Abstract
Nonsense-mediated RNA decay (NMD) is a highly conserved and selective RNA turnover pathway that has been subject to intense scrutiny. NMD identifies and degrades subsets of normal RNAs, as well as abnormal mRNAs containing premature termination codons. A core factor in this pathway—UPF3B—is an adaptor protein that serves as an NMD amplifier and an NMD branch-specific factor. UPF3B is encoded by an X-linked gene that when mutated causes intellectual disability and is associated with neurodevelopmental disorders, including schizophrenia and autism. Neu-Yilik et al. now report a new function for UPF3B: it modulates translation termination. Using a fully reconstituted in vitro translation system, they find that UPF3B has two roles in translation termination. First, UPF3B delays translation termination under conditions that mimic premature translation termination. This could drive more efficient RNA decay by allowing more time for the formation of RNA decay-stimulating complexes. Second, UPF3B promotes the dissociation of post-termination ribosomal complexes that lack nascent peptide. This implies that UPF3B could promote ribosome recycling. Importantly, the authors found that UPF3B directly interacts with both RNA and the factors that recognize stop codons—eukaryotic release factors (eRFs)—suggesting that UPF3B serves as a direct regulator of translation termination. In contrast, a NMD factor previously thought to have a central regulatory role in translation termination—the RNA helicase UPF1—was found to indirectly interact with eRFs and appears to act exclusively in post-translation termination events, such as RNA decay, at least in vitro. The finding that an RNA decay-promoting factor, UFP3B, modulates translation termination has many implications. For example, the ability of UPF3B to influence the development and function of the central nervous system may be not only through its ability to degrade specific RNAs but also through its impact on translation termination and subsequent events, such as ribosome recycling.
Keywords: Nonsense-mediated RNA decay, RNA, UPF3B, translation termination
Introduction
Gene expression is typically thought to be controlled at the level of RNA synthesis (transcription), but equally critical for determining steady-state RNA levels is the rate of RNA turnover. Among the best-characterized eukaryotic RNA turnover mechanisms is nonsense-mediated RNA decay (NMD), a highly conserved and selective pathway that detects and degrades mRNAs with translation stop codons positioned in specific (often suboptimal) contexts 1– 5. Originally regarded as a quality-control mechanism that rapidly degrades aberrant mRNAs with premature termination codons (PTCs), it is now clear that NMD also degrades many normal RNAs 5. Just as transcription factors regulate the rate of RNA synthesis from subsets of normal genes, NMD regulates the stability of subsets of normal mRNAs. While the physiological significance of this is not certain, increasing evidence strongly suggests that NMD is a highly regulated pathway that degrades subsets of mRNAs to influence specific biological events, including during development and in response to stress 3, 5.
Many proteins have been identified that function in NMD, including those that act in specific branches of the NMD pathway 2. One of these branch-specific factors is UPF3B, which has been shown to promote the decay of specific mRNAs in a cell type-specific manner 6– 10. In a recent article in the EMBO Journal, Neu-Yilik et al. 11 report that UPF3B has a new role. They demonstrate that UPF3B modulates specific steps in translation termination ( Figure 1). Remarkably, they provide evidence that UPF3B has a central role in translation termination, as it interacts with ribosomes, it binds to RNA, and it directly interacts with release factors (RFs), the proteins that recognize stop codons in order to trigger translation termination ( Figure 2 and Supplemental Figure 1). These discoveries significantly alter the view we have of UPF3B and they have implications for how we view the multiple steps of gene expression. The authors’ findings deepen our understanding of the underlying mechanism of translation termination and have potential consequences in terms of biology and neural disease, as we detail below.
Figure 1. UPF3B regulates translation termination.
In eukaryotes, translation termination proceeds through four consecutive stages. Using a reconstituted in vitro translation termination system, Neu-Yilik et al. 11 provided evidence that UPF3B modulates all four of these stages. They obtained evidence that UPF3B delays the middle stages, which may serve to regulate translation termination in RNAs with stop codons in ‘premature’ contexts. By contrast, UPF3B accelerates ribosome recycling, which may serve many functions, including facilitating the exposure of the mRNA body to nuclease attack during NMD. Of note, steps 2 and 3 are difficult to distinguish and thus it is possible that UPF3B acts on only one of these two steps. eRF, eukaryote release factor.
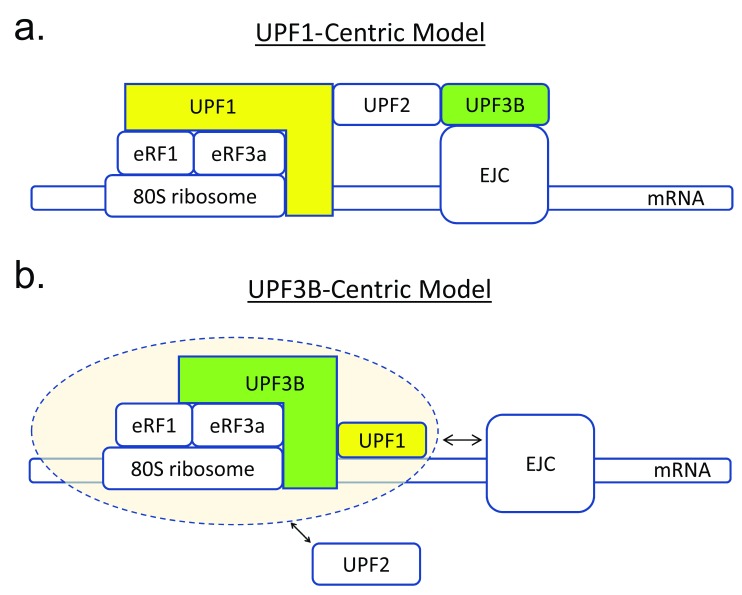
Figure 2. Two models that explain coupling of translation termination and nonsense-mediated RNA decay (NMD).
( a) In one model, UPF1 is the major factor molecularly connecting translation termination with NMD 1, 22. UPF1 interacts (directly or indirectly) with release factors (eRF1 and eRF3A) bound to the terminating ribosome stalled at a premature stop codon (PTC), based on co-immunoprecipitation analysis. UPF1 then recruits the adaptor protein, UPF2, which, in turn, binds to UPF3B, which is recruited to RNA by the exon junction complex (EJC) bound at splice junctions. UPF2 induces a conformational change in UPF1, which activates it for subsequent events that lead to RNA decay. ( b) In the UPF3B-centric model from Neu-Yilik et al. 11, UPF3B serves as an active assembly mediator that brings the translation termination machinery and the NMD machinery together (pink oval). Neu-Yilik et al. 11 found that UPF3B directly binds to RNA and interacts with ribosomes, which, according to their model, allows it to be loaded directly onto translated RNAs. At a PTC, UPF3B recruits the eRF1-eRF3-GTP complex, which leads to delayed translation termination. Neu-Yilik et al. 11 also demonstrated that UPF3B directly binds to UPF1 (without the necessity for UPF2 to serve as an adaptor), providing a mechanism by which UPF2-independent NMD occurs 28. It remains for future studies to determine how UPF2 and the EJC functionally interact (double-head arrows) with the NMD and translation termination machineries.
