Abstract
Adeno-associated virus (AAV) has shown promising therapeutic efficacy with a good safety profile in a wide range of animal models and human clinical trials. With the advent of clustered regulatory interspaced short palindromic repeat (CRISPR)-based genome-editing technologies, AAV provides one of the most suitable viral vectors to package, deliver, and express CRISPR components for targeted gene editing. Recent discoveries of smaller Cas9 orthologues have enabled the packaging of Cas9 nuclease and its chimeric guide RNA into a single AAV delivery vehicle for robust in vivo genome editing. Here, we discuss how the combined use of small Cas9 orthologues, tissue-specific minimal promoters, AAV serotypes, and different routes of administration has advanced the development of efficient and precise in vivo genome editing and comprehensively review the various AAV-CRISPR systems that have been effectively used in animals. We then discuss the clinical implications and potential strategies to overcome off-target effects, immunogenicity, and toxicity associated with CRISPR components and AAV delivery vehicles. Finally, we discuss ongoing non-viral-based ex vivo gene therapy clinical trials to underscore the current challenges and future prospects of CRISPR/Cas9 delivery for human therapeutics.
Keywords: CRISPR/Cas9 complex, genome editing, adeno-associated virus
Introduction
CRISPR (clustered regulatory interspaced short palindromic repeat)/Cas9-based RNA-guided DNA endonuclease is transforming biomedical science research and has quickly become the preferred genome-editing platform for interrogating endogenous gene function in vivo 1, 2. The CRISPR-based genome-editing tool has revolutionized the gene-editing technique because of its simplicity in target design, affordability, high efficiency, versatility, and multiplexing capability 3. The commonly used CRISPR system can be implemented in mammalian cells by co-expressing Cas9 nuclease along with chimeric guide RNA (gRNA), which is derived from a synthetic fusion of the CRISPR RNA array (crRNA) and trans-activating crRNA (tracrRNA) 3. The target site sequence of gRNA needs to be immediately followed by an optimal protospacer adjacent motif (PAM) sequence according to the species-derived Cas9 nuclease used at the 3′ end 4, 5. The background of and the recent developments in the CRISPR-based targeted genome-editing toolboxes have been extensively reviewed recently 1, 2, 6. Various applications of CRISPR technologies for genome engineering and medical research have also been reviewed recently 2, 6, 7.
The CRISPR/Cas9 complex can be introduced into the cell in the forms of DNA, messenger RNA (mRNA), or protein 8. The DNA encoding Cas9 and gRNA can be delivered into the cell using the plasmid and viral expression vectors 8. Through microinjection, liposome-mediated transfection, electroporation, or nucleofection 9, several recent studies have shown that the delivery format of active Cas9 protein/gRNA ribonucleoprotein (RNP) complex has lower off-target effects due to rapid clearance of RNPs from the cell 8– 10 and rapid gene editing as they cleave chromosomal DNA almost immediately after delivery 9, 10 as compared with plasmid DNA transfection. However, the delivery formats of mRNA and protein pose certain technical challenges in vivo. For example, many of the genetic brain disorders affect very large brain areas and more than a single structure in the brain 11. Therefore, global gene delivery to the central nervous system is a key to achieve effective therapies for neurological disorders. Although a recent study has successfully demonstrated genome editing in the mouse brain by local delivery of RNPs 12, it is technically challenging to deliver the RNPs globally to the central nervous system. Thus, the viral-based in vivo genome editing remains a popular choice to achieve stable or elevated expression of Cas9 and single-guide RNA (sgRNA) required for establishing animal disease models and therapeutic gene editing in animals. Indeed, it has been demonstrated that systemic delivery of adeno-associated virus (AAV) vectors enabled uniform and broad vector distribution, and subsequently led to extensive and widespread transgene expression in the adult mouse central nervous system 13– 15.
Animal models are preferred over cell models, as they help understand disease mechanisms at the physiological and systemic levels. Somatic mutagenesis via in vivo genome editing provides an ideal platform to accelerate the generation of transgenic animals for rapid exploration of human diseases and correcting genetic defects in gene therapy. In vivo genome editing also avoids laborious germline targeting and high costs of maintaining transgenic neonates through adulthood in animals with a long life span. Obviously, somatic mutagenesis in adult animals has proven technically more challenging than gene editing of one-cell-stage embryos or zygotes or pluripotent stem cells. The major hurdle is to efficiently deliver the CRISPR components in vivo. To date, various viral, non-viral (for example, lipid nanoparticles), and physical (for example, hydrodynamic injection) based delivery approaches of the CRISPR/Cas9 complex have been adopted for in vivo genome editing. The pros and cons of these delivery methods have been extensively reviewed recently 16– 18.
Given the great potential of viral vectors in gene and cell therapy, five major classes of viral vectors—retroviruses 19, lentiviruses 20, 21, adenoviruses 22, 23, AAVs 24, 25, and baculoviruses 26, 27—have been employed to deliver CRISPR components into mammalian cells for targeted genome editing. The advantages and disadvantages of using these viral vectors for in vivo delivery of the CRISPR transgenes have been extensively reviewed 24, 28– 30. In Table 1, we listed general characteristics and applications of various viral delivery vectors. Among these, the AAV vector is the overall focus of this review. The AAV system provides major advantages for research and therapeutics, including a very mild immune response and toxicity elicited by AAV in animal models. Moreover, AAVs remain primarily episomal upon transduction, avoiding random integration of the viral genetic materials into the host genome that can disrupt neighboring gene function and cause insertional mutagenesis 31. Indeed, there has been no reported case of disease caused by AAV in humans 31. Additionally, AAV can exist long-term as concatemers in non-dividing cells for stable transgene expressions 31. Given a good safety profile of AAV and therapeutic efficacy in a wide range of animal models and human clinical trials (ClinicalTrials.gov), AAV is thought to be one of the most suitable viral vectors for gene therapeutic applications and gene transfer in vivo. Furthermore, there is a wide range of AAV serotypes that can be selected to infect specific tissues in vivo. For these reasons, here, we provide an overview of the state of the art of various AAV-CRISPR systems as well as their principal vector designs for in vivo genome editing in animals. Senís and colleagues were amongst the first to exploit and demonstrate the use of the AAV vectors to package, deliver, and express CRISPR components for targeted gene editing in hard-to-transfect cells and the liver of adult mice 32. Subsequently, an increasing number of the studies used AAV vectors to deliver the CRISPR components into animals for in vivo genome editing. Of note, for certain applications such as packaging of a very large transgene 29, 33, genome-wide screening 20, 34, 35, and production of vaccines and recombinant proteins 36, 37, the other viral vectors may be a better option. However, in-depth discussions of the other viral delivery vehicles for the CRISPR components are beyond the scope of this review.
Table 1. Viral delivery vectors for the CRISPR/Cas9 complex.
| Characteristics for a typical
vector |
Retrovirus 19, 38, 39 | Lentivirus 20, 21, 38, 40, 41 | Adenovirus 22, 42– 48 | Adeno-associated
virus 30, 32, 49– 52 |
Baculovirus 27, 29, 53 |
|---|---|---|---|---|---|
| Common viral type | γ-retroviruses | HIV-1 | Ad5 | AAV2 | AcMNPV |
| Viral envelope | Yes | Yes | No | No | Yes |
| Nucleocapsid shape | Icosahedral | Icosahedral | Icosahedral | Icosahedral | Rod |
| Viral size | 80–130 nm | 80–130 nm | 70–105 nm | 18–26 nm | 250–300 nm
30–60 nm |
| Viral genome structure | Linear ssRNA | Linear ssRNA | Linear dsDNA | Linear ssDNA | Circular dsDNA |
| Viral genome size | 8.3 kb | 9.7 kb | 36 kb | 4.7 kb | 80–180 kb |
| Packaging capacity | <8.0 kb | <8.0 kb | <30 kb | <4.5 kb | >38 kb |
| Transgene is flanked by | LTRs | LTRs | ITRs | ITRs | Tn7s |
| Viral generation approach | Triple-plasmid transfection | Triple-plasmid transfection | Homologous recombination | Triple-plasmid transfection | Site-specific transposition |
| Competent cell used | Stbl3 | Stbl3 | AdEasier-1 | Stbl3 | DH10Bac |
| Host cells used | HEK293T | HEK293T | HEK293T or HER911 | HEK293T | Sf9 or Sf21 |
| Cells infected | Dividing | Dividing or non-dividing | Dividing or non-dividing | Dividing or non-dividing | Dividing or non-dividing |
| Transduction efficiency | Moderate | High | Very high | High | High |
| Transgene expression | Stable | Stable | Transient | Transient | Transient |
| Immune response | Moderate | Low | High | Very low | Very low |
| Toxicity | High | Moderate | High | Low | Low |
| Random genome integration | Yes | Yes | No | Generally no
(recombinant AAV has a low frequency of host genome integration events) |
No |
| Biosafety levels | BSL-2 | BSL-2 | BSL-2 | BSL-1 | BSL-1 |
| Common applications | Generating stable cell and
gene transfer, cancer and stem cell research |
Transduce difficult-to-
transfect cell, genome- wide screens |
Vaccine production, cancer
immune therapy |
Gene delivery
in vivo,
optogenetics |
Recombinant proteins
and vaccine production |
| Clinical trials | Very popular | Very popular | Popular | Increasing popularity | Growing interest |
| First
ex vivo gene transfer
clinical trial started (disease, ClinicalTrials.gov ID) |
1990
(severe combined immunodeficiency, NCT00001255) |
2007
(lymphoma, NCT00569985) |
2000
(hepatocellular carcinoma, NCT00300521) |
None | None |
| First
in vivo gene transfer
clinical trial started (disease, ClinicalTrials.gov ID) |
None | 2014
(sickle cell anemia, NCT02186418) |
1993
(cystic fibrosis, NCT00004779) |
1999
(cystic fibrosis, NCT00004533) |
None |
AcMNPV, autographa californica multicapsid nucleopolyhedrovirus; HIV, human immunodeficiency virus; ITR, inverted terminal repeat; LTR, long terminal repeat; Tn7, transposon 7.
Here, we aim to review various AAV-CRISPR systems recently demonstrated in mice and to discuss how the combined use of tissue-specific minimal promoters, AAV serotypes, different routes of administration, and small Cas9 orthologues has enabled investigators to achieve maximal efficiency and specificity for in vivo genome editing. In addition, we discuss the clinical implications and potential strategies to overcome off-target effects, immunogenicity, and toxicity associated with CRISPR components and AAV delivery vehicles. Finally, we discuss the promises and hurdles associated with ongoing ex vivo gene therapy clinical trials.
AAV-CRISPR-based in vivo genome editing in mice
At least 33 published studies successfully used CRISPR/Cas9 alongside AAV vectors for in vivo genome editing in mice ( Table 2). The AAV-CRISPR system is particularly useful for editing disease-associated genes in the brain or central nervous system of mice because of the inability of most cationic nanocarriers to cross the blood-brain barrier, high transduction efficiency of AAV vectors in the brain, and non-dividing properties of neurons for long-term therapeutic effects. The AAV-CRISPR system has also been successfully used to restore the gene function in muscle-associated diseases such as Duchenne muscular dystrophy 54– 57 and diseases associated with the eye 5, 58– 60, liver 4, 49, 61, heart 62– 64, and lung 65. This AAV-CRISPR-mediated gene editing in mice provided proof-of-principle studies for human disease modeling, gene therapy, or gene functional characterizations. We discuss recent developments and advancements in four critical aspects of the AAV-CRISPR system that have enabled efficient and precise in vivo genome editing in mice.
