SUMMARY
The actin cytoskeleton—a collection of actin filaments with their accessory and regulatory proteins—is the primary force-generating machinery in the cell. It can produce pushing (protrusive) forces through coordinated polymerization of multiple actin filaments or pulling (contractile) forces through sliding actin filaments along bipolar filaments of myosin II. Both force types are particularly important for whole-cell migration, but they also define and change the cell shape and mechanical properties of the cell surface, drive the intracellular motility and morphogenesis of membrane organelles, and allow cells to form adhesions with each other and with the extracellular matrix.
1. Introduction
The ability of cells to migrate as a whole, or move subcellular components, is essential for many unicellular organisms and all multicellular organisms. During embryonic development, cellular movements establish the body plan and form organs and tissues. For example, neuronal precursors migrate long distances to the site of differentiation and then extend long processes to make functional neuronal circuitry. Migration and shape changes of individual cells are also crucial for the functionality of body systems in adults. Muscle contraction generates macroscopic movements of animals and their organs. Fast motility of immune cells allows them to survey tissues, find and destroy pathogens, and initiate immune responses. Subtle movements of tiny projections from neuronal processes underlie the formation and loss of synapses (e.g., during learning and memory loss). Even nonmigratory cells acquire motile behavior upon tissue injury in order to close wounds and rebuild tissues. Movements of subcellular components are essential for cell growth and proliferation, the import and export of nutrients and signaling intermediates, degradation and renewal of cellular structures, communication with the environment, and many other aspects of normal cell physiology.
Cell motility also contributes to disease. Cell motility enhances invasion and metastasis of tumor cells. Migration of immune cells into tissues contributes to chronic inflammatory diseases. Additionally, some microbial pathogens manipulate motility mechanisms of the host cell to avoid immune surveillance and facilitate their own cell-to-cell spread.
Forces generated by the actin cytoskeleton power these diverse motility processes. The main component of the actin cytoskeleton is actin filaments, which are polar linear polymers of the abundant cytoplasmic protein actin. Many cellular actin filaments turn over constantly to remodel actin-based structures according to changing needs. Regulatory proteins control all aspects of actin filament dynamics in time and space, such as actin filament nucleation, elongation, and disassembly (reviewed by Pollard 2016). In cells, actin-binding proteins assemble most actin filaments into networks and bundles adapted to specific tasks. Additional accessory proteins allow actin filaments to act in association with cellular membranes.
Here, we review how the actin cytoskeleton produces pushing, pulling, and resistance forces responsible for multiple cell-motility events (Fig. 1). Whole-cell migration serves as a useful experimental system to decipher the molecular mechanisms of cell motility. Cells move by repeating cycles of protrusion and attachment of the cell front, followed by detachment and retraction of the rear (Fig. 1). Coordinated polymerization of multiple actin filaments produces protrusive forces that drive the extension of the plasma membrane at the cell leading edge (Pollard and Borisy 2003). Similar mechanisms drive propulsion of membrane-enclosed organelles and promote apposition of membranes during formation of cell–cell junctions (Chhabra and Higgs 2007). Contractile forces produced by myosin motors pulling on actin filaments retract the trailing end in migrating cells, a mechanism analogous to muscle contraction (Huxley and Hanson 1954; Huxley and Niedergerke 1954). A similar contractile mechanism separates daughter cells during cytokinesis (reviewed in Glotzer 2016), reinforces adhesion sites between cells or between a cell and the extracellular matrix, maintains and changes the cell shape, and defines the mechanical properties of the cell surface.
Figure 1.
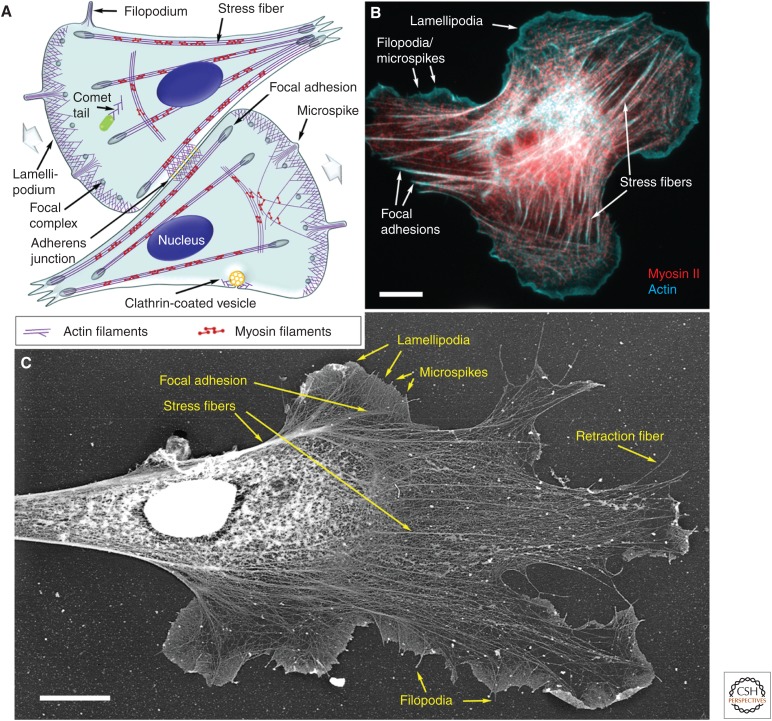
Components of the actin cytoskeleton in migrating cells. (A) Illustration of the components of the actin cytoskeleton in representative fibroblast-like cells. The direction of cell migration is indicated by wide gray arrows. (B) Fluorescence micrograph of a rat embryo fibroblast showing actin filaments (cyan) and myosin II (red). (C) Electron micrograph of the cytoskeleton of a Xenopus laevis fibroblast prepared by platinum shadowing after detergent extraction and critical point drying. Individual components of the actin cytoskeleton are marked in all panels. Scale bars, 10 µm. (C, Adapted from Svitkina and Borisy 1999.)
2. The actin cytoskeleton in protrusion
2.1. General Concept
To generate a pushing force for protrusion, the cell uses the energy of actin polymerization, as reviewed elsewhere (Pollard 2016). This general concept emerged from studies of the acrosomal reaction in invertebrate sperm by Tilney and coworkers (Tilney 1975). Although this discovery was made in a specialized cellular system, the concept has general significance and applies, in particular, to protrusion of the leading edge of migrating cells.
The directionality of pushing force produced by actin polymerization originates from the structural polarity of actin filaments (Huxley 1963), in which one end (barbed end or “plus end”) polymerizes faster than the other (pointed end or “minus end”) (Woodrum et al. 1975). In cells, polymerizing actin filaments are uniformly oriented with their barbed ends toward the load, commonly, the plasma membrane. Whereas elongating barbed ends push on the load, disassembly by severing and depolymerization occurs closer to the pointed ends to release monomers for recycling. As reviewed elsewhere (Pollard 2016), proteins of the Ena/VASP (vasodilator-stimulated phosphoprotein) and formin families increase the rate and duration of barbed end elongation by binding multiple profilin–actin complexes and rapidly transferring actin onto the barbed end (Fig. 2B,C) (Dominguez 2010). They also compete with capping proteins that could terminate barbed end elongation and keep elongating ends near to the membrane to increase the efficiency of pushing.
Figure 2.
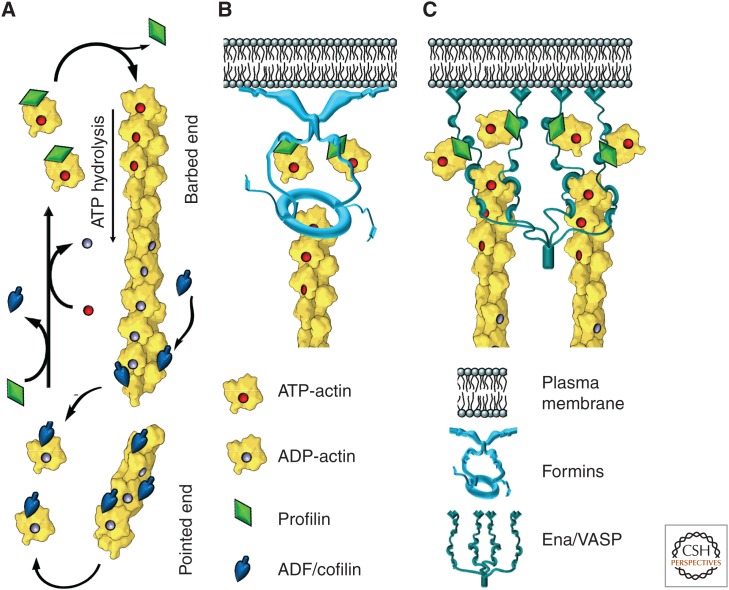
Accelerated dynamics of actin filaments. (A) Polymerization of the actin filament preferentially occurs at the barbed end from ATP-actin–profilin complexes; after incorporation of an actin monomer into the filament, profilin dissociates. Polymerization-triggered ATP hydrolysis and subsequent release of inorganic phosphate from actin subunits make filaments more susceptible to depolymerization and increase their affinity for actin-depolymerizing factor (ADF)/cofilin. ADF/cofilin severs filaments, promoting their depolymerization. Released actin subunits bind to profilin, which competes off ADF/cofilin and promotes nucleotide exchange in the actin monomer, thus producing new ATP-actin–profilin complexes. (B) Formins and (C) Ena/VASP (vasodilator-stimulated phosphoprotein) proteins associate with actin filament barbed ends, promote their elongation by recruiting actin–profilin complexes and protecting barbed ends from capping; they also anchor barbed ends to the membrane.
Polymerization of individual actin filaments produces piconewton forces (Kovar and Pollard 2004), and filaments are organized into parallel bundles in filopodia or branched networks in lamellipodia to produce large forces (Fig. 1). Filopodia are slender protrusions that can extend far beyond the cell edge and reach or sense distant targets. Lamellipodia are planar protrusions that generate large pushing forces that are mainly responsible for propelling the cell front. The balance of protrusion by filopodia and lamellipodia varies broadly among cell types and might reflect the relative needs for fast locomotion versus precise navigation. Distinct, but overlapping, sets of accessory proteins regulate actin polymerization in filopodia and lamellipodia.
2.2. Protrusion of Parallel Bundles
Parallel bundles of long actin filaments mediate protrusion of thin, long extensions of the plasma membrane and maintain their asymmetric shape. Different types of these finger-like protrusions share the basic mechanism of formation but vary in their functions, protein composition, and dynamics.
2.2.1. Acrosomal Processes
The discovery that actin polymerization generates force during the acrosome reaction in echinoderm sperm was also the first example of membrane protrusion driven by elongating actin filament bundles (Tilney et al. 1973). An initial contact of a sperm cell with an egg leads to explosive extension of a long acrosomal process from the sperm head. Growth of this process helps the sperm penetrate the egg jelly. Polymerization of a tight bundle of actin filaments inside the process drives its extension. These filaments are uniformly oriented with their barbed ends toward the growing tip of the process. Actin monomers in a complex with profilin are stored in the sperm head. Rapid addition of monomers to the barbed ends drives the elongation of the process until all the monomers are consumed.
2.2.2. Leading Edge Filopodia
Filopodia are finger-like protrusions formed at the leading edge of migrating cells, such as fibroblasts, or cells extending long processes, such as neurons (Fig. 3A). In contrast to acrosomal processes, filopodia, in theory, can protrude indefinitely because actin filaments turn over continuously by releasing monomers from the rear for reuse at the front. However, most filopodia buckle and retract after a period of elongation. The turnover of the actin bundles is best studied in filopodia at the leading edge.
Figure 3.
