Actin polymerization through the ER-bound formin INF2 stimulates mitochondrial division by promoting Drp1 recruitment. Chakrabarti et al. extend this work to show that INF2-mediated actin polymerization stimulates a second mitochondrial response independent of Drp1: increased ER–mitochondrial calcium transfer via MCU and inner mitochondrial membrane constriction, which requires electron transport chain activity.
Abstract
Mitochondrial division requires division of both the inner and outer mitochondrial membranes (IMM and OMM, respectively). Interaction with endoplasmic reticulum (ER) promotes OMM division by recruitment of the dynamin Drp1, but effects on IMM division are not well characterized. We previously showed that actin polymerization through ER-bound inverted formin 2 (INF2) stimulates Drp1 recruitment in mammalian cells. Here, we show that INF2-mediated actin polymerization stimulates a second mitochondrial response independent of Drp1: a rise in mitochondrial matrix calcium through the mitochondrial calcium uniporter. ER stores supply the increased mitochondrial calcium, and the role of actin is to increase ER–mitochondria contact. Myosin IIA is also required for this mitochondrial calcium increase. Elevated mitochondrial calcium in turn activates IMM constriction in a Drp1-independent manner. IMM constriction requires electron transport chain activity. IMM division precedes OMM division. These results demonstrate that actin polymerization independently stimulates the dynamics of both membranes during mitochondrial division: IMM through increased matrix calcium, and OMM through Drp1 recruitment.
Introduction
Contacts between the ER and mitochondrion are now widely appreciated as important for communication in several respects, including lipid synthesis and calcium transfer (Phillips and Voeltz, 2016). For calcium transfer, close ER–mitochondrial contacts (<30 nm) enable a disproportionate amount of stimulus-induced calcium release from the ER to be taken up by mitochondria instead of being released into the cytosol (Rizzuto et al., 1998; Csordás et al., 2006, 2010; Giacomello et al., 2010). This uptake is mediated by the mitochondrial calcium uniporter (MCU; Baughman et al., 2011; De Stefani et al., 2011). Moderate increases in mitochondrial calcium stimulate oxidative catabolism through activation of several dehydrogenases (Denton, 2009), whereas excessive mitochondrial calcium can trigger apoptosis (Baffy et al., 1993). Although several proteins have been shown to mediate ER–mitochondrial contacts (Phillips and Voeltz, 2016), mechanisms controlling these contacts during cell stimulation are unclear.
ER–mitochondrial contact also stimulates mitochondrial division (Friedman et al., 2011), which is required for diverse aspects of normal cellular physiology, including proper distribution of mitochondrial genomes (Lewis et al., 2016), metabolic adaptation (Mishra and Chan, 2016), mitophagy (Youle and van der Bliek, 2012; Burman et al., 2017), and immune response (Pernas and Scorrano, 2016). Defects in mitochondrial division have been linked to multiple pathologies, particularly neurodegenerative diseases (Nunnari and Suomalainen, 2012). Most mechanistic focus has been on outer mitochondrial membrane (OMM) division, with the dynamin GTPase Drp1 being a key factor (Labrousse et al., 1999; Labbé et al., 2014). Drp1 oligomerizes into a ring encircling the OMM, and GTP hydrolysis by Drp1 drives OMM constriction, leading to division. We have shown that one effector of ER-stimulated mitochondrial division in mammals is ER-bound inverted formin 2 (INF2), with INF2-mediated actin polymerization playing a key role in Drp1 recruitment to and oligomerization at division sites (Korobova et al., 2013; Ji et al., 2015). INF2 is linked to two human diseases: the neuropathy Charcot–Marie–Tooth disease (Boyer et al., 2011) and the kidney disease focal segmental glomerulosclerosis (Brown et al., 2010). Myosin II is also required for this process (DuBoff et al., 2012; Korobova et al., 2014), as well as the mitochondrially bound actin polymerization factor Spire1C (Manor et al., 2015). Other mechanisms of actin polymerization might also stimulate mitochondrial division (Li et al., 2015; Moore et al., 2016). Recent work shows that dynamin 2 accumulates on the OMM subsequent to Drp1 and acts at later stages of mitochondrial division (Lee et al., 2016).
Comparatively little is known about inner mitochondrial membrane (IMM) division, which must also occur for successful mitochondrial division. Early results in Caenorhabditis elegans showed that Drp1-deficient animals had an overall mitochondrial division defect but that matrix markers segregated (Labrousse et al., 1999). Drp1-independent mitochondrial constrictions have been observed (Lee and Yoon, 2014), and these constrictions appear to be through direct effects on the IMM (Cho et al., 2017). The constrictions occur at ER–mitochondrial contact sites, depend on increased intramitochondrial calcium, and precede full mitochondrial division (Cho et al., 2017).
In this paper, we show that INF2-mediated actin polymerization on the ER is necessary for mitochondrial calcium increase upon stimulation with either histamine or ionomycin. This calcium increase requires MCU, which is also required for stimulus-induced IMM contractions and mitochondrial division. INF2-mediated actin polymerization stimulates ER-to-mitochondrial calcium transfer by enhancing close ER–mitochondrial contact. During stimulus-induced mitochondrial division, the IMM divides before OMM division.
Results
Stimulus-induced actin polymerization enhances mitochondrial Ca2+
Studies from our laboratory and others show that a variety of stimuli causing increased cytosolic calcium, including ionomycin and histamine, trigger a transient cytosolic actin polymerization “burst” (Ji et al., 2015; Shao et al., 2015; Wales et al., 2016). These stimuli operate by distinct mechanisms to raise cytosolic calcium, with ionomycin requiring extracellular calcium and histamine relying only on calcium from intracellular stores (Fig. S1 A). We previously showed that the ionomycin-induced actin burst stimulates Drp1 oligomerization and subsequent mitochondrial division (Ji et al., 2015). Histamine stimulation also induces Drp1 oligomerization and mitochondrial division, albeit to lower levels than ionomycin (Fig. S1, B–D).
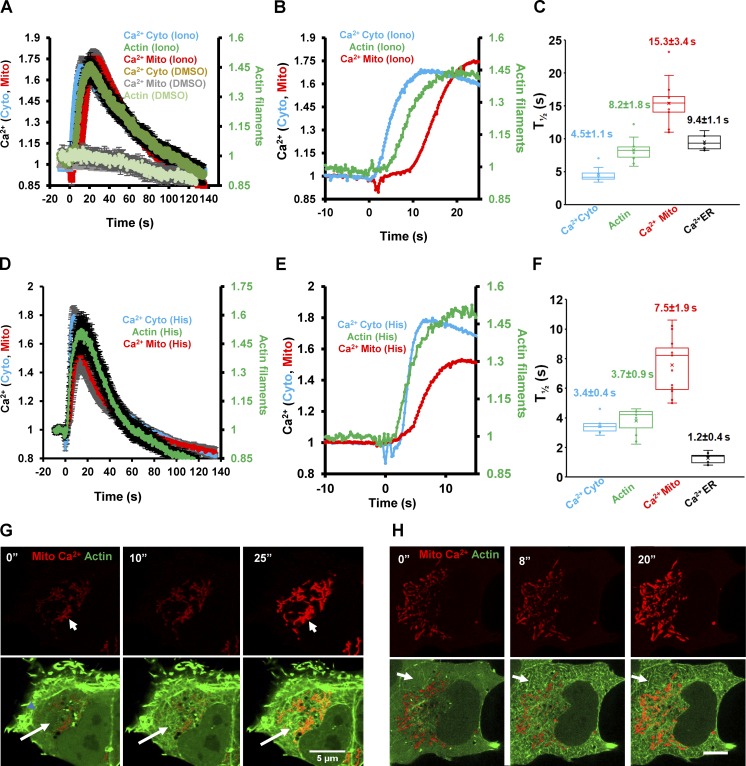
Because both ionomycin and histamine also induce an increase in mitochondrial matrix calcium (Rizzuto et al., 1993; Abramov and Duchen, 2003), we investigated the relative kinetics of changes in cytoplasmic calcium and mitochondrial calcium in relation to the actin burst using live-cell imaging. Both ionomycin and histamine trigger rapid increases in cytoplasmic calcium, with a t1/2 of 4.5 ± 1.1 and 3.4 ± 0.4 s, respectively (Fig. 1, A–F; and Videos 1 and 2). The actin burst lags significantly upon ionomycin stimulation (8.3 ± 1.8 s), but its histamine-stimulated rate is indistinguishable from that of cytoplasmic calcium (3.8 ± 0.9 s; Fig. 1, A–F; and Videos 1 and 2). The mitochondrial calcium increase occurs after the actin burst for both stimuli, with a t1/2 of 15.4 ± 3.4 for ionomycin and 7.5 ± 1.9 s for histamine (Fig. 1, A–F; and Videos 1 and 2). This result is confirmed by two-color imaging in the same cell for both ionomycin (Fig. 1 G and Video 3) and histamine (Fig. 1 H and Video 4). All responses are transient, returning to near baseline within 200 s for both stimuli. Further information on ionomycin stimulation is provided in Materials and methods and Fig. S5 (D and E).
Figure 1.
The stimulus-induced actin burst precedes the mitochondrial calcium spike. (A and D) Quantification of cytoplasmic calcium (Cyto-R-GECO), cytoplasmic actin (GFP-Ftractin), or mitochondrial calcium (Mito-R-GECO) in U2OS cells after 4 µM ionomycin (A) or 100 µM histamine (D) stimulation. Rapid acquisition mode (4.8 frames/s). Values on the y axis represent the value at time X normalized to the value at time 0 (F/F0); n = 10–16 cells (also given in C). Error bars represent SEM. Corresponds to Videos 1 and 2. (B and E) Zoom of early time points from graphs in A (ionomycin) and D (histamine) showing the distinct kinetics of cytoplasmic calcium, actin, and mitochondrial calcium. Error bars have been removed for clarity. (C and F) Box-and-whiskers plots of stimulation half-times based on the curves in A and D. Number of cells for each reading is 14 (cytoplasmic calcium, ionomycin), 10 (actin, ionomycin), 14 (mitochondrial calcium, ionomycin), 12 (ER calcium, ionomycin), 14 (cytoplasmic calcium, histamine), 11 (actin, histamine), 16 (mitochondrial calcium, histamine), and 10 (ER calcium, histamine). Each point represents one cell. Error bars represent SD (G and H) Time-lapse montages of actin (green, GFP-Ftractin) and mitochondrial calcium (red, mito-R-GECO) changes in the same cell after 4 µM ionomycin (G) or 100 µM histamine (H) stimulation. Mitochondrial calcium alone shown in the top panels and merge in the bottom panels; taken at one frame per 1.03 s (G) and 1.2 s (H). Arrows in the merged panel indicate regions of the cytoplasm displaying the cytoplasmic actin burst. Bars: (G) 5 µm; (H) 10 µm. Corresponds to Videos 3 and 4.
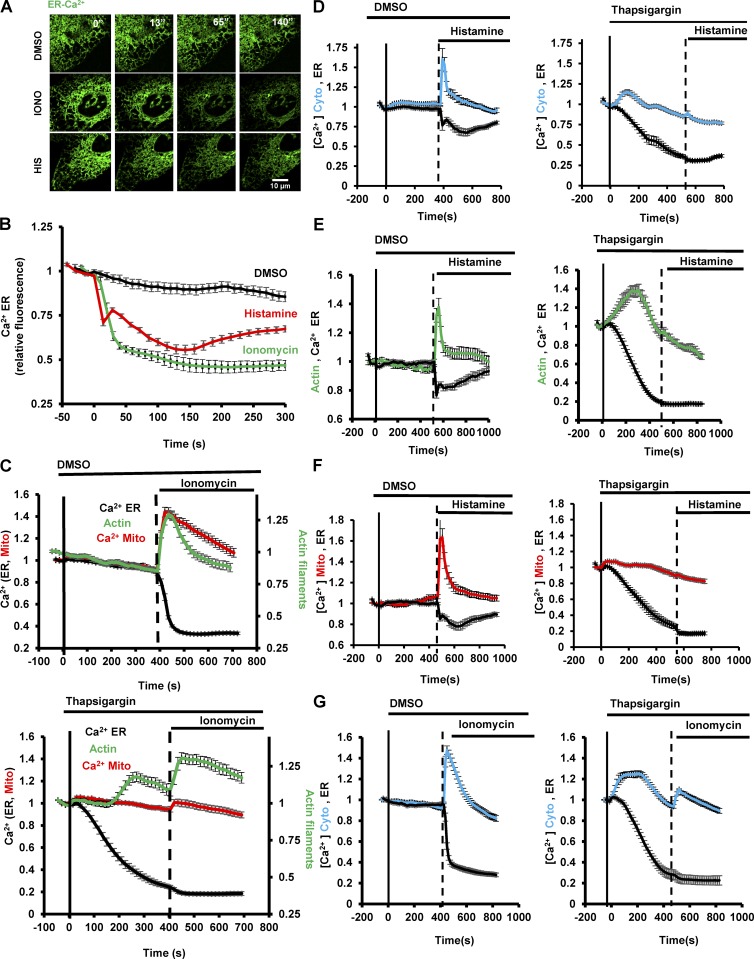
We probed in more detail the calcium source for the mitochondrial calcium spike. Because mitochondrial calcium responses often occur at sites of close ER contact (De Stefani et al., 2016), we directly measured changes in ER calcium upon ionomycin or histamine stimulation using a low-affinity calcium probe for ER, ER-GCaMP6-150 (150 µM Kd for Ca2+; de Juan-Sanz et al., 2017). As shown previously (Montero et al., 1997, 2003), histamine induces a rapid decrease in ER calcium that occurs in two phases (Fig. 2, A and B; and Video 5). The first phase precedes both the actin burst and the overall cytosolic calcium increase (t1/2 1.27 ± 0.36 s; Fig. 1 F), whereas the second phase occurs 20–60 s after stimulation. Interestingly, ionomycin also induces a rapid decrease in ER calcium (Fig. 2, A and B; and Video 5) with a t1/2 of 9.6 ± 1.1 s (Fig. 1 C). This result had been suggested previously, but not directly measured (Morgan and Jacob, 1994; Caridha et al., 2008), and implies that a significant component of the ionomycin-induced cytoplasmic calcium increase derives from intracellular stores, likely through calcium-mediated calcium release (Endo, 2009).