Table 2. CRISPR/Cas9-based in vivo genome editing in mice with tissue-specific promoters and AAV variants.
| Target tissue in
mouse (disease or phenotype) |
Promoter –
regulates CRISPR or reporter |
AAV
serotype |
Route of
administration (injection site) |
Application
(gene) |
Phenotypic impact or
therapeutic outcome |
Remark | Reference |
|---|---|---|---|---|---|---|---|
| Brain
(study gene function) |
pMecp2 promoter - SpCas9
hSyn1 promoter - GFP, KASH U6 promoter - gRNA |
AAV1 | Stereotactic injection
(dentate gyrus) |
Gene knockout
( Mecp2, Dnmt1, Dnmt3a, and Dnmt3b) |
Impaired contextual memory
and memory formation |
Can edit multiple
genes simultaneously |
25 |
| Brain
(study brain circuit) |
hSyn1 promoter - SaCas9,
Cre recombinase, GCaMP6f calcium sensor CAG promoter - tdTomato U6 promoter - Grna |
AAV2-retro | Stereotactic injection
(pontine nucleus, dorsal striatum) |
Gene knockout
( tdTomato) |
Enabled efficient mapping,
monitoring, and manipulation of projection neurons |
Modified AAV2
capsid with a 7-mer peptide |
68 |
| Brain
(Huntington disease) |
CMV promoter - SpCas9, eGFP
U6 promoter - gRNA H1 promoter - gRNA |
AAV1 | Stereotactic injection
(striata) |
SNP-
dependent editing ( Htt) |
Reduced expression of mutant
Htt allele in mouse brain |
Use transgenic
Huntington disease model |
76 |
| Brain
(schizophrenia) |
EF1a promoter - tdTomato
CBh promoter - ScGFP hSyn1 promoter - ScGFP GFAP promoter - ScGFP U6 promoter - gRNA |
AAV2g9 | Stereotaxic injections
(intracerebroventricular, cerebrospinal fluid) |
Gene deletion
( pre-miR137) |
Displayed preferential, robust,
and widespread neuronal transduction within the brain |
Use Cas9 mice and
an AAV chimeric derived from AAV2 and AAV9 |
80 |
| Brain
(inducible genome editing) |
Dox inducible Tight promoter
- SpCas9 TRE3G promoter - SpCas9 pMecp2 - SpCas9 CMV promoter - TetR, GFP, KASH H1/TO promoter - gRNA U6/TO promoter - gRNA |
AAV-DJ
AAV-DJ/8 |
Stereotaxic injection
(basal and lateral amygdala) |
Gene induction
( Tet2) |
Edited the genomes of
neurons in vivo within the mouse brain in a Dox- dependent manner |
Doxycycline-
dependent gRNA expression |
81 |
| Brain
(study gene function) |
CBh promoter - Cre
recombinase hSyn1 promoter - GFP, KASH U6 promoter - gRNA |
AAV1 | Stereotactic injection
(prefrontal cortex) |
Gene knock-in
( NeuN) |
NeuN protein depletion only in
the injected region |
Use Cre-dependent
SpCas9 Rosa26 knock-in mice |
65 |
| Brain
(glioblastoma) |
U6 promoter - gRNA
GFAP promoter - Cre recombinase |
AAV9 | Stereotactic injection | Gene mutation
( Trp53, Nf1, or Rb1) |
Induced tumor formation
that recapitulates human glioblastoma |
Use LSL-Cas9 mice | 79 |
| Brain
(precise genome editing) |
EFS promoter - SpCas9
U6 promoter - gRNA |
AAV1 | Intraventricular and
stereotactic injections |
Gene knock-in
( Camk2a, Erk2, Actb) |
vSLENDR enabled efficient
homology-directed repair in post-mitotic neurons in developing, adult, aged, and pathological brains |
Use wild-type and
Cas9 mice |
82 |
| Brain
(study gene function) |
U6 promoter - gRNA
CBh promoter - Cre recombinase |
AAV | Stereotactic injection
(pyramidal neurons and microglia in hippocampus) |
Gene
disruption ( Cnr2) |
Decreased contextual fear
memory, enhanced spatial working memory |
Use Camk2a-Cas9,
Gad2-Cas9, and Cx3cr1-Cas9 mice |
83 |
| Central nervous
system (amyotrophic lateral sclerosis) |
CMV promoter - SpCas9
U6 promoter - gRNA |
scAAV9 | Intrathecal injection | Gene
knockdown ( Igf1) |
Decreased D-amino acid
oxidase and increased D-serine, and caspase9 activation |
Use hSOD1G93A
ALS mouse model |
84 |
| Muscle
(Duchenne muscular dystrophy) |
CMV promoter - SaCas9
EFS promoter - SaCas9 CAGGS promoter - tdTomato U6 promoter - gRNA |
AAV9 | Intramuscular
(tibialis anterior) and intraperitoneal (intraperitoneal space) injections |
Exon deletion
( Dmd) |
Partially recovered muscle
functional deficiencies |
Use mdx mouse
model of DMD |
54 |
| Muscle
(Duchenne muscular dystrophy) |
CMV promoter - SaCas9
U6 promoter - gRNA |
AAV8 | Intramuscular (tibialis
anterior), intraperitoneal (intraperitoneal space), and intravenous (tail vein) injections |
Exon deletion
( Dmd) |
Improved muscle function | Use mdx mouse
model of DMD |
55 |
| Muscle
(Duchenne muscular dystrophy) |
CMV promoter - SpCas9
U6 promoter - gRNA |
AAV9 | Intraperitoneal,
intramuscular, and retro-orbital (venous sinus) injections |
Exon deletion
( Dmd) |
Enhanced skeletal muscle
function |
Use mdx mouse
model of DMD |
56 |
| Muscle
(Duchenne muscular dystrophy) |
CK8 promoter - SpCas9,
SaCas9 CMV promoter - mCherry U6 promoter - gRNA |
AAV6 | Intramuscular (tibialis
anterior) and systemic (retro-orbital) injection |
Exon deletion
and gene knock-in ( Dmd) |
Improved muscle function | Use mdx4cv mouse
model of DMD |
57 |
| Muscle
(congenital muscular dystrophy type 1A) |
CMV promoter - SaCas9
U6 promoter - gRNA |
AAV9 | Intraperitoneal injection | Gene
correction ( Lama2) |
Improved muscle histopathology
and function |
Use dy2J/dy2J mice | 69 |
| Retina
(age-related macular degeneration) |
EFS promoter - CjCas9
Spc512 promoter - CjCas9 U6 promoter - gRNA |
AAV9 | Intramuscular (tibialis
anterior) and intravitreal injection |
Gene knockout
( Rosa26, Vegfa, and Hif1a) |
Reduced the size of
laser-induced choroidal neovascularization |
Smallest Cas9
orthologue (smaller than SaCas9) |
5 |
| Retina
(Leber Congenital Amaurosis 10) |
CMV promoter - SpCas9,
SaCas9 hSyn1 promoter - eGFP GFAP promoter - eGFP U6 promoter - gRNA |
AAV5 | Subretinal injection | Gene deletion
( Cep290) |
Effectively removed intronic
mutation in Cep290 with minimized immune response |
Use self-limiting
CRISPR/Cas9 system |
58 |
| Retina
(retinal gene editing) |
pMecp2 promoter - SpCas9
hSyn1 promoter - mCherry, KASH U6 promoter - gRNA |
AAV2 | Intravitreal (intraocular)
injection |
Gene knockout
( YFP) |
Effective gene knockout without
affecting retinal function |
Use Thy1-YFP
transgenic mice |
59 |
| Retina
(retinal degeneration) |
CMV promoter - SpCas9,
tdTomato U6 promoter - gRNA |
AAV8 | Subretinal injection | Gene knockout
( Nrl) |
Prevented retinal degeneration,
improved rod survival, and preserved cone function |
Use mouse models of
retinal degeneration |
60 |
| Retina
(angiogenesis) |
pICAM2 - SpCas9
U6 promoter - gRNA |
AAV1 | Intravitreal injection | Gene silencing
( VEGFR2) |
Abrogated angiogenesis | Use mouse models
of oxygen-induced retinopathy and laser-induced choroid neovascularization |
85 |
| Liver
(ornithine transcarbamylase) |
TBG promoter - SaCas9
U6 promoter - gRNA |
AAV8 | Intravenous injection
(temporal vein) |
Gene knock-in
( Otc) |
Increased survival in mice
challenged with a high-protein diet |
Use adult OTC-
deficient mice |
49 |
| Liver
(hereditary tyrosinemia) |
EF1a promoter - GFP
U6 promoter - gRNA |
AAV8 | Systemic (tail vein)
injection |
Gene knock-in
( Fah) |
Rescued disease symptoms
such as weight loss and liver damage |
Use a mouse model
of human hereditary tyrosinemia |
61 |
| Liver
(total cholesterol) |
TBG promoter - SaCas9
U6 promoter - gRNA |
AAV8 | Systemic (tail vein)
injection |
Gene knockout
( Pcsk9) |
Reduced serum Pcsk9 and
total cholesterol levels |
Smaller Cas9
orthologue than SpCas9 |
4 |
| Liver
(LDL cholesterol) |
CB promoter - EmGFP
U6 promoter - gRNA |
AAV8 | Intraperitoneal injection | Gene knockout
( Ldlr, Apob) |
Resulted in severe
hypercholesterolemia and atherosclerosis |
Use Cas9 targeted
transgenic mice |
77 |
| Liver
(hemophilia B) |
HCRhAATp promoter - SaCas9
U6 promoter - gRNA |
AAV8 | Systemic (tail vein)
injection |
Gene knock-in
( F9) |
Restored hemostasis | Use F9-mutated mice | 71 |
| Heart
(PRKAG2 cardiac syndrome) |
CMV promoter - SpCas9
U6 promoter - gRNA |
AAV9 | Systemic injection | Gene knockout
( Prkag2) |
Restored the morphology
and function of the heart after disrupting the mutant Prkag2 allele |
Use H530R
Prkag2
transgenic knock-in mice |
62 |
| Heart
(cardiac disease modeling) |
Myh6 promoter - SpCas9, GFP,
TdTomato CMV promoter - ZsGreen U6 promoter - gRNA |
AAV9 | Intraperitoneal injection | Gene deletion
( Myh6) |
Displayed severe
cardiomyopathy and loss of cardiac function |
Use postnatal
cardiac-Cas9 transgenic mice |
63 |
| Heart
(cardiac myocyte maturation) |
cTNT promoter - Cre
recombinase U6 promoter - gRNA |
AAV9 | Subcutaneous injection | Gene knockout
( Jph2, Ryr2) |
Disrupted T-tubule structure
and maturation |
Use RosaCas9GFP/
Cas9GFP neonatal mice |
78 |
| Heart
(dystrophic cardiomyopathy) |
CK7-miniCMV promoter - SaCas9
U6 promoter - gRNA |
AAV rh74 | Systemic (retro-orbital,
intraperitoneal) injection |
Gene excision
( Dmd) |
Restored dystrophin expression
and cardiac function |
Use mdx/Utr
+/
−
dystrophic mice |
70 |
| Heart
(cardiac gene function) |
CB promoter - SpCas9
U6 promoter - gRNA |
AAV9 | Intracardiac | Gene disruption
( Myh6, Sav1, and Tbx20) |
Resulted in mosaic pattern of
gene disruption |
Use Myh6-Cre
transgenic mice |
64 |
| Lung
(lung adenocarcinoma) |
EFS promoter - Renilla
luciferase, Cre recombinase U6 promoter - gRNA |
AAV9 | Intranasal (nostril) and
intratracheal (trachea) injections |
Gene knock-in
( p53, Lkb1, and Kras) |
Led to macroscopic tumors
of lung adenocarcinoma pathology |
Use Cre-dependent
SpCas9 Rosa26 knock-in mice |
65 |
| Liver, heart, muscle
(host immune response) |
SMVP promoter - SpCas9,
SpCas9-VPR CASI promoter - SpCas9 CAG promoter - tdTomato U6 promoter - gRNA |
AAV9 | Intramuscular and
intraperitoneal injections |
Gene
activation ( Mstn, Fst, Pd-l1, and Cd47) |
Modest activation of
Pd-l1 and
Cd47 |
Use AAV-split-Cas9
system |
50 |
| Circulating
lymphocytes, spleen, liver, heart, lung, and kidney (eradication of HIV-1 DNA) |
CMV promoter - SaCas9
U6 promoter - gRNA |
AAV9 | Systemic (tail vein)
injection and retro- orbital inoculation |
Gene knockout
(HIV-1 DNA) |
Eradication of HIV-1 DNA | Use transgenic
mice and rats encompassing the HIV-1 genome |
86 |
| Spleen, lungs, heart,
colon, and brain (HIV-1 proviral DNA excision) |
CMV promoter - SaCas9
U6 promoter - gRNA |
AAV-DJ/8 | Systemic (tail vein)
injection |
Gene deletion
(HIV-1 DNA) |
Induced efficient excision of
HIV-1 proviral DNA |
Use HIV-1 Tg26
transgenic mice and humanized BLT mice with chronic HIV-1 infection |
72 |
AAV, adeno-associated virus; ALS, amyotrophic lateral sclerosis; Apob, apolipoprotein B; BLT, bone marrow/liver/thymus; CAG, a hybrid of CMV early enhancer and chicken beta-actin promoter; CAGGS, a hybrid of promoter composed of the CMV immediate-early enhancer, CBA promoter, and CBA intron 1/exon 1; Camk2a, calcium/calmodulin-dependent protein kinase II alpha; CASI, a hybrid of CMV enhancer and chicken β-actin promoter followed by a splice donor and splice acceptor; CB, chicken beta actin promoter; CBh, a hybrid form of the CBA promoter; Cd47, integrin-associated signal transducer; Cep290, centrosomal protein 290; CjCas9, Campylobacter jejuni Cas9; CK7, CK8, striated muscle-restricted promoter; CMV, cytomegalovirus; Cnr2, cannabinoid receptor 2; CRISPR, clustered regularly interspaced short palindromic repeat; cTNT, cardiac troponin T promoter; Cx3cr1, chemokine (C-X3-C motif) receptor 1; Dmd, Duchenne muscular dystrophy; Dnmt, DNA methyltransferase; EF1a, elongation factor-1 alpha; EFS, EF1a short; eGFP, enhanced green fluorescent protein; Erk2, mitogen-activated protein kinase 1; F9, coagulation factor IX; Fah, fumarylacetoacetate hydrolase; Fst, follistatin; Gad2, glutamic acid decarboxylase 2; GCaMP6f, green fluorescent calcium indicator; GFAP, glial fibrillary acidic protein; H1, human polymerase III RNA promoter; HCRhAATp, an enhancer element of the hepatic control region of the Apo E/C1 gene and the human anti-trypsin promoter; Hif1a, hypoxia-inducible factor 1 alpha; HIV, human immunodeficiency virus; hSyn1, human synapsin I; Htt, huntingtin; Igf1, insulin-like growth factor 1; Jph2, junctophilin-2; KASH, nuclear transmembrane domain; Kras, Kirsten rat sarcoma viral oncogene homolog; Lama2, laminin alpha 2; LDL, low-density lipoprotein; Ldlr, low-density lipoprotein receptor; Lkb1, serine/threonine kinase 11; mdx, dystrophin-deficient; Mecp2, methyl CpG binding protein 2; Mstn, myostatin; Myh6, myosin heavy polypeptide 6; NeuN, neuronal nuclear antigen; Nf1, neurofibromin 1; Nrl, neural retina-specific leucine zipper protein; Otc, ornithine transcarbamylase; Pcsk9, proprotein convertase subtilisin/kexin type 9; Pd-l1, programmed death-ligand 1; pICAM2, intercellular adhesion molecule 2 promoter; Prkag2, protein kinase AMP-activated non-catalytic subunit gamma 2; p53, transformation related protein 53; Rb1, RB transcriptional corepressor 1; Roa26, a safe harbor locus in mouse; Ryr2, ryanodine receptor 2; SaCas9, Staphylococcus aureus Cas9; Sav1, salvador family WW domain containing 1; SMVP, a hybrid of SV40 enhancer–CMV–promoter–chimeric intron; SNP, single-nucleotide polymorphism; SpCas9, Streptococcus pyogenes Cas9; Spc512, muscle-specific promoter; TBG, thyroxine binding globulin; Tbx20, T-box 20; Tet2, tet methylcytosine dioxygenase 2; Thy1, thymus cell antigen 1 theta; TRE3G, Tet-On 3G inducible promoter; TO, tetracycline operator; U6, human U6 small nuclear promoter; Vegfa, vascular endothelial growth factor A; VEGFR2, kinase insert domain protein receptor; VPR, a fusion of VP64–p65–Rta; vSLENDR, virus-mediated single-cell labeling of endogenous proteins via HDR; YFP, yellow fluorescent protein.