NMD
NMD is a translation-dependent pathway triggered by in-frame stop codons in specific contexts. The core components of the NMD pathway—UPF1, UPF2, and UPF3 ( Figure 2)—are conserved in all known eukaryotes. UPF1 is an RNA helicase that processively remodels RNA and ribonucleoproteins (RNPs) 12. UPF2 serves as an adaptor protein that bridges UPF1 with UPF3 1. In vertebrates, there are two UPF3 proteins with different functions: UPF3B is an NMD activator encoded by an X-linked gene that promotes a specific branch of the NMD pathway 2, 6, 13– 15, while UPF3A is a weak NMD factor encoded by an autosomal gene that recently was shown to be predominantly an NMD repressor in many cell types 14, 16, 17. In mammals, UPF3B interact with the exon junction complex (EJC), a set of proteins deposited just upstream of exon-exon junctions after RNA splicing 13. The EJC is thought to be responsible for the fact that in-frame stop codons more than about 50 nucleotides upstream of the last exon-exon junction almost always trigger NMD. The likely mechanism responsible for this “-50 boundary rule” is that EJCs bound at exon-exon junctions more than about 50 nucleotides downstream of the stop codon are not displaced by ribosomes (given the footprint lengths of the EJC and ribosome) and thus such bound EJCs are able to recruit factors, including UPF3B, which elicit RNA decay through the NMD pathway 3, 13.
Translation termination
The termination of translation in eukaryotes requires the release factors (that is, eukaryotic RF1 (eRF1) and eRF3). When a translating ribosome reaches a stop codon on an mRNA, the ribosome recruits eRF1 in a complex with the GTPase, eRF3, bound by GTP. eRF1 recognizes the stop codon by virtue of its mimicking the shape of tRNA. Stop codon recognition triggers eRF3 to hydrolyse GTP, leading to a conformational change of eRF1, peptidyl-tRNA hydrolysis, and dissociation of the nascent polypeptide and eRF3 from the ribosome. The essential ATP-binding cassette protein, ABCE1, then splits 80S ribosomes into free 60S subunits and 40S subunits 18, 19, followed by the release of the ribosomal subunits from the mRNA 20.
The NMD factor, UPF3B, functions in translation termination
While normal translational termination is well understood, the mechanisms underlying premature translation termination remain relatively obscure. Prevailing models for the mechanics of premature translation termination ascribe a critical role for the NMD factor UPF1 21– 23. In support, it has been found that genetic ablation of UPF1 in yeast leads to increased stop codon suppression 24, 25 and RNA interference–mediated depletion of UPF1 in human cells reduces stop codon read-through 26. In support of a direct role, UPF1 was found to interact with both eRF1 and eRF3A, based on co-immunoprecipitation (co-IP) experiments ( Figure 2a) 25– 27. These and other findings support a model in which UPF1 recruits eRFs to ribosomes that are stalled at a premature termination codon in an early phase of termination and then UPF1 promotes ribosome disassembly in a late phase of termination via its ATPase function, activated by UPF2 and UPF3 binding 21, 22, 29 ( Figure 2a).
While an attractive model, it has not been possible to experimentally test this model because there is no reliable in vivo termination assay. Thus, to test the validity of this model, Neu-Yilik et al. 11 chose instead to use a fully reconstituted in vitro mammalian system that recapitulates all phases of translation termination 30– 32. Using this in vitro system, these investigators found that neither UPF1 nor UPF1’s biochemical functions (such as ATP binding or hydrolysis), nor its phosphorylation, plays a discernible role in early or late phases of translation termination. Furthermore, they found that UPF1 does not measurably bind eRF1 and eRF3A directly, and thus the previously described interactions between UPF1 and eRFs (identified in co-IP experiments) are likely to be indirect. The data from Neu-Yilik et al. 11 suggest that UPF1 is functionally dispensable during translation termination in vitro and that the essential role of UPF1 (as well as UPF2) is exerted exclusively in the post-termination phase of translation, such as the rapid RNA decay elicited by NMD. Intriguingly, Neu-Yilik et al. 11 obtained evidence that another NMD factor—UPF3B—is instead critical for translation termination. Under conditions simulating premature translation termination, UPF3B delays termination and inhibits peptide release. UPF3B also has a second function: it promotes dissociation of post-termination ribosomal complexes after release of the nascent peptide. As we discuss below, the discovery of these new UPF3B functions alters how we view this NMD factor and lends further support to the growing view that translation termination and RNA decay are coupled.
UPF3B modulates translation termination
Translation termination at a PTC is thought to be less efficient than when it occurs at a normal stop codon. To examine the role of the UPFs in this process, Neu-Yilik et al. 11 used a fully reconstituted translation termination system that had previously been shown to mimic translation termination 33. In this system, ribosome subunits, amioacylated tRNAs, and a complete set of purified individual translational factors are assembled on a model mRNA encoding a tetrapeptide followed by a UAA stop codon. Neu-Yilik et al. 11 introduced various combinations of purified NMD factors to test their roles in this system. To simulate the inefficient translation termination at PTCs, the authors used limiting amounts of RFs.
Under this set of conditions, the authors found that introduction of UPF3B delayed translation termination, as assayed by a toeprinting assay. This assay showed that UPF3B dramatically reduced the ability of pre-termination complexes (preTCs) to convert into post-termination complexes (postTCs). A UPF3B mutant lacking the EJC-interaction domain that is defective in NMD was fully competent in inhibiting translation termination. This is an important finding from a mechanistic perspective and also has practical applications. For example, this mutant may allow one to tease out the relative role of UPF3B’s NMD activity from its UPF3B’s translation termination activity in different biological scenarios.
Interestingly, UPF2, which had no effect on its own, reversed the inhibitory role of UPF3B. Thus, UPF3B and UPF2 appear to act in a ‘yin-yang manner’ to modulate translation termination, perhaps for regulatory purposes.
To determine whether UPF3B downmodulates translation termination by inhibiting peptidyl-tRNA hydrolysis, Neu-Yilik et al. 11 tracked the progress of peptide release by using 35S-labeled initiator-tRNA in a reconstituted termination reaction. They found that the addition of UPF3B reduced the efficiency of peptidyl-tRNA hydrolysis under the same reaction conditions in which UPF3B inhibited translation termination. These data are consistent with UPF3B inhibiting translation termination by weakening peptidyl-tRNA hydrolysis.
