Abstract
Objective
The objective of this study was to determine the frequency of the antimicrobial resistance and genes encoding virulence factors of enterococci isolated in hospitalized burn patients in a major burn center in Ahvaz, southwest of Iran. A total of 340 bacterial isolates were collected from the burn center from February 2014 to February 2015. The antimicrobial susceptibility and MIC of vancomycin were determined using the disk diffusion and micro-agar dilution techniques. The genus and species-specific genes, potential virulence genes, and vanA and vanB genes were detected by polymerase chain reaction.
Results
According to our results, out of the 340 bacterial isolates, 16.4% (n = 56) were identified as enterococci. Out of the 56 enterococcal isolates, 35 (62.5%) were Enterococcus faecalis and 21 (37.5%) were Enterococcus faecium. More than 20% (n = 5) of E. faecium demonstrated resistance to vancomycin. The gelE and asa genes were the most prevalent virulence genes in E. faecalis (48.5%) and E. faecium (43%) isolates. The emergence of vancomycin resistant E. faecium strains which have several virulence factors should be considered as a major cause of concern for burn centers. Control and management of infections induced by enterococci should be regarded as highly important in burn patients.
Keywords: Enterococci, Vancomycin resistant, Virulence genes, Burn
Introduction
Nosocomial infections are the most common factors in increasing morbidity, mortality, long-term treatment, and cost in hospitalized patients [1, 2]. Burn patients are more at the risk of complications of nosocomial infections due to their weakened immune system [2]. In many countries, particularly developing countries including Iran, burn infections are one of the most serious issues in the field of health [3]. Organisms associated with nosocomial infections in burn patients include the patient’s own normal flora organisms and the hospital environment or the staff. Bacterial pathogens are the most common cause of burn infections such as wounds in burn patients. In addition to wound infections, bacterial pneumonia and bloodstream infections are also common causes of death in burn patients [4].
Among Gram-positive and Gram-negative bacteria, Staphylococcus aureus and Pseudomonas aeruginosa are considered as the most important factors causing burn wound infections [5]. According to reports published in recent years, enterococci have become important factors in acquiring nosocomial infections [6–8]. These Gram-positive bacteria are part of the normal flora of the human digestive system and were considered to be commensal bacteria in the past [6]. Since enterococci have the ability to cause infection in wounds, including burn wounds, and are intrinsically resistant to many antibiotics due to their ability to acquire antibiotic resistance genes, they are considered as one of the most important causes of nosocomial infections. Enterococci are now regarded as the third most common cause of hospital infections. Enterococci can cause urinary tract, intra-abdominal, pelvic, wound, and super infections (including those caused by burns) in patients [6, 9]. The emergence of multidrug-resistant enterococci strains (resistant to two or more classes of antibiotics) or MDR has caused numerous problems for patients in hospitals all over the world [10]. In Enterococcus species, E. faecium and E. faecalis are regarded as important nosocomial bacterial pathogens. The published reports of some countries indicated the prevalence of burn wound infections caused by VRE strains in hospitals, particularly in the intensive care unit (ICU) [11]. Studies on bacteria and burn infections have been conducted in Taleghani Hospital of Ahvaz as the main burn center in Khuzestan Province, southwest of Iran [12, 13]. Only a few studies have been published on the role of enterococci in burn patients in Iran, and given the increasing importance of VRE-related infections, the aim of the present study was to evaluate the frequency of enterococcal infections, virulence factors, and antibiotic resistance patterns in enterococci strains isolated from clinical samples of burn patients hospitalized in one of the largest burn centers in the southwest of Iran.
Main text
Methods
The present study was ethically approved by Ahvaz Jundishapur University of Medical Sciences, Institutional Review Board (Code No. 93146).