Figure 2.
Calcium release from the ER triggered by ionomycin and histamine. (A) Time-lapse montages of U2OS cells transfected with the ER calcium probe (ER-GCaMP6-150) and treated with DMSO (top), 4 µM ionomycin (middle), or 100 µM histamine (bottom). Time in seconds. Bar, 10 µm. Corresponds to Video 5. (B) Graph quantifying changes in ER calcium upon the treatments described in A; n = 12, 15, and 15 cells for DMSO, ionomycin, and histamine, respectively. Error bars represent SEM. (C) Effect of thapsigargin on ER calcium (black curves), actin burst (green curves), and mitochondrial calcium (red curves) before and after ionomycin stimulation. U2OS cells transfected with ER calcium probe and mApple-Ftractin. DMSO (top graph) or 1 µM thapsigargin (bottom graph) was applied at 0 s (bold line) and 4 µM ionomycin was applied at the time indicated by the dashed line; n = 10 cells (DMSO) or 13 cells (thapsigargin). Error bars represent SEM. (D) Effect of thapsigargin on histamine-induced cytoplasmic calcium and ER calcium changes. U2OS cells transfected with ER calcium probe and Cyto-R-GECO; n = 15 cells (DMSO) or 15 cells (thapsigargin). (E) Effect of thapsigargin on histamine-induced actin burst and ER calcium. U2OS cells were transfected with ER calcium probe and mApple-Ftractin; n = 10 cells (DMSO) or 11 cells (thapsigargin). Error bars represent SEM. (F) Effect of thapsigargin on histamine-induced mitochondrial calcium and ER calcium. U2OS cells transfected with ER calcium probe and mito-R-GECO; n = 12 cells (DMSO) or 14 cells (Thapsigargin). Error bars represent SEM. (G) Effect of thapsigargin on ionomycin-induced cytoplasmic calcium and ER calcium. U2OS cells transfected with both the Cyto-R-Geco and ER-GCaMP6-150; n = 17 cells (DMSO) or 16 cells (thapsigargin). Error bars represent SEM.
We used thapsigargin pretreatment to test whether ER calcium release was necessary for the mitochondrial calcium increase upon either histamine or ionomycin stimulation. Thapsigargin treatment depletes ER calcium significantly within 10 min (Fig. 2 C). The resulting rise in cytoplasmic calcium is sufficient to activate actin polymerization, but this activation is significantly slower than the acute activation caused by ionomycin or histamine (Fig. 2 C), with a t1/2 of >200 s. No mitochondrial calcium increase occurs upon thapsigargin stimulation. For histamine, stimulation after thapsigargin-induced depletion of ER calcium results in no increase in cytoplasmic calcium or mitochondrial calcium, similar to past experiments (Diarra and Sauvé, 1992), and histamine does not induce an actin burst (Fig. 2, D–F). For ionomycin stimulation, thapsigargin pretreatment reduces the increases in cytoplasmic calcium and actin polymerization, and the increase in mitochondrial calcium is eliminated (Fig. 2, C and G). These results suggest that the mitochondrial calcium increase results largely from ER calcium for both histamine and ionomycin stimulation.
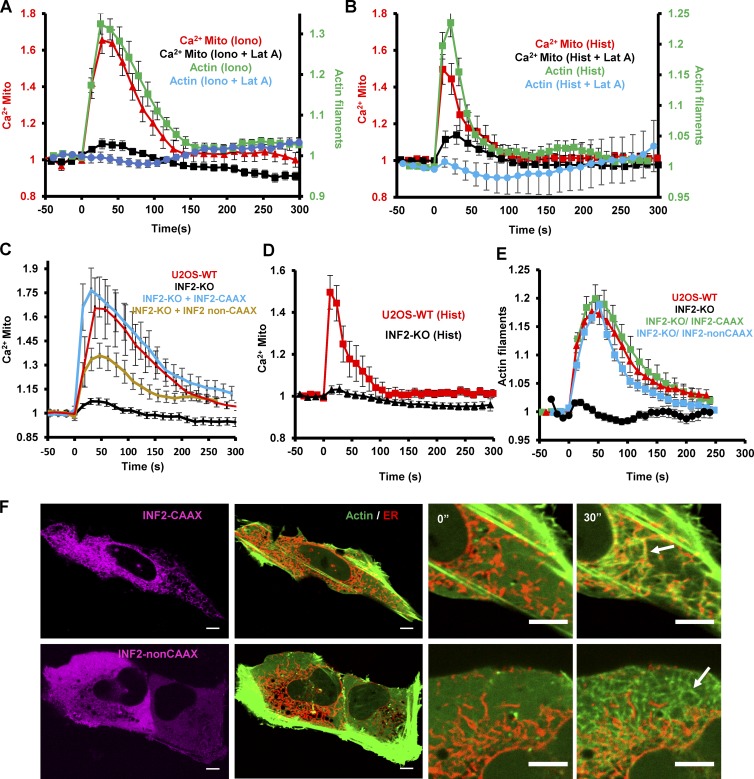
Because the actin burst precedes the mitochondrial calcium spike, we asked whether actin polymerization was necessary for increased mitochondrial calcium. Treatment with the actin sequestering molecule latrunculin A (LatA) strongly inhibits the mitochondrial calcium spike upon either ionomycin or histamine stimulation (Fig. 3, A and B), whereas increasing the magnitude of the cytosolic calcium increase (Fig. S2 A). These results suggest that actin polymerization is required for efficient stimulus-induced mitochondrial calcium entry.
Figure 3.
Actin requirement for stimulus-induced mitochondrial calcium spike. (A and B) Effect of 2 μM LatA treatment on ionomycin-induced (A) or histamine-induced (B) actin polymerization burst and mitochondrial calcium spike in U2OS cells; n = 10–15 cells. Error bars represent SEM. (C and D) Mitochondrial calcium spike after ionomycin (C) or histamine (D) stimulus in U2OS-WT, U2OS-INF2-KO, or INF2-KO cells reexpressing GFP-INF2-CAAX or GFP-INF2-nonCAAX. Control U2OS-INF2-KO cells are expressing GFP-Sec61β as an ER marker instead of INF2. Error bars represent SEM. (E) Actin polymerization burst after ionomycin stimulation in U2OS-WT and U2OS-INF2-KO cells. Rescue by transfection of GFP-INF2-CAAX or GFP-INF2-nonCAAX is also shown; n = 15–18 cells. Error bars represent SEM. (F) Ionomycin-induced actin morphology in INF2-KO cells reexpressing GFP-INF2-CAAX or GFP-INF2-nonCAAX. Whole-cell view shown prestimulation. Insets shown prestimulation (0”, left) and after 30 s stimulation (30”, right). Taken in a single confocal plane in an apical region of the cell body, which reduces actin background from stress fibers but causes the ER to appear fragmented. Arrows show examples of actin filament accumulation. Bars: (main) 5 µm; (inset) 2 µm. Corresponds to Video 6.
Actin polymerization on the ER enhances ER-to-mitochondrial Ca2+ transfer
We previously showed that an ER-bound isoform of the formin INF2, INF2-CAAX, plays a role in mitochondrial division (Korobova et al., 2013) and that siRNA-mediated INF2 suppression eliminates the ionomycin-induced actin burst (Ji et al., 2015). We therefore asked whether INF2-CAAX plays a role in the mitochondrial calcium spike. CRISPR-mediated knockout (KO) of INF2 in U2OS cells (Fig. S2, B and C) eliminates both the actin burst and the mitochondrial calcium spike upon either ionomycin or histamine stimulation (Fig. 3, C and D; and Fig. S2 D) while causing an increase in cytoplasmic calcium (Fig. S2 A). Reexpression of INF2-CAAX or INF2-nonCAAX as GFP fusions restores the actin burst (Fig. 3 E), but the resulting actin morphology differs between the two isoforms. INF2-CAAX–induced actin enriches around the ER, whereas INF2-nonCAAX–induced actin is not ER enriched (Fig. 3 F and Video 6). Interestingly, INF2-CAAX restores the mitochondrial calcium spike to a greater degree than INF2-nonCAAX (Figs. 3 C and S2 E), suggesting that ER localization is important.
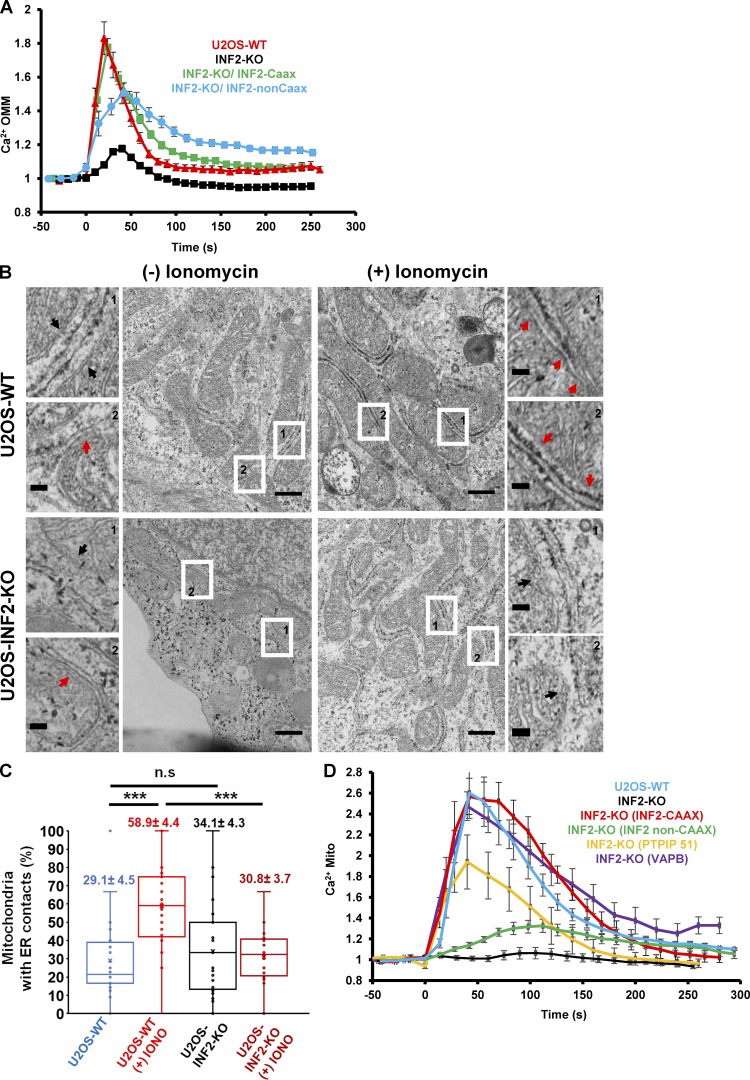
These results suggest that INF2-mediated actin polymerization enhances ER-to-mitochondrial calcium transfer. One mechanism to mediate this enhancement is by promoting close contact between ER and mitochondria. Evidence for ER–mitochondrial contact has been obtained previously using low-affinity calcium sensors tethered to the cytoplasmic face of the OMM, showing that increases in ER–mitochondrial contact cause increases in calcium concentration near the OMM that are in excess of the bulk cytoplasmic calcium concentration (Csordás et al., 2010; Giacomello et al., 2010). We used a similar low-affinity calcium probe (LAR-GECO-1.2, Kd of 12 µM for calcium; Wu et al., 2014) tethered to the cytoplasmic face of the OMM through MAS70. Stimulation of WT U2OS cells by ionomycin causes an increase in the OMM-calcium signal in a time course similar to the mitochondrial calcium increase, and this OMM-calcium increase is greatly attenuated in INF2-KO cells (Fig. 4 A). Re-expression of INF2-CAAX rescues the ionomycin-stimulated OMM-calcium increase fully, whereas reexpression of INF2-nonCAAX causes only partial rescue (Fig. 4 A).
Figure 4.
INF2-mediated actin polymerization is required for stimulation of ER–mitochondrial contact. (A) Change in calcium levels at the cytoplasmic face of the OMM upon 4 µM ionomycin stimulation in U2OS-WT and INF2-KO cells. Cells were transfected with the Mass70-LAR-GECO1.2 construct, a low-affinity calcium probe (Kd of 12 µM) tethered to the cytoplasmic face of the OMM. For rescue, INF2-KO cells were transfected with plasmid expressing GFP-INF2-CAAX or GFP-INF2-nonCAAX. Ionomycin was added at 0 s in n = 24 cells (WT), 19 cells (INF2-KO), 22 cells (INF2-CAAX), and 16 cells (INF2-non CAAX). Error bars represent SEM. (B) Electron micrographs showing examples of the relationship between the ER and mitochondria in WT cells (top images) and INF2-KO cells (bottom images) in either the unstimulated condition [(−) ionomycin, left images] or after 60 s stimulation with 4 µM ionomycin [(+) ionomycin, right images]. Two zooms shown for each panel (regions indicated by numbered arrows on the main panels). Examples of ER–mitochondrial contacts of <30 nm are indicated by red arrows, whereas examples of more distant contacts indicated by black arrows. Bars: (main micrographs) 500 nm; (zooms) 100 nm. (C) Quantification from electron micrographs of the percentage of mitochondria with close ER contacts in WT cells (n = 244 mitochondria), WT cells stimulated with ionomycin (n = 176 mitochondria), INF2-KO cells (n = 245 mitochondria), and INF2-KO cells stimulated with ionomycin (n = 204 mitochondria). Values represent mean ± SEM; ***, P < 0.001 (unpaired Student’s t test). Each point represents one imaged field. (D) Rescue of mitochondrial calcium response in INF2-KO cells by overexpression of ER–mitochondrial tethers. INF2-KO U2OS cells were transfected with a mitochondrial matrix calcium probe (Mito-R-GECO) along with either CFP-VAPB or GFP-PTPIP51 and then stimulated with 4 µM ionomycin. The effects of either GFP-INF2-CAAX or GFP-INF2-nonCAAX reexpression are shown for comparison; n = 10 cells (WT), 8 cells (INF2-KO), 11 cells (INF2 non-CAAX), 15 cells (INF2-CAAX), 10 cells (PTPIP51), or 14 cells (VAPB). Error bars represent SEM.