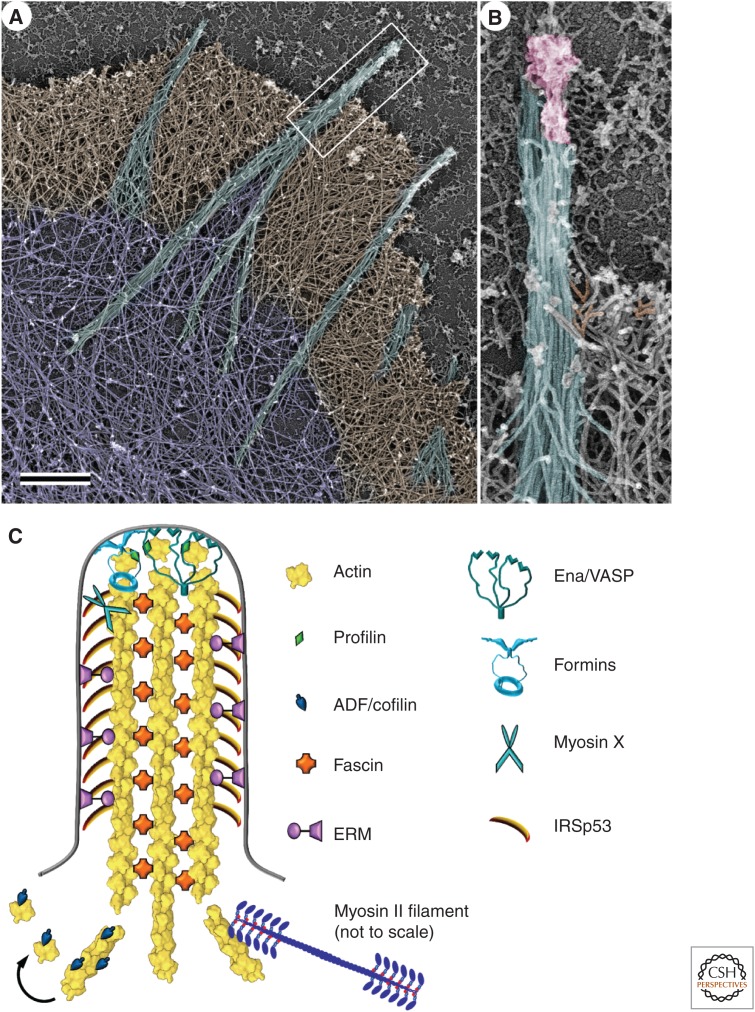
The organization of actin in filopodia and microspikes. (A) Platinum-replica electron microscopy of the cytoskeleton at the leading edge of a cultured mouse melanoma cell; actin filament bundles (cyan) that form the cytoskeleton of filopodia extend beyond the cell leading edge, span the dense actin filament network in lamellipodia (brown), and sometimes penetrate into the sparser actin network in the lamella (purple). Actin bundles in microspikes are mostly positioned within lamellipodia. Scale bar, 1 µm. (B) Enlarged boxed region from A shows long parallel actin filaments (cyan) and a complex of regulatory proteins (pink) at the filopodial tip. Some branched actin filaments in the adjacent lamellipodium are shaded orange. (C) Molecular organization of filopodia. Actin filaments are oriented with their barbed ends toward the filopodial tip. They are cross-linked into a bundle by fascin and laterally attached to the plasma membrane by ezrin-radixin-moesin (ERM) proteins. Formin and Ena/VASP (vasodilator-stimulated phosphoprotein) proteins associate with the actin filament barbed ends, anchor them to the membrane at the filopodial tip, and promote their elongation by protecting them from capping and recruiting actin–profilin complexes. Unconventional myosin X moves along actin filaments and accumulates at the filopodial tips. I-BAR-domain-containing proteins, such as IRSp53, help to maintain the tubular shape of the filopodial plasma membrane. Actin-depolymerizing factor (ADF)/cofilin and myosin II sever actin filaments at the filopodial base to stimulate actin filament depolymerization and monomer recycling.
2.2.2.1. Structure and dynamics
The internal actin filament bundle of leading edge filopodia is organized like that of an acrosomal process. Individual filaments span the entire length of the filopodium (Fig. 3B) and are uniformly oriented with their barbed ends toward the filopodial tip (Small et al. 1978). During protrusion, actin subunits are added at the filopodium tip, move away from the tip as a part of the filament lattice, and are released at the rear of filopodium (Wang 1985; Mallavarapu and Mitchison 1999). Introduction of fiducial marks into the actin filament bundle in filopodia showed that, although the filopodial tip grows forward, the entire bundle slides backward (Mallavarapu and Mitchison 1999). The balance between rates of protrusion and retrograde flow varies among individual filopodia. Retrograde flow can even completely counterbalance polymerization, so that the filopodium remains stationary, although actin keeps polymerizing at the tip. The amount of energy of polymerization that is converted into protrusion depends, at least in part, on the strength of linkages between the filopodium and the substrate. Accessory proteins regulate multiple aspects of actin dynamics in leading edge filopodia by controlling actin filament elongation, cross-linking, disassembly, and interaction with other structures (Fig. 3C).
2.2.2.2. Elongation
Formation of long actin filaments, like those in filopodia, in the cytoplasm of motile cells is a challenging task because of the presence of abundant capping proteins capable of binding barbed ends and terminating their elongation. Accordingly, Ena/VASP proteins and some formins are enriched at filopodial tips, where they protect barbed ends from capping proteins (Svitkina et al. 2003; Yang et al. 2007). Ena/VASP proteins are particularly important for formation of filopodia in neuronal growth cones (Lebrand et al. 2004), but share this function with formins in other cell types (Homem and Peifer 2009).
Another challenge for elongation of actin filaments in filopodia is delivery of actin monomers and other structural proteins from pools in the cell body to the filopodial tip, where polymerization takes place. The long thin shape of filopodia is unfavorable for component delivery by simple diffusion (Mogilner and Rubinstein 2005). Unconventional myosin X has been found to promote filopodium elongation and is thought to deliver actin subunits and/or regulators of actin polymerization, such as Ena/VASP, to filopodial tips by using its motor activity (Kerber and Cheney 2011).
2.2.2.3. Bundling and cross-linking
Fast elongation of actin filaments at filopodial tips allows for protrusion, but the force produced by elongation buckles actin filaments longer than 0.7 µm (Kovar and Pollard 2004), limiting their pushing ability. Cells overcome this problem by cross-linking filopodial filaments along their length, thus increasing their collective stiffness (Vignjevic et al. 2006). The major bundling protein in leading edge filopodia is fascin, a relatively small bivalent monomeric protein that makes tight cross-links between filaments. In vitro, it induces rigid needle-shaped bundles of actin filaments. Fascin turns over rapidly within filopodial bundles by constantly dissociating and rebinding to actin filaments. This property might help the cell to release stresses and torques within the bundle while it grows and encounters obstacles.
2.2.2.4. Depolymerization
The small number (<30; Mogilner and Rubinstein 2005) of actin filaments in filopodia means that very few pointed ends are available to release actin subunits. The rate of ADP-actin dissociation from pointed ends (0.1/sec) is slower than the rate of barbed end elongation, but severing of pieces of the filament can accelerate turnover. Both ADF/cofilin (Breitsprecher et al. 2011) and myosin II (Medeiros et al. 2006) are candidates to promote severing.
2.2.2.5. Shape maintenance by bundling and membrane interactions
The actin filament bundle within a filopodium functions as a scaffold to maintain the highly asymmetric shape of the filopodium. Notably, some filopodia contain a loose network of actin filaments with mixed polarity instead of a parallel bundle, suggesting a different mechanism of protrusion and shape maintenance (Korobova and Svitkina 2010). Lateral binding of actin filaments to the plasma membrane contributes to the stiffness of the filopodium. This linkage is likely mediated by proteins of the ezrin-radixin-moesin (ERM) family (Niggli and Rossy 2008). These ERM proteins concentrate in filopodia and interact simultaneously with actin filaments and membrane components, such as phospholipids and transmembrane proteins. Other membrane-binding proteins support the tubular geometry of filopodial membranes. When bound to the membrane through their membrane-bending I-BAR (inverted Bin/Amphiphysin/Rvs) or IF-BAR (inverted Fes/CIP4 homology BAR) domains, these proteins induce negative membrane curvature (away from the cytoplasm) (Suetsugu and Gautreau 2012). For example, the I-BAR domain of protein IRSp53 stabilizes the tubular shape of the plasma membrane in leading edge filopodia.
2.2.2.6. Interaction with the extracellular environment
Because of their elongated shape, filopodia are believed to function as “fingers” to reach, sense, and grab a target. By establishing communication with other cells or extracellular molecules, filopodia can guide cell locomotion during normal tissue morphogenesis or cancer metastasis, or form cell–cell junctions in epithelia and neurons (Mattila and Lappalainen 2008). Filopodia can also capture various particles, such as pathogens, for subsequent internalization (Romero et al. 2011). Consistent with these functions, tips of leading edge filopodia are enriched with adhesion and signaling proteins that could initiate adhesions when contacting adhesive surfaces, and trigger cellular responses when the filopodium finds an appropriate target.
2.2.3. Other Parallel Actin Bundles
Parallel bundles of actin filaments oriented with their barbed ends toward the plasma membrane are also present in other cell-surface extensions, such as microspikes (Hoglund et al. 1980) and retraction fibers (Svitkina et al. 1997) in migrating cells, microvilli in intestinal epithelia (Mooseker and Tilney 1975), and hair-cell stereocilia in the inner ear (Tilney et al. 1980).
2.2.3.1. Microspikes and retraction fibers
Microspikes and retraction fibers are rod-shaped bundles of actin filaments closely related to leading edge filopodia. In fact, all three structures form at the edge of migrating cells (Figs. 1 and 3) and can interconvert (Svitkina et al. 2003). Microspikes do not extend significantly beyond the cell leading edge but are embedded in and protrude at the same rate as the leading edge actin network. Retraction fibers are rod-shaped cell-surface extensions like filopodia, but they form during retraction of the cell edge, rather than by their own protrusion. Nevertheless, the tips of some retraction fibers elongate.
2.2.3.2. Microvilli
Microvilli are nonmotile finger-like protrusions from the apical surface of intestinal epithelial cells that increase the cell surface area for efficient absorption of nutrients. Microvilli are uniform in length (1–2 µm) and diameter (∼200 nm) and tightly packed next to each other in the so-called brush border. The bundle of approximately 20 to 30 parallel actin filaments inside a microvillus is structurally similar to filopodia (Brown and McKnight 2010), with the barbed ends at the tips embedded into material of unknown composition, which probably regulates the dynamics of actin filament barbed ends. These actin filaments turn over severalfold slower than those in filopodia (Loomis et al. 2003).
The actin bundle is laterally attached to the plasma membrane by myosin Ia, which binds to actin filaments by its motor domain and to acidic phospholipids in the plasma membrane by its tail (Nambiar et al. 2010). Myosin Ia is a fully functional motor and is believed to regulate membrane tension around microvilli. The main actin filament cross-linkers in the brush border microvilli are fimbrin and villin.
Microvilli are also present in lower densities on the surfaces of other cells types, such as other epithelial cells, lymphocytes, and various sensory cells. The main cross-linkers in sensory cell microvilli are fimbrin and espin, whereas villin is specific for various epithelial cells (Sekerkova et al. 2006).
2.2.3.3. Stereocilia
Stereocilia are specialized mechanosensory microvilli involved in hearing and the sense of balance. They differ from microvilli in size, shape, dynamics, and molecular composition. The lengths of stereocilia vary from 1 to 100 µm, but are tightly controlled to match the frequency of sound waves to be detected. Stereocilia have approximately 10-fold more actin filaments in the core bundle than microvilli. They undergo negligible dynamics (Zhang et al. 2012), suggesting that actin bundles play mostly a structural role in these cylindrical cellular extensions. Fimbrin and espin cross-link actin filaments in stereocilia (Sekerkova et al. 2006). Stereocilia taper toward their bases, which allows them to pivot rigidly in response to vibration. Myosins Ia, Ic, IIIa, VI, VIIa, and XVa have been functionally implicated in hearing by contributing to stereocilia structure, such as length regulation and membrane dynamics (Manor and Kachar 2008).
2.3. Protrusion by Branched Networks
Assembly of branched actin filament networks provides pushing forces for multiple cellular events, the best studied of which are protrusion of lamellipodia at the leading edge of migrating cells and propulsion of certain intracellular pathogens. In addition, branched networks promote various endocytic events, motility and biogenesis of intracellular organelles, and the formation of diverse cell–cell junctions, including neuronal and immune synapses.
2.3.1. Lamellipodia
Lamellipodia are flat undulating cellular protrusions at the leading edge of the cell that were named by Abercrombie and colleagues (Abercrombie et al. 1970). Lamellipodia can generate much greater protrusive forces than filopodia and serve as the major cellular engine to propel the leading edge forward. Lamellipodia also function as navigation devices for guiding a cell around obstacles, sensing soluble guidance cues, and probing the chemical and mechanical properties of the substratum. Although lamellipodia have mostly been studied using cells cultured on a flat solid surface, cells migrating in more physiological conditions, such as a three-dimensional matrix with physiological stiffness, also use lamellipodia for protrusion. However, the dimensions and overall morphology of lamellipodia depend on the geometry of the available adhesive surface and a balance of intracellular signaling pathways (Petrie and Yamada 2012).