A total of 340 bacterial strains were isolated from clinical samples of hospitalized burn patients (wound biopsies, blood, and urine) in a burn center in Ahvaz from February 2014 to February 2015. All the enterococci isolates were identified in the genus and species levels by Gram staining, catalase reaction, growth in 6.5% NaCl solution, motility assessment, use of arabinose, bile, and esculin hydrolysis tests [14]. Furthermore, a PCR-based study was conducted using specific primers (ddl E. faecium and ddl E. faecalis) for all E. faecium and E. faecalis isolates [15]. Antimicrobial susceptibility test for enterococci isolates was performed against vancomycin (30 μg), teicoplanin (30 μg), gentamicin (120 μg), ampicillin (10 μg), ciprofloxacin (5 μg), tetracycline (30 μg), chloramphenicol (30 μg), and linezolid (30 μg) (Mast, UK) by the disk diffusion technique. Vancomycin minimum inhibitory concentration (MICs) was detected by micro-agar dilution method based on the CLSI guidelines [17]. DNAs from different enterococcal isolates were extracted using an appropriate DNA Extraction Kit (Cinagene, Iran). Specific primers of ddl E. faecium, ddl E. faecalis vancomycin resistant genes (vanA and vanB), and virulence genes including hyaluronidase (hyl), enterococci surface protein (esp), haemolysin activator (cyl), gelatinase (gelE), and aggregation substance asaI (Metaboin, Germany) were used for PCR amplification as described previously [14, 16, 18].
Results and discussion
Prevalence of enterococci isolates
Enterococci were isolated from 16.4% (n = 56) of clinical specimens of burn hospitalized patients. The frequency of enterococci in wound, blood, and urine specimens were 37.5% (n = 21), 30.3% (n = 17), and 35% (n = 18), respectively. Among the 56 enterococci strains, 62.5% (n = 35) were identified as E. faecalis and 37.5% (n = 21) were identified as E. faecium. Table 1 shows the comparison of the frequency distribution of E. faecium and E. faecalis in different clinical specimens. Among Enterococcus species, E. faecalis was identified as the major cause of enterococcal infections. E. faecalis caused enterococcal infections approximately ten times more than other Enterococcus species [19]. E. faecalis has more virulence factors than E. faecium; consequently, it has higher pathogenicity [20]. Nevertheless, in recent years a microbial shift from E. faecium to E. faecalis has been observed which could be due to the emergence of MDR strains resistant to vancomycin (VRE) species in hospitals [19, 21]. One of the influential factors in increasing nosocomial infections resulted from enterococci is the emergence of VREs in hospitals. According to the studies conducted in Iran, nosocomial infections caused by enterococci, particularly the resistant strains are highly prevalent [10, 21]. In 2001, the National Nosocomial Infection Surveillance System (NNISS) reported that the frequency of burn wound infections caused by Enterococcus was 11% [2] which is close to the results of the current study (16%). There have been few studies investigating the existence of infection caused by enterococci in burn patients. Our results showed an increase in the contribution of E. faecium to infections which is consistent with our results on the reported frequency ratio of E. faecium to E. faecalis in Tehran’s hospitals in 2014. Although these results are different from other results reported in Iran and other countries, more than 80% of burn wound infections caused by Enterococcus was due to E. faecalis [6, 22–24].
Table 1.
Frequency distribution of E. faecalis and E. faecium isolates in clinical specimens of burn patients
| Urine | Wound | Blood | |
|---|---|---|---|
| E. faecalis (n = 35) | 26% (9) | 37% (13) | 37% (13) |
| E. faecium (n = 21) | 33% (7) | 47% (10) | 19% (4) |
Antimicrobial resistance patterns
Figure 1 presents the comparison of the results of antimicrobial susceptibility analysis of E. faecium and E. faecalis. More than 20% (n = 5) of E. faecium exhibited resistance to vancomycin and teicoplanin. This can be the reason for the increased presence of E. faecium which is resistant to vancomycin and can survive in the hospital environment. E. faecium strains exhibited higher levels of resistance to antibiotics. Higher levels of resistance to tetracycline and ampicillin were observed in 71% (n = 15) and 66% (n = 14) of E. faecium isolates, respectively. According to the findings, this can be an alarm for burn patients owing to the fact that more than 50% of E. faecium strains and 100% of VRE strains were resistant to ampicillin and gentamicin. No resistance to linezolid and vancomycin was observed in E. faecalis isolates. The most frequent resistance pattern was simultaneous resistance to ampicillin/tetracycline/gentamicin (AMP/TET/GEM) in 43% of E. faecium and tetracylin/gentamicin (TET/GEM) in 46% of E. faecalis isolates. All vancomycin resistant E. faecium (VREfm) possessed vanA gene, while no vanB gene was observed in VREfm strains. On the other hand, all VRE strains which possessed vanA gene showed resistance both to vancomycin and teicoplanin. MIC values for vancomycin ranged from 32 to 1024 µg/ml. In addition, there was no correlation between the presence of vancomycin resistance, virulence factors, and the clinical samples. So far, no documented results for the presence of VRE in burn patients have been published in Iran which may be attributed to the lack of expensive and purposive studies on enterococci in the burn units. However, some countries have reported VREs in burn units [25–28]. According to the results of this study and similar studies, linezolid was identified as the most effective antibiotic against enterococcal infections [10, 22, 23].
