Abstract
Many cytokines and all interferons activate members of a small family of kinases (the Janus kinases [JAKs]) and a slightly larger family of transcription factors (the signal transducers and activators of transcription [STATs]), which are essential components of pathways that induce the expression of specific sets of genes in susceptible cells. JAK-STAT pathways are required for many innate and acquired immune responses, and the activities of these pathways must be finely regulated to avoid major immune dysfunctions. Regulation is achieved through mechanisms that include the activation or induction of potent negative regulatory proteins, posttranslational modification of the STATs, and other modulatory effects that are cell-type specific. Mutations of JAKs and STATs can result in gains or losses of function and can predispose affected individuals to autoimmune disease, susceptibility to a variety of infections, or cancer. Here we review recent developments in the biochemistry, genetics, and biology of JAKs and STATs.
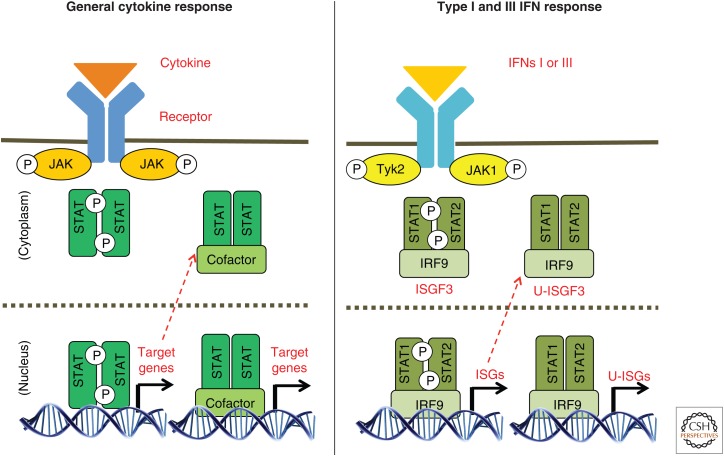
Because the basic biochemistry of Janus kinase–signal transducers and activators of transcription (JAK-STAT) signaling pathways has been frequently and extensively reviewed (see, for example, Stark and Darnell 2012; Cai et al. 2015; O’Shea et al. 2015; Villarino et al. 2015), we present here only a brief summary. After a cytokine or interferon binds to its specific receptor, the receptor forms homodimers, heterodimers, or trimers, depending on the cytokine, thus activating the tightly bound JAKs to cross-phosphorylate each other. The activated JAKs then phosphorylate specific tyrosine residues in the cytoplasmic domains of the receptors, providing binding sites for the STATs through their highly conserved SH2 domains. The receptor-bound STATs are phosphorylated, each on a highly conserved tyrosine residue, after which they leave the receptor as homo- or heterodimers whose association is strengthened by SH2-phosphotyrosine interactions. The phosphorylated STAT dimers are then transported to the nucleus, where they bind to and activate specific promoters. The basic outline of JAK-STAT signaling (Fig. 1) shows that interferon (IFN)-γ (type II IFN) and all of the cytokines primarily drive the formation of specific STAT homodimers, which then bind to DNA directly. However, in some cases, heterodimers involving STAT1 and STAT3 or STAT5A and STAT5B can also form. In contrast, IFN-β and the subtypes of IFN-α (collectively type I IFNs) and the subtypes of IFN-λ (collectively type III IFNs) drive the formation of STAT1-STAT2 heterodimers, which then associate with the DNA-binding interferon regulatory protein 9 (IRF9) to form interferon-stimulated gene factor 3 (ISGF3). The DNA sequences to which STAT homodimers bind (γ-activated sequences [GAS] elements) are related to, but distinct from, the sequences to which IRF9 binds (interferon-stimulated regulatory elements [ISREs]). The consensus GAS sequences are 5′TTCCNGGAA3′ for STAT1, 3, 4, and 5 and 5′TTCCNNGGAA for STAT6. Note that these motifs are palindromic, consistent with the need for each STAT subunit of the dimer to contact DNA. The consensus ISRE sequence is RRTTTCNNTTTCY (Decker and Kovarik 1999). Also shown in Figure 1 are STAT transcription factors that form and function without tyrosine phosphorylation (unphosphorylated STATs [U-STATs]). The genes encoding STAT3, STAT1, STAT2, and IRF9 are themselves transcriptional targets of the primary signals generated by phosphorylated STAT3 and ISGF3, respectively, leading to secondary increases in the concentrations of these U-STATs in response to the initial signal, driving STAT-STAT association and promoter binding even in the absence of tyrosine phosphorylation. This aspect is discussed further below.
Figure 1.
Outline of Janus kinase–signal transducers and activators of transcription (JAK-STAT) signaling shows that interferon (IFN)-γ (type II IFN) and all of the cytokines primarily drive the formation of specific STAT homodimers, which then bind to DNA directly. However, in some cases, heterodimers involving STAT1 and STAT3 or STAT5A and STAT5B can also form. In contrast, IFN-β and the subtypes of IFN-α (collectively type I IFNs), and the subtypes of IFN-λ (collectively type III IFNs) drive the formation of STAT1-STAT2 heterodimers, which then associate with the DNA-binding interferon regulatory protein 9 (IRF9) to form interferon-stimulated gene factor 3 (ISGF3). Also shown are STAT transcription factors that form and function without tyrosine phosphorylation (unphosphorylated STATs [U-STATs]).
As summarized in Table 1, many cytokines and all IFNs use JAK tyrosine kinases and STAT transcription factors to connect their cell-surface receptors to the activation of their specific gene targets. The assignments of specific JAKs and STATs to specific cytokines and IFNs in the table should be understood to represent the major pathways only. In reality, the situation is much more complex, and a single ligand–receptor pair may activate more than one STAT. The ratio of activated STATs can depend on their relative intracellular concentrations (specific examples for interleukin (IL)-6, IL-21, and IL-27 are cited below), on the specific cell type, on whether or not the cell has received prior signals (“priming”), and probably on other variables as well. As one example of cell-type-specific responses, van Boxel-Dezaire et al. (2010) found that different primary human leukocyte subsets respond quite differently to IFN-β. In B cells and CD4+ T cells, IFN-β activates STAT3 and STAT5 primarily, with biological effects opposite from those driven by the “canonical” activation of STAT1 and STAT2 in the other leukocyte subtypes that were studied.
Table 1.
Summary of cytokines and IFNs that use JAK tyrosine kinases and STAT transcription factors to connect their cell-surface receptors to the activation of specific gene targets
| STATs | JAKs | Major cytokines |
|---|---|---|
| STAT1 | JAK1, JAK2, TYK2 | Type I, II, and III IFNs |
| STAT2 | JAK1, TYK2 | Type I and III IFNs |
| STAT3 | JAK1, JAK2, JAK3, TYK2 | IL-6 family cytokines, IL-10, IL-27, IL-21 |
| STAT4 | JAK2, TYK2 | IL-12, IL-23 |
| STAT5A/B | JAK1, JAK2, JAK3 | IL-2, IL-7, IL-9, IL-15, EPO, TPO, GM-CSF, GH, PRL |
| STAT6 | JAK1, JAK2, JAK3, TYK2 | IL-4, IL-13 |
STATs, Signal transducers and activators of transcription; JAKs, Janus kinases; IFN, interferon; IL, interleukin; EPO, erythropoietin; TPO, thrombopoietin; GM-CSF, granulocyte macrophage colony-stimulating factor; GH, growth hormone; PRL, prolactin.
FUNCTIONALLY IMPORTANT CHEMICAL MODIFICATIONS OF THE STATs
The STATs are substrates for phosphorylation, methylation, and other posttranslational modifications that facilitate both positive and negative fine-tuning of the transcriptional responses.
Carboxy-Terminal Serine Phosphorylations
All the STATs except STAT2 share a functionally important serine phosphorylation site within a P(M)SP motif located near their carboxyl termini. Carboxy-terminal serine phosphorylation is stimulated by many different cytokines and growth factors, and is mediated by many different kinases, including extracellular signal-regulated kinase (ERK), p38, c-Jun amino-terminal kinase (JNK), mechanistic target of rapamycin (mTOR), nemo-like kinase (NLK), calcium/calmodulin-dependent protein kinase II (CaMKII), IκB kinase ɛ (IKKɛ), and protein kinase C δ (PKC-δ) (Schindler et al. 2007). Phosphorylation increases the transactivation potential of these proteins. The most intensively investigated phosphorylated serine residues are S727 in both STAT1 and STAT3. Phosphorylation of S727 of STAT1 and STAT3 is necessary for full activation of transcription in response to IFNs and IL-6 family cytokines, but the phosphorylation of S727 in either STAT1 or STAT3 is not associated with increased tyrosine phosphorylation (Wen et al. 1995). The serine to alanine mutation of S727 in STAT1 or STAT3 leads to reduction of the cytokine-induced transcription of specific genes, the extent of which is likely to vary in different cellular contexts. In addition to altering the transcriptional activation of STATs, serine phosphorylation of S727 in STAT1 and STAT3 has been correlated with enhanced DNA-binding ability (Eilers et al. 1995; Zhang et al. 1995; Ng and Cantrell 1997). Visconti et al. (2000) showed that mutating S721 of STAT4 decreased its transcriptional activity in IL-12-treated cells. Phosphorylation of both S725 and S779 of STAT5A and of S730 of STAT5B negatively regulate transactivation in response to stimulation of mammary glands with prolactin (Yamashita et al. 1998; Benitah et al. 2003). Wang et al. (2004) showed that IL-4 and IL-13 promote STAT6 phosphorylation on S756 in human T cells. However, the contribution of this phosphorylation to function is not yet clear.