2.3.1.1. Structure and dynamics
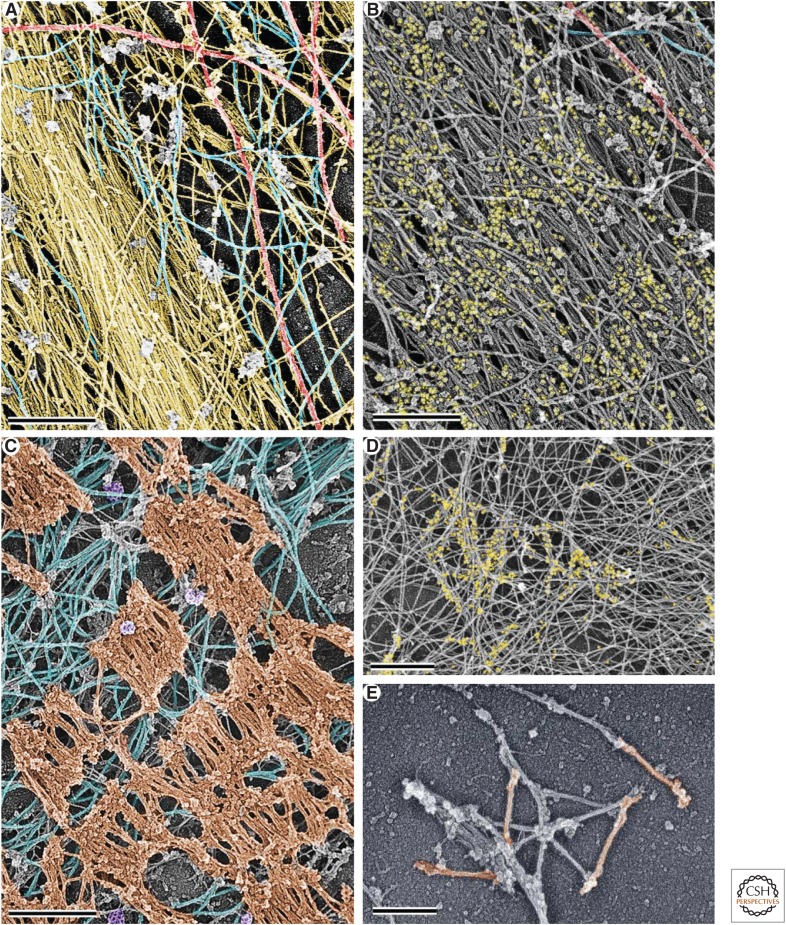
Early electron-microscopic studies of the lamellipodial cytoskeleton revealed that the lamellipodium is filled with a network of long diagonal actin filaments oriented with their barbed ends toward the leading edge (Small et al. 1978). Further scrutiny of the actin network architecture revealed multiple branched actin filaments formed by attachment of the pointed end of one filament to the side of another filament, with the formation of an ∼70° angle between the barbed ends of two filaments (Fig. 4A) (Svitkina et al. 1997; Svitkina and Borisy 1999) identical to branches formed by the Arp2/3 complex (Mullins et al. 1998). Specific structural parameters of these branched (or dendritic) actin networks in lamellipodia, such as the density, length, and orientation of actin filaments and frequency and distribution of branches, can vary among cell types and with the state of protrusion. Thus, lamellipodia containing a low density of long branched filaments protrude faster, but the filaments are more prone to buckling, which leads to ruffling and retraction of lamellipodia. Lamellipodia with a high density of short filaments are more persistent, although they protrude more slowly (Bear et al. 2002).
Figure 4.
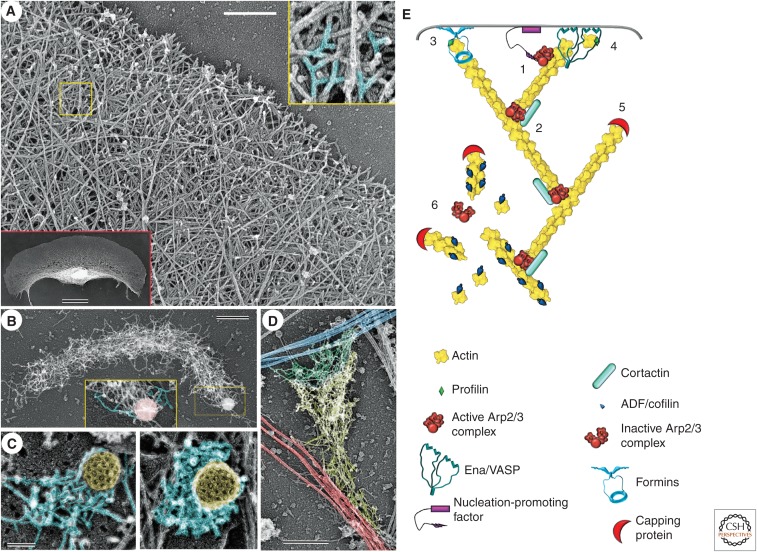
Branched actin filament networks. (A) Organization of actin filaments in the lamellipodium of a fish epidermal keratocyte revealed by platinum-replica electron microscopy. A region outlined by the yellow box is enlarged in the yellow-framed inset to show branched actin filaments (highlighted in cyan). The red-framed inset shows the entire keratocyte moving upward. (B) Branched actin network in a comet tail assembled on ActA-coated latex beads in a cytoplasmic extract from Xenopus oocytes. A region outlined by the yellow box is enlarged in the yellow-framed inset to show branched actin filaments (highlighted in cyan) at the surface of the bead (pink). (C) Patches of branched actin filaments (cyan) assembled in the vicinity of clathrin-coated vesicles (yellow). (Reprinted from Collins et al. 2011.) (D) Cytoskeleton of an excitatory synapse in a cultured hippocampal neuron. A branched actin network in the dendritic spine (yellow) resides on dendritic microtubules (red) and forms a junction with the branched actin network in the apposing presynaptic bouton (green) associated with axonal microtubules (blue). The position of the junction is approximate. Scale bars, 500 nm (A,B,D); 100 nm (C); and 10 µm (red inset in A). (E) Diagram showing molecular organization of branched actin networks. The Arp2/3 complex is cooperatively activated by a membrane-targeted nucleation-promoting factor and a preexisting “mother” actin filament (1). On activation, the Arp2/3 complex nucleates a new “daughter” actin filament at the side of the mother filament and remains associated with the branchpoint; branchpoints are further stabilized by cortactin (2). The nascent filament elongates with its barbed end oriented toward the membrane. Its elongation is promoted by formins (3) and/or Ena/VASP proteins (4). After a period of elongation, the barbed end is capped by capping protein and lags behind the protruding leading edge (5). Disassembly at the rear of the lamellipodial network occurs through dissociation of branches and ADF/cofilin-mediated severing (6).
Measurements of actin dynamics using fiducial marks in the lamellipodial actin network (Wang 1985; Theriot and Mitchison 1991) showed that actin subunits add near the plasma membrane, move away from the membrane, and dissipate from the network by the time they reach the lamellipodium base. As in filopodia, the energy of actin polymerization is distributed between protrusion and retrograde flow. However, actin turns over much faster in lamellipodia than in filopodia, and disassembly occurs throughout the lamellipodium (Theriot and Mitchison 1991), not only at the base, as in filopodia (Mallavarapu and Mitchison 1999).
The mechanism of actin turnover in lamellipodia (Fig. 4E) includes continuous nucleation of new “daughter” actin filaments at the side of preexisting “mother” filaments by Arp2/3 complex, a process called dendritic nucleation (Mullins et al. 1998). Daughter filaments then elongate and push on the plasma membrane. After a short period of elongation, their barbed end is capped and elongation is terminated. Disassembly of the network occurs through a combination of debranching and severing of actin filaments, followed by depolymerization of filament fragments. Thus, individual filaments in the network do not treadmill, but are “born” at a branchpoint, grow at the barbed end, become capped, and later “die” by depolymerization (Pollard and Borisy 2003). However, the array of branched filaments in lamellipodia undergoes treadmilling as a whole by assembling at the front and disassembling throughout its body.
The protrusive mechanism based on the dendritic nucleation principle has several advantages for generation of pushing forces, as compared with continuous elongation of individual filaments. First, short, stiff filaments are more efficient at pushing because they do not buckle. Second, being anchored to the mother filament and through it to the entire lamellipodial network, daughter filaments can efficiently transform the energy of polymerization into useful work, rather than slip backward in the absence of traction. Third, repetitive branching allows the network to easily expand or reorient itself by simply adjusting the rates and sites of nucleation and capping. Furthermore, according to the elastic Brownian ratchet model, filaments push more effectively when oriented at an angle to the membrane (Mogilner and Oster 1996, 2003). Indeed, an angled filament requires only tiny thermal fluctuations to allow for addition of an actin subunit but retains enough elasticity to produce force after recoil against a barrier such as the membrane. However, barbed end–associated proteins might also facilitate addition of actin subunits to the filament ends despite close membrane proximity and regardless of filament orientation (Dickinson and Purich 2006). These advantages can explain why the dendritic nucleation mechanism has emerged during evolution.
2.3.1.2. Molecular machinery
The molecular machinery controlling protrusion of dendritic networks includes a large number of essential and accessory molecules. Essential components besides actin itself include Arp2/3 complex, capping protein, and ADF/cofilin, all reviewed in detail elsewhere (Pollard 2016). A mixture of just these three proteins and an activator of Arp2/3 complex can generate a treadmilling actin network (Loisel et al. 1999).
Arp2/3 complex (Machesky et al. 1994; Welch et al. 1997) nucleates a daughter filament from the side of a mother filament, links mother and daughter filaments at the defined angle of 70° and caps the pointed end of the daughter filament (Mullins et al. 1998). Arp2/3 complex requires activation by a nucleation-promoting factor and binding to a mother filament to nucleate a branch (Machesky et al. 1999). The most important nucleation-promoting factors belong to the Wiskott–Aldrich syndrome protein (WASp) family, which in mammals include WASp, N-WASp, WAVE1–WAVE3, and some other proteins (Burianek and Soderling 2013). The heteropentameric WAVE regulatory complex typically activates Arp2/3 complex in lamellipodia. Signaling events and guidance cues recruit and activate nucleation-promoting factors at the plasma membrane to nucleate branched filaments that generate force on the plasma membrane exactly where it is needed.
Constant production of new filaments by the Arp2/3 complex requires concurrent termination of the barbed end growth to prevent exponential expansion of the lamellipodial network and unproductive consumption of monomers. This task in lamellipodia is predominantly performed by heterodimeric capping protein, which tightly binds the actin filament barbed end. Other barbed end capping proteins, such as Eps8 and gelsolin, are also present in lamellipodia and might contribute to the dynamics of branched networks (Zigmond 2004).
Disassembly of branched actin networks throughout the lamellipodium depends on severing by ADF/cofilin. Severing depends on hydrolysis of ATP bound to polymerized actin and dissociation of the γ-phosphate. The protein Aip1 promotes severing by ADF/cofilin (Okada et al. 1999). In cells moving rapidly and persistently in culture, such as isolated frog epidermal keratocytes, little ADF/cofilin binds to newly assembled ATP–actin filaments at the cell front. In other cells, such as fibroblasts, that show more-exploratory behavior, ADF/cofilin can bind actin filaments in anterior regions of the lamellipodial network, probably, because these regions contain mixtures of filaments with bound ADP or ATP (Svitkina and Borisy 1999). Actin filament severing by ADF/cofilin can also help the cell to start a new lamellipodium by creating new uncapped barbed ends (Ghosh et al. 2004).
Dissociation of a daughter filament from the mother filament also contributes to disassembly of the branched actin network. ADF/cofilin promotes debranching by changing the conformation of the mother filament and reducing the affinity for Arp2/3 complex (Chan et al. 2009). Cofilin-related GMF proteins (Gandhi et al. 2010) cause debranching by binding to Arp2/3 complex. In contrast, the actin-binding protein cortactin stabilizes branches, probably by forming an additional link between the Arp2/3 complex and the actin filament (Weaver et al. 2001).
Formins (Yang et al. 2007) and Ena/VASP proteins (Bear et al. 2002) produce long, unbranched actin filaments within the branched network by enhancing filament elongation and counterbalancing the activity of capping proteins. The long filaments can be bundled by fascin to produce filopodia (Svitkina et al. 2003) or buckled under the membrane and produce ruffles (Bear et al. 2002). Actin cross-linkers, having relatively long spacers between actin-binding sites, such as α-actinin and filamin A, are also present in lamellipodia, where they are thought to consolidate the entire network.