Fig. 1.
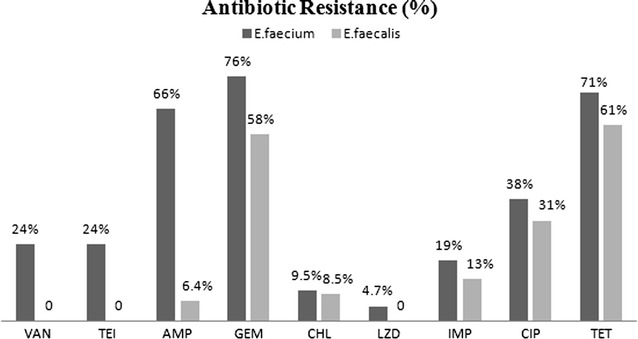
The comparison of antibiotic resistance (%) in E. faecium and E. faecalis isolates in burn patients
Distribution of virulence genes
Table 2 presents the frequency and distribution of virulence genes among clinical specimens. The gelE and asa genes were the most frequent virulence genes in E. faecalis (48.5%) and E. faecium (43%) isolates, respectively. This is consistent with the results reporting gelE as the most frequent virulence factor in E. faecalis strains [23, 25, 29], while some reports have indicated the absence or low rate of gelE gene in enterococcal isolates [25, 30, 31]. Some reports have indicated the absence or low rate of asa gene in E. faecium strains [31, 32]. The cyl gene was not detected in any E. faecium isolates. The hyl gene was found in 1.7% (n = 1) of E. faecium isolates. A total of 68.5% (n = 24) of E. faecalis isolates and 42.8% (n = 9) of E. faecium isolates were positive for more than one virulence gene. There was no relationship between virulence genes and clinical specimens.
Table 2.
Frequency distribution of virulence gene in E. faecalis and E. faecium isolates
| esp | gel | asa | hyl | cyl | |
|---|---|---|---|---|---|
| E. faecium/E. faecalis | E. faecium/E. faecalis | E. faecium/E. faecalis | E. faecium/E. faecalis | E. faecium/E. faecalis | |
| Blood | 4.7% (1): 16% (5) | 0%: 26% (9) | 14% (3): 8.5% (3) | 0%: 17% (6) | 0%: 0% |
| Urine | 9.5% (2): 13% (4) | 14% (3): 6% (2) | 14% (3): 20% (7) | 0%: 8.5% (3) | 0: 2.8% (1) |
| Wound | 4.7% (1): 8.5% (3) | 19% (4): 17% (6) | 14% (3): 20% (7) | 4.7% (1): 6% (2) | 0: 2.8% (1) |
| Total | 19% (4): 34% (12) | 33% (7): 48.5% (17) | 43% (9): 48.5% (17) | 4.7% (1): 31% (11) | 0: 5.6% (2) |
In conclusion, based on the results of the current study, it was demonstrated that the detection of enterococci in clinical samples of burn patients is highly important. In addition, the presence of E. faecium strains resistant to vancomycin with several virulence factors can be a source of alarm for burn centers. Considering the possible transmission of antibiotic resistance genes among enterococci, and staphylococci, the control and management of infections caused by enterococci as well as the appropriate administration of antibiotics in burn patients can be highly effective in treating nosocomial infections in burn centers.