A potential caveat with the above experiments is that the authors used excess amounts of UPF3B and other NMD proteins to saturate their interactions with other components of the translation termination machinery. Although this saturating approach successfully illuminated new functions for UPF3B, it will be important, in the future, to perform dose-response experiments to determine the functions of UPF3B when present at limiting concentrations.
UPF3B forms a complex with release factors
To assess whether UPF3B might function directly in translation termination, Neu-Yilik et al. 11 examined whether UPF3B forms complexes with RFs in vivo. Through co-IP experiments (in the presence of RNases to ablate RNA-dependent interactions), they found that UPF3B interacts with eRF3A ( Figure 2b and Supplemental Figure 1a). In contrast, UPF3B did not measurably interact with eRF1 ( Supplemental Figure 1a). However, UPF3B inhibited the ability of UPF1 to interact with eRF1, providing evidence that UPF3B does have an active role in eRF1 complexes. This negative regulatory role is consistent with UPF3B’s ability to inhibit translation.
To elucidate whether UPF3B directly interacts with eRFs, Neu-Yilik et al. 11 performed in vitro pull-down experiments. This revealed that UPF3B directly binds with both eRF1 and eRF3A ( Figure 2b and Supplemental Figure 1a, 1b). Surprisingly, this assay showed that other core NMD factors, including UPF1, did not directly interact with eRFs. This goes against the widely held assumption that UPF1 has a direct and integral role in translation termination (see the Introduction and below). Together, these data raised the possibility that, instead, UPF3B is the NMD factor that plays the most direct role in regulating translation termination.
Follow-up experiments using size exclusion chromatography (SEC) under physiological buffer conditions provided evidence that UPF3B forms a stable trimeric complex with both eRF1 and eRF3A ( Figure 2b and Supplemental Figure 1a, 1b). In experiments in which UPF3B was mixed with eRF1 and eRF3A individually, UPF3B was found to bind strongly with eRF3A but only weakly (probably transiently) with eRF1 ( Supplemental Figure 1a), the latter of which is consistent with the lack of measurable interaction of UPF3B with eRF1, as measured by co-IP ( Supplemental Figure 1a). SEC analysis also provided evidence that the stoichiometric relationship of eRF1, eRF3A, and UPF3B is about 1:1:1. Both in vitro pull-down and SEC assays demonstrated that this trimeric complex is stable in vitro ( Figure 2b and Supplemental Figure 1b) and its formation is independent of the eRF3A substrate GTP. However, this does not rule out that GTP hydrolysis by eRF3A could modulate the interactions between itself and eRF1 and UPF3B, an interesting question to address in the future.
Neu-Yilik et al. 11 then went on to perform mutational analysis to understand how UPF3B interacts with eRF3A. They found that UPF3B likely interacts with the amino (N)-terminus of eRF3A, based on the inability of an N-terminal eRF3A mutant to interact with UPF3B, both in vitro and in vivo. This is consistent with a regulatory role for UPF3B, as eRF3A does not require the N-terminus for its function in translation termination 34. Using both in vitro pull-down and co-IP analyses, they showed that eRF3A binds to UPF3B in a middle domain between UPF3B’s UPF2- and EJC-interaction domains ( Figure 3). Thus, this study defined a new functional domain in UPF3B. The finding that UPF3B uses different binding sites for eRF3A, UPF2, and the EJC suggests that the interactions between UPF3B and these factors are not mutually exclusive ( Figure 2b).
Figure 3. The interaction sites on human UPF3B for its cofactors.
The numbers denote the positions of amino acids in human UPF3B. The RNA recognition motif (RRM)-like domain overlaps with the UPF2-interacting domains 35. While Neu-Yilik et al. 11 established that UPF3B directly binds to RNA, whether the UPF3B RRM domain is involved is not known. EBM, exon junction complex binding motif.
UPF3B triggers disassembly of post-termination complexes
To address the role of UPF3B in normal translation termination, Neu-Yilik et al. 11 performed experiments with saturating amounts of eRFs. Under this condition, they found that UPF3B not only delayed translation termination but destabilized the postTCs; that is, it triggered their release from the mRNA. This activity was independent of UPF1 and prevented by UPF2. Analysis of a battery of UPF3B mutants showed that UPF3B’s EJC-interaction domain was dispensable, and that neither the UPF2-interaction domain nor the eRF3A-interaction (middle) domain was sufficient for this function. Subsequent experiments indicated that UPF3B dissociates postTCs after both GTP and peptidyl-tRNA hydrolysis. In contrast, UPF3B did not measurably impact either preTCs or postTCs before peptide hydrolysis. UPF3B was found to dissociate postTCs regardless of whether they are bound by eRF3A, implying that UPF3B interactions with eRF3A are dispensable for UPF3B to promote the dissociation of the ribosome from the mRNA.
Model
To understand how UPF3B might regulate translation termination, Neu-Yilik et al. 11 examined whether it might interact with ribosomes. Using a co-sedimentation assay, they found that UPF3B shares with UPF1 the ability to associate with ribosomes ( Figure 2b and Supplemental Figure 1a, 1b). In addition, they found that UPF3B interacts directly with RNA in vitro ( Figure 2b and Supplemental Figure 1a), consistent with the RNA-binding activity of UPF3B previously identified by individual-nucleotide resolution cross-linking and immunoprecipitation (iCLIP) analysis 36. These findings raise the exciting possibility that UPF3B can independently bind the ribosome and the mRNA without requiring additional bridging by other NMD proteins or the EJC. Thus, like UPF1, UPF3B is able to directly load onto actively translated RNAs, which has important implications for devising models explaining how UPF3B acts in translation termination and NMD.
An outstanding question is how UPF3B binds to RNA. UPF3B was previously shown to have a domain strongly reminiscent of an RNA-recognition motif (RRM), but this was shown to be responsible for binding to UPF2, not RNA 35.
The findings from Neu-Yilik et al. 11 led the authors to propose a new model in which UPF3B has a central role in translation termination and NMD ( Figure 1). In this model, UPF3B binds in the vicinity of the A site of the ribosome and assists in the recruitment of eRF1-eRF3A during the initial slow phase of termination at a PTC ( Figure 1, phase 1). UPF3B then delays termination by sterically impeding stop codon recognition and peptide release by eRF1 ( Figure 1, phase 2). After a retarded phase of peptide release ( Figure 1, phase 3), UPF3B contributes to the rescue of ribosomes stalled at a PTC by promoting the release of postTCs ( Figure 1, phase 4). According to their model, this last phase is independent of eRF binding and is promoted by interactions of UPF3B with the ribosome subunit interface. Subsequently, UPF1, UPF2, and UPF3B drive 3′ untranslated region (UTR) remodeling and the recruitment of enzymes responsible for the decay phase of NMD.