3. The actin cytoskeleton in membrane trafficking
Normal cell physiology depends on the presence of a functional intracellular transportation and delivery system that sorts, moves, and directs newly synthesized, preexisting, and imported molecules to specific destinations within the cell by using membrane vesicles as “parcels.” Actin polymerization is one of multiple mechanisms used by the cell to accomplish these tasks. Furthermore, some intracellular pathogens exploit vesicular transport mechanisms to access cellular resources, avoid immune surveillance in the extracellular environment, and facilitate their own spread from cell to cell (Haglund and Welch 2011). Although this microbial mimicry has adverse effects on human health, experiments with bacteria and viruses have been instrumental for deciphering the molecular mechanisms of protrusion driven by branched actin filament networks (Loisel et al. 1999). Our advanced understanding of the biochemical, biophysical, and structural mechanisms of actin-polymerization-driven motility now permits explicit mathematical modeling of this process by using quantitative parameters measured in live cells and in vitro systems. Computer simulations derived from these models accurately reproduce the behavior of the leading edge or bacterial “comet tails” but also help reveal hidden gaps in our knowledge that would remain undetectable at a qualitative level (Mogilner 2009).
3.1. Phagocytosis and Macropinocytosis
Internalization of a solid particle (phagocytosis) (Groves et al. 2008) or a portion of extracellular fluid (macropinocytosis) (Kerr and Teasdale 2009) depends on broad two-dimensional protrusions of the cell surface homologous to lamellipodia. Both processes evolved in amoebas and are used by immune cells of metazoa, such as macrophages and dendritic cells. Dendritic cells use macropinocytosis to take up extracellular fluids containing foreign soluble materials and initiate an immune response. Macrophages and neutrophils use phagocytosis to engulf and destroy pathogens and debris in the body. Various signaling pathways can trigger phagocytosis and macropinocytosis by nonimmune cells. For example, certain intestinal pathogens activate signaling pathways leading to Arp2/3 complex activation and formation of protrusions that engulf the bacterium. Once inside the cell, pathogens use additional molecular “tricks” to escape from the phagocytic vacuole and avoid proteolytic degradation in lysosomes (Haglund and Welch 2011).
3.2. Comet Tail Motility
The bacteria Listeria monocytogenes and Shigella flexneri—intracellular pathogens that cause severe intestinal disorders—were the first examples of bacteria that can exploit cellular resources to induce dendritic actin nucleation to facilitate their cell-to-cell spread (Bernardini et al. 1989; Tilney and Portnoy 1989). After accessing the cytoplasm of intestinal epithelial cells through induced phagocytosis, one pole of these bacteria assembles a comet-like tail of branched actin filaments. This tail propels the bacterium through the cytoplasm and into a cellular protrusion, which then can be ingested by a neighboring cell. Listeria has evolved a surface protein ActA that directly activates Arp2/3 complex by mimicking cellular nucleation-promoting factors (Welch et al. 1998). When adsorbed to plastic beads, ActA can induce comet tail formation in cytoplasmic extracts (Fig. 4B) or a solution of purified proteins (Loisel et al. 1999). Shigella has a protein named IcsA that recruits the cellular nucleation-promoting factor N-WASp (Egile et al. 1999). In both cases, the comet is made exclusively of cytoplasmic proteins of the host cell.
The comet-tail-driven motility of Listeria and Shigella is thought to represent an exaggerated version of membrane trafficking in animal cells, which depends on different nucleation-promoting factors to activate the Arp2/3 complex (Burianek and Soderling 2013). N-WASp is involved in the phagocytosis and intracellular motility of endosomes. WASH, another member of WASp family, is thought to participate in scission of recycling vesicles from endosomes. A further nucleation-promoting factor—WHAMM—participates in endoplasmic reticulum (ER)-to-Golgi trafficking, as well as in Golgi morphogenesis. The details of the mechanism underpinning the actin polymerization machinery during these membrane trafficking events are currently under investigation.
3.3. Clathrin-Mediated Endocytosis
Clathrin-mediated endocytosis is the major endocytic pathway used by cells to take up exogenous molecules and cell-surface components in a highly selective way (McMahon and Boucrot 2011). The formation of an endocytic vesicle from the plasma membrane involves local membrane invagination followed by constriction of the bud neck and, finally, vesicle scission. All these events are energetically unfavorable and require force-generating machinery. Pushing forces generated by dendritic actin networks make important contributions to this process (Mooren et al. 2012). Actin polymerization is absolutely essential for clathrin-mediated endocytosis in yeast and is also important for mammalian endocytosis. Dendritic actin networks in the form of small three-dimensional patches assemble at the clathrin-coated structures toward the end of the endocytic internalization cycle (Fig. 4C) (Collins et al. 2011). The pushing force generated by these patches is thought to facilitate invagination of the clathrin-coated pit, constriction of the bud neck, and vesicle scission.
4. The role of actin at cell–cell junctions
Actin polymerization by Arp2/3 complex is involved in formation of various cell–cell junctions, such as adherens junctions in epithelial cells (Michael and Yap 2013), synapses in neurons (Racz and Weinberg 2013) and immune cells (Billadeau and Burkhardt 2006), and prefusion junctions in myoblasts (Sens et al. 2010). Although branched actin networks have been visualized only in some of these situations, the involvement of a dendritic nucleation mechanism has been deduced from localization of essential components of the dendritic nucleation machinery at these junctions and their functional contribution to junction formation. Pushing forces generated by branched networks at cell–cell junctions most likely promote apposition of the contacting plasma membranes of two neighboring cells.
4.1. Neuronal Synapses
Excitatory synapses between neurons in the central nervous system are formed between presynaptic boutons filled with synaptic vesicles on axons and postsynaptic mushroom-shaped protrusions called dendritic spines on dendrites (Fig. 4D). The two plasma membranes at the synapse are connected by adhesion molecules and separated by a narrow cleft. Neurotransmitters secreted by the axonal bouton diffuse through the cleft and activate receptors on the head of the spines. The head of each dendritic spine is filled with a dense three-dimensional network of branched actin filaments, including Arp2/3 complex, capping protein, and VASP (Korobova and Svitkina 2010; Lin et al. 2010). Presynaptic boutons of cultured hippocampal neurons also contain a less elaborate branched actin network (Korobova and Svitkina 2010). The presynaptic and postsynaptic networks probably press against each other to promote interaction of cell–cell adhesion receptors at the synapse.
4.2. Adherens Junctions
Adherens junctions are the main type of cell–cell adhesions that depend on the actin cytoskeleton. Adherens junctions in epithelial sheets are essential for maintaining tissue integrity. Cell–cell interactions at adherens junctions are mediated primarily by members of the cadherin family of adhesion receptors and stabilized by the actin cytoskeleton. Ultrastructurally, adherens junctions in mature epithelial sheets appear to be associated mostly with actin filament bundles (Yonemura et al. 1995). However, other approaches have revealed that the integrity of adherens junctions depends on dynamic actin filaments and the Arp2/3 complex (Michael and Yap 2013). The contribution of branched actin networks is most apparent during the formation of adherens junctions, when colliding lamellipodia make the first cell–cell contacts (McNeill et al. 1993; Krendel and Bonder 1999; Hoelzle and Svitkina 2012). Networks of actin in two adjacent cells might press the membranes together and facilitate engagement of adhesion molecules.
4.3. Myoblast Fusion
Muscle cells in most animals are syncytia formed by fusion of thousands of myocytes during embryonic development. The fusion process is best studied in embryos of the fruitfly Drosophila melanogaster (Abmayr and Pavlath 2012), where muscles are formed by the asymmetric fusion of a founder cell with several fusion-competent myoblasts. Soon after a junction is formed between these two cell types, the fusion-competent myoblast forms an actin-rich protrusive “finger” that pushes into the founder cell (Sens et al. 2010). Formation of the finger depends on two Drosophila nucleation-promoting factors—WASp and Scar (homolog of WAVE)—and thus is likely to depend on Arp2/3 complex. In the founder cell, a thin layer of actin formed in a Scar-dependent manner at the contact site counteracts the pushing force from the finger and promotes juxtaposition of the contacting membranes. Next, a fusion pore forms at the junction site and expands until the two cells completely fuse. Less-well-characterized fusion occurs between other cell types, such as germ cells during fertilization, macrophages making giant cells, and placental trophoblasts.
5. The actin cytoskeleton in contraction
5.1. General Concept
The actin cytoskeleton generates pulling forces through mutual sliding of actin and myosin filaments, as originally established for skeletal muscles (Huxley and Hanson 1954; Huxley and Niedergerke 1954; Weber and Franzini-Armstrong 2002). As reviewed elsewhere (Sweeney and Hammers 2016), the contractile machinery in skeletal muscles is more highly ordered and stable than in other cells, but the mechanism of contraction in muscle cells is applicable to nonmuscle cells.
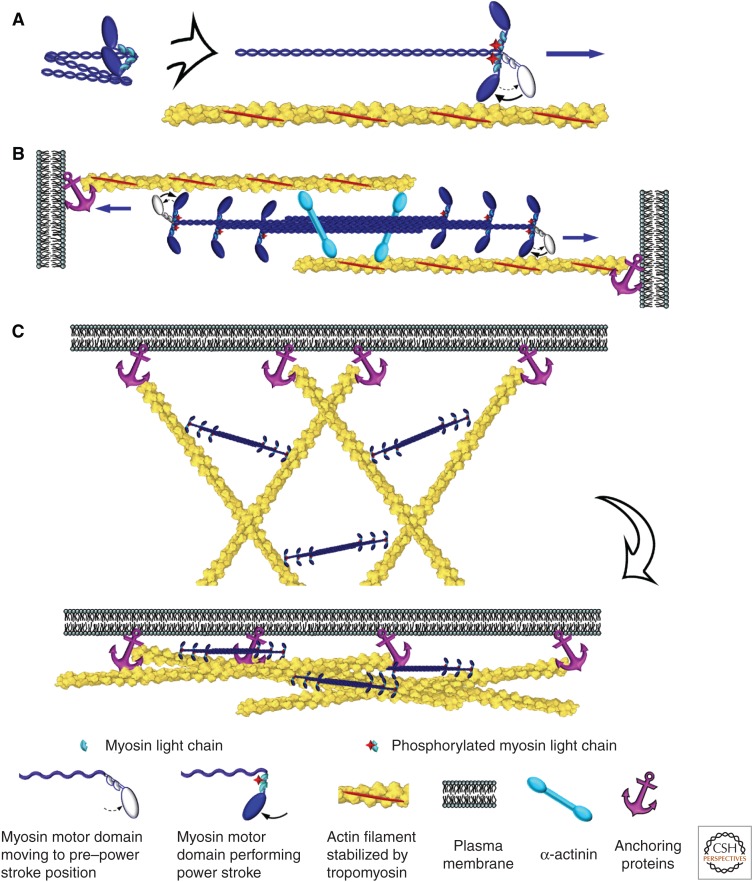
Myosins, a large superfamily of motor proteins (Syamaladevi et al. 2012), use the energy of ATP hydrolysis to move along actin filaments (reviewed by Sweeney and Holzbaur 2016). Class II myosins are specifically designed for contraction (Fig. 5). The amino-terminal motor domain of the heavy chain (“head”) has the ATPase and actin-binding activities, whereas the “tail” mediates parallel dimerization of heavy chains through coiled-coil interactions. Each heavy chain binds two different light chains—essential and regulatory—at the “neck,” a region between the head and the tail. Myosin II polymerizes into bipolar filaments through interactions of the tails, so the motor domains are located at the filament ends. These bipolar myosin filaments pull actin filaments together, causing contraction. Different members of class II myosins have different polymerization properties. Sarcomeric myosin II forms longer (1.5–2 µm), more stable bipolar filaments than smooth muscle and nonmuscle myosin II isoforms (∼0.3–0.4 µm) (Pollard 1975).
Figure 5.
Contractile activity of nonmuscle myosin II. (A) A hexameric molecule of nonmuscle myosin II has a folded conformation in an inactive state (left). Phosphorylation of the myosin regulatory light chain causes unfolding of the myosin II molecule and restoration of motor activity; this activated myosin II can move toward the barbed end of the actin filament by swinging its motor domain in an ATP-dependent manner (right). Contraction of actin–myosin-II bundles (B) or networks (C) is mediated by bipolar filaments of myosin II that move along oppositely oriented (B) or nonaligned (C) actin filaments anchored at their barbed ends to the plasma membrane or other cellular structures.
Actin filaments participating in contraction are stabilized by tropomyosins, a large group of elongated coiled-coil proteins that bind actin filaments along the length (Nevzorov and Levitsky 2011). Capping proteins stabilize both filament ends in striated muscles (Littlefield and Fowler 2008). However, actin filament ends appear to remain uncapped in nonmuscle contractile systems.
Organizing actin filaments into bundles makes contraction more efficient. The main actin filament cross-linking protein in contractile systems is α-actinin, an antiparallel homodimer with an actin-binding site at each end (Sjoblom et al. 2008). The spacer between actin-binding sites separates the actin filaments sufficiently to allow interaction with myosin filaments, but close enough to align filaments into bundles and maximize the actin–myosin-II interaction.
