Woodruff previews work from Huang et al. proposing how phase separtaion of a scaffold protein could facilitate spindle assembly.
Abstract
The spindle matrix has been proposed to facilitate mitotic spindle assembly. In this issue, Huang et al. (2018. J. Cell Biol. https://doi.org/10.1083/jcb.201706103) show that the spindle matrix protein BuGZ is sufficient to form micron-scale compartments that recruit and activate Aurora A, a critical kinase for spindle assembly.
Somatic cells assemble a proteinacious structure called the mitotic spindle to properly segregate replicated DNA during cell division. The mitotic spindle is a micron-scale supramolecular structure largely composed of microtubules that emanate from two opposite poles and connect to chromosomes via kinetochores. Numerous microtubule-associated proteins (MAPs) such as motor proteins, microtubule polymerases, and microtubule depolymerases organize the spindle microtubules and determine mitotic spindle size. Adding to its complexity, the mitotic spindle is incredibly dynamic and regulated: it is assembled, elongated, and summarily disassembled each cell cycle. A looming question in the field of cell biology has been how cells coordinate mitotic spindle assembly with the cell cycle.
Mitotic spindle assembly has been classically described as the collaboration of three pathways (Petry, 2016). In the first pathway, called search-and-capture, centrosomes (or nuclear pole bodies in yeast) nucleate microtubules that then grow outward and connect to kinetochores anchored on the chromosomes. In the second pathway, the chromosomes nucleate microtubules, which through the help of MAPs, self-organize into a bipolar structure. In the third pathway, called branching, the microtubules within the spindle secure γ-tubulin complexes that nucleate additional microtubules.
Emerging evidence suggests that a fourth spindle assembly pathway may exist that utilizes molecules called spindle matrix proteins. In most eukaryotes, the mitotic spindle assembles a few minutes after nuclear envelope breakdown. In the interim between nuclear envelope fenestration and spindle assembly, tubulin often rushes into the nucleoplasm and concentrates there above normal cytosolic levels, suggesting the existence of a nucleoplasmic scaffold that binds tubulin (Fig. 1; Yao et al., 2012). It was thought that this scaffold could assist mitotic spindle assembly, and thus it earned the name spindle matrix. Proteins were then identified that supported this concept: megator and chromator (Drosophila melanogaster) as well as BuGZ (Xenopus laevis and human) surrounded the spindle without appearing to be part of it, and they remained organized in a subnuclear compartment after drug-induced spindle disassembly (Ding et al., 2009; Lince-Faria et al., 2009; Ma et al., 2009). Importantly, depletion of these proteins impaired mitotic spindle assembly. How these proteins assist spindle assembly and whether they actually form a distinct matrix remained unclear.
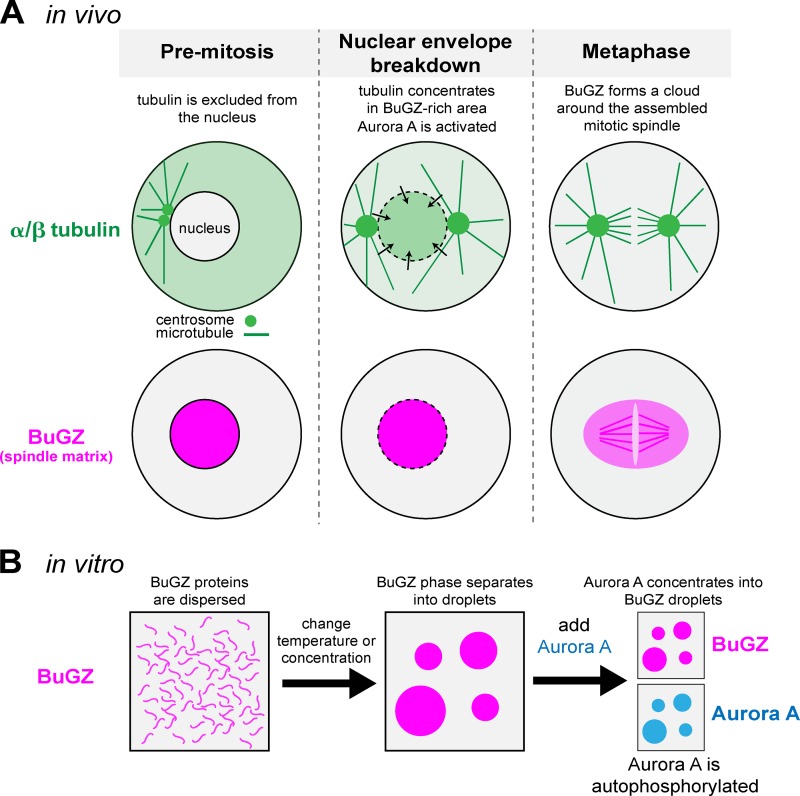
Figure 1.
