Abstract
Background
Long non-coding RNA PCAT7 has been revealed to participate in tumorigenesis of various cancers. However, the mechanism of PCAT7 in non-small cell lung cancer (NSCLC) has not been identified. Hence, this study aimed to determine the function of PCAT7 in NSCLC.
Material/Methods
The expression level of PCAT7 in 96 pairs of NSCLC tissues and 6 cell lines was detected by qRT-PCR. Proliferation assay, flow cytometric analysis, transwell/migration assay, and Western blotting assay were performed to detect the relation between PCAT7 and malignant behaviors of NSCLC cells in vitro, including cell proliferation, apoptosis, migration, invasion, and epithelial-to-mesenchymal transition (EMT). Rescue assays were carried out to confirm the contribution of PCAT7 to the progression of NSCLC cells by targeting miR-134-5p.
Results
PCAT7 was found to be overexpressed in NSCLC tissues (compared with corresponding non-tumor tissues) and NSCLC cells (compared with normal cell line 16-HBE). Overexpression of PCAT7 resulted in the promotion of tumor cell proliferation, inhibition of cells apoptosis, facilitation of cells metastasis, and formation of EMT phenotype, while PCAT7 expression deletion remarkably prohibited cell proliferation, accelerated their apoptosis, weakened metastasis, and reversed EMT to MET. miR-134-5P, as a target gene of PCAT7, restored the effects of down-regulation of PCAT7.
Conclusions
These findings demonstrate that PCAT7 participates in tumor progression in NSCLC by inhibiting miR-134-5p.
MeSH Keywords: Carcinogenesis; Carcinoma, Non-Small-Cell Lung; MicroRNAs; RNA, Long Noncoding
Background
Lung cancer remains the most common cause of cancer-related deaths in the United States [1]. Generally, lung cancer can be classified into 2 categories: small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC). NSCLC is the most common type of lung cancer, accounting for 80% of all cases [2]. Although treatments for NSCLC have increased gradually, such as surgery, chemotherapy, and radiotherapy, improvements in patient prognosis remain limited. One of the most crucial reasons for this situation is the lack of a detailed understanding of the pathogenetic mechanism. Therefore, it is necessary to further explore the pathogenetic mechanism of NSCLC for developing effective diagnosis and treatment.
lncRNAs (long non-coding RNAs) can be transcribed as transcripts over 200 nucleotides long, with no evidence of protein coding potential by the human genome [3]. There has been ample evidence suggesting that dysregulated lncRNAs are closely related to carcinogenesis [4]. Recently, many NSCLC-related lncRNAs have been identified, such as HOTAIR, PANDAR, and TUG1 [5–7], which can participate in tumorigenesis and cancer progression of NSCLC by affecting cellular homeostasis. These findings have indicated the potential of lncRNAs as novel biomarkers for prognosis or therapeutic targets. PCAT7 (prostate cancer-associated transcript 7) is a 1937-bp lncRNA that maps at chromosome 9q22.32 [8]. An increasing number of studies have reported that PCAT7 may be related to tumorigenesis. Recently, Liu et al. have reported that PCAT7 contributes to tumor progression in nasopharyngeal carcinoma by functioning as a ceRNA to sponge miR-134-5p [8]. Horie et al. indicated that PCAT7 is overexpressed in NSCLC [9]. However, it still needs to be determined whether PCAT7 is involved in tumor progression in NSCLC.
This study demonstrates that PCAT7 is overexpressed in NSCLC tissues and cell lines. PCAT7 negatively influenced the miR-134-5p and then promoted cell proliferation, inhibited cell apoptosis, accelerated cell migration/invasion, and induced EMT. PCAT7/miR-134-5p has potential to be a novel approach for the treatment of NSCLC.
Material and Methods
Patients and clinical samples collection
We obtained 96 NSCLC tissues and corresponding tumor tissues from the Department of Thoracic Surgery in the First Affiliated Hospital of Sun Yat-sen University between April 2012 and October 2016. These tissues were stored in liquid nitrogen before examination. All patients signed the inform consent.
Cell cultures
We purchased 6 NSCLC cell lines (A549, H292, H460, H1703, H358, and H1975) and a normal human lung bronchial epithelial cell line (16-HBE) from the American Type Culture Collection (ATCC, USA) and cultured them in RPMI-1640 Medium (Gibco BRL, Gaithersburg, MD) with 10% fetal bovine serum, ampicillin, and streptomycin in a humidified atmosphere of 95% air and 5% CO2, with the temperature at 37°C.