We also measured changes in ER–mitochondrial contact directly by thin-section EM, defining regions of <30 nm distance as ER–mitochondrial contact sites (Csordás et al., 2006). Before ionomycin stimulation, WT and INF2-KO cells contained a similar percentage of mitochondria with close ER contacts (Fig. 4, B and C). Upon ionomycin stimulation, the percentage of mitochondria with close ER contacts increased twofold in WT cells but did not increase in INF2-KO cells (Fig. 4, B and C). These results suggest that INF2-mediated actin polymerization increases ER–mitochondrial contact.
We next asked whether overexpression of ER–mitochondrial tethering proteins could compensate for the lack of ionomycin-induced mitochondrial calcium in INF2-KO cells. For this purpose, we chose VAPB (on the ER) and PTPIP51 (on mitochondria), because overexpression of either of these tether partners individually has been shown to increase ER–mitochondrial tethering (Gomez-Suaga et al., 2017). Indeed, expression of either protein significantly restores the ionomycin-induced mitochondrial calcium spike in INF2 KO cells (Figs. 4 D and S2 E). Collectively, these results suggest that actin polymerization by INF2-CAAX enhances ER-mitochondrial contact, leading to increased stimulus-induced mitochondrial calcium.
We have also reported that myosin IIA suppression by siRNA reduces ionomycin-triggered mitochondrial division while not affecting the actin burst (Ji et al., 2015). MyoIIA knockdown (KD) also attenuates the mitochondrial calcium spike for both ionomycin and histamine stimulation while having similar levels of actin burst in both cases (Fig. S3, A–C). Collectively, these results suggest that both INF2-mediated actin polymerization and myosin IIA activity are necessary for stimulus-induced mitochondrial calcium entry.
Role for mitochondrial calcium in mitochondrial division
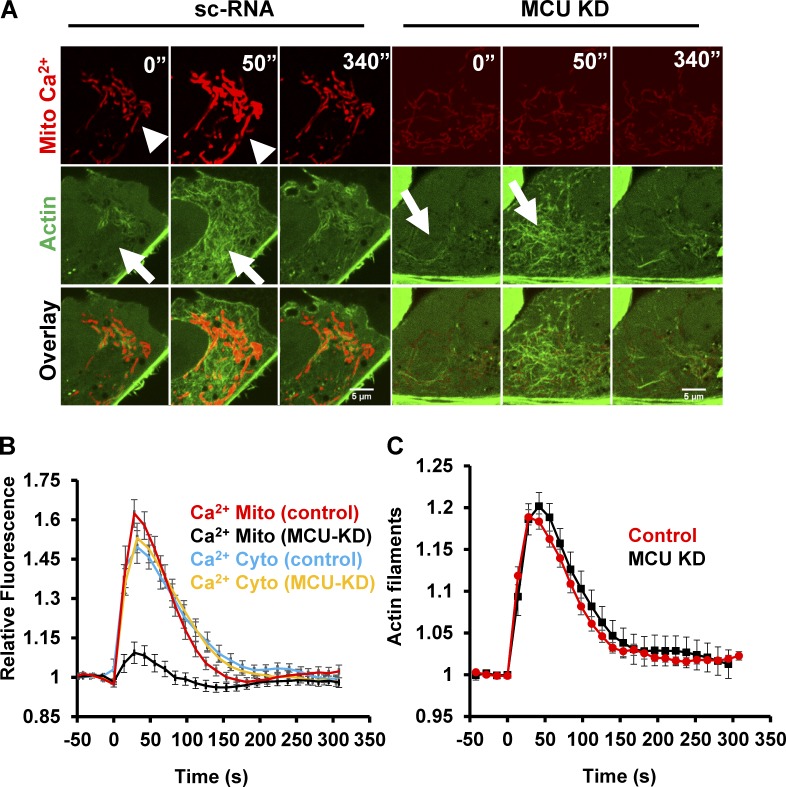
We next asked whether the mitochondrial calcium spike plays a role in mitochondrial division. MCU is the major IMM channel for mitochondrial calcium and accounts for most histamine-induced mitochondrial calcium entry (Baughman et al., 2011; De Stefani et al., 2011). In U2OS cells, MCU-targeted siRNA (MCU-KD; Fig. S3 D) eliminates the ionomycin-induced mitochondrial calcium spike without affecting the cytosolic calcium spike or the magnitude and duration of the actin burst (Fig. 5). MCU-KD also decreases baseline fluorescence of mito-R-GECO by 2.5-fold (Fig. S3, G and H), suggesting a decrease in basal mitochondrial calcium. Re-expression of an siRNA-resistant GFP-MCU construct rescues the ionomycin-induced mitochondrial calcium spike (Fig. S3, F and G).
Figure 5.
MCU suppression inhibits the mitochondrial calcium spike without inhibiting the actin burst. U2OS cells were transfected with either scrambled siRNA (control) or siRNA against MCU (MCU KD). After 48 h, cells were transfected with mito-R-GECO (mitochondrial calcium) and GFP-Ftractin (polymerized actin) or cyto-R-GECO (cytoplasmic calcium) and imaged at 72 h after siRNA treatment. (A) Confocal image montages of cells at indicated times after 4 µM ionomycin stimulation. For MCU KD, the mitochondrial calcium signal is enhanced to reveal the faint mitochondrial outline. Arrowheads show mitochondrial calcium rise, and arrows denote polymerized actin increases. Bars, 5 µm. (B) Quantification of the mitochondrial and cytosolic calcium spikes upon stimulation with 4 µM ionomycin (at time 0); n = 10 cells (Mito calcium control), 18 cells (Mito calcium MCU KD), 10 cells (Cyto calcium control), or 15 cells (Cyto calcium MCU KD). Error bars represent SEM. (C) Quantification of the actin burst upon stimulation with 4 µM ionomycin (at time 0); n = 10 cells (control) or 15 cells (MCU KD). Error bars represent SEM.
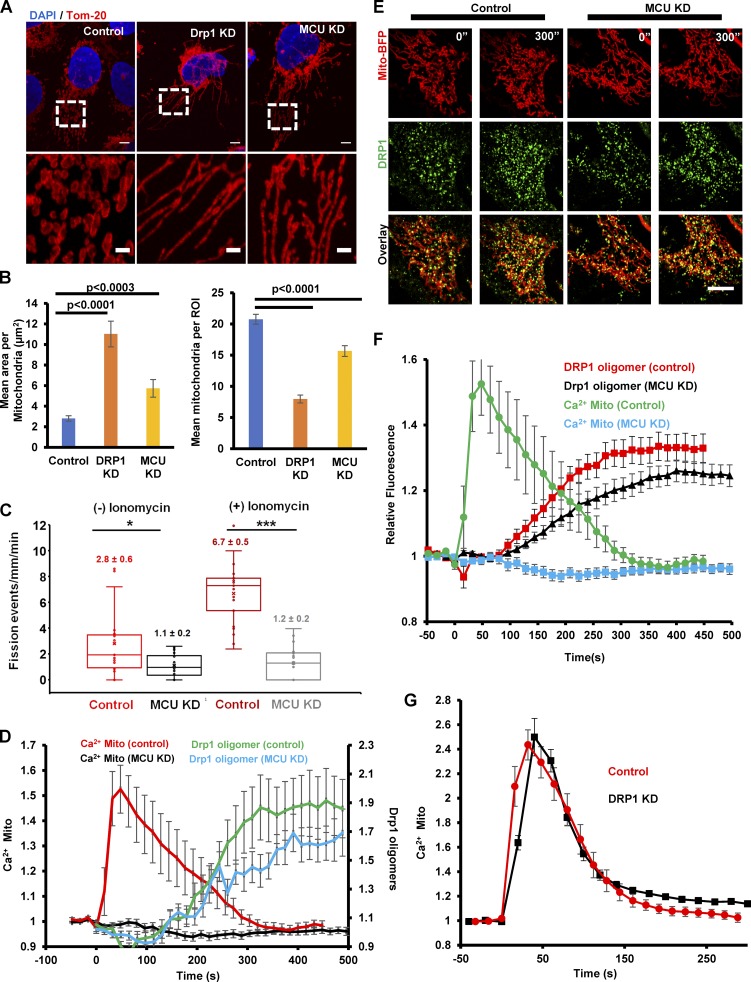
We used MCU-KD cells to ask whether mitochondrial calcium plays a role in mitochondrial division. Using a recently established morphometric assay (Lee et al., 2016), MCU-KD causes an increase in the mean area of individual mitochondria that is intermediate between control cells and Drp1-KD cells (Fig. 6, A and B). Given the possibility that the increase in length could be caused by either decreased division or increased fusion, we also used a live-cell assay to measure mitochondrial division rate directly. In unstimulated cells, MCU-KD causes a 2.5-fold decrease in the division rate (Fig. 6 C). Ionomycin stimulation induced a threefold increase in mitochondrial division rate for WT cells but no significant increase in MCU KD cells (Fig. 6 C). These results suggest that mitochondrial calcium entry plays a role in mitochondrial division, particularly in division stimulated by elevated calcium, and agrees with recent findings in neurons (Cho et al., 2017).
Figure 6.
MCU suppression inhibits mitochondrial division, but not Drp1 oligomerization. (A) U2OS cells were transfected with scrambled siRNA (control), MCU siRNA (MCU KD), and Drp1 siRNA (Drp1 KD) for 72 h Cells were then fixed and mitochondria stained using anti-Tom20 (red) and DAPI (blue). ROIs of fixed dimension were analyzed for mitochondrial length and number as described in Materials and methods. Images of control (left), Drp1 KD (middle), or MCU KD cells (right) are shown. Bars: (main) 5 µm; (insets) 2 µm. (B) Mean mitochondrial length quantification represented as area (square micrometers) per mitochondrion (left) and mean mitochondrial number quantification (right) for 80, 53, and 50 cells for control, MCU KD, and Drp1 KD cells, respectively. Errors bars represent SEM. P-values were obtained from an unpaired Student’s t test. (C) Quantification of mitochondrial division rate in control and MCU KD U2OS cells by measuring the number of division events in peripheral ROIs from live-cell videos of mitochondrial matrix marker (mito-BFP), either in unstimulated cells or cells in the first 10 min of 4 µM ionomycin stimulation; n = 19 cells [control (−) ionomycin], 21 cells [control (+) ionomycin], 20 cells [MCU KD (−) ionomycin], or 22 cells [MCU KD (+) ionomycin]. Each point represents one ROI per cell. P-values were obtained from an unpaired Student’s t test. *, P < 0.015; ***, P < 0.0001. Error bars represent SD. (D) Quantification of mitochondrially associated Drp1 oligomers and mitochondrial calcium in response to 4 µM ionomycin. GFP-Drp1 knock-in cells were treated with scrambled siRNA or MCU siRNA for 48 h and then transfected with mito-R-GECO and mito-BFP. Cells were imaged at 72 h after siRNA treatment; n = 20 cells in each case. Ionomycin addition at 0 s. Error bars represent SEM. (E) Micrographs from GFP-Drp1 knock-in U2OS cells (control and MCU siRNA treated) transfected with mito-BFP (red) and treated with 4 µM ionomycin as in B. Prestimulation (0”) and 300-s stimulations are shown. Bar, 10 µm. (F) Drp1 oligomerization kinetics in MCU KD cells, as measured in the whole cell. GFP-Drp1 knock-in U2OS cells were treated with control siRNA or MCU siRNA for 72 h and then stimulated with 4 µM ionomycin while acquiring GFP images. The amount of oligomerized Drp1 was assessed as GFP signal above the cytoplasmic background signal, as described in Materials and methods; n = 20 cells in each case. Error bars represent SEM. (G) Quantification of mitochondrial calcium increase in control or Drp1 KD U2OS cells upon 4 µM ionomycin stimulation; n = 14 cells in each case. Error bars represent SEM.
Because Drp1 is a key mitochondrial division factor, and because we previously showed that ionomycin causes increased Drp1 oligomerization (Ji et al., 2015), we tested the effect of MCU-KD on Drp1 oligomerization. In a CRISPR-derived GFP-Drp1 knock-in cell line (Ji et al., 2017), MCU suppression does not significantly alter ionomycin-induced Drp1 oligomerization on mitochondria (Fig. 6, D and E). Because Drp1 can oligomerize on peroxisomes and ER independently of mitochondria (Ji et al., 2017), we also assessed total cellular Drp1 oligomers and found similar ionomycin-induced oligomerization kinetics between control and MCU KD cells (Fig. 6 F). In addition, Drp1 suppression does not alter the ionomycin-induced mitochondrial calcium increase (Fig. 6 G). These results suggest that the role of mitochondrial calcium in mitochondrial division is not related to Drp1 recruitment.
Interestingly, siRNA of either Drp1 or MCU causes increased expression of the other protein, with MCU KD causing a twofold increase in Drp1 and Drp1 KD causing a twofold increase in MCU (Fig. S3, D and E). Furthermore, siRNA of INF2 causes approximately twofold increases in both Drp1 and MCU (Fig. S3, D and E). These results may suggest compensatory changes to maintain mitochondrial division upon depletion of key components.
Drp1-independent IMM constriction
Mitochondria in unstimulated U2OS cells periodically constrict simultaneously along their length (Fig. S4 A), as reported previously for other cell types (Lee and Yoon, 2014; Cho et al., 2017). Only rarely do these constrictions undergo division. Interestingly, constrictions accompany a transient rise in matrix calcium (Fig. S4 B). The distance between constrictions is ∼2 µm (Fig. S4 C).