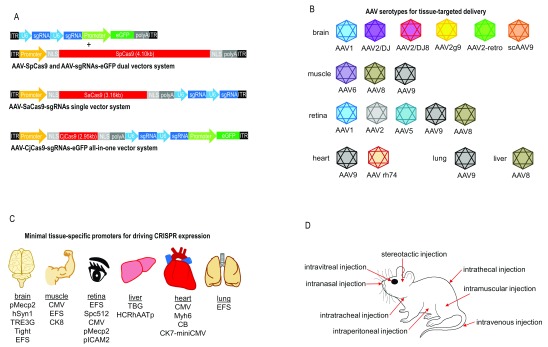
Small Cas9 orthologues
Even though the AAV vector is an extremely attractive delivery vehicle for CRISPR, its relatively small viral genome-packaging capacity has limited its use for delivering large transgenes. Using dual-vector AAV system or smaller Cas9 orthologues along with truncated regulatory elements (promoter and polyadenylation signal) has greatly circumvented the transgene packaging issue ( Figure 1A). Owing to a small AAV viral genome-packaging capacity (~4.8 kb), it has been technically challenging to co-package Streptococcus pyogenes-derived Cas9 (SpCas9) (4.1 kb) and multiple sgRNAs into all-in-one AAV vectors for multiplex genome editing. Recently, it has been demonstrated that when very small promoters were used (for example, miCMV promoter and H1 promoter to drive the expressions of SpCas9 and its gRNA, respectively), it was possible to package both SpCas9 and its gRNA into a single vector 32. However, the number of promoters that can be selected for tissue-specific transduction is limited. To circumvent this issue, the previously reported dual-vector system was adopted to express SpCas9 nuclease from one vector and to express its gRNAs together with a fluorescent reporter gene from another vector 25. Recently, St1Cas9 (~3.3 kb) from Streptococcus thermophilus 66 and a rationally designed truncated form of SpCas9 67 were developed to fit SpCas9 and its gRNA into a single AAV construct. Unfortunately, St1Cas9 requires a more complex PAM sequence that limits the number of targetable loci 66, and truncated SpCas9 has a much lower efficiency than its wild-type counterpart 67.
Figure 1. CRISPR/Cas9-based in vivo genome editing by using small Cas9 orthologues, different routes of AAV administration, tissue-specific minimal promoters, and AAV serotypes.
( A) AAV-CRISPR vectors. Campylobacter jejuni-derived Cas9 (CjCas9) is the smallest Cas9 orthologue (984 amino acids, 2.95 kb). Staphylococcus aureus-derived Cas9 (SaCas9) (1,053 amino acids, 3.16 kb) is smaller than the most widely used Cas9 derived from Streptococcus pyogenes (SpCas9) (1,368 amino acids, 4.10 kb). Owing to the large size of SpCas9, the SpCas9 gene and two gRNAs are packaged into two separate AAV vectors. The SpCas9-based dual-vector system enables one vector to express SpCas9 and another to express multiple gRNAs and a fluorescent reporter gene. Two gRNAs can be packaged with the SaCas9 into a single AAV vector if minimal promoter and polyadenylation signal are used. In the all-in-one vector system, CjCas9 enables packaging with two gRNAs and a fluorescent reporter gene, enhanced green fluorescent protein ( eGFP), into a single AAV vector. ( B) AAV serotypes for tissue-targeted delivery. ( C) Tissue-specific minimal promoters for driving CRISPR expression. ( D) Different routes of AAV administration into various tissues of a mouse. AAV, adeno-associated virus; CRISPR, clustered regulatory interspaced short palindromic repeat; gRNA, guide RNA.
To overcome these drawbacks, other recently discovered small Cas9 orthologues, including Staphylococcus aureus-derived Cas9 (SaCas9, 3.16 kb) 4 and Campylobacter jejuni-derived Cas9 (CjCas9, 2.95 kb) 5, have been used to package the Cas9 and its gRNA into a single AAV delivery vehicle for in vivo genome editing. To date, at least 11 independent in vivo studies have used the AAV-SaCas9 system to edit disease-associated genes in a variety of tissues, including brain 68, muscle 54, 55, 57, 69, retina 58, heart 70, and liver 4, 49, 71. More recently, the quadruplex gRNAs/SaCas9 vector consisting of SaCas9 and multiplex sgRNAs was successfully delivered using AAV-DJ/8 for in vivo excision of HIV-1 proviral DNA in various solid tissues/organs via a single intravenous injection in humanized bone marrow/liver/thymus (BLT) mice with chronic HIV-1 infection 72. In the all-in-one vector system, CjCas9 can be packaged with multiple gRNAs and even a fluorescent reporter gene into a single AAV vector 5. Even though CjCas9 represents the smallest Cas9 orthologue characterized to date, only one in vivo study has demonstrated the use of AAV vectors to deliver CjCas9 into the mouse’s retina for the treatment of age-related macular degeneration because of the recent development of CjCas9 5. Excitingly, AAV-CjCas9 offers the possibility of editing multiplex genes and tracking the expression of fluorescent reporter genes at high efficiency by simply introducing a single type of AAV vector into the mouse body. The CRISPR-mediated genome-editing specificities can be further improved by adopting truncated gRNAs 73 and small chemical molecules to regulate Cas9 activity 74 or to enhance double-stranded break-induced homologous recombination efficiency 75 without sacrificing on-target genome-editing efficiencies.
Natural and engineered AAV serotypes
Multiple naturally occurring AAV serotypes have been harnessed to deliver the CRISPR/Cas9 complex into different tissues and organs in mice ( Figure 1B). AAV1 is particularly efficient to drive CRISPR transgene expression in the brain 25, 65, 76, whereas AAV8 appears well suited for the transduction of liver 4, 49, 61, 71, 77 and muscle 55 in mice. AAV9 has the general ability to transduce all major tissues, including muscle 54, 56, 69, retina 5, heart 62– 64, 78, and lung 65, in mice. Recently, the AAV9-CRISPR system has been successfully used to identify functional tumor suppressors in glioblastoma by performing high-throughput mutagenesis in the brain of conditional-Cas9 mice to recapitulate human glioblastoma 79. AAV2 59 and AAV5 58 have also been used to transduce retina, whereas AAV6 57 enables CRISPR-mediated gene editing in muscle tissue.
In order to alter tropism, reduce blockage by neutralizing antibodies, or enhance transduction efficiency, AAV capsids have been chemically or genetically modified to produce hybrid capsids by combining the properties of multiple serotypes, capsid shuffling, directed evolution, rational mutagenesis, or carrying peptide insertions that introduce the novel receptor-binding activity. AAV variants with engineered capsids (AAV2-retro, AAV2g9, AAV-DJ, and AAV-DJ/8) have been used to package CRISPR for transduction in the mouse brain. An in vivo-directed evolution-engineered AAV variant, rAAV2-retro, permits robust retrograde access to projection neurons in functionally connected and highly distributed networks with a high efficiency for neural circuit dissection and in vivo genome editing 68. An AAV chimera derived from AAV2 and AAV9, AAV2g9, enables preferential, robust, and widespread neuronal transduction within the mouse brain for CRISPR/Cas9-mediated gene deletion for the treatment of neurological disorders 80. To provide substantially higher infectivity rates across a broad range of tissues than any known native serotypes, the AAV-DJ was engineered by DNA family shuffling to create a hybrid capsid from eight different native serotypes 87. AAV-DJ/8 was created by introducing two point mutations in the heparin-binding domain of AAV-DJ to increase uptake in brain tissue in vivo, leading to a similar ability to infect heart and brain tissues with AAV8 and AAV9 87, 88. In fact, AAV-DJ/8 has been successfully used alongside the CRISPR/Cas9 system with doxycycline-dependent gRNA expression for inducible genome editing of neurons in vivo within the mouse brain 81. AAV-DJ/8 has also been successfully used to package and deliver the CRISPR/SaCas9 vectors for in vivo excision of HIV-1 proviral DNA in animal models 72.
Recently, the capsids of AAV9, which is the most efficient native serotype characterized to date for in vivo transduction of the brain, have been further engineered for better performance in transductions and tissue targeting specificity. These include AAV-PHP.B (a chimeric AAV9 variant generated by inserting a 7-mer sequence, TLAVPFK, on AAV9 capsid) 13, AAV-AS (a chimeric AAV9 variant generated by inserting a poly-alanine peptide on AAV9 capsid) 14, and AAV9/3 (double tyrosine-mutant form of AAV9) 89, all of which were shown to be more efficient in transductions of the adult mouse central nervous system after intravenous injection compared with the wild-type AAV9. Furthermore, some of these engineered AAV capsids could de-target the native AAV tropism without affecting vector production, capsid assembly, infectivity, and gene transfer. Therefore, these novel engineered vectors offer promising options to package the CRISPR/Cas9 complex to efficiently edit disease-associated genes in the mouse brain or any other tissues. Additionally, self-complementary AAV9 (scAAV9) can be used to significantly decrease the time required for the onset of CRISPR expression in the central nervous system of mice 84. Second-strand synthesis is a rate-limiting step for efficient transduction by recombinant AAV vectors 31. The use of double-stranded self-complementary AAV (scAAV) has improved the transduction efficiencies by bypassing the second-strand synthesis step, but cloning capacity was further reduced to accommodate the complementary strand 90.
Tissue-specific minimal promoters
A tissue-specific promoter is important to ensure the expression of the CRISPR transgene in a tissue- or organ-specific manner. Various truncated or minimal tissue-specific promoters have been used to drive the expression of CRISPR in specific tissues upon injection of the AAV vectors into the mouse body ( Figure 1C). For example, the 235 base pair (bp) mouse pMecp2 (methyl CpG binding protein 2) and 485 bp human hSyn1 (human synapsin I) promoters have been used to efficiently and specifically drive the expression of CRISPR and other transgenes in the mouse brain 25 and retina 59. A striated muscle-restricted promoter, CK8, enabled specific expression of SpCas9 and SaCas9 for gene editing in the muscle of the mdx mouse model of Duchenne muscular dystrophy upon intramuscular (tibialis anterior) or systemic (retro-orbital) injection of AAV6 vectors 57. The liver-specific TBG (thyroxine binding globulin) 4, 49 and cardiac-specific Myh6 (myosin heavy polypeptide 6) 63 promoters have been successfully used to drive the expression of CRISPR for in vivo genome editing in mouse liver and heart, respectively. More general promoters such as CMV (cytomegalovirus) 54, 58, 60, 62 and EFS (elongation factor-1 alpha short) 5, 65 can be used to express the CRISPR in multiple tissues, including muscle, retina, heart, and lung, for simultaneous gene editing of various tissues in mice. Compared with the CMV promoter, the elongation factor-1 alpha (EF1a) promoter tends to give more consistent and prolonged expression regardless of cell type or physiology 91, 92. EF1a is more stable in long-term culture because it maintains high transgene expression and stability and the copy number of episomal vectors 91. Thus, despite a stronger gene expression driven by the CMV promoter in many mammalian cell types, EF1a is preferred for efficient transgene expression in mouse embryonic stem cell lines 93 and for engineering stable tumor cell lines 92. The library of minimal tissue-specific promoters alongside AAV vectors used for controlling the expression of CRISPR or other transgenes in vivo is expanding with the development of more tightly regulated synthetic hybrid promoters, such as widely used CAG, CAGGS, CASI, CBh, GFAP, TRE3G, SMVP, Spc512, H1/T0, CK7-miniCMV, pICAM2, HCRhAATp, and Tight promoters ( Table 2).