RNA extraction and quantitative real-time PCR
Total RNA extracted from tissues and cells was isolated with Trizol reagent (Invitrogen, Grand Island, CA) in accordance with the manufacturer’s instructions. The isolated RNA was first reversely transcribed to cDNA using the PrimeScript RT reagent kit (Takara, Japan) in compliance with the manufacturer’s protocol. qRT-PCR was performed with SYBR Prime Script RT-PCR Kits (Takara, Japan) based on the manufacturer’s protocol. The primers were:
PCAT7, Forward 5′-AAACAAGCCAACCGCACAAT-3′,
Reverse 5′-CCTGCTTGCTGTGTTACTGC-3′;
miR-134-5p, forward, 5′-ACACTGCATCCTGGCAATTC-3′;
reverse, 5′-CGTGGTGAATCGAGACTCAC-3′;
U6, forward, 5′-CTCGCTTCGGCAGCACA-3′ and
reverse, 5′-AACGCTTCACGAATTTGCGT-3′.
All assays were performed in triplicate. The relative expression levels were first calculated using the 2-ΔΔCt method, and then normalized to expression of GAPDH mRNA.
Cell transfection
Plasmid complementary DNA PCAT7 and miR-134-5p were constructed by amplification and introduction of PCAT7 and miR-134-5P cDNA sequence into the pGLV5 vector (ABM, Canada). The siRNA sequences targeting PCAT7 and si-NC were purchased from Genepharma Co., Ltd (Shanghai). According to the manufacturer’s protocol, the Lipofectamine 2000 kit (Invitrogen) was used to perform cell transfection.
Western bolt analysis
After being separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), cell protein lysates were first transferred to polyvinylidene fluoride membranes (Roche) and then incubated with specific rabbit anti-human antibodies, including E-cadherin (1: 5000 dilution), N-cadherin (1: 5000 dilution), Vimentin (1: 5000 dilution), caspase-3 (1: 5000 dilution), caspase-9 (1: 5000 dilution), and GAPDH (1: 3000 dilution). Afterwards, they were stored overnight at 4°C, followed by treatment with secondary antibodies, where ECL chromogenic substrate was used for quantification by densitometry (Quantity One software; Bio-Rad, Hercules, CA).
CCK-8 assay
The CCK-8 kit (Dojindo Laboratories, Kumamoto, Japan) was used to assess the viability of the cells, which were then seeded in a 96-well plate at a density of 1×104 cells per well. After being cultured for 24 h, the corresponding PCAT7 and siRNA were transfected and cultured in normal media. After adding the CCK-8 solution at 0, 24, 48, and 72 h after transfection, the relative number of cells was assessed by measuring at OD 460 nm. All assays were performed in triplicate.
Colony formation assay
After being placed in 6-well (300 cells/well) plates, the cells were incubated in RPMI 1640 with 10% FBS at 37°C for 2 weeks, then fixed and stained with 0.1% crystal violet (E. Merck, AG, Darmstadt, BRD), and then were photographed. Next, the number of visible colonies was counted manually.
Cell apoptosis assays
According to the manufacturer’s instructions, the cells were washed with PBS and the apoptosis was performed using flow cytometric analyses with Annexin V: 7-AAD Apoptosis Detection Kits (BD Biosciences, USA). After incubation, the samples were analyzed using flow cytometry (FACSCalibur, BD Biosciences, San Jose, CA). All samples were assayed in triplicate.
Cell migration and invasion assays
Transwell chambers (8-μm pore membrane filters; Costar, Corning, NY, USA) and Matrigel invasion (Becton Dickinson) were used to measure NSCLC cell migration and invasion. These cells, which had been cultivated in serum-free medium, were transferred into the upper chamber without or with 10 ug/ml Matrigel 48 h after transfection. Medium with 10% FBS was added into the lower chamber. The remaining cells on the upper surface of the filter were wiped off with a cotton swab, while those that had migrated were fixed in methanol, stained with 0.1% crystal violet, and counted using a microscope in 3 random fields per filter after incubation for 24 h.
Wound healing assay
After been transfected for 48 h, these cells were seeded into a 24-well plate, where they would be scraped into a single stripe with a sterile plastic 10-μl micropipette tip only if the monolayer cell reached approximately 90% confluence. After removing the detached cells, the plate was continuously cultured in RPMI 1640 without FBS. A series of pictures were taken using an inverted microscope in the same field at 0 h, 12 h, and 24 h. The wound width (μm) was measured using Image pro-plus 6.0 software. Wound closure rate=wound width (at 0 h, 12 h, or 24 h)/wound width (0 h)×100%.
Statistical analysis
All statistical analyses were conducted using the SPSS 22.0 software (SPSS Inc., Chicago, IL). Two-group comparisons were made using the t test and a one-way ANOVA, and the chi-square test was used to analyze multiple group comparisons. Spearman correlation analysis was used to detect the correlation between PACT7 expression levels in NSCLC tissues. Multivariate analysis of prognostic parameters in NSCLC patients was conducted using the Cox proportional hazards model, where P<0.05 was considered statistically significant.