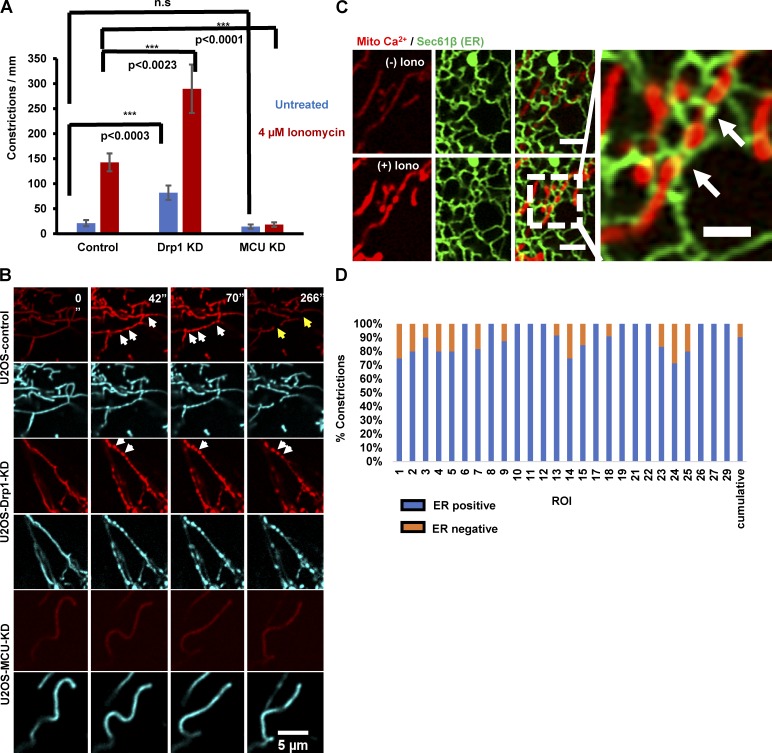
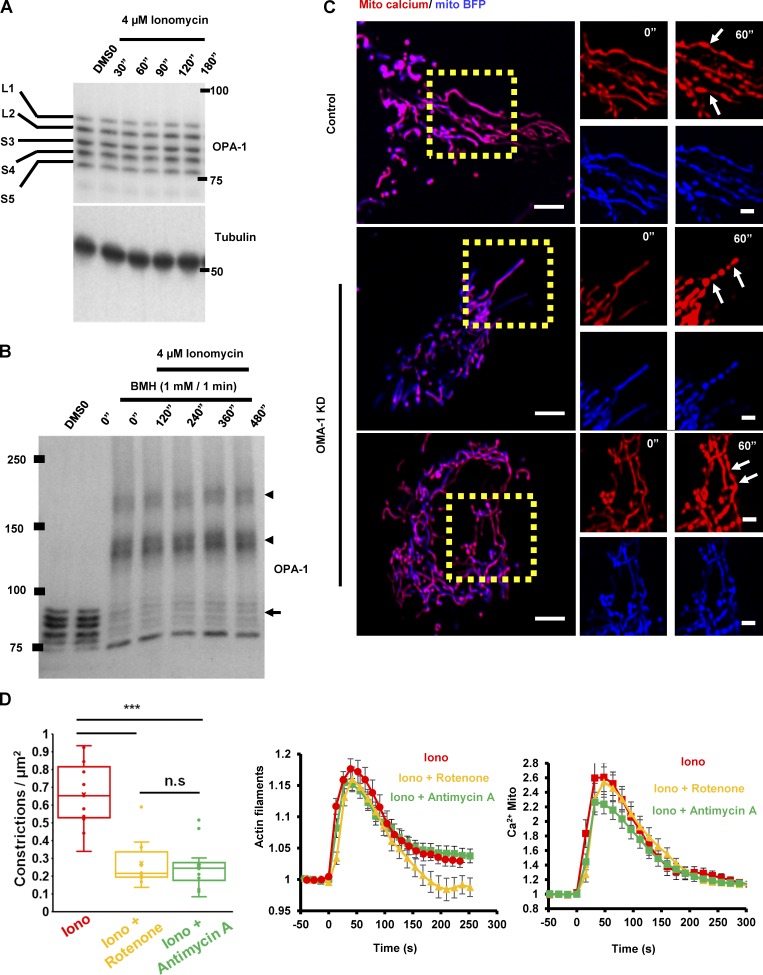
Ionomycin causes a 6.7-fold increase in mitochondrial constrictions, on a time scale similar to the mitochondrial calcium increase (Fig. 7, A and B; and Video 7). By time-lapse Airyscan microscopy, 90.4 ± 1.9% of the constrictions correspond to sites of close ER association (Fig. 7, C and D; and Video 8). We used siRNA suppression to ask whether constrictions were caused by Drp1-induced OMM constriction. Drp1-KD causes an increase in mitochondrial constrictions for both the unstimulated and ionomycin-stimulated conditions (Fig. 7, A and B; and Video 7), similar to a recent study showing enhancement of constrictions by dominant-negative Drp1 in neurons (Cho et al., 2017). These constrictions can be so exaggerated that aberrant bulges appear at adjacent regions. We next used MCU-KD cells to test whether increased mitochondrial calcium was necessary for these constrictions. MCU-KD results in a significantly reduced constriction number for both the unstimulated and ionomycin-stimulated conditions (Fig. 7, A and B; and Video 7). These findings suggest that the mitochondrial constrictions result from mitochondrial calcium influx and not Drp1 activity, as has recently been found in neurons (Cho et al., 2017).
Figure 7.
Mitochondrial constrictions are MCU dependent and Drp1 independent. (A) Quantification of mitochondrial constriction frequency in the unstimulated (blue) or ionomycin-stimulated (red) conditions, represented as constrictions per mm mitochondrial length in U2OS-control, Drp1-KD, and MCU-KD cells. P-values were obtained from an unpaired Student’s t test. Error bars represent SEM. Control:, 16 ROI, 2,508 µm mitochondrial length; Drp1 KD, 10 ROI, 1,000 µm; MCU KD: 20 ROI, 2,600 µm. (B) Representative confocal montages of control, Drp1-KD, and MCU-KD U2OS cells transfected with mito-R-GECO (mitochondrial calcium, red) and mitoBFP (mitochondrial matrix, blue) and then treated with 4 µM ionomycin at time 0. Time in seconds. White arrows point to constrictions and yellow arrows to fission events. Bar, 5 µm. Corresponds to Video 8. (C) U2OS cell transfected with GFP-Sec61β (green, ER marker) and mito-R-GECO (red, mitochondrial calcium) and imaged live before ionomycin stimulation (top) and at 35 s after 4 µM ionomycin addition (bottom). Image on the right is an expanded view of the (+) ionomycin condition. Bars: (regular panels) 2 µm; (expanded panel) 1 µm. Arrows denote ER presence at mitochondrial constrictions. Corresponds to Video 9. (D) Quantification of percent mitochondrial constrictions corresponding to ER contact from live-cell confocal microscopy of ionomycin-stimulated cells such as in C and Video 9. 204 constrictions from 26 ROIs from 13 cells assessed, totaling a 461-µm2 mitochondrial area.
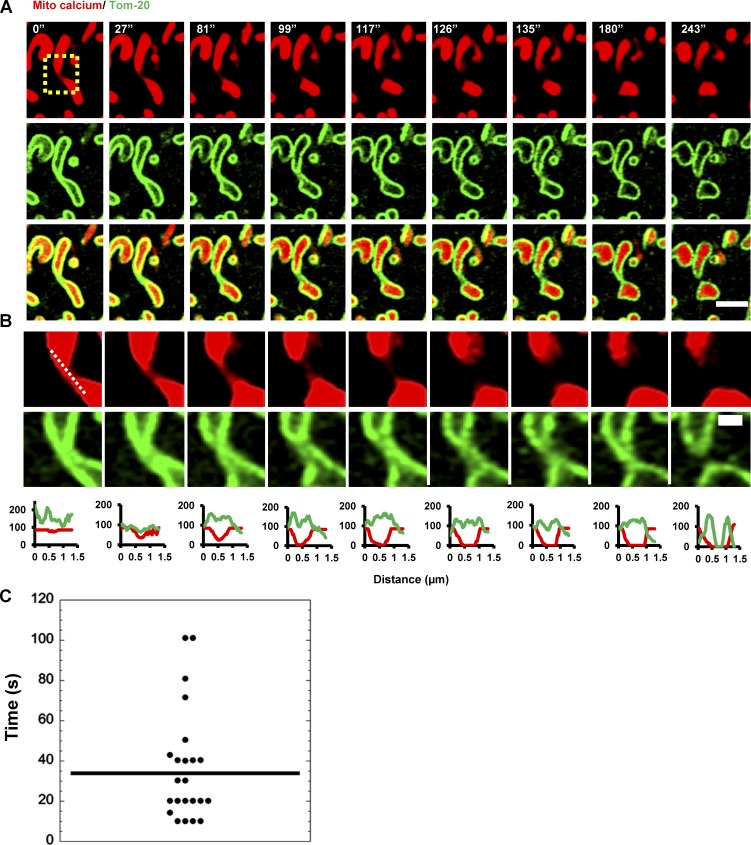
These results suggest that mitochondrial constrictions are caused by IMM dynamics. To examine dynamics of both IMM and OMM, we used Airyscan microscopy of cells expressing mito-R-GECO and GFP-Tom20 (an OMM marker). During ionomycin-stimulated mitochondrial division events, a consistently observable intermediate stage exists in which no discernable matrix marker is visible whereas the OMM marker presents as two parallel lines (Fig. 8, A and B; Video 9; and Fig. S4 D). Similar results occur on a comparatively slower time scale upon treatment with CGP37157 (Fig. S4 E and Video 10), an inhibitor of the mitochondrial Na+/Ca+ antiporter, a major route of mitochondrial calcium exit (Kaddour-Djebbar et al., 2010; Choudhary et al., 2011). From the CGP37157 data, the mean lag time between apparent matrix separation and apparent OMM separation for 29 measured events is 32.3 ± 29.1 s (29 events; Fig. 8 C).
Figure 8.
IMM division before OMM division during mitochondrial division. (A) U2OS cells transfected with mito-R-GECO (mitochondrial matrix calcium, red) and GFP-Tom20 (OMM, green) were imaged live by Airyscan microscopy at 9-s intervals after 4 µM ionomycin treatment. Representative time-lapse montage of a mitochondrial division event shown here. Bar, 2 µm. (B) Zoom of the division site (boxed in A). The mito-R-GECO levels have been enhanced to detect the existence of any thin matrix tether. Line scans indicate that the OMM tether persists in the absence of a matrix tether. Bar, 500 nm. (C) Quantification of time between apparent matrix separation and apparent OMM separation in mitochondrial division events after CGP37157 stimulation (example in Fig. S4 E). 29 division events from 23 cells. Mean time, 32.3 ± 29.1 s (SD). Each point represents one division event.
We probed the mechanism by which increased matrix calcium leads to IMM constriction by testing the role for Opa1, a dynamin GTPase that is attached to the IMM. Opa1 is well known to mediate IMM fusion, but targeted proteolysis releases Opa1 from the IMM and may allow Opa1 to participate in IMM division (Anand et al., 2014; MacVicar and Langer, 2016). One study showed evidence that Opa1 is not required for constrictions (Lee and Yoon, 2014), whereas another study showed that Oma1 processing of Opa1 is necessary for constrictions (Cho et al., 2017). We probed for changes in Opa1 proteolytic processing upon ionomycin stimulation and found no apparent change during the time period of increased IMM constrictions (Fig. 9 A). Using a covalent cross-linking assay adapted from others (Otera et al., 2016; Fig. S5 A), we found no apparent change in Opa1 oligomerization upon ionomycin stimulation (Fig. 9 B). In addition, ionomycin-induced constrictions were still present in U2OS cells upon Oma1 suppression (Fig. 9 C), despite effectively blocking Opa1 proteolytic processing (Fig. S5B, C). These results suggest that Opa1 processing by Oma1 is not required to induce IMM constrictions in U2OS cells.
Figure 9.
IMM constrictions do not require Oma1 activity but do require the ETC. (A) Western blot of Opa1 isoform pattern in U2OS cells during 4 µM ionomycin stimulation during the period of optimum calcium-induced constrictions. Mass is in kilodaltons. L1, L2, and S3–5 refer to Opa1 isoforms are as defined previously (Anand et al., 2014; Otera et al., 2016). (B) Western blot of Opa1 after live-cell cross-linking with BMH to reveal Opa1 oligomerization changes during 4 µM ionomycin stimulation. Oligomers are labeled with arrowheads and monomers with an arrow. Mass is in kilodaltons. (C) Constrictions in U2OS cells that had been transfected with Oma1 siRNA for 72 h, followed by transfection of mito-R-GECO and mitoBFP for 24 h and stimulation by 4 µM ionomycin for 60 s. Bars: (main) 5 µm; (insets) 2 µm. (D) Effect of 5 µM antimycin A or 2.5 µM rotenone on (left to right) mitochondrial constrictions, actin burst, and mitochondrial calcium spike induced by 4 µM ionomycin. Antimycin A or rotenone was added simultaneously to ionomycin. For mitochondrial constriction assessment: n = 10 ROIs, 200 µm2 mitochondrial area (Iono), 11 ROIs, 136 µm2 (Iono + rotenone); and 16 ROIs, 300 µm2 (Iono + Antimycin A). For actin filament assessment, 10 cells were used for each condition. For mitochondrial calcium assessment, 15–20 cells were used for each condition. P-values are from an unpaired Student’s t test. ***, P < 0.0011. Error bars represent SD (constriction quantification) or SEM (actin filament and mitochondrial calcium assessment).
We also asked whether a calcium-mediated increase in electron transport chain (ETC) activity was necessary for IMM constrictions, because calcium activates dehydrogenases associated with the ETC. Treatment with rotenone (complex I inhibitor) or antimycin A (complex III inhibitor) significantly decreases ionomycin-induced mitochondrial constrictions without decreasing the actin burst or mitochondrial calcium spike (Fig. 9 D). Because the ETC inhibitors are added simultaneous to the ionomycin stimulation, these results suggest that a calcium-mediated increase in ETC activity is required for IMM constrictions.