UPF1 does not measurably impact translation termination
The work of Neu-Yilik et al. 11 is important not only because of the new roles it defines for UPF3B but also for the insights it provides into the role of UPF1. As described in the Introduction, several previous lines of evidence had suggested that this central NMD factor has direct roles in translation termination. Thus, it was a surprise when Neu-Yilik et al. 11 found that recombinant UPF1 had no measurable effect on two steps regulated by UPF3B: (i) the preTC-to-postTC transition and (ii) release of the postTC from the mRNA. Furthermore, UPF1 failed to have a measurable effect regardless of whether there were limiting or saturating amounts of eRFs. While the authors confirmed previous reports 26, 27, 37 that UPF1 interacts with eRFs (through co-IP experiments), they found, using an in vitro pull-down assay, that UPF1 does not measurably directly bind with either eRF1 or eRF3A ( Supplemental Figure 1a). Thus, UPF1 interacts indirectly with eRFs. At first glance, the most obvious mechanism for this indirect binding is through the adaptor protein UPF2, as UPF1 directly binds to UPF2, and UPF2 directly binds to the eRF-binding protein UPF3B 13. However, this seems unlikely, as Neu-Yilik et al. 11 found that UPF2 did not measurably interact with eRF1 and only minimally interacted with eRF3A ( Supplemental Figure 1a). This raised the possibility that instead UPF1 directly binds to UPF3B, which they tested by the pull-down assay and found to be the case ( Figure 2b and Supplemental Figure 1a). Based on SEC analysis, UPF1 and UPF3B bound to each other with a stoichiometry of about 1:1. Of note, an earlier study concluded that UPF3B does not directly interact with UPF1 38, but this study used a truncated UPF1 variant.
Because ATP binding and hydrolysis are essential for UPF1’s role in NMD 1, Neu-Yilik et al. 11 also examined whether these biochemical steps impact translation termination. They found that translation termination in vitro is independent of both ATP binding and UPF1 ATP hydrolysis, as shown in toeprinting experiments by adding ATP or its non-hydrolyzable analog AMPPNP. Neu-Yilik et al. 11 also investigated the role of UPF1 phosphorylation, as this post-translation modification is considered an integral step in NMD 39– 41. According to current models, UPF1 phosphorylation occurs through formation of a SMG1-UPF1-eRF1-eRF3 (SURF) complex composed of UPF1, eRF1, eRF3, the UPF1 kinase SMG1, and two NMD factors (SMG8 and SMG9) known promote UPF1 phosphorylation 13. This SURF complex is thought to assemble at the site of translation termination and phosphorylate UPF1, leading to the recruitment of RNA decay-promoting factors 1. To test the role of UPF factors on SURF function, Neu-Yilik et al. 11 examined the effect of SMG1/8/9 on translation termination, in the presence or absence of UPF1, but found no measurable effect. The authors also examined UPF1 phosphorylation and found that UPF3B modestly inhibited the addition of this post-translational modification. In contrast, when UPF3B and UPF2 were added together, UPF1 phosphorylation was strongly inhibited. It is tempting to speculate that the repression of UPF1 phosphorylation by the other UPFs influences post-translation termination mechanisms such as NMD.
Collectively, the results from Neu-Yilik et al. 11 raise the possibility that UPF1 is dispensable for translation termination, and hence the function of UPF1 may be exclusively for post-termination events, such as the decay of transcripts through the NMD pathway. This notion is consistent with previous studies showing that UPF1 phosphorylation and its ATPase and helicase activities are important for UPF1’s functions in 3′-UTR mRNP remodeling and the recruitment of mRNA decay factors in metazoans 12, 42– 44.
A potential caveat with the experiments performed by Neu-Yilik et al. 11 is that even though limiting amounts of RFs were used to mimic premature termination, translation termination was relatively efficient, as indicated by the toeprinting assay. Thus, the conditions used by the authors could have masked a stimulatory effect of UPF1 on translation termination. Indeed, this possibility is supported by recent experiments in Saccharomyces cerevisiae showing that Upf1p promote translation termination and ribosome release at PTCs 45. While it remains to be determined if yeast and mammalian UPF1 differ in their ability to stimulate translation termination, it would not be surprising if they do differ, as the mammalian NMD pathway appears to be more complex than the NMD pathway in lower organisms. Mammalian NMD requires a large number of factors, appears to have several branches, and is controlled by diverse regulatory mechanisms, whereas yeast NMD appears to require fewer factors and is a single linear pathway.
Perspective
Physiological implications of UPF3B’s ability to regulate translation termination
The finding by Neu-Yilik et al. 11 that UPF3B is not only an RNA decay-promoting factor but influences translation termination was unexpected. What might be the physiological relevance of this? For example, what benefit would be provided by UPF3B’s ability to delay translation termination? One possibility is that this activity serves to stimulate the decay of RNAs targeted by the NMD pathway. Delayed translation termination would likely increase the time required to assemble NMD-promoting complexes, thereby increasing the probability that such an RNA would be degraded by NMD. In addition, paused ribosomes would be predicted to lead to reduced translation of ribosomes upstream, which could leave the RNA more exposed to nucleases. Support for this notion comes from S. cerevisiae, where it has been shown that slower translation elongation rates reduce translation initiation, which, in turn, stimulates mRNA decapping and degradation 46– 48.
What might be the physiological relevance of UPF3B stimulating the release of postTCs? The most obvious benefit conferred by this activity would be an enhancement of ribosome recycling. This would not only be energetically favorable for cells, but it would likely increase the efficiency of translation as well as translation-dependent activities, including NMD. Another possible benefit of the UPF3B-dependent quick release of postTCs stalled at PTCs is facilitation of mRNA exposure to ribonucleases for complete degradation. Indeed, recent studies in yeast support the notion that postTC release promotes RNA decay 45.
The finding that UPF3B regulates translation termination is intriguing in light of the fact that all known vertebrates harbor a second copy of the UPF3 gene. Might this other UPF3 paralog— UPF3A—encode a protein that also functions in translation termination? One possibility is that UPF3A promotes UPF3B’s translation termination functions. This follows from two findings: (i) the ability of both UPF3A and UPF3B to directly bind to the NMD factor, UPF2, and (ii) the discovery by Neu-Yilik et al. 11 that UPF2 inhibits UPF3B’s translation termination functions. By binding and thereby presumably sequestering UPF2, UPF3A could relieve UPF2’s inhibitory role and thus promote UPF3B-mediated effects on translation termination.
Does UPF3B influence translation termination in cells?