Additional Serine and Threonine Phosphorylations
Recent advances in mass spectrometry have revolutionized the analysis of protein phosphorylation by allowing rapid identification of the sites modified with precision and sensitivity. Several additional serine and threonine modifications sites on STATs have been found.
STAT1
Phosphorylation of S708 by TRIM6-activated IKKɛ regulates STAT1 homodimerization but not ISGF3 formation in response to IFN (Tenoever et al. 2007; Rajsbaum et al. 2014). Ultimately, the phosphorylation of S708 facilitates the induction of a subset of ISGs whose protein products are essential for the antiviral response in vitro and in vivo. Phosphorylation of S744/S747 has also been reported, but the binding of ISGF3 to ISREs was unaffected by a carboxy-terminal deletion of STAT1 that removed both of these residues (Tenoever et al. 2007).
STAT2
The first serine phosphorylation of STAT2 (S287) was revealed by the work of Steen et al. (2013). Phosphorylation-defective mutants of S287 of STAT2 enhanced the ability of ISGF3 to bind to DNA, revealing that this phosphorylation is a negative regulatory event. We have shown that the phosphorylation of T387 regulates the ability of ISGF3 to bind to DNA (Wang et al. 2017). This phosphorylation negatively regulates the expression of most genes induced by type I IFN, inhibiting the ability of IFN to protect cells against virus infection and to inhibit cell growth. In most untreated cell types, the great majority of STAT2 is phosphorylated on T387 constitutively. T387 lies in a cyclin-dependent kinase (CDK) consensus sequence, and CDK inhibitors decrease T387 phosphorylation markedly. Using CDK inhibitors to reverse the constitutive inhibitory phosphorylation of T387 of STAT2 might enhance the efficacy of type I IFNs.
STAT3
Waitkus et al. (2014) have provided evidence that GSK3 α/β directly phosphorylates STAT3, simultaneously on T714 and S727, and that these modifications are required for STAT3-dependent gene expression in response to simultaneous activation of epidermal growth factor receptor (EGFR) and protease-activated receptor 1 (PAR-1) in endothelial cells. Levels of both T714 and S727 phosphorylation of STAT3 are significantly elevated in renal tumor tissues, suggesting that the GSK3-activated STAT3 signaling may be important in this disease.
STAT5A
S127/S128 phosphorylation of STAT5A is required for ERBB4-induced Y694 phosphorylation and has a substantial impact on ERBB4-dependent regulation of STAT5A activity (Clark et al. 2005). The expression of a STAT5 mutant in which the S725 and S779 phosphorylation sites were altered prohibited transformation and induced apoptosis in bone marrow cells (Pircher et al. 1999; Xue et al. 2002; Friedbichler et al. 2010; Berger et al. 2014). S779 is phosphorylated by p21-activated kinase (PAK) in human myeloid malignancies.
STAT5B
STAT5B constitutively phosphorylated on S193 has been found in hematopoietic cancers (Mitra et al. 2012). This phosphorylation is dependent on the mTOR signaling pathway and positively regulates STAT5B DNA binding and transcriptional activity.
STAT6
Phosphorylation of STAT6 S707 is triggered by the virus infection–responsive protein STING, which is located in the endoplasmic reticulum. Homodimers of STAT6 phosphorylated on Y641 and S407 then activate specific target genes in the nucleus that mediate immune cell homing (Chen et al. 2011). S707 is phosphorylated by JNK, which can be activated in response to cellular stress or IL-1β. Phosphorylation of S707 is a negative regulatory event that decreases the DNA-binding ability of STAT6 following its activation by IL-4 (Shirakawa et al. 2011).
Lysine and Arginine Modifications
The lysine residues of proteins can be modified by acetylation or by the addition of one, two, or three methyl groups, and the arginine residues can be modified by methylation. Furthermore, these reactions are reversible, providing rich opportunities to modify function. Many enzymes carry out the reversible acetylation and methylation of histones, providing the chemical basis of the modification of chromatin structure and function known as the histone code (Allis and Jenuwein 2016). In many cases, the specifically modified lysine or arginine residues provide docking sites for the binding of accessory proteins that modulate function. As summarized in Table 2, several different lysine and arginine residues of STAT1 and STAT3 are methylated or acetylated, but we are not aware of reports of these modifications for any other STAT. In every case, the enzymes responsible for STAT1 or STAT3 modification were previously known to modify histones. One of the earliest papers reports the dimethylation of R31 of STAT1, which facilitates IFN-dependent gene expression by inhibiting the association of STAT1 with the negative regulator PIAS1 (Mowen et al. 2001). It is very interesting that the effects of gain-of-function mutations of STAT1 that cause disseminated yeast infections in patients can be ameliorated by reducing the level of PIAS1 or by facilitating STAT1 arginine methylation (Sampaio et al. 2013).
Table 2.
Summary of lysine and arginine modifications of STAT1 and STAT3
| STATs | Modifications | Sites |
|---|---|---|
| STAT1 | Methylation | R31me1 (Zhu et al. 2002) |
| R31me2 (Mowen et al. 2001) | ||
| Acetylation | K410/413ac (Kramer et al. 2006; Antunes et al. 2011; Kotla and Rao, 2015), controversial | |
| Sumoylation | K703sm (Ungureanu et al. 2003; Ungureanu et al. 2005; Gronholm et al. 2012) | |
| STAT3 | Methylation | R31me1 (Iwasaki et al. 2010) |
| K49me2 (Dasgupta et al. 2015b) | ||
| K140 (Yang et al. 2010) | ||
| Acetylation | K49/87ac (Ray et al. 2005; Hou et al. 2008; Nie et al. 2009) | |
| K679ac (Nie et al. 2009) | ||
| K685ac (Yuan et al. 2005; Lee et al. 2009, 2016; Dasgupta et al. 2014; Kang et al. 2015) | ||
| K707ac (Nie et al. 2009) | ||
| Sumoylation | K451sm (Zhou et al. 2016b) |
Listed posttranslational modifications (PTMs) can be found at www.phosphosite.org.
STATs, Signal transducers and activators of transcription.
We have described the reversible dimethylation of K140 of STAT3, which regulates STAT3-dependent gene expression negatively (Yang et al. 2010). In this case, the reaction is catalyzed by the histone lysine methyltransferase SET9 and occurs only after STAT3 has been bound to a promoter. The docking site for SET9 is provided by the phosphorylated S727 residue of STAT3, because the S727A mutant of STAT3 fails to recruit SET9 to the promoter. We reviewed several additional examples of the lysine methylation of promoter-bound transcription factors (Stark et al. 2011). Our working hypothesis, which needs to be tested further, is that the promoter-bound factor provides a docking site for a histone-modifying enzyme that then catalyzes functionally important modifications, not only of the transcription factor but potentially also of local histones and the transcriptional machinery itself. Another important modification of STAT3 is the dimethylation of K49, carried out by the lysine methyltransferase EZH2 (Dasgupta et al. 2015b). Failure to carry out this reaction inhibits the ability of STAT3 to activate the expression of a substantial fraction of its target genes by an as-yet-unknown mechanism.
SUMOylation, Glycosylation, and Additional Tyrosine Phosphorylation
Glycosylation (Gewinner et al. 2004) and tyrosine phosphorylation of STATs at additional sites, including STAT2, human, Y631 (Scarzello et al. 2007), STAT5A, mouse, Y682/683 (Schaller-Schonitz et al. 2014), and STAT5B, rat, Y679 (Kabotyanski and Rosen 2003), have been reported, but the functional importance of these modifications is not yet well established. On the other hand, SUMOylation of STAT1 on K703 is a modification of great functional importance (references in Table 2). This modification, catalyzed by PIAS1 (Rogers et al. 2003; Ungureanu et al. 2003), leads to inhibition of STAT1 function (Rogers et al. 2003; Ungureanu et al. 2005), and modulation of the response to IFN-γ is facilitated by the SUMOylation of STAT1 (Begitt et al. 2011; Maarifi et al. 2015).
NEGATIVE REGULATION
Failure to regulate cytokine-stimulated responses leads to catastrophic hyperinflammatory responses, and therefore elaborate mechanisms exist to achieve the necessary negative regulation. A major mechanism involves the suppressor of cytokine signaling (SOCS) proteins (reviewed by Kazi et al. 2014). These potent negative regulators are typically induced in response to acute exposure to specific cytokines, and they typically function by inhibiting STAT activation at the receptors (see Babon et al. 2014 for an example of the SOCS3 and the IL-6 family of cytokines). Defective SOCS3 function has been reported to contribute to many diseases, including allergy, autoimmune diseases such as rheumatoid arthritis, vascular inflammatory diseases, insulin resistance, and cancer, as reviewed by Yin et al. (2015). Many additional mechanisms contribute to adequate regulation of the induction of and responses to IFNs, including the PIAS proteins, which inhibit the function of activated STATs in the nucleus and also catalyze the inhibitory SUMOylation of STAT1, and protein tyrosine phosphatases, which inactivate the STATs (Hertzog and Williams 2013; Porritt and Hertzog 2015). Although the STAT3-induced expression of SOCS3 is important for dampening acute responses to IL-6 and other gp130-linked cytokines, tumor cells use an IL-6-induced association between the IL-6 receptor:gp130 complex and the EGFR to nullify the inhibitory effect of SOCS3 and thus to sustain STAT3 activation constitutively (Wang et al. 2013). It seems likely that specific mechanisms to prevent negative regulation of STAT activation will be used whenever sustained STAT activation is necessary in normal physiology.
