Here, Joo et al. provide new insights into the function of transcription factor TFIID. Their findings suggest a new model in which TAFs function as reinitiation factors, accounting for the differential responses of promoters to various transcription factor mutations.
Keywords: TFIID, TBP-associated factors, transcription activation, bromodomain, histone acetylation, Taf1
Abstract
TFIID binds promoter DNA to recruit RNA polymerase II and other basal factors for transcription. Although the TATA-binding protein (TBP) subunit of TFIID is necessary and sufficient for in vitro transcription, the TBP-associated factor (TAF) subunits recognize downstream promoter elements, act as coactivators, and interact with nucleosomes. In yeast nuclear extracts, transcription induces stable TAF binding to downstream promoter DNA, promoting subsequent activator-independent transcription reinitiation. In vivo, promoter responses to TAF mutations correlate with the level of downstream, rather than overall, Taf1 cross-linking. We propose a new model in which TAFs function as reinitiation factors, accounting for the differential responses of promoters to various transcription factor mutations.
Transcription by RNA polymerase II (RNAPII) requires a set of basal initiation factors that recognize core promoter sequences, position the polymerase appropriately, catalyze promoter melting, and mediate the transition from preinitiation complex (PIC) to stable elongation complex (EC) (Thomas and Chiang 2006). Building on decades of biochemistry and molecular genetics, recent structural studies are providing insight into how these factors fit and function together (Hahn and Buratowski 2016). However, some aspects of initiation factor function remain unclear. In particular, the basal factor TFIID has been the subject of much study and debate.
Early template commitment and DNase I footprinting experiments identified TFIID as the factor that first binds promoter DNA. It contacts the TATA box as well as DNA downstream near the transcription start site (TSS) and beyond (Nakajima et al. 1988). A single small protein, designated TATA-binding protein (TBP), is necessary and sufficient for PIC assembly and TFIID transcription activity in vitro (Buratowski et al. 1988, 1989). However, some fraction of TBP in cells is associated with roughly a dozen additional proteins, called TBP-associated factors (TAFs) (for review, see Thomas and Chiang 2006). While not required for in vitro transcription, TAFs are generally essential for cell and organism viability. Multiple activities have been proposed for TAFs.
One TAF function is to contact promoter DNA. Very short elements around the TSS and 20–30 base pairs (bp) downstream can contribute to promoter strength (Kadonaga 2012). These may affect RNAPII, TFIIH, or TFIID binding, but the effect of these elements is strongest when there is a weak nonconsensus TATA box. The TFIID DNase I footprint on some promoters extends much further downstream than TBP alone, and several TAFs cross-link at the TSS and beyond (Horikoshi et al. 1988a,b; Sypes and Gilmour 1994; Burke and Kadonaga 1997; Auty et al. 2004; Lee et al. 2005). A recent cryo-electron microscopy (cryo-EM) structure shows Taf1, Taf2, and Taf7 directly interacting with the TSS and +20 to +30 regions (Louder et al. 2016). Therefore, a subset of TAFs likely mediates extended promoter recognition.
TAFs have also been implicated in transcription activation. In vitro transcription with TBP supports basal transcription, but response to activators is often potentiated by TFIID. At some promoters, DNase I protection by TFIID downstream from the TATA box is seen only in the presence of an activator, suggesting that TFIID not only is a target for activators but may undergo activator-induced conformation changes (Horikoshi et al. 1988a,b; Chi and Carey 1996). The TAF “coactivator” model became controversial when early experiments in yeast showed that some genes continued expressing after TAFs were inactivated (summarized in Hahn 1998). In addition, compelling genetic and biochemical data showed that many activators function by contacting and recruiting other factors. Activator targets include histone acetyltransferases (HATs) and chromatin remodelers that make promoter DNA more accessible as well as the Mediator–RNAPII complex that facilitates PIC assembly. Nevertheless, both in vitro and in vivo experiments argue that some activators function via direct contacts with specific TAF subunits (Albright and Tjian 2000; Mencía et al. 2002; Papai et al. 2010; Chen et al. 2013).
Over time, extensive analyses produced a consensus that at least 90% of yeast genes show reduced expression upon TAF inactivation (Lee et al. 2000; Huisinga and Pugh 2004; Warfield et al. 2017). The most affected promoters tend to have weaker, nonconsensus TATA elements and are more likely to be constitutively expressed. The small minority of promoters that are relatively resistant to TAF mutations is strongly affected by mutations in the histone H3 acetyltransferase SAGA, although SAGA is clearly a general coactivator (Baptista et al. 2017). These genes generally have a strong consensus TATA and are more likely to be highly induced under specific conditions. While often oversimplified to categorize each gene as dependent on only one or the other complex, the data actually show combinatorial effects of double mutants at essentially all promoters (Huisinga and Pugh 2004; Bonnet et al. 2014; Grünberg et al. 2016; Baptista et al. 2017; Warfield et al. 2017). To reflect this subtlety, promoters are often referred to as TFIID-dominant or SAGA-dominant, although this categorization does not fully reflect the continuum of promoter responses. Importantly, many activators can function at both promoter classes, arguing that it is not simply the specific activator that determines response to TAFs (de Jonge et al. 2017).
TAFs also appear to mediate communication between TFIID and nucleosomes. Several TAFs contain histone fold domains, although it remains unclear whether they form a structure resembling a nucleosome within TFIID or interact with actual histone proteins (Selleck et al. 2001; Bieniossek et al. 2013). The C-terminal half of Taf1 in higher eukaryotes has two bromodomains and an extended C-terminal domain; i.e., a BET module. In Saccharomyces cerevisiae, the BET module is encoded by the BDF1 gene rather than being fused to Taf1 (Matangkasombut et al. 2000). The Bdf1 bromodomains bind specifically to acetylated histone H4 tails (Ladurner et al. 2003; Matangkasombut and Buratowski 2003). It remains unclear whether and how this interaction affects transcription.
Current models for TAF function are premised on TFIID acting before transcription initiation. However, while carrying out a proteomic analysis of RNAPII ECs, we unexpectedly discovered a striking transcription-dependent recruitment of TAFs, but not TBP or other basal factors, to DNA well downstream from the TATA box. In vitro transcription reactions suggest that this post-initiation TAF complex promotes additional rounds of transcription independently of an upstream activator. The in vivo relevance of these downstream interactions is supported by ChIP-exo (chromatin immunoprecipitation [ChIP] using λ exonuclease to digest transcription factor-unbound DNA after ChIP) data showing a downstream-extended TFIID footprint specifically on TFIID-dominant genes. Based on these data, we propose a new function for the TAF complex as an activator-independent reinitiation factor. This model explains some earlier confusing data about TFIID function and provides a simple explanation in which the level of TFIID dominance is determined by how efficiently a promoter supports TAF-dependent reinitiation.
Results
Transcription-dependent recruitment of TAFs to downstream DNA
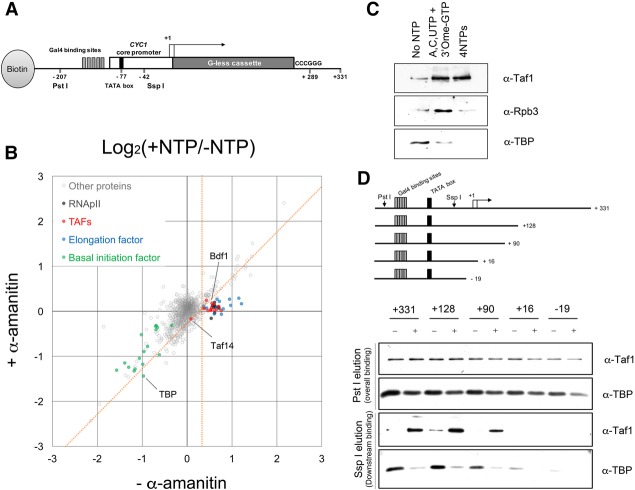
We and others have used quantitative mass spectrometry to analyze RNAPII initiation complexes assembled in vitro using bead-immobilized templates (Ranish et al. 2004; Sikorski et al. 2012). We extended this approach to ECs (YJ Joo, SB Ficarro, Y Chun, JA Marto, and S Buratowski, in prep.). DNA templates containing five Gal4-binding sites and the CYC1 core promoter upstream of a G-less cassette were immobilized on beads and incubated with nuclear extract. In the presence of ATP, UTP, CTP, and the chain terminator 3′-O-me-GTP, ECs extend to the end of the G-less cassette and stall. Digestion with restriction enzyme Ssp I cuts 30 bp downstream from the TATA box to release ECs bound to downstream DNA, while any remaining PIC components stay associated with upstream DNA on the bead (Fig. 1A).
Figure 1.
Transcription-mediated TAF association with downstream DNA. (A) Schematic of the transcription template used to isolate RNAPII ECs as described in the Materials and Methods. Downstream bound proteins are eluted using Ssp I endonuclease, which cuts 35 nucleotides downstream from the TATA box but upstream of the first TSS. Total proteins are eluted with Pst I (see Supplemental Fig. S1D). (B) Scatter plot comparing NTP-dependent protein binding to downstream DNA (log2 of +NTP/−NTP ratio) in the presence (Y-axis) or absence (X-axis) of α-amanitin. Each circle represents one protein (1511 total), with the value calculated from the sum of all peptide signals identified for that protein. Proteins in specific subgroups are color-coded as noted. Proteins specifically dependent on transcription are defined as those at the right of the vertical orange line (95% confidence level for enrichment with NTPs) on the X-axis and below the diagonal orange line (95% confidence level cutoff for reduction of NTP enrichment by α-amanitin) (see also Supplemental Fig. S1A–C; Supplemental Table S1). (C) Immunoblot of proteins eluted from immobilized templates with Ssp I before treatment with NTPs or after 15 min of incubation with A, C, and UTP plus 3′O-meGTP (to stall ECs) or all four NTPs to allow RNAPII runoff (see also Supplemental Fig. S2B). (D) TBP and Taf1 binding on immobilized templates of different lengths. The top panel shows a schematic of the template series analyzed as in B. The middle panels show immunoblots of total bound proteins eluted with Pst I, while the bottom panels show downstream binding proteins eluted with Ssp I (see also Supplemental Fig. S2A,C).