Neu-Yilik et al. 11 provide strong evidence that UPF3B influences premature translation termination in vitro. Does it also have this activity in live cells? Consistent with this possibility is the evidence that ribosome association is prolonged at premature stop codons in S. cerevisiae and mammalian cells 49, 50. Indeed, some evidence suggests that the kinetics of termination discriminates between normal termination codons and PTCs in vivo 51, 52. The ability of UPF3B to promote the disassembly of post-termination complexes is consistent with the finding that deletion of UPF3, as well as the other UPF genes, causes defects in ribosome release in yeast, both in vitro and in vivo 53.
To more directly test whether UPF3B functions in translation termination in cells, one could use ribosome profiling (Riboseq), a powerful technique that monitors translation genome-wide in vivo by deep sequencing of ribosome-protected mRNA fragments 54– 56. Riboseq has been successfully applied to analyze translation termination defects and ribosome recycling defects in many different contexts 54. If UPF3B promotes ribosome recycling, this predicts that cells from Upf3b-null mice 57 would show increased ribosomes stalling or queuing upstream of in-frame stop codons. This Riboseq analysis would also distinguish between UPF3B acting as a general ribosome release factor vs. a selective ribosome release factor targeting only specific RNAs, such as those ultimately degraded by NMD.
Potential implications of the new functional domain in UPF3B
Another important finding of Neu-Yilik et al. 11 is their identification of a new functional domain in UPF3B. They found that its middle region, which previously had no known function, serves as a direct binding site for eRF3A ( Figure 3). Interestingly, mutations in this particular region of UPF3B are found in patients with intellectual disability (ID); indeed, mutations in this region have been shown—through human pedigree analysis—to likely cause ID and even some neuro-developmental disorders such as autism spectrum disorder (ASD) and schizophrenia (SCZ) 58– 60. This region of UPF3B appears to be critical for mature neuron functions, based on a study by Alrahbeni et al. 61. These authors investigated the functional effect of missense mutations in this region from several different male patients with ID, some of whom also displayed ASD or SCZ. Using a gain-of-function tethering assay, the authors found that the mutant UPF3B proteins produced as a result of these missense mutations caused dramatically reduced NMD activity in both neural and non-neural cell lines. To test whether these mutant proteins influenced neuronal maturation, they transiently overexpressed them in neural cells cultured under differentiation conditions. They found that these mutant UPF3B proteins dramatically decreased neurite arborization, the process by which neurons generate branch-like dendritic processes in order to enhance neural connectivity with neighboring cells. This arborization defect was specific, as overexpression of wild-type UPF3B had no significant effect on arborization. The discovery that specific UPF3B mutations lead to reduced neurite arborization is significant, as both mouse models and post-mortem human studies have linked neuronal branching defects to neurodevelopmental disorders 62– 64.
These results from Alrahbeni et al. support a model in which UPF3B’s NMD function is critical for neuron arborization and consequent neural processes required for normal intellect and behavior. However, might UPF3B’s role in translation termination be responsible instead? Such an alternative model predicts that human ID patients with UPF3B missense mutations in the middle domain will express a form of UPF3B unable to bind to eRF3A and unable to regulate translation termination. If this model proves to be correct, it could potentially resolve a paradoxical finding made by another group—Jolly et al. 65. Studying post-mitotic hippocampal neurons, Jolly et al. found that short hairpin RNA-mediated depletion of UPF3B caused a significant increase in arborization of both axons and dendrites 66. Thus, the data of Jolly et al. support the notion that UPF3B inhibits arborization, whereas Alrahbeni et al. obtained evidence that UPF3B promotes arborization 61, 65. How can their opposite results be reconciled? One possibility is that UPF3B’s two functions—NMD and translation termination—exert opposite roles in arborization and are differentially affected by UPF3B depletion 66 and mutant UPF3B overexpression 61.
One means to dissect the biological roles of UPF3B’s different biochemical activities is to examine the effect of UPF3B mutants that lose one activity but retain another activity. In this regard, Neu-Yilik et al. 11 identified a UPF3B mutant (lacking the EJC-interaction domain) that has no NMD-promoting activity but is fully competent in inhibiting translation termination. By performing rescue experiments with this mutant versus wild-type UPF3B, one can begin to dissect the importance of UPF3B’s NMD versus UPF3B’s translation termination functions. By analogy, SMG6 mutants lacking either NMD function or telomerase maintenance function were used in rescue experiments to define the role of SMG6 in mouse embryonic stem cell differentiation 63, 66.
A simple prediction of the finding that UPF3B interacts with the N-terminus of eRF3A is that an N-terminal truncation protein of eRF3A would not be susceptible to UPF3B-mediated translation termination inhibition. Surprisingly, Neu-Yilik et al. 11 found that this was not the case. In addition, they found that the interaction between UPF3B and eRF3A interaction is dispensable for the ability of UPF3B to promote postTC dissociation. These results suggest that UPF3B modulates translation termination events independently of its ability to interact with eRF3A.
Poly-(A) binding protein (PABP) also binds the N-terminal region of eRF3A 67, 68. This is interesting given that PABP has the opposite functions as UPF3B, as PABP stimulates translation termination 68 and represses NMD 69. It is thus tempting to speculate that PABP and UPF3B compete for eRF3A binding as a means to determine whether an RNA is stabilized and translated or undergoes translation termination and is degraded. What mechanism might control this putative competition? One model is that the type of the stop codon determines the probability of whether eRF3A binds PABP or UPF3B. In this model, binding of PABP to eRF3A might be favored at a normal stop codon, leading to ribosome recycling, RNA stabilization, and further rounds of translation. In contrast, binding of UPF3B to eRF3A might be favored at a PTC, leading to delayed translation termination, activation of NMD, and consequent RNA decay.
Elucidating the molecular choreography guiding how UPF factors interact with RFs
To understand the molecular mechanisms by which UPF3B collaborates with eRFs and other UPF proteins in translation termination, it will be critical to solve the crystal structures of UPF3B-containing complexes. Neu-Yilik et al. 11 obtained in vitro evidence for two stable UPF3B-containing complexes—UPF3B/eRF3A/eRF1 and UPF3B/eRF3A/UPF1—both of whose structures will be intriguing to decipher. The structure of the UPF3B/eRF3A/eRF1 complex would likely represent a snapshot of early events in translation termination whereas the UPF3B/eRF3A/UPF1 complex would likely represent later events, such as the transition between translation termination and NMD. Solving such structures would not only reveal how UPF3B and eRF3A interact, but they might suggest how eRF1 and UPF1 function in these complexes. Ultimately, structural analysis could help decipher how translation termination and NMD are coordinated.