Chimeric gRNA expression is frequently driven by an RNA polymerase III promoter, most often by the human U6 small nuclear (U6) promoter. The human U6 promoter has been shown to be more efficient than its murine homolog for short hairpin RNA (shRNA) expression in both human and murine cells 94. The human U6 promoter was also proven more efficient than the human H1 promoter in driving shRNA expression for the long-term inhibition of gene expression in vitro and in vivo 95. However, the use of the U6 promoter to overexpress shRNAs in vivo can induce severe hepatotoxicity and inflammatory side effects due to abundant shRNA production 96. The weaker H1 promoter may offer a safer option in this case, but its therapeutic efficacy is dependent on the sequence of the shRNA 96. Some promoters require a particular nucleotide as the transcription start site (TSS) to initiate transcription. For instance, the human or mouse U6 promoters require a guanine nucleotide (G) at the 5′ end of the transcript for effective transcription 97. To use the U6 promoter for driving gRNA expression, the first nucleotide of the transcribed gRNA should be a G to maximize U6 promoter activity 97. If the G is absent at the TSS of the gRNA sequence, the human RNA polymerase III promoter H1 can be used to drive the expression of the gRNA 76. The use of H1 promoter-expressed gRNAs could enhance the versatility of the CRISPR technology by expanding the targetable genome 98. For multiplex CRISPR/Cas9-based genome editing such as large genomic DNA deletion or synergistically enhanced gene activation and repression, four independent polymerase III promoters (human U6 promoter, murine U6 promoter, 7SK, and H1) can be used to express four gRNAs in a single expression cassette 21. The use of distinct polymerase III promoters can avoid the problem of genetic recombination in a viral vector and the loss of the proximal gRNA due to identical DNA sequences of the promoters 99. The doxycycline-dependent promoters such as H1/TO and U6/TO can also be used for inducible in vivo genome editing by supplying animals with Dox-containing food to regulate the expression of the gRNA in a Dox-dependent manner 81. The introduction of Dox inhibits Tet repressor (TetR) binding and induces gRNA expression 81.
Local and global delivery of AAV vectors
The route of administration of AAV-CRISPR vectors into the mouse body also determines the efficacy and specificity of gene editing for in vivo therapeutics ( Figure 1D). Given the inability of the most native AAV serotypes (except for AAV9 100, AAVrh8 101, and AAVrh10 102) to cross the blood-brain barrier, stereotactic 25, 65, 76 and intrathecal 84 injections are still the preferred routes to systemic injection in order to introduce the AAV-CRISPR vectors into specific regions of the mouse brain or central nervous system for therapeutic gene editing. For the stereotactic injection, AAV-CRISPR vectors are intracranially injected into the brain by using a stereotaxic apparatus and the injection micropipette. In the intrathecal injection approach, AAV-CRISPR vectors are injected into the spinal canal or subarachnoid space to reach cerebrospinal fluid. For efficient therapeutic gene editing in the mouse muscle tissue, intramuscular (via tibialis anterior), intraperitoneal (via intraperitoneal space), or intravenous (via the tail vein, temporal vein, and retro-orbital) injections are the preferred methods to introduce AAV-CRISPR vectors, as have been demonstrated for the mdx mouse model of Duchenne muscular dystrophy 55– 57. Subretinal 58, 60 or intravitreal 59, 85 injections are used to deliver AAV-CRISPR vectors into the mouse retina for gene editing. As most of the AAV vectors will accumulate in the mouse liver tissue upon systemic injection, intravenous injection by tail vein or temporal vein can efficiently edit the genes associated with liver diseases 4, 49, 71. Systemic injection remains preferred to local injection in the liver tissue because the delivery is more uniform and the distribution of viral vectors is broader. It is also a good delivery mode for gene editing in multiple tissues, as it is almost non-invasive to the body. On the other hand, intracardiac 64, subcutaneous 78, intraperitoneal 63, or systemic 62, 70 injection has been used to introduce AAV9 vectors carrying CRISPR transgene into the mouse heart for gene editing. Intranasal and intratracheal injections are used to locally deliver the AAV-CRISPR vehicles into the mouse lung tissue 65. Therefore, the selection of an effective route of injections is dependent on the target tissue, AAV serotypes, and tissue-specific promoters used.
CRISPR/Cas9-based human gene and cell therapies
In August 2016, the first human clinical trial using a CRISPR-based gene-editing technique was started for cancer immunotherapy of metastatic non-small cell lung cancer 103. Subsequently, 10 more early phase clinical trials are underway (ClinicalTrials.gov). One of these clinical trials has proceeded to phase 2 for the treatment of advanced esophageal cancer. Six of these early phase clinical trials are using the CRISPR/Cas9 system to create genetically altered immune cells, which are infused back into patients with an advanced stage of lung, bladder, prostate, renal, gastric, esophageal, or nasopharyngeal carcinoma or lymphoma to target and eradicate cancer cells ( Table 3). In general, the programmed cell death protein 1 gene ( PDCD1) will be knocked out in autologous T cells. These engineered T cells then will be selected and expanded ex vivo before infusion back into the patients. PDCD1, more commonly known as PD-1, is an inhibitory cell surface receptor involved in the regulation of T-cell function during immune response and tolerance. Knockout of PD-1 in T cells extracted from the patient’s blood could prevent cells’ immune response from switch-off after re-introduction of the gene-edited cells into the patient’s bloodstream and attack and defeat the cancer cells 104. In fact, antibodies (for example, nivolumab and pembrolizumab) that neutralize PD-1 with high response and low adverse effect rates have been successfully used for cancer immunotherapy in human clinical trials 105, 106. Gene editing is expected to inhibit PD-1 with a greater certainty, and by multiplying the cells in the laboratory, scientists can enhance the efficacy of triggering an immune response against tumors 104.
Table 3. Ongoing human clinical trials involving CRISPR/Cas9-based gene and cellular therapies.
| Condition | Intervention | Phase | Type | Primary objective and study
design |
Principle | Start date | Finish date | ClinicalTrials.
gov identifier |
|---|---|---|---|---|---|---|---|---|
| Metastatic non-
small cell lung cancer |
Biological: CRISPR/Cas9-
mediated PD-1 knockout T cells from autologous origin Drug: cyclophosphamide, interleukin-2 |
Phase 1 | Ex vivo | A dose-escalation study to
evaluate the safety of ex vivo knocked-out, expanded, and selected PD-1 knockout engineered T cells that are infused back into the patient for the treatment of metastatic non- small cell lung cancer |
Target cancer
cell |
August 2016 | April 2018 | NCT02793856 |
| Muscle-invasive
bladder cancer stage IV |
Biological: CRISPR/Cas9-
mediated PD-1 knockout T cells from autologous origin Drug: cyclophosphamide, interleukin-2 |
Phase 1 | Ex vivo | A dose-escalation study of
ex vivo knocked-out, expanded, and selected PD-1 knockout engineered T cells that are infused back into the patient for the treatment of muscle-invasive bladder cancer |
Target cancer
cell |
September
2016 |
September
2019 |
NCT02863913 |
| Hormone-refractory
prostate cancer |
Biological: CRISPR/Cas9-
mediated PD-1 knockout T cells from autologous origin Drug: cyclophosphamide, interleukin-2 |
Phase 1 | Ex vivo | A dose-escalation study of
ex vivo knocked-out, expanded, and selected PD-1 knockout engineered T cells that are infused back into the patient for the treatment of castration- resistant prostate cancer |
Target cancer
cell |
November
2016 |
December
2020 |
NCT02867345 |
| Metastatic renal
cell carcinoma |
Biological: CRISPR/Cas9-
mediated PD-1 knockout T cells from autologous origin Drug: cyclophosphamide, interleukin-2 |
Phase 1 | Ex vivo | A dose-escalation study of
ex vivo knocked-out, expanded, and selected PD-1 knockout engineered T cells that are infused back into the patient for the treatment of metastatic advanced renal cancer |
Target cancer
cell |
November
2016 |
November
2020 |
NCT02867332 |
| Advanced
esophageal cancer |
Biological: CRISPR/Cas9-
mediated PD-1 knockout T cells from autologous origin Drug: cyclophosphamide, interleukin-2 |
Phase 2 | Ex vivo | Evaluate the safety of
ex vivo
knocked-out, expanded, and selected PD-1 knockout T cells that are infused back into the patient for the treatment of advanced esophageal cancer |
Target cancer
cell |
March 2017 | December
2018 |
NCT03081715 |
| Gastric carcinoma
stage IV, nasopharyngeal carcinoma stage IV, T-cell lymphoma stage IV, adult Hodgkin lymphoma stage IV, diffuse large B-cell lymphoma stage IV |
Biological: CRISPR/Cas9-
mediated PD-1 knockout T cells from autologous origin Drug: fludarabine, cyclophosphamide, interleukin-2 |
Phase 1/2 | Ex vivo | Evaluate the safety and clinical
response of cell therapy using CRISPR-Cas9-mediated PD-1 knockout EBV-CTL cells for the treatment of advanced-stage EBV-associated malignancies |
Target EBV-
associated cancer cell |
April 2017 | March 2022 | NCT03044743 |
| HIV-1-infection | Biological: CRISPR/Cas9-
mediated CCR5 modified CD34 + hematopoietic stem/ progenitor cells from donors Drug: anti-retroviral therapy |
Phase 1 | Ex vivo | Evaluate the safety and
feasibility of allotransplantation with CRISPR/Cas9 CCR5 gene modified CD34 + hematopoietic stem/progenitor cells in HIV-infected patients with hematological malignances |
Target CCR5-
positive immune cell |
May 2017 | May 2021 | NCT03164135 |
| B-cell leukemia,
B-cell lymphoma |
Biological: gene-disrupted
allogeneic CD19-directed BBζ CAR-T cells (termed UCART019) will be generated by combining the lentiviral delivery of CAR and CRISPR RNA electroporation to disrupt endogenous TCR and B2M genes |
Phase 1/2 | Ex vivo | Evaluate the feasibility, safety,
and in vivo persistence of UCART019 adoptively transferred T cells in patients with relapsed or refractory CD19 + leukemia and lymphoma |
Target cancer
cell |
June 2017 | May 2022 | NCT03166878 |
| Human
papillomavirus- related malignant neoplasm |
Biological: TALEN and
CRISPR/Cas9 |
Phase 1 | In vivo | An open-label and triple-cohort
study to evaluate the safety and efficacy of TALEN and CRISPR/ Cas9 plasmids for the treatment of HPV persistency and HPV- related cervical intraepithelial neoplasia |
Disrupt HPV
E6/E7 DNA |
January 2018 | January
2019 |
NCT03057912 |
| Neurofibromatosis
type 1 |
Biological: establish isogenic
NF1 wild-type ( NF1 +/ +), NF1 heterozygous ( NF1 +/ −), and NF1 homozygous ( NF1 −/ −) patient-specific iPSC lines using CRISPR/Cas9 technology |
Phase 1 | Ex vivo | Establish an iPSC bank for disease
phenotypic characterization, drug screening, and identification that can reverse or alleviate the disease phenotypes |
Collection of
stem cells |
November
2017 |
June 2019 | NCT03332030 |
| Gastrointestinal
infection |
Biological: knockout CRISPR
and gain-of-function CRISPR SAM Procedure: duodenal biopsy |
Phase 1 | Ex vivo | Identify and establish a list of
host cellular proteins that mediate norovirus infection in a stem cell- derived human intestinal enteroid model |
Genome-
wide genetic screening |
January 2018 | December
2020 |
NCT03342547 |
| Sickle cell disease | Overall genetic literacy,
CRISPR-specific literacy, and general attitudes and beliefs toward CRISPR |
Observational | Cross-
sectional |
Study the attitudes, beliefs, and
opinions of those with SCD, parents of those with SCD, and providers on the use of CRISPR/ Cas9 gene-editing |
– | October 2017 | June 2018 | NCT03167450 |
Based on ClinicalTrials.gov database on human clinical trials performed in the US and worldwide. B2M, beta-2-microglobulin; CAR, chimeric antigen receptor; CCR5, C-C chemokine receptor type 5; CTL, cytotoxic T-lymphocyte; EBV, Epstein-Barr virus; HPV, human papillomavirus; iPSC, induced pluripotent stem cell; NF1, neurofibromatosis type 1; PD-1, programmed cell death protein 1 gene; SCD, sickle cell disease; TALEN, transcription activator-like effector nuclease; TCR, T-cell receptor.
These interventional studies aimed to evaluate the clinical response, safety, and maximal tolerant dose of the CRISPR-mediated PD-1 knockout T cells in treating advanced-stage malignancies upon being infused back into patients. An anti-cancer chemotherapy drug, cyclophosphamide, will be administered for a few days to modify the immune micro-environment before cell infusion or to deplete regulatory T cells (Tregs) before collecting peripheral blood. After cell infusion, the interleukin-2 cytokine that has both immune-modulating and anti-tumor properties will be administered to sustain the survival of infused T cells. For cancer immunotherapy of lymphoma or leukemia, an anti-metabolite anti-cancer chemotherapy drug, fludarabine, will be co-administered with the cyclophosphamide before cell infusion. Biomarkers and immunological markers will be closely monitored after cell infusion to determine whether the injections are causing any serious adverse effects and to evaluate whether the patients will receive benefits from the treatment.