Quantitative mass spectrometry revealed multiple proteins associated with downstream DNA specifically in the presence of NTPs. To distinguish which were dependent on transcription rather than simply NTP hydrolysis, a parallel reaction with the elongation inhibitor α-amanitin was analyzed. Statistically significant enrichment (Fig. 1B; Supplemental Fig. S1A–C; Supplemental Table S1) of RNAPII subunits (Fig. 1B, black circles; Supplemental Fig. S1A–C, black circles) and known elongation factors (which include PAF subunits, Spt5, Spt6–Spn1, and Set2) (Fig. 1B, blue circles; Supplemental Fig. S1A–C, blue circles) showed that bona fide ECs were isolated. A detailed description of these proteomic results will appear elsewhere (YJ Joo, SB Ficarro, Y Chun, JA Marto, and S Buratowski, in prep.).
We were surprised to also detect very clear transcription-dependent enrichment of all known TAF proteins (Fig. 1B, red circles), including Bdf1, on the released downstream DNA. The one exception was Taf14/Tfg3, a YEATS domain protein also found associated with several other yeast transcription factors but not mammalian or Drosophila TFIID (Thomas and Chiang 2006). Enrichment of all Tafs except Taf14 and Taf10 (a small protein in both TFIID and SAGA) was blocked by α-amanitin (Fig. 1B; Supplemental Fig. S1C). Surprisingly, TBP behavior differed from that of TAFs. The signals for downstream binding of TBP and other basal transcription factors (likely PICs formed at weak cryptic TATAs in the G-less cassette) were strongly reduced by NTPs even when transcription was inhibited (Fig. 1B, green circles).
Many results argue that TAFs do not associate with ECs. First, cross-linking experiments detect TAFs near promoters but not downstream (Rhee and Pugh 2012). Second, the curated literature (http://www.yeastgenome.org) contains no reports of TAF mutants exhibiting 6-aza-uracil or mycophenolic acid sensitivity—phenotypes often associated with elongation defects. Finally, affinity purifications of TAFs and elongation factors failed to reveal physical association (Gavin et al. 2006; Krogan et al. 2006). To verify that TAFs are not associated with ECs in our system, we compared a reaction with U, C, and ATP plus 3′-O-me-GTP (where ECs stall at the end of the G-less cassette) with a reaction with all four NTPs, allowing elongation off the end of the template (Fig. 1C). In contrast to RNAPII, Taf1 remains associated under both conditions, showing TFIID independence from the EC.
Additional experiments further characterized the post-initiation TAF binding. Similar downstream TAF enrichment was seen on naked and chromatinized templates (Supplemental Fig. S1E). Increased TAF binding was also seen, albeit usually less pronounced, when the entire chimeric CYC1/G-less cassette template was released by Pst I digestion (Fig. 1D; Supplemental Fig. S1D) or when total proteins were eluted by boiling (Supplemental Fig. S2A). Therefore, while some TAFs likely transfer from upstream promoter regions, some may also be recruited de novo. Similar Taf1 increases were seen on several natural RNAPII transcribed promoters but not on the RNAPI transcribed ribosomal DNA (rDNA) promoter (Supplemental Fig. S2A). Importantly, NTPs produced far more Taf1 binding than ATP alone, further suggesting transcription-dependent TAF recruitment (Supplemental Fig. S2B).
A series of template deletions was tested to map which sequences were important. To rule out artifactual association of TAFs with the G-less cassette, we tested 5′ deletions, removing promoter sequences up to the SspI site or to the beginning of the G-less cassette (Supplemental Fig. S2C). Both truncations completely abrogated downstream TAF recruitment, showing promoter dependence. Finally, a series of shorter immobilized templates showed that 90 nucleotides downstream from the TSS supported post-transcription TAF binding, while 16 was insufficient (Fig. 1D). This is consistent with footprinting assays showing that TAFs interact with the TSS and regions just downstream. Altogether, our biochemical data suggest a post-initiation conformation of TFIID stably bound to downstream promoter DNA even after separation from the TATA box.
In vivo evidence for downstream TAF binding
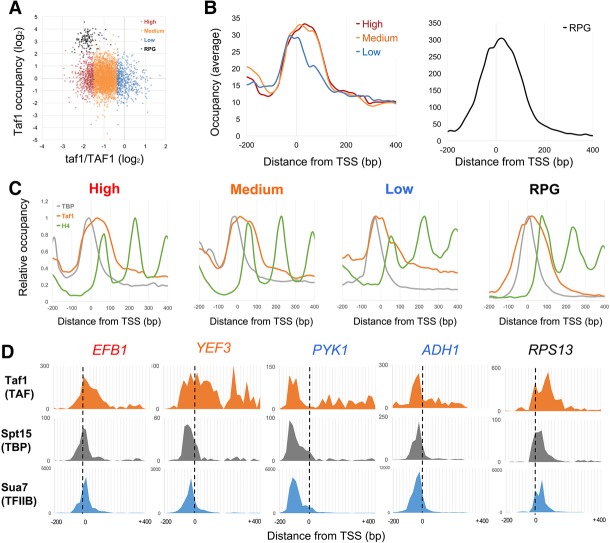
To determine the physiological relevance of downstream TAF binding, we looked for correlations between TAF occupancy and TAF dependence. TAF occupancy was defined as the number of Taf1 ChIP-exo reads (Rhee and Pugh 2012) from −110 to +290 relative to the TSS. TAF dependence was defined as the difference in expression in two different taf1 temperature-sensitive alleles (Supplemental Fig. S3A) versus TAF1 (Huisinga and Pugh 2004). The data showed that ribosomal protein genes (RPGs) were well separated from the other mRNA genes (Fig. 2A). While RPGs have very high TAF occupancy and strong TAF dependence, there was surprisingly little correlation for the other mRNA genes. Therefore, we instead used standard deviation (SD) from the mean to put genes into four categories by their response to taf1 temperature inactivation: RPGs (Fig. 2A, black), high (top 825 genes) (Fig. 2A, red), medium (middle 4327 genes) (Fig. 2A, orange), or low (bottom 729 genes) (Fig. 2A, blue). Note that these categories are not meant to imply clearly discrete groups but only represent extremes along a spectrum.
Figure 2.
Taf1 downstream binding correlates with its transcription function. (A) Scatter plot of Taf1 transcription dependence versus total occupancy. For 4348 scored mRNA genes, the taf1/TAF1 mRNA expression level ratios from Huisinga and Pugh (2004) were plotted in log2 scale on the X-axis. For each promoter, total Taf1 occupancy was plotted by summing ChIP-exo counts from −100 bp to +300 bp relative to the TSS (Rhee and Pugh 2012), dividing by the average for all mRNA gene promoters, and plotting in log2 scale on the Y-axis. Excluding the 137 RPGs, high TAF dependence genes (825) were defined as those with taf1/TAF1 expression ratios below −1 SD (SD = 0.57) from the mean of all gene ratios (−0.95). Medium dependence genes (4327) had ratios between −1 and +1 SD, and low dependence genes (729) had more than +1 SD from the mean of ratios. Average ratios in high, medium, low, and RPG groups were −1.73, −0.92, −0.08, and −1.82, respectively (see also Supplemental Fig. S3A). (B) Averaged Taf1 occupancy for the four groups was plotted relative to the TSS (high [red], medium [orange], low [blue], and RPG [black]) using ChIP-exo data from Rhee and Pugh (2012) (see also Supplemental Fig S3B,C). (C) Occupancies for TBP, Taf1, and nucleosomes were plotted for the four groups separately to compare peak positions. (Gray) TBP; (orange) Taf1; (green) histone H4. For each curve, data were normalized by setting the maximum value to 1.0. (D) Individual gene examples of Taf1 (TAF), Spt15 (TBP), and Sua7 (TFIIB) profiles at EFB1 (high TAF1 dependence, −2.02), YEF3 (medium TAF1 dependence, −1.21), PYK1 (low TAF1 dependence, 0.02), ADH1 (low TAF1 dependence, 0.35), and RPS13 (RPG; TAF1 dependence, −1.00).
In vivo evidence for differential downstream promoter interactions of TAFs came from re-examining high-resolution ChIP-exo data from Rhee and Pugh (2012). They sorted genes by Taf1 occupancy and observed that low occupancy genes had Taf1 cross-linking patterns similar to the narrow peak seen for TBP and TFIIB upstream of the TSS. In contrast, high occupancy genes showed additional cross-linking overlapping the +1 nucleosome. We repeated this analysis using the four categories defined above. In plots of averaged values, Taf1 cross-links appear to have three peaks: one overlapping TBP and TFIIB, one near the TSS, and one overlapping the +1 nucleosome (Fig. 2B,C). Downstream peaks are also seen for Taf2 and Bdf1 but not several other TAFs (Supplemental Fig. S3B). Notably, Drosophila Taf2 also cross-links predominantly downstream from the TSS (Shao and Zeitlinger 2017). These results are consistent with a recent structural model of TFIID showing a downstream lobe consisting of Taf1, Taf2, and Taf7 (Supplemental Fig. S3B; Louder et al. 2016).