Discussion
The combination of work performed here and in past studies (Korobova et al., 2013; Ji et al., 2015) demonstrates that INF2-mediated actin polymerization on the ER stimulates mitochondrial division by two independent mechanisms: (1) mitochondrial calcium uptake, leading to IMM constriction; and (2) Drp1 oligomerization, leading to OMM constriction. Actin stimulates the mitochondrial calcium increase by enhancing ER–mitochondrial interaction (this study), allowing more efficient flow of calcium from ER to mitochondrion. Independently, actin stimulates Drp1 oligomerization through direct binding (Ji et al., 2015; Hatch et al., 2016). Mitochondrial calcium uptake is rapid, with a t1/2 of <20 s, and triggers constriction of the IMM in a manner requiring the ETC. Drp1 oligomerization occurs on a slower time course, with a t1/2 of ∼100 s.
INF2 is activated by increased cytoplasmic calcium (Shao et al., 2015; Wales et al., 2016). We postulate that an initial rise in cytoplasmic calcium (stimulated by ionomycin or histamine here) causes INF2 activation, with the resulting ER-localized actin polymerization driving ER–mitochondrial contact, which then allows efficient calcium transfer to mitochondria through MCU. The relative kinetics of cytoplasmic calcium, actin polymerization, and mitochondrial calcium in response to these stimuli support this mechanism.
This is the first study demonstrating a role for actin polymerization in ER-to-mitochondrial calcium transfer. A past study showed that 3-h treatment with actin depolymerizing drugs reduced ER calcium release from hippocampal neurons (Wang et al., 2002), but the length of treatment raised the possibility of indirect effects. Here, we show that ionomycin- or histamine-stimulated mitochondrial calcium increase is blocked by LatA applied simultaneously with the stimulus or by INF2 KO. These stimuli activate calcium dynamics by two distinct mechanisms. Histamine activates ER calcium efflux through IP3 receptors (Berridge et al., 2003). Ionomycin causes initial calcium entry from the extracellular medium, which activates a secondary release of calcium from the ER. This “calcium-induced calcium release” from the ER is presumably largely through ryanodine receptors (Smith et al., 1986). Although ryanodine receptors are best characterized in muscle or neuronal systems (Endo, 2009), they are also widely expressed in nonexcitable cells (Giannini et al., 1995; Bennett et al., 1996; Querfurth et al., 1998; Park et al., 2014). It is known that a disproportionate amount of ER-released calcium enters the mitochondrion, instead of dissipating into the bulk cytosol. Close contacts of <30 nm between the ER and mitochondrion are thought to allow preferential mitochondrial calcium entry (Rizzuto et al., 1998; Csordás et al., 2006, 2010; Giacomello et al., 2010), and several proteins that mediate ER–mitochondrial tethering might be important in this interaction (Phillips and Voeltz, 2016).
It is not clear what population of actin filaments enhances ER–mitochondrial interaction. The INF2-mediated actin “burst” occurs throughout the cytosol (Ji et al., 2015; Shao et al., 2015; Wales et al., 2016), but we have shown that the ER-bound INF2 isoform is specifically associated with mitochondrial dynamics, and we have shown here and elsewhere (Ramabhadran et al., 2013) that this isoform stimulates actin polymerization on the ER. One model is that INF2 assembles actin directly at ER–mitochondrial contacts, perhaps in association with mitochondrially bound Spire1C (Manor et al., 2015). Short filamentous structures have been observed at ER–mitochondrial close contacts by thin-section EM (Csordás et al., 2006), but their molecular identity is unknown. The fact that even relatively stable actin-based structures are destroyed during traditional EM processing (Lehrer, 1981; Maupin and Pollard, 1983) suggests that the short actin filaments required to span ER–mitochondrial contacts (10–20 subunits) are likely to be largely eliminated during EM investigation of these contacts. Another model is that actin exerts its effect on ER–mitochondrial contact from a distance, perhaps by decreasing motility of each organelle, as we have observed for mitochondria (Korobova et al., 2013). In addition to actin polymerization directly at mitochondrial division sites, we have observed polymerization parallel to the mitochondrion (Ji et al., 2015), which could serve such a purpose.
Regardless of the mechanism, other molecules contribute to actin-mediated enhancement of ER–mitochondrial contact. The fact that myosin IIA is required for the mitochondrial calcium increase suggests that it is providing contractile tension for ER–mitochondrial contact. The aforementioned Spire1C has been shown to work with INF2 in other aspects of mitochondrial division (Manor et al., 2015). Other studies have shown evidence for actin “clouds” assembling around mitochondria, and these clouds correspond to increased mitochondrial division (Li et al., 2015; Moore et al., 2016). It is unclear whether these structures constitute the same process as that mediated by INF2, because the morphology of the actin filaments differs significantly.
In light of the bulk of our results, it is intriguing that thapsigargin treatment successfully elicits a cytoplasmic actin burst but fails to stimulate an increase in mitochondrial calcium. One difference is that the thapsigargin-induced actin burst is significantly slower than those elicited by ionomycin or histamine, the former requiring ∼100 s and the latter peaking within 10 s. This difference in kinetics may speak to the specific actin pool that must be activated to enhance ER–mitochondrial contact rather than the bulk cytoplasmic actin pool. Alternately, activation of other molecules, such as myosin IIA, through phosphorylation (Heissler and Sellers, 2016) might be insufficient in the slower thapsigargin response. More detailed examination of specific actin and myosin populations should shed light on this question.
The demonstration that IMM constricts before OMM division during mitochondrial division, both here and in a recently published study (Cho et al., 2017), suggests that specific mechanisms for IMM constriction must exist. In this study, we find that constrictions are strongly suppressed by ETC inhibitors (antimycin and rotenone). We apply these inhibitors simultaneous to the constriction stimulus, suggesting that an acute metabolic increase is necessary. Increased matrix calcium is known to activate pyruvate dehydrogenase and isocitrate dehydrogenase (Denton, 2009), linking the actin-stimulated mitochondrial calcium rise to IMM constrictions through increased metabolic flux.
At present, the mechanism by which increased metabolic flux drives IMM constriction is unclear. Cristae remodeling is an attractive possibility. A major player in cristae remodeling is the dynamin GTPase Opa1, which is bound to the IMM and mediates mitochondrial fusion but might also mediate IMM division upon proteolytic processing that releases it from the IMM (Anand et al., 2014; MacVicar and Langer, 2016). Opa1 is also involved in the maintenance of cristae morphology, as well as regulating OMM/IMM tethering through the mitochondrial contact site complex (Cogliati et al., 2016; Otera et al., 2016; Quintana-Cabrera et al., 2017). Findings from two studies differ on the importance of Opa1 to mitochondrial constrictions. One study (Lee and Yoon, 2014) shows that spontaneous mitochondrial constrictions do not require Opa1, but that Opa1 is required for subsequent IMM proton leak. Another study (Cho et al., 2017) shows that Oma1 or Opa1 suppression inhibits IMM constrictions in cortical neuron cultures. Possible explanations for these differences include distinct constriction mechanisms based on cell type or stimulus. A second Opa1-processing protease, Yme1 (Anand et al., 2014; MacVicar and Langer, 2016) has not yet been tested for its importance in constrictions and, whereas Oma1 is the relevant protease in neurons (Cho et al., 2017), there may be redundancy in other cells.
We show for the first time that apparent IMM division precedes OMM division. We use the term “apparent” because it is possible that a thin matrix tether persists that is undetectable in our studies, even though overprocessing of the matrix signal fails to detect such a tether. Despite this caveat, we observe a period of tens of seconds in which there is no detectable matrix and the OMM appears parallel at the eventual division site, which is strongly suggestive of IMM division before OMM division. However, we have never observed stable IMM division in the absence of eventual OMM division, suggesting that the two events are coupled. A repeated observation in the field has been that a Drp1-independent “preconstriction” occurs before full mitochondrial division (Labrousse et al., 1999; Legesse-Miller et al., 2003). Calcium-stimulated IMM constriction/division may be the origin of such preconstriction.
Materials and methods
Plasmids and siRNA oligonucleotides
Mito-R-GECO1, Cyto-R-GECO1 (Kd = 0.48 µM for calcium), and Mito-LAR-GECO1.2 (Kd = 12 µM for calcium) constructs were gifts from Y.M. Usachev (University of Iowa Carver College of Medicine, Iowa City, IA), are on the pcDNA3.1(−) backbone (Thermo Fisher Scientific), and have been described previously (Wu et al., 2014). The OMM-calcium probe was a gift from S. Strack (University of Iowa Carver College of Medicine, Iowa City, IA) and was constructed from the Mito-LAR-GECO1.2 plasmid by replacing the matrix targeting sequence with the OMM targeting sequence from Saccharomyces cerevisiae Mas70 (amino acids 1–25). mApple-F-tractin and GFP-F-tractin plasmid were gifts from C. Waterman and A. Pasapera (National Institutes of Health, Bethesda, MD) and were on a GFP-N1 backbone (Clontech), as described previously (Johnson and Schell, 2009). Mito-DsRed and mito-BFP (GFP-N1 backbone) constructs were previously described (Korobova et al., 2014) and consist of amino acids 1–22 of S. cerevisiae COX4 N terminal to the respective fusion protein. ER-tagRFP (modified GFP-N1 backbone) was a gift from E. Snapp (Albert Einstein College of Medicine, New York, NY), with prolactin signal sequence at 5′ of the fluorescent protein and KDEL sequence at 3′. GFP-hSec61β (pAc-GFP-C1 backbone; Clontech) was a gift from T. Rapoport (Harvard Medical School, Boston, MA). Tom20-GFP was made by restriction digest of Tom20 from Tom20-mCherry (a gift from A. York, National Institutes of Health, Bethesda, MD) with NheI and BamHI and cloning into eGFP-N1 (Clontech). ER-GCaMP6-150 (Kd = 150 μM for calcium) on an FCK(1.3)GW backbone (22217; Addgene) was described previously (de Juan-Sanz et al., 2017) and is available from Addgene (86918). CFP-tagged VAPB (in pECFP-C1; Clontech) and eGFP-tagged PTPIP51 (in pAcGFP-N1; Clontech) were gifts from C.C.J. Miller (King’s College, London, England, UK; Gomez-Suaga et al., 2017). The pDEST47-GFP-MCU (in pcDNA6.2/C-EmGFP-DEST) plasmid for rescue experiments was from Addgene (31732) and has been described previously (Baughman et al., 2011). The GFP-INF2-CAAX and nonCAAX plasmids (in eGFP-C1; Clontech) used for rescue were similar to those reported previously (Ramabhadran et al., 2013), except they were gene edited to correct errors. To gene edit, the plasmids were digested with Sal1/HindIII, and the gene-edited sequence containing bases 796–2541 (synthesized by IDT, Inc.) was ligated in. The gene-edited insert is 5′-GTCGACATGAGCAGCCACCAGGAGGTGTTTGCCTCCTTGTTCCACAAAGTGTCTTGCTCTCCTGTTTCTGCCCAGCTGCTGAGTGTACTGCAGGGCCTTCTGCACCTCGAACCTACTTTGAGGTCTTCACAGCTCCTGTGGGAGGCTCTGGAATCCCTCGTCAACCGGGCAGTTCTTCTCGCCTCCGATGCCCAGGAGTGTACCCTCGAAGAGGTGGTCGAGCGATTGCTTAGTGTGAAAGGTCGCCCTCGGCCTTCACCACTGGTGAAAGCGCATAAGAGTGTTCAAGCTAACCTTGACCAATCTCAGAGGGGTAGCTCTCCCCAGAATACCACCACTCCAAAGCCCTCAGTTGAAGGTCAGCAACCAGCCGCAGCCGCCGCCTGCGAACCGGTGGATCACGCTCAGAGTGAGTCTATCCTGAAAGTAAGCCAGCCAAGGGCCCTGGAGCAGCAAGCATCTACACCACCCCCACCTCCCCCACCACCTTTGCTCCCAGGAAGTTCCGCTGAACCTCCACCCCCTCCCCCCCCGCCTCCTTTGCCATCTGTAGGAGCTAAGGCTTTGCCCACAGCACCACCACCACCGCCCTTGCCTGGACTGGGAGCAATGGCTCCTCCTGCACCTCCTTTGCCTCCCCCTCTCCCAGGATCCTGTGAATTTCTGCCTCCTCCACCTCCCCCCCTGCCCGGTCTGGGGTGCCCTCCTCCCCCTCCGCCACTGCTCCCGGGCATGGGGTGGGGTCCCCCACCACCGCCCCCCCCACTCTTGCCATGTACATGCTCCCCTCCCGTCGCGGGCGGAATGGAAGAGGTTATCGTTGCTCAGGTAGATCACGGCTTGGGAAGCGCCTGGGTCCCCTCTCACCGGCGGGTCAACCCCCCTACCCTGAGAATGAAAAAACTTAACTGGCAGAAGCTGCCTAGTAATGTGGCACGGGAGCACAACAGCATGTGGGCGAGTCTTTCATCACCCGACGCCGAAGCCGTGGAGCCCGATTTCTCCTCAATCGAGAGACTCTTTTCCTTCCCCGCTGCGAAACCTAAAGAGCCAACAATGGTTGCTCCAAGAGCTCGGAAGGAGCCCAAGGAGATCACCTTCCTGGATGCGAAAAAGAGTCTGAACCTCAACATTTTTCTGAAACAATTCAAATGCTCTAATGAGGAGGTGGCCGCCATGATAAGAGCAGGAGACACCACTAAATTTGACGTGGAGGTTCTGAAGCAGCTCCTTAAACTTCTCCCCGAAAAGCACGAGATCGAGAATCTCAGGGCGTTCACCGAAGAGCGGGCGAAGCTCGCTTCTGCCGACCATTTCTATCTCCTGCTTCTGGCAATCCCTTGTTACCAGTTGCGCATCGAGTGCATGCTCCTGTGTGAAGGAGCAGCTGCAGTTCTGGATATGGTGCGCCCAAAAGCCCAGCTGGTGCTCGCCGCCTGTGAAAGTTTGCTTACTTCAAGACAGTTGCCCATATTCTGTCAATTGATTCTGAGAATTGGCAACTTCCTTAACTACGGATCTCATACGGGGGACGCGGACGGGTTCAAAATTTCAACACTTCTGAAACTTACTGAGACGAAGAGCCAACAGAATAGAGTGACTTTGCTGCACCACGTGCTTGAGGAGGCTGAGAAAAGCCACCCTGACCTGCTCCAGTTGCCTCGGGACTTGGAGCAGCCAAGCCAGGCCGCGGGGATCAACCTGGAAATAATTAGATCAGAAGCAAGCTCTAACCTCAAAAAGCTT-3′.
