periphery as a potential contributor to premature, and possibly normal, aging. Notably, expression of lamin A variants has been detected in physiological models of aging such as extensively passaged normal human fibroblasts and vascular smooth muscle cells, fibroblasts from centenarians, aged-patient skin biopsies, and sections from coronary arteries (Cao et al., 2007; McClintock et al., 2007; Rodriguez et al., 2009; Olive et al., 2010; Ragnauth et al., 2010; Lattanzi et al., 2014). In addition, many of the cellular defects observed in HGPS, such as elevated levels of DNA damage and telomere dysfunction, are the primary causes of other premature aging disorders (see text box), potentially placing the nuclear periphery upstream of these processes. Several observations in multiple premature aging disorders suggest that sequestration of proteins at the nuclear periphery, or loss thereof, may be responsible for prominent defects associated with premature and physiological aging. In particular, peripheral regulation appears to affect several key features of aging including impaired cell stemness, dysregulated cytotoxicity, and increased senescence, as well as aberrant chromatin structure and telomere stability.
Premature aging disorders and the nucleus
Most human premature aging disorders are caused by mutations in the cellular machinery involved in DNA metabolism (i.e., DNA replication, epigenetic regulation, telomere maintenance, and DNA damage signaling and repair). For example, mutations in LMNA cause HGPS and atypical Werner syndrome, mutation of ZMPSTE24 causes restrictive dermopathy, and mutations in BANF1 cause Nestor–Guillermo progeria syndrome (see Premature aging and the nuclear periphery). In addition, mutations in the RecQ helicases WRN, BLM, and RecQL4 cause the premature aging disorders Werner syndrome, Bloom syndrome, and Rothmund-Thomson syndrome, respectively. These helicases have conserved as well as nonredundant functions. WRN is primarily involved in DNA repair via nonhomologous end joining (NHEJ) and in DNA replication, H3K9 methylation, and telomere regulation (Croteau et al., 2014; Zhang et al., 2015b; de Renty and Ellis, 2017). BLM and RecQL4 are necessary for proper homologous recombination (HR) and DNA replication (Smeets et al., 2014; de Renty and Ellis, 2017). Together the RecQ diseases highlight how variations in DNA maintenance create distinct pathologies. Whereas ablation of RecQ function irrespective of isoform increases susceptibility to cancer, the differences in aging phenotypes are more nuanced (i.e., premature onset vs. developmental abnormalities).
Similar to NHEJ and HR, mutations in the nucleotide excision DNA damage repair pathway (NER) result in the premature aging disorders xeroderma pigmentosum (mutations in global NER), Cockayne syndrome (mutations in transcription-coupled NER; Mitchell et al., 2015), and trichothiodystrophy (mutations in the transcription factor IIH complex; Stefanini et al., 2010). How defects in NER cause progeroid and neurological pathologies as well as a reduced lifespan are poorly understood (Stefanini et al., 2010; Wilson et al., 2016). Nevertheless, the decreased life expectancy resulting from mutations in the NER pathway and the RecQ helicases strongly implicate DNA damage repair as a contributor to the aging phenotype. However, a caveat is revealed by defects in components of the DNA damage signaling and repair cascade such as ATM protein kinase (McKinnon, 2012) and the Fanconi anemia complex (Taniguchi et al., 2002) which lead to cancer, neurodegeneration, or microcephaly, but not premature aging. Together this set of disorders highlight that defective DNA damage signaling is insufficient to cause progeria, but the type and extent of damage are important determinants of disease pathology.
In support of an involvement of telomere maintenance in aging, mutations in several telomere regulatory proteins (i.e., TERT, TERC, DKC1, and TINF2, among others) lead to dyskeratosis congenita (Opresko and Shay, 2017). This monogenic telomere disease is hallmarked by genetic anticipation, where the lifespans of mutation carriers decrease with succeeding generations. Furthermore, telomere length has repeatedly been correlated with human morbidity in large cohort studies examining physiological aging (Blackburn et al., 2015). In addition, persistent DNA damage can disproportionally affect telomeres and telomere-associated repair foci increase with age (Hewitt et al., 2012). This telomere-specific damage has been reported to be irreparable, driving cellular senescence, and presumably contributing to organismal aging (Fumagalli et al., 2012). Intriguingly, fibroblasts derived from patients with dyskeratosis congenita have been shown to express progerin, further establishing the link between telomere attrition, senescence, and regulation of the nuclear lamina (Cao et al., 2011a).
An exception to the nuclear-based progeroid diseases is a mouse model with spontaneous mutations in mitochondrial DNA induced by homozygous expression of a proofreading-deficient catalytic subunit of mtDNA polymerase (POLG; Trifunovic et al., 2004; Kujoth et al., 2005; Bratic and Larsson, 2013). These mice have respiratory chain dysfunction and mitochondrial damage, coinciding with a shortened lifespan along with premature occurrence of alopecia, anemia, cardiomyopathy, kyphosis, reduced fertility, as well as hearing, hair, and weight loss. Although no corresponding naturally occurring human mutations are known, increased mitochondrial mutations are thought to occur during human aging (Bratic and Larsson, 2013). Current evidence indicates that stem cells are particularly sensitive to mitochondrial dysfunction (Hämäläinen et al., 2015), implying that rapid embryonic stem cell depletion may contribute to the premature aging phenotype (Ahlqvist et al., 2012, 2015). Curiously, mice with mutations in the mitochondrial DNA helicase Twinkle also display an accumulation of mtDNA mutations but do not exhibit premature aging symptoms (Tyynismaa et al., 2005). Hence, despite the dramatic phenotype, how mtDNA polymerase dysfunction causes premature aging remains poorly understood. One avenue may be through a number of mechanisms for mitochondrion-nucleus cross talk (Sahin et al., 2011; Cagin and Enriquez, 2015; Li et al., 2016; Lionaki et al., 2016; Quirós et al., 2016). However, if nuclear (dys)function contributes to the pathology in these models has not been examined.
Cell stemness
The regenerative potential of many tissues declines with age, contributing to dysfunction and degeneration (López-Otín et al., 2013). Although the progressive loss of stem cell function by a number of different stimuli has been associated with aging, HGPS models have highlighted that dysregulation of the pathways governing stem cell fate can likewise contribute to stem cell exhaustion and the aging phenotype (Scaffidi and Misteli, 2008; Rosengardten et al., 2011).
Notch
Notch activation regulates proliferation, migration, and differentiation and is critical in stem cell homeostasis (Conboy et al., 2003, 2005; Bjornson et al., 2012; Balistreri et al., 2016; Bray, 2016). Activation of the Notch receptor induces cleavage of the intracellular domain of the transmembrane receptor and translocation into the nucleus, where it interacts with transcriptional activators and repressors (Balistreri et al., 2016; Bray, 2016). In physiological aging, loss of Notch signaling prevents stem cell renewal in muscle and contributes to cardiovascular anomalies, whereas elevated Notch signaling can lead to cancer and promotes inflammation via nuclear factor-κB, MAPK, and TGFβ (Balistreri et al., 2016).
Progerin expression in human MSCs causes activation of Notch target genes and results in the loss of stem cell identity and differentiation defects (Scaffidi and Misteli, 2008). Interestingly, gene activation occurs in the absence of induction of the upstream portions of the pathway, consistent with the nuclear progerin protein being the driver of these effects. Mechanistically, progerin expression reduces levels of nuclear receptor corepressor (NCOR) and increases levels of the Notch pathway coactivator, SKI-interacting protein (SKIP; Zhou et al., 2000; Scaffidi and Misteli, 2008). NCOR and SKIP are anchored to the nuclear periphery in normal cells (Zhang et al., 2003; Demmerle et al., 2012; Fig. 2 A). NCOR is activated when sequestered to the nuclear periphery (Demmerle et al., 2012), whereas SKIP may be restricted from binding to Notch-dependent promoters. The increase in Notch signaling upon progerin expression coincides with SKIP relocalization from the periphery and into the nucleoplasm (Scaffidi and Misteli, 2008). Release of SKIP is thought to allow promoter binding, leading to the observed increases in Notch pathway activation and stem cell depletion (Scaffidi and Misteli, 2008). However, it is uncertain to what extent changes in the INM localization of other Notch cofactors, e.g., sirtuin 1 (SIRT1) or polycomb repressive complex (PRC), alter signaling (Bray, 2016). In support of a role for Notch dysregulation in HGPS, mice expressing progerin have altered wound healing and reduced stem cell function (Rosengardten et al., 2011), indicative of premature stem cell differentiation. Hence, nuclear defects that accrue with aging may hasten stem cell depletion and enrich for a Notch-insensitive population by altering peripheral regulation of NCOR/SKIP.
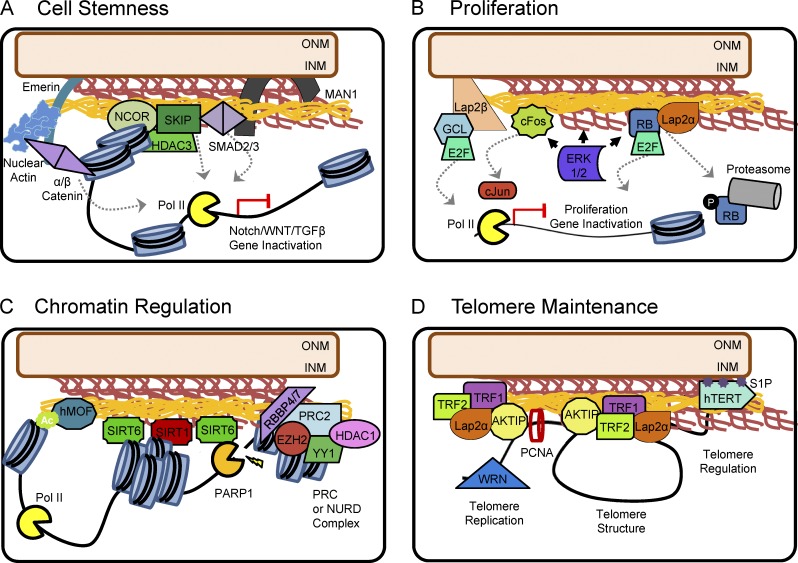
Figure 2.
Mechanisms of protein sequestration at the nuclear periphery. (A) The INM regulates cell stemness via sequestration of Notch, Wnt, and TGFβ pathway effectors. The Notch signaling corepressor NCOR and coactivator SKIP are anchored to the nuclear periphery in normal cells (Zhang et al., 2003; Demmerle et al., 2012). Although NCOR is activated at the nuclear periphery, SKIP may be restricted from binding to Notch-dependent promoters. SKIP has also been shown to interact with SMAD2/3 to regulate TGFβ-dependent transcription (Leong et al., 2001). Similarly, SMAD2/3 is bound to MAN1. The interaction with MAN1 sequesters SMAD2/3, attenuating heterodimerization with Smad4 and suppressing TGFβ-induced transcription (Lin et al., 2005; Pan et al., 2005). Wnt pathway regulation involves emerin-mediated nuclear export of β-catenin as well as the possible stabilization of β-catenin by α-catenin and nuclear actin (Markiewicz et al., 2006; Holaska and Wilson, 2007; Tilgner et al., 2009; Stubenvoll et al., 2015; Serebryannyy et al., 2017). (B) Proliferation is regulated by the interaction of the nuclear lamina with AP-1, Rb, and ERK1/2. Hypophosphorylated cFos and Rb are sequestered by the nuclear lamina, priming a readily available population of transcription factors for rapid cell cycle regulation (González et al., 2008; Rodríguez et al., 2010). Activation and nuclear translocation of ERK1/2 contribute to the phosphorylation of Rb and cFos. Phosphorylated Rb is degraded, freeing E2F to activate transcription. Similarly, phosphorylation of cFos facilitates dimerization with cJun and promoter binding. E2F is also sequestered to the nuclear periphery via its interaction with Germ cell-less (GCL) and Lap2β (Nili et al., 2001). Nuclear ERK1/2 can bind to lamin A/C as well (Rodríguez et al., 2010), potentially regulating its activity and turnover. (C) Lamin A is required for the proper localization and enzymatic activity of SIRT1 and SIRT6, regulating chromatin condensation and poly(ADP-ribose) polymerase 1 (PARP1) activity (Liu and Zhou, 2013; Ghosh et al., 2015). hMOF localization is also dependent on lamin A/C (Füllgrabe et al., 2013), implicating the nuclear lamina in both histone acetylation and deacetylation. In addition, lamin A binds the chromatin remodeling complexes PRC and NuRD, establishing a repressive heterochromatin state at the INM (Pegoraro et al., 2009; Cesarini et al., 2015). (D) The shelterin complex components TRF1/2 and AKTIP regulate telomere replication, length, and stability (Ludérus et al., 1996; Dechat et al., 2004; Wood et al., 2014; Chojnowski et al., 2015). Whereas TRF1 may bind lamin B (Crabbe et al., 2012), TRF2 stabilization of telomeres at the nuclear periphery is dependent on binding to lamin A/C and Lap2α (Chojnowski et al., 2015). Similarly, AKTIP interacts with lamin A/C, lamin B, and PCNA to regulate telomere replication and stability (Burla et al., 2016). The INM-incorporated lipid moiety S1P is also able to promote telomere stability by preventing the degradation of hTERT (Panneer Selvam et al., 2015).
Wnt
In conjunction with Notch, the Wnt pathway regulates the balance between stem cell proliferation and differentiation (Brack et al., 2007, 2008; Clevers and Nusse, 2012; Borggrefe et al., 2016). Wnt hyperactivation triggers accelerated cellular senescence (Liu et al., 2007), and aberrant Wnt signaling leads to cancer (Clevers and Nusse, 2012). In the cytoplasm, β-catenin is continuously degraded or maintained at adherens junctions. Upon Wnt receptor activation, β-catenin translocates into the nucleus, where it regulates transcription via lymphoid enhancer binding factor (LEF), T cell factor, and chromatin remodeling (Lien and Fuchs, 2014; McCrea and Gottardi, 2016). The nuclear β-catenin complex also mediates the DNA damage response (Zhang et al., 2011a; Serebryannyy et al., 2017) and oxidative stress sensitivity (Essers et al., 2005) and maintains telomerase levels (Hoffmeyer et al., 2012). Expression of progerin or a truncated form of lamin A reduces nuclear β-catenin/LEF1 levels, contributing to defective ECM synthesis and proliferation arrest (Hernandez et al., 2010). Similarly, mice knocked out for Zmpste24 have a defect in proliferation caused by decreased β-catenin levels (Espada et al., 2008), and mice deficient for LEF1 have defects in regulating bone formation (Noh et al., 2009), supporting a role for the Wnt pathway in HGPS and aging.
Although lamin A expression and nuclear β-catenin levels are correlated (Bermeo et al., 2015), how lamin A regulates β-catenin remains unclear. One mechanism may be through the INM protein emerin (Fig. 2 A), which facilitates β-catenin export from the nucleus (Markiewicz et al., 2006; Tilgner et al., 2009; Stubenvoll et al., 2015). Both lamin A and progerin interact with emerin (Kubben et al., 2010; Chojnowski et al., 2015). Although lamin A, nesprin-2, and protein 4.1R retain emerin at the INM (Zhang et al., 2005; Kubben et al., 2010; Meyer et al., 2011; Wu et al., 2014), its enrichment at the nuclear periphery appears decreased in progerin-expressing cells (Kubben et al., 2010; Eisch et al., 2016). It is tempting to speculate that mislocalization of emerin may impede proper β-catenin signaling. Alternatively, emerin binding to β-catenin at the INM may prevent β-catenin degradation. Both emerin and β-catenin also interact with nuclear actin (Holaska and Wilson, 2007; de Lanerolle and Serebryannyy, 2011; Daugherty et al., 2014; Serebryannyy et al., 2017; Fig. 2 A), and the interaction with nuclear actin may tether and stabilize the complex selectively to lamin A but not progerin (Simon et al., 2010). It is also possible that β-catenin/LEF1 may be regulated indirectly via other INM-associated proteins such as Yin Yang 1 (YY1), which binds lamin A/C and positions chromatin at the nuclear periphery (Harr et al., 2015). YY1 also directly binds the LEF1 promoter and represses its transcription (Yokoyama et al., 2010). Therefore, changes to the nuclear periphery may alter YY1 activity and consequently LEF1 levels. Regardless of the mechanism, these observations suggest that the nuclear periphery regulates the effectiveness of Wnt pathway signaling.
