Klaips et al. outline the pathways and molecular mechanisms of cellular protein homeostasis, or proteostasis, and discuss how a decline in proteostasis during aging contributes to disease.
Abstract
Ensuring cellular protein homeostasis, or proteostasis, requires precise control of protein synthesis, folding, conformational maintenance, and degradation. A complex and adaptive proteostasis network coordinates these processes with molecular chaperones of different classes and their regulators functioning as major players. This network serves to ensure that cells have the proteins they need while minimizing misfolding or aggregation events that are hallmarks of age-associated proteinopathies, including neurodegenerative disorders such as Alzheimer’s and Parkinson’s diseases. It is now clear that the capacity of cells to maintain proteostasis undergoes a decline during aging, rendering the organism susceptible to these pathologies. Here we discuss the major proteostasis pathways in light of recent research suggesting that their age-dependent failure can both contribute to and result from disease. We consider different strategies to modulate proteostasis capacity, which may help develop urgently needed therapies for neurodegeneration and other age-dependent pathologies.
Introduction
Human cells express more than ∼10,000 different proteins at any given time (Kulak et al., 2017), the majority of which must fold (and often assemble) to well-defined, three-dimensional structures to allow a myriad of cellular functions. Although the native conformation of a given protein is encoded by its amino acid sequence (Anfinsen, 1973), in the cell many proteins require assistance by molecular chaperones and other factors to fold efficiently and at a biologically relevant time scale (Balchin et al., 2016). Moreover, proteins often need to retain structural flexibility or contain significant unstructured regions to function, leaving them at risk of misfolding and aggregation (Chiti and Dobson, 2017). Even otherwise stably folded proteins may unfold and possibly aggregate under stress conditions, such as elevated temperatures. Finally, as proteins become terminally misfolded, or are no longer functionally required, they must be degraded to avoid damaging effects of their continued presence. Maintaining an intact proteome (proteostasis) thus requires not only strict control of the initial production and folding of a protein but also its conformational maintenance, control of abundance and subcellular localization, and finally, disposal by degradation.
A complex proteostasis network (PN) acts at each of these steps to maintain a balanced proteome linked by molecular chaperones of different classes as central players. These factors ensure de novo folding in a crowded cellular environment and maintain proteins in a soluble, nonaggregated state. Moreover, in conditions that disfavor folding or solubility, certain chaperones act to target misfolded proteins for degradation or spatial sequestration, thus protecting the rest of the proteome from aberrant interactions (Balchin et al., 2016; Sontag et al., 2017).
Here, we describe the major pathways of cellular proteostasis and outline the challenges they face during aging and disease. We exemplify these processes using mainly the proteostasis pathways operating in the cytosol, where most cellular proteins are produced. The major exceptions are the proteins associated with the endomembrane system and secretory proteins. These polypeptides generally fold and assemble in the ER. Although the environment of the ER is oxidizing and differs in several aspects from the reducing cytosol, the core principles governing overall proteostatic balance apply (Skach, 2009; Gidalevitz et al., 2013). Rather than focusing on specific disease states, we discuss common themes that have been shown to be relevant across multiple systems, suggesting a conserved and intimate linkage of proteostasis with the aging process and associated pathologies.
Organization of the PN
Because of the astronomically large number of possible conformations a polypeptide chain can adopt, the folding process is inherently error prone (Dobson et al., 1998; Bartlett and Radford, 2009). Production of misfolded proteins is further increased by stochastic errors of protein biogenesis occurring at the level of transcription and mRNA maturation and translation (Sachidanandam et al., 2001; Ng and Henikoff, 2006; Drummond and Wilke, 2008). Such failed protein products must be recognized and degraded to avoid aberrant interactions, making it a challenge to maintain a healthy proteome even under normal conditions. This challenge is exacerbated in the case of disease-associated mutations, environmental stress, and aging and if left unresolved can lead to the formation of toxic aggregate species. Thus, to maintain proteostasis, cells have evolved a wide variety of molecular chaperones and protein quality-control factors that are functionally linked with protein degradation machineries. This system is referred to as the PN (Balch et al., 2008; Fig. 1 A).
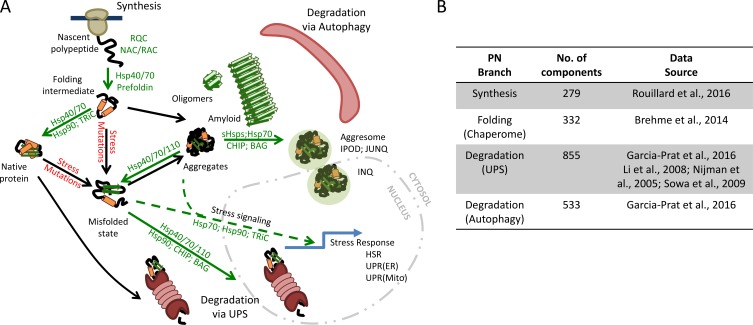
Figure 1.
The PN. (A) The flux of proteins through the PN relies on chaperones at many stages. PN capacity is sufficient to fold, often via intermediates, most newly synthesized polypeptides as they exit the ribosome. When a protein is no longer needed, it can be efficiently targeted for degradation either in the cytosol or nucleus via the UPS. Proteins that cannot be folded are also targeted for degradation via the UPS. During stress, the cell can increase PN capacity by activating a stress response. In aged or diseased cells, there is an increase in overall protein misfolding and aggregation, owing to increases in mutations and an overall decrease in PN capacity. These misfolded species may aggregate and/or be sequestered into large structures (aggresomes). A subset of misfolded species may form amyloid fibers that can further interfere with cellular processes. Chaperone factors that enhance particular steps are shown in green. Adapted from Hipp et al. (2014) and Kim et al. (2013). (B) Numbers for the major branches of the human PN, including synthesis, folding, and maintenance, and degradation branches, are shown. Datasets for generating these values were collected from the sources shown. The data were then arranged into nonoverlapping groups to represent the major PN branches. BAG, Bcl-2–associated athanogene; NAC, nascent chain-associated complex; RAC, ribosome-associated complex; RQC, ribosome quality control.
Defining the exact composition of the PN has proved difficult given the complexity of the human proteome. The PN has previously been proposed to consist of ∼1,000–1,400 components on the basis of our initial understanding of its organization (Balch et al., 2008; Powers et al., 2009; Kim et al., 2013; Hipp et al., 2014). Based on current annotations in databases and several large-scale genomic studies, we estimate that the PN comprises ∼2,000 factors that act in concert to maintain cellular proteostasis (Fig. 1 B). With increasing availability of functional annotations for the biological pathways in the human genome, these numbers will be further refined.
Operationally, the PN can be divided into three branches composed of factors belonging to major processes: (1) protein synthesis, (2) folding and conformational maintenance (often coupled to transport and/or assembly), and (3) protein degradation (the ubiquitin–proteasome system [UPS] and autophagy–lysosome system; Fig. 1). Molecular chaperones and their regulatory cofactors act as liaisons connecting all these processes.
A set of ∼280 components participate directly in nascent polypeptide chain synthesis (Wolff et al., 2014; Rouillard et al., 2016; Fig. 1 B). Apart from the core constituents of the translational machinery, several chaperones act on the ribosome to prevent premature misfolding of the nascent chain and assist in cotranslational folding. Quality-control factors of the UPS interface with protein synthesis to remove defective and stalled nascent chains as part of ribosomal quality-control pathways (Brandman and Hegde, 2016).
Newly synthesized proteins may fold cotranslationally or may rapidly complete their folding on release from the ribosome. Folding, and in some cases assembly to oligomeric complexes, is mediated by molecular chaperones, often involving sequential interactions with members of different chaperone classes (Langer et al., 1992; Frydman et al., 1994; Balchin et al., 2016). The repertoire of human chaperones (the “chaperome”) contains ∼330 members of several functionally distinct gene families, which cater to diverse substrate clients (Brehme et al., 2014; Sala et al., 2017; Fig. 1 B).
Misfolded and aggregated proteins must be removed from the system by proteolytic degradation to prevent the accumulation of toxic species. Eukaryotic cells invest extensively in protein degradation machineries, with the two major pathways of the UPS and autophagy comprising ∼850 and ∼500 different components, respectively (Nijman et al., 2005; Li et al., 2008; Sowa et al., 2009; Varshavsky, 2012; García-Prat et al., 2016; Fig. 1 B). The UPS mainly serves to target individual proteins to the proteasome, whereas the autophagy system clears larger aggregates or membrane-associated proteins (Menzies et al., 2015; Ciechanover and Kwon, 2017).
These branches of the PN are functionally coordinated by various signaling cascades, which sense and respond to imbalances in proteostasis (Fig. 1 A). In this way, cells constantly monitor and adjust their proteome status in response to internal and external changes. The PN not only enables this adjustment, but is itself adaptive to the needs of specific cell types. In “simpler” organisms such as yeast, the basic organization of the PN may be rather constant, only tuning itself to fluctuations in environmental conditions. However, in metazoans (Guisbert et al., 2013), especially in complex mammalian systems (Uhlén et al., 2015), tissue-specific proteomes and regulatory programs imply that there must be a marked heterogeneity in aspects of proteostasis across diverse cell types, suggesting the existence of tissue-specific PNs (Sala et al., 2017) with differing contributions of the three branches.
Many diseases, including type II diabetes and the major neurodegenerative pathologies, are associated with a reduced function of the PN, which may be caused by mutations in PN components (Kakkar et al., 2014) or by interference of toxic aggregate species with PN function (Hipp et al., 2014). Importantly, as shown in model organisms, aging is also associated with a general decrease in PN capacity and a corresponding increase in protein aggregation, which manifests as a functional decline in many cellular pathways (Taylor and Dillin, 2011; Labbadia and Morimoto, 2015a). In the following sections, we discuss the roles of the major pathways within the PN.
Protein synthesis
Although the production of individual proteins is regulated by specific factors and pathways, the levels of bulk protein synthesis must be adjusted to the protein folding capacity of the cell to avoid the accumulation of misfolded proteins. Indeed, in key lifespan extension pathways such as caloric restriction, increased proteostasis capacity is conferred, at least in part, by a general decrease in protein translation (Hansen et al., 2007; Taylor and Dillin, 2011). Translation attenuation is also critical in relieving PN overload in conditions of conformational stress. This is typically mediated by inhibition of translation initiation factor 2α (eIF2α). For example, on activation of the unfolded protein response (UPR) to the accumulation of misfolded proteins in the ER, protein kinase RNA-like ER kinase (PERK) in the ER membrane phosphorylates eIF2α, thereby attenuating its function in translation (Harding et al., 2001).
Protein folding and aggregation
Polypeptide chains fold by sequestering hydrophobic residues and forming stabilizing intramolecular interactions to achieve a low free-energy state (Fig. 2). Rather than sampling all potential folding states, a process that would take an insurmountable amount of time, polypeptides proceed toward their native conformation by increasingly forming local and long-range contacts between amino acid residues, thereby limiting the conformational space that must be explored (Dinner et al., 2000; Bartlett and Radford, 2009). In this way, many small proteins can achieve their proper fold quickly and efficiently in vitro. However, once placed in the highly crowded cellular environment, proteins often face significant challenges during folding, because partially folded states with exposed hydrophobic amino acids residues are in danger of misfolding and aggregating. Aberrant folding may occur during de novo synthesis or in conditions of conformational stress, where preexisting proteins may fail to maintain their folded states. Destabilizing mutations or the presence of intrinsically unstructured regions can also predispose polypeptides to misfolding (Dunker et al., 2008; Gershenson et al., 2014).