The following oligonucleotides for all siRNA used were synthesized by IDT: myosin IIA, 5′-GCCACGCCCAGAAGAACGAGAAUGC-3′; Drp1, 5′-GGAACGCAGAGCAGCGGAAAGAGCT-3′; MCU, 5′-UAAUUGACACUUUAGAUUAUCUCTT-3′ (#siRNA1); 5′-GUAACAUACUUCAUCACUUAUGGAA-3′ (#siRNA2), 5′-AUUGACAGAGUUGCUAUCUAUUCAC-3′ (siRNA3); and OMA1, 5′-AGGAAGGAGCAAGGUACCAAUAAUAGCUUGCU-3′. Silencer Negative Control (Ambion) was used as a negative control (5′-CGUUAAUCGCGUAUAAUACGCGUAT-3′).
Antibodies and Western blotting
Anti–myosin IIA (rabbit; 3403S; Cell Signaling Technology) was used at 1:1,000. Anti-MCU (rabbit; 14997; Cell Signaling Technology) was used at 1:1,000. Anti-Drp1 (rabbit; 8570; Cell Signaling Technology,) was used at 1:1,000. Anti-GAPDH (G-9, mouse; Santa Cruz Biotechnology) was used at 1:1,500. Anti-tubulin (DM1-α, mouse; Sigma-Aldrich,) was used at 1: 10,000 dilutions. Anti-actin (mouse; mab1501R; Millipore) used at 1:1,000. Anti-INF2 was described previously (Ramabhadran et al., 2011; 941-1249 antibody used, rabbit). Anti-Opa1 (mouse; 612606; BD Biosciences) was used at 1:2,000. Anti-Mff (rabbit; 17090-1-AP; Proteintech,) was used at 1:1,000. Anti-Fis1 (rabbit; 10956-1-AP; Proteintech) was used at 1:1,000. Anti-Mid51 (rabbit; 20164-1-AP; Proteintech) used at 1:500. Anti-OMA1 (sc-515788; Santa Cruz Biotechnology) was used at 1:1,000. Anti-Tom20 (rabbit; sc-145; Santa Cruz Biotechnology) was used at 1:500 for immunofluorescence. Anti–rabbit Texas red secondary antibody (goat; Tl1000; Vector Laboratories) was used at 1:500 for immunofluorescence. For Western blotting, cells were grown on a 6-well plate, trypsinized, washed with PBS, and resuspended 50 µl PBS. This solution was mixed with 34 µl 10% SDS and 1 µl of 1 M DTT, boiled 5 min, and cooled to 23°C, and then 17 µl of 300 mM freshly made N-ethylmaleimide (Thermo Fisher Scientific) in water was added. Just before SDS-PAGE, the protein sample was mixed 1:1 with 2xDB (250 mM Tris-HCl, pH 6.8, 2 mM EDTA, 20% glycerol, 0.8% SDS, 0.02% bromophenol blue, 1000 mM NaCl, and 4 M urea). Proteins were separated by 7.5% SDS-PAGE and transferred to a PVDF membrane (polyvinylidine difluoride membrane; Millipore). The membrane was blocked with TBS-T (20 mM Tris-HCl, pH 7.6, 136 mM NaCl, and 0.1% Tween-20) containing 3% BSA (Research Organics) for 1 h, then incubated with the primary antibody solution at 4°C overnight. After washing with TBS-T, the membrane was incubated with horseradish peroxidase (HRP)-conjugated secondary antibody (goat; 0300-0136 and 5196-2504; Bio-Rad) for 1 h at room temperature. Signals were detected by Chemiluminescence (Pierce).
Cell culture, transfection, and drug treatments
Human osteosarcoma U2OS cells (HTB96; American Type Culture Collection) were grown in DMEM (Invitrogen) supplemented with 10% calf serum (Atlanta Biologicals). The GFP-Drp1 KI U2OS cell line made by CRISPR-Cas9 is described elsewhere (Ji et al., 2017). In brief, we used the GeCKO system (Zhang laboratory, Massachusetts Institute of Technology, Cambridge, MA; http://genome-engineering.org/gecko/). The donor plasmid contained eGFP (A206K mutant) flanked by 445 bases upstream of the hDrp1 start codon and 308 bases downstream of the start (synthesized by IDT). The target guide sequence (5′-CATTCATTGCCGTGGCCGGC-3′) was predicted using the GeCKO website program and made by IDT. Donor and guide plasmids were transfected into U2OS cells at a 3:1 molar ratio using Lipofectamine 2000 (Invitrogen). Cells were put under puromycin selection, and clones were selected by FACS and single-cell cloning and then verified by immunofluorescence and Western blotting. The cell line expresses GFP-Drp1 at 50% of the total Drp1 level, with the remaining Drp1 being unmodified (overall Drp1 level unchanged from control cells).
To prepare the INF2-KO-U2OS cell line, the guide sequence to INF2 (5′-TCCGTGGGGTCCGAATCCTG-3′) was predicted and cloned into LentiCRISPRv2 vector by following the protocol found at http://genome-engineering.org/gecko/. The resulting guide plasmid, along with helper plasmids psPAX2 and pMD2.G, was transfected into HEK293 cells using Lipofectamine LTX (Invitrogen). Media containing transfection reagent was aspirated the next day and replaced with fresh media. Supernatant containing lentivirus was collected 48 h posttransfection and filtered through a 0.45-µm filter. Fresh virus was used to infect U2OS cells. After 48 h, media containing virus was removed and replaced with fresh media containing 2 µg/ml puromycin. Cells were put under puromycin selection, and clones were selected by FACS into 96-well plates and verified by immunofluorescence and Western blotting.
For transfection, cells were seeded at 4 × 105 cells per well in a 6-well dish ∼16 h before transfection. Plasmid transfections were performed in OPTI-MEM media (Invitrogen) with 2 µl Lipofectamine 2000 (Invitrogen) per well for 6 h, followed by trypsinization and replating onto concanavalin A (C5275; Sigma/Aldrich)–coated glass-bottom MatTek dishes (P35G-1.5-14-C) at ∼3.5 × 105 cells per well (coverslips were treated for ∼2 h with 100 g/ml concanavalin A in water at room temperature). Cells were imaged in live cell media (21063-029; Life Technologies) ∼16 to 24 h after transfection.
For all experiments, the following amounts of DNA were transfected per well (individually or combined for cotransfection): 500 ng for Mito-R-GECO1, Cyto-R-GECO1, Mito-LAR-GECO1.2, mt-DsRed, GFP-F-Tractin, and mito-BFP; 600 ng for GFP-Tom20; 750 ng for ER-GCaMP6-150; 1,000 ng for ER-tagRFP and GFP-Sec61β; 600 ng for CFP-VAPB and GFP-PTPIP51; and 500 ng for the OMM-Mito calcium construct.
For siRNA transfections, 105 cells were plated on 6-well plates, and 2 µl RNAimax (Invitrogen) and 63 pg siRNA were used per well. Cells were analyzed 72 to 84 h posttransfection for suppression.
For ionomycin (I0634; Sigma-Aldrich) or histamine (H7125; Sigma-Aldrich), cells were treated with 4 µM ionomycin (from a 1 mM stock in DMSO) or 100 µM histamine (from 100 mM stock in DMSO) at the fifth frame (∼1 min, depending on time interval used) during imaging and continued for another 5–10 min. Medium was preequilibrated for temp and CO2 content before use. DMSO was used as the negative control. Of note, the ionomycin treatment used here differs markedly from those in most other studies in that we conduct our stimulation in the presence of serum. This treatment results in the transient changes in cytoplasmic and mitochondrial calcium shown, with both returning to baseline in 120 s In contrast, most other studies treat in the absence of serum. In our hands, serum removal causes a higher and prolonged mitochondrial calcium increase, in addition to extensive fragmentation of both mitochondria and ER (Fig. S5 D).
For LatA (428021; Calbiochem) coupled with ionomycin treatment, cells were treated with live cell medium containing LatA (2 µM; added from a 2 mM DMSO stock) and ionomycin simultaneously at frame 5 (∼1 min, depends on time intervals) with DMSO used as the negative control. For CGP17357 (C8874; Sigma-Aldrich), cells were treated with 80 µM (from 10 mM stock in DMSO) at frame 10 (∼1 min depending on the time intervals used) during imaging. For thapsigargin (T7458; Life Technologies), cells were treated with 1 µM (from 1 mM stock in DMSO) at frame 4 (∼1 min depending on the time interval used) during imaging. ETC inhibitors (antimycin A, 5 µM final [A8674; Sigma-Aldich]-, Rotenone, 2.5 µM final [557368; Sigma-Aldrich]) were added simultaneously with ionomycin after 4 frames of imaging (∼1 min depending on frame speed used).
Live imaging by confocal and Airyscan microscopy
All imaging was conducted in DMEM (21063-029 with 4.5 g/liter d-glucose, l-glutamine, and 25 mM Hepes; Gibco) supplemented with 10% NCS (SH30118.03; HyClone).
MatTek dishes were imaged on a Wave FX spinning disk confocal microscope (Quorum Technologies, Inc.) on an Eclipse Ti microscope (Nikon) equipped with Hamamatsu ImageM EM CCD cameras and Bionomic Controller (20/20 Technology, Inc.) temperature-controlled stage set to 37°C. After equilibrating to temperature for 10 min, cells were imaged with the 60× 1.4 NA Plan-Apo objective (Nikon) using the 403-nm laser and 450/50 filter for BFP, 491 nm and 525/20 for GFP, 561 nm and 593/40 for RFP. Images acquired using Metamorph software (Molecular Devices, Inc.). Images in Figs. 2 A, 4 B, S1 (C and D), S4 (A and B), and S5 D were collected with Wave FX microscope.
For high-time-resolution imaging, a Dragonfly 302 spinning disk confocal (Andor Technology, Inc.) on a Nikon Ti-E base and equipped with an iXon Ultra 888 EMCCD camera, a Zyla 4.2 Mpixel sCMOS camera, and a Tokai Hit stage-top incubator was used. A solid-state 405 smart diode 100-mW laser, solid-state 488 OPSL smart laser 50-mW laser, solid-state 560 OPSL smart laser 50-mW laser, and solid-state 637 OPSL smart laser 140-mW laser were used (objective: 60× 1.4 NA CFI Plan Apo; Nikon). Images were acquired using Fusion software (Andor Technology, Inc.). Images in Figs. 1 (G and H), 3 F, 5 A, 6 (A and E), 9 C, S2 E, and S3 E were taken on the Dragonfly microscope.
Airyscan images were acquired on LSM 880 equipped with 63×/1.4 NA plan Apochromat oil objective using the Airyscan detectors (Carl Zeiss Microscopy). The Airyscan uses a 32-channel array of GaAsP detectors configured as 0.2 airy units per channel to collect the data that is subsequently processed using Zen2 software. After equilibrating to 37°C for 30 min, cells were imaged with the 405-nm laser and 450/30 filter for BFP, 488 nm and 525/30 for GFP, and 561 nm and 595/25 for RFP. Images in Figs. 7 C, 8 (A and B), and S4 (D and E) were taken using an LSM 880.
Measurements of calcium changes and actin burst in live cells
For the following experiments, imaging was conducted at either 0.21-s intervals (high time resolution) or 12- to 15-s intervals (low time resolution), as indicated. All imaging was in live cell media (DMEM + 4.5 g/liter glucose, l-glutamine, and Hepes [21063–029]; Life Technologies) containing 10% fetal calf serum.
Calcium measurements
Cells were seeded at 4 × 105 confluency and transfected with the indicated mitochondrial/cytosolic GECO probes or the ERGCaMP6-150 probe (ER calcium) before the day of imaging. 24 h after transfections, these cells were imaged for 1 min to establish baseline fluorescence (F0) at 488 nm excitation. Cells were then perfused with the indicated compounds by addition of equal volume of a 2× stock in medium with continuous imaging (F-each time point). Mean fluorescence was calculated for each cell using ImageJ (National Institutes of Health). Fluorescence values for each time point after drug treatment (F) were normalized with the mean initial fluorescence (first five or six frames − F0) and plotted against time as F/F0. The rapid mito-R-GECO response is not caused by a change in matrix volume, because the fluorescence intensity of cotransfected mito-BFP does not change appreciably.
Actin burst measurements
Cells were seeded at 4 × 105 confluency and transfected with GFP-Ftractin before the day of imaging. 24 h after transfections, these cells were imaged (at a vertical plane above the basal surface, to avoid actin stress fibers) for 1 min to establish baseline fluorescence at 488 nm excitation. Cells were then perfused with the indicated drugs with continuous imaging. Mean fluorescence values for each time point were calculated from four regions of interest (ROIs) per cell selected in the perinuclear region using ImageJ. Fluorescence values for each time point after drug treatment (F) were normalized with the mean initial fluorescence (first five or six frames − F0) and plotted against time as F/F0.
Mitochondrially associated Drp1 puncta
Drp1-KI cells transiently transfected with mitochondrial markers were imaged live by spinning disc confocal fluorescence microscopy for 10 min at 3-s intervals in a single focal plane. ROIs with readily resolvable mitochondria and Drp1 were processed as described previously (Ji et al., 2015). We thresholded mitochondrially associated Drp1 puncta by using the Colocalization ImageJ plugin with the following parameters: ratio 50% (0–100%); threshold channel 1: 30 (0–255); threshold channel 2: 30 (0–255); display value: 255 (0–255). Mitochondrially associated Drp1 puncta were further analyzed by Trackmate as described previously (Ji et al., 2015) to separate into low-threshold and high-threshold categories. The number of Drp1 puncta in each category was automatically counted frame by frame using the Find Stack Maxima ImageJ macro.