UNPHOSPHORYLATED STATs
Many studies have shown that U-STATs, which lack phosphorylation of their highly conserved tyrosine residues, are located in nuclei, bind to promoters, and activate gene expression (Chatterjee-Kishore et al. 2000; Yang et al. 2005, 2007; Cui et al. 2007; Cheon and Stark 2009; Cheon et al. 2013; Park et al. 2015). Whether the phosphorylation of other residues affects the function of U-STATs is not clear, but K685 of U-STAT3 is acetylated, and mutation of this residue results in loss of expression of many U-STAT3-induced genes (Dasgupta et al. 2014). U-STATs 1, 2, 3, 5, and 6 regulate gene expression through their interactions with partner cofactors. The expression of U-STATs 1, 2, and 3 is increased in response to cytokines that induce their tyrosine phosphorylation, but how the expression of the other U-STATs is regulated is not yet known. The roles of U-STATs in regulating gene expression were originally controversial, but it is now generally accepted that U-STATs are critical transcription factors that are involved in many biological events in both normal and pathological situations.
U-STATs as Positive Regulators of Gene Expression
The levels of the U-STAT1, U-STAT2, and U-STAT3 proteins are induced in response to signals that lead to the tyrosine phosphorylation of each STAT, because each phosphorylated STAT binds to the promoter of its own gene to induce expression (Yang et al. 2005; Cheon et al. 2013). The U-STAT proteins then accumulate, translocate into nuclei, bind to target gene promoters together with their cofactors, and induce the expression of these genes. U-STATs contribute to the steady-state constitutive expression of specific genes, while tyrosine-phosphorylated STATs induce rapid and transient responses in response to cytokine stimulation. Many target genes of U-STATs 1, 2, 3, and 6 encode proteins that promote cell survival and resistance to cell death, suggesting that the U-STAT system helps to sustain the survival of cells in stressful environments.
U-STAT1 induces its target genes as U-ISGF3, a tripartite complex with U-STAT2 and IRF9, and the expression of all three U-ISGF3 components is increased in response to either type I or type III IFN (Cheon et al. 2013; Sung et al. 2015). U-STAT1 does not activate gene expression as a homodimer, because high levels of U-STAT1 do not induce target gene expression if U-STAT2 and IRF9 are not present in sufficient quantities (Cheon and Stark 2009; Cheon et al. 2013). U-ISGF3 induces a subset of ISGs that are also induced in the initial response to IFNs, resulting in their prolonged expression. The phenotypes of cells that express high levels of only the U-ISGF3-induced genes are different from those of cells treated with high levels of IFNs, which express all of the ISGs in response to phosphorylated ISGF3. Cancer cells that express high levels of U-ISGF3 are more resistant to DNA damage, while IFNs inhibit cancer cell proliferation and increase their apoptosis (Borden et al. 2007). Similarly, hepatocytes expressing high levels of U-ISGF3 are more resistant to IFN-α therapy, although U-ISGF3 itself suppresses viral replication by inducing antiviral genes (Sung et al. 2015). Phosphorylated STAT3 mediates the induction of U-STAT3 in response to IL-6 and other cytokines that activate gp130-linked receptors (Yang et al. 2005). However, in strong contrast to the situation for U-STAT1, U-STAT3 induces the expression of a set of genes that is completely different from the set induced by phosphorylated STAT3 (Yang et al. 2005). Some of the genes specifically induced by U-STAT3 are important oncogenes, including SRC, MET, and MRAS. The mechanism by which U-STAT3 induces expression of target genes is only partially understood. However, for the expression of a subset of the induced genes, including RANTES, IL6, and IL8, U-STAT3 forms a complex with nuclear factor κB (NF-κB), which then binds to κB elements in the promoters of a small fraction of NF-κB target genes (Yang et al. 2007). Cui et al. (2007) show that U-STAT6 constitutively activates the expression of the COX2 gene. Similarly to the other U-STATs, U-STAT6 also forms a complex with a cofactor protein, p300, facilitating binding to the COX2 promoter.
U-STATs as Negative Regulators of Gene Expression
In contrast to their ability to regulate gene expression positively, some U-STATs repress gene expression instead. ChIP-seq data showing genome-wide distribution reveals that U-STAT5 and phosphorylated STAT5 bind to different cis-acting elements in the genome, and differently regulate gene expression (Park et al. 2015). In this study, phosphorylated STAT5 was shown to bind to GAS elements in canonical STAT5-induced promoters in response to thrombopoietin (TPO) in mouse hematopoietic stem cells. In the absence of TPO, however, a large portion of U-STAT5 occupies binding sites for early growth response (EGR), an activator that promotes the expression of megakaryocytic genes, thus repressing EGR-induced gene expression. Only a small portion of U-STAT5 binds to promoters that are occupied by phosphorylated STAT5 in response to TPO, but the role of U-STAT5 is not known in that situation. Differently from U-STAT5, U-ISGF3 and phosphorylated ISGF3 bind to similar ISRE elements in the promoters of ISGs that are induced by both transcription factors (Cheon et al. 2013). Using a ChIP-on-chip analysis, Testoni et al. (2011) showed that U-STAT2, possibly as a component of U-ISGF3, binds to more than half of the ISG promoters investigated before IFN-α treatment, but the effect on the expression of ISGs is not clear. Although U-ISGF3 itself increases the expression of some ISGs in the absence of IFN treatment, we do not yet know whether or not U-ISGF3 inhibits the expression of other ISGs in response to IFN.
Nongenomic Activity of U-STAT3 in Mitochondria
U-STAT3 plays important roles not only in the nucleus but also in mitochondria (Garama et al. 2016). The mitochondrial activity of STAT3 is not dependent on Y705 phosphorylation, but phosphorylation of S727 is necessary (Gough et al. 2009, 2014). Mitochondrial U-STAT3 regulates the activity of the electron transport chain, which is required for adenosine triphosphate (ATP) production, and the opening of the mitochondrial permeability transition pore (Gough et al. 2009; Wegrzyn et al. 2009). In RAS-transformed cells, the MEK–ERK pathway drives the phosphorylation of STAT3 on S727, and RAS-transformed cells carrying a mutation of S727 are partially resistant to inhibitors of the ERK pathway (Gough et al. 2013).
STRUCTURES AND INTRACELLULAR LOCATIONS OF STATS
Phosphorylated STATs are translocated into the nucleus after cytokine stimulation, but U-STATs shuttle constitutively between cytoplasm and nucleus (Meyer and Vinkemeier 2004; Pranada et al. 2004; Liu et al. 2005; Iyer and Reich 2007; Vogt et al. 2011). U-STATs 1, 3, and 5 form antiparallel dimers, whereas phosphorylated STAT dimers are in a parallel conformation that is stabilized by phosphotyrosine–SH2 domain interactions, allowing both DNA-binding domains to contact the GAS sequences simultaneously, resulting in strong binding (Mao et al. 2005; Neculai et al. 2005; Wenta et al. 2008; Timofeeva et al. 2012; Nkansah et al. 2013). In addition, phosphorylated homodimers of STAT1 bind to each other to form tetramers, facilitating gene expression in response to type II (but not type I) IFNs. Consistently, the F77A mutation of murine STAT1, which disrupts dimer–dimer interactions, blunts signaling in response to type II IFNs in mice (Begitt et al. 2014). Interestingly, Droescher et al. (2011) found that all activated STATs can form paracrystalline arrays in the nuclei of cytokine-stimulated cells, with important biological consequences. However, for STAT1 only, this phenomenon can be prevented by modification of the protein by SUMO (Begitt et al. 2011; Droescher et al. 2011). STAT2 forms a stable antiparallel heterodimer with either the unphosphorylated or tyrosine-phosphorylated forms of STAT1; these heterodimers are not transported to the nucleus and have no transcriptional activity, so STAT2 is a pervasive negative regulator of STAT1-dependent functions (Ho et al. 2016). However, when IRF9 is present in sufficient quantity, antiparallel STAT1-STAT2 heterodimers will be converted to U-ISGF3 (Cheon et al. 2013) or to hemiphosphorylated ISGF3 (Morrow et al. 2011), in which the relative orientation of the two STATs becomes parallel. They are then transported to the nucleus and bind to DNA to activate transcription. Thus, the availability of free STAT1 to signal in response, for example, to IFN-γ or IL-27 (Ho et al. 2016), will depend on the relative concentrations of both STAT2 and IRF9.