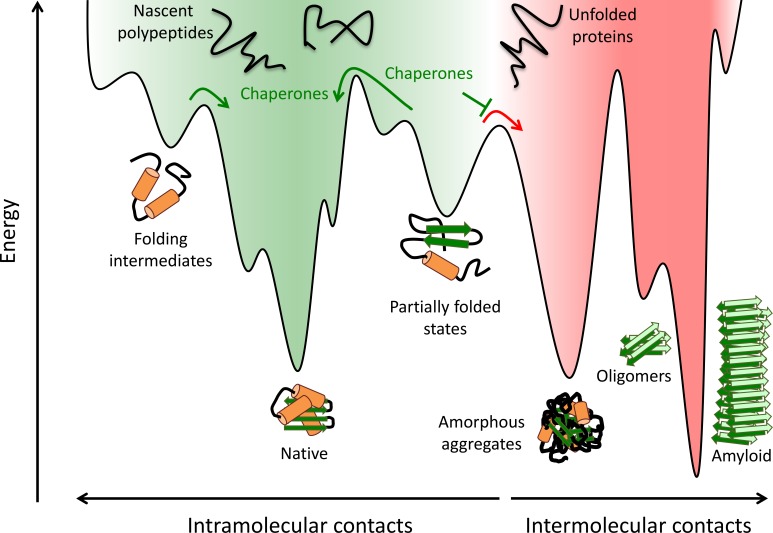
Figure 2.
Protein folding and aggregation. Nascent polypeptides fold by sampling various conformations and sequestering hydrophobic amino acid residues. Partially folded intermediates, both on- and off-pathway, can become trapped in localized energy minima. These species are at risk of aggregation by forming aberrant intermolecular contacts, which can lead to the formation of oligomers, amorphous aggregates, and amyloid fibrils. Molecular chaperones promote the formation of the native species by lowering free-energy barriers between kinetically stable intermediates, smoothing the protein folding landscape (green arrows), and preventing aberrant intermolecular interactions (red arrow). Adapted from Balchin et al. (2016), Hartl et al. (2011), and Kim et al. (2013).
Unlike in vitro folding studies (Anfinsen, 1973; Bartlett and Radford, 2009), which start from complete proteins, in the cell proteins are synthesized vectorially on the ribosome, which means that the structural information necessary for folding becomes available gradually and not all at once. Translation is slow relative to rates of folding, allowing for the possibility of partial structure formation, both native or misfolded, before the completion of protein synthesis. Cotranslational folding limits the amount of time nascent chains populate potentially vulnerable, nonnative states (Balchin et al., 2016). Some very small proteins (∼50 amino acids in length) even fold to completion within the widening exit portal of the ribosome (Holtkamp et al., 2015; Marino et al., 2016). However, the major part of the ribosomal exit channel is too narrow to allow structure formation (Wilson and Beckmann, 2011), and thus the nascent chains of larger proteins must first emerge from the ribosome before they can fold, which puts them at risk of misfolding and aberrant interactions. The ribosomal surface may influence the folding process (Kaiser et al., 2011; Cabrita et al., 2016). Moreover, the topology of ribosomes in the context of polysomes, where translating ribosomes may approach each other closely, is optimized to reduce the risk of interactions between nascent chains. The ribosomes adopt a staggered “pseudohelical” arrangement, in which their polypeptide exit sites are at maximal distance from each other (Brandt et al., 2009). Multidomain proteins often fold their domains sequentially during translation, thereby avoiding nonnative interactions between concomitantly folding domains (Netzer and Hartl, 1997; Frydman et al., 1999).
Average proteome and protein sizes have increased dramatically during evolution from bacteria to eukarya (Balchin et al., 2016; Fig. 3 A). Proteins of more than ∼100 amino acids in length constitute the vast majority of proteins in all domains of life and typically fold via intermediate states with incompletely buried hydrophobic residues (Dinner et al., 2000; Brockwell and Radford, 2007; Fig. 2). Such intermediates may be kinetically stable and may be highly aggregation prone (Gershenson et al., 2014), particularly in the crowded environment of the cell (∼300 g of protein per liter), where macromolecular interactions are enhanced compared with dilute solution (Ellis and Minton, 2006). Although the majority of resulting aggregates are amorphous (i.e., lacking long-range structural order), a subset of mostly smaller proteins, often containing unstructured regions (Dunker et al., 2008), can form ordered fibrillar aggregates, referred to as amyloid or amyloid-like, and which are characterized by β-strands running perpendicular to the fibril axis (cross-β-structure; Chiti and Dobson, 2017; Fig. 2). Such amyloid aggregates form insoluble deposits and are the hallmark of several age-dependent proteinopathies, including Alzheimer’s, Parkinson’s, and Huntington’s diseases (Ross and Poirier, 2004). More proteins transition to an insoluble, aggregated state when they exceed their normal cellular abundance or when imbalances occur between subunits of oligomeric complexes (Vendruscolo et al., 2011; Ciryam et al., 2013; Chiti and Dobson, 2017), a phenomenon that becomes more prevalent during aging, as shown in the nematode Caenorhabditis elegans (Walther et al., 2015).
Figure 3.
Proteome complexity increases from prokaryotes to eukaryotes. (A) The average protein length (in number of amino acids) and total number of chaperones in the proteomes of bacteria (Escherichia coli), yeast (Saccharomyces cerevisiae), and humans (Homo sapiens) are shown. The increase in proteome complexity with increasing organismal complexity, from prokaryotes to eukaryotes, and from single-cell to multicellular organisms is accompanied by an increase in chaperone number. Comparative chaperomes were generated by UniProtKB queries by using identical keyword searches for chaperones for each organism. Differences between the total human chaperome number and previously reported numbers (Brehme et al., 2014; Fig. 1 B) arise from a more-stringent definition here to ensure appropriate comparisons between organisms. (B) The numbers of members of each of the major chaperone classes are shown for different organisms with selective cochaperones shown for comparison (Genevaux et al., 2007; Vos et al., 2008; Brehme et al., 2014; Rizzolo et al., 2017). Protein folding in the bacterial (C) and eukaryotic (D) cytosol. Most proteins use folding assistance on exit from the ribosome. Ribosome-associated factors include Trigger factor in bacteria and the nascent chain-associated complex (NAC) and ribosome-associated complex (RAC) in eukaryotes. Downstream Hsp70s (DnaK in bacteria) work with their cofactors Hsp40 (DnaJ in bacteria) and nucleotide exchange factors (NEFs; GrpE in bacteria) in protein folding. Some proteins must be transferred to the chaperonin class of chaperones for further folding (GroEL/ES in bacteria and TRiC in eukaryotes). In eukaryotes, prefoldin can transfer some substrates directly to TRiC. Cofactors such as Hop (Hsp70-Hsp90 organizing protein) can mediate interactions with Hsp90 (HtpG in bacteria), which also acts downstream of Hsp70 in the folding of a subset of proteins mainly engaged in cell signaling. Adapted from Balchin et al. (2016).
Molecular chaperones—central organizers of the PN
To overcome the challenges to protein folding and solubility, cells have evolved molecular chaperones (Fig. 3 A). We define a molecular chaperone as a protein that assists in the folding, assembly, conformational maintenance, or regulation of another protein without becoming part of its final structure (Hartl, 1996). The chaperones that participate broadly in de novo folding recognize generic structural features of nonnative proteins, primarily exposed hydrophobic amino acid residues, and promote folding by kinetic partitioning of nonnative states (Kim et al., 2013; Fig. 2). Many chaperones are induced under conditions of stress, such as heat shock, and in addition to their functions in de novo folding are also involved in protein refolding, disaggregation, oligomeric assembly, trafficking, and degradation (Balchin et al., 2016; Fig. 1 A). Although the core chaperone machineries (heat-shock protein [Hsp] 70s, Hsp90s, chaperonins, and small Hsps [sHsps]) are already present in prokaryotes, a strong expansion in the number of their regulatory cofactors (Hsp40s, tetratricopeptide repeat proteins) is notable as eukaryotic proteomes increase in complexity (Genevaux et al., 2007; Vos et al., 2008; Brehme et al., 2014; Rizzolo et al., 2017; Fig. 3 B).
A first tier of chaperones interacts directly with the ribosome close to the polypeptide exit site. These components are typically not stress-inducible (Albanèse et al., 2006) and include Trigger factor in bacteria and specialized chaperone complexes, such as nascent chain-associated complex and ribosome-associated complex, in eukaryotes (Fig. 3, C and D). They interact with exposed hydrophobic sequences of the emerging nascent chain and act to prevent premature (mis)folding, maintaining the polypeptide in a nonaggregated, folding competent state until sufficient structural elements for productive folding are available (Agashe et al., 2004; Kaiser et al., 2006; Preissler and Deuerling, 2012; Nilsson et al., 2016). Although most proteins may only require these chaperones to achieve their native fold, proteins with complex domain topologies and multidomain proteins need assistance by additional chaperone classes that act downstream.
Such proteins may next interact cotranslationally or posttranslationally with a member of the Hsp70 chaperone family (DnaK in prokaryotes; Fig. 3, C and D), a ubiquitous class of ATP-dependent chaperones of ∼70 kD with a hub position in the PN. The Hsp70 C-terminal domain binds short hydrophobic peptide sequences of about seven residues that are exposed by nascent and nonnative protein substrates (Rüdiger et al., 1997; Mayer et al., 2000; Clerico et al., 2015). The affinity of the C-terminal domain for protein substrate is allosterically regulated by ATP binding and hydrolysis in the N-terminal ATPase domain. Hsp70s rely on regulatory chaperone cofactors of the Hsp40 class (also known as J-proteins), which typically bind first to exposed hydrophobic patches on nonnative proteins and recruit Hsp70 (Kampinga and Craig, 2010; Nillegoda et al., 2017). These factors then stimulate the hydrolysis of Hsp70-bound ATP, thereby catalyzing the closing of the Hsp70 peptide binding cleft (Clerico et al., 2015). There are ∼50 different Hsp40 proteins in human cells (Fig. 3 B), which fall into three structural subtypes and have different subcellular localizations (Kampinga and Craig, 2010). They confer broad functionality to the Hsp70 system, allowing these chaperones not only to participate in the initial folding of nascent chains but also in conformational maintenance (Mashaghi et al., 2016), disaggregation (Diamant et al., 2000; Ben-Zvi et al., 2004; Rampelt et al., 2012), and the targeting of terminally misfolded proteins for degradation (Kettern et al., 2010). Nucleotide exchange factors are necessary to allow Hsp70 to perform cycles of substrate binding and release (Laufen et al., 1999; Mayer and Bukau, 2005; Winkler et al., 2012).
Proteins that are unable to fold through such Hsp70 cycles may be transferred to the chaperonin class of chaperones (Hsp60s), which includes GroEL/GroES in bacteria, Hsp60 in mitochondria, and TRiC/CCT in the eukaryotic cytosol (Fig. 3, C and D). These chaperonin proteins form multimeric, cylindrical complexes that function by transiently encapsulating individual nonnative proteins so they can fold, unimpaired by aggregation (Lopez et al., 2015; Hayer-Hartl et al., 2016). The opening and closing of the folding chamber is regulated by the ATPase of the chaperonin, either in conjunction with a separate lid-like cofactor of the GroES-type (for GroEL and mitochondrial Hsp60) or lid-structure built into the chaperonin complex (for TRiC/CCT). Although only ∼10% of the proteome requires a chaperonin to fold, substrates include essential and highly abundant proteins, such as actin and tubulin. Accordingly, deletion or mutation of TRiC is toxic and has been implicated in disease. Neurodegenerative disorders affecting myelination, spastic paraplegia, and leukodystrophy are caused by autosomally inherited mutations in mitochondrial Hsp60 (Hansen et al., 2002; Magen et al., 2008).