There are many other intriguing questions regarding the role of UPF3B in translation termination. For instance, among its various binding partners, which one does UPF3B interact with first? Among the many biochemical steps that UPF3B could theoretically act in, which pathway does UPF3B preferentially take? Which reaction steps are rate-limiting and thus dictate the magnitude of UPF3B-mediated effects on translation termination? Does UPF3B reduce the affinity of eRFs for the terminating ribosome to inhibit translation termination? Or does UPF3B repress the ability of eRF3A to undergo GTP binding or hydrolysis? To address these questions, one could employ the strategy recently used to study factors involved in translation initiation 70. In this study, thermodynamic and kinetic analyses were performed on five essential single components (three RNA-binding proteins, RNA and ATP), coupled with computational simulation, to yield a comprehensive view of the molecular choreography driving translation initiation. To study the role of UPF3B in translation termination, a similar analysis could be done with all the components involved, including the three UPF factors, the two eRFs, the ribosome, RNA, and relevant small molecules (ATP and ADP bound by UPF1 and GTP and GDP bound by eRF3A). Elucidation of the molecular choreography by which UPF3B regulates translation termination could also serve as a model to understand other UPF-dependent post-transcriptional mechanisms, such as NMD itself.
Coda
The unexpected discovery that a well-established factor with RNA decay-promoting activity has roles in translation leads to the question of which activity came first. Was UPF3B’s primordial role in NMD? Or did UPF3B first function in translation? Its primordial activity likely arose over 500 million years ago, as UPF3 is found throughout the phylogenetic scale, from yeast to humans. We know that UPF3 acts as an NMD factor in all species so far tested, but whether it has a role in translation termination remains to be determined for most species. Regardless of its evolutionary origin, the ability of UPF3B to function in both biosynthesis (translational regulation) and decay (RNA turnover) provides another example of how biology is driven by forces that simultaneously synthesize and degrade. In the words of T.S. Eliot: “April is the cruelest month, breeding Lilacs out of the dead land, mixing memory and desire, stirring dull roots with spring rain”.
Abbreviations
ASD, autism spectrum disorder; co-IP, co-immunoprecipitation; EJC, exon junction complex; eRF, eukaryote release factor; ID, intellectual disability; NMD, nonsense-mediated RNA decay; N-terminus, amino terminus; PABP, poly-(A) binding protein; postTC, post-termination complex; preTC, pre-termination complex; PTC, premature termination codon; RF, release factor; Riboseq, ribosome profiling; RNP, ribonucleoprotein; SCZ, schizophrenia; SEC, size exclusion chromatography; SURF, SMG1–UPF1–eRF1–eRF3; UPF, up-frameshift; UTR, untranslated region.
Editorial Note on the Review Process
F1000 Faculty Reviews are commissioned from members of the prestigious F1000 Faculty and are edited as a service to readers. In order to make these reviews as comprehensive and accessible as possible, the referees provide input before publication and only the final, revised version is published. The referees who approved the final version are listed with their names and affiliations but without their reports on earlier versions (any comments will already have been addressed in the published version).
The referees who approved this article are:
J. Robert Hogg, Biochemistry and Biophysics Center, National Heart, Lung, and Blood Institute, National Institutes of Health, Bethesda, MD, USA
Marc Graille, Laboratoire de Biochimie, Ecole polytechnique, CNRS, Université Paris-Saclay, Palaiseau, France
Elena Alkalaeva, Engelhardt Institute of Molecular Biology, The Russian Academy of Sciences, Moscow, Russian Federation
Funding Statement
This work was supported by National Institutes of Health grant RO1 GM111838
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; referees: 3 approved]
Supplementary material
Supplemental Figure 1. Summary of the evidence for the interactions between eRF1, eRF3A, UPF3B, UPF2L, UPF1, mRNA, and the 80S ribosome, as reported in Neu-Yilik et al., 2017 11. (a) Two-component interactions detected by the indicated assays. ‘+’ and ‘−‘ denote interactions detected and not detected, respectively. The discrepancy between the in vivo co-immunoprecipitation (co-IP) and other in vitro assays likely reflects either indirect interactions meditated by other cofactors or through post-translational modifications. (b) Three-component complexes detected by the indicated assays. Blue denotes components in the examined complexes. PD, in vitro pull-down; SEC, size exclusion chromatography; SED, ribosome co-sedimentation.
References
- 1. Lykke-Andersen S, Jensen TH: Nonsense-mediated mRNA decay: an intricate machinery that shapes transcriptomes. Nat Rev Mol Cell Biol. 2015;16(11):665–77. 10.1038/nrm4063 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 2. Huang L, Wilkinson MF: Regulation of nonsense-mediated mRNA decay. Wiley Interdiscip Rev RNA. 2012;3(6):807–28. 10.1002/wrna.1137 [DOI] [PubMed] [Google Scholar]
- 3. Goetz AE, Wilkinson M: Stress and the nonsense-mediated RNA decay pathway. Cell Mol Life Sci. 2017;74(19):3509 –3531. 10.1007/s00018-017-2537-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Karousis ED, Nasif S, Mühlemann O: Nonsense-mediated mRNA decay: novel mechanistic insights and biological impact. Wiley Interdiscip Rev RNA. 2016;7(5):661–82. 10.1002/wrna.1357 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 5. Karam R, Wengrod J, Gardner LB, et al. : Regulation of nonsense-mediated mRNA decay: implications for physiology and disease. Biochim Biophys Acta. 2013;1829(6–7):624–33. 10.1016/j.bbagrm.2013.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chan WK, Huang L, Gudikote JP, et al. : An alternative branch of the nonsense-mediated decay pathway. EMBO J. 2007;26(7):1820–30. 10.1038/sj.emboj.7601628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Huang L, Lou CH, Chan W, et al. : RNA homeostasis governed by cell type-specific and branched feedback loops acting on NMD. Mol Cell. 2011;43(6):950–61. 10.1016/j.molcel.2011.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Karam R, Lou CH, Kroeger H, et al. : The unfolded protein response is shaped by the NMD pathway. EMBO Rep. 2015;16(5):599–609. 10.15252/embr.201439696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lou CH, Dumdie J, Goetz A, et al. : Nonsense-Mediated RNA Decay Influences Human Embryonic Stem Cell Fate. Stem Cell Reports. 2016;6(6):844–57. 10.1016/j.stemcr.2016.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Huang L, Shum EY, Jones SH, et al. : A Upf3b-mutant mouse model with behavioral and neurogenesis defects. Mol Psychiatry. 2017. 10.1038/mp.2017.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Neu-Yilik G, Raimondeau E, Eliseev B, et al. : Dual function of UPF3B in early and late translation termination. EMBO J. 2017;36(20):2968–86. 10.15252/embj.201797079 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 12. Fiorini F, Bagchi D, Le Hir H, et al. : Human Upf1 is a highly processive RNA helicase and translocase with RNP remodelling activities. Nat Commun. 2015;6: 7581. 10.1038/ncomms8581 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 13. Le Hir H, Saulière J, Wang Z: The exon junction complex as a node of post-transcriptional networks. Nat Rev Mol Cell Biol. 2016;17(1):41–54. 10.1038/nrm.2015.7 [DOI] [PubMed] [Google Scholar]