CHROMATIN REMODELING BY STATs
There are a few intriguing reports that STATs function to affect chromatin structure independently of their ability to activate transcription. U-STAT92E (the only Drosophila STAT) stabilizes heterochromatin in association with heterochromatin protein 1 (HP1) (Shi et al. 2008). Phosphorylation of U-STAT92E causes heterochromatin instability and promotes gene expression. Human U-STAT5A binds to HP1α, repressing the expression of multiple oncogenes (Hu et al. 2013). The overexpression of STAT5A Y704F, which cannot be phosphorylated at this site, has effects on global gene expression similar to the effects of over expressing HP1α. The phosphorylation of STAT1 remodels chromatin to generate a local environment appropriate for the activation of target gene expression. When human histocompatibility (MHC) genes are activated by IFN-γ, phosphorylated STAT1, which binds to specific elements of the target gene promoter and recruits the chromatin remodeling enzyme BRG1, causes the release of entire MHC locus loops from compacted chromatin (Christova et al. 2007).
JAK AND STAT MUTATIONS
Naturally Occurring STAT Mutations
Because the STATs have essential roles in infectious disease, immunity, and cancer, many naturally occurring mutations have been discovered that affect human health, providing a rich source of structural and functional information. All seven STATs form homo- and heterodimers following phosphorylation of their tyrosine residues, and thus there is great potential for dominant effects in which only one partner is mutated. These mutations can affect many different protein–protein interactions, leading to dominant phenotypes that reflect either loss of function (LOF) or gain of function (GOF), as summarized recently for STAT3 (Chandrasekaran et al. 2016). However, STAT mutations will be manifest in human disease only if they lead to a discernable biological phenotype and, although many dominant mutations have been described for STAT1 and STAT3, far fewer have emerged for the other STATs. Because the structures of the STAT dimers are similar, it seems likely that germline dominant mutations will occur with similar frequencies for all the STATs, but with far less frequent phenotypic consequences for STATs 2, 4, 5, and 6. In their summary of all human primary immunodeficiencies, Boisson et al. (2015) point out that the known autosomal-recessive deficiencies are all caused by alleles with LOF, and that 44 out of 61 autosomal-dominant defects are caused by LOF mutations. Negative regulation of cytokine-dependent signaling is vital to prevent overstimulation of immune responses, so that, within the 17 examples of GOF-dominant mutations, those affecting STAT1 lead to infection, autoimmunity, and malignancy, whereas GOF mutations of STAT3 affect autoimmunity, allergy, and autoinflammation. Y705, S727, and K49 mutations of STAT3 have not yet been reported, presumably because they would not be consistent with survival.
Gain-of-Function Somatic Mutations in Cancer
The constitutive activation of JAK-STAT signaling pathways in cancer cells, through somatic mutation and other mechanisms, drives proliferation and resistance to stresses, and helps to overcome barriers to perpetual cell growth. Because this topic has been well reviewed recently (O’Shea et al. 2015; Pilati and Zucman-Rossi 2015; Thomas et al. 2015), we present only a few illustrative examples here. As recent examples for the STATs, activating somatic mutations of STAT5B lead to leukemia (Rajala et al. 2013) and activating mutations of STAT6 lead to follicular lymphoma (Yildiz et al. 2015). For JAK2, the activating V617F mutation causes polycythemia vera and other myeloproliferative diseases (Spivak 2010).
The Gain-of-Function STAT1 Paradox in Infectious Disease
Resolution of infection depends heavily on IFN responses, and specific STAT1 and JAK mutations that lead to increased susceptibility to infection have been extensively reviewed (Casanova et al. 2012; Boisson et al. 2015). Because STAT1 is required for all known responses to all three IFN subtypes, dominant STAT1 mutations lead to increased susceptibility to a wide range of infectious agents. For example, some dominant LOF mutations of STAT1 underlie chronic infections, such as candidiasis (van de Veerdonk et al. 2011) and disseminated mycobacterial disease (Sampaio et al. 2012). It is surprising that germline GOF mutations that lead to constitutive STAT1 activity predispose affected individuals to diseases resulting from infection with a variety of mycobacteria and fungi (Sampaio et al. 2013; Uzel et al. 2013; Kumar et al. 2014). As summarized by Zerbe et al. (2016), “STAT1 LOF mutations are associated with viral, mycobacterial … and bacterial infections, while GOF mutations are associated with mucocutaneous and invasive fungal infections and viral infections.” STAT1 GOF mutations also cause failure to control persistent JC virus infections in the central nervous system, leading to progressive multifocal leukoencelphalopathy in humans (Zerbe et al. 2016). Furthermore, persistent activation of IFN signaling, which depends on STAT1 activation, facilitates persistent lymphocytic choriomeningitis virus (LCMV) infection in mice (Teijaro et al. 2013; Oldstone 2015). Why are we betrayed by STAT1-dependent systems whose primary roles should be to protect us from infection? As summarized by Michael Oldstone (2015) for the example of persistent LCMV infection in mice, production of type I IFN leads to the expression of IL-10 and PD-1/PD-L1, which in turn cause loss of antiviral T-cell function, so that inhibiting the IFN response helps to cure the persistent infection! The general problem is that the up-regulation of strong inhibitory responses to acute or persistent infections must be balanced by the need to limit these responses, to avoid killing uninfected cells, and to avoid autoimmunity, resulting in a complex system that is poised on a veritable knife-edge and thus susceptible to misregulation.
There are a few examples of human germline mutations in STATs other than STAT1 or STAT3 that affect infection or inflammation. Patients with an abnormally low level of STAT2 expression are susceptible to virus infections (Shahni et al. 2015) but, very surprisingly, the complete loss of STAT2 expression does not seem to eliminate completely the host defense against many viruses, but does sensitize affected individuals to measles (Hambleton et al. 2013). For STAT4, some polymorphisms influence the risk of developing juvenile arthritis (Fan et al. 2015).
STATs AND IFNs
Mechanisms of Misregulation of IFN Signaling
The complexity of how negative regulators fine-tune IFN responses and the consequences of failure to regulate these responses effectively have been well reviewed recently (Hertzog and Williams 2013; Porritt and Hertzog 2015). In hepatitis C virus (HCV) infections, the virus persists even though many IFN-induced proteins are expressed at a high level in the livers of chronically infected patients. However, the levels of tyrosine-phosphorylated STAT1 and STAT2 are low, and the patients respond poorly to exogenous IFN (Shin et al. 2016). The underlying mechanism is complex, revealing some important general principles (Sung et al. 2015). The levels of U-ISGF3 are high in these livers because of chronic exposure to IFN-λ and, as a result, the downstream gene ISG15 is constitutively activated. Increased concentrations of the ISG15 protein stabilize USP18, a negative regulator of the response to type I IFNs (Zhang et al. 2015). Negative regulators play an essential role in modulating IFN responses that, if not well controlled, are extremely deleterious. The expected phenotype of a cell with a low level of IFN-dependent signaling, as a result of either chronic exposure to a low level of IFN or to a GOF mutation of STAT1, is failure to respond effectively to the high level of IFN that would be produced in response to an infection. As examples, we note GOF STAT1 mutations that lead to increased constitutive STAT1 tyrosine phosphorylation and STAT1-dependent gene expression, but decreased ability of the affected cells to respond to restimulation by IFN (Sampaio et al. 2013; Uzel et al. 2013). We anticipate that increased expression of prominent negative regulators, such as the SOCS and PIAS proteins in response to GOF STAT mutations, will help to explain why cells bearing these mutations fail to respond well to a high level of the relevant cytokine, especially IFN. We also suspect that U-ISGF3, which activates some promoters (Cheon et al. 2013), also binds to other promoters without activating them, thus competing with phosphorylated ISGF3 to inhibit the IFN-induced responses of these promoters.
It is fascinating to observe that not all mammals use the IFN system in the same way. Zhou et al. (2016a) found that at least one species of bats expresses type I IFNs constitutively in the absence of exogenous stimulation, leading to the constitutive expression of U-ISGF3 and providing resistance to viruses that are pathogenic in other mammals. Some bats are especially sensitive to fungal infections (Verant et al. 2014), and it seems possible that desensitization to increased production of IFN on infection may contribute to this situation. It is also relevant that constitutive IFN signaling at very low levels in normal individuals modulates profound biological effects (Gough et al. 2012), including the regulation of basal STAT1 expression, as shown by the failure of Y701F STAT1 to sustain STAT1 gene expression in transgenic mice (Majoros et al. 2016). Another possibility for the deleterious effects of GOF mutations in STAT1 and STAT3 is the competition of these two proteins for activation by specific receptors, including the receptors for IL-21 (Wan et al. 2015), IL-6 (Costa-Pereira et al. 2002), and IFN-γ (Qing and Stark 2004). In the case of IL-21, STAT1 and STAT3 have opposing roles in regulating the function of CD4+ T cells (Wan et al. 2015). The relative concentrations of STAT1 and STAT3 are determined not only by endogenous factors but also by the actions of cytokines, because activated STAT3 drives the expression of the STAT3 gene (Yang et al. 2007) and ISGF3 drives the expression of not only the STAT1 gene but also the STAT2 and IRF9 genes (Cheon et al. 2013). Therefore, GOF mutations in STAT1 or STAT3 are likely to alter the steady-state levels of these two proteins and thus affect the biological responses to cytokines such as IL-21.