The highly conserved Hsp90 chaperone system also functions downstream of Hsp70 in maintaining a variety of signaling pathways via the folding and conformational regulation of their signal-transduction molecules (Sharma et al., 2012; Taipale et al., 2012). Hsp90 is active as a homodimer and mediates protein folding via ATP-dependent structural changes in cooperation with a multitude of cofactors (Wandinger et al., 2008). Hsp90 can bind substrates that are near native, thereby stabilizing metastable clients, such as kinases and steroid receptor molecules, in a conformation poised for activation by ligand binding (Fig. 1 A and Fig. 3, B and C). Because of its role in the folding of many disease-relevant proteins, pharmacologic inhibition of Hsp90 is being considered as a strategy in the treatment of many diseases from cancer to viruses (Whitesell et al., 1994; Balch et al., 2008; Geller et al., 2013; Mbofung et al., 2017).
Maintaining the metastable proteome
After initial folding, many proteins continue to require chaperone surveillance to maintain their functional form. Proteins are often active under conditions just at the cusp of stability, and their functional conformational states may be challenged under stress conditions. Additionally, many proteins contain intrinsically unstructured regions or sequences of low amino acid complexity important for their function, including up to 75% of signaling molecules in mammalian cells (Dunker et al., 2008). These proteins may acquire a stable structure only on binding to a ligand or other macromolecular surface. It has become clear in recent years that a hallmark of cellular aging is a gradual loss of proteome balance and accumulation of protein aggregates. This is thought to be due to at least in part an increase in the accumulation of errors in translation, splicing, or molecular misreading and to an increased production of oxidized and carbonylated proteins (Aguilaniu et al., 2003; López-Otín et al., 2013). The proteins of the PN are not exempt from such modifications. Indeed, studies in C. elegans and other model systems have shown that aged organisms have a markedly reduced ability to maintain metastable proteins in their soluble states (Morley et al., 2002; David et al., 2010; Gupta et al., 2011; Walther et al., 2015). In worms, this decline in PN capacity is tied to development, suggestive of a regulated program of aging (Ben-Zvi et al., 2009; Labbadia and Morimoto, 2015b).
A healthy chaperone network is thus required to maintain the metastable proteome and to prevent the accumulation of toxic aggregate species. The Hsp70 system and the so-called sHsps, the latter functioning as multidisperse chaperone oligomers (Haslbeck et al., 2005), are particularly important in this regard. Upon acute stress, such as heat exposure, 10–30% of cytosolic proteins are potential clients of sHsps, indicating an important role in the maintenance of proteome stability (Haslbeck et al., 2004; Mymrikov et al., 2017). When the system is overburdened, however, misfolded species will form and may aggregate. In these cases the association of sHsps with the aggregates themselves has been shown to aid in their resolution by the cell during recovery from conformational stress. Association of sHsps and chaperone cofactors with aggregates enables downstream processing (Ben-Zvi et al., 2004; Malinovska et al., 2012; Rampelt et al., 2012; Żwirowski et al., 2017) and eventual disaggregation by Hsp70/Hsp40/Hsp110 machineries (Mogk et al., 2003; Nillegoda and Bukau, 2015). In yeast, the disaggregation capacity of cells is further enhanced by a specialized AAA+ ATPase called Hsp104, which along with its Hsp70/Hsp40 cofactors has been shown to disaggregate many amyloid aggregates (Parsell et al., 1994; Glover and Lindquist, 1998; Wegrzyn et al., 2001).
Disposal by degradation
Proteins that are unable to fold or refold, despite intervention by chaperones, must be disposed of to prevent the accumulation of potentially toxic aggregate species. Such terminally misfolded proteins undergo proteolytic degradation mainly by the UPS (Varshavsky, 2012; Ciechanover and Kwon, 2017) or by chaperone-mediated lysosomal degradation (Kettern et al., 2010; Cuervo and Wong, 2014). The Hsp70 and Hsp90 chaperone systems are intimately involved in these processes, because the E3 ubiquitin ligase Chip binds the C terminus of these chaperones and ubiquitylates misfolded chaperone-bound proteins (Esser et al., 2004; Fig. 1 A). As shown recently, surplus proteins that fail to assemble with their partner molecules are recognized by a specific E3 ligase (UBE20; Yanagitani et al., 2017).
A subset of proteins that misfolds in the cytosol undergoes chaperone-mediated transport into the nucleus to be degraded by nuclear proteasomes (Heck et al., 2010; Prasad et al., 2010; Park et al., 2013; Shibata and Morimoto, 2014; Fig. 1 A). Most of the proteins known to use this pathway are ectopically expressed secretory proteins. The extent to which endogenous, misfolded proteins are degraded in the nucleus remains to be established. Given that an abundance of proteasomes is found in the nucleus (Russell et al., 1999; von Mikecz, 2006), it is tempting to speculate that compartmentalizing synthesis/folding and degradation provides an evolutionary advantage by preventing premature degradation. The same principle would apply to the process of ER-mediated degradation, wherein misfolded proteins are retrotranslocated from the ER to the cytosol for disposal by the proteasome (Vembar and Brodsky, 2008).
Aggregates may be resolved by the Hsp70/Hsp40/Hsp110 machinery before transfer into the proteasome (Hjerpe et al., 2016). Certain aggregate species resistant to disaggregation may be cleared directly by selective autophagy and lysosomal degradation (Lamark and Johansen, 2012), processes that also target a variety of additional substrates including membrane bound organelles (Mizushima, 2007).
Many cell types show a decline in UPS activity and autophagy during aging (Rubinsztein et al., 2011; Cuervo and Wong, 2014), contributing to the widespread protein aggregation that is observed in postmitotic cells, such as muscle and neurons, and predisposing the latter for certain neurodegenerative diseases (David et al., 2010; Hamer et al., 2010; Walther et al., 2015). Because disease-associated proteins tend to be metastable, a slight increase in their abundance as clearance systems decline can have dramatic effects on their aggregation propensity (Ciryam et al., 2013; Kundra et al., 2017). Aging cells are also less able to cope with and dispose of amyloid-like aggregates (Morley et al., 2002), as exemplified by the fact that cellular aggregate deposits persist although they are typically associated with ubiquitin (Lowe et al., 1988; Bence et al., 2001; Waelter et al., 2001). These aggregates often sequester important components of the PN, which leads to further proteostatic impairment with buildup of damaged protein species and increased risk of aggregation (Bennett et al., 2005; Hipp et al., 2014; Itakura et al., 2016).
Compartmentalization of damaged proteins
If attempts to prevent, refold, or degrade aberrant protein species fail, a final line of defense against their interference with cellular processes is their controlled sequestration into more benign aggregate deposits or inclusion bodies (Sontag et al., 2017). Depending on the properties of the misfolded proteins and their ability for eventual resolubilization, such deposits can be directed to several different localizations within the cytosol or nucleus and are referred to as an IPOD (for insoluble protein deposit), JUNQ (for juxtanuclear quality-control compartment), or INQ (for intranuclear quality-control compartment) in yeast and as an aggresome in mammalian cells (Johnston et al., 1998; Kaganovich et al., 2008; Miller et al., 2015; Fig. 1 A). Their formation is itself dependent on several quality-control components including chaperones (Malinovska et al., 2012; Escusa-Toret et al., 2013; Wolfe et al., 2013). In addition to providing an environment in which aggregates may be shielded and thus prevented from engaging in potentially toxic interactions, in dividing cells the inclusions also serve as a way to minimize the amount of aberrant proteins that are passed on to daughter cells (Hill et al., 2017). Like other proteostasis pathways, the ability of a cell to maintain spatial quality control also declines with age (Escusa-Toret et al., 2013), and cells that lack this ability show accelerated aging (Erjavec et al., 2007).
Toxicity caused by aggregation
Proteins have an intrinsic capacity to convert from their native state to intractable fibrillar aggregates, but under normal physiological conditions this tendency is resisted by cellular proteostasis mechanisms (Chiti and Dobson, 2017). However, the propensity to form amyloid-like aggregates is more pronounced for certain metastable proteins, including those associated with disease, especially when exceeding the cellular concentrations at which they are soluble (Ciryam et al., 2013). Dysregulation of protein abundance and protein stoichiometries may occur in an age-dependent manner, as observed in nematodes and other model organisms (Walther et al., 2015). Indeed, recent research shows that the formation of insoluble protein deposits in neurodegenerative syndromes such as Alzheimer’s disease occurs concomitantly with the aggregation of a large set of highly expressed and aggregation-prone proteins that constitute a metastable subproteome (Kundra et al., 2017). The metastable subproteome includes many RNA-binding proteins that contain unstructured or low-complexity sequences. As shown recently, such proteins often have the ability to undergo liquid–liquid phase transitions (Feric et al., 2016), forming dynamic droplet-like compartments in the nucleus and cytosol that participate in RNA metabolism, ribosome biogenesis, cell signaling, and other processes (Banani et al., 2017). However, the normally dynamic behavior of these condensates is highly sensitive to changes in the physicochemical environment of cells, and aberrant phase transition behavior, leading to fibril formation, has been linked with aging and diseases such as amyotrophic lateral sclerosis (Alberti and Hyman, 2016). These recent observations help explain how the age-dependent decline in protein homeostasis favors the stochastic manifestation of neurodegenerative aggregation. Importantly, even in dominantly inherited neurodegenerative disorders, such as those caused by expanded polyglutamine sequences (Scherzinger et al., 1999; Gusella and MacDonald, 2006), manifestation is age dependent and triggered by PN decline (Morley et al., 2002; Gidalevitz et al., 2006).
Aggregation in disease typically causes gain-of-function toxicity, which means that the cytotoxic effects are largely unrelated to the normal function of the disease protein (Ross and Poirier, 2004; Chiti and Dobson, 2017). However, the presence of large fibrillar aggregate deposits does not always correlate with disease onset or severity (Kayed et al., 2003; Leitman et al., 2013; Chiti and Dobson, 2017). Indeed, work over the past years revealed that the most toxic aggregate species may be soluble oligomers and small insoluble species with little or no fibrillar content (Chiti and Dobson, 2017). Such oligomers expose hydrophobic residues and unpaired polypeptide backbone structures, features that render them highly interactive with other proteins, including proteins enriched in low-complexity sequences and critical factors of the PN, and with membranes (Kayed et al., 2003; Olzscha et al., 2011; Winner et al., 2011; Park et al., 2013; Kim et al., 2016b; Woerner et al., 2016).
Although the exact nature of the most-toxic species remains a subject of discussion, in many cases the larger amyloid aggregates may exert a relative protective effect by sequestering the more-toxic oligomers and by having a reduced surface-to-volume ratio (Saudou et al., 1998; Arrasate et al., 2004; Douglas et al., 2008; Kim et al., 2016b). However, key cellular factors, including PN components, that interact aberrantly with soluble oligomers may also be sequestered in the insoluble deposits, contributing to cellular dysfunction (Olzscha et al., 2011; Park et al., 2013; Hipp et al., 2014; Ripaud et al., 2014; Yu et al., 2014; Woerner et al., 2016). Moreover, large intracellular aggregate deposits sterically displace membrane structures and may cause their fragmentation, as recently shown for inclusions of polyglutamine expansion proteins by cryo-electron tomography (Bäuerlein et al., 2017). In many model systems, exogenous expression of individual chaperone components, such as Hsp70, or up-regulation of multiple chaperones by pharmacologic induction of the stress response has been shown to either prevent toxic aggregation or to direct the formation of less-toxic but still aggregated species (Muchowski et al., 2000; Sittler et al., 2001; Holmes et al., 2014; Nagy et al., 2016).
