Hu et al. discuss challenges in studying aging and offer a perspective on the use of organoids as a model to understand the biology of aging.
Abstract
The biology of aging is challenging to study, particularly in humans. As a result, model organisms are used to approximate the physiological context of aging in humans. However, the best model organisms remain expensive and time-consuming to use. More importantly, they may not reflect directly on the process of aging in people. Human cell culture provides an alternative, but many functional signs of aging occur at the level of tissues rather than cells and are therefore not readily apparent in traditional cell culture models. Organoids have the potential to effectively balance between the strengths and weaknesses of traditional models of aging. They have sufficient complexity to capture relevant signs of aging at the molecular, cellular, and tissue levels, while presenting an experimentally tractable alternative to animal studies. Organoid systems have been developed to model many human tissues and diseases. Here we provide a perspective on the potential for organoids to serve as models for aging and describe how current organoid techniques could be applied to aging research.
Introduction
Aging is widely held to be the consequence of the accumulation of cellular damage over time (López-Otín et al., 2013). In humans, aging is marked by declining tissue function and increased susceptibility to diseases such as cancer, diabetes, cardiovascular disease, dementia, arthritis, sarcopenia, and renal dysfunction. Aging-associated diseases and phenotypes frequently manifest on the tissue level yet have their roots in molecular and cellular damage (López-Otín et al., 2013).
Most aging research is motivated by the need to understand the process of aging in humans. However, studying aging requires observing and perturbing molecules, cells, tissues, and systems in organisms over time—requirements that are not feasible in humans. Instead, aging is studied in model systems, each of which balances a desire for biological relevance on one hand and practicality on the other. Thus, most models of aging suffer from key limitations depending on where on this spectrum they lie. Organoid culture, especially human organoid culture, has the potential to address many of the limitations of traditional aging models. By providing organotypic structure in an in vitro model, organoids enable the study of aging in tissues in a higher-throughput, experimentally accessible manner. These potential advantages may be particularly useful for studying emergent signs of aging at the tissue level, clearing the way for future human therapies.
What is an organoid?
An organoid is a miniature, simplified organ that recreates physiological 3D tissue structure and cellular composition in vitro. Examples include a variety of 3D in vitro culture systems: cultured tissue fragments, tissues reconstituted from cultured cells, and tissues grown from stem cells, usually in the presence of ECM protein. In this perspective, we have broadly included examples and influences from several different organotypic culture systems. The use of organoids reflects the philosophy that the structure and composition of a tissue is relevant to its function (Simian and Bissell, 2017). In practice, organoids are characterized by some if not all of the following features:
• A physiologically relevant microenvironment
• Cells derived from primary tissues, primary cells, or stem cells, rather than immortalized cell lines
• Cells correctly organized with respect to one another and their ECM
• Multiple cell types from the original organ
Models of aging
The spectrum of aging models
The trade-off between relevance to humans and ease of experimentation is a limitation in any model, particularly when seeking to develop interventions to limit the effects of human aging. Organoids provide a powerful and physiologically relevant means to perform in vitro experiments on tissue and on human tissue in particular.
Nonhuman organisms and traditional cell culture have been invaluable tools for the study of fundamental aging processes, including the Nobel Prize-winning mechanisms of telomere regulation and DNA repair (Sancar and Rupp, 1983; Greider and Blackburn, 1987; Lindahl, 1993). However, model organisms may age in ways not applicable to humans. For example, Caenorhabditis elegans has a dauer stage, a stress- and aging-resistant larval form. Induction of dauer-like states may alter lifespan with questionable correlation to human aging (McElwee et al., 2006). In mice, the most common mammalian model organism, important features that affect aging, such as immune response, DNA repair, telomerase regulation, and telomere length, differ significantly between mice and humans (Rangarajan and Weinberg, 2003; Seok et al., 2013). Even naked mole rats and blind mole rats, two rodents both renowned for their sevenfold-longer lives than laboratory mice, owe their longevity to differing molecular mechanisms (Azpurua and Seluanov, 2013). Thus, different species, even within the same order, have developed distinct mechanisms for controlling lifespan and health span in relatively brief evolutionary time. These differences have practical implications. Some of the most prominent diseases of human aging, such as epithelial cancers, cardiovascular disease, osteoporosis, and Alzheimer’s disease, are rare or nonexistent in aged mice, who instead predominantly suffer from mesenchymal cancers (Rangarajan and Weinberg, 2003; Vanhooren and Libert, 2013). Consequently, interventions that reduce susceptibility to human age-related diseases may not affect mouse lifespan, and vice versa.
Ultimately, studies in humans are necessary to validate therapies for human aging, but ethical, logistical, and financial challenges render routine human aging studies infeasible. A widely practiced alternative is cell culture, but the correlations to human aging may be weak because of the lack of physiological context. Many signs of aging, namely those that manifest at the tissue and organ level, may be best represented by a more complex system. These signs of aging include a breakdown in tissue homeostasis, stem cell dysfunction, and susceptibility to tissue-specific diseases and cancer (López-Otín et al., 2013).
Although more technically challenging than traditional cell culture, organoids provide increased relevance to human health via a microenvironment supportive of tissue-specific function and physiological cell–cell and cell–matrix interactions. These factors contribute to increased viability of difficult-to-culture cells, including primary cells and prostate tumors (Gao et al., 2014). Moreover, 3D culture improves cell type–specific function and gene expression in cells such as chondrocytes, hepatocytes, and mammary and kidney epithelial cells (Ben-Ze’ev et al., 1988; Astashkina et al., 2012; Caron et al., 2012; Simian and Bissell, 2017). As a result, organoids may be useful for modeling age-related declines in tissue-specific function, such as chondrocyte matrix deposition (Barbero et al., 2004). Furthermore, age-related pathologies intrinsically linked to the ECM, such as osteoarthritis, are well-suited to study in a 3D system (Lozito et al., 2013). Organoids may be the most practical way to perform aging experiments at this level of complexity, particularly on human tissues.
Practical experiments with organoid models
Organoids provide a practical alternative to whole-organism studies in mammals. Patient-derived organoids have been scaled to high-throughput screens against cystic fibrosis and cancer, which would be cost-prohibitive in mice or humans (Dekkers et al., 2016; Pauli et al., 2017; Schütte et al., 2017). Demonstrating the scalability and high-throughput potential of organoids, Pauli et al. (2017) developed patient-derived tumor organoids for screening a library of >100 drugs at different concentrations and combinations, and Danahay et al. (2015) screened nearly 5,000 proteins for their effects on basal cell fate in human lung organoids.
Improved throughput is made possible not only by larger sample sizes but also simpler assays. Some molecular signs of aging, such as protein aggregation, oxidation, and glycation, can be assayed by nondestructive, high-throughput microscopy (Meerwaldt et al., 2004; Chaudhuri et al., 2006; Jung et al., 2010). Organoids are far more amenable to live-cell imaging than whole organisms, such as mice, and can complement mouse studies with imaging and perturbations that would be otherwise impractical. High-resolution live imaging of development, stem cell dynamics, and cancer invasion has been demonstrated in several organoid systems (Sato et al., 2011; Nguyen-Ngoc et al., 2015).
Finally, manipulating microenvironmental cues and specific cell populations is more straightforward in organoids than model organisms. Organoids are typically embedded in a matrix of the investigators’ choice, allowing for increased control over its source, composition, and mechanical properties. Cells acquired from tissue can be sorted, modified to delete or express specific genes, and expanded before assembly into organoids, allowing investigators to build organoids using defined building blocks from the bottom up. These organoids enable otherwise infeasible studies, such as tissues bearing gene knockouts that would be embryonically lethal. Additionally, this strategy allows the construction of organoids composed of specific proportions of cell subpopulations. For example, Sato et al. (2011) assembled intestinal organoids from stem cells with or without Paneth cells to identify Paneth cells as essential components of the intestinal stem cell (ISC) niche, a result previously obscured by incomplete lineage ablation. Moreover, viral vectors or transfection allows genetic manipulation of organoids with far less time and expense than developing a transgenic mouse (Koo et al., 2011). As a result, organoid approaches greatly enhance our capacity to study specific cell populations, phenotypes, and mutations within the context of a tissue. Organoid models of aging are in their earliest stages, but promising work, discussed in this perspective, has demonstrated their potential applicability to aging research.
Studying aging in organoids
Molecular and cellular processes
The genome can be damaged through several mechanisms, leading to the acquisition of genetic and epigenetic changes with age. DNA damage activates repair pathways but can also trigger mutations, apoptosis, or senescence. These outcomes can lead to aberrant behavior and deleterious effects on neighboring cells, such as inflammatory senescence-associated signaling (Parrinello et al., 2005). Epigenetically, aging cells acquire regional hyper- and hypomethylation, as well as increased variance in epigenetic marks (Jones et al., 2015). These cell-intrinsic aspects of aging are preserved in primary cells and tissue-derived organoids (Blokzijl et al., 2016).
Glycation, oxidation, and misfolding are foremost among molecular damage to proteins during aging. In particular, ECM proteins such as collagen, elastin, and crystallin are susceptible to nonenzymatic glycation, leading to stiffening of the ECM (Singh et al., 2001). Age-associated glycation, as well as altered structure, composition, and cross-linking of the ECM, has been associated with functional declines in a variety of tissues (Phillip et al., 2015). As matrix is a major component of organoid culture, organoids are a natural model for the effects of aged ECM on tissue function.
Organoid assays can enable researchers to experimentally link aging-associated changes in ECM structure to functional consequences. ECM substrates and gels for cell culture have been isolated from human and animal tissue (Voytik-Harbin et al., 2007; Zhang et al., 2009; O’Brien et al., 2010). Decreased cell proliferation and stem cell function have been observed on matrices derived from older donors, including Bruch’s membrane and cardiac and skeletal muscle ECM (Gullapalli et al., 2005; Williams et al., 2014; Stearns-Reider et al., 2017). More complex phenotypes can also be linked to ECM source by using organoid models. The Schedin group (Lyons et al., 2011) has shown that ECM from rats in different reproductive states can instruct morphogenesis and invasion of human mammary and breast cancer organoids. In addition, several groups have experimentally modified the ECM via advanced glycation end product–mediated cross-linking to mimic ECM aging, demonstrating aging phenotypes such as skin and cartilage stiffening and increased invasiveness (Verzijl et al., 2002; Pageon and Asselineau, 2005; Rodriguez-Teja et al., 2015).