TGFβ
TGFβ signaling promotes stem cell proliferation and renewal in a context-dependent manner (Watabe and Miyazono, 2009). Canonical TGFβ activation causes the phosphorylation and nuclear translocation of Smad2/3 transcriptional modulators, heterodimerization with Smad4, and chromatin binding (Watabe and Miyazono, 2009; Zhang, 2009). However, Smad proteins also signal via other pathways. In bone marrow–derived adult human MSCs, Smad3 activation induces nuclear translocation of β-catenin to promote proliferation and prevent differentiation (Jian et al., 2006). In differentiated mouse embryonic fibroblasts, TGFβ inhibits proliferation by dephosphorylating retinoblastoma protein (Rb) in a lamin A/C-protein phosphatase 2A (PP2A)–dependent manner (Van Berlo et al., 2005). Lamin A/C also facilitates dephosphorylation of Smad2 by PP2A, suggesting that the nuclear periphery mediates TGFβ-induced proliferation (Van Berlo et al., 2005).
Although it is not known how these interactions are altered in HGPS, patients with mutations in another INM protein, MAN1, are symptomatic for patches of abnormally increased bone density and disseminated patches of connective tissue including osteopoikilosis, Buschke–Ollendorff syndrome, and melorheostosis, which are associated with increased TGFβ signaling (Hellemans et al., 2004). Similar to lamin A/C, MAN1 binds phosphorylated Smad2/3 (Lin et al., 2005; Pan et al., 2005; Fig. 2 A). Binding to MAN1 reduces Smad2/3 phosphorylation, prevents heterodimerization with Smad4, alters the nuclear localization of activated Smad2/3, and ultimately suppresses TGFβ-induced transcription. Intriguingly, microarray analysis suggests that TGFβ signaling in fibroblasts derived from old individuals closely resembles that of cells derived from HGPS patients (Aliper et al., 2015).
The reactive oxygen species (ROS) response
With age, mitochondria lose their respiratory activity because of a buildup of oxidative damage, propagating a cycle of increasing oxidative stress and mitochondrial dysfunction (Bratic and Larsson, 2013; López-Otín et al., 2013; Ahlqvist et al., 2015). Accumulation of ROS can trigger loss of protein function, aberrant activation of signaling pathways, and the accumulation of DNA damage (Bratic and Larsson, 2013). As in physiological aging models, HGPS cells exhibit signs of mitochondrial dysfunction and increased ROS levels (Kubben et al., 2016; Xiong et al., 2016), implicating signaling from the nuclear periphery in mitochondria homeostasis.
Nuclear factor erythroid 2–related factor 2 (Nrf2)
Nrf2 is a transcriptional activator of a global cytoprotective response to reduce chemical and oxidative stress (Ma, 2013). Nrf2 is continuously degraded by the actin-bound Keap1/Cul3 ubiquitin ligase complex. When redox-sensitive cysteine residues in Keap1 are modified, Nrf2 is released and translocates into the nucleus to transcriptionally regulate autophagy, inflammasome activation, ER stress, and the unfolded protein response, as well as mitochondrial redox signaling (Ma, 2013). Nrf2 levels change with age in a tissue- and model-dependent manner (Zhang et al., 2015a). Yet it is poorly understood whether Nrf2 levels are increased in response to higher ROS damage with age or decreased as the capacity to respond to stress is impaired with age. This is further complicated by regulation of Nrf2 by multiple signaling factors including Notch (Wakabayashi et al., 2014), c-Myc, and DNA damage proteins (Zhang et al., 2015a). Nevertheless, given the long-standing hypothesis that unregulated ROS production contributes to physiological aging (Harman, 1956), Nrf2 may be a major contributor to the aging phenotype.
In HGPS fibroblasts, Nrf2 is improperly sequestered to the nuclear periphery by progerin, reducing its nucleoplasmic concentration and impairing Nrf2-dependent transcription (Kubben et al., 2016). In contrast, mutations in lamin A that cause various muscular dystrophies induce Nrf2 nuclear translocation and transcriptional activation (Dialynas et al., 2015). Hence, although some lamin A mutants are able to induce Nrf2 signaling, progerin impairs the Nrf2-mediated response to oxidative stress by sequestration. This finding explains why HGPS models have increased ROS levels and mitochondrial dysfunction (Viteri et al., 2010; Peinado et al., 2011; Richards et al., 2011; Rivera-Torres et al., 2013; Kubben et al., 2016; Xiong et al., 2016). Because this effect may be stressor dependent (Hashimoto et al., 2017), it will be important to determine whether Nrf2 is sequestered at the nuclear lamina in physiological aging.
Octamer-binding transcription factor 1 (Oct-1)
Oct-1 regulates several transcriptional pathways, including the cellular stress response, and represses aging-related collagenase genes (Imai et al., 1997; Malhas et al., 2009; Zhao, 2013). Similar to Nrf2, cellular stress stabilizes Oct-1 and promotes cell survival (Zhao, 2013). A fraction of nuclear Oct-1 associates with lamin B1 at the nuclear periphery (Kim et al., 1996; Malhas et al., 2009; Columbaro et al., 2013). Intriguingly, increasing the passage number of a preimmortalized cell line results in the loss of lamina-retained Oct-1 and increased collagenase gene expression (Imai et al., 1997). The JNK-dependent association of Oct-1 with lamin B1 has been estimated to differentially regulate 57 genes, many of which are involved in the oxidative stress response (Malhas et al., 2009; Boubriak et al., 2017). Knockdown of either Oct-1 or lamin B1 increases ROS accumulation and sensitivity to oxidative stress, whereas co-knockdown increases cell survival (Malhas et al., 2009; Shimi et al., 2011). Conversely, duplication of the LMNB1 gene in adult-onset autosomal dominant leukodystrophy, a disease characterized by the progressive degradation of myelin, increases sequestration of Oct-1 at the nuclear periphery (Columbaro et al., 2013). Therefore, retention of Oct-1 by lamin B1 may be a means to restrict Oct-1 binding to chromatin. Alternatively, peripheral Oct-1 may aid in recruiting genomic regions to the nuclear lamina; however, Oct-1 knockout does not appear to affect the interaction between lamin B1 and chromatin (Meuleman et al., 2013). Further studies should determine whether loss of lamin B during senescence or in HGPS affects the localization and activity of Oct-1, and what downstream impact this interaction has on ROS accumulation and the aging phenotype.
Proliferation
The inhibition of proliferation and accumulation of senescent cells has been strongly suggested to cause an age-associated decline in function via triggering of secretion of a set of inflammatory cytokines, growth factors, and interleukins, referred to as senescence-associated inflammatory/secretory phenotype, as well as the depletion of stem and progenitor cells (van Deursen, 2014; Childs et al., 2015; Baker et al., 2016). The nuclear periphery, in concert with a series of intricate checkpoints and signaling pathways, regulates the proliferative potential of the cell, as evidenced by the induction of senescence in HGPS models (Varela et al., 2005).
Rb
Cellular stressors such as DNA damage, hypoxia, and ROS activate the tumor suppressor protein p53 (Lasry and Ben-Neriah, 2015). p53 subsequently activates p21, and unresolved stress leads to downstream activation of p16INK4A and inhibition of the kinases CDK2, CDK4, and CDK6 (Childs et al., 2015; Lasry and Ben-Neriah, 2015). Inhibition of these cyclin-dependent kinases prevents Rb phosphorylation, facilitating the interaction of unphosphorylated Rb with and inhibition of the E2F transcription factors. This regulatory mechanism prevents damaged cells from proliferating by inhibiting cell cycle progression, alters transcription of a large number of gene targets including the RecQ helicases (Yamabe et al., 1998; Dimova and Dyson, 2005; Liu et al., 2008), and induces cellular senescence (Lasry and Ben-Neriah, 2015).
Hypophosphorylated Rb is sequestered via lamin A/C and Lap2α (Mittnacht and Weinberg, 1991; Templeton et al., 1991; Mancini et al., 1994; Kennedy et al., 2000; Markiewicz et al., 2002; Johnson et al., 2004; Pekovic et al., 2007; Fig. 2 B). The lamin–Rb interaction prevents complex degradation in a SUMO-dependent manner (Johnson et al., 2004; Sharma and Kuehn, 2016) and promotes further TGFβ-mediated Rb dephosphorylation (Van Berlo et al., 2005). It remains uncertain whether sequestration of unphosphorylated Rb also withdraws E2F to prevent transcription or whether Rb sequestration increases E2F signaling by impeding the E2F–Rb interaction (Nitta et al., 2006; Pekovic et al., 2007); however, current evidence suggests that lamin A/C acts to restrict proliferation (Johnson et al., 2004; Nitta et al., 2006; Naetar et al., 2008; Rodríguez et al., 2010). Furthermore, the lamin–Rb complex may exist in the nucleoplasm bound to Lap2α as well as at the periphery (Naetar et al., 2008; Rodríguez et al., 2010), and the function of these two pools remains a point of interest. Regardless, the nuclear retention of lamin A/C and stabilization of hypophosphorylated Rb likely prime a readily available population of Rb for rapid cell cycle regulation (Nitta et al., 2006). Similar to Rb, another E2F repressor termed Germ cell-less is tethered to INM-localized Lap2β. Germ cell-less is proposed to bind and recruit E2F to the periphery, preventing E2F-dependent transcription (Nili et al., 2001; Fig. 2 B).
In fibroblasts derived from HGPS patients, cell cycle dysfunction correlates with altered Rb signaling (Dechat et al., 2007; Marji et al., 2010), despite hyperactivation of p53 (Varela et al., 2005; Liu et al., 2006; Kudlow et al., 2008; Wheaton et al., 2017). Similar to HGPS, fibroblasts derived from normally aged elderly individuals exhibit signs of increased progerin expression and a respective decrease in Rb levels (Han et al., 2008; Marji et al., 2010). Therefore, progerin, unlike lamin A/C, may not protect Rb from degradation, causing the Rb pools to be exhausted too quickly. Furthermore, whereas Rb binding to the nuclear lamina regulates proliferation, release of Rb from the lamina may facilitate inhibitory binding of Rb with E2F on promoters, causing global changes in chromatin conformation and inducing senescence-associated heterochromatin foci (Narita et al., 2003). This process may be mediated via loss of lamin B1 (Shimi et al., 2011; Sadaie et al., 2013) and partially parallels the changes in chromatin organization that occur in HGPS (Chandra et al., 2015). Although multiple studies have confirmed the lamin–Rb interaction, and it is clear that Rb plays a critical role in proliferation, how the lamin–Rb complex contributes to aging warrants further study.
Activating protein 1 (AP-1)
The transcription factors Fos and Jun heterodimerize to form the AP-1 complex (Eferl and Wagner, 2003). c-Fos and c-Jun respond to mitogenic signals to translocate into the nucleus and promote cell cycle progression by activating transcription of cyclins and repressing p53 and p16INK4A (Eferl and Wagner, 2003). AP-1 is regulated by many of the pathways discussed here, including Notch (Chu et al., 2002), Wnt (Lien and Fuchs, 2014), TGFβ (Verrecchia et al., 2001), and NRF2 (Zhang et al., 2015a). As with Rb, lamin A/C is able to bind dephosphorylated c-Fos at the nuclear periphery, preventing heterodimerization with c-Jun and chromatin binding (Ivorra et al., 2006; González et al., 2008; Fig. 2 B). In response to mitogenic signals, ERK1/2 is phosphorylated and, in turn, increases expression of c-Fos and c-Jun (Eferl and Wagner, 2003). ERK1/2 also phosphorylates c-Fos to release it from the nuclear periphery, activating AP-1 transcription (González et al., 2008).
Notably, ERK1/2 interacts with lamin A/C as well, which may facilitate efficient activation of cotethered c-Fos, or lamin A/C may sequester ERK1/2 in a dormant state at the periphery. In support of the latter hypothesis, decreased lamin A/C expression correlates with increased ERK1/2 activation (Muchir et al., 2009b), and a mutation in the N terminus of lamin A/C that leads to Emery–Dreifuss muscular dystrophy exhibits increased ERK1/2 signaling (Muchir et al., 2007, 2009a). Interestingly, lamin A/C binds ERK1/2 and Rb in a mutually exclusive manner: activation of ERK1/2 dislodges lamin A/C–bound Rb at the nuclear periphery, promoting Rb phosphorylation, degradation, and cell cycle activation (Rodríguez et al., 2010; Fig. 2 B). In this way, as increasing ERK enters the nucleus, more Rb is dislodged from lamin A/C and degraded (Rodríguez et al., 2010). Similarly, more c-Fos is phosphorylated and released from lamin A/C (González et al., 2008). Both processes work together to induce proliferation. As lamin A/C becomes unbound from Rb and c-Fos, increasing amounts of ERK could be tethered to the lamina as a feedback mechanism to attenuate ERK signaling.
Proliferating cell nuclear antigen (PCNA)
PCNA is a necessary component of the DNA replication machinery and has roles in DNA damage repair and telomere stability (Vannier et al., 2013; Choe and Moldovan, 2017). PCNA expression decreases with age (Goukassian et al., 2000), and delayed induction of PCNA in aged rats may contribute to the accumulation of DNA damage (Kaneko et al., 2002). Indeed, a homozygous missense mutation in PCNA causes premature aging, likely induced by increased DNA damage sensitivity (Baple et al., 2014). Furthermore, PCNA interacts with lamins (Shumaker et al., 2008), BAF (Montes de Oca et al., 2009), and the helicase WRN (Lebel et al., 1999). A- and B-type lamins are thought to aid in the localization of PCNA (Shumaker et al., 2008), and disruption of the nuclear lamina using lamin mutants or progerin causes sequestration and aggregation of PCNA, interfering with replication (Spann et al., 1997; Moir et al., 2000; Hilton et al., 2017; Wheaton et al., 2017). Likewise, expression of prelamin A or progerin causes PCNA dysfunction, leading to stalled replication forks that become double-stranded breaks upon collapse (Cobb et al., 2016; Hilton et al., 2017; Wheaton et al., 2017). The impaired proliferation, increased DNA damage, and destabilized telomeres found in HGPS, physiological aging, and PCNA dysfunction implicate PCNA as a potential mediator of both physiological and pathological aging.
Chromatin remodeling
Changes in histone-modifying enzymes have been observed to correlate with lifespan in a wide range of organisms (López-Otín et al., 2013; Zane et al., 2014). Yet little is known regarding the relationships between the array of different chromatin remodeling enzymes and their effects on longevity. Nevertheless, given the importance of the nuclear lamina in genome organization, it is unsurprising that HGPS and senescing cells undergo a drastic remodeling of the chromatin landscape that correlates with changes in the nuclear lamina (Shah et al., 2013; Chandra et al., 2015).