Limitations
In this study, no significant differences exist between the presence of virulence genes and resistance to vancomycin. One of the reasons for this result may be the low number of vancomycin resistant strains. Therefore, more samples must be collected from hospitals in general and from burn centers in particular.
Authors’ contributions
LS and AE conceived the study. LS and ML conducted the experiments and analyzed the results. LA and LS drafted the manuscript and made substantial contributions to the design of the study. LA, AE, and SMA critically reviewed the manuscript. LS, SMA and ML, participated in data analysis. All authors read and approved the final manuscript.
Acknowledgements
We would like to thank all members of microbiology laboratory of Taleghani Hospital in Ahvaz.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
All the information supporting our conclusions and appropriate references are included in the manuscript.
Consent for publication
Not applicable.
Ethics approval and consent to participate
The present study was ethically approved by the Ahvaz Jundishapur University of Medical Sciences, Institutional Review Board (Code No. 93146). The Samples were accessed retrospectively. Permission was granted from the hospital and Ahvaz Jundishapur University of Medical Sciences to access the samples.
Funding
This research was funded by a grant from infection and tropical disease research centers, Jundishapur University of Medical Sciences, Ahvaz.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- MIC
minimum inhibition concentration
- E. faecium
Enterococcus faecium
- E. faecalis
Enterococcus faecalis
- MDR
multi-drug resistant
- VRE
vancomycin resistant enterococci
- PCR
polymerase chain reaction
- hyl
hyaluronidase
- esp
enterococci surface protein
- cyl
haemolysin activator
- gleE
gelatinase
- capD
capsular polysaccharides biosynthesis protein
- asaI
aggregation substance
- VAN
vancomycin
- TEI
teicoplanin
- AMP
ampicillin
- GEM
gentamicin
- CHL
chloramphenicol
- LZD
linezolid
- IMP
imipenem
- CIP
ciprofloxacin
- TET
tetracycline
Contributor Information
Leili Shokoohizadeh, Email: shokoohizadeh@yahoo.com.
Alireza Ekrami, Phone: + 98983373825, Phone: +989166064072, Email: aekrami@yahoo.com, Email: ekrami@ajums.ac.ir.
Maryam Labibzadeh, Email: labibzadeh4@yahoo.com.
Liaqat Ali, Email: liaqatbiotech@gmail.com.
Seyed Mohammad Alavi, Email: alavi1329dr@yahoo.com.
References
- 1.Haas JP. Measurement of infection control department performance: state of the science. Am J Infect Control. 2006;34(9):543–549. doi: 10.1016/j.ajic.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 2.Mayhall GC. The epidemiology of burn wound infections: then and now. Clin Infect Dis. 2003;37:543–550. doi: 10.1086/376993. [DOI] [PubMed] [Google Scholar]
- 3.Azimi L, Motevallian A, Ebrahimzadeh Namvar A, Asghari B, Lari AR. Nosocomial infections in burned patients in Motahari Hospital, Tehran, Iran. Dermatol Res Pract. 2011. [DOI] [PMC free article] [PubMed]
- 4.D’Avignon LC, Hogan BK, Murray CK, Loo FL, Hospenthal DR, Cancio LC, et al. Contribution of bacterial and viral infections to attributable mortality in patients with severe burns: an autopsy series. Burns. 2010;36:73. doi: 10.1016/j.burns.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 5.Lawrence J. Burn bacteriology during the last 50 years. Burns. 1994;18:23–29. doi: 10.1016/0305-4179(92)90066-4. [DOI] [PubMed] [Google Scholar]
- 6.Sood S, Malhotra M, Das BK, Kapi A. Enterococcal infections & antimicrobial resistance. Indian J Med Res. 2008;128:111–121. [PubMed] [Google Scholar]