Attachment of actin filaments to a load allows for conversion of the myosin-driven actin filament translocation into mechanical work. Actin filaments are typically attached to a load near their barbed ends. Myosin II moves toward the barbed end so it can pull on the actin filament. In striated muscles, barbed ends are attached to the Z-disk that forms a boundary between sarcomeres. In smooth muscles, actin filament barbed ends are embedded in dense bodies. In nonmuscle cells, barbed ends are typically anchored at the plasma membrane (Fig. 5C). These associations of actin filaments with a load are system specific and usually involve multiple proteins.
5.2. Contractile Systems in Nonmuscle Cells
The contractile forces generated by nonmuscle cells are used for many purposes, including migration, cytokinesis, cell-shape changes, organization of the extracellular matrix, and formation of cell–cell and cell–matrix junctions (Heissler and Manstein 2013). Nonmuscle cells exert pulling forces by using various arrays of actin and myosin II filaments. The organization of these arrays varies from quasi-sarcomeric bundles to mixed networks of actin and myosin II with imperfect alignment (Fig. 1). Nonmuscle contractile systems are much less ordered and more dynamic than striated muscle sarcomeres, allowing nonmuscle cells to build and dismantle a contractile system in response to changing conditions. Specific regulatory mechanisms, as well as the biochemical and biophysical properties of nonmuscle myosin II, underlie the flexibility of nonmuscle contractility.
5.2.1. Nonmuscle Myosin II
The assembly of bipolar filaments of myosin II in nonmuscle cells is reversible, allowing a cell to assemble a contractile system when and where needed. Phosphorylation of the myosin regulatory light chain (Adelstein and Conti 1975) regulates both the assembly and ATPase activity of myosin II in nonmuscle cells of vertebrates. When the regulatory light chains are dephosphorylated, myosin II folds into an inactive, autoinhibited conformation (Craig et al. 1983), unable to polymerize or bind to actin filaments. When the light chain is phosphorylated on one (Ser19) or two (Ser18 and Thr18) residues, nonmuscle myosin II unfolds into a linear conformation, restoring its motor activity and ability to polymerize (Fig. 5A). Phosphorylation of, or ligand binding to, the extreme carboxyl terminus of the heavy chain inhibits polymerization, but not the motor activity, of nonmuscle myosin II (Clark et al. 2007). This regulation might contribute to recycling nonmuscle myosin II molecules.
Mammals encode three isoforms (A, B, and C) of the nonmuscle myosin heavy chain (Wang et al. 2011; Heissler and Manstein 2013). Nonmuscle myosin IIA and IIB are expressed widely, whereas myosin IIC has restricted expression. The isoforms have both unique and overlapping functions in cells and the organism. Accordingly, they show some differences in their biophysical parameters of motor activity and dynamics of bipolar filament turnover (De La Cruz and Ostap 2004). The turnover differences depend on the intrinsic properties of the heavy chain tails, as well as on variations in the regulation of their polymerization and depolymerization (Sandquist and Means 2008).
Among three mammalian nonmuscle myosin II isoforms, myosin IIA is the fastest motor with the lowest duty ratio (a fraction of the ATP cycle, when myosin is strongly attached to the actin filament). Therefore, myosin IIA is well suited for fast movement along actin filaments and generation of contraction. Myosin IIA also shows fast cycles of polymerization and depolymerization, so that it can quickly change its subcellular distribution. Nonmuscle myosin IIB is the slowest motor, with the highest duty ratio among nonmuscle myosin II isoforms. Moreover, resistive load significantly increases the duty ratio of myosin IIB, more than that of other nonmuscle myosin II isoforms (Kovacs et al. 2007). These properties suggest that nonmuscle myosin IIB preferentially functions to maintain tension for extended periods of time, rather than to produce active contraction. Accordingly, the turnover rate of bipolar filaments of nonmuscle myosin IIB is relatively slow.
5.2.2. Contractile Actin–Myosin Bundles
Early studies, in the 1930s, of cultured cells by phase-contrast microscopy revealed so-called “stress fibers”—straight or slightly curved phase-dark lines that span the cell and appear to be under tension (Buckley and Porter 1967). Subsequent analyses showed that stress fibers represent mixed bundles of actin and bipolar filaments of nonmuscle myosin II that contract or exert isometric tension (Burridge and Wittchen 2013).
5.2.2.1. Structure
The structural organization of stress fibers in nonmuscle cells shows a slight resemblance to striated muscle sarcomeres. Immunofluorescence staining showed that nonmuscle myosin II is arranged in a semiperiodic fashion along stress fibers (Fig. 1) (Weber and Groeschel-Stewart 1974). Segments of the stress fiber with nonmuscle myosin II are also enriched in tropomyosin, whereas intervening areas are enriched in α-actinin (Lazarides 1976). Electron microscopy showed that myosin II in stress fibers forms bipolar filaments ∼300 nm in length (Langanger et al. 1986; Svitkina et al. 1989), which aligned in parallel stacks analogous to the skeletal muscle myosin II filaments in sarcomeres (Fig. 6A–C) (Verkhovsky et al. 1995). Bipolar myosin II filaments aligned with the stress fiber axis were also detected by dual-color fluorescence microscopy, with different probes for myosin II heads and tails, as triplets of alternating colors (Ebrahim et al. 2013; Beach et al. 2014).
Figure 6.
Contractile actin structures in nonmuscle cells. (A) A stress fiber fragment (extends from upper left to lower middle) and a network of other cytoskeletal components (upper right half) in a rat fibroblast. Color coding: yellow, actin filaments; red, microtubules; cyan, intermediate filaments. (Reprinted from Svitkina et al. 1995.) (B) Immunogold labeling of a stress fiber (extends from upper left to lower right) with an antibody to nonmuscle myosin II. Distribution of colloidal gold particles shows semiperiodic arrangement of myosin II along the actin filament bundle. Color coding: yellow, gold particles; red, microtubules; cyan, intermediate filaments. (C) Dissolution of actin filaments by treatment of the cytoskeleton with the actin-severing protein gelsolin reveals stacks of bipolar myosin II filaments separated by irregular intervals. The diagonal alignment of myosin II filaments suggests that stacks belong to the same stress fiber running in an upper left to lower right direction. Color coding: brown, myosin II filaments; cyan, intermediate filaments; purple, clathrin-coated pits. (D) Contractile actin–myosin-II network in the lamella of a fish epidermal keratocyte. Immunogold staining of myosin II shows linear sets of gold particles (colored in yellow) representing myosin II filaments among the network of actin filaments. The orientation of actin and myosin II filaments changes from the front (right) to the rear (left) of the cell, reflecting network contraction during keratocyte migration. (E) A cluster of nonoriented bipolar filaments of nonmuscle myosin II in the lamella of a fish epidermal keratocyte visualized after dissolution of actin filaments by gelsolin treatment. Filaments form chains (upper right), networks (center), and stacks (lower left). Several individual bipolar filaments are colored in brown. Scale bars, 500 nm (A–C) and 250 nm (D,E).
Although these features of stress fibers resemble striated muscle sarcomeres, the two systems do have significant differences. Stacks of bipolar filaments are unevenly spaced along the stress fiber, often staggered and not perfectly aligned with each other, and can fuse and interdigitate with each other to the extent that they become poorly discernable (Fig. 6B,C).
The organization of actin filaments in stress fibers also differs significantly from striated muscle sarcomeres in which the filaments have defined lengths, strictly alternating polarities, and interact only with one set of myosin II filaments (Huxley 1957). Fluorescence labeling of actin filaments shows that they are continuously distributed along the stress fiber length (Lazarides and Weber 1974), contrasting with their periodic distribution in striated muscle. Electron microscopy showed that the actin filaments in stress fibers have mixed polarity and show variable lengths, so that oppositely oriented filaments often overlap. Moreover, some actin filaments in the stress fibers are so long that they pass through several myosin-rich regions (Fig. 6A,B) (Sanger and Sanger 1980). These features could be due to the absence of capping proteins, as neither barbed end capping proteins nor the pointed end capper tropomodulin have been detected in stress fibers.
Proteins promoting barbed end elongation—VASP and formins—participate in the assembly of actin filaments in stress fibers. VASP is present in stress fibers in a punctate pattern (Reinhard et al. 1995), suggesting that elongating barbed ends are scattered throughout the actin filament bundle. VASP becomes particularly enriched in regions where a stress fiber is ruptured, where it promotes actin filament elongation to repair the damage (Smith et al. 2010). The actin-polymerizing activity of the formin mDia1 is also used during stress fiber formation, although mDia1 has not been detected in stress fibers (Watanabe et al. 1999). Thus, in contrast to the capped barbed ends in striated muscle, actin filament barbed ends in stress fibers of nonmuscle cells can elongate, and seem to be protected from capping.
5.2.2.2. Interaction with the load
Stress fibers are attached at their ends (focal adhesions) to the plasma membrane, and, through the plasma membrane, to the extracellular matrix. This transmembrane linkage involves members of the integrin family of transmembrane adhesion receptors (Hu and Luo 2013). Integrins directly bind the extracellular matrix outside the cell and use a set of adaptor proteins to bind actin filaments inside the cell (Wehrle-Haller 2012). Adaptor proteins, such as talin and vinculin, are highly enriched in focal adhesions. Stress fiber actin filaments in the vicinity of a focal adhesion are uniformly oriented with their barbed ends toward the plasma membrane (Sanger and Sanger 1980). These barbed ends are not capped and can incorporate actin subunits, likely owing to the activity of VASP that is highly enriched in focal adhesions. The occurrence of barbed end elongation at focal adhesions might help to maintain the connection of actin filaments to the plasma membrane in the presence of pulling forces exerted by myosin II. Regions of stress fibers associated with focal adhesions are enriched in α-actinin but rarely contain myosin II. In addition, focal adhesions contain a very large number of signaling proteins, which transmit information about the stiffness and chemistry of the underlying surface into the cell, so that the cell can adapt its behavior to external conditions (Wehrle-Haller 2012).
In addition to classical straight stress fibers that are associated with focal adhesions, bundles of actin and myosin II filaments can be bound to another load and/or acquire different shapes. For example, a stress fiber can have one end attached to the plasma membrane and the other end incorporated into another stress fiber or cytoskeletal structure. Migrating cells produce arc-shaped stress fibers with both ends enmeshed in surrounding actin networks and bundles (Naumanen et al. 2008). These arcs can be connected to focal adhesions through other stress fibers that radiate out of the arc and terminate in focal adhesions.
As reviewed elsewhere (Glotzer 2016), contractile rings of actin filaments (Schroeder 1969) and myosin II (Fujiwara and Pollard 1976) drive constriction of the cleavage furrow during cytokinesis. In the slime mold Dictyostelium discoideum, attachment of the contractile ring to the plasma membrane requires the activity of the focal adhesion protein talin (Tsujioka et al. 2012). Members of the ERM protein family of actin filament–membrane linkers (Niggli and Rossy 2008) might also contribute to the interaction of the contractile ring with the plasma membrane.
Bundles of actin filaments and myosin II are also associated with sites of cell–cell interaction at intercellular adherens junctions (Huveneers and de Rooij 2013). Adhesion receptors of the cadherin family make homophilic interactions between two cells and use adaptor proteins to bind to actin filaments in the cell interior. The adaptor proteins include adherens-junction-specific proteins, such as α-catenins and β-catenins, as well as proteins functioning in focal adhesions, such as α-actinin and vinculin.
In the cohesive layers of epithelial cells that compartmentalize tissues in an organism, adherens junctions form a continuous circumferential belt around the apical cell surface. Adherens junctions are linked to circular actin–myosin II bundles, which can transmit tangential tension to the adhesion belt. The nature of the mechanism attaching circumferential actin–myosin II bundles to the plasma membrane in continuous or “linear” adherens junctions remains uncertain. Local accumulations of cadherins also form intermittent cell–cell contacts. These “focal” adherens junctions are typically associated with the tips of stress fibers. As focal and linear adherens junctions use slightly distinct sets of adaptor proteins on the cytoplasmic side, the mechanisms of interaction between actin and the plasma membrane might have both shared and distinct features in these two types of adherens junctions.
5.2.2.3. Mechanism of contraction
In highly ordered striated muscle sarcomeres, bipolar myosin II filaments interdigitate with actin filaments with opposite polarities in two halves of the sarcomere. The sarcomere shortens and the cell contracts when myosin II heads pull the actin filaments toward the middle of the sarcomere (Huxley and Hanson 1954; Huxley and Niedergerke 1954).