Morphological analysis of mitochondrial size
Following methods described previously (Lee et al., 2016) and adapted for U2OS cells, U2OS-WT cells (plated on MatTek dishes) were treated with siRNAs against MCU or Drp1 for 72 h. Cells were then fixed with 1% glutaraldehyde in PBS for 10 min and treated with sodium borohydride in PBS to quench autofluorescence. These cells were then stained with Tom 20 antibody (mitochondria), anti-rabbit secondary antibody and DAPI (nucleus) in PBS + 10% calf serum, and imaged in PBS on the MatTek dish to prevent flattening due to mounting. z-stacks (20 × 0.4 µm) were acquired to obtain the total cell volume. Images were analyzed in a blinded manner (coded by one investigator and analyzed by a second investigator). Maximum intensity projections were generated from z-stacks acquired and background subtracted using FIJI (rolling ball 20.0). A 225-µm2 area of resolvable mitochondria was selected (one ROI/cell in the spread region of the cell) and analyzed using “Analyze Particle” plugin in FIJI to obtain number of mitochondrial fragments and the area of each fragment per ROI. Images were then decoded to generate the respective graphs.
Mitochondrial division rate
Mitochondrial division rate was described in detail previously (Ji et al., 2015). Suitable ROIs were selected for analysis based on whether individual mitochondria were resolvable and did not leave the focal plane. One ROI per cell selected. Files of these ROIs were assembled, coded and scrambled by one investigator, and analyzed for division by a second investigator in a blinded manner as to the treatment condition. The second investigator scanned the ROIs frame by frame manually for division events and determined mitochondrial length within the ROI using the ImageJ macro, Mitochondrial Morphology. The results were then given back to the first investigator for decoding.
Mitochondrial constriction quantification
Confocal time courses were collected of U2OS cells (control, Drp1-KD, or MCU-KD) transfected with mito-BFP and mito-R-GECO, recording at 12-s intervals for 1 min before ionomycin stimulation, and then for 300 s after ionomycin stimulation (4 µM in the presence of serum). Suitable ROIs were selected for analysis based on whether individual mitochondria were resolvable and did not leave the focal plane (one ROI per cell). Constrictions were scored visually as near-diffraction limited reduction in mito-BFP signal within mitochondria. For each ROI, constrictions were scored for each of the five frames before stimulation and averaged to give the unstimulated value and for 10 consecutive frames surrounding the 2-min mark poststimulation to give the ionomycin-induced value. Number of ROIs and the total length of mitochondria scanned for each set given in the figure legend.
EM and analysis of ER–mitochondrial contact
U2OS cells (control, INF2-KO) were seeded at 40,000 cells/cm2 onto glass-bottom MatTek dishes and cultured overnight in full growth medium. The next day, the cells were either left untreated or stimulated for 1 min with 4 µM ionomycin. Cells were fixed (1 h at room temp, two changes of fixative) in 2% glutaraldehyde, 3% paraformaldehyde in 0.1 M sodium cacodylate, pH 7.2, postfixed (1 h on ice, in the dark) in 1% OsO4 in 0.1 M sodium cacodylate, pH 7.2, rinsed (2 × 10min at room temp) in 0.1 M sodium cacodylate, pH 7.2, incubated (overnight, at RT, in the dark) in 2% aqueous uranyl acetate (EMS), dehydrated in graded ethanol (50%, 70%, 95%, 2 × 100%), and embedded in LX-112 resin (Ladd Research) using standard protocols (Stan et al., 2012). Sections parallel to the cellular monolayer were obtained using a Leica Ultracut 6 ultramicrotome, mounted on carbon/formvar–coated 300-mesh copper grids (Ted Pella), stained with uranyl acetate and lead citrate (EMS), examined under a Jeol 1010 electron microscope, and imaged using a bottom-mounted AMT CCD camera (2,048 × 2,048 pixels). TIFF images were acquired by one investigator (R.V. Stan) scrambled by another investigator (H.N. Higgs) and analyzed by a third investigator (R. Chakrabarti) using FIJI. Mitochondria and ER were located based on their respective morphology and were traced by hand. The distance between the organelles was measured using the “measure” tool in Fiji along the traced-out contact region. Mitochondria with ER within 30 nm at any point on the mitochondrial circumference were scored as a percentage of total mitochondria in the field. The values were decoded by the second investigator and the graphs generated.
Opa1 cross-linking
Cross-linking assay using bismaleimidohexane (BMH; Thermo Fisher Scientific) was modified from the protocols described previously (Patten et al., 2014; Otera et al., 2016). The first paper used a low concentration (50 µM) for 30 min, and the second paper used high concentration (1 mM) for 20 min. We first tested both concentrations for varying time (Fig. S5 A) and decided to use 1 mM BMH for 1 min to accommodate the rapid changes induced by ionomycin.
U2OS cells were treated with 4 µM ionomycin for the indicated times. At 1 min before termination, 2× BMH solution (2 mM) in medium was added. The medium was quickly aspirated and replaced with modified SDS-PAGE extraction buffer (250 mM Tris-HCl, pH 6.8, 2 mM EDTA, 20% glycerol, 0.8% SDS, 0.02% bromophenol blue, 1,000 mM NaCl, and 4 M urea) to extract protein for Western blotting.
Statistical analysis
All statistical analyses and p-value determinations were done using GraphPad Prism. To determine p-values, an unpaired Student’s t tests was performed between two groups of data, comparing full datasets stated in the figure legends.
Online supplemental material
Fig. S1 shows a comparison of ionomycin and histamine stimulation for cytoplasmic calcium increase (with and without EGTA in medium), Drp1 oligomer number, and mitochondrial division rate. Fig. S2 shows characterization of the CRISPR-mediated INF2-KO cell line and rescue of ionomycin-stimulated mitochondrial calcium increase by INF2 isoforms and ER–mitochondrial tethers. Fig. S3 shows the effects of myosin IIA KD on ionomycin- and histamine-stimulated actin burst and mitochondrial calcium increase; quantification of protein levels for KDs of Drp1, INF2, MCU, and myosin IIA; rescue of ionomycin-stimulated mitochondrial calcium increase in MCU-KD cells by re-expression of siRNA-resistant MCU; and the low basal mitochondrial calcium levels in MCU-KD cells. Fig. S4 shows spontaneous mitochondrial constrictions, as well as examples of IMM division before OMM division for ionomycin-stimulated and CGP37157-stimulated cells. Fig. S5 shows Opa1 oligomer assay pilot experiment, the effects of Oma1-KD on Opa1 band patterns, and comparison of ionomycin stimulation in the presence and absence of serum. Videos 1 and 2 correspond to Fig. 1, A and D, respectively, and show changes in cytoplasmic calcium, actin polymerization, and mitochondrial calcium in response to ionomycin (Video 1) and histamine (Video 2). Videos 3 and 4 correspond to Figs. 1, G and H, respectively, and simultaneously show changes in actin polymerization and mitochondrial calcium in response to ionomycin (Video 3) or histamine (Video 4). Video 5 corresponds to Fig. 2 A, and shows changes in ER calcium in response to ionomycin or histamine. Video 6 corresponds to Fig. 3 F and shows actin polymerization in cells expressing either INF2-CAAX or INF2-nonCAAX in response to ionomycin. Video 7 corresponds to Fig. 7 B and shows ionomycin-induced mitochondrial matrix constrictions in Drp1-KD or MCU-KD cells. Video 8 corresponds to Fig. 7 C and shows mitochondrial constrictions at ER contact sites after ionomycin stimulation. Videos 9 and 10 correspond to Fig. 8 A and Fig. S4 E, respectively, and show matrix division before OMM division in cells stimulated with ionomycin (Video 9) or CGP37157 (Video 10).
Supplementary Material
Acknowledgments
We thank Tom Blanpied, Sai Divakaruni, Minerva Contreras, Mike Hoppa, Stefan Strack, Charles Barlowe, Bill Wickner, Anna Hatch, Mu A, Lorna Young, Laura McCormick, and Lori Schoenfeld for advice and preliminary experiments that strengthened this work, as well as A. Lavanway for frequent microscopy help and Uma Clic for the ability to rise when asked.
This work was supported by the National Institutes of Health (grants GM069818, GM106000, P20 GM113132, and DK88826 to H.N. Higgs and grant NS036942 to T.A. Ryan). The purchase of the Airyscan microscope was made possible through the National Institute of General Medical Sciences (supplement to GM109965), the Provost and Dean of Sciences at Dartmouth College, and the Norris Cotton Cancer Center. The purchase of the Dragonfly microscope was made possible through support from the William Ruger Senior Fund at Dartmouth (560118) and the Department of Biochemistry and Cell Biology.
The authors declare no competing financial interests.
Author contributions: R. Chakrabarti and H.N. Higgs conceived of or designed the research and wrote the manuscript. R. Chakrabarti, R. Stan, and W.-K. Ji performed the experiments. J. de Juan-Sanz and T.A. Ryan designed ER Ca2+ experiments.
References
- Abramov A.Y., and Duchen M.R.. 2003. Actions of ionomycin, 4-BrA23187 and a novel electrogenic Ca2+ ionophore on mitochondria in intact cells. Cell Calcium. 33:101–112. 10.1016/S0143-4160(02)00203-8 [DOI] [PubMed] [Google Scholar]
- Anand R., Wai T., Baker M.J., Kladt N., Schauss A.C., Rugarli E., and Langer T.. 2014. The i-AAA protease YME1L and OMA1 cleave OPA1 to balance mitochondrial fusion and fission. J. Cell Biol. 204:919–929. 10.1083/jcb.201308006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baffy G., Miyashita T., Williamson J.R., and Reed J.C.. 1993. Apoptosis induced by withdrawal of interleukin-3 (IL-3) from an IL-3-dependent hematopoietic cell line is associated with repartitioning of intracellular calcium and is blocked by enforced Bcl-2 oncoprotein production. J. Biol. Chem. 268:6511–6519. [PubMed] [Google Scholar]
- Baughman J.M., Perocchi F., Girgis H.S., Plovanich M., Belcher-Timme C.A., Sancak Y., Bao X.R., Strittmatter L., Goldberger O., Bogorad R.L., et al. 2011. Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature. 476:341–345. 10.1038/nature10234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett D.L., Cheek T.R., Berridge M.J., De Smedt H., Parys J.B., Missiaen L., and Bootman M.D.. 1996. Expression and function of ryanodine receptors in nonexcitable cells. J. Biol. Chem. 271:6356–6362. 10.1074/jbc.271.11.6356 [DOI] [PubMed] [Google Scholar]
- Berridge M.J., Bootman M.D., and Roderick H.L.. 2003. Calcium signalling: dynamics, homeostasis and remodelling. Nat. Rev. Mol. Cell Biol. 4:517–529. 10.1038/nrm1155 [DOI] [PubMed] [Google Scholar]
- Boyer O., Nevo F., Plaisier E., Funalot B., Gribouval O., Benoit G., Huynh Cong E., Arrondel C., Tête M.J., Montjean R., et al. 2011. INF2 mutations in Charcot-Marie-Tooth disease with glomerulopathy. N. Engl. J. Med. 365:2377–2388. 10.1056/NEJMoa1109122 [DOI] [PubMed] [Google Scholar]
- Brown E.J., Schlöndorff J.S., Becker D.J., Tsukaguchi H., Tonna S.J., Uscinski A.L., Higgs H.N., Henderson J.M., and Pollak M.R.. 2010. Mutations in the formin gene INF2 cause focal segmental glomerulosclerosis. Nat. Genet. 42:72–76. 10.1038/ng.505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burman J.L., Pickles S., Wang C., Sekine S., Vargas J.N.S., Zhang Z., Youle A.M., Nezich C.L., Wu X., Hammer J.A., and Youle R.J.. 2017. Mitochondrial fission facilitates the selective mitophagy of protein aggregates. J. Cell Biol. 216:3231–3247. 10.1083/jcb.201612106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caridha D., Yourick D., Cabezas M., Wolf L., Hudson T.H., and Dow G.S.. 2008. Mefloquine-induced disruption of calcium homeostasis in mammalian cells is similar to that induced by ionomycin. Antimicrob. Agents Chemother. 52:684–693. 10.1128/AAC.00874-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho B., Cho H.M., Jo Y., Kim H.D., Song M., Moon C., Kim H., Kim K., Sesaki H., Rhyu I.J., et al. 2017. Constriction of the mitochondrial inner compartment is a priming event for mitochondrial division. Nat. Commun. 8:15754 10.1038/ncomms15754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary V., Kaddour-Djebbar I., Lakshmikanthan V., Ghazaly T., Thangjam G.S., Sreekumar A., Lewis R.W., Mills I.G., Bollag W.B., and Kumar M.V.. 2011. Novel role of androgens in mitochondrial fission and apoptosis. Mol. Cancer Res. 9:1067–1077. 10.1158/1541-7786.MCR-10-0445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogliati S., Enriquez J.A., and Scorrano L.. 2016. Mitochondrial Cristae: Where Beauty Meets Functionality. Trends Biochem. Sci. 41:261–273. 10.1016/j.tibs.2016.01.001 [DOI] [PubMed] [Google Scholar]