Average values of Taf1 overlapping the TBP peak are roughly equal in high, medium, or low TAF dependence classes (Fig. 2B). However, promoters with medium or high TAF dependence show the highest levels of Taf1 cross-linking near and downstream from the TSS in averaged plots (Fig. 2C) as well as when ratios of downstream to upstream cross-linking are calculated for individual genes (Supplemental Fig. S3C). Although RPG promoters have roughly 10-fold higher overall Taf1 cross-linking, their normalized profile is most similar to the high TAF dependence promoters. These trends can also be seen in individual gene traces (Fig. 2D). Therefore, the response of promoters to TAF1 mutations is correlated not with the absolute level of Taf1 binding but instead with downstream promoter interactions of TFIID.
Downstream TFIID binding correlates with Bdf1 and acetylated histone H4
We previously isolated Bdf1 as a Taf7-interacting protein corresponding to the BET module of mammalian Taf1 (Matangkasombut et al. 2000). Bdf1 was also identified as a subunit of the SwrC complex, which replaces H2A with H2A.Z/Htz1 (Krogan et al. 2003; Kobor et al. 2004; Mizuguchi et al. 2004). The Bdf1 bromodomains interact with acetylated H4 tails (Ladurner et al. 2003; Matangkasombut and Buratowski 2003). As histone H4 acetylation (H4ac) is highest at +1 nucleosomes of active promoters, Bdf1 and other BET proteins are thought to recruit their associated complexes to TSS regions.
Since Bdf1 behaved like TAFs, but not SwrC, in our immobilized template system, we asked which complex its behavior most closely resembled in vivo. ChIP-seq (ChIP combined with high-throughput sequencing) for TBP, Taf1, histone H3, and H4ac was performed. Consistent with their high transcription rate, RPG promoters have the lowest H3 occupancy but the highest H4ac (Supplemental Fig. S4A). Average H3 levels in the other three gene classes were similar, and low TAF dependence genes had notably less H4ac than high or medium dependence genes. These results were aligned (Fig. 3A) with previously published data sets for TFIIB, Bdf1, Swr1, and Htz1 (Rhee and Pugh 2012; Yen et al. 2013; Gu et al. 2015). Genes were sorted into the four classes of TAF dependence and then further within each category by the Taf1 downstream cross-linking signal from +30 to +170 relative to the TSS. Bdf1 correlated with Taf1 and TBP better than Swr1 and Htz1 (Supplemental Fig. S4B). Taf1 and Bdf1 were particularly high at RPGs, while Swr1 and Htz1 were largely absent (Fig. 3A; Supplemental Fig. S4B). H4ac correlated best with Bdf1, with both Swr1 and Taf1 also showing some correlation. Importantly, TBP correlated well with Taf1 and Bdf1 on TAF-dependent genes, while genes with low TAF dependence had high TBP levels even when Taf1 levels were low (Fig. 3A; Supplemental Fig. S4B).
Figure 3.
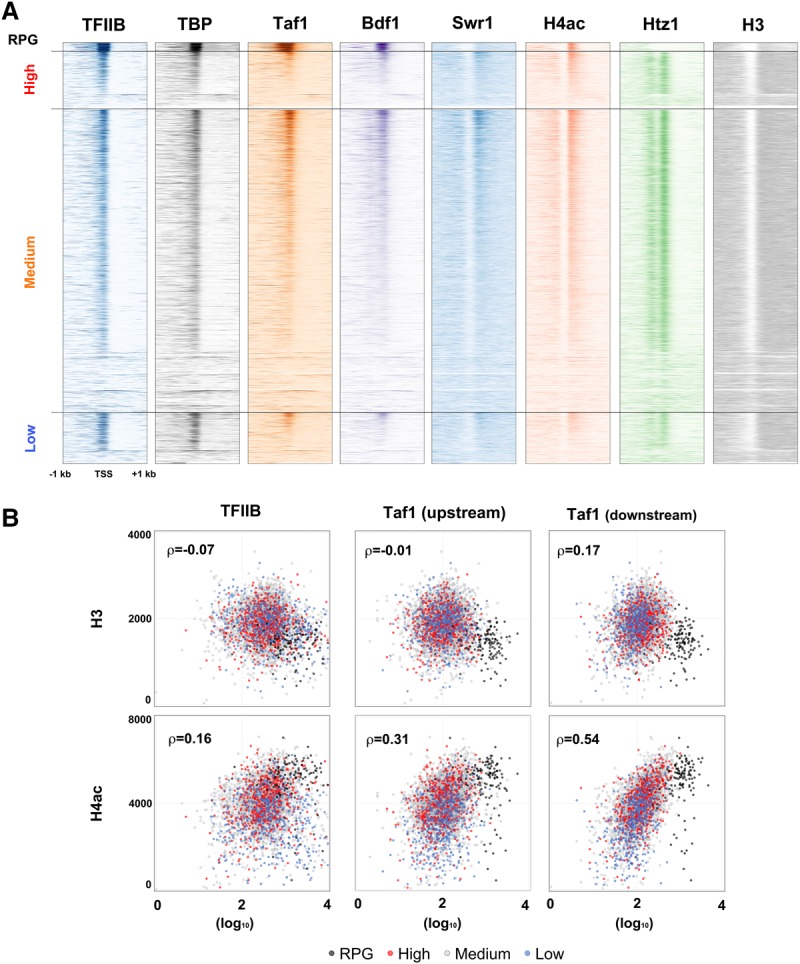
Downstream Taf1 binding correlates with +1 nucleosome H4ac. (A) ChIP patterns of TFIIB, TBP, Taf1, Bdf1, Swr1, H4ac, Htz1, and H3 at individual genes were plotted as horizontal lines to generate heat maps, with darker color signifying higher levels. Genes were sorted into the four TAF1 dependence groups and then within each group by Taf1 downstream occupancy (ChIP-exo reads from +20 bp to +160 bp relative to the TSS). Together with our newly generated data, raw data for TFIIB (Wong et al. 2014), Bdf1 (Rhee and Pugh 2012), Swr1 (Yen et al. 2013), and Htz1 (Gu et al. 2015) were reanalyzed as in the Materials and Methods (see also Supplemental Fig. S4A). (B) H3 and H4ac levels were plotted versus TFIIB and Taf1 binding. Occupancies of H3 and H4ac were determined as the sum of ChIP-seq reads from +30 bp to +170 bp relative to the TSS, encompassing the +1 nucleosome. Occupancies of Taf1 and TFIIB were determined as total ChIP-exo reads (Rhee and Pugh 2012) from −140 bp to −1 bp for the upstream core promoter and +20 bp to +160 bp for the downstream promoter. Each dot represents one mRNA gene (total 4720). For each graph, Spearman's rank correlation coefficient (ρ) is shown (see also Supplemental Fig. S4B).
Taf7 binds to both downstream Taf1/Taf2 (Louder et al. 2016) and Bdf1 (Matangkasombut et al. 2000), presumably positioning the Bdf1 bromodomains similarly to those found in metazoan Taf1. If so, bromodomain interactions with H4ac could promote the downstream DNA-binding conformation of TFIID. In agreement with this prediction, H4ac correlates better with Taf1 cross-linking to downstream promoter regions (+30 to +170) than to upstream Taf1 (−150 to −10) or the PIC, as assayed by TFIIB signal (Fig. 3B; Supplemental Fig. S4B). These results are consistent with a model in which Bdf1 (or the metazoan Taf1 bromodomains) binds H4ac to help tether TFIID to the +1 nucleosome and downstream promoter region.
TAFs and transcription promote activator-independent reinitiation
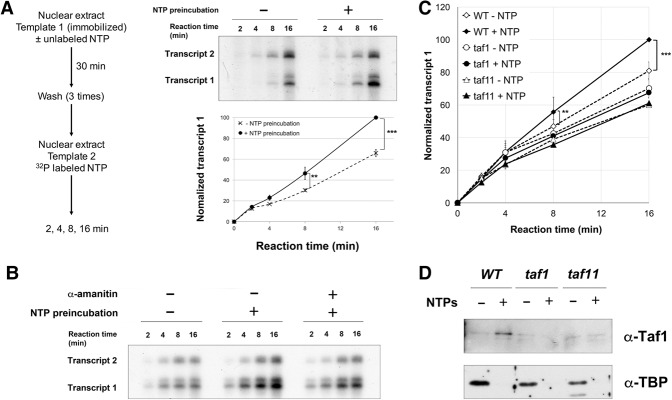
One likely function for stable post-transcription interaction of TAFs with downstream promoter DNA is to promote reinitiation. To test this idea, nuclear extracts were prepared from isogenic wild-type (TAF1) or temperature sensitive taf1 strains (the taf145 ts-1 strain in Walker et al. 1996). Single-round transcription was assayed by either limiting NTP exposure to 3 min (Fig. 4A; Supplemental Fig. S5A) or a runoff assay in which PICs were assembled on immobilized templates followed by extensive washing and then NTP addition in the absence of any additional factors that could support reinitiation (Supplemental Fig. S5B). Single-round transcription in the absence of activator was similar, and Gal4-vp16 produced a twofold to threefold increase in both the taf1 mutant and wild-type extracts (Fig. 4A; Supplemental Fig. S5A,B). Increased transcript levels were similarly produced by multiround conditions (45 min with NTPs) in the presence of activator. In contrast, the taf1 extract produced only about half as much transcription in the multiround reaction in the absence of activator (Fig. 4B,C; Supplemental Fig. S5A).
Figure 4.