Good and Bad IFNs in Cancer
Cancers are constitutively exposed to IFNs that are produced by immune cells in the tumor microenvironment, especially macrophages and dendritic cells. In addition, cancer cells make type I IFNs themselves in response to endogenous or induced DNA damage and in response to the enhanced expression of double-stranded RNAs that are encoded by endogenous retrovirus-like DNA sequences, following reduction in the extent of the DNA methylation that normally suppresses their expression (reviewed by Cheon et al. 2014 and Borden 2017). Acute exposure to endogenous or exogenous (therapeutic) IFN often leads to the arrest or death of cancer cells (Borden 2017). In particular, the IFN that is induced in response to DNA damage facilitates the arrest or killing of cancer cells (Widau et al. 2014; Yu et al. 2015). On the other hand, chronic exposure to a low level of IFN leads to the expression of genes comprising the IFN-related DNA damage-resistance signature (IRDS) (Weichselbaum et al. 2008), which is virtually identical to the pattern of gene expression observed in response to U-ISGF3 (see above). The IRDS phenotype in cancer is characterized by resistance to DNA damage, very likely because of the action of one or more IFN-induced protein. Cancer cells have to survive the toxic effects of endogenous IFNs that arise from constitutive DNA damage and the formation of endogenous double-stranded RNA (Leonova et al. 2013) and exogenous IFNs produced by immune cells in the tumor microenvironment. They do this by desensitizing the full response to IFNs, which otherwise would induce the expression of many cytotoxic or cytostatic proteins while retaining the partial response to IFN that is driven by U-ISGF3, which induces the expression of proteins that provide protection against DNA damage. How the cancer cells manage to achieve such a selective response to IFNs remains to be elucidated. Important factors are likely to be the amounts and types of IFN that are present and the modulatory effects of the many other signaling pathways that are activated by other cytokines in the tumor microenvironment.
SUMMARY AND CONCLUSIONS
Many complex mechanisms are required for appropriate control of how cells respond to cytokines and IFNs, including multiple posttranslational modifications of the STATs and inhibition by many constitutive and induced negative regulators. Furthermore, all the responses are time-dependent, with kinetics that are regulated in many ways as well, including changing levels of STAT expression and modulation of the effects of the negative regulators. How a cell responds to a specific cytokine or IFN is determined not only by the cell type but also by the experience of that cell, in which all the signals that the cell is receiving from the environment are integrated into a specific pattern of behavior. Defects in control become evident in patients with rare germline defects in the STATs and in the abnormal responses of cancer cells to extracellular signals. We can anticipate many more years of important new discoveries as the many layers of this amazing system are exposed to our view by ongoing research.
Footnotes
Editors: Warren J. Leonard and Robert D. Schreiber
Additional Perspectives on Cytokines available at www.cshperspectives.org
REFERENCES
- Allis CD, Jenuwein T. 2016. The molecular hallmarks of epigenetic control. Nat Rev Genet 17: 487–500. [DOI] [PubMed] [Google Scholar]
- Antunes F, Marg A, Vinkemeier U. 2011. STAT1 signaling is not regulated by a phosphorylation-acetylation switch. Mol Cell Biol 31: 3029–3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babon JJ, Varghese LN, Nicola NA. 2014. Inhibition of IL-6 family cytokines by SOCS3. Semin Immunol 26: 13–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begitt A, Droescher M, Knobeloch KP, Vinkemeier U. 2011. SUMO conjugation of STAT1 protects cells from hyperresponsiveness to IFNγ. Blood 118: 1002–1007. [DOI] [PubMed] [Google Scholar]
- Begitt A, Droescher M, Meyer T, Schmid CD, Baker M, Antunes F, Knobeloch KP, Owen MR, Naumann R, Decker T, et al. 2014. STAT1-cooperative DNA binding distinguishes type 1 from type 2 interferon signaling. Nat Immunol 15: 168–176. [DOI] [PubMed] [Google Scholar]
- Benitah SA, Valeron PF, Rui H, Lacal JC. 2003. STAT5a activation mediates the epithelial to mesenchymal transition induced by oncogenic RhoA. Mol Biol Cell 14: 40–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger A, Hoelbl-Kovacic A, Bourgeais J, Hoefling L, Warsch W, Grundschober E, Uras IZ, Menzl I, Putz EM, Hoermann G, et al. 2014. PAK-dependent STAT5 serine phosphorylation is required for BCR-ABL-induced leukemogenesis. Leukemia 28: 629–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisson B, Quartier P, Casanova JL. 2015. Immunological loss-of-function due to genetic gain-of-function in humans: Autosomal dominance of the third kind. Curr Opin Immunol 32: 90–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borden EC. 2017. Interferon signaling in cancer therapy: New opportunities. Nat Rev Drug Discov (in press). [Google Scholar]
- Borden EC, Sen GC, Uze G, Silverman RH, Ransohoff RM, Foster GR, Stark GR. 2007. Interferons at age 50: Past, current and future impact on biomedicine. Nat Rev Drug Discov 6: 975–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai B, Cai JP, Luo YL, Chen C, Zhang S. 2015. The specific roles of JAK/STAT signaling pathway in sepsis. Inflammation 38: 1599–1608. [DOI] [PubMed] [Google Scholar]
- Casanova JL, Holland SM, Notarangelo LD. 2012. Inborn errors of human JAKs and STATs. Immunity 36: 515–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekaran P, Zimmerman O, Paulson M, Sampaio EP, Freeman AF, Sowerwine KJ, Hurt D, Alcantara-Montiel JC, Hsu AP, Holland SM. 2016. Distinct mutations at the same positions of STAT3 cause either loss or gain of function. J Allergy Clin Immunol 138: 1222–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee-Kishore M, Wright KL, Ting JPY, Stark GR. 2000. How Stat1 mediates constitutive gene expression: A complex of unphosphorylated Stat1 and IRF1 supports transcription of the LMP2 gene. EMBO J 19: 4111–4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Sun H, You F, Sun W, Zhou X, Chen L, Yang J, Wang Y, Tang H, Guan Y, et al. 2011. Activation of STAT6 by STING is critical for antiviral innate immunity. Cell 147: 436–446. [DOI] [PubMed] [Google Scholar]
- Cheon H, Stark GR. 2009. Unphosphorylated STAT1 prolongs the expression of interferon-induced immune regulatory genes. Proc Natl Acad Sci 106: 9373–9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheon H, Holvey-Bates EG, Schoggins JW, Forster S, Hertzog P, Imanaka N, Rice CM, Jackson MW, Junk DJ, Stark GR. 2013. IFNβ-dependent increases in STAT1, STAT2, and IRF9 mediate resistance to viruses and DNA damage. EMBO J 32: 2751–2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheon H, Borden EC, Stark GR. 2014. Interferons and their stimulated genes in the tumor microenvironment. Semin Oncol 41: 156–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christova R, Jones T, Wu PJ, Bolzer A, Costa-Pereira AP, Watling D, Kerr IM, Sheer D. 2007. P-STAT1 mediates higher-order chromatin remodelling of the human MHC in response to IFN. J Cell Sci 120: 3262–3270. [DOI] [PubMed] [Google Scholar]
- Clark DE, Williams CC, Duplessis TT, Moring KL, Notwick AR, Long W, Lane WS, Beuvink I, Hynes NE, Jones FE. 2005. ERBB4/HER4 potentiates STAT5A transcriptional activity by regulating novel STAT5A serine phosphorylation events. J Biol Chem 280: 24175–24180. [DOI] [PubMed] [Google Scholar]
- Costa-Pereira AP, Tininini S, Strobl B, Alonzi T, Schlaak JF, Is’harc H, Gesualdo I, Newman SJ, Kerr IM, Poli V. 2002. Mutational switch of an IL-6 response to an interferon-γ-like response. Proc Natl Acad Sci 99: 8043–8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Zhang L, Luo J, Rajasekaran A, Hazra S, Cacalano N, Dubinett SM. 2007. Unphosphorylated STAT6 contributes to constitutive cyclooxygenase-2 expression in human non-small cell lung cancer. Oncogene 26: 4253–4260. [DOI] [PubMed] [Google Scholar]