Chemical damage can also lead to degradation and aggregation of proteins and other biomolecules, such as lipids. Protein damage accumulates in the form of carbonylation, fragmentation, and cross-linking, whereas lipid peroxidation compromises membranes and leads to the accumulation of oxidized fatty acids. Oxidation products not only have reduced function but are also prone to cross-linking. Together with misfolding, age-associated protein oxidation results in proteolysis-resistant aggregates both intra- and extracellularly. This feature is highly conserved across species; one of the most universal signs of aging is the accumulation of intracellular lipofuscin lipid-protein aggregates (Jung et al., 2010). Although their causal role in pathogenesis is disputed, protein and lipid-protein aggregates are prominently associated with central nervous system dysfunction, such as the infamous tangles, amyloids, and Lewy bodies in neurodegenerative disease, and the retinal drusen in age-related macular degeneration (Crabb et al., 2002; Caughey and Lansbury, 2003). These late-stage manifestations of neurodegenerative pathology may be more apparent in 3D culture than in traditional cell culture, where secreted proteins diffuse freely into culture media rather than accumulate near cells. In a human stem cell–derived model of familial Alzheimer’s disease, 3D culture was found to not only improve neuronal maturation but also promote tauopathy though the accumulation of β-amyloid aggregates in the ECM, observations that did not occur in 2D culture or mice (Choi et al., 2014). Similarly, scaffold-free 3D neural organoids, but not 2D culture, were shown to accumulate amyloid aggregates and model tauopathy. Significantly, these signs of human neurodegeneration were spontaneous in patient-derived organoids but are often only observed in mice after the expression of multiple human transgenes (Raja et al., 2016).
Tissue-level processes
At the tissue level, accumulated molecular damage can manifest in a variety of ways. These include changes in tissue function, susceptibility to cancer and other diseases, changes in stem cell function, and alterations in the immune system (Aw et al., 2007; López-Otín et al., 2013; Blau et al., 2015). Encouragingly, a variety of organoid models have already been explored to study aspects of each of these age-related phenomena.
One feature of aging is a progressive decline in physiological function. Various signs of aging, such as lower glomerular filtration rate, reduced cardiac output, and increased susceptibility to disease, can often be tied to tissue-specific changes that may be more apparent in 3D tissues than 2D culture. For example, a semi-enclosed cardiac organoid chamber allows for the measurement of pressure and ejection fraction (Lee et al., 2008). Barrier function can be measured across a kidney- or neural-endothelial interface (Vernetti et al., 2017), which could be used to model aging-associated declines in glomerular filtration rate and blood-brain barrier function (Tan et al., 2010; Elahy et al., 2015). Additionally, gastrointestinal organoids have been used extensively to study host-microbe interactions (Bartfeld et al., 2015; Engevik et al., 2015). They would be well-suited to studying the aging microbiome and age-related susceptibility to disease caused by ordinarily asymptomatic microbes, such as Clostridium difficile (Htwe et al., 2007; Heintz and Mair, 2014). Age-associated changes in circadian rhythm may also be modeled in organoids, considering that intestinal explants and stem cell–derived organoids, but not 2D culture, have been demonstrated to spontaneously produce robust, synchronized circadian oscillations (Myers and Badia, 1995; Moore et al., 2014). In general, organotypic cultures can represent more complex and accurate phenotypes than 2D culture and are thus ideal for modeling their decline in aging.
Organoids are widespread models for cancer progression that have been used to study tumor-initiating mutations and processes, such as cancer cell dissemination (Li et al., 2014; Nguyen-Ngoc et al., 2015). A natural extension of this work would be to use organoids from donors of different ages to understand how normal, aged tissue responds to in vitro oncogenic transformation. For example, transformed human mammary epithelial cells develop phenotypes dependent on donor age. Aged cells more frequently adopted a luminal-like phenotype and expressed estrogen receptor α in organoid culture, suggesting that age is a determinant of cancer subtype (Lee et al., 2015). In addition, organoid culture permits study of the tumor microenvironment, including neighboring cells and the ECM. Age-related changes to the microenvironment may play a role in altered susceptibility to cancer. Experimental glycation of the ECM increased invasiveness and decreased epithelial differentiation in nonmalignant prostate acini (Rodriguez-Teja et al., 2015). Similarly, the accumulation of senescent cells with age may exert tumor-promoting effects on neighboring cells. By coculturing fibroblasts with nonmalignant mammary organoids, Parrinello et al. (2005) demonstrated that senescent fibroblasts secreted factors that increased invasion and proliferation, and decreased epithelial differentiation.
Organoids are excellent models for stem cell maintenance and tissue repair, because organoids can recreate in vitro the minimal unit of a stem cell niche: cells and ECM in an organotypic structure (Murrow et al., 2017). Stem cell exhaustion, or reduced proliferation and biased differentiation of stem cells, contributes to aging in several tissues, and stem cell function is a common readout in aging studies (Brack et al., 2007; Pang et al., 2011; Garbe et al., 2012). For example, mouse intestinal organoids have been used to study the proliferation and differentiation of ISCs in the context of aging research. In one study of ISC function, which declines with age, Nalapareddy et al. (2017) used organoids from aged mice to determine that the aged ISC niche was deficient in Wnt signaling. Exogenous Wnt3a supplementation in organoid culture was sufficient to restore organoid formation and lobe growth of aged ISCs. Other investigators have used organoid-formation assays to study caloric restriction, an intervention shown to have anti-aging effects in several model organisms, including nonhuman primates (Mattison et al., 2017). By combining the Paneth cells and ISCs of ad libitum-fed and calorie-restricted mice, Yilmaz et al. (2012) demonstrated that Paneth cells from calorie-restricted mice were responsible for increasing ISC organoid formation and size. The capacity to separate and perturb the contributions of individual niche components, such as growth factors and accessory cells, during aging and anti-aging interventions in a rapid, systematic manner is an important advantage that organoids provide over model organisms.
Aging of the immune system is characterized by changing proportions of immune cells, declining innate immune function, and increased chronic inflammation (Aw et al., 2007). To study how these factors intersect with age-associated diseases such as cancer and osteoarthritis, aging researchers can take advantage of existing methods for studying the effects of immune cell co-culture and pro-inflammatory stimuli in organoids. For example, co-culture of primary mouse mammary epithelial organoids and immune cells demonstrated how immune cells regulate branching morphogenesis (Plaks et al., 2015). Systemic inflammation has also been studied in organoids by adding inflammatory cytokines or antiinflammatory drugs to the culture medium. Anti-inflammatory treatment of intestinal organoids suppressed the senescence-associated inflammatory response and reduced inflammation-mediated tumorigenesis (Pribluda et al., 2013). Sun et al. (2011) used both cytokines and macrophage-conditioned media to simulate immune involvement in a 3D human chondrocyte model of osteoarthritis. In response to inflammation, the cartilage scaffolds demonstrated phenotypic features of early osteoarthritis, such as increased and atypical matrix synthesis and degradation. These examples highlight systems that integrate organoids with immune cells and inflammatory cytokines.
Sourcing organoids
Organoids can be derived from several sources, most notably primary cells and tissue or pluripotent stem cells (PSCs). Cell lines are less suitable sources for most applications of organoids to aging research, because of immortalization, evasion of cellular senescence, or tumor origins. In contrast, tissues from donors of different ages are a more suitable model of the phenotypes of aging. These samples are becoming more readily available to the basic science research community. A handful of untransformed primary human cell types and tissues, such as keratinocytes, stromal and dermal fibroblasts, and mammary epithelial cells, are available from cell banks. Skin, blood, marrow, fat, breast, and muscle are relatively abundant tissues that can be acquired from healthy donors undergoing elective surgery. In addition, primary human material can be obtained from pathologically normal tumor-peripheral tissue, surgical discards, tissues ineligible for transplant, and trimmings from transplanted tissues (Barkauskas et al., 2017).
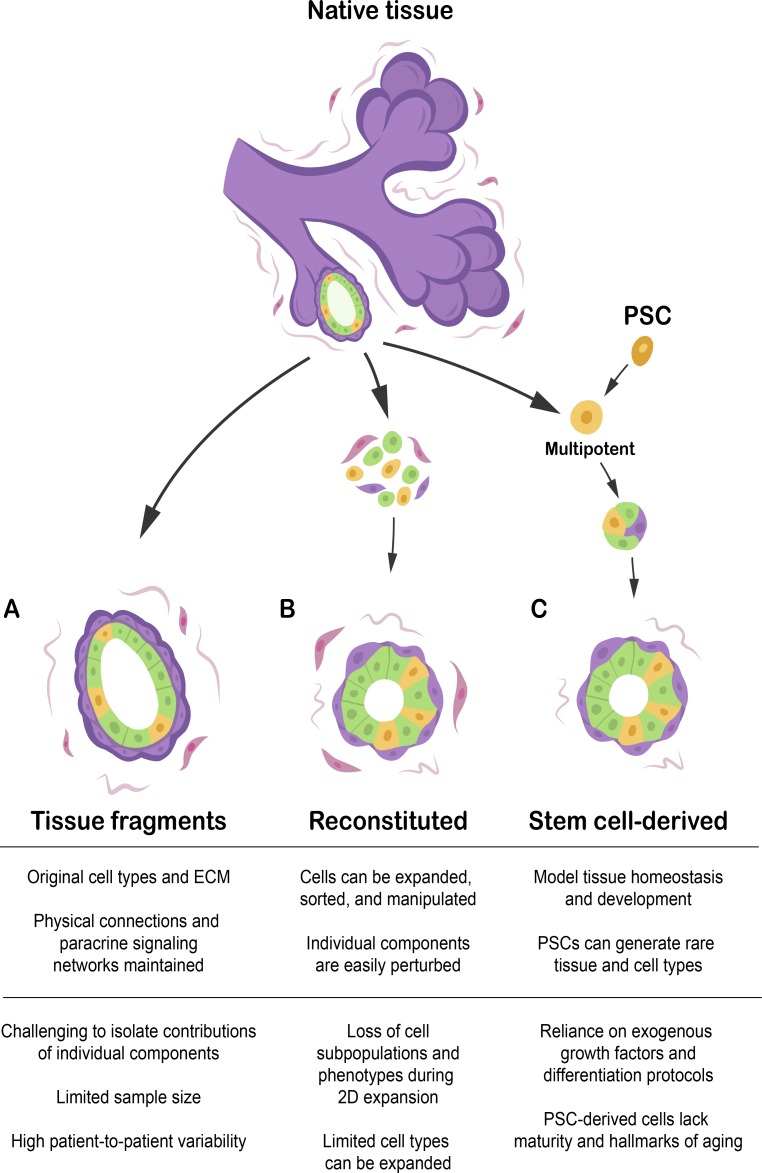
Fresh, primary tissue explants or fragments can be cultured with minimal processing, often retaining multiple cell types and the surrounding donor-derived ECM and stroma (Fig. 1 A; Tanos et al., 2013; Nguyen-Ngoc et al., 2015; Li et al., 2016). Some, such as arteries, skin, and central nervous system, are typically cultured as thin slices; alternatively, organoids may be derived from fragments of digested minced tissue. Both strategies maintain original tissue structure, minimize disruption to cell state, and may include ECM and accessory cells. The maintenance of intact paracrine signaling networks contributes to the overall function of tissue fragment-derived organoids. For example, the stromal compartment in intestinal organoids can maintain the stem cell niche without exogenous growth factors (Ootani et al., 2009). Similarly, human mammary organoids retaining stromal and ECM elements maintain hormone responsiveness and proliferate without exogenous growth factors (Tanos et al., 2013). These organoids represent the most physiologically relevant in vitro models and can be particularly valuable for studying complex, intercellular signaling networks in aging tissue. However, sample size and tissue types are limited, and it can be difficult to specifically perturb one component or cell type in the system.