SIRTs
SIRTs are a family of NAD-dependent deacetylases, which have diverse roles in metabolism, cancer, and aging (López-Otín et al., 2013). SIRT1 and SIRT6 regulate longevity via their roles in genomic stability, metabolic regulation, and chromatin modification (Liu and Zhou, 2013). SIRT1 binding to lamin A, but not progerin or prelamin A, properly activates SIRT1 (Liu and Zhou, 2013; Fig. 2 C). SIRT1 is also responsible for autophagy-mediated deacetylation of histone H4K16 (Füllgrabe et al., 2013), linking a full autophagy response to lamin A expression. Whereas SIRT1 decreases H4K16ac levels, hMOF, an acetyl transferase that increases H4K16ac, is down-regulated upon autophagy induction (Krishnan et al., 2011; Füllgrabe et al., 2013). hMOF has also been shown to bind to lamin A but not prelamin A, and this interaction facilitates localization to the nuclear periphery (Krishnan et al., 2011). Although the role of the hMOF–lamin A interaction in autophagy is unknown, lamin A may coordinate proper H4K16 acetylation. Similar to aged cycling fibroblasts (Zane et al., 2014), prelamin A–expressing cells are hypoacetylated at H4K16 (Krishnan et al., 2011), which correlates with defects in DNA damage repair and premature senescence. Determining how the balance between SIRT1 and hMOF activity is regulated by the nuclear lamina may give valuable insights into the peripheral regulation of heterochromatin and euchromatin.
SIRT6 deacetylates several histone lysines on histone H3, including on aa position K9, K27, K18, and K56, to aid in DNA damage repair (Zane et al., 2014) as well as prime telomeres for WRN binding (Tasselli et al., 2017). SIRT6 also represses expression of embryonic stem cell transcription factors, in addition to targets of the AP-1 complex, and coactivates NRF2 (Sundaresan et al., 2012; Pan et al., 2016; Tasselli et al., 2017). Like SIRT1, SIRT6 binding to lamin A, but not progerin, increases enzymatic activity (Ghosh et al., 2015). Lamin A is necessary for SIRT6 recruitment to DNA damage and proper chromatin interaction (Fig. 2 C). However, in HGPS models, SIRT6 activity and expression are reduced, which may contribute to the defects in DNA damage repair, chromatin organization, and telomere maintenance (Endisha et al., 2015; Ghosh et al., 2015). Although the role of peripheral versus nucleoplasmic lamin A is still in question, these studies demonstrate that controlled expression of lamin A is critical for proper function of SIRT1 and SIRT6.
Histone methylation complexes
Retinoblastoma binding protein (Rbbp)4/7 are members of the nucleosome remodeling and deacetylase (NuRD) complex and the polycomb repressive complex 2 (PRC2; Laugesen and Helin, 2014). These complexes are generally responsible for heterochromatin maintenance and both Rbbp4 and 7 bind lamin A and BAF, but not progerin (Montes de Oca et al., 2009; Pegoraro et al., 2009; Fig. 2 C). Indeed, cells derived from HGPS patients or fibroblasts from elderly individuals show reductions in Rbbp4/7, HP1γ, and HDAC1 levels. In HGPS cells, these reductions correlate with loss of heterochromatin (H3K9me3) and increased susceptibility to DNA damage (Pegoraro et al., 2009). Rbbp4 also regulates nuclear import, and loss of Rbbp4 induces senescence (Tsujii et al., 2015). Therefore, lamin A may maintain heterochromatin at the nuclear periphery by stabilizing the Rbbp4/7 complex, facilitating proper chromatin organization and nuclear function. (Figs. 1 A and 2 C).
PRC2 has also been found to interact with lamin A, and this interaction is necessary for its proper localization (Cesarini et al., 2015). For example, in undifferentiated myoblast cells, PRC2 is recruited with the Msx1 homeoprotein and MyoD to the nuclear periphery (Wang et al., 2011). PRC2 relocation increases H3K27me3 marks at the periphery and represses the associated muscle-specific differentiation factors. In contrast, HGPS fibroblasts exhibit altered H3K27me3 deposition, loss of heterochromatin, and changes in gene expression comparable to those of senescent cells (Shumaker et al., 2006; Bracken et al., 2007; McCord et al., 2013; Shah et al., 2013; Chandra et al., 2015). The decrease in histone methylation is likely a result of changes to the peripheral lamin A–PRC/EZH2 interaction (Wang et al., 2011; McCord et al., 2013; Harr et al., 2015). Along the same paradigm as NCOR/SKIP regulation, these studies suggest that chromatin remodeling factors are recruited to the nuclear periphery to invoke a repressive environment, whereas transcription factors are synergistically sequestered at the periphery to limit their activity.
Telomere maintenance
The well-established correlation between telomere length and longevity suggests that progressive attrition of telomeres with iterative cycles of replication causes DNA ends to become exposed, triggering DNA damage and cellular senescence (López-Otín et al., 2013). Telomeres in lower eukaryotes localize to the nuclear periphery (Gonzalez-Suarez et al., 2009), but only a subpopulation of telomeres are peripheral in human cells, and this localization may be sensitive to the proliferative state as well as lamin organization (Ludérus et al., 1996; Gonzalez-Suarez et al., 2009; Arnoult et al., 2010; Crabbe et al., 2012; Chojnowski et al., 2015; Guidi et al., 2015). Although the factors governing telomere positioning in humans remain poorly understood, models of HGPS indicate that the nuclear lamina is important for regulating telomere stability, whereas loss of telomeres may trigger progerin expression (Kudlow et al., 2008; Gonzalez-Suarez et al., 2009; Ottaviani et al., 2009; Cao et al., 2011a; Crabbe et al., 2012; Chojnowski et al., 2015).
Telomere repeat–binding factor (TRF) 2
In vertebrates, TRF1 and 2 bind duplex telomeric DNA as homodimers within the shelterin protein complex to ensure telomere integrity (Wood et al., 2014). TRF1 negatively regulates telomere length and may be localized by lamin B1 to the nuclear periphery during postmitotic nuclear assembly (Crabbe et al., 2012). TRF2 forms protective telomere DNA loops at chromosome ends and at interstitial telomeric sequences. Stabilization of telomeres by TRF2 at the nuclear periphery is dependent on binding to lamin A/C and Lap2α (Ludérus et al., 1996; Dechat et al., 2004; Wood et al., 2014; Chojnowski et al., 2015; Fig. 2 D). However, TRF2 does not interact with progerin, and cells lacking lamin A/C or cells derived from HGPS patients lose their telomeres (Wood et al., 2014). It remains unclear whether TRF2 no longer binds progerin because of its integration into the periphery or whether there is specific protein–protein disruption. Notably, Lap2α preferentially interacts with lamin A/C over progerin (Dechat et al., 2000; Kubben et al., 2010), and the levels of Lap2α (Scaffidi and Misteli, 2006; Vidak et al., 2015) as well as nucleoplasmic lamin A/C are reduced in models of HGPS (Pekovic et al., 2007; Naetar et al., 2008; Chojnowski et al., 2015). The reduction in Lap2α levels upon progerin expression may inhibit TRF2 binding to lamin A/C. While progerin expression promotes telomere attrition via dysregulated TRF2, expression of a dominant-negative TRF2 protein induces uncapping of telomeres and correlates with increased progerin production (Cao et al., 2011a). Additionally, TRF2 expression is reduced in response to DNA damage in models of adult-onset progeroid syndromes caused by LMNA mutations (Saha et al., 2013), potentially propagating DNA damage and p53-mediated senescence. These data suggest a model in which proper interactions with the nuclear periphery are important for TRF2 function and telomere stability, whereas TRF2 expression may be inhibited across aging disorders by unregulated DNA damage, potentially triggering progerin expression.
AKT-interacting protein (AKTIP)
AKTIP, a recently discovered component of the shelterin complex, binds TRF1, TRF2, and PCNA to facilitate telomeric DNA replication as well as regulate telomere stability (Burla et al., 2015). AKTIP is strongly localized at the nuclear periphery in a lamin A–dependent manner, where it transiently interacts with telomeres (Burla et al., 2015, 2016; Fig. 2 D). In HGPS models, the peripheral localization of AKTIP is lost (Burla et al., 2016). Depletion of AKTIP causes cells to senesce, and senescence corresponds with increased prelamin A expression and nuclear deformities (Burla et al., 2015, 2016). This regulatory loop sheds new light on how changes in the nuclear periphery lead to telomere dysfunction, and conversely, how shortened telomeres affect the nuclear periphery.
Sphingosine-1-phosphate (S1P)
Not only is the protein composition of the INM important for telomere maintenance, but nuclear lipid moieties may also play a role (Panneer Selvam et al., 2015). S1P binds human telomerase reverse transcription (hTERT) at the nuclear periphery, preventing hTERT degradation and in turn promoting telomere stability (Fig. 2 D). Expression of an hTERT mutant that is unable to bind S1P prevents hTERT localization to the nuclear periphery, and consequently this mutant is rapidly degraded. S1P stability is correlated with delayed senescence in primary cells and increased tumor growth in a xenograft model. Whereas S1P specifically binds and prevents hTERT degradation (Panneer Selvam et al., 2015), TRF2 stabilizes the telomere structure (Wood et al., 2014) and AKTIP facilitates proper telomere replication (Burla et al., 2015). Irrespective of these distinct mechanisms, the nuclear periphery appears to play a protective role in telomere regulation.
Concluding remarks
The disproportionate number of aging disorders caused by mutations in nuclear pathways is striking and illustrates the importance of nuclear homeostasis for the aging process. One general mechanism for how the cell nucleus contributes to aging appears to be the integrity of the nuclear periphery, which is negatively affected by aberrant lamin expression in both pathological and physiological aging. As demonstrated in the cited examples, a likely mechanism for how nuclear function contributes to aging is by altered sequestration of cellular factors at the nuclear periphery. Disrupting the nuclear periphery by mutant lamina proteins has wide-ranging consequences at the structural, signaling, and transcriptional levels. Not only is peripheral regulation important in aging as noted here, but these principles may also apply to cancer, nuclear envelopathies, and other diseases. Although the pathways highlighted in our discussion suggest a role for nuclear sequestration as a basic regulatory principle, many questions remain as to how this process mechanistically inhibits protein function and to what degree it may be generalized. It will be important to determine a complete proteomic description of lamina-associated proteins in various diseases and, although many of the relevant observations are based on correlation, it will be essential to demonstrate that sequestration or loss thereof has functional consequences in aging-relevant pathways.
Acknowledgments
Work in the Misteli laboratory is supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, and Center for Cancer Research and by the Progeria Research Foundation.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- AKTIP
- AKT-interacting protein
- HGPS
- Hutchinson–Gilford progeria syndrome
- hTERT
- human telomerase reverse transcription
- INM
- inner nuclear membrane
- LEF
- lymphoid enhancer binding factor
- MSC
- mesenchymal stem cell
- PCNA
- proliferating cell nuclear antigen
- Rb
- retinoblastoma protein
- ROS
- reactive oxygen species
- S1P
- sphingosine-1-phosphate
- TRF
- telomere repeat–binding factor
References
- Ahlqvist K.J., Hämäläinen R.H., Yatsuga S., Uutela M., Terzioglu M., Götz A., Forsström S., Salven P., Angers-Loustau A., Kopra O.H., et al. . 2012. Somatic progenitor cell vulnerability to mitochondrial DNA mutagenesis underlies progeroid phenotypes in Polg mutator mice. Cell Metab. 15:100–109. 10.1016/j.cmet.2011.11.012 [DOI] [PubMed] [Google Scholar]
- Ahlqvist K.J., Suomalainen A., and Hämäläinen R.H.. 2015. Stem cells, mitochondria and aging. Biochim. Biophys. Acta. 1847:1380–1386. 10.1016/j.bbabio.2015.05.014 [DOI] [PubMed] [Google Scholar]
- Aliper A.M., Csoka A.B., Buzdin A., Jetka T., Roumiantsev S., Moskalev A., and Zhavoronkov A.. 2015. Signaling pathway activation drift during aging: Hutchinson-Gilford progeria syndrome fibroblasts are comparable to normal middle-age and old-age cells. Aging (Albany NY). 7:26–37. 10.18632/aging.100717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnoult N., Schluth-Bolard C., Letessier A., Drascovic I., Bouarich-Bourimi R., Campisi J., Kim S.H., Boussouar A., Ottaviani A., Magdinier F., et al. . 2010. Replication timing of human telomeres is chromosome arm-specific, influenced by subtelomeric structures and connected to nuclear localization. PLoS Genet. 6:e1000920 10.1371/journal.pgen.1000920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker D.J., Childs B.G., Durik M., Wijers M.E., Sieben C.J., Zhong J., Saltness R.A., Jeganathan K.B., Verzosa G.C., Pezeshki A., et al. . 2016. Naturally occurring p16(Ink4a)-positive cells shorten healthy lifespan. Nature. 530:184–189. 10.1038/nature16932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balistreri C.R., Madonna R., Melino G., and Caruso C.. 2016. The emerging role of Notch pathway in ageing: Focus on the related mechanisms in age-related diseases. Ageing Res. Rev. 29:50–65. 10.1016/j.arr.2016.06.004 [DOI] [PubMed] [Google Scholar]
- Baple E.L., Chambers H., Cross H.E., Fawcett H., Nakazawa Y., Chioza B.A., Harlalka G.V., Mansour S., Sreekantan-Nair A., Patton M.A., et al. . 2014. Hypomorphic PCNA mutation underlies a human DNA repair disorder. J. Clin. Invest. 124:3137–3146. 10.1172/JCI74593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrowman J., and Michaelis S.. 2009. ZMPSTE24, an integral membrane zinc metalloprotease with a connection to progeroid disorders. Biol. Chem. 390:761–773. 10.1515/BC.2009.080 [DOI] [PubMed] [Google Scholar]
- Benson E.K., Lee S.W., and Aaronson S.A.. 2010. Role of progerin-induced telomere dysfunction in HGPS premature cellular senescence. J. Cell Sci. 123:2605–2612. 10.1242/jcs.067306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermeo S., Vidal C., Zhou H., and Duque G.. 2015. Lamin A/C acts as an essential factor in mesenchymal stem cell differentiation through the regulation of the dynamics of the Wnt/β-catenin pathway. J. Cell. Biochem. 116:2344–2353. 10.1002/jcb.25185 [DOI] [PubMed] [Google Scholar]