BuGZ self-assembly facilitates mitotic spindle formation. (A) Diagram depicting the localization of α/β tubulin and the spindle matrix protein BuGZ in a vertebrate cell entering mitosis. Before mitosis, tubulin is largely excluded from the nucleus. During nuclear envelope breakdown, α/β tubulin dimers enter the nucleoplasmic space and concentrate there above cytoplasmic levels presumably by interacting with BuGZ. The interaction of BuGZ with Aurora A kinase leads to Aurora A activation and mitotic spindle assembly. During metaphase, BuGZ binds weakly to microtubules and also forms a cloud surrounding the mitotic spindle. (B) Diagram depicting BuGZ assembly in vitro. In low temperatures or low protein concentrations, BuGZ exists in an unassembled state. Increasing the temperature or protein concentration triggers phase separation of BuGZ into micron-scale droplets. BuGZ droplets concentrate purified Aurora A kinase and enhance its autophosphorylation, which activates the kinase.
A significant breakthrough in understanding the spindle matrix came when Jiang et al. (2015) found that BuGZ is sufficient to assemble into micron-scale droplets in vitro. BuGZ droplets were sufficient to concentrate tubulin ∼10-fold over background, and mutations that prevented BuGZ assembly or tubulin recruitment inhibited spindle assembly in a Xenopus extract system. Intriguingly, BuGZ droplets were spherical and would often wet the surface of microtubules; similar liquid-like properties have been seen in dynamic biomolecular condensates such as germ granules and Tau droplets (Brangwynne et al., 2009; Hernández-Vega et al., 2017). These results suggested that BuGZ assembly through liquid–liquid phase separation could form the basis of the spindle matrix, which would then promote spindle assembly by concentrating tubulin.
In this issue, Huang et al. suggest that the spindle matrix assists spindle assembly by locally enhancing the activity of Aurora A kinases. Aurora A is activated primarily in mitosis and promotes spindle assembly by phosphorylating centrosome proteins and MAPs that regulate microtubule growth (e.g., TPX2 and TACC3) and shrinkage (e.g., MCAK). One mechanism of Aurora A activation involves trans-autophosphorylation of its T-loop (threonine 295 or threonine 288 in Xenopus or human Aurora A, respectively). However, the interaction between two Aurora A proteins is very weak (Kd > 300 µM), suggesting that most Aurora A would be inactive in physiological conditions, where concentrations exist in the low-micromolar range (e.g., Aurora A has a concentration of ∼1.2 µM in Caenorhabditis elegans embryos; Saha et al., 2016). Huang et al. (2018) demonstrate that BuGZ droplets are sufficient to concentrate purified Aurora A kinase and increase T-loop phosphorylation in vitro (Fig. 1). They then identified a zinc finger domain on BuGZ that is necessary and sufficient to bind Aurora A. Replacement of endogenous BuGZ with the zinc finger mutant reduced Aurora A activation in Xenopus egg extracts as judged by T-loop phosphorylation.