- 14. Shum EY, Jones SH, Shao A, et al. : The Antagonistic Gene Paralogs Upf3a and Upf3b Govern Nonsense-Mediated RNA Decay. Cell. 2016;165(2):382–95. 10.1016/j.cell.2016.02.046 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 15. Tarpey PS, Raymond FL, Nguyen LS, et al. : Mutations in UPF3B, a member of the nonsense-mediated mRNA decay complex, cause syndromic and nonsyndromic mental retardation. Nat Genet. 2007;39(9):1127–33. 10.1038/ng2100 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 16. Lykke-Andersen J, Shu MD, Steitz JA: Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell. 2000;103(7):1121–31. 10.1016/S0092-8674(00)00214-2 [DOI] [PubMed] [Google Scholar]
- 17. Kunz JB, Neu-Yilik G, Hentze MW, et al. : Functions of hUpf3a and hUpf3b in nonsense-mediated mRNA decay and translation. RNA. 2006;12(6):1015–22. 10.1261/rna.12506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Heuer A, Gerovac M, Schmidt C, et al. : Structure of the 40S-ABCE1 post-splitting complex in ribosome recycling and translation initiation. Nat Struct Mol Biol. 2017;24(5):453–60. 10.1038/nsmb.3396 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 19. Jackson RJ, Hellen CU, Pestova TV: The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11(2):113–27. 10.1038/nrm2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dever TE, Green R: The elongation, termination, and recycling phases of translation in eukaryotes. Cold Spring Harb Perspect Biol. 2012;4(7):a013706. 10.1101/cshperspect.a013706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Celik A, Kervestin S, Jacobson A: NMD: At the crossroads between translation termination and ribosome recycling. Biochimie. 2015;114:2–9. 10.1016/j.biochi.2014.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 22. He F, Jacobson A: Nonsense-Mediated mRNA Decay: Degradation of Defective Transcripts Is Only Part of the Story. Annu Rev Genet. 2015;49:339–66. 10.1146/annurev-genet-112414-054639 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 23. Kervestin S, Jacobson A: NMD: a multifaceted response to premature translational termination. Nat Rev Mol Cell Biol. 2012;13(11):700–12. 10.1038/nrm3454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Keeling KM, Lanier J, Du M, et al. : Leaky termination at premature stop codons antagonizes nonsense-mediated mRNA decay in S. cerevisiae. RNA. 2004;10(4):691–703. 10.1261/rna.5147804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang W, Czaplinski K, Rao Y, et al. : The role of Upf proteins in modulating the translation read-through of nonsense-containing transcripts. EMBO J. 2001;20(4):880–90. 10.1093/emboj/20.4.880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ivanov PV, Gehring NH, Kunz JB, et al. : Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J. 2008;27(5):736–47. 10.1038/emboj.2008.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Singh G, Rebbapragada I, Lykke-Andersen J: A competition between stimulators and antagonists of Upf complex recruitment governs human nonsense-mediated mRNA decay. PLoS Biol. 2008;6(4):e111. 10.1371/journal.pbio.0060111 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 28. Gehring NH, Kunz JB, Neu-Yilik G, et al. : Exon-junction complex components specify distinct routes of nonsense-mediated mRNA decay with differential cofactor requirements. Mol Cell. 2005;20(1):65–75. 10.1016/j.molcel.2005.08.012 [DOI] [PubMed] [Google Scholar]
- 29. Brogna S, McLeod T, Petric M: The Meaning of NMD: Translate or Perish. Trends Genet. 2016;32(7):395–407. 10.1016/j.tig.2016.04.007 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 30. Fan-Minogue H, Du M, Pisarev AV, et al. : Distinct eRF3 requirements suggest alternate eRF1 conformations mediate peptide release during eukaryotic translation termination. Mol Cell. 2008;30(5):599–609. 10.1016/j.molcel.2008.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pisarev AV, Unbehaun A, Hellen CU, et al. : Assembly and analysis of eukaryotic translation initiation complexes. Meth Enzymol. 2007;430:147–77. 10.1016/S0076-6879(07)30007-4 [DOI] [PubMed] [Google Scholar]
- 32. Pisareva VP, Pisarev AV, Komar AA, et al. : Translation initiation on mammalian mRNAs with structured 5'UTRs requires DExH-box protein DHX29. Cell. 2008;135(7):1237–50. 10.1016/j.cell.2008.10.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Alkalaeva EZ, Pisarev AV, Frolova LY, et al. : In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell. 2006;125(6):1125–36. 10.1016/j.cell.2006.04.035 [DOI] [PubMed] [Google Scholar]
- 34. Zhouravleva G, Schepachev V, Petrova A, et al. : Evolution of translation termination factor eRF3: is GSPT2 generated by retrotransposition of GSPT1's mRNA? IUBMB Life. 2006;58(4):199–202. 10.1080/15216540600686862 [DOI] [PubMed] [Google Scholar]
- 35. Kadlec J, Izaurralde E, Cusack S: The structural basis for the interaction between nonsense-mediated mRNA decay factors UPF2 and UPF3. Nat Struct Mol Biol. 2004;11(4):330–7. 10.1038/nsmb741 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 36. Hauer C, Sieber J, Schwarzl T, et al. : Exon Junction Complexes Show a Distributional Bias toward Alternatively Spliced mRNAs and against mRNAs Coding for Ribosomal Proteins. Cell Rep. 2016;16(6):1588–603. 10.1016/j.celrep.2016.06.096 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 37. Kashima I, Yamashita A, Izumi N, et al. : Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 2006;20(3):355–67. 10.1101/gad.1389006 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 38. Chamieh H, Ballut L, Bonneau F, et al. : NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat Struct Mol Biol. 2008;15(1):85–93. 10.1038/nsmb1330 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 39. Wilkinson MF: The cycle of nonsense. Mol Cell. 2003;12(5):1059–61. 10.1016/S1097-2765(03)00448-9 [DOI] [PubMed] [Google Scholar]
- 40. Ohnishi T, Yamashita A, Kashima I, et al. : Phosphorylation of hUPF1 induces formation of mRNA surveillance complexes containing hSMG-5 and hSMG-7. Mol Cell. 2003;12(5):1187–200. 10.1016/S1097-2765(03)00443-X [DOI] [PubMed] [Google Scholar]