- Bjornson C.R., Cheung T.H., Liu L., Tripathi P.V., Steeper K.M., and Rando T.A.. 2012. Notch signaling is necessary to maintain quiescence in adult muscle stem cells. Stem Cells. 30:232–242. 10.1002/stem.773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn E.H., Epel E.S., and Lin J.. 2015. Human telomere biology: A contributory and interactive factor in aging, disease risks, and protection. Science. 350:1193–1198. 10.1126/science.aab3389 [DOI] [PubMed] [Google Scholar]
- Borggrefe T., Lauth M., Zwijsen A., Huylebroeck D., Oswald F., and Giaimo B.D.. 2016. The Notch intracellular domain integrates signals from Wnt, Hedgehog, TGFβ/BMP and hypoxia pathways. Biochim. Biophys. Acta. 1863:303–313. 10.1016/j.bbamcr.2015.11.020 [DOI] [PubMed] [Google Scholar]
- Boubriak I.I., Malhas A.N., Drozdz M.M., Pytowski L., and Vaux D.J.. 2017. Stress-induced release of Oct-1 from the nuclear envelope is mediated by JNK phosphorylation of lamin B1. PLoS One. 12:e0177990 10.1371/journal.pone.0177990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brack A.S., Conboy M.J., Roy S., Lee M., Kuo C.J., Keller C., and Rando T.A.. 2007. Increased Wnt signaling during aging alters muscle stem cell fate and increases fibrosis. Science. 317:807–810. 10.1126/science.1144090 [DOI] [PubMed] [Google Scholar]
- Brack A.S., Conboy I.M., Conboy M.J., Shen J., and Rando T.A.. 2008. A temporal switch from notch to Wnt signaling in muscle stem cells is necessary for normal adult myogenesis. Cell Stem Cell. 2:50–59. 10.1016/j.stem.2007.10.006 [DOI] [PubMed] [Google Scholar]
- Bracken A.P., Kleine-Kohlbrecher D., Dietrich N., Pasini D., Gargiulo G., Beekman C., Theilgaard-Mönch K., Minucci S., Porse B.T., Marine J.C., et al. . 2007. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev. 21:525–530. 10.1101/gad.415507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratic A., and Larsson N.G.. 2013. The role of mitochondria in aging. J. Clin. Invest. 123:951–957. 10.1172/JCI64125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray S.J. 2016. Notch signalling in context. Nat. Rev. Mol. Cell Biol. 17:722–735. 10.1038/nrm.2016.94 [DOI] [PubMed] [Google Scholar]
- Burke B., and Stewart C.L.. 2013. The nuclear lamins: Flexibility in function. Nat. Rev. Mol. Cell Biol. 14:13–24. 10.1038/nrm3488 [DOI] [PubMed] [Google Scholar]
- Burla R., Carcuro M., Raffa G.D., Galati A., Raimondo D., Rizzo A., La Torre M., Micheli E., Ciapponi L., Cenci G., et al. . 2015. AKTIP/Ft1, a new shelterin-interacting factor required for telomere maintenance. PLoS Genet. 11:e1005167 10.1371/journal.pgen.1005167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burla R., Carcuro M., Torre M.L., Fratini F., Crescenzi M., D’Apice M.R., Spitalieri P., Raffa G.D., Astrologo L., Lattanzi G., et al. . 2016. The telomeric protein AKTIP interacts with A- and B-type lamins and is involved in regulation of cellular senescence. Open Biol. 6:160103 10.1098/rsob.160103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabanillas R., Cadiñanos J., Villameytide J.A., Pérez M., Longo J., Richard J.M., Alvarez R., Durán N.S., Illán R., González D.J., and López-Otín C.. 2011. Néstor-Guillermo progeria syndrome: A novel premature aging condition with early onset and chronic development caused by BANF1 mutations. Am. J. Med. Genet. A. 155A:2617–2625. 10.1002/ajmg.a.34249 [DOI] [PubMed] [Google Scholar]
- Cagin U., and Enriquez J.A.. 2015. The complex crosstalk between mitochondria and the nucleus: What goes in between? Int. J. Biochem. Cell Biol. 63:10–15. 10.1016/j.biocel.2015.01.026 [DOI] [PubMed] [Google Scholar]
- Campisi J. 2013. Aging, cellular senescence, and cancer. Annu. Rev. Physiol. 75:685–705. 10.1146/annurev-physiol-030212-183653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao K., Capell B.C., Erdos M.R., Djabali K., and Collins F.S.. 2007. A lamin A protein isoform overexpressed in Hutchinson-Gilford progeria syndrome interferes with mitosis in progeria and normal cells. Proc. Natl. Acad. Sci. USA. 104:4949–4954. 10.1073/pnas.0611640104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao K., Blair C.D., Faddah D.A., Kieckhaefer J.E., Olive M., Erdos M.R., Nabel E.G., and Collins F.S.. 2011a Progerin and telomere dysfunction collaborate to trigger cellular senescence in normal human fibroblasts. J. Clin. Invest. 121:2833–2844. 10.1172/JCI43578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao K., Graziotto J.J., Blair C.D., Mazzulli J.R., Erdos M.R., Krainc D., and Collins F.S.. 2011b Rapamycin reverses cellular phenotypes and enhances mutant protein clearance in Hutchinson-Gilford progeria syndrome cells. Sci. Transl. Med. 3:89ra58 10.1126/scitranslmed.3002346 [DOI] [PubMed] [Google Scholar]
- Capanni C., Cenni V., Haraguchi T., Squarzoni S., Schüchner S., Ogris E., Novelli G., Maraldi N., and Lattanzi G.. 2010. Lamin A precursor induces barrier-to-autointegration factor nuclear localization. Cell Cycle. 9:2600–2610. 10.4161/cc.9.13.12080 [DOI] [PubMed] [Google Scholar]
- Cesarini E., Mozzetta C., Marullo F., Gregoretti F., Gargiulo A., Columbaro M., Cortesi A., Antonelli L., Di Pelino S., Squarzoni S., et al. . 2015. Lamin A/C sustains PcG protein architecture, maintaining transcriptional repression at target genes. J. Cell Biol. 211:533–551. 10.1083/jcb.201504035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra T., Ewels P.A., Schoenfelder S., Furlan-Magaril M., Wingett S.W., Kirschner K., Thuret J.Y., Andrews S., Fraser P., and Reik W.. 2015. Global reorganization of the nuclear landscape in senescent cells. Cell Reports. 10:471–483. 10.1016/j.celrep.2014.12.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W., Worman H.J., and Gundersen G.G.. 2015. Accessorizing and anchoring the LINC complex for multifunctionality. J. Cell Biol. 208:11–22. 10.1083/jcb.201409047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Lee L., Kudlow B.A., Dos Santos H.G., Sletvold O., Shafeghati Y., Botha E.G., Garg A., Hanson N.B., Martin G.M., et al. . 2003. LMNA mutations in atypical Werner’s syndrome. Lancet. 362:440–445. 10.1016/S0140-6736(03)14069-X [DOI] [PubMed] [Google Scholar]
- Chen Z., Chang W.Y., Etheridge A., Strickfaden H., Jin Z., Palidwor G., Cho J.H., Wang K., Kwon S.Y., Doré C., et al. . 2017. Reprogramming progeria fibroblasts re-establishes a normal epigenetic landscape. Aging Cell. 16:870–887. 10.1111/acel.12621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs B.G., Durik M., Baker D.J., and van Deursen J.M.. 2015. Cellular senescence in aging and age-related disease: From mechanisms to therapy. Nat. Med. 21:1424–1435. 10.1038/nm.4000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe K.N., and Moldovan G.L.. 2017. Forging ahead through darkness: PCNA, still the principal conductor at the replication fork. Mol. Cell. 65:380–392. 10.1016/j.molcel.2016.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chojnowski A., Ong P.F., Wong E.S., Lim J.S., Mutalif R.A., Navasankari R., Dutta B., Yang H., Liow Y.Y., Sze S.K., et al. . 2015. Progerin reduces LAP2α-telomere association in Hutchinson-Gilford progeria. eLife. 4:e07759 10.7554/eLife.07759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu J., Jeffries S., Norton J.E., Capobianco A.J., and Bresnick E.H.. 2002. Repression of activator protein-1-mediated transcriptional activation by the Notch-1 intracellular domain. J. Biol. Chem. 277:7587–7597. 10.1074/jbc.M111044200 [DOI] [PubMed] [Google Scholar]
- Chung H.Y., Cesari M., Anton S., Marzetti E., Giovannini S., Seo A.Y., Carter C., Yu B.P., and Leeuwenburgh C.. 2009. Molecular inflammation: Underpinnings of aging and age-related diseases. Ageing Res. Rev. 8:18–30. 10.1016/j.arr.2008.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H., and Nusse R.. 2012. Wnt/β-catenin signaling and disease. Cell. 149:1192–1205. 10.1016/j.cell.2012.05.012 [DOI] [PubMed] [Google Scholar]
- Cobb A.M., Murray T.V., Warren D.T., Liu Y., and Shanahan C.M.. 2016. Disruption of PCNA-lamins A/C interactions by prelamin A induces DNA replication fork stalling. Nucleus. 7:498–511. 10.1080/19491034.2016.1239685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Columbaro M., Mattioli E., Maraldi N.M., Ortolani M., Gasparini L., D’Apice M.R., Postorivo D., Nardone A.M., Avnet S., Cortelli P., et al. . 2013. Oct-1 recruitment to the nuclear envelope in adult-onset autosomal dominant leukodystrophy. Biochim. Biophys. Acta. 1832:411–420. 10.1016/j.bbadis.2012.12.006 [DOI] [PubMed] [Google Scholar]
- Conboy I.M., Conboy M.J., Smythe G.M., and Rando T.A.. 2003. Notch-mediated restoration of regenerative potential to aged muscle. Science. 302:1575–1577. 10.1126/science.1087573 [DOI] [PubMed] [Google Scholar]
- Conboy I.M., Conboy M.J., Wagers A.J., Girma E.R., Weissman I.L., and Rando T.A.. 2005. Rejuvenation of aged progenitor cells by exposure to a young systemic environment. Nature. 433:760–764. 10.1038/nature03260 [DOI] [PubMed] [Google Scholar]
- Conneely K.N., Capell B.C., Erdos M.R., Sebastiani P., Solovieff N., Swift A.J., Baldwin C.T., Budagov T., Barzilai N., Atzmon G., et al. . 2012. Human longevity and common variations in the LMNA gene: a meta-analysis. Aging Cell. 11:475–481. 10.1111/j.1474-9726.2012.00808.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbe L., Cesare A.J., Kasuboski J.M., Fitzpatrick J.A., and Karlseder J.. 2012. Human telomeres are tethered to the nuclear envelope during postmitotic nuclear assembly. Cell Reports. 2:1521–1529. 10.1016/j.celrep.2012.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croteau D.L., Popuri V., Opresko P.L., and Bohr V.A.. 2014. Human RecQ helicases in DNA repair, recombination, and replication. Annu. Rev. Biochem. 83:519–552. 10.1146/annurev-biochem-060713-035428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csoka A.B., Cao H., Sammak P.J., Constantinescu D., Schatten G.P., and Hegele R.A.. 2004. Novel lamin A/C gene (LMNA) mutations in atypical progeroid syndromes. J. Med. Genet. 41:304–308. 10.1136/jmg.2003.015651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl K.N., Scaffidi P., Islam M.F., Yodh A.G., Wilson K.L., and Misteli T.. 2006. Distinct structural and mechanical properties of the nuclear lamina in Hutchinson-Gilford progeria syndrome. Proc. Natl. Acad. Sci. USA. 103:10271–10276. 10.1073/pnas.0601058103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugherty R.L., Serebryannyy L., Yemelyanov A., Flozak A.S., Yu H.J., Kosak S.T., deLanerolle P., and Gottardi C.J.. 2014. α-Catenin is an inhibitor of transcription. Proc. Natl. Acad. Sci. USA. 111:5260–5265. 10.1073/pnas.1308663111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies B.S., Fong L.G., Yang S.H., Coffinier C., and Young S.G.. 2009. The posttranslational processing of prelamin A and disease. Annu. Rev. Genomics Hum. Genet. 10:153–174. 10.1146/annurev-genom-082908-150150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechat T., Korbei B., Vaughan O.A., Vlcek S., Hutchison C.J., and Foisner R.. 2000. Lamina-associated polypeptide 2alpha binds intranuclear A-type lamins. J. Cell Sci. 113:3473–3484. [DOI] [PubMed] [Google Scholar]
- Dechat T., Gajewski A., Korbei B., Gerlich D., Daigle N., Haraguchi T., Furukawa K., Ellenberg J., and Foisner R.. 2004. LAP2alpha and BAF transiently localize to telomeres and specific regions on chromatin during nuclear assembly. J. Cell Sci. 117:6117–6128. 10.1242/jcs.01529 [DOI] [PubMed] [Google Scholar]
- Dechat T., Shimi T., Adam S.A., Rusinol A.E., Andres D.A., Spielmann H.P., Sinensky M.S., and Goldman R.D.. 2007. Alterations in mitosis and cell cycle progression caused by a mutant lamin A known to accelerate human aging. Proc. Natl. Acad. Sci. USA. 104:4955–4960. 10.1073/pnas.0700854104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker M.L., Chavez E., Vulto I., and Lansdorp P.M.. 2009. Telomere length in Hutchinson-Gilford progeria syndrome. Mech. Ageing Dev. 130:377–383. 10.1016/j.mad.2009.03.001 [DOI] [PubMed] [Google Scholar]
- de Lanerolle P., and Serebryannyy L.. 2011. Nuclear actin and myosins: Life without filaments. Nat. Cell Biol. 13:1282–1288. 10.1038/ncb2364 [DOI] [PubMed] [Google Scholar]
- de la Rosa J., Freije J.M., Cabanillas R., Osorio F.G., Fraga M.F., Fernández-García M.S., Rad R., Fanjul V., Ugalde A.P., Liang Q., et al. . 2013. Prelamin A causes progeria through cell-extrinsic mechanisms and prevents cancer invasion. Nat. Commun. 4:2268 10.1038/ncomms3268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delbarre E., Tramier M., Coppey-Moisan M., Gaillard C., Courvalin J.C., and Buendia B.. 2006. The truncated prelamin A in Hutchinson-Gilford progeria syndrome alters segregation of A-type and B-type lamin homopolymers. Hum. Mol. Genet. 15:1113–1122. 10.1093/hmg/ddl026 [DOI] [PubMed] [Google Scholar]
- Demmerle J., Koch A.J., and Holaska J.M.. 2012. The nuclear envelope protein emerin binds directly to histone deacetylase 3 (HDAC3) and activates HDAC3 activity. J. Biol. Chem. 287:22080–22088. 10.1074/jbc.M111.325308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Renty C., and Ellis N.A.. 2017. Bloom’s syndrome: Why not premature aging?: A comparison of the BLM and WRN helicases. Ageing Res. Rev. 33:36–51. 10.1016/j.arr.2016.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Sandre-Giovannoli A., Bernard R., Cau P., Navarro C., Amiel J., Boccaccio I., Lyonnet S., Stewart C.L., Munnich A., Le Merrer M., and Lévy N.. 2003. Lamin a truncation in Hutchinson-Gilford progeria. Science. 300:2055 10.1126/science.1084125 [DOI] [PubMed] [Google Scholar]
- Dialynas G., Shrestha O.K., Ponce J.M., Zwerger M., Thiemann D.A., Young G.H., Moore S.A., Yu L., Lammerding J., and Wallrath L.L.. 2015. Myopathic lamin mutations cause reductive stress and activate the nrf2/keap-1 pathway. PLoS Genet. 11:e1005231 10.1371/journal.pgen.1005231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Micco A., Frera G., Lugrin J., Jamilloux Y., Hsu E.T., Tardivel A., De Gassart A., Zaffalon L., Bujisic B., Siegert S., et al. . 2016. AIM2 inflammasome is activated by pharmacological disruption of nuclear envelope integrity. Proc. Natl. Acad. Sci. USA. 113:E4671–E4680. 10.1073/pnas.1602419113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimova D.K., and Dyson N.J.. 2005. The E2F transcriptional network: old acquaintances with new faces. Oncogene. 24:2810–2826. 10.1038/sj.onc.1208612 [DOI] [PubMed] [Google Scholar]
- Dittmer T.A., and Misteli T.. 2011. The lamin protein family. Genome Biol. 12:222 10.1186/gb-2011-12-5-222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmer T.A., Sahni N., Kubben N., Hill D.E., Vidal M., Burgess R.C., Roukos V., and Misteli T.. 2014. Systematic identification of pathological lamin A interactors. Mol. Biol. Cell. 25:1493–1510. 10.1091/mbc.E14-02-0733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doubaj Y., De Sandre-Giovannoli A., Vera E.V., Navarro C.L., Elalaoui S.C., Tajir M., Lévy N., and Sefiani A.. 2012. An inherited LMNA gene mutation in atypical progeria syndrome. Am. J. Med. Genet. A. 158A:2881–2887. 10.1002/ajmg.a.35557 [DOI] [PubMed] [Google Scholar]