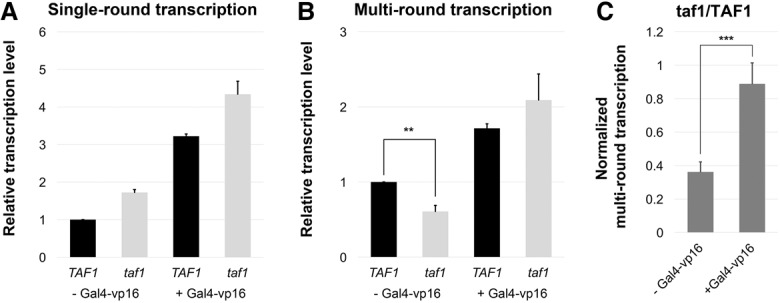
Taf1 promotes activator-independent multiround transcription. In vitro transcription was performed with TAF1 (YF157/YSW87; black) or taf1 (YF158/YSW90; gray) yeast nuclear extracts. NTPs were added for 3 min in single-round reactions (A) or 45 min for multiround transcription (B) in the presence or absence of transcription activator Gal4-vp16, as indicated. Transcripts were quantified, normalized, and plotted as the average value from three independent reactions. (C) The ratio of multiround transcripts in taf1 versus TAF1 extracts was divided by the same ratio in single-round reactions (see also Supplemental Fig. S5). Error bars show SD. (**) P = 0.014; (***) P = 0.006; (unmarked pairs) P > 0.01.
To test whether TAF-dependent stimulation of multiround transcription is connected to the transcription-dependent TAF deposition described above, a two-step reaction scheme was used (Fig. 5A). In the first step, nuclear extract was mixed with template 1 immobilized on magnetic beads, exactly as in the proteomics experiments in Figure 1. Preincubation with unlabeled NTPs allows transcription, or instead transcription can be blocked by omitting NTPs or adding α-amanitin with the NTPs. The complexes associated with template 1 were then washed extensively and added to a second reaction containing nuclear extract, a second template 2 as an internal control for de novo initiation, and radioactive NTPs for quantitation of transcription from both templates. Stimulation of reinitiation manifests as faster accumulation of transcripts from template 1 relative to template 2. As reinitiations also occur on template 2 after the initial round, the increase will not be cumulative over time and will have a maximum value of twofold (i.e., if every template 1 reinitiates once before any template 2 initiation in the second reaction).
Figure 5.
Taf1 promotes transcription reinitiation. (A) A schematic of two-step sequential reactions used to analyze transcription reinitiation is at the left. Note that no transcription activator was used in these experiments. Yeast nuclear extract was preincubated with immobilized template 1 (as in Fig. 1A) in the presence or absence of NTPs (400 µM each). After three washes, nuclear extract, 32P-labeled NTPs, and the longer G-less cassette template 2 (pG5CG-D2/F916) were added. (Top right panel) After 2, 4, 8, and 16 min, labeled transcripts were isolated and analyzed by gel electrophoresis and autoradiography. (Bottom right panel) Quantitation from three independent reactions plots transcript 1 levels over time, normalized by setting the 16-min maximum to 100 and using transcript 2 to correct for variations in recovery or reaction conditions. Error bars indicate SD (see also Supplemental Fig. S6A). (**) P = 0.01; (***) P = 7.5 × 10−5. (B) The sequential transcription assay was performed with NTPs and 10 µg/mL α-amanitin in the preincubation, as indicated. (C) Sequential transcription assay comparing wild-type (YSW87) with mutant taf1 (YSW90) or taf11 (YSB1732) nuclear extracts in the first preincubation reaction. After washes, the second transcription reaction was performed with wild-type nuclear extract prepared from strain BJ2168. Quantitation from three (taf11) or five (taf1) independent replicates is shown. (**) P = 0.08; (***) P = 5.2 × 10−5. All other differences were not statistically significant (see also Supplemental Fig. S6C). (D) Immobilized template reactions as in Figure 1 were performed with elution by SspI digestion and immunoblotting for Taf1 and TBP, comparing wild-type (YSW87) with mutant taf1 (YSW90) or taf11 (YSB1732) nuclear extracts. Note that Taf1 levels were roughly similar in all three extracts, as can be seen from the basal signal in the −NTP lanes.
When wild-type extract was used for both steps, a marked increase in template 1 transcription was seen when NTPs were included in the first reaction (Fig. 5A). Stimulation was strongly decreased by inclusion of α-amanitin in the first reaction (Fig. 5B), arguing that elongation by RNAPII, and not some other NTP function, is needed for enhanced reinitiation. Interestingly, the reinitiation effect was not seen when Gal4-vp16 was included in the reaction (Supplemental Fig. S6A), possibly because a high level of activator-induced de novo initiation swamps the reinitiation signal. Finally, when taf1 or taf11 mutant extract was used for the first reaction, no stimulation of reinitiation was observed even though wild-type extract was used for the second reaction (Fig. 5C; Supplemental Fig. S6C). Loss of reinitiation stimulation coincided with loss of increased downstream Taf1 binding in the mutant extracts (Fig. 5D).
Together, our in vitro results reveal the existence of a reinitiation intermediate that is transcription-dependent, TAF-dependent, and activator-independent. While superficially similar to the “Scaffold” complex defined by Yudkovsky et al. (2000), Scaffold can be formed with ATP alone and is activator-dependent. In our assay, preferential reinitiation is activator-independent (Supplemental Fig. S6A) and not seen with ATP alone (Supplemental Fig. S6B). Therefore, Scaffold cannot account for our results (see also the Discussion). These reinitiation intermediates are not mutually exclusive, and the residual stimulation that we observed with α-amanitin in the first reaction (Fig. 5B) could represent Scaffold-stimulated reinitiation.
In vivo evidence for two TFIID states
Based on the observations above, we developed the following model (also see the Discussion). First, a transcription activator promotes a pioneer round of transcription by recruiting coactivators such as SAGA and Swi/Snf to move promoter-occluding nucleosomes. A PIC then assembles, stimulated by activator–Mediator interactions. In this initial round of transcription, TAFs may be tethered to TBP without downstream contacts, rearranged by activator interaction (Chi and Carey 1996), or possibly even absent. After initiation and promoter clearance by RNAPII, the TAFs can then bind downstream, where they could promote one or more cycles of reinitiation. This model predicts at least two states of TFIID, distinguished by whether the Taf1/Taf2/Taf7/Bdf1 module is bound to the downstream promoter or not.
This idea was tested using Anchor Away depletion (Haruki et al. 2008) of Rpb1 or Bdf1 from nuclei. ChIP-seq of Taf1 and TBP was performed before and after a rapid 60-min depletion of the indicated factor. The relative change was plotted by heat map of individual promoters (Fig. 6A) or by gene-averaged plots (Supplemental Fig. S7). Depletion of Rpb1 led to a widespread drop in TBP cross-linking, presumably because lack of RNAPII limits PIC formation. Taf1 also dropped throughout the entire length of RPG and low TAF dependence promoters but behaved quite differently on high and medium promoters (Fig. 6B). Within this group, despite the drop in TBP, Taf1 cross-linking decreased upstream while at the same time increasing downstream. This shift is consistent with a TAF complex binding downstream of the TSS after departure of RNAPII.
Figure 6.
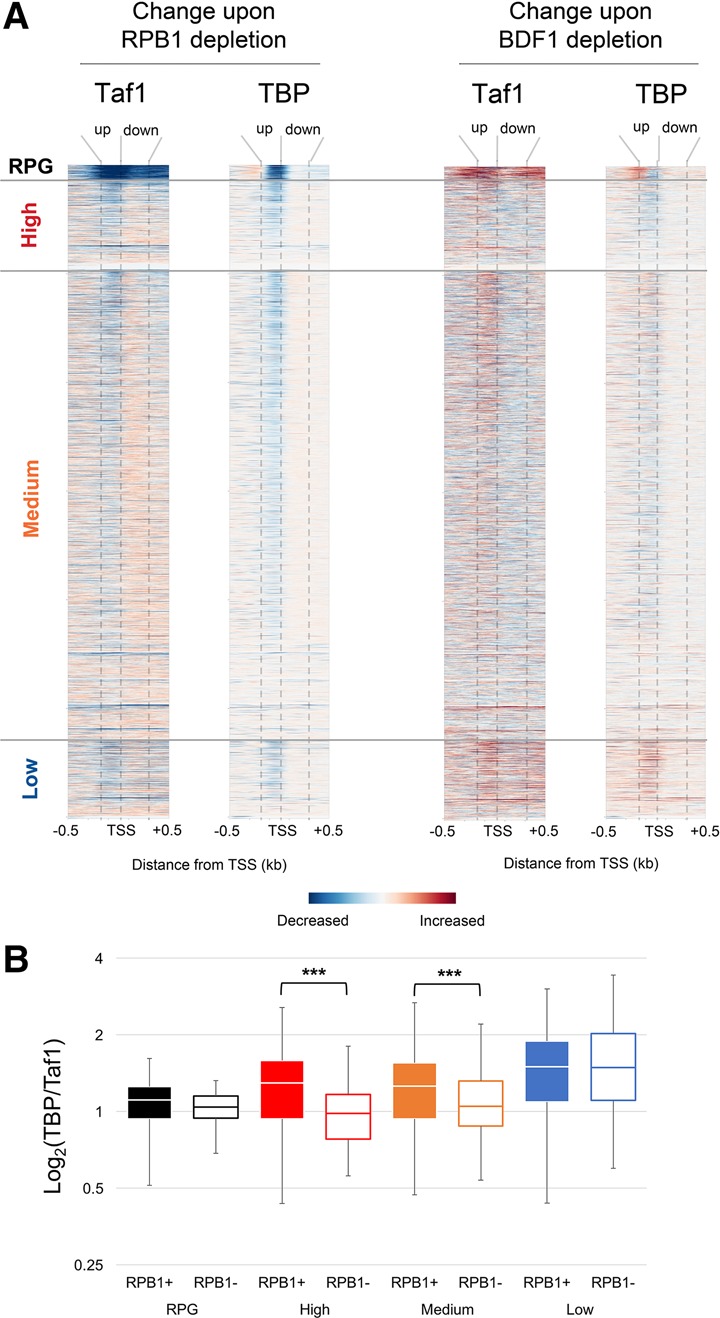
Depletion of Rpb1 or Bdf1 alters Taf1 binding in vivo. (A) ChIP-seq for Taf1 or TBP was performed from cells before or after a 60-min depletion of Rpb1 or Bdf1 by Anchor Away. Normalized reads (reads per million) were mapped, and differences were plotted as heat maps, showing individual genes sorted by the classes defined in Figure 2 (see also Supplemental Fig. S7A,B). (B) The ratios of total TBP and Taf1 reads for each promoter were calculated and plotted as quartile box and whisker plots.