- Dasgupta M, Unal H, Willard B, Yang J, Karnik SS, Stark GR. 2014. Critical role for lysine 685 in gene expression mediated by transcription factor unphosphorylated STAT3. J Biol Chem 289: 30763–30771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta A, Chen KH, Tian L, Archer SL. 2015a. Gone fission: An asymptomatic STAT2 mutation elongates mitochondria and causes human disease following viral infection. Brain 138: 2802–2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta M, Dermawan JK, Willard B, Stark GR. 2015b. STAT3-driven transcription depends upon the dimethylation of K49 by EZH2. Proc Natl Acad Sci 112: 3985–3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker T. 2016. Emancipation from transcriptional latency: Unphosphorylated STAT5 as guardian of hematopoietic differentiation. EMBO J 35: 555–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker T, Kovarik P. 1999. Transcription factor activity of STAT proteins: Structural requirements and regulation by phosphorylation and interacting proteins. Cell Mol Life Sci 55: 1535–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droescher M, Begitt A, Marg A, Zacharias M, Vinkemeier U. 2011. Cytokine-induced paracrystals prolong the activity of signal transducers and activators of transcription (STAT) and provide a model for the regulation of protein solubility by small ubiquitin-like modifier (SUMO). J Biol Chem 286: 18731–18746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers A, Georgellis D, Klose B, Schindler C, Ziemiecki A, Harpur AG, Wilks AF, Decker T. 1995. Differentiation-regulated serine phosphorylation of STAT1 promotes GAF activation in macrophages. Mol Cell Biol 15: 3579–3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan ZD, Wang FF, Huang H, Huang N, Ma HH, Guo YH, Zhang YY, Qian XQ, Yu HG. 2015. STAT4 rs7574865 G/T and PTPN22 rs2488457 G/C polymorphisms influence the risk of developing juvenile idiopathic arthritis in Han Chinese patients. PLoS ONE 10: e0117389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedbichler K, Kerenyi MA, Kovacic B, Li G, Hoelbl A, Yahiaoui S, Sexl V, Mullner EW, Fajmann S, Cerny-Reiterer S, et al. 2010. Stat5a serine 725 and 779 phosphorylation is a prerequisite for hematopoietic transformation. Blood 116: 1548–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garama DJ, White CL, Balic JJ, Gough DJ. 2016. Mitochondrial STAT3: Powering up a potent factor. Cytokine 87: 20–25. [DOI] [PubMed] [Google Scholar]
- Gewinner C, Hart G, Zachara N, Cole R, Beisenherz-Huss C, Groner B. 2004. The coactivator of transcription CREB-binding protein interacts preferentially with the glycosylated form of Stat5. J Biol Chem 279: 3563–3572. [DOI] [PubMed] [Google Scholar]
- Gough DJ, Corlett A, Schlessinger K, Wegrzyn J, Larner AC, Levy DE. 2009. Mitochondrial STAT3 supports Ras-dependent oncogenic transformation. Science 324: 1713–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough DJ, Messina NL, Clarke CJ, Johnstone RW, Levy DE. 2012. Constitutive type I interferon modulates homeostatic balance through tonic signaling. Immunity 36: 166–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough DJ, Koetz L, Levy DE. 2013. The MEK–ERK pathway is necessary for serine phosphorylation of mitochondrial STAT3 and Ras-mediated transformation. PLoS ONE 8: e83395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough DJ, Marie IJ, Lobry C, Aifantis I, Levy DE. 2014. STAT3 supports experimental K-RasG12D-induced murine myeloproliferative neoplasms dependent on serine phosphorylation. Blood 124: 2252–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronholm J, Vanhatupa S, Ungureanu D, Valiaho J, Laitinen T, Valjakka J, Silvennoinen O. 2012. Structure-function analysis indicates that sumoylation modulates DNA-binding activity of STAT1. BMC Biochem 13: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hambleton S, Goodbourn S, Young DF, Dickinson P, Mohamad SM, Valappil M, McGovern N, Cant AJ, Hackett SJ, Ghazal P, et al. 2013. STAT2 deficiency and susceptibility to viral illness in humans. Proc Natl Acad Sci 110: 3053–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzog PJ, Williams BR. 2013. Fine tuning type I interferon responses. Cytokine Growth Factor Rev 24: 217–225. [DOI] [PubMed] [Google Scholar]
- Ho J, Pelzel C, Begitt A, Mee M, Elsheikha HM, Scott DJ, Vinkemeier U. 2016. STAT2 is a pervasive cytokine regulator due to its inhibition of STAT1 in multiple signaling pathways. PLoS Biol 14: e2000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou T, Ray S, Lee C, Brasier AR. 2008. The STAT3 NH2-terminal domain stabilizes enhanceosome assembly by interacting with the p300 bromodomain. J Biol Chem 283: 30725–30734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Dutta P, Tsurumi A, Li J, Wang J, Land H, Li WX. 2013. Unphosphorylated STAT5A stabilizes heterochromatin and suppresses tumor growth. Proc Natl Acad Sci 110: 10213–10218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki H, Kovacic JC, Olive M, Beers JK, Yoshimoto T, Crook MF, Tonelli LH, Nabel EG. 2010. Disruption of protein arginine N-methyltransferase 2 regulates leptin signaling and produces leanness in vivo through loss of STAT3 methylation. Circ Res 107: 992–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer J, Reich NC. 2007. Constitutive nuclear import of latent and activated STAT5a by its coiled coil domain. FASEB J 22: 391–400. [DOI] [PubMed] [Google Scholar]
- Kabotyanski EB, Rosen JM. 2003. Signal transduction pathways regulated by prolactin and Src result in different conformations of activated Stat5b. J Biol Chem 278: 17218–17227. [DOI] [PubMed] [Google Scholar]
- Kang HJ, Yi YW, Hou SJ, Kim HJ, Kong Y, Bae I, Brown ML. 2015. Disruption of STAT3-DNMT1 interaction by SH-I-14 induces re-expression of tumor suppressor genes and inhibits growth of triple-negative breast tumor. Oncotarget 10.18632/oncotarget.4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazi JU, Kabir NN, Flores-Morales A, Ronnstrand L. 2014. SOCS proteins in regulation of receptor tyrosine kinase signaling. Cell Mol Life Sci 71: 3297–3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotla S, Rao GN. 2015. Reactive oxygen species (ROS) mediate p300–dependent STAT1 protein interaction with peroxisome proliferator-activated receptor (PPAR)-γ in CD36 protein expression and foam cell formation. J Biol Chem 290: 30306–30320. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kramer OH, Baus D, Knauer SK, Stein S, Jager E, Stauber RH, Grez M, Pfitzner E, Heinzel T. 2006. Acetylation of Stat1 modulates NF-κB activity. Genes Dev 20: 473–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N, Hanks ME, Chandrasekaran P, Davis BC, Hsu AP, Van Wagoner NJ, Merlin JS, Spalding C, La Hoz RM, Holland SM, et al. 2014. Gain-of-function signal transducer and activator of transcription 1 (STAT1) mutation-related primary immunodeficiency is associated with disseminated mucormycosis. J Allergy Clin Immunol 134: 236–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JL, Wang MJ, Chen JY. 2009. Acetylation and activation of STAT3 mediated by nuclear translocation of CD44. J Cell Biol 185: 949–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC, Min HY, Jung HJ, Park KH, Hyun SY, Cho J, Woo JK, Kwon SJ, Lee HJ, Johnson FM, et al. 2016. Essential role of insulin-like growth factor 2 in resistance to histone deacetylase inhibitors. Oncogene 35: 5515–5526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonova KI, Brodsky L, Lipchick B, Pal M, Novototskaya L, Chenchik AA, Sen GC, Komarova EA, Gudkov AV. 2013. p53 cooperates with DNA methylation and a suicidal interferon response to maintain epigenetic silencing of repeats and noncoding RNAs. Proc Natl Acad Sci 110: E89–E98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine RL, Gilliland DG. 2008. Myeloproliferative disorders. Blood 112: 2190–2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, McBride KM, Reich NC. 2005. STAT3 nuclear import is independent of tyrosine phosphorylation and mediated by importin-3. Proc Natl Acad Sci 102: 8150–8155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maarifi G, Maroui MA, Dutrieux J, Dianoux L, Nisole S, Chelbi-Alix MK. 2015. Small ubiquitin-like modifier alters IFN response. J Immunol 195: 2312–2324. [DOI] [PubMed] [Google Scholar]
- Majoros A, Platanitis E, Szappanos D, Cheon H, Vogl C, Shukla P, Stark GR, Sexl V, Schreiber R, Schindler C, et al. 2016. Response to interferons and antibacterial innate immunity in the absence of tyrosine-phosphorylated STAT1. EMBO Rep 17: 367–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X, Ren Z, Parker GN, Sondermann H, Pastorello MA, Wang W, McMurray JS, Demeler B, Darnell JE, Chen X. 2005. Structural bases of unphosphorylated STAT1 association and receptor binding. Mol Cell 17: 761–771. [DOI] [PubMed] [Google Scholar]
- Meyer T, Vinkemeier U. 2004. Nucleocytoplasmic shuttling of STAT transcription factors. Eur J Biochem 271: 4606–4612. [DOI] [PubMed] [Google Scholar]