Results
PCAT7 is up-regulated in human NSCLC tissues and cell lines
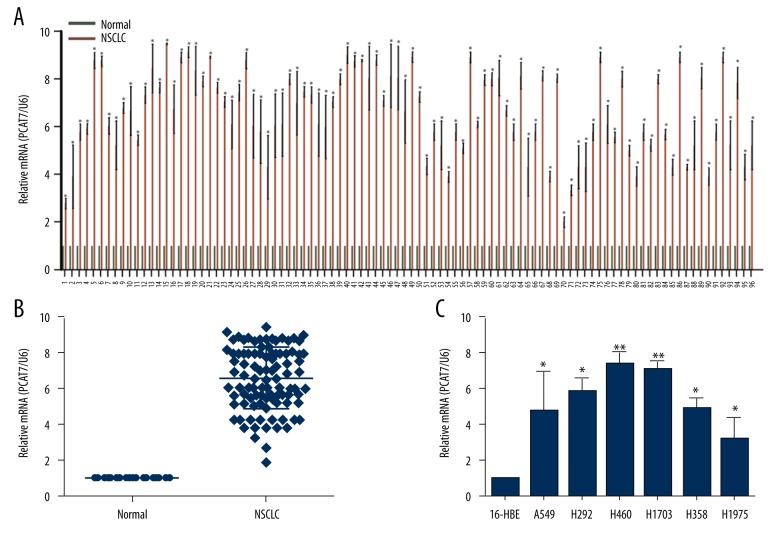
The expression level of PCAT7 was detected using qRT-PCR in 96 NSCLC lung tissues and pair-matched normal lung tissues. Compared with matched normal tissues, PCAT7 expression level in tumor tissues was extremely high (Figure 1A, 1B) (p<0.01). Subsequently, PCAT7 expressions in 6 NSCLC cell lines (A549, H292, H460, H1703, H358, and H1975), as well as in human bronchial epithelial (16HBE) cells, were determined. As shown in Figure 1C, PCAT7 expression in NSCLC cell lines was significantly increased compared with that in 16HBE cells.
Figure 1.
PCAT7 is overexpressed in human NSCLC tissues and cell lines. (A, B) Relative expression of PCAT7 expression in NSCLC tissues (n=96) and in paired adjacent normal tissues (n=96) by qRT-PCR, and normalized to U6 expression. (C) Relative expression of PCAT7 expression in 6 NSCLC cell lines and a normal oral epidermal cell by qRT-PCR. * P<0.05. ** P<0.01. Data are presented as mean ± standard error based on at least 3 independent experiments.
PCAT7 expression correlates with tumor progression and poor prognosis in NSCLC patients
Data from Table 1 show that PCAT7 expression level is remarkably correlated with TNM stage (p=0.035), histological grade (p=0.009), and lymph node metastasis (p=0.030). There was no connection between PCAT7 expression and sex, age, tumor size, or smoking history (p>0.05). Multivariate analysis of prognostics showed that PCAT7 expression (P=0.002) was negatively correlated with the prognosis of NSCLC patients. TNM stage, histological grade, and lymph node metastasis (Table 2, where P=0.029, 0.012, and 0.041, respectively) had the same negative correlation with the prognosis of NSCLC patients. These results suggest that PCAT7 is an adverse biomarker of prognosis in NSCLC.
Table 1.
Correlation between the PCAT7 and clinical charateristic of NSCLC patients.
| Characteristics | Expression | χ2 | P | |
|---|---|---|---|---|
| Low | High | |||
| Gender | ||||
| Male | 16 | 26 | 0.069 | 0.793 |
| Female | 22 | 32 | ||
| Age (years) | ||||
| <60 | 16 | 20 | 0.569 | 0.451 |
| ≥60 | 22 | 38 | ||
| Tumor size | ||||
| ≤3 cm | 18 | 17 | 3.232 | 0.072 |
| >3 cm | 20 | 41 | ||
| TNM stage | ||||
| I and II | 17 | 41 | 4.455 | 0.035* |
| III and IV | 21 | 44 | ||
| Histological grade | ||||
| Low or middle | 25 | 21 | 6.852 | 0.009* |
| High | 13 | 34 | ||
| Smoking history | ||||
| Never smokers | 14 | 31 | 0.590 | 0.442 |
| Smokers | 30 | 49 | ||
| Lymph node metastasis | ||||
| Negative | 14 | 10 | 4.704 | 0.030* |
| Positive | 24 | 48 | ||
Low/high by the sample median.
Statistically significant (P<0.05);
statistically significant (P<0.01).
Table 2.
Multivariate analysis of prognostic parameters in NSCLC patients by Cox proportional hazards model.
| Variable | Category | HR (95% CI) | P value |
|---|---|---|---|
| Gender | Male | 0.902 (0.374–1.488)) | 0.692 |
| Female | |||
| Age ≥60 (years) | <60 | 1.226 (0.533–1.891) | 0.358 |
| ≥60 | |||
| Tumor size (≥3 cm) | ≤3 cm | 2.259 (0.964–3.710) | 0.053 |
| >3 cm | |||
| TNM stage | I–II | 2.585 (1.145–4.677) | 0.029* |
| III–IV | |||
| Histological grade | Middle or low | 2.762 (1.456–5.889) | 0.012* |
| High | |||
| Smoking history | Never smokers | 0.952 (0.677–1.601) | 0.414 |
| Smokers | |||
| Lymphnode metastasis | Positive | 1.989 (1.094–3.552) | 0.041* |
| Negative | |||
| PCAT7 expression | High | 3.780 (1.372–7.220) | 0.002** |
| Low |
Statistically significant (P<0.05);
statistically significant (P<0.01).