- 41. Durand S, Franks TM, Lykke-Andersen J: Hyperphosphorylation amplifies UPF1 activity to resolve stalls in nonsense-mediated mRNA decay. Nat Commun. 2016;7: 12434. 10.1038/ncomms12434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lou CH, Shum EY, Wilkinson MF: RNA degradation drives stem cell differentiation. EMBO J. 2015;34(12):1606–8. 10.15252/embj.201591631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kurosaki T, Li W, Hoque M, et al. : A post-translational regulatory switch on UPF1 controls targeted mRNA degradation. Genes Dev. 2014;28(17):1900–16. 10.1101/gad.245506.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Okada-Katsuhata Y, Yamashita A, Kutsuzawa K, et al. : N- and C-terminal Upf1 phosphorylations create binding platforms for SMG-6 and SMG-5:SMG-7 during NMD. Nucleic Acids Res. 2012;40(3):1251–66. 10.1093/nar/gkr791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Serdar LD, Whiteside DL, Baker KE: ATP hydrolysis by UPF1 is required for efficient translation termination at premature stop codons. Nat Commun. 2016;7:14021. 10.1038/ncomms14021 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 46. Harigaya Y, Parker R: Codon optimality and mRNA decay. Cell Res. 2016;26(12):1269–70. 10.1038/cr.2016.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Radhakrishnan A, Chen YH, Martin S, et al. : The DEAD-Box Protein Dhh1p Couples mRNA Decay and Translation by Monitoring Codon Optimality. Cell. 2016;167(1):122–132.e9. 10.1016/j.cell.2016.08.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sweet T, Kovalak C, Coller J: The DEAD-box protein Dhh1 promotes decapping by slowing ribosome movement. PLoS Biol. 2012;10(6):e1001342. 10.1371/journal.pbio.1001342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Amrani N, Ganesan R, Kervestin S, et al. : A faux 3'-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature. 2004;432(7013):112–8. 10.1038/nature03060 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 50. Peixeiro I, Inácio Â, Barbosa C, et al. : Interaction of PABPC1 with the translation initiation complex is critical to the NMD resistance of AUG-proximal nonsense mutations. Nucleic Acids Res. 2012;40(3):1160–73. 10.1093/nar/gkr820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hilleren P, Parker R: Mechanisms of mRNA surveillance in eukaryotes. Annu Rev Genet. 1999;33:229–60. 10.1146/annurev.genet.33.1.229 [DOI] [PubMed] [Google Scholar]
- 52. Zünd D, Mühlemann O: Recent transcriptome-wide mapping of UPF1 binding sites reveals evidence for its recruitment to mRNA before translation. Translation (Austin). 2013;1(2): e26977. 10.4161/trla.26977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ghosh S, Ganesan R, Amrani N, et al. : Translational competence of ribosomes released from a premature termination codon is modulated by NMD factors. RNA. 2010;16(9):1832–47. 10.1261/rna.1987710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ingolia NT, Ghaemmaghami S, Newman JR, et al. : Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324(5924):218–23. 10.1126/science.1168978 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 55. Brar GA, Weissman JS: Ribosome profiling reveals the what, when, where and how of protein synthesis. Nat Rev Mol Cell Biol. 2015;16(11):651–64. 10.1038/nrm4069 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 56. Ingolia NT: Ribosome Footprint Profiling of Translation throughout the Genome. Cell. 2016;165(1):22–33. 10.1016/j.cell.2016.02.066 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 57. Huang L, Shum EY, Jones SH, et al. : A Upf3b-mutant mouse model with behavioral and neurogenesis defects. Mol Psychiatry. 2017. 10.1038/mp.2017.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Addington AM, Gauthier J, Piton A, et al. : A novel frameshift mutation in UPF3B identified in brothers affected with childhood onset schizophrenia and autism spectrum disorders. Mol Psychiatry. 2011;16(3):238–9. 10.1038/mp.2010.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lynch SA, Nguyen LS, Ng LY, et al. : Broadening the phenotype associated with mutations in UPF3B: two further cases with renal dysplasia and variable developmental delay. Eur J Med Genet. 2012;55(8–9):476–9. 10.1016/j.ejmg.2012.03.010 [DOI] [PubMed] [Google Scholar]
- 60. Szyszka P, Sharp SI, Dedman A, et al. : A nonconservative amino acid change in the UPF3B gene in a patient with schizophrenia. Psychiatr Genet. 2012;22(3):150–1. 10.1097/YPG.0b013e32834accbe [DOI] [PubMed] [Google Scholar]
- 61. Alrahbeni T, Sartor F, Anderson J, et al. : Full UPF3B function is critical for neuronal differentiation of neural stem cells. Mol Brain. 2015;8:33. 10.1186/s13041-015-0122-1 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 62. Belichenko PV, Oldfors A, Hagberg B, et al. : Rett syndrome: 3-D confocal microscopy of cortical pyramidal dendrites and afferents. Neuroreport. 1994;5(12):1509–13. 10.1097/00001756-199407000-00025 [DOI] [PubMed] [Google Scholar]
- 63. Baj G, Patrizio A, Montalbano A, et al. : Developmental and maintenance defects in Rett syndrome neurons identified by a new mouse staging system in vitro. Front Cell Neurosci. 2014;8:18. 10.3389/fncel.2014.00018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Krey JF, Paşca SP, Shcheglovitov A, et al. : Timothy syndrome is associated with activity-dependent dendritic retraction in rodent and human neurons. Nat Neurosci. 2013;16(2):201–9. 10.1038/nn.3307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Jolly LA, Homan CC, Jacob R, et al. : The UPF3B gene, implicated in intellectual disability, autism, ADHD and childhood onset schizophrenia regulates neural progenitor cell behaviour and neuronal outgrowth. Hum Mol Genet. 2013;22(23):4673–87. 10.1093/hmg/ddt315 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 66. Li T, Shi Y, Wang P, et al. : Smg6/Est1 licenses embryonic stem cell differentiation via nonsense-mediated mRNA decay. EMBO J. 2015;34(12):1630–47. 10.15252/embj.201489947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hoshino S, Imai M, Kobayashi T, et al. : The eukaryotic polypeptide chain releasing factor (eRF3/GSPT) carrying the translation termination signal to the 3'-Poly(A) tail of mRNA. Direct association of erf3/GSPT with polyadenylate-binding protein. J Biol Chem. 1999;274(24):16677–80. 10.1074/jbc.274.24.16677 [DOI] [PubMed] [Google Scholar]
- 68. Ivanov A, Mikhailova T, Eliseev B, et al. : PABP enhances release factor recruitment and stop codon recognition during translation termination. Nucleic Acids Res. 2016;44(16):7766–76. 10.1093/nar/gkw635 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 69. Behm-Ansmant I, Gatfield D, Rehwinkel J, et al. : A conserved role for cytoplasmic poly(A)-binding protein 1 (PABPC1) in nonsense-mediated mRNA decay. EMBO J. 2007;26(6):1591–601. 10.1038/sj.emboj.7601588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Gao Z, Putnam AA, Bowers HA, et al. : Coupling between the DEAD-box RNA helicases Ded1p and eIF4A. eLife. 2016;5: pii: e16408. 10.7554/eLife.16408 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.