Stress response pathways
Although the protein quality-control networks ensure proteostasis under basal conditions, on conformational stress, such as increases in temperature or exposure to oxidative agents, many additional proteins become prone to misfolding, with proteins comprising the metastable subproteome being particularly vulnerable. Cells adapt to such conditions by activating stress-response pathways to increase PN components, decrease substrate load, and resolve misfolded or aggregated species (Fig. 4). In metazoans the stress-response pathways additionally underlie cell nonautonomous regulation, allowing coordination within and between tissues and organs (Taylor et al., 2014; Sala et al., 2017).
Figure 4.
Healthy and aged proteostasis. (Left) The PN ensures that most proteins fold to a stable native state. When these proteins are no longer needed or errors in folding occur, they are efficiently targeted for degradation, primarily via the UPS. On transient stress (middle), otherwise healthy cells activate a stress response. Chaperone sensors bind to misfolded species and trigger the appropriate transcriptional program leading to a general increase in protein-folding capacity, increase in protein turnover, and decrease in the production of additional substrates via attenuation of general protein translation. In cases of chronic stress, such as during disease or aging (right), sequestration of key PN components by aggregates can lead to aberrant transcriptional programs, a deficit in folding capacity due to a lack of functionally available chaperones, and a buildup of misfolded species due to a decline in proteasomal capacity. This can lead to a chronic inability to restore PN balance and further accumulation of misfolded species.
The cytosolic stress response is regulated primarily by heat-shock transcription factor 1 (Hsf1), which is maintained in an inactive state by association with chaperones including Hsp90 (Zou et al., 1998) and Hsp70 (Zheng et al., 2016). The current model suggests that on heat stress, these chaperones are titrated away from Hsf1 by binding to denatured proteins. Hsf1 is then free to induce the transcription of a wide range of proteostasis components (Zou et al., 1998; Zheng et al., 2016), although general protein translation is decreased, reducing the load on the chaperone machinery. Concurrently, expression of chaperones and other quality-control elements, such as proteasomal components, is increased to prevent and resolve misfolded proteins and aggregation. Finally, once the stressor is removed, a negative-feedback loop on Hsf1 activity ensures a return to stasis within the system (Akerfelt et al., 2010; Gomez-Pastor et al., 2017).
Similar stress-response pathways include the UPR in the ER and mitochondria (Walter and Ron, 2011; Jovaisaite et al., 2014; Schulz and Haynes, 2015; Frakes and Dillin, 2017). The UPR of the ER has been studied extensively and is highly conserved from fungi to mammalian cells. The accumulation of misfolded proteins in the ER is sensed by three transmembrane signaling proteins, IRE-1, PERK and ATF6, which constitute distinct arms of the UPR and function to activate transcription factors for the production of a multiplicity of proteostasis components. PERK activation also leads to phosphorylation of eIF2α and thus to attenuation of general translation (Walter and Ron, 2011; Frakes and Dillin, 2017). Concomitantly, proteasome biogenesis is up-regulated by a recently discovered signaling pathway that adjusts cellular degradation capacity to demand (Rousseau and Bertolotti, 2016). Although the exact mode of activation of the UPR is distinct from that of the cytosolic heat-shock response, the overall goals are similar: an up-regulation of quality-control components and a decrease in potentially misfolded substrates though transient attenuation of translation.
Increasing evidence supports the existence of significant crosstalk between the various cellular stress-response pathways, with protein misfolding in the ER resulting in the aggregation of metastable proteins in the cytosol (Hamdan et al., 2017). This is consistent with ER stress triggering a partially protective cytosolic stress response when components of the UPR are defective (Liu and Chang, 2008). A link between mitochondrial stress and the cytosolic stress-response pathway has also been identified that can protect cells from disease associated aggregates (Kim et al., 2016a).
Chronic stress response
Although the up-regulation of protein quality-control components allows cells to resolve stress-induced misfolded proteins and aggregates that formed as the result of acute environmental stress, the amyloid aggregates associated with age-dependent diseases appear to be largely resistant to these rescue mechanisms (Klaips et al., 2014; Zaarur et al., 2015). The resulting chronic exposure of cells to misfolded species can have detrimental effects on the PN. For example, expression of model polyglutamine aggregates interferes with ER-associated protein degradation and leads to a prolonged activation of the ER stress response (Duennwald and Lindquist, 2008; Leitman et al., 2013).
On chronic production of certain misfolded or aggregated proteins, as may occur in disease or during aging, the stress response becomes activated but unable to clear the offending species (Lamech and Haynes, 2015; Fig. 4). This maladaptive stress response leaves cells vulnerable not only because the aggregates persist but also because the cells become refractive to additional stressors (Roth et al., 2014), consistent with aged cells and organisms being less responsive to stress insults (Ben-Zvi et al., 2009). A recent study in nematodes and mammalian cells revealed an interesting relationship among aging, chronic protein folding stress, and PN capacity (Tawo et al., 2017). These authors observed that normal turnover of the insulin-like growth factor receptor (Daf2 in C. elegans) involves the E3 ubiquitin ligase Chip. Both aging and the accumulation of protein aggregates were found to interfere with the degradation of insulin receptor, because Chip becomes increasingly engaged by misfolded proteins (Tawo et al., 2017). The resulting increase in Daf2 levels inhibits the Daf16 transcription factor (FOXO in mammals), causing a down-regulation of PN components and reduced lifespan.
Modulation of the PN
The persistence of disease-associated protein aggregates would suggest that the cellular PN is generally unable to cope with such substrates. However, cells may be able to adequately handle aberrant protein species for long periods, sometimes decades, as suggested by the fact that even the inherited forms of neurodegenerative disease, such as Huntington’s disease, do not present until advanced age.
Indeed, specific modulation of PN components can impact both aggregate morphology and lifespan in model systems, paving the way for therapeutic intervention (Balch et al., 2008; Powers et al., 2009). Expression of chaperones and cochaperones of different classes have consistently resulted in a decrease in disease-aggregate toxicity and even increased lifespan (Auluck et al., 2002; Hoshino et al., 2011; Chafekar et al., 2012). Analogous to the cellular stress responses, strategies for therapeutic interventions in aggregate-associated neurodegenerative diseases have focused on preventing further production of misfolded species, stabilization of properly folded species, and clearance of existing aggregates (Balch et al., 2008; Calamini et al., 2011). Toward these ends, small molecules have been identified that prolong translation attenuation on stress (Tsaytler et al., 2011); stabilize mutant proteins, such as transthyretin against aggregation; target folding and trafficking defects in specific disease associated proteins, such as mutant cystic fibrosis transmembrane conductance regulator (Baranczak and Kelly, 2016); and increase the clearance of toxic protein species through activation of the UPS (Lee et al., 2010) or autophagy (Sarkar et al., 2009). Because of the broad range of components sequestered by protein aggregates, enhancement of the endogenous stress-response pathways themselves has been particularly fruitful in extending lifespan and proteostatic health (Sittler et al., 2001; Mu et al., 2008; Akerfelt et al., 2010; Kumsta et al., 2017).
Concluding remarks
Work over the past decades has uncovered a remarkable ability of cells to maintain proteostasis under a variety of challenging conditions. The importance of this ability is underlined by our increasing understanding that many neurodegenerative and aging-associated diseases are caused by a breakdown in this process. In recent years we have gained considerable insight as to why PN capacity may decline with age. Although in some cases the buildup of stochastic mutations and damage can certainly contribute to disease onset, this seems insufficient to explain the universal age-dependent decline in PN health observed across species. Work in metazoans such as worms suggests that this decline may instead be a regulated process. Consistent with the basic tenet of the “disposable soma” theory (Kirkwood and Holliday, 1979), organisms may sacrifice their own proteostatic fitness to divert resources toward reproduction. This is a plausible explanation, especially for short-lived species such as C. elegans, in which proteostasis decline occurs abruptly in early adulthood, and the lifespan extension gained by activation of stress-response factors comes at the cost of reduced fecundity (Ben-Zvi et al., 2009). However, the mechanisms underlying the gradual deterioration of proteostasis in long-lived mammals are clearly more complex. Hopefully, a better understanding of the network connectivity in healthy cells and tissues and its changes during aging and disease will allow us to harness aspects of the PN to combat aggregation disorders and increase health span.
Acknowledgments
Work in the authors’ laboratory is supported by the European Commission (grant FP7 GA ERC-2012-SyG_318987–ToPAG) and the Munich Cluster for Systems Neurology (grant EXC 010 SyNergy). C.L. Klaips is the recipient of a postdoctoral fellowship from the Alexander von Humboldt Foundation.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- Hsp
- heat-shock protein
- PERK
- protein kinase RNA-like ER kinase
- PN
- proteostasis network
- sHsp
- small Hsp
- UPR
- unfolded protein response
- UPS
- ubiquitin–proteasome system
References
- Agashe V.R., Guha S., Chang H.C., Genevaux P., Hayer-Hartl M., Stemp M., Georgopoulos C., Hartl F.U., and Barral J.M.. 2004. Function of trigger factor and DnaK in multidomain protein folding: increase in yield at the expense of folding speed. Cell. 117:199–209. 10.1016/S0092-8674(04)00299-5 [DOI] [PubMed] [Google Scholar]
- Aguilaniu H., Gustafsson L., Rigoulet M., and Nyström T.. 2003. Asymmetric inheritance of oxidatively damaged proteins during cytokinesis. Science. 299:1751–1753. 10.1126/science.1080418 [DOI] [PubMed] [Google Scholar]