PCAT7 facilitates the proliferation of NSCLC cells
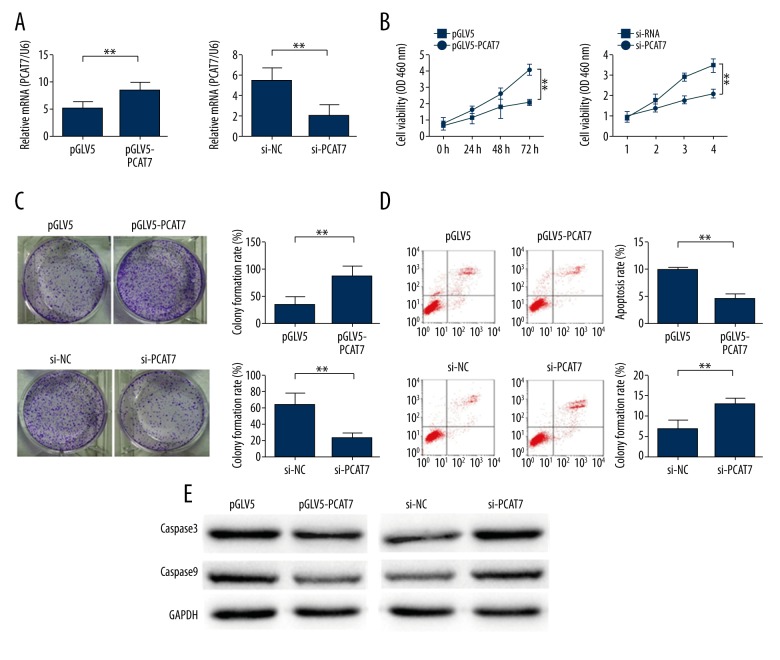
To further understand the oncogenic activity of PCAT7 in NSCLC cells, the PCAT7 expression level in A549 cell was up-regulated or down-regulated through transfection with pGLV5/PCAT7 or PCAT7-specific siRNA, respectively, with the empty vector or control siRNA as a negative control. Satisfactory efficiency was detected after transfecting A549 for 48 h (Figure 2A). As shown in CCK8 assay results, compared with cells transfected with pGLV5 vector, the proliferation ability of PCAT7-transfected cells was increased (p<0.01; Figure 2B). However, compared with pGLV5-transfected cells, poor proliferation ability was detected in PACT7-down-regulated cells (p<0.01; Figure 2B). Furthermore, results from colony formation assays showed that the proliferation of PCAT7-transfected cells was higher than that of pGLV5-transfected cells (p<0.05; Figure 2C). On the contrary, the proliferation in PACT7-down-regulated cells was suppressed compared with NC-transfected cells (p<0.05; Figure 2C). Subsequently, the effect of PCAT7 on apoptosis was assessed using flow cytometry. As shown in Figure 2D, the apoptosis in PCAT7-over-regulated cells was significantly decreased compared with negative controls (p<0.01). In contrast, PCAT7-down-regulation increased apoptosis compared with the NC group. To further understand the effect of PCAT7 on the apoptosis of NSCLC cells, the levels of apoptosis-related proteins (caspase3 and caspase9 proteins) were detected using Western blot analysis. As shown in Figure 2E, forcing the expression of PCAT7 resulted in decreased levels of caspase3 and caspase9, while the down-regulation of PCAT7 significantly increased the expression level of these 2 proteins. Therefore, the apoptosis might decelerate after down-regulated PCAT7 travels through the caspase-3-dependent pathway. These results show that PCAT7 might accelerate proliferation while reducing apoptosis through active caspase-3 in NSCLC cells.
Figure 2.
PCAT7 facilitate the proliferation of NSCLC cells. (A) The relative expression of PCAT7 48h in cells transfected with PCAT7 expression vector pGLV5/PCAT7 (or control) and PCAT7-specific sIRNA (or sIRNA control) detected by qRT-PCR. (B, C) CCK8 and colony formation assays on cells transfected with PCAT7 expression vector pGLV5/PCAT7 (or control) and PCAT7-specific sIRNA (or sIRNA control). (D) Flow cytometry of cells transfected with PCAT7 expression vector pGLV5/PCAT7 (or control) and PCAT7-specific siRNA (or siRNA control). (E) Western blot analysis of caspase-3 and capase-9 levels. * P<0.05. ** P<0.01. Data are presented as mean ± standard error based on at least 3 independent experiments.