In less-well-organized stress fibers, myosin II filaments are positioned among actin filaments with mixed polarity (Burridge and Wittchen 2013) and might be able to interact preferentially with actin filaments having the appropriate polarity to contract rather than lengthen the bundle. It is plausible that actin filaments that are properly oriented relative to myosin II and attached to a load are tensioned. The duty ratio of nonmuscle myosin II is increased under resisting load, which allows the motor to remain bound to these actin filaments for a longer time, favoring contraction (Kovacs et al. 2007). Moreover, actin filaments under tension have higher affinity for myosins (Uyeda et al. 2011), further increasing the probability of interaction between actin and myosin II.
5.2.2.4. Functions
In migrating cells, contractile forces generated by stress fibers are typically applied to focal adhesions. These contractile forces can rupture the adhesion and, thus, induce local retraction of the cell edge. Alternatively, they can strengthen the adhesion, which then can serve as a launchpad for the next round of protrusion (Wolfenson et al. 2011). In directionally moving cells, adhesion strengthening typically occurs at the cell front, whereas adhesion rupture is biased toward the cell rear. This differential response helps the cell to maintain its direction of migration. This behavior is regulated, at least in part, by microtubules (Vasiliev et al. 1970; Kaverina et al. 2002), although the biochemical mechanism underlying this is not clear (Stehbens and Wittmann 2012). Another important function of contractile actin–myosin bundles is to maintain isometric tension that supports the shape of the cells and defines the mechanical properties of the cell surface. For example, circumferential bundles of actin and myosin II associated with linear adherens junctions generate a tangential tension that is necessary to maintain junction integrity.
Cells migrating on a solid surface in two-dimensional culture have exaggerated stress fibers and focal adhesions, as compared with cells moving in vivo (Wong et al. 1983; Geraldo et al. 2012), in three-dimensional culture (Kubow et al. 2013), or on soft surfaces with physiologically relevant stiffness (Pelham and Wang 1997; Engler et al. 2004). The dependence of stress fiber formation on the physical properties of the substratum could reflect the fact that cells in tissues might use stress fibers and focal adhesions mostly for remodeling of the extracellular matrix. Indeed, stress fibers and focal adhesions are especially characteristic for fibroblasts—mesenchymal cells that secrete and organize the extracellular matrix in the organism. When grown in a collagen gel in cell culture, fibroblasts use focal adhesions and stress fibers to attach to and pull on collagen fibers, so that they can move and align these fibers. Thus, when grown on a stiff surface in a culture dish, fibroblasts build up contractile force by increasing the number and size of stress fibers, as if desperately trying to deform this noncompliant substratum.
5.2.3. Contractile Actin–Myosin Networks
5.2.3.1. Structure and mechanism of contraction
To mediate contraction, actin and myosin II filaments do not have to be aligned, as in sarcomeres or stress fibers, but can be arranged into a network in which individual filaments are oriented at a range of angles (Verkhovsky et al. 1999). In most cases, such organization is inferred from low-resolution fluorescence microscopy images of cells stained with probes for actin filaments and nonmuscle myosin II. With this technique, actin–myosin-II networks appear as a diffuse distribution of actin that contains scattered puncta of myosin II. This staining pattern is characteristic, for example, of the “cell cortex,” an actin–myosin-II layer underlying the plasma membrane in many cells (Zigmond et al. 1979). In some cases, the high-resolution organization of actin–myosin-II networks has been determined. For example, electron microscopy of the thin lamellae of migrating cells (Verkhovsky et al. 1995; Svitkina et al. 1997) or of the growth cones of cultured neurons (Bridgman 2002) has revealed bipolar filaments of nonmuscle myosin II, either individual ones or in the form of clusters of variable size, associating with a nonaligned network of relatively long actin filaments. Actin–myosin-II networks in the cell cortex and other locations might also have a similar basic organization, although the densities of actin and myosin II filaments and their relative abundance most likely vary from case to case.
Although actin–myosin-II networks have even less-ordered organization than stress fibers, they still can generate contractile forces, as shown decades ago when mixtures of actin and myosin in a test tube contracted upon addition of ATP (Szent-Gyorgyi 1950, 2004). In lamellae of migrating cells, interactions of the two ends of a bipolar myosin II filament with two actin filaments oriented at different angles causes rotation of actin and myosin II filaments around the anchor points until they all align between the attachment sites. Such mutual alignment of actin and myosin II filaments is accompanied by network contraction in the orthogonal direction (Fig. 5C) (Svitkina et al. 1997; Verkhovsky et al. 1999).
5.2.3.2. Interaction with the load
The molecular connections between the plasma membrane and actin–myosin-II networks are not yet well understood. In leading lamellae of migrating cells, actin–myosin-II networks appear to be associated with focal complexes (Shutova et al. 2012). These small and dynamic cell–matrix adhesions generate traction for cell migration or can serve as precursors for larger and more-stable focal adhesions (Rottner et al. 1999; Choi et al. 2008). They use a similar set of proteins to connect actin to the plasma membrane, as mature focal adhesions. The adhesive structures linking other actin–myosin-II networks to the plasma membrane might depend on additional transmembrane and adaptor proteins. Perhaps these molecules are widely distributed at low density over the plasma membrane. ERM-family proteins are candidates (Niggli and Rossy 2008) because their depletion decreases membrane-to-cortex attachment (Diz-Munoz et al. 2010). Many other proteins that can bind both actin and the membrane, such as class I myosins, formins, Ena/VASP proteins, and talin, might also contribute to this linkage.
5.2.3.3. Functions
In fast migrating cells such as keratocytes and neutrophils, contraction of actin–myosin-II networks brings forward the cell body, so that it keeps up with the advancing leading edge (Svitkina et al. 1997). In other cell types, network contraction leads to stress fiber formation through the alignment of actin and myosin II filaments (Verkhovsky et al. 1995) that mature into stress fibers with semiperiodic myosin II distribution (Stachowiak et al. 2012). The contraction of actin–myosin-II networks might represent a primordial mechanism to exert pulling forces, which evolved into more efficient contractile systems, such as stress fibers, contractile rings, and sarcomeres.
During migration, cells need traction to prevent slippage during protrusion. Traction is largely provided by focal adhesions. Counterintuitively, small and dynamic focal complexes associated with actin–myosin-II networks are more efficient in the generation of traction than large and stable focal adhesions (Beningo et al. 2001). Adhesion formation, in general, depends on force (Burridge and Wittchen 2013) produced by actin–myosin-II systems and transmitted to integrins, which activates integrins and traction generation. The forces required to form focal complexes are fairly small and can be produced by contraction of actin–myosin-II networks or derived from actin filament cross-linking (Choi et al. 2008) or a drag from actin retrograde flow (Alexandrova et al. 2008; Gardel et al. 2008).
Contraction of actin–myosin networks also leads to changes in cell shapes and remodels tissues. For example, the invagination of epithelial sheets during embryonic development drives gastrulation, neural tube formation, gland formation, and other processes (Kasza and Zallen 2011). During invagination, a fraction of epithelial cells acquires a conical shape by constricting their apical surface, leading to local bending of the epithelial sheet (Wessells et al. 1971). During gastrulation in Drosophila embryos, contraction of an actin–myosin-II network located underneath the apical surface drives apical constriction of epithelial cells, which is stabilized by the circumferential belt of actin filaments and myosin II associated with apical adherens junctions (Martin 2010).
The cortical actin–myosin-II network underlying the plasma membrane defines the mechanical properties of the cell surface, generates surface tension, which allows cells to resist external insults, and produces internal pressures (Salbreux et al. 2012). If the cortex is locally damaged or detached from the plasma membrane, internal pressure causes formation of blebs—balloon-like expansions of the plasma membrane. Reformation of the cortex in the bleb leads to withdrawal of the bleb through contraction of the cortical actin–myosin-II network. Blebs often form during apoptosis, possibly, as precursors of apoptotic bodies. They are common during cytokinesis, in which they help to release excessive intracellular pressure in daughter cells. An amoeboid mode of cell migration, especially when cells move in a spatially constrained environment, involves extensive blebbing (Liu et al. 2015; Ruprecht et al. 2015). In this case, blebs are thought to advance the cell front instead of lamellipodia and filopodia (Charras and Paluch 2008).
6. Conclusion
The activity of the actin cytoskeleton supports the vast majority of motile events in eukaryotic cells. A large set of accessory proteins controls the assembly of a common pool of actin, as well as the spatial organization, subcellular localization, and interactions of the filaments with other structures. This immense complexity has evolved for the benefit of cells and organisms and continues to offer interesting challenges to researchers aiming to understand how the actin cytoskeleton works.
ACKNOWLEDGMENTS
This work has been supported by National Institutes of Health grant R01 GM 095977. I thank Maria Shutova for the image used in Figure 1B and Farida Korobova for the image used in Figure 4D.
Footnotes
Editors: Thomas D. Pollard and Robert D. Goldman
Additional Perspectives on The Cytoskeleton available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Abercrombie M, Heaysman JE, Pegrum SM. 1970. The locomotion of fibroblasts in culture. II. ‘Ruffling’. Exp Cell Res 60: 437–444. [DOI] [PubMed] [Google Scholar]
- Abmayr SM, Pavlath GK. 2012. Myoblast fusion: Lessons from flies and mice. Development 139: 641–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adelstein RS, Conti MA. 1975. Phosphorylation of platelet myosin increases actin-activated myosin ATPase activity. Nature 256: 597–598. [DOI] [PubMed] [Google Scholar]
- Alexandrova AY, Arnold K, Schaub S, Vasiliev JM, Meister JJ, Bershadsky AD, Verkhovsky AB. 2008. Comparative dynamics of retrograde actin flow and focal adhesions: Formation of nascent adhesions triggers transition from fast to slow flow. PLoS ONE 3: e3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach JR, Shao L, Remmert K, Li D, Betzig E, Hammer JA. 3rd 2014. Nonmuscle myosin II isoforms coassemble in living cells. Curr Biol 24: 1160–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear JE, Svitkina TM, Krause M, Schafer DA, Loureiro JJ, Strasser GA, Maly IV, Chaga OY, Cooper JA, Borisy GG, et al. 2002. Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell 109: 509–521. [DOI] [PubMed] [Google Scholar]
- Beningo KA, Dembo M, Kaverina I, Small JV, Wang YL. 2001. Nascent focal adhesions are responsible for the generation of strong propulsive forces in migrating fibroblasts. J Cell Biol 153: 881–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardini ML, Mounier J, d’Hauteville H, Coquis-Rondon M, Sansonetti PJ. 1989. Identification of icsA, a plasmid locus of Shigella flexneri that governs bacterial intra- and intercellular spread through interaction with F-actin. Proc Natl Acad Sci 86: 3867–3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billadeau DD, Burkhardt JK. 2006. Regulation of cytoskeletal dynamics at the immune synapse: New stars join the actin troupe. Traffic 7: 1451–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitsprecher D, Koestler SA, Chizhov I, Nemethova M, Mueller J, Goode BL, Small JV, Rottner K, Faix J. 2011. Cofilin cooperates with fascin to disassemble filopodial actin filaments. J Cell Sci 124: 3305–3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridgman PC. 2002. Growth cones contain myosin II bipolar filament arrays. Cell Motil Cytoskeleton 52: 91–96. [DOI] [PubMed] [Google Scholar]
- Brown JW, McKnight CJ. 2010. Molecular model of the microvillar cytoskeleton and organization of the brush border. PLoS ONE 5: e9406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley IK, Porter KR. 1967. Cytoplasmic fibrils in living cultured cells. A light and electron microscope study. Protoplasma 64: 349–380. [DOI] [PubMed] [Google Scholar]