- Akerfelt M., Morimoto R.I., and Sistonen L.. 2010. Heat shock factors: integrators of cell stress, development and lifespan. Nat. Rev. Mol. Cell Biol. 11:545–555. 10.1038/nrm2938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albanèse V., Yam A.Y., Baughman J., Parnot C., and Frydman J.. 2006. Systems analyses reveal two chaperone networks with distinct functions in eukaryotic cells. Cell. 124:75–88. 10.1016/j.cell.2005.11.039 [DOI] [PubMed] [Google Scholar]
- Alberti S., and Hyman A.A.. 2016. Are aberrant phase transitions a driver of cellular aging? BioEssays. 38:959–968. 10.1002/bies.201600042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anfinsen C.B. 1973. Principles that govern the folding of protein chains. Science. 181:223–230. 10.1126/science.181.4096.223 [DOI] [PubMed] [Google Scholar]
- Arrasate M., Mitra S., Schweitzer E.S., Segal M.R., and Finkbeiner S.. 2004. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 431:805–810. 10.1038/nature02998 [DOI] [PubMed] [Google Scholar]
- Auluck P.K., Chan H.Y., Trojanowski J.Q., Lee V.M., and Bonini N.M.. 2002. Chaperone suppression of alpha-synuclein toxicity in a Drosophila model for Parkinson’s disease. Science. 295:865–868. 10.1126/science.1067389 [DOI] [PubMed] [Google Scholar]
- Balch W.E., Morimoto R.I., Dillin A., and Kelly J.W.. 2008. Adapting proteostasis for disease intervention. Science. 319:916–919. 10.1126/science.1141448 [DOI] [PubMed] [Google Scholar]
- Balchin D., Hayer-Hartl M., and Hartl F.U.. 2016. In vivo aspects of protein folding and quality control. Science. 353:aac4354 10.1126/science.aac4354 [DOI] [PubMed] [Google Scholar]
- Banani S.F., Lee H.O., Hyman A.A., and Rosen M.K.. 2017. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18:285–298. 10.1038/nrm.2017.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranczak A., and Kelly J.W.. 2016. A current pharmacologic agent versus the promise of next generation therapeutics to ameliorate protein misfolding and/or aggregation diseases. Curr. Opin. Chem. Biol. 32:10–21. 10.1016/j.cbpa.2016.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett A.I., and Radford S.E.. 2009. An expanding arsenal of experimental methods yields an explosion of insights into protein folding mechanisms. Nat. Struct. Mol. Biol. 16:582–588. 10.1038/nsmb.1592 [DOI] [PubMed] [Google Scholar]
- Bäuerlein F.J.B., Saha I., Mishra A., Kalemanov M., Martínez-Sánchez A., Klein R., Dudanova I., Hipp M.S., Hartl F.U., Baumeister W., and Fernández-Busnadiego R.. 2017. In Situ Architecture and Cellular Interactions of PolyQ Inclusions. Cell. 171:179–187. 10.1016/j.cell.2017.08.009 [DOI] [PubMed] [Google Scholar]
- Bence N.F., Sampat R.M., and Kopito R.R.. 2001. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 292:1552–1555. 10.1126/science.292.5521.1552 [DOI] [PubMed] [Google Scholar]
- Bennett E.J., Bence N.F., Jayakumar R., and Kopito R.R.. 2005. Global impairment of the ubiquitin-proteasome system by nuclear or cytoplasmic protein aggregates precedes inclusion body formation. Mol. Cell. 17:351–365. 10.1016/j.molcel.2004.12.021 [DOI] [PubMed] [Google Scholar]
- Ben-Zvi A., De Los Rios P., Dietler G., and Goloubinoff P.. 2004. Active solubilization and refolding of stable protein aggregates by cooperative unfolding action of individual hsp70 chaperones. J. Biol. Chem. 279:37298–37303. 10.1074/jbc.M405627200 [DOI] [PubMed] [Google Scholar]
- Ben-Zvi A., Miller E.A., and Morimoto R.I.. 2009. Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. Proc. Natl. Acad. Sci. USA. 106:14914–14919. 10.1073/pnas.0902882106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandman O., and Hegde R.S.. 2016. Ribosome-associated protein quality control. Nat. Struct. Mol. Biol. 23:7–15. 10.1038/nsmb.3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt F., Etchells S.A., Ortiz J.O., Elcock A.H., Hartl F.U., and Baumeister W.. 2009. The native 3D organization of bacterial polysomes. Cell. 136:261–271. 10.1016/j.cell.2008.11.016 [DOI] [PubMed] [Google Scholar]
- Brehme M., Voisine C., Rolland T., Wachi S., Soper J.H., Zhu Y., Orton K., Villella A., Garza D., Vidal M., et al. 2014. A chaperome subnetwork safeguards proteostasis in aging and neurodegenerative disease. Cell Rep. 9:1135–1150. 10.1016/j.celrep.2014.09.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockwell D.J., and Radford S.E.. 2007. Intermediates: ubiquitous species on folding energy landscapes? Curr. Opin. Struct. Biol. 17:30–37. 10.1016/j.sbi.2007.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrita L.D., Cassaignau A.M.E., Launay H.M.M., Waudby C.A., Wlodarski T., Camilloni C., Karyadi M.-E., Robertson A.L., Wang X., Wentink A.S., et al. 2016. A structural ensemble of a ribosome-nascent chain complex during cotranslational protein folding. Nat. Struct. Mol. Biol. 23:278–285. 10.1038/nsmb.3182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calamini B., Silva M.C., Madoux F., Hutt D.M., Khanna S., Chalfant M.A., Saldanha S.A., Hodder P., Tait B.D., Garza D., et al. 2011. Small-molecule proteostasis regulators for protein conformational diseases. Nat. Chem. Biol. 8:185–196. 10.1038/nchembio.763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chafekar S.M., Wisen S., Thompson A.D., Echeverria A., Walter G.M., Evans C.G., Makley L.N., Gestwicki J.E., and Duennwald M.L.. 2012. Pharmacological Tuning of Heat Shock Protein 70 Modulates Polyglutamine Toxicity and Aggregation. ACS Chem. Biol. 7:1556–1564. 10.1021/cb300166p [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiti F., and Dobson C.M.. 2017. Protein Misfolding, Amyloid Formation, and Human Disease: A Summary of Progress Over the Last Decade. Annu. Rev. Biochem. 86:27–68. 10.1146/annurev-biochem-061516-045115 [DOI] [PubMed] [Google Scholar]
- Ciechanover A., and Kwon Y.T.. 2017. Protein Quality Control by Molecular Chaperones in Neurodegeneration. Front. Neurosci. 11:185 10.3389/fnins.2017.00185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciryam P., Tartaglia G.G., Morimoto R.I., Dobson C.M., and Vendruscolo M.. 2013. Widespread aggregation and neurodegenerative diseases are associated with supersaturated proteins. Cell Rep. 5:781–790. 10.1016/j.celrep.2013.09.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerico E.M., Tilitsky J.M., Meng W., and Gierasch L.M.. 2015. How hsp70 molecular machines interact with their substrates to mediate diverse physiological functions. J. Mol. Biol. 427:1575–1588. 10.1016/j.jmb.2015.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo A.M., and Wong E.. 2014. Chaperone-mediated autophagy: roles in disease and aging. Cell Res. 24:92–104. 10.1038/cr.2013.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David D.C., Ollikainen N., Trinidad J.C., Cary M.P., Burlingame A.L., and Kenyon C.. 2010. Widespread protein aggregation as an inherent part of aging in C. elegans. PLoS Biol. 8:e1000450 10.1371/journal.pbio.1000450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamant S., Ben-Zvi A.P., Bukau B., and Goloubinoff P.. 2000. Size-dependent disaggregation of stable protein aggregates by the DnaK chaperone machinery. J. Biol. Chem. 275:21107–21113. 10.1074/jbc.M001293200 [DOI] [PubMed] [Google Scholar]
- Dinner A.R., Šali A., Smith L.J., Dobson C.M., and Karplus M.. 2000. Understanding protein folding via free-energy surfaces from theory and experiment. Trends Biochem. Sci. 25:331–339. 10.1016/S0968-0004(00)01610-8 [DOI] [PubMed] [Google Scholar]
- Dobson C.M., Sali A., and Karplus M.. 1998. Protein folding: a perspective from theory and experiment. Angew. Chem. Int. Ed. Engl. 37:868–893. [DOI] [PubMed] [Google Scholar]
- Douglas P.M., Treusch S., Ren H.Y., Halfmann R., Duennwald M.L., Lindquist S., and Cyr D.M.. 2008. Chaperone-dependent amyloid assembly protects cells from prion toxicity. Proc. Natl. Acad. Sci. USA. 105:7206–7211. 10.1073/pnas.0802593105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond D.A., and Wilke C.O.. 2008. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell. 134:341–352. 10.1016/j.cell.2008.05.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duennwald M.L., and Lindquist S.. 2008. Impaired ERAD and ER stress are early and specific events in polyglutamine toxicity. Genes Dev. 22:3308–3319. 10.1101/gad.1673408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunker A.K., Silman I., Uversky V.N., and Sussman J.L.. 2008. Function and structure of inherently disordered proteins. Curr. Opin. Struct. Biol. 18:756–764. 10.1016/j.sbi.2008.10.002 [DOI] [PubMed] [Google Scholar]
- Ellis R.J., and Minton A.P.. 2006. Protein aggregation in crowded environments. Biol. Chem. 387:485–497. 10.1515/BC.2006.064 [DOI] [PubMed] [Google Scholar]
- Erjavec N., Larsson L., Grantham J., and Nyström T.. 2007. Accelerated aging and failure to segregate damaged proteins in Sir2 mutants can be suppressed by overproducing the protein aggregation-remodeling factor Hsp104p. Genes Dev. 21:2410–2421. 10.1101/gad.439307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escusa-Toret S., Vonk W.I.M., and Frydman J.. 2013. Spatial sequestration of misfolded proteins by a dynamic chaperone pathway enhances cellular fitness during stress. Nat. Cell Biol. 15:1231–1243. 10.1038/ncb2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esser C., Alberti S., and Höhfeld J.. 2004. Cooperation of molecular chaperones with the ubiquitin/proteasome system. Biochim. Biophys. Acta. 1695:171–188. 10.1016/j.bbamcr.2004.09.020 [DOI] [PubMed] [Google Scholar]
- Feric M., Vaidya N., Harmon T.S., Mitrea D.M., Zhu L., Richardson T.M., Kriwacki R.W., Pappu R.V., and Brangwynne C.P.. 2016. Coexisting Liquid Phases Underlie Nucleolar Subcompartments. Cell. 165:1686–1697. 10.1016/j.cell.2016.04.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frakes A.E., and Dillin A.. 2017. The UPRER: Sensor and Coordinator of Organismal Homeostasis. Mol. Cell. 66:761–771. 10.1016/j.molcel.2017.05.031 [DOI] [PubMed] [Google Scholar]
- Frydman J., Nimmesgern E., Ohtsuka K., and Hartl F.U.. 1994. Folding of nascent polypeptide chains in a high molecular mass assembly with molecular chaperones. Nature. 370:111–117. 10.1038/370111a0 [DOI] [PubMed] [Google Scholar]
- Frydman J., Erdjument-Bromage H., Tempst P., and Hartl F.U.. 1999. Co-translational domain folding as the structural basis for the rapid de novo folding of firefly luciferase. Nat. Struct. Biol. 6:697–705. 10.1038/10754 [DOI] [PubMed] [Google Scholar]
- García-Prat L., Martínez-Vicente M., Perdiguero E., Ortet L., Rodríguez-Ubreva J., Rebollo E., Ruiz-Bonilla V., Gutarra S., Ballestar E., Serrano A.L., et al. 2016. Autophagy maintains stemness by preventing senescence. Nature. 529:37–42. 10.1038/nature16187 [DOI] [PubMed] [Google Scholar]
- Geller R., Andino R., and Frydman J.. 2013. Hsp90 inhibitors exhibit resistance-free antiviral activity against respiratory syncytial virus. PLoS One. 8:e56762 10.1371/journal.pone.0056762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genevaux P., Georgopoulos C., and Kelley W.L.. 2007. The Hsp70 chaperone machines of Escherichia coli: a paradigm for the repartition of chaperone functions. Mol. Microbiol. 66:840–857. 10.1111/j.1365-2958.2007.05961.x [DOI] [PubMed] [Google Scholar]