PCAT7 promotes metastasis and enhances the epithelial-to-mesenchymal transition of NSCLC cells
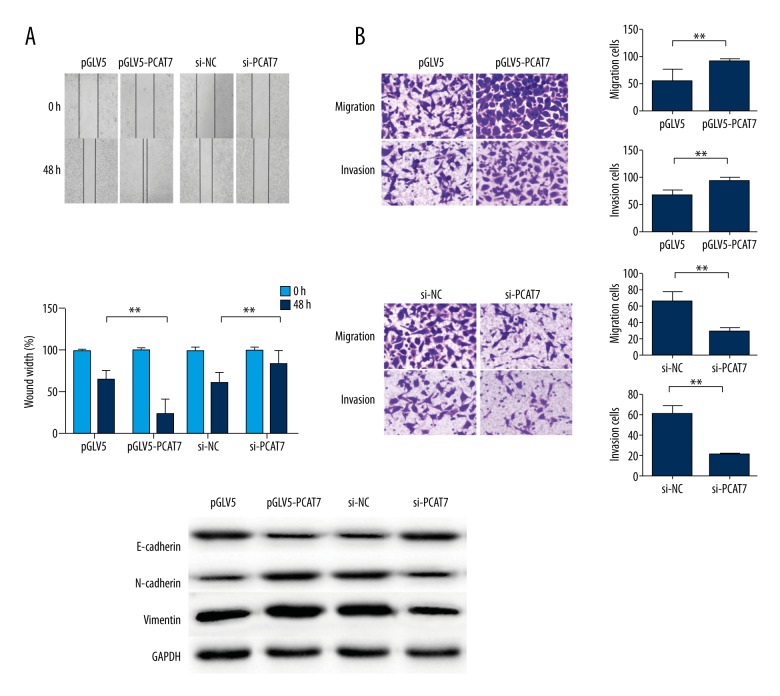
Metastasis is an important characteristic of tumor malignancy. Wound healing assay, as well as migration and invasion assays, were performed to investigate the relationship between PCAT7 and cell motility of NSCLC cell lines. Compared with negative controls, the wound healing ability was observed to accelerate in PCAT7-overexpressed cells, but weaken in PCAT7-underexpressed cells (Figure 3A). Moreover, enhanced migration/invasion capacity was observed in PCAT7-over-regulated NSCLC cells but not the control group (Figure 3B). On the contrary, down-regulation of the expression of PCAT7 reduced the migration/invasion ability (Figure 3B). Furthermore, Western blot assays revealed that a high level of PCAT7 reduced the expression of E-cadherin protein but increased the expression of N-cadherin protein and Vimentin protein, and the down-expression of PCAT7 reversed the EMT process to MET (Figure 3C). These data demonstrate that PCAT7 has a strong connection with NSCLC cell metastasis.
Figure 3.
PCAT7 promotes metastasis and enhances the epithelial-tomesenchymal transition of NSCLC cells. (A) Wound healing assay showed that PACT7 accerlated migration ability (×50); (B) Transwell migration assay and Matrigel invasion assay showed that NY-SAR-35 promoted migration/invasion capacity (×50); (C) Western blot analysis of E-cadherin, N-cadherin, and Vimentin expression levels. The columns show the mean for 3 separate experiments; bars, sd. * P<0.05. ** P<0.01
miR-134-5p is a target of PCAT7 and regulates the function of PCAT7 in NSCLC cells
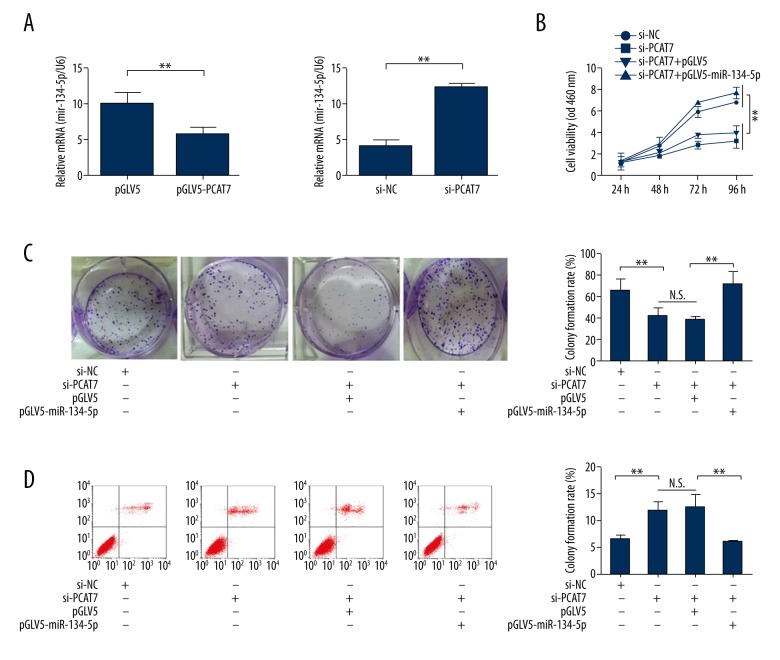
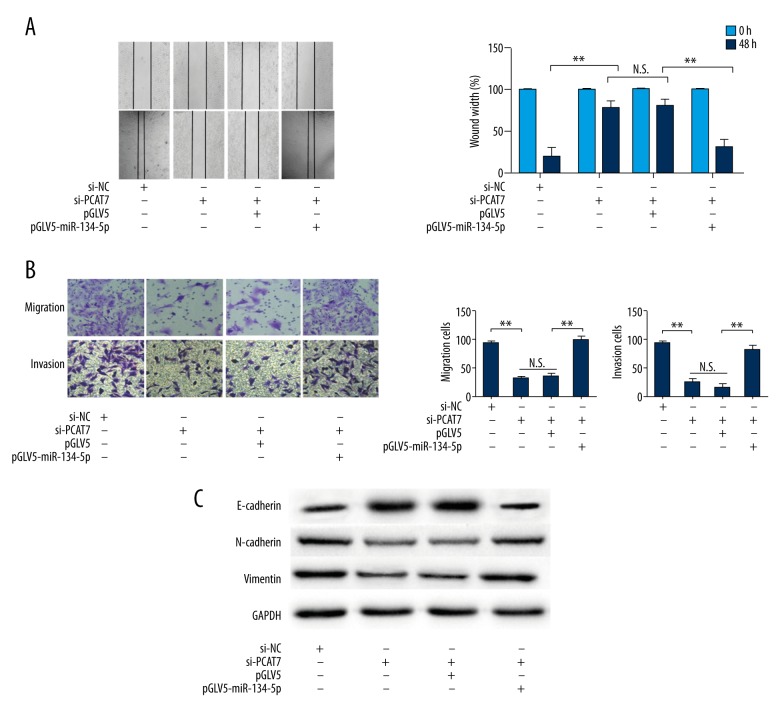
Since it has been made clear that miR-134-5P is a target of PCAT7 [8], the expression level of miR-134-5p can be measured in response to the level of PCAT7. Decreased expression of miR-134-5p was detected in PACT7-over-regulated NSCLC cells, while down-regulated PACT7 increased the expression of miR-134-5p (Figure 4A). Subsequently, rescue assays were performed to reveal the moderating effect of miR-134-5p on PCAT7 in NSCLC cells. The rescued proliferation ability of NSCLC cells after co-transfection with si-PCAT7 and miR-134-5P was detected by CCK8 assays and colony formation assays (Figure 4B, 4C). In addition, the rescued pro-apoptosis function of si-PCAT7 in NSCLC cells was found by flow cytometry analyses with introduction of miR-134-5p (Figure 4D). Additionally, as shown in Figure 5A, 5B, miR-134-5P partially rescued the anti-metastasis function of si-PCAT7. Furthermore, the MET induced by si-PCAT7 was reversed to EMT after introduction of miR-134-5P (Figure 5C). These data indicate that miR-134-5P regulates the function of PCAT7 in NSCLC cells.