Figure 1.
Types of organoids. (A) Tissue fragments and explants are generated from native tissue with little disruption to cell and ECM organization and no intermediate period in 2D culture. This limits total sample size and manipulability but is unmatched in mimicking the original tissue. (B) Reconstituted organoids are assembled in vitro from cultured or sorted cells. This permits cell expansion and modular assembly of components. However, some subpopulations or phenotypes, such as senescent and hormone-responsive cells, may be lost during culture. (C) Stem cell–derived organoids are differentiated from multipotent adult tissue stem cells or PSCs under the appropriate differentiation and maintenance conditions. PSCs specifically can generate rare tissue and cell types, but have limited maturity and lose the hallmarks of aging.
In contrast, primary human cells can be dissociated, optionally expanded in culture, and then reconstituted into organoids. These reconstituted organoids allow for expanded cell numbers and considerably more control over constituent cells and ECM, including the opportunity to perturb specific components (Fig. 1 B). Although tissue composition and cell state change rapidly on 2D culture, suitable conditions for preserving lineage-specific markers during expansion have been identified in a variety of tissues, including keratinocytes, fibroblasts, and mammary and lung epithelial cells (Wilson et al., 1992; Stampfer et al., 2013; Barkauskas et al., 2017). Lung epithelial cells can be expanded in culture and give rise to branched bronchial or alveolar organoids (Barkauskas et al., 2017). Human mammary epithelial cells are a notable aging resource established from surgical discards of donors of different ages and disease status, including healthy women and cancer patients (Stampfer et al., 2013). Nonimmortal mammary epithelial cells can be expanded in vitro for several passages, retain transcriptional signatures of their source tissue, and recapitulate the bilayered structure of the mammary gland in reconstituted organoids (Garbe et al., 2012; Cerchiari et al., 2015). One caveat is that expansion in culture selects for proliferative cells, potentially excluding cells with short telomeres, terminally differentiated nonproliferating cells, and cells undergoing senescence. Furthermore, 2D culture also causes loss of cell type–specific transcriptional profiles; some of this dedifferentiation can be reversed or ameliorated on transitioning to organotypic culture (Caron et al., 2012).
Primary tissue is also a source of multipotent adult tissue stem cells that can generate organoids in the presence of specific growth factors; stem cell–derived organoids are also generated from PSCs (Fig. 1 C). Organoids from multiple epithelial tissues, including stomach, small intestine, colon, liver, pancreas, breast, and prostate, have been expanded from single primary stem or progenitor cells (Table 1). However, an issue with this method is that their maintenance requires the inclusion of stem cell niche–maintaining soluble factors, which may obscure phenotypes of aging that originate in growth factor signaling (Broutier et al., 2016; Nalapareddy et al., 2017). In contrast, PSCs, including induced PSCs (iPSCs) and embryonic stem cells allow for a greater range of key human tissue types than can be practicably supplied by surgical discards, notably brain and cardiac muscle. These organoids frequently contain multiple cell types and self-organizing tissue structures that recapitulate embryonic development. For example, human iPSC-derived brain organoids have been used to model early stage and developmental disorders (Qian et al., 2016) and could become excellent models for neurodegenerative diseases such as Alzheimer’s disease. Raja et al. (2016) demonstrated that iPSC organoids derived from patients with familial Alzheimer’s disease develop robust pathology, including amyloid aggregation, hyperphosphorylated tau protein, and endosome abnormalities. Caution must be exercised when applying stem cell studies to aging research. Although familial disease–linked mutations are widely studied, genetic disease may differ mechanistically from sporadic, age-related disease. Furthermore, PSCs begin in a fetal-like state and require additional maturation. Many hallmarks of aging, aside from somatic mutations, are reverted on iPSC reprogramming, including DNA damage foci, epigenetic marks, oxidative damage, senescence markers, and telomere length (Horvath, 2013; Cornacchia and Studer, 2017).
Table 1. Examples of rodent and human tissues cultured as organoids.
Organoid culture has now been extended to a variety of tissues, including many from adult human primary tissue or cells. h, human; m, mouse; MSC, mesenchymal stem cell.
Outlook
Advances in organotypic culture techniques ultimately seek to obtain relevant phenotypes in vitro. Remarkable progress has been made in developing organoid techniques and culture systems, but several limitations remain to be addressed or validated. Furthermore, there are many opportunities to improve on the reproducibility and complexity of organoids, particularly by the inclusion of tissue engineering and microfluidics. These areas of current research are summarized in Table 2.
Table 2. Summary of current limitations and areas for development in organoid culture for aging.
| Goals | Approaches | Challenges |
|---|---|---|
| Increase developmental maturity of pluripotent stem cell–derived organoids | Long-term culture (Nicholas et al., 2013; Takasato et al., 2016; Tzatzalos et al., 2016) | Stem cells are highly sensitive to culture conditions, impeding robust protocols |
| In vivo maturation (Huch et al., 2013; Watson et al., 2014; Takebe et al., 2015) | ||
| Mechanical or electrical conditioning of muscle and cartilage (Ruan et al., 2016) | ||
| Improved culture conditions and differentiation protocols | ||
| Acceleration by small molecules (Chambers et al., 2012) | ||
| Source culturable, age-varied human cells | Surgical discards from elective surgery, transplant trimmings, and tissue peripheral to tumors | Hard to source certain tissues, especially healthy, culturable adult cells |
| Cells adapt to culture and are not infinitely renewable | ||
| Nontumor peripheral tissue may differ from healthy tissue | ||
| In vitro aging to model the effects of specific age-related lesions and provide a source of artificially aged cells | Induced senescence by DNA damage or environmental stress (Busuttil et al., 2003; Parrinello et al., 2005; Nasto et al., 2013) | Controversial which treatments best phenocopy aging |
| Long-term culture (Dos Santos et al., 2015) | ||
| Progeria mutations (Liu et al., 2011; Zhang et al., 2011; Miller et al., 2013) | ||
| Direct reprogramming of aged cells (Mertens et al., 2015) | ||
| Mimic the effects of the aged ECM in vitro | ECM from aged donors (Gullapalli et al., 2005; Williams et al., 2014; Stearns-Reider et al., 2017) | Controversial which treatments best phenocopy aging |
| Glycation crosslinking (Rodriguez-Teja et al., 2015) | ECM extraction from tissues can alter its mechanical properties, microstructure, and composition | |
| Enzymatic crosslinking (Levental et al., 2009) | ||
| Develop aging-relevant experimental readouts | Epigenetic clock (Hannum et al., 2013; Horvath, 2013) | Unclear which signs of aging are most important |
| Mutational analysis (Blokzijl et al., 2016) | Require signs of aging that change appreciably across the span of an experiment | |
| Protein/DNA oxidation | ||
| Protein aggregation | ||
| Tissue-specific functional assays | ||
| Long-term maintenance of organoids in a stable, growth-arrested state | Improved culture media | Relatively few published maintenance conditions |
| Improved ECM and bioreactors | Vetting culture conditions is lengthy | |
| Complete modeling of whole organs and physiological systems | Co-culture with immune cells, stromal cells, and microbiome bacteria (Parrinello et al., 2005; Engevik et al., 2015; Plaks et al., 2015) | Few good techniques for sophisticated organoid construction |
| Increase throughput and reproducibility | Vascularization (Auger et al., 2013) | Powerful techniques are often cumbersome and low-throughput |
| Innervation (Workman et al., 2017) | ||
| In vivo implantation (Watson et al., 2014; Takebe et al., 2015) | ||
| Microfluidic access to apical/basal fluid reservoirs and fluid transport between organ systems (Vernetti et al., 2017) | ||
| Organ-specific ECM (Voytik-Harbin et al., 2007; Zhang et al., 2009; O’Brien et al., 2010) | ||
| Defined artificial ECMs may decrease lot-to-lot variability (Gjorevski et al., 2016) | ||
| Growth factor distribution within ECM gels for spatial control over growth and differentiation (Wylie et al., 2011) | ||
| Cell patterning for control over initial organoid shape and composition (Nelson et al., 2008; Todhunter et al., 2015) | ||
| 3D-printed gels amenable to perfusion with control over large-scale tissue structure (Kolesky et al., 2016) | ||
| Morphological screening and sorting to enrich for correctly formed organoids (Arora et al., 2017) |
Although pluripotent stem cells are a significant source for organoids, one concern is the immaturity of cells and tissues derived from these cells, because PSCs begin in a fetal-like state. Functional and developmental maturation of stem cells is an active area of research and has been pursued by techniques such as long-term culture, altered concentrations of growth factors and inhibitors, direct differentiation, and mechanical or electrical stimulation (Chambers et al., 2012; Nicholas et al., 2013; Mertens et al., 2015; Ruan et al., 2016; Takasato et al., 2016; Tzatzalos et al., 2016). In vivo maturation, typically by implantation into a mouse, has also been demonstrated to increase organoid maturity and can be used to generate adult-like human tissue and organoids (Huch et al., 2013; Watson et al., 2014; Takebe et al., 2015).