- Meyer T, Begitt A, Lödige I, van Rossum M, Vinkemeier U. 2002. Constitutive and IFN-γ-induced nuclear import of STAT1 proceed through independent pathways. EMBO J 21: 344–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra A, Ross JA, Rodriguez G, Nagy ZS, Wilson HL, Kirken RA. 2012. Signal transducer and activator of transcription 5b (Stat5b) serine 193 is a novel cytokine-induced phospho-regulatory site that is constitutively activated in primary hematopoietic malignancies. J Biol Chem 287: 16596–16608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow AN, Schmeisser H, Tsuno T, Zoon KC. 2011. A novel role for IFN-stimulated gene factor 3II in IFN-γ signaling and induction of antiviral activity in human cells. J Immunol 186: 1685–1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowen KA, Tang J, Zhu W, Schurter BT, Shuai K, Herschman HR, David M. 2001. Arginine methylation of STAT1 modulates IFNα/β-induced transcription. Cell 104: 731–741. [DOI] [PubMed] [Google Scholar]
- Neculai D, Neculai AM, Verrier S, Straub K, Klumpp K, Pfitzner E, Becker S. 2005. Structure of the unphosphorylated STAT5a dimer. J Biol Chem 280: 40782–40787. [DOI] [PubMed] [Google Scholar]
- Ng J, Cantrell D. 1997. STAT3 is a serine kinase target in T lymphocytes. Interleukin 2 and T cell antigen receptor signals converge upon serine 727. J Biol Chem 272: 24542–24549. [DOI] [PubMed] [Google Scholar]
- Nie Y, Erion DM, Yuan Z, Dietrich M, Shulman GI, Horvath TL, Gao Q. 2009. STAT3 inhibition of gluconeogenesis is downregulated by SirT1. Nat Cell Biol 11: 492–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nkansah E, Shah R, Collie GW, Parkinson GN, Palmer J, Rahman KM, Bui TT, Drake AF, Husby J, Neidle S, et al. 2013. Observation of unphosphorylated STAT3 core protein binding to target dsDNA by PEMSA and X-ray crystallography. FEBS Lett 587: 833–839. [DOI] [PubMed] [Google Scholar]
- Oldstone MB. 2015. A Jekyll and Hyde profile: Type 1 interferon signaling plays a prominent role in the initiation and maintenance of a persistent virus infection. J Infect Dis 212: S31–S36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shea JJ, Schwartz DM, Villarino AV, Gadina M, McInnes IB, Laurence A. 2015. The JAK-STAT pathway: Impact on human disease and therapeutic intervention. Annu Rev Med 66: 311–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HJ, Li J, Hannah R, Biddie S, Leal-Cervantes AI, Kirschner K, Flores Santa Cruz D, Sexl V, Gottgens B, Green AR. 2015. Cytokine-induced megakaryocytic differentiation is regulated by genome-wide loss of a uSTAT transcriptional program. EMBO J 35: 580–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilati C, Zucman-Rossi J. 2015. Mutations leading to constitutive active gp130/JAK1/STAT3 pathway. Cytokine Growth Factor Rev 26: 499–506. [DOI] [PubMed] [Google Scholar]
- Pircher TJ, Petersen H, Gustafsson JA, Haldosen LA. 1999. Extracellular signal-regulated kinase (ERK) interacts with signal transducer and activator of transcription (STAT) 5a. Mol Endocrinol 13: 555–565. [DOI] [PubMed] [Google Scholar]
- Porritt RA, Hertzog PJ. 2015. Dynamic control of type I IFN signalling by an integrated network of negative regulators. Trends Immunol 36: 150–160. [DOI] [PubMed] [Google Scholar]
- Pranada AL, Metz S, Herrmann A, Heinrich PC, Muller-Newen G. 2003. Real time analysis of STAT3 nucleocytoplasmic shuttling. J Biol Chem 279: 15114–15123. [DOI] [PubMed] [Google Scholar]
- Qing Y, Stark GR. 2004. Alternative activation of STAT1 and STAT3 in response to interferon-γ. J Biol Chem 279: 41679–41685. [DOI] [PubMed] [Google Scholar]
- Rajala HL, Eldfors S, Kuusanmaki H, van Adrichem AJ, Olson T, Lagstrom S, Andersson EI, Jerez A, Clemente MJ, Yan Y, et al. 2013. Discovery of somatic STAT5b mutations in large granular lymphocytic leukemia. Blood 121: 4541–4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajsbaum R, Versteeg GA, Schmid S, Maestre AM, Belicha-Villanueva A, Martinez-Romero C, Patel JR, Morrison J, Pisanelli G, Miorin L, et al. 2014. Unanchored K48-linked polyubiquitin synthesized by the E3-ubiquitin ligase TRIM6 stimulates the interferon-IKKɛ kinase-mediated antiviral response. Immunity 40: 880–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray S, Boldogh I, Brasier AR. 2005. STAT3 NH2-terminal acetylation is activated by the hepatic acute-phase response and required for IL-6 induction of angiotensinogen. Gastroenterology 129: 1616–1632. [DOI] [PubMed] [Google Scholar]
- Rogers RS, Horvath CM, Matunis MJ. 2003. SUMO modification of STAT1 and its role in PIAS-mediated inhibition of gene activation. J Biol Chem 278: 30091–30097. [DOI] [PubMed] [Google Scholar]
- Sampaio EP, Bax HI, Hsu AP, Kristosturyan E, Pechacek J, Chandrasekaran P, Paulson ML, Dias DL, Spalding C, Uzel G, et al. 2012. A novel STAT1 mutation associated with disseminated mycobacterial disease. J Clin Immunol 32: 681–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampaio EP, Hsu AP, Pechacek J, Bax HI, Dias DL, Paulson ML, Chandrasekaran P, Rosen LB, Carvalho DS, Ding L, et al. 2013. Signal transducer and activator of transcription 1 (STAT1) gain-of-function mutations and disseminated coccidioidomycosis and histoplasmosis. J Allergy Clin Immunol 131: 1624–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarzello AJ, Romero-Weaver AL, Maher SG, Veenstra TD, Zhou M, Qin A, Donnelly RP, Sheikh F, Gamero AM. 2007. A mutation in the SH2 domain of STAT2 prolongs tyrosine phosphorylation of STAT1 and promotes type I IFN-induced apoptosis. Mol Biol Cell 18: 2455–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller-Schonitz M, Barzan D, Williamson AJ, Griffiths JR, Dallmann I, Battmer K, Ganser A, Whetton AD, Scherr M, Eder M. 2014. BCR-ABL affects STAT5A and STAT5B differentially. PLoS ONE 9: e97243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler C, Levy DE, Decker T. 2007. JAK-STAT signaling: From interferons to cytokines. J Biol Chem 282: 20059–20063. [DOI] [PubMed] [Google Scholar]
- Shahni R, Cale CM, Anderson G, Osellame LD, Hambleton S, Jacques TS, Wedatilake Y, Taanman JW, Chan E, Qasim WV. et al. 2015. Signal transducer and activator of transcription 2 deficiency is a novel disorder of mitochondrial fission. Brain 138: 2834–2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S, Larson K, Guo D, Lim SJ, Dutta P, Yan SJ, Li WX. 2008. Drosophila STAT is required for directly maintaining HP1 localization and heterochromatin stability. Nat Cell Biol 10: 489–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin EC, Sung PS, Park SH. 2016. Immune responses and immunopathology in acute and chronic viral hepatitis. Nat Rev Immunol 16: 509–523. [DOI] [PubMed] [Google Scholar]
- Shirakawa T, Kawazoe Y, Tsujikawa T, Jung D, Sato S, Uesugi M. 2011. Deactivation of STAT6 through serine 707 phosphorylation by JNK. J Biol Chem 286: 4003–4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spivak JL. 2010. Narrative review: Thrombocytosis, polycythemia vera, and JAK2 mutations: The phenotypic mimicry of chronic myeloproliferation. Ann Intern Med 152: 300–306. [DOI] [PubMed] [Google Scholar]
- Stark GR, Darnell JE Jr. 2012. The JAK-STAT pathway at twenty. Immunity 36: 503–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark GR, Wang Y, Lu T. 2011. Lysine methylation of promoter-bound transcription factors and relevance to cancer. Cell Res 21: 375–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen HC, Nogusa S, Thapa RJ, Basagoudanavar SH, Gill AL, Merali S, Barrero CA, Balachandran S, Gamero AM. 2013. Identification of STAT2 serine 287 as a novel regulatory phosphorylation site in type I interferon-induced cellular responses. J Biol Chem 288: 747–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung PS, Cheon H, Cho CH, Hong SH, Park DY, Seo HI, Park SH, Yoon SK, Stark GR, Shin EC. 2015. Roles of unphosphorylated ISGF3 in HCV infection and interferon responsiveness. Proc Natl Acad Sci 112: 10443–10448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teijaro JR, Ng C, Lee AM, Sullivan BM, Sheehan KC, Welch M, Schreiber RD, de la Torre JC, Oldstone MB. 2013. Persistent LCMV infection is controlled by blockade of type I interferon signaling. Science 340: 207–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenoever BR, Ng SL, Chua MA, McWhirter SM, Garcia-Sastre A, Maniatis T. 2007. Multiple functions of the IKK-related kinase IKKɛ in interferon-mediated antiviral immunity. Science 315: 1274–1278. [DOI] [PubMed] [Google Scholar]