Figure 4.
miR-134-5p was the target of PCAT7 and induced the proliferation in NSCLC cells. (A) The relative expression of miR-134-5p in response to the level of PCAT7 was detected by qRT-PCR. (B, C) CCK8 and colony formation assays on cells co-transfected with si-PCAT7 and miR-134-5p. (D) Flow cytometry of cells co-transfected with si-PCAT7 and miR-134-5p. Error bars represent the mean ±SEM of at least 3 independent experiments. * p<0.05, ** p<0.01.
Figure 5.
miR-134-5p was the target of PCAT7 and induced the metastasis in NSCLC cells. (A) Wound healing assay showed that miR-134-5p inhibited migration ability induced by si-PCAT7(×50); (B) Transwell migration assay and Matrigel invasion assay showed that N miR-134-5p restored migration ability restrained by si-PCAT7 (×50); (C) Western blot analysis of E-cadherin, N-cadherin, and Vimentin expression levels. Error bars represent the mean ±SEM of at least 3 independent experiments. * p<0.05, ** p<0.01.
Discussion
Lung cancer is a major cause of death worldwide, accounting for 19.4% of all cancer-related deaths [10]. NSCLC accounts for 80% of lung cancers [10] and the majority of cancer-related deaths worldwide. Recently, numerous studies have confirmed that lncRNAs, as a member of the non-coding RNAs family, shows great potential in regulation of biological processes in lung cancer [11]. However, the relationship between lncRNAs and NSCLC tumorigenesis is still basically unknown. This research is the first to analyze the expression level of P7CAT in NSCLC tissues and paired normal lung tissues, indicating that the expression level of PCAT7 was dramatically increased in NSCLC tissues and cell lines compared with normal lung tissues and 16-HBE, respectively. Aberrant expression of PCAT7 was closely associated with TNM stage (p=0.035), histological grade (P=0.009), and lymph node metastasis (p=0.03), and high PCAT7 expression was an adverse prognostic factor (P=0.002). It was also found that PCAT7, as a carcinogen in NSCLC, can promote cell proliferation, inhibit cell apoptosis, accelerate cell migration/invasion, and induce EMT. Our study shows that PCAT7 exert its function on NSCLC mainly by targeting miR-134-5P. PCAT7 may act as an oncogene and a key regulator in the prognosis and treatment of NSCLC.