Similarly, accelerated aging in vitro will be necessary for PSC-derived organoids and useful for studying the functional consequences of age-associated damage. Accelerated aging experiments typically involve cells derived from progeric individuals (Liu et al., 2011; Miller et al., 2013) or expose cells to aging-associated stress, such as reactive oxygen species, inflammation, or radiation, which appear to phenocopy aspects of aging (Nasto et al., 2013; Cornacchia and Studer, 2017). With regard to aging experiments, organoid culture, as opposed to cell culture, has the advantage of increased stability. This stability has allowed culture periods as long as 60 wk (Hibiya et al., 2017), which can be used to study chronological aging in vitro. For example, 120 d of culture of reconstituted human epidermis led to increased cellular senescence and morphological changes resembling those of chronological skin aging in vivo (Dos Santos et al., 2015). Furthermore, genomic stability has been reported to be higher in organoids than in traditional cell culture, as demonstrated by the use of organoid culture to clonally expand stem cells before mutational analysis (Behjati et al., 2014; Huch et al., 2015; Blokzijl et al., 2016). Notably, organoid clonal expansion was also used to quantify the accumulation of mutations in adult ISCs with age (Blokzijl et al., 2016).
Primary cell and organoid culture remains highly sensitive to culture conditions. Support of the ISC niche, for example, was a revolutionary advance, permitting long-term expansion of intestinal organoids that once could be cultured for only a couple of weeks (Ootani et al., 2009). Other tissues, such as artery or ovary, can still only be cultured for only short times. Additionally, aging studies may be particularly sensitive to culture conditions. Numerous culture parameters that can phenocopy aspects of aging, such as replicative aging, oxygen tension, and free glucose, must be carefully controlled for. Finally, long-term culture and studying tissue homeostasis require the maintenance of growth-arrested organoids resembling the resting state of adult tissues. Identifying microenvironmental conditions to transition organoid tissues from a proliferative to a more mature, growth-arrested state would represent an important contribution to aging research.
As in vitro models increasingly seek to recapitulate complex tissue- and organ-level phenotypes, transport of oxygen and nutrients by diffusion becomes limiting. Air–liquid interfaces have been used to culture larger explants, but in general, vasculature will be necessary for the survival of most organoids over a couple hundred micrometers wide (Auger et al., 2013), as well as useful for the study of aging-relevant phenotypes, such as metastasis and deterioration of the blood-brain barrier and glomerulus (Tan et al., 2010; Elahy et al., 2015). Furthermore, vascularization has the potential to network organoids into systemic, multiorgan contexts that allow the exchange of hormones, cytokines, and metabolites, which have been implicated in aging in mice (Villeda et al., 2014). Incorporation into a systemic environment presents an opportunity to complement the strengths of organoid and in vivo platforms: organoids generated and manipulated in vitro can be implanted into mice, where they mature and integrate into the host vasculature (Watson et al., 2014; Takebe et al., 2015). Such systems could be powerful tools to study the effects of an aged systemic environment on human tissues.
Similarly, microfluidics could increase the complexity of in vitro organoid systems while improving throughput and reproducibility. Multiorgan systems have been developed for the study of organ interactions via an in vivo-like sequential, organ-to-organ transfer of media, with a particular interest in drug interactions (Vernetti et al., 2017). These devices could provide insight into how aging tissues affect other parts of the body. Additionally, microfluidic devices permit lumen access and control over millimeter-scale morphology. Although complex tissue structures, such as intestinal crypts and kidney nephrons, are readily generated by organoid self-organization (Morizane et al., 2015), the process can be highly heterogeneous, making assay readout complex or impossible. For example, reproducible kidney organoids with a channel-accessible central duct would enable key functional assays, namely permeability and urea concentration. Methods for micron-scale organization of cells within a tissue or microfluidic chamber (Nelson et al., 2008; Todhunter et al., 2015; Kolesky et al., 2016) could be used to create reproducible, high-throughput organoid assays for aging and other diseases.
Organoids are a powerful emerging tool for the study of human health and disease. Aging research has long needed a model with the practical advantages of simpler organisms and traditional cell culture, as well as the complexity required to model the tissue-level phenotypes that accompany aging. As researchers develop organotypic cultures of increasing complexity, it is becoming clear that organoids will enable the study of tissue-level, and even systems-level, phenotypes of aging. Human organoids have the potential to model the human body with unmatched fidelity. With time, new models will be developed and validated, significantly accelerating human aging research and the search for therapies that will improve human health and lifespan.
Acknowledgments
We thank Drs. Hao Li and Saul Villeda for their constructive feedback and discussion.
J.L. Hu is supported by the National Science Foundation Graduate Research Fellowship Program. M.A. LaBarge and M.E. Todhunter are supported by the Era of Hope Scholar Award from the Congressionally Directed Medical Research Programs Breast Cancer Research Program (BC141351), the National Institute on Aging (R01AG040081), and the City of Hope Center for Cancer and Aging. Z.J. Gartner is a Chan Zuckerberg Biohub investigator and is supported by an Era of Hope Scholar Award (BC123047), the Center for Cellular Construction (NSF DBI-1548297), and the National Institutes of Health (DP2HD080351).
The authors declare no competing financial interests.
References
- Antonica F., Kasprzyk D.F., Opitz R., Iacovino M., Liao X.-H., Dumitrescu A.M., Refetoff S., Peremans K., Manto M., Kyba M., and Costagliola S.. 2012. Generation of functional thyroid from embryonic stem cells. Nature. 491:66–71. 10.1038/nature11525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora N., Imran Alsous J., Guggenheim J.W., Mak M., Munera J., Wells J.M., Kamm R.D., Asada H.H., Shvartsman S.Y., and Griffith L.G.. 2017. A process engineering approach to increase organoid yield. Development. 144:1128–1136. 10.1242/dev.142919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astashkina A.I., Mann B.K., Prestwich G.D., and Grainger D.W.. 2012. Comparing predictive drug nephrotoxicity biomarkers in kidney 3-D primary organoid culture and immortalized cell lines. Biomaterials. 33:4712–4721. 10.1016/j.biomaterials.2012.03.001 [DOI] [PubMed] [Google Scholar]
- Auger F.A., Gibot L., and Lacroix D.. 2013. The pivotal role of vascularization in tissue engineering. Annu. Rev. Biomed. Eng. 15:177–200. 10.1146/annurev-bioeng-071812-152428 [DOI] [PubMed] [Google Scholar]
- Aw D., Silva A.B., and Palmer D.B.. 2007. Immunosenescence: emerging challenges for an ageing population. Immunology. 120:435–446. 10.1111/j.1365-2567.2007.02555.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azpurua J., and Seluanov A.. 2013. Long-lived cancer-resistant rodents as new model species for cancer research. Front. Genet. 3:319 10.3389/fgene.2012.00319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbero A., Grogan S., Schäfer D., Heberer M., Mainil-Varlet P., and Martin I.. 2004. Age related changes in human articular chondrocyte yield, proliferation and post-expansion chondrogenic capacity. Osteoarthritis Cartilage. 12:476–484. 10.1016/j.joca.2004.02.010 [DOI] [PubMed] [Google Scholar]
- Barkauskas C.E., Cronce M.J., Rackley C.R., Bowie E.J., Keene D.R., Stripp B.R., Randell S.H., Noble P.W., and Hogan B.L.M.. 2013. Type 2 alveolar cells are stem cells in adult lung. J. Clin. Invest. 123:3025–3036. 10.1172/JCI68782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkauskas C.E., Chung M.-I., Fioret B., Gao X., Katsura H., and Hogan B.L.M.. 2017. Lung organoids: current uses and future promise. Development. 144:986–997. 10.1242/dev.140103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N., Huch M., Kujala P., van de Wetering M., Snippert H.J., van Es J.H., Sato T., Stange D.E., Begthel H., van den Born M., et al. . 2010. Lgr5(+ve) stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell. 6:25–36. 10.1016/j.stem.2009.11.013 [DOI] [PubMed] [Google Scholar]
- Bartfeld S., Bayram T., van de Wetering M., Huch M., Begthel H., Kujala P., Vries R., Peters P.J., and Clevers H.. 2015. In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology. 148:126–136.e6. 10.1053/j.gastro.2014.09.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behjati S., Huch M., van Boxtel R., Karthaus W., Wedge D.C., Tamuri A.U., Martincorena I., Petljak M., Alexandrov L.B., Gundem G., et al. . 2014. Genome sequencing of normal cells reveals developmental lineages and mutational processes. Nature. 513:422–425. 10.1038/nature13448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmin J., Bernard C., Corman B., Merval R., Esposito B., and Tedgui A.. 1995. Increased production of tumor necrosis factor and interleukin-6 by arterial wall of aged rats. Am. J. Physiol. 268:H2288–H2293. [DOI] [PubMed] [Google Scholar]