- 7.Lindenstrau AG, Pavlovic M, Bringmann A, Behr J, Ehrmann MA, Vogel RF. Comparison of genotypic and phenotypic cluster analyses of virulence determinants and possible role of CRISPR elements towards their incidence in Enterococcus faecalis and Enterococcus faecium. Syst Appl Microbiol. 2011;34:553–560. doi: 10.1016/j.syapm.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 8.Arshadi M, Douraghi M, Shokoohizadeh L, Moosavian SM, Pourmand MR. High prevalence of diverse vancomycin resistance Enterococcus faecium isolates in clinical and environmental sources in ICU wards in southwest of Iran. Microb Pathog. 2017;111:212–217. doi: 10.1016/j.micpath.2017.08.045. [DOI] [PubMed] [Google Scholar]
- 9.Willems RJ, Hanage WP, Bessen DE, Feil EJ. Population biology of Gram-positive pathogens: high-risk clones for dissemination of antibiotic resistance. FEMS Microbiol Rev. 2011;35(8):72–900. doi: 10.1111/j.1574-6976.2011.00284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shokoohizadeh L, Mobarez AM, Zali MR, Ranjbar R, Alebouyeh M, Sakinc T, et al. High frequency distribution of heterogeneous vancomycin resistant Enterococcus faecium (VREfm) in Iranian hospitals. Diagn pathol. 2013;8:163. doi: 10.1186/1746-1596-8-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.May AK, Melton SM, McGwin G, Cross JM, Moser SA, Rue LW. Reduction of vancomycin-resistant enterococcal infections by limitation of broad-spectrum cephalosporin use in a trauma and burn intensive care unit. Shock. 2000;14:259–264. doi: 10.1097/00024382-200014030-00003. [DOI] [PubMed] [Google Scholar]
- 12.Ekrami A, Hemadi A, Kalantar E, Latifi M, Kayedani A. Epidemiology of hospitalized burn patients during 5 years in Khuzestan province, Iran. Iran J Clin Infect Dis. 2010;5(1):40–44. [Google Scholar]
- 13.Ekrami A, Montazeri EA, Kaydani GA, Shokoohizadeh L. Methicillin resistant staphylococci: prevalence and susceptibility patterns in a burn center in Ahvaz from 2013–2014. Iran J Microbiol. 2015;7(4):208. [PMC free article] [PubMed] [Google Scholar]
- 14.Falkman RR, Collin MD. Identification of Enterococcus species isolated from human infections by a conventional test scheme. J Clin Microbiol. 1989;27:731–734. doi: 10.1128/jcm.27.4.731-734.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kariyama R, Mitsuhata R, Chow JW, Clewell DB, Kumon H. Simple and reliable multiplex PCR assay for surveillance, isolates of vancomycin resistant enterococci. J Clin Microbiol. 2000;38:3092–3095. doi: 10.1128/jcm.38.8.3092-3095.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Performance standards for antimicrobial susceptibility testing, vol. 31. 21th ed. Wayne: Clinical and Laboratory Standards Institute; 2011. p. 84–7.
- 17.Eaton TJ, Gasson MJ. Molecular screening of Enterococcus virulence determinants and potential for genetic exchange between food and medical isolates. Appl Environ Microbiol. 2001;67:1628–1635. doi: 10.1128/AEM.67.4.1628-1635.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vankerckhoven V, Van Autgaerden T, Vael C, Lammens C, Chapelle S, Rossi R, et al. Development of a multiplex PCR for the detection of asa1, gelE, cylA, esp, and hyl genes in enterococci and survey for virulence determinants among European hospital isolates of Enterococcus faecium. J Clin Microbiol. 2004;42:4473–4479. doi: 10.1128/JCM.42.10.4473-4479.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mundy LM, Sahm DF, Gilmore M. Relationships between enterococcal virulence and antimicrobial resistance. Clin Microbiol Rev. 2000;13:513–522. doi: 10.1128/CMR.13.4.513-522.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharifi Y, Hasani A, Ghotaslou R, Varshochi M, Hasani A, Aghazadeh M, Milani M. Survey of virulence determinants among vancomycin resistant Enterococcus faecalis and Enterococcus faecium isolated from clinical specimens of hospitalized patients of northwest of Iran. Open Microbiol J. 2012;6:34–39. doi: 10.2174/1874285801206010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lester CH, Sandvang D, Olsen SS, Schønheyder HC, Jarløv JO, Bangsborg J. Emergence of ampicillin-resistant Enterococcus faecium in Danish hospitals. J Antimicrob Chemother. 2008;62:1203–1206. doi: 10.1093/jac/dkn360. [DOI] [PubMed] [Google Scholar]