Recent research demonstrates how subcellular compartmentalization can be achieved via demixing of proteins from the cytoplasm, a process termed protein phase separation (Banani et al., 2017). The work of Huang et al. (2018) suggests that BuGZ phase separates to form such a compartment in the nucleoplasm, which could be the elusive spindle matrix. This compartment would in theory help initiate spindle assembly by (A) attracting elements needed for spindle assembly, such as tubulin and Aurora A, and (B) activating Aurora A. Exactly how the condensed BuGZ phase could activate Aurora A is still unclear. The simplest mechanism would be Aurora A concentration, which would enhance Aurora A dimerization and trans-autophosphorylation. Although this may in part explain Aurora A activation in vitro, it likely doesn’t explain Aurora A activation in vivo, as BuGZ depletion did not change the levels of Aurora A on the spindle relative to spindle microtubules (Huang et al., 2018). If the condensed BuGZ phase is more crowded or viscous than the surrounding cytoplasm, diffusion of Aurora A could be affected; this scenario would favor trans-autophosphorylation, as it is likely a transition state–limited reaction. The condensed BuGZ phase might also exclude phosphatases that dephosphorylate the T-loop on Aurora A.
Determining the mechanism by which BuGZ activates Aurora A is a clear next step in understanding spindle assembly. However, a more important step will be to determine whether the spindle matrix in vivo behaves like a phase-separated compartment as predicted from in vitro experiments using purified BuGZ.
Acknowledgments
J.B. Woodruff was supported by funding from the Max Planck Research Network in Synthetic Biology.
The author declares no competing financial interests.
References
- Banani S.F., Lee H.O., Hyman A.A., and Rosen M.K.. 2017. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18:285–298. 10.1038/nrm.2017.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne C.P., Eckmann C.R., Courson D.S., Rybarska A., Hoege C., Gharakhani J., Jülicher F., and Hyman A.A.. 2009. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 324:1729–1732. 10.1126/science.1172046 [DOI] [PubMed] [Google Scholar]
- Ding Y., Yao C., Lince-Faria M., Rath U., Cai W., Maiato H., Girton J., Johansen K.M., and Johansen J.. 2009. Chromator is required for proper microtubule spindle formation and mitosis in Drosophila. Dev. Biol. 334:253–263. 10.1016/j.ydbio.2009.07.027 [DOI] [PubMed] [Google Scholar]
- Hernández-Vega A., Braun M., Scharrel L., Jahnel M., Wegmann S., Hyman B.T., Alberti S., Diez S., and Hyman A.A.. 2017. Local Nucleation of Microtubule Bundles through Tubulin Concentration into a Condensed Tau Phase. Cell Reports. 20:2304–2312. 10.1016/j.celrep.2017.08.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Li1 T., Ems-McClung S.C., Walczak C.E., Prigent C., Zhu X., Zhang X., and Zheng Y.. 2018. Aurora A activation in mitosis promoted by BuGZ. J. Cell Biol. 217:93–102. 10.1083/jcb.201706103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Wang S., Huang Y., He X., Cui H., Zhu X., and Zheng Y.. 2015. Phase transition of spindle-associated protein regulate spindle apparatus assembly. Cell. 163:108–122. 10.1016/j.cell.2015.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lince-Faria M., Maffini S., Orr B., Ding Y., Florindo C., Sunkel C.E., Tavares A., Johansen J., Johansen K.M., and Maiato H.. 2009. Spatiotemporal control of mitosis by the conserved spindle matrix protein Megator. J. Cell Biol. 184:647–657. 10.1083/jcb.200811012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Tsai M.-Y., Wang S., Lu B., Chen R., Yates J.R. III, Zhu X., and Zheng Y.. 2009. Requirement for Nudel and dynein for assembly of the lamin B spindle matrix. Nat. Cell Biol. 11:247–256. 10.1038/ncb1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petry S. 2016. Mechanisms of Mitotic Spindle Assembly. Annu. Rev. Biochem. 85:659–683. 10.1146/annurev-biochem-060815-014528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S., Weber C.A., Nousch M., Adame-Arana O., Hoege C., Hein M.Y., Osborne-Nishimura E., Mahamid J., Jahnel M., Jawerth L., et al. 2016. Polar Positioning of Phase-Separated Liquid Compartments in Cells Regulated by an mRNA Competition Mechanism. Cell. 166:1572–1584. 10.1016/j.cell.2016.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao C., Rath U., Maiato H., Sharp D., Girton J., Johansen K.M., and Johansen J.. 2012. A nuclear-derived proteinaceous matrix embeds the microtubule spindle apparatus during mitosis. Mol. Biol. Cell. 23:3532–3541. 10.1091/mbc.E12-06-0429 [DOI] [PMC free article] [PubMed] [Google Scholar]