In another CRISPR/Cas9-based ex vivo gene therapy clinical trial, the CRISPR/Cas9 complex will be used to disrupt the C-C chemokine receptor type 5 ( CCR5) gene in the CD34 + hematopoietic stem or progenitor cells from donors (NCT03164135). Then, these CCR5 modified CD34 + cells from donors will be infused into the HIV-infected patients with AIDS and hematological malignancies. In principle, HIV-1 virus requires CCR5 receptor on the surface of host immune cells (for example, T cells) to infect the cell; hence, disrupting or blocking the CCR5 receptor may make the immune cell resistant to the virus infection. When the CCR5-modified immune cells from the donor are mixed inside the body with the CCR5-positive immune cells from HIV-infected patients, the HIV-1 virus will infect and kill the CCR5-positive immune cells and the CCR5-negative immune cells will survive and eventually take over the immune function. The primary objectives of this clinical trial are to determine the safety of the allotransplantation of these CD34 + cells and to evaluate the resistance to the HIV-1 virus in HIV-infected patients after infusion of these CD34 + cells. Prior to the cell infusion, the patients will be treated with anti-retroviral therapy to achieve undetectable HIV-1 virus in the peripheral blood. During the follow-up, HIV-1 viral load, CD4 + T cells, and CCR5-negative cell counts in the peripheral blood will be monitored in HIV-infected patients.
Another promising ex vivo gene therapy clinical trial is the combined use of CRISPR/Cas9 gene-editing technology and chimeric antigen receptor (CAR)-based cancer immunotherapy (NCT03166878). The CAR-expressed T cells can be engineered to recognize leukemia antigens such as CD19 on B cells for the treatment of relapsed or refractory B-cell malignancies. However, the cost and technical difficulties in the production and expansion of CAR-expressed T cells have hindered the wide application of personalized autologous CAR–T cell therapy. Thus, universal CAR–T cells derived from healthy unrelated donors are developed to overcome some of these drawbacks. To evade host-mediated immunity and deliver anti-leukemic effects without graft-versus-host disease in patients with relapsed or refractory B-cell malignancies, gene-disrupted allogeneic CD19-directed BBζ CAR–T cells (known as UCART019) are developed by combining the lentiviral delivery of CAR and CRISPR mRNA electroporation to disrupt both of the endogenous T-cell receptor ( TCR) and beta-2 microglobulin ( B2M) genes. The TCR and B2M play important roles in triggering the immune response; hence, disrupting both of these genes could minimize immunogenicity associated with allotransplantation. The primary goal of these allogeneic CD19 CAR–T cells is to evaluate the feasibility, safety, and in vivo persistence of UCART019 adoptively transferred T cells in patients with relapsed or refractory CD19 + leukemia and lymphoma. The amount of UCART019 cells (engraftment), humoral host immunity, and anti-tumor response upon UCART019 cell infusions will be monitored.
Owing to the tissue complexities in humans, toxicity concern, and potential complications such as host immune response following a high dose of AAV vectors used to achieve significant therapeutic efficacy 107, the AAV-CRISPR viral delivery system is restricted to ex vivo gene editing or genetic manipulation in animals. One of the promising strategies to overcome this issue was to mutagenize the surface-exposed tyrosine residues on the AAV capsid in order to avoid AAV degradation by the host cell proteasome machinery and to improve AAV intracellular trafficking to the nucleus, which can lead to high transduction efficiency at lower vector doses 107. The combined use of capsid-modified and genome-modified next-generation AAV vectors has allowed higher transduction efficiency and transgene expression at further reduced doses 108. To date, the AAV-CRISPR gene-editing system was tested in non-human animals only, but the first in vivo gene therapy using CRISPR/Cas9 technology will be carried out in human clinical trials soon. In January 2018, an open-label and triple-cohort study will be conducted to evaluate the safety of therapeutic doses and the dosing regimen of CRISPR/Cas9 plasmid to treat human papillomavirus (HPV) persistency and HPV-related cervical intraepithelial neoplasia. Given the important roles that E6 and E7 play in HPV-driven carcinogenesis, CRISPR/Cas9-mediated disruption of HPV16 and HPV18 E6/E7 DNA could be an attractive approach for therapeutic interventions by significantly downregulating the expression of E6/E7 in order to induce HPV-associated cell apoptosis and to inhibit cell growth.
Promises and hurdles associated with the AAV-CRISPR system for future clinical applications
An increasing number of studies in mice have clearly demonstrated that the combined use of tissue-specific minimal promoters, natural and engineered AAV serotypes, different routes of administration, and small Cas9 orthologues enables efficient packaging and precise delivery of AAV-CRISPR vectors for targeted in vivo genome editing in specific tissues with minimized side effects. Nevertheless, special considerations are required when selecting tissue-specific promoters, natural and engineered AAV serotypes, or routes of administration to avoid non-specific delivery and transgene expression of the AAV. In addition, the off-target effects, toxicity, and immunogenicity associated with CRISPR/Cas9 delivery remain to be fully resolved.
Non-specific delivery and transgene expression
Tissue-specific promoters and AAV serotypes might still be able to transduce and induce transgene expression in healthy tissues or other non-target organs if a strong promoter or a high dose of viral vectors is introduced into the body by intravenous injection. Despite a significant improvement in the efficiency and specificity of newly discovered natural and engineered AAV variants for systemic delivery in mice, the toxicity and side effects associated with the non-specific delivery of the transgenes to the non-target tissues and organs remain a concern 13, 89. It is easier to get delivery vehicles taken up by liver than other organs upon systemic injection. Therefore, it is more challenging to specifically target non-liver organs than liver via a systemic delivery approach. For instance, recently discovered synthetic vectors, AAV-PHP.B 13 and tyrosine-mutant AAV 89, were found to transduce the adult mouse central nervous system more efficiently and widely than the natural AAV9 after intravenous injection. However, these synthetic vectors can also transduce the liver and other peripheral organs upon systemic delivery 13, 89. In this case, local injections such as stereotaxic injection into the brain can minimize the possible complications or adverse side effects associated with non-specific expression of CRISPR in healthy tissues or other non-target organs.
Off-target effects
Even though CRISPR/Cas9 predominantly recognizes the intended target sites, a series of high-throughput genome-wide methods such as multiplex Digenome-seq 109, 110, ChIP-seq 111, 112, GUIDE-seq 113, and whole-genome sequencing 114 as well as targeted deep sequencing 112, Cas9 toxicity screens 115, and SITE-seq biochemical methods 116 have revealed evidence of off-target effects due to target mismatch tolerance of CRISPR/Cas9. As inter-individual natural genetic variation can affect CRISPR/Cas9 specificity 117, a recently developed CIRCLE-seq approach could be used to identify genome-wide off-target mutations of CRISPR/Cas9 that are associated with cell type-specific single-nucleotide polymorphisms to provide personalized specificity profiles 118. Notably, given the same target sequence of gRNA, off-target sites of Cas9 before and after being fused to a catalytic enzyme (for example, cytidine deaminase base editor and chromatin modifiers) could be different; therefore, independent assessment of their genome-wide specificities is recommended 119. Imprecise repair of Cas9-induced DNA double-stranded breaks can give rise to deleterious structural chromosomal rearrangements such as deletions, inversions, and translocations, which in turn may activate oncogenes or cause genome instability 120. Hence, high-throughput screenings of CRISPR/Cas9 off-target activity and further improvement in the fidelity of CRISPR/Cas9 on-target activity are essential for safety in clinical gene transfer applications.
There are many ways to minimize CRISPR/Cas9 off-target effects in the human genome. For example, a recently developed RNA-targeting Cas9 (RCas9) system could avoid permanent off-target genetic lesions in DNA-mediated CRISPR-based therapeutics 121. The RCas9 system consists of a fusion of rationally truncated dCas9 protein and PilT N-terminus (PIN) domain that can be packaged into the AAV vector for eliminating toxic microsatellite repeat expansion RNA or reducing repetitive RNA level without targeting the DNA 121. With a similar strategy, systemic delivery of dCas9/gRNA by AAV9 significantly reduced pathological RNA foci, rescued chloride channel 1 protein expression, and decreased myotonia in myotonic dystrophy mice by impeding the transcription of expanded microsatellite repeats 122. Another way is to deliver purified Cas9 RNPs rather than plasmid expression vectors 10, 123. Despite some technical constraints in non-viral delivery methods for in vivo administration, the use of Cas9 RNPs can limit the duration of Cas9 expression and decrease the chance of Cas9 nuclease cleaving at non-specific sites in a genome because of the rapid clearance from the cell 10, 123. Complementing the CRISPR-based editing capability with conditional genetic manipulation tools such as photoactivatable Cas9 124, 125, chemical-inducible Cas9 126, 127, or multiple inputs logic gate genetic circuits 128, 129 enables the precise spatial and temporal control of Cas9 activity inside the cell, which in turn leads to the reduction in off-target activity. Alternatively, a pair of Cas9 nickases 130– 132, dCas9-FokI 133, or high-fidelity Cas9 variants such as SpCas9-HF1 134, eSpCas9 135, and HypaCas9 136 can be used to minimize undesired off-target mutagenesis.
The use of a scarless genome-editing strategy for targeted point mutation knock-in can also minimize unwanted mutation formations to favor the desired clean base editing outcomes 137, 138. For a point mutation knock-in, the efficiency of precise sequence replacement by CRISPR-mediated homology-directed repair (HDR) could be significantly increased by using asymmetric donor DNA 139, HDR enhancer 140, or short ssDNA donor oligonucleotides as a donor template instead of long plasmid donor 139 or by inhibiting non-homologous end joining (NHEJ) activity 141. Also, the artificial chimeric RNAs 142—truncated 73, 113 and chemically modified 143 gRNAs—were shown to have lower off-target activity than the original gRNAs. A number of bioinformatics analysis tools such as GuideScan 144, CRISPRdirect 145, and Cas-OFFinder 146 permit the specific design of gRNAs to avoid binding at the non-intended target sites in the human genome. The specificity of the gRNA designed can be improved by selecting a target sequence with two Gs at the 5′ terminus 131 and avoiding potential mismatches at the seed sequence or base-pairing adjacent to the PAM 147. Another innovative way to improve the specificity and reduce the toxicity of the CRISPR/Cas9 is co-delivery of DNA decoys or competitive inhibitor oligonucleotides bearing all possible off-target sequences that can sequester and prevent the CRISPR/Cas9 from binding to the off-target sites within a host genome. A similar concept has been successfully demonstrated to alleviate the off-target effects of RNA interference by using RNA decoys to reduce the sense strand activity of shRNAs 148, while artificial microRNA (miRNA) sponges have been used to inhibit miRNA function and its ability to regulate natural mRNAs 149. In addition, a recently developed miRNA-responsive CRISPR/Cas9 switch could be useful for cell type-specific genome editing by sensing endogenous miRNA activities 150.
Immunogenicity
Owing to the exogenous nature of AAV and CRISPR components, host immune responses can attenuate therapeutic effects and cause side effects. Thus, the toxicity and immunogenicity associated with AAV and CRISPR components should be circumvented for safer and higher efficacy in clinical gene therapy applications. Even though the AAV generally elicits a very mild immune response and does not induce extensive cellular damage in vivo, certain AAV serotypes such as AAV9 may evoke humoral immune responses, as indicated by the presence of capsid-specific antibodies 50. While transient immunosuppression is one of the possible ways to mitigate the host immune response following the delivery of AAV vectors 151, it is not feasible for long-term therapeutic treatments of chronic diseases and is prone to adverse complications such as infections and malignancies. Empty capsid mutants can be used as decoys to overcome pre-existing humoral immunity by adsorbing antibodies in the bloodstream upon systemic delivery of both empty and functional AAVs 152. Alternatively, the AAV capsids can be genetically engineered or mutated to reduce the binding affinity and the neutralizing effects of AAV antibodies 15, 153– 155. In addition to AAVs, the presence of Cas9-specific antibodies in Cas9-exposed animals indicated that Cas9 could evoke deleterious cellular and humoral immune responses in vivo 50. Expression of Cas9 in vivo could affect the transcription of the genes associated with the immunological processes. This in turn may destabilize the host immune system, elicit significant cellular infiltration or expansion, and induce enlargement of the draining lymph nodes with increased immune cell counts 50. The Cas9-responsive T-cell clonotype described previously could serve as a distinctive biomarker to assess Cas9-specific immunity before clinical implementation of the CRISPR system 50. To minimize the immune response due to prolonged expression of Cas9, conditional genome editing with the self-limiting CRISPR/Cas9 system 58 or light-inducible 124, 125 or chemical-inducible 126, 127 Cas9 can be used to minimize the duration of Cas9 expression in the body.