- 22.Sharifi Y, Hasani A, Ghotaslou R, Varshochi M, Hasani A, Soroush MH, Aghazadeh M, Milani M. Vancomycin-resistant enterococci among clinical isolates from north-west Iran: identification of therapeutic surrogates. J Med Microbiol. 2012;61:600–602. doi: 10.1099/jmm.0.036780-0. [DOI] [PubMed] [Google Scholar]
- 23.Heidari H, Emaneini M, Dabiri H, Jabalameli F. Virulence factors, antimicrobial resistance pattern and molecular analysis of Enterococcal strains isolated from burn patients. Microb Pathog. 2016;90:93–97. doi: 10.1016/j.micpath.2015.11.017. [DOI] [PubMed] [Google Scholar]
- 24.Emaneini MO, Aligholi MA, Aminshahi MA. Characterization of glycopeptides, aminoglycosides and macrolide resistance among Enterococcus faecalis and Enterococcus faecium isolates from hospitals in Tehran. Pol J Microbiol. 2008;57:173–178. [PubMed] [Google Scholar]
- 25.Sharifi Y, Hasani A, Ghotaslou R, Naghili B, Aghazadeh M, Milani M, Bazmany A. Virulence and antimicrobial resistance in enterococci isolated from urinary tract infections. Adv Pharm Bull. 2013;3:197–201. doi: 10.5681/apb.2013.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altoparlak U, Koca O, Ozkurt Z, Akcay MN. Incidence and risk factors of vancomycin-resistant enterococcus colonization in burn unit patients. Burns. 2011;37:149–153. doi: 10.1016/j.burns.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 27.Wibbenmeyer L, Williams I, Ward M, Xiao X, Light T, Latenser B, et al. Risk factors for acquiring vancomycin-resistant Enterococcus and methicillin-resistant Staphylococcus aureus on a burn surgery step-down unit. J Burn Res. 2010;31:269–279. doi: 10.1097/BCR.0b013e3181d0f479. [DOI] [PubMed] [Google Scholar]
- 28.Falk PS, Winnike J, Woodmansee C, Desai M, Mayhall CG. Outbreak of vancomycin-resistant enterococci in a burn unit. Infect Control Hosp Epidemiol. 2000;21:575–582. doi: 10.1086/501806. [DOI] [PubMed] [Google Scholar]
- 29.Sabia C, De Niederhäusern S, Guerrieri E, Messi P, Anacarso I, Manicardi G, et al. Detection of bacteriocin production and virulence traits in vancomycin-resistant enterococci of different sources. J Appl Microbiol. 2008;104(4):970–979. doi: 10.1111/j.1365-2672.2007.03612.x. [DOI] [PubMed] [Google Scholar]
- 30.Waar K, Muscholl-Silberhorn AB, Willems RJ, Slooff MJ, Harmsen HJ, Degener JE. Genogrouping and incidence of virulence factors of Enterococcus faecalis in liver transplant patients differ from blood culture and fecal isolates. J Infect Dis. 2002;185(8):1121–1127. doi: 10.1086/339682. [DOI] [PubMed] [Google Scholar]
- 31.Hällgren A, Claesson C, Saeedi B, Monstein HJ, Hanberger H, Nilsson LE. Molecular detection of aggregation substance, enterococcal surface protein, and cytolysin genes and in vitro adhesion to urinary catheters of Enterococcus faecalis and E. faecium of clinical origin. Int J Med Microbiol. 2009;299(5):323–332. doi: 10.1016/j.ijmm.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 32.Billström H, Sullivan Å, Lund B. Cross-transmission of clinical Enterococcus faecium in relation to esp and antibiotic resistance. J Appl Microbiol. 2008;15:2115–2122. doi: 10.1111/j.1365-2672.2008.03983.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the information supporting our conclusions and appropriate references are included in the manuscript.