- Burianek LE, Soderling SH. 2013. Under lock and key: Spatiotemporal regulation of WASP family proteins coordinates separate dynamic cellular processes. Semin Cell Dev Biol 24: 258–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge K, Wittchen ES. 2013. The tension mounts: Stress fibers as force-generating mechanotransducers. J Cell Biol 200: 9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C, Beltzner CC, Pollard TD. 2009. Cofilin dissociates Arp2/3 complex and branches from actin filaments. Curr Biol 19: 537–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charras G, Paluch E. 2008. Blebs lead the way: How to migrate without lamellipodia. Nat Rev Mol Cell Biol 9: 730–736. [DOI] [PubMed] [Google Scholar]
- Chhabra ES, Higgs HN. 2007. The many faces of actin: Matching assembly factors with cellular structures. Nat Cell Biol 9: 1110–1121. [DOI] [PubMed] [Google Scholar]
- Choi CK, Vicente-Manzanares M, Zareno J, Whitmore LA, Mogilner A, Horwitz AR. 2008. Actin and α-actinin orchestrate the assembly and maturation of nascent adhesions in a myosin II motor-independent manner. Nat Cell Biol 10: 1039–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark K, Langeslag M, Figdor CG, van Leeuwen FN. 2007. Myosin II and mechanotransduction: A balancing act. Trends Cell Biol 17: 178–186. [DOI] [PubMed] [Google Scholar]
- Collins A, Warrington A, Taylor KA, Svitkina T. 2011. Structural organization of the actin cytoskeleton at sites of clathrin-mediated endocytosis. Curr Biol 21: 1167–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig R, Smith R, Kendrick-Jones J. 1983. Light-chain phosphorylation controls the conformation of vertebrate non-muscle and smooth muscle myosin molecules. Nature 302: 436–439. [DOI] [PubMed] [Google Scholar]
- De La Cruz EM, Ostap EM. 2004. Relating biochemistry and function in the myosin superfamily. Curr Opin Cell Biol 16: 61–67. [DOI] [PubMed] [Google Scholar]
- Dickinson RB, Purich DL. 2006. Diffusion rate limitations in actin-based propulsion of hard and deformable particles. Biophys J 91: 1548–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diz-Munoz A, Krieg M, Bergert M, Ibarlucea-Benitez I, Muller DJ, Paluch E, Heisenberg CP. 2010. Control of directed cell migration in vivo by membrane-to-cortex attachment. PLoS Biol 8: e1000544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez R. 2010. Structural insights into de novo actin polymerization. Curr Opin Struct Biol 20: 217–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahim S, Fujita T, Millis BA, Kozin E, Ma X, Kawamoto S, Baird MA, Davidson M, Yonemura S, Hisa Y, et al. 2013. NMII forms a contractile transcellular sarcomeric network to regulate apical cell junctions and tissue geometry. Curr Biol 23: 731–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egile C, Loisel TP, Laurent V, Li R, Pantaloni D, Sansonetti PJ, Carlier MF. 1999. Activation of the CDC42 effector N-WASP by the Shigella flexneri IcsA protein promotes actin nucleation by Arp2/3 complex and bacterial actin-based motility. J Cell Biol 146: 1319–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler AJ, Griffin MA, Sen S, Bonnemann CG, Sweeney HL, Discher DE. 2004. Myotubes differentiate optimally on substrates with tissue-like stiffness: Pathological implications for soft or stiff microenvironments. J Cell Biol 166: 877–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara K, Pollard TD. 1976. Fluorescent antibody localization of myosin in the cytoplasm, cleavage furrow, and mitotic spindle of human cells. J Cell Biol 71: 848–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi M, Smith BA, Bovellan M, Paavilainen V, Daugherty-Clarke K, Gelles J, Lappalainen P, Goode BL. 2010. GMF is a cofilin homolog that binds Arp2/3 complex to stimulate filament debranching and inhibit actin nucleation. Curr Biol 20: 861–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardel ML, Sabass B, Ji L, Danuser G, Schwarz US, Waterman CM. 2008. Traction stress in focal adhesions correlates biphasically with actin retrograde flow speed. J Cell Biol 183: 999–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraldo S, Simon A, Elkhatib N, Louvard D, Fetler L, Vignjevic DM. 2012. Do cancer cells have distinct adhesions in 3D collagen matrices and in vivo? Eur J Cell Biol 91: 930–937. [DOI] [PubMed] [Google Scholar]
- Ghosh M, Song X, Mouneimne G, Sidani M, Lawrence DS, Condeelis JS. 2004. Cofilin promotes actin polymerization and defines the direction of cell motility. Science 304: 743–746. [DOI] [PubMed] [Google Scholar]
- *.Glotzer M. 2016. Cytokinesis in metazoa and fungi. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a022343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves E, Dart AE, Covarelli V, Caron E. 2008. Molecular mechanisms of phagocytic uptake in mammalian cells. Cell Mol Life Sci 65: 1957–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haglund CM, Welch MD. 2011. Host-pathogen interactions: Pathogens and polymers: Microbe-host interactions illuminate the cytoskeleton. J Cell Biol 195: 7–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heissler SM, Manstein DJ. 2013. Nonmuscle myosin-2: Mix and match. Cell Mol Life Sci 70: 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoelzle MK, Svitkina T. 2012. The cytoskeletal mechanisms of cell–cell junction formation in endothelial cells. Mol Biol Cell 23: 310–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoglund AS, Karlsson R, Arro E, Fredriksson BA, Lindberg U. 1980. Visualization of the peripheral weave of microfilaments in glia cells. J Muscle Res Cell Motil 1: 127–146. [DOI] [PubMed] [Google Scholar]
- Homem CC, Peifer M. 2009. Exploring the roles of diaphanous and enabled activity in shaping the balance between filopodia and lamellipodia. Mol Biol Cell 20: 5138–5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu P, Luo BH. 2013. Integrin bi-directional signaling across the plasma membrane. J Cell Physiol 228: 306–312. [DOI] [PubMed] [Google Scholar]
- Huveneers S, de Rooij J. 2013. Mechanosensitive systems at the cadherin-F-actin interface. J Cell Sci 126: 403–413. [DOI] [PubMed] [Google Scholar]
- Huxley HE. 1957. The double array of filaments in cross-striated muscle. J Biophys Biochem Cytol 3: 631–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huxley HE. 1963. Electron microscope studies on the structure of natural and synthetic protein filaments from striated muscle. J Mol Biol 7: 281–308. [DOI] [PubMed] [Google Scholar]
- Huxley H, Hanson J. 1954. Changes in the cross-striations of muscle during contraction and stretch and their structural interpretation. Nature 173: 973–976. [DOI] [PubMed] [Google Scholar]
- Huxley AF, Niedergerke R. 1954. Structural changes in muscle during contraction: Interference microscopy of living muscle fibres. Nature 173: 971–973. [DOI] [PubMed] [Google Scholar]
- Kasza KE, Zallen JA. 2011. Dynamics and regulation of contractile actin-myosin networks in morphogenesis. Curr Opin Cell Biol 23: 30–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaverina I, Krylyshkina O, Small JV. 2002. Regulation of substrate adhesion dynamics during cell motility. Int J Biochem Cell Biol 34: 746–761. [DOI] [PubMed] [Google Scholar]
- Kerber ML, Cheney RE. 2011. Myosin-X: A MyTH-FERM myosin at the tips of filopodia. J Cell Sci 124: 3733–3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr MC, Teasdale RD. 2009. Defining macropinocytosis. Traffic 10: 364–371. [DOI] [PubMed] [Google Scholar]
- Korobova F, Svitkina T. 2010. Molecular architecture of synaptic actin cytoskeleton in hippocampal neurons reveals a mechanism of dendritic spine morphogenesis. Mol Biol Cell 21: 165–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs M, Thirumurugan K, Knight PJ, Sellers JR. 2007. Load-dependent mechanism of nonmuscle myosin 2. Proc Natl Acad Sci 104: 9994–9999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar DR, Pollard TD. 2004. Insertional assembly of actin filament barbed ends in association with formins produces piconewton forces. Proc Natl Acad Sci 101: 14725–14730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krendel MF, Bonder EM. 1999. Analysis of actin filament bundle dynamics during contact formation in live epithelial cells. Cell Motil Cytoskeleton 43: 296–309. [DOI] [PubMed] [Google Scholar]
- Kubow KE, Conrad SK, Horwitz AR. 2013. Matrix microarchitecture and myosin II determine adhesion in 3D matrices. Curr Biol 23: 1607–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langanger G, Moeremans M, Daneels G, Sobieszek A, De Brabander M, De Mey J. 1986. The molecular organization of myosin in stress fibers of cultured cells. J Cell Biol 102: 200–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarides E. 1976. Actin, α-actinin, and tropomyosin interaction in the structural organization of actin filaments in nonmuscle cells. J Cell Biol 68: 202–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarides E, Weber K. 1974. Actin antibody: The specific visualization of actin filaments in non-muscle cells. Proc Natl Acad Sci 71: 2268–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebrand C, Dent EW, Strasser GA, Lanier LM, Krause M, Svitkina TM, Borisy GG, Gertler FB. 2004. Critical role of Ena/VASP proteins for filopodia formation in neurons and in function downstream of netrin-1. Neuron 42: 37–49. [DOI] [PubMed] [Google Scholar]
- Lin WH, Nebhan CA, Anderson BR, Webb DJ. 2010. Vasodilator-stimulated phosphoprotein (VASP) induces actin assembly in dendritic spines to promote their development and potentiate synaptic strength. J Biol Chem 285: 36010–36020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlefield RS, Fowler VM. 2008. Thin filament length regulation in striated muscle sarcomeres: Pointed-end dynamics go beyond a nebulin ruler. Semin Cell Dev Biol 19: 511–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YJ, Le Berre M, Lautenschlaeger F, Maiuri P, Callan-Jones A, Heuze M, Takaki T, Voituriez R, Piel M. 2015. Confinement and low adhesion induce fast amoeboid migration of slow mesenchymal cells. Cell 160: 659–672. [DOI] [PubMed] [Google Scholar]
- Loisel TP, Boujemaa R, Pantaloni D, Carlier MF. 1999. Reconstitution of actin-based motility of Listeria and Shigella using pure proteins. Nature 401: 613–616. [DOI] [PubMed] [Google Scholar]
- Loomis PA, Zheng L, Sekerkova G, Changyaleket B, Mugnaini E, Bartles JR. 2003. Espin cross-links cause the elongation of microvillus-type parallel actin bundles in vivo. J Cell Biol 163: 1045–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky LM, Atkinson SJ, Ampe C, Vandekerckhove J, Pollard TD. 1994. Purification of a cortical complex containing two unconventional actins from Acanthamoeba by affinity chromatography on profilin-agarose. J Cell Biol 127: 107–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky LM, Mullins RD, Higgs HN, Kaiser DA, Blanchoin L, May RC, Hall ME, Pollard TD. 1999. Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc Natl Acad Sci 96: 3739–3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallavarapu A, Mitchison T. 1999. Regulated actin cytoskeleton assembly at filopodium tips controls their extension and retraction. J Cell Biol 146: 1097–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manor U, Kachar B. 2008. Dynamic length regulation of sensory stereocilia. Semin Cell Dev Biol 19: 502–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AC. 2010. Pulsation and stabilization: Contractile forces that underlie morphogenesis. Dev Biol 341: 114–125. [DOI] [PubMed] [Google Scholar]
- Mattila PK, Lappalainen P. 2008. Filopodia: Molecular architecture and cellular functions. Nat Rev Mol Cell Biol 9: 446–454. [DOI] [PubMed] [Google Scholar]
- McMahon HT, Boucrot E. 2011. Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat Rev Mol Cell Biol 12: 517–533. [DOI] [PubMed] [Google Scholar]
- McNeill H, Ryan TA, Smith SJ, Nelson WJ. 1993. Spatial and temporal dissection of immediate and early events following cadherin-mediated epithelial cell adhesion. J Cell Biol 120: 1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros NA, Burnette DT, Forscher P. 2006. Myosin II functions in actin-bundle turnover in neuronal growth cones. Nat Cell Biol 8: 215–226. [DOI] [PubMed] [Google Scholar]
- Michael M, Yap AS. 2013. The regulation and functional impact of actin assembly at cadherin cell–cell adhesions. Semin Cell Dev Biol 24: 298–307. [DOI] [PubMed] [Google Scholar]
- Mogilner A. 2009. Mathematics of cell motility: Have we got its number? J Math Biol 58: 105–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogilner A, Oster G. 1996. Cell motility driven by actin polymerization. Biophys J 71: 3030–3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogilner A, Oster G. 2003. Force generation by actin polymerization II: The elastic ratchet and tethered filaments. Biophys J 84: 1591–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogilner A, Rubinstein B. 2005. The physics of filopodial protrusion. Biophys J 89: 782–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooren OL, Galletta BJ, Cooper JA. 2012. Roles for actin assembly in endocytosis. Annu Rev Biochem 81: 661–686. [DOI] [PubMed] [Google Scholar]