Conclusions and perspectives
The AAV-CRISPR system holds enormous translational potential to develop curative therapeutic options for patients with severe and life-threatening genetic diseases by permanently editing disease-causing or risk genes in the human body. The delivery, efficacy, and safety issues in treating complex heritable and somatic disorders have to be fully resolved to fulfill this promise. Thus far, the ex vivo approach has been adopted to overcome the technical challenges associated with the in vivo delivery of the AAV-CRISPR viral vectors in humans. The ex vivo gene-editing approach is commonly used for the therapeutic treatment of blood disorders, cancers, and immune-related diseases. In the near future, in vivo genome editing is expected to offer better avenues to treat a wide range of human hereditary diseases in adults. In vivo gene therapy in humans provides several advantages over the ex vivo approach, and sometimes a combination of the two is necessary to achieve a good therapeutic outcome. For example, in vivo photoreceptor cell rescue can be used to halt retinal degeneration to preserve existing vision, while ex vivo photoreceptor cell replacement can be used to restore lost vision in patients with retinal dystrophy 156. After in vivo photoreceptor cell rescue, the retinal environment may become more permissive for transplanted photo-receptor survival and de novo synaptogenesis via the ex vivo approach 156.
The AAV vector already has a long history of success in clinical trials in vivo, but owing to the relatively recent arrival of CRISPR technology, the AAV-CRISPR system has yet to be tested in vivo for human gene therapy trials. Nevertheless, three ongoing human clinical trials (NCT03041324, NCT02702115, and NCT02695160) have used AAV vectors to deliver zinc finger nucleases, an earlier and well-established gene-editing tool, to the liver tissue for the treatment of hemophilia B and mucopolysaccharidosis. Similarly, the AAV vectors may be used for CRISPR delivery for human clinical trials in the future. Excitingly, the first in vivo human gene therapy trials using CRISPR/Cas9 plasmid administration to treat human HPV-related malignant neoplasms will be initiated in January 2018 (NCT03057912).
Although it is more feasible to deliver the transgene ex vivo than in vivo, the in vivo approach eliminates the need for cell transplants. Therefore, in vivo therapy can avoid potential graft-versus-host disease and immunosuppression-related complications such as infections and malignancies, as seen in cellular therapy. Recently, various genetically engineered AAV variants or structure-guided derivations of AAV mutants have been developed to significantly improve the efficiency and specificity for in vivo delivery 157. Compared with existing AAV serotypes, these newly engineered AAV capsids enabled higher transduction efficiency at a targeted tissue by altering the native tropism of the AAV capsid 13– 15, 158 and had low immunogenicity by evading pre-existing anti-AAV capsid neutralizing antibodies in the human body 159, 160. Given the encouraging results obtained with the next-generation synthetic AAV capsids in animals, this synthetic AAV vector may also be used to deliver CRISPR/Cas9 transgene in humans for in vivo genome editing in the near future. Further improvement in the performance of engineered AAV variants and CRISPR components is necessary to realize the full potential of the AAV-CRISPR system for in vivo genome editing.
Editorial Note on the Review Process
F1000 Faculty Reviews are commissioned from members of the prestigious F1000 Faculty and are edited as a service to readers. In order to make these reviews as comprehensive and accessible as possible, the referees provide input before publication and only the final, revised version is published. The referees who approved the final version are listed with their names and affiliations but without their reports on earlier versions (any comments will already have been addressed in the published version).
The referees who approved this article are:
Dirk Grimm, Department of Infectious Diseases, Virology, Heidelberg University Hospital, Heidelberg, Germany
Jin-Soo Kim, Department of Chemistry, Seoul National University, Seoul, 151-742, Korea, South
Funding Statement
This work was funded by National Institutes of Health grants AG017242, GM104459, and CA180126 (to Yousin Suh).
[version 1; referees: 2 approved]
References
- 1. Lau CH, Suh Y: Genome and Epigenome Editing in Mechanistic Studies of Human Aging and Aging-Related Disease. Gerontology. 2017;63(2):103–17. 10.1159/000452972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barrangou R, Doudna JA: Applications of CRISPR technologies in research and beyond. Nat Biotechnol. 2016;34(9):933–41. 10.1038/nbt.3659 [DOI] [PubMed] [Google Scholar]
- 3. Cong L, Ran FA, Cox D, et al. : Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–23. 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 4. Ran FA, Cong L, Yan WX, et al. : In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520(7546):186–91. 10.1038/nature14299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kim E, Koo T, Park SW, et al. : In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat Commun. 2017;8:14500. 10.1038/ncomms14500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Komor AC, Badran AH, Liu DR: CRISPR-Based Technologies for the Manipulation of Eukaryotic Genomes. Cell. 2017;168(1–2):20–36. 10.1016/j.cell.2016.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 7. Xiong X, Chen M, Lim WA, et al. : CRISPR/Cas9 for Human Genome Engineering and Disease Research. Annu Rev Genomics Hum Genet. 2016;17:131–54. 10.1146/annurev-genom-083115-022258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kouranova E, Forbes K, Zhao G, et al. : CRISPRs for Optimal Targeting: Delivery of CRISPR Components as DNA, RNA, and Protein into Cultured Cells and Single-Cell Embryos. Hum Gene Ther. 2016;27(6):464–75. 10.1089/hum.2016.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liang X, Potter J, Kumar S, et al. : Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. J Biotechnol. 2015;208:44–53. 10.1016/j.jbiotec.2015.04.024 [DOI] [PubMed] [Google Scholar]
- 10. Kim S, Kim D, Cho SW, et al. : Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24(6):1012–9. 10.1101/gr.171322.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fornito A, Zalesky A, Breakspear M: The connectomics of brain disorders. Nat Rev Neurosci. 2015;16(3):159–72. 10.1038/nrn3901 [DOI] [PubMed] [Google Scholar]
- 12. Staahl BT, Benekareddy M, Coulon-Bainier C, et al. : Efficient genome editing in the mouse brain by local delivery of engineered Cas9 ribonucleoprotein complexes. Nat Biotechnol. 2017;35(5):431–4. 10.1038/nbt.3806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Deverman BE, Pravdo PL, Simpson BP, et al. : Cre-dependent selection yields AAV variants for widespread gene transfer to the adult brain. Nat Biotechnol. 2016;34(2):204–9. 10.1038/nbt.3440 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 14. Choudhury SR, Harris AF, Cabral DJ, et al. : Widespread Central Nervous System Gene Transfer and Silencing After Systemic Delivery of Novel AAV-AS Vector. Mol Ther. 2016;24(4):726–35. 10.1038/mt.2015.231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Choudhury SR, Fitzpatrick Z, Harris AF, et al. : In vivo Selection Yields AAV-B1 Capsid for Central Nervous System and Muscle Gene Therapy. Mol Ther. 2016;24(7):1247–57. 10.1038/mt.2016.84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yin H, Kauffman KJ, Anderson DG: Delivery technologies for genome editing. Nat Rev Drug Discov. 2017;16(6):387–99. 10.1038/nrd.2016.280 [DOI] [PubMed] [Google Scholar]
- 17. Song M: The CRISPR/Cas9 system: Their delivery, in vivo and ex vivo applications and clinical development by startups. Biotechnol Prog. 2017;33(4):1035–45. 10.1002/btpr.2484 [DOI] [PubMed] [Google Scholar]
- 18. He ZY, Men K, Qin Z, et al. : Non-viral and viral delivery systems for CRISPR-Cas9 technology in the biomedical field. Sci China Life Sci. 2017;60(5):458–67. 10.1007/s11427-017-9033-0 [DOI] [PubMed] [Google Scholar]
- 19. Williams MR, Fricano-Kugler CJ, Getz SA, et al. : A Retroviral CRISPR-Cas9 System for Cellular Autism-Associated Phenotype Discovery in Developing Neurons. Sci Rep. 2016;6:25611. 10.1038/srep25611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Koike-Yusa H, Li Y, Tan EP, et al. : Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat Biotechnol. 2014;32(3):267–73. 10.1038/nbt.2800 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 21. Kabadi AM, Ousterout DG, Hilton IB, et al. : Multiplex CRISPR/Cas9-based genome engineering from a single lentiviral vector. Nucleic Acids Res. 2014;42(19):e147. 10.1093/nar/gku749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang D, Mou H, Li S, et al. : Adenovirus-Mediated Somatic Genome Editing of Pten by CRISPR/Cas9 in Mouse Liver in Spite of Cas9-Specific Immune Responses. Hum Gene Ther. 2015;26(7):432–42. 10.1089/hum.2015.087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cheng R, Peng J, Yan Y, et al. : Efficient gene editing in adult mouse livers via adenoviral delivery of CRISPR/Cas9. FEBS Lett. 2014;588(21):3954–8. 10.1016/j.febslet.2014.09.008 [DOI] [PubMed] [Google Scholar]
- 24. Schmidt F, Grimm D: CRISPR genome engineering and viral gene delivery: a case of mutual attraction. Biotechnol J. 2015;10(2):258–72. 10.1002/biot.201400529 [DOI] [PubMed] [Google Scholar]
- 25. Swiech L, Heidenreich M, Banerjee A, et al. : In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat Biotechnol. 2015;33(1):102–6. 10.1038/nbt.3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mansouri M, Ehsaei Z, Taylor V, et al. : Baculovirus-based genome editing in primary cells. Plasmid. 2017;90:5–9. 10.1016/j.plasmid.2017.01.003 [DOI] [PubMed] [Google Scholar]
- 27. Mansouri M, Bellon-Echeverria I, Rizk A, et al. : Highly efficient baculovirus-mediated multigene delivery in primary cells. Nat Commun. 2016;7:11529. 10.1038/ncomms11529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen X, Gonçalves MA: Engineered Viruses as Genome Editing Devices. Mol Ther. 2016;24(3):447–57. 10.1038/mt.2015.164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kwang TW, Zeng X, Wang S: Manufacturing of AcMNPV baculovirus vectors to enable gene therapy trials. Mol Ther Methods Clin Dev. 2016;3:15050. 10.1038/mtm.2015.50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Epstein BE, Schaffer DV: Combining Engineered Nucleases with Adeno-associated Viral Vectors for Therapeutic Gene Editing. Adv Exp Med Biol. 2017;1016:29–42. 10.1007/978-3-319-63904-8_2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mingozzi F, High KA: Therapeutic in vivo gene transfer for genetic disease using AAV: progress and challenges. Nat Rev Genet. 2011;12(5):341–55. 10.1038/nrg2988 [DOI] [PubMed] [Google Scholar]
- 32. Senís E, Fatouros C, Große S, et al. : CRISPR/Cas9-mediated genome engineering: an adeno-associated viral (AAV) vector toolbox. Biotechnol J. 2014;9(11):1402–12. 10.1002/biot.201400046 [DOI] [PubMed] [Google Scholar]
- 33. Zhu H, Lau C, Goh S, et al. : Baculoviral transduction facilitates TALEN-mediated targeted transgene integration and Cre/LoxP cassette exchange in human-induced pluripotent stem cells. Nucleic Acids Res. 2013;41(19):e180. 10.1093/nar/gkt721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Joung J, Konermann S, Gootenberg JS, et al. : Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat Protoc. 2017;12(4):828–63. 10.1038/nprot.2017.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Klann TS, Black JB, Chellappan M, et al. : CRISPR-Cas9 epigenome editing enables high-throughput screening for functional regulatory elements in the human genome. Nat Biotechnol. 2017;35(6):561–8. 10.1038/nbt.3853 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 36. Morris SJ, Turner AV, Green N, et al. : Laboratory-Scale Production of Replication-Deficient Adenovirus Vectored Vaccines. Methods Mol Biol. 2016;1349:121–35. 10.1007/978-1-4939-3008-1_8 [DOI] [PubMed] [Google Scholar]
- 37. Garnier A, Côté J, Nadeau I, et al. : Scale-up of the adenovirus expression system for the production of recombinant protein in human 293S cells. Cytotechnology. 1994;15(1–3):145–55. 10.1007/BF00762389 [DOI] [PubMed] [Google Scholar]
- 38. Fricano-Kugler CJ, Williams MR, Salinaro JR, et al. : Designing, Packaging, and Delivery of High Titer CRISPR Retro and Lentiviruses via Stereotaxic Injection. J Vis Exp. 2016; (111). 10.3791/53783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yi Y, Noh MJ, Lee KH: Current advances in retroviral gene therapy. Curr Gene Ther. 2011;11(3):218–28. 10.2174/156652311795684740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dropulić B: Lentiviral vectors: their molecular design, safety, and use in laboratory and preclinical research. Hum Gene Ther. 2011;22(6):649–57. 10.1089/hum.2011.058 [DOI] [PubMed] [Google Scholar]
- 41. Mátrai J, Chuah MK, VandenDriessche T: Recent advances in lentiviral vector development and applications. Mol Ther. 2010;18(3):477–90. 10.1038/mt.2009.319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li C, Guan X, Du T, et al. : Inhibition of HIV-1 infection of primary CD4 + T-cells by gene editing of CCR5 using adenovirus-delivered CRISPR/Cas9. J Gen Virol. 2015;96(8):2381–93. 10.1099/vir.0.000139 [DOI] [PubMed] [Google Scholar]