- Csordás G., Renken C., Várnai P., Walter L., Weaver D., Buttle K.F., Balla T., Mannella C.A., and Hajnóczky G.. 2006. Structural and functional features and significance of the physical linkage between ER and mitochondria. J. Cell Biol. 174:915–921. 10.1083/jcb.200604016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csordás G., Várnai P., Golenár T., Roy S., Purkins G., Schneider T.G., Balla T., and Hajnóczky G.. 2010. Imaging interorganelle contacts and local calcium dynamics at the ER-mitochondrial interface. Mol. Cell. 39:121–132. 10.1016/j.molcel.2010.06.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Juan-Sanz J., Holt G.T., Schreiter E.R., de Juan F., Kim D.S., and Ryan T.A.. 2017. Axonal Endoplasmic Reticulum Ca(2+) Content Controls Release Probability in CNS Nerve Terminals. Neuron. 93:867–881.e6. 10.1016/j.neuron.2017.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denton R.M. 2009. Regulation of mitochondrial dehydrogenases by calcium ions. Biochim. Biophys. Acta. 1787:1309–1316. 10.1016/j.bbabio.2009.01.005 [DOI] [PubMed] [Google Scholar]
- De Stefani D., Raffaello A., Teardo E., Szabò I., and Rizzuto R.. 2011. A forty-kilodalton protein of the inner membrane is the mitochondrial calcium uniporter. Nature. 476:336–340. 10.1038/nature10230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Stefani D., Rizzuto R., and Pozzan T.. 2016. Enjoy the Trip: Calcium in Mitochondria Back and Forth. Annu. Rev. Biochem. 85:161–192. 10.1146/annurev-biochem-060614-034216 [DOI] [PubMed] [Google Scholar]
- Diarra A., and Sauvé R.. 1992. Effect of thapsigargin and caffeine on Ca2+ homeostasis in HeLa cells: implications for histamine-induced Ca2+ oscillations. Pflugers Arch. 422:40–47. 10.1007/BF00381511 [DOI] [PubMed] [Google Scholar]
- DuBoff B., Götz J., and Feany M.B.. 2012. Tau promotes neurodegeneration via DRP1 mislocalization in vivo. Neuron. 75:618–632. 10.1016/j.neuron.2012.06.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo M. 2009. Calcium-induced calcium release in skeletal muscle. Physiol. Rev. 89:1153–1176. 10.1152/physrev.00040.2008 [DOI] [PubMed] [Google Scholar]
- Friedman J.R., Lackner L.L., West M., DiBenedetto J.R., Nunnari J., and Voeltz G.K.. 2011. ER tubules mark sites of mitochondrial division. Science. 334:358–362. 10.1126/science.1207385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomello M., Drago I., Bortolozzi M., Scorzeto M., Gianelle A., Pizzo P., and Pozzan T.. 2010. Ca2+ hot spots on the mitochondrial surface are generated by Ca2+ mobilization from stores, but not by activation of store-operated Ca2+ channels. Mol. Cell. 38:280–290. 10.1016/j.molcel.2010.04.003 [DOI] [PubMed] [Google Scholar]
- Giannini G., Conti A., Mammarella S., Scrobogna M., and Sorrentino V.. 1995. The ryanodine receptor/calcium channel genes are widely and differentially expressed in murine brain and peripheral tissues. J. Cell Biol. 128:893–904. 10.1083/jcb.128.5.893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Suaga P., Paillusson S., Stoica R., Noble W., Hanger D.P., and Miller C.C.. 2017. The ER-Mitochondria Tethering Complex VAPB-PTPIP51 Regulates Autophagy. Curr. Biol. 27:371–385. 10.1016/j.cub.2016.12.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch A.L., Ji W.K., Merrill R.A., Strack S., and Higgs H.N.. 2016. Actin filaments as dynamic reservoirs for Drp1 recruitment. Mol. Biol. Cell. 27:3109–3121. 10.1091/mbc.E16-03-0193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heissler S.M., and Sellers J.R.. 2016. Various Themes of Myosin Regulation. J. Mol. Biol. 428(9, 9 Pt B):1927–1946. 10.1016/j.jmb.2016.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji W.K., Hatch A.L., Merrill R.A., Strack S., and Higgs H.N.. 2015. Actin filaments target the oligomeric maturation of the dynamin GTPase Drp1 to mitochondrial fission sites. eLife. 4:e11553 10.7554/eLife.11553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji W.K., Chakrabarti R., Fan X., Schoenfeld L., Strack S., and Higgs H.N.. 2017. Receptor-mediated Drp1 oligomerization on endoplasmic reticulum. J. Cell Biol. 10.1083/jcb.201610057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson H.W., and Schell M.J.. 2009. Neuronal IP3 3-kinase is an F-actin-bundling protein: role in dendritic targeting and regulation of spine morphology. Mol. Biol. Cell. 20:5166–5180. 10.1091/mbc.E09-01-0083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaddour-Djebbar I., Choudhary V., Brooks C., Ghazaly T., Lakshmikanthan V., Dong Z., and Kumar M.V.. 2010. Specific mitochondrial calcium overload induces mitochondrial fission in prostate cancer cells. Int. J. Oncol. 36:1437–1444. [DOI] [PubMed] [Google Scholar]
- Korobova F., Ramabhadran V., and Higgs H.N.. 2013. An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2. Science. 339:464–467. 10.1126/science.1228360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korobova F., Gauvin T.J., and Higgs H.N.. 2014. A role for myosin II in mammalian mitochondrial fission. Curr. Biol. 24:409–414. 10.1016/j.cub.2013.12.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbé K., Murley A., and Nunnari J.. 2014. Determinants and functions of mitochondrial behavior. Annu. Rev. Cell Dev. Biol. 30:357–391. 10.1146/annurev-cellbio-101011-155756 [DOI] [PubMed] [Google Scholar]
- Labrousse A.M., Zappaterra M.D., Rube D.A., and van der Bliek A.M.. 1999. C. elegans dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane. Mol. Cell. 4:815–826. 10.1016/S1097-2765(00)80391-3 [DOI] [PubMed] [Google Scholar]
- Lee H., and Yoon Y.. 2014. Transient contraction of mitochondria induces depolarization through the inner membrane dynamin OPA1 protein. J. Biol. Chem. 289:11862–11872. 10.1074/jbc.M113.533299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.E., Westrate L.M., Wu H., Page C., and Voeltz G.K.. 2016. Multiple dynamin family members collaborate to drive mitochondrial division. Nature. 540:139–143. 10.1038/nature20555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legesse-Miller A., Massol R.H., and Kirchhausen T.. 2003. Constriction and Dnm1p recruitment are distinct processes in mitochondrial fission. Mol. Biol. Cell. 14:1953–1963. 10.1091/mbc.E02-10-0657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrer S.S. 1981. Damage to actin filaments by glutaraldehyde: Protection by tropomyosin. J. Cell Biol. 90:459–466. 10.1083/jcb.90.2.459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis S.C., Uchiyama L.F., and Nunnari J.. 2016. ER-mitochondria contacts couple mtDNA synthesis with mitochondrial division in human cells. Science. 353:aaf5549 10.1126/science.aaf5549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Xu S., Roelofs B.A., Boyman L., Lederer W.J., Sesaki H., and Karbowski M.. 2015. Transient assembly of F-actin on the outer mitochondrial membrane contributes to mitochondrial fission. J. Cell Biol. 208:109–123. 10.1083/jcb.201404050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacVicar T., and Langer T.. 2016. OPA1 processing in cell death and disease - the long and short of it. J. Cell Sci. 129:2297–2306. 10.1242/jcs.159186 [DOI] [PubMed] [Google Scholar]
- Manor U., Bartholomew S., Golani G., Christenson E., Kozlov M., Higgs H., Spudich J., and Lippincott-Schwartz J.. 2015. A mitochondria-anchored isoform of the actin-nucleating spire protein regulates mitochondrial division. eLife. 4:4 10.7554/eLife.08828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maupin P., and Pollard T.D.. 1983. Improved preservation and staining of HeLa cell actin filaments, clathrin-coated membranes, and other cytoplasmic structures by tannic acid-glutaraldehyde-saponin fixation. J. Cell Biol. 96:51–62. 10.1083/jcb.96.1.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra P., and Chan D.C.. 2016. Metabolic regulation of mitochondrial dynamics. J. Cell Biol. 212:379–387. 10.1083/jcb.201511036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero M., Barrero M.J., and Alvarez J.. 1997. [Ca2+] microdomains control agonist-induced Ca2+ release in intact HeLa cells. FASEB J. 11:881–885. [DOI] [PubMed] [Google Scholar]
- Montero M., Lobatón C.D., Gutierrez-Fernández S., Moreno A., and Alvarez J.. 2003. Modulation of histamine-induced Ca2+ release by protein kinase C. Effects on cytosolic and mitochondrial [Ca2+] peaks. J. Biol. Chem. 278:49972–49979. 10.1074/jbc.M308378200 [DOI] [PubMed] [Google Scholar]
- Moore A.S., Wong Y.C., Simpson C.L., and Holzbaur E.L.. 2016. Dynamic actin cycling through mitochondrial subpopulations locally regulates the fission-fusion balance within mitochondrial networks. Nat. Commun. 7:12886 10.1038/ncomms12886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan A.J., and Jacob R.. 1994. Ionomycin enhances Ca2+ influx by stimulating store-regulated cation entry and not by a direct action at the plasma membrane. Biochem. J. 300:665–672. 10.1042/bj3000665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunnari J., and Suomalainen A.. 2012. Mitochondria: in sickness and in health. Cell. 148:1145–1159. 10.1016/j.cell.2012.02.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otera H., Miyata N., Kuge O., and Mihara K.. 2016. Drp1-dependent mitochondrial fission via MiD49/51 is essential for apoptotic cristae remodeling. J. Cell Biol. 212:531–544. 10.1083/jcb.201508099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H.K., Lee J.E., Lim J., and Kang B.H.. 2014. Mitochondrial Hsp90s suppress calcium-mediated stress signals propagating from mitochondria to the ER in cancer cells. Mol. Cancer. 13:148 10.1186/1476-4598-13-148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patten D.A., Wong J., Khacho M., Soubannier V., Mailloux R.J., Pilon-Larose K., MacLaurin J.G., Park D.S., McBride H.M., Trinkle-Mulcahy L., et al. 2014. OPA1-dependent cristae modulation is essential for cellular adaptation to metabolic demand. EMBO J. 33:2676–2691. 10.15252/embj.201488349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernas L., and Scorrano L.. 2016. Mito-Morphosis: Mitochondrial Fusion, Fission, and Cristae Remodeling as Key Mediators of Cellular Function. Annu. Rev. Physiol. 78:505–531. 10.1146/annurev-physiol-021115-105011 [DOI] [PubMed] [Google Scholar]
- Phillips M.J., and Voeltz G.K.. 2016. Structure and function of ER membrane contact sites with other organelles. Nat. Rev. Mol. Cell Biol. 17:69–82. 10.1038/nrm.2015.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querfurth H.W., Haughey N.J., Greenway S.C., Yacono P.W., Golan D.E., and Geiger J.D.. 1998. Expression of ryanodine receptors in human embryonic kidney (HEK293) cells. Biochem. J. 334:79–86. 10.1042/bj3340079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana-Cabrera R., Mehrotra A., Rigoni G., and Soriano M.E.. 2017. Who and how in the regulation of mitochondrial cristae shape and function. Biochem. Biophys. Res. Commun. 10.1016/j.bbrc.2017.04.088 [DOI] [PubMed] [Google Scholar]
- Ramabhadran V., Korobova F., Rahme G.J., and Higgs H.N.. 2011. Splice variant-specific cellular function of the formin INF2 in maintenance of Golgi architecture. Mol. Biol. Cell. 22:4822–4833. 10.1091/mbc.E11-05-0457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramabhadran V., Hatch A.L., and Higgs H.N.. 2013. Actin monomers activate inverted formin 2 by competing with its autoinhibitory interaction. J. Biol. Chem. 288:26847–26855. 10.1074/jbc.M113.472415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzuto R., Brini M., Murgia M., and Pozzan T.. 1993. Microdomains with high Ca2+ close to IP3-sensitive channels that are sensed by neighboring mitochondria. Science. 262:744–747. 10.1126/science.8235595 [DOI] [PubMed] [Google Scholar]
- Rizzuto R., Pinton P., Carrington W., Fay F.S., Fogarty K.E., Lifshitz L.M., Tuft R.A., and Pozzan T.. 1998. Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science. 280:1763–1766. 10.1126/science.280.5370.1763 [DOI] [PubMed] [Google Scholar]
- Shao X., Li Q., Mogilner A., Bershadsky A.D., and Shivashankar G.V.. 2015. Mechanical stimulation induces formin-dependent assembly of a perinuclear actin rim. Proc. Natl. Acad. Sci. USA. 112:E2595–E2601. 10.1073/pnas.1504837112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J.S., Coronado R., and Meissner G.. 1986. Single channel measurements of the calcium release channel from skeletal muscle sarcoplasmic reticulum. Activation by Ca2+ and ATP and modulation by Mg2+. J. Gen. Physiol. 88:573–588. 10.1085/jgp.88.5.573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stan R.V., Tse D., Deharvengt S.J., Smits N.C., Xu Y., Luciano M.R., McGarry C.L., Buitendijk M., Nemani K.V., Elgueta R., et al. 2012. The diaphragms of fenestrated endothelia: gatekeepers of vascular permeability and blood composition. Dev. Cell. 23:1203–1218. 10.1016/j.devcel.2012.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wales P., Schuberth C.E., Aufschnaiter R., Fels J., García-Aguilar I., Janning A., Dlugos C.P., Schäfer-Herte M., Klingner C., Wälte M., et al. 2016. Calcium-mediated actin reset (CaAR) mediates acute cell adaptations. eLife. 5:e19850 10.7554/eLife.19850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Mattson M.P., and Furukawa K.. 2002. Endoplasmic reticulum calcium release is modulated by actin polymerization. J. Neurochem. 82:945–952. 10.1046/j.1471-4159.2002.01059.x [DOI] [PubMed] [Google Scholar]
- Wu J., Prole D.L., Shen Y., Lin Z., Gnanasekaran A., Liu Y., Chen L., Zhou H., Chen S.R., Usachev Y.M., et al. 2014. Red fluorescent genetically encoded Ca2+ indicators for use in mitochondria and endoplasmic reticulum. Biochem. J. 464:13–22. 10.1042/BJ20140931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle R.J., and van der Bliek A.M.. 2012. Mitochondrial fission, fusion, and stress. Science. 337:1062–1065. 10.1126/science.1219855 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.