- Dreesen O., and Stewart C.L.. 2011. Accelerated aging syndromes, are they relevant to normal human aging? Aging (Albany NY). 3:889–895. 10.18632/aging.100383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreesen O., Chojnowski A., Ong P.F., Zhao T.Y., Common J.E., Lunny D., Lane E.B., Lee S.J., Vardy L.A., Stewart C.L., and Colman A.. 2013. Lamin B1 fluctuations have differential effects on cellular proliferation and senescence. J. Cell Biol. 200:605–617. 10.1083/jcb.201206121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreger M., Bengtsson L., Schöneberg T., Otto H., and Hucho F.. 2001. Nuclear envelope proteomics: Novel integral membrane proteins of the inner nuclear membrane. Proc. Natl. Acad. Sci. USA. 98:11943–11948. 10.1073/pnas.211201898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eferl R., and Wagner E.F.. 2003. AP-1: A double-edged sword in tumorigenesis. Nat. Rev. Cancer. 3:859–868. 10.1038/nrc1209 [DOI] [PubMed] [Google Scholar]
- Egesipe A.-L., Blondel S., Cicero A.L., Jaskowiak A.-L., Navarro C., De Sandre-Giovannoli A., Levy N., Peschanski M., and Nissan X.. 2016. Metformin decreases progerin expression and alleviates pathological defects of Hutchinson–Gilford progeria syndrome cells. NPJ Aging Mech. Dis. 2:16026 10.1038/npjamd.2016.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisch V., Lu X., Gabriel D., and Djabali K.. 2016. Progerin impairs chromosome maintenance by depleting CENP-F from metaphase kinetochores in Hutchinson-Gilford progeria fibroblasts. Oncotarget. 7:24700–24718. 10.18632/oncotarget.8267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endisha H., Merrill-Schools J., Zhao M., Bristol M., Wang X., Kubben N., and Elmore L.W.. 2015. Restoring SIRT6 expression in Hutchinson-Gilford progeria syndrome cells impedes premature senescence and formation of dysmorphic nuclei. Pathobiology. 82:9–20. 10.1159/000368856 [DOI] [PubMed] [Google Scholar]
- Enyedi B., Jelcic M., and Niethammer P.. 2016. The cell nucleus serves as a mechanotransducer of tissue damage-induced inflammation. Cell. 165:1160–1170. 10.1016/j.cell.2016.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson M., Brown W.T., Gordon L.B., Glynn M.W., Singer J., Scott L., Erdos M.R., Robbins C.M., Moses T.Y., Berglund P., et al. . 2003. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature. 423:293–298. 10.1038/nature01629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espada J., Varela I., Flores I., Ugalde A.P., Cadiñanos J., Pendás A.M., Stewart C.L., Tryggvason K., Blasco M.A., Freije J.M., and López-Otín C.. 2008. Nuclear envelope defects cause stem cell dysfunction in premature-aging mice. J. Cell Biol. 181:27–35. 10.1083/jcb.200801096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers M.A., de Vries-Smits L.M., Barker N., Polderman P.E., Burgering B.M., and Korswagen H.C.. 2005. Functional interaction between beta-catenin and FOXO in oxidative stress signaling. Science. 308:1181–1184. 10.1126/science.1109083 [DOI] [PubMed] [Google Scholar]
- Fernandez P., Scaffidi P., Markert E., Lee J.H., Rane S., and Misteli T.. 2014. Transformation resistance in a premature aging disorder identifies a tumor-protective function of BRD4. Cell Reports. 9:248–260. 10.1016/j.celrep.2014.08.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Füllgrabe J., Lynch-Day M.A., Heldring N., Li W., Struijk R.B., Ma Q., Hermanson O., Rosenfeld M.G., Klionsky D.J., and Joseph B.. 2013. The histone H4 lysine 16 acetyltransferase hMOF regulates the outcome of autophagy. Nature. 500:468–471. 10.1038/nature12313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli M., Rossiello F., Clerici M., Barozzi S., Cittaro D., Kaplunov J.M., Bucci G., Dobreva M., Matti V., Beausejour C.M., et al. . 2012. Telomeric DNA damage is irreparable and causes persistent DNA-damage-response activation. Nat. Cell Biol. 14:355–365. 10.1038/ncb2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S., Liu B., Wang Y., Hao Q., and Zhou Z.. 2015. Lamin A Is an endogenous SIRT6 activator and promotes SIRT6-mediated DNA repair. Cell Reports. 13:1396–1406. 10.1016/j.celrep.2015.10.006 [DOI] [PubMed] [Google Scholar]
- Goldberg M.W., Huttenlauch I., Hutchison C.J., and Stick R.. 2008. Filaments made from A- and B-type lamins differ in structure and organization. J. Cell Sci. 121:215–225. 10.1242/jcs.022020 [DOI] [PubMed] [Google Scholar]
- Goldman R.D., Shumaker D.K., Erdos M.R., Eriksson M., Goldman A.E., Gordon L.B., Gruenbaum Y., Khuon S., Mendez M., Varga R., and Collins F.S.. 2004. Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson-Gilford progeria syndrome. Proc. Natl. Acad. Sci. USA. 101:8963–8968. 10.1073/pnas.0402943101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González J.M., Navarro-Puche A., Casar B., Crespo P., and Andrés V.. 2008. Fast regulation of AP-1 activity through interaction of lamin A/C, ERK1/2, and c-Fos at the nuclear envelope. J. Cell Biol. 183:653–666. 10.1083/jcb.200805049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Suarez I., Redwood A.B., Perkins S.M., Vermolen B., Lichtensztejin D., Grotsky D.A., Morgado-Palacin L., Gapud E.J., Sleckman B.P., Sullivan T., et al. . 2009. Novel roles for A-type lamins in telomere biology and the DNA damage response pathway. EMBO J. 28:2414–2427. 10.1038/emboj.2009.196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goukassian D., Gad F., Yaar M., Eller M.S., Nehal U.S., and Gilchrest B.A.. 2000. Mechanisms and implications of the age-associated decrease in DNA repair capacity. FASEB J. 14:1325–1334. 10.1096/fj.14.10.1325 [DOI] [PubMed] [Google Scholar]
- Guidi M., Ruault M., Marbouty M., Loïodice I., Cournac A., Billaudeau C., Hocher A., Mozziconacci J., Koszul R., and Taddei A.. 2015. Spatial reorganization of telomeres in long-lived quiescent cells. Genome Biol. 16:206 10.1186/s13059-015-0766-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hämäläinen R.H., Ahlqvist K.J., Ellonen P., Lepistö M., Logan A., Otonkoski T., Murphy M.P., and Suomalainen A.. 2015. mtDNA mutagenesis disrupts pluripotent stem cell function by altering redox signaling. Cell Reports. 11:1614–1624. 10.1016/j.celrep.2015.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X., Feng X., Rattner J.B., Smith H., Bose P., Suzuki K., Soliman M.A., Scott M.S., Burke B.E., and Riabowol K.. 2008. Tethering by lamin A stabilizes and targets the ING1 tumour suppressor. Nat. Cell Biol. 10:1333–1340. 10.1038/ncb1792 [DOI] [PubMed] [Google Scholar]
- Harman D. 1956. Aging: A theory based on free radical and radiation chemistry. J. Gerontol. 11:298–300. 10.1093/geronj/11.3.298 [DOI] [PubMed] [Google Scholar]
- Harr J.C., Luperchio T.R., Wong X., Cohen E., Wheelan S.J., and Reddy K.L.. 2015. Directed targeting of chromatin to the nuclear lamina is mediated by chromatin state and A-type lamins. J. Cell Biol. 208:33–52. 10.1083/jcb.201405110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto K., Majumdar R., and Tsuji Y.. 2017. Nuclear lamins and progerin are dispensable for antioxidant Nrf2 response to arsenic and cadmium. Cell. Signal. 33:69–78. 10.1016/j.cellsig.2017.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellemans J., Preobrazhenska O., Willaert A., Debeer P., Verdonk P.C., Costa T., Janssens K., Menten B., Van Roy N., Vermeulen S.J., et al. . 2004. Loss-of-function mutations in LEMD3 result in osteopoikilosis, Buschke-Ollendorff syndrome and melorheostosis. Nat. Genet. 36:1213–1218. 10.1038/ng1453 [DOI] [PubMed] [Google Scholar]
- Hernandez L., Roux K.J., Wong E.S., Mounkes L.C., Mutalif R., Navasankari R., Rai B., Cool S., Jeong J.W., Wang H., et al. . 2010. Functional coupling between the extracellular matrix and nuclear lamina by Wnt signaling in progeria. Dev. Cell. 19:413–425. 10.1016/j.devcel.2010.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzer M.W. 2010. The nuclear envelope. Cold Spring Harb. Perspect. Biol. 2:a000539 10.1101/cshperspect.a000539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt G., Jurk D., Marques F.D., Correia-Melo C., Hardy T., Gackowska A., Anderson R., Taschuk M., Mann J., and Passos J.F.. 2012. Telomeres are favoured targets of a persistent DNA damage response in ageing and stress-induced senescence. Nat. Commun. 3:708 10.1038/ncomms1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton B.A., Liu J., Cartwright B.M., Liu Y., Breitman M., Wang Y., Jones R., Tang H., Rusinol A., Musich P.R., and Zou Y.. 2017. Progerin sequestration of PCNA promotes replication fork collapse and mislocalization of XPA in laminopathy-related progeroid syndromes. FASEB J. 31:3882–3893. 10.1096/fj.201700014R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisama F.M., Lessel D., Leistritz D., Friedrich K., McBride K.L., Pastore M.T., Gottesman G.S., Saha B., Martin G.M., Kubisch C., and Oshima J.. 2011. Coronary artery disease in a Werner syndrome-like form of progeria characterized by low levels of progerin, a splice variant of lamin A. Am. J. Med. Genet. A. 155A:3002–3006. 10.1002/ajmg.a.34336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmeyer K., Raggioli A., Rudloff S., Anton R., Hierholzer A., Del Valle I., Hein K., Vogt R., and Kemler R.. 2012. Wnt/β-catenin signaling regulates telomerase in stem cells and cancer cells. Science. 336:1549–1554. 10.1126/science.1218370 [DOI] [PubMed] [Google Scholar]
- Holaska J.M., and Wilson K.L.. 2007. An emerin “proteome”: Purification of distinct emerin-containing complexes from HeLa cells suggests molecular basis for diverse roles including gene regulation, mRNA splicing, signaling, mechanosensing, and nuclear architecture. Biochemistry. 46:8897–8908. 10.1021/bi602636m [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim M.X., Sayin V.I., Akula M.K., Liu M., Fong L.G., Young S.G., and Bergo M.O.. 2013. Targeting isoprenylcysteine methylation ameliorates disease in a mouse model of progeria. Science. 340:1330–1333. 10.1126/science.1238880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S., Nishibayashi S., Takao K., Tomifuji M., Fujino T., Hasegawa M., and Takano T.. 1997. Dissociation of Oct-1 from the nuclear peripheral structure induces the cellular aging-associated collagenase gene expression. Mol. Biol. Cell. 8:2407–2419. 10.1091/mbc.8.12.2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivorra C., Kubicek M., González J.M., Sanz-González S.M., Alvarez-Barrientos A., O’Connor J.E., Burke B., and Andrés V.. 2006. A mechanism of AP-1 suppression through interaction of c-Fos with lamin A/C. Genes Dev. 20:307–320. 10.1101/gad.349506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian H., Shen X., Liu I., Semenov M., He X., and Wang X.F.. 2006. Smad3-dependent nuclear translocation of beta-catenin is required for TGF-beta1-induced proliferation of bone marrow-derived adult human mesenchymal stem cells. Genes Dev. 20:666–674. 10.1101/gad.1388806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson B.R., Nitta R.T., Frock R.L., Mounkes L., Barbie D.A., Stewart C.L., Harlow E., and Kennedy B.K.. 2004. A-type lamins regulate retinoblastoma protein function by promoting subnuclear localization and preventing proteasomal degradation. Proc. Natl. Acad. Sci. USA. 101:9677–9682. 10.1073/pnas.0403250101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko T., Tahara S., Tanno M., and Taguchi T.. 2002. Age-related changes in the induction of DNA polymerases in rat liver by gamma-ray irradiation. Mech. Ageing Dev. 123:1521–1528. 10.1016/S0047-6374(02)00119-7 [DOI] [PubMed] [Google Scholar]
- Katta S.S., Smoyer C.J., and Jaspersen S.L.. 2014. Destination: Inner nuclear membrane. Trends Cell Biol. 24:221–229. 10.1016/j.tcb.2013.10.006 [DOI] [PubMed] [Google Scholar]
- Kennedy B.K., Barbie D.A., Classon M., Dyson N., and Harlow E.. 2000. Nuclear organization of DNA replication in primary mammalian cells. Genes Dev. 14:2855–2868. 10.1101/gad.842600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.K., Lesoon-Wood L.A., Weintraub B.D., and Chung J.H.. 1996. A soluble transcription factor, Oct-1, is also found in the insoluble nuclear matrix and possesses silencing activity in its alanine-rich domain. Mol. Cell. Biol. 16:4366–4377. 10.1128/MCB.16.8.4366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korfali N., Wilkie G.S., Swanson S.K., Srsen V., Batrakou D.G., Fairley E.A., Malik P., Zuleger N., Goncharevich A., de Las Heras J., et al. . 2010. The leukocyte nuclear envelope proteome varies with cell activation and contains novel transmembrane proteins that affect genome architecture. Mol. Cell. Proteomics. 9:2571–2585. 10.1074/mcp.M110.002915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korfali N., Wilkie G.S., Swanson S.K., Srsen V., de Las Heras J., Batrakou D.G., Malik P., Zuleger N., Kerr A.R., Florens L., and Schirmer E.C.. 2012. The nuclear envelope proteome differs notably between tissues. Nucleus. 3:552–564. 10.4161/nucl.22257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan V., Chow M.Z., Wang Z., Zhang L., Liu B., Liu X., and Zhou Z.. 2011. Histone H4 lysine 16 hypoacetylation is associated with defective DNA repair and premature senescence in Zmpste24-deficient mice. Proc. Natl. Acad. Sci. USA. 108:12325–12330. 10.1073/pnas.1102789108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubben N., and Misteli T.. 2017. Molecular and cellular roots of pre-mature ageing and ageing-associated diseases. Nat. Rev. Mol. Cell Biol. 18:595–609. 10.1038/nrm.2017.68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubben N., Voncken J.W., Demmers J., Calis C., van Almen G., Pinto Y., and Misteli T.. 2010. Identification of differential protein interactors of lamin A and progerin. Nucleus. 1:513–525. 10.4161/nucl.1.6.13512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubben N., Adriaens M., Meuleman W., Voncken J.W., van Steensel B., and Misteli T.. 2012. Mapping of lamin A- and progerin-interacting genome regions. Chromosoma. 121:447–464. 10.1007/s00412-012-0376-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubben N., Zhang W., Wang L., Voss T.C., Yang J., Qu J., Liu G.H., and Misteli T.. 2016. Repression of the antioxidant NRF2 pathway in premature aging. Cell. 165:1361–1374. 10.1016/j.cell.2016.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudlow B.A., Stanfel M.N., Burtner C.R., Johnston E.D., and Kennedy B.K.. 2008. Suppression of proliferative defects associated with processing-defective lamin A mutants by hTERT or inactivation of p53. Mol. Biol. Cell. 19:5238–5248. 10.1091/mbc.E08-05-0492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujoth G.C., Hiona A., Pugh T.D., Someya S., Panzer K., Wohlgemuth S.E., Hofer T., Seo A.Y., Sullivan R., Jobling W.A., et al. . 2005. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 309:481–484. 10.1126/science.1112125 [DOI] [PubMed] [Google Scholar]