- 43. Gwiazda KS, Grier AE, Sahni J, et al. : High Efficiency CRISPR/Cas9-mediated Gene Editing in Primary Human T-cells Using Mutant Adenoviral E4orf6/E1b55k "Helper" Proteins. Mol Ther. 2016;24(9):1570–80. 10.1038/mt.2016.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. St George JA: Gene therapy progress and prospects: adenoviral vectors. Gene Ther. 2003;10(14):1135–41. 10.1038/sj.gt.3302071 [DOI] [PubMed] [Google Scholar]
- 45. Kochanek S: High-capacity adenoviral vectors for gene transfer and somatic gene therapy. Hum Gene Ther. 1999;10(15):2451–9. 10.1089/10430349950016807 [DOI] [PubMed] [Google Scholar]
- 46. Lusky M: Good manufacturing practice production of adenoviral vectors for clinical trials. Hum Gene Ther. 2005;16(3):281–91. 10.1089/hum.2005.16.281 [DOI] [PubMed] [Google Scholar]
- 47. Volpers C, Kochanek S: Adenoviral vectors for gene transfer and therapy. J Gene Med. 2004;6(Suppl 1):S164–71. 10.1002/jgm.496 [DOI] [PubMed] [Google Scholar]
- 48. McConnell MJ, Imperiale MJ: Biology of adenovirus and its use as a vector for gene therapy. Hum Gene Ther. 2004;15(11):1022–33. 10.1089/hum.2004.15.1022 [DOI] [PubMed] [Google Scholar]
- 49. Yang Y, Wang L, Bell P, et al. : A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nat Biotechnol. 2016;34(3):334–8. 10.1038/nbt.3469 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 50. Chew WL, Tabebordbar M, Cheng JK, et al. : A multifunctional AAV-CRISPR-Cas9 and its host response. Nat Methods. 2016;13(10):868–74. 10.1038/nmeth.3993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. During MJ: Adeno-associated virus as a gene delivery system. Adv Drug Deliv Rev. 1997;27(1):83–94. 10.1016/S0169-409X(97)00024-0 [DOI] [PubMed] [Google Scholar]
- 52. Kotterman MA, Schaffer DV: Engineering adeno-associated viruses for clinical gene therapy. Nat Rev Genet. 2014;15(7):445–51. 10.1038/nrg3742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Airenne KJ, Hu YC, Kost TA, et al. : Baculovirus: an insect-derived vector for diverse gene transfer applications. Mol Ther. 2013;21(4):739–49. 10.1038/mt.2012.286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tabebordbar M, Zhu K, Cheng JKW, et al. : In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science. 2016;351(6271):407–11. 10.1126/science.aad5177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nelson CE, Hakim CH, Ousterout DG, et al. : In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science. 2016;351(6271):403–7. 10.1126/science.aad5143 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 56. Long C, Amoasii L, Mireault AA, et al. : Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science. 2016;351(6271):400–3. 10.1126/science.aad5725 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 57. Bengtsson NE, Hall JK, Odom GL, et al. : Muscle-specific CRISPR/Cas9 dystrophin gene editing ameliorates pathophysiology in a mouse model for Duchenne muscular dystrophy. Nat Commun. 2017;8: 14454. 10.1038/ncomms14454 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 58. Ruan GX, Barry E, Yu D, et al. : CRISPR/Cas9-Mediated Genome Editing as a Therapeutic Approach for Leber Congenital Amaurosis 10. Mol Ther. 2017;25(2):331–41. 10.1016/j.ymthe.2016.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 59. Hung SS, Chrysostomou V, Li F, et al. : AAV-Mediated CRISPR/Cas Gene Editing of Retinal Cells In vivo. Invest Ophthalmol Vis Sci. 2016;57(7):3470–6. 10.1167/iovs.16-19316 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 60. Yu W, Mookherjee S, Chaitankar V, et al. : Nrl knockdown by AAV-delivered CRISPR/Cas9 prevents retinal degeneration in mice. Nat Commun. 2017;8: 14716. 10.1038/ncomms14716 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 61. Yin H, Song CQ, Dorkin JR, et al. : Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat Biotechnol. 2016;34(3):328–33. 10.1038/nbt.3471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Xie C, Zhang YP, Song L, et al. : Genome editing with CRISPR/Cas9 in postnatal mice corrects PRKAG2 cardiac syndrome. Cell Res. 2016;26(10):1099–111. 10.1038/cr.2016.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Carroll KJ, Makarewich CA, McAnally J, et al. : A mouse model for adult cardiac-specific gene deletion with CRISPR/Cas9. Proc Natl Acad Sci U S A. 2016;113(2):338–43. 10.1073/pnas.1523918113 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 64. Johansen AK, Molenaar B, Versteeg D, et al. : Postnatal Cardiac Gene Editing Using CRISPR/Cas9 With AAV9-Mediated Delivery of Short Guide RNAs Results in Mosaic Gene Disruption. Circ Res. 2017;121(10):1168–81. 10.1161/CIRCRESAHA.116.310370 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 65. Platt RJ, Chen S, Zhou Y, et al. : CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159(2):440–55. 10.1016/j.cell.2014.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 66. Kleinstiver BP, Prew MS, Tsai SQ, et al. : Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523(7561):481–5. 10.1038/nature14592 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 67. Nishimasu H, Ran FA, Hsu PD, et al. : Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156(5):935–49. 10.1016/j.cell.2014.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 68. Tervo DG, Hwang B, Viswanathan S, et al. : A Designer AAV Variant Permits Efficient Retrograde Access to Projection Neurons. Neuron. 2016;92(2):372–82. 10.1016/j.neuron.2016.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 69. Kemaladewi DU, Maino E, Hyatt E, et al. : Correction of a splicing defect in a mouse model of congenital muscular dystrophy type 1A using a homology-directed-repair-independent mechanism. Nat Med. 2017;23(8):984–9. 10.1038/nm.4367 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 70. El Refaey M, Xu L, Gao Y, et al. : In vivo Genome Editing Restores Dystrophin Expression and Cardiac Function in Dystrophic Mice. Circ Res. 2017;121(8):923–9. 10.1161/CIRCRESAHA.117.310996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ohmori T, Nagao Y, Mizukami H, et al. : CRISPR/Cas9-mediated genome editing via postnatal administration of AAV vector cures haemophilia B mice. Sci Rep. 2017;7(1):4159. 10.1038/s41598-017-04625-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Yin C, Zhang T, Qu X, et al. : In vivo Excision of HIV-1 Provirus by saCas9 and Multiplex Single-Guide RNAs in Animal Models. Mol Ther. 2017;25(5):1168–86. 10.1016/j.ymthe.2017.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 73. Fu Y, Sander JD, Reyon D, et al. : Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32(3):279–84. 10.1038/nbt.2808 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 74. Davis KM, Pattanayak V, Thompson DB, et al. : Small molecule-triggered Cas9 protein with improved genome-editing specificity. Nat Chem Biol. 2015;11(5):316–8. 10.1038/nchembio.1793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Yu C, Liu Y, Ma T, et al. : Small molecules enhance CRISPR genome editing in pluripotent stem cells. Cell Stem Cell. 2015;16(2):142–7. 10.1016/j.stem.2015.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 76. Monteys AM, Ebanks SA, Keiser MS, et al. : CRISPR/Cas9 Editing of the Mutant Huntingtin Allele In Vitro and In Vivo. Mol Ther. 2017;25(1):12–23. 10.1016/j.ymthe.2016.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Jarrett KE, Lee CM, Yeh YH, et al. : Somatic genome editing with CRISPR/Cas9 generates and corrects a metabolic disease. Sci Rep. 2017;7: 44624. 10.1038/srep44624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Guo Y, VanDusen NJ, Zhang L, et al. : Analysis of Cardiac Myocyte Maturation Using CASAAV, a Platform for Rapid Dissection of Cardiac Myocyte Gene Function In Vivo. Circ Res. 2017;120(12):1874–88. 10.1161/CIRCRESAHA.116.310283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Chow RD, Guzman CD, Wang G, et al. : AAV-mediated direct in vivo CRISPR screen identifies functional suppressors in glioblastoma. Nat Neurosci. 2017;20(10):1329–41. 10.1038/nn.4620 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 80. Murlidharan G, Sakamoto K, Rao L, et al. : CNS-restricted Transduction and CRISPR/Cas9-mediated Gene Deletion with an Engineered AAV Vector. Mol Ther Nucleic Acids. 2016;5(7):e338. 10.1038/mtna.2016.49 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 81. de Solis CA, Ho A, Holehonnur R, et al. : The Development of a Viral Mediated CRISPR/Cas9 System with Doxycycline Dependent gRNA Expression for Inducible In vitro and In vivo Genome Editing. Front Mol Neurosci. 2016;9:70. 10.3389/fnmol.2016.00070 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 82. Nishiyama J, Mikuni T, Yasuda R: Virus-Mediated Genome Editing via Homology-Directed Repair in Mitotic and Postmitotic Cells in Mammalian Brain. Neuron. 2017;96(4):755–768.e5. 10.1016/j.neuron.2017.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Li Y, Kim J: Distinct roles of neuronal and microglial CB2 cannabinoid receptors in the mouse hippocampus. Neuroscience. 2017;363:11–25. 10.1016/j.neuroscience.2017.08.053 [DOI] [PubMed] [Google Scholar]
- 84. Lin H, Hu H, Duan W, et al. : Intramuscular Delivery of scAAV9-hIGF1 Prolongs Survival in the hSOD1G93A ALS Mouse Model via Upregulation of D-Amino Acid Oxidase. Mol Neurobiol. 2016;1–14. 10.1007/s12035-016-0335-z [DOI] [PubMed] [Google Scholar]
- 85. Huang X, Zhou G, Wu W, et al. : Genome editing abrogates angiogenesis in vivo. Nat Commun. 2017;8(1):112. 10.1038/s41467-017-00140-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Kaminski R, Bella R, Yin C, et al. : Excision of HIV-1 DNA by gene editing: a proof-of-concept in vivo study. Gene Ther. 2016;23(8–9):690–5. 10.1038/gt.2016.41 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 87. Grimm D, Lee JS, Wang L, et al. : In vitro and in vivo gene therapy vector evolution via multispecies interbreeding and retargeting of adeno-associated viruses. J Virol. 2008;82(12):5887–911. 10.1128/JVI.00254-08 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 88. Hashimoto H, Mizushima T, Chijiwa T, et al. : Efficient production of recombinant adeno-associated viral vector, serotype DJ/8, carrying the GFP gene. Virus Res. 2017;238:63–8. 10.1016/j.virusres.2017.05.017 [DOI] [PubMed] [Google Scholar]
- 89. Iida A, Takino N, Miyauchi H, et al. : Systemic delivery of tyrosine-mutant AAV vectors results in robust transduction of neurons in adult mice. Biomed Res Int. 2013;2013: 974819. 10.1155/2013/974819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. McCarty DM, Monahan PE, Samulski RJ: Self-complementary recombinant adeno-associated virus (scAAV) vectors promote efficient transduction independently of DNA synthesis. Gene Ther. 2001;8(16):1248–54. 10.1038/sj.gt.3301514 [DOI] [PubMed] [Google Scholar]
- 91. Wang X, Xu Z, Tian Z, et al. : The EF-1α promoter maintains high-level transgene expression from episomal vectors in transfected CHO-K1 cells. J Cell Mol Med. 2017;21(11):3044–54. 10.1111/jcmm.13216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Teschendorf C, Warrington KH, Jr, Siemann DW, et al. : Comparison of the EF-1 alpha and the CMV promoter for engineering stable tumor cell lines using recombinant adeno-associated virus. Anticancer Res. 2002;22(6A):3325–30. [PubMed] [Google Scholar]
- 93. Chung S, Andersson T, Sonntag KC, et al. : Analysis of different promoter systems for efficient transgene expression in mouse embryonic stem cell lines. Stem Cells. 2002;20(2):139–45. 10.1634/stemcells.20-2-139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Roelz R, Pilz IH, Mutschler M, et al. : Of mice and men: human RNA polymerase III promoter U6 is more efficient than its murine homologue for shRNA expression from a lentiviral vector in both human and murine progenitor cells. Exp Hematol. 2010;38(9):792–7. 10.1016/j.exphem.2010.05.005 [DOI] [PubMed] [Google Scholar]
- 95. Mäkinen PI, Koponen JK, Kärkkäinen AM, et al. : Stable RNA interference: comparison of U6 and H1 promoters in endothelial cells and in mouse brain. J Gene Med. 2006;8(4):433–41. 10.1002/jgm.860 [DOI] [PubMed] [Google Scholar]
- 96. Sun CP, Wu TH, Chen CC, et al. : Studies of efficacy and liver toxicity related to adeno-associated virus-mediated RNA interference. Hum Gene Ther. 2013;24(8):739–50. 10.1089/hum.2012.239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Larson MH, Gilbert LA, Wang X, et al. : CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat Protoc. 2013;8(11):2180–96. 10.1038/nprot.2013.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Ranganathan V, Wahlin K, Maruotti J, et al. : Expansion of the CRISPR-Cas9 genome targeting space through the use of H1 promoter-expressed guide RNAs. Nat Commun. 2014;5:4516. 10.1038/ncomms5516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Vidigal JA, Ventura A: Rapid and efficient one-step generation of paired gRNA CRISPR-Cas9 libraries. Nat Commun. 2015;6:8083. 10.1038/ncomms9083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Saraiva J, Nobre RJ, Pereira de Almeida L: Gene therapy for the CNS using AAVs: The impact of systemic delivery by AAV9. J Control Release. 2016;241:94–109. 10.1016/j.jconrel.2016.09.011 [DOI] [PubMed] [Google Scholar]