- Ben-Ze’ev A., Robinson G.S., Bucher N.L.R., and Farmer S.R.. 1988. Cell-cell and cell-matrix interactions differentially regulate the expression of hepatic and cytoskeletal genes in primary cultures of rat hepatocytes. Proc. Natl. Acad. Sci. USA. 85:2161–2165. 10.1073/pnas.85.7.2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhumiratana S., Eton R.E., Oungoulian S.R., Wan L.Q., Ateshian G.A., and Vunjak-Novakovic G.. 2014. Large, stratified, and mechanically functional human cartilage grown in vitro by mesenchymal condensation. Proc. Natl. Acad. Sci. USA. 111:6940–6945. 10.1073/pnas.1324050111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blau H.M., Cosgrove B.D., and Ho A.T.V.. 2015. The central role of muscle stem cells in regenerative failure with aging. Nat. Med. 21:854–862. 10.1038/nm.3918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blokzijl F., de Ligt J., Jager M., Sasselli V., Roerink S., Sasaki N., Huch M., Boymans S., Kuijk E., Prins P., et al. . 2016. Tissue-specific mutation accumulation in human adult stem cells during life. Nature. 538:260–264. 10.1038/nature19768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brack A.S., Conboy M.J., Roy S., Lee M., Kuo C.J., Keller C., and Rando T.A.. 2007. Increased Wnt signaling during aging alters muscle stem cell fate and increases fibrosis. Science. 317:807–810. 10.1126/science.1144090 [DOI] [PubMed] [Google Scholar]
- Broutier L., Andersson-Rolf A., Hindley C.J., Boj S.F., Clevers H., Koo B.-K., and Huch M.. 2016. Culture and establishment of self-renewing human and mouse adult liver and pancreas 3D organoids and their genetic manipulation. Nat. Protoc. 11:1724–1743. 10.1038/nprot.2016.097 [DOI] [PubMed] [Google Scholar]
- Busuttil R.A., Rubio M., Dollé M.E.T., Campisi J., and Vijg J.. 2003. Oxygen accelerates the accumulation of mutations during the senescence and immortalization of murine cells in culture. Aging Cell. 2:287–294. 10.1046/j.1474-9728.2003.00066.x [DOI] [PubMed] [Google Scholar]
- Caron M.M.J., Emans P.J., Coolsen M.M.E., Voss L., Surtel D.A.M., Cremers A., van Rhijn L.W., and Welting T.J.M.. 2012. Redifferentiation of dedifferentiated human articular chondrocytes: comparison of 2D and 3D cultures. Osteoarthritis Cartilage. 20:1170–1178. 10.1016/j.joca.2012.06.016 [DOI] [PubMed] [Google Scholar]
- Caughey B., and Lansbury P.T.. 2003. Protofibrils, pores, fibrils, and neurodegeneration: separating the responsible protein aggregates from the innocent bystanders. Annu. Rev. Neurosci. 26:267–298. 10.1146/annurev.neuro.26.010302.081142 [DOI] [PubMed] [Google Scholar]
- Cerchiari A.E., Garbe J.C., Jee N.Y., Todhunter M.E., Broaders K.E., Peehl D.M., Desai T.A., LaBarge M.A., Thomson M., and Gartner Z.J.. 2015. A strategy for tissue self-organization that is robust to cellular heterogeneity and plasticity. Proc. Natl. Acad. Sci. USA. 112:2287–2292. 10.1073/pnas.1410776112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers S.M., Qi Y., Mica Y., Lee G., Zhang X.J., Niu L., Bilsland J., Cao L., Stevens E., Whiting P., et al. . 2012. Combined small-molecule inhibition accelerates developmental timing and converts human pluripotent stem cells into nociceptors. Nat. Biotechnol. 30:715–720. 10.1038/nbt.2249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.H., and Davies J.A.. 2012. An improved method of renal tissue engineering, by combining renal dissociation and reaggregation with a low-volume culture technique, results in development of engineered kidneys complete with loops of Henle. Nephron, Exp. Nephrol. 121:e79–e85. 10.1159/000345514 [DOI] [PubMed] [Google Scholar]
- Chaudhuri A.R., de Waal E.M., Pierce A., Van Remmen H., Ward W.F., and Richardson A.. 2006. Detection of protein carbonyls in aging liver tissue: A fluorescence-based proteomic approach. Mech. Ageing Dev. 127:849–861. 10.1016/j.mad.2006.08.006 [DOI] [PubMed] [Google Scholar]
- Choi S.H., Kim Y.H., Hebisch M., Sliwinski C., Lee S., D’Avanzo C., Chen H., Hooli B., Asselin C., Muffat J., et al. . 2014. A three-dimensional human neural cell culture model of Alzheimer’s disease. Nature. 515:274–278. 10.1038/nature13800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornacchia D., and Studer L.. 2017. Back and forth in time: Directing age in iPSC-derived lineages. Brain Res. 1656:14–26. 10.1016/j.brainres.2015.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabb J.W., Miyagi M., Gu X., Shadrach K., West K.A., Sakaguchi H., Kamei M., Hasan A., Yan L., Rayborn M.E., et al. . 2002. Drusen proteome analysis: an approach to the etiology of age-related macular degeneration. Proc. Natl. Acad. Sci. USA. 99:14682–14687. 10.1073/pnas.222551899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danahay H., Pessotti A.D., Coote J., Montgomery B.E., Xia D., Wilson A., Yang H., Wang Z., Bevan L., Thomas C., et al. . 2015. Notch2 is required for inflammatory cytokine-driven goblet cell metaplasia in the lung. Cell Reports. 10:239–252. 10.1016/j.celrep.2014.12.017 [DOI] [PubMed] [Google Scholar]
- Dekkers J.F., Berkers G., Kruisselbrink E., Vonk A., de Jonge H.R., Janssens H.M., Bronsveld I., van de Graaf E.A., Nieuwenhuis E.E.S., Houwen R.H.J., et al. . 2016. Characterizing responses to CFTR-modulating drugs using rectal organoids derived from subjects with cystic fibrosis. Sci. Transl. Med. 8:344ra84 10.1126/scitranslmed.aad8278 [DOI] [PubMed] [Google Scholar]
- Dos Santos M., Metral E., Boher A., Rousselle P., Thepot A., and Damour O.. 2015. In vitro 3-D model based on extending time of culture for studying chronological epidermis aging. Matrix Biol. 47:85–97. 10.1016/j.matbio.2015.03.009 [DOI] [PubMed] [Google Scholar]
- Dye B.R., Hill D.R., Ferguson M.A.H., Tsai Y.-H., Nagy M.S., Dyal R., Wells J.M., Mayhew C.N., Nattiv R., Klein O.D., et al. . 2015. In vitro generation of human pluripotent stem cell derived lung organoids. eLife. 4:e05098 10.7554/eLife.05098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiraku M., Takata N., Ishibashi H., Kawada M., Sakakura E., Okuda S., Sekiguchi K., Adachi T., and Sasai Y.. 2011. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature. 472:51–56. 10.1038/nature09941 [DOI] [PubMed] [Google Scholar]
- Elahy M., Jackaman C., Mamo J.C., Lam V., Dhaliwal S.S., Giles C., Nelson D., and Takechi R.. 2015. Blood-brain barrier dysfunction developed during normal aging is associated with inflammation and loss of tight junctions but not with leukocyte recruitment. Immun. Ageing. 12:2 10.1186/s12979-015-0029-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engevik M.A., Engevik K.A., Yacyshyn M.B., Wang J., Hassett D.J., Darien B., Yacyshyn B.R., and Worrell R.T.. 2015. Human Clostridium difficile infection: inhibition of NHE3 and microbiota profile. Am. J. Physiol. Gastrointest. Liver Physiol. 308:G497–G509. 10.1152/ajpgi.00090.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman B.S., Brooks C.R., Lam A.Q., Fu H., Morizane R., Agrawal V., Saad A.F., Li M.K., Hughes M.R., Werff R.V., et al. . 2015. Modelling kidney disease with CRISPR-mutant kidney organoids derived from human pluripotent epiblast spheroids. Nat. Commun. 6:8715 10.1038/ncomms9715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D., Vela I., Sboner A., Iaquinta P.J., Karthaus W.R., Gopalan A., Dowling C., Wanjala J.N., Undvall E.A., Arora V.K., et al. . 2014. Organoid cultures derived from patients with advanced prostate cancer. Cell. 159:176–187. 10.1016/j.cell.2014.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbe J.C., Pepin F., Pelissier F.A., Sputova K., Fridriksdottir A.J., Guo D.E., Villadsen R., Park M., Petersen O.W., Borowsky A.D., et al. . 2012. Accumulation of multipotent progenitors with a basal differentiation bias during aging of human mammary epithelia. Cancer Res. 72:3687–3701. 10.1158/0008-5472.CAN-12-0157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjorevski N., Sachs N., Manfrin A., Giger S., Bragina M.E., Ordóñez-Morán P., Clevers H., and Lutolf M.P.. 2016. Designer matrices for intestinal stem cell and organoid culture. Nature. 539:560–564. 10.1038/nature20168 [DOI] [PubMed] [Google Scholar]
- Greider C.W., and Blackburn E.H.. 1987. The telomere terminal transferase of Tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell. 51:887–898. 10.1016/0092-8674(87)90576-9 [DOI] [PubMed] [Google Scholar]
- Gullapalli V.K., Sugino I.K., Van Patten Y., Shah S., and Zarbin M.A.. 2005. Impaired RPE survival on aged submacular human Bruch’s membrane. Exp. Eye Res. 80:235–248. 10.1016/j.exer.2004.09.006 [DOI] [PubMed] [Google Scholar]
- Hannum G., Guinney J., Zhao L., Zhang L., Hughes G., Sadda S., Klotzle B., Bibikova M., Fan J.B., Gao Y., et al. . 2013. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol. Cell. 49:359–367. 10.1016/j.molcel.2012.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen D.V., Lui J.H., Parker P.R.L., and Kriegstein A.R.. 2010. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature. 464:554–561. 10.1038/nature08845 [DOI] [PubMed] [Google Scholar]