- Testoni B, Vollenkle C, Guerrieri F, Gerbal-Chaloin S, Blandino G, Levrero M. 2011. Chromatin dynamics of gene activation and repression in response to interferon (IFN ) reveal new roles for phosphorylated and unphosphorylated forms of the transcription factor STAT2. J Biol Chem 286: 20217–20227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas SJ, Snowden JA, Zeidler MP, Danson SJ. 2015. The role of JAK/STAT signalling in the pathogenesis, prognosis and treatment of solid tumours. Br J Cancer 113: 365–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timofeeva OA, Chasovskikh S, Lonskaya I, Tarasova NI, Khavrutskii L, Tarasov SG, Zhang X, Korostyshevskiy VR, Cheema A, Zhang L, et al. 2012. Mechanisms of unphosphorylated STAT3 transcription factor binding to DNA. J Biol Chem 287: 14192–14200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungureanu D, Vanhatupa S, Kotaja N, Yang J, Aittomaki S, Janne OA, Palvimo JJ, Silvennoinen O. 2003. PIAS proteins promote SUMO-1 conjugation to STAT1. Blood 102: 3311–3313. [DOI] [PubMed] [Google Scholar]
- Ungureanu D, Vanhatupa S, Gronholm J, Palvimo JJ, Silvennoinen O. 2005. SUMO-1 conjugation selectively modulates STAT1-mediated gene responses. Blood 106: 224–226. [DOI] [PubMed] [Google Scholar]
- Uzel G, Sampaio EP, Lawrence MG, Hsu AP, Hackett M, Dorsey MJ, Noel RJ, Verbsky JW, Freeman AF, Janssen E, et al. 2013. Dominant gain-of-function STAT1 mutations in FOXP3 wild-type immune dysregulation-polyendocrinopathy-enteropathy-X-linked-like syndrome. J Allergy Clin Immunol 131: 1611–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Boxel-Dezaire AH, Zula JA, Xu Y, Ransohoff RM, Jacobberger JW, Stark GR. 2010. Major differences in the responses of primary human leukocyte subsets to IFN-β. J immunol 185: 5888–5899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Veerdonk FL, Plantinga TS, Hoischen A, Smeekens SP, Joosten LA, Gilissen C, Arts P, Rosentul DC, Carmichael AJ, Smits-van der Graaf CA, et al. 2011. STAT1 mutations in autosomal dominant chronic mucocutaneous candidiasis. N Engl J Med 365: 54–61. [DOI] [PubMed] [Google Scholar]
- Verant ML, Meteyer CU, Speakman JR, Cryan PM, Lorch JM, Blehert DS. 2014. White-nose syndrome initiates a cascade of physiologic disturbances in the hibernating bat host. BMC Physiol 14: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarino AV, Kanno Y, Ferdinand JR, O’Shea JJ. 2015. Mechanisms of Jak/STAT signaling in immunity and disease. J Immunol 194: 21–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visconti R, Gadina M, Chiariello M, Chen EH, Stancato LF, Gutkind JS, O’Shea JJ. 2000. Importance of the MKK6/p38 pathway for interleukin-12-induced STAT4 serine phosphorylation and transcriptional activity. Blood 96: 1844–1852. [PubMed] [Google Scholar]
- Vogt M, Domoszlai T, Kleshchanok D, Lehmann S, Schmitt A, Poli V, Richtering W, Muller-Newen G. 2011. The role of the N-terminal domain in dimerization and nucleocytoplasmic shuttling of latent STAT3. J Cell Sci 124: 900–909. [DOI] [PubMed] [Google Scholar]
- Waitkus MS, Chandrasekharan UM, Willard B, Tee TL, Hsieh JK, Przybycin CG, Rini BI, Dicorleto PE. 2014. Signal integration and gene induction by a functionally distinct STAT3 phosphoform. Mol Cell Biol 34: 1800–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan CK, Andraski AB, Spolski R, Li P, Kazemian M, Oh J, Samsel L, Swanson PA 2nd, McGavern DB, Sampaio EP, et al. 2015. Opposing roles of STAT1 and STAT3 in IL-21 function in CD4+ T cells. Proc Natl Acad Sci 112: 9394–9399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Malabarba MG, Nagy ZS, Kirken RA. 2004. Interleukin 4 regulates phosphorylation of serine 756 in the transactivation domain of Stat6. Roles for multiple phosphorylation sites and Stat6 function. J Biol Chem 279: 25196–25203. [DOI] [PubMed] [Google Scholar]
- Wang Y, van Boxel-Dezaire AH, Cheon H, Yang J, Stark GR. 2013. STAT3 activation in response to IL-6 is prolonged by the binding of IL-6 receptor to EGF receptor. Proc Natl Acad Sci 110: 16975–16980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Nan J, Willard B, Wang X, Yang J, Stark GR. 2017. Negative regulation of type I IFN signaling by phosphorylation of STAT2 on T387. EMBO J 36: 202–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegrzyn J, Potla R, Chwae YJ, Sepuri NBV, Zhang Q, Koeck T, Derecka M, Szczepanek K, Szelag M, Gornicka A, et al. 2009. Function of mitochondrial Stat3 in cellular respiration. Science 323: 793–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichselbaum RR, Ishwaran H, Yoon T, Nuyten DS, Baker SW, Khodarev N, Su AW, Shaikh AY, Roach P, Kreike B, et al. 2008. An interferon-related gene signature for DNA damage resistance is a predictive marker for chemotherapy and radiation for breast cancer. Proc Natl Acad Sci 105: 18490–18495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Z, Zhong Z, Darnell JE Jr. 1995. Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell 82: 241–250. [DOI] [PubMed] [Google Scholar]
- Wenta N, Strauss H, Meyer S, Vinkemeier U. 2008. Tyrosine phosphorylation regulates the partitioning of STAT1 between different dimer conformations. Proc Natl Acad Sci 105: 9238–9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widau RC, Parekh AD, Ranck MC, Golden DW, Kumar KA, Sood RF, Pitroda SP, Liao Z, Huang X, Darga TE, et al. 2014. RIG-I-like receptor LGP2 protects tumor cells from ionizing radiation. Proc Natl Acad Sci 111: E484–E491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue HH, Fink DW Jr, Zhang X, Qin J, Turck CW, Leonard WJ. 2002. Serine phosphorylation of Stat5 proteins in lymphocytes stimulated with IL-2. Int Immunol 14: 1263–1271. [DOI] [PubMed] [Google Scholar]
- Yamashita H, Xu J, Erwin RA, Farrar WL, Kirken RA, Rui H. 1998. Differential control of the phosphorylation state of proline-juxtaposed serine residues Ser725 of Stat5a and Ser730 of Stat5b in prolactin-sensitive cells. J Biol Chem 273: 30218–30224. [DOI] [PubMed] [Google Scholar]
- Yang J, Chatterjee-Kishore M, Staugaitis SM, Nguyen H, Schlessinger K, Levy DE, Stark GR. 2005. Novel roles of unphosphorylated STAT3 in oncogenesis and transcriptional regulation. Cancer Res 65: 939–947. [PubMed] [Google Scholar]
- Yang J, Liao X, Agarwal MK, Barnes L, Auron PE, Stark GR. 2007. Unphosphorylated STAT3 accumulates in response to IL-6 and activates transcription by binding to NFκB. Genes Dev 21: 1396–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Huang J, Dasgupta M, Sears N, Miyagi M, Wang B, Chance MR, Chen X, Du Y, Wang Y, et al. 2010. Reversible methylation of promoter-bound STAT3 by histone-modifying enzymes. Proc Natl Acad Sci 107: 21499–21504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz M, Li H, Bernard D, Amin NA, Ouillette P, Jones S, Saiya-Cork K, Parkin B, Jacobi K, Shedden K, et al. 2015. Activating STAT6 mutations in follicular lymphoma. Blood 125: 668–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Liu W, Dai Y. 2015. SOCS3 and its role in associated diseases. Hum Immunol 76: 775–780. [DOI] [PubMed] [Google Scholar]
- Yu Q, Katlinskaya YV, Carbone CJ, Zhao B, Katlinski KV, Zheng H, Guha M, Li N, Chen Q, Yang T, et al. 2015. DNA-damage-induced type I interferon promotes senescence and inhibits stem cell function. Cell Rep 11: 785–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan ZL, Guan YJ, Chatterjee D, Chin YE. 2005. Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science 307: 269–273. [DOI] [PubMed] [Google Scholar]
- Zerbe CS, Marciano BE, Katial RK, Santos CB, Adamo N, Hsu AP, Hanks ME, Darnell DN, Quezado MM, Frein C, et al. 2016. Progressive multifocal leukoencephalopathy in primary immune deficiencies: Stat1 gain of function and review of the literature. Clin Infect Dis 62: 986–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Blenis J, Li HC, Schindler C, Chen-Kiang S. 1995. Requirement of serine phosphorylation for formation of STAT-promoter complexes. Science 267: 1990–1994. [DOI] [PubMed] [Google Scholar]
- Zhang X, Bogunovic D, Payelle-Brogard B, Francois-Newton V, Speer SD, Yuan C, Volpi S, Li Z, Sanal O, Mansouri D, et al. 2015. Human intracellular ISG15 prevents interferon-α/β over-amplification and auto-inflammation. Nature 517: 89–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Tachedjian M, Wynne JW, Boyd V, Cui J, Smith I, Cowled C, Ng JH, Mok L, Michalski WP, et al. 2016a. Contraction of the type I IFN locus and unusual constitutive expression of IFN-α in bats. Proc Natl Acad Sci 113: 2696–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Wang M, Li J, Xiao M, Chin YE, Cheng J, Yeh ET, Yang J, Yi J. 2016b. SUMOylation and SENP3 regulate STAT3 activation in head and neck cancer. Oncogene 35: 5826–5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Mustelin T, David M. 2002. Arginine methylation of STAT1 regulates its dephosphorylation by T cell protein tyrosine phosphatase. J Biol Chem 277: 35787–35790. [DOI] [PubMed] [Google Scholar]