- 101. Vagner T, Dvorzhak A, Wójtowicz AM, et al. : Systemic application of AAV vectors targeting GFAP-expressing astrocytes in Z-Q175-KI Huntington's disease mice. Mol Cell Neurosci. 2016;77:76–86. 10.1016/j.mcn.2016.10.007 [DOI] [PubMed] [Google Scholar]
- 102. Tanguy Y, Biferi MG, Besse A, et al. : Systemic AAVrh10 provides higher transgene expression than AAV9 in the brain and the spinal cord of neonatal mice. Front Mol Neurosci. 2015;8:36. 10.3389/fnmol.2015.00036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Cyranoski D: CRISPR gene-editing tested in a person for the first time. Nature. 2016;539(7630):479. 10.1038/nature.2016.20988 [DOI] [PubMed] [Google Scholar]
- 104. Cyranoski D: Chinese scientists to pioneer first human CRISPR trial. Nature. 2016;535(7613):476–7. 10.1038/nature.2016.20302 [DOI] [PubMed] [Google Scholar]
- 105. Costa R, Carneiro BA, Agulnik M, et al. : Toxicity profile of approved anti-PD-1 monoclonal antibodies in solid tumors: a systematic review and meta-analysis of randomized clinical trials. Oncotarget. 2017;8(5):8910–20. 10.18632/oncotarget.13315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Zhang T, Xie J, Arai S, et al. : The efficacy and safety of anti-PD-1/PD-L1 antibodies for treatment of advanced or refractory cancers: a meta-analysis. Oncotarget. 2016;7(45):73068–79. 10.18632/oncotarget.12230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Zhong L, Li B, Mah CS, et al. : Next generation of adeno-associated virus 2 vectors: point mutations in tyrosines lead to high-efficiency transduction at lower doses. Proc Natl Acad Sci U S A. 2008;105(22):7827–32. 10.1073/pnas.0802866105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Ling C, Li B, Ma W, et al. : Development of Optimized AAV Serotype Vectors for High-Efficiency Transduction at Further Reduced Doses. Hum Gene Ther Methods. 2016;27(4):143–9. 10.1089/hgtb.2016.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Kim D, Kim S, Kim S, et al. : Genome-wide target specificities of CRISPR-Cas9 nucleases revealed by multiplex Digenome-seq. Genome Res. 2016;26(3):406–15. 10.1101/gr.199588.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Kim D, Bae S, Park J, et al. : Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat Methods. 2015;12(3):237–43, 1 p following 243. 10.1038/nmeth.3284 [DOI] [PubMed] [Google Scholar]
- 111. Kuscu C, Arslan S, Singh R, et al. : Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat Biotechnol. 2014;32(7):677–83. 10.1038/nbt.2916 [DOI] [PubMed] [Google Scholar]
- 112. O'Geen H, Henry IM, Bhakta MS, et al. : A genome-wide analysis of Cas9 binding specificity using ChIP-seq and targeted sequence capture. Nucleic Acids Res. 2015;43(6):3389–404. 10.1093/nar/gkv137 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 113. Tsai SQ, Zheng Z, Nguyen NT, et al. : GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. 2015;33(2):187–97. 10.1038/nbt.3117 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 114. Yang L, Grishin D, Wang G, et al. : Targeted and genome-wide sequencing reveal single nucleotide variations impacting specificity of Cas9 in human stem cells. Nat Commun. 2014;5: 5507. 10.1038/ncomms6507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Morgens DW, Wainberg M, Boyle EA, et al. : Genome-scale measurement of off-target activity using Cas9 toxicity in high-throughput screens. Nat Commun. 2017;8: 15178. 10.1038/ncomms15178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Cameron P, Fuller CK, Donohoue PD, et al. : Mapping the genomic landscape of CRISPR-Cas9 cleavage. Nat Methods. 2017;14(6):600–6. 10.1038/nmeth.4284 [DOI] [PubMed] [Google Scholar]
- 117. Scott DA, Zhang F: Implications of human genetic variation in CRISPR-based therapeutic genome editing. Nat Med. 2017;23(9):1095–101. 10.1038/nm.4377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Tsai SQ, Nguyen NT, Malagon-Lopez J, et al. : CIRCLE-seq: a highly sensitive in vitro screen for genome-wide CRISPR-Cas9 nuclease off-targets. Nat Methods. 2017;14(6):607–14. 10.1038/nmeth.4278 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 119. Kim D, Lim K, Kim ST, et al. : Genome-wide target specificities of CRISPR RNA-guided programmable deaminases. Nat Biotechnol. 2017;35(5):475–80. 10.1038/nbt.3852 [DOI] [PubMed] [Google Scholar]
- 120. Koo T, Lee J, Kim JS: Measuring and Reducing Off-Target Activities of Programmable Nucleases Including CRISPR-Cas9. Mol Cells. 2015;38(6):475–81. 10.14348/molcells.2015.0103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Batra R, Nelles DA, Pirie E, et al. : Elimination of Toxic Microsatellite Repeat Expansion RNA by RNA-Targeting Cas9. Cell. 2017;170(5):899–912.e10. 10.1016/j.cell.2017.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 122. Pinto BS, Saxena T, Oliveira R, et al. : Impeding Transcription of Expanded Microsatellite Repeats by Deactivated Cas9. Mol Cell. 2017;68(3):479–490.e5. 10.1016/j.molcel.2017.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 123. Kim K, Park SW, Kim JH, et al. : Genome surgery using Cas9 ribonucleoproteins for the treatment of age-related macular degeneration. Genome Res. 2017;27(3):419–26. 10.1101/gr.219089.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Nihongaki Y, Kawano F, Nakajima T, et al. : Photoactivatable CRISPR-Cas9 for optogenetic genome editing. Nat Biotechnol. 2015;33(7):755–60. 10.1038/nbt.3245 [DOI] [PubMed] [Google Scholar]
- 125. Polstein LR, Gersbach CA: A light-inducible CRISPR-Cas9 system for control of endogenous gene activation. Nat Chem Biol. 2015;11(3):198–200. 10.1038/nchembio.1753 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 126. Dow LE, Fisher J, O'Rourke KP, et al. : Inducible in vivo genome editing with CRISPR-Cas9. Nat Biotechnol. 2015;33(4):390–4. 10.1038/nbt.3155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Liu KI, Ramli MN, Woo CW, et al. : A chemical-inducible CRISPR-Cas9 system for rapid control of genome editing. Nat Chem Biol. 2016;12(11):980–7. 10.1038/nchembio.2179 [DOI] [PubMed] [Google Scholar]
- 128. Liu Y, Zeng Y, Liu L, et al. : Synthesizing AND gate genetic circuits based on CRISPR-Cas9 for identification of bladder cancer cells. Nat Commun. 2014;5:5393. 10.1038/ncomms6393 [DOI] [PubMed] [Google Scholar]
- 129. Weinberg BH, Pham NT, Caraballo LD, et al. : Large-scale design of robust genetic circuits with multiple inputs and outputs for mammalian cells. Nat Biotechnol. 2017;35(5):453–62. 10.1038/nbt.3805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Shen B, Zhang W, Zhang J, et al. : Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat Methods. 2014;11(4):399–402. 10.1038/nmeth.2857 [DOI] [PubMed] [Google Scholar]
- 131. Cho SW, Kim S, Kim Y, et al. : Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2014;24(1):132–41. 10.1101/gr.162339.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Ran FA, Hsu PD, Lin CY, et al. : Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154(6):1380–9. 10.1016/j.cell.2013.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 133. Guilinger JP, Thompson DB, Liu DR: Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat Biotechnol. 2014;32(6):577–82. 10.1038/nbt.2909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Kleinstiver BP, Pattanayak V, Prew MS, et al. : High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529(7587):490–5. 10.1038/nature16526 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 135. Slaymaker IM, Gao L, Zetsche B, et al. : Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351(6268):84–8. 10.1126/science.aad5227 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 136. Chen JS, Dagdas YS, Kleinstiver BP, et al. : Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature. 2017;550(7676):407–10. 10.1038/nature24268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Komor AC, Kim YB, Packer MS, et al. : Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533(7603):420–4. 10.1038/nature17946 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 138. Kim K, Ryu SM, Kim ST, et al. : Highly efficient RNA-guided base editing in mouse embryos. Nat Biotechnol. 2017;35(5):435–7. 10.1038/nbt.3816 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 139. Richardson CD, Ray GJ, DeWitt MA, et al. : Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat Biotechnol. 2016;34(3):339–44. 10.1038/nbt.3481 [DOI] [PubMed] [Google Scholar]
- 140. Song J, Yang D, Xu J, et al. : RS-1 enhances CRISPR/Cas9- and TALEN-mediated knock-in efficiency. Nat Commun. 2016;7:10548. 10.1038/ncomms10548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Maruyama T, Dougan SK, Truttmann MC, et al. : Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol. 2015;33(5):538–42. 10.1038/nbt.3190 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 142. Cho SW, Kim S, Kim JM, et al. : Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31(3):230–2. 10.1038/nbt.2507 [DOI] [PubMed] [Google Scholar]
- 143. Hendel A, Bak RO, Clark JT, et al. : Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat Biotechnol. 2015;33(9):985–9. 10.1038/nbt.3290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Perez AR, Pritykin Y, Vidigal JA, et al. : GuideScan software for improved single and paired CRISPR guide RNA design. Nat Biotechnol. 2017;35(4):347–9. 10.1038/nbt.3804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Naito Y, Hino K, Bono H, et al. : CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics. 2015;31(7):1120–3. 10.1093/bioinformatics/btu743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Bae S, Park J, Kim JS: Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014;30(10):1473–5. 10.1093/bioinformatics/btu048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Zhang XH, Tee LY, Wang XG, et al. : Off-target Effects in CRISPR/Cas9-mediated Genome Engineering. Mol Ther Nucleic Acids. 2015;4(11):e264. 10.1038/mtna.2015.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Mockenhaupt S, Grosse S, Rupp D, et al. : Alleviation of off-target effects from vector-encoded shRNAs via codelivered RNA decoys. Proc Natl Acad Sci U S A. 2015;112(30):E4007–16. 10.1073/pnas.1510476112 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 149. Tay FC, Lim JK, Zhu H, et al. : Using artificial microRNA sponges to achieve microRNA loss-of-function in cancer cells. Adv Drug Deliv Rev. 2015;81:117–27. 10.1016/j.addr.2014.05.010 [DOI] [PubMed] [Google Scholar]
- 150. Hirosawa M, Fujita Y, Parr CJC, et al. : Cell-type-specific genome editing with a microRNA-responsive CRISPR-Cas9 switch. Nucleic Acids Res. 2017;45(13):e118. 10.1093/nar/gkx309 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 151. Manning WC, Zhou S, Bland MP, et al. : Transient immunosuppression allows transgene expression following readministration of adeno-associated viral vectors. Hum Gene Ther. 1998;9(4):477–85. 10.1089/hum.1998.9.4-477 [DOI] [PubMed] [Google Scholar]
- 152. Mingozzi F, Anguela XM, Pavani G, et al. : Overcoming preexisting humoral immunity to AAV using capsid decoys. Sci Transl Med. 2013;5(194):194ra92. 10.1126/scitranslmed.3005795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Huttner NA, Girod A, Perabo L, et al. : Genetic modifications of the adeno-associated virus type 2 capsid reduce the affinity and the neutralizing effects of human serum antibodies. Gene Ther. 2003;10(26):2139–47. 10.1038/sj.gt.3302123 [DOI] [PubMed] [Google Scholar]
- 154. Arbetman AE, Lochrie M, Zhou S, et al. : Novel caprine adeno-associated virus (AAV) capsid (AAV-Go.1) is closely related to the primate AAV-5 and has unique tropism and neutralization properties. J Virol. 2005;79(24):15238–45. 10.1128/JVI.79.24.15238-15245.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Maheshri N, Koerber JT, Kaspar BK, et al. : Directed evolution of adeno-associated virus yields enhanced gene delivery vectors. Nat Biotechnol. 2006;24(2):198–204. 10.1038/nbt1182 [DOI] [PubMed] [Google Scholar]
- 156. Bakondi B: In vivo versus ex vivo CRISPR therapies for retinal dystrophy. Expert Rev Ophthalmol. 2016;11(6):397–400. 10.1080/17469899.2016.1251316 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 157. Grimm D, Büning H: Small But Increasingly Mighty: Latest Advances in AAV Vector Research, Design, and Evolution. Hum Gene Ther. 2017;28(11):1075–86. 10.1089/hum.2017.172 [DOI] [PubMed] [Google Scholar]
- 158. Paulk NK, Pekrun K, Zhu E, et al. : Bioengineered AAV Capsids with Combined High Human Liver Transduction In vivo and Unique Humoral Seroreactivity. Mol Ther. 2017; pii: S1525-0016(17)30437-9. 10.1016/j.ymthe.2017.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Tse LV, Klinc KA, Madigan VJ, et al. : Structure-guided evolution of antigenically distinct adeno-associated virus variants for immune evasion. Proc Natl Acad Sci U S A. 2017;114(24):E4812–E4821. 10.1073/pnas.1704766114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Li C, Wu S, Albright B, et al. : Development of Patient-specific AAV Vectors After Neutralizing Antibody Selection for Enhanced Muscle Gene Transfer. Mol Ther. 2016;24(1):53–65. 10.1038/mt.2015.134 [DOI] [PMC free article] [PubMed] [Google Scholar]