- Gershenson A., Gierasch L.M., Pastore A., and Radford S.E.. 2014. Energy landscapes of functional proteins are inherently risky. Nat. Chem. Biol. 10:884–891. 10.1038/nchembio.1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidalevitz T., Ben-Zvi A., Ho K.H., Brignull H.R., and Morimoto R.I.. 2006. Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science. 311:1471–1474. 10.1126/science.1124514 [DOI] [PubMed] [Google Scholar]
- Gidalevitz T., Stevens F., and Argon Y.. 2013. Orchestration of secretory protein folding by ER chaperones. Biochim. Biophys. Acta. 1833:2410–2424. 10.1016/j.bbamcr.2013.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover J.R., and Lindquist S.. 1998. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell. 94:73–82. 10.1016/S0092-8674(00)81223-4 [DOI] [PubMed] [Google Scholar]
- Gomez-Pastor R., Burchfiel E.T., and Thiele D.J.. 2017. Regulation of heat shock transcription factors and their roles in physiology and disease. Nat. Rev. Mol. Cell Biol. 10.1038/nrn.2017.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guisbert E., Czyz D.M., Richter K., McMullen P.D., and Morimoto R.I.. 2013. Identification of a tissue-selective heat shock response regulatory network. PLoS Genet. 9:e1003466 10.1371/journal.pgen.1003466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R., Kasturi P., Bracher A., Loew C., Zheng M., Villella A., Garza D., Hartl F.U., and Raychaudhuri S.. 2011. Firefly luciferase mutants as sensors of proteome stress. Nat. Methods. 8:879–884. 10.1038/nmeth.1697 [DOI] [PubMed] [Google Scholar]
- Gusella J.F., and MacDonald M.E.. 2006. Huntington’s disease: seeing the pathogenic process through a genetic lens. Trends Biochem. Sci. 31:533–540. 10.1016/j.tibs.2006.06.009 [DOI] [PubMed] [Google Scholar]
- Hamdan N., Kritsiligkou P., and Grant C.M.. 2017. ER stress causes widespread protein aggregation and prion formation. J. Cell Biol. 216:2295–2304. 10.1083/jcb.201612165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamer G., Matilainen O., and Holmberg C.I.. 2010. A photoconvertible reporter of the ubiquitin-proteasome system in vivo. Nat. Methods. 7:473–478. 10.1038/nmeth.1460 [DOI] [PubMed] [Google Scholar]
- Hansen J.J., Dürr A., Cournu-Rebeix I., Georgopoulos C., Ang D., Nielsen M.N., Davoine C.-S., Brice A., Fontaine B., Gregersen N., and Bross P.. 2002. Hereditary spastic paraplegia SPG13 is associated with a mutation in the gene encoding the mitochondrial chaperonin Hsp60. Am. J. Hum. Genet. 70:1328–1332. 10.1086/339935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M., Taubert S., Crawford D., Libina N., Lee S.-J., and Kenyon C.. 2007. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell. 6:95–110. 10.1111/j.1474-9726.2006.00267.x [DOI] [PubMed] [Google Scholar]
- Harding H.P., Novoa I., Bertolotti A., Zeng H., Zhang Y., Urano F., Jousse C., and Ron D.. 2001. Translational regulation in the cellular response to biosynthetic load on the endoplasmic reticulum. Cold Spring Harb. Symp. Quant. Biol. 66:499–508. 10.1101/sqb.2001.66.499 [DOI] [PubMed] [Google Scholar]
- Hartl F.U. 1996. Molecular chaperones in cellular protein folding. Nature. 381:571–579. 10.1038/381571a0 [DOI] [PubMed] [Google Scholar]
- Hartl F.U., Bracher A., and Hayer-Hartl M.. 2011. Molecular chaperones in protein folding and proteostasis. Nature. 475:324–332. 10.1038/nature10317 [DOI] [PubMed] [Google Scholar]
- Haslbeck M., Braun N., Stromer T., Richter B., Model N., Weinkauf S., and Buchner J.. 2004. Hsp42 is the general small heat shock protein in the cytosol of Saccharomyces cerevisiae. EMBO J. 23:638–649. 10.1038/sj.emboj.7600080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslbeck M., Franzmann T., Weinfurtner D., and Buchner J.. 2005. Some like it hot: the structure and function of small heat-shock proteins. Nat. Struct. Mol. Biol. 12:842–846. 10.1038/nsmb993 [DOI] [PubMed] [Google Scholar]
- Hayer-Hartl M., Bracher A., and Hartl F.U.. 2016. The GroEL-GroES Chaperonin Machine: A Nano-Cage for Protein Folding. Trends Biochem. Sci. 41:62–76. 10.1016/j.tibs.2015.07.009 [DOI] [PubMed] [Google Scholar]
- Heck J.W., Cheung S.K., and Hampton R.Y.. 2010. Cytoplasmic protein quality control degradation mediated by parallel actions of the E3 ubiquitin ligases Ubr1 and San1. Proc. Natl. Acad. Sci. USA. 107:1106–1111. 10.1073/pnas.0910591107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill S.M., Hanzén S., and Nyström T.. 2017. Restricted access: spatial sequestration of damaged proteins during stress and aging. EMBO Rep. 18:377–391. 10.15252/embr.201643458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hipp M.S., Park S.-H., and Hartl F.U.. 2014. Proteostasis impairment in protein-misfolding and -aggregation diseases. Trends Cell Biol. 24:506–514. 10.1016/j.tcb.2014.05.003 [DOI] [PubMed] [Google Scholar]
- Hjerpe R., Bett J.S., Keuss M.J., Solovyova A., McWilliams T.G., Johnson C., Sahu I., Varghese J., Wood N., Wightman M., et al. 2016. UBQLN2 Mediates Autophagy-Independent Protein Aggregate Clearance by the Proteasome. Cell. 166:935–949. 10.1016/j.cell.2016.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes W.M., Klaips C.L., and Serio T.R.. 2014. Defining the limits: Protein aggregation and toxicity in vivo. Crit. Rev. Biochem. Mol. Biol. 49:294–303. 10.3109/10409238.2014.914151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtkamp W., Kokic G., Jäger M., Mittelstaet J., Komar A.A., and Rodnina M.V.. 2015. Cotranslational protein folding on the ribosome monitored in real time. Science. 350:1104–1107. 10.1126/science.aad0344 [DOI] [PubMed] [Google Scholar]
- Hoshino T., Murao N., Namba T., Takehara M., Adachi H., Katsuno M., Sobue G., Matsushima T., Suzuki T., and Mizushima T.. 2011. Suppression of Alzheimer’s disease-related phenotypes by expression of heat shock protein 70 in mice. J. Neurosci. 31:5225–5234. 10.1523/JNEUROSCI.5478-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E., Zavodszky E., Shao S., Wohlever M.L., Keenan R.J., and Hegde R.S.. 2016. Ubiquilins Chaperone and Triage Mitochondrial Membrane Proteins for Degradation. Mol. Cell. 63:21–33. 10.1016/j.molcel.2016.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston J.A., Ward C.L., and Kopito R.R.. 1998. Aggresomes: A cellular response to misfolded proteins. J. Cell Biol. 143:1883–1898. 10.1083/jcb.143.7.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovaisaite V., Mouchiroud L., and Auwerx J.. 2014. The mitochondrial unfolded protein response, a conserved stress response pathway with implications in health and disease. J. Exp. Biol. 217:137–143. 10.1242/jeb.090738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaganovich D., Kopito R., and Frydman J.. 2008. Misfolded proteins partition between two distinct quality control compartments. Nature. 454:1088–1095. 10.1038/nature07195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser C.M., Chang H.C., Agashe V.R., Lakshmipathy S.K., Etchells S.A., Hayer-Hartl M., Hartl F.U., and Barral J.M.. 2006. Real-time observation of trigger factor function on translating ribosomes. Nature. 444:455–460. 10.1038/nature05225 [DOI] [PubMed] [Google Scholar]
- Kaiser C.M., Goldman D.H., Chodera J.D., Tinoco I. Jr., and Bustamante C.. 2011. The ribosome modulates nascent protein folding. Science. 334:1723–1727. 10.1126/science.1209740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakkar V., Meister-Broekema M., Minoia M., Carra S., and Kampinga H.H.. 2014. Barcoding heat shock proteins to human diseases: looking beyond the heat shock response. Dis. Model. Mech. 7:421–434. 10.1242/dmm.014563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampinga H.H., and Craig E.A.. 2010. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 11:579–592. 10.1038/nrm2941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayed R., Head E., Thompson J.L., McIntire T.M., Milton S.C., Cotman C.W., and Glabe C.G.. 2003. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 300:486–489. 10.1126/science.1079469 [DOI] [PubMed] [Google Scholar]
- Kettern N., Dreiseidler M., Tawo R., and Höhfeld J.. 2010. Chaperone-assisted degradation: multiple paths to destruction. Biol. Chem. 391:481–489. 10.1515/bc.2010.058 [DOI] [PubMed] [Google Scholar]
- Kim H.E., Grant A.R., Simic M.S., Kohnz R.A., Nomura D.K., Durieux J., Riera C.E., Sanchez M., Kapernick E., Wolff S., and Dillin A.. 2016a Lipid Biosynthesis Coordinates a Mitochondrial-to-Cytosolic Stress Response. Cell. 166:1539–1552. 10.1016/j.cell.2016.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.E., Hipp M.S., Bracher A., Hayer-Hartl M., and Hartl F.U.. 2013. Molecular chaperone functions in protein folding and proteostasis. Annu. Rev. Biochem. 82:323–355. 10.1146/annurev-biochem-060208-092442 [DOI] [PubMed] [Google Scholar]
- Kim Y.E., Hosp F., Frottin F., Ge H., Mann M., Hayer-Hartl M., and Hartl F.U.. 2016b Soluble Oligomers of PolyQ-Expanded Huntingtin Target a Multiplicity of Key Cellular Factors. Mol. Cell. 63:951–964. 10.1016/j.molcel.2016.07.022 [DOI] [PubMed] [Google Scholar]
- Kirkwood T.B.L., and Holliday R.. 1979. The evolution of ageing and longevity. Proc. R. Soc. Lond. B Biol. Sci. 205:531–546. 10.1098/rspb.1979.0083 [DOI] [PubMed] [Google Scholar]
- Klaips C.L., Hochstrasser M.L., Langlois C.R., and Serio T.R.. 2014. Spatial quality control bypasses cell-based limitations on proteostasis to promote prion curing. Elife. 3:e04288 10.7554/eLife.04288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulak N.A., Geyer P.E., and Mann M.. 2017. Loss-less Nano-fractionator for High Sensitivity, High Coverage Proteomics. Mol. Cell. Proteomics. 16:694–705. 10.1074/mcp.O116.065136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumsta C., Chang J.T., Schmalz J., and Hansen M.. 2017. Hormetic heat stress and HSF-1 induce autophagy to improve survival and proteostasis in C. elegans. Nat. Commun. 8:14337 10.1038/ncomms14337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundra R., Ciryam P., Morimoto R.I., Dobson C.M., and Vendruscolo M.. 2017. Protein homeostasis of a metastable subproteome associated with Alzheimer’s disease. Proc. Natl. Acad. Sci. USA. 114:E5703–E5711. 10.1073/pnas.1618417114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbadia J., and Morimoto R.I.. 2015a The biology of proteostasis in aging and disease. Annu. Rev. Biochem. 84:435–464. 10.1146/annurev-biochem-060614-033955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbadia J., and Morimoto R.I.. 2015b Repression of the Heat Shock Response Is a Programmed Event at the Onset of Reproduction. Mol. Cell. 59:639–650. 10.1016/j.molcel.2015.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamark T., and Johansen T.. 2012. Aggrephagy: selective disposal of protein aggregates by macroautophagy. Int. J. Cell Biol. 2012:736905 10.1155/2012/736905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamech L.T., and Haynes C.M.. 2015. The unpredictability of prolonged activation of stress response pathways. J. Cell Biol. 209:781–787. 10.1083/jcb.201503107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer T., Lu C., Echols H., Flanagan J., Hayer M.K., and Hartl F.U.. 1992. Successive action of DnaK, DnaJ and GroEL along the pathway of chaperone-mediated protein folding. Nature. 356:683–689. 10.1038/356683a0 [DOI] [PubMed] [Google Scholar]