At promoters with high or medium TAF dependence, Bdf1 depletion had the opposite effect on Taf1 binding when compared with Rpb1 depletion. Supporting a role for Bdf1 in anchoring TFIID downstream, Taf1 cross-linking decreased downstream while remaining the same or increasing upstream. TBP was unaffected or slightly decreased. In contrast, TBP and Taf1 in both regions increased on low TAF dependence promoters, perhaps due to redistribution of TFIID from the other promoters. Taf1 also increased on RPG promoters, although less so in the region just downstream from the TSS. While unexpected, it should be noted that cells lacking Bdf1 grow very slowly yet are viable due to the presence of its homolog, Bdf2. The Bdf2 bromodomains do not show a preference for acetylated H4 (Matangkasombut and Buratowski 2003), so depletion of Bdf1 may result in retargeting of TFIID by Bdf2.
In summary, the differential response of Taf1 cross-linking to regions upstream of and downstream from the TSS is consistent with independent binding of TFIID modules. Bdf1 promotes downstream interactions, while RNAPII antagonizes them.
Discussion
In the process of isolating and characterizing RNAPII ECs, we unexpectedly discovered that transcription initiation results in stable association of TAFs with downstream promoter regions, independent of TBP bound to the TATA box. The downstream TAF complex may create a “memory” or “bookmark” of recent transcription, thereby promoting reinitiation. This transcription-dependent deposition of a TAF complex is reminiscent of the exon junction complex, which is deposited just upstream of splice junctions by the spliceosome to mark recent intron removal (Moore and Proudfoot 2009). Incorporating our data with earlier observations, we propose the following model (Fig. 7A).
Figure 7.
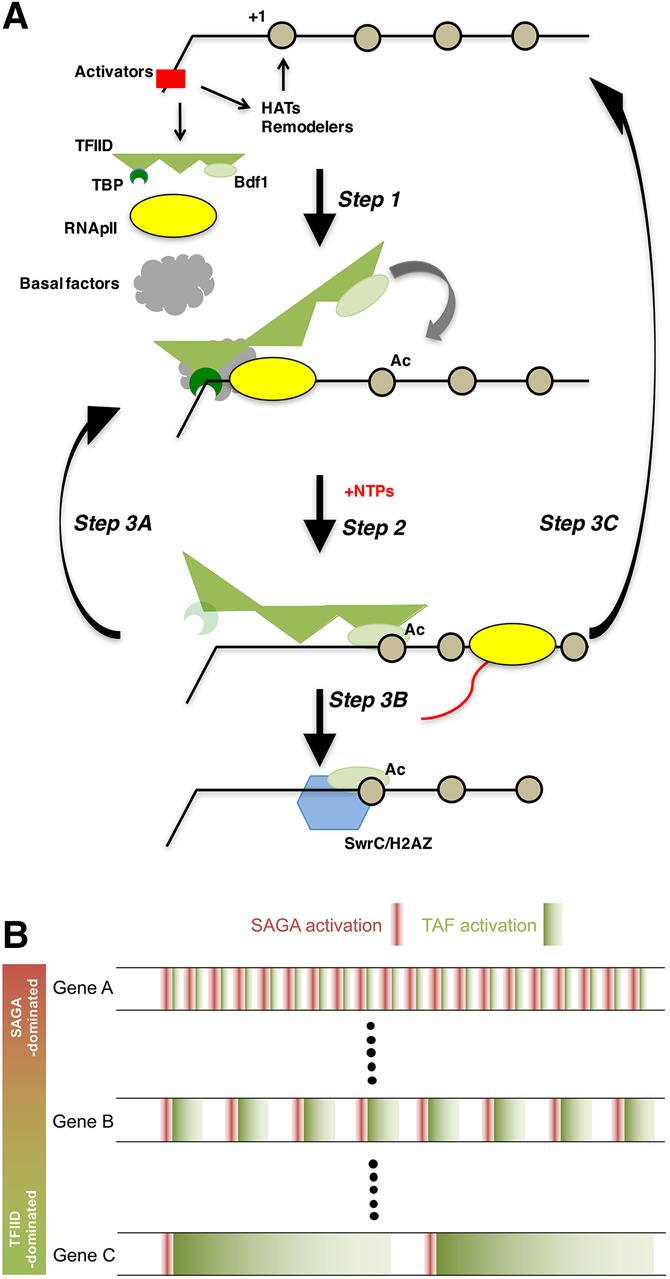
Mechanism of TAF-mediated RNAPII transcription. (A) Model for how TAFs may promote reinitiation (see the Discussion for details). TFIID is shown as TBP (dark green), TAFs (three flexibly linked triangles in medium green), and Bdf1 (light-green oval). (Black line) DNA; (beige circles) nucleosomes; (red rectangle) transcription activators; (gray cloud) basal factors; (yellow oval) RNAPII; (blue hexagon) SwrC; (red line) the RNA transcript. The lighter version of TBP after step 2 is to signify that it may be partially or completely dissociated after initiation. (B) A reinitiation model of how two promoters of similar strength can exhibit differential responses to TAF or activator/SAGA mutants (Huisinga and Pugh 2004; de Jonge et al. 2017). Each horizontal trace represents behavior over time. An initial activator- and SAGA-stimulated round of transcription is represented in red, while subsequent TAF-stimulated reinitiations are represented in green.
Step 1: pioneer round of PIC assembly
Regulatory transcription factors recruit HATs (SAGA and NuA4 in yeast) and ATP-dependent chromatin remodelers to move occluding nucleosomes from the core promoter (Cairns 2009; Weake and Workman 2010). Several mechanisms for TBP delivery have been described, and which is used may depend on which activators are present at the promoter. It has been proposed that TBP can arrive at the promoter via its association with SAGA (Eisenmann et al. 1992; Mohibullah and Hahn 2008), as free TBP (Buratowski et al. 1988), or as part of TFIID. If the latter, TFIID may be recruited by specific promoter sequences (Kadonaga 2012) or activators (Albright and Tjian 2000; Mencía et al. 2002; Papai et al. 2010; Chen et al. 2013). Once TBP is in place, the other basal factors, including Mediator–RNAPII, assemble into the PIC.
Step 2: initiation and establishment of downstream TAF interactions
NTPs trigger promoter melting and synthesis of RNA. Upon promoter escape by RNAPII, some factors may remain associated to facilitate reinitiation. An early in vitro study with purified mammalian basal factors found only TFIID remaining on immobilized templates after transcription (Zawel et al. 1995). Using yeast extracts, Yudkovsky et al. (2000) described a “Scaffold” complex containing TFIID, TFIIA, Mediator, TFIIH, and possibly TFIIE persisting at the promoter. However, recent ChIP-seq and ChEC-seq (chromatin endogenous cleavage [ChEC] with high-throughput sequencing) experiments argue that Mediator resides primarily at upstream activator-binding sites and only transiently at the basal promoter (Grünberg et al. 2012; Jeronimo and Robert 2014; Wong et al. 2014), so Scaffold may actually be two complexes: an activator–Mediator complex at the UAS/enhancer and a subset of basal factors at the core promoter.
Our data suggest a reinitiation intermediate in which TAFs interact with downstream promoter DNA and the acetylated +1 nucleosome. This complex is clearly distinct from Scaffold. First, Scaffolds were defined as what remains after extensively washed PICs were treated with NTPs or ATP only. In contrast, our initial round of transcription occurs in extracts, allowing post-initiation recruitment, as seen with TAFs (Supplemental Fig. S1D). Whereas Scaffold is dependent on activator, our reinitiation effect does not require activator (Fig. 4; Supplemental Fig. S6A). Finally, while Scaffold can be generated with ATP alone, our reinitiation intermediate requires a full set of NTPs for actual transcription (Figs. 1, 5B; Supplemental Fig. S6B).
In vivo evidence for downstream interactions in our reinitiation intermediate comes from patterns of Taf1 cross-linking (Figs. 2, 3). At TFIID-dominant promoters, downstream cross-linking increases after rapid RNAPII depletion even as upstream cross-linking drops (Fig. 6A). Interestingly, recent ChEC-seq work from Grünberg et al. (2016) showed that depletion of Mediator subunit Med14 caused increased Taf1-mediated cleavage downstream from the TSS. Other groups showed that TBP inactivation unexpectedly increased TAF cross-linking at some promoters (Mencía et al. 2002; Shen et al. 2003). Originally interpreted as TBP-independent binding of TAFs, our model suggests that these TAFs may instead remain from previous rounds of transcription.
Transcription-dependent TAF deposition was seen in vitro on both naked and chromatinized templates (Fig. 1; Supplemental Fig. S1E). However, interactions between the Taf1/Bdf1 bromodomains and the acetylated +1 nucleosome are also likely important for TFIID function in vivo. A recent TFIID structure lacks the bromodomains but shows a Taf1/Taf2/Taf7 module binding the promoter 50–60 bp downstream from the TATA box (Louder et al. 2016). Bdf1 specifically interacts with Taf7 (Matangkasombut et al. 2000), consistent with Taf1/Bdf1 bromodomains mediating communication between the +1 nucleosome and TFIID. Supporting this idea, acetylated H4 correlates better with downstream than upstream Taf1 cross-linking (Fig. 3B; Supplemental Fig. S4B). Even more compellingly, depletion of Bdf1 results in a reduction of Taf1 cross-linking specifically downstream from the TSS (Fig. 6B).