- Mooseker MS, Tilney LG. 1975. Organization of an actin filament-membrane complex. Filament polarity and membrane attachment in the microvilli of intestinal epithelial cells. J Cell Biol 67: 725–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins RD, Heuser JA, Pollard TD. 1998. The interaction of Arp2/3 complex with actin: Nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc Natl Acad Sci 95: 6181–6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambiar R, McConnell RE, Tyska MJ. 2010. Myosin motor function: The ins and outs of actin-based membrane protrusions. Cell Mol Life Sci 67: 1239–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumanen P, Lappalainen P, Hotulainen P. 2008. Mechanisms of actin stress fibre assembly. J Microsc 231: 446–454. [DOI] [PubMed] [Google Scholar]
- Nevzorov IA, Levitsky DI. 2011. Tropomyosin: Double helix from the protein world. Biochemistry 76: 1507–1527. [DOI] [PubMed] [Google Scholar]
- Niggli V, Rossy J. 2008. Ezrin/radixin/moesin: Versatile controllers of signaling molecules and of the cortical cytoskeleton. Int J Biochem Cell Biol 40: 344–349. [DOI] [PubMed] [Google Scholar]
- Okada K, Obinata T, Abe H. 1999. XAIP1: A Xenopus homologue of yeast actin interacting protein 1 (AIP1), which induces disassembly of actin filaments cooperatively with ADF/cofilin family proteins. J Cell Sci 112: 1553–1565. [DOI] [PubMed] [Google Scholar]
- Pelham RJ Jr, Wang Y. 1997. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc Natl Acad Sci 94: 13661–13665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie RJ, Yamada KM. 2012. At the leading edge of three-dimensional cell migration. J Cell Sci 125: 5917–5926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD. 1975. Electron microscopy of synthetic myosin filaments. Evidence for cross-bridge. Flexibility and copolymer formation. J Cell Biol 67: 93–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Pollard TD. 2016. Actin and actin-binding proteins. Cold Spring Harb Perspect Biol 8: a018226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Borisy GG. 2003. Cellular motility driven by assembly and disassembly of actin filaments. Cell 112: 453–465. [DOI] [PubMed] [Google Scholar]
- Racz B, Weinberg RJ. 2013. Microdomains in forebrain spines: An ultrastructural perspective. Mol Neurobiol 47: 77–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhard M, Jouvenal K, Tripier D, Walter U. 1995. Identification, purification, and characterization of a zyxin-related protein that binds the focal adhesion and microfilament protein VASP (vasodilator-stimulated phosphoprotein). Proc Natl Acad Sci 92: 7956–7960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero S, Grompone G, Carayol N, Mounier J, Guadagnini S, Prevost MC, Sansonetti PJ, Tran Van Nhieu G. 2011. ATP-mediated Erk1/2 activation stimulates bacterial capture by filopodia, which precedes Shigella invasion of epithelial cells. Cell Host Microbe 9: 508–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottner K, Hall A, Small JV. 1999. Interplay between Rac and Rho in the control of substrate contact dynamics. Curr Biol 9: 640–648. [DOI] [PubMed] [Google Scholar]
- Ruprecht V, Wieser S, Callan-Jones A, Smutny M, Morita H, Sako K, Barone V, Ritsch-Marte M, Sixt M, Voituriez R, et al. 2015. Cortical contractility triggers a stochastic switch to fast amoeboid cell motility. Cell 160: 673–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salbreux G, Charras G, Paluch E. 2012. Actin cortex mechanics and cellular morphogenesis. Trends Cell Biol 22: 536–545. [DOI] [PubMed] [Google Scholar]
- Sandquist JC, Means AR. 2008. The C-terminal tail region of nonmuscle myosin II directs isoform-specific distribution in migrating cells. Mol Biol Cell 19: 5156–5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger JM, Sanger JW. 1980. Banding and polarity of actin filaments in interphase and cleaving cells. J Cell Biol 86: 568–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder TE. 1969. The role of ‘contractile ring’ filaments in dividing Arbacia egg. Biol Bull 137: 413–414. [Google Scholar]
- Sekerkova G, Zheng L, Loomis PA, Mugnaini E, Bartles JR. 2006. Espins and the actin cytoskeleton of hair cell stereocilia and sensory cell microvilli. Cell Mol Life Sci 63: 2329–2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sens KL, Zhang S, Jin P, Duan R, Zhang G, Luo F, Parachini L, Chen EH. 2010. An invasive podosome-like structure promotes fusion pore formation during myoblast fusion. J Cell Biol 191: 1013–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shutova M, Yang C, Vasiliev JM, Svitkina T. 2012. Functions of nonmuscle myosin II in assembly of the cellular contractile system. PLoS ONE 7: e40814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjoblom B, Salmazo A, Djinovic-Carugo K. 2008. α-actinin structure and regulation. Cell Mol Life Sci 65: 2688–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small JV, Isenberg G, Celis JE. 1978. Polarity of actin at the leading edge of cultured cells. Nature 272: 638–639. [DOI] [PubMed] [Google Scholar]
- Smith MA, Blankman E, Gardel ML, Luettjohann L, Waterman CM, Beckerle MC. 2010. A zyxin-mediated mechanism for actin stress fiber maintenance and repair. Dev Cell 19: 365–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachowiak MR, McCall PM, Thoresen T, Balcioglu HE, Kasiewicz L, Gardel ML, O’Shaughnessy B. 2012. Self-organization of myosin II in reconstituted actomyosin bundles. Biophys J 103: 1265–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehbens S, Wittmann T. 2012. Targeting and transport: How microtubules control focal adhesion dynamics. J Cell Biol 198: 481–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetsugu S, Gautreau A. 2012. Synergistic BAR-NPF interactions in actin-driven membrane remodeling. Trends Cell Biol 22: 141–150. [DOI] [PubMed] [Google Scholar]
- Svitkina TM, Borisy GG. 1999. Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J Cell Biol 145: 1009–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkina TM, Surguchova IG, Verkhovsky AB, Gelfand VI, Moeremans M, De Mey J. 1989. Direct visualization of bipolar myosin filaments in stress fibers of cultured fibroblasts. Cell Motil Cytoskeleton 12: 150–156. [DOI] [PubMed] [Google Scholar]
- Svitkina TM, Verkhovsky AB, Borisy GG. 1995. Improved procedures for electron microscopic visualization of the cytoskeleton of cultured cells. J Struct Biol 115: 290–303. [DOI] [PubMed] [Google Scholar]
- Svitkina TM, Verkhovsky AB, McQuade KM, Borisy GG. 1997. Analysis of the actin-myosin II system in fish epidermal keratocytes: Mechanism of cell body translocation. J Cell Biol 139: 397–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkina TM, Bulanova EA, Chaga OY, Vignjevic DM, Kojima S, Vasiliev JM, Borisy GG. 2003. Mechanism of filopodia initiation by reorganization of a dendritic network. J Cell Biol 160: 409–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Sweeney HL, Hammers DW. 2016. Muscle contraction. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a023200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Sweeney HL, Holzbaur ELF. 2016. Motor proteins. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a021931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syamaladevi DP, Spudich JA, Sowdhamini R. 2012. Structural and functional insights on the Myosin superfamily. Bioinform Biol Insights 6: 11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szent-Gyorgyi A. 1950. Actomyosin and muscular contraction. Biochim Biophys Acta 4: 38–41. [DOI] [PubMed] [Google Scholar]
- Szent-Gyorgyi AG. 2004. The early history of the biochemistry of muscle contraction. J Gen Physiol 123: 631–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theriot JA, Mitchison TJ. 1991. Actin microfilament dynamics in locomoting cells. Nature 352: 126–131. [DOI] [PubMed] [Google Scholar]
- Tilney LG. 1975. The role of actin in nonmuscle cell motility. Soc Gen Physiol Ser 30: 339–388. [PubMed] [Google Scholar]
- Tilney LG, Portnoy DA. 1989. Actin filaments and the growth, movement, and spread of the intracellular bacterial parasite, Listeria monocytogenes. J Cell Biol 109: 1597–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilney LG, Hatano S, Ishikawa H, Mooseker MS. 1973. The polymerization of actin: Its role in the generation of the acrosomal process of certain echinoderm sperm. J Cell Biol 59: 109–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilney LG, Derosier DJ, Mulroy MJ. 1980. The organization of actin filaments in the stereocilia of cochlear hair cells. J Cell Biol 86: 244–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujioka M, Yumura S, Inouye K, Patel H, Ueda M, Yonemura S. 2012. Talin couples the actomyosin cortex to the plasma membrane during rear retraction and cytokinesis. Proc Natl Acad Sci 109: 12992–12997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyeda TQ, Iwadate Y, Umeki N, Nagasaki A, Yumura S. 2011. Stretching actin filaments within cells enhances their affinity for the myosin II motor domain. PloS ONE 6: e26200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiliev JM, Gelfand IM, Domnina LV, Ivanova OY, Komm SG, Olshevskaja LV. 1970. Effect of colcemid on the locomotory behaviour of fibroblasts. J Embryol Exp Morphol 24: 625–640. [PubMed] [Google Scholar]
- Verkhovsky AB, Svitkina TM, Borisy GG. 1995. Myosin II filament assemblies in the active lamella of fibroblasts: Their morphogenesis and role in the formation of actin filament bundles. J Cell Biol 131: 989–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkhovsky AB, Svitkina TM, Borisy GG. 1999. Network contraction model for cell translocation and retrograde flow. Biochem Soc Symp 65: 207–222. [PubMed] [Google Scholar]
- Vignjevic D, Kojima S, Aratyn Y, Danciu O, Svitkina T, Borisy GG. 2006. Role of fascin in filopodial protrusion. J Cell Biol 174: 863–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YL. 1985. Exchange of actin subunits at the leading edge of living fibroblasts: Possible role of treadmilling. J Cell Biol 101: 597–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A, Ma X, Conti MA, Adelstein RS. 2011. Distinct and redundant roles of the non-muscle myosin II isoforms and functional domains. Biochem Soc Trans 39: 1131–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe N, Kato T, Fujita A, Ishizaki T, Narumiya S. 1999. Cooperation between mDia1 and ROCK in Rho-induced actin reorganization. Nat Cell Biol 1: 136–143. [DOI] [PubMed] [Google Scholar]
- Weaver AM, Karginov AV, Kinley AW, Weed SA, Li Y, Parsons JT, Cooper JA. 2001. Cortactin promotes and stabilizes Arp2/3-induced actin filament network formation. Curr Biol 11: 370–374. [DOI] [PubMed] [Google Scholar]
- Weber A, Franzini-Armstrong C. 2002. Hugh E. Huxley: Birth of the filament sliding model of muscle contraction. Trends Cell Biol 12: 243–245. [DOI] [PubMed] [Google Scholar]
- Weber K, Groeschel-Stewart U. 1974. Antibody to myosin: The specific visualization of myosin-containing filaments in nonmuscle cells. Proc Natl Acad Sci 71: 4561–4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehrle-Haller B. 2012. Structure and function of focal adhesions. Curr Opin Cell Biol 24: 116–124. [DOI] [PubMed] [Google Scholar]
- Welch MD, DePace AH, Verma S, Iwamatsu A, Mitchison TJ. 1997. The human Arp2/3 complex is composed of evolutionarily conserved subunits and is localized to cellular regions of dynamic actin filament assembly. J Cell Biol 138: 375–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch MD, Rosenblatt J, Skoble J, Portnoy DA, Mitchison TJ. 1998. Interaction of human Arp2/3 complex and the Listeria monocytogenes ActA protein in actin filament nucleation. Science 281: 105–108. [DOI] [PubMed] [Google Scholar]
- Wessells NK, Spooner BS, Ash JF, Bradley MO, Luduena MA, Taylor EL, Wrenn JT, Yamada K. 1971. Microfilaments in cellular and developmental processes. Science 171: 135–143. [DOI] [PubMed] [Google Scholar]
- Wolfenson H, Bershadsky A, Henis YI, Geiger B. 2011. Actomyosin-generated tension controls the molecular kinetics of focal adhesions. J Cell Sci 124: 1425–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong AJ, Pollard TD, Herman IM. 1983. Actin filament stress fibers in vascular endothelial cells in vivo. Science 219: 867–869. [DOI] [PubMed] [Google Scholar]
- Woodrum DT, Rich SA, Pollard TD. 1975. Evidence for biased bidirectional polymerization of actin filaments using heavy meromyosin prepared by an improved method. J Cell Biol 67: 231–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Czech L, Gerboth S, Kojima S, Scita G, Svitkina T. 2007. Novel roles of formin mDia2 in lamellipodia and filopodia formation in motile cells. PLoS Biol 5: e317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonemura S, Itoh M, Nagafuchi A, Tsukita S. 1995. Cell-to-cell adherens junction formation and actin filament organization: Similarities and differences between non-polarized fibroblasts and polarized epithelial cells. J Cell Sci 108: 127–142. [DOI] [PubMed] [Google Scholar]
- Zhang DS, Piazza V, Perrin BJ, Rzadzinska AK, Poczatek JC, Wang M, Prosser HM, Ervasti JM, Corey DP, Lechene CP. 2012. Multi-isotope imaging mass spectrometry reveals slow protein turnover in hair-cell stereocilia. Nature 481: 520–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigmond SH. 2004. Beginning and ending an actin filament: Control at the barbed end. Curr Top Dev Biol 63: 145–188. [DOI] [PubMed] [Google Scholar]
- Zigmond SH, Otto JJ, Bryan J. 1979. Organization of myosin in a submembranous sheath in well-spread human fibroblasts. Exp Cell Res 119: 205–219. [DOI] [PubMed] [Google Scholar]