- Hegab A.E., Arai D., Gao J., Kuroda A., Yasuda H., Ishii M., Naoki K., Soejima K., and Betsuyaku T.. 2015. Mimicking the niche of lung epithelial stem cells and characterization of several effectors of their in vitro behavior. Stem Cell Res. (Amst.). 15:109–121. 10.1016/j.scr.2015.05.005 [DOI] [PubMed] [Google Scholar]
- Heintz C., and Mair W.. 2014. You are what you host: microbiome modulation of the aging process. Cell. 156:408–411. 10.1016/j.cell.2014.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibiya S., Tsuchiya K., Hayashi R., Fukushima K., Horita N., Watanabe S., Shirasaki T., Nishimura R., Kimura N., Nishimura T., et al. . 2017. Long-term inflammation transforms intestinal epithelial cells of colonic organoids. J. Crohn’s Colitis. 11:621–630. 10.1093/ecco-jcc/jjw186 [DOI] [PubMed] [Google Scholar]
- Hohwieler M., Illing A., Hermann P.C., Mayer T., Stockmann M., Perkhofer L., Eiseler T., Antony J.S., Müller M., Renz S., et al. . 2017. Human pluripotent stem cell-derived acinar/ductal organoids generate human pancreas upon orthotopic transplantation and allow disease modelling. Gut. 66:473–486. 10.1136/gutjnl-2016-312423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S. 2013. DNA methylation age of human tissues and cell types. Genome Biol. 14:R115 10.1186/gb-2013-14-10-r115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Htwe T.H., Mushtaq A., Robinson S.B., Rosher R.B., and Khardori N.. 2007. Infection in the elderly. Infect. Dis. Clin. North Am. 21:711–743: ix. 10.1016/j.idc.2007.07.006 [DOI] [PubMed] [Google Scholar]
- Huch M., Bonfanti P., Boj S.F., Sato T., Loomans C.J.M., van de Wetering M., Sojoodi M., Li V.S.W., Schuijers J., Gracanin A., et al. . 2013. Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. EMBO J. 32:2708–2721. 10.1038/emboj.2013.204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huch M., Gehart H., van Boxtel R., Hamer K., Blokzijl F., Verstegen M.M.A., Ellis E., van Wenum M., Fuchs S.A., de Ligt J., et al. . 2015. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell. 160:299–312. 10.1016/j.cell.2014.11.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M.J., Goodman S.J., and Kobor M.S.. 2015. DNA methylation and healthy human aging. Aging Cell. 14:924–932. 10.1111/acel.12349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung T., Höhn A., and Grune T.. 2010. Lipofuscin: detection and quantification by microscopic techniques. In Advanced Protocols in Oxidative Stress II. Armstrong D., editor. Humana Press, Totowa, NJ: 173–193. 10.1007/978-1-60761-411-1_13 [DOI] [PubMed] [Google Scholar]
- Karthaus W.R., Iaquinta P.J., Drost J., Gracanin A., van Boxtel R., Wongvipat J., Dowling C.M., Gao D., Begthel H., Sachs N., et al. . 2014. Identification of multipotent luminal progenitor cells in human prostate organoid cultures. Cell. 159:163–175. 10.1016/j.cell.2014.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesky D.B., Homan K.A., Skylar-Scott M.A., and Lewis J.A.. 2016. Three-dimensional bioprinting of thick vascularized tissues. Proc. Natl. Acad. Sci. USA. 113:3179–3184. 10.1073/pnas.1521342113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo B.K., Stange D.E., Sato T., Karthaus W., Farin H.F., Huch M., van Es J.H., and Clevers H.. 2011. Controlled gene expression in primary Lgr5 organoid cultures. Nat. Methods. 9:81–83. 10.1038/nmeth.1802 [DOI] [PubMed] [Google Scholar]
- Kwong J., Chan F.L., Wong K.K., Birrer M.J., Archibald K.M., Balkwill F.R., Berkowitz R.S., and Mok S.C.. 2009. Inflammatory cytokine tumor necrosis factor alpha confers precancerous phenotype in an organoid model of normal human ovarian surface epithelial cells. Neoplasia. 11:529–541. 10.1593/neo.09112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- L’Heureux N., Pâquet S., Labbé R., Germain L., and Auger F.A.. 1998. A completely biological tissue-engineered human blood vessel. FASEB J. 12:47–56 https://doi.org/0892-6638/97/0012-0047. [DOI] [PubMed] [Google Scholar]
- Lang S.H., Stark M., Collins A., Paul A.B., Stower M.J., and Maitland N.J.. 2001. Experimental prostate epithelial morphogenesis in response to stroma and three-dimensional matrigel culture. Cell Growth Differ. 12:631–640. [PubMed] [Google Scholar]
- Lee E.J., Kim D.E., Azeloglu E.U., and Costa K.D.. 2008. Engineered cardiac organoid chambers: toward a functional biological model ventricle. Tissue Eng. Part A. 14:215–225. 10.1089/tea.2007.0351 [DOI] [PubMed] [Google Scholar]
- Lee J.K., Garbe J.C., Vrba L., Miyano M., Futscher B.W., Stampfer M.R., and LaBarge M.A.. 2015. Age and the means of bypassing stasis influence the intrinsic subtype of immortalized human mammary epithelial cells. Front. Cell Dev. Biol. 3:13 10.3389/fcell.2015.00013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levental K.R., Yu H., Kass L., Lakins J.N., Egeblad M., Erler J.T., Fong S.F.T., Csiszar K., Giaccia A., Weninger W., et al. . 2009. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 139:891–906. 10.1016/j.cell.2009.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Nadauld L., Ootani A., Corney D.C., Pai R.K., Gevaert O., Cantrell M.A., Rack P.G., Neal J.T., Chan C.W.-M., et al. . 2014. Oncogenic transformation of diverse gastrointestinal tissues in primary organoid culture. Nat. Med. 20:769–777. 10.1038/nm.3585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Ootani A., and Kuo C.. 2016. An air–liquid interface culture system for 3D organoid culture of diverse primary gastrointestinal tissues. In Gastrointestinal Physiology and Diseases: Methods and Protocols. Ivanov A.I., York S.N., and York N., editors. Humana Press, New York: 33–40. 10.1007/978-1-4939-3603-8_4 [DOI] [PubMed] [Google Scholar]
- Lindahl T. 1993. Instability and decay of the primary structure of DNA. Nature. 362:709–715. 10.1038/362709a0 [DOI] [PubMed] [Google Scholar]
- Linnemann J.R., Meixner L.K., Miura H., and Scheel C.H.. 2011. An organotypic 3D assay for primary human mammary epithelial cells that recapitulates branching morphogenesis. In 3D Cell Culture: Methods and Protocols. Koledova Z., editor. Humana Press, New York: 125–137. [DOI] [PubMed] [Google Scholar]
- Liu G.H., Barkho B.Z., Ruiz S., Diep D., Qu J., Yang S.-L., Panopoulos A.D., Suzuki K., Kurian L., Walsh C., et al. . 2011. Recapitulation of premature ageing with iPSCs from Hutchinson-Gilford progeria syndrome. Nature. 472:221–225. 10.1038/nature09879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Otín C., Blasco M.A., Partridge L., Serrano M., and Kroemer G.. 2013. The hallmarks of aging. Cell. 153:1194–1217. 10.1016/j.cell.2013.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozito T.P., Alexander P.G., Lin H., Gottardi R., Cheng A.W., and Tuan R.S.. 2013. Three-dimensional osteochondral microtissue to model pathogenesis of osteoarthritis. Stem Cell Res. Ther. 4(Suppl 1):S6 10.1186/scrt367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons T.R., O’Brien J., Borges V.F., Conklin M.W., Keely P.J., Eliceiri K.W., Marusyk A., Tan A.-C., and Schedin P.. 2011. Postpartum mammary gland involution drives progression of ductal carcinoma in situ through collagen and COX-2. Nat. Med. 17:1109–1115. 10.1038/nm.2416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madden L., Juhas M., Kraus W.E., Truskey G.A., and Bursac N.. 2015. Bioengineered human myobundles mimic clinical responses of skeletal muscle to drugs. Elife. 4:e04885 10.7554/eLife.04885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciniak A., Cohrs C.M., Tsata V., Chouinard J.A., Selck C., Stertmann J., Reichelt S., Rose T., Ehehalt F., Weitz J., et al. . 2014. Using pancreas tissue slices for in situ studies of islet of Langerhans and acinar cell biology. Nat. Protoc. 9:2809–2822. 10.1038/nprot.2014.195 [DOI] [PubMed] [Google Scholar]
- Matsui T., Nieto-Estévez V., Kyrychenko S., Schneider J.W., and Hsieh J.. 2017. Retinoblastoma protein controls growth, survival, and neuronal migration in human cerebral organoids. Development. 15:1025–1034. 10.1242/dev.143636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattison J.A., Colman R.J., Beasley T.M., Allison D.B., Kemnitz J.W., Roth G.S., Ingram D.K., Weindruch R., de Cabo R., and Anderson R.M.. 2017. Caloric restriction improves health and survival of rhesus monkeys. Nat. Commun. 8:14063 10.1038/ncomms14063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElwee J.J., Schuster E., Blanc E., Thornton J., and Gems D.. 2006. Diapause-associated metabolic traits reiterated in long-lived daf-2 mutants in the nematode Caenorhabditis elegans. Mech. Ageing Dev. 127:458–472. 10.1016/j.mad.2006.01.006 [DOI] [PubMed] [Google Scholar]
- Meerwaldt R., Graaff R., Oomen P.H.N., Links T.P., Jager J.J., Alderson N.L., Thorpe S.R., Baynes J.W., Gans R.O.B., and Smit A.J.. 2004. Simple non-invasive assessment of advanced glycation endproduct accumulation. Diabetologia. 47:1324–1330. 10.1007/s00125-004-1451-2 [DOI] [PubMed] [Google Scholar]