- Laufen T., Mayer M.P., Beisel C., Klostermeier D., Mogk A., Reinstein J., and Bukau B.. 1999. Mechanism of regulation of hsp70 chaperones by DnaJ cochaperones. Proc. Natl. Acad. Sci. USA. 96:5452–5457. 10.1073/pnas.96.10.5452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B.-H., Lee M.J., Park S., Oh D.-C., Elsasser S., Chen P.-C., Gartner C., Dimova N., Hanna J., Gygi S.P., et al. 2010. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 467:179–184. 10.1038/nature09299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitman J., Ulrich Hartl F., and Lederkremer G.Z.. 2013. Soluble forms of polyQ-expanded huntingtin rather than large aggregates cause endoplasmic reticulum stress. Nat. Commun. 4:2753 10.1038/ncomms3753 [DOI] [PubMed] [Google Scholar]
- Li W., Bengtson M.H., Ulbrich A., Matsuda A., Reddy V.A., Orth A., Chanda S.K., Batalov S., and Joazeiro C.A.. 2008. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle’s dynamics and signaling. PLoS One. 3:e1487 10.1371/journal.pone.0001487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., and Chang A.. 2008. Heat shock response relieves ER stress. EMBO J. 27:1049–1059. 10.1038/emboj.2008.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez T., Dalton K., and Frydman J.. 2015. The Mechanism and Function of Group II Chaperonins. J. Mol. Biol. 427:2919–2930. 10.1016/j.jmb.2015.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Otín C., Blasco M.A., Partridge L., Serrano M., and Kroemer G.. 2013. The hallmarks of aging. Cell. 153:1194–1217. 10.1016/j.cell.2013.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe J., Blanchard A., Morrell K., Lennox G., Reynolds L., Billett M., Landon M., and Mayer R.J.. 1988. Ubiquitin is a common factor in intermediate filament inclusion bodies of diverse type in man, including those of Parkinson’s disease, Pick’s disease, and Alzheimer’s disease, as well as Rosenthal fibres in cerebellar astrocytomas, cytoplasmic bodies in muscle, and mallory bodies in alcoholic liver disease. J. Pathol. 155:9–15. 10.1002/path.1711550105 [DOI] [PubMed] [Google Scholar]
- Magen D., Georgopoulos C., Bross P., Ang D., Segev Y., Goldsher D., Nemirovski A., Shahar E., Ravid S., Luder A., et al. 2008. Mitochondrial hsp60 chaperonopathy causes an autosomal-recessive neurodegenerative disorder linked to brain hypomyelination and leukodystrophy. Am. J. Hum. Genet. 83:30–42. 10.1016/j.ajhg.2008.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinovska L., Kroschwald S., Munder M.C., Richter D., and Alberti S.. 2012. Molecular chaperones and stress-inducible protein-sorting factors coordinate the spatiotemporal distribution of protein aggregates. Mol. Biol. Cell. 23:3041–3056. 10.1091/mbc.E12-03-0194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino J., von Heijne G., and Beckmann R.. 2016. Small protein domains fold inside the ribosome exit tunnel. FEBS Lett. 590:655–660. 10.1002/1873-3468.12098 [DOI] [PubMed] [Google Scholar]
- Mashaghi A., Bezrukavnikov S., Minde D.P., Wentink A.S., Kityk R., Zachmann-Brand B., Mayer M.P., Kramer G., Bukau B., and Tans S.J.. 2016. Alternative modes of client binding enable functional plasticity of Hsp70. Nature. 539:448–451. 10.1038/nature20137 [DOI] [PubMed] [Google Scholar]
- Mayer M.P., and Bukau B.. 2005. Hsp70 chaperones: cellular functions and molecular mechanism. Cell. Mol. Life Sci. 62:670–684. 10.1007/s00018-004-4464-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer M.P., Rüdiger S., and Bukau B.. 2000. Molecular basis for interactions of the DnaK chaperone with substrates. Biol. Chem. 381:877–885. 10.1515/BC.2000.109 [DOI] [PubMed] [Google Scholar]
- Mbofung R.M., McKenzie J.A., Malu S., Zhang M., Peng W., Liu C., Kuiatse I., Tieu T., Williams L., Devi S., et al. 2017. HSP90 inhibition enhances cancer immunotherapy by upregulating interferon response genes. Nat. Commun. 8:451 10.1038/s41467-017-00449-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzies F.M., Fleming A., and Rubinsztein D.C.. 2015. Compromised autophagy and neurodegenerative diseases. Nat. Rev. Neurosci. 16:345–357. 10.1038/nrn3961 [DOI] [PubMed] [Google Scholar]
- Miller S.B., Ho C.T., Winkler J., Khokhrina M., Neuner A., Mohamed M.Y., Guilbride D.L., Richter K., Lisby M., Schiebel E., et al. 2015. Compartment-specific aggregases direct distinct nuclear and cytoplasmic aggregate deposition. EMBO J. 34:778–797. 10.15252/embj.201489524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N. 2007. Autophagy: process and function. Genes Dev. 21:2861–2873. 10.1101/gad.1599207 [DOI] [PubMed] [Google Scholar]
- Mogk A., Deuerling E., Vorderwülbecke S., Vierling E., and Bukau B.. 2003. Small heat shock proteins, ClpB and the DnaK system form a functional triade in reversing protein aggregation. Mol. Microbiol. 50:585–595. 10.1046/j.1365-2958.2003.03710.x [DOI] [PubMed] [Google Scholar]
- Morley J.F., Brignull H.R., Weyers J.J., and Morimoto R.I.. 2002. The threshold for polyglutamine-expansion protein aggregation and cellular toxicity is dynamic and influenced by aging in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 99:10417–10422. 10.1073/pnas.152161099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu T.W., Ong D.S., Wang Y.J., Balch W.E., Yates J.R. III, Segatori L., and Kelly J.W.. 2008. Chemical and biological approaches synergize to ameliorate protein-folding diseases. Cell. 134:769–781. 10.1016/j.cell.2008.06.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchowski P.J., Schaffar G., Sittler A., Wanker E.E., Hayer-Hartl M.K., and Hartl F.U.. 2000. Hsp70 and hsp40 chaperones can inhibit self-assembly of polyglutamine proteins into amyloid-like fibrils. Proc. Natl. Acad. Sci. USA. 97:7841–7846. 10.1073/pnas.140202897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mymrikov E.V., Daake M., Richter B., Haslbeck M., and Buchner J.. 2017. The Chaperone Activity and Substrate Spectrum of Human Small Heat Shock Proteins. J. Biol. Chem. 292:672–684. 10.1074/jbc.M116.760413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy M., Fenton W.A., Li D., Furtak K., and Horwich A.L.. 2016. Extended survival of misfolded G85R SOD1-linked ALS mice by transgenic expression of chaperone Hsp110. Proc. Natl. Acad. Sci. USA. 113:5424–5428. 10.1073/pnas.1604885113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netzer W.J., and Hartl F.U.. 1997. Recombination of protein domains facilitated by co-translational folding in eukaryotes. Nature. 388:343–349. 10.1038/41024 [DOI] [PubMed] [Google Scholar]
- Ng P.C., and Henikoff S.. 2006. Predicting the effects of amino acid substitutions on protein function. Annu. Rev. Genomics Hum. Genet. 7:61–80. 10.1146/annurev.genom.7.080505.115630 [DOI] [PubMed] [Google Scholar]
- Nijman S.M., Luna-Vargas M.P., Velds A., Brummelkamp T.R., Dirac A.M., Sixma T.K., and Bernards R.. 2005. A genomic and functional inventory of deubiquitinating enzymes. Cell. 123:773–786. 10.1016/j.cell.2005.11.007 [DOI] [PubMed] [Google Scholar]
- Nillegoda N.B., and Bukau B.. 2015. Metazoan Hsp70-based protein disaggregases: emergence and mechanisms. Front. Mol. Biosci. 2:57 10.3389/fmolb.2015.00057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nillegoda N.B., Stank A., Malinverni D., Alberts N., Szlachcic A., Barducci A., De Los Rios P., Wade R.C., and Bukau B.. 2017. Evolution of an intricate J-protein network driving protein disaggregation in eukaryotes. Elife. 6:e24560 10.7554/eLife.24560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson O.B., Müller-Lucks A., Kramer G., Bukau B., and von Heijne G.. 2016. Trigger Factor Reduces the Force Exerted on the Nascent Chain by a Cotranslationally Folding Protein. J. Mol. Biol. 428:1356–1364. 10.1016/j.jmb.2016.02.014 [DOI] [PubMed] [Google Scholar]
- Olzscha H., Schermann S.M., Woerner A.C., Pinkert S., Hecht M.H., Tartaglia G.G., Vendruscolo M., Hayer-Hartl M., Hartl F.U., and Vabulas R.M.. 2011. Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell. 144:67–78. 10.1016/j.cell.2010.11.050 [DOI] [PubMed] [Google Scholar]
- Park S.H., Kukushkin Y., Gupta R., Chen T., Konagai A., Hipp M.S., Hayer-Hartl M., and Hartl F.U.. 2013. PolyQ proteins interfere with nuclear degradation of cytosolic proteins by sequestering the Sis1p chaperone. Cell. 154:134–145. 10.1016/j.cell.2013.06.003 [DOI] [PubMed] [Google Scholar]
- Parsell D.A., Kowal A.S., Singer M.A., and Lindquist S.. 1994. Protein disaggregation mediated by heat-shock protein Hsp104. Nature. 372:475–478. 10.1038/372475a0 [DOI] [PubMed] [Google Scholar]
- Powers E.T., Morimoto R.I., Dillin A., Kelly J.W., and Balch W.E.. 2009. Biological and chemical approaches to diseases of proteostasis deficiency. Annu. Rev. Biochem. 78:959–991. 10.1146/annurev.biochem.052308.114844 [DOI] [PubMed] [Google Scholar]
- Prasad R., Kawaguchi S., and Ng D.T.W.. 2010. A nucleus-based quality control mechanism for cytosolic proteins. Mol. Biol. Cell. 21:2117–2127. 10.1091/mbc.E10-02-0111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preissler S., and Deuerling E.. 2012. Ribosome-associated chaperones as key players in proteostasis. Trends Biochem. Sci. 37:274–283. 10.1016/j.tibs.2012.03.002 [DOI] [PubMed] [Google Scholar]
- Rampelt H., Kirstein-Miles J., Nillegoda N.B., Chi K., Scholz S.R., Morimoto R.I., and Bukau B.. 2012. Metazoan Hsp70 machines use Hsp110 to power protein disaggregation. EMBO J. 31:4221–4235. 10.1038/emboj.2012.264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripaud L., Chumakova V., Antonin M., Hastie A.R., Pinkert S., Körner R., Ruff K.M., Pappu R.V., Hornburg D., Mann M., et al. 2014. Overexpression of Q-rich prion-like proteins suppresses polyQ cytotoxicity and alters the polyQ interactome. Proc. Natl. Acad. Sci. USA. 111:18219–18224. 10.1073/pnas.1421313111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzolo K., Huen J., Kumar A., Phanse S., Vlasblom J., Kakihara Y., Zeineddine H.A., Minic Z., Snider J., Wang W., et al. 2017. Features of the Chaperone Cellular Network Revealed through Systematic Interaction Mapping. Cell Rep. 20:2735–2748. 10.1016/j.celrep.2017.08.074 [DOI] [PubMed] [Google Scholar]
- Ross C.A., and Poirier M.A.. 2004. Protein aggregation and neurodegenerative disease. Nat. Med. 10(7, Suppl):S10–S17. 10.1038/nm1066 [DOI] [PubMed] [Google Scholar]
- Roth D.M., Hutt D.M., Tong J., Bouchecareilh M., Wang N., Seeley T., Dekkers J.F., Beekman J.M., Garza D., Drew L., et al. 2014. Modulation of the maladaptive stress response to manage diseases of protein folding. PLoS Biol. 12:e1001998 10.1371/journal.pbio.1001998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouillard A.D., Gundersen G.W., Fernandez N.F., Wang Z., Monteiro C.D., McDermott M.G., and Ma’ayan A.. 2016. The harmonizome: a collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database (Oxford). 2016:baw100 10.1093/database/baw100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau A., and Bertolotti A.. 2016. An evolutionarily conserved pathway controls proteasome homeostasis. Nature. 536:184–189. 10.1038/nature18943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein D.C., Mariño G., and Kroemer G.. 2011. Autophagy and aging. Cell. 146:682–695. 10.1016/j.cell.2011.07.030 [DOI] [PubMed] [Google Scholar]