Step 3: a TAF-mediated branch point
Depending on the particular promoter sequences and nearby chromatin modifications, three outcomes can be envisioned: (1) assembly of a new PIC for reinitiation; (2) dissociation of TAFs but not Bdf1, leading to SwrC recruitment and H2A.Z incorporation at the +1 nucleosome; or (3) TFIID dissociation and reversion to the nucleosome-occluded state.
Option 3A: TAF-dependent reinitiation
The TAF complex could stimulate reinitiation through at least two mechanisms. First, it can sterically block ingression of the +1 nucleosome to an occluding position, bypassing the need for activator-dependent recruitment of HATs and remodelers as in step 1. Second, the TAF complex can stabilize TBP or perhaps recruit a new TBP molecule. This might be particularly significant at promoters with weak TATA boxes.
At SAGA-dominant but not TFIID-dominant promoters, the +1 nucleosome overlaps the PIC (Rhee and Pugh 2012). Two recent studies showed that the +1 nucleosome markedly shifts downstream when promoters with low TAF occupancy become active (Nocetti and Whitehouse 2016; Zhou et al. 2016). In contrast, promoters that are highly expressed or with high TAF occupancy have a more static +1 nucleosome at the downstream-shifted position. Finally, Reja et al. (2015) showed that transcription inhibition causes the +1 nucleosome to shift upstream over TFIID-dominant RPG promoters. Together, these observations support a model in which TAFs help maintain the +1 nucleosome at a downstream position.
As TAF-dominant promoters tend to have weaker, nonconsensus TATA boxes, downstream TAF interactions can also help retain or rebind TBP at the TATA box. Supporting a retention model, an in vivo epitope-switching experiment (van Werven et al. 2009) found the highest TBP turnover at SAGA-dominant promoters and the lowest TBP turnover at promoters with high Taf1 levels.
Importantly, the TAF mutant extracts used here support single-round transcription as efficiently as wild-type extracts but have much less activity in multiround reactions (Figs. 4, 5). Similarly, TAF depletion from mammalian extracts did not affect activator-induced transcription in single-round transcription reactions but strongly reduced transcription in multiround reactions (Oelgeschläger et al. 1998). Whether the post-initiation TFIID complex assembles a new PIC by reusing basal factors from the Scaffold or recruits them de novo, this new cycle of transcription bypasses the need for the activator-dependent step 1.
Option 3B: recruitment of SwrC and H2A.Z
In yeast, if TAFs dissociate from downstream DNA before a new PIC can assemble, the acetylated +1 nucleosome might retain Bdf1. This would create a window for Bdf1 to recruit the SwrC complex for incorporation of H2A.Z/Htz1 at the +1 nucleosome. Htz1 has been proposed to act as a memory of recent transcription to allow rapid reinitiation. In agreement, Htz1 levels are highest at inactive promoters with acetylated +1 nucleosomes (Li et al. 2005; Zhang et al. 2005; Altaf et al. 2010).
Option 3C: return to the inactive state
TFIID dissociation could allow the promoter to revert to the repressed state at the beginning of step 1. The lifetime of post-initiation TFIID at promoters may vary depending on particular DNA sequence elements, TAF-recruiting activators, and +1 nucleosome acetylation.
The relative probability of TAF-dependent reinitiation versus returning to the repressed state provides a simple explanation for differential responses of promoters to SAGA and TAF mutations (Fig. 7B). Promoters with weak TFIID retention may need to continuously acetylate and remodel the +1 nucleosome, creating stronger SAGA dependence. In contrast, those with strongest TFIID retention will have more reinitation cycles and therefore appear more TAF-dependent. This model accommodates the growing consensus that SAGA and TFIID work redundantly or cooperatively at most promoters (Huisinga and Pugh 2004; Bonnet et al. 2014; Grünberg et al. 2016; Baptista et al. 2017; Warfield et al. 2017).
One prediction of our model is that SAGA-dominant promoters will have a stronger requirement for the continued presence of activator. In striking agreement, de Jonge et al. (2017) showed recently that nuclear depletion of the Hsf1 transcription activator causes rapid loss of transcription at its SAGA-dominant, but not its TFIID-dominant, target promoters. This could be explained by the persistence of activator-independent reinitiations in vivo.
Our model also suggests that TFIID could affect “transcription burst” properties of promoters by minimizing intervals of repressed transcription. Transcription bursting in yeast is most common at SAGA-dominant promoters with strong TATA boxes, while TAF-dominant promoters generally have lower levels of noise (Raser and O'Shea 2004; Blake et al. 2006). Interestingly, Taf1 reduces burst size and duration at mammalian promoters with strong TATA boxes (Pennington et al. 2013), perhaps reflecting competition between TAFs and RNAPII for downstream promoter sequences (see below).
TFIID conformations and transcription-dependent TAF interactions
An important future goal will be to determine the physical basis for transcription-dependent TAF binding. A large body of evidence, including experiments here (Fig. 6), suggests TFIID conformation changes in which TAFs can swing on and off the downstream promoter DNA (Fig. 7A). Early cryo-EM studies of TFIID noted substantial flexibility (Elmlund et al. 2009; Papai et al. 2010; Cianfrocco et al. 2013). The most recent model for TFIID bound to DNA (Louder et al. 2016) reveals specific downstream promoter–TAF interactions, but it is noteworthy that glutaraldehyde fixing was necessary to produce this structure. This TFIID model can be docked into a separate model of the PIC containing TBP only, but steric clashes led Louder et al. (2016) to surmise that downstream TAFs must release to accommodate RNAPII. Similarly, careful kinetic experiments by Yakovchuk et al. (2010) indicate that TAFs actually retard PIC assembly and that TFIID downstream promoter contacts rearrange or release as RNAPII forms a closed complex. Finally, Zhang et al. (2015) discovered a Taf2-binding chemical that locks the downstream TAFs onto the promoter, thereby blocking assembly of RNAPII into the PIC.
Therefore, the post-initiation TAF binding reported here may simply reflect competition between TAFs and RNAPII, with TAFs clamping down on the promoter after polymerase release into elongation. Alternatively, it is possible that some event or modification during promoter clearance actively places TAFs downstream. Some activators also trigger downstream TFIID conformation changes to promote initial PIC formation (Chi and Carey 1996; Horikoshi et al. 1988a,b), and these activation mechanisms are likely analogous to the activator-independent effects reported here.
Previous models presumed that TBP and TAFs act in concert as a single promoter recognition factor. Our model suggests that TBP and TAF binding to DNA can occur more independently at different stages of the transcription cycle. Interestingly, based on comparisons of free and DNA-bound TFIID structures, Nogales and colleagues (Cianfrocco et al. 2013) speculated that there could be a TFIID conformation having downstream TAF–DNA contacts, while TBP and more upstream TAFs release from the TATA box. Such a complex would be consistent with the reinitiation intermediate suggested here. The ability of a TBP–TAF complex to independently release upstream or downstream contacts may also resemble events during RNAPIII transcription, where TFIIIB and TFIIIC subunits interact with promoter elements downstream from the TSS to assemble the PIC yet somehow do not interfere with elongation. The TAF reinitiation function described here provides a new model for thinking about TFIID functions.
Materials and methods
Strains, plasmids, and oligonucleotides
S. cerevisiae strains, oligonucleotides, and plasmids used in this study are listed in Supplemental Tables S2–S4.
Yeast nuclear extract preparation
Nuclear extracts were prepared as described previously (Sikorski et al. 2012). Wild-type extracts for mass spectrometry analysis were prepared from strain BY4741 or BJ2168/YF4. For in vitro transcription assays, nuclear extracts were prepared from wild-type (YSW87/YF157), taf1 ts-1 (YSW93/YF158), or taf11-3000 (YSB1732) strains (Walker et al. 1996; Komarnitsky et al. 1999).
Immobilized template binding and quantitative mass spectrometry
Immobilized template-binding assay was performed as described (Sikorski et al. 2012) with some modifications. Biotinylated linear templates were amplified by PCR from pUC18-G5CYC1 G-(SB649) with the primers listed in Supplemental Table S3. Where noted, templates were chromatinized with yeast histone octamers as described (Sikorski et al. 2012). Chromatinized or naked templates (880 ng of DNA) were immobilized by binding to 200 µg of streptavidin beads (Dynabeads Streptavidin T1, Invitrogen) for 1 h at room temperature. Beads were isolated by magnetic concentration and then blocked in 600 µL of blocking solution [1× transcription buffer (100 mM K-acetate, 20 mM HEPES-KOH at pH 7.6, 1 mM EDTA, 5 mM Mg-acetate) with 60 mg/mL casein, 5 mg/mL polyvinylpyrrolidone, 2.5 mM dithiothreitol (DTT)] for 25 min at room temperature. Transcription activator Gal4-vp16 (800 ng) was incubated with templates for 5 min at room temperature. Yeast nuclear extracts (∼1.5 mg) were added in 240 µL of 1× transcription buffer complemented with 80 U of RNAsin, 0.44 U of creatine phosphokinase, 307.2 µg of phosphocreatine, 0.03% NP-40, 2 µg of tRNA, 160 µM S-adenosylmethionine, 20 µM acetyl-CoA, and 4 µg of Hae III-digested Escherichia coli DNA for 30 min at room temperature. Transcription was started by adding NTP mix (400 µM each ATP, CTP, UTP, and 3′-OmeGTP) to the mixture. Where indicated, α-amanitin was added at 10 µg/mL. The reaction was stopped after 15 min by washing twice in 200 µL of wash buffer (1× transcription buffer complemented with 0.05% NP-40 and 2.5 mM DTT). Downstream DNA and bound proteins were released with 120 U of SspI-HF or PstI-HF digestion (New England Biolabs) in 160 µL of 1× transcription buffer for 30 min at room temperature. Sample preparation for quantitative mass spectrometry and subsequent data analysis were performed as described (Sikorski et al. 2012). For relative quantitation, iTRAQ reporter signal intensity values of all unique peptide scans for a given protein were summed prior to calculation of ratios. All iTRAQ ratios were normalized to total reporter signal. Only proteins that were identified in multiple replicates were used. Statistical significance of enrichment was calculated by t-test (Supplemental Fig. S1A) and mixed model (Supplemental Fig. S1B,C) analyses.