- Kumaran R.I., Muralikrishna B., and Parnaik V.K.. 2002. Lamin A/C speckles mediate spatial organization of splicing factor compartments and RNA polymerase II transcription. J. Cell Biol. 159:783–793. 10.1083/jcb.200204149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasry A., and Ben-Neriah Y.. 2015. Senescence-associated inflammatory responses: Aging and cancer perspectives. Trends Immunol. 36:217–228. 10.1016/j.it.2015.02.009 [DOI] [PubMed] [Google Scholar]
- Lattanzi G., Ortolani M., Columbaro M., Prencipe S., Mattioli E., Lanzarini C., Maraldi N.M., Cenni V., Garagnani P., Salvioli S., et al. . 2014. Lamins are rapamycin targets that impact human longevity: A study in centenarians. J. Cell Sci. 127:147–157. 10.1242/jcs.133983 [DOI] [PubMed] [Google Scholar]
- Laugesen A., and Helin K.. 2014. Chromatin repressive complexes in stem cells, development, and cancer. Cell Stem Cell. 14:735–751. 10.1016/j.stem.2014.05.006 [DOI] [PubMed] [Google Scholar]
- Lebel M., Spillare E.A., Harris C.C., and Leder P.. 1999. The Werner syndrome gene product co-purifies with the DNA replication complex and interacts with PCNA and topoisomerase I. J. Biol. Chem. 274:37795–37799. 10.1074/jbc.274.53.37795 [DOI] [PubMed] [Google Scholar]
- Lee S.J., Jung Y.S., Yoon M.H., Kang S.M., Oh A.Y., Lee J.H., Jun S.Y., Woo T.G., Chun H.Y., Kim S.K., et al. . 2016. Interruption of progerin-lamin A/C binding ameliorates Hutchinson-Gilford progeria syndrome phenotype. J. Clin. Invest. 126:3879–3893. 10.1172/JCI84164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong G.M., Subramaniam N., Figueroa J., Flanagan J.L., Hayman M.J., Eisman J.A., and Kouzmenko A.P.. 2001. Ski-interacting protein interacts with Smad proteins to augment transforming growth factor-beta-dependent transcription. J. Biol. Chem. 276:18243–18248. 10.1074/jbc.M010815200 [DOI] [PubMed] [Google Scholar]
- Li Y., Hassinger L., Thomson T., Ding B., Ashley J., Hassinger W., and Budnik V.. 2016. Lamin mutations accelerate aging via defective export of mitochondrial mRNAs through nuclear envelope budding. Curr. Biol. 26:2052–2059. 10.1016/j.cub.2016.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L., Zhang H., and Gu X.. 2009. Homozygous LMNA mutation R527C in atypical Hutchinson-Gilford progeria syndrome: Evidence for autosomal recessive inheritance. Acta Paediatr. 98:1365–1368. 10.1111/j.1651-2227.2009.01324.x [DOI] [PubMed] [Google Scholar]
- Lien W.H., and Fuchs E.. 2014. Wnt some lose some: Transcriptional governance of stem cells by Wnt/β-catenin signaling. Genes Dev. 28:1517–1532. 10.1101/gad.244772.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F., Morrison J.M., Wu W., and Worman H.J.. 2005. MAN1, an integral protein of the inner nuclear membrane, binds Smad2 and Smad3 and antagonizes transforming growth factor-beta signaling. Hum. Mol. Genet. 14:437–445. 10.1093/hmg/ddi040 [DOI] [PubMed] [Google Scholar]
- Lionaki E., Gkikas I., and Tavernarakis N.. 2016. Differential protein distribution between the nucleus and mitochondria: Implications in aging. Front. Genet. 7:162 10.3389/fgene.2016.00162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., and Zhou Z.. 2013. Activation of SIRT1 by resveratrol requires lamin A. Aging (Albany NY). 5:94–95. 10.18632/aging.100532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G.H., Barkho B.Z., Ruiz S., Diep D., Qu J., Yang S.L., Panopoulos A.D., Suzuki K., Kurian L., Walsh C., et al. . 2011. Recapitulation of premature ageing with iPSCs from Hutchinson-Gilford progeria syndrome. Nature. 472:221–225. 10.1038/nature09879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Fergusson M.M., Castilho R.M., Liu J., Cao L., Chen J., Malide D., Rovira I.I., Schimel D., Kuo C.J., et al. . 2007. Augmented Wnt signaling in a mammalian model of accelerated aging. Science. 317:803–806. 10.1126/science.1143578 [DOI] [PubMed] [Google Scholar]
- Liu Y., Rusinol A., Sinensky M., Wang Y., and Zou Y.. 2006. DNA damage responses in progeroid syndromes arise from defective maturation of prelamin A. J. Cell Sci. 119:4644–4649. 10.1242/jcs.03263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., El-Naggar S., Clem B., Chesney J., and Dean D.C.. 2008. The Rb/E2F pathway and Ras activation regulate RecQ helicase gene expression. Biochem. J. 412:299–306. 10.1042/BJ20070975 [DOI] [PubMed] [Google Scholar]
- Loi M., Cenni V., Duchi S., Squarzoni S., Lopez-Otin C., Foisner R., Lattanzi G., and Capanni C.. 2016. Barrier-to-autointegration factor (BAF) involvement in prelamin A-related chromatin organization changes. Oncotarget. 7:15662–15677. 10.18632/oncotarget.6697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi M.L., and Lammerding J.. 2011. Keeping the LINC: The importance of nucleocytoskeletal coupling in intracellular force transmission and cellular function. Biochem. Soc. Trans. 39:1729–1734. 10.1042/BST20110686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Otín C., Blasco M.A., Partridge L., Serrano M., and Kroemer G.. 2013. The hallmarks of aging. Cell. 153:1194–1217. 10.1016/j.cell.2013.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludérus M.E., van Steensel B., Chong L., Sibon O.C., Cremers F.F., and de Lange T.. 1996. Structure, subnuclear distribution, and nuclear matrix association of the mammalian telomeric complex. J. Cell Biol. 135:867–881. 10.1083/jcb.135.4.867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y.B., Mitrpant C., Johnsen R.D., Fabian V.A., Fletcher S., Mastaglia F.L., and Wilton S.D.. 2013. Investigation of age-related changes in LMNA splicing and expression of progerin in human skeletal muscles. Int. J. Clin. Exp. Pathol. 6:2778–2786. [PMC free article] [PubMed] [Google Scholar]
- Ly D.H., Lockhart D.J., Lerner R.A., and Schultz P.G.. 2000. Mitotic misregulation and human aging. Science. 287:2486–2492. 10.1126/science.287.5462.2486 [DOI] [PubMed] [Google Scholar]
- Ma Q. 2013. Role of nrf2 in oxidative stress and toxicity. Annu. Rev. Pharmacol. Toxicol. 53:401–426. 10.1146/annurev-pharmtox-011112-140320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makatsori D., Kourmouli N., Polioudaki H., Shultz L.D., McLean K., Theodoropoulos P.A., Singh P.B., and Georgatos S.D.. 2004. The inner nuclear membrane protein lamin B receptor forms distinct microdomains and links epigenetically marked chromatin to the nuclear envelope. J. Biol. Chem. 279:25567–25573. 10.1074/jbc.M313606200 [DOI] [PubMed] [Google Scholar]
- Malhas A.N., Lee C.F., and Vaux D.J.. 2009. Lamin B1 controls oxidative stress responses via Oct-1. J. Cell Biol. 184:45–55. 10.1083/jcb.200804155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini M.A., Shan B., Nickerson J.A., Penman S., and Lee W.H.. 1994. The retinoblastoma gene product is a cell cycle-dependent, nuclear matrix-associated protein. Proc. Natl. Acad. Sci. USA. 91:418–422. 10.1073/pnas.91.1.418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariño G., Ugalde A.P., Salvador-Montoliu N., Varela I., Quirós P.M., Cadiñanos J., van der Pluijm I., Freije J.M., and López-Otín C.. 2008. Premature aging in mice activates a systemic metabolic response involving autophagy induction. Hum. Mol. Genet. 17:2196–2211. 10.1093/hmg/ddn120 [DOI] [PubMed] [Google Scholar]
- Marji J., O’Donoghue S.I., McClintock D., Satagopam V.P., Schneider R., Ratner D., Worman H.J., Gordon L.B., and Djabali K.. 2010. Defective lamin A-Rb signaling in Hutchinson-Gilford progeria syndrome and reversal by farnesyltransferase inhibition. PLoS One. 5:e11132 10.1371/journal.pone.0011132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markiewicz E., Dechat T., Foisner R., Quinlan R.A., and Hutchison C.J.. 2002. Lamin A/C binding protein LAP2alpha is required for nuclear anchorage of retinoblastoma protein. Mol. Biol. Cell. 13:4401–4413. 10.1091/mbc.E02-07-0450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markiewicz E., Tilgner K., Barker N., van de Wetering M., Clevers H., Dorobek M., Hausmanowa-Petrusewicz I., Ramaekers F.C., Broers J.L., Blankesteijn W.M., et al. . 2006. The inner nuclear membrane protein emerin regulates beta-catenin activity by restricting its accumulation in the nucleus. EMBO J. 25:3275–3285. 10.1038/sj.emboj.7601230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock D., Ratner D., Lokuge M., Owens D.M., Gordon L.B., Collins F.S., and Djabali K.. 2007. The mutant form of lamin A that causes Hutchinson-Gilford progeria is a biomarker of cellular aging in human skin. PLoS One. 2:e1269 10.1371/journal.pone.0001269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCord R.P., Nazario-Toole A., Zhang H., Chines P.S., Zhan Y., Erdos M.R., Collins F.S., Dekker J., and Cao K.. 2013. Correlated alterations in genome organization, histone methylation, and DNA-lamin A/C interactions in Hutchinson-Gilford progeria syndrome. Genome Res. 23:260–269. 10.1101/gr.138032.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCrea P.D., and Gottardi C.J.. 2016. Beyond β-catenin: Prospects for a larger catenin network in the nucleus. Nat. Rev. Mol. Cell Biol. 17:55–64. 10.1038/nrm.2015.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinnon P.J. 2012. ATM and the molecular pathogenesis of ataxia telangiectasia. Annu. Rev. Pathol. 7:303–321. 10.1146/annurev-pathol-011811-132509 [DOI] [PubMed] [Google Scholar]
- Meaburn K.J. 2016. Spatial genome organization and its emerging role as a potential diagnosis tool. Front. Genet. 7:134 10.3389/fgene.2016.00134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehmood S., Marcoux J., Gault J., Quigley A., Michaelis S., Young S.G., Carpenter E.P., and Robinson C.V.. 2016. Mass spectrometry captures off-target drug binding and provides mechanistic insights into the human metalloprotease ZMPSTE24. Nat. Chem. 8:1152–1158. 10.1038/nchem.2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merideth M.A., Gordon L.B., Clauss S., Sachdev V., Smith A.C., Perry M.B., Brewer C.C., Zalewski C., Kim H.J., Solomon B., et al. . 2008. Phenotype and course of Hutchinson-Gilford progeria syndrome. N. Engl. J. Med. 358:592–604. 10.1056/NEJMoa0706898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuleman W., Peric-Hupkes D., Kind J., Beaudry J.B., Pagie L., Kellis M., Reinders M., Wessels L., and van Steensel B.. 2013. Constitutive nuclear lamina-genome interactions are highly conserved and associated with A/T-rich sequence. Genome Res. 23:270–280. 10.1101/gr.141028.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer A.J., Almendrala D.K., Go M.M., and Krauss S.W.. 2011. Structural protein 4.1R is integrally involved in nuclear envelope protein localization, centrosome-nucleus association and transcriptional signaling. J. Cell Sci. 124:1433–1444. 10.1242/jcs.077883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.D., Ganat Y.M., Kishinevsky S., Bowman R.L., Liu B., Tu E.Y., Mandal P.K., Vera E., Shim J.W., Kriks S., et al. . 2013. Human iPSC-based modeling of late-onset disease via progerin-induced aging. Cell Stem Cell. 13:691–705. 10.1016/j.stem.2013.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell S.J., Scheibye-Knudsen M., Longo D.L., and de Cabo R.. 2015. Animal models of aging research: Implications for human aging and age-related diseases. Annu. Rev. Anim. Biosci. 3:283–303. 10.1146/annurev-animal-022114-110829 [DOI] [PubMed] [Google Scholar]
- Mittnacht S., and Weinberg R.A.. 1991. G1/S phosphorylation of the retinoblastoma protein is associated with an altered affinity for the nuclear compartment. Cell. 65:381–393. 10.1016/0092-8674(91)90456-9 [DOI] [PubMed] [Google Scholar]
- Moir R.D., Spann T.P., Herrmann H., and Goldman R.D.. 2000. Disruption of nuclear lamin organization blocks the elongation phase of DNA replication. J. Cell Biol. 149:1179–1192. 10.1083/jcb.149.6.1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montes de Oca R., Shoemaker C.J., Gucek M., Cole R.N., and Wilson K.L.. 2009. Barrier-to-autointegration factor proteome reveals chromatin-regulatory partners. PLoS One. 4:e7050 10.1371/journal.pone.0007050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulson C.L., Go G., Gardner J.M., van der Wal A.C., Smitt J.H., van Hagen J.M., and Miner J.H.. 2005. Homozygous and compound heterozygous mutations in ZMPSTE24 cause the laminopathy restrictive dermopathy. J. Invest. Dermatol. 125:913–919. 10.1111/j.0022-202X.2005.23846.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulson C.L., Fong L.G., Gardner J.M., Farber E.A., Go G., Passariello A., Grange D.K., Young S.G., and Miner J.H.. 2007. Increased progerin expression associated with unusual LMNA mutations causes severe progeroid syndromes. Hum. Mutat. 28:882–889. 10.1002/humu.20536 [DOI] [PubMed] [Google Scholar]
- Muchir A., Pavlidis P., Bonne G., Hayashi Y.K., and Worman H.J.. 2007. Activation of MAPK in hearts of EMD null mice: Similarities between mouse models of X-linked and autosomal dominant Emery Dreifuss muscular dystrophy. Hum. Mol. Genet. 16:1884–1895. 10.1093/hmg/ddm137 [DOI] [PubMed] [Google Scholar]
- Muchir A., Shan J., Bonne G., Lehnart S.E., and Worman H.J.. 2009a Inhibition of extracellular signal-regulated kinase signaling to prevent cardiomyopathy caused by mutation in the gene encoding A-type lamins. Hum. Mol. Genet. 18:241–247. 10.1093/hmg/ddn343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchir A., Wu W., and Worman H.J.. 2009b Reduced expression of A-type lamins and emerin activates extracellular signal-regulated kinase in cultured cells. Biochim. Biophys. Acta. 1792:75–81. 10.1016/j.bbadis.2008.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musich P.R., and Zou Y.. 2011. DNA-damage accumulation and replicative arrest in Hutchinson-Gilford progeria syndrome. Biochem. Soc. Trans. 39:1764–1769. 10.1042/BST20110687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naetar N., Korbei B., Kozlov S., Kerenyi M.A., Dorner D., Kral R., Gotic I., Fuchs P., Cohen T.V., Bittner R., et al. . 2008. Loss of nucleoplasmic LAP2alpha-lamin A complexes causes erythroid and epidermal progenitor hyperproliferation. Nat. Cell Biol. 10:1341–1348. 10.1038/ncb1793 [DOI] [PubMed] [Google Scholar]
- Naetar N., Ferraioli S., and Foisner R.. 2017. Lamins in the nuclear interior: Life outside the lamina. J. Cell Sci. 130:2087–2096. 10.1242/jcs.203430 [DOI] [PubMed] [Google Scholar]