- Mertens J., Paquola A.C.M., Ku M., Hatch E., Böhnke L., Ladjevardi S., McGrath S., Campbell B., Lee H., Herdy J.R., et al. . 2015. Directly reprogrammed human neurons retain aging-associated transcriptomic signatures and reveal age-related nucleocytoplasmic defects. Cell Stem Cell. 17:705–718. 10.1016/j.stem.2015.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messner S., Agarkova I., Moritz W., and Kelm J.M.. 2013. Multi-cell type human liver microtissues for hepatotoxicity testing. Arch. Toxicol. 87:209–213. 10.1007/s00204-012-0968-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.D., Ganat Y.M., Kishinevsky S., Bowman R.L., Liu B., Tu E.Y., Mandal P.K., Vera E., Shim J.W., Kriks S., et al. . 2013. Human iPSC-based modeling of late-onset disease via progerin-induced aging. Cell Stem Cell. 13:691–705. 10.1016/j.stem.2013.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore K.A., Polte T., Huang S., Shi B., Alsberg E., Sunday M.E., and Ingber D.E.. 2005. Control of basement membrane remodeling and epithelial branching morphogenesis in embryonic lung by Rho and cytoskeletal tension. Dev. Dyn. 232:268–281. 10.1002/dvdy.20237 [DOI] [PubMed] [Google Scholar]
- Moore S.R., Pruszka J., Vallance J., Aihara E., Matsuura T., Montrose M.H., Shroyer N.F., and Hong C.I.. 2014. Robust circadian rhythms in organoid cultures from PERIOD2:LUCIFERASE mouse small intestine. Dis. Model. Mech. 7:1123–1130. 10.1242/dmm.014399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morizane R., Lam A.Q., Freedman B.S., Kishi S., Valerius M.T., and Bonventre J.V.. 2015. Nephron organoids derived from human pluripotent stem cells model kidney development and injury. Nat. Biotechnol. 33:1193–1200. 10.1038/nbt.3392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrow L.M., Weber R.J., and Gartner Z.J.. 2017. Dissecting the stem cell niche with organoid models: an engineering-based approach. Development. 144:998–1007. 10.1242/dev.140905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers B.L., and Badia P.. 1995. Changes in circadian rhythms and sleep quality with aging: mechanisms and interventions. Neurosci. Biobehav. Rev. 19:553–571. 10.1016/0149-7634(95)00018-6 [DOI] [PubMed] [Google Scholar]
- Nalapareddy K., Nattamai K.J., Kumar R.S., Karns R., Wikenheiser-Brokamp K.A., Sampson L.L., Mahe M.M., Sundaram N., Yacyshyn M.B., Yacyshyn B., et al. . 2017. Canonical Wnt signaling ameliorates aging of intestinal stem cells. Cell Reports. 18:2608–2621. 10.1016/j.celrep.2017.02.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasto L.A., Robinson A.R., Ngo K., Clauson C.L., Dong Q., St Croix C., Sowa G., Pola E., Robbins P.D., Kang J., et al. . 2013. Mitochondrial-derived reactive oxygen species (ROS) play a causal role in aging-related intervertebral disc degeneration. J. Orthop. Res. 31:1150–1157. 10.1002/jor.22320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson C.M., Inman J.L., and Bissell M.J.. 2008. Three-dimensional lithographically defined organotypic tissue arrays for quantitative analysis of morphogenesis and neoplastic progression. Nat. Protoc. 3:674–678. 10.1038/nprot.2008.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevalainen M.T., Härkönen P.L., Valve E.M., Ping W., Nurmi M., and Martikainen P.M.. 1993. Hormone regulation of human prostate in organ culture. Cancer Res. 53:5199–5207. [PubMed] [Google Scholar]
- Nguyen-Ngoc K., Shamir E.R., Huebner R.J., Beck J.N., Cheung K.J., and Ewald A.J.. 2015. 3D culture assays of murine mammary branching morphogenesis and epithelial invasion. In Tissue Morphogenesis: Methods and Protocols. Nelson C.M., editor. Springer, New York: 135–162. 10.1007/978-1-4939-1164-6_10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas C.R., Chen J., Tang Y., Southwell D.G., Chalmers N., Vogt D., Arnold C.M., Chen Y.J.J., Stanley E.G., Elefanty A.G., et al. . 2013. Functional maturation of hPSC-derived forebrain interneurons requires an extended timeline and mimics human neural development. Cell Stem Cell. 12:573–586. 10.1016/j.stem.2013.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi T.K., Ninomiya N., Sekine M., Komazaki S., Wang P.-C., Asashima M., and Kurisaki A.. 2015. Generation of stomach tissue from mouse embryonic stem cells. Nat. Cell Biol. 17:984–993. 10.1038/ncb3200 [DOI] [PubMed] [Google Scholar]
- O’Brien J., Fornetti J., and Schedin P.. 2010. Isolation of mammary-specific extracellular matrix to assess acute cell-ECM interactions in 3D culture. J. Mammary Gland Biol. Neoplasia. 15:353–364. 10.1007/s10911-010-9185-x [DOI] [PubMed] [Google Scholar]
- Ootani A., Li X., Sangiorgi E., Ho Q.T., Ueno H., Toda S., Sugihara H., Fujimoto K., Weissman I.L., Capecchi M.R., and Kuo C.J.. 2009. Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche. Nat. Med. 15:701–706. 10.1038/nm.1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pageon H., and Asselineau D.. 2005. An in vitro approach to the chronological aging of skin by glycation of the collagen: the biological effect of glycation on the reconstructed skin model. Ann. N. Y. Acad. Sci. 1043:529–532. 10.1196/annals.1333.060 [DOI] [PubMed] [Google Scholar]
- Pang W.W., Price E.A., Sahoo D., Beerman I., Maloney W.J., Rossi D.J., Schrier S.L., and Weissman I.L.. 2011. Human bone marrow hematopoietic stem cells are increased in frequency and myeloid-biased with age. Proc. Natl. Acad. Sci. USA. 108:20012–20017. 10.1073/pnas.1116110108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrinello S., Coppe J.-P., Krtolica A., and Campisi J.. 2005. Stromal-epithelial interactions in aging and cancer: senescent fibroblasts alter epithelial cell differentiation. J. Cell Sci. 118:485–496. 10.1242/jcs.01635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli C., Hopkins B.D., Prandi D., Shaw R., Fedrizzi T., Sboner A., Sailer V., Augello M., Puca L., Rosati R., et al. . 2017. Personalized in vitro and in vivo cancer models to guide precision medicine. Cancer Discov. 7:462–477. 10.1158/2159-8290.CD-16-1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillip J.M., Aifuwa I., Walston J., and Wirtz D.. 2015. The mechanobiology of aging. Annu. Rev. Biomed. Eng. 17:113–141. 10.1146/annurev-bioeng-071114-040829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaks V., Boldajipour B., Linnemann J.R., Nguyen N.H., Kersten K., Wolf Y., Casbon A.J., Kong N., van den Bijgaart R.J.E., Sheppard D., et al. . 2015. Adaptive immune regulation of mammary postnatal organogenesis. Dev. Cell. 34:493–504. 10.1016/j.devcel.2015.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pribluda A., Elyada E., Wiener Z., Hamza H., Goldstein R.E., Biton M., Burstain I., Morgenstern Y., Brachya G., Billauer H., et al. . 2013. A senescence-inflammatory switch from cancer-inhibitory to cancer-promoting mechanism. Cancer Cell. 24:242–256. 10.1016/j.ccr.2013.06.005 [DOI] [PubMed] [Google Scholar]
- Qian X., Nguyen H.N., Song M.M., Hadiono C., Ogden S.C., Hammack C., Yao B., Hamersky G.R., Jacob F., Zhong C., et al. . 2016. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell. 165:1238–1254. 10.1016/j.cell.2016.04.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raja W.K., Mungenast A.E., Lin Y.-T., Ko T., Abdurrob F., Seo J., and Tsai L.-H.. 2016. Self-organizing 3D human neural tissue derived from induced pluripotent stem cells recapitulate Alzheimer’s disease phenotypes. PLoS One. 11:e0161969 10.1371/journal.pone.0161969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangarajan A., and Weinberg R.A.. 2003. Opinion: Comparative biology of mouse versus human cells: modelling human cancer in mice. Nat. Rev. Cancer. 3:952–959. 10.1038/nrc1235 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Teja M., Gronau J.H., Breit C., Zhang Y.Z., Minamidate A., Caley M.P., McCarthy A., Cox T.R., Erler J.T., Gaughan L., et al. . 2015. AGE-modified basement membrane cooperates with Endo180 to promote epithelial cell invasiveness and decrease prostate cancer survival. J. Pathol. 235:581–592. 10.1002/path.4485 [DOI] [PubMed] [Google Scholar]
- Ruan J.-L., Tulloch N.L., Razumova M.V., Saiget M., Muskheli V., Pabon L., Reinecke H., Regnier M., and Murry C.E.. 2016. Mechanical stress conditioning and electrical stimulation promote contractility and force maturation of induced pluripotent stem cell-derived human cardiac tissue. Circulation. 134:1557–1657. 10.1161/CIRCULATIONAHA.114.014998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A., and Rupp W.D.. 1983. A novel repair enzyme: UVRABC excision nuclease of Escherichia coli cuts a DNA strand on both sides of the damaged region. Cell. 33:249–260. 10.1016/0092-8674(83)90354-9 [DOI] [PubMed] [Google Scholar]
- Sato T., van Es J.H., Snippert H.J., Stange D.E., Vries R.G., van den Born M., Barker N., Shroyer N.F., van de Wetering M., and Clevers H.. 2011. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 469:415–418. 10.1038/nature09637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schütte M., Risch T., Abdavi-Azar N., Boehnke K., Schumacher D., Keil M., Yildiriman R., Jandrasits C., Borodina T., Amstislavskiy V., et al. . 2017. Molecular dissection of colorectal cancer in pre-clinical models identifies biomarkers predicting sensitivity to EGFR inhibitors. Nat. Commun. 8:14262 10.1038/ncomms14262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seok J., Warren H.S., Cuenca A.G., Mindrinos M.N., Baker H.V., Xu W., Richards D.R., McDonald-Smith G.P., Gao H., Hennessy L., et al. . 2013. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc. Natl. Acad. Sci. USA. 110:3507–3512. 10.1073/pnas.1222878110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simian M., and Bissell M.J.. 2017. Organoids: A historical perspective of thinking in three dimensions. J. Cell Biol. 216:31–40. 10.1083/jcb.201610056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Barden A., Mori T., and Beilin L.. 2001. Advanced glycation end-products: a review. Diabetologia. 44:129–146. 10.1007/s001250051591 [DOI] [PubMed] [Google Scholar]