Increasing numbers of studies have confirmed that lncRNAs, which were initially thought to be “transcriptional noise” [12], regulate gene expression through numerous mechanisms in lung cancer. Specifically, metastasis, which is associated with lung adenocarcinoma transcript 1 (MALAT1), as an oncogene, may achieve its effect not only by modulating activity of the downstream mediator P53 [13], but also by interacting with polycomb 2 (PC2) [14]. Hox transcript antisense intergenic RNA (HOTAIR) has been reported to be able to link with LSD1/CoREST/REST complexes and PRC2 to drive them to occupy their gene target regions, which results in target gene silencing [15,16]. It also has been reported that HOTAIR can regulate downstream genes via epigenetic modification [17]. Moreover, Horie et al. confirmed that PCAT7 plays a key role in hypomethylation in NSCLC cell lines [18]. Among all these mechanisms, the main function of lncRNA is to regulate the degradation of miRNA targets by acting as a ceRNA sponge [19,20]. Fu et al. reported that HOTAIR can mediate hepatocarcinogenesis through suppressing miRNA-218 expression and activating P14 and P16 signaling [19]. Li et al. found that LncRNA MALAT1 can exert oncogenic effects on lung adenocarcinoma by targeting miR-204 [21]. Our results identified that miR-134-5P, as the potential target gene of PCAT7, can positively regulate migration and invasion in si-PCAT7-transfected cells, while reversing the MET progress. These results indicate that miR-134-5P may serve as a potential target gene of PCAT7 and regulate metastasis in NSCLC.
EMT has a pivotal role in the initiation of metastasis, a process in which epithelial cells lose adhesion and cytoskeletal components, concomitant with a gain of mesenchymal components and the initiation of a migratory phenotype [22,23]. Recent publications have showed that BRAF and BC087858 unequivocally augment lung cancer cell motility and invasiveness in vitro [24,25]. In addition, LINC01133, a long non-coding RNA, can inhibit metastasis and epithelial-to-mesenchymal transition in colorectal cancer by interacting with its target gene SRSF6 [26]. This study detected the changes of some most representative EMT markers in cells that are transfected with pGLV5-PCAT7 (pGLV5 as control) and si-PCAT7 (siRNA as control), which indicate that PCAT7 can promote cell migration and invasion in NSCLC cells, partly by regulating the EMT pathway.
Although we have revealed that miR-134-5p is a potential target gene of PCAT7 in NSCLC cells, our research still has limitations. We need to define the exact regulatory mechanism between miR-134-5p and PCAT7 in NSCLC. In addition, we need investigate the relationship of PACT7 and chemo-resistance in NSCLC cells, and our ultimate goal is finding a new therapeutic target in NSCLC.
In summary, our results reveal the potential of PCAT7 as a new prognostic indicator in NSCLC. PCAT7, which has been over-regulated in NSCLC tissues and cell lines, can accelerate proliferation, prohibit apoptosis, and promote cell migration/invasion by regulating the EMT pathway. We also found that miR-134-5p is a potential target gene of PCAT7 in NSCLC cells. These results suggest that PCAT7 has potential to be applied in the prognosis and treatment of NSCLC.
Conclusions
Results of this study reveal the potential of PCAT7 as a new prognostic indicator in NSCLC, which can induce poor prognosis and promote tumorigenesis by inhibiting miR-134-5p in NSCLC.