- Rüdiger S., Buchberger A., and Bukau B.. 1997. Interaction of Hsp70 chaperones with substrates. Nat. Struct. Biol. 4:342–349. 10.1038/nsb0597-342 [DOI] [PubMed] [Google Scholar]
- Russell S.J., Steger K.A., and Johnston S.A.. 1999. Subcellular localization, stoichiometry, and protein levels of 26 S proteasome subunits in yeast. J. Biol. Chem. 274:21943–21952. 10.1074/jbc.274.31.21943 [DOI] [PubMed] [Google Scholar]
- Sachidanandam R., Weissman D., Schmidt S.C., Kakol J.M., Stein L.D., Marth G., Sherry S., Mullikin J.C., Mortimore B.J., Willey D.L., et al. International SNP Map Working Group . 2001. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 409:928–933. 10.1038/35057149 [DOI] [PubMed] [Google Scholar]
- Sala A.J., Bott L.C., and Morimoto R.I.. 2017. Shaping proteostasis at the cellular, tissue, and organismal level. J. Cell Biol. 216:1231–1241. 10.1083/jcb.201612111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S., Ravikumar B., Floto R.A., and Rubinsztein D.C.. 2009. Rapamycin and mTOR-independent autophagy inducers ameliorate toxicity of polyglutamine-expanded huntingtin and related proteinopathies. Cell Death Differ. 16:46–56. 10.1038/cdd.2008.110 [DOI] [PubMed] [Google Scholar]
- Saudou F., Finkbeiner S., Devys D., and Greenberg M.E.. 1998. Huntingtin acts in the nucleus to induce apoptosis but death does not correlate with the formation of intranuclear inclusions. Cell. 95:55–66. 10.1016/S0092-8674(00)81782-1 [DOI] [PubMed] [Google Scholar]
- Scherzinger E., Sittler A., Schweiger K., Heiser V., Lurz R., Hasenbank R., Bates G.P., Lehrach H., and Wanker E.E.. 1999. Self-assembly of polyglutamine-containing huntingtin fragments into amyloid-like fibrils: implications for Huntington’s disease pathology. Proc. Natl. Acad. Sci. USA. 96:4604–4609. 10.1073/pnas.96.8.4604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz A.M., and Haynes C.M.. 2015. UPR(mt)-mediated cytoprotection and organismal aging. Biochim. Biophys. Acta. 1847:1448–1456. 10.1016/j.bbabio.2015.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma K., Vabulas R.M., Macek B., Pinkert S., Cox J., Mann M., and Hartl F.U.. 2012. Quantitative proteomics reveals that Hsp90 inhibition preferentially targets kinases and the DNA damage response. Mol. Cell. Proteomics. 11:M111.014654 10.1074/mcp.M111.014654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y., and Morimoto R.I.. 2014. How the nucleus copes with proteotoxic stress. Curr. Biol. 24:R463–R474. 10.1016/j.cub.2014.03.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sittler A., Lurz R., Lueder G., Priller J., Lehrach H., Hayer-Hartl M.K., Hartl F.U., and Wanker E.E.. 2001. Geldanamycin activates a heat shock response and inhibits huntingtin aggregation in a cell culture model of Huntington’s disease. Hum. Mol. Genet. 10:1307–1315. 10.1093/hmg/10.12.1307 [DOI] [PubMed] [Google Scholar]
- Skach W.R. 2009. Cellular mechanisms of membrane protein folding. Nat. Struct. Mol. Biol. 16:606–612. 10.1038/nsmb.1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontag E.M., Samant R.S., and Frydman J.. 2017. Mechanisms and Functions of Spatial Protein Quality Control. Annu. Rev. Biochem. 86:97–122. 10.1146/annurev-biochem-060815-014616 [DOI] [PubMed] [Google Scholar]
- Sowa M.E., Bennett E.J., Gygi S.P., and Harper J.W.. 2009. Defining the human deubiquitinating enzyme interaction landscape. Cell. 138:389–403. 10.1016/j.cell.2009.04.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale M., Krykbaeva I., Koeva M., Kayatekin C., Westover K.D., Karras G.I., and Lindquist S.. 2012. Quantitative analysis of HSP90-client interactions reveals principles of substrate recognition. Cell. 150:987–1001. 10.1016/j.cell.2012.06.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawo R., Pokrzywa W., Kevei E., Akyuz M.E., Balaji V., Adrian S., Hohfeld J., and Hoppe T.. 2017. The Ubiquitin Ligase CHIP Integrates Proteostasis and Aging by Regulation of Insulin Receptor Turnover. Cell. 169:470–482. 10.1016/j.cell.2017.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor R.C., and Dillin A.. 2011. Aging as an event of proteostasis collapse. Cold Spring Harb. Perspect. Biol. 3:a004440 10.1101/cshperspect.a004440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor R.C., Berendzen K.M., and Dillin A.. 2014. Systemic stress signalling: understanding the cell non-autonomous control of proteostasis. Nat. Rev. Mol. Cell Biol. 15:211–217. 10.1038/nrm3752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsaytler P., Harding H.P., Ron D., and Bertolotti A.. 2011. Selective inhibition of a regulatory subunit of protein phosphatase 1 restores proteostasis. Science. 332:91–94. 10.1126/science.1201396 [DOI] [PubMed] [Google Scholar]
- Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A., et al. 2015. Proteomics. Tissue-based map of the human proteome. Science. 347:1260419 10.1126/science.1260419 [DOI] [PubMed] [Google Scholar]
- Varshavsky A. 2012. The ubiquitin system, an immense realm. Annu. Rev. Biochem. 81:167–176. 10.1146/annurev-biochem-051910-094049 [DOI] [PubMed] [Google Scholar]
- Vembar S.S., and Brodsky J.L.. 2008. One step at a time: endoplasmic reticulum-associated degradation. Nat. Rev. Mol. Cell Biol. 9:944–957. 10.1038/nrm2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendruscolo M., Knowles T.P.J., and Dobson C.M.. 2011. Protein solubility and protein homeostasis: a generic view of protein misfolding disorders. Cold Spring Harb. Perspect. Biol. 3:a010454 10.1101/cshperspect.a010454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Mikecz A. 2006. The nuclear ubiquitin-proteasome system. J. Cell Sci. 119:1977–1984. 10.1242/jcs.03008 [DOI] [PubMed] [Google Scholar]
- Vos M.J., Hageman J., Carra S., and Kampinga H.H.. 2008. Structural and functional diversities between members of the human HSPB, HSPH, HSPA, and DNAJ chaperone families. Biochemistry. 47:7001–7011. 10.1021/bi800639z [DOI] [PubMed] [Google Scholar]
- Waelter S., Boeddrich A., Lurz R., Scherzinger E., Lueder G., Lehrach H., and Wanker E.E.. 2001. Accumulation of mutant huntingtin fragments in aggresome-like inclusion bodies as a result of insufficient protein degradation. Mol. Biol. Cell. 12:1393–1407. 10.1091/mbc.12.5.1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P., and Ron D.. 2011. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 334:1081–1086. 10.1126/science.1209038 [DOI] [PubMed] [Google Scholar]
- Walther D.M., Kasturi P., Zheng M., Pinkert S., Vecchi G., Ciryam P., Morimoto R.I., Dobson C.M., Vendruscolo M., Mann M., and Hartl F.U.. 2015. Widespread Proteome Remodeling and Aggregation in Aging C. elegans. Cell. 161:919–932. (Published erratum appears in Cell 2017. 168:944) 10.1016/j.cell.2015.03.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandinger S.K., Richter K., and Buchner J.. 2008. The Hsp90 chaperone machinery. J. Biol. Chem. 283:18473–18477. 10.1074/jbc.R800007200 [DOI] [PubMed] [Google Scholar]
- Wegrzyn R.D., Bapat K., Newnam G.P., Zink A.D., and Chernoff Y.O.. 2001. Mechanism of prion loss after Hsp104 inactivation in yeast. Mol. Cell. Biol. 21:4656–4669. 10.1128/MCB.21.14.4656-4669.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitesell L., Mimnaugh E.G., De Costa B., Myers C.E., and Neckers L.M.. 1994. Inhibition of heat shock protein HSP90-pp60v-src heteroprotein complex formation by benzoquinone ansamycins: essential role for stress proteins in oncogenic transformation. Proc. Natl. Acad. Sci. USA. 91:8324–8328. 10.1073/pnas.91.18.8324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D.N., and Beckmann R.. 2011. The ribosomal tunnel as a functional environment for nascent polypeptide folding and translational stalling. Curr. Opin. Struct. Biol. 21:274–282. 10.1016/j.sbi.2011.01.007 [DOI] [PubMed] [Google Scholar]
- Winkler J., Tyedmers J., Bukau B., and Mogk A.. 2012. Hsp70 targets Hsp100 chaperones to substrates for protein disaggregation and prion fragmentation. J. Cell Biol. 198:387–404. 10.1083/jcb.201201074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winner B., Jappelli R., Maji S.K., Desplats P.A., Boyer L., Aigner S., Hetzer C., Loher T., Vilar M., Campioni S., et al. 2011. In vivo demonstration that alpha-synuclein oligomers are toxic. Proc. Natl. Acad. Sci. USA. 108:4194–4199. 10.1073/pnas.1100976108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woerner A.C., Frottin F., Hornburg D., Feng L.R., Meissner F., Patra M., Tatzelt J., Mann M., Winklhofer K.F., Hartl F.U., and Hipp M.S.. 2016. Cytoplasmic protein aggregates interfere with nucleocytoplasmic transport of protein and RNA. Science. 351:173–176. 10.1126/science.aad2033 [DOI] [PubMed] [Google Scholar]
- Wolfe K.J., Ren H.Y., Trepte P., and Cyr D.M.. 2013. The Hsp70/90 cochaperone, Sti1, suppresses proteotoxicity by regulating spatial quality control of amyloid-like proteins. Mol. Biol. Cell. 24:3588–3602. 10.1091/mbc.E13-06-0315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff S., Weissman J.S., and Dillin A.. 2014. Differential scales of protein quality control. Cell. 157:52–64. 10.1016/j.cell.2014.03.007 [DOI] [PubMed] [Google Scholar]
- Yanagitani K., Juszkiewicz S., and Hegde R.S.. 2017. UBE2O is a quality control factor for orphans of multiprotein complexes. Science. 357:472–475. 10.1126/science.aan0178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu A., Shibata Y., Shah B., Calamini B., Lo D.C., and Morimoto R.I.. 2014. Protein aggregation can inhibit clathrin-mediated endocytosis by chaperone competition. Proc. Natl. Acad. Sci. USA. 111:E1481–E1490. 10.1073/pnas.1321811111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaarur N., Xu X., Lestienne P., Meriin A.B., McComb M., Costello C.E., Newnam G.P., Ganti R., Romanova N.V., Shanmugasundaram M., et al. 2015. RuvbL1 and RuvbL2 enhance aggresome formation and disaggregate amyloid fibrils. EMBO J. 34:2363–2382. 10.15252/embj.201591245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Krakowiak J., Patel N., Beyzavi A., Ezike J., Khalil A.S., and Pincus D.. 2016. Dynamic control of Hsf1 during heat shock by a chaperone switch and phosphorylation. Elife. 5:e18638 10.7554/eLife.18638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J., Guo Y., Guettouche T., Smith D.F., and Voellmy R.. 1998. Repression of heat shock transcription factor HSF1 activation by HSP90 (HSP90 complex) that forms a stress-sensitive complex with HSF1. Cell. 94:471–480. 10.1016/S0092-8674(00)81588-3 [DOI] [PubMed] [Google Scholar]
- Żwirowski S., Kłosowska A., Obuchowski I., Nillegoda N.B., Piróg A., Ziętkiewicz S., Bukau B., Mogk A., and Liberek K.. 2017. Hsp70 displaces small heat shock proteins from aggregates to initiate protein refolding. EMBO J. 36:783–796. 10.15252/embj.201593378 [DOI] [PMC free article] [PubMed] [Google Scholar]