- Narita M., Nunez S., Heard E., Narita M., Lin A.W., Hearn S.A., Spector D.L., Hannon G.J., and Lowe S.W.. 2003. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 113:703–716. 10.1016/S0092-8674(03)00401-X [DOI] [PubMed] [Google Scholar]
- Navarro C.L., De Sandre-Giovannoli A., Bernard R., Boccaccio I., Boyer A., Geneviève D., Hadj-Rabia S., Gaudy-Marqueste C., Smitt H.S., Vabres P., et al. . 2004. Lamin A and ZMPSTE24 (FACE-1) defects cause nuclear disorganization and identify restrictive dermopathy as a lethal neonatal laminopathy. Hum. Mol. Genet. 13:2493–2503. 10.1093/hmg/ddh265 [DOI] [PubMed] [Google Scholar]
- Navarro C.L., Cadiñanos J., De Sandre-Giovannoli A., Bernard R., Courrier S., Boccaccio I., Boyer A., Kleijer W.J., Wagner A., Giuliano F., et al. . 2005. Loss of ZMPSTE24 (FACE-1) causes autosomal recessive restrictive dermopathy and accumulation of Lamin A precursors. Hum. Mol. Genet. 14:1503–1513. 10.1093/hmg/ddi159 [DOI] [PubMed] [Google Scholar]
- Navarro C.L., Esteves-Vieira V., Courrier S., Boyer A., Duong Nguyen T., Huong T.T., Meinke P., Schröder W., Cormier-Daire V., Sznajer Y., et al. . 2014. New ZMPSTE24 (FACE1) mutations in patients affected with restrictive dermopathy or related progeroid syndromes and mutation update. Eur. J. Hum. Genet. 22:1002–1011. 10.1038/ejhg.2013.258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nili E., Cojocaru G.S., Kalma Y., Ginsberg D., Copeland N.G., Gilbert D.J., Jenkins N.A., Berger R., Shaklai S., Amariglio N., et al. . 2001. Nuclear membrane protein LAP2beta mediates transcriptional repression alone and together with its binding partner GCL (germ-cell-less). J. Cell Sci. 114:3297–3307. [DOI] [PubMed] [Google Scholar]
- Nitta R.T., Jameson S.A., Kudlow B.A., Conlan L.A., and Kennedy B.K.. 2006. Stabilization of the retinoblastoma protein by A-type nuclear lamins is required for INK4A-mediated cell cycle arrest. Mol. Cell. Biol. 26:5360–5372. 10.1128/MCB.02464-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh T., Gabet Y., Cogan J., Shi Y., Tank A., Sasaki T., Criswell B., Dixon A., Lee C., Tam J., et al. . 2009. Lef1 haploinsufficient mice display a low turnover and low bone mass phenotype in a gender- and age-specific manner. PLoS One. 4:e5438 10.1371/journal.pone.0005438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive M., Harten I., Mitchell R., Beers J.K., Djabali K., Cao K., Erdos M.R., Blair C., Funke B., Smoot L., et al. . 2010. Cardiovascular pathology in Hutchinson-Gilford progeria: Correlation with the vascular pathology of aging. Arterioscler. Thromb. Vasc. Biol. 30:2301–2309. 10.1161/ATVBAHA.110.209460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opresko P.L., and Shay J.W.. 2017. Telomere-associated aging disorders. Ageing Res. Rev. 33:52–66. 10.1016/j.arr.2016.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottaviani A., Schluth-Bolard C., Rival-Gervier S., Boussouar A., Rondier D., Foerster A.M., Morere J., Bauwens S., Gazzo S., Callet-Bauchu E., et al. . 2009. Identification of a perinuclear positioning element in human subtelomeres that requires A-type lamins and CTCF. EMBO J. 28:2428–2436. 10.1038/emboj.2009.201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D., Estévez-Salmerón L.D., Stroschein S.L., Zhu X., He J., Zhou S., and Luo K.. 2005. The integral inner nuclear membrane protein MAN1 physically interacts with the R-Smad proteins to repress signaling by the transforming growth factor-beta superfamily of cytokines. J. Biol. Chem. 280:15992–16001. 10.1074/jbc.M411234200 [DOI] [PubMed] [Google Scholar]
- Pan H., Guan D., Liu X., Li J., Wang L., Wu J., Zhou J., Zhang W., Ren R., Zhang W., et al. . 2016. SIRT6 safeguards human mesenchymal stem cells from oxidative stress by coactivating NRF2. Cell Res. 26:190–205. 10.1038/cr.2016.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panneer Selvam S., De Palma R.M., Oaks J.J., Oleinik N., Peterson Y.K., Stahelin R.V., Skordalakes E., Ponnusamy S., Garrett-Mayer E., Smith C.D., and Ogretmen B.. 2015. Binding of the sphingolipid S1P to hTERT stabilizes telomerase at the nuclear periphery by allosterically mimicking protein phosphorylation. Sci. Signal. 8:ra58 10.1126/scisignal.aaa4998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegoraro G., Kubben N., Wickert U., Göhler H., Hoffmann K., and Misteli T.. 2009. Ageing-related chromatin defects through loss of the NURD complex. Nat. Cell Biol. 11:1261–1267. 10.1038/ncb1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado J.R., Quiros P.M., Pulido M.R., Marino G., Martinez-Chantar M.L., Vazquez-Martinez R., Freije J.M., Lopez-Otin C., and Malagon M.M.. 2011. Proteomic profiling of adipose tissue from Zmpste24−/− mice, a model of lipodystrophy and premature aging, reveals major changes in mitochondrial function and vimentin processing. Mol. Cell. Proteomics. 10:M111.008094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekovic V., Harborth J., Broers J.L., Ramaekers F.C., van Engelen B., Lammens M., von Zglinicki T., Foisner R., Hutchison C., and Markiewicz E.. 2007. Nucleoplasmic LAP2alpha-lamin A complexes are required to maintain a proliferative state in human fibroblasts. J. Cell Biol. 176:163–172. 10.1083/jcb.200606139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrini S., Borghi R., D’Oria V., Restaldi F., Moreno S., Novelli A., Bertini E., and Compagnucci C.. 2017. Aged induced pluripotent stem cell (iPSCs) as a new cellular model for studying premature aging. Aging (Albany NY). 9:1453–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasilova M., Chattopadhyay C., Pal P., Schaub N.A., Buechner S.A., Mueller H., Miny P., Ghosh A., and Heinimann K.. 2004. Homozygous missense mutation in the lamin A/C gene causes autosomal recessive Hutchinson-Gilford progeria syndrome. J. Med. Genet. 41:609–614. 10.1136/jmg.2004.019661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puente X.S., Quesada V., Osorio F.G., Cabanillas R., Cadiñanos J., Fraile J.M., Ordóñez G.R., Puente D.A., Gutiérrez-Fernández A., Fanjul-Fernández M., et al. . 2011. Exome sequencing and functional analysis identifies BANF1 mutation as the cause of a hereditary progeroid syndrome. Am. J. Hum. Genet. 88:650–656. 10.1016/j.ajhg.2011.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quirós P.M., Mottis A., and Auwerx J.. 2016. Mitonuclear communication in homeostasis and stress. Nat. Rev. Mol. Cell Biol. 17:213–226. 10.1038/nrm.2016.23 [DOI] [PubMed] [Google Scholar]
- Ragnauth C.D., Warren D.T., Liu Y., McNair R., Tajsic T., Figg N., Shroff R., Skepper J., and Shanahan C.M.. 2010. Prelamin A acts to accelerate smooth muscle cell senescence and is a novel biomarker of human vascular aging. Circulation. 121:2200–2210. 10.1161/CIRCULATIONAHA.109.902056 [DOI] [PubMed] [Google Scholar]
- Richards S.A., Muter J., Ritchie P., Lattanzi G., and Hutchison C.J.. 2011. The accumulation of un-repairable DNA damage in laminopathy progeria fibroblasts is caused by ROS generation and is prevented by treatment with N-acetyl cysteine. Hum. Mol. Genet. 20:3997–4004. 10.1093/hmg/ddr327 [DOI] [PubMed] [Google Scholar]
- Rivera-Torres J., Acín-Perez R., Cabezas-Sánchez P., Osorio F.G., Gonzalez-Gómez C., Megias D., Cámara C., López-Otín C., Enríquez J.A., Luque-García J.L., and Andrés V.. 2013. Identification of mitochondrial dysfunction in Hutchinson-Gilford progeria syndrome through use of stable isotope labeling with amino acids in cell culture. J. Proteomics. 91:466–477. 10.1016/j.jprot.2013.08.008 [DOI] [PubMed] [Google Scholar]
- Rodriguez S., Coppedè F., Sagelius H., and Eriksson M.. 2009. Increased expression of the Hutchinson-Gilford progeria syndrome truncated lamin A transcript during cell aging. Eur. J. Hum. Genet. 17:928–937. 10.1038/ejhg.2008.270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez J., Calvo F., González J.M., Casar B., Andrés V., and Crespo P.. 2010. ERK1/2 MAP kinases promote cell cycle entry by rapid, kinase-independent disruption of retinoblastoma-lamin A complexes. J. Cell Biol. 191:967–979. 10.1083/jcb.201004067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosengardten Y., McKenna T., Grochová D., and Eriksson M.. 2011. Stem cell depletion in Hutchinson-Gilford progeria syndrome. Aging Cell. 10:1011–1020. 10.1111/j.1474-9726.2011.00743.x [DOI] [PubMed] [Google Scholar]
- Sadaie M., Salama R., Carroll T., Tomimatsu K., Chandra T., Young A.R., Narita M., Pérez-Mancera P.A., Bennett D.C., Chong H., et al. . 2013. Redistribution of the Lamin B1 genomic binding profile affects rearrangement of heterochromatic domains and SAHF formation during senescence. Genes Dev. 27:1800–1808. 10.1101/gad.217281.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha B., Zitnik G., Johnson S., Nguyen Q., Risques R.A., Martin G.M., and Oshima J.. 2013. DNA damage accumulation and TRF2 degradation in atypical Werner syndrome fibroblasts with LMNA mutations. Front. Genet. 4:129 10.3389/fgene.2013.00129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin E., Colla S., Liesa M., Moslehi J., Müller F.L., Guo M., Cooper M., Kotton D., Fabian A.J., Walkey C., et al. . 2011. Telomere dysfunction induces metabolic and mitochondrial compromise. Nature. 470:359–365. 10.1038/nature09787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaffidi P., and Misteli T.. 2005. Reversal of the cellular phenotype in the premature aging disease Hutchinson-Gilford progeria syndrome. Nat. Med. 11:440–445. 10.1038/nm1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaffidi P., and Misteli T.. 2006. Lamin A-dependent nuclear defects in human aging. Science. 312:1059–1063. 10.1126/science.1127168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaffidi P., and Misteli T.. 2008. Lamin A-dependent misregulation of adult stem cells associated with accelerated ageing. Nat. Cell Biol. 10:452–459. 10.1038/ncb1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer E.C., and Gerace L.. 2005. The nuclear membrane proteome: Extending the envelope. Trends Biochem. Sci. 30:551–558. 10.1016/j.tibs.2005.08.003 [DOI] [PubMed] [Google Scholar]
- Schirmer E.C., Florens L., Guan T., Yates J.R. III, and Gerace L.. 2003. Nuclear membrane proteins with potential disease links found by subtractive proteomics. Science. 301:1380–1382. 10.1126/science.1088176 [DOI] [PubMed] [Google Scholar]
- Sebastiani P., Solovieff N., Dewan A.T., Walsh K.M., Puca A., Hartley S.W., Melista E., Andersen S., Dworkis D.A., Wilk J.B., et al. . 2012. Genetic signatures of exceptional longevity in humans. PLoS One. 7:e29848 10.1371/journal.pone.0029848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serebryannyy L.A., Yemelyanov A., Gottardi C.J., and de Lanerolle P.. 2017. Nuclear α-catenin mediates the DNA damage response via β-catenin and nuclear actin. J. Cell Sci. 130:1717–1729. 10.1242/jcs.199893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shachar S., and Misteli T.. 2017. Causes and consequences of nuclear gene positioning. J. Cell Sci. 130:1501–1508. 10.1242/jcs.199786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P.P., Donahue G., Otte G.L., Capell B.C., Nelson D.M., Cao K., Aggarwala V., Cruickshanks H.A., Rai T.S., McBryan T., et al. . 2013. Lamin B1 depletion in senescent cells triggers large-scale changes in gene expression and the chromatin landscape. Genes Dev. 27:1787–1799. 10.1101/gad.223834.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P., and Kuehn M.R.. 2016. SENP1-modulated sumoylation regulates retinoblastoma protein (RB) and Lamin A/C interaction and stabilization. Oncogene. 35:6429–6438. 10.1038/onc.2016.177 [DOI] [PubMed] [Google Scholar]
- Shimi T., Pfleghaar K., Kojima S., Pack C.G., Solovei I., Goldman A.E., Adam S.A., Shumaker D.K., Kinjo M., Cremer T., and Goldman R.D.. 2008. The A- and B-type nuclear lamin networks: Microdomains involved in chromatin organization and transcription. Genes Dev. 22:3409–3421. 10.1101/gad.1735208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimi T., Butin-Israeli V., Adam S.A., Hamanaka R.B., Goldman A.E., Lucas C.A., Shumaker D.K., Kosak S.T., Chandel N.S., and Goldman R.D.. 2011. The role of nuclear lamin B1 in cell proliferation and senescence. Genes Dev. 25:2579–2593. 10.1101/gad.179515.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimi T., Kittisopikul M., Tran J., Goldman A.E., Adam S.A., Zheng Y., Jaqaman K., and Goldman R.D.. 2015. Structural organization of nuclear lamins A, C, B1, and B2 revealed by superresolution microscopy. Mol. Biol. Cell. 26:4075–4086. 10.1091/mbc.E15-07-0461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumaker D.K., Dechat T., Kohlmaier A., Adam S.A., Bozovsky M.R., Erdos M.R., Eriksson M., Goldman A.E., Khuon S., Collins F.S., et al. . 2006. Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proc. Natl. Acad. Sci. USA. 103:8703–8708. 10.1073/pnas.0602569103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumaker D.K., Solimando L., Sengupta K., Shimi T., Adam S.A., Grunwald A., Strelkov S.V., Aebi U., Cardoso M.C., and Goldman R.D.. 2008. The highly conserved nuclear lamin Ig-fold binds to PCNA: Its role in DNA replication. J. Cell Biol. 181:269–280. 10.1083/jcb.200708155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon D.N., Zastrow M.S., and Wilson K.L.. 2010. Direct actin binding to A- and B-type lamin tails and actin filament bundling by the lamin A tail. Nucleus. 1:264–272. 10.4161/nucl.11799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeets M.F., DeLuca E., Wall M., Quach J.M., Chalk A.M., Deans A.J., Heierhorst J., Purton L.E., Izon D.J., and Walkley C.R.. 2014. The Rothmund-Thomson syndrome helicase RECQL4 is essential for hematopoiesis. J. Clin. Invest. 124:3551–3565. 10.1172/JCI75334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoyer C.J., Katta S.S., Gardner J.M., Stoltz L., McCroskey S., Bradford W.D., McClain M., Smith S.E., Slaughter B.D., Unruh J.R., and Jaspersen S.L.. 2016. Analysis of membrane proteins localizing to the inner nuclear envelope in living cells. J. Cell Biol. 215:575–590. 10.1083/jcb.201607043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somech R., Shaklai S., Amariglio N., Rechavi G., and Simon A.J.. 2005. Nuclear envelopathies—Raising the nuclear veil. Pediatr. Res. 57:8R–15R. 10.1203/01.PDR.0000159566.54287.6C [DOI] [PubMed] [Google Scholar]
- Spann T.P., Moir R.D., Goldman A.E., Stick R., and Goldman R.D.. 1997. Disruption of nuclear lamin organization alters the distribution of replication factors and inhibits DNA synthesis. J. Cell Biol. 136:1201–1212. 10.1083/jcb.136.6.1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spann T.P., Goldman A.E., Wang C., Huang S., and Goldman R.D.. 2002. Alteration of nuclear lamin organization inhibits RNA polymerase II-dependent transcription. J. Cell Biol. 156:603–608. 10.1083/jcb.200112047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanini M., Botta E., Lanzafame M., and Orioli D.. 2010. Trichothiodystrophy: From basic mechanisms to clinical implications. DNA Repair (Amst.). 9:2–10. 10.1016/j.dnarep.2009.10.005 [DOI] [PubMed] [Google Scholar]