- Squires G.R., Okouneff S., Ionescu M., and Poole A.R.. 2003. The pathobiology of focal lesion development in aging human articular cartilage and molecular matrix changes characteristic of osteoarthritis. Arthritis Rheum. 48:1261–1270. 10.1002/art.10976 [DOI] [PubMed] [Google Scholar]
- Stampfer M.R., Labarge M.A., Garbe J.C., and Schatten H.. 2013. An integrated human mammary epithelial cell culture system for studying carcinogenesis and aging. In Cell and Molecular Biology of Breast Cancer. Schatten H., editor. Humana Press, New York: 323–361. 10.1007/978-1-62703-634-4_15 [DOI] [Google Scholar]
- Stearns-Reider K.M., D’Amore A., Beezhold K., Rothrauff B., Cavalli L., Wagner W.R., Vorp D.A., Tsamis A., Shinde S., Zhang C., et al. . 2017. Aging of the skeletal muscle extracellular matrix drives a stem cell fibrogenic conversion. Aging Cell. 16:518–528. 10.1111/acel.12578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojadinovic O., Minkiewicz J., Sawaya A., Bourne J.W., Torzilli P., de Rivero Vaccari J.P., Dietrich W.D., Keane R.W., and Tomic-Canic M.. 2013. Deep tissue injury in development of pressure ulcers: a decrease of inflammasome activation and changes in human skin morphology in response to aging and mechanical load. PLoS One. 8:e69223 10.1371/journal.pone.0069223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Wang X., and Kaplan D.L.. 2011. A 3D cartilage - inflammatory cell culture system for the modeling of human osteoarthritis. Biomaterials. 32:5581–5589. 10.1016/j.biomaterials.2011.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasato M., Er P.X., Chiu H.S., Maier B., Baillie G.J., Ferguson C., Parton R.G., Wolvetang E.J., Roost M.S., Lopes S.M., and Little M.H.. 2016. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature. 536:238 10.1038/nature17982 [DOI] [PubMed] [Google Scholar]
- Takebe T., Enomura M., Yoshizawa E., Kimura M., Koike H., Ueno Y., Matsuzaki T., Yamazaki T., Toyohara T., Osafune K., et al. . 2015. Vascularized and complex organ buds from diverse tissues via mesenchymal cell-driven condensation. Cell Stem Cell. 16:556–565. 10.1016/j.stem.2015.03.004 [DOI] [PubMed] [Google Scholar]
- Tan J.C., Busque S., Workeneh B., Ho B., Derby G., Blouch K.L., Sommer F.G., Edwards B., and Myers B.D.. 2010. Effects of aging on glomerular function and number in living kidney donors. Kidney Int. 78:686–692. 10.1038/ki.2010.128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanos T., Sflomos G., Echeverria P.C., Ayyanan A., Gutierrez M., Delaloye J.-F., Raffoul W., Fiche M., Dougall W., Schneider P., et al. . 2013. Progesterone/RANKL is a major regulatory axis in the human breast. Sci. Transl. Med. 5:182ra55 10.1126/scitranslmed.3005654 [DOI] [PubMed] [Google Scholar]
- Toda S., Watanabe K., Yokoi F., Matsumura S., Suzuki K., Ootani A., Aoki S., Koike N., and Sugihara H.. 2002. A new organotypic culture of thyroid tissue maintains three-dimensional follicles with C cells for a long term. Biochem. Biophys. Res. Commun. 294:906–911. 10.1016/S0006-291X(02)00561-2 [DOI] [PubMed] [Google Scholar]
- Todhunter M.E., Jee N.Y., Hughes A.J., Coyle M.C., Cerchiari A., Farlow J., Garbe J.C., LaBarge M.A., Desai T.A., and Gartner Z.J.. 2015. Programmed synthesis of three-dimensional tissues. Nat. Methods. 12:975–981. 10.1038/nmeth.3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turco M.Y., Gardner L., Hughes J., Cindrova-Davies T., Gomez M.J., Farrell L., Hollinshead M., Marsh S.G.E., Brosens J.J., Critchley H.O., et al. . 2017. Long-term, hormone-responsive organoid cultures of human endometrium in a chemically defined medium. Nat. Cell Biol. 19:568–577. 10.1038/ncb3516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzatzalos E., Abilez O.J., Shukla P., and Wu J.C.. 2016. Engineered heart tissues and induced pluripotent stem cells: Macro- and microstructures for disease modeling, drug screening, and translational studies. Adv. Drug Deliv. Rev. 96:234–244. 10.1016/j.addr.2015.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhooren V., and Libert C.. 2013. The mouse as a model organism in aging research: usefulness, pitfalls and possibilities. Ageing Res. Rev. 12:8–21. 10.1016/j.arr.2012.03.010 [DOI] [PubMed] [Google Scholar]
- van Vliet E., Stoppini L., Balestrino M., Eskes C., Griesinger C., Sobanski T., Whelan M., Hartung T., and Coecke S.. 2007. Electrophysiological recording of re-aggregating brain cell cultures on multi-electrode arrays to detect acute neurotoxic effects. Neurotoxicology. 28:1136–1146. 10.1016/j.neuro.2007.06.004 [DOI] [PubMed] [Google Scholar]
- Vernetti L., Gough A., Baetz N., Blutt S., Broughman J.R., Brown J.A., Foulke-Abel J., Hasan N., In J., Kelly E., et al. . 2017. Functional coupling of human microphysiology systems: intestine, liver, kidney proximal tubule, blood-brain barrier and skeletal muscle. Sci. Rep. 7:42296 10.1038/srep42296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verzijl N., DeGroot J., Ben Z.C., Brau-Benjamin O., Maroudas A., Bank R.A., Mizrahi J., Schalkwijk C.G., Thorpe S.R., Baynes J.W., et al. . 2002. Crosslinking by advanced glycation end products increases the stiffness of the collagen network in human articular cartilage: a possible mechanism through which age is a risk factor for osteoarthritis. Arthritis Rheum. 46:114–123. [DOI] [PubMed] [Google Scholar]
- Villadsen R., Fridriksdottir A.J., Rønnov-Jessen L., Gudjonsson T., Rank F., LaBarge M.A., Bissell M.J., and Petersen O.W.. 2007. Evidence for a stem cell hierarchy in the adult human breast. J. Cell Biol. 177:87–101. 10.1083/jcb.200611114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeda S.A., Plambeck K.E., Middeldorp J., Castellano J.M., Mosher K.I., Luo J., Smith L.K., Bieri G., Lin K., Berdnik D., et al. . 2014. Young blood reverses age-related impairments in cognitive function and synaptic plasticity in mice. Nat. Med. 20:659–663. 10.1038/nm.3569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voytik-Harbin S.L., Brightman A.O., Waisner B.Z., Robinson J.P., and Lamar C.H.. 2007. Small intestinal submucosa: A tissue-derived extracellular matrix that promotes tissue-specific growth and differentiation of cells in vitro. Tissue Eng. 4:157–174. 10.1089/ten.1998.4.157 [DOI] [Google Scholar]
- Watson C.L., Mahe M.M., Múnera J., Howell J.C., Sundaram N., Poling H.M., Schweitzer J.I., Vallance J.E., Mayhew C.N., Sun Y., et al. . 2014. An in vivo model of human small intestine using pluripotent stem cells. Nat. Med. 20:1310–1314. 10.1038/nm.3737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C., Quinn K.P., Georgakoudi I., and Black L.D. III. 2014. Young developmental age cardiac extracellular matrix promotes the expansion of neonatal cardiomyocytes in vitro. Acta Biomater. 10:194–204. 10.1016/j.actbio.2013.08.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson J.L., Dollard S.C., Chow L.T., and Broker T.R.. 1992. Epithelial-specific gene expression during differentiation of stratified primary human keratinocyte cultures. Cell Growth Differ. 3:471–483. [PubMed] [Google Scholar]
- Workman M.J., Mahe M.M., Trisno S., Poling H.M., Watson C.L., Sundaram N., Chang C.-F., Schiesser J., Aubert P., Stanley E.G., et al. . 2017. Engineered human pluripotent-stem-cell-derived intestinal tissues with a functional enteric nervous system. Nat. Med. 23:49–59. 10.1038/nm.4233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W., Booth J.L., Duggan E.S., Wu S., Patel K.B., Coggeshall K.M., and Metcalf J.P.. 2010. Innate immune response to H3N2 and H1N1 influenza virus infection in a human lung organ culture model. Virology. 396:178–188. 10.1016/j.virol.2009.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wylie R.G., Ahsan S., Aizawa Y., Maxwell K.L., Morshead C.M., and Shoichet M.S.. 2011. Spatially controlled simultaneous patterning of multiple growth factors in three-dimensional hydrogels. Nat. Mater. 10:799–806. 10.1038/nmat3101 [DOI] [PubMed] [Google Scholar]
- Xiang Z., Hrabetova S., Moskowitz S.I., Casaccia-Bonnefil P., Young S.R., Nimmrich V.C., Tiedge H., Einheber S., Karnup S., Bianchi R., and Bergold P.J.. 2000. Long-term maintenance of mature hippocampal slices in vitro. J. Neurosci. Methods. 98:145–154. 10.1016/S0165-0270(00)00197-7 [DOI] [PubMed] [Google Scholar]
- Yilmaz Ö.H., Katajisto P., Lamming D.W., Gültekin Y., Bauer-Rowe K.E., Sengupta S., Birsoy K., Dursun A., Yilmaz V.O., Selig M., et al. . 2012. mTORC1 in the Paneth cell niche couples intestinal stem-cell function to calorie intake. Nature. 486:490–495. 10.1038/nature11163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Lian Q., Zhu G., Zhou F., Sui L., Tan C., Mutalif R.A., Navasankari R., Zhang Y., Tse H.-F., et al. . 2011. A human iPSC model of Hutchinson Gilford Progeria reveals vascular smooth muscle and mesenchymal stem cell defects. Cell Stem Cell. 8:31–45. 10.1016/j.stem.2010.12.002 [DOI] [PubMed] [Google Scholar]
- Zhang Y., He Y., Bharadwaj S., Hammam N., Carnagey K., Myers R., Atala A., and Van Dyke M.. 2009. Tissue-specific extracellular matrix coatings for the promotion of cell proliferation and maintenance of cell phenotype. Biomaterials. 30:4021–4028. 10.1016/j.biomaterials.2009.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]