Footnotes
Source of support: This study was supported by the Medical Scientific Technical Research Foundation of Guangdong Province, China (A2017355)
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. Cancer J Clin. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Han W, Du X, Wang J, et al. SNHG16 indicates a poor prognosis and affects cell proliferation, migration and invasion in non-small cell lung cancer. Exp Cell Res. 2017 doi: 10.1016/j.yexcr.2017.09.027. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 3.Ramos AD, Attenello FJ, Lim DA. Uncovering the roles of long noncoding RNAs in neural development and glioma progression. Neurosci Lett. 2016;625:70–79. doi: 10.1016/j.neulet.2015.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21(11):1253–61. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 5.Wang R, Chen X, Xu T, et al. MiR-326 regulates cell proliferation and migration in lung cancer by targeting phox2a and is regulated by HOTAIR. Am J Cancer Res. 2016;6(2):173–86. [PMC free article] [PubMed] [Google Scholar]
- 6.Li J, Li Z, Zheng W, et al. PANDAR: A pivotal cancer-related long non-coding RNA in human cancers. Mol Biosyst. 2017;13(11):2195–201. doi: 10.1039/c7mb00414a. [DOI] [PubMed] [Google Scholar]
- 7.Li Z, Shen J, Chan MT, Wu WK. TUG1: A pivotal oncogenic long non-coding RNA of human cancers. Cell Prolif. 2016;49(4):471–75. doi: 10.1111/cpr.12269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Y, Tao Z, Qu J, et al. Long non-coding RNA PCAT7 regulates ELF2 signaling through inhibition of miR-134-5p in nasopharyngeal carcinoma. Biochem Biophys Res Commun. 2017;491(2):374–81. doi: 10.1016/j.bbrc.2017.07.093. [DOI] [PubMed] [Google Scholar]
- 9.Horie M, Kaczkowski B, Ohshima M, et al. Integrative CAGE and DNA methylation profiling identify epigenetically regulated genes in NSCLC. Mol Cancer Res. 2017;15(10):1354–65. doi: 10.1158/1541-7786.MCR-17-0191. [DOI] [PubMed] [Google Scholar]
- 10.Sang H, Liu H, Xiong P, Zhu M. Long non-coding RNA functions in lung cancer. Tumour Biol. 2015;36(6):4027–37. doi: 10.1007/s13277-015-3449-4. [DOI] [PubMed] [Google Scholar]
- 11.Liz J, Esteller M. lncRNAs and microRNAs with a role in cancer development. Biochim Biophys Acta. 2016;1859(1):169–76. doi: 10.1016/j.bbagrm.2015.06.015. [DOI] [PubMed] [Google Scholar]
- 12.Ponjavic J, Ponting CP, Lunter G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 2007;17(5):556–65. doi: 10.1101/gr.6036807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tripathi V, Shen Z, Chakraborty A, et al. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS Genet. 2013;9(3):e1003368. doi: 10.1371/journal.pgen.1003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gutschner T, Hämmerle M, Eissmann M, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73(3):1180–89. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang J, Zhang P, Wang L, et al. Long non-coding RNA HOTAIR in carcinogenesis and metastasis. Acta Biochim Biophys Sin (Shanghai) 2014;46(1):1–5. doi: 10.1093/abbs/gmt117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsai MC, Manor O, Wan Y, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329(5992):689–93. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qiu MT, Hu JW, Yin R, Xu L. Long noncoding RNA: An emerging paradigm of cancer research. Tumour Biol. 2013;34(2):613–20. doi: 10.1007/s13277-013-0658-6. [DOI] [PubMed] [Google Scholar]
- 18.Horie M, Kaczkowski B, Ohshima M, et al. Integrative CAGE and DNA methylation profiling identify epigenetically regulated genes in NSCLC. Mol Cancer Res. 2017;15(10):1354–65. doi: 10.1158/1541-7786.MCR-17-0191. [DOI] [PubMed] [Google Scholar]
- 19.Fu WM, Zhu X, Wang WM, et al. Hotair mediates hepatocarcinogenesis through suppressing miRNA-218 expression and activating P14 and P16 signaling. J Hepatol. 2015;63(4):886–95. doi: 10.1016/j.jhep.2015.05.016. [DOI] [PubMed] [Google Scholar]
- 20.Ma MZ, Chu BF, Zhang Y, et al. Long non-coding RNA CCAT1 promotes gallbladder cancer development via negative modulation of miRNA-218-5p. Cell Death Dis. 2015;6:e1583. doi: 10.1038/cddis.2014.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li J, Wang J, Chen Y, et al. LncRNA MALAT1 exerts oncogenic functions in lung adenocarcinoma by targeting miR-204. Am J Cancer Res. 2016;6(5):1099–107. [PMC free article] [PubMed] [Google Scholar]
- 22.Yang P, Huo Z, Liao H, Zhou Q, et al. Cancer/testis antigens trigger epithelial-mesenchymal transition and genesis of cancer stem-like cells. Curr Pharm Des. 2015;21(10):1292–300. doi: 10.2174/1381612821666141211154707. [DOI] [PubMed] [Google Scholar]
- 23.Yao D, Peng S, Dai C. The role of hepatocyte nuclear factor 4alpha in metastatic tumor formation of hepatocellular carcinoma and its close relationship with the mesenchymal-epithelial transition markers. BMC Cancer. 2013;13:432. doi: 10.1186/1471-2407-13-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pan H, Jiang T, Cheng N, et al. Long non-coding RNA BC087858 induces non-T790M mutation acquired resistance to EGFR-TKIs by activating PI3K/AKT and MEK/ERK pathways and EMT in non-small-cell lung cancer. Oncotarget. 2016;7(31):49948–60. doi: 10.18632/oncotarget.10521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun M, Liu XH, Wang KM, et al. Downregulation of BRAF activated non-coding RNA is associated with poor prognosis for non-small cell lung cancer and promotes metastasis by affecting epithelial-mesenchymal transition. Mol Cancer. 2014;13:68. doi: 10.1186/1476-4598-13-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kong J, Sun W, Li C, et al. Long non-coding RNA LINC01133 inhibits epithelial-mesenchymal transition and metastasis in colorectal cancer by interacting with SRSF6. Cancer Lett. 2016;380(2):476–84. doi: 10.1016/j.canlet.2016.07.015. [DOI] [PubMed] [Google Scholar]