- Stubenvoll A., Rice M., Wietelmann A., Wheeler M., and Braun T.. 2015. Attenuation of Wnt/β-catenin activity reverses enhanced generation of cardiomyocytes and cardiac defects caused by the loss of emerin. Hum. Mol. Genet. 24:802–813. 10.1093/hmg/ddu498 [DOI] [PubMed] [Google Scholar]
- Sundaresan N.R., Vasudevan P., Zhong L., Kim G., Samant S., Parekh V., Pillai V.B., Ravindra P.V., Gupta M., Jeevanandam V., et al. . 2012. The sirtuin SIRT6 blocks IGF-Akt signaling and development of cardiac hypertrophy by targeting c-Jun. Nat. Med. 18:1643–1650. 10.1038/nm.2961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi H., and Rünger T.M.. 2013. Longwave UV light induces the aging-associated progerin. J. Invest. Dermatol. 133:1857–1862. 10.1038/jid.2013.71 [DOI] [PubMed] [Google Scholar]
- Tang H., Hilton B., Musich P.R., Fang D.Z., and Zou Y.. 2012. Replication factor C1, the large subunit of replication factor C, is proteolytically truncated in Hutchinson-Gilford progeria syndrome. Aging Cell. 11:363–365. 10.1111/j.1474-9726.2011.00779.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi T., Garcia-Higuera I., Xu B., Andreassen P.R., Gregory R.C., Kim S.T., Lane W.S., Kastan M.B., and D’Andrea A.D.. 2002. Convergence of the fanconi anemia and ataxia telangiectasia signaling pathways. Cell. 109:459–472. 10.1016/S0092-8674(02)00747-X [DOI] [PubMed] [Google Scholar]
- Tasselli L., Zheng W., and Chua K.F.. 2017. SIRT6: Novel mechanisms and links to aging and disease. Trends Endocrinol. Metab. 28:168–185. 10.1016/j.tem.2016.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton D.J., Park S.H., Lanier L., and Weinberg R.A.. 1991. Nonfunctional mutants of the retinoblastoma protein are characterized by defects in phosphorylation, viral oncoprotein association, and nuclear tethering. Proc. Natl. Acad. Sci. USA. 88:3033–3037. 10.1073/pnas.88.8.3033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thul P.J., Åkesson L., Wiking M., Mahdessian D., Geladaki A., Ait Blal H., Alm T., Asplund A., Björk L., Breckels L.M., et al. . 2017. A subcellular map of the human proteome. Science. 356:eaal3321 10.1126/science.aal3321 [DOI] [PubMed] [Google Scholar]
- Tilgner K., Wojciechowicz K., Jahoda C., Hutchison C., and Markiewicz E.. 2009. Dynamic complexes of A-type lamins and emerin influence adipogenic capacity of the cell via nucleocytoplasmic distribution of beta-catenin. J. Cell Sci. 122:401–413. 10.1242/jcs.026179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran J.R., Chen H., Zheng X., and Zheng Y.. 2016. Lamin in inflammation and aging. Curr. Opin. Cell Biol. 40:124–130. 10.1016/j.ceb.2016.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifunovic A., Wredenberg A., Falkenberg M., Spelbrink J.N., Rovio A.T., Bruder C.E., Bohlooly-Y M., Gidlöf S., Oldfors A., Wibom R., et al. . 2004. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 429:417–423. 10.1038/nature02517 [DOI] [PubMed] [Google Scholar]
- Tsujii A., Miyamoto Y., Moriyama T., Tsuchiya Y., Obuse C., Mizuguchi K., Oka M., and Yoneda Y.. 2015. Retinoblastoma-binding protein 4-regulated classical nuclear transport is involved in cellular senescence. J. Biol. Chem. 290:29375–29388. 10.1074/jbc.M115.681908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turgay Y., Eibauer M., Goldman A.E., Shimi T., Khayat M., Ben-Harush K., Dubrovsky-Gaupp A., Sapra K.T., Goldman R.D., and Medalia O.. 2017. The molecular architecture of lamins in somatic cells. Nature. 543:261–264. 10.1038/nature21382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyynismaa H., Mjosund K.P., Wanrooij S., Lappalainen I., Ylikallio E., Jalanko A., Spelbrink J.N., Paetau A., and Suomalainen A.. 2005. Mutant mitochondrial helicase Twinkle causes multiple mtDNA deletions and a late-onset mitochondrial disease in mice. Proc. Natl. Acad. Sci. USA. 102:17687–17692. 10.1073/pnas.0505551102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungricht R., Klann M., Horvath P., and Kutay U.. 2015. Diffusion and retention are major determinants of protein targeting to the inner nuclear membrane. J. Cell Biol. 209:687–703. 10.1083/jcb.201409127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Berlo J.H., Voncken J.W., Kubben N., Broers J.L., Duisters R., van Leeuwen R.E., Crijns H.J., Ramaekers F.C., Hutchison C.J., and Pinto Y.M.. 2005. A-type lamins are essential for TGF-beta1 induced PP2A to dephosphorylate transcription factors. Hum. Mol. Genet. 14:2839–2849. 10.1093/hmg/ddi316 [DOI] [PubMed] [Google Scholar]
- van Deursen J.M. 2014. The role of senescent cells in ageing. Nature. 509:439–446. 10.1038/nature13193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannier J.B., Sandhu S., Petalcorin M.I., Wu X., Nabi Z., Ding H., and Boulton S.J.. 2013. RTEL1 is a replisome-associated helicase that promotes telomere and genome-wide replication. Science. 342:239–242. 10.1126/science.1241779 [DOI] [PubMed] [Google Scholar]
- Varela I., Cadiñanos J., Pendás A.M., Gutiérrez-Fernández A., Folgueras A.R., Sánchez L.M., Zhou Z., Rodríguez F.J., Stewart C.L., Vega J.A., et al. . 2005. Accelerated ageing in mice deficient in Zmpste24 protease is linked to p53 signalling activation. Nature. 437:564–568. 10.1038/nature04019 [DOI] [PubMed] [Google Scholar]
- Varela I., Pereira S., Ugalde A.P., Navarro C.L., Suárez M.F., Cau P., Cadiñanos J., Osorio F.G., Foray N., Cobo J., et al. . 2008. Combined treatment with statins and aminobisphosphonates extends longevity in a mouse model of human premature aging. Nat. Med. 14:767–772. 10.1038/nm1786 [DOI] [PubMed] [Google Scholar]
- Verrecchia F., Chu M.L., and Mauviel A.. 2001. Identification of novel TGF-beta/Smad gene targets in dermal fibroblasts using a combined cDNA microarray/promoter transactivation approach. J. Biol. Chem. 276:17058–17062. 10.1074/jbc.M100754200 [DOI] [PubMed] [Google Scholar]
- Verstraeten V.L., Broers J.L., van Steensel M.A., Zinn-Justin S., Ramaekers F.C., Steijlen P.M., Kamps M., Kuijpers H.J., Merckx D., Smeets H.J., et al. . 2006. Compound heterozygosity for mutations in LMNA causes a progeria syndrome without prelamin A accumulation. Hum. Mol. Genet. 15:2509–2522. 10.1093/hmg/ddl172 [DOI] [PubMed] [Google Scholar]
- Verstraeten V.L., Ji J.Y., Cummings K.S., Lee R.T., and Lammerding J.. 2008. Increased mechanosensitivity and nuclear stiffness in Hutchinson-Gilford progeria cells: Effects of farnesyltransferase inhibitors. Aging Cell. 7:383–393. 10.1111/j.1474-9726.2008.00382.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidak S., and Foisner R.. 2016. Molecular insights into the premature aging disease progeria. Histochem. Cell Biol. 145:401–417. 10.1007/s00418-016-1411-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidak S., Kubben N., Dechat T., and Foisner R.. 2015. Proliferation of progeria cells is enhanced by lamina-associated polypeptide 2α (LAP2α) through expression of extracellular matrix proteins. Genes Dev. 29:2022–2036. 10.1101/gad.263939.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viteri G., Chung Y.W., and Stadtman E.R.. 2010. Effect of progerin on the accumulation of oxidized proteins in fibroblasts from Hutchinson Gilford progeria patients. Mech. Ageing Dev. 131:2–8. 10.1016/j.mad.2009.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi N., Skoko J.J., Chartoumpekis D.V., Kimura S., Slocum S.L., Noda K., Palliyaguru D.L., Fujimuro M., Boley P.A., Tanaka Y., et al. . 2014. Notch-Nrf2 axis: Regulation of Nrf2 gene expression and cytoprotection by notch signaling. Mol. Cell. Biol. 34:653–663. 10.1128/MCB.01408-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Kumar R.M., Biggs V.J., Lee H., Chen Y., Kagey M.H., Young R.A., and Abate-Shen C.. 2011. The Msx1 homeoprotein recruits polycomb to the nuclear periphery during development. Dev. Cell. 21:575–588. 10.1016/j.devcel.2011.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Das A., Xiong Z.M., Cao K., and Hannenhalli S.. 2015. Phenotype-dependent coexpression gene clusters: Application to normal and premature ageing. IEEE/ACM Trans. Comput. Biol. Bioinformatics. 12:30–39. 10.1109/TCBB.2014.2359446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Lichter-Konecki U., Anyane-Yeboa K., Shaw J.E., Lu J.T., Östlund C., Shin J.Y., Clark L.N., Gundersen G.G., Nagy P.L., and Worman H.J.. 2016. A mutation abolishing the ZMPSTE24 cleavage site in prelamin A causes a progeroid disorder. J. Cell Sci. 129:1975–1980. 10.1242/jcs.187302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watabe T., and Miyazono K.. 2009. Roles of TGF-beta family signaling in stem cell renewal and differentiation. Cell Res. 19:103–115. 10.1038/cr.2008.323 [DOI] [PubMed] [Google Scholar]
- Wheaton K., Campuzano D., Ma W., Sheinis M., Ho B., Brown G.W., and Benchimol S.. 2017. Progerin-induced replication stress facilitates premature senescence in Hutchinson-Gilford progeria syndrome. Mol. Cell. Biol. 37:e00659-16 10.1128/MCB.00659-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie G.S., Korfali N., Swanson S.K., Malik P., Srsen V., Batrakou D.G., de las Heras J., Zuleger N., Kerr A.R., Florens L., and Schirmer E.C.. 2011. Several novel nuclear envelope transmembrane proteins identified in skeletal muscle have cytoskeletal associations. Mol. Cell. Proteomics. 10:M110.003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson B.T., Stark Z., Sutton R.E., Danda S., Ekbote A.V., Elsayed S.M., Gibson L., Goodship J.A., Jackson A.P., Keng W.T., et al. . 2016. The Cockayne Syndrome Natural History (CoSyNH) study: Clinical findings in 102 individuals and recommendations for care. Genet. Med. 18:483–493. 10.1038/gim.2015.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood A.M., Rendtlew Danielsen J.M., Lucas C.A., Rice E.L., Scalzo D., Shimi T., Goldman R.D., Smith E.D., Le Beau M.M., and Kosak S.T.. 2014. TRF2 and lamin A/C interact to facilitate the functional organization of chromosome ends. Nat. Commun. 5:5467 10.1038/ncomms6467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D., Flannery A.R., Cai H., Ko E., and Cao K.. 2014. Nuclear localization signal deletion mutants of lamin A and progerin reveal insights into lamin A processing and emerin targeting. Nucleus. 5:66–74. 10.4161/nucl.28068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W., Chojnowski A., Boudier T., Lim J.S., Ahmed S., Ser Z., Stewart C., and Burke B.. 2016. A-type lamins form distinct filamentous networks with differential nuclear pore complex associations. Curr. Biol. 26:2651–2658. 10.1016/j.cub.2016.07.049 [DOI] [PubMed] [Google Scholar]
- Xiong Z.M., Choi J.Y., Wang K., Zhang H., Tariq Z., Wu D., Ko E., LaDana C., Sesaki H., and Cao K.. 2016. Methylene blue alleviates nuclear and mitochondrial abnormalities in progeria. Aging Cell. 15:279–290. 10.1111/acel.12434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamabe Y., Shimamoto A., Goto M., Yokota J., Sugawara M., and Furuichi Y.. 1998. Sp1-mediated transcription of the Werner helicase gene is modulated by Rb and p53. Mol. Cell. Biol. 18:6191–6200. 10.1128/MCB.18.11.6191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama N.N., Pate K.T., Sprowl S., and Waterman M.L.. 2010. A role for YY1 in repression of dominant negative LEF-1 expression in colon cancer. Nucleic Acids Res. 38:6375–6388. 10.1093/nar/gkq492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zane L., Sharma V., and Misteli T.. 2014. Common features of chromatin in aging and cancer: Cause or coincidence? Trends Cell Biol. 24:686–694. 10.1016/j.tcb.2014.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.E. 2009. Non-Smad pathways in TGF-beta signaling. Cell Res. 19:128–139. 10.1038/cr.2008.328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Dowd D.R., Staal A., Gu C., Lian J.B., van Wijnen A.J., Stein G.S., and MacDonald P.N.. 2003. Nuclear coactivator-62 kDa/Ski-interacting protein is a nuclear matrix-associated coactivator that may couple vitamin D receptor-mediated transcription and RNA splicing. J. Biol. Chem. 278:35325–35336. 10.1074/jbc.M305191200 [DOI] [PubMed] [Google Scholar]
- Zhang D.Y., Wang H.J., and Tan Y.Z.. 2011a Wnt/β-catenin signaling induces the aging of mesenchymal stem cells through the DNA damage response and the p53/p21 pathway. PLoS One. 6:e21397 10.1371/journal.pone.0021397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Davies K.J.A., and Forman H.J.. 2015a Oxidative stress response and Nrf2 signaling in aging. Free Radic. Biol. Med. 88(Pt B):314–336. 10.1016/j.freeradbiomed.2015.05.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Lian Q., Zhu G., Zhou F., Sui L., Tan C., Mutalif R.A., Navasankari R., Zhang Y., Tse H.F., et al. . 2011b A human iPSC model of Hutchinson Gilford progeria reveals vascular smooth muscle and mesenchymal stem cell defects. Cell Stem Cell. 8:31–45. 10.1016/j.stem.2010.12.002 [DOI] [PubMed] [Google Scholar]
- Zhang Q., Ragnauth C.D., Skepper J.N., Worth N.F., Warren D.T., Roberts R.G., Weissberg P.L., Ellis J.A., and Shanahan C.M.. 2005. Nesprin-2 is a multi-isomeric protein that binds lamin and emerin at the nuclear envelope and forms a subcellular network in skeletal muscle. J. Cell Sci. 118:673–687. 10.1242/jcs.01642 [DOI] [PubMed] [Google Scholar]
- Zhang W., Li J., Suzuki K., Qu J., Wang P., Zhou J., Liu X., Ren R., Xu X., Ocampo A., et al. . 2015b Aging stem cells. A Werner syndrome stem cell model unveils heterochromatin alterations as a driver of human aging. Science. 348:1160–1163. 10.1126/science.aaa1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Cook P.C., Zindy E., Williams C.J., Jowitt T.A., Streuli C.H., MacDonald A.S., and Redondo-Muñoz J.. 2016. Integrin α4β1 controls G9a activity that regulates epigenetic changes and nuclear properties required for lymphocyte migration. Nucleic Acids Res. 44:3031–3044. 10.1093/nar/gkv1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F.Q. 2013. Octamer-binding transcription factors: Genomics and functions. Front. Biosci. (Landmark Ed.). 18:1051–1071. 10.2741/4162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S., Fujimuro M., Hsieh J.J., Chen L., Miyamoto A., Weinmaster G., and Hayward S.D.. 2000. SKIP, a CBF1-associated protein, interacts with the ankyrin repeat domain of NotchIC to facilitate NotchIC function. Mol. Cell. Biol. 20:2400–2410. 10.1128/MCB.20.7.2400-2410.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]