ChIP and ChIP-seq analysis
For Figure 3, strain BY4741 was used. For nuclear depletion of Rpb1 (YSB3202) or Bdf1 (YSB3323) in Figure 6, the Anchor Away system (Haruki et al. 2008) was used, with 1 µg/mL rapamycin added for 1 h to cells containing the appropriate FRB fusion. Cells were grown to OD600 = 0.5 and then cross-linked with 1% formaldehyde for 20 min at room temperature. Cross-linking was stopped with 125 mM glycine for 5 min, and cells were collected and washed. Sheared chromatin (100∼300 bp) was prepared using a Misonix Sonicator 300 with cup horns.
Cross-linked chromatin was precipitated by standard methods with specific antibodies against Taf1 (provided by Joe Reese, Pennsylvania State University), Spt15 (TBP), H3 (Abcam, ab1791), or H4ac (Millipore, 06-598). Libraries for multiplex sequencing were generated as described previously (Wong et al. 2013) and sequenced on an Illumina 2500 machine by the Harvard Bauer Center Genomics Core Facility. Sequenced reads were aligned to the yeast S288C reference genome (http://www.yeastgenome.org, with annotations provided by Burak Alver, Harvard Medical School) using Bowtie 1 (Langmead et al. 2009). MACS (version 2) was used to visualize peaks from sequence read distribution (Feng et al. 2012). Custom Python scripts for analyzing and graphing data (from L. Soares) are available at https:// github.com/Buratowski/NGS. Data have been deposited in Gene Expression Omnibus (GEO; accession no. GSE94851). In order to reanalyze genome-wide occupancy, raw data for TFIIB (Wong et al. 2014), Taf1, Bdf1 (Rhee and Pugh 2012), Htz1 (Gu et al. 2015), and Swr1 (Yen et al. 2013) were adapted and reanalyzed in parallel as above (GEO accession nos. SRR1197807 [TFIIB], SRR396796 [Taf1], SRR397550 [Bdf1],SRR1118262 [Htz1], and SRR948403 [Swr1]).
In vitro transcription
In vitro transcription was performed as described (Sikorski et al. 2012). After preincubation of template and extract, NTPs were added for 3 min for single-round transcription and for 45 min for multiround reactions. For the two-step transcription analysis in Figure 5, NTP preincubation was performed as in the immobilized template-binding assay as described above, except Gal4-vp16 was not added. Template 1, shown in Figure 1A, was incubated with wild-type or taf1 or taf11 mutant extract and unlabeled NTPs for 30 min. Beads were then washed three times in 200 µL of wash buffer. For the second reaction, template 2 (pG5CG-D2 from Pardee et al. 2003), ∼1.5 mg of wild-type yeast nuclear extract, and 32P-labeled NTPs (250 µM ATP, 250 µM CTP, 10 µM UTP, 1.2 µCi of 32P-labeled UTP) in 240 µL of 1× transcription buffer were added to the template 1-bound beads. At each indicated time point, 60 µL of the mix was collected, and the reaction was stopped with 200 µL of RNase T1 mixture (1000 U; Thermo Scientific). Transcripts were recovered and analyzed by standard gel electrophoresis, autoradiography, and phosphorimager.
Supplementary Material
Acknowledgments
We thank Joe Reese for Taf1 antibodies and strains; Fred Ponticelli and Steve Hahn for plasmids; and Kevin Struhl, Fred Winston, Steve Hahn, Frank Holstege, Mike Carey, and members of the Buratowski and Marto laboratories for helpful discussions. This work was supported by Dana-Farber Cancer Institute/Blais Proteomics Center support to J.A.M., and National Institutes of Health grant GM46498 to S.B.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.306324.117.
References
- Albright SR, Tjian R. 2000. TAFs revisited: more data reveal new twists and confirm old ideas. Gene 242: 1–13. [DOI] [PubMed] [Google Scholar]
- Altaf M, Auger J, Brodeur J, Piquet S, Cramet M, Bouchard N, Lacoste N, Utley RT, Gaudreau L, et al. 2010. NuA4-dependent acetylation of nucleosomal histones H4 and H2A directly stimulates incorporation of H2A.Z by the SWR1 complex. J Biol Chem 285: 15966–15977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auty R, Steen H, Myers LC, Persinger J, Bartholomew B , Gygi SP, Buratowski S. 2004. Purification of active TFIID from Saccharomyces cerevisiae. J Biol Chem 279: 49973–49981. [DOI] [PubMed] [Google Scholar]
- Baptista T, Grünberg S, Minoungou N, Koster MJE, Timmers HTM, Hahn S, Devys D, Tora L. 2017. SAGA is a general cofactor for RNA polymerase II. Mol Cell 68: 130–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieniossek C, Papai G, Schaffitzel C, Garzoni F, Chaillet M, Scheer E, Papadopoulos P, Tora L, Schultz P, Berger I. 2013. The architecture of human general transcription factor TFIID core complex. Nature 493: 699–702. [DOI] [PubMed] [Google Scholar]
- Blake WJ, Balázsi G, Kohanski MA, Isaacs FJ, Murphy KF, Kuang Y, Cantor CR, Walt DR, Collins JJ. 2006. Phenotypic consequences of promoter-mediated transcriptional noise. Mol Cell 24: 853–865. [DOI] [PubMed] [Google Scholar]
- Bonnet J, Wang CY, Baptista T, Vincent SD, Hsiao WC, Stierle M, Kao CF, Tora L, Devys D. 2014. The SAGA coactivator complex acts on the whole transcribed genome and is required for RNA polymerase II transcription. Genes Dev 28: 1999–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S, Hahn S, Sharp PA, Guarente L. 1988. Function of a yeast TATA element-binding protein in a mammalian transcription system. Nature 334: 37–42. [DOI] [PubMed] [Google Scholar]
- Buratowski S, Hahn S, Guarente L, Sharp PA. 1989. Five intermediate complexes in transcription initiation by RNA polymerase II. Cell 56: 549–561. [DOI] [PubMed] [Google Scholar]
- Burke TW, Kadonaga JT. 1997. The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila. Genes Dev 11: 3020–3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns BR. 2009. The logic of chromatin architecture and remodelling at promoters. Nature 461: 193–198. [DOI] [PubMed] [Google Scholar]
- Chen WY, Zhang J, Geng H, Du Z, Nakadai T, Roeder RG. 2013. A TAF4 coactivator function for E proteins that involves enhanced TFIID binding. Genes Dev 27: 1596–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi T, Carey M. 1996. Assembly of the isomerized TFIIA–TFIID–TATA ternary complex is necessary and sufficient for gene activation. Genes Dev 10: 2540–2550. [DOI] [PubMed] [Google Scholar]
- Cianfrocco MA, Kassavetis GA, Grob P, Fang J, Juven-Gershon T, Kadonaga JT, Nogales E. 2013. Human TFIID binds to core promoter DNA in a reorganized structural state. Cell 152: 120–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jonge WJ, O'Duibhir E, Lijnzaad P, van Leenen D, Groot Koerkamp MJ, Kemmeren P, Holstege FC. 2017. Molecular mechanisms that distinguish TFIID housekeeping from regulatable SAGA promoters. EMBO J 36: 274–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenmann DM, Arndt KM, Ricupero SL, Rooney JW, Winston F. 1992. SPT3 interacts with TFIID to allow normal transcription in Saccharomyces cerevisiae. Genes Dev 6: 1319–1331. [DOI] [PubMed] [Google Scholar]
- Elmlund H, Baraznenok V, Linder T, Szilagyi Z, Rofougaran R, Hofer A, Hebert H, Lindahl M, Gustafsson CM. 2009. Cryo-EM reveals promoter DNA binding and conformational flexibility of the general transcription factor TFIID. Structure 17: 1442–1452. [DOI] [PubMed] [Google Scholar]
- Feng J, Liu T, Qin B, Zhang Y, Liu XS. 2012. Identifying ChIP-seq enrichment using MACS. Nat Protoc 7: 1728–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dümpelfeld B, et al. 2006. Proteome survey reveals modularity of the yeast cell machinery. Nature 440: 631–636. [DOI] [PubMed] [Google Scholar]
- Grünberg S, Warfield L, Hahn S. 2012. Architecture of the RNA polymerase II preinitiation complex and mechanism of ATP-dependent promoter opening. Nat Struct Mol Biol 19: 788–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grünberg S, Henikoff S, Hahn S, Zentner GE. 2016. Mediator binding to UASs is broadly uncoupled from transcription and cooperative with TFIID recruitment to promoters. EMBO J 35: 2435–2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu M, Naiyachit Y, Wood TJ, Millar CB. 2015. H2A.Z marks antisense promoters and has positive effects on antisense transcript levels in budding yeast. BMC Genomics 16: 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn S. 1998. The role of TAFs in RNA polymerase II transcription. Cell 95: 579–582. [DOI] [PubMed] [Google Scholar]
- Hahn S, Buratowski S. 2016. Structural biology: snapshots of transcription initiation. Nature 533: 331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruki H, Nishikawa J, Laemmli UK. 2008. The anchor-away technique: rapid, conditional establishment of yeast mutant phenotypes. Mol Cell 31: 925–932. [DOI] [PubMed] [Google Scholar]
- Horikoshi M, Carey MF, Kakidani H, Roeder RG. 1988a. Mechanism of action of a yeast activator: direct effect of GAL4 derivatives on mammalian TFIID-promoter interactions. Cell 54: 665–669. [DOI] [PubMed] [Google Scholar]
- Horikoshi M, Hai T, Lin YS, Green MR, Roeder RG. 1988b. Transcription factor ATF interacts with the TATA factor to facilitate establishment of a preinitiation complex. Cell 54: 1033–1042. [DOI] [PubMed] [Google Scholar]
- Huisinga KL, Pugh BF. 2004. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol Cell 13: 573–585. [DOI] [PubMed] [Google Scholar]
- Jeronimo C, Robert F. 2014. Kin28 regulates the transient association of Mediator with core promoters. Nat Struct Mol Biol 21: 449–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadonaga JT. 2012. Perspectives on the RNA polymerase II core promoter. Wiley Interdiscip Rev Dev Biol 1: 40–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobor MS, Venkatasubrahmanyam S, Meneghini MD, Gin JW, Jennings JL, Link AJ, Madhani HD, Rine J. 2004. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol 2: e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarnitsky PB, Michel B, Buratowski S. 1999. TFIID-specific yeast TAF40 is essential for the majority of RNA polymerase II-mediated transcription in vivo. Genes Dev 13: 2484–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Keogh MC, Datta N, Sawa C, Ryan OW, Ding H, Haw RA, Pootoolal J, Tong A, Canadien V, et al. 2003. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol Cell 12: 1565–1576. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, et al. 2006. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440: 637–643. [DOI] [PubMed] [Google Scholar]
- Ladurner AG, Inouye C, Jain R, Tjian R. 2003. Bromodomains mediate an acetyl-histone encoded antisilencing function at heterochromatin boundaries. Mol Cell 11: 365–376. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Causton HC, Holstege FC, Shen WC, Hannett N, Jennings EG, Winston F, Green MR, Young RA. 2000. Redundant roles for the TFIID and SAGA complexes in global transcription. Nature 405: 701–704. [DOI] [PubMed] [Google Scholar]
- Lee DH, Gershenzon N, Gupta M, Ioshikhes IP, Reinberg D, Lewis BA. 2005. Functional characterization of core promoter elements: the downstream core element is recognized by TAF1. Mol Cell Biol 25: 9674–9686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Pattenden SG, Lee D, Gutiérrez J, Chen J, Seidel C, Gerton J, Workman JL. 2005. Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc Natl Acad Sci 102: 18385–18390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louder RK, He Y, López-Blanco JR, Fang J, Chacón P, Nogales E. 2016. Structure of promoter-bound TFIID and model of human pre-initiation complex assembly. Nature 531: 604–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matangkasombut O, Buratowski S. 2003. Different sensitivities of bromodomain factors 1 and 2 to histone H4 acetylation. Mol Cell 11: 353–363. [DOI] [PubMed] [Google Scholar]
- Matangkasombut O, Buratowski RM, Swilling NW, Buratowski S. 2000. Bromodomain factor 1 corresponds to a missing piece of yeast TFIID. Genes Dev 14: 951–962. [PMC free article] [PubMed] [Google Scholar]
- Mencía M, Moqtaderi Z, Geisberg JV, Kuras L, Struhl K. 2002. Activator-specific recruitment of TFIID and regulation of ribosomal protein genes in yeast. Mol Cell 9: 823–833. [DOI] [PubMed] [Google Scholar]
- Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C. 2004. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303: 343–348. [DOI] [PubMed] [Google Scholar]
- Mohibullah N, Hahn S. 2008. Site-specific cross-linking of TBP in vivo and in vitro reveals a direct functional interaction with the SAGA subunit Spt3. Genes Dev 22: 2994–3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Proudfoot NJ. 2009. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell 136: 688–700. [DOI] [PubMed] [Google Scholar]
- Nakajima N, Horikoshi M, Roeder RG. 1988. Factors involved in specific transcription by mammalian RNA polymerase. Mol Cell Biol 8: 4028–4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nocetti N, Whitehouse I. 2016. Nucleosome repositioning underlies dynamic gene expression. Genes Dev 30: 660–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelgeschläger T, Tao Y, Kang YK, Roeder RG. 1998. Transcription activation via enhanced preinitiation complex assembly in a human cell-free system lacking TAFIIs. Mol Cell 1: 925–931. [DOI] [PubMed] [Google Scholar]
- Papai G, Tripathi MK, Ruhlmann C, Layer JH, Weil PA, Schultz P. 2010. TFIIA and the transactivator Rap1 cooperate to commit TFIID for transcription initiation. Nature 465: 956–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardee TS, Ghazy MA, Ponticelli AS. 2003. Yeast and Human RNA polymerase II elongation complexes: evidence for functional differences and postinitiation recruitment of factors. Eukaryot Cell 2: 318–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennington KL, Marr SK, Chirn GW, Marr MT. 2013. Holo-TFIID controls the magnitude of a transcription burst and fine-tuning of transcription. Proc Natl Acad Sci 110: 7678–7683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranish JA, Hahn S, Lu Y, Yi EC, Li XJ, Eng J, Aebersold R. 2004. Identification of TFB5, a new component of general transcription and DNA repair factor IIH. Nat Genet 36: 707–713. [DOI] [PubMed] [Google Scholar]
- Raser JM, O'Shea EK. 2004. Control of stochasticity in eukaryotic gene expression. Science 304: 1811–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reja R, Vinayachandran V, Ghosh S, Pugh BF. 2015. Molecular mechanisms of ribosomal protein gene coregulation. Genes Dev 29: 1942–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee HS, Pugh BF. 2012. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature 483: 295–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selleck W, Howley R, Fang Q, Podolny V, Fried MG, Buratowski S, Tan S. 2001. A histone fold TAF octamer within the yeast TFIID transcriptional coactivator. Nat Struct Biol 8: 695–700. [DOI] [PubMed] [Google Scholar]
- Shao W, Zeitlinger J. 2017. Paused RNA polymerase II inhibits new transcriptional initiation. Nature Gen 49: 1045–1051. [DOI] [PubMed] [Google Scholar]
- Shen WC, Bhaumik SR, Causton HC, Simon I, Zhu X, Jennings EG, Wang T-H, Young RA, Green MR. 2003. Systematic analysis of essential yeast TAFs in genome-wide transcription and preinitiation complex assembly. EMBO J 22: 3395–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski TW, Joo YJ, Ficarro SB, Askenazi M, Buratowski S, Marto JA. 2012. Proteomic analysis demonstrates activator- and chromatin-specific recruitment to promoters. J Biol Chem 287: 35397–35408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sypes MA, Gilmour DS. 1994. Protein/DNA crosslinking of a TFIID complex reveals novel interactions downstream of the transcription start. Nucleic Acids Res 22: 807–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas MC, Chiang CM. 2006. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol 41: 105–178. [DOI] [PubMed] [Google Scholar]
- van Werven FJ, van Teeffelen HA, Holstege FC, Timmers HT. 2009. Distinct promoter dynamics of the basal transcription factor TBP across the yeast genome. Nat Struct Mol Biol 16: 1043–1048. [DOI] [PubMed] [Google Scholar]
- Walker SS, Reese JC, Apone LM, Green MR. 1996. Transcription activation in cells lacking TAFIIS. Nature 383: 185–188. [DOI] [PubMed] [Google Scholar]
- Warfield L, Ramachandran S, Baptista T, Devys D, Tora L, Hahn S. 2017. Transcription of nearly all yeast RNA polymerase II-transcribed genes is dependent on transcription factor TFIID. Mol Cell 68: 118–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weake VM, Workman JL. 2010. Inducible gene expression: diverse regulatory mechanisms. Nat Rev Genet 11: 426–437. [DOI] [PubMed] [Google Scholar]
- Wong KH, Jin Y, Moqtaderi Z. 2013. Multiplex Illumina sequencing using DNA barcoding. Curr Protoc Mol Biol 101: 7.11.1–7.11.11. [DOI] [PubMed] [Google Scholar]
- Wong KH, Jin Y, Struhl K. 2014. TFIIH phosphorylation of the Pol II CTD stimulates mediator dissociation from the preinitiation complex and promoter escape. Mol Cell 54: 601–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakovchuk P, Gilman B, Goodrich JA, Kugel JF. 2010. RNA polymerase II and TAFs undergo a slow isomerization after the polymerase is recruited to promoter-bound TFIID. J Mol Biol 397: 57–68. [DOI] [PubMed] [Google Scholar]
- Yen K, Vinayachandran V, Pugh BF. 2013. SWR-C and INO80 chromatin remodelers recognize nucleosome-free regions near +1 nucleosomes. Cell 154: 1246–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yudkovsky N, Ranish JA, Hahn S. 2000. A transcription reinitiation intermediate that is stabilized by activator. Nature 408: 225–229. [DOI] [PubMed] [Google Scholar]
- Zawel L, Kumar KP, Reinberg D. 1995. Recycling of the general transcription factors during RNA polymerase II transcription. Genes Dev 9: 1479–1490. [DOI] [PubMed] [Google Scholar]
- Zhang H, Roberts DN, Cairns BR. 2005. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell 123: 219–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Boskovic Z, Hussain MM, Hu W, Inouye C, Kim HJ, Abole AK, Doud MK, Lewis TA, Koehler AN, et al. 2015. Chemical perturbation of an intrinsically disordered region of TFIID distinguishes two modes of transcription initiation. Elife 4: e07777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Blocker AW, Airoldi EM, O'Shea EK. 2016. A computational approach to map nucleosome positions and alternative chromatin states with base pair resolution. Elife 5: 572. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.